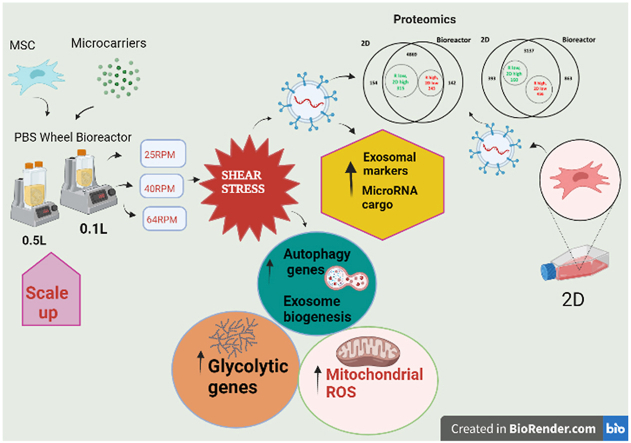
Abstract
Human mesenchymal stromal cells (hMSCs) are mechanically sensitive undergoing phenotypic alterations when subjected to shear stress, cell aggregation, and substrate changes encountered in 3D dynamic bioreactor cultures. However, little is known about how bioreactor microenvironment affects the secretion and cargo profiles of hMSC-derived extracellular vesicles (EVs) including the subset, “exosomes”, which contain therapeutic proteins, nucleic acids, and lipids from the parent cells. In this study, bone marrow–derived hMSCs were expanded on 3D Synthemax II microcarriers in the PBS mini 0.1L Vertical-Wheel bioreactor system under variable shear stress levels at 25, 40, and 64 RPM (0.1–0.3 dyn/cm2). The bioreactor system promotes EV secretion from hMSCs by 2.5-fold and upregulates the expression of EV biogenesis markers and glycolysis genes compared to the static 2D culture. The microRNA cargo was also altered in the EVs from bioreactor culture including the upregulation of miR-10, 19a, 19b, 21, 132, and 377. EV protein cargo was characterized by proteomics analysis, showing upregulation of metabolic, autophagy and ROS-related proteins comparing with 2D cultured EVs. In addition, the scalability of the Vertical-Wheel bioreactor system was demonstrated in a 0.5L bioreactor, showing similar or better hMSC-EV secretion and cargo content compared to the 0.1L bioreactor. This study advances our understanding of bio-manufacturing of stem cell-derived EVs for applications in cell-free therapy towards treating neurological disorders such as ischemic stroke, Alzheimer's disease, and multiple sclerosis.
Keywords: Human mesenchymal stromal cells, Extracellular vesicles, Cargo profile, 3D microcarriers, Shear stress, Vertical-Wheel bioreactor
Graphical abstract
Highlights
-
•
The bioreactor system promotes exosome secretion of hMSCs by 2.5-fold and upregulates the exosome biogenesis markers.
-
•
The microRNA cargo is also promoted in the exosome from bioreactor culture.
-
•
The exosome protein cargo shows high similarity to exosome of 3D hMSC aggregates.
-
•
The scalability of the Vertical-Wheel bioreactor system is demonstrated in 0.5L bioreactor.
-
•
This study advances our understanding of bio-manufacturing of stem cell-derived exosomes.
1. Introduction
The number of publications and clinical trials for human mesenchymal stromal cells (hMSCs) and their secreted extracellular vesicles (EVs) have been growing exponentially over the last two decades as their potential for facilitating tissue repair and treating autoimmune disorders has been realized [[1], [2], [3], [4], [5], [6], [7], [8], [9]]. As a direct result, the demand for therapeutically potent hMSCs and the derived EVs has risen dramatically highlighting the need for xeno-free bioreactor culture systems that are able to deliver high cell densities without compromising inherent hMSC properties underpinned by energy metabolism [5,10].
Recent studies have demonstrated hMSCs having limited migration capacity and poor survival at lesion sites post transplantation, leading to the conclusion that the main therapeutic benefits of hMSCs are mechanistically occurring through paracrine action and not through direct engraftment or replenishment of damaged tissue [[11], [12], [13], [14]]. Of particular interest are hMSC-secreted EVs including a subset of smaller vesicles, exosomes (30–150 nm), composed of a lipid bilayer encompassing cell-specific proteins, nucleic acids, and lipids [15]. Once secreted from the parental cells, exosomes and target cell interactions occur through receptor-ligand interactions, direct fusion of exosomes with the plasma membrane, or through endocytosis [16]. Direct delivery via exosomes of cell-specific cargos as well as EVs loaded with drugs/therapeutics post isolation are gaining interest due to low toxicity, low immunogenicity, high engineering flexibility, and ability to target specific cell types and diseased cells [17,18]. Stem cell-derived exosomes have already been demonstrated to inhibit tumor cell growth, treat Parkinson's disease, and ameliorate ischemic injuries [[19], [20], [21]]. While not fully understood, there is evidence of significant crosstalk between EV biogenesis and autophagy [[22], [23], [24]]. Exosomes are formed initially as intraluminal vesicles (ILVs) by inward budding of late endosomes and multivesicular bodies (MVBs). Following formation of ILVs, MVBs can fuse with the lysosome where the contents are degraded in autophagy, or fuse with the plasma membrane where the ILVs are secreted from the cells as exosomes [24]. Proteins canonically regarded as critical for autophagy (ATG5, ATG16L1, etc.) play significant roles in the destination of the ILVs [23].
Recent studies have shown that biomechanical forces such as shear stress in 3D microenvironment can be exploited to induce hMSC differentiation [25,26] and enhance anti-inflammatory mediators and chemokine production [27], which are both underpinned through shifts in cellular metabolism to meet the changing energy demands of the cells. Multiple studies have suggested that EV production and parent cell metabolism are inherently linked [28,29]. Three of the top ten proteins identified in exosomes on EVpedia are glycolytic enzymes [29], and the enhanced metabolic activity leads to increased exosome production [30]. Higher levels of glycolysis have been shown to correlate with higher production of exosomes in cancer cells [31]. EV biogenesis and autophagy are linked through the endolysomal pathway with significant crosstalk and may act synergistically to relieve cell stress [32]. hMSC-derived EVs have been used to ameliorate ischemia by replenishing ATP and NADH levels, which is thought due to the addition of glycolytic enzymes after EV phagocytosis by damaged cells [33]. Therapeutic efficacy of hMSCs may depend on the protein levels secreted through EVs. hMSC-EV bio-manufacturing for clinical use has been reported using T flasks and CellFactory [3,34,35]. Yet, upscaling production of hMSC-EVs in dynamic bioreactor microenvironments is necessary to produce EVs of clinically relevant quantities. Our previous study has characterized EVs secreted by 3D hMSC aggregates cultured under wave motion [36]. However, the influence of 3D dynamic bioreactor microenvironment on EV biogenesis and cargo profiles is still not fully understood.
Vertical-Wheel bioreactors have recently been demonstrated for the production of hMSCs on microcarriers and human pluripotent stem cell (hPSC) aggregates as well as cerebellar spheroids [[37], [38], [39], [40], [41], [42], [43]]. But their use for EV production from human stem cells has not been demonstrated. With increased attention to the importance of hMSC secretome on their therapeutic efficacy, this study seeks to link the metabolic status of the hMSCs [44] to EV production and their protein cargo profiles determined by proteomics as well as selected microRNA cargo, based on the methods established in our previous investigations [[45], [46], [47], [48]]. In this study, it is hypothesized that shear stress and 3D microenvironment in the PBS Vertical-Wheel Bioreactors alter cellular energy metabolism and lead to an increase in cell signaling modulating EV biogenesis. It is further speculated that the interplay between reactive oxygen species (ROS), autophagy, and EV biogenesis in the bioreactor system could have significant impacts on hMSC-derived EV production and cargo profiles. The scalability of the hMSC and EV production in the Vertical-Wheel bioreactor was also demonstrated from 0.1L to 0.5L bioreactors. This study advances the bio-manufacturing process for hMSC-EV therapy towards treating neurological disorders such as ischemic stroke, Alzheimer's disease, and multiple sclerosis.
2. Materials and methods
2.1. hMSC 2-D planar cultures
Bone marrow-derived hMSCs were isolated from human bone marrow mononuclear cells (BM-hMNCs, purchased from Lonza Walkersville Inc.) from a de-identified healthy donor. hMSCs were derived from BM-hMNCs according to the manufacturer's instruction [49]. hMSCs cryogenically preserved in CryoStor CS10 (StemCell Technologies, Vancouver, Canada) were thawed and plated into Corning CellBIND T-25 cm2, T-75 cm2, and T-175 cm2 Rectangular Canted Neck Cell Culture Flasks with Vent Cap (Corning, Corning, NY, USA). Cells were seeded at a density of 2500 cells/cm2 in RoosterNourish-MSC-XF (RoosterBio, Frederick, MD, USA) and cultured under standard condition (5% CO2, 37 °C). Media were exchanged on day 3 and cells were harvested on day 4 after incubation for 5 min with Trypsin-EDTA (0.05%) (Thermo Fisher Scientific Inc., Waltham, MA, USA). Cells were cultured through 4 passages when they were harvested for inoculation of the Vertical-Wheel bioreactors and 2D control. hMSCs from two donors were compared (Supplementary Fig. S1) and the cells with better expansion were selected in the following experiments.
2.2. Microcarrier-based hMSC expansion in PBS Vertical-Wheel bioreactors
Corning Low Concentration Synthemax II Microcarriers (Corning Incorporated, Corning, NY, USA) were added to the PBS Vertical-Wheel bioreactors (PBS Biotech Inc., Camarillo, CA, USA) [50] at a concentration of 20 g/L of working volume (Supplementary Fig. S2). The vessels were filled up to the seeding volume with RoosterNourish-MSC-XF media (60 mL and 300 mL for the 0.1L and 0.5L vessels respectively). Vessels were then inoculated at 22,500 cells/mL (relative to working volume) and maintained at the lowest possible agitation to suspend the microcarriers (25 RPM and 19 RPM for the 0.1L and 0.5L vessels respectively) for 24 h for cell attachment. On day one, vessels were brought up to the full working volume (100 mL and 500 mL for the 0.1L and 0.5L vessels respectively) and bioreactor agitation was adjusted to experimental values (25 RPM, 40 RPM, and 64 RPM for the shear stress portion of the study; maintained at the lowest possible agitation for suspension in scaling up portion of the study). On day three, 80% of media were exchanged in each bioreactor. On day five, 100% of the media were exchanged with RoosterCollect-EV (RoosterBio, Frederick, MD, USA) for EV collection at day 8.
In parallel to bioreactors, Corning CellBIND T-25 cm2, T-75 cm2, and T-175 cm2 Rectangular Canted Neck Cell Culture Flasks with Vent Cap were inoculated with hMSCs at 1250 cells/cm2 and grown in standard culture conditions. Media were exchanged on day three of culture and were replaced with RoosterCollect-EV on day 5 for EV collection at day 8.
2.3. Cell number and metabolite measurements
For sampling, each bioreactor was set to 50 RPM and 10 mL of samples were taken out for cell count and metabolite analysis on day 1, 3, 5 and 8. Samples were strained with a 70 μm pluriStrainer (pluriSelect, El Cajon, CA, USA) to remove cell-adhered microcarriers from media. Media were immediately analyzed with a BioProfile Flex2 (Nova Biomedical, Waltham, MA, USA) analyzer for metabolite and gas analysis. Cells were washed out of the strainer with 2 mL of Dulbecco's Phosphate-Buffered Saline (DPBS) (Corning Cellgro, Corning, NY, USA) and lysed for 30 min with the addition of an equal volume of Reagent A100 (ChemoMetec, Bohemia, NY, USA). After lysis, nuclei were stabilized with Reagent B (ChemoMetec, Bohemia, NY, USA) with addition of a volume equal to Reagent A. The solution was well mixed and loaded into a Via1-Cassette (ChemoMetec, Bohemia, NY, USA) which was subsequently loaded into a NucleoCounter NC-200 (ChemoMetec, Bohemia, NY, USA) that determined cell concentration via DAPI staining. For 2D controls, hMSCs were washed three times with DPBS and trypsinized for 5 min at 37 °C. Cells were loaded into a Via1-Cassette for NucleoCounter NC-200 counting. The spent media were collected and immediately analyzed with a BioProfile Flex2 analyzer.
2.4. Cell harvest and EV media collection
For bioreactors, cells and microcarriers were allowed to settle for 10 min without agitation. Conditioned media were then collected and stored at −80 °C. Bioreactors were filled to 50% working volume with DPBS and agitated at 25 RPM for 90 s. Cell-adhered microcarriers were again allowed to settle for 10 min and supernatants were removed. This process was repeated for a total of three washes. After the final washing step, 50% of DPBS was removed and replaced with an equal volume of Trypsin-EDTA (0.05%) and incubated under 25 RPM agitation at 37 °C for 10 min. Bioreactor agitation was then increased to 80 RPM and incubated for another 5 min. An equal volume of media was added to dilute Trypsin-EDTA and the microcarriers were allowed to settle. Supernatants containing cells were then collected and an additional rinse of DPBS (25% of working volume) was added to vessels agitated at 25 RPM for 90 s. Microcarriers were allowed to settle and supernatants were pooled together for centrifugation. Cells were pelleted and either stored at −80 °C or used immediately for characterizations. For 2D controls, conditioned media were collected and stored at −80 °C. hMSCs were washed three times with DPBS and trypsinized for 5 min at 37 °C. Cells were pelleted and either stored at −80 °C or used immediately for characterization.
2.5. EV isolation
To isolate the hMSC-EVs, the differential ultracentrifugation method was performed followed by characterization using nanoparticle tracking analysis [45,46]. Briefly, the conditioned media were centrifuged at 500 g for 5 min at 4 °C. The supernatants were collected and centrifuged again at 2000 g for 10 min. The collected supernatants were then centrifuged at 10,000 g for 30 min. EVs were isolated using an inexpensive polyethylene glycol (PEG)-based method as reported previously [45,46]. Briefly, after centrifugation at 10,000 g for 30 min, supernatants were collected and mixed with PEG solution (16% wt/vol in 1 M NaCl) at a 1:1 vol and incubated at 4 °C overnight. The mixed solutions were centrifuged at 3000 g for 1 h. The crude EV pellets were resuspended in particle-free PBS and then ultracentrifuged at 100,000 g for 70 min. Purified EV pellets were resuspended in 100 μL PBS and stored in −80 °C until further use.
2.6. Nanoparticle tracking analysis (NTA)
NTA was performed on the isolated EV samples in triplicate to determine size distribution and particle concentration. NTA was performed on a Nanosight LM10-HS instrument (Malvern Instruments, Malvern, UK) configured with a blue (488 nm) laser and sCMOS camera [45]. The EV samples were diluted to 1–2 μg protein per mL in PBS. For each replicate, three videos of 60 s were acquired with camera shutter speed fixed at 30.00 ms. To ensure accurate and consistent detection of small particles, camera level was set to 13, and detection threshold was maintained at 5. The laser chamber was cleaned thoroughly with particle-free water between each sample reading. The collected videos were analyzed using NTA3.0 software to obtain the mode and mean size distribution, as well as the concentration of particles per mL of solution. Compared to the mean size, the mode size is usually a more accurate representation because the vesicle aggregates may affect the value of mean size.
2.7. Western blotting for EV markers
EV and cell samples were lysed in radio-immunoprecipitation assay (RIPA) buffer (150 mM sodium chloride, 1.0% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 50 mM Tris, pH 8.0 and 1X Thermo Scientific Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific Inc., Waltham, MA, USA). Samples were then lysised for 20 min on ice, and spun down at 14,000 RPM for 20 min. The supernatant was collected and a Bradford assay was carried out to determine the protein concentration. Protein lysate concentration was normalized, and denatured at 100 °C in 2X Laemmli Sample buffer for 5 min. About 3–10 μg of proteins were loaded into each well. Proteins were separated by 12% Bis-Tris-SDS gels and transferred onto a nitrocellulose membrane (Bio-rad, Hercules, CA. USA). For the detection of non-phosphorylated proteins, the membranes were blocked for 1 h in 1% skim milk (w/v) in Tris-buffered saline (10 mM Tris-HCl, pH 7.5, and 150 mM NaCl) with 0.1% Tween 20 (v/v) (TBST). Membranes were incubated overnight in the presence of the primary antibodies (Supplementary Table S1) diluted in blocking buffer at 4 °C. Afterward, the membranes were washed four times with TBST and then incubated with an IR secondary (LI-COR, Lincoln, NE) at 1:5000 for 180 min at room temperature. Blots were washed another four times with TBST and processed using the LI-COR Odyssey (LI-COR).
2.8. Reverse transcription and quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from different cell samples using the RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. The samples were further treated using DNA-Free RNA Kit (Zymo, Irvine, CA, USA) [51]. Reverse transcription was carried out according to the manufacturer's instructions using 2 μg of total mRNA, anchored oligo-dT primers (Operon, Huntsville, AL), and Superscript III (Invitrogen, Carlsbad, CA, USA). The software Oligo Explorer 1.2Primers (Genelink, Hawthorne, NY, USA) was used to design the primers specific for target genes (Supplementary Table S2). For normalization of expression levels, β-actin was used as an endogenous control. Using SYBR1 Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA), real-time reactions were performed on an ABI7500 instrument (Applied Biosystems). The amplification reactions were performed as follows: 2 min at 50 °C, 10 min at 95 °C, and 40 cycles of 95 °C for 15 s and 55 °C for 30 s, and 68 °C for 30 s. The Ct values of the target genes were firstly normalized to the Ct values of the endogenous control selected as β-actin (over GAPDH and B2M-1). The corrected Ct values were then compared for the bioreactor conditions to the 2D control, or between the compared samples. Fold changes in gene expression was calculated using the comparative Ct method: to obtain the relative expression levels.
2.9. RT-qPCR analysis for microRNA expression
Total microRNA (miRNA) was isolated from different EV samples using the miRNeasy Micro Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Reverse transcription was carried out using commercial qScript miR cDNA synthesis kit (discontinued, Quantabio, Beverly, MA). The PerfeCTa Universal/miR-specific PCR Primer (QuantaBio) has been designed and validated to work specifically with PerfeCTa SYBR Green SuperMix using miRNA cDNA produced. The levels of miRs were determined with U6 or SNORD44 as a housekeeping gene for normalization of miR expression levels (Primer sequences are shown in Supplementary Table S3). Real-time RT-qPCR reactions were performed on an Applied Biosystems Quantstudio 7 flex (Applied Biosystems, Foster City, CA), using PerfeCTa SYBR Green SuperMix. The amplification reactions were performed as follows: 2 min at 95 °C, and 40 cycles of 95 °C for 5 s and 60 °C for 15 s, and 70 °C for 15 s. Fold variation in gene expression was quantified by means of the comparative Ct method: , which is based on the comparison of expression of the target gene (normalized to U6 or SNORD44) between the compared samples.
2.10. Transmission electron microscopy (TEM)
TEM was performed to confirm the morphology of EVs according to Lasser et al. [52] and also as shown in our previous publication [46]. Briefly, EV isolates were resuspended in 50–100 μL of sterile filtered PBS. For each sample preparation, intact EVs (5 μL) were dropped onto Parafilm. A carbon coated 400 Hex Mesh Copper grid (Electron Microscopy Sciences, EMS) was positioned using forceps with coating side down on top of each drop for 1 h. Grids were washed with sterile filtered PBS three times and then the EV samples were fixed for 10 min in 2% PFA (EMS, EM Grade). After washing, the grids were transferred on top of a 20 μL drop of 2.5% glutaraldehyde (EMS, EM Grade) and incubated for 10 min at room temperature. Grid samples were stained for 10 min with 2% uranyl acetate (EMS grade). Then the samples were embedded for 10 min with 0.13% methyl cellulose and 0.4% uranyl acetate. The coated side of the grids were left to dry before imaging on the CM120 Biotwin electron microscope [52].
2.11. Total and mitochondrial ROS, autophagosomes, and phenotyping by flow cytometry
For ROS measurement, cell suspension was incubated with 25 μM carboxy-H2DCFDA (Molecular Probe) at 37 °C for 30 min and total ROS was determined using flow cytometry. For mitochondrial ROS measurement, cell suspension was incubated with 5 μM MitoSOX Red (Molecular Probe) at 37 °C for 10 min and analyzed using flow cytometry. For autophagy measurement, cell suspension was incubated with 20 μM Cyto-ID Green (Enzo Life Sciences, Farmingdale, NY, USA), a fluorescent dye that selectively labels accumulated autophagic vacuoles, at 37 °C for 30 min, and analyzed by flow cytometry and calculated according to the manufacturer's instructions. For NAD+ dependent enzymes, hMSCs were harvested with 0.25% trypsin-EDTA solution, washed in PBS, and then fixed in 4% paraformaldehyde (PFA) at RT for 15 min. Cells were then permeabilized in 0.2% Triton X-100 for 10 min. Non-specific binding sites were blocked with 1% bovine serum albumin in PBS for 15 min. After washing, cells were incubated with specific primary antibodies (Supplementary Table S1) for human Sirt-1, Sirt-3, NAMPT, CD38, and Parp1 (Santa Cruz Biotechnology, Dallas, TX, USA) at room temperature for 2 h, followed by incubation with corresponding secondary antibody (Molecular Probe). Labeled samples were acquired using BD FACSCanto II flow cytometer (Becton Dickinson) along with isotype control. The results were analyzed using FlowJo software.
2.12. Human fibroblast (hFB) growth and in vitro wound healing model
Primary human dermal fibroblasts (containing mitochondria) were purchased from American Type Culture Collection (PCS-201-012, ATCC, Manassas, VA, USA) and subcultured up to passage 15. All reagents were purchased from Sigma Aldrich (St. Louis, MO, USA) unless otherwise noted. In vitro wound healing assay was modified to evaluate the effects of EVs [53]. Briefly, hFBs were seeded (0.2 × 106 cell/per well) onto a tissue culture treated 24-well plate and grown overnight in DMEM plus 10% FBS. An artificial wound was introduced with a 200 μL pipette tip and images were captured with an Olympus IX70 inverted microscope for 0–24 h. EVs then were added at the concentration 1 × 108 EV/mL medium for next 24 h. Cell growth was then recorded and analyzed by ImageJ to calculate the wound closure rate.
2.13. Proteomics analysis for hMSCs and the secreted EVs
The hMSC-EVs (from the 2D and the bioreactor-25 RPM group) were isolated using ExtraPEG and then extracted for proteins, together with the parent cells. Based on protein quantification result, up to 25-μg proteins were isolated on S-trap micro column (Protifi, K02-micro). The isolated proteins (triplicate for each group) were alkylated and digested on column based on manufacturer's instructions. All the eluted peptides were fractionated by Pierce high pH reverse phase peptide fractionation kit (Thermo, 84868) into 5 fractions for each sample. Then all the samples were vacuumed dried and submitted to FSU Translational Science Laboratory. The samples were analyzed on the Thermo Q Exactive HF as previously described [54,55]. Briefly, resulting raw files were searched with Proteome Discoverer 2.4 using SequestHT, Mascot and Amanda as search engines. Scaffold (version 5.0) was used to validate the protein and peptide identity. Peptide identity was accepted if Scaffold Local false discovery rate (FDR) algorithm demonstrated a probability greater that 99.0%. Likewise, protein identity was accepted if the probability level was greater than 99.0% and contained a minimum of two recognized peptides. Gene Ontology (GO) annotation was carried out by g:Profiler.
2.14. Statistical analysis
Data were analyzed in Graphpad Prism software. Data shown are represented as mean and standard deviation with the exception of the RT-qPCR data expressed in box and whisker plots. Statistical analysis for RT-qPCR data were performed with multiple Mann-Whitney tests and significance was accepted at p ≤ 0.05. All other statistical analyses were performed with a Student's t-test and significance was accepted at p ≤ 0.05.
3. Results
3.1. Effects of agitation speeds on hMSC expansion and metabolism
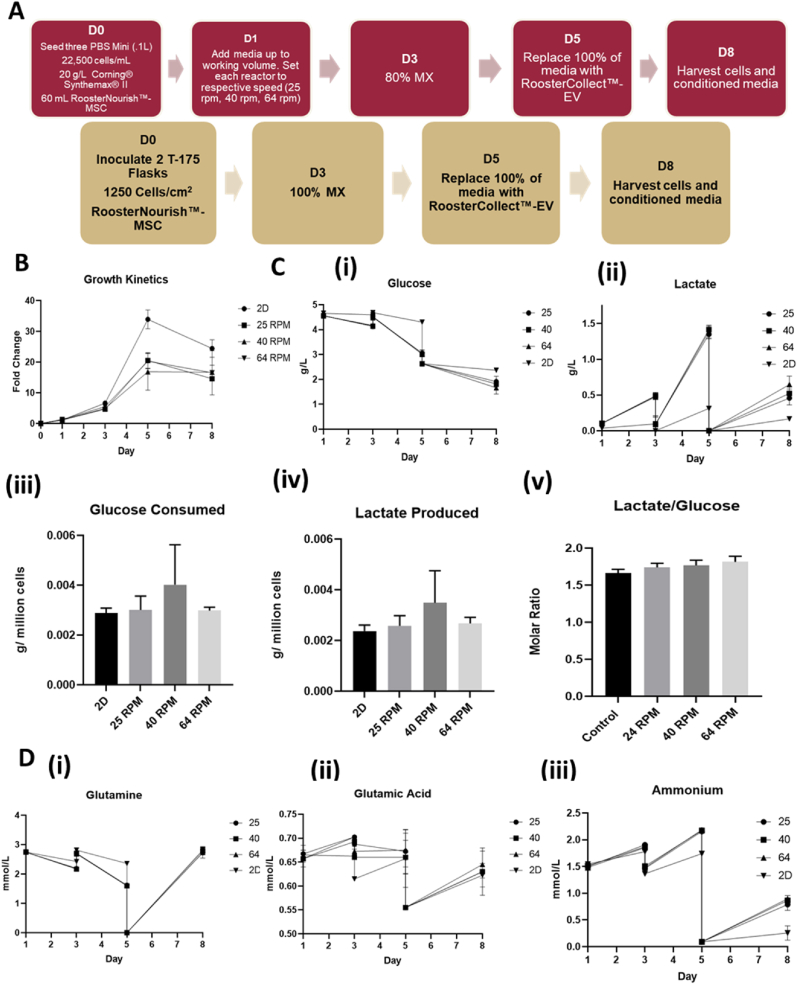
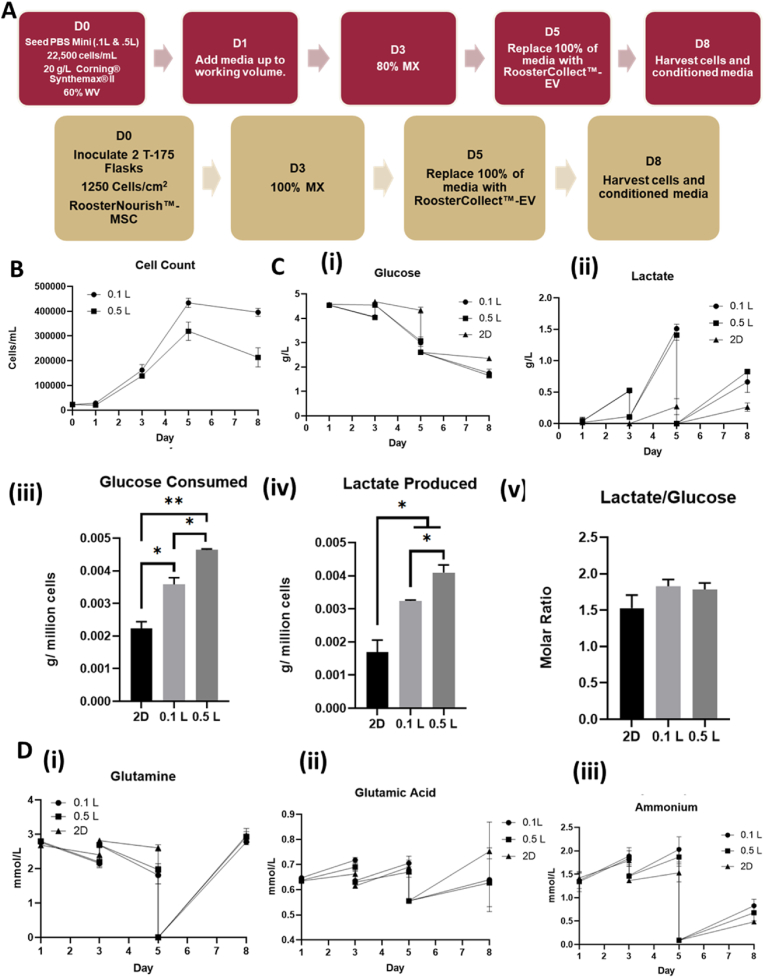
PBS 0.1L bioreactor systems and Corning CellBIND T-flasks were inoculated with hMSCs and cultured as shown in Fig. 1A. To study the effects of shear stress on hMSC metabolism and EV production, hMSCs were expanded at 25, 40, and 64 RPM agitations (i.e., 0.12, 0.2, and 0.3 dyn/cm2 respectively) in the 3D bioreactor system corresponding to a 2.5-fold linear increase in shear stress at each localized measured point (Supplementary Figs. S3, S4, S5). Following the typical lag phase, hMSC expansion was robust in each bioreactor with 20-fold increase on day 5 (Fig. 1B and Supplementary Figs. S6 and S7). Switching the hMSC expansion media to EV collection media shows no growth during day 5–8. The 2D control group followed a similar expansion pattern achieving >30-fold increase on day 5. The 24, 40, and 64 RPM conditions showed similar hMSC expansion with average cell densities of 4–5 × 105 cells/mL on day 5. Glucose and lactate concentration measurements showed similar changes for the three bioreactor groups, indicating that shear stress increase did not significantly alter hMSC proliferation and metabolism while the 2D group had higher glucose and lower lactate concentrations (Fig. 1C). Glucose consumption and lactate production normalized to cell number, as well as the molar ratio of lactate/glucose during the exponential phase (day 3–5), showed similar values for the four groups. Other metabolites that supplement energy, protein synthesis, and nucleic acid synthesis, (i.e., glutamine and glutamic acid) showed similar changes for the three bioreactor groups, but the 2D group had lower glutamine consumption and ammonia production (Fig. 1D and Supplementary Fig. S7). Glutamine consumption normalized cell number was comparable for all the four groups, but ammonium production normalized to cell number was higher for the 2D control than the bioreactor groups. The pH value, oxygen and CO2 saturation values were similar for the three bioreactor groups (Supplementary Fig. S7).
Fig. 1.
hMSC expansion and metabolism on Synthemax II microcarriers in PBS Vertical-Wheel Bioreactors at 25, 40, and 64 RPM. (A) Process flow for hMSC expansion on Synthemax II microcarrers in PBS Vertical-Wheel Bioreactors; (B) hMSC growth kinetics during expansion and EV collection stages; (C) Glucose and Lactate metabolism: (i) Glucose concentration kinetics; (ii) Lactate concentration kinetics. (iii) Glucose consumed normalized to cell number; (iv) Lactate produced normalized to cell number; (v) Lactate production to glucose consumption ratios. (D) Glutamine metabolism: (i) Glutamine concentration kinetics. The extracellular vesicle (EV) collection medium is formulated with Glutamax. Therefore, the glutamine concentration at day 5 is 0. (ii) Glutamic acid concentration kinetics; (iii) Ammonia concentration kinetics.
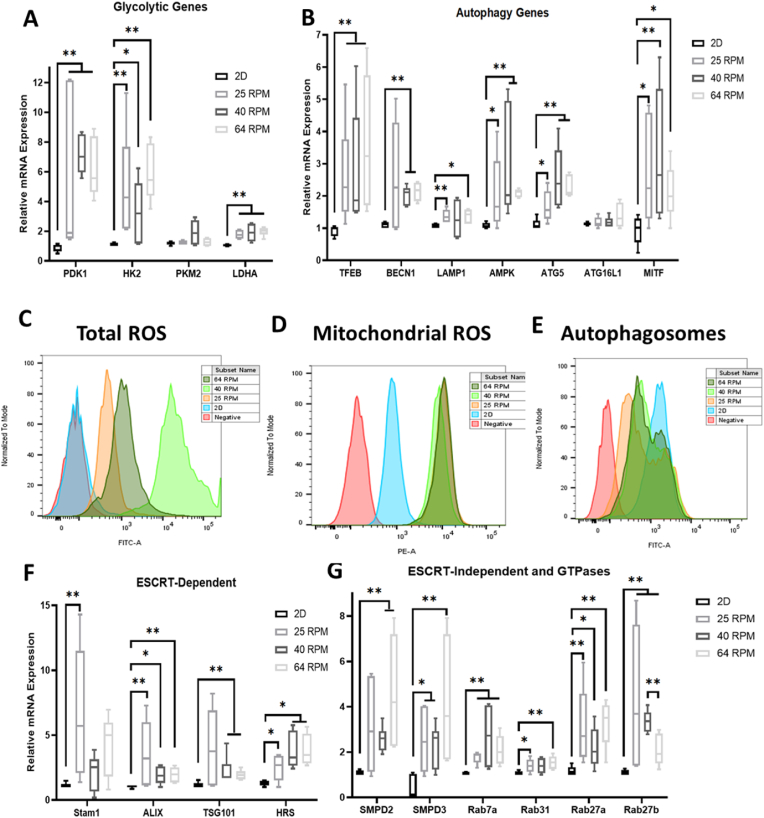
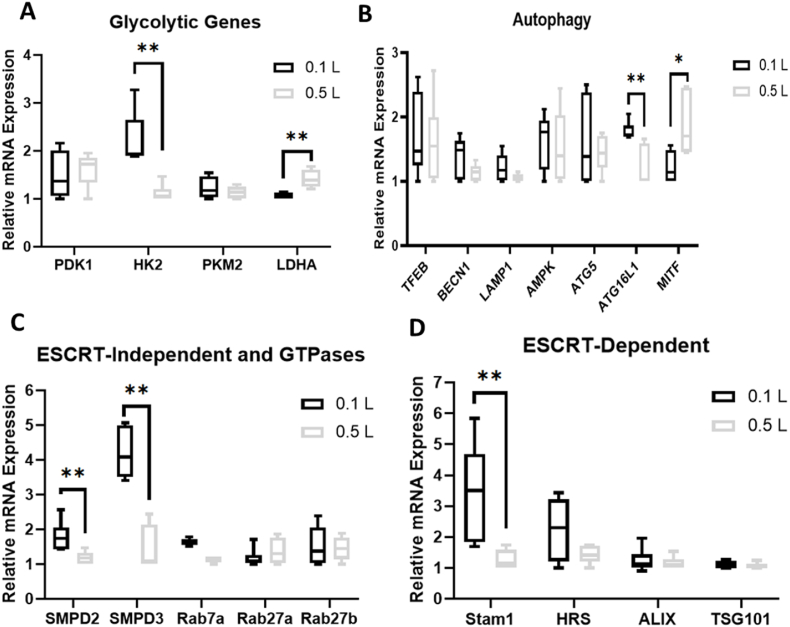
The bioreactor-expanded hMSCs were measured for mRNA expression of genes responsible for activating and regulating key steps in glycolysis, in comparison to the 2D control Three endogenous controls were compared and ACTB shows more stable expression than GAPDH and B2M-1, and thus was used in following experiments (Supplementary Fig. S8). PDK1, HK2, and LDHA mRNA expression was significantly upregulated (8–12 fold, 5–8 fold, and 2-fold respectively) for the bioreactor groups compared to the 2D control, but not PKM2 expression (Fig. 2A). However, no discernible distinctions could be made amongst the three bioreactor groups. mRNA levels integral to key functions of autophagy were also significantly upregulated in the bioreactor groups compared to the 2D controls (Fig. 2B). Specifically, transcription factors TFEB (2–6 fold) and MITF (2–5 fold) involved in boosting lysosomal biogenesis were upregulated in each of the bioreactor groups. AMPK (2–5 fold), an upstream initiator shown to induce autophagy activation, was also upregulated. mRNA levels for proteins that are key in autophagic vesicle formation, lysosomal integrity, and membrane trafficking (i.e., BECN1, LAMP1, and ATG5) were also upregulated in the bioreactor groups (2–4 fold, 1.5–2 fold, and 2–3 fold respectively). Of all autophagy related markers tested, only ATG16L1 showed no statistically significant change.
Fig. 2.
Metabolic and extracellular vesicle (EV) biogenesis pathways and reactive oxygen species (ROS) analysis for hMSCs in PBS Vertical-Wheel Bioreactors at 25, 40, and 64 RPM. Various genes were measured by RT-qPCR on the mRNAs isolated from the harvested cells at the end of EV collection (n = 6, results were from two independent bioreactor runs), relative to 2D control. (A) Glycolytic genes; (B) Autophagy genes; Flow cytometry histograms for (C) Total ROS; (D) Mitochondrial ROS; and (E) Autophagosomes. Red line: negative control; blue line: 2D culture control; orange line: 25 RPM bioreactor culture; light green: 40 RPM, and dark green: 64 RPM. (F) ESCRT-dependent EV biogenesis markers; (G) ESCRT-independent EV biogenesis markers. * indicates p < 0.05; ** indicates p < 0.01, *** indicates p < 0.001.
Both total and mitochondrial ROS were upregulated in the bioreactor groups compared with the 2D control (Fig. 2C and D, and Supplementary Fig. S9A). Expression of mitochondrial ROS was similar in each bioreactor group. However, the 40 RPM group had the highest total ROS while the 25 RPM and 64 RPM groups showed similar expression to each other. Lower autophagosomes accumulation was observed in bioreactor groups compared to 2D culture (Fig. 2E). Flow cytometry analysis of NAD-pathway related proteins [44,56] was performed for the bioreactor-expanded cells and the 2D control (Supplementary Fig. S9B). For Sirt-1, lower expression of the 2D control than the bioreactor groups was observed, and the three agitation speeds showed similar expression levels. A similar trend was observed for Nampt. However, there is no difference between the 2D control and the bioreactor groups for Sirt-3 and CD38. For Parp1, the 64 RPM condition showed lower expression than the other conditions.
mRNA levels of the endosomal sorting complexes required for transport (ESCRT)-dependent exosomal markers Alix (2–5 fold), TSG101 (2–7 fold), and HRS (2–5 fold) were significantly upregulated in the bioreactor groups compared to the 2D control (Fig. 2F and Supplementary Fig. S10). Stam1 was upregulated in the 25 RPM group but not statistically significant for the other two bioreactor groups. mRNA levels for SMPD2 and SMPD3 (2–7 fold), hydrolyzers of sphingomyelin that generate EVs in an ESCRT-independent manner, were also upregulated in the bioreactor groups as well as the mRNA levels for GTPases Rab7a (2–4 fold), Rab27a (2–4 fold) and Rab27b (2–7 fold) that have functions in multivesicular endosomal docking to the plasma membrane (Fig. 2G). Slight increase in Rab31 expression in the bioreactor was observed.
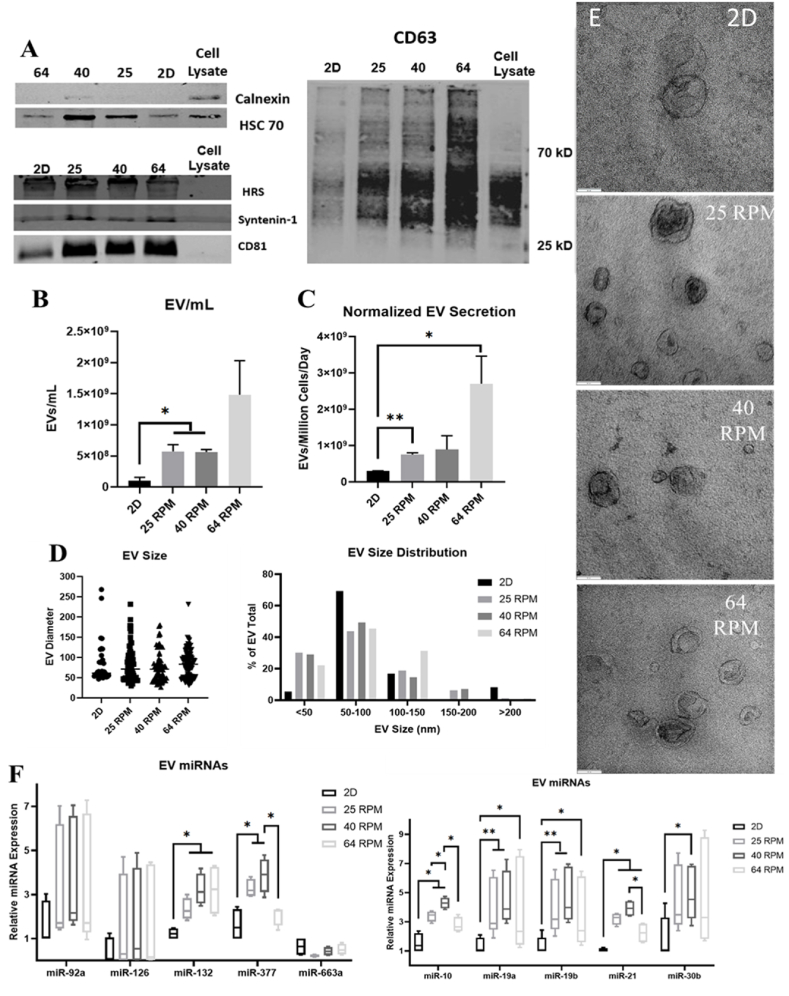
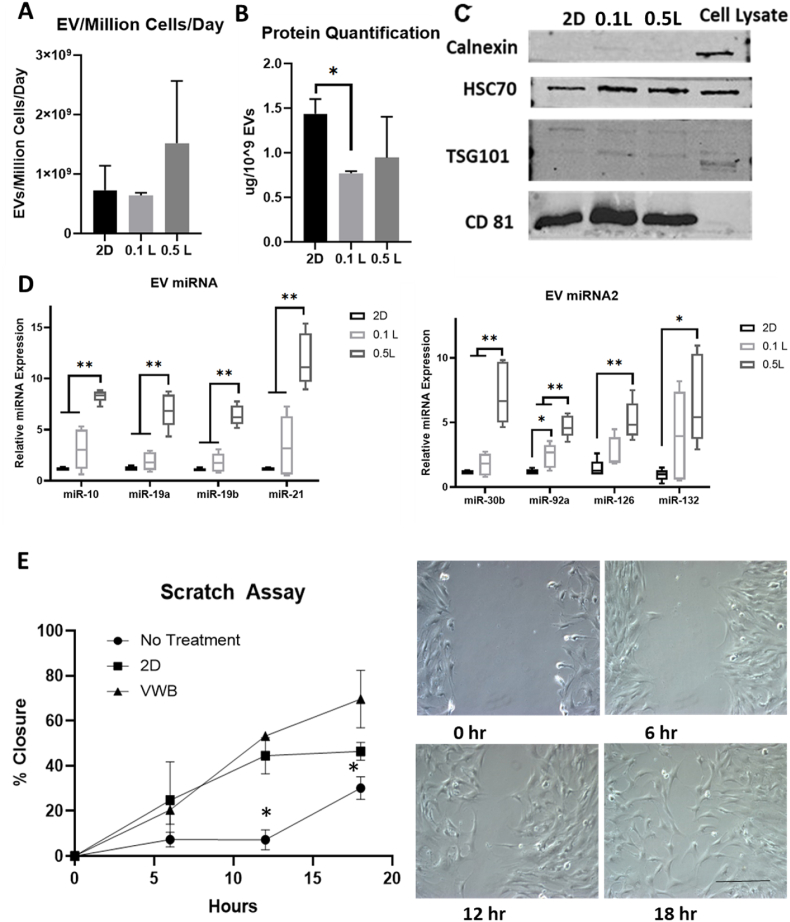
3.2. Effects of agitation speeds on hMSC-EV generation and miRNA cargo
After processing the conditioned media of bioreactor and 2D cultures, EVs were isolated and known exosomal markers were measured by Western blot (Fig. 3A and Supplementary Fig. S11A). The expression of HRS, HSC70, syntenin-1, CD81, and CD63 was confirmed in each EV group, while bioreactor groups showed higher expression of HSC70, HRS, CD81 and CD63. Negative marker calnexin was run in parallel to prove no cell debris contamination. As expected, EV number/mL yield increased more than 5.5-fold in bioreactor groups compared to the 2D cultures (Fig. 3B). When normalized to cell number, EV secretion in the bioreactor groups was increased more than 2.5-fold over the 2D culture (Fig. 3C). In both cases, the three bioreactor groups did not show statistically significant differences in EV secretion between each other. EV size and morphology were examined via TEM with all groups showing classic exosomal cup shaped morphologies, suggesting shear stress from bioreactor culture does not impact membrane integrity (Fig. 3D and E) [57]. Using ImageJ software to quantify EV size, no statistically significant changes were observed among EV diameter in any of the groups. However, the bioreactor groups did have a greater proportion of EVs less than 50 nm in diameter (between 22 and 30%) in comparison to the 2D group (5.5%) (Fig. 3D). There is no statistical difference in protein content normalized to EV number for all the four conditions (Supplementary Fig. S11B). But bioreactor culture generates more total EV protein content due to the higher EV number than 2D culture.
Fig. 3.
Characterizations of extracellular vesicles (EVs) secreted by 0.1L bioreactor-expanded hMSCs. (A) Western Blot of exosomal marker expression, including HSC70, TSG101, CD81, Calnexin (negative marker); HRS, Syntenoin-1, CD81, and CD63. (B) Comparison of EV yield for the number of EVs per mL spent medium (n = 3), (C) Number of EVs normalized to the cell number (n = 3). (D) EV size and distribution determined based on TEM images (n = 50–80). (E) TEM images of EVs for various groups. Scale bars: 100 nm. (F) miRNA expression in the isolated EVs determined by RT-qPCR (n = 6). The value shows ΔΔCt. Results were combined from two independent bioreactor runs. *p < 0.05; **p < 0.01, ***p < 0.001.
EVs were lysed to determine relative quantities of miRNAs previously shown to respond in cell populations to shear stress in 3D microenvironment, known as “mechano-miRNAs” [58]. Of the 10 miRNAs tested, 7 miRs (i.e., miR-132, miR-377, miR-10, miR-19a, miR-19b, miR-21, and miR-30b) were significantly upregulated (3–7 fold) in the EVs isolated from the bioreactor groups compared to the 2D group with miR-92a, miR-126, and miR-663a showing non-statistically significant upregulation (Fig. 3F and Supplementary Fig. S11C). Varying the shear stress levels encountered by the cells in the 3D bioreactors showed little effects on miRNA levels in the EVs among the three bioreactor groups.
3.3. Proteomic analysis of parent hMSCs and the secreted EVs
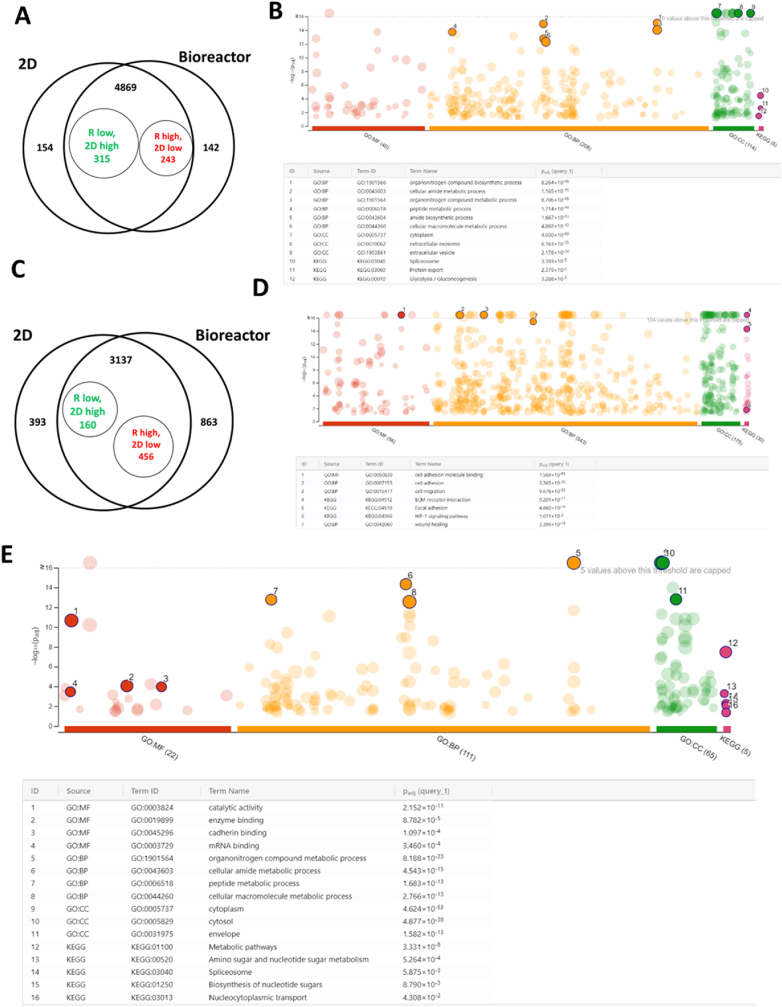
To determine the protein cargo of the EVs, proteomics analysis was performed for the 2D EV, bioreactor (25 RPM) EV, as well as the two groups of the corresponding parent cells, using equal amounts of proteins (25 μg) per sample. The two EV samples for proteomics were characterized by NTA and protein assay (Supplementary Fig. S12), showing the comparable size and the protein content. Proteomics analysis of the parent cells showed 4869 shared differentially expressed proteins (DEPs) for the bioreactor and 2D cultures (Fig. 4A and Data file S1). 315 DEPs were upregulated in 2D culture and 243 DEPs were upregulated in bioreactor culture. There were 154 DEPs exclusively expressed in 2D culture and 142 exclusively expressed in bioreactor culture. Gene Ontology (GO) annotation shows that the DEPs (Fig. 4B and Supplementary Table S4, Fig. S12) are enriched in various types of cellular metabolic and biosynthesis processes, glycolysis, cell adhesion/cadherin binding, extracellular vesicles, extracellular exosome, protein export etc. For the proteomic analysis of hMSC-EVs, among 3137 shared DEPs, 160 DEPs were upregulated in 2D EVs and 456 DEPs were upregulated in bioreactor EVs (Fig. 4C). There were 393 DEPs exclusively expressed in 2D EVs and 863 exclusively expressed in bioreactor EVs. GO annotation shows that the enriched DEPs are involved in cell adhesion, cell migration, ECM-receptor interaction, focal adhesion, HIF-1 signaling pathway, and wound healing (Fig. 4D and Supplementary Fig. S12). GO annotation for the DEPs in the EVs for bioreactor only subset (863 DEPs) reveals that the enriched pathways are related to metabolic processes (peptides, macromolecules, etc.) with some of them having catalytic activities (Fig. 4E and Supplementary Fig. S12). The comparison of DEPs in parent cells and in the secreted EVs was also performed (Table 1). Almost all DEPs upregulated in the parent cells were also upregulated in the secreted EVs. The DEPs downregulated in the parent cells showed a mixture of upregulation and downregulation in the EVs, indicating that the protein content of the parent cells contributes significantly to the protein cargo of the secreted EVs.
Fig. 4.
Bioreactor-expanded hMSCs and the secreted EV protein cargo analysis by proteomics. (A) Venn diagram of the differentially expressed proteins (DEPs) from hMSCs of bioreactor culture vs. 2D culture. (B) GO annotation of the hMSC DEPs (558 as in A) for the enriched pathways. (C) Venn diagram of the DEPs for hMSC-EVs. (D) GO annotation of the hMSC-EV DEPs (616 as in C) for the enriched pathways. (E) GO annotation of the DEPs (863 as in C) for the Bioreactor only subset of hMSC-EV protein cargo.
Table 1.
Relation between hMSC cell and EV DEPs.
| ID | Fold change in Cell | Fold change in EV | ID | Fold change in Cell | Fold change in EV |
|---|---|---|---|---|---|
| CRIP1 | INF | INF | NIBAN2 | 0.9 | 2.5 |
| PLAT | INF | 5.7 | EHD4 | 0.8 | 1.7 |
| TAOK2 | 8.4 | INF | MYO1B | 0.8 | 8.1 |
| COL4A1 | 7.2 | 2.7 | TP53BP1 | 0.5 | INF |
| MYO15B | 3.8 | INF | AHNAK2 | 0.7 | 4.3 |
| FABP3 | 3.3 | 3.9 | FERMT2 | 0.7 | 5.6 |
| SERPINE1 | 3.1 | 1.6 | IQGAP1 | 0.7 | 3.0 |
| ABCC1 | 3.0 | INF | MMP14 | 0.7 | 1.9 |
| MGLL | 2.8 | INF | NQO1 | 0.7 | 1.7 |
| STX2 | 2.8 | INF | PARVA | 0.7 | 2.9 |
| MYADM | 2.6 | 2.0 | PLIN3 | 0.7 | 5.1 |
| LDHAL6B | 2.3 | INF | RAB18 | 0.7 | 8.0 |
| HELZ2 | 2.2 | INF | ENPP1 | 0.4 | 1.5 |
| RAB22A | 2.2 | 5.2 | ADAM9 | 0.6 | 1.8 |
| RAB23 | 2.2 | 3.3 | CHMP1A | 0.6 | INF |
| FKBP1A | 2.1 | 2.2 | CTNNB1 | 0.6 | 3.8 |
| S100A10 | 2.1 | 3.2 | EGFR | 0.6 | 2.0 |
| CAPN2 | 1.9 | 5.6 | NIBAN1 | 0.6 | 21.0 |
| PI4K2A | 1.9 | INF | PDCD6IP | 0.6 | 1.3 |
| AK3 | 1.6 | INF | ITGB5 | 0.4 | 2.8 |
| CBR1 | 1.6 | 8.6 | ZYX | 0.6 | 4.5 |
| CYB5R3 | 1.6 | INF | AHNAK | 0.5 | 4.3 |
| NME2 | 1.6 | 2.1 | CTSK | 0.5 | 5.7 |
| SLC12A4 | 1.6 | 3.0 | VTA1 | 0.4 | 4.3 |
| ANXA5 | 1.5 | 1.9 | KPNB1 | 0.5 | 14.0 |
| ARF6 | 1.5 | 2.1 | MYO1D | 0.5 | 3.1 |
| CAP1 | 1.5 | 2.1 | RFTN1 | 0.3 | 2.0 |
| PGK1 | 1.5 | 1.6 | PDGFRB | 0.2 | 1.5 |
| UGP2 | 1.5 | 3.5 | THBS2 | 0.5 | 0.5 |
| WDFY1 | 1.5 | INF | POSTN | 0.5 | 0.3 |
| FAH | 1.4 | 7.1 | THBS1 | 0.5 | 0.4 |
| FKBP5 | 1.4 | INF | GTF2I | 0.4 | 0.0 |
| GPI | 1.4 | 5.6 | AKR1D1 | 0.3 | 0.4 |
| HSD17B12 | 1.4 | INF | COL12A1 | 0.3 | 0.2 |
| MYOF | 1.4 | 4.2 | COL14A1 | 0.3 | 0.2 |
| TRPV2 | 1.4 | INF | AKAP8 | 0.2 | 0.0 |
| ANXA1 | 1.3 | 2.9 | UBTF | 0.2 | 0.3 |
| ANXA2 | 1.3 | 2.1 | FEN1 | 0.1 | 0.5 |
| FAP | 1.3 | 2.4 | HBG2 | 0.0 | 0.2 |
| FASN | 1.3 | 1.7 | PIWIL2 | 0.0 | 0.0 |
| HPCAL1 | 1.3 | 2.4 | PSMD2 | 0.8 | 0.4 |
| PKM | 1.3 | 3.2 | SFPQ | 0.7 | 0.1 |
| UGDH | 1.3 | 2.5 | SDF4 | 0.6 | 0.2 |
| IDH1 | 1.2 | 6.2 | FTH1 | 0.5 | 0.6 |
| HSPA4 | 1.1 | 8.0 | |||
| GIT1 | 2.8 | 0.0 |
Potential mechanisms that could be contributing to enhanced EV biogenesis in bioreactors were examined through the DEPs in the parent cells (Table 2). The DEPs related to autophagy, glucose and lipid metabolic process, as well as the protein carbolic process mostly showed upregulation in the bioreactor group. Similarly, DEPs related to cellular responses to ROS were also upregulated for the bioreactor group (Table 2). DEPs related to EV biogenesis and miRNA sorting showed upregulation in the secreted EVs of the bioreactor group, including VPS4A that is critical to miRNA sorting (Supplementary Fig. S13). The DEPs related to wound healing were mostly comparable for the bioreactor vs. 2D cell groups, with a few showing the upregulation, including ANXA1, SERPINE1, and VIL1.
Table 2.
Function related identified hMSC proteins.
| Function | Protein ID | P value | Fold Change |
|---|---|---|---|
| Autophagy | NPC1_HUMAN | 0.01 | 3.7 |
| LAMP2_HUMAN | 0.012 | 2.4 | |
| RAB8A_HUMAN | 0.043 | 1.7 | |
| CLH1_HUMAN | 0.018 | 1.2 | |
| LRSM1_HUMAN | 0 | INF | |
| PK3C3_HUMAN | 0.024 | 2.7 | |
| VTI1A_HUMAN | 0.039 | 2.2 | |
| ATG5_HUMAN | 0.2 | 0.7 | |
| LAMP1_HUMAN | 0 | 1.9 | |
| AAPK2_HUMAN |
0.7 |
1.6 |
|
| glucose metabolic process | CLN5_HUMAN | 0.011 | 0.5 |
| PGK1_HUMAN | 0.046 | 1.5 | |
| APOD_HUMAN | 0.002 | 20 | |
| G6PI_HUMAN | 0.01 | 1.4 | |
| 4F2_HUMAN | 0.019 | 0.6 | |
| KPYM_HUMAN | 0.02 | 1.3 | |
| ACOC_HUMAN | 0.043 | 1.5 | |
| FABP5_HUMAN | 0.019 | 0.7 | |
| LDH6B_HUMAN | 0.005 | 2.3 | |
| HXK1_HUMAN | 0.8 | 1 | |
| HXK2_HUMAN | 0.1 | 1.6 | |
| HXK3_HUMAN | 0.5 | 3.2 | |
| LDHA_HUMAN |
0.043 |
1.4 |
|
| lipid metabolic process | TPP1_HUMAN | 0.001 | 2.5 |
| ACSL1_HUMAN | 0.009 | 1.8 | |
| LACTB_HUMAN | 0.007 | 1.9 | |
| LRP1_HUMAN | 0.038 | 0.8 | |
| PLCD3_HUMAN |
0.034 |
5 |
|
| protein catabolic process | MMP2_HUMAN | 0.018 | 0.5 |
| CAN2_HUMAN | 0 | 1.9 | |
| DPP4_HUMAN | 0.007 | 6.3 | |
| SEPR_HUMAN | 0.025 | 1.3 | |
| CNDP2_HUMAN | 0.02 | 0.8 | |
| NEUL_HUMAN | 0.018 | 0.4 | |
| XPP3_HUMAN | 0.016 | 1.9 | |
| DPP2_HUMAN |
0.012 |
2 |
|
| cellular response to reactive oxygen species | TPM4_HUMAN | 0.016 | 0.7 |
| NDUS3_HUMAN | 0.003 | 1.3 | |
| EGFR_HUMAN | 0.01 | 0.6 | |
| SODM_HUMAN | 0.001 | 6.2 | |
| CBR1_HUMAN | 0.019 | 1.6 | |
| MK14_HUMAN |
0.02 |
1.8 |
|
| wound healing | ACTG_HUMAN | 0.6 | 1.1 |
| ANXA1_HUMAN | 0.026 | 1.3 | |
| B4GT1_HUMAN | 0.2 | 2.9 | |
| CSKP_HUMAN | 0.1 | INF | |
| CD109_HUMAN | 0.2 | 0.6 | |
| CLAP1_HUMAN | 0.8 | 1.1 | |
| CRK_HUMAN | 0.5 | 0.9 | |
| FERM2_HUMAN | 0.006 | 0.7 | |
| CXA1_HUMAN | 0.8 | 0.9 | |
| HMGB1_HUMAN | 0.1 | 0.6 | |
| ITB1_HUMAN | 0.8 | 0.9 | |
| KANK1_HUMAN | 0.8 | 0.9 | |
| MTOR_HUMAN | 1 | 1 | |
| MYLK_HUMAN | 0.9 | 1 | |
| PAI1_HUMAN | 0.001 | 3.1 | |
| SMAD3_HUMAN | 0.02 | 0.3 | |
| SRSF6_HUMAN | 0.008 | 0.7 | |
| VILI_HUMAN | 0.2 | 3.1 |
The DEPs in the EVs of the bioreactor vs. the 2D group were compared to our previous publication about the characterizations of EVs from 3D hMSC aggregates (Supplementary Fig. S14 and Data file S2). High similarity of EV protein cargo was observed between the PBS bioreactor group and the EVs from the 3D hMSC aggregates (showing function in immunomodulation and neuroprotection) in our previous publication [36], but not the EVs from the 2D hMSC culture.
3.4. Scale up effects of the PBS bioreactor on hMSC expansion and metabolism
PBS 0.1L and 0.5L bioreactor systems were inoculated in parallel alongside Corning CellBIND T-flasks (Fig. 5A). After the seeding phase, the 0.1L and 0.5L systems were filled to their working volumes of 100 mL and 500 mL respectively in order to study the effects of scaling on hMSC-EV production. While seeding parameters and feeding schedules were kept constant, sample counts deviated around day 5 with the 0.1L bioreactor having 4.3 × 105 cells/mL (about 20-fold expansion) while the 0.5L bioreactor achieved a cell density of 3.2 × 105 cells/mL (about 14-fold expansion, which may be due to the sampling error as cell densities following trypsinization were all in the 3.5 × 105–4.2 × 105 range) (Fig. 5B and Supplementary Fig. S15). The 2D group reached >30-fold increase on day 5, probably due to the lower seeding density. Media analysis revealed similar values in the 0.1L and 0.5L bioreactors for glucose and lactate concentrations, while the 2D culture showed higher glucose and lower lactate concentration values due to lower cell density (Fig. 5C). Glucose consumption and lactate production in spent media of bioreactors when normalized to cell number were found to be significantly higher than the 2D group, and the 0.5L bioreactor showed higher consumption/production per million cells than the 0.1L bioreactor. However, the molar ratio of lactate produced to glucose consumed stayed constant in the 1.5–1.8 range for the three groups. Other measured metabolites (i.e., glutamine, glutamic acid, and ammonium) only showed minor deviations for the three groups (Fig. 5D). The oxygen saturation in the 0.5L bioreactor was lower than the 0.1L bioreactor, and the CO2 saturation was higher in the 0.5L bioreactor than the 0.1L bioreactor. The pH value was comparable for the two groups (Supplementary Fig. 15). The total cell harvesting efficiency for the 0.5L bioreactor was 71%–81%, higher than the 0.1L bioreactor (54%–62%). And the harvested cell density for the 0.5L bioreactor (3.6–4.1 × 105 cells/mL) was similar to the 0.1L bioreactor (4.0–4.2 × 105 cells/mL).
Fig. 5.
hMSC expansion and metabolism on Synthemax II microcarriers in 0.5L PBS Vertical-Wheel Bioreactors. (A) Process flow for hMSC expansion on Synthemax II microcarrers in PBS Vertical-Wheel Bioreactors; (B) hMSC numbers during expansion and EV collection stages; (C) Glucose and Lactate metabolism: (i) Glucose concentration kinetics; (ii) Lactate concentration kinetics. (iii) Glucose consumed normalized to cell number; (iv) Lactate produced normalized to cell number; (v) Lactate production to glucose consumption ratios. (D) Glutamine metabolism: (i) Glutamine concentration kinetics. The EV collection medium is formulated with Glutamax. (ii) Glutamic acid concentration kinetics; (iii) Ammonia concentration kinetics.
Upon scale up from the 0.1L to the 0.5L bioreactor, mRNA levels for genes regulating glycolysis, autophagy, and EV biogenesis were determined. mRNA levels of HK2 was downregulated (0.4-fold) in the 0.5L group, yet LDHA was upregulated (1.5-fold) in the 0.5L group where higher levels of lactate production per cell was observed (Fig. 6A and Supplementary Fig. S16). PDK1 and PKM2 had no change. For genes responsible for autophagy, only ATG16L1 (0.5-fold) and MITF (2-fold) showed statistically significant changes for the 0.5L bioreactor compared to the 0.1L bioreactor (Fig. 6B). For EV biogenesis, SMPD2 and SMPD3 both showed downregulation (0.25–0.5 fold) in the 0.5L group, none of the GTPases (i.e., Rab7a, Rab27a, Rab27b) showed significant changes (Fig. 6C). Of the ESCRT-dependent EV biogenesis markers, only STAM1 showed upregulation (0.25-fold) in the 0.5L group (Fig. 7D). HRS, ALIX, and TSG101 remained at comparable levels for the 0.5L bioreactor compared to the 0.1L bioreactor group.
Fig. 6.
Metabolic and extracellular vesicle biogenesis pathways for hMSCs in 0.5L PBS Vertical-Wheel Bioreactors. Various genes were measured by RT-qPCR on the mRNAs isolated from the harvested cells at the end of EV collection (n = 6, results were from two independent bioreactor runs), relative to 0.1L bioreactor control. (A) Glycolytic genes; (B) Autophagy genes; (C) ESCRT-dependent EV biogenesis markers; (D) ESCRT-independent EV biogenesis markers. * indicates p < 0.05; ** indicates p < 0.01, *** indicates p < 0.001.
Fig. 7.
Characterizations of extracellular vesicle secreted by 0.5 L bioreactor-expanded hMSCs. (A) Comparison of EV production yield as the number of EVs normalized to the cell number. (B) Protein content of isolated EVs normalized to EV number. (C) Western Blot of exosomal marker expression. (D) miRNA expression in the isolated EVs determined by RT-qPCR (n = 6, results were from two independent bioreactor runs). (E) In vitro wound healing assay for the human fibroblasts that received the 2D and bioreactor hMSC-EVs. Scale bar: 100 μm. VWB: vertical-wheel bioreactor. * indicates p < 0.05; ** indicates p < 0.01, *** indicates p < 0.001.
3.5. Scale up effects on hMSC-EV production and EV miRNA cargo
Upon scaling up from the 0.1L to the 0.5L bioreactor, the yields of EV number per mL spent media increased about 3.5-fold for the 0.5L bioreactor group compared to the 2D cultures (Supplementary Fig. S17). Interestingly, when normalized to cell number, EV secretion in the 0.1L bioreactor group was comparable to the 2D group while the 0.5L bioreactor group was 2.75-fold higher (Fig. 7A). Protein quantification of EVs when normalized to EV number for the 0.1L bioreactor showed about 50% less protein compared to the EVs isolated from 2D culture (Fig. 7B), while the difference for the 0.5L bioreactor group was not statistically significant from the 2D group. Western blots for known exosomal markers were run to confirm the positive expression of HSC70 and CD81, with the bioreactor groups having higher expression than the 2D group. The TSG101 expression was weak for all the groups including the cell lysates, which may require higher protein loading during the assay. Negative marker calnexin was run in parallel to prove no cell debris contamination (Fig. 7C). Analysis of EV miRNA cargo showed upregulation in all miRNAs (i.e., miR-10, 19a, 19b, 21, 30b, 92a, 126, and 132) for the 0.5L bioreactor group in comparison to the 2D group as well as the 0.1L bioreactor group (with the exception of miR-126 and 132). (Fig. 7D and Supplementary Fig. S18). The in vitro wound healing assay was then performed to evaluate the bioreactor-produced hMSC-EV function (Fig. 7E). As expected, the hMSC-EVs promoted wound healing and the bioreactor EV group showed faster scratch closure rates than the 2D EV group.
4. Discussion
External stressors such as thermal fluctuations, hypoxia, medium pH, substrate stiffness, cytoskeletal alterations, and drug stimulation have been reported to increase EV production [59,60] and affect EV composition and cargo [5,61]. Specifically, it has been postulated that biophysical force induced by bioreactor shear stress and 3D microenvironment alter the cytoskeletal function of stem cells and promote EV biogenesis [59]. However, the shear stress encountered by hMSCs in 3D bioreactor microenvironments has yet to be thoroughly investigated. This study demonstrates the xeno-free and scalable production of hMSCs and the secreted EVs in the PBS 0.1L and 0.5L Vertical-Wheel bioreactors. Furthermore, EV quantities isolated from the bioreactor cultures showed more than 2.5-fold increase for the EV number per cell secretion over traditional 2D culture. mRNA levels of genes regulating glycolysis, autophagy, and EV biogenesis were all altered in the bioreactor culture compared to the 2D culture.
There are several differences in the 3D Vertical-Wheel bioreactor cultures compared to the 2D cultures, including the presence of shear stress, substrate materials, curvature (and thus cell shape or 3D microenvironment), and nutrient availability (due to agitation and differences in cell density). For example, softer hydrogels with lower integrand ligand density has been reported to secrete roughly 10-fold more EVs from hMSCs than rigid substrates [62]. However, in Supplementary Fig. S14 and our previous publication [36], proteomics analysis showed high similarity between microcarrier-based Vertical-Wheel bioreactor-generated UC-hMSC-EVs and the dynamic 3D hMSC aggregate-generated EVs (not the 2D culture) indicating that the presence of shear stress and the 3D culture microenvironment may contribute more to the EV protein cargo than the microcarriers (or substrate materials). EV secretion from good manufacturing practice (GMP)-cultured cells on different coating surfaces (gelatin and Synthemax) showed that there was no difference on EV size distribution and total protein level [63].
hMSCs are highly sensitive to biomechanical forces, including shear stress generated through the suspension of 3D microcarriers in dynamic bioreactor systems [64]. Using COMSOL to simulate the velocity profile and shear stress distribution in the Vertical-Wheel bioreactor, the average shear stress increases roughly 3-fold (0.1–0.3 dyn/cm2) over the range of 25–64 RPM (Supplementary Figs. S3–S5). Among the bioreactor groups in this study, varying shear stress via agitation speed did not cause significant changes in gene expression of the cells nor did it impact EV biogenesis. However, significant differences were identified and quantified between the bioreactor and static 2D cultures.
Shear stress has been reported to induce hMSC differentiation and enhance anti-inflammatory mediators (e.g., PGE2) and chemokine production [[25], [26], [27]]. When endothelial cells are subjected to shear stress, ROS production is increased primarily through NADPH oxidases (NOXs) [65,66]. In this study, both total ROS as well as mitochondrial ROS were upregulated in bioreactor culture compared to 2D culture suggesting potentially conserved cellular mechanisms of ROS generation though NOXs. hMSC secretion activity as well as apoptosis are heavily dependent upon ROS acting as cell signaling intermediates on downstream signaling cascades (e.g., Akt/mTOR, Wnt, FOXO, etc.) [[67], [68], [69]]. While mechanistically cross talk between ROS and autophagic machinery is well documented, it has been proposed that ROS induce autophagy as a cellular antioxidant defense to decrease ROS concentrations [[70], [71], [72]]. Our RT-qPCR analysis showed upregulation of genes responsible for regulation of autophagy (TFEB, BECN1, LAMP1, AMPK, ATG5, and MITF) in bioreactors, potentially counteracting ROS accumulation. The genes responsible for regulating glycolysis PDK1, HK2, and LDHA also showed upregulation in the bioreactor. Bioreactor-expanded hMSCs showed upregulation of Sirt1 protein expression which could modulate an increase in autophagy through Sirt1 deacytlyation of FOXO1, ATG5, and ATG7 in a redox-dependent manner as previously reported [65].
Overall, upregulation of GTPases and ESCRT-independent/dependent genes critical in EV biogenesis was observed in Vertical-Wheel bioreactor culture translating to a 2.5-fold increase in EV secretion per cell. While it is still unclear if ESCRT-independent/dependent EV biogenesis pathways operate synergistically or in parallel, there is emerging evidence that EV biogenesis and autophagy are intimately linked through shared molecular machinery in the endolysosomal pathway [22,23,32,73]. For example, ATG5 inhibits the vacuolar proton pump on MVBs preventing the acidification and degradation of the ILV contents and redirecting the MVB to the plasma membrane where the ILV are secreted as exosomes [23]. Interestingly, Vertical-Wheel bioreactor culture increased ATG5 expression which may be redirecting MVB away from lysosomal degradation and upregulating exosomal secretion. Similarly, bioreactor culture upregulates MITF, a transcription factor that binds to the promoter region of lysosomal genes encoding hydrolases, lysosomal membrane permeases, and lysosome-associated proteins [74]. Furthermore, MITF is a direct transcription factor for GTPase Rab27a, which is critical in the downstream MVB docking and subsequent EV release at the plasma membrane [75,76]. Indeed, our results did show an increase in Rab27a and Rab27b expression along with other small GTPases (Rab31 and Rab7a) implicated in ESCRT-independent exosomal release [77]. Further investigations are needed in order to determine if altered autophagy in bioreactor culture is affecting EV biogenesis. It is also noticed that CD63, which recently was confirmed as a predominate small EV (exosome) marker, was upregulated in all bioreactor conditions, and EV size distribution showed consistent results with the increased small particle (less than 50 nm) ratio.
MiRNA cargo has been recognized as the major contributor to the therapeutic effects of hMSC-derived exosomes [7,78,79]. Shear stress is noted to play an essential role in stimulating angiogenesis and remodeling of the vasculature [[80], [81], [82], [83], [84]]. There is also evidence to show that miRNAs can be induced in cells exposed to biophysical forces, including miR-21 and miR-132 [[85], [86], [87]]. Therefore, EV miRNA cargo that have implications in angiogenesis and wound healing were investigated in this study. miR-10, miR-21, miR-19a, and miR-132, which have all been shown to promote or facilitate angiogenic behavior, were upregulated in bioreactor culture compared to the 2D control. miR-10 sequesters vascular endothelial growth factor (VEGF) through the regulation of cell surface ligands [88]. miR-21 promotes angiogenesis through targeting and inhibition of the Fas ligand protein [89]. miR-19a promotes cell proliferation as well as angiogenesis through the inhibition of the antiangiogenic factor TSP-1 and the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) signaling pathway [90]. miR-132 has been shown to act an “angiogenic switch” in the endothelium inducing vascularization [91]. Contrarily, the upregulation of miR-19b, miR-30b, and miR-377 was observed in this study, which have been shown to play roles in inhibiting or suppressing angiogenesis. miR-19b may potentially inhibit angiogenesis via VEGF receptor targeting in the endocytosis signaling pathway [92], while miR-30b and miR-377 inhibitions promoted angiogenesis [93,94]. Together, these miRNAs that have a regulatory mechanism in angiogenesis and wound healing may be impacted by bioreactor culture. It needs to be noted that miR-377 and miR-132 showed differential expression across different culture conditions, indicating that their expression is dependent on the shear stress level. miR-132 expression reached a plateau (after 40 RPM) with shear stress increases, while miR-377 expression is the highest under the 40 RPM condition and the level decreases under the 64 RPM condition. This is consistent with previous literature demonstrating that miR-132 increases stepwise with fluid shear stress (0–15 dyn/cm2) in periodontal ligament cells [95].
Besides angiogenesis, the miRNAs upregulated in bioreactor-produced EVs also have neuroprotective effects (e.g., miR-10, 19a, 19b, 21, 132, and 377). miR-19a can inhibit the Cyclin D1 expression, regulate cell cycle and its interactions with Wnt signaling pathway, and exhibit anti-apoptotic effects [96,97]. miR-21 has potent neuroprotective effects, alleviates blood-brain barrier (BBB) disruption in ischemic stroke, and promotes neurite outgrowth by regulating programmed cell death protein 4 [[98], [99], [100], [101]]. miR-21 was also identified as a long-term memory keeper of the fibrogenic program in hMSCs regulated through the acutely mechanosensitive myocardin-related transcription factor-A (MRTF-A/MLK-1) [87]. miR-132 has been reported to play an important role in maintaining the BBB integrity as well as regulating neurite outgrowth and neuronal transmission [102]. Blocking the miR-132 delivery from neurons to cerebral endothelial cells can induce BBB leakage.
Additionally, very high levels of cell and microcarrier aggregation around day three and five of culture were observed (Supplementary Fig. S6). Similar 3D aggregate culture has been reported to have significant impacts on EV proteins and miRNA cargo [103,104]. Our previous study demonstrated that 3D hMSC aggregates promote EV biogenesis and modulate EV protein cargo with potential to enhance immunomodulatory potential compared to the 2D culture [36], possibly due to the cytoskeleton reorganization and the reconfigured cell metabolism. In this study, proteomics analysis reveals that the DEPs in the parent cells may contribute to the DEPs in the secreted EVs, and the altered cellular metabolism in PBS bioreactor microenvironment (e.g., exposure to shear stress, the 3D microcarrier culture) may contribute to the enhanced EV biogenesis. The protein cargo of EVs of PBS bioreactor-expanded hMSCs has similar profile to the EVs of 3D hMSC aggregates, which has been well characterized in our previous studies, including in vitro functional analysis and in vivo therapeutic effects on Alzheimer's disease animal models [36,105]. The DEPs in the bioreactor-EVs are enriched in cell adhesion, cell migration, ECM-receptor interaction, focal adhesion, and HIF-1 signaling pathway. The DEPs in the EVs in bioreactor only subset shows the enrichment of proteins related to metabolic processes and cadherin binding. While the protein expression of bioreactor-EVs possesses similar profiles to EVs from 3D hMSC aggregates, in vivo functional analysis needs to be investigated in the future to confirm similar therapeutic benefits.
Increasing production of hMSCs and their derived EVs to meet growing demands as cellular therapeutics requires progressive scaling into larger bioreactor systems that ensure the therapeutic efficacy [106]. Our study found scaling hMSC and EV production resulted in similar expansion in the 0.5L Vertical-Wheel bioreactor compared to the 0.1L bioreactor. Each bioreactor was run at the lowest possible agitation speed to suspend microcarriers (25 and 19 RPM for the 0.1L and the 0.5L bioreactor respectively). Lower shear stress may have resulted in larger aggregation of hMSCs and microcarriers in the 0.5L bioreactor on days 5 and 8, during high confluency, resulting in non-homogenous mixtures during sampling. Overall, mRNA levels for genes modulating glycolysis, autophagy, and EV biogenesis in the 0.5L vessel showed only minor changes from the 0.1L bioreactor. The increased yields of EV production per cell and per mL media over 2D culture were retained, suggesting that scaling does not have a significant impact on hMSCs and the derived EVs. Yet, there was a significant increase in miRNA cargo in EVs from the 0.5L vessel. The interface of the media with the gas in the headspace of each bioreactor is of similar sizes, therefore, the near equivalent flux of oxygen into the media may not be able to replenish increased oxygen consumption in the 0.5L vessel and lower oxygen tension may increase miRNA levels [107]. Introduction of sparging or a steady flow of oxygen into the headspace are proven strategies at replenishing oxygen levels [108]. The Vertical-Wheel bioreactor system has potential for the scaled production up to 15–80L.
5. Conclusion
This study demonstrates that in PBS bioreactor microenvironment (i.e., the presence of shear stress, 3D microcarrier culture) hMSCs shift metabolic processes and ROS accumulation is increased. hMSC microenvironment in a Vertical-Wheel bioreactor significantly increases EV yields and promotes EV biogenesis markers compared to traditional 2D culture. Increasing shear stress doesn't impact hMSC proliferation and EV production over tested range (25–64 RPM). Bioreactor microenvironment alters miRNA cargo (upregulated miRNAs with therapeutic potential) and protein cargo, which shows the similarity to the EVs from 3D hMSC aggregates. The scale up potential is also demonstrated in 0.5L bioreactor. This study demonstrates the feasibly of scalable production of hMSC-EVs in bioreactors with defined cargo and provides the possible mechanisms that contribute to the promoted EV biogenesis in bioreactors. Together, the scale up potential of hMSC-EVs in the Vertical-Wheel bioreactor was demonstrated in this study.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding authors on reasonable request.
Ethics approval and consent to participate
Not applicable.
CRediT authorship contribution statement
Richard Jeske: Conceptualization, Investigation, Formal analysis, Data curation, Writing – original draft, Writing – review & editing, Visualization. Chang Liu: Investigation, Validation. Leanne Duke: Methodology, Investigation. Maria L. Canonicco Castro: Software, Formal analysis. Laureana Muok: Validation. Peggy Arthur: Investigation, Validation. Mandip Singh: Investigation, Supervision. Sunghoon Jung: Conceptualization, Resources, Supervision, Project administration. Li Sun: Conceptualization, Methodology, Investigation, Data curation, Writing – review & editing, Visualization, Supervision, Project administration. Yan Li: Conceptualization, Methodology, Investigation, Resources, Data curation, Writing – original draft, Writing – review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgement
The authors would like to thank Dr. Brian K. Washburn and Kristina Poduch in Florida state university (FSU) Department of Biological Sciences for their help with RT-qPCR analysis. The authors would also thank for the support by FSU Flow Cytometry Core facility and FSU College of Medicine Translational Science laboratory for proteomics. TEM at the Biological Science Imaging Resource (BSIR) is supported by Florida State University. LM held McKnight Doctorial Fellowship and was also supported in part by FAMU title III funding. This work is supported by National Science Foundation (CBET-1743426 and CBET-1917618). Research reported in this publication was also partially supported by the National Institutes of Health (USA) under Award Number R01NS125016. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2022.07.004.
Contributor Information
Richard Jeske, Email: rjj15@my.fsu.edu.
Chang Liu, Email: cl20ev@my.fsu.edu.
Leanne Duke, Email: leanne.duke@med.fsu.edu.
Maria L. Canonicco Castro, Email: mlc16m@my.fsu.edu.
Laureana Muok, Email: laureana1.muok@famu.edu.
Peggy Arthur, Email: PEGGY1.ARTHUR@famu.edu.
Mandip Singh, Email: mandip.sachdeva@famu.edu.
Sunghoon Jung, Email: sjung@pbsbiotech.com.
Li Sun, Email: li.sun@med.fsu.edu.
Yan Li, Email: yli4@fsu.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Han C., Sun X., Liu L., Jiang H., Shen Y., Xu X., Li J., Zhang G., Huang J., Lin Z., Xiong N., Wang T. Exosomes and their therapeutic potentials of stem cells. Stem Cell. Int. 2016;2016 doi: 10.1155/2016/7653489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jarmalavičiūtė A., Pivoriūnas A. Exosomes as a potential novel therapeutic tools against neurodegenerative diseases. Pharmacol. Res. 2016;113:816–822. doi: 10.1016/j.phrs.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Gimona M., Pachler K., Laner-Plamberger S., Schallmoser K., Rohde E. Manufacturing of human extracellular vesicle-based therapeutics for clinical use. Int. J. Mol. Sci. 2017;18(6) doi: 10.3390/ijms18061190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reiner A.T., Witwer K.W., van Balkom B.W.M., de Beer J., Brodie C., Corteling R.L., Gabrielsson S., Gimona M., Ibrahim A.G., de Kleijn D., Lai C.P., Lotvall J., Del Portillo H.A., Reischl I.G., Riazifar M., Salomon C., Tahara H., Toh W.S., Wauben M.H.M., Yang V.K., Yang Y., Yeo R.W.Y., Yin H., Giebel B., Rohde E., Lim S.K. Concise review: developing best-practice models for the therapeutic use of extracellular vesicles. Stem Cells Transl Med. 2017;6(8):1730–1739. doi: 10.1002/sctm.17-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel D.B., Santoro M., Born L.J., Fisher J.P., Jay S.M. Towards rationally designed biomanufacturing of therapeutic extracellular vesicles: impact of the bioproduction microenvironment. Biotechnol. Adv. 2018;36(8):2051–2059. doi: 10.1016/j.biotechadv.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiklander O.P.B., Brennan M.A., Lotvall J., Breakefield X.O., El Andaloussi S. Advances in therapeutic applications of extracellular vesicles. Sci. Transl. Med. 2019;11(492) doi: 10.1126/scitranslmed.aav8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Z.G., Buller B., Chopp M. Exosomes - beyond stem cells for restorative therapy in stroke and neurological injury. Nat. Rev. Neurol. 2019;15(4):193–203. doi: 10.1038/s41582-018-0126-4. [DOI] [PubMed] [Google Scholar]
- 8.Feng R., Ullah M., Chen K., Ali Q., Lin Y., Sun Z. Stem cell-derived extracellular vesicles mitigate ageing-associated arterial stiffness and hypertension. J. Extracell. Vesicles. 2020;9(1) doi: 10.1080/20013078.2020.1783869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim H.Y., Kim T.J., Kang L., Kim Y.J., Kang M.K., Kim J., Ryu J.H., Hyeon T., Yoon B.W., Ko S.B., Kim B.S. Mesenchymal stem cell-derived magnetic extracellular nanovesicles for targeting and treatment of ischemic stroke. Biomaterials. 2020;243 doi: 10.1016/j.biomaterials.2020.119942. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X., Zhang H., Gu J., Zhang J., Shi H., Qian H., Wang D., Xu W., Pan J., Santos H.A. Engineered extracellular vesicles for cancer therapy. Adv. Mater. 2021;33(14) doi: 10.1002/adma.202005709. [DOI] [PubMed] [Google Scholar]
- 11.Ranganath S.H., Levy O., Inamdar M.S., Karp J.M. Harnessing the mesenchymal stem cell secretome for the treatment of cardiovascular disease. Cell Stem Cell. 2012;10(3):244–258. doi: 10.1016/j.stem.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linero I., Chaparro O. Paracrine effect of mesenchymal stem cells derived from human adipose tissue in bone regeneration. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0107001. e107001-e107001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H., Rong P., Ma X., Nie W., Chen C., Yang C., Zhang J., Dong Q., Wang W. Paracrine effect of mesenchymal stem cell as a novel therapeutic strategy for diabetic nephropathy. Life Sci. 2018;215:113–118. doi: 10.1016/j.lfs.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Berglund A.K., Fortier L.A., Antczak D.F., Schnabel L.V. Immunoprivileged no more: measuring the immunogenicity of allogeneic adult mesenchymal stem cells. Stem Cell Res. Ther. 2017;8(1) doi: 10.1186/s13287-017-0742-8. 288-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skotland T., Sagini K., Sandvig K., Llorente A. An emerging focus on lipids in extracellular vesicles. Adv. Drug Deliv. Rev. 2020;159:308–321. doi: 10.1016/j.addr.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y., Liu Y., Liu H., Tang W.H. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. 2019;9(1):19. doi: 10.1186/s13578-019-0282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang Y., Duan L., Lu J., Xia J. Engineering exosomes for targeted drug delivery. Theranostics. 2021;11(7):3183–3195. doi: 10.7150/thno.52570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bunggulawa E.J., Wang W., Yin T., Wang N., Durkan C., Wang Y., Wang G. Recent advancements in the use of exosomes as drug delivery systems. J. Nanobiotechnol. 2018;16(1):81. doi: 10.1186/s12951-018-0403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenberger L., Ezquer M., Lillo-Vera F., Pedraza P.L., Ortúzar M.I., González P.L., Figueroa-Valdés A.I., Cuenca J., Ezquer F., Khoury M., Alcayaga-Miranda F. Stem cell exosomes inhibit angiogenesis and tumor growth of oral squamous cell carcinoma. Sci. Rep. 2019;9(1):663. doi: 10.1038/s41598-018-36855-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H.-X., Liang F.-C., Gu P., Xu B.-L., Xu H.-J., Wang W.-T., Hou J.-Y., Xie D.-X., Chai X.-Q., An S.-J. Exosomes derived from mesenchymal stem cells repair a Parkinson's disease model by inducing autophagy. Cell Death Dis. 2020;11(4):288. doi: 10.1038/s41419-020-2473-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang X., Lew K.S., Chen Q., Richards A.M., Wang P. Human mesenchymal stem cell-derived exosomes reduce ischemia/reperfusion injury by the inhibitions of apoptosis and autophagy. Curr. Pharmaceut. Des. 2018;24(44):5334–5341. doi: 10.2174/1381612825666190119130441. [DOI] [PubMed] [Google Scholar]
- 22.Xing H., Tan J., Miao Y., Lv Y., Zhang Q. Crosstalk between exosomes and autophagy: a review of molecular mechanisms and therapies. J. Cell Mol. Med. 2021;25(5):2297–2308. doi: 10.1111/jcmm.16276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salimi L., Akbari A., Jabbari N., Mojarad B., Vahhabi A., Szafert S., Kalashani S.A., Soraya H., Nawaz M., Rezaie J. Synergies in exosomes and autophagy pathways for cellular homeostasis and metastasis of tumor cells. Cell Biosci. 2020;10(1):64. doi: 10.1186/s13578-020-00426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baixauli F., López-Otín C., Mittelbrunn M. Exosomes and autophagy: coordinated mechanisms for the maintenance of cellular fitness. Front. Immunol. 2014;5 doi: 10.3389/fimmu.2014.00403. 403-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yourek G., McCormick S.M., Mao J.J., Reilly G.C. Shear stress induces osteogenic differentiation of human mesenchymal stem cells. Regen. Med. 2010;5(5):713–724. doi: 10.2217/rme.10.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henderson K., Sligar A.D., Le V.P., Lee J., Baker A.B. Biomechanical regulation of mesenchymal stem cells for cardiovascular tissue engineering. Adv. Healthc. Mater. 2017;6(22) doi: 10.1002/adhm.201700556. [DOI] [PubMed] [Google Scholar]
- 27.Diaz M.F., Vaidya A.B., Evans S.M., Lee H.J., Aertker B.M., Alexander A.J., Price K.M., Ozuna J.A., Liao G.P., Aroom K.R., Xue H., Gu L., Omichi R., Bedi S., Olson S.D., Cox C.S., Jr., Wenzel P.L. Biomechanical forces promote immune regulatory function of bone marrow mesenchymal stromal cells. Stem Cell. 2017;35(5):1259–1272. doi: 10.1002/stem.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.K. Göran Ronquist, Extracellular Vesicles and Energy Metabolism, (1873-3492 (Electronic)). [DOI] [PubMed]
- 29.I. Lazar, E. Clement, C. Attane, C. Muller, L. Nieto, A New Role for Extracellular Vesicles: How Small Vesicles Can Feed Tumors' Big Appetite, (1539-7262 (Electronic)). [DOI] [PMC free article] [PubMed]
- 30.Wang J., Bonacquisti E.E., Brown A.D., Nguyen J. Boosting the biogenesis and secretion of mesenchymal stem cell-derived exosomes. Cells. 2020;9(3):660. doi: 10.3390/cells9030660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei Y., Wang D., Jin F., Bian Z., Li L., Liang H., Li M., Shi L., Pan C., Zhu D., Chen X., Hu G., Liu Y., Zhang C.-Y., Zen K. Pyruvate kinase type M2 promotes tumour cell exosome release via phosphorylating synaptosome-associated protein 23. Nat. Commun. 2017;8(1) doi: 10.1038/ncomms14041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu J., Camfield R., Gorski S.M. The interplay between exosomes and autophagy – partners in crime. J. Cell Sci. 2018;131(15) doi: 10.1242/jcs.215210. [DOI] [PubMed] [Google Scholar]
- 33.Arslan F., Lai R.C., Smeets M.B., Akeroyd L., Choo A., Aguor E.N.E., Timmers L., van Rijen H.V., Doevendans P.A., Pasterkamp G., Lim S.K., de Kleijn D.P. Mesenchymal stem cell-derived exosomes increase ATP levels, decrease oxidative stress and activate PI3K/Akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury. Stem Cell Res. 2013;10(3):301–312. doi: 10.1016/j.scr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 34.Pachler K., Lener T., Streif D., Dunai Z.A., Desgeorges A., Feichtner M., Oller M., Schallmoser K., Rohde E., Gimona M. A Good Manufacturing Practice-grade standard protocol for exclusively human mesenchymal stromal cell-derived extracellular vesicles. Cytotherapy. 2017;19(4):458–472. doi: 10.1016/j.jcyt.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Colao I.L., Corteling R., Bracewell D., Wall I. Manufacturing exosomes: a promising therapeutic platform. Trends Mol. Med. 2018;24(3):242–256. doi: 10.1016/j.molmed.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 36.Yuan X., Sun L., Jeske R., Nkosi D., York S., Liu Y., Grant S.C., Meckes D.G.J., Li Y. Engineering extracellular vesicles by three-dimensional dynamic culture of human mesenchymal stem cells. J. Extracell. Vesicles. 2022;11(6) doi: 10.1002/jev2.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nogueira D.E.S., Rodrigues C.A.V., Carvalho M.S., Miranda C.C., Hashimura Y., Jung S., Lee B., Cabral J.M.S. Strategies for the expansion of human induced pluripotent stem cells as aggregates in single-use Vertical-Wheel bioreactors. J. Biol. Eng. 2019;13:74. doi: 10.1186/s13036-019-0204-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sousa M.F., Silva M.M., Giroux D., Hashimura Y., Wesselschmidt R., Lee B., Roldao A., Carrondo M.J., Alves P.M., Serra M. Production of oncolytic adenovirus and human mesenchymal stem cells in a single-use, Vertical-Wheel bioreactor system: impact of bioreactor design on performance of microcarrier-based cell culture processes. Biotechnol. Prog. 2015;31(6):1600–1612. doi: 10.1002/btpr.2158. [DOI] [PubMed] [Google Scholar]
- 39.Lembong J., Kirian R., Takacs J.D., Olsen T.R., Lock L.T., Rowley J.A., Ahsan T. Bioreactor parameters for microcarrier-based human MSC expansion under xeno-free conditions in a vertical-wheel system. Bioengineering. 2020;7(3) doi: 10.3390/bioengineering7030073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Sousa Pinto D., Bandeiras C., de Almeida Fuzeta M., Rodrigues C.A.V., Jung S., Hashimura Y., Tseng R.J., Milligan W., Lee B., Ferreira F.C., Lobato da Silva C., Cabral J.M.S. Scalable manufacturing of human mesenchymal stromal cells in the vertical-wheel bioreactor system: an experimental and economic approach. Biotechnol. J. 2019;14(8) doi: 10.1002/biot.201800716. [DOI] [PubMed] [Google Scholar]
- 41.Borys B.S., So T., Colter J., Dang T., Roberts E.L., Revay T., Larijani L., Krawetz R., Lewis I., Argiropoulos B., Rancourt D.E., Jung S., Hashimura Y., Lee B., Kallos M.S. Optimized serial expansion of human induced pluripotent stem cells using low-density inoculation to generate clinically relevant quantities in vertical-wheel bioreactors. Stem Cells Transl Med. 2020;9(9):1036–1052. doi: 10.1002/sctm.19-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva T.P., Fernandes T.G., Nogueira D.E.S., Rodrigues C.A.V., Bekman E.P., Hashimura Y., Jung S., Lee B., Carmo-Fonseca M., Cabral J.M.S. Scalable generation of mature cerebellar organoids from human pluripotent stem cells and characterization by immunostaining. JoVE. 2020;160 doi: 10.3791/61143. [DOI] [PubMed] [Google Scholar]
- 43.Silva T.P., Sousa-Luis R., Fernandes T.G., Bekman E.P., Rodrigues C.A.V., Vaz S.H., Moreira L.M., Hashimura Y., Jung S., Lee B., Carmo-Fonseca M., Cabral J.M.S. Transcriptome profiling of human pluripotent stem cell-derived cerebellar organoids reveals faster commitment under dynamic conditions. Biotechnol. Bioeng. 2021;18(7):2781–2803. doi: 10.1002/bit.27797. [DOI] [PubMed] [Google Scholar]
- 44.Yuan X., Liu Y., Bijonowski B., Tsai A.C., Fu Q., Logan T.M., Ma T., Li Y. NAD+/NADH redox alterations reconfigure metabolism and rejuvenate senescent human mesenchymal stem cells in vitro. Commun. Biol. 2020;3:774. doi: 10.1038/s42003-020-01514-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rider M.A., Hurwitz S.N., Meckes D.G., Jr. ExtraPEG: a polyethylene glycol-based method for enrichment of extracellular vesicles. Sci. Rep. 2016;6 doi: 10.1038/srep23978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marzano M., Bejoy J., Cheerathodi M., Sun L., York S., Zhao J., Kanekiyo T., Bu G., Meckes D.G., Jr., Li Y. Differential effects of extracellular vesicles of lineage-specific human pluripotent stem cells on cellular behaviours of isogenic cortical spheroids. Cells. 2019;8:993–1014. doi: 10.3390/cells8090993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cone A.S., Hurwitz S.N., Lee G., Yuan X., Zhou Y., Li Y., Meckes D.G., Jr. Alix and Syntenin-1 traffic amyloid precursor protein and amyloid beta into extracellular vesicles. BMC Mol. Cell Biol. 2020;21:58–78. doi: 10.1186/s12860-020-00302-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marzano M., Bou-Dargham M.J., Cone A.S., York S., Helsper S., Grant S.C., Meckes D.G., Jr., Sang Q.X., Li Y. Biogenesis of extracellular vesicles produced from human stem cell-derived cortical spheroids exposed to iron oxides. ACS Biomater. Sci. Eng. 2021;7(3):1111–1122. doi: 10.1021/acsbiomaterials.0c01286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jung S., Sen A., Rosenberg L., Behie L.A. Human mesenchymal stem cell culture: rapid and efficient isolation and expansion in a defined serum-free medium. J Tissue Eng. Regen. Med. 2012;6(5):391–403. doi: 10.1002/term.441. [DOI] [PubMed] [Google Scholar]
- 50.Yuan X., Chen X., Zeng C., Meckes D.G., Jr., Li Y. Extracellular vesicle collection from human stem cells grown in suspension bioreactors. Methods Mol. Biol. 2022;2436:193–204. doi: 10.1007/7651_2021_416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song L., Wang K., Li Y., Yang Y. Nanotopography promoted neuronal differentiation of human induced pluripotent stem cells. Colloids Surf. B Biointerfaces. 2016;148:49–58. doi: 10.1016/j.colsurfb.2016.08.041. [DOI] [PubMed] [Google Scholar]
- 52.Lasser C., Eldh M., Lotvall J. Isolation and characterization of RNA-containing exosomes. JoVE. 2012;59 doi: 10.3791/3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roney K.E., O'Connor B.P., Wen H., Holl E.K., Guthrie E.H., Davis B.K., Jones S.W., Jha S., Sharek L., Garcia-Mata R., Bear J.E., Ting J.P. Plexin-B2 negatively regulates macrophage motility, Rac, and Cdc42 activation. PLoS One. 2011;6(9) doi: 10.1371/journal.pone.0024795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hurwitz S.N., Meckes D.G., Jr. An adaptable polyethylene glycol-based workflow for proteomic analysis of extracellular vesicles. Methods Mol. Biol. 2017;1660:303–317. doi: 10.1007/978-1-4939-7253-1_25. [DOI] [PubMed] [Google Scholar]
- 55.Hurwitz S.N., Sun L., Cole K.Y., Ford C.R., Olcese J.M., 3rd, Meckes D.G., Jr. An optimized method for enrichment of whole brain-derived extracellular vesicles reveals insight into neurodegenerative processes in a mouse model of Alzheimer's disease. J. Neurosci. Methods. 2018;307:210–220. doi: 10.1016/j.jneumeth.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeske R., Lewis S., Tsai A.C., Sanders K., Liu C., Yuan X., Li Y. Agitation in a microcarrier-based spinner bioreactor modulates homeostasis of human mesenchymal stem cells. Biochem. Eng. J. 2021;168 doi: 10.1016/j.bej.2021.107947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willms E., Johansson H.J., Mäger I., Lee Y., Blomberg K.E.M., Sadik M., Alaarg A., Smith C.I.E., Lehtiö J., El Andaloussi S., Wood M.J.A., Vader P. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016;6(1) doi: 10.1038/srep22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar S., Kim C.W., Simmons R.D., Jo H. Role of flow-sensitive microRNAs in endothelial dysfunction and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2014;34(10):2206–2216. doi: 10.1161/ATVBAHA.114.303425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu C., Su C. Design strategies and application progress of therapeutic exosomes. Theranostics. 2019;9(4):1015–1028. doi: 10.7150/thno.30853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jeyaram A., Lamichhane T.N., Wang S., Zou L., Dahal E., Kronstadt S.M., Levy D., Parajuli B., Knudsen D.R., Chao W., Jay S.M. Enhanced loading of functional miRNA cargo via pH gradient modification of extracellular vesicles. Mol. Ther. 2019;28(3):975–985. doi: 10.1016/j.ymthe.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patel D.B., Luthers C.R., Lerman M.J., Fisher J.P., Jay S.M. Enhanced extracellular vesicle production and ethanol-mediated vascularization bioactivity via a 3D-printed scaffold-perfusion bioreactor system. Acta Biomater. 2018;95:236–244. doi: 10.1016/j.actbio.2018.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lenzini S., Debnath K., Joshi J.C., Wong S.W., Srivastava K., Geng X., Cho I.S., Song A., Bargi R., Lee J.C., Mo G.C.H., Mehta D., Shin J.-W. Cell–matrix interactions regulate functional extracellular vesicle secretion from mesenchymal stromal cells. ACS Nano. 2021;15(11):17439–17452. doi: 10.1021/acsnano.1c03231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andriolo G., Provasi E., Lo Cicero V., Brambilla A., Soncin S., Torre T., Milano G., Biemmi V., Vassalli G., Turchetto L., Barile L., Radrizzani M. Exosomes from human cardiac progenitor cells for therapeutic applications: development of a GMP-grade manufacturing method. Front. Physiol. 2018;9:1169. doi: 10.3389/fphys.2018.01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsai A.-C., Jeske R., Chen X., Yuan X., Li Y. Influence of microenvironment on mesenchymal stem cell therapeutic potency: from planar culture to microcarriers. Front. Bioeng. Biotechnol. 2020;8:640. doi: 10.3389/fbioe.2020.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu J., Bi X., Chen T., Zhang Q., Wang S.X., Chiu J.J., Liu G.S., Zhang Y., Bu P., Jiang F. Shear stress regulates endothelial cell autophagy via redox regulation and Sirt1 expression. Cell Death Dis. 2015;6(7) doi: 10.1038/cddis.2015.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsieh H.-J., Liu C.-A., Huang B., Tseng A.H.H., Wang D.L. Shear-induced endothelial mechanotransduction: the interplay between reactive oxygen species (ROS) and nitric oxide (NO) and the pathophysiological implications. J. Biomed. Sci. 2014;21(1):3. doi: 10.1186/1423-0127-21-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Atashi F., Modarressi A., Pepper M.S. The role of reactive oxygen species in mesenchymal stem cell adipogenic and osteogenic differentiation: a review. Stem Cell. Dev. 2015;24(10):1150–1163. doi: 10.1089/scd.2014.0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu Y., Yuan X., Muñoz N., Logan T.M., Ma T. Commitment to aerobic glycolysis sustains immunosuppression of human mesenchymal stem cells. Stem Cells Transl. Med. 2019;8(1):93–106. doi: 10.1002/sctm.18-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu C., Zhao L., Peng C., Li L. Regulation of the mitochondrial reactive oxygen species: strategies to control mesenchymal stem cell fates ex vivo and in vivo. J. Cell Mol. Med. 2018;22(11):5196–5207. doi: 10.1111/jcmm.13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Azad M.B., Chen Y., Gibson S.B. Regulation of autophagy by reactive oxygen species (ROS): implications for cancer progression and treatment. Antioxidants Redox Signal. 2009;11(4):777–790. doi: 10.1089/ars.2008.2270. [DOI] [PubMed] [Google Scholar]
- 71.Filomeni G., De Zio D., Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 2015;22(3):377–388. doi: 10.1038/cdd.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fang C., Gu L., Smerin D., Mao S., Xiong X. The interrelation between reactive oxygen species and autophagy in neurological disorders. Oxid. Med. Cell. Longev. 2017;2017 doi: 10.1155/2017/8495160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hessvik N.P., Llorente A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018;75(2):193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perera R.M., Di Malta C., Ballabio A. MiT/TFE family of transcription factors, lysosomes, and cancer. Annu. Rev. Cell Biol. 2019;3:203–222. doi: 10.1146/annurev-cancerbio-030518-055835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chiaverini C., Beuret L., Flori E., Busca R., Abbe P., Bille K., Bahadoran P., Ortonne J.P., Bertolotto C., Ballotti R. Microphthalmia-associated transcription factor regulates RAB27A gene expression and controls melanosome transport. J. Biol. Chem. 2008;283(18):12635–12642. doi: 10.1074/jbc.M800130200. [DOI] [PubMed] [Google Scholar]
- 76.Ostrowski M., Carmo N.B., Krumeich S., Fanget I., Raposo G., Savina A., Moita C.F., Schauer K., Hume A.N., Freitas R.P., Goud B., Benaroch P., Hacohen N., Fukuda M., Desnos C., Seabra M.C., Darchen F., Amigorena S., Moita L.F., Thery C. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010;12(1):19–30. doi: 10.1038/ncb2000. ;, sup pp 1-13. [DOI] [PubMed] [Google Scholar]
- 77.Wei D., Zhan W., Gao Y., Huang L., Gong R., Wang W., Zhang R., Wu Y., Gao S., Kang T. RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res. 2021;31(2):157–177. doi: 10.1038/s41422-020-00409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.S. Sart, X. Yuan, R. Jeske, Y. Li, Engineering exosomal MicroRNAs in human stem cells, Adv. Stem Cell Biol. A chapter to “Induced Pluripotent Stem Cells – Novel Concepts” (2021) In Press.
- 79.Chen X., Liang H., Zhang J., Zen K., Zhang C.Y. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22(3):125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 80.Fisher A.B., Chien S., Barakat A.I., Nerem R.M. Endothelial cellular response to altered shear stress. Am. J. Physiol. Lung Cell Mol. Physiol. 2001;281(3):L529–L533. doi: 10.1152/ajplung.2001.281.3.L529. [DOI] [PubMed] [Google Scholar]
- 81.Ueda A., Koga M., Ikeda M., Kudo S., Tanishita K. Effect of shear stress on microvessel network formation of endothelial cells with in vitro three-dimensional model. Am. J. Physiol. Heart Circ. Physiol. 2004;287(3):H994–H1002. doi: 10.1152/ajpheart.00400.2003. [DOI] [PubMed] [Google Scholar]
- 82.Galie P.A., Nguyen D.-H.T., Choi C.K., Cohen D.M., Janmey P.A., Chen C.S. Fluid shear stress threshold regulates angiogenic sprouting. Proc. Natl. Acad. Sci. U. S. A. 2014;111(22):7968–7973. doi: 10.1073/pnas.1310842111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Buchanan C.F., Verbridge S.S., Vlachos P.P., Rylander M.N. Flow shear stress regulates endothelial barrier function and expression of angiogenic factors in a 3D microfluidic tumor vascular model. Cell Adhes. Migrat. 2014;8(5):517–524. doi: 10.4161/19336918.2014.970001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kolluru G.K., Sinha S., Majumder S., Muley A., Siamwala J.H., Gupta R., Chatterjee S. Shear stress promotes nitric oxide production in endothelial cells by sub-cellular delocalization of eNOS: a basis for shear stress mediated angiogenesis. Nitric Oxide. 2010;22(4):304–315. doi: 10.1016/j.niox.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 85.Ni C.W., Qiu H., Jo H. MicroRNA-663 upregulated by oscillatory shear stress plays a role in inflammatory response of endothelial cells. Am. J. Physiol. Heart Circ. Physiol. 2011;300(5):H1762–H1769. doi: 10.1152/ajpheart.00829.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Qi L., Zhang Y. The microRNA 132 regulates fluid shear stress-induced differentiation in periodontal ligament cells through mTOR signaling pathway. Cell. Physiol. Biochem. 2014;33(2):433–445. doi: 10.1159/000358624. [DOI] [PubMed] [Google Scholar]
- 87.Li C.X., Talele N.P., Boo S., Koehler A., Knee-Walden E., Balestrini J.L., Speight P., Kapus A., Hinz B. MicroRNA-21 preserves the fibrotic mechanical memory of mesenchymal stem cells. Nat. Mater. 2017;16(3):379–389. doi: 10.1038/nmat4780. [DOI] [PubMed] [Google Scholar]
- 88.Hassel D., Cheng P., White M.P., Ivey K.N., Kroll J., Augustin H.G., Katus H.A., Stainier D.Y.R., Srivastava D. MicroRNA-10 regulates the angiogenic behavior of zebrafish and human endothelial cells by promoting vascular endothelial growth factor signaling. Circ. Res. 2012;111(11):1421–1433. doi: 10.1161/CIRCRESAHA.112.279711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Du X., Hong L., Sun L., Sang H., Qian A., Li W., Zhuang H., Liang H., Song D., Li C., Wang W., Li X. miR-21 induces endothelial progenitor cells proliferation and angiogenesis via targeting FASLG and is a potential prognostic marker in deep venous thrombosis. J. Transl. Med. 2019;17(1):270. doi: 10.1186/s12967-019-2015-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gollmann-Tepeköylü C., Pölzl L., Graber M., Hirsch J., Nägele F., Lobenwein D., Hess M.W., Blumer M.J., Kirchmair E., Zipperle J., Hromada C., Mühleder S., Hackl H., Hermann M., Al Khamisi H., Förster M., Lichtenauer M., Mittermayr R., Paulus P., Fritsch H., Bonaros N., Kirchmair R., Sluijter J.P.G., Davidson S., Grimm M., Holfeld J. miR-19a-3p containing exosomes improve function of ischaemic myocardium upon shock wave therapy. Cardiovasc. Res. 2020;116(6):1226–1236. doi: 10.1093/cvr/cvz209. [DOI] [PubMed] [Google Scholar]
- 91.Anand S., Majeti B.K., Acevedo L.M., Murphy E.A., Mukthavaram R., Scheppke L., Huang M., Shields D.J., Lindquist J.N., Lapinski P.E., King P.D., Weis S.M., Cheresh D.A. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat. Med. 2010;16(8):909–914. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yin R., Guo L., Gu J., Li C., Zhang W. Over expressing miR-19b-1 suppress breast cancer growth by inhibiting tumor microenvironment induced angiogenesis. Int. J. Biochem. Cell Biol. 2018;97:43–51. doi: 10.1016/j.biocel.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 93.Zhang Q., Liu S., Zhang J., Ma X., Dong M., Sun B., Xin Y. Roles and regulatory mechanisms of miR-30b in cancer, cardiovascular disease, and metabolic disorders (Review) Exp. Ther. Med. 2021;21(1):44. doi: 10.3892/etm.2020.9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fan Y., Ding S., Sun Y., Zhao B., Pan Y., Wan J. MiR-377 regulates inflammation and angiogenesis in rats after cerebral ischemic injury. J. Cell. Biochem. 2018;119(1):327–337. doi: 10.1002/jcb.26181. [DOI] [PubMed] [Google Scholar]
- 95.Qi L., Zhang Y. The microRNA 132 regulates fluid shear stress-induced differentiation in periodontal ligament cells through mTOR signaling pathway. Cell. Physiol. Biochem. 2014;33(2):433–445. doi: 10.1159/000358624. [DOI] [PubMed] [Google Scholar]
- 96.Song J.L., Nigam P., Tektas S.S., Selva E. microRNA regulation of Wnt signaling pathways in development and disease. Cell. Signal. 2015;27(7):1380–1391. doi: 10.1016/j.cellsig.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nie X., Liu Y., Chen W.D., Wang Y.D. Interplay of miRNAs and canonical Wnt signaling pathway in hepatocellular carcinoma. Front. Pharmacol. 2018;9:657. doi: 10.3389/fphar.2018.00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang T., Ni S., Luo Z., Lang Y., Hu J., Lu H. The protective effect of microRNA-21 in neurons after spinal cord injury. Spinal Cord. 2019;57(2):141–149. doi: 10.1038/s41393-018-0180-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yao X., Wang Y., Zhang D. microRNA-21 confers neuroprotection against cerebral ischemia-reperfusion injury and alleviates blood-brain barrier disruption in rats via the MAPK signaling pathway. J. Mol. Neurosci. 2018;65(1):43–53. doi: 10.1007/s12031-018-1067-5. [DOI] [PubMed] [Google Scholar]
- 100.Li S., Liang Z., Xu L., Zou F. MicroRNA-21: a ubiquitously expressed pro-survival factor in cancer and other diseases. Mol. Cell. Biochem. 2012;360(1–2):147–158. doi: 10.1007/s11010-011-1052-6. [DOI] [PubMed] [Google Scholar]
- 101.Jiang Y., Zhao S., Ding Y., Nong L., Li H., Gao G., Zhou D., Xu N. MicroRNA21 promotes neurite outgrowth by regulating PDCD4 in a rat model of spinal cord injury. Mol. Med. Rep. 2017;16(3):2522–2528. doi: 10.3892/mmr.2017.6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xu B., Zhang Y., Du X.F., Li J., Zi H.X., Bu J.W., Yan Y., Han H., Du J.L. Neurons secrete miR-132-containing exosomes to regulate brain vascular integrity. Cell Res. 2017;27(7):882–897. doi: 10.1038/cr.2017.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Villasante A., Marturano-Kruik A., Ambati S.R., Liu Z., Godier-Furnemont A., Parsa H., Lee B.W., Moore M.A., Vunjak-Novakovic G. Recapitulating the size and cargo of tumor exosomes in a tissue-engineered model. Theranostics. 2016;6(8):1119–1130. doi: 10.7150/thno.13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rocha S., Carvalho J., Oliveira P., Voglstaetter M., Schvartz D., Thomsen A.R., Walter N., Khanduri R., Sanchez J.C., Keller A., Oliveira C., Nazarenko I. 3D cellular architecture affects MicroRNA and protein cargo of extracellular vesicles. Adv. Sci. 2019;6(4) doi: 10.1002/advs.201800948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cone A.S., Yuan X., Sun L., Duke L.C., Vreones M.P., Carrier A.N., Kenyon S.M., Carver S.R., Benthem S.D., Stimmell A.C., Moseley S.C., Hike D., Grant S.C., Wilber A.A., Olcese J.M., Meckes D.G.J. Mesenchymal stem cell-derived extracellular vesicles ameliorate Alzheimer's disease-like phenotypes in a preclinical mouse model. Theranostics. 2021;11(17):8129–8142. doi: 10.7150/thno.62069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cherian D.S., Bhuvan T., Meagher L., Heng T.S.P. Biological considerations in scaling up therapeutic cell manufacturing. Front. Pharmacol. 2020;11(654) doi: 10.3389/fphar.2020.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shen G., Li X., Jia Y.-f., Piazza G.A., Xi Y. Hypoxia-regulated microRNAs in human cancer. Acta Pharmacol. Sin. 2013;34(3):336–341. doi: 10.1038/aps.2012.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lembong J., Kirian R., Takacs J.D., Olsen T.R., Lock L.T., Rowley J.A., Ahsan T. Bioreactor parameters for microcarrier-based human MSC expansion under xeno-free conditions in a vertical-wheel system. Bioengineering. 2020;7(3):73. doi: 10.3390/bioengineering7030073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding authors on reasonable request.