Abstract
Objective
Haematogenous dissemination is a prevalent route of colorectal cancer (CRC) metastasis. However, as the gatekeeper of vessels, the role of tumour pericytes (TPCs) in haematogenous metastasis remains largely unknown. Here, we aimed to investigate the heterogeneity of TPCs and their effects on CRC metastasis.
Design
TPCs were isolated from patients with CRC with or without liver metastases and analysed by single-cell RNA sequencing (scRNA-seq). Clinical CRC specimens were collected to analyse the association between the molecular profiling of TPCs and CRC metastasis. RNA-sequencing, chromatin immunoprecipitation-sequencing and bisulfite-sequencing were performed to investigate the TCF21-regulated genes and mechanisms underlying integrin α5 on TCF21 DNA hypermethylation. Pericyte-conditional Tcf21-knockout mice were constructed to investigate the effects of TCF21 in TPCs on CRC metastasis. Masson staining, atomic force microscopy, second-harmonic generation and two-photon fluorescence microscopy were employed to observe perivascular extracellular matrix (ECM) remodelling.
Results
Thirteen TPC subpopulations were identified by scRNA-seq. A novel subset of TCF21high TPCs, termed ‘matrix–pericytes’, was associated with liver metastasis in patients with CRC. TCF21 in TPCs increased perivascular ECM stiffness, collagen rearrangement and basement membrane degradation, establishing a perivascular metastatic microenvironment to instigate colorectal cancer liver metastasis (CRCLM). Tcf21 depletion in TPCs mitigated perivascular ECM remodelling and CRCLM, whereas the coinjection of TCF21high TPCs and CRC cells markedly promoted CRCLM. Mechanistically, loss of integrin α5 inhibited the FAK/PI3K/AKT/DNMT1 axis to impair TCF21 DNA hypermethylation in TCF21high TPCs.
Conclusion
This study uncovers a previously unidentified role of TPCs in haematogenous metastasis and provides a potential diagnostic marker and therapeutic target for CRC metastasis.
Keywords: cancer, colorectal cancer, liver metastases, extracellular matrix
Significance of this study
What is already known on this subject?
Colorectal cancer (CRC) metastasises mainly to the liver, which results from haematogenous dissemination.
Tumour pericytes (TPCs) are the major components of tumour vessels; their heterogeneity and functions in CRC metastasis remain largely unknown.
What are the new findings?
The heterogeneity of TPCs derived from patients with CRC was dissected by single-cell RNA sequencing.
This study elucidated an undefined role of TPCs in promoting CRC metastasis.
We identified a novel TPC-related prometastatic signalling pathway for CRC metastasis.
How might it impact on clinical practice in the foreseeable future?
This study provides new insights into the heterogeneity of TPCs and their prometastatic effects on CRC.
Therapeutic targeting of the prometastatic TCF21high TPCs may represent a new paradigm for cancer therapy.
Introduction
Colorectal cancer (CRC) is the third most common malignant tumour worldwide, and colorectal cancer liver metastasis (CRCLM) counts as the leading cause of disease mortality.1 CRCLM results from haematogenous dissemination,2 which refers to tumour cell intravasation into circulation and extravasation from blood vessels, followed by the formation of metastatic foci in liver.3 Tumour intravasation is a critical and rate-limiting step of haematogenous metastasis, during which tumour cells invade the perivascular extracellular matrix (ECM), breach the endothelial barrier and enter blood circulation.4 5 During intravasation, the invading tumour cells contact endothelial and immune cells to form a tumour microenvironment of metastasis (TMEM).6 Pericytes are contractile cells embedded within the capillary walls and wrapped around endothelial cells, where they act as the supervisors of endothelial cells to regulate vessel stabilisation and vascular permeability.7 However, the role of tumour pericytes (TPCs) in haematogenous metastasis remains controversial.
TPCs attached to endothelial cells act as physiological barriers to inhibit tumour cell intravasation. Genetic depletion of NG2+ or PDGFRβ+ pericytes or pharmaceutical inhibition of pericyte recruitment has been shown to enhance vascular permeability and intratumoral hypoxia, which promotes tumour cell epithelial-to-mesenchymal transition (EMT) and tumour metastasis.8–10 Tumor-derived PDGF-BB induces pericyte–fibroblast transition (PFT), leading to cell detachment from blood vessels or secretion of interleukin-33 to recruit tumor-associated macrophages, enhancing haematogenous metastasis.11 12 Tumor-derived exosomes activate KLF4 in pericytes, resulting in cell detachment and abundant deposition of fibronectin in secondary organs, thereby establishing a premetastatic niche to facilitate haematogenous metastasis.13 However, TPCs also play a prometastatic role. CD45-VLA-1bri or endosialin-expressing pericytes in primary tumours facilitate haematogenous metastasis by promoting tumour cell intravasation in a cell contact-dependent manner without altering the tumour vasculature structure or permeability.14 15 These contradictory effects of TPCs on haematogenous metastasis may be associated with their heterogeneity,16 which has not been determined.
TCF21, a member of the basic helix–loop–helix family of transcription factors, is critical for embryogenesis. Deficiency of TCF21 leads to abnormalities in multiple organs and neonatal lethality.17 High expression of TCF21 in coronary artery smooth muscle cells inhibits their differentiation and promotes their migration and proliferation, contributing to stable atherosclerotic plaques and reducing the risk of coronary artery disease.18 19 TCF21 also drives the expression of inflammatory genes and deposition of collagen IV in visceral adipose stem cells.20 TCF21 acts as a tumour suppressor in various types of tumours, which is low expressed in tumour cells owing to its promoter hypermethylation.17 However, the expression profile and function of TCF21 in TPCs remain unknown.
Here, we employed single-cell RNA sequencing (scRNA-seq) to dissect the heterogeneity of TPCs derived from patients with CRC with or without liver metastases, and 13 distinct TPC subpopulations were identified. Among them, a novel subset of TCF21high TPCs, termed matrix–pericytes, was correlated with CRCLM. TCF21 in TPCs increased perivascular collagen deposition and rearrangement and basement membrane degradation, facilitating the establishment of the perivascular metastatic microenvironment (PMM) to enhance tumour cell intravasation. Pericyte-specific knockout of Tcf21 inhibited CRCLM by reducing perivascular ECM remodelling, maintaining the integrity of the basement membrane, and decreasing circulating tumour cells (CTCs). Moreover, the expression of TCF21 in matrix–pericytes was regulated by the loss of integrin α5, which conferred to the DNA hypermethylation of TCF21 via the FAK/PI3K/AKT-DNMT1 axis.
Materials and methods
The detailed materials and methods can be found in the online supplemental materials.
Results
Identification of a distinct subpopulation of TPCs associated with CRC metastasis
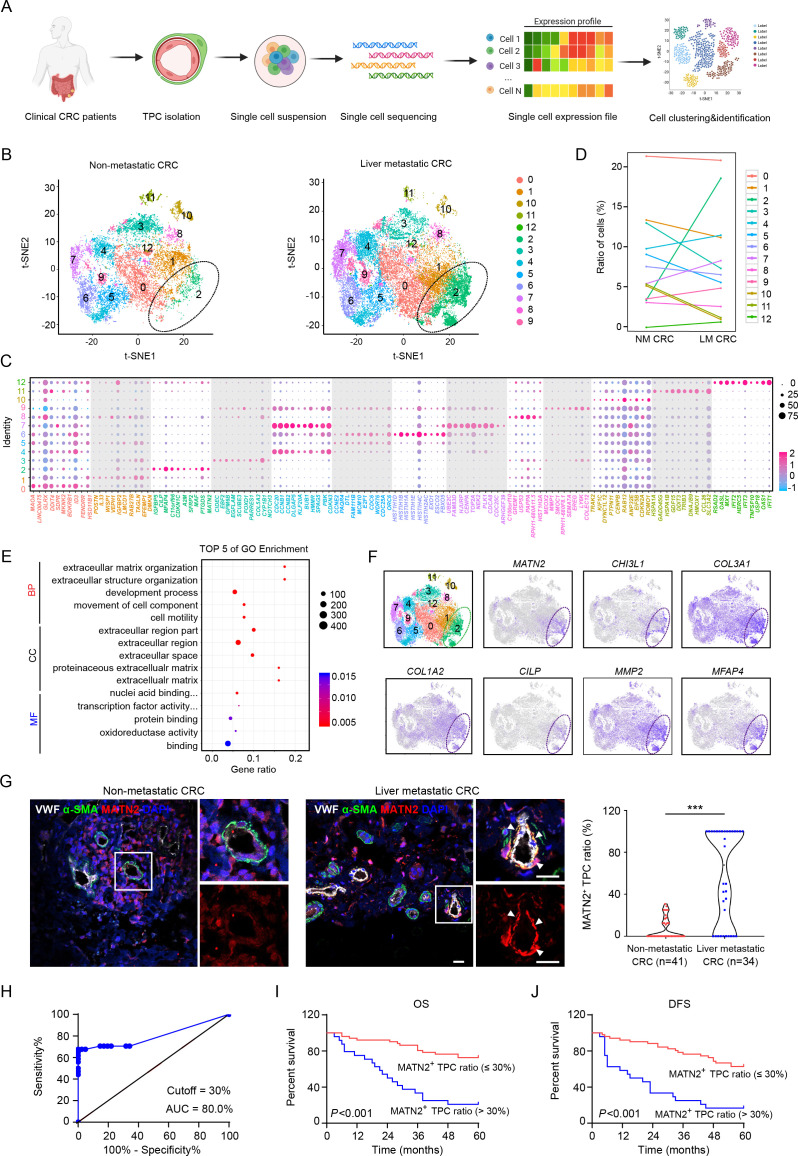
To dissect the heterogeneity of TPCs and assess their contribution to CRC metastasis, TPCs were isolated from primary tumour tissues of patients with CRC with or without liver metastases and were analysed by scRNA-seq using the 10× chromium platform. Single-cell transcriptomes were generated from 50 000 cells, which were juxtaposed in t-stochastic neighbour embedding (t-SNE) space to identify distinct clusters (figure 1A). The gene expression profiles of TPCs were classified into 13 distinct clusters (figure 1B), and their origins were determined using known pericyte markers (online supplemental figures 1A, B). The top 10 cluster-specific genes are shown in figure 1C . The proportion of cells in cluster 2 showed the greatest difference among all subsets between patients with CRC with (18.5%) and without (3.4%) liver metastases (figure 1D), indicating that this subset was associated with CRCLM. Gene Ontology (GO) enrichment analysis indicated that the upregulated genes in samples from patients with CRC with liver metastases were associated with several GO terms related to the ECM (figure 1E), including MATN2, CHI3L1, COL3A1, COL1A2, CILP, MMP2, MFAP4, FBLN1 and FBLN2 (figure 1F and online supplemental figure 2). Therefore, cells in cluster 2 were defined as matrix–pericytes. One of the genes exclusively enriched in cluster 2, MATN2, which encodes the protein matrilin-2 (MATN2), was selected as a biomarker for matrix–pericytes. The ratio of MATN2+ TPCs was higher in tumour sections derived from patients with CRC with liver metastases than in those without liver metastases (figure 1G and online supplemental table 1). In addition, receiver operating characteristic (ROC) curve analysis showed that the optimal cut-off percentage of MATN2+ TPCs to predict CRCLM with high sensitivity and specificity was 30% (figure 1H). Kaplan-Meier survival analysis showed that overall survival (OS) (figure 1I) and disease-free survival (DFS) (figure 1J) were shorter in patients with a high (>30%) ratio of MATN2+ TPCs. Furthermore, the correlation analysis between the MATN2+ TPC ratio and clinicopathological parameters showed that the MATN2+ TPC ratio had a positive correlation with the TNM stage and liver metastasis of patients with CRC (online supplemental table 2). Taken together, these data indicate that matrix–pericytes are notably associated with CRC metastasis.
Figure 1.
Transcriptomic characterisation of matrix-pericytes in TPCs derived from CRC patients. (A) Schematic diagram of scRNA-seq for TPCs. (B) t-SNE visualisation of TPC subsets derived from primary tumour tissues of CRC patients with (n=2) or without (n=2) liver metastasis. (C) Dot plots of the top ten marker genes in each TPC subset. (D) Percentage of each subset of TPCs. (E) GO analysis of the upregulated genes in cluster 2. (F) t-SNE visualisation of TPCs at all stages is combined, overlaid with the expression of indicated genes. (G) Immunofluorescence analysis and quantification of MATN2+ matrix-pericyte ratio in primary tumour tissues from CRC patients with (n=34) or without (n=41) liver metastasis. white arrows indicate the staining of MATN2 in TPCs. scale bar, 20 µm. each sample on the violin plots represents individual patient data. ***P < 0.001 by two-tailed Mann-Whitney test. (H) ROC curve analysis of MATN2+ TPC ratio in CRC patients (n=75). (I, J) Kaplan-Meier analysis of the OS (I) or DFS (J) in CRC patients with high or low MATN2+ matrix-pericyte ratio (based on 30% cut-off, n=75). P<0.001 by Log-rank (Mantel-Cox) test. BP, biological process; CC, cellular component; CRC, colorectal cancer; DAPI, 4',6-diamidino-2-phenylindole; DFS, disease-free survival; GO, Gene Ontology; LM CRC, liver metastatic colorectal cancer; MF, molecular function; NM CRC, non-metastatic colorectal cancer; OS, overall survival; ROC, receiver operating characteristic; TPC, tumour pericyte; t-SNE, t-stochastic neighbour embedding; VWF, von willebrand factor.
gutjnl-2022-327913supp002.pdf (3.4MB, pdf)
gutjnl-2022-327913supp001.xlsx (19KB, xlsx)
TCF21 in TPCs is associated with CRC metastasis
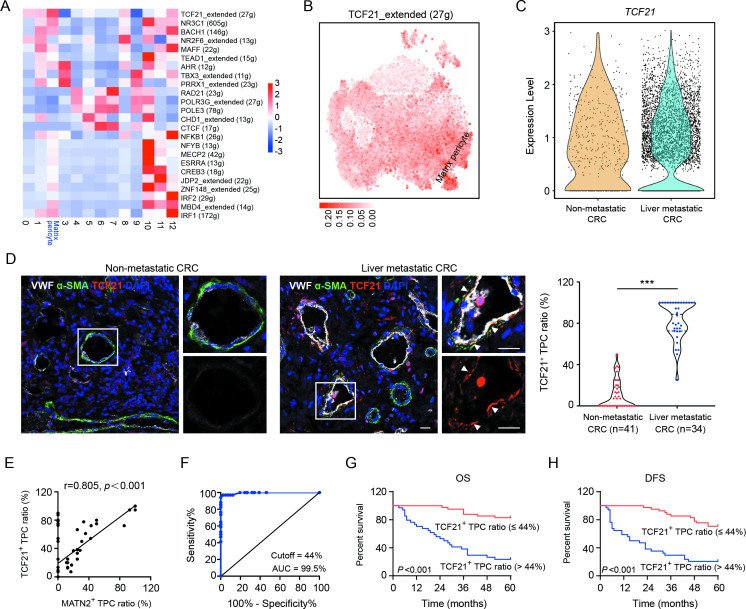
Key regulators of matrix–pericytes were identified using the single-cell regulatory network inference and clustering (SCENIC) pipeline, which connects cis-regulatory sequence information with scRNA-seq data.21 SCENIC analysis showed that the regulon activity of TCF21 was the highest in matrix–pericytes (figure 2A), which was confirmed in the t-SNE space of all cells (figure 2B). In addition, the number of TCF21high TPCs was markedly increased in samples from patients with CRC with liver metastases compared with those without liver metastases (figure 2C). In addition, the ratio of TCF21+ TPCs was increased in tumour sections from patients with CRC with liver metastases (figure 2D and online supplemental table 1). However, TCF21 was undetectable in TPCs in hepatic metastatic tumours derived from patients with CRC (online supplemental figure 3). Pearson’s correlation coefficient analysis showed a positive correlation between the ratio of TCF21+ TPCs and MATN2+ matrix–pericytes (r=0.805, P<0.001; figure 2E). Additionally, flow cytometry analysis showed that the proportion of TCF21+MATN2+ TPCs was significantly increased in TPCs derived from patients with CRC without metastases (TPCNM) infected with lentivirus harbouring TCF21-overexpressing plasmid (TPCNM TCF21) compared with those harbouring vector (TPCNM Vector) (online supplemental figure 4A). In contrast, knockdown of TCF21 in TPCs derived from patients with CRC with liver metastases (TPCLM) showed opposite effects (online supplemental figure 4B). These data indicate that TCF21+ TPCs are closely associated with matrix–pericytes. ROC curve analysis revealed that the optimal percentage of TCF21+ TPCs for predicting liver metastases in patients with CRC was 44% (figure 2F). Kaplan-Meier survival analysis suggested that OS (figure 2G) and DFS (figure 2H) were significantly shorter in patients with higher (>44%) ratio of TCF21+ TPCs. Furthermore, the TCF21+ TPC ratio was notably correlated with TNM stage and liver metastasis in patients with CRC (online supplemental table 3). These findings indicate that TCF21 in TPCs can serve as a predictive biomarker for CRC metastasis.
Figure 2.
TCF21 in matrix–pericytes is positively associated with liver metastasis of CRC. (A) Heat map analysis of the regulon activities in distinct subsets of TPCs. (B) t-SNE visualisation of TCF21 regulon modulus in all TPC subsets. (C) Expression of TCF21 in matrix–pericytes derived from CRC patients with or without liver metastasis. (D) Immunofluorescence staining and quantification of TCF21+ TPC ratio in tumour sections from CRC patients with (n=34) or without (n=41) liver metastases. White arrows indicate TCF21 staining in TPCs. Scale bar, 20 µm. Each sample on the violin plots represents individual patient data. ***P<0.001 by two-tailed Mann-Whitney test. (E) Pearson’s correlation analysis of TCF21+ TPC ratio and MATN2+ matrix–pericyte ratio (n=75). (F) ROC curve analysis of TCF21+ TPC ratio in CRC patients (n=75). (G, H) Kaplan-Meier analysis of OS (G) or DFS (H) in CRC patients with high or low TCF21+ TPC ratio (based on the 44% cut-off, n=75). P<0.001 by log-rank (Mantel-Cox) test. CRC, colorectal cancer; DAPI, 4',6-diamidino-2-phenylindole; DFS, disease-free survival; OS, overall survival; ROC, receiver operating characteristic; TPC, tumour pericyte; t-SNE, t-stochastic neighbour embedding; VWF, von willebrand factor.
TCF21 in TPCs contributes to the phenotype transition of matrix–pericytes and the remodelling of perivascular ECM
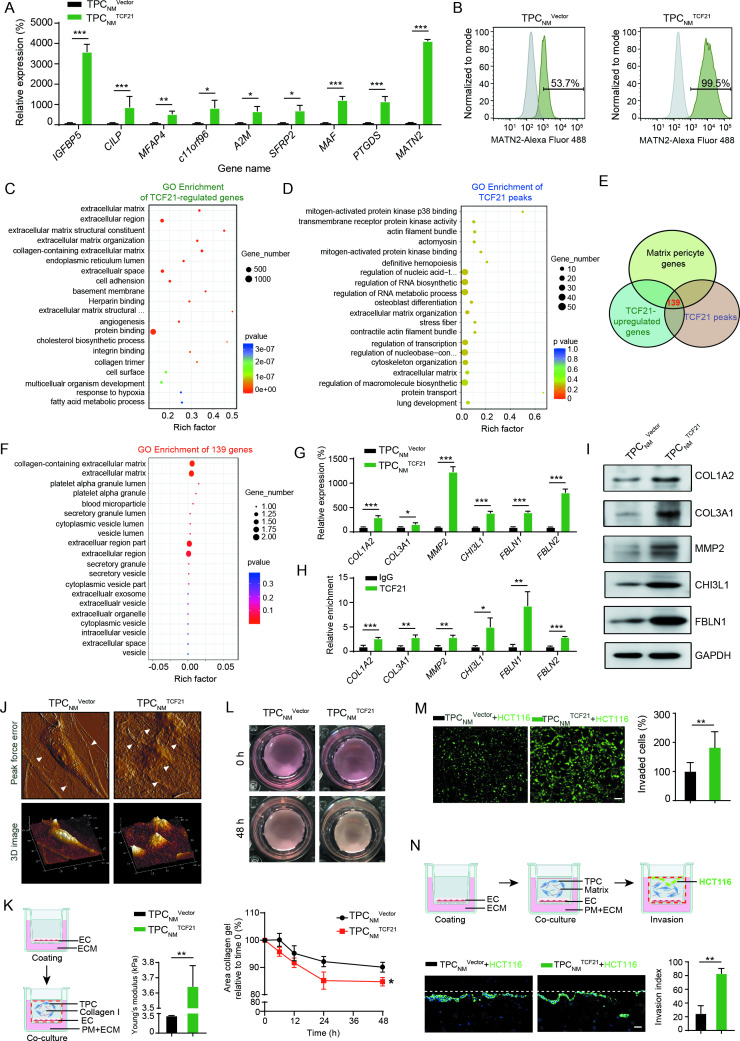
The positive correlation between TFC21 upregulation in TPCs and CRCLM suggested that TCF21 may be involved in the activation of TPCs and may promote the phenotype transition of matrix–pericytes. Depletion or overexpression of TCF21 in TPCs had negligible effects on cell proliferation, adhesion and migration (online supplemental figure 5). The levels of nine genes in matrix–pericytes, including IGFBP5, CILP, MFAP4, c11orf96, A2M, SFRP2, MAF, PTGDS and MATN2, were significantly higher in TPCNM TCF21 than in TPCNM Vector (figure 3A). Conversely, the levels of these genes were significantly lower in TPCLM transfected with siTCF21 (TPCLM siTCF21) than in those transfected with negative control siRNA (TPCLM siNC) (online supplemental figure 6A). In addition, the percentage of MATN2+ matrix–pericytes was increased by overexpression of TCF21 in TPCNM and decreased by the knockdown of TCF21 in TPCLM (figure 3B and online supplemental figure 6B).
Figure 3.
TCF21 is essential for the formation of matrix–pericytes and facilitates CRC cell invasion by ECM remodelling. (A) RT-qPCR analysis for matrix–pericyte-related genes in TPCNM Vector or TPCNM TCF21 (n=3). (B) FCM analysis for MATN2 expression in TPCNM Vector and TPCNM TCF21 (n=3). (C) GO terms of differentially expressed genes derived from RNA-seq in TPCNM TCF21 compared with those in TPCNM Vector. (D) GO terms of TCF21 peaks derived from ChIP-Seq in TPCNM TCF21. (E) Venn diagram showing the number of overlapped genes derived from TCF21-upregulated genes, TCF21 peaks and matrix–pericyte genes. (F) GO analysis for the intersected genes derived from E. (G) RT-qPCR analysis for the indicated genes in TPCNM Vector and TPCNM TCF21 (n=3). (H) ChIP-qPCR analysis for the binding of TCF21 with the promoter of indicated genes in TPCNM TCF21 (n=3). (I) Western blotting analysis for the expression of indicated proteins in TPCNM Vector and TPCNM TCF21 (n=3). (J) AFM imaging of topography and roughness of the collagen fibres incubated with TPCNM Vector or TPCNM TCF21 (n=3). White arrows indicate the collagen fibres. (K) Young’s modulus detection of the stiffness of collagen gels (n=3). Schematics of the 3D coculture model (left) and Young’s modulus detection (right) are shown. Red dotted line indicates the removed part. (L) Representative images and quantification of collagen contraction caused by TPCNM Vector or TPCNM TCF21 (n=3). (M) Representative images and quantification of invaded HCT116 cells (green) (n=3). Scale bar, 50 µm. (N) Schematics of organotypical culture system for studying the invasion of tumour cells (upper). Representative images and quantification of HCT116 cells that invaded into the TPC-containing matrix (bottom) (n=3). Scale bar, 20 µm. Red dotted line indicates the removed part; white lines indicate the initiation. Data are presented as mean±SEM. *P<0.05, **P<0.01, ***P<0.001 by two-tailed unpaired t-test. AFM, atomic force microscopy; ChIP-qPCR, chromatin immunoprecipitation-quantitative PCR; CRC, colorectal cancer; EC, endothelial cell; ECM, endothelial culture medium; GO, Gene Ontology; RT-qPCR, real-time quantitative PCR; TPC, tumour pericyte.
Genes acting downstream of TCF21 were investigated in TPCs using whole-transcriptome RNA-seq and ChIP-seq assays (online supplemental figure 6C, D). RNA-seq assays showed that 1276 genes were upregulated, and 1397 genes were downregulated in TPCNM TCF21 compared with TPCNM Vector (online supplemental figure 6E). A heatmap of the RNA-seq results showed that the expression of matrix–pericyte-specific genes, including MFAP4, CILP, MATN2, MMP2, COL3A1, COL1A2, CHI3L1 and FBLN1, was significantly higher in TPCNM TCF21 (online supplemental figure 6F). GO enrichment analysis of the RNA-seq and ChIP-seq results showed that genes regulated by TCF21 were associated with the categories ‘ECM organisation’ and ‘ECM’ (figure 3C, D), which were similar to the expression profiles in matrix–pericytes. Moreover, comparison of the TCF21 peaks detected by ChIP-seq analysis, TCF21-induced genes detected by RNA-seq and matrix–pericyte genes detected by scRNA-seq identified 139 overlapping genes (figure 3E). GO analysis of these overlapping genes showed their associations with the categories ‘collagen-containing ECM’ and ECM (figure 3F). These results were confirmed by RT-qPCR and ChIP-qPCR, and the binding of TCF21 to the promoter of matrix–pericyte-specific genes was significantly upregulated in TCF21-overexpressing TPCNM (figure 3G, H, and online supplemental figure 6G and H), which were confirmed by western blotting (figure 3I and online supplemental figure 6I). These results indicate that TCF21 is critical for the generation of matrix–pericytes.
ECM remodelling is crucial for tumour metastasis,22 which is represented by aberrant collagen production and cross-linking leading to tissue stiffness, thus promoting tumour metastasis.23 24 Moreover, degradation of the vascular basement membrane by proteases such as matrix metalloproteinases facilitates the escape of tumour cells from primary tumour sites.25 We proposed that TCF21 may play a role in perivascular ECM remodelling and CRC metastasis. Neither overexpression nor knockdown of TCF21 in TPCs altered tumour cell proliferation, migration, EMT or endothelial cell tube formation in vitro (online supplemental figure 7A–H). Atomic force microscopy (AFM) revealed that TPCNM TCF21 possessed greater ability than TPCNM Vector in inducing local alignment of collagen, reorganisation of curly and sheet-like collagen fibres into radial and bundling structures, and increased roughness (figure 3J). Moreover, TPCNM TCF21 was superior to TPCNM Vector in stiffening the mechanical properties of collagen (figure 3K) and increasing collagen contraction (figure 3L), thereby enhancing ECM stiffness. To further assess the effects of TCF21-mediated perivascular ECM remodelling on the transmigration of CRC cells, PKH-67-labelled HCT116 or DLD-1 cells mixed with TPCNM Vector, TPCNM TCF21, TPCLM siNC or TPCLM siTCF21 were seeded into Matrigel. TPCNM TCF21 was better than TPCNM Vector in facilitating CRC cell invasion through the Matrigel-coated transwell membranes (figure 3M and online supplemental figure 8A), whereas TPCLM siTCF21 reduced the invasion of CRC cells compared with TPCLM siNC (online supplemental figure 8B, C). The experiments performed on an organotypical culture system showed that the number of invaded HCT116 cells was significantly increased when matrix, consisting of collagen I and Matrigel, was premixed with TPCNM TCF21 compared with TPCNM Vector (figure 3N). Collectively, these results indicate that TCF21 in matrix–pericytes induces perivascular ECM remodelling and facilitates CRC cell invasion through the perivascular matrix.
We further investigated whether MATN2 exerted prometastatic effects similar to those of TCF21. Given that TCF21high TPCs facilitated tumour metastasis through ECM remodelling, the levels of genes encoding ECM proteins, such as COL1A2, COL3A1, MMP2, CHI3L1 and FBLN1, were examined by RT-qPCR in TPCs with overexpression or knockdown of MATN2 (online supplemental figure 9A, B). Our results showed that MATN2 in TPCs had negligible effects on the levels of these ECM-related genes (online supplemental figure 9C, D). Additionally, MATN2 in TPCs had negligible effects on tumour cell invasion (online supplemental figure 9E, F). These data indicate that MATN2+ TPCs could not give metastatic phenotypes to tumour cells similar to TCF21high TPCs, which may merely serve as a characteristic marker for matrix–pericytes.
Knockout of Tcf21 in TPCs inhibits CRC metastasis
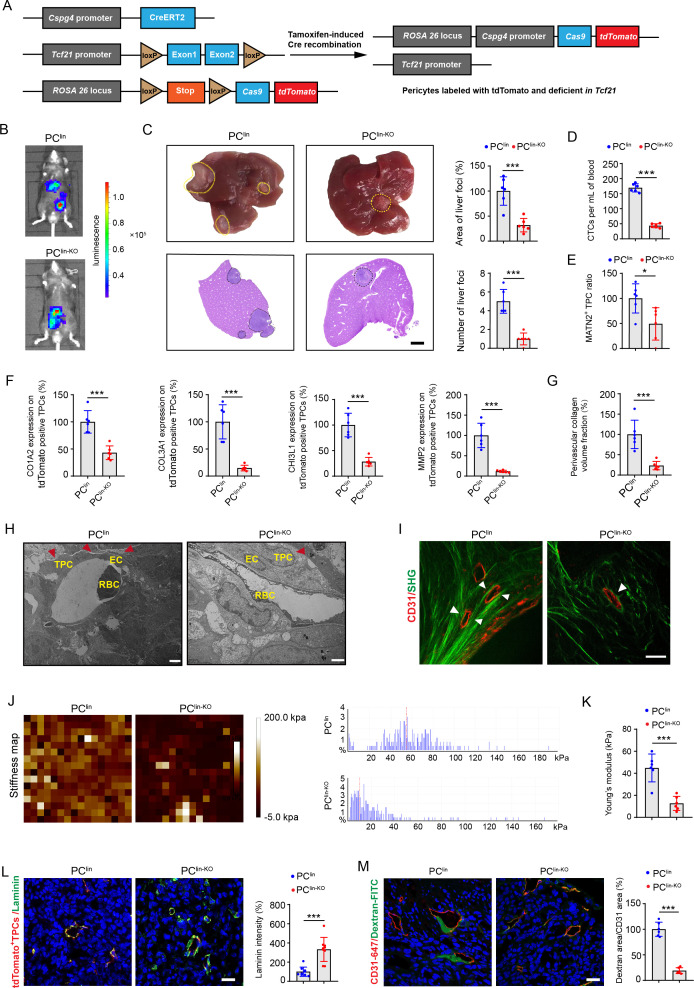
To determine whether TCF21 in TPCs is essential for CRC metastasis in vivo, pericyte lineage-tracing mice (PClin) and tamoxifen-inducible Cspg4-driven pericyte-specific Tcf21 knockout mice (PClin-KO) were generated (figure 4A and online supplemental figure 10A, B). Tamoxifen administration to both PClin and PClin-KO mice resulted in the permanent labelling of pericytes with tdT fluorescence, and the knockout of Tcf21 in PClin-KO mice. To examine the effects of TCF21 in TPCs on CRC metastasis, PClin and PClin-KO mice were orthotopically injected with luciferase-labelled tumour cells (MC38-luc-LM3), followed by treatment with tamoxifen for 1 week (online supplemental figure 11A). Tamoxifen treatment had negligible effects on the non-tumour tissues (online supplemental figure 11B) but induced Cre activity in TPCs, as indicated by tdT fluorescence in both PClin and PClin-KO mice and loss of TCF21 in PClin-KO mice (online supplemental figure 11C, D). In vivo, the TPC-specific deletion of Tcf21 significantly inhibited the formation of liver metastases (figure 4B). The area and number of liver metastatic foci were significantly decreased in PClin-KO mice compared with those in PClin mice (figure 4C). Furthermore, the number of EpCAM+CD45− CTCs was significantly lower in PClin-KO mice than that in PClin mice (figure 4D and online supplemental figure 12A, B). Knockout of Tcf21 in TPCs had negligible effects on primary tumour growth and EMT, as indicated by equal levels of E-cadherin, vimentin, and Ki67 in tumour sections from PClin and PClin-KO mice (online supplemental figure 13A, B). These data indicate that TCF21 in TPCs promotes CRC metastasis, independent on tumour growth and EMT.
Figure 4.
Pericyte-specific knockout of Tcf21 inhibits the remodelling of perivascular ECM and CRC metastasis. (A) schematic diagram of the construction of Cspg4 (NG2) lineage-tracing Tcf21 inducible knockout mice. (B) Representative images and quantification of bioluminescence signals of mice orthotopically injected with MC38-luc-LM3 cells (n=6). (C) Representative images and H&E analysis of livers derived from MC38-luc-LM3 xenograft-bearing mice (n=6). Yellow and black dotted lines indicate the liver metastatic loci. Scale bar, 2 mm. (D) Quantification of CTCs (n=6). (E) Quantification of MATN2+ TPC ratio in primary tumour sections (n=6). (F) Quantification of the expression of COL3A1, MMP2, COL1A1 and CHI3L1 on TPCs (tdTomato) in primary tumour sections (n=6). (G) Quantification of perivascular collagen in primary tumour sections (n=6). (H) Representative TEM images of collagen fibres surrounding TPCs (n=6). Red arrowheads indicate the perivascular collagen fibres. Scale bar, 2 µm. (I) Representative images of perivascular collagen organisation in tumours imaged by SHG (green) and CD31 (red) (n=6). White arrowheads indicate the perivascular collagen fibres. Scale bar, 50 µm. (J, K) Representative AFM images and quantification of the stiffness of the perivascular area in primary tumour sections (n=6). (L) Immunofluorescence staining and quantification of laminin (green) surrounding TPCs (tdTomato) in primary tumour sections (n=6). Scale bar, 20 µm. (M) Immunofluorescence staining and quantification of FITC–dextran 40 kD (green) surrounding vessels labelled with CD31 in primary tumour sections (n=6). Scale bar, 20 µm. Data are presented as mean±SEM. *P< 0.05, ***P <0.001 by two-tailed unpaired t-test. AFM, atomic force microscopy; CRC, colorectal cancer; CTC, circulating tumour cell; EC, endothelial cell; ECM, extracellular matrix; RBC, red blood cell; SHG, second-harmonic generation; TEM, transmission electron microscopy; TPC, tumour pericyte.
The effects of TCF21 in TPCs on vascular structure and function were further evaluated by assessing pericyte coverage and the percentage of MATN2+ matrix–pericytes in tumour vessels. Immunofluorescence staining showed that deletion of Tcf21 in TPCs had negligible effects on total pericyte coverage (online supplemental figure 13C), whereas the number of MATN2+ matrix–pericytes in primary tumour tissues of MC38-luc-LM3 CRCLM xenografts was lower in PClin-KO mice than in PClin mice (figure 4E and online supplemental figure 12C). In addition, the expression of ECM remodelling-related proteins, including MMP2, COL1A2, COL3A1 and CHI3L1, in TPCs from PClin mice was significantly higher than those from PClin-KO mice (figure 4F and online supplemental figure 12D). Masson staining showed that the density of fibrillar collagen components was significantly decreased in tumour sections from PClin-KO mice compared with that in PClin mice (figure 4G and online supplemental figure 12E). Transmission electron microscopy analysis indicated that the collagen bundles at the perivascular region were thinner in tumour sections from PClin-KO than in those from PClin mice (figure 4H). Collagen deposition is required for tissue stiffness and the local invasiveness of tumour cells, and changes in collagen orientation also contribute to tumour metastasis.26 The structure of perivascular collagen was further examined by second-harmonic generation and two-photon excited fluorescence. Perivascular collagen fibres in PClin-KO mice were arranged irregularly around the tumour vessels with curly and unfixed outlines, whereas perivascular collagen in PClin mice had an almost parallel orientation and were tightly embraced surrounded the tumour vessels (figure 4I). The stiffness of perivascular areas was determined by AFM and analysed with Young’s modulus patterns, and we found that the perivascular areas in PClin-KO mice were significantly softer than those in PClin mice (figure 4J, K). As the basement membrane serves as a physical barrier for tumour cell intravasation, degradation of the basement membrane could weaken its barrier function and increase vessel permeability.27 Our results showed that the expression of laminin, the most abundant component in the vascular basement membrane, was significantly higher in PClin-KO mice than that in PClin mice (figure 4L), and deletion of Tcf21 in TPCs resulted in decreased vessel permeability in PClin-KO mice compared with PClin mice (figure 4M). Taken together, TCF21 in matrix–pericytes might be a key regulator of perivascular ECM remodelling, which establishes a PMM to facilitate CRC cell intravasation.
Loss of integrin α5 attenuates DNA methylation of TCF21 promoter and increases TCF21 expression in TPCs
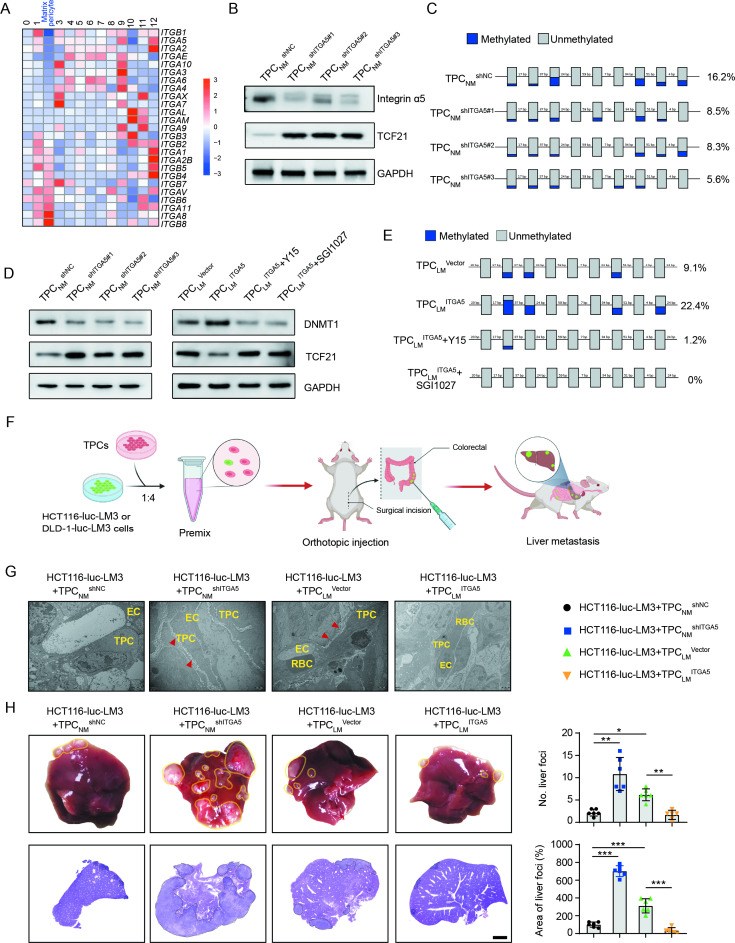
Integrins, the key receptors of ECM components, function as mechanical transducers to facilitate tumour metastasis.28 Our results showed that ITGA2, ITGA5 and ITGB1 were significantly decreased in matrix–pericytes among the 13 clusters of TPCs (figure 5A). Integrin α2 and integrin β1 were unrelated to TCF21, whereas integrin α5 expression was inversely correlated with TCF21 expression in TPCs (online supplemental figure 14A, B). Gain-of-function and loss-of-function experiments showed that TCF21 was negatively regulated by integrin α5 at both the mRNA and protein levels (figure 5B and online supplemental figure 14C-E), whereas integrin α2 and integrin β1 had negligible effects on TCF21 expression (online supplemental figure 14F).
Figure 5.
Loss of integrin α5 inhibits hypermethylation of TCF21 promoter and upregulates the expression of TCF21 in TPCs. (A) Heat map of the expression of integrins in all subsets of TPCs. (B) Western blotting analysis of integrin α5 and TCF21 in integrin α5-knockdown TPCNM (n=3). (C) BSP analysis for TCF21 promotor region in integrin α5-knockdown TPCNM (n=3). Blue and grey circles represent the methylated and unmethylated CpGs, respectively. The percentage of total methylated CpGs is given on the right. (D) Western blotting analysis of DNMT1 and TCF21 in integrin α5-knockdown (left) or integrin α5-overexpressing (right) TPCs treated with or without FAK inhibitor (Y15) or DNMT1 inhibitor (SGI1027) (n=3). (E) BSP analysis for the methylation and unmethylation levels of TCF21 in TPCLM treated with or without Y15 and SGI1027 (n=3). (F) Schematic diagram of the in vivo experiments. (G) Representative TEM images of collagen fibres surrounding TPCs in primary HCT116 orthotopic xenografts (n=6). Red arrowheads indicate perivascular collagen fibres. Scale bar, 2 µm. (H) Representative images of the whole liver and H&E analysis of liver metastatic foci (n=6). Yellow and black dotted lines indicate the metastatic loci. Scale bar, 2 mm. Data are presented as mean±SEM. *P<0.05, **P<0.01, ***P<0.001 by one-way analysis of variance followed by Tukey’s post hoc test. BSP, bisulfite sequencing PCR; CRC, colorectal cancer; EC, endothelial cell; RBC, red blood cell; TEM, transmission electron microscopy; TPC, tumour pericyte.
Hypermethylation of TCF21 DNA suppresses TCF21 expression in various types of tumour cells, including non-small cell lung cancer, head and neck squamous cell carcinoma, and CRC.29–31 Evaluation of the methylation of CpG islands within the promoter of TCF21 in TPCs showed that TCF21 expression in TPCs was inversely correlated with its methylation status, as indicated by the decreased 5-mC level of TCF21 promoter of TPCLM compared with TPCNM (online supplemental figure 14G). In addition, integrin α5 positively regulated TCF21 DNA hypermethylation, and loss of integrin α5 in TPCs attenuated DNA hypermethylation of TCF21 promoter in TPCs (figure 5C and online supplemental figure 14H).
DNMT1 has been reported to regulate the DNA hypermethylation of TCF21 in lung cancers.32 Our results showed that depletion of integrin α5 reduced the expression of DNMT1 in TPCNM by inhibiting the FAK/PI3K/AKT axis, whereas overexpression of integrin α5 in TPCLM exerted opposite effects, which could be reversed by the FAK inhibitor Y15 (figure 5D and online supplemental figure 14I). Furthermore, either Y15 or DNMT inhibitor SGI1027 decreased the expression of DNMT1 and suppressed the DNA hypermethylation of TCF21 (figure 5E), followed by an increase in the expression of TCF21 in TPCLM ITGA5 (figure 5D). These results indicate that the loss of integrin α5 upregulates TCF21 in TPCs by suppressing the DNA hypermethylation of TCF21.
The effects of integrin α5 in TPCs on CRC metastasis in vivo were investigated in an orthotopic xenograft model generated by the co-injection of HCT116-luc-LM3 or DLD1-luc-LM3 cells and integrin α5-overexpressing or -knockdown TPCs (figure 5F). Compared with TPCNM shNC, TPCNM shITGA5 increased collagen density in perivascular areas of tumour tissues (figure 5G and online supplemental figure 15A, B), facilitated perivascular basement membrane degradation (online supplemental figure 15C, D) and significantly increased the number and area of metastatic foci in the liver (figure 5H and online supplemental figure 15E). Conversely, co-injection of tumour cells and TPCLM ITGA5 showed the opposite effects compared with those with TPCLM Vector (figure 5G, H, and online supplemental figure 15A–E). These data indicate that loss of integrin α5 in TPCs promotes CRC metastasis.
TCF21high matrix–pericytes correlate with perivascular ECM remodelling and liver metastasis in patients with CRC
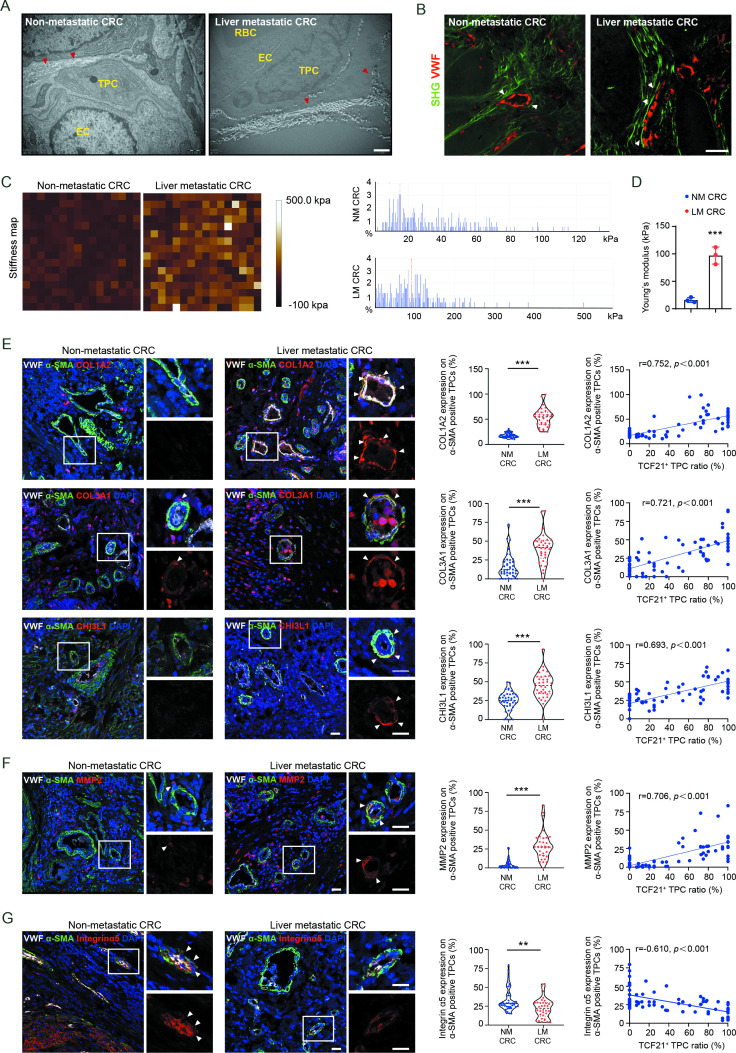
To determine whether the above findings were applicable to patients with CRC, perivascular collagen deposition and alignment were assessed in patients with CRC with and without liver metastases. Compared with patients with CRC without liver metastases, the perivascular collagen abundance, and the reorientation of perivascular collagen fibres into a radial alignment were more prominent in patients with CRC with liver metastases (figure 6A, B, and online supplemental figure 16A, B). In addition, the stiffness of perivascular areas was significantly enhanced in tumour tissues from patients with CRC with liver metastases compared with those without liver metastases (figure 6C, D). Moreover, the expression of COL1A2, COL3A1 and CHI3L1 (figure 6E), as well as MMP2, a key protease involved in the degradation of basement membrane (figure 6F), was higher in TPCs from patients with CRC with liver metastases than those without liver metastases. Correspondently, the expression of integrin α5 was lower in TPCs from patients with CRC with liver metastases than in those without liver metastases (figure 6G). Pearson correlation analysis indicated that the levels of MMP2, COL1A2, COL3A1 and CHI3L1 in TPCs were positively correlated, whereas the level of integrin α5 in TPCs was negatively correlated with the ratio of TCF21high TPCs (figure 6E–G). These clinical data indicate that integrin α5 loss-induced upregulation of TCF21 is an important regulator of perivascular ECM remodelling and the establishment of the PMM, thus facilitating CRC metastasis.
Figure 6.
TCF21 in matrix–pericytes is associated with the remodelling of perivascular ECM in primary tumours derived from patients with CRC. (A) Representative TEM images of collagen fibres surrounding TPCs in primary tumour tissues from patients with CRC (n=3). Red arrowheads indicate the perivascular collagen fibres. Scale bar, 1 µm. (B) Representative images of perivascular collagen structure and density in patients with CRC imaged by SHG (green) and VWF (red) (n=3). White arrowheads indicate perivascular collagen fibres. Scale bar, 50 µm. (C, D) Representative AFM images and quantification of stiffness in perivascular area in primary tumour tissues from patients with CRC. Data are presented as mean±SEM (n=3).***P<0.001 by two-tailed unpaired t-test. (E–G) Immunofluorescence staining and quantification of COL1A2, COL3A1, CHI3L1 (E), MMP2 (F) and integrin α5 expression (G) in αSMA+ TPCs in primary tumour sections from patients with CRC (n=75), and Pearson’s correlation analysis for COL1A2, COL3A1, CHI3L1, MMP2 or integrin α5 expression in αSMA+ TPCs with TCF21+ TPC ratio (%) are shown (n=75). White arrowheads indicate the staining of indicated proteins in TPCs. Scale bar, 20 µm. Each sample on the violin plots represents individual patient data. **P<0.01, ***P<0.001 by two-tailed Mann-Whitney test. AFM, atomic force microscopy; DAPI, 4',6-diamidino-2-phenylindole; EC, endothelial cell; ECM, extracellular matrix; LM CRC, liver metastatic colorectal cancer; NM CRC, non-metastatic colorectal cancer; RBC, red blood cell; SHG, second-harmonic generation; TEM, transmission electron microscopy; TPC, tumour pericyte; VWF, von willebrand factor.
Discussion
Cellular heterogeneity in the tumour microenvironment (TME) may result in distinct pathological phenotypes and different responses to cancer therapy.33 The heterogeneity of tumour cells,34 tumor-infiltrating immune cells,35 cancer-associated fibroblasts (CAFs),36 and endothelial cells37 has been extensively investigated by scRNA-seq, which has also been employed to analyse pericytes derived from tumours, enteritis, and normal GI tract.38–41 However, the specific phenotype and function of pericytes in tumour progression are largely unknown. Therefore, evaluating the heterogeneity of TPCs may provide new insights into the mechanisms underlying haematogenous metastasis. Here, the heterogeneity of TPCs was first revealed by scRNA-seq. By comparing the subpopulations of TPCs in our study with those in previous studies,39 42 four novel subpopulations of TPCs were specifically identified in our work (online supplemental figure 17A–C).
Loss of NG2 is an important hallmark of PFT, during which pericytes shed from the blood vessels where they were originally attached.11 We found that the expression of NG2 in clusters 10, 11 and 12 was extremely low, suggesting that PFT may have occurred in these subpopulations. However, the expression of NG2 in cluster 2 was not significantly decreased, indicating that these TPCs retained perivascular characteristics despite being activated. Furthermore, trajectory analysis indicated that cluster 2 originated from cluster 9 and later evolved into cluster 12 (online supplemental figure 18A, B), further demonstrating that cluster 2 was a newly identified subset of TPCs. Therefore, our findings improve understanding of the heterogeneity of TPCs.
To initiate metastasis, tumour cells must break through the collagen-enriched ECM by matrix deposition and degradation, which generate ‘tracks’ or ‘tunnels’ that help tumour cells pass.43 On reaching blood vessels, these tumour cells interact with proangiogenic TIE2high/VEGFhigh macrophages and luminal endothelial cells to construct a TMEM, which supports the transendothelial migration of tumour cells into blood circulation.6 During intravasation, tumour cells physically contact with tumour endothelial cells via juxtracrine and paracrine signalling.5 Although TPCs attach to endothelial cells, their roles in tumour cell intravasation and the underlying mechanisms remain unclear. Our study demonstrates that matrix–pericytes are one of the TPC subpopulations promote CRC metastasis by establishment of PMM, providing an unidentified function of TPCs in haematogenous metastasis.
TCF21 in tumour cells is considered to inhibit tumour growth and metastasis,17 whereas the expression and function of TPCs in haematogenous metastasis remain largely unknown. In contrast to its suppressive role in tumour cells and CAFs,44 TCF21 promoted phenotypical transition of TPCs into matrix–pericytes, which facilitated the haematogenous metastasis of CRC. The distinct functions of TCF21 in TPCs in haematogenous metastasis may be caused by the different methylation levels of the TCF21. DNA methylation of TCF21 was lower in matrix–pericytes than in tumour cells, which increased the expression of TCF21 in TPCs from patients with CRC with liver metastases. DNA methylation of TCF21 was regulated by the FAK/AKT/DNMT1 signalling pathway, whereas the expression and activation of FAK in TPCs are negatively correlated with tumour angiogenesis, tumour growth, and metastasis.45 However, the mechanism by which cancer cell regulates integrin α5 and TCF21 expression in TPCs has not been revealed. Metastatic CRC cells could rely on gene mutations46 including TP53, BRAF and KRAS or extracellular vehicles (EVs)47 to promote tumour metastasis. We found that the TP53, BRAF and KRAS mutations were not independent predictors of CRCLM (online supplemental table 4), and the above gene mutations were not associated with the TCF21+ TPC ratio or the expression of integrin α5 in TPCs (online supplemental figure 19A and B). Interestingly, we found that metastatic CRC cells could reduce the expression of integrin α5 while increase the expression of TCF21 in an EV-dependent manner (online supplemental figure 19C, D). Nevertheless, the mechanisms underlying tumor-derived EVs and other factors involved in the regulation of TPCs require further investigation.
In conclusion, this study used scRNA-seq to reveal the heterogeneity of TPCs in patients with CRC and to identify a novel subpopulation of TCF21high TPCs associated with CRCLM. These findings revealed the effects and mechanisms of TCF21high TPCs in the construction of the PMM to facilitate CRC metastasis by remodelling the perivascular ECM and provided a potential diagnostic marker for haematogenous metastasis.
Acknowledgments
We are grateful to Guangzhou Genedenovo Biotechnology Co. for assistance with sequencing and/or bioinformatics analysis of single-cell RNA sequencing. We thank Guangzhou LC-Bio Technology Co. for their assistance with RNA-seq and CHIP-seq. We thank GL for assistance with atomic force microscopy and data analysis.
Footnotes
XL, JP and TL contributed equally.
Correction notice: This article has been corrected since it published Online First. Figure one and the supplementary files have been corrected.
Contributors: DZ, WY, MC and YC designed and supervised the experiments, revised the manuscript and were responsible for the drafts of the manuscript, as well as all requested revisions. QQ, MH, LD and PM-KT critically revised the manuscript. MC, XL, JP and TL wrote the manuscript and analysed the data. XL, MC, WY, QM, ZZ and SQ performed experiments. JP, DH, RD and YZ collected human colorectal cancer tissues, reviewed the pathological sections and assessed preclinical and clinical samples. YG and WZ directed the second-harmonic generation and two-photon-excited fluorescence experiments. HL directed the Young modulus measurements.
Funding: This work was supported by the National Natural Science Foundation of China (81973340, 81803566, 81973341, 81773758 and U1801287), Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program (2017BT01Y036), Natural Science Foundation of Guangdong Province (2019A1515010144, 2019A1515110543, A1515110543, 2021A1515110242, 2019A1515011934 and 2020A1515010071), Ministry of Science and Technology of China (2018ZX09711001-008-008), National High-Level Personnel of Special Support Program (Zhang Dongmei), National Key R&D Program of China (2017YFC1703800), Key-Area Research and Development Program of Guangdong Province (2020B1111110004, 2021B1111110004), Science and Technology Program of Guangzhou (202002030010), Guangdong Province Key Laboratory of Pharmacodynamic Constituents of TCM and New Drugs Research, College of Pharmacy (2020B1212060076), Science and Technology Projects in Guangzhou (202102070001), Young S&T Talent Training ProgramProgramme of Guangdong Provincial Association for S&T, China (SKXRC202216).
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, conduct, reporting or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. scRNA-seq data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through the GEO accession number GSE199726. RNA-seq and ChIP-seq data were deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession numbers GSE200064 and GSE200065, respectively.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
Not applicable.
References
- 1. Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2021. CA Cancer J Clin 2021;71:7–33. 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 2. Enquist IB, Good Z, Jubb AM, et al. Lymph node-independent liver metastasis in a model of metastatic colorectal cancer. Nat Commun 2014;5:3530. 10.1038/ncomms4530 [DOI] [PubMed] [Google Scholar]
- 3. Welch DR, Hurst DR. Defining the hallmarks of metastasis. Cancer Res 2019;79:3011–27. 10.1158/0008-5472.CAN-19-0458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell 2017;168:670–91. 10.1016/j.cell.2016.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reymond N, d'Água BB, Ridley AJ. Crossing the endothelial barrier during metastasis. Nat Rev Cancer 2013;13:858–70. 10.1038/nrc3628 [DOI] [PubMed] [Google Scholar]
- 6. Harney AS, Arwert EN, Entenberg D, et al. Real-Time Imaging Reveals Local, Transient Vascular Permeability, and Tumor Cell Intravasation Stimulated by TIE2hi Macrophage-Derived VEGFA. Cancer Discov 2015;5:932–43. 10.1158/2159-8290.CD-15-0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Armulik A, Genové G, Betsholtz C. Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev Cell 2011;21:193–215. 10.1016/j.devcel.2011.07.001 [DOI] [PubMed] [Google Scholar]
- 8. Cooke VG, LeBleu VS, Keskin D, et al. Pericyte depletion results in hypoxia-associated epithelial-to-mesenchymal transition and metastasis mediated by Met signaling pathway. Cancer Cell 2012;21:66–81. 10.1016/j.ccr.2011.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xian X, Håkansson J, Ståhlberg A, et al. Pericytes limit tumor cell metastasis. J Clin Invest 2006;116:642–51. 10.1172/JCI25705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Keskin D, Kim J, Cooke VG, et al. Targeting vascular pericytes in hypoxic tumors increases lung metastasis via angiopoietin-2. Cell Rep 2015;10:1066–81. 10.1016/j.celrep.2015.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hosaka K, Yang Y, Seki T, et al. Pericyte-fibroblast transition promotes tumor growth and metastasis. Proc Natl Acad Sci U S A 2016;113:E5618–27. 10.1073/pnas.1608384113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang Y, Andersson P, Hosaka K, et al. The PDGF-BB-SOX7 axis-modulated IL-33 in pericytes and stromal cells promotes metastasis through tumour-associated macrophages. Nat Commun 2016;7:11385. 10.1038/ncomms11385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murgai M, Ju W, Eason M, et al. Klf4-Dependent perivascular cell plasticity mediates pre-metastatic niche formation and metastasis. Nat Med 2017;23:1176–90. 10.1038/nm.4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sinha D, Chong L, George J, et al. Pericytes promote malignant ovarian cancer progression in mice and predict poor prognosis in serous ovarian cancer patients. Clin Cancer Res 2016;22:1813–24. 10.1158/1078-0432.CCR-15-1931 [DOI] [PubMed] [Google Scholar]
- 15. Viski C, König C, Kijewska M, et al. Endosialin-Expressing pericytes promote metastatic dissemination. Cancer Res 2016;76:5313–25. 10.1158/0008-5472.CAN-16-0932 [DOI] [PubMed] [Google Scholar]
- 16. Lyle LT, Lockman PR, Adkins CE, et al. Alterations in pericyte subpopulations are associated with elevated blood-tumor barrier permeability in experimental brain metastasis of breast cancer. Clin Cancer Res 2016;22:5287–99. 10.1158/1078-0432.CCR-15-1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ao X, Ding W, Zhang Y, et al. Tcf21: a critical transcription factor in health and cancer. J Mol Med 2020;98:1055–68. 10.1007/s00109-020-01934-7 [DOI] [PubMed] [Google Scholar]
- 18. Nagao M, Lyu Q, Zhao Q, et al. Coronary Disease-Associated Gene TCF21 Inhibits Smooth Muscle Cell Differentiation by Blocking the Myocardin-Serum Response Factor Pathway. Circ Res 2020;126:517–29. 10.1161/CIRCRESAHA.119.315968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wirka RC, Wagh D, Paik DT, et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat Med 2019;25:1280–9. 10.1038/s41591-019-0512-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Akama T, Chun T-H. Transcription factor 21 (TCF21) promotes proinflammatory interleukin 6 expression and extracellular matrix remodeling in visceral adipose stem cells. J Biol Chem 2018;293:6603–10. 10.1074/jbc.RA117.000456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Suo S, Zhu Q, Saadatpour A, et al. Revealing the critical regulators of cell identity in the mouse cell atlas. Cell Rep 2018;25:1436–45. 10.1016/j.celrep.2018.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Winkler J, Abisoye-Ogunniyan A, Metcalf KJ, et al. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat Commun 2020;11:5120. 10.1038/s41467-020-18794-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shen Y, Wang X, Lu J, et al. Reduction of liver metastasis stiffness improves response to bevacizumab in metastatic colorectal cancer. Cancer Cell 2020;37:800–17. 10.1016/j.ccell.2020.05.005 [DOI] [PubMed] [Google Scholar]
- 24. Reuten R, Zendehroud S, Nicolau M, et al. Basement membrane stiffness determines metastases formation. Nat Mater 2021;20:892–903. 10.1038/s41563-020-00894-0 [DOI] [PubMed] [Google Scholar]
- 25. Koikawa K, Ohuchida K, Ando Y, et al. Basement membrane destruction by pancreatic stellate cells leads to local invasion in pancreatic ductal adenocarcinoma. Cancer Lett 2018;425:65–77. 10.1016/j.canlet.2018.03.031 [DOI] [PubMed] [Google Scholar]
- 26. Provenzano PP, Eliceiri KW, Campbell JM, et al. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med 2006;4:38. 10.1186/1741-7015-4-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kalluri R. Assembly and role in tumour angiogenesis. Nat Rev Cancer 2003;3:422–33. [DOI] [PubMed] [Google Scholar]
- 28. Paszek MJ, Zahir N, Johnson KR, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell 2005;8:241–54. 10.1016/j.ccr.2005.08.010 [DOI] [PubMed] [Google Scholar]
- 29. Smith LT, Lin M, Brena RM, et al. Epigenetic regulation of the tumor suppressor gene TCF21 on 6q23-q24 in lung and head and neck cancer. Proc Natl Acad Sci U S A 2006;103:982–7. 10.1073/pnas.0510171102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen B, Zeng C, Ye Y, et al. Promoter methylation of TCF21 may repress autophagy in the progression of lung cancer. J Cell Commun Signal 2018;12:423–32. 10.1007/s12079-017-0418-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dai Y, Duan H, Duan C, et al. Tcf21 functions as a tumor suppressor in colorectal cancer through inactivation of PI3K/Akt signaling. Onco Targets Ther 2017;10:1603–11. 10.2147/OTT.S118151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu H, Zhou J, Zeng C, et al. Curcumin increases exosomal TCF21 thus suppressing exosome-induced lung cancer. Oncotarget 2016;7:87081–90. 10.18632/oncotarget.13499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marusyk A, Janiszewska M, Polyak K. Intratumor heterogeneity: the Rosetta stone of therapy resistance. Cancer Cell 2020;37:471–84. 10.1016/j.ccell.2020.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim C, Gao R, Sei E, et al. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell 2018;173:879–93. 10.1016/j.cell.2018.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zheng C, Zheng L, Yoo J-K, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 2017;169:1342–56. 10.1016/j.cell.2017.05.035 [DOI] [PubMed] [Google Scholar]
- 36. Muhl L, Genové G, Leptidis S, et al. Single-Cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nat Commun 2020;11:3953. 10.1038/s41467-020-17740-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goveia J, Rohlenova K, Taverna F, et al. An integrated gene expression landscape profiling approach to identify lung tumor endothelial cell heterogeneity and angiogenic candidates. Cancer Cell 2020;37:21–36. 10.1016/j.ccell.2019.12.001 [DOI] [PubMed] [Google Scholar]
- 38. Zhao Q, Eichten A, Parveen A, et al. Single-Cell transcriptome analyses reveal endothelial cell heterogeneity in tumors and changes following antiangiogenic treatment. Cancer Res 2018;78:2370–82. 10.1158/0008-5472.CAN-17-2728 [DOI] [PubMed] [Google Scholar]
- 39. Kinchen J, Chen HH, Parikh K. Data from: composition of the colonic mesenchyme and the nature of its plasticity in inflammatory bowel disease. Gene Expression Omnibus 2018. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE114374 [Google Scholar]
- 40. Kim J-E, Fei L, Yin W-C, et al. Single cell and genetic analyses reveal conserved populations and signaling mechanisms of gastrointestinal stromal niches. Nat Commun 2020;11:334. 10.1038/s41467-019-14058-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Teuwen L-A, De Rooij LPMH, Cuypers A, et al. Tumor vessel co-option probed by single-cell analysis. Cell Rep 2021;35:109253. 10.1016/j.celrep.2021.109253 [DOI] [PubMed] [Google Scholar]
- 42. Rao M, Oh K, Moffitt R. Single-Cell RNA-seq analysis of Metastasis-Primary tumor dissimilarity in a patient with gastrointestinal neuroendocrine cancer. Gene Expression Omnibus 2020. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi [Google Scholar]
- 43. Gaggioli C, Hooper S, Hidalgo-Carcedo C, et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol 2007;9:1392–400. 10.1038/ncb1658 [DOI] [PubMed] [Google Scholar]
- 44. Hussain A, Voisin V, Poon S, et al. Distinct fibroblast functional states drive clinical outcomes in ovarian cancer and are regulated by TCF21. J Exp Med 2020;217:e20191094. 10.1084/jem.20191094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lechertier T, Reynolds LE, Kim H, et al. Pericyte FAK negatively regulates Gas6/Axl signalling to suppress tumour angiogenesis and tumour growth. Nat Commun 2020;11:2810. 10.1038/s41467-020-16618-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bartolini A, Cardaci S, Lamba S, et al. BCAM and Lama5 mediate the recognition between tumor cells and the endothelium in the metastatic spreading of KRAS-mutant colorectal cancer. Clin Cancer Res 2016;22:4923–33. 10.1158/1078-0432.CCR-15-2664 [DOI] [PubMed] [Google Scholar]
- 47. Zhao S, Mi Y, Zheng B, et al. Highly-metastatic colorectal cancer cell released miR-181a-5p-rich extracellular vesicles promote liver metastasis by activating hepatic stellate cells and remodelling the tumour microenvironment. J Extracell Vesicles 2022;11:e12186. 10.1002/jev2.12186 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2022-327913supp002.pdf (3.4MB, pdf)
gutjnl-2022-327913supp001.xlsx (19KB, xlsx)
Data Availability Statement
Data are available in a public, open access repository. scRNA-seq data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through the GEO accession number GSE199726. RNA-seq and ChIP-seq data were deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession numbers GSE200064 and GSE200065, respectively.