Abstract
Objective
Time‐restricted eating (TRE) restores circadian rhythms in mice, but the evidence to support this in humans is limited. The objective of this study was to investigate the effects of TRE on 24‐hour profiles of plasma metabolites, glucoregulatory hormones, and the subcutaneous adipose tissue (SAT) transcriptome in humans.
Methods
Men (n = 15, age = 63 [4] years, BMI 30.5 [2.4] kg/m2) were recruited. A 35‐hour metabolic ward stay was conducted at baseline and after 8 weeks of 10‐hour TRE. Assessment included 24‐hour profiles of plasma glucose, nonesterified fatty acid (NEFA), triglyceride, glucoregulatory hormones, and the SAT transcriptome. Dim light melatonin onset and cortisol area under the curve were calculated.
Results
TRE did not alter dim light melatonin onset but reduced morning cortisol area under the curve. TRE altered 24‐hour profiles of insulin, NEFA, triglyceride, and glucose‐dependent insulinotropic peptide and increased transcripts of circadian locomotor output cycles protein kaput (CLOCK) and nuclear receptor subfamily 1 group D member 2 (NR1D2) and decreased period circadian regulator 1 (PER1) and nuclear receptor subfamily 1 group D member 1 (NR1D1) at 12:00 am. The rhythmicity of 450 genes was altered by TRE, which enriched in transcripts for transcription corepressor activity, DNA‐binding transcription factor binding, regulation of chromatin organization, and small GTPase binding pathways. Weighted gene coexpression network analysis revealed eigengenes that were correlated with BMI, insulin, and NEFA.
Conclusions
TRE restored 24‐hour profiles in hormones, metabolites, and genes controlling transcriptional regulation in SAT, which could underpin its metabolic health benefit.
Study Importance.
What is already known?
Time‐restricted eating (TRE) restores circadian rhythms in the animal model.
TRE improves glucose metabolism in humans.
TRE alters the circadian rhythm of amino acid metabolism in skeletal muscle.
What does this study add?
TRE restored circadian rhythms in metabolites and glucoregulatory hormones.
TRE altered clock gene expression and restored 24‐hour profiles in genes controlling transcriptional regulation in human adipose tissue.
TRE may promote healthy adipose tissue function and metabolism.
How might these results change the direction of research or the focus of clinical practice?
The study provides the first evidence that TRE restores 24‐hour profiles to genes in adipose tissue in humans.
Providing potential mechanisms of TRE in regulating metabolic health in humans could be critical to translating TRE into clinical practice.
INTRODUCTION
The circadian system controls multiple behavioral and metabolic processes [1]. At a molecular level, the system is controlled by transcriptional‐translational feedback loops. Period (PER1/2/3) and cryptochrome (CRY1/2) circadian regulator genes are positively regulated by the transcription factors circadian locomotor output cycles protein kaput and aryl hydrocarbon receptor nuclear translocator‐like protein 1 complex (CLOCK:BMAL1/ARNTL) and are repressed by their own translation products. Additionally, nuclear activators and repressors (nuclear receptors RORα/β/γ, nuclear receptor subfamily 1 group D member 1 and 2 [NR1D1 and NR1D2]) regulate BMAL1 transcription [1].
Nutrient‐signaling molecules are strong regulators of clock genes in peripheral tissues. Feeding results in the activation of the insulin‐phosphorylated protein kinase B(pAKT)‐mammalian target of rapamycin (mTOR) pathway, which increases the stability and translation of PER [2, 3], whereas fasting activates AMP‐activated protein kinase (AMPK) and nicotinamide phosphoribosyltransferase (NAMPT), reducing the stability and transcription of CRY and PER [4, 5]. Therefore, meal timing is an entraining cue for peripheral clocks, and mistimed eating with shortened overnight fasting periods dampens peripheral clocks in mice [6, 7].
Time‐restricted eating (TRE) extends the overnight fasting period by limiting the contiguous eating duration to 6 to 10 hours [6, 8, 9, 10, 11]. In mice with diet‐induced obesity, TRE reduces body weight gain and body fat accumulation, improves glucose tolerance, and restores amplitude of core clock genes [6, 7, 11, 12]. In humans, TRE decreases body weight [9, 13] and improves cardiovascular end points, insulin sensitivity, and β‐cell responsiveness [8]. TRE also regulates the rhythmicity of transcriptional profiles involved in amino acid transport but it does not alter clock genes in human skeletal muscle [14].
The effect of TRE on 24‐hour adipose tissue transcriptomic profiles in humans remains untested. Individuals with obesity and type 2 diabetes mellitus (T2DM) have a flattened amplitude in the adipose tissue transcriptome, with <2% of expressed genes showing a diurnal rhythm compared with 8% in lean individuals [15]. Adipose tissue also plays a vital role in the body's adaptive response to fasting by inhibiting de novo fatty acid synthesis and adipogenesis and stimulating lipolysis, thereby elevating circulating nonesterified fatty acid (NEFA) [16]. In human adipose tissue, acute fasting also alters the transcripts involved in circadian clocks [16, 17]. Accordingly, we hypothesized that TRE would restore 24‐hour rhythms in metabolites, glucoregulatory hormones, and the subcutaneous adipose tissue (SAT) transcriptome in humans with obesity.
METHODS
Participants
A total of 15 men (aged 40‐70 years) with obesity (BMI = 30.5 [2.4] kg/m2; waist circumference = 113 [4] cm) participated. The study Consolidated Standards of Reporting Trials (CONSORT) diagram, sample‐size calculation, and other clinical characteristics of the study participants have been previously reported [18].
Design
This study was designed as an open‐label, pre‐post trial, including 2‐week baseline monitoring and an 8‐week TRE intervention. Individuals following a prolonged daily eating duration lifestyle (>12 h/d) during the baseline monitoring period were recruited to participate in the study in the South Australia Health and Medical Research Institute in Adelaide. First enrollment of participants was on July 17, 2018, and the trial was completed on April 3, 2019. The study was approved by the Central Adelaide Local Health Network Human Research Ethics Committee, University of Adelaide, and the University of South Australia, and it was registered with ClinicalTrials.gov (NCT03590158). Written informed consent was obtained from each participant prior to the enrollment.
In the 2 weeks prior to baseline and the week‐8 metabolic testing visit, activity levels and sleep patterns were measured by ActiGraph (wGT3X‐BT). All eating and drinking events for the entire 10‐week study duration were photographed and annotated by photograph‐based smartphone application “myCircadianClock” (https://mycircadianclock.org/, Satchidananda Panda, Salk Institute, La Jolla, California). Prior to the metabolic testing visit, standardized foods were provided at 100% of calculated total daily energy requirements (50% of carbohydrate, 30% fat, 20% protein) for 3 days. Whole‐body composition was assessed by dual‐energy x‐ray absorptiometry.
Dietary intervention
Participants were instructed to eat their habitual diets within a self‐selected consistent 10‐hour time frame each day for 8 weeks (the latest eating occasion was to be completed by 7:30 pm). Water and energy‐free beverage consumption was allowed ad libitum. No other dietary instructions were provided, but participants were asked to maintain their usual daily physical activity and sleep patterns.
Procedures during the metabolic ward stay
Metabolic testing was performed in a light‐, noise‐, and temperature‐controlled sleep and chronobiology laboratory (Figure 1A). Four participants per run arrived at 4:30 pm on the afternoon prior to testing. Dinner was provided at 6:30 pm, and an 8‐hour sleep opportunity was provided at 10:00 pm and was measured via polysomnography. For periods of wakefulness, light intensity was <50 lux and <0.03 lux during sleep. On the metabolic testing day, three identical meals and two identical snacks were provided at calculated energy balance based on the sedentary activity levels. The timing of meals differed between baseline and week 8, when snack 2 was either consumed between a 14‐hour prolonged eating window of 8:00 am to 10:00 pm (10:00 pm at baseline) or a 10‐hour restricted eating window of 8:00 am to 6:00 pm (10:30 am at week 8). Water was allowed ad libitum.
FIGURE 1.
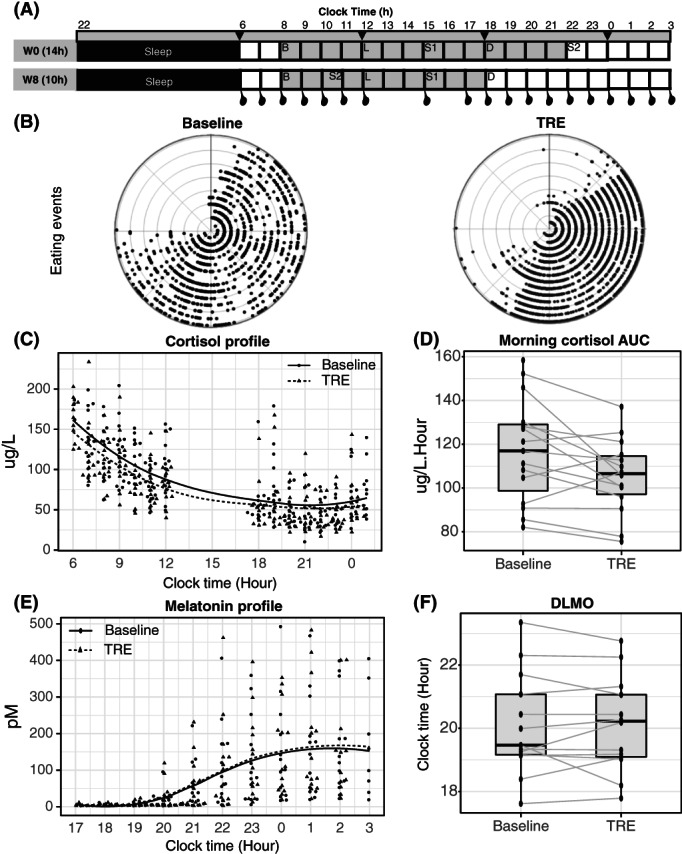
(A) Schematic of the study design: participants attended the sleep laboratory lab for a 35‐hour metabolic testing visit at baseline and week 8. Identical meals were provided within a 14‐hour time frame at W0 (upper panel, 8:00 am‐10:00 pm) and 10‐hour time frame at W8 (lower panel, 8:00 am‐6:00 pm; gray area). Abdominal SAT biopsies were performed every 6 hours from 6:00 am (black arrows), and blood samples were collected every 3 hours from 6:00 am to 3:00 am hourly from 6:00 am to 12:00 pm and 5:00 pm to 3:00 am (black dots). (B) Polar plot of eating events of each individual plotted against time of the day in each concentric circle (each circle represents a different participant, n = 15) during the baseline monitoring period (black) and during TRE intervention (gray). (C,D) Levels of cortisol (n = 15) from 6:00 am to 12:00 pm and from 6:00 pm to 1:00 am in hourly plasma samples collected during the metabolic ward stay and morning cortisol area under the curve. (E,F) Levels of melatonin (n = 13) from 5:00 pm to 3:00 am in hourly samples collected during the metabolic ward stay and DLMO at 10 pm. DLMO, dim light melatonin onset; SAT, subcutaneous adipose tissue; TRE, time‐restricted eating.
The metabolic testing day started at 6:00 am followed by an overnight fast and completed at 3:00 am the following day (i.e., for a 24‐hour period) to obtain a total of four SAT biopsies every 6 hours, as previously described [19], and eight blood samples every 3 hours from 6:00 am (Figure 1A). Blood samples collected every 3 hours were used to measure glucose, glucoregulatory hormones (insulin, ghrelin, glucagon‐like peptide‐1 [GLP‐1], gastric inhibitory polypeptide [GIP]), and metabolites (triglycerides and NEFA). A total of 20 μL of 200mM 4‐(2‐aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF, Gold Biotechnology; Cat#: A‐540‐5, CAS 30827‐99‐7) and 10 μL of dipeptidyl peptidase‐IV (DPP‐4) inhibitor (Sigma‐Aldrich; Cat#: 634867) were added to 1 mL of 3‐hour whole‐blood samples for the measurement of glucoregulatory hormones (ghrelin, GLP‐1, GIP). Blood samples were also taken every hour from 6:00 am to 12:00 am and from 5:00 pm to 3:00 am to measure morning cortisol levels and evening melatonin and cortisol levels, respectively (Figure 1A).
Plasma hormone and metabolite measurements
Blood glucose and triglycerides were measured using commercially available enzymatic kits on an AU480 clinical analyzer (Beckman Coulter, Inc.). NEFA was measured using a colorimetric assay (Randox Laboratories Ltd.). Plasma insulin (EMD), cortisol (DIAsource ImmunoAssays S.A.), and melatonin (Bühlmann Laboratories AG) were measured by radioimmunology assay, as described elsewhere [20, 21, 22]. Plasma ghrelin, GLP‐1, and GIP were measured by magnetic bead‐based quantitative immunoassay (MilliporeSigma). Samples from each participant were analyzed within the same run to minimize interassay variation.
Adipose tissue RNA sequencing
Total RNA was extracted from 50 to 100 mg of SAT using the RNeasy Lipid Tissue Mini Kit (Qiagen) according to the manufacturer's recommendations. RNA integrity number was verified with Agilent 2200 TapeStation system (Agilent Technologies). RNA‐seq libraries (complementary DNA [cDNA] libraries from polyA mRNA) and sequencing were performed using Illumina's TruSeq RNA library Preparation Kit according to manufacturer's instructions (Illumina), starting with 200 ng of total RNA. Libraries were pooled into groups of 12 to a lane and sequenced using an Illumina HiSeq 2500 with 50‐base pair (bp) single‐read chemistry.
Statistical methods
Baseline measurements and plasma values
The dim light melatonin onset (DLMO) was calculated as the absolute threshold > 10 pM for each participant [23], which was used for determining the endogenous circadian phase. Melatonin and cortisol data were plotted relative to clock time. Data were grouped into time bins: 3 hours for blood metabolites and glucoregulatery hormones and 6 hours for adipose tissue transcriptome (6:00 am to 12:00 am). Paired Student t test (two‐sided) or Wilcoxon signed rank test was applied to analyze the DLMO, morning and evening cortisol area under the curve (AUC), cortisol meal response, and objective sleep measurement. The correlation between the change in morning cortisol AUC and the change in glycated hemoglobin (HbA1c) was tested via Pearson correlation analysis. Analysis of all temporal profiles of metabolites was first carried out using a linear mixed‐effect model fit by restricted maximum likelihood, with the time of day (6:00 am, 9:00 am, 12:00 pm, 3:00 pm, 6:00 pm, 9:00 pm, 12:00 am, 3:00 am), TRE (baseline, week 8), and time of day by TRE interaction as the fixed factors and participant ID as a random factor. Data were log‐transformed if the skewness or heteroscedasticity in the residuals was observed. Statistical analysis was performed using R software (version 3.6.1; The R Foundation for Statistical Computing).
RNA‐seq quality control and analysis
Sequence images were transformed FastQ files through Illumina software (BaseCaller). The quality check was done by using FastQC, version 0.11.5 (Babraham Bioinformatics). A total of 97% of bases had a quality score greater than or equal to Q30 with a mean quality score of 36.53. Sequence alignments were performed using Star version 2.5.3a, and data were preprocessed and analyzed in the R/Bioconductor environment. Sequenced reads were mapped to the GRCh37 version of the human genome assembly (also known as hg19) [24], as downloaded from the University of California Santa Cruz Genome FTP site (http://hgdownload.cse.ucsc.edu/goldenpath/hg19/bigZips/hg19.fa.gz) using Star v2.5.3a. Gene‐level read counts were generated using Homer (v4.10) and hg19 annotation (ftp://hgdownload.soe.ucsc.edu/goldenPath/hg19/bigZips/genes/). After removing genes with <10 counts combined across all samples, the remaining genes were considered expressed and normalized using trimmed mean of M‐values normalization and their dispersion was estimated using edgeR [25].
Analysis of differentially expressed genes by edgeR
Analysis of differentially expressed genes was carried out via edgeR [25], with designs that accounted for pre‐ and postintervention at each time point. A genewise quasi‐likelihood negative binomial generalized log‐linear model was fit to the count data, and statistically differentially expressed gene expression was assessed using a likelihood ratio test and corrected for multiple hypotheses testing using the Benjamini‐Hochberg method (false discovery rate [FDR] < 0.05).
Spline regression analysis
Rhythmic analysis was not applicable for this data set, as our adipose tissue biopsy specimens were sampled at only four time points [3]. Therefore, time‐series samples were treated independently, and a natural cubic spline regression model (NCSRM) [26] with three degrees of freedom for an experimental two‐way design with one treatment factor and time as a continuous variable was fitted to the edgeR normalized count data to identify the transcripts that followed the different 24‐hour profiles after TRE versus baseline using the R package splineTimeR [27]. Differences in the fitted NCSRM coefficient values were assessed using an empirical Bayes method and were corrected for multiple hypotheses testing using the Benjamini‐Hochberg method (FDR < 0.05).
Enrichment analysis
Enriched ontology terms were found using WebGestaltR to perform overrepresentation analysis against the Molecular Signatures Database and Gene Ontology databases, using all expressed genes as the background [28]. An FDR of 5% was used to filter significantly enriched terms.
Construction of weighted gene coexpression network and identification of significant modules and hub genes
The R package WGCNA (version 1.70‐3) was used for coexpression analysis [29]. Network construction and module detection were performed on the 450 genes with variable profiles identified from the spline analysis, as previously described [29]. In brief, a signed network was constructed, and Pearson correlations were used. The correlation matrix was transferred to the adjacency matrix through the adjacency function from the WGCNA package with power β (calculated using pickSoftThreshold, 21). One minus the Topological Overlap Measure (1 − TOM) was used to calculate a dissimilarity measure and assign genes into modules based on coexpression [30], using the dynamic tree‐cutting method (minimum cluster size 30, deepSplit 2) [30]. The minimum number of genes in each module was set to 30.
The module eigengene (MEs) identified for each module is a robust representative of the character of the closely coexpressed genes that form each module. The module‐trait relationships were calculated using Pearson correlation coefficient between each ME and the clinical traits. Clinical traits included BMI, percentage of body fat mass, glucose, NEFA, triglycerides, insulin, GIP, and GLP‐1. The significance of the Pearson correlation between modules and traits was determined using an asymptotic t test [31].
Correlation between the MEs and the gene expression profiles was calculated to identify the most central element in each module's network, the “hub gene” of the module. Enrichment analysis of each module's co‐expressed genes was conducted according to the previously described methods.
RESULTS
Participants
The clinical characteristics are briefly summarized. A total of 15 men (mean [SD], age: 63 [4] years; waist circumference: 113 [4] cm, BMI: 30.5 [2.4]; body mass: 95.2 [12.2] kg; body fat percentage: 34.3% [1.2%]) started and completed the study without adverse events. Photograph‐based food diary records showed excellent adherence to TRE, with a significant reduction in eating hours per day (14.6 [1.0] hours vs. 10.6 [1.0] hours, p < 0.0001; Figure 1B, polar plot).
TRE reduced stress hormone and increased rapid eye movement sleep stage but did not alter melatonin rhythm
TRE reduced morning cortisol AUC by −17.4 (9.4) μg/L/h (p = 0.02; Figure 1C,D) but did not significantly alter evening cortisol AUC (change = −4.4 [4.5] μg/L/h, p = 0.35; Figure 1C). The exploratory analysis showed that the change in morning cortisol AUC was positively correlated with the change in HbA1c (r = 0.60, p = 0.02), but the significance was lost after multiple adjustment (Supporting Information Table S1). TRE also significantly reduced the meal‐induced cortisol response after dinner by −10.7 μg/L/h (p = 0.04). TRE did not alter DLMO (p = 0.86; Figure 1E,F) but increased the percentage of rapid eye movement sleep without changing sleep onset latency, total sleep time, sleep efficiency, and percentage of sleep stage 1, 2, and 3 (Supporting Information Table S2).
TRE altered 24‐hour profiles of plasma insulin, NEFA, triglycerides, and GIP concentrations
A time of day by TRE interaction was observed in 24‐hour profiles of insulin (F = 2.39, p < 0.02; Figure 2A), NEFA (F = 4.76, p < 0.001; Figure 2B), triglycerides (F = 3.71, p < 0.001; Figure 2C), and GIP (F = 5.55, p < 0.001; Figure 2D). Post hoc analysis showed that, in response to TRE, insulin was increased at midday (p = 0.02) and decreased at midnight (p = 0.01; Figure 2A). NEFA concentrations were decreased at 9:00 am and midday (p = 0.04 and 0.03, respectively) and increased at midnight (p < 0.001; Figure 2B), whereas triglycerides were lower at midnight and 3:00 am (p < 0.001 and 0.003, respectively; Figure 2C). There were no changes in the concentrations of NEFA and triglycerides at other time points. Only GIP was higher at 9:00 am, 12:00 pm, and 6:00 pm (p = 0.05, 0.003, and 0.01, respectively) and lower at midnight (p < 0.001; Figure 2D). There were no changes in GLP‐1 (Figure 2E), glucose (Figure 2F), and ghrelin (Supporting Information Figure S1 and Table S3).
FIGURE 2.
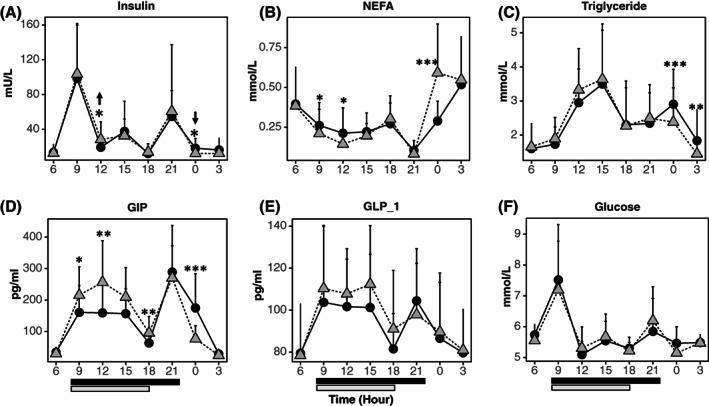
The impact of TRE on 24‐hour profiles of plasma metabolites and glucoregulatory hormones. Linear mixed‐effect model fit with the time of day (6:00 am, 9:00 am, 12:00 pm, 3:00 pm, 6:00 pm, 9:00 pm, 12:00 am, 3:00 am), TRE (baseline, week 8), and time of day by TRE interaction as the fixed factors and participant ID as a random factor (n = 15, mean ± SD). Time of day by TRE effect, post hoc test: *p < 0.05, **p < 0.01, ***p < 0.001. Solid line represents baseline, dash line represents TRE. Black bar and gray bar at the bottom represent the eating window at baseline (8:00 am to 10:00 pm) and after TRE (8:00 am to 6:00 pm). GIP, glucose‐dependent insulinotropic peptide; GLP‐1, glucagon‐like peptide‐1; NEFA, nonesterified fatty acid; TRE, time‐restricted eating.
TRE altered SAT transcriptional profiles
RNA sequencing of SAT samples detected that 319, 610, and 3407 transcripts were differentially expressed by TRE at 6:00 am, 6:00 pm, and 12:00 am, respectively (Supporting Information Tables S4‐S6, respectively). Only five genes were altered at all of these time points, namely, zinc finger protein 587 (ZNF587), carboxyl‐terminal domain phosphatase subunit 1(CTDP1), retinoic acid‐induced 1 (RAI1), chromobox 4 (CBX4), and transforming growth factor beta‐like stimulated clone 22 domain family member 4 (TSC22D4).
First, we examined the 24‐hour profiles of the known core clock genes. TRE increased CLOCK (FDR = 0.054) and NR1D2 (FDR = 0.011) gene expression and decreased the expression of PER1 (FDR = 0.023) and NR1D1 (FDR = 0.016) at 12:00 am (Figure 3). To analyze the impact of TRE on the temporal profile of adipose tissue transcripts, we used spline regression models and identified 450 genes (Figure 4 and Supporting Information Table S7). The changes in the 24‐hour profiles of core clock genes were not detected by spline analysis. Pathway analysis yielded four gene ontology pathways (Table 1): 15 genes were enriched in transcription corepressor activity pathway (FDR = 0.045; Supporting Information Table S8); 19 genes were enriched in DNA‐binding transcription factor binding pathway (FDR < 0.001; Supporting Information Table S8); 13 genes were enriched in the regulation of chromatin organization pathway (FDR < 0.001; Supporting Information Table S8), and 24 genes were enriched in small GTPase binding (FDR = 0.008; Supporting Information Table S8).
FIGURE 3.
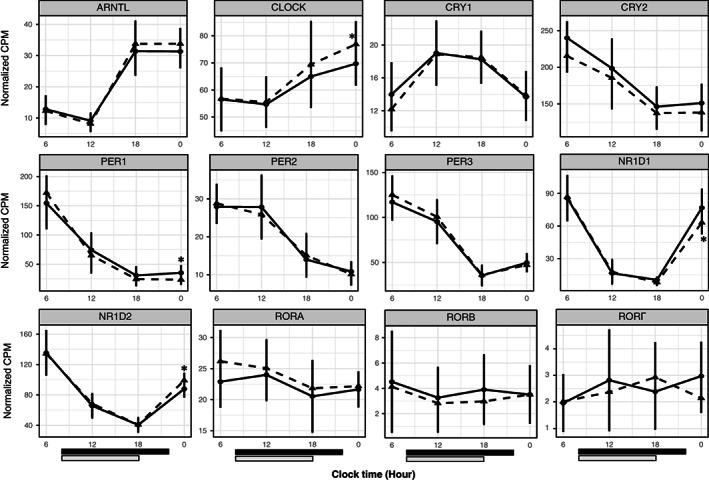
Twenty‐four‐hour profiles of core clock genes. Average (SD, n = 12) normalized counts of core clock genes in subcutaneous adipose tissue at different times of the day. The solid line represents baseline; the dash line represents time‐restricted eating (TRE). Black bar and gray bar at the bottom represent the eating window at baseline (8:00 am to 10:00 pm) and after TRE (8:00 am to 6:00 pm). *False discovery rate < 0.05.
FIGURE 4.
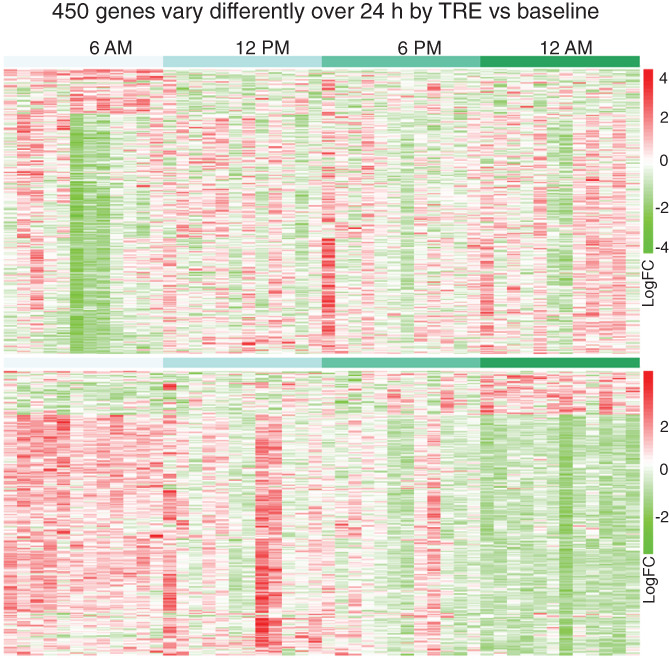
Spline regression analysis identified 450 profiles of genes that displayed different rhythmicity by TRE (lower panel) versus baseline (upper panel; n = 12 at each time point). TRE, time‐restricted eating. [Color figure can be viewed at wileyonlinelibrary.com]
TABLE 1.
GO enrichment analysis of 450 genes that showed altered rhythmicity in response to TRE
| ID | Category | Gene set term | Overlap | Enrichment ratio | p value | FDR |
|---|---|---|---|---|---|---|
| GO_MF | GO:0031267 | GO_SMALL_GTPASE_BINDING | 24 | 2.538 | 0.000 | 0.008 |
| GO_BP | GO:0006325 | GO_REGULATION_OF_CHROMATIN_ORGANIZATION | 13 | 3.367 | 0.000 | 0.025 |
| GO_ MF | GO:0140297 | GO_DNA_BINDING_TRANSCRIPTION_FACTOR_BINDING | 19 | 2.599 | 0.000 | 0.026 |
| GO_ MF | GO:0003714 | GO_TRANSCRIPTION_COREPRESSOR_ACTIVITY | 15 | 2.834 | 0.000 | 0.045 |
Abbreviations: BP, biological process; FDR, false discovery rate; GO, gene ontology; MF, molecular function; TRE, time‐restricted eating.
Network analysis indicated gene alteration in response to TRE in human adipose tissue
Four distinct coexpression MEs were identified from spline analysis by using WGCNA (Figure 5A). The calculating module‐trait correlation is presented as a heatmap (Figure 5B). Genes that clustered in the gray module (n = ) were negatively correlated with BMI, percentage of body fat mass, insulin, and GIP and positively correlated with NEFA. Genes in the brown (n = 43) and blue (n = 105) modules were positively correlated with BMI, percentage of body fat, and insulin, and negatively correlated with NEFA. The top hub gene for the blue module was fibrosin (FBRS); for brown, it was Rho guanine nucleotide exchange factor 1 (ARHGEF1); for turquoise, it was methyl‐CpG binding domain protein (MBD3); and for gray, it was heat shock protein 90 alpha family class B member 1 (HSP90AB1) (Figure 5C). To further explore the key regulatory nodes, we applied pathway analysis of genes in each module. Only the genes in the gray model, showing the strongest correlations with the clinical traits, were enriched in specific pathways. These pathways were chaperone‐mediated protein complex assembly, response to topologically incorrect protein, posttranslational protein modification, and RNA splicing while maximizing gene coverage (Figure 5D).
FIGURE 5.
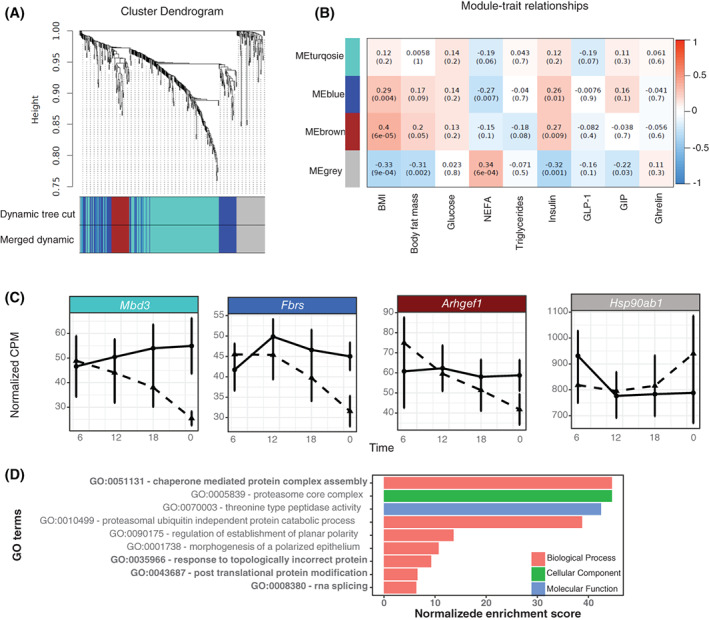
Weighted gene coexpression network analysis (WGCNA) and target modules and hub genes screening. (A) Network analysis of 450 genes generated from spline analysis to identify distinct modules of coexpression genes. (B) Pearson correlation coefficient between the eigengene of modules and the sample feature vector. Numbers represent the correlation coefficients and numbers in brackets indicate the corresponding p values. (C) Top hub gene in each eigengene of module. (D) Significant GO terms that were enriched in the gray module (FDR < 0.05). Bolded four GO terms represent the top gene sets while maximizing gene coverage. ARHGEF1, Rho/Rac guanine nucleotide exchange factor 18; FBRS, fibrosin; FDR, false discovery rate; GO, gene ontology; HSP90AB1, heat shock protein 90 alpha family class B member 1; MBD3, methyl‐CpG binding domain protein. [Color figure can be viewed at wileyonlinelibrary.com]
DISCUSSION
TRE improves metabolic health by restoring circadian rhythms in rodent models [6, 7]. However, the evidence that TRE alters 24‐hour rhythms in humans is currently limited. In this study, TRE decreased morning cortisol and midnight levels of insulin, GIP, and triglycerides and increased NEFA. Four out of twelve core clock genes were altered in adipose tissue, and four hundred and fifty genes that were arrhythmic at baseline became rhythmic in response to TRE. Overrepresentation analysis indicated that the pathways altered by TRE were transcription corepressor activity, DNA‐binding transcription factor binding, regulation of chromatin organization, and small GTPase binding. WGCNA found the correlations among three eigengenes and insulin, NEFA, and BMI.
TRE improves insulin sensitivity and increases NEFA suppression in response to meals given at breakfast and over 24 hours in humans [8, 18, 32]. The present study extends these findings by showing changes in the 24‐hour profiles of insulin, NEFA, triglycerides, and GIP in a manner consistent with improved insulin sensitivity and metabolic health. Parr et al. also reported that TRE altered the 24‐hour profiles of insulin, NEFA, GIP, and GLP‐1 [32]. TRE did not alter DLMO, which is primarily regulated by the light‐dark cycle [33] and has been reported previously [34]. Interestingly, rapid eye movement sleep was increased by TRE (~2.1%, 9 min), indicating an improvement in sleep health, which is reverse to what happens during Ramadan fasting, when food intake occurs later in the evening [35]. Contrary to expectations [36, 37], evening cortisol was not reduced by TRE, potentially because of the small size of the late evening snack in this study (~200 kcal). However, morning cortisol was lowered by TRE, which could have contributed to the lower rise in morning glucose and HbA1c [38].
The molecular machinery of the circadian clock Is a combination of transcriptional activators and repressors coordinately acting at thousands of sites in the chromatin fiber, driving a highly specific program of gene expression in a circadian manner to maintain metabolic homeostasis [1, 39]. Obesity is associated with a dampening in the amplitude of core clock genes, including CLOCK, PER1, NR1D1, and NR1D2, in adipose tissue of mice and humans [15], which is restored by TRE in mice [6, 7, 11]. In the present study, TRE increased CLOCK and NR1D2 and decreased PER1 and NR1D1 transcripts at midnight. Only one study has previously performed serial time point sampling over 24 hours under unrestricted versus time‐restricted conditions in skeletal muscle [14]. In that study, TRE altered the rhythmicity of genes involved in amino acid transport but did not alter clock genes [14]. Interestingly, muscle clocks are known to be less responsive to food cues than the liver or adipose tissue clocks in mice [40]. Two other human studies have reported that TRE induced changes in clock genes at two four time points. Early TRE decreased PER1 at 8:00 pm and increased CRY1/2 and RORΑ at 8:00 am and 8:00 pm [41]. Increased amplitude in BMAL1, CRY1, PER2, and RORα was also reported in white blood cells of patients with T2DM who ate three meals in 12 hours versus six meals in 15 hours [42].
In human adipose tissue, we show for the first time, to our knowledge, a rhythmic induction of 450 genes following TRE [17]. An advance occurs as transcripts positively related to PER1 are reinforced by temporal feeding, whereas genes negatively related to PER1 are reinforced by temporal fasting [17]. Insulin has recently emerged as a mediator of clocks that induces translation of PER2 and PER1 in human stem cell‐derived adipocytes, mouse 3 T3‐L1 cells, and adipose tissue explants from mPer2Luc knockin mice [2, 3]. PER2 and PER3 transcripts were also upregulated, and NR1D2 was downregulated by insulin versus saline in human adipose tissue [3]. In total, ~2% of transcripts were altered by insulin versus saline, including AMPK, phosphatidylinositol, and mTOR pathways [3]. We speculate that TRE induced reductions in evening insulin may have driven the alterations in clocks and downstream genes, which was supported by a correlation between insulin and mEs. Two eigengene modules had a positive relationship with insulin, and one module showed a negative relationship with insulin. Pathway analysis of the latter module showed enrichment in chaperone‐mediated protein complex assembly, post‐translational protein modification, response to topologically incorrect protein, and RNA splicing. The hub gene was HSP90AB1, a molecular co‐chaperone isoform of heat shock protein 90 that is involved in maintaining cellular levels of BMAL1 [43], and, therefore, potentially another key factor in TRE induced restoration in adipose tissue rhythms.
Having healthy adipose tissue is essential for optimal metabolic functioning. Circadian timing is a crucial aspect in adipose tissue health, promoting the accumulation of lipids during periods of energy intake and lipid mobilization during times of fasting. In the 450‐gene‐set analysis, the pathways in adipose tissue that were altered by TRE were those involved in the regulation of chromatin organization (e.g., sterol regulatory element binding transcription factor 1 [SREBF1]; ribosomal protein S6 kinase A4 [RPS6KA4]; tripartite Motif Containing 28 [TRIM28]; C‐terminal binding protein 1 [CTBP1]; lysine methyltransferase 2B [KMT2B]; and H1 histone family member X [H1FX]), transcription corepressor activity, and DNA transcription factor binding (e.g., nuclear receptor corepressor 2 [NCOR2], CTBP1, CBX4, and transcription factor 3 [TCF3]). Previously, pathways encoding histone deacetylation and transcriptional regulation, including DNA‐binding transcription repressor, protein serine/threonine phosphatase, and transcription corepressor/coregulatory/cofactors, were altered by TRE in human muscle [14]. SREBF1 is a transcription factor involved in the regulation of lipogenesis in adipose tissue. SREBF1 activity is regulated by nutrient availability through the insulin‐mTOR complex1 signaling pathway, peaking during the nocturnal feeding period in mice [44] and lowered by fasting [45]. TRE also altered SREBF1 in the liver of CRY1−/−, CRY2−/−, and BMAL1−/− knockout mice [44, 46]. Therefore, TRE could indicate restoration in the rhythmic regulation of de novo lipogenesis in SAT, which may promote insulin sensitization of adipocytes [47]. This was supported by downregulated SREBF1 targeted pathways at 6:00 am enriched by gene‐set enrichment analysis (data are not shown), such as LDL receptor (LDLR), ELOVL fatty acid elongase 6 (ELOVL6), acetyl‐CoA carboxylase alpha (ACACA), malic enzyme 1 (ME1), 3‐hydroxy‐3‐methylglutaryl‐CoA synthase 1 (HMGCS1), acyl‐CoA synthetase short chain family member 2 (ACSS2), ATP citrate lyase (ACLY), acetoacetyl‐CoA synthetase (AACS), and sterol‐C5‐desaturase (SC5D). Small GTPase binding regulates vesicular trafficking, and the temporal profile of this pathway was altered by TRE. Changes in Rho GTPases (Rho/Rac guanine nucleotide exchange factor 17 or 18 [ARHGEF17 or 18]; Rho GDP dissociation inhibitor alpha [ARHGDIα]; Pleckstrin homology and Rho GEF domain containing G3 [PLEKHG3]; ARHGEF1; Rac family small GTPase 1 [RAC1]; CDC42 effector protein 1 [CDC42EP1]) and Rab GTPases (RAB11 family interacting protein 3 [RAB11FIP3]), which promote glucose uptake through increasing glucose transport type 4 (GLUT4) translocation to cell surface [48, 49], were observed. The rhythmicity in the small GTPases contribute to known daily variation in insulin sensitivity in peripheral tissues [49], and we speculate that TRE could restore diurnal variation in insulin‐mediated glucose uptake in adipose tissue.
Finally, cell proliferation in adipose tissue is diurnal, with a loss in rhythm following high‐fat diet and unrestricted eating [50]. The present study shows that TRE induced changes in the temporal regulation of transcription factors involved in adipogenesis (including TRIM28, disheveled segment polarity protein [DVL], and TCF3) and adipose tissue browning, including cAMP‐regulated transcriptional coactivator (CRTC) family [51], CTBP1 (a corepressor of peroxisome proliferator‐activated receptor γ) [52], and CBX4 (a polycomb group protein for maintaining the stability of transcriptional coactivator PRDM16) [53]. Therefore, TRE could restore the temporal regulation in the cellular fate of preadipocytes in humans, which could also influence healthy adipose tissue expansion and function; however, this requires further study.
The study has several limitations. This was an uncontrolled, pre‐post study with a small sample size, solely in White men in their 60s with obesity. As such, our findings cannot be directly extended to men from different age groups, women, individuals with normal weight, those with established metabolic disorders such as T2DM and metabolic syndrome, and people from other races. Because sampling was not performed at more than four time points, we did not perform a cosine model to predict the mesor, amplitude, and phase shift of the 24‐hour profiles of the adipose tissue transcriptome and glucoregulatory hormones. The changes in the adipose tissue transcriptome could have partially resulted from different lengths of fasting prior to the adipose tissue biopsy at midnight between baseline and TRE. Spline analysis displayed a group of altered gene expression profiles, and we are assuming the rhythmicity of these genes may be restored by TRE; however, this needs to be further confirmed in the constant routine protocol.
CONCLUSION
TRE restored 24‐hour profiles in glucoregulatory factors and adipose tissue transcripts in a manner that was suggestive of healthy adipose tissue function and metabolism.
AUTHOR CONTRIBUTIONS
Leonie K. Heilbronn, Amy T. Hutchison, Siobhan Banks, Gary A. Wittert, and Lijun Zhao designed the research. Leonie K. Heilbronn, Amy T. Hutchison, Bo Liu, and Lijun Zhao collected data. Lijun Zhao and Leonie K. Heilbronn analyzed plasma biomarkers and Lijun Zhao extracted high‐quality mRNA from adipose tissue. Gary A. Wittert and Campbell H. Thompson provided clinical support and supervision, Leanne Nguyen and John Au provided clinical support. Emily N.C. Manoogian and Satchidananda Panda supplied the myCircadianClock application. Emily N.C. Manoogian, Satchidananda Panda, and Hiep D. Le prepared adipose tissue for RNA sequencing. April E. Williams and Lijun Zhao performed adipose tissue RNA‐seq data analysis. Siobhan Banks supervised metabolic test days during the ward stay at the sleep and chronobiology laboratory. Lijun Zhao and Andrew Vincent performed statistical analysis and data visualization. All authors contributed to the data interpretation and preparation of the manuscript. Leonie K. Heilbronn had full access to the data and had primary responsibility for the final publication.
FUNDING INFORMATION
The research was funded by Diabetes Australia (grant number Y18G‐HEIL). Lijun Zhao is supported by a Beacon of Enlightenment Scholarship from The University of Adelaide.
CONFLICT OF INTEREST
Gary A. Wittert has received research funding for testosterone pharmacology studies (Weight Watchers, Bayer) as well as speaking (Besins, Bayer, World Obesity), expert testimony (Australian Health Practitioners Regulation Agency), International Advisory Board (Bayer), and consultancy (Elsevier) fees. Satchidananda Panda is the author of the books The Circadian Code and The Circadian Diabetes Code. The other authors declared no conflict of interest.
CLINICAL TRIAL REGISTRATION
ClinicalTrials.gov identifier NCT03590158.
Supporting information
Figure S1 Supporting Information.
Table S1 Supporting Information
ACKNOWLEDGMENTS
The research in the SP lab is supported by NIH (grant number: DK115214 and DK118278) and The Robert Wood Johnson Foundation (grant number 76014). The authors also thank Mark Salkeld and Scott Standfield for the technical support in melatonin extraction and measurement. Open access publishing facilitated by The University of Adelaide, as part of the Wiley ‐ The University of Adelaide agreement via the Council of Australian University Librarians.
Zhao L, Hutchison AT, Liu B, et al. Time–restricted eating alters the 24‐hour profile of adipose tissue transcriptome in men with obesity. Obesity (Silver Spring). 2023;31(Suppl. 1):63‐74. doi: 10.1002/oby.23499
Funding information Beacon of Enlightenment Scholarship from The University of Adelaide; Diabetes Australia Research Trust, Grant/Award Number: Y18G‐HEIL; Robertwood Johnson Foundation, Grant/Award Number: 76014; National Institutes of Health, Grant/Award Numbers: DK118278, DK115214
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available upon reasonable request for academic use. The raw sequencing data are deposited in the NCBI Gene Expression Omnibus (GEO) repository (accession number GSE168705). The statistical analysis code are also available upon request.
REFERENCES
- 1. Sinturel F, Petrenko V, Dibner C. Circadian clocks make metabolism run. J Mol Biol. 2020;432:3680‐3699. [DOI] [PubMed] [Google Scholar]
- 2. Crosby P, Hamnett R, Putker M, et al. Insulin/IGF‐1 drives PERIOD synthesis to entrain circadian rhythms with feeding time. Cell. 2019;177:896‐909.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tuvia N, Pivovarova‐Ramich O, Murahovschi V, et al. Insulin directly regulates the circadian clock in adipose tissue. Diabetes. 2021;70:1985‐1999. [DOI] [PubMed] [Google Scholar]
- 4. Lamia KA, Sachdeva UM, DiTacchio L, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437‐440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone‐Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK‐SIRT1. Science. 2009;324:654‐657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hatori M, Vollmers C, Zarrinpar A, et al. Time‐restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high‐fat diet. Cell Metab. 2012;15:848‐860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chaix A, Lin T, Le HD CMW, Panda S. Time‐restricted feeding prevents obesity and metabolic syndrome in mice lacking a circadian clock. Cell Metab. 2018;29:303‐319.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sutton EF, Beyl R, Early KS, Cefalu WT, Ravussin E, Peterson CM. Early time‐restricted feeding improves insulin sensitivity, blood pressure, and oxidative stress even without weight loss in men with prediabetes. Cell Metab. 2018;27:1212‐1221.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cienfuegos S, Gabel K, Kalam F, et al. Effects of 4‐ and 6‐h time‐restricted feeding on weight and cardiometabolic health: a randomized controlled trial in adults with obesity. Cell Metab. 2020;32:366‐378.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wilkinson MJ, Manoogian ENC, Zadourian A, et al. Ten‐hour time‐restricted eating reduces weight, blood pressure, and atherogenic lipids in patients with metabolic syndrome. Cell Metab. 2019;31:92‐104.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chaix A, Zarrinpar A, Miu P, Panda S. Time‐restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell Metab. 2014;20:991‐1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Regmi P, Chaudhary R, Page AJ, et al. Early or delayed time‐restricted feeding prevents metabolic impact of obesity in mice. J Endocrinol. 2021;248:75‐86. [DOI] [PubMed] [Google Scholar]
- 13. Gabel K, Hoddy KK, Haggerty N, et al. Effects of 8‐hour time restricted feeding on body weight and metabolic disease risk factors in obese adults: a pilot study. Nutr Healthy Aging. 2018;4:345‐353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lundell LS, Parr EB, Devlin BL, et al. Time‐restricted feeding alters lipid and amino acid metabolite rhythmicity without perturbing clock gene expression. Nat Commun. 2020;11:4643. doi: 10.1038/s41467-020-18412-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stenvers DJ, Jongejan A, Atiqi S, et al. Diurnal rhythms in the white adipose tissue transcriptome are disturbed in obese individuals with type 2 diabetes compared with lean control individuals. Diabetologia. 2019;62:704‐716. [DOI] [PubMed] [Google Scholar]
- 16. Defour M, Michielsen CCJR, O'Donovan SD, Afman LA, Kersten S. Transcriptomic signature of fasting in human adipose tissue. Physiol Genomics. 2020;52:451‐467. [DOI] [PubMed] [Google Scholar]
- 17. Loboda A, Kraft WK, Fine B, et al. Diurnal variation of the human adipose transcriptome and the link to metabolic disease. BMC Med Genomics. 2009;2:7. doi: 10.1186/1755-8794-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao L, Hutchison AT, Liu B, et al. Time‐restricted eating improves glycemic control and dampens energy‐consuming pathways in human adipose tissue. Nutrition. 2022;96:111583. doi: 10.1016/j.nut.2021.111583 [DOI] [PubMed] [Google Scholar]
- 19. Tam CS, Viardot A, Clément K, et al. Short‐term overfeeding may induce peripheral insulin resistance without altering subcutaneous adipose tissue macrophages in humans. Diabetes. 2010;59:2164‐2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Riad‐Fahmy D, Read GF, Gaskell SJ, Dyas J, Hindawi R. A simple, direct radioimmunoassay for plasma cortisol, featuring a 125I radioligand and a solid‐phase separation technique. Clin Chem. 1979;25:665‐658. [PubMed] [Google Scholar]
- 21. Fraser S, Cowen P, Franklin M, Lewy AJ. Direct radioimmunoassay and gas chromatography‐mass spectrometry compared for determination of melatonin in plasma. Clin Chem. 1983;29:1703‐1704. [PubMed] [Google Scholar]
- 22. Morgan CR, Lazarow A. Immunoassay of insulin: two antibody system: plasma insulin levels of normal, subdiabetic and diabetic rats. Diabetes. 1963;12:115‐126. [Google Scholar]
- 23. Benloucif S, Burgess HJ, Klerman EB, et al. Measuring melatonin in humans. J Clin Sleep Med. 2008;4:66‐69. [PMC free article] [PubMed] [Google Scholar]
- 24. Heinz S, Benner C, Spann N, et al. Simple combinations of lineage‐determining transcription factors prime cis‐regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576‐589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen Y, Lun ATL, Smyth GK. From reads to genes to pathways: differential expression analysis of RNA‐Seq experiments using Rsubread and the edgeR quasi‐likelihood pipeline. F1000Res. 2016;5:1438. doi: 10.12688/f1000research.8987.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Michna A, Braselmann H, Selmansberger M, et al. Natural cubic spline regression modeling followed by dynamic network reconstruction for the identification of radiation‐sensitivity Gene Association networks from time‐course transcriptome data. PLoS One. 2016;11:e0160791. doi: 10.1371/journal.pone.0160791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Michna A. Package ‘splineTimeR’ Time‐course differential gene expression data analysis using spline regression models followed by gene association network reconstruction. Published July 24, 2022. https://www.bioconductor.org/packages/devel/bioc/manuals/splineTimeR/man/splineTimeR.pdf.
- 28. Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199‐W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Horvath S, Zhang B, Carlson M, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci USA. 2006;103:17402‐17407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakamura H, Fujii K, Gupta V, et al. Identification of key modules and hub genes for small‐cell lung carcinoma and large‐cell neuroendocrine lung carcinoma by weighted gene co‐expression network analysis of clinical tissue‐proteomes. PLoS One. 2019;14:e0217105. doi: 10.1371/journal.pone.0217105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang B, Horvath S. A general framework for weighted gene co‐expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128 [DOI] [PubMed] [Google Scholar]
- 32. Parr EB, Devlin BL, Radford BE, Hawley JA. A delayed morning and earlier evening time‐restricted feeding protocol for improving glycemic control and dietary adherence in men with overweight/obesity: a randomized controlled trial. Nutrients. 2020;12:505. doi: 10.3390/nu12020505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zeitzer JM, Dijk DJ, Kronauer RE, Brown EN, Czeisler CA. Sensitivity of the human circadian pacemaker to nocturnal light: melatonin phase resetting and suppression. J Physiol. 2000;526(Pt 3):695‐702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wehrens SMT, Christou S, Isherwood C, et al. Meal timing regulates the human circadian system. Curr Biol. 2017;27:1768‐1775.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Almeneessier AS, BaHammam AS. How does diurnal intermittent fasting impact sleep, daytime sleepiness, and markers of the biological clock? Current insights. Nat Sci Sleep. 2018;10:439‐452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gu C, Brereton N, Schweitzer A, et al. Metabolic effects of late dinner in healthy volunteers –a randomized crossover clinical trial. J Clin Endocrinol Metab. 2020;105:2789‐2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Richter J, Herzog N, Janka S, Baumann T, Kistenmacher A, Oltmanns KM. Twice as high diet‐induced thermogenesis after breakfast vs dinner on high‐calorie as well as low‐calorie meals. J Clin Endocrinol Metab. 2020;105:dgz311. doi: 10.1210/clinem/dgz311 [DOI] [PubMed] [Google Scholar]
- 38. Ortiz R, Kluwe B, Odei JB, et al. The association of morning serum cortisol with glucose metabolism and diabetes: the Jackson Heart Study. Psychoneuroendocrinology. 2019;103:25‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhu Q, Belden WJ. Molecular regulation of circadian chromatin. J Mol Biol. 2020;432:3466‐3482. [DOI] [PubMed] [Google Scholar]
- 40. Yasumoto Y, Hashimoto C, Nakao R, et al. Short‐term feeding at the wrong time is sufficient to desynchronize peripheral clocks and induce obesity with hyperphagia, physical inactivity and metabolic disorders in mice. Metabolism. 2016;65:714‐727. [DOI] [PubMed] [Google Scholar]
- 41. Jamshed H, Beyl R, Della Manna D, Yang E, Ravussin E, Peterson C. Early time‐restricted feeding improves 24‐hour glucose levels and affects markers of the circadian clock, aging, and autophagy in humans. Nutrients. 2019;11:1234. doi: 10.3390/nu11061234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jakubowicz D, Landau Z, Tsameret S, et al. Reduction in glycated hemoglobin and daily insulin dose alongside circadian clock upregulation in patients with type 2 diabetes consuming a three‐meal diet: a randomized clinical trial. Diabetes Care. 2019;42:2171‐2180. [DOI] [PubMed] [Google Scholar]
- 43. Schneider R, Linka RM, Reinke H. HSP90 affects the stability of BMAL1 and circadian gene expression. J Biol Rhythms. 2014;29:87‐96. [DOI] [PubMed] [Google Scholar]
- 44. Gilardi F, Migliavacca E, Naldi A, et al. Genome‐wide analysis of SREBP1 activity around the clock reveals its combined dependency on nutrient and circadian signals. PLoS Genet. 2014;10:e1004155. doi: 10.1371/journal.pgen.1004155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Le Martelot G, Claudel T, Gatfield D, et al. REV‐ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA. 2009;106:21453‐24158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Song Z, Xiaoli AM, Yang F. Regulation and metabolic significance of de novo lipogenesis in adipose tissues. Nutrients. 2018;10:1383. doi: 10.3390/nu10101383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Z, Oh E, Clapp DW, Chernoff J, Thurmond DC. Inhibition or ablation of p21‐activated kinase (PAK1) disrupts glucose homeostatic mechanisms in vivo. J Biol Chem. 2011;286:41359‐41367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gendaszewska‐Darmach E, Garstka MA, Błażewska KM. Targeting small GTPases and their Prenylation in diabetes mellitus. J Med Chem. 2021;64:9677‐9710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ribas‐Latre A, Santos RB, Fekry B, et al. Cellular and physiological circadian mechanisms drive diurnal cell proliferation and expansion of white adipose tissue. Nat Commun. 2021;12:3482. doi: 10.1038/s41467-021-23770-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sonntag T, Ostojić J, Vaughan JM, et al. Mitogenic signals stimulate the CREB coactivator CRTC3 through PP2A recruitment. iScience. 2019;11:134‐145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vernochet C, Peres SB, Davis KE, et al. C/EBPalpha and the corepressors CtBP1 and CtBP2 regulate repression of select visceral white adipose genes during induction of the brown phenotype in white adipocytes by peroxisome proliferator‐activated receptor gamma agonists. Mol Cell Biol. 2009;29:4714‐4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen Q, Huang L, Pan D, Zhu LJ, Wang YX. Cbx4 Sumoylates Prdm16 to regulate adipose tissue thermogenesis. Cell Rep. 2018;22:2860‐2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Supporting Information.
Table S1 Supporting Information
Data Availability Statement
The data that support the findings of this study are available upon reasonable request for academic use. The raw sequencing data are deposited in the NCBI Gene Expression Omnibus (GEO) repository (accession number GSE168705). The statistical analysis code are also available upon request.


