Summary
SEQUIN is a web-based application (app) that allows fast and intuitive analysis of RNA sequencing data derived for model organisms, tissues, and single cells. Integrated app functions enable uploading datasets, quality control, gene set enrichment, data visualization, and differential gene expression analysis. We also developed the iPSC Profiler, a practical gene module scoring tool that helps measure and compare pluripotent and differentiated cell types. Benchmarking to other commercial and non-commercial products underscored several advantages of SEQUIN. Freely available to the public, SEQUIN empowers scientists using interdisciplinary methods to investigate and present transcriptome data firsthand with state-of-the-art statistical methods. Hence, SEQUIN helps democratize and increase the throughput of interrogating biological questions using next-generation sequencing data with single-cell resolution.
Keywords: RNA sequencing, transcriptome analysis, gene expression, single-cell analysis, R/Shiny app, iPSC profiler, t-SNE, UMAP, dimensionality reduction, data visualization
Graphical abstract
Highlights
-
•
SEQUIN enables RNA-seq data analysis for users without bioinformatic expertise
-
•
SEQUIN empowers users to analyze bulk and single-cell transcriptome data firsthand
-
•
SEQUIN enables data visualization and generation of publication-ready figures
-
•
iPSC Profiler helps measure and compare pluripotent and differentiated cell types
Motivation
Bulk and single-cell RNA sequencing are the most widely used technologies to study gene expression patterns at the population or single-cell level. The size of datasets generated by these analyses raise computational challenges in data analysis that typically require expertise in bioinformatic methods for data visualization. Rapidly evolving sequencing techniques and statistical methods create a bottleneck for scientists who want to analyze RNA-seq datasets but do not have in-depth coding knowledge to learn a new programming language or software tool. Here, we present a web-based application that gives scientists without expertise in bioinformatics the ability to upload bulk and single-cell RNA-seq datasets and rapidly obtain data visualization and differential gene expression analysis.
Weber et al. present SEQUIN, a web-based application for analysis of bulk and single-cell RNA-seq data, including quality control, gene set enrichment, data visualization, dimensionality reduction, differential gene expression analysis, and generation of publication-ready figures and tables. Users may upload datasets for comparison of gene expression within or across experiments.
Introduction
Over the past decade, RNA sequencing (RNA-seq) has become the method of choice for gene expression profiling and single-cell analysis (single-cell RNA-seq [scRNA-seq]).1 Massively parallel next-generation sequencing is increasingly accessible and affordable for the broader scientific community. However, as new sequencing technologies and statistical methods are developed and rapidly evolve,2,3,4 each new approach requires testing and learning a new programming language or software tool. This is a particular challenge for scientists that want to take advantage of RNA-seq but do not have special expertise or training in data analysis and bioinformatics. Other challenges include the need to merge independent sequencing libraries that are generated de novo or downloaded from public repositories containing vast amounts of data from the published literature. For instance, to draw meaningful biological conclusions, batch correction is of great relevance for reproducible data comparisons and hypothesis generation.5 Furthermore, visualizing and presenting the data are integral parts of the scientific process and inform data interpretation, flexible decision making, and project planning. Confidence in the robustness and reproducibility of any data analysis workflow is critical for saving time and resources, thereby enabling high-throughput multi-scale experiments. Addressing these challenges and considering the complex and multifaceted nature of analyzing sequencing data, we established a platform that we named SEQUIN. Specifically, we used the recommended National Institutes of Health (NIH) principles of scientific data management, including findability, accessibility, interoperability, and reusability.6 SEQUIN represents a versatile R/Shiny app for real-time analysis and visualization of bulk and scRNA-seq raw count and metadata. Among the unique advantages of SEQUIN is that it enables novice users to perform complex data analysis, interactive exploration, and generation of publication-ready figures in one place. SEQUIN is available as open source and is currently the most comprehensive platform for web browser-based gene expression analysis (https://sequin.ncats.io/app/).
Results
Workflow for bulk RNA-seq data analysis
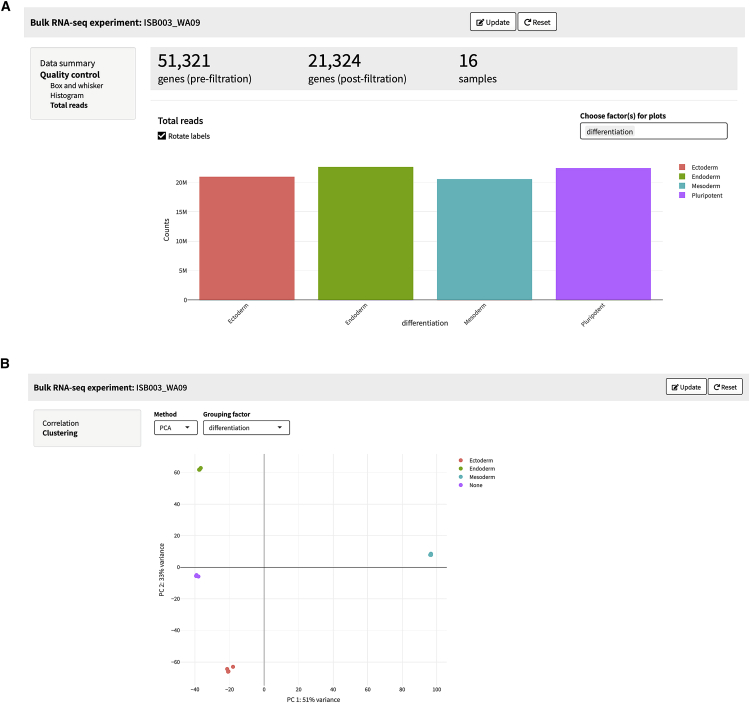
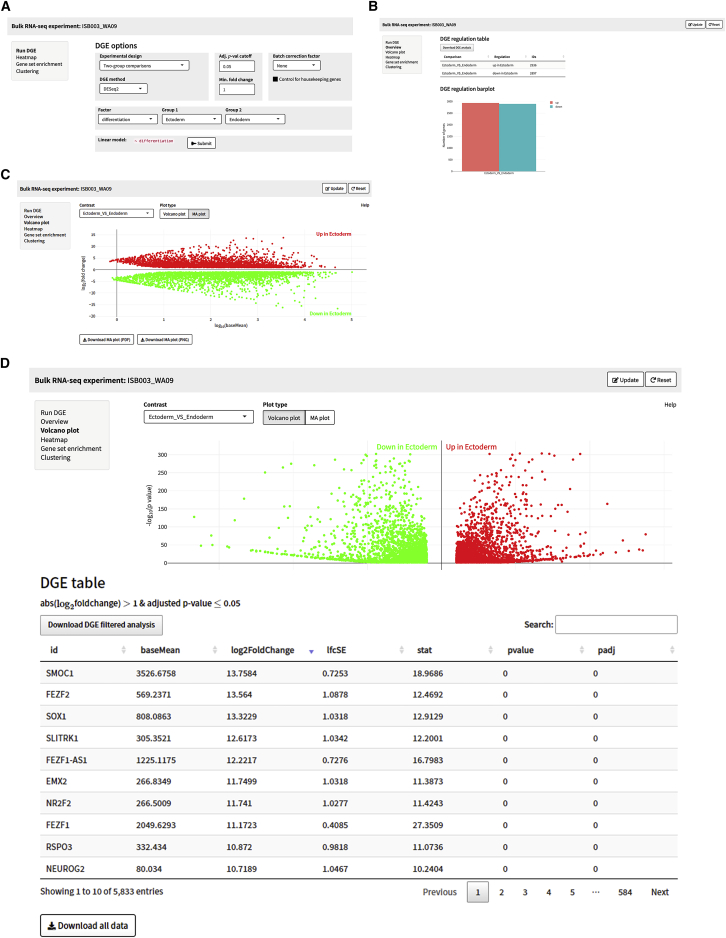
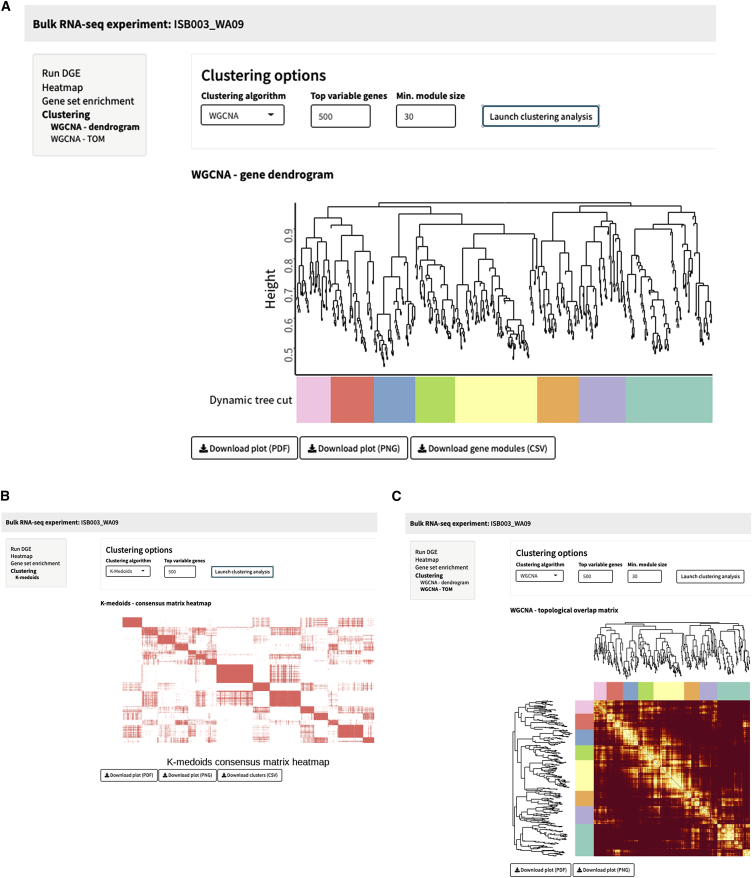
SEQUIN is a fully integrated, user-friendly, and scalable approach for analyzing RNA-seq data (Figure 1). To illustrate a typical workflow in SEQUIN, we first analyzed RNA-seq data derived from a well-established in vitro model system, which is based on controlled differentiation of human embryonic stem cells (hESCs; WA09 cell line) into the primary embryonic germ layers. In these standardized experiments, hESCs were efficiently differentiated into lineage-committed ectodermal, mesodermal, and endodermal precursor cells under chemically defined conditions. Following differentiation, we generated both bulk and single-cell RNA-seq data for analysis using SEQUIN to confirm efficient differentiation of the WA09 cell line into the three germ layers. Detailed information is provided in our previously published study,7 with brief data interpretation below. We uploaded the entire dataset from these experiments (bulk samples, dataset named ISB003_WA09) and removed pseudogenes, ribosomal and mitochondrial genes, and genes with row sums less than 10 and transformed the data for normalization (log2[count + 1]; Figure 2A). These are the default settings recommended for routine use or novice users. After outlier detection and/or optional downsampling, users can visualize a portion of the count matrix, the metadata, and visualize the distribution of total reads per sample (data summary step). A total of 16 samples were analyzed (n = 4 for pluripotent and lineage-committed cells), and 21,324 genes remained after filtering. The samples had relatively balanced counts per cell, with an average of 21 million counts per sample (quality control step; Figure 3A). Principal-component analysis (PCA) in the data structure step revealed clustering of replicates by sample with 51% and 33% of the variance explained by principal component 1 (PC1) and PC2, respectively (Figure 3B). At this point, data are prepared for either differential gene expression (DGE) analysis, or one can skip forward to use the iPSC Profiler, which will be discussed below. We performed DGE analysis using pairwise two-group comparisons with DESeq2 with a minimum fold change of 1 and an adjusted p value of 0.05 (Figure 2B). Of note, several other modeling options are available in the app including edgeR and limma-voom. These choices automatically generate a preview of the linear model design next to the “Submit” button (Figure 4A). Because no batch effect across these samples was observed in this particular experiment, we did not batch correct (see STAR Methods). The output for DGE is a table and plot of the total number of genes that are up- or downregulated (Figure 4B). The volcano plot section provides options to view volcano or MA (ratio intensity) plots as well as a significant filtered DGE table (Figures 4C and 4D). A volcano plot shows log2 fold change versus −log10 p values per gene, while an MA plot depicts log fold change versus mean expression values between two samples or groups. Guided by pairwise DGE comparisons, we explored upregulated differentially expressed (DE) genes and confirmed the expression of lineage-specific genes. Cell type-specific DE genes were significantly upregulated in pluripotent and differentiated cells. As mentioned above, more detailed information on experimental results is provided in our previous publication.7 Using such RNA-seq datasets, it is possible to view a heatmap of the top 100 DE genes by contrast or by selecting a custom gene list. Cells can also be clustered by gene expression using two unsupervised approaches: weighted correlation network analysis (WGCNA)8 or k-medoids9 (Figure 5). These options are also available for scRNA-seq, as described below.
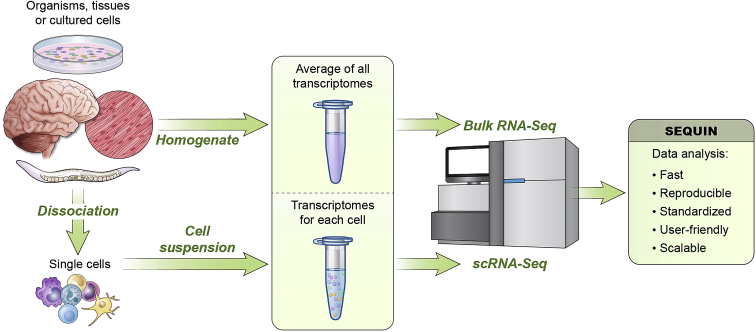
Figure 1.
Versatility of SEQUIN for bulk and single-cell RNA-seq
Overview of experimental models and next-generation sequencing that generate transcriptomic datasets. Two paths of data generation are shown: “bulk” populations of cells from homogenized organisms, tissues, or cultured cells or single-cell suspension after dissociation. The first path leads to averaged gene expression values of the transcriptomes, while the second creates a transcriptome library of each cell. Either sequencing data format can be input to SEQUIN for analysis, which is a free and fully featured R/Shiny application for both types of data.
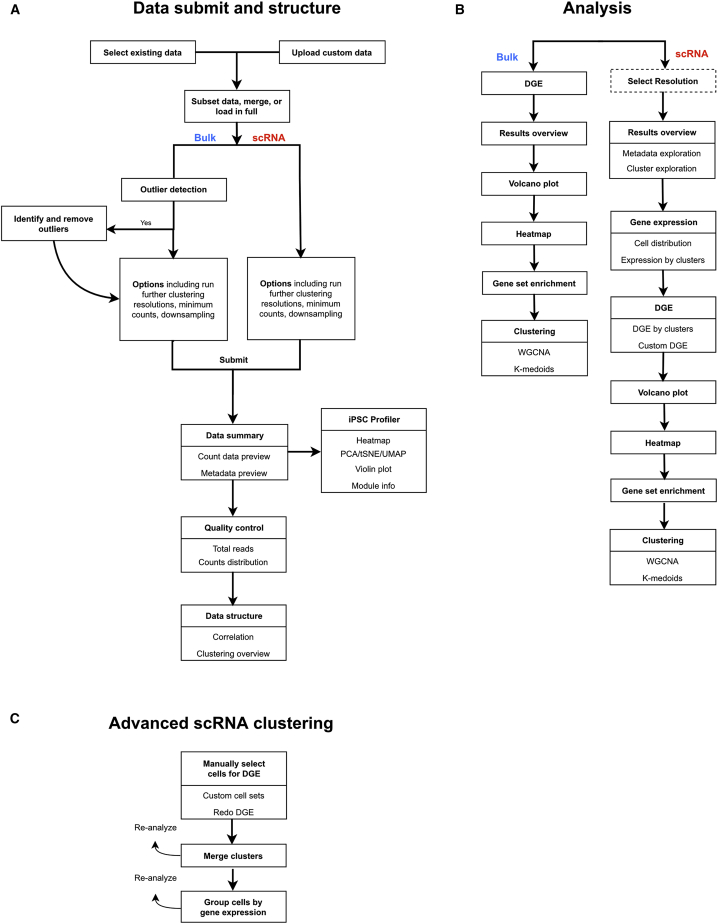
Figure 2.
Overview of workflows describing SEQUIN
(A) Diagram depicting the workflow for the bulk and single-cell RNA (scRNA) sequencing data submit options, data summary, quality control and data structure sections of the app.
(B) Workflow for analysis sections specific to bulk or scRNA. The select resolution (in dashed box) is only available when the user selects “Run multiple resolutions using Seurat.”
(C) Workflow for advanced scRNA clustering with key features and options.
Figure 3.
Bulk RNA-seq analysis
(A) Quality control step showing total reads by differentiation stage, which are consistently on average 21 million total reads by differentiation stage of dataset ISB003 (WA09).
(B) PCA plot of samples and replicates showing that PC1 accounts for 51% and PC2 33% of the variance in the data, respectively. Samples and replicates separate strongly by PC1 and PC2 clustering depending on lineage specification.
Figure 4.
Example analyses of DE genes
(A) DGE analysis options for the bulk RNA-seq analysis of ISB003 (WA09) showing two-group comparisons by differentiation using DESeq2. The adjusted p value cutoff is 0.05, with a minimum fold change of 1 and the linear model ∼differentiation.
(B) Total DE genes by linear model showing the total number of genes up- and downregulated in the ectoderm versus endoderm comparison.
(C) MA plot of total up- and downregulation DE genes for the ectoderm versus endoderm comparison.
(D) Volcano plot of total up- and downregulation DE genes for the ectoderm and endoderm comparison. Test p values and adjusted p values come from DESeq2 default statistical methods, which are the Wald test and Benjamini-Hochberg multiple test correction.
Figure 5.
WGCNA and k-medoid clustering
(A and B) Weighted gene co-expression network analysis (WGCNA) clustering of ISB003 (WA09). The WGCNA gene dendrogram for ISB003 (WA09) is based on the hierarchical clustering of all genes. Colors below the row and column dendrograms are dynamic tree cuts, which indicate size (total number of cells per cluster). The WGCNA topological matrix plot is the correlation between pairs of genes and pairs of gene modules. A k-medoids consensus matrix heatmap based on all genes and samples from the same dataset (B).
(C) k-medoids heatmap reflects an estimate of the similarity between pairs of genes.
Analyzing scRNA-seq data
To perform scRNA-seq data analysis, we again used a recently published dataset (IS006_WA09) that includes well-characterized human pluripotent and differentiated cells generated by standardized methods.7 Prior to data submission to the server, the full set of cells was randomly downsampled from 16,582 to 10,000 cells, and all rRNA, mitochondrial, and pseudogenes were filtered out. The option to downsample was implemented to reduce count matrix load time and the RAM constraints in R. If the user prefers not to downsample and is willing to wait longer, the entire dataset can be uploaded. Of note, data are stored only for the duration of the app session. Hence, all user-uploaded data from a session is immediately destroyed once the session ends.
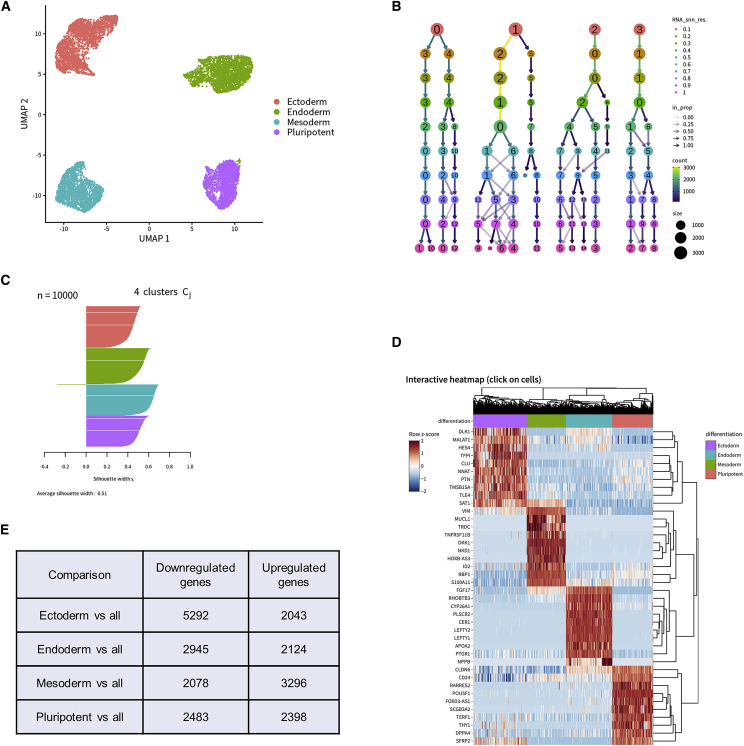
The widely used standard Seurat workflow10 is incorporated into SEQUIN with options to adjust the number of PCA dimensions either by setting a cumulative percentage of variance in the PCs or using the default of 75% and the range of clustering resolutions. Prior to performing downstream analyses, the user can optionally view a snapshot of the count matrix, inspect the metadata, or preview the distribution of cells or samples after the dataset has been fully loaded. The samples analyzed here had relatively balanced counts per cell (median 10,000) and the threshold for minimum counts removed poor quality or empty cells. Overall variance between samples can be visualized. Moreover, review of dimensionality reduction by selected metadata factor is possible with PCA, t-stochastic neighbor embedding (tSNE), and uniform manifold approximation projection (UMAP). Four distinct cell clusters were identified and are depicted in the UMAP plot (Figure 6A).
Figure 6.
Analysis of iPSC differentiation into embryonic germ layers
(A) UMAP clearly separates clusters by differentiation stage.
(B) Clustree flowchart identifies how cells sort into clusters at various resolutions.
(C) Silhouette plot cleanly separates clusters at 0.1 resolution.
(D) Interactive heatmap with a custom set of lineage-specific genes.
(E) Total up- and downregulated DE genes by differentiation stage compared with the rest of the clusters.
Next, we submitted the scRNA-seq dataset with varied resolutions from 0.1 to 1 by steps of 0.1. As expected, higher resolutions generated more clusters. Four to 16 clusters were created for resolutions 0.1 to 1, respectively. On the basis of the UMAP, Clustree plot (Figure 6B), and silhouette plot (Figure 6C) we concluded that 0.1 was the optimal resolution for clustering the different developmental lineages (Figures 6D and 6E). Clustree is helpful to identify the best resolution for cell clustering on the basis of the distribution.11 Each resolution is represented by a row, with circle sizes corresponding to the total number of cells, and arrow thickness indicating the proportion of cells that flow from one cluster to another. Over-clustering occurs when there is frequent crossover of cells from one cluster to another at higher resolutions, and the user should choose a stable resolution lower than this point (Figure 6B, rows 5–10). In the silhouette plot, the positive silhouette widths indicate that a given cell is closer to other cells within that cluster than to other clusters, which is ideal.12 Together with the dimensionality reduction and Clustree plots, we confirmed that 0.1 resolution clearly separated different cells. The UMAP revealed that four clusters separate cells on the basis of their developmental stage and lineage specification, which further supports the 0.1 resolution for accurate clustering. Furthermore, the metadata can be explored in various other ways as presented (Figures S1A and S1B). Considering the UMAP, Clustree plot, and silhouette plot together, we confirmed good clustering of the different developmental lineages, indicating initial pluripotency and subsequent efficient differentiation of the human iPSC line (WA09).
Prior to performing DGE, we inspected lineage-specific genes using the selection from the drop-down menu of available genes. Gene expression can be displayed by cluster or sample name in the form of bar plots and in PCA, UMAP, and tSNE. Example UMAPs are shown for POU5F1, HES4, DKK1, and PTGR1 (Figures S2A–S2H). SEQUIN also provides alternative options for DGE analysis. The user can compare a given cluster to the rest or select a factor in the metadata and the desired comparisons (Figure S3). Using an interactive UMAP, tSNE, or PCA plot, the user can manually lasso-select cells to perform DGE and gene set enrichment analysis (GSEA) and download these tables. Lasso selection of cells to define new, custom clusters on the basis of the user’s knowledge is a powerful feature of SEQUIN. To identify the uniquely DE genes per cluster, Custom DGE was performed (Figure 6E) and lineage-specific results were consistent with a recent report.7 Several thousand statistically significant genes were up- or downregulated for pluripotent and differentiated cells. The user can explore pairwise DE genes in a volcano plot or by generating a DE genes table on the basis of log fold change and adjusted p value thresholds. Although we provide core differential expression analysis, there are many options to further explore these genes. The user can generate a heatmap of the top 50 DE genes or by specifying a custom list of genes across samples. A selected metadata grouping factor for cells can be overlaid onto the heatmap samples. For instance, we targeted a set of genes involved in cell differentiation that showed scaled, normalized gene expression varied greatly by lineage (Figure 6D). Subsequently, we performed GSEA on the statistically significant DE genes that were upregulated in each respective cluster (lineage) versus the rest of the clusters (the top 100 DE genes upregulated in the selected cluster). GSEA calls the Enrichr API13 with a mirror of all gene set libraries available from that resource, including the popular Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology, and ARCHS4.14 To our knowledge, the unified integration of DGE and GSEAs in one R/Shiny app is unique to SEQUIN.
Next, a valuable app feature is the ability to merge existing clusters or cluster cells on the basis of the mean, median, or sum of gene(s) expression (Figures 2C and S4). A resolution of 0.1 created four distinct clusters, clearly separated by tissue lineage. Alternatively, a resolution of 0.3 was able to further separate the endoderm and ectoderm clusters to give a total of six clusters. If after exploring the Clustree and other plots, it becomes clear that a different clustering would be optimal, the user can backtrack to merge clusters, which can then be named and given a comment to describe the merge. By clicking “Save to database,” the updated metadata will be saved in the Relational Database Service (RDS) and can be reloaded with the count data for downstream analysis (Figures S5A–S5D). Only metadata changes to existing experiments will be saved; custom uploaded data will not be stored in the RDS.
iPSC profiler
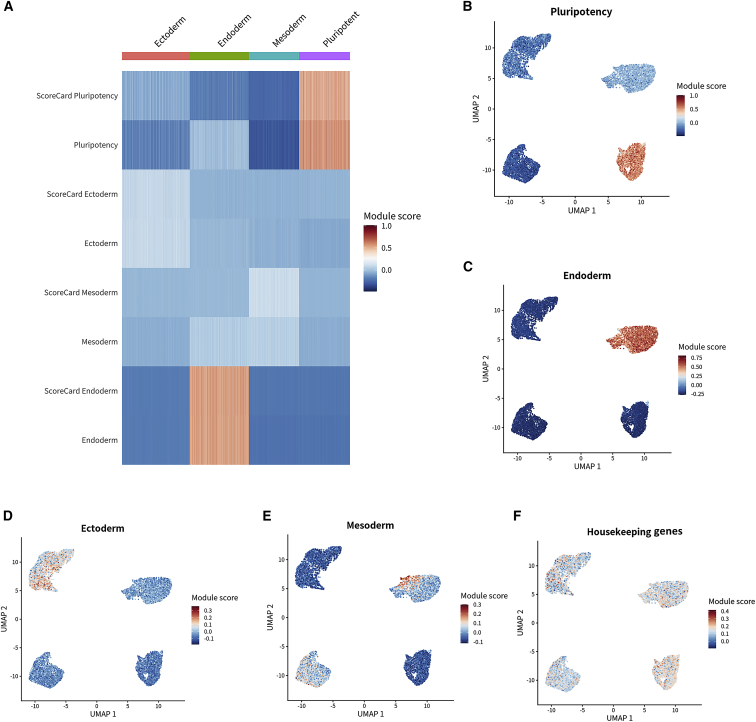
The iPSC Profiler is a special feature of SEQUIN and was developed to characterize cell identities representing human pluripotent and lineage-committed cells. It can be used to complement and extend PluriTest and ScoreCard, which are previously established resources to measure pluripotency and embryoid body differentiation on the basis of microarray technology and qPCR.15,16,17 The iPSC Profiler was established as a gene module scoring tool for both bulk and scRNA-seq data using the Seurat AddModuleScore function, which classifies single cells or whole samples as pluripotent, ectodermal, mesodermal, or endodermal (Figure 2A).16 Module scores differ from the simple average of a gene set in that they are directly comparable across set sizes and contents. We ran the iPSC Profiler for an exemplary dataset (IS006_WA09) and found that module scores were well matched to the ScoreCard modules for pluripotency, ectoderm, mesoderm, and endoderm (Figure 7A). We then generated a UMAP of the pluripotency module (Figure 7B) and a reference UMAP colored by expected cell lineage (Figure 6A). Module scores were highest for endoderm and pluripotency gene sets (Figures 7B and 7C). Ectoderm and mesoderm cells scored relatively low for their gene sets, with only a subset of the cells having high scores for these two modules (Figures 7D and 7E). The housekeeping gene module scores were consistent across all lineages (Figure 7F). Altogether, the iPSC Profiler is a practical tool to compare pluripotent and lineage-committed cells.
Figure 7.
iPSC Profiler module scores
(A) Heatmap module scores from both ScoreCard and iPSC Profiler indicating similar scores for pluripotency, ectoderm, mesoderm, and endoderm.
(B) UMAP plot colored by pluripotency module scores reveals high scores in hESCs (WA09).
(C–E) Endoderm module scores are highest in the endoderm cluster. Similarly, the ectoderm and mesoderm clusters are represented by their respective modules.
(F) Module for housekeeping genes yields comparable results across all cell clusters analyzed.
Comparison with other data analysis methods
Although there are many software applications or platforms with a graphical user interface that broadly work with RNA-seq data, each typically addresses only a few steps in a full analysis pipeline, may or may not be hosted on a public server or are not freely available, among other considerations (Table 1). We evaluated features in popular commercial and open-source tools that are similar in scope to SEQUIN.18,19,20,21 We only chose tools that begin with a raw gene count by samples table and a metadata table, and result in outputs of tables and figures, including DE genes, heatmaps, and clustering plots. Overall, SEQUIN was unique in that it is a fully featured resource, for both bulk and single cell data, and is freely available without server start-up overhead, while offering rigorous statistical methods. Among the apps evaluated, only two could handle both kinds of data with start-to-finish analysis in one place, and they are commercial products (Partek Flow and Illumina BaseSpace DRAGEN). However, those platforms do not include all features that are available in SEQUIN, and when comparing across all platforms, none offered cell and gene module scoring similar to the iPSC Profiler. Outlier detection and removal in bulk data were nearly completely absent in other apps. Critically, in order to accomplish certain SEQUIN-equivalent tasks, it was sometimes required to do manual or command-line work outside of the app such as subsetting samples, library merging, or DE model customization. This latter feature was noticeably missing or modestly featured in other free apps, perhaps out of simplicity, but with the result that complex experimental designs would be impossible to accurately model (such as “time × treatment × cell line”). Also, display of data or model results varied widely. The level of plotting customization options was heterogeneous in terms of the plot type, content, and graphical design, even in the commercial products. Plot types commonly used in the workflow to show gene expression levels were not offered, such as violin, dot plot, tSNE, or UMAP. We provide all the previously mentioned plot types and analysis features in one interactive environment. This cross-platform benchmarking is currently the best estimation for comparable apps; however, each app is unique and specific needs are user-dependent.
Table 1.
Comparison of SEQUIN with other available apps
| Feature | SEQUIN | IRIS-EDA | Partek Flow | 10X Genomics Cellranger and Cell Loupe Browser | TIBCO SpotFire OmicsOffice | Illumina BaseSpace and DRAGEN | FlowJo SeqGeq | TCC-GUI | RNfuzzyApp | FGCZ | RNAseq DRaMA | SCHNAPPs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Free | ✓ | ✓ | ✗ | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Server is already hosted | ✓ | ✓ | ✓ | ✓ | ✗ | ✓ | ✓ | ✓ | ✗ | ✓ | ✗ | ✗ |
| Sever stability/disconnects | ✓ | ✗ | ✓ | ✓ | N/A | ✓ | ✓ | ✗ | N/A | ✓ | N/A | N/A |
| Analyze both bulk and single-cell RNA-seq | ✓ | ✗ | ✓ | ✗ | ✗ | ✓ | ✓ | ✗ | ✗ | ✓a,b | ✗ | ✗ |
| Subset samples | ✓ | ✗ | ✓ | ✗ | ✓a | ✗ | ✗ | ✗ | ✗ | ✓ | ✗ | ✗ |
| Library merging | ✓ | ✗ | ✓ | ✓a | ✓a | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ |
| Batch correction (bulk) | ✓ | ✗ | ✓ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ |
| Sample outlier detection/removal | ✓ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✓ | ✗ |
| QC reporting (library size) | ✓ | ✓ | ✓ | ✓ | ✗ | ✓ | ✓ | ✓ | ✓ | ✗ | ✓ | ✓ |
| Sample correlation | ✓ | ✓ | ✓ | ✗ | ✗ | ✓ | ✗ | ✗ | ✗ | ✗ | ✓ | ✗ |
| Sample clustering (distance) | ✓ | ✓ | ✓ | ✗ | ✗ | ✓ | ✗ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Cell clustering (distance) | ✓ | ✓ | ✓ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✓ |
| Cell clustering (2D or 3D) | ✓ | ✓ | ✓ | ✓ | ✗ | ✗ | ✓ | ✗ | ✗ | ✗ | ✗ | ✓ |
| DE analysis | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✗c | ✓ |
| DE model customization | high | medium | medium | none | none | low | medium | none | none | none | nonea | low |
| tSNE | ✓ | ✓ | ✓ | ✓ | ✗ | ✗ | ✓ | ✗ | ✗ | ✗ | ✗ | ✓ |
| UMAP | ✓ | ✗ | ✓ | ✗ | ✗ | ✓ | ✗ | ✗ | ✗ | ✗ | ✗ | ✓ |
| PCA | ✓ | ✓ | ✓ | ✗ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Violin plot | ✓ | ✗ | ✓ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✓ |
| Dot plot | ✓ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ |
| Boxplot | ✓ | ✗ | ✓ | ✗ | ✓ | ✗ | ✗ | ✓ | ✓ | ✓ | ✗ | ✓ |
| Scatterplot | ✓ | ✓ | ✓ | ✗ | ✓ | ✗ | ✓ | ✗ | ✗ | ✗ | ✗ | ✓ |
| Volcano/MA plot | ✓ | ✓ | ✓ | ✗ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Heatmap | ✓ | ✓ | ✓ | ✗ | ✓ | ✗ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Plot customization of content or aesthetics | high | low | high | none | high | low | medium | high | low | low | medium | medium |
| Gene set enrichment analysis | ✓ | ✗d | ✓ | ✗ | ✓ | ✓ | ✓ | ✗ | ✓ | ✗ | ✓ | ✗ |
| Custom cell clustering including merging clusters | ✓ | ✗ | ✓ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✓ |
| Cell gene module scoring | ✓ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ |
Key features in SEQUIN were compared with those in other available apps for presence or absence or strength in customization options for the feature. 2D, two-dimensional; DE, differential expression; QC, quality control.
Requires more complicated manual or command line work to accomplish the task.
In separate R/Shiny apps with limited functions.
Only visualizes pre-computed DE results.
Only provides DE gene results table for export outside of the app (for GSEA).
In-app benchmarking
To help users understand the processing time for running SEQUIN, we time-stamped each step using scRNA-seq datasets of varying sizes from 90 to 19,759 cells (which are the datasets example_sc and IS006_11, respectively; Table S1). With pre-computed cell clusters and using all cells without downsampling, time to load data was between 8 s and 29 min. The most computationally intensive step was cell clustering when multiple resolutions were requested (0.4–2.8 in steps of 0.4), which took 13 s to 1.5 h for the smallest and largest datasets mentioned. A relatively long clustering step is expected for any single cell analysis algorithm and users are allowed to proceed given a notice that it will take time. We adjusted the R/Shiny server timeout to allow up to two hours of processing wait. After clustering, the remaining steps took between one second and 3 min 45 s to run (custom DGE on the largest dataset). Users will experience shorter wait times when default data load settings are used and/or they downsample cells to at least 10,000.
Discussion
Analyzing and integrating single-cell and bulk RNA-seq data has become a powerful approach in biomedical research. Practical cost-efficient strategies are required to handle large datasets, compare disparate and diverse biological systems, identify biomarkers, and measure relevant endpoints and signatures in molecular medicine. To democratize and standardize this process, we developed SEQUIN as a standalone data analysis and visualization platform. Using exemplary datasets derived from a well-defined human stem cell model, we studied multilineage differentiation of hESCs and iPSCs and performed comparisons of cell type-specific and differentially regulated genes. These experiments yielded consistent and reproducible results and were used as case demonstrations to introduce SEQUIN. Moreover, we developed the iPSC Profiler, a convenient gene module scoring tool that provided quick confirmation of successful differentiation of human pluripotent cells into the three embryonic germ layers. Because transcription factor-based reprogramming of somatic cells into iPSCs is a widely used technology, many new cell lines have been or are in the process of being generated. However, standardized RNA-seq data analysis for iPSCs has not been reported so far. The iPSC Profiler can aid in characterizing new cell lines including determining successful reprogramming, assessing cellular heterogeneity, and multilineage differentiation potential. In the future, we envisage that gene modules for other terminally differentiated cell types (e.g., hepatocytes, cardiomyocytes, neuronal subtypes) can be incorporated into SEQUIN. In summary, SEQUIN empowers users from different backgrounds and levels of bioinformatics expertise to perform customizable analyses of bulk and single-cell RNA-seq in real time and in one location. We propose that SEQUIN sets a standard for RNA-seq analysis within the R/Shiny environment.
Limitations of the study
Despite the user-friendly interface and capability to perform rapid analyses of bulk and single-cell RNA-seq datasets, SEQUIN does not allow for indefinite data storage. This limitation would require the users to upload data for each session, as data are not retained in this application. Another limitation is inherent to the R and R/Shiny software environment: data are stored in memory, so it can be time consuming to wait for new data to load and be ready to use. These limitations are surmountable if the user installs a local instance of the application with sufficiently powerful hardware.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| scRNA-seq dataset (IS006_WA09) | Tristan et al.7 | NCBI SRA PRJNA657268 |
| Bulk RNA-seq dataset (ISB003_WA09) | Tristan et al.7 | NCBI SRA PRJNA657268 |
| Bulk RNA-seq dataset (ISB003_WA09) | Chen et al.22 | NCBI SRA PRJNA552890 |
| Bulk RNA-seq dataset (example_bulk) | Walker et al.23 | NCBI GEO GSE110021 |
| Software and algorithms | ||
| SEQUIN | This paper | https://sequin.ncats.io/app/, https://doi.org/10.5281/zenodo.7554907 |
| IRIS-EDA | Monier et al.18 | https://bmbls.bmi.osumc.edu/IRIS/ |
| Partek Flow | Kanehisa Laboratories and Pathway Solutions, Inc. | https://www.partek.com/partek-flow/ |
| 10X Genomics Cellranger and Cell Loupe Browser | 10X Genomics, Inc. |
https://www.10xgenomics.com/products/loupe-browser https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation |
| TIBCO SpotFire OmicsOffice | PerkinElmer, Inc. | https://perkinelmerinformatics.com/products/exclusive-reseller/tibco-spotfire |
| Illumina BaseSpace and DRAGEN | Illumina, Inc. | https://www.illumina.com/products/by-type/informatics-products/basespace-sequence-hub/apps.html |
| FlowJo SeqGeq | BD Biosciences, Inc. | https://www.flowjo.com/learn/flowjo-university/seqgeq |
| TCC-GUI | Su et al.19 | https://github.com/swsoyee/TCC-GUI |
| RNfuzzyApp | Haering et al.20 | https://gitlab.com/habermann_lab/rna-seq-analysis-app |
| FGCZ | Functional Genomics Center Zurich | https://github.com/fgcz |
| RNAseq DRaMa | HSS David Z. Rosensweig Genomics Research Center | https://hssgenomics.shinyapps.io/RNAseq_DRaMA/ |
| SCHNAPPS | Jagla et al.21 | https://www.rna-seqblog.com/schnapps-single-cell-shiny-applications/ |
Resource availability
Lead contact
Further information and resource requests should be directed to Ilyas Singeç (ilyassingec@gmail.com).
Materials availability
This study did not generate new unique reagents.
Method details
Dataset workflow
The primary workflow is as follows: loading data from the existing bulk database or custom upload, Data structure, Analysis, and the iPSC Profiler (Figures 1 and 2). Extending from two previous frameworks, IRIS-EDA and scClustViz, there are many feature enhancements for analysis that can be finely adjusted by the user in a highly interactive fashion.12,18
We included two main additional features for bulk RNA-Seq: removing outliers and batch correction. Outliers can be identified by selecting “Subset data” then “Identify outliers” which creates a PCA. The user can select specific samples or technical replicates to remove. If bulk samples from different sequencing libraries are selected, batch correction is run by default using the RUVSeq RUVg algorithm with an empirical set of housekeeping genes.24 We chose RUVg batch correction because it does not dampen biological signals as aggressively as other methods, such as ComBat-seq.25 RUVg does not require a balanced experimental design with the same samples in each batch, making it highly convenient for merging disparate sequencing libraries. If batch correction was performed, the weighting variable is included first in the design as recommended by Risso et al.24
We included five main features for single-cell data: 1. calculate multiple nearest-neighbor resolutions to find ideal cluster assignment, 2. improved visualizations using clustree for resolution selection, with converted scClustViz plots to ggplot2 and lasso selection for Seurat-based plots, 3. GSE, 4. options to manually combine nearest-neighbor clusters and create updated clusters based on the expression of selected gene(s), and 5. the iPSC Profiler tool. For both bulk and scRNA data, the user can merge samples across multiple experiments in the database or their own uploaded data, allowing comparisons to previously published data. For bulk data, we have also included batch correction, which allows for rapid comparison across datasets with reduced technical confounding factors. To exemplify the utility of SEQUIN, we describe bulk and single-cell analysis of human embryonic stem cells (WA09, WiCell, Madison, WI) and iPSCs (LiPSC-GR1.1, NIH Common Fund) that were differentiated into ectoderm, endoderm, and mesoderm using standardized kits (STEMCELL Technologies) as previously described.7
As the number of cells increases for scRNA experiments, so do the limitations within the R environment. While it is possible to load upwards of 100,000 cells into R, this will cause problems in R/Shiny. While we can confirm that we are able to load and analyze 55,074 cells in SEQUIN, it comes with a significant wait time for loading plots, tables, and complete analyses. As scRNA datasets become even more prevalent and larger, this will also be a limitation of the app.
Although we included the option to batch correct bulk RNA-seq datasets, we did not include batch correction for scRNA datasets. Differential gene expression tests are sensitive to batch effects, but most approaches can only control for simple batch structure.26 More complex experimental designs such as unbalanced samples or uneven total cell counts across datasets can be problematic. We tested several batch corrections approaches on various scRNA datasets as well as simulated data, and we concluded that the majority of these methods completely dampened true biological signal rather than simply removing batch effects.
The iPSC Profiler was developed as a tool to quickly visualize the expression of gene sets in samples or cells or the score for a given gene module. The score is directly computed from Seurat’s AddModuleScore function, developed by Tirosh et al.27 Since our data focuses on stem cells and iPSCs, genes from the ScoreCard were included in addition to longer literature and internal experimental based gene lists for the three primary tissue lineages.16 The user may choose either the canonical or expanded modules for fast assessment of early lineage attainment, as indicated by module name. For a brief comparison of the module score differences for multilineage differentiation, see Figure 7A.
Quantification and statistical analysis
The application was built on several existing statistical packages in R. Namely, the following were used in whole or part with adaptations described previously: DESeq2,28 limma-voom,29 and edgeR30 for bulk RNA-Seq data analysis; Seurat,10 including the AddModuleScore function developed by Tirosh et al.27 in the iPSC Profiler tool; RUV-Seq batch correction methods for bulk RNA-Seq by Risso et al.24; scClustViz12 and clustree11 converted to ggplot2 plots for scRNA-Seq clustering; enrichR for gene set enrichment. Exact steps, settings, and significance thresholds used in SEQUIN for scRNA-Seq and bulk RNA-Seq data analysis were described in the previous sections, “Workflow for bulk RNA-seq data analysis” and “Analyzing scRNA-seq data”, respectively. Sample counts for the bulk datasets pre-loaded in the application are: “ISB003_WA09”: 16; “ISB008”: 6; “example_bulk”: 48. Cell counts for the pre-loaded scRNA datasets are: “example_sc”: 90; “IS020_WA09”: 10058; “IS018”: 3875; “IS006_WA09”: 16582; “IS006_11”: 19759.
Acknowledgments
This study was supported by the NIH Common Fund (Regenerative Medicine Program) and in part by the National Center for Advancing Translational Sciences (NCATS) Division of Preclinical Innovation (DPI) Intramural Research Program. We thank Rancho Biosciences and the ITRB and Informatics Group at NCATS. We acknowledge the contributions of all Stem Cell Translation Laboratory (SCTL) scientists for suggesting feature enhancements and testing SEQUIN. This project used the computational resources of the NIH High-Performance Computing (HPC) Biowulf cluster for R pipeline development (http://hpc.nih.gov). The graphical abstract was created using BioRender.com.
Author contributions
Conceptualization, C.W., M.B.H., and P.-H.C.; methodology, investigation, and data curation, C.W., M.B.H., B.E., and N.J.S.; writing – original draft and review & editing, C.W., M.B.H., H.M.B., C.A.T., and I.S.; funding acquisition, I.S.; resources, K.M.W. and K.W.; supervision, C.A.T., H.M.B., and I.S.
Declaration of interests
The authors declare no conflicts of interest.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: March 6, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2023.100420.
Contributor Information
Carlos A. Tristan, Email: carlos.tristan@nih.gov.
Ilyas Singeç, Email: ilyassingec@gmail.com.
Supplemental information
Data and code availability
-
•
This paper analyzes existing, publicly available data. The accession numbers for the datasets are listed in the key resources table. Six publicly available experimental RNA-seq datasets are included in the app for training and demonstration purposes. The datasets generated by the Stem Cell Translation Laboratory (SCTL) were derived from hESCs (WA09) and iPSCs (LiPSC-GR1.1): “ISB003” (WA09 differentiated into three germ layers);7 bulk RNA-seq data “ISB008” (WA09 and LiPSC-GR1.1) from Chen et al.22 and four scRNA datasets from Tristan et al.7: “IS020” (WA09 cultured manually and robotically); “IS018” (WA09 differentiated into neurons), “IS006_WA09” (WA09 differentiated into three germ layers); “IS006_11” (LiPSC-GR1.1 differentiated into germ layers). A small RNA-Seq dataset “example_bulk” is from WI-38 fibroblasts with and without TGF-β treatment on days 1 and 20 from Walker et al.23 This dataset was selected because it is from human-derived samples, has a simple and balanced experimental design, and includes sufficient metadata to demonstrate all features and statistical models available for bulk RNA-seq in SEQUIN. A small scRNA-seq dataset “example_sc” was obtained from the IRIS-EDA GitHub repository (https://github.com/OSU-BMBL/IRIS) and consists of human preimplantation embryos and hESCs at different passages.18 This dataset was selected because it is from human-derived samples and is small but sufficient for quickly demonstrating all features available for scRNA-seq in SEQUIN. A publicly hosted version with example datasets from SCTL and the literature is available at https://sequin.ncats.io/app/.
-
•
All original code and a stand-alone package that can run locally is available here: https://github.com/ncats/public_sequin. All original code has been deposited at Zenodo and is publicly available as of the date of publication under https://doi.org/10.5281/zenodo.7554907.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Wang Y.J., Schug J., Lin J., Wang Z., Kossenkov A., Kaestner K.H. Comparative analysis of commercially available single-cell RNA sequencing platforms for their performance in complex human tissues. bioRxiv. 2019 doi: 10.1101/541433. Preprint at. [DOI] [Google Scholar]
- 2.Adossa N., Khan S., Rytkönen K.T., Elo L.L. Computational strategies for single-cell multi-omics integration. Comput. Struct. Biotechnol. J. 2021;19:2588–2596. doi: 10.1016/j.csbj.2021.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simoneau J., Dumontier S., Gosselin R., Scott M.S. Current RNA-seq methodology reporting limits reproducibility. Brief. Bioinform. 2021;22:140–145. doi: 10.1093/bib/bbz124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stupple A., Singerman D., Celi L.A. The reproducibility crisis in the age of digital medicine. NPJ Digit. Med. 2019;2:2. doi: 10.1038/s41746-019-0079-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeng S.L., Chi Y.C., Ma M.C., Chan S.H., Sun H.S. Gene expression analysis of combined RNA-seq experiments using a receiver operating characteristic calibrated procedure. Comput. Biol. Chem. 2021;93:107515. doi: 10.1016/j.compbiolchem.2021.107515. [DOI] [PubMed] [Google Scholar]
- 6.Wilkinson M.D., Dumontier M., Aalbersberg I.J.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.W., da Silva Santos L.B., Bourne P.E., et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tristan C.A., Ormanoglu P., Slamecka J., Malley C., Chu P.H., Jovanovic V.M., Gedik Y., Jethmalani Y., Bonney C., Barnaeva E., et al. Robotic high-throughput biomanufacturing and functional differentiation of human pluripotent stem cells. Stem Cell Rep. 2021;16:3076–3092. doi: 10.1016/j.stemcr.2021.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Budiaji W., Leisch F. Simple K-medoids partitioning algorithm for mixed variable data. Algorithms. 2019;12:177. [Google Scholar]
- 10.Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zappia L., Oshlack A. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. GigaScience. 2018;7:giy083. doi: 10.1093/gigascience/giy083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Innes B.T., Bader G.D. scClustViz - single-cell RNAseq cluster assessment and visualization. F1000Res. 2018;7 doi: 10.12688/f1000research.16198.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lachmann A., Torre D., Keenan A.B., Jagodnik K.M., Lee H.J., Wang L., Silverstein M.C., Ma'ayan A. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018;9:1366. doi: 10.1038/s41467-018-03751-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Miguel M.P., Fuentes-Julián S., Alcaina Y. Pluripotent stem cells: origin, maintenance and induction. Stem Cell Rev. Rep. 2010;6:633–649. doi: 10.1007/s12015-010-9170-1. [DOI] [PubMed] [Google Scholar]
- 16.Tsankov A.M., Akopian V., Pop R., Chetty S., Gifford C.A., Daheron L., Tsankova N.M., Meissner A. A qPCR ScoreCard quantifies the differentiation potential of human pluripotent stem cells. Nat. Biotechnol. 2015;33:1182–1192. doi: 10.1038/nbt.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Müller F.J., Brandl B., Loring J.F. StemBook. 2012. Assessment of human pluripotent stem cells with PluriTest. [DOI] [PubMed] [Google Scholar]
- 18.Monier B., McDermaid A., Wang C., Zhao J., Miller A., Fennell A., Ma Q. IRIS-EDA: an integrated RNA-Seq interpretation system for gene expression data analysis. PLoS Comput. Biol. 2019;15:e1006792. doi: 10.1371/journal.pcbi.1006792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su W., Sun J., Shimizu K., Kadota K. TCC-GUI: a Shiny-based application for differential expression analysis of RNA-Seq count data. BMC Res. Notes. 2019;12:133. doi: 10.1186/s13104-019-4179-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haering M., Habermann B.H. RNfuzzyApp: an R shiny RNA-seq data analysis app for visualisation, differential expression analysis, time-series clustering and enrichment analysis. F1000Res. 2021;10 doi: 10.12688/f1000research.54533.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jagla B., Libri V., Chica C., Rouilly V., Mella S., Puceat M., Hasan M. SCHNAPPs - single Cell sHiNy APPlication(s) J. Immunol. Methods. 2021;499:113176. doi: 10.1016/j.jim.2021.113176. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y., Tristan C.A., Chen L., Jovanovic V.M., Malley C., Chu P.H., Ryu S., Deng T., Ormanoglu P., Tao D., et al. A versatile polypharmacology platform promotes cytoprotection and viability of human pluripotent and differentiated cells. Nat. Methods. 2021;18:528–541. doi: 10.1038/s41592-021-01126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker E.J., Heydet D., Veldre T., Ghildyal R. Transcriptomic changes during TGF-beta-mediated differentiation of airway fibroblasts to myofibroblasts. Sci. Rep. 2019;9:20377. doi: 10.1038/s41598-019-56955-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Risso D., Ngai J., Speed T.P., Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y., Parmigiani G., Johnson W.E. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform. 2020;2:lqaa078. doi: 10.1093/nargab/lqaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kharchenko P.V. The triumphs and limitations of computational methods for scRNA-seq. Nat. Methods. 2021;18:723–732. doi: 10.1038/s41592-021-01171-x. [DOI] [PubMed] [Google Scholar]
- 27.Tirosh I., Izar B., Prakadan S.M., Wadsworth M.H., 2nd, Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G., et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper analyzes existing, publicly available data. The accession numbers for the datasets are listed in the key resources table. Six publicly available experimental RNA-seq datasets are included in the app for training and demonstration purposes. The datasets generated by the Stem Cell Translation Laboratory (SCTL) were derived from hESCs (WA09) and iPSCs (LiPSC-GR1.1): “ISB003” (WA09 differentiated into three germ layers);7 bulk RNA-seq data “ISB008” (WA09 and LiPSC-GR1.1) from Chen et al.22 and four scRNA datasets from Tristan et al.7: “IS020” (WA09 cultured manually and robotically); “IS018” (WA09 differentiated into neurons), “IS006_WA09” (WA09 differentiated into three germ layers); “IS006_11” (LiPSC-GR1.1 differentiated into germ layers). A small RNA-Seq dataset “example_bulk” is from WI-38 fibroblasts with and without TGF-β treatment on days 1 and 20 from Walker et al.23 This dataset was selected because it is from human-derived samples, has a simple and balanced experimental design, and includes sufficient metadata to demonstrate all features and statistical models available for bulk RNA-seq in SEQUIN. A small scRNA-seq dataset “example_sc” was obtained from the IRIS-EDA GitHub repository (https://github.com/OSU-BMBL/IRIS) and consists of human preimplantation embryos and hESCs at different passages.18 This dataset was selected because it is from human-derived samples and is small but sufficient for quickly demonstrating all features available for scRNA-seq in SEQUIN. A publicly hosted version with example datasets from SCTL and the literature is available at https://sequin.ncats.io/app/.
-
•
All original code and a stand-alone package that can run locally is available here: https://github.com/ncats/public_sequin. All original code has been deposited at Zenodo and is publicly available as of the date of publication under https://doi.org/10.5281/zenodo.7554907.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.