Abstract
Metalation, the acquisition of metals by proteins, must avoid mis‐metalation with tighter binding metals. This is illustrated by four selected proteins that require different metals: all show similar ranked orders of affinity for bioavailable metals, as described in a universal affinity series (the Irving–Williams series). Crucially, cellular protein metalation occurs in competition with other metal binding sites. The strength of this competition defines the intracellular availability of each metal: its magnitude has been estimated by calibrating a cells' set of DNA‐binding, metal‐sensing, transcriptional regulators. This has established that metal availabilities (as free energies for forming metal complexes) are maintained to the inverse of the universal series. The tightest binding metals are least available. With these availabilities, correct metalation is achieved.
Keywords: cobalt, copper, iron, Irving–Williams series, magnesium, manganese, metal sensor, metalation, nickel, zinc
Metalation of proteins is logical within defined intracellular metal availabilities. Mis‐metalation is avoided because weaker binding metals are more available than tighter binding ones.
Abbreviation
SodA, superoxide dismutase
Metal affinities and mis‐metalation
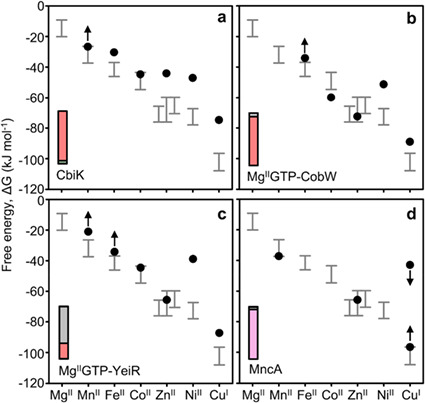
Metalation is crucial for metalloproteins to achieve their proper enzymatic activity and/or structure. Mis‐metalation occurs in part because proteins are flexible and metal binding is non‐conservative: a wrong metal can use a sub‐set of ligands from the bona fide site, recruit additional adventitious ligands, and/or distort the native geometry. With such limited constraint, it is anticipated that most metalloproteins are at risk of mis‐metalation of their nascent binding sites. It is estimated that 47% of enzymes contain metals [1, 2]. How do cells overcome this pervasive challenge to enable enzymes to bind metal(s) with the correct chemical properties, and not simply those that bind most tightly? To exemplify the challenge of mis‐metalation, Fig. 1 shows how tightly essential metals bind to the four selected proteins: Namely, a chelatase (CbiK) that acquires CoII for a molecular cofactor (vitamin B12), a CoII metallochaperone (CobW), a homologous ZnII delivery protein (YeiR) and a MnII enzyme that entraps metal during folding (MncA) [3, 4, 5]. The tightness of binding is represented as differences in free energies for forming the respective metal complexes. Affinities were measured for what is considered the exchangeable available form of each metal in the cytosol, that is divalent except for copper which is monovalent (albeit MncA ratios were estimated for CuII as described later) [6]. For all four proteins, the tightest binding metal is not the one required for activity (Fig. 1). The orders of binding follow, or tend towards, the Irving–Williams series: MgII < MnII < FeII < CoII < NiII < CuII (CuI) > ZnII, from weakest to tightest [6, 7]. If surplus metals were allowed to inter‐compete, three proteins would be mis‐metalated with CuI, and one inferred to become mis‐metalated with ZnII (represented as black and grey insets in Fig. 1).
Fig. 1.
Metal binding to four proteins exemplifies the risk of mis‐metalation. Tightness of binding to the four proteins is shown for the available and exchangeable forms of metals in the cytosol (or CuII for periplasmic MncA). Values (black circles) are free energies for forming complexes calculated via the standard relationship ∆G = −RTlnK A (∆G free energy change, R molar gas constant, T temperature in kelvin, K A association constant). Note that values are logarithmically related to binding constants. The more negative the value the tighter the binding. Arrows indicate values that were at the minimum or maximum limits of the respective determinations of metal affinity. CbiK is a CoII chelatase for vitamin B12 biosynthesis [3], MgIIGTP‐CobW is a CoII metallochaperone from an alternative vitamin B12 biosynthetic pathway, MgIIGTP‐YeiR is analogous to MgIIGTP‐CobW but implicated in handling ZnII [5], MncA is a MnII cupin [4]. Values for MncA are assigned based upon competition between metals (details noted in the text). Insets show percentage occupancies with copper (black) and ZnII (grey) as a proportion of total metal occupancy. The correct metals are not the tightest binding metals and the orders of binding tend to follow the Irving–Williams series [6, 7].
Examples of delivery proteins and the synthesis of molecular cofactors
Some proteins bind metals in pre‐assembled cofactors such as iron in haem or iron sulfur clusters, nickel in cofactor F430 or cobalt in vitamin B12, as examples. However, the correct metal must still partition onto the cofactor assembly pathway in the first place. CbiK is a chelatase that inserts CoII into corrin in one of two pathways for the synthesis of vitamin B12 [8, 9, 10]. Fig. 1 shows that CuI binds tightly to CbiK, with ZnII and NiII comparable to CoII, highlighting the question as to how tight‐binding non‐cognate metals are avoided.
Metals are supplied to some proteins by delivery pathways involving metallochaperones and here the final metalation step is aided by selective protein–protein interactions [11, 12, 13, 14]. CobW supplies CoII to a different chelatase, not CbiK, in an alternative pathway for vitamin B12 biosynthesis [5, 8, 15]. CobW has a predicted GTPase domain and when bound to MgIIGTP, CobW forms a tight complex with CoII but crucially it forms an even tighter complex with CuI [5]. As an aside, binding of NiII to MgIIGTP‐CobW is relatively weak, departing from the Irving–Williams series (Fig. 1). This is probably because association with MgIIGTP pre‐organises the CoII site of CobW into a tetrahedral geometry thereby limiting the ability of NiII to distort the site into its preferred planar geometry [5]. YeiR is analogous to CobW but is implicated in the delivery of ZnII rather than CoII [5, 16, 17]. In common with CobW, when bound to MgIIGTP, YeiR forms tighter complexes with CuI than with its cognate metal. How do the correct metals somehow partition onto these, and other, delivery pathways inside cells to avoid mis‐metalation, or blocked metalation, of the proteins that they supply?
Example of kinetic trapping in a folded protein
Some metals become kinetically trapped within folded proteins. However, the folding pathway may still preferentially entrap the wrong metals. This is illustrated by the MnII cupin MncA. An experiment in which MncA was folded in vitro in the presence of an equimolar surplus of CuI and MnII led to the wrong metal, CuI, being entrapped implying that CuI binds MncA more than 10 times more tightly. Similar experiments competing MnII against either ZnII or CuII again led to entrapment of the wrong metal [4]. Presumably, some nascent flexible site along the folding pathway tends to follow the Irving–Williams series. Competition experiments with increasing molar excesses of MnII versus either CuII or ZnII yielded full loading of MncA with MnII at 10 000 and 100 000‐fold surpluses of MnII respectively [4]. Notably, this suggests a slightly tighter affinity for ZnII than CuII, but the experiment used Tris buffers, which may have formed unaccounted CuII‐complexes. Affinities for ZnII and CuII binding to a nascent site in MncA have been assigned in Fig. 1D to reflect the measured differences relative to MnII.
Mis‐metalation within cells
The MnII form of Escherichia coli superoxide dismutase (SodA) is commonly mis‐metalated with FeII and inactive [18]. The detection of reactive oxygen species by a sensor, OxyR, triggers expression of a manganese importer (MntH) which in turn leads to nascent superoxide dismutase SodA being correctly metalated with MnII [19]. Small molecule cofactors can also become mis‐metalated. For example, exposure to elevated levels of CoII leads to mis‐metalation of iron sulfur clusters with CoII [20]. Furthermore, some forms of iron deficiency cause the accumulation of zinc protoporphyrin IX in place of haeme [21]. The copper cupin CucA is found in the same cyanobacterial periplasmic compartments as MnIIMncA, but CucA does acquire copper using the same ligands within the same fold as MncA [4]. Importantly, while CucA is secreted via the sec‐pathway to fold within the periplasm where it acquires copper, MncA is a TAT‐substrate which folds in the cytoplasm where it entraps MnII. This suggests that the location of protein folding can determine the specificity of metalation, and moreover that MnII must be significantly more available than either CuI or ZnII in the cytosol, with the latter ratio being at least 100 000‐fold. These observations reveal the crucial contributions of metal availabilities at the sites of protein folding to the avoidance of mis‐metalation [4].
Metalation in cells
Cells have a diversity of mechanisms that maintain metal availabilities within tolerable ranges [22, 23, 24, 25, 26, 27, 28, 29, 30]. For example, importers acquire more of metals that are deficient while storage proteins and exporters sequester or remove those in surplus. Mechanisms also exist to sustain optimal metal availabilities within intracellular compartments and to maintain extracellular systemic metal supply in multicellular organisms. These mechanisms are controlled by a variety of metal sensors. DNA‐binding metal sensing transcriptional regulators have been especially well characterised in bacterial cells: They include metal‐dependent de‐repressors [31], metal‐dependent co‐repressors [32], and metal‐dependent activators [33, 34], of gene transcription. The allosteric mechanisms of these sensors have evolved to couple metal binding to DNA binding, and to respond within the ranges of intracellular metal availabilities over which their cognate metals fluctuate in viable cells [35, 36]. When sensitivities are adjusted to lie outside the vital range, sensors become unresponsive to changes in metal levels [35]. These sensors offer a route to read‐out the ranges over which intracellular metal availabilities fluctuate.
Calibrating metal sensors to decode metalation
A set of DNA‐binding metal sensors of the three types outlined above, have been characterised in detail from Salmonella enterica serovar Typhimurium (hereafter Salmonella), for the purpose of calibrating their responses to intracellular metal availabilities [3]. The Salmonella sensors are almost identical to those of E. coli [5, 37]: They include MnII‐responsive co‐repressor MntR, FeII‐responsive co‐repressor Fur, CoII‐responsive de‐repressor RcnR, NiII‐responsive co‐repressor NikR, ZnII‐responsive activator ZntR, ZnII‐responsive co‐repressor ZntR and CuI‐responsive activator CueR [3]. After confirming their cognate metals, the metal affinities, DNA affinities of apo‐ and metal‐bound forms of each sensor, along with the number of promoter binding sites and the number of sensor molecules per cell (in high and in low metal) were all determined [3]. These values were used to calculate response curves that relate the state of each sensor to the intracellular availability of the cognate metal. The states (on or off DNA, with or without metal) that exist at different metal availabilities form coupled thermodynamic cycles [38, 39]. Before resolving these cycles mathematically in order to generate response curves, there needs to be prior consideration of the nature of available metals inside cells.
A ZnII‐buffered in vitro transcription assay previously established that ZnII sensors of E. coli respond at femtomolar ZnII concentrations [40]. It was noted that one atom per cell volume (approximately a femtolitre) equates to nanomolar concentrations, suggesting that the sensors detect the transition from ZnII deficiency to excess at a million times less than one (hydrated) atom per cell. An explanation is that there are surplus binding sites in the intracellular milieu such that all ZnII atoms are bound, leading to a suggestion that ZnII might be delivered to its destinations by dedicated proteins analogous to copper metallochaperones [40, 41]. There is evidence of some ZnII delivery proteins [42], albeit this deflects attention towards the specificity of metal acquisition by the delivery proteins themselves. Moreover, it is anticipated that ZnII‐metallochaperones are exceptional, with a multitude of ZnII‐proteins acquiring metal directly. Importantly, metal transfer by associative ligand exchange, analogous to transfer from metallochaperones [13, 41, 43, 44], may also occur from small molecule ligands such as glutathione or free histidine [35, 45, 46, 47, 48, 49, 50, 51, 52].
Cytosolic metal buffering by small molecules, metallochaperones and other labile sites, coupled with associative metal‐transfer, has several implications: (a) It becomes mathematically possible to resolve the coupled thermodynamic cycles to calculate sensor states at different metal availabilities when the metal is buffered (because metal binding to the sensor does not alter the available metal concentration, thus removing an otherwise dependent variable); (b) metal transfer by associative ligand exchange is rapid because it is not limited by the slow rate of release to the hydrated state; (c) an equilibrium state will better approximate an in vivo state if metal transfer to and from sensors (along with other proteins) is associative and fast; (d) an almost non‐existent pool of hydrated metal ions is not a limitation if metal transfer is associative; (e) the concentration of the negligible hydrated pool (equating to one ZnII atom per million cells at any given instant or an atom in every cell one millionth of the time, in the earlier example) enables the strength of competition from the intracellular milieu to be calculated as an activity or difference in free energy. The range of internal metal availabilities over which each Salmonella sensor transitions between its off‐DNA states and its DNA‐bound states, or in the case of the activators, DNA‐metal‐bound state, were thus calculated [3]. Notably, the activators distort DNA to align critical nucleotide sequences only in their metal‐bound state. In contrast, repression is mediated by apo‐ or metalated sensors, albeit the proportions associated with DNA differ for co‐repressors versus de‐repressors. The calculated response curves were seen to depart from those generated from the metal affinities of sensors alone [3]. For example, some metal sensors are autoregulatory and a change in sensor abundance with metal concentration introduces hysteresis. Also, because metal binding and DNA binding are allosterically coupled, metal binding alters DNA affinity but reciprocally DNA binding alters metal affinity, and this influences the metal response curves.
Metal availability follows the inverse of the Irving–Williams series
The grey bars in each panel of Fig. 2 show the availabilities over which the sensor(s) for each metal are calculated to respond, ranging from 10% to 90% cognate DNA occupancy with sensor, or with solely metalated sensor for activators [3]. Availabilities are shown as free energies for forming complexes that would be 50% metalated at the respective metal concentration. Metal availability is the inverse of the Irving–Williams series. The more competitive metals are maintained at the lowest availabilities. This has been a long‐standing hypothesis which is now experimentally supported by the calculated sensitivities of a cells' detectors/controllers of metal availabilities [3, 6]. These data provide estimates of the magnitude of competition from labile binding sites in the intracellular milieu [3]. In turn, this provides a frame of reference against which it becomes possible to re‐interpret metalation, for example, of the four proteins shown in Fig. 1, in a biological context.
Fig. 2.
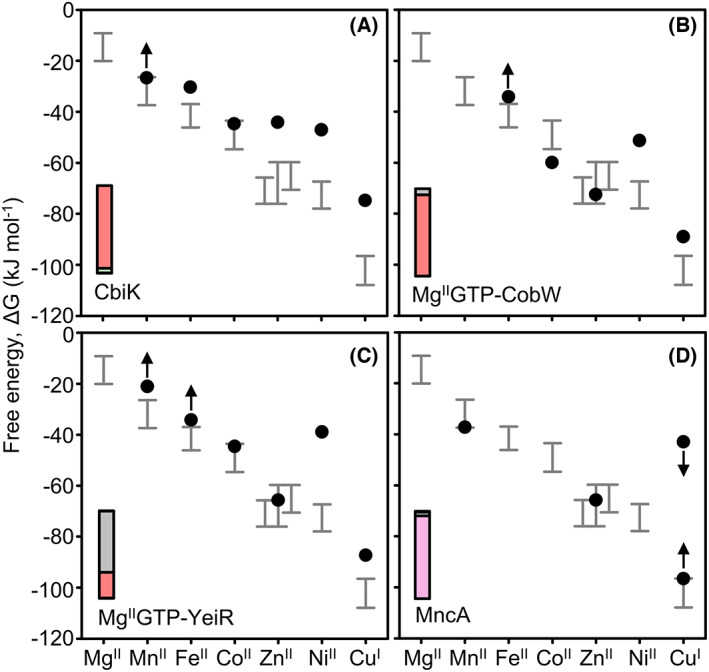
Metal availability is the inverse of the Irving–Williams series and decodes correct metalation. Grey bars show the ranges from 10% to 90% of the transcriptional responses of the cognate sensors for each metal as free energies for forming complexes that would be 50% saturated at the respective availability [3]. Metal availability is inverse to the Irving–Williams series. The ranges indicate the range of strengths of competition from exchangeable cytosolic binding sites against which the sensors have evolved to compete to sustain optimal metal availabilities. Metalation of other proteins similarly involves competition with these exchangeable metal binding sites. Black circles and arrows replicate metal binding data from Fig. 1, except for limits to CuI binding to MncA where the weakest value is derived from a competition experiment and the tightest inferred to give negligible (1%) CuI occupancy. The four cognate metals become apparent when binding is considered in relation to availability, as shown in the insets with CoII (salmon red), ZnII (grey), MnII (pink) [3, 5]. These proportional metal occupancies are calculated for idealised cells in which the sensors are at the mid‐points of their ranges. Total calculated metal occupancies are 16% CbiK, 99% MgIIGTP‐CobW, 36% MgIIGTP‐YeiR and (using the selected K A MnII) 91% MncA, implying substantial amounts of apo‐CbiK and apo‐YeiR exist under these conditions. Online metalation calculators similarly decode metal occupancies in the context of defined metal availabilities (https://mib‐nibb.webspace.durham.ac.uk/metalation‐calculators/) [53].
Decoding metalation
The insets in Figs 1 and 2 depict fractional occupancies of each protein with metals: copper black, ZnII grey, CoII salmon red, FeII green and MnII pink. Once competition against the intracellular milieu is considered, calculated metalation switches from the black and grey insets in Fig. 1 (representing the tightest but wrong metals copper and ZnII) to the colours in Fig. 2 [representing the correct metals CoII (MgIIGTP‐CobW), ZnII (MgIIGTP‐YeiR), CoII (CbiK) and MnII (MncA)]. These occupancies have been estimated for an idealised cell in which metal‐availabilities match the mid‐points of the ranges for the respective sensor(s) [3, 4, 5]. They have been calculated from the difference in free energy for forming the respective metal complex with the protein of interest, versus that inferred for the competing intracellular milieu. Metalation can occur if the gradient favours transfer to the protein [3, 5]. The gradient may be favourable for more than one metal and hence values have been inter‐competed to ensure that occupancy of a single site does not exceed a stoichiometry of one [5]. Intriguingly, in the absence of nucleotide or with MgIIGDP, the gradient disfavours transfer of CoII to CobW, whereas MgIIGTP‐CobW favours CoII‐transfer, providing insight into the mechanistic cycle for this metallochaperone [5]. Because metals are trapped within MncA it is difficult to determine the affinities of the nascent site at which binding occurs during folding and, as noted earlier, values have only been estimated for the relative affinities of three metals [4]. Fig. 2D therefore, shows the free energies for forming ZnII complexes (and limits for CuI complexes) with the nascent site in MncA relative to an assigned MnII value.
Metalation calculators have been created which perform analogous calculations. They enable simulations of intracellular metalation of proteins of interest from inputted metal affinities and either by using default metal availabilities originally estimated for Salmonella, or by inputting known or simulated metal availabilities for other organisms (such as E. coli) [53].
New frontiers in protein metalation
Questions and methods
To what extent does metal‐protein speciation depart from predictions based on differences in free energies for complex formation relative to competing intracellular sites? Constraints and uses of the approach described here have been catalogued [53]. Additional factors that could influence metalation include kinetic contributions such as proximity to sites of metal import (where availability departs from that averaged over the compartment as a whole) and selective interactions with metal buffering molecules, including metallochaperones as extreme examples. The scale of such additional contributions could become evident from the extent to which observed metalation departs from the predictions of metalation calculators. The disparities may be relatively small, and hence at risk of being dismissed, but viewed in the context of the landscape of competition from intracellular‐binding sites their crucial contributions to correct metalation might become evident.
To what extent do the metal‐binding preferences of some proteins depart from the Irving–Williams series? A cautionary note is that metal affinities of proteins can be challenging to measure and many reported values are not correct [54, 55]. We have already discussed how the formation of adducts with other molecules such as MgIIGTP can pre‐organise a binding site to introduce steric selection [5]. It is known that cooperativity at di‐metal sites can similarly improve selectivity, for example, in favour of MnII relative to FeII in a class Ib diMnII ribonucleotide reductase and in the MnII/FeII oxidase R2lox [56, 57]. Change in oxidation state post‐binding, away from that of the labile pool, can favour retention of a selected metal [58]. Synthetic proteins have been generated with metal preferences that depart from the Irving–Williams series [59]. If better metal selectivity could have evolved, why has not it? Perhaps, because greater selectivity comes at a price such as reduced flexibility at the active site diminishing the catalytic repertoire [57, 59]. Perhaps because there has been limited pressure for greater selectivity when evolution has occurred within the thermodynamic landscape for metal availabilities shown in Fig. 2. Evolution of metal homeostasis, rather than adaptation of the vast complement of metal sites, has probably offered the more parsimonious solution when metal supply has changed over time.
By how much does metal availability vary in different compartments and organisms? It is anticipated that availability is the inverse of the Irving–Williams series in the compartments of most cells (e.g. albeit CuI may be substantially more available in the trans‐Golgi network of eukaryotic cells). Existing metalation calculators could initially be extrapolated to simulations for other cell types. However, modest change in availabilities of two metals, but in opposing directions (one more available, one less available), could switch the specificity of metalation. Thus, bespoke calculators should ideally be generated by substituting availabilities determined for the respective compartment and growth condition, albeit using the same understandings and web‐based template. However, it took about a decade to calibrate the sensors of Salmonella and in many compartments and species it is less clear which cellular sensors could be used to replicate this approach. The metal affinities of sites that modulate the trafficking or processing of metal‐transporters, change the stability or translatability of transcripts encoding proteins of metal homeostasis, or modulate other post‐transcriptional mechanisms, offer a possible route to define metal availability in idealised cellular compartments. However, it is less clear how these sites could be used to read out availabilities in conditional cells. An attractive idea is to use artificial intelligence to predict availabilities based on global surveys of protein metal affinities. However, a preponderance of erroneous affinities in the literature could confound the signal to noise ratio. Better yet, metal‐responsive probes, including cell permeable chromogenic molecules, have been developed [60, 61, 62]. There is uncertainty about what some probes read‐out in a biological context: But these uncertainties seem resolvable such that the probes could be calibrated to read‐out the free energies of available metal. Furthermore, these probes could be cross‐correlated inside Salmonella by comparison with values obtained from the characterised DNA‐binding metal sensors. This latter approach may allow the generation of bespoke metalation calculators for a variety of cells and compartments to be more swiftly created.
Applications
The term nutritional immunity encompasses mechanisms by which pathogens are subjected to metal excess or deficiency as part of the host defences [23, 30, 63, 64]. This includes the sequestration of metals by calprotectin released from neutrophils [65, 66], the depletion of metals in macrophage phagosomes by natural resistance‐associated macrophage protein one [67, 68], the elevation of copper in the same compartment [69], the sequestration of iron scavenging siderophores by siderocalins [70], amongst others. There is a history of using metals and chelants to limit the growth of pathogens in medicine, in agriculture and in consumer goods [71]. Knowledge of the activities of available metals in pathogens should enable the identification of proteins that are liable to mis‐metalation, creating opportunities to tailor antimicrobials to subvert metalation, for example of enzymes involved in antimicrobial resistance.
Mis‐metalation occurs in some diseases [72]. This might be a primary cause or a secondary symptom. Metalation could be simulated for proteins associated with such diseases to identify those liable to mis‐metalation, perhaps informing future treatments. Such simulations could use metalation calculators in which availabilities have been entered that match determined free energies of available metals in the respective compartment of human cells. The latter might be determined using cell permeable chromogenic probes as discussed earlier. In plants, the generation of analogous calculators for their varied compartments has the potential to assist approaches to improve the nutritional supply of metals associated with hidden hunger [73].
The sources of some technology‐critical metals required in electronic devices and batteries are at risk [74]. This generates a need for targeted metal recovery and sustainable recycling. Sensors are known that detect several non‐essential metals and there is scope to identify more [33, 34, 75, 76, 77, 78]. These sensors could be calibrated to monitor and quantify sub‐lethal intracellular availabilities of the critical elements. In turn, this knowledge would assist the engineering of accumulation and bio‐recovery of technology‐critical metals.
In synthetic biology, heterologous (introduced) proteins may be mis‐matched to metal availabilities in the engineered cells. There is scope to use metalation calculators for organisms such as E. coli, and in future yeast, to optimise metalation in support of sustainable industrial biotechnology. The heterologous proteins might be products of in vitro evolution or of targeted engineering. Encouragingly, these proteins need not be engineered for the tightest binding metal to be the correct metal, but merely to meet the more attainable goal of acquiring the correct metal in the context of the prevailing intracellular availabilities as illustrated in Fig. 2. The pathway for synthesis of cofactor F430 stalled at the point of NiII insertion; likewise, that for vitamin B12 stalled at CoII insertion, when introduced into E. coli, which does not naturally produce either molecule [5, 79]. The latter vitamin B12 pathway involved CobW as in Figs 1B and 2B. Calibrated qPCR with E. coli transcripts indicated that intracellular CoII availability in cells grown in LB medium was below 10% of the range for CoII‐sensing RcnR, while ZnII approximated to the mid‐point for ZnII‐sensing ZntR and Zur, equating to idealised cells for ZnII but not for CoII [5]. Under these conditions, MgIIGTP‐CobW is predicted to be ~75% mis‐metalated with ZnII [5]. Supplementation of culture media with 10 μm cobalt was estimated (via qPCR) to raise the intracellular free energy of available CoII sufficiently to reverse mis‐metalation of the metallochaperone and indeed under these conditions, vitamin B12 synthesis proceeded, matching calculated loading of MgIIGTP‐CobW with CoII [5]. This presents opportunities to optimise the bioprocess for manufacturing vitamin B12 by manipulating the supply of CoII or ZnII via supplementation and chelation or by further engineering metal homeostasis. Importantly, vitamin B12 is neither made nor used by plants with the vegan society recommending supplements [80, 81]. As individuals adopt more plant‐based diets to reduce environmental demand for food production, efficient bio‐manufacture of vitamin B12 may gain in importance. Web‐based metalation calculators are now available for E. coli strains grown under specified culture conditions with plans to iteratively update the resource (https://mib‐nibb.webspace.durham.ac.uk/metalation‐calculators/) [53]. With an estimated half of the reactions of life requiring metals, optimisation of metalation informed by metalation calculators, promises to assist the transition to more sustainable manufacturing.
Acknowledgements
In memory of Deenah Morton (née Osman). We thank members of the research group, Tessa Young, Sophie Clough, William Michaels, and Arthur Glasfeld for valuable insights. Metalation and mis‐metalation research, plus the generation of metalation calculators, is supported by Biotechnology and Biological Sciences Research Council awards BB/V006002/1 and BB/W015749/1.
Edited by Martin Högbom
Data accessibility
Data sharing is not applicable as no new data were created or analysed in this review.
References
- 1. Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: from enzyme databases to general principles. J Biol Inorg Chem. 2008;13(8):1205–18. [DOI] [PubMed] [Google Scholar]
- 2. Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460(7257):823–30. [DOI] [PubMed] [Google Scholar]
- 3. Osman D, Martini MA, Foster AW, Chen J, Scott AJP, Morton RJ, et al. Bacterial sensors define intracellular free energies for correct enzyme metalation. Nat Chem Biol. 2019;15(3):241–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tottey S, Waldron KJ, Firbank SJ, Reale B, Bessant C, Sato K, et al. Protein‐folding location can regulate manganese‐binding versus copper‐ or zinc‐binding. Nature. 2008;455(7216):1138–42. [DOI] [PubMed] [Google Scholar]
- 5. Young TR, Martini MA, Foster AW, Glasfeld A, Osman D, Morton RJ, et al. Calculating metalation in cells reveals CobW acquires CoII for vitamin B12 biosynthesis while related proteins prefer ZnII. Nat Commun. 2021;12(1):1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Da Silva JF, Williams RJP. The biological chemistry of the elements: the inorganic chemistry of life. Oxford: Oxford University Press; 2001. [Google Scholar]
- 7. Irving H, Williams R. Order of stability of metal complexes. Nature. 1948;162(4123):746–7. [Google Scholar]
- 8. Osman D, Cooke A, Young TR, Deery E, Robinson NJ, Warren MJ. The requirement for cobalt in vitamin B12: a paradigm for protein metalation. Biochim Biophys Acta Mol Cell Res. 2021;1868(1):118896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Raux E, Thermes C, Heathcote P, Rambach A, Warren MJ. A role for Salmonella typhimurium cbiK in cobalamin (vitamin B12) and siroheme biosynthesis. J Bacteriol. 1997;179(10):3202–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schubert HL, Raux E, Wilson KS, Warren MJ. Common chelatase design in the branched tetrapyrrole pathways of heme and anaerobic cobalamin synthesis. Biochemistry. 1999;38(33):10660–9. [DOI] [PubMed] [Google Scholar]
- 11. Lacasse MJ, Douglas CD, Zamble DB. Mechanism of selective nickel transfer from HypB to HypA, Escherichia coli [NiFe]‐hydrogenase accessory proteins. Biochemistry. 2016;55(49):6821–31. [DOI] [PubMed] [Google Scholar]
- 12. Watanabe S, Kawashima T, Nishitani Y, Kanai T, Wada T, Inaba K, et al. Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni‐metallochaperone HypA and its enhancer. Proc Natl Acad Sci U S A. 2015;112(25):7701–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Banci L, Bertini I, Cantini F, Felli IC, Gonnelli L, Hadjiliadis N, et al. The Atx1‐Ccc2 complex is a metal‐mediated protein‐protein interaction. Nat Chem Biol. 2006;2(7):367–8. [DOI] [PubMed] [Google Scholar]
- 14. Yang X, Li H, Lai TP, Sun H. UreE‐UreG complex facilitates nickel transfer and preactivates GTPase of UreG in Helicobacter pylori . J Biol Chem. 2015;290(20):12474–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crouzet J, Levy‐Schil S, Cameron B, Cauchois L, Rigault S, Rouyez MC, et al. Nucleotide sequence and genetic analysis of a 13.1‐kilobase‐pair Pseudomonas denitrificans DNA fragment containing five cob genes and identification of structural genes encoding Cob(I)alamin adenosyltransferase, cobyric acid synthase, and bifunctional cobinamide kinase‐cobinamide phosphate guanylyltransferase. J Bacteriol. 1991;173(19):6074–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blaby‐Haas CE, Flood JA, Crécy‐Lagard V, Zamble DB. YeiR: a metal‐binding GTPase from Escherichia coli involved in metal homeostasis. Metallomics. 2012;4(5):488–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haas CE, Rodionov DA, Kropat J, Malasarn D, Merchant SS, de Crécy‐Lagard V. A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life. BMC Genomics. 2009;10(1):470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Privalle CT, Fridovich I. Transcriptional and maturational effects of manganese and iron on the biosynthesis of manganese‐superoxide dismutase in Escherichia coli . J Biol Chem. 1992;267(13):9140–5. [PubMed] [Google Scholar]
- 19. Anjem A, Varghese S, Imlay JA. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli . Mol Microbiol. 2009;72(4):844–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ranquet C, Ollagnier‐de‐Choudens S, Loiseau L, Barras F, Fontecave M. Cobalt stress in Escherichia coli. The effect on the iron‐sulfur proteins. J Biol Chem. 2007;282(42):30442–51. [DOI] [PubMed] [Google Scholar]
- 21. Labbé RF, Vreman HJ, Stevenson DK. Zinc protoporphyrin: a metabolite with a mission. Clin Chem. 1999;45(12):2060–72. [PubMed] [Google Scholar]
- 22. Bird AJ. Cellular sensing and transport of metal ions: implications in micronutrient homeostasis. J Nutr Biochem. 2015;26(11):1103–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chandrangsu P, Rensing C, Helmann JD. Metal homeostasis and resistance in bacteria. Nat Rev Microbiol. 2017;15(6):338–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clemens S. The cell biology of zinc. J Exp Bot. 2021;73(6):1688–98. [DOI] [PubMed] [Google Scholar]
- 25. Ma Z, Jacobsen FE, Giedroc DP. Coordination chemistry of bacterial metal transport and sensing. Chem Rev. 2009;109(10):4644–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Merchant SS, Helmann JD. Elemental economy: microbial strategies for optimizing growth in the face of nutrient limitation. Adv Microb Physiol. 2012;60:91–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nevitt T, Öhrvik H, Thiele DJ. Charting the travels of copper in eukaryotes from yeast to mammals. Biochim Biophys Acta Mol Cell Res. 2012;1823(9):1580–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Palmer CM, Guerinot ML. Facing the challenges of Cu, Fe and Zn homeostasis in plants. Nat Chem Biol. 2009;5(5):333–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Podar D, Maathuis FJM. Primary nutrient sensors in plants. iScience. 2022;25(4):104029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Murdoch CC, Skaar EP. Nutritional immunity: the battle for nutrient metals at the host–pathogen interface. Nat Rev Microbiol. 2022. 10.1038/s41579-022-00745-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morby AP, Turner JS, Huckle JW, Robinson NJ. SmtB is a metal‐dependent repressor of the cyanobacterial metallothionein gene smtA: identification of a Zn inhibited DNA‐protein complex. Nucleic Acids Res. 1993;21(4):921–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hantke K. Cloning of the repressor protein gene of iron‐regulated systems in Escherichia coli K12. Mol Gen Genet. 1984;197(2):337–41. [DOI] [PubMed] [Google Scholar]
- 33. Lund PA, Ford SJ, Brown NL. Transcriptional regulation of the mercury‐resistance genes of transposon Tn501. J Gen Microbiol. 1986;132(2):465–80. [DOI] [PubMed] [Google Scholar]
- 34. O'Halloran T, Walsh C. Metalloregulatory DNA‐binding protein encoded by the merR gene: isolation and characterization. Science. 1987;235(4785):211–4. [DOI] [PubMed] [Google Scholar]
- 35. Foster AW, Pernil R, Patterson CJ, Scott AJP, Pålsson LO, Pal R, et al. A tight tunable range for Ni(II) sensing and buffering in cells. Nat Chem Biol. 2017;13(4):409–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Osman D, Foster AW, Chen J, Svedaite K, Steed JW, Lurie‐Luke E, et al. Fine control of metal concentrations is necessary for cells to discern zinc from cobalt. Nat Commun. 2017;8:1884. 10.1038/s41467-017-02085-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413(6858):852–6. [DOI] [PubMed] [Google Scholar]
- 38. VanZile ML, Chen X, Giedroc DP. Allosteric negative regulation of smt O/P binding of the zinc sensor, SmtB, by metal ions: a coupled equilibrium analysis. Biochemistry. 2002;41(31):9776–86. [DOI] [PubMed] [Google Scholar]
- 39. Pennella MA, Arunkumar AI, Giedroc DP. Individual metal ligands play distinct functional roles in the zinc sensor Staphylococcus aureus CzrA. J Mol Biol. 2006;356(5):1124–36. [DOI] [PubMed] [Google Scholar]
- 40. Outten CE, O'Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292(5526):2488–92. [DOI] [PubMed] [Google Scholar]
- 41. Robinson NJ, Winge DR. Copper metallochaperones. Annu Rev Biochem. 2010;79(1):537–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weiss A, Murdoch CC, Edmonds KA, Jordan MR, Monteith AJ, Perera YR, et al. Zn‐regulated GTPase metalloprotein activator 1 modulates vertebrate zinc homeostasis. Cell. 2022;185(12):2148–2163.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lamb AL, Torres AS, O'Halloran TV, Rosenzweig AC. Heterodimeric structure of superoxide dismutase in complex with its metallochaperone. Nat Struct Biol. 2001;8(9):751–5. [DOI] [PubMed] [Google Scholar]
- 44. Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O'Halloran TV. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science. 1999;284(5415):805–8. [DOI] [PubMed] [Google Scholar]
- 45. Foster AW, Young TR, Chivers PT, Robinson NJ. Protein metalation in biology. Curr Opin Chem Biol. 2022;66:102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ma Z, Chandrangsu P, Helmann TC, Romsang A, Gaballa A, Helmann JD. Bacillithiol is a major buffer of the labile zinc pool in Bacillus subtilis. Mol Microbiol. 2014;94(4):756–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Murphy JT, Bruinsma JJ, Schneider DL, Collier S, Guthrie J, Chinwalla A, et al. Histidine protects against zinc and nickel toxicity in Caenorhabditis elegans . PLoS Genet. 2011;7(3):e1002013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nairn B, Lonergan ZR, Wang J, Braymer JJ, Zhang Y, Calcutt MW, et al. The response of Acinetobacter baumannii to zinc starvation. Cell Host Microbe. 2016;19(6):826–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Morgan MT, Nguyen LAH, Hancock HL, Fahrni CJ. Glutathione limits aquacopper(I) to sub‐femtomolar concentrations through cooperative assembly of a tetranuclear cluster. J Biol Chem. 2017;292(52):21558–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brawley HN, Lindahl PA. Low‐molecular‐mass labile metal pools in Escherichia coli: advances using chromatography and mass spectrometry. J Biol Inorg Chem. 2021;26(4):479–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stewart LJ, Ong CY, Zhang MM, Brouwer S, McIntyre L, Davies MR, et al. Role of glutathione in buffering excess intracellular copper in Streptococcus pyogenes . MBio. 2020;11(6):e02804–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hider RC, Kong XL. Glutathione: a key component of the cytoplasmic labile iron pool. Biometals. 2011;24(6):1179–87. [DOI] [PubMed] [Google Scholar]
- 53. Foster AW, Clough SE, Aki Z, Young TR, Clarke AR, Robinson NJ. Metalation calculators for E. coli strain JM109 (DE3): aerobic, anaerobic and hydrogen peroxide exposed cells cultured in LB media. Metallomics. 2022;14(9):mfac058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xiao Z, Wedd AG. The challenges of determining metal‐protein affinities. Nat Prod Rep. 2010;27(5):768–89. [DOI] [PubMed] [Google Scholar]
- 55. Young TR, Xiao Z. Principles and practice of determining metal–protein affinities. Biochem J. 2021;478(5):1085–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Grāve K, Griese JJ, Berggren G, Bennett MD, Högbom M. The Bacillus anthracis class Ib ribonucleotide reductase subunit NrdF intrinsically selects manganese over iron. J Biol Inorg Chem. 2020;25(4):571–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kisgeropoulos EC, Griese JJ, Smith ZR, Branca RMM, Schneider CR, Högbom M, et al. Structural motifs balance metal binding and oxidative reactivity in a heterobimetallic Mn/Fe protein. J Am Chem Soc. 2020;142(11):5338–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cotruvo JA, Stubbe J. Escherichia coli Class Ib ribonucleotide reductase contains a dimanganese(III)‐tyrosyl radical cofactor in vivo. Biochemistry. 2011;50(10):1672–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Choi TS, Tezcan FA. Overcoming universal restrictions on metal selectivity by protein design. Nature. 2022;603(7901):522–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zastrow ML, Huang Z, Lippard SJ. HaloTag‐based hybrid targetable and ratiometric sensors for intracellular zinc. ACS Chem Biol. 2020;15(2):396–406. [DOI] [PubMed] [Google Scholar]
- 61. Chung CY‐S, Posimo JM, Lee S, Tsang T, Davis JM, Brady DC, et al. Activity‐based ratiometric FRET probe reveals oncogene‐driven changes in labile copper pools induced by altered glutathione metabolism. Proc Natl Acad Sci. 2019;116(37):18285–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fahrni CJ. Synthetic fluorescent probes for monovalent copper. Curr Opin Chem Biol. 2013;17(4):656–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Antelo GT, Vila AJ, Giedroc DP, Capdevila DA. Molecular evolution of transition metal bioavailability at the host–pathogen interface. Trends Microbiol. 2021;29(5):441–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Weiss G, Carver PL. Role of divalent metals in infectious disease susceptibility and outcome. Clin Microbiol Infect. 2018;24(1):16–23. [DOI] [PubMed] [Google Scholar]
- 65. Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319(5865):962–5. [DOI] [PubMed] [Google Scholar]
- 66. Zygiel EM, Nelson CE, Brewer LK, Oglesby‐Sherrouse AG, Nolan EM. The human innate immune protein calprotectin induces iron starvation responses in Pseudomonas aeruginosa . J Biol Chem. 2019;294(10):3549–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vidal SM, Malo D, Vogan K, Skamene E, Gros P. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell. 1993;73(3):469–85. [DOI] [PubMed] [Google Scholar]
- 68. Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, et al. Cloning and characterization of a mammalian proton‐coupled metal‐ion transporter. Nature. 1997;388(6641):482–8. [DOI] [PubMed] [Google Scholar]
- 69. Osman D, Waldron KJ, Denton H, Taylor CM, Grant AJ, Mastroeni P, et al. Copper homeostasis in Salmonella is atypical and copper‐CueP is a major periplasmic metal complex. J Biol Chem. 2010;285(33):25259–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432(7019):917–21. [DOI] [PubMed] [Google Scholar]
- 71. Lemire JA, Harrison JJ, Turner RJ. Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat Rev Microbiol. 2013;11(6):371–84. [DOI] [PubMed] [Google Scholar]
- 72. Adlard PA, Bush AI. Metals and Alzheimer's disease. J Alzheimers Dis. 2006;10:145–63. [DOI] [PubMed] [Google Scholar]
- 73. Assunção AGL, Cakmak I, Clemens S, González‐Guerrero M, Nawrocki A, Thomine S. Micronutrient homeostasis in plants for more sustainable agriculture and healthier human nutrition. J Exp Bot. 2022;73(6):1789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Royal Society of Chemistry . Elements in danger. 2022. [cited 2022 July 28]. Available from: https://www.rsc.org/new‐perspectives/sustainability/elements‐in‐danger/
- 75. Borremans B, Hobman JL, Provoost A, Brown NL, van der Lelie D. Cloning and functional analysis of the pbr lead resistance determinant of Ralstonia metallidurans CH34. J Bacteriol. 2001;183(19):5651–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pontel LB, Audero MEP, Espariz M, Checa SK, Soncini FC. GolS controls the response to gold by the hierarchical induction of Salmonella‐specific genes that include a CBA efflux‐coding operon. Mol Microbiol. 2007;66(3):814–25. [DOI] [PubMed] [Google Scholar]
- 77. Xu C, Shi W, Rosen BP. The chromosomal arsR gene of Escherichia coli encodes a trans‐acting metalloregulatory protein (*). J Biol Chem. 1996;271(5):2427–32. [DOI] [PubMed] [Google Scholar]
- 78. Endo G, Silver S. CadC, the transcriptional regulatory protein of the cadmium resistance system of Staphylococcus aureus plasmid pI258. J Bacteriol. 1995;177(15):4437–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Moore SJ, Sowa ST, Schuchardt C, Deery E, Lawrence AD, Ramos JV, et al. Elucidation of the biosynthesis of the methane catalyst coenzyme F430. Nature. 2017;543(7643):78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Pawlak R, Parrott SJ, Raj S, Cullum‐Dugan D, Lucus D. How prevalent is vitamin B(12) deficiency among vegetarians? Nutr Rev. 2013;71(2):110–7. [DOI] [PubMed] [Google Scholar]
- 81. The Vegan Society . What every vegan should know about vitamin B12. [cited 2022 July 27]. Available from: https://www.vegansociety.com/resources/nutrition‐and‐health/nutrients/vitamin‐b12/what‐every‐vegan‐should‐know‐about‐vitamin‐b12
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable as no new data were created or analysed in this review.


