Abstract
RNA 2′O-methylation is a ‘self’ epitranscriptomic modification allowing discrimination between host and pathogen. Indeed, human immunodeficiency virus 1 (HIV-1) induces 2′O-methylation of its genome by recruiting the cellular FTSJ3 methyltransferase, thereby impairing detection by RIG-like receptors. Here, we show that RNA 2′O-methylations interfere with the antiviral activity of interferon-stimulated gene 20-kDa protein (ISG20). Biochemical experiments showed that ISG20-mediated degradation of 2′O-methylated RNA pauses two nucleotides upstream of and at the methylated residue. Structure-function analysis indicated that this inhibition is due to steric clash between ISG20 R53 and D90 residues and the 2′O-methylated nucleotide. We confirmed that hypomethylated HIV-1 genomes produced in FTSJ3-KO cells were more prone to in vitro degradation by ISG20 than those produced in cells expressing FTSJ3. Finally, we found that reverse-transcription of hypomethylated HIV-1 was impaired in T cells by interferon-induced ISG20, demonstrating the direct antagonist effect of 2′O-methylation on ISG20-mediated antiviral activity.
Graphical Abstract
Graphical Abstract.
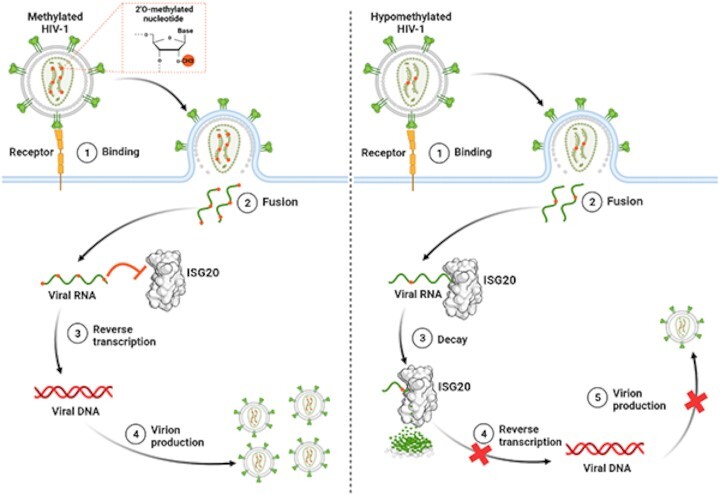
Role of HIV-1 RNA 2′O-methylation in limiting ISG20 antiviral action. The presence of methyl on the HIV-1 RNA promotes virus replication by preventing recognition and viral RNA decay by ISG20.
INTRODUCTION
RNA structure and functions are regulated by post-transcriptional modifications (PTMs). To date, more than 140 chemical epitranscriptomic modifications have been described and they regulate most RNA-mediated cellular processes (e.g. splicing, nuclear export, microRNA biogenesis, localization, translation mechanisms, degradation and sensing by innate immunity) (1). Despite the increasing knowledge on PTM roles in RNA biological functions, the viral RNA (vRNA) epitranscriptomic modification landscape is still poorly characterized (2,3), and the role of these chemical modifications in virus replication remains poorly understood. RNA capping is one of the best characterized PTMs in the viral world, which consists of a guanosine moiety linked via a 5′-5′ triphosphate to the first nucleotide (N1) at the 5′ end of most vRNAs (GpppN1) (4). This cap structure is methylated at the N7 position of the guanosine residue by an S-adenosyl-l-methionine-dependent N7-methyltransferase (MTase), forming the cap-0 (mGpppN1) structure. In some cases, the first nucleotide (N1) of the cap-0 structure is methylated on the ribose 2′OH by a virally encoded 2′O-MTase, leading to the formation of a cap-1 structure (mGpppNm) (5). This cap structure plays critical role in protecting RNA against 5′ exonucleases such as XRN1 (6) and DXO (7) and favors the recognition of Eif4E, thus promoting mRNA translation (8). Moreover, the cap 2′O-methylation is sensed by the host cytosolic retinoic acid-inducible gene I protein (RIG-I) as a hallmark of the self (9,10). The detection of ‘non-self’ RNAs (i.e. vRNAs without cap-1 structure) by RIG-like receptors initiates a cascade of events, leading to the secretion of type I interferon (IFN-I) and proinflammatory cytokines (10). In addition, it has been shown that some IFN-stimulated genes (ISG), such as IFN-induced proteins with tetratricopeptide repeats (IFIT) 1/3, can specifically discriminate capped RNA without the N1 2′O-methylation mark (11). The modality of recognition of this RNA structure as ‘non-self’ by these restriction factors has been established by X-ray crystallography (12). By sequestering ‘non-self’ RNAs, IFIT1/3 block their translation into proteins (13,14).
In the viral world, several MTases involved in cap methylation have been characterized biochemically and structurally (15–19). The activity of these MTases depends on their ability to recruit and accommodate specific substrates in their catalytic pocket. Besides their functional role in RNA capping, the MTases of Flaviviruses and Filoviruses can methylate internal adenosines at the 2′O position within the vRNA (20–23). These internal epitranscriptomic marks have been mapped in the genome of some RNA viruses using RiboMethSeq (24), Nm-seq (25) and sensitive ‘agnostic’ mass spectrometry techniques (2). The presence of 2′O-methylated nucleotides (Nm) has been demonstrated within the genome of Zika virus (ZIKV), Dengue virus (DENV) (2), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (25). Strikingly, internal Nm have been identified also in the genomes of polioviruses and hepatitis C viruses (2), although they do not encode any known MTase. This suggests that host 2′O-MTases might be recruited to promote these epitranscriptomic vRNA modifications. In line with these findings, type 1 human immunodeficiency virus (HIV-1) recruits the complex composed of a transactivation-responsive RNA-binding protein (TRBP) and the cellular 2′O-MTase FTSJ3 to catalyze the 2′O-methylation of 17 internal nucleotides in its genome (26). These modifications have a ‘proviral’ effect, at least by limiting the sensing by the RIG-like receptor MDA5 (26). Indeed, the production of HIV-1 in FTSJ3 knock-out (KO) cells leads to the release of hypomethylated progeny viruses that trigger the expression of IFN-I upon the specific activation of the cytoplasmic MDA5 sensor (26). Although the exact molecular mechanism whereby MDA5 discriminates self and non-self has not been fully characterized, it has been suggested that the internal 2′O-methylation might prevent MDA5 interaction with the vRNA (27). Moreover, internal RNA 2′O-methylation might also confer resistance towards cellular restriction factors, including the IFN-induced exonucleases RNase L (28) and ISG20 (29,30).
In this study, we focused on ISG20, an interferon-induced 3′ to 5′ exonuclease that inhibits the replication of a broad range of RNA viruses, including Flaviviruses (Yellow fever, West Nile, DENV, bovine viral diarrhea virus, and hepatitis C) (31), Togaviruses (Sindbis, Chikungunya, and Venezuelan equine encephalitis viruses), Rhabdoviruses (vesicular stomatitis virus), Orthomyxoviruses (influenza virus), Bunyaviruses, Hepadnaviruses (hepatitis B) (32) and Retroviruses (HIV-1) (33). ISG20 is a 20-kDa protein that is enriched in the nucleolus and Cajal bodies (34). It contains three conserved exonuclease motifs (Exo I, II and III) and belongs to the DEDDh box 3′-5′ exonucleases. Members of this superfamily are usually involved in ssRNA and ssDNA degradation, but ISG20 might preferentially target RNA substrates (35,36). This hypothesis is supported by the crystal structure of ISG20 in complex with uridine 5′ monophosphate (UMP) showing a specific hydrogen bond between residues M14 and R53 of ISG20 and the 2′OH moiety of the nucleotide ribose (35) that might stabilize the RNA in the catalytic pocket. Here, we produced and purified human ISG20 to precisely characterize its exonuclease activity on RNA substrates, and 2′O-methylation role in ISG20 RNase activity. We confirmed our biochemical results in an infectious cell model and demonstrated that the HIV-1 genome, which is naturally 2′O-methylated by the host FTSJ3, is more resistant to ISG20-mediated degradation than hypomethylated vRNA. In addition, using pseudotyped HIV-1 particles, we demonstrated that the vRNA stability and luciferase reporter gene expression are reduced in the absence of 2′O-methylation. We further confirmed that ISG20 interferes with the retro-transcription of hypomethylated HIV-1 within T cells. Together, our data shed light on a new proviral role of HIV-1 RNA 2′O-methylation by FTSJ3 in protecting the viral transcripts against ISG20-mediated degradation during the early stages of infection.
MATERIALS AND METHODS
Cloning of ISG20 coding sequence for recombinant protein expression
The coding sequence of human ISG20 (amino acids 1–181) followed by a C-terminal hexa-histidine tag was cloned into the pDEST14 vector (Thermo Fisher Scientific) using the Gateway cloning system following the manufacturer's instruction. Mutations were introduced in the expression plasmid by PCR using the QuickChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's instructions. Primers are listed in Supplementary Table S1.
ISG20 expression and purification
ISG20 was produced in Escherichia coli BL21-CodonPlus (DE3)-RIPL bacteria (Agilent). Cells were grown in LB medium containing 1% glucose, 100 μg/ml of ampicillin, and 34 μg/ml chloramphenicol at 37°C. Expression was induced at OD600nm = 0.7 with 0.2 mM isopropyl-thiogalactopyranoside (IPTG) and stopped after 4h. Cells were harvested by centrifugation at 8000 × g at 4°C for 20 min, and pellets were stored at –80°C until purification. Pellets were thawed on ice and resuspended in lysis buffer (50 mM Tris pH 7, 50 mM NaCl, 1 mM β-mercaptoethanol, 1 mM PMSF and 10% glycerol). The suspension was sonicated and centrifuged at 30 000 × g 4°C for 30 min. After clarification, 0.3% (w/v) of polyethyleneimine was slowly added to the lysate with gentle shaking at 4°C for 30 min, followed by another centrifugation step. The supernatant was collected and supplemented with 5 mM imidazole pH 7.9 and 500 mM NaCl. Soluble ISG20 was purified on NiNTA resin, the column was washed with buffer W1 (50 mM Tris pH 7.9, 500 mM NaCl, 1 mM β-mercaptoethanol, 25 mM imidazole), W2 (50 mM Tris pH 7.9, 1 M NaCl, 25 mM imidazole, 1 mM β-mercaptoethanol), W3 (50 mM Tris pH 7.9, 100 mM NaCl, 25 mM imidazole, 1 mM β-mercaptoethanol), and W1. Proteins were eluted with buffer E (50 mM Tris pH 7.9, 250 mM NaCl, 250 mM imidazole and 1 mM β-mercaptoethanol), then dialyzed against 50 mM Tris pH 7, 150 mM NaCl, 1 mM β-mercaptoethanol and 0.1 mM dithiothreitol (DTT). Finally, proteins were concentrated on Amicon Ultra 5 kDa, and stored in 50% glycerol at -20°C.
Synthesis of RNA and DNA substrates
The synthetic RNAs (Biomers) and DNAs (Eurofins Genomics) used in this study are listed in Table 1 and S2. Synthetic RNAs and DNAs (HPLC grade) were purchased. A27 with an N6-methyl adenosine at position 7 from the 3′ end (A20AN6mA6) was synthesized as previously described (37). Synthesis was performed at the 1 mmol scale on an ABI 394 DNA synthesizer using commercially available 5′-O-DMTr-2′-O-pivaloyloxymethyl-3′-O-(O-cyanoethyl-N,N-diisopropylphosphoramidite N-6-phenoxyacetyl adenosine (Chemgenes Corp). The N6-methyl adenosine (AN6m) was introduced in the sequence using the 5′-O-DMTr-2′-O-pivaloyloxymethyl-3′-O-(O-cyanoethyl-N,N-diisopropylphosphoramidite N6-methyl adenosine synthesized by us. After deprotection and release from the solid support upon basic conditions (DBU then aqueous ammonia treatment at 37°C for 4 h), A20AN6mA6 was purified by IEX-HPLC to high purity.
Table 1.
List of the RNA substrates used for the exonuclease assays with ISG20
| Name | Sequence from 5′ to 3′ | ΔG (kcal/mol) | Modifications |
|---|---|---|---|
| A27 | AAAAAAAAAAAAAAAAAAAAAAAAAAA | –0.00 | No |
| A26Am | AAAAAAAAAAAAAAAAAAAAAAAAAAAm | –0.00 | 2′O-methylation |
| A26Um | AAAAAAAAAAAAAAAAAAAAAAAAAAUm | –0.00 | 2’O-methylation |
| A26Cm | AAAAAAAAAAAAAAAAAAAAAAAAAACm | –0.00 | 2′O-methylation |
| A26Gm | AAAAAAAAAAAAAAAAAAAAAAAAAAGm | –0.00 | 2′O-methylation |
| A24AmA2 | AAAAAAAAAAAAAAAAAAAAAAAAAmAA | –0.00 | 2′O-methylation |
| A20AmA6 | AAAAAAAAAAAAAAAAAAAAAmAAAAAA | –0.00 | 2′O-methylation |
| A20UmA6 | AAAAAAAAAAAAAAAAAAAAUmAAAAAA | –0.00 | 2′O-methylation |
| A20CmA6 | AAAAAAAAAAAAAAAAAAAACmAAAAAA | –0.00 | 2′O-methylation |
| A20GmA6 | AAAAAAAAAAAAAAAAAAAAGmAAAAAA | –0.00 | 2′O-methylation |
| RNA4-A27 | AGAUCGUGAACUUACAAACUAUACAAA | –0.00 | No |
| RNA4-A27m | AGAUCGUGAACUUACAAACUAUACAAAm | –0.43 | 2′O-methylation |
| RNA4-C27 | AGAUCGUGAACUUACAAACUAUACAAC | –0.44 | No |
| RNA4-C27m | AGAUCGUGAACUUACAAACUAUACAACm | –0.44 | 2’O-methylation |
32Pradiolabeling of the 5′ of substrates
The non-fluorescent substrates were radiolabeled at their 5′ end using T4 polynucleotide kinase (New England) and [γ32P]ATP (Perkin Elmer) according to the manufacturer's instructions.
Exonuclease assays
Nuclease activity assays were performed by mixing 2 nM of recombinant ISG20 with 500 nM of radiolabeled substrate in optimized buffer (50 mM Tris–HCl pH 7, 2.5 mM MnCl2, 1 mM β-mercaptoethanol, 0.1 mM DTT and 0.1% Triton X-100). Reaction was performed at 37°C and at each time point (0, 1, 5, 15, 30 and 60 min), 5 μl of suspension was taken and a double volume of loading buffer containing 96% formamide and 10 mM EDTA was added to stop the reaction. Then, the digested products were loaded on 7 M urea-containing 14% (wt/vol) polyacrylamide gels (acrylamide/bisacrylamide ratio, 19:1) buffered with 0.5× Tris-NH2-taurine-EDTA and run at 65 W. Results were visualized by phosphor imaging using a Typhoon-9410 variable-mode scanner (GE Healthcare). ISG20 exonuclease activity was quantified using the FujiImager and Image Gauge analysis software. The nuclease activity of NP1 (New England Biolabs) and PDE (Sigma-Aldrich) was tested according to manufacturer recommendation at 37°C. All assays were done at least twice.
Structural modeling
2′O-Methylated UMP was designed using PubChem Sketcher V2.4 (https://pubchem.ncbi.nlm.nih.gov//edit3/index.html) then aligned with the UMP of the ISG20 structure (1WLJ) via UCSF Chimera (38) to visualize the molecular basis of ISG20 inhibition at N0. Modeling of ISG20 with an RNA was done by superimposing the previously described structure of this nuclease (PDB:1WLJ) with that of SDN1 (PDB: 5Z9X) in ChimeraX (39). Methyl groups were added to the RNA of this model at the 2′O of riboses at positions N–2 and N0 using the chimera build structure option that guaranties proper angles and bond distances.
Cell lines and culture conditions
The cell lines used in this study are: parental HEK293T (HEK-WT), FTSJ3 knock-out HEK293T (HEK-FTSJ3-KO), Jurkat and HeLa P4C5 cells. The HEK-FTSJ3-KO cells (26) were kindly provided by Y. Bennasser (IGH, Montpellier, France) and were used to produce viral or pseudoviruses particles containing a hypomethylated genome. HeLa P4C5 is HeLa-derived cell line expressing CD4 and CCR5 (40). The HeLa P4C5, HEK-WT and HEK-FTSJ3-KO cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco), whereas the Jurkat (clone E6-1) cells were cultured in Roswell Park Memorial Institute medium (RPMI,Gibco). Both media were supplemented with 10% FCS, supplemented with 1% penicillin–streptomycin (Thermo Fisher Scientific). When indicated, HeLa P4C5 and Jurkat cells were treated with recombinant human IFN-α2a (R&D Systems).
Plasmids and siRNA
A codon-optimized plasmid encoding for a 3xFlag WT or R53A/D90A ISG20 was purchased from TWIST bioscience for eukaryotic cell expression. siRNA directed against human ISG20 were purchased from Horizon as pools of four individual siRNAs (ON-TARGETplus SMARTpool). The non-targeting control siRNA are directed against human PPIB (CypB), also as an ON-TARGETplus SMARTpool (Horizon). siRNA transfections into Jurkat cells were performed by nucleofection, using the SE Cell Line 4D-Nucleofector™ X Kit (Lonza) in a 4D-Nucleofector apparatus, following the manufacturer's recommendations. For transfections of HeLa P4C5 cells, siRNA and plasmids were transfected simultaneously using Lipofectamine 3000 (Invitrogen) following the manufacturer's instructions.
Production of WT and hypomethylated HIV-1 particles
The HIV-1 plasmid pNL4-3.Luc.R-E (41) was kindly provided by N. Landau (Aaron Diamond AIDS Research Center, The Rockefeller University, New York, USA). VSV-G pseudotyped luciferase-encoding HIV-1 particles were produced by transient transfection of HEK293T (WT or FTSJ3-KO) with pNL4-3.Luc.R-E and pVSV-G using calcium phosphate, and were concentrated by ultracentrifugation for 1 h at 22 000 × g (Beckman Coulter) at 4°C (26). Replication-competent, infectious HIV-1 particles were produced by transient transfection of HEK293T (WT or FTSJ3-KO) with pNL4-3 and were concentrated using Lenti-X Concentrator (Takara). Virus yields were measured by p24 enzyme-linked immunosorbent assay following the manufacturer's instructions (Lenti-X p24 Rapid Titer Kit, Takara). All pseudotyped and infectious HIV-1 particles were treated with 250 U/ml benzonase (Merck) at 37°C for 20 min before being used in the experiments.
Western blot analyses
Cells were lysed in a buffer containing 50 mM Tris–HCl (pH7.4), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 and EDTA-free protease inhibitor cocktail (Roche). Protein extracts were analyzed on 12% SDS-PAGE gels, and transferred onto nitrocellulose membranes. Membranes were incubated with primary antibodies against FTSJ3 (rabbit; Bethyl Laboratories), Flag (clone M2; Sigma-Aldrich), ISG20 (rabbit, Proteintech) and β-actin (mouse, Sigma-Aldrich), followed by anti-mouse and anti-rabbit HRP-conjugated antibodies. Chemiluminescent bands were imaged with a ChemidocTM MP Imager and analyzed for quantification using the Image LabTM desktop software (Bio-Rad Laboratories).
RT-qPCR analyses
Total RNA was extracted using the RNeasy Mini kit (Qiagen) and subjected to deoxyribonuclease (DNase) treatment, following the manufacturer's instructions. RNA concentration and purity were evaluated by spectrophotometry (NanoDrop 2000c, Thermo Fisher Scientific). Cellular mRNA were reverse-transcribed with both oligo(dT) and random primers using a PrimeScript RT Reagent Kit (Perfect Real Time, Takara) in a 10 μl reaction.
HIV-1 RNA (50 ng) was reverse-transcribed using an oligo(dT) primer (located at the 3′ end of HIV-1 RNA) or the M661 primer (in the Gag gene) (Zack et al., 1990) and the PrimeScript RT Reagent Kit (Perfect Real Time, Takara) in a 20 μl reaction. Real-time PCR amplifications were performed in triplicate using Takyon Rox SYBRMasterMix dTTP Blue (Eurogentec) on an Applied Biosystems QuantStudio 5 instrument (Thermo Fischer Scientific). RT products were quantified using the M667/AA55 primers that amplify the R/U5 region of HIV-1, and the following program: 3 min at 95°C followed by 40 cycles of 15 s at 95°C, 20 s at 60°C, and 20 s at 72°C. Endogenous ISG20 transcripts were quantified with ISG20-F and ISG20-R primers, whereas transfected ISG20 (WT or R53A/D90A), which has been codon-optimized, was quantified using ISG20co-F and ISG20co-R primers. Ct values for ISG20 and ISG20co were normalized to expression levels of RPL13A (60S ribosomal protein L13a), using the 2-ΔΔCt method. Primers are listed in Supplementary Table S1.
Quantification of HIV-1 RT products by qPCR
Cells were either transduced with VSV-G-pseudotyped HIV-1 or infected with replication-competent HIV-1 (produced in HEK-WT or HEK-FTSJ3-KO cells). Cells were collected 6 h post-infection and total DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen). Real-time PCR reactions were performed as described above. RT products were quantified using M667/M661 primers and results were normalized to GAPDH expression level. Primers are listed in Supplementary Table S1.
RESULTS
Internal 2′O-methylation of RNA limits ISG20 activity at N–2 and N0 of the methylated residue
Recombinant human ISG20 was purified (Supplementary Figure S1A) and its exonuclease activity was assessed using synthetic 27-nucleotide RNAs (A27, C27, U27) (Supplementary Figure S1B, Supplementary Table S2) after optimization of the reaction conditions (Supplementary Figure S1C, D). The laddering degradation profile confirmed ISG20 distributive 3′ exonuclease activity. Sequence analysis showed that ISG20 contains conserved ‘DEDDh’ motifs (Supplementary Figure S2, Supplementary Table S3). After alanine substitution of residues in the Exo I (D11A, E13A), Exo II (D94A) and Exo III (H149A, D154A) motifs, the five recombinant ISG20 mutants were purified (Supplementary Figure S3A). The exonuclease activity of wild type (WT) ISG20 was confirmed using synthetic A27 RNA, whereas that of the five mutants was strongly impaired (Supplementary Figure S3B). Moreover, assessment of ISG20 nuclease activity using ssRNA1, ssRNA2 that forms a 3′ hairpin structure, and ssDNA1 (Supplementary Table S2) showed that the presence of an RNA secondary structure stalled ISG20 exonuclease activity and that the enzyme was inactive on ssDNA (Supplementary Figure S3C). Lastly, ISG20 also degraded RNA involved in RNA/DNA heteroduplexes (Supplementary Figure S3D).
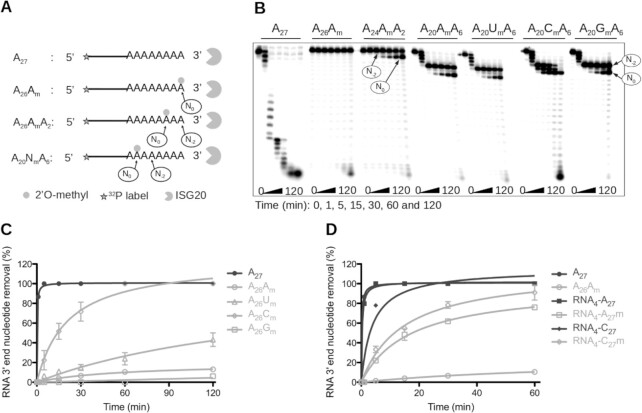
Next, to investigate whether the internal 2′O-methylation mark conferred resistance to ISG20-mediated degradation, radiolabeled A27 fragments carrying a 2′O-methyl group at different positions were incubated with ISG20 (Figure 1A), and their decay analyzed on urea-PAGE. A27 that contained one Nm at the 3′ end (A26Am) or two nucleotides upstream of its 3′ end (A24AmA2) was almost completely protected from ISG20-mediated degradation (Figure 1B). Conversely, if the methylation was 7 nucleotides upstream of the 3′ end (A20AmA6), the RNA remained sensitive to ISG20 exonuclease activity, but hydrolysis paused before further degradation of the substrate. The stalling occurred at two distinct positions. The first main pause was detected two nucleotides upstream of the Nm (N–2) and the second at the Nm (N0). This inhibition is likely to be ISG20-specific because the 3′ exonuclease activity of two other 3′-5′ exonucleases, namely nuclease P1 (NP1) and phosphodiesterase I (PDE), was barely affected by the presence of Nm (Supplementary Figure S4). Then, the same experiment was repeated to evaluate whether 2′O-methylation on ribonucleotides other than ‘A’ led to the same degradation pattern. A20UmA6 and A20GmA6 displayed a similar degradation profile as A20AmA6 (Figure 1B). However, the pause observed at N-2 and N0 seemed to be less pronounced with A20CmA6, and ISG20 overcame the Nm after a longer incubation period (2 h). The 2′O-methylation mark also protected heteropolymeric RNAs from degradation (Supplementary Figure S5), but less efficiently than the homopolymeric A27 (Figure 1B). These findings indicated that ISG20 exonuclease activity pauses two nucleotides upstream of the Nm, and at the Nm and that the length of these pauses depends on the Nm nature and its sequence context.
Figure 1.
Impact of RNA 2′O-methylation on ISG20 nuclease activity. (A–D) Recombinant human ISG20 was incubated with different 5′-radiolabeled RNAs carrying 2′O-methylated residues at various positions in a time-course experiment, and the substrate hydrolysis was followed by PAGE. The degradation kinetics of various substrates in (C and D) were monitored by PAGE separation. Quantification of nucleotide removal, relative to the total RNA length, was done using the FujiImager and Image Gauge analysis software. (A) Schematic representation of the experimental procedure of panel (B). (B) Effect of RNA 2′O-methylation on ISG20 exonuclease activity. The assay was performed using non-methylated A27, and 3′ end 2′O-methylated A26Am, A24AmA2, A20AmA6, A20UmA6, A20CmA6, and A20GmA6. (C) Quantification of non-methylated (A27) and 3′ end 2′O-methylated RNA (A26Am, A26Um, A26Cm, A26Gm) degradation by ISG20. (D) Quantification of non-methylated RNA4-A27 and RNA4-C27 and 3′-end 2′O-methylated RNA4-A27m, RNA4-C27m degradation by ISG20. The results in panel B and C correspond to the mean values and standard deviation of three independent experiments.
2′O-Methylation of poly(A) at its 3′end suppresses degradation by ISG20
To further characterize the stalling observed at the Nm, ISG20 was incubated with a poly(A) carrying Am, Gm, Cm or Um at its 3′ end for different times. Following PAGE separation, the hydrolysis of RNA substrates was quantified by densitometry. This showed that the 3′ to 5′ excision kinetics of A26Nm was dependent on the nature of the Nm at the 3′ end. Indeed, ISG20 fully hydrolyzed non-methylated A27 in ∼5 min (Figure 1C). After 120 min ∼100% of A26Cm, ∼43% of A26Um, and 13% of A27m were degraded, conversely A26Gm was almost completely protected from ISG20 degradation (6%) (Figure 1C). These results confirm that the presence of a methyl group on the 2′O of the last nucleotide strongly reduces RNA decay and increases its lifespan to a different extent in function of the methylated residue (G > A>>U>>C). Again, ISG20 was more active on heteropolymeric linear ssRNAs that contained an Nm at their 3′ end, as indicated by the degradation of ∼75% and 95% of RNA4-A27m and RNA4-C27m RNA, respectively, after 60 min of incubation (Figure 1D).
2′O-Methylated ribonucleotides limit ISG20 RNase activity through steric hindrance
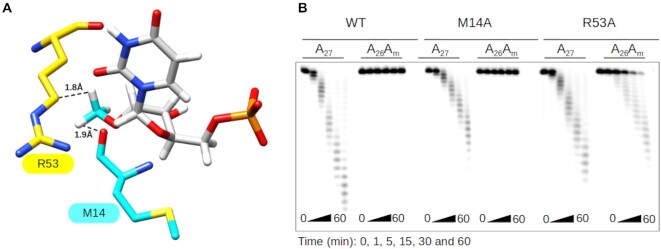
Next, the previously described 3D structure of ISG20 in complex with UMP (32) was used to determine whether ISG20 exonuclease activity inhibition resulted from a direct inhibition of the catalytic reaction or from altered substrate recognition. A methyl group was modeled on the 2′O of the UMP ribose to determine its impact on the catalytic/binding pocket of ISG20. This modification led to a potential steric clash of the methyl group with the M14 and R53 residues of ISG20 (Figure 2A), suggesting that UMP would need to be repositioned within the catalytic pocket to accommodate the methyl group. To test this hypothesis, these two ISG20 residues were substituted with the less bulky alanine residue (Supplementary Figure S6) and the activity of each mutant was evaluated using A27 and A26Am (as substrate to analyze the pause at N0). The exonuclease activity of these two ISG20 mutants toward A27 was slightly reduced compared with WT ISG20, confirming the role of these residues in RNA stabilization in ISG20 catalytic pocket (Figure 2B). When incubated with A26Am, WT ISG20 and the M14A mutant did not show any nuclease activity. Conversely, the R53A mutant could degrade A26Am, indicating that this mutant can overcome the stop at N0 (Figure 2B). Therefore, it can be inferred that the stop at N0 results from a steric hindrance mechanism with residue R53 of ISG20 (Supplementary Figure S7A).
Figure 2.
Molecular basis of ISG20 inhibition by 2′O-methylation at N0. (A) Structural model of ISG20 showing the steric clash between residues R53 (yellow surface) and M14 (cyan surface), and 2′O-methylated UMP (UMP is in grey and 2′O-methyl in cyan). The methylated UMP was built using PubChem Sketcher V2.4 and superimposed on the UMP present in the catalytic pocket of the ISG20 structure (PDB: 1WLJ). (B) The exonuclease activity of wild-type (WT) and M14A and R53A mutant ISG20 on methylated and non-methylated A27 RNAs was monitored by PAGE analysis as in Figure 1.
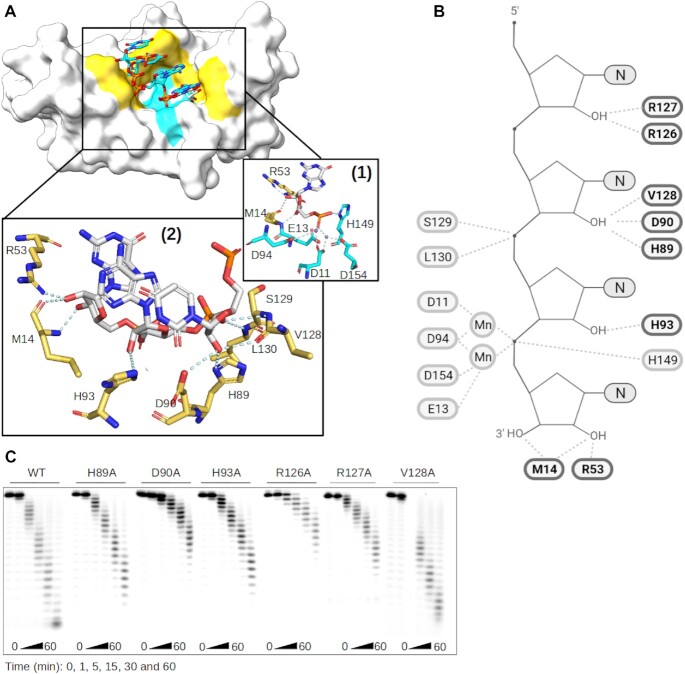
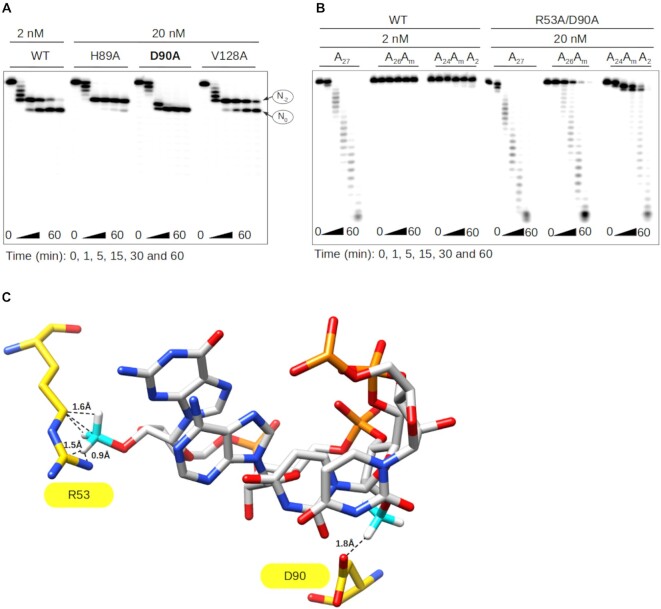
Then, the molecular mechanism of the N-2 stop was investigated using the structural information of two ISG20 homologs (SDN1 and ExoX) in complex with a nucleic acid chain (42,43). Superimposition of ISG20 with SDN1 and ExoX showed a good fit of their catalytic sites with a Root-Mean-Square Deviation of 0.836 Å and 1.148 Å, respectively, and the superposition of the nucleic acid chains with the UMP of ISG20 (Supplementary Figure S8). Based on the structural alignment, putative candidate residues of the RNA binding domain (RBD) were selected: M14, R53, H89, D90, H93, R126, R127, V128, S129 and L130 (Figure 3A, B). To experimentally test the structural model, alanine scanning was carried out on residues that were most likely to affect the binding (H89A, D90A, H93A, R126A, R127A, V128A) and the different proteins were produced and purified (Supplementary Figure S6) for RNA nuclease assays. Although most ISG20 mutants conserved their exonuclease activity on A27 (Figure 3C), the D90A, R126A and R127A mutants showed reduced hydrolytic capacity that may result from disruption of the interaction between ISG20 and the RNA substrate (Figure 3B). According to our structural model, the stop at N-2 could implicate residue(s) H89, D90 and/or V128 that are supposed to interact with the 2′OH of N0 in the RNA and could be struggling to accommodate the methylated nucleotide (Supplementary Figure S7B). To confirm this hypothesis, the effect of 2′O-methylation on the ISG20 H89A, D90A, and V128A mutant activity on A20AmA6 was assessed at a concentration of 20 nM instead of 2 nM to overcome their decreased enzymatic activity. All mutants showed the same activity profile as WT ISG20, including the two intermediate degradation products detected at N-2 and N0, with the exception of the D90A mutant that only stopped at N0 (Figure 4A). These results confirmed that D90 is the key player in the N-2 stop, which is likely due to a steric clash with the Nm (Supplementary Figure S7B).
Figure 3.
Structural activity relationship of ISG20 RNA binding Domain. (A) Model of ISG20 in interaction with an RNA substrate was built based on superimposition of ISG20 (PDB: 1WLJ) and SDN1 in complex with RNA (PDB: 5Z9X). Surface representation of ISG20 (off-white) containing an RNA from SDN1 structure (sticks). Highlighted in yellow are the residues of the RNA binding domain (RBD) and in cyan the conserved residues of the exonuclease domain. Zoom on the ISG20 catalytic pocket showing the interaction (cyan) between the (1) residues of its catalytic domain (cyan), manganese (purple), and the nucleotide to be excised (grey), (2) residues of its RBD (yellow) and the RNA (grey). (B) Cartoon representing the ISG20 model in interaction with an RNA substrate performed using Biorender in which the residues of ISG20 RBD interacting with the RNA 2′OH are highlighted in bold. (C) Mutagenesis analysis of the residues highlighted in panel (B). The different ISG20 mutants were produced and their exonuclease activity was followed as in Figure 1 using non-methylated A27.
Figure 4.
The D90 and R53 residues play a key role in ISG20 inhibition by 2′O-methylation. (A) Exonuclease activity of WT ISG20 and the H89A, D90A, and V128A mutants was assessed on A20AmA6, and the RNA degradation products were separated on PAGE and analyzed by autoradiography, as in Figure 1. (B) Exonuclease activity of WT ISG20 and the double mutant R53A/D90A was assessed using the A27, A26Am and A24AmA2 RNA substrates. Degradation was monitored by PAGE and analyzed by autoradiography, as described in Figure 1. (C) Structural model of ISG20 catalytic site (PDB: 1WLJ) with a methylated RNA (the methyl groups were added to the RNA using the chimera build structure option). Residues D90 and R53 are in yellow, the RNA is in grey, the RNA 2′O-methyl moieties in cyan. The model shows the steric hindrance between the distinct methylated nucleotides and the R53 and D90 residues of ISG20.
A double mutant R53A/D90A was next generated. Despite partial loss of its nuclease activity on substrates at 2 nM, the double mutant digested both methylated (A26Am and A24AmA2) and non-methylated A27 in a time-dependent manner at 20 nM (Figure 4B). Although the hydrolytic activity of the mutant was reduced, it could bypass the N-2 and N0 stops, unlike WT ISG20. Altogether, these results give insights into the molecular basis of the hydrolysis blockage that is caused by two consecutive steric hindrances between the D90 and R53 residues of ISG20 and the methylated nucleotide (Figure 4C & Supplementary Figure S7). The specificity of the 2′O-methylation-linked inhibition was confirmed by introducing another PTM, adenosine N6-methylation (AN6m), in the RNA substrate. This methylation did not interact with the ISG20 RBD in the structural model (Supplementary Figure S9A) nor affected ISG20 exonuclease activity (Supplementary Figure S9B).
Internal 2′O-methylation limits HIV-1 RNA degradation by ISG20
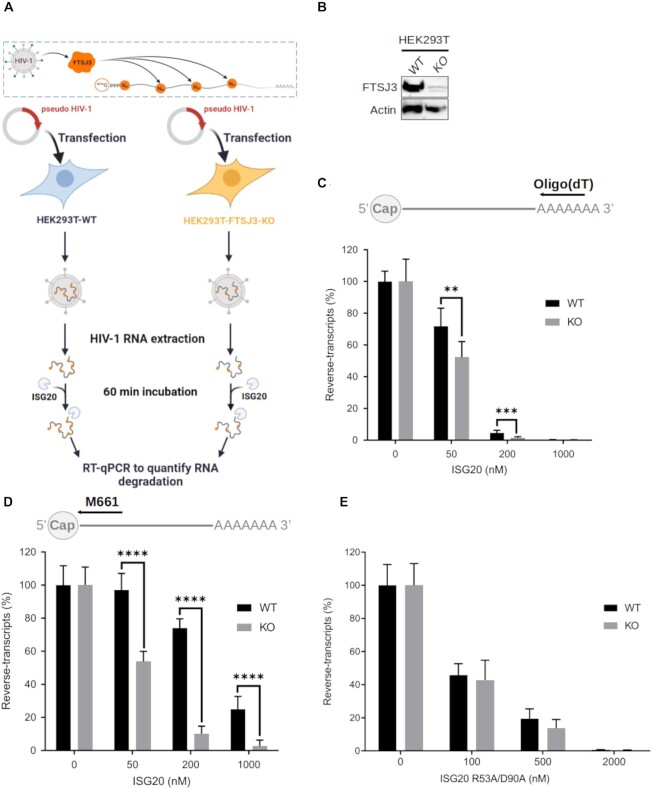
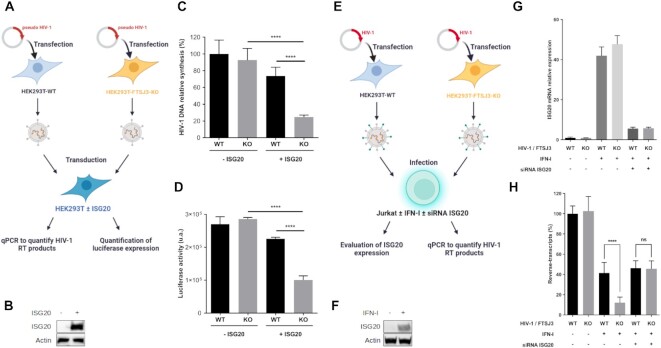
In the HIV-1 genome, internal 2′O-methylation is catalyzed by the cellular 2′O-MTase FTSJ3 (26). Therefore, parental and FTSJ3-KO HEK293T cells (Figure 5A & B) were used to produce HIV-1 virions. After extraction, naturally methylated (from parental cells) and hypomethylated (from FTSJ3-KO cells) full-length vRNAs were incubated with increasing concentrations of recombinant human ISG20 (Figure 5A). After 1 h incubation, the exonuclease reaction was stopped by heating, and the remaining HIV-1 vRNAs were reverse-transcribed using primers that anneal to the 5′ (M661, in the Gag gene) or the 3′ end (oligo(dT)) of the HIV-1 genome (Figure 5C, D). Quantification of the reverse-transcription (RT) products by qPCR showed that HIV-1 RNA could be degraded by ISG20 and that the 5′ end of the genome was less prone to RNA degradation, as expected for 3′ to 5′ exonuclease activity. Moreover, hypomethylated HIV-1 RNA was more sensitive to ISG20-mediated degradation than methylated RNA. Then, methylated and hypomethylated HIV-1 RNAs were incubated with the ISG20 double mutant R53A/D90A, the activity of which is barely affected by RNA 2′O-methylation (Figure 4B). Methylated and hypomethylated HIV-1 RNAs were degraded to a similar extent (Figure 5E), confirming that Nm presence in the HIV-1 genome does not alter the activity of the ISG20 double mutant. These results prompted us to evaluate the antiviral effect in ISG20-expressing cells during infection. To this aim, luciferase-encoding VSV-G pseudotyped viruses were produced in parental and FTSJ3-KO HEK293T cells to generate HIV-1 pseudoparticles that contained methylated and hypomethylated vRNAs, respectively (Figure 6A). Then, HEK293T cells that overexpress or not Flag-tagged human ISG20 (Figure 6B) were transduced with these pseudoviruses and the reverse-transcription of HIV-1 RNA in infected cells was monitored by qPCR (Figure 6A). Quantification of HIV-1 DNA products at 6 h post-infection showed a decreased in RT efficiency of hypomethylated viruses in ISG20-expressing cells (Figure 6C). This observation suggests an increased sensitivity of hypomethylated vRNA to ISG20-mediated degradation in infected cells. This was further supported by the lower luciferase expression observed in ISG20-expressing cells infected with hypomethylated compared to naturally methylated pseudotyped viruses (Figure 6D, FTSJ3-KO). Collectively, these results suggest that RNA 2′O-methylation induced by FTSJ3 impairs the antiviral activity of ISG20 by limiting the degradation of HIV-1 genomes.
Figure 5.
Hypomethylated HIV-1 RNA is sensitive to in vitro ISG20-mediated degradation. (A–D) Parental and FTSJ3-KO HEK293T cells were transfected with HIV-1 molecular clone pNL4-3.Luc.R-E in order to produce HIV-1 virions containing naturally methylated (WT) and hypomethylated (KO) genome, respectively. Upon recovery of viral particles, full-length vRNAs were extracted and incubated with recombinant human ISG20 (WT or double mutant R53A/D90A) for 60 min. Finally, vRNAs degradation by ISG20 was monitored by RT-qPCR. (A) Schematic representation of the experimental procedure. (B) FTSJ3 expression in parental (WT) and FTSJ3-KO (KO) HEK293T cells was assessed by western blotting before transfection of the HIV-1 molecular clone pNL4.3. (C, D) 50 ng of RNA extracted from HIV-1 particles produced in parental (WT) or FTSJ3-KO HEK293T cells was incubated with ISG20 at the indicated concentrations at 37°C for 1h. After ISG20 inactivation (20 min at 70°C), RNA products were reverse-transcribed using a (C) oligo(dT) or (D) the M661 primer, and the reverse-transcripts quantified by qPCR. The relative amount of HIV-1 reverse-transcripts in each condition is shown. (E) The exonuclease activity of ISG20 R53A/D90A was evaluated using HIV-1 RNA extracted from viral particles produced in parental (WT) or FTSJ3-KO HEK293T cells. RNA degradation products were reverse-transcribed using the M661 primer, and the reverse-transcripts quantified by qPCR as in (D). Data are the mean ± standard deviation of three independent experiments performed in triplicate. **P < 0.01, ***P < 0.001 and **** P < 0.0001 (Student's t-test).
Figure 6.
Restriction of the hypomethylated HIV-1 replication by ISG20. (A–D) ISG20 restricts the reverse-transcription of hypomethylated VSV-G pseudotyped HIV-1. WT and FTSJ3-KO HEK293T cells were used to produce VSV-G-pseudotyped HIV-1 particles containing WT and hypomethylated (KO) genome, respectively. Extracted virions were used to infect 48 h WT HEK293T cells over-expressing or not Flag-tagged human ISG20. At 6 h post-infection, total cell DNA was extracted and HIV-1 reverse-transcripts were quantified by qPCR and HIV-1 infection was estimated by measuring luciferase activity in cell extracts 24h post-infection. (A) Schematic representation of the experimental procedure. (B) HEK293T cells were transfected with an empty plasmid or with a plasmid encoding Flag-tagged human ISG20. The expression of ISG20 was analyzed by western blotting using an anti-Flag antibody. (C) Quantification of HIV-1 reverse-transcripts by qPCR. (D) HIV-1 infection was estimated by measuring luciferase activity in cell extracts 24 h post-infection. (C, D) Data are the mean ± standard deviation of three independent experiments performed in triplicate; **P < 0.01, ***P < 0.001 and **** P < 0.0001 (Student's t-test). (E–H) Jurkat cells were treated or not with 1000 IU/ml human IFN-I for 18 h and transfected with a siRNA control (−) or targeting ISG20 (+), as indicated. At 48 h post-transfection, cells were infected with HIV-1 NL4-3 produced in WT or in FTSJ3-KO HEK293T cells. At 6 h post-infection, cells were harvested and total RNA and DNA were extracted for specific qPCR analysis. (E) Schematic representation of the experimental procedure. (F) Jurkat cells were treated or not with 1000 IU/ml human IFN-I for 18h and ISG20 expression was assessed by western-blot. (G) Evaluation of endogenous ISG20 expression by RT-qPCR. (H) Quantification of HIV-1 RT products by qPCR. (G, H) Data represents the mean of three independent experiments performed in duplicates ± SD. ****P < 0.0001, as determined by one-way ANOVA with Bonferroni post hoc test. ns, non-significant.
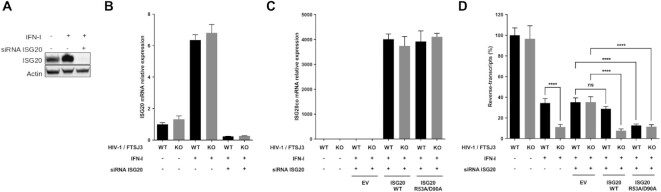
We next sought to determine whether ISG20 expression would affect the reverse-transcription of replication-competent HIV-1 in its main target cells, CD4+ T lymphocytes. To this end, we evaluated the antiviral effect of ISG20 in human Jurkat cells infected with HIV-1 particles either containing naturally methylated (WT) or a hypomethylated genome (Figure 6E). In these cells, the constitutive expression of ISG20 is undetectable, but can be significantly increased following type I interferon (IFN-I) treatment (Figure 6F, G). We therefore assessed the ability of WT and hypomethylated HIV-1 to perform RT in Jurkat cells following IFN-I treatment, with or without silencing the expression of ISG20 by a specific siRNA. Interestingly, IFN-I treatment led to a significantly greater inhibition of the reverse-transcription of hypomethylated HIV-1 compared to the WT viruses (Figure 6H). However, this effect was lost upon ISG20 knockdown, suggesting that hypomethylated HIV-1 was more sensitive than WT virus to ISG20-mediated decay (Figure 6H). Moreover, we wanted to determine whether the overexpression of WT ISG20 or the double mutant R53A/D90A could revert the effect of ISG20 silencing on replication-competent HIV-1. However, since Jurkat cells proved to be very difficult to transfect and that we could not use HIV-1 vectors to transduce them, we decided to perform the next experiments in HeLa P4C5 cells, which express HIV-1 receptors and are easy to transfect. Unlike Jurkat cells, HeLa cells constitutively express ISG20, and its expression can be further increased by IFN-I treatment (Figure 7A, B). In order to evaluate the sensitivity to ISG20 of infectious HIV-1 containing naturally methylated or hypomethylated genome, cells were first pre-treated or not with IFN-I, then co-transfected with a siRNA control (−) or targeting ISG20 (+) and an empty vector (EV) or a plasmid encoding either WT or the R53A/D90A ISG20. Then, the cells were infected with HIV-1 produced in WT or FTSJ3-KO HEK293T cells and the amount of RT products was quantified by RT-qPCR 6 h post-infection. In this experimental setup, we observed that the level of IFN-I induced ISG20 expression was lower than that observed in Jurkat cells, probably due to its relatively high constitutive expression in HeLa cells (Figures 6F, G and 7A, B). In addition, the siRNA-mediated ISG20 silencing in this experimental system was very efficient (Figure 7B) and overexpression of ISG20-encoding plasmids, which are not sensitive to ISG20 siRNA, was assessed by RT-qPCR (Figure 7C). As expected and concordantly with the results obtained in Jurkat cells, the RT of hypomethylated viruses was more affected by IFN-I treatment than that of WT viruses (Figure 7D). Interestingly, the RT of hypomethylated viruses was restored to WT levels following ISG20 knockdown, which once again supports the conclusion that difference in IFN-I sensitivity results from an increased sensitivity of hypomethylated HIV-1 to ISG20 restriction (Figure 7D). To further confirm these observations, we compensated ISG20-depleted HeLa cells with ectopic ISG20 expression. We thus observed a similar profile as in IFN-I treated cells, illustrated by a significant inhibition of the RT of hypomethylated viruses, but not of WT viruses (Figure 7D). The sensitivity of ISG20 to viral RNA 2′O-methylations was further demonstrated using the ISG20 double mutant R53A/D90A, which, unlike WT ISG20, inhibited the RT process of both viruses in a similar manner (Figure 7D). Altogether, our in cellula assays showed that ISG20 interferes with the retro-transcription of hypomethylated viruses to a greater extent than that of the WT viruses. However, it should be noted that this ISG20-induced inhibition of HIV-1 is less pronounced than that observed in our in vitro experiments. This could be due to various factors such as the accessibility of the vRNA to the cellular ISG20, the methylation level of viral genomic RNA, the position of the Nm within the vRNA and the sequence context of Nm. Despite this, all of our results highlight the antagonistic effect 2′O-methylation on ISG20 nuclease-mediated antiviral activity and reveal a new proviral role for the 2′O-methylations of the HIV-1 genome.
Figure 7.
Restriction profile of ISG20 WT and the R53A/D90A mutant on replication competent HIV-1. (A) HeLa P4C5 cells were treated or not with 1000 IU/ml human IFN-α2a for 18h and transfected with a siRNA control (−) or targeting ISG20 (+), as indicated. At 48h post-transfection, ISG20 expression was assessed by western-blot using anti-ISG20 antibodies. (B–D) HeLa P4C5 cells were treated or not with 1000 IU/ml human IFN-I for 18 h and transfected with a siRNA control (−) or targeting ISG20 (+), as indicated. At 48h post-transfection, cells were infected with HIV-1 NL4-3 produced in WT or in FTSJ3-KO cells. At 6 h post-infection, cells were harvested and total RNA and DNA were extracted for specific qPCR analysis. (B) The expression of endogenous ISG20 upon IFN-I induction was evaluated by RT-qPCR. (C) Quantification of ectopic expression of transfected ISG20 WT and double mutant by RT-qPCR. (D) The amount of HIV-1 RT products was estimated by qPCR. Data represent the mean of three independent experiments performed in duplicate ± SD. ****P < 0.0001, as determined by one-way ANOVA with Bonferroni post hoc test. ns, non-significant.
DISCUSSION
ISG20 is an IFN-regulated protein with antiviral activity against a broad range of RNA viruses (31–33,44). Although it has been suggested that its antiviral activity is linked to its exonuclease activity, ISG20 also inhibits the translation of vRNA (45). Several recent studies have shown that the genome of many RNA viruses contains PTM, including Nm (1–3,25,26). Here, we found that this epitranscriptomic modification can inhibit ISG20 exonuclease activity in vitro and thereby limit the host antiviral response mediated by ISG20. Using synthetic RNAs carrying 2′O-methylation at different positions, we demonstrated that ISG20 nuclease activity is specifically impaired by this epitranscriptomic mark. We also observed that this inhibition is modulated by several factors, such as the nature of the methylated nucleotide moiety (Gm > Am > Um > Cm>>A), its position in the RNA sequence, and the substrate sequence (Figure 1 and Supplementary Figure S5). We found that ISG20 activity was strongly impaired when the 2′O-methylation occurred in the context of homopolymeric RNA, such as poly(A), and at the 3′ end of the sequences than in heteropolymeric RNA substrates (Figure 1 and Supplementary Figure S5). Together, these data indicate that Nm can regulate RNA decay and that synthetic poly(A) is quite resistant to ISG20-mediated degradation when modified at its 3′ end. However, the proportion of methylated A residues in viral poly(A) tails has not been investigated yet, to our knowledge.
In addition, we showed that the degradation of RNA carrying internal 2′O-methylation (A20NmA6) by ISG20 was paused at N-2 and N0 from the Nm (Figure 1A). Through a structure-guided analysis, we selected key residues of ISG20 RBD that may be involved in this inhibition (M14, R58, H89, D90, and V128; Figure 3A, B). Alanine mutagenesis of these residues revealed that the pauses at N–2 and N0 were overcome by ISG20 harboring the D90A and R53A mutation, respectively (Figure 2 and 4A, and Supplementary Figure S7), indicating that this inhibition occurs through steric hindrance. This was further confirmed by the finding that the double mutant R53A/D90A can hydrolyze methylated RNA without pausing at N–2 and N0 (Figure 4B) and highlights the role of residues R53 and D90 as 2’O-methyl group sensors within the ISG20 catalytic pocket. Moreover, this inhibition seems to be specific to RNA 2′O-methylation because N6-methylation, another important epitranscriptomic mark present on vRNA, barely affected ISG20 exonuclease activity (Supplementary Figure S9B). This observation is also supported by the structural model of ISG20 in the presence of an RNA substrate showing that N6-methylation is outside the RNA binding pocket (Supplementary Figure S9A). This is consistent with the study by Imam et al. demonstrating that ISG20 targets HBV transcripts containing AN6m (46). Therefore, it is likely that ISG20 inhibition is specifically related to the presence of RNA 2′O-methylation, suggesting that viruses that induce such internal PTM, using their own (21–23) or the host MTase (26), might limit ISG20 antiviral activity. This possible role of internal 2′O-methylation is reminiscent of other viral strategies to overcome the cell defenses, such as cap 2′O-methylation that prevents vRNA sensing by RIG-I (9,10) and its subsequent sequestration by IFIT1/3 restriction factors (11).
Although the viral epitranscriptome is still poorly characterized for most viruses, the recent development of sequencing-based methods to identify modified nucleotides and their location has boosted the knowledge on the mechanisms and functions of 2′O-methylation in RNA viruses (1–3,25,26). For instance, HIV-1 exploits the cell machinery to catalyze 2′O-methylation of its own genome through a tightly regulated process. Indeed, HIV-1 recruits FTSJ3, a cellular MTase, to catalyze the 2′O-methylation of 17 specific residues of its own RNA. This prevents the sensing of vRNA by MDA5, which in turn limits the production of IFN-I (26). Therefore, we asked whether internal methylation of HIV-1 RNA might also influence its resistance to ISG20 restriction. We addressed this question by comparing in vitro the sensitivity to ISG20 of HIV-1 RNAs extracted from viruses produced in parental HEK293T cells or cells that lack FTSJ3. As expected, we found that recombinant ISG20 was able to degrade HIV-1 genome and that internal 2′O-methylation could counteract this degradation (Figure 5C, D). However, the protection conferred by vRNA 2′O-methylation was less drastic than observed in the in vitro experiments performed on 2′O-methylated homopolymeric synthetic RNA. This might be due to the fact that only 17 confident methylations were detected inside the HIV RNA (26) and are distributed along the viral genome. In addition, it is noteworthy that the methylations detected in the HIV genome are in heteropolymeric regions which are more susceptible to ISG20 in vitro degradation (Figure 1D and Supplementary Figure S5) and we do not know yet whether the poly(A) tail of HIV-1 contains Nm. We next confirmed this result in cell lines (HEK293T, Jurkat, HeLaP4C5) infected with naturally methylated or hypomethylated VSV-G pseudotyped HIV-1 or with replication-competent HIV-1 virus. Using a VSV-G pseudotyped HIV-1 encoding luciferase, we first showed that ectopically expressed ISG20 reduced the RT and subsequent luciferase expression of HIV-1 vectors containing a hypomethylated genome. (Figure 6C, D). Using replication-competent HIV-1, we also found that IFN-induced ISG20 reduced the RT level of hypomethylated HIV-1 in Jurjat cells (Figure 6H). Finally, we showed that the ectopic expression of ISG20 in HeLa-CD4 cells, only interfered with the RT of hypomethylated HIV-1 viruses (Figure 7D), whereas the R53A/D90A ISG20 mutant inhibited the RT of both WT and hypomethylated viruses at the same extent, as expected (Figure 7D). It should be noted that the levels of HIV RT inhibition by ISG20 in infected cells were rather modest compared to the results obtained in vitro. This observation illustrates the fact that vRNA is protected by a core during the early stages of infection, presumably making it difficult for ISG20 to access. However, the fact that viruses with hypomethylated genomes are partly susceptible to ISG20 restriction suggests that the vRNA remains relatively accessible. Further experiments will be required to determine whether this relative accessibility of the HIV-1 genome only occurs at certain stages of the cycle (e.g. RT initiation or decapsidation) and whether ISG20 degrades RNA in the cytoplasm or nucleus. In any case, contrary to the study by Espert et al. (33), we did not observe any susceptibility of WT HIV-1 to ISG20 restriction.
Collectively, our results highlight the role of internal 2′O-methylations in the early phases of HIV-1 infection, preventing ISG20 antiviral activity. Indeed, 2′O-methylation is a viral countermeasure to the cellular antiviral response originally identified as the subversion of ‘self’ sensing to impair type 1 IFN expression. Our study also demonstrates that at least for HIV-1, 2′O-methylation also directly interferes with the antiviral effect of ISG20 by protecting the vRNA from its exonuclease activity. The importance of 2′O-methylation in viral replication has been shown in various RNA viruses, including Nipah and Hendra viruses (47), but the dual function observed in HIV-1 (limiting vRNA sensing and degradation) must be investigated in other viruses to determine whether this dual mechanism is universal.
The analysis of SARS-CoV-2 epitranscriptome also showed the presence of internal 2′O-methylation marks (25). However, the implicated MTase remains unknown. These Nm marks, were mainly detected in the 5′ and 3′ untranslated regions (UTR) and in the non-coding regions of the vRNA (25) that are enriched in hairpin structures. Therefore, these methylation marks might prevent the detection of vRNA by MDA5, as observed for HIV-1. Their impact on ISGs that target double stranded RNA, such as 2′-5′-oligoadenylate synthetase 1 (48) and the RNase L, is questionable. Moreover, as a recent study revealed a transient upregulation of ISG20 expression upon SARS-CoV-2 infection (49), the resistance to ISG20-mediated restriction could result from the adoption of countermeasures (e.g. hyper-2′O-methylation at the 3′UTR) (25) evolved by the virus. More studies are needed to confirm these hypotheses in the context of SARS-CoV-2 infection. Internal 2′O-methylation has also been described in flaviviruses, and in vitro studies showed that the NS5 MTases of DENV (22) and ZIKV (21) specifically methylate adenosines. However, the epitranscriptome analysis of both viruses highlighted the presence of 2′O-methylation marks on all four ribonucleotides (2). This implies that the host 2′O-MTases might also participate in these methylation processes. This is consistent with the binding of fibrillarin, a nucleolar 2′O-MTase, in the DENV and ZIKV genomes (50). These observations suggest that there is synergy and fine-tuning in the 2′O-methylation performed by viral and cellular MTases to shield vRNAs from innate immune censors and the IFN response.
Furthermore, our experimental data raise an important question as how cellular RNAs escape the exonuclease activity of ISG20. A recent study showed that ISG20 can discriminate self from non-self RNA (45), but the molecular signatures involved in this process are not yet identified. Our results suggest that epitranscriptomic modifications such as 2′O-methylation of ribonucleotides in the poly(A) might play a role in such process, although the mechanisms governing the 2′O-methylation of cellular RNA are still poorly characterized. Despite the fact that numerous cellular RNA MTases have been identified, their involvement in self-marking is still an open question. The mechanism governing specific RNA recognition is barely described except for Fibrilarin, which is involved in RNA guided methylation and was particularly studied in the case of ribosomal RNA (51–53).
The identification of the functions associated with internal 2′O-methylation on MDA5 sensing and ISG20 exonuclease activity may also be considered in the context of improving mRNA-based therapeutics. Indeed treatment with exogenous RNA can be hampered by the rapid degradation of RNA molecules and/or its capacity to induce inappropriate innate immunity stimulation. Various PTMs are used to overcome these drawbacks. For instance, addition of an RNA cap structure (54–56), a 5′ and 3′ untranslated region (57), or Poly-A (56,58) promotes mRNA stabilization and increases protein translation. In addition, modified nucleosides, such as AN6m, 5-methylcytidine, and pseudo-uridine, can be used to avoid innate immunity overstimulation and to increase RNA translation (59,60). The results from this study and earlier work (26) highlight the role of 2′O-methylation in RNA sensing and suggest that this modification may be used to improve the half-life of exogenous RNAs, acting as a protection against ISG20-mediated degradation and the cellular response against non-self RNA.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Yamina Bennasser (IGH, Montpellier, France) for kindly providing HEK293T cells knocked-out for FTSJ3 (HEK-FTSJ3-KO). The luciferase-encoding HIV-1 plasmid (pNL4-3.Luc.R-E) was kindly provided by Nathaniel R Landau (Aaron Diamond AIDS Research Center, The Rockefeller University, New-York, USA). We thank Bhawna Sama and Coralie Valle for their careful critical reading of this manuscript.
Author contributions: Conceptualization: P.E.K. and E.D.; Methodology: P.E.K., B.Co., S.N. and E.D.; Investigation: P.E.K., N.R., C.C., L.P., F.F., N.S.; Formal analysis: P.E.K., F.F., S.N. and E.D.; Validation: P.E.K. and E.D.; Resources: N.R., F.D., B.Co., N.S., B.Ca. and E.D.; Data curation: P.E.K., F.F., N.S., Writing – original draft: P.E.K, and E.D; Writing – review & editing: P.E.K., N.R., L.P., C.C., F.F, F.D., B.Ca., D.M., B.Co, S.N. and E.D.; Visualization: P.E.K., N.S.; Project administration: P.E.K. and E.D.; Funding acquisition: B.Ca., S.N., D.M. and E.D.
Contributor Information
Priscila El Kazzi, AFMB, CNRS, Aix-Marseille University, UMR 7257, Case 925, 163 Avenue de Luminy, 13288 Marseille Cedex 09, France.
Nadia Rabah, AFMB, CNRS, Aix-Marseille University, UMR 7257, Case 925, 163 Avenue de Luminy, 13288 Marseille Cedex 09, France; Université de Toulon, 83130 La Garde, France.
Célia Chamontin, IRIM, CNRS UMR9004, Université de Montpellier, Montpellier, France.
Lina Poulain, AFMB, CNRS, Aix-Marseille University, UMR 7257, Case 925, 163 Avenue de Luminy, 13288 Marseille Cedex 09, France.
François Ferron, AFMB, CNRS, Aix-Marseille University, UMR 7257, Case 925, 163 Avenue de Luminy, 13288 Marseille Cedex 09, France; European Virus Bioinformatics Center, Leutragraben 1, 07743 Jena, Germany.
Françoise Debart, IBMM, UMR 5247 CNRS, Université de Montpellier, ENSCM, Montpellier, France.
Bruno Canard, AFMB, CNRS, Aix-Marseille University, UMR 7257, Case 925, 163 Avenue de Luminy, 13288 Marseille Cedex 09, France.
Dorothée Missé, MIVEGEC, Univ. Montpellier, CNRS, IRD, Montpellier, France.
Bruno Coutard, Unité des Virus Émergents (UVE: Aix-Marseille Univ-IRD 190-Inserm 1207), Marseille, France.
Sébastien Nisole, IRIM, CNRS UMR9004, Université de Montpellier, Montpellier, France.
Etienne Decroly, AFMB, CNRS, Aix-Marseille University, UMR 7257, Case 925, 163 Avenue de Luminy, 13288 Marseille Cedex 09, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
P.E.K. was supported by the Foundation Méditerranée infection; Agence nationale de la recherche [ANR-20-CE11-0024-02, ANR-17-CE15-0029]; Foundation pour la recherche médicale [FRM-REPLI80C/U160]; Agence Nationale de la Recherche sur le SIDA et les Hépatites virales [ANRS-ECTZ80C/U160]. Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1. Netzband R., Pager C.T.. Epitranscriptomic marks: emerging modulators of RNA virus gene expression. WIREs RNA. 2020; 11:e1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McIntyre W., Netzband R., Bonenfant G., Biegel J.M., Miller C., Fuchs G., Henderson E., Arra M., Canki M., Fabris D.et al.. Positive-sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Res. 2018; 46:5776–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kennedy E.M., Courtney D.G., Tsai K., Cullen B.R.. Viral epitranscriptomics. J. Virol. 2017; 91:e02263-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Decroly E., Ferron F., Lescar J., Canard B.. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 2012; 10:51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Decroly E., Canard B.. Biochemical principles and inhibitors to interfere with viral capping pathways. Curr. Opin. Virol. 2017; 24:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang C.-T., Muthukumar S., Weber R., Levdansky Y., Chen Y., Bhandari D., Igreja C., Wohlbold L., Valkov E., Izaurralde E.. A low-complexity region in human XRN1 directly recruits deadenylation and decapping factors in 5′-3′ messenger RNA decay. Nucleic Acids Res. 2019; 47:9282–9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jurado A.R., Tan D., Jiao X., Kiledjian M., Tong L.. Structure and function of pre-mRNA 5′-end capping quality control and 3′-end processing. Biochemistry. 2014; 53:1882–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lazaris-Karatzas A., Montine K.S., Sonenberg N.. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990; 345:544–547. [DOI] [PubMed] [Google Scholar]
- 9. Devarkar S.C., Wang C., Miller M.T., Ramanathan A., Jiang F., Khan A.G., Patel S.S., Marcotrigiano J.. Structural basis for m7G recognition and 2′-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schuberth-Wagner C., Ludwig J., Bruder A.K., Herzner A.-M., Zillinger T., Goldeck M., Schmidt T., Schmid-Burgk J.L., Kerber R., Wolter S.et al.. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N1-2′O-Methylated self RNA. Immunity. 2015; 43:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.-Y., Schneller S., Zust R., Dong H.et al.. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010; 468:452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abbas Y.M., Pichlmair A., Górna M.W., Superti-Furga G., Nagar B.. Structural basis for viral 5′-PPP-RNA recognition by human IFIT proteins. Nature. 2013; 494:60–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kumar P., Sweeney T.R., Skabkin M.A., Skabkina O.V., Hellen C.U.T., Pestova T.V.. Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′-terminal regions of cap0-, cap1- and 5′ppp- mRNAs. Nucleic Acids Res. 2014; 42:3228–3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson B., VanBlargan L.A., Xu W., White J.P., Shan C., Shi P.-Y., Zhang R., Adhikari J., Gross M.L., Leung D.W.et al.. Human IFIT3 modulates IFIT1 RNA binding specificity and protein stability. Immunity. 2018; 48:487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shuman S., Hurwitz J.. Mechanism of mRNA capping by vaccinia virus guanylyltransferase: characterization of an enzyme–guanylate intermediate. Proc. Natl. Acad. Sci. U.S.A. 1981; 78:187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Egloff M.-P., Benarroch D., Selisko B., Romette J.-L., Canard B.. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002; 21:2757–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G.et al.. Crystal structure and functional analysis of the SARS-Coronavirus RNA cap 2′-O-Methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011; 7:e1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paesen G.C., Collet A., Sallamand C., Debart F., Vasseur J.-J., Canard B., Decroly E., Grimes J.M.. X-ray structure and activities of an essential mononegavirales L-protein domain. Nat. Commun. 2015; 6:8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Valle C., Martin B., Ferron F., Roig-Zamboni V., Desmyter A., Debart F., Vasseur J.-J., Canard B., Coutard B., Decroly E.. First insights into the structural features of ebola virus methyltransferase activities. Nucleic Acids Res. 2021; 49:1737–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barral K., Sallamand C., Petzold C., Coutard B., Collet A., Thillier Y., Zimmermann J., Vasseur J.-J., Canard B., Rohayem J.et al.. Development of specific dengue virus 2′-O- and N7-methyltransferase assays for antiviral drug screening. Antiviral Res. 2013; 99:292–300. [DOI] [PubMed] [Google Scholar]
- 21. Coutard B., Barral K., Lichière J., Selisko B., Martin B., Aouadi W., Lombardia M.O., Debart F., Vasseur J.-J., Guillemot J.C.et al.. Zika virus methyltransferase: structure and functions for drug design perspectives. J. Virol. 2017; 91:e02202-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dong H., Chang D.C., Hua M.H.C., Lim S.P., Chionh Y.H., Hia F., Lee Y.H., Kukkaro P., Lok S.-M., Dedon P.C.et al.. 2′-O Methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog. 2012; 8:e1002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martin B., Coutard B., Guez T., Paesen G.C., Canard B., Debart F., Vasseur J.-J., Grimes J.M., Decroly E.. The methyltransferase domain of the sudan ebolavirus l protein specifically targets internal adenosines of RNA substrates, in addition to the cap structure. Nucleic Acids Res. 2018; 46:7902–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ayadi L., Motorin Y., Marchand V.. Quantification of 2′-O-Me residues in RNA using next-generation sequencing (Illumina ribomethseq protocol). Methods Mol. Biol. Clifton NJ. 2018; 1649:29–48. [DOI] [PubMed] [Google Scholar]
- 25. Yang S.L., DeFalco L., Anderson D.E., Zhang Y., Aw J.G.A., Lim S.Y., Lim X.N., Tan K.Y., Zhang T., Chawla T.et al.. Comprehensive mapping of SARS-CoV-2 interactions in vivo reveals functional virus-host interactions. Nat. Commun. 2021; 12:5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ringeard M., Marchand V., Decroly E., Motorin Y., Bennasser Y.. FTSJ3 is an RNA 2′-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature. 2019; 565:500–504. [DOI] [PubMed] [Google Scholar]
- 27. Wu B., Peisley A., Richards C., Yao H., Zeng X., Lin C., Chu F., Walz T., Hur S.. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013; 152:276–289. [DOI] [PubMed] [Google Scholar]
- 28. Silverman R.H. Viral encounters with 2′,5′-Oligoadenylate synthetase and RNase l during the interferon antiviral response. J. Virol. 2007; 81:12720–12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Espert L., Degols G., Gongora C., Blondel D., Williams B.R., Silverman R.H., Mechti N.. ISG20, a new interferon-induced RNase specific for single-stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. J. Biol. Chem. 2003; 278:16151–16158. [DOI] [PubMed] [Google Scholar]
- 30. Weiss C.M., Trobaugh D.W., Sun C., Lucas T.M., Diamond M.S., Ryman K.D., Klimstra W.B.. The interferon-induced exonuclease ISG20 exerts antiviral activity through upregulation of type i interferon response proteins. Msphere. 2018; 3:e00209-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jiang D., Weidner J.M., Qing M., Pan X.-B., Guo H., Xu C., Zhang X., Birk A., Chang J., Shi P.-Y.et al.. Identification of five interferon-induced cellular proteins that inhibit west nile virus and dengue virus infections. J. Virol. 2010; 84:8332–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu H., Lei Y., Zhong S., Peng F.-Y., Zhou Z., Li K., Ren H.. Antiviral activities of ISG20 against hepatitis C virus. Chin. J. Hepatol. 2013; 21:33–37. [DOI] [PubMed] [Google Scholar]
- 33. Espert L., Degols G., Lin Y.-L., Vincent T., Benkirane M., Mechti N.. Interferon-induced exonuclease ISG20 exhibits an antiviral activity against human immunodeficiency virus type 1. J. Gen. Virol. 2005; 86:2221–2229. [DOI] [PubMed] [Google Scholar]
- 34. Espert L., Eldin P., Gongora C., Bayard B., Harper F., Chelbi-Alix M.K., Bertrand E., Degols G., Mechti N.. The exonuclease ISG20 mainly localizes in the nucleolus and the cajal (Coiled) bodies and is associated with nuclear SMN protein-containing complexes. J. Cell. Biochem. 2006; 98:1320–1333. [DOI] [PubMed] [Google Scholar]
- 35. Horio T., Murai M., Inoue T., Hamasaki T., Tanaka T., Ohgi T.. Crystal structure of human ISG20, an interferon-induced antiviral ribonuclease. FEBS Lett. 2004; 577:111–116. [DOI] [PubMed] [Google Scholar]
- 36. Nguyen L.H., Espert L., Mechti N., Wilson D.M.. The human Interferon- and Estrogen-Regulated ISG20/HEM45 gene product degrades single-stranded RNA and DNA in vitro. Biochemistry. 2001; 40:7174–7179. [DOI] [PubMed] [Google Scholar]
- 37. Mauer J., Luo X., Blanjoie A., Jiao X., Grozhik A.V., Patil D.P., Linder B., Pickering B.F., Vasseur J.-J., Chen Q.et al.. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. 2017; 541:371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 39. Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E.. UCSF chimerax: structure visualization for researchers, educators, and developers. Protein Sci. Publ. Protein Soc. 2021; 30:70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Amara A., Gall S.L., Schwartz O., Salamero J., Montes M., Loetscher P., Baggiolini M., Virelizier J.-L., Arenzana-Seisdedos F.. HIV coreceptor downregulation as antiviral principle: SDF-1α–dependent internalization of the chemokine receptor CXCR4 contributes to inhibition of HIV replication. J. Exp. Med. 1997; 186:139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. He J., Choe S., Walker R., Di Marzio P., Morgan D.O., Landau N.R.. Human immunodeficiency virus type 1 viral protein r (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 1995; 69:6705–6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen J., Liu L., You C., Gu J., Ruan W., Zhang L., Gan J., Cao C., Huang Y., Chen X.et al.. Structural and biochemical insights into small RNA 3′ end trimming by arabidopsis SDN1. Nat. Commun. 2018; 9:3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang T., Sun H.-L., Cheng F., Zhang X.-E., Bi L., Jiang T.. Recognition and processing of double-stranded DNA by ExoX, a distributive 3′–5′ exonuclease. Nucleic Acids Res. 2013; 41:7556–7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Deymier S., Louvat C., Fiorini F., Cimarelli A.. ISG20: an enigmatic antiviral RNase targeting multiple viruses. FEBS Open Bio. 2022; 1096–1111.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu N., Nguyen X.-N., Wang L., Appourchaux R., Zhang C., Panthu B., Gruffat H., Journo C., Alais S., Qin J.et al.. The interferon stimulated gene 20 protein (ISG20) is an innate defense antiviral factor that discriminates self versus non-self translation. PLoS Pathog. 2019; 15:e1008093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Imam H., Kim G.-W., Mir S.A., Khan M., Siddiqui A.. Interferon-stimulated gene 20 (ISG20) selectively degrades N6-methyladenosine modified hepatitis b virus transcripts. PLoS Pathog. 2020; 16:e1008338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Deffrasnes C., Marsh G.A., Foo C.H., Rootes C.L., Gould C.M., Grusovin J., Monaghan P., Lo M.K., Tompkins S.M., Adams T.E.et al.. Genome-wide siRNA screening at biosafety level 4 reveals a crucial role for fibrillarin in henipavirus infection. PLoS Pathog. 2016; 12:e1005478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wickenhagen A., Sugrue E., Lytras S., Kuchi S., Noerenberg M., Turnbull M.L., Loney C., Herder V., Allan J., Jarmson I.et al.. A prenylated dsRNA sensor protects against severe COVID-19. Science. 2021; 374:eabj3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hammoudeh S.M., Hammoudeh A.M., Bhamidimarri P.M., Al Safar H., Mahboub B., Künstner A., Busch H., Halwani R., Hamid Q., Rahmani M.et al.. Systems immunology analysis reveals the contribution of pulmonary and extrapulmonary tissues to the immunopathogenesis of severe COVID-19 patients. Front. Immunol. 2021; 12:2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Flynn R.A., Belk J.A., Qi Y., Yasumoto Y., Wei J., Alfajaro M.M., Shi Q., Mumbach M.R., Limaye A., DeWeirdt P.C.et al.. Discovery and functional interrogation of SARS-CoV-2 RNA-host protein interactions. Cell. 2021; 184:2394–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Barros-Silva D., Klavert J., Jenster G., Jerónimo C., Lafontaine D.L.J., Martens-Uzunova E.S.. The role of oncosnornas and ribosomal RNA 2′-O-methylation in cancer. RNA Biol. 2021; 18:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tomkuvienė M., Ličytė J., Olendraitė I., Liutkevičiūtė Z., Clouet-d’Orval B., Klimašauskas S.. Archaeal fibrillarin-Nop5 heterodimer 2′-O-methylates RNA independently of the C/D guide RNP particle. RNA. 2017; 23:1329–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Massenet S., Bertrand E., Verheggen C.. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 2017; 14:680–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Martin S.A., Paoletti E., Moss B.. Purification of mRNA guanylyltransferase and mRNA (guanine-7-) methyltransferase from vaccinia virions. J. Biol. Chem. 1975; 250:9322–9329. [PubMed] [Google Scholar]
- 55. Stepinski J., Waddell C., Stolarski R., Darzynkiewicz E., Rhoads R.E.. Synthesis and properties of mRNAs containing the novel ‘anti-reverse’ cap analogs 7-methyl(3′-O-methyl)GpppG and 7-methyl (3′-deoxy)GpppG. RNA. 2001; 7:1486–1495. [PMC free article] [PubMed] [Google Scholar]
- 56. Jaïs P.H., Decroly E., Jacquet E., Le Boulch M., Jaïs A., Jean-Jean O., Eaton H., Ponien P., Verdier F., Canard B.et al.. C3P3-G1: first generation of a eukaryotic artificial cytoplasmic expression system. Nucleic Acids Res. 2019; 47:2681–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ross J., Sullivan T.D.. Half-lives of beta and gamma globin messenger RNAs and of protein synthetic capacity in cultured human reticulocytes. Blood. 1985; 66:1149–1154. [PubMed] [Google Scholar]
- 58. Gallie D.R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991; 5:2108–2116. [DOI] [PubMed] [Google Scholar]
- 59. Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D.. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. J. Am. Soc. Gene Ther. 2008; 16:1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Karikó K., Buckstein M., Ni H., Weissman D.. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005; 23:165–175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.