Synopsis
Generalist coral species may play an important role in predicting, managing, and responding to the growing coral reef crisis as sea surface temperatures are rising and reef wide bleaching events are becoming more common. Pocilloporids are amongst the most widely distributed and studied of generalist corals, characterized by a broad geographic distribution, phenotypic plasticity, and tolerance of sub-optimal conditions for coral recruitment and survival. Emerging research indicates that microbial communities associated with Pocilloporid corals may be contributing to their persistence on coral reefs impacted by thermal stress; however, we lack detailed information on shifts in the coral–bacterial symbiosis during bleaching events across many of the reef habitats these corals are found. Here, we characterized the bacterial communities of healthy and bleached Pocillopora damicornis corals during the bleaching events that occurred during the austral summer of 2020 on Heron Island, on the southern Great Barrier Reef, and the austral summer of 2019 on Lord Howe Island, the most southerly coral reef in Australia. Regardless of reef location, significant differences in α and β diversities, core bacterial community, and inferred functional profile of the bleached microbiome of P. damicornis were not detected. Consistent with previous reports, patterns in the Pocilloporid coral microbiome, including no increase in pathogenic taxa or evidence of dysbiosis, are conserved during bleaching responses. We hypothesize that the resilience of holobiont interactions may aid the Pocilloporids to survive Symbiodiniaceae loss and contribute to the success of Pocilloporids.
Introduction
Anthropogenically induced climate change is driving record warm temperatures and rates of warming culminating in large-scale bleaching and mortality events across the world's coral reefs (Hughes et al. 2017; van Woesik and Kratochwill 2022). Bleaching events are becoming increasingly frequent and severe (Hughes et al. 2017, 2018; Eakin et al. 2019; Sully et al. 2019). For example, on Australia's Great Barrier Reef (GBR), anomalously warm temperatures have triggered six mass coral bleaching events since the late 1990’s, four of which have occurred since 2016 (2016, 2017, 2020, and 2022; and GBRMPA 2017, GBRMPA, AIMS, & CSIRO Reef Snapshot 2022). Mean coral cover in the central section of the GBR was also affected by the repeated bleaching events in 2016 and 2017, declining from 22% coverage in 2016 to 14% coverage in 2018 (AIMS 2018), but increasing to 36% in 2022 (AIMS 2022). Bleaching events have also impacted high-latitude reefs over the last two decades, including at Sodwana Bay, South Africa (Celliers and Schleyer 2002), Lord Howe Island (LHI), Australia (Dalton et al. 2011, 2020; Harrison et al. 2011), Rottnest Island, Australia (Thomson et al. 2011), the Houtman Abrolhos Islands, Australia (Abdo et al. 2012; Smale et al. 2012), and Norfolk Island, Australia (Parks Australia Report 2021). Thus, the consequences of ocean warming are evident on coral reefs worldwide, including those located at high latitude reefs that have been hypothesized as global warming refuges (Kim et al. 2019).
Studies of coral bleaching induced mortality events are increasingly assessing changes in the composition and functional role of the microbial communities of the corals that undergo a loss of symbiotic algal partners (Symbiodiniaceae) due to bleaching partial mortality and colony mortality (Cziesielski et al. 2019; Gardner et al. 2019; van Oppen and Blackall 2019). It is clear from this research that changes in community composition in response to bleaching are not consistent across corals, reef locations or bleaching events. Studies have shown that the composition of the bacterial microbiome associated with corals can either change (Mouchka et al. 2010; Tout et al. 2015; McDevitt-Irwin et al. 2017) or remain unchanged during a thermal stress event (Tracy et al. 2015; Hadaidi et al. 2017; Epstein et al. 2019; van Oppen and Blackall 2019; Bergman et al. 2021). In some studies, thermal stress has been shown to correlate with shifts in the microbial community through an increase of opportunistic pathogens (Acropora millepora [Bourne et al. 2008] and Acropora, Porites, and Pocillopora [Maher et al. 2019]) and that pathogens found in the communities of heat-stressed corals resemble those found in diseased corals (Porites compressa [Thurber et al. 2009] and P. damicornis [Tout et al. 2015]). Similarly, some bleached corals have been shown to host higher proportions of Vibrio and Acidobacteria than healthy corals (Mouchka et al. 2010; Morrow et al. 2018). Furthermore, the “Anna Karenina Principle” (AKP) of dysbiosis suggests that bacterial microbiome changes induced by perturbations are stochastic and lead to unstable community states instead of a specific microbiome configuration associated with stress (Zaneveld et al. 2017). However, many studies are also increasingly pointing to stability or resilience of the microbiome while the host corals are undergoing Symbiodiniaceae loss, bleaching, and/or thermal stress. For example, in many studies of Pocilloporid corals, the microbiome has been shown to remain stable throughout thermal stress (P. acuta [Epstein et al. 2019], P. damicornis [Brener-Raffalli et al. 2018; Bergman et al. 2021], and the P. damicornis species complex [Ziegler et al. 2017; Pogoreutz et al. 2018]). Stability of the microbiome is defined here as no significant increase in diversity of bacterial communities of the host coral during heat stress, such as that observed during two degree heating weeks of thermal stress in P. acuta (Epstein et al. 2019). While it has been suggested that a stable microbiome may contribute to resilience against dynamic environmental conditions (Ziegler et al. 2017; Grottoli et al. 2018), alternative hypotheses suggest this may also prevent the colonization of beneficial microbes (Reshef et al. 2006) and contribute to a breakdown of the meta-organism relationship among host, symbionts, and microbiome (Tracy et al. 2015). The diversity of documented coral microbial community responses to thermal stress also suggests that the response of the coral microbiome to thermal stress will vary under certain degrees of stress, extent of bleaching, or past history of bleaching. Clearly, the coral holobiont and microbial community responses are complex and therefore likely vary across the substantial diversity of Scleractinia, coral reef habitats, and the severity of heat stress and bleaching impacts to the coral host.
One of the most broadly distributed families of corals are the Pocilloporids, a widespread taxonomic group of generalist scleractinian corals. Generalist coral species thrive across a breadth of environmental conditions and are generally considered more tolerant of sub-optimal conditions than species existing within a narrow environmental niche (Richmond et al. 2005; Clavel et al. 2011; Darling et al. 2012). The Pocilloporids include P. damicornis, shown to be one of the most geographically cosmopolitan and generalist species of scleractinian coral in the pan Pacific, which has also often been characterized as sensitive to bleaching (Dalton et al.; Loya et al. 2001; van Woesik et al. 2011; Claar and Baum 2019). However, generalists are regarded to be more successful than specialists in the current climate crisis due to extensive plasticity and tolerance of sub-optimal environmental conditions (Clavel et al. 2011; Chichorro et al. 2019). P. damicornis has been characterized as bleaching-sensitive on the GBR, despite containing higher density of Symbiodiniaceae than more bleaching-resistant species (Ulstrup et al. 2006); however, its ability to rely on heterotrophic feeding may contribute to its persistence in a changing environment whilst undergoing bleaching responses (Todd 2008; Schmidt-Roach et al. 2014). The cosmopolitan distribution of P. damicornis, as well as its potential for success in changing environmental conditions as a generalist, make it an ideal study species for examining trends in coral physiology and stress response in a range of environmental conditions.
Here, we therefore investigated the microbiome of the generalist coral species P. damicornis during reef wide coral bleaching events that occurred in 2019 and 2020 on a subtropical and tropical shallow coral reef lagoonal ecosystem (Fig. 1). P. damicornis colonies were sampled during the onset of reef wide bleaching on the tropical reef lagoons of Heron Island (HI) on the southern GBR in summer of 2020 and LHI in summer of 2019. This study characterizes the microbiome composition and inferred bacterial functions of P. damicornis during the bleaching events on these reefs to investigate the characteristics of a bleached coral microbiome for the generalist coral species of two different coral reef habitats.
Fig. 1.
Conceptual map showing study site locations. (A) HI and (B) LHI.
Methods
Sample collection and DNA extraction
Heron Island (HI). Single branch fragments per colony (∼4 cm) of P. damicornis were collected on snorkel using needle-nose pliers sterilized between samples with 70% molecular-grade ethanol from the shallow reef flat at 1–2 m depth in the research zone of HI in March 2020. Samples were transported to Heron Island Research Station in individual WhirlPak© bags. Bleaching of P. damicornis colonies was recorded during ecological monitoring of the reef wide bleaching event (Ainsworth et al. 2021) and colonies were selected for sampling based on visual signs of bleaching (white, n = 5) and stability with algal symbionts (e.g., no clear visual indication of bleaching stress, herein referred to as healthy, n = 5). Samples were collected from the center of colonies >3 m apart and separated by distinct sand patches to reduce the likelihood that colonies were clonal (Permit: G19/41974.1). Samples were immediately snap-frozen in WhirlPak© bags dipped in liquid nitrogen and stored at −80°C until processing for DNA extraction following the protocol outlined below. Each entire coral fragment, including the coral skeleton, (1.34 ± 0.14 g, mean ± SE, n = 10) was added to a 2 mL tube containing 1.4 mm ceramic spheres (Matrix E, MP Biomedicals). DNA extraction followed manufacturer protocols using a QIAGEN QIAmp DNA Mini Kit (cat. #56304) with minor adjustments as briefly outlined here. Digestion buffers were doubled for all protocols, a step which greatly increased DNA yields following overnight digestion. A FastPrep-24 5G homogenizer (MP Biomedicals, Irvine, CA, USA) was programed to run three rounds of 20 s each (6.0 m/s) to homogenize the sample. Following homogenization, all samples were incubated overnight (18–24 hours) at 56°C. Samples were then centrifuged (3 min at 6000 x g) to pellet calcium carbonate remaining from the coral skeleton. The resulting supernatant was transferred to a new 2 mL tube, centrifuged again to pellet out any remaining calcium carbonate (3 min at 20,000 x g), and supernatant was transferred to a new 2 mL tube. Following the remainder of the manufacturer's protocol, AL buffer and 100% molecular-grade ethanol were doubled to increase in proportion to the increased ATL and Proteinase K buffer volume prior to vortexing and adding to the collection column (QIAmp DNA Mini Kit, Qiagen). The manufacturer's protocol was followed for the remainder of the procedure and DNA eluted to 60 µL.
Lord Howe Island (LHI). Single branch fragments per colony (∼4 cm) of P. damicornis were collected on snorkel using needle-nose pliers sterilized between samples with 70% molecular-grade ethanol from the shallow fringing Lagoon reef at 1–2 m depth on the western side of LHI in March 2019 (Permit MEAA19/206). Bleached (n = 5) and healthy (n = 5) colonies were selected based on the visual health surveys outlined in Steinberg et al. (2022) during the reef wide bleaching event in March 2019. Sterile 16% paraformaldehyde ampules (Electron Microscopy Sciences, cat # 50980487) were used for the preparation of 4% paraformaldehyde (PFA) by a 1:3 dilution with phosphate buffered saline (PBS) solution (PBS tablets [Invitrogen, Waltham, MA, USA]) in UltraPure DNA/RNA-Free Distilled Water (ThermoFisher Scientific, Waltham, MA, USA). Samples were then added to 50 mL conical tubes immediately following collection and covered with the 4% PFA preservative solution. After 14 hours, PFA solution was removed and replaced with the DNA/RNA free PBS for storage. Samples were stored at 4°C for ∼3–6 months. DNA was extracted from coral fragments collected from LHI (1.29 ± 0.16 g, mean ± SE, n = 10) by adding each entire fragment to a 2 mL tube containing 1.4 mm ceramic spheres (Matrix E, MP Biomedicals). DNA extraction followed manufacturer protocols using a RecoverAll™ Total Nucleic Acid Isolation Kit for FFPE (cat #AM1975) protocol with minor adjustments as briefly outlined here. 400 µL of digestion buffer and 8 µL of Proteinase K were added prior to bead-beating as described previously. Following homogenization, all samples were incubated overnight (18–24 hours) at 50°C and centrifuged twice (3 min at 6000 x g and 3 min at 20,000 x g) to pellet and remove calcium carbonate remaining from the coral skeleton. Isolation additive and 100% molecular-grade ethanol were doubled to increase in proportion to the increased digestion buffer and Proteinase K buffer volume prior to vortexing and adding to the collection column. The manufacturer's protocol was followed for the remainder of the procedure and DNA eluted to 60 µL.
16S rRNA gene amplicon sequencing and analysis. For all samples, extracted DNA concentration and purity were quantified using a Qubit Fluorometer and Qubit dsDNA broad-spectrum assay kits (Life Technologies, Thornton, NSW, Australia). Extracted DNA was stored at −20°C prior to PCR amplification and sequencing. DNA extraction, amplification, and sequencing were performed on all samples as well as on two negative controls (no sample template, to account for contamination introduced during DNA extraction) prepared per site (n = 20 samples plus four negative controls in total). Sequencing was performed by MR DNA (Molecular Research LP, Shallowater, TX, USA) on the Illumina MiSeq platform following manufacturer's guidelines. The 16S rRNA gene V1–V3 regions PCR primers 27F/519R, commonly used in studies of the coral microbiome (e.g., Glasl et al. 2019; Marchioro et al. 2020) were used in a 35 cycle PCR using the HotStarTaq Plus Master Mix Kit (Qiagen, Germantown, MD, USA) under the following conditions: 95°C for 5 minutes, followed by 35 cycles of 95°C for 30 seconds, 53°C for 40 seconds, and 72°C for 1 minute, after which a final elongation step at 72°C for 10 minutes was performed. Samples were multiplexed using unique dual indices, pooled together in equal proportions based on molecular weight and DNA concentrations, and purified using calibrated Ampure XP beads. The pooled and purified PCR product was then used to prepare an Illumina DNA library and sequenced on the Illumina MiSeq (2 × 300 bp-paired end reads) following the manufacturer's guidelines.
Data analysis
Sequence data were analyzed using Quantitative Insights Into Microbial Ecology version 2 (QIIME2, (Bolyen et al. 2019). After denoising and primer removal using the DADA2 pipeline (Callahan et al. 2016), taxonomy was assigned to amplicon sequence variants (ASVs) in QIIME2 using a naïve Bayes classifier trained on SILVA 138 reference sequence and taxonomy files pre-formatted for use with QIIME2 using RESCRIPt (Robeson et al. 2021). ASVs assigned as “chloroplasts,” “mitochondria,” or unassigned (classification absent at a phylum level) were removed and excluded from the final ASV table. Contaminant removal was conducted in R based on contaminants identified in two negative controls per site/extraction protocol (version 4.1.2) using the package decontam at a threshold of 0.5, which implements a statistical classification procedure that identifies contaminants in sequencing data (Davis et al. 2018). Data analysis was conducted in R using the packages phyloseq (McMurdie and Holmes 2013) and ampvis2 (Andersen et al. 2018). Linear discriminant analysis effect size tests (LEfSe, [Segata et al. 2011]) were conducted using the package microbiomeMarker (Cao 2020) on unrarefied data, using an LDA cutoff of 1. At a higher LDA cutoff, no differences were observed in bleached samples. The functional composition of bacterial communities was inferred using both software platforms PICRUSt2 (Douglas et al. 2020) and Tax4Fun2 (Wemheuer et al. 2020), and overlapping KEGG Orthologs (KO) between PICRUSt2 and Tax4Fun2 outputs were used for final data visualization with PICRUSt2 abundance values. The coral core microbiome was determined for species-specific community taxa at a prevalence threshold of 30, 60, and 90% using the “occupancy method” defined by Custer et al. (2023), e.g., present in equal to or greater than X% of samples (Ainsworth et al. 2015; Hernandez-Agreda et al. 2018a; Ricci et al. 2022).
Statistical analysis
All statistical analyses were performed in R, using the package vegan (Dixon 2003) for multivariate statistics and ggplot2 (Wickham and Chang 2016) for data visualization. Alpha diversity metrics were analyzed using separate one-way ANOVAs for each treatment (healthy and bleached) using unrarefied and rarefied data (Shannon, Chao1, and Inverse Simpson). In cases where residuals were not normally distributed (determined via Shapiro test), the non-parametric Kruskall–Wallis test was used to analyze differences in alpha diversity metrics between healthy and bleached corals at each location. For beta diversity, Euclidean distance matrices on centered log-ratio (clr)transformed data were analyzed between healthy and bleached corals at each location. Homogeneity of dispersion around group centroids was assessed for each beta diversity metric between bleached and healthy corals using PERMDISP (betadisper function in vegan). A permutational multivariate analysis of variance (PERMANOVA, n = 9999, adonis function in vegan) was performed to test for dissimilarities in microbial community composition between healthy and bleached samples.
Prior to assessing the beta diversity of healthy and bleached corals at each site, we evaluated the effectiveness of different normalization methods on our dataset. A common challenge in interpreting data from 16S rRNA ecological datasets is finding a normalization technique that best fits data characteristics. As 16S rRNA data is compositional, the starting point for analysis used a ratio transformation of the data (Gloor and Reid 2016; Gloor et al. 2017). A common problem with ASV tables is the high degree of zero inflation of the proportional data (up to 90%) (Paulson et al. 2013). Following the recommendations of Gloor et al. (2017), a + 1 count was added to the ASV tables and we used a clr transformation prior to undertaking community composition analysis (as used by Gloor et al. 2017). The benefit of a clr transformation (Aitchison 1982) is that ratio transformations can capture the relationships between features in the dataset regardless of if the data are counts or proportions. clr-transformed values are scale-invariant, which means that the same ratio will be obtained from samples with high or low read counts. Given the low read counts within coral microbiome samples (due to high DNA concentration of host and dinoflagellate DNA in the mixed holobiont sample), we also compared a clr transformation and Aitchison distance matrix to the widely used method of data rarefaction. For DNA amplified coral samples from HI to rarefaction was at a depth of 733 (90% of the minimum sequence count) resulting in 633 taxa identified across the 10 samples, removing 41% of taxa. It is important to note that for DNA amplified from coral samples from LHI rarefaction was at a depth of 8352 (90% of the minimum sequence count) resulting in 326 taxa across 10 samples, removing ∼10% of taxa. We therefore assessed community composition using the clr-transformed data and the Aitchison distance matrix, as no taxa were lost using this method.
Results
Sequencing statistics. In total, 311,748 sequences from 20 samples and 4 negative controls were generated within the study. Quality control and removal of chloroplasts, mitochondria, unassigned ASVs (classification absent at a phylum level), and potential contaminants resulted in the retention of 304,678 sequences with a mean of 15,233 ± 2,437 (± SE) reads per sample, ranging from a minimum of 815 reads to a maximum of 41,209 reads (Table S1). Clustering at ASV level and removing negative controls yielded 1427 distinct ASVs for analysis of the microbial community. 1085 ASVs were identified in coral samples from HI (n = 10 samples, 815–40,635 reads per sample), and 365 ASVs were identified in coral samples from LHI (n = 10 samples, 9280–33,218 reads per sample).
HI coral bacterial communities
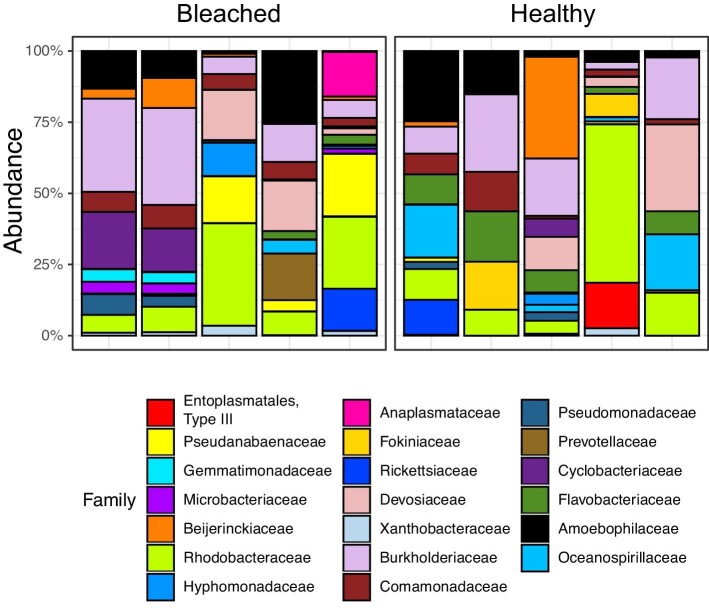
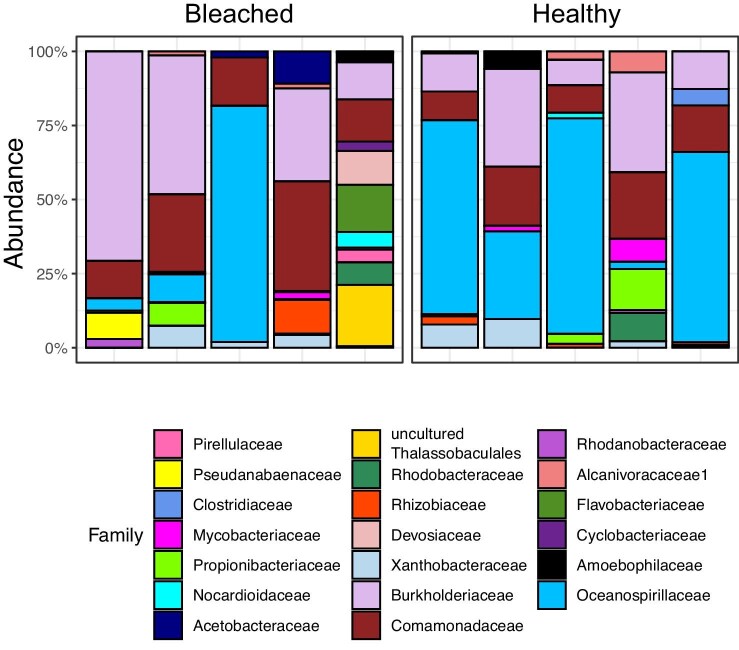
Characteristics of the bleached coral microbiome from HI. Phylum-level assignment of bacteria ASVs indicated the dominance of Proteobacteria in both healthy (83% of taxa) and bleached (64% of taxa) samples. Family-level assignment of bacterial ASVs indicated the dominance of Rhodobacteraceae (15% of taxa) in healthy samples from HI, followed by Sphingomonadaceae (5% of taxa) and Colwelliaceae (4% of taxa). However, the families “Type III,” Burkholderiaceae, and Amoebophilaceae had the highest average relative abundances in healthy samples (Fig. 2). Rhodobacteraceae (19% of taxa) was also the most prevalent family in bleached HI samples, followed by Pseudanabaenaceae (5% of taxa) and Colwelliacae (4% of taxa). The families with the highest relative abundance in bleached HI samples were Burkholderiaceae, Anaplasmataceae, and Amoebophilaceae (Fig. 2), similar to the community observed from the control samples.
Fig. 2.
Relative % abundance of ASVs in the HI community microbiome, agglomerated down to top 20 families and sorted by health status.
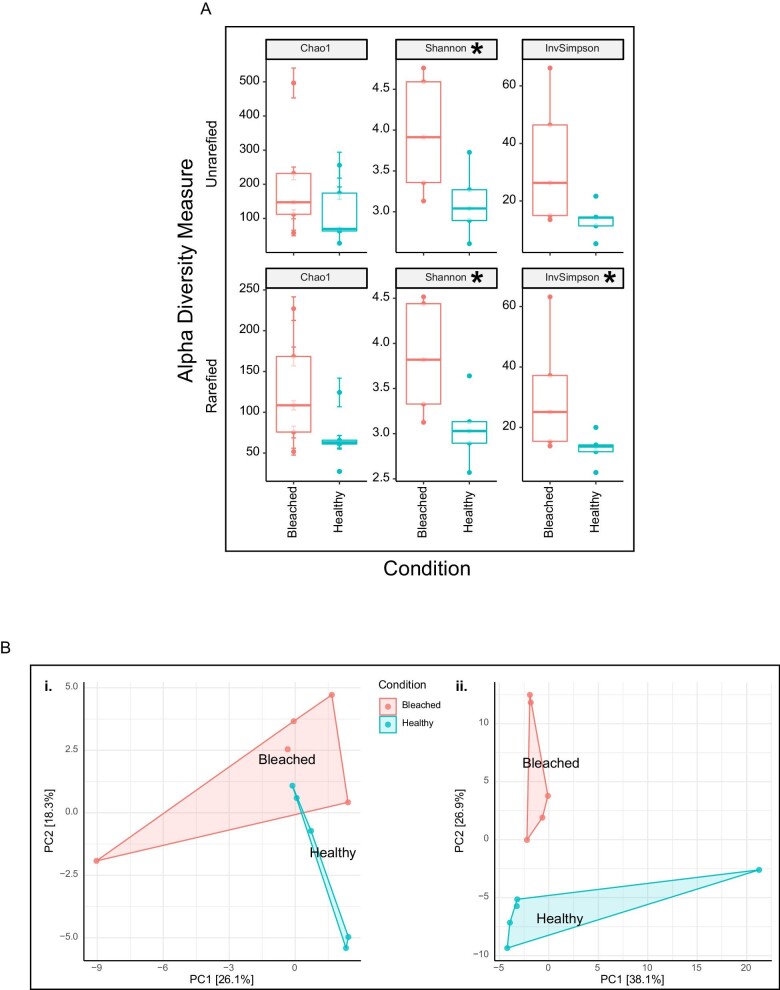
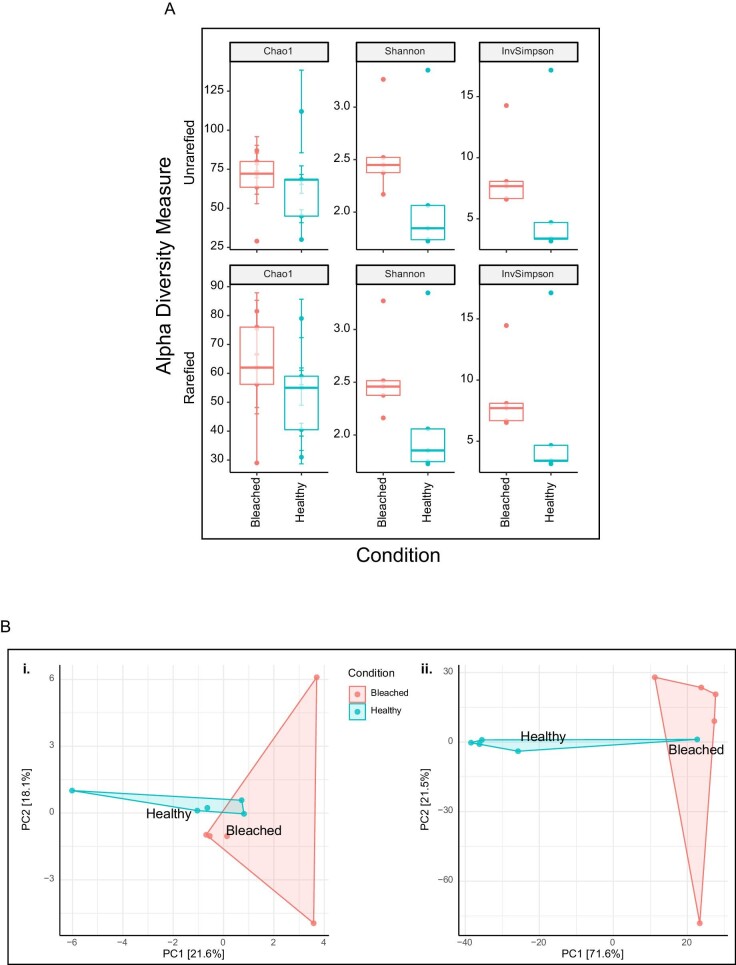
Beta diversity did not differ between bleached or healthy bacterial communities from HI, but Shannon diversity did. Shannon diversity differed between healthy and bleached corals (χ2 = 3.938, p = 0.047), but there was no statistical difference between healthy and bleached communities from HI samples in ASV richness (Chao1, χ2 = 0.534, p = 0.465) and dominance (Simpson, χ2 = 3.153, p = 0.076) (Fig. 3A). Beta diversity did not differ between healthy or bleached bacterial communities of corals from HI during the 2020 bleaching event (PERMANOVA on clr-transformed Euclidean distances, F = 1.195, p = 0.093; Fig. 3B).
Fig. 3.
Community-level analyses for HI. (A) Alpha diversity metrics on both unrarefied and rarefied data comparing healthy and bleached samples. Significant differences are indicated by an *. (B) Principal component analyses (PCoA) comparing clr-transformed (1) and rarefied (2) Euclidean distance matrices. No significant differences were observed in beta diversity metrics.
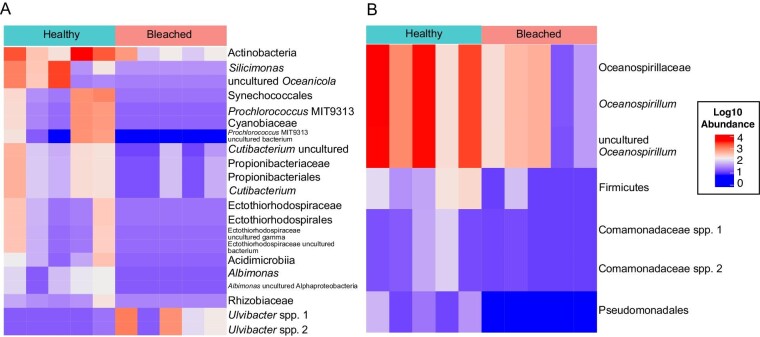
Differentially abundant ASVs in bleached and healthy coral samples from HI do not include pathogenic taxa. Employing linear discriminant analysis effect size (LeFSe) analysis, we identified members of the coral microbiome at an ASV level that were differentially abundant in bleached or healthy samples. In corals from HI, Ulvibacter spp. were found in higher relative abundance in healthy coral tissues (Fig. 4A). More taxa were found to be higher in relative abundance in bleached corals than healthy, with individual ASVs from the families Rhodobacteraceae, Cyanobiaceae, Propionibacteriaceae, Ectothiorhodospiraceae, and Rhizobiaceae found at a higher abundance in bleached samples (Fig. 4A). However, potentially disease-associated taxa (e.g Vibrio) were not found at a higher abundance in bleached coral samples than in healthy coral samples from HI (Fig. 4A). In healthy coral samples, an uncultured Vibrio ASV was present in one sample at 0.15% relative abundance. In bleached coral samples, one V. neocaledonicus ASV was present in one sample at 0.01% relative abundance. There was also no differential abundance of Endozoicomonas ASVs in either healthy or thermally stressed samples, a common bacterial associate in Pocilloporid corals (Pogoreutz et al. 2018; Epstein et al. 2019; Voolstra and Ziegler 2020; Ricci et al. 2022). There was a very low relative abundances of Endozoicomonas ASVs in one healthy sample (0.11%) and none detected in bleached samples.
Fig. 4.
Heatmap resulting from LEfSe analysis, showing differentially abundant ASVs in bleached and healthy corals for (A) HI and (B) LHI. Abundances were log(10) transformed for visualization. Taxa are listed to the lowest level of classification identified.
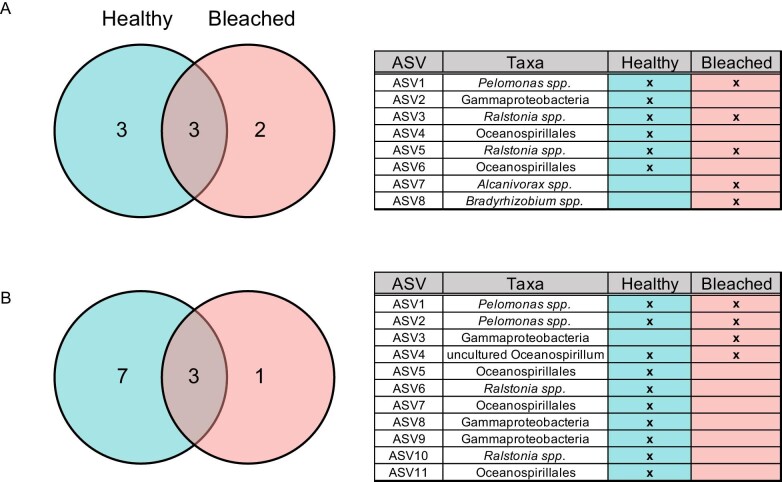
Coral core microbiome in corals from HI. The core microbiome was determined at HI for both bleached and healthy corals at a 90% threshold, as this would capture taxa present across all samples (n = 5 per health status, per site, sensu Hernandez-Agreda et al. 2017). The majority of bacterial ASVs (healthy: 98.3% of ASVs, bleached: 99.3% of ASVs were not present across individuals with the same health status (Fig. 5A). In corals from HI, six ASVs were found in healthy samples and five ASVs were found in bleached samples. The core ASVs found associated with healthy samples were all in the phylum Proteobacteria, and included: an unidentified Gammaproteobacteria, two Ralstonia spp., a Pelomonas spp., and two unidentified Oceanospirillales. The core ASVs found associated with bleached samples were all in the phylum Proteobacteria as well, and included: an Alcanivorax spp., a Bradyrhizobium spp., two Ralstonia spp., and a Pelomonas spp. Of these, three ASVs were found in both healthy and bleached samples: two Ralstonia spp. and one Pelomonas spp. The presence of only three commonly identified taxa across all samples at 90% prevalence, as well as the majority of bacterial ASV's not found to be consistently present across at least 90% of samples, indicates that ASVs affiliated with rare bacterial taxa dominated the coral microbiome in the present study.
Fig. 5.
Venn diagrams showing common bacterial phylotypes shared between bleached and healthy P. damicornis colonies at 90% prevalence at (A) HI and (B) LHI. Venn diagrams are accompanied by a table listing core ASVs at the lowest level of classification identified.
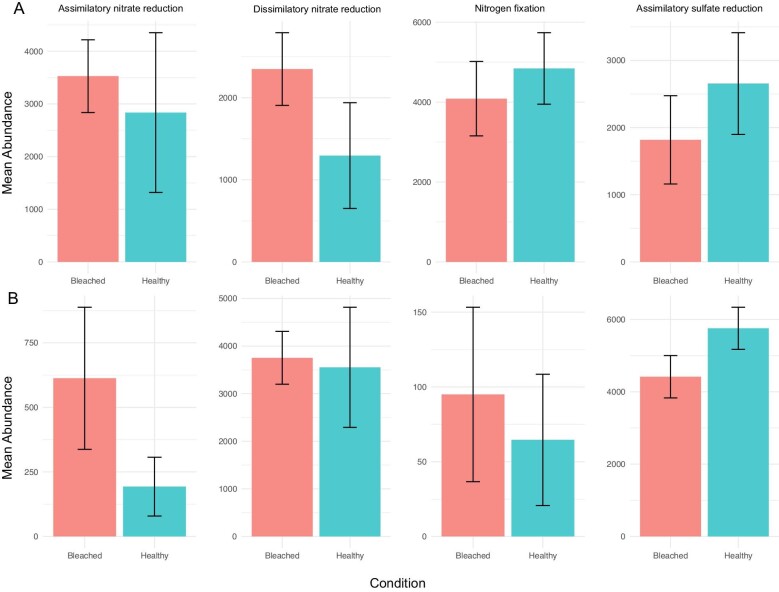
Predicted microbial functional profiles do not differ between bleached and healthy corals at HI. Herein focusing exclusively on nitrogen and sulphur metabolism, four KEGG pathways were identified in the microbial communities of bleached and healthy corals from HI: assimilatory nitrate reduction, dissimilatory nitrate reduction, nitrogen fixation, and assimilatory sulfate reduction. There was no clear enrichment of either nitrogen or sulphur metabolism KEGG pathways between healthy and bleached corals from HI (Fig. 6A).
Fig. 6.
Barplot of mean abundance for key nitrate and sulfur metabolism pathways identified in consensus between PICRUSt2 and Tax4Fun2 between bleached and healthy corals at (A) HI and (B) LHI. No significant differences were observed between functions.
LHI coral bacterial communities
Characteristics of the bleached coral microbiome from LHI. Phylum-level assignment of bacteria ASVs indicated the dominance of Proteobacteria in both healthy (76% of taxa) and bleached (82% of taxa) samples. The most prevalent family in healthy samples from LHI was Comamonadaceae (9% of taxa), followed by Rhodobacteraceae (7% of taxa) and Beijerinckiaceae (5% of taxa) (Fig. 7). However, the families Oceanospirillaceae, Burkholderiaceae, and Propionibacteriaceae had the highest average relative abundances (Fig. 8A). Thalassospiraceae (11% of taxa) was the most prevalent family in bleached LHI samples, followed by Comamonadaceae (6% of taxa) and an uncultured bacterium in the order Thalassobaculales (5% of taxa). The families with the highest relative abundance in bleached LHI samples were Burkholderiaceae, Flavobacteriaceae, and Comamonadaceae (Fig. 7).
Fig. 7.
Relative % abundance of ASVs in the LHI community microbiome, agglomerated down to top 20 families and sorted by health status.
Fig. 8.
Community-level analyses for LHI. (A) Alpha diversity metrics on both unrarefied and rarefied data comparing healthy and bleached samples. Significant differences are indicated by an *. (B) Principal component analyses (PCoA) comparing clr-transformed (1) and rarefied (2) Euclidean distance matrices. No significant differences were observed in beta diversity metrics.
No difference in alpha and beta diversity was found between the microbial communities of bleached and healthy samples from LHI. There was no difference between healthy and bleached samples in Shannon (F = 1.307, p = 0.286), Chao1 (F = 0.008, p = 0.475), or Simpson (F = 0.561, p = 0.929) diversity metrics (Fig. 8A). Beta diversity did not differ between healthy or bleached bacterial communities of corals from LHI during the 2019 bleaching event (PERMANOVA on clr-transformed Euclidean distances, F = 1.157, p = 0.119; Fig. 8B).
Differentially abundant ASVs in bleached and healthy coral samples from LHI do not include pathogenic taxa. LeFSe analyses on LHI samples showed individual ASVs from the orders Oceanospirillales, Burkholderiales, and Pseudomonadales were higher in healthy samples. One ASV from the phylum Firmicutes was also higher in healthy samples. No members of the coral microbiome were higher in the bleached samples (Fig. 4B). Bleaching sensitivity is paralleled by the emergence of opportunistic bacterial species, but similarly to bleached samples from HI, there was no marked increase in relative abundance of Vibrio taxa. One Vibrionales species, Photobacterium spp., was present in one sample from LHI at <0.01% relative abundance, and no Vibrio taxa were present in bleached samples. Endozoicomonadaceae composed 3% of the taxa in bleached LHI samples, but this was driven by presence in a single sample at 0.002% relative abundance.
Coral core microbiome in corals from LHI. At a 90% threshold for core microbiome, 10 ASVs were found in healthy samples and 4 ASVs in bleached samples from LHI. The majority of bacterial ASVs were not present across individuals with the same health status (Fig. 5B). The core ASVs found associated with healthy samples were all in the phylum Proteobacteria, and included: two Pelomonas spp., three unidentified Oceanspirillales, two Ralstonia spp., two unidentified Gammaproteobacteria, and an uncultured Oceanospirillum. The core ASVs found associated with bleached samples were all in the phylum Proteobacteria as well, and included: two Pelomonas spp., an unidentified Gammaproteobacteria, and an uncultured Oceanospirillum. Of these, three ASVs were shared between both healthy and bleached samples: two Pelomonas spp. and an uncultured Oceanospirillum. At a 90% threshold, 94.6% of taxa were not present in healthy samples from LHI (188 taxa) and 98.1% of taxa were not present in bleached samples from LHI (213 taxa).
Predicted microbial functional profiles do not differ between bleached and healthy corals at LHI. Herein focusing exclusively on the fundamental properties of nitrogen and sulphur metabolism, four KEGG pathways were identified in the microbial communities of bleached and healthy corals from LHI: assimilatory nitrate reduction, dissimilatory nitrate reduction, nitrogen fixation, and assimilatory sulfate reduction. There was no clear enrichment of either nitrogen or sulphur metabolism KEGG pathways between healthy and bleached corals from LHI (Fig. 6).
Discussion
In this study, we investigated the characteristics of the bleached P. damicornis coral microbiome during a reef wide bleaching event at two distinct coral reef locations, to determine the microbiome composition and inferred microbial functioning of the generalist coral. We found that stability in the coral microbiome, herein described as both (1) no change in beta diversity between healthy and bleached corals and (2) no increase in pathogenic taxa in bleached corals, was consistent in both coral reef lagoonal environments. Some taxa associated with both healthy and bleached corals at HI and LHI, such as Rhodobacteraceae and Flavobacteriia, were similar to what has previously been observed in P. acuta from Havannah Island and Pandora Reef on the GBR (Botté et al. 2022) and P. damicornis from HI (van Oppen et al. 2018). Ricci et al. (2022) found a high abundance of Pseudoalteromonas species in the tissues and skeleton of Pocilloporidae from the GBR that was not observed in the present study, possibly due to different bacterial communities associated with different sample collection methods targeting different regions of the holobiont (Bergman et al. 2022) or potentially seasonal difference in the microbiome which need to be assessed in future studies (Sharp et al. 2017; Yu et al. 2021b). To date, the P. damicornis bacterial microbiome at LHI has yet to be characterized, making this study the first to do so. In both locations, similar patterns in the microbial community in response to bleaching were observed.
Several patterns of microbial stability during bleaching emerged that were consistent in both distinct locations. These trends differ from two commonly presented microbial community hypotheses to heat stress. First, the AKP of dysbiosis, which suggests that microbial changes induced by perturbations lead to unstable community states and are stochastic (Zaneveld et al. 2017). Here, the stable microbiome composition observed (no significant difference between the microbiome of bleached and healthy corals from HI and LHI) is also similar to what has been reported during reef wide P. acuta thermal stress (Epstein et al. 2019) and ex situ simulated bleaching in P. damicornis (Bergman et al. 2021). Both Bergman et al. 2021 and Epstein et al. 2019 reported significant differences in beta diversity at the sequence variant level throughout P. damicornis thermal stress response (Epstein et al. 2019) and bleaching (Bergman et al. 2021), but further analysis revealed that no time points were driving this significance and led both studies to conclude overall bacterial stability in bleached colonies over time during a thermal stress event. Epstein et al. (2019) remark that in some coral species even significant thermal stress may not result in visible signs of bleaching; however, in both studies similar patterns of no marked increase in beta diversity was observed in the microbial communities of either bleached or heat stressed corals, and no clear pattern in beta diversity was identified to correlate to either bleaching or heat stress responses (Epstein et al. 2019; Bergman et al. 2021). Similarly, in the present study, variation in one metric of alpha diversity between healthy and bleached samples was identified (e.g., Shannon diversity in coral samples from HI), but no marked increases in beta diversity and Chao1/Simpson measures of alpha diversity were observed between healthy and bleached samples at both locations. Epstein et al. (2019) has shown that if heat stress does not result in severe bleaching or bleaching induced coral mortality, then the beta diversity of the coral microbiome is not altered. However, in the current study where large-scale coral bleaching and bleaching-induced mortality occurred across both reefs studied, community changes to the microbiome were not apparent at the time of sampling. These results may indicate that the timing of sampling within the bleaching event, reflective of the degree of heat stress and progression to mortality of the coral hosts, is potentially critical in understanding any correlation between bleaching, the microbiome, and bleaching outcomes for the coral colonies.
The complementary hypothesis to the AKP is a shift toward a pathogenic community similar to that found in diseased corals, which has been reported in heat-stressed corals (Bourne and Munn 2005; Thurber et al. 2009; Tout et al. 2015), but was also not found in the present study. V. coralliilyticus, a temperature-dependent pathogen of P. damicornis (Ben-Haim et al. 2003), and other Vibrio taxa were not found here. A lack of significant increase in Vibrio-affiliated sequences has also been observed in the P. verrucosa microbiome during a bleaching event in the South China Sea (Yang et al. 2021). Similar to Yang et al. (2021), the corals in the present study were collected during the reef wide bleaching event and outside of controlled aquaria conditions. Increases in the relative abundance of Rhodobacteraceae in bleached corals (as seen in the present study) has been associated with parallel increases in Vibrio-affiliated sequences previously in P. damicornis (Tout et al. 2015), but not in bleached P. lutea colonies (Pootakham et al. 2019) or in P. damicornis from HI undergoing thermal stress (Bergman et al. 2021). A caveat is that Pootakham et al. (2019) and Bergman et al. (2021), and the present study all used different reverse primers (519R or 1492R) to that used by Tout et al. 2015a) (1392R), so primer selection is also an important consideration.
Additionally, for corals collected from HI, the microbiome was almost entirely lacking Endozoicomonas, a common bacterial associate in Pocilloporid corals (Pogoreutz et al. 2018; Epstein et al. 2019; Voolstra and Ziegler 2020; Ricci et al. 2022), in both healthy and bleached/heat stressed samples. Ricci et al. (2022) characterized the microbiome of P. damicornis collected from HI at 0–1 m depth in January 2020, prior to bleaching events of 2020, and reported a high abundance of Endozoicomonas (uncultured species) associated with the coral tissues. The difference between our results and Ricci et al. (2022) suggests that warming water may disrupt the coral–Endozoicomonas association, an interpretation also supported by Botté et al. (2022) who reported low abundances of Endozoicomonas in P. acuta samples from Pandora Reef and Havannah Island on the GBR during a bleaching event and suggested that the onset of coral bleaching (3.5–5.6°C-weeks) represents a tipping point for Endozoicomonas species in P. acuta. As a loss of Endozoicomonas is often recorded in bleached or diseased corals (Bayer et al. 2013; Meyer et al. 2014; Glasl et al. 2016), an additional possibility suggested by Botté et al. (2022) is that repetitive and severe bleaching on the GBR has greatly reduced populations of coral tissue-associated Endozoicomonas over time. Epstein et al. (2019) found Endozoicomonas present in the majority of P. acuta samples from Orpheus Island collected in Feb—May 2016, prior to the repetitive mass bleaching events recorded from 2017, but found no variation in relative abundances between thermally stressed and healthy samples. As Epstein et al. (2019) also sampled corals during an in situ bleaching event, one interesting possibility is that all corals sampled in both Epstein et al. (2019) and the present study may have been environmentally stressed by reef wide warming regardless of bleaching status at the time of sample collection. This presents an alternative hypothesis to explain the lack of both Endozoicomonas and increased dysbiosis observed herein. In either instance, if Endozoicomonas populations do not return to their original abundances in a coral host following a bleaching event, then the coral host's resistance to stress may also decrease. However, further studies are needed to determine the cause of low abundances of Endozoicomas on the bacterial communities of corals following repetitive mass bleaching and sampled from the GBR in 2020, as in the present study.
In the current study, taxa that were found to be differentially abundant between bleached and healthy corals had varying functions. In healthy coral samples from HI, Ulvibacter spp. were found in a higher relative abundance. Ulvibacter spp. have been identified in seaweeds as a polysaccharide utilizer (Nedashkovskaya et al. 2016), which suggests that the organic carbon (e.g., cell wall of Symbiodiniaceae) in the coral holobiont may be utilized by Ulvibacter (Gong et al. 2020). In healthy corals, higher relative abundance of Ulvibacter could be aiding the coral holobiont in carbon uptake. More taxa were found to be at higher relative abundances in bleached corals than in healthy corals from HI, with individual ASVs from the families Rhodobacteraceae, Cyanobiaceae, Propionibacteriaceae, Ectothiorhodospiraceae, and Rhizobiaceae found at a higher abundance in bleached samples. Similar increases in the relative abundances of opportunistic Rhizobiales and Rhodobacteriales have been observed in P. lutea bacterial communities during an in situ bleaching event in the Andaman Sea (Pootakham et al. 2018), although similar to the results of the present study, no marked increase in relative abundances of disease-associated taxa (e.g., Vibrio) accompanied increases in Rhodobacteriales in heat-stressed corals (Pootakham et al. 2019). In healthy samples from LHI, individual ASVs from the orders Oceanospirillales and Burkholderiales were present in a higher abundance than in bleached samples, along with Pseudomonadales and the phylum Firmicutes. However, no ASVs were present at a higher abundance in bleached samples than healthy samples from LHI. A dominance of Oceanospirillales (47%) and Burkholderiales (8.5%) has previously been observed in many healthy coral species, including P. damicornis from HI (Tout et al. 2015; Ricci et al. 2022), in Stylophora pistillata from the Red Sea (Bayer et al. 2013), and P. acuta from Orpheus Island (Epstein et al. 2019). At LHI, both the healthy and bleached P. damicornis microbiome therefore show resemblance to a healthy coral microbiome, further supporting that no dysbiosis was observed in bleached P. damicornis samples.
At both HI and LHI, the majority of bacterial ASVs were not present across individuals with the same health status. One of the ASVs identified in both healthy and bleached samples from HI, as well as in healthy samples from LHI, belonged to Ralstonia spp. Ralstonia phylotypes have been commonly reported in studies of stony and soft corals (Sunagawa et al. 2010; Williams et al. 2015; Woo et al. 2017; Yu et al. 2021a). A Pelomonas spp. ASV was also observed in both healthy and bleached coral core microbiomes from both HI and LHI. Pelomonas is ubiquitous in coral microbiome studies (Röthig et al. 2017), but its role in the coral microbiome remains equivocal. It has been previously identified in the core microbiome of other corals such as the deep-sea coral Eguchipsammia fistula at a 100% threshold (Röthig et al. 2017) and P. damicornis at an 80% threshold (Bergman et al. 2021), removed as a contaminant (Pogoreutz et al. 2017), and associated with A. hemprichii samples from polluted sites (Ziegler et al. 2019). 1–6% of ASVs were shared across all members of a particular treatment group at either site, which is similar to a recent characterization of the P. damicornis microbiome that found 5–7% of all OTUs to be contained in the core microbiome (Ostria-Hernández et al. 2022). While one possibility is that widespread generalist species may not maintain a true coral microbiome (e.g., only one phylotype found persistent across three depth generalist coral species, Hernandez-Agreda et al. 2018b), the high specificity of coral core microbes also (Hernandez-Agreda et al. 2017, 2018b) suggests that variation in the abundance of the core microbes provides insight into the response of the coral host to stress. This is supported by Ostria-Hernández et al. (2022) finding that the relative abundance of the core microbiome of P. damicornis differed among anthropogenic stress levels. However, the core microbes that varied were not known coral pathogens (e.g., Vibrio species), which further supports microbial stability throughout bleaching in P. damicornis.
An important finding of the current study is that inferred functional profiles of the coral microbiome also did not differ between bleached and healthy corals at either location for the KEGG pathways of nitrogen and sulphur metabolism. Four functions were identified in bleached and healthy corals from HI and LHI: assimilatory nitrate reduction, dissimilatory nitrate reduction, nitrogen fixation, and assimilatory sulfate reduction. At both sites, there was no clear enrichment of either nitrogen or sulphur metabolism functions between healthy and bleached corals (Fig. 6). However, in interpreting these findings, acknowledging the resolution limitation of amplicon-based functional predictive tools is important as rare environment-specific functions may not be identified or maybe below the current detection limits, so these interpretations are made with caution (Douglas et al. 2020). A previous study comparing predicted functional profiles of the microbial communities of P. damicornis tissues and mucus reported 19 pathways to differ significantly between bleached and healthy microbial communities; however, similarly none of the altered functions were identified as nitrogen and sulfur metabolism (Zou et al. 2022). Additionally, for P. verrucosa during a bleaching event in the South China Sea, no difference in predicted functional profiles were found between bleached and healthy corals for nitrogen and sulfur metabolism pathways (Sun et al. 2022).
Overall, the characteristics of the bleached P. damicornis microbiome were similar in two distinct coral reef locations. Despite indications of bleaching, the bacterial communities and predicted functional roles of the bleached P. damicornis coral microbiome remained unchanged. It has been suggested that the role of P. damicornis and closely related members of its species complex (Schmidt-Roach et al. 2014) as environmental generalists may contribute to microbial stability throughout bleaching (Bergman et al. 2021), as similar stability has been observed in heat-stressed P. damicornis (Brener-Raffalli et al. 2018), P. acuta (Epstein et al. 2019; Botté et al. 2022), and P. verrucosa (Pogoreutz et al. 2018; Ziegler et al. 2019). Interestingly, a similar study comparing the microbiome of bleached and healthy coral species during a natural bleaching event in the Seychelles found stability was evident regardless of whether the coral species sampled was considered a “winner” or a “loser” in thermal tolerance (Gardner et al. 2019). This suggests that Symbiodiniaceae densities and bleaching response may be uncoupled from bacterial community, as seen in studies where the coral microbiome remains stable despite bleaching (Hadaidi et al. 2017). Overall, a conserved lack of differences in microbial community response to heat stress observed in the present study reflects a combination of host physiology and thermal stress responses that could be uncoupled from changes in the bacterial microbiome.
Finally, the conserved microbial response observed herein is also interesting given the different thermal regimes of the two bleaching events at HI and LHI. In a study of coral microbial assemblages from tropical and subtropical corals in the South China Sea, coral microbiome composition was found to vary across thermal regimes, with greater heterogeneity in corals from tropical reefs than subtropical possibly indicative of the microbial network transitioning from a stable to unstable state at bleaching threshold temperatures (Gong et al. 2020). In 2020, heat stress across the GBR (including HI) with max temperatures up to 37°C in 2020 (Ainsworth et al. 2021) highlighted the high temperatures experienced by the region on a cyclical basis. LHI has also experienced several bleaching events, documented in 2010, 2011, and 2019, with lower max temperatures of up to 25°C (Dalton et al. 2020; Steinberg et al. 2022). Additionally, seawater and sediments are one of the main drivers affecting microbiome composition, with more than 30% of ASVs shared between surrounding seawater, sediment, and P. damicornis on the GBR at a point in time (Ricci et al. 2022). Different seawater and sediment microbial communities are likely to differ between sites and therefore contribute to the differences in microbial community seen between sites herein. Future studies should therefore also examine seawater and sediment microbial communities, in order to understand if the taxa observed are site-specific to the coral, sediment, or seawater environment.
Natural variation between factors in corals in situ may also contribute to masking changes in the coral microbiome and are addressed here. For example, in the present study, different colonies on the reef flat were sampled at each timepoint. This contrasts with Epstein et al. (2019), who sampled the same colony throughout thermal stress events. While Epstein et al. (2019) found no change in the bacterial microbiome of the same colonies over time, it's possible that in the present study the microbiome of a single colony may have changed following bleaching but that natural variability is masked by colony-specific variation. In addition, natural variation in time since onset of bleaching (the caveat of an in situ experiment vs. a controlled ex situ experiment) could be an important factor explaining the breadth of variation observed amongst the bleached colonies, which would also mask any bleaching-specific variation. This raises the question that while we recorded photophysiological decline and symbiont loss concomitant with bleaching in the present study, it's not possible to know the length of time since the onset of bleaching or if corals were eventually pushed to mortality in situ. Both of these would affect the eventual development of dysbiosis in the coral microbiome and contribute to the variation within groups observed in the present study. With the small sample size in the present study, variation within groups could also potentially mask differences in diversity or composition among groups. We did not detect significant differences between groups in the present study, but future studies not limited by sample size could have increased statistical power to disentangle variability within groups from variability among groups. These factors are all caveats that should be considered when conducting an experiment in situ and in the interpretation of the results herein.
In conclusion, by examining the microbial communities of healthy and bleached corals during reef wide bleaching events of two distinct coral reef locations, we find consistent bleaching responses of the bacterial community between tropical (HI) and subtropical (LHI) Pocilloporid corals. While differences in the bacterial taxa of the holobiont confirm other studies reporting distinct bacterial communities associated with coral reef habitats (Hernandez-Agreda et al. 2016), an overall consistently stable microbial community response (e.g., no increase in diversity or shift toward a dysbiotic state) suggests that there is the possibility of a generalized host-specific or environment-specific influence on the holobiont of P. damicornis bleaching responses. Microbial community stability conserved across environments may contribute to the role of P. damicornis as a widespread environmental generalist species. Structurally stable microbiomes, presumed to be strongly selected, may host bacterial communities with a specialized set of functions (Ley et al. 2006; McDevitt-Irwin et al. 2017; Ziegler et al. 2017). It has been suggested that stable members play important roles in coral health, whereas transient members may vary with environmental conditions or perturbances (McDevitt-Irwin et al. 2017; Hernandez-Agreda et al. 2018b). Our results highlight the need for the inclusion of a broad range of (1) sites with varying thermal regimes and (2) species with different functional traits (e.g., specialist vs. generalist) to comprehensively characterize key members of a bleached and healthy coral microbiome.
Supplementary Material
Acknowledgements
This research was conducted at Heron Island Research Station, Lord Howe Island, and the University of New South Wales. We thank A. Fordyce, T. Moriarty, C. Page, and R. Steinberg for assistance in both the field and in the lab.
Contributor Information
J L Bergman, Biological, Earth, and Environmental Sciences, University of New South Wales, Sydney, NSW, 2052, Australia.
F Ricci, Biological, Earth, and Environmental Sciences, University of New South Wales, Sydney, NSW, 2052, Australia; School of BioSciences, University of Melbourne, Melbourne, VIC, 3010, Australia.
W Leggat, School of Environmental and Life Sciences, University of Newcastle, Callaghan, NSW, 2308, Australia.
T D Ainsworth, Biological, Earth, and Environmental Sciences, University of New South Wales, Sydney, NSW, 2052, Australia.
Funding
This work was supported by an Australian Research Council Discovery Project Grant [DP 180103199] and an UNSW Scientia Funding to T.D. Ainsworth. J.L. Bergman was also supported by the UNSW Scientia PhD Scholarship, the Australian Coral Reef Society's Student Research Award, and the Holsworth Wildlife Research Endowment—Equity Trustees Charitable Foundation & the Ecological Society of Australia.
Conflict of interest
The authors declare no competing interests.
Author contributions
J.L. Bergman and T.D. Ainsworth planned the experiments. J.L. Bergman conducted the experiments. J.L. Bergman analyzed the data and wrote the manuscript with advice, contributions, review, and editing from F. Ricci, W. Leggat, and T.D. Ainsworth.
Data availability
The sequencing data underlying this article are available in an NCBI repository accessible at https://www.ncbi.nlm.nih.gov/, PRJNA802894.
References
- Abdo DA, Bellchambers LM, Evans SN. 2012. Turning up the heat: increasing temperature and coral bleaching at the high latitude coral reefs of the Houtman Abrolhos Islands. PLoS One 7:e43878. 10.1371/journal.pone.0043878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AIMS . 2018. Annual summary report of coral reef condition 2018. Townsville: Australian Institute of Marine Science. [Google Scholar]
- AIMS . 2022. Annual summary report of coral reef condition 2021/2022. Townsville: Australian Institute of Marine Science. [Google Scholar]
- Ainsworth TD, Krause L, Bridge T, Torda G, Raina J-B, Zakrzewski M, Gates RD, Padilla-Gamiño JL, Spalding HL, Smith Cet al. 2015. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J 9:2261–74. 10.1038/ismej.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsworth TD, Leggat W, Silliman BR, Lantz CA, Bergman JL, Fordyce AJ, Page CE, Renzi JJ, Morton J, Eakin CMet al. 2021. Rebuilding relationships on coral reefs: coral bleaching knowledge-sharing to aid adaptation planning for reef users. Bioessays 43:1–9. 10.1002/bies.202100048. [DOI] [PubMed] [Google Scholar]
- Aitchison J. 1982. The statistical analysis of compositional data. J R Stat Soc Series B Stat Methodol 44:139–60. 10.1111/j.2517-6161.1982.tb01195.x. [DOI] [Google Scholar]
- Andersen K, Kirkegaard R, Karst S, Albertsen M. 2018. ampvis2: an R package to analyse and visualise 16S rRNA amplicon data. Biorxiv (doi: 10.1101/299537). [DOI] [Google Scholar]
- Bayer T, Neave MJ, Alsheikh-Hussain A, Aranda M, Yum LK, Mincer T, Hughen K, Apprill A, Voolstra CR. 2013. The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue-associated endozoicomonas bacteria. Appl Environ Microbiol 79:4759–62. 10.1128/AEM.00695-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarz VN, Grover R, Maguer J-F, Fine M, Ferrier-Pagès C. 2017. The assimilation of diazotroph-derived nitrogen by scleractinian corals depends on their metabolic status. Am Soc Microbiol 8:e02058. 10.1128/mBio.02058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Haim Y, Thompson FL, Thompson CC, Cnockaert MC, Hoste B, Swings Jet al. 2003. Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int J Syst Evol Microbiol 53:309–15. [DOI] [PubMed] [Google Scholar]
- Bergman JL, Leggat W, Ainsworth TD. 2021. The meta-organism response of the environmental generalist Pocillopora damicornis exposed to differential accumulation of heat stress. Front Mar Sci 8:1819. [Google Scholar]
- Bergman JL, Shaw T, Egan S, Ainsworth TD. 2022. Assessing the coral microbiome at the scale of tissue-specific habitats within the coral meta-organism. Front Mar Sci 9:985456. 10.3389/fmars.2022.985496. [DOI] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar Fet al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–7. 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botté ES, Cantin NE, Mocellin VJL, O'brien PA, Rocker MM, Frade PR, Webster NS. 2022. Reef location has a greater impact than coral bleaching severity on the microbiome of Pocillopora acuta. Coral Reefs 41:63–79. 10.1007/s00338-021-02201-y. [DOI] [Google Scholar]
- Bourne DG, Munn CB. 2005. Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ Microbiol 7:1162–74. 10.1111/j.1462-2920.2005.00793.x. [DOI] [PubMed] [Google Scholar]
- Bourne D, Iida Y, Uthicke S, Smith-Keune C. 2008. Changes in coral-associated microbial communities during a bleaching event. ISME J 2:350–63. 10.1038/ismej.2007.112. [DOI] [PubMed] [Google Scholar]
- Brener-Raffalli K, Clerissi C, Vidal-Dupiol J, Adjeroud M, Bonhomme F, Pratlong M, Aurelle D, Mitta G, Toulza E. 2018. Thermal regime and host clade, rather than geography, drive symbiodinium and bacterial assemblages in the scleractinian coral Pocillopora damicornis sensu lato. Microbiome 6:39. 10.1186/s40168-018-0423-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, Mcmurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–3. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y. 2020. microbiomeMarker: microbiome biomarker analysis. R package version 1.2.2 (https://github.com/yiluheihei/microbiomeMarker) Accessed 20 Nov 2022.
- Celliers L, Schleyer MH. 2002. Coral bleaching on high-latitude marginal reefs at Sodwana Bay, South Africa. Mar Pollut Bull 44:1380–7. 10.1016/S0025-326X(02)00302-8. [DOI] [PubMed] [Google Scholar]
- Chichorro F, Juslén A, Cardoso P. 2019. A review of the relation between species traits and extinction risk. Biol Conserv 237:220–9. 10.1016/j.biocon.2019.07.001. [DOI] [Google Scholar]
- Claar DC, Baum JK. 2019. Timing matters: survey timing during extended heat stress can influence perceptions of coral susceptibility to bleaching. Coral Reefs 38:559–65. 10.1007/s00338-018-01756-7. [DOI] [Google Scholar]
- Clavel J, Julliard R, Devictor V. 2011. Worldwide decline of specialist species: toward a global functional homogenization? Front Ecol Environ 9:222–8. 10.1890/080216. [DOI] [Google Scholar]
- Custer GF, Gans M, Van Diepen LTA, Dini-Andreote F, Buerkle CA. 2023. Comparative analysis of core microbiome assignments: implications for ecological synthesis. Msystems 8:e01066–22. 10.1128/msystems.01066-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cziesielski MJ, Schmidt-Roach S, Aranda M. 2019. The past, present, and future of coral heat stress studies. Ecol Evol 9:10055–66. 10.1002/ece3.5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton SJ, Carroll AG. 2011. Monitoring coral health to determine coral bleaching response at high latitude eastern Australian reefs: an applied model for a changing climate. Diversity 3:592–610. 10.3390/d3040592. [DOI] [Google Scholar]
- Dalton SJ, Carroll AG, Sampayo E, Roff G, Harrison PL, Entwistle K, Huang Z, Salih A, Diamond SL. 2020. Successive marine heatwaves cause disproportionate coral bleaching during a fast phase transition from El Niño to La Niña. Sci Total Environ 715:136951. 10.1016/j.scitotenv.2020.136951. [DOI] [PubMed] [Google Scholar]
- Darling ES, Alvarez-Filip L, Oliver TA, Mcclanahan TR, Côté IM. 2012. Evaluating life-history strategies of reef corals from species traits. Ecol Lett 15:1378–86. 10.1111/j.1461-0248.2012.01861.x. [DOI] [PubMed] [Google Scholar]
- Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. 2018. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6:226. 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon P. 2003. VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–30. 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MGI. 2020. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol 38:685–8. 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eakin CM, Sweatman HPA, Brainard RE. 2019. The 2014–2017 global-scale coral bleaching event: insights and impacts. Coral Reefs 38:539–45. 10.1007/s00338-019-01844-2. [DOI] [Google Scholar]
- Epstein HE, Torda G, Van Oppen MJH. 2019. Relative stability of the Pocillopora acuta microbiome throughout a thermal stress event. Coral Reefs 38:373–86. 10.1007/s00338-019-01783-y. [DOI] [Google Scholar]
- Gardner SG, Camp EF, Smith DJ, Kahlke T, Osman EO, Gendron G, Hume BCC, Pogoreutz C, Voolstra CR, Suggett DJ. 2019. Coral microbiome diversity reflects mass coral bleaching susceptibility during the 2016 El Niño heat wave. Ecol Evol 9:938–56. 10.1002/ece3.4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBRMPA . 2017. Final report: 2017 coral bleaching event on the Great Barrier Reef. Townsville: Great Barrier Reef Marine Park Authority. [Google Scholar]
- GBRMPA, AIMS, and CSIRO . 2022. Reef snapshot: summer 2021–22. Townsville: Great Barrier Reef Marine Park Authority. [Google Scholar]
- Glasl B, Herndl GJ, Frade PR. 2016. The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J 10:2280–92. 10.1038/ismej.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasl B, Smith CE, Bourne DG, Webster NS. 2019. Disentangling the effect of host-genotype and environment on the microbiome of the coral Acropora tenuis. PeerJ 7:e6377. 10.7717/peerj.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor GB, Macklaim JM, Pawlowsky-Glahn V, Egozcue JJ. 2017. Microbiome datasets are compositional: and this is not optional. Front Microbiol 8:2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor GB, Reid G. 2016. Compositional analysis: a valid approach to analyze microbiome high-throughput sequencing data. Can J Microbiol 62:692–703. 10.1139/cjm-2015-0821. [DOI] [PubMed] [Google Scholar]
- Gong S, Jin X, Ren L, Tan Y, Xia X. 2020. Unravelling heterogeneity of coral microbiome assemblages in tropical and subtropical corals in the South China Sea. Microorganisms 8:604. 10.3390/microorganisms8040604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grottoli AG, Dalcin Martins P, Wilkins MJ, Johnston MD, Warner ME, Cai W-J, Melman TF, Hoadley KD, Pettay DT, Levas Set al. 2018. Coral physiology and microbiome dynamics under combined warming and ocean acidification. PLoS One 13:e0191156. 10.1371/journal.pone.0191156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadaidi G, Röthig T, Yum LK, Ziegler M, Arif C, Roder C, Burt J, Voolstra CR. 2017. Stable mucus-associated bacterial communities in bleached and healthy corals of Porites lobata from the Arabian Seas. Sci Rep 7:1–11. 10.1038/srep45362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PL, Dalton SJ, Carroll AG. 2011. Extensive coral bleaching on the world's southernmost coral reef at Lord Howe Island, Australia. Coral Reefs 30:775. 10.1007/s00338-011-0778-7. [DOI] [Google Scholar]
- Hernandez-Agreda A, Gates RD, Ainsworth TD. 2017. Defining the core microbiome in corals’ microbial soup. Trends Microbiol 25:125–40. 10.1016/j.tim.2016.11.003. [DOI] [PubMed] [Google Scholar]
- Hernandez-Agreda A, Leggat W, Ainsworth TD. 2018a. A comparative analysis of microbial DNA preparation methods for use with massive and branching coral growth forms. Front Microbiol 9:2146. 10.3389/fmicb.2018.02146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Agreda A, Leggat W, Bongaerts P, Ainsworth TD. 2016. The microbial signature provides insight into the mechanistic basis of coral success across reef habitats. mBio 7:e00560–16. 10.1128/mBio.00560-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Agreda A, Leggat W, Bongaerts P, Herrera C, Ainsworth TD. 2018b. Rethinking the coral microbiome: simplicity exists within a diverse microbial biosphere. mBio 9:e00812–18. 10.1128/mBio.00812-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TP, Kerry JT, Álvarez-Noriega M, Álvarez-Romero JG, Anderson KD, Baird AH, Babcock RC, Beger M, Bellwood DR, Berkelmans Ret al. 2017. Global warming and recurrent mass bleaching of corals. Nature 543:373–7. 10.1038/nature21707. [DOI] [PubMed] [Google Scholar]
- Hughes TP, Kerry JT, Simpson T. 2018. Large-scale bleaching of corals on the Great Barrier Reef. Ecology 99:501. 10.1002/ecy.2092. [DOI] [PubMed] [Google Scholar]
- Kim SW, Sampayo EM, Sommer B, Sims CA, Gómez-Cabrera MDC, Dalton SJ, Beger M, Malcolm HA, Ferrari R, Fraser Net al. 2019. Refugia under threat: mass bleaching of coral assemblages in high-latitude eastern Australia. Glob Chang Biol 25:3918–31. 10.1111/gcb.14772. [DOI] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837–48. 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Loya Y, Sakai K, Yamazato K, Nakano Y, Sambali H, Van Woesik R. 2001. Coral bleaching: the winners and the losers. Ecol Lett 4:122–31. 10.1046/j.1461-0248.2001.00203.x. [DOI] [Google Scholar]
- Maher RL, Rice MM, Mcminds R, Burkepile D E, Vega Thurber R. 2019. Multiple stressors interact primarily through antagonism to drive changes in the coral microbiome. Sci Rep 9:1–12. 10.1038/s41598-019-43274-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchioro GM, Glasl B, Engelen AH, Serrão EA, Bourne DG, Webster NS, Frade PR. 2020. Microbiome dynamics in the tissue and mucus of acroporid corals differ in relation to host and environmental paragmeteres. PeerJ 8:e9644. 10.7717/peerj.9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcdevitt-Irwin JM, Baum JK, Garren M, Vega Thurber RL. 2017. Responses of coral-associated bacterial communities to local and global stressors. Front Mar Sci 4:262. 10.3389/fmars.2017.00262. [DOI] [Google Scholar]
- Mcmurdie PJ, Holmes S. 2013. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8 . e61217. 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JL, Paul VJ, Teplitski M. 2014. Community shifts in the surface microbiomes of the coral Porites astreoides with unusual lesions. PLoS One 9:e100316. 10.1371/journal.pone.0100316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow KM, Muller E, Lesser MP. 2018. “How does the coral microbiome cause, respond to, or modulate the bleaching process?” In: Coral bleaching. Cham: Springer, p. 153–88. 10.1007/978-3-319-75393-5_7. [DOI] [Google Scholar]
- Mouchka ME, Hewson I, Harvell CD. 2010. Coral-associated bacterial assemblages: current knowledge and the potential for climate-driven impacts. Integr Comp Biol 50:662–74. 10.1093/icb/icq061. [DOI] [PubMed] [Google Scholar]
- Nedashkovskaya OI, Kukhlevskiy AD, Zhukova N, Kim SB. 2016. Amylibacter ulvae sp. nov., a new alphaproteobacterium isolated from the Pacific green alga ulva fenestrata. Arch Microbiol 198:251–6. [DOI] [PubMed] [Google Scholar]
- Ostria-Hernández ML, Hernández-Zulueta J, Vargas-Ponce O, Díaz-Pérez L, Araya R, Rodríguez-Troncoso AP, Ríos-Jara E, Rodríguez-Zaragoza FA. 2022. Core microbiome of corals Pocillopora damicornis and Pocillopora verrucosa in the northeastern tropical Pacific. Mar Ecol 43:e12729. 10.1111/maec.12729. [DOI] [Google Scholar]
- Parks Australia . 2021. Norfolk Island lagoonal reef ecosystem health assessment 2020/2021. Norfolk Island, NSW. [Google Scholar]
- Paulson JN, Stine OC, Bravo HC, Pop M. 2013. Differential abundance analysis for microbial marker-gene surveys. Nat Methods 10:1200–2. 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogoreutz C, Rädecker N, Cárdenas A, Gärdes A, Wild C, Voolstra CR. 2017. Dominance of endozoicomonas bacteria throughout coral bleaching and mortality suggests structural inflexibility of the Pocillopora verrucosa microbiome. Ecol Evol 8:2240–52. 10.1002/ece3.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogoreutz C, Rädecker N, Cárdenas A, Gärdes A, Wild C, Voolstra CR. 2018. Dominance of endozoicomonas bacteria throughout coral bleaching and mortality suggests structural inflexibility of the Pocillopora verrucosa microbiome. Ecol Evol 8:2240–52. 10.1002/ece3.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pootakham W, Mhuantong W, Putchim L, Yoocha T, Sonthirod C, Kongkachana W, Sangsrakru D, Naktang C, Jomchai N, Thongtham Net al. 2018. Dynamics of coral-associated microbiomes during a thermal bleaching event. Microbiologyopen 7:e00604. 10.1002/mbo3.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pootakham W, Mhuantong W, Yoocha T, Putchim L, Jomchai N, Sonthirod C, Naktang C, Kongkachana W, Tangphatsornruang S. 2019. Heat-induced shift in coral microbiome reveals several members of the Rhodobacteraceae family as indicator species for thermal stress in Porites lutea. Microbiologyopen 8:e935. 10.1002/mbo3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshef L, Koren O, Loya Y, Zilber-Rosenberg I, Rosenberg E. 2006. The coral probiotic hypothesis. Environ Microbiol 8:2068–73. 10.1111/j.1462-2920.2006.01148.x. [DOI] [PubMed] [Google Scholar]
- Ricci F, Tandon K, Black JR, Lê Cao K-A, Blackall LL, Verbruggen H. 2022. Host traits and phylogeny contribute to shaping coral-bacterial symbioses. mSystems 7:e00044–22. 10.1128/msystems.00044-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond CE, Breitburg DL, Rose KA. 2005. The role of environmental generalist species in ecosystem function. Ecol Modell 188:279–95. 10.1016/j.ecolmodel.2005.03.002. [DOI] [Google Scholar]
- Robeson MS, O'rourke DR, Kaehler BD, Ziemski M, Dillon MR, Foster JT, Bokulich NA. 2021. RESCRIPt: reproducible sequence taxonomy reference database management. PLoS Comput Biol 17:e1009581. 10.1371/journal.pcbi.1009581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röthig T, Roik A, Yum LK, Voolstra CR. 2017. Distinct bacterial microbiomes associate with the deep-sea coral Eguchipsammia fistula from the red sea and from aquaria settings. Front Mar Sci 4:259. 10.3389/fmars.2017.00259. [DOI] [Google Scholar]
- Schmidt-Roach S, Miller KJ, Lundgren P, Andreakis N. 2014. With eyes wide open: a revision of species within and closely related to the Pocillopora damicornis species complex (Scleractinia; Pocilloporidae) using morphology and genetics. Zool J Linn Soc 170:1–33. 10.1111/zoj.12092. [DOI] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:1–18. 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp KH, Pratte ZA, Kerwin AH, Rotjan RD, Stewart FJ. 2017. Season, but not symbiont state, drives microbiome structure in the temperate coral astrangia poculata. Microbiome 5:120. 10.1186/s40168-017-0329-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale DA, Wernberg T. 2012. Ecological observations associated with an anomalous warming event at the Houtman Abrolhos Islands, Western Australia. Coral Reefs 31:441. 10.1007/s00338-012-0873-4. [DOI] [Google Scholar]
- Steinberg RK, Ainsworth TD, Moriarty T, Bednarek T, Dafforn KA, Johnston EL. 2022. Bleaching susceptibility and resistance of octocorals and anemones at the world's southern-most coral reef. Front Physiol 0:726. 10.3389/fphys.2022.804193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sully S, Burkepile D E, Donovan MK, Hodgson G, Van Woesik R. 2019. A global analysis of coral bleaching over the past two decades. Nat Commun 10:1264. 10.1038/s41467-019-09238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Yang H, Shi Q, Wang G. 2022. Changes in coral bacterial communities during a natural bleaching event linked to El Niño in the South China Sea. Reg Stud Mar Sci 53:102383. 10.1016/j.rsma.2022.102383. [DOI] [Google Scholar]
- Sunagawa S, Woodley CM, Medina M. 2010. Threatened corals provide underexplored microbial habitats. PLoS One 5:e9554. 10.1371/journal.pone.0009554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson DP, Bearham D, Graham F, Eagle JV. 2011. High latitude, deeper water coral bleaching at Rottnest Island, Western Australia. Coral Reefs 30:1107. 10.1007/s00338-011-0811-x. [DOI] [Google Scholar]
- Thurber RV, Willner-Hall D, Rodriguez-Mueller B, Desnues C, Edwards RA, Angly F, Dinsdale E, Kelly L, Rohwer F. 2009. Metagenomic analysis of stressed coral holobionts. Environ Microbiol 11:2148–63. 10.1111/j.1462-2920.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- Todd PA. 2008. Morphological plasticity in scleractinian corals. Biol Rev 83:315–37. 10.1111/j.1469-185X.2008.00045.x. [DOI] [PubMed] [Google Scholar]
- Tout J, Siboni N, Messer LF, Garren M, Stocker R, Webster NS, Ralph PJ, Seymour JR. 2015. Increased seawater temperature increases the abundance and alters the structure of natural Vibrio populations associated with the coral Pocillopora damicornis. Front Microbiol 6:432. 10.3389/fmicb.2015.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy AM, Koren O, Douglas N, Weil E, Harvell CD. 2015. Persistent shifts in Caribbean coral microbiota are linked to the 2010 warm thermal anomaly. Environ Microbiol Rep 7:471–9. 10.1111/1758-2229.12274. [DOI] [PubMed] [Google Scholar]
- Ulstrup K, Berkelmans R, Ralph P, Van Oppen M. 2006. Variation in bleaching sensitivity of two coral species across a latitudinal gradient on the Great Barrier Reef: the role of zooxanthellae. Mar Ecol Prog Ser 314:135–48. 10.3354/meps314135. [DOI] [Google Scholar]
- Van Oppen MJH, Blackall LL. 2019. Coral microbiome dynamics, functions and design in a changing world. Nat Rev Microbiol 17:557–67. 10.1038/s41579-019-0223-4. [DOI] [PubMed] [Google Scholar]
- Van Oppen MJH, Bongaerts P, Frade P, Peplow LM, Boyd SE, Nim HT, Bay LK. 2018. Adaptation to reef habitats through selection on the coral animal and its associated microbiome. Mol Ecol 27:2956–71. 10.1111/mec.14763. [DOI] [PubMed] [Google Scholar]
- Van Woesik R, Kratochwill C. 2022. A global coral-bleaching database, 1980–2020. Scientific Data 2022 9:1 9:1–7. 10.1038/s41597-022-01121-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Woesik R, Sakai K, Ganase A, Loya Y. 2011. Revisiting the winners and the losers a decade after coral bleaching. Mar Ecol Prog Ser 434:67–76. 10.3354/meps09203. [DOI] [Google Scholar]
- Voolstra CR, Ziegler M. 2020. Adapting with microbial help: microbiome flexibility facilitates rapid responses to environmental change. Bioessays 42:2000004. 10.1002/bies.202000004. [DOI] [PubMed] [Google Scholar]
- Wemheuer F, Taylor JA, Daniel R, Johnston E, Meinicke P, Thomas T, Wemheuer B. 2020. Tax4Fun2: prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ Microbiomes 15:1–12. 10.1186/s40793-020-00358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H, Chang W. 2016. Package “ggplot2.” (https://cran.microsoft.com/snapshot/2015-01-06/web/packages/ggplot2/ggplot2.pdf) [Accessed August 30, 2022].
- Williams AD, Brown BE, Putchim L, Sweet MJ. 2015. Age-related shifts in bacterial diversity in a reef coral. PLoS One 10:e0144902. 10.1371/journal.pone.0144902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo S, Yang S-H, Chen H-J, Tseng Y-F, Hwang S-J, De Palmas S, Denis V, Imahara Y, Iwase F, Yum Set al. 2017. Geographical variations in bacterial communities associated with soft coral Scleronephthya gracillimum. PLoS One 12:e0183663. 10.1371/journal.pone.0183663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Zhang Y, Ahmad M, Ling J, Zhou W, Zhang Y, Dong J. 2021. Microbial community structure shifts and potential symbiodinium partner bacterial groups of bleaching coral Pocillopora verrucosa in South China Sea. Ecotoxicol 30:966–74. 10.1007/s10646-021-02380-y. [DOI] [PubMed] [Google Scholar]
- Yu X, Yu K, Chen B, Liao Z, Liang J, Yao Q, Qin Z, Wang H, Yu J. 2021a. Different responses of scleractinian coral acropora pruinosa from Weizhou Island during extreme high temperature events. Coral Reefs 40:1697–711. 10.1007/s00338-021-02182-y. [DOI] [Google Scholar]
- Yu X, Yu K, Liao Z, Chen B, Deng C, Yu J, Yao Q, Qin Z, Liang J. 2021b. Seasonal fluctuations in symbiotic bacteria and their role in environmental adaptation of the scleractinian coral acropora pruinosa in high-latitude coral reef area of the South China Sea. Sci Total Environ 792:148438. 10.1016/j.scitotenv.2021.148438. [DOI] [PubMed] [Google Scholar]
- Zaneveld JR, Mcminds R, Vega Thurber R. 2017. Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat Microbiol 2:17121. 10.1038/nmicrobiol.2017.121. [DOI] [PubMed] [Google Scholar]
- Ziegler M, Grupstra CGB, Barreto MM, Eaton M, Baomar J, Zubier K, Al-Sofyani A, Turki AJ, Ormond R, Voolstra CR. 2019. Coral bacterial community structure responds to environmental change in a host-specific manner. Nat Commun 10:1–11. 10.1038/s41467-019-10969-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler M, Seneca FO, Yum LK, Palumbi SR, Voolstra CR. 2017. Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat Commun 8:1–8. 10.1038/ncomms14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Chen Y, Wang L, Zhang S, Li J. 2022. Differential responses of bacterial communities in coral tissue and mucus to bleaching. Coral Reefs 41:951–60. 10.1007/s00338-022-02261-8. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data underlying this article are available in an NCBI repository accessible at https://www.ncbi.nlm.nih.gov/, PRJNA802894.