Abstract
The expansion of intratumoral stem-like/progenitor exhausted CD8+ T (Tstem/Tpex) cells provides a potential approach to improve the therapeutic efficacy of immune checkpoint blockade (ICB). Thus, here we demonstrate a strategy to facilitate Tstem/Tpex cell expansion by combining an alarmin high-mobility group nucleosome binding domain 1 (HMGN1) peptide with programmed death-ligand 1 (PD-L1) blockade. The antitumor effects of HMGN1, anti–PD-L1, and their combined treatment were monitored in the B16F10, LLC, Colon26, or EO771 tumor–bearing mice. The comprehensive immunologic analyses, such as high-dimensional flow cytometry, transcriptome analysis, and single-cell RNA-sequencing (scRNA-seq), were used to investigate the cellular and molecular mechanisms of antitumor immune responses after treatments. We identified the immunostimulatory domain (EPKRR SARLS AKPPA KVEAK PKK) on HMGN1 and synthesized this domain as a therapeutic peptide (minP1). Combined treatment with minP1 and PD-L1 blockade induced durable tumor regression in tumor-bearing mice. minP1 increased the number of intratumoral mature DCs enriched in immunoregulatory molecules (mregDC) and enhanced their MHC class I antigen–presenting program. minP1 also synergized with PD-L1 blockade in augmenting intratumoral Tstem/Tpex cell number. Analysis of our scRNA-seq dataset by CellPhonDB suggested potential interactions between mregDCs and Tstem/Tpex cells in tumors. Our results indicate that HMGN1 peptide (minP1) serves as an immunoadjuvant to promote effective anti–PD-L1 immunotherapy with increased Tstem/Tpex cells in tumors.
Introduction
Blocking programmed cell death 1 and its ligand programmed death ligand 1 (PD-1/PD-L1) can increase survival in patients with cancer by reinvigorating the functions of tumor-reactive CD8+ T cells (1-5); however, not all patients respond equally well to PD-1/PD-L1 blockade. The overall response rate to PD-1/PD-L1 blockade alone is below 30% in patients with melanoma, non–small cell lung carcinoma, ovarian cancer, and renal cell cancer (6, 7). Low response rates to immune checkpoint blockade (ICB) therapies are associated with inadequate antigen presentation and insufficient generation of tumor-reactive CD8+ T cells (8, 9). To address these issues, researchers have attempted to combine various types of immune stimulators with PD-1/PD-L1 blockade to strengthen the antigen-presenting function of intratumoral dendritic cells (DC) and accelerate the generation of tumor-reactive CD8+ T cells, such as R848 (a TLR-7 agonist; ref. 10), CpG oligodeoxynucleotide (ODN; a TLR-9 agonist; ref. 11), or SD-101 (a novel class of CpG-ODN TLR9 agonist; ref. 12). Here, we explored the possibility of combining an immune stimulator called high-mobility nucleosome binding domain 1 (HMGN1) with PD-L1 blockade.
HMGN1 contains two functional domains, a nucleosome binding domain (NBD) and a chromatin unfolding domain (CHUD; refs. 13, 14), and functions as both an intracellular and extracellular regulatory protein (15). Intracellular HMGN1 is a transcriptional factor that supports higher order chromatin folding and gene regulation (13, 14). Extracellular HMGN1 acts as an alarmin, which is released from apoptotic or necrotic cells, to promote DC activation via MYD88 innate immune signal transduction adaptor (MYD88)– and toll-like receptor adaptor molecule 1 (TRIF)–dependent signaling pathways (16-20). Currently, recombinant HMGN1 is considered to act as a DC adjuvant for T-cell–mediated cancer therapy due to its benefits of promoting CD8+ T-cell expansion (17-19, 21). As reported previously, an in vitro assay demonstrated that recombinant HMGN1 can promote DC-dependent CD8+ T-cell expansion in cocultured Pmel-1 CD8+ T cells with gp100-pulsed bone marrow (BM)–derived DCs (21). An in vivo study also demonstrated that HMGN1 synergizes anti-CD4–depleting antibody treatment in enhancing the expansion of intratumoral CD8+ T cells in mice (21). Thus, we speculated that combining HMGN1 with PD-L1 blockade may facilitate intratumoral CD8+ T-cell expansion.
Intratumoral CD8+ T-cell expansion is a promising strategy to improve the therapeutic efficacy of ICB, particularly if it results in the expansion of a newly defined progenitor exhausted CD8+ T (Tpex) cell subset with proliferation and differentiation capacities (22, 23), which acts as resource cells to mediate tumor control in response to ICB therapy (23-25). The Tpex subset is also known as precursor-exhausted (25), stem-like (24, 26, 27), or memory-like CD8+ T cells (27, 28). However, because there is no specific term for this T-cell subset, here we used stem-like/progenitor exhausted CD8+ T (Tstem/Tpex) to satisfy a wide range of readers. There is increasing evidence that the Tstem/Tpex subset is characterized by high expression of SLAM family member 6 (SLAMF6, also known as Ly108), and transcriptional factor TCF1 (encoded by the Tcf7 gene); the intermediate expression of PD-1; low expression of T-cell immunoglobulin and mucin domain 3 (TIM-3; also known as hepatitis A virus cellular receptor 2, HAVCR2) and lymphocyte-activation gene 3 (LAG-3); and high proliferative capacity (22-24, 29, 30). Patients with melanoma who had increased numbers of Tstem/Tpex cells display durable tumor regression and longer progression-free survival after ICB therapy (23, 31). Conversely, a lack of adequate Tstem/Tpex cells or an abundance of terminally exhausted CD8+ T (Tex) cells restricts the therapeutic efficacy of ICB therapy and limits the survival benefits to both tumor-bearing mice and patients with cancer (31).
Here, we demonstrate a new strategy to facilitate Tstem/Tpex cell expansion by combining a HMGN1-derived immunostimulatory peptide with PD-L1 blockade. The HMGN1 peptide synergized with PD-L1 blockade to increase the number and antigen-presenting ability of tumor-infiltrating mature DCs enriched in immunoregulatory molecules (mregDC), which correlated with the expansion of Tstem/Tpex cells in tumors. Using single-cell RNA-sequencing (scRNA-seq), we predicted the cell–cell interactions among mregDC, Tstem/Tpex, and Tex subsets by CellPhoneDB. The regulatory molecules on mregDCs may support Tstem/Tpex cells but not Tex-cell activation and expansion.
Materials and Methods
Identification and generation of immunostimulatory HMGN1 peptides
Recombinant HMGN1 proteins were produced in E. coli system. The expression pET-17b plasmid with murine HMGN1 coding region (122–412) or human HMGN1 coding region (205–507) was transformed into BL21 Star (DE3) E. coli (Thermo Fisher Scientific) by heat shock method. The single colony was picked up and precultivated in an LB/ampicillin medium until the OD600 absorbance reaches 0.6. Added isopropyl β-D-1-thiogalactopyranoside (IPTG, final concentration equal to 1 mmol/L) into culture medium and cultivated the cells for 4 hours. The cell pellet was collected by centrifugation at 15,000 × g for 5 minutes, and then the soluble proteins were extracted from the cell pellet by sonication. The recombinant HMGN1 proteins were then purified using sequential fractionation by heparin affinity column, ion exchange column, and reverse-phase column as previously described (21). The purity of HMGN1 (>99%) was confirmed by SDS-PAGE. The absolute endotoxin concentration of murine HMGN1 (0.0008 endotoxin unit per mg (EU/mg) and human HMGN1 (0.001 EU/mg) were assessed by Endospecy ES-50M Kit (Seikagaku Corporation). The exact amount of endotoxin remained in the range of 0.08 μg to 0.4 μg for HMGN1 per injection is in the ranged from <8 × 10−8 ng to 4 × 10−7 ng. The protein sequence of HMGN1 was identified by Applied Biosystems Procise 492 HT (Thermo Fisher Scientific). To negate the possibility of endotoxin and/or additional bacterial component contamination in HMGN1 peptide preparation, all HMGN1 peptides (Supplementary Table S1) were chemically synthesized by Eurofins Genomics.
Mice
Seven-week-old female BALB/c and C57BL/6 were purchased from Japan SLC, Inc. The IL12-YFP reporter C57BL/6 mice (B6.129-Il12btm1.1Lky/J) were purchased from the Jackson Laboratory. For tumor growth experiments, each group contained 8 mice except where otherwise specified. All animal experiments were conducted in accordance with institutional guidelines with the approval of the Animal Care and Use Committee of The University of Tokyo and Tokyo University of Science.
Cell lines and tumor models
The murine tumor cell line Colon26 was obtained from the Cell Resource Center for Biomedical Research (RRID: CVCL_0240; Cell Banker, RIKEN BRC, Japan). The Lewis lung carcinoma (LLC) cell line was provided from Dr. F. Abe (RRID: CVCL_4358; Nipponkayaku). The cell line B16F10 was obtained from the American Type Culture Collection (ATCC; RRID: CVCL_0159). The cell line EO771 was obtained from CH3 BioSystems (CVCL_GR23). Colon26, LLC, and EO771 cell lines were cultured in RPMI1640 medium supplemented with 10% heat-inactivated FBS, 1% penicillin/streptomycin, and 10 mmol/L HEPES. B16F10 cell line was cultured in DMEM supplemented with 10% heat-inactivated FBS, 1% penicillin/streptomycin, and 10 mmol/L HEPES. All cell lines were tested for Mycoplasma, cultured at 37 °C in a humidified 5% CO2 incubator, and maintained by subculturing every 2 to 3 days for 2 weeks before use. Colon26 (2 × 105 cells), LLC (2 × 105 cells), and B16F10 (5 × 105) cells were subcutaneously inoculated into the right flank of mice (BALB/c mice for Colon26; C57BL/6 mice for LLC and B16F10). EO771 (2 × 105 cells) cells were implanted into the inguinal mammary fat pad of C57BL/6 mice. Tumor diameter was measured twice weekly and used to calculate tumor volume (V, mm3) using the formula V = L × W × W/2; L is tumor length and W is tumor width. The maximum tumor volume did not exceed 2,000 mm3 for mice, which is the endpoint for each tumor growth analysis. For tumor rechallenge, the parental tumor cell line or a different tumor cell line was subcutaneously inoculated into the right flank of mice that cleared tumors after treatments. Intraperitoneal injections of anti–PD-L1 (200 μg/injection on days 4, 8, 14, and 18; clone 10F.9G2; BioXcell), and intraperitoneal injections of HMGN1 and its derived peptides (Supplementary Table S1), including minP1 (0.08 μg/injection on days 9, 14, 17, and 20), were given to tumor-bearing mice. For dose–response experiments, anti–PD-L1 and various doses (0.008 to 2 μg/mL) of HMGN1 proteins and peptides were administered intraperitoneally to Colon26- or B16F10-bearing mice.
Immune cell isolation
Each tumor was cut into small pieces and digested for 45 minutes at 37°C with 0.1% collagenase (FUJIFILM Wako) at 180 rpm in a shaker. The total cells went through density separation with 35% and 65% Percoll PLUS (Cytiva), and leukocytes were recovered from the middle layer. Ammonium chloride potassium (ACK) buffer was used to lyse red blood cells. The live cell number was counted using Flow-Count fluorospheres (Beckman Coulter) plus propidium iodide (PI; Biolegend) and a CytoFLEX flow cytometer (Beckman Coulter). Flow cytometric data were analyzed using Flowjo software (version 10.5; BD Biosciences). For enrichment of CD8+ T cells, an anti-CD8-biotin and a Streptavidin MicroBeads (Miltenyi Biotec Inc) were used to perform magnetic separation of CD8+ T cells from the tumor. Finally, the enriched CD8+ T cells (85% to 90% purity) were further sorted by using FACSAria III Cell Sorter (BD Biosciences). The purity of sorted CD8+ T cells or sorted Ly108+TIM-3−, Ly108+TIM-3+, and Ly108−TIM-3+ CD8+ T-cell subsets were consistently over 95%. For the bulk RNA-sequencing (RNA-seq) library preparation and transcriptome analysis, the sorted total CD8+ T cell and its subsets were used.
Flow cytometry
Unless otherwise stated, total cell suspensions from the tumor tissues of Colon26 or LLC tumor–bearing mice on day 12 (after two rounds of minP1 and anti–PD-L1 treatments) and 16 (after three rounds of minP1 and anti–PD-L1 treatments) after tumor inoculation were collected. Three minutes before collecting tissues, intravascular leukocytes were stained by intravenous injection of fluorescein isothiocyanate (FITC)–conjugated antibody (3 μg/mouse) against CD45 (32). Total cell suspensions were prepared by enzymatic and mechanical dissociation of tissues, as described previously (33). Flow-Count Fluorospheres (Beckman Coulter) were used to determine cell numbers. Cells were pretreated with Fc blocking reagent (anti-mouse CD16/CD32 mAb, clone 2.4G2; BioXcell), then stained with a mixture of fluorophore-conjugated anti-mouse antibodies (Supplementary Table S1). Data were acquired on a CytoFlex flow cytometer (Beckman Coulter) and analyzed by using Flowjo 10.5 software (Flowjo, LLC). The forward and side scatter profiles were first used to exclude doublets and debris first, and dead cells were then removed by PI or 7-amino-actinomycin D (7-AAD; Biolegend) staining. For intracellular cytokine detection, enriched tumor-infiltrating CD8+ T cells were restimulated with ionomycin (IM, 1 μg/mL; Sigma-Aldrich) and phorbol myristate acetate (PMA, 25 ng/mL; Sigma-Aldrich) in the presence of GolgiPlug (BD Biosciences) for 4 hours at 37°C. The restimulated CD8+ T cells were stained with surface antigens, and then stained for intracellular cytokines using a Cytofix/Cytoperm kit (BD Biosciences), according to the manufacturer’s instructions.
Classification of immune cell population by t-distributed stochastic neighbor embedding
Multicolor flow cytometric data was visualized by t-distributed stochastic neighbor embedding (t-SNE) analysis (34-36), which was used to classify different immune cell populations in various organs in tumor-bearing mice. Nine days after tumor inoculation, the total cell suspensions from tumor, draining lymph node (dLN), spleen (SP), and BM were first stained with a mixture of 12 fluorophores (7-AAD, Ly6G-PE-594, and other 10 parameters as mentioned below) at 4° C for 25 minutes, then a total of 1 × 106 cells were incubated with FITC-conjugated minP1 (FITC-minP1, 0.5 μg/mL) at 37°C in a humidified 5% CO2 incubator for 30 minutes, Using Flowjo software, live cells were gated on the 7-AAD negative population and minimized to 3 × 104 cells per sample. Triplicate samples were concatenated into one group and each group underwent analysis by t-SNE analysis using Flowjo software (version 10.5; BD Biosciences). To identify immune cell populations in the tumors, dLN, and SP, ten parameters (B220-PE-Cy7, CD3-BV500, CD4-BV421, CD8-Alexa700, CD11b-BV605, CD11c-APC, CD19-PerCP-Cy5.5, I-A/I-E-BV786, NK1.1-PE, and Ly6C-APC-Cy7) were used to separate B-cell, CD4+ T-cell, CD8+ T-cell, NK-cell, NKT-cell, monocyte, macrophage/monocyte (monocyte-to-macrophage transition), and DC populations. To identify immune system precursor cells in the BM, ten parameters (B220-PE-Cy7, CD11b-BV510, CD24-PE-594, CD115-PE, CD117-APC-Cy7, CD135-PerCP-Cy5.5, Ly6C-Alexa700, Ly6G-APC, CX3CR1-BV-605, Sca-1-BV421) were used to separate cell populations, including B220+ B-cell progenitors, Ly6G+ granulocyte progenitors, monocyte progenitors (MP), monocyte-DC progenitors (MDP), common DC progenitors (CDP), Ly6C+ monocytes, and Ly6C− monocytes.
Competitive protein binding assay
Nine days after LLC tumor inoculation, total cell suspensions were prepared by enzymatic and mechanical dissociation of tumor tissues, as described previously (33). Total cell suspensions were first stained with an antibody mixture of 12 fluorophores (anti-B220, -CD3, -CD4, -CD8, -CD11b, -CD11c, -CD19, -I-A/I-E, -NK1.1, and -Ly6C) at 4°C for 25 minutes. For dose-dependent competitive assay, cells were then incubated with FITC-minP1 (0.5 μg/mL) and a range of concentrations (0, 0.5, 1, 2, 4, 8, 16, 32, and 64 mg/mL) of unlabeled-minP1 (at 37°C in a humidified 5% CO2 incubator for 30 minutes), with the FITC-minP1 as the competitive protein and unlabeled-P2 as the uncompetitive control. Flowjo Software was used to measure the mean fluorescence intensity (MFI) of FITC-minP1 for each cell population. To compare the competitive and uncompetitive control groups, the FITC-minP1 binding affinity and the half-maximal inhibitory concentration (IC50) were calculated by using 6-parameter logistic regression.
Bulk RNA-seq library preparation and analysis
Total RNA was lysed from 30,000 total cells of each tumor tissue or 10,000 sorted CD8+ T cells by lysis/storage buffer [(1% Lithium Lauryl sulfate (NACALAI TESQUE), 100 mmol/L pH 7. Tris-HCl (NIPPON GENE), 500 mmol/L LiCl (Sigma-Aldrich), and 10 mmol/L EDTA (NIPPON GENE)] and stored in the same lysis/storage buffer. mRNA was isolated from total RNA by Dynabeads M-270 streptavidin with biotin-oligo (dT)25 (Thermo Fisher Scientific). First- and second-strand cDNA synthesis was performed by Superscript reverse transcriptase IV (Thermo Fisher Scientific) and Kapa HiFi DNA polymerase (Roche). While capturing on the beads, cDNA was digested with fragmentase (0.5 μL for sample; NEW ENGLAND BioLabs). Each 3′end of cDNA fragment (100 ng) was ligated to an adapter carrying Ion-Barcode-common sequence 1 (CS1; 1.5 μmol/L). Finally, an 8-cycle PCR step with a PCR mixture (10 μmol/L of trP1 primer, 10 μmol/L Ion_barcode, and 1x KAPA Hifi Hotstart ReadyMix) was performed to enrich for the desired cDNA library molecules by using MiniAmp Thermal Cycler (Thermo Fisher Scientific), and cDNA library products were purified by size selection using AMPure XP beads (Beckman Coulter). Products were confirmed using the Agilent High Sensitivity DNA kit (Agilent Technologies).
The above 5′end Serial Analysis of Gene Expression (SAGE)–sequencing libraries (input library concentration was 65 pmol/L) were used for sequencing by an Ion 540 Chef kit, an Ion 540 Chip kit, and an Ion S5 Sequencer (Thermo Fisher Scientific) according to the manufacturer’s instructions. Adapter trimming and quality filtering of sequencing data were performed by using Cutadpat-v2.4 (37) and Trimommatic-v0.36 (38). The filtered reads were mapped on Refseq mm10 using Bowtie2–2.2.5 (parameters: -t -p 11 -N 1 -D 200 -R 20 -L 20 -i S, 1, 0.50 –nofw). The mapped reads per gene (raw tag counts) were be quantified as gene expression. Between-sample normalization of gene expression was performed against raw count data by using R 3.6.1 (https://cran.r-project.org/) with DESeq2 (39) and pheatmap (40) packages. Genes with adjusted P value less than 0.05 and a fold change of ≥2 between at least three samples were identified as statistically significant differentially expressed genes (DEG). Raw data from the experiment have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) under accession GSE139291.
Functional analysis of DEGs was performed by using Cytoscape 3.7.1 with ClueGO plugin (v2.5.4; refs. 41, 42). Significantly enriched Gene Ontology (GO) terms (ref. 43; GO-immune system process, GO levels: 3–8, version: February 27, 2019) and Kyoto Encyclopedia of Genes and Genomes (KEGG; version: February 27, 2019) pathway terms (44) in DEGs were explored and grouped, and a term network was constructed based on the overlap of their elements (kappa score = 0.4). Leading terms within each group were defined as the most significantly enriched term in each group. Terms not connected with any other term were excluded.
scRNA-seq library preparation and analysis
For scRNA-seq, we sorted 30,000 CD45+ cells (with 90% purify) from Colon26 tumors by using anti-CD45-APC with an anti-APC MicroBeads (Miltenyi Biotec Inc), and AutoMACS (Miltenyi Biotec). Sorted cells were stained with TotalSeq anti-mouse Hashtag antibodies (clone M1/42, 30-F11; A0301, A0302, A0303, A0304, A0305, A0306, A0307, A0308, A0309, A0310, A0311, A0312, A0313, and A0314; BioLegend, USA). 10,000 labeled cells were trapped and reverse-transcribed using BD Rhapsody (BD) according to the manufacturer’s instructions. For scRNA-seq, cDNA libraries and hashtag libraries were prepared by an optimized process similar to our magnetic bead-based bulk RNA-seq method (45). Sequencing libraries were generated using the NEBNext UltraII FS Library Prep Kit (New England BioLabs), and QC of cDNA and final libraries was performed by MultiNA and qPCR library quantification assay (KAPA). Samples were sequenced on an Illumina Novaseq 6000 S4 flow cell (67 bp read 1 and 140 bp read 2; Illumina) to a depth of approximately 100,000 reads per cell. For mouse data, after adaptor removal by Cutadpat-v2.10 (37), gene-expression libraries were aligned to the mm10 reference transcriptome by Bowtie2–2.3.4.1, and count matrices were generated using the home-built shell scripts and the modified python script of BD Rhapsody workflow. Valid cell barcodes were identified as cell barcodes above inflection threshold of knee-plot of total read counts of each cell barcode identified by DropletUtils package (46). Sample origins and doublets were identified based on fold change of the normalized read counts of the hashtags. Data were log transformed [log(TPM + 1)] for all downstream analyses, including “CreatSeuratObject()”, “AddMetaData()”, “NormalizeData()”, and “ScaleData()” were performed using the R software package Seurat v2.3.4 (ref. 47; http://satijalab.org/seurat) to analyze immune cell populations. An overview of the experimental data has been deposited in the NCBI GEO (http://www.ncbi.nlm.nih.gov/geo) under accession GSE167192.
For each cell, three quality control metrics were calculated: (i) the total detected number of genes, (ii) the proportion of ribosome encoded transcripts, and (iii) the proportion of mitochondrially encoded transcripts. Cells were excluded from downstream analysis if fewer than 200 genes were detected. There was an expression matrix of 7,613 cells by 22,833 genes. Each gene expression measurement was normalized by total expression within the corresponding cell and multiplied by a scaling factor of 1,000,000. A subset of features that exhibit high cell-to-cell variation (high variable genes) in the dataset were calculated by “FindVariableFeatures()” function in the Seurat software package (v2.3.4, Satija Labs, New York Genome Center) and used for principal components analysis. Principal components were determined to be significant (P < 0.05) using the jackstraw method, and Fit-SNE (48) was performed on these key principal components. Unsupervised clustering was performed using a shared nearest neighbor modularity optimization-based algorithm, as described previously (49). The reclustering analysis was used to remove contaminating subpopulations. Differential gene expression and signature enrichment analyses were performed by a Wilcoxon rank-sum test. To perform an analysis of cell–cell interactions among DC and T-cell subsets, we analyzed our scRNA-seq data by CellPhoneDB (ref. 50; www.CellPhoneDB.org). As previously reported (51), CellPhoneDB can calculate the means of the average expression of each receptor–ligand pair in each pairwise comparison between two cell types, and provide a P value for each receptor–ligand pair. We then determined the order of interactions that were highly enriched between cell types based on the number of significant pairs, and manually selected biologically genes relevant to the costimulatory/coinhibitory molecules between T-cell and DC subsets. The means of the average expression of interacting molecule 1 in cell cluster 1 and interacting molecule 2 in cell cluster 2 were indicated by color, and P values were indicated by circle size, scale. Assays were carried out at the mRNA level but were extrapolated to protein interactions.
Immunofluorescent staining
Acetone-fixed, 8-μm tumor sections of LLC tumor–bearing IL12-YFP reporter C57BL/6 mice on day 14 after tumor inoculation were prestained with anti-mouse MHC class II (clone M5/114.15.2, BioLegend) and incubated with PI. Sections were mounted with ProLong Gold Antifade Mountant (Thermo Fisher Scientific) and examined under a TCS SP5 confocal microscope (Leica Microsystems). Distribution of IL12b+ and MHC class II+ DCs in the tumor were scored by IL12b (green—YFP) MHC class II (red—Alexa Fluor 647) coexpression, and nuclear staining for PI (blue—Alexa Fluor 514) with a 200 μm scale bar.
Quantification and statistical analysis
Data were analyzed using Prism 8.0 software (GraphPad Software). For comparisons among groups in the in vivo study, we used one-way ANOVA with the Dunnett test. For comparisons between the means of two variables, we used two-sided unpaired Student t test. For correlation, we used a two-sided Pearson correlation with coefficient r. All statistical analyses were presented as mean with SEM and conducted with a significance level of α = 0.05 (P< 0.05).
Results
Combined HMGN1 and PD-L1 blockade induces durable tumor regression
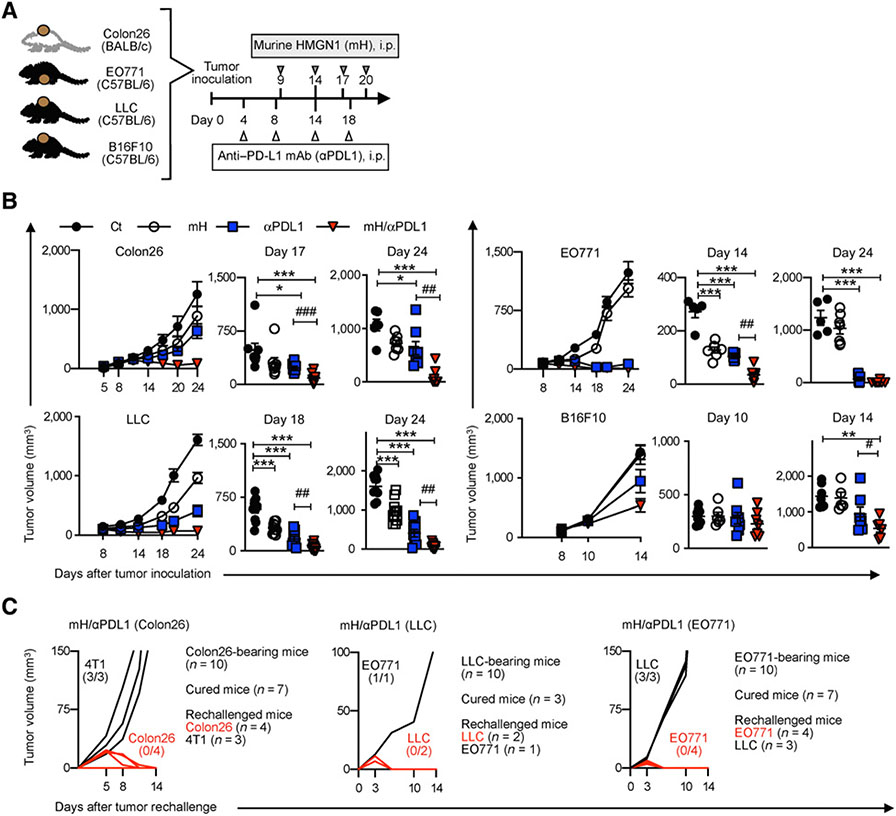
To determine the optimal dosage of murine HMGN1 (mH) in combination with an anti–PD-L1, we first performed a dose-response assessment of combined treatment in B16F10 or Colon26 tumor-bearing mice. The best antitumor effect was at 0.08 μg mH in the B16F10 model, and at a dose range of 0.08 to 0.4 μg mH in the Colon26 model. A 200-μg anti–PD-L1 dose was followed by or combined with mH per treatment (Supplementary Fig. S1A). We, therefore, used the optimized dose of 0.08 μg mH and 200 μg anti–PD-L1 in the following experiments (Fig. 1A). We next evaluated the antitumor effects of mH/anti–PD-L1 treatment in Colon26, LLC, EO771, and B16F10 models. Compared with anti–PD-L1 alone, combined mH/anti–PD-L1 showed significant improvement in tumor growth inhibition in all four different tumor models: Colon26 (day 17, P < 0.001; day 24, P < 0.01); LLC (day 18, P < 0.01; day 24, P < 0.01); EO771 (day 14, P < 0.01); and B16F10 (day 14, P < 0.05; Fig. 1B). These results demonstrated that combining HMGN1 with PD-L1 blockade induces durable tumor regression.
Figure 1.
Combined treatment with HMGN1 and PD-L1 blockade induces durable tumor regression. A, The optimized protocol for HMGN1/anti–PD-L1 treatment in Colon26, LLC, EO771, and B16F10 tumor–bearing mice. B, Tumor growth analysis during mH/anti–PD-L1 treatment and tumor volume of Colon26 (on days 17 and 24), LLC (on days 18 and 24), EO771 (on days 14 and 24), and B16F10 (on days 10 and 14). C, Tumor rechallenge. One week after tumor clearance, mice that cleared Colon26, LLC, and EO771 tumors were rechallenged with 4T1 and Colon26 tumor cells, with EO771 and LLC tumor cells or with LLC and EO771 tumor cells, respectively. Tumor growth is representative of three independent experiments with at least eight mice/group. Data are presented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 determined using Dunnett post hoc test (compared with control); #, P < 0.05; ##, P < 0.01; ###, P < 0.001 determined using Student t test (comparing mH/anti–PD-L1–treated and anti–PD-L1–treated groups). Ct, control.
We observed that 70% of Colon26 (7/10), 30% of LLC (3/10), and 70% of EO771 (7/10) tumor–bearing mice cleared primary tumors after mH/anti–PD-L1 treatment, but did not observe this in untreated or single treatment groups. To confirm that this tumor rejection was due to the tumor-specific immune responses, we performed a rechallenge experiment. We found that tumor-regressed mice resisted rechallenge of primary tumor cells but failed to control primary challenge with another tumor type, confirming that the robust antitumor effects of mH/anti–PD-L1 treatment were associated with tumor-specific immunologic memory (Fig. 1C).
minP1 derived from the HMGN1 NBD retains its antitumor effects
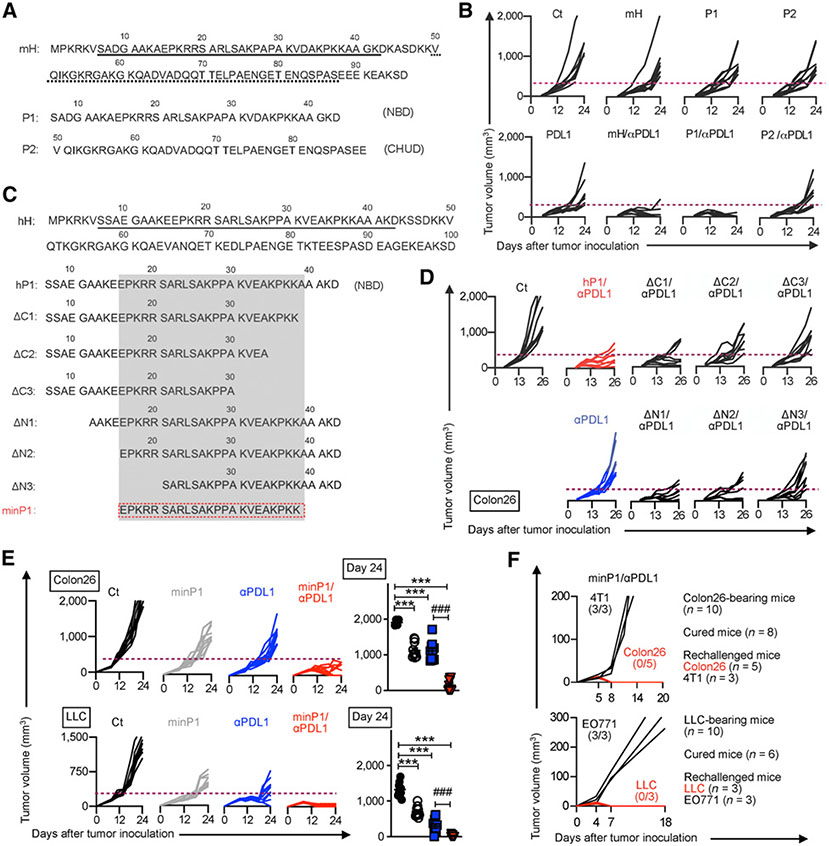
As clinical agents, synthetic peptides have advantages over recombinant proteins because they are quickly and easily synthesized without the risk of endotoxin contamination, and are relatively inexpensive; therefore, we tried to identify the immunostimulatory domain on HMGN1 and used the synthesized HMGN1-derived immunostimulatory domain as a therapeutic peptide in combination with anti–PD-L1 treatment. To determine the immunostimulatory domain on HMGN1, we first synthesized the murine HMGN1 NBD peptide (P1) and CHUD peptide (P2) and evaluated the antitumor effects of P1/anti–PD-L1 and P2/anti–PD-L1 treatments in the Colon26 model, respectively (Fig. 2A). P1/anti–PD-L1 treatment, but not P2/anti–PD-L1 treatment, showed synergistic antitumor effects equivalent to mH/anti–PD-L1 treatment (Fig. 2B), suggesting that the HMGN1 immunostimulatory domain remains within the NBD.
Figure 2.
minP1, a minimized immunostimulatory peptide derived from the HMGN1 NBD, retains its antitumor effects. A, The amino acid sequence of murine HMGN1 (mH) and its NBD (peptide 1, P1) and CHUD (peptide 2, P2). B, Tumor growth for mice treated with mH, P1, or P2 in combination with anti–PD-L1 treatment in each mouse from day 0 to day 24. C, The amino acid sequence of the human HMGN1 (hH) NBD and derived peptides with various lengths. D, Tumor growth after treatment with HMGN1 NBD-derived peptides in combination with anti–PD-L1 treatment in each mouse. E, Tumor growth after treatment with the minP1 (human HMGN1 NBD-derived immunostimulatory peptide) in combination with anti–PD-L1 treatment in both Colon26 and LLC models from day 0 to day 24, and tumor volume at day 24. F, Tumor rechallenge. One week after tumor clearance, mice that cleared Colon26 or LLC tumors were rechallenged with 4T1 and Colon26 tumor cells or with EO771 and LLC tumor cells, respectively. Tumor growth is representative of three independent experiments with at least eight mice/group. Data are presented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 using Dunnett post hoc test (compared with control); #, P < 0.05; ##, P < 0.01; ###, P < 0.001 using Student t test (comparing minP1/anti–PD-L1–treated and anti–PD-L1–treated groups). Ct, control.
Next, to minimize the region of immunostimulatory domain within the HMGN1 NBD, we further selected human HMGN1 [which showed equal antitumor efficacy to mH in our previous report (21)] and synthesized a human HMGN1 NBD peptide (hP1) and six hP1-derived N-terminally or C-terminally truncated peptides (ΔN1, ΔN2, ΔN3, ΔC1, ΔC2, ΔC3; Fig. 2C). In the Colon26 model, hP1, ΔN1, ΔN2, and ΔC1 displayed similar synergistic antitumor effects in combination with anti–PD-L1 treatment, but ΔC2, ΔC3, and ΔN3 peptides failed, indicating that the C-terminal region from K31 to K38 and the N-terminal region from E16 to R20 were required for antitumor function (Fig. 2D). Thus, we hypothesized that this minimal peptide (minP1, spanning 23-amino acid residues from E16 to K38, EPKRR SARLS AKPPA KVEAK PKK) retained the HMGN1-induced antitumor effects.
minP1 exhibited similar antitumor efficacy to mH, P1, and hP1 in the Colon26 model (Supplementary Fig. S1B and S1C). Combined treatment with minP1 and anti–PD-L1 showed significant improvement in tumor growth inhibition compared with anti–PD-L1 alone in both Colon26 and LLC models (Fig. 2E; Supplementary Fig. S1D). We also observed that 80% of Colon26 (8/10), 60% of LLC (6/10), and 40% of EO771 (4/10) tumor–bearing mice achieved complete regression of primary tumors only in the minP1/anti–PD-L1 treatment group, which showed similar antitumor efficacy to mH/anti–PD-L1. With tumor rechallenge, mice that cleared tumors resisted primary tumor rechallenge (Fig. 2F; Supplementary Fig. S1E), consistent with the tumor rechallenge results of mH/anti–PD-L1 treatment (Fig. 1C). Taken together, these data demonstrated that minP1 retains the HMGN1 immunostimulatory function and shows the equal antitumor effects to full-length HMGN1 protein.
minP1 preferentially binds intratumoral DC populations
To further understand how minP1 regulates antitumor immunity in tumor-bearing mice, we first tried identifying the potential target cell populations of minP1 therapy in tumors and primary and secondary lymphoid organs. Nine days after LLC tumor inoculation, total cell suspensions from the tumor, dLN, SP, and BM were prestained with cell surface antibodies, and then incubated with FITC-minP1 to detect cell populations that bind to minP1. To identify immune cell populations in the tumors, dLN, and spleen, ten parameters (B220, CD3, CD4, CD8, CD11b, CD11c, CD19, I-A/I-E, NK1.1, and Ly6C) were used to separate B-cell, CD4+ T-cell, CD8+ T-cell, NK-cell, NKT-cell, monocyte, macrophage, and DC populations (Supplementary Fig. S2A). To identify immune system precursor cells in the BM, ten parameters (B220, CD11b, CD24, CD115, CD117, CD135, Ly6C, Ly6G, CX3CR1, Sca-1) were used to separate cell populations, including B220+ immature B cells, Ly6G+ granulocyte progenitors, MPs, MDPs, CDPs, Ly6C+ monocytes, and Ly6C− monocytes (Supplementary Fig. S2A). Next, we clustered the immune cell populations using t-SNE analysis (Supplementary Fig. S2B). We detected higher FITC-minP1 intensities on monocyte, macrophage, and DC clusters in tumors, but not on any cell cluster in dLN, SP, or BM (Supplementary Fig. S2B). These results suggest that minP1 preferentially binds to intratumoral immune cells, including monocytes, macrophages, and DCs.
To confirm the results of t-SNE analysis, we evaluated the FITC-minP1 binding affinity to intratumoral immune cell populations by competitive protein binding assay, which involves incubating intratumoral immune cell populations with FITC-minP1 and a range of concentrations of unlabeled-minP1 or -P2 (unlabeled minP1 served as the competitive protein and unlabeled-P2 as the uncompetitive control). Using an unlabeled-minP1 as competitive protein, we observed that monocyte, macrophage, and DC clusters had decreased FITC-minP1 intensities unlike the CD4+ T-, CD8+ T-, or NK-cell clusters (Supplementary Fig. S2C). In a dose-dependent competitive protein binding assay, as the concentration of unlabeled minP1 (competitive protein) increased, the amounts of FITC-minP1 bound to macrophage and DC clusters decreased, but the decreasing amounts of FITC-minP1 was not observed for the monocyte cluster (Supplementary Fig. S2D). The half-maximal inhibitory concentration (IC50) values of the DC (69.96 μg/mL) cluster was lower than those for macrophages (192.98 μg/mL), monocyte (>200 μg/mL), and other immune cell populations (>200 μg/mL) in tumors (Supplementary Fig. S2D). Taken together, these results demonstrated that minP1 preferentially binds on intratumoral DC populations and may further regulate their biological functions.
minP1 upregulates MHC class I antigen presentation program on intratumoral mregDC population
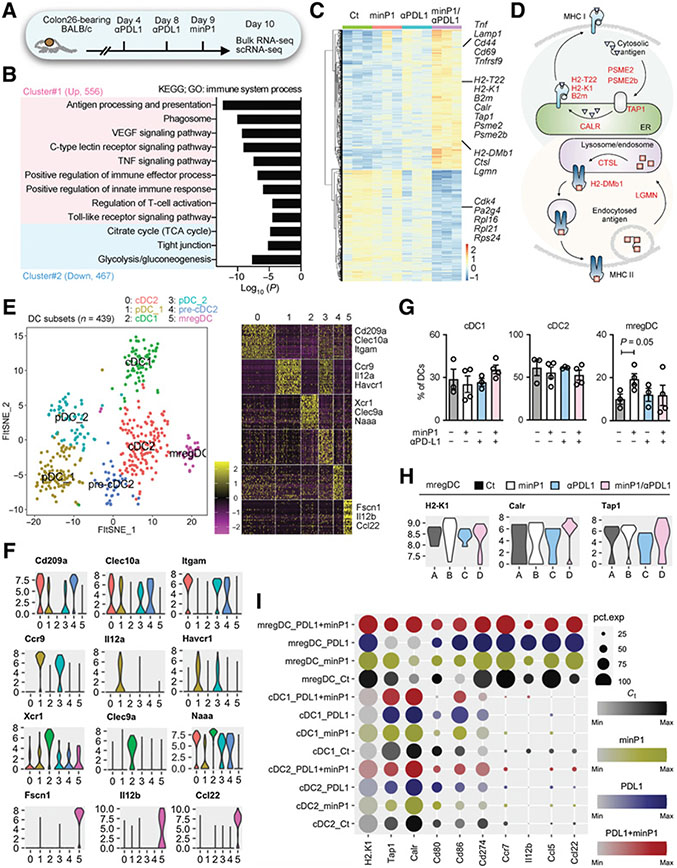
To examine the role of how minP1 shapes antitumor immunity in the tumor microenvironment and how minP1 regulates the biological functions of intratumoral DC populations, we analyzed the transcriptomic changes among the minP1, anti–PD-L1, and minP1/anti–PD-L1 treatment groups in the Colon26 model by using bulk RNA-seq on whole tumor tissues and scRNA-seq on CD45+ immune cell populations, respectively (Fig. 3A). We found a total of 1,023 DEGs (with adjusted P < 0.05 and a fold change ≥2 among groups) between anti–PD-L1 and minP1/anti–PD-L1 treatment groups. GO analysis yielded a large cluster (cluster #1) containing 556 upregulated genes involved in the positive regulation of innate immune responses and immune effector processes, which included genes involved in antigen processing and presentation, phagocytosis, the C-type lectin receptor signaling pathway, the TNF signaling pathway, T-cell activation, and the toll-like signaling pathway (Fig. 3B). Many upregulated genes in the minP1/anti–PD-L1 treatment group, such as histocompatibility 2, class II, locus Mb1 (H2-DMb1), histocompatibility 2, T region locus 22 (H2-T22), histocompatibility 2, K1, K region (H2-K1), beta-2 microglobulin (B2m), transporter 1, ATP-binding cassette, subfamily B (MDR/TAP; Tap1), calreticulin (Calr), cathepsin L (Ctsl), legumain (Lgmn), proteasome activator subunit 2 (PA28 beta; Psme2), and Psme2b, were involved in the antigen processing and presentation pathway (Fig. 3C and D). Upregulated genes after combination treatment, including tumor necrosis factor (Tnf), lysosomal-associated membrane protein 1 (Lamp1), Cd44, Cd69, TNF receptor superfamily, member 9 (Tnfrsf9), and IFN gamma receptor 1 (Ifngr1), were involved in regulation of T-cell activation (Fig. 3C). Conversely, some downregulated genes after combination treatment, including cyclin-dependent kinase 4 (Cdk4), proliferation-associated 2G4 (Pa2g4), and ribosomal proteins 16, 21, and S24 (Rpl16, Rpl21, Rps24), were involved in immunosuppression and T-cell exclusion (Fig. 3C; ref. 52). Our bulk RNA-seq demonstrated that minP1 shaped antitumor immunity by enhancing antigen presentation and T-cell activation programs after anti–PD-L1 treatment in the tumor microenvironment.
Figure 3.
minP1 transcriptionally upregulates the MHC class I antigen presentation program in intratumoral mregDCs. A, Schema for the preparation of tumor cell suspensions from Colon26 tumor–bearing mice for bulk RNA-seq (n = 12, three mice for each treatment groups) and scRNA-seq (n = 14, three mice for untreated control and anti–PD-L1 groups and four mice for minP1 and minP1/PD-L1 treatment groups) on day 10. B, GO analysis of DEGs upregulated (cluster #1) and downregulated (cluster #2) in the minP1/anti–PD-L1 treatment group. C, Heatmap of the 1,023 DEGs. Each column represents a group, and each row represents an individual gene. The Z-scores of module groups are shown at the bottom-right corner of the heatmap. D, Summary diagram of the antigen processing and presentation pathway components based on the KEGG pathway map04612 (https://www.genome.jp/dbget-bin/www_bget?path:map04612). E, A t-SNE projection and corresponding heatmap for scRNA-seq profiling of 439 DCs in tumors of Colon26 tumor–bearing mice. DC clusters are distinct colors. cDC1 (Xcr1, Clec9a, Naaa), cDC2 (Cd209a, Clec10a, Itgam), pDC (Ccr9, Il12ra, Havcr1), and mregDC (Fscn1, Il12b, Ccl22) were profiled. F, Violin plots showing the expression distribution of selected genes in different DC clusters. The y-axis represents log-normalized expression values. G, Proportion of DCs for cDC1, cDC2, and mregDC clusters in different treatment groups. H, Violin plots showing the expression distribution of selected genes (H2-K1, Calr, Tap1) in different treatment groups. The y-axis represents log-normalized expression values. I, Dot plots showing expression of antigen presentation (H2-K1, Tap1, Calr), DC immunostimulatory (Cd80, Cd86, Cd274), and cytokine and chemokine (Ccr7, Il12b, Ccl5, Ccl22) genes among DC clusters in different treatment groups. Node size is proportional to the expression frequency in a cell cluster. Node color max to min is proportional to the gene enrichment score in each cluster; black represents control (Ct) group. Yellow represents minP1 treatment group. Blue represents anti–PD-L1 treatment group. Red represents anti–PD-L1+minP1 treatment group.
scRNA-seq showed a total of 27 unsupervised cell clusters from tumors were profiled and characterized by unique gene signatures of each cell populations (Supplementary Fig. S3A-S3C). To gain insight about how different treatments (untreated, minP1, anti–PD-L1, and minP1/anti–PD-L1) impacted the biological functions of intratumoral DC populations, we selected DC clusters for analysis. With reference to the previously defined DC subsets in literature (53, 54), we characterized a total of six DC clusters by their unique representative gene signatures, including conventional DC type1 (cDC1; Xcr1, Clec9a, and Naaa), conventional DC type 2 and precursors (cDC2 and pre-cDC2; Cd209a, Clec10a, and Itgam), plasmacytoid DCs (pDC_1 and pDC_2; Ccr9, Il12a, and Havcr1), and mregDC (Fscn1, Il12b, and Ccl22; Fig. 3E and F). Among the DC clusters, minP1 treatment significantly increased the proportion of mregDCs (relative to the untreated group; Fig. 3G), and also enhanced the expression of MHC class I antigen presentation-related genes, H2-K1, Calr, and Tap1, on mregDCs (Fig. 3H and I). We did not observe a percentage change or expression change in cDC1 or cDC2 clusters after minP1 treatment (Fig. 3H and I). We also observed that the upregulated genes in the minP1/anti–PD-L1 treatment group, such as Cd70, Tnfrsf14, Calr, Fadd, Iqsec1, Kif5c, Cyth2, influenced the positive regulation of T-cell-mediated cytotoxicity, antigen processing and presentation, and endocytosis (Supplementary Fig. S4A and S4B). Conversely, downregulated genes, including Actr3, Fscn1, and Rac1, contributed to the positive regulation of lamellipodium organization and assembly (Supplementary Fig. S4A and S4B). Taken together, our scRNA-seq data further suggested that minP1 treatment results in an increasing proportion of mregDCs in the tumor microenvironment and also transcriptionally upregulates their MHC class I antigen presentation program.
minP1 increases the number of intratumoral mregDCs and enhances their MHC class I expression
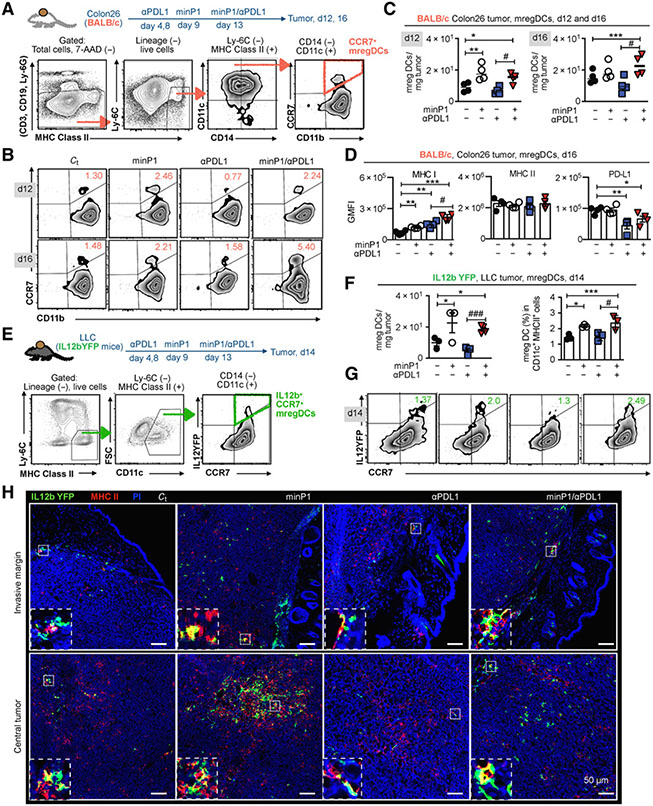
To confirm our scRNA-seq results and assess whether the proportion and number of mregDCs changed with different treatments (untreated, minP1, anti–PD-L1, and minP1/anti–PD-L1), we prepared total cell suspensions from the tumor tissues of Colon26 tumor-bearing mice on day 12 (after two rounds of minP1 and anti–PD-L1 treatments) and 16 (after three rounds of minP1 and anti–PD-L1 treatments) after tumor inoculation. Using flow cytometry, we identified a CCR7+CD11b+CD11c+CD14−Ly6C−MHC class II+ cell population as CCR7+ mregDCs from a lineage-negative cell population (CD3−, CD19−, Ly6G−; Fig. 4A). Increasing numbers of intratumoral CCR7+ mregDCs were observed in Colon26 tumor–bearing mice treated with minP1 or minP1/anti–PD-L1 on days 12 and 16 compared with untreated or anti–PD-L1 treatment, respectively (P < 0.05; Fig. 4B and C). In the minP1/anti–PD-L1 treatment group, increased CCR7+ mregDCs exhibited higher MHC class I and lower PD-L1 expression compared with those from the untreated or anti–PD-L1 treatment group, respectively (P < 0.05); however, their MHC class II expression did not exhibit any difference (Fig. 4D), which is consistent with our scRNA-seq results and suggests that minP1 might enhance the MHC class I antigen–presenting ability of intratumoral CCR7+ mregDCs. Consistent with the results in Colon26 tumor–bearing mice, the increasing proportion and the number of intratumoral IL12b+CCR7+ mregDCs (55) were also observed in LLC tumor–bearing IL12b-YFP mice treated with minP1 or minP1/anti–PD-L1 compared to untreated or anti–PD-L1 treatment, respectively (P < 0.05; Fig. 4E-G).
Figure 4.
minP1 increases the number of intratumoral mregDCs at both the invasive margin and central tumor and enhances their MHC class I expression. A, Schema on the preparation of tumor immune cells from Colon26 tumor–bearing BALB/c mice and flow cytometry gating of CCR7+ mregDCs. B and C, The frequency and number of CCR7+ mregDCs in the tumors on days 12 and 16 after tumor inoculation. D, The expression of MHC class I, MHC class II, and PD-L1 on CCR7+ mregDCs on day16. GMFI, geometric MFI. E, Schema on the preparation of tumor immune cells from LLC tumor–bearing IL12b-YFP reporter C57BL/6 mice and flow cytometry gating of IL12b+CCR7+ mregDCs. F and G, Frequency and number of the IL12b+CCR7+ mregDCs in tumors from LLC tumor–bearing IL12b-YFP reporter C57BL/6 mice treated with combination treatment on day 14 after tumor inoculation. H, The number and localization of IL12b+CCR7+ mregDCs in LLC tumor sections identified by coexpression of IL12b-YFP (green) and MHC class II (red), with a 200 μm scale bar. Bottom panels indicate zoomed and merged images of costained mregDCs. Each result is representative of three independent experiments with at least four mice/group. Data are presented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 using Dunnett post hoc test (compared with control); #, P < 0.05; ##, P < 0.01; ###, P < 0.001 using Student t test (comparing with minP1/anti–PD-L1–treated and anti–PD-L1–treated groups).
To determine where these increased intratumoral IL12b+CCR7+ mregDCs localize, we investigated the invasive margins and the central tumors of LLC tumor–bearing IL12-YFP mice. Using immunofluorescence staining, we identified the IL12b-YFP (green)+ and MHC class II (red)+ cell population as mregDC in tumors. Increasing numbers of IL12b+CCR7+ mregDCs were observed in both the invasive margins and the central tumors of minP1- and minP1/anti–PD-L1–treated mice (Fig. 4H). Taken together, these results indicated that minP1 enhances the MHC class I antigen–presenting ability of mregDCs and increases the number of mregDCs at both the invasive margin and central tumor.
Expansion of intratumoral Tstem/Tpex cells by minP1/αPD-L1 treatment associates with increased tumoral mregDCs
Our bulk RNA-seq suggested that minP1/anti–PD-L1 treatment shaped antitumor immunity by promoting regulation of T-cell activation in tumors (Fig. 3B). To confirm our bulk RNA-seq results, we prepared total cell suspensions from the tumor tissues of Colon26 or LLC tumor–bearing mice on day 12 (after two minP1 treatments) and analyzed intratumoral CD8+ T cells in two different type of tumors by flow cytometry (Fig. 5A). Significantly increasing numbers of intratumoral CD8+ T cells were found in minP1, anti–PD-L1, and minP1/anti–PD-L1 treatment groups in both models (P < 0.01; relative to the untreated control group, Fig. 5B). We also observed that minP1/anti–PD-L1 synergistically increased the numbers of intratumoral CD8+ T cells (P < 0.01; relative to anti–PD-L1 treatment, Fig. 5B). A concomitant increase in numbers of mregDCs and CD8+ T cells was also observed in the minP1/anti–PD-L1 treatment groups of both Colon26 and LLC models (Fig. 5C).
Figure 5.
Expansion of intratumoral stem-like/progenitor exhausted CD8+ T cells by minP1/anti–PD-L1 treatment is associated with an increased number of mregDCs in tumors. A, Schema on the preparation of tumor immune cells from Colon26 or LLC tumor–bearing mice. FCM, flow cytometry. B, The number of CD8+ T cells in the tumors of Colon26 or LLC tumor–bearing mice on day 12 after tumor inoculation. CD45IVS, intravenous CD45 staining. C, Scatter plots of mregDCs and CD8+ T-cell numbers in tumors. The symbol color reflects different treatment groups. D, Flow cytometry gating of intratumoral Tstem/Tpex (Ly108+TIM-3−CD8+) and Tex (Ly108−TIM-3+CD8+) cells. The tSNE defines the Tstem/Tpex and Tex populations among CD8+ T cells and displays the fluorescence intensity of CD44, PD-1, Ly108, and TIM-3 in this population. GMFI, geometric MFI. E, Expression of the intracellular cytokines IFNg, IL2, and TNFα. Cell populations 1 to 3 correspond to populations indicated in D. F, Heatmap of the top 15 DEGs among the Tstem/Tpex, Ttran, and Tex cell subsets. G and H, The compartments, frequencies, and numbers of Tstem/Tpex (Ly108+TIM-3−CD8+), Ttran (Ly108+TIM-3+CD8+), and Tex (Ly108−TIM-3+CD8+) cells in the tumors of LLC or Colon26 tumor–bearing mice on day 12 after tumor inoculation. I, Scatter plots of mregDCs and Tstem/Tpex cell numbers or mregDCs and Tex cell numbers in tumors. The symbol color reflects different treatment groups. Each result is representative of three independent experiments with at least four mice/group. Data are presented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 using Dunnett post hoc test (compared with control); #, P < 0.05; ##, P < 0.01; ###, P < 0.001 using Student t test (comparing between minP1/anti–PD-L1–treated and anti–PD-L1–treated groups).
Next, we investigated which intratumoral CD8+ T-cell subsets increased after treatments. Thus, we first profiled different intratumoral CD8+ T-cell subsets from CD44highPD-1+CD3+ CD8+ T cells by their expression of Ly108 and TIM-3 (Fig. 5D). The Ly108+TIM-3− subset exhibited high IL2, intermediate TNFα, and low IFNγ compared with Ly108+TIM-3+ and Ly108−TIM-3+ subsets (Fig. 5E). To verify subset-specific gene signatures, we then performed bulk RNA-seq on sorted Ly108+TIM-3−, Ly108+TIM-3+, and Ly108−TIM-3+ subsets from tumors of LLC tumor–bearing mice on day 15 (Supplementary Fig. S5A and S5B). The Ly108+TIM-3− subset had specific gene signatures (e.g., Slamf6, Tcf7, Sell, Id3) related to the Tstem/Tpex subset; the Ly108− TIM-3+ subset had specific gene signatures (e.g., Havcr2, Gzmb, Entpd1) related to the Tex subset; and the Ly108+ TIM-3+ subset had intermediate expression of genes (Havcr2, Gzmb, Slamf6) related to the transitory exhausted (Ttran) subset (Fig. 5F; Supplementary S5A and S5B), consistent with the previous reports (22-24, 29, 30).
In the minP1/anti–PD-L1 treatment group, increasing proportion and number of Tstem/Tpex cells were observed in both Colon26 and LLC models compared to anti–PD-L1 alone (Fig. 5G and 5H, left). No significant differences were found in Ttran cell numbers among the treatment groups in both Colon26 and LLC models (Fig. 5H, middle). Conversely, the proportion and number of Tex cells decreased in both models after minP1/anti–PD-L1 treatment (relative to anti–PD-L1 treatment; Fig. 5H, right). We also observed a concomitant increase in mregDCs and Tstem/Tpex cell numbers in the minP1/anti–PD-L1 treatment group of both Colon26 and LLC models, but not Tex cells (Fig. 5I). Taken together, these results suggested that minP1/anti–PD-L1 treatment exerts a synergistic effect on the concomitant increase in Tstem/Tpex cell and mregDC numbers in tumors; however, the mechanism remains unclear.
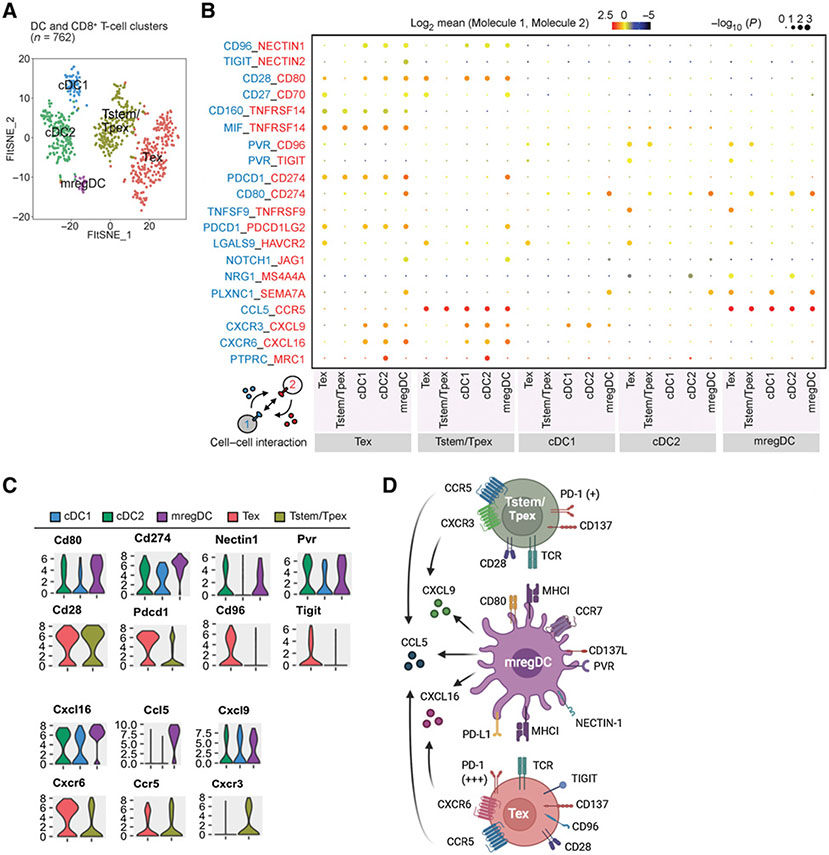
CellPhonDB reveals the potential interactions between mregDCs, Tstem/Tpex cells, and Tex cells in tumors
As previously reported, an increasing number of IL12+CCR7+ mregDCs correlates with the activation and expansion of intratumoral CD8+ T cells, owing to (i) the chemokine receptor CCR7, which enables mregDC to transport antigens from tumor to lymph nodes and then support T-cell priming and expansion (53, 56-58); (ii) the cytokine IL12, which enables mregDCs to augment CD8+ T-cell activation via T cell–DC cross-talk beyond the cytokines IFNγ and IL12 (55); and (iii) immunoregulatory molecules (PD-L1, PD-L2, CD80, etc.), which enable mregDCs control T-cell activation (53). To understand relationship among mregDCs, Tstem/Tpex, and Tex cells in our study, we used CellPhoneDB (50) to predict the relevant interacting ligand–receptor partners from our scRNA-seq data (Fig. 6A and B). The mregDC subset is defined as FSCN1 (Fscn1)+ IL12 (Il12b)+ CCL22 (Ccl22)+ (Fig. 3F); the Tstem/Tpex cell subset, as CD62 L (Sell)− CD44+ Scal (Ly6a)− CD95 (Fas)− PD-1 (Pdcd1)lo TIM-3 (Havarc2)− CD38− TCF1 (Tcf1)+; and the Tex cell subset, as CD62 L (Sell)− CD44+ Scal (Ly6a)− CD95 (Fas)− PD-1 (Pdcd1)high TIM-3 (Havarc2)+ CD38+ TCF1 (Tcf7)− (Supplementary Fig. S5C and S5D). We first analyzed chemokine/chemokine interactions and found that mregDCs expressed high Ccl5, Cxcl9, and Cxcl16, whereas Tstem/Tpex cells expressed Ccr5 and Cxcr3, the receptors for Ccl5 and Cxcl9, respectively. In contrast, Tex cells expressed Ccr5 and Cxcr6, the receptors for Ccl5 and Cxcl16, respectively (Fig. 6C).
Figure 6.
CellPhonDB reveals the potential interactions between mregDCs, Tstem/Tpex cells, and Tex cells in tumors. A, A t-SNE projection of scRNA-seq profiling from 762 T cells and DCs in tumors of Colon26 tumor–bearing mice. T-cell and DC clusters are represented as distinct colors. B, Overview of select ligand–receptor interactions; P values are indicated by circle size [scale on right (permutation test)]. The means of the average expression of interacting molecule 1 in cell cluster 1 and interacting molecule 2 in cell cluster 2 are indicated by color. Assays were carried out at the mRNA level but were extrapolated to protein interactions. C, Violin plots exhibiting the expression distribution of selected genes in T-cell and DC clusters. The y-axis represents log-normalized expression values. D, Summary diagram of the main receptors and ligands expressed on the mregDC, Tstem/Tpex cell, and Tex cell clusters that were involved in cellular recruitment and costimulation/coinhibition. Created with BioRender.com.
Next, we analyzed costimulatory/coinhibitory molecule interactions. MregDCs expressed higher immunostimulatory molecules, such as Cd80 and Tnfsf9 (known as CD137 ligand), and immunoregulatory molecules, such as Cd274 (known as PD-L1), Nectin1, and Pvr, compared to cDC1 and cDC2 subsets (Fig. 6C). The corresponding receptors or ligands to those immunoregulatory molecules, such as Pdcd1, Cd96, and Tigit, were highly expressed on Tex cells but not Tstem/Tpex cells. By contrast, Tstem/Tpex cells expressed slightly higher Cd28 (Fig. 6C). These results suggested that mregDCs provide a CD80–CD28 costimulatory cross-talk for both Tstem/Tpex cells and Tex cells, but multiple coinhibitory cross-talks (i.e., PD-L1–PD-1, NECTIN1–CD96, and PVR-TIGIT) likely occur between mregDCs and Tex cells (Fig. 6D).
Discussion
The expansion of intratumoral Tstem/Tpex cells provides a potential approach to improving the efficacy of ICB therapy. Our team has previously demonstrated that HMGN1 promotes DC activation via the MYD88 pathway and facilitates DC-dependent CD8+ T-cell expansion (16, 21). We here explored the possibility of combining the HMGN1 (minP1) with PD-L1 blockade. minP1 treatment increased the number of intratumoral mregDCs and enhanced their MHC class I antigen–presenting program; however, high PD-L1 expression observed on mregDCs may attenuate T-cell expansion (53, 59). minP1 treatment enhanced the effects of blocking the PD-1/PD-L1 axis in PD-L1high mregDCs and resulted in the expansion of intratumoral CD8+ T cells, particularly the Tstem/Tpex subset.
Notably, the Tstem/Tpex subset (TCF1+ PD-1+) with stem-like properties and long-lived potential act as a resource cell population, differentiating into Ttran/Tex subsets and mediating tumor control in response to ICB therapy. Although the Ttran/Tex subsets may show increased cytotoxicity, their short-lived features highlight again the essential of the Tstem/Tpex pool that contributes to a longer duration of response to ICB therapy and robust antitumor effect (23, 31). Tstem/Tpex cells mainly reside near a high density of antigen-presenting cell (APC) niches within tumor, and require the support from APCs to maintain their function and differentiation (60). For example, an adequate number of DCs reinvigorates intratumoral CD8+ T cells from exhaustion and support CD8+ T-cell expansion in tumors by CD28-CD80 costimulation (29, 61). On the other hand, DCs are one of the major PD-L1+ immune cells within tumors and are among the significant immune targets of PD-L1 blockade, which may shape the therapeutic efficacy of PD-1/PD-L1 blockade (62, 63). Here, we found that the minP1 synergizes with PD-L1 blockade in increasing the number of mregDCs, enhancing their antigen-presenting function with a moderate increase in PD-L1 expression. The upregulation of antigen presentation–related molecules and PD-L1 expression on mregDCs may be due to the increasing IFNγ derived from tumor-infiltrating CD8+ T cells after PD-L1 treatment. IFNγ upregulates PD-L1 expression (59), promotes the noncanonical NF-kB pathway (55), and induces the secretion of IL12 from tumor-infiltrating DCs. Thus, CD8+ T-cell–derived IFNγ may drive the mregDC program to enhance antitumor immunity (53).
It has been reported that the mregDC subset is a conserved state of tumor-infiltrating DCs after tumor antigen capture, and it may play distinct immunoregulatory roles within tumors to regulate the functions of tumor-infiltrating T cells due to their high expression of both costimulatory molecules (such as CD40, CD80, CD86) and coinhibitory molecules (such as PD-L1, PD-L2, CD200; refs. 53, 54). However, mregDCs resemble activated CCR7+ migratory DCs [also known as DC3s (64), LAMP3+ DCs (65), CCR7+IL12+ DCs (58), and BATF3+ DCs (66)] and act as primary APCs after migrating into the lymph nodes, where they can activate tumor antigen-specific T cells (58). Based on our scRNA-seq data analysis by CellPhoneDB, mregDCs may favor the association with Tstem/Tpex cells via CXCL9–CXCR3 and expand through CD28–CD80 interactions. Alternatively, mregDCs may favor the association with Tex cells via CXCL16–CXCR6 and regulate through multiple coinhibitory interactions (such as PD-1–PD-L1, NECTIN1–CD96, and PVR–TIGIT).
The higher MHC class I expression on mregDCs after combination treatment (minP1/anti–PD-L1) and the increased Tstem/Tpex cells but decreased Tex cells are puzzling. One possibility is the high expression of multiple coinhibitory molecules (such as PD-1, CD96, TIGIT, TIM-3) observed on Tstem/Tpex cells and the receptor/ligand to these coinhibitory molecules on mregDCs. Once the PD-1/PD-L1 axis is blocked, mregDCs may selectively suppress Tex cell proliferation via the remaining coinhibitory molecules. The combined interaction of mregDCs with Tstem/Tpex cells and Tex cells in the tumor may not be the only mechanisms. The supply of Tstem/Tpex cells from the dLN via circulation may also dictate the equilibrium of Tstem/Tpex and Tex cells (59, 67). These possibilities to account for the observation of increased mregDCs with more Tstem/Tpex cells but lower Tex cells after combination treatment will be examined in future studies.
Our work highlights three important implications for both basic and clinical research. First, we have designed an effective strategy for combining the HMGN1 peptide (minP1) with PD-L1 blockade to improve the efficacy of ICB therapy. The use of peptides as therapeutics has advantages of standardized synthesis protocols, low toxicity, and good efficacy. Compared with the HMGN1, minP1 shows a lower risk of endotoxin contamination during synthesis. It could be produced in larger quantities within a short time period and has increasing potential to use as an adjuvant for cancer immunotherapies in the clinic. Second, the transcriptome signature of tumor tissues after minP1/anti–PD-L1 treatment could be used as an indicator to evaluate clinical response and help determine prognosis of patients with cancer. The gene cluster including H2-DMb1, H2-T22, H2-K1, B2m, Tap1, Calr, Ctsl, Lgmn, Psme2, and Psme2b was involved in antigen processing and presentation, whereas the cluster including Tnf, Lamp1, Cd44, Cd69, Tnfrsf9, and Ifngr1 was involved in regulation of T-cell activation, and these genes were all upregulated after combination treatment. Conversely, the gene cluster including Cdk4, Rpl16, Rpl21, Rps24, and Pa2g4 was involved in immunosuppression and T-cell exclusion and was downregulated after combination treatment. These gene clusters could be used as indicators to predict and monitor the efficacy of combination treatment. Third, we identified the domain responsible for the immunostimulatory functions of HMGN1. Although previous studies identified the HMGN1 NBD and CHUD based on their distinct intracellular functions (13, 14), our previous work has further demonstrated that HMGN1 also has an extracellular function, inducing an immunostimulatory response that results in synergistic antitumor effects (16-21). Nevertheless, the area of the protein mediating the immunostimulatory function was unclear. Here, we found a 23-amino acid peptide within the HMGN1 NBD that retains the immunostimulatory responses and synergistic antitumor effects of HMGN1. This finding will enable in-depth examination of this immunostimulatory domain to understand how HMGN1 binds and interacts with its candidate receptors, such as lymphocyte antigen 96 (LY96; also known as MD2; ref. 16) or Gαi protein coupled receptor (GiPCR; ref. 20). Overall, our study provides the rationale for combining an HMGN1 immunostimulatory peptide with PD-L1 blockade for cancer immunotherapy.
Supplementary Material
Acknowledgments
The authors thank Shin Aoki, Shunichi Fujita, and Ai Yamashita (The University of Tokyo, Tokyo, Japan) for help with maintenance of laboratory and animal facility, Rina Matsukiyo and Kazushige Shiraishi (Tokyo University of Science, Chiba, Japan) for help with cell sorting, and Junko Yasuda (Tokyo University of Science, Chiba, Japan) for help with RNA-seq and analysis. C.-Y. Chen was supported by The University of Tokyo Fellowship. Research work in the laboratory of K. Matsushima is funded by Grant-in-Aid for Scientific Research on Innovative Areas (grant number: 17929397).
Footnotes
Authors’ Disclosures
C.-Y. Chen reports a patent for HMGN partial peptide and cancer therapy using same (patent no. WO2019–172358). S. Ueha reports grants from Japan Society for the Promotion of Science (17H06392 and 20281832) during the conduct of the study; other support from ImmunoGeneTeqs, Inc. (stock), IDAC Theranostics, Inc., and ImmunoGeneTeqs, Inc. (advisory role) outside the submitted work; and a patent for HMGN partial peptide and cancer therapy using the same pending. S. Shichino reports personal fees from ImmunoGeneTeqs, Inc. during the conduct of the study. S. Shibayama reports a patent for HMGN partial peptide and cancer therapy using the same pending. K. Matsushima reports a patent for HMGN partial peptide and cancer therapy using same (patent no. WO2019–172358). No disclosures were reported by the other authors.
Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
References
- 1.Iwai Y, Ishida M, Tanaka Y, Okazaki T, Honjo T, Minato N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc Natl Acad Sci U S A 2002;99:12293–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strome SE, Dong H, Tamura H, Voss SG, Flies DB, Tamada K, et al. B7-H1 blockade augments adoptive T-cell immunotherapy for squamous cell carcinoma. Cancer Res 2003;63:6501–5. [PubMed] [Google Scholar]
- 3.Blank C, Gajewski TF, Mackensen A. Interaction of PD-L1 on tumor cells with PD-1 on tumor-specific T cells as a mechanism of immune evasion: implications for tumor immunotherapy. Cancer Immunol Immunother 2005;54:307–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, Anderson AC. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J Exp Med 2010;207:2187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: mechanisms, response biomarkers, and combinations. Sci Transl Med 2016;8:328rv4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012;366:2455–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science 2015;348:69–74. [DOI] [PubMed] [Google Scholar]
- 9.Jenkins RW, Barbie DA, Flaherty KT. Mechanisms of resistance to immune checkpoint inhibitors. Br J Cancer 2018;118:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nie Y, Yang D, Trivett A, Han Z, Xin H, Chen X, et al. Development of a curative therapeutic vaccine (TheraVac) for the treatment of large established tumors. Sci Rep 2017;7:14186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salmon H, Idoyaga J, Rahman A, Leboeuf M, Remark R, Jordan S, et al. Expansion and activation of CD103(+) dendritic cell progenitors at the tumor site enhances tumor responses to therapeutic PD-L1 and BRAF inhibition. Immunity 2016;44:924–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ribas A, Medina T, Kummar S, Amin A, Kalbasi A, Drabick JJ, et al. SD-101 in combination with pembrolizumab in advanced melanoma: results of a phase Ib, multicenter study. Cancer Discov 2018;8:1250–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bustin M Chromatin unfolding and activation by HMGN(*) chromosomal proteins. Trends Biochem Sci 2001;26:431–7. [DOI] [PubMed] [Google Scholar]
- 14.Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev 2005;15:496–506. [DOI] [PubMed] [Google Scholar]
- 15.Hock R, Furusawa T, Ueda T, Bustin M. HMG chromosomal proteins in development and disease. Trends Cell Biol 2007;17:72–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang D, Postnikov YV, Li Y, Tewary P, de la Rosa G, Wei F, et al. High-mobility group nucleosome-binding protein 1 acts as an alarmin and is critical for lipopolysaccharide-induced immune responses. J Exp Med 2012;209:157–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei F, Yang D, Tewary P, Li Y, Li S, Chen X, et al. The alarmin HMGN1 contributes to antitumor immunity and is a potent immunoadjuvant. Cancer Res 2014;74:5989–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang D, Bustin M, Oppenheim JJ. Harnessing the alarmin HMGN1 for anticancer therapy. Immunotherapy 2015;7:1129–31. [DOI] [PubMed] [Google Scholar]
- 19.Nie Y, Yang D, Oppenheim JJ. Alarmins and antitumor immunity. Clin Ther 2016;38:1042–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang D, Han Z, Alam MM, Oppenheim JJ. High-mobility group nucleosome binding domain 1 (HMGN1) functions as a Th1-polarizing alarmin. Semin Immunol 2018;38:49–53. [DOI] [PubMed] [Google Scholar]
- 21.Chen CY, Ueha S, Ishiwata Y, Yokochi S, Yang De, Oppenheim JJ, et al. Combined treatment with HMGN1 and anti-CD4 depleting antibody reverses T cell exhaustion and exerts robust anti-tumor effects in mice. J Immunother Cancer 2019;7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paley MA, Kroy DC, Odorizzi PM, Johnnidis JB, Dolfi DV, Barnett BE, et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 2012;338:1220–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 2019;20:326–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 2016;537:417–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kallies A, Zehn D, Utzschneider DT. Precursor exhausted T cells: key to successful immunotherapy? Nat Rev Immunol 2020;20:128–36. [DOI] [PubMed] [Google Scholar]
- 26.Vodnala SK, Eil R, Kishton RJ, Sukumar M, Yamamoto TN, Ha NH, et al. T cell stemness and dysfunction in tumors are triggered by a common mechanism. Science 2019;363:eaau0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu T, Ji Y, Moseman EA, Xu HC, Manglani M, Kirby M, et al. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci Immunol 2016;1:eaai8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snell LM, MacLeod BL, Law JC, Osokine I, Elsaesser HJ, Hezaveh K, et al. CD8(+) T cell priming in established chronic viral infection preferentially directs differentiation of memory-like cells for sustained immunity. Immunity 2018;49:678–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hashimoto M, Kamphorst AO, Im SJ, Kissick HT, Pillai RN, Ramalingam SS, et al. CD8 T cell exhaustion in chronic infection and cancer: opportunities for interventions. Annu Rev Med 2018;69:301–18. [DOI] [PubMed] [Google Scholar]
- 30.McLane LM, Abdel-Hakeem MS, Wherry EJ. CD8 T cell exhaustion during chronic viral infection and cancer. Annu Rev Immunol 2019;37:457–95. [DOI] [PubMed] [Google Scholar]
- 31.Siddiqui I, Schaeuble K, Chennupati V, Fuertes Marraco SA, Calderon-Copete S, Ferreira DP, et al. Intratumoral Tcf1(+)PD-1(+)CD8(+) T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 2019;50:195–211. [DOI] [PubMed] [Google Scholar]
- 32.Anderson KG, Mayer-Barber K, Sung H, Beura L, James BR, Taylor JJ, et al. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat Protoc 2014;9:209–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ueha S, Yokochi S, Ishiwata Y, Ogiwara H, Chand K, Nakajima T, et al. Robust antitumor effects of combined anti-CD4-depleting antibody and anti-PD-1/PD-L1 immune checkpoint antibody treatment in mice. Cancer Immunol Res 2015;3:631–40. [DOI] [PubMed] [Google Scholar]
- 34.van der Maaten L Accelerating t-SNE using tree-based algorithms. J Mach Learn Res 2014;15:3221–45. [Google Scholar]
- 35.van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res 2008;9:2579–605. [Google Scholar]
- 36.Simoni Y, Becht E, Fehlings M, Loh CY, Koo SL, Teng KWW, et al. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 2018;557:575–9. [DOI] [PubMed] [Google Scholar]
- 37.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 2011;17:10–2. [Google Scholar]
- 38.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolde RR: pheatmap: Pretty Heatmaps. 2015. [Google Scholar]
- 41.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009;25:1091–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, et al. A travel guide to Cytoscape plugins. Nat Methods 2012;9:1069–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 2013;8:1551–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353–D61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shichino S, Ueha S, Hashimoto S, Otsuji M, Abe J, Tsukui T, et al. Transcriptome network analysis identifies protective role of the LXR/SREBP-1c axis in murine pulmonary fibrosis. JCI Insight 2019;4:e122163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Griffiths JA, Richard AC, Bach K, Lun ATL, Marioni JC. Detection and removal of barcode swapping in single-cell RNA-seq data. Nat Commun 2018;9:2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Linderman GC, Rachh M, Hoskins JG, Steinerberger S, Kluger Y. Fast interpolation-based t-SNE for improved visualization of single-cell RNA-seq data. Nat Methods 2019;16:243–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015;161:1202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 2020;15:1484–506. [DOI] [PubMed] [Google Scholar]
- 51.Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 2018;563:347–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su MJ, Melms JC, et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell 2018;175:984–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maier B, Leader AM, Chen ST, Tung N, Chang C, LeBerichel J, et al. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature 2020;580:257–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gerhard GM, Bill R, Messemaker M, Klein AM, Pittet MJ. Tumor-infiltrating dendritic cell states are conserved across solid human cancers. J Exp Med 2021;218:e20200264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garris CS, Arlauckas SP, Kohler RH, Trefny MP, Garren S, Piot C, et al. Successful anti-PD-1 cancer immunotherapy requires T cell-dendritic cell cross-talk involving the cytokines IFN-gamma and IL-12. Immunity 2018;49:1148–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hirao M, Onai N, Hiroishi K, Watkins SC, Matsushima K, Robbins PD, et al. CC chemokine receptor-7 on dendritic cells is induced after interaction with apoptotic tumor cells: critical role in migration from the tumor site to draining lymph nodes. Cancer Res 2000;60:2209–17. [PubMed] [Google Scholar]
- 57.Ohl L, Mohaupt M, Czeloth N, Hintzen G, Kiafard Z, Zwirner J, et al. CCR7 governs skin dendritic cell migration under inflammatory and steady-state conditions. Immunity 2004;21:279–88. [DOI] [PubMed] [Google Scholar]
- 58.Roberts EW, Broz ML, Binnewies M, Headley MB, Nelson AE, Wolf DM, et al. Critical role for CD103(+)/CD141(+) dendritic cells bearing CCR7 for tumor antigen trafficking and priming of T cell immunity in melanoma. Cancer Cell 2016;30:324–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng Q, Qiu X, Zhang Z, Zhang S, Zhang Y, Liang Y, et al. PD-L1 on dendritic cells attenuates T cell activation and regulates response to immune checkpoint blockade. Nat Commun 2020;11:4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jansen CS, Prokhnevska N, Master VA, Sanda MG, Carlisle JW, Bilen MA, et al. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature 2019;576:465–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kamphorst AO, Wieland A, Nasti T,Yang S, Zhang R, Barber DL, et al. Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science 2017;355:1423–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin H, Wei S, Hurt EM, Green MD, Zhao L, Vatan L, et al. Host expression of PD-L1 determines efficacy of PD-L1 pathway blockade-mediated tumor regression. J Clin Invest 2018;128:805–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Curiel TJ, Wei S, Dong H, Alvarez X, Cheng P, Mottram P, et al. Blockade of B7-H1 improves myeloid dendritic cell-mediated antitumor immunity. Nat Med 2003;9:562–7. [DOI] [PubMed] [Google Scholar]
- 64.Zilionis R, Engblom C, Pfirschke C, Savova V, Zemmour D, Saatcioglu HD, et al. Single-Cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity 2019;50:1317–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao R, et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell 2019;179:829–45. [DOI] [PubMed] [Google Scholar]
- 66.Zhang L, Li Z, Skrzypczynska KM, Fang Q, Zhang W, O’Brien SA, et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell 2020;181:442–59. [DOI] [PubMed] [Google Scholar]
- 67.Dammeijer F, van Gulijk M, Mulder EE, Lukkes M, Klaase L, van den Bosch T, et al. The PD-1/PD-L1-checkpoint restrains T cell immunity in tumor-draining lymph nodes. Cancer Cell 2020;38:685–700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.