Abstract
A growing body of evidence shows that circular RNAs (circRNAs) participate in tumor growth and metastasis and also play crucial roles in the treatment and prognosis of various cancers. In this article, we identified a novel circRNA, circSOBP (has_circ_0001633), based on the results of high-throughput RNA sequencing, and its expression was subsequently validated via quantitative reverse transcription polymerase chain reaction in bladder cancer (BCa) tissues and cell lines. The association between circSOBP expression and the clinicopathologic features and prognosis of 56 recruited BCa patients was then analyzed, and the biological roles of circSOBP were assessed by in vitro cloning formation, wound healing, transwell, CCK-8, and in vivo xenograft mouse models. Next, the competitive endogenous RNA mechanism was explored through fluorescence in situ hybridization, RNA pull-down, luciferase reporter, bioinformatics analysis, and rescue experiments. Western blot and immunohistochemistry detected the expression of downstream mRNA, and we were able to determine that circSOBP was downregulated in BCa tissues and cell lines and that lower circSOBP expression was associated with more advanced pathological stage, larger tumor size, and poorer overall survival with BCa patients. Overexpressed circSOBP suppressed cell proliferation, migration, and invasion both in vitro and in vivo. Mechanistically, competitive interactions between circSOBP and miR-200a-3p enhanced target gene PTEN expression. In addition, we found a significant correlation between higher expression of circSOBP in BCa patients after immunotherapy than before and a better treatment outcome, indicating that circSOBP might regulate the programmed death 1/programmed death ligand 1 pathway. Overall, circSOBP inhibits BCa tumorigenesis and metastasis by a novel miR-200a-3p/PTEN axis, which makes it an excellent biomarker and therapeutic target for treating BCa.
Keywords: circSOBP, miR-200a-3p, PTEN, proliferation, bladder cancer
Introduction
Bladder cancer (BCa) is the 10th most commonly diagnosed cancer worldwide, with approximately 573,000 new cases and 213,000 deaths in 2021 1 . The primary standard of treatment for BCa includes surgery, radiation, chemotherapy, immunotherapy, and targeted therapy, but tumoral heterogeneity often contributes to drug resistance, relapse, and metastasis after therapy, resulting in poor survival outcomes 2 . Currently, there are no effective biomarkers for BCa for diagnosis, therapy, or prognosis. Therefore, in this article we set out to investigate the molecular mechanism that drives BCa development and progression in the hopes of potentially identifying promising new therapeutic targets.
Circular RNAs (circRNAs) are a kind of noncoding RNAs that are produced by connecting the terminal 5′ and 3′ ends of RNAs through alternative back-splicing 3 . circRNAs are widespread in eukaryotic cells and are usually highly conserved and stably expressed across species 4 . Currently, with the development of next-generation throughput sequencing techniques, mounting evidence shows that abnormal circRNA expression is closely associated with the pathogenesis of various malignant tumors and conduces essential effects in the regulation of gene expression, cell proliferation, migration, invasion, apoptosis, and cell cycle progression5,6. circRNAs have several biological properties, but their “miRNA sponge” property is the main regulatory mechanism due to the presence of microRNA (miRNA)-binding sites. circRNAs can also operate as competitive endogenous RNAs (ceRNAs) to bind miRNA for moderation of the expression of target genes (circRNA/miRNA/mRNA axis) 7 . In addition, circRNAs have recently been found to regulate the programmed death 1 (PD-1)/programmed death ligand 1 (PD-L1) pathway, meaning that they thus also participate in the body’s immune response and in immunotherapy8–10. Taken together, circRNAs have many properties of superior molecular biomarkers and therapeutic targets.
We identified 87 remarkably differentially expressed circRNAs using RNA sequencing from five pairs of BCa patients and their adjacent tissues in our research group 11 , including the top 40 upregulated and 47 downregulated circRNAs. Among them, circSOBP was the most downregulated in BCa tissues and cell lines and exerted the greatest tumor-suppressive function on BCa proliferation and metastasis by sponging miR-200a-3p to regulate the expression PTEN protein. Clinically, we found that low-expressed circSOBP was positively correlated with earlier pathological stage, smaller tumor size, and better prognosis. Moreover, we also discovered a positive correlation between changes in circSOBP expression in BCa patients before/after immunotherapy and outcome of treatment, indicating that circSOBP might regulate the PD-1/PD-L1 pathway, meaning that it participates in the body’s immune response and in immunotherapy. Our findings shed new light on the role of circSOBP in BCa development.
Materials and Methods
Clinical Tissue Specimens
The 61 pairs of BCa tissues and matched adjacent normal urothelial (MANU) tissues including five pairs of samples for RNA sequencing (RNA-Seq) that were used in this study were obtained at the Second Hospital of Tianjin Medical University from October 2020 to October 2021. All patients underwent robot-assisted or laparoscopic radical resection for BCa, and they did not receive any preoperative chemotherapy, radiotherapy, or immunotherapy. In addition, 23 BCa patients performed immunotherapy combined with albumin-paclitaxel, and tumor samples were obtained to conduct transcriptome sequencing before and after treatment. These fresh tissues were immediately frozen in liquid nitrogen after surgical removal and stored at −80°C. This study was approved and authorized by the Ethics Committee of the Second Hospital of Tianjin Medical University, and each patient signed written informed consent forms. In addition, RNA-seq data of 433 BCa tissues, miRNA-seq data of 437 BCa tissues, data on 19 adjacent normal tissues, and data on 9 GTEx for BCa were also downloaded from The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov) database.
Cell Cultures
Human cell lines T24, EJ, 253J, and 5637 and the immortalized normal uroepithelium cell line SV-HUC-1 came from our laboratory (Key Laboratory of Urology, Second Hospital of Tianjin Medical University). Among them, T24 was cultivated in McCoy’s 5A medium (BIOIND, Kibbutz Beit Haemek, Israel), EJ was maintained in F12K medium (BOSTER, Wuhan, China), and 253J and 5637 were fostered in RPMI 1640 medium (BIOIND). All media were complemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin, and all cell lines were cultured in a humidified incubator at 37°C with 5% CO2.
Nucleic Acid Purification and qRT-PCR
Total RNA and genomic DNA were extracted from the tissues and cells using an HP Total RNA Kit (Omega Bio-Tek, Norcross, GA, USA) and Tissue DNA Kit (Omega Bio-Tek), respectively. miRNA was extracted using an miRNA isolation Kit (R6842-01; Omega Bio-Tek), and total RNA and miRNA were reverse-transcribed into complementary DNA (cDNA) with a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher, Waltham, MA, USA) and an mir-XTM miRNA First-Strand Synthesis Kit (Takara, Tokyo, Japan), respectively. Real-time quantitative polymerase chain reaction (qRT-PCR) and the 2−ΔΔCT method were used to analyze the relative gene expression data. qRT-PCR was performed on a Quantagene q225 system (Kubo Tech, Beijing, China) using TOROGreen qPCR Master Mix (Toroivd, shanghai, China), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as internal reference to measure the expression level of circRNA and mRNA. U6 snRNA was used for miRNA. The forward primers of all miRNAs were designed by adding the Poly (A) tail method and the universal reverse primer supported by Sangon Biotech (Shanghai, China). All other related primers are shown in Supplemental Table S1.
Cell Transfection
The lentiviruses of overexpressed and negative control circSOBP were constructed by Hanbio Biotechnology (Shanghai, China). Briefly, T24 and 253J cell lines were seeded into six-well plates the day before transfection at 60% to 70% confluency. Cell transfections were then implemented according to the manufacturer’s instructions, and we placed them into their respective normal media after 8 h of infection. Stable expression cells were then selected in a medium containing puromycin 4 μg/ml (10mg/ml; Biosharp, Hefei, China). Finally, the transfection efficiency was validated through cell fluorescence and qRT-PCR.
CCK-8 Assay and Cloning Formation Assay
The transfected T24 and 253J cells were plated in 96-well plates at a density of 1,500 cells per well after cell counting. At 0-, 24-, 48-, 72-, and 96-h time points after seeding, the cells were treated with 10 µl of Cell Counting Kit-8 solution (CCK-8; BOSTER) + 90 µl of serum-free medium and incubated at 37°C for 2 h. Cell proliferation capability was then evaluated through the detection of light absorbance at 450 nm of each well. For cloning formation assay, 400 post-transfection cells were seeded into six-well plates and added into 1640/McCoy’s 5A medium containing 10% FBS. After 7 days, the old medium was replaced with new, and after 2 weeks, cell fixation was performed in 4% paraformaldehyde (PFA) for 30 min, and the samples were stained with 0.1% crystal violet for 10 min prior to imaging and quantification.
Wound-Healing Assay
Different groups of transfected cells were planted in six-well plates and grew until 100%. Then, a straight wound in the cell layer was drawn in three different positions using sterile 200 µl pipette tips that had been washed gently two to three times with phosphate-buffered saline (PBS), and the cells were then cultured in serum-free medium. The cell migration images were photographed at 0 and 12 h after scratching using an optical microscope.
Transwell Assay
Transwell chambers of 8-µm pore size polycarbonate membrane (SORFA, Beijing, China) and Matrigel (BD BioCoat, CA, USA) were used for various assays. For migration assay, 1.5 × 104 cells were suspended in 200 µl of serum-free medium and seeded into upper chambers. However, for invasion assay, the upper chambers were coated with 50 µl of Matrigel. A total of 800 µl medium containing 20% FBS was placed into the lower chambers. Following an incubation period (24 h), the cells that had migrated onto the lower membrane surface were fixed with PFA and stained with crystal violet solution. We then randomly selected three fields to photograph and counted the number of cells using Image J.
RNA Fluorescence In Situ Hybridization
RNA fluorescence in situ hybridization (RNA FISH) assay was applied to identify the subcellular location of circSOBP and miR-200a-3p in BCa cells and BCa tissues. A FISH Kit was purchased from GenePharma (Shanghai, China). Following the manufacturer’s instructions, cell climbing piece and tissue paraffin sections were hybridized with specific Cy3-labeled circSOBP probes and a Cy5-labeled miR-200a-3p probe (GenePharma) at 37°C away from light overnight (12–16 h). The cell nuclei were then stained with 4′6-diamidino-2-phenylindole (DAPI), and all images were captured by a fluorescence inverted microscope (Olympus, Tokyo, Japan). The probe sequences are presented in Supplemental Table S2.
Actinomycin D Assay
Cells (50 ×104) were seeded into six-well plates for 24 h and treated with actinomycin D (2 µg/ml) to disturb the transcriptional process. Total RNA was then extracted at five time points (0, 4, 8, 12, and 24 h) for qRT-PCR analysis.
Dual-Luciferase Reporter Assay
We seeded HEK-293T cells (10 × 104/well) in 24-well plates and grew them until 60%–70%. Then, the cells were cotransfected with wide/mute-type circSOBP or PTEN 3′-UTR (untranslated region) report plasmid that had been synthesized and cloned into pSI-Check2 vector and miRNA mimics (Hanbio). After 48 h of transfection, firefly and Renilla luciferase activity were measured using the Dual-Luciferase Reporter Assay System (E1910; Promega, Madison, WI, USA) and GloMax 20/20 luminometer (Promega). miRNA sequences are listed in Supplemental Table S3.
RNA Pull-Down Assay
For RNA pull-down assay, cells were lysed with RIPA lysis buffer (BL504A; Biosharp) and then heat-denatured. Next, biotinylated circSOBP probe and Oligo probe (GenePharma) were added to the cell lysates and hybridized overnight at 4°C. Streptavidin magnetic beads were then added as well and incubated at room temperature for 2 h to generate probe-coated bead pull-down for enrichment. The magnetic beads were then washed, and total RNA and miRNAs were extracted and analyzed by qRT-PCR (Supplemental Table S4).
Western Blot Analysis
Proteins were extracted for Western blot analysis using RIPA lysis buffer and were quantified using a bicinchoninic acid (BCA) kit (Thermo Fisher). To accomplish this, equal amounts of proteins were electrophoresed by 10% SDS-PAGE (sodium dodecyl sulphate-polyacrylamide gel electrophoresis) and transferred onto polyvinylidene difluoride membranes. After blocking for 1 h with 5% skim milk powder at room temperature, the membranes were incubated overnight at 4°C with primary antibodies specific for PTEN (1:1,000; BOSTER) and GAPDH (1:5,000; BOSTER). The membranes were then incubated with horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h, and the bands were visualized using ImmobilonTM Western Chemiluminescent HRP Substrate (Millipore, Billerica, MA, USA).
Tumor Xenografts and Metastasis Models
Four-week-old balb/c male nude mice were purchased from SPF Biotechnology (Beijing, China), and all animal experiments were approved by the animal ethics committee of the Second Hospital of Tianjin Medical University. For subcutaneous tumor formation assays, 10 mice were randomly divided into two groups (n = 5 for each group). T24 cell suspension that had been stably transfected with circSOBP and control lentivirus was prepared at a density of 200 × 104/100 µl, mixed with 100 µl of Matrigel, and subsequently injected into the region of the axilla. Tumors were measured on alternate days by caliper, and ultimate tumor volume was recorded (volume = (width) 2 × length/2). The mice were euthanized 4 weeks later and the tumors were weighed after harvesting for hematoxylin and eosin (HE) staining and immunohistochemistry (IHC) analysis. For metastasis assays, T24-luc cells stably transfected with circSOBP and control lentivirus were prepared at a density of 1 × 105/100 µl, and 100-µl cells suspensions were intravenously injected into the tail veins of the nude mice (n = 5 for each group). We then conducted bioluminescence imaging after 6 weeks. Finally, the lungs of these nude mice were excised postmortem for HE staining.
Statistical Analysis
All measurements were expressed as mean ± standard deviation (SD) from at least three independent experiments. GraphPad prism 9.0 software and R version 3.6.3 R were used to conduct all statistical analyses. Student t tests and χ2 tests (depending on the data type) were used to calculate the statistical differences between groups. Survival analysis was performed using Kaplan-Meier (KM) survival curve analysis with a log-rank test. Differences were said to be statistically significant as indicated by the following: *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
circSOBP Is Downregulated in BCa and Closely Associated With Clinicopathological Features
To select BCa-related circRNAs, five pairs of fresh frozen BCa tissues and adjacent normal tissues underwent RNA-seq of the entire transcriptome by our team 11 . Differently expressed circRNAs were identified under the criteria of|Log2FC|≥2 and P value <0.05, and are shown in a heatmap (Fig. 1A, Supplemental Table S5). Among these downregulated circRNAs, circSOBP showed obviously lower expression (logFC = −4.114966, P = 0.023934) in BCa tissues compared with paracarcinoma tissues, but the detailed biological function of circSOBP has not been studied and reported in BCa previously. Thus, circSOBP was chosen for further examination.
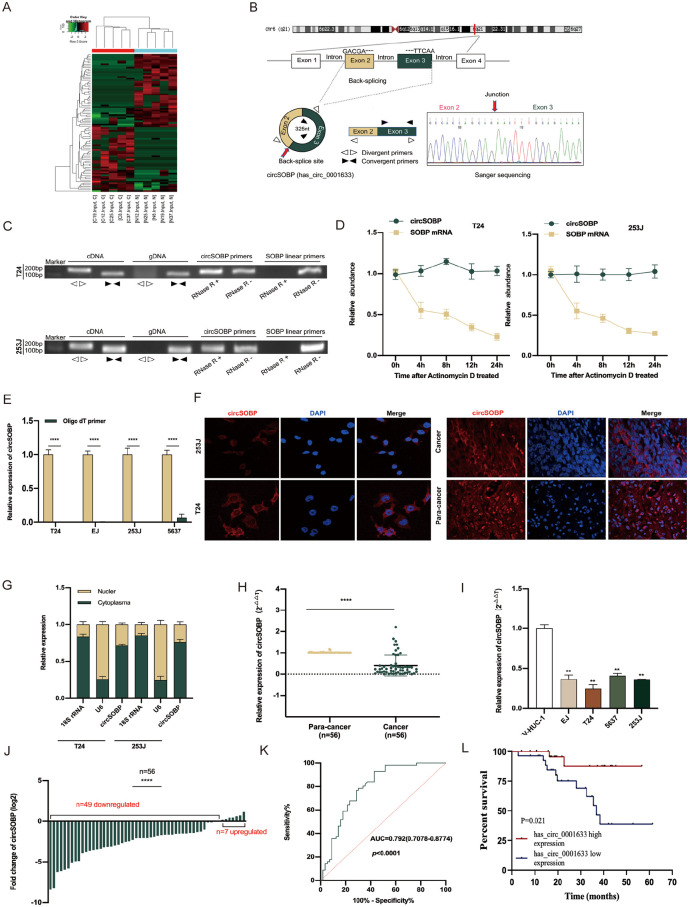
Figure 1.
The validation and characteristics of circSOBP in BCa tissues and cells. (A) The heatmap plots demonstrated the dysregulated circRNAs in five pairs of BCa tissues and corresponding adjacent normal tissues. Red represents upregulated and green downregulated. (B) The SOBP gene and circSOBP genomic regions. Sanger sequencing demonstrated head-to-hail splicing. (C) The PCR products of circSOBP and linear SOBP were tested by gel electrophoresis. CircSOBP could only be amplified from cDNA, not gDNA using divergent primers. The expression of circSOBP and SOBP mRNA was measured by qRT-PCR with or without RNase R. (D) qRT-PCR analysis of circSOBP and SOBP mRNA after treatment with actinomycin D at the indicated time points. (E) qRT-PCR analysis of circSOBP in the reverse transcription products using random primers or oligo dT primers. (F) FISH confirmed that circSOBP was predominantly located in cytoplasm both in BC cells and in tissues (red). Nuclei were stained with DAPI (blue). (G) Nuclear and cytoplasmic fraction assay further confirmed the same result. (H) qRT-PCR analysis of circSOBP in 56 paired BCa and adjacent noncancerous tissue. (I) qRT-PCR analysis of circSOBP in BC cell lines and normal urothelial epithelium. (J) The percentage of upregulated or downregulated expression of circSOBP in 56 BCa patients. (K) The ROC curve for distinguishing BCa and ANP tissues based on circSOBP expression. (L) Patients with BCa were divided into two groups according to the median level of circSOBP expression. Overall survival was assessed by the Kaplan-Meier method with the log-rank test. BCa: blood cancer; PCR: polymerase chain reaction; FISH: fluorescence in situ hybridization; DAPI: 4′6-diamidino-2-phenylindole; ROC: receiver operating characteristic; AUC: area under the curve; qRT-PCR: quantitative reverse transcription polymerase chain reaction; cDNA: complementary DNA; gDNA: genomic DNA; circRNA: circular RNA; ANP: adjacent normal paracancerous.
According to the circBank database (http://www.circbank.cn/) annotation, circSOBP (circbase ID: hsa_circ_0001633) stems from exon 2 and exon 3 of its host gene (SOBP), which is located on human 6q21, chr6:107824860-107827631, and its circular transcript length is 325 nt. The total sequence and qPCR product of circSOBP in agarose gel electrophoresis are presented in Supplemental Figure S1. circSOBP’s head-to-tail splicing was confirmed by Sanger sequencing (Fig. 1B), and we also designed divergent and convergent primers to demonstrate the head-to-tail splicing of circSOBP rather than trans-splicing or genomic rearrangements.
Agarose gel electrophoresis results showed that circSOBP was only detected in cDNA but not in genomic DNA (gDNA). In addition, the circular isoform of circSOBP was resistant to RNase R (a processive 3′ to 5′ exoribonuclease). Nevertheless, the linear transcript was easily digested (Fig. 1C). To check the stability of circSOBP, T24 and 253J cells were treated by actinomycin D (an inhibitor of RNA synthesis), and qRT-PCR showed that the expression of circSOBP hardly changed but that SOBP linear RNA had obviously degraded within 48 h (Fig. 1D). Due to circRNAs’ lack of 3′ polyadenylated tail structure, total RNA was reversed into cDNA using oligo dT primers and random primers, and the targeted fragment of circSOBP was amplified by qRT-PCR (Fig. 1E). The results indicated that circSOBP is a highly stable circRNA. Furthermore, the subcellular location of circSOBP was analyzed with RNA FISH and nucleus-cytoplasmic fractionation assays, and these results illustrated the prominent cytoplasmic localization of circSOBP in BCa cells. In addition, the expression of circSOBP in carcinoma tissues was lower than in adjacent normal tissues (Fig. 1F, G).
Next, circSOBP expression was detected in tissue specimens and BCa cell lines, where qRT-PCR revealed that circSOBP expression was significantly lower in BCa tissues (Fig. 1H) and BC cells (Fig. 1I) than in paracarcinoma tissues and immortalized normal uroepithelium cells, which was consistent with the RNA-seq results. Among 56 paired specimens, 7.14% (4/56) were found not to be statistically different (P > 0.05), and 12.5% (7/56) showed a contrary tendency (Fig. 1J). In addition, the receiver operating characteristic (ROC) curve was also constructed to evaluate the different circSOBP expression between BCa and normal tissues. The area under the curve (AUC) was 0.792 (sensitivity = 0.929, 1 − specificity = 0.429) (Fig. 1K), indicating that circSOBP may be an excellent diagnostic biomarker.
We also collected the detailed clinicopathologic parameters of the BCa patients and classified them into relatively low- and high-circSOBP expression groups based on the median expression of circSOBP. As shown in Table 1, elevated circSOBP expression was positively associated with tumor size and pathology stage. Further, Kaplan-Meier survival curves suggested that higher circSOBP expression predicted longer patients’ overall survival (Fig. 1L). Briefly, the above results imply that circSOBP is a tumor suppressor in BCa and that higher expression of circSOBP predicts a better prognosis.
Table 1.
Correlations Between circSOBP Expression and Clinicopathological Characteristics in BCa.
| Parameter | Case | circSOBP expression | P value | |
|---|---|---|---|---|
| Low (28) | High (28) | |||
| Gender | 1.000 | |||
| Female | 7 | 3 (5.4%) | 4 (7.1%) | |
| Male | 49 | 25 (44.6%) | 24 (42.9%) | |
| Age | 0.469 | |||
| <60 | 9 | 3 (5.4%) | 6 (10.7%) | |
| ≥60 | 47 | 25 (44.6%) | 22 (39.3%) | |
| Tumor size | 0.015* | |||
| <3cm | 24 | 7 (12.5%) | 17 (30.4%) | |
| ≥3cm | 32 | 21 (37.5%) | 11 (19.6%) | |
| Pathology stage | 0.007* | |||
| pTa-pT1 | 31 | 10 (17.9%) | 21 (37.5%) | |
| pT2-pT4 | 25 | 18 (32.1%) | 7(12.5%) | |
| Grade, n (%) | 0.491 | |||
| Low | 2 | 2 (3.6%) | 0 (0%) | |
| high | 54 | 26 (46.4%) | 28 (50%) | |
| Lymphatic metastasis status | 0.329 | |||
| N0 | 44 | 24 (42.9%) | 20 (35.7%) | |
| N1 | 12 | 4 (7.1%) | 8 (14.3%) | |
| Distant metastasis status | 1.000 | |||
| M0 | 55 | 28 (50%) | 27 (48.2%) | |
| M1 | 1 | 0 (0%) | 1 (1.8%) | |
BCa: bladder cancer.
P < 0.05.
circSOBP Inhibits the Proliferation, Migration, and Invasion of BCa In Vitro
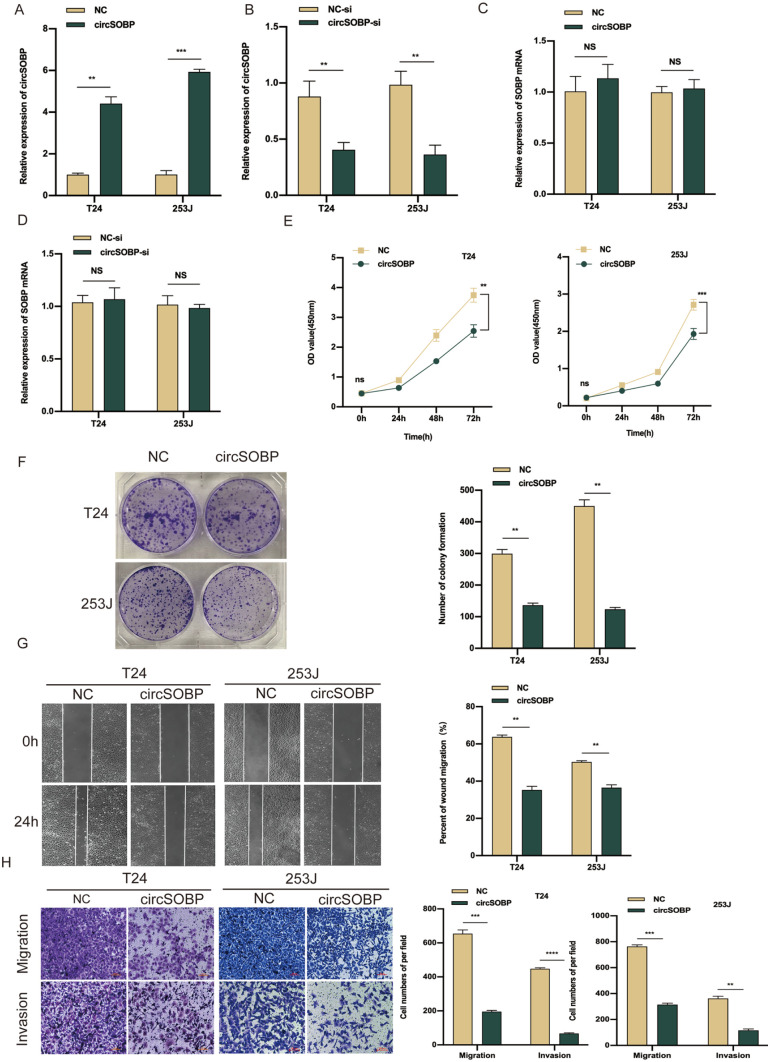
Gain-of-function and loss-of function methods were used to explore the possible biological function of circSOBP in BCa, and to do this, circSOBP overexpression lentivirus and circSOBP-si vectors were constructed and transfected in T24 and 253J cells, respectively. The overexpression and knockdown efficiency of circSOBP were then detected via qRT-PCR, where we found that the expression of circSOBP was remarkably increased or decreased, respectively (Fig. 2A, B). We also observed that SOBP mRNA was downregulated in our RNA-seq data (Fig. S2A), whereas SOBP mRNA expression was unaltered with a change in circSOBP (Fig. 2C, D), which excludes any potential effect of SOBP mRNA on the biological functions of BCa. The CCK-8 and colony formation assays showed that the overexpression of circSOBP remarkably suppressed the proliferation capability of BCa cells as well (Fig. 2E, F). Accordingly, wound-healing and transwell assays showed that the ability of migration invasion of BCa cells was significantly inhibited after circSOBP overexpression (Fig. 2G, H), and circSOBP knockdown restrained the above phenotypes (Fig. S2B–E).
Figure 2.
Overexpression of circSOBP suppresses BCa cell proliferation, migration, and invasion in vitro. (A, B) The verification of circSOBP overexpression and knockdown efficiency via qRT-PCR. (C, D) qRT-PCR analysis of SOBP mRNA in BCa cells after circSOBP was overexpression or knockdown. (E, F) Effect of circSOBP overexpression on the proliferative capacity of BCa cells by CCK-8 and colony formation assays. (G, H) Effect of circSOBP overexpression on migration and invasion abilities of BCa cells by wound-healing and transwell assays. BCa: bladder cancer; qRT-PCR: real-time quantitative polymerase chain reaction; PCR: polymerase chain reaction; CCK-8: Cell Counting Kit-8; NC: negative control.
circSOBP Functions as an miRNA Sponge for miR-200a-3p
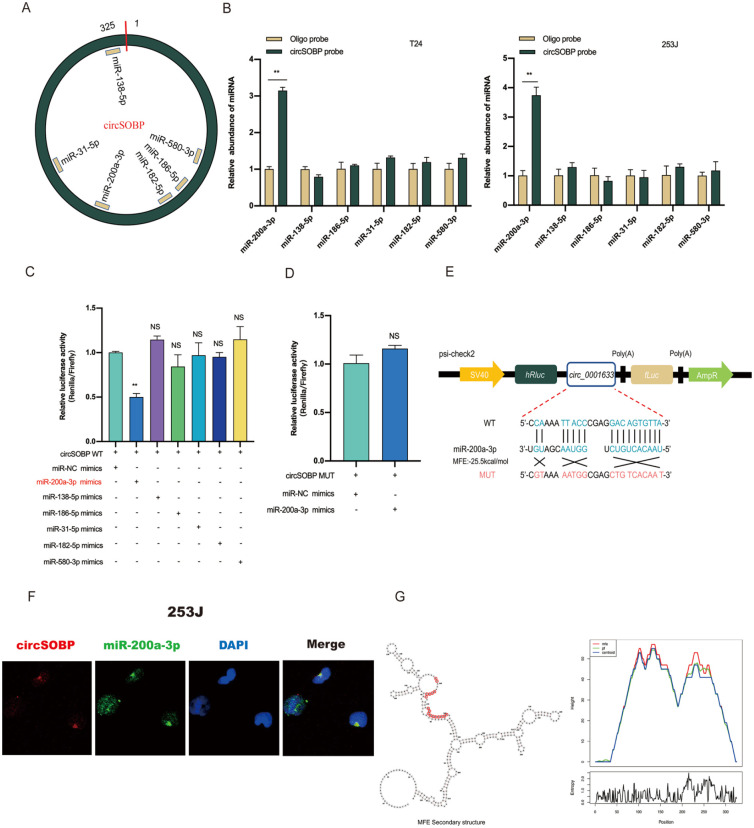
One of the main functions of circRNAs is to sponge miRNAs. Due to the cytoplasmic localization of circSOBP, it occupies the same space with miRNAs. We thus speculate that circSOBP may perform miRNA sponge functions. To test this, circRNA-miRNA interactions were predicted by bioinformatics analysis on circBank (http://www.circbank.cn/index.html), CircInteractome (https://circinteractome.nia.nih.gov), and RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/) databases, and six candidate miRNAs (miR-31-5p, miR-138-5p, miR-186-5p, miR-200a-3p, miR-580-3p, and miR-182-5p) were filtered (Fig. 3A). To validate our findings, we performed a pull-down assay using a biotinylated probe specific to circSOBP. Among the six candidate miRNAs, only miR-200a-3p could be abundantly pulled down by circSOBP probe in both T24 and 253J cells (Fig. 3B). Furthermore, dual-luciferase reporter assays were used to test whether these miRNAs directly target circSOBP. We cotransfected circSOBP wild-type (WT) plasmid and miRNA mimics in HEK-293T cells and found that miR-200a-3p mimics decreased the luciferase activity remarkably but not with the mutant-type (MUT) circSOBP vector (Fig. 3C, D). The binding site between circSOBP and miR-200a-3p is shown in Fig 3E.
Figure 3.
circSOBP serves as an miR-200a-3p sponge in BCa cells. (A) Six potential target miRNAs of circSOBP were predicted by circBank, circinteractome, and RNAhybrid. (B) Relative expression levels of six miRNAs in T24 and 253J lysates pulled down by biotinylated circSOBP probe or oligo probe. (C, D) Dual-luciferase reporter assay in HEK-293T cell was used to analyze the direct interaction between circSOBP and six predicted miRNAs. (E) Sequence alignment of miR-200a-3p with circSOBP wild-type and mutated plasmid. (F) The colocalization between circSOBP and miR-200a-3p was shown by FISH in 253J cells. Nuclei were stained with DAPI. (G) The secondary structure of circSOBP was predicted by ViennaRNA Web Services, and the red region indicates binding sites with miR-200a-3p. Minimum free energy (MFE) = −25.5. BCa: bladder cancer; miRNA: microRNA; FISH: fluorescence in situ hybridization; DAPI: 4′6-diamidino-2-phenylindole; MUT: mutant-type; WT: wild-type.
The RNA FISH assay indicated the presence the colocalization of circSOBP and miR-200a-3p in 253J cells (Fig. 3F). Here, an interesting phenomenon was observed: we predicted the secondary structure of circSOBP on ViennaRNA Web Services (http://rna.tbi.univie.ac.at) and found that the predictive binding position of miR-200a-3p partly located in the loop region of circSOBP demonstrated the minimum free energy (MFE) compared with other miRNAs and indicated lower base-pair probabilities of circSOBP itself but higher base-pair probabilities with miRNAs (Fig. 3G). This is consistent with our experimental results.
The Antitumor Effects of circSOBP Depends on miR-200a-3p
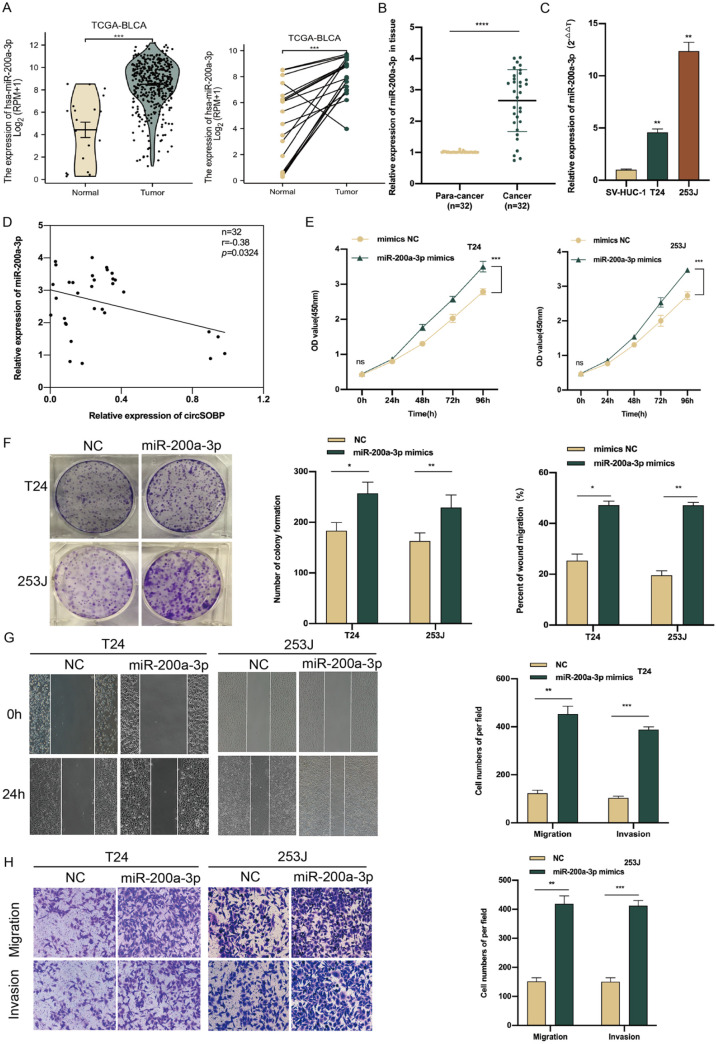
Given the interaction between circSOBP and miR-200a-3p, we next assessed the potential biological function of miR-200a-3p in BCa. First, we analyzed the expression of miR-200a-3p on the TCGA-BLCA RNA-seq database and found that miR-200a-3p was significantly upregulated in BCa tissues compared with normal tissues whether in paired or unpaired samples (Fig. 4A). Then, we detected the expression level of miR-200a-3p by qRT-PCR in 32 pairs of the same population of BCa patients and found that the expression of miR-200a-3p was also upregulated (Fig. 4B). The same result for differential expression was further validated in BCa cell lines via qRT-PCR (Fig. 4C). Furthermore, Pearson correlation analysis on the expression of circSOBP and miR-200a-3p in the same BCa cohort showed that they were negatively correlated (r = −0.38, P = 0.0324) (Fig. 4D), and increased expression of miR-200a-3p substantially accelerated T24 and 253J cell proliferation, migration, and invasion compared with mimics in the negative control (NC) group (Fig. 4E–H).
Figure 4.
miR-200a-3p promotes BCa cell proliferation, migration, and invasion in vitro. (A) The expression of miR-200a-3p in BCa and normal tissues in TCGA database. (B) The expression of miR-200a-3p in the 32 pairs of the same BCa cohort. (C) The relative expression of miR-200a-3p in BCa cell lines. (D) Pearson correlation analysis showed the connection between circSOBP and miR-200a-3p expression in 32 pairs of the same population of BCa patients. (E, F) Effect of miR-200a-3p mimics on proliferative capacity of BCa cells by CCK-8 and colony formation assays. (G, H) Effect of miR-200a-3p mimics on migration and invasion abilities of BCa cells by wound healing and transwell assays. BCa: bladder cancer; TCGA: The Cancer Genome Atlas; CCK-8: Cell Counting Kit-8; BLCA: bladder urothelial carcinoma; NC: negative control; SV-HUC-1: human ureteral immortalized cells.
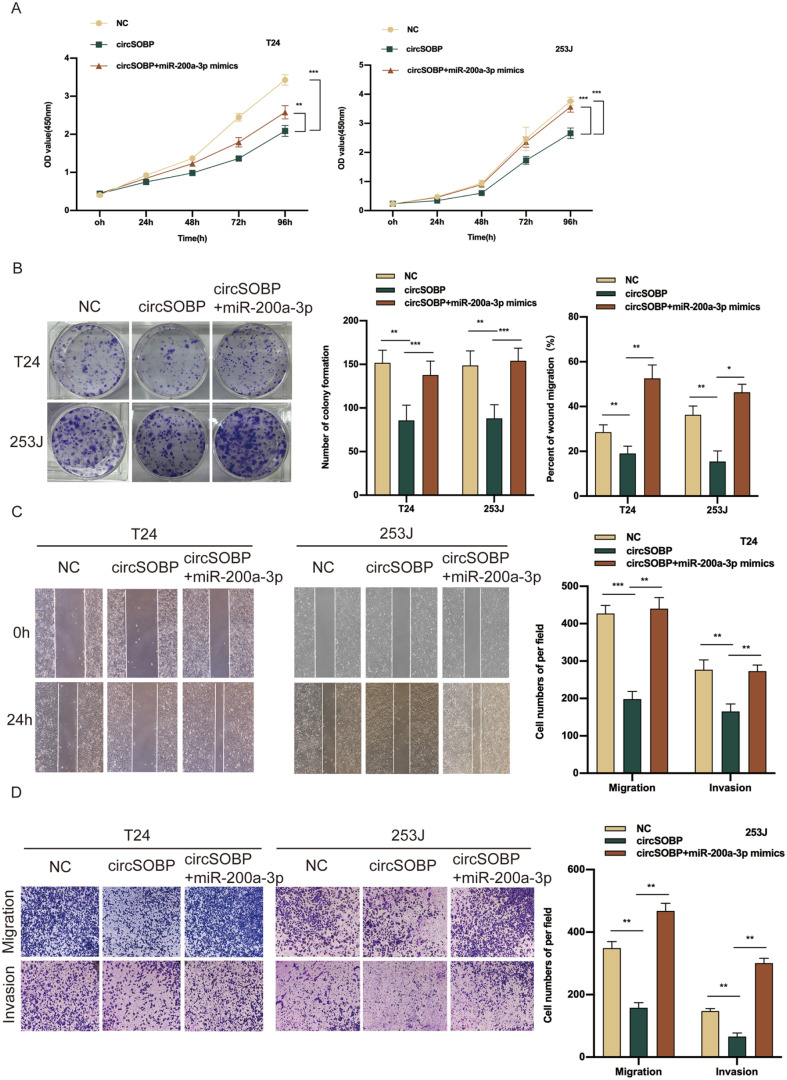
However, the above phenotypes were also drastically inhibited in T24 and 253J cells transfected with miR-200a-3p inhibitors (Supplemental Fig. S3A–D). These findings reveal that miR-200a-3p is highly expressed in BCa and plays an oncogenic role in BCa progression. To evaluate whether circSOBP restrains the malignant phenotype of BCa cells by targeting miR-200a-3p, rescue experiments were implemented by cotransfecting circSOBP and miR-200a-3p mimics into T24 and 253J cells. After this, only the inhibitory abilities of proliferation, migration, and invasion with the BCa cells transfected with circSOBP alone were markedly reversed by the cotransfected system with circSOBP and miR-200a-3p mimics (Fig. 5A–D). Jointly, these results demonstrate that circSOBP suppresses the oncogenic phenotype of BCa cells, at least to some degree, by sponging miR-200a-3p.
Figure 5.
miR-200a-3p reverses the inhibitory effect of circSOBP on BCa cells in vitro. (A, B) CCK-8 and colony formation assays indicated that the cell proliferation ability of BCa cells transfected with circSOBP was reversed when cotransfected with miR-200a-3p mimics. (C, D) Wound-healing and transwell assays demonstrated that the cell migration and invasion abilities of BCa cells transfected with circSOBP were reversed when cotransfected with miR-200a-3p mimics. BCa: bladder cancer; CCK8: Cell Counting Kit-8; NC: negative control.
circSOBP Competitively Binds to miR-200a-3p as a ceRNA to Upregulate PTEN Protein Expression
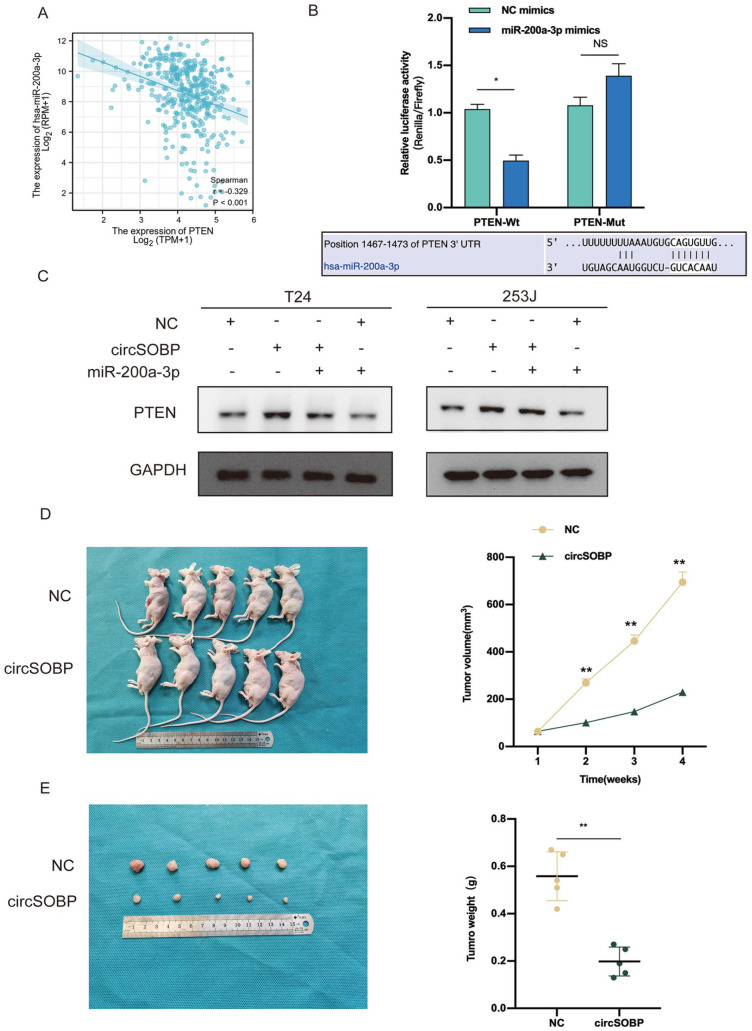
PTEN was identified as the downstream target gene of miR-200a-3p from the following three pieces of evidence: (1) four databases including miRWALK, starbase, Target gene, and miRDB co-predicted that miR-200a-3p can target the 3′-UTR of PTEN mRNA via bioinformatics analysis; (2) PTEN protein was found to be downregulated in BCa tissues in a previous study 12 ; (3) Spearman correlation analysis showed that the expression level of PTEN was negatively correlated with miR-200a-3p in BCa tissues according to TCGA data (r = −0.329, P < 0.001) (Fig. 6A). Subsequently, we explored the interaction between miR-200a-3p and PTEN mRNA 3′-UTR by dual-luciferase reporter assay. Here, the above conclusion was supported by our results that miR-200a-3p could bind to PTEN 3′-UTR (Fig. 6B). Furthermore, Western blot showed that when BCa cells were cotransfected with circSOBP lentivirus and miR-200a-3p mimics, PTEN protein expression was considerably lower than when the cells were transfected with circSOBP alone (Fig. 6C). These results thus show that circSOBP can sponge miR-200a-3p and regulate PTEN expression to inhibit BCa development.
Figure 6.
circSOBP competitively binds to miR-200a-3p as a ceRNA to upregulate PTEN protein expression. Meanwhile, circSOBP inhibits the growth of BCa tumor in vivo. (A) Spearman correlation between miR-200a- 3p level and PTEN level in TCGA-BLCA dataset. (B) Dual-luciferase reporter assay in HEK-293T cells was used to analyze the direct interaction between miR-200a-3p and PTEN. Binding site of PTEN 3′-UTR and miR- 200a-3p was predicted by Interactome. (C) Western blot suggested that miR-200a-3p could partly decrease the protein expression level of PTEN, which was promoted by circSOBP. (D) Hypodermic injection of T24 cells stably transfected with circSOBP or NC control into nude mice established subcutaneous xenograft tumors (n = 5). (E) Representation picture of tumor formation of xenograft in nude mice (n = 5). GAPDH: glyceraldehyde 3-phosphate dehydrogenase; BCa: bladder cancer; UTR: untranslated region; ceRNA: competitive endogenous RNA; BLCA: bladder urothelial carcinoma; NC: negative control.
circSOBP Inhibits the Growth and Metastasis of BCa Cells In Vivo
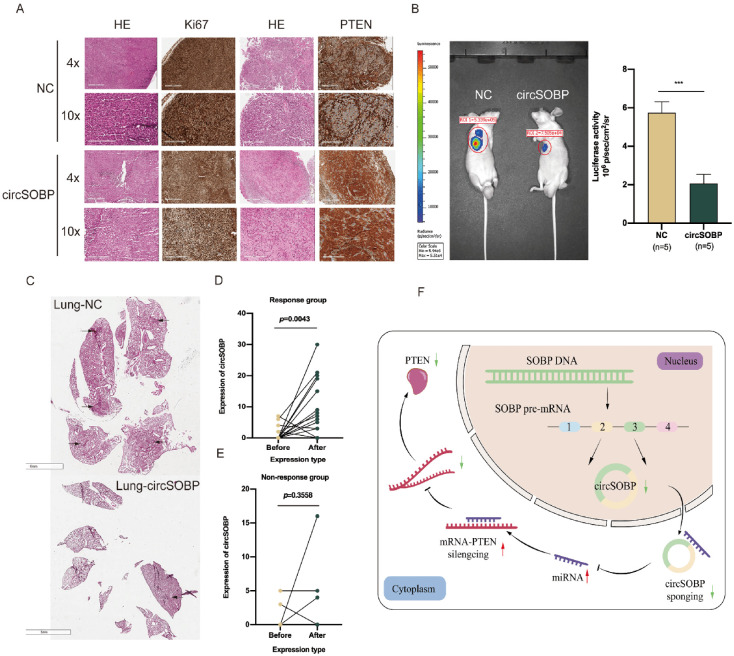
In our in vivo experiments, subcutaneous tumor formation assays revealed that the circSOBP overexpression group had an observably smaller growth rate of tumors and tumor weights than controls (Fig. 6D, E). The results of HE and IHC analysis are shown in Fig. 7A. In the overexpressed circSOBP group, the expression of Ki67 and PTEN protein was decreased and elevated, respectively. In addition, in metastasis assays, live imaging of nude mice showed that circSOBP overexpression demonstrated fewer metastatic foci in the lungs of the treated mice than in the NC group. The representative images are shown in Fig. 7B. Finally, HE staining validated the fact that the number of metastatic foci was also decreased in the lungs compared with the control groups (Fig. 7C).
Figure 7.
circSOBP suppresses the metastasis of BCa tumor in vivo. (A) HE staining and IHC analysis of Ki67 and PTEN protein expression in subcutaneous xenograft tumors. (B, C) Representative image (fluorescence and HE) of pulmonary metastatic nodules in lung metastasis models. (D, E) Expression of circSOBP between before-after immunotherapy in response or nonresponse groups with BCa patients. (F) Schematic illustration of the mechanism of circSOBP/miR-200a-3p/PTEN axis in BCa progression and metastasis. BCa: bladder cancer; HE: hematoxylin and eosin; IHC: immunohistochemistry; NC: negative control.
Changes in circSOBP Expression Positively Correlate With the Response to Immunotherapy Combined With Chemotherapy
Previous research suggests that there is an association between circRNAs and immune checkpoint proteins such as PD-1 and PD-L1. However, few relevant reports have focused on BCa. To explore whether circSOBP expression is related to immune response, whole-transcriptome sequencing of tumor samples was performed in 23 BCa patients before and after immunotherapy. The treatment option at our center was tislelizumab 200 mg and albumin-paclitaxel 200 mg every 3 weeks for one cycle. After four therapy cycles, an efficacy assessment was performed using enhanced computed tomography and pathology examination, and all patients received a disease response evaluation. There were five complete responses (CRs), eight pathological complete responses (pCRs), four partial responses (PRs), one progressive disease (PD), and five stable diseases (SDs). The detailed results are shown in Table S6. In the responder group (CR + pCR + PR), expressions of circSOBP after treatment were significantly elevated compared with baseline before treatment (P = .0043) (Fig. 7D). Nevertheless, in the nonresponder group (SD + PD), we found no statistical difference before and after treatment (P = 0.3558) (Fig. 7E).
Discussion
The last few years have witnessed the research value of circRNAs transform from “waste” to “treasure.” circRNAs with abnormal expressions are found in virtually all cancers and play critical roles in cancer etiology as oncogenes and tumor suppressors13–16. circRNAs have also been discovered to regulate the growth, metastasis, apoptosis, autophagy, metabolism, stemness, tumor immunology, and drug resistance of cancer cells and genome instability through various mechanisms17–21. These RNAs could serve as potentially targetable markers in clinical treatments due to their unique properties such as tissue and cell developmental stage-specificity, high stability, and longer half-lives and their extensive presence in various body fluids. However, it is unclear whether circRNAs mediate BCa cell invasion and migration, and the possible molecular regulatory mechanism of circRNA in mediating anticancer processes in BCa is also still unclear.
In this article, we identified a novel downregulated expression of target circRNA (circSOBP) using high-throughput sequencing and qRT-PCR in tissues and cells. Upregulated expression of circSOBP was shown to be positively linked with advanced clinical stage and tumor size when the clinicopathological parameters of 56 BCa patients were analyzed. Specifically, our cohort of 56 patients underwent up to 4 years of follow-up wherein we observed that more highly expressed circSOBP tended to lead to a better prognosis. Combined with a higher AUC, circSOBP showed excellent diagnostic and prognostic value for BCa as well.
Similar findings have been reported in the literature. For example, Yang et al. 22 showed that circNSD2 expression was dramatically increased in osteosarcoma (OS) and that higher circNSD2 expression correlated with more advanced clinical stage, larger tumor size, higher incidence of distant metastases, and poorer overall survival in OS patients. Yan et al. 23 similarly demonstrated that patients with lower levels of circEVI5 expression in their BCa tissues had a poorer rate of relapse-free survival (RFS) and larger tumors.
In vitro and in vivo experiments have also indicated that circSOBP functions as a tumor suppressor to inhibit the growth and aggressiveness of BCa cells. The subcellular localization of RNAs determines the underlying functioning mechanism 24 , and the majority of circRNAs have been reported to reside in the cytoplasm and to act as miRNA sponges, regulating tumor growth via a ceRNA mechanism 25 . Some circRNAs are primarily produced in the nucleus to interact with RNA-binding proteins (RBPs) and regulate their function, however. For example, circACTN4 might combine with FUBP1 to promote the occurrence and development of breast cancer by enhancing the expression of MYC 26 . In our study, we found that circSOBP was located predominantly in the cytoplasm via FISH, suggesting that circSOBP might be an miRNA sponge. Next, bioinformatics tools, RNA pull-down, and dual-luciferase assay results agreed that circSOBP and miR-200a-3p had intermolecular interactions. Previous research has found that miR-200a-3p is elevated in a variety of cancers, including ovarian cancer and non-small-cell lung cancer27,28. In addition, our results illustrated that miR-200a-3p was overexpressed in BCa tissues and cells and facilitated malignant phenotypes, in agreement with a previous study 29 .
To search for downstream target genes of miR-200a-3p, four databases were used to discover that 3′-UTR untranslated regions bind to an miR-200a-3p seed region, and here the PTEN gene attracted our attention. In human malignancies, PTEN has become well known as one of the most frequent tumor suppressor genes, and it is a negative regulator of the phosphatidylinositol-3-kinase (PI3K)/AKT signaling cascade, which affects cell proliferation, invasiveness, survival, and metabolism due to mutation, deletion, or aberrant expression30,31. In previous studies, PTEN protein has been shown to be downregulated in paired BCa specimens compared with normal adjacent tissue samples 12 . Moreover, PTEN knockout can strengthen the migration and invasion of BCa cells, and cause resistance to chemotherapy and adverse prognosis patients 32 . In our study, we found that there were targeted site interactions between miR-200a-3p and PTEN mRNA via the dual-luciferase assay. Furthermore, miR-200a-3p expression was negatively associated with PTEN in the TCGA database. Western blot experiments also made it clear that circSOBP favorably regulated PTEN protein levels to suppress malignant phenotypes of BCa dependent on miR-200a-3p. Therefore, a novel ceRNA mechanism was elucidated.
Currently, immunotherapy with checkpoint inhibitors has been shown to be effective in treating BCa 33 and lung cancer 34 , and many circRNAs have been reported to affect the tumor microenvironment via influencing the PD-1/PD-L1 pathway. Huang et al. 35 showed that circMET induces hepatocellular carcinoma development, immune tolerance, and anti-PD1 treatment resistance through Mir-30-5p/Snail/ DPP4/CXCL10 axis, and Luo et al. 8 illustrated that hsa_circ_0000190 may be used as a novel biomarker for the effectiveness of systemic and immunotherapy in advanced lung cancers.
Finally, chemotherapy can enhance T-cell response and boost antitumor activity, which allows chemotherapy and immune checkpoint inhibitors to work together synergistically to achieve better therapeutic outcomes36,37. An increasing body of scientific and clinical evidence also suggests that chemotherapy regimens can cause immunogenic cell death; however, the exact mechanism is still being investigated. For this study, our center used a novel therapeutic strategy that set it apart from conventional single-agent immunotherapy, and we attained a high rate of response. The mechanism of this treatment regimen is still being investigated, and as stated above, circSOBP variations were observed before and after treatment to test whether this novel circRNA plays a role in the mechanism. Indeed, the detailed molecular mechanisms of circSOBP-involved modulation of the PD-1/PD-L1 pathway remain to be further explored.
Conclusion
In summary, we identified a new circular RNA, circSOBP, that is linked to prognosis and immune response in BCa patients and that inhibits the progression and metastasis of BCa via the miR-200a-3p/PTEN axis. These findings elucidate a novel regulatory network that may shed light on new clinical biomarkers, therapeutic agents, and drug targets for BCa.
Supplemental Material
Supplemental material, sj-docx-1-cll-10.1177_09636897231165874 for circSOBP Inhibits Bladder Cancer Proliferation and Metastasis by Regulating the miR-200a-3p/PTEN Axis and Participating in the Immune Response by Yinglang Zhang, Zhi Li, Yu Zhang, Chong Shen, Zhe Zhang, Xiaolei Ren, Changgang Guo, Shaobo Yang, Zejin Wang and Hailong Hu in Cell Transplantation
Supplemental material, sj-png-2-cll-10.1177_09636897231165874 for circSOBP Inhibits Bladder Cancer Proliferation and Metastasis by Regulating the miR-200a-3p/PTEN Axis and Participating in the Immune Response by Yinglang Zhang, Zhi Li, Yu Zhang, Chong Shen, Zhe Zhang, Xiaolei Ren, Changgang Guo, Shaobo Yang, Zejin Wang and Hailong Hu in Cell Transplantation
Supplemental material, sj-png-3-cll-10.1177_09636897231165874 for circSOBP Inhibits Bladder Cancer Proliferation and Metastasis by Regulating the miR-200a-3p/PTEN Axis and Participating in the Immune Response by Yinglang Zhang, Zhi Li, Yu Zhang, Chong Shen, Zhe Zhang, Xiaolei Ren, Changgang Guo, Shaobo Yang, Zejin Wang and Hailong Hu in Cell Transplantation
Supplemental material, sj-png-4-cll-10.1177_09636897231165874 for circSOBP Inhibits Bladder Cancer Proliferation and Metastasis by Regulating the miR-200a-3p/PTEN Axis and Participating in the Immune Response by Yinglang Zhang, Zhi Li, Yu Zhang, Chong Shen, Zhe Zhang, Xiaolei Ren, Changgang Guo, Shaobo Yang, Zejin Wang and Hailong Hu in Cell Transplantation
Supplemental material, sj-xlt-5-cll-10.1177_09636897231165874 for circSOBP Inhibits Bladder Cancer Proliferation and Metastasis by Regulating the miR-200a-3p/PTEN Axis and Participating in the Immune Response by Yinglang Zhang, Zhi Li, Yu Zhang, Chong Shen, Zhe Zhang, Xiaolei Ren, Changgang Guo, Shaobo Yang, Zejin Wang and Hailong Hu in Cell Transplantation
Acknowledgments
The authors thank AiMi Academic Services (www.aimieditor.com) for English language editing and review services.
Footnotes
Author Contributions: Yinglang Zhang, Chong Shen, and Hailong Hu designed this study. Yinglang Zhang wrote the manuscript. Yinglang Zhang, Zhi li, Yu Zhang, Zhe Zhang, Shaobo Yang, and Zejin Wang performed the experiments. Xiaolei Ren and Changgang Guo analyzed the corresponding data. All authors read and approved the final manuscript. Yinglang Zhang, Zhi Li, and Yu Zhang contributed equally to this work.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article and its supplementary information files.
Ethical Approval: This study was approved by our institutional review board.
Statement of Human and Animal Rights: This article does not contain any studies with human or animal subjects.
Statement of Informed Consent: There are no human subjects in this article and informed consent is not applicable.
Research Ethics and Patient Consent: The current study was approved by the Medical Ethics Committee of the Second Hospital of Tianjin Medical University (NO.KY2020K063). Written informed consent was obtained from each patient for their anonymized information to be published in this article.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the Natural Science Foundation of Inner Mongolia (2022QN08020) and Technology Project of Tianjin Binhai New Area Health Commission (Grant No. 2019BWKY026).
ORCID iDs: Yinglang Zhang  https://orcid.org/0000-0003-0205-2878
https://orcid.org/0000-0003-0205-2878
Hailong Hu  https://orcid.org/0000-0001-8675-5524
https://orcid.org/0000-0001-8675-5524
Supplemental Material: Supplemental material for this article is available online.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 Countries. CA Cancer J Clin. 2021;71(3): 209–49. [DOI] [PubMed] [Google Scholar]
- 2.Meeks JJ, Al-Ahmadie H, Faltas BM, Taylor JA, 3rd, Flaig TW, DeGraff DJ, Christensen E, Woolbright BL, McConkey DJ, Dyrskjøt L. Genomic heterogeneity in bladder cancer: challenges and possible solutions to improve outcomes. Nat Rev Urol. 2020;17(5): 259–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19(3): 188–206. [DOI] [PubMed] [Google Scholar]
- 4.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2): 141–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang D, Yang S, Wang H, Wang J, Zhang Q, Zhou S, He Y, Zhang H, Deng F, Xu H, Zhong S, et al. The progress of circular RNAs in various tumors. Am J Transl Res. 2018;10(6): 1571–82. [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao W, Dong M, Pan J, Wang Y, Zhou J, Ma J, Liu S. Circular RNAs: a novel target among non-coding RNAs with potential roles in malignant tumors (Review). Mol Med Rep. 2019;20(4): 3463–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441): 384–88. [DOI] [PubMed] [Google Scholar]
- 8.Luo YH, Yang YP, Chien CS, Yarmishyn AA, Ishola AA, Chien Y, Chen YM, Huang TW, Lee KY, Huang WC. Plasma level of circular RNA hsa_circ_0000190 correlates with tumor progression and poor treatment response in advanced lung cancers. Cancers. 2020;12(7): 1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang W, Pan S, Chen X, Wang ZW, Zhu X. The role of lncRNAs and circRNAs in the PD-1/PD-L1 pathway in cancer immunotherapy. Mol Cancer. 2021;20(1): 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding L, Lu S, Li Y. Regulation of PD-1/PD-L1 pathway in cancer by noncoding RNAs. Pathol Oncol Res. 2020;26(2): 651–63. [DOI] [PubMed] [Google Scholar]
- 11.Shen C, Wu Z, Wang Y, Gao S, Da L, Xie L, Qie Y, Tian D, Hu H. Downregulated hsa_circ_0077837 and hsa_circ_0004826, facilitate bladder cancer progression and predict poor prognosis for bladder cancer patients. Cancer Med. 2020;9(11): 3885–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Q, Liu T, Feng H, Yang R, Zhao X, Chen W, Jiang B, Qin H, Guo X, Li L, Guo H. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18(1): 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Sheng W, Xu T, Xu J, Gao R, Zhang Z. CircRNA hsa_circ_0110102 inhibited macrophage activation and hepatocellular carcinoma progression via miR-580-5p/PPARα/CCL2 pathway. Aging. 2021;13(8): 11969–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang X, Guo S, Wang S, Zhang Y, Chen H, Wang Y, Liu R, Niu Y, Xu Y. EIF4A3-induced circARHGAP29 promotes aerobic glycolysis in docetaxel-resistant prostate cancer through IGF2BP2/c-Myc/LDHA signaling. Cancer Res. 2022;82(5): 831–45. [DOI] [PubMed] [Google Scholar]
- 15.Zheng X, Huang M, Xing L, Yang R, Wang X, Jiang R, Zhang L, Chen J. The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Mol Cancer. 2020;19(1): 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu C, Cheng Z, Cui S, Mao X, Li B, Fu Y, Wang H, Jin H, Ye Q, Qin W. circFOXM1 promotes proliferation of non-small cell lung carcinoma cells by acting as a ceRNA to upregulate FAM83D. J Exp Clin Cancer Res. 2020;39(1): 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, Shan G. CircRNA in cancer: fundamental mechanism and clinical potential. Cancer Lett. 2021;505:49–57. [DOI] [PubMed] [Google Scholar]
- 18.Zhong C, Wu K, Wang S, Long Z, Yang T, Zhong W, Tan X, Wang Z, Li C, Mao X. Autophagy-related circRNA evaluation reveals hsa_circ_0001747 as a potential favorable prognostic factor for biochemical recurrence in patients with prostate cancer. Cell Death Dis. 2021;12(8): 726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi P, Li Y, Guo Q. Circular RNA circPIP5K1A contributes to cancer stemness of osteosarcoma by miR-515-5p/YAP axis. J Transl Med. 2021;19(1): 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang B, Teng F, Chang L, Wang J, Liu DL, Cui YS, Li GH. Tumor-derived exosomal circRNA_102481 contributes to EGFR-TKIs resistance via the miR-30a-5p/ROR1 axis in non-small cell lung cancer. Aging. 2021;13(9): 13264–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu G, Yang Z, Peng T, Lv Y. Circular RNAs: rising stars in lipid metabolism and lipid disorders. J Cell Physiol. 2021;236(7): 4797–806. [DOI] [PubMed] [Google Scholar]
- 22.Yang B, Li L, Tong G, Zeng Z, Tan J, Su Z, Liu Z, Lin J, Gao W, Liu Q, Lin L. Circular RNA circ_001422 promotes the progression and metastasis of osteosarcoma via the miR-195-5p/FGF2/PI3K/Akt axis. J Exp Clin Cancer Res. 2021;40(1): 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan M, Niu L, Liu J, Yao Y, Li H. circEVI5 acts as a miR-4793-3p sponge to suppress the proliferation of gastric cancer. Cell Death Dis. 2021;12(8): 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buxbaum AR, Haimovich G, Singer RH. In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol. 2015;16(2): 95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang Q, Hann SS. Biological roles and mechanisms of circular RNA in human cancers. Onco Targets Ther. 2020;13:2067–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Xing L, Yang R, Chen H, Wang M, Jiang R, Zhang L, Chen J. The circACTN4 interacts with FUBP1 to promote tumorigenesis and progression of breast cancer by regulating the expression of proto-oncogene MYC. Mol Cancer. 2021;20(1): 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi C, Yang Y, Zhang L, Yu J, Qin S, Xu H, Gao Y. MiR-200a-3p promoted the malignant behaviors of ovarian cancer cells through regulating PCDH9. Onco Targets Ther. 2019;12:8329–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei S, Wang K, Huang X, Zhao Z, Zhao Z. LncRNA MALAT1 contributes to non-small cell lung cancer progression via modulating miR-200a-3p/programmed death-ligand 1 axis. Int J Immunopathol Pharmacol. 2019;33:2058738419859699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wan P, Chen Z, Huang M, Jiang H, Wu H, Zhong K, Ding G, Wang B. miR-200a-3p facilitates bladder cancer cell proliferation by targeting the A20 gene. Transl Androl Urol. 2021;10(11): 4262–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee YR, Chen M, Pandolfi PP. The functions and regulation of the PTEN tumor suppressor: new modes and prospects. Nat Rev Mol Cell Biol. 2018;19(9): 547–62. [DOI] [PubMed] [Google Scholar]
- 31.Luongo F, Colonna F, Calapà F, Vitale S, Fiori ME, De Maria R. PTEN tumor-suppressor: the dam of stemness in cancer. Cancers. 2019;11(8): 1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashrafizadeh M, Zarrabi A, Samarghandian S, Najafi M. PTEN: what we know of the function and regulation of this onco-suppressor factor in bladder cancer? Eur J Pharmacol. 2020;881:173226. [DOI] [PubMed] [Google Scholar]
- 33.Hussain SA, Birtle A, Crabb S, Huddart R, Small D, Summerhayes M, Jones R, Protheroe A. From clinical trials to real-life clinical practice: the role of immunotherapy with PD-1/PD-L1 inhibitors in advanced urothelial carcinoma. Eur Urol Oncol. 2018;1(6): 486–500. [DOI] [PubMed] [Google Scholar]
- 34.Yang KN, Han W, Qin YJ, Chen LN. Effects of different levels of soluble PD-L1 protein on the growth of Lewis lung cancer transplanted tumor. J Biol Regul Homeost Agents. 2019;33(2): 537–42. [PubMed] [Google Scholar]
- 35.Huang XY, Zhang PF, Wei CY, Peng R, Lu JC, Gao C, Cai JB, Yang X, Fan J, Ke AW, Shi GM. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Molecular Cancer. 2020;19(1): 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z, Zhao J, Ma Z, Cui J, Shu Y, Liu Z, Cheng Y, Leaw SJ, Wu Y, Ma Y, Tan W, et al. A phase 2 study of tislelizumab in combination with platinum-based chemotherapy as first-line treatment for advanced lung cancer in Chinese patients. Lung Cancer. 2020;147:259–68. [DOI] [PubMed] [Google Scholar]
- 37.Zhang P, Ma Y, Lv C, Huang M, Li M, Dong B, Liu X, An G, Zhang W, Zhang J, Zhang L, et al. Upregulation of programmed cell death ligand 1 promotes resistance response in non-small-cell lung cancer patients treated with neo-adjuvant chemotherapy. Cancer Sci. 2016;107(11): 1563–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-cll-10.1177_09636897231165874 for circSOBP Inhibits Bladder Cancer Proliferation and Metastasis by Regulating the miR-200a-3p/PTEN Axis and Participating in the Immune Response by Yinglang Zhang, Zhi Li, Yu Zhang, Chong Shen, Zhe Zhang, Xiaolei Ren, Changgang Guo, Shaobo Yang, Zejin Wang and Hailong Hu in Cell Transplantation
Supplemental material, sj-png-2-cll-10.1177_09636897231165874 for circSOBP Inhibits Bladder Cancer Proliferation and Metastasis by Regulating the miR-200a-3p/PTEN Axis and Participating in the Immune Response by Yinglang Zhang, Zhi Li, Yu Zhang, Chong Shen, Zhe Zhang, Xiaolei Ren, Changgang Guo, Shaobo Yang, Zejin Wang and Hailong Hu in Cell Transplantation
Supplemental material, sj-png-3-cll-10.1177_09636897231165874 for circSOBP Inhibits Bladder Cancer Proliferation and Metastasis by Regulating the miR-200a-3p/PTEN Axis and Participating in the Immune Response by Yinglang Zhang, Zhi Li, Yu Zhang, Chong Shen, Zhe Zhang, Xiaolei Ren, Changgang Guo, Shaobo Yang, Zejin Wang and Hailong Hu in Cell Transplantation
Supplemental material, sj-png-4-cll-10.1177_09636897231165874 for circSOBP Inhibits Bladder Cancer Proliferation and Metastasis by Regulating the miR-200a-3p/PTEN Axis and Participating in the Immune Response by Yinglang Zhang, Zhi Li, Yu Zhang, Chong Shen, Zhe Zhang, Xiaolei Ren, Changgang Guo, Shaobo Yang, Zejin Wang and Hailong Hu in Cell Transplantation
Supplemental material, sj-xlt-5-cll-10.1177_09636897231165874 for circSOBP Inhibits Bladder Cancer Proliferation and Metastasis by Regulating the miR-200a-3p/PTEN Axis and Participating in the Immune Response by Yinglang Zhang, Zhi Li, Yu Zhang, Chong Shen, Zhe Zhang, Xiaolei Ren, Changgang Guo, Shaobo Yang, Zejin Wang and Hailong Hu in Cell Transplantation