Abstract
Microglia is considered the central nervous system (CNS) resident macrophages that establish an innate immune response against pathogens and toxins. However, the recent studies have shown that microglial gene and protein expression follows a circadian pattern; several immune activation markers and clock genes are expressed rhythmically without the need for an immune stimulus. Furthermore, microglia responds to an immune challenge with different magnitudes depending on the time of the day. This review examines the circadian control of microglia function and the possible physiological implications. For example, we discuss that synaptic prune is performed in the cortex at a certain moment of the day. We also consider the implications of daily microglial function for maintaining biological rhythms like general activity, body temperature, and food intake. We conclude that the developmental stage, brain region, and pathological state are not the only factors to consider for the evaluation of microglial functions; instead, emerging evidence indicates that circadian time as an essential aspect for a better understanding of the role of microglia in CNS physiology.
Keywords: biological rhythms, circadian rhythms, microglia, microglial polarization, neuroimmune response
Main Points
Microglia are rhythmic cells with an oscillatory expression in clock genes, cytokines, and other microglial markers.
Microglial circadian rhythms determine their physiological function and immune responses.
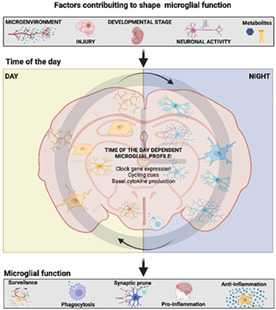
1. INTRODUCTION
Physiological processes are studied under the precept that every bodily function is temporally organized to adapt to the daily alternation between day and night. Every cell of the body is subjected to this daily change whereby the intrinsic capacity to present a 24‐h oscillatory activity pattern (circadian rhythms) came about as an adaptation to this geophysical phenomenon (Bollinger & Schibler, 2014).
Circadian rhythms are generated by biological clocks. In mammals, the suprachiasmatic nucleus (SCN) of the hypothalamus, is considered the master clock. This specialized nucleus shows self‐sustained rhythms in its neuronal activity that is adjusted to the 24‐h light–dark cycle. Then, the SCN coordinates whole‐body circadian rhythms by transmitting its cyclical message to other brain regions as well as peripheral clocks, thereby modulating distinct physiological processes such as hormone secretion, glucose production, autonomic output, alertness, among others (Buijs et al., 2019). In this way, several brain regions show daily oscillations in neuronal activity.
This capacity is provided by clock genes that have been identified to codify transcription factors which activity is regulated by negative feedback loops operating within a ~ 24 h period. Specifically, one of the products of these clock genes, the CLOCK protein forms a heterodimer with BMAL1, another clock gene product, acting as a transcriptional activator that is active during the day and promotes the expression of the Per gene as well as other output genes whose expression oscillate in a circadian fashion. In the cytoplasm, PER forms a dimer with CRY which stabilizes it; the PER/CRY complex then moves to the nucleus where it represses its own expression by inhibiting CLOCK/BMAL1 activity (Albrecht, 2002; Cermakian et al., 2013; Cox & Takahashi, 2019; Ko & Takahashi, 2006)
Recent evidence has shown that besides neurons, glial cells in the SCN also have fluctuating functions essential for maintaining whole‐body circadian rhythms (Brancaccio et al., 2017; Sueviriyapan et al., 2020) indicating that neurons are not the only oscillating cells in the central nervous system (CNS). Specifically, it has been demonstrated that microglia, the resident macrophages of the CNS, also have circadian rhythms in clock gene expression, macrophage markers, cytokine production and secretion (Wang et al., 2021) and in the magnitude of their response to immune stimuli (Fonken et al., 2015). The circadian fluctuation in microglial function may also place this cell population as a new player regulating the circadian system since it was recently shown that depleting microglial cells induces arrhythmicity in several complex physiological processes (De Luca et al., 2019).
The present review will discuss the studies describing the circadian rhythm of microglia functions. We will also discuss the possible cues that provide a temporal timestamp to these cells and the physiological implications of this circadian regulation. Finally, we will propose circadian time as a novel factor to be considered when studying microglial activity under physiological and pathological conditions.
2. MICROGLIAL PHYSIOLOGICAL FUNCTIONS
In the last decade, microglial studies have focused on the molecular signature of microglia determining that their molecular signature is subjected to the individual's developmental stage, brain region, pathological condition, sex, species and even whether these cells are studied in vivo or in vitro. The results derived from transcriptomics suggest that microglia is a more diverse dell population than previously thought and may acquire different functionalities depending in several factors.
One of the most relevant functions of microglia is their surveillance capacity. Two‐photon imaging have shown that microglial processes are in constant motility, which is necessary for monitoring the brain parenchyma (Davalos et al., 2005; Nimmerjahn et al., 2005). This brain surveillance and directed process motility are aimed to monitor the activity and conditions of synapses (Nimmerjahn et al., 2005) to prevent hyper‐excitability of activated brain areas under physiological conditions (Merlini et al., 2021), and to phagocytize synapses during development (Schafer et al., 2012; Stevens et al., 2007) and even in adulthood by motoring the status of said synapses. The movement of microglial processes is also directed to either mechanically damaged tissue, dying neurons and invading microorganisms (Haynes et al., 2006).
Previously, it was considered that upon stimulation with an immune challenge microglial cells could polarized into one of two states; a “classical” pro‐inflammatory (M1) profile that expresses inducible nitric oxide synthase (Paxinos) for nitric oxide production, pro‐inflammatory cytokines (TNF‐α, interleukin [IL]‐ 1β, IL‐12) (Gordon & Taylor, 2005; Villalta et al., 2009),and an “alternative” anti‐inflammatory (M2) state that expresses arginase 1 (ARG1), anti‐ inflammatory cytokines (Il4, Il10, Il13, and TGF‐β), CD206, among others (Colton, 2009; Novak & Koh, 2013; Shechter & Schwartz, 2013; Stein et al., 1992). Nonetheless, the concepts of M1 and M2 polarization were coined based on data suggesting that macrophages could activate as T‐cells are activated into T helper 1 and 2 (TH1 and TH2). Furthermore, the fact that macrophage surface markers can elicit one of these two T‐cell phenotypes, helped to establish that macrophages would adopt the same nomenclature to describe its activation state. However, the recent studies have challenged this dichotomous classification (Ransohoff, 2016). As it was noted, the only studies that have been able to obtain the M1 and M2 phenotypes were done by exposing cells in vitro to immune challenges including LPS or diverse combinations of cytokines, while in vivo microglial phenotypes are only described with one or two markers that do not demonstrate a pro or anti‐inflammatory function.
Furthermore, in 2013, Butovsky et al. demonstrated that the transcriptomic profile of freshly isolated microglia and that of the BV2 cell line was significantly different and that many microglial markers were not even present in any of the cell lines employed (Butovsky et al., 2014). Suggesting that microglial immune response and physiological functions most be analyzed considering all the previously mentioned factors (brain region, sex, developmental stage, etc.). In the next sections we will propose the time of the day as another factor to be taken into consideration while studying microglial functions, that is, the time of the day.
3. EVIDENCE OF THE CIRCADIAN OSCILLATIONS OF MICROGLIAL CLOCK GENES
In mammals, the molecular clock consists of interlinked auto‐regulatory transcriptional–translational feedback loops that drive rhythmic ~24‐h protein expression patterns in individual cells (Reppert & Weaver, 2002). The main loop, called the core clock, includes the following genes: circadian locomotor output cycles kaput (clock), brain and muscle ARNT like protein1 (bmal1), period (per 1, 2, and 3), and cryptocrome (cry 1 and 2). As stated above, CLOCK and BMAL1 form a dimer that promotes the transcription of per and cry (1 and 2). As these genes are expressed, their protein products dimerize to form a complex that represses clock and bmal1, inhibiting their own transcription (Albrecht, 2002). In addition, the gene npas2 is an ortholog of clock, and can substitute for clock in clock‐mutant mice (DeBruyne et al., 2007; Reick et al., 2001).
Additional loops are formed by retinoic acid‐related orphan nuclear receptors Rev‐Erba and RORs, along with D‐box binding protein (DBP), which fine‐tune the oscillations generated by the core clock and contribute to the strength of the molecular clock circuitry (Albrecht, 2002; Cox & Takahashi, 2019). Clock proteins also regulate other target genes known as clock‐controlled genes (CCGs) through the binding of the CLOCK: BMAL1 dimer to the E‐box element in their promoter region.
One of the first studies demonstrating the presences of clock genes in microglial cells was published in 2011, by Nakazato and colleagues showing that microglia isolated from neonatal mouse brains and the microglial cell line BV‐2 expressed: Per1, Per2, Per3, Cry1‐2, Bmal1, Clock Dec1‐2, and Npas2 (Nakazato et al., 2011). Later, the circadian oscillations of BMAL, CLOCK, CRY1 and 2, PER 1 and 2, and Nr1d1 (which encodes the Rev‐Erba protein), and DBP proteins was described in the BV‐2 microglial cell line (Wang et al., 2020), confirming that microglia do not only express clock genes, but also they actually have an oscillating molecular clock in vitro.
It was not until 2013, which Hayashi and colleagues demonstrated that Per1 and Per2 present the highest levels within the first 2 h of the night (the active phase in nocturnal rodents) and that Rev‐erbα showed peaked in the middle of the dark phase. In the same study. In contrast, the acrophase of the core transcription factor Bmal1 was observed at the beginning of the day (the resting phase). Later the work of Fonken et al. demonstrated the presence of clock genes in rat hippocampal microglia (Fonken et al., 2015) (Figure 1).
FIGURE 1.
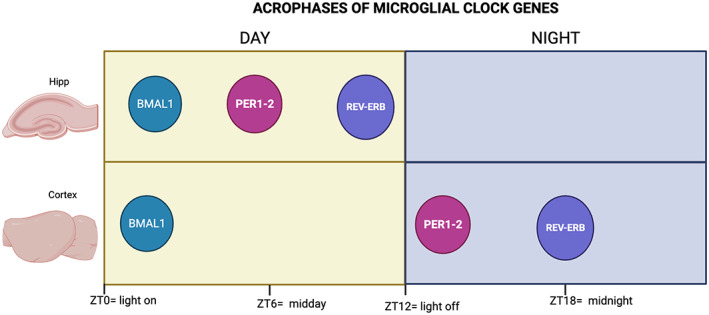
Time of maximal clock gene expression map for BMAL1, PER1 and 2 and REV‐ERB in the hippocampus (Hipp) and the cortex. Yellow panels indicate the light phase, and blue panels show the dark phase. ZT refers to Zeitgeber time, which literally means “provider of time” and refers to an environmental signal that indicates time, like the onset of light (ZT = 0)
In chronobiology, to determine if a particular oscillation, either physiological, behavioral, or genetic, is endogenous and not the product of changes in the environment, it is necessary to evaluate if the temporal changes persist even in the absence of time cues like the daily alternation between light and dark periods. Studies determined that the circadian oscillations of microglial clock genes persisted in mice kept in constant darkness for a full 24 h cycle, indicating that these rhythms are intrinsic (Hayashi, Koyanagi, et al., 2013). Accordingly, microglia isolated from the cortex of clock‐mutant mice does not show a significant time‐of‐day difference in the expression of clock genes in cortical microglia (Fonken et al., 2015; Hayashi, 2013). Together, these data demonstrate that microglial cells in at least some brain areas express the molecular machinery to generate circadian oscillations. More studies are necessary to further characterize the circadian expression of microglial clock genes in the entire brain and to determine if these oscillations are indeed linked to oscillatory functions and are necessary to maintain normal microglial function.
Importantly, circadian oscillations in microglial clock genes could provide the basis for the oscillations of microglial function. Hippocampal microglia expresses both Bmal‐1 and pro‐inflammatory cytokines at relatively high levels during the light phase (Fonken et al., 2015). Bmal1 deficiency reduced mRNA levels of IL‐1β, TNF‐α, and IL‐6 in response to LPS administration in the microglial BV‐2 cell line. In contrast, IL‐10 mRNA levels were increased (Wang et al., 2020). In primary microglia from the neocortex of Bmal‐1 deficient mice, LPS administration induced a lower IL‐6 expression than in wild‐type mice (Nakazato et al., 2017). Together, these data suggest that Bmal‐1 promotes the expression of pro‐inflammatory cytokines and decrease the expression of anti‐inflammatory cytokines in physiological conditions and upon LPS administration. The molecular pathways of this interaction are still unknown, but Bmal‐1 modulation of cytokine production could be carried out through the NRF2 transcription factor, which is known to promote IL‐1β expression in macrophages after Bmal‐1 binds to its E‐box (Early et al., 2018).
The relationship of Bmal‐1 with cytokine expression has also been studied in APP‐KI mice, which over‐express amyloid precursor protein. Microglia of these mice showed malfunctioning of the CLOCK/BMAL1 feedback loops. A reduced expression of BMAL1 in these mice was related to increase TNF‐α and IL‐1β levels in cortical microglia (Ni et al., 2019). Furthermore, Bmal1‐deficient B6.129‐Arntltm1Bra/J mice exhibit increased gene expression of anti‐oxidative and anti‐inflammatory factors in microglia, and Bmal1 knock‐down microglial BV‐2 cell showed reduced responses to LPS, suggesting that the different elements of the molecular clock are crucial for the correct expression of microglial physiological and immune functions (Wang et al., 2020) (Table 1).
TABLE 1.
Circadian oscillation of clock genes in microglia
| Model | Brain region | Circadian peak | Reference | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Per1 | Per2 | Per3 | Cry1 | Cry 2 | Bmal1 | Clock | Npas2 | Nr1d1 | Dbp | RevErb‐α | |||
| Neonatal mice | Whole brain | P | P | P | P | P | P | P | P | ‐ | P | ‐ | Nakazato et al. (2011) |
| BV‐2 microglia cell line | ‐ | P | P | P | P | P | P | A | P | ‐ | P | ‐ | Nakazato et al. (2011) |
| BV‐2 microglia cell line synchronized with dexamethasone a | ‐ | 4 | 4 | 24 and 8 | 8 | 4 | 4 | ‐ | 16 | ‐ | ‐ | ‐ | Nakazato et al. (2017) |
| 8–10 week old mice | Cortex | 14 | 14 | ‐ | ‐ | ‐ | 2 | ‐ | ‐ | ‐ | ‐ | 18 | Hayashi, Koyanagi, et al. (2013) |
| 2‐ month old mice | Cortex | 18 | 18 | ‐ | ‐ | ‐ | 2 | ‐ | ‐ | ‐ | ‐ | 10 | Ni et al. (2019) |
| MG6 microglia cell line synchronized with dexamethasone a | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | 18 | ‐ | ‐ | ‐ | ‐ | 24 | Ni, Wu, Stoka, et al. (2019) |
| Sprague–Dawley rats | Hippocampus | 6 | 6 | ‐ | ‐ | ‐ | 24 | NR | ‐ | ‐ | ‐ | 12 | Fonken et al. (2015) |
| Fischer‐Brown Norway rats | Hippocampus | 6 | 12 | ‐ | ‐ | ‐ | 24 | ‐ | ‐ | ‐ | ‐ | 12 | Fonken, Kitt, et al. (2016) and Fonken, Weber, et al. (2016) |
| BV‐2 microglia cell line | ‐ | 24 | 8 | ‐ | 4 | NR | 4 | 8 | ‐ | 16 | 8 | ‐ | Wang et al. (2020) |
| 557BL/6 J 10‐ week‐old mice | Whole brain | 13.5 | 15 | ‐ | 15 | 13.5 | 4.5 | 7.5 | ‐ | 12 | 12 | ‐ | Wang et al. (2021) |
Abbreviations: P, the presence of the gene was measured but not its oscillation; ‐, not measured; a, absent; NR, non‐rhythmic.
ZT is considered as the moment of adding dexamethasone to the medium.
Oscillating clock genes provides cells with the molecular mechanism to oscillate in a circadian manner, but the following question remains: Besides clock genes: are there other microglial genes that present circadian oscillations? In the next section, we will discuss the studies that report daily changes in genes or proteins involved known as microglial markers or functional products.
4. OTHER MICROGLIAL GENES AND PROTEINS THAT EXPRESS A CIRCADIAN RHYTHM
Although clock genes are rhythmically expressed in microglial cells, it is unclear whether they promote oscillation of genes that determine the function in these macrophages. Nevertheless, reports show that other microglial proteins besides clock genes also display circadian oscillations.
Under physiological conditions several microglial markers exhibit a rhythm. The expression levels of the macrophage marker, the ionized calcium‐binding adapter molecule 1 (Iba‐1) varies according to the time of the day in the hippocampus of male mice, with significantly more Iba‐1 positive cells in the day, which corresponds to the resting phase in rodents (Griffin et al., 2019). Although, Iba‐1 is not considered a functional marker for macrophages, we can hypothesize that circadian changes in its expression might be indicative of an oscillatory pattern in microglial functions.
Besides Iba‐1, other microglial markers in the hippocampus exhibit a circadian rhythm for example; the cluster of differentiation 68 (CD68) which peaks during the day (Choudhury et al., 2020). Although pathological models such as stroke, have proposed CD68 as a general marker of phagocytic macrophages (Perego et al., 2011). CD68 is a marker related to lysosomal glycoproteins transported through vesicles between lysosomes, endosomes, and the plasma membrane thus, its circadian expression could be related to either phagocytosis, autophagy or even the secretion of lytic enzymes.
Choudhury et al, reported circadian changes in matrix metalloproteinases (MMPs) 2, 13, and 14 in the prefrontal cortex of adult mice (PFC) that could be involved in microglial migration and invasion (Lively & Schlichter, 2013; Nakanishi, 2003; Sacks et al., 2018). In addition, the expression of CD11b and CD45 was also increased during the sleeping phase as compared to the active phase. The microglial cells expressing CD11b in the day time also presented larger cell bodies and contained more synapsin I protein (Choudhury et al., 2020), suggesting that these cells are performing phagocytosis, this possible functional implication of microglial rhythmicity will be discussed in detail in the later section.
Another region that exhibits daily changes in the expression of microglial markers is the medio‐basal hypothalamus (MBH) of male mice. The number of Iba‐1‐positive cells, the number of ramifications per microglial cell, and TNF‐α expression is higher at night than in the day. Importantly, this difference is lost after 24 h of fasting (Yi et al., 2017), suggesting that the signal conveying rhythmicity to hypothalamic microglia is related to food intake. Supporting this data, obese mice fed a high‐fat diet (HFD) also show an arrythmic Iba‐1 and TNF‐α expression throughout the day (Yi et al., 2017), suggesting that the rhythm of these cells in the hypothalamus is determined by metabolic signals.
Furthermore, the same research group also reported that CD68 levels in the MBH are also higher during the day, and that this expression is under the control of clock genes, since microglial knock‐down of Bmal‐1 up‐regulates CD68 along the entire light–dark cycle (Wang et al., 2021). Altogether, these results suggest that some microglial markers might be clock controlled genes, further studies should assess this hypothesis.
Besides the circadian oscillation in macrophage markers, microglial cytokine expression also exhibits circadian changes under basal conditions. Wang et al. described a rhythm in the expression of the pro‐inflammatory cytokines IL1‐β, and IL‐6 in primary microglial cultures with the highest expression in the light phase (Wang et al., 2020). In addition, Hippocampal microglia in rats show a daily rhythm in the expression of IL1‐β, IL‐6, and TNF‐α and the major histocompatibility complex II (MHC‐II), each reaching their maximal expression at different time points of the light phase (Fonken et al., 2015) while cortical isolated microglia also display daily differences in the mRNA expression of TNF‐α, IL1‐β, and the myeloid differentiation primary response 88 (Myd88), an adaptor for pro‐inflammatory pathways located downstream of IL1‐β (Milanova et al., 2019). All together these data demonstrate that the basal cytokine levels oscillate daily under physiological conditions without the need of an immune stimulation. Moreover, it is possible that microglial physiological functions might employ cytokines secretion to maintain parenchymal homeostasis in a circadian manner. More studies are necessary in order to corroborate this.
The relationship between microglial immune products and clock genes has also been described, so that deletion of clock genes induces a higher production of inflammatory cytokines and chemokines (Sato et al., 2014; Vieira et al., 2020),
Bmal1 deficiency reduces mRNA levels of IL‐1β, TNF‐α, and IL‐6, and increases IL‐10 in response to LPS administration in the microglial cell line BV‐2 (Wang et al., 2020). Additionally, the primary microglia cultures from the neocortex of Bmal‐1 deficient mice treated with LPS, secrete less IL‐6 than the microglia isolated from wild‐type mice (Nakazato et al., 2017), suggesting that Bmal‐1 deficiency decreases the inflammatory response of cortical microglia.
Furthermore, Rev‐erbα is considered one of the links between circadian rhythms and immune functions as it modulates the expression of genes involved in innate immunity, such as Il6, Il19, Cxcl6, Cxcl11, and Ccl2 in immune cells including macrophages (Gibbs et al., 2012; Sato et al., 2014). In accordance, Rev‐erbα‐deficient mice lose the daily oscillation of hippocampal Iba‐1 and CD68 and present constantly high levels throughout the day (Griffin et al., 2019).These data suggest that Rev‐erbα deficiency is necessary to maintain the proper oscillations in hippocampal microglial markers. Future studies should assess the functional implications of the lack of oscillations in the hippocampal microglia.
The circadian differences in the expression of cytokines, clock genes, and functional microglial markers suggest that these cells change their cell function in accordance with the time of the day and most importantly they do so in the absence of pathogens or injury.
If microglial cells are able to modulate their function in accordance with the time of the day, then what are the mechanisms that synchronize them to the light–dark cycle? In the next section, we will discuss how these cells may acquire an oscillatory profile.
5. POSSIBLE MECHANISMS IMPOSING A TIME STAMP ON MICROGLIA
Until now, there is no evidence that the SCN, the biological clock, may directly regulate the circadian oscillations in microglia since there are no anatomical reports of SCN neurons contacting these macrophages in the different nuclei in which they present daily changes in gene expression. One possibility is that oscillating changes in the levels of metabolites, neurotransmitters and hormones known to modulate microglial function could provide an indirect temporal signal to these cells (Desale & Chinnathambi, 2020; Fonken et al., 2015; Fonken, Kitt, et al., 2016; Fonken, Weber, et al., 2016; Logiacco et al., 2021).
Neuronal activity is known to vary throughout the day in several nuclei (Caba et al., 2014; Iyer et al., 2014; Kononen et al., 1990). Furthermore, circulating metabolites like free fatty acids (Panda et al., 2002) and glucose (Hsieh et al., 2019; Kalsbeek et al., 2014) or hormones like leptin (Gao et al., 2018; Schoeller et al., 1997; Yildiz et al., 2004), and glucocorticoids (Buijs et al., 2019; Halberg et al., 1959; Leliavski et al., 2015; Oster et al., 2006) display circadian rhythms that may potentially synchronize microglial function within a 24 h period (Desale & Chinnathambi, 2020; Fonken et al., 2015; Fonken, Kitt, et al., 2016; Fonken, Weber, et al., 2016; Logiacco et al., 2021).
Hormones like insulin, glucocorticoids and leptin are known to reach areas like the hypothalamus, hippocampus, amygdala, and the prefrontal cortex (Banisadr et al., 2005; McGregor & Harvey, 2018; Thundyil et al., 2012; Trujeque‐Ramos et al., 2018; van Swieten et al., 2014; Weisz et al., 2017). Although, it is unknown whether their brain levels may oscillate along the light–dark cycle, there are some reports suggesting that glucocorticoids might act as a synchronizing signal for microglial cells.
The oscillation of glucocorticoids in plasma have one of the best described circadian rhythms, with the highest levels just before the waking hours and lower levels while organisms are resting (Buijs et al., 2019; Halberg et al., 1959; Leliavski et al., 2015; Oster et al., 2006). Fonken et al. demonstrated that corticosterone stimulation restored Per1 expression in hippocampal microglia isolated from aged rats. Interestingly, the authors employed low doses of corticosterone (10 or 100 nM) for only 2 h, suggesting that this hormone may synchronize microglial function under physiological conditions. Importantly, microglia isolated from adrenalectomized rats maintain the oscillation of clock and inflammatory genes, indicating that glucocorticoids are not the only signal sustaining circadian gene expression but also may convey an entraining signal (Fonken et al., 2015; Fonken, Kitt, et al., 2016; Fonken, Weber, et al., 2016).
Plasmatic glucose levels also oscillate daily (Kalsbeek et al., 2014). This is an essential metabolite for microglial function that in their quiescent state mainly express the glucose transporter 5 (Glut5), a transporter with low glucose affinity, while in response to an immune challenge like LPS, microglia over‐express the high affinity glucose transporter 1 (Glut1) thus, elevating their intracellular glucose levels and promoting an anaerobic glycolytic metabolism (Bernier et al., 2020; Lauro & Limatola, 2020; Orihuela et al., 2016).
Fluctuating glucose levels in the milieu can also influence microglial functions. Hsieh et al. demonstrated that increasing glucose concentrations in murine BV‐2 cultures promotes cell proliferation and TNF‐α production (Hsieh et al., 2019; Wang et al., 2020). Furthermore, hypothalamic microglial cells rearrange around neuropeptide y (NPY) neurons during hypoglycemia (Winkler et al., 2019), suggesting that glucose levels may modulate microglial surveillance state in response to glucose.
In addition, there are several brain regions that show a circadian rhythm in glucose utilization (Crane et al., 1980; Newman et al., 1992). Recently, Rodríguez‐Cortes et al. demonstrated a circadian rhythm in the entry of glucose into the hypothalamic arcuate nucleus (Rodríguez‐Cortés et al., 2022), suggesting that the daily variations in glucose levels in different brain areas could also be a cyclic signal modulating microglial function.
The peak of microglial oscillations depends on the brain region, and therefore, it is unlikely that a single cycling cue may be able to entrain every microglial cell in the brain. It is also improbable that neurotransmitters, metabolites, and hormones present the same rhythm among the different brain areas. Therefore, we believe that the characteristics of each brain region, together with the patterns of neuronal activity and the access of metabolites and hormones, might provide a specific timestamp to the daily functions of microglia. Future studies should address if the daily changes observed in microglia result from the circadian oscillations in blood‐borne hormones and metabolites.
We have presented evidence showing that microglial function is controlled in a circadian manner, but why do these cells oscillate every day? The following section will discuss the possible physiological implications of the circadian rhythm in microglial activity.
6. PHYSIOLOGICAL IMPLICATIONS OF MICROGLIAL CIRCADIAN CONTROL
As previously discussed, several microglial genes oscillate but the physiological implication of these cyclic patterns remains poorly understood.
Synaptic prune is one of the main functions that microglial cells perform (Hong & Stevens, 2016; Paolicelli et al., 2014; Sacks et al., 2018). One possibility for the circadian changes observed in microglial gene and protein expression might be related with the reduction in the number of synapses widely reported during the resting phase (Acsády & Harris, 2017; de Vivo et al., 2017; Diering et al., 2017; Vyazovskiy et al., 2008).
The microglia‐specific protease cathepsin S show a circadian pattern (Hayashi, Koyanagi, et al., 2013) and deletion of CatS causes an arrhythmic phenotype in locomotor activity and inhibits the diurnal variations in the synaptic activity and spine density in cortical neurons (Hayashi, Koyanagi, et al., 2013; Takayama et al., 2017). Furthermore, Choudhury et al, reported the possible phagocytic synaptic elimination in the PFC during the resting phase. In Choudhury's study, synaptophysin‐immunoreactive synapses were found co‐localizing with complement C3 immunoreactivity, which were presumably inside of phagosomes in the microglial soma (Choudhury et al., 2020). These data suggest that microglia is involved in the remotion of synapses during the resting phase in the PFC. Future studies should asses if this is a functional implication for the daily changes in gene and protein expression reported in the microglia of areas like the hippocampus and the medio basal hypothalamus (Figure 2).
FIGURE 2.
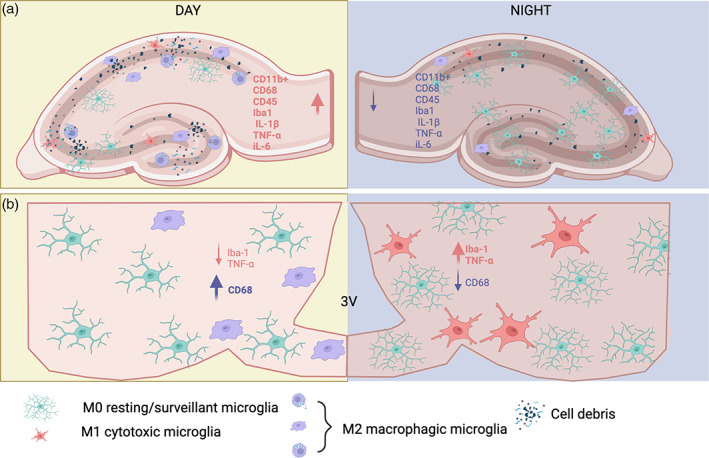
Circadian rhythm of microglia in the hippocampus and hypothalamus. Under physiological conditions, microglial genes are rhythmically expressed in the hippocampus and hypothalamus of rodents. In the hippocampus (a) CD11B+, CD68, CD45, Iba‐1, IL‐1β, TNF‐α, and IL‐6, are higher in the day than at night. Conversely, the microglial profile in the mediobasal hypothalamus (b) is inverted, with higher levels of Iba‐1, and TNF‐α during the night, in contrast, CD6 increases during the day
The circadian variations of immune functions are ubiquitous in immunology, and microglial daily oscillations might be important for the modulation of their immune response (Wang et al., 2022). Although the exact physiological significance behind the time‐dependent sensitivity to immune challenges is unclear, the daily differences in response to immune challenges might be one of the most illustrative examples of microglial rhythms.
In two different studies, Fonken et al. reported higher mRNA levels of IL‐1β, TNFα, and IL‐6 in response to ex vivo LPS administration in hippocampal microglia isolated from rats during the day as compared to those isolated during the night
(Fonken et al., 2015; Fonken, Kitt, et al., 2016; Fonken, Weber, et al., 2016). In contrast, IL‐10 mRNA levels were higher after LPS in hippocampal microglia isolated in the middle of the night. Furthermore, infecting cultured MG6 microglial cell line with P. gingivalis (a pathogenic bacterium that causes periodontitis) induced process extension in cortical microglia that were significantly greater when the immune challenge was given during the day (Takayama et al., 2016). Together, these results suggest that the elevated pro‐inflammatory cytokine expression in the hippocampus and cortex observed during the resting phase (light) could favor microglial pro‐inflammatory response at this particular time of the day (Figure 3) (Fonken et al., 2015).
FIGURE 3.
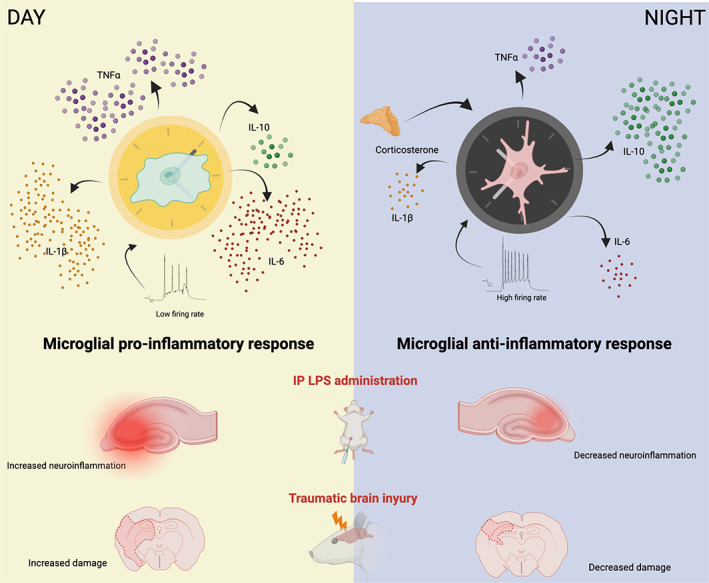
Circadian differences in the microglial response to immune challenges. Hypothetical outcomes of an intraperitoneal (IP) lipopolysaccharide (LPS) administration and a traumatic brain injury (TBI) performed either during the light (resting) phase or at night (activity). In the resting phase, microglia expresses higher levels of pro‐inflammatory cytokines which could contribute to the exacerbation of inflammation after an immune challenge. Instead, during the active phase microglia mainly express IL‐10, which probably ameliorates the damage caused by either an LPS administration or a TBI
Time‐of‐day dependent responses to stress have been also reported in microglia exposed to an inflammatory stimulus (Fonken, Kitt, et al., 2016; Fonken, Weber, et al., 2016). Rats exposed to tail shocks during the day have increased ex vivo microglial activation in response to an LPS challenge than those exposed during the night (Weber et al., 2015). In addition, hippocampal microglia isolated from rats that underwent inescapable stress during the day showed increased mRNA expression of the inflammatory cytokines IL‐1β, IL‐6, and TNF‐α. In contrast, the inflammatory response of the microglia isolated from rats that were subjected to a stress challenge during the night was similar to that of non‐stressed animals (Fonken, Kitt, et al., 2016; Fonken, Weber, et al., 2016). These studies suggest that the particular time of the day in which an immune stimulus is given will determine how microglia will respond to an infection or injury.
A clear example can be observed in a study by Martinez‐Tapia et al. that demonstrated that the outcome of a traumatic brain injury (TBI) differs depending on the moment of the light–dark cycle in which the injury occurs. In this study, the rats subjected to a TBI protocol at midnight had better behavioral outcome than those subjected during the light phase (Martinez‐Tapia et al., 2020). Coincidentally the moment of the worst behavioral outcome coincides with the time of the day in which microglia has the highest levels of pro‐inflammatory cytokines in the cortex (Ni et al., 2019). Importantly the moment of the worst behavioral outcome coincides with the time of the day in which microglia has the highest levels of pro‐inflammatory cytokines in the cortex (Ni et al., 2019). Therefore, the nocturnal low pro‐inflammatory cytokine levels could explain why cortical microglia might be less reactive to TBI event at this moment, leading to a better outcome. Future studies should assess the gene expression profile of cortical microglia before and after the TBI inductions to determine the implication of the time of this type of microglial immune response.
Besides immune challenges, microglial function seems to respond to neuronal activity as well by increasing its surveillance state in response to neurotransmitter release when neurons are most active (Logiacco et al., 2021). Studies have shown that cortical microglia rearrange their filopodia towards firing neurons; this re‐direction of microglial processes is necessary to prevent neuronal network hyper‐excitability (Li et al., 2013; Merlini et al., 2021).
Similarly, hypothalamic neuronal activity in response to acute hypoglycemia rapidly induces the activation of NPY positive neurons thus, promoting the reorganization of surrounding microglial cells and increasing their Iba‐1+ immunoreactivity (Winkler et al., 2019).
Conversely, activating the microglial toll‐like receptor 2 (TLR2) by its ligand Pam3CSK4 (a synthetic triacylated lipopeptide) also induces the rearrangement of hypothalamic microglia toward proopiomelanocortin (POMC) neurons in the hypothalamus, which correlates with increased glutamatergic inputs towards these neurons (Jin et al., 2016). All together, these studies indicate that microglial processes move towards actively firing neurons, that probably, as Merlini et. al, demonstrated could help prevent hyper‐synchronization and hyper excitability of neuronal circuits that consequently could lead either to neuronal death or pathologies (Merlini et al., 2021). The daily microglial activity might contribute to the maintenance of the local milieu and the orchestration of complex physiological processes.
Finally, daily microglial activity might also be involved in the orchestration of complex physiological processes in the overall physiology as demonstrated by Sominsky et al. which employed a genetic rat model of microglial depletion using DTR expression driven by the CX3CR1 promoter; these rats upon the administration of the diphtheria toxin underwent a significant microglial depletion, resulting in a significant loss in the circadian organization of body temperature, energy expenditure, and locomotor activity (De Luca et al., 2019; Sominsky et al., 2021).
Furthermore, Lorea‐Hernández et al. showed that the inhibition or depletion of microglia reduces the respiratory rhythm, suggesting that the microglial release of excitatory modulators such as cytokines might be involved in the daily respiratory rhythm (Camacho‐Hernández et al., 2019). These results suggest that microglia is not only necessary for the maintenance of local brain homeostasis but is also important for ensuring the proper physiological and behavioral output of the central nervous system.
7. CONCLUSIONS AND FUTURE DIRECTIONS
One of the main questions arising from the present review is why the time of the day also shapes microglial functions? Moreover, why should we consider this circadian variation in future studies of microglial functions? The answers might reside in the results of recent single‐cell studies demonstrating that microglial transcriptional profiles vary depending on a wide variety of factors such as; the developmental stage, brain region, and health status of an individual (Bennett et al., 2016; Gosselin et al., 2017; Keren‐Shaul et al., 2017; Li et al., 2019; Masuda et al., 2019).
In 2019, Hammond et al. described the transcriptomic profiles of developing microglia; they found the highest diversity during early development and demonstrated that specialized subpopulations of microglia exist within the brain during development (Hammond et al., 2019). Later, Matsuda and collabs described distinct subclasses of microglial populations in both mice and humans, revealing four major clusters of microglial cells in healthy humans similar to the genetic profiles of mouse microglia (Masuda et al., 2019). In the same study, microglial cells derived from the cerebellum, spinal cord, forebrain, midbrain, cortex, hippocampus, corpus callosum, and facial nucleus were further classified into different sub‐clusters; therefore, various spatiotemporal subclasses of microglial populations were defined (Masuda et al., 2019). Another study by Zheng et al. also determined that cortical and spinal microglia differ, finding three subtypes in the cortex and two in the spinal cord (Zheng et al., 2021). Given the daily changing physiology of the body and the brain, the study of microglial subtypes and their immune responses might benefit from considering microglial circadian rhythms as another factor influencing microglial function and immune response. Nowadays, chronobiologists are convinced that it is crucial to understand the daily changes of every system in the body, and the brain is no exception. Therefore, it is vital to consider the perspective of chronobiology to elucidate whether pharmacological therapies aimed to regulate microglial function will provide the optimal outcome (Corsi et al., 2021).
AUTHOR CONTRIBUTIONS
Mara A. Guzmán‐Ruiz, Natalí N. Guerrero‐Vargas, designed the scope and drafted the manuscript. Mara A. Guzmán‐Ruiz, Natalí N. Guerrero‐Vargas, Alejandra Lagunes‐Cruz, Shellye González‐González, Jesús Enrique García‐Aviles, Gabriela Hurtado‐Alvarado, Rebeca Mendez‐Hernández, wrote original draft, collected data and formatted tables, visualized figures conceptualized the manuscript and supervised the study and edited the original draft. Anahí Chavarría‐Krauser, Jean‐Pascal Morin, Virginia Arriaga‐Avila, Ruud M Buijs, and Rosalinda Guevara‐Guzmán critically reviewed the manuscript. All authors reviewed and approved the final version of the manuscript.
FUNDING INFORMATION
This study was supported by grants DGAPA‐PAPIIT IA204121 to MAGR, DGAPA‐PAPIIT IN221819 to RGG, DGAPA‐PAPIIT IG201321 to RMB and DGAPA‐PAPIIT IA206620 to NNGV, DGAPA‐PAPIIT IN214821 to ACh, CONACyT Ciencia de Frontera‐2019 1783 to ACh.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGMENTS
The authors want to thank Dr. Alfredo Cárdenas‐Rivera and Jorge Marquez for their input and Professor Josefina Bolado for proof‐reading. Rebeca Mendez‐Hernández is a doctoral student from the Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México (UNAM). Graphical abstract and figures were constructed using BioRender (https://biorender.com). This work was supported by grants: DGAPA‐PAPIIT IA204121 to MAGR, DGAPA‐PAPIIT IA206620 to NNGV, DGAPA‐PAPIIT IN214821 to ACK, CONACyT Ciencia de Frontera‐2019 1783 to ACK, DGAPA‐PAPIIT IG201321 to RMB, and DGAPA‐PAPIIT IN221819 to RGG.
Guzmán‐Ruiz, M. A. , Guerrero‐Vargas, N. N. , Lagunes‐Cruz, A. , González‐González, S. , García‐Aviles, J. E. , Hurtado‐Alvarado, G. , Mendez‐Hernández, R. , Chavarría‐Krauser, A. , Morin, J.‐P. , Arriaga‐Avila, V. , Buijs, R. M. , & Guevara‐Guzmán, R. (2023). Circadian modulation of microglial physiological processes and immune responses. Glia, 71(2), 155–167. 10.1002/glia.24261
Mara A. Guzmán‐Ruiz and Natalí N. Guerrero‐Vargas equally contributed to the elaboration of this work and share the 1rs authorship.
Funding information ACh, Grant/Award Numbers: DGAPA‐PAPIIT IN214821, CONACyT Ciencia de Frontera‐2019 1783; NNGV, Grant/Award Number: DGAPA‐PAPIIT IA206620; RMB, Grant/Award Number: DGAPA‐PAPIIT IG201321; RGG, Grant/Award Number: DGAPA‐PAPIIT IN221819; MAGR, Grant/Award Number: DGAPA‐PAPIIT IA204121
Contributor Information
Mara A. Guzmán‐Ruiz, Email: marda1808@gmail.com.
Rosalinda Guevara‐Guzmán, Email: rguevara@unam.mx.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Acsády, L. , & Harris, K. D. (2017). Synaptic scaling in sleep. Science, 355(6324), 457. 10.1126/science.aam7917 [DOI] [PubMed] [Google Scholar]
- Albrecht, U. (2002). Invited review: Regulation of mammalian circadian clock genes. Journal of Applied Physiology (Bethesda, MD: 1985), 92(3), 1348–1355. 10.1152/japplphysiol.00759.2001 [DOI] [PubMed] [Google Scholar]
- Banisadr, G. , Gosselin, R. D. , Mechighel, P. , Rostène, W. , Kitabgi, P. , & Mélik Parsadaniantz, S. (2005). Constitutive neuronal expression of CCR2 chemokine receptor and its colocalization with neurotransmitters in normal rat brain: Functional effect of MCP‐1/CCL2 on calcium mobilization in primary cultured neurons. The Journal of Comparative Neurology, 492(2), 178–192. 10.1002/cne.20729 [DOI] [PubMed] [Google Scholar]
- Bennett, M. L. , Bennett, F. C. , Liddelow, S. A. , Ajami, B. , Zamanian, J. L. , Fernhoff, N. B. , Mulinyawe, S. B. , Bohlen, C. J. , Adil, A. , Tucker, A. , Weissman, I. L. , Chang, E. F. , Li, G. , Grant, G. A. , Hayden Gephart, M. G. , & Barres, B. A. (2016). New tools for studying microglia in the mouse and human CNS. Proceedings of the National Academy of Sciences of the United States of America, 113(12), E1738–E1746. 10.1073/pnas.1525528113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier, L. P. , York, E. M. , Kamyabi, A. , Choi, H. B. , Weilinger, N. L. , & MacVicar, B. A. (2020). Microglial metabolic flexibility supports immune surveillance of the brain parenchyma. Nature Communications, 11(1), 1559. 10.1038/s41467-020-15267-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger, T. , & Schibler, U. (2014). Circadian rhythms ‐ from genes to physiology and disease. Swiss Medical Weekly, 144, w13984. 10.4414/smw.2014.13984 [DOI] [PubMed] [Google Scholar]
- Brancaccio, M. , Patton, A. P. , Chesham, J. E. , Maywood, E. S. , & Hastings, M. H. (2017). Astrocytes control circadian timekeeping in the Suprachiasmatic nucleus via glutamatergic signaling. Neuron, 93(6), 1420–1435. 10.1016/j.neuron.2017.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buijs, R. M. , Guzmán Ruiz, M. A. , Méndez Hernández, R. , & Rodríguez Cortés, B. (2019). The suprachiasmatic nucleus; a responsive clock regulating homeostasis by daily changing the setpoints of physiological parameters. Autonomic Neuroscience, 218, 43–50. 10.1016/j.autneu.2019.02.001 [DOI] [PubMed] [Google Scholar]
- Butovsky, O. , Jedrychowski, M. P. , Moore, C. S. , Cialic, R. , Lanser, A. J. , Gabriely, G. , Koeglsperger, T. , Dake, B. , Wu, P. M. , Doykan, C. E. , Fanek, Z. , Liu, L. , Chen, Z. , Rothstein, J. D. , Ransohoff, R. M. , Gygi, S. P. , Antel, J. P. , & Weiner, H. L. (2014). Identification of a unique TGF‐β‐dependent molecular and functional signature in microglia. Nature Neuroscience, 17(1), 131–143. 10.1038/nn.3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caba, M. , Pabello, M. , Moreno, M. L. , & Meza, E. (2014). Main and accessory olfactory bulbs and their projections in the brain anticipate feeding in food‐entrained rats. Chronobiology International, 31(8), 869–877. 10.3109/07420528.2014.918625 [DOI] [PubMed] [Google Scholar]
- Camacho‐Hernández, N. P. , Lorea‐Hernández, J. J. , & Peña‐Ortega, F. (2019). Microglial modulators reduce respiratory rhythm long‐term facilitation in vitro. Respiratory Physiology & Neurobiology, 265, 9–18. 10.1016/j.resp.2018.07.012 [DOI] [PubMed] [Google Scholar]
- Cermakian, N. , Lange, T. , Golombek, D. , Sarkar, D. , Nakao, A. , Shibata, S. , & Mazzoccoli, G. (2013). Crosstalk between the circadian clock circuitry and the immune system. Chronobiology International, 30(7), 870–888. 10.3109/07420528.2013.782315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury, M. E. , Miyanishi, K. , Takeda, H. , Islam, A. , Matsuoka, N. , Kubo, M. , Matsumoto, S. , Kunieda, T. , Nomoto, M. , Yano, H. , & Tanaka, J. (2020). Phagocytic elimination of synapses by microglia during sleep. Glia, 68(1), 44–59. 10.1002/glia.23698 [DOI] [PubMed] [Google Scholar]
- Colton, C. A. (2009). Heterogeneity of microglial activation in the innate immune response in the brain. Journal of Neuroimmune Pharmacology, 4(4), 399–418. 10.1007/s11481-009-9164-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi, G. , Picard, K. , di Castro, M. A. , Garofalo, S. , Tucci, F. , Chece, G. , … Limatola, C. (2021). Microglia modulate hippocampal synaptic transmission and sleep duration along the light/dark cycle. Glia, 70, 89–105. 10.1002/glia.24090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, K. H. , & Takahashi, J. S. (2019). Circadian clock genes and the transcriptional architecture of the clock mechanism. Journal of Molecular Endocrinology, 63(4), R93–R102. 10.1530/jme-19-0153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane, P. D. , Braun, L. D. , Cornford, E. M. , Nyerges, A. M. , & Oldendorf, W. H. (1980). Cerebral cortical glucose utilization in the conscious rat: Evidence for a circadian rhythm. Journal of Neurochemistry, 34(6), 1700–1706. 10.1111/j.1471-4159.1980.tb11263.x [DOI] [PubMed] [Google Scholar]
- Davalos, D. , Grutzendler, J. , Yang, G. , Kim, J. V. , Zuo, Y. , Jung, S. , … Gan, W. B. (2005). ATP mediates rapid microglial response to local brain injury in vivo. Nature Neuroscience, 8(6), 752–758. 10.1038/nn1472 [DOI] [PubMed] [Google Scholar]
- De Luca, S. N. , Sominsky, L. , Soch, A. , Wang, H. , Ziko, I. , Rank, M. M. , & Spencer, S. J. (2019). Conditional microglial depletion in rats leads to reversible anorexia and weight loss by disrupting gustatory circuitry. Brain, Behavior, and Immunity, 77, 77–91. 10.1016/j.bbi.2018.12.008 [DOI] [PubMed] [Google Scholar]
- de Vivo, L. , Bellesi, M. , Marshall, W. , Bushong, E. A. , Ellisman, M. H. , Tononi, G. , & Cirelli, C. (2017). Ultrastructural evidence for synaptic scaling across the wake/sleep cycle. Science, 355(6324), 507–510. 10.1126/science.aah5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBruyne, J. P. , Weaver, D. R. , & Reppert, S. M. (2007). CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nature Neuroscience, 10(5), 543–545. 10.1038/nn1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desale, S. E. , & Chinnathambi, S. (2020). Role of dietary fatty acids in microglial polarization in Alzheimer's disease. Journal of Neuroinflammation, 17(1), 93. 10.1186/s12974-020-01742-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diering, G. H. , Nirujogi, R. S. , Roth, R. H. , Worley, P. F. , Pandey, A. , & Huganir, R. L. (2017). Homer1a drives homeostatic scaling‐down of excitatory synapses during sleep. Science, 355(6324), 511–515. 10.1126/science.aai8355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early, J. O. , Menon, D. , Wyse, C. A. , Cervantes‐Silva, M. P. , Zaslona, Z. , Carroll, R. G. , Palsson‐McDermott Eva M., Angiari Stefano, Ryan Dylan G., Corcoran Sarah E., Timmons George, Geiger Sarah S., Fitzpatrick Darren J., O'Connell Daniel, Xavier Ramnik J., Hokamp Karsten, Luke A. J O'Neill, Curtis, A. M. (2018). Circadian clock protein BMAL1 regulates IL‐1β in macrophages via NRF2. Proceedings of the National Academy of Sciences of the United States of America, 115(36), E8460–E8468. doi: 10.1073/pnas.1800431115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonken, L. K. , Frank, M. G. , Kitt, M. M. , Barrientos, R. M. , Watkins, L. R. , & Maier, S. F. (2015). Microglia inflammatory responses are controlled by an intrinsic circadian clock. Brain, Behavior, and Immunity, 45, 171–179. 10.1016/j.bbi.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonken, L. K. , Kitt, M. M. , Gaudet, A. D. , Barrientos, R. M. , Watkins, L. R. , & Maier, S. F. (2016). Diminished circadian rhythms in hippocampal microglia may contribute to age‐related neuroinflammatory sensitization. Neurobiology of Aging, 47, 102–112. 10.1016/j.neurobiolaging.2016.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonken, L. K. , Weber, M. D. , Daut, R. A. , Kitt, M. M. , Frank, M. G. , Watkins, L. R. , & Maier, S. F. (2016). Stress‐induced neuroinflammatory priming is time of day dependent. Psychoneuroendocrinology, 66, 82–90. 10.1016/j.psyneuen.2016.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y. , Vidal‐Itriago, A. , Milanova, I. , Korpel, N. L. , Kalsbeek, M. J. , Tom, R. Z. , Kalsbeek, A. , Hofmann, S. M. , & Yi, C. X. (2018). Deficiency of leptin receptor in myeloid cells disrupts hypothalamic metabolic circuits and causes body weight increase. Molecular Metabolism, 7, 155–160. 10.1016/j.molmet.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs, J. E. , Blaikley, J. , Beesley, S. , Matthews, L. , Simpson, K. D. , Boyce, S. H. , … Loudon, A. S. (2012). The nuclear receptor REV‐ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proceedings of the National Academy of Sciences of the United States of America, 109(2), 582–587. 10.1073/pnas.1106750109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, S. , & Taylor, P. R. (2005). Monocyte and macrophage heterogeneity. Nature Reviews. Immunology, 5(12), 953–964. 10.1038/nri1733 [DOI] [PubMed] [Google Scholar]
- Gosselin, D. , Skola, D. , Coufal, N. G. , Holtman, I. R. , Schlachetzki, J. C. M. , Sajti, E. , … Glass, C. K. (2017). An environment‐dependent transcriptional network specifies human microglia identity. Science, 356(6344), eaal3222. 10.1126/science.aal3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin, P. , Dimitry, J. M. , Sheehan, P. W. , Lananna, B. V. , Guo, C. , Robinette, M. L. , Hayes, M. E. , Cedeño, M. R. , Nadarajah, C. J. , Ezerskiy, L. A. , Colonna, M. , Zhang, J. , Bauer, A. Q. , Burris, T. P. , & Musiek, E. S. (2019). Circadian clock protein Rev‐erbα regulates neuroinflammation. Proceedings of the National Academy of Sciences of the United States of America, 116(11), 5102–5107. 10.1073/pnas.1812405116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halberg, F. , Albrecht, P. G. , & Bittner, J. J. (1959). Corticosterone rhythm of mouse adrenal in relation to serum corticosterone and sampling. The American Journal of Physiology, 197, 1083–1085. 10.1152/ajplegacy.1959.197.5.1083 [DOI] [PubMed] [Google Scholar]
- Hammond, T. R. , Dufort, C. , Dissing‐Olesen, L. , Giera, S. , Young, A. , Wysoker, A. , Walker, A. J. , Gergits, F. , Segel, M. , Nemesh, J. , Marsh, S. E. , Saunders, A. , Macosko, E. , Ginhoux, F. , Chen, J. , RJM, F. , Piao, X. , & Stevens, B. (2019). Single‐cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell‐state changes. Immunity, 50(1), 253–271. 10.1016/j.immuni.2018.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, Y. , Koyanagi, S. , Kusunose, N. , Okada, R. , Wu, Z. , Tozaki‐Saitoh, H. , Ukai, K. , Kohsaka, S. , Inoue, K. , Ohdo, S. , & Nakanishi, H. (2013). The intrinsic microglial molecular clock controls synaptic strength via the circadian expression of cathepsin S. Scientific Reports, 3, 2744. 10.1038/srep02744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes, S. E. , Hollopeter, G. , Yang, G. , Kurpius, D. , Dailey, M. E. , Gan, W. B. , & Julius, D. (2006). The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nature Neuroscience, 9(12), 1512–1519. 10.1038/nn1805 [DOI] [PubMed] [Google Scholar]
- Hong, S. , & Stevens, B. (2016). Microglia: Phagocytosing to clear, sculpt, and eliminate. Developmental Cell, 38(2), 126–128. 10.1016/j.devcel.2016.07.006 [DOI] [PubMed] [Google Scholar]
- Hsieh, C. F. , Liu, C. K. , Lee, C. T. , Yu, L. E. , & Wang, J. Y. (2019). Acute glucose fluctuation impacts microglial activity, leading to inflammatory activation or self‐degradation. Scientific Reports, 9(1), 840. 10.1038/s41598-018-37215-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, R. , Wang, T. A. , & Gillette, M. U. (2014). Circadian gating of neuronal functionality: A basis for iterative metaplasticity. Frontiers in Systems Neuroscience, 8, 164. 10.3389/fnsys.2014.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, S. , Kim, J. G. , Park, J. W. , Koch, M. , Horvath, T. L. , & Lee, B. J. (2016). Hypothalamic TLR2 triggers sickness behavior via a microglia‐neuronal axis. Scientific Reports, 6, 29424. 10.1038/srep29424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsbeek, A. , la Fleur, S. , & Fliers, E. (2014). Circadian control of glucose metabolism. Molecular Metabolism, 3(4), 372–383. 10.1016/j.molmet.2014.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren‐Shaul, H. , Spinrad, A. , Weiner, A. , Matcovitch‐Natan, O. , Dvir‐Szternfeld, R. , Ulland, T. K. , … Amit, I. (2017). A unique microglia type associated with restricting development of Alzheimer's disease. Cell, 169(7), 1276–1290. 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- Ko, C. H. , & Takahashi, J. S. (2006). Molecular components of the mammalian circadian clock. Hum Mol Genet, 15 Spec No 2, R271–R277. doi: 10.1093/hmg/ddl207 [DOI] [PubMed] [Google Scholar]
- Kononen, J. , Koistinaho, J. , & Alho, H. (1990). Circadian rhythm in c‐fos‐like immunoreactivity in the rat brain. Neuroscience Letters, 120(1), 105–108. 10.1016/0304-3940(90)90179-d [DOI] [PubMed] [Google Scholar]
- Lauro, C. , & Limatola, C. (2020). Metabolic reprograming of microglia in the regulation of the innate inflammatory response. Frontiers in Immunology, 11, 493. 10.3389/fimmu.2020.00493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leliavski, A. , Dumbell, R. , Ott, V. , & Oster, H. (2015). Adrenal clocks and the role of adrenal hormones in the regulation of circadian physiology. Journal of Biological Rhythms, 30(1), 20–34. 10.1177/0748730414553971 [DOI] [PubMed] [Google Scholar]
- Li, Q. , Cheng, Z. , Zhou, L. , Darmanis, S. , Neff, N. F. , Okamoto, J. , Gulati, G. , Bennett, M. L. , Sun, L. O. , Clarke, L. E. , Marschallinger, J. , Yu, G. , Quake, S. R. , Wyss‐Coray, T. , & Barres, B. A. (2019). Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single‐cell RNA sequencing. Neuron, 101(2), 207–223. 10.1016/j.neuron.2018.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Du, X. F. , & Du, J. L. (2013). Resting microglia respond to and regulate neuronal activity in vivo. Communicative & Integrative Biology, 6(4), e24493. 10.4161/cib.24493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lively, S. , & Schlichter, L. C. (2013). The microglial activation state regulates migration and roles of matrix‐dissolving enzymes for invasion. Journal of Neuroinflammation, 10, 75. 10.1186/1742-2094-10-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logiacco, F. , Xia, P. , Georgiev, S. V. , Franconi, C. , Chang, Y. J. , Ugursu, B. , Sporbert, A. , Kühn, R. , Kettenmann, H. , & Semtner, M. (2021). Microglia sense neuronal activity via GABA in the early postnatal hippocampus. Cell Reports, 37(13), 110128. 10.1016/j.celrep.2021.110128 [DOI] [PubMed] [Google Scholar]
- Martinez‐Tapia, R. J. , Estrada‐Rojo, F. , Lopez‐Aceves, T. G. , Rodríguez‐Mata, V. , Perez‐Torres, A. , Barajas‐Martinez, A. , Garcia‐Velasco, S. , Ugalde‐Muñiz, P. , & Navarro, L. (2020). Diurnal variation induces neurobehavioral and neuropathological differences in a rat model of traumatic brain injury. Frontiers in Neuroscience, 14, 564992. 10.3389/fnins.2020.564992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda, T. , Sankowski, R. , Staszewski, O. , Böttcher, C. , Amann, L. , Sagar, Scheiwe, C. , Nessler, S. , Kunz, P. , van Loo, G. , Coenen, V. A. , Reinacher, P. C. , Michel, A. , Sure, U. , Gold, R. , Grün, D. , Priller, J. , Stadelmann, C. , & Prinz, M. (2019). Spatial and temporal heterogeneity of mouse and human microglia at single‐cell resolution. Nature, 566(7744), 388–392. 10.1038/s41586-019-0924-x [DOI] [PubMed] [Google Scholar]
- McGregor, G. , & Harvey, J. (2018). Regulation of hippocampal synaptic function by the metabolic hormone, leptin: Implications for health and neurodegenerative disease. Frontiers in Cellular Neuroscience, 12, 340. 10.3389/fncel.2018.00340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini, M. , Rafalski, V. A. , Ma, K. , Kim, K. Y. , Bushong, E. A. , Rios Coronado, P. E. , Yan, Z. , Mendiola, A. S. , Sozmen, E. G. , Ryu, J. K. , Haberl, M. G. , Madany, M. , Sampson, D. N. , Petersen, M. A. , Bardehle, S. , Tognatta, R. , Dean, T., Jr. , Acevedo, R. M. , Palop, J. J. , … Akassoglou, K. (2021). Microglial G(i)‐dependent dynamics regulate brain network hyperexcitability. Nature Neuroscience, 24(1), 19–23. 10.1038/s41593-020-00756-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milanova, I. V. , Kalsbeek, M. J. T. , Wang, X. L. , Korpel, N. L. , Stenvers, D. J. , Wolff, S. E. C. , de Goede, P. , Heijboer, A. C. , Fliers, E. , la Fleur, S. E. , Kalsbeek, A. , & Yi, C. X. (2019). Diet‐induced obesity disturbs microglial Immunometabolism in a time‐of‐day manner. Frontiers in Endocrinology (Lausanne), 10, 424. 10.3389/fendo.2019.00424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi, H. (2003). Microglial functions and proteases. Molecular Neurobiology, 27(2), 163–176. 10.1385/mn:27:2:163 [DOI] [PubMed] [Google Scholar]
- Nakazato, R. , Hotta, S. , Yamada, D. , Kou, M. , Nakamura, S. , Takahata, Y. , … Takarada, T. (2017). The intrinsic microglial clock system regulates interleukin‐6 expression. Glia, 65(1), 198–208. 10.1002/glia.23087 [DOI] [PubMed] [Google Scholar]
- Nakazato, R. , Takarada, T. , Yamamoto, T. , Hotta, S. , Hinoi, E. , & Yoneda, Y. (2011). Selective upregulation of Per1 mRNA expression by ATP through activation of P2X7 purinergic receptors expressed in microglial cells. Journal of Pharmacological Sciences, 116(4), 350–361. 10.1254/jphs.11069fp [DOI] [PubMed] [Google Scholar]
- Newman, G. C. , Hospod, F. E. , Patlak, C. S. , & Moore, R. Y. (1992). Analysis of in vitro glucose utilization in a circadian pacemaker model. The Journal of Neuroscience, 12(6), 2015–2021. 10.1523/jneurosci.12-06-02015.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, J. , Wu, Z. , Meng, J. , Saito, T. , Saido, T. C. , Qing, H. , & Nakanishi, H. (2019). An impaired intrinsic microglial clock system induces neuroinflammatory alterations in the early stage of amyloid precursor protein knock‐in mouse brain. Journal of Neuroinflammation, 16(1), 173. 10.1186/s12974-019-1562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn, A. , Kirchhoff, F. , & Helmchen, F. (2005). Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science, 308(5726), 1314–1318. 10.1126/science.1110647 [DOI] [PubMed] [Google Scholar]
- Novak, M. L. , & Koh, T. J. (2013). Macrophage phenotypes during tissue repair. Journal of Leukocyte Biology, 93(6), 875–881. 10.1189/jlb.1012512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orihuela, R. , McPherson, C. A. , & Harry, G. J. (2016). Microglial M1/M2 polarization and metabolic states. British Journal of Pharmacology, 173(4), 649–665. 10.1111/bph.13139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster, H. , Damerow, S. , Kiessling, S. , Jakubcakova, V. , Abraham, D. , Tian, J. , … Eichele, G. (2006). The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metabolism, 4(2), 163–173. 10.1016/j.cmet.2006.07.002 [DOI] [PubMed] [Google Scholar]
- Panda, S. , Antoch, M. P. , Miller, B. H. , Su, A. I. , Schook, A. B. , Straume, M. , … Hogenesch, J. B. (2002). Coordinated transcription of key pathways in the mouse by the circadian clock. Cell, 109(3), 307–320. 10.1016/s0092-8674(02)00722-5 [DOI] [PubMed] [Google Scholar]
- Paolicelli, R. C. , Bisht, K. , & Tremblay, M. (2014). Fractalkine regulation of microglial physiology and consequences on the brain and behavior. Frontiers in Cellular Neuroscience, 8, 129. 10.3389/fncel.2014.00129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego, C. , Fumagalli, S. , & De Simoni, M. G. (2011). Temporal pattern of expression and colocalization of microglia/macrophage phenotype markers following brain ischemic injury in mice. Journal of Neuroinflammation, 8, 174. 10.1186/1742-2094-8-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff, R. M. (2016). A polarizing question: Do M1 and M2 microglia exist? Nature Neuroscience, 19(8), 987–991. 10.1038/nn.4338 [DOI] [PubMed] [Google Scholar]
- Reick, M. , Garcia, J. A. , Dudley, C. , & McKnight, S. L. (2001). NPAS2: An analog of clock operative in the mammalian forebrain. Science, 293(5529), 506–509. 10.1126/science.1060699 [DOI] [PubMed] [Google Scholar]
- Reppert, S. M. , & Weaver, D. R. (2002). Coordination of circadian timing in mammals. Nature, 418(6901), 935–941. 10.1038/nature00965 [DOI] [PubMed] [Google Scholar]
- Rodríguez‐Cortés, B. , Hurtado‐Alvarado, G. , Martínez‐Gómez, R. , León‐Mercado, L. A. , Prager‐Khoutorsky, M. , & Buijs, R. M. (2022). Suprachiasmatic nucleus‐mediated glucose entry into the arcuate nucleus determines the daily rhythm in blood glycemia. Current Biology, 32, 796–805. 10.1016/j.cub.2021.12.039 [DOI] [PubMed] [Google Scholar]
- Sacks, D. , Baxter, B. , Campbell, B. C. V. , Carpenter, J. S. , Cognard, C. , Dippel, D. , Eesa, M. , Fischer, U. , Hausegger, K. , Hirsch, J. A. , Hussain, M. S. , Jansen, O. , Jayaraman, M. V. , Khalessi, A. A. , Kluck, B. W. , Lavine, S. , & Vorwerk, D. (2018). Multisociety consensus quality improvement revised consensus statement for endovascular therapy of acute ischemic stroke. International Journal of Stroke, 13(6), 612–632. 10.1177/1747493018778713 [DOI] [PubMed] [Google Scholar]
- Sato, S. , Sakurai, T. , Ogasawara, J. , Takahashi, M. , Izawa, T. , Imaizumi, K. , Taniguchi, N. , Ohno, H. , & Kizaki, T. (2014). A circadian clock gene, rev‐erbα, modulates the inflammatory function of macrophages through the negative regulation of Ccl2 expression. Journal of Immunology, 192(1), 407–417. 10.4049/jimmunol.1301982 [DOI] [PubMed] [Google Scholar]
- Schafer, D. P. , Lehrman, E. K. , Kautzman, A. G. , Koyama, R. , Mardinly, A. R. , Yamasaki, R. , Ransohoff, R. M. , Greenberg, M. E. , Barres, B. A. , & Stevens, B. (2012). Microglia sculpt postnatal neural circuits in an activity and complement‐dependent manner. Neuron, 74(4), 691–705. 10.1016/j.neuron.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeller, D. A. , Cella, L. K. , Sinha, M. K. , & Caro, J. F. (1997). Entrainment of the diurnal rhythm of plasma leptin to meal timing. The Journal of Clinical Investigation, 100(7), 1882–1887. 10.1172/jci119717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter, R. , & Schwartz, M. (2013). CNS sterile injury: Just another wound healing? Trends in Molecular Medicine, 19(3), 135–143. 10.1016/j.molmed.2012.11.007 [DOI] [PubMed] [Google Scholar]
- Sominsky, L. , Dangel, T. , Malik, S. , De Luca, S. N. , Singewald, N. , & Spencer, S. J. (2021). Microglial ablation in rats disrupts the circadian system. The FASEB Journal, 35(2), e21195. 10.1096/fj.202001555RR [DOI] [PubMed] [Google Scholar]
- Stein, M. , Keshav, S. , Harris, N. , & Gordon, S. (1992). Interleukin 4 potently enhances murine macrophage mannose receptor activity: A marker of alternative immunologic macrophage activation. The Journal of Experimental Medicine, 176(1), 287–292. 10.1084/jem.176.1.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, B. , Allen, N. J. , Vazquez, L. E. , Howell, G. R. , Christopherson, K. S. , Nouri, N. , Micheva, K. D. , Mehalow, A. K. , Huberman, A. D. , Stafford, B. , Sher, A. , Litke, A. M. , Lambris, J. D. , Smith, S. J. , John, S. W. , & Barres, B. A. (2007). The classical complement cascade mediates CNS synapse elimination. Cell, 131(6), 1164–1178. 10.1016/j.cell.2007.10.036 [DOI] [PubMed] [Google Scholar]
- Sueviriyapan, N. , Tso, C. F. , Herzog, E. D. , & Henson, M. A. (2020). Astrocytic modulation of neuronal activity in the Suprachiasmatic nucleus: Insights from mathematical modeling. Journal of Biological Rhythms, 35(3), 287–301. 10.1177/0748730420913672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama, F. , Hayashi, Y. , Wu, Z. , Liu, Y. , & Nakanishi, H. (2016). Diurnal dynamic behavior of microglia in response to infected bacteria through the UDP‐P2Y6 receptor system. Scientific Reports, 6, 30006. 10.1038/srep30006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama, F. , Zhang, X. , Hayashi, Y. , Wu, Z. , & Nakanishi, H. (2017). Dysfunction in diurnal synaptic responses and social behavior abnormalities in cathepsin S‐deficient mice. Biochemical and Biophysical Research Communications, 490(2), 447–452. 10.1016/j.bbrc.2017.06.061 [DOI] [PubMed] [Google Scholar]
- Thundyil, J. , Pavlovski, D. , Sobey, C. G. , & Arumugam, T. V. (2012). Adiponectin receptor signalling in the brain. British Journal of Pharmacology, 165(2), 313–327. 10.1111/j.1476-5381.2011.01560.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujeque‐Ramos, S. , Castillo‐Rolón, D. , Galarraga, E. , Tapia, D. , Arenas‐López, G. , Mihailescu, S. , & Hernández‐López, S. (2018). Insulin regulates GABA(a) receptor‐mediated tonic currents in the prefrontal cortex. Frontiers in Neuroscience, 12, 345. 10.3389/fnins.2018.00345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Swieten, M. M. , Pandit, R. , Adan, R. A. , & van der Plasse, G. (2014). The neuroanatomical function of leptin in the hypothalamus. Journal of Chemical Neuroanatomy, 61‐62, 207–220. 10.1016/j.jchemneu.2014.05.004 [DOI] [PubMed] [Google Scholar]
- Vieira, E. , Mirizio, G. G. , Barin, G. R. , de Andrade, R. V. , Nimer, N. F. S. , & La Sala, L. (2020). Clock genes, inflammation and the immune system‐implications for diabetes, obesity and neurodegenerative diseases. International Journal of Molecular Sciences, 21(24), 9743. 10.3390/ijms21249743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalta, S. A. , Nguyen, H. X. , Deng, B. , Gotoh, T. , & Tidball, J. G. (2009). Shifts in macrophage phenotypes and macrophage competition for arginine metabolism affect the severity of muscle pathology in muscular dystrophy. Human Molecular Genetics, 18(3), 482–496. 10.1093/hmg/ddn376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyazovskiy, V. V. , Cirelli, C. , Pfister‐Genskow, M. , Faraguna, U. , & Tononi, G. (2008). Molecular and electrophysiological evidence for net synaptic potentiation in wake and depression in sleep. Nature Neuroscience, 11(2), 200–208. 10.1038/nn2035 [DOI] [PubMed] [Google Scholar]
- Wang, C. , Lutes, L. K. , Barnoud, C. , & Scheiermann, C. (2022). The circadian immune system. Science Immunology, 7(72), eabm2465. 10.1126/sciimmunol.abm2465 [DOI] [PubMed] [Google Scholar]
- Wang, X. L. , Kooijman, S. , Gao, Y. , Tzeplaeff, L. , Cosquer, B. , Milanova, I. , … Yi, C. X. (2021). Microglia‐specific knock‐down of Bmal1 improves memory and protects mice from high fat diet‐induced obesity. Molecular Psychiatry, 26, 6336–6349. 10.1038/s41380-021-01169-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. L. , Wolff, S. E. C. , Korpel, N. , Milanova, I. , Sandu, C. , Rensen, P. C. N. , Kooijman, S. , Cassel, J.‐C. , Kalsbeek, A. , Boutillier, A.‐L. , & Yi, C. X. (2020). Deficiency of the circadian clock gene Bmal1 reduces microglial Immunometabolism. Frontiers in Immunology, 11, 586399. 10.3389/fimmu.2020.586399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, M. D. , Frank, M. G. , Tracey, K. J. , Watkins, L. R. , & Maier, S. F. (2015). Stress induces the danger‐associated molecular pattern HMGB‐1 in the hippocampus of male Sprague Dawley rats: A priming stimulus of microglia and the NLRP3 inflammasome. The Journal of Neuroscience, 35(1), 316–324. 10.1523/jneurosci.3561-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisz, F. , Piccinin, S. , Mango, D. , Ngomba, R. T. , Mercuri, N. B. , Nicoletti, F. , & Nisticò, R. (2017). The role of adiponectin receptors in the regulation of synaptic transmission in the hippocampus. Synapse, 71(5). 10.1002/syn.21964 [DOI] [PubMed] [Google Scholar]
- Winkler, Z. , Kuti, D. , Polyák, Á. , Juhász, B. , Gulyás, K. , Lénárt, N. , Dénes, Á. , Ferenczi, S. , & Kovács, K. J. (2019). Hypoglycemia‐activated hypothalamic microglia impairs glucose Counterregulatory responses. Scientific Reports, 9(1), 6224. 10.1038/s41598-019-42728-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, C. X. , Walter, M. , Gao, Y. , Pitra, S. , Legutko, B. , Kälin, S. , Layritz, C. , García‐Cáceres, C. , Bielohuby, M. , Bidlingmaier, M. , Woods, S. C. , Ghanem, A. , Conzelmann, K. K. , Stern, J. E. , Jastroch, M. , & Tschöp, M. H. (2017). TNFα drives mitochondrial stress in POMC neurons in obesity. Nature Communications, 8, 15143. 10.1038/ncomms15143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz, B. O. , Suchard, M. A. , Wong, M. L. , McCann, S. M. , & Licinio, J. (2004). Alterations in the dynamics of circulating ghrelin, adiponectin, and leptin in human obesity. Proceedings of the National Academy of Sciences of the United States of America, 101(28), 10434–10439. 10.1073/pnas.0403465101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, J. , Ru, W. , Adolacion, J. R. , Spurgat, M. S. , Liu, X. , Yuan, S. , Liang, R. X. , Dong, J. , Potter, A. S. , Potter, S. S. , Chen, K. , Chen, R. , Varadarajan, N. , & Tang, S. J. (2021). Single‐cell RNA‐seq analysis reveals compartment‐specific heterogeneity and plasticity of microglia. iScience, 24(3), 102186. 10.1016/j.isci.2021.102186 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


