Abstract
The epigenetic landscape of oligodendrocyte lineage cells refers to the cell-specific modifications of DNA, chromatin, and RNA that define a unique gene expression pattern of functionally specialized cells. Here, we focus on the epigenetic changes occurring as progenitors differentiate into myelin-forming cells and respond to the local environment. First, modifications of DNA, RNA, nucleosomal histones, key principles of chromatin organization, topologically associating domains, and local remodeling will be reviewed. Then, the relationship between epigenetic modulators and RNA processing will be explored. Finally, the reciprocal relationship between the epigenome as a determinant of the mechanical properties of cell nuclei and the target of mechanotransduction will be discussed. The overall goal is to provide an interpretative key on how epigenetic changes may account for the heterogeneity of the transcriptional profiles identified in this lineage.
Keywords: brain, chromatin, development, histone, myelin, transcription
INTRODUCTION
While all the cells of an organism share the same genetic information, functional specialization is achieved by the establishment of a cell-specific transcriptional program. The spatial organization of the genome within nuclear territories, its relationship with the nuclear lamina, together with modifications of nucleic acids and histone proteins, and the state of compaction of chromatin, collectively define the epigenomic landscape of a cell. Parts of the genome containing genes not compatible with specialized cell functions are rendered inaccessible, while other regions regulating functional specificity are made accessible and become either poised for transcription or transcriptionally competent.
In the nuclei of eukaryotic cells, the DNA is wrapped around protein octamers called histones, which define the nucleosome, the basic unit of chromatin. The amino acids in the N terminal tails of these proteins undergo several post-translational modifications, including lysine acetylation, methylation or ubiquitination, serine phosphorylation, and arginine methylation or citrullination. These histone marks result from the concerted actions of enzymatic histone writers (responsible for the deposition of the modification) and erasers (responsible for the removal of the modification). The modified amino acids are recognized by specialized reader proteins via specific recognition domains, including bromodomains that recognize lysine acetylation or chromo-domains that recognize methylated residues. DNA can also be modified by the addition of a methyl group at the 5′ cytosine residue (5mC), resulting in transcriptional repression, or by the oxidation of the methyl group (5hmC), leading to hydroxymethylation, associated with transcriptional activation. The dynamic regulation of histone and DNA modifications is associated with distinct transcriptional states allowing for transcriptionally competent euchromatin or repressed heterochromatin. Eukaryotic cell nuclei also possess higher-order organization in large domains of repressed or active chromatin within specific topological domains.
DNA methylation masking transcription factor recognition sites, repressive histone modifications, chromatin compaction, and recruitment to the nuclear lamina act by hindering the access of transcription factors to DNA, reducing or diminishing the ability of cells to respond to external cues (Figure 1). Gene-poor or -silenced genomic regions are recruited to the nuclear periphery where they interact within the nuclear lamina, forming lamina-associated domains (LADs),1,2 while gene-rich transcriptionally active regions tend to arrange around transcription factors to form transcriptional “factories” and cluster into topologically associating domains (TADs)3–5 (Figure 2). While these domains and associated modifications are generally stable, they also have the potential to be reversed, and this implies that the epigenetic landscape is not static, but allows for adaptation to external conditions.
FIGURE 1.
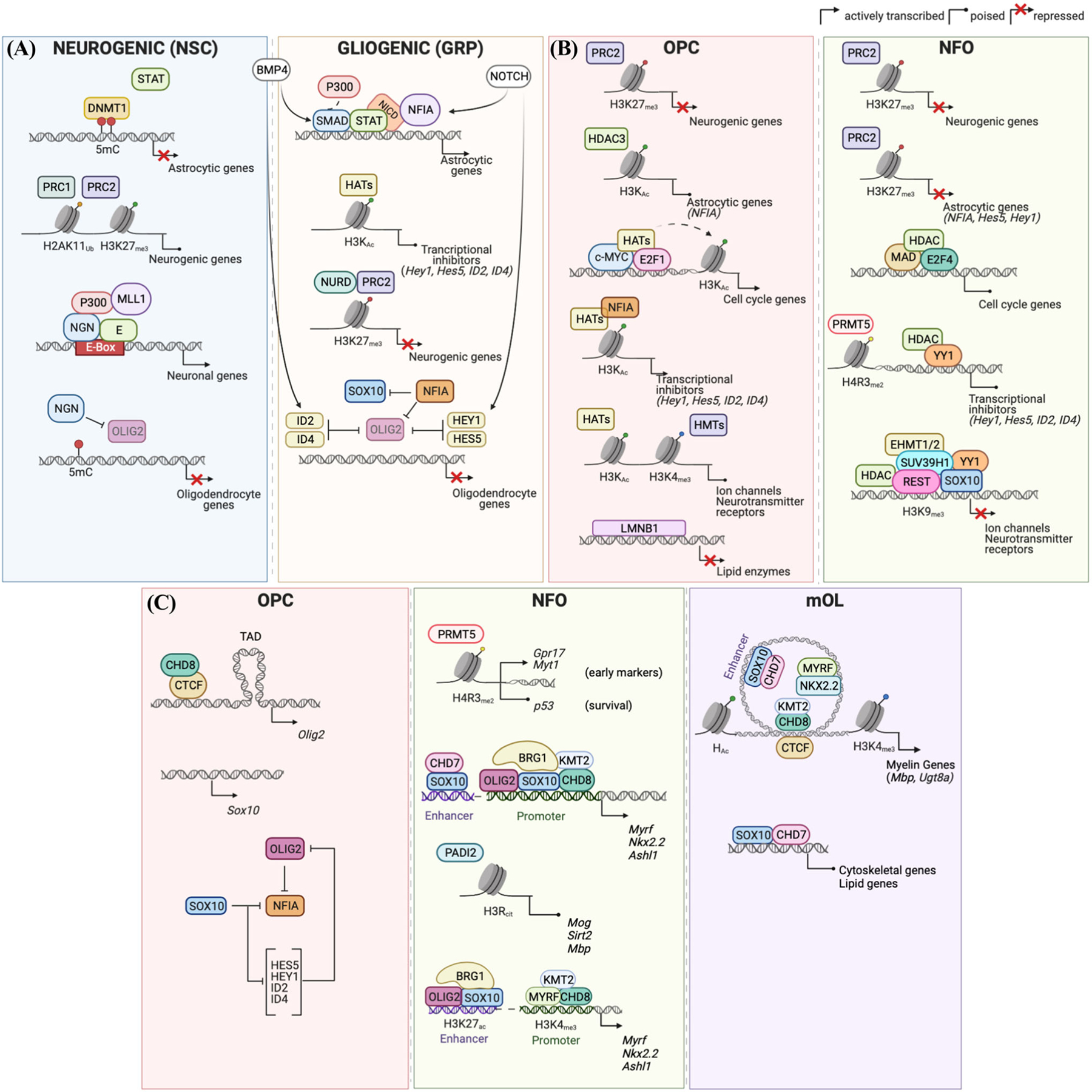
Regulation of gene expression during the major transitions from neural stem cell to oligodendrocyte. Histone and DNA modifications and relative enzymatic activities regulating the expression of the indicated gene categories during the transition from neurogenic to gliogenic stem cell (panel A). Repressive histone modifications and transcriptional inhibitors responsible for the maintenance of the undifferentiated, proliferative, and electrically active state of oligodendrocyte progenitor cells (OPCs) and for the transition to newly formed oligodendrocyte (NFO) (panel B). Schematic representation of the histone modifications and chromatin remodeling events regulating the differentiation from progenitor (OPC) to newly formed oligodendrocyte (NFO) to mature oligodendrocyte (mOL) (panel C). Created with BioRender.
FIGURE 2.
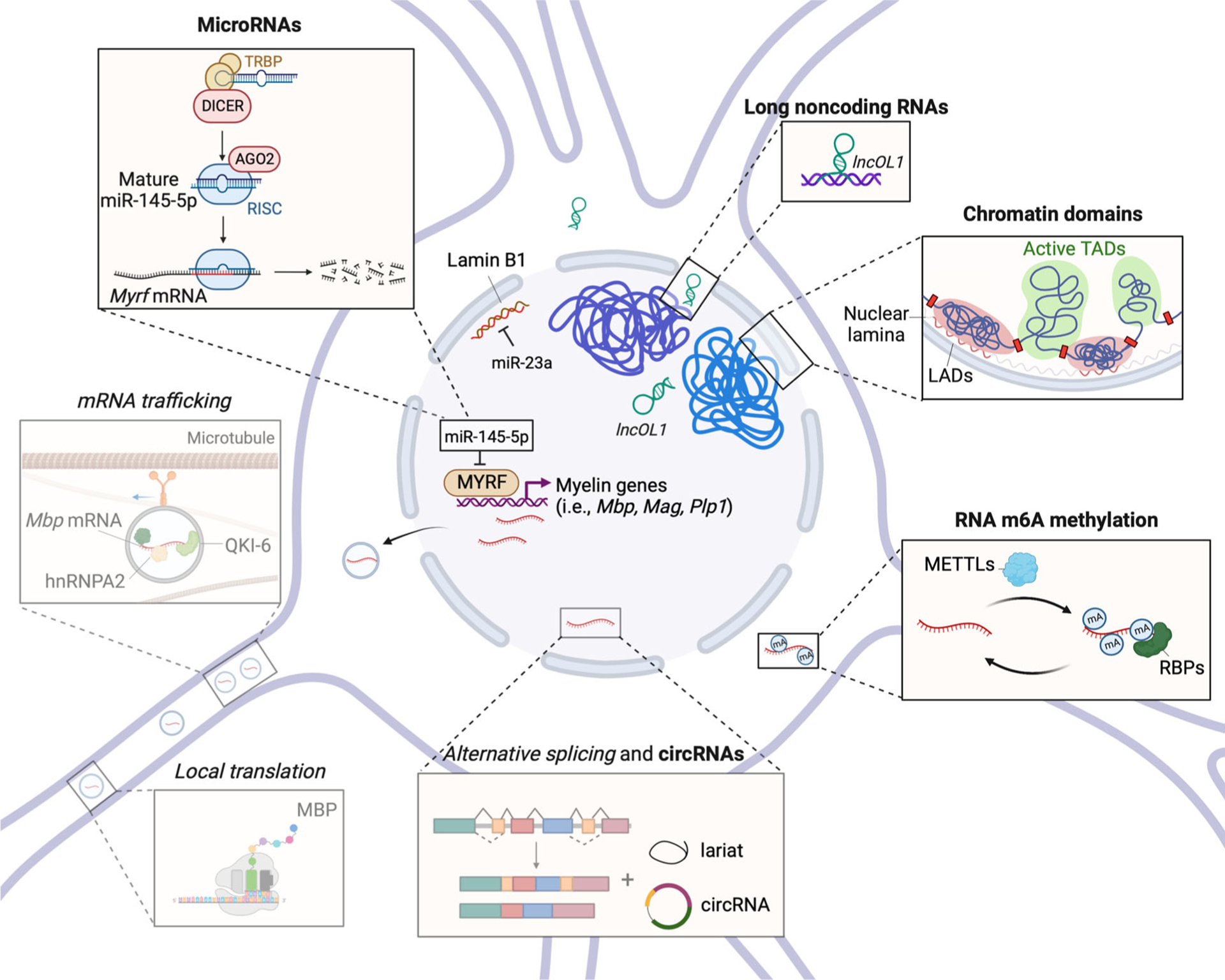
The epigenomic landscape of an oligodendrocyte. Depicted here is a schematic representation of the 3D nuclear organization of the genome, including lamina-associated domain (LAD) and topologically associating domain (TAD), which together with histone and DNA modifications and chromatin remodelers, define the cell-specific unique epigenomic landscape. RNA processing, modification, and trafficking are shown in shaded gray boxes. Epigenetic regulation of gene expression by microRNA, long noncoding RNA, and circRNAs is shown in separate boxes. Created with BioRender. Abbreviations: circRNA, circular RNA; mRNA, messenger RNA.
Here, we will first discuss some generalities on mechanisms of repression and activation, then address the role of histone and DNA modifications during oligodendrocyte lineage progression, followed by a discussion of their crosstalk with mechanisms of RNA processing and we conclude with a review of the role of nuclear chromatin and lamina in determining the mechanical properties of the nuclei and also as a point of convergence of physical forces in the regulation of gene expression.
General concepts of epigenetic regulation of gene expression
Acetylation of lysine residues in the histone tails is considered a modification permissive for transcription, as it neutralizes the positive charge of the amino acid, loosens the interaction with negatively charged DNA, and facilitates the access of transcription factors. This modification is modulated by the opposing actions of writer histone acetyltransferases (HATs) and eraser histone deacetylases (HDACs) and is, therefore, suitable for the dynamic regulation of genes.6,7 Histone methylation is a more complex event, catalyzed by enzymes that transfer one or more methyl groups on specific lysine or arginine residues.8 The transcriptional outcome is dependent on the number of methyl groups added to lysine residues (mono-, di-, or tri-methylation), as well as the position of a given lysine residue within the histone tails. For instance, lysine K4 in histone H3 is associated with transcriptional activation. In its monomethylated state (H3K4me1), this modification is found in active enhancers, while its trimethylated state (H3K4me3) marks transcriptionally active promoters.9,10 This modification is placed by the enzymatic writers MLL1/KMT2A and MLL4/KMT2B and reversed by the enzymatic erasers KDM5A/JARID1B and KDM5B/JARID1A. Dimethylation of arginine residues, catalyzed by protein methyl transferases PRMTs, has been shown to play an important role in transcriptional activation or repression, depending on the position of the arginine residue and whether the deposition of the methyl groups is symmetrically or asymmetrically placed.11 While specific erasers for arginine methylated marks have not been identified, the activity of deiminase enzymes (PADI) responsible for the conversion of arginine into citrulline has been proposed to reverse the transcriptional consequences of arginine methylation on histones.12
The trimethylation of K9 and K27 residues in histone H3 results in transcriptional repression. The enzymes responsible for H3K27me3 deposition are part of the Polycomb group PRC2 (e.g., EZH1 and EZH2), which deposit the mark at transcriptionally inactive promoters.13 Recognition of the H3K27me3 mark by distinct chromo-domain binding proteins can lead to poised expression or stable repression, depending on whether the recruited PRC1 complexes (RING1A and B) induce ubiquitination of specific lysine residues in histone H2A (and result in poised expression of genes) or recruit NURD repressive complexes and lead to heterochromatin formation.14 Thus, even though erasers for the H3K27me3 mark exist (i.e., UTX/KDM6A and JMJD3/KDM6B), the activity of the PRC2 complex results in long-term repression due to chromatin compaction.
H3K9me3 marks are deposited by specific histone writers (e.g., SUV39H1/2) and removed by erasers, such as KDM3A/JMJD1 and KDM4/JMJD2. Both writers and erasers are highly expressed in the oligodendrocyte lineage,15–17 thereby suggesting that this modification may play an essential role in regulating transient repression. In addition, H3K9me3 marks have been shown to be recognized by specific readers (such as HP1-α)18 and contribute to chromatin compaction and recruitment to the nuclear periphery.
DNA methylation catalyzed by methyltransferases (DNMTs) plays an essential role in many biological processes, including transcription and cellular differentiation. DNA methylation preferentially occurs at exon-boundaries, characterized by high CpG density.19,20 It has been shown that DNA methylation cooperates with other histone modifications to further stabilize repression. For instance, the 5mC mark is recognized by a family of proteins carrying conserved methyl-CpG binding domains (e.g., MECP2), which recruits other histone modifiers and further alters the local chromatin conformation.21–23 Importantly, MECP2 was reported to also regulate alternative exon splicing by recruiting HDACs and altering the kinetics of RNA pol II elongation.24 A relationship between DNA methylation and splicing was also reported in the oligodendrocyte lineage,25 as cell-specific deletion of Dnmt1 in progenitors impaired the alternative splicing of several transcripts regulating lipid metabolism, myelination, and cell cycle.25
While DNA methylation at promoters has been associated with transcriptional repression, and a true demethylase has not been identified, it has been shown that methylated cytosines are targeted by enzymes called ten-eleven translocases (TETs) and converted to hydroxymethyl cytosines which can be oxidized to formyl cytosine and then carboxyl cytosine, which is eventually excised by the DNA repair complex.26
Gene silencing is further stabilized by establishing contacts between heterochromatin and the nuclear lamina. These genomic regions forming stable interactions with the nuclear lamina have been identified as LADs.27,28 The nuclear lamina in oligodendrocyte lineage cells consists of intermediate filaments called lamins. Lamin A and lamin C are expressed in mature oligodendrocytes, and lamin B1 is expressed in oligodendrocyte progenitor cells (OPCs). These proteins can directly bind to chromatin via specific interactors and also associate with the cytoskeleton through the linker of nucleoskeleton and cytoskeleton (LINC) protein complex (see below), thereby enabling the direct transduction of physical forces from the exterior of the cell to its genome.29
HISTONE AND DNA MODIFICATIONS RESPONSIBLE FOR THE ACQUISITION OF THE OLIGODENDROCYTE LINEAGE IDENTITY
Although neural stem cells (NSCs) are multipotential and able to generate neurons and glia within the developing nervous system, they are neurogenic at mid-gestation and gliogenic at late gestation. This section aims to review the mechanisms regulating the switch from neurogenic to gliogenic transcriptional competence of NSCs and then delve into the transcriptional and epigenetic mechanisms responsible for the transition from oligodendrocyte progenitors to oligodendrocytes.
Epigenetic landscape of NSCs during the neurogenic to gliogenic transition
The neurogenic to gliogenic switch of NSC during embryonic development, despite the exposure to the same external factors, implies the involvement of epigenetic mechanisms regulating their responsiveness to these external cues. At least two mechanisms have been described: DNA methylation and histone post-translational modifications (Figure 1A). DNA methylation has been shown to prevent the binding of transcription factors to astrocytic genes during the neurogenic phase by altering their recognition site and keeping astrocytic genes repressed.30,31 At the same time, neurogenic genes are poised for transcription due to the coexistence of the transcriptionally competent histone H3K27 acetylation and repressive H2AK119 ubiquitination, mediated by the recruitment of the PRC1 complex (RING1B) to H3K27me3 marks at specific genomic sites.32
The switch to the gliogenic phase is characterized by the activation of astrocytic genes and stable ubiquitin-independent repression of neurogenic genes (Figure 1A). Late gestation is characterized by the expression of the transcription factor NFIA,33 previously identified as a critical astrogliogenic factor,34,35 which releases the DNA methylation block by dislodging the DNA methyltransferase DNMT1 and allows transcription factors like STATs and SMADs to recruit HATs and activate astrocytic genes.36 Consistent with this mechanism, mice with genetic ablation of Dnmt1 in NSC30,31 and those with forced expression of NFIA37 are characterized by precocious astrogenesis. Neurogenic genes lose the H3K27 acetylation and become stably repressed due to the concerted activity of the PRC2 complex, which imposes the repressive H3K27me3 mark and the PRC1 complex, containing PHC2, which promotes clustering and further recruits repressive NURD complexes containing HDAC1 and other repressors, ultimately effecting heterochromatin formation.32 In support of the importance of the PRC2 complex in lineage commitment, oligodendrocyte-lineage cells lacking PRC2 components, such as EZH2 or EED, were characterized by a gene expression pattern reminiscent of the astrocytic phenotype and severe hypomyelination.38,39 The decision of NSC and glial-restricted precursors to become either astrocytes or oligodendrocytes occurs in late embryogenesis when the epigenomic landscape organizes the cell-specific pattern of gene expression in response to external cues. The presence of Notch or BMP, for instance, promotes an astrocytic pattern of gene expression, whereas noggin or Sonic hedgehog favors the establishment of the oligodendrocytic transcriptome.40 Therefore, late embryonic NSCs are characterized by a remarkable susceptibility to external signals due to a receptive epigenomic configuration, underlying the importance of both extracellular and intracellular contexts for attaining specific developmental outcomes.41–43
Epigenetic landscape of OPCs: Repression of neurogenic genes
OPCs are electrically active cells44–47 that receive direct synaptic inputs from excitatory and inhibitory neurons48–51 and respond to neuronal activity with proliferation52–54 or differentiation.55,56 It is, therefore, not surprising that they retain the expression of several neurotransmitter receptors and ion channels, characteristically defined as neuronal genes. Despite the expression of these neuronal genes and initial reports that neurons can be generated from OPC in specific conditions57 and in selective brain regions,58 it is now clear that oligodendrocyte progenitors are not neurogenic59 as neurogenic genes are silenced in these cells, as described above (Figure 1B). The concept that OPCs behave as multipotential cells with the ability to generate neurons was suggested by initial in vitro studies, with cells cultured in the presence of growth factors60 or HDAC inhibitors.57 However, with the exception of a few reports in selected brain regions,58 in vivo fate mapping studies did not support OPCs as neurogenic cells.61–63 Consistent with the inability of OPCs to form neurons in vivo, their epigenomic landscape is characterized by the presence of widespread PRC2-dependent repressive H3K27me3 marks on neurogenic genes, which remain unaltered throughout the process of differentiation.64 The presence of these marks on neurogenic genes is also in agreement with the previously reported presence of the PRC2 enzymatic complex EZH2 on genes promoting neuronal fate, during the transition from embryonic stem cells to OPCs.65,66 Nevertheless, a more recent study showed that OLIG2+ cells lacking Ezh2 expression can still generate oligodendrocyte precursors, indicating that the absence of PRC2 activity per se is not sufficient to transition into a neuronal fate.38 Thus, the loss of neurogenic potential of OPCs is consistent with the silencing of neurogenic genes during embryonic development, using a mechanism of repression involving both PRC2-dependent histone methylation and PRC1-dependent recruitment of HDACs.67 In specific regions of the central nervous system (e.g., ventral areas of the neural tube), however, OPCs share the same precursors with interneurons.68 Due to this common origin, it is not surprising that their epigenome reflects this shared origin of interneuron-specific genes in OPCs that are mostly characterized by unmodified histones, which render them responsive to direct reprogramming into neuronal cells upon transfection of neurogenic transcription factors.69
As OPCs differentiate into oligodendrocytes, they progressively lose synaptic contacts and become electrically inert,49 a process which is accompanied by the downregulation of a large number of broadly defined neuronal genes, including neurotransmitter receptors and ion channels25,64 (Figure 1B).
Epigenetic landscape of OPCs: Repression of astrocytic genes
The prolonged exposure of cultured OPCs to morphogenic factors and mitogens70 or HDAC inhibitors60 favored the expression of astrocytic and neuronal genes in these cells. However, in vivo, only some OPCs in the ventral gray matter can generate astrocytes during the embryonic period.63 This suggests that the epigenomic landscape of cultured OPCs is different from that of brain OPCs, whose lineage becomes progressively more restricted over time. DNA methylation is not used as one of the mechanisms for lineage restriction, as the ablation of Dnmt1 in OPCs does not result in lineage switch or even misexpression of astrocytic or neuronal genes.25 Rather, it is accompanied by dysregulation of genes regulating cell division, subsequent defective proliferation, downregulation of transcription factors important for oligodendrogliogenesis (e.g., Ascl1, Sox10, and Myrf), and defective RNA splicing events (related to exon skipping) ultimately leading to decreased survival.25 In contrast, ablation of the enzymes responsible for PRC2-dependent repression in OPCs resulted in the upregulation of Notch pathway-related genes, including the astrogliogenic factor NFIA,38 and increased astrogliogenesis. Since the astrogliogenic function of NFIA in OPCs is antagonized by OLIG2,34,71 it is not surprising that increased generation of protoplasmic astrocytes in the dorsal forebrain during embryonic development could also be detected after deletion of Olig2 in OPCs. Importantly, the ability of OPCs to form astrocytes was not stable over time, but rather it became less effective with aging.72 NFIA astrogliogenic function in OPCs is also antagonized by SOX1035,71 and HDAC3 activity.73 In glial progenitor cells, NFIA cooperates with SOX9 to generate astrocytes.71,73,74 As OPCs become committed to the oligodendrocyte lineage, the increasing levels of SOX10 can act by directly antagonizing NFIA (Figure 1B) and indirectly upregulating the levels of the microRNA miR-338 reduces SOX9 levels.75 The genetic deletion of Sox10 also increases the generation of astrocytes at the expense of oligodendrocytes.71 Ablation of Hdac3 induces a very similar phenotype to that of Olig2 deletion,73 thereby suggesting the cooperation between transcription factors and epigenomic modulators in regulating the oligodendrocytic/astrocytic switch.
From NSC to oligodendrocyte lineage cells and back: The role of pioneer transcription factors
It is important to clarify here that progressive lineage restriction occurs during normal development and requires the establishment of repressive DNA methylation and chromatin compaction, as delineated above. However, although the transcriptional program defining cell identity is stable, it is possible, in specific circumstances, to reactivate silent genes by altering the epigenomic landscape. The discovery that somatic cells could be reprogrammed into pluripotent stem cells by the transfection of four transcription factors (e.g., OCT4, SOX2, KLF4, and MYC) suggested that their expression was sufficient to erase the epigenetic landscape responsible for cell specialization.76 A deeperunderstanding of this mechanism of action led to the concept of pioneer transcription factors. These proteins are characterized by the ability to bind only to partial transcription factor recognition motifs displayed on the surface of nucleosomes, and either alone or in tandem, they are capable of affecting the epigenetic landscape by inducing nucleosomal remodeling independent of ATP or affecting DNA methylation.77 Thus, specific transcription factors have the dual ability to both establish or dismantle the epigenetic landscape.
In an attempt to define the key transcription factors responsible for the transition from NSCs to OPCs, an elegant study78 focused on three transcription factors, differentially expressed between human NSCs (already expressing OLIG2) and OPCs (e.g., ASCL1, SOX10, and NKX2.2) and analyzed the transcriptional profile induced in NSCs by overexpressing each of them separately and in combination. While the combined expression led to oligodendrocyte lineage cell generation, the expression of ASCL1 induced both oligodendrocyte and neuronal genes.78 This result is consistent with the role of ASCL1 as a pioneering transcription factor with the ability to reactivate neuronal genes even within the context of a repressive chromatin state, and also explains previous studies on its role as both neurogenic and oligodendrogliogenic factors.79–81
Intriguingly, the expression of SOX10 alone was sufficient to induce a gene expression pattern that resembled that of primary human OPCs, in terms of repressed and activated genes.78 Since SOX10 is an HMG transcription factor and a member of the SOX family, it is conceivable that, like SOX2, it may play the role of “pioneer” in opening genes required for myelin production within the heterochromatic nuclei of differentiating OPCs. Consistent with its role as a pioneer transcription factor, a recent study reported that expression of OLIG2, ASCL1, SOX10, and NKX2.2 is sufficient to reprogram human dermal fibroblasts into oligodendrocyte lineage cells.82
Epigenetic landscape of differentiating oligodendrocyte progenitors: Repressive events regulating cell cycle, electrical properties, and transcriptional inhibitors of myelin genes
The transition from OPCs to mature oligodendrocytes is characterized by a series of functional changes. This section will summarize the current knowledge of the changes in the epigenetic landscape occurring during this transition (Figure 1B,C). They include (1) exit from the cell cycle; (2) loss of migratory capacity associated with morphological changes (from bipolar to multiprocess bearing cell); (3) loss of electrical responsiveness to neuronal stimulation; and (4) generation of large quantities of lipid and specialized proteins to be assembled into a membrane structure called myelin. These processes are not concurrent but rather sequential, with the early repressive events referring to regulatory RNA processes and downregulation of genes encoding for positive regulators of proliferation and negative regulators of myelin gene expression.83
We have previously discussed how OPCs are electrically active cells with the ability to respond to neurotransmitters by activating ionotropic receptors and voltage-gated ion channels.44,49,84,85 They receive synaptic neuronal contacts and are proliferative, migratory, electrically active cells whose genomic distribution of the repressive methylation mark H3K27me3 is localized on neurogenic genes and not present on genes regulating the expression of neurotransmitter receptors, ion channels, and synaptic transmission.64 However, as the OPCs start to differentiate into oligodendrocytes, eventually forming myelin to provide axonal insulation, it is of paramount importance that genes regulating their electrical properties are silenced.25,49 This occurs by the deposition of the repressive histone mark H3K9me3 at genomic loci responsible for the expression of ion channels and pathways related to synaptic transmission.64 Downregulation of the enzymatic writer for H3K9me3 (e.g., SUV39H1) during the differentiation process prevents the changes in the excitability of these cells.64 Thus, the OPCs adopt two distinct mechanisms of neuronal gene silencing: an early PRC2-dependent mechanism of repression of neurogenic genes and a later SUV39H1-dependent mechanism of inactivation of the electrical properties of the cells. H3K9me3 is also found on GABAergic genes as OPCs differentiate.64 The H3K9me3 mark is recognized by specific chromodomain binding proteins, such as HP1-α, a protein that we previously reported to be enriched in white matter tracts during developmental myelination.86 In nuclear domains characterized by transcriptionally silent chromatin, HP1-α has the ability to recruit genes to the nuclear periphery that are repressed due to this histone mark.
At the same time, additional mechanisms of transcriptional repression are in place to prevent the premature differentiation of OPCs into myelin-forming oligodendrocytes, as initially shown by our group first in cultured OPCs87 and then in developing and aging rodents86,88,89 and subsequently validated by several other groups.90–92 The initial discoveries identified reversible histone acetylation of specific lysine residues as histone marks in the nuclei of OPCs,86,87,89 which are favored by the presence of mitogens87 and differentially affected by astrogliogenic and oligodendrogliogenic signals.70 Additional crosstalk between epigenetic modifications, such as PRMT5-mediated symmetric arginine methylation and histone acetylation,93 and regulation of chromatin accessibility by arginine citrullination,94 further contribute to tighter regulation of this transcriptional network. The histone deacetylase HDAC1 was identified as the major enzyme responsible for the transcriptional repression occurring during the transition from OPCs to oligodendrocytes,86,95 and genetic approaches further validated this observation in mice and cultured cells.70,96
Repression occurs at genomic loci encoding for transcriptional repression of inhibitory bHLH transcription factors class V (e.g., ID2 and ID4) and class VI (e.g., HES5), which were shown to play a major role as inhibitors of myelin gene expression.97–100 Importantly, besides acetylation/deacetylation regulated by the opposing activity of HATs and HDACs, these genes were also shown to be regulated by PRMT5101 and complexes regulating the chromatin accessibility at their promoter region.102
It is worth noting that several of these genes, such as transcriptional inhibitors103 and cell cycle regulatory genes,25 are further silenced by additional mechanisms, including histone and DNA methylation and also recruitment to the nuclear lamina after being repressed by initial histone deacetylation.104
The relationship between cell cycle exit and differentiation in OPCs was shown to be tightly connected to the major transition in the epigenomic landscape, as transcription factors involved in cell cycle regulation, such as members of the E2F and MYC family, act as major transcriptional switches. For instance, E2F1 and MYC were shown to modulate the recruitment of HATs to genomic locations encoding for chromatin components and regulatory enzymes, as well as transcriptional inhibitors of differentiation.105,106
Conditions favoring histone acetylation at the expense of deacetylation prevent the differentiation of OPCs. Those conditions include treatment of OPCs with HDAC inhibitors,86,87 exposure to BMPs70 and genetic ablation of the histone arginine methyltransferase PRMT5,93 the histone deacetylase HDAC1,96 or one of the transcription factors responsible for their recruitment, such as YY1.98 Conversely, silencing E2F1106 or MYC,105 or blocking the activity of histone acetylation readers,107 are capable of inducing the progression of OPCs toward the first stages of oligodendrocyte differentiation but not sufficient to induce the myelinating terminally differentiated phenotype.
An additional mechanism preventing premature differentiation of OPCs into myelinating oligodendrocytes is the association of lipid metabolism genes to the nuclear lamina.104 Downregulation of LmnB1 as OPCs differentiate is guaranteed by miR23, and the inability to decrease the levels of this nuclear lamina protein results in nuclear structural defects, including the formation of atypical intranuclear membrane and decreased expression of myelin genes.108,109 Indeed, failure to decrease LMNB1 levels results in reduced Plp1 expression110 and overall decreased transcription of myelin genes.108,111 From a mechanistic perspective, the progressive decrease of LMNB1 during differentiation results in increased expression levels of genes encoding for key lipid enzymes, resulting from the release from the nuclear periphery.104 This also explained the dramatic dysmyelinating phenotype that was reported in transgenic mice overexpressing Lmnb1.112
Epigenetic landscape of oligodendrocyte progenitors differentiating into oligodendrocytes: Transcriptional activation of genes regulating cytoskeleton, lipids, and myelin proteins
The transcriptional signature of differentiated oligodendrocytes includes the activation of several genes necessary for the synthesis of myelin lipids and proteins as well as cytoskeletal remodelers necessary for axon wrapping, membrane extension, and transporters of ions and metabolites, which allow for the exchange of solutes at the neuro–glial interface. While the mechanisms of derepression defined above address the timing of myelin protein gene expression, a number of additional gene activation mechanisms need to occur to achieve the identity of myelinating oligodendrocytes. The program of activation includes the recruitment of ATP chromatin remodelers (such as BRG1) to enhancer regions containing SOX10 and OLIG2 binding sites in OPCs and to OLIG2 binding sites near the transcriptional start site for early differentiation genes (e.g., Cnp, Mbp, and Sirt2) and genes encoding for cytoskeletal elements in newly formed oligodendrocytes.113 The role of chromatin remodeling complexes at later stages of differentiation, however, appears less prominent, as indicated by the modest phenotype in mice with genetic ablation of Brg1 in differentiating oligodendrocytes.114 The binding of SOX10 to enhancer regions of genes related to the actin cytoskeleton and metabolic regulation115 is essential for the progression of OPCs to oligodendrocytes, and SOX10 acts as an orchestrator of both repressive64 and activating marks characterizing the epigenomic landscape as well as to aid in the recruitment of the chromodomain helicase DNA-binding (CHD) family of ATP-dependent chromatin remodelers. Among those, CHD8 was shown to open chromatin at promoter regions around the transcriptional start site and allow either for repression or activation of gene expression, the latter favored by the recruitment of the histone methyltransferase KMT2102 and responsible for the deposition of the activating H3K4me3 histone mark at the promoters of genes associated with oligodendrocyte differentiation (e.g., Olig1, Sox10, Nkx2.2, Tcf7l2, Myrf, Mbp, and Ugt8). CHD7, in contrast, was identified at later stages of differentiation to be recruited at SOX10 binding sites of genes modulating lipid homeostasis, cytoskeletal reorganization, myelin, and axonal ensheathment (e.g., Olig1, Nkx2.2, Myrf, and Sip1).116 An additional mechanism regulating chromatin accessibility in differentiated oligodendrocytes involves the removal of repressive arginine methylation mediated by PADI2, the enzyme responsible for converting arginine residues into citrulline at loci encoding for oligodendrocyte differentiation genes.94
Finally, DNA hydroxymethylation, mediated by TET enzymes, has been shown to induce the activation of genes regulating the crosstalk between myelinating oligodendrocytes and neurons. The global levels of brain hydroxymethylation increase during development and decline with age. This pattern correlates with decreased levels of the TET1, but not of the TET2 isoform in oligodendrocytes, thereby suggesting TET1 as the main enzyme responsible for DNA hydroxymethylation in the oligodendroglial lineage.117 The functional significance of TET1 was inferred by characterizing the phenotype of mice with lineage-specific ablation of this gene,117,118 which revealed important phenotypic similarities but also significant differences, possibly due to the different regions of targeted ablation. While mice with lineage-specific ablation of Tet1 catalytic domain (encoded by exons 11–13, Tet1Δ11–13) showed defects in developmental myelination, cell-cycle progression, OPC differentiation, and adult myelin repair,118 mice with cell-specific ablation of exon 4 (Tet1Δ4) did not display defective differentiation but were characterized by impaired adult repair of demyelinated lesions due to misexpression of genes regulating axo–glial interaction.117 These findings are intriguing, as they suggest that the N-terminal domain of TET1 might exert a role which is independent of the enzymatic activity. In this regard, it is noted that in other cell types, the N-terminal domain of TET1 has been shown to modulate the levels of repressive histone mark H3K27me3 and repress the expression of developmental genes.119,120 Future studies are needed to provide a better understanding of the role of the distinct TET family members at the distinct stages of the oligodendrocyte lineage.
EPIGENETIC REGULATION OF GENE EXPRESSION MEDIATED BY RNAs
As OPCs differentiate into mature oligodendrocytes, their functional specialization is guaranteed by the presence of a set of diverse proteins encoded by the same gene due to alternative splicing of the pre-mRNA, which allows for exons and introns to be either retained or skipped121 and thereby allowing the developmental stage-specific expression of the same protein with different functional domains.122,123 These RNAs can also be reversibly modified by specific enzymes (e.g., METTL3 and METTL14) and transported to distal sides of the cell to allow for local translation in response to external stimuli. It is clear that RNA processing plays a prominent role in the oligodendrocytes,83 and the detection of RNA splicing defects in mice with lineage-specific ablation of histone or DNA modifiers25,93 further highlights the existence of crosstalk between epigenetics and RNA processing. Additional modalities of epigenetic regulation of gene expression by RNAs include the study of methylation of adenosine residues in RNA, regulatory microRNAs (miRNA) (responsible for fine-tuning of transcript levels in response to external conditions), long noncoding RNAs (lncRNAs) (which may bind to chromatin reorganize genomic architecture), and circular RNAs (circRNAs)124 (Figure 2).
mRNA methylation
N6-Methyladenosine (m6A) mRNA methylation has recently been identified as a post-transcriptional reversible chemical modification affecting gene expression.125 This mark is deposited by a multiprotein methyltransferase complex composed of METTL3 and METTL14 enzymes.126,127 While METTL14 acts as a scaffold for RNA-binding, the biochemical reaction is catalyzed by METTL3.
The m6A methyl RNA mark can be recognized by several reader proteins, including members of the YT521-B homology (YTH) domain family (such as YTHDC1), IGF2 binding proteins (IGF2BPs), eukaryotic elongation factor (eIF3), HNRNPA2/B1,128,129 and PRRC2A.130 The latter is of special interest, as gene ontology enrichment analysis of PRRC2A targeting m6A-methylated mRNAs revealed transcripts regulating various biological functions related to brain development, gliogenesis, oligodendrocyte differentiation, and myelination. Among the transcripts bearing the m6A modification and bound by PRRC2A is the Olig2 transcript, suggesting that the m6A RNA/PRCC2A pathway may play an important role in the regulation of this important transcription factor for the OL. Consistently, neural-cell-specific deletion of Prrc2a resulted in downregulation of gliogenesis and myelination-related genes, as well as hypomyelination and cognitive dysfunction.130 In addition, the PRRC2A paralogue, PRRC2C, has also been identified as an oligodendrocyte-specific substrate of the protein arginine methyltransferase PRMT5.131
Erasers of the m6A mark on RNA are specific m6A demethylases, such as ALKBH5 (the ALKB homolog H5) and FTO (Fat mass and obesity-associated),132,133 the latter with the ability to recognize and remove the m6A mark from the Olig2 mRNA, leading to its degradation.130 Thus, m6A mRNA methylation can affect the mRNA stability and favor the interaction with RNA-binding proteins resulting in the formation of condensates or membrane-less compartments regulated by phase separation, such as stress granules.134–138m6A mRNA profiling in the oligodendrocyte lineage identified thousands of differentially modified mRNAs between OPCs and mature oligodendrocytes,137 with only 23 transcripts showing the same pattern of m6A methylation at both stages. Oligodendrocyte lineage-specific ablation of Mettl14 did not affect the progenitor state, or the translation or subcellular distribution of Mbp transcripts.137 However, it impaired their differentiation of OPC into oligodendrocytes with consequent hypomyelination accompanied by aberrant splicing of numerous transcripts, including Ptprz1 (protein tyrosine phosphate receptor type Z1) and Nfasc (neurofascin).137
MicroRNAs
MiRNAs are short, small noncoding RNAs (ncRNA) that negatively regulate their targets in a sequence-specific manner. miRNAs are derived from primary miRNAs (pri-miRNAs) cleaved by the nuclear Drosha complex into shorter precursor miRNAs (pre-miRNAs). Then, these molecules are translocated into the cytoplasm, where they are further processed into mature miRNAs by the endoribonuclease Dicer and incorporated into the RNA-induced silencing complex (RISC), which regulates their interactions with the mRNA targets.139 The RISC–miRNA complex recognizes a complementary sequence in the 3’UTR of mRNA, inhibiting its translation with or without mRNA degradation.140
Ablation of Dicer in mice—generated by in vivo recombination—demonstrated the critical role of miRNAs in mouse development. Lineage-specific deletion of Dicer1 in the oligodendrocyte lineage resulted in delayed oligodendrocyte differentiation and myelin production in vivo and in vitro and late neurodegeneration.106,107,141–143
Several miRNAs were identified as differentially regulated at distinct stages of oligodendrocyte development.141,143–147 The levels of miR-219, miR-338, miR-138, miR-29, and miR-23, for instance, were shown to increase during the differentiation of OPCs and into oligodendrocytes.141,143,144,148,149 These miRNAs were found to decrease the differentiation inhibitors’ levels further, thereby resulting in a prodifferentiative effect.142 For instance, miR-219 mRNA targets included mitogen receptors and transcriptional inhibitors (e.g., Pdgfrα, Sox6, and Hes5) as well as inhibitors of myelination (e.g., Lingo1 and Etv5).150 The overexpression of miR-219 and miR-338 in cultured primary OPCs lacking Dicer1 partially rescued oligodendrocyte differentiation by inducing myelin gene expression, including Mbp, Cnp, and Mog,141,143,150 and reducing the levels of Sox9.75 In contrast, miR-145–5p was shown to be downregulated during oligodendrocyte differentiation and exert an overall proliferative effect150. Downregulation of miR-145–5p (anti-145) promoted differentiation of cultured OPCs,151 possibly due to the release of the brake on the levels of Myrf (Figure 2), a critical transcription factor regulating oligodendrocyte maturation, and also a downstream target of miR-145–5p.151 Additional miRNAs of functional relevance to the oligodendrocyte lineage include miR-23a, a prodifferentiation factor, which acts as a negative regulator of LmnB1108,146 while also favoring the expression of lncRNA (2700046G09Rik), and together with the lncRNA decrease PTEN levels.111
Long noncoding RNAs
LncRNAs are nonprotein-coding RNAs that are spliced, mostly polyadenylated, and longer than 200 nucleotides in length and are generally characterized by low expression levels and nuclear localization. Many lncRNAs exhibit cell type-specific expression and are primarily associated with distinct developmental stages.152 They modulate gene expression by binding to chromatin and changing the epigenetic landscape by modifying the genomic architecture. Although both human and mouse genomes encode thousands of lncRNAs, only a few lncRNAs have been functionally characterized.
Recent transcriptomic studies identified cell type-specific lncRNAs in eight mouse brain cell types,121,153,154 including 355 lncRNAs as differentially expressed between NSCs and OPCs. Of those transcripts, 254 were upregulated during oligodendrogenesis, and 88 of them were characterized by the presence of OLIG2 binding sites within 10 kb of their transcription start site. Among them, lnc-OPC levels increased during the transition from NSCs to OPCs,155 and its ablation in NSC impaired the generation of OPCs.153 Additional lncRNAs were identified at later stages of oligodendrocyte lineage progression.155
Another study analyzing lncRNAs differentially expressed in cultured primary mouse OPCs, immature oligodendrocytes, and mature oligodendrocytes identified 1342 unique gene loci named lncOLs characterized by the presence of SOX10 binding sites and activating H3K27ac and H3K4me3 histone marks and exhibiting conservation across species.154 A total of 301 lncRNAs were identified as differentially expressed in differentiating oligodendrocytes compared to OPCs. Of those, lncOL1 and lncOL4 were associated with neurogenesis and gliogenesis and their function was strongly linked with the expression of oligodendrocyte and myelin-specific genes, such as Nkx2-2, Mbp, Mog, and Plp1. Their expression was not detected in NSC at birth but peaked at postnatal day P14 as OPCs differentiated and then sharply declined. Targeted siRNA knockdown was sufficient to decrease Mbp, Plp1, and Cnp expression in cultured OPCs, while their overexpression increased the levels of Mbp, Mag, and Myrf.154 Consistent with the in vitro data, lncOL1-deficient mice were also characterized by a low number of mature oligodendrocytes in the spinal cord and exhibited severely impaired myelination at postnatal day 21.154 lncOL1 was detected both in cytoplasmic and nuclear compartments with the nuclear lncOL1 detected in discrete puncta in association with chromatin.154 This was of interest as the RNA–protein prediction algorithm identified SUZ12, a subunit of the PRC2 complex, as a potential interactor with lncOL1.154 The functional consequence of the interaction between lncOL1 and SUZ12, as shown by immunoprecipitation, was further validated by the detection of increased transcript levels of target genes in lncOL1 knockout mice.154 Collectively, these results suggest that lncRNAs modulate the epigenome by modifying chromatin transcriptional states in oligodendrocytes (Figure 2).
Circular RNAs
CircRNAs are single-stranded, stable, functional RNAs that have a closed-loop structure. They are produced by noncanonical splicing events known as back-splicing, where a downstream 5′ splice site is joined to an upstream 3′ splice site forming an exon-containing lariat precursor. CircRNAs are predominantly found in the cytoplasm, and the lack of a 5′ cap and 3′ tail makes these molecules escape degradation and have a longer half-life than linear RNAs. They play important roles in regulating miRNA activity, alternative splicing, RNA binding protein sequestration, and transcription through interactions with RNA polymerase II.156–159 Unlike miRNAs and lncRNAs, a subset of circRNAs is reported to be translated into proteins.
Similar to linear isoforms, multiple circRNAs variants can be generated from a single gene by alternative splicing, containing different combinations of exons and/or introns.160 For instance, circCFAP299 has two isoforms in human oligodendroglioma cells; the short isoform is favored during differentiation, suggesting a distinct functional role for the unique exon in the long isoform, including interactions with RNA-binding proteins.161 In addition to the isoform switch, the circular transcriptome profile also changes at different stages of differentiation in oligodendroglioma cells.161 For instance, circVPS13C, whose mutation results in mitochondrial dysfunction, is upregulated in differentiated cells.
MECHANOTRANSDUCTION ALLOWS CHROMATIN AND NUCLEAR LAMINA TO ADJUST IN RESPONSE TO PHYSICAL FORCES
Over the last decade, it is becoming well-accepted that mechanotransduction is very important for the epigenetic regulation of gene expression because cells respond to the stiffness or elasticity of substrates they come in contact with as well as strain and compression forces. Mechanotransduction can, therefore, modify histone marks and change the association of chromatin with the nuclear lamina.162 Several reports have highlighted the biological response of oligodendrocyte lineage cells exposed to distinct mechanical stimuli.163–167 In addition, the dynamic state of chromatin condensation has been related to changes in the mechanical properties of the nuclei of OPC as they differentiate into mature oligodendrocytes.168 The nuclei of OPCs are relatively elastic and oscillating,169 characterized by the presence of loose euchromatin and by a nuclear lamina predominantly formed by LMNB1, conferring a level of softness and elasticity, which is compatible with the migratory and proliferative ability of these cells. The nuclei of differentiated oligodendrocytes, in contrast, are much more rigid, characterized by high levels of heterochromatin especially distributed along the nuclear periphery, where the nuclear lamina is now characterized by the presence of LMNA. These features allow the protection of the genetic material of highly specialized cells and are compatible with the highly branched morphology of the mature oligodendrocytes that make contact with several axons and wrap them with their membrane, providing both insulation and metabolic support.168 In this section, we will first discuss general mechanisms of mechanotransduction and then provide an outlook of potential pathways connecting mechanosensing to the regulation of gene expression in oligodendrocyte lineage cells (Figure 3).
FIGURE 3.
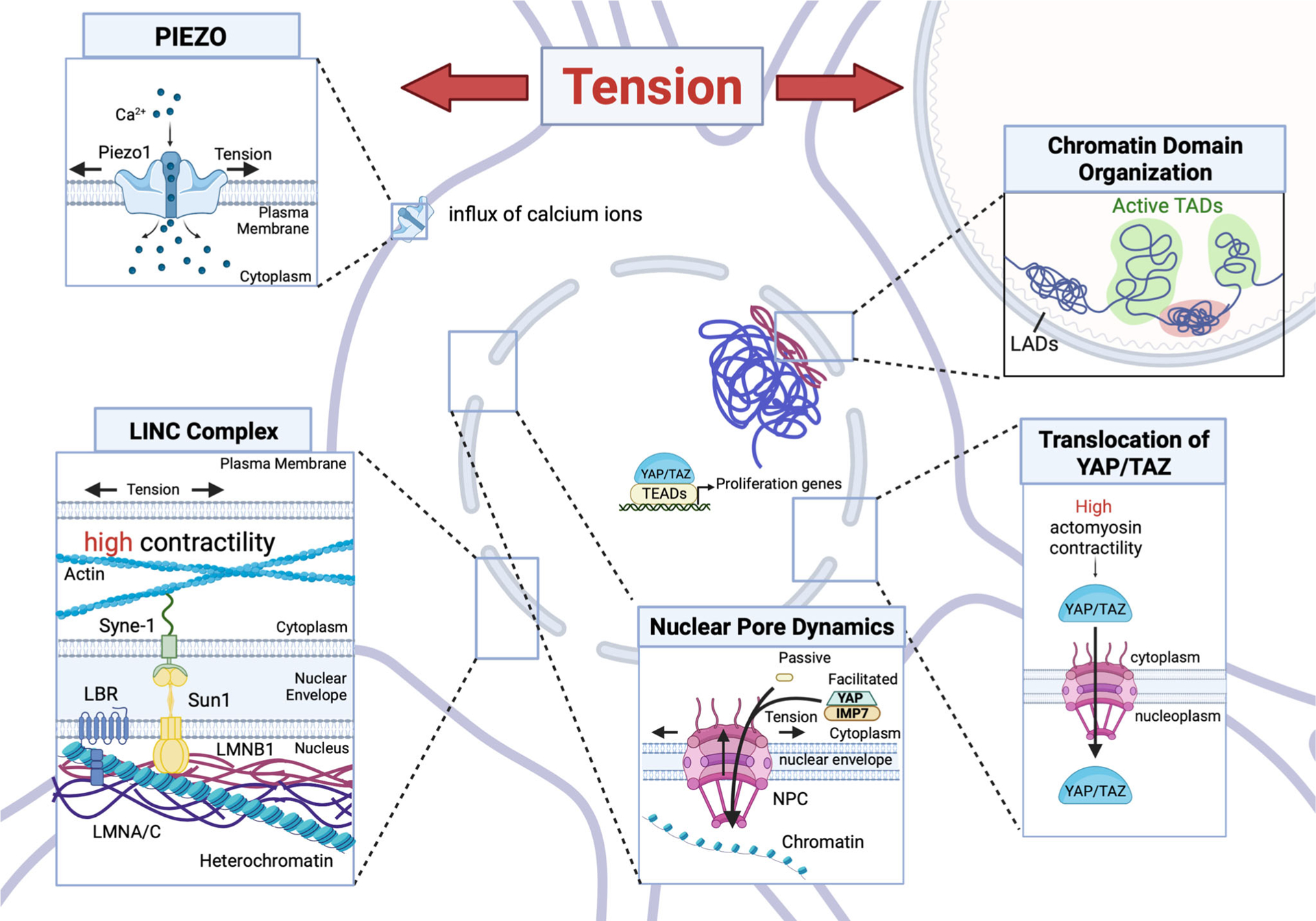
Major pathways of mechanotransduction signal epigenetic changes in response to tension. The figure shows the main pathways responsible for the transmission of the signal from the outside of the cell to the nucleus. Boxes represent modalities of response to tensile stretches. Created with BioRender. Abbreviations: LAD, lamina-associated domain; LINC, linker of nucleoskeleton and cytoskeleton; NPC, nuclear pore complex; TAD, topologicallly associating domain.
Chromatin remodeling complexes and HDACs respond to actomyosin contractility
Studies on the differentiation properties of stem cells plated on substrates with diverse ranges of softness and stiffness suggested that the mechanical properties of the substrate were capable of inducing specialized patterns of gene expression. For instance, very rigid surfaces induced expression of bone and ossification genes, soft surfaces induced brain-specific genes, and substrates of intermediate stiffness induced muscle-specific genes.170 Within the normal brain parenchyma, however, more subtle regional and temporal differences in stiffness between gray (softer) and white matter could be correlated to the amount of myelin in distinct regions.171–173 Gray matter stiffness increased from prenatal to postnatal periods (from 0.3 to 0.7 kPa), and white matter stiffness remained substantially higher (up to 10 kPa)174 and increased with aging.166,175 Fine-tuning of substrate elasticity using hydrogels allowed to define the range of stiffness promoting neuronal (0.1–0.5 kPa) versus glial (1–10 kPa) specification.176 Among the glial cells, OPCs were studied by several groups and were shown to respond by increasing proliferation and migration on stiffer substrates and differentiation on softer ones, suggesting that these cells respond to external mechanical stimuli.165–168
There is evidence that the growth of OPC on softer substrates reduced actomyosin contractility and led to myosin-dependent translocation of HDAC3 from the cytoplasm to the nucleus, leading to increased deacetylation and compaction of the chromatin177 and differentiation. Reduced actomyosin contractility also resulted in a reduction of polymerized actin and a shift from F to G actin, which is critical for the process of myelination.178,179 The role of deploy-merized nuclear actin as a modulator of chromatin in response to mechanical cues has been reported in HeLa cells since nuclear actin can bind to all three RNA polymerases,180 interact with transcription factors, and suppress class I HDAC activity.181 Globular actin in the nucleus also has the ability to bind to specific components of the BRG1 chromatin–remodeling complexes,182 thereby suggesting a direct link between cytoskeletal remodeling and transcriptional competence of chromatin.183 BRG1, in the SWI/SNF complex, has been shown to have spatiotemporal relevance to the development of oligodendrocytes in the early brain by regulating the expression of the key transcription factor, OLIG2,184 remodeling chromatin,113 and modulating differentiation.114
Mechanotransduction in response to tension: Piezo1, YAP, and regulation of nuclear pores
As the brain develops, oligodendrocyte lineage cells are also exposed to different types of tensile forces, including those created on migratory OPCs by blood pulsation, cerebrospinal fluid flow, axonal growth, and those affecting the several branches of newly differentiated oligodendrocytes—each contacting axons in all directions. The tensile strain was reported to decrease nuclear oscillations in OPCs and increase their rigidity.169 This was consistent with the increased deacetylation of histone H3K14185 and also with the results of studies supporting increased H3K9me3 and the formation of heterochromatin.164 The molecular players responsible for the relationship between tensile strain and epigenome modulators remain to be defined, but several potential mechanisms have been proposed, including the induction of transient calcium waves via the membrane protein Piezo1 and the nuclear accumulation of the coactivator Yes associated protein1 (YAP) and transcription factors, which are due to stretching of the nuclear pores. Piezo1 is a propeller-shaped trans-membrane protein that responds to changes in membrane tension by opening a cation channel that causes an influx of calcium ions into the cell.186–188 It is expressed in neurons, astrocytes, and microglial cells at low levels. In the oligodendrocyte lineage cells, Piezo1 is highly expressed in OPCs, where its activation decreases both proliferation and migration.166,189
The transcriptional coactivators YAP and TAZ with the PDZ domain encoded by the gene WWTR1 are part of the Hippo (named after the Hpo/MTS1/2 kinase) pathway cascade, which regulates organ size by modulating cell proliferation and apoptosis (Figure 3). In the unphosphorylated state, the coactivators YAP and TAZ are nuclear and favor proliferation due to the transcription of cell cycle genes. In conditions that favor low actomyosin contractility (such as softer substrates), YAP is phosphorylated by nuclear kinases LATS1/2 and becomes cytosolic and unable to activate transcription of proliferative genes, thereby contributing to cell cycle exit.190 Mechanical stimulation, including shear stress, or cytoskeletal tension results in the transmission of physical forces to the nuclear envelope and stretching of the nuclear pores191,192 to allow for YAP nuclear localization and accumulation and activation of proliferation genes.190,193
Chromatin at the nuclear envelope
A major hallmark of differentiation is the accumulation of constitutive and facultative heterochromatin at the nuclear periphery, which confers rigidity to the nucleus of differentiated oligodendrocytes. An intricate array of proteins play a major role in organizing the epigenome. These proteins include those that span the nuclear envelope (e.g., SYNE/nesprins and SUN proteins), a mesh of intermediate filaments called lamins (e.g., lamin B1 and lamin A/C) at the inner nuclear membrane, and proteins involved in regulating the organization of chromatin and its association with the nuclear envelope. Indeed, a large number of proteins containing lamina association domains, as well as readers of acetylated and methylated histone marks (e.g., LAP2A, LBR, and emerin) serve as bridge and anchor chromatin to the nuclear lamina. This recruitment allows the creation of lamin-associated domains consisting of heterochromatic regions characterized by low rates of gene expression and silenced genes.1 The transition from OPC to mature oligodendrocytes shows clear changes in the mechanical properties of the nucleus, from a softer nucleus to one that is more rigid,168 and this coincides with the progressive accumulation of peripheral heterochromatin. The mechanical properties of the nucleus also respond to changes in substrate rigidity by changes in the composition of the nuclear lamina via upregulation of specific lamins and phosphorylation of emerin.194,195 Oligodendrocyte progenitors, for instance, express LMNB1 and progressively upregulate the levels of LMNA during differentiation and when cultured on stiffer substrates, thereby differentially affecting heterochromatin recruitment to the nuclear envelope.104,194
As the brain grows and the skull sutures close, the oligodendrocyte lineage cells undergo rapid expansion to adjust to the extensive requirement for myelination across distinct brain regions. These events lead to an overall increased cell density, which is sensed by the cells via a complex that connects the plasma membrane through the nucleus via the LINC complex (Figure 3). In the adult brain, in inflammatory conditions characterized by increased blood–brain barrier permeability, immune cells can enter into the parenchyma creating local crowding conditions. These physical constraints exert physical forces on the cells, which can be studied by culturing cells in the presence of inert microspheres196 or using a compression device.164 Both conditions were shown to induce changes in histone repressive marks and heterochromatin recruitment to the nuclear envelope.164 These nuclear changes and the effect on differentiation have been shown to be mediated by the SYNE–SUN–actin complex, as silencing of the KASH-domain containing LINC complex component SYNE1 prevented the mechanisms of differentiation of OPCs induced by mechanotransduction, without altering biochemical-induced differentiation.164
Tethering of heterochromatin to the periphery of the nucleus has been shown in retinal cells, keratinocytes, and fibroblasts to depend upon both LBR and lamin A.197 Future studies will be needed to further clarify the role of nuclear lamins in regulating chromatin organization in oligodendrocyte lineage cells.
CONCLUDING REMARKS
This review highlights the complexity and current understanding of the epigenomic landscape in oligodendrocyte lineage cells, providing a perspective on the multiplicity of events that regulate gene expression and underlie the transcriptional heterogeneity of OPCs as described by many labs. Importantly, stable modifications of gene expression allow for the expression of genes responsible for the functional specialization of cells. However, chronic conditions and pathological states may allow for a reorganization of the landscape and allow for the expression of genes that are silenced in physiological conditions (e.g., immune genes). While some of the key eraser enzymes and pioneer transcription factors have been identified, it will be important to understand how distinct pathological conditions lead to pathological transcriptional states.
In addition, it is important to realize that we have just started to identify responsible histone, DNA, and RNA modifications, and much remains to be explored. Our current models are biased by the use of experimental genetic approaches using conditional mouse lines and developmental myelination as readout. This approach has been successful in the definition of enzymatic activities and transcriptional networks involved in the process. However, it has possibly led to over-generalization, and future work will need to take into consideration the functional role of time and stage of differentiation on topological domains. Novel approaches need to be considered in order to explore the relative role of protein readers of epigenetic marks and the relative contribution of the biophysical state of chromatin and membrane-less organelles at each stage of lineage progression.
ACKNOWLEDGMENTS
The authors would like to acknowledge funding from NIH-NINDS Grant R35 NS111604 to P.C. and thank all the members of the Casaccia lab for their critical reading of the manuscript.
Funding information
National Institute of Neurological Disorders and Stroke, Grant/Award Number: R35NS111604
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, & van Steensel B (2008). Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature, 453, 948–951. [DOI] [PubMed] [Google Scholar]
- 2.Towbin BD, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, & Gasser SM (2012). Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell, 150, 934–947. [DOI] [PubMed] [Google Scholar]
- 3.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, Gribnau J, Barillot E, Blüthgen N, Dekker J, & Heard E (2012). Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature, 485, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, & Cavalli G (2012). Three-dimensional folding and functional organization principles of the Drosophila genome. Cell, 148, 458–472. [DOI] [PubMed] [Google Scholar]
- 5.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, & Ren B (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature, 485, 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roth SY, Denu JM, & Allis CD (2001). Histone acetyltransferases. Annual Review of Biochemistry, 70(1), 81–120. [DOI] [PubMed] [Google Scholar]
- 7.Park S-Y, & Kim J-S (2020). A short guide to histone deacetylases including recent progress on class II enzymes. Experimental & Molecular Medicine, 52(2), 204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin C, & Zhang Y (2005). The diverse functions of histone lysine methylation. Nature Reviews Molecular Cell Biology, 6(11), 838–849. [DOI] [PubMed] [Google Scholar]
- 9.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Calcar SV, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, & Ren B (2007). Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genetics, 39(3), 311–318. [DOI] [PubMed] [Google Scholar]
- 10.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, … Ren B (2009). Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature, 459(7243), 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guccione E, & Richard S (2019). The regulation, functions and clinical relevance of arginine methylation. Nature Reviews. Molecular Cell Biology, 20(10), 642–657. [DOI] [PubMed] [Google Scholar]
- 12.Yu K, & Proost P (2022). Insights into peptidylarginine deiminase expression and citrullination pathways. Trends in Cell Biology, 32(9), 746–761. [DOI] [PubMed] [Google Scholar]
- 13.Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, & Simon JA (2002). Histone methyltransferase activity of a drosophila polycomb group repressor complex. Cell, 111(2), 197–208. [DOI] [PubMed] [Google Scholar]
- 14.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, & Brockdorff N (2004). Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Developmental Cell, 7(5), 663–676. [DOI] [PubMed] [Google Scholar]
- 15.Marques S, Zeisel A, Codeluppi S, van Bruggen D, Mendanha Falcão A, Xiao L, Li H, Häring M, Hochgerner H, Romanov RA, Gyllborg D, Muñoz-Manchado AB, La Manno G, Lönnerberg P, Floriddia EM, Rezayee F, Ernfors P, Arenas E, Hjerling-Leffler J, … Castelo-Branco G (2016). Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science, 352(6291), 1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma K, Schmitt S, Bergner CG, Tyanova S, Kannaiyan N, Manrique-Hoyos N, Kongi K, Cantuti L, Hanisch U-K, Philips M-A, Rossner MJ, Mann M, & Simons M (2015). Cell type– and brain region–resolved mouse brain proteome. Nature Neuroscience, 18(12), 1819–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, Bertagnolli D, Goldy J, Shapovalova N, Parry S, Lee C, Smith K, Bernard A, Madisen L, Sunkin SM, … Zeng H (2016). Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nature Neuroscience, 19(2), 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, & Kouzarides T (2001). Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410(6824), 120–124. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz S, Meshorer E, & Ast G (2009). Chromatin organization marks exon-intron structure. Nature Structural & Molecular Biology, 16(9), 990–995. [DOI] [PubMed] [Google Scholar]
- 20.Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Sung KWK, Rigoutsos I, Loring J, & Wei C-L (2010). Dynamic changes in the human methylome during differentiation. Genome Research, 20(3), 320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nan X, Ng H-H, Johnson CA, Laherty CD, Turner BM, Eisenman RN, & Bird A (1998). Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature, 393(6683), 386–389. [DOI] [PubMed] [Google Scholar]
- 22.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, & Wolffe AP (1998). Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genetics, 19(2), 187–191. [DOI] [PubMed] [Google Scholar]
- 23.Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, & Kouzarides T (2003). The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. The Journal of Biological Chemistry, 278(6), 4035–4040. [DOI] [PubMed] [Google Scholar]
- 24.Maunakea AK, Chepelev I, Cui K, & Zhao K (2013). Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Research, 23(11), 1256–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moyon S, Huynh JL, Dutta D, Zhang F, Ma D, Yoo S, Lawrence R, Wegner M, John GR, Emery B, Lubetzki C, Franklin RJM, Fan G, Zhu J, Dupree JL, & Casaccia P (2016). Functional characterization of DNA methylation in the oligodendrocyte lineage. Cell Reports, 15(4), 748–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohli RM, & Zhang Y (2013). TET enzymes, TDG and the dynamics of DNA demethylation. Nature, 502(7472), 472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rullens PMJ, & Kind J (2021). Attach and stretch: Emerging roles for genome–lamina contacts in shaping the 3D genome. Current Opinion in Cell Biology, 70, 51–57. [DOI] [PubMed] [Google Scholar]
- 28.Vahabikashi A, Adam SA, Medalia O, & Goldman RD (2022). Nuclear lamins: Structure and function in mechanobiology. APL Bioengineering, 6(1), 011503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellad JA, Warren DT, & Shanahan CM (2011). Nesprins LINC the nucleus and cytoskeleton. Current Opinion in Cell Biology, 23(1), 47–54. [DOI] [PubMed] [Google Scholar]
- 30.Fan G, Martinowich K, Chin MH, He F, Fouse SD, Hutnick L, Hattori D, Ge W, Shen Y, Wu H, ten Hoeve J, Shuai K, & Sun YE (2005). DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development, 132(15), 3345–3356. [DOI] [PubMed] [Google Scholar]
- 31.He F, Ge W, Martinowich K, Becker-Catania S, Coskun V, Zhu W, Wu H, Castro D, Guillemot F, Fan G, de Vellis J, & Sun YE (2005). A positive autoregulatory loop of Jak-STAT signaling controls the onset of astrogliogenesis. Nature Neuroscience, 8(5), 616–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsuboi M, Kishi Y, Yokozeki W, Koseki H, Hirabayashi Y, & Gotoh Y (2018). Ubiquitination-independent repression of PRC1 targets during neuronal fate restriction in the developing mouse neocortex. Developmental Cell, 47(6), 758–772.e5. [DOI] [PubMed] [Google Scholar]
- 33.Tiwari N, Pataskar A, Péron S, Thakurela S, Sahu SK, Figueres-Oñate M, Marichal N, López-Mascaraque L, Tiwari VK, & Berninger B (2018). Stage-specific transcription factors drive astrogliogenesis by remodeling gene regulatory landscapes. Cell Stem Cell, 23(4), 557–571.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, & Anderson DJ (2006). The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron, 52(6), 953–968. [DOI] [PubMed] [Google Scholar]
- 35.Kang P, Lee HK, Glasgow SM, Finley M, Donti T, Gaber ZB, Graham BH, Foster AE, Novitch BG, Gronostajski RM, & Deneen B (2012). Sox9 and NFIA Coordinate a Transcriptional Regulatory Cascade during the Initiation of Gliogenesis. Neuron, 74(1), 79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukuda S, Abematsu M, Mori H, Yanagisawa M, Kagawa T, Nakashima K, Yoshimura A, & Taga T (2007). Potentiation of astrogliogenesis by STAT3-mediated activation of bone morphogenetic protein-smad signaling in neural stem cells. Molecular and Cellular Biology, 27(13), 4931–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Namihira M, Kohyama J, Semi K, Sanosaka T, Deneen B, Taga T, & Nakashima K (2009). Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Developmental Cell, 16(2), 245–255. [DOI] [PubMed] [Google Scholar]
- 38.Wang W, Cho H, Kim D, Park Y, Moon JH, Lim SJ, Yoon SM, McCane M, Aicher SA, Kim S, Emery B, Lee JW, Lee S, Park Y, & Lee S-K (2020). PRC2 acts as a critical timer that drives oligodendrocyte fate over astrocyte identity by repressing the notch pathway. Cell Reports, 32(11), 108147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Yang L, Dong C, Wang J, Xu L, Qiu Y, Weng Q, Zhao C, Xin M, & Lu QR (2020). EED-mediated histone methylation is critical for CNS myelination and remyelination by inhibiting WNT, BMP, and senescence pathways. Science Advances, 6(33), eaaz6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McMahon JA, Takada S, Zimmerman LB, Fan C-M, Harland RM, & McMahon AP (1998). Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes & Development, 12(10), 1438–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao MS, & Mayer-Proschel M (1997). Glial-restricted precursors are derived from multipotent neuroepithelial stem cells. Developmental Biology, 188(1), 48–63. [DOI] [PubMed] [Google Scholar]
- 42.Gregori N, Pröschel C, Noble M, & Mayer-Pröschel M (2002). The tripotential glial-restricted precursor (GRP) cell and glial development in the spinal cord: Generation of bipotential oligodendrocyte-type-2 astrocyte progenitor cells and dorsal–ventral differences in GRP cell function. The Journal of Neuroscience, 22(1), 248–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herrera J, Yang H, Zhang S-C, Proschel C, Tresco P, Duncan ID, Luskin M, & Mayer-Proschel M (2001). Embryonic-derived glial-restricted precursor cells (GRP cells) can differentiate into astrocytes and oligodendrocytes in vivo. Experimental Neurology, 171(1), 11–21. [DOI] [PubMed] [Google Scholar]
- 44.Káradóttir R, Hamilton NB, Bakiri Y, & Attwell D (2008). Spiking and nonspiking classes of oligodendrocyte precursor glia in CNS white matter. Nature Neuroscience, 11(4), 450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagy B, Hovhannisyan A, Barzan R, Chen T-J, & Kukley M (2017). Different patterns of neuronal activity trigger distinct responses of oligodendrocyte precursor cells in the corpus callosum. PLOS Biology, 15(8), e2001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibson EM, Purger D, Mount CW, Goldstein AK, Lin GL, Wood LS, Inema I, Miller SE, Bieri G, Zuchero JB, Barres BA, Woo PJ, Vogel H, & Monje M (2014). Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science, 344(6183), 1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paez PM, & Lyons DA (2020). Calcium signaling in the oligodendrocyte lineage: Regulators and consequences. Annual Review of Neuroscience, 43, 163–186. [DOI] [PubMed] [Google Scholar]
- 48.Kukley M, Nishiyama A, & Dietrich D (2010). The fate of synaptic input to NG2 glial cells: Neurons specifically downregulate transmitter release onto differentiating oligodendroglial cells. The Journal of neuroscience : the official journal of the Society for Neuroscience, 30(24), 8320–8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Biase LM, Nishiyama A, & Bergles DE (2010). Excitability and synaptic communication within the oligodendrocyte lineage. Journal of Neuroscience, 30(10), 3600–3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin SC, & Bergles DE (2004). Synaptic signaling between GABAergic interneurons and oligodendrocyte precursor cells in the hippocampus. Nature Neuroscience, 7(1), 24–32. [DOI] [PubMed] [Google Scholar]
- 51.Bergles DE, Roberts JDB, Somogyi P, & Jahr CE (2000). Glutamatergic synapses on oligodendrocyte precursor cells in the hippocampus. Nature, 405(6783), 187–191. [DOI] [PubMed] [Google Scholar]
- 52.Venkatesh HS, Tam LT, Woo PJ, Lennon J, Nagaraja S, Gillespie SM, Ni J, Duveau DY, Morris PJ, Zhao JJ, Thomas CJ, & Monje M (2017). Targeting neuronal activity-regulated neuroligin-3 dependency in high-grade glioma. Nature, 549(7673), 533–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gallo V, Zhou J, McBain C, Wright P, Knutson P, & Armstrong R (1996). Oligodendrocyte progenitor cell proliferation and lineage progression are regulated by glutamate receptor-mediated K+ channel block. The Journal of Neuroscience, 16(8), 2659–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vautier F, Belachew S, Chittajallu R, & Gallo V (2004). Shaker-type potassium channel subunits differentially control oligodendrocyte progenitor proliferation. Glia, 48(4), 337–345. [DOI] [PubMed] [Google Scholar]
- 55.Ortiz FC, Habermacher C, Graciarena M, Houry P-Y, Nishiyama A, Oumesmar BN, & Angulo MC (2019). Neuronal activity in vivo enhances functional myelin repair. JCI Insight, 4(9), e123434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Demerens C, Stankoff B, Logak M, Anglade P, Allinquant B, Couraud F, Zalc B, & Lubetzki C (1996). Induction of myelination in the central nervous system by electrical activity. Proceedings of the National Academy of Sciences, 93(18), 9887–9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu A, Han YR, Li J, Sun D, Ouyang M, Plummer MR, & Casaccia-Bonnefil P (2007). The glial or neuronal fate choice of oligodendrocyte progenitors is modulated by their ability to acquire an epigenetic memory. Journal of Neuroscience, 27(27), 7339–7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rivers LE, Young KM, Rizzi M, Jamen F, Psachoulia K, Wade A, Kessaris N, & Richardson WD (2008). PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nature Neuroscience, 11(12), 1392–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richardson WD, Young KM, Tripathi RB, & McKenzie I (2011). NG2-glia as multipotent neural stem cells: Fact or fantasy? Neuron, 70(4), 661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kondo T, & Raff M (2000). Oligodendrocyte precursor cells reprogrammed to become multipotential CNS stem cells. Science (New York, N.Y.), 289(5485), 1754–1757. 10.1126/science.289.5485.1754 [DOI] [PubMed] [Google Scholar]
- 61.Dimou L, Simon C, Kirchhoff F, Takebayashi H, & Götz M (2008). Progeny of olig2-expressing progenitors in the gray and white matter of the adult mouse cerebral cortex. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 28(41), 10434–10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kang SH, Fukaya M, Yang JK, Rothstein JD, & Bergles DE (2010). NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron, 68(4), 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu X, Hill RA, Dietrich D, Komitova M, Suzuki R, & Nishiyama A (2011). Age-dependent fate and lineage restriction of single NG2 cells. Development (Cambridge, England), 138(4), 745–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu J, Magri L, Zhang F, Marsh NO, Albrecht S, Huynh JL, Kaur J, Kuhlmann T, Zhang W, Slesinger PA, & Casaccia P (2015). Chromatin landscape defined by repressive histone methylation dur ing oligodendrocyte differentiation. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 35(1), 352–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sher F, Rössler R, Brouwer N, Balasubramaniyan V, Boddeke E, & Copray S (2008). Differentiation of neural stem cells into oligodendrocytes: Involvement of the polycomb group protein Ezh2. Stem Cells (Dayton, Ohio), 26(11), 2875–2883. [DOI] [PubMed] [Google Scholar]
- 66.Sher F, Boddeke E, Olah M, & Copray S (2012). Dynamic changes in Ezh2 gene occupancy underlie its involvement in neural stem cell self-renewal and differentiation towards oligodendrocytes. PLoS One, 7(7), e40399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu J, & Casaccia P (2010). Epigenetic regulation of oligodendrocyte identity. Trends in Neurosciences, 33(4), 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boshans LL, Factor DC, Singh V, Liu J, Zhao C, Mandoiu I, Lu QR, Casaccia P, Tesar PJ, & Nishiyama A (2019). The chromatin environment around interneuron genes in oligodendrocyte precursor cells and their potential for interneuron reprograming. Serotonin Receptors in Neurobiology, 13, 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boshans LL, Soh H, Wood WM, Nolan TM, Mandoiu II, Yanagawa Y, Tzingounis AV, & Nishiyama A (2021). Direct reprogramming of oligodendrocyte precursor cells into GABAergic inhibitory neurons by a single homeodomain transcription factor Dlx2. Scientific Reports, 11(1), 3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu M, Hernandez M, Shen S, Sabo JK, Kelkar D, Wang J, O’Leary R, Phillips GR, Cate HS, & Casaccia P (2012). Differential modulation of the oligodendrocyte transcriptome by sonic hedgehog and bone morphogenetic protein 4 via opposing effects on histone acetylation. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 32(19), 6651–6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Glasgow SM, Zhu W, Stolt CC, Huang T-W, Chen F, LoTurco JJ, Neul JL, Wegner M, Mohila C, & Deneen B (2014). Mutual antagonism between Sox10 and NFIA regulates diversification of glial lineages and glioma subtypes. Nature Neuroscience, 17(10), 1322–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zuo H, Wood WM, Sherafat A, Hill RA, Lu QR, & Nishiyama A (2018). Age-dependent decline in fate switch from NG2 cells to astrocytes after olig2 deletion. The Journal of Neuroscience, 38(9), 2359–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang L, He X, Liu L, Jiang M, Zhao C, Wang H, He D, Zheng T, Zhou X, Hassan A, Ma Z, Xin M, Sun Z, Lazar MA, Goldman SA, Olson EN, & Lu QR (2016). Hdac3 interaction with p300 histone acetyltransferase regulates the oligodendrocyte and astrocyte lineage fate switch. Developmental Cell, 36(3), 316–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klum S, Zaouter C, Alekseenko Z, Björklund ÅK, Hagey DW, Ericson J, Muhr J, & Bergsland M (2018). Sequentially acting SOX proteins orchestrate astrocyte- and oligodendrocyte-specific gene expression. EMBO Reports, 19(11), e46635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reiprich S, Cantone M, Weider M, Baroti T, Wittstatt J, Schmitt C, Küspert M, Vera J, & Wegner M (2017). Transcription factor Sox10 regulates oligodendroglial Sox9 levels via microRNAs. Glia, 65(7), 1089–1102. [DOI] [PubMed] [Google Scholar]
- 76.Takahashi K, & Yamanaka S (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell, 126(4), 663–676. [DOI] [PubMed] [Google Scholar]
- 77.Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, & Zaret KS (2015). Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell, 161(3), 555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang J, Pol SU, Haberman AK, Wang C, O’Bara MA, & Sim FJ (2014). Transcription factor induction of human oligodendrocyte progenitor fate and differentiation. Proceedings of the National Academy of Sciences of the United States of America, 111(28), E2885–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parras CM, Galli R, Britz O, Soares S, Galichet C, Battiste J, Johnson JE, Nakafuku M, Vescovi A, & Guillemot F (2004). Mash1 specifies neurons and oligodendrocytes in the postnatal brain. The EMBO Journal, 23(22), 4495–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Battiste J, Helms AW, Kim EJ, Savage TK, Lagace DC, Mandyam CD, Eisch AJ, Miyoshi G, & Johnson JE (2007). Ascl1 defines sequentially generated lineage-restricted neuronal and oligodendrocyte precursor cells in the spinal cord. Development, 134(2), 285–293. 10.1242/dev.02727 [DOI] [PubMed] [Google Scholar]
- 81.Nakatani H, Martin E, Hassani H, Clavairoly A, Maire CL, Viadieu A, Kerninon C, Delmasure A, Frah M, Weber M, Nakafuku M, Zalc B, Thomas J-L, Guillemot F, Nait-Oumesmar B, & Parras C (2013). Ascl1/Mash1 promotes brain oligodendrogenesis during myelination and remyelination. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 33(23), 9752–9768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tanabe K, Nobuta H, Yang N, Ang CE, Huie P, Jordan S, Oldham MC, Rowitch DH, & Wernig M (2022). Generation of functional human oligodendrocytes from dermal fibroblasts by direct lineage conversion. Development, 149(20), dev199723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Swiss VA, Nguyen T, Dugas J, Ibrahim A, Barres B, Androulakis IP, & Casaccia P (2011). Identification of a gene regulatory network necessary for the initiation of oligodendrocyte differentiation. PLoS ONE, 6(4), e18088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kukley M, Capetillo-Zarate E, & Dietrich D (2007). Vesicular glutamate release from axons in white matter. Nature Neuroscience, 10(3), 311–320. [DOI] [PubMed] [Google Scholar]
- 85.Ziskin JL, Nishiyama A, Rubio M, Fukaya M, & Bergles DE (2007). Vesicular release of glutamate from unmyelinated axons in white matter. Nature Neuroscience, 10(3), 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shen S, Li J, & Casaccia-Bonnefil P (2005). Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. Journal of Cell Biology, 169(4), 577–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marin-Husstege M, Muggironi M, Liu A, & Casaccia-Bonnefil P (2002). Histone deacetylase activity is necessary for oligodendrocyte lineage progression. The Journal of Neuroscience, 22(23), 10333–10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shen S, Sandoval J, Swiss VA, Li J, Dupree J, Franklin RJM, & Casaccia-Bonnefil P (2008). Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nature Neuroscience, 11(9), 1024–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shen S, Liu A, Li J, Wolubah C, & Casaccia-Bonnefil P (2008). Epigenetic memory loss in aging oligodendrocytes in the corpus callosum. Neurobiology of Aging, 29(3), 452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lyssiotis CA, Walker J, Wu C, Kondo T, Schultz PG, & Wu X (2007). Inhibition of histone deacetylase activity induces developmental plasticity in oligodendrocyte precursor cells. Proceedings of the National Academy of Sciences of the United States of America, 104(38), 14982–14987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Conway GD, O’Bara MA, Vedia BH, Pol SU, & Sim FJ (2012). Histone deacetylase activity is required for human oligodendrocyte progenitor differentiation. Glia, 60(12), 1944–1953. [DOI] [PubMed] [Google Scholar]
- 92.Douvaras P, Rusielewicz T, Kim KH, Haines JD, Casaccia P, & Fossati V (2016). Epigenetic modulation of human induced pluripotent stem cell differentiation to oligodendrocytes. International Journal of Molecular Sciences, 17(4), 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Scaglione A, Patzig J, Liang J, Frawley R, Bok J, Mela A, Yattah C, Zhang J, Teo SX, Zhou T, Chen S, Bernstein E, Canoll P, Guccione E, & Casaccia P (2018). PRMT5-mediated regulation of developmental myelination. Nature Communications, 9(1), 2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Falcão AM, Meijer M, Scaglione A, Rinwa P, Agirre E, Liang J, Larsen SC, Heskol A, Frawley R, Klingener M, Varas-Godoy M, Raposo AASF, Ernfors P, Castro DS, Nielsen ML, Casaccia P, & Castelo-Branco G (2019). PAD2-Mediated citrullination contributes to efficient oligodendrocyte differentiation and myelination. Cell Reports, 27(4), 1090–1102.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.He Y, Sandoval J, & Casaccia-Bonnefil P (2007). Events at the transition between cell cycle exit and oligodendrocyte progenitor differentiation: The role of HDAC and YY1. Neuron Glia Biology, 3(3), 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ye F, Chen Y, Hoang T, Montgomery RL, Zhao X, Bu H, Hu T, Taketo MM, van Es JH, Clevers H, Hsieh J, Bassel-Duby R, Olson EN, & Lu QR (2009). HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nature Neuroscience, 12(7), 829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Samanta J, & Kessler JA (2004). Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development, 131(17), 4131–4142. [DOI] [PubMed] [Google Scholar]
- 98.He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, Shi Y, Nave KA, & Casaccia-Bonnefil P (2007). The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron, 55(2), 217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marin-Husstege M, He Y, Li J, Kondo T, Sablitzky F, & Casaccia-Bonnefil P (2006). Multiple roles of Id4 in developmental myeli-nation: Predicted outcomes and unexpected findings. Glia, 54(4), 285–296. [DOI] [PubMed] [Google Scholar]
- 100.Liu A, Li J, Marin-Husstege M, Kageyama R, Fan Y, Gelinas C, & Casaccia-Bonnefil P (2006). A molecular insight of Hes5-dependent inhibition of myelin gene expression: Old partners and new players. The EMBO Journal, 25(20), 4833–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huang J, Vogel G, Yu Z, Almazan G, & Richard S (2011). Type II arginine methyltransferase PRMT5 regulates gene expression of inhibitors of differentiation/DNA binding Id2 and Id4 during glial cell differentiation. The Journal of Biological Chemistry, 286(52), 44424–44432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao C, Dong C, Frah M, Deng Y, Marie C, Zhang F, Xu L, Ma Z, Dong X, Lin Y, Koenig S, Nait-Oumesmar B, Martin DM, Wu LN, Xin M, Zhou W, Parras C, & Lu QR (2018). Dual requirement of CHD8 for chromatin landscape establishment and histone methyltransferase recruitment to promote CNS myelination and repair. Developmental Cell, 45(6), 753–768.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tiane A, Schepers M, Riemens R, Rombaut B, Vandormael P, Somers V, Prickaerts J, Hellings N, Hove D. van den, & Vanmierlo T (2021). DNA methylation regulates the expression of the negative transcriptional regulators ID2 and ID4 during OPC differentiation. Cellular and Molecular Life Sciences, 78(19–20), 6631–6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yattah C, Hernandez M, Huang D, Park H, Liao W, & Casaccia P (2020). Dynamic lamin B1-gene association during oligodendrocyte progenitor differentiation. Neurochemical Research, 45(3), 606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Magri L, Gacias M, Wu M, Swiss VA, Janssen WG, & Casaccia P (2014). C-Myc-dependent transcriptional regulation of cell cycle and nucleosomal histones during oligodendrocyte differentiation. Neuroscience, 276, 72–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Magri L, Swiss VA, Jablonska B, Lei L, Pedre X, Walsh M, Zhang W, Gallo V, Canoll P, & Casaccia P (2014). E2F1 coregulates cell cycle genes and chromatin components during the transition of oligodendrocyte progenitors from proliferation to differentiation. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 34(4), 1481–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gacias M, Gerona-Navarro G, Plotnikov AN, Zhang G, Zeng L, Kaur J, Moy G, Rusinova E, Rodriguez Y, Matikainen B, Vincek A, Joshua J, Casaccia P, & Zhou M-M (2014). Selective chemical modulation of gene transcription favors oligodendrocyte lineage progression. Chemistry & Biology, 21(7), 841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lin S-T, & Fu Y-H (2009). MiR-23 regulation of lamin B1 is crucial for oligodendrocyte development and myelination. Disease Models & Mechanisms, 2(3–4), 178–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Prüfert K, Vogel A, & Krohne G (2004). The lamin CxxM motif promotes nuclear membrane growth. Journal of Cell Science, 117(25), 6105–6116. [DOI] [PubMed] [Google Scholar]
- 110.Heng MY, Lin S-T, Verret L, Huang Y, Kamiya S, Padiath QS, Tong Y, Palop JJ, Huang EJ, Ptácχek LJ, & Fu Y-H (2013). Lamin B1 mediates cell-autonomous neuropathology in a leukodystrophy mouse model. The Journal of Clinical Investigation, 123(6), 2719–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lin S-T, Huang Y, Zhang L, Heng MY, Ptáček LJ, & Fu Y-H (2013). MicroRNA-23a promotes myelination in the central nervous system. Proceedings of the National Academy of Sciences, 110(43), 17468–17473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rolyan H, Tyurina YY, Hernandez M, Amoscato AA, Sparvero LJ, Nmezi BC, Lu Y, Estécio MRH, Lin K, Chen J, He RR, Gong P, Rigatti LH, Dupree J, Bayır H, Kagan VE, Casaccia P, & Padiath QS (2015). Defects of Lipid Synthesis Are Linked to the Age-Dependent Demyelination Caused by Lamin B1 Overexpression. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 35(34), 12002–12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, Liu L, Liu W, Wu LMN, Mao M, Chan JR, Wu J, & Lu QR (2013). Olig2 Targets Chromatin Remodelers to Enhancers to Initiate Oligodendrocyte Differentiation. Cell, 152(1–2), 248–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bischof M, Weider M, Küspert M, Nave K-A, & Wegner M (2015). Brg1-dependent chromatin remodelling is not essentially required during oligodendroglial differentiation. The Journal of Neuroscience, 35(1), 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lopez-Anido C, Sun G, Koenning M, Srinivasan R, Hung HA, Emery B, Keles S, & Svaren J (2015). Differential Sox10 genomic occupancy in myelinating glia. Glia, 63(11), 1897–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.He D, Marie C, Zhao C, Kim B, Wang J, Deng Y, Clavairoly A, Frah M, Wang H, He X, Hmidan H, Jones BV, Witte D, Zalc B, Zhou X, Choo DI, Martin DM, Parras C, & Lu QR (2016). Chd7 cooperates with sox10 and regulates the onset of CNS myelination and remyelination. Nature Neuroscience, 19(5), 678–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Moyon S, Frawley R, Marechal D, Huang D, Marshall-Phelps KLH, Kegel L, Bøstrand SMK, Sadowski B, Jiang Y-H, Lyons DA, Möbius W, & Casaccia P (2021). TET1-mediated DNA hydroxymethylation regulates adult remyelination in mice. Nature Communications, 12(1), 3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang M, Wang J, Zhang K, Lu G, Liu Y, Ren K, Wang W, Xin D, Xu L, Mao H, Xing J, Gao X, Jin W, Berry K, Mikoshiba K, Wu S, Lu QR, & Zhao X (2021). Ten-eleven translocation 1 mediated-DNA hydroxymethylation is required for myelination and remyelination in the mouse brain. Nature Communications, 12(1), 5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lejart A, Zentout S, Chapuis C, D’Augustin O, Smith R, Salbert G, & Huet S (2022). The N-terminal domain of TET1 promotes the formation of dense chromatin regions refractory to transcription. Chromosoma, 131(1–2), 47–58. [DOI] [PubMed] [Google Scholar]
- 120.Chrysanthou S, Tang Q, Lee J, Taylor SJ, Zhao Y, Steidl U, Zheng D, & Dawlaty MM (2022). The DNA dioxygenase Tet1 regulates H3K27 modification and embryonic stem cell biology independent of its catalytic activity. Nucleic Acids Research, 50(6), 3169–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, & Wu JQ (2014). An RNA-Sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. Journal of Neuroscience, 34(36), 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hobson GM, Huang Z, Sperle K, Sistermans E, Rogan PK, Garbern JY, Kolodny E, Naidu S, & Cambi F (2006). Splice-site contribution in alternative splicing of PLP1 and DM20: Molecular studies in oligodendrocytes. Human Mutation, 27(1), 69–77. [DOI] [PubMed] [Google Scholar]
- 123.Wang E, Dimova N, & Cambi F (2007). PLP/DM20 ratio is regulated by hnRNPH and F and a novel G-rich enhancer in oligodendrocytes. Nucleic Acids Research, 35(12), 4164–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ransohoff JD, Wei Y, & Khavari PA (2018). The functions and unique features of long intergenic non-coding RNA. Nature Reviews. Molecular Cell Biology, 19(3), 143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yue Y, Liu J, & He C (2015). RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes & Development, 29(13), 1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, Dai Q, Chen W, & He C (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nature Chemical Biology, 10(2), 93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, & Zhao JC (2014). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nature Cell Biology, 16(2), 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, & Soller M (2016). M6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature, 540(7632), 301–304. [DOI] [PubMed] [Google Scholar]
- 129.Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, & Tavazoie SF (2015). HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell, 162(6), 1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wu R, Li A, Sun B, Sun J-G, Zhang J, Zhang T, Chen Y, Xiao Y, Gao Y, Zhang Q, Ma J, Yang X, Liao Y, Lai W-Y, Qi X, Wang S, Shu Y, Wang H-L, Wang F, … Yuan Z (2019). A novel m6A reader Prrc2a controls oligodendroglial specification and myelination. Cell Research, 29(1), 23–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dansu DK, Liang J, Selcen I, Zheng H, Moore DF, & Casaccia P (2022). PRMT5 interacting partners and substrates in oligodendrocyte lineage cells. Frontiers in Cellular Neuroscience, 16, 820226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang C-M, Li CJ, Vågbø CB, Shi Y, Wang W-L, Song S-H, Lu Z, Bosmans RPG, Dai Q, Hao Y-J, Yang X, Zhao W-M, Tong W-M, Wang X-J, Bogdan F, … He C (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular Cell, 49(1), 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang Y-G, & He C (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nature Chemical Biology, 7(12), 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, Ben-Haim MS, Eyal E, Yunger S, Pinto Y, Jaitin DA, Viukov S, Rais Y, Krupalnik V, Chomsky E, … Hanna JH (2015). Stem cells. M6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science, 347(6225), 1002–1006. [DOI] [PubMed] [Google Scholar]
- 135.Roundtree IA, Luo G-Z, Zhang Z, Wang X, Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, He E, Shen B, & He C (2017). YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. ELife, 6, e31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Anders M, Chelysheva I, Goebel I, Trenkner T, Zhou J, Mao Y, Verzini S, Qian S-B, & Ignatova Z (2018). Dynamic m6A methylation facilitates mRNA triaging to stress granules. Life Science Alliance, 1(4), e201800113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xu H, Dzhashiashvili Y, Shah A, Kunjamma RB, Weng Y-L, Elbaz B, Fei Q, Jones JS, Li YI, Zhuang X, Ming G-L, He C, & Popko B (2020). M6A mRNA Methylation Is Essential for Oligodendrocyte Maturation and CNS Myelination. Neuron, 105(2), 293–309.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, Patil DP, Kwak H, Lee JH, & Jaffrey SR (2019). M6A enhances the phase separation potential of mRNA. Nature, 571(7765), 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bartel DP (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 116(2), 281–297. [DOI] [PubMed] [Google Scholar]
- 140.Wu L, & Belasco JG (2008). Let me count the ways: Mechanisms of gene regulation by miRNAs and siRNAs. Molecular Cell, 29(1), 1–7. [DOI] [PubMed] [Google Scholar]
- 141.Dugas JC, Cuellar TL, Scholze A, Ason B, Ibrahim A, Emery B, Zamanian JL, Foo LC, McManus MT, & Barres BA (2010). Dicer1 and miR-219 are required for normal oligodendrocyte differentiation and myelination. Neuron, 65(5), 597–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shin D, Shin J-Y, McManus MT, Ptáček LJ, & Fu Y-H (2009). Dicer ablation in oligodendrocytes provokes neuronal impairment in mice. Annals of Neurology, 66(6), 843–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhao X, He X, Han X, Yu Y, Ye F, Chen Y, Hoang T, Xu X, Li H, Xin M, Wang F, Appel B, & Lu QR (2010). MicroRNA-Mediated control of oligodendrocyte differentiation. Neuron, 65(5), 612–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kuypers NJ, Bankston AN, Howard RM, Beare JE, & Whittemore SR (2016). Remyelinating oligodendrocyte precursor cell miRNAs from the sfmbt2 cluster promote cell cycle arrest and differentiation. Journal of Neuroscience, 36(5), 1698–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, & Hudson LD (2008). Identification of dynamically regulated MicroRNA and mRNA networks in developing oligodendrocytes. The Journal of Neuroscience, 28(45), 11720–11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Letzen BS, Liu C, Thakor NV, Gearhart JD, All AH, & Kerr CL (2010). MicroRNA expression profiling of oligodendrocyte differentiation from human embryonic stem cells. PLoS ONE, 5(5), e10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhao X, Wu J, Zheng M, Gao F, & Ju G (2012). Specification and maintenance of oligodendrocyte precursor cells from neural progenitor cells: Involvement of microRNA-7a. Molecular Biology of the Cell, 23(15), 2867–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.de Faria O Jr., Cui Q-L, Bin JM, Bull S-J, Kennedy TE, Bar-Or A, Antel JP, Colman DR, & Dhaunchak AS (2012). Regulation of miRNA 219 and miRNA Clusters 338 and 17–92 in Oligodendrocytes. Frontiers in Genetics, 3. 10.3389/fgene.2012.00046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shin D, Howng SYB, Ptáček LJ, & Fu Y-H (2012). MiR-32 and its target SLC45A3 regulate the lipid metabolism of oligodendrocytes and myelin. Neuroscience, 213, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang H, Moyano AL, Ma Z, Deng Y, Lin Y, Zhao C, Zhang L, Jiang M, He X, Ma Z, Lu F, Xin M, Zhou W, Yoon SO, Bongarzone ER, & Lu QR (2017). MiR-219 cooperates with miR-338 in myelination and promotes myelin repair in the CNS. Developmental Cell, 40(6), 566–582.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kornfeld SF, Cummings SE, Fathi S, Bonin SR, & Kothary R (2021). MiRNA-145–5p prevents differentiation of oligodendrocyte progenitor cells by regulating expression of myelin gene regulatory factor. Journal of Cellular Physiology, 236(2), 997–1012. [DOI] [PubMed] [Google Scholar]
- 152.Liu SJ, Nowakowski TJ, Pollen AA, Lui JH, Horlbeck MA, Attenello FJ, He D, Weissman JS, Kriegstein AR, Diaz AA, & Lim DA (2016). Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biology, 17(1). 10.1186/s13059-016-0932-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dong X, Chen K, Cuevas-Diaz Duran R, You Y, Sloan SA, Zhang Y, Zong S, Cao Q, Barres BA, & Wu JQ (2015). Comprehensive identification of long non-coding RNAs in purified cell types from the brain reveals functional LncRNA in OPC fate determination. PLoS Genetics, 11(12), e1005669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.He D, Wang J, Lu Y, Deng Y, Zhao C, Xu L, Chen Y, Hu Y-C, Zhou W, & Lu QR (2017). LncRNA functional networks in oligodendrocytes reveal stage-specific myelination control by an lncOL1/Suz12 complex in the CNS. Neuron, 93(2), 362–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wei H, Dong X, You Y, Hai B, Duran RC-D, Wu X, Kharas N, & Wu JQ (2021). OLIG2 regulates lncRNAs and its own expression during oligodendrocyte lineage formation. BMC Biology, 19(1), 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, & Kadener S (2014). CircRNA biogenesis competes with pre-mRNA splicing. Molecular Cell, 56(1), 55–66. [DOI] [PubMed] [Google Scholar]
- 157.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, & Kjems J (2013). Natural RNA circles function as efficient microRNA sponges. Nature, 495(7441), 384–388. [DOI] [PubMed] [Google Scholar]
- 158.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, & Rajewsky N (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature, 495(7441), 333–338. [DOI] [PubMed] [Google Scholar]
- 159.Zhang X-O, Wang H-B, Zhang Y, Lu X, Chen L-L, & Yang L (2014). Complementary sequence-mediated exon circularization. Cell, 159(1), 134–147. [DOI] [PubMed] [Google Scholar]
- 160.Li X, Zhang B, Li F, Yu K, & Bai Y (2020). The mechanism and detection of alternative splicing events in circular RNAs. PeerJ, 8, e10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Li Y, Wang F, Teng P, Ku L, Chen L, Feng Y, & Yao B (2022). Accurate identification of circRNA landscape and complexity reveals their pivotal roles in human oligodendroglia differentiation. Genome Biology, 23(1), 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Stephens AD, Banigan EJ, Adam SA, Goldman RD, & Marko JF (2017). Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Molecular Biology of the Cell, 28(14), 1984–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Dansu DK, Sauma S, & Casaccia P (2021). Oligodendrocyte progenitors as environmental biosensors. Seminars in Cell & Developmental Biology, 116, 38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hernandez M, Patzig J, Mayoral SR, Costa KD, Chan JR, & Casaccia P (2016). Mechanostimulation promotes nuclear and epigenetic changes in oligodendrocytes. The Journal of Neuroscience, 36(3), 806–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lourenço T, Paes de Faria J, Bippes CA, Maia J, Lopes-da-Silva JA, Relvas JB, & Grãos M (2016). Modulation of oligodendrocyte differentiation and maturation by combined biochemical and mechanical cues. Scientific Reports, 6(1), 21563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Segel M, Neumann B, Hill MFE, Weber IP, Viscomi C, Zhao C, Young A, Agley CC, Thompson AJ, Gonzalez GA, Sharma A, Holmqvist S, Rowitch DH, Franze K, Franklin RJM, & Chalut KJ (2019). Niche stiffness underlies the ageing of central nervous system progenitor cells. Nature, 573(7772), 130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Urbanski MM, Kingsbury L, Moussouros D, Kassim I, Mehjabeen S, Paknejad N, & Melendez-Vasquez CV (2016). Myelinating glia differentiation is regulated by extracellular matrix elasticity. Scientific Reports, 6(1), 33751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jagielska A, Norman AL, Whyte G, Vliet KJV, Guck J, & Franklin RJM (2012). Mechanical environment modulates biological properties of oligodendrocyte progenitor cells. Stem Cells and Development, 21(16), 2905–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Makhija E, Jagielska A, Zhu L, Bost AC, Ong W, Chew SY, Shivashankar GV, & Vliet KJV (2018). Mechanical strain alters cellular and nuclear dynamics at early stages of oligodendrocyte differentiation. Frontiers in Cellular Neuroscience, 12, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Engler AJ, Sen S, Sweeney HL, & Discher DE (2006). Matrix elasticity directs stem cell lineage specification. Cell, 126(4), 677–689. [DOI] [PubMed] [Google Scholar]
- 171.Budday S, Nay R, de Rooij R, Steinmann P, Wyrobek T, Ovaert TC, & Kuhl E (2015). Mechanical properties of gray and white matter brain tissue by indentation. Journal of the Mechanical Behavior of Biomedical Materials, 46, 318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Weickenmeier J, de Rooij R, Budday S, Steinmann P, Ovaert TC, & Kuhl E (2016). Brain stiffness increases with myelin content. Acta Biomaterialia, 42, 265–272. [DOI] [PubMed] [Google Scholar]
- 173.Weickenmeier J, de Rooij R, Budday S, Ovaert TC, & Kuhl E (2017). The mechanical importance of myelination in the central nervous system. Journal of the Mechanical Behavior of Biomedical Materials, 76, 119–124. [DOI] [PubMed] [Google Scholar]
- 174.Chatelin S, Constantinesco A, & Willinger R (2010). Fifty years of brain tissue mechanical testing: From in vitro to in vivo investigations. Biorheology, 47(5–6), 255–276. [DOI] [PubMed] [Google Scholar]
- 175.Elkin BS, Ilankovan A, & Morrison B (2010). Age-dependent regional mechanical properties of the rat hippocampus and cortex. Journal of Biomechanical Engineering, 132(1), 011010. [DOI] [PubMed] [Google Scholar]
- 176.Saha K, Keung AJ, Irwin EF, Li Y, Little L, Schaffer DV, & Healy KE (2008). Substrate modulus directs neural stem cell behavior. Biophysical Journal, 95(9), 4426–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Jain N, Iyer KV, Kumar A, & Shivashankar GV (2013). Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proceedings of the National Academy of Sciences, 110(28), 11349–11354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Nawaz S, Sánchez P, Schmitt S, Snaidero N, Mitkovski M, Velte C, Brückner BR, Alexopoulos I, Czopka T, Jung SY, Rhee JS, Janshoff A, Witke W, Schaap IAT, Lyons DA, & Simons M (2015). Actin filament turnover drives leading edge growth during myelin sheath formation in the central nervous system. Developmental Cell, 34(2), 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zuchero JB, Fu M, Sloan SA, Ibrahim A, Olson A, Zaremba A, Dugas JC, Wienbar S, Caprariello AV, Kantor C, Leonoudakis D, Lariosa-Willingham K, Kronenberg G, Gertz K, Soderling SH, Miller RH, & Barres BA (2015). CNS myelin wrapping is driven by actin disassembly. Developmental Cell, 34(2), 152–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.de Lanerolle P, & Serebryannyy L (2011). Nuclear actin and myosins: Life without filaments. Nature Cell Biology, 13(11), 1282–1288. [DOI] [PubMed] [Google Scholar]
- 181.Serebryannyy LA, Cruz CM, & de Lanerolle P (2016). A role for nuclear actin in HDAC 1 and 2 regulation. Scientific Reports, 6(1), 28460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Park H-J, Tsai E, Huang D, Weaver M, Frick L, Alcantara A, Moran JJ, Patzig J, Melendez-Vasquez CV, Crabtree GR, Feltri ML, Svaren J, & Casaccia P (2022). ACTL6a coordinates axonal caliber recognition and myelination in the peripheral nerve. IScience, 25(4), 104132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chang L, Azzolin L, Biagio DD, Zanconato F, Battilana G, Xiccato RL, Aragona M, Giulitti S, Panciera T, Gandin A, Sigismondo G, Krijgsveld J, Fassan M, Brusatin G, Cordenonsi M, & Piccolo S (2018). The SWI/SNF complex is a mechanoregulated inhibitor of YAP and TAZ. Nature, 563(7730), 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Matsumoto S, Banine F, Feistel K, Foster S, Xing R, Struve J, & Sherman LS (2016). Brg1 directly regulates Olig2 transcription and is required for oligodendrocyte progenitor cell specification. Developmental Biology, 413(2), 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jagielska A, Lowe AL, Makhija E, Wroblewska L, Guck J, Franklin RJM, Shivashankar GV, & Vliet KJV (2017). Mechanical strain promotes oligodendrocyte differentiation by global changes of gene expression. Frontiers in Cellular Neuroscience, 11, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Coste B, Mathur J, Schmidt M, Earley TJ, Ranade S, Petrus MJ, Dubin AE, & Patapoutian A (2010). Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science, 330(6000), 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Saotome K, Murthy SE, Kefauver JM, Whitwam T, Patapoutian A, & Ward AB (2018). Structure of the mechanically activated ion channel Piezo1. Nature, 554(7693), 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Guo YR, & MacKinnon R (2017). Structure-based membrane dome mechanism for Piezo mechanosensitivity. ELife, 6, e33660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Velasco-Estevez M, Koch N, Klejbor I, Caratis F, & Rutkowska A (2022). Mechanoreceptor piezo1 is downregulated in multiple sclerosis brain and is involved in the maturation and migration of oligodendrocytes in vitro. Frontiers in Cellular Neuroscience, 16. 10.3389/fncel.2022.914985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Low BC, Pan CQ, Shivashankar GV, Bershadsky A, Sudol M, & Sheetz M (2014). YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS Letters, 588(16), 2663–2670. [DOI] [PubMed] [Google Scholar]
- 191.Andreu I, Granero-Moya I, Chahare NR, Clein K, Molina-Jordán M, Beedle AEM, Elosegui-Artola A, Abenza JF, Rossetti L, Trepat X, Raveh B, & Roca-Cusachs P (2022). Mechanical force application to the nucleus regulates nucleocytoplasmic transport. Nature Cell Biology, 24(6), 896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.García-García M, Sánchez-Perales S, Jarabo P, Calvo E, Huyton T, Fu L, Ng SC, Sotodosos-Alonso L, Vázquez J, Casas-Tintó S, Görlich D, Echarri A, & Del Pozo MA (2022). Mechanical control of nuclear import by Importin-7 is regulated by its dominant cargo YAP. Nature Communications, 13(1), 1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Shimizu T, Osanai Y, Tanaka KF, Abe M, Natsume R, Sakimura K, & Ikenaka K (2017). YAP functions as a mechanotransducer in oligodendrocyte morphogenesis and maturation. Glia, 65(2), 360–374. 10.1002/glia.23096 [DOI] [PubMed] [Google Scholar]
- 194.Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PCDP, Pinter J, Pajerowski JD, Spinler KR, Shin J-W, Tewari M, Rehfeldt F, Speicher DW, & Discher DE (2013). Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science, 341(6149), 1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Guilluy C, Osborne LD, Landeghem LV, Sharek L, Superfine R, Garcia-Mata R, & Burridge K (2014). Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nature Cell Biology, 16(4), 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Rosenberg SS, Kelland EE, Tokar E, De la Torre AR, & Chan JR (2008). The geometric and spatial constraints of the microenvironment induce oligodendrocyte differentiation. Proceedings of the National Academy of Sciences of the United States of America, 105(38), 14662–14667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, Herrmann H, Blum H, Engelkamp D, Stewart CL, Leonhardt H, & Joffe B (2013). LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell, 152(3), 584–598. [DOI] [PubMed] [Google Scholar]


