Abstract
Background: It is essential to develop better biomarkers for diagnosis, prediction, and treating glioma patients to ensure successful clinical outcomes. The clinical application of Insulin-Like Growth Factor Binding Proteins (IGFBPs) for glioma is yet to be investigated. Methods: Cohorts were obtained from TCGA, GTEx, CGGA, HPA, Oncomine, CancerSEA, TISCH, etc. The expressions, methylation, and survival association of IGFBPs were analyzed. The Least Absolute Shrinkage and Selection Operator (LASSO) regression was used to construct a prognostic model. The correlation of IGFBPs and immune cells or immune molecules was analyzed. The effect of IGFBPs on immune therapy and chemotherapy was analyzed. The top 500 correlated genes of IGFBPs were enriched in GO terms. Correlations between IGFBPs and functional states in glioma single-cells and correlations between IGFBPs and chemokines in samples were analyzed. Results: IGFBPs were overexpressed in glioma. Malignant cells were the major cell types that expressed IGFBPs. Higher-grade glioma had a higher expression of IGFBPs, which were associated with worse survival. A survival model of IGFBPs for glioma patients was trained and validated. A nomogram was generated. Generally, IGFBPs mRNA expression was negatively correlated with B cells and T cells. IGFBPs were associated with multiple immune molecules and drug sensitivity. Conclusion: IGFBPs were powerful diagnostic, prognostic, and therapeutic prediction biomarkers for glioma.
Keywords: IGFBPs, glioma, expression, survival, immune cells
Introduction
The incidence of glioma, one of the most common forms of malignant brain tumor, is on the rise globally [1,2]. It is categorized into four distinct grades based on histologic and clinical data; Grade 1, Grade 2, Grade 3, and Grade 4 [3]. Grade 1 glioma is typically benign, and is most commonly seen in children, with successful outcomes from surgical resection. Grade 2 and 3 gliomas are commonly referred to as low-grade gliomas (LGG) while Grade 4 glioma is known as the highest-grade glioma, and is also referred to as glioblastoma or glioblastoma multiforme (GBM) [4]. Glioma, especially grade 4 glioma, remains a fatal condition despite the evolution of treatments, with most cases having a very bad survival [4]. So far, the lack of effective diagnostic and prognostic strategies and the low response rate of glioma therapy prevent the improvement of glioma survival. As such, it is of critical importance to develop reliable biomarkers to improve the diagnosis, prognosis, and therapeutic prediction of gliomas.
Insulin-Like Growth Factor Binding Proteins (IGFBPs, gene symbols are IGFBP1-7, we also referred to as IGFBPs in this study), known as insulin-like growth factor (IGF) carrier proteins, are circulating bioactive molecules that play a role in the modulation of IGF [5]. There are seven IGFBPs (IGFBP1-7) that bind IGFs [6]. Recent studies revealed that IGFBPs were closely associated with cancer development. In lung cancer, IGFBPs have been suggested to be a biomarker for patients [7]. Reduction in IGFBP1 suppressed beta-element-inhibited cell proliferation of lung cancer cells [8]. IGFBP1 was found overexpressed in colorectal cancer and regulated tumor and metastasis [9]. IGFBP2 mediated many tumor functions in breast cancer [10].
The prognostic value of IGFBPs for many types of cancer has been reported [11-13]. A previous study investigated the immune association and clinical association of IGFBP2 in glioma [14]. Yet, so far, the clinical value of IGFBPs in glioma has not been studied comprehensively.
Methods
Data acquisition
Cohorts of glioma were obtained from The Cancer Genome Atlas (TCGA) [15], Genotype-Tissue Expression (GTEx) [16], and the Chinese Glioma Genome Atlas (CGGA) [17]. Immunohistochemistry images were accessed from the Human Protein Atlas (HPA) [18]. Data of expression comparison across analyses were accessed using Oncomine [19]. The chemotherapy response and expression data (n=454) were accessed from the ROC Plotter [20]. Single-cell sequencing cohorts were accessed from the Tumor Immune Single-cell Hub (TISCH) [21] and Cancer single-cell states atlas (CancerSEA) [22]. Data used in this study included TCGA [15], GETx [16], CGGA [17], GSE131928 [23], SRA PRJNA482620 [24], GSE84465 [25], and GSE31095 [26].
Data analysis and plotting
All the analyses of TCGA, GTEx, and CGGA cohorts were conducted by R v4.0.3 and ggplot2 v3.3.2. Principal component analysis (PCA) dimensionality reduction on samples was conducted using the GEPIA2 [27]. Functional prediction of genes and gene interaction networks was conducted using GeneMANIA [28]. The Estimating the Proportions of Immune and Cancer cells (EPIC) algorithms was used to estimate the immune cell infiltration level [29]. Potential immune checkpoint blockade (ICB) response was calculated using the Tumor Immune Dysfunction and Exclusion (TIDE) algorithm [30]. The GSCALite [31] was used to evaluate the area under the dose-response curve (AUC) values for drug association of IGFBPs in cancer cell lines. GDSC and CTRP data were integrated for investigation. Single-cell sequencing data were analyzed and plotted by the TISCH. The gene-miRNA-transcriptional factor coregulation network was constructed using RegNetwork [32]. T-test or One-way ANOVA with Tukey test was used to analyze the difference between groups. Spearman correlation was used to analyze correlations between two variates. Gene expression differences were compared using Wilcoxon or Kruskal-Wallis tests. Survival analysis was conducted using Kaplan-Meier analysis, log-rank tests and Cox regression tests, while Pearson’s correlation tests were used to evaluate the correlation of two variables except for drug sensitivity analysis. Statistical significance was set at P<0.05.
Results
Diagnostic value of IGFBPs for glioma
TCGA data suggested that the gene mutation rate of IGFBPs in glioma was low. Among 403 GBM samples, only 2% of GBM had IGFBP7 and IGFBP3 mutation and 1% of GBM had IGFBP1 mutation and less than 1% of GBM had mutations in IGFBP5 with no mutation in the other IGFBPs. Among 526 LGG samples, only 1% of LGG had IGFBP5 and IGFBP2 mutations with no mutation in the other IGFBPs (Figure 1A). Hence, we suggested that IGFBP mutations were not critical for glioma.
Figure 1.
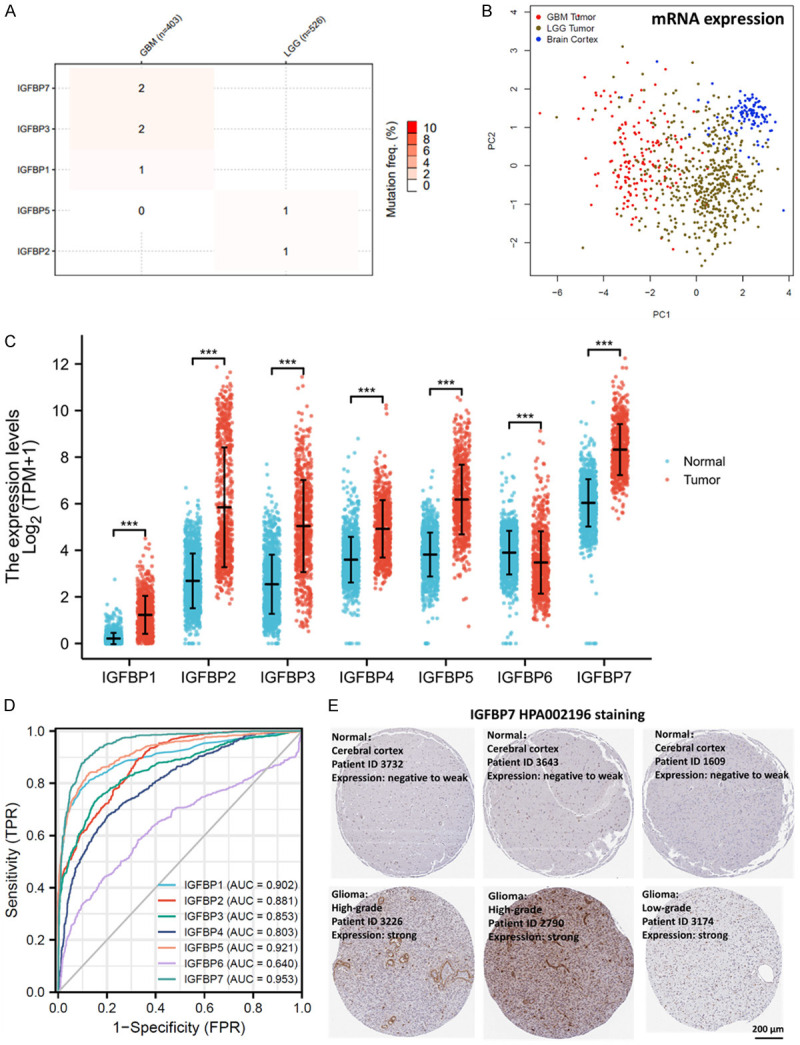
Diagnostic value of IGFBPs for glioma. A. Single-nucleotide variant percentage heatmap of glioma. TCGA (LGG+GBM) cohort was analyzed. B. PCA dimensionality reduction on the mRNA expression of IGFBPs from glioma and normal brain cortex tissues based on their expression of IGFBPs. TCGA (LGG+GBM) and GETx cohorts were compared. C. The mRNA expressions of IGFBPs in glioma (n=1157) and normal brain (n=689) tissues. TCGA (LGG+GBM) and GETx cohorts were compared. D. Diagnostic ROC curve of IGFBPs in glioma (LGG and GBM). TCGA (LGG+GBM) and GETx cohorts were analyzed. E. Representative immunohistochemical images of IGFBP protein in glioma and normal brain tissues (100X). Antibody staining of IGFBP7 protein in normal brain and glioma tissues were downloaded from the Human Protein Atlas and shown as representative images. *P<0.05, **P<0.01,***P<0.001. GBM: glioblastoma; LGG: low grade glioma; TPM: transcripts per million; ID: identity number; TPR: True Positive Rate; FPR: False Positive Rate; AUC: Area under the ROC Curve; ROC: Receiver operating characteristic.
To determine whether the expression of IGFBPs distinguished between glioma and normal brain, we conducted PCA dimensionality reduction on samples from glioma and brain based on the IGFBPs mRNA levels. The PCA plotting showed that the clusters of glioma samples, including GBM (grade 4 glioma) and LGG (grade 2-3 glioma) samples, considerably distinguished from the cluster of normal brain tissues (Figure 1B). This indicated that the expressions of IGFBPs were different between glioma and brain tissues. To further specify the difference, the expressions of IGFBPs in glioma and normal brain tissues were plotted. Statistical analysis showed that, compared to normal brain tissues, glioma expressed significantly higher IGFBPs except for IGFBP6 (Figure 1C). To validate the up-regulation of IGFBPs in glioma, the Oncomine was queried for the overexpression or copy number gain of IGFBPs in glioma. 14, 16, 20, 10, 15, 6, and 13 analyses of glioma were gathered for IGFBP1, IGFBP2, IGFBP3, IGFBP4, IGFBP5, IGFBP6, and IGFBP7 respectively (Supplementary Figure 1). All seven IGFBPs were significantly overexpressed or overcopied in glioma compared with normal brain tissues. IGFBP6 had fewer data sets supporting the overexpression in glioma, which was consistent with the TCGA data. In addition, correlation analysis of IGFBP mRNA expression revealed that IGFBPs were correlated with each other (Supplementary Figure 2).
To estimate the detection biomarker potential of IGFBPs for glioma, diagnostic ROC curves of IGFBPs in glioma were plotted. Results showed that the AUCs of IGFBPs were over 0.8 except for IGFBP6 (0.64) (Figure 1D). IGFBP6 had the lowest AUC, which was consistent with the expression plot. Therefore, IGFBPs showed very good diagnostic values, except for IGFBP6. The IGFBP7 had the highest AUC of 0.953, thus, we downloaded representative staining images of IGFBP7 protein to show the potential diagnostic use of this biomarker for cancer detection. The image data suggested glioma expressed higher IGFBP7 at the protein level (Figure 1E). These data revealed that IGFBPs may be diagnostic biomarkers for glioma. Thus we believe that IGFBPs have value in the diagnosis of glioma. It would be helpful to check whether the level of IGFBPs in more accessible tissues such as blood is consistent with the expression in tumor tissue. Thus, we analyzed the mRNA expression of IGFBPs in platelets from glioma patients and healthy donors. However, only IGFBP3 was significantly upregulated in platelets from glioma patients (Supplementary Figure 3). Yet, the analysis was rather preliminary since only 20 samples were included, thus the significance might be subjected to a low sample number. More studies will be required to confirm the results or distinguish the any differences.
IGFBP expression in glioma cell sublines
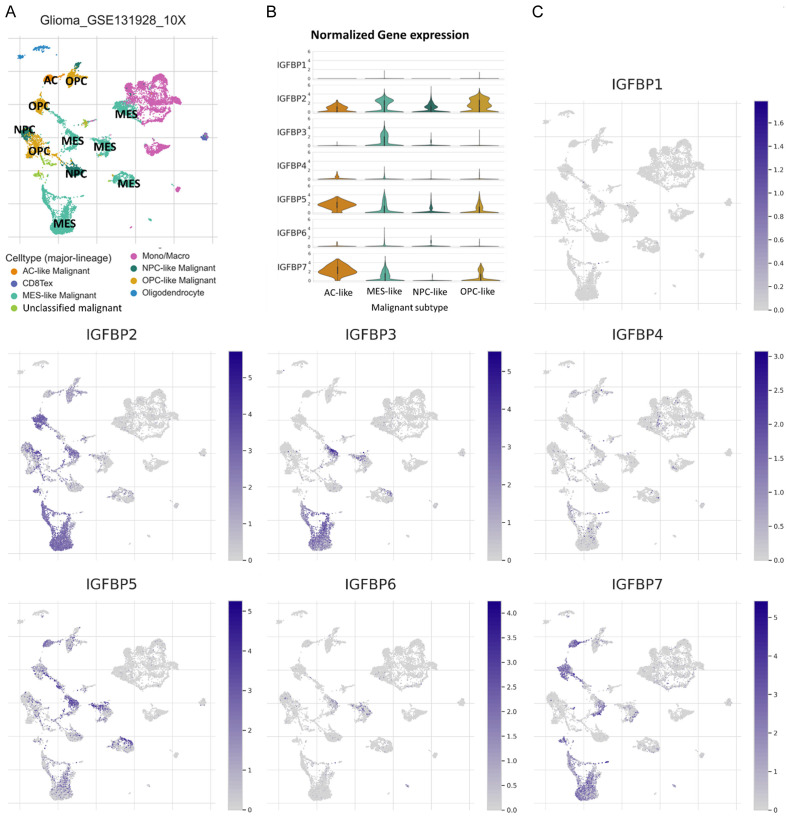
This study also analyzed single-cell mRNA data to specify the glioma cell sub-types that expressed IGFBP genes. In this study, single-cell data analysis revealed that IGFBP1, IGFBP4, and IGFBP6 had very low expression in all glioma cell subtypes, which was consistent with the above expression analysis showing that IGFBP2, IGFBP3, IGFBP5, and IGFBP7 had the four highest expressions of IGFBP genes (Figure 1B). Among all cell subtypes in glioma, malignant cells were the major cell subtype that expressed IGFBP genes. Malignant glioma cells had been classified into four sub-lines based on single cell expression profiles, including 1) neural-progenitor-like (NPC-like), 2) oligodendrocyte-progenitor-like (OPC-like), 3) astrocyte-like (AC-like), and 4) mesenchymal-like (MES-like) [23]. IGFBP2 was relatively evenly expressed in OPC-like, AC-like, MES-like, and NPC-like malignant cells. IGFBP3 was only highly found in MES-likecells. IGFBP5 and IGFBP7 were highly expressed in AC-like cells and slightly expressed in MES-like and OPC-like malignant cells (Figure 2).
Figure 2.
Identification of IGFBPs expressing cell sub-types in glioma. Single-cell mRNA expression cohort GSE131928 (Glioma_GSE131928_10X) was accessed and analyzed using the TISCH. Abbreviation: neural-progenitor-like (NPC-like), oligodendrocyte-progenitor-like (OPC-like), astrocyte-like (AC-like), and mesenchymal-like (MES-like), CD8Tex (exhausted CD8+T cells). A. Cell-subtype annotation. The gene expression level was displayed using UMAP (Dimensionality reduction by Uniform Manifold Approximation and Projection). The cell clustering was conducted and the cell-type annotation at the single-cell level was provided by the TISCH. B. Expressions of IGFBPs mRNA in cells of glioma samples, log2 (TPM+1). C. Violin plots of IGFBPs mRNA expressions in different subtypes of glioma cells, log2 (TPM+1).
Clinical association of IGFBPs
Next, we compared IGFBPs in different clinical subgroups, including WHO grades of glioma, IDH gene mutation status, the 1p/19q codeletion, primary therapy outcome, gender, age, race, and histology. Results showed that higher-grade glioma, which is more severe, expressed significantly higher IGFBPs than lower-grade glioma (Figure 3A). IDH mutated glioma usually has a better prognosis. Our analysis revealed that mutated IDH glioma expressed lower IGFBPs than IDH wild type glioma (Figure 3B). Another protective factor for glioma, 1p/19q co-deletion, was also associated with better survival in glioma. Data suggested that glioma with 1p/19q co-deletion reduced all IGFBP levels except for IGFBP1 (Figure 3C). As for the primary therapy outcome group, data suggested that, generally, the PD (progressive disease) group expressed higher IGFBPs, while the CR (complete response) group expressed relatively lower IGFBPs (Figure 3D). In addition, gender and race made no difference in IGFBPs (Figure 3E and 3G) but patients older than 60 expressed significantly higher IGFBPs (Figure 3F). Among the histologic groups, glioblastoma expressed a higher level of IGFBPs than astrocytoma, oligoastrocytoma, or oligodendroglioma (Figure 3H). Glioblastoma is the most aggressive glioma type compared to astrocytoma, oligoastrocytoma, and oligodendroglioma. These clinical association analysis results suggested that IGFBP might associate with a worse prognostic phenotype of glioma.
Figure 3.
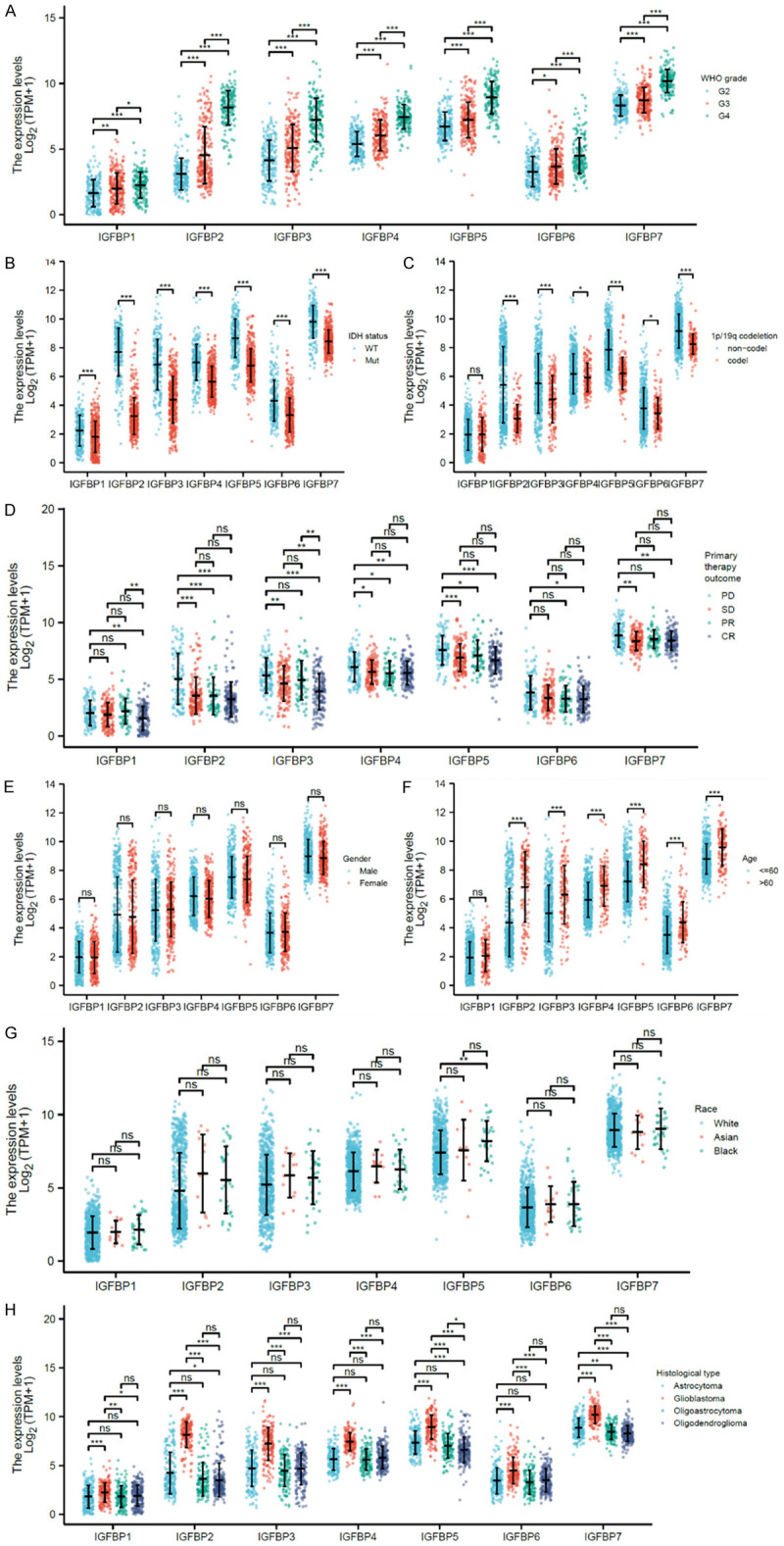
Clinical association of IGFBPs. TCGA (LGG+GBM) and GETx cohorts were analyzed for mRNA expression comparisons. A. WHO grades of glioma: Grade 2 (n=224), Grade 3 (n=243), and Grade 4 (n=168). B. IDH gene mutation status: wildtype (n=246) and mutated (n=440). C. The 1p/19q codeletion: non-co-deleted (n=518) and co-deleted (n=171). D. Primary therapy outcomes: PD (progressive disease) (n=112), SD (stable disease) (n=147), PR (partial response) (n=64), and CR (complete response) (n=139). E. Gender: male (n=398) and female (n=298). F. Age: ≤60 (n=553) and >60 (n=143). G. Race: White (n=637), Asian (n=13), and Black (n=33). H. Histology: astrocytoma (n=195), glioblastoma (n=168), oligoastrocytoma (n=134), and oligodendroglioma (n=199). *P<0.05, **P<0.01, ***P<0.001. Ns: not significant; WT: wild-type; mut: mutant; WHO: World Health Organization.
Regulation of IGFBP expression in glioma
This study concerned how IGFBP expression was regulated. First, epigenetic data analysis revealed that the methylation of IGFBPs in glioma might regulate their mRNA expression. Results showed that higher-grade glioma had significantly lower methylation of all seven IGFBP genes (Supplementary Figure 4A). Data also suggested that the methylation of IGFBPs in glioma was mostly negatively correlated with gene expression, especially for LGG data (Supplementary Figure 4B). The methylation level was also associated with overall survival, except for IGFBP3 (Supplementary Figure 4C). In addition, to identify possible miRNAs and transcriptional factors that might regulate IGFBP expression, a gene-miRNA-transcriptional factor coregulation network of IGFBPs was constructed (Supplementary Figure 5A). Data also revealed that most of the transcription factors and some of the miRNAs identified in the coregulation network were correlated with the expression of IGFBPs (Supplementary Figure 5B and 5C). This study further identified 32 transcriptional factors associated with glioma prognosis (Supplementary Figure 5D). Most of these transcriptional factors were expressed differentially in glioma compared to normal brains (Supplementary Figure 6A) or differently expressed depending on the grade of glioma (Supplementary Figure 6B), inferring that they may be associated with the development of glioma.
Survival prediction value of IGFBPs
Since we have demonstrated that IGFBPs may associate with a worse glioma phenotype, IGFBP expression may also associate with survival. To investigate the prognostic value of IGFBPs for glioma, this study conducted Kaplan-Meier (KM) plots and log-rank analysis using the TCGA (LGG+GBM) mRNA expression cohort. The analysis revealed that all of the IGFBPs were survival-associated (Supplementary Figure 7A). External validation log-rank analysis was conducted using CGGA mRNA expression data, which further confirmed that IGFBPs were significantly associated with worse survival (Supplementary Figure 7B). To estimate the survival prediction performance of IGFBPs for glioma, time-dependent receiver operating characteristic (ROC) curves were plotted. IGFBP2, IGFBP3, IGFBP4, IGFBP5, and IGFBP7 had AUCs of over 0.7, which indicated their considerably good value for prognosis. IGFBP1 had AUCs of 0.54-0.6 and IGFBP6 had AUCs of 0.62-0.74, which also had a possible prognostic value (Supplementary Figure 7C).
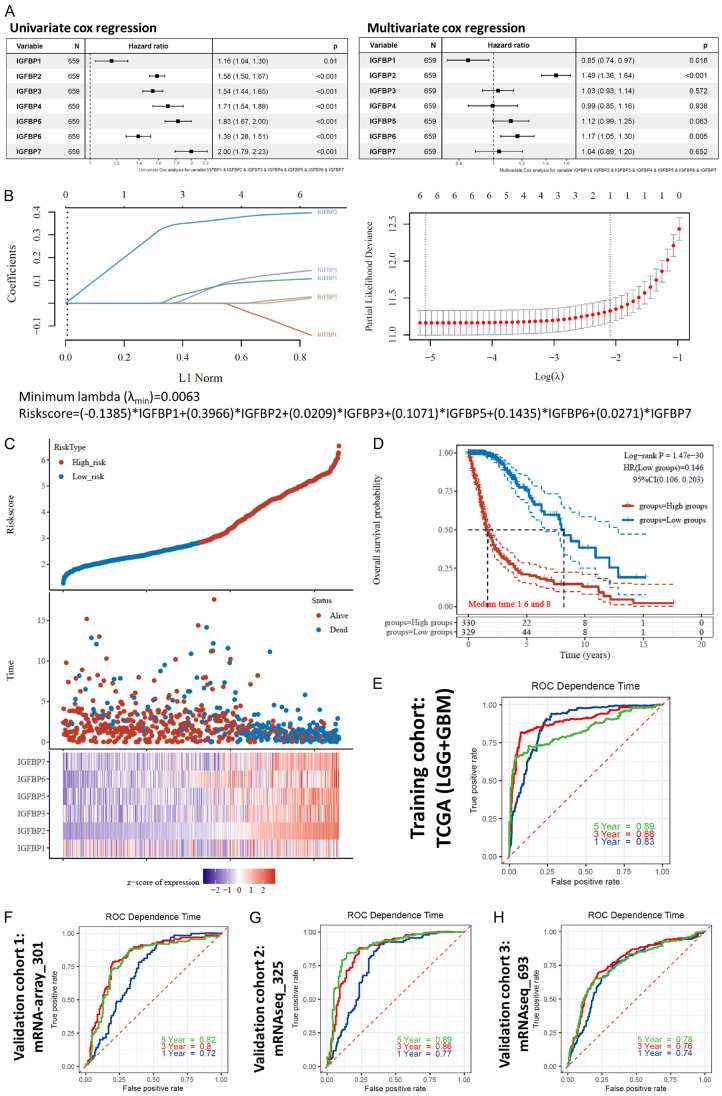
Univariate Cox analysis confirmed that each of the IGFBP genes was associated with survival (Figure 4A left panel). Multivariate Cox analysis revealed that the HR of IGFBP2 and IGFBP6 were still larger than 1and the HR of IGFBP1 was lower than 1 while the other IGFBPs had no significance (Figure 4A right panel). In univariate Cox regression, IGFBP1 was a risk factor, but after being adjusted for the other IGFBPs, multivariate Cox regression showed that IGFBP1 was a protective factor. This means that IGFBP1 can be used as a risk factor alone, but when IGFBP1 was used with the other IGFBPs, it can only be used as a protective factor. The multivariate Cox regression analysis of IGFBPs suggested that IGFBPs were not independent of each other in their survival association except for IGFBP1, IGFBP2, and IGFBP6. Therefore, to further optimize the prognostic application of IGFBPs, a gene signature model is required.
Figure 4.
Construction of a machine-learning prognostic model. TCGA (LGG+GBM) mRNA expression cohorts were used to train the model with the LASSO algorithm, while CGGA mRNA-array_301, mRNAseq_325, and mRNAseq_693 mRNA expression cohorts were used to validate the model. A. Overall survival Cox regression analysis of IGFBPs for glioma patients. B. left panel: Coefficients of IGFBPs shown by lambda parameter. Right panel: Partial likelihood deviance versus log (λ) drawn using LASSO (Least Absolute Shrinkage and Selection Operator) regression model. Bottom panel: The equation of the LASSO regression model. C. Prognostic analysis of the LASSO regression model. Top panel: The distribution of risk score. The dots represent the risk scores and were divided into low-risk and high-risk groups by median. Middle panel: Survival status of the patients. Bottom panel: Heatmap of the expression profiles of the prognostic genes in the LASSO regression model. D. Overall survival KM plots and log-rank analysis of risk score in glioma. HR was calculated using cox regression. Median survival times are shown. E. Overall survival time-dependent receiver operating characteristic (ROC) curves of the risk score for glioma patients in the training cohort. F-H. Overall survival time-dependent ROC curves of the risk score for glioma patients in three validation cohorts.
Hence, we constructed a machine-learning prognostic model based on IGFBPs (Figure 4B). The Least Absolute Shrinkage and Selection Operator (LASSO) was used to perform variable selection. The tuning parameter lambda was chosen by cross-validation. When lambda was small, the result is essentially the least-squares estimates and the model will have high accuracy without overfitting (Figure 4C). Results showed that this model had very high prognostic confidence for glioma patients in the training cohort (Figure 4D, 4E). Three external independent glioma cohorts were used for the validation of this model. The AUCs of ROC were all between 0.72-0.89 (Figure 4F-H). Consequently, based on this model, we constructed another prognostic nomogram for glioma patients (Supplementary Figure 8). These survival analyses suggested that IGFBPs had a very high prognostic value for glioma patients.
The predictive value of IGFBPs for glioma immunotherapy
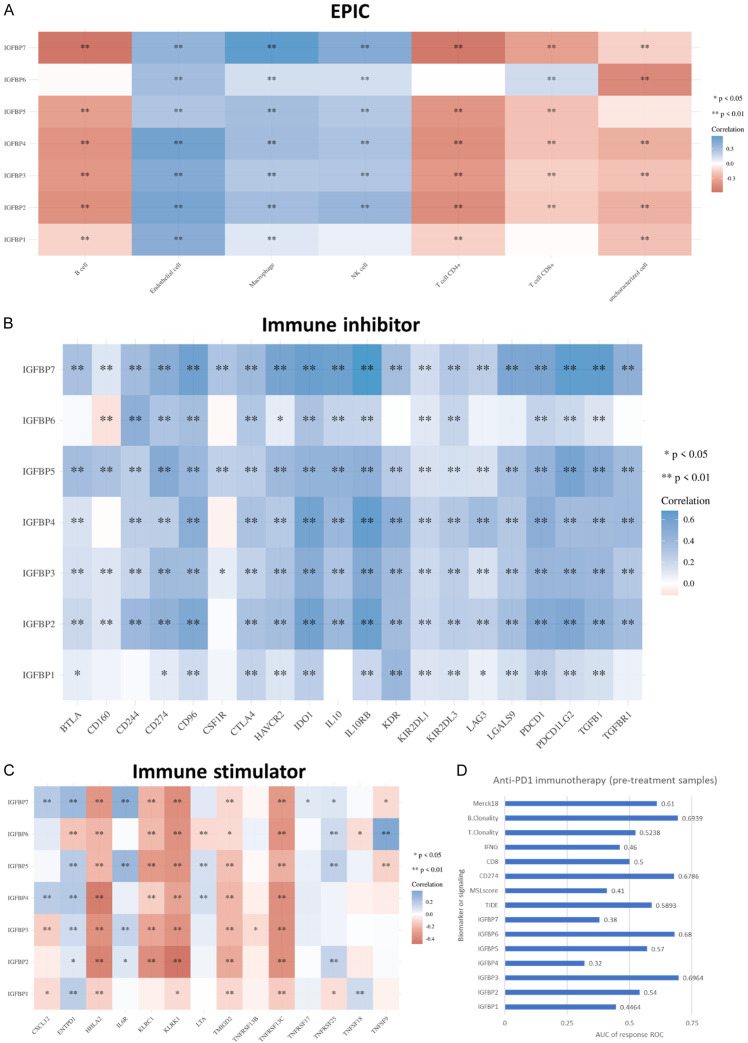
This study also investigated the predictive value of IGFBPs for glioma immunotherapy. The analysis revealed that IGFBPs were positively correlated with endothelial cells, macrophages, and NK cells. IGFBPs were negatively correlated with B cells, T cell CD4+, and T cell CD8+, except for B cell/IGFBP6, T cell CD4+/IGFBP6, T cell CD8+/IGFBP1, and T cell CD8+/IGFBP6. T cell CD8+ was positively correlated with IGFBP6 (Figure 5A). This study also analyzed the correlation between IGFBPs and immune inhibitors or stimulators, which were immune checkpoint blockade targets for immunotherapy. Results showed that, in brief, most of the immune inhibitors were positively correlated with the expression of IGFBPs (Figure 5B), while a majority of the immune stimulators analyzed were negatively correlated with IGFBPs (Figure 5C). These results indicated that IGFBPs might decrease B cell and T cell infiltration level, promote immune inhibition, and reduce immune stimulation. In addition, this study also revealed that IGFBP expression was positively correlated with most of the MHC molecules (Supplementary Figure 9), which might result from negative feedback of the low infiltration level of T cells.
Figure 5.
Immunity association of IGFBPs in glioma. A. Correlations of immune cell infiltration levels and expression of IGFBPs. The EPIC (Estimating the Proportions of Immune and Cancer cells) algorithms were used to estimate the immune cell infiltration levels. TCGA (LGG+GBM) mRNA expression cohort was analyzed. B. Correlations of expressions of immune inhibitors and expressions of IGFBPs. TCGA (LGG+GBM) mRNA expression cohort was analyzed. C. Correlations of expressions of immune stimulators and expressions of IGFBPs. TCGA (LGG+GBM) mRNA expression cohort was analyzed. D. AUC of anti-PD1 immunotherapy response ROC (responders n=8, non-responders n=7). SRA PRJNA482620 data were accessed and analyzed using the TIDE.
In order to further evaluate the predictive value of IGFBPs for glioma immune therapy, TIDE algorithms [30] were used to predict the immune checkpoint blockade (ICB) response of IGFBP-high (75-100%) glioma and IGFBP-low glioma (0-25%). Results showed that IGFBP-high gliomas had higher TIDE scores and were more likely to respond to ICB therapy (Supplementary Figure 10). Therefore, IGFBPs genes were signs of the immune microenvironment and predictive biomarkers for immune therapies in glioma. To compare the predictive value of IGFBPs with the current popular immunotherapy biomarkers/signals, we analyzed a PD1 blockade immunotherapy cohort. We compared the AUC of anti-PD1 immunotherapy response ROC of IGFBPs with 8 commently-used immunotherapy biomarkers/signals, including TIDE, microsatellite instability (MSI.score), CD274, CD8, interferon-gamma (IFNG), T cell clonality (T.Clonality), B cell clonality (B.Clonality), and Merck18 signature. Results revealed that IGFBP3 had the best performance for the prediction of anti-PD1 immunotherapy response with an AUC of 0.6964, which was higher than all of the immunotherapy biomarkers/signals. IGFBP6 was the second highest IGFBP, with an AUC of 0.68 which was higher than all of the commonly-used immunotherapy biomarkers/signals except for B.Clonality. The AUC of the other IGFBPs were all lower than 0.6 (Figure 5D). Thus, IGFBP3 and IGFBP6 had great predictive power in anti-PD1 immunotherapy response.
Predictive value of IGFBPs for glioma drug therapy
On the other hand, we studied drug therapy response. Results revealed that, in the GDSC database, IGFBP6, IGFBP4, IGFBP3, and IGFBP1 were positively correlated with the IC50 of most of the top 30 correlated drugs (Figure 6A). In the CTRP database, IGFBPs were positively correlated with the IC50 of most of the top 30 correlated drugs except for IGFBP2 (Figure 6B). The complete correlation results of all correlated drugs were shown in Supplementary Figures 11 and 12. These data suggested that IGFBPs were signs that cancer cells were insensitive to these drugs. However, these results were not based on glioma cell lines but based on all cancer cell lines in the database. Whether the drug-sensitive prediction value was specific to glioma cells requires further analysis.
Figure 6.
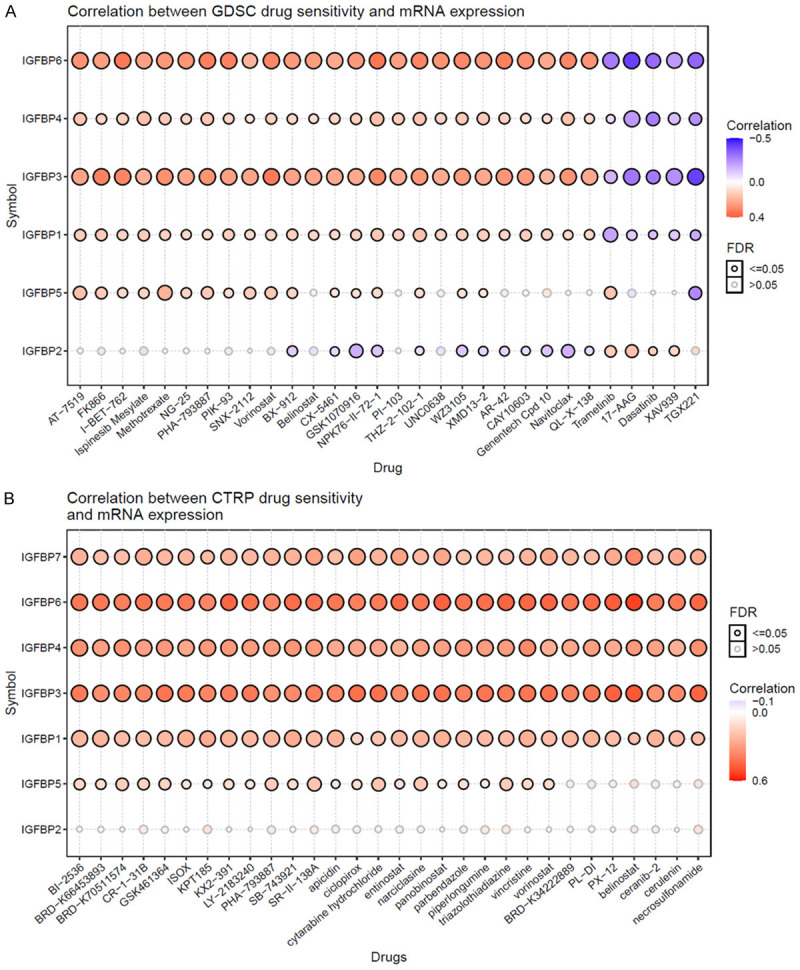
The predictive value of IGFBPs for drug therapy in cancers. A. Genomics of Drug Sensitivity in Cancer (GDSC) drugs. B. The Cancer Therapeutics Response Portal (CTRP) drugs. The GSCALite was used to evaluate the area under the dose-response curve (AUC) values for drugs and gene expression profiles of IGFBPs in different cancer cell lines. Drug sensitivity and gene expression profiling data of cancer cell lines in GDSC and CTRP are integrated for investigation. The expression of each gene in the gene set was performed by Spearman correlation analysis with the small molecule/drug sensitivity (IC50). Data of the top 30 drugs are shown.
Possible functions of IGFBPs in glioma
The GeneMANIA predicted the functions of IGFBPs. The predictions suggested that IGFBPs might have foundations in growth factor binding, glycosaminoglycan binding, sulfur compound binding, etc (Supplementary Figure 13). We also analyzed single-cell data to investigate the role of IGFBPs in glioma. Seven IGFBP genes were queried as a gene list. The single-cell data revealed that IGFBPs showed strong correlations to glioma cell metastasis, epithelial-mesenchymal transition (EMT), hypoxia, and angiogenesis (R>0.5). IGFBPs also had moderate correlations to glioma cell inflammation, invasion, apoptosis, quiescence, and differentiation (0.2<R<0.5) (Supplementary Figure 14). EMT was associated with cancer migration. The analysis revealed that IGFBP genes were positively associated with multiple chemokines and most of the chemokine receptors (Supplementary Figure 15). As chemokine signals are critical for cell migration, these results suggested that IGFBPs might be associated with glioma cell migration, which might be related to the correlation of IGFBPs and EMT. Surprisingly, as growth factor binding proteins, IGFBPs were significantly negatively correlated with glioma cell stemness (Supplementary Figure 14). To confirm a possible correlation of IGFBPs and glioma cell stemness, the one-class logistic regression (OCLR) algorithms [33] were used to estimate the stemness of TCGA samples. Results showed that all seven IGFBP genes were negatively associated with glioma stemness (Supplementary Figure 16).
Discussion
This study used bioinformatic analysis to support the clinical values of IGFBPs for glioma treatment. This study indicated that IGFBPs might be used for the diagnosis of glioma because they were overexpressed in glioma tissues. On the other hand, IGFBPs can be used for glioma prognosis as they were associated with patients’ survival. The prognostic use of IGFBPs has been reported often in other cancer types, such as ovarian cancer [12] and breast cancer [11,34]. IGFBP-4 expression was suggested to be adversely associated with lung cancer prognosis [13]. IGFBP5 was reported as a poor prognostic factor in patients with urothelial carcinomas of the upper urinary tracts and urinary bladder [35]. In addition, IGFBPs as biomarkers were also used for many other diseases besides cancer. For instance, circulating IGFBP3 was used for the prognosis of liver cirrhosis [36]. IGFBP7 was identified as a clinical biomarker for patients with dyspnea [37].
For glioma, IGFBP2 has been proposed as one of the critical preoperative diagnostic and prognostic biomarkers for GBM [38]. Patients with GBM who had higher levels of IGFBP2 expression had worse survival outcomes [14]. These previous papers were consistent with this study that IGFBP2 was one of the most striking biomarkers among all IGFBPs in general glioma. However, most of the previous studies on IGFBPs in cancer reported the use of one certain IGFBP separately. In this study, results revealed that IGFBPs were coexpressed with each other at different levels. Therefore, their diagnostic and prognostic indications might not be independent. To figure out this issue, this study applied a comprehensive prognostic model that considered their independent impacts and coexpressions. LASSO regression is widely applied to establish prognostic models for cancer. For example, a study used it to train a prognostic model of ferroptosis-related gene signatures for patients with hepatocellular carcinoma [39]. This method can reduce the estimation variance while providing an interpretable final model [40]. In the present study, the survival prediction models of IGFBPs for glioma patients were constructed with TCGA data as a training cohort and subsequently validated using three independent validation cohorts. Since the training and validation include a substantial number of clinical glioma patients and the models showed very high levels of confidence, this study concluded that using IGFBPs for glioma prognosis was reliable and practicable.
As for the regulation of IGFBPs expression in glioma, this study did studies or predictions in methylation, miRNA, and transcriptional factors. The methylation levels of IGFBPs in glioma were found negatively associated with the expression of IGFBPs. This indicated that methylation might be the major regulation of IGFBP expression. For the miRNA and transcriptional factors, this study only predicted candidates for future studies. But, these predictions were limited as they were mostly based on all tissues not specific for cancer or glioma.
The identification of glioma cell subtypes expressing IGFBPs sheds light on the regulation of IGFBPs. The four glioma cell sublines are differentially expressed during five different phases of malignant glioma development. A previous study has divided malignant glioma development into 4 phases, including T0, T1, T2, End phase, and EndSVZ phase according to single-cell molecular characteristics [41]. Their data suggested that glioma at the T0 phase highly expressed MES-like cells; glioma at the T1 phase highly expressed NPC-like cells; glioma at the T2 phase highly expressed AC-like cells; glioma at the End phase and the EndSVZ phase highly expressed OPC-like cells [41]. IGFBP2 was relatively evenly expressed in OPC-like, AC-like, MES-like, and NPC-like malignant cells, indicating that IGFBP2 might function widely in glioma. IGFBP3 was only highly expressed in MES-like malignant cells, indicating that it might function in the early stage of glioma. IGFBP5 and IGFBP7 were highly expressed in AC-like malignant cells, inferring that IGFBP5 and IGFBP7 might be involved in the regulation of later stages of glioma development.
IGFBPs have been suggested to regulate immune cells associated with cancer. IGFBP2 has been found to drive tumor growth in pancreatic ductal adenocarcinoma by shifting the polarization of macrophages. [42]. This study indicated that IGFBPs could serve as an indicator of glioma immunity. Results indicated that all the IGFBPs were highly associated with the infiltration of immune cells and the expression of immune molecules. Recently, immunotherapy has been developed as a form of cancer treatment and has been met with favorable results [43]. Immunotherapy, such as the blockade of programmed cell death protein 1 (PD-1), is an effective way of treating multiple cancer types by immune checkpoint blockade (ICB) [44-47]. The results of ICB therapy can differ depending on the individual patient [47]. The identification of differences in the response of different genomic subtypes to ICB treatment is a crucial challenge that needs to be addressed. Research has shown that IGFBP2, one of the IGFBPs, can regulate PD-L1 expression by activating the epidermal growth factor receptor-signal transducer and activator of the transcription 3 signaling pathway in malignant melanoma [48]. This study showed that IGFBPs could be used as an indicator of the glioma immune microenvironment, which is in line with previous research. The results of the ICB analysis indicated that gliomas with high levels of IGFBPs were more likely to be responsive to ICB therapy. In addition, IGFBP3, IGFBP5, and IGFBP7 might also be predictive biomarkers for chemotherapy. In fact, the effects of IGFBPs on cancer therapy were reported in previous literature. The Erk pathway was said to be activated by IGFBP-1 expression, leading to Tamoxifen resistance in breast cancer cells [49]. Another study reported that the overexpression of IGFBP5 promoted radiosensitivity in prostate cancer through the PI3K-AKT pathway [50].
The most surprising finding in this study was that IGFBPs were negatively correlated with glioma stemness. Individual IGFBPs have been shown to have IGF-independent actions [5]. The explanation of the negative correlation of IGFBPs and glioma stemness might be that, as growth factor binding proteins, IGFBPs bind to growth factors and thereby inhibit cancer stem cell proliferation. A prior study had demonstrated that eliminating IGFBP7 could foster the development of hepatocellular carcinoma, which is in agreement with our result [51]. Further studies are needed to determine the functional role of IGFBPs in glioma, as the functional predictions in this study were only speculative.
Conclusion
IGFBPs are diagnostic, prognostic, and therapeutic biomarkers for glioma.
Acknowledgements
The author thanks the support of Weifen Chen, Zongxiong Liu, and Yaqi Yang.
Disclosure of conflict of interest
None.
Abbreviations
- IGFBP
Insulin-Like Growth Factor Binding Protein
- IGFBPs
IGFBP1, IGFBP2, IGFBP3, IGFBP4, IGFBP5, IGFBP6, and IGFBP7
- LGG
low-grade glioma
- GBM
glioblastoma multiforme
- VGSCs
voltage-gated sodium channels
- ICB
immune checkpoint blockade
- TCGA
The Cancer Genome Atlas
- GTEx
Genotype-Tissue Expression
- CGGA
Chinese Glioma Genome Atlas
- TISCH
Tumor Immune Single-cell Hub
- CancerSEA
Cancer single cell states atlas
- ROC
receiver operating characteristic
- PRS
primary and recurrent status
- AUC
area under the ROC curve
- DEGs
differentially expressed genes
- CTL
cytotoxic T lymphocytes
- TIDE
Tumor Immune Dysfunction and Exclusion
- NPC
neural progenitor cell
- OPC
oligodendrocyte-progenitor
- AC
astrocyte
- MES
mesenchymal
Supporting Information
References
- 1.Weller M, Wick W, Aldape K, Brada M, Berger M, Pfister SM, Nishikawa R, Rosenthal M, Wen PY, Stupp R, Reifenberger G. Glioma. Nat Rev Dis Primers. 2015;1:15017. doi: 10.1038/nrdp.2015.17. [DOI] [PubMed] [Google Scholar]
- 2.Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh KM, Wrensch MR, Barnholtz-Sloan JS. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2014;16:896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claus EB, Walsh KM, Wiencke JK, Molinaro AM, Wiemels JL, Schildkraut JM, Bondy ML, Berger M, Jenkins R, Wrensch M. Survival and low-grade glioma: the emergence of genetic information. Neurosurg Focus. 2015;38:E6. doi: 10.3171/2014.10.FOCUS12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bach LA. IGF-binding proteins. J Mol Endocrinol. 2018;61:T11–T28. doi: 10.1530/JME-17-0254. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Zhang M, He T, Yang W, Wang L, Zhang L, Guo M. Epigenetic silencing of IGFBPL1 promotes esophageal cancer growth by activating PI3K-AKT signaling. Clin Epigenetics. 2020;12:22. doi: 10.1186/s13148-020-0815-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Q, Zhou Y, Ying K, Ruan W. IGFBP, a novel target of lung cancer? Clin Chim Acta. 2017;466:172–177. doi: 10.1016/j.cca.2017.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Zheng F, Tang Q, Zheng XH, Wu J, Huang H, Zhang H, Hann SS. Inactivation of Stat3 and crosstalk of miRNA155-5p and FOXO3a contribute to the induction of IGFBP1 expression by beta-elemene in human lung cancer. Exp Mol Med. 2018;50:1–14. doi: 10.1038/s12276-018-0146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JC, Ha YJ, Tak KH, Roh SA, Kim CW, Kim TW, Kim SK, Kim SY, Cho DH, Kim YS. Complex behavior of ALDH1A1 and IGFBP1 in liver metastasis from a colorectal cancer. PLoS One. 2016;11:e0155160. doi: 10.1371/journal.pone.0155160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du Y, Wang P. Upregulation of MIIP regulates human breast cancer proliferation, invasion and migration by mediated by IGFBP2. Pathol Res Pract. 2019;215:152440. doi: 10.1016/j.prp.2019.152440. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Luo XX, Tang YL, Xu JX, Zeng ZG. The prognostic values of insulin-like growth factor binding protein in breast cancer. Medicine (Baltimore) 2019;98:e15561. doi: 10.1097/MD.0000000000015561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng R, Chen W, Xia W, Zheng J, Zhou Q. The prognostic values of the insulin-like growth factor binding protein family in ovarian cancer. Biomed Res Int. 2020;2020:7658782. doi: 10.1155/2020/7658782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao Y, Zhu S, Yin W, Liu X, Hu Y. IGFBP-4 expression is adversely associated with lung cancer prognosis. Oncol Lett. 2017;14:6876–6880. doi: 10.3892/ol.2017.7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai J, Chen Q, Cui Y, Dong J, Chen M, Wu P, Jiang C. Immune heterogeneity and clinicopathologic characterization of IGFBP2 in 2447 glioma samples. Oncoimmunology. 2018;7:e1426516. doi: 10.1080/2162402X.2018.1426516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Jensen MA, Zenklusen JC. A practical guide to the cancer genome atlas (TCGA) Methods Mol Biol. 2016;1418:111–141. doi: 10.1007/978-1-4939-3578-9_6. [DOI] [PubMed] [Google Scholar]
- 16.GTEx Consortium. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Z, Zhang KN, Wang Q, Li G, Zeng F, Zhang Y, Wu F, Chai R, Wang Z, Zhang C, Zhang W, Bao Z, Jiang T. Chinese glioma genome atlas (CGGA): a comprehensive resource with functional genomic data from Chinese glioma patients. Genomics Proteomics Bioinformatics. 2021;19:1–12. doi: 10.1016/j.gpb.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 19.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menyhárt O, Fekete JT, Győrffy B. Gene expression-based biomarkers designating glioblastomas resistant to multiple treatment strategies. Carcinogenesis. 2021;42:804–813. doi: 10.1093/carcin/bgab024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun D, Wang J, Han Y, Dong X, Ge J, Zheng R, Shi X, Wang B, Li Z, Ren P, Sun L, Yan Y, Zhang P, Zhang F, Li T, Wang C. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021;49:D1420–D1430. doi: 10.1093/nar/gkaa1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan H, Yan M, Zhang G, Liu W, Deng C, Liao G, Xu L, Luo T, Yan H, Long Z, Shi A, Zhao T, Xiao Y, Li X. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019;47:D900–D908. doi: 10.1093/nar/gky939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, Richman AR, Silverbush D, Shaw ML, Hebert CM, Dewitt J, Gritsch S, Perez EM, Gonzalez Castro LN, Lan X, Druck N, Rodman C, Dionne D, Kaplan A, Bertalan MS, Small J, Pelton K, Becker S, Bonal D, Nguyen QD, Servis RL, Fung JM, Mylvaganam R, Mayr L, Gojo J, Haberler C, Geyeregger R, Czech T, Slavc I, Nahed BV, Curry WT, Carter BS, Wakimoto H, Brastianos PK, Batchelor TT, Stemmer-Rachamimov A, Martinez-Lage M, Frosch MP, Stamenkovic I, Riggi N, Rheinbay E, Monje M, Rozenblatt-Rosen O, Cahill DP, Patel AP, Hunter T, Verma IM, Ligon KL, Louis DN, Regev A, Bernstein BE, Tirosh I, Suvà ML. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178:835–849. e821. doi: 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao J, Chen AX, Gartrell RD, Silverman AM, Aparicio L, Chu T, Bordbar D, Shan D, Samanamud J, Mahajan A, Filip I, Orenbuch R, Goetz M, Yamaguchi JT, Cloney M, Horbinski C, Lukas RV, Raizer J, Rae AI, Yuan J, Canoll P, Bruce JN, Saenger YM, Sims P, Iwamoto FM, Sonabend AM, Rabadan R. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med. 2019;25:462–469. doi: 10.1038/s41591-019-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darmanis S, Sloan SA, Croote D, Mignardi M, Chernikova S, Samghababi P, Zhang Y, Neff N, Kowarsky M, Caneda C, Li G, Chang SD, Connolly ID, Li Y, Barres BA, Gephart MH, Quake SR. Single-cell RNA-seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 2017;21:1399–1410. doi: 10.1016/j.celrep.2017.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nilsson RJ, Balaj L, Hulleman E, van Rijn S, Pegtel DM, Walraven M, Widmark A, Gerritsen WR, Verheul HM, Vandertop WP, Noske DP, Skog J, Würdinger T. Blood platelets contain tumor-derived RNA biomarkers. Blood. 2011;118:3680–3683. doi: 10.1182/blood-2011-03-344408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Racle J, Gfeller D. EPIC: a tool to estimate the proportions of different cell types from bulk gene expression data. Methods Mol Biol. 2020;2120:233–248. doi: 10.1007/978-1-0716-0327-7_17. [DOI] [PubMed] [Google Scholar]
- 30.Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, Li Z, Traugh N, Bu X, Li B, Liu J, Freeman GJ, Brown MA, Wucherpfennig KW, Liu XS. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu CJ, Hu FF, Xia MX, Han L, Zhang Q, Guo AY. GSCALite: a web server for gene set cancer analysis. Bioinformatics. 2018;34:3771–3772. doi: 10.1093/bioinformatics/bty411. [DOI] [PubMed] [Google Scholar]
- 32.Liu ZP, Wu C, Miao H, Wu H. RegNetwork: an integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database (Oxford) 2015;2015:bav095. doi: 10.1093/database/bav095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lian H, Han YP, Zhang YC, Zhao Y, Yan S, Li QF, Wang BC, Wang JJ, Meng W, Yang J, Wang QH, Mao WW, Ma J. Integrative analysis of gene expression and DNA methylation through one-class logistic regression machine learning identifies stemness features in medulloblastoma. Mol Oncol. 2019;13:2227–2245. doi: 10.1002/1878-0261.12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mita K, Zhang Z, Ando Y, Toyama T, Hamaguchi M, Kobayashi S, Hayashi S, Fujii Y, Iwase H, Yamashita H. Prognostic significance of insulin-like growth factor binding protein (IGFBP)-4 and IGFBP-5 expression in breast cancer. Jpn J Clin Oncol. 2007;37:575–582. doi: 10.1093/jjco/hym066. [DOI] [PubMed] [Google Scholar]
- 35.Liang PI, Wang YH, Wu TF, Wu WR, Liao AC, Shen KH, Hsing CH, Shiue YL, Huang HY, Hsu HP, Chen LT, Lin CY, Tai C, Wu JY, Li CF. IGFBP-5 overexpression as a poor prognostic factor in patients with urothelial carcinomas of upper urinary tracts and urinary bladder. J Clin Pathol. 2013;66:573–582. doi: 10.1136/jclinpath-2012-201278. [DOI] [PubMed] [Google Scholar]
- 36.Correa CG, Colombo Bda S, Ronsoni MF, Soares E Silva PE, Fayad L, Silva TE, Wildner LM, Bazzo ML, Dantas-Correa EB, Narciso-Schiavon JL, Schiavon Lde L. Circulating insulin-like growth factor-binding protein 3 as prognostic biomarker in liver cirrhosis. World J Hepatol. 2016;8:739–748. doi: 10.4254/wjh.v8.i17.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ibrahim NE, Afilalo M, Chen-Tournoux A, Christenson RH, Gaggin HK, Hollander JE, Kastner P, Levy PD, Mang A, Masson S, Nagurney JT, Nowak RM, Pang PS, Peacock WF, Dipl-Stat VR, Walters EL, Januzzi JL Jr. Diagnostic and prognostic utilities of insulin-like growth factor binding protein-7 in patients with dyspnea. JACC Heart Fail. 2020;8:415–422. doi: 10.1016/j.jchf.2020.02.009. [DOI] [PubMed] [Google Scholar]
- 38.Gállego Pérez-Larraya J, Paris S, Idbaih A, Dehais C, Laigle-Donadey F, Navarro S, Capelle L, Mokhtari K, Marie Y, Sanson M, Hoang-Xuan K, Delattre JY, Mallet A. Diagnostic and prognostic value of preoperative combined GFAP, IGFBP-2, and YKL-40 plasma levels in patients with glioblastoma. Cancer. 2014;120:3972–3980. doi: 10.1002/cncr.28949. [DOI] [PubMed] [Google Scholar]
- 39.Liang JY, Wang DS, Lin HC, Chen XX, Yang H, Zheng Y, Li YH. A novel ferroptosis-related gene signature for overall survival prediction in patients with hepatocellular carcinoma. Int J Biol Sci. 2020;16:2430–2441. doi: 10.7150/ijbs.45050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tibshirani R. The lasso method for variable selection in the cox model. Stat Med. 1997;16:385–395. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, Zhou R, Xiong Y, Zhou L, Yan X, Wang M, Li F, Xie C, Zhang Y, Huang Z, Ding C, Shi K, Li W, Liu Y, Cao Z, Zhang ZN, Zhou S, Chen C, Zhang Y, Chen L, Wang Y. Sequential fate-switches in stem-like cells drive the tumorigenic trajectory from human neural stem cells to malignant glioma. Cell Res. 2021;31:684–702. doi: 10.1038/s41422-020-00451-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun L, Zhang X, Song Q, Liu L, Forbes E, Tian W, Zhang Z, Kang Y, Wang H, Fleming JB, Pasche BC, Zhang W. IGFBP2 promotes tumor progression by inducing alternative polarization of macrophages in pancreatic ductal adenocarcinoma through the STAT3 pathway. Cancer Lett. 2021;500:132–146. doi: 10.1016/j.canlet.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baxevanis CN, Perez SA, Papamichail M. Cancer immunotherapy. Crit Rev Clin Lab Sci. 2009;46:167–189. doi: 10.1080/10408360902937809. [DOI] [PubMed] [Google Scholar]
- 44.Xia L, Liu Y, Wang Y. PD-1/PD-L1 blockade therapy in advanced non-small-cell lung cancer: current status and future directions. Oncologist. 2019;24(Suppl 1):S31–S41. doi: 10.1634/theoncologist.2019-IO-S1-s05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahoney KM, Freeman GJ, McDermott DF. The next immune-checkpoint inhibitors: PD-1/PD-L1 blockade in melanoma. Clin Ther. 2015;37:764–782. doi: 10.1016/j.clinthera.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Li G. PD-1/PD-L1 blockade in cervical cancer: current studies and perspectives. Front Med. 2019;13:438–450. doi: 10.1007/s11684-018-0674-4. [DOI] [PubMed] [Google Scholar]
- 47.Wang X, Guo G, Guan H, Yu Y, Lu J, Yu J. Challenges and potential of PD-1/PD-L1 checkpoint blockade immunotherapy for glioblastoma. J Exp Clin Cancer Res. 2019;38:87. doi: 10.1186/s13046-019-1085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li T, Zhang C, Zhao G, Zhang X, Hao M, Hassan S, Zhang M, Zheng H, Yang D, Liu L, Mehraein-Ghomi F, Bai X, Chen K, Zhang W, Yang J. IGFBP2 regulates PD-L1 expression by activating the EGFR-STAT3 signaling pathway in malignant melanoma. Cancer Lett. 2020;477:19–30. doi: 10.1016/j.canlet.2020.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng Y, Sowers JY, Houston KD. IGFBP-1 expression promotes tamoxifen resistance in breast cancer cells via erk pathway activation. Front Endocrinol (Lausanne) 2020;11:233. doi: 10.3389/fendo.2020.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen X, Yu Q, Pan H, Li P, Wang X, Fu S. Overexpression of IGFBP5 enhances radiosensitivity through pi3k-akt pathway in prostate cancer. Cancer Manag Res. 2020;12:5409–5418. doi: 10.2147/CMAR.S257701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akiel M, Rajasekaran D, Gredler R, Siddiq A, Srivastava J, Robertson C, Jariwala NH, Fisher PB, Sarkar D. Emerging role of insulin-like growth factor-binding protein 7 in hepatocellular carcinoma. J Hepatocell Carcinoma. 2014;1:9–19. doi: 10.2147/JHC.S44460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.