Abstract
Purpose of the review
In this review, we aim to analyze the progress in understanding the genetic basis of the epilepsies, as well as ongoing efforts to define the increasingly diverse and novel presentations, phenotypes, and divergences from the expected that have continually characterized the field.
Recent Findings
A genetic workup is now considered to be standard of care for individuals with an unexplained epilepsy, due to mounting evidence that genetic diagnoses significantly influence treatment choices, prognostication, community support, and increasingly, access to clinical trials. As more individuals with epilepsy are tested, novel presentations of known epilepsy genes are being discovered, and more individuals with self-limited epilepsy are able to attain genetic diagnoses. Additionally, new genes causative of epilepsy are being uncovered through both traditional and novel methods, including large international data-sharing collaborations and massive sequencing efforts as well as computational methods and analyses driven by the Human Phenotype Ontology (HPO).
Summary
New approaches to gene discovery and characterization are advancing rapidly our understanding of the genetic and phenotypic architecture of the epilepsies. This review highlights relevant and groundbreaking studies published recently that have pushed forward the field of epilepsy genetics.
Keywords: Epilepsy, genetics, gene discovery, phenotypic characterization
INTRODUCTION
The majority of epilepsy is known to be attributable to genetic factors, as has been long established. Genetic variation is thought to explain up to two thirds of the overall liability to epilepsy in the general population, and genetic susceptibility can be attributed to monogenic, polygenic, and other multifactorial contributions (1). Vast progress has been made in the past twenty years in the identification of the genes causative of epilepsy, as well as the characterization of the phenotypic architecture of many of the genetic epilepsies. The first gene to be identified as causative of epilepsy, CHRNA4, was identified in 1995 (2) and now, over 15 years later, over 1,000 genes have been identified that have been associated with monogenic epilepsy. In addition, it has now been established as standard-of-care to complete a genetic workup for individuals with unexplained epilepsies, even individuals who have typical development and epilepsy that is well-controlled (3)**. Whole exome sequencing is now recommended as a first-line test in individuals with developmental delay and intellectual disability (4), conditions where genetic contributions are known to be highly comorbid with that of epilepsy. With this huge advancement in possible genes to identify, as well as people in whom these findings can be identified routinely, the field of epilepsy genetics has experienced an explosive growth in available knowledge and new questions to answer.
PROGRESS IN IDENTIFICATION OF GENETIC BASES OF THE EPILEPSIES
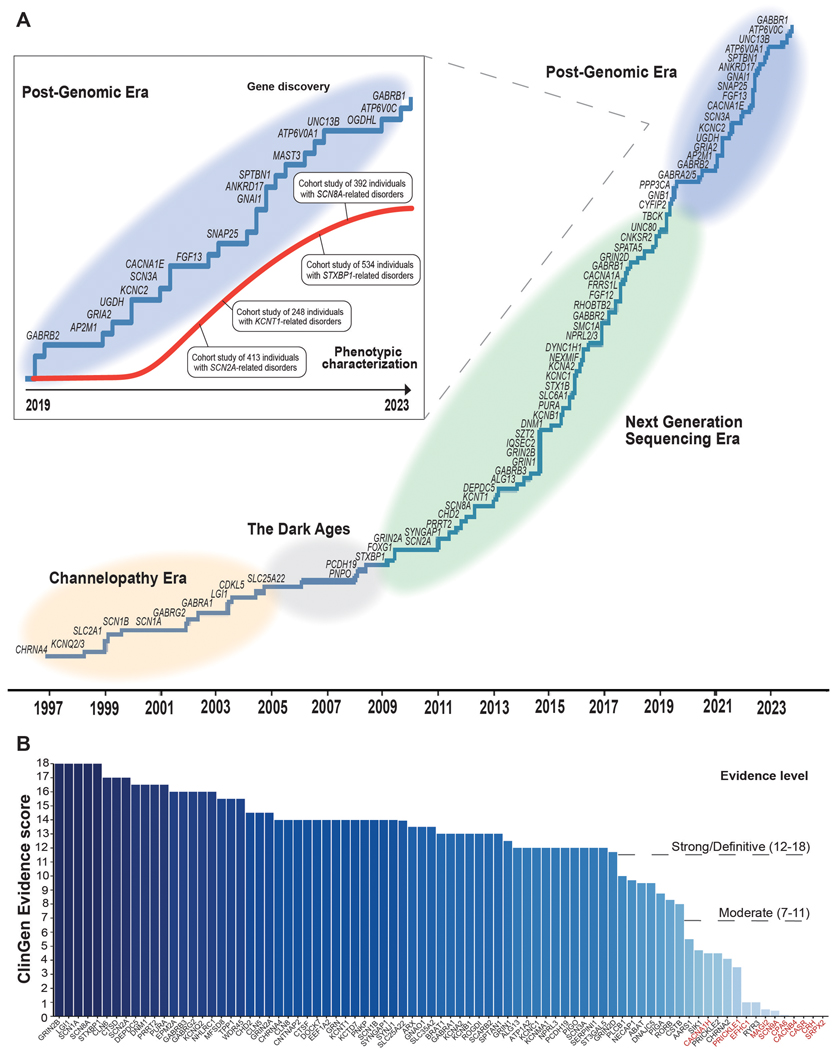
Ten years ago, the field was revolutionized by novel high-throughput methods that led to the the discovery of what is still the most common known cause of monogenic epilepsy, PRRT2 (5), which causes self-limited familial infantile-onset seizures, as well as the discoveries of the monogenic causes of Malignant Migrating Partial Seizures of Infancy (MMPSI) (6) and Autosomal Dominant Nocturnal Frontal Lobe Epilepsy (ADNFLE) (7), KCNT1. Genetic etiologies are still being discovered now as they were then, through large family studies and international “genematching” collaborations (8). Though, in addition, international data sharing collaborations and large exome sequencing studies performed on thousands of individuals, such as Epi4K and Epi25 Collaborative (9), the Deciphering Developmental Disorders (DDD) project (10)**, and cohorts analyzed from clinical testing labs (11), are creating massive datasets that have the power to allow new gene discoveries to rise to the surface. While gene identification is essential, it is just as imperative that evidence supporting gene-disease relationship emerge from a both genetic and experimental evidence. Over time, evidence has accumulated significantly to create definitive or strong disease-gene relationships for genes causative of epilepsy, as has been systematically analyzed by Clinical Genomics Resource Epilepsy Expert panel (Figure 1B).
Figure 1. A recent history of gene identification and phenotypic characterizations of the epilepsies.
A. The pace of gene discovery is continuing at a steady pace but slowing over time as we reach the limits of gene discovery power achievable with current exome studies. However, our knowledge of phenotypic characterization continues to rapidly increase, leveraged by large studies of many hundreds of individuals with genetic epilepsy. B. Evidence supporting the validity of gene-disease relationship is bolstered over time by repeated case series, natural history analyses, and evidence gained from functional testing. Importantly, mounting genetic and experimental evidence also leads to gene-disease relationships being refuted over time (as represented by genes highlighted in red).
NOVEL GENE DISCOVERY
Hundreds of new epilepsy genes have been discovered in the past decade that explain epilepsies that cross the spectrum of self-limited epilepsy to developmental and epileptic encephalopathy (Figure 1A). Diagnoses for children with DEEs now can be identified over 40% of the time, due to the steady advancement of gene discovery and characterization (12). Collaborative international networks have led to the discovery of further relatively common genes causative of DEEs such as SCN3A (13), which is causative of a severe childhood onset epilepsy, sometimes with malformations of cortical development.
In addition to SCN3A, further ion channel genes have been identified as causative genes for human epilepsies in the last three years, gradually unveiling the full landscape of epilepsy-related channelopathies. KCNC2, encoding a member of the shaw-related voltage gated potassium channel (KV3) family important for high-frequency repetitive firing of action potentials was identified as a rare cause of developmental and epileptic encephalopathies and generalized epilepsies (14, 15)*. Both gain- and loss-of-function variants were identified as disease-causing, and in contrast to phenotypes associated with the closely related KCNC1 gene, none of the reported individuals have been found to have progressive myoclonus epilepsy. In addition, bi-allelic loss-of-function variants in CACNA2D1, encoding the auxiliary α2δ subunit of voltage-gated calcium channels were identified as a rare cause of severe developmental and epileptic encephalopathies (16).
The synapse disorders represent a second gene group with constantly increasing number of disease genes implicated in human epilepsy. In addition to the discovery of AP2M1 (17), encoding the μ-subunit of the adaptor protein complex 2 involved in clathrin-mediated endocytosis and the synaptic vesicle cycle, SNAP25 (18) and UNC13B (19), and have been identified as genetic causes of human seizure disorders in the last three years. In contrast to UNC13B, which was found as a cause of a mild focal epilepsy, both AP2M1 and SNAP25 cause developmental and epileptic encephalopathies or complex neurodevelopmental disorders.
Other notable genes identified in the last three years include SPTBN1, encoding a neuronal βII-spectrin (20) given its relatedness to SPTAN1, one of the earliest identified genes for developmental and epileptic encephalopathies listed as developmental and epileptic encephalopathy 5 in OMIM (21). In parallel to SPTAN1, the phenotypic spectrum of SPTBN1 disease-causing variants include severe epilepsies and neurodevelopmental differences. An additional gene worth noting includes the SLC32A1 gene, encoding the vesicular inhibitory aminoacid cotransporter VGAT. The SLC32A1 gene is notable as it is one of the few recently identified genes for familial epilepsies inherited in an autosomal-dominant manner (22). Specifically, disease-causing variants were first identified in families with Genetic Epilepsy with Febrile Seizures Plus (GEFS+), followed by the identification of a broader phenotypic range due to de novo variants that includes developmental and epileptic encephalopathy with a prominent hyperkinetic movement disorder (23).
The genetic architecture of the epilepsies is built through up to 20 genes recurring commonly across many people and families, and then a “long tail” of remaining genes, each explaining no more than a fraction of a percent of genetic epilepsy on their own. Most genetic discoveries of recent identify individually rare, but collectively common conditions. There are the genes that can cause a spectrum of epilepsies, based on differing genetic variants and factors yet to be discovered, such as KCNC2, which can lead to both genetic generalized epilepsy presentations and DEE presentations (14, 15). NPRL3, a modulator of the GATOR complex, an mTOR regulator within the brain, has been identified as causative of a spectrum of focal epilepsies which can run in families with variable penetrance and expressivity (24). This gene has been found to explain a much higher proportion of familial and non-familial focal epilepsy than could have been predicted with the gene’s discovery in 2016. Perhaps this phenomenon could be explained by the increased testing available for individuals who have self-limited epilepsies, and underscores the importance of testing availability for individuals with self-limited and focal epilepsies.
Finally, new genes have been discovered through novel methods, such as phenotypic similarity analyses. While genes are consistently discovered through genotype-first approaches, such as massive sequencing efforts that allow for identification of rare and de novo variants in novel genes, it has been demonstrated that phenotype-first approaches can be used to identify new epilepsy genes. AP2M1, a gene causative of neurodevelopmental disorders with generalized epilepsy, especially myoclonic-atonic seizures, was discovered through analyzing phenotypic similarities constructed through phenotyping over 300 individuals with DEEs with the Human Phenotype Ontology (HPO) (17). These novel approaches to gene identification will continue to grow as the phenotypic variability of the epilepsies is further defined.
POLYGENIC AND OTHER GENETIC MECHANISMS
Novel disease mechanisms including pathogenic intronic variants and repeat expansions, and innovative integration of genetic data with polygenic risk scores have created paradigm shifts in the field. Non-coding variants within an intronic region of SCN1A were found to promote inclusion of a poison exon, causing Dravet Syndrome through reduced SCN1A expression (25). Intronic expansions in SAMD12 and other genes were identified as the cause of adult familial myoclonic epilepsy (FAME) (26, 27), a borderland condition between myoclonic epilepsy and tremor that was found to affect hundreds of individuals given the familial inheritance in this condition.
Since a large percentage of the genetic liability for generalized epilepsy is expected to be explained by common genetic variants, polygenic risk scores may reflect a significant contribution to the overall population risk. Despite providing valuable information about risk, polygenic risk scores do not indicate underlying biological causes of disease, and do not appear likely to ever do so, because the risk signals are spread broadly through the genome. For this reason, it appears unlikely that therapies can often be effectively targeted on the basis of individual polygenic risk scores. Polygenic risk assessment is also currently limited to specific populations. Recent progress has been made in understanding polygenic epilepsies through Genome-Wide Association (GWAS) and polygenic risk analyses. Recently, new genetic factors related to propensity towards febrile seizures have been identified, collectively explaining 2.8% of the variance in the liability to febrile seizures (28)*. Genetic factors involved in this propensity include those that regulate response to fever, as well as genes that influence neuronal excitability through ion channel (SCN1A, SCN2A, ANO3) and synaptic transmission (BSN, ERC2, GABRG2, HERC1). Over 7,000 cases and 83,000 controls were analyzed to attain these results, reflecting the massive numbers of research participants that will be required to further define and investigate polygenic risk. Complex inheritance of multiple, common variants in key genes also have been analyzed using data from international consortia for sporadically affected individuals, as well as people with familial epilepsy and their unaffected relatives (29). This analysis was able to demonstrate that individuals with familial epilepsies (OR 1.20) and sporadic epilepsies (OR 1.09) had an elevated polygenic risk compared to controls. PRS scores were most pronounced in familial cases of GGE (OR 1.58), emphasizing the importance of familial aggregation particularly in the generalized epilepsies.
NOVEL PRESENTATIONS OF KNOWN GENES
It has been repeatedly seen within the field of epilepsy genetics that monogenic explanations are found first for severe developmental and epileptic encephalopathies, and then mild and novel presentations are later attributed to the same gene. Both STXBP1 and KCNQ2-related developmental and epileptic encephalopathy (DEE) were first identified as causes of Ohtahara syndrome in neonates who went on to experience intractable seizures across their lifespan (30, 31); these conditions now are known to cause much more variable presentations of epilepsy and developmental phenotypes, including infantile spasms without prior neonatal seizures (32), and sometimes neurodevelopmental disorders without seizures (33, 34). As more individuals are analyzed through genetic workups, explanations for more mildly affected individuals with epilepsy are increasingly being identified. Mild variants in genes once thought to be causative of DEEs in all cases are being found in individuals with self-limited epilepsies and more mild impacts to their learning and development, as has now been reported rarely in STXBP1 (34)*.
Further, novel disease-causing mechanisms and variants within known epilepsy genes are being identified. DNM1-related disorder, a severe DEE characterized by profound hypotonia, highly abnormal hypssarrythmic EEGs, West syndrome evolving to Lennox-Gastaut syndrome, and severe to profound developmental delays, was first identified to be caused by missense gain-of-function variants within the ATPase and middle domains of the protein, interfering with the protein’s ability to heterodimerize and aid in vesicle recycling (35). Recently, the most recurrent described variant in DNM1 was identified, an apparent splice variant (c.1197–8G>A) in the canonical isoform of the protein, but one that actually acts as a non-apparent missense variant in the isoform of the protein most highly expressed in the pediatric brain (36)*. Additionally, genetic etiologies first characterized as operating through dominant negative or loss-of-function mechanisms have had novel variants identified that are proven to operate in a gain-of-function manner, resulting in novel presentations and phenotypes. Novel variants in SCN1A have recently been shown to cause a severe early-onset phenotype diverging from Dravet syndrome, characterized by epilepsy that onsets significantly prior to the classic 5–6 months of age and which results in a unique developmental profile (37, 38)*.
PROGRESS IN VARIANT ANALYSIS AND INTERPRETATION
Novel approaches to variant databasing, aggregation, functional testing, and interpretation are growing exponentially in recent years, which is critical step towards making data available and interpretable to those providing clinical care. Variant and functional data has been grouped into publicly available portals for the NMDA-receptor family of genes (39), SLC6A1 (40), and the sodium channel family of genes (41). These browsable resources allow for the combination of genetic, phenotypic, and functional data for improved variant interpretation and VUS resolution. Functional characterization of a variant’s effect on a protein is essential to establish the underlying pathobiology of the genetic condition to create opportunities for precision therapy, as well as to resolve variants of uncertain significance. Large, collaborative efforts within the NIH-funded Centers Without Walls (CWOW) have performed mass characterization of variants within multiple epilepsy genes, as was recently demonstrated in KCNQ2 (42). These studies create the basis of pathobiology for these variants, and at times identify paradoxical effects on channels that have led to the discovery of novel presentations of known epilepsy genes, such as that seen in SCN1A (37, 38). This increased knowledge of variant impact and functional analyses has led to increasingly mounting evidence proving gene-disease relationships.
PROGRESS IN UNDERSTANDING THE CLINICAL PICTURE OF GENETIC EPILEPSIES
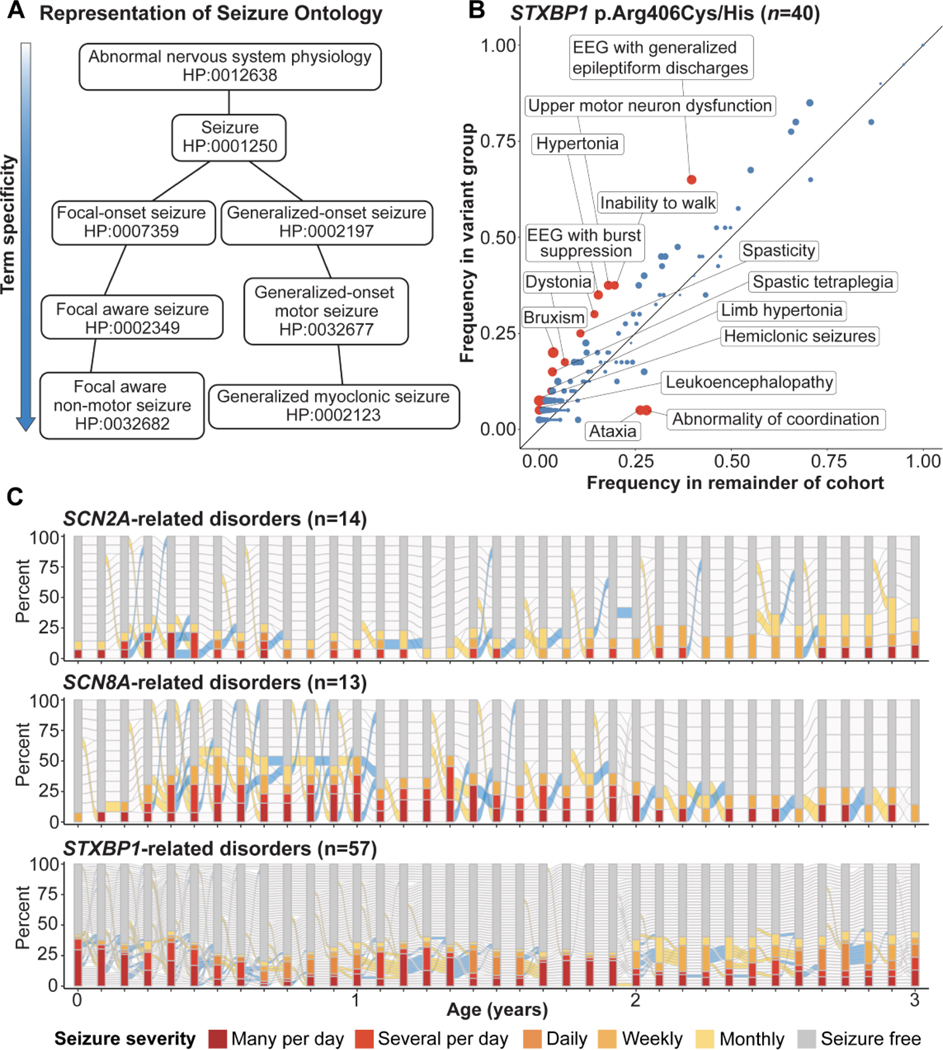
In parallel to the explosion of the understanding of genotypic data that has occurred over the last decade, it has become increasingly imperative to characterize the phenotypic architecture of genetic epilepsies being more routinely diagnosed (Figure 2). Our understanding of the genetic underpinnings of epilepsy has thus far outpaced our understanding of phenotypic architecture and variability. Outlining the natural history and progression of genetic epilepsies is a massive undertaking requiring a multi-pronged approach: including case series and large family studies, natural history studies that follow smaller groups of children affected by a genetic condition in granular and comprehensive detail, and, finally, the use of computational methods, such as phenotypic reconstruction through HPO and Electronic Medical Record (EMR) data to characterize genetic epilepsies based on thousands of years of patient data that already exists, waiting to be analyzed. Natural history studies for Rett syndrome, Angelman, Dravet, and CDKL5-related disorder have shown prime examples of deep phenotypic description and characterization of affected individuals over time. Parallel to deep phenotypic characterization in smaller groups of affected individuals are efforts to aggregate existing data from the literature in addition to data captured in routine clinical care and the EMR (43), which allow for enough power to bring new features and genotype/phenotype correlations to light, as has been seen in STXBP1, SCN2A, KCNT1, and SCN8A (34, 44–46). Finally, reconstruction of genetic diseases through HPO-based methods has allowed the field to place computational power behind static and longitudinal phenotypic description, leading to novel discoveries (47).This emerging field of phenomics allows for static and longitudinal disease reconstructions that will allow for identification of natural history study and trial endpoints in the future. These joint efforts are leading to a comprehensive understanding of the complex phenotypic architecture of the epilepsies over time.
Figure 2. Novel methods of phenotypic characterization.
A. Representation of the Human Phenotype Ontology (HPO), the system that has accelerated phenotypic characterization through computational methods. Example HPO terms with their associated standardized code are shown. B. Genotype/ phenotype correlations that emerge for STXBP1 related disorders when approached through methods of phenotype reconstruction (from Xian et al., 2021-open access paper). Red points indicated HPO terms with uncorrected P-values < 0.05, while blue points indicated HPO terms with uncorrected P-values ≥ 0.05 C. Longitudinal seizure reconstruction of three of the most common causes of genetic epilepsy, demonstrating the difference in seizure presentation in the first three years of life, which can be reconstructed from data already within the Electronic Medical Record (EMR).
PROGRESS IN PRECISION MEDICINE
With diagnoses being identified in an increasing number of people with epilepsy, the question remains as to the extent to which treatment is affected by genetic diagnosis. Through systematic analysis of this question, it has been found that genetic diagnosis can influence treatment in nearly 50% of cases (48)*, through the influence on treatment options including anti-seizure medication (ASM) choice and the facilitation of preventative and specialty care due to known risks associated with particular genetic diagnoses. Through the analysis of over 400 individuals, it was found that new medications were added or initiated in over 1/3 of individuals following genetic diagnosis. Many genetic diagnoses causative of epilepsy have an associated precision medicine therapy that is recommended or suggested post-diagnosis. For example, carbamazepine is recommended as a first-line medication for people with infantile seizures due to PRRT2, but also for adult-onset dyskinesias often present in this disorder (49). Sodium channel blockers are recommended to be avoided for loss-of-function variants with sodium channel genes such as SCN1A, SCN2A, and SCN8A, but they have been trialed with success in individuals with gain-of-function variants in these genes (37). Considering diagnostic rate of genetic testing in individuals with epilepsy across all testing modalities has been found to be 17%, and much higher in individuals with developmental and epileptic encephalopathy and neurodevelopmental disorders (50), a significant proportion of the pediatric and adult population with epilepsy has the potential to benefit from a clinical genetics evaluation. Though this potential exists, it should be stated that, for most genes causative of epilepsy, a precision medicine is not available, or has been found to be less effective than initially theorized. Future research will seek to compare effectiveness of known and novel epilepsy medications in individuals with genetic diagnoses.
In addition, the first gene and genetic therapies are now trialed for distinct genetic epilepsies, including for SCN1A-related Dravet, and KCNT1. However, at the writing of this review, it is critical to point out that none of these studies have been published, a caveat that is critical given the high and sometimes unrealistic expectations for novel treatments. Gene therapies are under development for several genetic epilepsies, though it is still uncertain as to how these novel approaches will be delivered safely to the central nervous system, and the robustness of an effect they might have (51).
THE ROLE OF ADVOCACY ORGANIZATIONS
As more rare genetic epilepsies are identified and more individuals diagnosed, the role of genetic epilepsy advocacy organizations has become increasingly critical. Recently, the Rare Epilepsy Landscape Analysis (RELA) performed a qualitative analysis of 44 rare genetic epilepsy organizations that have shared goals of improving care and outcomes in individuals with genetic epilepsy, to establish a national registry, and to increase funding for epilepsy research through new paradigms (52)*. The Rare Epilepsy Network (REN) registry, an engagement partnership initiative of The Epilepsy foundation, now includes representation from family foundations of more than forty rare genetic epilepsies. Advocacy organizations now have and must continue to be driving all steps of clinical and translational research. Disease concept model studies, often driven by family foundations, have identified novel symptoms and disease impacts not previously described in the scientific literature (53, 54). In addition, advocacy organizations are identifying and driving fast-paced efforts to push precision medicine into the clinic, as demonstrated by the Foundation for Angelman Syndrome Therapeutics (FAST) collaborative, as well as the work performed by the STXBP1 and SLC6A1 foundations to push forward drug repurposing efforts (55).
CONCLUSION
Through international collaboration between clinicians, scientists, and family advocacy, knowledge of the genetic epilepsies continues to grow exponentially. In the clinic, genetic testing for individuals with any unexplained epilepsy has been officially set as standard of care by the National Society of Genetic Counselors (NSGC) (3). Gene discovery continues at a steady pace, and we have a better understanding than ever of the natural history of genetic epilepsies, gained through gold-standard and innovative methods. We are seeing the first gene modulatory therapies being trialed in the genetic epilepsies, and vast amounts of research focused on precision medicine approaches. Continued efforts of collaboration, data-sharing and harmonization, and integration of the massive amounts of data that has been collected in the research and clinical domains over the past years will underpin our future discoveries and understanding.
KEY POINTS.
Genetic testing for all individuals with an unexplained epilepsy is now standard-of-care, with exome sequencing often being the most appropriate test of choice.
Increased genetic testing of individuals and huge multi-national exome sequencing and datasharing efforts have lead to further genotypic and phenotypic characterization in the epilepsies.
Collaboration with disease advocacy organizations has pushed the field of epilepsy genetics and precision medicine forward.
ACKNOWLEDGEMENTS
The authors would like to thank Lucy Brancazio with assistance in development of the article figures.
FINANCIAL SUPPORT AND SPONSORSHIP
I.H. was supported by The Hartwell Foundation (Individual Biomedical Research Award), NINDS (K02NS112600, U24NS120854-01, U54NS108874-04), the Eunice Kennedy Shriver National Institute of Child Health and Human Development through the Children’s Hospital of Philadelphia and the University of Pennsylvania (U54HD086984), the German Research Foundation (HE5415/3-1, HE5415/5-1, HE5415/6-1, HE5415/7-1), the National Center for Advancing Translational Sciences of the NIH (UL1TR001878), the Institute for Translational Medicine and Therapeutics’ (ITMAT) at the Perelman School of Medicine of the University of Pennsylvania, and by Children’s Hospital of Philadelphia through the Epilepsy NeuroGenetics Initiative (ENGIN). S.R. is supported by The STXBP1 Disorders foundation via an Individual Award.
Footnotes
CONFLICTS OF INTEREST
None of the authors have any conflicts of interest to declare.
References
- 1.Thomas RH, Berkovic SF. The hidden genetics of epilepsy-a clinically important new paradigm. Nat Rev Neurol. 2014;10(5):283–92. [DOI] [PubMed] [Google Scholar]
- 2.Steinlein OK, Mulley JC, Propping P, Wallace RH, Phillips HA, Sutherland GR, et al. A missense mutation in the neuronal nicotinic acetylcholine receptor alpha 4 subunit is associated with autosomal dominant nocturnal frontal lobe epilepsy. Nat Genet. 1995;11(2):201–3. [DOI] [PubMed] [Google Scholar]
- 3. Smith L, Malinowski J, Ceulemans S, Peck K, Walton N, Sheidley BR, et al. Genetic testing and counseling for the unexplained epilepsies: An evidence-based practice guideline of the National Society of Genetic Counselors. J Genet Couns. 2022. **This practice guideline provides the first evidence-based approach to genetic testing and counseling for individuals with unexplained epilepsy
- 4.Manickam K, McClain MR, Demmer LA, Biswas S, Kearney HM, Malinowski J, et al. Exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability: an evidence-based clinical guideline of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2021;23(11):2029–37. [DOI] [PubMed] [Google Scholar]
- 5.Heron SE, Grinton BE, Kivity S, Afawi Z, Zuberi SM, Hughes JN, et al. PRRT2 mutations cause benign familial infantile epilepsy and infantile convulsions with choreoathetosis syndrome. Am J Hum Genet. 2012;90(1):152–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barcia G, Fleming MR, Deligniere A, Gazula VR, Brown MR, Langouet M, et al. De novo gain-of-function KCNT1 channel mutations cause malignant migrating partial seizures of infancy. Nat Genet. 2012;44(11):1255–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heron SE, Smith KR, Bahlo M, Nobili L, Kahana E, Licchetta L, et al. Missense mutations in the sodium-gated potassium channel gene KCNT1 cause severe autosomal dominant nocturnal frontal lobe epilepsy. Nat Genet. 2012;44(11):1188–90. [DOI] [PubMed] [Google Scholar]
- 8.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36(10):928–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Epi KC. Epi4K: gene discovery in 4,000 genomes. Epilepsia. 2012;53(8):1457–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kaplanis J, Samocha KE, Wiel L, Zhang Z, Arvai KJ, Eberhardt RY, et al. Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature. 2020;586(7831):757–62. **Integrating genetic data from over 30,000 trio exomes from individuals with developmental disorders including epilepsy, evidence was found to support the discovery of many new genetic disorders. In addition, insight was generated suggesting that over 1,000 novel genetic etiologies still are yet to be discovered.
- 11.Lindy AS, Stosser MB, Butler E, Downtain-Pickersgill C, Shanmugham A, Retterer K, et al. Diagnostic outcomes for genetic testing of 70 genes in 8565 patients with epilepsy and neurodevelopmental disorders. Epilepsia. 2018;59(5):1062–71. [DOI] [PubMed] [Google Scholar]
- 12.Hamdan FF, Myers CT, Cossette P, Lemay P, Spiegelman D, Laporte AD, et al. High Rate of Recurrent De Novo Mutations in Developmental and Epileptic Encephalopathies. Am J Hum Genet. 2017;101(5):664–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaman T, Helbig I, Bozovic IB, DeBrosse SD, Bergqvist AC, Wallis K, et al. Mutations in SCN3A cause early infantile epileptic encephalopathy. Ann Neurol. 2018;83(4):703–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwarz N, Seiffert S, Pendziwiat M, Rademacher AV, Brunger T, Hedrich UBS, et al. Spectrum of Phenotypic, Genetic, and Functional Characteristics in Patients With Epilepsy With KCNC2 Pathogenic Variants. Neurology. 2022;98(20):e2046–e59. *In this cohort, KCNC2 was demonstrated to be explanatory for several unusual epilepsy phenotypes in unrelated individuals, including early onset absence epilepsy, Doose syndrome, and continuous spike and wave during slow sleep (CSWS), phenotypes that have not been demonstrated previously to be explained by the same gene in different individuals.
- 15.Vetri L, Cali F, Vinci M, Amato C, Roccella M, Granata T, et al. A de novo heterozygous mutation in KCNC2 gene implicated in severe developmental and epileptic encephalopathy. Eur J Med Genet. 2020;63(4):103848. [DOI] [PubMed] [Google Scholar]
- 16.Dahimene S, von Elsner L, Holling T, Mattas LS, Pickard J, Lessel D, et al. Biallelic CACNA2D1 loss-of-function variants cause early-onset developmental epileptic encephalopathy. Brain. 2022;145(8):2721–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helbig I, Lopez-Hernandez T, Shor O, Galer P, Ganesan S, Pendziwiat M, et al. A Recurrent Missense Variant in AP2M1 Impairs Clathrin-Mediated Endocytosis and Causes Developmental and Epileptic Encephalopathy. Am J Hum Genet. 2019;104(6):1060–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klockner C, Sticht H, Zacher P, Popp B, Babcock HE, Bakker DP, et al. De novo variants in SNAP25 cause an early-onset developmental and epileptic encephalopathy. Genet Med. 2021;23(4):653–60. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Qiao JD, Liu XR, Liu DT, Chen YH, Wu Y, et al. UNC13B variants associated with partial epilepsy with favourable outcome. Brain. 2021;144(10):3050–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cousin MA, Creighton BA, Breau KA, Spillmann RC, Torti E, Dontu S, et al. Pathogenic SPTBN1 variants cause an autosomal dominant neurodevelopmental syndrome. Nat Genet. 2021;53(7):1006–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Syrbe S, Harms FL, Parrini E, Montomoli M, Mutze U, Helbig KL, et al. Delineating SPTAN1 associated phenotypes: from isolated epilepsy to encephalopathy with progressive brain atrophy. Brain. 2017;140(9):2322–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heron SE, Regan BM, Harris RV, Gardner AE, Coleman MJ, Bennett MF, et al. Association of SLC32A1 Missense Variants With Genetic Epilepsy With Febrile Seizures Plus. Neurology. 2021;96(18):e2251–e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Platzer K, Sticht H, Bupp C, Ganapathi M, Pereira EM, Le Guyader G, et al. De Novo Missense Variants in SLC32A1 Cause a Developmental and Epileptic Encephalopathy Due to Impaired GABAergic Neurotransmission. Ann Neurol. 2022;92(6):958–73. [DOI] [PubMed] [Google Scholar]
- 24.Ricos MG, Hodgson BL, Pippucci T, Saidin A, Ong YS, Heron SE, et al. Mutations in the mammalian target of rapamycin pathway regulators NPRL2 and NPRL3 cause focal epilepsy. Ann Neurol. 2016;79(1):120–31. [DOI] [PubMed] [Google Scholar]
- 25.Carvill GL, Engel KL, Ramamurthy A, Cochran JN, Roovers J, Stamberger H, et al. Aberrant Inclusion of a Poison Exon Causes Dravet Syndrome and Related SCN1A-Associated Genetic Epilepsies. Am J Hum Genet. 2018;103(6):1022–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bennett MF, Oliver KL, Regan BM, Bellows ST, Schneider AL, Rafehi H, et al. Familial adult myoclonic epilepsy type 1 SAMD12 TTTCA repeat expansion arose 17,000 years ago and is present in Sri Lankan and Indian families. Eur J Hum Genet. 2020;28(7):973–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishiura H, Doi K, Mitsui J, Yoshimura J, Matsukawa MK, Fujiyama A, et al. Expansions of intronic TTTCA and TTTTA repeats in benign adult familial myoclonic epilepsy. Nat Genet. 2018;50(4):581–90. [DOI] [PubMed] [Google Scholar]
- 28. Skotte L, Fadista J, Bybjerg-Grauholm J, Appadurai V, Hildebrand MS, Hansen TF, et al. Genome-wide association study of febrile seizures implicates fever response and neuronal excitability genes. Brain. 2022;145(2):555–68. *This GWAS-based study provides evidence febrile seizures can be due to common variations in genes linked to liability to seizure as well as genes that regulate response to fever. Genetic variants identified in this study explain up to 2.8% of the variance in liability to febrile seizures in humans.
- 29.Oliver KL, Ellis CA, Scheffer IE, Ganesan S, Leu C, Sadleir LG, et al. Common risk variants for epilepsy are enriched in families previously targeted for rare monogenic variant discovery. EBioMedicine. 2022;81:104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saitsu H, Kato M, Okada I, Orii KE, Higuchi T, Hoshino H, et al. STXBP1 mutations in early infantile epileptic encephalopathy with suppression-burst pattern. Epilepsia. 2010;51(12):2397–405. [DOI] [PubMed] [Google Scholar]
- 31.Weckhuysen S, Mandelstam S, Suls A, Audenaert D, Deconinck T, Claes LR, et al. KCNQ2 encephalopathy: emerging phenotype of a neonatal epileptic encephalopathy. Ann Neurol. 2012;71(1):15–25. [DOI] [PubMed] [Google Scholar]
- 32.Millichap JJ, Miceli F, De Maria M, Keator C, Joshi N, Tran B, et al. Infantile spasms and encephalopathy without preceding neonatal seizures caused by KCNQ2 R198Q, a gain-of-function variant. Epilepsia. 2017;58(1):e10–e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miceli F, Millevert C, Soldovieri MV, Mosca I, Ambrosino P, Carotenuto L, et al. KCNQ2 R144 variants cause neurodevelopmental disability with language impairment and autistic features without neonatal seizures through a gain-of-function mechanism. EBioMedicine. 2022;81:104130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xian J, Parthasarathy S, Ruggiero SM, Balagura G, Fitch E, Helbig K, et al. Assessing the landscape of STXBP1-related disorders in 534 individuals. Brain. 2022;145(5):1668–83. *The first genotypephenotype correlations were elucidated for STXBP1-related disorders in this study, which used phenotypic reconstruction of every published individual, as well as many unpublished, to create a comprehensive overview of a common neurodevelopmental disorder.
- 35.von Spiczak S, Helbig KL, Shinde DN, Huether R, Pendziwiat M, Lourenco C, et al. DNM1 encephalopathy: A new disease of vesicle fission. Neurology. 2017;89(4):385–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Parthasarathy S, Ruggiero SM, Gelot A, Soardi FC, Ribeiro BFR, Pires DEV, et al. A recurrent de novo splice site variant involving DNM1 exon 10a causes developmental and epileptic encephalopathy through a dominant-negative mechanism. Am J Hum Genet. 2022;109(12):2253–69. *A paradoxical mechanism underlies a complex developmental and epileptic encephopathy, which is demonstrated through phenotypic data and gene expression data obtained from pediatric brain surgery samples.
- 37. Brunklaus A, Brunger T, Feng T, Fons C, Lehikoinen A, Panagiotakaki E, et al. The gain of function SCN1A disorder spectrum: novel epilepsy phenotypes and therapeutic implications. Brain. 2022;145(11):3816–31. *This study defined the phenotypic spectrum of variants in SCN1A that lead to an SCN1A-related epilepsy disorder entirely separate from Dravet syndrome. Variants were functionally analyzed, providing evidence for precision medicine within gain-of-function SCN1A-related disorder.
- 38.Clatot J, Parthasarathy S, Cohen S, McKee JL, Massey S, Somarowthu A, et al. SCN1A gain-of-function mutation causing an early onset epileptic encephalopathy. Epilepsia. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.GRIN Portal: BROAD Institute; [Available from: https://grin-portal.broadinstitute.org/.
- 40.SLC6A1 Portal: BROAD Institute; [Available from: https://slc6a1-portal.broadinstitute.org/.
- 41.SCN Portal: BROAD Institute; [Available from: https://scn-portal.broadinstitute.org/.
- 42.Vanoye CG, Desai RR, Ji Z, Adusumilli S, Jairam N, Ghabra N, et al. High-throughput evaluation of epilepsy-associated KCNQ2 variants reveals functional and pharmacological heterogeneity. JCI Insight. 2022;7(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewis-Smith D, Ganesan S, Galer PD, Helbig KL, McKeown SE, O’Brien M, et al. Phenotypic homogeneity in childhood epilepsies evolves in gene-specific patterns across 3251 patient-years of clinical data. Eur J Hum Genet. 2021;29(11):1690–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crawford K, Xian J, Helbig KL, Galer PD, Parthasarathy S, Lewis-Smith D, et al. Computational analysis of 10,860 phenotypic annotations in individuals with SCN2A-related disorders. Genet Med. 2021;23(7):1263–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johannesen KM, Liu Y, Koko M, Gjerulfsen CE, Sonnenberg L, Schubert J, et al. Genotype-phenotype correlations in SCN8A-related disorders reveal prognostic and therapeutic implications. Brain. 2022;145(9):2991–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonardi CM, Heyne HO, Fiannacca M, Fitzgerald MP, Gardella E, Gunning B, et al. KCNT1-related epilepsies and epileptic encephalopathies: phenotypic and mutational spectrum. Brain. 2021;144(12):3635–50. [DOI] [PubMed] [Google Scholar]
- 47.Lewis-Smith D, Parthasarathy S, Xian J, Kaufman MC, Ganesan S, Galer PD, et al. Computational analysis of neurodevelopmental phenotypes: Harmonization empowers clinical discovery. Hum Mutat. 2022;43(11):1642–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McKnight D, Morales A, Hatchell KE, Bristow SL, Bonkowsky JL, Perry MS, et al. Genetic Testing to Inform Epilepsy Treatment Management From an International Study of Clinical Practice. JAMA Neurol. 2022. *Genetic diagnosis is found to directly influence medical management of individuals with epilepsy through this systematic analysis.
- 49.Symonds JD, Zuberi SM, Stewart K, McLellan A, O’Regan M, MacLeod S, et al. Incidence and phenotypes of childhood-onset genetic epilepsies: a prospective population-based national cohort. Brain. 2019;142(8):2303–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheidley BR, Malinowski J, Bergner AL, Bier L, Gloss DS, Mu W, et al. Genetic testing for the epilepsies: A systematic review. Epilepsia. 2022;63(2):375–87. [DOI] [PubMed] [Google Scholar]
- 51.Lu Z, He S, Jiang J, Zhuang L, Wang Y, Yang G, et al. Base-edited cynomolgus monkeys mimic core symptoms of STXBP1 encephalopathy. Mol Ther. 2022;30(6):2163–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Miller IP. Raring for change: Confluence of scientific discovery and advocate alignment warrants vital new investments in The Epilepsies. Epilepsy Behav. 2020;111:107276. *In response to a massive rise of disease advocacy organizaitons in the epilepsies, The Rare Epilepsy Landscape Analysis (RELA) identified common challenges within the genetic epilepsies and issued recommendations for improved care as defined by patients and caregivers.
- 53.Nabbout R, Auvin S, Chiron C, Irwin J, Mistry A, Bonner N, et al. Development and content validation of a preliminary core set of patient- and caregiver-relevant outcomes for inclusion in a potential composite endpoint for Dravet Syndrome. Epilepsy Behav. 2018;78:232–42. [DOI] [PubMed] [Google Scholar]
- 54.Willgoss T, Cassater D, Connor S, Krishnan ML, Miller MT, Dias-Barbosa C, et al. Measuring What Matters to Individuals with Angelman Syndrome and Their Families: Development of a Patient-Centered Disease Concept Model. Child Psychiatry Hum Dev. 2021;52(4):654–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Phenylbutyrate for STXBP1 Encephalopathy and SLC6A1 Neurodevelopmental Disorder: clinicaltrials.gov; [Available from: https://clinicaltrials.gov/ct2/show/NCT04937062?cond=stxbp1&draw=2&rank=2.