Abstract
COVID-19 has been found to affect the expression profile of several mRNAs and miRNAs, leading to dysregulation of a number of signaling pathways, particularly those related to inflammatory responses. In the current study, a systematic biology procedure was used for the analysis of high-throughput expression data from blood specimens of COVID-19 and healthy individuals. Differentially expressed miRNAs in blood specimens of COVID-19 vs. healthy specimens were then identified to construct and analyze miRNA–mRNA networks and predict key miRNAs and genes in inflammatory pathways. Our results showed that 171 miRNAs were expressed as outliers in box plot and located in the critical areas according to our statistical analysis. Among them, 8 miRNAs, namely miR-1275, miR-4429, miR-4489, miR-6721-5p, miR-5010-5p, miR-7110-5p, miR-6804-5p and miR-6881–3p were found to affect expression of key genes in NF-KB, JAK/STAT and MAPK signaling pathways implicated in COVID-19 pathogenesis. In addition, our results predicted that 25 genes involved in above-mentioned inflammatory pathways were targeted not only by these 8 miRNAs but also by other obtained miRNAs (163 miRNAs). The results of the current in silico study represent candidate targets for further studies in COVID-19.
Keywords: COVID-19, MicroRNA (miRNA), Inflammation, JAK-STAT, MAPK, NF-κB
1. Introduction
Health care systems across the world have been noticeably influenced by Coronavirus disease 2019 (COVID-19) pandemic (Das et al., 2021; Rakhsha et al., 2020). COVID-19 mortalities and morbidities including post-recovery syndromes mainly result from immune-homeostasis disruption in infected people (Choudhury and Mukherjee, 2020). It has recently been found that spike glycoprotein, which is the major infectious surface protein of SARS-CoV-2, acts as a ligand for human TLR4 (Choudhury et al., 2022), and this protein–protein interaction activates transcription factors such as NF-κB, interferon regulatory factor and activator protein 1 (AP-1) that encode pro-inflammatory cytokines (Patra et al., 2021a) such as CC-chemokine ligand 2 (CCL2), interleukin 6 (IL-6), tumor necrosis factor (TNF), C-reactive protein (CRP), and CXC-chemokine ligand 10 (CXCL10) (Jiang et al., 2022). These inflammatory mediators are capable of activating various adaptive and innate immune cells, which in turn amplifies the inflammatory environment and leads to clear immunopathology, that is ‘cytokine storm’. Major organs such as heart, pancreas, and kidney are directly damaged by cytokine storm (Choudhury et al., 2021; Mukherjee, 2022; Patra et al., 2021b). Note that inflammatory molecules not only increase the progress of the disease early during viral infection but also remain in the patients' plasma after acute infection and may impact patients surviving COVID-19 in long term (Patel et al., 2021). It has been found that this hyper-inflammation, known as Multisystem Inflammatory Syndrome (MIS), affects patients 4 to 6 weeks after their infection (Yao et al., 2021) and leads to shock, gastrointestinal symptoms, cardiac dysfunction, and increased inflammatory markers (Mezochow et al., 2022).
Despite the existence of multiple vaccine options and treatments across the globe to prevent COVID-19 infection, the majority of these are not sufficiently efficient to target late-stage cytokine storm associated with COVID-19. Hence, in addition to the prevention or blocking of the entrance of the virus, greater attention should be paid to the reduction of hyperinflammation induced by COVID-19 (Zoulikha et al., 2022).
According to the literature, multiple physiological parameters change due to infection and cytokine storm, among which, microRNAs (miRNAs) can represent this poor state given their pivotal contribution to various essential biological processes (Garcia-Giralt et al., 2022). These processes include neuronal development, metabolic control, muscle differentiation, stem cell differentiation, cell cycle, and immune modulation. Thus, miRNA dysfunction can be linked to a diverse range of diseases, such as cancers, immune system disorders, and cardiovascular diseases (Condrat et al., 2020).
As small non-coding RNAs with an approximate length of 20 to 25 nucleotides, miRNAs are responsible for regulating the expression of several target genes via sequence-specific hybridization to the 3′ untranslated region (UTR) of messenger RNAs. These miRNAs can either prevent the translation or directly degrade their target messenger RNAs (Leichter et al., 2017). Regarding the fact that perfect complementarity is not needed for target recognition by miRNAs, a single miRNA is able to regulate several messenger RNAs. Despite the slight impact of miRNAs on each messenger RNA's target, the combined impact is considerable and generates notable phenotypic results. The fact that the majority of miRNAs are conserved across several species of animals indicates the evolutionary significant modulatory role of these molecules in essential biological processes and pathways (Christopher et al., 2016).
These targeting pathways of miRNAs in human diseases represent a novel and potentially strong therapeutic candidate for different pathological afflictions (Christopher et al., 2016). In addition, as reported in many research works, miRNAs can be released into extracellular fluids including plasma and serum, saliva, cerebrospinal fluid, breast milk, tears, urine, peritoneal fluid, colostrum, bronchial lavage, ovarian follicular fluid, and seminal fluid. Extracellular miRNAs, in contrast to cellular RNAs, demonstrate high stability, such that they can resist degradation when exposed to ambient temperature for up to four days and to harmful conditions including low or high pH, several freeze-thaw cycles, and boiling. Hence, extracellular miRNAs can be employed as biomarkers for various diseases(O'Brien et al., 2018).
According to the above discussion, the present in silico research was designed to discover the most relevant regulatory miRNAs of major inflammatory pathways (JAK-STAT, NF-kb, and MAPK) that are dysregulated in COVID-19 infection.
2. Methods
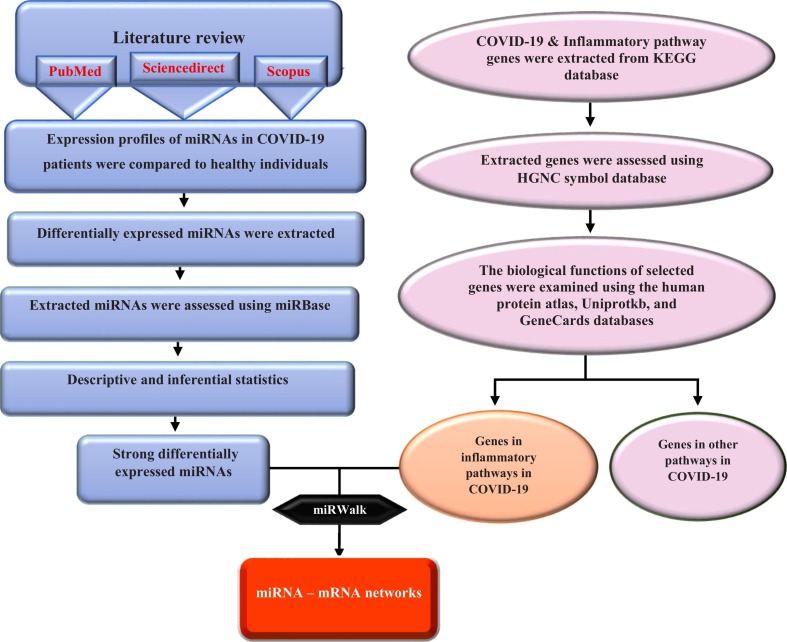
In the current study, a systematic biologic procedure was used for the analysis of high-throughput expression data from blood samples of COVID-19 and healthy individuals. Since it is only an in silico study, we did not include any patients or control samples in this study. We aimed to detect differentially expressed miRNAs in blood samples of COVID-19 vs. healthy specimens to construct and analyze miRNA–mRNA networks and predict key miRNAs and genes in inflammatory pathways. Fig. 1 briefly illustrates the steps applied in the in silico analysis (Fig. 1).
Fig. 1.
Study design flowchart.
2.1. miRNA expression profiling
Firstly, we searched web-based references in which the expression profiles of miRNAs in blood samples of COVID-19 patients are provided in comparison to healthy individuals. The search criteria included COVID-19 patients in all severity-based categories, mild, moderate as well as severe respiratory failure, diagnosed by means of available qPCR Detection Kits. Among all the available sources, miRNA expression profiles of 11 paired datasets which included the profile of COVID-19 patients versus healthy samples were selected for further investigation (Table 1 ). In the following steps, the expression profiles of the selected miRNAs were processed using statistical methods. To do so, we used Shapiro-Wilks and Kolmogorov-Smirnov tests with 0.01 and 0.001 significant levels for investigating normality of data. After that, some parametric-statistical tests were applied to determine the critical area of data in comparison to mean of data. To investigate how data are distributed, Box-Plot with test, frequency table, as well as some appropriate plots such as histogram and line plot, were depicted.
Table 1.
Summary of high-throughput research methodology to assess miRNA expression profiles in COVID-19 patients.
| Group | Sample | Method | Reference |
|---|---|---|---|
| COVID-19/control | Whole blood | High-throughput sequencing -Illumina HiseqX Ten platform | Li et al. (2020) |
| COVID-19/control | Whole blood Plasma cfRNA |
BGISEQ-500 sequencer | Yang et al. (2021) |
| COVID-19/control | Whole blood | Multi-transcriptome sequencing -Illumina Hiseq 4000 platform | Tang et al. (2020) |
| COVID-19/control | Peripheral blood/plasma | MiRCURY LNA miRNome qPCR panels | Fayyad-Kazan et al. (2021) |
| COVID-19/control | Peripheral blood | Whole-Transcriptome RNA Sequencing-Illumina HiSeq X Ten platform | Li et al. (2021) |
| COVID-19/control | Plasma sample | Small RNA sequencing- Illumina | Farr et al. (2021) |
| COVID-19/control | Whole blood/ Plasma | miRNA sequencing- HiSeq 2500 platform Illumina | Nicoletti et al. (2022) |
| COVID-19/control | Peripheral blood | miRNA sequencing- Illumina | Duecker et al., 2021 |
| COVID-19/control | Blood/plasma | HTG EdgeSeq miRNA Whole Transcriptome Assay | Akula et al. (2022) |
| COVID-19/control | Plasma | Small RNA sequencing- Illumina | Gutmann et al. (2022) |
| COVID-19/control | Plasma | Small RNA sequencing- Illumina | Fernández-Pato et al. (2022) |
2.2. Identification of key genes in inflammatory signaling pathways in COVID-19
KEGG pathway database (https://www.genome.jp/kegg/) was used to determine key genes involved in inflammatory signaling pathways in COVID-19 pathogenesis. This database provides a set of depicted pathway maps indicating molecular interaction and relation networks for genetic and environmental information processing, cellular processes, organismal systems, metabolism, human diseases, and drug development. Based on KEGG database, genes involved in common inflammatory pathways, i.e. JAK/STAT, NF-KB, and MAPK were obtained. We further assessed our selected genes using HGNC symbol database (https://www.genenames.org/), a web-based resource in which human gene nomenclatures are approved. After that, genes that not only participated in the above-mentioned inflammatory pathways but also made a unique contribution in COVID-19 pathogenesis were chosen for further investigation.
2.3. miRNA – mRNA networks prediction
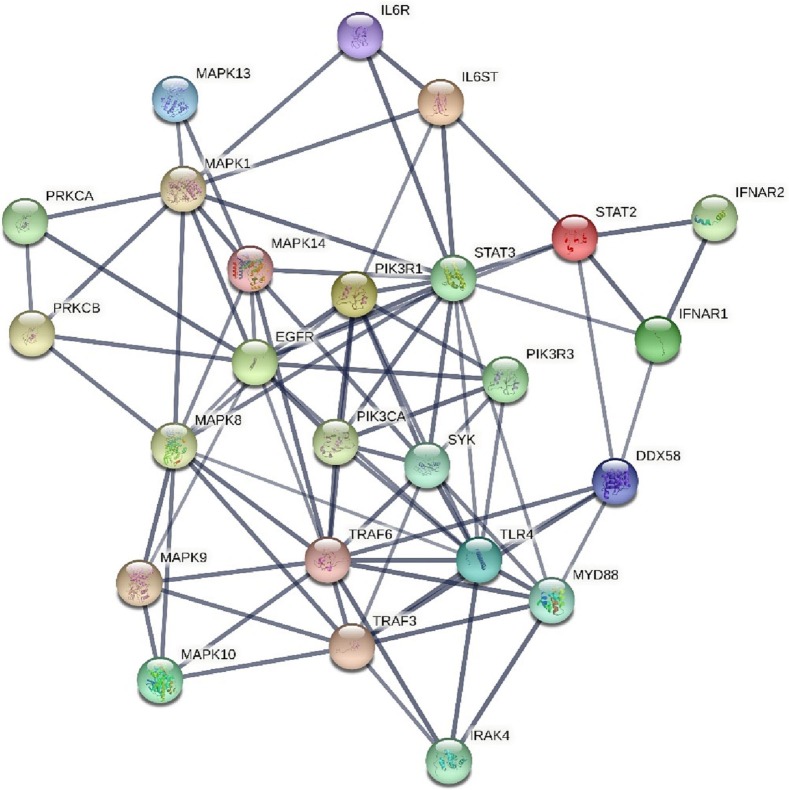
In this report, we applied the miRWalk (http://mirwalk.umm.uni-heidelberg.de/), an open-source high accuracy platform, to predict miRNAs of interest and inflammatory target gene interactions. This online tool provides the most comprehensive collection of predicted and experimentally verified miRNA-target gene interactions based on TarPmiR algorithm which utilizes a random-forest based learning approach to predict miRNA target sites as accurately as possible. Furthermore, miRWalk database is capable of extracting information from popular gene-miRNA prediction databases such as TargetScan (https://www.targetscan.org/vert_80/), miRDB (http://mirdb.org/), and miRTarBase (https://mirtarbase.cuhk.edu.cn/∼miRTarBase/miRTarBase_2022/php/index.php). To validate protein-protein interaction networks, we recruited STRING database (https://string-db.org/) in which all known and predicted physical and functional protein associations are integrated. For miRNA–mRNA networks construction and analysis, the Cytoscape software version 1.7.3 was used. To do so, the gained miRNA–protein interactions were chosen and depicted in this open source bioinformatics software.
2.4. Biological functions of genes potentially dysregulated in COVID-19 pathogenesis
Among all genes playing critical roles in COVID-19 pathogenesis, some potentially dysregulated genes were selected and further investigated in terms of molecular and biological functions. For this purpose, the human protein atlas database (https://www.proteinatlas.org/) providing the expression levels of mRNAs and proteins of human genes, as well as their biological roles, according to different genomics technologies such as transcriptomics and systems biology was employed. Moreover, other protein-based databases including UniProtKB (https://www.uniprot.org/help/uniprotkb) as well as GeneCards (https://www.genecards.org/) were utilized for further assessment.
3. Results
3.1. miRNA expression profile processing
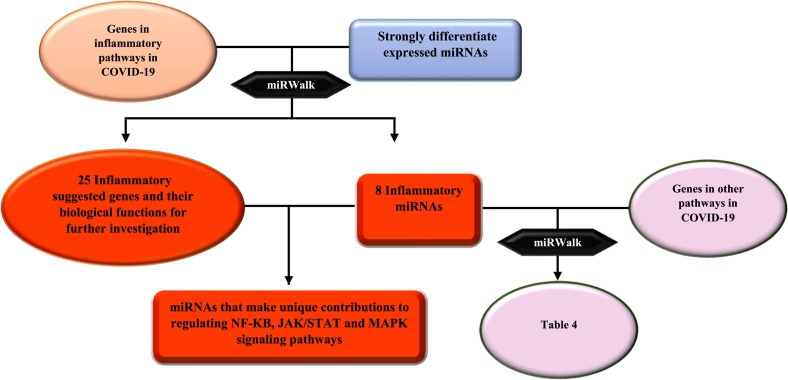
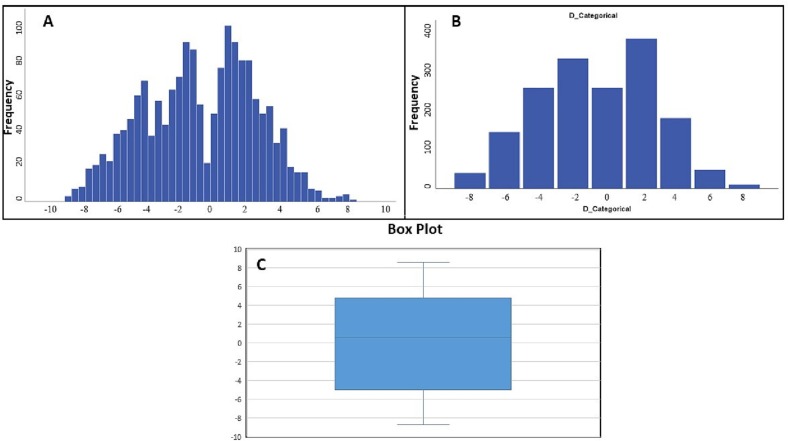
Figure 2 briefly illustrates the results obtained in this study (Fig. 2). At first, miRNA expression profiles in blood samples of COVID-19 vs. healthy specimens were obtained according to previously described criteria. The demographic characteristics of selected paired samples were succinctly recorded (Table 2 ). As shown in Table 2, the enrolled COVID-19 and healthy participants belonged to various sex and age-groups in all severity-based categories, i.e. mild, moderate, and severe respiratory failure. We selected differentially expressed miRNAs in blood samples of COVID-19 vs. healthy samples for further assessment. The statistical processing of the miRNA expression profiles was implemented using descriptive and inferential statistics. The normality of data was strongly confirmed via Shapiro–Wilk test in which the null hypothesis is a sample x1, …, xn coming from a normally distributed population. In different studies, confidence levels (1-α) % of 95 or 99% are frequently used for both tests, with the significance level (α)% equal to 1% or 5%. Considering the main and critical condition for the parametric test, normality, t-test with a confidence level of 95% was conducted, which is a statistical hypothesis test used to determine whether an unknown population mean is different from a specific value. The results indicated significant difference between the reported mean of miRNA expression and the mean of existing data. Confidence intervals for the expression levels of miRNA (log2 fold change) were regarded from −5 to 4.8. Based on the outliers in this box plot (Fig. 2.C), miRNAs expressed outside of this interval were in our critical areas of interest (more than max and less than min). Furthermore, Box-Plot with test results indicated that miRNAs expressed outside of the confidence intervals and entering quartile distance can be selected for further investigation. Our results showed that 171 miRNAs were expressed outside of the defined and entered quartile distance (Fig. 3 ).
Fig. 2.
Short description of obtained results in this study.
Table 2.
Demographic features of COVID-19 patients considered in this study.
| Group | Population | Sex(male/female) | Age (years) | Symptoms | Reference |
|---|---|---|---|---|---|
| COVID-19 | 10 | 4/6 | 44.90 ± 19.94 | Mild Moderate |
Li et al. (2020) |
| Control | 4 | 2/2 | 44.75 ± 11.84 | – | |
| COVID-19 | 5 | – | – | Covid-19 patient | Yang et al. (2021) |
| Control | 3 | – | – | – | |
| COVID-19 | 6 6 |
4/2 5/1 |
50–89 60–89 |
Moderate Severe |
Tang et al. (2020) |
| Control | 4 | 2/2 | 50–69 | – | |
| COVID-19 | 14 13 6 |
7/6 9/5 4/2 |
39.29 ± 7.477 46.31 ± 8.548 57.67 ± 2.16 |
Mild Moderate Severe |
Fayyad-Kazan et al. (2021) |
| Control | 10 | – | – | – | |
| COVID-19 | 10 | 4/6 | 44:90 ± 19:94 | – | Li et al. (2021) |
| Control | 4 | 1/3 | 34:75 ± 11:84 | – | |
| COVID-19 | 10 | 4/6 | 53.5 ± 17.2 | – | Farr et al. (2021) |
| Control | 10 | 4/6 | 53 ± 17.6 | – | |
| COVID-19 | 4 4 |
2/2 2/2 |
61.8 ± 11.7 64.0 ± 8.6 |
Mild/moderate severe/critical |
Nicoletti et al. (2022) |
| Control | 4 | 2/2 | 62.8 ± 14.9 | – | |
| COVID-19 | 11 10 |
9/2 8/2 |
76 (48–91) 69 (52–79) |
Moderate Severe |
Duecker et al. (2021) |
| Control | 8 | 5/3 | 68 (49–89) | – | |
| COVID-19 | 12 | 6/6 | 47.8 ± 9.8 | Moderate Severe |
Akula et al. (2022) |
| Control | 8 | 5/3 | 46 ± 7.3 | – | |
| COVID-19 | 18 18 |
56٪/44٪ 42٪/58٪ |
55.0 (36.0, 66.0) 58.0 (39.0, 66.0) |
Mild to Moderate Severe |
Gutmann et al. (2022) |
| Control | 11 | 42٪/58٪ | 40.0 (30.0, 46.0) | – | |
| COVID-19 | 32 52 12 |
23/9 26/26 4/8 |
63.4 (52.9–78.3) 59.4 (53.0–71.8) 66.2 (44.4–72.6) |
Severe Moderate Asymptomatic/Mild |
Fernández-Pato et al. (2022) |
| Control | 13 | 7/6 | 66.7 (57–68.9) | – |
Fig. 3.
A) Frequency distribution of miRNAs involved in COVID-19 pathogenesis. X-axis shows the miRNA expression profiling intervals based on fold changes reported for miRNA expression in high-throughput studies. Y-axis shows the frequency of miRNAs in determined intervals B) Frequency distribution of categorical miRNAs involved in COVID-19 pathogenesis. X-axis shows the miRNA expression profiling intervals based on fold changes reported for miRNA expression in high-throughput studies. Y-axis shows the frequency of miRNAs in determined intervals. Y-axis shows the frequency of miRNAs in determined intervals (0–400) C) Box-Plot with test of miRNAs involved in COVID-19 pathogenesis. miRNA expression profiling values (From bottom to top of the chart: Minimum, Quartile 1, Median, Quartile 3, Maximum).
At first, miRNA expression profiles in blood samples of COVID-19 vs. healthy specimens were obtained according to previously described criteria. The demographic characteristics of the selected paired samples were succinctly recorded (Table 2). As shown in Table 2, the enrolled COVID-19 and healthy participants were ranged in various sex and age-groups in all severity-based categories, i.e. mild, moderate as well as severe respiratory failure. We selected differentially expressed miRNAs in blood samples of COVID-19 vs. healthy specimens for further assessment. The statistical processing of the miRNA expression profiles was implemented using descriptive and inferential statistics. The normality of data was strongly confirmed. Considering the main and critical condition for parametric test, normality, t-test was conducted and the results indicated the significant difference between the reported mean of miRNA expression and the mean of existing data. Confidence intervals for the expression levels of miRNAs (log2 fold change) were regarded from −5 to 4.8. Based on the defined confidence intervals, miRNAs expressed out of this interval, were in our critical areas of interest. Furthermore, Box-Plot with test results indicated that miRNAs expressed out of enter quartile distance can be selected for further investigation. Our results have shown that 171 miRNAs are expressed out of defined confidence intervals and entered quartile distance (Fig. 3).
3.2. Potential capability of dysregulated miRNAs in regulating inflammatory signaling pathways in COVID-19
Looking into KEGG database, genes encoding proteins that are related to COVID-19 pathogenesis as well as genes involved in common inflammatory signaling pathways, JAK/STAT, NF-KB, and MAPK were gained. HGNC was recruited and extracted gene names were checked and corrected, accordingly. As a result, we obtained 74 genes playing roles in so-called inflammatory pathways in COVID-19 pathogenesis. To evaluate whether selected inflammatory genes are the genuine targets of determined miRNAs (miRNAs that were expressed outside of the defined confidence intervals and entered quartile distance), miRWalk database was used. Our results revealed that many components of inflammatory pathways associated with COVID-19 pathogenesis were the targetome of determined miRNAs. Considering predicted miRNA-gene interactions, 8 strong differentially expressed miRNAs, including miR-1275, miR-4429, miR-4489, miR-6721-5p, miR-5010-5p, miR-7110-5p, miR-6804-5p and miR-6881–3p, affecting about one third of inflammatory genes, were selected as candidates for further investigation.
3.3. miRNA – mRNA networks analysis and construction
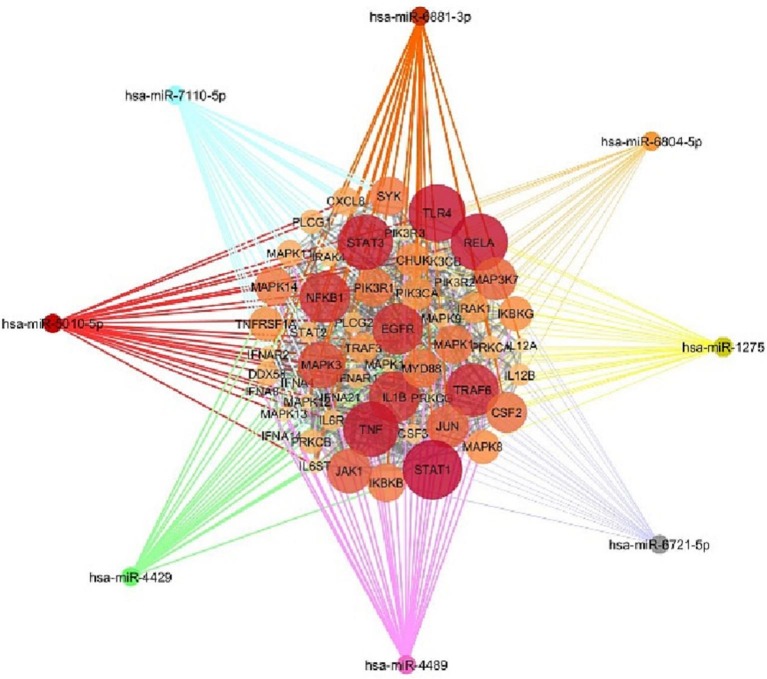
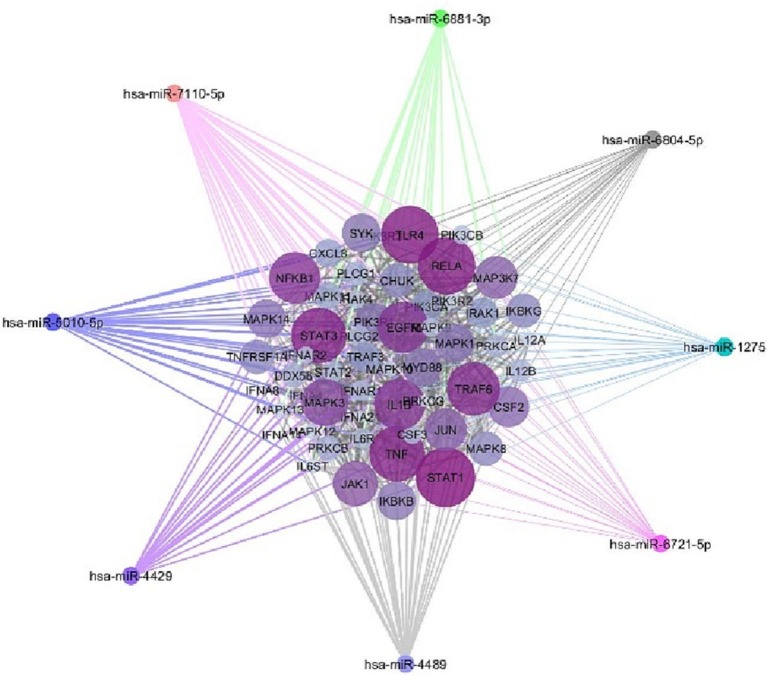
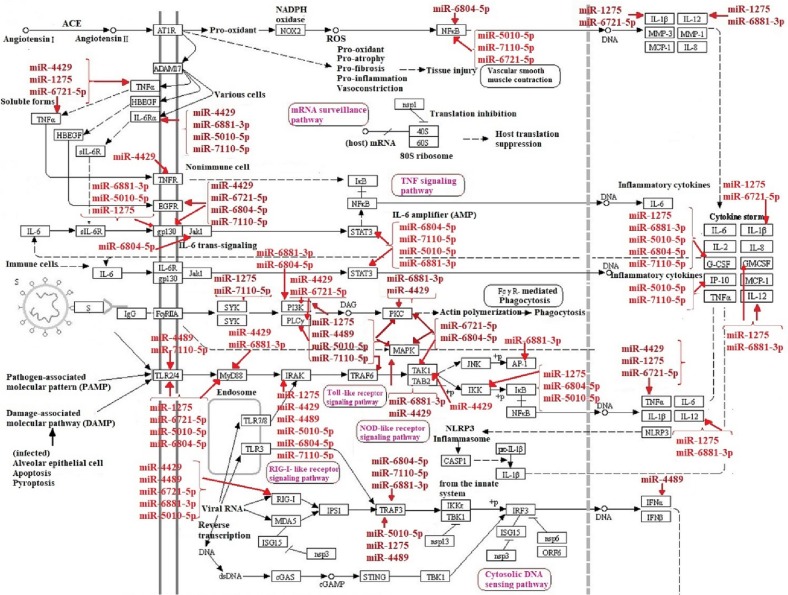
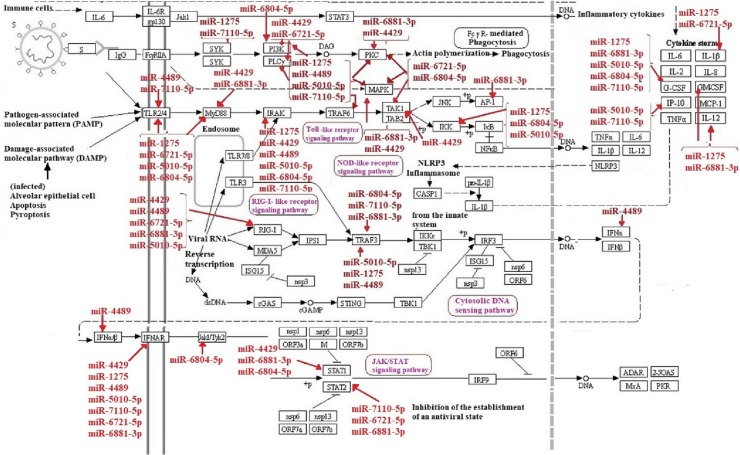
Altogether, miRWalk web tool predicted 202 interactions between inflammatory genes and miRNAs of interest. Protein-protein interaction network was gained using STRING database with confidence edges and high confidence score (0.700). In the following step, the complete list of the interactions of the determined miRNAs of interest and inflammatory genes along with protein-protein interactions was imported into Cytoscape software version 1.7.3, analyzed, and then visualized (Fig. 4 , Table 3 ). As shown in Fig. 4, genes that have more interactions are shown larger in size. Moreover, there is direct relation between the number of protein interactions and color intensity. Regarding the striking roles of miRNAs of interest in regulating inflammatory genes in COVID-19 pathogenesis, we focused on the effects of miRNAs of interest in regulating other genes associated with COVID-19 pathogenesis. Potential target genes of miRNAs of interest in other signaling pathways involved in COVID-19 pathogenesis are summarized in Table 4 . Accordingly, 252 miRNA-gene interactions were predicted and pictured in Cytoscape software (Fig. 5 ). Apart from this, the role of miRNAs of interest (miR-1275, miR-4429, miR-4489, miR-6721-5p, miR-5010-5p, miR-7110-5p, miR-6804-5p and miR-6881–3p) in regulating key genes in NF-KB, JAK/STAT and MAPK signaling pathways involved in COVID-19 pathogenesis was pictured based on KEGG data base (Fig. 6, Fig. 7 ). (See Fig. 8.)
Fig. 4.
The potential interactions between potential target genes of miRNAs of interest in NF-KB, JAK/STAT and MAPK signaling pathways participating in COVID-19 pathogenesis and miR-1275, miR-4429, miR-4489, miR-6721-5p, miR-5010-5p, miR-7110-5p, miR-6804-5p and miR-6881–3p were depicted by Cytoscape. Genes that have more interactions are indicated larger in size. Moreover, there is direct relation between the number of protein interactions and color intensity.
Table 3.
Potential targets of miRNAs involved in COVID-19 pathogenesis.
| Gene | miRNA |
|---|---|
| MAPK1 | miR-4429, miR-6721-5p, miR-5010-5p, miR-6804-5p, miR-4489, miR-7110-5p |
| MAPK3 | miR-1275, miR-6881–3p, miR-5010-5p, miR-6804-5p, miR-4489, miR-7110-5p |
| MAPK8 | miR-5010-5p, miR-6804-5p, miR-4489 |
| MAPK9 | miR-4429, miR-1275, miR-6721-5p, miR-5010-5p, miR-6804-5p, miR-4489, miR-7110-5p |
| MAPK10 | miR-4429, miR-1275, miR-6881–3p, miR-5010-5p, miR-6804-5p,miR-4489, miR-7110-5p, miR-6721-5p |
| MAPK11 | miR-4429, miR-6721-5p,miR-4489, miR-7110-5p |
| MAPK12 | miR-6881–3p, miR-7110-5p |
| MAPK13 | miR-4429, miR-1275, miR-6721-5p, miR-6881–3p, miR-6804-5p, miR-4489, miR-7110-5p, miR-5010-5p |
| MAPK14 | miR-4429, miR-1275, miR-6804-5p, miR-5010-5p, miR-4489 |
| MAP3K7 | miR-4429, miR-6721-5p, miR-6804-5p |
| PRKCG | miR-4429, miR-1275, miR-6721-5p, miR-5010-5p, miR-4489, miR-7110-5p |
| EGFR | miR-4429, miR-6721-5p, miR-6804-5p, miR-7110-5p |
| STAT1 | miR-4429, miR-6881–3p, miR-6804-5p |
| DDX58 | miR-4429, miR-6721-5p, miR-6881–3p, miR-5010-5p, miR-4489 |
| PLCG2 | miR-4429, miR-1275, miR-6721-5p, miR-4489, miR-7110-5p |
| PIK3CA | miR-4429, miR-1275, miR-6881–3p, miR-5010-5p, miR-4489 |
| IL6R | miR-4429, miR-6881–3p, miR-5010-5p, miR-7110-5p |
| TNF | miR-4429, miR-1275, miR-6721-5p |
| IFNAR1 | miR-4429, miR-1275, miR-5010-5p, miR-4489, miR-7110-5P |
| IRAK1 | miR-4429, miR-4489 |
| TNFRSF1A | miR-4429 |
| PIK3R3 | miR-4429, miR-1275, miR-6721-5p, miR-6881–3p, miR-5010-5p, miR-6804-5p, miR-4489, miR-7110-5p |
| IRAK4 | miR-4429, miR-1275, miR-5010-5p, miR-6804-5p, miR-7110-5p |
| MYD88 | miR-4429, miR-1275, miR-6721-5p, miR-6881–3p, miR-5010-5p, miR-6804-5p |
| IL6ST | miR-4429, miR-1275, miR-6721-5p, miR-6881–3p, miR-5010-5p, miR-6804-5p, miR-7110-5p |
| PIK3R1 | miR-4429, miR-1275, miR-6881–3p, miR-5010-5p, miR-6804-5p, miR-7110-5p |
| SYK | miR-1275, miR-7110-5p |
| IKBKB | miR-1275, miR-6721-5p, miR-6881–3p, miR-6804-5p, miR-4489 |
| TRAF3 | miR-1275, miR-5010-5p, miR-6804-5p, miR-4489, miR-7110-5p, miR-6881–3p |
| PIK3CB | miR-1275, miR-6721-5p, miR-7110-5p |
| CHUK | miR-1275, miR-5010-5p, miR-6804-5p |
| IL12B | miR-1275 |
| TLR4 | miR-1275, miR-6721-5p, miR-5010-5p, miR-6804-5p, miR-4489, miR-7110-5p |
| TRAF6 | miR-1275, miR-5010-5p, miR-4489, miR-7110-5p |
| IL1B | miR-1275, miR-6721-5p |
| CSF3 | miR-1275, miR-6881–3p, miR-5010-5p, miR-6804-5p, miR-7110-5p |
| IFNAR2 | miR-1275, miR-6721-5p, miR-6881–3p, miR-7110-5p |
| PRKCA | miR-6721-5p, miR-6881–3p, miR-5010-5p, miR-4489 |
| STAT2 | miR-6721-5p, miR-6881–3p, miR-7110-5p |
| STAT3 | miR-6804-5p, miR-7110-5p, miR-5010-5p, miR-6881–3p |
| JUN | miR-6881–3p |
| RELA | miR-5010-5p, miR-7110-5p, miR-6721-5p |
| IL12A | miR-6881–3p |
| PLCG1 | miR-7110-5p, miR-5010-5p |
| PRKCB | miR-4489, miR-7110-5p, miR-6804-5p |
| CXCL8 | miR-5010-5p, miR-7110-5p |
| JAK1 | miR-6804-5p |
| NFKB1 | miR-6804-5p |
| IKBKG | miR-4489, miR-7110-5p |
| IFNA8 | miR-4489 |
| IFNA14 | miR-4489 |
| IFNA21 | miR-4489 |
| CSF2 | miR-6881–3p,miR-1275 |
| PIK3R2 | miR-4489 |
| IFNA4 | miR-4489 |
Table 4.
Potential target genes of miRNAs of interest in other signaling pathways involved in COVID-19 pathogenesis.
| miRNA | Gene | Biological process |
|---|---|---|
| hsa-miR-5010-5P | ACE | Renin-Angiotensin Pathway |
| ADAR | Antiviral defense, Immunity, Innate immunity, mRNA processing, RNA-mediated gene silencing | |
| C1QC, C2 | Complement pathway, Immunity, Innate immunity | |
| CASP1 | Apoptosis | |
| CXCL10 | Chemotaxis, Inflammatory response | |
| CGAS | Antiviral defense, DNA damage, DNA repair, Host-virus interaction, Immunity, Innate immunity | |
| EIF2AK2 | Antiviral defense, Host-virus interaction, Immunity, Innate immunity, Transcription, Transcription regulation | |
| F13A1 | Blood coagulation, Hemostasis | |
| HBEGF | Immunity | |
| MAS1 | ACE Inhibitor Pathway, Peptide ligand-binding receptors | |
| MASP1 | Complement activation lectin pathway, Immunity, Innate immunity | |
| MAVS | Antiviral defense, Host-virus interaction, Immunity, Innate immunity | |
| MX2 | Antiviral defense, Immunity, Innate immunity, mRNA transport, Protein transport, Translocation, Transport | |
| NFKBIB | Immunity, Inflammatory response | |
| NLRP3 | Immunity, Inflammatory response, Innate immunity, Transcription, Transcription regulation | |
| OAS1,2 | Antiviral defense, Immunity, Innate immunity | |
| RPL7,10,13,14,22, | Host-virus interaction, rRNA processing, Translation, Gene expression | |
| 22 L1,27A,32,34, | ||
| 35A,37,38 | ||
| RPS4Y1,8,15A, | Host-virus interaction, rRNA processing, Translation | |
| 20,23,29, 27 L | ||
| TLR3 | Immunity, Inflammatory response, Innate immunity | |
| UBA52 | Host-virus interaction, Translation | |
| hsa-miR-6804-5P | ACE | Renin-Angiotensin Pathway |
| ADAR | Antiviral defense, Immunity, Innate immunity, mRNA processing, RNA-mediated gene silencing | |
| C7 | Complement pathway, Immunity, Innate immunity | |
| CYBB | Electron transport, Ion transport, Transport | |
| F13A1 | Blood coagulation, Hemostasis | |
| MAS1 | ACE Inhibitor Pathway, Peptide ligand-binding receptors | |
| MASP1 | Complement activation lectin pathway, Immunity, Innate immunity | |
| MAVS | Antiviral defense, Host-virus interaction, Immunity, Innate immunity | |
| OAS1,2 | Antiviral defense, Immunity, Innate immunity | |
| RPL10,13,14, | Host-virus interaction, rRNA processing, Translation, Gene expression | |
| 22 L1,28,32,37,37A | ||
| RPS15A,24,27 L | Host-virus interaction, rRNA processing, Translation | |
| TLR3 | Immunity, Inflammatory response, Innate immunity | |
| UBA52 | Host-virus interaction, Translation | |
| hsa-miR-7110-5P | ADAR | Antiviral defense, Immunity, Innate immunity, mRNA processing, RNA-mediated gene silencing |
| ADAM17 | Notch signaling pathway | |
| CASP1 | Apoptosis | |
| C1QA,C1QC | Complement pathway, Host-virus interaction, Immunity, Innate immunity | |
| C3AR,C5AR1 | Chemotaxis | |
| CXCL10 | Chemotaxis, Inflammatory response | |
| CGAS | Antiviral defense, DNA damage, DNA repair, Host-virus interaction, Immunity, Innate immunity | |
| C2,6,7,C8B | Complement pathway, Immunity, Innate immunity | |
| CYBB | Electron transport, Ion transport, Transport | |
| EIF2AK2 | Antiviral defense, Host-virus interaction, Immunity, Innate immunity, Transcription, Transcription regulation | |
| FGB, FGA | Adaptive immunity, Blood coagulation, Hemostasis, Immunity, Innate immunity | |
| HBEGF | Immunity | |
| MAS1 | ACE Inhibitor Pathway, Peptide ligand-binding receptors | |
| MASP1,2 | Complement activation lectin pathway, Immunity, Innate immunity | |
| MAVS | Antiviral defense, Host-virus interaction, Immunity, Innate immunity | |
| MX2 | Antiviral defense, Immunity, Innate immunity, mRNA transport, Protein transport, Translocation, Transport | |
| NFKBIB | Immunity, Inflammatory response | |
| OAS1,2,3 | Antiviral defense, Immunity, Innate immunity | |
| RPL10L,11,13, | Host-virus interaction, rRNA processing, Translation, Gene expression | |
| 14,22 L1,23,23A, | ||
| 27A,28,32, 36, | ||
| 37,37A,38, 41 | ||
| RPS3A,9,14, | Host-virus interaction, rRNA processing, Translation | |
| 15A,16,19,29,27 L | ||
| RPS10-NUDT3 | Transcription | |
| STING1 | Autophagy, Host-virus interaction, Immunity, Innate immunity | |
| SELP | Cell adhesion | |
| TMPRSS2 | Regulation of Androgen receptor activity, Host-virus interaction | |
| TLR2,3 | Immunity, Inflammatory response, Innate immunity | |
| UBA52 | Host-virus interaction, Translation | |
| hsa-miR-6881–3P | ACE | Renin-Angiotensin Pathway |
| CASP1 | Apoptosis | |
| C3AR1 | Chemotaxis | |
| CFD | Complement alternate pathway, Immunity, Innate immunity | |
| CYBB | Electron transport, Ion transport, Transport | |
| EIF2AK2 | Antiviral defense, Host-virus interaction, Immunity, Innate immunity, Transcription, Transcription regulation | |
| FCGR2A | Immunity | |
| MAS1 | ACE Inhibitor Pathway, Peptide ligand-binding receptors | |
| MAVS | Antiviral defense, Host-virus interaction, Immunity, Innate immunity | |
| NRP1 | Angiogenesis, Differentiation, Host-virus interaction, Neurogenesis | |
| NFKBIB | Immunity, Inflammatory response | |
| OAS1,3 | Antiviral defense, Immunity, Innate immunity | |
| RPS19,14 | Host-virus interaction, rRNA processing, Translation | |
| RPL13, 14,22, 36A,37,38 | Host-virus interaction, rRNA processing, Translation, Gene expression | |
| UBA52 | Host-virus interaction, Translation | |
| hsa-miR-6721-5P | ACE | Renin-Angiotensin Pathway |
| CFD | Complement alternate pathway, Immunity, Innate immunity | |
| EIF2AK2 | Antiviral defense, Host-virus interaction, Immunity, Innate immunity, Transcription, Transcription regulation | |
| FCGR2A | Immunity | |
| MASP1 | Complement activation lectin pathway, Immunity, Innate immunity | |
| MMP1 | Collagen degradation, Host-virus interaction | |
| MX1 | Antiviral defense, Immunity, Innate immunity | |
| MX2 | Antiviral defense, Immunity, Innate immunity, mRNA transport, Protein transport, Translocation, Transport | |
| OAS1,2 | Antiviral defense, Immunity, Innate immunity | |
| RPL4,7,10,13,22, | Host-virus interaction, rRNA processing, Translation, Gene expression | |
| 23,27A,28,31,32, | ||
| 37,37A | ||
| RPS3,4×,15A, | Host-virus interaction, rRNA processing, Translation | |
| 16,19,24,29 | ||
| RPS10-NUDT3 | Transcription | |
| RSL24D1 | RSL24D1 | |
| TLR2 | Immunity, Inflammatory response, Innate immunity | |
| hsa-miR-4429 | CYBB | Electron transport, Ion transport, Transport |
| C6,7 | Complement pathway, Immunity, Innate immunity | |
| FAU | Host-virus interaction, RNA processing, Transcription and Translation | |
| MAS1 | ACE Inhibitor Pathway, Peptide ligand-binding receptors | |
| MAVS | Antiviral defense, Host-virus interaction, Immunity, Innate immunity | |
| OAS2 | Antiviral defense, Immunity, Innate immunity | |
| RPL3,14,15,22 L1, | Host-virus interaction, rRNA processing, Translation, Gene expression | |
| 28,32,34,37,38 | ||
| RPS3,4×,19,26,29 | Host-virus interaction, rRNA processing, Translation | |
| RSL24D1 | Ribosome biogenesis | |
| STING1 | Autophagy, Host-virus interaction, Immunity, Innate immunity | |
| TLR7 | Immunity, Inflammatory response, Innate immunity | |
| UBA52 | Host-virus interaction, Translation | |
| hsa-miR-1275 | ACE | Renin-Angiotensin Pathway |
| ADAR | Antiviral defense, Immunity, Innate immunity, mRNA processing, RNA-mediated gene silencing | |
| C7 | Complement pathway, Cytolysis, Immunity, Innate immunity | |
| C1QC | Complement pathway, Immunity, Innate immunity | |
| CASP1 | Apoptosis | |
| HBEGF | Immunity | |
| IKBKE | DNA damage, Host-virus interaction | |
| MAS1 | ACE Inhibitor Pathway, Peptide ligand-binding receptors | |
| MASP2 | Complement activation lectin pathway, Immunity, Innate immunity | |
| MAVS | Antiviral defense, Host-virus interaction, Immunity, Innate immunity | |
| MBL2 | Complement activation lectin pathway, Complement pathway, Immunity, Innate immunity | |
| MX1 | Antiviral defense, Immunity, Innate immunity | |
| MX2 | Antiviral defense, Immunity, Innate immunity, mRNA transport, Protein transport, Translocation, Transport | |
| OAS2,3 | Antiviral defense, Immunity, Innate immunity | |
| RPL4,13,14, | Host-virus interaction, rRNA processing, Translation, Gene expression | |
| 23A,27A,28,31 | ||
| RPS3A,4×,19, | Host-virus interaction, rRNA processing, Translation | |
| 14,29 | ||
| RPS10-NUDT3 | Transcription | |
| STING1 | Autophagy, Host-virus interaction, Immunity, Innate immunity | |
| TMPRSS2 | Regulation of Androgen receptor activity, Host-virus interaction | |
| UBA52 | Host-virus interaction, Translation | |
| hsa-miR-4489 | ACE | Renin-Angiotensin Pathway |
| ACE2 | Host-virus interaction | |
| AGTR1 | Renin-Angiotensin Pathway | |
| C2,7 | Complement pathway, Immunity, Innate immunity | |
| EIF2AK2 | Antiviral defense, Host-virus interaction, Immunity, Innate immunity, Transcription, Transcription regulation | |
| FGB | Adaptive immunity, Blood coagulation, Hemostasis, Immunity, Innate immunity | |
| MAS1 | ACE Inhibitor Pathway, Peptide ligand-binding receptors | |
| MASP1 | Complement activation lectin pathway, Immunity, Innate immunity | |
| MX1,MX2 | Antiviral defense, Immunity, Innate immunity, mRNA transport, Protein transport, Translocation, Transport | |
| OAS1,3 | Antiviral defense, Immunity, Innate immunity | |
| RPL14,15,10,13A, | Host-virus interaction, rRNA processing, Translation, Gene expression | |
| 19,28,31,35A,37 | ||
| RPL36A-HNRNPH2 | Transcription | |
| RPS4X,19,14,29 | Host-virus interaction, rRNA processing, Translation | |
| TMPRSS2 | Regulation of Androgen receptor activity, Host-virus interaction | |
| UBA52 | Host-virus interaction, Translation |
Fig. 5.
Predicted interactions between target genes of miRNAs of interest in other signaling pathways involved in COVID-19 pathogenesis and miR-1275, miR-4429, miR-4489, miR-6721-5p, miR-5010-5p, miR-7110-5p, miR-6804-5p and miR-6881–3p were depicted via Cytoscape. There is direct relation between the number of protein interactions and color intensity.
Fig. 6.
The role of miRNAs of interest (miR-1275, miR-4429, miR-4489, miR-6721-5p, miR-5010-5p, miR-7110-5p, miR-6804-5p and miR-6881–3p) in regulating key genes in NF-KB and MAPK signaling pathways contributing to COVID-19 pathogenesis.
Fig. 7.
The role of miRNAs of interest (miR-1275, miR-4429, miR-4489, miR-6721-5p, miR-5010-5p, miR-7110-5p, miR-6804-5p and miR-6881–3p) in regulating key genes in JAK/STAT signaling pathway involved in COVID-19 pathogenesis.
Fig. 8.
Potential interactions between target genes of miRNAs of interest in other signaling pathways involved in COVID-19 pathogenesis and miR-1275, miR-4429, miR-4489, miR-6721-5p, miR-5010-5p, miR-7110-5p, miR-6804-5p and miR-6881–3p were depicted via Cytoscape. There is direct relation between the number of protein interactions and color intensity.
3.4. Predicting marked dysregulated genes in inflammatory signaling pathways participating in COVID-19 pathogenesis
Our results predicted that among many genes (509) of JAK/STAT, NF-KB, and MAPK signaling pathways, 25 genes were not only targeted by 171 miRNAs that were expressed outside of the defined confidence intervals and entered quartile distance but also strongly influenced by miRNAs of interest (miR-1275, miR-4429, miR-4489, miR-6721-5p, miR-5010-5p, miR-7110-5p, miR-6804-5p and miR-6881–3p). Potential target genes regulated by more than twenty miRNAs and their biological functions are represented in Table 5 . The role of 25 key genes in other inflammatory pathways and their interactions are shown in Table 6 and Fig. 7. It can be seen that a total of 11 out of 25 genes are targeted by 30 miRNAs: IL6R, IL6ST, MAPK9, MAPK10, MAPK13, TRAF3, PIK3R1, PIK3R3, TLR4, SYK, and IRAK4. Apart from this, our results highlighted that miR-5010-5p and miR-7110-5p make a unique contribution to regulating NF-KB signaling pathway. Strikingly, miR-6805-5p and miR-4489 strongly affected MAPK signaling pathway. Moreover, the components of JAK/STAT signaling pathway are significant targets of miR-7110-5p and miR-6881–3p. Based on this result, we found very significant candidates for prospective studies in COVID-19.
Table 5.
Strong possible target genes of investigated miRNAs.
| Gene symbol | Molecular function | Protein function |
|---|---|---|
| DDX58 | Helicase | Production of proinflammatory cytokines and IFN-I, RIG-I-mediated antiviral signaling, Transcription of antiviral immunological genes, Detection of both positive and negative strand RNA viruses |
| Hydrolase | ||
| RNA-binding | ||
| EGFR | Host cell receptor for virus entry | Activates downstream signaling cascades such as the RAS-RAF-MEK-ERK, PI3K /AKT, PLCγ1, STATs modules and NF-kappa-B |
| IFNAR1 | Receptor | A component of IFN-I receptor |
| Activation of JAK-STAT pathway | ||
| IFNAR2 | Receptor | A component of IFN-I receptor |
| Activation of JAK-STAT pathway | ||
| IL6ST | Receptor | A signal transduction molecule associated with IL6R and activates JAK-STAT3 and JAK-MAPK signaling pathways. |
| IL6R | Receptor | Part of the receptor for IL6 that associated with IL6ST and leads to the immune response regulation. |
| IRAK4 | Serine/threonine-protein kinase | Activation of NF-kappa-B in both TLR and TCR signaling pathways. |
| MAPK1 | Serine/threonine-protein kinase | MAPK1/ERK2 participates in the MAPK/ERK cascade. Depending on the cellular context, this cascade is involved in various biological processes such as cell growth, adhesion, survival and differentiation. Moreover, It plays key roles in apoptosis, cell cycle, and host-virus interaction. |
| MAPK8 | Serine/threonine-protein kinase | Stimulated MAPK8/JNK1 via extracellular stimuli such as proinflammatory cytokines or physical stress phosphorylates primarily components of AP-1 (JUN, JDP2 and ATF2), resulting in regulation of AP-1 transcriptional activity. |
| MAPK9 | Serine/threonine-protein kinase | Stimulated MAPK9/JNK2 via proinflammatory cytokines or physical stress phosphorylates primarily components of AP-1 (JUN, JDP2 and ATF2), resulting in regulates AP-1 transcriptional activity. |
| MAPK10 | Serine/threonine-protein kinase | Stimulated MAPK10/JNK3 via proinflammatory cytokines or physical stress phosphorylates primarily components of AP-1 (JUN, JDP2 and ATF2), resulting in regulates AP-1 transcriptional activity. |
| MAPK13 | Serine/threonine-protein kinase | Stimulated MAPK13 via proinflammatory cytokines or physical stress leads to direct activation of transcription factors such as ELK1 and ATF2. |
| MAPK14 | Serine/threonine-protein kinase | Stimulated MAPK14 via proinflammatory cytokines or physical stress leads to direct or indirect activation of transcription factors such as RPS6KA5/MSK1, RPS6KA4/MSK2, CREB1, ATF1, the NF-kappa-B isoform RELA/NFKB3, STAT1 and STAT3. |
| MYD88 | Cytosolic adapter | Adapter protein involved in the innate immune response, NF-kappa-B activation, cytokine production and inflammatory responses. |
| PIK3CA | Serine/threonine-protein kinase | Inflammation |
| PIK3R1 | Adaptor, Regulatory subunit | Host-virus interaction, IL-4 Signaling Pathways, IL-2 Pathways |
| Hypoxia-inducible factor regulation | ||
| PIK3R3 | Regulatory subunit | IL-4 Signaling Pathways, IL-2 Pathways |
| PRKCA | Serine/threonine-protein kinase | Positive and negative regulation of platelet function and inflammation |
| PRKCB | Serine/threonine-protein kinase | B-cell activation, Activation of the NF-kappa-B pathway |
| STAT2 | Signal transducer and transcription activator | Signal transducer and activator of transcription mediating signaling by type I interferons. Moreover, recruiting USP18 to the type I IFN receptor subunit IFNAR2, STAT2 has a negative regulatory role in the type I IFN signaling. |
| STAT3 | Signal transducer and transcription activator | It mediates cellular response to interleukins, KITLG/SCF, LEP and other growth factors. It is Involved in inflammatory response by regulating differentiation of naive CD4(+) T-cells into T-helper Th17 or regulatory T-cells. Moreover, STAT3 is capable of binding to the IL6-responsive elements in the promoter region of different acute-phase protein genes. |
| SYK | Non-receptor tyrosine kinase | It mediates signal transduction downstream of a variety of transmembrane receptors including BCR. It plays a key role in stimulation of neutrophil phagocytosis by IL15. It is involved in dendritic and mast cells activation as well as IL-3/IL3-mediated signaling in basophils. It strengthens integrin-mediated activation of neutrophils and macrophages and P-selectin receptor/SELPG-mediated leukocyte recruitment to inflammatory sites. |
| TRAF3 | Transferase | It regulates pathways resulting in the activation of NF-kappa-B and MAP kinases. It plays critical roles in B-cell survival, normal antibody isotype switching from IgM to IgG, T-cell dependent immune responses, antiviral responses as well as the production of cytokines and interferon. |
| TRAF6 | Hydrolase, Receptor | It mediates dendritic cells (DCs) maturation and/or activation as well as NF-kappa-B and JUN activation. It plays a role in signal transduction initiated via TNF receptor, IL-1 receptor and IL-17 receptor. TRAF6 and MAP3K8 regulate immunoglobulin production via CD40 signals that activate ERK in B-cells and macrophages. |
| TLR4 | Receptor | Receptor protein involved in the innate immune responses, activation of NF-kappa-B, production of cytokines and inflammatory responses |
Table 6.
The inflammatory pathways of strong potential target genes.
| Gene symbol | Inflammatory pathway |
|---|---|
| DDX58 | NF-KB |
| EGFR | JAK-STAT,MAPK |
| IFNAR1 | JAK-STAT |
| IFNAR2 | JAK-STAT |
| IL6ST | JAK-STAT |
| IL6R | JAK-STAT |
| IRAK4 | NF-KB |
| MAPK1 | MAPK |
| MAPK8 | MAPK |
| MAPK9 | MAPK |
| MAPK10 | MAPK |
| MAPK13 | MAPK |
| MAPK14 | MAPK |
| MYD88 | NF-KB |
| PIK3CA | JAK-STAT |
| PIK3R1 | JAK-STAT |
| PIK3R3 | JAK-STAT |
| PRKCA | MAPK |
| PRKCB | NF-KB, MAPK |
| STAT2 | JAK-STAT |
| STAT3 | JAK-STAT |
| SYK | NF-KB |
| TRAF3 | NF-KB |
| TRAF6 | NF-KB |
| TLR4 | NF-KB |
4. Discussion
Given the fact that mechanisms underlying the role of mis-expressed miRNAs in the regulation of inflammatory pathways in COVID-19 have not been fully understood, a comprehensive literature review and a bioinformatics approach were employed to predict miRNA–mRNA networks and identify potential key inflammatory miRNAs in the COVID-19 pathogenesis. This approach highlighted the importance of eight miRNAs, namely miR-1275, miR-4429, miR-4489, miR-6721-5p, miR-5010-5p, miR-7110-5p, miR-6804-5p, and miR-6881–3p, and 25 target genes in COVID-19 pathogenesis. Note that the expression of the above-mentioned miRNAs and their underlying mechanisms had not been completely investigated in COVID-19 patients. However, in line with the obtained results, the significant down-regulation of miR-1275 and miR-4429 in two separate COVID-19 patient cohorts had been previously confirmed (Duecker et al., 2021). In this study, we aimed to investigate the participation of these miRNAs in regulating inflammatory signaling pathways. The results of this study predicted the significant participation of the mentioned miRNAs in regulating inflammatory pathways.
The role of the JAK/STAT, NF-KB, and MAPK signaling pathways in generating inflammatory responses in COVID-19 was established. SARS-CoV-2 infection employs the JAK/STAT pathway to trigger an inflammatory response, which leads to serving dendritic cells, endothelial cells, monocytes, natural killer cells, lymphocytes, macrophages, and pneumocytes and proceeding towards cytokine storm. As a result of this process, different inflammatory markers are produced in the host that specify the severity of the disease. In addition, JAK/STAT pathway utilizes B cell and T cell differentiation to mediate immune responses (Satarker et al., 2021). For instance, an association was found between components of JAK/STAT signaling pathway such as JAK1–3 and STAT1–6 and different immune response roles, which include IL-12, IL-6, IL-4, IL-2 signaling, Th (T-helper)1, Th2, Treg cell and Th9, Th 17 cell differentiation, viral selective CD8+ T cell proliferation and B-cell lymphoma 2 (Bcl-2) (Banerjee et al., 2017; O'Shea and Plenge, 2013). Furthermore, NF-κB governs the expression of pro-inflammatory cytokines, which leads to an increased activation of NF-κB through a positive feedback loop (Chen and Greene, 2004). In COVID-19, pro-inflammatory cytokines with high expression levels further stimulate NF-KB (Neufeldt et al., 2022). The upregulation of NF-κB may contribute to inflammatory responses, as observed in patients with COVID-19. Intriguingly, some proteins belonging to SARS-CoV-2 are recruited as the NF-κB activators. For instance, as the most potent NF-kB inducer, ORF7a is thus a producer of pro-inflammatory cytokines (Su et al., 2021). However, by up-regulating the p38 MAPK signaling pathway, several pro-inflammatory cytokines, like IL-1β, TNF-α, and IL-6, are activated (Zarubin and Han, 2005). The lost activity of ACE2 upon the entrance of the virus, which converts Angiotensin II into Angiotensin 1–7, potentially enables the predomination of Ang II-mediated activation of p38 in the heart and lungs when Ang 1–7 is down-regulated. This enables unrestrained inflammation and generates a positive feedback loop since the activation of p38 also upregulates ADAM17, a protease found to cleave the ACE2 ectodomain to further decrease local ACE2 protective activity (Scott et al., 2011). In addition, SARS-CoV utilizes a viral protein to upregulate p38 activity directly, like other RNA respiratory viruses that may be capable of hijacking p38 activity to promote replication (Grimes and Grimes, 2020).
miRNA target databases indicate that miRNAs regulate most human genes. miRNAs also affect the expression levels of genes involved in the NF-KB, JAK/STAT, and MAPK signaling pathways participating in inflammatory responses in COVID-19. In line with the results of our study, studies have demonstrated that some miRNAs obtained in this study can affect signaling pathways genes involved in inflammatory responses. For example, the combined influence of miR-146a and miR-1275 as well as miR-132 resulted in mRNA degradation and protein inhibition of IRAK1 and MAPK3, respectively (Betáková and Švančarová, 2013). In addition, miR-1275 was implicated in the regulation of NF-κB and IL-6 (Plowman and Lagos, 2021). The involvement of miR-1275 down-regulation has been established in cancers, infections, and obesity (Zhou et al., 2017). MiR-1275 silences ELK1, an E-twenty-six-domain transcription factor related to adipocyte differentiation, and thus suppresses human visceral pre-adipocyte differentiation (Pang et al., 2016). Also, IL6 and TNFα can down-regulate miR-1275 expression in mature human adipocytes. Furthermore, it has been established that NF-κB participates in regulating miR-1275 transcription via attaching to its promoter region. Responding to TNF-α, NF-κB attached to the promoter region of miR-1275 and suppressed its transcription. This somewhat explains the miR-1275 reduction in mature human adipocytes in obesity (Zhou et al., 2017). Fawzy et al. (2015) demonstrated that IGF1R is the direct target of miR-1275 and that this miRNA is able to somewhat control the development of hepatocellular carcinoma by directly targeting IGF1R and IGF2BPs genes (Fawzy et al., 2015). By targeting IGF1R, AKT3, MAPK1, and PTEN genes involved in mTOR and PI3K/Akt signaling pathways, miR-4429 plays a role in the appearance of Biliary atresia (BA) disease. In this disease, the progressive inflammation, extrahepatic bile ducts and fibrosis of intrahepatic, resulting in cirrhosis were observed (Dong et al., 2016). Expression profiling study is the most cost-effective tool with greatest promise for identifying new biomarkers for diseases and drug responses (Gurwitz, 2013). Comparing 10 patients with COVID-19 and 10 healthy individuals as the control in a profiling study found several interesting differential miRNA expression signatures. The results indicated a notable down-regulation of miR-1275 in the COVID-19 patient cohort. Hence, miR-1275 can serve as a beneficial biomarker to identify an inflammatory response in COVID-19 (1, D5). Moreover, the expression profiling of miRNAs in sepsis secondary to pneumonia patients showed the up-regulated expression levels of miR-7110-5p in patients compared with healthy individuals. In addition, research showed the promise of these miRNAs for use as sepsis biomarkers (Zhang et al., 2019).
As far as we know, this is the first report to introduce a set of miRNAs regulating inflammatory signaling pathways by conducting a comprehensive profiling investigation on COVID-19 patients. Among these miRNAs, the role of miR-1275 in regulating inflammatory pathways has received the attention of many studies; this confirms the results of our study. Although the relationship of some of miRNAs obtained in this study with genes in inflammatory signaling pathways has not been investigated, bioinformatics predictions of this study indicate that miR-4429, miR-4489, miR-6721-5p, miR-5010-5p, miR-7110-5p, miR-6804-5p and miR-6881–3p can affect the expression of a significant number of genes involved in inflammatory pathways in COVID-19. It is possible to use these miRNAs as therapeutic targets and potential biomarkers for this and other inflammatory disorders.
Note that the results of this study were obtained via investigating high-throughput studies using in silico tools to predict potential miRNAs-mRNA interactions in inflammatory networks. Therefore, using the results, researchers can develop hypotheses for future experimental studies in the area of inflammation in relation to not only COVID-19 but also other inflammatory disease.
Ethics approval and consent to participant
Not applicable.
Consent of publication
Not applicable.
Authors' contributions
SGF and SHS wrote the draft and revised it. SHS, BMH and MDO designed and supervised the study. ZAF, PM, ZaAF, ShKh and NS collected the data and performed the bioinformatic analysis. All the authors read and approved the submitted version.
Author statement
The authors declare that all persons who meet authorship criteria are listed and actively participated in the design and writing of this work. We also declare that we have no any financial conflict of interest that could have appeared to influence the work.
Declaration of Competing Interest
The authors declare they have no conflict of interest.
Acknowledgement
This study is related to the project NO. 1400/65255 From Student Research Committe, Shahid Beheshti University of Medical Sciences, Tehran, Iran. We also appreciate the “Student Research Committee” and “Research & Technology Chancellor” in Shahid Beheshti University of Medical Sciences for their financial support of this study.
Edited by: Pradeep Kumar
References
- Akula S.M., Bolin P., Cook P.P. Cellular miR-150-5p may have a crucial role to play in the biology of SARS-CoV-2 infection by regulating nsp10 gene. RNA Biol. 2022;19:1–11. doi: 10.1080/15476286.2021.2010959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Biehl A., Gadina M., Hasni S., Schwartz D.M. JAK–STAT signaling as a target for inflammatory and autoimmune diseases: current and future prospects. Drugs. 2017;77:521–546. doi: 10.1007/s40265-017-0701-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betáková T., Švančarová P. Role and application of RNA interference in replication of influenza viruses. Acta Virol. 2013;57:97–104. doi: 10.4149/av_2013_02_97. [DOI] [PubMed] [Google Scholar]
- Chen L.F., Greene W.C. Shaping the nuclear action of NF-κB. Nat. Rev. Mol. Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- Choudhury A., Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J. Med. Virol. 2020;92:2105–2113. doi: 10.1002/jmv.25987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury A., Das N.C., Patra R., Mukherjee S. In silico analyses on the comparative sensing of SARS-CoV-2 mRNA by the intracellular TLRs of humans. J. Med. Virol. 2021;93:2476–2486. doi: 10.1002/jmv.26776. [DOI] [PubMed] [Google Scholar]
- Choudhury A., Sen Gupta P.S., Panda S.K., Rana M.K., Mukherjee S. Designing AbhiSCoVac - a single potential vaccine for all ‘corona culprits’: Immunoinformatics and immune simulation approaches. J. Mol. Liq. 2022;351 doi: 10.1016/j.molliq.2022.118633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher A., Kaur R., Kaur G., Kaur A., Gupta V., Bansal P. MicroRNA therapeutics: discovering novel targets and developing specific therapy. Perspect. Clin. Res. 2016;7:68. doi: 10.4103/2229-3485.179431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condrat C.E., Thompson D.C., Barbu M.G., Bugnar O.L., Boboc A., Cretoiu D., Suciu N., Cretoiu S.M., Voinea S.C. miRNAs as biomarkers in disease: latest findings regarding their role in diagnosis and prognosis. Cells. 2020;9 doi: 10.3390/cells9020276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das N.C., Labala R.K., Patra R., Chattoraj A., Mukherjee S. In silico identification of new anti-SARS-CoV-2 agents from bioactive Phytocompounds targeting the viral spike glycoprotein and human TLR4. Lett. Drug Des. Discov. 2021;19:175–191. doi: 10.2174/1570180818666210901125519. [DOI] [Google Scholar]
- Dong R., Shen Z., Zheng C., Chen G., Zheng S. Serum microRNA microarray analysis identifies miR-4429 and miR-4689 are potential diagnostic biomarkers for biliary atresia. Sci. Rep. 2016;6 doi: 10.1038/srep21084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duecker R.P., Adam E.H., Wirtz S., Gronau L., Khodamoradi Y., Eberhardt F.J., Donath H., Gutmann D., Vehreschild M.J.G.T., Zacharowski K., Kreyenberg H., Chiocchetti A.G., Zielen S., Schubert R. The mir-320 family is strongly downregulated in patients with covid-19 induced severe respiratory failure. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms221910351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr R.J., Rootes C.L., Rowntree L.C., Nguyen T.H.O., Hensen L., Kedzierski L., Cheng A.C., Kedzierska K., Au G.G., Marsh G.A., Vasan S.S., Foo C.H., Cowled C., Stewart C.R. Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawzy I.O., Hamza M.T., Hosny K.A., Esmat G., El Tayebi H.M., Abdelaziz A.I. MiR-1275: a single microRNA that targets the three IGF2-mRNA-binding proteins hindering tumor growth in hepatocellular carcinoma. FEBS Lett. 2015;589:2257–2265. doi: 10.1016/j.febslet.2015.06.038. [DOI] [PubMed] [Google Scholar]
- Fayyad-Kazan M., Makki R., Skafi N., El Homsi M., Hamade A., El Majzoub R., Hamade E., Fayyad-Kazan H., Badran B. Circulating miRNAs: potential diagnostic role for coronavirus disease 2019 (COVID-19) Infect. Genet. Evol. 2021;94 doi: 10.1016/j.meegid.2021.105020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Pato A., Virseda-Berdices A., Resino S., Ryan P., Martínez-González O., Pérez-García F., Martin-Vicente M., Valle-Millares D., Brochado-Kith O., Blancas R., Martínez A., Ceballos F.C., Bartolome-Sánchez S., Vidal-Alcántara E.J., Alonso D., Blanca-López N., Ramirez Martinez-Acitores I., Martin-Pedraza L., Jiménez-Sousa M.Á., Fernández-Rodríguez A. Plasma miRNA profile at COVID-19 onset predicts severity status and mortality. Emerg. Microbes Infect. 2022;11:676–688. doi: 10.1080/22221751.2022.2038021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Giralt N., Du J., Marin-Corral J., Bódalo-Torruella M., Blasco-Hernando F., Muñoz-Bermúdez R., Clarós M., Nonell L., Perera-Bel J., Fernandez-González M., Nogues X., Sorli-Redó L., Güerri-Fernández R. Circulating microRNA profiling is altered in the acute respiratory distress syndrome related to SARS-CoV-2 infection. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-10738-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes J.M., Grimes K.V. p38 MAPK inhibition: a promising therapeutic approach for COVID-19. J. Mol. Cell. Cardiol. 2020;144:63–65. doi: 10.1016/j.yjmcc.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurwitz D. Expression profiling: a cost-effective biomarker discovery tool for the personal genome era. Genome Med. 2013;5 doi: 10.1186/gm445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann C., Khamina K., Theofilatos K., Diendorfer A.B., Burnap S.A., Nabeebaccus A., Fish M., McPhail M.J.W., O’Gallagher K., Schmidt L.E., Cassel C., Auzinger G., Napoli S., Mujib S.F., Trovato F., Sanderson B., Merrick B., Roy R., Edgeworth J.D., Shah A.M., Hayday A.C., Traby L., Hackl M., Eichinger S., Shankar-Hari M., Mayr M. Association of cardiometabolic microRNAs with COVID-19 severity and mortality. Cardiovasc. Res. 2022;118:461–474. doi: 10.1093/cvr/cvab338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Zhao T., Zhou X., Xiang Y., Gutierrez-Castrellon P., Ma X. Inflammatory pathways in COVID-19: mechanism and therapeutic interventions. MedComm. 2022;3 doi: 10.1002/mco2.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichter A.L., Sullivan M.J., Eccles M.R., Chatterjee A. MicroRNA expression patterns and signalling pathways in the development and progression of childhood solid tumours. Mol. Cancer. 2017;16 doi: 10.1186/s12943-017-0584-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Hu X., Li L., Li J. Hui. Differential microRNA expression in the peripheral blood from human patients with COVID-19. J. Clin. Lab. Anal. 2020;34 doi: 10.1002/jcla.23590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.X., Chen J., Lv S.K., Li J.H., Li L.L., Hu X. Whole-transcriptome RNA sequencing reveals significant differentially expressed mRNAs, miRNAs, and lncRNAs and related regulating biological pathways in the peripheral blood of COVID-19 patients. Mediat. Inflamm. 2021;2021 doi: 10.1155/2021/6635925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezochow G., Zlotoff D., Brenner L., Nestor J., Bebell L.M. Multisystem inflammatory syndrome in adults (MIS-A) associated with COVID-19: a presentation of mixed shock. Ann. Intern. Med. Clin. Cases. 2022;1 doi: 10.7326/aimcc.2021.0066. [DOI] [Google Scholar]
- Mukherjee S. Toll-like receptor 4 in COVID-19: friend or foe? Futur. Virol. 2022;17:415–417. doi: 10.2217/fvl-2021-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeldt C.J., Cerikan B., Cortese M., Frankish J., Lee J.Y., Plociennikowska A., Heigwer F., Prasad V., Joecks S., Burkart S.S., Zander D.Y., Subramanian B., Gimi R., Padmanabhan S., Iyer R., Gendarme M., El Debs B., Halama N., Merle U., Boutros M., Binder M., Bartenschlager R. SARS-CoV-2 infection induces a pro-inflammatory cytokine response through cGAS-STING and NF-κB. Commun. Biol. 2022;5 doi: 10.1038/s42003-021-02983-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoletti A. de S., Visacri M.B., da Ronda C.R. da S.C., Vasconcelos P.E.D.N.S., Quintanilha J.C.F., de Souza R.N., Ventura D. de S., Eguti A., Silva L.F. de S., Perroud Junior M.W., Catharino R.R., Reis L.O., dos Santos L.A., Durán N., Fávaro W.J., Lancellotti M., da Costa J.L., Moriel P., Pincinato E. de C. Differentially expressed plasmatic microRNAs in Brazilian patients with coronavirus disease 2019 (COVID-19): preliminary results. Mol. Biol. Rep. 2022;49:6931–6943. doi: 10.1007/s11033-022-07338-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J., Hayder H., Zayed Y., Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne). 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea J.J., Plenge R. JAKs and STATs in Immunoregulation and immune-mediated disease. Immunity. 2013;36:542–550. doi: 10.1016/j.immuni.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang L., You L., Ji C., Shi C., Chen L., Yang L., Huang F., Zhou Y., Zhang J., Chen X., Guo X. miR-1275 inhibits adipogenesis via ELK1 and its expression decreases in obese subjects. J. Mol. Endocrinol. 2016;57:33–43. doi: 10.1530/JME-16-0007. [DOI] [PubMed] [Google Scholar]
- Patel P., Decuir J., Abrams J., Campbell A.P., Godfred-Cato S., Belay E.D. Clinical characteristics of multisystem inflammatory syndrome in adults: a systematic review. JAMA Netw. Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.26456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra R., Chandra Das N., Mukherjee S. Targeting human TLRs to combat COVID-19: a solution? J. Med. Virol. 2021;93:615–617. doi: 10.1002/jmv.26387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra R., Das N.C., Bhattacharya M., Shit P.K., Patra B.C., Mukherjee S. Applications of artificial intelligence (AI) protecting from COVID-19 pandemic: a clinical and socioeconomic perspective. EAI/Springer Innov. Commun. Comput. 2021;45–60 doi: 10.1007/978-3-030-68936-0_3. [DOI] [Google Scholar]
- Plowman T., Lagos D. Non-coding rnas in covid-19: emerging insights and current questions. Non-coding RNA. 2021;7 doi: 10.3390/ncrna7030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhsha A., Azghandi S., Taghizadeh-Hesary F. Decision on chemotherapy amidst COVID-19 pandemic: a review and a practical approach from Iran. Infect. Chemother. 2020;52:496–502. doi: 10.3947/IC.2020.52.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satarker S., Tom A.A., Shaji R.A., Alosious A., Luvis M., Nampoothiri M. JAK-STAT pathway inhibition and their implications in COVID-19 therapy. Postgrad. Med. 2021;133:489–507. doi: 10.1080/00325481.2020.1855921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A.J., O’Dea K.P., O’Callaghan D., Williams L., Dokpesi J.O., Tatton L., Handy J.M., Hogg P.J., Takata M. Reactive oxygen species and p38 mitogen-activated protein kinase mediate tumor necrosis factor α-converting enzyme (TACE/ADAM-17) activation in primary human monocytes. J. Biol. Chem. 2011;286:35466–35476. doi: 10.1074/jbc.M111.277434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C.M., Wang L., Yoo D. Activation of NF-κB and induction of proinflammatory cytokine expressions mediated by ORF7a protein of SARS-CoV-2. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-92941-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Hao, Gao Yuehan, Li Zhaohuai, Miao Yushan, Huang Zhaohao, Liu Xiuxing, Xie Lihui, Li He, Wen Wen, Zheng Yingfeng, Su Wenru. The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID-19. Clin. Transl. Med. 2020;10 doi: 10.1002/ctm2.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Zhao Y., Li J., Liu C., Zhu L., Zhang J., Yu Y., Wang W.J., Lei G., Yan J., Sun F., Bian C., Meng F., Xu Z., Bai C., Ye B., Guo Y., Shu L., Yuan X., Zhang N., Bi Y., Shi Y., Wu G., Zhang S., Gao G.F., Liu L., Liu W.J., Sun H.X. Downregulated miR-451a as a feature of the plasma cfRNA landscape reveals regulatory networks of IL-6/IL-6R-associated cytokine storms in COVID-19 patients. Cell. Mol. Immunol. 2021;18:1064–1066. doi: 10.1038/s41423-021-00652-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Q., Waley L., Liou N. Adult presentation of multisystem inflammatory syndrome (MIS) associated with recent COVID-19 infection: lessons learnt in timely diagnosis and management. BMJ Case Rep. 2021;14 doi: 10.1136/bcr-2021-243114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarubin T., Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 2005;15:11–18. doi: 10.1038/sj.cr.7290257. [DOI] [PubMed] [Google Scholar]
- Zhang W., Jia J., Liu Z., Si D., Ma L., Zhang G. Circulating microRNAs as biomarkers for Sepsis secondary to pneumonia diagnosed via Sepsis 3.0. BMC Pulm. Med. 2019;19 doi: 10.1186/s12890-019-0836-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.F., Fu Z.Y., Chen X.H., Cui Y., Ji C.B., Guo X.R. Tumor necrosis factor-α and interleukin-6 suppress microRNA-1275 transcription in human adipocytes through nuclear factor-κB. Mol. Med. Rep. 2017;16:5965–5971. doi: 10.3892/mmr.2017.7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoulikha M., Huang F., Wu Z., He W. COVID-19 inflammation and implications in drug delivery. J. Control. Release. 2022;346:260–274. doi: 10.1016/j.jconrel.2022.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]