Abstract
Background: N6-methyladenosine (m6A) is one of the most common RNA modifications in eukaryotes and has effects on RNA structure and stability. Recent studies have shown that m6A methylation is involved in human carcinogenesis. In the present study, we investigated the effects of m6A demethylases FTO and ALKBH5 on renal cell carcinoma (RCC) cell lines. Methods: The epithelial-mesenchymal in vitro knockdowns of FTO and ALKBH5 induced by antisense oligonucleotides (LNA-GapmeR system) were established in RCC cell lines. Their effects on migration and proliferation were investigated subsequently. The influence of FTO and ALKBH5 knockdown on key epithelial-mesenchymal transition (EMT) genes was analyzed. Results: Inactivation of FTO and ALKBH5 resulted in decreased proliferation and motility in all cell lines examined (ACHN, Caki-1, 769-P). Vimentin (VIM) was downregulated after the knockdown of FTO and ALKBH5, indicating an EMT switch. Conclusions: Knockdown of the m6A erasers FTO and ALKBH5 inhibits the malignant potential in the cell cultures studied by means of an EMT switch.
Keywords: m6A-erasers, clear cell renal cell carcinoma (ccRCC), epithelial-mesenchymal transition (EMT), FTO, ALKBH5
Introduction
Renal cell carcinoma (RCC) accounts for approximately 3% of all adult cancers and 2% of cancer-related deaths [1,2]. The prognosis of patients with locally advanced or metastatic RCC is poor. The 5-year survival rate of patients with metastatic RCC is approximately 5% [1,2]. Modern pharmacotherapy of RCC with tyrosine kinase inhibitors, anti-VEGF antibodies, or checkpoint immunotherapy has only moderately improved patient survival. Complete remission rates of 17% and median overall survival of 40 months are described [3,4].
RNA methylation was first described in the 1970s. m6A methylation is a dynamic, reversible modification involved in many physiologic processes of eukaryotic cells, such as mRNA localization, translational inhibition or activation, and miRNA processing [5]. M6A methylation affects several important biologic processes, such as tissue development, sex selection, and DNA damage response [6-8]. M6A methylation involves the binding, recognition, and removal of methyl groups from RNA molecules that act as binding proteins, reading proteins, and deletion proteins, respectively [6-8]. If this process is disrupted, abnormal gene expression can occur, leading to a variety of cancers such as leukemia, lung cancer, pancreatic cancer, glioblastoma, and breast cancer [9]. M6A methylation is regulated by enzymes that insert and remove these methylations and are referred to as methylation “writers” and “erasers”, respectively. The corresponding localization and activity of these enzymes on chromatin are regulated by chromatin “readers”, protein modules that recognize histone and DNA modifications [10,11]. To date, only two erasers, fat mass and obesity-associated protein (FTO) and AlKBH5 (AlkB family homolog 5), have been discovered [12,13]. FTO regulates adipogenesis and energy homeostasis [12]. ALKBH5 affects RNA nuclear export and metabolism as well as gene expression [13]. FTO and ALKBH5 are involved in the carcinogenesis of various tumors [14-16]. Little is known about the role of FTO and ALKBH5 in the genesis of renal cell carcinoma. Strick et al [17] demonstrated that ALKBH5 and FTO are prognostic biomarkers in ccRCC patients, but the functional role of these genes has not been investigated. Epithelial-mesenchymal transition (EMT) is a biologic process in which polarized epithelial cells lose their adhesive properties and adopt a mesenchymal cell phenotype [18]. This process is involved not only in normal embryonic development and tissue repair but is also closely related to the invasion and migration ability of carcinomas. In malignant tumors, EMT is a key process for migration, invasion, metastasis, angiogenesis, and tumorigenicity [18]. It is important to elucidate the regulatory mechanisms of EMT. By understanding these processes, drug therapies can be derived. This could be done by inhibiting tumor cell migration [18].
This study aims to investigate the effects of the knockdown of FTO and ALKBH5 in renal cancer cell lines and to explore the associated mechanisms during EMT and cell proliferation.
Materials and methods
Database analysis and RNAseq expression data
The expression values of FTO and ALKBH5 were obtained from The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov) and the cBioPortal for Cancer Genomics (https://www.cbioportal.org/) [19,20]. The log2-transformed RNAseq V2 expression data of ALKBH5 and FTO were compared in the available tumor entities via the cBioportal for Cancer Genomics. Data were processed and normalized using the RSEM method [19,20].
Cell culture and transfection
The renal carcinoma cell lines ACHN and 769P were purchased from the American Type Culture Collection (Manassas, VA, USA); Caki-1 was purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany). The cells were cultivated with RPMI Medium 1640 including 5% L-glutamine, 1% penicillin, and 10% fetal bovine serum (all: Gibco, Gaithersburg, MD, USA) at 37°C and 5% CO2. Cells were seeded in 6-well plates (2 × 105 cells/well). After 24 hours, the cells were transfected with antisense LNA GapmeRs against ALKBH5 or FTO; GapmeRs were designed and synthesized by Qiagen (Hilden, Germany); see Table S1. GapmeRs were labeled with 5’-FAM. Successful transfection was confirmed by fluorescence microscopy after transfection. Transfection was carried out with 10 µl GapmeR, 10 µl FuGENE HD transfection reagent (Promega Mannheim, Germany), and 1980 µl cell culture media. The experiments included an untreated, a negative control GapmeR and a GapmeR against GAPDH. All experiments were carried out in triplicate.
Quantitative real-time polymerase chain reaction (qRT-PCR)
The Total RNA Purification Mini Spin Kit (Genaxxon, Ulm, Germany) was used to isolate total RNA according to the manufacturer’s recommendation. Reverse transcription was performed with 1 µg of total RNA using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio, Tokyo, Japan). The expression levels of FTO and ALKBH5 were determined using a QuantStudio5 (Applied Biosystems, Waltham, MA, USA) real-time PCR cycler. cDNAs were amplified using SYBR Green Real-time PCR Master Mix (Takara Bio) and 0.4 µl (10 pmol/µl) of each primer pair. Amplification was performed according to the following protocol: Hot start at 95°C for 30 s, followed by 35 cycles (denaturation at 95°C for 5 s, extension at 60°C for 30 s, preheating at 79°C for 30 s), and concluded with melting curve analysis. All experiments were performed in triplicate on a 384-well plate. Relative expression values were calculated using QuantStudio Design & Analysis software V1.5.1 (Thermo Fisher Scientific, Carlsbad, CA, USA), and expression values were normalized to ACTB expression. Primer sequences are listed in Table S2.
Western blotting
Cell lysates (20 μL) were resolved by SDS-PAGE gel. The separated protein was transferred to the Nitrocellulose membranes (Thermo Fisher Scientific) and subjected to immunoblotting overnight at 4°C (see Table S3 for antibodies). Afterward, incubation with corresponding secondary antibodies (see Table S3) was performed at room temperature for 1 h. After washing with Tris-buffered saline containing 0.1% Tween 20, the blot was developed with WesternBright-ECL-Spray (Advansta, Menlo Park, CA, USA) and images were archived on a Fujifilm LAS-3000 (Fujifilm, Tokyo, Japan) digital photography system.
Cell viability assay
The EZ4U assay (Biomedica, Aschaffenburg, Germany) was used to determine cell viability according to the manufacturer’s recommendations. Absorbance was measured at 450 nm with 620 nm as reference using a photometer (Spectra III; Tecan, Crailsheim, Germany) after 3 hours of incubation at 37°C. The cell growth rate was calculated according to the following formula: Cell growth rate (%) = absorbance of the GapmeR group/absorbance of the negative control group × 100%. The cell growth rate of the negative control group was set to 1 (100%). Each cell group included four samples and each experiment was repeated three times. The significance of the treatment was calculated by a t-test using the mean and standard deviation of the growth rate.
Cell migration assay
We used 24-well plates with Falcon Cell Culture Inserts (Thermo Fisher Scientific) for cell migration experiments. Approximately 5 × 104 transfected cells were added to the top of the insert and 300 µl of RPMI Medium 1640 (2% FBS) was added to the specimen. 700 µl of RPMI medium 1640 (20% FBS) was added to the bottom wells to stimulate migration. The cells were incubated at 37°C and 5% CO2 for 24 hours.
After fixing the cells with 4% paraformaldehyde in phosphate buffer (AppliChem, St. Louis, MO, USA), the nuclei were stained with hematoxylin.
The cells that had not migrated were wiped on the inside of the insert. The migrated cells on the outside of the membrane were mounted on a slide. Photographs of each membrane were taken with a microscope (Olympus DP70, Tokyo, Japan) to evaluate migration. The migrated cells were quantified using QuPath [21]. The cell migration rate was calculated using the following formula: Migration rate (%) = cell number of the gapmeR group/cell number of the negative control group × 100%. The migration rate of the negative control group was set to 1 (100%). Each cell line was run at least 3 times.
Statistical analysis
All measured data were expressed by mean ± standard deviation (mean ± SD). An independent-samples t-test was used to evaluate the significance of the different groups. All data were analyzed using GraphadPrism8.0 software (GraphPad Software, San Diego, CA, USA). Counted data were expressed as % and analyzed by chi-square test. Differences with P < 0.05 were considered significant.
Results
First, we analyzed the FTO and ALKBH5 expression profiles in different cancer entities using the Cancer Genome Atlas Research Network (TCGA) datasets with the cBioportal software [19,20] (Figure 1). Of note, both FTO and ALKBH5 expressions were highly expressed in ccRCC. This indicates the functional importance of m6A demethylases in this particular tumor type.
Figure 1.
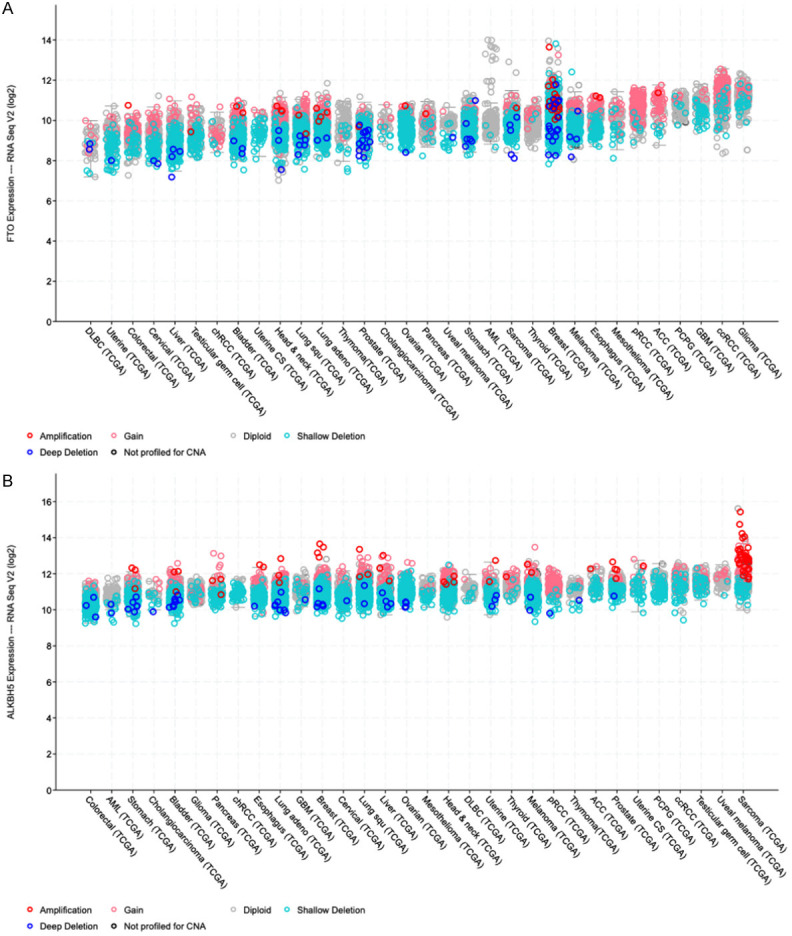
RNAseq expression data of cBioPortal database. A: Available RNA sequencing expression data from 32 cancer entities were studied, and high mRNA expression of FTO in ccRCC, in a pan-cancer analysis of the TCGA (The Cancer Genome Atlas) dataset via cBioportal (cBioPortal for Cancer Genomics, https://www.cbioportal.org), FTO showed the second-highest expression at the transcriptional level in ccRCC. B: Available RNA sequencing expression data from 32 cancer entities were studied, and high mRNA expression of ALKBH5 in ccRCC, in a pan-cancer analysis of the TCGA dataset via cBioportal, ALKBH5 showed the 4th highest expression at the transcriptional level in ccRCC.
To better understand the functional role of both highly expressed m6A demethylases, we performed knockdown experiments to analyze the functional influences of the deletion of FTO and ALKBH5 on parameters of malignancy. After 48 hours of incubation, efficient transfection of fluorescently labeled GapmeRs was evident in the ccRCC cell lines examined. High knockdown efficiency of FTO and ALKBH5 was induced in all cell lines with significantly decreased expression of both genes at both the mRNA and protein levels (see Figure 2). Cell proliferation and migration (see Figures 3 and 4) were significantly decreased after the knockdown of FTO and ALKBH5 in all cell lines examined. In our opinion, the impaired migratory ability of the cells due to the knockdown of FTO and ALKBH5 was due to the resulting disruption of the EMT signaling pathway.
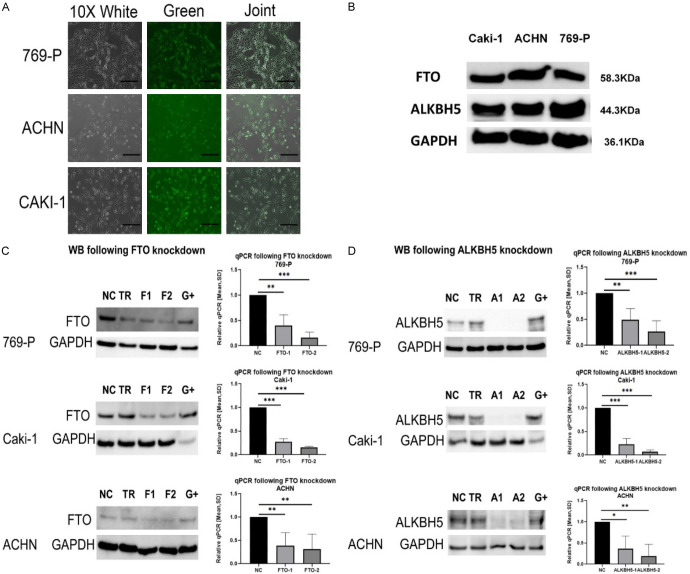
Figure 2.
Knockdown FTO and ALKBH5 successfully. NT = untreated cells; TR = transfection reagent FuGENE; NC = negative control GapmeR; F1 = FTO-1 GapmeR (Sequence 1); F2 = FTO-2 GapmeR (Sequence 2); A1 = ALKBH5-1 GapmeR (Sequence 1); A2 = ALKBH5-2 GapmeR (Sequence 2); G+ = GAPDH GapmeR (positive control, knock down), all treated cells were collected after transfection GapmeRs for 72 hours; A: All three cell lines, 769-P, Caki-1, and ACHN, show transfection efficiency for all the GapmeRs greater than 90%. The FITC-labeled GapmeRs were transfected with FuGENE HD transfection reagent and photos were taken by a fluorescence microscope. All cells were displayed in the White, green fluorescence GapmeR was observed in the Green and the overlay of both in the Joint picture. B: The protein expression of FTO and ALKBH5 for all three renal cell carcinoma cell lines are detectable and at the correct kD size in western blot analysis. GAPDH was used as a Housekeeper. C: The knockdown effect was determined by real-time PCR and Western blotting. The protein level decreased of FTO compared to Negative Control GapmeR, by at least 50%. The mRNA expression of FTO mRNA relative to the NC group were both significantly downregulated in the renal cancer cell lines (769-P, Caki-1 and ACHN) (ns = no statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). D: The knockdown effect was determined by real-time PCR and Western blotting. The protein level decreased of ALKBH5, compared to Negative Control GapmeR, by at least 50%. The mRNA expression of ALKBH5 mRNA relative to the NC group were both significantly downregulated in the renal cancer cell lines (769-P, Caki-1 and ACHN) (ns = no statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test).
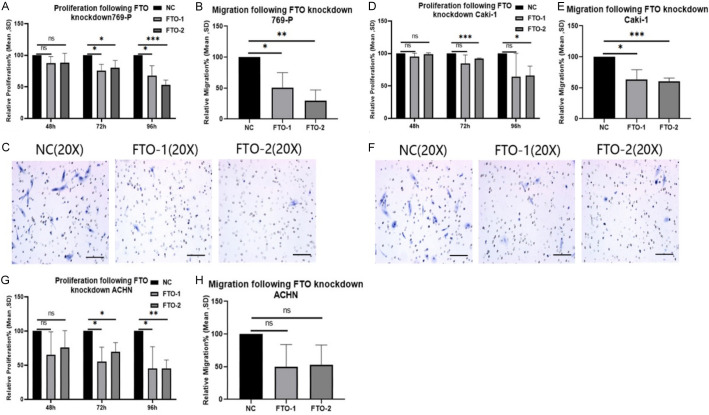
Figure 3.
Knockdown of FTO decreased migration and proliferation for all studied cell lines. A: Effects of knocking down FTO on 769-P cell lines respectively growth/proliferation by EZ4U test. (ns = not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). B: Effects of knocking down FTO on 769-P cell lines. Results show the migration capacity by cell count analysis on Boyden chamber membranes. (ns = not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). C: Pictures of cell count analysis on Boyden chamber membranes by 20 × microscopy to demonstrate migration after 24 h. The effect of FTO knocking down on 769-P cell lines. The ACHN showed no statistically significant difference, so we did not show the changes by 20 × microscopy. D: Effects of knocking down FTO on Caki-1 cell lines respectively growth/proliferation by EZ4U test. (ns = not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). E: Effects of knocking down FTO on Caki-1 cell lines respectively migration capacity by cell count analysis on Boyden chamber membranes. (ns = not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). F: Pictures of cell count analysis on Boyden chamber membranes by 20 × microscopy to demonstrate migration after 24 h. The effect of FTO knocking down on Caki-1 cell lines. The ACHN showed no statistical significance, so we do not show the changes of 20 × microscopy. G: Effects of knocking down FTO on ACHN cell lines respectively. Growth/proliferation by EZ4U test. (ns = not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). H: Effects of knocking down FTO on ACHN cell lines respectively migration capacity by cell count analysis on Boyden chamber membranes. (ns = not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test).
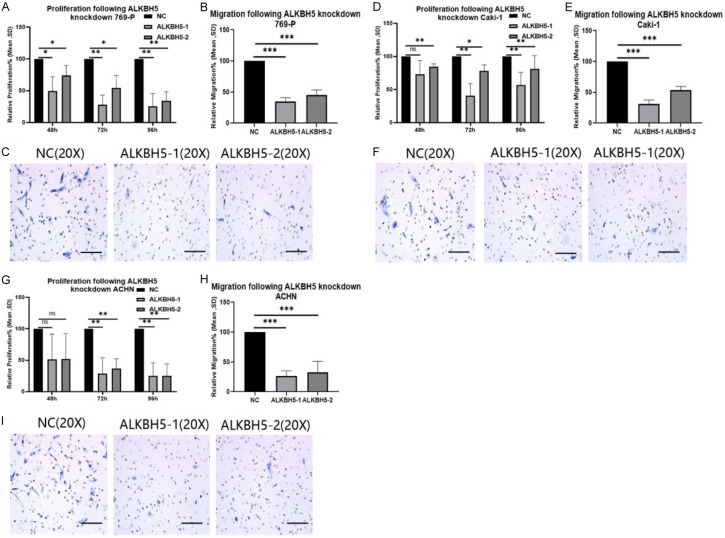
Figure 4.
The knockdown of ALKBH5 decreased migration and proliferation for all studied cell lines. A: Effects of knocking down ALKBH5 on 769-P cell lines respectively growth/proliferation by EZ4U test. (ns = not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). B: Effects of knocking down ALKBH5 on 769-P cell lines respectively migration capacity by cell count analysis on Boyden chamber membranes. (ns = not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). C: Pictures of 769-P cell lines count analysis on Boyden chamber membranes by 20 × microscopy to demonstrate migration after 24 h. D: Effects of knocking down ALKBH5 on Caki-1 cell lines respectively growth/proliferation by EZ4U test. (ns = not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). E: Effects of knocking down ALKBH5 on Caki-1 cell lines respectively migration capacity by cell count analysis on Boyden chamber membranes. (ns = not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). F: Pictures of Caki-1 cell lines count analysis on Boyden chamber membranes by 20 × microscopy to demonstrate migration after 24 h. G: Effects of knocking down ALKBH5 on ACHN cell lines respectively growth/proliferation by EZ4U test. (ns = not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). H: Effects of knocking down ALKBH5 on ACHN cell lines respectively migration capacity by cell count analysis on Boyden chamber membranes. (ns = not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). I: Pictures of ACHN cell lines counted analysis on Boyden chamber membranes by 20 × microscopy to demonstrate migration after 24 h.
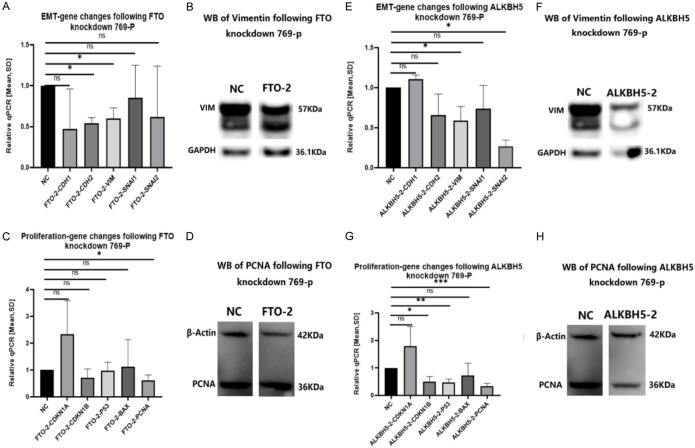
The GapmeRs of FTO-2 and ALKBH5-2 resulted in the best and most stable reduction of migration and proliferation of the 769-p cell line. Therefore, the GapmeRs (FTO-2 & ALKBH5-2) were selected first to test the RQ-mean of mRNA changes of EMT and proliferation genes of the 769-p cell line. Silencing of FTO or ALKBH5 expression resulted in changes in EMT and proliferation process, for EMT genes (VIM, CDH2, SNAI2) and proliferation genes (PCNA, CDKN1B, P53). Both were significantly downregulated after knockdown of ALKBH5 or FTO in RT-PCR. Hereafter, we tested the protein level changes of these six genes. Only VIM and PCNA showed significant results. After specific FTO or ALKBH5 knockdown, we examined VIM expression at the transcriptional and translational levels in 769-P cells.
Correlating with the impaired proliferation potential after FTO or ALKBH5 knockdown, the expression of the proliferation marker PCNA was suppressed (see Figure 5). In the ccRCC-TCGA cohort, both m6A demethylases were positively correlated with VIM and PCNA expression (see Figure 6), in agreement with our experimental results.
Figure 5.
Effects of EMT and proliferation genes changes by knockdown FTO or ALKBH5 on 769-P. Our GapmeRs of FTO-2 and ALKBH5-2 show the best and most stable results on migration and proliferation of the 769-p cell line, so we choose GapmeRs (FTO-2 & ALKBH5-2) to test the RQ-mean of mRNA changes on EMT and Proliferation Genes of 769-p cell line. Silencing FTO and ALKBH5 expression resulted in alterations in the EMT and proliferation process. A: From the RT-PCR results of EMT genes, we found Vimentin (VIM) and N-Cadherin (CDH2) downregulated with statistical significance after FTO knockdown. (ns = not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). B: The protein of VIM was downregulated as well in the western blot experiment. (ns = not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). C: The RT-PCR results of proliferation genes demonstrate a statistically significant downregulation of PCNA after FTO knockdown. (ns = not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). D: There was no significant downregulation of PCNA after FTO knockdown on the protein level of PCNA. (ns = not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). E: VIM and Snail Family Transcriptional Repressor 2 (SNAI2) were significantly downregulated after ALKBH5 knockdown in RT-PCR. (ns = not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). F: We used VIM to correlate this knockdown effect on the protein level. Change of Proliferation Genes for mRNA and proteins after Silencing FTO-2 and ALKBH5-2 in the 769-p cell line. The genes of CDKN1A, CDKN1B, p53, BAX, and PCNA were tested. (ns = not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). G: CDKN1B, p53 and PCNA mRNA were downregulated with statistic significance after ALKBH5 knockdown. (ns = not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). H: The protein of PCNA was also downregulated following ALKBH5 knockdown. (ns = not statistically significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test).
Figure 6.
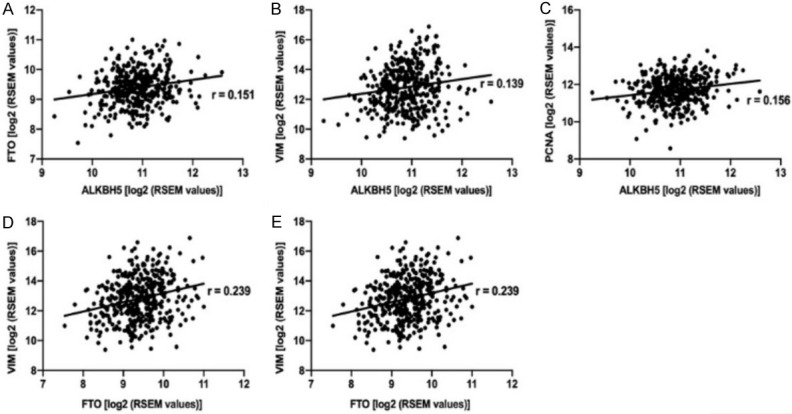
The cBioportal software is used to analyze the correlation between FTO and VIM and PCNA, we can find they are related from its Pearson’s R correlation coefficients. In the ccRCC-TCGA data set, a weak positive correlation was evident between ALKBH5 and FTO (A). Both ALKBH5 and FTO showed a weak positive correlation to VIM (B, D) and PCNA (C, E).
Discussion
M6A methylation is the most abundant posttranscriptional modification (> 50%) [22] with the development of high-throughput sequencing technology and advances in epigenetics research [23-26], renewed attention has focused on the function and role of m6A methylation in various biologic processes. M6A methylation differs from other mRNA internal modifications (such as 5-methylcytosine RNA methylation, m5C [27]) because it regulates the post-transcriptional expression level of the gene without altering the base sequence. This is mainly done by affecting the binding of the mRNA to the writing protein. This regulates the splicing, translation, and stabilization of the mRNA. This process is reversible and precisely controlled. This process involves 3 steps: ① Methylation, the enzyme that adds the methyltransferase (writer); ② Demethylation, i.e. the methyl group is removed, the methyl group is stripped from the carbocyclic ring by a demethylase (eraser); ③ Binding: the RNA base sites where the methylation is changed are recognized and bound.
Several studies have shown that aberrant m6A levels are associated with carcinogenesis in human malignancies [14-16]. Methylation “writers” (consisting of METTL3, METTL14, and WTAP) mediate m6A modification of RNA, while two distinct proteins (FTO and ALKBH5) mediate demethylation. So far, it is not known whether the only human m6A eraser proteins FTO and ALKBH5 have distinct or redundant functions [10-12], FTO and ALKBH5 are abnormally expressed in many tumors [14-16]. FTO plays a critical role in carcinogenesis as an m6A eraser. R-2HG targets the FTO/m6A/MYC/CEBPA axis and has antitumor activity in leukemia and brain tumors [9,14,28]. ALKBH5 is known to affect mRNA export and RNA metabolism. In the pathogenesis of glioblastoma and breast cancer, ALKBH5 acts as an oncoprotein by affecting the proliferation and self-renewal of cancer stem cells [15,16,29]. ALKBH5 expression is abnormally upregulated in gliomastem cells, and its increased expression is associated with poor disease outcome in patients with glioblastoma multiforme [15]. Suppression of ALKBH5 expression in hypoxic MCF-7 and MDA-MB-231 cells increased the m6A content of the total cellular RNA pool, indicating that ALKBH5 plays an important role in regulating RNA methylation in human breast cancer cells [16,29]. The function of m6A demethylases in RCC is poorly understood.
We observed that the expression levels of the two M6A demethylases studied were comparatively high in ccRCC, suggesting their possible importance in the pathogenesis of this tumor entity. We knocked down FTO and ALKBH5 in RCC cell lines (769-P, Caki-1, ACHN) using gapmeRs. A decrease in FTO and ALKBH5 gene expression inhibited proliferation and migration in the cell lines studied. Epithelial cells can undergo epithelial-mesenchymal transition (EMT) to transform into highly motile and invasive phenotypes and migrate from the original tissue. EMT is therefore a crucial step in cancer progression [18,30-33].
Suppression of FTO and ALKBH5 expression resulted in changes in the EMT (indicated by altered VIM mRNA/protein expression) and proliferation (altered PCNA mRNA/protein expression) axes. Accordingly, m6A demethylases affect ccRCC progression through changes in EMT and proliferation.
In summary, we demonstrated that the m6A demethylases FTO and ALKBH5 play an oncogenic role by promoting cell migration and proliferation through modulation of the epithelial-mesenchymal transition and cell cycle. The genes involved are VIM and PCNA. The mechanism by which other genes such as CDH2, SNAI2 among EMT genes and CDKN1B, and P53 regulating proliferation are involved in this process is currently unclear and needs further investigation. FTO and ALKBH5 are highly expressed in renal cancer and may represent therapeutic targets for the treatment of renal cancer.
Acknowledgements
WH was supported by grant No. 2019CFB743 from the Hubei Province Natural Science Foundation, China.
The experimental protocol was established, according to the ethical guidelines of the Helsinki Declaration and was approved by the Human Ethics Committee of University Hospital Bonn. Written informed consent was obtained from individual or guardian participants.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR, Winchester DP. The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin. 2017;67:93–99. doi: 10.3322/caac.21388. [DOI] [PubMed] [Google Scholar]
- 2.Jonasch E, Walker CL, Rathmell WK. Clear cell renal cell carcinoma ontogeny and mechanisms of lethality. Nat Rev Nephrol. 2021;17:245–261. doi: 10.1038/s41581-020-00359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choueiri TK, Motzer RJ. Systemic therapy for metastatic renal-cell carcinoma. N Engl J Med. 2017;376:354–366. doi: 10.1056/NEJMra1601333. [DOI] [PubMed] [Google Scholar]
- 4.Trott JF, Kim J, Abu Aboud O, Wettersten H, Stewart B, Berryhill G, Uzal F, Hovey RC, Chen CH, Anderson K, Graef A, Sarver AL, Modiano JF, Weiss RH. Inhibiting tryptophan metabolism enhances interferon therapy in kidney cancer. Oncotarget. 2016;7:66540–66557. doi: 10.18632/oncotarget.11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubin DT, Taylor RH. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975;2:1653–1668. doi: 10.1093/nar/2.10.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 2013;29:108–115. doi: 10.1016/j.tig.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu N, Pan T. N6-methyladenosine-encoded epitranscriptomics. Nat Struct Mol Biol. 2016;23:98–102. doi: 10.1038/nsmb.3162. [DOI] [PubMed] [Google Scholar]
- 8.Wang T, Kong S, Tao M, Ju S. The potential role of RNA N6-methyladenosine in cancer progression. Mol Cancer. 2020;19:88. doi: 10.1186/s12943-020-01204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng X, Su R, Feng X, Wei M, Chen J. Role of N(6)-methyladenosine modification in cancer. Curr Opin Genet Dev. 2018;48:1–7. doi: 10.1016/j.gde.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torres IO, Fujimori DG. Functional coupling between writers, erasers and readers of histone and DNA methylation. Curr Opin Struct Biol. 2015;35:68–75. doi: 10.1016/j.sbi.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, Lu Z, Bosmans RP, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia G, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG, He C. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, Qin X, Tang L, Wang Y, Hong GM, Huang H, Wang X, Chen P, Gurbuxani S, Arnovitz S, Li Y, Li S, Strong J, Neilly MB, Larson RA, Jiang X, Zhang P, Jin J, He C, Chen J. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, Majumder S, He C, Huang S. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606. e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, He X, Semenza GL. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A. 2016;113:E2047–2056. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strick A, von Hagen F, Gundert L, Klümper N, Tolkach Y, Schmidt D, Kristiansen G, Toma M, Ritter M, Ellinger J. The N(6)-methyladenosine (m(6)A) erasers alkylation repair homologue 5 (ALKBH5) and fat mass and obesity-associated protein (FTO) are prognostic biomarkers in patients with clear cell renal carcinoma. BJU Int. 2020;125:617–624. doi: 10.1111/bju.15019. [DOI] [PubMed] [Google Scholar]
- 18.Tretbar S, Krausbeck P, Müller A, Friedrich M, Vaxevanis C, Bukur J, Jasinski-Bergner S, Seliger B. TGF-β inducible epithelial-to-mesenchymal transition in renal cell carcinoma. Oncotarget. 2019;10:1507–1524. doi: 10.18632/oncotarget.26682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG, James JA, Salto-Tellez M, Hamilton PW. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 23.Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A, Helm M, Bujnicki JM. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 25.Trinh BN, Long TI, Laird PW. DNA methylation analysis by MethyLight technology. Methods. 2001;25:456–462. doi: 10.1006/meth.2001.1268. [DOI] [PubMed] [Google Scholar]
- 26.Schwartz S, Motorin Y. Next-generation sequencing technologies for detection of modified nucleotides in RNAs. RNA Biol. 2017;14:1124–1137. doi: 10.1080/15476286.2016.1251543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Motorin Y, Lyko F, Helm M. 5-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010;38:1415–1430. doi: 10.1093/nar/gkp1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, Ni T, Zhang ZS, Zhang T, Li C, Han L, Zhu Z, Lian F, Wei J, Deng Q, Wang Y, Wunderlich M, Gao Z, Pan G, Zhong D, Zhou H, Zhang N, Gan J, Jiang H, Mulloy JC, Qian Z, Chen J, Yang CG. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell. 2019;35:677–691. e10. doi: 10.1016/j.ccell.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang C, Zhi WI, Lu H, Samanta D, Chen I, Gabrielson E, Semenza GL. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget. 2016;7:64527–64542. doi: 10.18632/oncotarget.11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim T, Veronese A, Pichiorri F, Lee TJ, Jeon YJ, Volinia S, Pineau P, Marchio A, Palatini J, Suh SS, Alder H, Liu CG, Dejean A, Croce CM. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med. 2011;208:875–883. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strouhalova K, Přechová M, Gandalovičová A, Brábek J, Gregor M, Rosel D. Vimentin intermediate filaments as potential target for cancer treatment. Cancers (Basel) 2020;12:184. doi: 10.3390/cancers12010184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 2011;68:3033–3046. doi: 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin S, Chen FF, Yang GF. Vimentin immunohistochemical expression as a prognostic factor in gastric cancer: a meta-analysis. Pathol Res Pract. 2018;214:1376–1380. doi: 10.1016/j.prp.2018.07.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.