Abstract
This study shows that gain‐of‐function variants in KLHL24 causing EBS and DCM, do not only originate in the start‐codon and suggest that any nonsense‐inducing variant affecting nucleotides c.4_84 will likely cause the same effect on protein level and a similar potential lethal phenotype.
dear editor, KLHL24 is a ubiquitin ligase that mediates keratin 1 , 2 and desmin 3 degradation in the skin and heart, respectively. Previously, five start‐codon variants in KLHL24 caused gain‐of‐function, due to absence of 28 N‐terminal amino acids (KLHL24‐ΔN28) via loss of the translation‐initiation codon. 1 , 2 , 4 Currently, 48 patients with heterozygous start‐codon variants leading to epidermolysis bullosa simplex (EBS) with high risk for dilated cardiomyopathy (DCM) have been reported. 3 , 5 Here, we publish a four‐generation Australian family where 14 members suffer(ed) from EBS and five from DCM. Next‐generation sequencing and segregation analysis identified a novel heterozygous variant in KLHL24:c.22A>T [p.(Arg8*)] in the proband and several other affected members (Figure 1a). All EBS‐affected members had aplasia cutis congenita, mild skin fragility and variably developed hypopigmentation, dystrophic toenails, hypoplastic teeth and frequent dental caries, but no alopecia or oral defects. Skin biopsies revealed basal intraepidermal blister formation, but normal keratin‐14 staining (Figure 1b). Following the sudden cardiac death of patient III:6 at 16 years of age, where post‐mortem evaluation showed an enlarged heart with fibrosis and a reduction in desmin staining (Figure 1b), four other EBS‐affected members were diagnosed with DCM. Patient III:1, diagnosed at age 16 years, had chronic dyspnoea from a young age, limited exercise tolerance, palpitations and an ejection fraction of 31%. He received heart failure medication, but declined an implantable cardiac defibrillator (ICD) and died suddenly at 21 years of age. The proband II:2, diagnosed with DCM at age 46 years, carries an ICD and has a current ejection fraction of 25%, while patients II:4 and II:5 were diagnosed at 49 and 44 years of age, respectively. The latter died from end‐stage heart failure at age 57 years of age. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Figure 1.
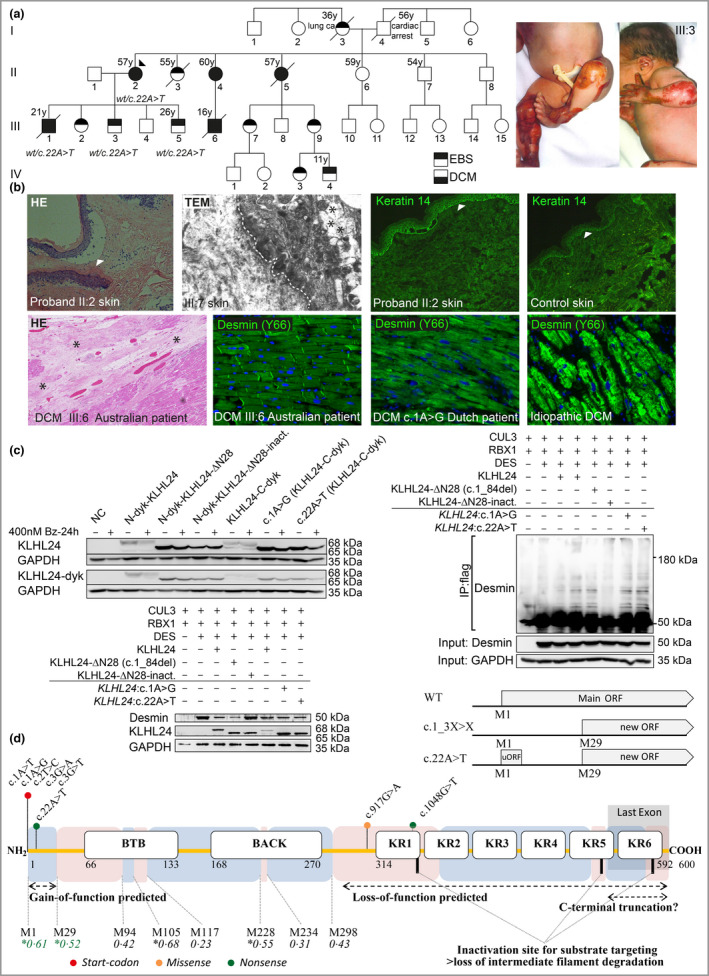
(a) Pedigree of an Australian family with epidermolysis bullosa simplex (EBS) and/or dilated cardiomyopathy (DCM), with the aplasia cutis congenita depicted on the right. The arrowhead indicates the proband and co‐segregation confirmation of variant KLHL24:c.22A>‐T is illustrated below family members. y, years. (b) Analysis of ex vivo skin and heart. Top left to right: haematoxylin and eosin staining (HE) of patient skin (×20), transmission electron microscopy of patient skin (*, cytolysis) (~ ×13 500) and immunofluorescence stainings of keratin‐14 (×20). Arrowheads/dotted line, basement membrane. Bottom left to right: HE of a postmortem heart (*, fibrosis) (×10) and immunofluorescence stainings of cardiac desmin (×20). (c) Western blot analysis of transfected HEK293A cells. Top left: the analysis of different KLHL24 clone transfections. Bz, Bortezomib; N, N‐terminal; C, C‐terminal; dyk, FLAG‐tagged; KLHL24‐ΔN28‐inactivated: mutagenesis at E355A, E535K and Y584C; NC, negative control; antibodies KLHL24‐dyk (M2) and KLHL24 (ab104089); KLHL24 = 68 kDa; KLHL24‐ΔN28 = 65 kDa. Bottom left: the functional desmin levels (50 kDa) after different co‐transfections (desmin: KLHL24>1:1 transfection ratio). Right: immunoprecipitated desmin levels (> 50 kDa) after different co‐transfections (IP desmin‐dyk M2; desmin: KLHL24>5:1 transfection ratio). (d) Schematic representation of KLHL24, including the clinically reported variants, protein domains [BTB, BACK and Kelch Repeats (KR)], in‐frame start‐codons (M1, M29..) with predicted translation‐initiation sites, based on the Kozak sequence (unlikely: 0·5 > x > *0·5:likely), with the confirmed active translation‐initiation sites in green; (u)ORF, (upstream) open reading frame.
Variant KLHL24:c.22A>T was absent from controls in the Genome Aggregation Population Database and had not been reported in ClinVar or HGMD variation databases. While predicted to result in loss‐of‐function, we hypothesized that this nonsense variant also causes N‐terminal truncation (KLHL24‐ΔN28) via translation re‐initiation, resulting in gain‐of‐function and excessive degradation of intermediate filaments. To prove this, we cloned start‐codon variant c.1A>G and variant c.22A>T into the pcDNA3·1(+)‐C‐DYK backbone containing the wildtype KLHL24 sequence. These plasmids encode different KLHL24 wildtype and variant proteins, fused to a C‐terminal FLAG‐tag. After overexpression in HEK293A cells, we compared the abundance and weight of the resulting KLHL24 protein with our validated wildtype and KLHL24‐ΔN28 mutants. 3 Transfection experiments confirmed that variant c.22A>T results in a protein, equal in abundancy and weight as the protein pathology of the previously reported start‐codon variants (Figure 1c). These results imply that KLHL24:c.22A>T generates a nonfunctional upstream open reading frame (uORF; Met1_Arg8), 6 causing skipping of translation of Leu9_Glu28, due to the stop‐codon at position 8. Translation re‐initiating takes place at Met29, leading to protein levels similar to the validated KLHL24‐ΔN28 mutant. To prove that the c.22A>T mutant is functionally as active as the c.1A>G and KLHL24‐ΔN28 mutants, we cloned DES and KRT14 1 into the pcDNA3·1(+)‐N‐DYK backbone. Co‐transfections were performed using either DES or KRT14, with different combinations of ubiquitin‐ligase factors RBX1, CUL3 and KLHL24 clones. Desmin levels were indeed excessively reduced in combination with KLHL24:c.22A>T, comparable with the c.1A>G and KLHL24‐ΔN28 mutant (Figure 1c). Immunoprecipitation studies substantiated that the KLHL24:c.22A>T/RBX1/CUL3–ubiquitin‐ligase complex leads to equally excessive ubiquitination levels of desmin as the KLHL24:c.1A>G/RBX1/CUL3–ubiquitin‐ligase complex (Figure 1c). Meanwhile, (ubiquitinated) keratin‐14 levels remained unaltered in all transfection conditions.
KLHL24 contains nine exons (main ORF: exons 3–9) (Figure 1d). Two homozygous variants previously led to loss‐of‐function, 7 implying that homozygous nonsense‐inducing variants after Met298 will result in loss‐of‐function, causing hypertrophic cardiomyopathy without skin fragility. Upstream of position 299, KLHL24 contains eight methionines that could serve as potential start‐codons. Here, we report the first variant downstream of the start‐codon results in gain‐of‐function. This is remarkable and our analyses indicate that this is the result of translation re‐initiation, as the resulting KLHL24 protein has the same size as a variant that lacks the first translation‐initiation codon. Re‐initiation of translation after a short uORF 6 is not uncommon and it is likely that other nonsense‐inducing variants between nucleotides c.4_84, can also result in the truncated protein KLHL24‐ΔN28. Of note, we detected similar amounts of desmin degradation by the KLHL24:c.22A>T/RBX1/CUL3 and KLHL24:c.1A>G/RBX1/CUL3–ubiquitin‐ligase complex, but could not detect KLHL24‐mediated degradation of keratin‐14 in transfected HEK293A cells. This is in line with follow‐up studies to the initial study on KLHL24‐related EBS from Lin et al. 2 , 3 , 4 Although, further studies to elucidate the KLHL24‐induced activity towards keratin‐14 are necessary, this may be related to ageing. 8 To conclude, this study shows that gain‐of‐function variants in KLHL24 causing EBS and DCM do not originate only in the start‐codon and suggest that any nonsense‐inducing variant affecting nucleotides c.4_84 will likely cause the same effect on protein level and a similar potential lethal phenotype.
Author contributions
Mathilde C.S.C. Vermeer: Conceptualization (equal); data curation (equal); formal analysis (equal); investigation (equal); methodology (equal); validation (equal); visualization (equal); writing – original draft (equal); writing – review and editing (equal). Mohammad Al‐shinnag: Conceptualization (equal); data curation (equal); investigation (equal); writing – original draft (supporting); writing – review and editing (supporting). Herman Sillje: Conceptualization (equal); methodology (equal); supervision (equal); writing – review and editing (equal). Antonio Gaytan: Data curation (equal); formal analysis (equal); investigation (equal); writing – review and editing (supporting). Dedee F Murrell: Data curation (equal); investigation (equal); methodology (equal); validation (equal); writing – review and editing (supporting). Julie McGaughran: Investigation (equal); writing – review and editing (supporting). wei melbourne: Data curation (equal); writing – review and editing (supporting). Timothy Cowan: Data curation (equal); investigation (equal); writing – review and editing (supporting). Peter C. van den Akker: Writing – review and editing (supporting). Karin van Spaendonck: Investigation (equal); writing – review and editing (supporting). Peter van der Meer: Conceptualization (equal); funding acquisition (equal); supervision (equal); writing – review and editing (equal). Marieke Bolling: Conceptualization (equal); funding acquisition (equal); resources (equal); supervision (equal); validation (equal); writing – review and editing (equal).
Acknowledgments
We thank the family members for their participation in this study. Patients and participating patients gave their oral or written informed consent, respectively.
M.C.S.C.V., M.A‐S. and H.H.W.S. contributed equally.
Funding sources: This work was funded by the Human Frontier Science Program (grant number RGY 0071/2014 to P.v.d.M.), Vlinderkind (no grant number; patient organization funding to M.C.B.) and the European Research Counsel [STOP‐HF (StG); grant number 715732, ERC‐2016‐STG to P.v.d.M.]. None of the funders had a role in the design and conduct of the study; collection, management, analysis and interpretation of the data; preparation, review or approval of the manuscript; and decision to submit the manuscript for publication.
Conflicts of interest: the authors declare they have no conflicts of interest.
Data availability: The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Lin Z, Li S, Feng C et al. Stabilizing mutations of KLHL24 ubiquitin ligase cause loss of keratin 14 and human skin fragility. Nat Genet 2016; 48:1504–16. [DOI] [PubMed] [Google Scholar]
- 2. He Y, Maier K, Leppert J et al. Monoallelic mutations in the translation initiation codon of KLHL24 cause skin fragility. Am J Hum Genet 2016; 99:1395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vermeer MCSC, Bolling MC, Bliley JM et al. Gain‐of‐function mutation in ubiquitin‐ligase KLHL24 causes desmin degradation and dilatation in hiPSC‐derived engineered heart tissues. J Clin Invest 2021; 131:e140615. doi: 10.1172/JCI140615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee JYW, Liu L, Hsu C‐K et al. Mutations in KLHL24 add to the molecular heterogeneity of epidermolysis bullosa simplex. J Invest Dermatol 2017; 137:1378–80. [DOI] [PubMed] [Google Scholar]
- 5. Schwieger‐Briel A, Fuentes I, Castiglia D et al. Epidermolysis bullosa simplex with KLHL24 mutations is associated with dilated cardiomyopathy. J Invest Dermatol 2019; 139:244–9. [DOI] [PubMed] [Google Scholar]
- 6. Barbosa C, Peixeiro I, Romão L. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet 2013; 9:e1003529. doi: 10.1371/journal.pgen.1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hedberg‐Oldfors C, Abramsson A, Osborn DPS et al. Cardiomyopathy with lethal arrhythmias associated with inactivation of KLHL24. Hum Mol Genet 2019; 28:1919–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vermeer MCSC, Silljé HHW, Pas HH et al. K14 degradation and ageing in epidermolysis bullosa simplex due to KLHL24 gain‐of‐function mutations. J Invest Dermatol 2022; 142:2271–74.e6. doi: 10.1016/j.jid.2021.12.027. [DOI] [PubMed] [Google Scholar]


