Abstract
Diabetic foot ulcers (DFUs) remain a very prevalent and challenging complication of diabetes worldwide due to high morbidity, high risks of lower extremity amputation and associated mortality. Despite major advances in diabetes treatment in general, there is a paucity of FDA approved technologies and therapies to promote successful healing. Furthermore, accurate biomarkers to identify patients at risk of non-healing and monitor response-to-therapy are significantly lacking. To date, research has been slowed by a lack of coordinated efforts among basic scientists and clinical researchers and confounded by non-standardized heterogenous collection of biospecimen and patient associated data. Novel technologies, especially those in the single and ‘multiomics’ arena, are being used to advance the study of diabetic foot ulcers but require pragmatic study design to ensure broad adoption following validation. These high throughput analyses offer promise to investigate potential biomarkers across wound trajectories and may support information on wound healing and pathophysiology not previously well understood. Additionally, these biomarkers may be used at the point-of-care. In combination with national scalable research efforts, which seek to address the limitations and better inform clinical practice, coordinated and integrative insights may lead to improved limb salvage rates.
Keywords: Diabetic foot ulcers, Infection, Limb salvage, Osteomyelitis, Standard care
1. Problem statement
Diabetes mellitus (DM) may affect every third adult American by 2050.1 The estimated prevalence of DM among US adults increased from 5.3 % in 1976–1980 to 11.5 % in 2011–2014, and outpaced the global increase during the same period 2 and up to 35 % of patients with DM will develop a diabetic foot ulcer (DFU).3 Even when adherent to current standard care (e.g., wound debridement, dressing changes, offloading, DM control), less than half of DFU heal and up to 60 % may develop infection.4-6 These data suggest that additional factors are involved in the development of DFU infection and may also explain why many patients fail to respond to standard care and have a high rate of DFU reoccurrence. As five-year mortality rates range from 29 %–69 % following minor (partial foot) amputation and from 52 % to 80 % for patients with major lower extremity amputation,7-10 better understanding the various factors promoting DFU healing and non-healing will help to prevent all amputation.
Relevant biomarkers to identify individuals at-risk of developing complications from DFU are currently lacking. Traditional biomarkers (e.g., wound surface area and % wound granulation) used as surrogate markers to predict eventual wound healing and identify infections are reactive and require direct observation over multiple weeks to predict wound progression.
Development and implementation of cutting-edge technologies to prevent and to evaluate personal risk of complications from DFU at the point of care are needed. DFU biomarkers can be used to inform and alter care, when necessary, for an individual immediately upon evaluation rather than after several weeks of monitoring. Thus, it is critical to develop more intuitive biomarkers to advance standard care for DFU care.
2. Diabetic foot ulcers current state of play
Those providers who manage DFUs should be familiar with the four primary tenets of wound healing and are supported by multiple national and international clinical practice guidelines. These tenets are universally applied during initial and reassessment of DFU because normal wound healing is a dynamic continuum.
The first tenet is identification (and eradication) of infection. Diabetic foot infections can be acute, subacute, and chronic. Infection management may require tissue and/or bone cultures to guide therapy. Enhanced assessment of infection should include radiographs and enhanced modalities such as MRIs and bone scans, as needed. Management strategies include: medical management with antibiotics, or a variety of surgical procedures as a more definitive approach. How best to eradicate infection must be weighed against patient medical status and quality of life. The second tenet is assessment of a patient's lower extremity vascular status. The delivery of antibiotics and essential nutrients for healing can only occur if the patient's blood flow is adequate. Assessment of vascular status include measures of ankle-brachial index, toe-brachial index, transcutaneous oxygen pressure (tcPO2), or use of duplex ultrasound, as needed. Outside of traditional pharmacological therapy (e.g., antiplatelet, statin, and antihypertensive agents), interdisciplinary management may require endovascular or surgical revascularization despite greater risk of adverse perioperative events in this patient population. Hyperbaric oxygen therapy also remains an adjunctive treatment to increase oxygen delivery and promotion of healing. Third, wound debridement is essential for wound healing. There is a saying in the wound healing world, “what you take off of the wound is more important than what you put on the wound.” Removing devitalized tissue deceases the bioburden and allows for angiogenesis and the stimulation of growth factors key in for the wound healing process to progress. There are many advanced dressings, but they are only effective if applied to a healthy wound bed. Studies have unequivocally demonstrated wound debridement has qualitative and quantitative benefits which promote wound healing.11,12 Finally, a fourth tenet for wound healing of DFU is pressure relief or ‘offloading’. Pressure relief allows a DFU the ability to heal without the constant pressure of one's own body weight and is essential.
Patient and provider adherence to this offloading tenet is not universal and the delivery of usual care varies. It is well documented the “gold-standard” for offloading DFU is total contact casting (TCC). TCC can reduce pressure on the plantar foot by up to 84 % to 92 % at the ulcer site, and heals most DFUs within six to eight weeks.16 Bus and coworkers conducted a systematic review and found that non-removable offloading devices (i.e., TCC) are more effective than removable devices at healing neuropathic foot ulcers.17 Better offloading adherence predicted smaller DFU size at 6 weeks but adherence to offloading recommendation was estimated to be under 30 % with removable-type offloading devices.13 Despite this knowledge, in a large multi-center study published by Wu et al., among 895 centers involved in the treatment of DFUs across the United States, only 1.7 % used TCC for the majority of cases.14 18 The authors noted several contributing factors to the minimal use of TCC, including patient tolerance, time needed to apply and remove the cast, materials cost, and reimbursement challenges.18
2.1. A lack of informative time-sensitive biomarkers to be used at point-of care
Currently, there are no effective tools in clinical wound practice that will readily identify and differentiate healing from non-healing patients beside tracking wound surface area progression over time. Surface area measurements including percent change in area, logarithmic (log) healing rate,14 or log wound surface area ratio14 can be used as surrogate endpoints for later complete wound reepithelization.15-17 It is well established that DFU which do not have a 40 % wound surface area reduction by 4-weeks undergoing standard care will be unlikely to heal by 12-weeks by similar therapy.16 More specifically, DFU reaching 60 % wound surface area reduction by 4-weeks with standard care therapy demonstrate a 77 % probability of healing by 16-weeks.18 However, these measures have up to a 45 % error rate and are confounded further by an intra-rater reliability of only 30 % and an inter-rater reliability of 60 %.19
Additional wound parameters, such as wound color (e.g., % wound granulation), have been shown to have prognostic value. Wounds that have a higher % of granulation tissue heal more readily and it is often used as an outcome in DFU clinical trials.20-22 Prior work from Valenzueula-Silva et al., found that wound base granulation tissue >50 % at 2 weeks, >75 % at the end of treatment, and 16 % reduction in area had >70 % positive predictive value for healing.23 In this same study, wound granulation tissue was estimated by the provider in infected DFU (Wagner grade 3 or 4)23 thereby introducing an element of bias and excluding many DFUs which are chronic but non-infected. Therefore, while useful in infected DFU, the demonstrated utility of % granulation tissue as a biomarker in non-infected DFU is transitive, not causal or proven.
Even still, receiver operator curves (ROC) for various DFU predictive models, those which predict whether a wound will ultimately heal or not, leave much to be desired. An early model incorporated wound clinical features (e.g., age of wound, size of wound, extent of infection) to predict DFUs that will not heal. The ROC for this approach was limited with area under the receiver operating characteristic curve (AUROC) of 0.66–0.70.24 An updated version in 2022 which assessed the clinical factors of wound surface area and wound duration (in days) combined with machine learning (ML) prediction had an improved discrimination capacity AUROC of 0.7212 for wound healing.25 Others have taken a systematic approach to evaluate patient-specific variables from a comprehensive literature review which were found to be associated with healing. Fife et al. created the ‘Wound Healing Index (WHI)’ to highlight ten of these factors; the WHI was able to generate a predictive AUROC of 0.648–0.668 for wound healing.26 Indeed, prognostic classification systems using multivariate analysis of perfusion- and wound-based predictors, like the Wound, Ischemia, and Foot infection (WiFi) classification system,27 have provided a useful tool to clinicians, but only in a subset of patients. Kaplan-Meier curve analyses demonstrated Wifi classification was a better predictor of diabetic wound healing than angiosome perfusion or pedal arch grade27; however, these data are only applicable in patients with critical limb ischemia undergoing revascularization. Kaplan-Meier analysis also support TcPO2 as a useful prognosis as patients with values >40 mmHg possess a significantly 25 % higher probability of wound closure compared to patients with <20 mmHg28; however, reproducibility and technical feasibility remain an ongoing dilemma for practical use of tcPO2. Additional models evaluated the ratio of matrix metalloproteinase-9 (MMP-9) to tissue inhibitor of MMPs (TIMP) and found a cutoff value of MMP-9/TIMP ratio at <0.395 best predicted a reduction in wound surface area of >80 % by 4 weeks.29 Unfortunately, the hailed biomarker suffered from relatively poor discriminative capacity (AUROC 0.65). Furthermore, these models failed to provide relevant time-to-event information and have not been widely adopted as a result.30
2.2. Major identified gaps limiting progress
As evidenced by worsening trends of non-traumatic lower extremity amputations,31 there are at least four major gaps which require actionable change to advance standard care and prevent additional amputations. First, and most problematic, is that diabetic foot complications do not receive broad public attention. The impact of DFU on individuals are enormous. This is demonstrated by total costs associated with diabetes-related lower extremity complications. In 2015, costs approached $80 billion and paralleled all cancer care costs in the US.32 To combat this, greater advocacy must occur more broadly through public service announcements. Emphasis can be placed on finding a cure for DM and to highlight the personalized approaches needed to heal DFU. For example, breast cancer advocacy groups have aligned with the National Football League (NFL) every October and promotes compassion for those suffering from breast cancer. DFU should be no different and more spirited representation of DM will increase awareness of this condition and its related complications.
Second, provider education about management topics in diabetic foot need to be emphasized in medical education and training. This includes providers, allied health professionals, and anyone who interacts with a patient with DM. DFU is estimated to be the most common cause of diabetes-related admission to hospital33 and will be encountered in a medical career regardless of specialty training. Currently, this knowledge gap among medical professionals is persistent and is best demonstrated by the diabetic Charcot foot. In a survey of >400 providers at a major tertiary care facility, approximately 67 % of physicians self-reported poor or no knowledge about the diabetic Charcot foot.34 It suggests teaching of general management strategies (in diabetic foot) are needed. Thus, the burden to improve care for diabetic foot complications remains on the medical community and this gap may not be only due to provider knowledge limitations, but also health system limitations in team care integration. Structured and dedicated limb salvage services are needed at every institution to triage and appropriately manage all diabetes-related lower extremity complications. This concerted effort will further support the multidisciplinary care team required to care for these patients longitudinally.
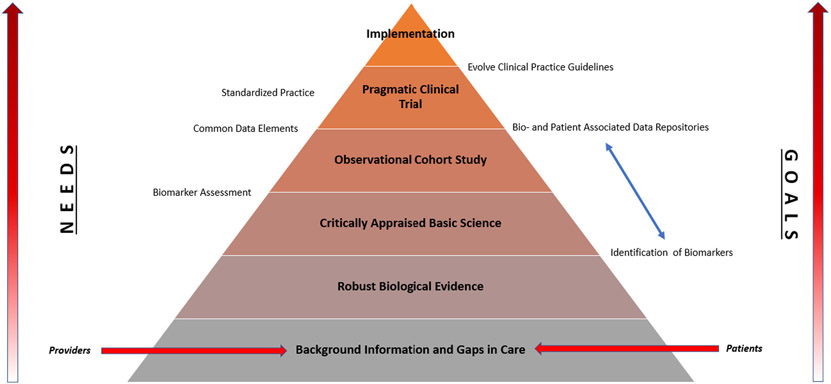
Third, and which highlights the former, is a requirement for clinical trials to be pragmatically designed and evaluate novel wound products under real-life routine practice conditions. Historically, standard care involved the application of a wet to dry (WTD) dressing to a wound. When WTD is removed, it performed non-selective debridement and was re-applied as prescribed. This standard care remains a benchmark which novel wound care products are compared against. However, for clinical practitioners, an effectiveness study, which emphasizes whether or not a treatment works pragmatically in the real-world, is more appropriate.35 We must uphold this standard rather than a historic one which is grossly oversimplified with respect to the DFU question. A ladder of opportunity to advance standard care practice is presented in Fig. 1.
Fig. 1.
Opportunities to advance standard care for diabetic foot. Providers and patients provide clinical evidence of known gaps in care. The process to advance care becomes progressively more demanding at each level. Through focused planning, common data collection, and execution of pragmatic trials in scientific networks, rigorous assessment of new evidence will lead to broad implementation of evidence into regular care of DFU patients. “NEEDS” and “GOALS” at each level are highlighted on the left and right, respectively.
Finally, access to standardized clinical data to better understand best practices is lacking. While requirements for ethical billing are well defined in local care determination (LCD) documents, provider notes are largely not easily shared and remain prohibitively inaccessible to interested researchers. A registry like the “Diabetic Foot Ulcer Registry” (ClinicalTrials.gov identifier: NCT02813161) represents a collaborative effort between the American Podiatric Medical Association (APMA) and the U.S. Wound Registry to create a longitudinal real-world registry of DFUs abstracted from electronic health record (EHR) data obtained in the course of clinical care.36 The richness in data produced by clinical care documentation is an untapped research tool which can inform and monitor best clinical practice through big data analyses. Until there is honest, unified documentation standards and outcome reporting in the DFU space, practitioners and patients alike will continue to be subjected to customary practice rather than evidence-based medicine.
2.3. Innovative new methods
To fill knowledge gaps, molecular techniques offer promise to investigate potential DFU biomarkers at various clinical stages and may support information on wound trajectory. Biomarker assessment can be categorized according to biological material studied. Inclusion of inflammatory, genomic, proteomic, metabolic, epigenetic, microbial, and imaging biomarkers is necessary as they are interrelated with emerging technologies and can be integrated with standard care practices. Multiomic data provides a more granular understanding of the spatial and temporal relationships of cells involved in the wound healing continuum. Below are emerging technologies which may contribute significantly to progressing knowledge of DFU biomarkers at a molecular level. The techniques are highlighted with available clinically relevant data provided in each subsection.
2.4. Single-cell RNA-sequencing (scRNA-seq)
Single-cell RNA-sequencing (scRNA-Seq) analysis provides deep insight into cell function and disease pathophysiology.37 It profiles the transcriptome landscape of individual cells in heterogenous tissues, disease or normal. In cancer biology, scRNA-Seq is used to plot cancer microenvironments and evaluate molecular mechanisms which may contribute to the (specific) disease state. Evaluating the microenvironment allows investigators to identify potential therapeutic targets. This approach has only recently been conducted in DFU tissue.37
An initial study of DFU resected during foot surgery was used for single cell-preparation and compared to non-diabetic skin and non-DFU tissue. Analysis demonstrated different fibroblast clusters were expressive in distinctive patterns and associated with an “distinctive injury response-associated gene expression profile” thought to be related DFU chronic inflammation in healers versus non-healers. However, these measurements were cross-sectional and qualified healers and non-healers at 12-weeks following intervention. Regardless, having the ability to map transitory states of healing during longitudinal follow-up encounters may prove valuable when assessing patients across wound healing trajectories. Further research is needed to better inform this technology.
In a murine model, RNA sequencing was used to evaluate expressional profile differences in epidermal, dermal, and subcutaneous layers of skin directly adjacent to wounded skin. The authors noted key wound signaling pathway involvement in specific layers. Additionally, dressing material and/or the placement of negative pressure wound therapy (NPWT) demonstrated the capacity to modulate early wound signaling in the skin layers.38 This study provided valuable information on medical material interactions with wound tissue and/or NPWT. It also provided information about genomic changes occurring early in the wound healing process that was previously not well elucidated. In summary, the authors noted the experimental data offered new perspective and methodology for characterizing wound tissue responses to wounding and provided new insight into localizing expressional patterns and genomic profiles within different wound regions.38
The level of additional detail provided by a layered approach may ultimately help improve clinical therapies. It highlights the growing utility of RNA seq to evaluate the wound healing environment at various time intervals.
2.5. Spatial transcriptomics (ST)
A component lost during cellular assessment via scRNA-Seq is the spatial relationship of molecular information.39 Because of cell emulsification methods used to perform scRNA-Seq, visualization and quantification of the molecular transcriptome is lost. However, spatial transcriptomics (ST) is a more recent method that enables the visualization and quantitation of the transcriptome in individual tissue sections.
Traditional measures of cell surface profiles which measure tissue depth and define cell lineages40 are insufficient to identify cell subtype by region within a wound. Using a combination of ST techniques, including scRNA-seq with single cell chromatin accessibility (scATAC-seq), investigators have shown in murine models that traditional cell surface markers are insufficient to characterize regional heterogeneity among wound healing fibroblasts.41 Specifically, they found there were no differences in fibroblast subpopulations based on region of the wound (i.e., inner and outer wound) when using traditional measures of cell lineage.41
Moreover, other ST platforms exist and include RNA-scope and multiplexed error-robust fluorescence in situ hybridization (MER-FISH).42,43 First, RNA-scope is a novel in situ hybridization assay for detection of target RNA within intact cells while MERFISH allows for simultaneously measuring the copy number and spatial distribution of RNA species from individual cells. These methods can facilitate improved understanding of spatial resolution of RNA transcripts within the variety of subcellular populations involved throughout the phases of wound healing. However, at the time of this review, ST techniques have not been utilized in DFU research. Use of ST to interpret the transcriptome as it relates to biogeography in DFU will be important to elucidate cellular differences with enhanced temporospatial resolution. The level of detail may allow a more tailored wound product application to specific part of a wound (e.g., areas of increased depth versus superficial margins) and promote wound healing.
2.6. Proteomics
Proteomics enables characterization of protein expression, immunophenotype, and functionality. It can be especially valuable in immunology and wound healing where a myriad of cellular processes is ongoing and in coordination with one another. A tool called spatial proteomics or CO-detection by indexing (CODEX), supports the localization of proteins, and their dynamic interactions, at the subcellular level; allowing for a more complete description of tissue architecture.44,45 Furthermore, newer studies have harnessed the power of comparative spatial proteomics as a discovery tool as well. The DFU landscape is no exception.
For example, to elucidate how DFU healing occurs while using negative pressure wound therapy (NPWT), the authors analyzed granulation tissue from debrided DFUs using label-free quantitative mass spectrometry prior to- and following NPWT for one week.46 The authors discovered that 33 proteins were upregulated and 3 proteins were downregulated; NPWT altered multiple proteins within the granulation tissue of these ulcers, mostly associated with detoxification and antioxidation, regulation of inflammation, complement, and coagulation cascades, and modification of lipid metabolism and extracellular matrix.46 Specifically, their findings highlighted certain proteins (e.g., PRDX2, PROS1, CTSS and ITIH4)46 that are supported by congruous reports47,48 and which could act as future potential biomarkers to assess the efficacy of treatment response to NPWT in DFU.
2.7. Immunophenotyping and epigenetics
Impaired wound healing in DM is thought to occur due to defects in the inflammatory response, epithelialization, angiogenesis, and migration of epithelial cells. Although many immune and structural cells are altered in diabetic wounds, one of the most well-studied immune cells specifically related to the inflammatory process is the monocyte/macrophage. In most of the wound healing literature to date, it is well described that in normal wound repair, monocyte/macrophages demonstrate high plasticity and convert from a pro-inflammatory macrophage to a reparative macrophage during the course of healing. While significant strides have been made in recognizing the contribution of blood monocyte-derived macrophages to tissue repair, it remains challenging to study monocyte/macrophage phenotypes in vivo due to a lack of consensus on nomenclature, cell-marker definitions, and debate over effector-cell origin.49,50 For example, M1-like (pro-inflammatory) consensus genes (Arg1, Socs1, Nos2, Marco, Il27, Il23a, Il12a, Il6, Nfkbiz, Tnf, and Irf5) demonstrate a resemblance between in vivo Ly6CHi wound monocyte/macrophages and M1[LPS/IFNγ] in vitro macrophages. Further, M2-like (reparative) consensus genes (Arg1, Chil3, Retnla, Nos2, Ccl24, Marco, Irf4, Ccl22, Ccl17, Alox15 and Socs2) demonstrate genes shared by both subsets, including Arg1 and Nos2, were most strongly expressed by in vivo Ly6C wound monocyte/macrophages. In fact, many of the genes that were distinct to the M2 subset were not highly expressed by any in vivo subset.51 Cytometry by time of flight (CyTOF) is one method where mass cytometry is capable of quantifying over 40 labeled targets on the surface of single cells, allowing for high-throughput interrogation of immune cell subpopulations. When coupled with proteomic stabilizing techniques, which preserve the integrity of single cell structure and proteome, researchers can characterize both extracellular and intracellular protein signatures and shifts in protein production at single cell resolution. What has come of the advanced sequencing data is the conclusion that there are multiple monocyte-macrophage phenotypes in tissue that exist along a spectrum and thus, prior conventional descriptions are not valid in vivo. These findings help to promote more accurate immunophenotyping of monocyte/macrophages in human tissue and may identify future targets for interventional therapy.
Similarly, newly published work by Wolf et al. depict epigenetic regulation of inflammation in keratinocytes during wound repair which was informed by previous ChIP-seq/RNA-seq findings. The team showed histone methyltransferase mixed-lineage leukemia 1 (MLL1) regulates IFNk in keratinocytes. Impaired IFNk production was evident in both human and murine diabetic wounds. Indeed, when IFNk was ablated, aberrant early phase inflammation was established with concomitant perturbation in wound healing. Upon keratinocyte specific deletion of MLL1, a similar decrease in IFNk expression was seen. These data strongly suggest that keratinocytes of the DM patient undergo epigenetic regulation of IFNk gene expression. The exact upstream mechanisms regulating MLL1-mediated Ifnk gene expression in keratinocytes remain unclear but MLL1/IFNk i in keratinocytes could be a novel approach to target inflammation in wounds. These studies demonstrate advancement of bench science informs our clinical understanding of wound healing.
2.8. Metagenomic assessment
Infection in DFU, including osteomyelitis, remain a diagnostic and therapeutic challenge. Consensus guidelines depend upon conventional culture techniques. Studies which evaluate culture-independent techniques often lack standardized assessment of a sentinel DFU. Numerous validated diabetic foot risk classification systems are described in the literature53-56 but focus on individuals who are at-risk to develop DFU and do not assess the risk of the DFU itself. Nearly all diabetic foot infections are preceded by DFU; the prevalence of these infections have been reported to range between 25 and 60 %.4-6 Patients who develop an infection have a 155-fold increased risk of amputation compared to those who do not.57 Studies evaluating the development of infection are much less common and this represents a critical gap in pathogenesis of DFU.
Culture-independent techniques such as next generation sequencing (NGS) or metagenomics next-generation sequencing (mNGS) can facilitate rapid interpretation of pathogen without the delay in time normally seen in conventional cultures. Present in all living organisms is the highly conserved 16S rRNA gene; (m)NGS examines the 16S rRNA gene's nine hypervariable regions to identify bacterial taxa58,59 from a sample. These approaches have shown promise in research settings but remain labor-intensive and bioinformatically intensive limiting their clinical value.60 Loesche et al. demonstrated, by evaluating dynamic microbiota changes longitudinally, that increased DFU microbiota diversity was associated with a faster (DFU) healing rate.61 Longitudinal sampling of DFU via wound swabs has also shown Staphylococcus aureus (S. aureus) strain diversity to be associated with outcome and effective wound debridement shifts the microbiome in an outcome-dependent manner.62 However, employment of amplicon-based sequencing failed to distinguish individual species making detection of microbial community assembly difficult.63,64 Moreover, the use of wound swabs for microbial evaluation is opposed by both the Infectious Disease Society of American (IDSA) and International Wound Group Diabetic Foot (IWGDF)65,66 as swabs demonstrate discordance with increased wound depth and are not consistent with good clinical practice.67
Still, early data from culture-independent techniques provides valuable insight into microbiome analysis of a DFU as care is provided. In fact, when ideal standard care is applied which includes the use of optimized offloading, the microbiome shifts dynamically. In a study by Issac et al., DFU patients were treated with regular wound debridement and application of a TCC weekly. NGS swab testing was performed at each visit, following debridement, and demonstrated significant variance in distribution of organisms from week to week. Specifically, the authors note initial abundances of S. aureus, Corynebacterium spp., and Acinetobacter spp. were most prevalent.68 As the wound followed a wound healing trajectory, the abundance diversity [of varied organisms] decreased. Additionally, genes which coded for beta-lactamase and tetracycline resistance also decreased as the wound followed a healing trajectory.68 Conversely, it mirrors research supporting negative DFU outcomes infected with drug resistant organisms.69-71
Lacking from NGS studies are clinical trials and translational applications. The major barrier to clinical implementation of these culture-independent tools is the technology's inability to distinguish between viable and nonviable microbes. The biohistory and biogeography of DFU require more well-controlled investigations to determine the importance of type and (relative or absolute) abundance of microbes for both infected- and non-infected clinical states. This foundational work is required to identify risk factors for a DFU to become infected. For example, bacteria in acutely infected DFU prioritize motility over biofilm and demonstrate greater pathogenicity and mechanisms as compared to a chronic wound.72 Only once more specific bacteria-related information is elucidated from culture-independent techniques will focused targeted clinical trials be able to establish their role in therapeutic trials in DFU with or without clinical infection. For now, this remains aspirational.
2.9. Wound image assessment
Obtaining DFU imaging biomarkers require refinement and advanced image processing in conjunction with machine learning (ML) methods to facilitate development of novel imaging biomarkers directly extractable from clinically obtained images. Several cutting-edge image technologies have been developed for management of DFU. These include imaging technologies that use a spectrum of imaging devices, ranging from specialized 3D cameras to regular smartphones, for image capturing coupled with computational methods for analysis of wound area determination and healing score evaluation.1,3
Deep learning approaches have been utilized in several analyses of DFU images.15,73,74 Advanced image processing and ML methods for assessment of images captured by smartphones and tablets have been developed recently.19,75 From here, processors are then able to extract clinically relevant biomarkers from the wound photographs. The imaging features are then combined with available clinical attributes obtained from the EHR to predict the chance and timing of healing of an index DFU.
To make this feasible, and as an example, our wound segmentation method processes input images (e.g., digital photos of wounds), removes any artifacts, and is then fed into a Convolutional Neural Network (CNN) to produce a probability map. The generated probability maps are then processed to extract the wound region and is then post-processed to reduce false positives.76 When compared with Support Vector Machines (SVM) and a patch-based CNN using Dice score, among others (Table 1), as the main metrics to assess the quality of wound segmentation, it was demonstrated our method is vastly superior for any of the image processing quality metrics.
Table 1.
Comparison between proposed method and other machine learning methods for wound segmentation after removal of image artifacts and producing a Convolutional Neural Network (CNN) probability map. When the proposed method is compared with Support Vector Machines (SVM) and a patch-based CNN, vastly superior precision, sensitivity, specificity, pixel accuracy, Dice score, and Matthew's Correlation Coefficient (MCC) are noted. Thus, image analysis supports a high likelihood of success in prediction biomarker development.
| Method | Precision | Sensitivity | Specificity | Pixel accuracy | Mean IoU | Dice | MCC |
|---|---|---|---|---|---|---|---|
| Patch-based CNN | 0.722 | 0.9 | 0.947 | 0.934 | 0.660 | 0.770 | 0.753 |
| SVM (RBG) | 0.564 | 0.806 | 0.896 | 0.877 | 0.472 | 0.596 | 0.594 |
| Proposed method | 0.830 | 0.917 | 0.973 | 0.966 | 0.761 | 0.845 | 0.839 |
With advances in wound segmentation processes, it enables interrogation and quantification of biomarkers available in wound images. These biomarkers may not always be discernable to the naked eye. Image biomarkers could be as simple as percent wound granulation tissue, as noted previously, or more involved and may include shape-based features, such as “wound solidity” or the density of a wound region. This can be quantified using image technologies and ML. Thus, the advanced wound segmentation method supports the extraction of imaging biomarkers from the now-processed clinically obtained image.
Besides deep learning-based features that are becoming highly popular in many other biomedical imaging systems, work continues to incorporate important imaging features that are meaningful, handcrafted, and have high clinical relevance. However, several challenges exist. First, diversity of data is lacking. Most wound image data exists from a single locale or academic center and definitive conclusions cannot be drawn from the data because of insufficient generalizability. The second reason, which further highlights the concern with diversity of data, is a lack of standardization in digital photography of wounds. For example, external factors such as angle of an image, positioning of the foot with respect to the camera, and lighting, all are not consistently defined. The resulting methodologies make data extraction and biomarker analysis highly dependent on the quality of a single image. Thus, the interpretation of data is also highly susceptible to image quality. Thirdly, reliable ML is predicated on integration of clinical phenotypic information. Datasets which include comprehensive patient-specific data to be used to integrate with wound image analysis is challenging. Finally, transparent ML is needed to inform clinical incorporation. ML is well known to be able to predict a clinical outcome, but these outcomes may be of little clinical value. Because the eventual outcome prediction is generally opaque and nontransparent, clinical utility of any extracted biomarkers would therefore remain low. With standardized approaches and richness in data, ML could provide reasoning behind rules and assignments for outcomes, which would be more user friendly for providers. It is therefore vital for the newest technology to perform standardized wound image analysis as a part of standard care at any point in care of a DFU to increase its clinical value.
3. New directions to address the problem with a look towards the future
To address these issues, at least one scalable national effort is established to further investigate care of individuals with a DFU. The NIDDK-funded Diabetic Foot Consortium (DFC) is the first-ever research network which seeks to tackle diabetic foot complications. The DFC aims to lay the foundation for a clinical trial network to test how to improve diabetic wound healing and prevent amputations among the 34 million, and growing, American adults with DM. Initially, the DFC was focused on identifying novel valid biomarkers to implement in clinical care. Additionally, studies now include a standardized tissue collection and preservation biorepository of biospecimens from individuals enrolled in the DFC's various studies (Clinicaltrials.gov: NCT05092620) and a fully integrated Ancillary Studies arm. The DFC is a fertile ground for validating potentially relevant DFU research with its access to robust clinical and research infrastructure critically dedicated to supporting cross-sectional and longitudinal DFU research. The DFC includes a Data Coordinating Center for access to robust patient demographic and phenotypic information as well.
Moreover, challenges remain after sample procurement has occurred in the form of data integration from ‘-omics’ datasets. First, to take the data from a single-omics study and combine into a multi-omics approach is dependent on appropriate gene- and feature selection. Gene selection methods are divided into five subsets: embedded, ensemble, hybrid, filter, and wrapper. Each has their benefits and drawbacks, with substantial bioinformatic expertise required alongside increased computational costs. These data-intensive demands exist because genetic products can have multiple relationships with their microenvironment. Second, in multi-omics studies involving various cancers, multiple cellular relationships exist and interfere with final data synthesis.77 The DFU environment is similarly complex. Any tissue- or cell related associations must be closely considered prior to data integration to provide the most reliable and most stable results. Likewise, feature selection in the single -omics data must also be reliable to inform feature selection in the multi-omics approach. Without this, the final performance of data integration may be endangered.
Thus, advances of the techniques being employed in the DFU field need to occur, but also in the reduction of the bioinformatic requirements to evaluate the data derived and the way we study novel clinical applications. Only then will the understanding of wound healing advance to alter standard care practice.
4. Final recommendations
In summary, we present a comprehensive overview of current knowledge gaps, the newest technologies and associated biomarkers which can be assessed to advance our understanding of wound healing biology in DFU and infection. Understanding the origin, activation, and differentiation trajectories of wound healing cells through a combination of these techniques is necessary to develop therapeutic strategies to promote optimal tissue repair. Incorporation of new knowledge gained from standardized clinical data and biospecimens from patients with DFUs will inform and enrich pragmatic clinical trial implementation.
Future clinical studies should include an active comparator, leverage the available DFC to promote larger sample size studies to limit the historical (over-)reliance on cross-sectional studies, and incorporate standardized assessments of DFU and related outcomes. This will assist in fine tuning appropriate gene- and feature selection to identify biomarkers involved in various phases of wound healing. Ultimately, these implementations will better support ongoing advancements in data integration and further foster the bench-to-bedside process.
Acknowledgements
The “Frontiers in Diabetes Complications - from Biology to Technologies” Conference, workshops and white papers were supported through a grant from NIDDK/Diacomp DK076169 and DK115255.
RPB is also supported by U01DK119083, U01DK119083-03S1, R01DK107956, and the JDRF center of excellence.
BS is supported by U01DK119083, 1K23DK131261, and U01DK119083-03S1.
CH is supported by U01DK119083.
KN is supported by U01DK119083.
KG is supported by U01DK119083.
Footnotes
Declaration of competing interest
None.
References
- 1.Boyle JP, Thompson TJ, Gregg EW, Barker LE, Williamson DF. Projection of the year 2050 burden of diabetes in the US adult population: dynamic modeling of incidence, mortality, and prediabetes prevalence. Popul Health Metr. 2010;8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L, Li X, Wang Z, et al. Trends in prevalence of diabetes and control of risk factors in diabetes among US adults, 1999–2018. JAMA. 2021;326:704–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong DG, Boulton AJ, Bus SA. Diabetic foot ulcers and their recurrence. N EngL J Med. 2017;376:2367–2375. [DOI] [PubMed] [Google Scholar]
- 4.Pickwell K, Siersma V, Kars M, et al. Predictors of lower-extremity amputation in patients with an infected diabetic foot ulcer. Diabetes Care. 2015;38:852–857. [DOI] [PubMed] [Google Scholar]
- 5.Prompers L, Huijberts M, Apelqvist J, et al. High prevalence of ischaemia, infection and serious comorbidity in patients with diabetic foot disease in Europe. Baseline results from the eurodiale study. Diabetologia. 2007;50:18–25. [DOI] [PubMed] [Google Scholar]
- 6.Holman N, Young B, Stephens H, Jeffcoate W, Group motNFCAS. Pilot study to assess measures to be used in the prospective audit of the management of foot ulcers in people with diabetes. Diabetic Medicine. 2015;32:78–84. [DOI] [PubMed] [Google Scholar]
- 7.Thorud JC, Plemmons B, Buckley CJ, Shibuya N, Jupiter DC. Mortality after nontraumatic major amputation among patients with diabetes and peripheral vascular disease: a systematic review. J Foot Ankle Surg. 2016;55:591–599. [DOI] [PubMed] [Google Scholar]
- 8.Rastogi A, Goyal G, Kesavan R, et al. Long term outcomes after incident diabetic foot ulcer: multicenter large cohort prospective study (EDI-FOCUS investigators) epidemiology of diabetic foot complications study. Diabetes Res Clin Pract. 2020; 108113. [DOI] [PubMed] [Google Scholar]
- 9.Pop-Busui R, Boulton AJ, Feldman EL, et al. Diabetic neuropathy: a position statement by the American Diabetes Association. Diabetes Care. 2017;40:136–154. 10.2337/dc16-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt B, Holmes C, Ye W, Pop-Busui R. A tale of two eras: mining big data from electronic health records to determine limb salvage rates with podiatry. Curr Diabetes Rev. 2018;16(6):497–502. 10.2174/1573399814666181017104818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardinal M, Eisenbud DE, Armstrong DG, et al. Serial surgical debridement: a retrospective study on clinical outcomes in chronic lower extremity wounds. Wound Repair Regen. 2009;17:306–311. [DOI] [PubMed] [Google Scholar]
- 12.Wilcox JR, Carter MJ, Covington S. Frequency of debridements and time to heal: a retrospective cohort study of 312 744 wounds. JAMA Dermatol. 2013;149: 1050–1058. [DOI] [PubMed] [Google Scholar]
- 13.Crews RT, Shen BJ, Campbell L, et al. Role and determinants of adherence to offloading in diabetic foot ulcer healing: a prospective investigation. Diabetes Care. 2016;39:1371–1377. 10.2337/dc15-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu SC, Jensen JL, Weber AK, Robinson DE, Armstrong DG. Use of pressure offloading devices in diabetic foot ulcers: do we practice what we preach? Diabetes Care. 2008;31:2118–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Margolis DJ, Gelfand JM, Hoffstad O, Berlin JA. Surrogate end points for the treatment of diabetic neuropathic foot ulcers. Diabetes Care. 2003;26:1696–1700. [DOI] [PubMed] [Google Scholar]
- 16.Sheehan P, Jones P, Caselli A, Giurini JM, Veves A. Percent change in wound area of diabetic foot ulcers over a 4-week period is a robust predictor of complete healing in a 12-week prospective trial. Diabetes Care. 2003;26:1879–1882. [DOI] [PubMed] [Google Scholar]
- 17.Snyder RJ, Cardinal M, Dauphinée DM, Stavosky J. A post-hoc analysis of reduction in diabetic foot ulcer size at 4 weeks as a predictor of healing by 12 weeks. Ostomy Wound Manage. 2010;56:44. [PubMed] [Google Scholar]
- 18.Lavery LA, Barnes SA, Keith MS, Seaman JW, Armstrong DG. Prediction of healing for postoperative diabetic foot wounds based on early wound area progression. Diabetes Care. 2008;31:26–29. [DOI] [PubMed] [Google Scholar]
- 19.Sprigle S, Nemeth M, Gajjala A. Iterative design and testing of a hand-held, non-contact wound measurement device. J Tissue Viability. 2012;21:17–26. [DOI] [PubMed] [Google Scholar]
- 20.Armstrong DG, Lavery LA, Consortium DFS. Negative pressure wound therapy after partial diabetic foot amputation: a multicentre, randomised controlled trial. Lancet. 2005;366:1704–1710. [DOI] [PubMed] [Google Scholar]
- 21.Blume PA, Walters J, Payne W, Ayala J, Lantis J. Comparison of negative pressure wound therapy using vacuum-assisted closure with advanced moist wound therapy in the treatment of diabetic foot ulcers: a multicenter randomized controlled trial. Diabetes Care. 2008;31:631–636. [DOI] [PubMed] [Google Scholar]
- 22.Sepúlveda G, Espíndola M, Maureira M, et al. Negative-pressure wound therapy versus standard wound dressing in the treatment of diabetic foot amputation. A randomised controlled trial. Cirugía Española (English Edition). 2009;86:171–177. [DOI] [PubMed] [Google Scholar]
- 23.Valenzuela-Silva CM, Tuero-Iglesias ÁD, García-Iglesias E, et al. Granulation response and partial wound closure predict healing in clinical trials on advanced diabetes foot ulcers treated with recombinant human epidermal growth factor. Diabetes Care. 2013;36:210–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margolis DJ, Allen-Taylor L, Hoffstad O, Berlin JA. Diabetic neuropathic foot ulcers: predicting which ones will not heal. Am J Med. 2003;115:627–631. [DOI] [PubMed] [Google Scholar]
- 25.Margolis DJ, Mitra N, Malay DS, et al. Further evidence that wound size and duration are strong prognostic markers of diabetic foot ulcer healing. Wound Repair Regen. 2022. 10.1111/wrr.13019. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fife CE, Horn SD, Smout RJ, Barrett RS, Thomson B. A predictive model for diabetic foot ulcer outcome: the wound healing index. Adv Wound Care. 2016;5:279–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weaver ML, Hicks CW, Canner JK, et al. The Society for Vascular Surgery Wound, ischemia, and foot infection (Wifi) classification system predicts wound healing better than direct angiosome perfusion in diabetic foot wounds. J Vasc Surg. 2018; 68:1473–1481. [DOI] [PubMed] [Google Scholar]
- 28.Ladurner R, Kuper M, Konigsrainer I, et al. Predictive value of routine transcutaneous tissue oxygen tension (tcpO2) measurement for the risk of non-healing and amputation in diabetic foot ulcer patients with non-palpable pedal pulses. Med Sci Monit. 2010; 16, CR273–7. [PubMed] [Google Scholar]
- 29.Li Z, Guo S, Yao F, Zhang Y, Li T. Increased ratio of serum matrix metalloproteinase-9 against TIMP-1 predicts poor wound healing in diabetic foot ulcers. J Diabetes Complications. 2013;27:380–382. [DOI] [PubMed] [Google Scholar]
- 30.Fife CE, Eckert KA, Carter MJ. Publicly reported wound healing rates: the fantasy and the reality. Adv Wound Care. 2018;7:77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geiss LS, Li Y, Hora I, Albright A, Rolka D, Gregg EW. Resurgence of diabetes-related nontraumatic lower-extremity amputation in the young and middle-aged adult US population. Diabetes Care. 2019;42:50–54. [DOI] [PubMed] [Google Scholar]
- 32.Armstrong DG, Swerdlow MA, Armstrong AA, Conte MS, Padula WV, Bus SA. Five year mortality and direct costs of care for people with diabetic foot complications are comparable to cancer. J Foot Ankle Res. 2020;13:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richard J-L, Sotto A, Lavigne J-P. New insights in diabetic foot infection. World J Diabetes. 2011;2:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt BM, Wrobel JS, Holmes CM. Physician knowledge of a rare foot condition - influence of diabetic patient population on self-described knowledge and treatment. Clin Diabetes Endocrinol. 2017;3:2. 10.1186/s40842-017-0041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gottrup F, Apelqvist J, Price P. Outcomes in controlled and comparative studies on non-healing wounds: recommendations to improve the quality of evidence in wound management. J Wound Care. 2010;19:237–268. [DOI] [PubMed] [Google Scholar]
- 36.Fife CE, Carter MJ, Walker D, Thomson B, Eckert KA. Diabetic foot ulcer off-loading: The gap between evidence and practice. Data from the US Wound Registry. Adv Skin Wound Care. 2014;27:310–316. [DOI] [PubMed] [Google Scholar]
- 37.Theocharidis G, Thomas BE, Sarkar D, et al. Single cell transcriptomic landscape of diabetic foot ulcers. Nat Commun. 2022;13:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hodge JG, Pistorio AL, Neal CA, et al. Novel insights into negative pressure wound healing from an in situ porcine perspective. Wound Repair Regen. 2022;30:64–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merritt CR, Ong GT, Church SE, et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat Biotechnol. 2020;38:586–599. [DOI] [PubMed] [Google Scholar]
- 40.Driskell RR, Lichtenberger BM, Hoste E, et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. 2013;504:277–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Foster DS, Januszyk M, Yost KE, et al. Integrated spatial multiomics reveals fibroblast fate during tissue repair. Proceedings of the National Academy of Sciences. 2021; 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang G, Moffitt JR, Zhuang X. Multiplexed imaging of high density libraries of RNAs with MERFISH and expansion microscopy. Sci Rep. 2018;8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia C, Fan J, Emanuel G, Hao J, Zhuang X. Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proc Natl Acad Sci. 2019;116:19490–19499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lundberg E, Borner GH. Spatial proteomics: a powerful discovery tool for cell biology. Nat Rev Mol Cell Biol. 2019;20:285–302. [DOI] [PubMed] [Google Scholar]
- 45.Goltsev Y, Samusik N, Kennedy-Darling J, et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell. 2018;174:968–981. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jia Z, Liu L, Zhang S, et al. Proteomics changes after negative pressure wound therapy in diabetic foot ulcers. Mol Med Rep. 2021;24:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma Y, Li R, Wang J, et al. ITIH4, as an inflammation biomarker, mainly increases in bacterial bloodstream infection. Cytokine. 2021;138, 155377. [DOI] [PubMed] [Google Scholar]
- 48.El Eter E, Al Masri A, Habib S, et al. Novel links among peroxiredoxins, endothelial dysfunction, and severity of atherosclerosis in type 2 diabetic patients with peripheral atherosclerotic disease. Cell Stress and Chaperones. 2014;19:173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murray PJ, Allen JE, Biswas SK, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willenborg S, Lucas T, van Loo G, et al. CCR2 recruits an inflammatory macrophage subpopulation critical for angiogenesis in tissue repair. Blood. 2012;120:613–625. 10.1182/blood-2012-01-403386. [DOI] [PubMed] [Google Scholar]
- 53.Boulton AJ, Armstrong DG, Albert SF, et al. Comprehensive foot examination and risk assessment: a report of the task force of the foot care interest group of the American Diabetes Association, with endorsement by the American Association of Clinical Endocrinologists. Diabetes Care. 2008;31:1679–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boyko EJ, Ahroni JH, Cohen V, Nelson KM, Heagerty PJ. Prediction of diabetic foot ulcer occurrence using commonly available clinical information: the Seattle diabetic foot study. Diabetes Care. 2006;29:1202–1207. [DOI] [PubMed] [Google Scholar]
- 55.Lavery LA, Peters EJ, Williams JR, Murdoch DP, Hudson A, Lavery DC. Reevaluating the way we classify the diabetic foot: restructuring the diabetic foot risk classification system of the international working group on the diabetic foot. Diabetes Care. 2008;31:154–156. [DOI] [PubMed] [Google Scholar]
- 56.Leese GP, Reid F, Green V, et al. Stratification of foot ulcer risk in patients with diabetes: a population-based study. Int J Clin Pract. 2006;60:541–545. [DOI] [PubMed] [Google Scholar]
- 57.Lavery LA, Armstrong DG, Wunderlich RP, Mohler MJ, Wendel CS, Lipsky BA. Risk factors for foot infections in individuals with diabetes. Diabetes Care. 2006;29: 1288–1293. [DOI] [PubMed] [Google Scholar]
- 58.Van de Peer Y, Chapelle S, De Wachter R. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 1996;24:3381–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmidt BM. Emerging diabetic foot ulcer microbiome analysis using cutting edge technologies. J Diabetes Sci Technol. 2022;16:353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmidt BM, Erb-Downward J, Ranjan P, Dickson R. Metagenomics to identify pathogens in diabetic foot ulcers and the potential impact for clinical care. Curr Diab Rep. 2021;21:1–8. [DOI] [PubMed] [Google Scholar]
- 61.Loesche M, Gardner SE, Kalan L, et al. Temporal stability in chronic wound microbiota is associated with poor healing. J Investig Dermatol. 2017;137:237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalan LR, Meisel JS, Loesche MA, et al. Strain- and species-level variation in the microbiome of diabetic wounds is associated with clinical outcomes and therapeutic efficacy. Cell Host Microbe. 2019;25:641–655.e5. 10.1016/j.chom.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meisel JS, Hannigan GD, Tyldsley AS, et al. Skin microbiome surveys are strongly influenced by experimental design. J Investig Dermatol. 2016;136:947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lipsky BA, Berendt AR, Cornia PB, et al. 2012 Infectious Diseases Society of America clinical practice guideline for the diagnosis and treatment of diabetic foot infections. Clin Infect Dis. 2012;54:e132–e173. [DOI] [PubMed] [Google Scholar]
- 65.Lipsky BA, Berendt AR, Cornia PB, et al. 2012 Infectious Diseases Society of America clinical practice guideline for the diagnosis and treatment of diabetic foot infections. Clin Infect Dis. Jun 2012;54:e132–e173. 10.1093/cid/cis346. [DOI] [PubMed] [Google Scholar]
- 66.Lipsky BA, Aragón-Sánchez J, Diggle M, et al. IWGDF guidance on the diagnosis and management of foot infections in persons with diabetes. Diabetes Metab Res Rev. 2016;32:45–74. [DOI] [PubMed] [Google Scholar]
- 67.Ertugrul MB, Baktiroglu S, Salman S, et al. Pathogens isolated from deep soft tissue and bone in patients with diabetic foot infections. J Am Podiatr Med Assoc. 2008;98: 290–295. [DOI] [PubMed] [Google Scholar]
- 68.Isaac ALTM, Colwell R, Armstrong DG. Metagenomics of diabetic foot ulcer undergoing treatment with total contact casting. J Wound Care. 2022;31(9). 10.12968/jowc.2022.31.Sup9.S45. [DOI] [PubMed] [Google Scholar]
- 69.Schmidt Brian YW, Shiwei Zhou. Multi-Drug Resistant Organism Predicts Ulcer Recurrence following Surgical Management of Diabetic Foot Osteomyelitis. International wound journal. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dang C, Prasad Y, Boulton A, Jude E. Methicillin-resistant Staphylococcus aureus in the diabetic foot clinic: a worsening problem. Diabet Med. 2003;20:159–161. [DOI] [PubMed] [Google Scholar]
- 71.Saltoglu N, Ergonul O, Tulek N, et al. Influence of multidrug resistant organisms on the outcome of diabetic foot infection. Int. J. Infect. Dis. May 2018;70:10–14. 10.1016/j.ijid.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 72.Malone M, Radzieta M, Peters TJ, et al. Host-microbe metatranscriptome reveals differences between acute and chronic infections in diabetes-related foot ulcers. APMIS. 2021. 10.1111/apm.13200. In press. [DOI] [PubMed] [Google Scholar]
- 73.Rastogi A, Goyal G, Kesavan R, et al. Long term outcomes after incident diabetic foot ulcer: multicenter large cohort prospective study (EDI-FOCUS investigators) epidemiology of diabetic foot complications study: epidemiology of diabetic foot complications study. Diabetes Res Clin Pract. 2020;162, 108113. [DOI] [PubMed] [Google Scholar]
- 74.Schmidt BM, Holmes CM, Ye W, Pop-Busui R. A tale of two eras: mining big data from electronic health records to determine limb salvage rates with podiatry. Curr Diabetes Rev. 2019;15:497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim RB, Gryak J, Mishra A, et al. Utilization of smartphone and tablet camera photographs to predict healing of diabetes-related foot ulcers. Comput Biol Med. 2020;126, 104042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cui C, Thurnhofer-Hemsi K, Soroushmehr R, et al. Diabetic wound segmentation using convolutional neural networks. IEEE. 2019:1002–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Momeni Z, Hassanzadeh E, Abadeh MS, Bellazzi R. A survey on single and multi omics data mining methods in cancer data classification. J Biomed Inform. 2020;107, 103466. [DOI] [PubMed] [Google Scholar]