Abstract
Aim
The pathogenesis of acute diverticulitis (AD) remains incompletely understood, despite it being one of the most common gastrointestinal conditions worldwide. The aim of this study was to investigate the role of the colonic microbiome in the pathogenesis of AD.
Method
A prospective case–control study was performed, comparing the microbiome of AD patients with that of controls, using 16S rRNA sequencing of rectal swab samples.
Results
The microbiome of individuals with AD showed lower diversity than that of controls. There were significant compositional differences observed, with a lower abundance of commensal bacterial families and genera such as Lachnospiraceae, Ruminococcus and Faecalibacterium in AD patients compared with controls, and there was an increase in several genera with known pathogenic roles including Fusobacteria, Prevotella and Paraprevotella.
Conclusion
This is the largest study to date to examine the microbiota of AD patients, and adds evidence to the proposed hypothesis that alterations in the colonic microbiome play a role in the pathogenesis of AD.
Keywords: acute diverticulitis, colonic Mircobiome treatment, microbiom
What does this paper add to the literature?
This paper adds evidence for the proposal hypothesizing that alterations in the colonic microbiome play a role in the pathogenesis of acute diverticulitis (AD). While this is a challenging area to study, improved understanding of the potential role of the microbiota in the pathogenesis of AD has significant clinical importance and warrants further investigation.
INTRODUCTION
Acute diverticulitis (AD) is a disease with a significant impact on global health and resources [1]. This condition is characterized by peri‐colonic inflammation arising in an outpouching (diverticulum) in the colonic wall. Complicated AD, defined by the modified Hinchey classification (Stages Ib–IV) [2] as the presence of perforation, abscess, fistula or purulent or faeculent peritonitis, can lead to systemic sepsis and the requirement for operative intervention [3]. Aging populations, global uptake of a Western‐style diet and lifestyles, and increasing incidence in younger people are driving a surge in incidence [4]. The requisite precursor for AD, diverticulosis, is an asymptomatic condition with a lifetime prevalence of up to 72% [5]. However, fewer than 5% of patients with diverticulosis go on to develop AD, and the reasons why this small proportion progress are unclear [6]. Recent research has indicated a genetic contribution, while environmental risk factors, including diet, sedentary lifestyle, obesity, smoking and medications, have been repeatedly demonstrated in large correlational studies [7, 8, 9, 10, 11, 12]. The exact pathogenic mechanism through which these risk factors exert their effect is again unknown, but current theory suggests that these factors impact the gut microbiota and its interplay with the human host [13].
Research findings over the last 10 years have demonstrated the importance of the gut microbiota and its impact on human health. The gut microbiota influences colonic mucosal defences and local and systemic inflammation. A role in the pathogenesis of gastrointestinal diseases, including Clostridium difficile colitis, inflammatory bowel disease and irritable bowel syndrome, has been confirmed, while several small studies have indicated a role of the microbiota in AD [14]. Dysbiosis of the microbiome, either locally within a diverticulum or throughout the colon, could lower the threshold for an initiating stimulus, such as impaction of a faecolith, to trigger the pathway to AD.
The aim of this study is to compare the gut microbiota of individuals with AD with that of controls. Demonstrating a link between AD and the gut microbiota would advance our understanding of this disease and has the potential to provide an opportunity for prevention of both primary and recurrent disease and aid clinical decision‐making with regard to elective colonic resection to prevent future attacks.
METHOD
Study design
A prospective, single‐centre case–control study was performed comparing the microbiome of patients with AD (Hinchey Ia–IV) with that of controls without signs of diverticulitis. All adult (>18 years) patients admitted to Christchurch Hospital, New Zealand between 1 February and 31 August 2020 with a diagnosis of AD were assessed for eligibility for the study. Diagnosis of AD was confirmed by CT scan. The decision to admit to hospital was at the discretion of the clinical team but generally followed existing literature and guidelines. Patients with uncomplicated disease (Hinchey 1a), absence of comorbidities or immunocompromised state, with the ability to tolerate oral intake and an adequate social situation, were considered for outpatient management. All others were admitted and were eligible for inclusion. Exclusion criteria were those patients who had antibiotics in the 6 weeks preceding admission (excepting antibiotics commenced at the current admission, within 24 h of obtaining the rectal swab), mechanical bowel preparation (MBP) in the preceding 6 weeks, known or subsequent diagnosis of colorectal cancer or inflammatory bowel disease or patients unable to consent. Subgroup analysis of uncomplicated (Hinchey Ia) and complicated AD (Hinchey Ib–IV) was done.
Controls were adults without a previous history of AD. This cohort included people from a previous feasibility study run by our group and also individuals who were admitted to hospital with a diagnosis that was unrelated to the gastrointestinal system, for example those admitted for hernia repair or trauma‐related injury.
Rectal samples were taken from cases and controls using DNA/RNA Shield Collection tubes with swabs (Zymo Research). These swabs were placed in a DNA/RNA Shield Collection tube, prefilled with a solution that preserves DNA and RNA at ambient temperature.
DNA isolation and amplification
DNA for microbiome analysis was extracted from the swabs using a ZymoBIOMICS DNA Kit (Zymo Research) according to the manufacturer’s instructions. DNA concentration and purity were assessed using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific).
16S rRNA gene sequencing
Microbiome analysis was carried out using 16S rRNA sequencing. A Nextera® XT DNA Index kit (Illumina) was used for library preparation using a single‐step polymerase chain reaction (PCR) library preparation method that has dual‐index PCR primers that flank the V3–V4 hyper‐variable region of the 16S rRNA gene (16SF_V3: 5′‐TATG GTAATTGGCCTACGGGAGGCAGCAG‐3′ and 16SR_V4: 5′‐AGTCAGTCAGCCGGACTACHVGGGTWTCTAAT‐3′), and Illumina sequencing adaptors and barcodes were added using limited‐cycle PCR. The libraries were pooled by equal molarity before loading on the Illumina MiSeq platform with PhiX as 20% of the library and paired‐end reads of length 250 bp were generated.
Bioinformatics analyses
Quality control
DADA2 (v.1.18.0) was used to filter, trim, join sequencing reads and remove chimeras to obtain amplicon sequence variants [15]. The parameters for filtering and trimming are shown in Table S1. The SILVA database (v.132) was utilized to assign taxonomy to the amplicon sequence variants, with the tryRC option set to True and utilizing the silva_sp and addSpecies commands to identify species‐level taxonomy, where possible [16]. The taxonomy, sample metadata and sequences were combined into a phyloseq object for subsequent analysis [17]. The associated code is available at https://gitlab.com/alsulit08/uoc_diverticulitis.
Diversity analyses
For alpha diversity, we rarefied all samples to the smallest library size of 11,659 reads. We used phyloseq’s estimate richness function for Observed (richness) and Shannon (richness and evenness) measures, and compared differences in the alpha diversity measures by Wilcoxon tests. A p‐value <0.05 was considered significant. We also used principal component analysis to test for overall differences in the microbiome between groups. We normalized our samples using centred‐log ratios through the microbiome R package, and performed ordination using phyloseq’s ordinate function with the ‘RDA’ option [18]. Significance was then calculated through Permanova using the adonis() function of the vegan package [19], and betadisper() to compare dispersions between groups.
Differential abundance
We used DESEq2 to obtain differentially abundant genera between the following comparisons: (1) AD versus control, (2) complicated AD versus uncomplicated AD, (3) complicated AD versus control and (4) uncomplicated AD versus control [20]. Sequencing of samples was performed in two batches, and we therefore used batch as a covariate in our design formula. We considered a fold‐change between comparisons to be significant if it has a p‐adjust value <0.05.
Detailed analysis of the sequences may be accessed at https://gitlab.com/alsulit08/ uoc_diverticulitis.
Metadata analysis
To evaluate differences in baseline patient characteristics, Fisher’s exact and chi‐square tests were used for categorical data, with the Mann–Whitney U‐test and t‐test used for continuous data where appropriate. A p‐value of <0.05 was considered statistically significant.
RESULTS
Patient characteristics
During the study period, 120 patients were admitted with confirmed AD. A total of 65 patients with AD were included, 21 of whom had complicated disease. The reasons for exclusion were antibiotic use, either in the preceding 6 weeks (n = 16) or for more than 24 h for the current illness (n = 18), the patient declining to participate (n = 14), the patient was unable to consent (n = 2), immunosuppression (n = 2) and recent bowel preparation (n = 1). One individual with complicated AD agreed to participate but was excluded from analysis as insufficient DNA was extracted from the rectal swab. A total of 27 controls were included. The baseline demographics for each group are shown in Table 1 and for subgroups of uncomplicated and complicated AD in Table 2.
TABLE 1.
Baseline patient characteristics: acute diverticulitis compared with controls
| Acute diverticulitis (n = 65) | Control group (n = 27) | p‐value | |
|---|---|---|---|
| Female (%) | 27 (42) | 13 (48) | 0.28 |
| Age (years) (range) | 58 (27–91) | 46 (26–88) | <0.01 |
| Smoker (%) | 10 (15) | 2 (7) | 0.25 |
TABLE 2.
Baseline patient characteristics of the acute diverticulitis group compared by disease classification
| Uncomplicated AD (n = 44) | Complicated AD (n = 21) | p‐value | |
|---|---|---|---|
| Age (years) (range) | 56 (27–91) | 57 (39–78) | 0.52 |
| Female (%) | 23 (52.3%) | 5 (23.8%) | 0.04 |
| Mean BMI (kg/m2) (SD) | 30.7 (5.0) | 29.0 (4.0) | 0.50 |
| Comorbidity | |||
| ASA grade | |||
| 1 | 17 (38.6%) | 7 (33.3%) | 0.56 |
| 2 | 22 (50.0%) | 13 (61.9%) | |
| 3 | 5 (11.4%) | 1 (4.8%) | |
| 4 | 0 (0%) | 0 (0%) | |
| Modified Hinchey classification | |||
| Ia | 44 (100%) | 0 (0%) | N/A |
| Ib | 0 (0%) | 19 (90.4%) | |
| II | 0 (0%) | 1 (4.8%) | |
| III | 0 (0%) | 1 (4.8%) | |
| IV | 0 (0%) | 0 (0%) | |
| Current smoker | 6 (13.6%) | 4 (19.0%) | 0.72 |
| Ischaemic heart disease | 6 (13.6%) | 1 (4.8%) | 0.41 |
| Pulmonary disease | 5 (11.4%) | 3 (14.3%) | 0.70 |
| Renal impairment | 1 (2.3%) | 0 (0%) | 1.00 |
| Diabetes | 4 (9.1%) | 1 (4.8%) | 1.00 |
| Immunosuppression | 1 (2.3%) | 3 (14.3%) | 0.10 |
| Previous colonic resection | 1 (2.3%) | 0 (0%) | 1.00 |
| Previous appendicectomy | 6 (13.6%) | 5 (23.8%) | 0.31 |
| Mean CRP at admission (SD) | 72.5 (49.5) | 132.5 (83.2) | <0.01 |
| Mean WCC at admission (SD) | 12.2 (3.2) | 13.7 (3.1) | 0.09 |
| Duration of symptoms (days) (range) | 2 (1–14) | 2 (1–40) | 0.49 |
| Previous episode of AD | 9 (20.5%) | 8 (38.1%) | 0.15 |
Abbreviations: AD, acute diverticulitis; ASA, American Society of Anesthesiologists; BMI, body mass index; CRP, serum C‐reactive protein (mg/L); WCC, white blood cell count (×109/L).
Alpha diversity
We compared Observed and Shannon measures of alpha diversity between control and AD samples. There was a statistically significant decrease in Observed alpha diversity and Shannon diversity index, p = <0.001 and p = 0.019, respectively, in AD compared with controls (Figure 1).
FIGURE 1.
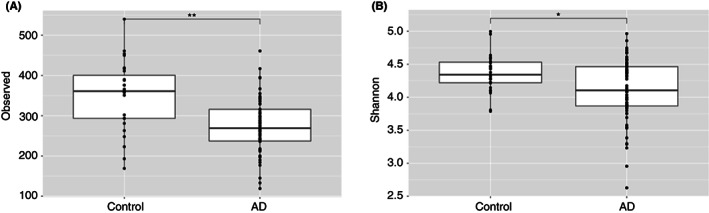
Observed (A) and Shannon (B) alpha diversities between acute diverticulitis and controls. p‐value: *<0.05; **≤0.01; ***≤0.001.
Both uncomplicated and complicated AD showed a statistically significant decrease in alpha diversity compared with controls (Figure 2).
FIGURE 2.
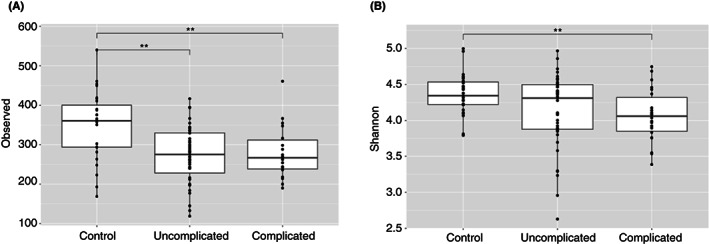
Observed (A) and Shannon (B) alpha diversities between uncomplicated, complicated and control groups. p‐value: *<0.05; **≤0.01; ***≤0.001.
Principal component analysis
Principal components (PCs) 1 and 2 accounted for 6% and 4% of the total variation, respectively, with the remaining components contributing significantly less to the variation seen. These two PCs were used to generate Figure 3.
FIGURE 3.
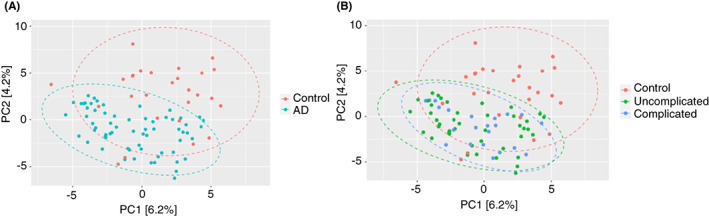
Principal component (PC) analysis using PC1 and PC2 showing separation between (A) acute diverticulitis (AD) and control groups and (B) complicated, uncomplicated, and control groups.
This analysis shows clear separation between all AD cases and controls (Figure 3A), but no separation between complicated and uncomplicated AD (Figure 3B). Permanova showed that our samples grouped significantly between disease states (AD versus control) but this grouping only explains 2.9% of the variance (adonis: R 2 = 0.029, p = 0.001). We also showed that there was a significant difference between within‐group dispersions (p = 0.001) possibly affecting PERMANOVA analysis, although our ordination supports the separation between AD and control group states.
Microbiome results
When comparing patients with AD (including subgroup comparisons of uncomplicated AD and complicated AD) with the control group using Wilcoxon tests there was a significant difference between the percentage abundances of the phyla Actinobacteria (AD versus control p < 0.001; complicated AD versus control p‐adjust = 0.001; uncomplicated AD versus control p‐adjust = 0.011) and Proteobacteria (AD versus control p < 0.0001; complicated AD versus control p‐adjust <0.001; uncomplicated AD versus control p‐adjust <0.001). Figure 4(A) shows the relative abundance of phyla between AD and controls and Figure 4(B) that between complicated AD, uncomplicated AD and controls.
FIGURE 4.
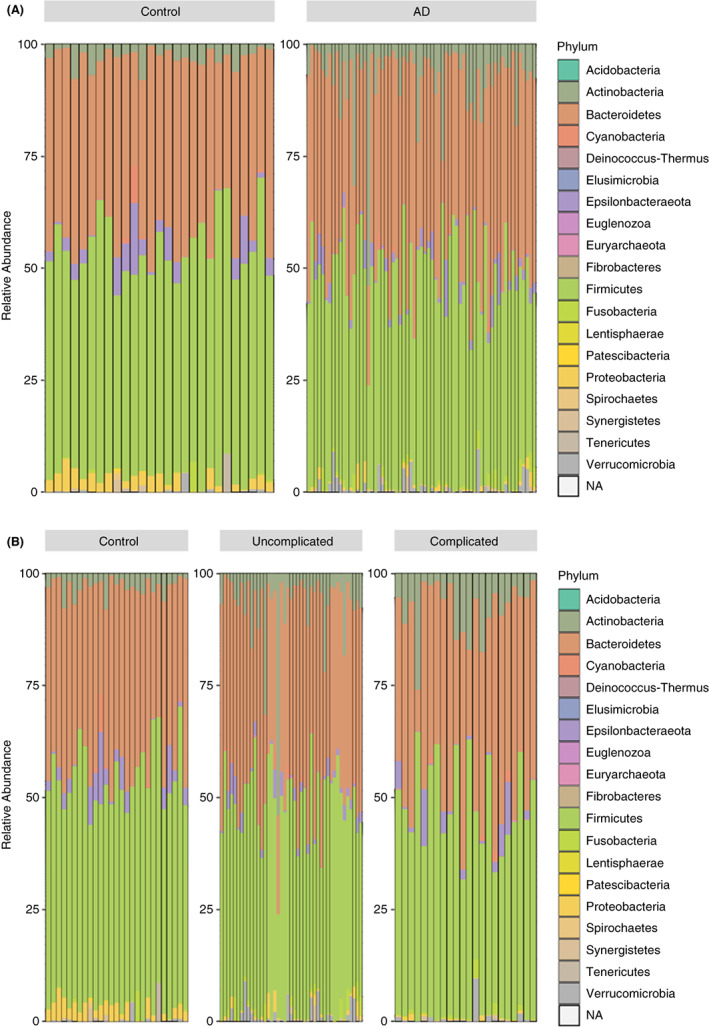
Relative abundance of phyla (A) between acute diverticulitis (AD) and controls and (B) between complicated AD, uncomplicated AD and controls, expressed as a percentage of the total taxonomic composition for each individual included in the study.
Differential abundance of genera
The heatmap in Figure 5 shows the differentially abundant genera between AD patients and controls. The heatmap on the left shows the log2 fold‐changes between AD and controls ((+) value = abundant in AD; (−) value = abundant in controls), while the heatmap on the right shows the abundance of these significant genera across samples with AD or from controls.
FIGURE 5.
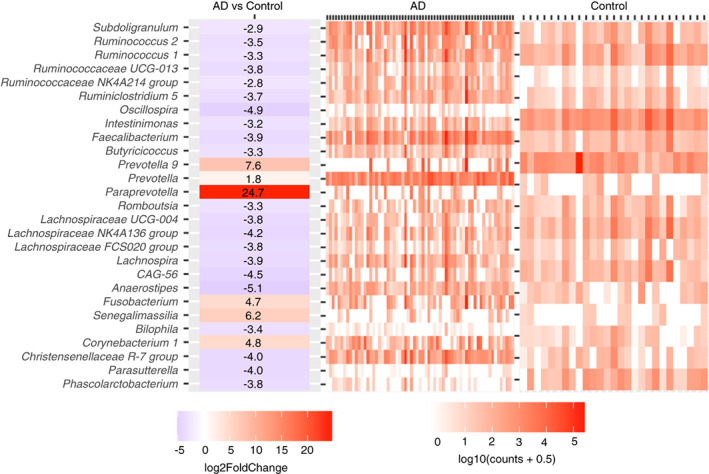
Log2 fold‐change of differentially abundant genera between acute diverticulitis (AD) and controls (left), and normalized abundances per sample in a group (right). Counts are DESeq2 normalized counts with an additional pseudocount of 0.5.
There were significant differences in microbiome composition noted for AD overall compared with controls. Twenty‐one genera were found to be significantly decreased in abundance in AD compared with controls, and were predominantly known commensal bacterial families and genera, such as Lachnospiraceae, Ruminococcus and Faecalibacterium, and contain species producing short‐chain fatty acids (SCFAs) [21, 22]. Only six genera were significantly more abundant in AD than in controls, and these included genera with known pathogenic roles, including Fusobacteria, Prevotella and Paraprevotella [23, 24, 25, 26]. Significance values for all differentially abundant taxa are included in Table S2.
Differences between uncomplicated AD and complicated AD were also observed, albeit less marked. In subgroup analysis, greater abundance of genera including Prevotella, Fusicatenibacter and Faecalibacterium were observed in the complicated group compared with uncomplicated AD (Figure 6).
FIGURE 6.
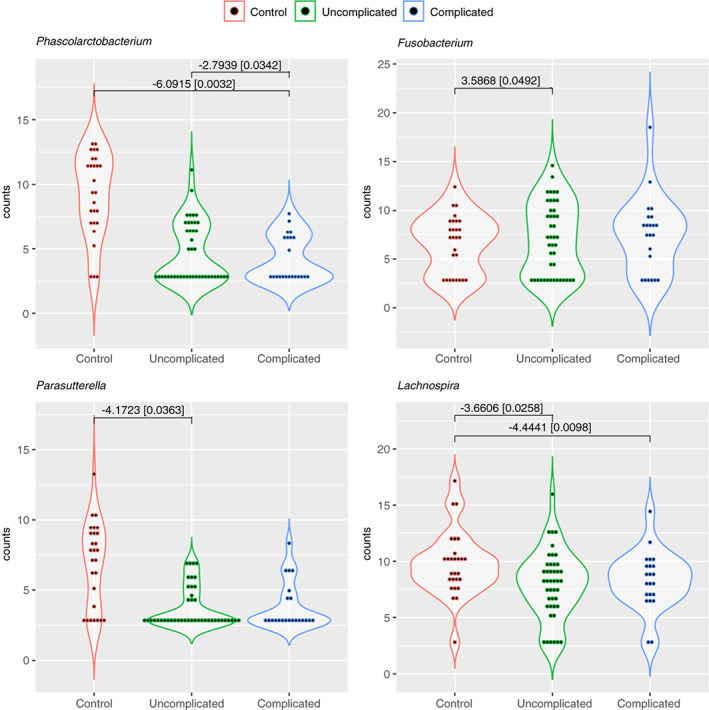
Violin plot demonstrating examples of differentially abundant genera comparing controls, uncomplicated acute diverticulitis (AD) and complicated AD with log2 fold‐change [p‐adjusted values].
DISCUSSION AND CONCLUSIONS
This study demonstrates distinct compositional changes in the microbiome of AD, with decreased diversity and decreased abundance of commensal taxa in patients with AD compared with controls. In addition, several known pathogenic taxa were increased in abundance in AD compared with controls.
There is little published literature examining the microbiota of AD, with differing methodologies limiting direct comparison. Daniels et al. compared the microbiota from rectal swabs of 31 CT‐confirmed cases of uncomplicated AD with 27 controls [27]. In contrast to the current study, they observed increased alpha diversity in AD compared with controls, predominantly as a result of differences in the phylum Proteobacteria. This may be due to a confounding effect as a result of study design – while rectal swabs in the AD group were taken in the emergency department before administration of antibiotics, swabs in the control group were taken at the time of colonoscopy, implying use of MBP. The use of MBP has been shown to significantly affect the composition of the microbiota, notably reducing diversity [28, 29]. Furthermore, inclusion of patients with disease states including inflammatory bowel disease and benign and malignant neoplasms in the control group is again likely to bias results, as these states are associated with decreased diversity [30, 31].
Gueimonde et al. compared Bifodobacterium species from distal colonic mucosal biopsies from 21 individuals with colorectal cancer, nine with diverticulitis and four with inflammatory bowel disease [32]. There were distinct differences between the three groups, again indicating a role of the microbiota in gastrointestinal disease; however, the study was limited by its sample size and by only evaluating a single species of the microbiota. The colonic microbiota is complex and disease states are likely to result from whole microbiota dysbiosis. With the exception of a few solitary disease‐causing pathogens, studying single species can be misleading [33].
The microbiome of symptomatic uncomplicated diverticular disease (SUDD) has been examined. This entity, characterized by abdominal symptoms (pain, diarrhoea, bloating) in an individual with diverticulosis but without overt inflammation is associated in one study with decreased abundance of Clostridium cluster IX, Fusobacterium and Lactobacillaceae compared with controls [26]. In a second study, increased abundance of Akkermansia muciniphila was noted in both asymptomatic diverticulosis patients and SUDD compared with controls [34].
It is uncertain if the microbiome is altered in individuals with diverticulosis. Two studies have been published, both of which found no difference between those with diverticulosis and those without [35, 36]. These studies have limitations – both were single centre and both analysed the microbiome of individuals undergoing screening colonoscopy. This presents a possible source of bias through the affect of MBP on microbial makeup. There is also potential for selection bias in the study by Jones et al. [35] because participants were undergoing screening colonoscopy, which in the USA requires health insurance and therefore this population may not be representative of the general population.
Disease states related to microbiome changes arise through two mechanisms, firstly through the presence (or increased abundance) of disease‐causing microbiota and secondly through decreased abundance and reduced diversity of commensal microbiota [33]. In the current study, patients with AD had a decreased abundance of recognized commensal genera, including those that produce SCFAs (propionate, acetate and butyrate). SCFAs provide 70% of the energy requirement of colonocytes; abundance of these molecules supports the metabolic demands of the cell, enabling it to maintain the intestinal barrier through improved tight‐junction integrity and mucin production [37, 38]. The luminal concentration of SCFAs has been directly correlated with the thickness of the mucous layer in animal studies [39]. SCFAs also enhance cellular metabolism in lymphocytes, with proven increases in lymphocytes, supporting activation, plasma‐cell differentiation and antibody production [40]. Loss of commensal microbiota genera and their by‐products leads to impairment of the intestinal barrier, leading to increased microbiota–immune cell interactions. Combined with downregulation of anti‐inflammatory processes, a proinflammatory state can ensue.
There were several taxa with increased abundance in AD identified in the current study, including known pathogens. Prevotella is a large genus with over 40 species cultured, at least three of which are known to reside in the gut. These species are generally commensal, being especially predominant in individuals who consume diets high in plant‐based carbohydrates. However, these microbes are highly adaptable, responding to niches within the human body by modulating gene repertoires [41]. This fact, in combination with significant inter‐individual variability in microbe–host interactions, leads to differences in perceived interaction, from beneficial to detrimental. Prevotella copri has been shown to increase susceptibility to colitis in mice, and with increased cytokine production and chronic colonic inflammation in individuals with HIV [23, 24]. There was increased abundance of Prevotella in AD patients in the current study, and particularly in patients with complicated AD. Another genus with observed increased abundance was Fusobacterium, which is correlated with the pathogenesis of colorectal cancer through activation of host inflammatory responses [25, 42]. While the current study cannot provide evidence of causation, the authors propose the mechanism of pathogenic involvement as an alteration of the protective and pathogenic interactions between the microbiota, the colonic cells and the immune system. This alteration contributes, along with other established factors, to an individual’s propensity to develop diverticulitis. While these changes in the microbiota may exist throughout the colon, it is in the sigmoid colon where diverticula are predominantly found (in the West), where microbial density is higher and where stool transit time is slower, thus allowing for more microbiota–host interactions.
Evaluation of colonic flora has traditionally been obtained through faecal sampling; however, alternative means include mucosal biopsy and rectal swab. The optimum method is debated. There are differences in the microbiome present in the bowel lumen versus the mucous layer of the bowel wall. Some authors argue that only the mucosal microbiome is of clinical relevance, as most of the host–bacteria interactions occur at this level [43, 44, 45]. Analysis of the mucosal microbiome is more challenging as generally this is done with flexible endoscopy. This procedure is invasive, and usually requires MBP, which can in itself alter the composition of the microbiome [28, 29]. Rectal swab was used in the current study as it was minimally invasive and could be used at point of care, prior to the administration of antibiotics. Rectal swabs have been shown to correlate with faecal samples in previous studies [46, 47, 48, 49] and in a feasibility study undertaken by our group [50].
The strengths of the current study include the requirement for imaging confirmation of AD and the large sample size relative to published data. A limitation of the study is the single time‐point analysis, which provides only a snapshot in time. Observed changes in the microbiota may not reflect an individual’s long‐term microbiota, and may reflect a response to, rather than a contribution to, the pathogenesis of AD. However, a longitudinal study to resolve this would be impractical given the rarity of progression of diverticulosis to AD. We were unable to control for potential confounding due to comorbidity and for the presence of diverticulosis in the control group which could introduce bias. While this study is the largest in the field, the numbers are still relatively small, particularly for subgroup analysis, and from a single centre. A further limitation lies in 16S rRNA gene sequencing, which is currently limited to providing genus‐level information.
The pathogenesis of AD is complex with many protective and pathogenic factors identified. The current study has added to the evidence that the microbiota is another contributor, and has a similarly complex role. While this is a challenging area to study, an improved understanding of the potential role of the microbiota in the pathogenesis of AD has significant clinical importance and this subject warrants further investigation.
AUTHOR CONTRIBUTIONS
Michael O’Grady: Study formulation, collection and processing of microbiome specimens, manuscript writing. Greg Turner: Study formulation, collection and processing of microbiome specimens, manuscript writing. Arielle Sulit: Statistical analysis. Frank Frizelle: Study formulation, supervision, manuscript writing. Rachel Purcell: Study formulation, supervision, manuscript writing.
CONFLICT OF INTEREST
No conflict of interest declared for any author.
FUNDING INFORMATION
Richard Stewart Scholarship 2020, Dunedin Basic Medical Sciences Trust.
ETHICS STATEMENT
This study was approved by the University of Otago Human Ethics Committee (Health), reference H20/009 and registered with the Canterbury District Health Board Research Office, reference RO#20008. Written informed consent was obtained from patients.
Supporting information
Table S1
Table S2
ACKNOWLEDGEMENTS
Our sincere thanks goes to all patients who participated in the study, and to staff of the Christchurch Hospital Surgical Wards and Department of Surgery, University of Otago, Christchurch. Open access publishing facilitated by University of Otago, as part of the Wiley ‐ University of Otago agreement via the Council of Australian University Librarians.
MJ O, Turner GA, A S, Frizelle FA, R P. Distinct changes in the colonic microbiome associated with acute diverticulitis. Colorectal Dis. 2022;24:1591–1601. 10.1111/codi.16271
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in UOC_diverticulitis at https://gitlab.com/alsulit08/uoc_diverticulitis, reference number 32064025.
REFERENCES
- 1. O’Grady M, Turner G, Currie W, Yi M, Frizelle F, Purcell R. Acute diverticulitis: an ongoing economic burden on the health system. ANZ J Surg. 2020;90(10):2046–9. [DOI] [PubMed] [Google Scholar]
- 2. Miller AS, Boyce K, Box B, Clarke MD, Duff SE, Foley NM, et al. The Association of Coloproctology of Great Britain and Ireland consensus guidelines in emergency colorectal surgery. Colorectal Dis. 2012;23(2):476–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sharma PV, Eglinton T, Hider P, Frizelle F. Systematic review and meta‐analysis of the role of routine colonic evaluation after radiologically confirmed acute diverticulitis. Ann Surg. 2014;259(2):263–72. [DOI] [PubMed] [Google Scholar]
- 4. Turner GA, O’Grady MJ, Purcell RV, Frizelle FA. Acute diverticulitis in young patients: a review of the changing epidemiology and etiology. Dig Dis Sci. 2021;67:1156–62. [DOI] [PubMed] [Google Scholar]
- 5. Peery AF, Keku TO, Galanko JA, Sandler RS. Sex and race disparities in diverticulosis prevalence. Clin Gastroenterol Hepatol. 2020;18(9):1980–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shahedi K, Fuller G, Bolus R, Cohen E, Vu M, Shah R, et al. Long‐term risk of acute diverticulitis among patients with incidental diverticulosis found during colonoscopy. Clin Gastroenterol Hepatol. 2013;11(12):1609–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Granlund J, Svensson T, Olén O, Hjern F, Pedersen NL, Magnusson PKE, et al. The genetic influence on diverticular disease – a twin study. Aliment Pharmacol Ther. 2012;35(9):1103–7. [DOI] [PubMed] [Google Scholar]
- 8. Strate LL, Erichsen R, Baron JA, Mortensen J, Pedersen JK, Riis AH, et al. Heritability and familial aggregation of diverticular disease: a population‐based study of twins and siblings. Gastroenterology. 2013;144(4):736–42. [DOI] [PubMed] [Google Scholar]
- 9. Schafmayer C, Harrison JW, Buch S, Lange C, Reichert MC, Hofer P, et al. Genome‐wide association analysis of diverticular disease points towards neuromuscular, connective tissue and epithelial pathomechanisms. Gut. 2019;68(5):854–65. [DOI] [PubMed] [Google Scholar]
- 10. Aune D, Sen A, Norat T, Riboli E. Dietary fibre intake and the risk of diverticular disease: a systematic review and meta‐analysis of prospective studies. Eur J Nutr. 2019;59(2):421–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aune D, Sen A, Leitzmann MF, Tonstad S, Norat T, Vatten LJ. Tobacco smoking and the risk of diverticular disease – a systematic review and meta‐analysis of prospective studies. Colorectal Dis. 2017;19(7):621–33. [DOI] [PubMed] [Google Scholar]
- 12. Hjern F, Wolk A, Håkansson N. Obesity, physical inactivity, and colonic diverticular disease requiring hospitalization in women: a prospective cohort study. Am J Gastroenterol. 2012;107(2):296–302. [DOI] [PubMed] [Google Scholar]
- 13. Jeganathan NA, Davenport ER, Yochum GS, Koltun WA. The microbiome of diverticulitis. Curr Opin Physiol. 2021;22:100452. [Google Scholar]
- 14. Skowron KB, Shogan BD, Rubin DT, Hyman NH. The new frontier: the intestinal microbiome and surgery. J Gastrointest Surg. 2018;22(7):1277–85. [DOI] [PubMed] [Google Scholar]
- 15. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high‐resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web‐based tools. Nucleic Acids Res. 2013;41(database issue):D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lahti L, Shetty S. Introduction to the microbiome R package. [cited 13 Dec 2021]. Available from: https://microbiome.github.io/tutorials/
- 19. Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: Community Ecology Package. 2019. [cited 16 Sep 2019]. Available from: https://CRAN.R‐project.org/package=vegan
- 20. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ze X, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 2012;6(8):1535–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Louis P, Young P, Holtrop G, Flint HJ. Diversity of human colonic butyrate‐producing bacteria revealed by analysis of the butyryl‐CoA:acetate CoA‐transferase gene. Environ Microbiol. 2010;12(2):304–14. [DOI] [PubMed] [Google Scholar]
- 23. Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;5(2):e01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dillon SM, Lee EJ, Kotter CV, Austin GL, Gianella S, Siewe B, et al. Gut dendritic cell activation links an altered colonic microbiome to mucosal and systemic T‐cell activation in untreated HIV‐1 infection. Mucosal Immunol. 2016;9(1):24–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Purcell RV, Visnovska M, Biggs PJ, Schmeier S, Frizelle FA. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci Rep. 2017;7(1):11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barbara G, Scaioli E, Barbaro MR, Biagi E, Laghi L, Cremon C, et al. Gut microbiota, metabolome and immune signatures in patients with uncomplicated diverticular disease. Gut. 2016;66(7):1252–61. [DOI] [PubMed] [Google Scholar]
- 27. Daniels L, Budding AE, de Korte N, Eck A, Bogaards JA, Stockmann HB, et al. Fecal microbiome analysis as a diagnostic test for diverticulitis. Eur J Clin Microbiol Infect Dis. 2014;33(11):1927–36. [DOI] [PubMed] [Google Scholar]
- 28. Nagata N, Tohya M, Fukuda S, Suda W, Nishijima S, Takeuchi F, et al. Effects of bowel preparation on the human gut microbiome and metabolome. Sci Rep. 2019;9(1):4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Harrell L, Wang Y, Antonopoulos D, Young V, Lichtenstein L, Huang Y, et al. Standard colonic lavage alters the natural state of mucosal‐associated microbiota in the human colon. PLoS One. 2012;7(2):e32545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zuo T, Kamm MA, Colombel J‐F, Ng SC. Urbanization and the gut microbiota in health and inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2018;15(7):440–52. [DOI] [PubMed] [Google Scholar]
- 31. Ai D, Pan H, Li X, Gao Y, Liu G, Xia LC. Identifying gut microbiota associated with colorectal cancer using a zero‐inflated lognormal model. Front Microbiol. 2019;24(10):826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gueimonde M. Qualitative and quantitative analyses of the bifidobacterial microbiota in the colonic mucosa of patients with colorectal cancer, diverticulitis and inflammatory bowel disease. World J Gastroenterol. 2007;13(29):3985–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Duvallet C, Gibbons SM, Gurry T, Irizarry RA, Alm EJ. Meta‐analysis of gut microbiome studies identifies disease‐specific and shared responses. Nat Commun. 2017;8(1):1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tursi A, Mastromarino P, Capobianco D, Elisei W, Miccheli A, Capuani G, et al. Assessment of fecal microbiota and fecal metabolome in symptomatic uncomplicated diverticular disease of the colon. J Clin Gastroenterol. 2016;50(suppl 1):S9–12. [DOI] [PubMed] [Google Scholar]
- 35. Jones RB, Fodor AA, Peery AF, Tsilimigras MCB, Winglee K, McCoy A, et al. An aberrant microbiota is not strongly associated with incidental colonic diverticulosis. Sci Rep. 2018;8(1):4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van Rossen TM, Ooijevaar RE, Kuyvenhoven JP, Eck A, Bril H, Buijsman R, et al. Microbiota composition and mucosal immunity in patients with asymptomatic diverticulosis and controls. PLoS One. 2021;16(9):e0256657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 1990;70(2):567–90. [DOI] [PubMed] [Google Scholar]
- 38. Ríos‐Covián D, Ruas‐Madiedo P, Margolles A, Gueimonde M, de los Reyes‐Gavilán CG, Salazar N. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol. 2016;7:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shortt C, Hasselwander O, Meynier A, Nauta A, Fernández EN, Putz P, et al. Systematic review of the effects of the intestinal microbiota on selected nutrients and non‐nutrients. Eur J Nutr. 2018;57(1):25–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe‐derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–50. [DOI] [PubMed] [Google Scholar]
- 41. Ley RE. Prevotella in the gut: choose carefully. Nat Rev Gastroenterol Hepatol. 2016;13(2):69–70. [DOI] [PubMed] [Google Scholar]
- 42. Kelly D, Yang L, Pei Z. Gut microbiota, fusobacteria, and colorectal cancer. Diseases. 2018;6(4):109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tang MS, Poles J, Leung JM, Wolff MJ, Davenport M, Lee SC, et al. Inferred metagenomic comparison of mucosal and fecal microbiota from individuals undergoing routine screening colonoscopy reveals similar differences observed during active inflammation. Gut Microbes. 2015;6(1):48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zoetendal EG, von Wright A, Vilpponen‐Salmela T, Ben‐Amor K, Akkermans ADL, de Vos WM. Mucosa‐associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68(7):3401–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schieffer KM, Sabey K, Wright JR, Toole DR, Drucker R, Tokarev V, et al. The microbial ecosystem distinguishes chronically diseased tissue from adjacent tissue in the sigmoid colon of chronic, recurrent diverticulitis patients. Sci Rep. 2017;7(1):8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Budding AE, Grasman ME, Eck A, Bogaards JA, Vandenbroucke‐Grauls CMJE, van Bodegraven AA, et al. Rectal swabs for analysis of the intestinal microbiota. PLoS One. 2014;9(7):e101344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reyman M, van Houten MA, Arp K, Sanders EAM, Bogaert D. Rectal swabs are a reliable proxy for faecal samples in infant gut microbiota research based on 16S‐rRNA sequencing. Sci Rep. 2019;9(1):16072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bassis CM, Moore NM, Lolans K, Seekatz AM, Weinstein RA, Young VB, et al. Comparison of stool versus rectal swab samples and storage conditions on bacterial community profiles. BMC Microbiol. 2017;17(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang N, Li T‐Z, Zheng K, Mou D‐L, Liang L‐C, Zhang T, et al. Use of rectal swab samples for analysis of the intestinal microbiome in children. Chin Med J (Engl). 2018;131(4):492–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Turner G, O’Grady M, Hudson D, Morgan X, Frizelle F, Purcell R. Rectal swabs are a reliable method of assessing the colonic microbiome. Int J Med Microbiol. 2022;312(2):151549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2
Data Availability Statement
The data that support the findings of this study are openly available in UOC_diverticulitis at https://gitlab.com/alsulit08/uoc_diverticulitis, reference number 32064025.


