Abstract
In CRISPR/Cas9 genome editing, the tight and persistent target binding of Cas9 provides an opportunity for efficient genetic and epigenetic modification on genome. In particular, technologies based on catalytically dead Cas9 (dCas9) have been developed to enable genomic regulation and live imaging in a site-specific manner. While post-cleavage target residence of CRISPR/Cas9 could alter the pathway choice in repair of Cas9-induced DNA double strand breaks (DSBs), it is possible that dCas9 residing adjacent to a break may also determine the repair pathway for this DSB, providing an opportunity to control genome editing. Here, we found that loading dCas9 onto a DSB-adjacent site stimulated homology-directed repair (HDR) of this DSB by locally blocking recruitment of classical non-homologous end-joining (c-NHEJ) factors and suppressing c-NHEJ in mammalian cells. We further repurposed dCas9 proximal binding to increase HDR-mediated CRISPR genome editing by up to 4-fold while avoiding exacerbation of off-target effects. This dCas9-based local inhibitor provided a novel strategy of c-NHEJ inhibition in CRISPR genome editing in place of small molecule c-NHEJ inhibitors, which are often used to increase HDR-mediated genome editing but undesirably exacerbate off-target effects.
INTRODUCTION
Since its induction, the clustered regularly interspaced short palindromic repeat (CRISPR) system has become a powerful, revolutionary genome editing tool with broad application in biology, agriculture and medicine (1,2). In CRISPR genome editing, site-specific DNA double strand breaks (DSBs) induced by CRISPR-associated (Cas) nucleases in eukaryotic cells are repaired mainly by two evolutionarily conserved DSB repair mechanisms, homology-directed repair (HDR) and non-homologous end joining (NHEJ), generating the desired DNA edits among varieties of repair products (3). In mammalian cells, the HDR pathway is restricted to the S/G2 phase of the cell cycle where the primary homologous template for HDR is provided by sister chromatids. The DNA ends of a DSB are resected to form the Rad51 filament for homology search and pairing in HDR of the DSB. In contrast, the NHEJ pathway operates throughout the cell cycle and is generally a faster process. NHEJ can be further divided into at least two sub-pathways, the primary classical NHEJ (c-NHEJ) and alternative end joining (a-EJ) (4,5). C-NHEJ requires the core NHEJ factors such as DNA-PK catalytic subunit (DNA-PKcs), Ku70/Ku80 and XRCC4/DNA ligase 4 to catalyze ligation of DNA ends. Upon Cas nuclease-induced DSBs, the Ku70/Ku80 heterodimer, the most abundant end-binding proteins in mammalian cells, binds to the DSB ends along with DNA-PKcs and recruits XRCC4/DNA ligase 4 for end ligation while protecting the ends from an attack by end processing enzymes. Previous studies have demonstrated that c-NHEJ is intrinsically accurate in repair of Cas nuclease-induced DSBs, the ends of which are readily ligatable (6–8). A-EJ is considered more error-prone and employed to re-ligate the ends if either of the core NHEJ factors is deficient or not engaged.
Given the slower process of HDR, the availability of homologous templates, the restriction of cell cycle stages and predominant c-NHEJ competition, HDR is generally much less efficient than c-NHEJ in mammalian cells. The low level of HDR often limits application of CRISPR genome editing. One major effort in CRISPR genome editing is to develop approaches to increase the HDR efficiency (3,9). Increasing local concentration of homologous templates at the DSB site has been attempted for enhancing HDR in CRISPR genome editing (3,9–11). Fusion of HDR facilitators to the widely used Cas nuclease Streptococcus pyogenes Cas9 (SpCas9) and arresting the cell cycle at the S/G2 phase by chemicals have improved HDR-mediated CRISPR genome editing (3,9,12–18). Chemical inhibition or genetic inactivation of c-NHEJ, the most dominant competing pathway against HDR, could channel DSBs that are supposedly repaired by c-NHEJ to HDR, thereby increasing the efficiency of HDR in repair of Cas nuclease-induced DSBs (3,8,9). Indeed, inhibition of c-NHEJ is often used to promote HDR-mediated genome editing (3,9,19–22). However, due to global inhibition of c-NHEJ, our recent study demonstrated that this approach unavoidably exacerbates off-target effect (8). We reason this off-target problem could be solved by local inhibition of c-NHEJ; but such a strategy has yet to be developed.
Binding of SpCas9 as well as many other Cas nucleases to its target is mediated by the base pairing between the sgRNA spacer and DNA target strand and by the interactions between the Cas protein and target DNA prior to DSB induction (23,24). These interactions entail strong and persistent binding of the Cas9–sgRNA complex to its target and help maintain its target residence for hours even after Cas9-induced DNA cleavage at some sites (25,26). Cas9-induced DSBs are exposed after target dissociation of Cas9–sgRNA. As exposure of Cas9-induced DSBs is prerequisite for engaging DSB repair, target residence of Cas9–sgRNA adds a layer of control on pathway choices in repair of Cas9-induced DSBs, contributing to the heterogeneity of CRISPR/Cas9 genome editing (8,24). The similarly persistent binding of nuclease dead SpCas9 (dSpCas9) to its target is capable to slow or block DNA replication and transcription in cells (27–29). It is also conceivable that tight target binding and persistent target residence of dCas9 enable efficient application of dCas9-based platforms in transcription regulation, epigenetic modification, genomic imaging, base editing and prime editing (30). Additionally, dSpCas9 proximal binding increases DNA cleavage by other Cas nucleases, thus enhancing both NHEJ- and HDR-mediated genome editing (31). In vitro assays indicated that dSpCas9 proximal binding modulated neighboring chromatin dynamics and increased the accessibility of Lachnospiraceae bacterium Cas12a (LbCas12a) target for DNA cleavage (32). Considering tight binding and persistent residence of Cas9–sgRNA at many target sites, dCas9–sgRNA targeting to a site adjacent to a DSB could compete with Ku70/Ku80 for binding to DNA ends of the DSB. Thus, dCas9 proximal binding might preclude the access of DNA ends to Ku70/Ku80, the end binding of which is necessary for efficient recruitment of DNA-PKcs and XRCC4/DNA ligase 4 and for efficient c-NHEJ repair of the DSB. This prompts us to hypothesize that dCas9 proximal binding may sufficiently and locally promotes HDR by suppressing c-NHEJ.
Here, we tested the hypothesis above and found that dSpCas9 loaded onto a site adjacent to a break stimulate HDR of this DSB. This HDR stimulation requires the presence of c-NHEJ factors in the cells. Further investigation revealed that dSpCas9 proximal binding blocked recruitment of the core NHEJ factors to the ends of the neighboring DSBs, thus locally suppressing c-NHEJ. We repurposed dSpCas9 proximal binding to increase HDR-mediated CRISPR genome editing by up to 4-fold and extended this strategy to catalytically dead Staphylococcus aureus Cas9 (dSaCas9). Unlike NHEJ inhibition by chemical or genetic approaches that exacerbate off-target effect in CRISPR genome editing (8), dCas9 proximal binding promotes HDR locally while avoiding exacerbation of off-target effect, thus providing an improved strategy of c-NHEJ inhibition in HDR-mediated CRISPR genome editing.
MATERIALS AND METHODS
Plasmids and single-stranded oligodeoxynucleotides (ssODN)
The U6-sgRNA plasmid for LbCas12a was generated by cloning CACCGAATTTCTACTAAGTGTAGAT(BbsI) aagtcttcgaattcgaagacgg (BbsI) TTTTTT into the BbsI sites of the U6-sgRNA vector for SpCas9 and the 22-nt spacer between the two newly inserted BbsI sites can be replaced. The sgRNA target sequences and respective mismatch mutations for SpCas9, SaCas9 and LbCas12a are listed in Supplementary Table S1. The expression plasmids for truncated and mismatched sgRNAs were constructed as described previously (33), and the expression plasmids for SpCas9 variants eSpCas9 or SpCas9-HF1 were described before (34,35). dSaCas9 was generated by site-directed mutation using KOD Plus-Neo Kit (TOYOBO) (36).
The donor plasmid containing truncated green fluorescent protein (GFP) for GFP correction experiments was derived from the HDR reporter plasmid by deleting the I-SceI-GFP cassette. The HDR reporter plasmid was previously constructed (37). The ssODN donor contained about 150-nt GFP homology flanking the I-SceI site on the HDR reporter and was synthesized by TsingKe Biological Technology (Supplementary Table S2). The donor plasmid for targeted GFP knock-in at the Rosa26 locus of mouse embryonic stem cells (mESCs) was generated by placing a homology arm to both sides of a PGK-GFP expression cassette. The donor sequences including homology arms are listed in Supplementary Table S2.
Cell lines
mESCs containing a single copy of the NHEJ reporter or the HDR reporter and U2OS cells containing a single copy of the HDR reporter were previously established as described before (8,37–40). mESCs were grown in medium supplied with 20% fetal bovine serum (Gibco), 1% penicillin-streptomycin (Gibco), 2 mM L-glutamine (Gibco), 0.1 mM β-mercaptoethanol (Sigma), 0.1 mM non-essential amino acid (Gibco), 1 mM sodium pyruvate (Gibco) and 1000 U/ml leukemia inhibitory factor (Millipore) on either MEF feeders or gelatinized plates. Human U2OS cells were cultured in high glucose DMEM containing 10% fetal bovine serum, 1% penicillin-streptomycin and 2 mM L-glutamine. Isogenic XRCC4+/+ and XRCC4–/– mESCs containing the HDR reporter and DNA-PKcs–/– and Ku80–/– HDR reporter mESCs along with isogenic wild-type clones were generated previously (8,41).
Generation of the I-SceI-GFP correction reporter
To generate I-SceI-GFP correction reporter mESC clones by the paired Cas9–sgRNA approach previously established (6), 2 × 105 mESCs harboring HDR reporter were transfected with the expression plasmids for Cas9 and paired sgRNAs targeting TrGFP cassette in a 24-well plate, and were seeded on mouse embryonic fibroblast (MEF) feeder cells at 10-cm plate at 2 days (d) post transfection for single clones without any antibiotic selection. Single clones were picked at 7–14 d post-transfection, expanded and verified by PCR along with Sanger sequencing. Similarly, to generate I-SceI-GFP correction reporter U2OS clones, 1.0 × 105 U2OS cells harboring HDR reporter were transfected with the expression plasmids for Cas9 and paired sgRNAs targeting TrGFP cassette in a 24-well plate. After 3 days, 200–400 cells were seeded onto a 100-mm plate. Single clones were picked after 14 d, expanded and verified by PCR along with Sanger sequencing. Primers for PCR were listed on (Supplementary Table S2).
Transfection and DSB repair reporter assays
Transfection of mESCs was done with Lipofectamine 2000 (Invitrogen) in 24-well plates as previously described (42). Total 2 × 105 mESCs harboring the HDR/NHEJ reporter were transfected with 0.5μg total DNA. For U2OS cells transfection, 1.0 × 105 cells were seeded on a 24-well plate and grown to 80–95% confluence. 0.8 μg total DNA were transfected by Lipofectamine 2000. Cells harboring the NHEJ or HDR reporter were transfected with pcDNA3β-I-SceI or the expression plasmids for SpCas9–sgRNA, LbCas12a-sgRNA or SaCas9–sgRNA as previously described (8).
In dSpCas9–sgRNA blockage experiments, cells were co-transfected with the expression plasmids for I-SceI, the LbCas12a-sgRNA complex or the SaCas9–sgRNA complex, together with the expression plasmids for dSpCas9–sgRNA. The ratio of I-SceI, LbCas12a-sgRNA or SaCas9–sgRNA to dSpCas9–sgRNA in transfection amount is 1:1. If necessary, cells were treated with the DNA-PKcs inhibitor NU7441 (TopScience Cat# T6276) or Nocodazole (Sigma-Aldrich Cat# M1404) at 6 h post-transfection. NU7441 was replaced with a fresh addition of the drug the next day. Nocodazole was withdrawn after 12 h and replaced by fresh medium for the rest of the experiment. GFP+ cells were determined by fluorescence-activated cell sorting (FACS) using Beckmann Coulter CytoFLEX at 72 h post-transfection. The frequencies of NHEJ, HDR and genome editing were calculated after being corrected with background readings and normalized with transfection efficiencies as described before (8).
In dSaCas9–sgRNA blockage experiments, cells were co-transfected with the expression plasmids for I-SceI or the SpCas9–sgRNA complex, together with the expression plasmids for dSaCas9–sgRNA. The ratio of I-SceI or SpCas9–sgRNA to dSaCas9–sgRNA in transfection amount is 1:1. GFP+ cells were determined by FACS at 72 h post-transfection.
HDR-based GFP correction and knock-in editing experiments
For HDR-based GFP correction, cells containing the single copy of inactive I-SceI-GFP were co-transfected with 0.25 μg plasmid DNA or 0.125 μg ssODN as HDR donor templates, the expression plasmids for I-SceI, SaCas9–sgRNA or LbCas12a-sgRNA, and/or the expression plasmids for dSpCas9–sgRNA, and treated with NU7441 and Nocodazole as needed. At 3 d post-transfection, GFP+ events were measured by FACS to determine the percentage of HDR-based gene correction events. To detect the GFP– cells, cells were determined by FACS at 4 days post-transfection. For HDR-based GFP knock-in at the mRosa26 locus, mESCs were transfected with a pCMV-β-globin intron-GFP plasmid donor and the expression plasmids for LbCas12a-sgRNA or SaCas9–sgRNA, together with the expression plasmids for dSpCas9–sgRNA. The percentages of HDR events were determined 10 d post-transfection.
In dSaCas9-mediated HDR stimulation experiments, donor templates including 0.25μg plasmid DNA or 0.125μg ssODN were transfected with the expression plasmids for I-SceI or SpCas9–sgRNA variants (i.e. eSpCas9–sgRNA, SpCas9-HF1-sgRNA and SpCas9-T17) and the expression plasmids for dSaCas9–sgRNA. GFP+ cells were determined by FACS at 72 h post-transfection.
Electrophoretic mobility shift assay
The in vitro DNA binding and electrophoretic mobility shift assay (EMSA) were performed as described previously (8). The dSpCas9 nuclease (PC1351, 0.5 μg/μl) was purchased from Inovogen Biotech. All sgRNAs used for dSpCas9 were synthesized by GenScript Biotech and were dissolved in RNA-free water to 1 μM before use. The primers labeled with either 5′-DyLight-680 were purchased from Takara BioMed (Supplementary Table S2). PCR was performed to generate 600–700 bp fluorescence-labeled DNA fragments. For the dSpCas9–sgRNA binding reaction, dSpCas9–sgRNA complex was pre-assembled by mixture of 0.5 pmol dSpCas9 with sgRNA and its variants for 1 h, then add 0.1 pmol target DNA to incubate for 1 h or 24 h. The samples were resolved on 4–20% SurePAGE non-denatured gel (GenScript) in 0.5× TBE buffer at 200 V for 150 min in 4°C cooling water for fluorescence-imaging analysis. The fluorescence imaging of gel electrophoresis was captured by Licor Odyssey infrared scanner and quantified by ImageJ. The percentages of unbound DNA were calculated as the ratios of the intensity of unbound DNA bands to the combined intensity of total bound and unbound DNA.
For the competition assay, 1 pmol dSpCas9 and 1 pmol sgRNA or sgRNA variants were incubated with 0.1 pmol target dsDNA for 2 h. We then added 1 pmol indicated SpCas9-20-nt sgRNA into the reaction solution to cleave dissociated DNA from preassembled dSpCas9–sgRNA–DNA for 6–24 h at 37°C. The reaction was quenched by the addition of 2 μl of denatured loading dye and the cleaved DNA was resolved by 2% agarose gel electrophoresis.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed using SimpleChIP® Plus Enzymatic Chromatin IP Kit (#9003, CST) following the manufacturer's instructions. Briefly, 2 × 105 HDR reporter or NHEJ reporter mESCs were transfected with expression plasmids for LbCas12a-sgRNAs, together with the expression plasmids for dSpCas9–sgRNAs. Total 107 cells were collected from at least three 24-well plates at 24 h after transfection, fixed with 1% formaldehyde at 37°C for 10 min and quenched with 0.125 M glycine for 5 min at room temperature. Cell pellets were washed and suspended in 1 ml Buffer A to separate the nuclei. The nuclei were then resuspended in 100 μl Buffer B and treated with 0.5 μl Micrococcal nuclease for 15 min to digest the DNA to length of 150–600 bp. The digest reaction was stopped with 10 μl 0.5M EDTA, the digested nuclei was pelleted by centrifugation at 12 000 × g at 4°C for 1 min, and the supernatant was removed. The nuclei pellets were suspended in 100 μl ChIP buffer and sonicated with several pulses to break the nuclear membrane. The membrane pellets were removed by centrifugation and 100 μl supernatant was diluted into 400 μl ChIP buffer. The chromatin supernatant was incubated with antibodies against Ku80 (#2753, CST), Mre11 (ab109623, Abcam) and Flag (#14793, CST), as well as nonspecific IgG antibody for 12 h. 20 μl Protein G magnetic beads was added to pull down chromatin fragments. After the crosslinked DNA-protein complex in the chromatin fragments was de-crosslinked, DNA were purified with spin columns from the ChIP Kit (#9003, CST). Specifically, 750 μl DNA Binding Buffer was added to 150 μl de-crosslinked DNA–protein solution. The mixture was transferred to the spin columns for centrifugation at 12 000 rpm for 1 min. The spin columns were washed twice with 750 μl DNA Wash Buffer. 50 μl DNA Elution Buffer was added to each spin column and the purified DNA were collected into a new tube by centrifugation at 12000 rpm for 1 min.
The primer pairs in Supplementary Table S2 were used to detect the enrichment of DNA fragments by qRT-PCR on CFX 96 Thermocycler (Bio-Rad). To perform qRT-PCR, 2 μl purified DNA were added to 18 μl reaction solution including 10 μl SYBR Green mix (Vazyme) and 1 μl primer pairs (10 μM) as indicated. The target DNA was amplified as follows: denaturation first at 95°C for 10 min and 40 cycles then at 95°C for 15 s, 55°C for 15 s and 72°C for 15 s. The fold enrichment of Ku80 and Flag-dSpCas9 at each genomic position relative to the negative IgG background was determined using the following equation: Fold enrichment = 2−(Ct genomic fragment with antibody of interest–Ct genomic fragment with IgG).
Genomic DNA extraction, PCR amplification and illumina deep sequencing
For analysis of targeted genome editing at endogenous genome loci, cells were collected after NHEJ induced by Cas9–sgRNAs. These cells were also transfected with pcDNA3β-GFP for transfection efficiencies. Genomic DNA (gDNA) was isolated from these cells using a gDNA purification kit (Axygen). The targeted regions were PCR-amplified with respective primers listed in Supplementary Table S2. PCR products purified with PCR Clean-up kit (Axygen) were end-repaired, adenylated at 3′ ends, ligated with adapters, purified, and amplified by the second round of PCR to incorporate the P7 and P5 Illumina adapters according to the manufacturer's protocols with Hieff NGS Ultima DNA Library Prep Kit for Illumina (Yeasen). The Illumina deep sequencing was performed at Novogene Co. Ltd and sequences were analyzed to identify edited events with different indels at repair junctions using DBS-Aligner as described previously (43).
Off-target analysis
Potential off-target sites were identified using the latest version of the CRISPR Off-Target prediction website (http://crispor.tefor.net/). All potential sites were ranked by an off-target hit score, and high-ranked potential sites were selected. Off-target sites were amplified by PCR with primers listed in Supplementary Table S2 after gDNA extraction from cells transfected with Cas9–sgRNA at 3 days post-transfection. Off-target editing efficiency was determined by Illumina deep sequencing. The off-target rate was determined as the ratio of off-target to on-target mutagenesis levels.
RESULTS
Target binding of dSpCas9 adjacent to a DSB promotes HDR of the DSB
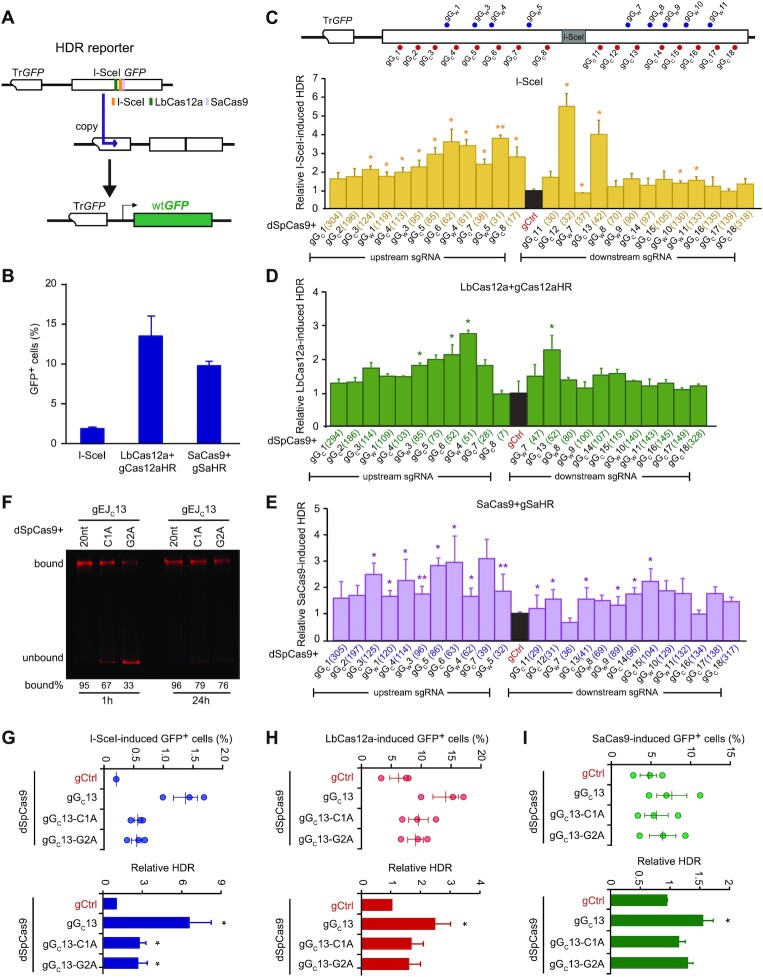
Previously, we found that SpCas9 with strong target binding and long residence at cleaved target affected DSB repair pathway choice (8). We speculated that the repair pathway choice for a given DSB could also be affected by dSpCas9 with such strong target binding and long target residence tethered to the DSB. Because dSpCas9 does not share its sgRNAs with LbCas12a or SaCas9, dSpCas9 can be used simultaneously with LbCas12a or SaCas9 to test the effect of dSpCas9 tethered to the DSB ends on HDR of the DSBs induced by LbCas12a or SaCas9. In contrast, both dSpCas9 and SpCas9 can use the same sgRNAs if both are present at the same time, interfering the activity of each other. As a result, dSpCas9 should not be used for SpCas9-induced DSBs. Thus, using mESCs harboring a single-copy HDR reporter (37,40), we tethered dSpCas9–sgRNA to the sites near a DSB induced by I-SceI, LbCas12a or SaCas9 in the HDR reporter and analyzed the effect on HDR of the DSB. The HDR reporter contains two copies of inactive GFP, the first truncated at 5’-end (i.e. TrGFP) and the second with the insertion of the 18-bp I-SceI site (i.e. I-SceI-GFP). Upon site-specific DNA breakage induced by I-SceI, LbCas12a-gCas12aHR or SaCas9-gSaHR around the I-SceI site, HDR of this DSB could use TrGFP of the sister chromatid as a homologous template to generate wild-type GFP (wtGFP), making cells GFP+ (Figure 1A). While I-SceI, LbCas12a-gCas12aHR or SaCas9-gSaHR induced GFP+ cells with the frequency at ∼1.8%, 12% and 10% in the reporter, respectively (Figure 1B), the level of HDR induced by I-SceI, LbCas12a-gCas12aHR or SaCas9-gSaHR was quantified as the frequency of induced GFP+ cells. The difference in these frequencies induced by I-SceI, LbCas12a and SaCas9 is likely due to different cutting efficiency of these nucleases. Among 25 sgRNAs tested for tethering dSpCas9 to the DSB induced by I-SceI, LbCas12a-gCas12aHR or SaCas9-gSaHR, some stimulated I-SceI-induced HDR by up to 5.5-fold, LbCas12a-induced HDR by about 2.5-fold and SaCas9-induced HDR by up to 3-fold whereas some others had little effect (Figure 1C–E, Supplementary Figure S1A–C and Supplementary Figure S2A–C). In the tested ranges, although SpCas9–sgRNAs at these tested sites efficiently mediated knock-out of the GFP gene, the HDR stimulation by dSpCas9 tethered to the DSB ends was negatively correlated with the distance of dSpCas9 from the DSBs induced by I-SceI and LbCas12a, but not by SaCas9 (Supplementary Figure S3A–D). This suggests that dSpCas9–sgRNAs loaded farther away from the breaks induced by I-SceI and LbCas12a tend to be less effective in stimulating HDR.
Figure 1.
dSpCas9–sgRNA proximal binding promotes HDR. (A) Schematic of the HDR reporter. Repair of I-SceI-, LbCas12a- and SaCas9-induced DSBs by HDR between sister chromatids can generate GFP+ cells. (B) The HDR efficiency induced by I-SceI, LbCas12a-gCas12aHR and SaCas9-gSaHR in HDR reporter mESCs. (C–E) Effect of dSpCas9–sgRNAs tethered adjacent to a DSB on HDR in HDR reporter mESCs. As indicated on the schematic of the reporter, the DSB was induced by I-SceI (C), LbCas12a-gCas12aHR (D) or SaCas9-gSaHR (E), and dSpCas9–sgRNAs were tethered to DNA sequences flanking the DSB. The distance of individual dSpCas9–sgRNA from the DSB was defined between the third PAM-proximal nucleotide of each dSpCas9–sgRNA binding site and the break point by I-SceI, LbCas12a or SaCas9 and indicated in parenthesis. After FACS measurement of nuclease-induced GFP+ cells, relative nuclease-induced HDR was calculated by normalizing ‘dSpCas9 + gCtrl’ control to 1.0. Columns indicate the mean ± standard error of the mean (S.E.M.) of at least three independent experiments, each in triplicates. Error bars indicate S.E.M. Significance was determined by Student's t-test between ‘gCtrl’ and each ‘dSpCas9–sgRNA’ and indicated by *P< 0.05 and **P< 0.01. (F) Target residence of dSpCas9-gGC13 and its sgRNA mismatch variants. dSpCas9–sgRNAs were incubated with fluorescence-labeled target DNAs for 1 and 24 h. DNAs bound with dSpCas9–sgRNAs or not were resolved by 4–20% native PAGE gel. The intensity ratio of bound DNA to total DNA were shown in percentages under each DNA gel. (G–I) Effect of dSpCas9–sgRNA target-binding affinity on HDR induced by I-SceI (G), LbCas12a-gCas12aHR (H) or SaCas9-gSaHR (I). Frequencies of GFP+ cells induced by I-SceI, LbCas12a and SaCas9 (top) were determined by FACS, and relative HDR (bottom) was calculated by normalizing ‘dSpCas9 + gCtrl’ treatment to 1.0. Each circle indicates one independent experiment, each in triplicates, and the mean of these independent experiments is also shown. Error bars indicate S.E.M. Columns indicate the mean of relative HDR. Significance was detected by Student's t-test between ‘dSpCas9 + gCtrl’ and ‘dSpCas9 + gGC13’ (*P< 0.05).
Among the four sgRNAs (i.e. gGC11, gGC12, gGW7 and gGC13) located adjacent to the break site, dSpCas9-gGW7 did not stimulate HDR induced by I-SceI, LbCas12a-gCas12aHR or SaCas9-gSaHR (Figure 1C–E). As SpCas9-gGW7 appeared to mediate target cleavage as efficient as the other three (Supplementary Figure S3A), it is possible that the target residence of dSpCas9-gGW7 would be shorter than other dSpCas9–sgRNA complex, thus failing to stimulate HDR. We thus performed the in vitro competition assays to compare the target residence of dSpCas9 complexed with gGC11, gGC12, gGW7 and gGC13 (Supplementary Figure S4A). After 2-h incubation of the dSpCas9–sgRNA complexes with their 620-bp target DNA, their respective SpCas9–sgRNAs were added into the reaction to cleave target DNA newly released from the dSpCas9–sgRNA–DNA ternary complexes. While SpCas9 complexed with the four sgRNAs cleaved their target DNA with similar efficiency in 6-h reaction, cleaved DNA detected the most at 12 h and 24 h were from the dSpCas9-gGW7-DNA complex (Supplementary Figure S4B). In other words, target DNA is more quickly dissociated from the dSpCas9–gGW7–DNA complex as compared to the other three dSpCas9–gRNA–DNA ternary complexes, indicating the shorter target residence for dSpCas9-gGW7.
To further determine whether target binding and residence were important for dSpCas9–sgRNA at the sites proximal to the DSB to stimulate HDR, we tested dSpCas9-gGC13, among the best that stimulated HDR, and its sgRNA variants C1A and G2A for their effects on HDR induced by I-SceI, LbCas12a-gCas12aHR and SaCas9-gSaHR. As expected, the mismatches reduced the target binding affinity of dSpCas9-gGC13 upon 1-h or 24-h incubation in vitro (Figure 1F). While dSpCas9-gGC13 strongly enhanced HDR induced by I-SceI, LbCas12a-gCas12aHR and SaCas9-gSaHR, both dSpCas9-gGC13 C1A and G2A lost most of this HDR stimulation (Figure 1G-I). Taken together, these results suggest that HDR stimulation by dSpCas9–sgRNAs is determined by their target binding ability and target residence adjacent to the break.
Local stimulation of HDR by dSpCas9 proximal binding is impaired in cells deficient for c-NHEJ
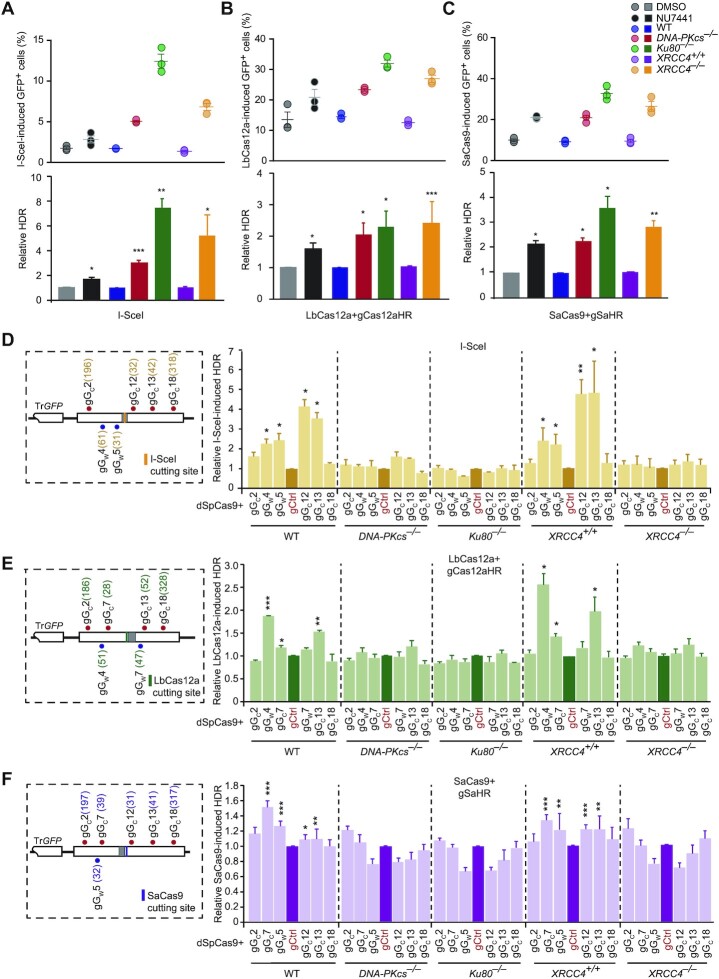
Previous studies have shown that inactivation of c-NHEJ would stimulate HDR (44–46). We wondered whether HDR stimulation by dSpCas9–sgRNAs was mediated by inactivation of c-NHEJ. We thus analyzed the effect of dSpCas9–sgRNAs on HDR in HDR reporter mESC where c-NHEJ was inactivated. Consistently, the DNA-PKcs inhibitor NU7441 increased HDR induced by I-SceI, LbCas12a-gCas12aHR and SaCas9-gSaHR by up to 100% (Figure 2A–C). Deletion of Ku80, DNA-PKcs or XRCC4 in HDR reporter mESC even more significantly stimulated HDR induced by I-SceI, LbCas12a-gCas12aHR and SaCas9-gSaHR (Figure 2A–C). However, while dSpCas9 in complex with gGW4, gGW5, gGC12 and gGC13 strongly stimulated I-SceI-induced HDR in wild-type mESC (WT and XRCC4+/+ mESC), no local stimulation was observed in isogenic DNA-PKcs–/–, Ku80–/– and XRCC4–/– mESC (Figure 2D and Supplementary Figure S5A). Similarly, as opposed to significant HDR stimulation in wild-type cells, LbCas12a- and SaCas9-induced HDR was not stimulated by dSpCas9 respectively loaded at the gGW4, gGC7 or gGC13 sites and at the gGC7, gGW5 or gGC13 sites adjacent to the break in isogenic DNA-PKcs–/–, Ku80–/– and XRCC4–/– mESC (Figure 2E, F and Supplementary Figure S5B, C). This indicates that the presence of c-NHEJ factors is required for local HDR stimulation by dSpCas9–sgRNA in cells.
Figure 2.
HDR stimulation by dSpCas9 proximal binding requires core NHEJ factors. (A–C) Effect on HDR of DNA-PKcs inhibition and DNA-PKcs, Ku80 or XRCC4 deficiency in mESCs transfected with expression plasmids for I-SceI (A), LbCas12a-gCas12aHR (B) or SaCas9-gSaHR (C). Frequencies of GFP+ cells induced by I-SceI, LbCas12a and SaCas9 (top) were determined by FACS at 3 days post-transfection and relative HDR (bottom) calculated by normalizing DMSO control, isogenic WT cells and isogenic XRCC4+/+ cells to 1.0. Each circle indicates one independent experiment, each in triplicates, and the mean of these independent experiments is also shown. Columns indicate the mean of relative HDR. Error bars indicate S.E.M. Significance was detected by Student's t-test and indicated by *P< 0.05, **P< 0.01 and ***P< 0.001. (D, E) Effect of dSpCas9 proximal binding on HDR induced by I-SceI (D), LbCas12a-gCas12aHR (E) or SaCas9-gSaHR (F) in mESCs deficient for DNA-PKcs, Ku80 or XRCC4 as compared to isogenic WT or XRCC4+/+ mESCs. HDR reporter mESCs were transfected with expression plasmids for I-SceI, LbCas12a-gCas12aHR or SaCas9-gSaHR, along with expression plasmids for dSpCas9–sgRNAs. Left: The HDR reporter was indicated with the cleavage site for I-SceI, LbCas12a-gCas12aHR or SaCas9-gSaHR and the target sites for dSpCas9–sgRNA target tethering. The distance between the third PAM-proximal nucleotide of each dSpCas9–sgRNA binding site and the DSB was shown in parenthesis. Right: Frequencies of GFP+ cells were measured by FACS at 3 days post-transfection and relative HDR was determined by normalizing control treatment (i.e. ‘gCtrl’) to 1.0. Columns indicate the mean ± S.E.M. of at least three independent experiments, each in triplicates. Error bars indicate S.E.M. Significance was detected by Student's t-test between ‘gCtrl’ and each ‘dSpCas9–sgRNA’ and indicated by *P< 0.05, **P< 0.01 and ***P< 0.001.
dSpCas9 tethered adjacent to a DSB inhibits c-NHEJ of the DSB
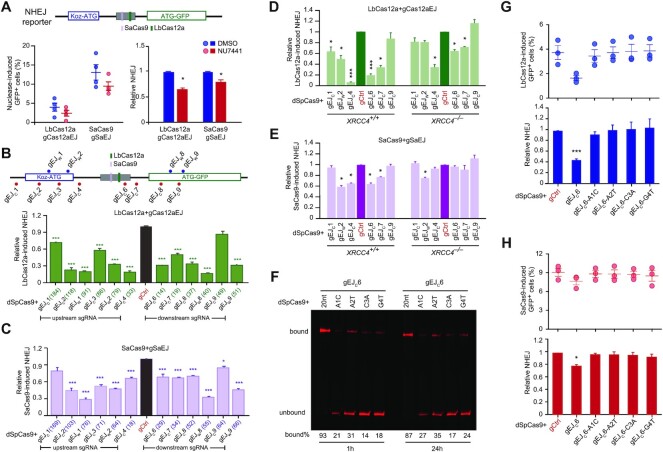
Proximal target binding of dSpCas9 may block binding of c-NHEJ factors to the ends of a DSB induced by I-SceI, LbCas12a or SaCas9, thereby suppressing c-NHEJ and stimulating HDR. To directly evaluate the effect of proximal dSpCas9–sgRNA target binding on c-NHEJ, we thus used an NHEJ reporter to quantitatively measure NHEJ and to compare the effect of DNA-PKcs inhibition and dSpCas9–sgRNA on NHEJ induced by LbCas12a and SaCas9 (Figure 3A and Supplementary Figure S6A). In this NHEJ reporter previously developed (38), an upstream, out-of-frame translation start site (‘Koz-ATG’) inactivates GFP (Supplementary Figure S6A). Upon DSBs induced by Cas nucleases at a 34-bp intervening sequence between ‘Koz-ATG’ and ‘ATG-GFP’ cassette, repair by mutagenic NHEJ (mNHEJ) with net loss of ‘3n + 1’ bp or net addition of ‘3n + 2’ bp at the repair junction could correct the reading frame of GFP, making cells GFP+. Thus, the frequency of GFP+ cells would represent the level of NHEJ in cells.
Figure 3.
Strong proximal binding of dSpCas9–sgRNA inhibits c-NHEJ. (A) Effect of DNA-PKcs inhibition by NU7441 on NHEJ induced by LbCas12a-gCas12aEJ and SaCas9-gSaEJ in NHEJ reporter mESCs. Cells were transfected with expression plasmids for LbCas12a-gCas12aEJ and SaCas9-gSaEJ, and site-specific DSBs were induced at the sites indicated in the NHEJ reporter. Frequencies of nuclease-induced GFP+ cells (left) were measured by FACS and relative NHEJ (right) was calculated by normalizing DMSO treatment to 1.0. Each circle indicates one independent experiment, each in triplicates, and the means of these independent experiments are also indicated. Error bar denotes S.E.M. Columns indicate the mean ± S.E.M. of relative NHEJ. Significance was analyzed by Student's t-test between ‘DMSO’ and ‘NU7441’ and indicated by *P< 0.05. (B, C) Effect of dSpCas9–sgRNA tethered adjacent to a DSB on NHEJ in NHEJ reporter mESCs. As indicated on the schematic of the reporter, the DSB was induced by LbCas12a-gCas12aEJ (B) or SaCas9-gSaEJ (C) at or around the I-SceI site, and dSpCas9–sgRNA was tethered to DNA sequences flanking the DSB. Frequencies of GFP+ cells were measured by FACS and relative NHEJ was calculated by normalizing ‘dSpCas9-gCtrl’ control (i.e. ‘gCtrl’) to 1.0. Columns indicate the mean ± S.E.M. of at least three independent experiments, each in triplicates. Error bars indicate S.E.M. The number in parenthesis indicated the distance between the third PAM-proximal nucleotide of each dSpCas9–sgRNA and the break point by LbCas12a or SaCas9. Significance was determined by Student's t-test between ‘gCtrl’ and each ‘dSpCas9–sgRNA’ and indicated by *P< 0.05, **P< 0.01 and ***P< 0.001. (D, E) Effect of dSpCas9 proximal binding on NHEJ induced by LbCas12a-gCas12aEJ (D) or SaCas9-gSaEJ (E) in XRCC4+/+ and XRCC4–/– mESCs. NHEJ reporter mESCs were transfected with expression plasmids for LbCas12a-gCas12aEJ or SaCas9-gSaEJ, along with expression plasmids for dSpCas9–sgRNAs. Frequencies of GFP+ cells were measured by FACS at 3 d post-transfection and relative NHEJ was determined by normalizing control treatment (i.e. ‘gCtrl’) to 1.0. Columns indicate the mean ± S.E.M. of at least three independent experiments, each in triplicates. Error bars indicate S.E.M. Significance was detected by Student's t-test between ‘gCtrl’ and each ‘dSpCas9–sgRNA’ and indicated by *P< 0.05, **P< 0.01 and ***P< 0.001. (F) Target residence of dSpCas9-gEJC6 and its sgRNA mismatch variants. dSpCas9–sgRNAs were incubated with fluorescence-labeled target DNAs for 1 and 24 h. DNAs bound with dSpCas9–sgRNAs or not were resolved by 4–20% native PAGE gel. The intensity ratio of bound DNA to total DNA were shown in percentages under each DNA gel. (G, H) Effect of dSpCas9–sgRNA target-binding affinity on NHEJ induced by LbCas12a (G) or SaCas9 (H). Frequencies of GFP+ cells induced by LbCas12a and SaCas9 (top) were determined by FACS, and relative NHEJ (bottom) was calculated by normalizing ‘dSpCas9 + gCtrl’ treatment to 1.0. Each circle indicates one independent experiment, each in triplicates, and the mean of these independent experiments is also shown. Error bars indicate S.E.M. Columns indicate the mean of relative NHEJ. Significance was detected by Student's t-test between ‘dSpCas9 + gCtrl’ and ‘dSpCas9 + gEJC6’ (*P< 0.05 and ***P< 0.001).
NU7441 reduced mNHEJ induced by LbCas12a-gCas12aEJ and SaCas9-gSaEJ in wild-type mESC by 40% and 25%, respectively (Figure 3A). As shown previously (8,47), c-NHEJ induced by LbCas12a and SaCas9 is mostly mutagenic due to enrichment of mNHEJ events after repeated cleavage of accurate NHEJ products by LbCas12a and SaCas9 at their target sites. Inactivation of c-NHEJ by NU7441 thus reduced LbCas12a- and SaCas9-induced NHEJ represented by the level of mNHEJ. We targeted dSpCas9 in complexed with gEJC6, gEJC7, gEJC8 or gEJC9 to the sites adjacent to LbCas12a- or SaCas9-induced DSBs and found that similar to NU7441, dSpCas9–sgRNAs bound to their targets upstream or downstream of the LbCas12a-gCas12aEJ and SaCas9-gSaEJ target sites suppressed LbCas12a- and SaCas9-induced mNHEJ by 40–80% (Figure 3B, C and Supplementary Figure S6B, C). These data suggest that target binding of dCas9–sgRNA adjacent to a DSB inhibits c-NHEJ of the DSB as does chemical inhibition of DNA-PKcs by NU7441.
Similar to the effect of DNA-PKcs inhibition by NU7441 on LbCas12a- and SaCas9-induced NHEJ, deletion of XRCC4 also reduced mNHEJ induced by LbCas12a-gCas12aEJ and SaCas9-gSaEJ (Figure 3D, E and Supplementary Figure S6D, E). However, while dSpCas9-gEJW2, dSpCas9-gEJC4, dSpCas9-gEJC6 and dSpCas9-gEJC7 inhibited LbCas12a- or SaCas9-induced mNHEJ in XRCC4+/+ mESC, this local inhibition of LbCas12a- and SaCas9-induced mNHEJ was reduced or even abolished in XRCC4–/– mESC (Figure 3D, E and Supplementary Figure S6D, E). This demonstrated that presence of c-NHEJ factors is required for local NHEJ inhibition by dSpCas9 proximal binding.
To determine whether target binding or residence was important for dSpCas9–sgRNA to suppress c-NHEJ, we tested dSpCas9-gEJC6, among the best that locally suppresses c-NHEJ, and its mismatch sgRNA mutants A1C, A2T, C3A and G4T for their effects on c-NHEJ induced by LbCas12a and SaCas9. As dSpCas9 complexed with either of these gEJC6 mismatch mutants was less able to bind their DNA substrate as opposed to dSpCas9-gEJC6 (Figure 3F), it was not surprised that dSpCas9 complexed with these mismatch sgRNA mutants lost the ability to suppress c-NHEJ induced by LbCas12a-gCas12aEJ and SaCas9-gSaEJ (Figure 3G, H). Taken together, these results supported that local inhibition of c-NHEJ by dSpCas9–sgRNAs is reliant upon the target binding ability and target residence of dSpCas9–sgRNAs adjacent to the break.
Target binding of dSpCas9 adjacent to a DSB blocks local recruitment of c-NHEJ factors
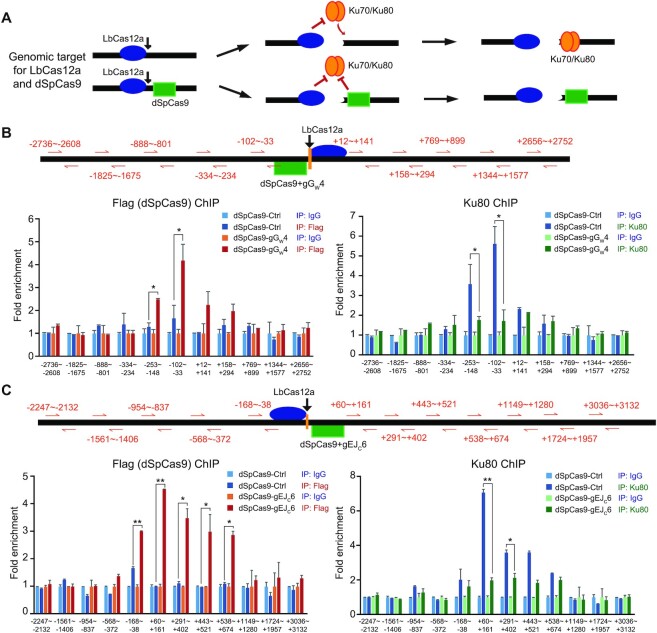
It is possible that proximal target binding of dSpCas9 may block binding of c-NHEJ factors to the ends of a DSB, thereby suppressing c-NHEJ and stimulating HDR in repair of this DSB. To examine this possibility, we performed the ChIP assays using HDR reporter and NHEJ reporter mESCs to directly determine whether dSpCas9 loaded onto its target sites near a DSB induced by LbCas12a could block recruitment of the core NHEJ factor Ku80 to the ends of the DSB (Figure 4A). Upon LbCas12a-induced DNA cleavage, the PAM-distal end is free and the PAM-proximal end remained bound with LbCas12a-sgRNA (Figure 4A) (48–51). We thus selected a target site within the free end for dSpCas9-gGW4 loading at -54bp ∼ -31bp in the HDR reporter and for dSpCas9-gEJC6 loading at +2 ∼ +25 in the NHEJ reporter (Figure 4B, C). DNA fragments were indeed enriched by Flag-tagged dSpCas9 at –253 ∼ –148 and –102 ∼ –33 in the HDR reporter and at –168 ∼ –38, +60 ∼ +161, +291 ∼ +402, +443 ∼ +521 and + 538 ∼ +674 in the NHEJ reporter near the free end of LbCas12a-induced DSB in cells transfected with dSpCas9-gGW4 (Figure 4B, C). In the absence of dSpCas9-gGW4 expression, DNA fragments were highly enriched by Ku80 at the free end much more than at the end bound with LbCas12a (i.e. –253 ∼ –148 and –102 ∼ –33 versus +12 ∼ +141 and +158 ∼ +294 in the HDR reporter, and +60 ∼ +161 and +291 ∼ +402 versus –168 ∼ –38 and –568 to –372 in the NHEJ reporter) (Figure 4B, C). However, this Ku80 recruitment was abolished by dSpCas9-gGW4 expression and subsequent enrichment of dSpCas9-gGW4 at its target site within the free end (Figure 4B, C). This suggested that tethering dSpCas9 to a target near a DSB blocks recruitment of c-NHEJ factors to the DSB and helped explain how proximal target binding of dSpCas9 locally suppresses c-NHEJ and stimulates HDR in DSB repair.
Figure 4.
dSpCas9–sgRNA proximal binding blocks recruitment of Ku80. (A) Schematic of the competitive binding between dSpCas9 (green square) and Ku70/Ku80 (orange tandem oval) at the end of DSBs induced by LbCas12a (blue oval). While the Ku70/Ku80 heterodimer is able to bind the free ends of LbCas12a-induced DSBs, it is expected that this recruitment could be blocked by post-cleavage target residence of LbCas12a at the PAM-proximal end and the target binding of dSpCas9 to a site within the free end. (B, C) Binding of Flag-dSpCas9-gGW4 or Flag-dSpCas9-gGC6 and Ku80 to DSBs induced by LbCas12a-gCas12aHR in the HDR reporter (B) and by LbCas12a-gCas12aEJ in the NHEJ reporter (C). Reporter mESCs were transfected with expression plasmids for LbCas12a-gCas12aHR or LbCas12a-gCas12aEJ, along with expression plasmids for Flag-dSpCas9-gGW4 or Flag-dSpCas9-gGC6. Binding of Flag-dSpCas9 and Ku80 to the DSBs induced by LbCas12a was detected by ChIP analysis performed with anti-Flag antibody (left) and anti-Ku80 antibody (right) as well as with anti-IgG background control. Fold enrichment of Flag-dSpCas9 and Ku80 at each position was assessed by real-time qPCR amplification using primer pairs located at varying distance away from the LbCas12a cleavage site as indicated in orange arrows and numbers and calculated relative to 1 for fold enrichment with the negative IgG antibody control. The mean and S.E.M for four independent experiments were shown. Significance as indicated was detected by Student's t-test: *P< 0.05 and **P< 0.01.
Proximal binding of dSpCas9 enhances HDR-mediated LbCas12a/SaCas9 genome editing
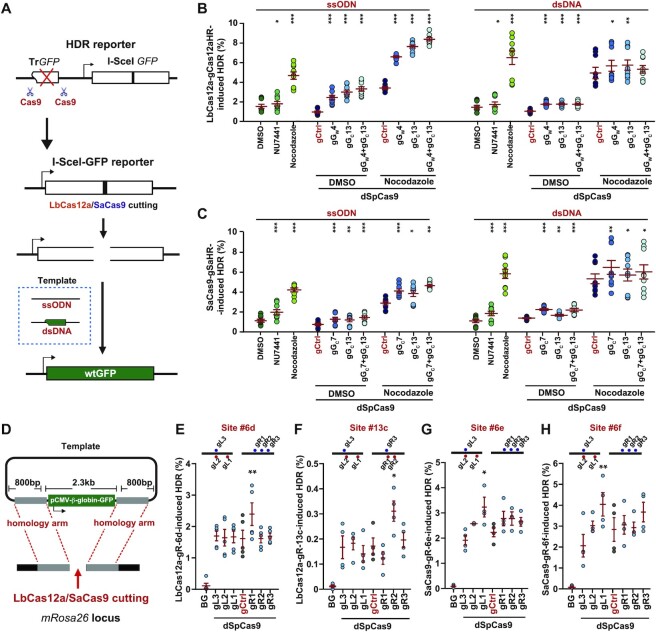
Given that dSpCas9–sgRNA loaded onto locations adjacent to the break facilitates HDR by suppressing c-NHEJ, we wondered whether this proximal binding of dSpCas9–sgRNA could improve the HDR-based knock-in or gene correction in CRISPR genome editing. To address this question, we first used SpCas9–sgRNA to precisely delete the TrGFP cassette from the single-copy chromosomal HDR reporter in our HDR reporter mESCs and U2OS cells and generated an I-SceI-GFP gene correction reporter (Figure 5A). After induction of a site-specific DSB in the reporter by LbCas12a-gCas12aHR or SaCas9-gSaHR, the inactive I-SceI-GFP gene could be corrected to WT GFP with either ssODN containing the correct GFP sequences overlapping the I-SceI site or a double-stranded DNA (dsDNA) plasmid harboring the TrGFP copy as the homologous template (Figure 5A). In fact, with either of homologous donors, LbCas12a- and SaCas9-induced HDR respectively converted ∼1–2% and 0.5–1% GFP– cells into GFP+ cells (Figure 5B, C). Treatment with NU7441 showed a modest enhancing effect on LbCas12a- or SaCas9-induced gene correction while Nocodazole at 0.25 μM increased the LbCas12a-induced gene correction frequencies by about 2.5-fold with the ssODN template and by about 4-fold with the plasmid DNA template and the SaCas9-induced gene correction frequencies by about 4-fold with the ssODN template and by about 6-fold with the plasmid DNA template (Figure 5B, C).
Figure 5.
dSpCas9–sgRNA proximal binding stimulates HDR with ssODN and dsDNA donors. (A) Schematic for generating the I-SceI-GFP correction reporter. The TrGFP copy of the original HDR reporter was deleted by paired Cas9–sgRNA in HDR reporter mESCs to generate the I-SceI-GFP correction reporter. Upon LbCas12a- or SaCas9-induced DNA breakage in the modified HDR reporter, wtGFP can be generated by HDR with exogenous ssODN or plasmid dsDNA homologous templates. (B, C) HDR stimulation by dSpCas9–sgRNA tethered adjacent to a break in I-SceI-GFP reporter mESCs. A site-specific DSB was induced by LbCas12a-gCas12aHR (B) or SaCas9-gSaHR (C), and GFP+ cells were generated by HDR of the DSB with ssODN or plasmid dsDNA homologous templates. Frequency of GFP+ cells representing the level of HDR was determined by FACS. During HDR, reporter cells were treated with NU7441 or Nocodazole alone, or with expression of individual dSpCas9–sgRNA or dSpCas9-gCtrl together with DMSO or Nocodazole as indicated. Each circle indicated one independent experiment, each in triplicates, and the mean of at least six independent experiments was also indicated. Error bars indicated S.E.M. Significance was analyzed by Student's t-test between each control group (‘DMSO’ and ‘gCtrl’) and each sample group and indicated by * for P< 0.05, **P< 0.01 and ***P< 0.001. (D) Schematic for HDR-mediated knock-in (KI) of a GFP gene into the mouse Rosa26 locus targeted by LbCas12a or SaCas9. The targeting vector contains the CMV promoter-β-globin intron-GFP (pCMV-β-globin-GFP) cassette flanked by ∼800 bp Rosa26 homology arms on either side. The sgRNAs adjacent to the breakpoint were generated to guide dSpCas9 binding. (E–H) Enhancement of HDR-mediated KI by dSpCas9–sgRNA tethered adjacent to the site of KI. mESCs were co-transfected with expression plasmids for LbCas12a-gR-6d (E), LbCas12a-gR-13c (F), SaCas9-gR-6e (G) or SaCas9-gR-6f (H), together with the pCMV-β-globin-GFP KI template and expression plasmids for 6 individual dSpCas9–sgRNAs or control vector (gCtrl). Cells transfected with the pCMV-β-globin-GFP KI template alone without DSB induction at the KI site served as background (BG). Frequencies of GFP+ cells were analyzed by FACS at 10 d post-transfection and the level of HDR induced by LbCas12a or SaCas9, representing the HDR-mediated KI efficiency, was corrected by the BG level and normalized with transfection efficiency. Each circle indicated one independent experiment, each in triplicates, and the mean of at least three independent experiments was also indicated. Error bars indicated S.E.M. Significance was analyzed by Student's t-test between ‘dSpCas9-gCtrl’ and each sample group and indicated by *P< 0.05 and **P< 0.01.
Because LbCas12a- and SaCas9-induced HDR in the HDR reporter were respectively stimulated by the proximal binding of dSpCas9-gGW4 or dSpCas9-gGC13 near LbCas12a-induced DSBs and by the proximal binding of dSpCas9-gGC7 or dSpCas9-gGC13 near SaCas9-induced DSBs (Figures 1D, E and 2E, F), we analyzed the effect of the dSpCas9 proximal binding on the gene correction mediated by LbCas12a and SaCas9. Targeting dSpCas9 with gGW4, gGC13 and gGW4/gGC13 to locations near the break increased LbCas12a-induced gene correction by more than 2-fold with the ssODN template or by nearly 2-fold with the plasmid dsDNA template (Figure 5B). The proximal target binding of dSpCas9 enhanced LbCas12a-induced gene correction further in the presence of Nocodazole, more than 2-fold over Nocodazole alone and up to 8-fold in comparison to the DMSO mock treatment (Figure 5B). Similarly, targeting dSpCas9 with gGC7, gGC13 and gGC7/gGC13 increased SaCas9-induced gene correction by about 2-fold with the ssODN template or with the plasmid dsDNA template, and in combination with Nocodazole, dSpCas9 proximal binding further elevated SaCas9-induced gene correction (Figure 5C). In human U2OS cells, dSpCas9 proximal binding also elevated the LbCas12a- and SaCas9-induced gene correction frequencies with the ssODN template or with the plasmid dsDNA template, and such elevation was further stimulated in combination with Nocodazole treatment (Supplementary Figure S7A, B).
We also tested whether dSpCas9 proximal binding would improve targeted knock-in of a larger insert at a natural genomic site. We loaded dSpCas9–sgRNA onto the locations near LbCas12a- or SaCas9-induced site-specific DSBs at the Rosa26 targeting sites and analyzed the efficiency of targeting 2.3-kb GFP expression cassette along with 800-bp homologous sequences on either side of a donor plasmid into the Rosa26 targets (Figure 5D). Gene targeting mediated by LbCas12a and SaCas9 was highly efficient at three sites with 1–3% of GFP+ frequency and less efficient at 1 site with 0.2% of GFP+ frequency (Figure 5E–H). Of note, without induction of a DSB at the targeting site, the random integration of donor plasmid is negligible (Figure 5E–H). Among sgRNAs guiding dSpCas9 proximal binding, many had little stimulatory effect on gene targeting induced by LbCas12a or SaCas9. However, the efficiency of gene targeting induced by LbCas12a-gR-6d, LbCas12a-gR-13c, SaCas9-gR-6e and SaCas9-gR-6f was elevated from 1.6% to 2.3% by dSpCas9-gR1, from 0.15% to 0.3% by dSpCas9-gR2, from 2.1% to 3.2% by dSpCas9-gL1, and from 2.6% to 4.0% by dSpCas9-gL1, respectively (Figure 5E–H). This suggests that dSpCas9 proximal binding could be designed to locally increase the efficiency of HDR-mediated gene targeting.
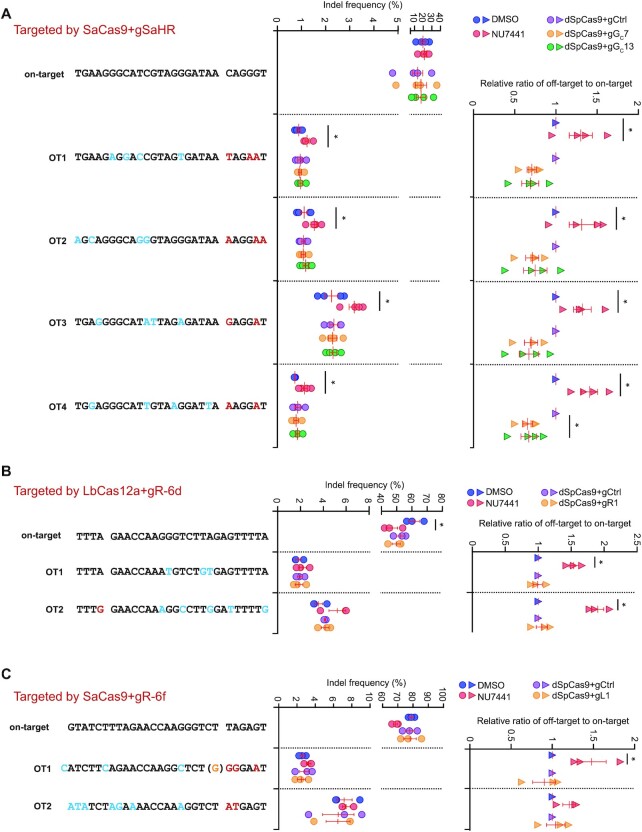
Proximal binding of dSpCas9 does not exacerbate off-target effects
Inactivation of c-NHEJ by chemical or genetic approaches is often used to enhance HDR-mediated CRISPR genome editing; however, our previous study revealed that this strategy often caused stronger off-target effects due to its global impact (8). In contrast, due to its localized action, dSpCas9 proximal target binding was expected to limit its influence on off-target sites. To test this possibility, we analyzed the frequencies of indels at on-target and off-target sites of SaCas9-gSaHR after respective treatment with NU7441 and dSpCas9 proximal target binding, both of which stimulate HDR induced by SaCas9-gSaHR (Figure 5B). NU7441 did not alter the on-target indel efficiency but increased mutagenesis significantly relative to DMSO at four different gSaHR off-target sites (Figure 6A). Using the ratio of off-target to on-target indel levels as a metric of off-target effect, we observed that NU7441 caused significant reduction in target specificity as anticipated (Figure 6A). In contrast, neither gGC7 nor gGC13 in complex with dSpCas9 enhanced mutagenesis as compared to the sgRNA control at off-target sites (Figure 6A).
Figure 6.
Proximal binding of dSpCas9–sgRNA adjacent to a DSB for HDR stimulation induces no exacerbation of off-target effect for SaCas9 with gSaHR (A), LbCas12a with gR-6d (B) and SaCas9 with gR-6f (C). HDR reporter mESCs were transfected with expression plasmids for SaCas9-gSaHR, LbCas12a-gR-6d and SaCas9-gR-6f, and then either treated with DMSO or NU7441, or co-transfected with expression plasmids for dSpCas9–sgRNA tethered adjacent to the on-target cutting site. The indel frequency at on-target and selected off-target sites as indicated were measured by amplicon deep sequencing and defined as the ratio of edited reads to total reads normalized by transfection efficiency. The ratio of off-target frequency to on-target frequency, i.e. ratio of off-target to on-target, indicated off-target effect. Relative ratio of off-target to on-target was determined by normalizing control treatment (i.e. ‘DMSO’ or ‘dSpCas9-gCtrl’) to 1.0. Each circle or triangle indicated one independent experiment, each in triplicates, and the mean of these independent experiments was also shown. Error bars indicated S.E.M. Statistical significance was detected by Student's t-test between ‘DMSO’ and ‘NU7441’ and indicated by *P< 0.05.
Because NU7441, dSpCas9-gR1 and dSpCas9-gL1 improved gene targeting induced by LbCas12a-gR-6d and SaCas9-gR-6f at the natural genomic site (Figure 5E, H), we also analyzed NU7441, dSpCas9-gR1 and dSpCas9-gL1 for their effect on the off-target activities of LbCas12a-gR-6d and SaCas9-gR-6f. We found that both NU7441 and dSpCas9 binding slightly reduced on-target editing of two sites by about 20–30%, suggesting repeated cleavage by LbCas12a and SaCas9 (Figure 6B, C). At off-target sites, NU7441 increased the mutagenesis whereas neither gR1 nor gL1 in complex with dSpCas9 did (Figure 6B, C). These results indicate that local dSpCas9 proximal binding, unlike NU7441, avoid exacerbating off-target effect while enhancing HDR-mediated genome editing.
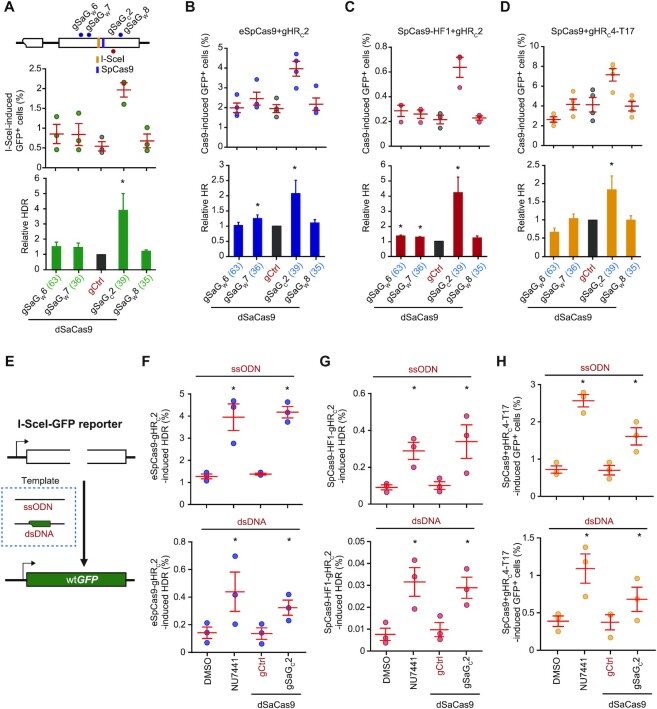
Proximal binding of dSaCas9 enhances HDR-mediated SpCas9 genome editing
Several SpCas9–sgRNA variants including eSpCas9, SpCas9-HF1 and truncated sgRNA have been engineered to improve the specificity of SpCas9–sgRNA and reduce off-target effect in genome editing; However, this improvement is often offset by reduced efficiency of on-target editing (8,33–35). Because of a reduction in their target interaction, chemical inhibition of c-NHEJ could efficiently stimulate HDR induced by these SpCas9–sgRNA variants as we previously showed (8). Undesirably, off-target effects of these SpCas9–sgRNA variants are also expected to increase as those of LbCas12a or SaCas9 do. The dCas9 proximal binding strategy could thus be applied to improve the efficiency of HDR-mediated genome editing by these more target-specific SpCas9–sgRNA variants while avoiding exacerbation of any off-target effect. Because dSpCas9 shares the same sgRNAs with SpCas9–sgRNA variants, dSpCas9 cannot be used for DSBs induced by the SpCas9–sgRNA variants. Instead, the catalytically dead SaCas9 (dSaCas9), which does not share the same sgRNAs with the SpCas9–sgRNA variants, could be used. Thus, using dSaCas9 as previously reported (36), we started to analyze the effect of dSaCas9 proximal binding on HDR by SpCas9-HF1, eSpCas9 and SpCas9-truncated 17-nt sgRNA (i.e. T17). We first tethered dSaCas9–sgRNA to the region within 63 bp of I-SceI-induced DSB in the HDR reporter (Figure 7A). Among the 4 sgRNAs, gSaGC2 stimulated I-SceI-induced HDR by near 4-fold while the other three gSaGW6, gSaGW7 and gSaGW8 had little stimulation (Figure 7A), likely due to different distance to the break and different target binding affinity of dSaCas9 in complex with either of these four sgRNAs (Supplementary Figure S8). Similarly, among the 4 sgRNAs, gSaGC2 elicited 2-, 4- and 2-fold stimulation of HDR induced by eSpCas9-gHRC2, SpCas9-HF1-gHRC2 and SpCas9-gHRC4-T17, respectively when dSaCas9 was loaded adjacent to the DSB by gSaGC2 (Figure 7B–D). Together, these data indicated dSaCas9 proximal target binding to the break site also functioned as a local c-NHEJ inhibitor to promote HDR.
Figure 7.
dSaCas9–sgRNA proximal binding stimulates HDR. (A–D) Effect of dSaCas9–sgRNA tethered adjacent to a DSB on HDR induced by I-SceI (A), eSpCas9-gHRC2 (B), SpCas9-HF1-gHRC2 (C), SpCas9-gHRC4-T17 (D) in HDR reporter mESCs. Upon DNA breakage induced by I-SceI and SpCas9 as indicated on the schematic of the reporter, 4 dSaCas9–sgRNAs were individually tethered to DNA sequences flanking the DSB to influence HDR. Frequency of GFP+ cells was measured by FACS and relative HDR was calculated relative to 1.0 for ‘dSaCas9 + gCtrl’ control. Columns indicated the mean ± S.E.M. of at least three independent experiments, each in triplicates. Error bars indicated S.E.M. The number in parenthesis following each sgRNA indicated the distance between the closest point of the PAM-containing 27-nt target site of each dSaCas9–sgRNA to the break point by I-SceI, eSpCas9-gHRC2, SpCas9-HF1-gHRC2, SpCas9-gHRC4-T17. Significance was determined by Student't-test between ‘gCtrl’ and each ‘dSaCas9–sgRNA’ and indicated by *P< 0.05 and **P< 0.01. (E) Schematic for the I-SceI-GFP correction reporter. Upon site-specific DNA breakage in the modified HDR reporter, wtGFP could be generated by HDR with exogenous ssODN or plasmid dsDNA homologous templates. (F–H) HDR stimulation by dSaCas9–sgRNA tethered adjacent to a DSB induced by eSpCas9-gHRC2 (F), SpCas9-HF1-gHRC2 (G), SpCas9-gHRC4-T17 (H) in I-SceI-GFP reporter mESCs. GFP+ cells were generated by HDR of the DSB with ssODN or plasmid dsDNA homologous templates. Frequency of GFP+ cells was determined by FACS. During HDR, reporter cells were treated with NU7441 alone, or with expression of individual dSaCas9-gSaGC2 or dSaCas9-gCtrl together with DMSO as indicated. Each circle indicates one independent experiment, each in triplicates, and the mean of at least six independent experiments is also indicated. Error bars indicate S.E.M. Significance was analyzed by Student's t-test between each sample group and its respective control group (‘DMSO’ and ‘gCtrl’), and indicated by *P< 0.05, **P< 0.01 and *** P< 0.001.
We further applied the dSaCas9-based strategy to HDR-mediated CRISPR gene correction in mESCs containing the I-SceI-GFP correction reporter (Figure 7E). SpCas9 variants (i.e. eSpCas9- gHRC2 and SpCas9-HF1-gHRC2) and SpCas9 with truncating sgRNA (i.e. SpCas9-gHRC4-T17) were chosen for DSB introduction with either ssODN or a dsDNA plasmid as homologous template. Like NU7441, dSaCas9-gSaGC2 effectively stimulated eSpCas9- and SpCas9-HF1-mediated HDR by up to 4-fold with either ssODN or plasmid DNA (Figure 7F, G). Using truncated gHRC4-T17, dSaCas9 binding at proximal region also significantly stimulated the SpCas9-induced HDR by about 2-fold, a smaller stimulation than 3-fold with NU7441 (Figure 7H). These results demonstrated the proximal dSaCas9 binding was effective for stimulating HDR induced by high-fidelity SpCas9–sgRNA variants and expanded the application of the dCas9-based local NHEJ inhibitor strategy.
Although high-fidelity SpCas9–sgRNA variants cause less off-target effect, both NU7441 and dSaCas9-gSaGC2 could still increase off-target activities of these variants while stimulating HDR for these variants. We thus examined the effects of NU7441 and dSaCas9-gSaGC2 on the off-target activities of SpCas9-HF1-gHRC2, eSpCas9-gHRC2 and SpCas9-gHRC4-T17. Both NU7441 and dSaCas9-gSaGC2 reduced on-target indel efficiency of these SpCas9–sgRNA variants (Supplementary Figure 9A-C). This is likely due to the major use of c-NHEJ in generation of on-target indels induced by these SpCas9–sgRNA variants as by SpCas9–sgRNAs (Figure 3A–C) (8). However, NU7441 stimulated off-target activities of these SpCas9–sgRNA variants (Supplementary Figure 9A–C). As a result, NU7441 caused significant reduction in target specificity as anticipated (Supplementary Figure 9A–C). In contrast, gSaGC2 in complex with dSaCas9 enhanced mutagenesis as compared to the sgRNA control at off-target sites (Supplementary Figure 9A–C). These results indicate that unlike NU7441, local dSaCas9 proximal binding as well as dSpCas9 proximal binding did not cause any increase of off-target effect while enhancing HDR-mediated genome editing.
DISCUSSION
Precise editing by HDR has a broad application in genome and cell engineering by CRISPR genome editing but is often limited by the low efficiency of HDR (1,3,52). Many approaches have been taken to improve HDR-mediated genome editing (3,9). Targeting Cas9-mediated DNA cleavage or exposure of DNA breaks in the S/G2 phase of the cell cycle can be used to increase HDR because the HDR machinery is evolved to act in the S/G2 phase in mammalian cells (3,9,15–17). HDR can also be promoted by local enrichment of homologous templates at the repair site (9–11). Recent studies show that non-integrating rAAV6 can deliver a high level of homologous templates to break sites for more efficient HDR (53,54). In addition, many studies have employed Cas9 fusion with a HDR facilitator involving key steps of HDR to locally enhance HDR-mediated correction (3,9,12–18). Another approach to promote HDR is suppression of the competing pathway c-NHEJ by chemical inhibitors, genetic deletion of genes encoding c-NHEJ factors and fusing Cas9 with c-NHEJ suppressor proteins such as 53BP1 domain (3,9,13,19–22). Last but not least, target cleavage could be enhanced by more efficient Cas nucleases and by more accessible chromatin with active transcription, epigenetic modifications or forced unwrapping of nucleosomes, thus leading to more HDR and NHEJ products (3,9,27,31,32,55–57).
Among these strategies, many work in a global manner and have a potential to exacerbate off-target effects in CRISPR genome editing. In particular, our previous study has demonstrated that inactivation of c-NHEJ by chemical or genetic approaches increases the frequency of indels at off-target sites where Cas9 binding is generally weaker and lasts shorter, thus causing stronger off-target effects (8). Off-target effects are a serious problem in CRISPR genome editing and have greatly limited clinical use of this technology; however, the stimulation of off-target effect by chemical or genetic inhibition of c-NHEJ was often ignored in CRISPR genome editing (8,58). Therefore, a strategy is urgently needed to inhibit c-NHEJ while causing no additional off-target effects in CRISPR genome editing. After having demonstrated c-NHEJ inhibition and HDR stimulation by dCas9 proximal binding, this study established dCas9 proximal binding as a strategy of local c-NHEJ inhibitor to address this need.
Upon DSBs induced by SpCas9 at many sites, spontaneous dissociation of SpCas9–sgRNAs from the cleaved targets exposes the DSB ends, which can be readily recognized and bound by the DNA-PKcs/Ku70/Ku80 (i.e. DNA-PK) holoenzyme and ligated by XRCC4/DNA ligase 4. The binding affinity of Ku70/Ku80 to DNA ends, each molecule in close contact with 13–21 bp of DNA, is generally high with the dissociation constant (Kd) at 0.15–0.4 nM and this strong binding is necessary for efficient recruitment of DNA-PKcs and XRCC4/DNA ligase 4 (59). After being loaded onto DNA ends, the Ku70/Ku80 heterodimer may be pushed inwards along DNA, allowing DNA-PKcs to bind the DNA ends (59). The DNA-PK holoenzyme interacts with ∼37 bp of DNA as Ku70/Ku80 binds DNA next to DNA-PKcs (60). The DNA-PK complex may further translocate away from the DSB ends so that XRCC4/DNA ligase 4 can bind 12–13 bp of DNA at the DSB ends for end ligation (61). As dSpCas9 binds to its targets with the Kd approximately at 0.2–4 nM (25,26,62), it is possible that dSpCas9 proximal binding can directly compete with Ku70/Ku80 for DNA binding or prevent the inward movement of Ku70/Ku80 along DNA. In fact, an in vitro assay has demonstrated that SpCas9 residing at the cleaved target cannot be displaced by 100-fold molar excess of the Ku70/Ku80 complex (27). It is conceivable that the c-NHEJ apparatus may not be properly assembled for c-NHEJ after dSpCas9 proximal binding. This possibility prompts us to devise the strategy of dSpCas9 proximal binding to suppress c-NHEJ at a given site. Acting as a c-NHEJ inhibitor, dSpCas9 tethered adjacent to a DSB may preclude the access of DSB ends by Ku70/Ku80 for end binding or block the inward sliding of Ku70/Ku80 along DNA for recruitment of DNA-PKcs and XRCC4/DNA ligase 4 and stimulate HDR by locally suppressing c-NHEJ that depends upon DNA-PKcs/Ku70/Ku80 and XRCC4/DNA ligase 4. Direct competition with Ku70/Ku80 for end binding and blockage of Ku70/Ku80 end sliding may disrupt different c-NHEJ steps that require different c-NHEJ factors. As loss of different c-NHEJ factors results in different a-EJ outcomes and different levels of HDR stimulation as we and others have demonstrated before (8,44,63–65), this helps explain why dCas9 could still inhibit mutagenic NHEJ, albeit to less extent, in XRCC4–/– cells (Figure 3D, E).
Unlike the global c-NHEJ inhibition induced by chemicals or genetic ablation, the c-NHEJ inhibition imposed by dSpCas9 proximal binding rarely occurs simultaneously at the off-target sites for DNA cleavage by other Cas nucleases, therefore not exacerbating the off-target effects. While dSpCas9 proximal binding promotes HDR induced by different CRISPR/Cas systems such as LbCas12a and SaCas9, HDR induced by SpCas9–sgRNA variants such as eSpCas9, SpCas9-HF1 and truncated sgRNAs can also be facilitated by dSaCas9 tethered near the repair site. The SpCas9 variants eSpCas9 and SpCas9-HF1 and truncated sgRNAs have been developed to improve the specificity of CRISPR/Cas9 genome editing and reduce off-target effects by removing the excessive target binding of Cas9–sgRNA. This improvement is however often offset by a reduction in target cleavage. The application of these Cas9–sgRNA variants could be particularly helped by dSaCas9 proximal binding that enhances the efficiency of genome editing mediated by these variants without increasing their off-target activities.
Previous studies have suggested that dSpCas9 proximal binding can induce unwrapping of neighboring nucleosomes and increase the accessibility of nucleosomal DNA target for Cas12a-mediated cleavage (31,32). This model would help explain the HDR stimulation by dCas9 proximal binding; it is however inconsistent with requirement of c-NHEJ factors for the HDR stimulation, dCas9-mediated inhibition of c-NHEJ or reduced recruitment of Ku80 to DNA ends in our study. It is worth noting that the HDR stimulation by dSpCas9 proximal binding and the underlying mechanisms may vary considerably between targets, between cell types and between cell cycle stages. For example, when SpCas9-induced DSBs are exposed by DNA replication forks at many sites, c-NHEJ is little or not even engaged in repair of these DSBs (8,66). Thus, like chemical inhibitors of c-NHEJ, which often generate mixed results in stimulating SpCas9-induced HDR (3,8), dCas9 proximal binding may not suppress c-NHEJ of SpCas9-induced DSBs at these sites in favor of HDR. Instead, if dCas9 proximal binding still stimulates HDR in this case, it is not mediated by locally suppressing c-NHEJ, but possibly by dCas9-mediated alteration of chromatin dynamics (31,32).
Upon binding to DSB ends, Ku70/Ku80 protect the ends from end processing and promote c-NHEJ that is innately accurate in joining readily ligatable ends (6–8). This study demonstrated that dCas9 proximal binding locally suppressed c-NHEJ and stimulated HDR by blocking end binding or end sliding of Ku70/Ku80 at specific sites. However, it remains unclear how exactly the HDR factors are engaged to the DSB ends where dCas9 resides. Likely, after preventing end binding or end sliding of Ku70/Ku80, dCas9 may be subsequently released from its target sites due to the dynamic of its target residence and local DNA metabolism, allowing late engagement of HDR. In addition, studies have shown that removal of bound proteins such as Ku70/Ku80 and other blocks from the ends is s a critical step for short-range end resection and requires Mre11 endonuclease activity that cleaves the 5’-terminated strand at positions up to 300–400 nt away from the ends (67–74). It is possible that, like Ku70/Ku80 or other protein blocks bound to the ends, dCas9 tethered to DNA ends could induce recruitment of Mre11 to create a nick at a position up to 300–400 nt away from the neighboring ends, initiate short-range end resection to dislodge dCas9 from the ends and facilitate HDR. However, using Mre11 ChIP analysis, we did not find significant enrichment of Mre11 at position up to 300–400 nt away from the ends in the presence of dSpCas9 proximal binding (Supplementary Figure S10A, B). Instead, it appeared that Mre11 was enriched further away from the ends at the regions from -801 nt to -2736 nt; but the reason is unclear. In addition, if Mre11 is indeed recruited to initiate short-range end resection at positions up to 300–400 nt away from the ends for HDR, some of the resected ends might fail to engage HDR and could be repaired by NHEJ, generating large deletions in indel-based NHEJ products. However, junction analysis by targeted PCR amplicon deep sequencing revealed that like NU7441, dSpCas9 or dSaCas9 tethered to a DSB end did not increase large deletions while generally having modest effect on the length distribution of deletions within 150 bp (Supplementary Figure S11A–F). Considering that NHEJ events with such large deletions are in a much smaller portion compared to all indel-based NHEJ products, this result is not surprising. Moreover, targeted PCR amplicon deep sequencing in this study is only applicable for PCR products of 300 bp and may be unsuitable for detecting large deletions. A better approach is needed to properly and systemically analyze the effect of dSpCas9 proximal binding on end resection in the future.
Like any other dCas9-based platforms, the efficiency of dCas9-based local c-NHEJ inhibitor is determined by several factors: the distance of dCas9 proximal binding to the DSB ends, the chromatin state at the site for dCas9 proximal binding, and the binding affinity and residence duration of dCas9 at its target (24,30). While the distance of dCas9 proximal binding to the DSB ends is easy to control, the latter two are hard to predict. Preassembled nucleosome at the dCas9 target may prevent dCas9 proximal binding, making ineffective this strategy of dCas9-based local c-NHEJ inhibition (32,75,76). In addition, dCas9 proximal binding may require a strong binding affinity and persistent residence to block the binding of Ku70/Ku80 to DSB ends. However, the binding affinity and residence duration of dCas9 may vary significantly from target to target and from cell type to cell type. Some are rather strong and some others quite weak. While reducing excessive target interaction of Cas9 is a useful strategy to minimize off-target effect in CRISPR genome editing, there remains a need for engineered dCas9 variants with a stronger and more persistent binding ability to improve the effect of dCas9-based platforms including this local c-NHEJ inhibition (24).
DATA AVAILABILITY
Deep sequencing raw data are available in the Sequence Read Archive (SRA) under accession number PRJNA851524 (https://www.ncbi.nlm.nih.gov/sra/ PRJNA851524). Flow cytometry raw data for this study has also been deposited at the Zenodo, where it is directly accessible at https://doi.org/10.5281/zenodo.7131697 and https://doi.org/10.5281/zenodo.7133853. Source data for the figures and supplementary figures are provided as a Source Data file with this paper.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Xie lab for helpful discussions. We also thank Bi Chao and Hong Xiaoli from the Core Facilities, Zhejiang University School of Medicine for their technical support. We thank J. Hu at Peking University for the gift of expression plasmids for Cas9 variants.
Contributor Information
Yi-Li Feng, Innovation Center for Minimally Invasive Technique and Device, Department of General Surgery, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang 310019, P.R. China; Institute of Translational Medicine, Zhejiang University School of Medicine and Zhejiang University Cancer Center, Hangzhou, Zhejiang 310029, P.R. China.
Si-Cheng Liu, Innovation Center for Minimally Invasive Technique and Device, Department of General Surgery, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang 310019, P.R. China; Institute of Translational Medicine, Zhejiang University School of Medicine and Zhejiang University Cancer Center, Hangzhou, Zhejiang 310029, P.R. China.
Ruo-Dan Chen, Innovation Center for Minimally Invasive Technique and Device, Department of General Surgery, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang 310019, P.R. China; Institute of Translational Medicine, Zhejiang University School of Medicine and Zhejiang University Cancer Center, Hangzhou, Zhejiang 310029, P.R. China.
Xiu-Na Sun, Innovation Center for Minimally Invasive Technique and Device, Department of General Surgery, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang 310019, P.R. China; Institute of Translational Medicine, Zhejiang University School of Medicine and Zhejiang University Cancer Center, Hangzhou, Zhejiang 310029, P.R. China.
Jing-Jing Xiao, Innovation Center for Minimally Invasive Technique and Device, Department of General Surgery, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang 310019, P.R. China; Institute of Translational Medicine, Zhejiang University School of Medicine and Zhejiang University Cancer Center, Hangzhou, Zhejiang 310029, P.R. China.
Ji-Feng Xiang, Innovation Center for Minimally Invasive Technique and Device, Department of General Surgery, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang 310019, P.R. China; Institute of Translational Medicine, Zhejiang University School of Medicine and Zhejiang University Cancer Center, Hangzhou, Zhejiang 310029, P.R. China; Department of General Surgery, Chongqing General Hospital, Chongqing 400013, China.
An-Yong Xie, Innovation Center for Minimally Invasive Technique and Device, Department of General Surgery, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang 310019, P.R. China; Institute of Translational Medicine, Zhejiang University School of Medicine and Zhejiang University Cancer Center, Hangzhou, Zhejiang 310029, P.R. China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31870806 to A.Y.X., 32071439 to Y.L.F.]; Department of Science and Technology of Hangzhou [202204A05 to A.Y.X., 202204B08 to Y.L.F.]; Natural Science Foundation of Zhejiang Province [LZ22C050001 to Y.L.F., LQ20C050004 to S.C.L.]. Funding for open access charge: Department of Science and Technology of Hangzhou.
Conflict of interest statement. None declared.
REFERENCES
- 1. Doudna J.A. The promise and challenge of therapeutic genome editing. Nature. 2020; 578:229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nambiar T.S., Baudrier L., Billon P., Ciccia A.. CRISPR-based genome editing through the lens of DNA repair. Mol. Cell. 2022; 82:348–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ramsden D.A., Carvajal-Garcia J., Gupta G.P.. Mechanism, cellular functions and cancer roles of polymerase-theta-mediated DNA end joining. Nat. Rev. Mol. Cell. Biol. 2021; 10:1038. [DOI] [PubMed] [Google Scholar]
- 5. Stinson B.M., Loparo J.J.. Repair of DNA double-strand breaks by the nonhomologous end joining pathway. Annu. Rev. Biochem. 2021; 90:137–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guo T., Feng Y.-L., Xiao J.-J., Liu Q., Sun X.-N., Xiang J.-F., Kong N., Liu S.-C., Chen G.-Q., Wang Y.et al.. Harnessing accurate non-homologous end joining for efficient precise deletion in CRISPR/Cas9-mediated genome editing. Genome Biol. 2018; 19:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bétermier M., Bertrand P., Lopez B.S.. Is non-homologous end-joining really an inherently error-prone process. PLoS Genet. 2014; 10:e1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu S.-C., Feng Y.-L., Sun X.-N., Chen R.-D., Liu Q., Xiao J.-J., Zhang J.-N., Huang Z.-C., Xiang J.-F., Chen G.-Q.et al.. Target residence of Cas9–sgRNA influences DNA double-strand break repair pathway choices in CRISPR/Cas9 genome editing. Genome Biol. 2022; 23:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang H., Ren S., Yu S., Pan H., Li T., Ge S., Zhang J., Xia N.. Methods favoring homology-directed repair choice in response to CRISPR/Cas9 induced-double strand breaks. Int. J. Mol. Sci. 2020; 21:E6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carlson-Stevermer J., Abdeen A.A., Kohlenberg L., Goedland M., Molugu K., Lou M., Saha K.. Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA aptamer for precise gene editing. Nat. Commun. 2017; 8:1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Savic N., Ringnalda F.C., Lindsay H., Berk C., Bargsten K., Li Y., Neri D., Robinson M.D., Ciaudo C., Hall J.et al.. Covalent linkage of the DNA repair template to the CRISPR-Cas9 nuclease enhances homology-directed repair. Elife. 2018; 7:e33761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Charpentier M., Khedher A.H.Y., Menoret S., Brion A., Lamribet K., Dardillac E., Boix C., Perrouault L., Tesson L., Geny S.et al.. CtIP fusion to Cas9 enhances transgene integration by homology-dependent repair. Nat. Commun. 2018; 9:1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jayavaradhan R., Pillis D.M., Goodman M., Zhang F., Zhang Y., Andreassen P.R., Malik P.. CRISPR-Cas9 fusion to dominant-negative 53BP1 enhances HDR and inhibits NHEJ specifically at Cas9 target sites. Nat. Commun. 2019; 10:2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nambiar T.S., Billon P., Diedenhofen G., Hayward S.B., Taglialatela A., Cai K., Huang J.-W., Leuzzi G., Cuella-Martin R., Palacios A.et al.. Stimulation of CRISPR-mediated homology-directed repair by an engineered RAD18 variant. Nat. Commun. 2019; 10:3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin S., Staahl B.T., Alla R.K., Doudna J.A.. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014; 3:e04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang D., Scavuzzo M.A., Chmielowiec J., Sharp R., Bajic A., Borowiak M.. Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases. Sci. Rep. 2016; 6:21264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wienert B., Nguyen D.N., Guenther A., Feng S.J., Locke M.N., Wyman S.K., Shin J., Kazane K.R., Gregory G.L., Carter M.A.M.et al.. Timed inhibition of CDC7 increases CRISPR-Cas9 mediated templated repair. Nat. Commun. 2020; 11:2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reint G., Li Z., Labun K., Keskitalo S., Soppa I., Mamia K., Tolo E., Szymanska M., Meza-Zepeda L.A., Lorenz S.et al.. Rapid genome editing by CRISPR-Cas9-POLD3 fusion. Elife. 2021; 10:e75415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L.. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015; 33:538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kühn R.. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015; 33:543–548. [DOI] [PubMed] [Google Scholar]
- 21. Robert F., Barbeau M., Éthier S., Dostie J., Pelletier J.. Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Med. 2015; 7:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Riesenberg S., Maricic T.. Targeting repair pathways with small molecules increases precise genome editing in pluripotent stem cells. Nat. Commun. 2018; 9:2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang F., Doudna J.A.. CRISPR–Cas9 structures and mechanisms. Annu. Rev. Biophys. 2017; 46:505–529. [DOI] [PubMed] [Google Scholar]
- 24. Feng Y., Liu S., Chen R., Xie A.. Target binding and residence: a new determinant of DNA double-strand break repair pathway choice in CRISPR/Cas9 genome editing. J. Zhejiang Univ. Sci. B. 2021; 22:73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A.. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014; 507:62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E.. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016; 34:339–344. [DOI] [PubMed] [Google Scholar]
- 27. Clarke R., Heler R., MacDougall M.S., Yeo N.C., Chavez A., Regan M., Hanakahi L., Church G.M., Marraffini L.A., Merrill B.J.. Enhanced bacterial immunity and mammalian genome editing via RNA-polymerase-mediated dislodging of Cas9 from double-strand DNA breaks. Mol. Cell. 2018; 71:42–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A.. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013; 152:1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Whinn K.S., Kaur G., Lewis J.S., Schauer G.D., Mueller S.H., Jergic S., Maynard H., Gan Z.Y., Naganbabu M., Bruchez M.P.et al.. Nuclease dead Cas9 is a programmable roadblock for DNA replication. Sci. Rep. 2019; 9:13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang H., La Russa M., Qi L.S.. CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 2016; 85:227–264. [DOI] [PubMed] [Google Scholar]
- 31. Chen F., Ding X., Feng Y., Seebeck T., Jiang Y., Davis G.D.. Targeted activation of diverse CRISPR-Cas systems for mammalian genome editing via proximal CRISPR targeting. Nat. Commun. 2017; 8:14958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Strohkendl I., Saifuddin F.A., Gibson B.A., Rosen M.K., Russell R., Finkelstein I.J.. Inhibition of CRISPR-Cas12a DNA targeting by nucleosomes and chromatin. Sci. Adv. 2021; 7:eabd6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K.. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014; 32:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K.. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016; 529:490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F.. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016; 351:84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Friedland A.E., Baral R., Singhal P., Loveluck K., Shen S., Sanchez M., Marco E., Gotta G.M., Maeder M.L., Kennedy E.M.et al.. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 2015; 16:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chandramouly G., Kwok A., Huang B., Willis N.A., Xie A., Scully R.. BRCA1 and CtIP suppress long-tract gene conversion between sister chromatids. Nat. Commun. 2013; 4:2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xie A., Kwok A., Scully R.. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 2009; 16:814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Feng Y.-L., Liu Q., Chen R.-D., Liu S.-C., Huang Z.-C., Liu K.-M., Yang X.-Y., Xie A.-Y.. DNA nicks induce mutational signatures associated with BRCA1 deficiency. Nat. Commun. 2022; 13:4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rass E., Chandramouly G., Zha S., Alt F.W., Xie A.. Ataxia telangiectasia mutated (ATM) is dispensable for endonuclease I-SceI-induced homologous recombination in mouse embryonic stem cells. J. Biol. Chem. 2013; 288:7086–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xie A., Hartlerode A., Stucki M., Odate S., Puget N., Kwok A., Nagaraju G., Yan C., Alt F.W., Chen J.et al.. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol. Cell. 2007; 28:1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xie A., Puget N., Shim I., Odate S., Jarzyna I., Bassing C.H., Alt F.W., Scully R.. Control of sister chromatid recombination by histone H2AX. Mol. Cell. 2004; 16:1017–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Feng Y.-L., Xiang J.-F., Liu S.-C., Guo T., Yan G.-F., Feng Y., Kong N., Li H.-D., Huang Y., Lin H.et al.. H2AX facilitates classical non-homologous end joining at the expense of limited nucleotide loss at repair junctions. Nucleic Acids Res. 2017; 45:10614–10633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pierce A.J., Hu P., Han M., Ellis N., Jasin M.. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001; 15:3237–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Allen C., Kurimasa A., Brenneman M.A., Chen D.J., Nickoloff J.A.. DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:3758–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Delacôte F., Han M., Stamato T.D., Jasin M., Lopez B.S.. An xrcc4 defect or Wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nucleic Acids Res. 2002; 30:3454–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yin J., Lu R., Xin C., Wang Y., Ling X., Li D., Zhang W., Liu M., Xie W., Kong L.et al.. Cas9 exo-endonuclease eliminates chromosomal translocations during genome editing. Nat. Commun. 2022; 13:1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Singh D., Mallon J., Poddar A., Wang Y., Tippana R., Yang O., Bailey S., Ha T.. Real-time observation of DNA target interrogation and product release by the RNA-guided endonuclease CRISPR Cpf1 (Cas12a). Proc. Natl. Acad. Sci. U.S.A. 2018; 115:5444–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jeon Y., Choi Y.H., Jang Y., Yu J., Goo J., Lee G., Jeong Y.K., Lee S.H., Kim I.-S., Kim J.-S.et al.. Direct observation of DNA target searching and cleavage by CRISPR-Cas12a. Nat. Commun. 2018; 9:2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Strohkendl I., Saifuddin F.A., Rybarski J.R., Finkelstein I.J., Russell R.. Kinetic basis for DNA target specificity of CRISPR-Cas12a. Mol. Cell. 2018; 71:816–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Swarts D.C., Jinek M.. Mechanistic insights into the cis- and trans-acting DNase activities of Cas12a. Mol. Cell. 2019; 73:589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rezalotfi A., Fritz L., Förster R., Bošnjak B.. Challenges of CRISPR-based gene editing in primary T cells. Int. J. Mol. Sci. 2022; 23:1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dever D.P., Bak R.O., Reinisch A., Camarena J., Washington G., Nicolas C.E., Pavel-Dinu M., Saxena N., Wilkens A.B., Mantri S.et al.. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016; 539:384–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lattanzi A., Camarena J., Lahiri P., Segal H., Srifa W., Vakulskas C.A., Frock R.L., Kenrick J., Lee C., Talbott N.et al.. Development of β-globin gene correction in human hematopoietic stem cells as a potential durable treatment for sickle cell disease. Sci. Transl. Med. 2021; 13:eabf2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takayama K., Igai K., Hagihara Y., Hashimoto R., Hanawa M., Sakuma T., Tachibana M., Sakurai F., Yamamoto T., Mizuguchi H.. Highly efficient biallelic genome editing of human ES/iPS cells using a CRISPR/Cas9 or TALEN system. Nucleic Acids Res. 2017; 45:5198–5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu B., Chen S., Rose A.L., Chen D., Cao F., Zwinderman M., Kiemel D., Aïssi M., Dekker F.J., Haisma H.J.. Inhibition of histone deacetylase 1 (HDAC1) and HDAC2 enhances CRISPR/Cas9 genome editing. Nucleic Acids Res. 2020; 48:517–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li G., Zhang X., Wang H., Liu D., Li Z., Wu Z., Yang H.. Increasing CRISPR/Cas9-mediated homology-directed DNA repair by histone deacetylase inhibitors. Int. J. Biochem. Cell Biol. 2020; 125:105790. [DOI] [PubMed] [Google Scholar]
- 58. Kim D., Luk K., Wolfe S.A., Kim J.-S.. Evaluating and enhancing target specificity of gene-editing nucleases and deaminases. Annu. Rev. Biochem. 2019; 88:191–220. [DOI] [PubMed] [Google Scholar]
- 59. Dynan W.S., Yoo S.. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998; 26:1551–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen X., Xu X., Chen Y., Cheung J.C., Wang H., Jiang J., de Val N., Fox T., Gellert M., Yang W.. Structure of an activated DNA-PK and its implications for NHEJ. Mol. Cell. 2021; 81:801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ochi T., Wu Q., Chirgadze D.Y., Grossmann J.G., Bolanos-Garcia V.M., Blundell T.L.. Structural insights into the role of domain flexibility in human DNA ligase IV. Structure. 2012; 20:1212–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jinek M., Jiang F., Taylor D.W., Sternberg S.H., Kaya E., Ma E., Anders C., Hauer M., Zhou K., Lin S.et al.. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014; 343:1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chang H.H.Y., Pannunzio N.R., Adachi N., Lieber M.R.. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell. Biol. 2017; 18:495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schulte-Uentrop L., El-Awady R.A., Schliecker L., Willers H., Dahm-Daphi J.. Distinct roles of XRCC4 and Ku80 in non-homologous end-joining of endonuclease- and ionizing radiation-induced DNA double-strand breaks. Nucleic Acids Res. 2008; 36:2561–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Guirouilh-Barbat J., Rass E., Plo I., Bertrand P., Lopez B.S.. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:20902–20907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Richardson C.D., Kazane K.R., Feng S.J., Zelin E., Bray N.L., Schäfer A.J., Floor S.N., Corn J.E.. CRISPR-Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat. Genet. 2018; 50:1132–1139. [DOI] [PubMed] [Google Scholar]
- 67. Cejka P., Symington L.S.. DNA end resection: mechanism and control. Annu. Rev. Genet. 2021; 55:285–307. [DOI] [PubMed] [Google Scholar]
- 68. Paiano J., Wu W., Yamada S., Sciascia N., Callen E., Paola Cotrim A., Deshpande R.A., Maman Y., Day A., Paull T.T.et al.. ATM and PRDM9 regulate SPO11-bound recombination intermediates during meiosis. Nat. Commun. 2020; 11:857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mimitou E.P., Yamada S., Keeney S.. A global view of meiotic double-strand break end resection. Science. 2017; 355:40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chanut P., Britton S., Coates J., Jackson S.P., Calsou P.. Coordinated nuclease activities counteract Ku at single-ended DNA double-strand breaks. Nat. Commun. 2016; 7:12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Deshpande R.A., Myler L.R., Soniat M.M., Makharashvili N., Lee L., Lees-Miller S.P., Finkelstein I.J., Paull T.T.. DNA-dependent protein kinase promotes DNA end processing by MRN and CtIP. Sci. Adv. 2020; 6:eaay0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Myler L.R., Gallardo I.F., Soniat M.M., Deshpande R.A., Gonzalez X.B., Kim Y., Paull T.T., Finkelstein I.J.. Single-molecule imaging reveals how Mre11-Rad50-Nbs1 initiates DNA break repair. Mol. Cell. 2017; 67:891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Reginato G., Cannavo E., Cejka P.. Physiological protein blocks direct the Mre11-Rad50-Xrs2 and Sae2 nuclease complex to initiate DNA end resection. Genes Dev. 2017; 31:2325–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang W., Daley J.M., Kwon Y., Krasner D.S., Sung P.. Plasticity of the Mre11-Rad50-Xrs2-Sae2 nuclease ensemble in the processing of DNA-bound obstacles. Genes Dev. 2017; 31:2331–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Horlbeck M.A., Witkowsky L.B., Guglielmi B., Replogle J.M., Gilbert L.A., Villalta J.E., Torigoe S.E., Tjian R., Weissman J.S.. Nucleosomes impede Cas9 access to DNA in vivo and in vitro. Elife. 2016; 5:e12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Isaac R.S., Jiang F., Doudna J.A., Lim W.A., Narlikar G.J., Almeida R.. Nucleosome breathing and remodeling constrain CRISPR-Cas9 function. Elife. 2016; 5:e13450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deep sequencing raw data are available in the Sequence Read Archive (SRA) under accession number PRJNA851524 (https://www.ncbi.nlm.nih.gov/sra/ PRJNA851524). Flow cytometry raw data for this study has also been deposited at the Zenodo, where it is directly accessible at https://doi.org/10.5281/zenodo.7131697 and https://doi.org/10.5281/zenodo.7133853. Source data for the figures and supplementary figures are provided as a Source Data file with this paper.