Abstract
Background
The aim of this study was to evaluate oral bacteria‐ and interleukin (IL)‐1β‐induced protein and mRNA expression profiles of monocyte chemoattractant protein‐1‐induced protein (MCPIP)‐1 and mucosa‐associated lymphoid tissue lymphoma translocation protein (MALT)‐1 in human gingival keratinocyte monolayers and organotypic oral mucosal models.
Methods
Human gingival keratinocyte (HMK) monolayers were incubated with Porphyromonas gingivalis, Fusobacterium nucleatum, P. gingivalis lipopolysaccharide (LPS) and IL‐1β. The protein levels of MCPIP‐1 and MALT‐1 were examined by immunoblots and mRNA levels by qPCR. MCPIP‐1 and MALT‐1 protein expression levels were also analyzed immunohistochemically using an organotypic oral mucosal model. One‐way analysis of variance followed by Tukey correction was used in statistical analyses.
Results
In keratinocyte monolayers, MCPIP‐1 protein expression was suppressed by F. nucleatum and MALT‐1 protein expression was suppressed by F. nucleatum, P. gingivalis LPS and IL‐1β. P. gingivalis seemed to degrade MCPIP‐1 and MALT‐1 at all tested time points and degradation was inhibited when P. gingivalis was heat‐killed. MCPIP‐1 mRNA levels were increased by P. gingivalis, F. nucleatum, and IL‐1β, however, no changes were observed in MALT‐1 mRNA levels.
Conclusion
Gingival keratinocyte MCPIP‐1 and MALT‐1 mRNA and protein expression responses are regulated by infection and inflammatory mediators. These findings suggest that periodontitis‐associated bacteria‐induced modifications in MCPIP‐1 and MALT‐1 responses can be a part of periodontal disease pathogenesis.
Keywords: infections, inflammation, periodontal diseases
1. INTRODUCTION
Periodontitis is an infection‐induced chronic inflammatory disease of teeth‐supporting tissues. 1 Porphyromonas gingivalis, a keystone pathogen for the development of dysbiosis, suppresses toll like receptor (TLR)‐4 activation, prevents nuclear factor‐κB (NF‐κB) p65 homodimer nuclear translocation, inhibits interleukin (IL)‐8 expression, stimulates of IL‐1β secretion, and eventually induces alveolar bone loss and periodontitis. 2 , 3 F. nucleatum, a bridge organism between early late colonizers of the oral biofilms, induces proinflammatory cytokine (e.g., IL‐8, IL‐1β) expression, and acts as an opportunistic pathogen during the pathogenesis of periodontitis. 4 P. gingivalis lipopolysaccharide (LPS) causes disruptions on the innate immune response through host receptors, for example through TLR‐4, leading to modifications in the proinflammatory cytokine responses. 5 , 6 , 7 , 8
Monocyte chemoattractant protein‐induced protein‐1 (MCPIP‐1), also called Regnase‐1, is a host inflammatory response cytokine RNase. 9 It was recently demonstrated that P. gingivalis can degrade MCPIP‐1 by its gingipains, which leads to the overactivation of the NF‐κB signaling pathway and increase the transcripts of proinflammatory cytokines. 10 MCPIP‐1 acts as a broad suppressor of miRNA activity and biogenesis. 11 Its involvement in the deubiquitination process provides inhibition of LPS and IL‐1‐induced NF‐kB signaling pathway, whereas its RNase activity suppresses proinflammatory cytokine (IL‐6, IL‐1β, or IL‐8) mRNA activity. 12 , 13 , 14 , 15 , 16 Mucosa‐associated lymphoid tissue lymphoma translocation protein 1 (MALT‐1) is a cysteine protease that cleaves the inflammatory signaling suppressors and eventually activates immune cells. MALT‐1 suppression can cause inflammatory genes to synthesize cytokines (MCPIP‐1 included) and inflammation develops in the absence of MALT‐1. Indeed, suppression of MALT‐1 increases MCPIP‐1 activity. 17 It was hypothesized that MCPIP‐1 depletion in periodontitis can be an outcome of MALT‐1 activation, however, this was not yet proven. 10 Indeed, simultaneous regulations of MCPIP‐1 and MALT‐1 gene and protein expressions by periodontitis‐associated bacteria and inflammatory mediators have not been elucidated as well.
In the present study, we hypothesized that MCPIP‐1 and its inhibitor MALT‐1 expressions of human gingival keratinocytes are regulated by periodontitis‐associated bacteria and by proinflammatory cytokines. Therefore, this study aimed to analyze the MCPIP‐1 and MALT‐1 gene and protein expression regulation by P. gingivalis, P. gingivalis LPS, Fusobacterium nucleatum (F. nucleatum), and IL‐1β, using both gingival keratinocyte monolayer and organotypic oral mucosal models.
2. MATERIALS AND METHODS
For the flowchart see Figure S1 in online Journal of Periodontology.
2.1. HMK cell line
Spontaneously immortalized nontumorigenic gingival keratinocyte cell lines, HMK cells, were used in all experiments. 18 HMK cells (passages of 7‒13) were sustained in a 75 cm 2 cell culture flask with a keratinocyte serum‐free media,* containing human recombinant epidermal growth factor, bovine pituitary extract, and antibiotics (penicillin‐streptomycin, 10,000 U/ml) at 37°C and 5% CO2. Keratinocyte growth media were replaced with fresh media three times per week, and the HMK cells were passaged weekly until they reached an 80–90% confluence.
2.2. Bacterial strains
P. gingivalis (ATCC 33277) and F. nucleatum (ATCC 25586) were obtained from the Anaerobe Reference Laboratory of the Finnish Institute for Health and Welfare (previously National Public Health Institute), Helsinki, Finland. Strains were revived from skimmed milk stocks and were grown on Brucella blood agar plates supplemented with hemin (5 mg L−1) and vitamin K1 (10 mg L−1) in an anaerobic chamber† (10% H2, 5% CO2, and 85% N2) at 37°C for 72 h. In all experiments, bacteria were suspended in phosphate buffer solution (PBS) and the optical density of the suspension was adjusted to 0.6 at 670 nm for P. gingivalis (3×108 CFU/ml) and to 0.6 at 620 nm for F. nucleatum (6×108 CFU/ml).
2.3. Stock solutions of P. gingivalis LPS and IL‐1β
P. gingivalis LPS ultra‐pure stock solution (1 mg/ml) was prepared by dissolving 1 mg of ultra‐pure P. gingivalis LPS‡ in 1 ml of endotoxin‐free water.
IL‐1β stock solution (100 μg/ml) was prepared by dissolving 100 μg of IL‐1β§ in 1 ml of endotoxin‐free water.
2.4. Incubation of HMK monolayers with bacteria, P. gingivalis LPS, and IL‐1β
HMK cells (3×105) were cultured in 12‐well plates and allowed to grow until 80%‒90% confluency. HMK cells were incubated with P. gingivalis ATCC 33277 and F. nucleatum ATCC 25586 at a multiplicity of infections (MOI) of 1:50 (2×105 HMK cells to 1×107 CFU bacteria) and 1:100 (2×105 HMK cells to 2×107 CFU bacteria), P. gingivalis LPS (1 μl/ml) and IL‐1β (15 ng/ml). In addition, HMK cells were also cultured with heat‐killed (95°C for 5 min) P. gingivalis (MOI of 1:100) and with P. gingivalis suspension containing serine and cysteine protease inhibitor (50 mg/ 7 ml).** HMK cells, which were incubated for 2, 6, and 24 h in the absence of bacteria, P. gingivalis LPS, or IL‐1β, served as control groups. After incubating the cell cultures in a CO2 incubator (10% H2, 5% CO2, and 85% N2) at 37°C for 2, 6, and 24 h, cells were first scraped mechanically and then lysed with a lysis buffer (150 μl, 50 mM Tris‐HCl, 150 mM NaCl, 1% Triton‐x.100, pH 8.0, on a shaker (250 rpm) for 10 min). The lysates were collected, sonicated, and stored at ‐70°C until they were used for immunoblots. Another group of cells were first scraped mechanically and lysed with 1 ml of trizol.†† Lysates were stored at ‐70°C until used for mRNA isolation.
A parallel set of cell cultures were performed to determine the effects of P. gingivalis, F. nucleatum, P. gingivalis LPS, and IL‐1β on HMK cell viability using a proliferation assay.‡‡
2.5. MCPIP‐1 and MALT‐1 protein expression determination and immunoblotting
Protein levels of each sample were determined using the Bradford method.§§
Western blot was used to analyze MCPIP‐1 and MALT‐1 levels in cell lysates. Same amount of protein for each sample was mixed with Laemmli buffer and mercaptoethanol, and then heated at 95°C for 5 min. Sodium dodecyl sulfate (SDS)‐polyacrylamide gels (15%) were used for the separation of proteins. Afterwards, the proteins were transferred to membranes*** and incubated overnight with either MCPIP‐1 (1:750 dilution)††† or MALT‐1 (1:1000 dilution)‡‡‡ antibody. Finally, the membranes were incubated with goat anti‐Rabbit IgG (H+L) secondary antibody (1:10000 dilution)§§§ for 1 h. The detection of HRP was performed using a chemiluminescent substrate,**** and the bands were detected with a digital imaging equipment.†††† Blots were stained with anti‐β ‐Actin antibody (1:10000)‡‡‡‡ to confirm equal loading. A non‐commercial software§§§§ was used for intensity analysis. All experiments were performed in triplicate and repeated at least three times at independent time points.
2.6. MCPIP‐1 and MALT‐1 gene expression determination
Total RNA was extracted from the cell lysates with trizol. A microvolume spectrophotometer***** was used to determine the purity and concentration of extracted RNA. First‐strand cDNA was synthesized from 500 ng of total extracted RNA using the cDNA synthesis kit.║║║ RT‐qPCR reactions were performed in 20‐μl volumes with 2 μl of template cDNA and 1 μl of gene‐specific primer‐probe mix¶¶¶ and 10 μl DNA probes master mix.### GAPDH was used as a control. The reaction was performed in a real‐time PCR system and consisted of initial denaturation at 95°C for 10 min, and 45 cycles of 95°C for 30s, and 60°C for 30s. All experiments were performed in triplicates and expression levels were analyzed using delta Ct method.
2.7. Constructing the organotypic oral mucosal model
A modified organotypic oral mucosal model was constructed as described previously. 19 , 20 , 21 The isolation and the characterization of cell types used were described previously (gingival keratinocytes by Mäkelä et al. 18 and gingival fibroblasts by Oksanen et al. 2002 19 ). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal calf serum, 1% essential amino acids, and supplemented with antibiotics at 37°C in 5% CO2. To construct the multilayer culture model, human gingival fibroblasts were suspended at a density of 3×105/ml in collagen solution††††† and plated in 10 mm culture inserts.‡‡‡‡‡ Then, the inserts were placed in 12‐well tissue culture plates for 24 h to solidify. The gingival keratinocytes were seeded on collagen‐fibroblast gels at a density of 8×105/ml, and when the cells reached confluence, the inserts were placed on metal grids to obtain the air‒liquid interface. The model was allowed to grow for an additional 14 days. The cultures were grown at 37°C in a humidified atmosphere, containing 95% air and 5% CO, 2 for 7 days before the experiments.
2.8. Incubation of the organotypic oral mucosal model cells with bacteria, P. gingivalis LPS and IL‐1β
Cellular responses of the organotypic oral mucosal model were stimulated as described previously. 22 The model was activated with P. gingivalis (3×106 CFU), F. nucleatum (3×106 CFU), P. gingivalis LPS (10 ng), and IL‐1β (150 pg) using a nitrocellulose filter as a scaffold. An organotypic model with only the membrane filter formed the control group. After 2 h of incubation, models were fixed overnight in phosphate buffered neutral 10% formalin at ‐20°C and later embedded in paraffin.
2.9. Immunohistochemical examination of organotypic oral mucosal model
Paraffin‐embedded samples of the organotypic oral mucosa model were cut into 5‐μm thick sections and deparaffinized. The sections were immunostained with hematoxylin and eosin (H&E), thymidine blue, Ki‐67, MCPIP‐1, and MALT‐1 using an automated immunostainer.§§§§§ Briefly, the sections underwent antigen retrieval in 1 mmol/L citrate buffer (pH 6.0) in a microwave twice for 5 min and incubated with 3% H2O2 to block endogenous peroxidase activity. The primary antibodies—MCPIP‐1 Antibody (1:200 dilution) and MALT‐1(1:100 dilution)—were detected with biotinylated secondary antibody. The membranes have been incubated for 1 h with goat anti‐Rabbit IgG (H+L) secondary antibody (1:10000 dilution)****** and streptavidin–horseradish peroxidase and visualized with 3,3’ diaminobenzidine tetrahydrochloride in horseradish peroxidase buffer. Human lung (for MALT‐1) and human kidney (for MCPIP‐1) tissues were used as positive staining controls. Extra sets of staining omitting primary antibodies were performed as negative controls.
The immunohistochemical stainings were evaluated under a light microscope†††††† and high‐resolution images were captured to analyze signal intensities‡‡‡‡‡‡ with software.§§§§§§ Intensities of the staining were determined from both epithelium and connective tissue.
2.10. Statistical analysis
A commercial statistical analysis program******* was used in all analyses. In all figures bar charts present values of means and standard deviations. One‐way analysis of variance followed by post‐hoc Tukey test was used to analyze levels of differences. A p‐value ˂0.05 was defined as statistically significant.
3. RESULTS
3.1. Effects of P. gingivalis, F. nucleatum, P. gingivalis LPS and IL‐1β on the viability of HMK monolayers
Viabilities of HMK monolayers after 2, 6, and 24 h of incubation with P. gingivalis (ATCC 33277), F. nucleatum (ATCC 25586), P. gingivalis LPS and IL‐1β were presented in Figure 1. After 2 h of incubation with F. nucleatum the HMK proliferation increased (MOI 1:50 P = 0.005 and MOI 1:100 P = 0.003). P. gingivalis suppressed HMK proliferation at 6 and 24 h (P<0.001). P. gingivalis LPS and IL‐1β incubation did not affect the proliferation of HMK cells.
FIGURE 1.
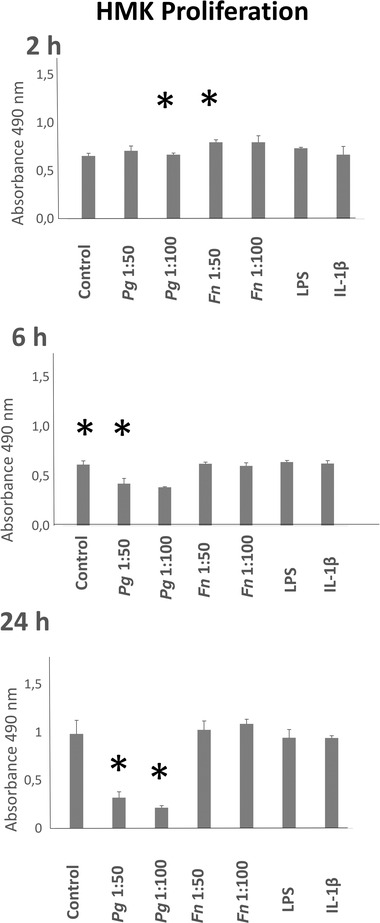
HMK viability levels after 2, 6, and 24 h of incubations with P. gingivalis (ATCC 33277), F. nucleatum (ATCC 25586), P. gingivalis LPS, and IL‐1β. * Statistical difference (P < 0.005) with control. Bars indicate mean values and standard deviations of triplicate assays
3.2. Effects of P. gingivalis, F. nucleatum, P. gingivalis LPS and IL‐1β on MCPIP‐1 protein expression of HMK monolayers
MCPIP‐1 protein expressions of HMK monolayers after 2, 6, and 24 h of incubation with P. gingivalis (ATCC 33277), F. nucleatum (ATCC 25586), P. gingivalis LPS, and IL‐1β were presented in Figure 2. P. gingivalis seemed to degrade MCPIP‐1 at all time points. MCPIP‐1 protein expression was suppressed by F. nucleatum (MOI 1:100) after 2 h incubation (P = 0.003). After 24 h of incubation, MCPIP‐1 protein expressions were suppressed by F. nucleatum (MOI 1:50) (P<0.001), F. nucleatum (MOI 1:100) (p<0.001), P. gingivalis LPS (P<0.001), and by IL‐1β (P = 0.003). No changes in MCPIP‐1 protein levels were observed after 2 and 6 h of incubations with F. nucleatum (MOI 1:50), P. gingivalis LPS, and IL‐1β (Figure 2).
FIGURE 2.
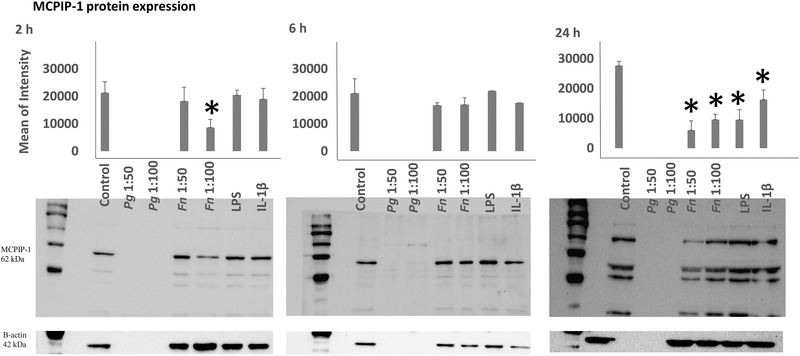
MCPIP‐1 protein expression levels after 2, 6, and 24 h of incubation with P. gingivalis (ATCC 33277), F. nucleatum (ATCC 25586), P. gingivalis LPS, and IL‐1β. *Statistical difference (P <0.005) with control. Bars indicate mean values and standard deviations. (Fifteen μg of protein were loaded to 2 and 6 h gels and 40 μg of protein to 24 h gels)
3.3. Effects of P. gingivalis, F. nucleatum, P. gingivalis LPS and IL‐1β on MALT‐1 protein expression of HMK monolayers
MALT‐1 protein expressions of HMK monolayers after 2, 6, and 24 h of incubation with P. gingivalis (ATCC 33277), F. nucleatum (ATCC 25586), P. gingivalis LPS, and IL‐1β were presented in Figure 3. MALT‐1 protein expression was suppressed by F. nucleatum (MOI 1:50 P = 0.011), F. nucleatum (MOI 1:100 P<0.001), P. gingivalis LPS (P = 0.016), and IL‐1β (P = 0.022) after 2 h of incubation. P. gingivalis seemed to degrade MALT‐1 at all time points. No changes were observed in MALT‐1 levels after 6 and 24 h of incubation (Figure 3).
FIGURE 3.
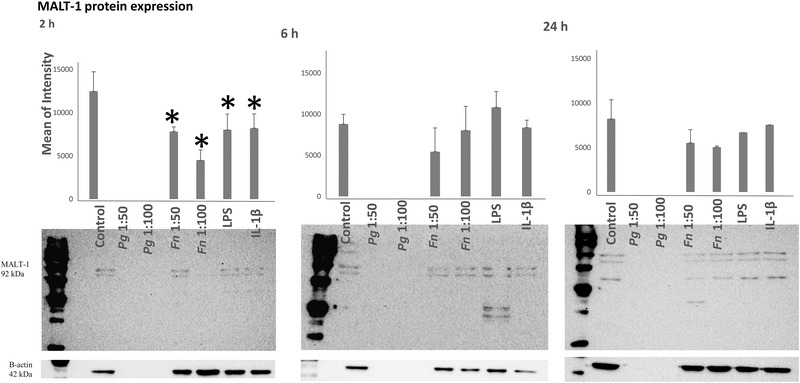
MALT‐1 protein expression levels after 2, 6, and 24 h of incubation with P. gingivalis, F. nucleatum, P. gingivalis LPS, and IL‐1β. *Statistical difference (P<0.005) with control. Bars indicate mean values and standard deviations
3.4. Effects of P. gingivalis, F. nucleatum, P. gingivalis LPS, and IL‐1β on MCPIP‐1 and MALT‐1 mRNA expressions of HMK monolayers
MCPIP‐1 mRNA expression levels after 2, 6, and 24 h of incubation with P. gingivalis, F. nucleatum, P. gingivalis LPS and IL‐1β were given in Figure 4. MCPIP‐1 mRNA synthesis was increased by F. nucleatum (MOI 1:50 P = 0.007 and MOI 1:100 P = 0.005) after 2 h incubation. After 6 h of incubation, MCPIP‐1 mRNA syntheses were increased by P. gingivalis (MOI 1:100 P = 0.01), F. nucleatum (MOI 1:50 P = 0.046 and MOI 1:100 P = 0.013), and IL‐1β (P = 0.022). After 24 h after incubation, MCPIP‐1 mRNA synthesis was increased by F. nucleatum (MOI 1:50 P = 0.02 and 1:100 P = 0.010) (Figure 4).
FIGURE 4.
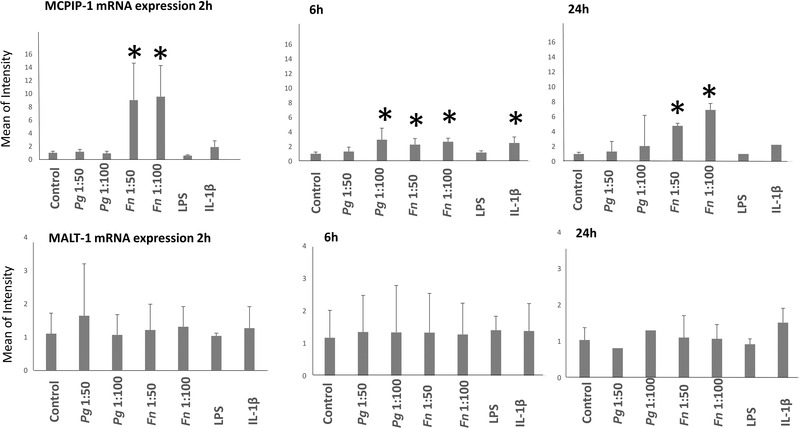
mRNA expression levels of MCPIP‐1 and MALT‐1 after 2, 6, and 24 h of incubation with P. gingivalis, F. nucleatum, P. gingivalis LPS, and IL‐1β. *Statistical difference with control. Bars indicate mean values and standard deviations
MALT‐1 mRNA expression levels after 2, 6, and 24 h of incubation with P. gingivalis, F. nucleatum, P. gingivalis LPS, and IL‐1β are presented in Figure 4. No significant changes in MALT‐1 mRNA were detected at any time points.
3.5. MCPIP‐1 and MALT‐1 protein expressions of HMK monolayers incubated with heat‐killed P. gingivalis and/or P. gingivalis with protease inhibitor
MCPIP‐1 and MALT‐1 expression levels of HMK monolayers were analyzed after 2, 6, and 24 h of incubation with P. gingivalis in the presence of protease inhibitor or with heat‐killed P. gingivalis (MOI 1:100) (Figure 5; see Figure S2 in online Journal of Periodontology for full size membranes). MCPIP‐1 and MALT‐1 proteins were detected at all time points when P. gingivalis was heat‐killed. Protease inhibitor alone did not inhibit MCPIP‐1 or MALT‐1 degradation.
FIGURE 5.
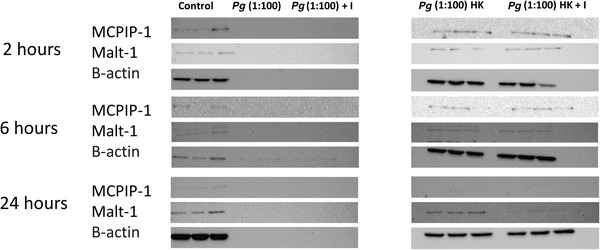
MCPIP‐1 and MALT‐1 protein expression profiles after 2, 6, and 24 h of incubation with P. gingivalis, P. gingivalis and protease inhibitor, heat killed P. gingivalis, and heat killed P. gingivalis with protease inhibitor. Letter I indicates incubation media containing enzyme inhibitors and letters HK indicate heat‐killed P. gingivalis groups (see Figure S2 in online Journal of Periodontology for full size membranes)
3.6. Effects of P. gingivalis, F. nucleatum, P. gingivalis LPS, and IL‐1β on MCPIP‐1 and MALT‐1 protein expression of the organotypic oral mucosal model
MCPIP‐1 and MALT‐1 protein expressions of the organotypic oral mucosal model after 2 h of incubation with P. gingivalis (ATCC 33277), F. nucleatum (ATCC 25586), P. gingivalis LPS, and IL‐1β were presented in Figure 6. No statistical difference was observed in MCPIP1 and MALT‐1 protein expression levels between the test groups (Figure 6).
FIGURE 6.
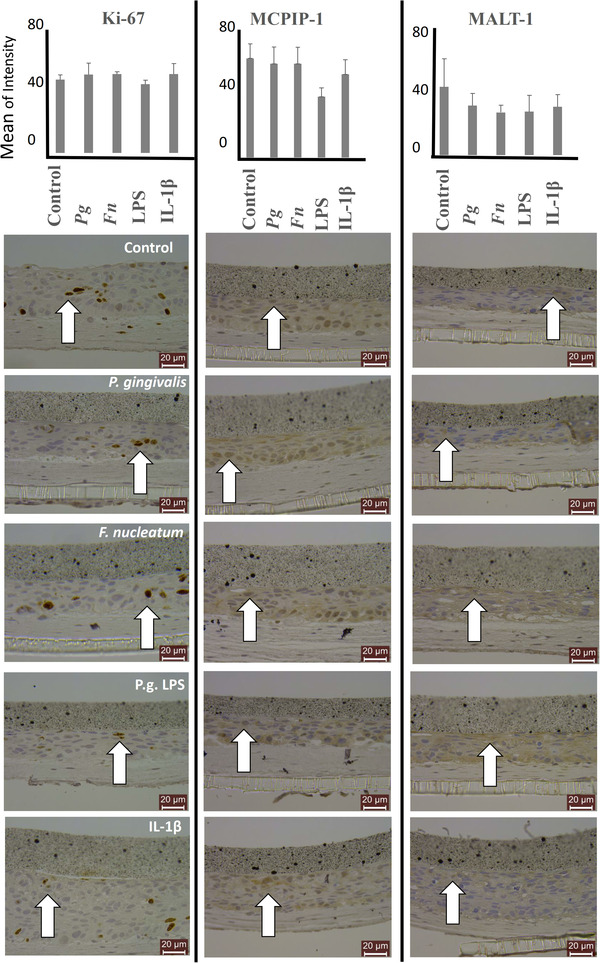
MCPIP‐1 and MALT‐1 protein expression levels of organotypic oral mucosal models after incubation with P. gingivalis, F. nucleatum, P. gingivalis LPS, and IL‐1β. Cellular proliferation is detected with Ki‐67 stainings. Bars indicate mean values and standard deviations. White arrows indicate representative positively stained cells
4. DISCUSSION
To our knowledge, the present study is the first to demonstrate the suppressive effect of F. nucleatum, IL‐1β, and P. gingivalis LPS, on MCPIP‐1 protein expressions of gingival keratinocyte monolayers. P. gingivalis, on the other hand, seemed to induce degradation of both MCPIP‐1 and MALT‐1 proteins; however, this outcome was reversed when P. gingivalis was heat‐killed. These findings indicate that oral bacteria may have both posttranscriptional and posttranslational modifications on MCPIP‐1 and MALT‐1, which supported our hypothesis. In addition, we also demonstrated the expressions of MCPIP‐1 and MALT‐1 in an organotypic oral mucosa model, which mimics the oral mucosa better than traditional on‐a‐plastic monolayer models.
One major strength of the present study was the simultaneous analysis of mRNA and protein expressions of MCPIP‐1 and MALT‐1 in gingival keratinocyte monolayers. We also applied different stimulation intervals to follow time‐dependent changes. Due to the shifts in cell numbers with time, different amounts of proteins were loaded in immunoblots for each time point. Thus, present results do not allow us to follow the time‐dependent changes. Another major strength was the use of organotypic oral mucosal culture models. Organotypic oral mucosal model contains gingival keratinocytes and gingival fibroblasts and allows to implement cellular interactions between these two cell types into the study design. One limitation of the applied organotypic oral mucosa model was with its short test period in comparison to the monolayer model. The present organotypic oral mucosa‐infection model forces bacteria to be in direct contact with the air. This is significantly different from the monolayer model, in which the human cells and bacteria are in a liquid environment. Due to its strict anaerobic character, P. gingivalis cannot survive in the presence of oxygen. On the other hand, as we observed in our previous studies, F. nucleatum can survive and increase its numbers in aerobic environments. 20 Due to their distinct characteristics in terms of oxygen tolerance, we limited the culture time of the organoid oral mucosa‐infection model with F. nucleatum and P. gingivalis for 2 h.
Previous studies indicated that proinflammatory mediators, such as LPS, IL‐1β, and bacteria can activate MCPIP‐1 gene expression. 23 , 24 According to the present results, P. gingivalis seems to induce the breakdown of MCPIP‐1. It was very recently demonstrated that P. gingivalis cleaves gingival keratinocyte MCPIP‐1 with its gingipain activity. 10 Our study indicated that the MCPIP‐1 degradation can be inhibited by heat‐killing of P. gingivalis, but not with the use of protease inhibitors. According to previous studies’ results, serine and cysteine protease inhibitions were not sufficient to stop the degradative activity of P. gingivalis. 10 However, heat‐killing of P. gingivalis inhibited its actin degradation ability, suppressed cytokine response activation, and also downregulated its IL‐1β stimulation capability from host cells. 25 Structural actin is also a target for P. gingivalis gingipains, and P. gingivalis ATCC 33277 and W50 strains can degrade actins as early as 1 h. 26 As demonstrated in our results, MCPIP‐1, MALT‐1, and β‐actin degradations were inhibited when P. gingivalis was heat‐killed. On the other hand, the altered recognition of heat‐killed bacteria may change innate immune responses therefore this method has limitations to follow the cellular responses of human cells against bacteria. 27 Moreover, MCPIP‐1 breakdown was not observed in the organotypic oral mucosal models, indicating that the stratification of keratinocytes allows them to defend themselves against bacterial proteolytic activities by limiting the adherence and invasion of bacteria and keeping bacteria and its proteases away from the proliferating basal cells. 28 An interesting finding of the present study was that the incubation of HMK monolayers with F. nucleatum suppressed MCPIP‐1 protein expression, while at the same time MCPIP‐1 mRNA levels elevated at all tested time points. An explanation of this phenomenon can be the degradation of MCPIP‐1 by F. nucleatum, as described previously. 29 , 30 , 31 Inverse correlations between mRNA and protein expressions due to post‐ or transcriptional modifications involved in turning mRNA into protein or proteolytic degradation of the proteins had been described previously. 32 Yet, in the limits of this study, the reason behind this inverse relationship is left unexplained.
Based on our results, P. gingivalis degrades HMK MALT‐1 proteins and this outcome can be inhibited by heat‐killing of P. gingivalis. F. nucleatum, P. gingivalis LPS, and IL‐1β suppress MALT‐1 protein levels only after short (2 h) incubation. It was demonstrated that MALT‐1 regulates the expression of proinflammatory cytokines in keratinocytes and MALT‐1 protease activity is an essential part of innate and adaptive responses. 33 , 34 , 35 , 36 MALT‐1 activity is controlled by antigen receptor ligation and monoubiquitination; however, contribution of inflammatory cytokines or bacterial mediators to MALT‐1 activity is unknown. 37 , 38 , 39 , 40 Moreover, to our knowledge, there is no study to show the modulation of MALT‐1 expression or activity by periodontal bacteria or in periodontitis. Yet, early (2 h) suppression of MALT‐1 and late (24 h) suppression of MCPIP‐1 protein expressions by inflammatory and infection mediators may indicate that the activation of these two inter‐related proteins is not simultaneous. The present study limited itself to three time points (2, 6, and 24 h) It was previously demonstrated that the bacteria‐induced IL‐1β and IL‐8 expressions of gingival epithelial cells reach their peak levels after 6‒12 h incubations and stay steady after 24 h. 41 , 42 Moreover it was also shown that the degradation of MCPIP‐1 by P. gingivalis starts as early as 30 min. 10 Further studies with longer incubation periods can be beneficial to demonstrate the time‐dependent changes in MCPIP‐1, MALT‐1, and proinflammatory cytokine interactions.
5. CONCLUSIONS
F. nucleatum, P. gingivalis, and IL‐1β dysregulate gingival keratinocyte MCPIP‐1 and MALT‐1 protein expression profiles. While it is difficult to clinically interpret the results of this in vitro study, our findings may propose that periodontitis‐associated bacteria‐induced modifications in MCPIP‐1 and MALT‐1 responses can be a part of periodontal disease pathogenesis. Well‐optimized MALT‐1/MCPIP‐1 gene silencing or knockout models will elucidate the contributions of MALT‐1 and MCPIP‐1 on cellular responses of gingival keratinocytes and will portray causal relationships between MALT‐1 activity and MCPIP‐1 degradation during infection and inflammation.
CONFLICT OF INTEREST
The authors have no conflicts of interest related to this study.
AUTHOR CONTRIBUTIONS
Yigit Firatli performed the laboratory and data analysis, and manuscript writing. Erhan Firatli was responsible for the supervision, data interpretation, and critical editing of the manuscript. Vuokko Loimaranta and Samira Elmanfi performed the laboratory analysis and critical editing of the manuscript. Ulvi K. Gursoy was responsible for the supervision, funding acquisition, project administration, and critical editing of the manuscript.
Supporting information
Supporting Information
Supporting Information
ACKNOWLEDGMENTS
This study is supported by Finnish National Agency for Education (EDUFI), Helsinki, Finland (Dr. Yigit Firatli ‐ Finnish Government Scholarship Pool KM‐20‐11377); University of Turku Joint Research Grant Fund, Turku, Finland (Dr. Yigit Firatli); SHS The Finnish Dental Society Apollonia, Helsinki, Finland (Dr. Ulvi K. Gursoy); and Minerva Foundation, Helsinki, Finland (Dr. Ulvi K. Gursoy). The authors are grateful for the skillful technical assistance of Katja Sampalahti, Oona Hällfors, and Tatjana Peskova from the University of Turku.
Firatli Y, Firatli E, Loimaranta V, Elmanfi S, Gürsoy UK. Regulation of gingival keratinocyte monocyte chemoattractant protein‐1‐induced protein (MCPIP)‐1 and mucosa‐associated lymphoid tissue lymphoma translocation protein (MALT)‐1 expressions by periodontal bacteria, lipopolysaccharide, and interleukin‐1β. J Periodontol. 2023;94:130–140. 10.1002/JPER.22-0093
Footnotes
SFM‐X, Gibco, Thermo Fisher Scientific, Waltham, MA, USA
Whitley A35 Anaerobic Workstation, Don Whitley Scientific, West Yorkshire, UK
Cat #tlrl‐ppglps Invivogen, San Diego, CA, USA
Recombinant Human IL‐1β cat # RH‐P0168, Genemed Synthesis Inc., San Antonio, TX, USA
Cat. # 11836170001, Roche Diagnostics, Mannheim, Germany
Invitrogen Cat. No 15596‐018, CA, USA
CellTiter 96 AQueous Non‐Radioactive Cell Proliferation Assay, Promega, WI, USA
Bio‐Rad, Hercules, CA, USA
Trans‐Blot Turbo Transfer System, Bio‐Rad, Hercules, CA, USA
Cat. # PA5‐24458, Thermo Fisher, USA
Cat. # PA5‐79622, Thermo Fisher, China
Cat. # 31460, Thermo Fisher, USA
Novex ECL Chemiluminescent Substrate Reagent Kit, Invitrogen, CA, USA
ChemiDoc MP Imaging System, Bio‐Rad, Hercules, CA, USA
Cat. # PA1 16889, Thermo Fisher, USA
ImageJ, National Institutes of Health, Bethesda, MD, USA
NanoDrop Lite Spectrophotometer, Thermo Fisher Scientific, Vantaa, Finland
Vitrogen, Cohesion Technologies, Palo Alto, CA, USA
ThinCert, Greiner Bio‐One, LA, USA
The Lab Vision Autostainer automated immunostaining system, Lab Vision Corporation, Europe
Cat. # 31460, Thermo Fisher, USA
Leica DMLB, Leica, Wetzlar, Germany
Leica DC 300 V 2.0, Leica, Wetzlar, Germany
ImageJ version 1.46c with immunohistochemistry image analysis toolbox plugin version 2; Rasband WS, National Institutes of Health, Bethesda, MD, USA
SPSS Statistics, version 26, IBM, Armonk, NY, USA
REFERENCES
- 1. Könönen E, Gursoy M, Gursoy UK. Periodontitis: A multifaceted disease of tooth‐supporting tissues. J Clin Med. 2019;8(8):1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hajishengallis G, Lamont RJ. Breaking bad: manipulation of the host response by Porphyromonas gingivalis . Eur J Immunol. 2014;44(2):328‐338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hajishengallis G, Diaz PI. Porphyromonas gingivalis: Immune subversion activities and role in periodontal dysbiosis. Curr Oral Health Rep. 2020;7(1):12‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peyyala R, Emecen‐Huja P, Ebersole JL. Environmental lead effects on gene expression in oral epithelial cells. J Periodontal Res. 2018;53(6):961‐971. [DOI] [PubMed] [Google Scholar]
- 5. Brian WB, Darveau R. Porphyromonas gingivalis lipopolysaccharide: an unusual pattern recognition receptor ligand for the innate host defense system. Acta Odonta Scand. 2001;59(3), 131‐138. [DOI] [PubMed] [Google Scholar]
- 6. Kornman KS, Page RC, Tonetti MS. The host response to the microbial challenge in periodontitis: assembling the players. Periodontol 2000. 1997;14:33‐53. [DOI] [PubMed] [Google Scholar]
- 7. Nakayama M, Ohara N. Molecular mechanisms of Porphyromonas gingivalis‐host cell interaction on periodontal diseases. Jpn Dent Sci Rev. 2017;53:134‐140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khan SA, Kong EF, Meiller TF, MA Jabra‐Rizk. Periodontal diseases: bug induced, host promoted. PLoS Pathog. 2015;11(7):e1004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fischer M, Weinberger T, Schulz C. The immunomodulatory role of regnase family RNA‐binding proteins. RNA Biology. 2020;17:1721‐1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gasiorek A, Dobosz E, Potempa B, et al. Subversion of lipopolysaccharide signaling in gingival keratinocytes via MCPIP‐1 degradation as a novel pathogenic strategy of inflammophilic pathobionts. mBio 2021;12:e00502‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Suzuki HI, Arase M, Matsuyama H, et al. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol Cell 2011;44:424‐436. [DOI] [PubMed] [Google Scholar]
- 12. Liang J, Saad Y, Lei T, et al. MCP‐induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF‐kappaB signaling. J Exp Med. 2010;207(13):2959‐2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jura J, Skalniak L, Koj A. Monocyte chemotactic protein‐1‐induced protein‐1 (MCPIP1) is a novel multifunctional modulator of inflammatory reactions. Biochim Biophys Acta. 2012;1823(10):1905‐1913. [DOI] [PubMed] [Google Scholar]
- 14. Dobosz E, Wadowska M, Kaminska M, et al. MCPIP‐1 restricts inflammation via promoting apoptosis of neutrophils. Front Immunol. 2021;12:627922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jin Z, Zheng E, Sareli C, Kolattukudy PE, Niu J. Monocyte chemotactic protein‐induced protein 1 (MCPIP‐1): A key player of host defense and immune regulation. Front Immunol. 2021;12:727861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Y, Huang X, Huang S, et al. Central role of myeloid MCPIP1 in protecting against LPS‐induced inflammation and lung injury. Signal Transduct Target Ther. 2017;2:17066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Uehata T, Iwasaki H, Vandenbon A, et al. Malt1‐induced cleavage of regnase‐1 in CD41 helper T cells regulates immune activation. Cell 2013;153:1036–1049. [DOI] [PubMed] [Google Scholar]
- 18. Mäkelä M, Larjava H, Pirilä E, et al. Matrix metalloproteinase 2 (gelatinase A) is related to migration of keratinocytes. Exp Cell Res. 1999;251:67‐78. [DOI] [PubMed] [Google Scholar]
- 19. Oksanen J, Hormia M. An organotypic in vitro model that mimics the dento‐epithelial junction. J. Periodontol. 2002;73:86–93. [DOI] [PubMed] [Google Scholar]
- 20. Gursoy UK, Pöllänen M, Könönen E, Uitto VJ. Biofilm formation enhances the oxygen tolerance and invasiveness of Fusobacterium nucleatum in an oral mucosa culture model. J Periodontol. 2010;81:1084–1091. [DOI] [PubMed] [Google Scholar]
- 21. Pöllänen MT, Gursoy UK, Könönen E, Uitto VJ. Fusobacterium nucleatum biofilm induces epithelial migration in an organotypic model of dento‐gingival junction. J Periodontol. 2012;83:1329‐1335. [DOI] [PubMed] [Google Scholar]
- 22. Gursoy UK, Salli K, Söderling E, Gursoy M, Hirvonen J, Ouwehand AC. Regulation of hBD‐2, hBD‐3, hCAP18/LL37, and proinflammatory cytokine secretion by human milk oligosaccharides in an organotypic oral mucosal model. Pathogens. 2021;11:10‐739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mizgalska D, Wegrzyn P, Murzyn K, et al. Interleukin‐1‐inducible MCPIP protein has structural and functional properties of RNase and participates in degradation of IL‐1beta mRNA. FEBS J. 2009;276(24):7386‐7399. [DOI] [PubMed] [Google Scholar]
- 24. Blazusiak E, Florczyk D, Jura J, Potempa J, Koziel J. Differential regulation by Toll‐like receptor agonists reveals that MCPIP1 is the potent regulator of innate immunity in bacterial and viral infections. J Innate Immun. 2013;5(1):15‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stathopoulou PG, Benakanakere MR, Galicia JC, Kinane DF. Epithelial cell pro‐inflammatory cytokine response differs across dental plaque bacterial species. J Clin Periodontol. 2010;37(1):24‐29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kinane JA, Benakanakere MR, Zhao J, Hosur KB, Kinane DF. Porphyromonas gingivalis influences actin degradation within epithelial cells during invasion and apoptosis. Cell Microbiol. 2012;14(7):1085‐1096. [DOI] [PubMed] [Google Scholar]
- 27. Strunk T, Richmond P, Prosser A, et al. Method of bacterial killing differentially affects the human innate immune response to Staphylococcus epidermidis . Innate Immun. 2011;17(6):508‐516. [DOI] [PubMed] [Google Scholar]
- 28. Groeger SE, Meyle J. Epithelial barrier and oral bacterial infection. Periodontol 2000. 2015;69:46‐67. [DOI] [PubMed] [Google Scholar]
- 29. Musson R, Szukała W, Jura J. MCPIP1 RNase and its multifaceted role. Int J Mol Sci. 2020;21:7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tefiku U, Popovska M, Cana A, et al. Determination of the role of Fusobacterium nucleatum in the pathogenesis in and out the mouth. Prilozi, 2020;41:87–99. [DOI] [PubMed] [Google Scholar]
- 31. Kang W, Jia Z, Tang D, Zhang Z, Gao H, He K, Feng Q. Fusobacterium nucleatum facilitates apoptosis, ROS generation, and inflammatory cytokine production by activating AKT/MAPK and NF‐κB signaling pathways in human gingival fibroblasts. Oxid Med Cell Longev. 2019;13;1681972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schmitt A, Grondona P, Maier T, et al. MALT1 Protease activity controls the expression of inflammatory genes in keratinocytes upon zymosan stimulation. J Invest Dermatol. 2016;136(4):788‐797. [DOI] [PubMed] [Google Scholar]
- 34. Zhang S, Wang M, Wang C, et al. Intrinsic abnormalities of keratinocytes initiate skin inflammation through the IL‐23/T17 axis in a MALT1‐dependent manner. J Immunol. 2021;15;206(4):839‐848. [DOI] [PubMed] [Google Scholar]
- 35. Li Y, Huang S, Huang X, et al. Pharmacological inhibition of MALT1 protease activity suppresses endothelial activation via enhancing MCPIP1 expression. Cell Signal. 2018;50:1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yu JW, Hoffman S, Beal AM, Dykon A, et al. MALT1 protease activity is required for innate and adaptive immune responses. PLoS One. 2015;12:10(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McAllister‐Lucas LM, Finally Lucas PC., MALT1 is a protease! Nat Immunol. 2008;9:231‐233. [DOI] [PubMed] [Google Scholar]
- 38. Qin H, Wu T, Liu J, et al. MALT‐1 inhibition attenuates the inflammatory response of ankylosing spondylitis by targeting NF‐κB activation. Injury 2021;52(6):1287‐1293. [DOI] [PubMed] [Google Scholar]
- 39. Pelzer C, Cabalzar K, Wolf A, Gonzalez M, Lenz G, Thome M. The protease activity of the paracaspase MALT1 is controlled by monoubiquitination. Nat Immunol. 2013;14(4):337‐345. [DOI] [PubMed] [Google Scholar]
- 40. Monajemi M, Pang YCF, Bjornson S, Menzies SC, van Rooijen N, Sly LM. Malt1 blocks IL‐1β production by macrophages in vitro and limits dextran sodium sulfate‐induced intestinal inflammation in vivo. J Leukoc Biol. 2018;104:557‐572. [DOI] [PubMed] [Google Scholar]
- 41. Sfakianakis A, Barr CE, Kreutzer D. Mechanisms of Actinobacillus actinomycetemcomitans‐induced expression of interleukin‐8 in gingival epithelial cells. J Periodontol. 2001;72:1413‐1419. [DOI] [PubMed] [Google Scholar]
- 42. Giacaman RA, Asrani AC, Ross KF, Herzberg MC. Cleavage of protease‐activated receptors on an immortalized oral epithelial cell line by Porphyromonas gingivalis gingipains. Microbiology (Reading). 2009;155:3238‐3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information


