Abstract
Psyllids are phloem‐feeding insects that can transmit plant pathogens such as phytoplasmas, intracellular bacteria causing numerous plant diseases worldwide. Their microbiomes are essential for insect physiology and may also influence the capacity of vectors to transmit pathogens. Using 16S rRNA gene metabarcoding, we compared the microbiomes of three sympatric psyllid species associated with pear trees in Central Europe. All three species are able to transmit ‘Candidatus Phytoplasma pyri’, albeit with different efficiencies. Our results revealed potential relationships between insect biology and microbiome composition that varied during psyllid ontogeny and between generations in Cacopsylla pyri and C. pyricola, as well as between localities in C. pyri. In contrast, no variations related to psyllid life cycle and geography were detected in C. pyrisuga. In addition to the primary endosymbiont Carsonella ruddii, we detected another highly abundant endosymbiont (unclassified Enterobacteriaceae). C. pyri and C. pyricola shared the same taxon of Enterobacteriaceae which is related to endosymbionts harboured by other psyllid species from various families. In contrast, C. pyrisuga carried a different Enterobacteriaceae taxon related to the genus Sodalis. Our study provides new insights into host–symbiont interactions in psyllids and highlights the importance of host biology and geography in shaping microbiome structure.
1. INTRODUCTION
Insects establish numerous associations with viruses, bacteria and fungi and can act as vectors of pathogens that cause severe diseases of humans and plants. In particular, some hemipteran insects are devastating pests of annual and perennial crops, mainly due to their ability to transmit numerous phytopathogenic bacteria and viruses, causing serious diseases and leading to significant agricultural losses (Alma et al., 2008; Wilkinson, 2012). The vector competence, that is, the ability of an insect to transmit pathogens, depends on the biology of both the insect vector and the pathogen, as well as on their interaction (Gonella et al., 2019; Orlovskis et al., 2015; Perilla‐Henao & Casteel, 2016; Tamborindeguy et al., 2017). Despite rapid progress in insect‐borne pathogen research in the last decade, it is still poorly understood which factors could favour or impede pathogen transmission.
Recent studies on insect microbiomes have revealed the essential role of the microbiome in the development, physiology and evolution of their hosts (Brucker & Bordenstein, 2013; Newell & Douglas, 2014; Shin et al., 2011; Storelli et al., 2011; van Opstal & Bordenstein, 2019). Insect symbionts are known to provide their hosts with important benefits (synthesizing essential amino acids and vitamins, or protection against predators and pathogens) (Douglas, 2015; Hypša & Nováková, 2008; Jaenike, 2012; Moran et al., 2008; Wilkinson, 2012) but can also manipulate their host's reproduction or shorten its lifespan to spread and persist within host populations (Engelstädter & Hurst, 2009; Hurst & Frost, 2015; Macke et al., 2017; McCutcheon et al., 2019; McMeniman et al., 2009). Moreover, some insect symbionts are known to affect the host's ability to transmit pathogens, which may have a direct impact on the dynamics of vector‐borne diseases (Bourtzis et al., 2014; Chu et al., 2016; Fagen et al., 2012; Gonella et al., 2019; Gottlieb et al., 2010; McMeniman et al., 2009). For instance, the presence of the bacterial endosymbiont Hamiltonella in the whitefly Bemisia tabaci (Hemiptera: Aleyrodidae) was shown to increase the acquisition and transmission efficiency of the Tomato yellow leaf curl virus (TYLCV) in this vector species (Gottlieb et al., 2010; Su et al., 2013). Similarly, Wolbachia may mitigate or impede pathogen acquisition and transmission in the Asian citrus psyllid Diaphorina citri (Hemiptera: Liviidae) (Fagen et al., 2012; Hosseinzadeh et al., 2019; Kruse et al., 2017; Song et al., 2019). Furthermore, Wolbachia was shown to inhibit the transmission of a plant virus by Nilaparvata lugens (Hemiptera: Delphacidae) (Gong et al., 2020). These observations indicate that certain members of insect microbiomes can have a strong impact on vector‐borne disease transmission.
The composition of an insect‐associated bacterial community can vary along an insect's life cycle and shift in response to changes in environmental conditions and/or insect biology (Colman et al., 2012; Fromont et al., 2017; Macke et al., 2017; Meng et al., 2019; Nováková et al., 2017; Yun et al., 2014). For instance, a shift in abundant bacterial taxa was observed across different life stages of D. citri (Meng et al., 2019). Likewise, seasonal changes affected microbial abundance and microbiome composition in mosquitoes and fleas (Cohen et al., 2015; Nováková et al., 2017). Furthermore, prolonged larval diapause in the parasitoid wasp Nasonia vitripennis modified microbiome composition in adults after termination of diapause (Dittmer & Brucker, 2021). Notably, Wolbachia does not survive prolonged larval diapause in this species (Perrot‐Minnot et al., 1996). This variability could potentially affect the vector competence of an insect host (Duguma et al., 2015; Ramsey et al., 2017; Rodríguez‐Ruano et al., 2018), for instance, when a bacterial antagonist of the vectored pathogen is lost from the microbiome due to insect ontogeny or changes in environmental conditions.
Psyllids (Insecta: Hemiptera: Psylloidea) are among the most economically important vectors of bacterial pathogens (Jing et al., 2014; Wilkinson, 2012). Among the major psyllid vectors, D. citri and Bactericera cockerelli (Triozidae) transmit ‘Candidatus Liberibacter’, which causes Huanglongbing (citrus greening) and zebra chip disease of potato (Bové, 2006; Hall et al., 2012; Hansen et al., 2008). In addition, psyllids transmit various strains of ‘Candidatus Phytoplasma’, intracellular bacterial pathogens which cause diseases of fruit trees with high economic importance in Europe and worldwide, such as apple proliferation on apples, stone fruit yellows on apricots and plums and Pear Decline on pears and peaches (Jarausch et al., 2019). The psyllid vectors of phytoplasmas comprise several Palearctic species from the genus Cacopsylla (Psyllidae) (Jarausch et al., 2019; Tamborindeguy et al., 2017). Among them, Cacopsylla pyri, C. pyricola and C. pyrisuga are sympatric in Central Europe and represent the most common psyllid species associated with pear trees. Cacopsylla pyri and C. pyricola are important vectors of ‘Candidatus Phytoplasma pyri causing Pear Decline and thus belong to the most harmful pests of pears in Europe and North America where they were introduced (Jarausch et al., 2019; Tougeron et al., 2021). Both vectors produce up to 3–5 generations per year and show resistance to insecticides, making it difficult to control population outbreaks (Civolani, 2012). In contrast to C. pyri and C. pyricola, C. pyrisuga had been considered a non‐vector species until recently, when its vector competence was experimentally proven in Austria (Riedle‐Bauer et al., 2022). However, it is not known to what extent this species acts as a vector in other parts of Europe. Importantly, C. pyri and C. pyricola differ from C. pyrisuga in numerous aspects of their biology, that is, in their life cycles, voltinism and feeding behaviour (Figure 1). C. pyri and C. pyricola represent closely related, seasonally polymorphic, polyvoltine species (Cho et al., 2020; Conci et al., 1993; Hodkinson, 2009; Soroker et al., 2013). They complete their life cycles on their host‐plants (Pyrus spp.), where they also predominantly overwinter (Hodkinson, 2009; Lauterer, 1999) (Figure 1). Interestingly, in its invasive range in North America, where C. pyricola was introduced from Europe, a number of individuals were found overwintering on conifers or other shelter plants (e.g., in apple orchards), while only a fraction of the population was associated solely with pear trees during an entire year (Horton et al., 1992, 1994). This suggests that C. pyricola individuals may overwinter on conifers in North America, but this association does not appear to be obligate. Both C. pyri and C. pyricola species produce several long‐day summer generations (with small‐sized, light‐coloured morphs) and usually one morphologically different short‐day overwintering generation (with a large‐sized, dark‐coloured morph) (Lauterer, 1999; Soroker et al., 2013; Figure S1). A new annual cycle begins in spring when the overwintered adults lay eggs, which give rise to the first summer generation (Figure S2A) (Hodkinson, 2009). In central Europe, the overwintering generation starts at the beginning of autumn with the eggs laid by adults of the last summer‐form generation. Later in autumn, the adults from the overwintering generation undergo a reproductive diapause which ends in the middle of winter, followed by a very slow reproductive development, and in spring a new annual cycle starts over (Figure S2A) (Hodkinson, 2009; Horton et al., 1998; Lauterer, 1999). In contrast, C. pyrisuga (Figures 1 and S1) produces only a single generation per year. It is also monophagous on pear trees (i.e., its immatures cannot develop on any other plant taxa), but in contrast to C. pyri and C. pyricola, this species has an obligate overwintering association with conifers (Figure 1) where it undergoes a reproductive diapause as adults (Hodkinson, 2009). In spring, overwintered adults of C. pyrisuga move back to pear trees where they lay their eggs (Figure S2B) (Lauterer, 1999). In summer, young adults migrate to shelters on occasional plants and afterwards on conifers. In light of the differences in host biology and possibly also vector competence (Figure 1), C. pyri, C. pyricola and C. pyrisuga represent an interesting study system to investigate the potential links between host biology, vector competence and the microbiome.
FIGURE 1.
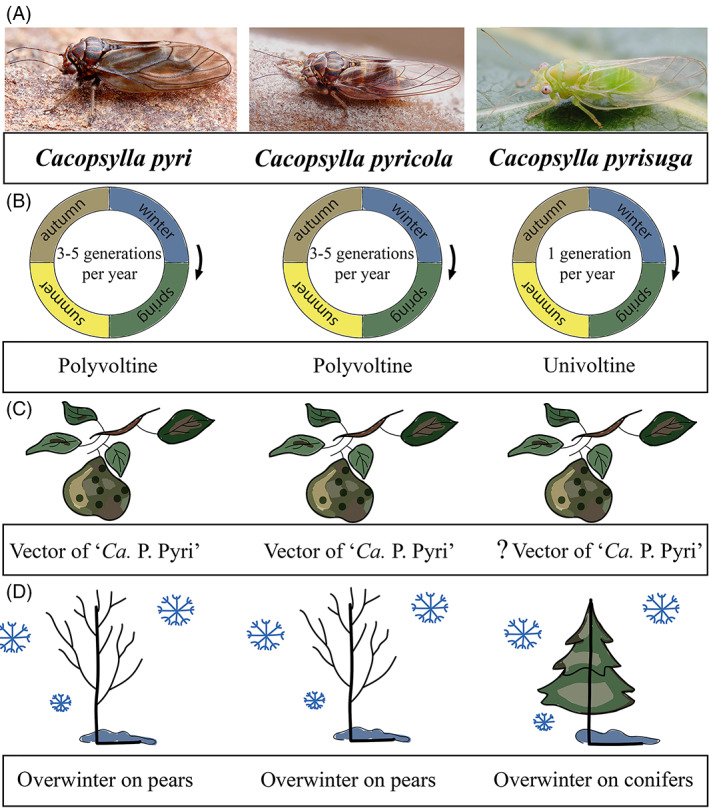
Schematic overview of the biology of the three pear‐associated Cacopsylla species. (A) Adult individuals, (B) differences in voltinism (generations per year), (C) vector competence, and (D) differences in overwintering strategies. The vector competence of Cacopsylla pyrisuga is marked with a question mark (?) because its vector capacity has been demonstrated for Austrian populations but is uncertain for CZ.
Like other groups of sap‐feeding insects, psyllids have established obligatory relationships with a primary endosymbiont which supplies the insect with essential amino acids lacking from its plant‐sap diet. The primary endosymbiont of psyllids is ‘Candidatus Carsonella ruddii’ (hereafter Carsonella), which is present in all psyllid species studied to date (e.g., Cooper et al., 2017; Cooper, Garczynski, & Horton, 2015; Morrow et al., 2017; Overholt et al., 2015; Song et al., 2019; Thao et al., 2000). Carsonella is localized in specialized host cells, bacteriocytes, which constitute a tissular structure called a bacteriome. In addition, numerous other secondary endosymbionts were detected in various psyllids. Depending on the psyllid species, these belong to variable bacterial groups, notably Alphaproteobacteria (Wolbachia), Betaproteobacteria (Profftella) and Enterobacteriaceae (Arsenophonus, Sodalis, unclassified Enterobacteriaceae) (Fromont et al., 2016, 2017; Fu et al., 2020; Morrow et al., 2017, 2020; Nakabachi et al., 2013; Nakabachi, Malenovský, et al., 2020; Nakabachi, Piel, et al., 2020). Some of these symbionts can be found in various host tissues, including the bacteriome, fat body, hemolymph, salivary glands and reproductive organs (Pontes et al., 2008; Wilkinson, 2012).
The aim of this work was to investigate and compare the microbiomes of the three psyllid species sympatric on pear trees in Central Europe, C. pyri, C. pyricola and C. pyrisuga. Our specific aims were to (i) characterize the microbiomes of various populations of C. pyri, C. pyricola and C. pyrisuga, (ii) investigate potential differences in microbiome composition between the three pear psyllid species, and (iii) investigate the impact of host ontogeny and seasonal generations on psyllid microbiome. To this end, our dataset comprises several populations from various geographical localities across different life stages and different generations for the polyvoltine species. Our results provide insights into the potential effects of host biology and geography on the microbiomes of pear psyllids.
2. MATERIALS AND METHODS
2.1. Sample collection
Most individuals of Cacopsylla pyri, C. pyricola and C. pyrisuga were selected from samples collected throughout an entire year from February 2020 until February 2021 in a pear orchard in Starý Lískovec (Brno, Czech Republic) (Tables 1 and S1; Figure S2). The sampling effort in this orchard was standardized for the collection of adults, with a time interval of 7–10 days between each visit. At each visit, psyllids were collected from five branches of five randomly chosen pear trees. Adults were collected using entomological sweeping nets and a beating tray, while eggs and 4–5th instar immatures were collected with a camelhair brush. Adults of C. pyricola were selected for sequencing based on the abundance peaks during the sampling year, representing different generations of this species (Figure S2A). In contrast to C. pyricola, the population of C. pyri did not show clear seasonal peaks but specimens were present at a constant but low density throughout the sampling year. Therefore, specimens of C. pyri likely representing different generations were selected for sequencing based on the information on its life cycle published for the sampling region (Lauterer, 1999). Regarding the univoltine C. pyrisuga, specimens were selected for sequencing from its only seasonal peak in late spring and early summer (Figure S2B). We also included C. pyrisuga specimens collected at the beginning of spring, corresponding to adults from the previous year that had overwintered on conifers and migrated back to pear trees in early spring (remigrants). In addition to Starý Lískovec, specimens of C. pyri were also collected in two other localities in the Czech Republic and one locality in Italy, specimens of C. pyricola in three other localities in the Czech Republic and one locality in France, and additional specimens of C. pyrisuga were obtained in another locality in the Czech Republic (Tables 1 and 1S). There is no information on the phytoplasma infection status of the pear orchards where we collected psyllids. Psyllids were identified to species based on adult and immature morphology (Burckhardt, 2010; Ossiannilsson, 1992).
TABLE 1.
List of analysed psyllid samples with information on their developmental stage, sex, number of pooled specimens (N), sampling and locality data with abbreviations used in the figures. CZ = Czech Republic
| Psyllid species | Developmental stage, gender | N samples | Date | Locality with abbreviations | Latitude | Longitude |
|---|---|---|---|---|---|---|
| Cacopsylla pyri | Adult, ♂ | 5 | 04 December 2019 | CZ, Litenčice = CZ1 | −49.2056 | −17.2131 |
| Adult, ♀ | 5 | 04 December 2019 | CZ, Litenčice = CZ1 | −49.2056 | −17.2131 | |
| Immature | 5 | 04 December 2019 | CZ, Litenčice = CZ1 | −49.2056 | −17.2131 | |
| Adult, ♂ | 1 | 15 February 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 | |
| Adult, ♀ | 1 | 15 February 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 | |
| Immature | 2 | 17 May 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 | |
| Adult, ♂ | 2 | 07 July 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 | |
| Adult, ♀ | 2 | 07 July 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 | |
| Immature | 5 | 07 July 2020 | CZ, Staré Město (Svitavy) = CZ3 | −49.7857 | −16.6882 | |
| Adult, ♂ | 4 | 08 May 2020 | Italy, Merano, Plaus = IT | −46.6545 | −11.0421 | |
| Adult, ♀ | 4 | 08 May 2020 | Italy, Merano, Plaus = IT | −46.6545 | −11.0421 | |
| Cacopsylla pyricola | Adult, ♂ | 5 | 23 February 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 |
| Adult, ♀ | 5 | 23 February 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 | |
| Adult, ♂ | 5 | 10 May 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 | |
| Adult, ♀ | 5 | 10 May 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 | |
| Immature | 5 | 10 May 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 | |
| Adult, ♂ | 5 | 02 August 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 | |
| Adult, ♀ | 5 | 02 August 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 | |
| Immature | 3 | 13 September 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 | |
| Immature | 2 | 27 September 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 | |
| Adult, ♂ | 4 | 18 October 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 | |
| Adult, ♀ | 4 | 18 October 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 | |
| Adult, ♂ | 4 | 06 July 2020 | CZ, Božanov = CZ4 | |||
| Adult, ♀ | 5 | 06 July 2020 | CZ, Božanov = CZ4 | |||
| Adult, ♂ | 5 | 12 July 2020 | CZ, Hradec Králové = CZ5 | |||
| Adult, ♀ | 5 | 12 July 2020 | CZ, Hradec Králové = CZ5 | |||
| Adult, ♂ | 4 | 14 August2020 | CZ, Staré Hutě = CZ6 | −49.1394 | −17.2867 | |
| Adult, ♀ | 3 | 14 August 2020 | CZ, Staré Hutě = CZ6 | −49.1394 | −17.2867 | |
| Adult, ♂ | 2 | 01 August 2018 | France, Haut‐Rhin, Wittelsheim Rothmoos = FR | −47.7782 | −7.2386 | |
| Adult, ♀ | 3 | 01 August 2018 | France, Haut‐Rhin, Wittelsheim Rothmoos = FR | −47.7782 | −7.2386 | |
| Cacopsylla pyrisuga | Adult, ♂ | 3 | 29 March 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 |
| Adult, ♀ | 5 | 29 March 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 | |
| Immature | 5 | 10 May 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 | |
| Adult, ♂ | 3 | 02 June 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 | |
| Adult, ♀ | 5 | 02 June 2020 | CZ, Starý Lískovec (Brno) = CZ2 | −49.1623 | −16.5868 | |
| Adult, ♂ | 5 | 09 May 2020 | CZ, Staré Město (Svitavy) = CZ3 | −46.5000 | −11.3626 | |
| Adult, ♀ | 5 | 09 May 2020 | CZ, Staré Město (Svitavy) = CZ3 | −46.5000 | −11.3626 | |
| Immature | 5 | 09 May 2020 | CZ, Staré Město (Svitavy) = CZ3 | −46.5000 | −11.3626 | |
| Egg | 3 | 09 May 2020 | CZ, Staré Město (Svitavy) = CZ3 | −46.5000 | −11.3626 |
2.2. DNA extraction and 16S rRNA gene amplicon sequencing
DNA was extracted from 151 psyllid individuals (C. pyri [N = 36], C. pyricola [N = 79], C. pyrisuga [N = 39]), as well as three egg samples (5 egg specimens per sample) from C. pyrisuga (Table S1) using E.Z.N.A.® Tissue DNA Kit (Omega Bio‐tek). The specimens represented three categories of psyllid life stages: adults (N = 119), immatures (N = 32) and eggs (N = 3). Two‐step‐PCR amplification was performed using KAPA2G Robust HotStart Polymerase (Merck) (Pafčo et al., 2018). In the first step, the V3–V4 variable region of the 16S rRNA gene was targeted using primer pair 341F and 805F (Klindworth et al., 2013) containing ‘tails’ serving as priming sites for the second PCR. The thermal conditions were as follows: 95°C for 3 min, 30 cycles (95°C for 15 s, 55.5°C for 15 s, and 72°C for 15 s) and 72°C for 3 min. In the second step, Nextera primers with sample‐specific barcodes and sequencing adaptors were used under the following conditions: 95°C for 3 min, 16 cycles (95°C for 15 s, 55°C for 30 s, and 72°C for 30 s), and 72°C for 3 min. Each sample was prepared in duplicate with a unique barcode to minimize the presence of putative chimeras or other kinds of non‐biological variants (Pafčo et al., 2018). Six negative controls were also included from extraction to sequencing, which was performed on an Illumina MiSeq platform using MiSeq Reagent Kit v3 (2 × 300 bp) chemistry.
Five immature specimens of C. pyri collected from the site CZ1 in winter (Table S1) were checked for parasitoid COI sequences due to their unusual microbiome composition (e.g., missing Carsonella and unclassified Enterobacteriaceae, Figure S4). PCR amplification was performed using DreamTaq PCR Master Mix (Thermo Fisher) and the primer pair LCO and HCO (Folmer et al., 1994) with the following thermal conditions: 35 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 60 s. Parasitoid DNA was amplified from two out of the five individuals. The two amplified parasitoid COI sequences were identical and therefore only one sequence from a single specimen (P18) was submitted to GenBank with the accession number OK561501. Based on BLAST search, the obtained parasitoid sequences were identified as Syrphophagus aphidivorus (Hymenoptera: Encyrtidae) with 98.84% identity. Ten adult specimens of C. pyri were additionally checked for the presence of parasitoid DNA using two primer sets, first using the LCO and HCO primer set (Folmer et al., 1994) (the same we used earlier for the detection of parasitoid DNA in immatures), which did not produce any PCR products. Then, we used the Cacopsylla‐specific primer set VPm_COI_F2 and VPm_COI_R4 (Oettl & Schlink, 2015) and successfully amplified psyllid DNA from all tested individuals using following PCR conditions: 35 cycles of 95°C for 30 s, 46°C for 30 s, and 72°C for 60 s.
2.3. Microbiome analyses
The obtained reads were quality‐checked using FastQC (Andrews, 2010) and trimmed to a final length of 250 bp using Trimmomatic implemented on Galaxy (Afgan et al., 2018) to remove low‐quality bases at the ends of the reads. Paired reads were then joined using the join_paired_ends.py script from QIIME (v1.9) (Caporaso et al., 2010). The joined reads were denoized and assigned to ASVs using the dada2 plugin in QIIME2 (Bolyen et al., 2019). Taxonomy was assigned to the representative sequence of each ASV based on the Silva database (v132) (Yilmaz et al., 2014) using the RDP classifier (Table S2). ASVs identified as mitochondria and chloroplasts as well as low‐abundance ASVs with <3 reads were removed from the ASV table. All ASVs amplified in the negative controls (N = 193) were removed from the ASV table to eliminate potential contaminants from downstream analyses.
All statistical analyses and graphics were performed in QIIME (v1.9) and R (v3.6.3), using the following packages: vegan, MASS, ggplot2, edgeR and indicspecies. Bacterial species richness and diversity were determined using the species richness estimator Chao 1 and the Shannon index of diversity after rarefaction of all samples to an even sampling depth of 10,000 reads. Alpha diversity indices were compared (i) between host species, and (ii) within each species, between different developmental stages, seasons, localities and between males and females). This was done using pairwise t‐tests after 1000 Monte Carlo permutations, as implemented in the QIIME script compare_alpha_diversity.py. Global differences in beta diversity between the three psyllid species were analysed using Principal Coordinates Analysis (PCoA) based on Bray–Curtis distances and ANOSIM after 10,000 permutations using the QIIME script compare_categories.py. Finer scale differences in microbiome composition depending on psyllid host developmental stage, generations/seasonality, locality, and sex were investigated within each species using nonmetric multidimensional scaling (NMDS) based on Bray–Curtis distances. The EnvFit function from the R package ‘vegan’ was then applied to fit the host variables onto the NMDS ordination scores and to identify significant correlations between microbiome composition and host traits. For C. pyri and C. pyricola, development and seasonal generations were analysed both separately and together, due to their potential biological interplay. For C. pyrisuga, which produces only one generation per year, microbiome composition was compared between overwintered adults (remigrants) and adults of a new summer generation. To compare the microbiome composition between males and females, samples representing immatures and eggs, for which the sex could not be identified, were removed from the dataset. A Venn diagram indicating the amount of shared versus species‐specific ASVs was produced using the Bioconductor package ‘limma’ using the data from the only locality (CZ2) where all three Cacopsylla species were present at the adult stage.
Bacterial genera that were mainly responsible for the differences in microbiomes between the three psyllid species were determined using two different approaches: (i) The indval function from the R package ‘indicspecies’. This function performs Indicator Species Analysis by calculating an Indicator Value (IndVal) for each bacterial taxon (De Caceres & Legendre, 2009). The bacterial genera with a p‐value < 0.034 were considered significant. (ii) The R package ‘edgeR’ for differential gene expression analysis (Robinson et al., 2010). Bacterial abundance was based on log CPM (counts per million) values (Table S4). Only genera present in at least 15 samples were included in the analysis and bacterial genera were considered differentially abundant with a log2‐fold change ≥1 and with an FDR‐adjusted p‐value ≤ 0.05.
2.4. Phylogenetic reconstruction of unclassified Enterobacteriaceae endosymbionts
To understand the phylogenetic relationships of ASVs identified as unclassified Enterobacteriaceae between the three Cacopsylla species, a phylogenetic analysis of these ASVs was performed. This analysis included (1) the seven most dominant ASVs out of 10 found in Cacopsylla pyri, (2) all 20 ASVs found in C. pyricola, and (3) the 10 most dominant ASVs out of 44 found in C. pyrisuga (11,914–139,391 reads per ASV) (Table S2). The 16S rRNA gene sequences of several endosymbionts from other psyllid species, namely C. myrthi (AF263559), C. burckhardti (TAAB01000016), C. jukyungi (TAAB01000018), A. mori (AB013087) and D. citri (AB038366) from the family Psyllidae, as well as two species from Aphalaridae, C. fiscella (KY428658) and C. maniformis (KY428659), were included in the analysis. As an out group, we used Pseudomonas entomophila (MK214821). The sequences of the endosymbionts from C. burckhardti C. myrthi, C. jukyungi and A. mori were included in the dataset as they are the closest known species to the ASVs of C. pyri, C. pyricola and C. pyrisuga based on a BLAST search. The maximum likelihood (ML) analysis was performed using IQ‐TREE (Trifinopoulos et al., 2016). Nodal support was evaluated using a standard nonparametric bootstrap (BS) with 1000 replicates. Clades recovered with BS > 70% were considered significantly supported. The ML tree was visualized with iTOL (Letunic & Bork, 2021).
3. RESULTS
3.1. 16S rRNA gene amplicon sequencing
The microbiomes of 151 Cacopsylla individuals (Cacopsylla pyri, C. pyricola and C. pyrisuga), as well as three egg clutches from C. pyrisuga, were sequenced on the Illumina MiSeq platform. After raw data processing, amplicon sequencing yielded 2352–59,068 high‐quality reads per sample (mean = 24,080), which were clustered into 4–72 amplicon sequence variants (ASVs) per sample (mean = 15.88) (Table S1). Rarefaction curves reached a plateau, indicating that our sequencing effort was sufficient to capture the bacterial diversity of these communities (Figure S3).
After an initial analysis, five immature specimens of C. pyri showed very different microbiomes from all other specimens, which led us to test for parasitoid DNA. We detected parasitoid DNA in two out of the five immature individuals of C. pyri collected in winter from the site CZ1 (Table S1, highlighted in yellow; Figure S4). Since the microbiomes of the other three immature individuals from the same collection date and site were similar to the microbiomes of the two confirmed parasitized individuals, all five immature specimens of C. pyri were discarded from the analyses. No parasitoid DNA was detected in adult specimens of C. pyri.
3.2. Pear psyllid species harbour different endosymbionts taxa
PCoA based on Bray–Curtis distances discriminated the three Cacopsylla species from each other, showing that the three pear psyllid species harbour different microbiomes (Figure 2A). Differences in the bacterial species composition among the three Cacopsylla species were significant based on ANOSIM (R = 0.927, p = 0.0001) and explained 40.50% of the variation between the bacterial communities. When comparing the microbiomes of the three Cacopsylla species from a locality where all species co‐occurred (CZ2), only 7 out of 1046 ASVs were found in all three Cacopsylla species (Figure 2B). The species which shared the most ASVs were Cacopsylla pyricola and C. pyrisuga (18), whereas C. pyri and C. pyricola shared only 2 ASVs, and C. pyri and C. pyrisuga had no ASVs in common. Hence, even when co‐occurring in the same locality, the three species maintain distinct bacterial communities. Despite the clear differences in microbiome composition, we found no differences in bacterial species richness (Chao 1) between the three species (C. pyri vs. C. pyricola: t‐test = −0.056, p = 1.000; C. pyricola vs. C. pyrisuga: t‐test = −1.088, p = 0.859; and C. pyri vs. C. pyrisuga: t‐test = −1.194, p = 0.701) (Figure 2C). However, the bacterial diversity based on the Shannon index was significantly higher in C. pyri compared to both C. pyricola (t‐test = −3.267, p = 0.006), and C. pyrisuga (t‐test = −2.523, p = 0.042) (Figure 2D). Specifically, bacterial diversity reached a mean Shannon index of 2.39 (range = 1.41–3.13) in C. pyri, compared to 2.02 (range = 1.17–3.88) in C. pyricola and 2.14 (range = 1.36–3.00) in C. pyrisuga (Table S1).
FIGURE 2.
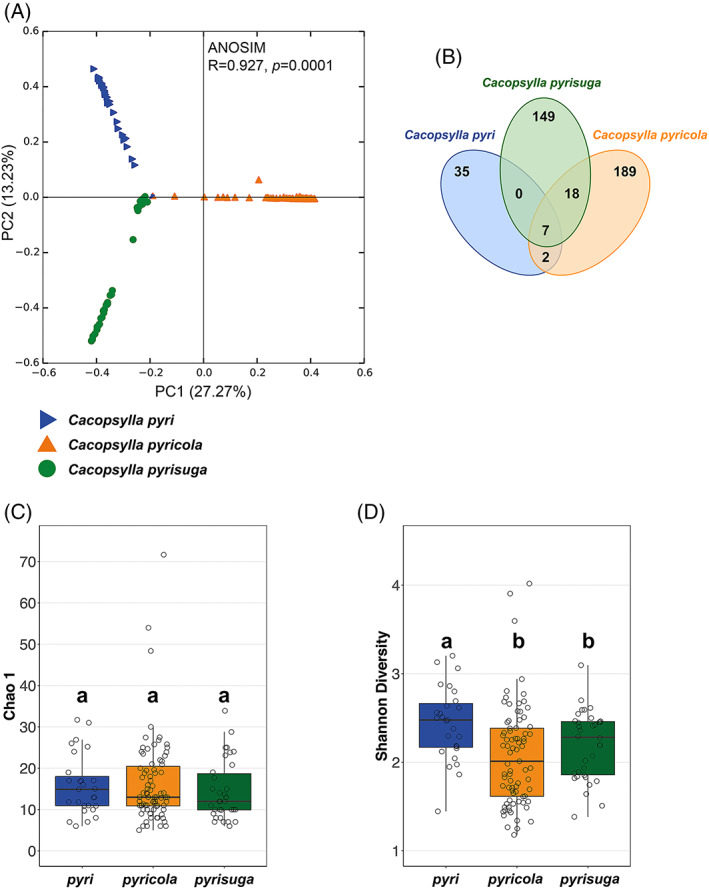
(A) Principal coordinates analysis based on bray–Curtis dissimilarity metrics showing differences in microbiome composition between the three Cacopsylla species. (B) Venn diagram showing the shared and specific bacterial ASVs between adult specimens of all three species co‐occurring in the same locality (CZ2). (C) Comparison of bacterial species richness (Chao 1) and (D) diversity (Shannon index) between individuals of the three Cacopsylla species.
In this study, Carsonella reads represented only 0%–21.8% of the reads per specimen. The primer set used (341F and 805R) was designed to amplify a wide range of bacteria but contained several mismatches with Carsonella in the forward primer (Klindworth et al., 2013; Morrow et al., 2017; Nakabachi et al., 2022). This led to an inefficient amplification of Carsonella, but favoured the amplification of other bacterial taxa (consistent with Morrow et al., 2017, 2020 and Kwak et al., 2021).
Apart from Carsonella, the microbiomes of the studied psyllid species were dominated by very few bacterial taxa. Hence, only 9–20 ASVs per psyllid species corresponded to an abundance >1% of the reads for the given species: 15 ASVs for C. pyri, 9 ASVs for C. pyricola and 20 ASVs for C. pyrisuga. Specifically, all individuals harboured abundant bacteria classified as Enterobacteriaceae (Figure 3A). Both C. pyri and C. pyricola harboured several ASVs of this endosymbiont, which was given for convenience the following provisional names in our study: ‘Group1 pyri’ (58.90% of all reads from C. pyri) for ASVs found in C. pyri and ‘Group1 pyricola’ (89.90% of all reads from C. pyricola) for those found in C. pyricola. A maximum likelihood phylogenetic analysis of these ASVs and 16S rRNA sequences from other psyllid‐associated endosymbionts showed that Group 1 pyri and Group 1 pyricola formed a strongly supported clade together with the endosymbiont of Cacopsylla jukyungi (BS = 97), which was well‐supported as a sister clade to the endosymbionts of C. myrthi and Anomoneura mori (BS = 100) (Figure 3B). Likewise, the close relationship with Enterobacteriaceae endosymbionts in C. myrthi and A. mori had been observed also in other Cacopsylla species (Morrow et al., 2020; Schuler et al., 2022), suggesting that these ASVs represent an endosymbiont clade widespread in the subfamily of Psyllinae (Psyllidae). In contrast, the most abundant ASVs assigned to Enterobacteriaceae in C. pyrisuga were referred to as “Group2 pyrisuga” in our study (87.56% of all reads from C. pyrisuga). Group2 pyrisuga showed its close relatedness to the Sodalis endosymbionts of Cacopsylla burckhardti and Cardiaspina maniformis (Aphalaridae) (BS = 96) (Figure 3B). Together, Group2 pyrisuga and the Sodalis endosymbionts of C. burckhardti and C. maniformis were placed as a sister clade to the one formed by Group1 pyri, Group1 pyricola and the endosymbionts of C. jukyungi, C. myrthi and A. mori (BS = 74). Last, Arsenophonus endosymbionts of C. fiscella (Aphalaridae) and D. citri (Psyllidae) established a well‐supported monophyletic group (BS = 97) that was placed as a sister to all analysed bacterial taxa. Taken together, our results indicate that the three studied psyllid species each harbour a highly abundant endosymbiont and that C. pyrisuga harbours a different taxon compared to the endosymbiont of C. pyri and C. pyricola.
FIGURE 3.
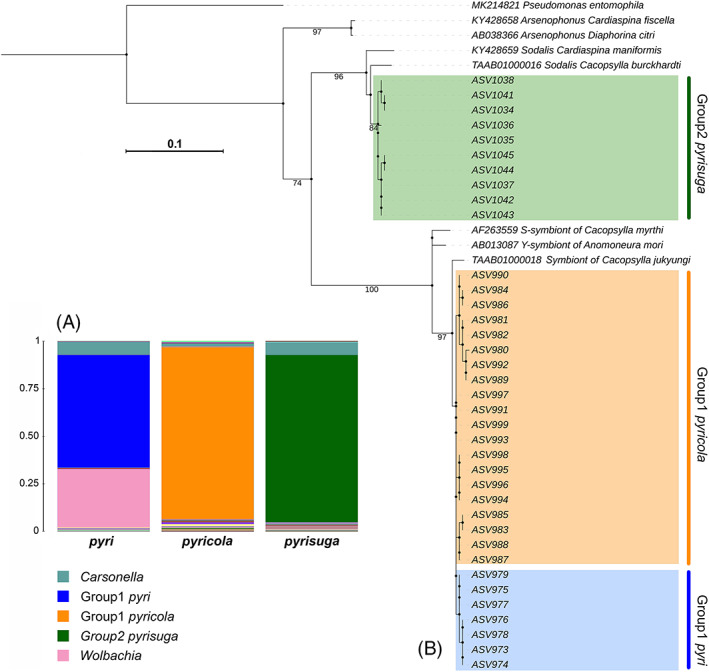
(A) Microbiome composition of the three Cacopsylla species. The most abundant bacterial genera are represented in the legend. (B) Maximum likelihood tree of 16S rRNA gene ASV sequences belonging to the Enterobacteriaceae endosymbionts of pear psyllids. The tree displays the relationships between the endosymbionts Group1 pyri, Group1 pyricola and Group2 pyrisuga, harboured by Cacopsylla pyri, C. pyricola and C. pyrisuga, respectively.
To better understand which bacterial taxa were responsible for the differences between the microbiomes of the three Cacopsylla species, we determined indicator species (using the R function IndVal) and differentially abundant bacterial taxa (edgeR). The IndVal analysis detected 10 bacterial taxa as potential indicator taxa which were predominantly present in only one psyllid species: six taxa in C. pyri, and two taxa each for C. pyricola and C. pyrisuga. These corresponded to Group1 pyri, Wolbachia, Streptococcus, Pediococcus, Microbacterium and Fimbriimonas for C. pyri; Group 1 pyricola and Gluconobacter for C. pyricola; and Group 2 pyrisuga and Rickettsia for C. pyrisuga (Figure 4). Five additional bacterial taxa were identified as differentially abundant between the three psyllid species. Specifically, Bradyrhizobium and Methylobacterium were more abundant in C. pyricola compared to C. pyri and C. pyrisuga, and Sphingomonas and Variovorax were more abundant in C. pyricola compared only to C. pyrisuga. Pajaroellobacter was more abundant in C. pyri and C. pyricola compared to C. pyrisuga (Figure 4).
FIGURE 4.
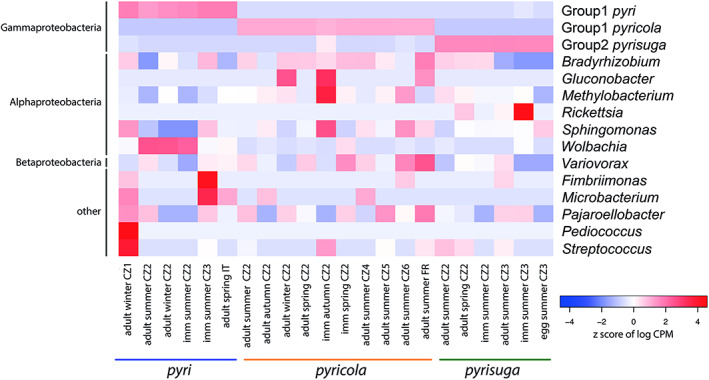
Heatmap showing the abundance (log counts per million) of 15 bacterial genera identified as either indicator species and/or differentially abundant between the three Cacopsylla species. Each column represents several psyllid individuals that are grouped together based on their developmental stage, generation (seasonality) and geographic location. Bacterial genera are ordered based on class level designations represented on the left of the heatmap. Psyllid species are indicated by the coloured bar below the heatmap.
3.3. Impact of host biology and geography on psyllid microbiome
The effect of different developmental stages, generations for polyvoltine species, seasonality for univoltine species, sex and different geographical localities on microbiome structure were tested for each of the three Cacopsylla species. Correlations between these factors and differences in microbiome composition (beta diversity) were evaluated using EnvFit (Table 2) applied to NMDS analyses based on Bray–Curtis distances (Figures 5A–D). The species richness and diversity indices (Chao 1 and Shannon, respectively) were used to detect differences in alpha diversity depending on the above‐mentioned factors (Table S3; Figures 5E–F). All analyses were performed for each psyllid species separately due to the substantial differences in their biology and sampling sites. There were no differences in microbiome composition between males and females in any of the studied psyllid species (Table 2; Figures 6C, 7C and 8E).
TABLE 2.
EnvFit results for each Cacopsylla species applied to nonmetric multidimensional scaling (NMDS) based on Bray–Curtis distances, with the following categories examined: Developmental stage, generation (seasonality), locality and sex. Significant p‐values are in bold
| Psyllid species | Developmental stage | Generation (seasonality) | Developmental stage + generation | Locality | Sex |
|---|---|---|---|---|---|
| Cacopsylla pyri | r 2 = 0.053, p = 0.191, stress = 0.165 | r 2 = 0.089, p = 0.263, stress = 0.165 | r 2 = 0.243, p = 0.004, stress = 0.165 | r 2 = 0.330, p = 0.001, stress = 0.165 | r 2 = 0.054, p = 0.319, stress = 0.140 |
| Cacopsylla pyricola | r 2 = 0.089, p = 0.002, stress = 0.206 | r 2 = 0.040, p = 0.453, stress = 0.206 | r 2 = 0.164, p = 0.005, stress = 0.206 | r 2 = 0.032, p = 0.775, stress = 0.206 | r 2 = 0.011, p = 0.478, stress = 0.208 |
| Cacopsylla pyrisuga | r 2 = 0.036, p = 0.614, stress = 0.144 | r 2 = 0.073, p = 0.065, stress = 0.144 | r 2 = 0.006, p = 0.833, stress = 0.144 | r 2 = 0.075, p = 0.143, stress = 0.124 |
FIGURE 5.
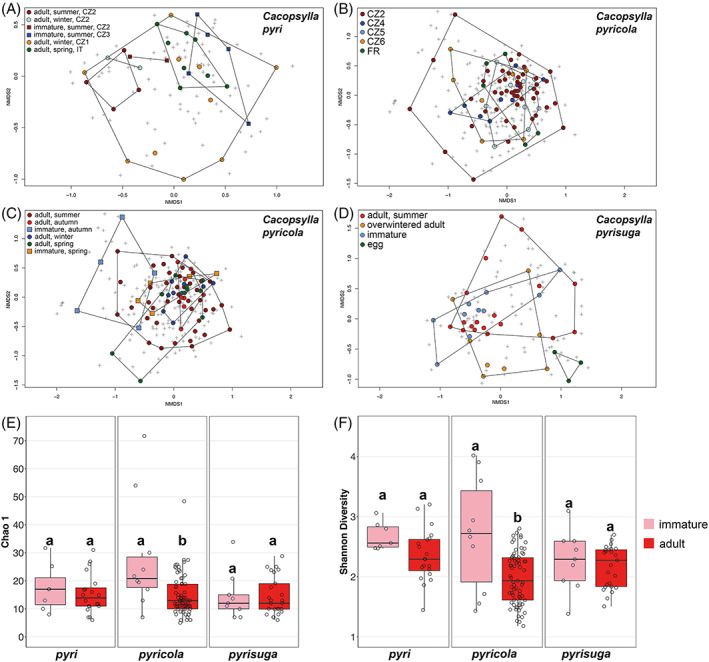
Impact of host life history traits on the bacterial communities of the three Cacopsylla species visualized with nonmetric multidimensional scaling. Datapoints are coloured based on the tested life history traits. Group boundaries were drawn using the ordihull function (R package ‘vegan’). (A) Microbiome composition of C. pyri individuals along host ontogeny, generations and localities. (B) Microbiome composition of C. pyricola individuals from different localities. (C) Microbiome composition of C. pyricola individuals along host ontogeny and generations. (D) Microbiome composition of C. pyrisuga individuals along host ontogeny and seasonality. Comparison of bacterial species richness (E) and diversity (F) between immature and adult individuals of the three Cacopsylla species. Different letters indicate significant differences based on t‐tests.
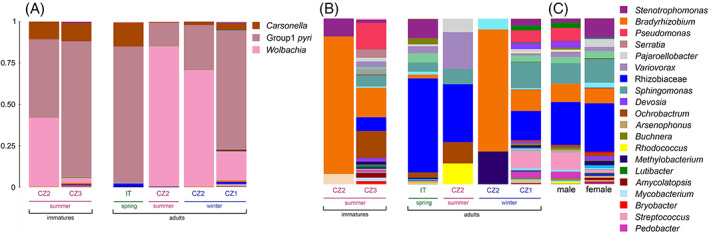
FIGURE 6.
Microbiome composition of Cacopsylla pyri depending on developmental stages, generations and localities. The most abundant bacterial genera are represented in the legend. (A) Microbiome composition including the most abundant taxa Carsonella, Group1 pyri and Wolbachia and (B) after removing ASVs of Carsonella, Group1 pyri and Wolbachia. (C) Comparison of microbiome composition between males and females after removing ASVs belonging to Carsonella, Group1 pyri and Wolbachia. Detailed information on all samples is provided in Table S1.
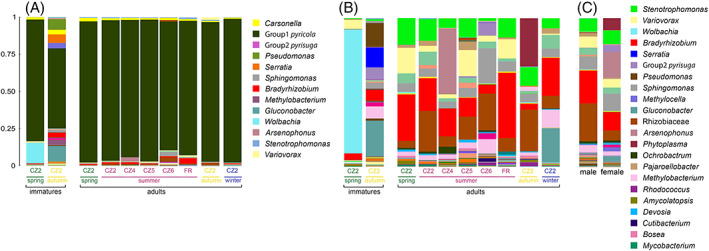
FIGURE 7.
Microbiome composition of Cacopsylla pyricola depending on developmental stages, generations and localities. The most abundant bacterial genera are represented in the legend. (A) Microbiome composition including the most abundant taxa Carsonella and Group 1 pyricola and (B) after removing ASVs of Carsonella and Group 1 pyricola. (C) Comparison of microbiome composition between males and females after removing ASVs belonging to Carsonella and Group1 pyricola. Detailed information on all samples is provided in Table S1.
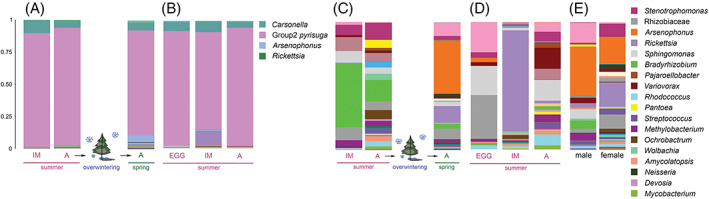
FIGURE 8.
Microbiome composition of Cacopsylla pyrisuga depending on developmental stages, seasonality and localities. The most abundant bacterial genera are represented in the legend. (A) Microbiome composition across developmental stages of C. pyrisuga from a single locality (CZ2) including Carsonella and Group 2 pyrisuga and (C) after removing ASVs of Carsonella and Group2 pyrisuga. (B) Microbiome composition across developmental stages of C. pyrisuga from a single locality (CZ3) including Carsonella and Group 2 pyrisuga and (D) after removing ASVs of Carsonella and Group 2 pyrisuga. (E) Comparison of microbiome composition between males and females after removing ASVs of Carsonella and Group 2 pyrisuga. Detailed information on all samples is provided in Table S1.
3.3.1. The microbiome of C. pyri varied across development, seasonal generations and geography
We found significant differences in microbiome composition between different developmental stages, generations and localities for C. pyri (Table 2; Figure 5A). Notably, the interaction between host developmental stages and generations was significant (EnvFit: r 2 = 0.243, p = 0.004), while both factors were not significant when tested independently (Table 2). The effect of geographical localities on the microbiome was tested for four populations of C. pyri collected in two countries (Czech Republic, Italy) (Tables 1 and S1). The locality was the most significant factor shaping microbiome composition, even when the most distant population collected in Italy (IT) was excluded from the analysis to evaluate only the differences between the Czech populations (EnvFit: r 2 = 0.330, p = 0.001). A major difference between the different localities was that individuals from the Czech populations CZ1, CZ2 and CZ3 harboured Wolbachia at varying relative abundances (20.19%, 65.42% and 3.02%, respectively), whereas only traces of Wolbachia (0.10%) were found in the IT population (Figures 6A and S4).
In total, the microbiome of C. pyri contained 20 ASVs assigned to Wolbachia (Table S2). Eight were present in all specimens (P1–P8) collected at the site CZ2 (Figure S4A), which represented different developmental stages and generations (Table S1). The relative abundance of Wolbachia varied with host ontogeny when comparing specimens collected from the same locality (CZ2) (Figure 6A). Wolbachia was more abundant in adults from the summer (84.43%) and overwintering generation (70.29%) compared to summer‐form immatures (41.54%). At the site CZ1, Wolbachia was found in a single male individual (P20) which exhibited a very high Wolbachia abundance (91.47% of all reads from this specimen) represented by 3 ASVs identical to the ones harboured by the individuals from CZ2 (Table S2; Figure S4A). In contrast, Wolbachia from the site CZ3 comprised a single ASV different from those found at the sites CZ1 and CZ2 (Table S2). To obtain a better picture of the more abundant bacterial taxa apart from the predominating endosymbionts Group1 pyri, Carsonella and Wolbachia, their ASVs were removed from the dataset (Figures 6B and S4B). This revealed several abundant taxa assigned either to the genus Bradyrhizobium or to the family Rhizobiaceae. Both ASV groups were present in most individuals but fluctuated in relative abundance between different generations and localities (Figures 6B and S4B).
3.3.2. The microbiome of Cacopsylla pyricola varied across development and seasonal generations but not geography
In Cacopsylla pyricola, geography had no significant effect on its bacterial community structure despite being tested on five populations collected in two countries (Czech Republic, France) (Tables 2 and S4; Figure 5B). However, as for C. pyri, in C. pyricola the interaction between host developmental stages and generations was correlated with changes in microbiome composition (EnvFit: r 2 = 0.164, p = 0.005) (Table 2; Figure 5C). Notably, host developmental stages had a significant impact on microbiome composition on its own (EnvFit: r 2 = 0.089, p = 0.002) (Table 2; Figure 5C). In addition, bacterial species richness and diversity were higher in immatures than in adults (Chao 1: t‐test = −3.966, p = 0.001; Shannon: t‐test = −4.191, p = 0.002) (Figures 5E–F). In contrast, the effect of different generations was significant only in combination with host developmental stages (Table 2; Figure 5C), suggesting that changes in microbiome composition between different generations occurred in only one developmental stage, either immatures or adults, but not in both. Considering the NMDS pattern, this seems more likely for immatures, since immatures collected in spring and autumn do not cluster together (Figure 5C). This suggests that differences in microbiome composition were most strongly correlated with host development in this species. This pattern could be explained by the high abundance of Wolbachia only in spring‐form immatures (83.19% of reads after removing the ASVs of the predominant endosymbionts), whereas no Wolbachia was found in autumn‐form immatures from the same site, nor in adults of C. pyricola (Figures 7A–B). Contrary to spring‐forms (Figures 7A–B), autumn‐form immatures harboured several other abundant bacterial taxa, such as Gluconobacter (24.01%), Pseudomonas (15.58%) and Serratia (13.46%). Bacterial species richness and diversity were also significantly higher in autumn‐form compared to spring‐form immatures (mean Chao 1 = 35.998, mean Shannon = 3.130 for autumn‐form immatures; and mean Chao 1 = 14.975, mean Shannon = 2.139 for spring‐form immatures) (Table S1). In contrast to immatures, Gluconobacter, Pseudomonas and Serratia were not detected in autumn‐form adults which, however, carried higher relative abundances of unidentified Rhizobiaceae (27.86%) and Phytoplasma (32.46%) (Figure 7B). The latter was found in a single female where Phytoplasma reached an abundance of 93% of all reads (Figure S5B). Surprisingly, adults collected in winter showed a similarly high relative abundance of Gluconobacter as immatures (22.96%), but no Pseudomonas and Serratia (Figure 7B). Taken together, these results suggest that most changes in microbiome composition in C. pyricola occur in immatures, whose microbiomes differed between seasons and were different from the microbiomes of adults from the same season.
3.3.3. Developmental stage, seasonality and geography had no effect on microbiome of Cacopsylla pyrisuga
In C. pyrisuga, neither host developmental stages, seasonality nor geography were significantly correlated with changes in bacterial species richness, diversity and microbiome composition (Tables 2 and S4, Figures 5E,F), suggesting that the bacterial community of C. pyrisuga remains relatively stable during the psyllid life cycle and between populations from different localities. Nonetheless, there may be a trend towards a different microbiome composition in eggs compared to other developmental stages, as seen by the clustering of the egg microbiome when visualized employing the NMDS ordination (Figure 5D). Apart from Group2 pyrisuga and Carsonella (Figures 8A–B and S6A), several bacterial genera reached relatively high relative abundances in this species, namely Bradyrhizobium, Arsenophonus, Rickettsia, Sphingomonas and unclassified Rhizobiaceae (Figures 8C,D and S6B). The latter two taxa reached high proportions (22.86% and 34.40%, respectively) in the three egg samples from the site CZ3 (Figure 8D). Compared to the eggs, a high relative abundance of Rickettsia (79.35%), represented by three ASVs, was found in immatures from the same locality (Table S2). During the host ontogeny from immature to adult, Rickettsia declined drastically, while Variovorax (16.55%) and Sphingomonas (16.32%) increased in relative abundance (Figure 8D). At the immature stage, these genera were either absent or present at low relative abundance. Minor compositional differences were detected between immatures and summer‐form adults from site CZ2 (Figure 8C). Interestingly, only overwintered adults collected on pears from the same site in spring harboured Arsenophonus (41.74%), followed by Rickettsia (13.32%) (Figure 8C). The latter occurred in a single female, reaching 93.15% relative abundance in this particular specimen (Figure S6B).
4. DISCUSSION
Our study characterized and compared the microbiomes of the three psyllid species Cacopsylla pyri, C. pyricola and C. pyrisuga associated with pear trees in Central Europe. The microbiomes of the three species were clearly distinct from one another, which led us to investigate whether differences in host life cycle, developmental stages, sex and geography could lead to changes in the bacterial community structure of the insect host (Gonella et al., 2019; McLean et al., 2019). We also examined the phylogenetic relationships of the prevailing endosymbionts within the studied psyllid species. Overall, our metabarcoding approach captured three different types of psyllid‐associated bacteria: (i) Bacteriome‐associated primary endosymbionts which provide the host with essential nutrients, (ii) facultative intracellular bacteria which can be localized in various host tissues and may have beneficial, neutral or detrimental effects on the host, and (iii) extracellular bacteria, notably within the insect gut.
4.1. Impact of host biology, ontogeny and geography on pear psyllid microbiomes
We found that the microbiomes of C. pyri and C. pyricola were more variable compared to the microbiome of C. pyrisuga. This was most pronounced in C. pyri, whose microbiome composition varied between different localities but also along host ontogeny and between different seasonal generations. In C. pyricola, the microbiome underwent major changes during development towards a lower species richness and diversity at the adult stage. Similarly, the bacterial diversity in the Asian Citrus psyllid D. citri showed a declining tendency towards the adult stage (Meng et al., 2019), and the bacterial richness in the tobacco whitefly Bemisia tabaci was found to be highest in first instar nymphs (Indiragandhi, 2010). We can only hypothesize about the mechanisms underlying the shifts in microbiome structure throughout the ontogenetic development of psyllids. For example, following recent studies on feeding behaviour (George et al., 2018; Görg & Gross, 2021), immatures of psyllids spend more time ingesting phloem than adults. This longer feeding time on nutrient‐rich phloem might explain the higher bacterial richness and diversity found in immatures of C. pyricola. Another plausible explanation of varying bacterial richness and diversity along host life cycle might be differences in host physiology (feeding behaviour, immune response, maturation of the reproductive organs) between different life stages (Arp et al., 2016; Inoue et al., 2009; Kandasamy et al., 2022; Nishide et al., 2019).
Cacopsylla pyri and C. pyricola are two species with very similar biology, but different from C. pyrisuga: both species produce several generations per year and overwinter predominantly on pear trees. Interestingly, the studies by Horton et al. (1992, 1994) on the biology of C. pyricola from North America, where this species was introduced from Europe, demonstrated that some individuals overwinter on conifers or in apple orchards, while other individuals from the same population stay in pear orchards throughout an entire year. In contrast to C. pyri and C. pyricola, C. pyrisuga is a univoltine species that has an obligate overwintering association with conifers (Hodkinson, 2009; Lauterer, 1999). This suggests that the presence of only one generation in C. pyrisuga or its obligate overwintering on shelter plants may contribute to the relative stability of microbiome of C. pyrisuga.
In contrast to C. pyri and C. pyricola, in C. pyrisuga we found no changes in the bacterial community structure depending on the tested factors (developmental stages, seasonality and geography). C. pyrisuga had not been considered a competent vector of ‘Ca. Phytoplasma mali’ (Jarausch et al., 2019; Jarausch & Jarausch, 2010), until Riedle‐Bauer et al. (2022) recently demonstrated experimentally that remigrants of C. pyrisuga are able to transmit ‘Ca. Phytoplasma mali’ to pear trees in Austria. Although naturally infected individuals of C. pyrisuga were previously found in the Czech Republic, their ability to transmit phytoplasma has not yet been proven there (Kučerová et al., 2007). In a similar manner, the apple psyllid Cacopsylla melanoneura was shown to transmit ‘Ca. Phytoplasma mali’ in Northern Italy but not in Germany (Mayer et al., 2009; Tedeschi et al., 2012). Thus, the capability to transmit phytoplasma might vary across regions. Therefore, transmission trials are needed to validate a putative vector competence of C. pyrisuga across its distributional range. Future research should also test if the stability in the microbiome structure of C. pyrisuga from Czech populations or the presence of Arsenophonus or a Sodalis‐like potential co‐primary endosymbiont in the microbiome could contribute to its vector incompetence/competence in this area.
4.2. Major facultative symbionts of the studied Cacopsylla species
In our study, Wolbachia was found at a high relative abundance only in Cacopsylla pyri, particularly at the adult stage. This is consistent with results from other studies on psyllid microbiomes, demonstrating that Wolbachia abundance tends to increase towards the adult stage (Meng et al., 2019; Ramsey et al., 2017). This is thought to be associated with high nutritional demands and organogenesis during psyllid development (Ramsey et al., 2017). Wolbachia is a common bacterium, infecting 40%–52% of terrestrial arthropods, and is mostly maternally inherited (Kaur et al., 2021; Weinert et al., 2015). Although Wolbachia maintains nutritional mutualistic associations with some hemipteran hosts (Hosokawa et al., 2010), most studies on insects demonstrated Wolbachia‐induced reproductive manipulations (Engelstädter & Hurst, 2009; Schuler et al., 2016), and/or protection against pathogens (Hedges et al., 2008; Teixeira et al., 2008). In psyllids, the role of Wolbachia has been explored only in several pest species, despite Wolbachia's ubiquitous distribution in this group (Fromont et al., 2016, 2017; Fu et al., 2020; Morrow et al., 2017, 2020; Nakabachi et al., 2022; Overholt et al., 2015). Particularly, previous studies on D. citri demonstrated a link between Wolbachia infection and ‘Ca. Liberibacter asiaticus’ titers, indicating that Wolbachia may both mitigate and impede the transmission of the pathogen in D. citri (Fagen et al., 2012; Hosseinzadeh et al., 2019; Kruse et al., 2017; Song et al., 2019). Likewise, Wolbachia strains that might induce cytoplasmatic incompatibility were detected in the potato psyllid B. cockerelli (Cooper, Swisher, et al., 2015; Fu et al., 2020; Hail et al., 2012; Nachappa et al., 2011). Considering the major biological roles that Wolbachia plays in other insect taxa (Bourtzis et al., 2014; Chrostek et al., 2013; Landmann, 2019; Nikoh et al., 2014; Weinert et al., 2015), its impact on biology and vector competence of C. pyri is of great interest.
Arsenophonus was observed in this study in high relative abundance only in overwintered adults (remigrants) of C. pyrisuga, although it was also detected at low abundance in several individuals of C. pyri and C. pyricola. Like other univoltine species that overwinter on conifers, that is, Cacopsylla pruni and C. picta (Barthel et al., 2020; Gallinger & Gross, 2018), C. pyrisuga can probably feed on conifer sap. The additional food source broadens the potential bacterial influx. In theory, both the conifer sap and the pear tree phloem (Gallinger & Gross, 2018) might mediate horizontal transmission of different bacteria among various insects associated with these plants. The genus Arsenophonus is known for its diversity and frequent horizontal transmissions between distantly related insect species (Jousselin et al., 2013; Mouton et al., 2012; Russell et al., 2003). Its broad range of symbiotic relationships encompasses both beneficial and manipulative associations (Ferree et al., 2008; Gherna et al., 1991; Hansen et al., 2007; McCutcheon et al., 2019; Nováková et al., 2009; Šochová et al., 2017). While Arsenophonus was previously detected in various psyllid species (Hall et al., 2016; Morrow et al., 2017; Nakabachi et al., 2022; Schuler et al., 2022; Sloan & Moran, 2012; Subandiyah et al., 2000; Thao et al., 2000), potentially as an obligate or at least beneficial symbiont (Hall et al., 2016; Hansen et al., 2007; Morrow et al., 2017; Subandiyah et al., 2000), its role in C. pyrisuga, particularly in remigrants that could transmit phytoplasma, remains to be elucidated.
To date, only a few studies have reported Rickettsia in psyllids (Cooper et al., 2021; Morrow et al., 2020; Nakabachi et al., 2022; Pilgrim et al., 2021; Schuler et al., 2022). We showed both immatures and overwintered adults of C. pyrisuga to harbour Rickettsia. Previously, mostly known as vector‐borne pathogens of vertebrates, Rickettsia are intracellular bacteria associated with a broad range of arthropods including phloem‐feeding hemipterans (Pilgrim et al., 2021; Wang et al., 2020; Weinert et al., 2015). Some hemipteran strains of Rickettsia have profound effects on insect biology and vector–pathogen interactions (Kliot et al., 2014; Morrow et al., 2020; Pilgrim et al., 2021; Wang et al., 2020). For instance, it was demonstrated that Rickettsia manipulates the transmission efficiency of the Tomato yellow leaf curl virus by the whitefly Bemisia tabaci (Kliot et al., 2014). This highlights the importance of future studies on potential effects of Rickettsia and Arsenophonus associates pathogen transmission abilities of C. pyrisuga.
Apart from these facultative intracellular symbionts, we also detected numerous other bacterial taxa, many of which likely represented extracellular bacteria from the insect gut. Despite being captured only at low abundance compared to the predominating intracellular symbionts, some of these were identified as species‐specific (Fimbriimonas, Gluconobacter, Microbacterium, Pediococcus and Streptococcus) or differentially abundant taxa (Bradyrhizobium, Methylobacterium, Pajaroellobacter, Sphingomonas and Variovorax), thereby contributing to the differences between the microbiomes of the three species. However, to date nothing is known regarding the potential roles of these bacteria in psyllids.
4.3. Potential co‐primary endosymbionts of pear psyllids and their phylogenetic relationships
Like other groups of sap‐feeding insects, psyllids maintain an obligatory association with the primary endosymbiont ‘Candidatus Carsonella ruddii’, which synthesizes essential amino acids and thus compensates for their deficiency in the phloem sap (Nakabachi et al., 2006; Sloan & Moran, 2012; Tamames et al., 2007). Carsonella resides in specialized host cells, bacteriocytes, which constitute a tissular structure called a bacteriome. It appears to be universally present in psyllids and represents a case of strict host–symbiont co‐speciation, as a result of vertical transmission and long‐term evolutionary co‐divergence between the endosymbiont and its insect host (Fromont et al., 2016; Hall et al., 2016; Thao et al., 2000). In addition to the obligate primary (P‐) endosymbiont Carsonella, numerous other secondary endosymbionts were detected in various psyllids. Depending on the psyllid species, these belong to various bacterial groups, notably Betaproteobacteria (Profftella) and Enterobacteriaceae (Arsenophonus, Sodalis, unclassified Enterobacteriaceae) (Fromont et al., 2016, 2017; Morrow et al., 2017, 2020;Nakabachi et al., 2013 ; Nakabachi, Malenovský, et al., 2020 ; Nakabachi, Piel, et al., 2020). Interestingly, some of these endosymbionts were shown to occur in the syncytium of the bacteriome, that is, they co‐inhabit the bacteriome together with Carsonella and can therefore be vertically transmitted (Fukatsu & Nikoh, 1998; Nakabachi et al., 2013; Sloan & Moran, 2012; Subandiyah et al., 2000). Hence, these secondary endosymbionts may in fact act as co‐primary (CP‐) nutritional and/or defensive endosymbionts with reduced genomes due to their obligate host‐associated lifestyle (Nakabachi et al., 2013; Nakabachi, Piel, et al., 2020; Sloan & Moran, 2012). In contrast to Carsonella, other endosymbionts such as Arsenophonus and Sodalis exhibit patterns of past horizontal transmission, suggesting more recent independent acquisitions and/or occasional host shifts (Fromont et al., 2016; Hall et al., 2016).
In our study, we detected Enterobacteriaceae in all analysed specimens of the three Cacopsylla species. These endosymbionts were recovered at a very high relative abundance and were identified as unclassified Enterobacteriaceae that are not yet formally described. Similarly, Morrow et al. (2017, 2020), Kwak et al. (2021), Nakabachi et al. (2022) and Schuler et al., 2022 found endosymbionts referred to as unclassified Enterobacteriaceae in several Cacopsylla species. Altogether, unclassified Enterobacteriaceae were detected in six out of seven currently recognized psyllid families (cf. Burckhardt et al., 2021), namely Aphalaridae, Calophyidae, Carsidaridae, Liviidae, Psyllidae and Triozidae (Fromont et al., 2017; Kwak et al., 2021; Morrow et al., 2017, 2020; Nakabachi et al., 2022; Overholt et al., 2015). Our study showed that C. pyri and C. pyricola share similar Enterobacteriaceae endosymbionts, closely related to the endosymbionts of C. jukyungi (the latter species is also associated with pear trees but distributed in eastern Asia; Cho et al., 2017, 2020), C. myrthi and A. mori, while the endosymbionts of C. pyrisuga and C. burckhardti are more closely related to the genus Sodalis. Nakabachi et al. (2022) identified the endosymbiont of C. burckhardti as Sodalis given that its ASVs showed 94.6%–97.9% identity to previously identified Sodalis from various insect groups. Given that C. pyrisuga and C. burckhardti are two closely related species, both associated with pear trees (Cho et al., 2017, 2020), it is interesting to note that their endosymbionts also seem to be closely related and might represent a new Sodalis‐like lineage. However, because our results on the phylogenetic relationships of Enterobacteriaceae endosymbionts of the studied Cacopsylla species were based on short fragments of the 16S rRNA gene, this should be interpreted with caution. Considering the occurrence of Enterobacteriaceae endosymbionts at high abundance in all analysed psyllid specimens as well as in other psyllid taxa (e.g., Kwak et al., 2021; Morrow et al., 2017, 2020; Nakabachi et al., 2022; Overholt et al., 2015; Schuler et al., 2022), their presence throughout the entire psyllid life cycle and relatedness to the endosymbionts of C. myrthi and A. mori, which are known to be localized in the bacteriome together with Carsonella, it is likely that the Enterobacteriaceae endosymbionts recovered from C. pyri, C. pyricola and C. pyrisuga are vertically transmitted, obligate co‐primary (CP‐) endosymbionts. However, to validate our hypothesis, the metabolic potential and tissular localization of the Enterobacteriaceae endosymbionts still need to be elucidated.
Based on the phylogenomic study of Psylloidea by Percy et al. (2018), all Cacopsylla species associated with Rosaceae, including C. pyri, C. pyricola and C. pyrisuga, belong to one clade. In turn, C. myrthi associated with the plant family Rhamnaceae is a member of a different Cacopsylla species group, whereas A. mori belongs to a more distantly related genus of Psyllidae. Considering this pattern of phylogenetic relationships within Psyllidae (Cho et al., 2020; Percy et al., 2018), the phylogeny of the Enterobacteriaceae endosymbionts does not fully match the phylogeny of their hosts. This is in line with previously observed incongruences between phylogenies of psyllids and their secondary endosymbionts (CP‐endosymbionts in the present study) (Hall et al., 2016; Thao et al., 2000; Thao & Baumann, 2004) and supports the hypothesis of past horizontal transfers of the secondary endosymbionts and their host shifts throughout the evolutionary history of psyllids.
According to our results, the most striking distinction between the studied pear psyllids lies in the presence of their dominant Enterobacteriaceae endosymbionts. As shown in several studies on insect microbiomes, various endosymbionts can play different functional roles in their hosts (Gottlieb et al., 2010; Hansen et al., 2007; Nakabachi et al., 2020b; Nikoh et al., 2014). For instance, a defensive role was demonstrated for Profftella, which co‐occurs with Carsonella in the bacteriome of Diaphorina spp. (Nakabachi et al., 2013; Nakabachi, Malenovský, et al., 2020; Nakabachi, Piel, et al., 2020). Future investigations of the biological role(s) of CP‐endosymbionts of pear psyllids using genomics would be necessary to better understand their functional and metabolic potential. This will also contribute to a better understanding of the relationships between CP‐endosymbionts and their psyllid hosts across the psyllid tree of life.
AUTHOR CONTRIBUTIONS
Liliya Štarhová Serbina and Hannes Schuler conceived the project; Liliya Štarhová Serbina, Domagoj Gajski and Igor Malenovský collected and identified the psyllid specimens; Liliya Štarhová Serbina, Ludek Zurek and Eva Nováková organized the data; Liliya Štarhová Serbina, Domagoj Gajski and Barbora Pafčo conducted molecular analyses; Jessica Dittmer supervised bioinformatics and statistical analyses; Liliya Štarhová Serbina and Jessica Dittmer performed data analysis; Liliya Štarhová Serbina prepared the first draft and wrote the manuscript together with Jessica Dittmer. All authors provided feedback that helped to improve the manuscript.
Supporting information
Figure S1 Habitus of adult Cacopsylla pyri – female (A), C. pyricola – female, summer morph (B) and C. pyrisuga – male, freshly emerged, early summer individual (C), and immature C. pyricola (D) and C. pyrisuga (E). The photos from the living specimens were taken by Ondřej Michálek.
Figure S2 The abundance distribution of adult Cacopsylla pyricola (A) and C. pyrisuga (B) collected on pears in Starý Lískovec (Brno, Czech Republic = CZ2) throughout an entire year from February 2020 to February 2021. The graphs start in May 2020 for convenience, to show the distribution of generations across different seasons. Black arrows indicate the abundance peaks from which adult psyllid specimens were selected for sequencing.
Figure S3 Rarefaction curves showing the number of observed ASVs depending on the number of reads at an even sampling depth of 10,000 reads per sample.
Figure S4 Microbiome composition in individuals of Cacopsylla pyri. (A) Including the most abundant taxa Carsonella, Group1 pyri and Wolbachia, and (B) after removing ASVs of Carsonella, Group1 pyri and Wolbachia. Samples with parasitoid DNA (P14–P18) are presented here but were discarded from the analysis.
Figure S5 Microbiome composition in individuals of Cacopsylla pyricola. (A) Including the most abundant taxa Carsonella and Group1 pyricola, and (B) after removing ASVs of Carsonella and Group1 pyricola.
Figure S6 Microbiome composition in individuals of Cacopsylla pyrisuga. (A) Including the most abundant taxa Carsonella and Group2 pyrisuga, and (B) after removing ASVs of Carsonella and Group2 pyrisuga.
Table S1 Metadata for all specimens analysed in this study. Samples with parasitoid DNA (P14–P18) that were discarded from the analyses are highlighted in yellow. Number of ASVs is given after removing ASVs containing <3 reads. Species richness (Chao 1) and diversity (Shannon) indices are based on an even sampling depth of 10,000 reads per sample. For specimens with <10,000 reads, the richness and diversity indices are not defined (ND).
Table S2 ASV table used for all microbiome analyses. For the specimen ID abbreviations, see Table S1.
Table S3 Bacterial species richness (Chao 1) and diversity (Shannon Index) in the three Cacopsylla species depending on developmental stages, generations (seasonality), localities and gender. Significant p‐values are highlighted in bold.
Table S4 Log CPM (counts per million) values for the 15 bacterial genera identified as either indicator species and/or differentially abundant by IndVal and edgeR, respectively. For information on locality abbreviations, see Table 1.
ACKNOWLEDGEMENTS
We are grateful to the anonymous reviewers for their constructive criticism and useful recommendations that helped to improve the original manuscript. We would like to thank Daniel Burckhardt for providing us with samples of Cacopsylla pyricola collected in France and for his valuable ideas and suggestions that helped to shape this study. LŠS is very grateful to her husband, Michal Štarha, for his enormous help in collecting psyllids in pear orchards. We are obliged to Stanislav Pekár for his support during the field work. We are also very grateful to Monica Pramatarova for collecting the specimens of C. pyricola in Božanov (Czech Republic), and Boris Tichý and Filip Pardy (CF Genomics CEITEC MU) for their support with obtaining sequencing data presented in this paper. We also thank Ondřej Michálek for taking photos from live specimens of C. pyri, C. pyricola and C. pyrisuga. This work was supported by the Autonomous Province of Bozen‐Bolzano to Liliya Štarhová Serbina and Hannes Schuler, and by a Brno PhD Talent Scholarship, funded by the Brno City Municipality, to Domagoj Gajsk. Open Access Funding provided by Libera Universita di Bolzano within the CRUI‐CARE Agreement.
Štarhová Serbina, L. , Gajski, D. , Pafčo, B. , Zurek, L. , Malenovský, I. , Nováková, E. et al. (2022) Microbiome of pear psyllids: A tale about closely related species sharing their endosymbionts. Environmental Microbiology, 24(12), 5788–5808. Available from: 10.1111/1462-2920.16180
Hannes Schuler and Jessica Dittmer contributed equally to this work.
[Corrections added on 28 September 2022, after first online publication: Affiliation 8 has been corrected in this version.]
Funding information Autonomous Province of Bozen‐Bolzano
DATA AVAILABILITY STATEMENT
All sequences were deposited into Sequence Read Archive (Bioproject PRJNA772591). The Sanger sequence was deposited into GenBank (accession number OK561501).
REFERENCES
- Afgan, E. , Baker, D. , Batut, B. , van den Beek, M. , Bouvier, D. , Čech, M. et al. (2018) The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Research, 46, 537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alma, A. , Daffonchio, D. , Gonella, E. & Raddadi, N. (2008) Symbiotic microorganisms in leafhopper and planthopper vectors of phytoplasmas in grapevine. In: Bourtzis, K. & Miller, T.A. (Eds.) Insect Symbiosis, Vol. 3. Boca Raton: CRC Press, pp. 339–359. [Google Scholar]
- Andrews, S. (2010) FastQC: a quality control tool for high throughput sequence data. URL https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Arp, A. , Hunter, W. & Pelz‐Stelinski, K. (2016) Annotation of the Asian citrus psyllid genome reveals a reduced innate immune system. Frontiers in Physiology, 7, 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel, D. , Schuler, H. , Galli, J. , Borruso, L. , Geier, J. , Heer, K. et al. (2020) Identification of plant DNA in adults of the phytoplasma vector Cacopsylla picta helps understanding its feeding behavior. Insects, 11(12), 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen, E. , Rideout, J.R. , Dillon, M.R. , Bokulich, N.A. , Abnet, C.C. , Al‐Ghalith, G.A. et al. (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology, 37, 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtzis, K. , Dobson, S.L. , Xi, Z. , Rasgon, J.L. , Calvitti, M. , Moreira, L.A. et al. (2014) Harnessing mosquito‐Wolbachia symbiosis for vector and disease control. Acta Tropica, 132, 150–163. [DOI] [PubMed] [Google Scholar]
- Bové, J. (2006) Huanglongbing: a destructive, newly emerging, century‐old disease of citrus. Journal of Plant Pathology, 88, 7–37. [Google Scholar]
- Brucker, R.M. & Bordenstein, S.R. (2013) The hologenomic basis of speciation: gut bacteria cause hybrid lethality in the genus Nasonia . Science, 341, 667–669. [DOI] [PubMed] [Google Scholar]
- Burckhardt, D. (2010) Psyllid‐key of Cacopsylla on Rosaceae . https://www.dlr.rlp.de/Psylliden-english
- Burckhardt, D. , Ouvrard, D. & Percy, D.M. (2021) An updated classification of the jumping plant‐lice (Hemiptera: Psylloidea) integrating molecular and morphological evidence. Eur J Taxon, 736, 137–182. [Google Scholar]
- Caporaso, J.G. , Kuczynski, J. , Stombaugh, J. , Bittinger, K. , Bushman, F.D. , Costello, E.K. et al. (2010) QIIME allows analysis of high‐throughput community sequencing data. Nature Methods, 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, G. , Burckhardt, D. , Inoue, H. , Luo, X. & Lee, S. (2017) Systematics of the east Palaearctic pear psyllids (Hemiptera: Psylloidea) with particular focus on the Japanese and Korean fauna. Zootaxa, 4362, 75–98. [DOI] [PubMed] [Google Scholar]
- Cho, G. , Malenovský, I. , Burckhardt, D. , Inoue, H. & Lee, S. (2020) DNA barcoding of pear psyllids (Hemiptera: Psylloidea: Psyllidae), a tale of continued misidentifications. Bulletin of Entomological Research, 110, 521–534. [DOI] [PubMed] [Google Scholar]
- Chrostek, E. , Marialva, M.S.P. , Esteves, S.S. , Weinert, L.A. , Martinez, J. , Jiggins, F.M. et al. (2013) Wolbachia variants induce differential protection to viruses in Drosophila melanogaster: a phenotypic and phylogenomic analysis. PLoS Genetics, 9(12), e1003896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, C.‐C. , Gill, T.A. , Hoffmann, M. & Pelz‐Stelinski, K.S. (2016) Inter‐population variability of endosymbiont densities in the Asian citrus psyllid (Diaphorina citri Kuwayama). Microbial Ecology, 71, 999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civolani, S. (2012) The past and present of pear protection against the pear psylla, Cacopsylla pyri L. In: Perveen, F. (Ed.) Insecticides—Pest Engineering. 17. London: IntechOpen. [Google Scholar]
- Cohen, C. , Toh, E. , Munro, D. , Dong, Q. & Hawlena, H. (2015) Similarities and seasonal variations in bacterial communities from the blood of rodents and from their flea vectors. The ISME Journal, 9, 1662–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman, D.R. , Toolson, E.C. & Takacs‐Vesbach, C.D. (2012) Do diet and taxonomy influence insect gut bacterial communities? Molecular Ecology, 21, 5124–5137. [DOI] [PubMed] [Google Scholar]
- Conci, C. , Rapisarda C., and & Tamanini, L. (1993) Annotated catalogue of the Italian Psylloidea. First part (Insecta Homoptera). Atti Accad roveretana Agiati vii 2B: 33–135. [Google Scholar]
- Cooper, W. , Horton, D. , Swisher‐Grimm, K. , Krey, K. & Wildung, M. (2021) Bacterial endosymbionts of Bactericera maculipennis and three mitochondrial haplotypes of B. cockerelli (Hemiptera: Psylloidea: Triozidae). Environl Entomol, 51, 94–107. [DOI] [PubMed] [Google Scholar]
- Cooper, W. , Swisher, K. , Garczynski, S. , Mustafa, T. , Munyaneza, J. & Horton, D. (2015) Wolbachia infection differs among divergent mitochondrial haplotypes of Bactericera cockerelli (Hemiptera: Triozidae). Annals of the Entomological Society of America, 108, 137–145. [Google Scholar]
- Cooper, W.R. , Garczynski, S.F. & Horton, D.R. (2015) Relative abundance of Carsonella ruddii (Gamma Proteobacterium) in females and males of Cacopsylla pyricola (Hemiptera: Psyllidae) and Bactericera cockerelli (Hemiptera: Triozidae). Insect Sci., 15, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, W.R. , Garczynski, S.F. , Horton, D.R. , Unruh, T.R. , Beers, E.H. , Peter, W.S. et al. (2017) Bacterial endosymbionts of the psyllid Cacopsylla pyricola (Hemiptera: Psyllidae) in the Pacific northwestern United States. Environmental Entomology, 46, 393–402. [DOI] [PubMed] [Google Scholar]
- De Caceres, M. & Legendre, P. (2009) Associations between species and groups of sites: indices and statistical inference. Ecology, 90, 3566–3574. [DOI] [PubMed] [Google Scholar]
- Dittmer, J. & Brucker, R.M. (2021) When your host shuts down: larval diapause impacts host‐microbiome interactions in Nasonia vitripennis . Microbiome, 9, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, A.E. (2015) Multiorganismal insects: diversity and function of resident microorganisms. Annual Review of Entomology, 60(60), 17–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duguma, D. , Hall, M.W. , Rugman‐Jones, P. , Stouthamer, R. , Terenius, O. , Neufeld, J.D. et al. (2015) Developmental succession of the microbiome of Culex mosquitoes. BMC Microbiology, 15, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelstädter, J. & Hurst, G.D.D. (2009) The ecology and evolution of microbes that manipulate host reproduction. Annual Review of Ecology, Evolution, and Systematics, 40, 127–149. [Google Scholar]
- Fagen, J. , Giongo, A. , Brown, C. , Davis‐Richardson, A. , Gano, K. & Triplett, E. (2012) Characterization of the relative abundance of the citrus pathogen ca. Liberibacter asiaticus in the microbiome of its insect vector, Diaphorina citri, using high throughput 16S rRNA sequencing. Open Microbiology Journal, 6, 29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferree, P.M. , Avery, A. , Azpurua, J. , Wilkes, T. & Werren, J.H. (2008) A bacterium targets maternally inherited centrosomes to kill males in Nasonia . Current Biology, 18, 1409–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmer, O. , Black, M. , Hoeh, W. , Lutz, R. & Vrijenhoek, R. (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology, 3, 294–299. [PubMed] [Google Scholar]
- Fromont, C. , Riegler, M. & Cook, J.M. (2016) Phylogeographic analyses of bacterial endosymbionts in fig homotomids (Hemiptera: Psylloidea) reveal codiversification of both primary and secondary endosymbionts. FEMS Microbiology Ecology, 92(12), fiw205. [DOI] [PubMed] [Google Scholar]
- Fromont, C. , Riegler, M. & Cook, J.M. (2017) Relative abundance and strain diversity in the bacterial endosymbiont community of a sap‐feeding insect across its native and introduced geographic range. Microbial Ecology, 74, 722–734. [DOI] [PubMed] [Google Scholar]
- Fu, Z. , Meier, A. , Epstein, B. , Bergland, A. , Castillo Carrillo, C. , Cooper, W. et al. (2020) Host plants and Wolbachia shape the population genetics of sympatric herbivore populations. Evolutionary Applications, 13, 2740–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu, T. & Nikoh, N. (1998) Two intracellular symbiotic bacteria from the mulberry psyllid Anomoneura mori (Insecta, Homoptera). Applied and Environmental Microbiology, 64, 3599–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallinger, J. & Gross, J. (2018) Unraveling the host plant alternation of Cacopsylla pruni—adults but not nymphs can survive on conifers due to phloem/xylem composition. Frontiers in Plant Science, 9, 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George, J. , Ammar, E.‐D. , Hall, D.G. , Shatters, R.G. & Lapointe, S.L. (2018) Prolonged phloem ingestion by Diaphorina citri nymphs compared to adults is correlated with increased acquisition of citrus greening pathogen. Scientific Reports, 8, 10352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherna, R.L. , Werren, J.H. , Weisburg, W. , Cote, R. , Woese, C.R. , Mandelco, L. et al. (1991) NOTES: Arsenophonus nasoniae gen. Nov., sp. nov., the causative agent of the son‐killer trait in the parasitic wasp Nasonia vitripennis . International Journal of Systematic Bacteriology, 41, 563–565. [Google Scholar]
- Gonella, E. , Tedeschi, R. , Crotti, E. & Alma, A. (2019) Multiple guests in a single host: interactions across symbiotic and phytopathogenic bacteria in phloem‐feeding vectors—a review. Entomologia Experimentalis et Applicata, 167, 171–185. [Google Scholar]
- Gong, J.‐T. , Li, Y. , Li, T.‐P. , Liang, Y. , Hu, L. , Zhang, D. et al. (2020) Stable introduction of plant‐virus‐inhibiting Wolbachia into planthoppers for rice protection. Current Biology, 30, 4837–4845. [DOI] [PubMed] [Google Scholar]
- Görg, L.M. & Gross, J. (2021) Influence of ontogenetic and migration stage on feeding behavior of Cacopsylla picta on ‘Candidatus Phytoplasma Mali’ infected and non‐infected apple plants. Journal of Insect Physiology, 131, 104229. [DOI] [PubMed] [Google Scholar]
- Gottlieb, Y. , Zchori‐Fein, E. , Mozes‐Daube, N. , Kontsedalov, S. , Skaljac, M. , Brumin, M. et al. (2010) The transmission efficiency of tomato yellow leaf curl virus by the whitefly Bemisia tabaci is correlated with the presence of a specific symbiotic bacterium species. Journal of Virology, 84, 9310–9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hail, D. , Dowd, S.E. & Bextine, B. (2012) Identification and location of symbionts associated with potato psyllid (Bactericera cockerelli) lifestages. Environmental Entomology, 41, 98–107. [DOI] [PubMed] [Google Scholar]
- Hall, A.A.G. , Morrow, J.L. , Fromont, C. , Steinbauer, M.J. , Taylor, G.S. , Johnson, S.N. et al. (2016) Codivergence of the primary bacterial endosymbiont of psyllids versus host switches and replacement of their secondary bacterial endosymbionts: evolution of psyllid endosymbionts. Environmental Microbiology, 18, 2591–2603. [DOI] [PubMed] [Google Scholar]
- Hall, D. , Richardson, M.L. , Ammar, E.‐D. & Halbert, S. (2012) Asian citrus psyllid, Diaphorina citri (Hemiptera: Psyllidae), vector of citrus huanglongbing disease. Entomologia Experimentalis et Applicata, 146, 207–222. [Google Scholar]
- Hansen, A. , Trumble, J. , Stouthamer, R. & Paine, T. (2008) A new huanglongbing species, “Candidatus Liberibacter psyllaurous,” found to infect tomato and potato, is vectored by the psyllid Bactericera cockerelli (Sulc). Applied and Environmental Microbiology, 74, 5862–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, A.K. , Jeong, G. , Paine, T.D. & Stouthamer, R. (2007) Frequency of secondary symbiont infection in an invasive psyllid relates to parasitism pressure on a geographic scale in California. Applied and Environmental Microbiology, 73, 7531–7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges, L.M. , Brownlie, J.C. , O'Neill, S.L. & Johnson, K.N. (2008) Wolbachia and virus protection in insects. Science, 322(5902), 702. [DOI] [PubMed] [Google Scholar]
- Hodkinson, I.D. (2009) Life cycle variation and adaptation in jumping plant lice (Insecta: Hemiptera: Psylloidea): a global synthesis. Journal of Natural History, 43, 65–179. [Google Scholar]
- Horton, D.R. , Broers, D.A. , Hinojosa, T. & Lewis, T. M. (1998) Ovarian development in overwintering pear psylla, Cacopsylla pyricola (Homoptera: Psyllidae): seasonality and effects of photoperiod. Can Entomol, 130, 859–867. [Google Scholar]
- Horton, D.R. , Higbee, B. & Krysan, J. (1994) Postdiapause development and mating status of pear psylla (Homoptera: Psyllidae) affected by pear and nonhost species. Annals of the Entomological Society of America, 87, 241–249. [Google Scholar]
- Horton, D.R. , Higbee, B.S. , Unruh, T.R. & Westigard, P.H. (1992) Spatial characteristic and effects of fall density and weather on overwintering loss of pear psylla (Homoptera: Psyllidae). Environmental Entomology, 21, 1319–1332. [Google Scholar]
- Hosokawa, T. , Koga, R. , Kikuchi, Y. , Meng, X.‐Y. , Fukatsu, T. & Moran, N.A. (2010) Wolbachia as a bacteriocyte‐associated nutritional mutualist. Proceedings of the National Academy of Sciences, 107, 769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseinzadeh, S. , Shams‐Bakhsh, M. , Mann, M. , Fattah‐Hosseini, S. , Bagheri, A. , Mehrabadi, M. et al. (2019) Distribution and variation of bacterial endosymbiont and “Candidatus Liberibacter asiaticus” titer in the huanglongbing insect vector, Diaphorina citri Kuwayama. Microbial Ecology, 78, 206–222. [DOI] [PubMed] [Google Scholar]
- Hurst, G. & Frost, C. (2015) Reproductive parasitism: maternally inherited symbionts in a biparental world. Cold Spring Harbor Perspectives in Biology, 7(5), a017699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hypša, V. & Nováková, E. (2008) Insect symbionts and molecular phylogenetics. In: Bourtzis, K. & Miller, T.A. (Eds.) Insect Symbiosis, Vol. 3. Boca Raton: CRC Press, pp. 8–25. [Google Scholar]
- Indiragandhi, P. (2010) Microbial communities in the developmental stages of B and Q biotypes of sweet potato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). Journal of Korean Society for Applied Biological Chemistry, 53, 605–617. [Google Scholar]
- Inoue, H. , Ohnishi, J. , Ito, T. , Tomimura, K. , Miyata, S. , Iwanami, T. et al. (2009) Enhanced proliferation and efficient transmission of Candidatus Liberibacter asiaticus by adult Diaphorina citri after acquisition feeding in the nymphal stage. Annals of Applied Biology, 155, 29–36. [Google Scholar]
- Jaenike, J. (2012) Population genetics of beneficial heritable symbionts. Trends in Ecology & Evolution, 27, 226–232. [DOI] [PubMed] [Google Scholar]
- Jarausch, B. & Jarausch, W. (2010) Psyllid vectors and their control. In: Weintraub, P.G. & Jones, P. (Eds.) Phytoplasmas: genomes, plant hosts and vectors. Wallingford: CABI, pp. 250–271. [Google Scholar]
- Jarausch, B. , Tedeschi, R. , Sauvion, N. , Gross, J. & Jarausch, W. (2019) Psyllid vectors: transmission and management of phytoplasma‐associated diseases. In: Bertaccini, A. , Weintraub, P.G. , Rao, G.P. & Mori, N. (Eds.) Phytoplasmas: plant pathogenic bacteria—II. Singapore: Springer, pp. 53–78. [Google Scholar]
- Jing, X. , Wong, A.C.‐N. , Chaston, J.M. , Colvin, J. , McKenzie, C.L. & Douglas, A.E. (2014) The bacterial communities in plant phloem‐sap‐feeding insects. Molecular Ecology, 23, 1433–1444. [DOI] [PubMed] [Google Scholar]
- Jousselin, E. , Cœur d'Acier, A. , Vanlerberghe‐Masutti, F. & Duron, O. (2013) Evolution and diversity of Arsenophonus endosymbionts in aphids. Molecular Ecology, 22, 260–270. [DOI] [PubMed] [Google Scholar]
- Kandasamy, T. , Ekbal, S. , Kumari, K. , Lohot, V. , Mohanasundaram, A. & Sharma, K. (2022) Unraveling bacterial diversity of the Indian lac insect Kerria lacca (Kerr) using next generation sequencing. International Journal of Tropical Insect Science, 42, 2365–2372. 10.1007/s42690-022-00758-x [DOI] [Google Scholar]
- Kaur, R. , Shropshire, J.D. , Cross, K.L. , Leigh, B. , Mansueto, A.J. , Stewart, V. et al. (2021) Living in the endosymbiotic world of Wolbachia: a centennial review. Cell Host & Microbe, 29, 879–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klindworth, A. , Pruesse, E. , Schweer, T. , Peplies, J. , Quast, C. , Horn, M. et al. (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next‐generation sequencing‐based diversity studies. Nucleic Acids Research, 41, e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliot, A. , Cilia, M. , Czosnek, H. & Ghanim, M. (2014) Implication of the bacterial endosymbiont Rickettsia spp. in interactions of the whitefly Bemisia tabaci with tomato yellow leaf curl virus. Journal of Virology, 88, 5652–5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse, A. , Fattah‐Hosseini, S. , Saha, S. , Johnson, R. , Warwick, E. , Sturgeon, K. et al. (2017) Combining 'omics and microscopy to visualize interactions between the Asian citrus psyllid vector and the Huanglongbing pathogen Candidatus Liberibacter asiaticus in the insect gut. PLoS One, 12(6), e0179531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kučerová, J. , Talácko, L. , Lauterer, P. , Navratil, M. & Fialová, R. (2007) Molecular tests to determine 'Candidatus Phytoplasma pyri' presence in psyllid vectors from a pear tree orchard in The Czech Republic—a preliminary report. Bulletin of Insectology, 60, 191–192. [Google Scholar]
- Kwak, Y. , Sun, P. , Meduri, V.R. , Percy, D.M. , Mauck, K.E. & Hansen, A.K. (2021) Uncovering symbionts across the psyllid tree of life and the discovery of a new Liberibacter species, “Candidatus Liberibacter capsica ”. Frontiers in Microbiology, 12, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmann, F. (2019) The Wolbachia endosymbionts. Microbiology Spectrum, 7(2), 1–15. 10.1128/microbiolspec.BAI-0018-2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterer, P. (1999) Results of the investigations on Hemiptera in Moravia, made by the Moravian museum (Psylloidea 2). Acta Musei Moraviae, Scientiae Biologicae, 84, 71–151. [Google Scholar]
- Letunic, I. & Bork, P. (2021) Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Research, 49, 293–296. 10.1093/nar/gkab301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macke, E. , Tasiemski, A. , Massol, F. , Callens, M. & Decaestecker, E. (2017) Life history and eco‐evolutionary dynamics in light of the gut microbiota. Oikos, 126, 508–531. [Google Scholar]
- Mayer, C. , Jarausch, B. , Jarausch, W. , Jelkmann, W. , Vilcinskas, A. & Gross, J. (2009) Cacopsylla melanoneura has no relevance as vector of apple proliferation in Germany. Phytopathology, 99, 729–738. [DOI] [PubMed] [Google Scholar]
- McCutcheon, J.P. , Boyd, B.M. & Dale, C. (2019) The life of an insect endosymbiont from the cradle to the grave. Current Biology, 29, 485–495. [DOI] [PubMed] [Google Scholar]
- McLean, A.H.C. , Godfray, H.C.J. , Ellers, J. & Henry, L.M. (2019) Host relatedness influences the composition of aphid microbiomes. Environmental Microbiology Reports, 11, 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMeniman, C.J. , Lane, R.V. , Cass, B.N. , Fong, A.W.C. , Sidhu, M. , Wang, Y.‐F. et al. (2009) Stable introduction of a life‐shortening Wolbachia infection into the mosquito Aedes aegypti . Science, 323, 141–144. [DOI] [PubMed] [Google Scholar]
- Meng, L. , Li, X. , Cheng, X. & Zhang, H. (2019) 16S rRNA Ggne sequencing reveals a shift in the microbiota of Diaphorina citri during the psyllid life cycle. Frontiers in Microbiology, 10, 1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, N. , McCutcheon, J. & Nakabachi, A. (2008) Genomics and evolution of heritable bacterial symbionts. Annual Review of Genetics, 42, 165–190. [DOI] [PubMed] [Google Scholar]
- Morrow, J.L. , Hall, A.A.G. & Riegler, M. (2017) Symbionts in waiting: the dynamics of incipient endosymbiont complementation and replacement in minimal bacterial communities of psyllids. Microbiome, 5, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow, J.L. , Om, N. , Beattie, G.A.C. , Chambers, G.A. , Donovan, N.J. , Liefting, L.W. et al. (2020) Characterization of the bacterial communities of psyllids associated with Rutaceae in Bhutan by high throughput sequencing. BMC Microbiology, 20, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouton, L. , Thierry, M. , Henri, H. , Baudin, R. , Gnankine, O. , Reynaud, B. et al. (2012) Evidence of diversity and recombination in Arsenophonus symbionts of the Bemisia tabaci species complex. BMC Microbiology, 12, S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachappa, P. , Levy, J. , Pierson, E. & Tamborindeguy, C. (2011) Diversity of endosymbionts in the potato psyllid, Bactericera cockerelli (Hemiptera: Triozidae), vector of zebra chip disease of potato. Current Microbiology, 62, 1510–1520. [DOI] [PubMed] [Google Scholar]
- Nakabachi, A. , Inoue, H. & Hirose, Y. (2022) Microbiome analyses of 12 psyllid species of the family Psyllidae identified various bacteria including Fukatsuia and Serratia symbiotica, known as secondary symbionts of aphids. BMC Microbiology, 22, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi, A. , Malenovský, I. , Gjonov, I. & Hirose, Y. (2020) 16S rRNA sequencing detected Profftella, Liberibacter, Wolbachia, and Diplorickettsia from relatives of the Asian citrus psyllid. Microbial Ecology, 80, 410–422. [DOI] [PubMed] [Google Scholar]
- Nakabachi, A. , Piel, J. , Malenovský, I. & Hirose, Y. (2020) Comparative genomics underlines multiple roles of Profftella, an obligate symbiont of psyllids: providing toxins, vitamins, and carotenoids. Genome Biology and Evolution, 12, 1975–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi, A. , Ueoka, R. , Oshima, K. , Teta, R. , Mangoni, A. , Gurgui, M. et al. (2013) Defensive bacteriome symbiont with a drastically reduced genome. Current Biology, 23, 1478–1484. [DOI] [PubMed] [Google Scholar]
- Nakabachi, A. , Yamashita, A. , Toh, H. , Ishikawa, H. , Dunbar, H.E. , Moran, N.A. et al. (2006) The 160‐kilobase genome of the bacterial endosymbiont Carsonella . Science, 314, 267–267. [DOI] [PubMed] [Google Scholar]
- Newell, P.D. & Douglas, A.E. (2014) Interspecies interactions determine the impact of the gut microbiota on nutrient allocation in Drosophila melanogaster . Applied and Environmental Microbiology, 80, 788–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh, N. , Hosokawa, T. , Moriyama, M. , Oshima, K. , Hattori, M. & Fukatsu, T. (2014) Evolutionary origin of insect‐Wolbachia nutritional mutualism. Proceedings of the National Academy of Sciences, 111, 10257–10262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishide, Y. , Kageyama, D. , Yokoi, K. , Jouraku, A. , Tanaka, H. , Futahashi, R. et al. (2019) Functional crosstalk across IMD and toll pathways: insight into the evolution of incomplete immune cascades. Proceedings of the Royal Society of London. Series B, Biological Sciences, 286, 20182207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková, E. , Hypša, V. & Moran, N.A. (2009) Arsenophonus, an emerging clade of intracellular symbionts with a broad host distribution. BMC Microbiology, 9, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková, E. , Woodhams, D.C. , Rodríguez‐Ruano, S.M. , Brucker, R.M. , Leff, J.W. , Maharaj, A. et al. (2017) Mosquito microbiome dynamics, a background for prevalence and seasonality of West Nile virus. Frontiers in Microbiology, 8, 526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettl, S. & Schlink, K. (2015) Molecular identification of two vector species, Cacopsylla melanoneura and Cacopsylla picta (Hemiptera: Psyllidae), of apple proliferation disease and further common psyllids of northern Italy. Journal of Economic Entomology, 108, 2174–2183. [DOI] [PubMed] [Google Scholar]
- Orlovskis, Z. , Canale, M.C. , Thole, V. , Pecher, P. , Lopes, J. & Hogenhout, S. (2015) Insect‐borne plant pathogenic bacteria: getting a ride goes beyond physical contact. Current Opinion in Insect Science, 9, 16–23. [DOI] [PubMed] [Google Scholar]
- Ossiannilsson, F. (1992) The psylloidea (Homoptera) of fennoscandia and denmark. In: Brill, E.J. (Ed.), Fauna Entomologica Scandinavica. [Google Scholar]
- Overholt, W.A. , Diaz, R. , Rosskopf, E. , Green, S.J. & Overholt, W.A. (2015) Deep characterization of the microbiomes of Calophya spp. (Hemiptera: Calophyidae) gall‐inducing psyllids reveals the absence of plant pathogenic bacteria and three dominant endosymbionts. PLoS One, 10(7), e0132248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pafčo, B. , Čížková, D. , Kreisinger, J. , Hasegawa, H. , Vallo, P. , Shutt, K. et al. (2018) Metabarcoding analysis of strongylid nematode diversity in two sympatric primate species. Scientific Reports, 8, 5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percy, D.M. , Crampton‐Platt, A. , Sveinsson, S. , Lemmon, A.R. , Lemmon, E.M. , Ouvrard, D. et al. (2018) Resolving the psyllid tree of life: phylogenomic analyses of the superfamily Psylloidea (Hemiptera). Systematic Entomology, 43, 762–776. [Google Scholar]
- Perilla‐Henao, L.M. & Casteel, C.L. (2016) Vector‐borne bacterial plant pathogens: interactions with hemipteran insects and plants. Frontiers in Plant Science, 7, 1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrot‐Minnot, M.J. , Guo, L.R. & Werren, J.H. (1996) Single and double infections with Wolbachia in the parasitic wasp Nasonia vitripennis: effects on compatibility. Genetics, 143, 961–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilgrim, J. , Thongprem, P. , Davison, H.R. , Siozios, S. , Baylis, M. , Zakharov, E.V. et al. (2021) Torix rickettsia are widespread in arthropods and reflect a neglected symbiosis. GigaScience, 10, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes, M.H. , Smith, K. , Smith, W. & Dale, C. (2008) Insect facultative symbionts: biology, culture, and genetic modification. In: Bourtzis, K. & Miller, T.A. (Eds.) Insect Symbiosis, Vol. 3. Boca Raton: CRC Press, pp. 377–396. [Google Scholar]
- Ramsey, J.S. , Chavez, J.D. , Johnson, R. , Hosseinzadeh, S. , Mahoney, J.E. , Mohr, J.P. et al. (2017) Protein interaction networks at the host–microbe interface in Diaphorina citri, the insect vector of the citrus greening pathogen. Royal Society Open Science, 4, 160545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedle‐Bauer, M. , Paleskić, C. , Schönhuber, C. , Staples, M. & Brader, G. (2022) Vector transmission and epidemiology of ‘Candidatus Phytoplasma pyri’ in Austria and identification of Cacopsylla pyrisuga as new pathogen vector. J Plant Dis Protect, 129, 375–386. 10.1007/s41348-021-00526-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D. , McCarthy, D.J. & Smyth, G.K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Ruano, S.M. , Škochová, V. , Rego, R.O.M. , Schmidt, J.O. , Roachell, W. , Hypša, V. et al. (2018) Microbiomes of north American Triatominae: the grounds for Chagas disease epidemiology. Frontiers in Microbiology, 9, 1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, J.A. , Latorre, A. , Sabater‐Munoz, B. , Moya, A. & Moran, N.A. (2003) Side‐stepping secondary symbionts: widespread horizontal transfer across and beyond the Aphidoidea. Molecular Ecology, 12, 1061–1075. [DOI] [PubMed] [Google Scholar]
- Schuler, H. , Dittmer, J. , Galli, J. , Borruso, L. , Fischnaller, S. , Anfora, G. et al. (2022) The microbial community of Cacopsylla spp. and its potential role in vector competence of phytoplasma. Environmental Microbiology, 1–16. 10.1111/1462-2920.16138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler, H. , Köppler, K. , Daxböck‐Horvath, S. , Rasool, B. , Krumböck, S. , Schwarz, D. et al. (2016) The hitchhiker's guide to Europe: the infection dynamics of an ongoing Wolbachia invasion and mitochondrial selective sweep in Rhagoletis cerasi . Molecular Ecology, 25, 1595–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, S.C. , Kim, S.H. , You, H. , Kim, B. , Kim, A.C. , Lee, K.A. et al. (2011) Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science, 334, 670–674. [DOI] [PubMed] [Google Scholar]
- Sloan, D.B. & Moran, N.A. (2012) Genome reduction and co‐evolution between the primary and secondary bacterial symbionts of psyllids. Molecular Biology and Evolution, 29, 3781–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šochová, E. , Husník, F. , Nováková, E. , Halajian, A. & Hypša, V. (2017) Arsenophonus and Sodalis replacements shape evolution of symbiosis in louse flies. PeerJ, 5, e4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X. , Peng, A. , Ling, J. , Cui, Y. , Cheng, B. & Zhang, L. (2019) Composition and change in the microbiome of Diaphorina citri infected with Candidatus Liberibacter asiaticus in China. International Journal of Tropical Insect Science, 39, 283–290. [Google Scholar]
- Soroker, V. , Alchanatis, V. , Harari, A. , Talebaev, S. , Anshelevich, L. , Reneh, S. et al. (2013) Phenotypic plasticity in the pear psyllid, Cacopsylla bidens (Šulc) (Hemiptera, Psylloidea, Psyllidae) in Israel. Israel Journal of Entomology, 43, 21–31. [Google Scholar]
- Storelli, G. , Defaye, A. , Erkosar, B. , Hols, P. , Royet, J. & Leulier, F. (2011) Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR‐dependent nutrient sensing. Cell Metabolism, 14, 403–414. [DOI] [PubMed] [Google Scholar]
- Su, Q. , Pan, H. , Liu, B. , Chu, D. , Xie, W. , Wu, Q. et al. (2013) Insect symbiont facilitates vector acquisition, retention and transmission of plant virus. Scientific Reports, 3, 1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subandiyah, S. , Nikoh, N. , Tsuyumu, S. , Somowiyarjo, S. & Fukatsu, T. (2000) Complex endosymbiotic microbiota of the citrus psyllid Diaphorina citri (Homoptera: Psylloidea). Zoological Science, 17, 983–989. [Google Scholar]
- Tamames, J. , Gil, R. , Latorre, A. , Peretó, J. , Silva, F.J. & Moya, A. (2007) The frontier between cell and organelle: genome analysis of Candidatus Carsonella ruddii . BMC Evolutionary Biology, 7, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamborindeguy, C. , Huot, O.B. , Ibanez, F. & Levy, J. (2017) The influence of bacteria on multitrophic interactions among plants, psyllids, and pathogen. Insect Science, 24, 961–974. [DOI] [PubMed] [Google Scholar]
- Tedeschi, R. , Baldessari, M. , Mazzoni, V. , Trona, F. , and & Angeli, G. (2012) Population dynamics of Cacopsylla melanoneura (Hemiptera: Psyllidae) in Northeast Italy and its role in the apple proliferation epidemiology in apple orchards. Journal of Economic Entomology 105: 322–328. [DOI] [PubMed] [Google Scholar]
- Teixeira, L. , Ferreira, A. & Ashburner, M. (2008) The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster . PLoS Biology, 6(12), e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao, M.L. & Baumann, P. (2004) Evidence for multiple acquisition of Arsenophonus by whitefly species (Sternorrhyncha: Aleyrodidae). Current Microbiology, 48, 140–144. [DOI] [PubMed] [Google Scholar]
- Thao, M.L. , Clark, M.A. , Baumann, L. , Brennan, E.B. , Moran, N.A. & Baumann, P. (2000) Secondary endosymbionts of psyllids have been acquired multiple times. Current Microbiology, 41, 300–304. [DOI] [PubMed] [Google Scholar]
- Tougeron, K. , Iltis, C. , Renoz, F. , Albittar, L. , Hance, T. , Demeter, S. et al. (2021) Ecology and biology of the parasitoid Trechnites insidiosus and its potential for biological control of pear psyllids. Pest Management Science, 77, 4836–4847. [DOI] [PubMed] [Google Scholar]
- Trifinopoulos, J. , Nguyen, L.‐T. , von Haeseler, A. & Minh, B.Q. (2016) W‐IQ‐TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research, 44, 232–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opstal, E.J. & Bordenstein, S.R. (2019) Phylosymbiosis impacts adaptive traits in Nasonia wasps. mBio, 10, e00887–e00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Lei, T. , Wang, X. , Maruthi, M.N. , Zhu, D. , Cameron, S.L. et al. (2020) A newly recorded rickettsia of the Torix group is a recent intruder and an endosymbiont in the whitefly Bemisia tabaci . Environmental Microbiology, 22, 1207–1221. [DOI] [PubMed] [Google Scholar]
- Weinert, L.A. , Araujo‐Jnr, E.V. , Ahmed, M.Z. & Welch, J.J. (2015) The incidence of bacterial endosymbionts in terrestrial arthropods. Proceedings of the Royal Society B: Biological Sciences, 282, 20150249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, T.L. (2012) Facultative tenants from the Enterobacteriaceae within phloem‐feeding insects. In: Zchori‐Fein, E. & Bourtzis, L. (Eds.) Manipulative tenants: bacteria associated with arthropods. Boca Raton: CRC Press, pp. 73–88. [Google Scholar]
- Yilmaz, P. , Parfrey, L.W. , Yarza, P. , Gerken, J. , Pruesse, E. , Quast, C. et al. (2014) The SILVA and “all‐species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Research, 42, 643–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun, J.‐H. , Roh, S.W. , Whon, T.W. , Jung, M.‐J. , Kim, M.‐S. , Park, D.‐S. et al. (2014) Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Applied and Environmental Microbiology, 80, 254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Habitus of adult Cacopsylla pyri – female (A), C. pyricola – female, summer morph (B) and C. pyrisuga – male, freshly emerged, early summer individual (C), and immature C. pyricola (D) and C. pyrisuga (E). The photos from the living specimens were taken by Ondřej Michálek.
Figure S2 The abundance distribution of adult Cacopsylla pyricola (A) and C. pyrisuga (B) collected on pears in Starý Lískovec (Brno, Czech Republic = CZ2) throughout an entire year from February 2020 to February 2021. The graphs start in May 2020 for convenience, to show the distribution of generations across different seasons. Black arrows indicate the abundance peaks from which adult psyllid specimens were selected for sequencing.
Figure S3 Rarefaction curves showing the number of observed ASVs depending on the number of reads at an even sampling depth of 10,000 reads per sample.
Figure S4 Microbiome composition in individuals of Cacopsylla pyri. (A) Including the most abundant taxa Carsonella, Group1 pyri and Wolbachia, and (B) after removing ASVs of Carsonella, Group1 pyri and Wolbachia. Samples with parasitoid DNA (P14–P18) are presented here but were discarded from the analysis.
Figure S5 Microbiome composition in individuals of Cacopsylla pyricola. (A) Including the most abundant taxa Carsonella and Group1 pyricola, and (B) after removing ASVs of Carsonella and Group1 pyricola.
Figure S6 Microbiome composition in individuals of Cacopsylla pyrisuga. (A) Including the most abundant taxa Carsonella and Group2 pyrisuga, and (B) after removing ASVs of Carsonella and Group2 pyrisuga.
Table S1 Metadata for all specimens analysed in this study. Samples with parasitoid DNA (P14–P18) that were discarded from the analyses are highlighted in yellow. Number of ASVs is given after removing ASVs containing <3 reads. Species richness (Chao 1) and diversity (Shannon) indices are based on an even sampling depth of 10,000 reads per sample. For specimens with <10,000 reads, the richness and diversity indices are not defined (ND).
Table S2 ASV table used for all microbiome analyses. For the specimen ID abbreviations, see Table S1.
Table S3 Bacterial species richness (Chao 1) and diversity (Shannon Index) in the three Cacopsylla species depending on developmental stages, generations (seasonality), localities and gender. Significant p‐values are highlighted in bold.
Table S4 Log CPM (counts per million) values for the 15 bacterial genera identified as either indicator species and/or differentially abundant by IndVal and edgeR, respectively. For information on locality abbreviations, see Table 1.
Data Availability Statement
All sequences were deposited into Sequence Read Archive (Bioproject PRJNA772591). The Sanger sequence was deposited into GenBank (accession number OK561501).


