Abstract
Recent advances in cancer characterization have consistently revealed marked heterogeneity, impeding the completion of integrated molecular and clinical maps for each malignancy. Here, we focus on chronic lymphocytic leukemia (CLL), a B cell neoplasm with variable natural history which is conventionally categorized into two subtypes distinguished by extent of somatic mutations in the heavy chain variable region of immunoglobulin genes (IGHV). To build the ‘CLL map,’ we integrated genomic, transcriptomic, and epigenomic data from 1148 patients. We identified 202 candidate genetic drivers of CLL (109 novel) and refined the characterization of IGHV subtypes, which revealed distinct genomic landscapes and leukemogenic trajectories. Discovery of new gene expression subtypes further subcategorized this neoplasm and proved to be independent prognostic factors. Clinical outcomes were associated with a combination of genetic, epigenetic, and gene expression features, further advancing our prognostic paradigm. Overall, this work reveals fresh insights into CLL oncogenesis and prognostication.
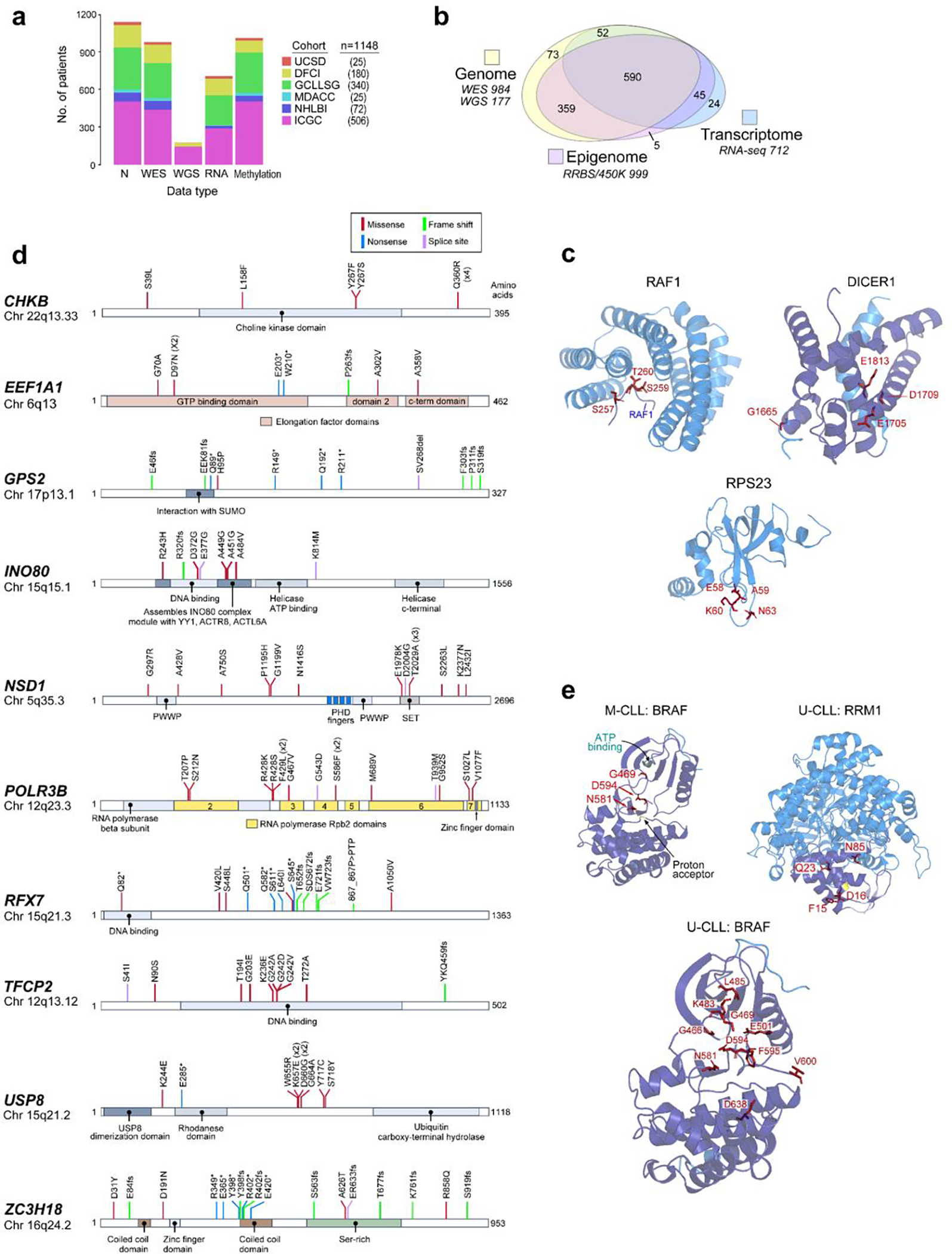
Previous analyses have provided only fragments of the ‘CLL map’, each focusing on particular patient populations or different data types1–9, but none have built a comprehensive atlas with sufficient power and resolution to fully characterize the whole bioclinical spectrum of the disease. We set out to assemble, from existing and newly generated data, the largest CLL dataset to date. This encompassed samples from 1095 CLL patients and 54 patients with monoclonal B cell lymphocytosis (MBL) from which whole-exome or -genome sequencing (WES/WGS) (n=1074), RNA-sequencing (RNA-seq) (n=712) and DNA methylation data (n=999) were analyzed (Extended Data Fig. 1a–b). Samples were collected during active surveillance (n=680), after treatment (n=52) or upon enrollment in therapeutic clinical trials1–3,10–13 (n=416; n=371 treatment-naive; n=45 relapsed/refractory) (Supplementary Table 1). This large dataset enabled more complete delineation of the biological underpinnings of CLL and its molecular subtypes.
RESULTS
Identification of novel CLL drivers
To generate a comprehensive catalogue of drivers, we first focused on the 984 CLL samples with WES. To ensure consistency and highest accuracy of the mutation calls, we reprocessed the data with an updated suite of tools, detecting somatic single nucleotide variants (sSNVs), short insertion/deletion mutations (indels), and copy number alterations (sCNAs). We also applied specialized tools for detecting recently described CLL driver events such as the g.3A>C mutation of the spliceosome-related small nuclear RNA U114 (U1) and the R110 mutation in the IGLV3–21 gene15,16 (IGLV3–21R110) (Methods). Our prior power estimates17 suggested that with ~1000 WES samples and somatic background mutation rate of ~1/Mb in CLL, we should be able to discover >90% of drivers mutated in 2% of patients, whereas with ~500 samples the power drops to 50%. To verify these estimates, we performed a down-sampling analysis and confirmed that the number of drivers almost doubled, increasing from an average of 38.8 with 500 cases to 74.5 with ~1000 cases, with the majority of new drivers mutated in <2% of patients (Fig. 1a, Methods). Likewise, increased cohort size enabled discovery of significantly recurrent sCNAs across all frequencies, with the steepest increase in lower frequency drivers (<3%, Fig. 1b).
Figure 1: Increased power enables CLL driver gene detection.
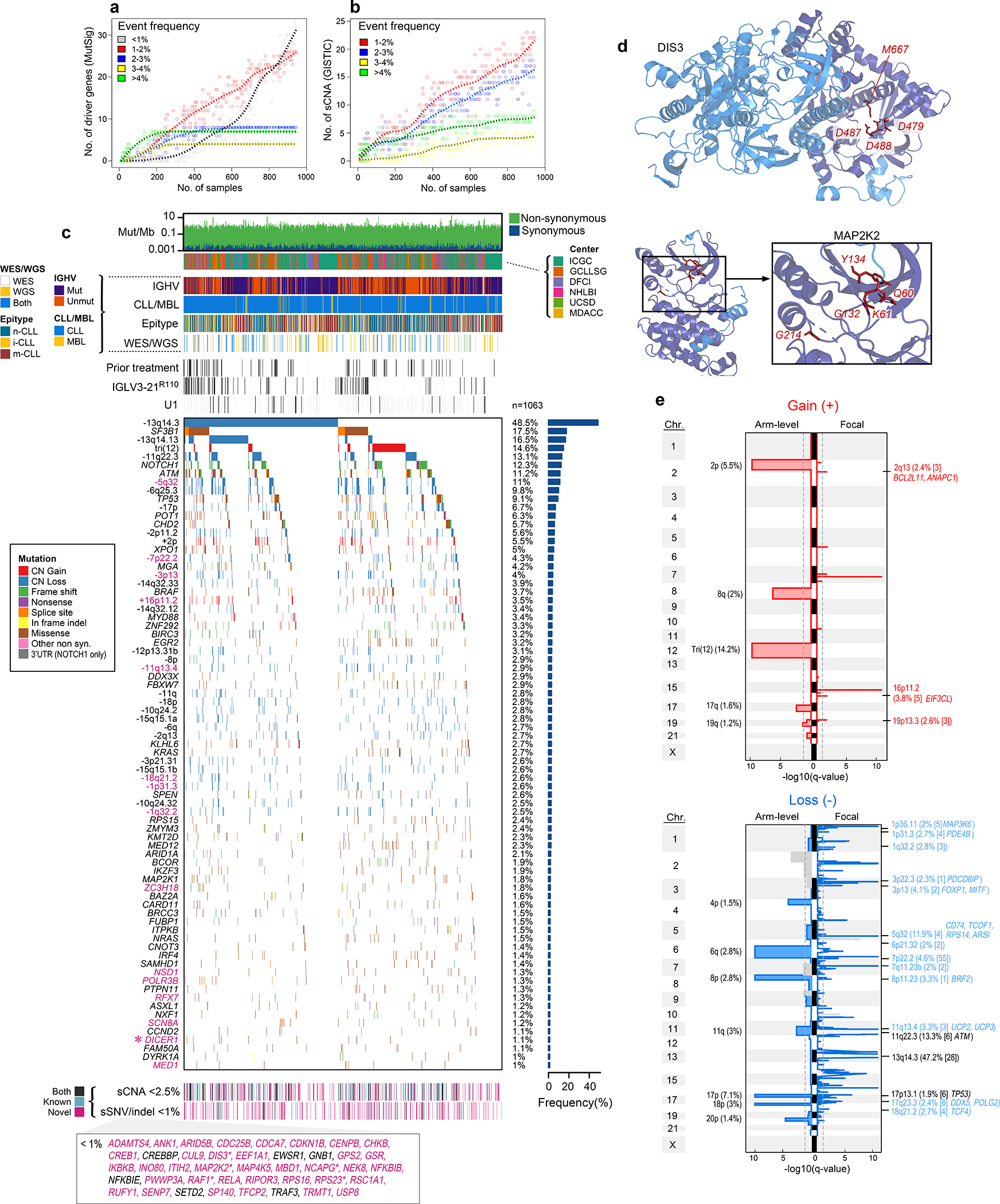
a-b. By down-sampling analysis, driver gene (a) and sCNA (b) discovery increases with additional samples. Points represent a random subset of samples with smoothed fit line; analysis separated by frequency.
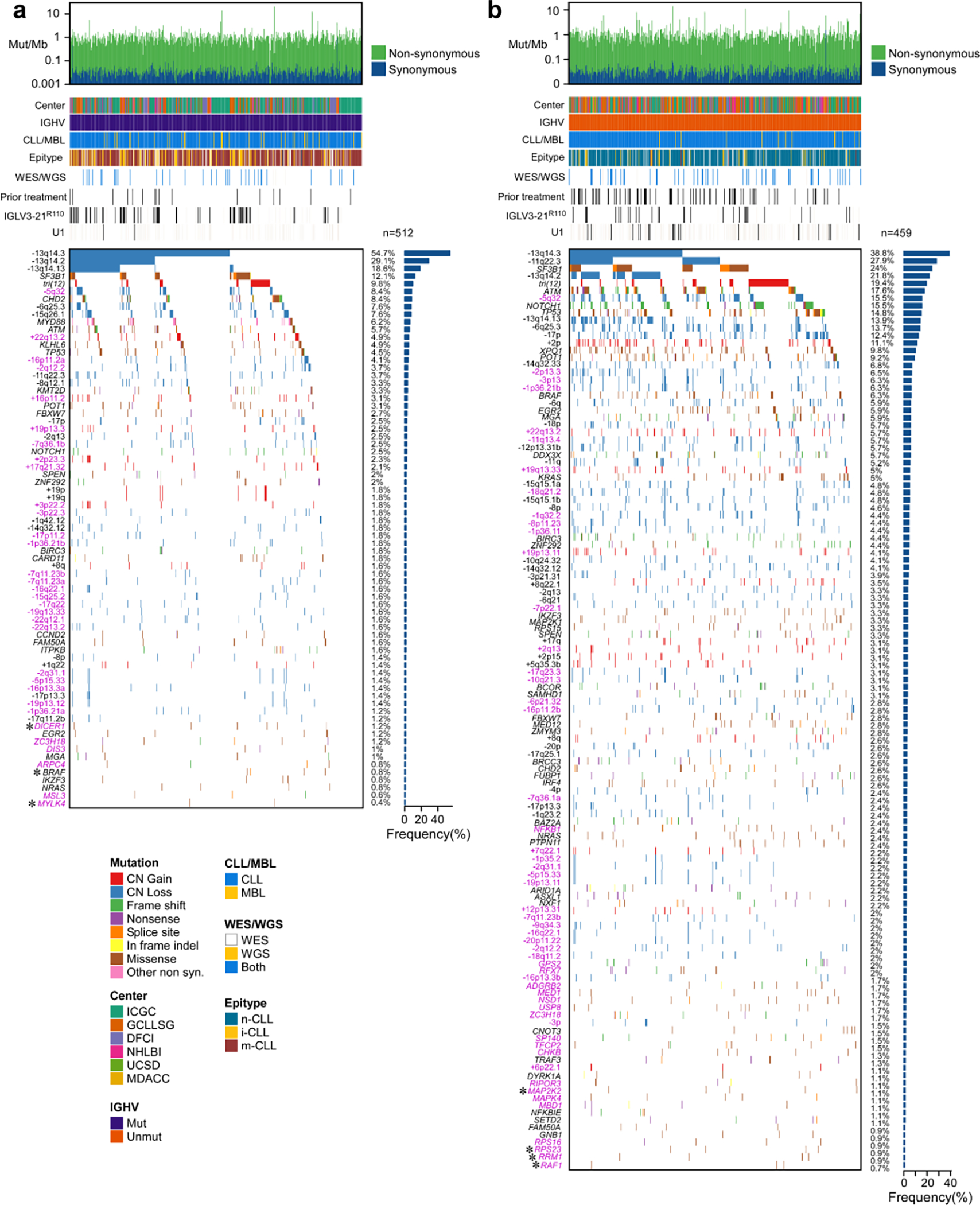
c. Landscape of genetic alterations in CLL with frequency of alterations (right, n=1063 patients). Header tracks - annotation of cohort, IGHV status, CLL or MBL sample, epigenetic subtype (epitype: naive-like, n-CLL; intermediate, i-CLL; memory-like, m-CLL), sequencing data type; prior treatment, U1 and IGLV3–21R110 mutations - black; magenta label - novel alterations. Asterisks - discovery by CLUMPS. Bottom tracks - Lower frequency sSNV/indels and sCNAs, designated as novel (magenta), known events (blue) or both (black). Bottom boxed inset - candidate driver genes, frequency <1%.
d. Representative genes identified by CLUMPS (see Supplementary Table 5). 3D protein structure of MAP2K2 and DIS3. Mutated residues (red labels) cluster in functional regions (purple).
e. Recurrent copy number gains (top) and losses (bottom) by GISTIC analysis showing arm level (left) and focal events (right). Chromosome number - vertical axis; dashed line - significance, q=0.1. Blacklisted regions - gray. Arm level events are labeled with cytoband and frequency (n=984). Focal events denote cytoband, frequency, number of genes encompassed in peak (bracketed), and genes of interest. Red/blue font: novel focal events with frequency >2%. Black font: previously known events (see Supplementary Table 7).
Our dataset revealed 82 putative CLL driver genes based on recurrent sSNV/indel mutations (q<0.1), of which 37 were not previously identified as significantly altered in CLL1,2,18–20 (Methods, Fig. 1c, Supplementary Table 2–4). Beyond the previously known CLL drivers, such as SF3B1, NOTCH1, ATM, and TP53 (mutated in 17.5%, 12.3%, 11.2%, and 9.1% of patients, respectively), as well as mutations in IGLV3–21R110 and U1 (mutated in 9.5% and 3.8%, respectively), the frequencies of the remaining events form a long, gradually decreasing tail (59 of 82 drivers mutated in <2% of patients). Although most newly discovered genes were mutated at low frequency, 24.2% of patients harbored at least one mutation in a novel putative driver. Notably, they were also the sole sSNV/indel driver in 4% of patients. Six additional putative drivers were discovered through spatial clustering of mutations in 3-D protein structures, using CLUMPS21, including MAP2K2, DIS3, and DICER1 (Fig. 1d, Extended Data Fig. 1c, Supplementary Table 5). Three MAP2K2 mutations were localized in the kinase domain, which activates ERK signaling and is functionally similar to MAP2K1, a previously identified CLL driver1. DIS3 encodes the catalytic subunit of a critical RNA exosome complex22 and is recurrently mutated in multiple myeloma23. Two of four altered sites in DIS3 were in cancer hotspots (D479 and D48824) and located in the catalytic domain25. Beyond sSNV/indels in coding regions, an analysis of 177 WGS did not reveal novel noncoding CLL drivers2,14 (Methods, Supplementary Table 3, Supplementary Note).
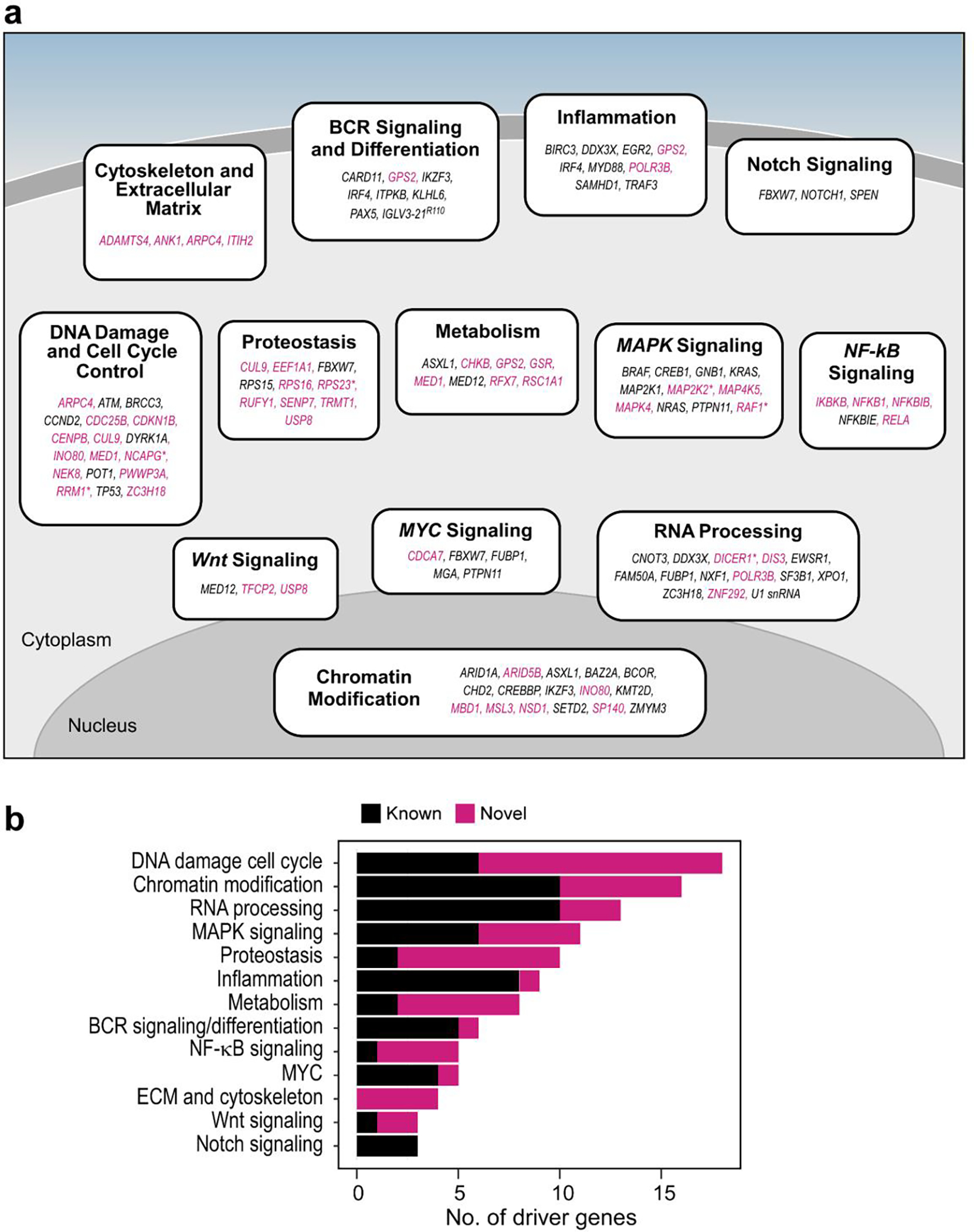
In support of the newly discovered drivers, we noted that 7 (18.9%) had mutations clustered in functional domains (Extended Data Fig. 1d). For example, mutations were identified in the DNA-binding domain of INO80, which encodes the catalytic subunit of a chromatin remodeling complex that regulates genome stability26 and is frequently mutated in hepatosplenic T cell lymphoma27. Additionally, 7 (18.9%) have a role in other mature B cell malignancies such as the tumor suppressor gene, RFX7, implicated in Burkitt lymphoma28 and diffuse large B cell lymphoma29. These candidate drivers were also enriched in biological pathways known to contribute to CLL pathogenesis such as DNA damage and chromatin modification1,2,14. However, they also identified processes not previously highlighted by driver genes like protein synthesis and stability as well as regulation of cytoskeletal proteins and the extracellular matrix (Extended Data Fig. 2a–b, Supplementary Table 6).
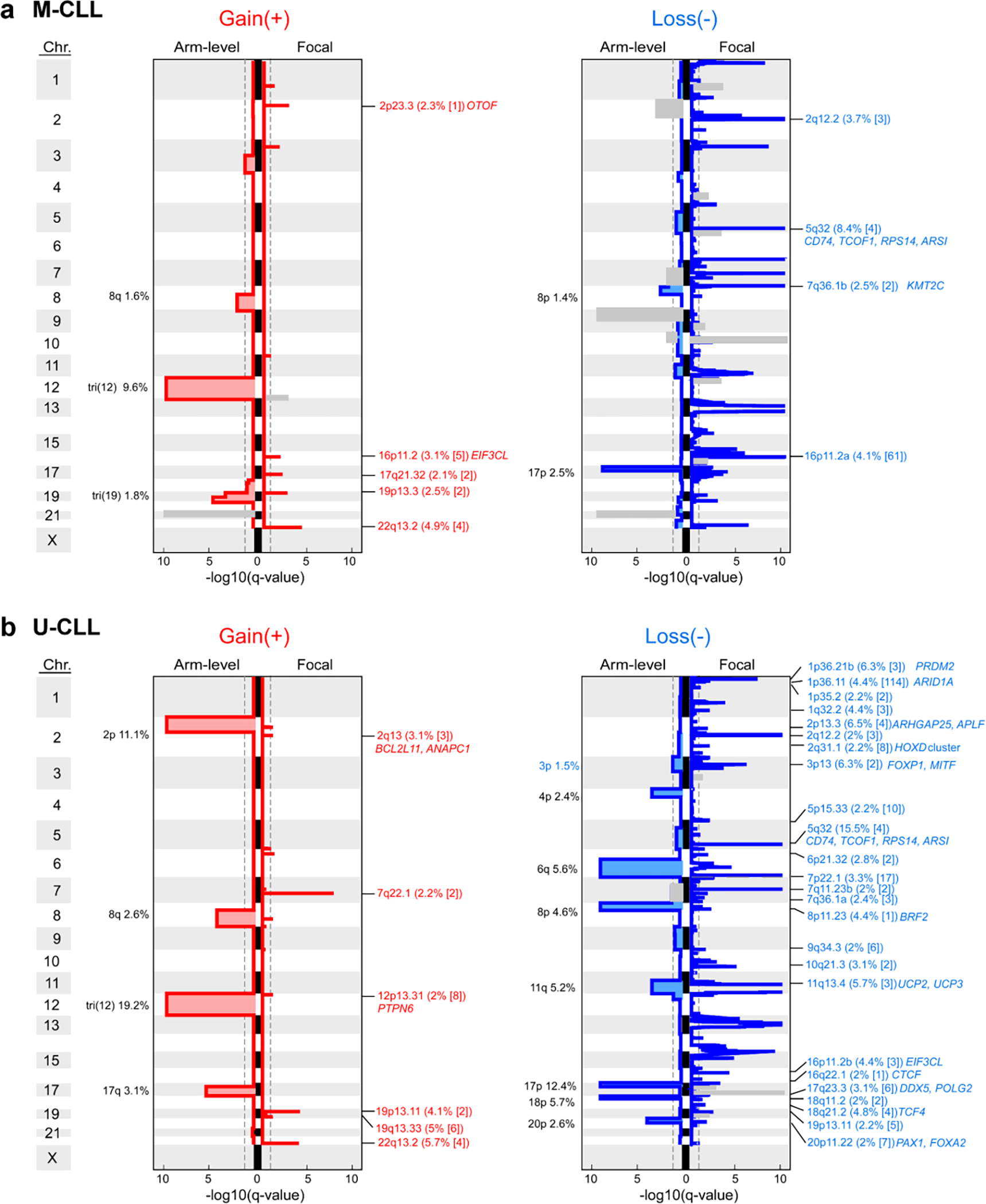
A striking finding provided by our increased statistical power was the abundance of yet unreported focal sCNAs associated with CLL, including 5 novel gains and 30 new losses (of 6 and 53 total, respectively)1,2,30–32 (Fig. 1e, Supplementary Table 7). One such deletion in 5q32 (11.9% of samples) encompassed ARSI, TCOF1, CD74, and RPS14, which is part of the common deleted region in 5q- syndrome, a low-risk subtype of myelodysplastic syndrome (MDS)33. Two of these genes, RPS14 and TCOF1, are involved in ribosome function or biogenesis and have been implicated in inflammatory Toll-like receptor signaling in MDS models34 and in maintaining genomic integrity after DNA damage35, respectively, suggesting that multiple genes in this region are associated with pathways involved in CLL oncogenesis. Other deletions contain UCP2 and UCP3 in 11q13.4 (3.3%), which encode mitochondrial uncoupling proteins that function as tumor suppressors altering redox homeostasis36,37 and multiple other regions that include known cancer associated genes38 (Supplementary Table 7). We were further enabled to identify rarely reported arm level sCNAs including 17q gain (1.6%) and 4p loss (1.5%)1,31. Altogether, our results vastly expand the map of CLL drivers and reveal convergent mechanisms through which cardinal cellular processes are altered in this disease.
Molecular profiles of IGHV subtypes
We leveraged our increased cohort size to discover distinct candidate driver genes, sCNAs, and structural variants (SVs) in 512 CLLs with mutated IGHV (M-CLLs) and 459 CLLs with unmutated IGHV (U-CLLs), expanding previous work that identified only a limited number of discrete molecular characteristics associated with IGHV status1,2,39 (Methods, Supplementary Table 8). The IGHV subtype-specific mutation analyses increased our sensitivity to identify 7 additional putative drivers that were not identified in the pan-CLL analysis (Extended Data Fig. 1e and 3, Supplementary Table 4–5). In U-CLL, this included NFKB1, a regulator of NFκB signaling40, and RRM1, which encodes the catalytic subunit of ribonucleotide reductase that is critical for DNA replication and repair as well as the target of nucleoside analogs including fludarabine41.
Although M-CLL and U-CLL had similar cohort sizes and comparable mutational burdens in coding regions (1.14/Mb vs. 1.11/Mb medians, respectively; Wilcoxon rank-sum test p=0.98; though the mean number of clonal mutations genome-wide was increased in M-CLL - 12.6 versus 9.6, p=6×10−14), the number of significant putative drivers was much higher in U-CLL (54 versus 25 genes, respectively; ratio 2.16, Binomial test p=0.0015). To ensure that this difference was not due to prior therapy, we compared only treatment-naive samples within each cohort (n=375; M-CLL was downsampled), and again found more drivers in U-CLL (ratio 2.82, one sample t-test p=5×10−11). Most drivers were significant in either M-CLL (n=9) or U-CLL (n=38) while only a minority were significant in both subgroups (n=16, 25.4% of total) (Fig. 2a). Of these shared drivers, 10 of the 16 were twice as frequent in U-CLL, consistent with increased driver frequency in this subtype.
Figure 2: M-CLL and U-CLL have unique genomic landscapes.
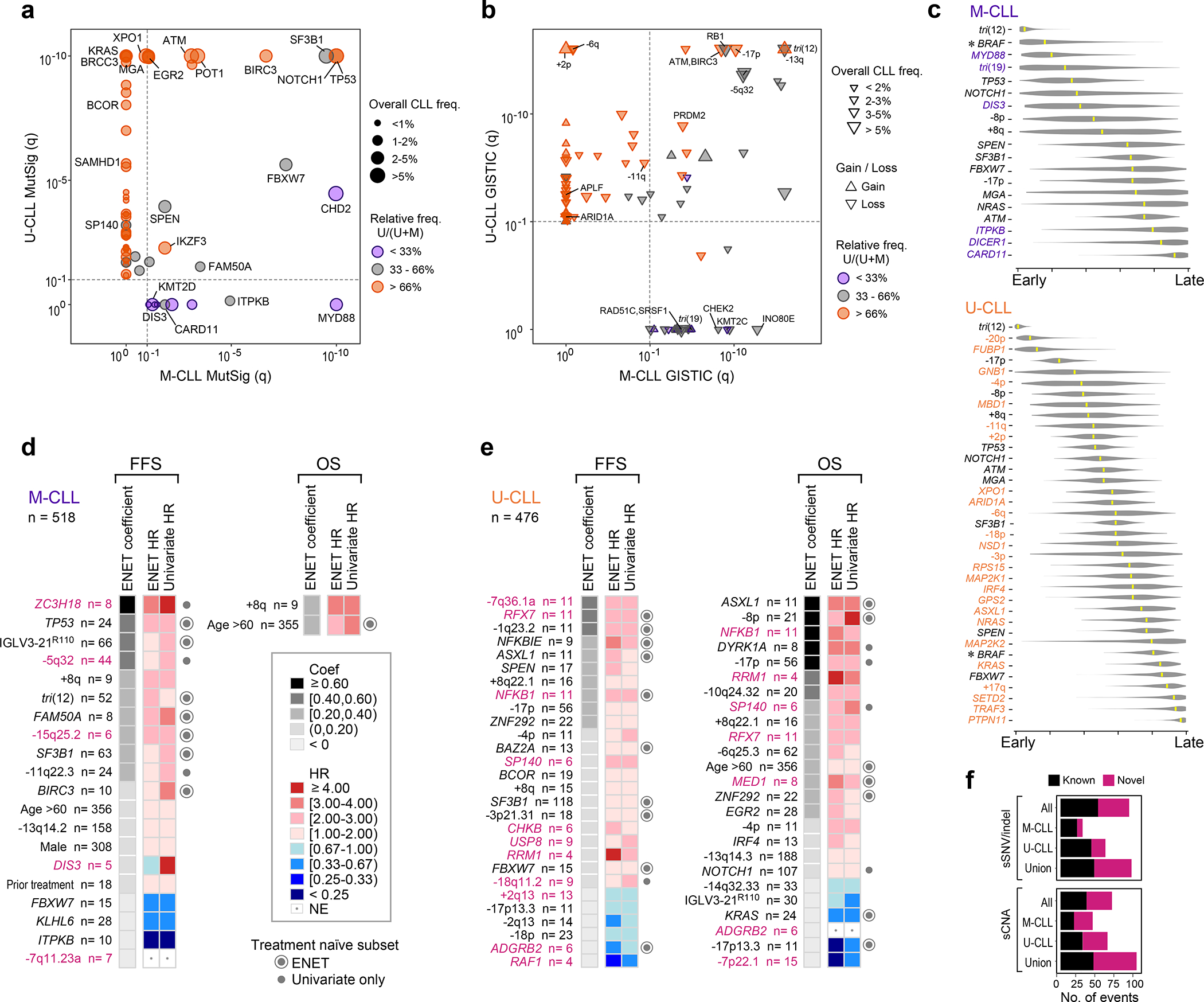
a-b. Comparison of candidate driver genes (a) or copy number gains/losses (up/down triangle, respectively, b) in U-CLL (y-axis, WES, n=459) vs. M-CLL (x-axis, WES, n=512) plotted by −log10(q-value). Significance - dashed line. Representative candidate drivers are annotated. Frequency in entire cohort (n=984) - size of circle (a) or triangle (b). Orange - drivers predominantly in U-CLL; purple - predominantly in M-CLL.
c. League model timing diagrams comparing acquisition of somatic mutation and arm level sCNAs in M-CLL (top, n=251) and U-CLL (bottom, n=354). Higher timing score (x-axis) denotes later event; median scores - yellow marks (95% confidence interval, gray). Purple - events significant in M-CLL; orange - events significant in U-CLL; black - events shared by M-CLL and U-CLL. Asterisks - significant difference in timing (q<0.1).
d-e. Somatic alterations associated with failure free survival (FFS) and overall survival (OS) in M-CLL (d, WES/WGS, n=518 and U-CLL (e, WES/WGS, n=476). Events ranked by elastic net (ENET) coefficients, which identifies variables to be included in the model, shrinking coefficients to 0 when excluded. Heatmap denotes hazard ratios (HR) for ENET and univariate Cox regressions. Events included by ENET model (concentric circle) or significant in univariate analysis only (closed circle) in treatment-naive, non-trial patients (M-CLL, n=393; U-CLL, n=247) annotated on right. Magenta label - novel alterations (see Supplementary Table 11).
f. Number of candidate drivers in three genomic driver detection analyses: entire cohort (All, n=984), M-CLL (n=512) and U-CLL (n=459). For each analysis set, sSNV/indel represents candidate driver genes from MutSig2CV and CLUMPS, while sCNA represents recurrent events from GISTIC. Union - total putative drivers identified in any of the three analysis sets.
IGHV subtypes were also distinguished by sCNA profiles (70 in either M-CLL or U-CLL vs. 20 shared) (Fig. 2b, Extended Data Fig. 4, Supplementary Table 7). Trisomy 19 (1.8%) was only observed in M-CLL, consistent with previous studies42. In contrast, 8 arm level events including 2p gain (11.1%) and loss of 6q (5.6%) were only significant in U-CLL. The majority of focal events distinguishing the IGHV subtypes were novel1,2,30,31, comprising 18 of 23 events enriched in M-CLL and 25 of 37 in U-CLL, and some provided orthogonal evidence for CLL driver genes discovered through mutation analysis. For example, loss of 1p36.11 (4.4%) contained ARID1A, a known driver gene2, and both this sCNA and sSNV were only significant in U-CLL. The sCNAs identified also emphasized underlying biology important in CLL leukemogenesis. In M-CLL, the region in 7q36.1b loss (2.5%) included KMT2C, a lysine-specific methyltransferase involved in epigenetic regulation43 (Fig. 2b, Extended Data Fig. 4). A related tumor suppressor, KMT2D, is a candidate driver44 also enriched specifically in M-CLL (Fig. 2a, Extended Data Fig. 3), demonstrating a convergence of different genetic alteration mechanisms on the same biologic pathway in this IGHV subtype.
We further identified differences between M-CLL and U-CLL on the basis of SVs. From 177 WGS (88 M-CLL, 87 U-CLL and 2 non-evaluable), we discovered 681 SV breakpoints in 141 (79.7%) patients (average of 4.8 per patient; Methods; Supplementary Table 9). Approximately 46% of SVs were clonal, supporting a potential role for SVs in CLL initiation (Supplementary Table 9, Methods). The most recurrent SVs involving the immunoglobulin (Ig) loci (as identified by IgCaller45, Methods) distinguished M-CLL from U-CLL (Extended Data Fig. 5a–b, Supplementary Table 9). We confirmed that the most common Ig translocation partner in M-CLL was BCL2 (5 of 88 cases, 5.7%)2. Conversely, a large 37-Mb deletion in chromosome 14 was identified in U-CLL (4 of 87 cases, 4.6%), which deletes candidate CLL drivers (DICER1, TRAF3) and directly perturbs ZFP36L1, a tumor suppressor gene that down-regulates NOTCH146. The rearrangement mechanism also differed between these events, with aberrant V(D)J recombination driving the BCL2 events in M-CLL and class-switch recombination (CSR) facilitating the ZFP36L1-associated deletions in U-CLL (Methods, Extended Data Fig. 5b), consistent with CSR events occurring prior to germinal cell commitment47,48. These different patterns and underlying mechanisms were confirmed in the WES cohort where IgCaller detected 9 additional cases with BCL2 translocations in M-CLL and only 1 in U-CLL (Supplementary Table 9, Extended Data Fig. 5c).
To evaluate possible differences in mechanisms of somatic mutation generation in M-CLL and U-CLL, we performed mutation signature analysis on 177 WGS and identified activity of 5 mutational processes (Extended Data Fig. 6a, Supplementary Note). In addition to confirming the presence of the aging, canonical activation-induced cytidine deaminase (c-AID) and non-canonical AID (nc-AID) related signatures in both clonal and subclonal mutations11, we also found evidence of signature SBS18, likely due to damage from reactive oxygen species, and splitting of the c-AID signatures (SBS84 and SBS85). Of note, clustered mutations in U-CLL were enriched in SBS84 relative to M-CLL, although non-significantly (Wilcoxon rank-sum test, p=0.19), whereas SBS85 was more prevalent in M-CLL, likely reflecting unique mutational processes arising from AID in each subtype (p=1.6×10−9, Extended Data Fig. 6b–c).
Further highlighting the differences between M-CLL and U-CLL, we detected distinct inferred timing of acquired sSNV/indels and arm level sCNAs when analyzed by PhylogicNDT49 (Methods, Fig. 2c). Trisomy 12 was an early event and shared drivers such as TP53 and NOTCH1 were intermediate in both CLL subtypes1. In contrast, acquisition of BRAF mutations was an early event in M-CLL but occurred late in U-CLL (q<0.1). Of those drivers specifically enriched per subtype, MYD88 was an early event in M-CLL whereas chromosome 20p loss and FUBP1 alterations may be initiating lesions in U-CLL. We separately assessed the temporal acquisition of sSNV/indels by analyzing their cancer cell fractions (CCF) (Extended Data Fig. 6d). Only 12 (12.4%) driver genes had predominantly clonal events with a median CCF>85%, and 6 of these 12 were novel, including MSL3 and USP8 identified in M-CLL and U-CLL, respectively. This panoply of genetic differences underscores M-CLL and U-CLL as distinct molecular entities and support their unique trajectories of leukemogenesis.
Given these differences, we analyzed the clinical impact of putative genetic drivers from each IGHV subtype (Methods, Fig. 2d–e, Table 1, Supplementary Table 10–11). Relative to M-CLL, U-CLL had more genetic changes associated with either failure-free survival (FFS) and/or overall survival (OS) (41 in U-CLL versus 18 in M-CLL, Binomial test p=0.004; Fig. 2d–e). Of these, 18 were novel events (5 of 18 in M-CLL and 13 of 41 in U-CLL; Fig. 2d–e). In M-CLL, ZC3H18 mutations and losses of 5q32 and 15q25.2 were novel alterations associated with risk of short FFS in addition to known factors such as TP53 and IGLV3–21R110 mutations. The prognostic impact of many of these novel putative drivers was also supported when the dataset was restricted to only treatment-naive, non-trial samples (n=393) (Table 1, Supplementary Table 10). Only two features were associated with reduced survival in M-CLL, which were age >60 years and gain of 8q, the chromosomal arm containing MYC. In U-CLL, RFX7 and NFKB1 were novel candidate drivers associated with poor FFS and OS, although only FFS was shorter in the treatment-naive subset (n=247, Supplementary Table 10). The prognostic impact of known but less frequent drivers, such as NFKBIE and ASXL1, was also evident in addition to verifying the known effects of more common features like 17p deletion. Of note, 17p deletion and TP53 mutations significantly co-occur1, which partially explains why only one was significant in our modeling. Further analysis of either alteration alone or in combination demonstrated that TP53 mutation in the absence of 17p deletion was not associated with adverse outcomes in U-CLL (Supplementary Table 10). This likely reflects the use of contemporary therapies such as ibrutinib and venetoclax where TP53 mutation alone has not been shown to influence prognosis50,51.
Table 1:
Patient characteristics in clinical analyses
| Overall* N (%) | IGHV mutated N (%) | IGHV unmutated N (%) | Treatment Naïveǂ N (%) | Treatment Naïveǂ IGHV mutated N (%) | Treatment Naïveǂ IGHV unmutated N (%) | Expression Cluster Cohort N (%) | Integrated Analysis N (%) | |
|---|---|---|---|---|---|---|---|---|
| N, Patients | 1009 | 518 | 476 | 640 | 393 | 247 | 603 | 506 |
| Site | ||||||||
| UCSD | 21 (2) | 8 (2) | 13 (3) | 21 (3) | 8 (2) | 13 (5) | 20 (3) | 17 (3) |
| DFCI | 172 (17) | 103 (20) | 69 (15) | 138 (22) | 96 (24) | 42 (17) | 105 (17) | 64 (13) |
| GCLLSG | 278 (28) | 107 (21) | 160 (34) | 0 (0) | 0 (0) | 0 (0) | 206 (34) | 172 (34) |
| MDACC | 22 (2) | 0 (0) | 21 (4) | 2 (<1) | 0 (0) | 2 (1) | 0 (0) | 0 (0) |
| NHLBI | 68 (7) | 23 (4) | 45 (9) | 46 (7) | 19 (5) | 27 (11) | 11 (2) | 10 (2) |
| ICGC | 448 (44) | 277 (53) | 168 (35) | 433 (68) | 270 (69) | 163 (66) | 261 (43) | 243 (48) |
| Treatment Naïve | 920 (91) | 500 (97) | 407 (86) | 640 (100) | 393 (100) | 247 (100) | 603 (100) | 0 (0) |
| Age at time of Sample yrs, median (range) | 63 (19, 94) | 65 (32, 90) | 61 (19, 94) | 65 (19, 94) | 66 (32, 90) | 62 (19, 94) | 63 (32, 91) | 63 (34, 91) |
| <60 yrs. | 375 (37) | 163 (31) | 208 (44) | 227 (35) | 118 (30) | 109 (44) | 226 (37) | 189 (37) |
| ≥60 yrs. | 634 (63) | 355 (69) | 268 (56) | 413 (65) | 275 (70) | 138 (56) | 377 (63) | 317 (63) |
| Sex | ||||||||
| Male | 655 (65) | 308 (59) | 336 (71) | 384 (60) | 218 (55) | 166 (67) | 405 (67) | 342 (68) |
| Female | 354 (35) | 210 (41) | 140 (29) | 256 (40) | 175 (45) | 81 (33) | 198 (33) | 164 (32) |
| Rai Stage at Dx | ||||||||
| 0 | 368 (36) | 250 (48) | 115 (24) | 347 (54) | 241 (61) | 106 (43) | 222 (37) | 185 (37) |
| 1 | 192 (19) | 74 (14) | 113 (24) | 105 (16) | 53 (13) | 52 (21) | 122 (20) | 101 (20) |
| 2 | 114 (11) | 47 (9) | 65 (14) | 30 (5) | 13 (3) | 17 (7) | 67 (11) | 56 (11) |
| 3 | 15 (1) | 4 (1) | 11 (2) | 7 (1) | 1 (<1) | 6 (2) | 9 (1) | 7 (1) |
| 4 | 31 (3) | 12 (2) | 19 (4) | 8 (1) | 4 (1) | 4 (2) | 18 (3) | 16 (3) |
| Unknown | 290 (29) | 132 (25) | 153 (32) | 143 (22) | 81 (21) | 62 (25) | 165 (27) | 141 (28) |
| IGHV | ||||||||
| mutated | 518 (51) | 518 (0) | 0 (0) | 394 (61) | 393 (100) | 0 (0) | 319 (53) | 272 (54) |
| unmutated | 476 (47) | 0 (0) | 476 (0) | 247 (39) | 0 (0) | 247 (100) | 272 (45) | 234 (46) |
| unknown | 15 (1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 12 (2) | 0 (0) |
| Expression Cluster | ||||||||
| EC-m1 | --- | --- | --- | --- | --- | --- | 53 (9) | 47 (9) |
| EC-u1 | --- | --- | --- | --- | --- | --- | 188 (31) | 152 (30) |
| EC-m2 | --- | --- | --- | --- | --- | --- | 48 (8) | 43 (9) |
| EC-o | --- | --- | --- | --- | --- | --- | 21 (3) | 19 (4) |
| EC-u2 | --- | --- | --- | --- | --- | --- | 64 (11) | 53 (10) |
| EC-m3 | --- | --- | --- | --- | --- | --- | 54 (9) | 47 (9) |
| EC-m4 | --- | --- | --- | --- | --- | --- | 113 (19) | 92 (18) |
| EC-i | --- | --- | --- | --- | --- | --- | 62 (10) | 53 (10) |
| Epitype (n=874)** | ||||||||
| memory | 342 (39) | --- | --- | --- | --- | --- | --- | 216 (43) |
| intermediate | 141 (16) | --- | --- | --- | --- | --- | --- | 79 (16) |
| naïve | 391 (45) | --- | --- | --- | --- | --- | --- | 211 (42) |
| Copy number alterations*** | ||||||||
| tri(12) | 149 (15) | 52 (10) | 94 (20) | 90 (14) | 34 (9) | 56 (23) | --- | 68 (13) |
| del(13q14.3) | 488 (48) | 293 (56) | 188 (40) | 306 (48) | 219 (56) | 87 (35) | --- | 255 (50) |
| del(11q) | 169 (17) | 24 (5) | 163 (34) | 83 (21) | 11 (3) | 72 (30) | --- | 87 (17) |
| del(17p) | 89 (9) | 13 (3) | 56 (12) | 30 (5) | 9 (2) | 21 (9) | --- | 31 (7) |
excluding patients sampled because they had enrolled on a treatment trial
Abbreviations: UCSD, University of California San Diego. DFCI, Dana-Farber Cancer Institute. GCLLSG, German CLL Study Group. MDACC, MD Anderson Cancer Center. NHLBI, National Heart Lung and Blood Institute. ICGC, International Cancer Genome Consortium.
with OS and sequencing data.
Epitype was not included in the genetics analyses, but it is included for descriptive purposes.
All copy number alterations were defined by GISTIC (Methods). del(17p) and del(11q) includes arm and focal events encompassing TP53 [del(17p) + del(17p13.1)] and ATM [del(11q) + del(11q22.3)].
--- Not analyzed in the cohort.
In summary, aggregation of three separate genomic analyses of the entire cohort (n=984), M-CLL (n=512), and U-CLL (n=459) revealed a total of 97 putative CLL driver genes and 105 sCNAs in addition to U1 and IGLV3–21R110 mutations (Fig. 2f). Our previous studies demonstrated that 8.9% of patients lacked an identifiable driver1,2. In our current analysis, the percent of patients lacking at least one potential driver was reduced to 3.8%. These patients without identifiable drivers were predominantly M-CLL (Fisher’s Exact test p=1.04×10−7; 6.6% relative to 0.6% in U-CLL), confirming yet another distinction between the IGHV subtypes2.
CLL subtypes based on epigenomic and transcriptomic features
In addition to subtypes based on IGHV status, genome-wide DNA methylation studies previously identified three epigenetic groups (epitypes), defined based on distinct methylation profiles of pre- and post-germinal center experienced B cells: naive-like CLL (n-CLL, predominantly U-CLL), intermediate CLL (i-CLL, mix of M-CLL and U-CLL), and memory-like CLL (m-CLL, predominantly M-CLL)6,7. Furthermore, cell division results in epigenetic imprints that correlate with the proliferative history of the cell. A mitotic clock score called epigenetically-determined cumulative mitoses (epiCMIT) has further delineated prognosis within epitypes where higher epiCMIT scores corresponded with worse prognosis52. Epitypes and epiCMIT were defined previously7,52 using 450k DNA methylation arrays (n=490), but we also developed and validated new methodologies to incorporate reduced representation bisulfite sequencing data (RRBS) (n=509) (Methods, Extended Data Fig. 7a–f, Supplementary Table 2 and 12). Evaluating the entire dataset (n=999), we found that the two main sources of variation in the CLL DNA methylome are explained by components of cellular memory: the cell of origin (epitype) and the proliferative history of the cell (epiCMIT) (Fig. 3a).
Figure 3: CLL subtypes based on epigenetic and transcriptomic features.
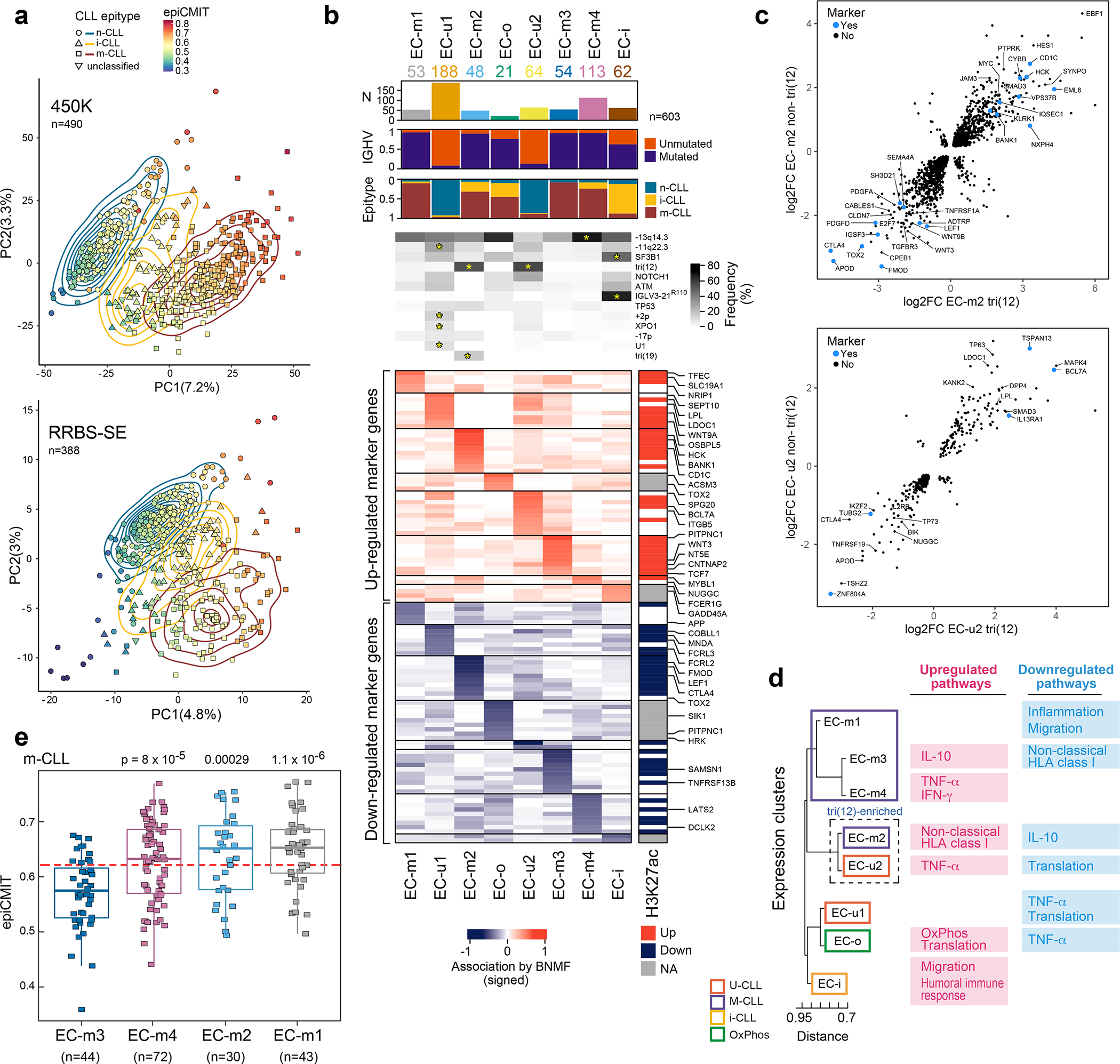
a. Main sources of variability in the DNA methylome are epitype and epiCMIT as determined by unsupervised principal component analysis in samples analyzed by 450k methylation array (top, n=490) or single-end reduced representation bisulfite sequencing (RRBS-SE, bottom, n=388).
b. Eight gene expression clusters (ECs, columns) were identified by Bayesian non-negative matrix factorization (BNMF) method in 603 treatment-naive samples. Heatmap demonstrates associated upregulated (red) and downregulated (blue) marker genes for each cluster (rows) with select genes (right, see Supplementary Table 13). Right vertical panel demonstrates upregulated (red) or downregulated (blue) histone 3 lysine 27 acetylation (H3K27ac) in regulatory regions for each marker gene; EC-o and EC-i H3K27ac was not assessed due to low sample size (NA, gray). Header - number of samples in ECs; association with IGHV subtype (M-CLL, purple; U-CLL, orange); epitype (n-CLL, blue; i-CLL, yellow; m-CLL, red). Frequency of common CLL alterations is shown for each EC. Significant associations - asterisks (q<0.1, curveball algorithm, Methods).
c. Differential gene expression of tri(12)-positive and -negative cases in EC-m2 (top) and EC-u2 (bottom) compared to all other M-CLL or U-CLLs, respectively (EC marker genes shown in blue).
d. Dendrogram of ECs with associated upregulated and downregulated biologic pathways determined by gene set enrichment analysis (see Extended Data Fig. 9b).
e. Cellular proliferative history, represented by epiCMIT, varied in ECs enriched with m-CLL epitype. EC-m3 had significantly lower epiCMIT relative to EC-m1, EC-m2, and EC-m4 (p-values by two-sided t-test; unadjusted). The dashed red line marks the mean epiCMIT in all m-CLLs (n=404). Boxplots: center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range.
While the overall DNA methylome mainly reflects the cellular past of each CLL, the present phenotypic state can be determined by investigating transcriptomes. By applying Bayesian non-negative matrix factorization for unsupervised clustering of RNA-seq data from 603 treatment-naive CLL samples, we identified 8 robust expression clusters (ECs) (Fig. 3b, Extended Data Fig. 8a–d, Supplementary Table 13, Supplementary Note). The ECs strongly associated with IGHV mutational status and/or epitype, revealing two subtypes of U-CLL/n-CLL (EC-u1, EC-u2) and four subtypes of M-CLL/m-CLL (EC-m1, EC-m2, EC-m3, and EC-m4) (Supplementary Table 13). EC-i was best defined by the i-CLL epitype whereas EC-o, the smallest cluster (n=21; 3.5%), was not significantly associated with any previously defined CLL group. Both EC-i and EC-o displayed borderline identity of somatic hypermutations in IGHV with germline, close to the 98% threshold distinguishing M-CLL from U-CLL (Extended Data Fig. 8e).
Although most ECs associated with IGHV status and epitype, expression-based clustering further refined and defined subsets within these conventional distinctions. However, 8% of samples had discordant IGHV status and EC assignment (i.e., M-CLLs included in EC-u clusters or vice versa). As an example of these discordant cases, we observed that 8 M-CLLs clustered in EC-u2, comprising 13% of this EC-u cluster. IGHV mutation rate for discordant cases was compared to those with concordant expression profiles, and while a small difference in mean percent identity in U-CLL was detected (t-test p=0.032, 99.65% versus 99.96% means, respectively), no difference was found among M-CLL cases (p=0.24, 93.96% versus 93.25%) (Extended Data Fig. 8f). Although correctly classified, some discordant cases had borderline IGHV status (97.5–98.5% IGHV identity; n=7) consistent with enrichment of the i-CLL epitype (17% in discordant vs. 8.3% in concordant samples, Fisher’s Exact test p=0.03). Interestingly, CHD2 alterations were overrepresented in discordant M-CLL cases where 45% had either CHD2 mutation or loss of 15q26.1 encompassing CHD2 (p=0.002).
We further explored whether the ECs were enriched with specific drivers. Indeed, EC-u1 was associated with loss of 11q22.3, gain 2p, and XPO1 and U1 mutations, whereas EC-u2 displayed enrichment of tri(12) (q<0.1) (Fig. 3b, Supplementary Table 13). EC-m2 was also associated with tri(12), occurring in 56%, as well as tri(19)53. SF3B1 and IGLV3–21R110 mutations were both enriched in EC-i (53% and 77%, respectively), which is consistent with previous work demonstrating their association with the i-CLL epitype54. Conversely, EC-m1 was enriched with driverless patients (24% of M-CLLs, Fisher’s Exact test q=0.004, odds-ratio 4.9; considering M-CLLs only). In addition to assessing genetic alterations, we analyzed which ECs displayed major stereotyped immunoglobulin genes, which are found in 13.5% of CLL and are divided into subsets that associate with clinical outcome55. All EC-m clusters had a lower proportion of major stereotyped B cell receptors (BCRs, 4–6%), whereas there was a higher incidence in the other ECs (14–20%) (Extended Data Fig. 8g). EC-i was associated with CLL stereotyped subset #2 and IGLV3–21 gene expression consistent with IGLV3–21R110 mutations previously described in this subset54 (Extended Data Fig. 8h–i).
Although genetic events were associated with most ECs, they cannot fully capture these expression phenotypes, which reflect an ensemble of genetic, epigenetic and other biological effects. EC-m2 and EC-u2, for example, were strongly associated with tri(12) events, but these occurred in only 56% and 67% of their samples, respectively. To delineate if a non-genetic unifying phenotype was present, we separately compared the tri(12)-positive and -negative subsets of EC-m2 and EC-u2 to M-CLL or U-CLL samples in other ECs, respectively (Fig. 3c). EC-m2 tri(12)-positive and -negative cases shared overexpression of HES1, MYC and EBF1, which encodes a regulator of B-cell differentiation previously associated with tri(12)9, as well as downregulation of Wnt signaling genes (WNT3, WNT9B, and LEF1). EC-u2 cases shared downregulation of pro-apoptotic genes, TP7356 and BIK57, and overexpression of MAPK4, which activates prosurvival pathways59. Thus, non-tri(12) samples ‘phenocopy’ the tri(12) samples within each of these clusters.
To further explore the biological differences among the ECs, we identified marker genes that were significantly upregulated or downregulated and which were respectively supported by increased or decreased histone 3 lysine 27 acetylation levels (H3K27ac, a mark of active regulatory elements) (Methods, Fig. 3b, Extended Data Fig. 8j–k, Supplementary Table 13). The top upregulated marker genes in EC-u1 included SEPT10 and LPL, which have been previously described in U-CLL and associated with poor prognosis58. Another upregulated EC-u1 gene, OSBPL5, was the top expression marker predicting shorter time to progression after treatment with fludarabine, cyclophosphamide, and rituximab59.
Differentially expressed genes in each EC reflected heterogeneity in biological pathways that was captured by gene set enrichment analysis (Methods, Fig. 3d, Extended Data Fig. 9a–b, Supplementary Table 13). Although EC-o was not associated with IGHV status or epitype, it was defined by enrichment in oxidative phosphorylation signaling (q=4.7×10−15). The EC-m clusters were distinguished by either upregulated or downregulated inflammatory signaling or antigen expression via nonclassical HLA class I. The EC-u clusters shared gene expression changes reflecting impaired protein translation, but differed in TNFα signaling. EC-i was enriched for pathways regulating migration and the humoral immune response, possibly reflecting autonomous BCR signaling by IGLV3–21R110. Finally, we compared the epiCMIT scores of the ECs within each epitype. In EC-m clusters, EC-m3 had the lowest epiCMIT, consistent with a lower proliferative history and suggestive of better patient outcomes (Fig. 3e).
To evaluate the robustness of EC classification and its potential application for prognostication in new samples, we built an EC classifier based on differentially expressed genes, which achieved ~80% overall accuracy (Methods). Performance was particularly high for EC-m3 and EC-i, which had perfect positive predictive value (PPV) at ~85% recall (Supplementary Table 13). By computing EC-specific precision-recall (PR) curves (average area under the curve = 0.88), we show that restricting predictions to the higher-confidence cases can improve performance (Supplementary Table 13, Extended Data Fig. 9c–f). Importantly, similar performance was achieved when training the models with only 26 genes (Extended Data Fig. 9g). Applying the classifier to samples that were excluded from the initial EC discovery (n=105; 44% were post-treatment) and to an external CLL cohort (n=136)60 showed comparable EC distributions per sample set and similar compositions of IGHV subtypes per EC, supporting the generalizability of these ECs (Extended Data Fig. 9h–i; Methods). Finally, by analyzing longitudinally sampled CLL specimens from 19 patients, we confirmed EC stability over years of disease in most cases (p<10−6 by permutation, Methods, Extended Data Fig. 9j). This provides further evidence that the ECs are generally a stable readout, with EC shifts potentially reflecting clonal evolution, both of which are useful for prognostication.
Integrative analysis predicts outcome
Multivariable analysis integrating clinical features and IGHV status confirmed independent prognostic impact of the ECs on FFS (n=603, p<0.001) and OS (p=0.007) (Methods, Table 1, Supplementary Table 11 and 14). The EC-u clusters had similarly short FFS and EC-i displayed intermediate FFS (Fig. 4a). However, outcomes in EC-m clusters were distinct where EC-m1, EC-m2, and EC-m4 demonstrated shorter FFS relative to EC-m3, the cluster with best prognosis and lowest epiCMIT score. Differentiation of EC-m clusters was also evident when evaluating OS (Fig. 4b). This confirmed ECs as an independent prognostic factor in CLL, particularly in distinguishing between EC-m clusters.
Figure 4: Expression clusters and integrated analysis predicts clinical outcome.
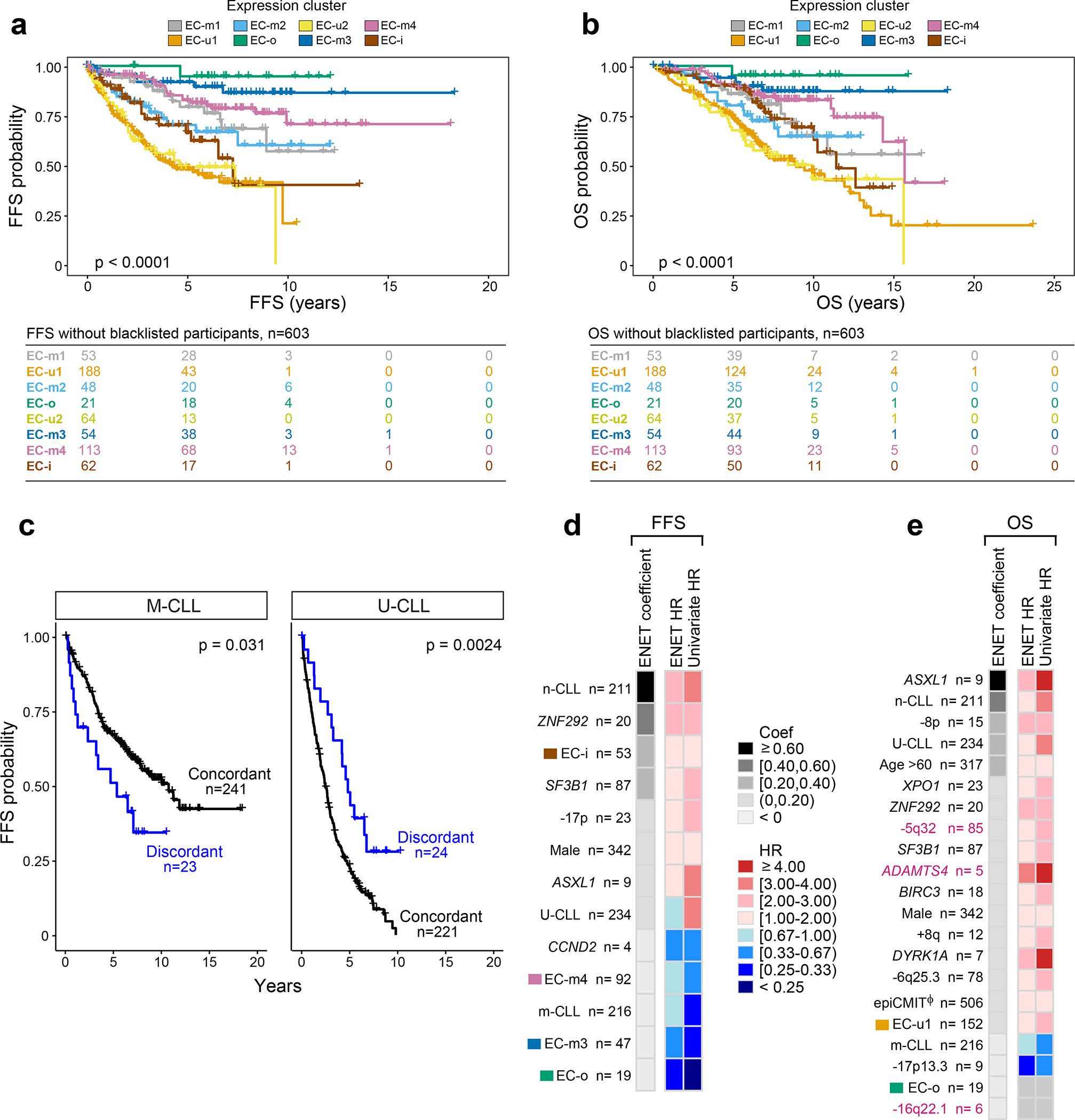
a-b. Kaplan Meier analysis of the impact of expression clusters on (a) failure free survival (FFS) and (b) overall survival (OS) probabilities in 603 treatment-naive samples (log-rank test).
c. Kaplan Meier analysis assessing the difference in FFS probability between samples with concordant IGHV status and ECs (e.g., M-CLLs in EC-m clusters) versus those that are discordant (e.g., M-CLLs in EC-u clusters). M-CLLs - left; U-CLLs - right. Log-rank test (two-sided; unadjusted p-values).
d-e. Genetic, epigenetic, and transcriptomic features associated with (d) FFS and (e) OS in treatment-naive samples (n=506). Events ranked by elastic net (ENET) coefficients, which identifies variables to be included in the model, shrinking coefficients to 0 when excluded. Heatmap denotes hazard ratios (HR) for ENET and univariate Cox regressions (see Supplementary Table 14). Continuous variable - Φ (epiCMIT).
Focusing on 47 cases for which there was discordance between their IGHV status and EC, we asked whether this discordance influenced outcome. FFS was shorter in discordant M-CLLs and longer in discordant U-CLLs relative to the concordant cases (log-rank test p=0.031 and p=0.0024, respectively) (Fig. 4c). For instance, median FFS of discordant M-CLLs (i.e., M-CLLs in EC-u clusters) was 5.3 years compared to 10.7 years in concordant cases (M-CLLs in EC-m clusters), thus revealing added prognostic value of the ECs relative to traditional classification.
To systematically assess the features contributing to outcome, we integrated IGHV subtype, genetic alterations, epitypes, epiCMIT and ECs in a multivariable model (Fig. 4d–e, Supplementary Table 14). The n-CLL epitype emerged as a strong predictor of FFS and OS, emphasizing the known importance of cell of origin. IGHV status and epiCMIT also influenced OS to a greater degree than FFS. A limited set of previously identified genetic alterations were associated with shorter FFS (ZNF292, SF3B1, ASXL1, and 17p deletion), but 11 adversely affected OS including novel events such as loss of 5q32. We noted the absence of known alterations, such as ATM and NOTCH1, which were significant by univariate analysis only. This likely reflects co-occurrence with other prognostic factors, similar to what we observed with TP53 and 17p deletion (Supplementary Table 14). Specific ECs were particularly informative in the model, with EC-i associated with adverse FFS and EC-o, EC-m3 and EC-m4 as protective. Altogether, this integrated model reveals a refined prognostic paradigm where genetics, epigenetics, and gene expression classification all contribute to clinical outcome.
DISCUSSION
Through integration of harmonized multiomic data, this work has expanded the molecular map of CLL and provided additional insights into its biological and clinical heterogeneity. The number of previously unrecognized putative drivers was doubled, thus achieving a more complete genetic basis for this cancer. These alterations highlight important cellular pathways not previously impacted by candidate drivers that may provide opportunities for development of new therapies in the future. Beyond cataloging the overall landscape, we delineated the distinction between its molecular subtypes by comprehensively analyzing the CLL genome, epigenome, and transcriptome. IGHV subtypes were enriched in unique genetic driver alterations leading to divergent clonal trajectories. We found a significant increase in genetic heterogeneity in U-CLL with more putative drivers relative to M-CLL. Notably, the driverless samples were almost exclusively M-CLL2, suggestive of alternative mechanisms of leukemogenesis in this subtype. Despite lower genetic complexity, M-CLL displayed increased transcriptional diversity segregating mainly into four ECs, which had different proliferative histories. Furthermore, the discovery of expression clusters expands our contemporary disease framework. While specific ECs were associated with IGHV status, epitypes, and genetic events, none of these previously defined groups completely captured the phenotypic diversity exhibited in the expression profiles. Additionally, identifying discordant cases with gene expression profiles inconsistent with their IGHV status was prognostic and CHD2 alterations may be contributing to this changed phenotype in M-CLL. This reveals the complex nature of CLL and provides the first version of a comprehensive molecular atlas of CLL that forms the basis for further exploration of unique mechanisms of pathogenesis.
By integrating these biological insights with patient outcomes, we highlighted the prognostic implications of even rare genetic events within IGHV subtypes, such as mutations in ZC3H18 and RFX7. Incorporating these data in a unified model revealed the importance of integrating multiple data layers in this disease. Critical components associated with outcome included the ECs, novel genetic alterations such as loss of 5q32 in addition to known factors including the cell of origin (IGHV status and epitype), proliferative history (epiCMIT), 17p deletion, and SF3B1 mutations. This refines our current disease paradigm and establishes a new spectrum of events contributing to leukemogenesis that may have implications beyond prognostication. In the future, this molecular foundation may allow for better prediction of response to therapy or provide the basis for rational combination of novel agents.
METHODS
Data Availability
The molecular data used in this study are publicly available and are included in the following patient cohorts (Table 1, Supplementary Table 1–2, Extended Data Fig. 1a): Dana-Farber Cancer Institute (DFCI), German CLL Study Group (GCLLSG), International Cancer Genome Consortium (ICGC), MD Anderson Cancer Center (MDACC), National Heart Lung and Blood Institute (NHLBI) and University of California San Diego (UCSD). Sequencing, expression, and genotyping is available at European Genome-Phenome Archive (EGA, http://www.ebi.ac.uk/ega/), which is hosted at the European Bioinformatics Institute (EBI), under accession numbers EGAS00000000092 (ICGC cohort) and in dbGaP under accession numbers: phs001473.v2.p1 (MDACC, NHLBI), phs000922.v2.p1 (GCLLSG), phs001431.v2.p1 (DFCI, UCSD), phs001091.v1.p1 (MDACC), phs000435.v3.p1 (DFCI), phs002297.v2.p1 (NHLBI), phs000879.v1.p1 (DFCI) and GEO accession number GSE143673 (GCLLSG). 450k array data is available at EGA under accession number EGAD00010001975 (ICGC). Project data portal: https://cllmap.org.
Code availability
Terra methods used in the study can be found at https://app.terra.bio/#workspaces/broad-firecloud-wupo1/CLLmap_Methods_Apr2021. Source code used in the study can be found at https://github.com/getzlab/CLLmap. The RFcaller pipeline is available at https://github.com/xa-lab/RFcaller. The new epiCMIT suitable for Illumina arrays and NGS approaches as well as the CLL epitype classifier can be found at https://github.com/Duran-FerrerM/CLLmap-epigenetics.
Human samples
The characteristics of the 1154 CLL/MBL samples from 1148 patients are described in Supplementary Table 1 and clinical characteristics of the 1009 CLL samples used in the clinical analysis are listed in Table 1. These included tumor and germline samples collected either during active surveillance (n=680), post-treatment (n=52)1–3,11, or at enrollment of a clinical trial prior to first cycle of therapy (n=416; treatment-naive n=371, relapsed/refractory n=45)1,10,12,13,61. Briefly, these trials included: (i) comparison of fludarabine and cyclophosphamide (FC) to FC-rituximab (FCR) in previously untreated patients (CLL8 trial, n=309)1,61; (ii) treatment-naive TP53 mutated patients within phase 2 CLL20 trial who all received alemtuzumab (n=31)62; (iii) ibrutinib or R-ibrutinib in relapsed/refractory (R/R) or untreated patients with 17p deletion, TP53 mutation, and/or 11q deletion (n=76; treatment-naive n=31; R/R n=45)10,12,13. Written informed consent was obtained from all patients. Samples were collected via protocols approved by institutional review boards or ethics and policy committees from the International Cancer Genome Consortium, German CLL Study Group, Dana-Farber Cancer Institute, the CLL Research Consortium, National Heart, Blood and Lung Institute, and MD Anderson Cancer Center. All clinical trials were conducted in accordance with the Declaration of Helsinki and International Conference on Harmonization Guidelines for Good Clinical Practice. If multiple samples were obtained from a patient, then the earliest collected sample was selected for analysis. Peripheral blood mononuclear cells were isolated and DNA and/or RNA were extracted and prepared with protocols varying between the different studies1–3,10–13,61. Briefly, either positive or negative immunomagnetic selection of CLL cells was performed in either all samples or those with low white blood cell counts, depending on the study. DNA was extracted using Qiagen kits (Qiagen Inc.) and RNA was obtained either using RNAeasy kit (Qiagen Inc.) or Trizol reagent (Invitrogen Life Technologies) per manufacturer’s instructions.
Molecular data retrieval and assembly
We retrieved previously reported sequencing data from CLL and MBL samples, including 984 whole-exome sequences1–3, 177 whole-genome sequences2,11, 448 RNA-seqs2,3,10,13,63, 490 methylation 450k arrays2 and 547 reduced-representation bisulfite sequencing64. Additionally, we sequenced 264 RNA-seq samples, and performed targeted DNA sequencing of the NOTCH1 3’ UTR for 293 samples (Supplementary Note). Single nucleotide polymorphism (SNP)-based fingerprinting comparisons within and between these sequencing data types were conducted with CrosscheckFingerprints65 for quality control to remove data redundancy and to verify patient-matched data, where appropriate.
Sequence data processing and analysis
All sequencing data (WES, WGS, RNA-seq, RRBS and targeted NOTCH1 sequencing) were processed and analyzed using methods implemented in the Broad Institute’s cloud-based Terra platform (https://app.terra.bio). The main Terra methods are available at https://app.terra.bio/#workspaces/broad-firecloud-wupo1/CLLmap_Methods_Apr2021 in addition to the detailed descriptions herein.
WES/WGS alignment and quality control
We processed all DNA sequence data through the Broad Institute’s data processing pipeline. For each sample, this pipeline combines data from multiple libraries and flow cell runs into a single BAM file. This file contains reads aligned to the human genome hg19 genome assembly (version b37) done by the Picard and Genome Analysis Toolkit (GATK)66 developed at the Broad Institute, a process that involves marking duplicate reads, recalibrating base qualities and realigning around indels. Reads were aligned to the hg19 genome assembly (version b37) using BWA-MEM (version 0.7.15-r1140).
Mutation calling
Prior to variant calling, the impact of oxidative damage (oxoG) to DNA during sequencing was quantified using DeToxoG67. The cross-sample contamination was measured with ContEst based on the allele fraction of homozygous SNPs68, and this measurement was used in the downstream mutation calling pipeline. From the aligned BAM files, somatic alterations were identified using a set of tools developed at the Broad Institute (www.broadinstitute.org/cancer/cga). The details of our sequencing data processing have been described elsewhere23,69. Briefly, for sSNVs/indel detection, high-confidence somatic mutation calls were made by applying MuTect70, MuTect271 and Strelka272 to WES/WGS sequencing data. Given that normal blood samples might also contain CLL cells, we used DeTiN73 to estimate tumor in normal (TiN) contamination in order to recover falsely rejected sSNVs/indels. Next, we applied four types of filters: (i) a realignment-based filter, which removes variants that can be attributed entirely to ambiguously mapped reads; (ii) an orientation bias filter, which removes possible oxoG and FFPE artifacts67; (iii) a ContEst filter, which removes variants that might come from other samples due to contamination; and (iv) an allele fraction specific panel-of-normals filter, which compares the detected variants to a large panel of normal exomes or genomes and removes variants that were observed in the two panel-of-normals (PoNs): one consists of 8,334 normal samples in TCGA while the other consists of 481 CLL-matched normal samples with TiN estimates of 0. All four filters together contributed to the exclusion of potential false-positive events (e.g., commonly occurring germline variants or sequencing artifacts), which ultimately yielded the final list of mutations. All filtered events in candidate CLL driver genes were also manually reviewed using the Integrated Genomics Viewer (IGV)74.
In order to increase the sensitivity and precision of mutation calls in candidate driver genes, an additional variant calling step was performed for the candidate driver gene loci using RFcaller (https://github.com/xa-lab/RFcaller), a pipeline that uses read-level features and extra trees/random forest algorithms for the detection of somatic mutations. This pipeline was run with default parameters for WES or WGS data, as well as for RNA-seq data for NOTCH1, which has low coverage in hotspot regions in some samples due to high GC content. All candidate mutations that passed filters and were detected by both pipelines were considered positives. Mutations detected by only one of the callers were visually inspected by a set of at least four expert curators, considering the following exclusion criteria: (i) low evidence due to limited number of reads supporting the mutation in the tumor sample or excessive mutant reads in the normal sample; (ii) low depth of coverage to rule out germline variant; (iii) low base quality region; (iv) low mapping quality region leading to multi-mapped reads; (v) calls supported by reads with a strong strand bias.
Identification of significantly mutated genes
To identify candidate cancer genes using our mutation calls from WES, we first used SignatureAnalyzer75 to identify mutational processes and potential artifact signatures. We discovered a signature likely due to the bleedthrough sequencing artifact and then filtered mutations with greater than 95% chance attributed to that bleedthrough signature. Next, we ran MutSig2CV76 to identify driver genes from the filtered WES Mutation Annotation Format (MAF) file. A stringent manual review was conducted using the IGV74 to review the mutations in the driver genes and further exclude low evidence calls. Then we reran MutSig2CV on the filtered set of mutation calls from WES to identify the final candidate driver genes. In addition, we also used CLUMPS21 (https://github.com/getzlab/getzlab-CLUMPS2) to identify driver genes based on clustering of mutations in the 3D structure of the protein product (see Supplementary Table 5). For CLUMPS, we applied two FDR corrections: one for all candidates and a second restricted hypothesis testing focused on genes in the COSMIC Cancer Gene Census38. Finally, for further stringency and to exclude candidates irrelevant to CLL biology, we discarded candidate genes that were not expressed in RNA-seq of 603 treatment-naive CLL samples, using a one-sided t-test testing for difference from 0 in TPM space. This discarded 15 candidate genes (Supplementary Table 4).
Copy number analysis
For detecting somatic copy number alterations (sCNAs) we used the GATK4 CNV pipeline (http://github.com/gatk-workflows/gatk4-somatic-cnvs), which involves the CalculateTargetCoverage, NormalizeSomaticReadCounts, and Circular Binary Segmentation (CBS) algorithms77 for genome segmentation. In order to identify candidate sCNA drivers (genomic regions that are significantly amplified or deleted), we then apply GISTIC 2.078. To exclude potential germline CNAs, we first ran GISTIC 2.0 on the matched normal samples and then concatenated the recurrent CNAs this outputted (q < 0.1) to the blacklisted regions. Then we ran GISTIC 2.0 on the tumor samples to produce a list of candidate sCNA driver regions. A force-calling process was applied to identify the presence/absence of each sCNA driver event across tumor samples (https://github.com/getzlab/GISTIC2_postprocessing). To further filter the potential false positive drivers, we only accepted sCNA drivers with population frequency greater than 1%. Finally, all filtered sCNA drivers were manually reviewed using IGV74 to exclude drivers that are based on sCNA events with low supporting evidence or that were localized close to centromeres. sCNA drivers were annotated by intersection with our list of CLL mutation driver genes and with genes in the COSMIC Cancer Gene Census38 (v90; Supplementary Table 7).
Structural variants calling
For structural variation (SV) detection, our pipeline (the Broad Institute’s Cancer Genome Analysis (CGA) SV pipeline)79 integrates evidence from three structural variation detection algorithms (Manta80, SvABA81 and dRanger23,69,82) to generate a list of structural variation events with high confidence. We followed the three SV detection tools with BreakPointer83 to pinpoint the exact breakpoint at base-level resolution. SVs calls were filtered if called by less than 2 tools or if they were identified in a panel of normal samples79. Next, breakpoint information was aggregated per sample to identify: (i) balanced translocations, which were defined as those with breakpoints on reverse strands within 1-kb of each other; (ii) inversions supported on both ends; (iii) complex events, based on the number of clustered events within 50-kb of each other (Supplementary Table 9). Breakpoints were annotated by intersection with our lists of CLL driver genes and significant sCNA regions, as well as with genes in the COSMIC Cancer Gene Census (v90)38 (Supplementary Table 9). These SV calls were compared to SVs called in Puente et al2, from which an additional 90 SVs were added after manual review. Clonal events were defined as those with cancer cell fraction (CCF) ≥ 0.75 and identified using the CGA SV pipeline algorithm (https://github.com/getzlab/REBC_tools;v1.1.3 )79. This method could be applied to the 569 SVs detected by the CGA SV pipeline, which provides the required information for CCF calculation, out of which we could successfully estimate CCF for 558 (98%)79. IgCaller45 (v1.1) was used to identify additional structural variants involving immunoglobulin genes (Supplementary Note).
Immunoglobulin (IG) gene characterization
The IG heavy (IGH) and light (IGL) chain gene rearrangements and mutational status were obtained from WGS/WES and RNA-seq using IgCaller (v1.1)45 and MiXCR (v.3.0.10)84, respectively. The rearrangements obtained were visually inspected in IGV74. The obtained sequences were used as input in IMGT/V-QUEST (v3.5.18; release 202018–4)85 to confirm gene annotations and mutational status. IGH gene rearrangements were complemented with Sanger sequencing available for 1076 cases. IGH and IGL characterization from the different sources were integrated and compared and used to infer IGLV3–21 R110 mutation status. See Supplementary Note and Supplementary Table 8.
RNA-seq analysis
RNA-seq data was processed in Terra using the GTEx V7 pipeline (https://github.com/broadinstitute/gtex-pipeline). Briefly, reads were aligned with STAR (v2.6.1d)86 to hg19 (b37) using the GENCODE v19 annotation, and quality control metrics and gene expression were computed with RNA-SeQC v2.3.6 (https://github.com/getzlab/rnaseqc)87. A collapsed version of the GENCODE annotation was used to quantify gene-level expression (available from gs://gtex-resources/GENCODE/gencode.v19.genes.v7.collapsed_only.patched_contigs.gtf). TPMs were used for sample clustering, while gene counts were used for differential gene expression, as required. See Supplementary Table 2 for sequencing and quality metrics.
RNA expression cluster detection
Gene-level TPMs were estimated with RNA-SeQC (v2.3.6) for RNA-seq from 603 treatment-naive CLL (https://github.com/getzlab/rnaseqc)87. Genes expressed at less than 0.1 TPM in 10% of samples were discarded, retaining 11,119 genes, which were batch corrected (as described below), followed by selection of the top 2,500 most varying genes. The clustering methodology combines consensus hierarchical clustering and Bayesian non-negative matrix factorization, as previously described88. Further details about the methodology and machine learning classifier are provided in Supplementary Note.
DNA methylation data processing
We analyzed DNA methylome data for a total of 1,037 samples, including 490 samples profiled with Illumina 450k array previously analyzed52 (EGA accession EGAD00010001975), and 547 samples profiled using reduced representation bisulfite sequencing (RRBS, with either single-end (SE), or paired-end (PE) approaches; Supplementary Table 2)64. We developed a pipeline in Terra to obtain the CpG methylation estimates from RRBS data (Supplementary Note). The epitype classifier and the epiCMIT mitotic clock were previously developed for Illumina 450K and EPIC array data52 and we therefore adapted the methods for the RRBS data (Supplementary Note).
Statistical Methods
Unless otherwise stated, two-sided t-test was used for mean comparison and multiple testing was corrected to compute false discovery rate (FDR, q) by the Benjamini-Hochberg procedure89. Categorical enrichments were computed using a two-sided Fisher’s Exact test unless otherwise stated.
Clinical outcome modeling
Failure-free survival (FFS) was calculated for treatment-naïve patients as the time from the date of the sequenced sample to the date of first treatment (“natural progression”), progression (if the patient was sampled at the time of enrollment on a clinical trial) or death, and censored at the last known event-free date. In the genetics-focused analysis (Supplementary Table 10), the first event was defined as time to next treatment in patients who received therapy within 30 days. Subset analysis included patients who were treatment-naïve at the time of the sequenced sample and not enrolled on a therapeutic clinical trial; in this analysis, time between sample and date of first treatment was used. Overall survival (OS) was calculated as the time from the date of the sequenced sample to the date of death and censored at the date last known alive. Patient characteristics and number included in each clinical outcome analysis are defined in Table 1. Univariate and multivariable Cox regression models were constructed for each subset of data. Final models were selected using the glmnet function for regularized Cox regression using an elastic net penalty within the Coxnet package in R. Ten-fold cross-validation using the cv.glmnet function with a partial-likelihood deviance metric to minimize λ was performed and the minimum CV-error model was used. The alpha was set to 1 corresponding to a Lasso penalty. The maximum iterations (maxit) parameter was set to 1000. Features identified as having non-zero coefficient values using elastic net and selected in the final model were then included in a Cox regression model to obtain the hazard ratios. These hazard ratios estimate the magnitude of effect but p-values and confidence intervals are not readily interpretable in the elastic net model and are therefore not reported. For the integrated analysis of all available datatypes (Supplementary Table 14), variables including expression cluster and epitype categories were dummy coded. Prognostic significance of expression cluster and IGHV status were also considered using a chi-squared test with the difference in −2log likelihood (−2logL) between models including sSNVs and sCNAs. The Breslow approximation was used for handling ties in survival time.
Extended Data
ED Fig 1. Dataset description and representative driver gene maps.
a. Full dataset (n=1148), with contributions by cohort and data type delineated (see Supplementary Table 1). b. Numbers of samples with genomic, epigenomic, and transcriptomic data. c. 3D protein structures of representative genes identified by CLUMPS in pan-CLL analysis (n=984, see Supplementary Table 5). Mutated residues - red labels. A peptide from RAF1 (designated at bottom-center, in complex with 14–3-3 zeta) shows clustered mutations around S259, whose phosphorylation regulates RAF1 activity and is a cancer mutational hotspot90 that, when mutated, perturbs the interaction with the 14–3-3 zeta and upregulates RAF1 kinase activity91,92. In DICER1, mutations occur in the RNase III domain (purple), including the cancer hotspot residue E181321,24. This region is critical for Mg2+ binding and is required for ribonuclease activity to process microRNAs and mediate post-transcriptional gene regulation93. RPS23 mutations are clustered in a conserved loop of the ribosomal decoding center, surrounding P62, whose post-translational hydroxylation affects translation termination accuracy94. These RPS23 mutations have a median CCF >80% (Extended Data Fig. 6d; Supplementary Table 3). d. Individual mutations maps of selected novel, putative driver genes. Mutation subtype and position are shown. e. Selected genes identified by CLUMPS in IGHV subtypes; mutated residues - red. Although BRAF was not identified as a potential M-CLL driver via MutSig2CV (see Extended Data Fig. 3, Methods), CLUMPS revealed three mutated sites clustered in the kinase domain (purple) that are cancer hotspots24, thus confirming BRAF as a shared driver (left). Mutated residues in BRAF in U-CLL (bottom) are shown for comparison, revealing a greater number of clustered mutations relative to M-CLL. In U-CLL, novel mutations were found in RRM1 (right). Somatic alterations were clustered in the N-terminal ATP-binding site (purple) and therefore have potential to impact enzymatic activity95.
ED Fig 2. CLL biological pathways affected by candidate driver genes.
a. Schema of CLL pathways containing previously identified (black) and novel (magenta) putative driver genes (see Supplementary Table 6). Novel drivers cluster in central processes driving CLL (e.g., DNA damage, chromatin modification, RNA processing)1,2, but also highlight new pathways not previously implicated by driver genes (e.g., cytoskeleton and extracellular matrix, proteostasis, metabolism). Asterisks - mutated genes discovered by CLUMPs. b. Stacked barplot ranked by the number of candidate driver genes per CLL pathway. Magenta bars show the number of newly identified drivers in each pathway.
ED Fig 3. Candidate driver alterations discovered in IGHV subtypes.
a-b. Landscape of putative driver genes and sCNAs in M-CLL (a, n=512) and U-CLL (b, n=459) with associated frequencies (rows, barplots). Header tracks annotate cohort, IGHV status (purple, M-CLL; orange, U-CLL), disease type (blue, CLL; yellow, MBL), epitype (blue, n-CLL; yellow, i-CLL; red, m-CLL), datatype (white, WES; yellow, WGS; blue, both); prior treatment, U1 and IGLV3–21R110 mutations are annotated in black; magenta label - novel alterations; asterisks - discovery by CLUMPS.
ED Fig 4. Chromosomal gains and losses identified in IGHV subtypes.
a-b. Recurrent copy number gains (left) and losses (right) by GISTIC analysis showing arm level (left per plot) and focal events (right per plot) in M-CLL (a, n=512) and U-CLL (b, n=459). Chromosomes are labeled along the vertical axis; dashed line - significance at q=0.1. Blacklisted regions are colored gray. All arm level events are labeled with cytoband arm and frequency in cohort. Focal events are annotated by cytoband, frequency, number of genes encompassed in peak (bracketed), and genes of interest. Red/blue font: novel focal events with frequency >2%. Black font: previously identified events (see Supplementary Table 7).
ED Fig 5. Landscape of driver alterations and chromosomal aberrations in IGHV subtypes.
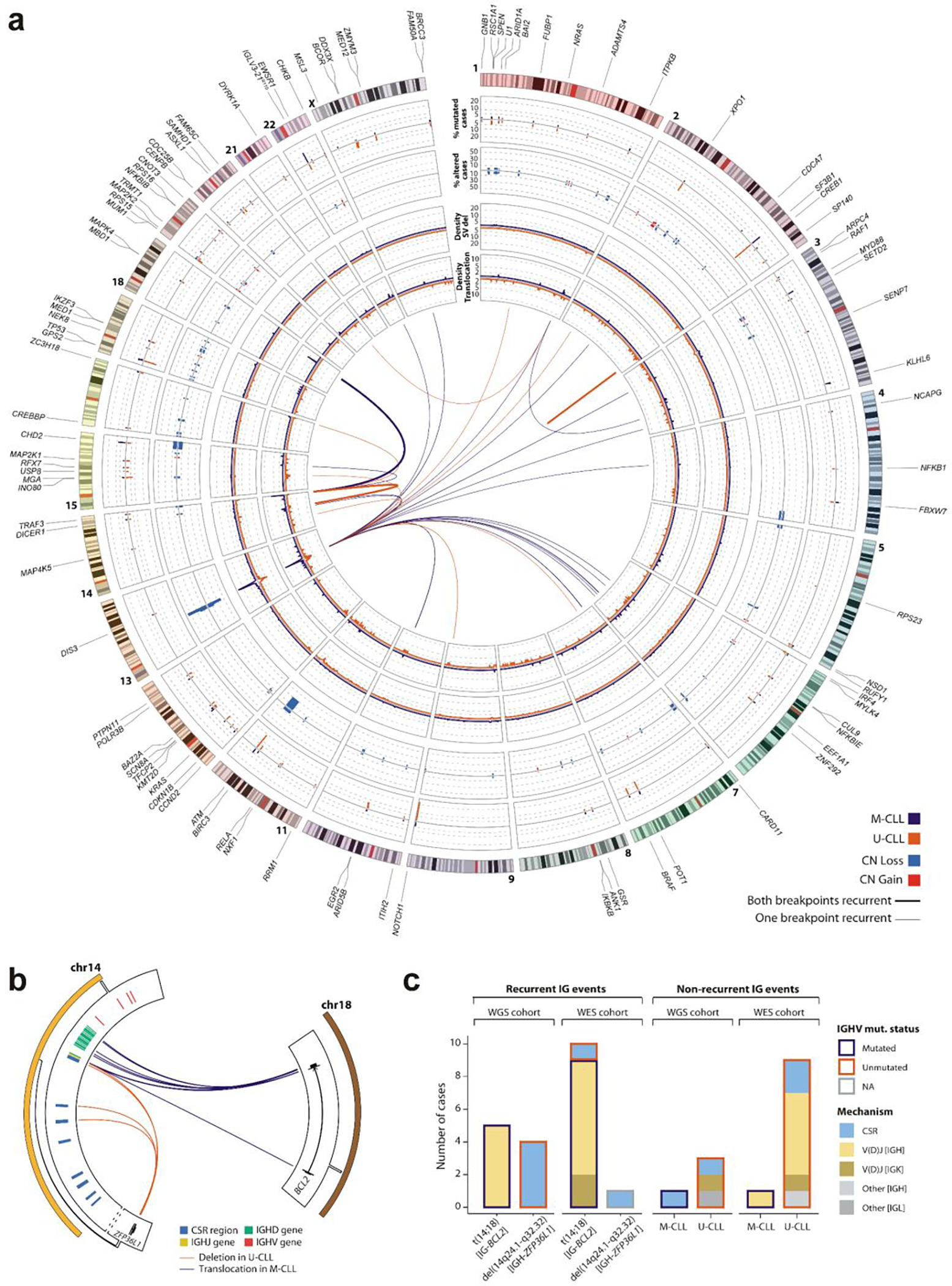
a. The genomic landscape of CLL IGHV subtypes. Driver genes, U1 and IGLV3–21R110 mutations are labeled according to their genomic location (outside ring, numbered by chromosome). The tracks show the frequency and locations of driver genes in M-CLL (purple) vs. U-CLL (orange) (track 1; outermost), focal sCNAs (track 2; gains, red; losses, blue), and density of SV breakpoints of deletions (track 3) and translocations (track 4) (M-CLL n=88; U-CLL n=87; WGS, windows of 1-Mb). Innermost plot highlights translocations in which either one or both breakpoints are recurrent in at least 3 cases (windows of 1-Mb considered to define recurrence) in M-CLL (purple) and U-CLL (orange). Deletions, inversions, and tandem duplications where both breakpoints were found in at least 2 cases and did not overlap with a driver sCNA are shown (Note: only focal deletion in SP140 in two U-CLL cases met this criterion. b. Schema of recurrent IG-BCL2 translocation and IGH-ZFP36L1 deletion in the WGS cohort. All 5 BCL2 translocations were in M-CLL with immunoglobulin (IG) breakpoints in J or D genes, suggesting mediation by aberrant V(D)J recombination. In contrast, 4 U-CLL cases carried IGH-ZFP36L1 truncating deletions, which were all clonal (CCF=1). Breakpoints in IGH class-switch regions suggested mediation by aberrant class-switch recombination (CSR). c. Immunoglobulin (IG) SVs in 177 WGS and 984 WES. In WES, 9 of 10 BCL2 translocations were in M-CLL and mediated by aberrant V(D)J recombination in IGH (n=7) or IGK (n=2). The sole BCL2 translocation in U-CLL was due to aberrant CSR. One CSR-mediated IGH-ZFP36L1 deletion was observed in a case with unclassified IGHV status due to presence of two populations (one M-CLL, one U-CLL; the latter was more prevalent). Of note, in WES, U-CLLs carry a higher number of non-recurrent IG events than M-CLL.
ED Fig 6. Mutational mechanisms and cancer cell fractions of candidate drivers.
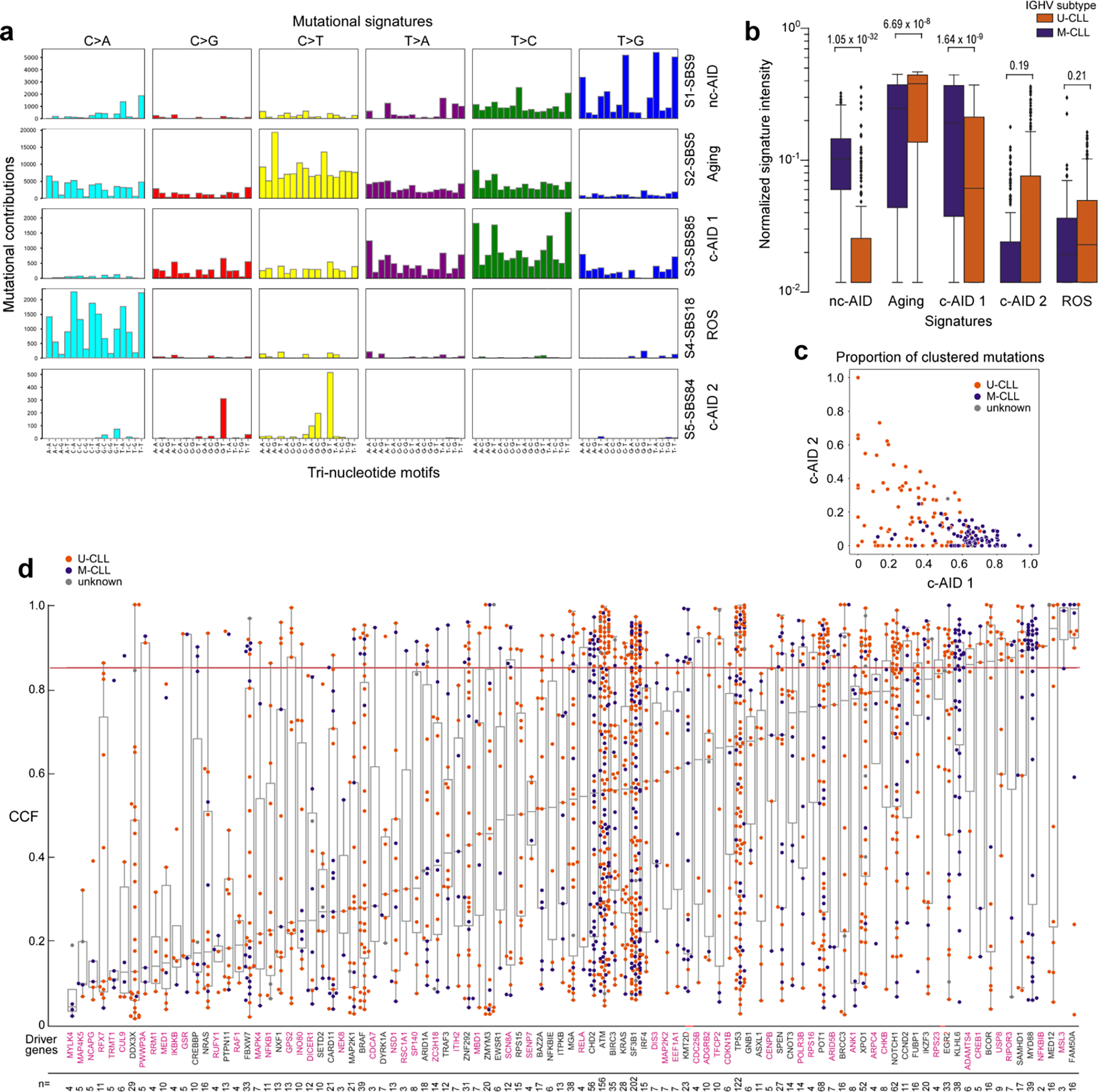
Eight mutational signatures were identified in 177 WGS, but 3 signatures corresponded to known artifacts and were therefore excluded (see Supplementary Note 2). Boxplots demonstrating mutation contribution for each of the 5 signatures are labeled with single-base substitution (SBS) number and identity (per COSMIC v3.1). b. Comparison of the normalized signature intensity of the mutational signatures in U-CLL (orange, n=87) vs. M-CLL (purple, n=88). The nc-AID and c-AID 1 signatures were enriched in M-CLL, whereas the aging signature was more prevalent in U-CLL. Although not significant, there was a trend of increased mutations due to the c-AID 2 signature in U-CLL. All p-values were calculated with Wilcoxon rank-sum test, two-sided. Boxplots: center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers. c. Proportions of clustered mutations contributed by the two c-AID related signatures (SBS84, c-AID 1 vs. SBS85, c-AID 2) for each IGHV subtype (M-CLL, purple; U-CLL, orange) d. Mean cancer cell fraction (CCF) for each non-silent mutation across all candidate driver genes identified in WES samples (n=984). Color of dots depicts the IGHV subtype (M-CLL, purple; U-CLL, orange). The horizontal red line is the threshold for clonality (CCF>85%). Magenta labels - newly identified putative driver genes. The number of non-silent mutations per driver gene is shown at the bottom. Boxplots: center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range.
ED Fig 7. Development and validation of epitype assignment and epiCMIT in RRBS data.
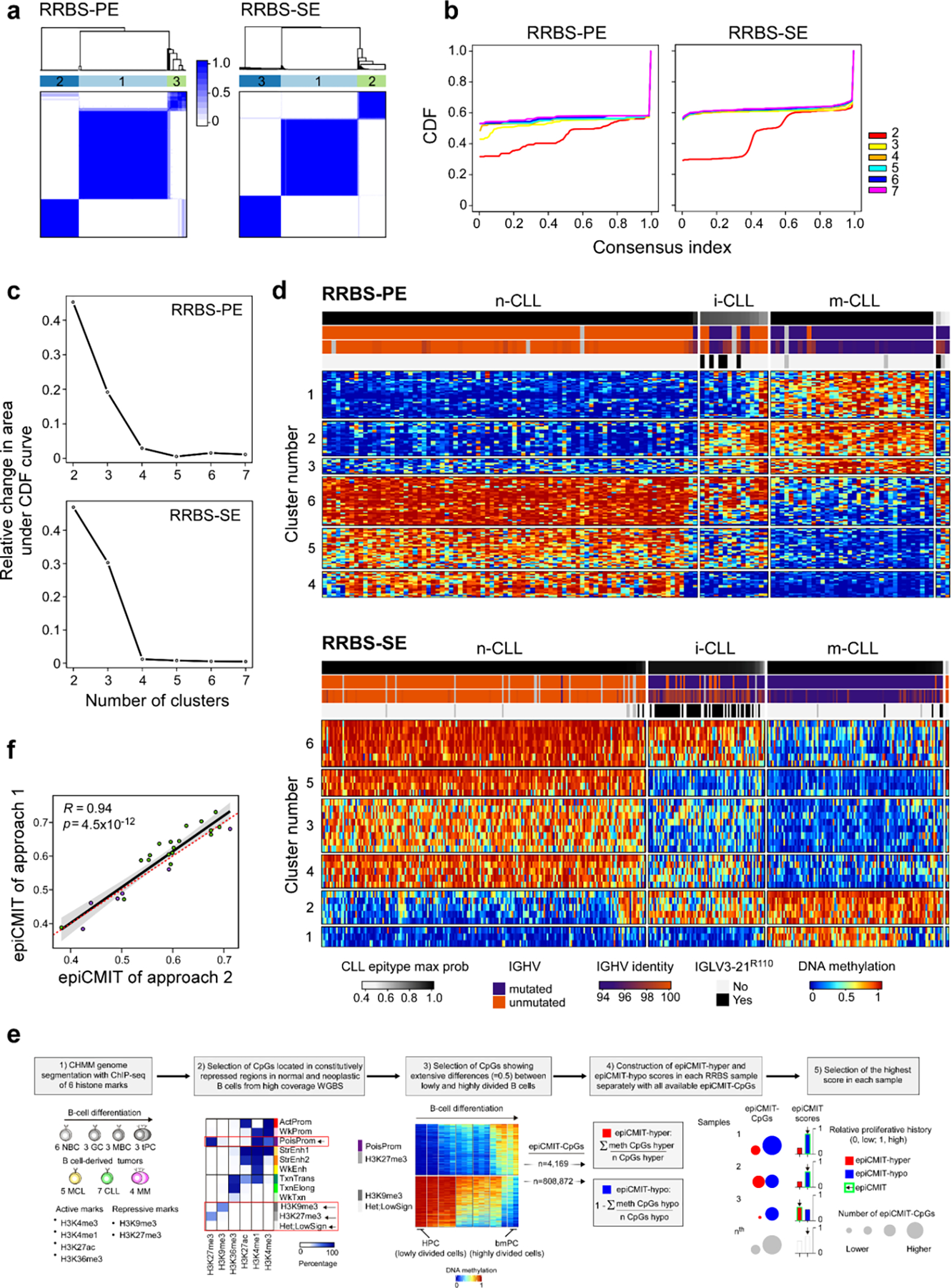
a. Consensus clustering matrices for K=3 groups for paired-end (n=136; 153 CpGs in consensus matrix) and single-end (n=388; 32 CpGs) RRBS data. (d). b. Empirical cumulative distribution functions (CDFs) for consensus matrices with K=2 to K=7. c. Relative change under the CDF for K=2 to K=7. d. Heatmaps of the CpGs used for consensus clustering in (a). Each sample (columns) is annotated by tracks: epitype max probability, IGHV status (M-CLL, purple; U-CLL, orange), IGHV percent identity, and presence of IGLV3–21R110 mutation (black). e. The development of the new epiCMIT methodology for RRBS data. The genome was segmented into Chromatin Hidden Markov Model (CHMM)96 states using ChIP-seq data to get repressed chromatin regions, where differential DNA methylation analyses was performed in high coverage whole-genome bisulfite sequencing (WGBS) data between the cells with the lowest and highest accumulated cell divisions in the B-cell lineage, namely the hematopoietic precursor cells (HPC) and bone-marrow plasma cells (bmPC). Only CPGs showing extensive differences were retained and constituted the epiCMIT-hyper CpGs or epiCMIT-hypo CpGs depending whether they gain or lose DNA methylation from 0.9 to ≤0.5 from HPC to bmPC, respectively. EpiCMIT-hyper and epiCMIT-hypo scores were calculated according to the available epiCMIT-CpGs per sample, and the higher score in each sample was then selected. f. epiCMIT values on the same samples profiled twice with different platforms. Approach 1 - profiled with Illumina-450k (green); approach 2 - profiled with RRBS-PE (violet). In samples profiled with Illumina 450k, the original epiCMIT-CpGs were used52. In samples profiled with RRBS, epiCMIT was calculated with all available epiCMIT-CpGs for the new catalog (e, Methods). P-value by Pearson correlation test, two-sided; Error band - 95% confidence intervals of the Pearson correlation coefficient.
ED Fig 8. Identification of expression clusters with associated biologic features.
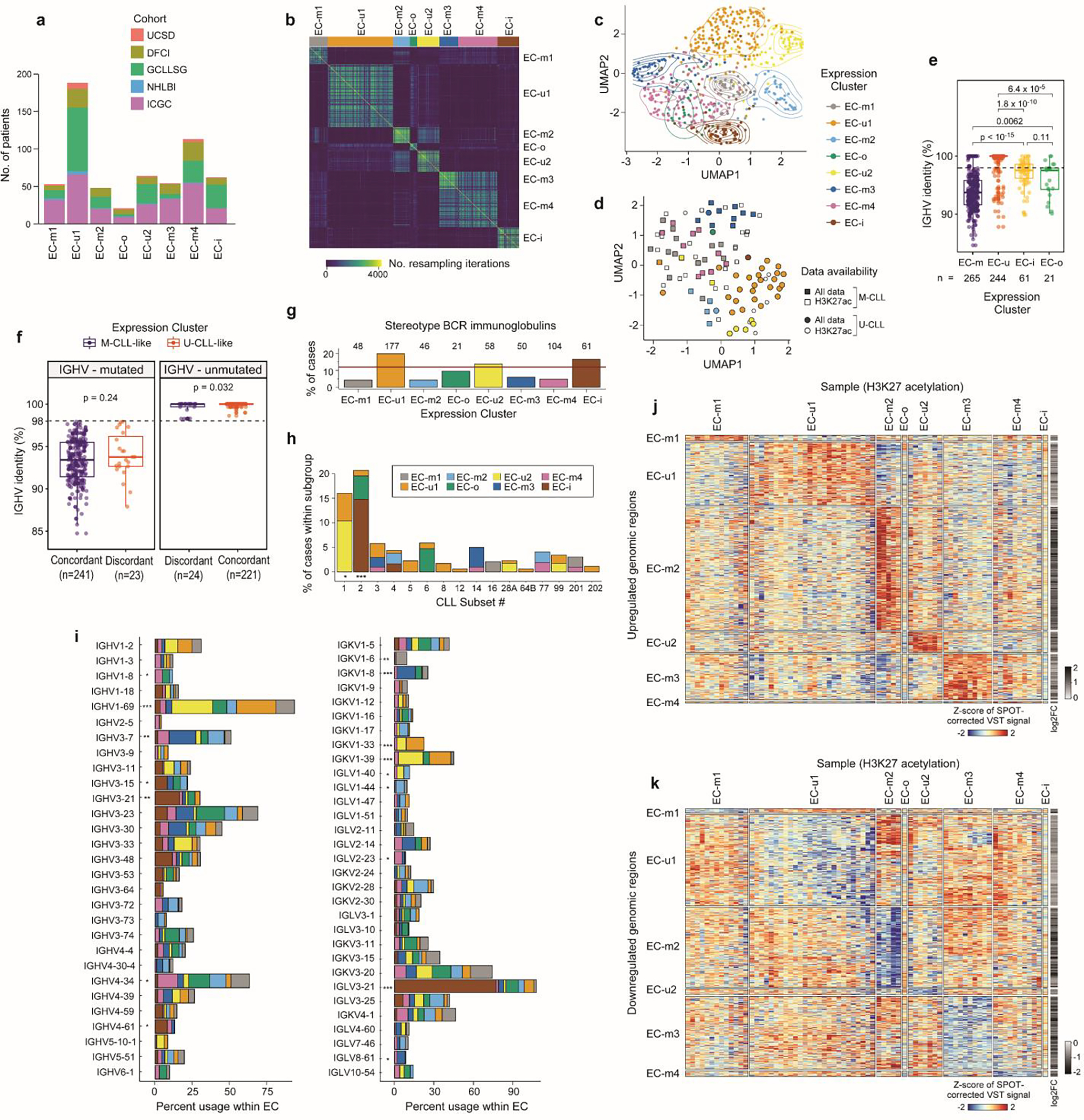
a. Cohort representation in each expression cluster. b. Consensus matrix for RNA expression profiles of 603 treatment-naive CLLs by repeated hierarchical clustering with 80% resampling and varying cutoffs for number of clusters, which is inputted to the BayesNMF procedure (Methods). c. Uniform manifold approximation and projection (UMAP) showing clustering of ECs (n=603; EC-u clusters (top), EC-m and EC-o (middle), EC-i (bottom)). Analysis was performed using the marker genes identified by BayesNMF. d. UMAP of H3K27ac profiles (n=104)8 denoting EC designation where available (colored points, n=73) and IGHV status. e. Comparison of the percent IGHV identity among ECs. Dotted line: 98% threshold defining M-CLL and U-CLL. P-values by two-sided t-tests. Boxplots: center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range. f. Comparison of the percent IGHV identity between those samples with concordant IGHV status and ECs (e.g., M-CLLs in EC-m clusters) versus the discordant samples (e.g., M-CLLs in EC-u clusters). IGHV mutated cases - left; IGHV unmutated samples - right. P-values by two-sided t-tests. Boxplots: center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range. g. Percentage of cases carrying stereotyped immunoglobulin genes within each EC. Red horizontal line: percentage of stereotyped cases in the whole cohort. h. Fraction of cases classified in each CLL stereotype subset according to their EC. i. Percentage of IGHV (left) and IG(K/L)V (right) gene usage within each EC. IGKV genes from proximal and distal clusters were merged for simplification. All p-values were calculated using Chi-squared tests corrected by the Benjamini-Hochberg procedure (q-values, q). q < 0.1; *, q < 0.05; **, q < 0.001; ***, q < 0.0001. j-k. Heatmaps showing upregulated (j) and downregulated (k) H3K27ac levels of EC marker genes and 2,000 bp upstream to capture regulatory regions (Methods).
ED Fig 9. EC differential gene expression, pathway activity, and classifier.
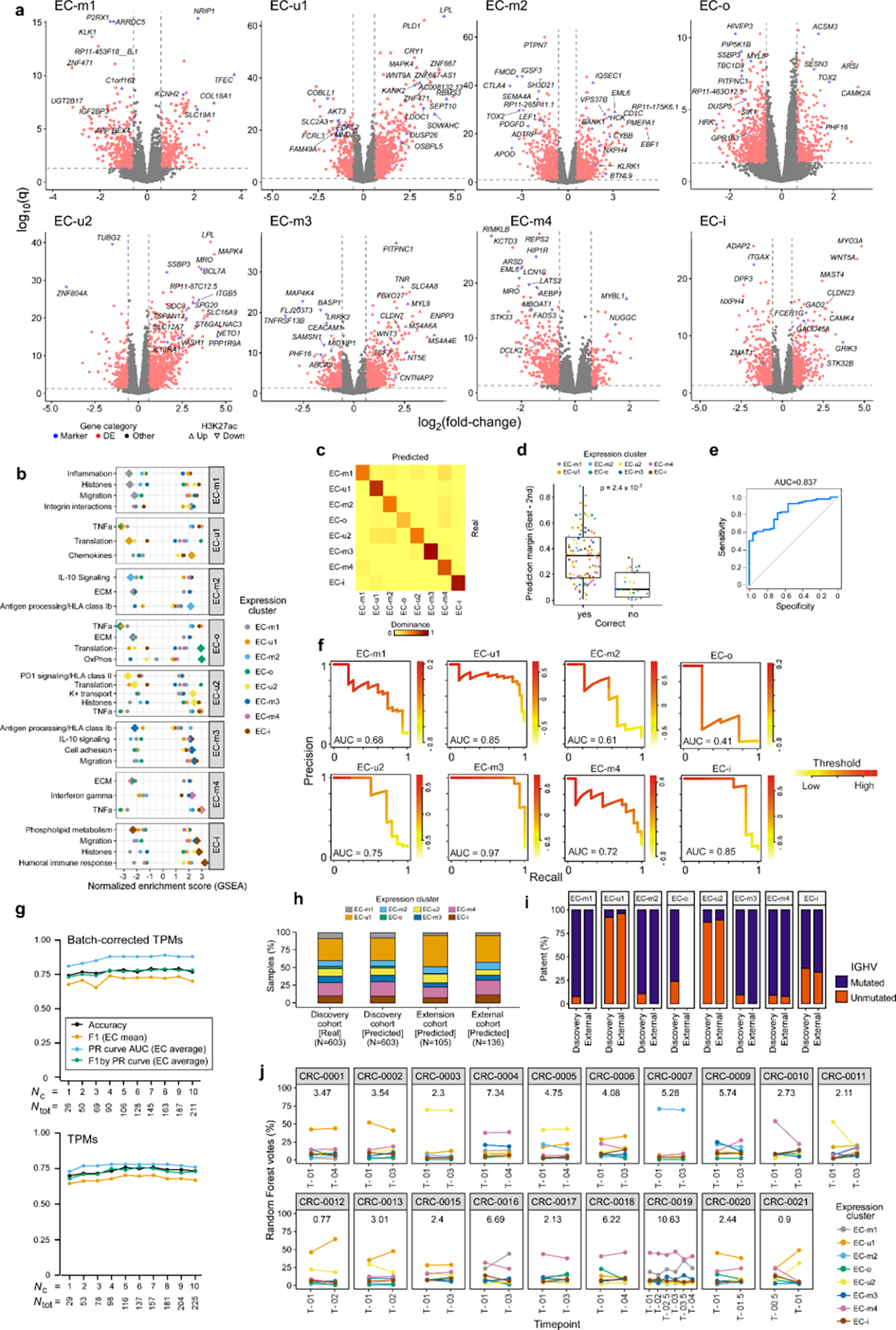
Differentially expressed genes per EC (red) using discovery set (n=603); EC marker genes by BayesNMF (blue). Significant up- or down-regulation of H3K27ac levels are directionally marked with triangles (ChIP-seq available for n=73; n=1 for EC-o and EC-i, thus unevaluable). b. EC gene set enrichment analysis (GSEA). Diamond denotes the EC compared to all others (circles). c. Confusion matrix for the EC classifier on the test set (“Dominance” defined in Methods). d. Confidence in correctly classified samples (n=95) is greater than for incorrectly classified samples (n=25; two-sided t-test). “Prediction margin” defined in Methods. Boxplots: center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range. e. Receiver-operator curve (ROC) showing the tradeoff between sensitivity and specificity for the range of cutoffs that can be applied based on the “prediction margin”, where samples under the cutoff are excluded from performance evaluation. AUC, area under curve. f. Precision-recall (PR) curves for EC classification performance on the test set (n=120), using the selected model (see Methods). The weighted average of AUC is 0.88. g. Performance metrics for models trained with differing amounts of input genes, demonstrating accuracy even with smaller gene sets. Metrics: Accuracy, overall; Average, weighted average across ECs (Methods). Nc, Ntot - number of genes (see Methods). h. EC distributions by BayesNMF compared to classifier predictions on the discovery cohort (n=603), an extension cohort not included discovery (n=105), and an external CLL cohort (n=136)60. i. IGHV status distributions per EC in discovery (n=603) and external (n=136) cohorts. The difference in IGHV-mutated samples per EC is 2–10% (p>0.05, Fisher’s Exact, Methods). j. Stability of the ECs over time in longitudinally sampled CLL samples3. Sample timepoints (x-axis); years between first and last sample (above curve).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Wandi Zhang, Satyen Gohil, Ignaty Leshchiner, Dimitri Livitz, Daniel Rosebrock, John Gribben, Kanti R. Rai, Michael J. Keating, Julian M. Hess, Nicholas J. Haradhvala, Armand Mohammed and Andi Gnirke for helpful discussions. We thank Candace Patterson, Sam Pollock, Kara Slowik, Oriol Olive, Conner J. Shaughnessy and Hayley Lyon for assistance in data collection and organization. We thank the patients, their families and the investigators of the clinical trials for providing samples and clinical data. This study was supported by NIH/NCI P01 CA206978 (to C.J.W. and G.G.) and the Broad/IBM Cancer Resistance Research Project (G.G. and L.P.). B.A.K. was supported by a long-term EMBO fellowship (ALTF 14-2018). C.K.H. was supported by the NHLBI Training Program in Molecular Hematology (T32HL116324). S.S. and E.T. were supported by the DFG (SFB1074, subproject B1, B2 and B10). A.W. and C.Su. were supported by the Intramural Research Program at NIH/NHLBI. J.A.B. was supported by MD Anderson’s Moon Shot Program in CLL, the CLL Global Research Foundation, and in part by the MD Anderson Cancer Center Support Grant CA016672. S.L. was supported by the NCI Research Specialist Award (R50CA251956). J.R.B. was supported by NIH R01 CA 213442, NIH/NCI P01 CA206978 and the Melton Family Foundation. X.S.P. acknowledges funding by the Spanish Ministerio de Economía y Competitividad (MINECO) (Grant No. SAF2017-87811-R and PID2020–117185RB-I00). A.D-N. was supported by the Department of Education of the Basque Government (PRE_2017_1_0100) and P.B-M. by a fellowship by MINECO. This study was supported by “la Caixa” Foundation (CLLEvolution- LCF/PR/HR17/52150017, Health Research 2017 Program “HR17-0022” to E.C.), the European Research Council under the European Union’s Horizon 2020 research and innovation program (Project BCLLATLAS, grant agreement 810287) (to J.I.M.-S. and E.C.), the Accelerator award CRUK/AIRC/AECC joint funder-partnership (to J.I.M.-S.), Generalitat de Catalunya Suport Grups de Recerca AGAUR 2017-SGR-1142 (to E.C.) and 2017-SGR-736 (to J.I.M.-S.), CERCA Programme / Generalitat de Catalunya. E.C. is an Academia Researcher of Catalan Institution for Research and Advanced Studies (ICREA).
Footnotes
COMPETING INTEREST DECLARATION
The authors declare the following conflicts related to the CLLmap project: C.J.W. receives research support from Pharmacyclics. E.C. has been a consultant for Illumina. G.G. receives research funds from IBM and Pharmacyclics; and is an inventor on patent applications related to SignatureAnalyzer-GPU. S.S. reports honoraria for consultancy, advisory board membership, speaker honoraria, research grants and travel support from AbbVie, Amgen, AstraZeneca, Celgene, Gilead, GSK, Hoffmann La-Roche, Janssen, Novartis. C.J.W., G.G., B.A.K., Z.L. and C.K.H. are inventors on a patent “Compositions, panels, and methods for characterizing chronic lymphocytic leukemia” (PCT/US21/45144). The following conflicts are unrelated to the CLLmap project: F.N. has received honoraria from Janssen for speaking at educational activities. E.T. declares research support by Abbvie and Roche; Advisory Boards and Speakers Bureau for Janssen, Abbvie and Roche. A.W. received research funding from Pharmacyclics, Acerta, Merck, Verastem, Genmab, Nurix. J.R.B. has served as a consultant for Abbvie, Acerta/Astra-Zeneca, Beigene, Bristol-Myers Squibb/Juno/Celgene, Catapult, Genentech/Roche, Janssen, MEI Pharma, Morphosys AG, Novartis, Pfizer, Rigel; received research funding from Gilead, Loxo/Lilly, Verastem/SecuraBio, Sun, TG Therapeutics; and served on the data safety monitoring committee for Invectys. J.A.B. received research support from AstraZeneca, BeiGene, Gilead, and Pharmacyclics; travel and speaker honoraria from Janssen. X.S.P. is a cofounder of and holds an equity stake in DREAMgenics. C.J.W. holds equity in BioNTech, Inc.. E.C. has been a consultant for Takeda and NanoString Technologies; has received honoraria from Janssen and Roche for speaking at educational activities; and is an inventor on a Lymphoma and Leukemia Molecular Profiling Project patent “Method for subtyping lymphoma subtypes by means of expression profiling” (PCT/US2014/64161). G.G. is an inventor on patent applications related to MSMuTect, MSMutSig, MSIDetect, and POLYSOLVER; and is a founder and consultant of and holds privately held equity in Scorpion Therapeutics. The other authors have no competing interests to declare.
REFERENCES
- 1.Landau DA et al. Mutations driving CLL and their evolution in progression and relapse. Nature 526, 525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puente XS et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature 526, 519 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Gruber M et al. Growth dynamics in naturally progressing chronic lymphocytic leukaemia. Nature 570, 474–479 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dvinge H et al. Sample processing obscures cancer-specific alterations in leukemic transcriptomes. Proceedings of the National Academy of Sciences 111, 16802–16807 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferreira PG et al. Transcriptome characterization by RNA sequencing identifies a major molecular and clinical subdivision in chronic lymphocytic leukemia. Genome Res. 24, 212–226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oakes CC et al. DNA methylation dynamics during B cell maturation underlie a continuum of disease phenotypes in chronic lymphocytic leukemia. Nat. Genet. 48, 253–264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulis M et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat. Genet. 44, 1236–1242 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Beekman R et al. The reference epigenome and regulatory chromatin landscape of chronic lymphocytic leukemia. Nat. Med. 24, 868–880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bloehdorn J et al. Multi-platform profiling characterizes molecular subgroups and resistance networks in chronic lymphocytic leukemia. Nat. Commun. 12, 5395 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landau DA et al. The evolutionary landscape of chronic lymphocytic leukemia treated with ibrutinib targeted therapy. Nat. Commun. 8, 2185 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasar S et al. Whole-genome sequencing reveals activation-induced cytidine deaminase signatures during indolent chronic lymphocytic leukaemia evolution. Nat. Commun. 6, 8866 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burger JA et al. Safety and activity of ibrutinib plus rituximab for patients with high-risk chronic lymphocytic leukaemia: a single-arm, phase 2 study. Lancet Oncol. 15, 1090–1099 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burger JA et al. Clonal evolution in patients with chronic lymphocytic leukaemia developing resistance to BTK inhibition. Nat. Commun. 7, 11589 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shuai S et al. The U1 spliceosomal RNA is recurrently mutated in multiple cancers. Nature 574, 712–716 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Minici C et al. Distinct homotypic B-cell receptor interactions shape the outcome of chronic lymphocytic leukaemia. Nat. Commun. 8, 15746 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maity PC et al. IGLV3–21*01 is an inherited risk factor for CLL through the acquisition of a single-point mutation enabling autonomous BCR signaling. Proc. Natl. Acad. Sci. U. S. A. 117, 4320–4327 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawrence MS et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleinstern G et al. Tumor mutational load predicts time to first treatment in chronic lymphocytic leukemia (CLL) and monoclonal B-cell lymphocytosis beyond the CLL international prognostic index. Am. J. Hematol. 95, 906–917 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leeksma AC et al. Clonal diversity predicts adverse outcome in chronic lymphocytic leukemia. Leukemia 33, 390–402 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landau DA et al. Evolution and Impact of Subclonal Mutations in Chronic Lymphocytic Leukemia. Cell 152, 714–726 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamburov A et al. Comprehensive assessment of cancer missense mutation clustering in protein structures. Proc. Natl. Acad. Sci. U. S. A. 112, E5486–95 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dziembowski A, Lorentzen E, Conti E & Séraphin B A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 14, 15–22 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Chapman MA et al. Initial genome sequencing and analysis of multiple myeloma. Nature 471, 467–472 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang MT et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat. Biotechnol. 34, 155–163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amblar M, Barbas A, Fialho AM & Arraiano CM Characterization of the Functional Domains of Escherichia coli RNase II. J. Mol. Biol. 360, 921–933 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Papamichos-Chronakis M, Watanabe S, Rando OJ & Peterson CL Global Regulation of H2A.Z Localization by the INO80 Chromatin-Remodeling Enzyme Is Essential for Genome Integrity. Cell 144, 200–213 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKinney M et al. The Genetic Basis of Hepatosplenic T-cell Lymphoma. Cancer Discov. 7, 369–379 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.López C et al. Genomic and transcriptomic changes complement each other in the pathogenesis of sporadic Burkitt lymphoma. Nat. Commun. 10, 1459 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber J et al. PiggyBac transposon tools for recessive screening identify B-cell lymphoma drivers in mice. Nat. Commun. 10, 1415 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edelmann J et al. High-resolution genomic profiling of chronic lymphocytic leukemia reveals new recurrent genomic alterations. Blood 120, 4783–4794 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Setlur SR et al. Comparison of familial and sporadic chronic lymphocytic leukaemia using high resolution array comparative genomic hybridization. Br. J. Haematol. 151, 336–345 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stilgenbauer S et al. Incidence and clinical significance of 6q deletions in B cell chronic lymphocytic leukemia. Leukemia 13, 1331–1334 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Boultwood J et al. Narrowing and genomic annotation of the commonly deleted region of the 5q− syndrome. Blood 99, 4638–4641 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Schneider RK et al. Rps14 haploinsufficiency causes a block in erythroid differentiation mediated by S100A8 and S100A9. Nat. Med. 22, 288–297 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ciccia A et al. Treacher Collins syndrome TCOF1 protein cooperates with NBS1 in the DNA damage response. Proceedings of the National Academy of Sciences 111, 18631–18636 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nowinski SM et al. Mitochondrial uncoupling links lipid catabolism to Akt inhibition and resistance to tumorigenesis. Nat. Commun. 6, 8137 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aguilar E et al. UCP2 Deficiency Increases Colon Tumorigenesis by Promoting Lipid Synthesis and Depleting NADPH for Antioxidant Defenses. Cell Rep. 28, 2306–2316.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sondka Z et al. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer 18, 696–705 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burns A et al. Whole-genome sequencing of chronic lymphocytic leukaemia reveals distinct differences in the mutational landscape between IgHVmut and IgHVunmut subgroups. Leukemia 32, 332–342 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Q, Lenardo MJ & Baltimore D 30 Years of NF-κB: A Blossoming of Relevance to Human Pathobiology. Cell 168, 37–57 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gandhi V & Plunkett W Cellular and Clinical Pharmacology of Fludarabine. Clin. Pharmacokinet. 41, 93–103 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Sellmann L et al. Trisomy 19 is associated with trisomy 12 and mutated IGHV genes in B-chronic lymphocytic leukaemia. Br. J. Haematol. 138, 217–220 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Shilatifard A The COMPASS Family of Histone H3K4 Methylases: Mechanisms of Regulation in Development and Disease Pathogenesis. Annu. Rev. Biochem. 81, 65–95 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleinstern G et al. Tumor mutational load predicts time to first treatment in chronic lymphocytic leukemia (CLL) and monoclonal B-cell lymphocytosis beyond the CLL international prognostic index. Am. J. Hematol. 95, 906–917 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nadeu F et al. IgCaller for reconstructing immunoglobulin gene rearrangements and oncogenic translocations from whole-genome sequencing in lymphoid neoplasms. Nat. Commun. 11, 3390 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hodson DJ et al. Deletion of the RNA-binding proteins ZFP36L1 and ZFP36L2 leads to perturbed thymic development and T lymphoblastic leukemia. Nat. Immunol. 11, 717–724 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oppezzo P et al. Chronic lymphocytic leukemia B cells expressing AID display dissociation between class switch recombination and somatic hypermutation. Blood vol. 101 4029–4032 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Roco JA et al. Class-Switch Recombination Occurs Infrequently in Germinal Centers. Immunity vol. 51 337–350.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leshchiner I et al. Comprehensive analysis of tumour initiation, spatial and temporal progression under multiple lines of treatment. bioRxiv 508127 (2019). [Google Scholar]
- 50.Tausch E et al. Prognostic and predictive impact of genetic markers in patients with CLL treated with obinutuzumab and venetoclax. Blood 135, 2402–2412 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Burger JA et al. Long-term efficacy and safety of first-line ibrutinib treatment for patients with CLL/SLL: 5 years of follow-up from the phase 3 RESONATE-2 study. Leukemia 34, 787–798 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duran-Ferrer M et al. The proliferative history shapes the DNA methylome of B-cell tumors and predicts clinical outcome. Nature Cancer 1, 1066–1081 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sellmann L et al. Trisomy 19 is associated with trisomy 12 and mutatedIGHVgenes in B-chronic lymphocytic leukaemia. British Journal of Haematology vol. 138 217–220 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Nadeu F et al. IGLV3–21R110 identifies an aggressive biological subtype of chronic lymphocytic leukemia with intermediate epigenetics. Blood (2020) doi: 10.1182/blood.2020008311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Agathangelidis A et al. Higher-order connections between stereotyped subsets: implications for improved patient classification in CLL. Blood 137, 1365–1376 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobbelstein M, Strano S, Roth J & Blandino G p73-induced apoptosis: a question of compartments and cooperation. Biochem. Biophys. Res. Commun. 331, 688–693 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Chinnadurai G, Vijayalingam S & Rashmi R BIK, the founding member of the BH3-only family proteins: mechanisms of cell death and role in cancer and pathogenic processes. Oncogene 27 Suppl 1, S20–9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bilban M et al. Deregulated expression of fat and muscle genes in B-cell chronic lymphocytic leukemia with high lipoprotein lipase expression. Leukemia 20, 1080–1088 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Herling CD et al. Time-to-progression after front-line fludarabine, cyclophosphamide, and rituximab chemoimmunotherapy for chronic lymphocytic leukaemia: a retrospective, multicohort study. Lancet Oncol. 20, 1576–1586 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dietrich S et al. Drug-perturbation-based stratification of blood cancer. J. Clin. Invest. 128, 427–445 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 61.Stilgenbauer S et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood 123, 3247–3254 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Stilgenbauer S et al. Alemtuzumab combined with dexamethasone, followed by alemtuzumab maintenance or Allo-SCT in ‘ultra high-risk’ CLL: Final results from the CLL2O phase II study. Blood 124, 1991–1991 (2014). [Google Scholar]
- 63.Wang L et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 365, 2497–2506 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Landau DA et al. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell 26, 813–825 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Javed N et al. Detecting sample swaps in diverse NGS data types using linkage disequilibrium. Nat. Commun. 11, 3697 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKenna A et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Costello M et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 41, e67 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cibulskis K et al. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics 27, 2601–2602 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berger MF et al. The genomic complexity of primary human prostate cancer. Nature 470, 214–220 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cibulskis K et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Benjamin D et al. Calling Somatic SNVs and Indels with Mutect2. bioRxiv 861054 (2019) doi: 10.1101/861054. [DOI] [Google Scholar]
- 72.Kim S et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods 15, 591–594 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Taylor-Weiner A et al. DeTiN: overcoming tumor-in-normal contamination. Nat. Methods 15, 531–534 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robinson JT et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim J et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat. Genet. 48, 600–606 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lawrence MS et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Olshen AB, Venkatraman ES, Lucito R & Wigler M Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 5, 557–572 (2004). [DOI] [PubMed] [Google Scholar]
- 78.Mermel CH et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morton LM et al. Radiation-related genomic profile of papillary thyroid carcinoma after the Chernobyl accident. Science (2021) doi: 10.1126/science.abg2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen X et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32, 1220–1222 (2016). [DOI] [PubMed] [Google Scholar]
- 81.Wala JA et al. SvABA: genome-wide detection of structural variants and indels by local assembly. Genome Res. 28, 581–591 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bass AJ et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat. Genet. 43, 964–968 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Drier Y et al. Somatic rearrangements across cancer reveal classes of samples with distinct patterns of DNA breakage and rearrangement-induced hypermutability. Genome Res. 23, 228–235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bolotin DA et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods 12, 380–381 (2015). [DOI] [PubMed] [Google Scholar]
- 85.Brochet X, Lefranc M-P & Giudicelli V IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36, W503–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Graubert A, Aguet F, Ravi A, Ardlie KG & Getz G RNA-SeQC 2: Efficient RNA-seq quality control and quantification for large cohorts. Bioinformatics (2021) doi: 10.1093/bioinformatics/btab135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Robertson AG et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 171, 540–556.e25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Benjamini Y & Hochberg Y Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300 (1995). [Google Scholar]
- 90.Pandit B et al. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat. Genet. 39, 1007–1012 (2007). [DOI] [PubMed] [Google Scholar]
- 91.Rommel C et al. Activated Ras displaces 14–3-3 protein from the amino terminus of c-Raf-1. Oncogene 12, 609–619 (1996). [PubMed] [Google Scholar]
- 92.Dhillon AS, Meikle S, Yazici Z, Eulitz M & Kolch W Regulation of Raf-1 activation and signalling by dephosphorylation. EMBO J. 21, 64–71 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Provost P et al. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 21, 5864–5874 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Loenarz C et al. Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc. Natl. Acad. Sci. U. S. A. 111, 4019–4024 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Qiu W, Zhou B, Darwish D, Shao J & Yen Y Characterization of enzymatic properties of human ribonucleotide reductase holoenzyme reconstituted in vitro from hRRM1, hRRM2, and p53R2 subunits. Biochem. Biophys. Res. Commun. 340, 428–434 (2006). [DOI] [PubMed] [Google Scholar]
- 96.Ernst J & Kellis M ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The molecular data used in this study are publicly available and are included in the following patient cohorts (Table 1, Supplementary Table 1–2, Extended Data Fig. 1a): Dana-Farber Cancer Institute (DFCI), German CLL Study Group (GCLLSG), International Cancer Genome Consortium (ICGC), MD Anderson Cancer Center (MDACC), National Heart Lung and Blood Institute (NHLBI) and University of California San Diego (UCSD). Sequencing, expression, and genotyping is available at European Genome-Phenome Archive (EGA, http://www.ebi.ac.uk/ega/), which is hosted at the European Bioinformatics Institute (EBI), under accession numbers EGAS00000000092 (ICGC cohort) and in dbGaP under accession numbers: phs001473.v2.p1 (MDACC, NHLBI), phs000922.v2.p1 (GCLLSG), phs001431.v2.p1 (DFCI, UCSD), phs001091.v1.p1 (MDACC), phs000435.v3.p1 (DFCI), phs002297.v2.p1 (NHLBI), phs000879.v1.p1 (DFCI) and GEO accession number GSE143673 (GCLLSG). 450k array data is available at EGA under accession number EGAD00010001975 (ICGC). Project data portal: https://cllmap.org.


