Abstract
Background
Impaired microbial development and decreased levels of short‐chain fatty acids, particularly butyrate, is suggested to have a role in the development of atopic dermatitis (AD).
Methods
Faecal microbiota composition, abundance of selected bacterial groups and fermentation metabolites were compared at 90, 180 and 360 days of life between 27 children who developed AD by age one (AD group), and 39 controls (non‐AD group) among the CARE (Childhood AlleRgy, nutrition and Environment) study cohort.
Results
Diversity within the Firmicutes and Bacteroidetes phyla in the faecal microbiota was lower in the AD group compared with the non‐AD group. Longitudinal analysis showed multiple amplicon sequence variants (ASV) within the same bacterial family to be differentially abundant. Namely, Ruminococcus bromii, a keystone primary starch degrader, and Akkermansia muciniphila, a mucin‐utilizer, had lower abundance among the AD group. Children with AD were less likely to have high levels of faecal butyrate at 360 days compared with those without AD (11.5% vs 34.2%). At 360 days, children with high abundance of R. bromii had higher level of butyrate as well as lower proportion of children with AD compared to children with low abundance of R. bromii (11.1–12.5% vs 44.4–52.5%), which was independent of the abundance of the major butyrate producers.
Conclusion
Our results suggested that R. bromii and other primary degraders might play an important role in the differences in microbial cross‐feeding and metabolite formation between children with and without AD, which may influence the risk of developing the disease.
Keywords: atopic dermatitis, butyrate, microbiota, resistant starch, short‐chain fatty acid
Children who developed atopic dermatitis (AD) in infancy were less likely to have high faecal butyrate levels at 1 year old, compared to children without AD. The abundance of Ruminococcus bromii, a keystone starch degrader, was lower in children who developed AD than in children without AD, while the abundance of major butyrate producers was similar. Abbreviations: AD, atopic dermatitis; CARE study, Childhood AlleRgy, nutrition and Environment study; R. bromii, Ruminococcus bromii

Abbreviations
- AD
-
atopic dermatitis
CARE study, Childhood Allergy, nutrition and Environment study
R. bromii, Ruminococcus bromii
SCFA
short‐chain fatty acid
1. INTRODUCTION
During the first year of life, the gut microbiome develops from a relatively simple composition to a more complex adult‐like pattern. 1 , 2 In the neonatal period, the genus Bifidobacterium including species that are characterized by their adaptation to utilize the non‐digestible carbohydrates in breast milk, such as human milk oligosaccharides (HMO), are prevalent. There is a compositional shift around the time of solid food introduction to a more diverse microbiota, dominated by species of the phyla Bacteroidetes or Firmicutes. 1 , 2 This change within the microbial community increases microbial functionality as shown by the changes in faecal levels of fermentation metabolites, such as short‐chain fatty acids (SCFA), which are mostly acetate, butyrate and propionate. The increase in the abundance of specialized primary degraders allows the breakdown of complex dietary and endogenous carbohydrates including resistant starch (e.g. Ruminococcus bromii) and complex carbohydrates associated with plant cell walls (e.g. members of Bacteroides and Bifidobacterium) or with endogenously produced mucin glycoproteins (e.g. Akkermansia muciniphila). 3 Primary degraders release carbohydrate mono, di‐ and oligomers and produce fermentation intermediates as well as final fermentation products, such as propionate and butyrate. The fermentation intermediates, acetate, succinate, lactate, formate and 1,2‐propanediol (1,2‐PD) can be utilized by other bacteria and further metabolized to butyrate and propionate. 4 Hence, this exchange of products between microbes, termed microbial cross‐feeding, may results in the overall increase in production of SCFAs.
Previous studies suggest that delayed gut microbial maturation is associated with the development of allergic diseases later in life. It has been shown that children with reduced microbial diversity or lower abundance of specific taxa characteristic for a particular age group, were more likely to develop asthma or atopic dermatitis (AD). 5 , 6 , 7 , 8 , 9 There is also increasing evidence suggesting that gut microbiota can affect the risk of developing allergic disease via indirect effects through the production of SCFAs. 10 , 11 Butyrate and propionate were shown to have protective effect by increasing differentiation of regulatory T cells (Tregs), which play an important role to promote and maintain tolerance. 12 , 13 , 14 In the PASTURE (Protection against Allergy Study in Rural Environments) cohort, infants who developed asthma had less “mature” gut microbiome, which was characterized by decreased abundance of taxa that are involved with butyrate production, compared with healthy children. 6 A Singaporean birth cohort study suggested that microbiota of infants with AD had a compromised developmental trajectory, with higher abundance of Escherichia coli and Klebsiella pneumoniae at 3 weeks of life followed by delayed accumulation of butyrate and propionate producers after 3 months of age compared with infants without AD. 7
These recent studies suggests that impaired microbiome development can lead to differences in metabolic activities including butyrate production, and development of allergic disease. However, there is still a gap in the knowledge on how and when this impairment occurs and on the other factors affecting butyrate production. Although dietary factors influence the gut microbiota composition and its metabolic activity, 15 , 16 few studies have investigated the effect of diet during the weaning period. The introduction of foods such as yoghurt, fish, fruit and vegetables during infancy was associated with increased level of faecal butyrate among the PASTURE cohort, suggesting the potential association between diet, development of infant microbiota and allergic disease. 17 It was therefore the aim of this study to gain a better understanding of the mechanism of how the microbiome can affect the development of AD in early life with a focus on butyrate.
Using data from a subset of the CARE (Childhood AlleRgy, nutrition and Environment) study cohort, we previously showed that Clostridium sensu stricto was among the butyrate‐forming pioneers in the gut during infancy. 18 This was followed by the emergence of groups with higher butyrate‐producing capacity, Eubacterium rectale/Roseburia spp., Faecalibacterium prausnitzii and Anaerobutyricum hallii, resulting in the increase in butyrate levels. For the current analysis, we compared the gut microbiota composition and metabolite profiles using the same data between children who developed AD in their first year of life and those who did not.
2. METHODS
2.1. Study design
CARE study is an ongoing prospective cohort study conducted in St. Gallen, Switzerland, with the aim to investigate the association between early life exposures and allergic diseases, especially AD. Healthy newborns are recruited at the Kantonsspital St. Gallen after birth and followed up by physical examination (4 months and 1 year) and questionnaires (at birth, 4 months and 1 year) to collect information on their symptoms of allergic diseases, demographic and environmental factors. Information on the child's diet is collected weekly throughout the first year of life. Written informed consent is obtained from the mother at recruitment. The study was approved by the Ethic Commission Ostschweiz (EKOS).
AD within the first year of life was defined as having either,
a doctor's diagnosis of AD or
itchy rash at specific locations reported by the parents in the 4‐month‐old or 1‐year‐old questionnaire, or
AD diagnosed by the research doctor at the 4‐month‐old or 1‐year‐old physical examination.
At the time the selection of participants was finalized in May 2019, there were 96 children who had complete follow‐up to 1 year old in the cohort. A total of 66 children, 39 children with no history of AD (non‐AD group) and 27 children with AD (AD group) in the first year of life had faecal samples available at the selected time points and were included in this nested case–control study. Demographic factors besides maternal history of allergy or the prevalence of AD were not different between children who were included in this study and those were not (data not shown). The onset of AD for the children included in this study was before 4 months for n = 11, and between 4 months and 1 year old for n = 16.
2.2. Faecal sample collection
Faecal samples collected at the following three time points, approximately at 90 days (mean ± SD, 94 ± 17 days: n = 62 for this time point), 180 days (181 ± 14 days, n = 66) and 360 days (362 ± 18 days, n = 66) were used in this project.
2.2.1. Microbiota and metabolite analysis
The method of faecal microbiota characterization has been described in our previous publication in detail. 18 Briefly, DNA was extracted from faecal samples using the Fast DNA Spin Kit for soil (MP Biomedicals, Switzerland) and analyzed using qPCR and 16S rRNA gene sequencing. qPCR was performed to determine the abundance of total bacteria and selected bacterial (functional) groups using the Roche LightCycle 480 System (Hoffmann‐La Roche, Switzerland), 18 or a CFX Connect (Biorad, Denmark). Clostridium sensu stricto, F. prausnitzii, Roseburia/E. rectale and E. hallii were previously identified as major butyrate‐producing groups in infants at 1 year of age and thus selected for our analysis. 19 Primers targeted the 16S rRNA or functional genes (Table S1). Briefly, each reaction comprised 5 μl iTaq Universal SYBR Green Supermix (Biorad, Denmark), 2 μl of forward and reverse primers (10 pmol), 1 μl of 10‐fold diluted DNA, and 2 μl of nuclease‐free water. The reaction was run using a CFX Connect instrument starting with heating at 95°C for 3 min followed by 40 cycles of denaturation at 95°C for 10 s and annealing and extension at 60°C for 30 s, and a melting curve analysis. Each run contained negative controls without template DNA. A ten‐fold dilution series of each standard was included in the runs to determine linear range and the limits of detection. For determining microbiota composition, gene sequencing targeting the 16S rRNA gene V4 hypervariable region (accession number PRJNA616703) was used. Each run included a negative control using water instead of the DNA template during library preparation. Negative controls recovered zero reads and were removed before further analysis. Sample processing is fully specified previously. 18 In short, raw reads were processed using the open‐source software package DADA2. 20 Reads with expected error rates higher than 3 and 4 for forward and reverse reads were removed. Chimeric sequences were identified and removed using the ‘consensus’ method included in the DADA2 pipeline. From n = 175 samples, we obtained 6,668,205 reads with a mean and median of 38,104 and 38,247 reads per sample respectively. Amplicon Sequence Variants (ASV) with less than 10 reads/dataset were removed before further analysis. For alpha‐diversity analysis, data was rarefied to 15,055 reads per sample to match the lowest number of reads found in a sample; richness and diversity indices were calculated at the ASV‐level. Cell counts, absolute abundance of ASVs and major bacterial families were calculated from relative abundance data and total bacteria cells counts determined using qPCR. Correction factors to account for multiple 16S rRNA gene copies were used to estimate cell numbers from gene copies as previously described. 18 We used the R package, MetaLonDa (Metagenomic Longitudinal Differential Abundance method) 21 to identify differentially abundant ASVs at different time intervals using the age in days of the child when the faecal sample was collected. This method employs a negative binomial distribution together with a semi‐parametric SS‐ANOVA to model the normalized count reads, then performs the significance testing based on unit time intervals using permutation testing procedure.
Fucose, fermentation intermediates formate, succinate, lactate, 1,2‐PD and acetate and final products propionate and butyrate present in faecal samples were quantified using high‐pressure liquid chromatography with refractive index detection. 18
2.3. Statistical analysis
Baseline characteristics were compared between children with and without AD using chi‐squared test or Fisher's exact test for categorical variables and t‐test for continuous variables. The number of observed species, Shannon index, relative and absolute abundance of major bacterial families, and the abundance of selected bacterial groups based on qPCR were compared between the two groups using Mann–Whitney U test.
Fucose and fermentation metabolites were compared between AD and non‐AD by the proportion of samples with detectable levels or with levels above the 75‐percentile value using chi‐squared test and by the observed level as continuous variables by Mann–Whitney U test at each time point. The correlations between butyrate, abundance of butyrate producers and R. bromii were assessed using Spearman correlation with Benjamini–Hochberg correction for multiple comparisons. Butyrate concentration and the proportion of children with AD by the abundance of R. bromii and butyrate producers were each compared using Dunn test with Benjamini–Hochberg adjustment and logistic regression. Chi‐squared test was used to analyze the association between demographic, dietary factors and the abundance of selected bacterial groups which were categorized at the 75‐percentile value. Statistical tests were carried out by R (http://www. R‐project.org) and figures were plotted using the ggplot package 22 and SigmaPlot 13 (Systat Sotfware Inc.).
3. RESULTS
3.1. Cohort characteristics
Children in the AD group were more likely to have history of paternal allergic disease but not maternal allergic disease compared with the non‐AD group (Table 1). There was no difference in sex, mode of delivery, antibiotics use up to 4 months, having siblings, having pets or dietary factors between the two groups.
TABLE 1.
Demographic and clinical characteristics of the children included in the analysis
| Children with no AD (non‐AD group, n = 39) % | Children with AD up to 1 year (AD group, n = 27) % | Total (n = 66) % | p‐value | ||||
|---|---|---|---|---|---|---|---|
| Sex | |||||||
| Boy | 20 | 51.3 | 13 | 48.1 | 33 | 50.0 | 0.80 |
| Girl | 19 | 48.7 | 14 | 51.9 | 33 | 50.0 | |
| Maternal history of allergic disease a | |||||||
| No | 15 | 38.5 | 9 | 33.3 | 24 | 36.4 | 0.67 |
| Yes | 24 | 61.5 | 18 | 66.7 | 42 | 63.6 | |
| Paternal history of allergic disease a | |||||||
| No | 26 | 66.7 | 10 | 37.0 | 36 | 54.5 | 0.02 |
| Yes | 13 | 33.3 | 17 | 63.0 | 30 | 45.5 | |
| Mode of delivery | |||||||
| Vaginal | 32 | 82.1 | 20 | 74.1 | 52 | 78.8 | 0.44 |
| Caesarean section | 7 | 17.9 | 7 | 25.9 | 14 | 21.2 | |
| Antibiotics use in pregnancy or up to 4 months | |||||||
| None | 33 | 84.6 | 23 | 85.2 | 56 | 84.8 | 0.95 |
| Once or more | 6 | 15.4 | 4 | 14.8 | 10 | 15.2 | |
| Any siblings | |||||||
| None | 24 | 61.5 | 20 | 74.1 | 44 | 66.7 | 0.29 |
| One or more | 15 | 38.5 | 7 | 25.9 | 22 | 33.3 | |
| Pet dog or cat during pregnancy | |||||||
| No | 28 | 71.8 | 24 | 88.9 | 52 | 78.8 | 0.10 |
| Yes | 11 | 28.2 | 3 | 11.1 | 14 | 21.2 | |
| Duration of breastfeeding (within 1 year) mean ± SD (weeks) | 34.9 ± 17.6 | 36.7 ± 16.2 | 0.67 | ||||
| Breastfeeding at 4 months b | |||||||
| No | 7 | 19.4 | 3 | 11.5 | 10 | 16.1 | 0.65 |
| Partially | 9 | 25.0 | 6 | 23.1 | 15 | 24.2 | |
| Exclusively | 20 | 55.6 | 17 | 65.4 | 37 | 59.7 | |
| Breastfeeding at 1 year b | |||||||
| No | 21 | 58.3 | 16 | 61.5 | 37 | 59.7 | 0.80 |
| yes | 15 | 41.7 | 10 | 38.5 | 25 | 40.3 | |
| Timing of solid introduction (mean ± SD, weeks) | 20.8 ± 3.0 | 19.6 ± 4.3 | 0.17 | ||||
| Numbers of food categories introduced within 1 year c (mean ± SD, max 23) | 17.2 ± 2.7 | 17.4 ± 3.3 | 0.82 | ||||
| Timing of faecal sample collection (mean ± SD, days) | |||||||
| 3‐month‐old sample (n = 62) | 92.0 ± 16.3 | 97.4 ± 18.0 | 0.23 | ||||
| 6‐month‐old sample | 180.0 ± 12.7 | 183.1 ± 15.4 | 0.37 | ||||
| 1‐year‐old sample | 361.0 ± 16.3 | 361.7 ± 19.8 | 0.86 | ||||
Note: p‐values were derived by Chi‐squared test for categorical variables and t‐test for continuous variables.
Abbreviations: AD, atopic dermatitis; SD, standard deviation.
Maternal and paternal history of allergic disease was defined as having report of atopic dermatitis, asthma, allergic rhinitis, or food allergy.
Numbers do not add up to total due to missing data for breastfeeding information (n = 4).
Foods introduced to the child's diet were categorized as the following: milk, butter, yoghurt, other dairy products, grains with and without gluten, 4 categories of vegetables, 2 categories of fruits, meat, eggs, fish or seafood, peanut, almond, other nuts, soy products, legumes, oils, sweets and spices.
3.2. Longitudinal comparison of the gut microbiota and AD
Comparison of beta‐diversity (between sample variation) between AD and non‐AD based on weighted Unifrac, which takes relative abundance into consideration, and Bray–Curtis distance metrics did not show any difference. There was a trend for a difference in unweighted Unifrac at 90 days and in Jaccard metric at 360 days, which are both methods that are more sensitive for differences in taxa with low abundance (Figure S1 and Table S1).
Alpha‐diversity (within‐sample diversity) of the total microbiota, based on the Shannon index, increased similarly among both groups over time (Figure 1). AD group had a lower diversity within the Firmicutes phylum and Bacteroidetes phylum at 180 days compared to non‐AD group, with a similar trend at 90 days for both phyla. Within the Firmicutes phylum, diversity of Clostridiaceae was lower in the AD group at 180 days with a similar trend at 90 days, and there was a trend for lower diversity of Lachnospiraceae at 360 days. The number of observed species was lower in AD compared with non‐AD for the total microbiota at 90 days (Table S3).
FIGURE 1.
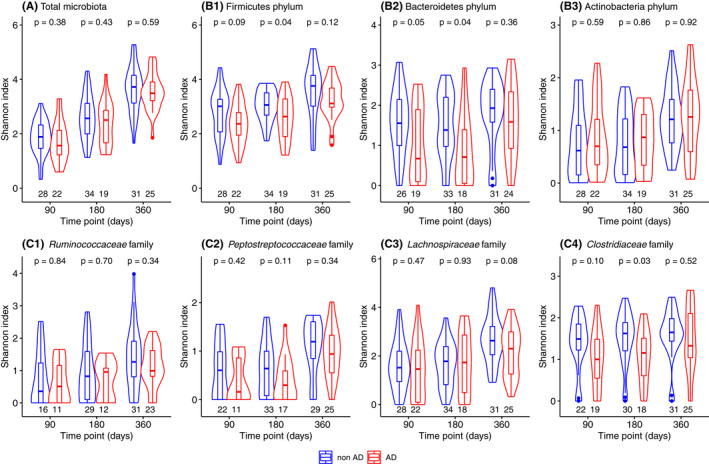
Shannon diversity index compared between children with and without atopic dermatitis in the first year of life for the (A) total microbiota, (B) phylum level: Firmicutes, Bacteroidetes and Actinobacteria, and (C) family level within the Firmicutes phylum: Ruminococcaceae, Peptostreptococcaceae, Lachnospiraceae and Clostridiaceae. Faecal microbiota composition was determined using 16S rRNA gene sequencing and samples with more than 15,000 reads were included in this analysis (n = 50, 53 and 56 at 90, 180 and 360 days, respectively). The violin plots describe the abundance with rotated kernel density plots, a marker and a box each indicating the median and interquartile range. The number on the x‐axis below each box plot describes the number of samples with species of the relevant taxa. p‐values were derived by Mann–Whitney U test
The difference in gut microbiota composition was also assessed by comparing the relative and absolute abundance of major bacterial families. Bifidobacteriaceae was the most abundant family in both groups with a relative abundance at around 70% at 90 days, which decreased to around 30% at 360 days (Figure S2). Relative abundance of Erysipelotrichaceae, Ruminococcaceae and Akkermansiaceae was lower in AD group at each 90, 180 and 360 days, respectively, compared with non‐AD (Table S4). Cell counts of Bifidobacteriaceae and Ruminococcaceae were lower in the AD group at 180 days (Table S5), with a similar trend at 360 days, while Coriobacteriaceae were lower at 360 days.
MetalonDa, which was used to assess differentially abundant ASV (which refers to single DNA sequences that are equivalent of strains) using a longitudinal method, identified ASVs of the phyla Firmicutes, Bacteroidetes and Verrumicrobia as being different in relative abundance during the first year of life between AD and non‐AD (Figure 2). Bacteroides ovatus ASV247, Lachnospiraceae ASV1315, Oscillibacter ASV1173 and Enterobacteriaceae ASV36 were dominant for more than 250 days in non‐AD, while Clostridium sensu stricto ASV914 and Terrisporobacter ASV799 were more abundant during the majority of the first year in AD. Akkermansia muciniphila ASV137 and Faecalibacterium ASV1053 became more abundant in the non‐AD group at around 350 days. R. bromii ASV1013 was more abundant in non‐AD at around 170 days and after 350 days. Several ASVs assigned to Enterococcus and Escherichia coli/Shigella were more abundant in AD group until around 300 days.
FIGURE 2.
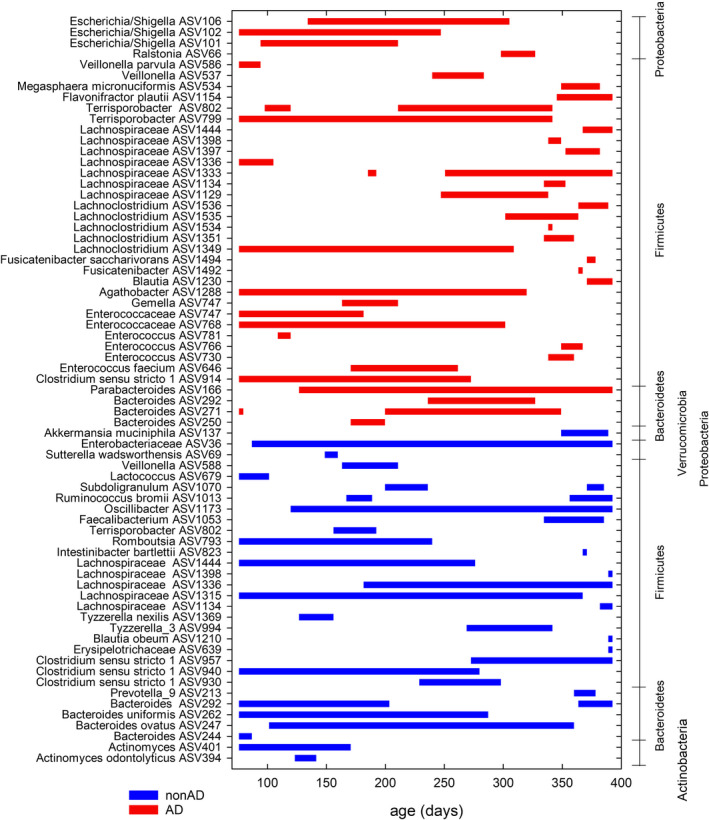
Time intervals of differentially abundant ASVs identified by MetaLonDa using the age of the samples in days. Faecal microbiota composition was determined using 16S rRNA gene sequencing (n = 168) and ASV with less than 50 reads were filtered out for the analysis. The blue lines represent the intervals where samples from children without atopic dermatitis (non‐AD group) had more reads and the red lines represent those where samples from children with atopic dermatitis (AD group) had more reads
3.3. Abundance of butyrate producers and key primary degraders using qPCR and AD
We used qPCR to quantify and compare abundance of selected butyrate producers, E. rectale/Roseburia spp., F. prausnitzii, A. hallii and Clostridium sensu stricto, since abundance levels were too low for consistent detection using 16S rRNA gene sequencing (Figure 3). Clostridium sensu stricto was the most abundant butyrate producer at 90 days and with little change in abundance at 180 and 360 days, while the abundance of the other three groups increased over time in both groups. There was a tendency of decrease in the abundance of E. rectale/Roseburia spp. in AD compared with non‐AD at 360 days but there was no difference in the abundance of other butyrate producers.
FIGURE 3.
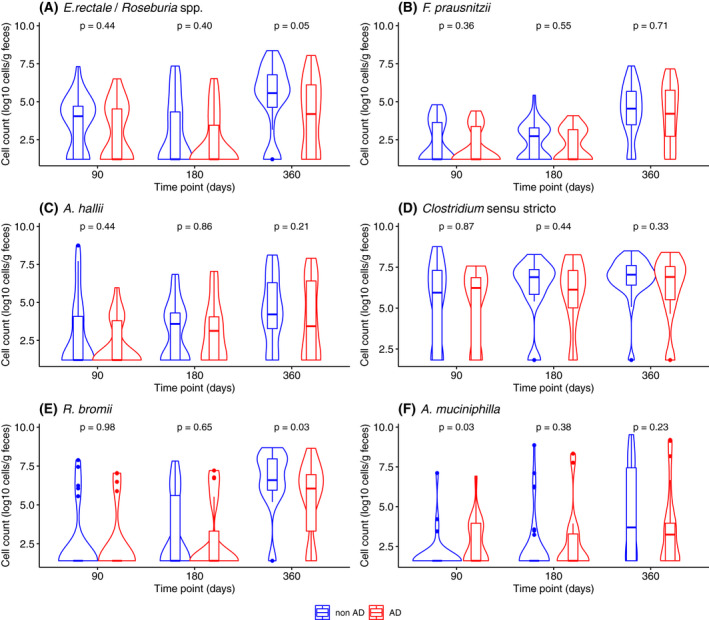
Abundance of major butyrate‐producing groups and cross‐feeders among children with and without atopic dermatitis during their first year of life quantified by qPCR. The number of samples included in this analysis was n = 48, 54 and 66 at 90, 180 and 360 days, respectively. The violin plots describe the abundance with rotated kernel density plots, a marker and a box each indicating the median and interquartile range. p‐values were derived by Mann–Whitney U test
We quantified R. bromii and A. muciniphila, which both had higher relative abundance around 360 days in non‐AD compared with AD in the MetaLonDa analysis (Figure 3). To assess the abundance of R. bromii, we quantified the genus Ruminococcus as a proxy, since R. bromii was the most abundant of this genus among the CARE cohort. The abundance of R. bromii increased during the first year of life in both groups but was lower in the AD group compared with non‐AD group at 360 days. The absolute abundance of A. muciniphila was higher in AD group at 90 days, but not different between the two groups at 180 or 360 days.
3.4. Gut microbial metabolites and AD
We compared the detectability and concentrations of propionate, butyrate and fermentation intermediates, succinate, lactate, formate, acetate and 1,2‐PD, as well as fucose (a major component of HMO and the source of 1,2‐PD) between AD and non‐AD (Figure S3 and Table S6). At 90 days, a higher proportion of samples had detectable 1,2‐PD among AD compared with non‐AD (92 vs 53%, supp table 6). There was no difference in the level of the three major SCFAs, butyrate, propionate, and acetate, or any of the other fermentation intermediates between AD and non‐AD when compared as a continuous variable. When categorized using the 75‐percentile value of the SCFAs at each time point, children in the AD group were less likely to have high level of butyrate (cut‐off at 18.86 μmol/g) at 360 days (11.5% vs 34.2%, Figure 4).
FIGURE 4.
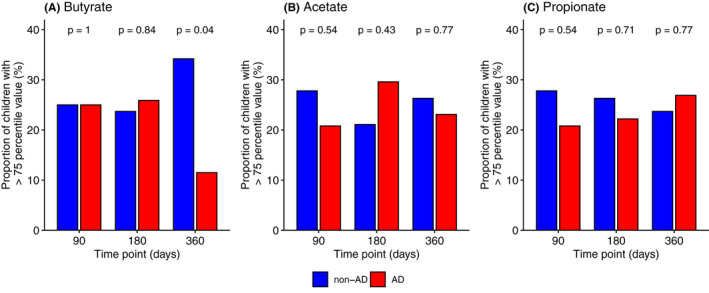
The association between the abundance of the three major short‐chain fatty acids and atopic dermatitis during the first year of life. Measurements were completed among n = 60, 65 and 64 samples at 90, 180 and 360 days, respectively. The proportion of children with high level of each butyrate, acetate and propionate was compared between the two groups using the 75‐percentile value (butyrate: 90 days 1.19 μmol/g, 180 days 4.79 μmol/g, 360 days 18.86 μmol/g, acetate: 90 days 144.31 μmol/g, 180 days 138.90 μmol/g, 360 days 120.96 μmol/g and propionate: 90 days 16.09 μmol/g, 180 days 25.29 μmol/g, 360 days 27.62 μmol/g) as a cut‐off at each time point. p‐values were derived by Chi‐squared test
3.5. Association between butyrate concentration, butyrate producers, R. bromii and AD
Based on the previous results, we explored the influence of R. bromii on the abundance of butyrate producers and butyrate levels among the 360 days sample (Figure 5). There was weak to moderate positive correlation (Spearman's rho, 0.3–0.49) between the abundance of the three major butyrate producers, E. rectale. F. prausnitzii and A. hallii, as well as with R. bromii and butyrate concentrations (Figure 5A). The abundance of Clostridium sensu stricto did not show similar correlation.
FIGURE 5.
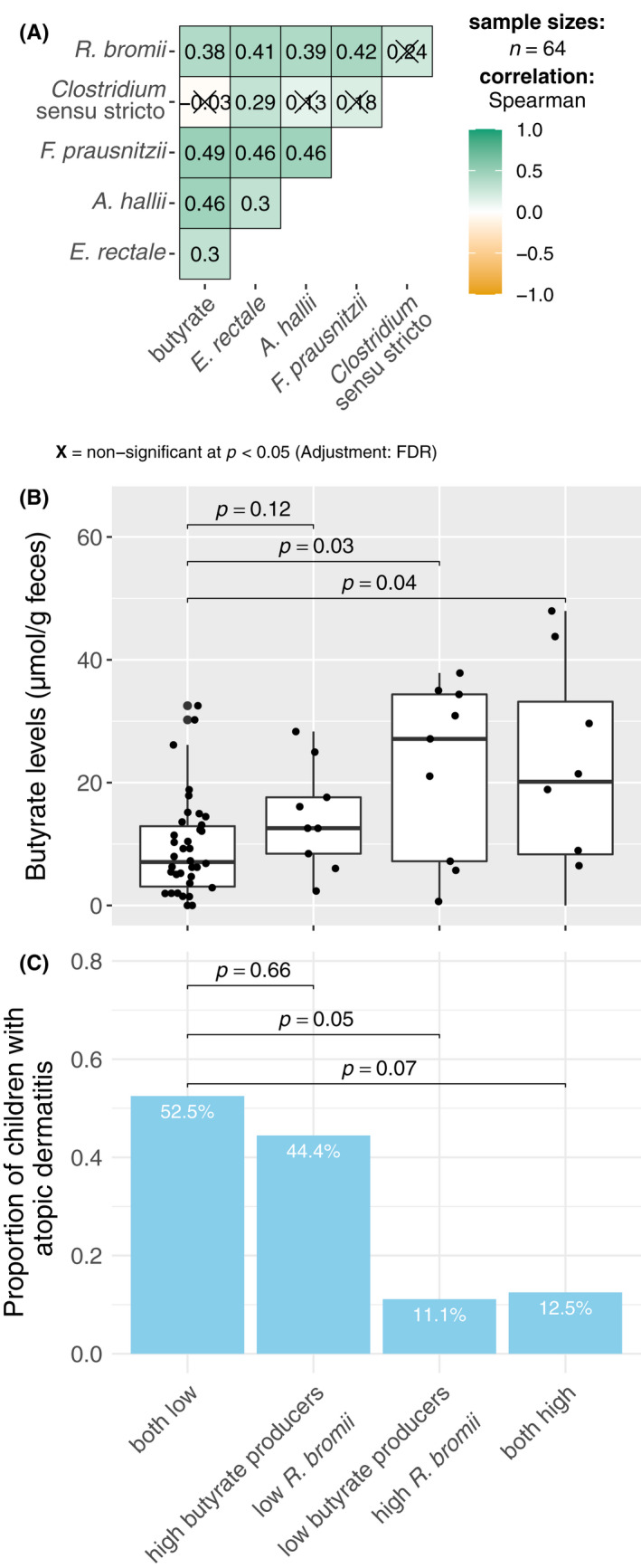
Association between butyrate level, abundance of the major butyrate producers (Clostridium sensu stricto, E. rectale, A. hallii and F. prausnitzii), R. bromii at 360 days and atopic dermatitis (AD) during the first year of life. (A) Correlation between the butyrate level, abundance based on qPCR of the major butyrate producers and R. bromii using Spearman correlation with Benjamini–Hochberg adjustment for multiple comparison (n = 64). (B) Butyrate level and the (C) proportion of children with AD by the combination of the abundance of the 3 major butyrate producers at 360 days (E. rectale, A. hallii and F. prausnitzii) and R. bromii. Children were categorized into mutually exclusive 4 groups; those with low abundance of both butyrate producers and R. bromii (n = 40), low R. bromii and high butyrate producers (n = 9), high R. bromii and low butyrate producers (n = 9), and a group having both at high levels (n = 8), using the 75‐percentile value of the abundance of the sum of the 3 butyrate producers and R. bromii (7.06 and 7.71 log10 cells/g feces, respectively) as a cut‐off. *p‐values for (B) were derived by Dunn test with Benjamini–Hochberg adjustment and for (C) were derived by logistic regression using the both low category as reference
To assess the potential interaction between the abundance of butyrate producers and R. bromii for the effect on butyrate level and risk of AD, we categorized the children into four mutually exclusive groups based on the 75‐percentile of the sum of the abundance of E. rectale. F. prausnitzii and A. hallii, and R. bromii (7.1 and 7.7 log10 cells/g feces, respectively); group 1: children with low abundance (<75‐percentile) of both butyrate producers and R. bromii (n = 40), group 2: high (>75 percentile) of butyrate producers and low R. bromii (n = 9), group 3: low butyrate producers and high R. bromii (n = 9) and group 4: children with high abundance of both (n = 8). Compared with group 1, the groups with high abundance of R. bromii (groups 3 and 4) showed higher level of butyrate, independent of the abundance of the butyrate producers (Figure 5B). Similarly, the proportion of children with AD was lower in the group with high abundance of R. bromii (group 3: 11.1% [1/9], p = 0.05 and group 4: 12.5% [1/8], p = 0.07), compared with group 1 (52.5% [21/40]), (Figure 5C).
3.6. Association between environmental factors, dietary factors and butyrate level, the abundance of butyrate producers and R. bromii
We explored the effect of several known environmental factors that affect the microbiota and dietary factors, butyrate level, the abundance of the three main butyrate producers (E. rectale. F. prausnitzii and A. hallii) and R. bromii at 360 days (Table S7 and Figure S4). Children with siblings were more likely to have higher butyrate concentration and a trend for increased abundance of A. hallii. Other environmental factors examined were not associated with any of the outcomes. Children with longer consumption of yoghurt or fish within the first year of life had a higher level of butyrate (Figure S4).
4. DISCUSSION
Our analysis of data from a longitudinal birth cohort study showed differences in faecal microbiota development and in fermentation activity between children with AD and those without AD during the first year of life.
The shift from the Bifidobacteriaceae‐dominated microbial community to a more complex composition occurred similarly in AD and non‐AD groups in terms of the overall microbial community composition (beta‐diversity) with a trend of difference found at several time points using diversity indexes that give more weight to taxa with low abundance. Furthermore, the abundance of several bacterial families such as Erysipelotrichaceae, Ruminococcaceae and Akkermansiaceae was lower in the AD group compared with the non‐AD group at different time points. Additionally, diversity (alpha‐diversity) within the Firmicutes and Bacteroidetes phyla was lower in the AD group.
Previous studies on the composition of gut microbiota during early childhood and allergic disease have shown inconsistent results, 23 which might be due to the difference in study population, timing, methods of sample analysis and the taxonomic levels reported. In fact, in our MetaLonDa analysis, multiple species within the same bacterial family were differentially abundant at various timings and durations. This suggests that the key taxa characterizing the microbiota of non‐AD or AD children are at the lower taxonomic level and for specific time intervals throughout the first year of life. Up to 300 days, infants with AD had increased abundance of several bacteria belonging to Enterococcus and Escherichia coli/Shigella, both classical pioneer species that create an anaerobic environment to promote subsequent colonization of other anaerobes. 2 Increased abundance of E. coli in the microbiota of children who develop AD has been reported 7 , 24 and was suggested to impact the successive establishment of other functional groups including butyrate or propionate producers. Colonization by E. coli has also been suggested to contribute to impairment of intestinal permeability and increased immune activation. 25 , 26
Children with AD in this study were less likely to have high butyrate levels at 360 days. This is in line with previous studies, among which several have also reported reduced abundance of butyrate producers in children with allergic diseases. 7 , 27 , 28 , 29 , 30 In our analysis, one of the major butyrate producers, E. rectale/Roseburia spp., was marginally decreased among AD children at 360 days. Additionally, we found several primary degraders with possible association to butyrate production, to be decreased in children with AD. Among these was the genus Ruminococcus, which was represented by R. bromii, that has also been reported to be less abundant in children who developed sensitization among a Canadian birth cohort. 27 E. rectale/Roseburia spp. and R. bromii are among the main bacteria that degrade resistant starch (RS) 31 , 32 and R. bromii is the key microbe in the initial breakdown of RS, which leads to microbial cross‐feeding and enhancing butyrate production. 32 , 33 , 34 We found higher level of butyrate and lower proportion of AD among children with high abundance of R. bromii (Figure 5), although it was among a small group of children (n = 17) and there was no clear interaction between the abundance of butyrate producers and R. bromii in relation to butyrate levels or the risk of AD. Nevertheless, our results suggested that the abundance of R. bromii is an important factor in determining faecal butyrate level and potentially the risk of developing AD. Other primary degraders that were less abundant among the AD group in the MetaLonDa analysis at around 360 days were A. muciniphila, a keystone species which degrades mucin glycans 35 and Bacteroides uniformis, a highly glycolytic and potential mucin degrader 36 up to around 300 days. Reduced abundance of mucin utilizers has been also reported among children with allergic disease. 37 , 38 Additionally, 1,2‐PD, a fermentation metabolite that originates from dietary fibre, fucosylated HMOs and host mucins, which are compositionally and structurally similar, and is converted to propanol and propionate, 39 was higher in AD than non‐AD in our study. Taken together, our results suggest some disturbance in the fermentation pathway of fucosylated HMO or mucin in children with AD. The abundance of A. muciniphilla based on qPCR was not different between AD and non‐AD or correlated with butyrate levels at 360 days (data not shown), when butyrate level was different between the two groups. However, we cannot exclude the possibility of other primary degraders besides R. bromii playing a role for the difference in butyrate production. Similarly, although the four bacterial groups we assessed have been reported to be the major butyrate‐producing bacteria among this age group, 19 it is possible that the reduced butyrate production in children with AD was affected via the difference in other butyrate producers or pathways and our results will need to be confirmed in future studies.
Many of the known risk factors for allergic disease, such as pet exposure, antibiotics use, birth order and mode of delivery, are known to affect the microbiota 40 , 41 and may be a potential confounder for the association between microbiota composition, butyrate level and AD. In this study, children with siblings were more likely to have higher butyrate concentration, similar to the results from the PASTURE study. 17 Although this was not associated with the outcome of AD in our study population, the potential confounding by these factors along with genetic factors such as family history of disease will need to be assessed in future studies with larger sample size. As for dietary factors, our analyses showed that children with longer duration of yoghurt or fish consumption had a higher level of butyrate, which is in line with previous reports. 17 , 42 We did not see a clear association with vegetable and fruit consumption, which may have been affected by the limited sample size. Based on the results from several intervention studies that have shown the abundance of R. bromii to increase in response of RS rich diet, 33 , 34 the increased abundance of these bacterial groups in the non‐AD group may have been due to the increased amount of RS intake. However, we were unable to assess this because we did not have this information. Notwithstanding the limitations of our exploratory analysis, our results suggest that this possible effect of several food items on butyrate level or its productivity is an important area of future research.
The strength of this analysis is that faecal samples were collected at multiple time points that allowed comparison of the composition and functionality of the microbiota throughout the first year of life. There are several limitations that need to be considered. Among the n = 27 children in the AD group, the onset was before 4 months for n = 11, and between 4 months and 1 year old for n = 16. We were limited in statistical power to detect a difference in the 90‐ and 180‐day sample including only children who developed AD between 4 months and 1 year old to assess the temporality of the associations. Furthermore, the differences we have shown in the abundance of several bacterial groups or butyrate level in our original analysis was only among the 360 days sample. So, it is possible that the differences that we have found are not causal but rather a consequence of the disease. The varying onset within the AD group may have also affected the comparison between AD and non‐AD by reducing the difference in the two groups and underestimating the effect of the exposures of interest. Most of the AD cases had mild disease and many with unknown sensitization status due to missing data, which may have also led to underestimation of the effect if there was a dose‐responsive association with the severity of AD or with concomitant sensitization. Finally, by conducting multiple comparisons we have increased the possibility of a type I error, and our findings will need to be confirmed in future studies.
In conclusion, the decreased abundance of several primary degraders and the accompanying difference in metabolic activity, namely the starch degrader, R. bromii and butyrate, might be a potential mechanistic explanation for the association between early life nutrition and the gut microbiome in the development of AD.
AUTHOR CONTRIBUTIONS
CS, RF, ReFCR and the CK‐CARE study group were involved in the conception of the study. CS, RF, ReF and CR designed the study. RuF, EB, CA, ReF and CR were involved in the collection of samples and clinical information. CS, ARG and QL have conducted the faecal microbial analysis. MS, CS, ARG and QL performed the statistical analysis. MS and CS wrote the manuscript. All authors contributed to data interpretation, critically appraised the manuscript and approved the final version.
CONFLICT OF INTEREST
All authors have no conflict of interest to declare in relation to this study.
Supporting information
Appendix S1
ACKNOWLEDGMENTS
We thank all the children and their parents for their participation. We are grateful to Olivia Appert and the CARE study team, Susanne Loeliger, Neeta Bühler, Bigna Fischer, Tanja Wachinger and Petra Schürmann for their contribution to the study. The CARE study is funded by the Kühne Foundation. Clarissa Schwab acknowledges funding from the Aarhus Universitets Forskningsfond. Part of the original data collection was supported by the Biostime Institute for Nutrition and Care ‐ Geneva Funding programs. Open access funding provided by Universitat Zurich.
APPENDIX A.
CK‐CARE study group
Thomas Bieber1,2; Peter Schmid‐Grendelmeier1,3; Claudia Traidl‐Hoffmann1,4,5; Marie‐Charlotte Brüggen1,3,6,7; Claudio Rhyner1,2,8
1Christine Kühne‐Center for Allergy Research and Education Davos (CK‐CARE), Davos, Switzerland
2Davos Biosciences, Davos, Switzerland
3Allergy Unit, Department of Dermatology, University Hospital of Zürich, Zürich, Switzerland
4Environmental Medicine, Faculty of Medicine, University of Augsburg, Augsburg, Germany
5Institute of Environmental Medicine, Helmholtz Zentrum Muenchen, German Research Center for Environmental Health, Augsburg, Germany
6Hochgebirgsklinik Davos, Davos, Switzerland
7Faculty of Medicine, University of Zürich, Zürich, Switzerland
8Swiss Institute of Allergy and Asthma Research (SIAF), Davos, Switzerland
Sasaki M, Schwab C, Ramirez Garcia A, et al. The abundance of Ruminococcus bromii is associated with faecal butyrate levels and atopic dermatitis in infancy. Allergy. 2022;77:3629‐3640. doi: 10.1111/all.15440
Mari Sasaki, Clarissa Schwab, Remo Frei and Caroline Roduit contributed equally to this work.
CK‐CARE study group members are listed in Appendix A.
REFERENCES
- 1. Moore RE, Townsend SD. Temporal development of the infant gut microbiome. Open Biol. 2019;9(9):190128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bäckhed F, Roswall J, Peng Y, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17(5):690‐703. [DOI] [PubMed] [Google Scholar]
- 3. Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3(4):289‐306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. 2017;19(1):29‐41. [DOI] [PubMed] [Google Scholar]
- 5. Stokholm J, Blaser MJ, Thorsen J, et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun. 2018;9(1):141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Depner M, Taft DH, Kirjavainen PV, et al. Maturation of the gut microbiome during the first year of life contributes to the protective farm effect on childhood asthma. Nat Med. 2020;26(11):1766‐1775. [DOI] [PubMed] [Google Scholar]
- 7. Ta LDH, Chan JCY, Yap GC, et al. A compromised developmental trajectory of the infant gut microbiome and metabolome in atopic eczema. Gut Microbes. 2020;12(1):1‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang M, Karlsson C, Olsson C, et al. Reduced diversity in the early fecal microbiota of infants with atopic eczema. J Allergy Clin Immunol. 2008;121(1):129‐134. [DOI] [PubMed] [Google Scholar]
- 9. Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol. 2012;129(2):434–440, 40.e1‐2. [DOI] [PubMed] [Google Scholar]
- 10. Sbihi H, Boutin RC, Cutler C, Suen M, Finlay BB, Turvey SE. Thinking bigger: how early‐life environmental exposures shape the gut microbiome and influence the development of asthma and allergic disease. Allergy. 2019;74(11):2103‐2115. [DOI] [PubMed] [Google Scholar]
- 11. Frei R, Lauener RP, Crameri R, O'Mahony L. Microbiota and dietary interactions: an update to the hygiene hypothesis? Allergy. 2012;67(4):451‐461. [DOI] [PubMed] [Google Scholar]
- 12. Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T‐cell generation. Nature. 2013;504(7480):451‐455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short‐chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569‐573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Noval Rivas M, Burton OT, Wise P, et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol. 2013;131(1):201‐212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leeming ER, Louca P, Gibson R, Menni C, Spector TD, Le Roy CI. The complexities of the diet‐microbiome relationship: advances and perspectives. Genome Med. 2021;13(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baxter NT, Schmidt AW, Venkataraman A, Kim KS, Waldron C, Schmidt TM. Dynamics of human gut microbiota and short‐chain fatty acids in response to dietary interventions with three fermentable fibers. MBio. 2019;10(1):e02566‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roduit C, Frei R, Ferstl R, et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy. 2019;74(4):799‐809. [DOI] [PubMed] [Google Scholar]
- 18. Appert O, Garcia AR, Frei R, et al. Initial butyrate producers during infant gut microbiota development are endospore formers. Environ Microbiol. 2020;22(9):3909‐3921. [DOI] [PubMed] [Google Scholar]
- 19. Vital M, Karch A, Pieper DH. Colonic butyrate‐producing communities in humans: an overview using omics data. mSystems. 2017;2(6):e00130‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high‐resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581‐583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Metwally AA, Yang J, Ascoli C, Dai Y, Finn PW, Perkins DL. MetaLonDA: a flexible R package for identifying time intervals of differentially abundant features in metagenomic longitudinal studies. Microbiome. 2018;6(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer‐Verlag New York; 2016. [Google Scholar]
- 23. Zimmermann P, Messina N, Mohn WW, Finlay BB, Curtis N. Association between the intestinal microbiota and allergic sensitization, eczema, and asthma: a systematic review. J Allergy Clin Immunol. 2019;143(2):467‐485. [DOI] [PubMed] [Google Scholar]
- 24. Penders J, Thijs C, van den Brandt PA, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA birth cohort study. Gut. 2007;56(5):661‐667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Secher T, Brehin C, Oswald E. Early settlers: which E. coli strains do you not want at birth? Am J Physiol Gastrointest Liver Physiol. 2016;311(1):G123‐G129. [DOI] [PubMed] [Google Scholar]
- 26. Secher T, Payros D, Brehin C, et al. Oral tolerance failure upon neonatal gut colonization with Escherichia coli producing the genotoxin colibactin. Infect Immun. 2015;83(6):2420‐2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cait A, Cardenas E, Dimitriu PA, et al. Reduced genetic potential for butyrate fermentation in the gut microbiome of infants who develop allergic sensitization. J Allergy Clin Immunol. 2019;144(6):1638‐1647.e3. [DOI] [PubMed] [Google Scholar]
- 28. Wopereis H, Sim K, Shaw A, Warner JO, Knol J, Kroll JS. Intestinal microbiota in infants at high risk for allergy: effects of prebiotics and role in eczema development. J Allergy Clin Immunol. 2018;141(4):1334‐1342.e5. [DOI] [PubMed] [Google Scholar]
- 29. Chiu CY, Cheng ML, Chiang MH, et al. Gut microbial‐derived butyrate is inversely associated with IgE responses to allergens in childhood asthma. Pediatr Allergy Immunol. 2019;30(7):689‐697. [DOI] [PubMed] [Google Scholar]
- 30. Nylund L, Nermes M, Isolauri E, Salminen S, de Vos WM, Satokari R. Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate‐producing bacteria. Allergy. 2015;70(2):241‐244. [DOI] [PubMed] [Google Scholar]
- 31. Ramsay AG, Scott KP, Martin JC, Rincon MT, Flint HJ. Cell‐associated alpha‐amylases of butyrate‐producing Firmicute bacteria from the human colon. Microbiology (Reading). 2006;152(Pt 11):3281‐3290. [DOI] [PubMed] [Google Scholar]
- 32. Ze X, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 2012;6(8):1535‐1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abell GC, Cooke CM, Bennett CN, Conlon MA, McOrist AL. Phylotypes related to Ruminococcus bromii are abundant in the large bowel of humans and increase in response to a diet high in resistant starch. FEMS Microbiol Ecol. 2008;66(3):505‐515. [DOI] [PubMed] [Google Scholar]
- 34. Walker AW, Ince J, Duncan SH, et al. Dominant and diet‐responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5(2):220‐230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Belzer C, Chia LW, Aalvink S, et al. Microbial metabolic networks at the mucus layer lead to diet‐independent butyrate and vitamin B(12) production by intestinal symbionts. MBio. 2017;8(5):e00770‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Benítez‐Páez A, Gómez Del Pulgar EM, Sanz Y. The glycolytic versatility of Bacteroides uniformis CECT 7771 and its genome response to oligo and polysaccharides. Front Cell Infect Microbiol. 2017;7:383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Demirci M, Tokman HB, Uysal HK, et al. Reduced Akkermansia muciniphila and Faecalibacterium prausnitzii levels in the gut microbiota of children with allergic asthma. Allergol Immunopathol (Madr). 2019;47(4):365‐371. [DOI] [PubMed] [Google Scholar]
- 38. Cani PD, de Vos WM. Next‐generation beneficial microbes: the case of Akkermansia muciniphila. Front Microbiol. 2017;8:1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cheng CC, Duar RM, Lin X, et al. Ecological importance of cross‐feeding of the intermediate metabolite 1,2‐propanediol between bacterial gut symbionts. Appl Environ Microbiol. 2020;86(11):e00190‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lambrecht BN, Hammad H. The immunology of the allergy epidemic and the hygiene hypothesis. Nat Immunol. 2017;18(10):1076‐1083. [DOI] [PubMed] [Google Scholar]
- 41. Tramper‐Stranders G, Ambrożej D, Arcolaci A, et al. Dangerous liaisons: bacteria, antimicrobial therapies, and allergic diseases. Allergy. 2021;76(11):3276‐3291. [DOI] [PubMed] [Google Scholar]
- 42. Makki K, Deehan EC, Walter J, Bäckhed F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe. 2018;23(6):705‐715. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1


