Summary
Light-sheet fluorescence microscopy (LSFM) can produce high-resolution tomograms of tissue vasculature with high accuracy. However, data processing and analysis is laborious due to the size of the datasets. Here, we introduce VesselExpress, an automated software that reliably analyzes six characteristic vascular network parameters including vessel diameter in LSFM data on average computing hardware. VesselExpress is ∼100 times faster than other existing vessel analysis tools, requires no user interaction, and integrates batch processing and parallelization. Employing an innovative dual Frangi filter approach, we show that obesity induces a large-scale modulation of brain vasculature in mice and that seven other major organs differ strongly in their 3D vascular makeup. Hence, VesselExpress transforms LSFM from an observational to an analytical working tool.
Keywords: light-sheet fluorescence microscopy, 3D vasculature, high-throughput analysis, open-source application
Graphical abstract
Highlights
-
•
An open-source software for analysis of vessel trees in 3D data of various organs
-
•
End-to-end automation and parallel processing enable high-throughput analysis
-
•
VesselExpress can be executed via a single command in the terminal or a GUI
-
•
The modular workflow design facilitates custom analysis
Motivation
The blood circulatory system represents a complex network of vessels with varying lengths and diameters that are precisely organized in space to allow proper tissue function. Light-sheet fluorescence microscopy (LSFM) can generate tomograms of tissue vasculature with high spatial accuracy. Yet, quantitative LSFM analysis is still cumbersome and available methods are restricted to single organs and advanced computing hardware. Here, we present VesselExpress, a software to analyze LSFM 3D data of blood vessel system rapidly and in a fully automated manner.
Spangenberg et al. present VesselExpress, an open-source software designed for high-throughput analysis of 3D vessel data. VesselExpress extracts nine characteristic vascular network parameters from raw data fully automatically without requiring user interaction.
Introduction
Vascular reorganization is a key process accompanying various pathophysiological conditions. For instance, sterile tissue inflammation induced by hypoxia-ischemia in stroke, myocardial infarction, or tumor development leads to massive vascular remodeling that critically determines tissue fate.1,2,3 Due to its unprecedented 3D visualization capacity, light-sheet fluorescence microscopy (LSFM), which has already been employed for analyzing a variety of organs,2,4,5,6,7 also allows a thorough exploration of disease-associated vessel reorganization. The labeling of blood vessels can be achieved by the injection of fluorescently labeled endothelial-specific antibodies,5 the perfusion of animals with fluorescent dyes,8 or the use of transgenic animals in which fluorescent reporter proteins are expressed under endothelial-specific promoters.9 After vessel labeling, vertebrate organs need to be optically cleared before LSFM.10,11,12 The power of LSFM enables high-resolution tomograms of whole cleared organs to be acquired quickly. The processing and analysis of the large (10–100 GB) datasets, however, can be laborious, which strongly inhibits the application of LSFM for large-scale quantitative studies of vessel systems.
Even though methods have been put forward that allow the quantification of blood vessel structures, even in large organs such as the murine brain,4,6 their application has only been demonstrated in the investigated organ, hence lacking general usability. Furthermore, they lack processing scalability, depend on advanced computing hardware, and even with advanced hardware, are too specific to allow for the systematic analysis of more complex pathophysiological settings by non-experts. Recently, efforts have been made to make 3D vessel analysis accessible for non-experts.13,14,15 However, the practicability of the proposed tools is limited in different ways. The approach by Bumgarner13 requires pre-segmented data and thus does not provide analysis end-to-end, i.e., from raw data to statistical endpoints. Another promising approach has been investigated through semi-automated random walk-based segmentations of blood vasculature.14,15 The approach is computationally highly efficient,16 yet limited to small sample numbers because it relies on substantial manual interference in an evaluation-edit-processing loop. Other existing solutions6 lack features such as computing platform independence, modular and extendable software design, or parallelizable workflow processing that are essential for medium or high throughput studies.17 In short, the lack of reliable and robust methods with modular and parallelizable workflow management for the quantification of phenotypic vessel features from the large 3D image stacks is a major limiting factor. Overcoming this limitation promises a paradigm to study disease processes and, potentially, to also screen compound libraries for effects on whole organ vascularization.18
Here, we present VesselExpress, a software to fully automatically analyze LSFM 3D data of blood vessel systems. It allows fast and reliable image analysis including image-processing methods and graph construction and analysis, which are assembled in a modularized software workflow. VesselExpress enables high-volume analyses, supported by corresponding algorithms, computational tools,19,20 and workflow management systems.17 With VesselExpress, we analyzed blood vessels from different murine organs, which were labeled either with fluorescein isothiocyanate (FITC)-albumin or fluorescent antibodies against CD31. Using a thresholding strategy with two cascaded Frangi filters for vessel segmentation, we were able to extract a comprehensive set of microvascular network characteristics that yield reliable information on microvascular length, branching, and diameter. Obtained measurements are well in line with conventional microscopic quantifications and match or exceed the performance of semi-automated analyses while operating orders of magnitude faster and requiring less hands-on time. Furthermore, VesselExpress is platform independent, available as open source, and provides a browser-based web interface that requires no specific computational expertise.
Results
VesselExpress software and workflow
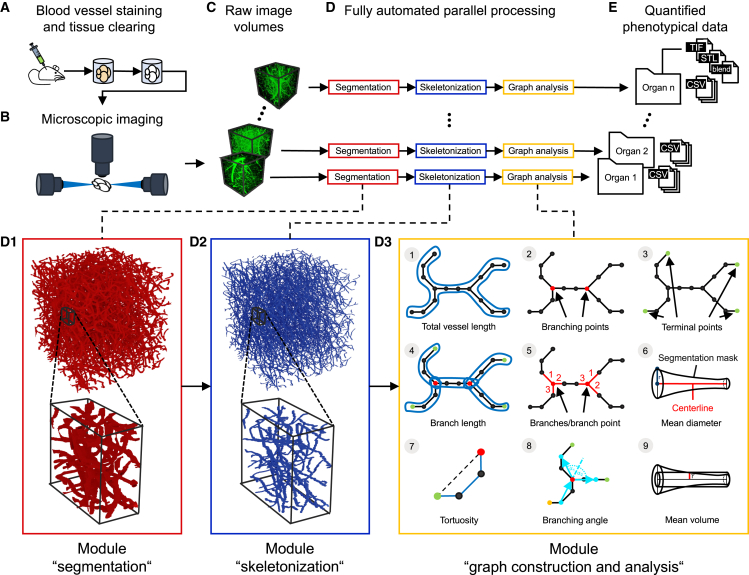
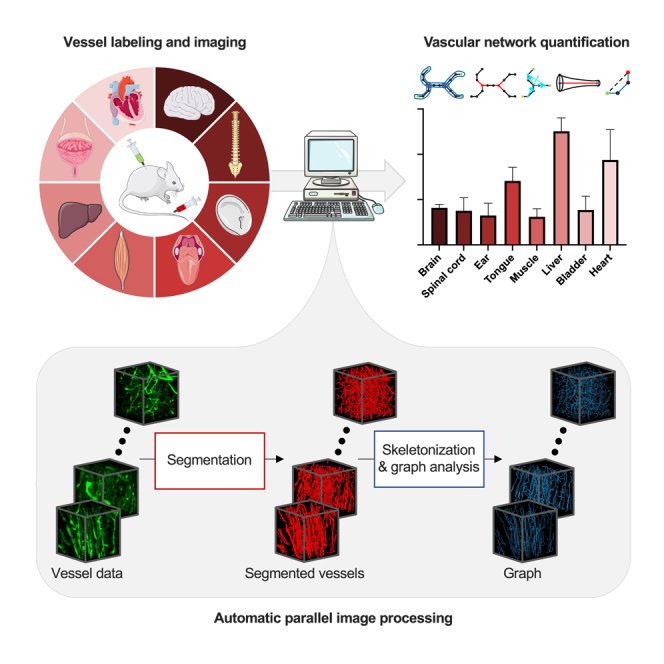
VesselExpress is an open-source software (https://github.com/RUB-Bioinf/VesselExpress) designed for rapid, fully automated, and scalable analysis of blood vessel trees in 3D datasets from LSFM sequences. The workflow is illustrated in Figure 1. VesselExpress accepts raw LSFM 3D images in the standard image file formats as input. Blood vessel staining and tissue clearing (Figure 1A) are required as sample preparation steps for the subsequent imaging of blood vessels in high resolution (Figure 1B). The processing pipeline takes raw image volumes (Figure 1C) as input and is fully automated without any user interaction (Figure 1D). The processing steps include segmentation (Figure 1D), skeletonization (Figure 1D) and graph construction with analysis (Figure 1D). In the first step, the raw images are segmented using workflows extended from Allen Cell and Structure Segmenter.21 The segmentation workflow involves certain parameters (Table S3) that need organ-specific adjustment. While default parameters for different murine organs are provided, VesselExpress includes an interactive software tool for parameter tuning based on the Napari22 platform. In the second step, the vessels’ centerlines are extracted from the binary images through the parallel thinning algorithm23 implemented in the scikit-image Python package.20 The centerlines are then transformed into undirected graphs by using the Python 3scan toolkit.24 Finally, the vessels are traced in a depth-first search (DFS), and nine phenotypic features (Figure 1E) are calculated.
Figure 1.
Pipeline for automatic quantitative blood vessel analysis
(A) Sample preparation includes blood vessel staining and tissue clearing.
(B–D) (B) Microscopic imaging of blood vessels. Generated blood vessel images (C) are processed via VesselExpress (D). In the segmentation, the raw images are binarized into foreground, representing the vessels, and background (D1). The segmentation mask is used to extract the vessels’ centerlines (D2). Undirected graphs are constructed from the skeleton mask and used for subsequent data analysis (D3).
(E) The results are quantified phenotypical data tables for statistical analysis and image masks for visual inspection of segmentation and skeletonization (E).
VesselExpress is implemented in Python and is deployed as a Docker19 container, which includes the necessary run environment and can be executed from all major operating systems via command line or the web interface integrated into the Docker image. VesselExpress processes raw microscopic images of blood vessels in parallel and produces quantified phenotypical data fully automated without requiring user interaction. All processing steps are automated in a pipeline integrated into the workflow management system Snakemake,17 which enables high scalability so that the run time of the software automatically benefits from larger RAM and more CPU cores without having to make any changes to the code. This allows comprehensive studies to be carried out within a short time. Due to the modular software design, each processing step can also be executed individually. Therefore, each function (“module”) can be easily exchanged with custom functions, if preferred, while maintaining the integrity of the pipeline. The pipeline outputs the nine quantified phenotypical features as text files, which can be directly statistically analyzed and visualized as required. Furthermore, 3D TIF images of the segmented and skeletonized vasculature as well as images of the graph, branching, and terminal points are provided. Optionally, the 3D vessel tree can automatically be rendered in Blender.25 Images of the rendered vessel tree can be provided along with the corresponding Blender project file that may be used for visual inspection and presentation.
The end-to-end automation from raw data to statistical output implemented in VesselExpress along with validation on multiple organs and microscopy modalities sets VesselExpress apart from previous approaches, as detailed in Figure S4A and Table S3. VesselExpress is tailored toward screening larger numbers of samples of identical tissue type. The usage scenario addressed by VesselExpress is to provide tools for calibrating a manageable number of parameters to a given tissue type and a given microscope setup and then to perform fully automated end-to-end analysis on large numbers of samples. This requires robust segmentation and graph analysis along with end-user suitable workflow management, which is not covered by any existing approach. A complete list of parameters used throughout segmentation and graph analysis is displayed in Table 1. As indicated, most parameters were used under default parameters throughout all experiments in our study. The main parameter that needs to be adjusted for different organs is the gamma parameter of the Frangi filter, which has a smoothing effect and can be adjusted to the specific noise level of a given dataset. For this calibration task, we provide the interactively assisted Napari plugin.
Table 1.
Parameters used by VesselExpress for segmentation and graph analysis
| Parameter | Type | Function | Default |
|---|---|---|---|
| Segmentation | |||
| smoothing | binary | if 1, edge preserving smoothing is performed | 1a |
| core_threshold | numeric | primary threshold to separate foreground from background as μ+k∘σ, where k is the core threshold parameter. 0.0 indicates no thresholding. | 3.0 |
| gamma_1 | numeric | the sensitivity of the Frangi filter to tubular shapes larger gamma for images with low SNR or vessels with low contrast, smaller gamma for images with high SNR or vessels with high contrast | 5.0 |
| sigma_1 | numeric | the kernel size of the Frangi filter; Should be set according to expected vessel diameter | 1.0 |
| cutoff_method_1 | categorical | thresholding method for secondary foreground-background separation (threshold_otsu, threshold_triangle, threshold_li) | threshold_triangle |
| core_vessel_2 | binary | if 1, a secondary Frangi filter is applied | 1 |
| gamma_2 | numeric | the sensitivity of the secondary Frangi filter to tubular shapes | 5.0 |
| sigma_2 | numeric | the kernel size of the secondary Frangi filter; should be set according to expected vessel diameter | 1.0 |
| cutoff_method_2 | categorical | same as cutoff_method_1 | threshold_triangle |
| post_closing | integer | voxel size of cube structuring element of closing operation | 5 |
| post_thinning | binary | indicator to perform topology-preserving-thinning | 1 |
| thin | integer | the number of voxels to be removed from the outer boundary of vessels toward the centerline | 1 |
| min_thickness | integer | half of minimum thickness to maintain under thinning | 1 |
| post_cleaning | integer | minimum voxel size of vessel object | 100 |
| Graph analysis | |||
| pixel_dimensions | string “z,y,x” | voxel size in μm specified according to image recording conditions | “1.0,1.0,1.0” |
| pruning_scale | numeric | all branches whose length is shorter than B·s are pruned, where B is half the vessel thickness at the branching point and s is the pruning_scale factor26 (see Figure S4C) | 1.5 |
| length_limit | numeric | all branches shorter than the specified length in μm will be removed | 3.0a |
| diameter_scale | numeric | all branches whose length is shorter than their diameter multiplied by this factor are removed | 2.0a |
| branching_threshold | percentage | percentage of segment length to use for vectorization of branches for branching angle calculation (see Figure S4E) | 0.25a |
| remove_border_end_pts | binary | if 1, endpoints located at the image border are excluded from quantification | 1a |
|
remove_end_pts_ from_small_filaments |
binary | if 1, endpoints from networks with less than five vessel segments are excluded from quantification | 1a |
| seg_interpolate | binary | if 1, segment interpolation (smoothing) is performed13 | 0a |
| spline_degree | integer | B-spline degree for smoothing | 3a |
| cut_neighbor_brpt_segs | binary | if 0, segments below the defined length limit that are connected through two branch points will not be removed | 1a |
Parameters that remained constant throughout all experiments throughout the paper in their default values.
Benchmarking and validation
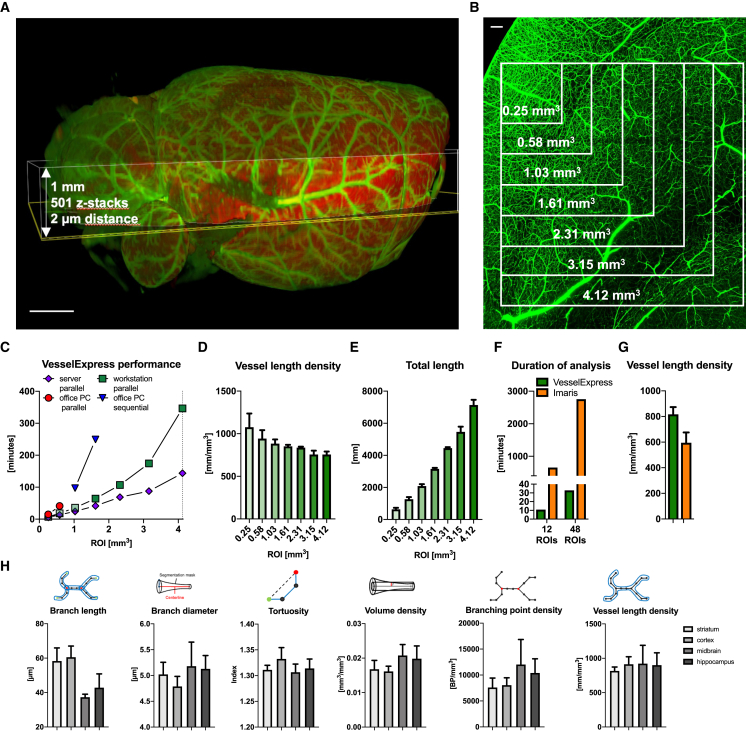
We generated high-resolution LSFM datasets of mouse brains with their blood vessels labeled by injection of FITC-albumin (Figures 2A and 2B) to benchmark VesselExpress against Imaris as the de facto industry standard tool for 3D vessel analysis. When these datasets were analyzed using VesselExpress on a workstation or multi-CPU (central processing unit) server with large amounts of RAM (random access memory), we observed a linear increase in computing time that was proportional to the region of interest (ROI) volume (Figure 2C). In example images from mouse brains, ROIs with a size of up to 0.25 mm3, i.e., 250 MB’s of 16-bit intensity images, could be analyzed in parallel on an office PC. Unlike other comparable analysis methods, the performance of VesselExpress benefits from the availability of RAM and the number of CPU cores, further supported by integration of Dask27 to overcome main memory limitations. Interestingly, the vessel length density (Figure 2D) and total length (Figure 2E) versus ROI volume showed only a slight deviation from constancy or linear growth respectively, with larger ROIs likely representing the highly heterogeneous capillary density in murine brains that becomes obvious at deeper regions of, e.g., the cortex.4 A head-to-head comparison of 12 or 48 different ROIs of the same brain regions showed that VesselExpress is about 70 or 95 times faster, respectively, compared to our reference system based on Imaris (Figure 2F). This is due to the capability for parallelized batch processing of arbitrarily large ROI sets in VesselExpress, whereas Imaris requires each ROI to be annotated manually and separately. Nevertheless, the quantifications obtained from both approaches differed only insignificantly (Figure 2G). We benchmarked the scalability of VesselExpress by investigating run times on an increasing number of 508 × 508 × 1,000 μm ROIs. Our experiments involved 1,024 ROIs on a workstation and high-performance computing hardware. Figure S1A confirms that VesselExpress scales linearly with respect to input size and concurrency.
Figure 2.
Analysis of FITC-albumin labeled brain vessels using automated pipeline VesselExpress
(A) Overview of brain vasculature in a mouse brain scanned with LSFM. Highlighted area represents region in which detail scans for vessel analyses in (C–G) were performed.
(B) Maximum intensity projection of detailed image from LSFM scan using 6.64× magnification and a step size of 2 μm. Inlets showing the region of interest used for vascular analysis in (C–E).
(C) Duration of vascular analysis in VesselExpress using four ROIs each in parallel with increasing image sizes. Analysis was performed on different devices. Using an office PC containing an Intel Core Duo i9 processor with 2.40 GHz, 8 cores, and 32 GB RAM, parallel analysis was possible with ROIs of a maximum size of 0.58 mm3 (red circles). Using one ROI per run, it was possible to analyze a ROI with 2.31 mm3, and total run times were summed up (blue triangle).
(D and E) Green squares represent a workstation equipped with an Intel Xeon W-1255 CPU with 3.30 GHz and 512 GB RAM, whereas violet diamonds display runtime of the very same ROIs on a server working with an Intel Xeon E7-8890 v4 CPU with 2.20GHz, 96 cores, and 1.97 TB RAM. Results from this analysis are displayed as vessel length density (D) and total vessel length (E).
(F and G) Duration (F) and vessel length density (G) from the analysis of either 12 ROIs or 48 ROIs with an image size of 0.25 mm3 using the automated pipeline VesselExpress or sequentially analysis using Imaris software (Bitplane, Oxford Instruments).
(H) Analysis of FITC-albumin-labeled vascular structure performed in 4–12 ROIs of striatum, cortex, midbrain, hippocampus, and corpus callosum of brains from up to six male mice using VesselExpress. Data in (D), (E), (G), and (H) are means ± standard deviation values. Scale bars represent 1 mm in (A) and 100 μm in (B).
We investigated the validity of VesselExpress outputs along several lines. The first evidence of the validity was provided by the insignificant deviation between VesselExpress and the Imaris-based workflow (Figure 2G). Since Imaris requires inspection and adjustment of intermediate results by the user, it can be considered a gold standard reference. The agreement between the semi-automated Imaris workflow and our highly automated VesselExpress was also supported by detailed visual overlays of specific branching points (Figure S1B). As further validation, we generated synthetic data that model blood vessels with known lengths and diameters as ground truth. As shown in Table S2, vessel lengths determined by VesselExpress deviated by less than 2%, while the average vessel diameter deviation was below 15%. Furthermore, we confirmed the robustness of VesselExpress in ROIs that involve widely varying vessel diameters (Figure S2C). We analyzed six typically used vesselness features provided by VesselExpress in different regions of the brain (Figure 2H) and found distinct values for every brain region that are well in line with published data.4,6 We observed that obtaining vessel diameters in line with published literature crucially relied on adequate image segmentation as the first analysis step. Our cascaded Frangi-filter approach yielded vessel diameters that very well matched with data from histological analyses, summing up to 1%–2% of fractional brain volume density (Figure 2H).28
Comparing VesselExpress with the semi-automated Imaris-based segmentation and the recently published VesselVio13 using the segmented data from VesselExpress produced highly similar results for the synthetic data (Table S2), whereas vessel length density in real brain data was ∼18% higher in VesselExpress than in VesselVio and Imaris (Figure S1C). This effect disappeared after vessel smoothing, which introduces an artificial shortening of vessel lengths, so we refrained from using it in further analyses. Smoothing is, however, implemented as an optional choice in VesselExpress. Importantly, VesselVio lacks the crucial step of image segmentation, so the comparison required input images that were pre-segmented using VesselExpress. Furthermore, compared to VesselVio, VesselExpress supports processing along a fully automated workflow (Figure S1D) and performs significantly faster. VesSAP in comparison with VesselExpress yields much more plausible vessel lengths and branching point counts (Figure S1E).
Obese mice show defects in brain vascularization
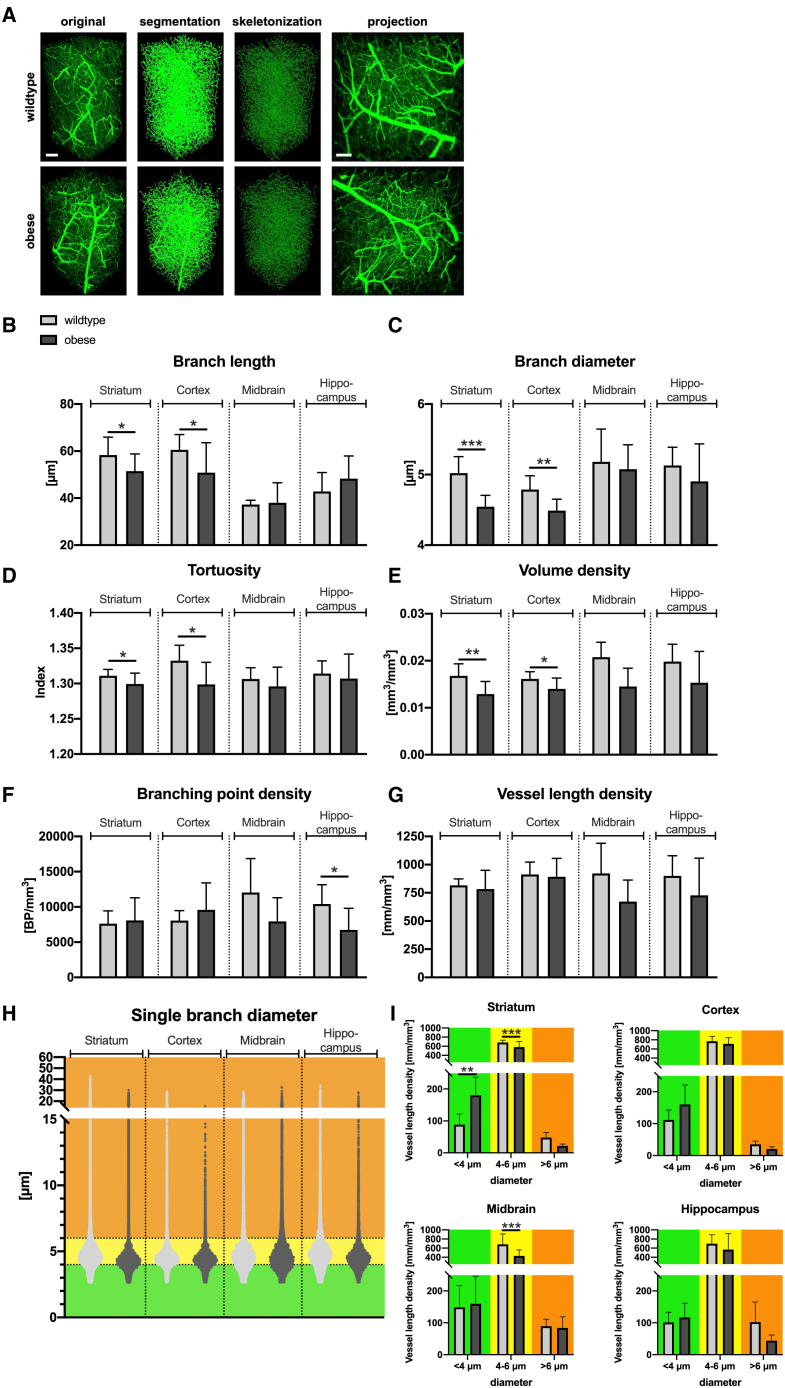
To demonstrate that VesselExpress enables automated analysis of datasets addressing a specific scientific question, we fluorescently labeled vessels in male 9- to 12-week-old C57BL/6 wild-type or homozygously leptin-deficient (ob/ob) mice of the same age by perfusion with FITC-albumin-containing hydrogel. A point mutation of the leptin gene in these animals leads to the development of hyperlipidemia and associated comorbidities such as hyperglycemia, hyperinsulinemia, and infertility.29 We then evaluated ROIs at different rostrocaudal levels of the brain including striatum, cortex, midbrain, and hippocampus using VesselExpress (Figure 3). Our structural data revealed a substantial impairment of microvascular network integrity in different brain regions of ob/ob compared to wild-type mice (Figures 3A–3G). In striatum and cortex of obese mice, we observed significantly lower branch lengths, branch diameters, and tortuosity indices resulting in lower vessel volumes compared to wild-type mice, while the total vessel length density was indistinguishable at all sites. A more detailed analysis of vessel length density in relation to small (<4 μm), medium (4–6 μm), and large diameters (>6 μm; Figure 3H) revealed that obese mice have more thin vessels (<4 μm) but fewer vessels larger than 4 μm (Figure 3I) in the striatum. Hence, LSFM followed by automated VesselExpress analysis can uncover subtle vascular changes in the brain associated with a metabolic disease. Interestingly, mice with a defect in a related protein, the leptin receptor (db/db), which are characterized by obesity and diabetes, were recently shown to reveal a biphasic vascular development with a juvenile hypovascularization followed by aberrant hypervascularization in later adulthood.30 To the best of our knowledge, the consequences of leptin deficiency for brain microvascular networks had not been studied by LSFM. Obesity and diabetes evoke multifaceted vascular changes in the brain.31 Obesity triggered by a high-fat diet induces a significant increase in vascular density, especially in the hippocampus under physiological conditions as assessed by LSFM,32 but compromises cerebral microvascular proliferation and remodeling post-ischemia.33 Importantly, it has been shown in humans that obesity negatively influences brain perfusion.34 Hence, LSFM data in mice appear to show the microvascular correlate of this finding.
Figure 3.
Vascular changes in brains of wild-type compared to obese mice
(A) Representative images of striatal brain regions showing a 3D view of original data, segmented pictures, and skeletonized images provided by VesselExpress as well as a maximum projection of stacked image (right).
(B–G) VesselExpress analysis of brain vessels reveals significant decrease in branch length (B), branch diameter (C), tortuosity (D), and branch volume density (E) in the striatum and cortex in obese mice compared to age-matched wild-type mice but not regarding branching point density (F) or vessel length density (G).
(H) Dot plot displaying single vessels according to diameter in different brain regions in wild-type (light gray) and obese mice (dark gray). Dashed lines represent thresholds used for the analysis of vessel length density in (I).
(I) Vessel length density in vessels with diameters <4 μm, 4–6 μm, or larger than 6 μm shows increased vessel length density of obese mice in smaller vessels and reduced vessel length density in larger vessels in the striatum.
Data are means ± SD values. ∗p ≤ 0.05, ∗∗p ≤ 0.01, and ∗∗∗p ≤ 0.001 compared with wild type (WT); six WT mice and five obese (ob) mice were used and ROIs from the left or right hemisphere were selected. The number of ROIs used was n = 12 (WT) and n = 10 (ob) in the striatum, n = 12 (WT) and n = 8 (ob) in the cortex, n = 4 (WT) and n = 10 (ob) in the midbrain, and n = 8 (WT) and n = 6 (ob) in the hippocampus; data were analyzed by students t test (in B–G) or two-way ANOVA using Sidaks multiple comparison test in (I). Scale bars represent 100 μm.
VesselExpress can analyze the vasculature of many organs
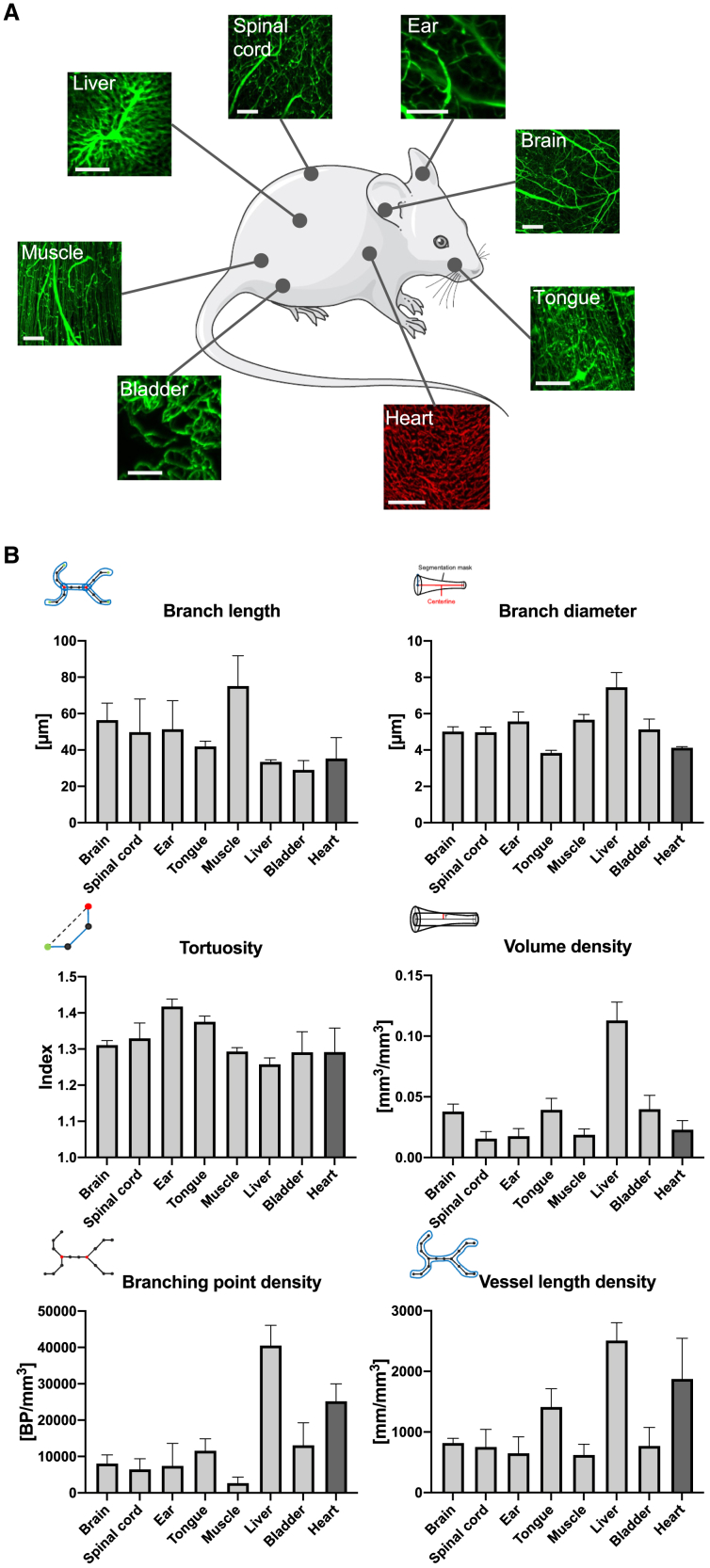
An obvious key requirement for a general-purpose vessel analysis tool is the transferability across different organs. Currently available tools, while certainly extremely powerful for their specific use in brain,4,6 are not able to analyze LSFM datasets of other organs. Hence, we validated VesselExpress for analyzing blood vessels of seven different organs and an additional heart dataset from a different series. We prepared six animals by injection of FITC-albumin, isolated the organs, and generated high-resolution datasets of blood vessels (Figure 4A). We found that VesselExpress could effectively extract complex vessel features from each organ. This analysis showed very similar values for vessel tortuosity, while all other features varied strongly between organs, thereby showing varying degrees of value spreading or homogeneity between samples. The liver turned out to be the most heavily vascularized organ with the highest vessel length and volume density (Figure 4B). Interestingly, close observation of the vessels by diameter revealed a different distribution depending on the organ. Thus, a predominant number of the vessels in the tongue and heart were thinner than in other organs, while vessels of the liver, muscle, and ear were thicker than those in the brain (Figure S2A). The distribution of the length of single vessels is comparable throughout the different organs, although brain and muscle reveal a bigger proportion of long vessels (Figure S2B). This demonstrates that LSFM datasets from various organs are ready to use when applying the provided organ-specific configurations in the pipeline without the need for further modifications. To test if VesselExpress could also be used for studying vascular features in animals exhibiting antibody-mediated labeling of endothelial cells, we finally examined microvascular network characteristics in murine hearts exposed to myocardial ischemia/reperfusion (I/R) injury, to which we intravenously delivered AF647-labeled anti-CD31 antibodies prior to animal sacrifice.2 Our study showed that VesselExpress was able to image microvessels in these samples with results very similar to previously published data2 (Figures 4A, 4B, and S3A,). Furthermore, VesselExpress also analyzed brain vessels labeled simultaneously using FITC-albumin hydrogel as well as AF647-CD31 antibody with very comparable results (Figures S3B and S3C). In addition, VesselExpress is not restricted to LSFM images. As shown in Figure S3D, results from VesselExpress in brain vasculature imaged with confocal microscopy yielded similar results regarding branch length, branch diameter, tortuosity, branch volume, and branching point density.
Figure 4.
VesselExpress is applicable for a variety of organs of mice
A total of six mice were used from which 1–4 ROIs per organ were selected. Mice were perfused with FITC-albumin gelatin hydrogel and indicated organs (A) were removed, dehydrated, and cleared. In a proof of evidence experiment, the heart was labeled separately by injection of AlexaFluor-647-labeled anti-CD31 into additional mice. LSFM was performed as described before, and vascular parameters including (B) branch length, branch diameter, tortuosity, branch volume density, branching point density, and vessel length density were calculated.
Scale bars represent 100 μm. The numbers of ROIs used from at least three different mice were n = 6 for brain, n = 6 for spinal cord, n = 8 for ear, n = 9 for tongue, n = 6 for muscle, n = 5 for liver, and n = 10 for bladder. In heart, we used an n = 3 using the very same ROIs already analyzed in Merz et al., 2019.2 Data are means ± SD values. ∗p ≤ 0.05, ∗∗p ≤ 0.01, and ∗∗∗p ≤ 0.001 analyzed by two-way ANOVA using Tukey’s multiple comparison test.
Discussion
With the development of advanced 3D imaging methods and LSFM in particular, there is a high demand for automated analysis of large datasets. Existing software is able to analyze individual 2D or 3D images,1,35 but currently the analysis especially of 3D data is disproportionately time-consuming and requires frequent and extensive hands-on time. A major bottleneck in existing solutions is the first and essential processing step of segmentation, which critically affects analysis outcome and thus often involves hands-on intervention. Our implementation of VesselExpress as an automated end-to-end processing pipeline facilitates a paradigm shift in the application of LSFM for comprehensive measurement campaigns since the analysis of the ensuing data is removed as a bottleneck. The workflow-based approach systematically automates data analysis, follows high-throughput paradigms of high-content image analysis approaches, and is both installable and executable by microscopists as users. Furthermore, the software design reduces the computational time by up to two orders of magnitude, thus making higher throughput studies and analyses possible on broadly available commodity hardware. With these considerations in mind, VesselExpress represents a fully automated analysis method for LSFM datasets with low barrier to entry. It is easily accessible via the provided Docker image operated by a simple command line or via an integrated web interface with few required configuration steps and does not need technically sophisticated hardware, specific operating systems, or advanced computer skills compared to other methods but produces comparable results.4,6 For example, analysis time for an ROIs with a volume of 0.25 mm3 is reduced to 3 min of computation time compared to an average of 54 min using the established industry standard. As we demonstrated, parallel processing capabilities of VesselExpress allows thousands of images to be processed in batches on high-performance computing hardware, facilitating analyses in VesselExpress that are impractical in comparable sequential processing pipelines.4,13
Automated data analysis pipelines such as VesselExpress almost inevitably involve parameters, which naturally raises the question of parameter robustness against data heterogeneity, including the degree of noise or artifacts. The broad range of tissues examined in our study allows practical conclusions for future studies: while a manageable number of parameters needs to be adjusted for each tissue type, parameter settings can remain constant across large numbers of measurements for the same type of tissue. This insight is crucial for utilizing automated vasculature analysis in the context of using LSFM as a screening tool. In this context, our publicly available data may serve as a reference and benchmark for future studies.
A downside of existing microvascular segmentation solutions is that calculated vessel diameters overestimate true microvessel diameters, resulting in, depending on the mode of image acquisition, image axis and data processing, mean brain capillary diameters >5 μm1 or, even significantly, >10 μm6. In confocal microscopic analyses, the diameter of capillary lumina in the cerebral cortex of C57Bl/6 mice and Wistar rats was shown to be 3.5–4.0 μm and 5.4 ± 1.5 μm, respectively,28,36 whereas that in the rat cerebral cortex was 4.2 ± 1.2 μm in Indian ink-stained corrosion casts.37 As a consequence of vessel diameter overestimations, the majority of existing LSFM studies including our studies refrained from statistically evaluating microvascular diameters.4,32,38,39 Here, we now made particular efforts to obtain reliable vessel diameters using an innovative strategy that combines statistics-based thresholding with up to two customized Frangi filter segmentations. By this approach, we computed mean microvessel diameters in the cerebral cortex of 4.8 ± 0.2 μm, summing up for the whole brain to 1%–2% of fractional volume density, which matches data from conventional histological analyses very well.28 Hence, we can extract comprehensive sets of network characteristics that comprise information on microvessel length, branching, and diameters. With this tool, we for the first time detected statistically significant differences in the vascular make-up of leptin-deficient ob/ob compared with wild-type mice, reflected by an increase of thin (<4 μm) microvessels.
Taken together, our approach enables the transition of LSFM from observational single-case studies to high content analyses that systematically and quantitatively evaluate organs and tissues. Our results demonstrate that LSFM can generate detailed vessel tomograms of multiple organs and thus could form the basis for a novel type of organ map that considers the 3D organization of higher-order structures. Since blood vessel densities and characteristics are important, e.g., for the efficiency of chemotherapy of solid cancers,40 VesselExpress also bears substantial potential for broad applicability.41
Limitations of the study
At present, VesselExpress analyses are optimized for vessels filled with a hydrogel containing a fluorescent dye. Labeling of vessels using endothelial-specific antibodies poses particular challenges, since endothelial labeling results in hollow tubes. In the case of the brain and heart, vascular analysis was similarly possible following endothelial labeling with anti-CD31 antibodies. We, so far, did not evaluate the utility of VesselExpress for studying CD31-labelled vascular networks in tissues with larger caliber vessels. Future studies will examine which pipeline adaptations are required to enable robust network analysis in such tissues. The modular design of VesselExpress would, however, allow it to easily expand the pipeline for required additional components, including the extension to whole-organ vasculature, which typically requires stitching,42 which is currently not supported by VesselExpress.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Alexa Fluor® 647 anti-mouse CD31 Antibody | BioLegend | Cat# 102515; RRID:AB_2161030 |
| Chemicals, peptides, and recombinant proteins | ||
| Albumin–fluorescein isothiocyanate conjugate | Sigma-Aldrich | Cat# A9771 |
| Tetrahydrofuran | Sigma-Aldrich | Cat# 401757 |
| Ethyl cinnamate | Sigma-Aldrich | Cat# 112372 |
| Deposited data | ||
| Raw LSFM, segmented and skeletonized images of all organs | Zenodo | https://doi.org/10.5281/zenodo.6025935 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6JOlaHsd | Envigo | Order code: 057; RRID:MGI:3042249 |
| Mouse: B6.Cg-Lepob/J | Jackson Laboratory | JAX: 000,632; RRID:IMSR_JAX:000632 |
| Software and algorithms | ||
| Python | Python Software Foundation | https://www.python.org/ |
| VesselExpress software | This paper |
https://github.com/RUB-Bioinf/VesselExpress https://zenodo.org/record/7677102 |
| Napari plugin for segmentation parameter tuning | This paper |
https://github.com/MMV-Lab/vessel-express-napari https://zenodo.org/record/7677102 |
| Prism 9.0 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Imaris 9.8.0 | Bitplane, Oxford Instruments | https://imaris.oxinst.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Axel Mosig (axel.mosig@rub.de).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animals
Animal experiments were performed in accordance with the regulations of the National Institute of Health Guidelines for the Care and Use of Laboratory Animals in compliance with ARRIVE guidelines and the permission of local authorities (Landesamt für Natur, Umwelt und Verbraucherschutz (LANUV) which is part of the Ministry for Environment, Agriculture, Conservation and Consumer Protection (MULNV) of the State of North Rhine-Westphalia). Organs of interest were removed in Ketamin/Xylazin anesthesia from 9 to 12 weeks old male C57BL/6J wildtype or leptin deficient (B6.Cg-Lepob/J) mice after perfusion with 40 mL 1 x PBS followed by 40 mL 4% PFA and 10 mL FITC-albumin containing gelatine hydrogel as described before.1 Mice were housed under specific-pathogen-free (SPF) conditions in a 12/12 h light/dark cycle with free access to water and food. For analysis of hearts, mice were subjected to a myocardial (I/R) injury in vivo, as reported.2 For this purpose, the animals’ rib cage was opened through a left lateral thoracotomy via which the left coronary artery (LCA) was ligated. After 45 min of ischemia, reperfusion was allowed for 5 days. For labeling of endothelial cells, these mice received 5 μg Alexa 647-coupled anti-CD31 antibody (Cat No. 102516, BioLegend) i.v. in a total volume of 150 μL PBS 10 min prior to sacrifice by cervical dislocation followed by animal perfusion with PBS.2 All tissues used in this study were dehydrated with increasing concentrations of tetrahydrofluran (THF) for 12 h each and cleared using ethylcinnamate (ECi), as outlined before.1
Method details
Light sheet data acquisition
Cleared organs were imaged using a light sheet Ultra-Microscope Blaze (LaVision BioTec (Miltenyi), Bielefeld, Germany) equipped with a supercontinuum white light laser source for bidirectional light sheet illumination, an sCMOS camera having a 2,048 × 2,048 chip of 6.5 μm pixel size, and three dipping objectives with magnification 1.1× (0.1NA), 4X (0.35NA) and 12X (0.53NA), which could be combined with additional tube lens magnification of 0.6×, 1x, 1.67x or 2.5x. Serial optical imaging was performed by exciting the FITC-albumin, tissue autofluorescence or AF647-anti-CD31 labeled vessels using excitation-emission bandpass filter combinations of 470/30–525/50, 560/40:620/60, and 630/30:680/30 respectively. Overview images were acquired at 1.1× magnification with 10 μm steps in the axial direction. Detailed images of blood vessels were acquired at 6.68× magnification (4× + 1.67X) with 2 μm steps in the axial direction. The highest lightsheet NA was used to give a sheet thickness at the focus of around 5μm. With excitation at 488 nm in ECi the actual sheet length can be estimated from twice the Rayleigh range for a focused Gaussian beam, which would correspond to a length of around 362 μm. Therefore, to obtain optimal axial resolution across the full image width of 1997 μm the dynamic focus function of the instrument was employed. For this the lightsheet focus was sequentially shifted across the image width in steps of approx. 250 μm. With the imaging parameters used the transverse image resolution was actually limited by Nyquist sampling to 1.94μm.
Confocal image acquisition
For confocal microscopy, 1 mm brain sections dehydrated and cleared by the CUBIC method43 were used. For this purpose, PFA-fixed brains from mice were incubated in CUBIC-1 reagent for 5 days with constant shaking while the solution was changed daily resulting in completely clear tissue. Subsequently, the image stacks from cubic cleared brain slices were acquired on a Leica TCS SP8 inverted confocal microscope equipped with a white light laser source combined with an acousto-optic tuneable filter for excitation wavelength selection and spectral detection on hybrid photodetectors via an acousto-optic beamsplitter. For image stack acquisition, an HCPL Fluotor 10X/0.3 air objective was used and the confocal scanner configured to acquire 266 image frames of 512 X 512 pixels (0.994 micron pixel dimension xy) in the z-direction with a z-stepping size of 4 microns. The FITC-albumin hydrogel filled brain vasculature was excited with a wavelength of 488 nm and the fluorescence emission detected from 500 nm to 580 nm.
VesselExpress analysis
Vascular analysis using the automated pipeline VesselExpress was performed in organs derived from the same group of mice perfused with FITC-albumin after transcardial perfusion with PBS and 4% PFA as described above. A detailed description of the Snakemake workflow, segmentation, skeletonization, graph construction and analysis, rendering and validation is described below. For a detailed analysis of vessels, image stacks acquired with 6.64× magnification and a step size of 2 μm in ventrodorsal direction were used. From each of these image stacks, ROIs covering representative vascular structures of each tissue were obtained. In the brain, identification of ROIs in the striatum was performed as described before.1 ROIs in cortex were selected directly adjacent to ROI in striatum at bregma level 0.74 mm. For the identification of ROIs in the hippocampus and midbrain, autofluorescence signal was used in Imaris to visualize the whole brain in 3D. Image dimensions of ROIs in cortex, midbrain and hippocampus were 305 × 305 × 600 μm, i.e. 0.056 mm3. The size of all ROIs is listed in Table S1. Files with a size of up to 3 or 4 GB in 16-bit TIF format were analyzed in parallel on a workstation containing an Intel Xeon W-1255 CPU with 3.30 GHz, 10 cores and 512 GB RAM or on a server equipped with an Intel Xeon CPU E7-8890 v4 with 2.20GHz, 96 cores and 1.97 TB RAM, respectively. Images with a file size of up to 1.5 GB were analyzed in parallel or sequentially on an office PC containing an Intel Core i9 CPU with 2.40 GHz, 8 cores and 32 GB RAM.
Vascular quantification using Imaris
For detailed vascular quantification image stacks acquired with 6.68× magnification and a step size of 2 μm in ventrodorsal direction were also used. From each of these image stacks, ROI covering representative vascular structures of each tissue. In these ROIs, microvasculature was analyzed after network modeling using the Imaris 3D rendering software filament tracer tool. The very same ROIs were used for VesselExpress analysis.
VesselExpress pipeline and validation
Snakemake workflow
VesselExpress consists of multiple modules which are automated in a pipeline with the workflow management system Snakemake.17 All modules of the pipeline are defined by rules in a Snakefile. A rule specifies input, output, environment, and the shell command to execute the corresponding Python script. Each module requires different software packages and contains parameters. The required software packages are defined in a YAML file. The default settings of all parameters are stored in a JSON configuration file which is also specified in the Snakefile. Thus, all parameters can be flexibly adjusted in the configuration file. Due to the modular software design, each processing step can also be executed individually. If preferred, custom functions can easily be integrated into VesselExpress by changing the rules in the Snakefile. All modules can be run by one terminal command (detailed instructions can be found on GitHub). Figure S4A illustrates the workflow for processing a 3D image including all steps.
The workflow is also accessible through a web-browser based graphical user interface (Video S1). The video also provides guidance on how to adjust the workflow parameters.
Segmentation
The segmentation method is extended from classic image segmentation workflows from the Allen Cell and Structure Segmenter,21 which is a 3-step workflow (pre-processing, core segmentation, post-processing) with a minimal number of parameters to tune and minimal number of functions to choose from. In the pre-processing step, we use the edge-preserving smoothing function in Segmenter. In the core segmentation step, we use a combination of a statistics-based thresholding method and customized Frangi filter-based segmentation. The statistical based thresholding method calculates the mean and standard deviation of the intensity of the whole image, denoted by m and std, respectively. The threshold value is set as m + S x std, where S is an empirically determined parameter (usually between 2.0 and 4.0, 3.0 was used for all organs). The customized Frangi filter based segmentation method takes three parameters: “sigma” (i.e., roughly representing the thickness of the vessels), “gamma” (i.e., the sensitivity to structures), and “cutoff method” (i.e., how to binarize the filter output into segmentation, options include Otsu, Li,44 and Triangle45). Different parameters are optimized and used for different organs. Users can either load our pre-defined parameters for different organs or further adjust these parameters within a Napari plugin.
The statistics-based thresholding is mainly used to capture those very thick vessels with much higher intensity than the rest (e.g., main artery), while the rest (e.g., microvessels) will be picked up by the customized Frangi filter based segmentation. In certain organs, the variation of the thickness of different microvessels is too high for one single Frangi filter to segment accurately all vessel calibers (recall that the parameter “sigma” needs to be set according to vessel thickness). To address this issue, we used an innovative strategy employing two different Frangi filters with different parameters (optimized using the Napari plugin) to obtain accurate segmentation of various microvessels. It is worth mentioning that this “2 Frangi filters” method is different from the original multi-scale Frangi filter, where multiple sigma values are used in one single filter. For example, we were able to use one Frangi filter with sigma = 1, gamma = 5, cutoff method = Li and another Frangi filter with sigma = 2, gamma = 100, cutoff method = Otsu. However, using the original multi-scale Frangi filter with two sigma values (e.g., sigma = 1, 2) is only comparable to our “2 Frangi filter” method when both Frangi filters have the same gamma value and the same cutoff method. Without the flexibility of using different gamma values and different cutoff methods in the two Frangi filters, the original multi-scale Frangi filter usually suffers from over-segmentation errors (i.e., a thin vessel segmented much thicker than it should be or two proximal thin vessels falsely segmented as a merged one). The “2 Frangi filters” strategy is the key ingredient for successfully segmenting all vessels with accurate thickness in different tissues.
Before post-processing, a logical OR operation is applied to the output from the statistical based thresholding and the customized Frangi filter-based segmentations to combine the results. At last, the post-processing step ensues with three different functions to choose from, depending on the specific organ and imaging quality. These functions include topology-preserving thinning21 to refine the thickness of the segmentation, morphological closing to connect potentially fragmented vessels and small holes, and a size filter to remove small segmented objects due to noise or imaging artifacts.
Skeletonization and graph construction
The vessels’ centerlines are extracted from the binary mask through the parallel thinning algorithm23 implemented in the scikit-image Python package.20 The centerlines are then transformed into undirected graphs by using the Python 3scan toolkit24 (Figure S4B). The graph construction consists of two main steps: first vertices are created for each image point (Figures S4B1 and S4B2). Vertices are defined by their image coordinates. Neighboring vertices are connected via edges. After this step, the graph typically contains cliques, i.e., subgraphs in which all vertices are connected, resulting in an over quantification of branching points and underdetection of terminal points (vertices with one neighbor). To avoid this, all cliques comprising three vertices are resolved by removing the longest edge (Figure S4B3). The final graph is stored as an adjacency list. For each vertex, there is an adjacency list that contains all neighboring vertices.
The graph obtained from skeletonization may contain spurious branches that do not represent the topology of the object. These occur when the binary image has bumps on the edge of the object. Therefore, these are branches that lie at the edge of the object and thus start from the center of the object in a branching point and end in a terminal point at the border of the object. These branches are identified and removed by the criteria defined by Montero and Lang42,26 found in Equation 1:
| (Equation 1) |
A branch with branching point bp and terminal point tp is removed exactly when the length of a branch is less than or equal to the distance of the branching point Bp to the nearest background point ƒ (Figure S4C). With the scaling factor s, this distance can be scaled arbitrarily. The higher the scaling factor, the more branches are removed (“pruned”). The nearest background point ƒ is determined using the Euclidean distance transformation from the scikit-image Python package.20,26
Graph analysis
The graph is used to extract relevant features of the vascular network. The features can be divided into filament and segment statistics. Each connected component of the graph is identified as one filament, and each direct path between branching or terminal points is considered as a segment. Figure S4D shows a connected graph, which consists of five segments. To identify segments, each connected graph is traversed in a depth-first search (DFS) starting from a randomly chosen terminal point. For each vertex, the number of neighbors is readout from the adjacency list to check if it is a branching or terminal point. Once a branching or terminal point is found, the segment is backtracked to the last terminal or branching point. If a previously visited vertex is found, it is designated a circle. Once a segment is found, all features of the segment are calculated. The features may be calculated of the raw segments or of the interpolated segments. Segments are interpolated by basis splines. The functions for segment interpolation are used from VesselVio. In a post-processing step edge points which are located at the border of an image are excluded from the terminal point quantification since they cannot be considered actual terminal points. Also segments with length or diameter below user-defined thresholds are removed from the analysis.
For each filament, the number of branching and terminal points is counted. In addition, the number of neighbors of each branching point is readout from the adjacency list. The total vessel length is calculated from the sum of all segment lengths. The length of a segment corresponds to the sum of the Euclidean distance of all points from starting point to terminal point e of the segment according to the pixel dimensions :
| (Equation 2) |
Furthermore, the tortuosity is calculated for each segment with the Equation 4. Equation 3 is used to calculate the Euclidean distance between the starting point and the terminal point for each dimension of a segment. VesselExpress outputs the quotient of the Euclidean distance and .
| (Equation 3) |
| (Equation 4) |
In addition, diameter and volume of the vessels are determined. The decisive value is the radius. This is calculated by the Euclidean distance transformation using Equation 5. The distance transformation is carried out on the binary image and replaces the pixel values with the Euclidean distance of the point to the nearest background point considering the dimensions. The diameter results from the average of all radii of the skeleton points of a segment from Equation 6.
| (Equation 5) |
| (Equation 6) |
The vessel volume is calculated by using the vessel’s average radius:
| (Equation 7) |
For segments that are connected to a predecessor via a branching point, branching angles are also determined. The angles are calculated with the standard angle equation for two vectors:
| (Equation 8) |
where corresponds to the considered segment and bicorresponds to the respective adjacent segment of the predecessor . A threshold value specifies the percentage length of the segment, from which the vector of a segment is formed.
Therefore, the value range of the threshold is between 0 and 1. If 0, the first neighboring node and if 1, the last node is selected as an approximation of the vessel orientation (Figure S4E).
Rendering
VesselExpress features an automated Blender25 integration, running on Version 2.83.5. Using the provided Cycles or Eevee rendering engines, VesselExpress can automatically generate high-definition images of the calculated segmentation as well as the skeleton.
This rendering makes full use of the Blender capabilities using the Principled BSDF material for the meshes and raytraced rendering on the GPU (if available). The mesh-, material-, lighting-, and color-properties, as well as camera angles can be freely customized beforehand.
The meshes for rendering are created via the scikit-image implementation of the Marching Cubes Algorithm.46,47 As an optional output, the generated meshes can be saved for further use in form of a.stl,.glb, and.blend file each.
Validation on synthetic ground truth dataset
To evaluate the accuracy against absolute ground truth, we created a synthetic dataset with each image containing six tubes of a certain size and diameter with varying intensities. Random Gaussian distributed noise with a mean of 0 and a standard deviation of 50 was added. The images were generated to replicate the resolution of the original light-sheet microscopic images as closely as possible by convolving the images with a Gaussian filter (sigma 0.85 in xy and 1.7 in z). For each image, we compared the mean length and diameter of the pipeline results (actual value) of VesselExpress or VesselVio with the expected target values. Table S2 shows the results obtained using parameters optimized for the synthetic images using the corresponding VesselExpress Napari plugin. This comparison reveals that lengths are precisely calculated (average deviation of 1.83% or 1.91%) as well as vessel diameters (average deviation of 5.1% or 7.55%) using VesselExpress or VesselVio, respectively. The average deviation is the mean of the absolute deviations of all tube images. Vessel lengths are systematically slightly underestimated, which can be explained by the thinning operation performed during each step of the iterative skeletonization procedure. Since the synthetic vessels are linear and do not end in branching points, one or few pixels may be missed at either end of the vascular-like linear segment, leading to a slight underestimation.
Comparison of VesselExpress to existing tools
We compared VesselExpress to VesselVio, Imaris, ClearMap and VesSAP by 18 features (Table S3). VesSAP and ClearMap are focused on analyzing the vascular network in the whole mouse brain using surface markers whereas VesselExpress is designed for large-scale studies in which the vascular network in specific regions of various organs should be analyzed. VesselExpress is the only tool which provides a fully automated pipeline from segmentation to feature extraction and enables parallel processing of images for high scalability. The other tools focus on parallelization within the single processing steps to make the runtime faster for processing large images sequentially. VesselExpress does not include stitching. However, available stitching tools like BigStitcher or TeraStitcher, which is used by the VesSAP authors, can easily be included into the pipeline due to the modular software design. While VesSAP relies on deep learning-based segmentation, VesselExpress relies on standard image processing with a dual-Frangi approach for segmentation. The model of VesSAP was trained with a large set of synthetic images using transfer-learning. Generating realistic synthetic images for transfer-learning is very difficult to achieve for most scenarios. Therefore, VesselExpress relies on calibrating a relatively small set of parameters for different types of images which can easily be done by the provided Napari plugin. With this approach we have analyzed images from multiple organs compared to others which analyzed images of a single organ.
Quantification and statistical analysis
Data are expressed as mean ± standard deviation (SD). In case of comparisons between 2 groups, two-tailed t tests were used. Details regarding number of samples and mice are provided in the respective figure legends. p values ≤ 0.05 were defined to indicate statistical significance. The statistical details are given in the figure legends. Statistical analyzes were performed using GraphPad Prism version 9.0 software.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (CRC TRR332 project C6 to D.M.H. and M.G.; RU/FOR5427 SP4, EN984/15-1, EN984/16-1, CRC/TR296 P09, CRC/TR332 A3, and CRC/TR332 Z1 to D.R.E.). Parts of the graphical abstract were drawn by using pictures from Servier Medical Art. Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License (https://creativecommons.org/licenses/by/3.0/).
Author contributions
Conceptualization, J.C., M.G., D.M.M., and A.M.; methodology, P.S., N.H., A.S., S.D.K., J.C., M.G., and A.M.; software, P.S., N.F., S.D.K., D.S., L.S., J.C., and A.M.; validation, P.S., A.S., J.C., M.G., D.M.M., and A.M., formal analysis, P.S., N.H., A.S., J.C., M.G., D.M.M., and A.M.; investigation, P.S., N.H., A.S., S.D.K., Y.Q., A.M.Y., J.W., L.K., S.K., Z.C., A.A.T., V.S., A.A.T., L.M.N.M., I.H., and J.C.; resources, P.S., A.S., S.D.K., Y.Q., A.M.Y., J.W., A.G., M.T., D.R.E., P.L., L.M.N.M., I.H., M.G., and D.M.M.; data curation, P.S., N.H., A.S., S.D.K., and D.S.; visualization, P.S., N.H., N.F., S.D.K., D.S., and J.C.; supervision, J.C., M.G., D.M.M., and A.M.; writing – original draft, P.S., N.H., J.C., M.G., D.M.M., and A.M., writing – review & editing, P.S., N.H., A.S., S.D.K., V.S., J.C., M.G., D.M.M., and A.M.
Declaration of interests
The authors declare no competing interests.
Published: March 17, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2023.100436.
Contributor Information
Matthias Gunzer, Email: matthias.gunzer@uk-essen.de.
Dirk M. Hermann, Email: dirk.hermann@uk-essen.de.
Axel Mosig, Email: axel.mosig@rub.de.
Supplemental information
Data and code availability
-
•
All data have been deposited at https://zenodo.org (Zenodo: https://doi.org/10.5281/zenodo.6025935) and are publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
All original code has been deposited at https://zenodo.org (Zenodo: https://doi.org/10.5281/zenodo.7677102) and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
References
- 1.Lugo-Hernandez E., Squire A., Hagemann N., Brenzel A., Sardari M., Schlechter J., Sanchez-Mendoza E.H., Gunzer M., Faissner A., Hermann D.M. 3D visualization and quantification of microvessels in the whole ischemic mouse brain using solvent-based clearing and light sheet microscopy. J. Cerebr. Blood Flow Metabol. 2017;37:3355–3367. doi: 10.1177/0271678X17698970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merz S.F., Korste S., Bornemann L., Michel L., Stock P., Squire A., Soun C., Engel D.R., Detzer J., Lörchner H., et al. Contemporaneous 3D characterization of acute and chronic myocardial I/R injury and response. Nat. Commun. 2019;10:2312. doi: 10.1038/s41467-019-10338-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong S.M., Jung D., Kiemen A., Gaida M.M., Yoshizawa T., Braxton A.M., Noë M., Lionheart G., Oshima K., Thompson E.D., et al. Three-dimensional visualization of cleared human pancreas cancer reveals that sustained epithelial-to-mesenchymal transition is not required for venous invasion. Mod. Pathol. 2020;33:639–647. doi: 10.1038/s41379-019-0409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirst C., Skriabine S., Vieites-Prado A., Topilko T., Bertin P., Gerschenfeld G., Verny F., Topilko P., Michalski N., Tessier-Lavigne M., Renier N. Mapping the fine-scale organization and plasticity of the brain vasculature. Cell. 2020;180:780–795.e25. doi: 10.1016/j.cell.2020.01.028. [DOI] [PubMed] [Google Scholar]
- 5.Klingberg A., Hasenberg A., Ludwig-Portugall I., Medyukhina A., Männ L., Brenzel A., Engel D.R., Figge M.T., Kurts C., Gunzer M. Fully automated evaluation of total glomerular number and capillary tuft size in nephritic kidneys using lightsheet microscopy. J. Am. Soc. Nephrol. 2017;28:452–459. doi: 10.1681/ASN.2016020232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Todorov M.I., Paetzold J.C., Schoppe O., Tetteh G., Shit S., Efremov V., Todorov-Völgyi K., Düring M., Dichgans M., Piraud M., et al. Machine learning analysis of whole mouse brain vasculature. Nat. Methods. 2020;17:442–449. doi: 10.1038/s41592-020-0792-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zundler S., Klingberg A., Schillinger D., Fischer S., Neufert C., Atreya I., Gunzer M., Neurath M.F. Three-Dimensional cross-sectional light-sheet microscopy imaging of the inflamed mouse gut. Gastroenterology. 2017;153:898–900. doi: 10.1053/j.gastro.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 8.Mohamud Yusuf A., Hagemann N., Schulten S., Rausch O., Wagner K., Hussner T., Qi Y., Totzeck M., Kleinschnitz C., Squire A., et al. Light sheet microscopy using FITC-albumin followed by immunohistochemistry of the same rehydrated brains reveals ischemic brain injury and early microvascular remodeling. Front. Cell. Neurosci. 2020;14:625513. doi: 10.3389/fncel.2020.625513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molbay M., Kolabas Z.I., Todorov M.I., Ohn T.L., Ertürk A. A guidebook for DISCO tissue clearing. Mol. Syst. Biol. 2021;17:e9807. doi: 10.15252/msb.20209807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ueda H.R., Ertürk A., Chung K., Gradinaru V., Chédotal A., Tomancak P., Keller P.J. Tissue clearing and its applications in neuroscience. Nat. Rev. Neurosci. 2020;21:61–79. doi: 10.1038/s41583-019-0250-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grüneboom A., Kling L., Christiansen S., Mill L., Maier A., Engelke K., Quick H.H., Schett G., Gunzer M. Next-generation imaging of the skeletal system and its blood supply. Nat. Rev. Rheumatol. 2019;15:533–549. doi: 10.1038/s41584-019-0274-y. [DOI] [PubMed] [Google Scholar]
- 12.Richardson D.S., Lichtman J.W. Clarifying tissue clearing. Cell. 2015;162:246–257. doi: 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bumgarner J.R., Nelson R.J. Open-source analysis and visualization of segmented vasculature datasets with VesselVio. Cell Rep. Methods. 2022;2:100189. doi: 10.1016/j.crmeth.2022.100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hägerling R., Drees D., Scherzinger A., Dierkes C., Martin-Almedina S., Butz S., Gordon K., Schäfers M., Hinrichs K., Ostergaard P., et al. VIPAR, a quantitative approach to 3D histopathology applied to lymphatic malformations. JCI Insight. 2017;2:e93424. doi: 10.1172/jci.insight.93424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirschnick N., Drees D., Redder E., Erapaneedi R., Pereira da Graca A., Schäfers M., Jiang X., Kiefer F. Rapid methods for the evaluation of fluorescent reporters in tissue clearing and the segmentation of large vascular structures. iScience. 2021;24:102650. doi: 10.1016/j.isci.2021.102650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drees D., Eilers F., Jiang X. Hierarchical random walker segmentation for large volumetric biomedical images. IEEE Trans. Image Process. 2022;31:4431–4446. doi: 10.1109/TIP.2022.3185551. [DOI] [PubMed] [Google Scholar]
- 17.Köster J., Rahmann S. Snakemake-a scalable bioinformatics workflow engine. Bioinformatics. 2018;34:3600. doi: 10.1093/bioinformatics/bty350. [DOI] [PubMed] [Google Scholar]
- 18.Tran T.C., Sneed B., Haider J., Blavo D., White A., Aiyejorun T., Baranowski T.C., Rubinstein A.L., Doan T.N., Dingledine R., Sandberg E.M. Automated, quantitative screening assay for antiangiogenic compounds using transgenic zebrafish. Cancer Res. 2007;67:11386–11392. doi: 10.1158/0008-5472.CAN-07-3126. [DOI] [PubMed] [Google Scholar]
- 19.Merkel D. Docker: lightweight linux containers for consistent development and deployment. Linux J. 2014;239:2. [Google Scholar]
- 20.van der Walt S., Schönberger J.L., Nunez-Iglesias J., Boulogne F., Warner J.D., Yager N., Gouillart E., Yu T., scikit-image contributors scikit-image: image processing in Python. PeerJ. 2014;2:e453. doi: 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J., Ding L., Viana M.P., Hendershott M.C., Yang R., Mueller I.A., Rafelski S.M. The Allen Cell Structure Segmenter: a new open source toolkit for segmenting 3D intracellular structures in fluorescence microscopy images. bioRxiv. 2018 doi: 10.1101/491035. Preprint at. [DOI] [Google Scholar]
- 22.Nicholas Sofroniew T.L., Evans K., Nunez-Iglesias J., Bokota G., Peña-Castellanos G., Winston P., Yamauchi K., Bussonnier M., Doncila Pop D., Liu Z., et al. 2021. napari/napari: 0.4.12rc2. Preprint at. [DOI] [Google Scholar]
- 23.Lee T.C., Kashyap R.L., Chu C.N. Building skeleton models via 3-D medial surface axis thinning algorithms. CVGIP Graph. Models Image Process. 1994;56:462–478. doi: 10.1006/cgip.1994.1042. [DOI] [Google Scholar]
- 24.Vemuri P., Pesavento M. 3D Image Skeletonization Tools. 2019. https://github.com/pranathivemuri/skeletonization
- 25.Blender Online Community Blender - a 3D modelling and rendering package. 2018. https://www.blender.org
- 26.Solís Montero A., Lang J. Skeleton pruning by contour approximation and the integer medial axis transform. Comput. Graph. 2012;36:477–487. [Google Scholar]
- 27.Dask . 2016. Dask: Library for Dynamic Task Scheduling.https://github.com/dask [Google Scholar]
- 28.Tsai P.S., Kaufhold J.P., Blinder P., Friedman B., Drew P.J., Karten H.J., Lyden P.D., Kleinfeld D. Correlations of neuronal and microvascular densities in murine cortex revealed by direct counting and colocalization of nuclei and vessels. J. Neurosci. 2009;29:14553–14570. doi: 10.1523/JNEUROSCI.3287-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindström P. The physiology of obese-hyperglycemic mice [ob/ob mice] Sci. World J. 2007;7:666–685. doi: 10.1100/tsw.2007.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y., Chen D., Smith A., Ye Q., Gao Y., Zhang W. Three-dimensional remodeling of functional cerebrovascular architecture and gliovascular unit in leptin receptor-deficient mice. J. Cerebr. Blood Flow Metabol. 2021;41:1547–1562. doi: 10.1177/0271678X211006596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hermann D.M., Kleinschnitz C. Modeling vascular risk factors for the development of ischemic stroke therapies. Stroke. 2019;50:1310–1317. doi: 10.1161/STROKEAHA.118.024673. [DOI] [PubMed] [Google Scholar]
- 32.Gruber T., Pan C., Contreras R.E., Wiedemann T., Morgan D.A., Skowronski A.A., Lefort S., De Bernardis Murat C., Le Thuc O., Legutko B., et al. Obesity-associated hyperleptinemia alters the gliovascular interface of the hypothalamus to promote hypertension. Cell Metabol. 2021;33:1155–1170.e10. doi: 10.1016/j.cmet.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zechariah A., ElAli A., Hagemann N., Jin F., Doeppner T.R., Helfrich I., Mies G., Hermann D.M. Hyperlipidemia attenuates vascular endothelial growth factor-induced angiogenesis, impairs cerebral blood flow, and disturbs stroke recovery via decreased pericyte coverage of brain endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2013;33:1561–1567. doi: 10.1161/ATVBAHA.112.300749. [DOI] [PubMed] [Google Scholar]
- 34.Amen D.G., Wu J., George N., Newberg A. Patterns of regional cerebral blood flow as a function of obesity in adults. J. Alzheimers Dis. 2020;77:1331–1337. doi: 10.3233/JAD-200655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rust R., Kirabali T., Grönnert L., Dogancay B., Limasale Y.D.P., Meinhardt A., Werner C., Laviña B., Kulic L., Nitsch R.M., et al. A practical guide to the automated analysis of vascular growth, maturation and injury in the brain. Front. Neurosci. 2020;14:244. doi: 10.3389/fnins.2020.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z.G., Zhang L., Tsang W., Soltanian-Zadeh H., Morris D., Zhang R., Goussev A., Powers C., Yeich T., Chopp M. Correlation of VEGF and angiopoietin expression with disruption of blood-brain barrier and angiogenesis after focal cerebral ischemia. J. Cerebr. Blood Flow Metabol. 2002;22:379–392. doi: 10.1097/00004647-200204000-00002. [DOI] [PubMed] [Google Scholar]
- 37.Miyoshi Y., Date I., Ohmoto T. Three-dimensional morphological study of microvascular regeneration in cavity wall of the rat cerebral cortex using the scanning electron microscope: implications for delayed neural grafting into brain cavities. Exp. Neurol. 1995;131:69–82. doi: 10.1016/0014-4886(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 38.Di Giovanna A.P., Tibo A., Silvestri L., Müllenbroich M.C., Costantini I., Allegra Mascaro A.L., Sacconi L., Frasconi P., Pavone F.S. Whole-brain vasculature reconstruction at the single capillary level. Sci. Rep. 2018;8:12573. doi: 10.1038/s41598-018-30533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gregorius J., Wang C., Stambouli O., Hussner T., Qi Y., Tertel T., Börger V., Mohamud Yusuf A., Hagemann N., Yin D., et al. Small extracellular vesicles obtained from hypoxic mesenchymal stromal cells have unique characteristics that promote cerebral angiogenesis, brain remodeling and neurological recovery after focal cerebral ischemia in mice. Basic Res. Cardiol. 2021;116:40. doi: 10.1007/s00395-021-00881-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takagi S., Inenaga R., Oya R., Nakamura S., Ikemura K. Blood vessel density correlates with the effects of targeted intra-arterial carboplatin infusion with concurrent radiotherapy for squamous cell carcinomas of the oral cavity and oropharynx. Br. J. Cancer. 2006;94:1580–1585. doi: 10.1038/sj.bjc.6603138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merz S.F., Jansen P., Ulankiewicz R., Bornemann L., Schimming T., Griewank K., Cibir Z., Kraus A., Stoffels I., Aspelmeier T., et al. High-resolution three-dimensional imaging for precise staging in melanoma. Eur. J. Cancer. 2021;159:182–193. doi: 10.1016/j.ejca.2021.09.026. [DOI] [PubMed] [Google Scholar]
- 42.Hörl D., Rojas Rusak F., Preusser F., Tillberg P., Randel N., Chhetri R.K., Cardona A., Keller P.J., Harz H., Leonhardt H., et al. BigStitcher: reconstructing high-resolution image datasets of cleared and expanded samples. Nat. Methods. 2019;16:870–874. doi: 10.1038/s41592-019-0501-0. [DOI] [PubMed] [Google Scholar]
- 43.Susaki E.A., Tainaka K., Perrin D., Yukinaga H., Kuno A., Ueda H.R. Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat. Protoc. 2015;10:1709–1727. doi: 10.1038/nprot.2015.085. [DOI] [PubMed] [Google Scholar]
- 44.Li C.H., Tam P.K.S. An iterative algorithm for minimum cross entropy thresholding. Pattern Recogn. Lett. 1998;19:771–776. doi: 10.1016/s0167-8655(98)00057-9. [DOI] [Google Scholar]
- 45.Zack G.W., Rogers W.E., Latt S.A. Automatic measurement of sister chromatid exchange frequency. J. Histochem. Cytochem. 1977;25:741–753. doi: 10.1177/25.7.70454. [DOI] [PubMed] [Google Scholar]
- 46.Lewiner T., Lopes H., Vieira A.W., Tavares G. Efficient implementation of marching cubes' cases with topological guarantees. J. Graph. Tool. 2012;8:1–15. doi: 10.1080/10867651.2003.10487582. [DOI] [Google Scholar]
- 47.Lorensen W.E., Cline H.E. Marching cubes: a high resolution 3D surface construction algorithm. SIGGRAPH Comput. Graph. 1987;21:163–169. doi: 10.1145/37402.37422. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data have been deposited at https://zenodo.org (Zenodo: https://doi.org/10.5281/zenodo.6025935) and are publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
All original code has been deposited at https://zenodo.org (Zenodo: https://doi.org/10.5281/zenodo.7677102) and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.