Abstract
Adult T‐cell leukemia/lymphoma (ATL) patients have a very poor prognosis. The humanized anti‐CCR4 therapeutic monoclonal antibody, mogamulizumab, is a key agent for ATL treatment. Our previous integrated molecular analysis demonstrated that among all the driver genes in ATL, CCR7 gene alterations were significantly associated with clinical response to mogamulizumab. Accordingly, here we investigated the detailed clinical impact of CCR7 alterations in a larger cohort of ATL patients. These CCR7 alterations, most of which lead to C‐terminus truncations, were observed in 27 of 223 patients (12%). For patients receiving mogamulizumab but not allogeneic hematopoietic stem cell transplantation (HSCT), CCR7 alterations were significantly associated with worse survival (median survival from the first dose of mogamulizumab of 0.7 years for 12 patients with CCR7 alterations vs. 1.6 years for 72 patients without, p = 0.020). On the other hand, the presence or absence of CCR7 alterations had no significant impact on survival in the entire cohort (median overall survival of 1.4 and 1.8 years, respectively, p = 0.901), or on the survival of patients receiving allogeneic HSCT (median survival from the day of transplantation of 0.9 years for 6 patients with CCR7 alterations and 1.4 years for 48 without, p = 0.543). Multivariate analysis indicated that patients with CCR4 alterations but lacking CCR7 alterations (n = 20) had significantly better survival after receiving mogamulizumab‐containing treatments (hazard ratio for survival, 0.437, 95% confidence interval, 0.192–0.994). This study contributes to the establishment of precision medicine for ATL.
Keywords: adult T‐cell leukemia/lymphoma, CCR4, CCR7, mogamulizumab
1. INTRODUCTION
Adult T‐cell leukemia/lymphoma (ATL) is a peripheral T‐cell neoplasm caused by human T‐cell lymphotropic virus type‐1. It has a very poor prognosis. 1 , 2 Allogeneic hematopoietic stem cell transplantation (HSCT) is generally considered to be the only curative treatment for ATL. Therefore, younger patients (≤70 years of age) and those with relatively well‐controlled ATL are recommended to receive this treatment, aiming for long‐term survival. 3 , 4 , 5 , 6 However, treatment‐related adverse events associated with allogeneic HSCT are generally severe compared to other treatments. In addition, many Japanese ATL patients are older (median age at diagnosis, 68 years). 7 Accordingly, the proportion of ATL patients who are candidates for allogeneic HSCT due to their age is decreasing year by year.
The humanized anti‐CCR4 monoclonal antibody mogamulizumab has a defucosylated Fc region, which enhances antibody‐dependent cellular cytotoxicity (ADCC). 8 , 9 , 10 It is approved in Japan for patients with newly diagnosed or relapsed/refractory ATL, 11 , 12 and offers clinical benefit for patients with this disease. 13 With respect to the relationship with allogeneic HSCT, pre‐transplantation treatment with mogamulizumab within approximately 2 months should be avoided, because it presumably result in increased severity and refractoriness of graft‐versus‐host disease after transplantation. 14 , 15 On the other hand, currently, most patients deemed unsuitable for allogeneic HSCT receive mogamulizumab‐containing treatment as first‐line therapy. 15 , 16 , 17 Accordingly, it is currently a key agent for ATL treatment. In this context, an integrated molecular analysis of ATL was conducted to identify genomic biomarkers of this antibody therapy. Among all the driver genes in ATL, only 2 alterations in the CCR4 and CCR7 genes were significantly associated with clinical response to mogamulizumab. Thus, the complete response rate of patients with CCR7 alterations was 29% (2/7) compared to 72% (40/56) of those without (p = 0.036). 18 This prompted us to focus on the clinical significance of CCR7 alterations, and here we report an investigation on a larger cohort of ATL patients.
2. METHODS
2.1. ATL patients
The present study included 223 ATL patients. The diagnosis and assignment of clinical subtypes of ATL was conducted according to the criteria recommended by the Japan Lymphoma Study Group. 2 The present study was approved by the Institutional Ethics Committees of Nagoya City University Graduate School of Medical Sciences (Nagoya, Japan), Imamura General Hospital (Kagoshima, Japan), Oita Prefectural Hospital (Oita, Japan), Fukuoka University (Fukuoka, Japan), National Hospital Organization Kyushu Medical Center (Fukuoka, Japan), and Nagoya University Graduate School of Medicine (Nagoya, Japan).
2.2. Nucleic acid extraction
We used AllPrep DNA/RNA from formalin‐fixed paraffin‐embedded (FFPE) Kits (80234, QIAGEN Inc., Germantown, MD) to extract genomic DNA and total RNA FFPE tissues in the 202 patients diagnosed with ATL by histopathology. In the remaining 21 ATL patients, AllPrep DNA/RNA Mini Kits (80204, QIAGEN Inc.) were used to extract genomic DNA and total RNA from peripheral blood mononuclear cells (PBMC) containing >30% abnormal lymphocytes.
2.3. Detection of CCR7 single nucleotide variants/insertion‐deletions by targeted next‐generation sequencing
DNA fragments encompassing codons 338–364 of CCR7 were amplified from genomic DNA using the primers CCR7‐F (5’‐GGCGTCAAGTTCCGCAAC‐3’) and CCR7‐R (5’‐GGCCTCCACACTCATGGA‐3’). The amplicons were then purified using Agencourt AMPure XP (Beckman Coulter Inc., Brea, CA) and barcoded using a Nextera XT Index Kit (Illumina Inc.). Subsequently, the library was re‐purified, quantified, and checked for purity using the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). An iSeq instrument (Illumina Inc.) was used for sequencing. All of these procedures were carried out according to Illumina's recommended methods (http://jp.support.illumina.com).
2.4. Detection of TP53, CD28, and CCR4 alterations
TP53 alterations including single nucleotide variants (SNVs)/Indels and copy number variations (CNVs) were analyzed, as previously described. 19 CD28 alterations including CD28‐related fusions, SNVs/Indels, and CNVs were also analyzed as previously described. 20 CCR4 SNVs/indels were analyzed by targeted next‐generation sequencing in samples from 46 patients. DNA fragments encompassing codons 322–348 of CCR4 were amplified from genomic DNA using primers CCR4‐F (5’‐CAAGTACATCCTACAGCTCTTC‐3’) and CCR4‐R (5’‐CATGATCCATGGTGGACTG‐3’). The subsequent procedures were the same as used for CCR7, as described above. For the remaining samples from 177 patients, CCR4 SNVs/indels in codons 322–348 were detected, as previously described. 21
2.5. Statistical analysis
Differences between two groups were examined with the Mann–Whitney U‐test or Fisher's exact test. Univariate and multivariate Cox proportional hazards regression models were used to identify prognostically relevant variables. The probability of survival was estimated by the Kaplan–Meier method, and survival times were compared using the log‐rank test. Overall survival (OS) was measured from the day when the tumor sample was obtained to death resulting from any cause. To evaluate the impact of allogeneic HSCT, survival was measured from the day of transplantation. When evaluating the impact of mogamulizumab‐containing treatment, survival was measured from the day of the first dose of antibody. All analyses were performed with SPSS Statistics 25 (IBM Corporation, Armonk, NY). In this study, p < 0.050 (two‐sided) was considered statistically significant.
3. RESULTS
3.1. Clinical characteristics of the ATL patients
The ATL patients enrolled in this study included 115 men and 108 women (age range, 36–90 years; median, 66 years) (Table 1). Tumor samples were obtained from each patient at the time of initial presentation at the participating hospital, and the clinical characteristics including clinical subtypes were recorded at that time. Treatments administered to these ATL patients varied because they were determined at the discretion of each clinical investigator.
TABLE 1.
Characteristics of Adult T‐cell leukemia/lymphoma (ATL) patients according to CCR7 alterations
| Characteristics | CCR7 alterations | ||
|---|---|---|---|
| Absent | Present | p value | |
| Number (%) | 196 (88) | 27 (12) | |
| Sex | 0.063 | ||
| Female | 90 (46) | 18 (67) | |
| Male | 106 (54) | 9 (33) | |
| Clinical subtype | 1.000 | ||
| Chronic, smoldering | 24 (12) | 3 (11) | |
| Acute, lymphoma | 172 (88) | 24 (89) | |
| ECOG PS* | 0.816 | ||
| 0, 1 | 144 (74) | 19 (70) | |
| 2, 3, 4 | 51 (26) | 8 (30) | |
| Serum sIL‐2R (U/ml)** | 1.000 | ||
| <20,000 | 111 (59) | 16 (62) | |
| >20,000 | 77 (41) | 10 (38) | |
| Serum Ca (mg/dl)***$ | 0.528 | ||
| <11.0 | 168 (88) | 21 (84) | |
| >11.0 | 23 (12) | 4 (16) | |
| Serum Albmin (g/dl)**** | 0.823 | ||
| >3.5 | 131 (69) | 17 (65) | |
| <3.5 | 60 (31) | 9 (35) | |
| Age (years) | 0.582 | ||
| Mean | 65 | 68 | |
| Median | 64 | 67 | |
| Range | 41–90 | 41–85 | |
| WBC (/ul)***** | 0.712 | ||
| Mean | 13,916 | 18,260 | |
| Median | 8185 | 7760 | |
| Range | 2500‐165,800 | 2800‐232,100 | |
| Hb (g/dl)***** | 0.412 | ||
| Mean | 12.9 | 12.6 | |
| Median | 13.2 | 13.1 | |
| Range | 6.0–17.1 | 2.9–15.0 | |
| Plt (×103/μl)***** | 0.445 | ||
| Mean | 228 | 221 | |
| Median | 222 | 190 | |
| Range | 4–622 | 111–444 | |
| CCR4 alterations | 0.660 | ||
| Absent | 134 (68) | 20 (74) | |
| Present | 62 (32) | 7 (26) | |
| CD28 alterations | 0.832 | ||
| Absent | 122 (62) | 18 (67) | |
| Present | 74 (38) | 9 (33) | |
| TP53 alterations | 0.059 | ||
| Absent | 120 (61) | 11 (41) | |
| Present | 76 (39) | 16 (59) | |
Abbreviations: ATL, adult T‐cell leukemia/lymphoma; Alb, albumin; Ca, calcium; CCR4, CC chemokine receptor 4; ECOG, Eastern Cooperative oncology Group; Hb, hemoglobin; PS, performance status; Plt, platelet count; sIL‐2R, soluble interleukin‐2 receptor; WBC, white blood cell count; $When serum Alb level was less than 4.0 g/dl, serum Ca was adjusted by the concentration of serum Alb as follows: adjusted Ca level (mg/dl) = measured Ca level (mg/dl) + [4‐Alb level (g/dl)]; *A patient's data was unknown; **Nine patients' data were unknown; ***Seven patients' data were unknown; ****Six patients' data were unknown; *****Five patients' data were unknown.
3.2. CCR7 gene alterations in ATL patients
CCR7 alterations, comprising one C346Y, three Q349*, two E350*, one E350 fs, four Q351*, four Q354*, nine W355*, one S357fs, and two R363 C CCR7 SNVs/indels, were detected in 27 of 223 patients (12%) (Figure 1).
FIGURE 1.
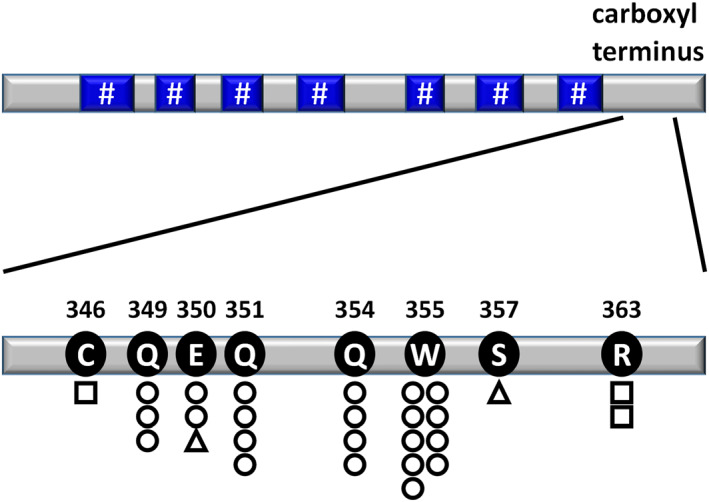
Positions, types, and frequencies of somatic alterations of CCR7 in Adult T‐cell leukemia/lymphoma (ATL) patients. DNA fragments encompassing codons 338–364 of CCR7 were amplified from genomic DNA. Circles, squares, and triangles indicate nonsense, missense, and frameshift single nucleotide variants (SNVs)/indels, respectively. The number of circles, squares, or triangles indicates the number of patients with alterations among the 223 ATL cases. A hash‐tag indicates a transmembrane domain of CCR7. CCR7 amino acid alterations were determined by reference to the NCBI protein sequence NP_001829.1
3.3. TP53, CD28 and CCR4 alterations in ATL patients
Sixty SNVs/indels of TP53 were identified in 49 ATL patients (22%), and six patients were found to harbor more than one of these. TP53 CNVs, such as homozygous and heterozygous deletions, were observed in 17 and 39 patients (8% and 17%), respectively, and thirteen patients harbored both TP53 SNVs/indels and CNVs. Collectively, TP53 alterations were observed in 92 patients (41%) (data not shown).
CD28 alterations were found in 83 patients (37%), including 35 CD28‐related fusions (5 CTLA4‐CD28 and 30 ICOS‐CD28), three activating SNVs (F51I, D124V, and D124E), and 53 CNVs (34 gains and 19 amplifications). Five patients simultaneously had a CD28‐related fusion and CNVs, two harbored two different types of CD28‐related fusions, and one had a CD28‐related fusion and SNV (data not shown).
CCR4 alterations were detected in 69 patients (31%), including one T321fs, four R323fs, two F326fs, 17 C329*, one C329fs, five Q330*, one Q330fs, 22 Y331*, four Y331fs, 8 Q336*, one I337fs, and three S345fs CCR4 SNVs/indels (data not shown).
3.4. Characteristics of ATL patients stratified by CCR7 alterations
Female patients tended to be more likely to harbor CCR7 alterations (18/108 [17%] vs. male, 9/115 [8%], p = 0.063). There were no significant differences between patients with or without CCR7 alterations in the other clinical characteristics recorded, such as clinical subtype (chronic, smoldering or acute, lymphoma), Eastern Cooperative Oncology Group (ECOG) performance status (PS), (0,1 vs. 2,3,4), serum soluble interleukin‐2 receptor (sIL‐2R) (≤20,000 U/ml vs. > 20,000 U/ml), serum adjusted calcium (Ca) (≤11.0 vs. > 11.0 mg/dl), serum albumin (≥3.5 vs. < 3.5 g/dl), age (years), white blood cell count (/uL), hemoglobin level (g/dl), or platelet count (x 103/ul). Patients with CCR7 alterations tended to be more likely to harbor TP53 alterations (16/27 [59%] vs. 76/196 [39%], respectively, p = 0.059). However, there were no significant differences between patients with or without CCR7 alterations in terms of the presence of CCR4 alterations (7/27 [26%] vs. 62/196 [32%], respectively, p = 0.660) or CD28 alterations (9/27 [33%] vs. 74/196 [38%], respectively, p = 0.832) (Table 1).
3.5. OS of ATL patients stratified by CCR7 alterations
The median OS of all patients enrolled in the present study was 1.7 years (Figure 2A). Patients aged >70 years had a significantly shorter OS than those aged ≤70 years (median OS, 1.2 vs. 1.9 years, respectively, p = 0.036) (data not shown). The OS of patients with an acute or lymphoma subtype was significantly shorter than of those with a chronic or smoldering subtype (median OS, 1.4 years vs. not reached [NR], respectively, p < 0.001) (data not shown). The OS of patients with TP53 alterations was significantly shorter than of those without (median OS, 0.9 years vs. Not reached, respectively, p < 0.001). The OS of patients with CD28 alterations was significantly shorter than of those without (median OS, 1.0 vs. 2.2 years, respectively, p = 0.010) (data not shown). Finally, there were no significant differences in OS between patients with and without CCR4 alterations (median OS, 1.4 vs. 1.8 years, respectively, p = 0.878) (data not shown), or CCR7 alterations (median OS, 1.4 vs. 1.8 years, respectively, p = 0.901) (Figure 2B).
FIGURE 2.
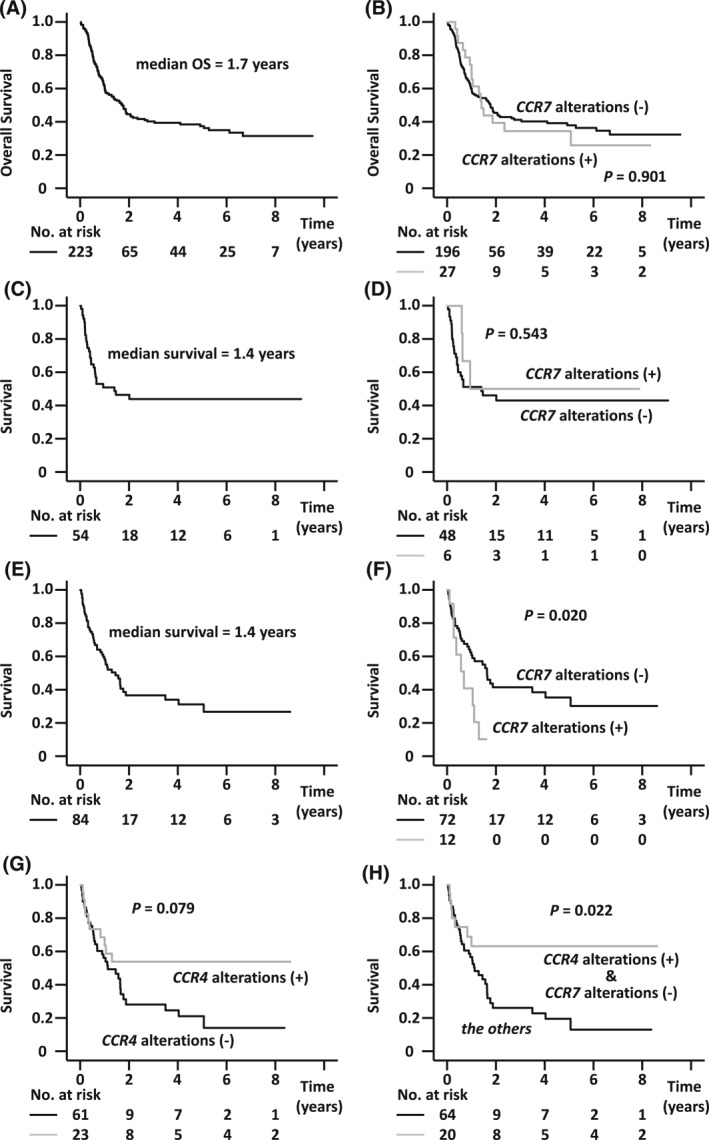
Survival of Adult T‐cell leukemia/lymphoma (ATL) patients stratified according to CCR7 gene alterations. (A) Overall survival (OS) of all ATL patients enrolled in the study (n = 223). The median OS was 1.8 years (95% confidence interval [CI], 1.3–2.1 years). (B) OS according to CCR7 alterations (with CCR7 alterations [+] compared with no alterations [−]; hazard ratio [HR], 1.034, 95% CI, 0.610–1.752). (C) Survival of ATL patients from the day of allogeneic hematopoietic stem cell transplantation (HSCT) (n = 54). The median survival was 1.4 years (95% CI, 0.0–2.9 years). (D) Survival from the day of allogeneic HSCT according to CCR7 alterations (with CCR7 alterations [+] compared to those without [−]; HR, 0.691, 95% CI, 0.208–2.293). (E) Survival of ATL patients receiving mogamulizumab but not allogeneic HSCT (n = 84). The median survival from the first dose of mogamulizumab was 1.4 years (95% CI, 0.8–2.1 years). (F) Survival from the first dose of mogamulizumab according to CCR7 alterations (with CCR7 alterations [+] vs. without [‐]; HR, 2.372, 95% CI, 1.119–5.027). (G) Survival from the first dose of mogamulizumab according to CCR4 alterations (with CCR4 alterations [+] compared to without [−]; HR, 0.538, 95% CI, 0.266–1.087). (H) Survival from the first dose of mogamulizumab according to CCR4 and CCR7 alterations (with CCR4 alterations [+] but without CCR7 alterations [−] compared with the others; HR, 0.400, 95% CI, 0.178–0.901)
3.6. Survival of ATL patients receiving allogeneic HSCT stratified by CCR7 alterations
The median survival from the day of allogeneic HSCT of all 54 transplanted patients was 1.4 years (Figure 2C). The median survival from the day of transplantation of patients with or without TP53 alterations was 0.4 years or NR, respectively (p = 0.002) (data not shown). In contrast, there were no significant differences in survival between patients with or without alterations of CD28 (median survival, 0.6 vs. 2.0 years, respectively, p = 0.447), CCR4 (median survival, respectively, 0.6 vs. 1.5 years, p = 0.952) (data not shown), or CCR7 alterations (median survival, 0.9 vs. 1.4 years, respectively, p = 0.543) (Figure 2D).
3.7. Survival of ATL patients receiving mogamulizumab but not allogeneic HSCT, stratified by CCR7 alterations
The median survival from the day of the first dose of mogamulizumab of 84 patients who did not receive allogeneic HSCT was 1.4 years (Figure 2E). There was a trend toward worse survival in patients with a higher age (>70 years vs. ≤ 70 years) at the first dose of mogamulizumab (median survival, 0.8 vs. 1.6 years, respectively, p = 0.095) (data not shown). There was no significant difference in survival between patients with an acute or lymphoma subtype and those with a chronic or smoldering subtype (median survival, 1.1 vs. 3.5 years, respectively, p = 0.179) (data not shown). In contrast, the difference in median survival from the first dose of antibody of patients with or without TP53 alterations was highly significant, at 0.7 versus 5.1 years, respectively (p < 0.001). The presence of CD28 alterations also had a significant negative impact, at 0.6 versus 1.6 years, respectively (p = 0.019) (data not shown). Survival from the first dose of antibody of patients with CCR7 alterations was also significantly worse than of those without (median survival, 0.7 vs. 1.6 years, respectively, p = 0.020) (Figure 2F). On the other hand, there was a trend toward better survival from the first dose of antibody in patients with CCR4 alterations relative to those without (median survival, NR vs. 1.1 years, respectively, p = 0.079) (Figure 2G). Finally, survival from the first dose of mogamulizumab was analyzed in patients stratified by both CCR4 and CCR7 alterations together. Thus, median survival in patients with CCR4 alterations but lacking CCR7 alterations was NR, which was significantly better than in the other patients (median survival, 1.1 years, p = 0.022) (Figure 2H).
3.8. Multivariate analysis of survival from the first dose of mogamulizumab
Multivariate analysis was performed using the following 5 variables: age at the first dose of mogamulizumab, clinical subtype, TP53 alterations, CD28 alterations, and a combination of CCR4 and CCR7 alterations. Of these, two variables including TP53 alterations, and presence of CCR4 alterations in the absence of CCR7 alterations, were significantly associated with survival (hazard ratio [HR], 2.685; 95% confidence interval [CI], 1.427–5.050, and HR, 0.437; 95% CI, 0.192–0.994, respectively) (Table 2).
TABLE 2.
Multivariate analysis for survival from the first dose of mogamulizumab
| Variables | Number | Hazard ratio | (95% CI) | p value |
|---|---|---|---|---|
| Age a , years | ||||
| <70 | 54 | 1.000 | ‐ | Reference |
| >70 | 30 | 1.837 | (0.979–3.447) | 0.058 |
| Clinical subtype | ||||
| Chronic, smoldering | 8 | 1.000 | ‐ | Reference |
| Acute, lymphoma | 76 | 2.457 | (0.742–8.133) | 0.141 |
| TP53 alterations | ||||
| Absent | 49 | 1.000 | ‐ | Reference |
| Present | 35 | 2.685 | (1.427–5.050) | 0.002 |
| CD28 alterations | ||||
| Absent | 51 | 1.000 | ‐ | Reference |
| Present | 33 | 1.820 | (0.959–3.456) | 0.067 |
| Both CCR4 alterations (+) and CCR7 alterations (−) | ||||
| No | 64 | 1.000 | ‐ | Reference |
| Yes | 20 | 0.437 | (0.192–0.994) | 0.048 |
Abbreviation: CI, confidence interval.
age at the first dose of mogamulizumab.
4. DISCUSSION
CCR7, a 7 transmembrane G‐protein‐coupled receptor similar to CCR4, is commonly expressed on ATL cells, and interactions with its ligands including CCL19 or CCL21 are important for tumor cell infiltration into or for residence in the lymphoid tissues. 22 , 23 CCR7 alterations, most of which were SNVs/indels leading to C‐terminus truncations, were observed in these patients. Such CCR7 alterations had gain‐of‐function effects leading to enhanced downstream signaling, 24 , 25 and have also been observed in other mature T‐cell neoplasms. 26
Analyzing the entire cohort of ATL patients examined here, those with CCR7 alterations did not appear to possess any specific clinical characteristics differentiating them from those without any alterations. However, there was a trend for patients with CCR7 alterations to be more likely to harbor TP53 alterations as well, which might be associated with the contribution of TP53 to genomic stability. 27 , 28 , 29 Regarding clinical outcomes, CCR7 alterations did not have a significant impact on OS, in contrast to TP53 or CD28 alterations, but similar to CCR4 alterations.
In general, because prognostic factors vary according to the treatment strategy, even in the same disease, we investigated the prognostic significance of CCR7 alterations in ATL patients stratified according to their treatment modality. CCR7 alterations, in the cohort of patients receiving allogeneic HSCT, did not have a significant impact on survival from the date of transplantation, in contrast to TP53 alterations, but similar to CD28 or CCR4 alterations. Next, we evaluated the impact of CCR7 alterations in patients receiving mogamulizumab but not allogeneic HSCT. The analysis focusing on this population is clinically important, in the context of the relationship between mogamulizumab and allogeneic HSCT, as mentioned in the Introduction and elsewhere. 3 , 4 , 14 , 15 In the present study, univariate analysis indicated that CCR7 alterations were significantly associated with a worse outcome in patients receiving mogamulizumab‐containing treatments. This finding is consistent with our earlier study identifying the relationship between CCR7 alterations and the clinical response to mogamulizumab. 18 We previously reported that the efficacy of mogamulizumab was lower in nodal ATL lesions compared to blood or skin ATL disease. 11 In this context, the enhanced capacity of ATL cells to remain resident in lymphoid tissues as a result of CCR7 alterations might contribute to refractoriness to mogamulizumab. However, mechanisms related to this phenomenon have not yet been clarified and further investigations are warranted. Regarding CCR4 alterations, findings in the present study were similar to those of previous reports indicating that they were associated with better survival in patients receiving mogamulizumab‐containing treatments. 18 , 21 When CCR4 and CCR7 alterations were assessed together, multivariate analysis indicated that the presence of CCR4 alterations combined with a lack CCR7 alterations was an independent predictor of significantly better survival.
In conclusion, the present study documents that the presence or absence of CCR7 alterations, especially in combination with CCR4 alterations, is a valuable biomarker to predict the clinical outcome of mogamulizumab‐containing treatments in ATL. The present study contributes to the establishment of precision medicine for patients with ATL.
AUTHOR CONTRIBUTIONS
Yuma Sakamoto, Takashi Ishida and Hiroshi Inagaki designed the research. Yuma Sakamoto, Takashi Ishida, Ayako Masaki, Takayuki Murase, Eiichi Ohtsuka, Morishige Takeshita, Reiji Muto, Hiromi Iwasaki, Asahi Ito, Shigeru Kusumoto, Nobuaki Nakano, Masahito Tokunaga, Kentaro Yonekura, Yukie Tashiro, Shinsuke Iida, Atae Utsunomiya, Ryuzo Ueda, and Hiroshi Inagaki performed the experiments. Takashi Ishida, Ryuzo Ueda, and Hiroshi Inagaki analyzed and interpreted data. All authors wrote and approved the manuscript.
CONFLICT OF INTEREST
Hiromi Iwasaki received research funding from Kyowa Kirin Co., Ltd. Shigeru Kusumoto received research funding from Chugai Pharmaceutical Co., Ltd., and Daiichi Sankyo Co., Ltd., and received honoraria from Chugai Pharmaceutical Co., Ltd. and Kyowa Kirin Co., Ltd. Nobuaki Nakano received honoraria from Novartis, Takeda Pharmaceutical Co., Ltd., Chugai Pharmaceutical Co., Ltd., Celgene, Otsuka Pharmaceutical Co., Ltd., Nippon Shinyaku Co., Ltd., Kyowa Kirin Co., Ltd., and Asahi Kasei Pharma Co., Ltd., and received consulting fee from JIMRO. Kentaro Yonekura received honoraria from AbbVie, Celgene, Daiichi Sankyo Co., Ltd., Eisai, Eli Lilly Japan, Janssen Pharmaceuticals, Kaken Pharmaceutical, Kyowa Kirin Co., Ltd., Maruho, Minophagen Pharmaceutical, Novartis, Sanofi, Taiho Pharmaceutical, Torii Pharmaceutical, and UCB Japan. Shinsuke Iida received honoraria from Janssen, Celgene, Ono, Takeda, Sanofi, and Daiichi Sankyo Co., Ltd., and received research funding from Sanofi, Chugai, Ono, Takeda, Kyowa Kirin Co., Ltd., Celgene, Janssen, Bristol‐Myers Squibb, Abbie, and Glaxo‐Smith‐Klein. Atae Utsunomiya received honoraria from Bristol‐Myers and Meiji Seika Pharma Co., Ltd., and received consulting fees from JIMRO, Meiji Seika Pharma Co., Ltd. and Otsuka Medical Devices Co., Ltd. Ryuzo Ueda received research funding from Kyowa Kirin Co., Ltd., Chugai Pharmaceutical Co., Ltd., and Ono Pharmaceutical Co., Ltd. The other authors have no COI to disclose.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/hon.3072.
ACKNOWLEDGMENTS
This work was supported by Grants‐in‐Aid for Early‐Career Scientists (20K16177, and 22K15426 to Yuma Sakamoto), a Grant‐in‐Aid for Challenging Research (Exploratory) (No 21K19900 to Takashi Ishida), a Grant‐in‐Aid for Scientific Research (B) (No 22H02918 to Takashi Ishida), and Grants‐in‐Aid from the Japan Agency for Medical Research and Development (No 20ae0101074h0001, 21ae0101074h0001, and 22ae0101074h0001 to Ryuzo Ueda).
Sakamoto Y, Ishida T, Masaki A, et al. CCR7 alterations associated with inferior outcome of adult T‐cell leukemia/lymphoma under mogamulizumab treatment. Hematol Oncol. 2022;40(5):876‐884. 10.1002/hon.3072
[Correction added on 23‐Sep‐2022, after first online publication: The author ORCID IDs were missing and have been added to this version.] [Correction added on 23‐Sep‐2022, after first online publication: Figure 2 contained minor typesetting errors and has been corrected in this version.]
Contributor Information
Takashi Ishida, Email: itakashi@med.nagoya-u.ac.jp.
Hiroshi Inagaki, Email: hinagaki@med.nagoya-cu.ac.jp.
DATA AVAILABILITY STATEMENT
The data that support these findings are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Uchiyama T, Yodoi J, Sagawa K, Takatsuki K, Uchino H. Adult T‐cell leukemia: clinical and hematologic features of 16 cases. Blood. 1977;50(3):481‐492. 10.1182/blood.v50.3.481.481 [DOI] [PubMed] [Google Scholar]
- 2. Shimoyama M. Diagnostic criteria and classification of clinical subtypes of adult T‐cell leukemia‐lymphoma. A report from the Lymphoma Study Group (1984‐1987). Br J Haematol. 1991;79(3):428‐437. 10.1111/j.1365-2141.1991.tb08051.x [DOI] [PubMed] [Google Scholar]
- 3. Cook LB, Fuji S, Hermine O, et al. Revised adult T‐cell leukemia‐lymphoma international consensus meeting report. J Clin Oncol. 2019;37(8):677‐687. 10.1200/jco.18.00501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Utsunomiya A. Progress in allogeneic hematopoietic cell transplantation in adult T‐cell leukemia‐lymphoma. Front Microbiol. 2019;10:2235. 10.3389/fmicb.2019.02235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ishida T, Hishizawa M, Kato K, et al. Allogeneic hematopoietic stem cell transplantation for adult T‐cell leukemia‐lymphoma with special emphasis on preconditioning regimen: a nationwide retrospective study. Blood. 2012;120(8):1734‐1741. 10.1182/blood-2012-03-414490 [DOI] [PubMed] [Google Scholar]
- 6. Ishida T, Hishizawa M, Kato K, et al. Impact of graft‐versus‐host disease on allogeneic hematopoietic cell transplantation for adult T cell leukemia‐lymphoma focusing on preconditioning regimens: nationwide retrospective study. Blood Marrow Transplant. 2013;19(12):1731‐1739. 10.1016/j.bbmt.2013.09.014 [DOI] [PubMed] [Google Scholar]
- 7. Nosaka K, Iwanaga M, Imaizumi Y, et al. Epidemiological and clinical features of adult T‐cell leukemia‐lymphoma in Japan, 2010‐2011: a nationwide survey. Cancer Sci. 2017;108(12):2478‐2486. 10.1111/cas.13398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ishida T, Utsunomiya A, Iida S, et al. Clinical significance of CCR4 expression in adult T‐cell leukemia/lymphoma: its close association with skin involvement and unfavorable outcome. Clin Cancer Res. 2003;9(10Pt1):3625‐3634. [PubMed] [Google Scholar]
- 9. Ishida T, Iida S, Akatsuka Y, et al. The CC chemokine receptor 4 as a novel specific molecular target for immunotherapy in adult T‐Cell leukemia/lymphoma. Clin Cancer Res. 2004;10(22):7529‐7539. 10.1158/1078-0432.ccr-04-0983 [DOI] [PubMed] [Google Scholar]
- 10. Ishii T, Ishida T, Utsunomiya A, et al. Defucosylated humanized anti‐CCR4 monoclonal antibody KW‐0761 as a novel immunotherapeutic agent for adult T‐cell leukemia/lymphoma. Clin Cancer Res. 2010;16(5):1520‐1531. 10.1158/1078-0432.ccr-09-2697 [DOI] [PubMed] [Google Scholar]
- 11. Ishida T, Joh T, Uike N, et al. Defucosylated anti‐CCR4 monoclonal antibody (KW‐0761) for relapsed adult T‐cell leukemia‐lymphoma: a multicenter phase II study. J Clin Oncol. 2012;30(8):837‐842. 10.1200/jco.2011.37.3472 [DOI] [PubMed] [Google Scholar]
- 12. Ishida T, Jo T, Takemoto S, et al. Dose‐intensified chemotherapy alone or in combination with mogamulizumab in newly diagnosed aggressive adult T‐cell leukaemia‐lymphoma: a randomized phase II study. Br J Haematol. 2015;169(5):672‐682. 10.1111/bjh.13338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shichijo T, Nosaka K, Tatetsu H, et al. Beneficial impact of first‐line mogamulizumab‐containing chemotherapy in adult T‐cell leukaemia‐lymphoma. Br J Haematol. 2022. PMID: 35607839. 10.1111/bjh.18281 [DOI] [PubMed] [Google Scholar]
- 14. Fuji S, Inoue Y, Utsunomiya A, et al. Pretransplantation anti‐CCR4 antibody mogamulizumab against adult T‐cell leukemia/lymphoma is associated with significantly increased risks of severe and corticosteroid‐refractory graft‐versus‐host disease, nonrelapse mortality, and overall mortality. J Clin Oncol. 2016;34(28):3426‐3433. 10.1200/jco.2016.67.8250 [DOI] [PubMed] [Google Scholar]
- 15. Yonekura K, Kusumoto S, Choi I, et al. Mogamulizumab for adult T‐cell leukemia‐lymphoma: a multicenter prospective observational study. Blood Adv. 2020;4(20):5133‐5145. 10.1182/bloodadvances.2020003053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ishida T, Utsunomiya A, Jo T, et al. Mogamulizumab for relapsed adult T‐cell leukemia‐lymphoma: updated follow‐up analysis of phase I and II studies. Cancer Sci. 2017;108(10):2022‐2029. 10.1111/cas.13343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ishida T, Jo T, Takemoto S, et al. Follow‐up of a randomised phase II study of chemotherapy alone or in combination with mogamulizumab in newly diagnosed aggressive adult T‐cell leukaemia‐lymphoma: impact on allogeneic haematopoietic stem cell transplantation. Br J Haematol. 2019;184(3):479‐483. 10.1111/bjh.15123 [DOI] [PubMed] [Google Scholar]
- 18. Tanaka N, Mori S, Kiyotani K, et al. Genomic determinants impacting the clinical outcome of mogamulizumab treatment for adult T‐cell leukemia/lymphoma. Haematologica. 2022. 10.3324/haematol.2021.280352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sakamoto Y, Ishida T, Masaki A, et al. Clinical significance of TP53 mutations in adult T‐cell leukemia/lymphoma. Br J Haematol. 2021;195(4):571‐584. 10.1111/bjh.17749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sakamoto Y, Ishida T, Masaki A, et al. Clinical significance of CD28 gene‐related activating alterations in adult T‐cell leukaemia/lymphoma. Br J Haematol. 2021;192(2):281‐291. 10.1111/bjh.17211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sakamoto Y, Ishida T, Masaki A, et al. CCR4 mutations associated with superior outcome of adult T‐cell leukemia/lymphoma under mogamulizumab treatment. Blood. 2018;132(7):758‐761. 10.1182/blood-2018-02-835991 [DOI] [PubMed] [Google Scholar]
- 22. Zlotnik A, Burkhardt AM, Homey B. Homeostatic chemokine receptors and organ‐specific metastasis. Nat Rev Immunol. 2011;11(9):597‐606. 10.1038/nri3049 [DOI] [PubMed] [Google Scholar]
- 23. Hasegawa H, Nomura T, Kohno M, et al. Increased chemokine receptor CCR7/EBI1 expression enhances the infiltration of lymphoid organs by adult T‐cell leukemia cells. Blood. 2000;95(1):30‐38. 10.1182/blood.v95.1.30.001k09_30_38 [DOI] [PubMed] [Google Scholar]
- 24. Kataoka K, Nagata Y, Kitanaka A, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015;47(11):1304‐1315. 10.1038/ng.3415 [DOI] [PubMed] [Google Scholar]
- 25. Nakagawa M, Schmitz R, Xiao W, et al. Gain‐of‐function CCR4 mutations in adult T cell leukemia/lymphoma. J Exp Med. 2014;211(13):2497‐2505. 10.1084/jem.20140987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Watatani Y, Sato Y, Miyoshi H, et al. Molecular heterogeneity in peripheral T‐cell lymphoma, not otherwise specified revealed by comprehensive genetic profiling. Leukemia. 2019;33(12):2867‐2883. 10.1038/s41375-019-0473-1 [DOI] [PubMed] [Google Scholar]
- 27. Levine AJ. p53: 800 million years of evolution and 40 years of discovery. Nat Rev Cancer. 2020;20(8):471‐480. 10.1038/s41568-020-0262-1 [DOI] [PubMed] [Google Scholar]
- 28. Mantovani F, Collavin L, Del Sal G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019;26(2):199‐212. 10.1038/s41418-018-0246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bykov VJN, Eriksson SE, Bianchi J, Wiman KG. Targeting mutant p53 for efficient cancer therapy. Nat Rev Cancer. 2018;18(2):89‐102. 10.1038/nrc.2017.109 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support these findings are available from the corresponding author upon reasonable request.


