Abstract
RNA-based therapeutics have shown great promise in treating a broad spectrum of diseases through various mechanisms including knockdown of pathological genes, expression of therapeutic proteins, and programmed gene editing. Due to the inherent instability and negative-charges of RNA molecules, RNA-based therapeutics can make the most use of delivery systems to overcome biological barriers and to release the RNA payload into the cytosol. Among different types of delivery systems, lipid-based RNA delivery systems, particularly lipid nanoparticles (LNPs), have been extensively studied due to their unique properties, such as simple chemical synthesis of lipid components, scalable manufacturing processes of LNPs, and wide packaging capability. LNPs represent the most widely used delivery systems for RNA-based therapeutics, as evidenced by the clinical approvals of three LNP-RNA formulations, patisiran, BNT162b2, and mRNA-1273. This review covers recent advances of lipids, lipid derivatives, and lipid-derived macromolecules used in RNA delivery over the past several decades. We focus mainly on their chemical structures, synthetic routes, characterization, formulation methods, and structure–activity relationships. We also briefly describe the current status of representative preclinical studies and clinical trials and highlight future opportunities and challenges.
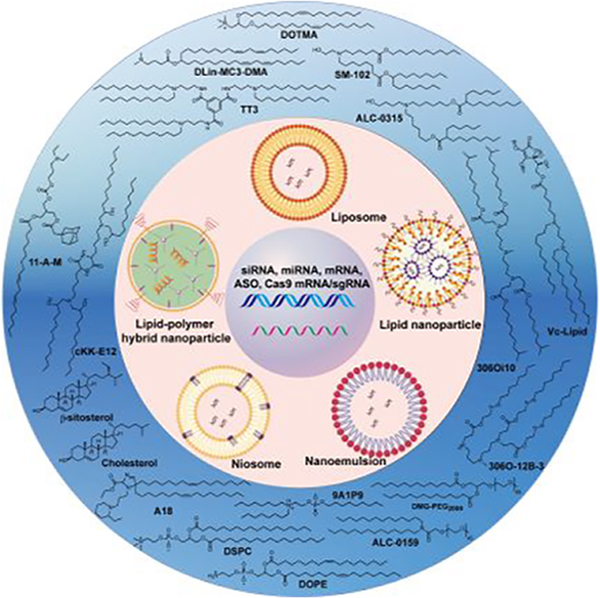
Graphical Abstract
1. INTRODUCTION
1.1. RNA-Based Therapeutics
RNA-based therapeutics have gained extensive interest in treating diverse diseases including those associated with “undruggable” targets.1–3 In the past decade, a series of RNA-based therapies have been approved for therapeutic applications in different types of diseases such as macular degeneration, spinal muscular atrophy, hypercholesterolemia, and TTR-mediated amyloidosis.4–6 Table 1 lists representative examples of RNA-based therapeutics for clinical use. Most recently, two formulations of lipid nanoparticles encapsulating mRNA, BNT162b2 and mRNA-1273, have obtained emergency use authorizations (EUA) from the FDA and EMA as SARS-CoV-2 vaccines for the prevention of coronavirus disease 2019 (COVID-19).7–9 RNA molecules mainly include antisense oligonucleotides (ASOs), small interfering RNA (siRNA), microRNA (miRNA), and mRNA (mRNA).
Table 1.
Representative RNA-Based Therapeutics Approved for Clinical Use
| RNA-based therapeutic products | Approval year | Therapeutic Indication |
|---|---|---|
| ASO | ||
| Mipomersen | 2013 | Familial hypercholesterolemia |
| Eteplirsen | 2016 | Duchenne muscular dystrophy |
| Nusinersen | 2016 | Spinal muscular atrophy |
| Inotersen | 2018 | Hereditary transthyretin amyloidosis |
| Golodirsen | 2019 | Duchenne muscular dystrophy |
| Volanesorsen | 2019 | Familial chylomicronaemia syndrome |
| siRNA | ||
| Patisiran | 2018 | Hereditary transthyretin amyloidosis |
| Givosiran | 2019 | Acute hepatic porphyria |
| Lumasiran | 2020 | Primary Hyperoxaluria Type 1 |
| Inclisiran | 2020 | Hypercholesterolemia or mixed dyslipidemia |
| mRNA | ||
| BNT162b2 | 2020 | COVID-19 vaccine |
| mRNA-1273 | 2020 | COVID-19 vaccine |
Short antisense oligonucleotides (ASOs) consist of single antisense stranded DNAs or RNAs with a sequence length of 8–50 nucleotides that can specifically bind to their target mRNA through complementary base pairing, leading to the degradation of mRNA by endogenous cellular RNase H10,11 or a functional blockade of mRNAs through steric effects.12,13 Chemical modification of ASOs, such as phosphorothioate ASOs, can increase the stability of ASOs and facilitate their interactions with targeted cells.14,15
In 1998, Fire et al. discovered the RNA interference (RNAi) pathway,16 which involves the formation of an RNA-induced silencing complex (RISC) in cell cytosol and subsequent decay of the target mRNA. These important findings led to the emergence of RNAi as a new type of RNA-based therapeutics.17 Small interfering RNA (siRNA) and microRNA (miRNA) are two major types of RNA molecules for RNA interference. siRNA, one of the most important classes of RNAi therapeutics, is typically a double-stranded RNA (dsRNA) molecule that consists of less than 30 base pairs. siRNA-based therapeutics have been investigated as potential therapies for diseases caused by abnormal expression or mutation such as cancers,18–21 viral infections,22,23 and genetic disorders.24 Following Onpattro (patisiran), the first approved RNAi therapeutic,25–28 three other RNAi therapeutics have been approved for clinical application, including givosiran,29,30 inclisiran,31,32 and lumasiran.33,34 Meanwhile, miRNA, usually an endogenous small noncoding RNA (ncRNA), negatively controls the expression of the target mRNA.35,36 Researchers have discovered numerous miRNA for the treatment of cancer37,38 and fibrosis.39 For instance, miR-34a was studied for the treatment of lung cancer.40
Messenger RNA (mRNA) carries genetic information transcribed from the genomic DNA in the nucleus to the sites of protein synthesis in the cytoplasm.41,42 The sequences of mRNA play important roles in coding a specific protein and modulating the post-translational modifications. Besides, mRNA has a relatively short half-life, which induces transient protein expression. Given these properties, mRNA has become a new class of therapeutics,43–45 which have shown considerable promise in vaccine development,46–57 allergy tolerization,58 and the treatment of a broad spectrum of diseases, including sepsis,59 hemophilia B,60,61 HIV,62 myocardial infarction,63 and several types of cancer.64,65 Theoretically, engineered mRNA can act as a vaccine platform to produce any emerging immunogen. Additionally, mRNA has been used for gene editing and genomic engineering.66,67 In recent years, gene editing systems have been a biotechnological breakthrough, providing a strategy for the treatment of various diseases. Specifically, the CRISPR/Cas system68 uses programmable DNA nucleases to permanently and precisely manipulate the genome.69 The codelivery of Cas9 mRNA and single-guide RNA (sgRNA) against a certain genomic target via base pairing between the sgRNA and the target DNA has been examined for gene editing in numerous genes.70–74 Additionally, RNA aptamer,75–78 RNA decoys,79 ribozymes,80,81 and circular RNA (cirRNA)82–85 have also been explored for biological and therapeutic applications, which have been well-summarized in other reviews.
1.2. Biological Barriers to RNA Delivery
Despite the great potential of RNA-based therapeutics for treating a variety of diseases, many barriers must be circumvented for the successful delivery of these therapeutic RNAs into targeted cell types. Biological obstacles to the effective delivery of RNAs include extracellular and intracellular barriers (Figure 1). The extracellular matrix (ECM) is the first barrier that protects the integrity of the cells from foreign agents, which can inhibit the transport of the RNA molecules from the extracellular environment to the target cells. Cell membrane and endosomal trapping are two major obstacles as intracellular barriers.86–88 As shown in Figure 1, once RNA-loading nanoparticles are administered into the bloodstream, they need to protect RNAs from rapid degradation by serum ribonucleases (RNase).89,90 Meanwhile, nanoparticles must evade phagocytosis, cross the vascular endothelial cells, and traverse the extracellular space to reach the target cells.91 Typical nanoparticles, with small particle size, can penetrate the vascular endothelial pores to pass the extracellular environment. Cellular uptake of RNA-loading nanoparticles into the cytoplasm involves many different pathways, such as clathrin-mediated endocytosis (CME),92,93 caveolae-mediated endocytosis (CvME), and macropinocytosis.94,95 Then, the nanoparticles must escape from the endosome before the lysosome formation which would result in the enzymatic degradation of the nanoparticles.96–98 It was reported that only 1–2% of lipid nanoparticles (LNPs) can escape the endosomes.99 Two main mechanisms have been proposed for the process of nanoparticles endosomal escape including the proton sponge effect and lipid flipping by fusogenic properties during nonlamellar phase transitions.100 During the process of endosomal maturation, the endosomal environment changes from neutral to slightly acidic (pH ~ 6.3 in early endosomes; pH ~ 5.5 in late endosomes; and pH ~ 4.7 in lysosomes),101,102 which makes the ionizable components of the nanoparticles become protonated. Protonation of the ionizable components destabilizes the anionic vesicular membrane and facilitates nanoparticle disassembly, leading to the release of RNA to the cytosol, where RNA elicits its functions.97,103 Upon acidification, lipid nanoparticles that contain protonated ionizable lipids or cationic lipids may adopt an inverted hexagonal (HII) phase and rapidly fuse with anionic endosomal membranes, resulting in the endosomal escape of nanoparticles.104 Incorporation of helper lipids, such as 1,2-dioleoyl-sn-glycerol-3-phosphatidylethanolamine (DOPE), can enhance endosomal fusion as well as endosomal escape of lipid nanoparticles, by undergoing a conformational change upon protonation and promoting an inverted hexagonal (HII) phase change.105–108
Figure 1.
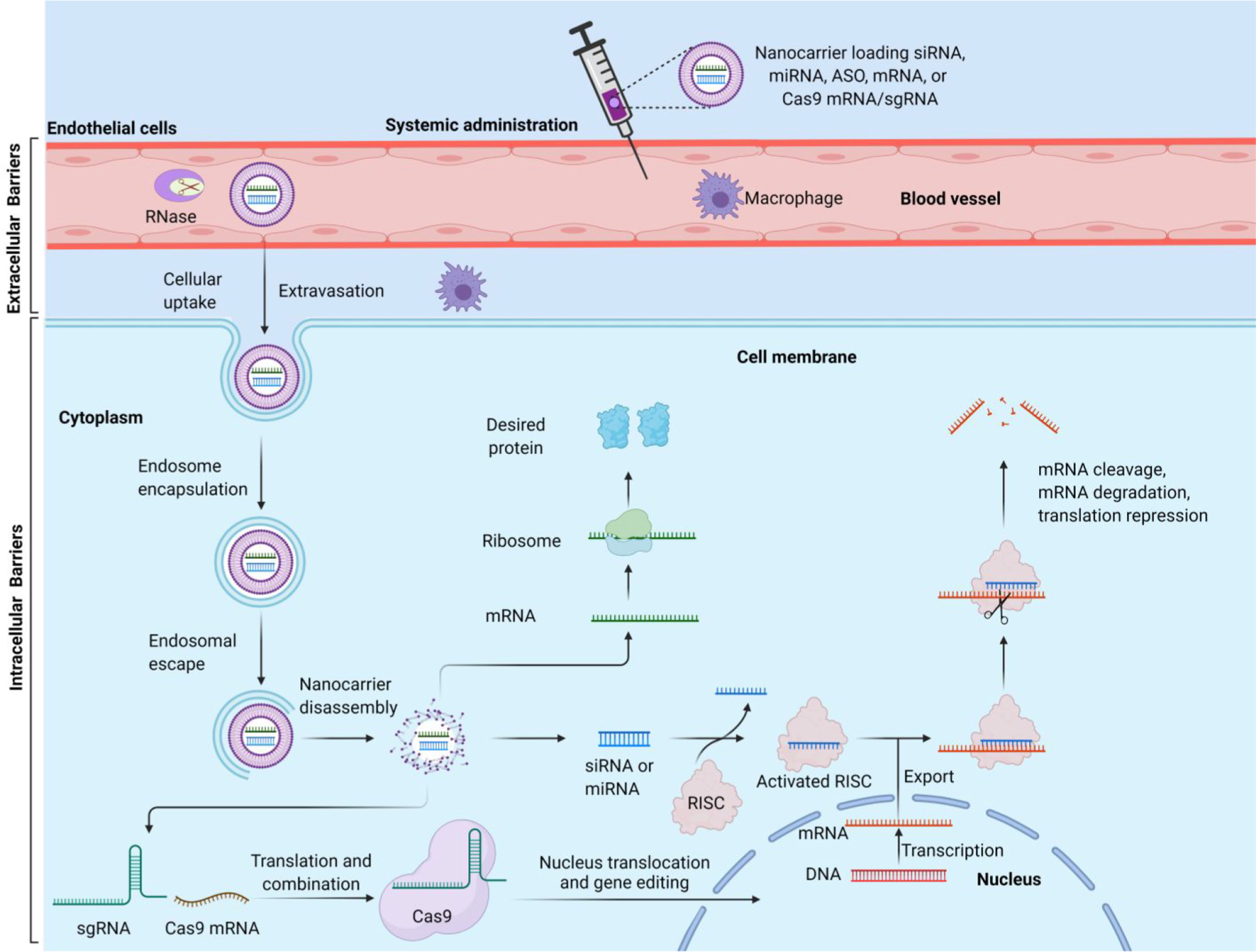
Schematic illustration of the extracellular and intracellular barriers to effective systemic delivery of RNAs and the mechanism of RNA-based therapeutics. Figure was created with BioRender.com.
1.3. Techniques for RNA Delivery
Effective delivery of RNA molecules into cells is a crucial step for successful RNA-based therapeutics. An ideal RNA delivery technique should have high delivery efficiency, low toxicity, as well as high cell specificity. Currently, techniques for RNA delivery can be divided into three types: physical methods, biological carriers, and synthetic approaches Table 2.
Table 2.
RNA Delivery Techniques
| RNA Delivery Techniques | refs | |
|---|---|---|
| Physical Methods | Microinjection | 111–113 |
| Electroporation | 114–116 | |
| Sonoporation | 117–119 | |
| Photoporation | 120, 121 | |
| Magnetofection | 122, 123 | |
| Hydroporation | 124 | |
| Microfluidic squeezing | 125–129 | |
| Biological Carriers | Extracellular vesicles (EVs) | 146–167 |
| Cell/cell membrane-based vectors | 135,168–171 | |
| Synthetic Approaches | Lipid-based nanocarriers | 175–177 |
| Polymer-based delivery systems | 206–217 | |
| Inorganic nanoparticles | 221–232 | |
| Nucleic acid nanostructures | 236, 238, 243, 246–253 | |
| Chemically conjugated RNAs | 261–278 | |
1.3.1. Physical Methods.
The physical methods for RNA delivery generally provide external forces, magnetic field or electrical field to the cells of interest, including microinjection, electroporation, sonoporation, magnetofection, photoporation, hydrodynamic delivery, and microfluidic squeezing.109,110 Microinjection involves direct injection of RNAs into the cytosol using a glass micropipette.111–113 Electroporation employs an electric field, transiently increasing cell membrane permeability, to import RNAs from extracellular compartments into cells.114–116 Sonoporation leads to a perforated cell membrane using ultrasound waves.117–119 Photoporation applies a focused laser beam to produce a submicron hole in cell membranes, which is most commonly used to treat single cells.120,121 Magnetofection involves attaching RNA with cationic magnetic nanoparticles. These nanoparticles are concentrated into target cells under a magnetic field.122,123 Hydroporation is the hydrodynamic capillary effect that can create pores in the cell membrane to allow entry of RNAs.124 Microfluidic squeezing is a microfluidic membrane deformation technique for delivering macromolecules in the surrounding medium into cells by forming transient pores in the plasma membrane.125–129 This technique shows low effects on the normal functions of cells and is broadly applicable for the cytosolic delivery of various macromolecules (e.g., RNA, carbon nanotubes, proteins, quantum dots) to different types of cells.130
1.3.2. Biological Carriers.
Biological carriers are delivery vehicles obtained from living organisms including extracellular vesicles (EVs) and cell/cell membrane-based vehicles, such as exosomes-based vehicles, red blood cells extracellular vesicles (RBCEVs), platelet membrane-coating vehicles, red blood cell (RBC) membrane coating nanoparticles, cancer cell membrane-coated nanoparticles, and macrophage-based vehicles.131–136 These carriers can protect RNA cargos from the degradation by RNase and early clearance by the immune system.137,138
Extracellular vesicles (EVs) are important mediators involved in intercellular communications, which are cell-derived membranous nanosized particles with a lipid bilayer membrane.139–142 Based on their size, surface markers, and mode of biogenesis, EVs are classified into three classes: exosomes (40–120 nm), microvesicles (100–500 nm), and apoptotic bodies (800–5000 nm).143–145 EVs have been applied as an RNA delivery system due to their characteristics such as high biocompatibility, long circulation time, and low toxicity.146–148 Exosomes are considered as “nature’s delivery system”, as it has been shown that exosomes naturally transport DNAs and RNAs between cells, inducing genetic modifications in both biological and pathogenic processes.149–151 Accumulating interests have been focused on harnessing exosomes as vehicles for siRNA152–155 and miRNA156–159 delivery to induce gene silencing.145,160–162 mRNA-loading exosomes have shown tumor-suppressor function in orthotopic phosphatase and tensin homologue (PTEN)-deficient glioma mouse models.163 EVs released from mature red blood cells (RBCEVs), for example, have been used as an RNA delivery system for miRNA inhibiting and CRISPR/Cas9 genome editing in xenograft mouse models.164 RBCs are selected to produce EVs for RNA delivery because mature RBCs lack both mitochondrial and nuclear DNA,165 so the risk of horizontal gene transfer is avoided. In previous studies, RBCEVs were loaded with ASOs, Cas9 mRNA, and sgRNAs or plasmids, respectively, and delivered these agents to target cells in both solid and liquid tumors.164 For example, RBCEVs encapsulating miRNA-125b ASO significantly silenced miRNA-125b and reduced infiltrated cancer cells in acute myeloid leukemia (AML) MOLM13 engrafted mice. Cas9 mRNA and sgRNAs can be codelivered to MOLM13 cells using RBCEVs, inducing genome editing effects.164 Platelet-derived microparticles (PMPs) are extracellular vesicles, 0.1–1 μm in diameter, that are involved in the enhancement of angiogenesis, invasion, and metastasis of tumors.166 PMPs have been shown to infiltrate solid tumors and deliver platelet-derived miRNA to tumor cells both in vivo and in vitro, resulting in gene silencing in tumor cells with broad tumor type specificity.167 Macrophages are appealing carriers for solid tumor targeting RNA delivery due to their inherent capacities to home to tumors at significant numbers throughout tumor progression.168,169 Moreover, macrophages can easily load and secrete nanoparticles into the surrounding microenvironment. Wayne et al. developed a macrophage-based targeted siRNA delivery system that delivered calcium integrin binding protein-1 (CIB1)-siRNA to MDA-MB-468 human breast cancer cells, leading to reduced expression of CIB1 and KI67 and decreased tumor growth.170 Zhang et al. prepared platelet membrane-camouflaged PLGA/DOTAP nanoparticles to deliver anti-Pcsk9 siRNA efficiently, resulting in ~28% reduction in the level of plasma LDL-C.171
1.3.3. Synthetic Approaches.
Synthetic approaches have constructed numerous types of natural or synthetic materials and formulations for delivering RNAs into cells, including lipid-based nanocarriers, polymer-based systems, inorganic nanoparticles, nucleic acid nanostructures, chemically modified RNAs, and many others.172–174
Among the various synthetic approaches for RNA-based therapeutics, lipid-based nanocarriers have been recognized as one of the most promising RNA delivery systems.175–177 These nanocarriers can be prepared in various forms such as cationic liposomes, ionizable lipid nanoparticles (LNPs), lipid–polymer hybrid nanoparticles (LPHNPs), lipid calcium phosphate (LCP) nanoparticles, niosomes, cationic nanoemulsions (CNEs), and neutral lipid nanoemulsions (NLEs).178–183 Examples of clinical trials of lipid-based RNA therapies are summarized in Table 3. In the 1960s, Bangham et al. reported that biocompatible lipid/phospholipid spontaneously formed closed phospholipid bilayer structures in an excess of water, which was termed as liposome.184,185 Liposomes contain an aqueous compartment that is surrounded by one or more phospholipid bilayers, which can serve as unique vehicles for the entrapment of hydrophilic drugs (e.g., Doxil)186 and DNA.187,188 Cationic liposomes are also among the earliest synthetic approaches used for RNA delivery.189–191 The positive charges of the cationic lipid-based liposomes can improve the RNA encapsulation efficiency as the result of electrostatic interactions between the negatively charged phosphate backbone of RNA molecules and the positively charged head groups of cationic lipids. Generally, the nitrogen/phosphate ratio (N/P) is modulated so that the liposome has a net positive charge, thus neutralizing RNA molecules and avoiding aggregation of liposomes. Besides, the excess positive charge facilitates the binding of liposomes to the negatively charged cell membranes.192 PEGylated cationic liposomes were developed to increase the circulation stability of liposomes, thus improving RNA delivery efficiency in vivo.192,193
Table 3.
Representative Clinical Trials of Lipid-Based RNA Therapies
| Name | Indication | RNA Payload | Delivery System | Delivery Route | Sponsoring Institution | Phase | Clinical Trial Identifier |
|---|---|---|---|---|---|---|---|
| siRNA-based therapeutics | |||||||
| ALN-TTR02 (Patisiran) | TTR-mediated Amyloidosis | siRNA against TTR | LNP (MC3/CHOL/DSPC/PEG2000-C-DMG) | IV | Alnylam Pharmaceuticals |
Approved | NCT03862807 |
| ALN-VSP02 | Solid tumor | siRNA targeting VEGF and KSP | LNP | IV | Alnylam NCT00882180 |
I |
NCT01158079 Pharmaceuticals |
| TKM-080301 | Tumor | siRNA targeting PLK1 | LNP | IV | Tekmira Pharmaceuticals |
I |
NCT01262235
NCT01437007 NCT02191878 |
| ALN-PCS02 | Hypercholesterolemia | siRNA against PCSK9 | LNP | IV | Alnylam Pharmaceuticals |
I | NCT01437059 |
| siRNA-EphA2-DOPC | Advanced malignant solid neoplasm | siRNA targeting EphA2 | Liposome (DOPC, Tween 20) | IP | M.D. Anderson Cancer Center | I | NCT01591356 |
| Atu027 | Pancreatic ductal carcinoma | siRNA targeting PKN3 | Liposome (AtuFECT01/PEG-DSPE/CHOL) | IV | Silence Therapeutics GmbH | I/II |
NCT01808638
NCT00938574 |
| ND-L02-s0201 | Hepatic fibrosis, Idiopathic pulmonary fibrosis | siRNA against HSP47 | LNP (DC-6-14/Chol/DOPE/Vitamin A) | IV | Bristol-Myers Squibb | II | NCT03538301 |
| ARB-001467 | Hepatitis B | siRNA against HBV gene | LNP | IV | Arbutus Biopharma | II | NCT02631096 |
| DCR-MYC | Hepatocellular carcinoma | siRNA targeting MYC | LNP | IV | Dicerna Pharmaceuticals, Inc. | I/II | NCT02314052 |
| DCR-HBVS | Hepatitis B | siRNA against HBV gene | LNP | IV | Dicerna Pharmaceuticals, Inc. | I | NCT03772249 |
| mRNA-based vaccinesagainst infection | |||||||
| mRNA-1273 | COVID-19 | mRNA encoding SARS-CoV-2 spike protein | LNP (SM-102/CHOL/DSPC/DMG-PEG2000) | IM | Moderna Therapeutics | Emergency Use Authorization (EUA) | NCT04470427 |
| BNT162b2 | COVID-19 | mRNA encoding SARS-CoV-2 spike protein | LNP (ALC-0315/CHOL/DSPC/ALC-0159) | IM | BioNTech/Pfizer | Emergency Use Authorization (EUA) | NCT04537949 |
| ARCoV | COVID-19 | mRNA encoding RBD of SARS-CoV-2 | LNP | IM | Abogen Biosciences | I | ChiCTR200003411 |
| ARCT-021 | COVID-19 | mRNA encoding SARS-CoV-2 spike protein | LNP (ATX/DSPC/CHOL/DMG-PEG2000) | IM | Arcturus Therapeutics | II | NCT04728347 |
| ChulaCov19 | COVID-19 | mRNA encoding SARS-CoV-2-specific antigen | LNP | IM | Chulalongkorn University | I | NCT04566276 |
| CVnCoV | COVID-19 | mRNA encoding SARS-CoV-2 spike protein | LNP | IM | CureVac | II/III | NCT04652102 |
| CV7202 | Rabies | mRNA encoding Rabies virus G glycoprotein | LNP | IM | CureVac | I | NCT03713086 |
| mRNA-1440 | Influenza H10N8 | mRNA encoding Hemagglutinin | LNP | ID or IM | Moderna Therapeutics | I | NCT03076385 |
| mRNA-1851 | Influenza H7N9 | mRNA encoding Hemagglutinin | LNP | ID or IM | Moderna Therapeutics | I | NCT03345043 |
| mRNA-1653 | hMPV/PIV3 | mRNA encoding fusion protein of hMPV and PIV3 | LNP | ID | Moderna Therapeutics | I | NCT03392389 |
| mRNA-1325 | Zika | mRNA encoding prM-E glycoproteins | LNP | IM | Moderna Therapeutics | I | NCT03014089 |
| mRNA-1893 | Zika | mRNA encoding prM-E glycoproteins | LNP | IM | Moderna Therapeutics | I | NCT04064905 |
| mRNA-1647 and mRNA-1443 | Cytomegalovirus | mRNA encoding Pentameric complex and B glycoprotein | LNP | ID | Moderna Therapeutics | I | NCT03392389 |
| mRNA-1388 | Chikungunya | mRNA encoding Chikungunya virus antigens | LNP | IM | Moderna Therapeutics | I | NCT03325075 |
| mRNA-1944 | Chikungunya | mRNA encoding Chikungunya virus antigens | LNP | IV | Moderna Therapeutics | I | NCT03829384 |
| mRNA-based vaccinesagainst infection | |||||||
| GSK 692342 | Tuberculosis | mRNA encoding immunogenic fusion protein (M72) of tuberculosis | LNP | IM. | GlaxoSmithKline | II | NCT01669096 |
| mRNA-based cancer immunotherapy | |||||||
| BNT111 | Melanoma | mRNA encoding TAAs | Liposome (DOTMA/DOPE) | IV | BioNTech RNA Pharmaceuticals GmbH | I | NCT04382898 |
| BNT112 | Prostate cancer | mRNA encoding TAAs | Liposome (DOTMA/DOPE) | IV | BioNTech RNA Pharmaceuticals GmbH | I | NCT03418480 |
| BNT113 | HPV16-positive cancers | mRNA encoding TAAs | Liposome (DOTMA/DOPE) | IV | BioNTech RNA Pharmaceuticals GmbH | I | NCT04534205 |
| BNT114 | Triple negative breast cancer | mRNA encoding TAAs | Liposome (DOTMA/DOPE) | IV | BioNTech RNA Pharmaceuticals GmbH | I | NCT02410733 |
| BNT115 | Ovarian cancer | mRNA encoding TAAs | Liposome (DOTMA/DOPE) | IM | BioNTech RNA Pharmaceuticals GmbH | I | NCT04163094 |
| RO7198457 (BNT122) | Locally advanced and metastatic tumors | mRNA encoding TAAs | Liposome | IV | Genentech, Inc. | II | NCT03815058 |
| mRNA-2752 | Solid tumors and lymphomas | mRNA encoding OX40L, IL-23, and IL-36γ | LNP | IT | Moderna Therapeutics | I | NCT03739931 |
| mRNA-2416 | Solid tumors lymphomas and ovarian cancer | mRNA encoding OX40L | LNP | IT | Moderna Therapeutics | I | NCT03323398 |
| mRNA-4157 | Bladder carcinoma, Melanoma | mRNA encoding TAAs | LNP | IM | Moderna Therapeutics | II | NCT03897881 |
| mRNA-4650 | Gastrointestinal cancer | mRNA encoding TAAs | LNP | IM | National Cancer Institute (NCI) | I/II | NCT03480152 |
| mRNA-5671/V941 | Nonsmall cell lung cancer, colorectal cancer, pancreatic adenocarcinoma | mRNA encoding KRAS antigens | LNP | IM | Merck Sharp & Dohme Corp. | I | NCT03948763 |
| HARE-40 | HPV positive cancers | mRNA encoding HPV oncoproteins E6 and E7 | LNP | ID | University of Southampton | I/II | NCT03418480 |
| SAR441000 (BNT131) | Solid tumors | mRNA encoding L-12sc, IL-15sushi, IFNα and GM-CSF | LNP | IT | Sanofi | I | NCT03871348 |
| W_ova1 | Ovarian cancer | mRNA encoding TAAs | Liposome | IV | University Medical Center Groningen | I | NCT04163094 |
| MEDI1191 | Solid tumors | mRNA encoding IL-12 | LNP | IT | MedImmune LLC. | I | NCT03946800 |
| mRNA-based therapeutics forgene disorders | |||||||
| mRNA-3704 | Isolated Methylmalonic Acidemia | mRNA encoding Methylmalonyl-CoA mutase | LNP | IV | Moderna Therapeutics | I/II | NCT03810690 |
| mRNA-3927 | Propionic academia | mRNA encoding Propionyl-CoA carboxylase | LNP | IV | Moderna Therapeutics | I/II | NCT04159103 |
| MRT5201 | Ornithine transcarbamylase deficiency | mRNA encoding Ornithine transcarbamylase | LNP | IV | Translate Bio, Inc. | I/II | NCT03767270 |
| MRT5005 | Human Cystic fibrosis | mRNA encoding CFTR | LNP | INH | Translate Bio, Inc. | I/II | NCT03375047 |
| NTLA-2001 | Transthyretin amyloidosis with polyneuropathy | CRISPR/Cas9 gene editing system | LNP | IV | Intellia Therapeutics | I | NCT04601051 |
| Other RNA-based therapeutics | |||||||
| MTL-CEBPA | Hepatocellular carcinoma | CEBPA-51 saRNA targeting CEBPA | Liposome (POPC/DOPE/MoChol/CHEMS) | IV | MiNA Therapeutics | I | NCT02716012 |
| BP1001 | AML, ALL, MDS, and CML | ASO targeting Grb2 mRNA | Liposome (DOPC/Tween 20) | IV | Bio-Path Holdings, Inc. | II | NCT02781883 |
| LErafAON-ETU | Advanced cancer | ASO targeting C-raf | LNP | IV | INSYS Therapeutics | I | NCT00100672 |
Later on, researchers synthesized ionizable lipids with apparent pKa values less than 7, which exhibit positive charges and interact with RNA molecules when protonated under acidic conditions, while they are neutral in physiologic conditions (pH = 7.4). Apart from ionizable lipids, PEG lipids and helper lipids are fundamental lipid components in the LNP formulations, such as DMG-PEG2000, 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), and cholesterol.194,195 Microfluidic mixing of solutions of lipid components in organic solvent with aqueous solutions is a readily scalable and precisely controlled technique for the preparation of LNPs.196–198 Different from cationic liposomes, typical LNPs only have a single phospholipid outer layer encapsulating the electron-dense core, where the ionizable lipids aggregate into inverted micelles around the entrapped RNA molecules.199–201 Additionally, under acidic conditions of the endosomes, these ionizable lipids are protonated and can bind to the negatively charged endosomal membranes, inducing endosome disruption and resulting in enhanced endosomal escape.202 These characteristics make lipid nanoparticles important materials for RNA delivery. In terms of clinical research, lipid nanoparticles (LNPs) are the most advanced synthetic approaches for RNA therapies for treating a range of diseases up to date.203 The approvals of patisiran,204,205 BNT162b2,8,9 and mRNA-12737,8 in the clinical application are milestones in the development of LNP-based RNA delivery.
In addition to lipid nanoparticles, both naturally derived and synthetic polymers have been utilized for RNA delivery, such as poly(ethyleneimine) (PEI),206,207 poly-l-lysine (PLL),208–210 poly(β-amino ester) (PBAE),211–213 chitosan,214–216 and polysaccharide.217 By tuning the physiochemical characteristics of polymers, efficient RNA delivery can be achieved in cell and animal models.218–220 A broad range of inorganic nanoparticles have also been explored as carriers for the controlled and targeted RNA delivery, such as gold nanoparticles,221–223 silica nanoparticles,224–226 calcium phosphate nanoparticles,227–229 and iron oxides nanoparticles.230–232 DNA and RNA strands are versatile building blocks for creating functional nucleic acid nanostructures with structural programmability, spatial addressability, molecular recognition capability, and biocompatibility.233–236 Over the past several decades, nucleic acid-based nanotechnology has made great achievements in various applications.237–246 For example, DNA and RNA nanostructures have been used in the delivery of ASOs and siRNA.236,238,243,246–253 Additionally, researchers have developed numerous chemical strategies of RNA conjugation, which can improve the RNA-binding affinity, thermostability, circulation time, and pharmacokinetic properties of RNA.254–260 A chemically conjugated RNA is a direct covalent conjugation of an RNA molecule and various moieties that promotes intracellular uptake, targets the drug to specific cells/tissues, or reduces clearance from the circulation. These moieties include lipids (e.g., cholesterol,261 α-Tocopherol262,263), peptides (e.g., cell-penetrating peptide264,265), aptamers,266–268 antibodies,269–271 and receptor ligand.272–276 The conjugation of siRNA and N-acetylgalactosamine (GalNAc) increases the cellular internalization in the liver through interactions of the GalNAc with the asialoglycoprotein receptor (ASGPR) on the surface of hepatocytes.259,277,278 This hepatocyte-specific delivery platform has led to the clinical use of givosiran,29,30 inclisiran,32 and lumasiran.33,34
In this review article, we focus on the chemical perspectives of lipids including a variety of lipid derivatives and lipid-derived macromolecules used in lipid-based RNA delivery systems over the past three decades. We summarize the advances of lipids, lipid derivatives, and lipopolymers regarding their chemical structures, synthetic routes, characterizations, and structure–activity relationships. We also briefly introduce the status of representative preclinical and clinical studies and highlight future opportunities and challenges.
2. CATIONIC, IONIZABLE, AND ZWITTERIONIC LIPIDS
Cationic or ionizable lipids are of great importance for the delivery of RNAs because their positively charged head groups under the formulation environment can interact with the negatively charged phosphate backbone of the RNA cargos.279,280 In 1978, Dimitriadis reported the delivery of rabbit globin mRNA into mouse lymphocytes ex vivo using phosphatidylserine-based unilamellar liposomes.189 In 1987, Felgner et al. synthesized the cationic lipid N-[1-(2,3-dioleyloxy)propyl]-N,N,N-trimethylammonium chloride (DOTMA, Figure 2) and used it for in vitro gene delivery.281 The encapsulation efficiency of DOTMA-based liposomes to pDNA is about 100%, and their pDNA delivery efficiency is 5–100 times higher than that of calcium phosphate or diethylaminoethyl-dextran.281 In 1989, Malone et al. developed DOTMA-based liposomes (Lipofection) for in vitro delivery of luciferase mRNA into NIH 3T3 mouse cells.191
Figure 2.
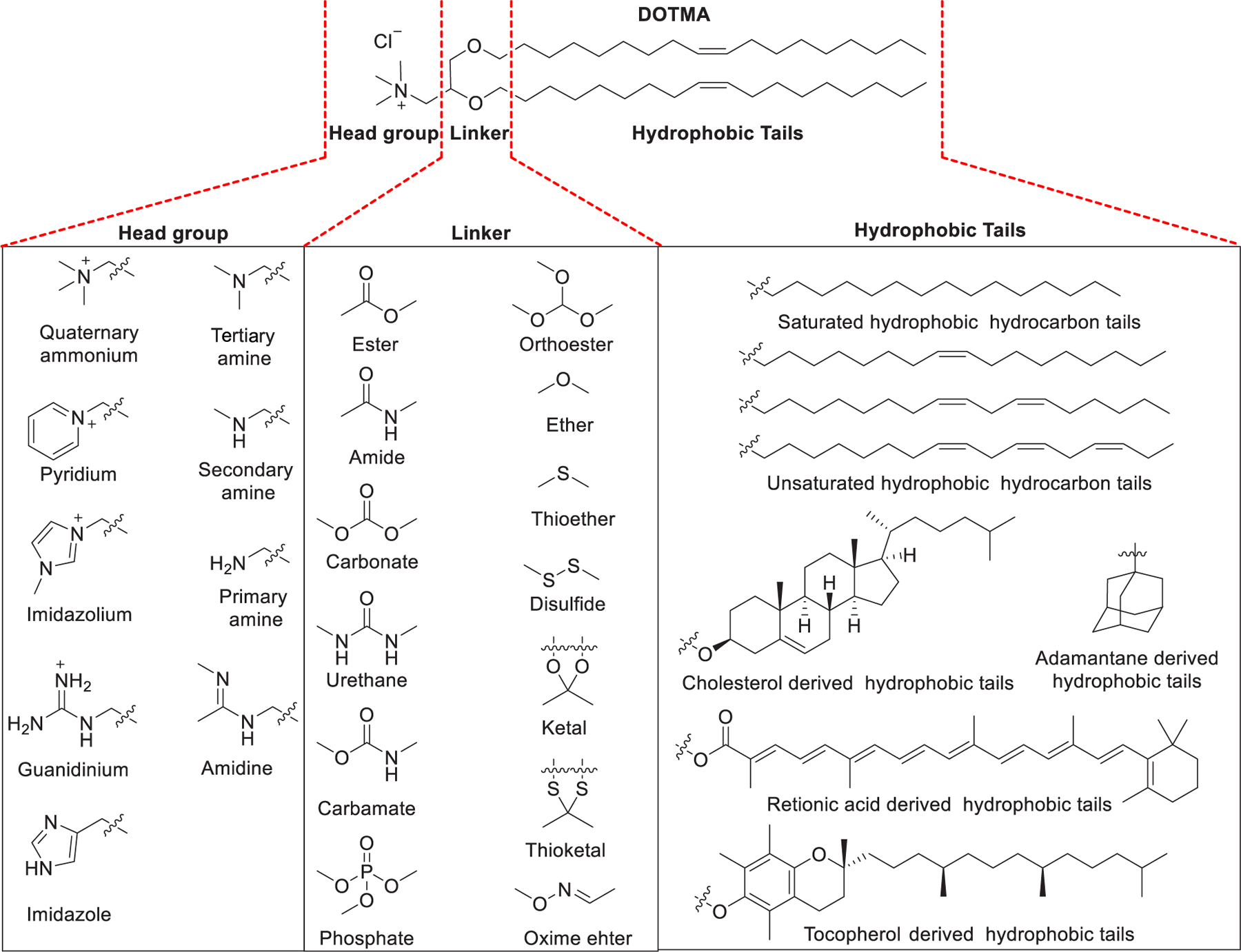
Chemical structures of the cationic lipid DOTMA and three segments of lipids.
Structurally, synthetic lipids usually contain three parts: (i) cationic or ionizable head groups, (ii) linker groups, and (iii) hydrophobic tails (Figure 2).282,283 The head groups exhibiting positive charge(s) can interact with the negatively charged RNA backbone through electrostatic attractions; in this way complexes containing condensed RNA are formed. The lipids and lipid derivatives can be classified into various categories based on the characteristics of their head groups: (i) cationic lipids, (ii) ionizable lipids, and (iii) zwitterionic lipids.282–285 The structure of the hydrophobic tails of lipids can affect their pKa, lipophilicity, transition temperature, and potency for RNA delivery.105,286 A cholesterol derivative or a hydrocarbon chain or even a tocopherol derivative can act as a hydrophobic component of lipids. The hydrocarbon tails are generally between 8 to 18 carbon units in length with various unsaturation degrees (e.g., oleoyl group, linoleoyl group), and symmetry is not necessary for them.280 Incorporation of unsaturated fatty acid as lipid tails has resulted in higher delivery efficiency in certain formulations, possibly owing to their low transition temperature and their influence on increasing membrane fluidity.287 An ideal linker group should be biodegradable and preserve strong circulation stability to survive in a biological environment. The commonly employed linker groups include ethers and esters, phosphate or phosphonate linker, glycerol-type moiety, or peptides. Carbamate and amide are also frequently used as linker, as both of them are chemically stable and biodegradable. Ester and ether are alternative linkers, which are chemically stable. The linker groups can be designed to be tunable; thus, they are stable enough for storage and have higher circulation stability but can be degraded rapidly at the target sites to facilitate the release of the RNA payload.
Geometry is an important characteristic of amphiphile lipids with regard to their application as RNA carriers. Amphiphile lipids form aggregates above a certain concentration in an aqueous environment, adopting various structures, including the micellar phase, hexagonal (HI) phase, lamellar phase, inverted micellar phase, and inverted hexagonal (HII) phase (Figure 3). These different types of structures can be predicted by the packing parameter P of the lipid, which is defined as the ratio of the amphiphile lipid volume (V) to its head group area (a), and the critical tail length (lc) .288–290 When P is less than 1/2, the conical-shaped amphiphile lipids assemble into micelles or a hexagonal (HI) phase. When 1/2 < P ≤ 1, cylindrical-shaped amphiphile lipids with a curvature close to 0 adopt the stable lamellar phase. The inclusion of lipids with a cylindrical shape, such as DSPC, increases the stability and circulation time of lipid-based nanoparticles.291 When P > 1, the structures formed by the inversed conical-shaped amphiphile lipids tend to adopt inverted micelles or inverted hexagonal (HII) phases. Thus, when P > 1, the inverted conical-shaped lipids (e.g., DOPE) can destabilize the endosomal membrane and allow the endosomal escape and release of the RNA payload into the cytosol of the target cells.286,292,293 This section describes a large number of lipid derivatives with their chemical structures, synthetic routes, and RNA delivery properties.
Figure 3.
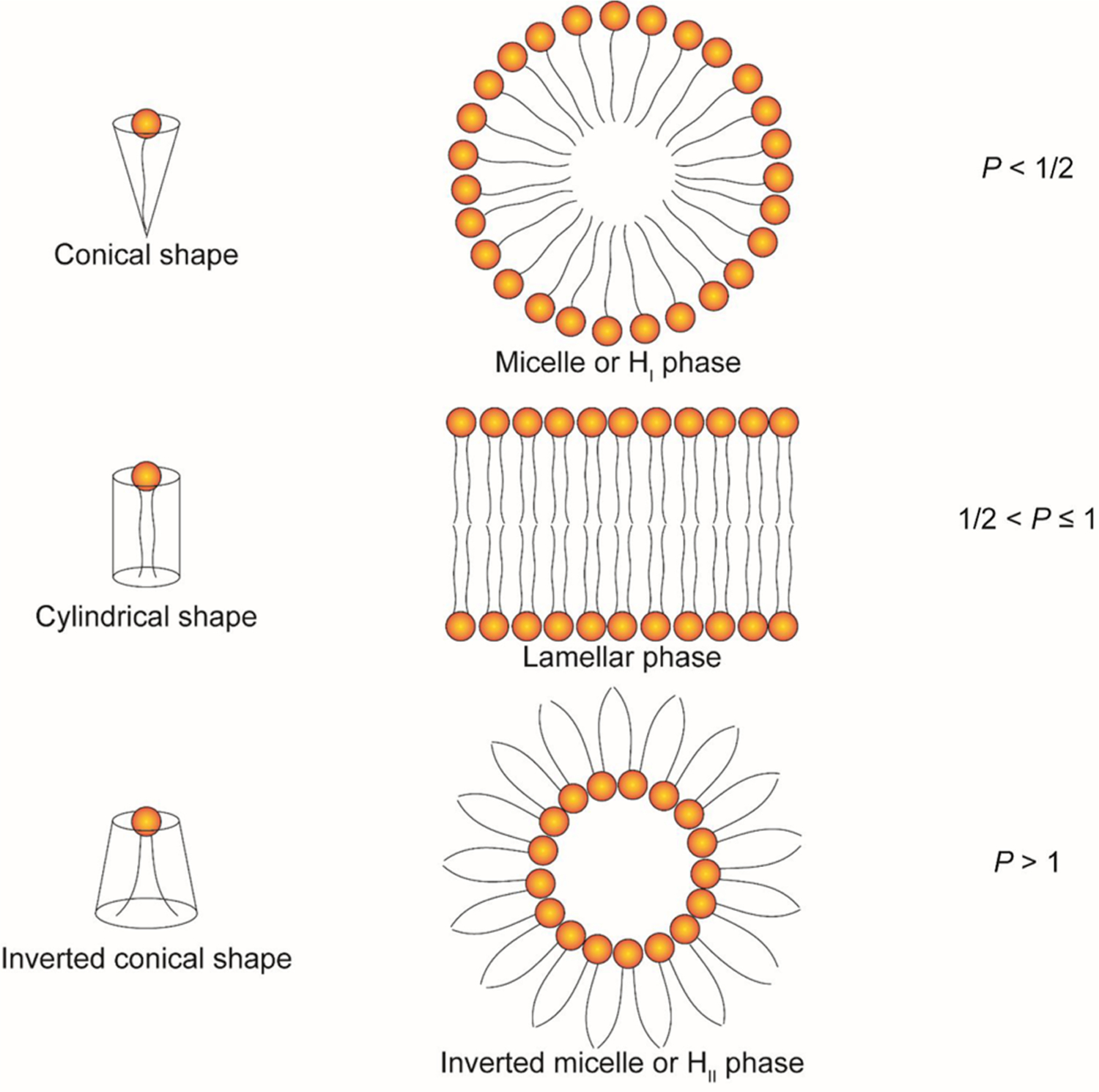
Schematic illustration of the shape structure concept of lipids.
2.1. Cationic Lipids
Cationic lipids refer to lipids with head groups bearing permanent positive charges. They have been well-explored for nucleic acids (DNA and RNA) delivery as components of liposomes and lipoplexes, due to their capability of encapsulating nucleic acids. According to the chemical structures of their head groups, they are grouped into four types in this part: quaternary ammonium lipids, guanidinium lipids, pyridinium lipids, and imidazolium lipids. Cationic lipids are permanently positively charged and are chemically stable even in the environment of strong oxidants and acids. However, the positive charges might lead to potential cytotoxicity, e.g., hemolytic and undesired immunostimulation.294,295 Cytotoxicity of cationic lipids may be related to the generation of reactive oxygen species (ROS) and the increase of cellular calcium levels.296,297 Besides, the positive charge of cationic lipids could result in their rapid plasma clearance and short circulation time.294,295 It is noteworthy that cationic lipids with delocalized positive charges, such as pyridinium, imidazolium, and guanidinium, showed lower cytotoxicity as compared to quaternary ammonium lipids.298–301 In certain cases, cationic lipids may act as vaccine adjuvants by taking advantage of their inflammatory effects.302
2.1.1. Quaternary Ammonium Lipids.
Ever since the 1980s, numerous cationic lipids with quaternary ammonium head groups have been developed for the delivery of DNA and RNA, such as DOTAP, DMRIE, and DODAB, which were reported to be effective for RNA delivery (Figure 4).303 As the purification of positively charged compounds is relatively challenging, the cationic head groups are preferentially formed via quaternization of the corresponding tertiary amines in the final step of the synthesis.304–306 The synthesis of DOTMA, for example, began with the combination of amino alcohol 1 with oleyl bromide 2 via ether bond formation; then quaternization of the resulting amine 3 with chloroform under reflux condition gave DOTMA.281 In 2016, Kranz et al. developed a DOTMA/DOPE LNPs-mRNA vaccine that specifically targeted dendritic cells (DCs) in vivo by changing the surface charge from positive to slightly positive or neutral. The optimized vaccine induced specific immune responses following intravenous administration.307 This DOTMA/DOPE LNPs formulation has also been used to deliver mRNA containing 1-methylpseudouridine (m1Ψ) instead of uracil for precision therapy of autoimmune diseases in mice.308 In 2020, Reinhard et al. engineered T cells by utilizing DOTMA-based LNPs to deliver mRNA encoding a single-chain variable fragment (scFv) that could specifically recognize the overexpressed cancer cell surface protein claudin 6 (CLDN6). The resulting CAR-T cells led to improved regression in mouse models of intractable tumors, such as CT26 colon carcinoma mouse models.309 In a recent phase 1 clinical trial of an mRNA vaccine for melanoma (melanoma FixVac), DOTAP-based LNPs were formulated to encapsulate mRNA encoding four tumor associated antigens (TAAs) that show high prevalence in melanoma; mRNA molecules were delivered into immature DCs in lymphoid tissues, driving TAA presentation on both MHC I and MHC II molecules after intravenous administration. Patients showed TLR activation, increased body temperature, elevated cytokines level in plasma, and specific response against at least one TAA after vaccination.310 In 2017, Cheng et al. used LNPs formulated with DOTAP/Cholesterol/eggPC/Tween 80 at a molar ratio of 25:20:50:5 to deliver G3139, an antisense oligonucleotide, into A549 lung cancer cells, resulting in 40% knockdown of bcl-2 mRNA and approximately 83% reduction of the bcl-2 protein level, respectively.311 In 2019, DOTAP LNPs encapsulating hARG1 mRNA were used to treat arginase deficiency in inherited metabolic liver disorder, achieving 54% of normal hepatic arginase 24 h after administration in mice.312 Cationic liposomes can act as immunomodulators that stimulate the innate immune response in some cases.313 For example, DOTAP-based cationic liposomes have been used as a vaccine adjuvant.302 The immunostimulatory effects of the components of cationic liposomes were related to the length and saturation degree of hydrophobic tails. Lipids possessing unsaturated tails or short saturated tails may be stronger immunomodulators than lipids with long saturated tails.313 Cationic liposomes formulated with DOTAP/DSPC/cholesterol were shown to be capable of activating Toll-like receptor 4 (TLR4), inducing a greater pro-inflammatory response with enhanced Th1 cytokines expression in mice compared with ionic liposomes formulated with DSPG/HSPC/cholesterol.314 DOTAP-based cationic nanoemulsions (CNEs) have also been used as vectors for mRNA delivery.315–320 For instance, CNEs formulated with DOTAP/sorbitan trioleate/polysorbate 80/squalene were reported to deliver an mRNA vaccine against several viral and bacterial infections in nonhuman primates with two doses of 75 μg.317 In a follow-up study, CNEs encapsulating the HIV Type 1 envelope protein mRNA were shown to be well-tolerated and immunogenic in nonhuman primates.318 Additionally, DOTAP was incorporated to prepare lipid–polymer hybrid nanoparticles (LPHNPs), which are composed of a biodegradable RNA-loaded polymer core surrounded by lipid/PEG-lipid layers.321,322 LPHNPs combine the unique strengths of liposomes and polymeric nanoparticles but exclude some of their limitations such as short circulation time and structural disintegration.323 Gao et al. formulated LPHNPs with DOTAP/DOPE/cholesterol (25:43:25) and poly(amidoamine)/siRNA for siRNA delivery. The resulting LPHNPs effectively delivered T7-modified anti-EGFR siRNA to an MCF-7 tumor xenograft murine model and inhibited tumor growth.324
Figure 4.
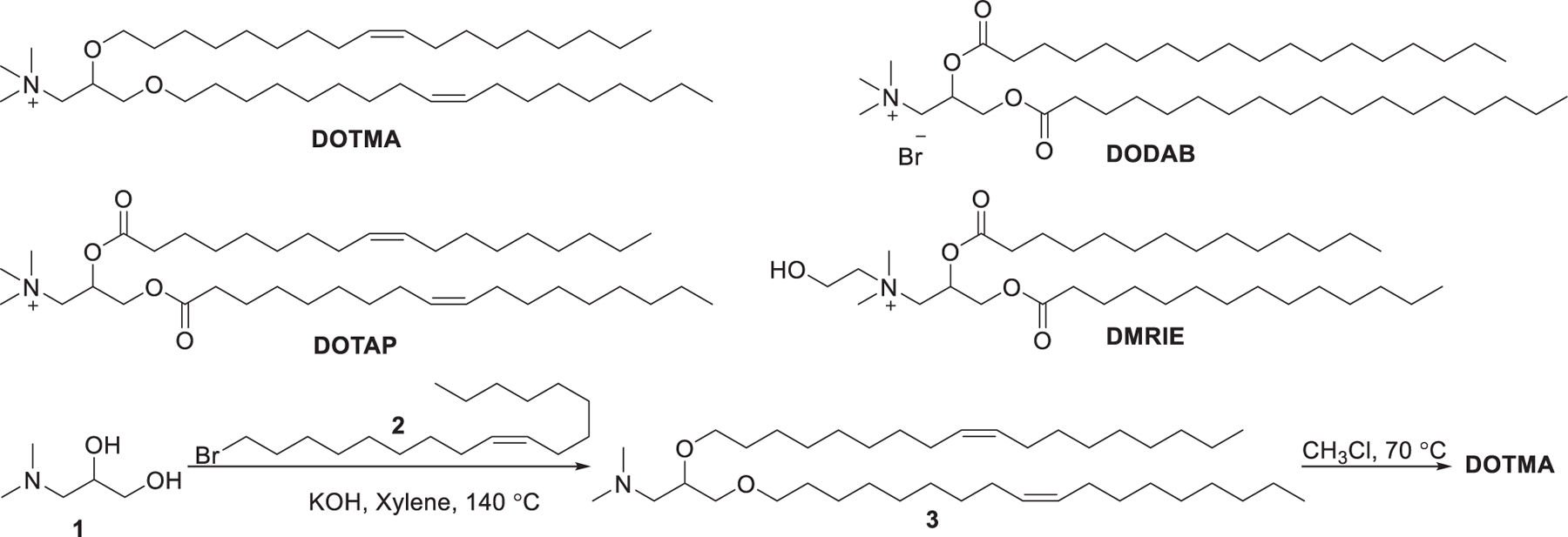
Chemical structures and synthesis of DOTMA and its analogs.
A hydroxyalkyl chain incorporated in the head group was capable of providing hydrogen bonding to neighboring head groups, thus decreasing the head group hydration and improving the encapsulation of nucleic acids via hydrogen bonding with the lipid. In the previous studies lipids used in DNA delivery, DORIE and DORI, were obtained by replacing one of the methyl groups in the head groups of DOTMA and DOTAP with a hydroxyethyl group, respectively, and both of them exhibited greater DNA delivery activity than DOTMA or DOTAP (Figure 5).325,326 DMDHP and MLRI were synthesized as analogs of DOTAP for mRNA delivery.327,328 MLRI, an asymmetric analog of DORI, contains a myristoyl group and a lauroyl group as the hydrophobic tails. Results indicated that mRNA/cationic lipid lipoplexes formulated with MLRI or DMDHP could protect mRNA from degradation by RNases in human cerebrospinal fluid (hCSF) for at least 4 h.327
Figure 5.
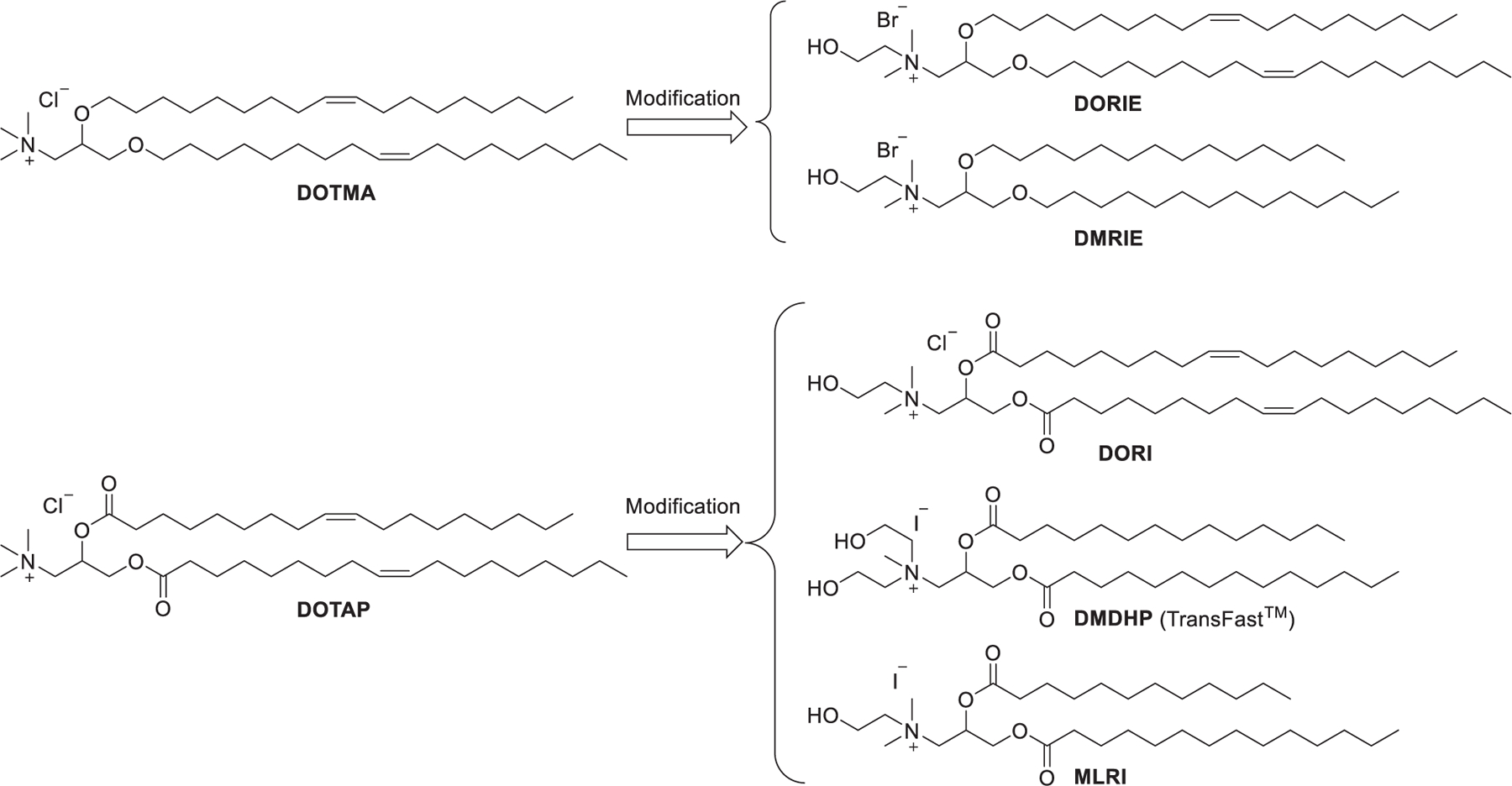
Chemical modifications of DOTMA and DOTAP by introducing hydroxyethyl groups
In 1999, Kikuchi et al. developed a series of quaternary ammonium lipids and identified DC-16-4 as the lead cationic lipid for delivering pDNA to human peritoneal disseminated tumors both in vitro and in vivo.329 As shown in Figure 6, the hydrophobic tails were installed via acylation of diol 4 with myristoyl chloride 5, and the head group was installed via an amide bond formation followed by quaternization of the tertiary amine group. In 2008, Sato et al. reported the delivery of gp46 siRNA using vitamin A-coupled DC-6-14 LNPs in rats, resulting in effective treatment of liver fibrosis and prolonged survival time by targeting hepatic stellate cells.330 ND-L02-s0201 is another vitamin A-coupled DC-6-14 LNP encapsulating siRNA targeting heat shock protein 47 (HSP47), which is involved in the fibrosis of the liver. Results of the phase I clinical study of ND-L02-s0201 showed that intravenous administration of ND-L02-s0201 at a siRNA dose of 90 mg for 3 weeks was well-tolerated in healthy adults.331
Figure 6.
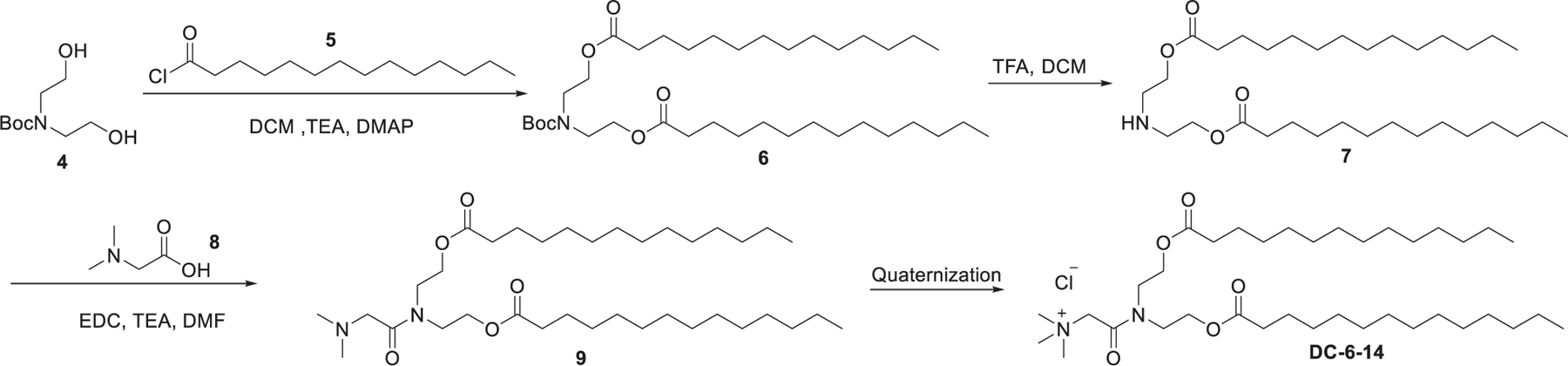
Chemical structure and synthetic route of DC-6-14.
In 1997, Gorman et al. reported that dimyristoyl-sn-glycero-3-ethylphosphocholine (EDMPC, 12), a phosphotriester derived from phosphocholine, was able to mediate efficient gene delivery.332 Then, Macdonald et al. found that phosphotriesters were slowly metabolized by intracellular phospholipases in endosomes and lysosomes and showed low cytotoxicity.333 In 1999, Macdonald et al. developed a series of alkyl phosphatidylcholine triesters, the cationic ethylphosphatidylcholines (ePCs), by introducing a third alkyloxy group, through substitution reaction between the phosphoric acid and ethyl trifluoromethylsulfonates (Figure 7).334 This method represents a straightforward way to convert zwitterionic phospholipids to cationic phospholipids via a simple substitution reaction. This transformation not only eliminates the negative-charge of phosphatidylcholines but also reduces their hydrogen bond accepting potential. This class of cationic lipids has shown effective delivery of pDNA both in vitro and in vivo for anticystic fibrosis and antitumor gene therapies.335–338 Dimyristoleoyl-ePC (EDMPC) 12 was identified as the most efficient ePC that could be used for delivering GFP siRNA into breast cancer cells.339 The structure–activity relationship of the ePCs showed that the high siRNA delivery efficiency of dimyristoleoyl-ePC (EDMPC) 12 stems from its high fusogenicity and ability to induce inverted hexagonal (HII) phases.339 In 2017, a lipopolyplex mRNA vaccine was prepared by trapping the mRNA/PBAE core in a bilayer lipid shell containing EDOPC/DOPE/DSPE-PEG. This mRNA vaccine showed adjuvant activity by stimulating the expression of INF-β and IL-12 in dendritic cells. Subcutaneous administration of this mRNA vaccine resulted in a reduction of tumor nodules by 90% in mice with lung metastatic B16-OVA tumors.340 In 2019, Zhang et al. prepared LNPs with dipalmitoyl-ePC 11 and cholesterol at a molar ratio of 70:30 for delivering mRNA.341 Results indicated that these LNPs could deliver mRNA encoding an anti-RAS antibody into a range of human cancer cells.341
Figure 7.
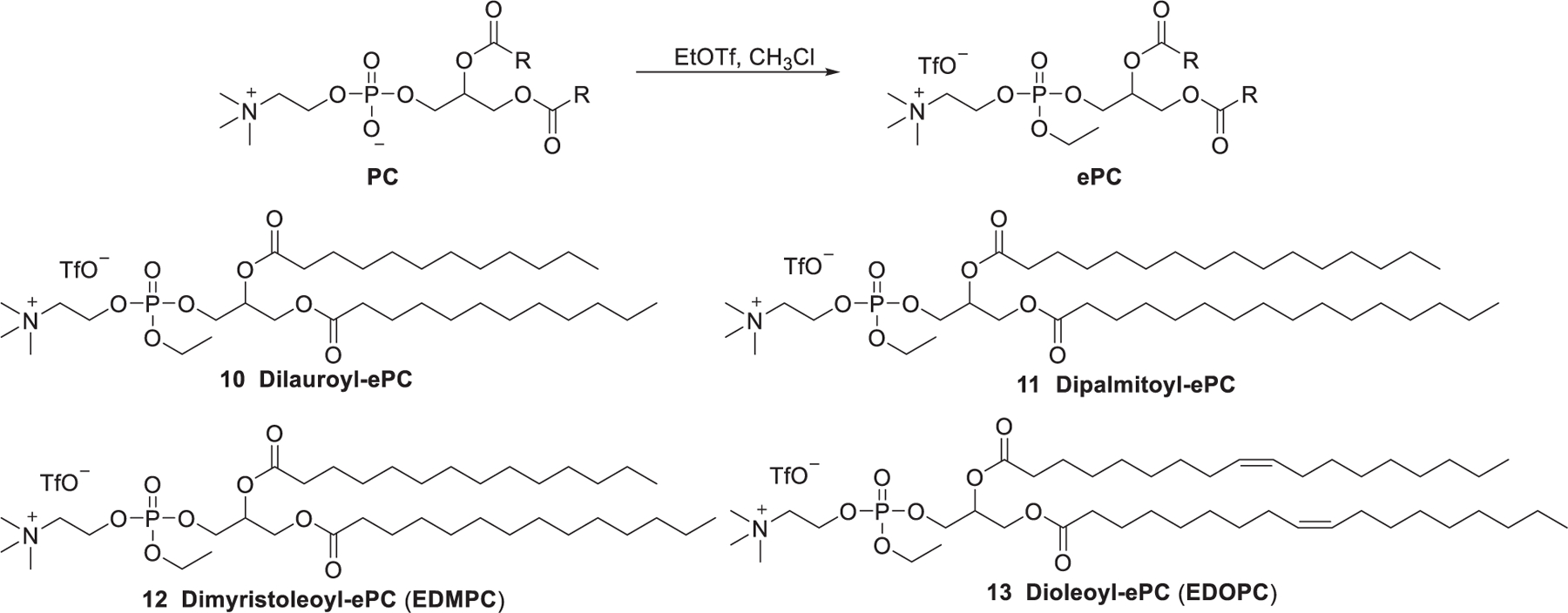
Chemical structures and general synthetic route of ethylphosphatidylcholines (ePCs).
The membrane-disruptive properties of detergent (e.g., Triton X-100 (TX100)) are considered for improving nucleic acids delivery. Pierrat et al. synthesized a panel of cationic phospholipid–detergent conjugates by covalently attaching a detergent molecule (such as Triton X-100 (TX100)) to DOPC (Figure 8).342 Conjugate 14 was able to deliver luciferase siRNA into mammalian cell cytosol without helper lipids, but its application was limited by its high toxicity. To address the toxicity issue, DOPC and Triton X-100 were conjugated through linker groups showing various chemical and biological stabilities (Figure 8).342 Results showed that conjugates 20 and 21 obtained by replacing the phosphoester bond of conjugate 14 with a phospho(alkyl)enecarbonate group showed no loss of siRNA delivery activity, whereas the cytotoxicities of conjugates 20 and 21 were significantly decreased. The conjugates incorporating the succinate moiety (conjugates 18 and 19) showed even lower toxicity along with a reduced siRNA delivery efficiency. The low siRNA delivery efficiency of conjugates 18 and 19 may be attributed to their more labile chemical structures. Another such series of conjugates were synthesized by coupling DOPC with low molecular weight alcohol or carboxylic acids.343 Lipids 30 and 31 efficiently delivered luciferase siRNA into U87 cells, provoking up to 80% of luciferase gene knockdown, whereas the other lipids (24–28) were inactive.343
Figure 8.
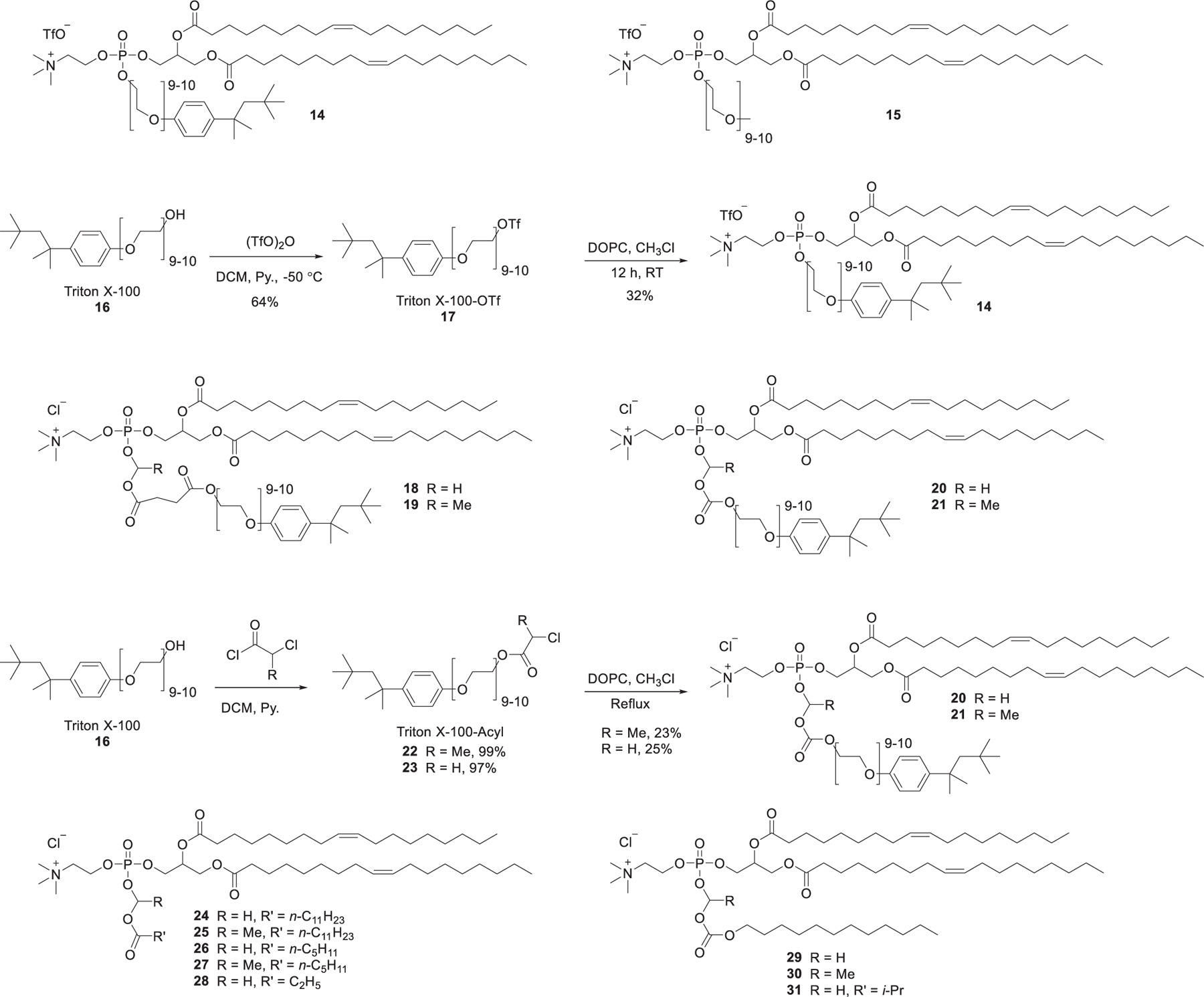
Representative chemical structures and synthetic routes of DOPC-based lipids.
Didodecyldimethylammonium bromide (DDAB) (Figure 9) was previously used to prepare nanoparticles as gene delivery vectors, such as cationic liposomes,344 niosomes,345 and LNPs.346 In 2011, lipid nanoparticles prepared with DDAB/MOG/siRNA showed effective siRNA delivery and induced gene silencing in vitro.347 DDAB-based liposomes have also been used in delivering siRNA to lung metastasized tumor following systemic injection348 and siRNA delivery targeting dendritic cells.349 The quaternary ammonium lipids can be further modified by introducing a second quaternary ammonium group, giving cationic gemini lipids (Figure 9).350,351 In 2001, Rosenzweig et al. developed a class of diquaternary ammonium lipids for DNA delivery via quaternization of their corresponding tetramethyldiamines with alkyl halides.352 A class of dimers of DODAC were synthesized by the Cullis group, among which TODMACS6 exhibited the best delivery ability. Results suggested that the second quaternary ammonium group may strengthen the interactions with DNA, and the delivery efficiency could be tuned by the length of the linker between the two head groups.353 Cardiolipin is a class of natural phospholipids that exists mainly in the heart and skeletal muscles.354 In 2005, Kasireddy et al. synthesized a series of gemini quaternary ammonium cardiolipin analogs (CCLAs) by replacing the two negatively charged phosphate groups of cardiolipin with two quaternary ammonium groups (Figure 9).355 The CCLA-based liposomes, formulated with CCLA/DOPA at a ratio of 1:2, delivered c-raf siRNA efficiently both in vitro and in vivo, inducing up to 62% of tumor growth repression in mice.356 A CCLA-based liposome encapsulating anti-Raf-1 siRNA, designated as NeoPhectin-AT, was shown to repress Raf-1 gene expression and concomitantly downregulate cyclin D1 gene expression.357
Figure 9.
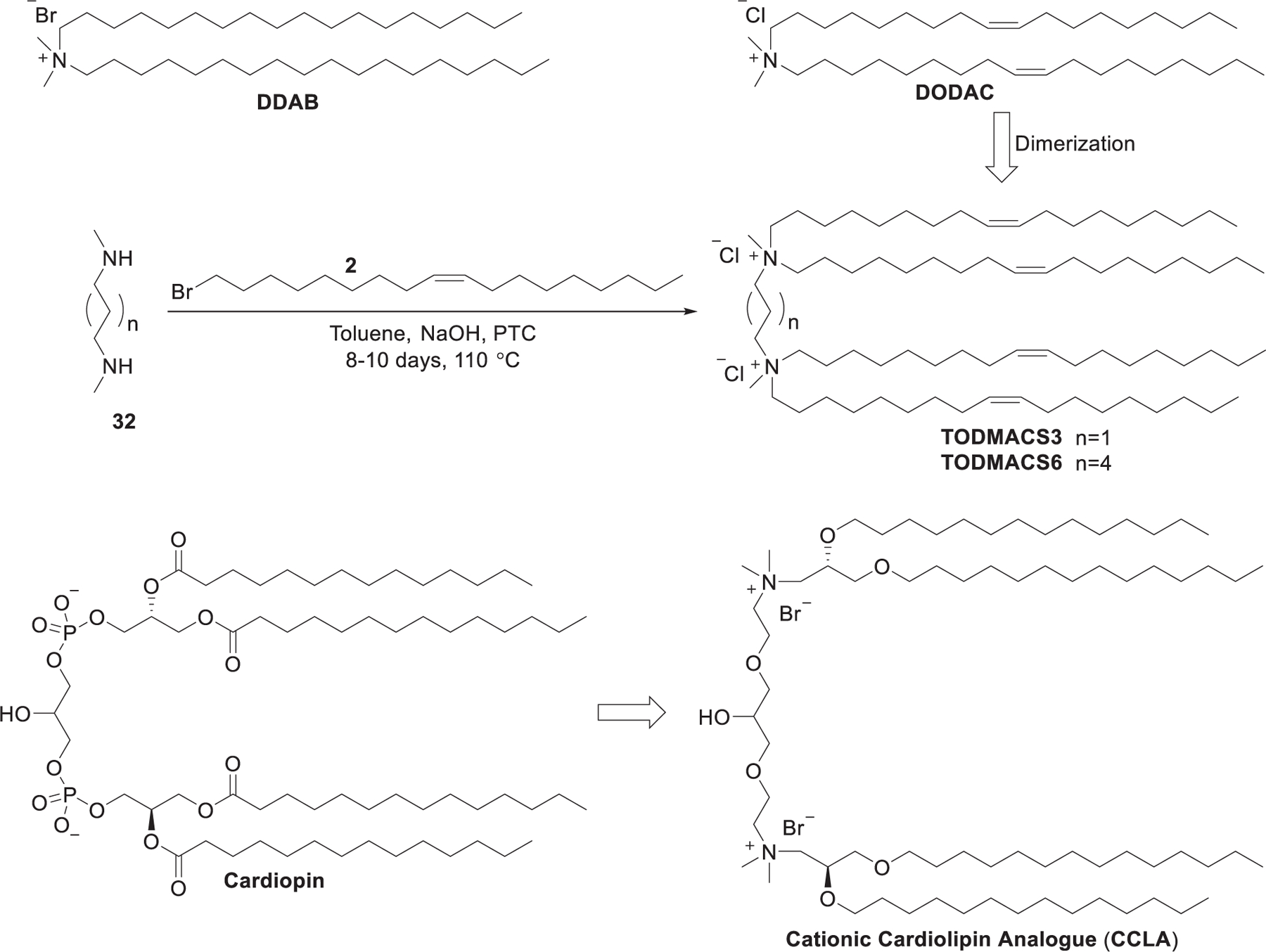
Synthesis of gemini diquaternary ammoniums TODMACS3, TODMACS 6, and CCLA.
Previous reports suggested that the incorporation of carbohydrate in cationic lipids increases the stability of DNA-loading cationic liposomes, decreases cytotoxicity, and enhances DNA delivery efficiency.306,358 Besides, cholesterol is biologically compatible and able to stabilize membranes and form stable liposomes. Maslov et al. synthesized a library of cholesterol-based cationic lipids with morpholinium, pyridinium, or imidazolium as their head groups (Figure 10). They also synthesized other cholesterol-based cationic lipids incorporating a carbohydrate residue with piperidinium, morpholinium, pyridinium, or imidazolium as the head groups.359,360 LNPs formulated with lipid 37/DOPE showed effective EGFP siRNA delivery in vitro and provided pronounced down-regulation of EGFP expression in BHK cells.348
Figure 10.
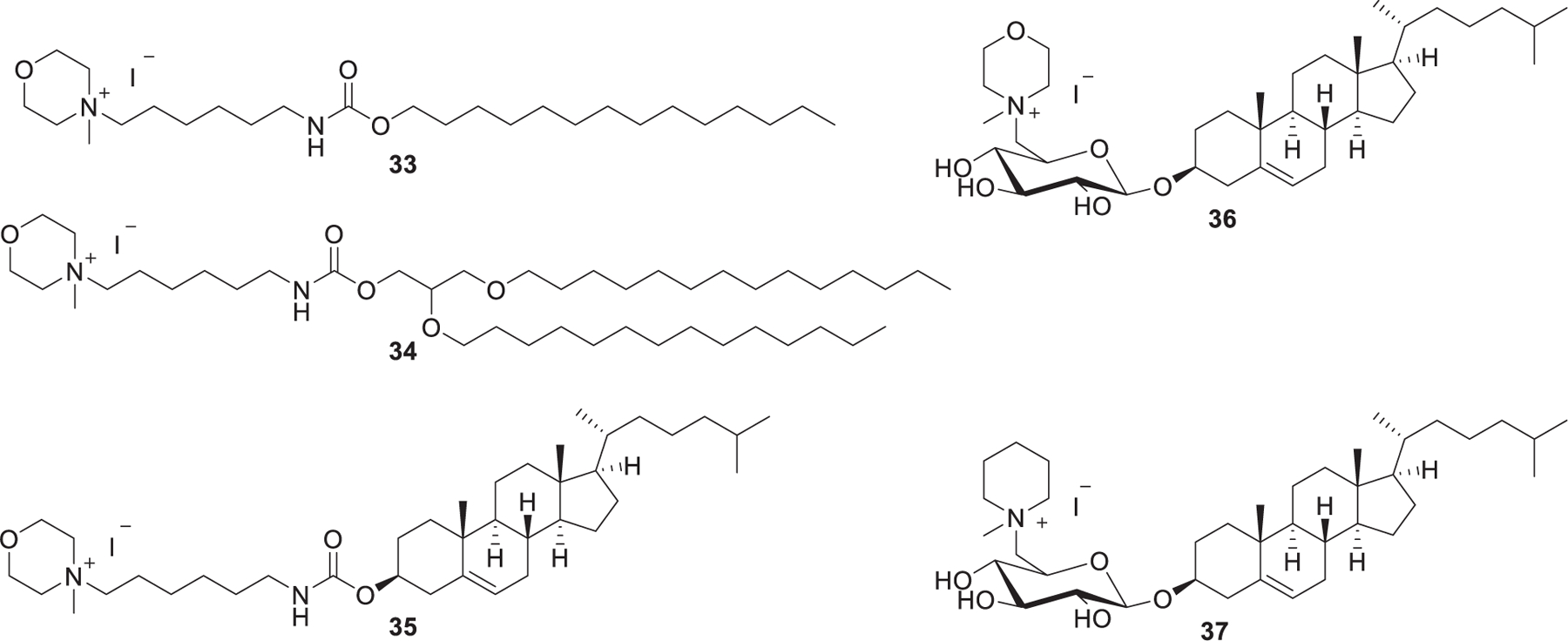
Chemical structures of piperidinium and morpholinium lipids.
Nucleolipids are amphiphilic compounds that possess head groups that can recognize nucleic acids and hydrophobic tails.361–364 Ceballos et al. developed a series of nucleoside lipids containing different bases attached at the anomeric position, a quaternary ammonium head group at the 5′, and two hydrophobic tails at the 2′ and 3′ positions. As for nucleolipids 38–43 (Figure 11), two oleoyls were installed to the 2′ and 3′ positions and 3-nitropyrrole, 5-nitroindole, or 4-nitroimidazole was attached at the anomeric carbon atom with different stereochemistry.365–367 Nucleolipids 44 and 45 are cationic lipids with hydrophobic domains connected to the nucleosides via a ketal linker (44) or an orthoester linker (45), respectively.367,368 The synthesis of nucleolipid 38 started with the selective protection of the hydroxy groups of d-ribose 46, giving protected sugar 47.369 Next, the protected sugar 47 was stereoselectively chlorinated with hexamethylphosphorus triamide (HMPT) and CCl4370 followed by attachment of 3-nitropyrrole.371 Removal of the TBS protecting group gave alcohol 49, which was tosylated, desilylated, and coupled with oleic acids to afford compound 50. The final quaternization reaction with trimethylamine afforded the nucleolipids 38. In the case of nucleolipid 45, the starting material 2′-deoxythymidine 53 was methanesulfonylated with methanesulfonyl chloride and the resulting compound underwent a coupling reaction with trihexadecyl orthoformate 52 promoted by pyridinium p-toluenesulfonate (PPTS).372 siRNA delivery by nucleolipid 38 LNPs resulted in protein knockdown in several cell lines, such as hamster ovarian cells, mouse fibroblast cells, and human liver cells. In the following studies based on lipids 40–42, results showed that both the stereochemistry at the anomeric carbon and the properties of the bases affected the formation of nucleolipids–siRNA complexes. Nucleolipid 45 LNPs were shown to be able to deliver the human RecQ helicase (RECQL4) siRNA into tumor cells. The cleavage of the orthoester group might promote endosome escape via the in situ generation of fatty alcohol.367,368
Figure 11.
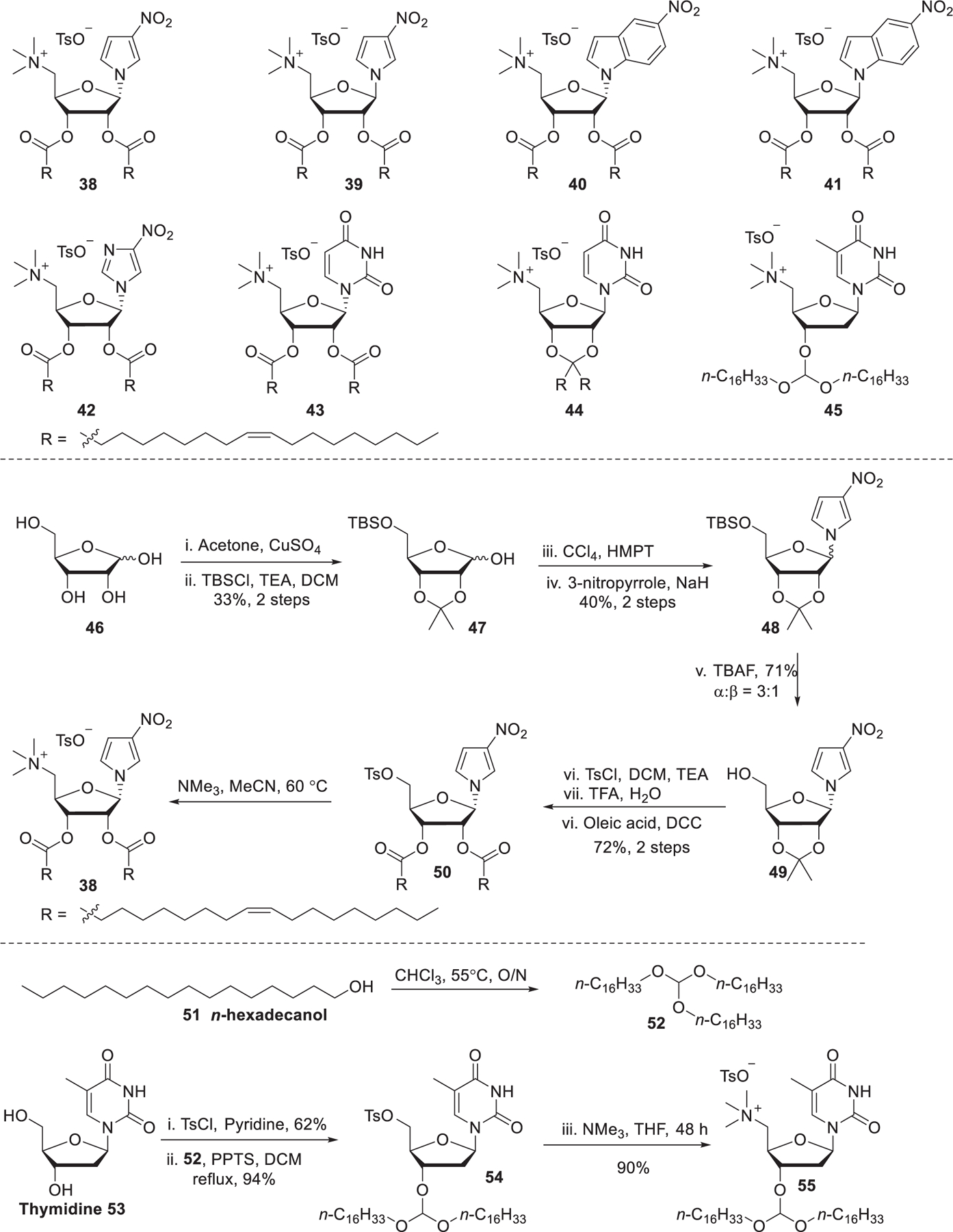
Chemical structures and synthetic routes of nucleoside derived lipids.
As mentioned in previous sections, ePCs are quaternary cationic lipids with two nonrigid hydrophobic tails, while DC-Chol is an example of a lipid with a rigid cholesterol component that has been reported to be efficient in DNA delivery (Figure 12).373,374 It was reported that rigid cationic lipids could self-assemble into tightly packed nanoparticles due to limited motional flexibility at the hydrophobic domain.375 In 2012, Pungente et al. developed five carotenoids-derived single-tailed rigid cationic lipids, which were expected to be capable of self-assembling and delivering siRNA efficiently.376 As shown in Figure 12, cationic lipids 56–60 contain the same rigid C30-carotenoid hydrophobic tails and different cationic head groups. The synthesis of these lipids started with the hydrolysis of ethyl-β-apo-8′-carotenoate 61, affording β-apo-8′-carotenoic acid 62. Then, these lipids were obtained via esterification followed by amination or quaternization. Results indicated that these single-tailed rigid cationic lipids were able to deliver siRNA to eukaryotic cells in vitro. Cationic lipids containing quaternary ammonium head groups with a hydroxyethyl moiety (lipids 58–60) showed enhanced siRNA delivery efficiency. Additionally, introducing a second hydroxyethyl group to the head group (lipids 59 and 60) increased the cytotoxicity without enhancing siRNA delivery efficiency.376
Figure 12.
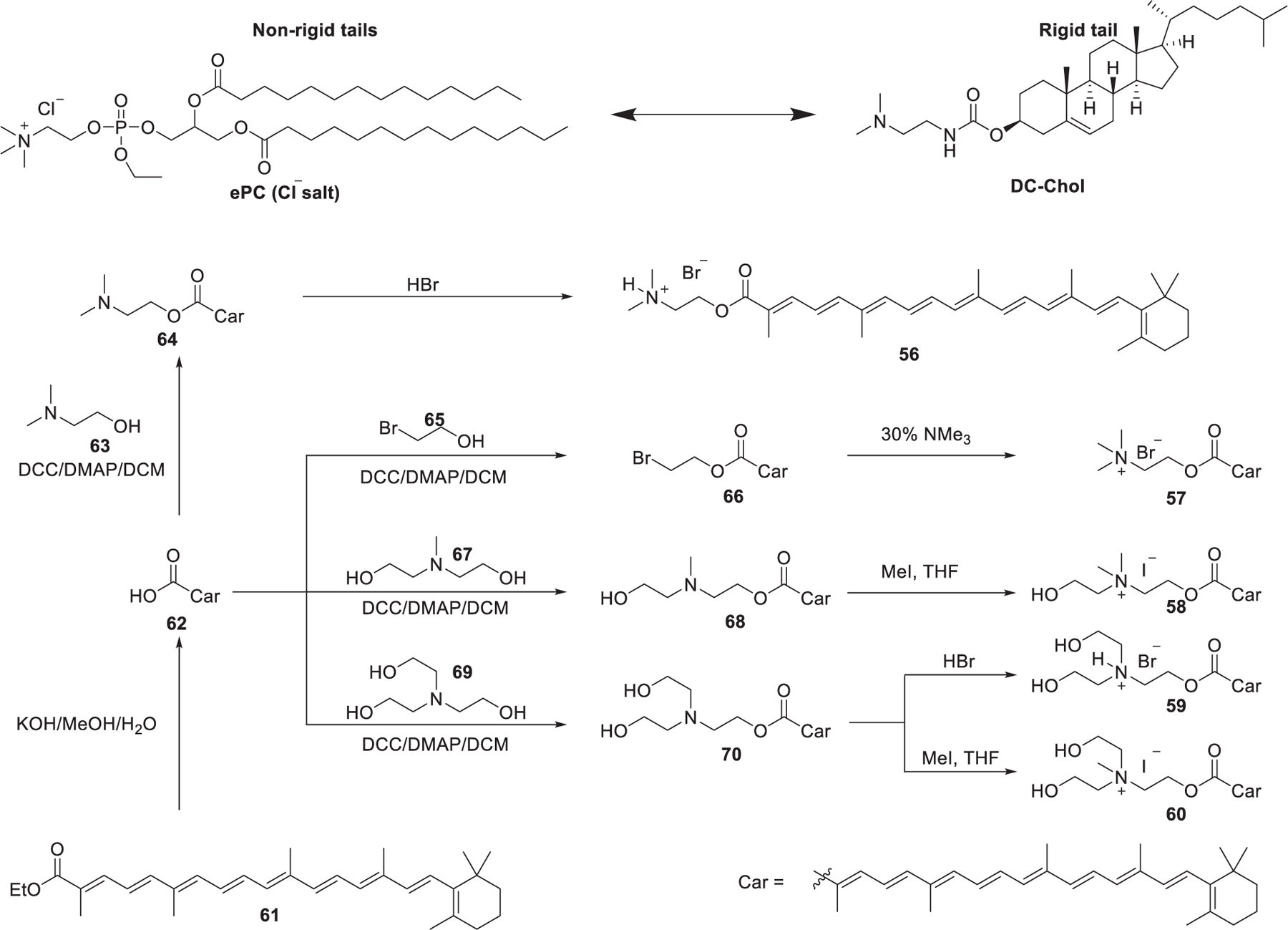
Chemical structures and synthetic route of carotenoid-derived lipids
It is known that sialic acid (SA) is overexpressed on the surface of a variety of cancer cells377,378 and phenylboronic acid (PBA) is an effective targeting ligand for improving cancer cell recognition and adhesion via interacting with SA.379–381 In 2019, Tang et al. synthesized a cationic lipid incorporating a phenylboronic acid ligand, designated as PBA-BADP,382 via quaternization of the bioreducible ionizable lipid BADP developed by Wang et al. (Figure 13).383 PBA-BADP LNPs showed an enhanced delivery of luciferase mRNA compared with BADP NPs in SA-overexpressing HeLa cervical cancer cells. Besides, PBA-BADP LNPs selectively delivered firefly luciferase (FLuc) mRNA to cancer cells including HeLa cells and DU145 cells rather than noncancer HK-2 and CCC-HPF-1 cells. PBA-BADP LNPs encapsulating Cas9 mRNA and HPV18E6 sgRNA induced 18.7% HPV18E6 gene knockout efficiency in HeLa cells.382
Figure 13.
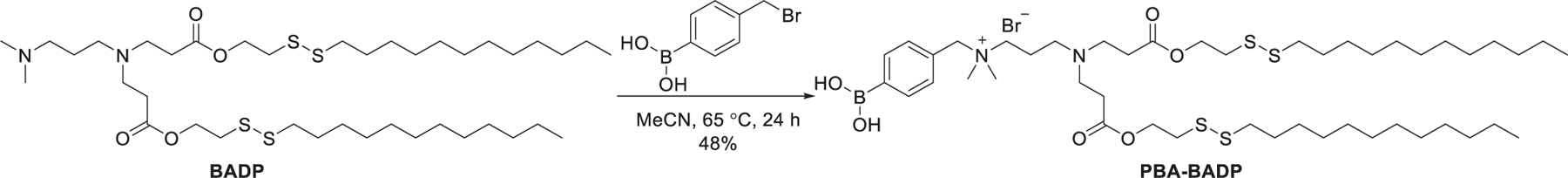
Chemical structure of PBA-BADP.
2.1.2. Guanidinium Lipids.
Potential toxicity is one of the challenges for the application of quaternary ammonium-based delivery materials.384,385 One strategy to overcome this issue is to delocalize the permanently positive charge of the cationic head group. Thus, cationic lipids with amidine, guanidinium, pyridinium, or imidazolium head groups are developed. Lipid diC14-amidine (Vectamidine) with an amidine head group is an early example of this class of lipids to delocalize the positive charge (Figure 14).386
Figure 14.
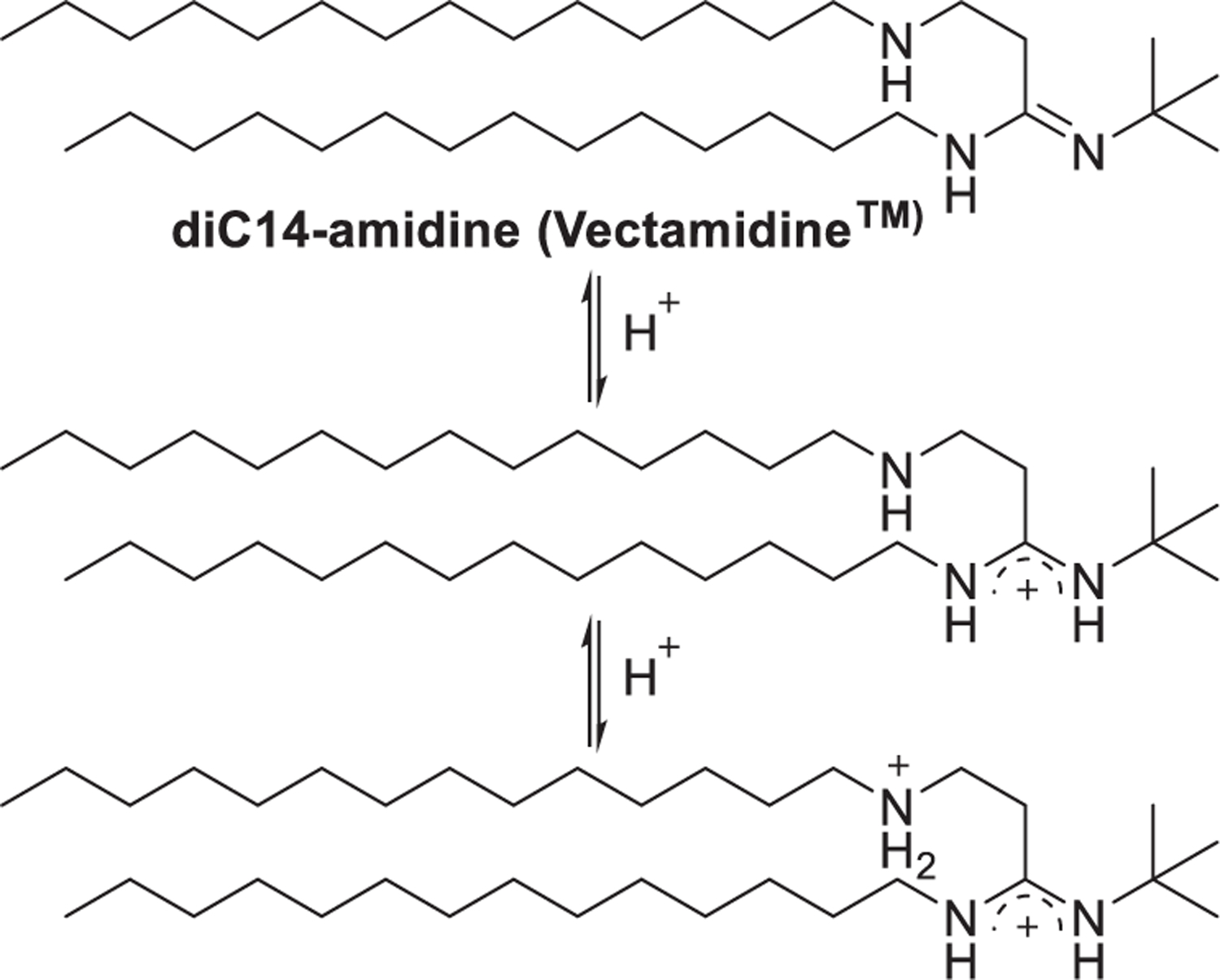
Chemical structure of vectamidine and the delocalization of the positive charge.
Cationic lipids incorporating a guanidinium functional group represent another option for RNA delivery because guanidinium has the following characteristics: (a) The guanidine is a basic functional group with a pKa value at about 13.5, so it can be protonated over a wide range of pH environments, resulting in a permanently positively charged guanidinium head group at physical pH. (b) The guanidinium group interacts with the phosphate backbone of nucleic acids, thus facilitating their encapsulation.387 (c) Hydrogen bonding between the guanidinium group and the RNA phosphate backbone can also enhance nucleic acids entrapment. (d) The guanidinium group binds with negatively charged proteoglycans on the cell membrane, thus enhancing cell uptake of the nanoparticles.388–390
Several groups have chosen arginine, a natural amino acid containing a guanidinium group, as the starting material to develop guanidinium type cationic lipids. In 2006, Santel et al. synthesized an arginine-derived guanidinium lipid AtuFECT01 (Figure 15).391 Formulated with commercially available helper lipids DPhyPE and DSPE-PEG, the resulting siRNA-Lipoplex was delivered to the tumor endothelial cell following intravenous administration, resulting in reduced Tie2 and CD31 expression in the vasculature of mice.391,392 Aleku et al. used Atu027 for the inhibition of protein kinase N3 (PKN3) in endothelial cells for the treatment of prostate and pancreatic cancers in mice;393,394 the favorable preclinical data led to the clinical trial of Atu027.395,396 A phase Ib/IIa study of synergistic therapy of pancreatic adenocarcinoma with Atu027 and gemcitabine showed this therapy for pancreatic carcinoma was safe and well-tolerated.396 In 2009, Chen et al. synthesized a series of guanidinium lipids which contain an l-lysine residue as well as a guanidinium functional group as the head group, such as DSGLA (Figure 15).397 siRNA encapsulated in the LNPs containing DSGLA showed enhanced cellular uptake and induced stronger gene silencing in H460 tumor cells both in vitro and in vivo as compared to that formulated with DOTAP.397 In their following work, they used liposome–polycation–DNA (LPD) nanoparticles containing DSAA to codeliver VEGF siRNA and Dox. Results showed that DSAA acted as an agent that increased the sensitivity of MDR cells to chemotherapy drugs and inhibited the expression of MDR transporters.398
Figure 15.
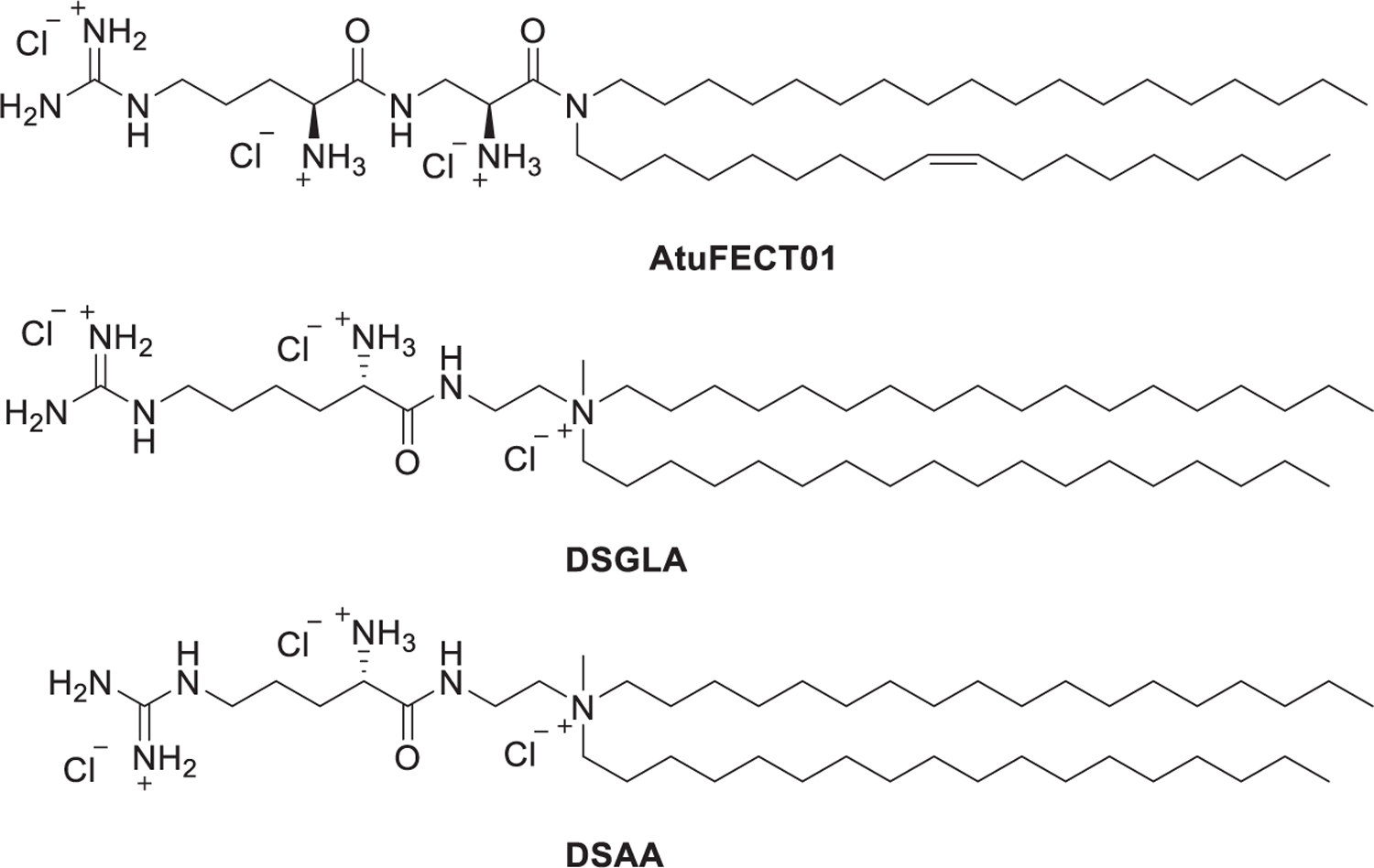
Chemical structures of AtuFECT01 and DSGLA.
In 2010, Mevel et al. reported the synthesis of several cationic lipids comprising a dialkyl glycyclamide or cholesteryl-moiety conjugated with a guanidinium head group (Figure 16).399 The synthesis of DODAG-9 was accomplished in three steps. The starting N-Boc-glycine 71 was coupled with dioctadecylamine 72 via amide bond formation to give amide 73, which was treated with TFA to remove the Boc protecting group, leading to the key glycine amide intermediate 74. Conjugation of intermediate 74 with the guanidinylation reagent, 1H-pyrazole-1-carboxamidine monohydrochloride 75,400 in ethanol afforded DODAG-9. DODAG-9 was used to deliver antihepatitis B virus (HBV) siRNAs to the murine liver in vivo.399
Figure 16.
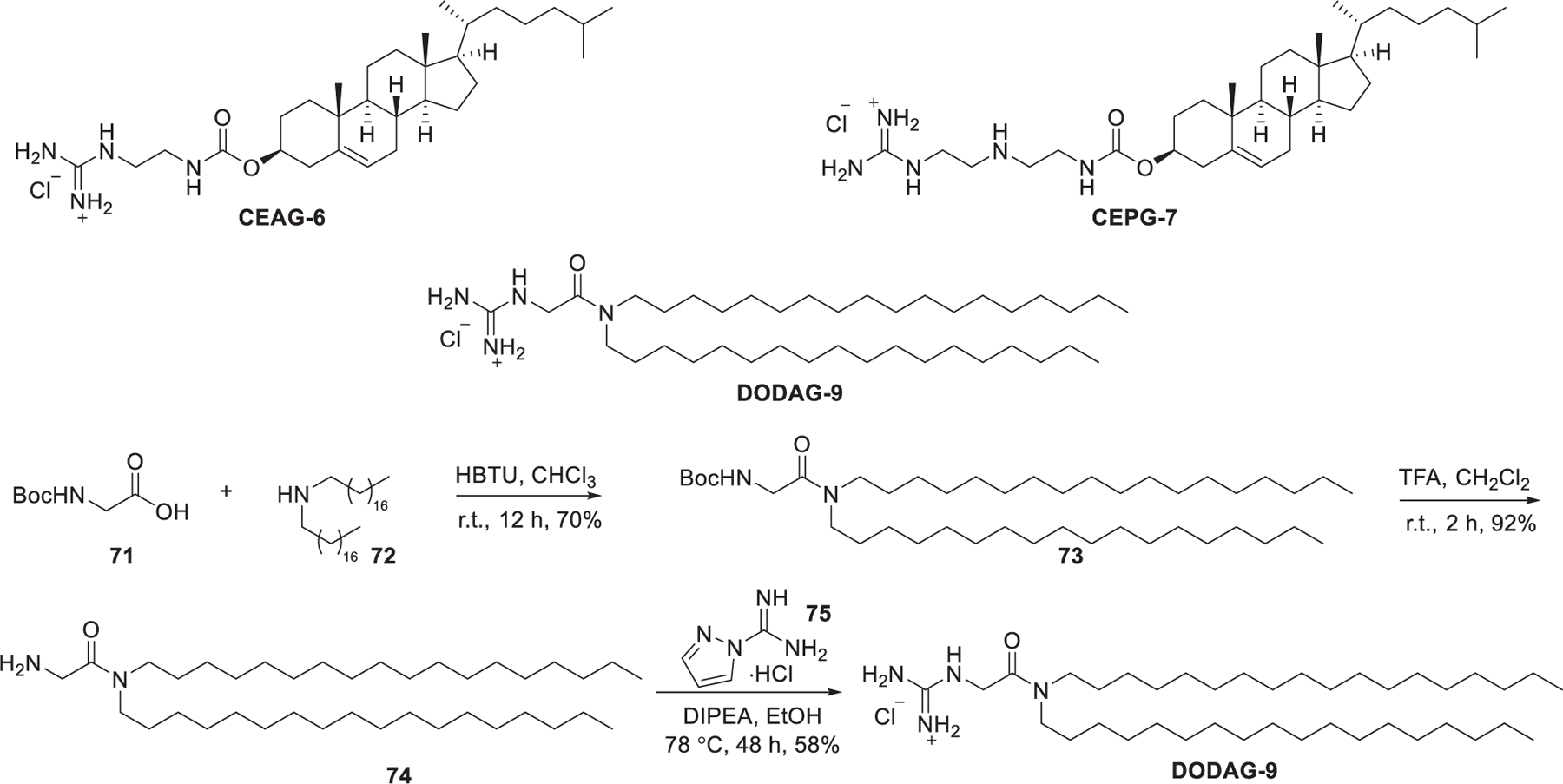
Chemical structures of guanidinium lipids and synthetic route of DODAG-9.
In 2011, Adami et al. created a library of guanidinium type lipids termed DiLA2 based on natural and modified arginine for siRNA delivery (Figure 17).401 The amino groups and carboxyl groups on these compounds are reaction sites for attaching hydrophobic tails with various unsaturation degrees and lengths. A series of DiLA2 analogs were synthesized based on nornorarginine, norarginine, l-arginine, and homoarginine; two sets of short hydrocarbon tails (C8 and C10) were attached to the α-amino group and the carboxyl group. In vitro screening of this panel of DiLA2 led to the identification of norarginine as the desirable amino acid to build the DiLA2 library. Then, a series of symmetric norarginine DiLA2 was synthesized by incorporating hydrocarbon tails from C12 to C18, and the asymmetric C18:1-norArg-C16 was synthesized by incorporating a C18:1 tail along with a C16 tail.401 C18:1-norArg-C16 was identified as the best-performing lipid in this library, which could deliver FVII siRNA efficiently, leading to 90% inhibition of FVII mRNA at a dose of 1 mg/kg after intravenous administration in mice.401 In 2020, Sanchez-Arribas et al. synthesized another arginine-based double-chained guanidinium type lipid, designated as C12ANHC18, which consists of the arginine residue linked to a 12-carbon atom alkyl chain and an unsaturated C18 alkyl chain.402 The C12ANHC18 LNPs lipoplexes could deliver GFP siRNA efficiently into HeLa cells and T731 cells in vitro.402
Figure 17.
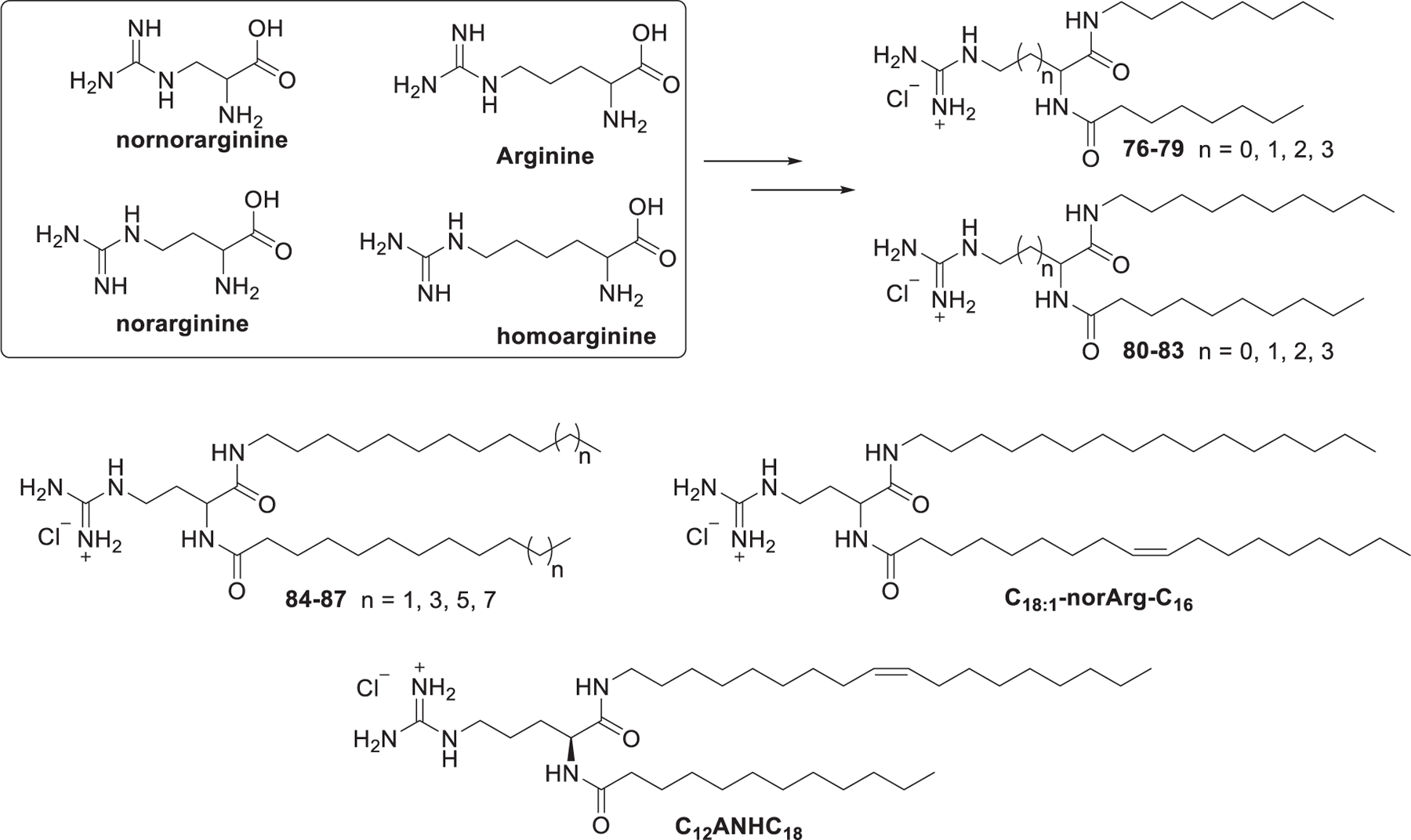
Chemical structures of guanidinium DiLA2 compounds and C12ANHC18.
In 2011, Metwally et al. synthesized four guanidinium derivatives of N4,N9-diacylated spermine,403 based on the symmetrical fatty acid amides of spermine (Figure 18).404 Starting from the selective protection of the two primary amino groups of spermine 92 with ethyl 1,3-dioxoisoindoline-2-carboxylate 93, followed by coupling of amine 94 with oleic acid 95, amides 96 was obtained. Removal of phthalimides protecting group with ammonium hydroxide gave diamine 97, which was treated with 1,3-di-Boc-2-(trifluoromethylsulfonyl)-guanidine 98 to install the protected guanidinium group, affording compound 99. Guanidinium lipid 90 was obtained after deprotection of 99 with TFA in DCM (Figure 18). These guanidinium lipids efficiently bound siRNA and formed the corresponding nanoparticles for siRNA delivery in HeLa cells. LNPs formulated with guanidinium lipid 90 were able to deliver GFP siRNA into HeLa cells, leading to a reduction of GFP expression by 26%.404
Figure 18.
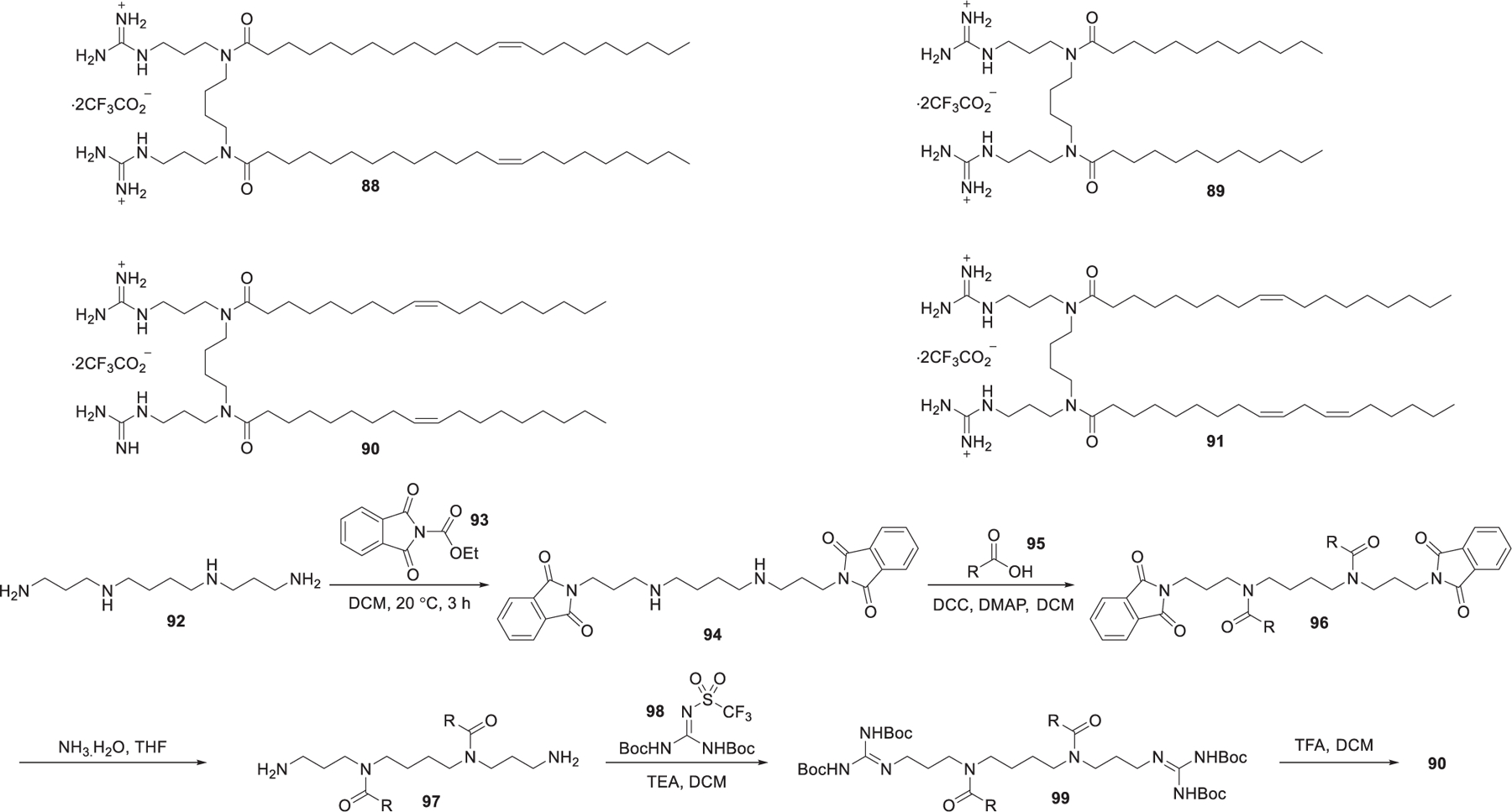
Chemical structures and synthesis of N1,N12-diamidino-N4,N9-diacylated spermines.
Squalene, a natural precursor for the synthesis of cholesterol, has low toxicity and is well-tolerated in animal tests.405 It has been used as a helper lipid, in substitution of cholesterol, in the preparation of cationic niosomes used in gene delivery.406 Bertrand et al. reported the delivery of anti-EWS/Fli-1 siRNA into A673 cells in vitro by taking advantage of squalene-derived guanidinium lipid 100 (SQ-NH(NH2)C=NH2+, AcO−) (Figure 19).407 Cholesterol-derived cationic lipid may function as not only a helper lipid but also a complexation agent of RNA in the preparation of LNPs encapsulating RNA molecules, as cholesterol is an important stabilizer in the preparation of lipid-based nanoparticles. In 2015, Jon et al. reported an arginine–cholesterol-derived guanidinium lipid named MA-Chol (Figure 19).408 MA-Chol was synthesized via the coupling of the protected form of arginine [Boc-Arg(Pbf)-OH] 101 and cholesterol followed by deprotection with TFA. Systemic administration of anti-PSK siRNA-loaded MA-Chol LNPs resulted in preferential accumulation of siRNA at the tumor site and ~81% suppression of tumor growth at a dose of 1 mg/kg in mice.408
Figure 19.
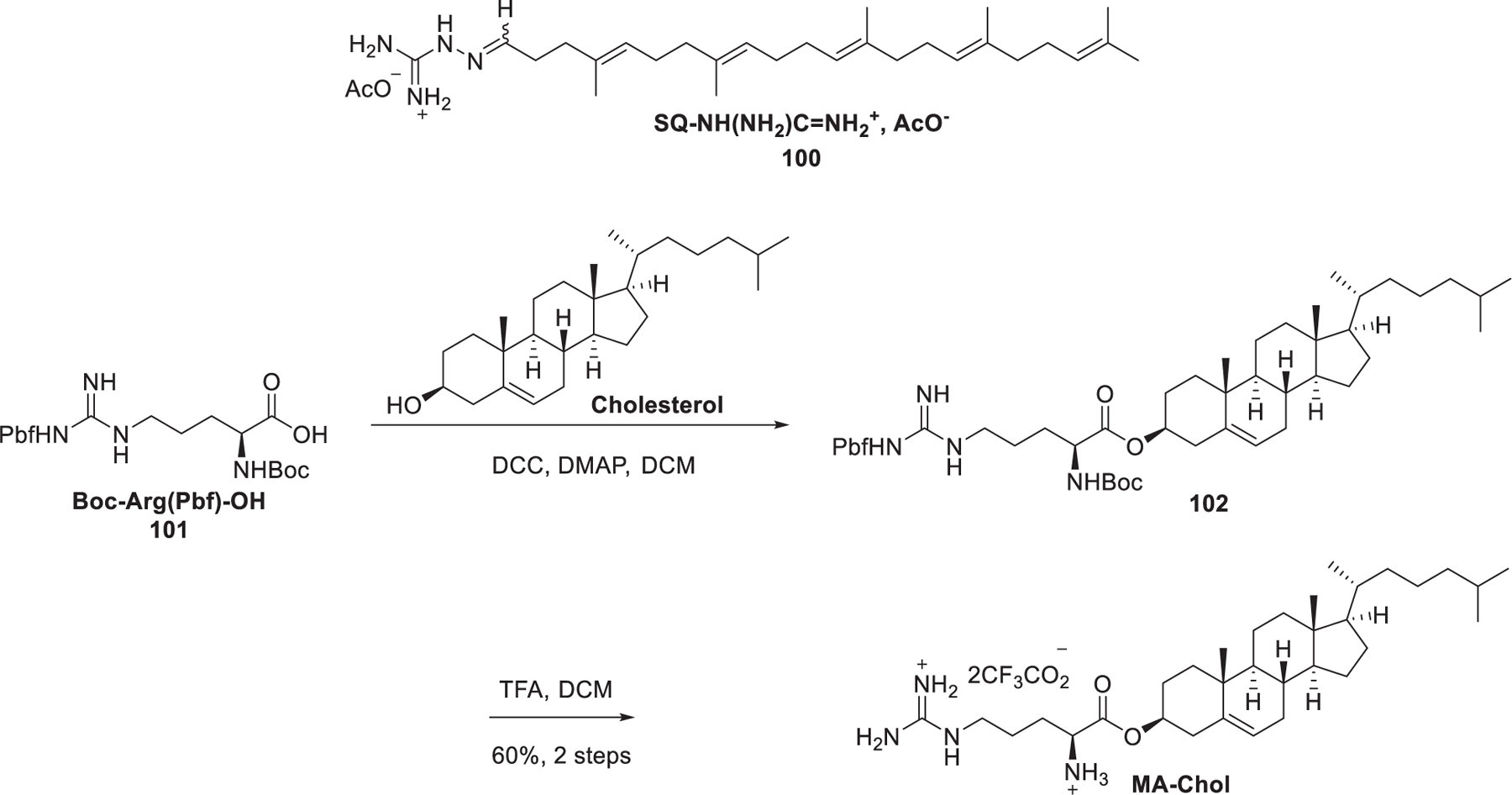
Chemical structures of squalene and cholesterol-derived guanidinium lipids.
In 2018, Bang et al. prepared a library of guanidinium lipids with arginine, oleyl amine, and cysteine as the building blocks (Figure 20).409 A cysteine was incorporated in the peptides because cysteine has been shown to increase intracellular delivery.410 These compounds were composed of two linear peptides (H-RCL and Bz-RCL) and two branched peptides (Tri-RCL and Di-RCL). Results showed that H-RCL and Bz-RCL were able to effectively deliver GAPDH siRNA in HeLa cells, while Di-RCL and Tri-RCL exhibited low delivery efficiency. The incorporation of the hydrophobic benzoyl group in Bz-RCL may improve its interaction with the cell membrane and consequently enhance siRNA delivery compared to H-RCL LNPs.409
Figure 20.
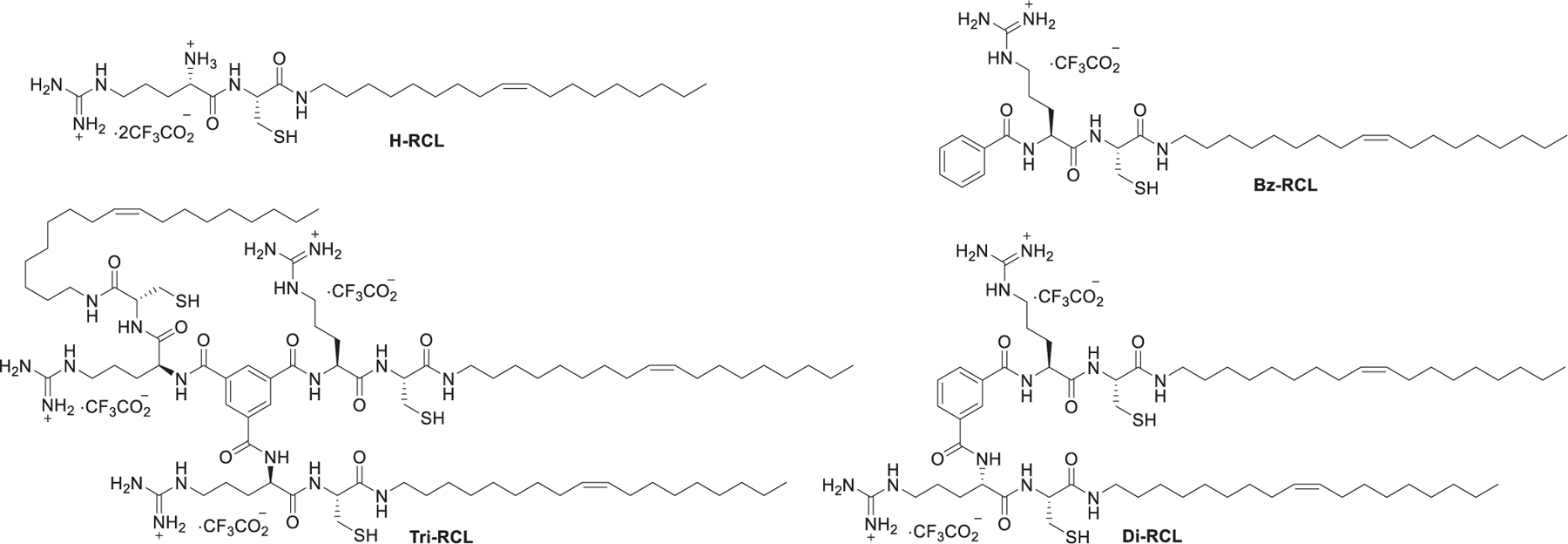
Chemical structures of arginine- and cysteine-derived guanidinium type lipids.
2.1.3. Pyridinium Lipids.
Apart from the guanidinium head group, the permanent positive charge can also be delocalized in heterocyclic rings, such as pyridinium rings and imidazolium rings. The pyridinium cationic lipids generally have lower cytotoxicity compared to lipids with quaternary ammonium head groups.300
In 1997, van der Woude et al. developed a class of pyridinium lipids, termed synthetic amphiphiles interdisciplinary (SAINT) (Figure 21).299 As shown in Figure 21, the synthesis of SAINT began with the reactions of dialkylation of 4-methylpyridine 103 and various alkyl bromides, giving alkylpyridine 104, which was quaternized with methyl iodide to afford N-alkylpyridinium iodide 105. Finally, N-alkylpyridinium iodide 105 was treated with ion exchange resins (Cl− form) to obtain the N-alkylpyridinium chloride salts.299 SAINT-C18 liposome (SAINT-O-Somes) could selectively and effectively deliver anti-VCAM-1 siRNA and anti-E-selectin siRNA into inflammation activated primary endothelial cells in vitro, inducing significant downregulation of target genes.411,412 In 2014, they further reported that specific PEGylated SAINT-C18 LNPs could deliver anti-VCAM-1 siRNA to activate endothelial cells in vivo, resulting in attenuation of VE-cadherin gene expression after intravenous administration.413
Figure 21.
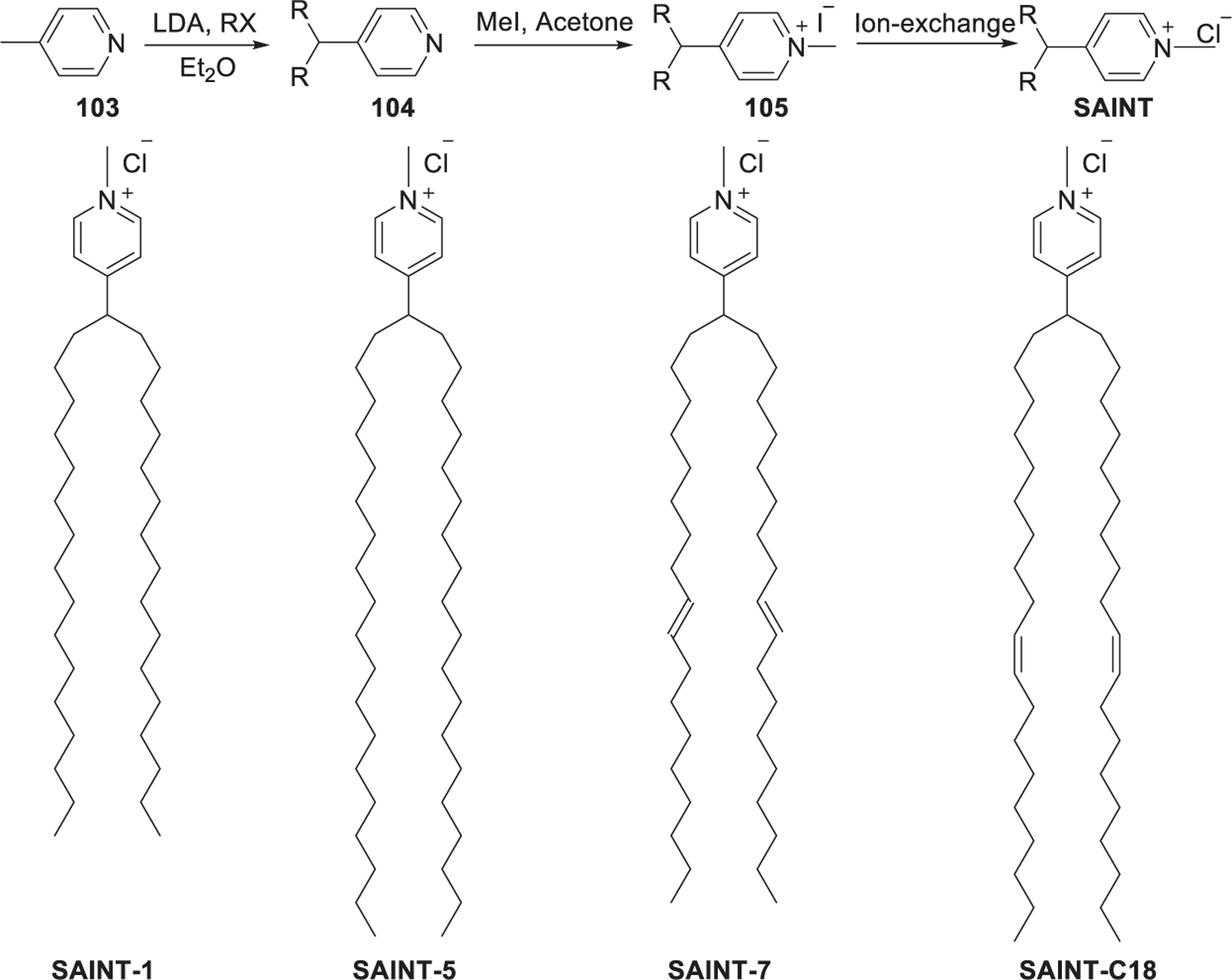
Chemical structure and synthesis of SAINT.
In 2010, Maslov et al. prepared cholesterol derived pyridinium lipid 111 (Figure 22), in which the pyridinium head group is attached to C-6 of the carbohydrate β-glucosyl spacer.359 Cationic lipids with imidazolium or morpholinium or piperidinium as the head groups were also synthesized. Condensation of acetobromoglucose 106 and cholesterol in the presence of Hg(CN)2 gave glucosides 107. Removal of the acetyl groups with sodium methoxide gave cholesteryl β-d-glucoside 108, of which the C-6 hydroxy group was regioselectively mesylated followed by acylation of the other hydroxy groups to afford compound 109. After direct quaternization of pyridine with 4 and deacylation, pyridinium lipid 111 was obtained. LNPs containing lipid 111 showed effective EGFP siRNA delivery and down-regulation of EGFP in BHK IR780 cells in vitro.359
Figure 22.
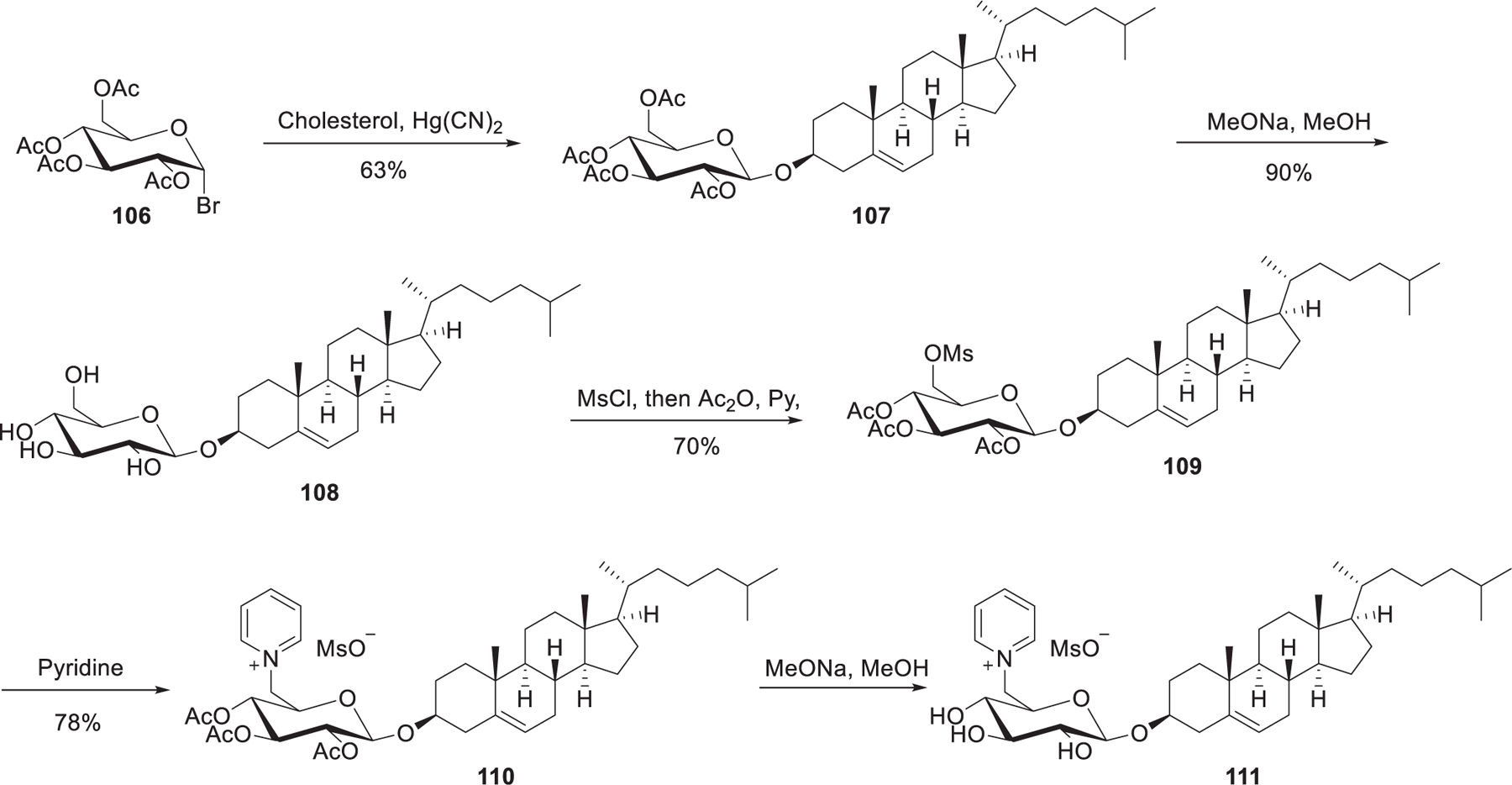
Synthesis of cholesterol-derived pyridinium lipids.
In 2013, Maslov et al. developed a series of pyridinium cationic lipids that contained various hydrophobic domains, including tetradecanol, dialkyl glycerol, and cholesterol (Figure 23).359,360 Carbamates 118 were obtained via coupling of 6-amino-1-hexanol 117 with tetradecanol 115 promoted by N,N′-carbonyldiimidazole (CDI). Bromination of the hydroxy group in compounds 118 followed by quaternization with pyridine gave cationic lipid 112. Results showed the type of hydrophobic tails determines the delivery activity of siRNA; LNPs containing lipid 113 exhibited better activity in siRNA delivery in vitro than that of 112 and 114.345
Figure 23.
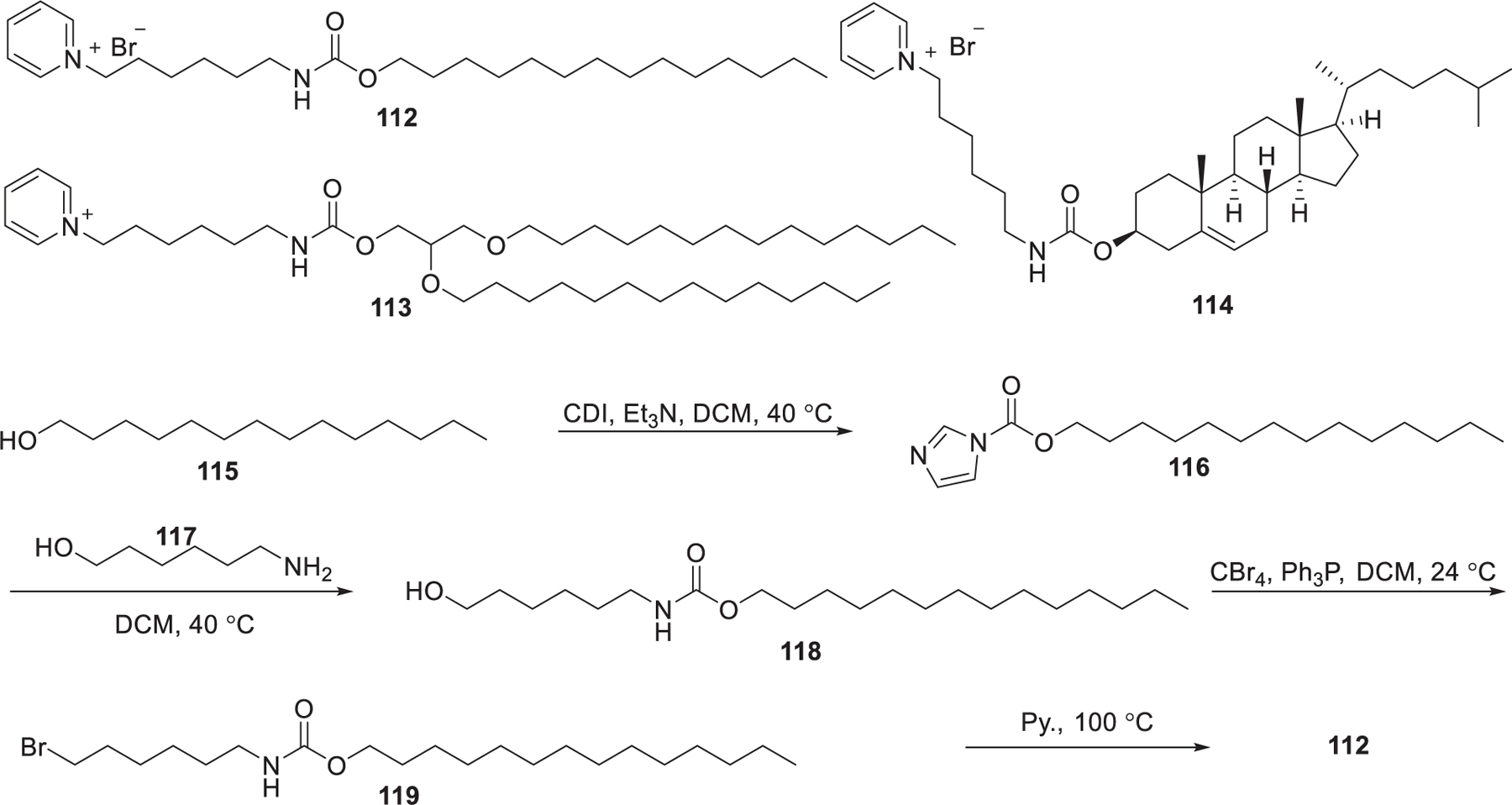
Chemical structures and synthesis of pyridinium lipids
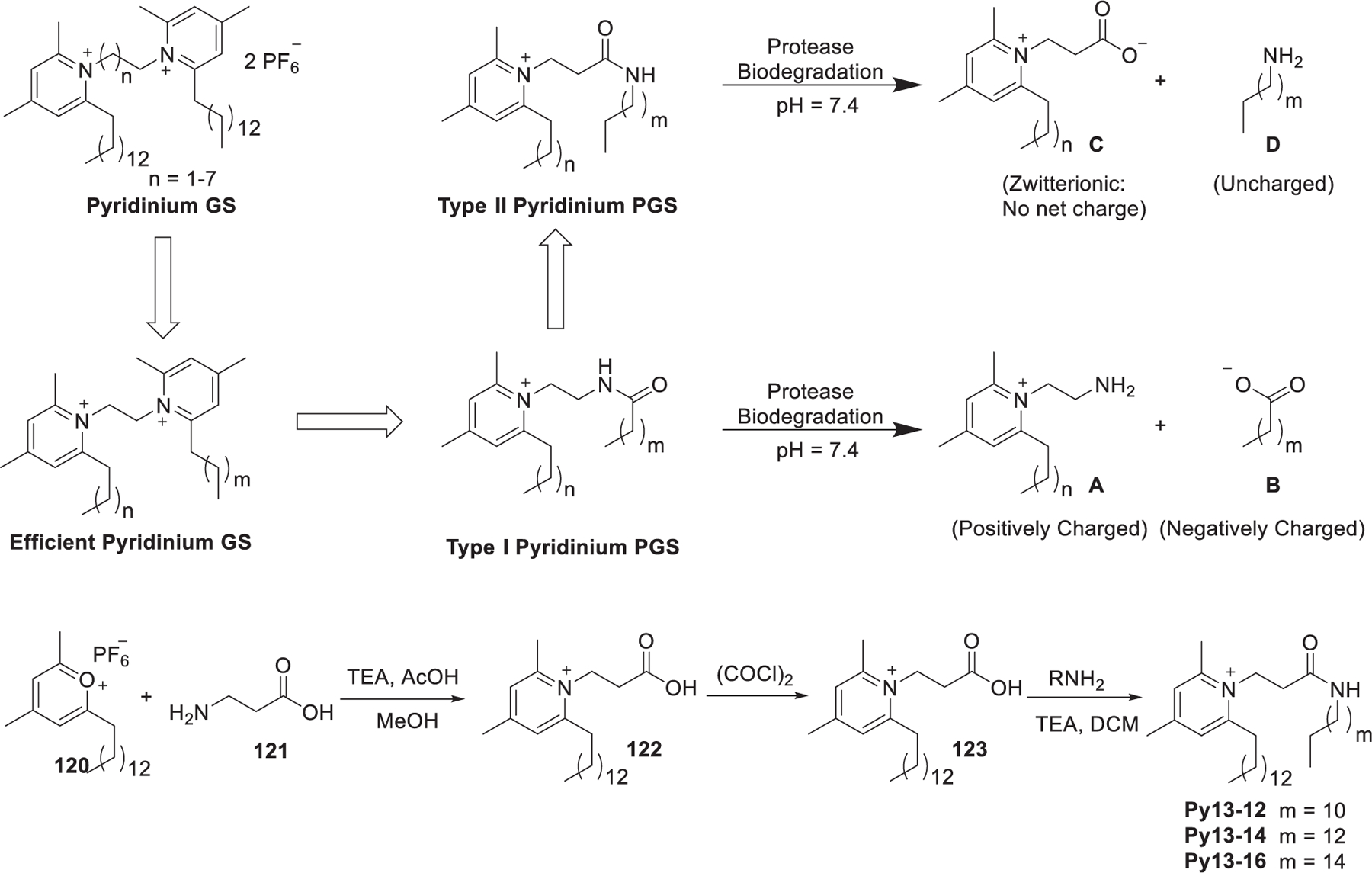
Pyridinium gemini surfactants (GSs), with a higher charge/mass ratio than pyridinium type lipids, can generate lipoplexes with smaller size.414,415 Structure–activity relationship studies of GSs in DNA delivery showed that the gene delivery efficiency of GSs was similar to or higher than that of their pyridinium lipid analogs.299,416,417 GSs with C2 spacing showed higher delivery efficiency than the other analogs. Although the relatively high molecular curvature of GSs is beneficial for increasing the delivery efficiency, it will increase their cytotoxicity.418–421 In 2017, Satyal et al. developed a class of analogs of GS, named pyridinium pseudogemini surfactants (PGS), in which one of the pyridinium head groups were replaced by a noncharged polar moiety that were capable of biodegradation and hydrogen bonding (Figure 24).422 The pyridinium pseudogemini surfactants (PGS) can mimic the tapered shape of pyridinium gemini surfactant (GS) with one positive charge.423 These pyridinium lipids can be hydrolyzed by amidase into neutral components, thus reducing their potential cytotoxicity. Type I PGSs showed efficient delivery of pDNA and siRNA toward several cell lines.416 To further reduce the cytotoxic effect, type II PGSs are designed in which the position of the amide was switched. Upon biodegradation, type II PGSs generate two species with no net charge and therefore display much lower cytotoxicity. The type II pyridinium PGSs were synthesized via the reaction of pyrylium salts 120 with the amino acid 121 to generate the substituted pyridinium head group followed by amide bond formation (Figure 24). Py13-16/DOPE was shown to be an efficient formulation for the delivery of pDNA, siRNA, and mRNA in vitro.422
Figure 24.
Design, synthesis, and proposed biodegradation pattern of pyridinium psudogemini surfactants.
2.1.4. Imidazolium Lipids.
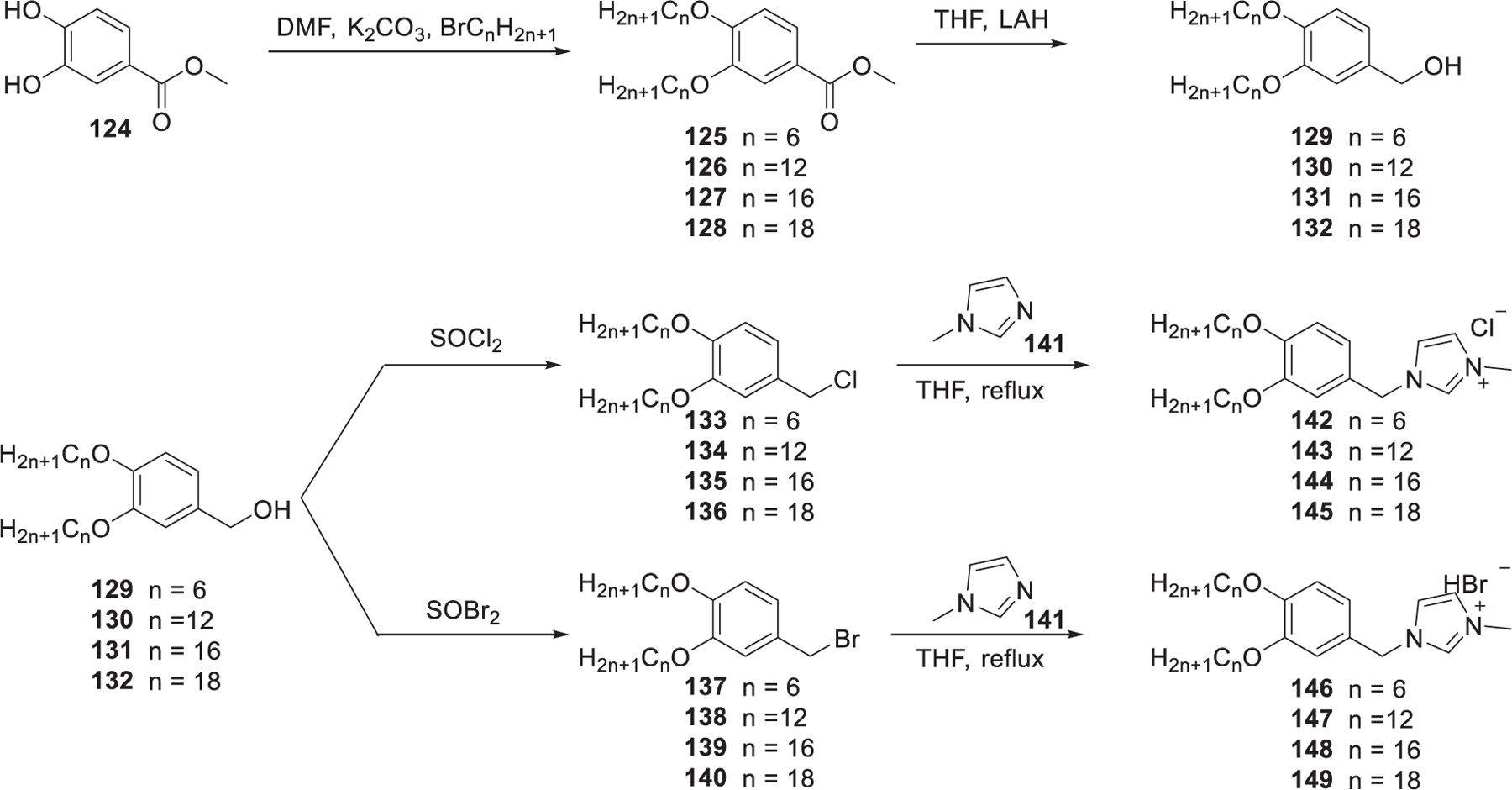
In 2009, Dobbs et al. developed two series of imidazolium lipids, chloride (142–145) and bromide (146–149) derivatives of 1-methyl-3-[3,4-bis(alkoxy)-benzyl]4H-imidazolium with different lengths of hydrophobic tails (Figure 25).424 These lipids were prepared in three steps, following a slightly modified literature procedure.425 Methyl 3,4-dihydroxybenzoate 124 was used as the starting material, which was etherified with 1-bromoalkanes 125–128 in the presence of potassium carbonate in DMF followed by reduction of the ester group, giving benzyl alcohols 129–132. Bromination or chlorination of benzyl alcohols 129–132 was carried out with SOBr2 or SOCl2 as the solvent, respectively; the resulting 3,4-bis(alkoxy)benzyl chlorides or bromides were finally converted to the desired imidazolium lipids via quaternization with 1-methylimidazole 141. Both 143 LNPs and 147 LNPs could induce 80% inhibition of the luciferase gene in A549-Luc cells at the anti-Luc siRNA concentration of 10 nM. Structure–activity relationship analysis showed that imidazolium lipids containing dodecyl tails (143, 147) showed enhanced siRNA encapsulation efficiency and higher siRNA delivery efficiency than other analogs.424
Figure 25.
Chemical structures and synthesis of imidazolium lipids.
In 2011, Perche et al. reported the preparation of histidylated LNPs by incorporating imidazole/imidazolium lipophosphoramidate lipids (Figure 26).426,427 Results showed that histidylated LNPs were an efficient delivery system for the tumor antigen mRNA delivery into splenic dendritic cells.426,427 In 2013, they developed siRNA-loading LNPs formulated with imidazole/imidazolium lipophosphoramidate and histidinylated polyethylenimine for siRNA delivery into HeLa cells in vitro.428
Figure 26.
Chemical structures of imidazole/imidazolium lipophosphoramidate lipids.
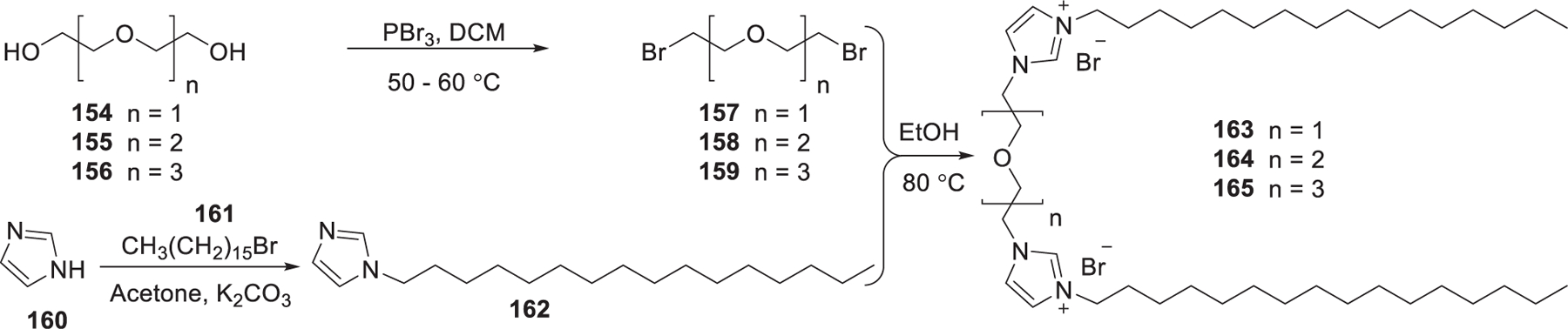
In 2012, Kumar et al. reported the synthesis of imidazolium gemini lipids and evaluated their application in siRNA delivery.429 The imidazolium gemini lipids were synthesized as shown in Figure 27. Bromination of the diol 154–156 with PBr3 in dichloromethane gave 157–159; alkylation of imidazole was realized via substitution reaction between imidazole 160 and 1-bromohexadecane 161 according to the reported procedure.430,431 The imidazolium gemini surfactants 163–165 were synthesized by refluxing the corresponding dibromoalkoxyalkanes 157–159 with N-n-hexadecyl imidazole 162 in ethanol at 80 °C for 3 days.429 In vitro biological analysis indicated that imidazolium gemini surfactants 163–165 yielded efficient siRNA delivery into HEK 293T cells, H1299 cells, and HeLa cells.429
Figure 27.
Synthesis of gemini imidazolium lipids 163−165.
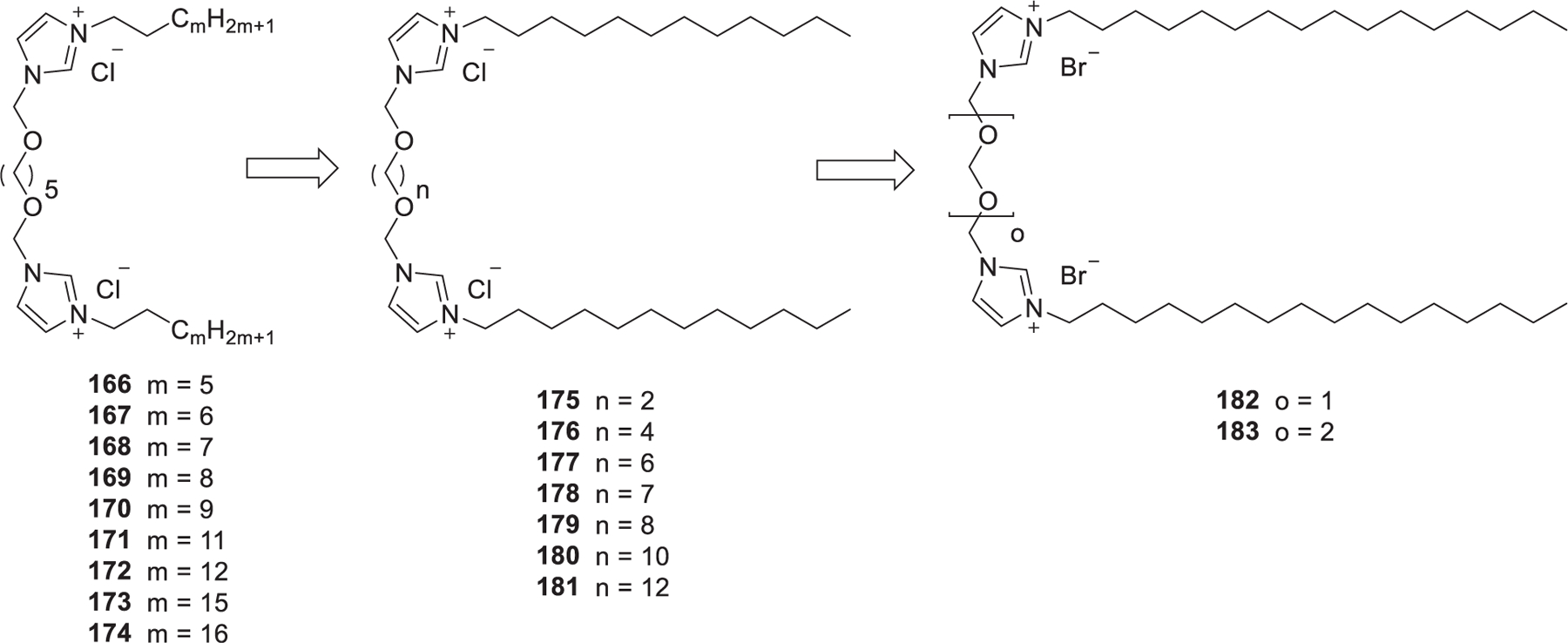
Cationic gemini lipids generally show greatly enhanced properties and lower cytotoxicity as compared to their corresponding monovalent counterparts.432–434 In 2013, Pietralik et al. prepared a library of imidazolium gemini lipids (166–174) with ether type linking groups and hydrophobic tails with different lengths (m = 5, 6, 7, 8, 9, 11, 12, 14, and 16) (Figure 28).435 Structure–activity relationship analysis showed that the hydrophobic tails should not be shorter than 11 carbon atoms (m > 9) to form complexes with 21 bp DNA and RNA. When the hydrophobic tail was 12 atoms (m = 12) in length, lipids exhibited the highest complexing activity with various nucleic acids.435 In 2016, Andrzejewska et al. developed another library of imidazolium gemini lipids (175–181) with variable lengths of dioxyalkyl linker groups and dodecyl tails.436 All of these gemini surfactants can effectively complex siRNA in a P/N ratio ranging from 1.5 to 10. Imidazolium gemini lipids containing dioxyethyl (n = 2, 175) and dioxyhexyl (n = 6, 177) spacer groups showed the strongest complexing with siRNA, and they also promoted the formation of an inverted hexagonal (HII) phase.436 In 2016, found that gemini lipid with shorter linking spacer (182, o = 1) showed higher anti-GFP siRNA delivery efficiency in vitro than gemini lipid with longer linking spacer (183, o = 2).437
Figure 28.
Optimization of imidazolium gemini surfactants.
2.2. Ionizable Lipids
In order to overcome the limitation of cationic lipids and further improve the RNA delivery efficiency, numerous ionizable lipids with ionizable amino head groups have been developed as a critical component for the formulation of LNPs.384,438–440 These ionizable lipids usually contain amino head groups and have an acid dissociation constant (pKa) less than 7;441,442 thus, they are protonated and positively charged at acidic pH (pH < 6.0) and neutral at physiological condition (pH = 7.4). They can form LNPs with an overall surface charge close to neutral, which exhibit reduced toxicity and prolonged circulation times as compared with cationic delivery systems after systemic administration, resulting in access to many tissues.443–445 In 2001, Semple et al. reported that ASO-loading LNPs formulated with DODAP/Chol/DSPC/PEG-CerC14 exhibited a half-life (t1/2) of 5–6 h, whereas ASO-loading LNPs containing DODAC/Chol/DSPC/PEG-CerC14 exhibited a half-life of only 15 min.444 Rapid plasma elimination of DODAC LNPs may be caused by the interactions between the cationic lipid DODAC and the anionic plasma proteins. In an acidified endosome, ionizable lipids are protonated and the resulting positively charged lipids can interact with negatively charged endosomal membranes, leading to the endosomal membrane destabilization and the release of RNA cargo into the cytosol.292 Optimization of ionizable lipids is explored by combining iterative screening and modification of lipids in one or more domains of their head groups, linkers, or hydrophobic tails.
2.2.1. Primary and Secondary Amino Lipids.
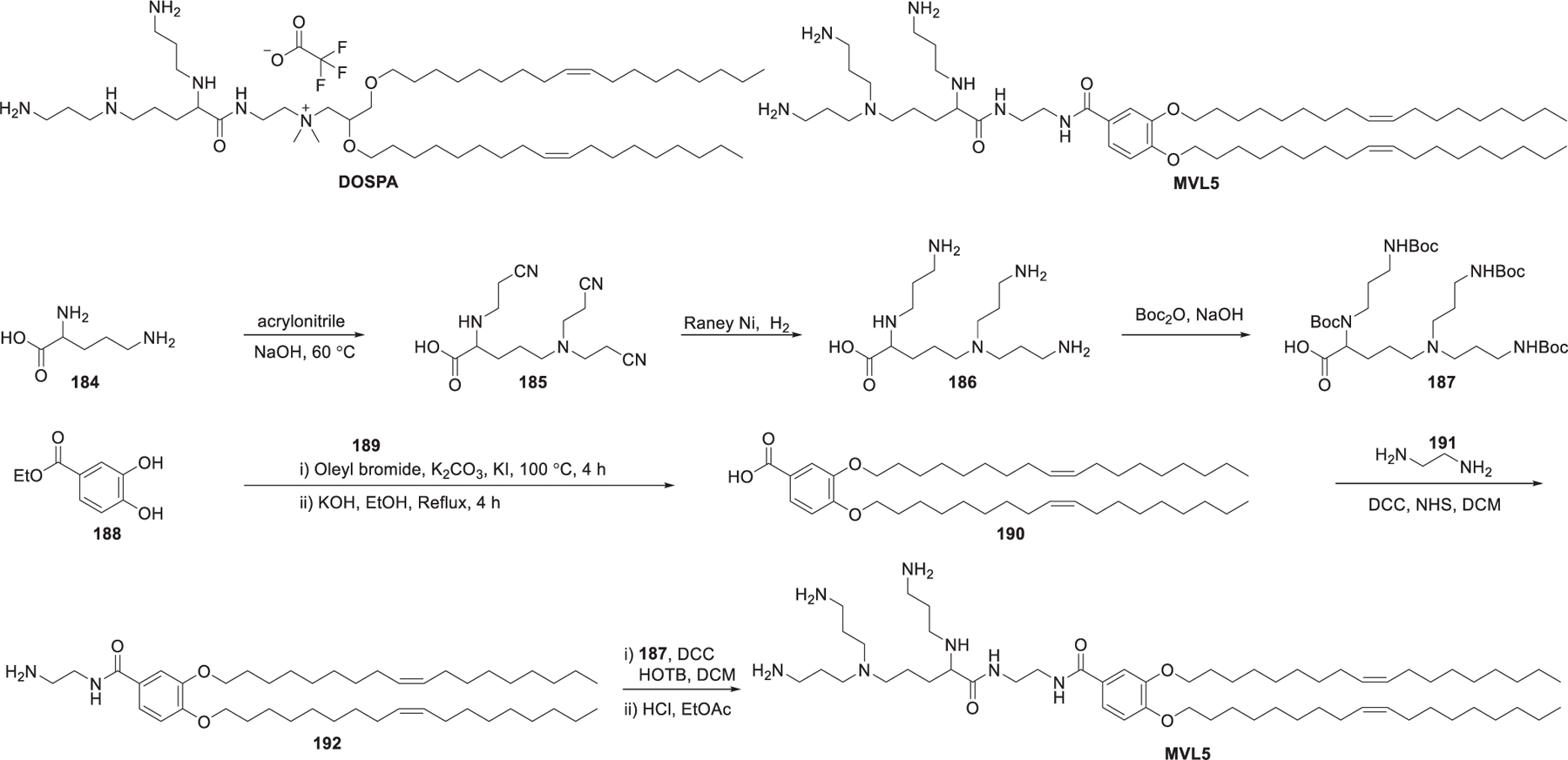
DOSPA is a multivalent lipid in which the spermine head group is linked to the hydrophobic domain that contains two oleyl tails via an amide bond (Figure 29).446 Lipofectamine is a widely used commercially available transfection reagent for pDNA or RNA delivery that contains DOSPA and DOPE at a molar ratio of 3:1.447–449 In 2002, Ewart et al. synthesized MVL5, a pentavalent ionizable lipid used for nucleic acid delivery.450 As depicted in Figure 29, the multivalent building block 186 was prepared by Michael addition of ornithine 184 to acrylonitrile followed by hydrogenation of the resulting nitrile moieties 185. Boc-protection of all amino groups in compound 186 yielded acid 187. Benzoic acid 190451 was synthesized by etherification of ethyl 3,4-dihydroxy benzoate 188 with oleyl bromide 189 followed by hydrolysis of the ethyl ester. Coupling of benzoic acid 190 and ethylenediamine 191 afforded amine 192, which was coupled with acid 187; the resulting product was deprotected with TFA to give MVL5. Compared to the monovalent cationic lipid DOTAP/DOPE, MVL5/DOPE exhibited lower toxicity and higher gene silencing efficiency in mammalian cells.452 As for a LNP formulated with MVL5 and monooleate glycerol (MOG), the high positive charge density of MVL5 and positive Gaussian curvature due to MOG facilitated endosomal escape, leading to efficient siRNA delivery and gene silencing in vitro.453
Figure 29.
Chemical structures of DOSPA and MVL5 and a synthetic route to lipid MVL5.
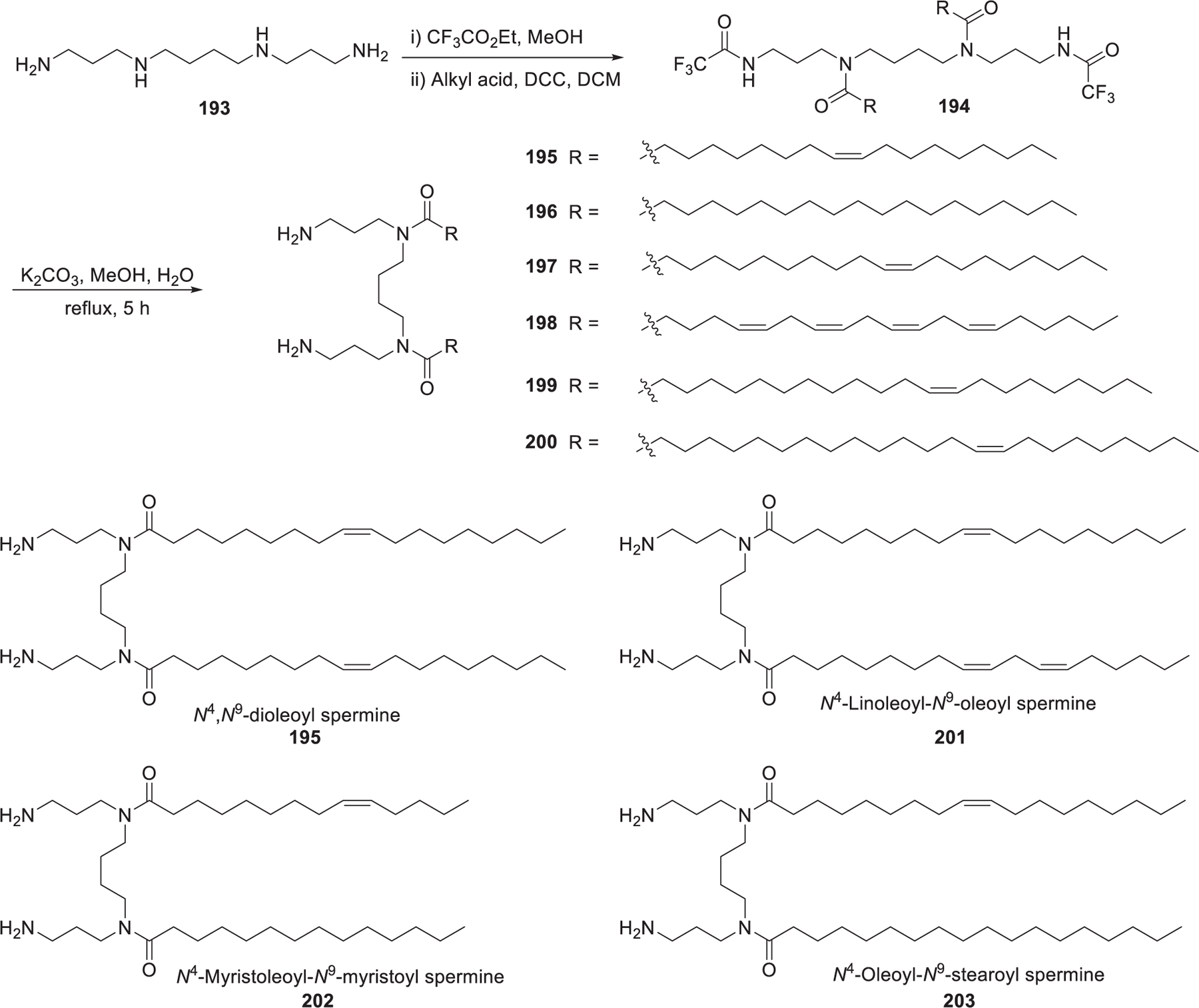
In 2006, Ghonaim et al. synthesized six symmetric N4,N9-diacyl spermines based on spermine, a naturally occurring polyamine, to evaluate the effects of the length and unsaturation degree of hydrocarbon tails on DNA and siRNA formulation.454 These N4,N9-diacyl polyamines with long chains454 (Figure 30) were synthesized through a three-step route based on their previous work.455,456 First, the two primary amino groups of spermine 193 were selectively protected as trifluoroacetamides via reaction with ethyl trifluoroacetate.457 Then aliphatic acyl chains were attached to the remaining secondary amino groups via amide bond formation. Lastly, the desired lipids were obtained after selective removal of the ditrifluoroacetyl protecting groups.458 By adding two mono-cis-unsaturated C20 or C22 chains, the resulting N4,N9-dieicosenoyl spermine 197 and N4,N9-dierucoyl spermine 199 were shown to be the lead lipids for siRNA delivery in FEK4 and HtTA cells in vitro.454 In 2012, Metwally et al. synthesized seven asymmetric N4,N9-diacyl spermines (e.g., 201 and 202), which contained two different hydrocarbon tails, varying in length from C18 to C24 with different unsaturation degrees.459 Results showed that C18 acyl tails with one or two unsaturation degrees induced effective EGFP gene silencing in HeLa cells. Besides, lipids that improved cell uptake of siRNA-loading LNPs did not necessarily show higher gene silencing activity.459 Among another six asymmetric N4,N9-diacyl spermines, N4-oleoyl-N9-stearoyl spermine 203 and N4-myristoleoyl-N9-myristoyl spermine 202 were effective in siRNA delivery, leading to 34% EGFP gene silencing in FEK4 primary skin cells in vitro. Structure–activity relationship analysis showed that the presence of an unsaturated bond in at least one of the hydrophobic tails is necessary for effective gene silencing.460
Figure 30.
Synthesis of spermine-derived lipids.
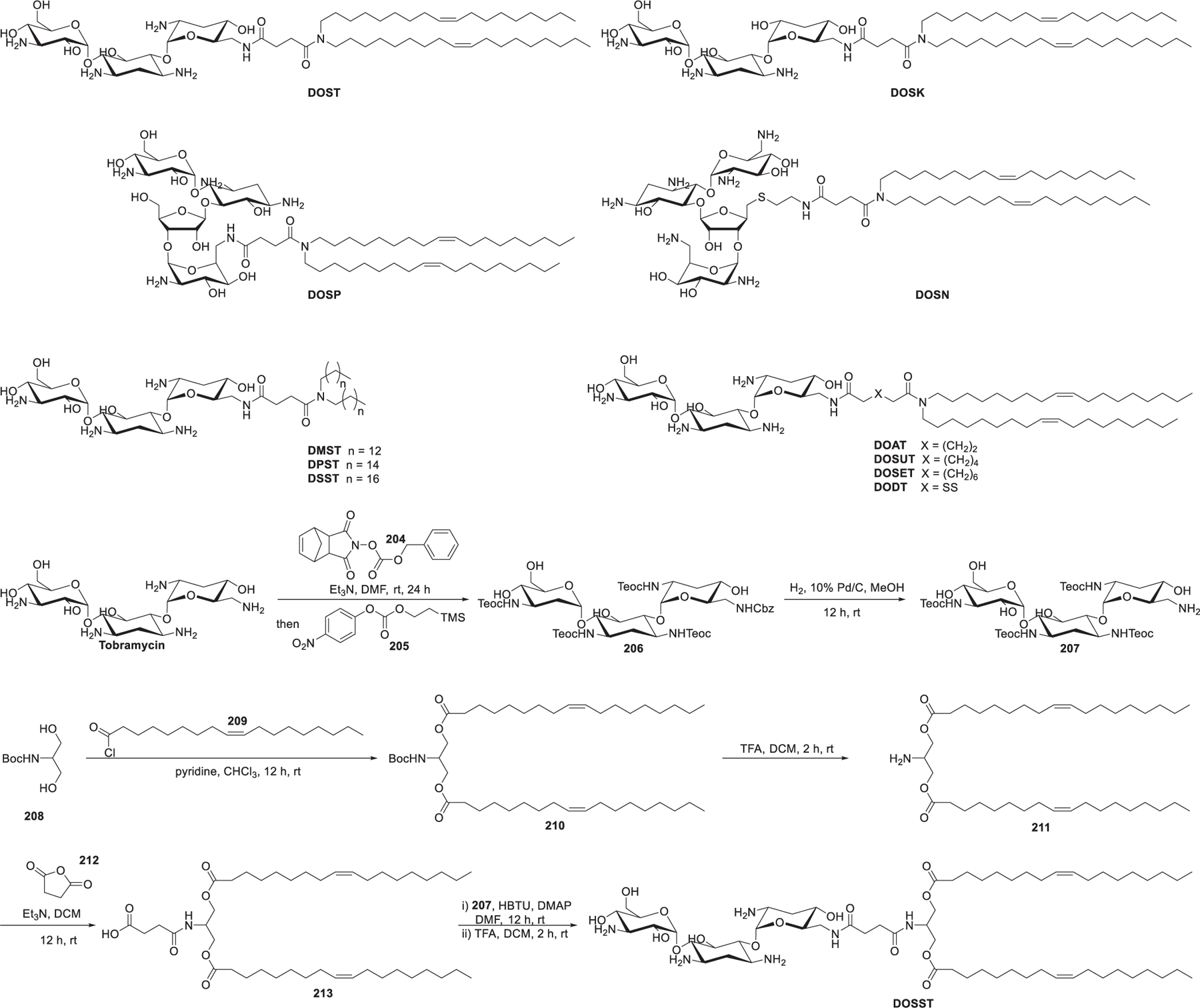
Aminoglycosides, broadly used as antibiotics, are applied for the synthesis of ionizable lipids for RNA delivery due to their natural affinity for RNA461–463 as well as their multifunctionality and structural variety. In 2007, Desigaux et al. synthesized a series of primary amino lipids by linking two dioleyl tails to various aminoglycoside head groups via a succinyl spacer (Figure 31).464 DOST and DOSK are derived from aminoglycosides which contain a 4,6-disubstituted 2-deoxystreptamine (4,6-DDS) ring, whereas DOSP and DOSN are derived from aminoglycosides with a disubstituted 2-deoxystreptamine (4,5-DDS) ring. Results showed that compared with the other three aminoglycoside-derived lipids,465 DOSP/siRNA complexes, with smaller particle size and higher colloidal stability, exhibited obvious GFP silencing in d2GFP cells and lamin A/C silencing in HEK293 and Hela cells in vitro. The flexibility of DOSP may enhance the endosomal escape of siRNA by forming lamellar microdomains, which can destabilize the endosomal membrane efficiently.464 Afterward, a structure-activity relationship study was performed to assess the importance of the hydrophobic tails, the spacer between the head group and the tails, and the behavior of stimuli-responsive linkers in delivering different DNA, siRNA, and mRNA.466 With DOST as the starting point, a set of another seven tobramycin-based lipids (DMST, DPST, DSST, DOAT, DOSUT, DOSET, DODT, and DOSST) were synthesized. As shown in Figure 31, starting with tobramycin, consecutively selective Cbz-protection of the less sterically hindered primary amine and Teoc-protection of the four remaining primary amines were realized by sequential addition of reagents 204 and 205 in one pot, giving the protected tobramycin derivative 206 in 71% yield. Hydrogenation of 206 allowed for the removal of the Cbz protecting group to provide 207. Diacylation of N-Boc serinol 208 with oleoyl chloride 209 yielded 210, which was treated with TFA to release the free amino group, resulting in amine 211. Then, amine 211 was reacted with succinic anhydride 212 to give the conjugate 213. Lastly, 213 and protected tobramycin 207 were coupled via the amide bond formation and afforded DOSST after deprotection of the Teoc groups. DOSST was an effective lipid for the delivery of mRNA, siRNA, and DNA. Structure-activity relationship studies showed that the length of the linker and the properties of the hydrophobic tails were important parameters to be considered in building efficient lipids, with the dioleyl tails suggested to be a better choice compared to shorter or saturated alkyl tails.466
Figure 31.
Structure of the aminoglycoside-derived ionizable lipids.
Ionizable lipids containing naturally occurring amino acids, such as lysine, with different hydrophobic tails were first reported to be used for DNA delivery.467,468 In 2009, Suh et al. synthesized an ionizable lipid N,N′-dioleylglutamide (DoGo1), which contains two oleylamine tails connecting to the two carboxylic acid groups of glutamic acid through amide bond linkages (Figure 32).469 Results showed that RFP-specific siRNA formulated with DoGo1 was effective in siRNA delivery in vivo, resulting in significant RFP gene knockdown in tumor tissues in mice.469 To minimize the toxicity of the amino head groups and increase the biostability of the resulting lipids, Xiao et al. synthesized a series of lipids incorporating peptidomimetics based on lysine and glutamine (Figure 32).470 N-Boc-glutamic acid 214 was coupled with oleylamine via amide bonds formation; the resulting amide was treated with trifluoroacetic acid to give DoGo1, which was attached to the Boc-protected diornithine peptide 217 followed by Boc removal to afford DoGo3. A single injection of siRNA-loading DoGO3 LNPs resulted in a 90% knockdown of apolipoprotein B (ApoB) mRNA and a 60% decrease in ApoB protein level in mouse liver at a siRNA dose of 7 mg/kg.470
Figure 32.
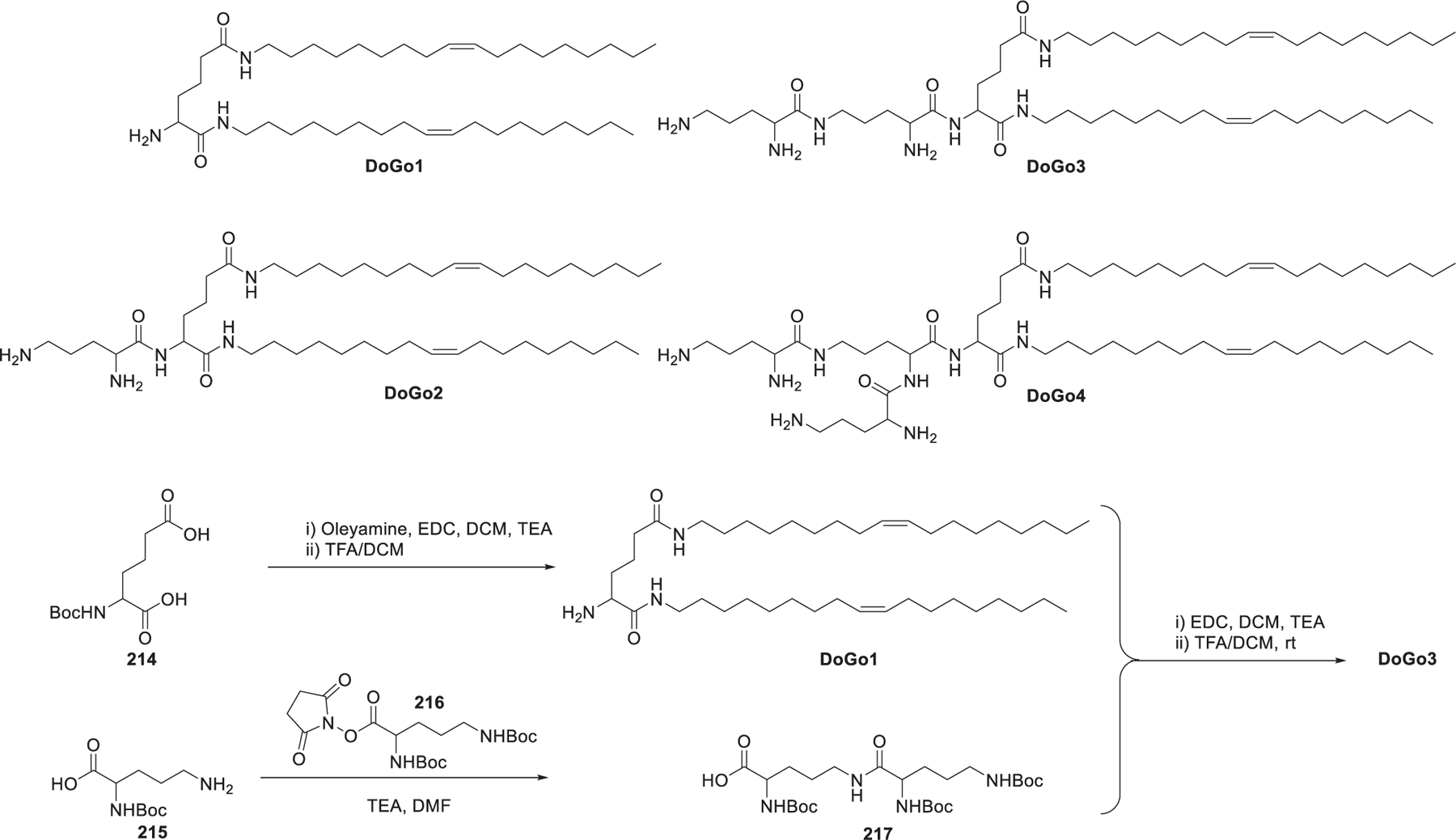
Chemical structure of lysine-derived lipids.
The Lu group developed a family of multifunctional ionizable lipids that are comprised of an ionizable head group, two distal hydrophobic tails, and two peptide-type linkers (Figure 33).471–473 In their first library, EHCO LNPs showed a high siRNA delivery efficiency into U87-Luc cells.456 Then they further modified EHCO and SHCO by removing the histidine residue and incorporating hydrophobic tails with various unsaturation degrees. In the secondary library, ECO and ECLn exhibited the highest luciferase gene silencing activity in HT29 cells and CHO cells.472 These lipids were synthesized using resin-based solid phase synthesis. For example, the synthesis of ECO started with the covalent attachment of one of the amine groups of ethylene diamine 219 to the solid material 218; then Michael addition of the remaining amine group to methyl acrylate 221 gave compound 222. Aminolysis of the esters of compound 222 with ethylene diamine 219 afforded diamine 223, which was coupled with Fmoc-Cys(Trt)-OH 224 via peptide bonds formation. The Fmoc group in the resulting compound was selectively removed with 20% piperidine to afford compound 225. Finally, the hydrophobic tails were attached to the free amino groups via amide bond formation and ECO was obtained after global deprotection with TFA (Figure 33).456 These lipids can form stable LNPs with siRNA without the inclusion of other helper lipids. Auto-oxidation of the thiol groups in the cysteine residue incorporated in the linker leads to the formation of intermolecular disulfide bonds, which can further stabilize the LNPs.474 The disulfide bonds in LNPs are relatively stable in the plasma and can be reduced by the endogenous glutathione to facilitate siRNA release. Besides, the thiol groups can also act as functional groups for modification of LNPs with biocompatible polymers and targeting ligands to minimize the immunogenicity and improve the siRNA delivery efficiency of the LNPs.475–477
Figure 33.
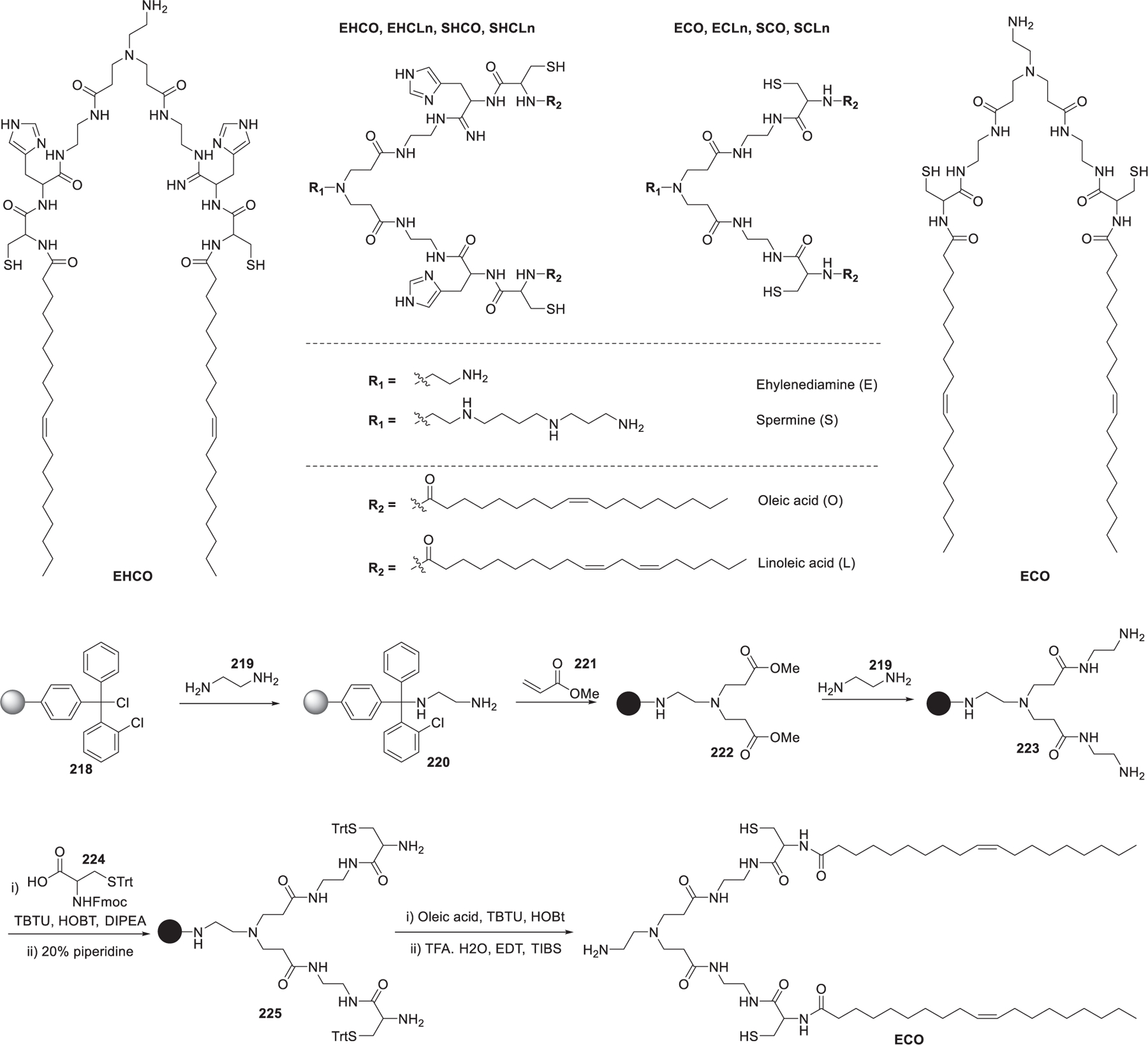
Chemical structures of multifunctional ionizable lipids.
In 2011, Asai et al. synthesized a dicetyl phosphate–tetraethylenepentamine conjugate (DCP-TEPA) that contained four ionizable amino groups attaching to the hydrophobic phosphate portions through a phosphoramide bond (Figure 34). In HT1080 human fibrosarcoma cells, the silencing activity of DCP-TEPA LNPs encapsulating siRNA was closely related to the N/P ratio of DCP-TEPA/siRNA.478 In 2019, Asai et al. synthesized similar lipids, a dioleylphosphate-diethylenetriamine conjugate (DOP-DETA) (Figure 34). The two oleyl tails offered DOP-DETA higher membrane fluidity and induced membrane fusion. EGFP siRNA-loading LNPs formulated with DOP-DETA/DPPC/cholesterol produced significant gene knockdown in HT1080-EGFP human fibrosarcoma cells in vitro. The molar ratio of DOP-DETA to siRNA is the determining factor of the gene knockdown efficiency.479 In 2019, Okamoto et al. studied the pKa of ionizable lipids by replacing hydrogen atom(s) with fluorine atom(s) and evaluated the influence of pKa on gene silencing efficiency.480 Two groups of lipids, the EtDA group and the DiETA group, were synthesized (Figure 34). The synthesis of ET-CH2F, for example, started with reductive amination of 2-fluoroethylamine 226 with N-Boc-2-aminoacetaldehyde 227; the resulting amine was treated with 4 M HCl to remove the Boc protecting group to give diamine 228. Chemoselective phosphorylation of the primary amine with dicetyl chlorophosphate 229 gave Et-CH2F (Figure 34). Due to the strong electron-withdrawing inductive effect of the fluorine atom,481 the pKa’s of the ionizable lipids decreased with the increase in the number of fluorine atoms. Results showed that the optimal lipid pKa for a high gene silencing effect varied according to the number of ionizable amines. LNPs containing Et-CH3 (pKa = 8.2) showed the highest gene knockdown efficiency in the EtDA group, and LNPs containing Di-CF3 (pKa1 = 7.1, pKa2 < 3.0) outperformed other lipids in the DiETA group. These results indicated that the balance between the number of ionizable amines and fluorine atoms was crucial to achieving high gene silencing activity.480
Figure 34.
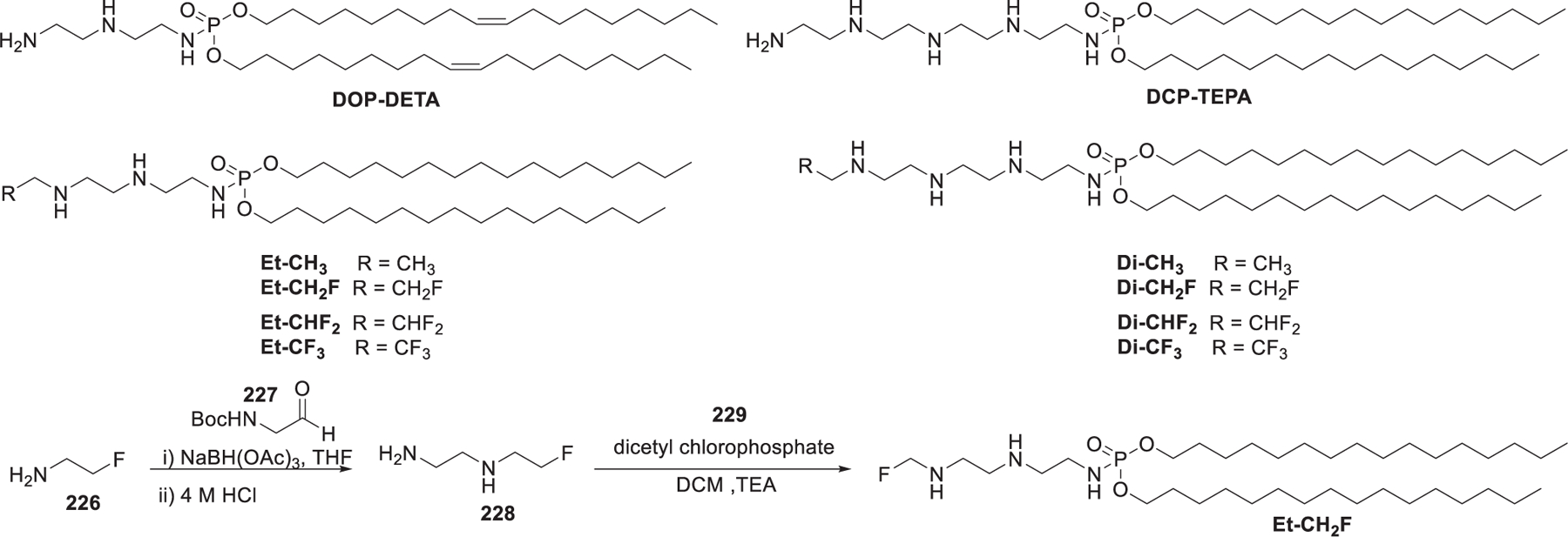
Chemical structures of dialkyl phosphate–polyamine conjugates and synthesis of Et-CH2F.
In 2011, Chang et al. developed a codelivery system of Mcl-1-specific anticancer siRNA and anticancer drugs mitoxantrone (MTO), in which lipids derived from anticancer drug acted as ionizable lipids that formed the LNPs for siRNA delivery.482 As shown in Figure 35, palmitoleic acid was conjugated to the mitoxantrone (MTO), generating two palmitoyl MTO (Pal-MTO) lipids: monopalmitoyl MTO 233 and dipalmitoyl MTO 232 (Figure 35). Nanoparticles containing monopalmitoyl MTO 233 and dipalmitoyl MTO 232 at a molar ratio of 1:1 showed effective Mcl-1 siRNA delivery in vitro, resulting in a reduction of B16F10-RFP tumor cell viability by 81%. This LNP-siRNA formulation showed stronger anticancer activity after intratumoral administration compared to LNPs alone.482
Figure 35.
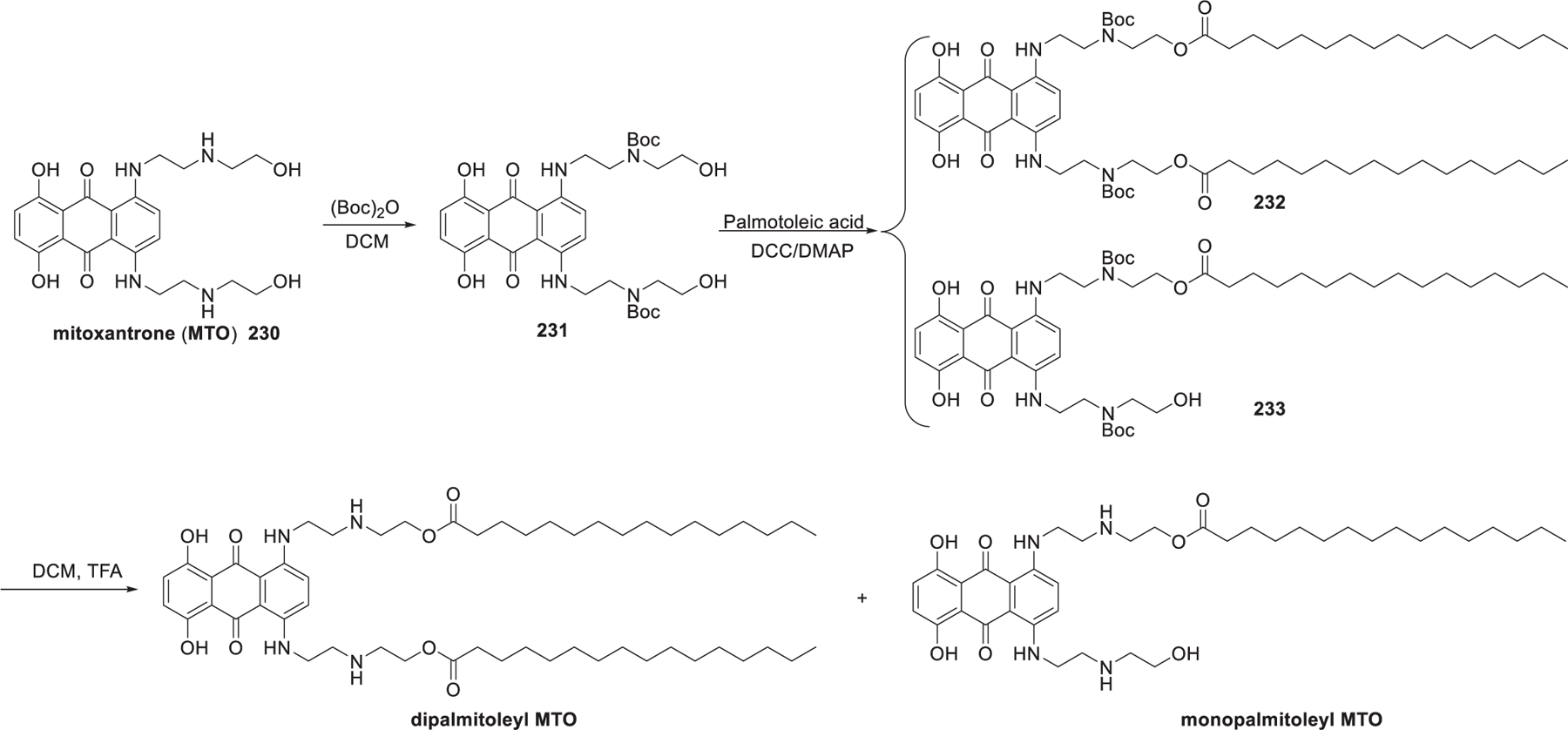
Synthesis of MTO-derived ionizable lipids.
2.2.2. Tertiary Amino Lipids.
Tertiary amines are compounds in which a nitrogen atom has three organic substituents. Generally, tertiary amines are less basic than secondary amines, because the steric hindrance of the attached alkyl or aryl groups hinders the protonation of the nitrogen atom. Sometimes primary and secondary amino lipids may exhibit different charge states in specific pH conditions compared to their tertiary amino counterparts. According to the characteristics of the chemical structures and moieties that are incorporated, tertiary amino lipids are divided into two major types in this session: (i) two-tailed amino lipids and (ii) bioactive molecules derived lipids.
2.2.2.1. Two-Tailed Amino Lipids.
In 2005, Heyes et al. synthesized two-tailed dimethyl amino lipids by modifying DOTMA.105 The trimethylammonium head group was replaced by a dimethylamino head group, and four hydrophobic tails with incremental degrees of saturation were conjugated via the ether linkers, leading to four amino lipids (DSDMA, DODMA, DLinDMA, and DLenDMA) (Figure 36). They showed that the unsaturation degree of the hydrophobic tails affected fusogenicity, lipid pKa, cellular uptake, and intracellular RNA delivery efficiency. The 31P-NMR analysis indicated that the increase of the saturation degree of hydrophobic tails from 2 to 0 double bonds compromised the fusogenicity of the LNPs. They also found that DLinDMA that contains two 18-carbon hydrocarbon tails with two unsaturated degrees showed the highest luciferase gene silencing effect in Nuro2A cells. They speculated that the linoleyl chains could produce a lipid with an inverted conical shape and tend to adopt the membrane-destabilizing inverted hexagonal (HII) phase.105 In 2006, LNPs formulated with DLinDMA/DSPC/cholesterol/PEG-C-DMA were used to deliver siRNAs, resulting in significant silencing of ApoB (>90%) when administrated systemically in nonhuman primates at a dose of 2.5 mg/kg.483 LNPs containing DODMA could also deliver CDK4 siRNA to MDAMB-468 cells and Hela cells, inducing a significant reduction of CDK4 protein expression.484 In 2013, DODMA-based LNP was used for systemic delivery of R-122 miRNA, a liver-specific tumor suppressor miRNA, leading to growth suppression of hepatocellular carcinoma (HCC) xenografts by 50% in mice.485 A-066 is an analog of DODMA that contains a pyrrole head group. A-066 LNPs encapsulating TetR siRNA could induce significant antitumor efficacy in orthotopic hepatocellular carcinoma models.486
Figure 36.
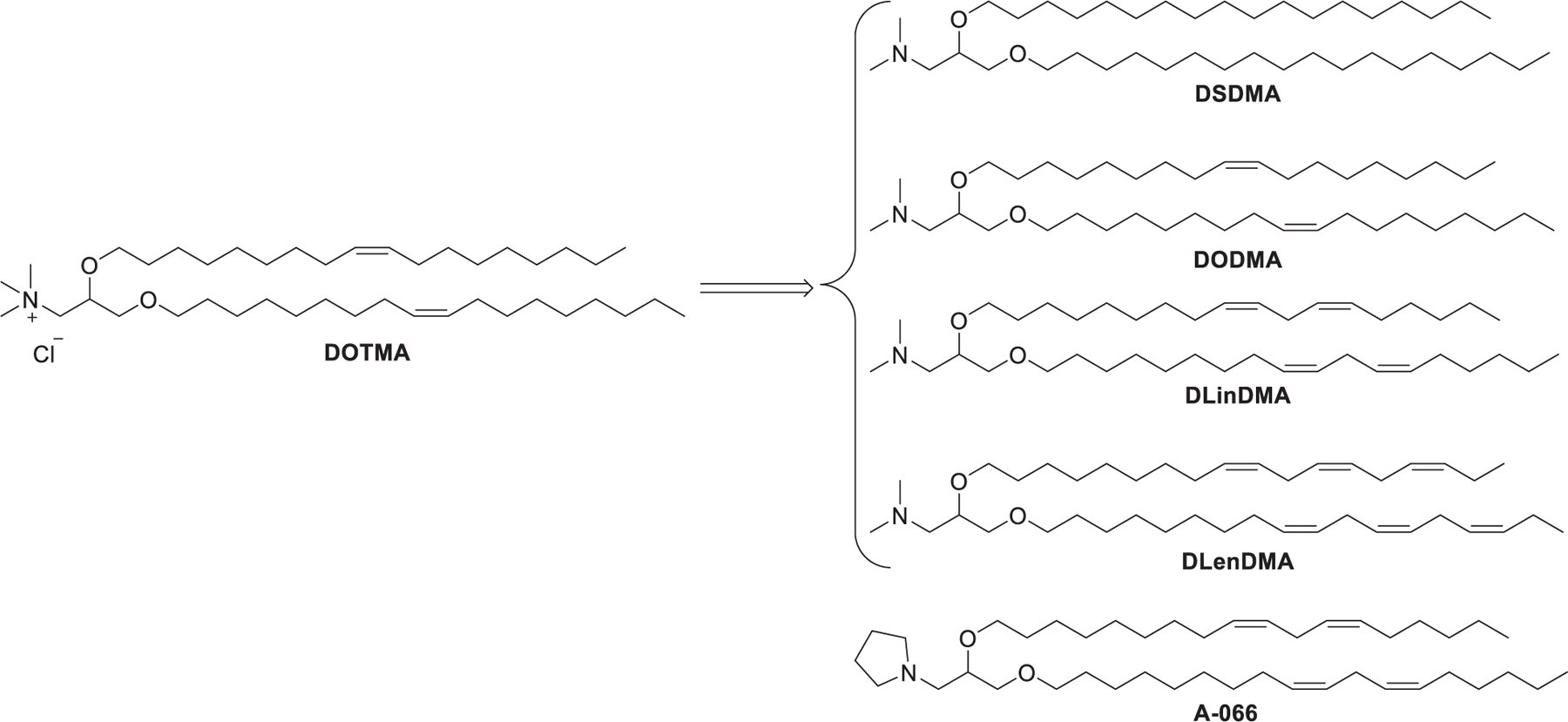
Chemical structures of the first generation of two-tailed amino lipids.
Researchers have shown that lipids containing asymmetric tails may improve delivery efficiency compared to lipids with symmetric tails, owing to the increased fusogenesity of lipids and improved membrane fluidity of LNPs.487–490 For example, Tao et al. developed a library of dimethylamino lipids with asymmetric tails that were a unique modification of DLinDMA (Figure 37).491 Different from DLinDMA, the CLinDMA series incorporated a cholesteryl ether hydrophobic domain in the lipid tail. They evaluated the siRNA delivery efficiency of these lipids to elucidate their structure–activity relationship and found that the interaction between ionizable lipid and biomembrane was closely related to the pKa value of the lipid and the environmental pH, and highly charged ionizable lipids lead to stronger interaction.492–495 Results also showed that the enhanced fusogenesity and membrane destabilizing capability of the ionizable lipids with asymmetric tails appeared to correlate with the overall lipid volume.496 Besides, the inclusion of a rigid cholesterol ether tail in lipids could produce LNPs that more mimic cell membrane and decrease protein adsorption to lipid membranes.497
Figure 37.
Chemical structures of two-tailed amino lipids with asymmetric tails.
Other modifications include replacing the ether linkers in DODMA and DLinDMA with esters, leading to DODAP and DLinDAP (Figure 38). 3-(Dimethylamino)propane-1,2-diyldioleate (DODAP), an ionizable variant of DOTAP with a pKa of 6.6, is one of the early biodegradable ionizable lipids used for the delivery of nucleic acids.498–500 In 2016, LNPs formulated with DODAP/DSPC/cholesterol/C16-PEG2000-Ceramide were used to deliver RVG-9r siRNA, leading to efficient silencing of mutant ataxin-3 gene and reduction in behavior deficits in two Machado–Joseph disease (MJD) mice following intravenous administration.501
Figure 38.
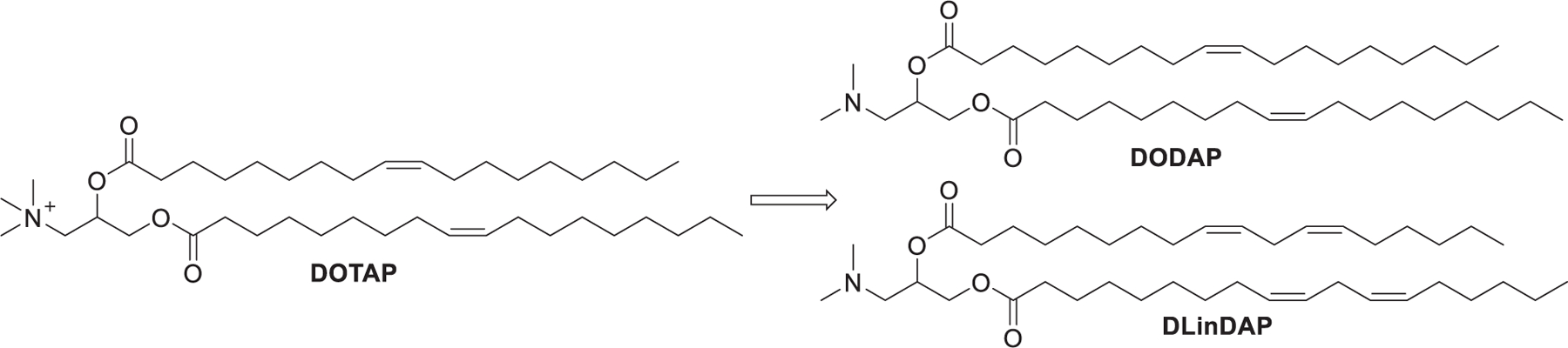
Chemical structures of DODAP and DLinDAP.
In 2010, Semple et al. described the synthesis of latently biodegradable cationic lipids by introducing a ketal ring into the linker domain, named as a class of DLin-K-DMA (Figure 39).502 As shown in Figure 39, at the beginning of the synthesis of DLin-KC2-DMA, linoleyl alcohol 234 was converted into linoleyl bromide 236 via methanesulfonylation followed by bromination. Then addition reaction between ethyl formate 237 and Grignard reagent prepared from 236 gave secondary alcohol 238, which was oxidized to afford ketone 239. Finally, the head group was attached via ketalization of ketone 239 with diol 240 to furnish DLin-KC2-DMA.502 These ionizable lipids, with pKa values in a range between 6.2 and 6.7, displayed potent gene silencing activity in vivo. Particularly, DLin-KC2-DMA was identified as the lead candidate among the four lipids for siRNA delivery. DLin-KC2-DMA showed tolerability in both rodent and nonhuman primates, LNPs formulated with DLinKC2-DMA/DSPC/cholesterol/PEG-lipid at a molecular ratio of 40:10:40:10 exhibited activity at anti-TTR siRNA doses as low as 0.01 mg/kg in rodent and 0.1 mg/kg in nonhuman primates, respectively.502 In 2012, a DLin-KC2-DMA formulation with an androgen receptor (AR) siRNA was developed as an AR inhibitor to treat prostate cancer. This formulation showed strong suppression of AR both in vitro and in vivo.503 In 2013, Lin et al. examined in vivo gene silencing activities of LNP-siRNA systems incorporating DLinDAP, DLinDMA, DLin-K-DMA, and DLinKC2-DMA and found that their activity varied over 3 orders of magnitude with the following order DLinKC2-DMA > DLin-K-DMA > DLinDMA ≫ DLinDAP.504 This tendency was in accordance with results of previous in vivo gene silencing studies in human prostate tumor tissue503 and primary antigen-presenting cells (APC) of the spleen and peritoneal cavity.505
Figure 39.
Chemical structures and synthesis of latently biodegradable two-tailed lipids.
Based on the studies on DLin-K-DMA, additional biodegradable two-tailed dimethyl amino lipids were then synthesized (Figure 40).441 The linker domain of the amino lipid was further modified by introducing biodegradable ester, amide, carbamate, or carbonate groups to modulate the lipid pKa (Figure 40a). 53 amino lipids with pKa values between 4.17 and 8.12 were prepared and complexed with siRNA to form LNPs. Interestingly, gene silencing potency was closely related with pKa value: ionizable lipids with pKa between 6.2 and 6.5 had the lowest ED50. The lead compound identified in this study was named DLin-MC3-DMA (MC3), which contains a dilinoleic acid tail and an ester linker group and enables potent hepatic gene silencing with an ED50 of 0.005 mg/kg for rodents and less than 0.03 mg/kg for nonhuman primates (a molecular ratio of 50:38.5:10:1.5 for MC3/cholesterol/DSPC/PEG-lipid).441,502 This LNPs formulation has been applied in Onpattro (patisiran), the first FDA approved LNP-based siRNA drug, which targets liver hepatocytes, the primary site for the synthesis of TTR protein.506 In 2016, Nabhan et al. used MC3 LNPs encapsulating human frataxin (FXN) mRNA for supplementing FXN protein in dorsal root ganglia in mice following intrathecal administration.507
Figure 40.
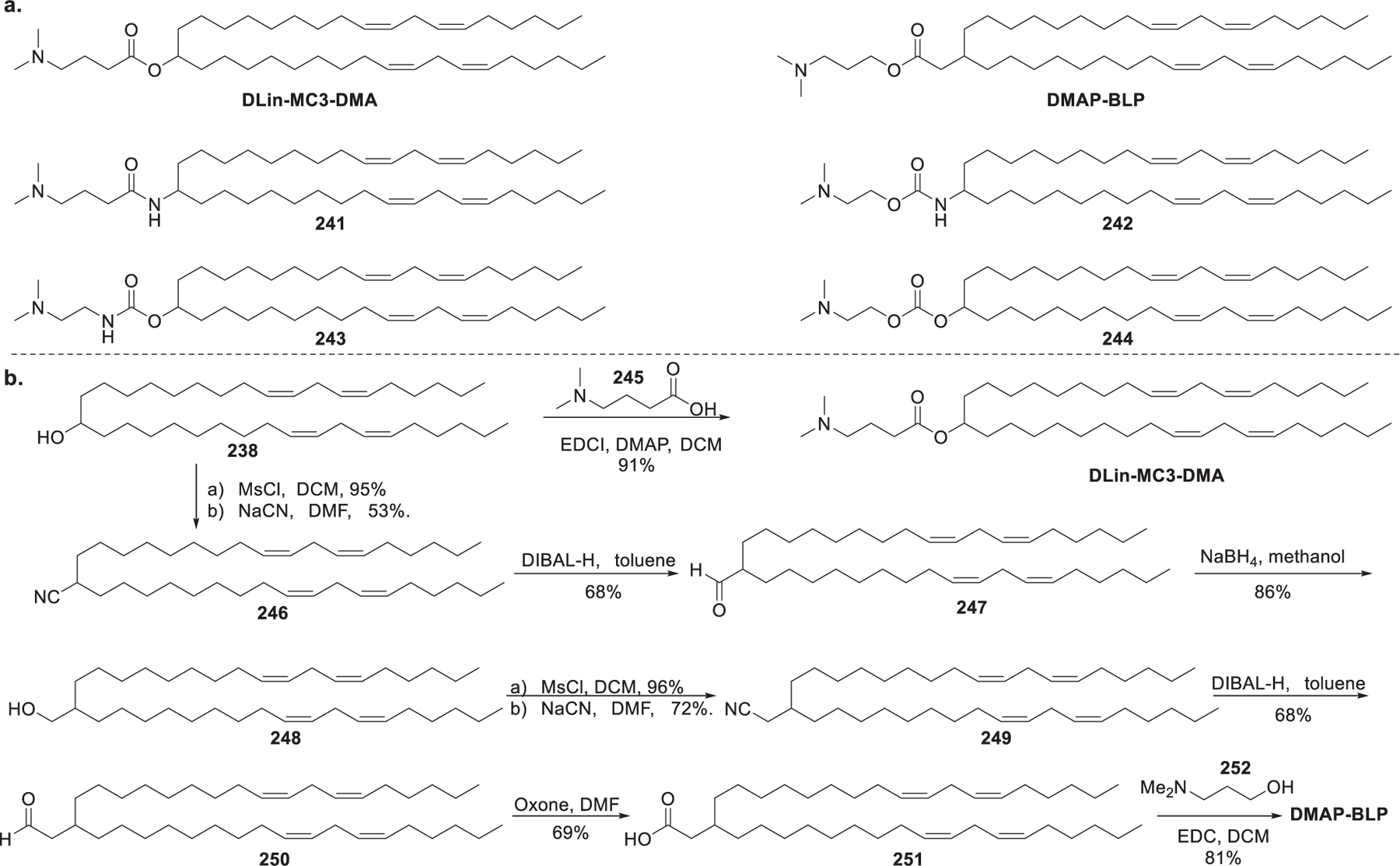
Representative chemical structures and synthetic routes to biodegradable two-tailed lipids.
In 2013, Rungta et al. synthesized 3-(dimethylamino)propyl-(12Z,15Z)-3-[(9Z,12Z)-octadeca-9,12-dien-1-yl]henicosa-12,15-dienoate (DMAP-BLP) (Figure 40b), an analog of DLin-MC3-DMA.508 The synthesis of DMAP-BLP began with sulfonylation of secondary alcohol 238 with methanesulfonyl chloride followed by SN2 reaction with sodium cyanide, giving cyanide 246. Cyanide 246 was reduced with DIBAL-H, and the resulting aldehyde 247 was treated with NaBH4 to afford primary alcohol 248, which was converted to cyanide 249. Then, reduction of cyanide 249 gave aldehyde 250, which was oxidized with oxone to generate acid 251. Finally, the combination of the head group with the hydrophobic tails via an esterification reaction between acid 251 and amino alcohol 252 promoted by EDCI provided DMAP-BLP (Figure 40).508 Rungta et al. used LNPs formulated with DMAP-BLP to deliver GRIN1 siRNA, resulting in efficient silencing of neuronal gene expression in vitro and selective reduction of synaptic N-methyl-d-aspartate receptor (NMDAR) currents in the brain in mice through intracranial injection.508 In 2016, DMAP-BLP-based LNPs were used for investigating the effect of particle size on the in vivo activity of siRNA-loaded LNPs. Results showed that siRNA-loading LNPs with diameters < 45 nm were considerably less potent in gene silencing as compared to LNPs in larger sizes. This is partially attributed to the rapid dissociation of lipid components which leads to a decrease in the stability of smaller LNPs.509 DMAP-BLP has also been used to formulate mRNA vaccines in several preclinical studies510,511 and clinical studies.510,512 In a series of influenza preclinical studies, DMAP-BLP LNP delivered nucleoside-modified mRNA encoding hemagglutinin (HA) immunogens intradermally, resulting in full protection of mice against a lethal challenge with a single dose of 0.4 μg per mouse.510 In a phase 1 trial using DMAP-BLP LNPs to deliver two distinct mRNA-encoded HA immunogens, all of the 23 participants had HAI titers > 1:40 at a dose of 100 μg via intramuscular injection.512 In 2017, a DMAP-BLP-based LNP formulated with DMAP-BLP/cholesterol/DSPC/PEG-lipid at a weight ratio of 50:38.5:10:1.5 was used for delivering IgEsig-prM-E mRNA encoding immunogen for the Zika virus. This LNP-mRNA formulation was capable of protecting mice lacking type I and II IFN signaling against a lethal challenge with a single dose of 10 μg or two 2 μg doses in a prime-boost approach.513,514 In 2018, Jhon et al. prepared DMAP-BLP LNPs encapsulating mRNAs encoding the human cytomegalovirus (CMV) glycoprotein B (gB). The DMAP-BLP/mRNA vaccine was efficiently delivered in mice and nonhuman primates (NHPs) following intramuscular injection, resulting in broadly neutralizing antibodies and potent immune responses.515
It is challenging to introduce biodegradable functionality in lipids and maintain high RNA delivery efficacy at the same time. In 2013, Maier et al. further modified DLin-MC3-DMA by incorporating a biodegradable primary ester linker in place of one of the double bonds in the hydrophobic tail (Figure 41).516 The synthesis of L-319 started with the addition of Grignard reagent prepared from 9-bromo-1-nonene 253 to ethyl formate, giving secondary alcohol 254, which underwent acylation with 4-bromobutanoyl chloride 255 to provide ester 256. Oxidization of the terminal alkene groups in 256 with RuO4 led to bis-acid 257. Then, the dimethylamino head group was installed via substitution of the bromine atom to afford amino bis-acid 258. L-319 was finally obtained via bis-esterification of bis-acid 258 with (Z)-2-nonen-1-ol 259 with EDCI as the carboxyl activating agent. Structure-activity relationship analysis showed that the position of the ester bond was critical to the delivery efficiency and clearance rate of lipids. Moving the ester bond closer toward the amine head group led to decreased delivery efficacy in vivo, which might be related to the decreased pKa. When the ester bond was positioned further away from the amine head group, the lipids were more resistant in the mouse liver. L-319 was identified as the lead biodegradable amino lipid from the screening, and L-319 LNPs had an FVII ED50 of less than 0.01 mg/kg in mouse studies. Results from pharmacokinetic studies in mice suggested that the resulting water-soluble metabolites from ester cleavage of L-319 were rapidly cleared from plasma and tissues without significant toxicity.516
Figure 41.
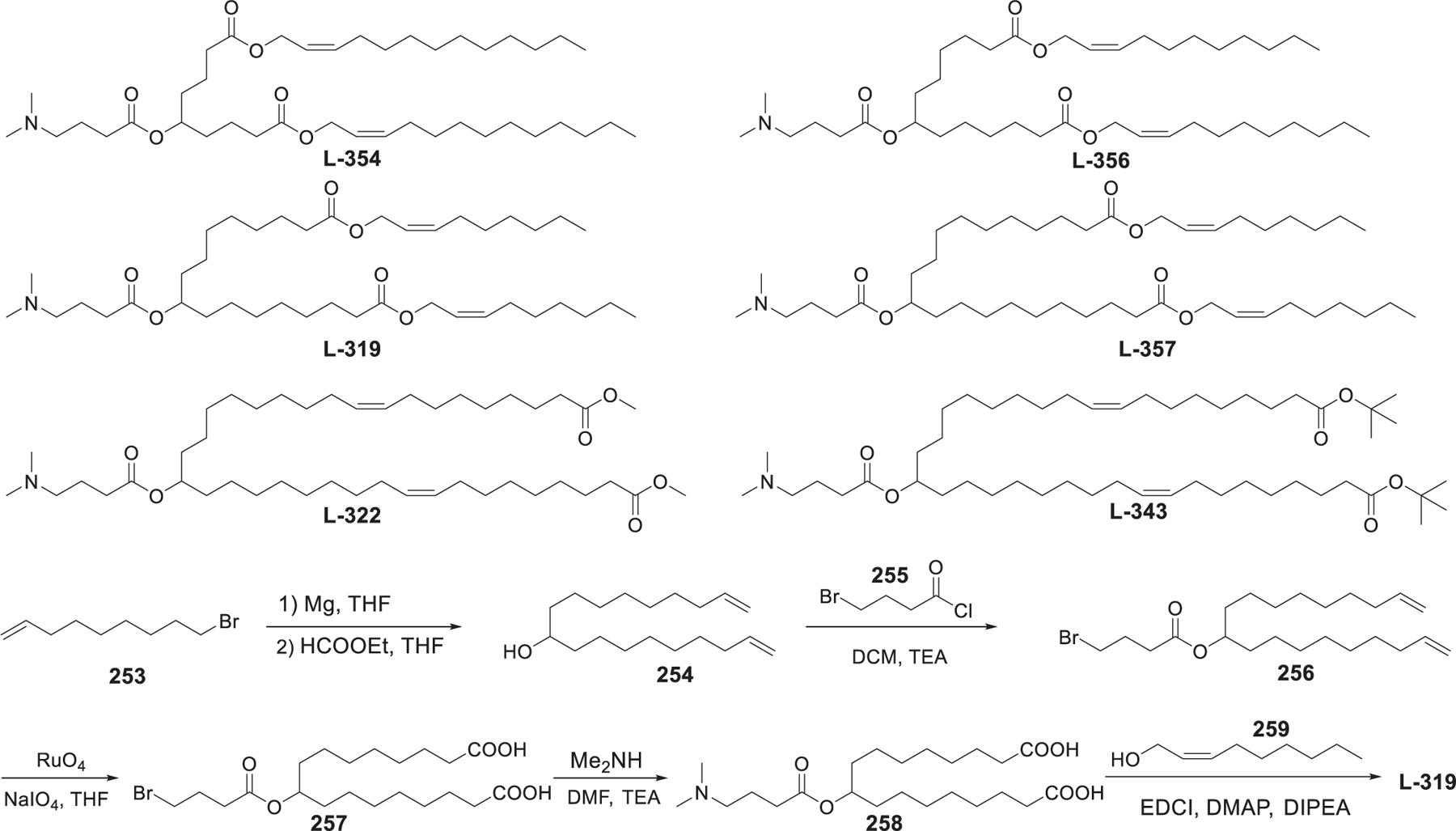
Synthesis of biodegradable dimethyl amino lipids.
In 2020, Miao et al. synthesized a library of biodegradable analogs of DLin-MC3-DMA by introducing ester and alkyne groups into the hydrophobic tails (Figure 42).517 They observed that the incorporation of alkyne groups in the hydrophobic tails of the ionizable lipids could enhance their fusion with the endosomal membrane, thus facilitating endosomal membrane destabilization, endosomal escape, and the release of the mRNA payload. Particularly, A6 showed an 8.5-fold higher production of human erythropoietin (hEPO) than MC3 LNPs in mice. The enhanced endosomal fusion might be caused by the tail protrusion and the lateral diffusion of the outer layer lipids into the endosomal membrane. Additionally, coformulation of A6 and cKK-E12518 synergistically boosted mRNA delivery into hepatocytes.517
Figure 42.
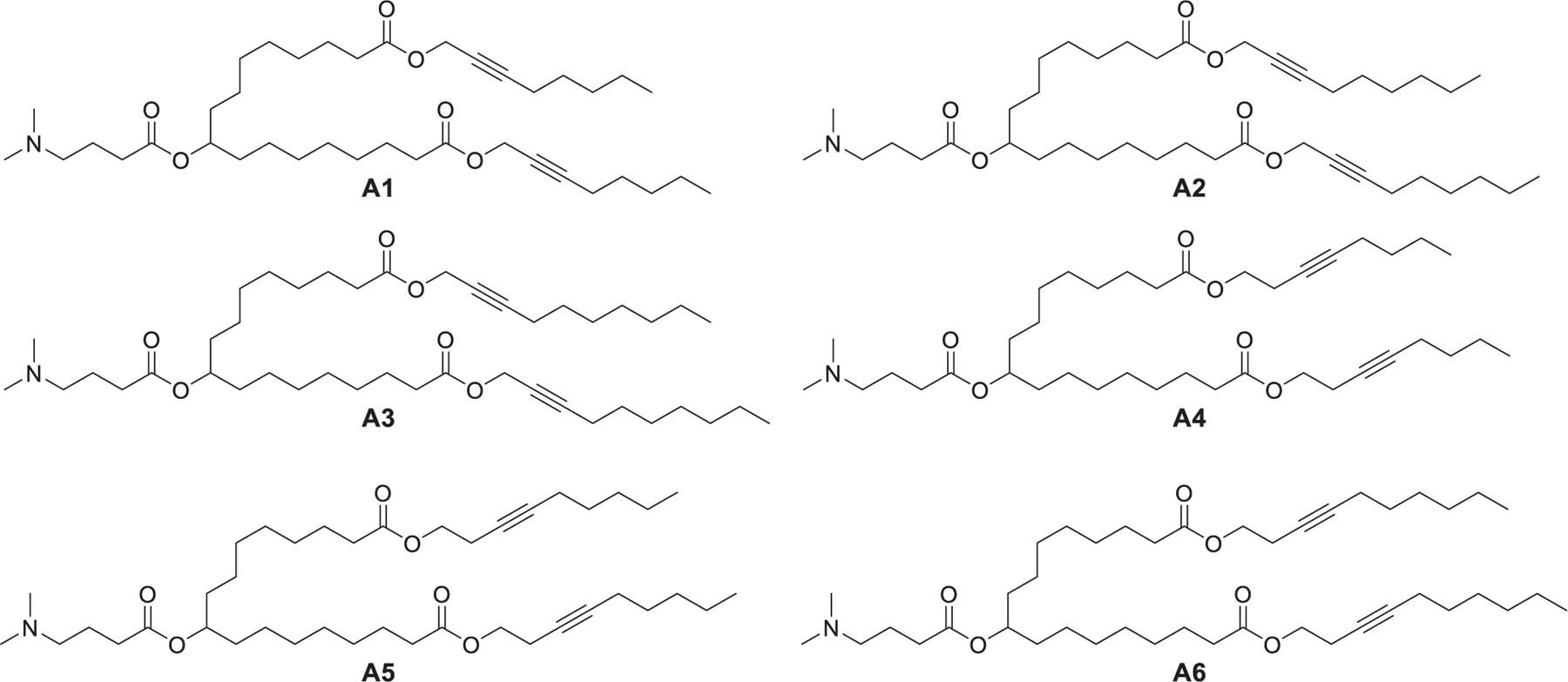
Chemical structures of biodegradable alkyne analogs of DLin-MC3-DMA.
Rajappan et al. synthesized a library of biodegradable ionizable lipids (referred to as ATX) that contain a central nitrogen atom connecting with two esterase-sensitive hydrophobic tails and the amino head group via a thiocarbamate union (Figure 43).519,520 ATX with diverse chemical structures were obtained by changing the acid chain length, altering the esterforming alcohol, and tuning the distance between the sulfur atom and the head group. The synthesis of ATX-2, for example, started with the combination of methyl-8-bromooctanoate 260 and benzylamine 261, giving benzylic amine 262, which was hydrogenated in the presence of Boc-anhydride, affording diester 263. Hydrolysis of bis-ester 263 led to diacid 264, which was bis-esterified with (Z)-2-nonenol 265 to provide diester 266. The Boc protection was removed, and the resulting Bisester 267 was treated with triphosgene and 2-(dimethylamino)-ethanethiol 268 in pyridine to yield ATX-2. The lipophilicity (indicated by cLogD) and basicity (indicated by pKa) of the lipids were measured, and lipids were formulated into LNPs encapsulating anti-FVII siRNA for evaluating their activities in the FVII knockdown assay in vivo. Results showed that the in vivo activity of lipids is related to a multivariable equation with strong ties to not only its pKa but also its lipophilicity. For example, ATX-131 LNP, with a measured pKa of 6.21, showed no FVII gene knockdown activity due to its low lipophilicity (cLogD < 10).519 In 2017, ATX-2 LNPs encapsulating human FIX (hFIX) mRNA were used for treating a Factor IX (FIX)-deficient mouse model of hemophilia B, resulting in 2-fold more hFIX protein expression than MC3 LNPs at an mRNA dose of 2 mg/kg following intravenous injection.61 One of the ATX was used to deliver mRNA in the development of the SARS-CoV-2 vaccine (LUNAR 36-COV19), of which a single dose of 2 μg per mouse protected mice from both mortality and infection following wild-type SARS-CoV-2 challenge.521
Figure 43.
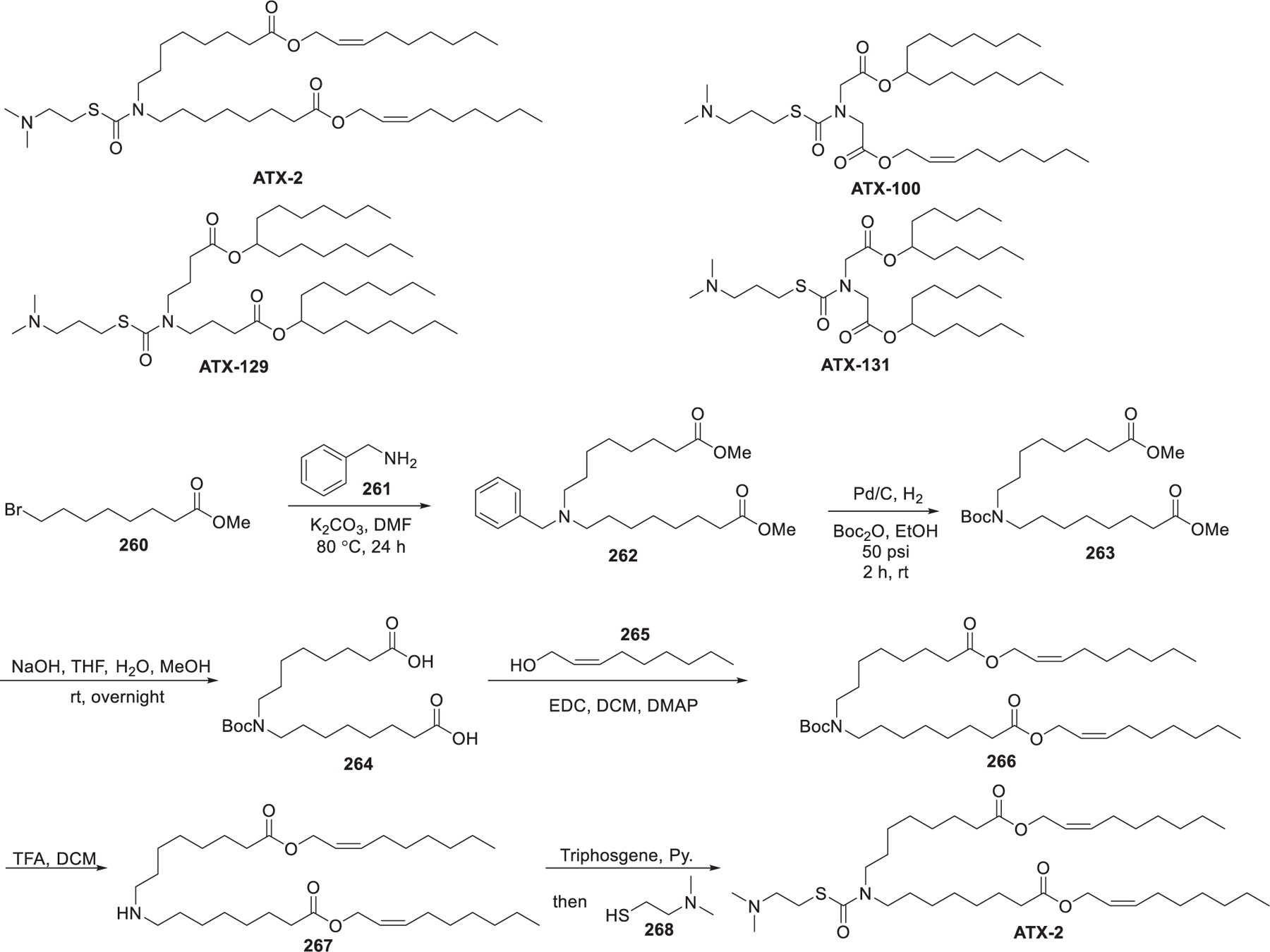
Chemical structures and synthesis of ATX lipids.
In 2020, Ramishetti et al. synthesized a library of analogs of MC3 that contained other linker groups such as hydrazine, ethanolamine, and hydroxylamine for gene silencing in leukocyte subsets (Figure 44).522 Lipids incorporating hydroxylamine and ethanolamine linkers were more efficient in gene silencing in CD8+ and CD4+ T leukocytes compared to that with hydrazine linkers. Additionally, lipids with a piperazine head group (lipid 270) accumulated more in the spleen than the liver, while lipids with a dimethylamino head group (lipid 269) resulted in more accumulation in the liver than the spleen. Moreover, lipids with a branched ester as the tail (lipids 271 and 272) were less effective for siRNA delivery than those with linoleyl chains (lipids 273 and 269). Systemic administration of lipid 269 LNPs encapsulating CD45 siRNA resulted in signification reduction of the CD45 level in both CD4+ and CD8+ T lymphocytes.522
Figure 44.
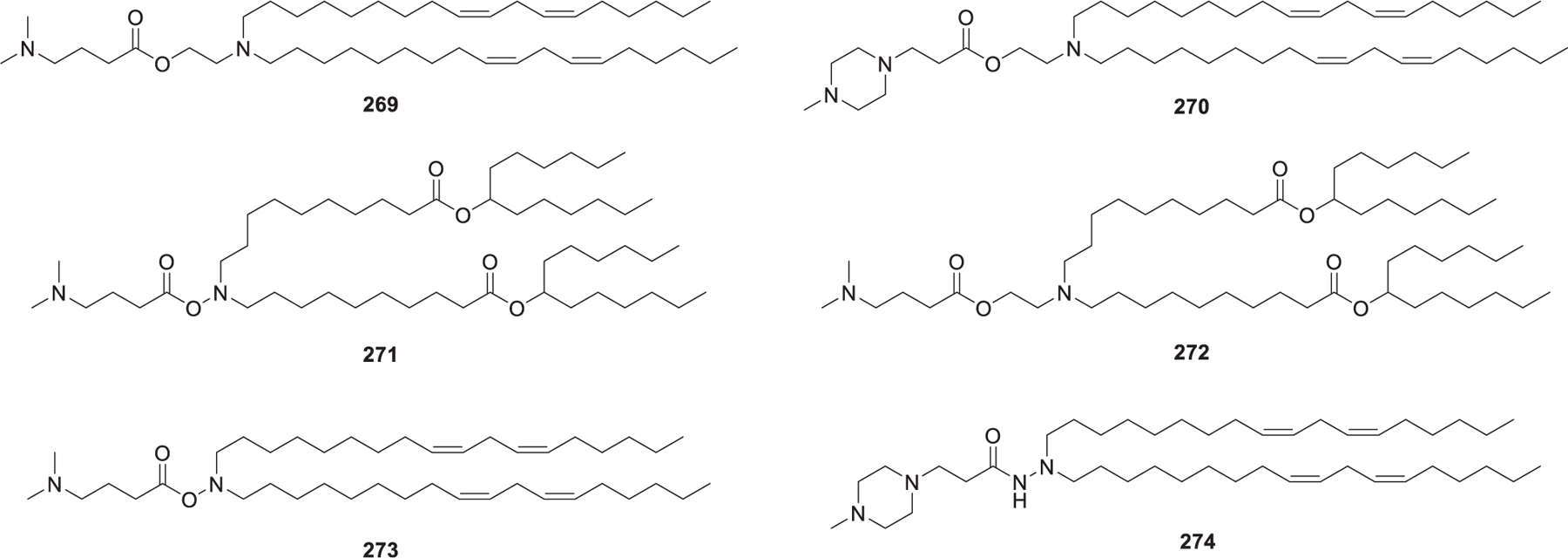
Chemical structures of two-tailed lipids with other linkers.
The results of clinical trials of BNT162b2523–525 and mRNA-1273526–528 vaccines successfully led to their approval in clinical application,7–9 providing a powerful approach to counteract the worldwide COVID-19 pandemic. Ionizable lipids used in the BNT162b2 and mRNA-1273 formulations were disclosed as ALC-0315529,530 and SM-102 (Lipid-H),530,531 respectively (Figure 45). ALC-0315 has two degradable branched ester tails, and SM-102 contains a primary degradable ester tail and a degradable branched ester tail. ALC-0315 LNPs showed approximately 6-fold higher firefly luciferase level than MC3 LNPs at a FLuc mRNA dose of 0.3 mg/kg in mice,529 whereas Lipid 5, an analog of SM-102 (Lipid-H), displayed a 3–6-fold increase in protein expression or immune responses in mice compared to MC3 when delivering an mRNA encoding influenza immunogen.531 These results indicated that the branching of tails may create a more cone-shaped structure and facilitate the endosomal escape of RNA molecules.532,533 BNT162b2 uses LNPs formulated with ALC-0315/cholesterol/DSPC/PEG-lipid to deliver mRNA encoding immunogen that is a diproline-stabilized, membrane-bound spike protein.525 mRNA-1273 uses SM-102 LNPs to deliver mRNA encoding transmembrane-anchored diproline-stabilized prefusion spike with a native furin cleavage site. SM-102 (lipid H)531 and lipid 5534 belong to a class of biodegradable ionizable lipids that were synthesized by incorporating greater branching rather than the dilinoleic alkyl tails of MC3.531,534 Both lipid 5534 and SM-102 (lipid H)531 contain an ethanolamine head group, a saturated linear tail with a primary ester linker and a second saturated branched tail with a less biodegradable secondary ester linker. Lipid 5 LNPs displayed luciferase expression 3-fold higher as compared to MC3 LNPs in mice and induced hEPO expression 5-fold higher than MC3 LNPs in nonhuman primates following intravenous administration. The enhanced protein expression may possibly be due to a high endosomal release of the mRNA cargo in lipid 5 LNPs. Pharmacokinetic studies revealed that lipid 5 LNPs decreased liver accumulation and were fully degraded within 24 h in both rats and nonhuman primates. SM-102 (lipid-H), which structurally differs from lipid 5 by a two-carbon displacement of the primary ester tail, was identified as the most promising lipid following intramuscular (IM) administration. The pKa of SM-102 LNPs is slightly higher than that of lipid 5 LNP (6.68 vs 6.56).531 Results of a phase 3 clinical trial showed that two 30 μg doses of BNT162b2 induced 95% protection against COVID-19 (95% credible interval, 90.3 to 97.6),525 and mRNA-1273 conferred 94.1% protection against COVID-19 illness after receiving two 100 μg doses in another phase 3 clinical trial.527 In 2021, Elia et al. used lipid 274 LNPs and lipid 275 LNPs to deliver mRNA encoding SARS-CoV-2 human Fc-conjugated receptor binding domain (RBD-hFc mRNA).57 While both lipid 274 LNP RBD-hFc mRNA and lipid 275 LNP RBD-hFc mRNA induced equal cellular and humoral responses in mice at an mRNA dose of 5 μg, only lipid 275 LNP RBD-hFc mRNA exhibited strong immunogenicity following intradermal administration. Both intradermal administration and intramuscular administration of lipid 275 LNPs could activate antigen presenting cells (APCs), thus inducing cellular responses.57
Figure 45.
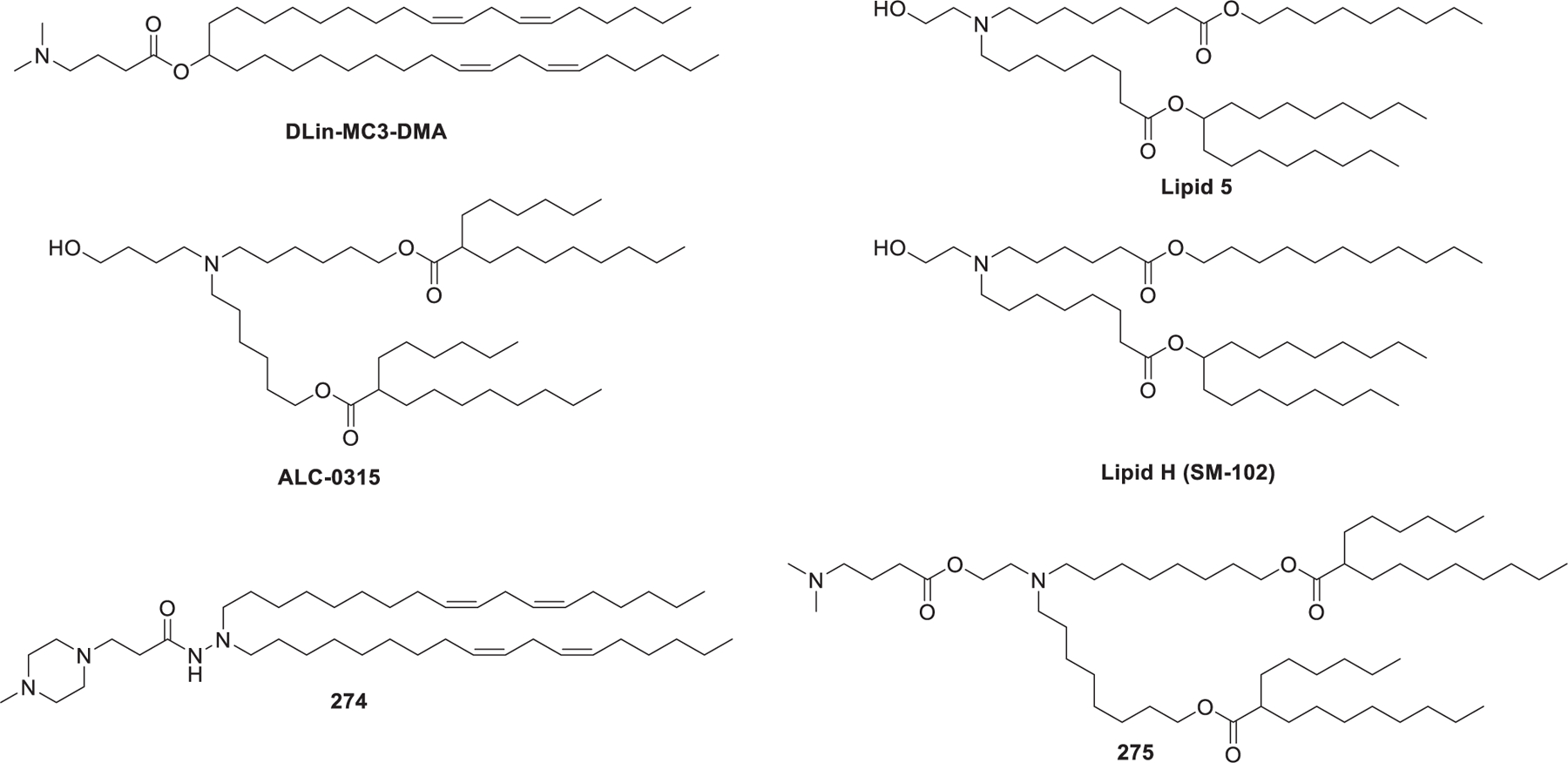
Chemical structures of ionizable lipids used in the development of COVID-19 mRNA vaccines.
The stability of LNP–RNA formulations is related to the properties of both RNA molecules and LNPs. For example, chemically modified siRNA molecules are double-stranded and relatively stable, while mRNA molecules are prone to be degraded via hydrolysis of the phosphodiester bonds535 and oxidation of the nucleobases.536 The chemical and physical features of LNPs can greatly affect their stability. The unsaturated hydrocarbon tails of lipids or cholesterol can be oxidized by various enzymes or agents.537,538 Aggregation and fusion can lead to physical destabilization of LNPs.539,540 Generally, PEG-lipids are incorporated in LNPs to prevent the aggregation of nanoparticles and to improve their circulation stability.541 At 2–8 °C, mRNA-1273 and BNT162b2 can be stored for 30 days and 5 days, respectively. Both of them need to be stored in a frozen state at extremely low temperature for long shelf life (6 months) and thawed before injection.542 The shelf life of the LNP–siRNA formulation of patisiran is as long as three-years when kept between 2 and 8 °C. The main difference of lipid components between the two LNP–mRNA vaccines and the LNP–siRNA formulation of patisiran lies in the ionizable lipids (SM-102, ALC-0315 vs MC3); therefore, it may be the stability of mRNA, rather than that of LNP, which determines the storage conditions and shelf life of LNP–mRNA formulations.543
In 2017, Fin et al. reported the simultaneous delivery of Cas9 mRNA and sgRNA for transthyretin gene editing in vivo by using LP-01 LNPs, resulting in a significant transthyretin gene editing in the liver in mice and a reduction in serum transthyretin levels by >97% after a single administration (Figure 46).544 In 2017, Dahlman et al. developed an efficient method to measure the biodistribution of many barcoded nanoparticles in a single mouse.545 This barcoded nanoparticle system can facilitate the characterizations of a large number of LNPs targeting specific tissues and cells and aid understanding of the structure–activity relationship of lipids in vivo. In 2019, they found that certain LNPs could deliver sgRNA and siRNA into splenic T cells in vivo.546 They synthesized a library of 16 constrained ionizable lipids containing small head groups (diethylamino head group or 1-pyrrodinyl head group), carbonate linkers, and two hydrophobic tails. As shown in Figure 46, the two hydrophobic tails were attached to two hydroxyl groups trimethylolmethane 276 via successive esterification, and the amino head group was conjugated to the remaining hydroxyl group via a carbonate linker to afford the desired lipids. 55 LNPs carrying anti-GFP siRNA and unique barcodes were formulated from 15 ionizable lipids, C14-PEG2000, cholesterol, and DSPC at four different ratios. Results showed that the top-performing 11-A-M LNPs were enriched more than other LNPs at a dose of 0.5 mg/kg in mice. The biodistribution of 11-A-M LNPs was in splenic CD8+ T cells, CD4+ T cells, and B cells. 11-A-M LNPs were also used to deliver chemically modified sgRNA targeting GFP to downregulate the expression of GFP in CD8+ and CD4+ T cells in mice.546
Figure 46.
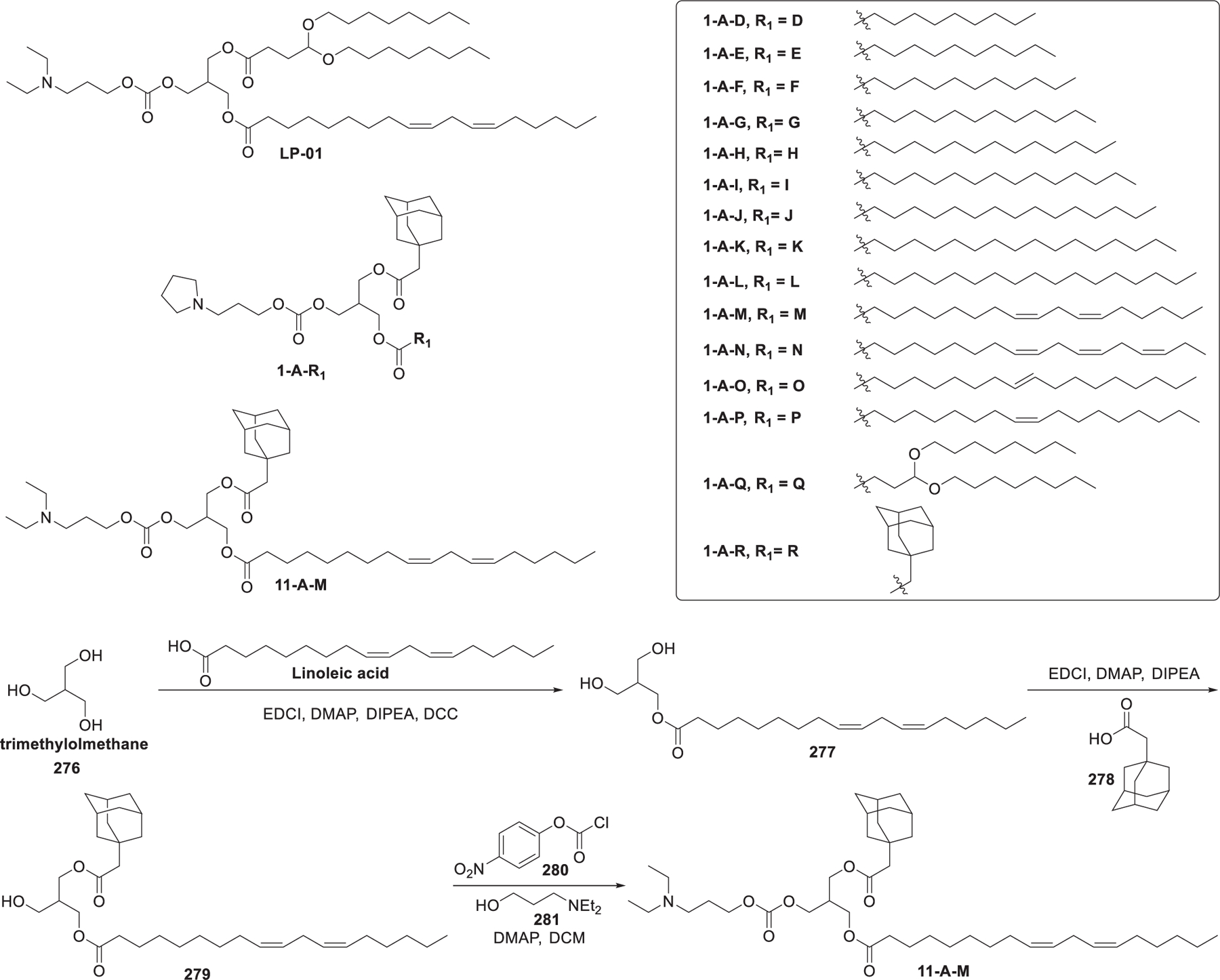
Chemical structures of constrained lipids with small head groups.
ssPalm are a class of gemini ionizable lipids, in which the two tertiary amino head groups are connected via a bioreducible disulfide linker and each amino head group is connected with a methyl group and hydrophobic tails (Figure 47).547–552 The starting material for the preparation of ssPalmM was 2,2′-dithioldiethanol 282, which was methanesulfonylated to give methanesulfonate ester 283. The head groups were installed via the substitution reaction between methanesulfonate ester 283 and 3-(methylamino)-1-propanol 284, affording diol 285. Diol 285 was esterified with myristic acid to give ssPalM.547 Prior studies reported that ssPalm was protonated after internalization of ssPalm LNPs into cells, thus facilitating the endosomal escape of ssPalm LNPs.504 Besides, the disulfide bridge cleavage induced by intracellular glutathione (GSH) in lysosomes facilitated the disassembly of LNPs and the release of the encapsulated RNA molecules.547,553 In 2015, Akita et al. found that ssPalmE LNPs could deliver FVII siRNA to hepatic cells more efficiently as compared to ssPalmM or ssPalmA after intravenous administration at a siRNA dose of 4 mg/kg in mice.554 ssPalmA LNPs were used to deliver siRNA against Col1a1, resulting in an inhibitory effect on hepatic fibrosis in mice following systemic injection, with an ED50 of 0.25 mg/kg.555 In 2015, they further modified ssPalmE by incorporating more flexible piperidine and systemically prolonging the distance between the hydrophobic domains and the ionizable nitrogen atoms, resulting in the pKa being increased from 6.03 to 6.18, thus allowing the tertiary amine groups more readily to be pronated. The increased pKa of the ssPalmE-P4-C2 led to the ED50 value for the ssPalm-P4-C2 LNPs in FVII knockdown as low as 0.035 mg/kg in mice.554 In 2018, by integrating the oleic acid scaffold into the ssPalm, Tanaka et al. prepared ssPalmO and its derivatives, which exhibited high mRNA delivery efficiency in the colon and less immune stimulation.556,557 In 2019, Tateshita et al. reported the ex vivo transfection of DCs using ssPalmE LNPs encapsulating ovalbumin (OVA) mRNA; the transfected DCs induced strong cytokines production in mouse bone marrow-derived dendritic cells (BMDCs).558 In 2020, Tanaka et al. synthesized a self-degradable analog of ssPalmO, ssPalmO-Phe, by introducing a phenyl ester linker. The self-degradation of ssPalmO-Phe involved in the mechanism of “hydrolysis accelerated by intra-particle enrichment of reactant (HyPER)”.550 In this reaction, disulfide bonds are cleaved under a reducing environment, generating enriched hydrophobic thiols, which can attack the phenyl ester linker group and break them down. Additionally, codelivery of Cas9 mRNA and sgRNA targeting TTR by ssPalmO-Phe LNPs allowed the editing of 55% of the TTR-encoding genome and led to a 95% reduction in serum TTR level after systemic administration in mice.550
Figure 47.
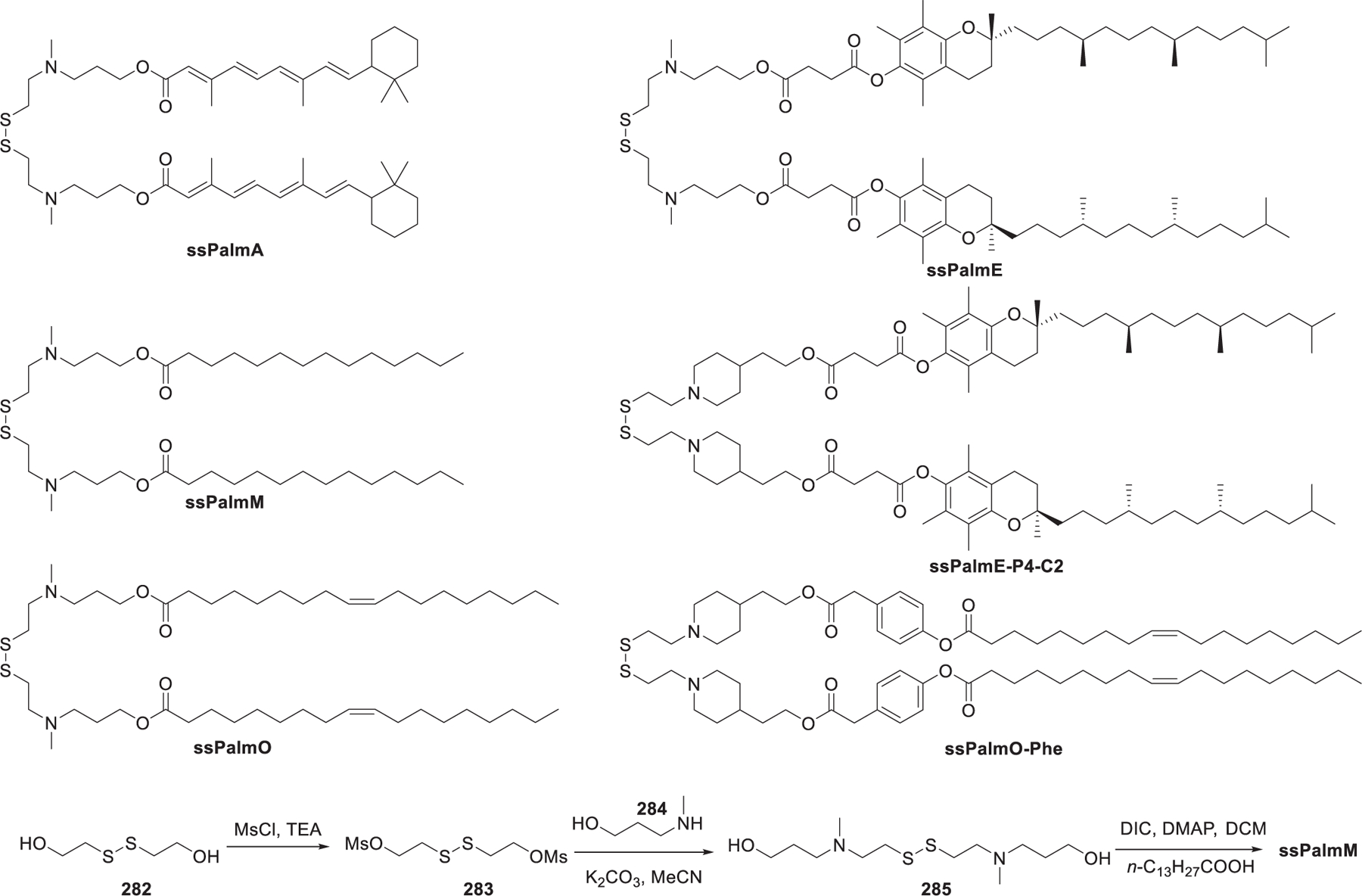
Chemical structures of representative ssPalm and a synthetic route to ssPalmM.
The Harashima group developed the YSK series of ionizable lipids; among them several lead lipids have been identified, such as the first generation YSK05,202 the second generation YSK13-C3,559,560 as well as the third generation YSK12-C4561 and CL4H6562 (Figure 48). In 2012, DODAP-based lipid YSK05 was developed, which contained an ionizable cyclic tertiary amino head group, a ketal linker, and two linoleyl chains. YSK05, with a pKa value of 6.4, could promote endosomal membrane fusion and endosomal escape. YSK05 LNPs encapsulating anti-PLK1 siRNA showed efficient and durable PLK1 gene silencing activity after intravenous administration in mice.202,563 YSK05 LNPs have also been used in siRNA delivery for the treatment of hepatitis C virus (HCV) infections in mice with humanized livers564 and the codelivery of sorafenib (SOR) and anti-midkine gene (anti-MK) siRNA to treat hepatocellular carcinoma (HCC).565 To enhance the tissue clearance as well as RNA delivery efficiency, biodegradable YSK13-C3 was then developed.559,560 The ED50 of gene silencing in mice following intravenous administration of YSK13-C3 LNPs encapsulating blood-clotting factor VII (FVII) siRNA was 0.015 mg/kg, which was about 4-fold more active than that of YSK05 LNPs.560 Intravenous injection of HBV siRNA-loading YSK13-C3 LNPs into mice could simultaneously reduce the levels of the HBV DNA and HBV antigens (HBsAg and HBeAg).560 In 2016, they developed YSK12-C4, which contained a hydroxy group near the amino head group. The use of YSK12-C4 LNPs encapsulating scavenger receptor class B type 1 (SR-B1) resulted in a gene silencing in mouse BMDCs, with an ED50 of 1.5 nM. Additionally, downregulation of the suppressor of cytokine signaling 1 led to an elevation of the cytokine level and significant inhibition of tumor growth.561 Based on the study of YSK12-C4, CL4H6, a lipid that contained oleic acid esters in the hydrophobic tails and had a pKa value of 6.25 was identified as another lead lipid (Figure 48).562 Structure–activity relationships showed that the structure of the head group was key to the apparent pKa’s of ionizable lipids and played an important role in the intrahepatic distribution and endosomal escape of LNPs. Intravenous administration of CL4H6 LNPs encapsulating FVII siRNA into mice resulted in significant FVII gene silencing with a hepatic ED50 of 0.0025 mg/kg.562 As shown in Figure 48, the starting material for the synthesis of CL4H6 was 6-bromo-1-hexanol 286, which was treated with tert-butyldimethylsilyl chloride (TBSCl) to protect the hydroxy group, giving compound 287. Grignard reagent prepared from compound 287 was reacted with δ-valerolactone 288 to afford diol 289. The amino head group was installed via chemical selective sulfonylation of the primary hydroxy group of diol 289 followed by a substitution reaction with dipropylamine 290, providing compound 291. Removal of the two TBS protecting groups led to triol 292, which underwent bis-esterification of the two primary hydroxy groups with oleic acid to obtain EL4H6 (Figure 48).562
Figure 48.
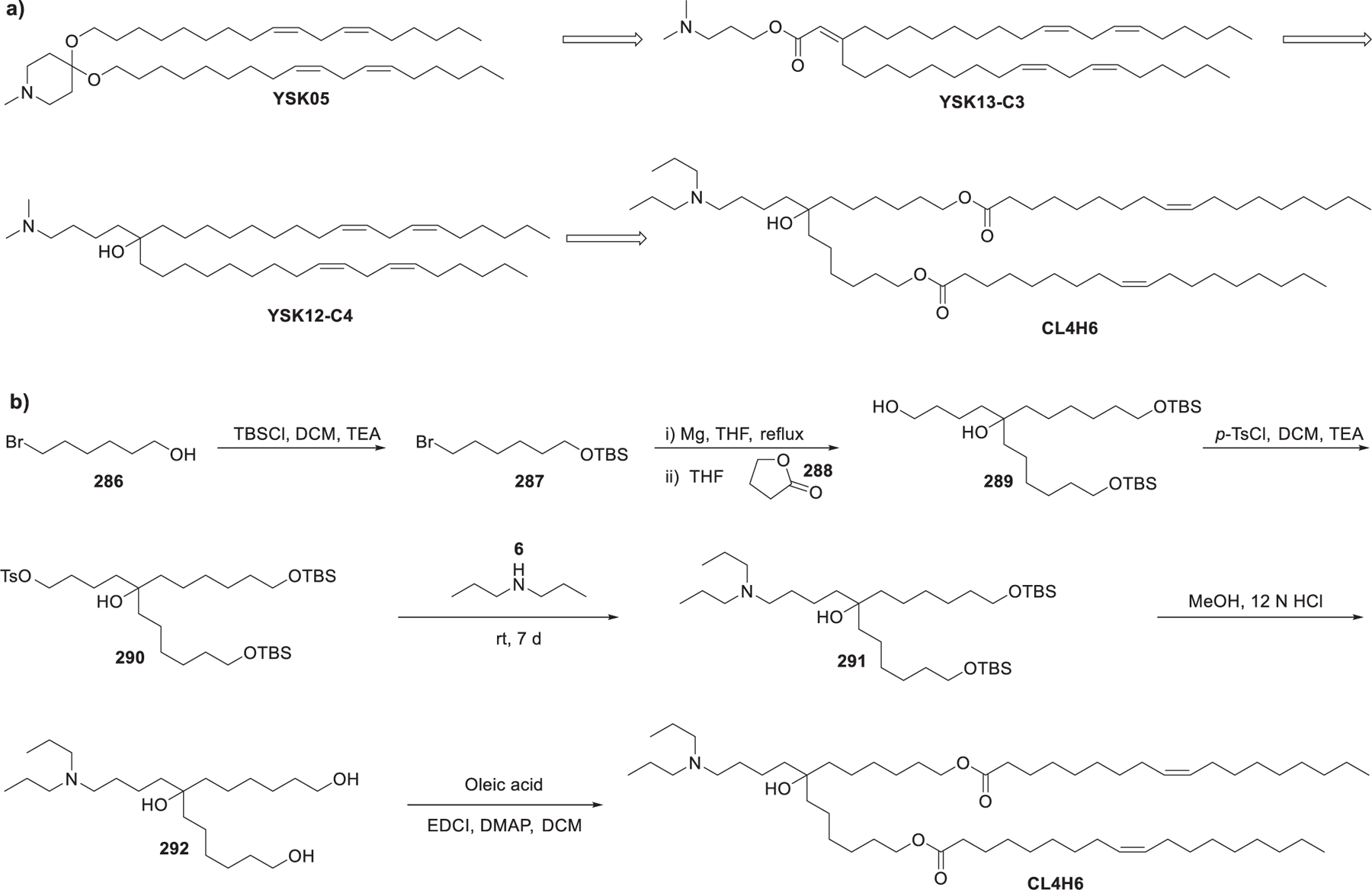
Chemical structures of YSK series lipids and a synthetic route to CL4H6.
In 2014, Gindy et al. reported that LNPs containing lipids with asymmetric tails were well-tolerated as they were rapidly cleared in vivo.566 Asymmetric lipid-formulated LNPs favor the adoption of destabilizing nonbilayer structures to facilitate the release of RNA cargo. However, these LNPs suffer from physical instability during storage, due to their weak hydrophobic interactions between the lipid components in LNPs, which may be driven by the asymmetric lipids.566 Based on this work, Suzuki et al. developed a series of ionizable lipids that contained two asymmetric hydrophobic tails (e.g., L021) (Figure 49). In vitro screening and structure–activity relationship studies identified L021 that contained an N-methylpiperdine head group and a C9 short tail, as the lead candidate for siRNA delivery (Figure 49).567 Intravenous injection of L021 LNPs encapsulating FVII siRNA induced significant FVII gene knockdown with an ED50 of ~0.02 mg/kg in mice. Additionally, L021 LNPs were stable to be stored at 4 °C for 1.5 years as a liquid without an increase in LNPs size or degradation of lipids. The cyclic N-methylpiperidine head group may increase hydrophobic interactions between the lipid components in the LNPs, thus enhancing the bilayer packing and conferring LNPs with sufficient stability.567 In 2016, they also revealed that both low-density lipoprotein (LDL) receptor and Apolipoprotein E (ApoE) played significant roles in in vivo intracellular uptake of L021 LNPs. Besides, in vivo biodistribution of L021 LNPs was significantly influenced by ApoE.568 In 2017, they synthesized a biodegradable analog of L021, L101, by introducing an ester linker into the long hydrophobic tail (Figure 49).569 As shown in Figure 49, monohydrolysis of diethyl decanedioate 294 gave decanedioic acid monoethyl ester 295, which was treated with oxalyl chloride to afford acyl chloride 296. Substitution reaction between acyl chloride 296 and in situ generated organocuprate reagent led to ketone 297. The biodegradable tail was installed via transesterification between 297 and (Z)-non-2-en-1-ol 298 catalyzed by Ti(Oi-Pr)4, giving compound 299. Finally, the ketone 299 was reduced and coupled with amino acid 301 to furnish lipid L101. L101 LNPs encapsulating anti-FVII siRNA induced significant FVII gene silencing in vivo in mice in a dose-dependent manner with an ED50 of 0.02 mg/kg in terms of siRNA. They also found that L101 was resistant to mouse or human serum esterase but could be degraded by intracellular hepatic enzymes (e.g., lysosomal esterases). Compared to biodegradable L101, nondegradable L021 showed a 100-fold more accumulation in mouse liver.569
Figure 49.
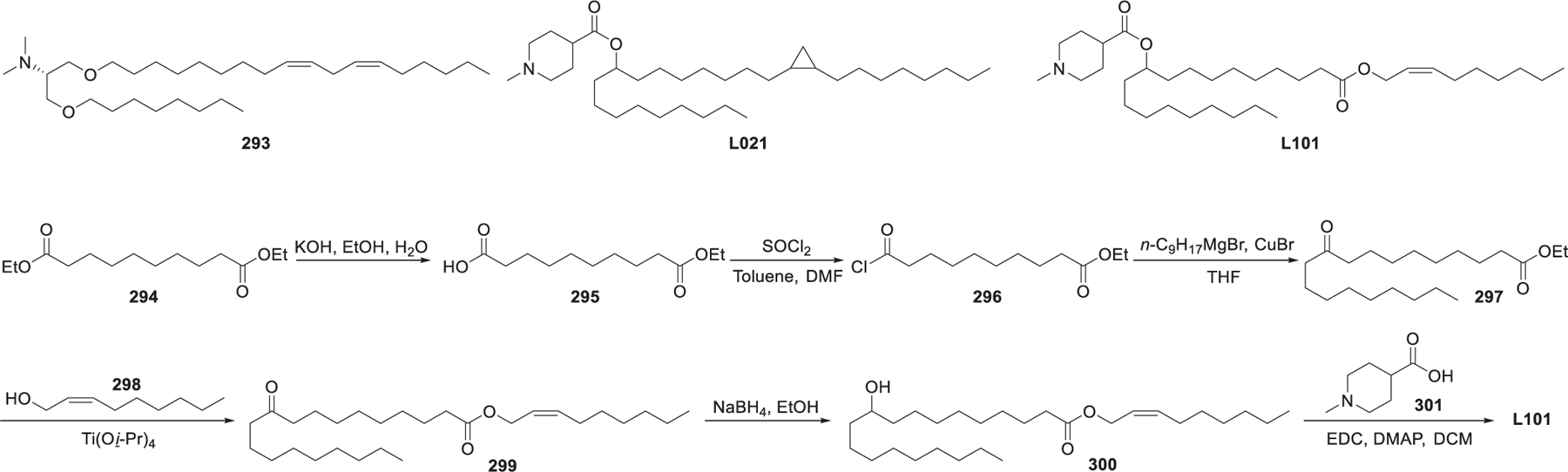
Chemical structures of lipid 293, L021, and L101 and a synthetic route to L101.
In 2017, Viricel et al. developed a series of ionizable switchable lipids that contained a tricyclic ionizable head group and two dodecyl hydrophobic tails.570 Various ionizable amino groups can be attached to the 4′-position of the central pyridine ring to modify the head group (Figure 50a). The synthesis of CSL3 began with a substitution reaction between 2,6-dibromopyridin-4-amine 302 and 2-iodo-N,N-dimethylethan-1-amine 303 followed by methylation of the resulting amine, giving amine 304. The tricyclic core 306 was formed via Suzuki coupling571,572 between compound 304 and phenylboronic acid 305. Finally, the hydrophobic tails were installed via Sonogashira coupling573 followed by hydrogenation of the carbon–carbon tribonds (Figure 50b). As reported in their previous work, lipids with this type of backbone can change their conformation upon protonation of the central pyridine at endosomal pH values (pH 5) (Figure 50c), and lipids with alkyl tails of 12 carbons showed the highest siRNA delivery efficiency in vivo.574 Once the central pyridine is protonated, intramolecular hydrogen bonding within the tricyclic core will lead to an orientation change of the two hydrophobic tails. Conformation change of the hydrocarbon tails destabilizes LNPs and prompts the release of the siRNA payload (Figure 50c).570,574 Results showed that siRNA-loading CSL2 LNPs and siRNA-loading CSL3 LNPs showed dose-dependent gene knockdown activity in vitro, whereas siRNA-loading CSL1 LNPs was ineffective. The failure of CSL1 LNPs in gene knockdown may be attributed to the bulky head group that can inhibit intramolecular hydrogen bond formation. CSL4, lacking the two methoxy groups, remained in endosomes as it could not undergo a conformational change.570
Figure 50.
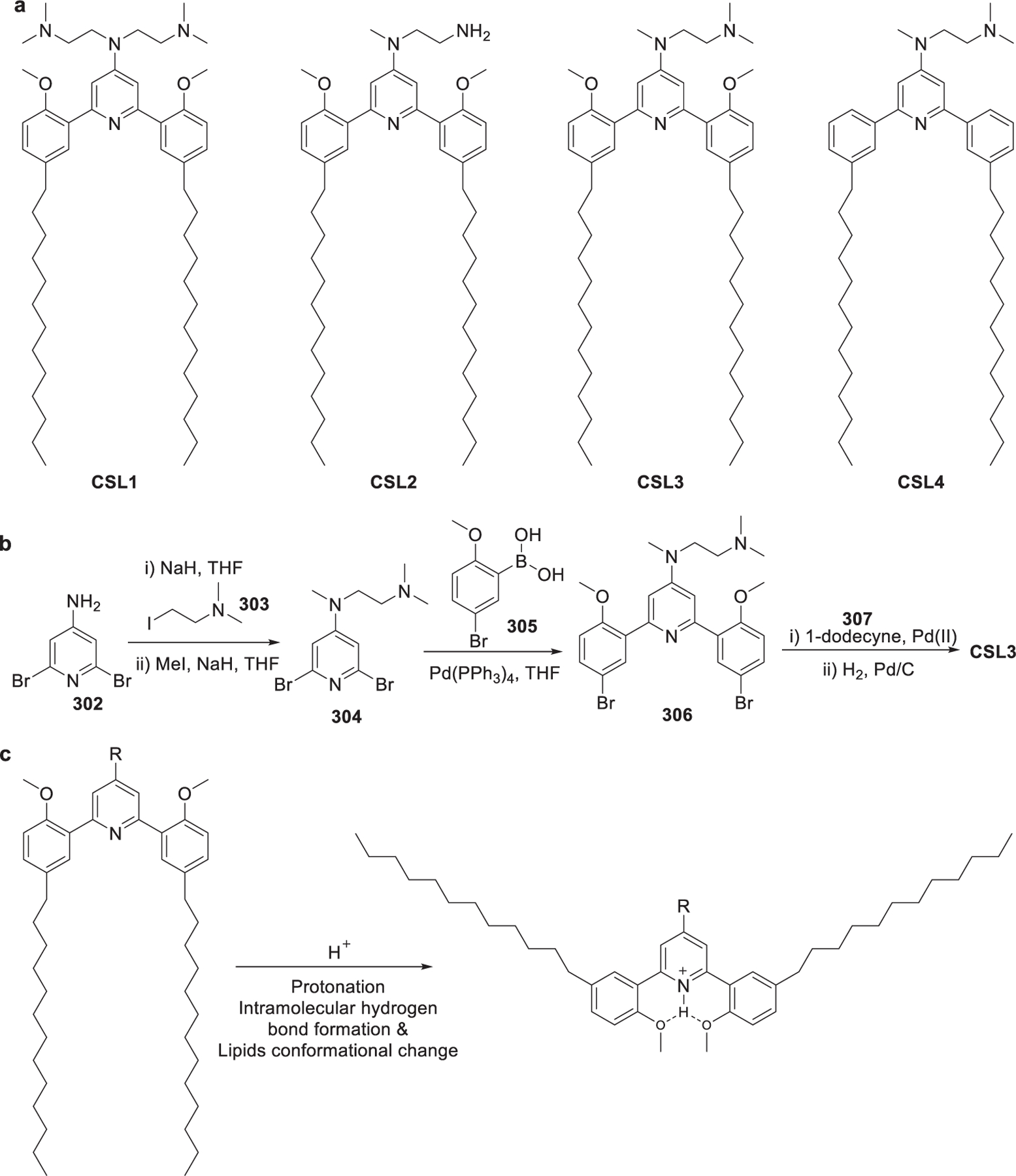
(a) Chemical structures of ionizable switchable lipids. (b) Synthetic route to CSL3;. (c) Protonation-induced conformational change of the ionizable switchable lipids.
2.2.2.2. Bioactive Molecules Derived Lipids.
Lipids incorporating various bioactive molecules, such as oligopeptides/amino acids, sugar,575 and small molecular drugs,59,576,577 have been developed as formulation components for RNA delivery. Cholesterol, for example, is biologically compatible and generally used as a helper lipid to stabilize the LNPs.578,579 Many cholesterol-based ionizable lipids have been synthesized and used in the applications of RNA delivery.496,580,581
In 1991, Gao et al. reported the synthesis of DC-Chol, a cholesterol-derived ionizable lipid in which the dimethylamine head group is attached to the cholesterol moiety via a biodegradable carbamate linker (Figure 51).581 In contrast to cationic liposome formulated with fully charged cationic lipids, such as DOTMA and DOTAP, LNPs formulated with DC-Chol/DOPE are only partially charged at pH 7.4,582 thus inhibiting the aggregation of LNPs.583 In 2011, Zhang et al. found that at a DC-chol/siRNA weight ratio as high as five or ten, DC-Chol/DOPE LNPs showed the highest siRNA delivery efficiency into SK-BR3 cells in vitro.584 In a mouse tumor model, PEGylated DC-Chol/DOPE LNPs containing kinesin spindle protein (KSP) siRNA led to a significant KSP gene silencing at tumor sites and a strong inhibition of tumor growth after systemic administration in mice at a siRNA dose of 1 mg/kg.585 The DC-Chol/DOPE LNPs have also been used in encapsulating alpha-1-antitrypsin (AAT) mRNA, resulting in high transfection efficiencies and sustained AAT protein expression in A549 cells in vitro.586 In 2016, Zhao et al. synthesized another cholesterol-based ionizable lipid, designated as DMAPA-chems, in which the head group and cholesterol moiety were connected through a 1,4-succinic acid linker with biodegradable ester and amide linkage bonds (Figure 51).587 DMAPA-chems LNPs encapsulating Notch1 siRNA showed high Notch1 gene knockdown efficiency in SKOV3 cells at an N/P ratio of 100.587
Figure 51.
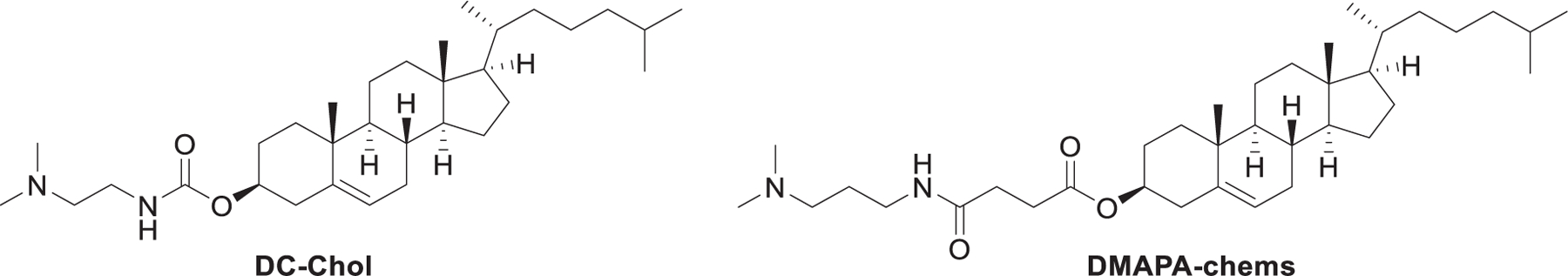
Chemical structures of cholesterol-derived ionizable lipids.
In 2019, Hou et al. reported a library of vitamin-derived lipids: VB3-Lipid, VC-Lipid, VD-Lipid, VE-Lipid, and VH-Lipid (Figure 52).59 These vitamin-derived lipids were prepared through the conjugation of vitamins or their derivatives with an amino lipid with a carboxylic acid group through ester or amide bond formation. LNPs formulated with VC-Lipid were optimal in delivering mRNA encoding an antimicrobial peptide IB367 along with a cathepsin B (CatB) via a cleavable linker into macrophages. The VC-LNPs engineered macrophages provide a promising therapeutic against multidrug-resistant bacteria-induced sepsis.59
Figure 52.
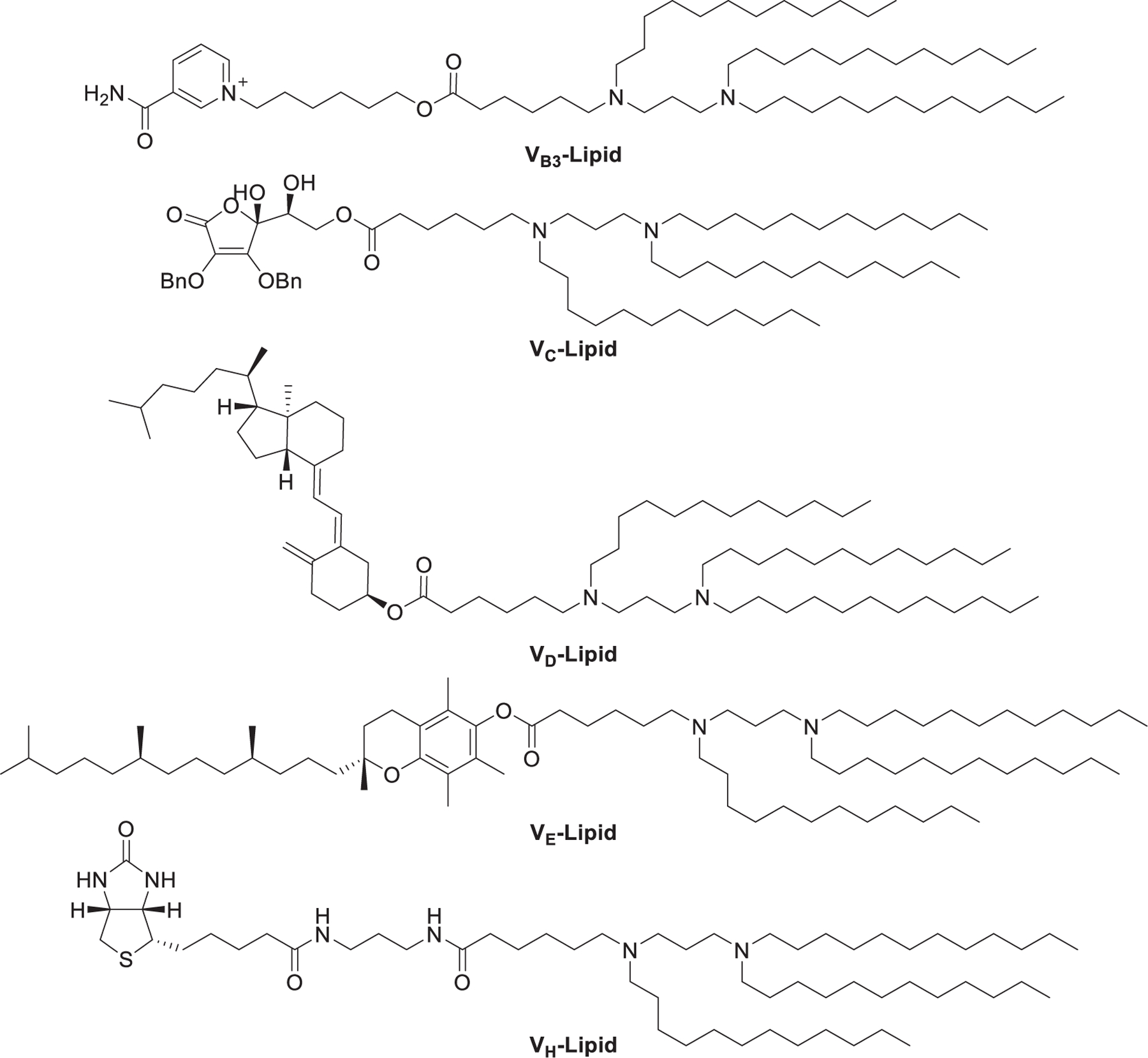
Chemical structures of vitamin-derived ionizable lipids.
Given the fact that phospholipids and glycolipids are natural components of the cell membrane, Zhang et al. synthesized a library of phospholipid and glycolipid-derived ionizable lipids (PLs and GLs) and used these materials to formulate biomimetic nanoparticles for mRNA delivery (Figure 53).588 The synthesis of PL1 started with phosphorylation of 3-bromo-1-propanol 309, giving phosphate triester 310, while the synthesis of GL1 began with glycosylation of β-d-galactose-pentaacetate 314 with 3-bromo-1-propanol 309 promoted by a boron trifluoride diethyl etherate complex. The remaining chemical conversions include the substitution of bromide in compounds 310 and 315 with monoprotected 1,3-propanediamine 311, removal of the Boc protecting group using TFA, and reductive amination reaction.
Figure 53.
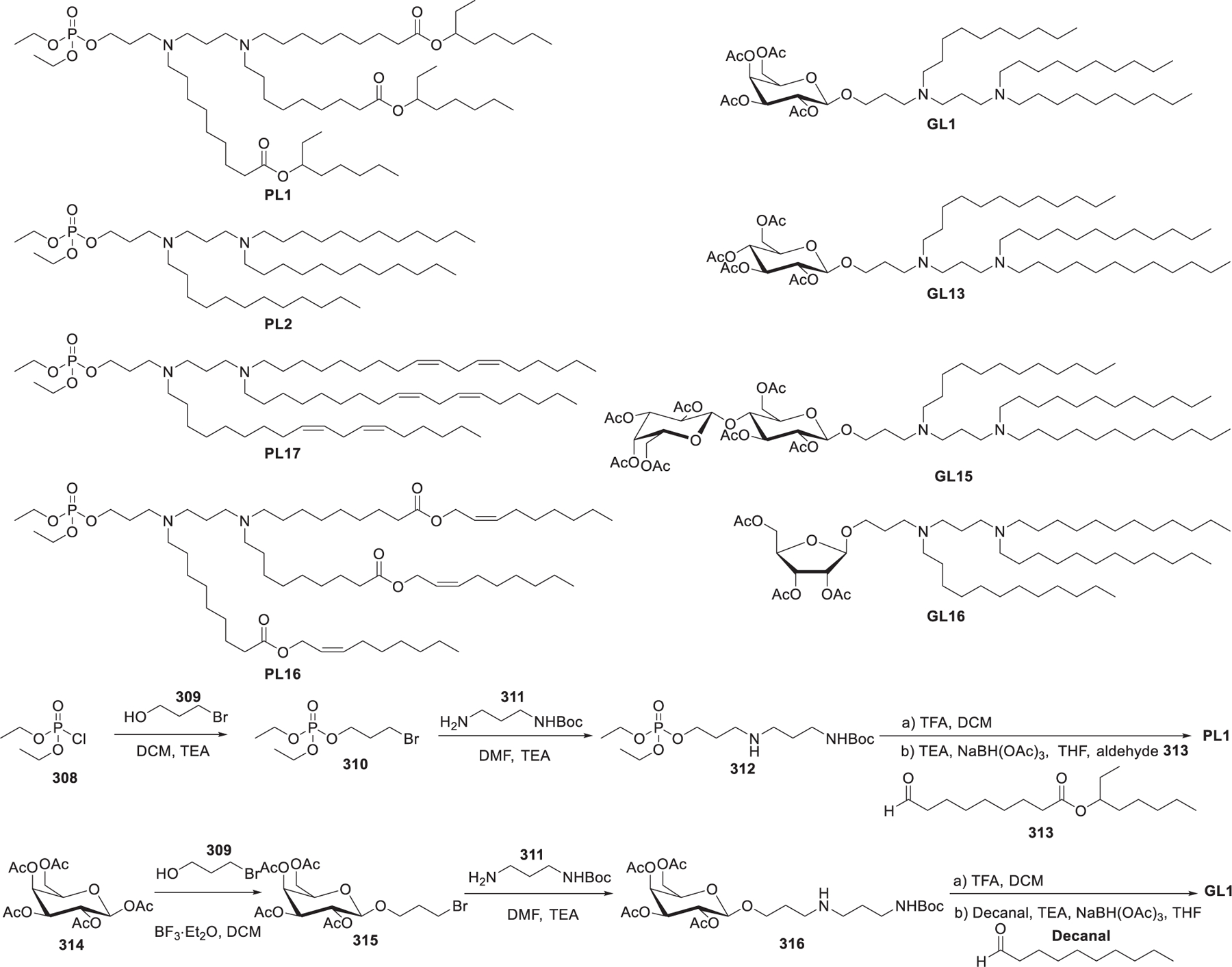
Chemical structures and synthesis of representative phospholipids and glycolipids.
2.2.3. Lipidoids.
Combinatorial chemistry approaches permit the rapid synthesis of large libraries of ionizable lipids, which are termed lipidoids or lipid-like compounds.518,589–592 An important advantage of combinatorial chemistry approaches is that a vast number of lipids with large diversities can be synthesized by assembling variant building blocks. The combination of this method with high-throughput screening can lead to the quick discovery of ionizable lipids with good RNA delivery efficiency.518,589–591 Many types of reactions can be applied to produce lipidoids, including aza-Michael addition of amines with acrylamides or acrylesters,589,593,594 ring-opening of epoxides with amines,590,595,596 and reductive amination of aldehydes597–599 (Figure 54). Thiol–yne click reaction600,601 and multicomponent reactions602–604 can also be used in the synthesis of lipidoids (Figure 54).
Figure 54.
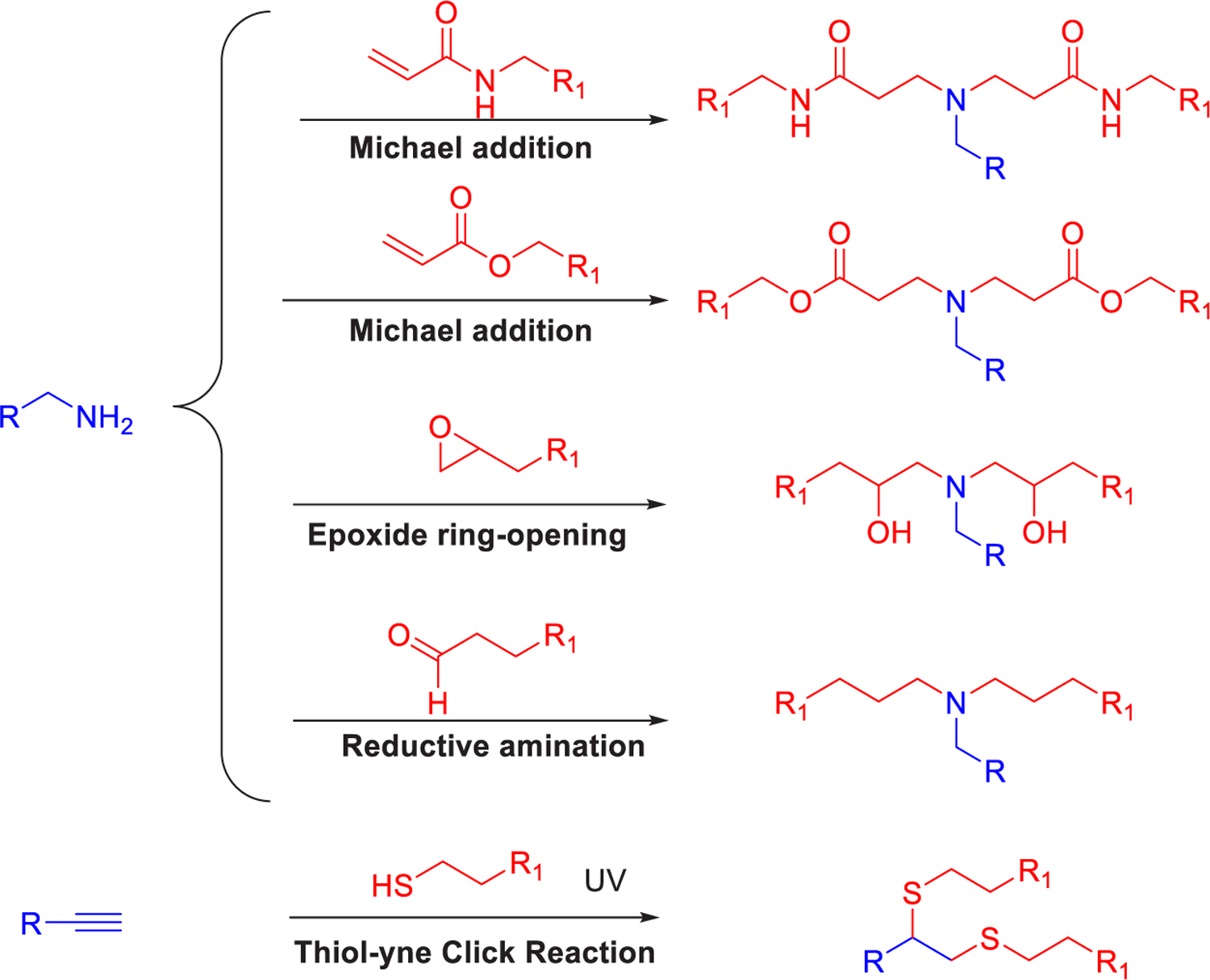
Representative reactions for the preparation of lipidoids.
2.2.3.1. Lipidoids Synthesized via Michael Addition.
In 2008, the Anderson and Langer group reported a library of lipidoids composed of 1200 lipids by conjugating 54 polyamines with 17 acrylamides and acryl ester with varying length of the alkyl group (between 9 to 18) (Figure 55).589 98N-12 with 5 tails attached (98N12-5) turned out to be a lead lipid when tested for gene silencing in HeLa cells. 98N12-5 LNPs encapsulating anti-FVII siRNA induced a 90% reduction of expression of the blood clotting Factor VII at a siRNA dose of 5 mg/kg in murine liver.589 In 2009, Nguyen et al. found that 98N12-5 LNPs encapsulating siRNA targeting the influenza nucleoprotein gene could lead to effective antiviral activity in vivo following systemic administration in mice.605 Pharmacodynamic studies of 98N12-5 LNPs showed they specifically targeted the liver following intravenous and intraperitoneal injection,606 and tissue biodistribution data indicated that >90% injected dose was distributed in the liver.607 PCSK9 siRNA-loading LNPs formulated with 98N12-5/cholesterol/DMG-mPEG2000 at a molar ratio of 42:48:10 led to significant silencing of PCSK9 gene in mice following systemic administration.608
Figure 55.
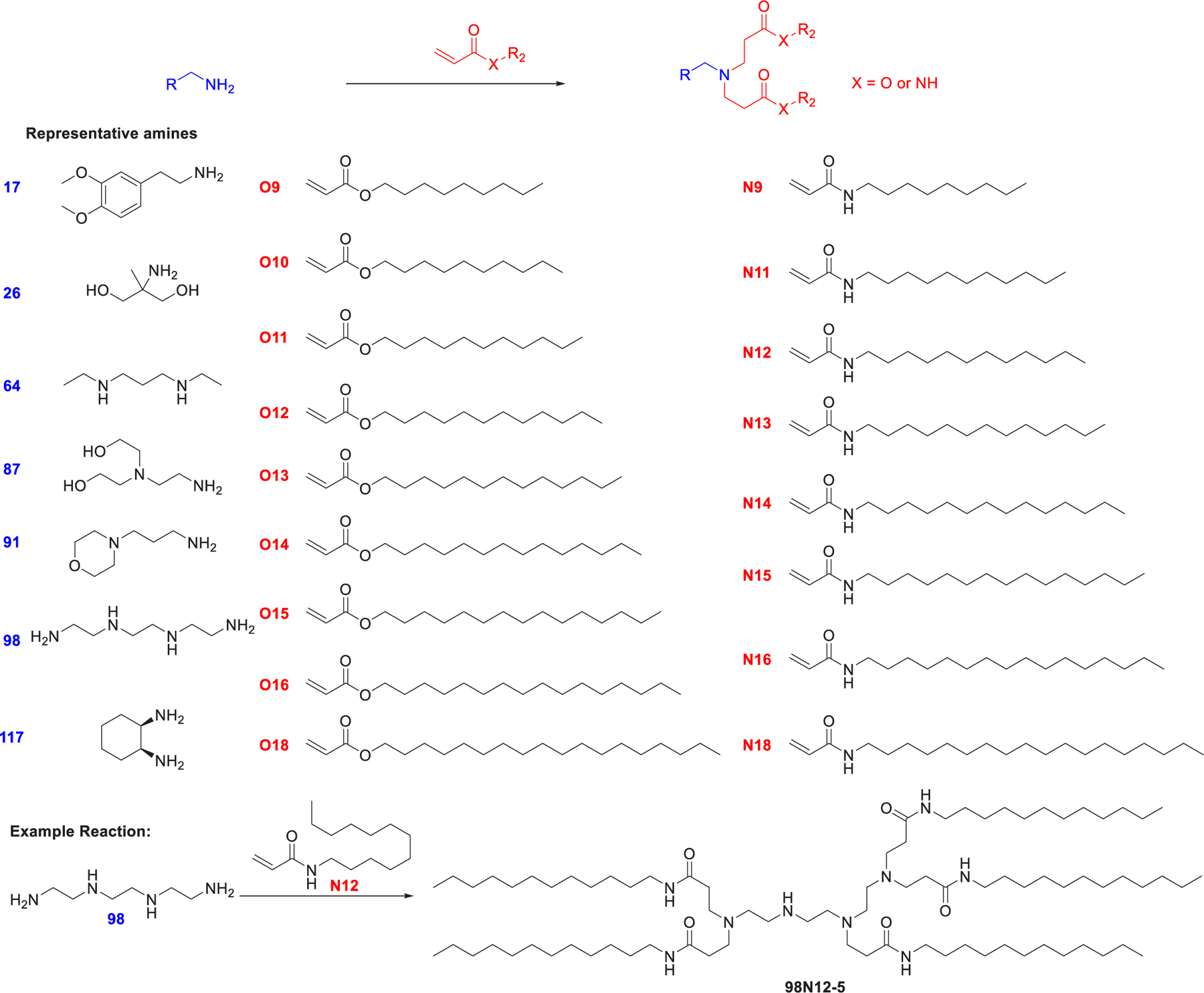
Synthesis of lipidoids via Michael addition of amines with acrylamides and acryl ester.
In 2014, Whitehead et al. evaluated the antifactor VII siRNA delivery efficacy of a library of 1400 lipids prepared from amines and acrylate tails and identified 304O13 as the lead candidate, which induced factor VII gene silencing with an ED50 of 0.01 mg/kg in mice (Figure 56).609 Structure–activity relationship studies showed that lipids containing three or more O13 tails and at least one tertiary amine may exhibit strong siRNA delivery efficacy. Then a secondary library of lipids was prepared from 12 polyamines and O13, and 503O13 was shown to be the best lipid for siRNA delivery in vivo.609 In 2016, the Whitehead group studied another library of lipidoids synthesized from the combination of 6 amines and 13 tails (Figure 56) and found that lipidoids synthesized from alkyl acrylate were shown to be more effective in siRNA delivery than lipidoids prepared from the methacrylate tail.610 Isodecyl acrylate (Oi10)-conjugated lipidoids exhibited the highest siRNA delivery efficiency in the whole library. The chemical structure of the tails significantly affected the surface pKa values of LNPs.610 In 2018, Ball et al. reported that 306Oi10-based LNPs can codeliver Factor VII siRNA and luciferase mRNA in vivo, which may be a potential method for treating diseases associated with both aberrant upregulation and downregulation of genes. Co-formulation of siRNA and mRNA in a single LNP substantially enhanced their delivery efficiency compared to LNPs encapsulating individual RNAs.611 In 2019, Hajj et al. found that 306Oi10, with a one-carbon branch, increased luciferase mRNA delivery 10-fold compared to the linear tailed 306O10 (Figure 56).612 The improvement may be due to the branch in the Oi10 tail that increases the distance between lipidoid molecules within the LNP bilayer, thus facilitating protonation of the lipidoid molecules at the late stage of the endosome.612 Additionally, 360Oi10 LNPs could deliver Cy5-labeled luciferase mRNA to hepatocytes, endothelial cells, and Kupffer cells in the liver following intravenous administration. Besides, 360Oi10 LNPs were capable of sufficiently encapsulating and codelivering functionally three different mRNAs (mCherry, firefly luciferase, and erythropoietin) in mice in vivo.613
Figure 56.
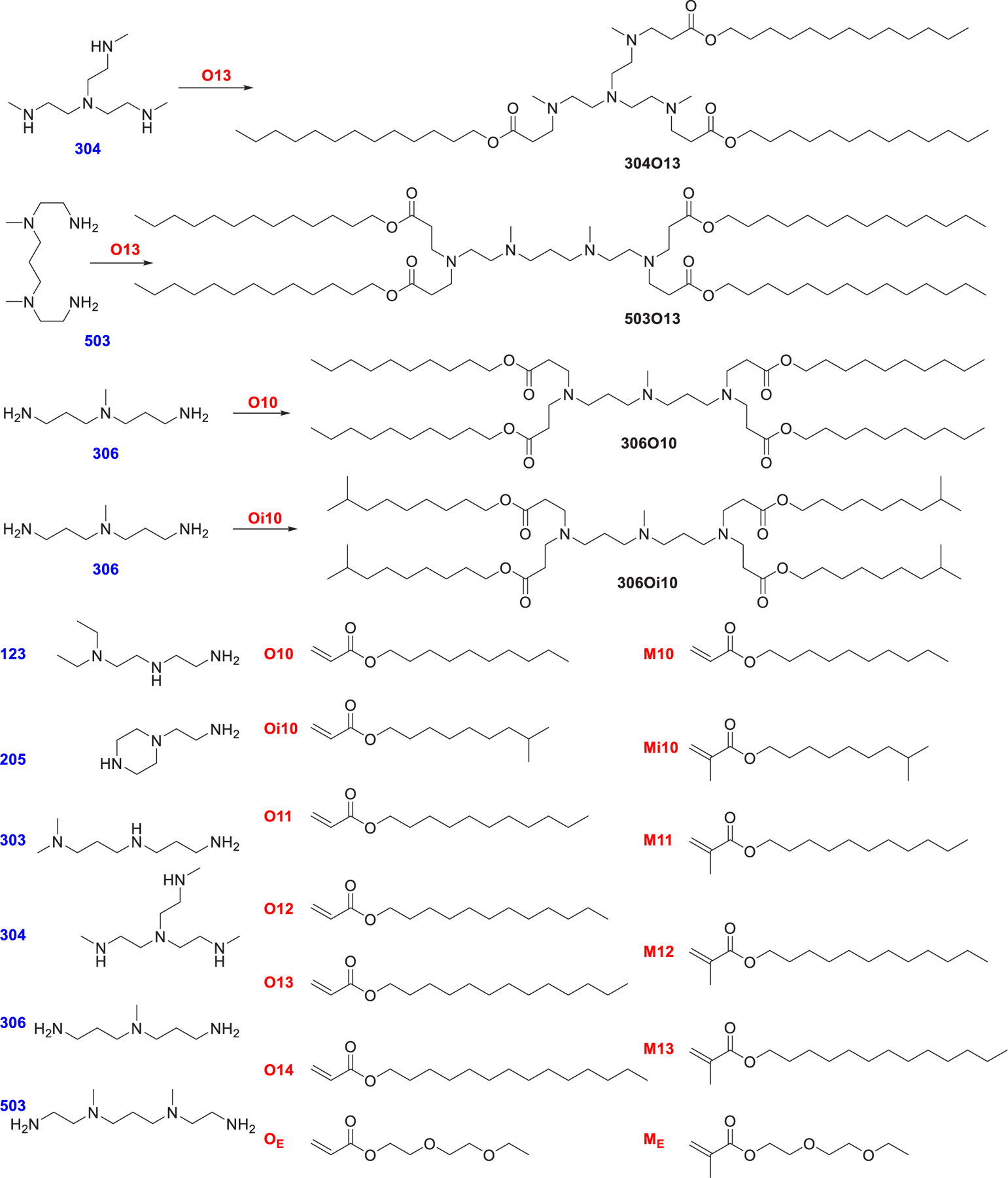
Lipidoids synthesized via Michael addition of alkyl-amines (blue) to alkyl-acrylate or methacrylate tails (red).
By utilizing the combinatorial strategy, the Xu group has developed many bioreducible ionizable lipids, of which the hydrophobic tails contain both disulfide bonds and ester bonds (Figure 57).614–621 Disulfide bonds are cleavable under the intracellular GSH condition, and esters can be hydrolyzed by esterase, thus facilitating degradation of lipidoids and RNA release. In 2014, 1-O16B was found as a lead candidate, which can deliver GFP-targeting siRNA to downregulate GFP in MDA-MB-231 cells.383 In 2016, by screening another library of bioreducible ionizable lipids, 8-O14B was shown to be capable of delivering Cas9 and sgRNA into GFP-HEK cells, resulting in greater than 70% knockdown of GFP expression with efficiency. Structure–activity relationship study suggested that lipids containing hydrophobic tails with a tail length between 14 to 18 carbons were more efficient in Cre protein delivery than lipids with 12-carbon tails.622 BAME-O16B LNPs were used in codelivery of Cas9 mRNA and sgRNA, leading to knockout of cellular GFP expression with efficiency up to 90% in human embryonic kidney cells and reduction of serum PCSK9 by 80% in C57BL/6 mice.618 In 2019, LNPs formulated with 306-O12B-3/cholesterol/DOPE/DSPE-PEG2000 at a ratio of 16:4:1:1 (w/w) were used to deliver ASOs targeting proprotein convertase subtilisin/kexin type 9 (PCSK9) in mice, resulting in significant PCSK9 silencing with the ED50 as low as 0.034 mg/kg following systemic administration.623 In 2020, Ma et al. synthesized a class of neurotransmitter-derived lipidoids (NT-lipidoids)615 for brain delivery of RNA, as neurotransmitters (e.g., tryptamine) can traverse the blood–brain barrier (BBB) effectively.624,625 LNPs formulated with 306-O12B-3 and NT1-O14B at a ratio of 7:3 (w/w) could cross the BBB and deliver Tau-ASO into neuronal cells, leading to a reduction of tau mRNA by ~50% and a reduction of tau protein by ~30% at a Tau-ASO dose of 20 μg following five times intravenous administration.615 In 2021, Zhao et al. reported two top lipidoid candidates, 93-O17S and 9322-O17S, for delivering mRNA into the primary T lymphocytes via a rough-to-detail screening approach.614 Structure–activity analysis showed that the amine head group, heteroatom substitution, spacer length, and linker type in the tail are all critical factors that influenced the RNA delivery efficiency. Both 93-O17S LNPs and 9322-O17S LNPs could deliver Fluc mRNA to the spleen efficiently in mice in vivo. Systemic delivery of Cre mRNA using 93-O17S LNPs could result in ~8.2% and ~6.5% of delivery efficacy into CD4+ and CD8+ T cells in mice, respectively.614
Figure 57.
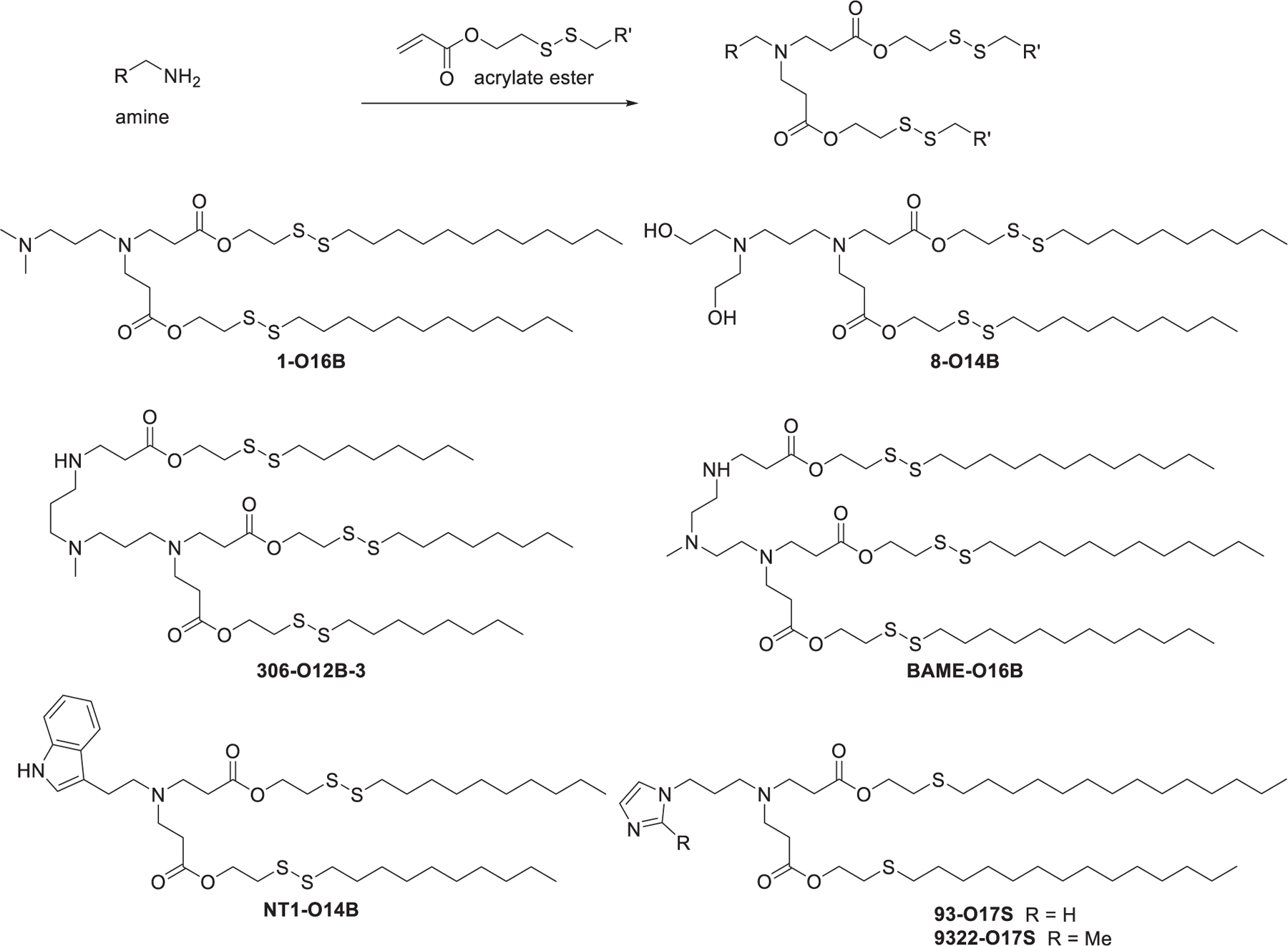
Chemical synthesis of bioreducible lipids.
In 2016, Zhou et al. prepared a library of over 1500 degradable dendrimer-like lipids via Michael addition between various amines and acrylate esters followed by thiol–ene reaction with alkyl thiols.626 5A2-SC8, a lipid with five nitrogen atoms and five short alkyl chains, was identified as the top-performing lipid in the library (Figure 58). LNPs encapsulating let-7g miRNA prepared from 5A2-SC8/DSPC/cholesterol/PEG-lipid induced strong let-7g gene silencing effects, inhibited tumor growth, and prolonged the survival time of mice with liver cancer at a miRNA dose of 1 mg/kg following weekly intravenous administration.626 In 2018, Zhou et al. optimized the formulation of 5A2-SC8 LNPs by lowering the mole fraction of 5A2-SC8 to 24% and using DOPE instead of DSPC; the optimized 5A2-SC8 LNPs showed strong luciferase mRNA delivery efficacy in mice.627 In FAH−/−mice, 5A2-SC8 LNPs encapsulating fumarylacetoacetate hydrolase (FAH) mRNA induced equivalent levels of ALT, TBIL, and AST compared to wild type mice following intravenous injection.627
Figure 58.
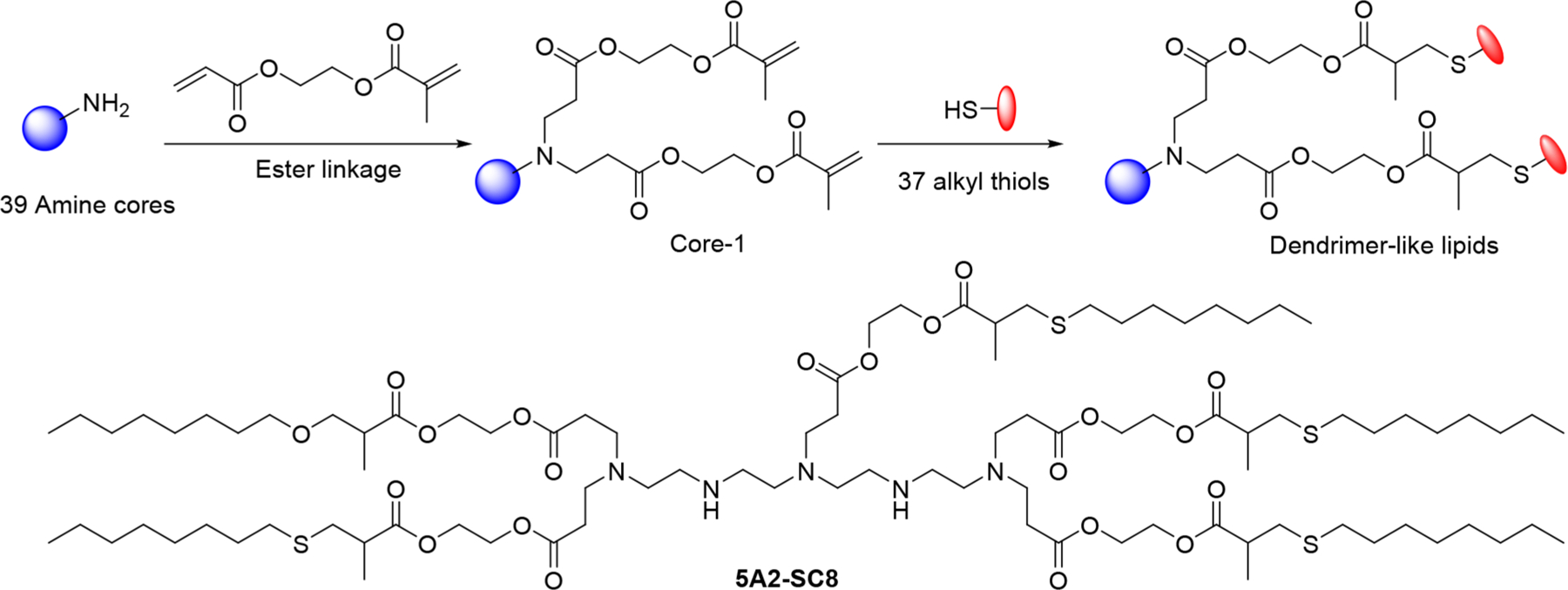
Modular strategy for the synthesis of dendrimer-like lipids and chemical structure of 5A2-SC8.
2.2.3.2. Lipidoids Synthesized via Epoxide Ring-Opening.
In 2010, Love et al. synthesized a library of amino alcohol-derived lipids via a ring-opening reaction of epoxides with varying tail lengths with amines (Figure 59a).590 C12-200 LNPs encapsulating FVII siRNA induced effective silencing of factor VII in mouse hepatocytes at low doses after intravenous administration.590 In 2013, Sahay et al. confirmed that siRNA delivery by C12-200 LNPs was mediated by Cdc42-dependent micropinocytosis.628 In a later study, a formulation of C12-200/DOPE/cholesterol/C14-PEG2000 at a molar ratio of 35:16:46.5:2.5 was used to prepare the optimized mRNA-loading C12-200 LNPs, which induced 7-fold more EPO protein expression than the initial formulation (C12-200/DSPC/cholesterol/C14-PEG2000).629 C12-200 LNPs prepared in this formulation have been used in the study for mRNA-mediated human α-galactosidase protein replacement therapy in mice and nonhuman primates.630 In a later study, direct intramyocardial injection of C14-113 LNPs encapsulating GFP mRNA in mice induced rapid and transient GFP expression in a dose-dependent manner with limited off-target biodistribution.631 In 2013, Xu et al. synthesized G0-C14632 via epoxide ring-opening of alkyl epoxide by generation 0 of poly(amidoamine) (PAMAM) (Figure 59b); this ionizable lipid has been used in the delivery of both siRNA632 and mRNA633,634 for the treatment of cancer in mice.
Figure 59.
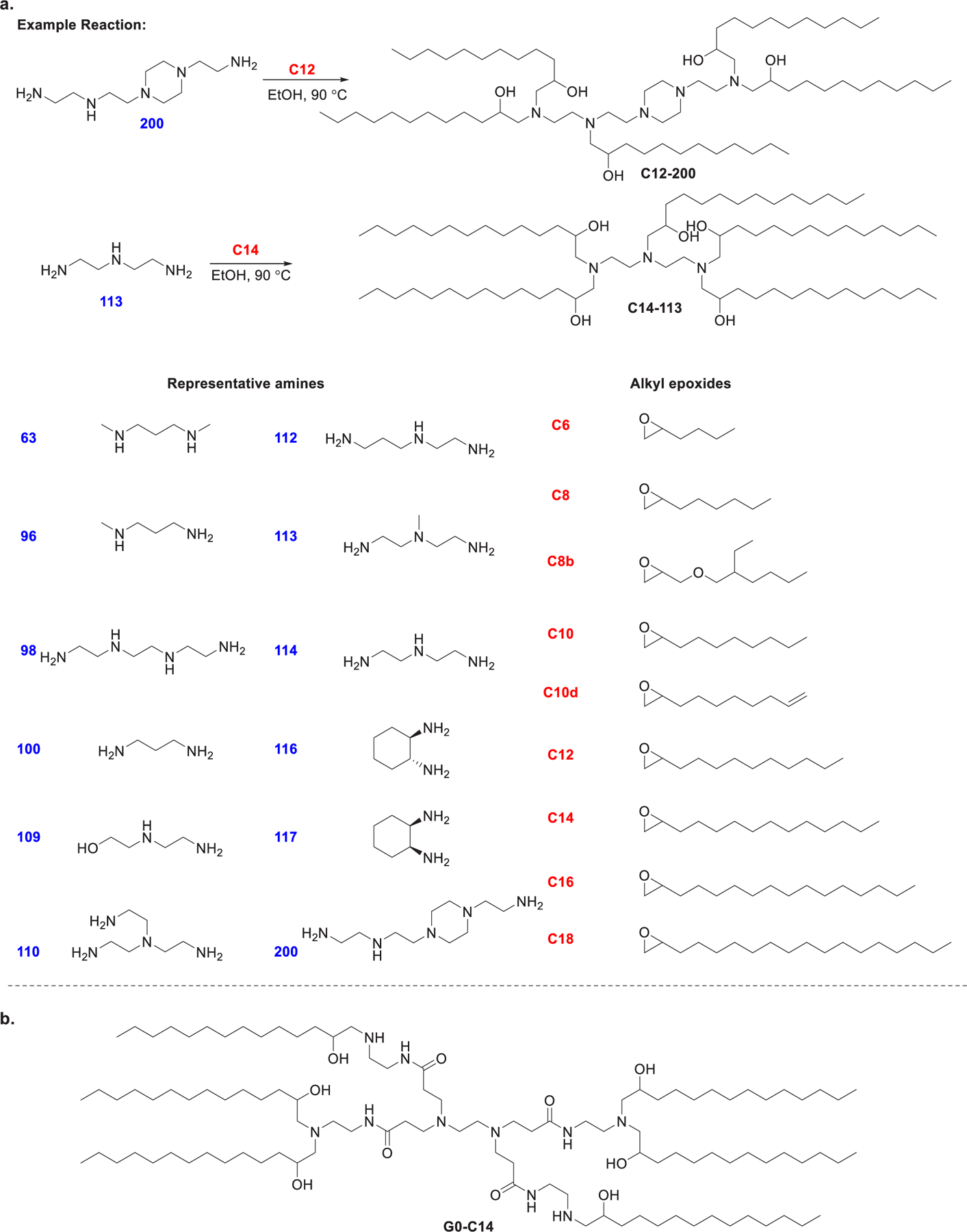
Synthesis of lipids from ring-opening reaction between amines and epoxides.
Aminoglycosides are a class of antibiotics that contain three to five amino-substituted sugars.635,636 The general mechanism of their antibacterial bioactivities is to selectively bind to bacterial 30S rRNA (rRNA).637,638 Besides, aminoglycosides are capable of penetrating the cell membrane of bacteria by disrupting the lipopolysaccharide components639 which might facilitate endosomal escape. In 2013, Zhang et al. synthesized a family of lipid-modified aminoglycosides for in vitro and in vivo siRNA delivery.640 Eight aminoglycosides were reacted with terminal epoxides bearing tail lengths of 10–16 carbons (Figure 60). Their results indicated that C11 and C12 tails were the preferred chain length for siRNA delivery, whereas lipids with tails longer than C13 were generally inefficient in siRNA delivery, which may be caused by their decreased solubility and lower fluidity. HG and GT are favored core scaffold aminoglycosides for the synthesis of lipids with high siRNA delivery efficiency, while AP-, PR-, NO-, and RB-derived lipids generally showed low siRNA delivery efficiency. HG-C11 LNPs encapsulating anti-FVII siRNA were able to induce effective Factor VII knockdown in mice with an ED50 of 0.04 mg/kg.640 In 2020, Yu et al. synthesized another library of aminoglycoside-derived lipids and evaluated their potency as mRNA formulation components.641 Four aminoglycosides, namely hygromycin, amikacin, gentamycin, and Geneticin, were reacted with seven epoxides bearing hydrocarbon tails and with five acrylic esters through epoxide ring-opening and Michael addition, respectively. GT-C10, obtained from the reaction of gentamycin and epoxide C10, was able to deliver luciferase mRNA at a dose of 0.05 mg/kg to produce a 107 average luminescence intensity in mouse liver following an intravenous administration.641
Figure 60.
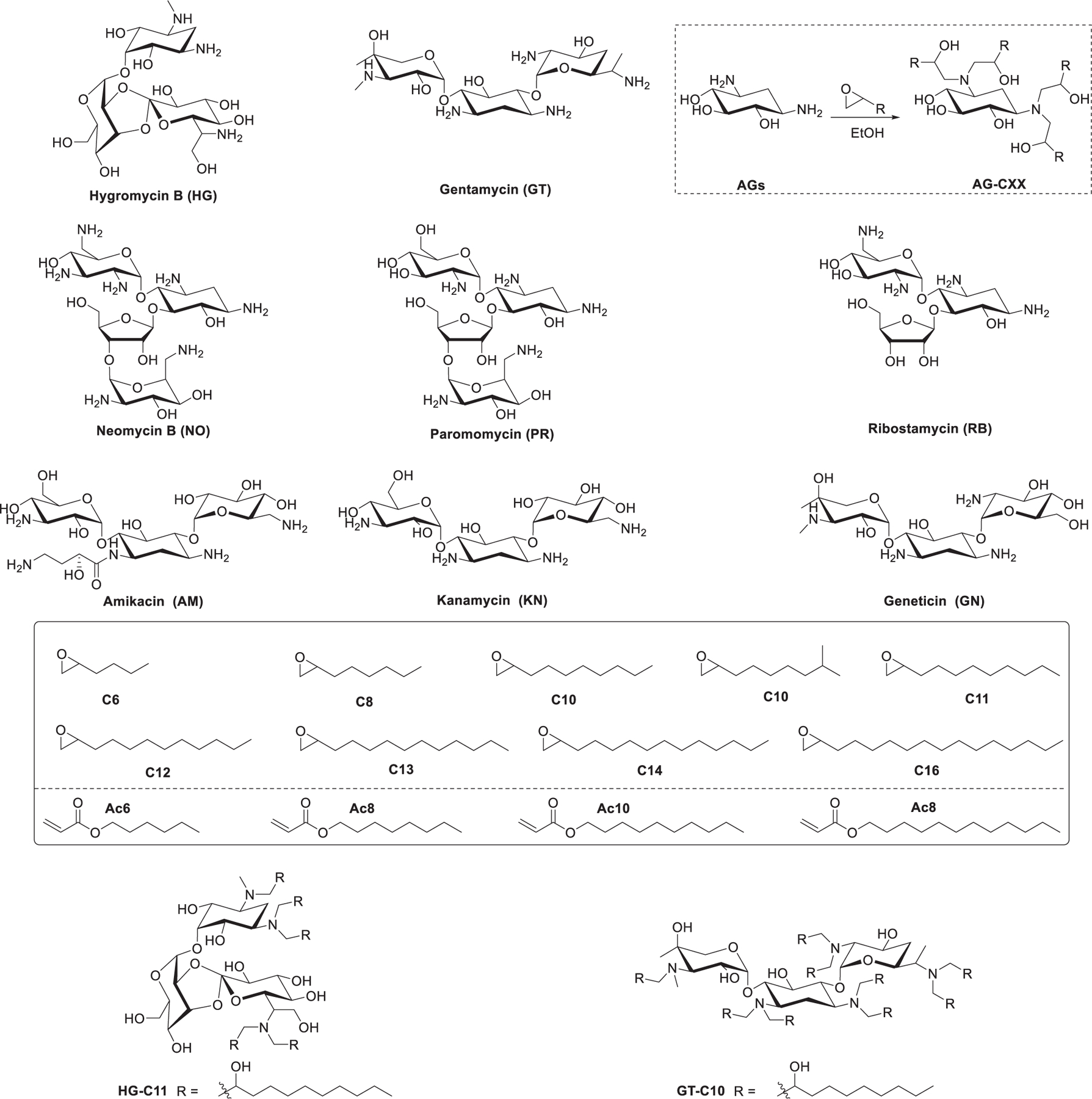
Chemical structures of aminoglycosides, epoxides, and acrylic esters and a representative schematic reaction between aminoglycoside and epoxide.
In 2014, Dong et al. developed a series of 1,3,5-triazinane-2,4,6-trione (TNT)-derived lipids that contained a six-membered core and three hydrophobic tails (Figure 61). TNT-4 was identified as a lead lipid, which showed efficient delivery of pDNA and siRNA both in vitro and in vivo.642 In 2016, Li et al. synthesized four analogs of TNT-4 by exchanging the positions of the tertiary amino group and secondary hydroxyl group relative to the TNT ring (Figure 61) and evaluated the effects of spacing around the trazinane trione (TNT) ring on mRNA delivery.643 The TNT-b10 luciferase mRNA LNPs induced 2-fold higher luciferase level than that of TNT-4 in vitro. Intravenous and intraperitoneal administration of TNT-b10 LNPs encapsulating luciferase mRNA induced the luciferase expression in the mouse spleen.643
Figure 61.
Chemical structures of TNT-4 and TNT-b10.
In 2014, Dong et al. developed a library of biomimetic ionizable lipids derived from amino acids, peptides, and polypeptides.518 These biomimetic lipids were synthesized via an addition reaction of amines to alkyl epoxide and acrylate esters as well as a reductive amination reaction between amines and alkyl aldehyde (Figure 62). cKK-E12 was identified as the lead lipid, which reduced FVII protein by 50% at a siRNA dose as low as 0.002 mg/kg in mice. Intravenous injection of cKK-E12 siRNA LNPs into nonhuman primates (NHP) at a siRNA dose of 0.3 mg/kg led to a reduction of TTR serum level by 95%. Structure–activity relationship analysis showed that lipids that contained a dilysine-derived diketopiperazine core and lipid tails between 12 and 14 carbon tail length were the most effective. cKK-E12, composed of a diketopiperazine core derived from lysine and four amino alcohol-based hydrophobic tails, can be synthesized via dimerization of Cbz-protected lysine, followed by removal of Cbz and epoxide ring-opening of the epoxide compound. cKK-E12 LNPs had also been used in codelivery of Cas9 mRNA and sgRNA to hepatocytes at clinical doses in mice, inducing >80% editing of PCSK9 gene in the liver.644 In 2019, Lokugamage et al. used cKK-E12 LNPs encapsulating mRNA to study the effects of toll-like receptor 4 (TLR4) on LNPs-mediated mRNA delivery.645
Figure 62.
Synthesis of amino acid-derived lipidoids.
In 2016, Fenton et al. synthesized four alkenyl α-amino alcohols (AAA)646,647 type analogs of cKK-E12 by conjugating the dilysine-derived diketopiperazine core of cKK-E12 with alkenyl epoxides derived from biologically relevant fatty acids (Figure 63a).648 Alkenyl epoxide precursors derived from the corresponding fatty acids were synthesized in four steps. First, a lithium aluminum hydride reduction of fatty acids followed by oxidation gave aldehydes 325. Then a one-pot α-chlorination/sodium borohydride reduction of aldehyde 325 provided the 1,2-chloroalcohols 326.649 The following basic dioxane promoted ring closure of these 1,2-chloroalcohols furnished the desired alkenyl epoxides AE-00 through AE-03 in 4 steps with only one silica gel chromatographic purification. Lastly, epoxide ring-opening of these alkenyl epoxides with the dilysine-derived diketopiperazine core 324 gave the desired alkenyl α-amino alcohol lipids OF-00 through OF-03. EPO mRNA-loading OF-02 LNPs outperformed EPO mRNA-loading cKK-E12 LNPs, with a 2-fold increase in EPO concentration to 14200 ± 1500 ng/mL when administrated intravenously at a dose of 0.75 mg/kg in C57BL/6 mice.648 In another report, Fenton et al. developed an OF-Deg-Lin LNPs mRNA delivery system that was able to induce functional protein expression in mouse B lymphocytes.650 OF-Deg-Lin is a biodegradable variant of OF-02 lipid with four degradable ester linkers attached to the diketopiperazine core and four doubly unsaturated tails; it can generate nontoxic linoleic acid upon hydrolytic cleavage (Figure 63b). The synthesis of OF-Deg-Lin started with the conjugation of the TBS-protected aldehyde 327 and the diketopiperazine core via reductive amination; then removal of the TBS protective group promoted by TBAF followed by esterification of the hydroxyl groups with linoleic acids gave OF-Deg-Lin. Much like nondegradable OF-02 Cy5 mRNA LNPs, intravenous administration of OF-Deg-Lin Cy5 mRNA LNPs in mice resulted in predominant accumulation of Cy5 mRNA in the liver. Systemic injection of nondegradable OF-02 LNPs encapsulating FLuc mRNA in mice resulted in protein expression mainly in the liver.442,651 In 2018, Fenton et al. further modified OF-Deg-Lin by prolonging the carbon linker length from a two carbon spacer to a four carbon spacer (Figure 63b); the resulting lipid OF-C4-Deg-Lin was shown to be more potent than OF-Deg-Lin for both anti-FLuc siRNA and FLuc mRNA delivery in HeLa cells in vitro. OF-C4-Deg-Lin LNPs encapsulating firefly luciferase mRNA induced the majority of FLuc protein expression in the mouse spleen following an intravenous administration.652
Figure 63.
Chemical structures and synthesis of OF-XX and OF-Deg-Lin.
In 2020, Billingsley et al. synthesized a library of 24 ionizable lipids for delivering mRNA to human T cells ex vivo.653 The 24 ionizable lipids were synthesized via epoxide ring-opening of three alkyl epoxides with eight polyamines (Figure 64), and their luciferase mRNA delivery efficiency in vitro was evaluated. Results showed that C14-4 LNPs were the top performers, which could deliver CAR mRNA efficiently into primary human T cells ex vivo, generating CAR T cells with enhanced antitumor ability.653
Figure 64.
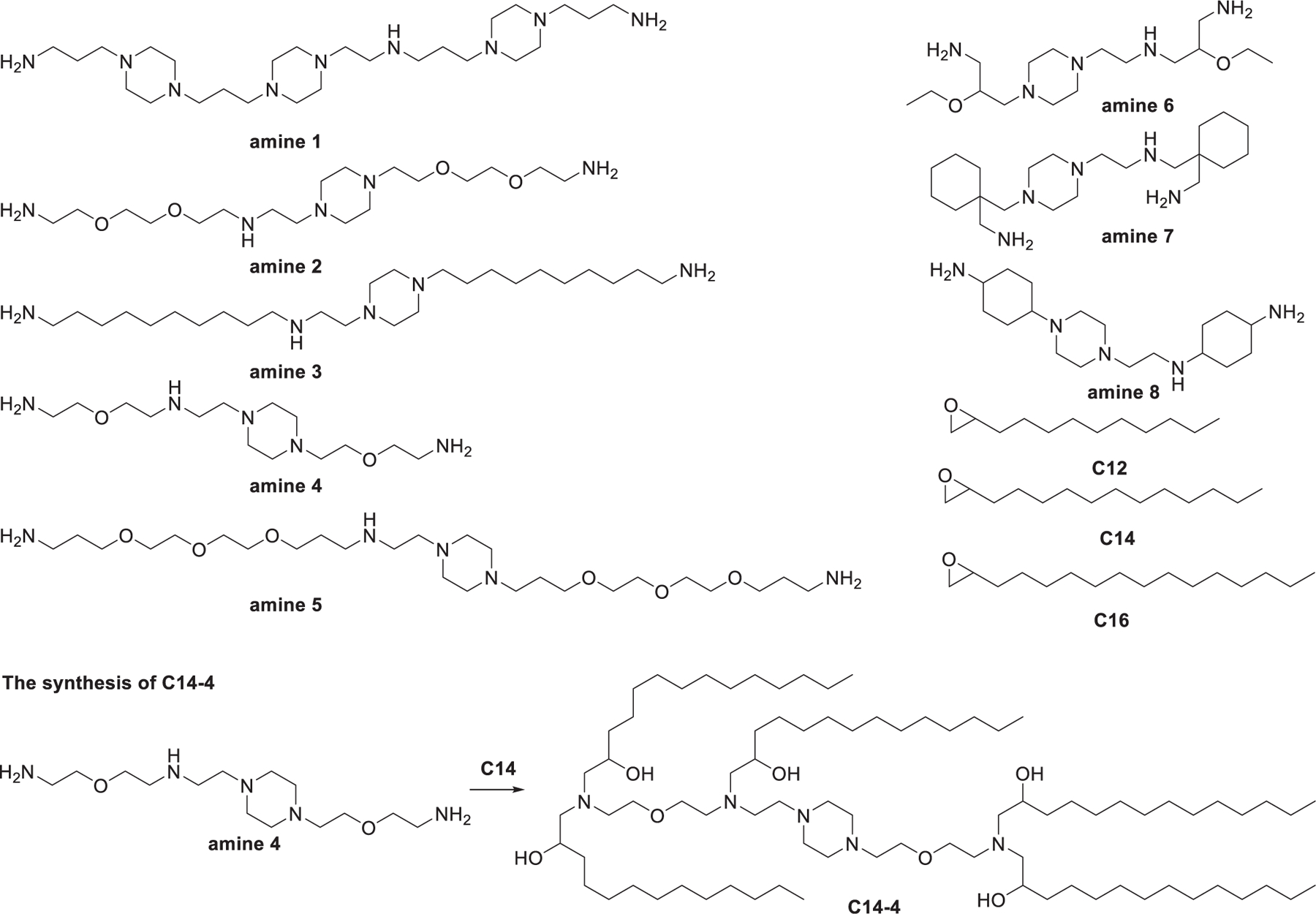
Synthesis of lipidoids via epoxide ring-opening of alkyl epoxides with polyamines.
2.2.3.3. Lipidoids Synthesized via Reductive Amination.
In 2016, Li et al. reported a library of ionizable lipids, designated as TTs, which consist of a phenyl ring, three amide linkers, and three hydrophobic hydrocarbon tails (Figure 65).598 TT2-TT8 lipids vary in the spacer length of the amide linker. In vitro screening identified TT3 as the lead lipid. PEGylated TT3 LNPs can efficiently deliver human factor IX (hFIX) mRNA in FIX-knockout mice in vivo after intravenous injection, resulting in the restoration of the level of hFIX.598 In 2020, TT3 LNPs were formulated to encapsulate engineered mRNA encoding various SARS-CoV-2 antigens.52 Vaccination of mice with this TT3 LNPs-mRNA formulation resulted in over 300-fold more specific antibody against anti-S1 as compared to that of MC3 LNPs-mRNA formulation. Additionally, antigen-specific antibodies induced by intramuscular administration of this formulation were 5-fold more than that of subcutaneous injection.52 In 2020, Zhang et al. further developed a library of ionizable lipids designated as FTT by combining the core of TT3 with various biodegradable tails (Figure 65).597 Results showed that LNPs formulated with FTT lipids with branched ester tails (e.g., FTT6) mainly delivered mRNAs into the liver and spleen, and their mRNA delivery efficiencies were higher than that of FTT lipids with linear ester tails (e.g., FTT10). Among all the FTT compounds, FTT5 was the lead lipid to deliver mRNA to the liver. Images of FTT5 LNPs captured by cryo-TEM revealed the spherical morphology of FTT5 LNPs. Intravenous injection of FTT5 LNPs encapsulating FVIII mRNA in both wild type mice and hemophilia A mice resulted in potent expression of hFVIII protein. Moreover, FTT5 LNPs were able to induce dramatic base editing of PCSK9 at a dose of 0.125 mg/kg in mice.597
Figure 65.
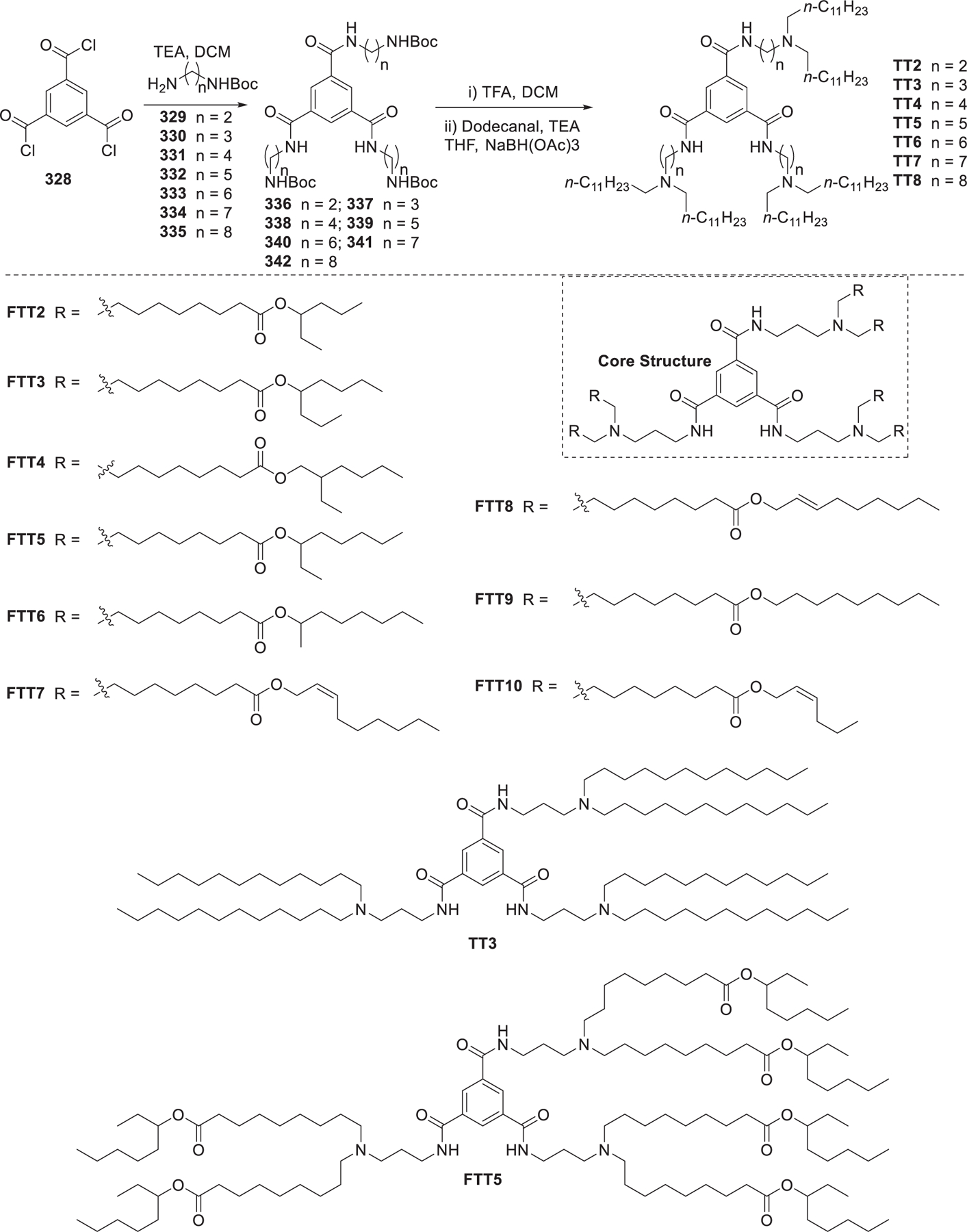
Synthesis of TT and FTT series of ionizable lipids.
2.2.3.4. Lipidoids Synthesized via Click Chemistry.
In 2012, Li et al. employed thiol–yne click chemistry654,655 to synthesize a library of more than 100 ionizable lipids (Figure 66a).601 These lipids were synthesized using eight alkyl thiols with varying length of the alkyl group (C6–C16), two alkynyl carboxylic acids, and seven amines. The synthesis was realized in two consecutive modular steps. First, a carboxylic acid with two hydrophobic tails was obtained via a thiol–yne click reaction between an alkyl thiol and an alkynyl carboxylic acid. Then the ionizable head group was installed through amide coupling, giving an ionizable thioether lipid (Figure 66a). Ionizable lipids with undecyl and dodecyl as hydrophobic tails showed stronger in vitro pDNA delivery efficiency compared to the other lipids, and A1C11 surpassed Lipo2000 in delivering siRNA into HEK293T cells in vitro.601 In 2013, Alabi et al. produced a library of 32 ionizable lipids using a similar method.600 In this study, lipids were synthesized via Michael addition between amines and propargyl acrylate followed by the thiol–yne click reaction with alkyl thiols in the presence of UV and a photocatalyst (Figure 66b).600 These lipids were then utilized to evaluate a multiparametric approach to screen LNPs. Results showed that the pKa of the whole LNPs, rather than the pKa of individual lipids, was a key determinant of LNPs function in vivo. LNPs with above 50% silencing had pKa values ranging from 6 to 7, while LNPs with pKa values below 5.8 exhibited no gene knockdown activities both in vitro and in vivo.600
Figure 66.
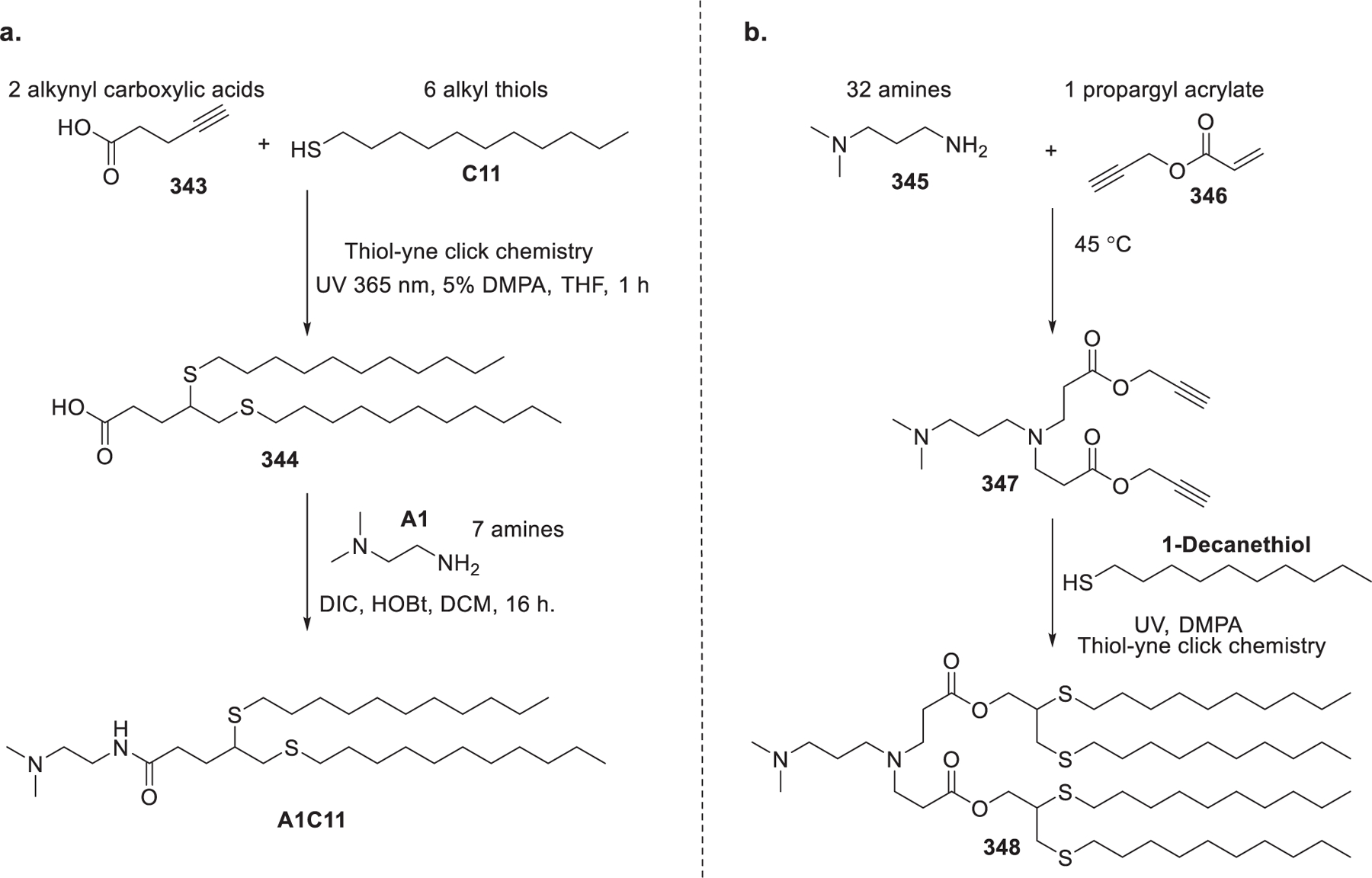
Synthetic routes to lipids via thiol–yne click chemistry.
2.2.3.5. Lipidoids Synthesized via Multicomponent Reactions.
Multicomponent reactions represent an efficient way to synthesize compounds with diversities.656 In 2018, Molla et al. reported the synthesis of a library of 288 ionizable lipids via a three-component thiolactone ring-opening reaction followed by a thiol–disulfide exchange reaction.604 Structurally, these lipids are composed of a head group with one or more ionizable tertiary amines, an amide linker, and two asymmetric alkyl tails, of which one contains a bioreducible disulfide bond (Figure 67). In vitro screening of these lipids identified some potent lipids for pDNA delivery in HEK-293T cells.604 In 2020, Molla et al. further evaluated the in vitro anti-GFP siRNA delivery efficiency of these lipids in HeLa-GFP cells.602 T16-PY12-A17, T18U-PY12-A17, and T18U-PY18-A4 (Figure 67) were shown to be effective in in vitro siRNA delivery, resulting in a reduction of GFP expression by 65%. Kdrl:EGFP Zebrafish embryos injected with T18U-PY18-A4 LNPs encapsulating anti-GFP siRNA showed a significant reduction in EGFP expression in blood vessels.602
Figure 67.
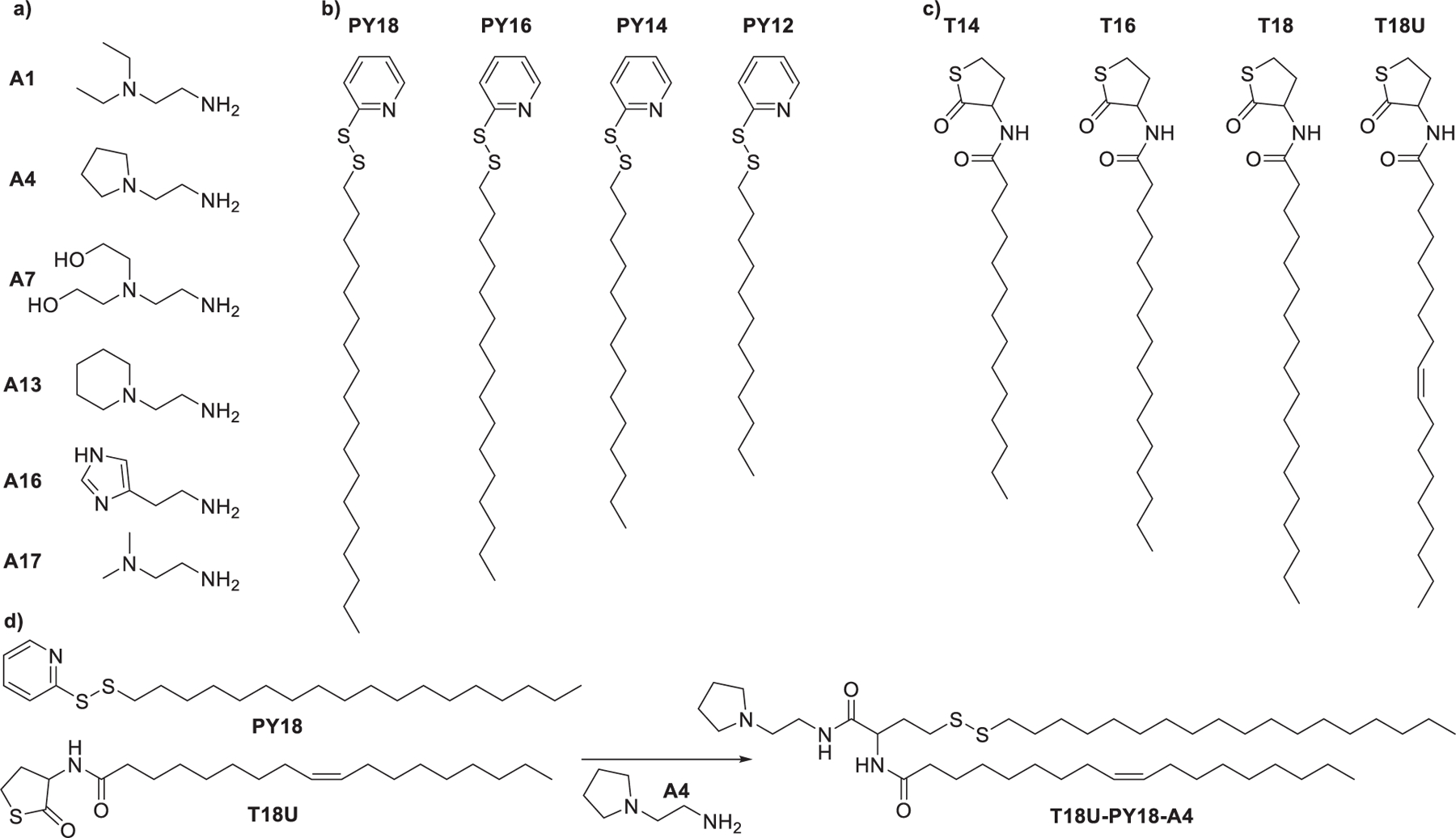
Synthesis of lipidoids via thiolactones ring-opening reaction followed by the thiol–disulfide exchange reaction. (a) Structures of amines. (b) Structures of pyridyl disulfide derivatives. (c) Structures of thiolactone derivatives. (d) Representative reaction scheme.
In 2019, Miao et al. developed a library of 1080 ionizable lipids603 via isocyanide-mediated three-component reaction (3-CR)657 that simultaneously coupled primary or secondary amines,658 ketones with different alkyl tail lengths and different degrees of saturation,648,659 and isocyanides or isocyanide derivatives (Figure 68).660 Results showed that lipids with longer, unsaturated tails showed higher mRNA delivery efficiency, and A2-Iso5-2DC18 (A2) and A12-Iso5–2DC18 (A12) were the top-performing lipids for mRNA delivery in bone marrow-derived dendritic cells (BMDCs), bone marrow-derived macrophages (BMDMs), and Hela cells. Both A2 and A12 contain two amines that are spaced three carbons apart and an ester group. A2 LNPs and A12 LNPs encapsulating Cre mRNA led to comparable levels of protein expression and could deliver mRNA into central APCs including CD11b+ and CD11c+ in the Ai14D reporter mouse model. A2 LNPs encapsulating mRNA encoding OVA induced much higher adaptive immune response and antitumor efficacy in the Ovalbumin (OVA)-expressing B16F10 mouse compared to A12 LNPs encapsulating mRNA encoding OVA. Rechallenge experiments indicated that only the A2 LNPs were able to induce strong antitumor immunity. Structurally, the main difference between A2 and A12 lies in their head groups: A2 has a heterocyclic amine head group whereas A12 contains a linear amine head group. They further developed a secondary library of lipids with different head groups to investigate the relationships between the head group (cyclic versus linear) and their immunogenicity (Figure 68). Lipids containing heterocyclic amine head groups induced higher expression of IFN-γ as compared to lipids with linear amine head groups after mRNA encoding OVA vaccination. In particular, A18 LNPs showed much higher IFN-γ-positive secretion than the other lipids, and lipid A18 also had intrinsic stimulatory effects. A18 was finally identified as the lead cyclic lipid candidate, which facilitated the mRNA delivery and induces a strong immune response partially mediated by the stimulator of the interferon gene (STING) pathway.
Figure 68.
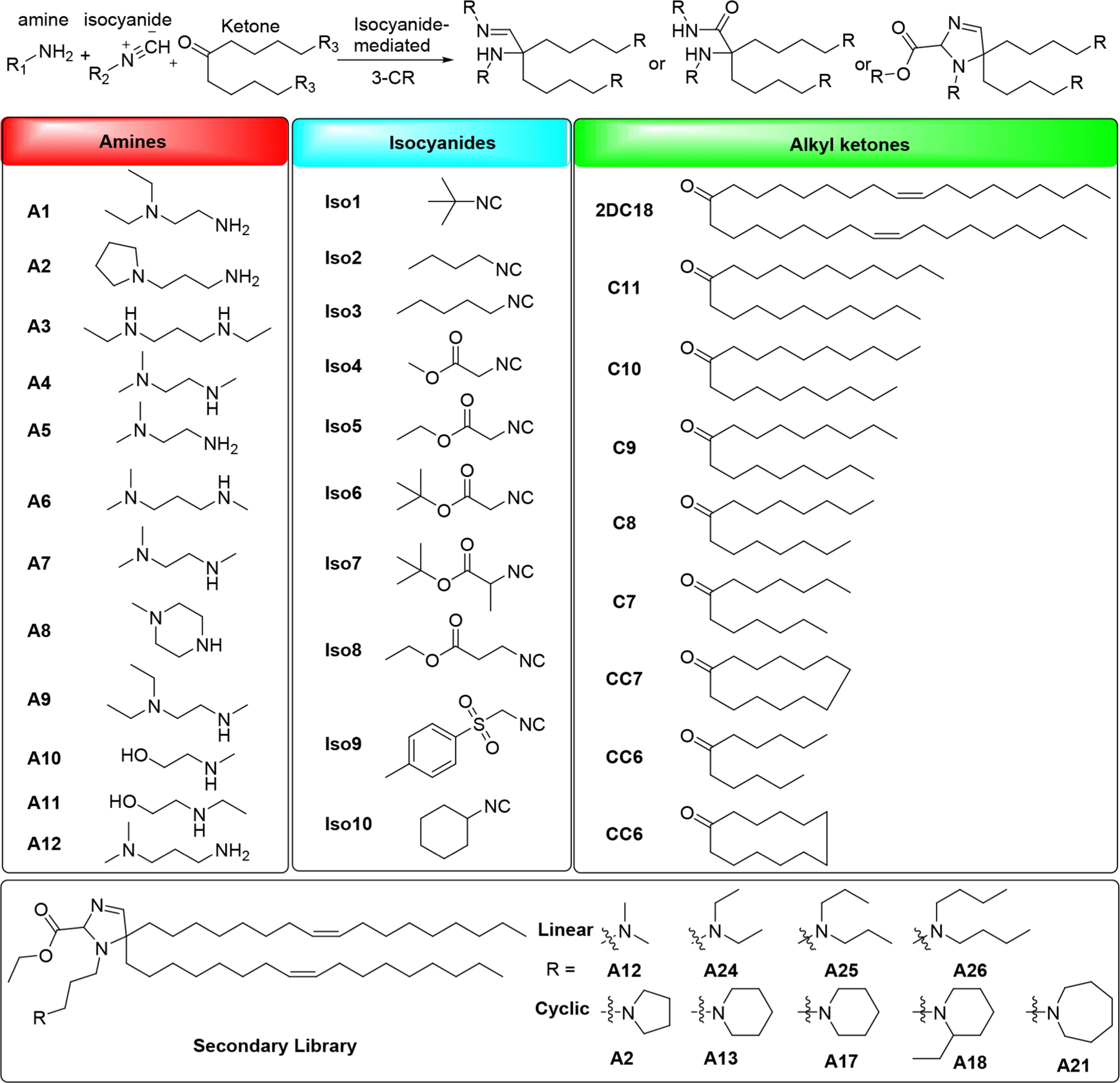
Isocyanide-mediated three-component reactions for the synthesis of lipidoids.
2.2.3. Zwitterionic Lipids
Zwitterionic lipids are lipids that contain covalently bonded cationic groups and anionic groups. In 2011, Sonoke et al. synthesized a galactose-modified zwitterionic lipid GDOPE by conjugating lactose with DOPE via reductive amination reactions (Figure 69).661 Compared to nongalactosylated LNPs, GDOPE-based LNPs showed enhanced delivery of siRNA to the liver in mice.
Figure 69.
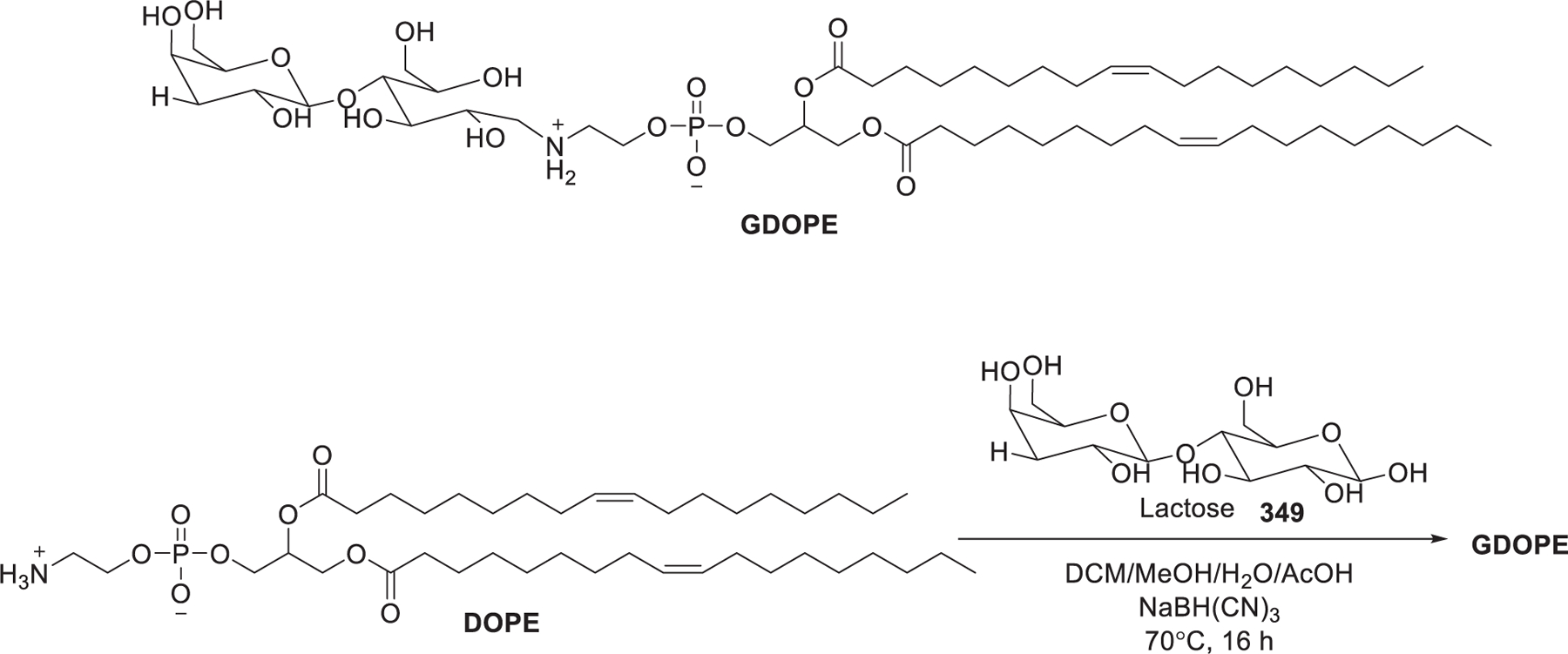
Chemical structure of zwitterionic lipid GDOPE.
In 2012, the Szoka group synthesized a class of zwitterionic lipids with head groups containing a tertiary amine or quaternary ammonium head group and carboxylate linked by various carbon spacers (Figure 70).662 LNPs containing these zwitterionic lipids showed efficient siRNA encapsulation when ionized.662 DOBAQ LNPs was used for the delivery of Cas9 mRNA to the back of the eye in mice.663
Figure 70.
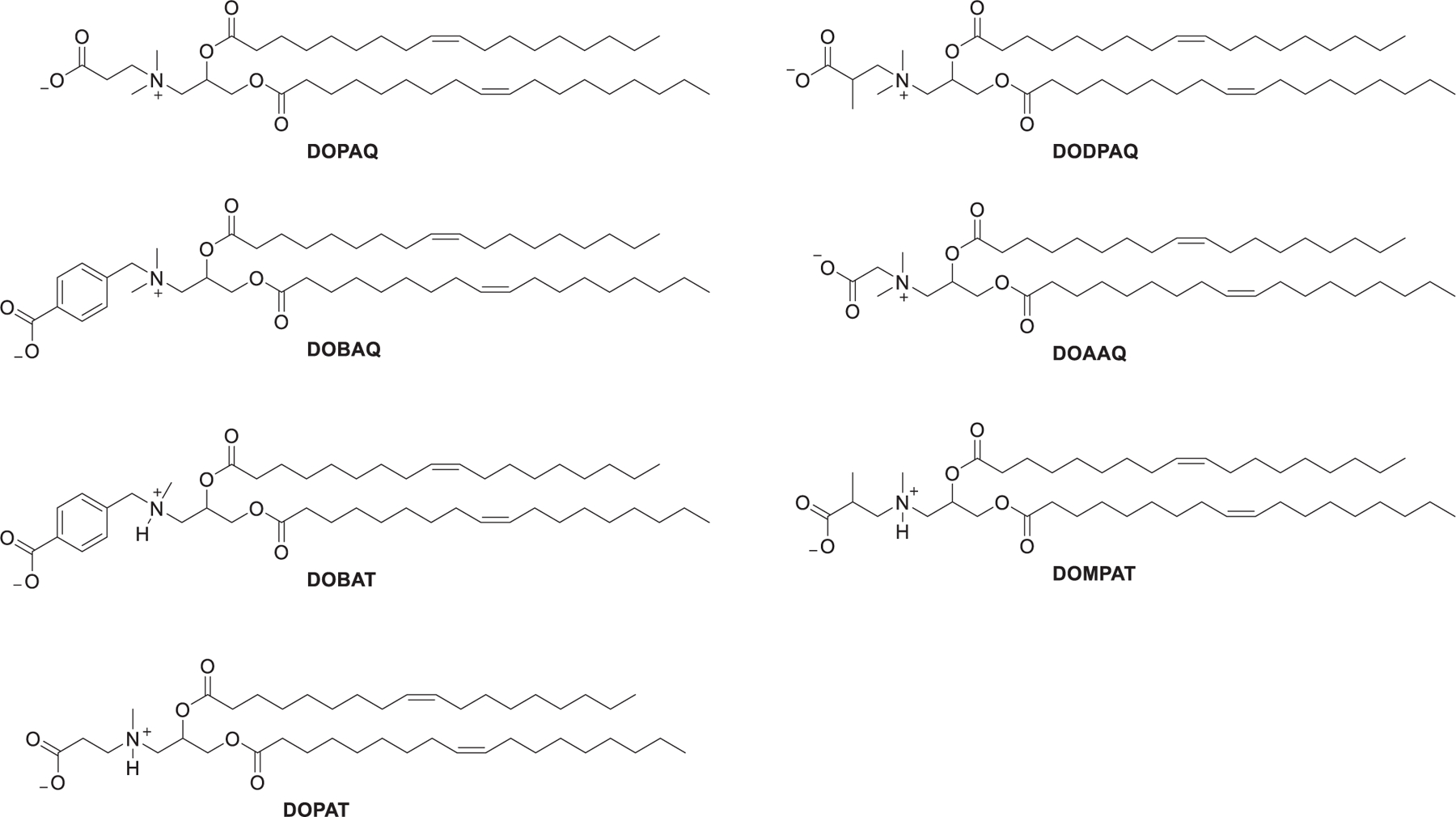
Chemical structures of zwitterionic lipids.
In 2013, Walsh et al. developed a series of lysine-based zwitterionic lipids termed ILL, which contained a zwitterionic lysine head group linked to dialkyl amines through an amide linker at the lysine α-amine (Figure 71).664 As shown in Figure 71, synthesis of LOA-LysC2 began with the coupling of linoleic acid 351 with oleylamine 350 promoted by EDC; the resulting amide 352 was treated with lithium aluminum hydride to give dialkylamine 353. The dialkylamine 353 was then functionalized via Michael addition reaction with tert-butyl acrylate 354 followed by deprotection of the resulting tert-butyl ester 355, affording β-amino acid 356. Compound 356 was coupled with protected lysine 357 via amide bond formation; the following global deprotection of the resulting compound yielded LOA-LysC2. Containing a primary amine, a tertiary amine, and a carboxylate, these ILLs exhibited a pH-dependent ionization property that varied with their chemical structures.664 ILLs form small-diameter LNPs that could efficiently entrap anti-Luc siRNA and deliver anti-Luc siRNA in HeLa cells, resulting in potent luciferase gene silencing.664
Figure 71.
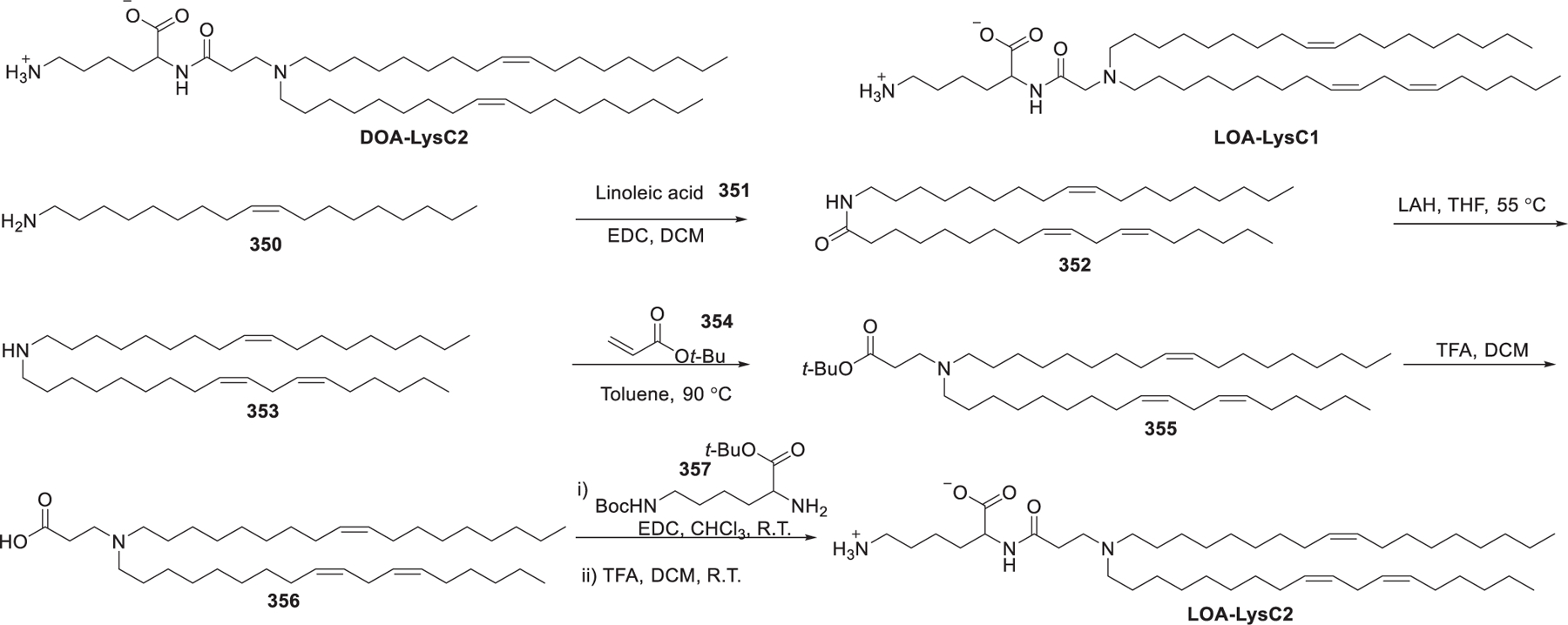
Chemical structures of lysine-derived zwitterionic lipids.
In 2017, Miller et al. developed a library of zwitterionic amino lipids (ZALs) that contained a zwitterionic sulfobetaine head group and an amine-rich core that were attached to various hydrophobic tails via Michael addition or epoxide ring-opening (Figure 72).665 Acrylation of N,N1-dimethylethane-1,2-diamine 359 gave 2-(dimethylamino) ethyl acrylamide 360, which underwent ring-opening reaction with 1,3-propane sultone 361 to afford the electrophilic precursor 362 (SBAm). Then a series of zwitterionic amines were prepared by conjugate addition of a library of polyamines to SBAm. The free amines of the resulting zwitterionic intermediate 364 were reacted with alkyl epoxides and alkyl acrylates to install hydrophobic tails and alcohol or ester groups, resulting in a 72-member ZALs library (Figure 72). Epoxide-based ZALs (ZAx-Epm) were more efficient than acrylate-based ZALs (ZAx-Acn; Figure 72) in siRNA delivery into HeLa cells. Intravenous administration of ZA3-EP10 LNPs containing Cas9 mRNA and LoxP sgRNA induced floxed tdTomato expression and enabled elimination of the STOP cassette in the liver, lungs, and kidneys of engineered mice.665
Figure 72.
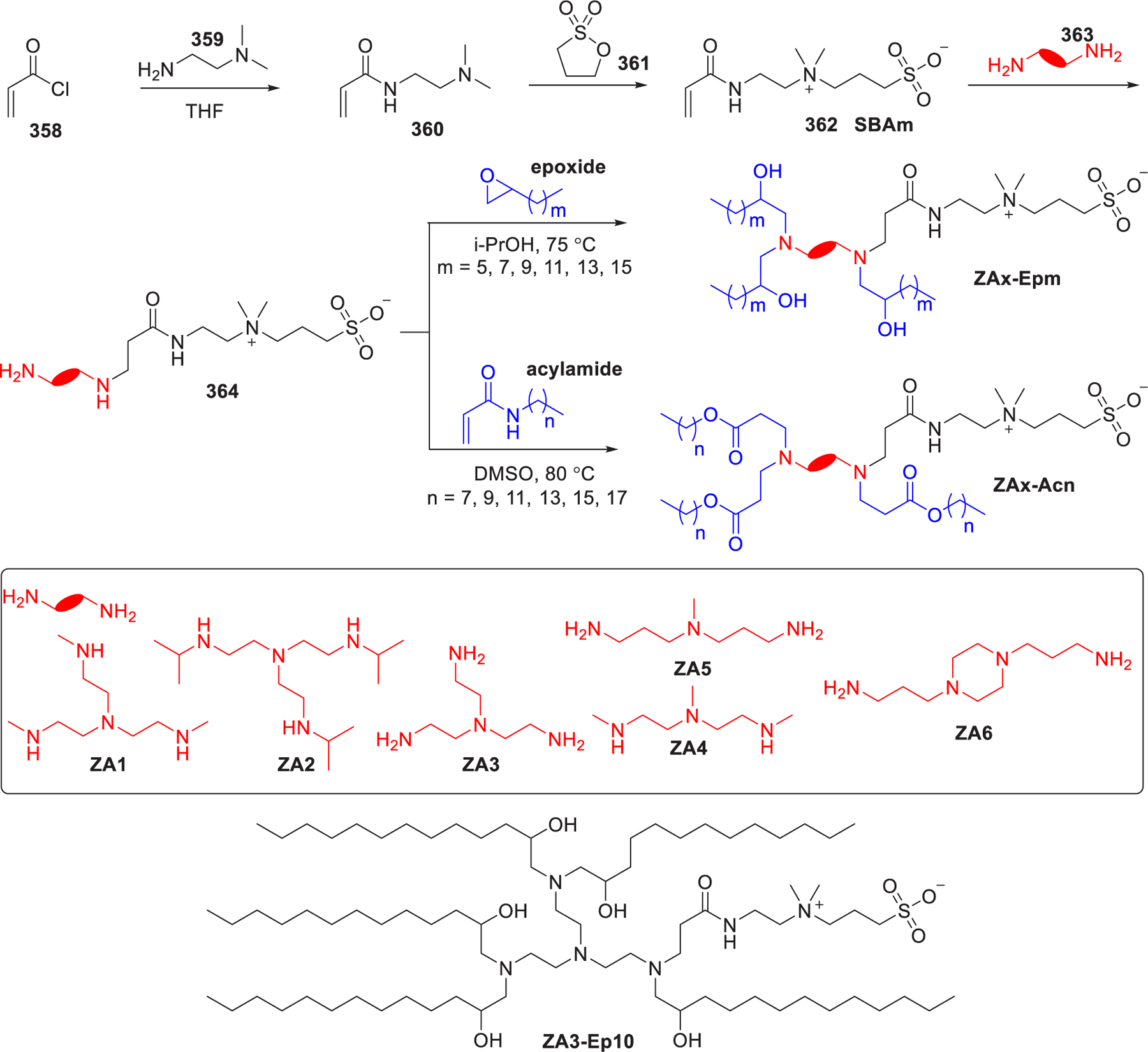
Chemical structures and the synthesis of zwitterionic lipid ZA3-Ep10.
In 2020, Hirai et al. synthesized a DOPE-derived charge-reversible lipid, dioleoylglycerophosphate–diethylenediamine conjugate (DOP-DEDA), which was composed of a negatively charged phosphoric acid, two ionizable amino groups, and two oleyl hydrocarbon tails.666 LNPs composed of DOP-DEDA/DPPC/cholesterol (DOP-DEDA LNPs) were stable in the physiological medium in the absence of PEG-conjugated lipid. The charge property of DOP-DEDA LNPs depends on pH: at pH 6.0 the DOP-DEDA LNP is protonated and positively charged, at physical pH (pH 7.4) it is neutral, and at pH 8.0 it is negatively charged (Figure 73). DOP-DEDA LNPs encapsulating polo-like kinase 1 (PLK1) siRNA-induced suppression on the PLK1 expression in a dose-dependent manner in MDA-MB-231 cells.666
Figure 73.
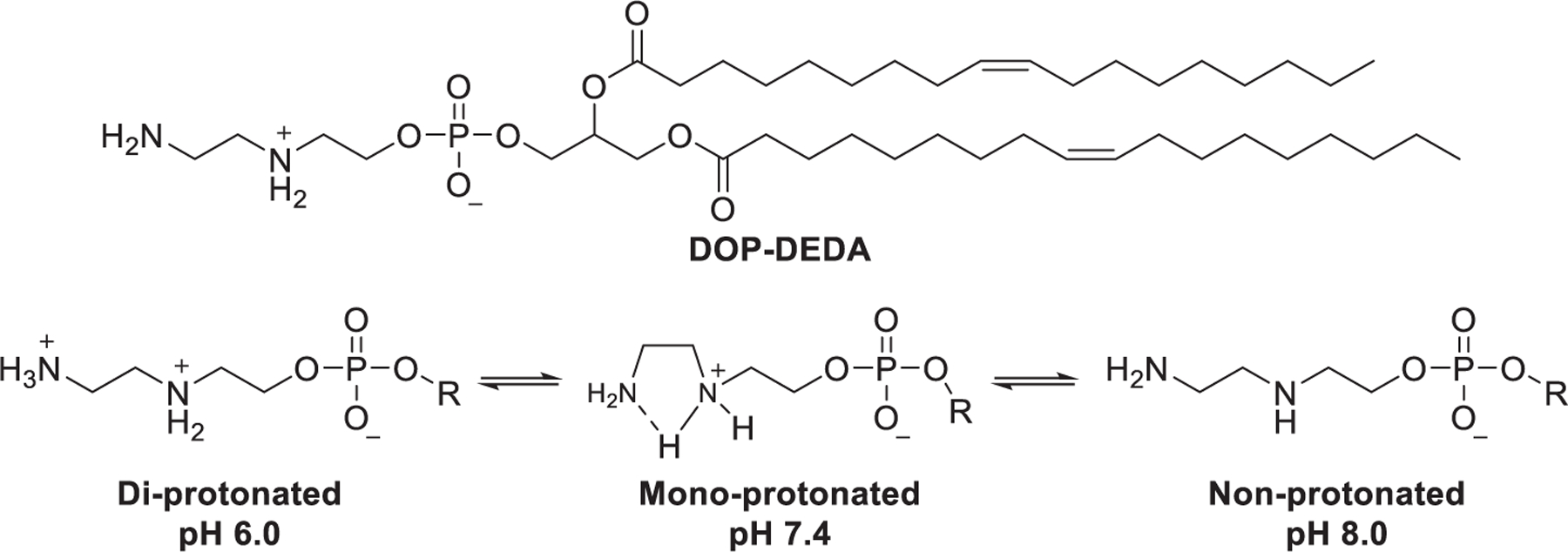
Chemical structure of DOP-DEDA and schematic illustration of the pH-responsive ability of DOP-DEDA.
To improve the endosomal escape of LNPs, Liu et al. synthesized a library of zwitterionic phospholipids (iPhos) that mimic natural phospholipids (e.g., DOPC and DOPE) through combinatorial chemistry in 2021.667 As shown in Figure 74, phosphorylation of variant aliphatic alcohols with different alkyl tail lengths gave the key intermediate alkylated dioxaphospholane oxide (Pm), which underwent ring-opening reaction668,669 when treated with variant amines (1A-28A), generating the library of 572 zwitterionic phospholipids (iPhos). Results of initial screening experiments for mRNA delivery in ovarian cancer cells showed that iPhos 7A1P4–13A1P16, containing an ionizable tertiary amine, a negatively charged phosphoric acid, and three hydrophobic hydrocarbon tails, were the top-performing phospholipids for mRNA delivery. 7A1P4–13A1P16, composed of the small head groups and large tails body, tend to adopt the inverted hexagonal (HII) phase, thus promoting membrane fusion, destabilizing the endosomal membrane, and allowing the endosomal escape of RNA molecules. iPhos 9A1P9 was shown to be the most active component of iPhos LNPs (iPLNPs); it exhibited 40 and 965 times higher in vivo mRNA delivery efficiency than DOPE and DSPC, respectively. Optimization of formulation of 9A1P9 LNPs was then carried out by introducing a supplemental selective organ-targeting (SORT)670,671 lipid. 9A1P9-5A2-SC8 LNPs mainly delivered Cre mRNA in the liver, while 9A1P9-DDAB LNPs mediated high accumulation of Cre mRNA in the lung. 9A1P9-5A2-SC8 LNPs and 9A1P9-DDAB LNPs were used for codelivering Cas9 mRNA and Tom1 sgRNA into Ai9 mice via intravenous administration at an mRNA dose of 0.75 mg/kg, resulting in specific Tom1 gene editing in the liver and the lung, respectively.667
Figure 74.
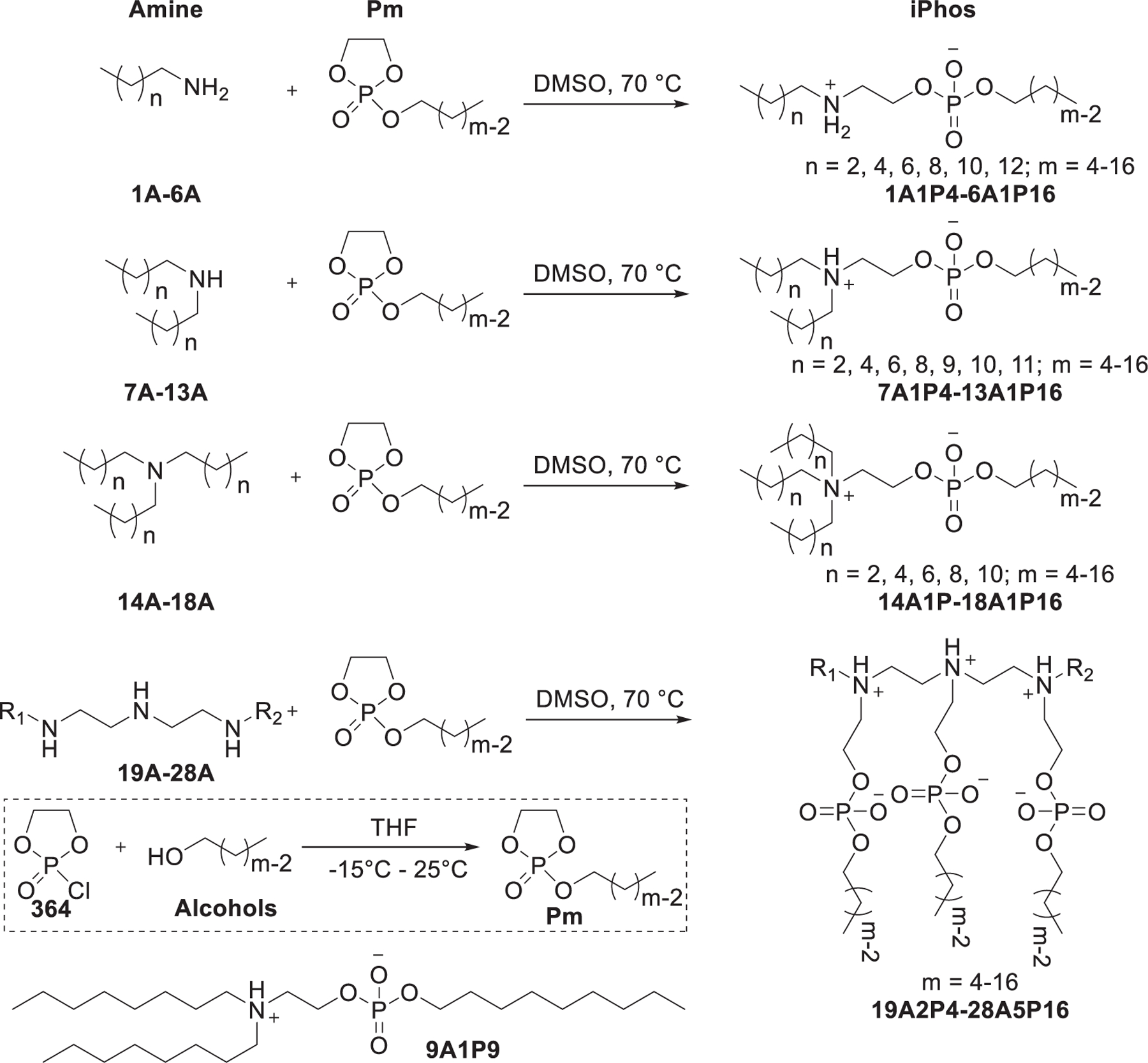
Chemical structures and synthesis of representative iPhos series lipids.
2.4. Other Lipids
In 2016, Kim et al. developed a class of coordinative amphiphiles (CAs) as transporters for siRNA delivery.672 These amphiphiles mimic cationic lipids, in which the cationic head group was replaced by zinc(II)–dipicolylamine complex (Zn/DPA) as an RNA phosphate backbone coordinating group, and a variety of membrane-directing groups were incorporated as substitutes of the hydrophobic tails (Figure 75). The CAs aggregate in aqueous solutions and the Zn/DPA head group coordinate with the negatively charged phosphate backbones of siRNAs, protecting siRNAs from degradation by RNase. The induction of different membrane-directing groups is necessary for enhanced siRNA delivery, as the Zn/DPA head group alone exhibits only moderate delivery efficiency.672
Figure 75.
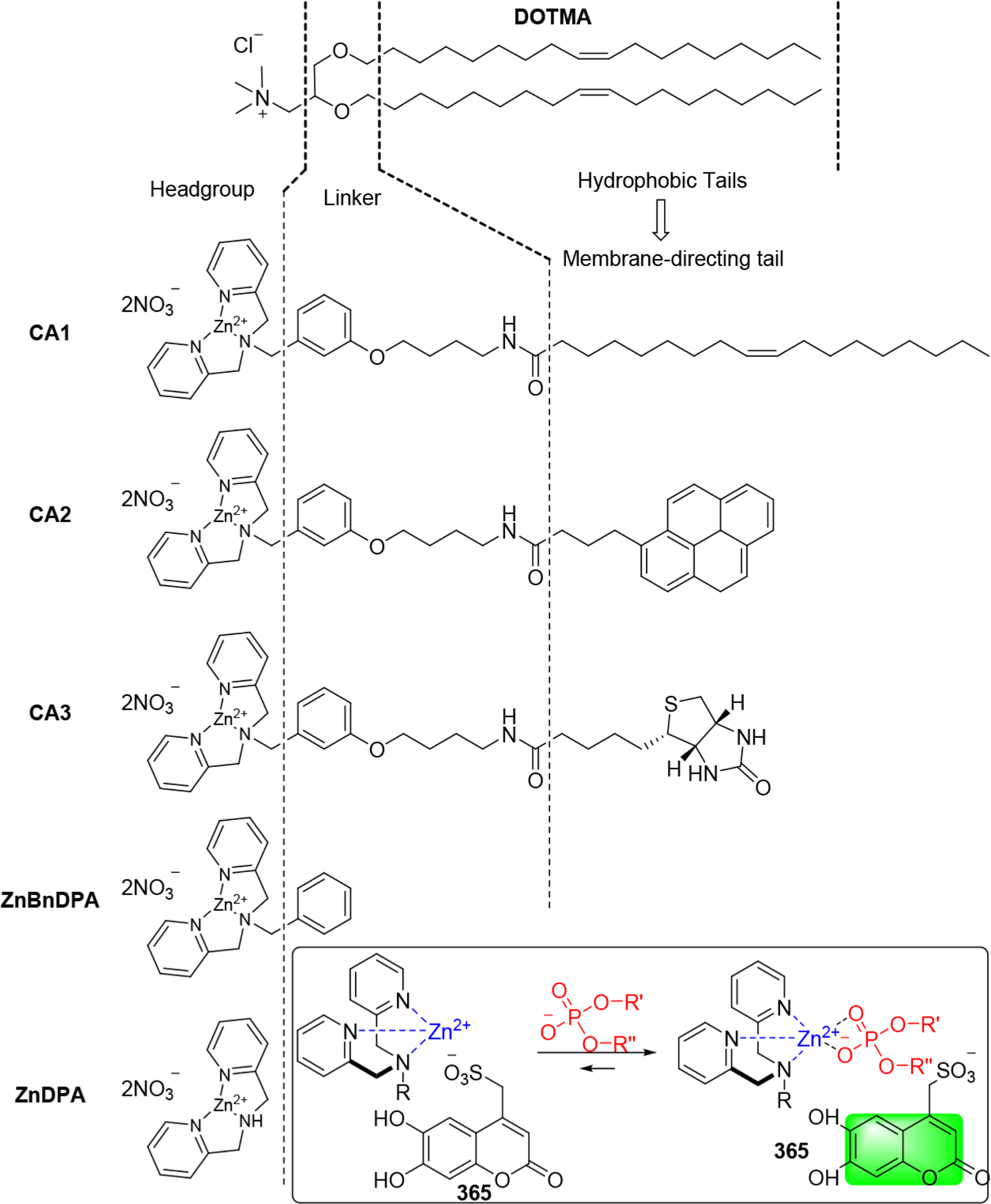
Schematic illustration of the interactions between Zn/DPA, sulfonic acid 365, and phosphate.
In 2018, Tai et al. developed a class of bifunctional chemical tags (366–371) that were capable of noncovalently binding and delivering siRNA into the cytosol directly (Figure 76).673 The bifunctional tags are composed of a siRNA-binding domain and a steroid region that can readily fuse with cell membrane. Compared to the conventional covalent siRNA–steroid conjugates, the noncovalently tagged siRNA is cell membrane-permeant and cytosol targeting, thus enabling effective siRNA delivery directly into PC-3 cells without involving the endocytic pathway.673
Figure 76.
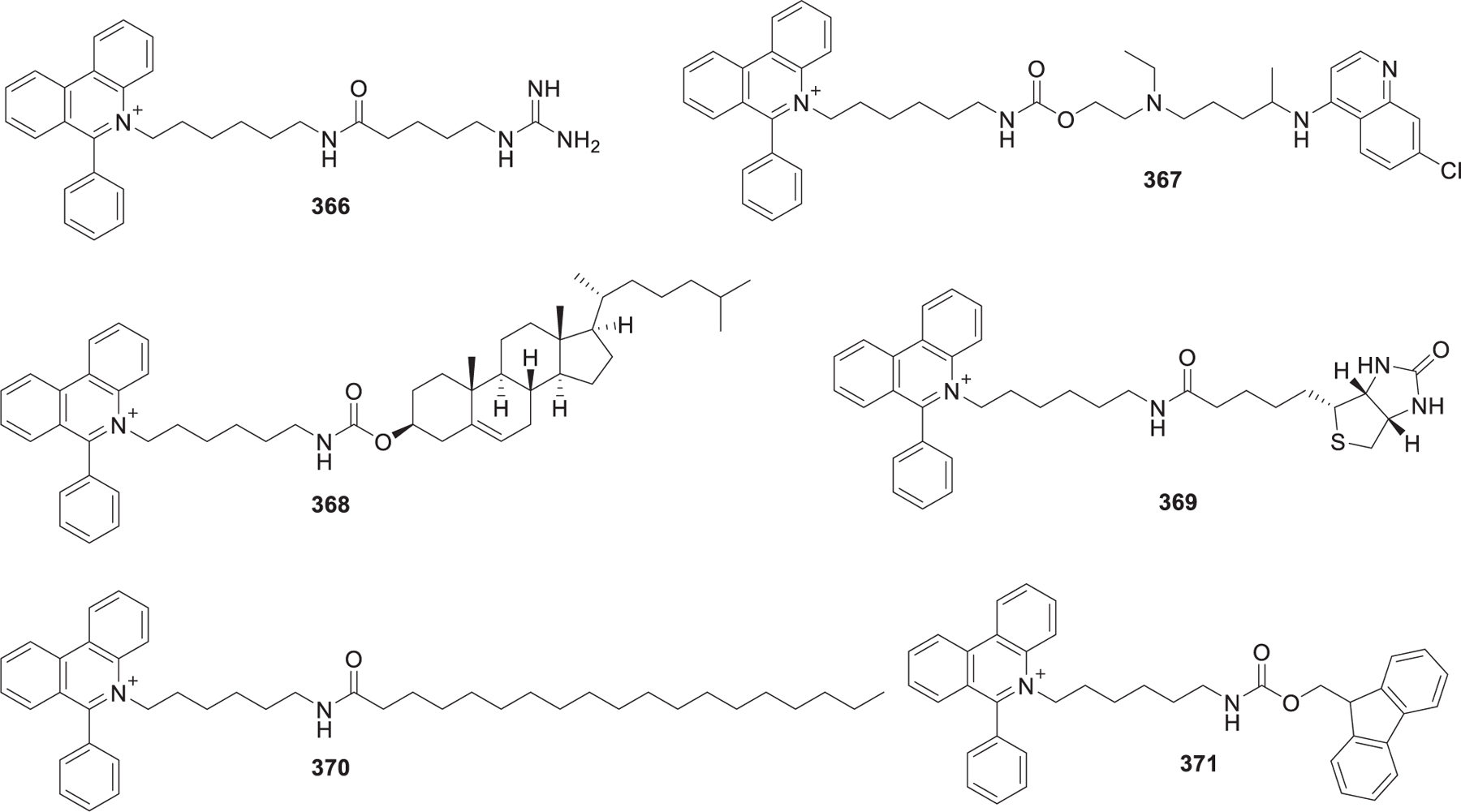
Chemical structures of representative coordinative amphiphiles (CAs).
3. HELPER LIPIDS
To stabilize the lipid-based RNA delivery system, many helper lipids, such as cholesterol, 1,2-dioctadecanoyl-sn-glycero-3-phosphocholine (DSPC), or 1,2-dioleoyl-sn-glycerol-3-phos-phoethanolamine (DOPE),288 have been included as the formulation components. These helper lipids may not only stabilize the particles but also enhance RNA delivery efficiency.194
3.1. Phospholipids
A typical phospholipid is composed of glycerol, two hydrophobic fatty acid tails, and a phosphate-linked head group (Figure 77). Due to their amphiphilic characteristic, they can form lipid bilayers as the main components of the cell membrane. Modifications of the phosphate group with the simple organic molecules choline, ethanolamine, or serine can give the corresponding phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylserine (PS). Phospholipids provide important structural components for the LNPs and may also aid the process of endosomal escape.107 Both synthetic and natural phospholipids can be used in the formulation of LNPs for RNA delivery.
Figure 77.
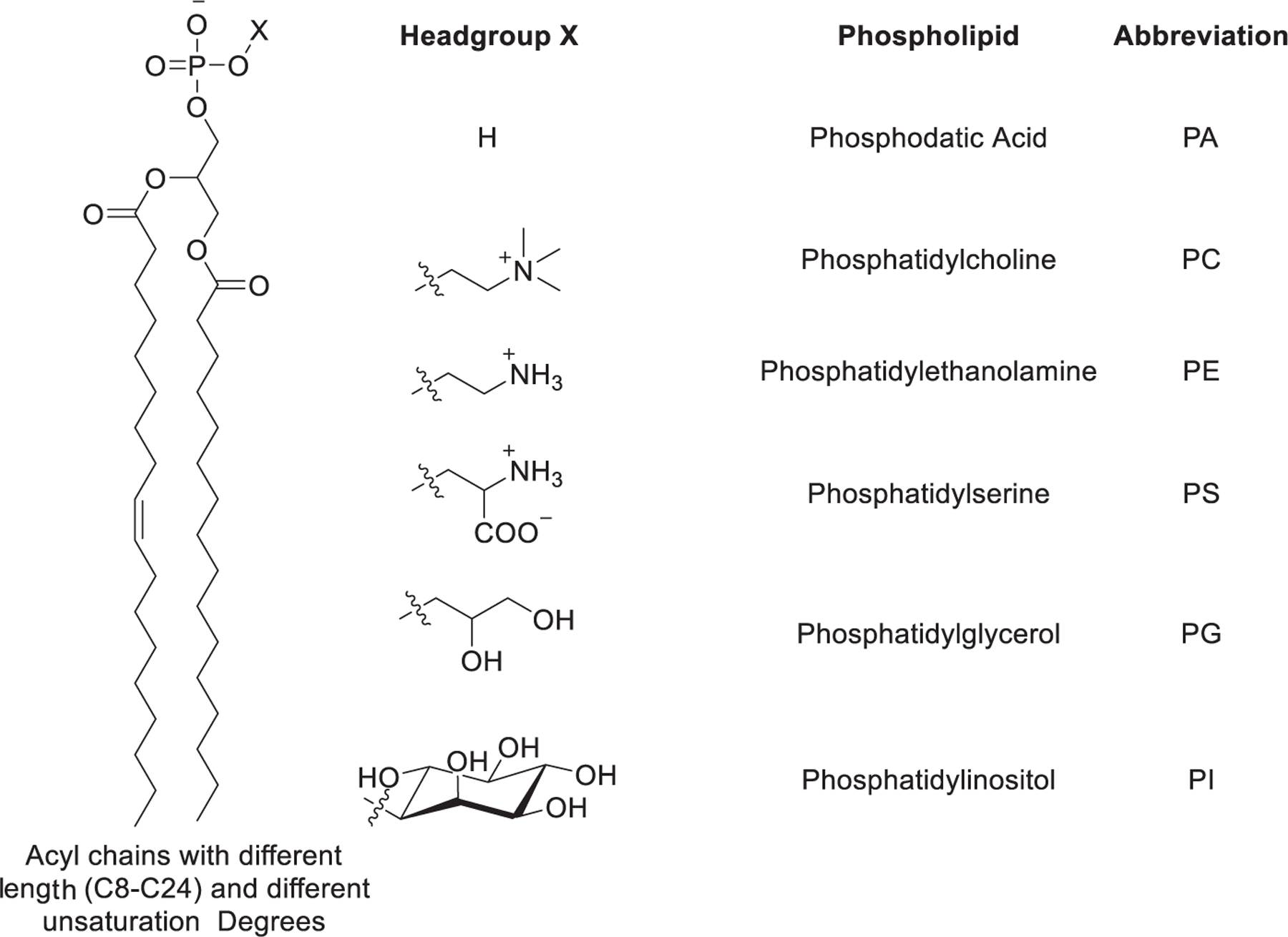
General chemical structure of phospholipids.
3.1.1. Phosphatidylcholines.
Phosphatidylcholines (PCs) are one of the major components (50%) of cell membranes. Due to the cylindrical geometry of PCs, they tend to adopt a bilayer phase (Figure 78),674 thus improving the stability of LNPs.675 PCs with saturated tails, such as HSPC (hydrogenated soybean PC) and DSPC (distearoylphosphatidylcholine), have high melting temperatures and are generally used to prepare highly stable LNPs. It is worth mentioning that DSPC is used as a helper lipid in patisiran, mRNA-1273, and BNT162b2.530 DOPC liposomes encapsulating EphA2 siRNA were shown to induce EphA2 gene silencing and repression of tumor growth in mice following either intravenous or intraperitoneal administration.676,677 Cyclo PC is found in the membrane of Escherichia coli.678 In 2019, the Cullis group studied the role of DSPC-cholesterol in LNPs formulation of siRNA and found that in empty LNP systems, DSPC-cholesterol resides in the outer layers of LNPs, whereas in siRNA-loaded LNPs, DSPC–cholesterol was partially internalized together with siRNA.195 DSPC can enhance the encapsulation of siRNA in the LNP–siRNA system by participating in the formation of siRNA–lipid complexes.195 In 2020, the Sahay group formulated LNPs with Cyclo PC instead of DSPC; the resulting Cyclo PC-LNPs showed enhanced delivery of mRNA into the cells compared to DSPC LNPs.679 They found that the structural differences between Cyclo PC and DSPC were not significant enough to influence in vitro delivery.679 This can be further supported by the report that alteration of symmetrical hydrophobic tails to asymmetrical hydrophobic tails did not significantly change the siRNA delivery efficiency of LNPs.568 DOPC has also been used in the formulation of neutral lipid emulsions (NLEs)288 along with Tween 20/squalene; the resulting NLEs selectively delivered R-34a miRNA to lung tumors, resulting in a 60% reduction of tumor area in mice after intravenous administraiton.680
Figure 78.
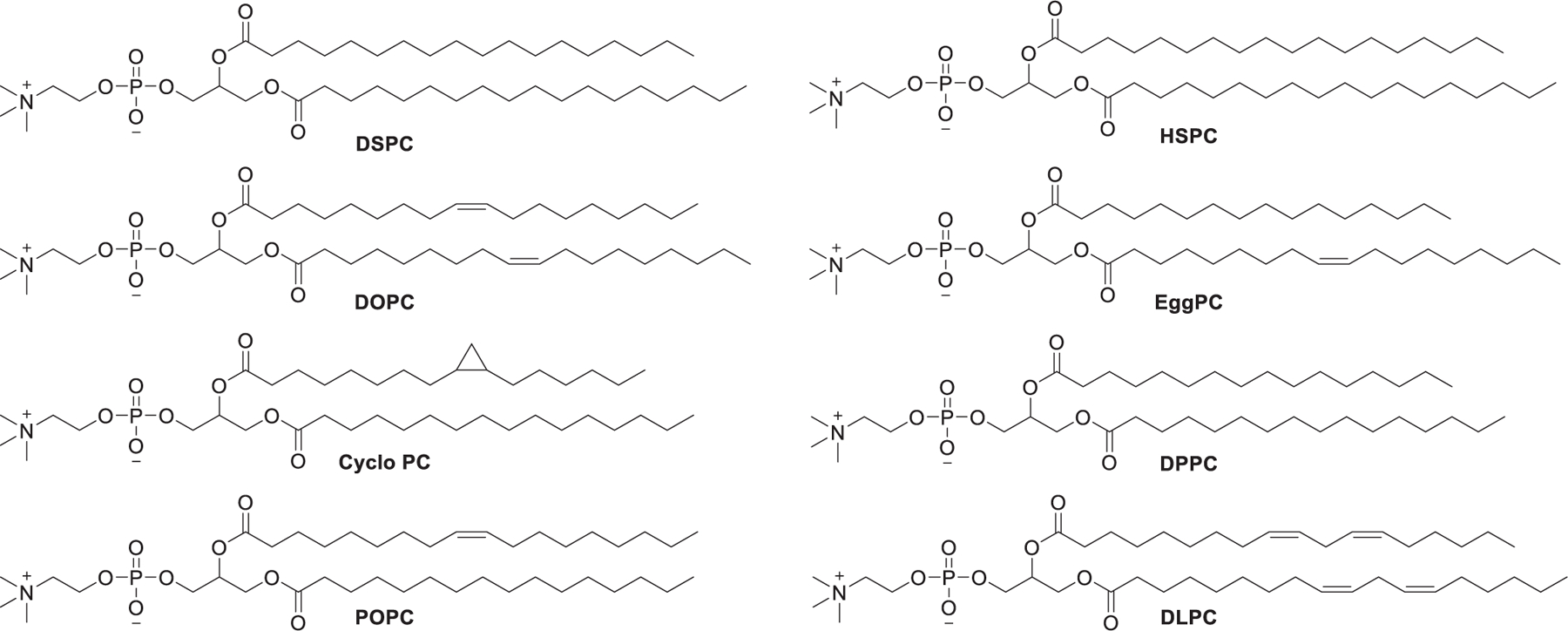
Chemical structures of representative phosphatidylcholines.
Adamantane is a diamondoid hydrocarbon that consists of three linked cyclohexane rings. When the adamantyl group is incorporated in small molecular drugs, not only can it influence the interactions between the small molecules and cell membrane,681 but it also can improve the pharmacokinetics of small molecule drugs.682 Besides, the constrained adamantyl group improves drug metabolic stability.683 In 2019, the Dahlman group reported that LNPs formulated with ionizable lipids that contain an adamantly group could deliver siRNA to splenic T cells without targeting ligands (Figure 79).546 Generally, PCs that are incorporated in LNPs contain unconstrained hydrocarbon tails. Based on the unique properties of constrained adamantyl groups, they developed a library of phosphatidylcholines containing an adamantyl group and further evaluated their mRNA delivery in vivo.684 They synthesized a series of constrained phosphatidylcholines, each of which contained a quaternary ammonium head group, a phosphodiester linkage, a constrained adamantly group, and an unconstrained hydrocarbon chain with varied length and saturation degree. The synthesis of these phosphatidylcholines started with the dibutyltin oxide-mediated chemoselective monoacylation of l-α-glyceryl phosphorylcholine 374 with adamantyl chloride 373.685 Then the hydrocarbon tail was attached to the remaining hydroxyl group via Steglich esterification,686 affording desired lipids A-10 through A-17-2Z (Figure 79). Then they utilized Fast Identification of Nanoparticle Delivery (FIND)687 to quantify mRNA delivery mediated by the 109 LNPs formulated with the constrained phosphatidylcholines in a single Ai14 mouse. Intravenous administration of A-11 LNPs encapsulating Cre mRNA in Ai14 mice at a dose of 0.5 mg/kg resulted in Cre mRNA accumulation in the liver Kupffer cells preferentially.685
Figure 79.
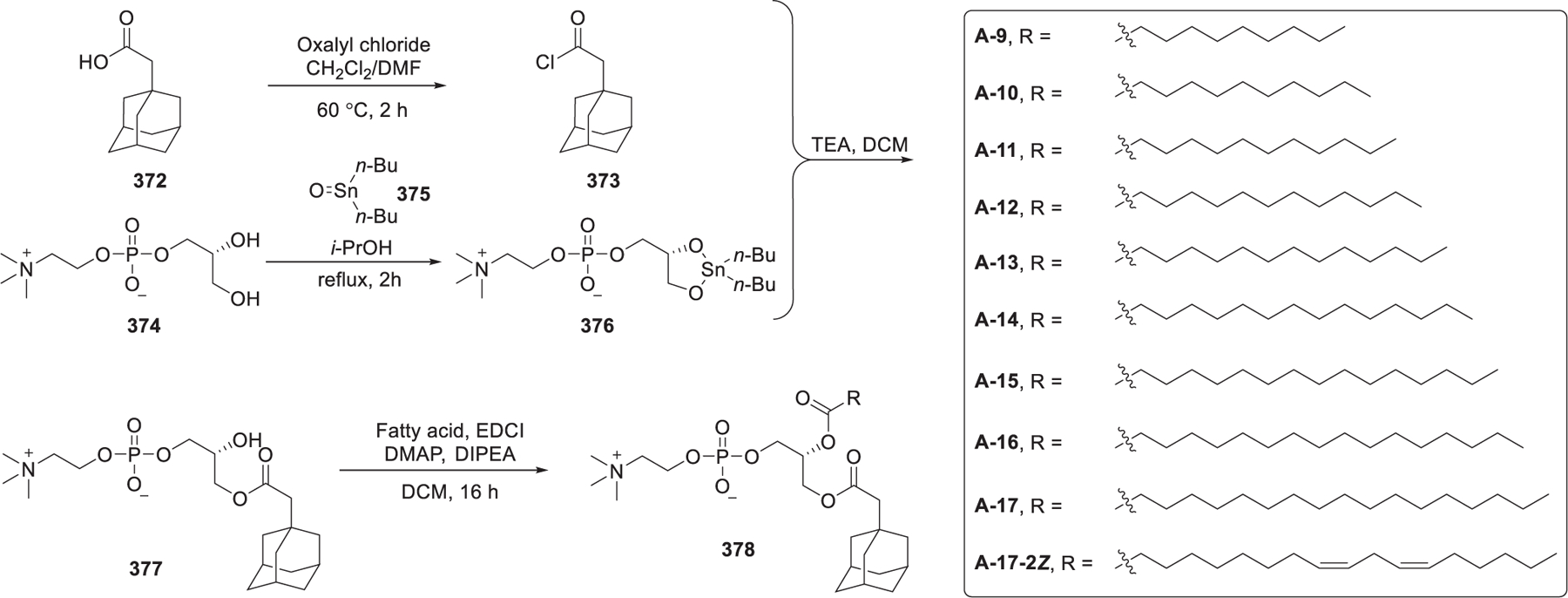
Synthesis of phosphatidylcholines incorporating adamantyl groups.
3.1.2. Phosphatidylethanolamines.
Previously, DOPE (Figure 80) was frequently used as a helper lipid in the formulation of cationic liposomes for DNA delivery for gene therapy.688 DOPE is composed of a primary amino head group, phosphoethanolamine, and two unsaturated oleoyl tails. With two unsaturated chains, DOPE has low melting temperatures (30 °C). In the physiological temperature, DOPE can induce the inverted hexagonal (HII) phase, facilitating membrane fusion and/or bilayer disruption.689
Figure 80.
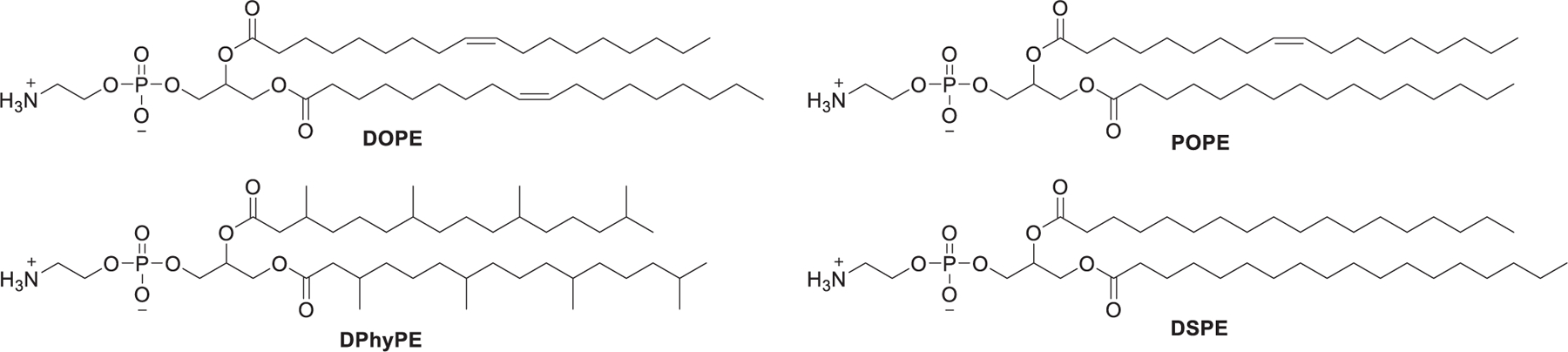
Chemical structures of representative phosphatidylethanolamines.
Phosphatidylcholines (PCs), containing a quaternary amine head group and two saturated hydrocarbon tails, have a P value less than 1, so they tend to aggregate into a lamellar structure. Phosphatidylethanolamine (PE), which contains a bit smaller primary amine head group than that of PCs and a P value > 1, favors inverted micelles or an inverted hexagonal (HII) phase.690 Kauffman et al. designed a series of experiments to determine the importance of LNP formulation on delivery efficacy and found that the use of the phospholipid DOPE instead of DSPC enhanced RNA delivery efficiency.629 In several studies, DOPE-formulated LNPs are more efficient than DSPC-formulated LNPs for mRNA delivery.598,611,627,629 The inclusion of DOPE during the formulation of LNPs may reduce membrane stability, thus facilitating endosomal escape.691,692 Another possible reason may be that the stronger complexation of mRNA to lipid in LNPs containing DSPC may hinder the decomplexation of mRNA from lipids in the cytosol, thus inhibiting the release and translation of mRNA payload. DSPC usually inhibits membrane fusion with the endosomal membrane, thus inhibiting endosomal escape, whereas DOPE, with unsaturated hydrophobic tails, can undergo a phase transition to an inverted hexagonal (HII) phase, thus facilitating membrane fusion-mediated endosomal escape.693,694 In 2006, Santel et al. prepared siRNA-lipoplex by formulating ATUFect01 with DPhyPE, a diphytanoyl zwitterionic phospholipid. Systemic administration of this siRNA-lipoplex led to downregulation of the corresponding mRNA and protein in vivo.391 In 2021, Zhang et al. investigated the influence of DOPE and DSPC on the interactions between LNPs and ApoE.695 Results of high-throughput in vivo screening of 96 LNPs showed that several LNPs incorporating DOPE (e.g., LNP 42) tended to accumulate in the liver, whereas LNPs formulated with DSPC (LNP 90) preferentially delivered RNA in the spleen. Results of QCM-D experiments showed that LNP containing DOPE (e.g., LNP 42) had stronger interactions with ApoE than LNP 90.695
3.1.3. Phosphatidylglycerols.
Phosphatidylglycerol (PG) is a non-pH sensitive anionic lipid that can provide balancing charges to an ionizable lipid (Figure 81). In 2012, Kapoor et al. developed effective DOPG-based anti-eGFP siRNA lipoplexes.696 In this work, anionic lipoplexes were prepared by complexing anionic liposomes formulated with DOPG and DOPE and siRNA using calcium ion bridges. The silencing activity of the anionic lipoplex composed of DOPG/DOPE was similar to that of Lipofectamine 2000. To inhibit the rapid growth of calcium phosphate (CaP) particles697 that encapsulate RNA, Zhang et al. developed a lipid-coated calcium phosphate (LCP) nanoparticle encapsulating cMyc siRNA, wherein the CaP core is coated with DOPA and DOTAP.698 The combination of chemo- and gene-therapeutics resulted in a dramatic suppression of tumor growth in H460 tumor-bearing mice following intravenous administration.698
Figure 81.
Chemical structures of representative phosphatidylglycerol.
3.1.4. Phosphatidylserines.
In 1978, Giorgos J. Dimitriadis prepared phosphatidylserine-based unilamellar liposomes to deliver rabbit globin mRNA into mouse lymphocytes ex vivo, resulting in functional protein expression.189 It was reported that anionic phosphatidylserine in the endosomal membrane could displace plasmids from the plasmids lipoplex, thus assisting in the release of plasmid after cell uptake of nanoparticles.699–701 In 2001, Hafez et al. found that coformulation of anionic phosphatidylserines (Figure 82) and cationic lipids preferentially adopts the inverted hexagonal (HII) phase, thus facilitating the release of nucleic acid payload into the cytoplasm.292 Besides, replacing 1,2-distearoyl-sn-glycero-3-phospho-l-serine (DSPS) with 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS) resulted in a clear inverted hexagonal (HII) phase when fusogenic cationic lipids were incorporated.292
Figure 82.
Chemical structures of representative phosphatidylserine.
3.2. Sterols
Steroids are a family of terpenoid lipids that contain four fused cyclic carbon rings, and sterols are steroids that contain a hydroxyl group, such as cholesterol.702 Cholesterol and related sterols are precursors for the synthesis of many vital steroids; they are naturally occurring lipids present in animal cell membranes. In the formulation of lipid-based nanoparticles, cholesterol acts as a helper lipid that can enhance nanoparticle stability and promotes the fusion of the nanoparticles with the cell membrane.176,703,704 Lipid nanoparticles without cholesterol may result in cholesterol shuttling from serum components to nanoparticles.705 Besides, cholesterol might reside on the surface of lipid nanoparticles in a crystalline form.706,707 It is reported that a cholesterol level increase by 7% improved the release of drug from 5% to 90%,708 which proves the essentiality of cholesterol in the delivery of siRNA.
Cholesterol variants derived from natural esterification and oxidation of cholesterol are differentially trafficked via lipoproteins to cells including endothelial cells, hepatocytes, and macrophage.709 In 2018, Paunovska et al. evaluated whether incorporation of different cholesterol variants (Figure 83) in LNPs would lead to different nanoparticle targeting in vivo.710 In vivo RNA delivery data points showed that modified cholesterols could affect the targeting ability of nanoparticles. Additionally, LNPs formulated with esterified cholesterol (e.g., cholesterol stearate) showed higher RNA delivery efficiency compared to LNPs containing regular or oxidized cholesterol (e.g., 7B-OH cholesterol) in mice. They also identified LNPs containing cholesteryl oleate as efficient nanocarriers for delivery of siRNA and sgRNA to liver endothelial cells in mice.710 This work also showed the possibility that rational design of the cholesterol analogs that closely mimic natural lipoproteins or interact with natural cholesterol trafficking pathways may enhance LNPs delivery efficiency.711
Figure 83.
Chemical structures of cholesterol variants formulated in LNPs.
In 2019, a library of nine cholesterol analogs was studied to evaluate the influence of chemical structures of sterol variants on mRNA delivery efficiency of LNPs.712 These side-chain or ringoxidized cholesterol analogs were obtained via treating cholesterol with enzymes (Figure 84). 125 FIND545,713 LNPs were formulated with the ionizable lipid cKK-E12,714 two PEG-lipids, the DOPE, and one of nine cholesterol variants. They observed that systematically oxidative modifications on the hydrocarbon tail (e.g., 20-hydroxycholesterol) were more potent than those with an oxidized B ring (e.g., 7-ketohydroxycholesterol). LNPs formulated with 20-hydroxycholesterol preferentially delivered Cre mRNA to hepatic cells and Kuffper cells rather than hepatocytes at a dose of 0.5 mg/kg after systemic administration in Ai14 mice. Oxidation of the tail attached to the sterol ring D may alter the interaction between LNPs and serum proteins, resulting in the biodistribution of LNPs in preference to the liver endothelial cells and Kupffer cells.712
Figure 84.
Chemical structures of sterol variants modified from cholesterol.
The Sahay group improved RNA delivery by using plant-based analogs of cholesterol instead.715–717 In 2020, Eygeris et al. found that when C-24-alkylated derivatives of cholesterol (e.g., β-sitosterol) were included in the mRNA-loading LNPs, enhanced mRNA delivery was observed.715 To further evaluate the effect of cholesterol analogs on RNA delivery efficacy, three groups of naturally occurring cholesterol analogs were selected based on structural resemblances (Figure 85).716 Group I contained vitamin D1, vitamin D2, and vitamin D3, group II was composed of C-24 α-alkyl sterols (e.g., β-sitosterol), and group III consisted of pentacyclic terpenoids (e.g., betulin). The screening result showed that group I analogs showed low mRNA delivery efficiency. Incorporation of the group II analog β-sitosterol in LNPs could result in mRNA translation efficiency improvement by 48-fold in cancer cells, whereas the inclusion of group III analogues led to ≥50% decrease in mRNA encapsulation efficiency and increased size of LNPs, resulting in poor mRNA delivery efficiency.718,719
Figure 85.
Chemical structures of three groups of cholesterol analogs.
The C-24 alkyl group of phytosterols induces crystal defects that are in proportion to the length of the C-24 alkyl group (cholesterol < campesterol < β-sitosterol).720 Three additional phytosterols, namely stigmastanol, fucosterol, and campesterol, were selected and formulated (Figure 86). The resulting LNPs exhibited high encapsulation (>90%) and an 11- to 211-fold improvement in mRNA delivery efficiency than cholesterol-based LNPs. Results of brassicasterol, ergosterol, and 9,11-dehydroergosterol (Figure 86) showed that reduced flexibility in the body and tail domain of cholesterol variants could inhibit RNA delivery. β-Sitosterol amino acid conjugates (polar) and β-sitosterol acetate (nonpolar) were also evaluated and showed that shielding the hydroxy group on ring A resulted in low or no RNA delivery efficiency. Lysosomal transporters recognize the hydroxyl group on cholesterol ring A and deliver cholesterol to the endoplasmic reticulum.721 The structure–activity relationship analysis of cholesterol analogs revealed that the high mRNA delivery efficiency was closely related to the flexibility of the sterol ring, the alkyl tail length, and the polarity associated with the hydroxyl group. The enhanced mRNA delivery efficacy may have been caused by morphological changes in the internal and external structure of mRNA-loading LNP.716,722 The structural analysis revealed that LNPs formulated with phytosterols had a polymorphic shape and exhibited different degrees of multilamellarity and rigidity. Unlike the smooth surface of LNPs containing cholesterol, LNPs containing β-sitosterol had a multifaceted surface, which might be caused by the changes in the surface lipid composition723 and could enhance the fusion of LNPs with membranes,724 thus leading to higher mRNA delivery efficiency. LNPs formulated with campesterol, stigmasterol, or β-sitosterol showed higher lamellarity and few internal defects as compared to LNPs containing cholesterol. The increased lamellarity may promote the fusion of LNPs with the endosomal membrane, while the internal structure of the LNPs may not be critical for the improved mRNA delivery efficiency of LNPs.715 They further found that LNPs formulated with vitamin D2 cannot cross the cell membrane due to the high fragility of their fluid lipid membrane, while the fucosterol-containing LNPs fail to deliver the mRNA payload due to their strong stability caused by the excessively rigid membrane. Moreover, live-cell imaging showed that LNP containing β-sitosterol showed extended retention in the endocytic vesicles, thus boosting the endosome escape of LNPs.717
Figure 86.
Structural features of C-24 alkyl derivatives of cholesterol.
3.3. Fatty Acids
When anionic lipids (Figure 87) are formulated into LNPs, they can interact with cationic lipids to form ion pairs, which can improve the pH-sensitivity of the LNPs and promote endosomal membrane destabilization, thus increasing nucleic acids delivery efficiency.725,726 Anionic lipids, such as fatty acids, can be protonated in the acidic endosome following cellular uptake. Protonation of the anionic lipids can cause the LNPs to be surface positively charged and/or induce lipid phase transition. Previous studies showed that if unsaturated fatty acids such as oleic acid (OA), linoleic acid (LA), or linolenic acid (LNA) are incorporated in LNPs, in place of PCs, the delivery efficiency of siRNA or miRNA may be significantly enhanced.194,727,728 DOTMA-based LNPs containing oleic acid (OA) were shown to be more efficient in microRNA-122 delivery than the commercially available Lipofectamine 2000.729 As linoleic acid (LA) is an essential fatty acid, which can participate in fatty acid metabolism of liver cells through plasma membrane fatty acid binding protein (FABPpm),730–733 the inclusion of LA can enhance hepatocyte uptake of LNPs. Yu et al. reported that their TRENL3-based LNPs formulated with anionic fatty acids had a much lower surface charge, thus leading to the improvement of the biodistribution of LNPs and enhanced uptake into hepatocytes in mice following intravenous administration. Besides, incorporating unsaturated fatty acids in LNPs led to a slight reduction of the mean diameter of LNPs.734 In the formulation of Smarticles LNPs, carboxylate-based anionic lipids (e.g., CHEMS) were incorporated to achieve pH-responsiveness. Smarticles LNPs have been used in the delivery of small activating RNA (saRNA) into HepG2 human hepatocellular carcinoma cells in vitro, resulting in activation of CEBPA mRNA and growth inhibition of liver cancer cell.735 Sodium dodecyl sulfate (SDS) is an anionic lipid that is not commonly used in the preparation of RNA delivery systems. In 2013, SDS-CTAB vesicles were developed by Russo et al., which could efficiently deliver CAT-A98 mRNA into the HEK-293 cells.736
Figure 87.
Chemical structures of anionic lipids.
He et al. synthesized an anionic helper lipid named DC and developed DC-based LNPs, termed nano-Transformers.737 Synthesis of DC began with the coupling of (9E)-octadec-9-enoic acid 379 with diol 380; the resulting ester 381 was treated with TFA to remove the Boc protecting group, giving amine 382. Ring-opening reaction of citraconic anhydride 383 with the primary amine group of 382 afforded DC (Figure 88). DC LNPs were negatively charged in physiological pH (pH = 7.4) and nearly neutral in endosome. Upon protonation at acidic pH, the positive surface charge of DC LNPs induces endosomal membrane fusion, thus facilitating the release of siRNA into the cytosol. DC LNPs delivered cyclin-dependent kinase 1 (CDK1)-siRNA efficiently, leading to up to 95% reduction of CDK1 mRNA in HepG2 cells in vitro, and significantly suppressed the HepG2 tumor growth in nude mice.737
Figure 88.
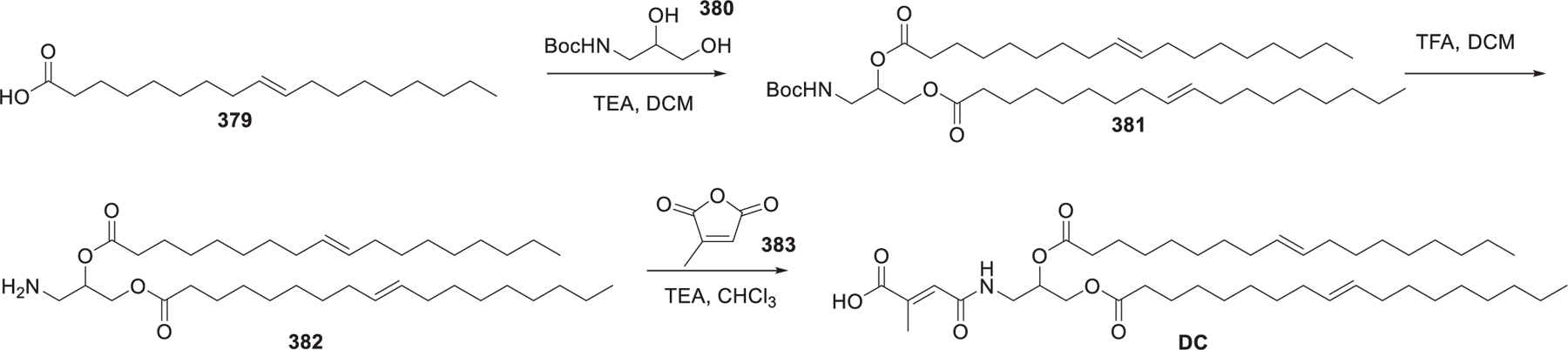
Synthesis procedures of DC.
3.4. Fatty Acid Esters
Both Span80738 and Tween80739 (Figure 89) have been used as surfactants in the formulation of cationic noisome for siRNA delivery. MOG can act as a helper lipid in LNPs-siRNA-mediated gene silencing because it can form gyroid inverted cubic structures that favor membrane fusion.347,437,740–744 DODAB/MOG (2:1) liposomes showed an efficient siRNA delivery.743,745,746 Cetyl palmitate is used as a component of some LNPs for RNA delivery, where the inclusion of cetyl palmitate can decrease the diameter of LNPs.747 Compritol 888 ATO is a mixture of glyceryl monobehenate (12–18% w/w), glyceryl dibehenate (45–54% w/w), and glyceryl tribehenate (28–32% w/w). Montana et al. reported delivery of siRNA via LNPs formulated with Compritol ATO 888 as matrix lipid.748
Figure 89.
Chemical structures of fatty acid esters for RNA delivery.
3.5. Other Helper Lipids
LNPs can be modified by incorporating lipids that contain various ligands to control their biological properties such as circulation stability and targeting ability.749 Antigen-presenting cells (APCs), such as dendritic cells and macrophages, express abundant mannose receptors on their cell surfaces; these mannose receptors can bind carbohydrates and their conjugates.750–753 To achieve site-specific targeting and enhance uptake of LNPs, mannose has been extensively investigated for modification of LNPs.575 Mannosylated lipid conjugate 384 (Figure 90) contains a d-mannose residue that is connected with dialkylglycerol via succinyl. The mannosylated LNPs incorporating mannosylated lipid conjugate 384 were used to deliver melanoma total RNA into DCs both ex vivo and in vivo and was shown to be more efficient in inducing CTL response compared to the control group.754 Mannosylated LNPs formulated with lipoconjugate 384 or 385 were efficient in delivering melanoma B16 RNA in vivo, inducing the generation of the melanoma B16-specific T-lymphocytes in mice and B16 cell apoptosis.755 Manchol, a mannose–cholesteryl amine conjugate, has also been used in the formulation of mannosylated LNPs loading self-amplifying mRNA (SAM) encoding hemagglutinin.756 Compared to the unglycosylated LNPs, mannosylated LNPs showed enhanced in vitro cellular uptake in BMDCs and induced a faster antibody response in mice, independent of the administration route.756 DGTS (Figure 90), a plant-derived structural lipid, is associated with lipid metabolism679 and cell survival in stress conditions.757,758 Kim et al. substituted DSPC in an LNP formulation, to a series of naturally occurring membrane lipids, especially DGTS.679 DGTS LNPs induced 20-fold lower luciferase expression compared to DSPC LNPs. However, DGTS led to an enhanced liver delivery of mRNA. This discrepancy may be due to the DGTS-induced LNP morphology modification, which changes both the stability and tolerance of LNPs.759 Both squalene760–762 and lycopene763,764 (Figure 90) can be used as neutral helper lipids in the preparation of noisome for RNA delivery.
Figure 90.
Chemical structures of other helper lipids for RNA delivery.
4. LIPID-DERIVED MACROMOLECULES
In addition to small molecular lipids and lipid derivatives, lipid-derived macromolecules have also been extensively explored as RNA delivery materials.722,765 The composition, structure type, and charge of lipid macromolecules have an important impact on the delivery efficiency of RNA.183 The lipid-derived macromolecules used for RNA delivery mainly include lipopolymers,766 lipopeptides,767 and lipoproteins.768 Since each type of macromolecule contains a large number of compounds, we generally highlight these lipid-derived macromolecules with an emphasis on their diverse chemical structures.
4.1. Lipopolymers
Lipopolymers have been widely used as a class of RNA delivery materials for decades.769,770 Lipopolymers are usually included in lipid nanoparticle formulations as auxiliary lipids or directly complexed with RNA molecules.771–773 They generally have the functions of enhancing the stability of lipid nanoparticles, promoting cellular uptake, and targeting diseased sites, thereby improving the efficiency of RNA delivery.774 From the perspective of chemical structures, lipopolymers are mainly composed of two parts: lipid fragments and polymer backbones. In this section, we mainly classify lipopolymers according to the types of polymer backbones including PEG-lipids, PEI-lipids, dendrimer-lipids, and other types of lipopolymers.
4.1.1. PEG-lipids.
PEG-lipids are important components usually incorporated on the surface of LNPs.195 PEG-lipids are composed of hydrophilic PEG conjugated to a hydrophobic alkyl chain through phosphate, glycerol, or other linkers. The PEG component can increase the stability of RNA-loaded LNPs and prolong their circulation time in the blood, thereby promoting the distribution and accumulation of nanoparticles in the diseased site.775 In this part, PEG-lipids are divided into two major types: general PEG-lipids and functionalized PEG-lipids.
4.1.1.1. General PEG-lipids.
PEG-lipids have been commonly applied in the construction of LNPs for drug delivery, such as DSPE-PEG2000 as one of the important components of Doxil LNPs.776,777 Later on, PEG-lipids are also used as an important ingredient of LNPs for RNA delivery.778 PEG2000-C-DMG is used as the PEG-lipid anchor in the Onpattro (patisiran) LNP formulation, which is the first FDA approved LNP-based siRNA drug for the treatment of hereditary transthyretin (hTTR) amyloidosis.779 Recently, DMG-PEG2000 and ALC-0159 are respectively used as one of the components of the mRNA-1273 and BNT162b2 vaccines against the COVID-19 pandemic.524 The PEG-lipids in the LNPs formulations of these three clinically used RNA drugs/vaccines have similar characteristics. These three PEG-lipids possess an mPEG2000 component as the hydrophilic chains and two alkyl chains with 14 carbon chains in length.
Since PEG-lipids have a great influence on the properties of LNPs, many studies have been conducted to investigate the effects of PEG-lipid on RNA LNPs.780,781 In 2008, Sonoke et al. synthesized several PEG-lipids containing acyl chains with 12 to 18 carbon atoms, which were used to prepare siRNA LNPs to investigate the influence of the alkyl chains length on LNPs.782 Each PEG-lipid/siRNA complex was intravenously injected in mice. Four hours post administration, the plasma concentration of 3H-labeled siRNA was over 10-fold higher in the group of C18-PEG2000 than that of the PEG-lipid/siRNA complex formed by shorter acyl chains (e.g., C-12 to C-16). In 2013, Mui et al. found that PEG-lipids with long alkyl chains (e.g., C-18) had a longer time associated with the LNPs than PEG-lipids with short alkyl chains (e.g., C-14).783 The dissociation of C-18 PEG-lipids from LNPs is approximately 0.2%/h in mice. Moreover, the molar ratio of PEG-lipid is another important factor.
In 2017, Oberli et al. found that the molecular weight of PEG (e.g., Mw: 350, 1000, 2000, 3000) and the length of the anchoring lipid (e.g., C-14, C-18) affected the particle size of LNPs (DMPE-PEG2000: ~67 nm).784 They applied DMPE-PEG2000 to prepare LNPs loaded with mRNA encoding various antigens as potential cancer vaccines. In the same year, Zhu et al. used the hybrid nanoparticle platform of a lipid-PEG shell and a PLGA/G0-C14 solid core to effectively achieve dePEGylation and control the delivery of siRNA.785 They found that the length of the PEG-lipids alkyl chains has a great influence on the dissociation rate of PEG from LNPs, and the dissociation rate of PEG on long alkyl chain lipids (e.g., DSG-PEG and DSPE-PEG) is slower than that on short alkyl chains (e.g., DMG-PEG and DMPE-PEG). Meanwhile, the unsaturated alkyl chain accelerates the dissociation of PEG (ceramide-PEG vs DPG-PEG), while the neutral and anionic PEG-lipids (DPG-PEG vs DPPE-PEG; DSG-PEG vs DSPE-PEG) with the same alkyl chain showed similar dissociation kinetics.785 This lipid–polymer hybrid RNA delivery system has also been used in delivering phosphatase and tensin homologue (PTEN) mRNA to restore tumor-growth suppression in mice.786
The preparation of acid-sensitive PEG-lipids is a method to improve the performance of LNPs. In 2012, Kulkarni et al. used the acid-sensitive benzylidene acetal bond to make Chol-PVA-PEG (Figure 91).787 The formulated Chol-PVA-PEG/siRNA nanoparticles showed a gene silencing activity comparable to bPEI and Lipofectamine 2000.
Figure 91.
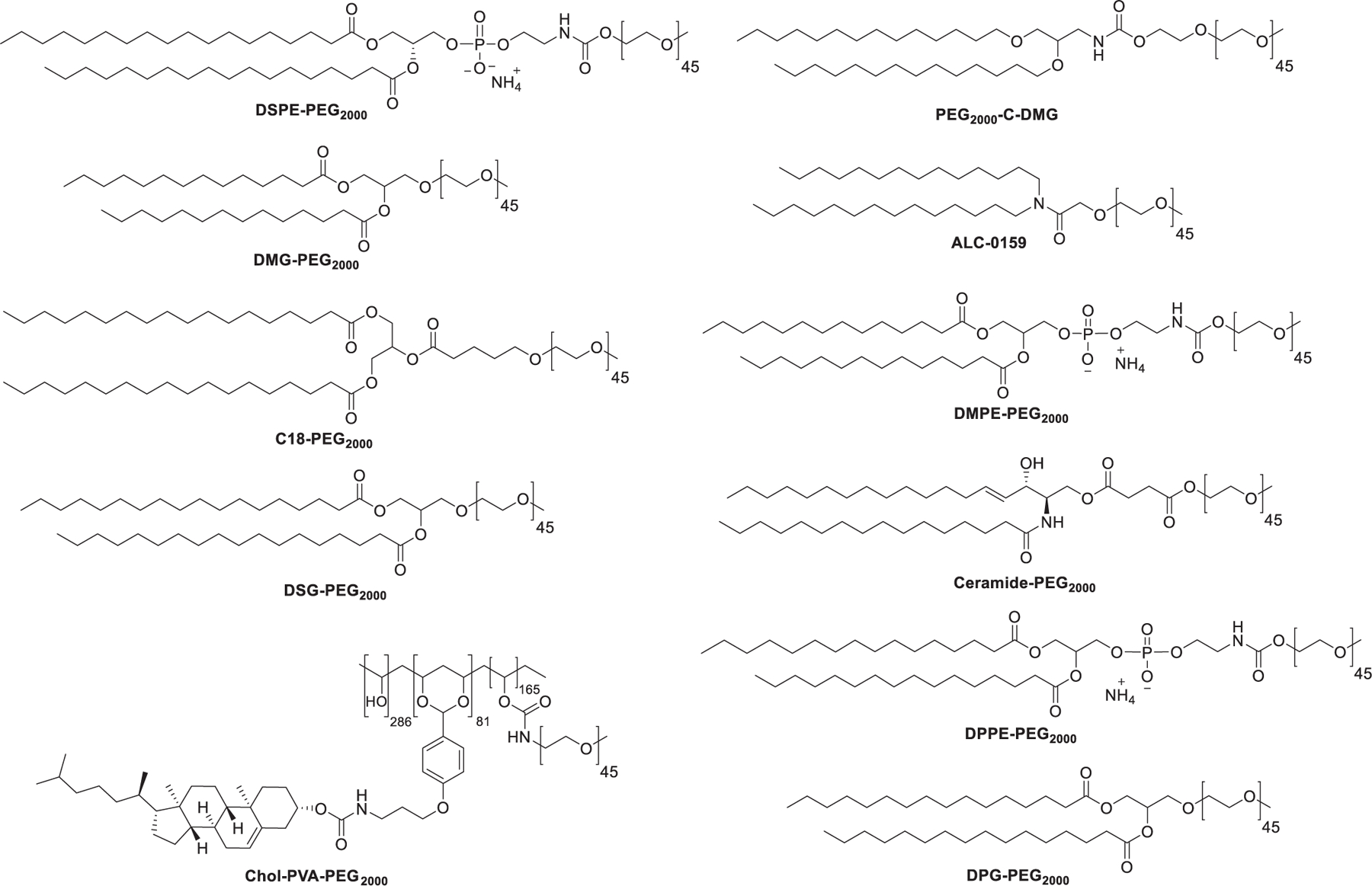
Chemical structures of PEG-lipids.
4.1.1.2. Functionalized PEG-lipids.
PEG can also be functionalized to incorporate specific functions such as cell targeting.788,789 In 2008, Li et al. linked anisamide to DSPE-PEG and prepared DSPE-PEG-anisamide that can target the sigma receptor on the B16F10 cancer cells.790 Then, DSPE-PEG-anisamide was used as a PEG-lipid to form a targeted LNPs-siRNA formulation. Compared to nontargeted LNPs, the targeted LNPs showed a 4-fold increase in cellular uptake in B16F10 cells. In 2010, Akinc et al. used targeting ligands containing multivalent N-acetylgalactosamine (GalNAc) clusters and linked them with DSG-PEG to prepare DSG-PEG-GalNAc that can be targeted to the liver hepatocytes.791 DSG-PEG-GalNAc, factor VII-targeting siRNA, and other components were formulated to LNPs, which were intravenously administered in mice at the siRNA doses of 0.022, 0.067, and 0.2 mg/kg. The LNPs-mediated factor VII in mice exhibited dose-dependent silencing. In 2016, Zang et al. conjugated anti-EphA10 antibody with Chol-SIB-PEG to prepare tumor-targeting Chol-SIB-PEG-Eph.792 In MCF-7/ADR cells, MDR1 siRNA LNPs prepared with Chol-SIB-PEG-Eph reduced the expression level of MDR1 protein. Furthermore, the LNPs containing the fluorescent molecule DIR were injected into the MCF-7/ADR xenograft tumor mouse model for fluorescence imaging. After 24 h of injection, the Chol-SIB-PEG-Eph-formulated LNPs displayed the highest accumulation in the tumor compared with the control group.792
In 2017, Krzysztoń et al. applied DSPE-PEG-FA as a folate-targeted PEG-lipid to form LNPs for siRNA delivery.793 These LNPs could significantly down-regulate luciferase activity compared with LNPs without a folate targeting group. In the same year, Santiwarangkool et al. synthesized DSG-PEG-GALA containing a polypeptide GALA that could target the lung endothelium for RNA delivery.794 Compared with LNPs composed of DSG-PEG, LNPs composed of DSG-PEG-GALA showed higher silencing activity in the mouse lungs, with an ED50 value of 0.21 mg/kg. In 2020, Qiao et al. synthesized vitamin A-modified PEG-lipid Chol-PEG-VA for RNA delivery (Figure 92).795 The cell uptake of Chol-PEG-VA containing LNPs was over 80% in HSC-T6. Compared with cells treated with single Col1α1 siRNA LNPs or TIMP-1 siRNA LNPs, double-siRNA nanoparticles Col1α1/TIMP-1 siRNA LNPs showed a higher inhibitory effect on collagen I accumulation. Among the various siRNA LNPs treatment groups, Col1α1/TIMP-1 siRNA LNPs treated mice led to the lowest collagen accumulation, which is similar to the level in normal mice.795
Figure 92.
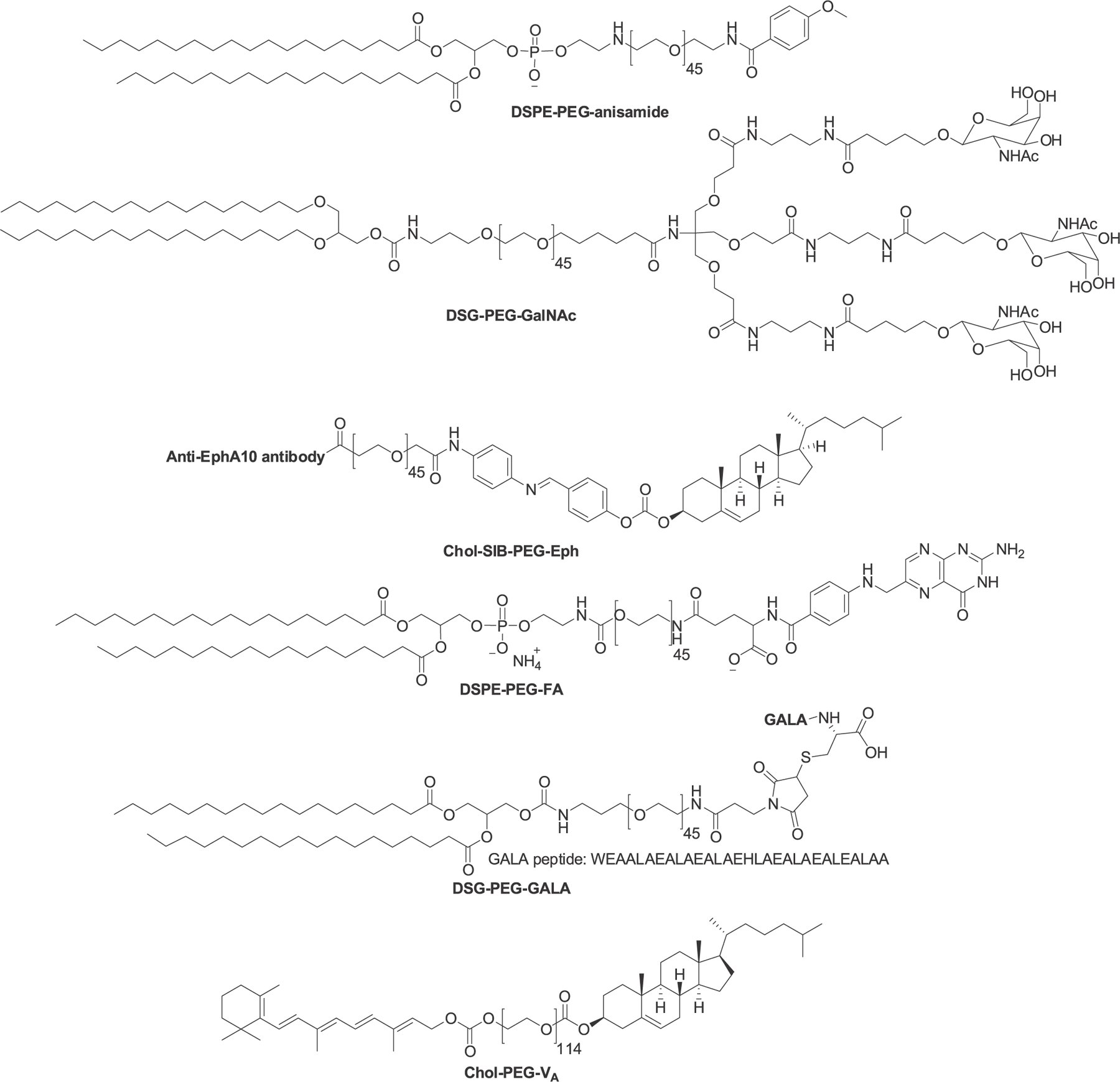
Chemical structures of functionalized PEG-lipids.
4.1.2. PEI-lipids.
PEI is another class of polymers used in RNA delivery.796,797 To improve the pharmaceutical properties of PEI, various lipids are conjugated with PEI.798,799 In 2014, Dahlman et al. synthesized a library of PEI analogs with diverse structures by conjugating small polyamines with alkyl tails of different lengths in different molar ratios.800 PEI-lipid 386 (Figure 93) was obtained by the epoxy-opening reaction between C15 epoxy-terminated lipid chains and PEI600 (14:1 molar ratio) and screened from the in vitro cell assays after formulation with PEG-lipid and siRNA. The lead material was able to silence multiple endothelial genes in mice.800 In the same year, Navarro et al. combined DOPE with PEI to obtain the lipopolymer 387.801 The DOPE-PEI effectively delivered GFP siRNA in c166 cells stably expressing GFP.801 In 2020, Fan et al. synthesized the Triton X-100-modified PEI (low molecular weight), which was then attached to dopamine grafted vitamin E (VEDA) and 4-carboxyphenylboronic acid (PBA) to obtain lipopolymer 388 (Figure 93).802 388-siEGFR complexes suppressed the protein expression of EGFR in A375 cells to 43.1% and reduced its EGFR mRNA level to 34.9% (Figure 93).
Figure 93.
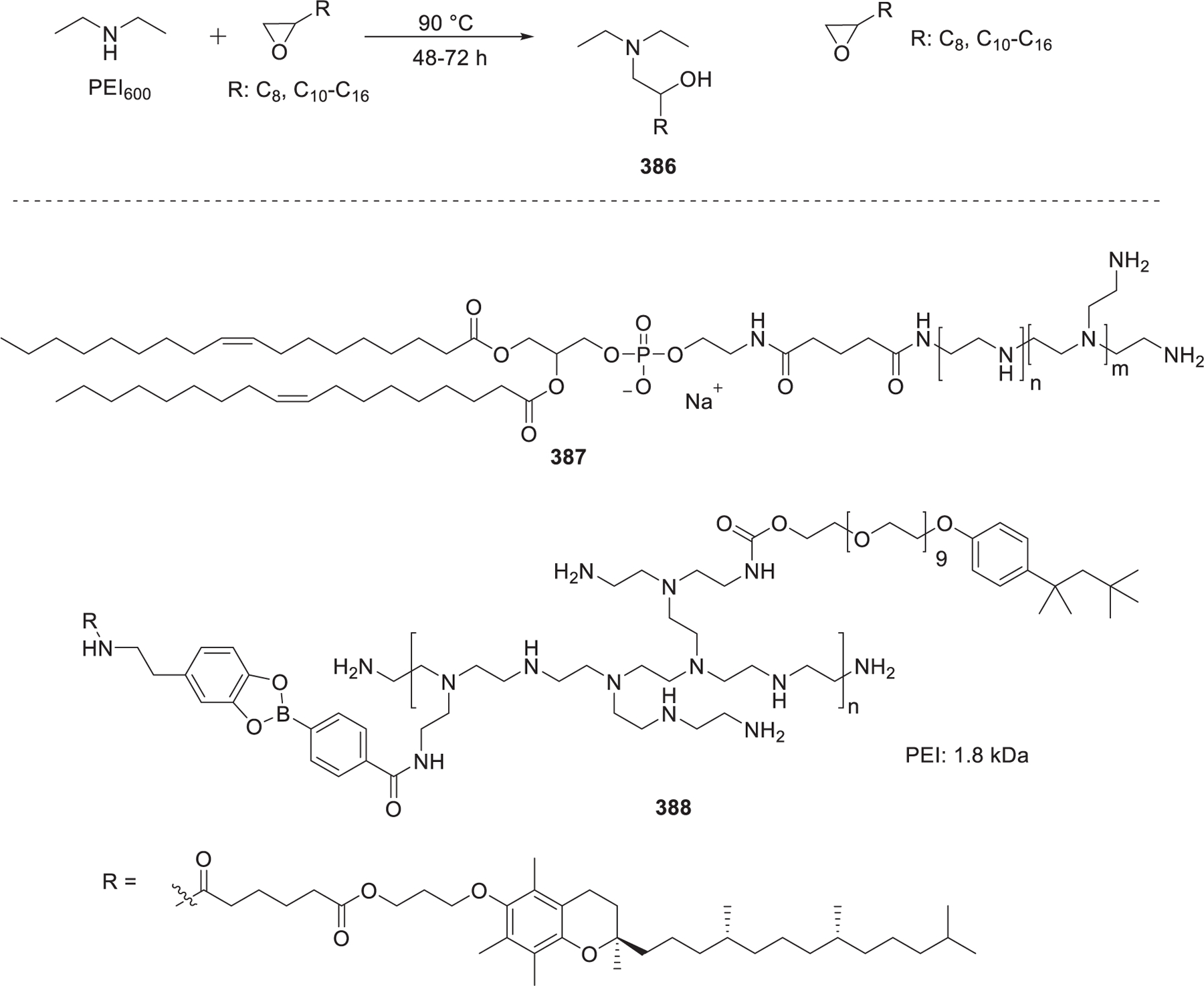
Chemical structures of PEI-lipids.
4.1.3. Dendrimer-lipids.
Dendrimers consist of three main parts: a central core, a growing branch unit (called generation (G)), and a large number of terminal functional groups (usually amino groups) on the surface.803 The terminal groups of the dendrimer can be lipid-modified to obtain the dendrimer-lipids.804–806 In 2012, Yu et al. synthesized a class of dendrimer-lipids with different alkyl chain lengths and dendrimer generations through click chemistry.807 The long alkyl chain of 389 (Figure 94) promotes the assembly of siRNA/carrier, can form a stable self-assembled complex with siRNA, and increases the stability of the complex through hydrophobic interactions. Moreover, the dendrimer-lipid 389/siRNA complex significantly downregulated Hsp27 and induced in a mouse prostate cancer model.807 In 2014, Khan et al. prepared LNPs with dendrimer-lipids 390 and 391 to deliver siRNA to the liver endothelium in vivo (Figure 94).808 In a hepatocellular tumor model, the formulated 390 and 391 showed 51% and 92% knockdown of Alpha-fetoprotein (AFP), respectively (siRNA dose: 1 mg/kg). In 2015, they used terminal epoxides with different alkyl chain lengths to react with free amines in poly(propylenimine) and poly(amidoamine) dendrimers to obtain dendrimer-lipids 390–394 (Figure 94).809 The nanoparticles of 393 loaded with Cy5.5-labeled siRNA showed greater fluorescence than other dendrimer-lipids in mouse lungs.
Figure 94.
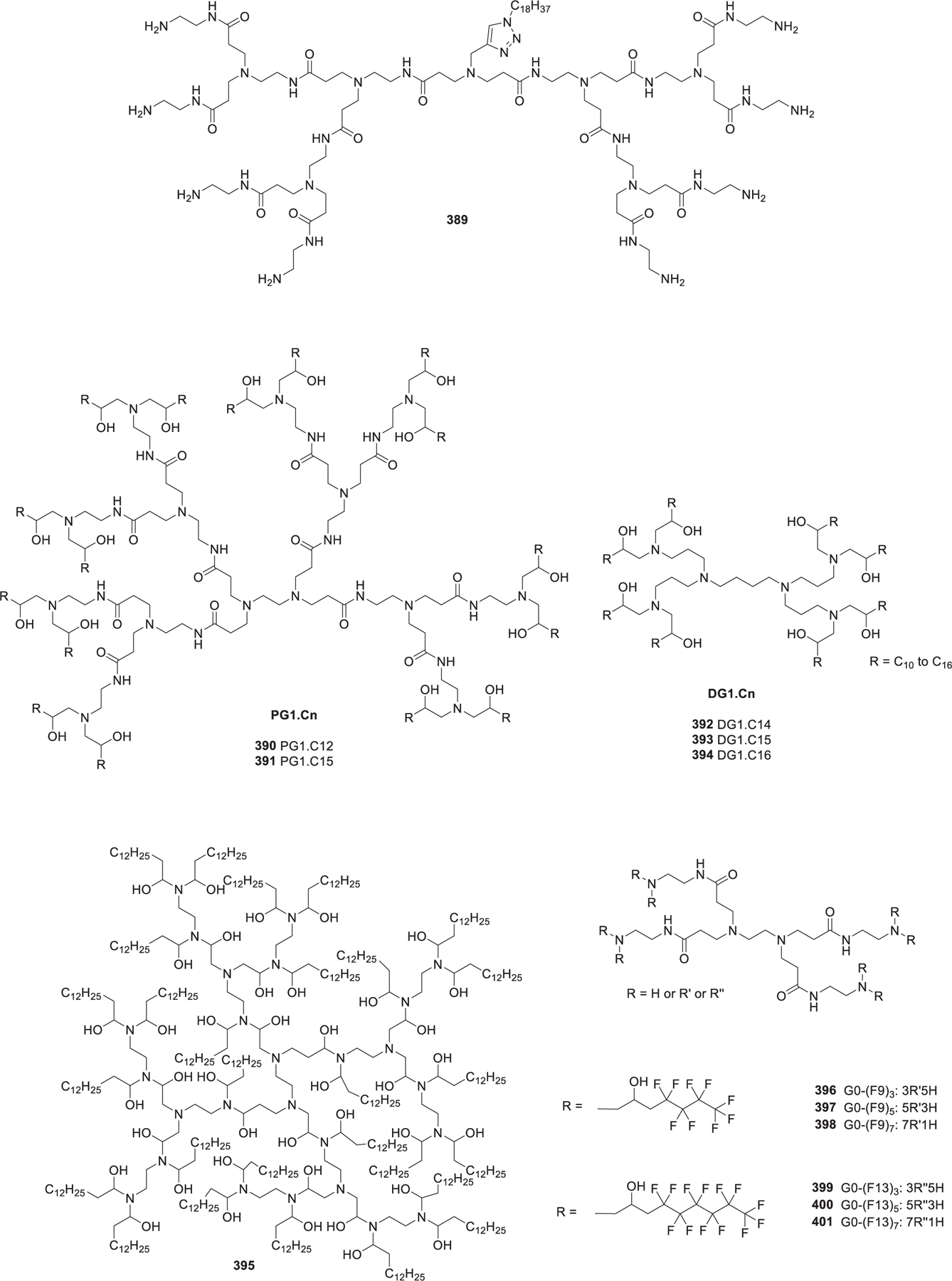
Chemical structures of dendrimer-lipids.
In 2016, Chahal et al. synthesized the modified dendrimer 395 and formulated it with PEG-lipids and self-amplifying RNA to form the nanoparticles (Figure 94).773 These nanoparticles were able to deliver many types of self-amplifying RNAs encoding viral antigens such as Ebola virus, Toxoplasma gondii, and H1N1 influenza. After a single-dose vaccination, the nanoparticles-treated mice developed effective CD8 T cells and antibody responses against specific pathogens.773
In 2020, Xiong et al. synthesized six low-molecular-weight epoxy-derived fluorine-substituted dendrimers (396–401) by reacting fluorine-containing alkyl propylene oxide with PAMAM-G0 (Figure 94).810 The formulation of dendrimer lipid 400 with Luc siRNA decreased over 50% of the luminescence signal at a siRNA concentration of 20 nM in HeLa-Luc cells. The siRNA targeting PHB1 (PHB1 siRNA) was also complexed with dendrimer lipid 400 to prepare the nanoparticles, which resulted in increased cell apoptosis rate in A549 lung carcinoma cells after treatment with this formulation.810
4.1.4. Other Types of Lipopolymers.
There are many other types of lipopolymers such as lipocationic polyesters, brush-like lipopolymers with poly(glycoamidoamine) (PGAAs) as a backbone, and charge-altering releasable transporters (CARTs) lipopolymers.811,812 In 2015, Hao et al. synthesized over one hundred lipocationic polyesters (404) using amino- or alkyl- valerolactone monomers (Figure 95).813 Six of these lipocationic polyester-formulated nanoparticles enabled over 90% silencing at a siRNA concentration of 38.4 nM in vitro. One lead material showed effective siRNA delivery in a MDA-MB-231-Luc xenograft mouse tumor model.813 In 2016, Dong et al. synthesized a series of lipopolymers (e.g., TarN3C10) through the reactions of poly(glycoamidoamine) (PGAAs) and epoxides (Figure 95).814 A number of the formulated PGAAs-lipids silenced over 50% of the FVII production.814 In 2017, Luo et al. continued to improve the brush lipopolymer based on the PGAA backbone and synthesized three additional PGAA lipid derivatives.815 The GluN4C10 nanoparticles showed more effective gene silencing of FVII compared to the TarN3C10 nanoparticles. In the same year, McKinlay et al. synthesized several amphiphilic charge-altering releasable transporters (CARTs) for RNA delivery (Figure 95).816 Among the different CARTs, CART-D13:A11 showed effective mRNA delivery in BALB/c mice. Later on, Benner et al. further developed Ser-CART 13b and evaluated its mRNA delivery via intramuscular or intravenous (iv) injection in mice.811
Figure 95.
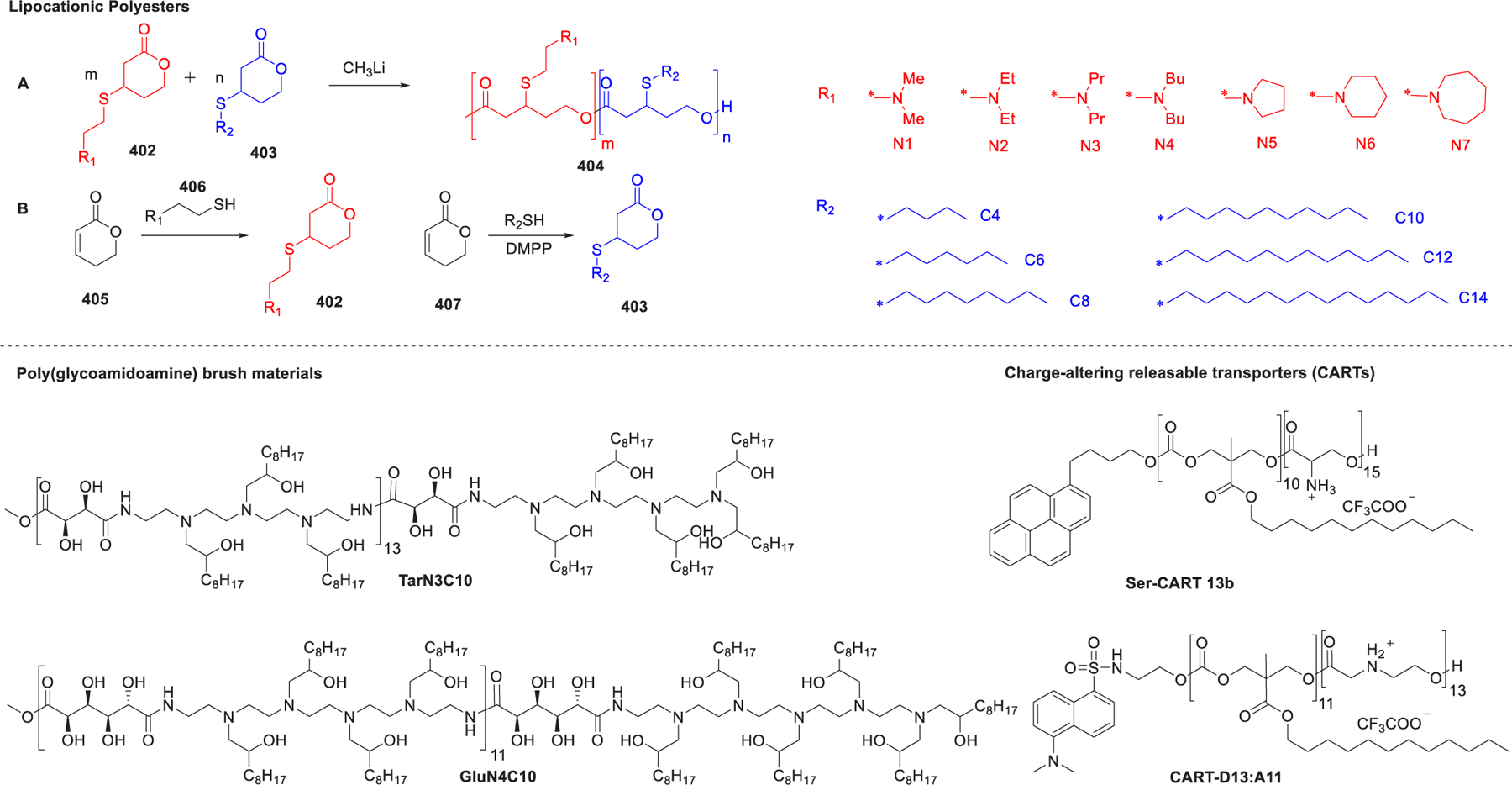
Chemical structures of other types of lipopolymers.
4.2. Lipopeptides
Lipopeptides possess the properties of both lipids and peptides.817–819 Lipopeptides not only have biological functions such as targeting effects but also take advantage of lipid features such as hydrophobic interactions.820,821 In 2006, Kim et al. conjugated oligoarginine with cholesteryl chloroformate to produce the oligomer Chol-R9 (Figure 96).822 The complex of Chol-R9 and siRNA against VEGF inhibited tumor growth after intratumoral injections in a CT-26 mouse tumor model. In 2011, Cline et al. conjugated cholic acid, spermidine, and lysine to prepare diwalled and tetrawalled umbrella compounds (Figure 96).823 Then, these umbrella compounds were reacted with the octaarginine peptide to obtain a variety of lipopeptides. Lipopeptide 408/siRNA complex showed similar silencing activity to that of Lipofectamine 2000.823 In 2017, Sharma et al. modified CGKRK, a tumor targeting peptide, with saturated and unsaturated fatty acids to synthesize the lipopeptides including 409.824 The oleic acid-CGKRK lipopeptide is a lead material for siRNA delivery found in cell studies. In 2020, Zhao et al. formulated cholesterol peptide (CP, 410, Figure 96), cabazitaxel, IKBKE siRNA, and cholesterol-hyaluronic acid (CHA) to assemble siRNA nanocomplex (CHA/CP/siRNA/cabazitaxel).817 This formulation could target CD44 overexpressed on triple-negative breast cancer (TNBC) cells. Moreover, this nanocomplex could codeliver IKBKE siRNA and cabazitaxel to TNBC cells, which significantly inhibited tumor growth in an orthotopic MDA-MB-231 TNBC mouse mode.817
Figure 96.
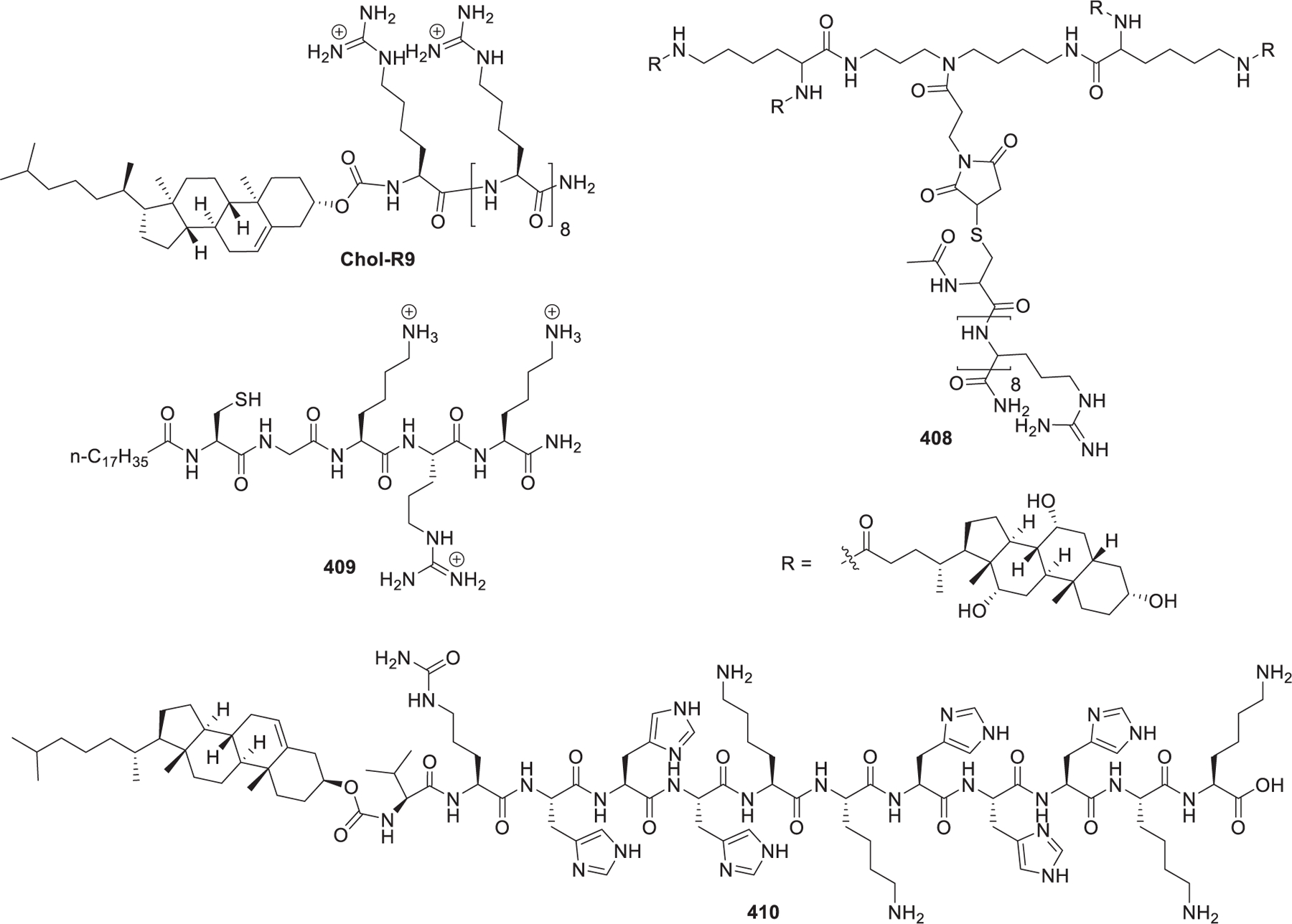
Chemical structures of lipopeptides.
4.3. Lipoproteins
Lipoproteins are endogenous nanocomplexes in the human body, which can be divided into chylomicrons (CM), very low density lipoproteins (VLDL), low density lipoproteins (LDL), and high density lipoproteins (HDL) based on their density.825,826 Lipoproteins serve as natural transporters for proteins, vitamins, and hormones. Lipoprotein particles are composed of surface components (apolipoproteins, phospholipids, and cholesterol) and core components (cholesterol esters and triglycerides).827–829 Researchers have applied lipoproteins to deliver small molecular drugs and nucleic acids for therapeutic applications.711,830–832 For example, Yang et al. prepared an HDL-mimicking peptide–phospholipid scaffold (HPPS) by integrating amphipathic α-helical peptides, cholesteryl oleate, and phospholipids; HPPS loaded with cholesterol-modified bcl-2 siRNA greatly reduced the expression of bcl-2 in KB cells.833 Based on this study, Cruz et al. constructed HPPS encapsulating siRNA against spalt-like transcription factor 4 (SALL4).834 The HPPS-siRNA nanoparticles showed significant inhibition of tumor growth in a hepatocellular carcinoma mouse model. In another study, McMahon et al. formulated templated lipoprotein particles (TLP) using gold nanoparticle, ApoA-I, two phospholipids, and cholesterol, which were then complexed with the DOTAP–RNA mixture to produce siRNA-TLP.835 These siRNA-TLP nanoparticles targeting the androgen receptor (AR) significantly inhibited tumor growth in a LNCaP xenograft mouse tumor model. In 2017, Huang et al. applied calcium phosphate, phospholipids, apolipoprotein E3, and siRNA to formulate siRNA-CaP-rHDL nanoparticles.836 These siRNA-CaP-rHDL promote the penetration of the blood–brain barrier and deliver siRNA to glioblastoma cells. In a C6 glioblastoma mouse model, the nanoparticles loaded with siRNA against activating transcription factor-5 (ATF5) reduced the ATF5 protein level and improved the mouse survival time. Recently, Jiang et al. developed SLNP, a lipoprotein-like nanoparticle using multiple components including calcium phosphate, phospholipids, stromal cell-derived factor 1 (SDF1) mimic peptides, and apolipoprotein E (apoE).829 These SLNP nanoparticles effectively delivered miR34a to glioma initiating cells (GICs) and extended overall survival in a patient-derived GICs glioma mouse model.
5. CONCLUSIONS AND OUTLOOK
Synthetic chemists and material scientists have spent over three decades on developing novel structures and formulations of lipids, lipid derivatives, and lipid-derived macromolecules for RNA delivery. With a better understanding of structural and biophysical properties for effective RNA delivery as well as the underlying mechanisms involved in the cellular uptake and endosomal release of the nanoparticles, more and more lipid-based RNA delivery systems are in different stages of clinical trials. In particular, MC3, ALC-0315, and SM-102-based lipid nanoparticles have been approved by the FDA and EMA for clinical use. These significant advances open up numerous opportunities for lipids and lipid derivatives and their related RNA-based therapeutics. (i) New chemical structures of lipids and lipid derivatives can be conceived and synthesized based on the current knowledge of lipid components such as cationic or ionizable head groups, linker groups, and hydrophobic tails. Many types of helper lipids and lipopolymers can be incorporated into nanoparticle formulations. (ii) A large number of chemical reactions such as Michael addition, epoxide-ring-opening, reductive amination, click chemistry, or one-pot multicomponent reactions permit the rapid synthesis of libraries of lipids with diversified chemical structures, paving a way to evaluate the structure–activity relationship. Incorporation of some biocompatible or targeting ligands, such as mannose, cell penetrating peptides, aptamers, or vitamins into the head group domains of cationic/ionizable lipids can improve RNA delivery efficiency with higher biocompatibility and lower immunogenicity. (iii) Detailed studies on LNPs properties such as physicochemical properties, enzymatic stability, morphology, and shelf life are needed. These studies will expand our understanding of the features of LNPs and provide useful design criteria for future LNPs development. (iv) Pharmaceutical properties of LNPs such as pharmacokinetics, pharmacodynamics, immunogenicity, and safety should also be carefully characterized. Knowledge obtained from these studies will not only elucidate the LNPs characteristics and their interactions with the host but also benefit the development of a wide variety of LNPs. (v) LNP-formulated RNA therapeutics can be broadly applicable to diverse diseases. Current clinical trials are investigating a series of LNPs-RNA candidates in cancer immunotherapy, protein replacement therapy, and gene editing. Importantly, LNPs-RNA formulations can potentially overcome “non-druggable” targets to treat diseases that cannot be addressed with current medicines, such as neurodegenerative disorders and genetic diseases. In summary, with the increasing chemical diversity and formulation number of lipids and lipid derivatives, more and more RNA-based therapeutics are translating from bench to bedside and improving the quality of life.
ACKNOWLEDGMENTS
Y.D. acknowledges the support of the Early Career Investigator Award from the Bayer Hemophilia Awards Program, Research Awards from the National PKU Alliance, New Investigator Grant from the AAPS Foundation, Maximizing Investigators’ Research Award R35GM119679 from the National Institute of General Medical Sciences, R01HL136652 grant from the National Heart, Lung, and Blood Institute, as well as the startup fund from the College of Pharmacy at The Ohio State University.
ABBREVIATIONS
- AML
acute myeloid leukemia
- APC
antigen-presenting cell
- ApoB
apolipoprotein B
- ApoE
apolipoprotein E
- ASGPR
asialoglycoprotein receptor
- ASO
antisense oligonucleotides
- BBB
blood–brain barrier
- Boc
tert-butyloxycarbonyl
- BMDC
bone marrow-derived dendritic cell
- BMDM
bone marrow-derived macrophage
- CAR-T cell
chimeric antigen receptor T cell
- Cas9
CRISPR associated protein 9
- Cbz
benzyloxycarbonyl
- CD
circular dichroism
- CDI
N,N′-carbonyldiimidazole
- cirRNA
circular RNA
- CME
clathrin-mediated endocytosis
- CMC
critical micelle concentration
- CNE
cationic nanoemulsion
- COVID-19
coronavirus disease 2019
- CRISPR
clusters of regularly interspaced short palindromic repeats
- CPP
cell penetrating peptide
- CTL
cytotoxic T lymphocyte
- CvME
caveolae-mediated endocytosis
- DC
dendritic cell
- DCC
N,N′-dicyclohexylcarbodiimide
- DCM
dichloromethane
- DIPEA
N,N′-diisopropylethylamine
- DMAP
4-dimethylaminopyridine
- DMF
dimethylformamide
- DOPE
1,2-dioleoyl-sn-glycerol-3-phosphatidylethanolamine
- DSPC
1,2-distearoyl-sn-glycero-3-phosphocholine
- dsRNA
double-stranded RNA
- EC
endothelial cell
- EVs
extracellular vesicles
- ECM
extracellular matrix
- ED50
median effective dose
- EDCI
1-ethyl-3-(3-(dimethylamino)propyl)-carbodiimide
- EGFP
enhanced green fluorescent protein
- ePC
ethylphosphatidylcholine
- EUA
emergency use authorization
- FIND
Fast Identification of Nanoparticle Delivery
- FVII
factor VII
- FXN
frataxin
- GalNAc
N-acetylgalactosamine
- GHS
glutathione
- HA
hemagglutinin
- HAI
hemagglutination inhibition
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- hCSF
human cerebrospinal fluid
- HDL
high density lipoprotein
- HMPT
hexamethylphosphorus triamide
- IDL
intermediate density lipoproteins
- IFN
interferon
- LDL
low density lipoprotein
- LNP
lipid nanoparticles
- LPHNP
lipid–polymer hybrid nanoparticle
- Luc
luciferase
- MHC I
major histocompatibility complex I
- MHC II
major histocompatibility complex II
- miRNA
micro RNA
- MOG
monooleoyl glycerol
- mRNA
mRNA
- MTO
mitoxantrone
- ncRNA
noncoding RNA
- NHP
nonhuman primate
- NLE
neutral lipid emulsion
- OVA
ovalbumin
- HIV
human immunodeficiency virus
- Pal
palmitoleyl
- PBA
phenylboronic acid
- pBAE
poly(β-amino ester)
- pDNA
plasmid DNA
- PEG
polyethylene glycol
- PEI
poly(ethylenimine)
- Ka
acidity constant
- PLGA
poly(lactic-co-glycolic acid)
- PLL
poly-l-lysine
- PPTS
p-toluenesulfonate
- PMP
platelet-derived microparticle
- RBC
red blood cell
- RBD
receptor binding domain
- RISC
RNA-induced silencing complex
- RNase
ribonucleases
- RNAi
RNA interference
- RVG
rabies virus glycoprotein
- SA
sialic acid
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- scFv
single-chain variable fragment
- siRNA
small interfering RNA
- sgRNA
single-guide RNA
- SORT
selective organ targeting
- SR-SAXS
small-angle X-ray scattering
- STING
stimulator of interferon genes
- TLP
templated lipoprotein particle
- TAA
tumor associated antigen
- TEA
triethylamine
- TFA
trifluoroacetic acid
- THF
tetrahydrofuran
- TTR
transthyretin
- VLDL
very low density lipoprotein
Biographies
Yuebao Zhang received his B.S. degree in pharmaceutical science from Zhengzhou University in 2011. In 2017, he received his Ph.D. degree in medicinal chemistry from Sichuan University under the supervision of Professor Zhenlei Song. He is currently a postdoctoral research associate in the lab of Dr. Yizhou Dong at The Ohio State University. His current research focuses on development of novel ionizable lipids and evaluation of lipids for their application in mRNA delivery.
Changzhen Sun received his bachelor degree in Pharmacy from Qingdao Agricultural University, China, in 2010. He obtained a master’s degree in pharmaceutical chemistry under the supervision of Professor Zhenlei Song in 2014, and a Ph.D. degree in organic chemistry under the guidance of Professor Bin He in 2017, Sichuan University, China. Then, he joined the Traditional Chinese Medicine Hospital Affiliated to Southwest Medical University. He is a postdoctoral research associate in the lab of Yizhou Dong at The Ohio State University. His research focuses on the construction and biological evaluation of novel RNA delivery systems.
Chang Wang is a postdoctoral associate in the College of Pharmacy at The Ohio State University. He received his B.Sc. in Pharmacy from Tianjin University, China, and his Ph.D. in Chemistry from Lehigh University, PA, where he investigated fundamental and translational studies of lipids and membranes. His Ph.D. work was focusing on a significant role unsaturated phospholipids play in the lipid rafts formation using a cell membrane model. Currently, under the mentorship of Dr. Yizhou Dong at The Ohio State University, he is devoted to developing multifunctional and biodegradable lipid-like molecules for mRNA delivery and genome editing.
Katarina E. Jankovic is a second-year undergraduate honors student in the program of neuroscience at The Ohio State University. She is an undergraduate researcher in the Dong group. Her research interests include cell specific drug delivery systems with a focus on mRNA therapeutics.
Yizhou Dong is an Associate Professor in the Division of Pharmaceutics and Pharmacology, College of Pharmacy at The Ohio State University. He received his B.S. in pharmaceutical sciences from Peking University, Health Science Center, and his M.S. in organic chemistry from Shanghai Institute of Organic Chemistry. In 2009, he received his Ph.D. degree in pharmaceutical sciences from the University of North Carolina at Chapel Hill (UNC-CH) under the supervision of Professor K.-H. Lee. From 2010 to 2014, he was a postdoctoral fellow in the laboratory of Professors Robert Langer and Daniel Anderson at Harvard Medical School and Massachusetts Institute of Technology. His research focuses on the design and development of nanomaterial and biotechnology platforms for the treatment of genetic disorders, infectious diseases, and cancers. He has authored over 70 papers and patents. Several of his inventions have been licensed and are currently under development as drug candidates for clinical trials. He serves as a board member of the Gene Delivery and Editing (GDE) Focus Group of the Controlled Release Society and an editorial board member for Bioactive Materials. He is a scientific advisory board member of Oncorus Inc. He is the recipient of Young Innovator in Cellular and Molecular Bioengineering from the Biomedical Engineering Society, Maximizing Investigators’ Research Award from the National Institute of General Medical Sciences (NIGMS), Ohio State Early Career Innovator of the Year, and American Association of Pharmaceutical Scientists (AAPS) Emerging Leader Award.
Footnotes
The authors declare the following competing financial interest(s): Y.D. is a scientific advisory board member of Oncorus Inc and serves as a consultant of Rubius Therapeutics.
Contributor Information
Yuebao Zhang, Division of Pharmaceutics & Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio 43210, United States.
Changzhen Sun, Division of Pharmaceutics & Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio 43210, United States.
Chang Wang, Division of Pharmaceutics & Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio 43210, United States.
Katarina E. Jankovic, Division of Pharmaceutics & Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio 43210, United States
Yizhou Dong, Division of Pharmaceutics & Pharmacology, College of Pharmacy and Department of Biomedical Engineering, The Center for Clinical and Translational Science, The Comprehensive Cancer Center, Dorothy M. Davis Heart & Lung Research Institute, Department of Radiation Oncology, The Ohio State University, Columbus, Ohio 43210, United States.
REFERENCES
- (1).DeWitt DE; Hirsch IB Outpatient Insulin Therapy in Type 1 and Type 2 Diabetes Mellitus. JAMA 2003, 289, 2254–2264. [DOI] [PubMed] [Google Scholar]
- (2).Akhtar MJ; Ahamed M; Kumar S; Siddiqui H; Patil G; Ashquin M; Ahmad I Nanotoxicity of Pure Silica Mediated Through Oxidant Generation Rather Than Glutathione Depletion in Human Lung Epithelial Cells. Toxicology 2010, 276, 95–102. [DOI] [PubMed] [Google Scholar]
- (3).Lin Y-X; Wang Y; Blake S; Yu M; Mei L; Wang H; Shi J RNA Nanotechnology-Mediated Cancer Immunotherapy. Theranostics 2020, 10, 281–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Naldini L Gene Therapy Returns to Centre Stage. Nature 2015, 526, 351–360. [DOI] [PubMed] [Google Scholar]
- (5).Weissman D; Karikó K mRNA: Fulfilling the Promise of Gene Therapy. Mol. Ther 2015, 23, 1416–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Ozcan G; Ozpolat B; Coleman RL; Sood AK; Lopez-Berestein G Preclinical and Clinical Development of siRNA-Based Therapeutics. Adv. Drug Delivery Rev 2015, 87, 108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).FDA. Fact Sheet for Healthcare Providers Administering Vaccine (Vaccination Providers) Emergency Use Authorization (EUA) of the Moderna COVID-19 Vaccine to Prevent Coronavirus Disease 2019 (COVID-19) https://www.fda.gov/media/144637/download (accessed 2021-05-19).
- (8).EMA. EMA Recommends COVID-19 Vaccine Moderna for Authorisation in the EU https://www.ema.europa.eu/en/news/ema-recommends-covid-19-vaccine-moderna-authorisation-eu (accessed on 2021-05-19).
- (9).FDA. FDA Takes Key Action in Fight Against COVID-19 by Issuing Emergency Use Authorization for First COVID-19 Vaccine https://www.fda.gov/news-events/press-announcements/fda-takes-key-action-fight-against-covid-19-issuing-emergency-use-authorization-first-covid-19 (accessed 2021-05-19).
- (10).Crooke ST Molecular Mechanisms of Antisense Oligonucleotides. Nucleic Acid Ther 2017, 27, 70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Vickers TA; Crooke ST Antisense Oligonucleotides Capable of Promoting Specific Target mRNA Reduction via Competing RNase H1-Dependent and Independent Mechanisms. PLoS One 2014, 9, No. e108625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kole R; Krainer AR; Altman S RNA Therapeutics: Beyond RNA Interference and Antisense Oligonucleotides. Nat. Rev. Drug Discovery 2012, 11, 125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Boiziau C; Kurfurst R; Cazenave C; Roig V; Thuong NT; Toulmé J-J Inhibition of Translation Initiation by Antisense Oligonucleotides via an RNase-H Independent Mechanism. Nucleic Acids Res 1991, 19, 1113–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Evers MM; Toonen LJA; van Roon-Mom WMC Antisense Oligonucleotides in Therapy for Neurodegenerative Disorders. Adv. Drug Delivery Rev 2015, 87, 90–103. [DOI] [PubMed] [Google Scholar]
- (15).Bennett CF; Swayze EE RNA Targeting Therapeutics: Molecular Mechanisms of Antisense Oligonucleotides as a Therapeutic Platform. Annu. Rev. Pharmacol. Toxicol 2010, 50, 259–293. [DOI] [PubMed] [Google Scholar]
- (16).Fire A; Xu S; Montgomery MK; Kostas SA; Driver SE; Mello CC Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis Elegans. Nature 1998, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- (17).Elbashir SM; Harborth J; Lendeckel W; Yalcin A; Weber K; Tuschl T Duplexes of 21-Nucleotide RNAs Mediate RNA Interference in Cultured Mammalian Cells. Nature 2001, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- (18).Devi GR siRNA-Based Approaches in Cancer Therapy. Cancer Gene Ther 2006, 13, 819–829. [DOI] [PubMed] [Google Scholar]
- (19).Zuckerman JE; Davis ME Clinical Experiences With Systemically Administered siRNA-Based Therapeutics in Cancer. Nat. Rev. Drug Discovery 2015, 14, 843–856. [DOI] [PubMed] [Google Scholar]
- (20).Mahmoodi Chalbatani G; Dana H; Gharagouzloo E; Grijalvo S; Eritja R; Logsdon CD; Memari F; Miri SR; Rad MR; Marmari V Small Interfering RNAs (siRNAs) in Cancer Therapy: a Nano-Based Approach. Int. J. Nanomed 2019, 14, 3111–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Mainini F; Eccles MR Lipid and Polymer-Based Nanoparticle siRNA Delivery Systems for Cancer Therapy. Molecules 2020, 25, 2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Qureshi A; Tantray VG; Kirmani AR; Ahangar AG A Review on Current Status of Antiviral siRNA. Rev. Med. Virol 2018, 28, No. e1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Levanova A; Poranen MM RNA Interference as a Prospective Tool for the Control of Human Viral Infections. Front. Microbiol 2018, 9, 2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Wittrup A; Lieberman J Knocking Down Disease: a Progress Report on siRNA Therapeutics. Nat. Rev. Genet 2015, 16, 543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Mullard A FDA Approves Landmark RNAi Drug. Nat. Rev. Drug Discovery 2018, 17, 613–613. [DOI] [PubMed] [Google Scholar]
- (26).Garber K Alnylam Launches Era of RNAi Drugs. Nat. Biotechnol 2018, 36, 777–778. [DOI] [PubMed] [Google Scholar]
- (27).Akinc A; Maier MA; Manoharan M; Fitzgerald K; Jayaraman M; Barros S; Ansell S; Du X; Hope MJ; Madden TD; et al. The Onpattro Story and the Clinical Translation of Nanomedicines Containing Nucleic Acid-Based Drugs. Nat. Nanotechnol 2019, 14, 1084–1087. [DOI] [PubMed] [Google Scholar]
- (28).Adams D; Gonzalez-Duarte A; O’Riordan WD; Yang C-C; Ueda M; Kristen AV; Tournev I; Schmidt HH; Coelho T; Berk JL; et al. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med 2018, 379, 11–21. [DOI] [PubMed] [Google Scholar]
- (29).Balwani M; Sardh E; Ventura P; Peiró PA; Rees DC; Stölzel U; Bissell DM; Bonkovsky HL; Windyga J; Anderson KE; et al. Phase 3 Trial of RNAi Therapeutic Givosiran for Acute Intermittent Porphyria. N. Engl. J. Med 2020, 382, 2289–2301. [DOI] [PubMed] [Google Scholar]
- (30).Scott LJ Givosiran: First Approval. Drugs 2020, 80, 335–339. [DOI] [PubMed] [Google Scholar]
- (31).Ray KK; Wright RS; Kallend D; Koenig W; Leiter LA; Raal FJ; Bisch JA; Richardson T; Jaros M; Wijngaard PLJ; et al. Two Phase 3 Trials of Inclisiran in Patients With Elevated LDL Cholesterol. N. Engl. J. Med 2020, 382, 1507–1519. [DOI] [PubMed] [Google Scholar]
- (32).Raal FJ; Kallend D; Ray KK; Turner T; Koenig W; Wright RS; Wijngaard PLJ; Curcio D; Jaros MJ; Leiter LA; et al. Inclisiran for the Treatment of Heterozygous Familial Hypercholesterolemia. N. Engl. J. Med 2020, 382, 1520–1530. [DOI] [PubMed] [Google Scholar]
- (33).Scott LJ; Keam SJ Lumasiran: First Approval. Drugs 2021, 81, 277–282. [DOI] [PubMed] [Google Scholar]
- (34).Jaklevic MC First drug approved for rare genetic disorder affecting kidneys. JAMA 2021, 325, 214–214. [DOI] [PubMed] [Google Scholar]
- (35).Broderick JA; Zamore PD microRNA Therapeutics. Gene Ther 2011, 18, 1104–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Christopher A; Kaur R; Kaur G; Kaur A; Gupta V; Bansal P microRNA Therapeutics: Discovering Novel Targets and Developing Specific Therapy. Perspect. Clin. Res 2016, 7, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Forterre A; Komuro H; Aminova S; Harada M A Comprehensive Review of Cancer microRNA Therapeutic Delivery Strategies. Cancers 2020, 12, 1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Chen Y; Gao D-Y; Huang L In Vivo Delivery of miRNAs for Cancer Therapy: Challenges and Strategies. Adv. Drug Delivery Rev 2015, 81, 128–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Li R-Q; Wu Y; Zhi Y; Yang X; Li Y; Xua F-J; Du J PGMA-Based Star-Like Polycations with Plentiful Hydroxyl Groups Act as Highly Efficient miRNA Delivery Nanovectors for Effective Applications in Heart Diseases. Adv. Mater 2016, 28, 7204–7212. [DOI] [PubMed] [Google Scholar]
- (40).Wiggins JF; Ruffino L; Kelnar K; Omotola M; Patrawala L; Brown D; Bader AG Development of a Lung Cancer Therapeutic Based on the Tumor Suppressor microRNA-34. Cancer Res 2010, 70, 5923–5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Gros F; Hiatt H; Gilbert W; Kurland CG; Risebrough RW; Watson JD Unstable Ribonucleic Acid Revealed by Pulse Labelling of Escherichia Coli. Nature 1961, 190, 581–585. [DOI] [PubMed] [Google Scholar]
- (42).Brenner S; Jacob F; Meselson M An Unstable Intermediate Carrying Information From Genes to Ribosomes for Protein Synthesis. Nature 1961, 190, 576–581. [DOI] [PubMed] [Google Scholar]
- (43).Sahin U; Karikó K; Türeci Ö mRNA-Based Therapeutics-Developing a New Class of Drugs. Nat. Rev. Drug Discovery 2014, 13, 759–780. [DOI] [PubMed] [Google Scholar]
- (44).Vallazza B; Petri S; Poleganov MA; Eberle F; Kuhn AN; Sahin U Recombinant Messenger RNA Technology and Its Application in Cancer Immunotherapy, Transcript Replacement Therapies, Pluripotent Stem Cell Induction, and Beyond. Wiley Interdiscip. Rev. RNA 2015, 6, 471–499. [DOI] [PubMed] [Google Scholar]
- (45).Xiong Q; Lee GY; Ding J; Li W; Shi J Biomedical Applications of mRNA Nanomedicine. Nano Res 2018, 11, 5281–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Pardi N; Hogan MJ; Porter FW; Weissman D mRNA Vaccines - a New Era in Vaccinology. Nat. Rev. Drug Discovery 2018, 17, 261–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Mascola JR; Fauci AS Novel Vaccine Technologies for the 21st Century. Nat. Rev. Immunol 2020, 20, 87–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Pardi N; Hogan MJ; Weissman D Recent Advances in mRNA Vaccine Technology. Curr. Opin. Immunol 2020, 65, 14–20. [DOI] [PubMed] [Google Scholar]
- (49).Batty CJ; Heise MT; Bachelder EM; Ainslie KM Vaccine Formulations in Clinical Development for the Prevention of Severe Acute Respiratory Syndrome Coronavirus 2 Infection. Adv. Drug Delivery Rev 2021, 169, 168–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Izda V; Jeffries MA; Sawalha AH COVID-19: A Review of Therapeutic Strategies and Vaccine Candidates. Clin. Immunol 2021, 222, 108634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Tai W; Zhang X; Drelich A; Shi J; Hsu JC; Luchsinger L; Hillyer CD; Tseng C-TK; Jiang S; Du L A Novel Receptor-Binding Domain (RBD)-Based mRNA Vaccine Against SARS-CoV-2. Cell Res 2020, 30, 932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Zeng C; Hou X; Yan J; Zhang C; Li W; Zhao W; Du S; Dong Y Leveraging mRNA Sequences and Nanoparticles to Deliver SARS-CoV-2 Antigens In Vivo. Adv. Mater 2020, 32, 2004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Zhang N-N; Li X-F; Deng Y-Q; Zhao H; Huang Y-J; Yang G; Huang W-J; Gao P; Zhou C; Zhang R-R; et al. A Thermostable mRNA Vaccine Against COVID-19. Cell 2020, 182, 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Laczkó D; Hogan MJ; Toulmin SA; Hicks P; Lederer K; Gaudette BT; Castaño D; Amanat F; Muramatsu H; Oguin TH; et al. A Single Immunization with Nucleoside-Modified mRNA Vaccines Elicits Strong Cellular and Humoral Immune Responses Against SARS-CoV-2 in Mice. Immunity 2020, 53, 724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Lu J; Lu G; Tan S; Xia J; Xiong H; Yu X; Qi Q; Yu X; Li L; Yu H; et al. A COVID-19 mRNA Vaccine Encoding SARS-CoV-2 Virus-Like Particles Induces a Strong Antiviral-Like Immune Response in Mice. Cell Res 2020, 30, 936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).McKay PF; Hu K; Blakney AK; Samnuan K; Brown JC; Penn R; Zhou J; Bouton CR; Rogers P; Polra K; et al. Self-Amplifying RNA SARS-CoV-2 Lipid Nanoparticle Vaccine Candidate Induces High Neutralizing Antibody Titers in Mice. Nat. Commun 2020, 11, 3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Elia U; Ramishetti S; Rosenfeld R; Dammes N; Bar-Haim E; Naidu GS; Makdasi E; Yahalom-Ronen Y; Tamir H; Paran N; et al. Design of SARS-CoV-2 hFc-Conjugated Receptor-Binding Domain mRNA Vaccine Delivered via Lipid Nanoparticles. ACS Nano 2021, DOI: 10.1021/acsnano.0c10180. [DOI] [PubMed] [Google Scholar]
- (58).Leader B; Baca QJ; Golan DE Protein Therapeutics: a Summary and Pharmacological Classification. Nat. Rev. Drug Discovery 2008, 7, 21–39. [DOI] [PubMed] [Google Scholar]
- (59).Hou X; Zhang X; Zhao W; Zeng C; Deng B; McComb DW; Du S; Zhang C; Li W; Dong Y Vitamin Lipid Nanoparticles Enable Adoptive Macrophage Transfer for the Treatment of Multidrug-Resistant Bacterial Sepsis. Nat. Nanotechnol 2020, 15, 41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).DeRosa F; Guild B; Karve S; Smith L; Love K; Dorkin JR; Kauffman KJ; Zhang J; Yahalom B; Anderson DG; et al. Therapeutic Efficacy in a Hemophilia B Model Using a Biosynthetic mRNA Liver Depot System. Gene Ther 2016, 23, 699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Ramaswamy S; Tonnu N; Tachikawa K; Limphong P; Vega JB; Karmali PP; Chivukula P; Verma IM Systemic Delivery of Factor IX Messenger RNA for Protein Replacement Therapy. Proc. Natl. Acad. Sci. U. S. A 2017, 114, E1941–E1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Pardi N; Secreto AJ; Shan X; Debonera F; Glover J; Yi Y; Muramatsu H; Ni H; Mui BL; Tam YK; et al. Administration of Nucleoside-Modified mRNA Encoding Broadly Neutralizing Antibody Protects Humanized Mice From HIV-1 Challenge. Nat. Commun 2017, 8, 14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Zangi L; Lui KO; von Gise A; Ma Q; Ebina W; Ptaszek LM; Später D; Xu H; Tabebordbar M; Gorbatov R; et al. Modified mRNA Directs the Fate of Heart Progenitor Cells and Induces Vascular Regeneration After Myocardial Infarction. Nat. Biotechnol 2013, 31, 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Tang X; Zhang S; Fu R; Zhang L; Huang K; Peng H; Dai L; Chen Q Therapeutic Prospects of mRNA-Based Gene Therapy for Glioblastoma. Front. Oncol 2019, 9, 1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Liang X; Li D; Leng S; Zhu X RNA-Based Pharmacotherapy for Tumors: From Bench to Clinic and Back. Biomed. Pharmacother 2020, 125, 109997. [DOI] [PubMed] [Google Scholar]
- (66).Wang H; Yang H; Shivalila CS; Dawlaty MM; Cheng AW; Zhang F; Jaenisch R One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell 2013, 153, 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Bernal JA RNA-Based Tools for Nuclear Reprogramming and Lineage-Conversion: Towards Clinical Applications. J. Cardiovasc. Transl. Res 2013, 6, 956–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Jinek M; Chylinski K; Fonfara I; Hauer M; Doudna JA; Charpentier E A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Cox DBT; Platt RJ; Zhang F Therapeutic Genome Editing: Prospects and Challenges. Nat. Med 2015, 21, 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Long C; Amoasii L; Mireault AA; McAnally JR; Li H; Sanchez-Ortiz E; Bhattacharyya S; Shelton JM; Bassel-Duby R; Olson EN Postnatal Genome Editing Partially Restores Dystrophin Expression in a Mouse Model of Muscular Dystrophy. Science 2016, 351, 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Ding Q; Strong A; Patel Kevin M; Ng S-L; Gosis Bridget S; Regan Stephanie N; Cowan Chad A; Rader Daniel J; Musunuru K Permanent Alteration of PCSK9 With In Vivo CRISPR-Cas9 Genome Editing. Circ. Res 2014, 115, 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Yang Y; Wang L; Bell P; McMenamin D; He Z; White J; Yu H; Xu C; Morizono H; Musunuru K; et al. A Dual AAV System Enables the Cas9-Mediated Correction of a Metabolic Liver Disease in Newborn Mice. Nat. Biotechnol 2016, 34, 334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Yin H; Xue W; Chen S; Bogorad RL; Benedetti E; Grompe M; Koteliansky V; Sharp PA; Jacks T; Anderson DG Genome Editing with Cas9 in Adult Mice Corrects a Disease Mutation and Phenotype. Nat. Biotechnol 2014, 32, 551–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Li B; Niu Y; Ji W; Dong Y Strategies for the CRISPR-Based Therapeutics. Trends Pharmacol. Sci 2020, 41, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Adachi T; Nakamura Y Aptamers: A Review of Their Chemical Properties and Modifications for Therapeutic Application. Molecules 2019, 24, 4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Nuzzo S; Brancato V; Affinito A; Salvatore M; Cavaliere C; Condorelli G The Role of RNA and DNA Aptamers in Glioblastoma Diagnosis and Therapy: A Systematic Review of the Literature. Cancers 2020, 12, 2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Keefe AD; Pai S; Ellington A Aptamers as Therapeutics. Nat. Rev. Drug Discovery 2010, 9, 537–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Kang K-N; Lee Y-S In Future Trends in Biotechnology; Zhong J-J, Ed.; Springer Berlin Heidelberg: Berlin, Heidelberg, 2013; pp 153–169; DOI: 10.1007/10_2012_136. [DOI] [Google Scholar]
- (79).Göpel Y; Görke B Lies and Deception in Bacterial Gene Regulation: the Roles of Nucleic Acid Decoys. Mol. Microbiol 2014, 92, 641–647. [DOI] [PubMed] [Google Scholar]
- (80).Balke D; Müller S Advances in Nucleic Acid Therapeutics; The Royal Society of Chemistry, 2019; pp 434–452; DOI: 10.1039/9781788015714-00434. [DOI] [Google Scholar]
- (81).Lewin AS; Hauswirth WW Ribozyme Gene Therapy: Applications for Molecular Medicine. Trends Mol. Med 2001, 7, 221–228. [DOI] [PubMed] [Google Scholar]
- (82).Lei B; Tian Z; Fan W; Ni B Circular RNA: a Novel Biomarker and Therapeutic Target for Human Cancers. Int. J. Med. Sci 2019, 16, 292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Li H-M; Ma X-L; Li H-G Intriguing Circles: Conflicts and Controversies in Circular RNA Research. Wiley Interdiscip. Rev.: RNA 2019, 10, No. e1538. [DOI] [PubMed] [Google Scholar]
- (84).Geng X; Jia Y; Zhang Y; Shi L; Li Q; Zang A; Wang H Circular RNA: Biogenesis, Degradation, Functions and Potential Roles in Mediating Resistance to Anticarcinogens. Epigenomics 2020, 12, 267–283. [DOI] [PubMed] [Google Scholar]
- (85).Li J; Yang J; Zhou P; Le Y; Zhou C; Wang S; Xu D; Lin H-K; Gong Z Circular RNAs in Cancer: Novel Insights Into Origins, Properties, Functions and Implications. Am. J. Cancer Res 2015, 5, 472–480. [PMC free article] [PubMed] [Google Scholar]
- (86).Sahay G; Querbes W; Alabi C; Eltoukhy A; Sarkar S; Zurenko C; Karagiannis E; Love K; Chen D; Zoncu R Efficiency of siRNA Delivery by Lipid Nanoparticles is Limited by Endocytic Recycling. Nat. Biotechnol 2013, 31, 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Wang T; Upponi JR; Torchilin VP Design of Multifunctional Non-Viral Gene Vectors to Overcome Physiological Barriers: Dilemmas and Strategies. Int. J. Pharm 2012, 427, 3–20. [DOI] [PubMed] [Google Scholar]
- (88).Yin H; Kanasty RL; Eltoukhy AA; Vegas AJ; Dorkin JR; Anderson DG Non-Viral Vectors for Gene-Based Therapy. Nat. Rev. Genet 2014, 15, 541–555. [DOI] [PubMed] [Google Scholar]
- (89).Sorrentino S Human Extracellular Ribonucleases: Multiplicity, Molecular Diversity and Catalytic Properties of the Major RNase Types. Cell. Mol. Life Sci 1998, 54, 785–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Semple SC; Chonn A; Cullis PR Interactions of Liposomes and Lipid-Based Carrier Systems With Blood Proteins: Relation to Clearance Behaviour In Vivo. Adv. Drug Delivery Rev 1998, 32, 3–17. [DOI] [PubMed] [Google Scholar]
- (91).Whitehead KA; Langer R; Anderson DG Knocking Down Barriers: Advances in siRNA Delivery. Nat. Rev. Drug Discovery 2009, 8, 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Rappoport JZ Focusing on Clathrin-Mediated Endocytosis. Biochem. J 2008, 412, 415–423. [DOI] [PubMed] [Google Scholar]
- (93).Le Roy C; Wrana JL Clathrin- and Non-Clathrin-Mediated Endocytic Regulation of Cell Signalling. Nat. Rev. Mol. Cell Biol 2005, 6, 112–126. [DOI] [PubMed] [Google Scholar]
- (94).Marchini C; Pozzi D; Montani M; Alfonsi C; Amici A; Amenitsch H; Candeloro De Sanctis S; Caracciolo G Tailoring Lipoplex Composition to the Lipid Composition of Plasma Membrane: A Trojan Horse for Cell Entry? Langmuir 2010, 26, 13867–13873. [DOI] [PubMed] [Google Scholar]
- (95).Zhao F; Zhao Y; Liu Y; Chang X; Chen C; Zhao Y Cellular Uptake, Intracellular Trafficking, and Cytotoxicity of Nanomaterials. Small 2011, 7, 1322–1337. [DOI] [PubMed] [Google Scholar]
- (96).Martens TF; Remaut K; Demeester J; De Smedt SC; Braeckmans K Intracellular Delivery of Nanomaterials: How to Catch Endosomal Escape in the Act. Nano Today 2014, 9, 344–364. [Google Scholar]
- (97).Vaidyanathan S; Orr BG; Banaszak Holl MM Role of Cell Membrane-Vector Interactions in Successful Gene Delivery. Acc. Chem. Res 2016, 49, 1486–1493. [DOI] [PubMed] [Google Scholar]
- (98).Varkouhi AK; Scholte M; Storm G; Haisma HJ Endosomal Escape Pathways for Delivery of Biologicals. J. Controlled Release 2011, 151, 220–228. [DOI] [PubMed] [Google Scholar]
- (99).Gilleron J; Querbes W; Zeigerer A; Borodovsky A; Marsico G; Schubert U; Manygoats K; Seifert S; Andree C; Stöter M; et al. Image-Based Analysis of Lipid Nanoparticle-Mediated siRNA Delivery, Intracellular Trafficking and Endosomal Escape. Nat. Biotechnol 2013, 31, 638–646. [DOI] [PubMed] [Google Scholar]
- (100).Rehman ZU; Zuhorn IS; Hoekstra D How Cationic Lipids Transfer Nucleic Acids Into Cells and Across Cellular Membranes: Recent Advances. J. Controlled Release 2013, 166, 46–56. [DOI] [PubMed] [Google Scholar]
- (101).Selby LI; Cortez-Jugo CM; Such GK; Johnston APR Nanoescapology: Progress Toward Understanding the Endosomal Escape of Polymeric Nanoparticles. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol 2017, 9, No. e1452. [DOI] [PubMed] [Google Scholar]
- (102).Casey JR; Grinstein S; Orlowski J Sensors and Regulators of Intracellular pH. Nat. Rev. Mol. Cell Biol 2010, 11, 50–61. [DOI] [PubMed] [Google Scholar]
- (103).Rehman ZU; Hoekstra D; Zuhorn IS Mechanism of Polyplex- and Lipoplex-Mediated Delivery of Nucleic Acids: Real-Time Visualization of Transient Membrane Destabilization without Endosomal Lysis. ACS Nano 2013, 7, 3767–3777. [DOI] [PubMed] [Google Scholar]
- (104).Koltover I; Salditt T; Rädler JO; Safinya CR An Inverted Hexagonal Phase of Cationic Liposome-DNA Complexes Related to DNA Release and Delivery. Science 1998, 281, 78–81. [DOI] [PubMed] [Google Scholar]
- (105).Heyes J; Palmer L; Bremner K; MacLachlan I Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Controlled Release 2005, 107, 276–287. [DOI] [PubMed] [Google Scholar]
- (106).Remaut K; Sanders NN; De Geest BG; Braeckmans K; Demeester J; De Smedt SC Nucleic Acid Delivery: Where Material Sciences and Bio-Sciences Meet. Mater. Sci. Eng., R 2007, 58, 117–161. [Google Scholar]
- (107).Zuhorn IS; Bakowsky U; Polushkin E; Visser WH; Stuart MCA; Engberts JBFN; Hoekstra D Nonbilayer phase of lipoplex-membrane mixture determines endosomal escape of genetic cargo and transfection efficiency. Mol. Ther 2005, 11, 801–810. [DOI] [PubMed] [Google Scholar]
- (108).Fattal E; Couvreur P; Dubernet C Smart” Delivery of Antisense Oligonucleotides by Anionic pH-Sensitive Liposomes. Adv. Drug Delivery Rev 2004, 56, 931–946. [DOI] [PubMed] [Google Scholar]
- (109).Ramamoorth M; Narvekar A Non Viral Vectors in Gene Therapy-an Overview. J. Clin. Diagn. Res 2015, 9, GE01–GE06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Herrero MJ; Sendra L; Miguel A; Aliño SF In Safety and Efficacy of Gene-Based Therapeutics for Inherited Disorders; Brunetti-Pierri N, Ed.; Springer International Publishing: Cham, 2017; pp 113–135; DOI: 10.1007/978-3-319-53457-2_6. [DOI] [Google Scholar]
- (111).Yang CH; Shen SC; Lee JC; Wu PC; Hsueh SF; Lu CY; Meng CT; Hong HS; Yang LC Seeing the Gene Therapy: Application of Gene Gun Technique to Transfect and Decolour Pigmented Rat Skin With Human Agouti Signalling Protein cDNA. Gene Ther 2004, 11, 1033–1039. [DOI] [PubMed] [Google Scholar]
- (112).Herweijer H; Wolff JA Progress and Prospects: Naked DNA Gene Transfer and Therapy. Gene Ther 2003, 10, 453–458. [DOI] [PubMed] [Google Scholar]
- (113).Zhang Y; Yu L-C Microinjection as a Tool of Mechanical Delivery. Curr. Opin. Biotechnol 2008, 19, 506–510. [DOI] [PubMed] [Google Scholar]
- (114).Yarmush ML; Golberg A; Serša G; Kotnik T; Miklavčič D Electroporation-Based Technologies for Medicine: Principles, Applications, and Challenges. Annu. Rev. Biomed. Eng 2014, 16, 295–320. [DOI] [PubMed] [Google Scholar]
- (115).Odani N; Ito K; Nakamura H Electroporation as an Efficient Method of Gene Transfer. Dev., Growth Differ 2008, 50, 443–448. [DOI] [PubMed] [Google Scholar]
- (116).Shi J; Ma Y; Zhu J; Chen Y; Sun Y; Yao Y; Yang Z; Xie J A Review on Electroporation-Based Intracellular Delivery. Molecules 2018, 23, 3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Chen Z-Y; Lin Y; Yang F; Jiang L; Ping Ge S Gene Therapy for Cardiovascular Disease Mediated by Ultrasound and Microbubbles. Cardiovasc. Ultrasound 2013, 11, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).Tomizawa M; Shinozaki F; Motoyoshi Y; Sugiyama T; Yamamoto S; Sueishi M Sonoporation: Gene Transfer Using Ultrasound. World J. Methodol 2013, 3, 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Escoffre J-M; Zeghimi A; Novell A; Bouakaz A In-Vivo Gene Delivery by Sonoporation: Recent Progress and Prospects. Curr. Gene Ther 2013, 13, 2–14. [DOI] [PubMed] [Google Scholar]
- (120).Hosokawa Y; Ochi H; Iino T; Hiraoka A; Tanaka M Photoporation of Biomolecules into Single Cells in Living Vertebrate Embryos Induced by a Femtosecond Laser Amplifier. PLoS One 2011, 6, No. e27677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Xiong R; Samal SK; Demeester J; Skirtach AG; De Smedt SC; Braeckmans K Laser-Assisted Photoporation: Fundamentals, Technological Advances and Applications. Adv. Phys.: X 2016, 1, 596–620. [Google Scholar]
- (122).Prosen L; Prijic S; Music B; Lavrencak J; Cemazar M; Sersa G Magnetofection: a Reproducible Method for Gene Delivery to Melanoma Cells. BioMed Res. Int 2013, 2013, 209452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Bi Q; Song X; Hu A; Luo T; Jin R; Ai H; Nie Y Magnetofection: Magic Magnetic Nanoparticles for Efficient Gene Delivery. Chin. Chem. Lett 2020, 31, 3041–3046. [Google Scholar]
- (124).Wells DJ Gene Therapy Progress and Prospects: Electroporation and Other Physical Methods. Gene Ther 2004, 11, 1363–1369. [DOI] [PubMed] [Google Scholar]
- (125).Sharei A; Cho N; Mao S; Jackson E; Poceviciute R; Adamo A; Zoldan J; Langer R; Jensen KF Cell Squeezing as a Robust, Microfluidic Intracellular Delivery Platform. J. Visualized Exp 2013, 81, No. e50980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (126).Han X; Liu Z; Jo MC; Zhang K; Li Y; Zeng Z; Li N; Zu Y; Qin L CRISPR-Cas9 Delivery to Hard-to-Transfect Cells via Membrane Deformation. Sci. Adv 2015, 1, No. e1500454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Ding X; Stewart MP; Sharei A; Weaver JC; Langer RS; Jensen KF High-Throughput Nuclear Delivery and Rapid Expression of DNA via Mechanical and Electrical Cell-Membrane Disruption. Nat. Biomed. Eng 2017, 1, 0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).Li J; Wang B; Juba BM; Vazquez M; Kortum SW; Pierce BS; Pacheco M; Roberts L; Strohbach JW; Jones LH; et al. Microfluidic-Enabled Intracellular Delivery of Membrane Impermeable Inhibitors to Study Target Engagement in Human Primary Cells. ACS Chem. Biol 2017, 12, 2970–2974. [DOI] [PubMed] [Google Scholar]
- (129).DiTommaso T; Cole JM; Cassereau L; Buggé JA; Hanson JLS; Bridgen DT; Stokes BD; Loughhead SM; Beutel BA; Gilbert JB; et al. Cell Engineering With Microfluidic Squeezing Preserves Functionality of Primary Immune Cells In Vivo. Proc. Natl. Acad. Sci. U. S. A 2018, 115, E10907–E10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Sharei A; Zoldan J; Adamo A; Sim WY; Cho N; Jackson E; Mao S; Schneider S; Han M-J; Lytton-Jean A; et al. A Vector-Free Microfluidic Platform for Intracellular Delivery. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 2082–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).Sil S; Dagur RS; Liao K; Peeples ES; Hu G; Periyasamy P; Buch S Strategies for the use of Extracellular Vesicles for the Delivery of Therapeutics. J. Neuroimmune Pharmacol 2020, 15, 422–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).O’Brien K; Breyne K; Ughetto S; Laurent LC; Breakefield XO RNA Delivery by Extracellular Vesicles in Mammalian Cells and Its Applications. Nat. Rev. Mol. Cell Biol 2020, 21, 585–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (133).Tan S; Wu T; Zhang D; Zhang Z Cell or Cell Membrane-Based Drug Delivery Systems. Theranostics 2015, 5, 863–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (134).Yurkin ST; Wang Z Cell Membrane-Derived Nanoparticles: Emerging Clinical Opportunities for Targeted Drug Delivery. Nanomedicine 2017, 12, 2007–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (135).Zhuang J; Gong H; Zhou J; Zhang Q; Gao W; Fang RH; Zhang L Targeted Gene Silencing In Vivo by Platelet Membrane-Coated Metal-Organic Framework Nanoparticles. Sci. Adv 2020, 6, No. eaaz6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (136).Wang S; Duan Y; Zhang Q; Komarla A; Gong H; Gao W; Zhang L Drug Targeting via Platelet Membrane-Coated Nanoparticles. Small Struct 2020, 1, 2000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (137).Kim KM; Abdelmohsen K; Mustapic M; Kapogiannis D; Gorospe M RNA in Extracellular Vesicles. Wiley Interdiscip. Rev. RNA 2017, 8, No. e1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (138).Hu C-MJ; Fang RH; Wang K-C; Luk BT; Thamphiwatana S; Dehaini D; Nguyen P; Angsantikul P; Wen CH; Kroll AV; et al. Nanoparticle biointerfacing by platelet membrane cloaking. Nature 2015, 526, 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Veziroglu EM; Mias GI Characterizing Extracellular Vesicles and Their Diverse RNA Contents. Front. Genet 2020, 11, 700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (140).Raposo G; Stahl PD Extracellular Vesicles: a New Communication Paradigm? Nat. Rev. Mol. Cell Biol 2019, 20, 509–510. [DOI] [PubMed] [Google Scholar]
- (141).Möller A; Lobb RJ The Evolving Translational Potential of Small Extracellular Vesicles in Cancer. Nat. Rev. Cancer 2020, 20, 697–709. [DOI] [PubMed] [Google Scholar]
- (142).van Niel G; D’Angelo G; Raposo G Shedding Light on the Cell Biology of Extracellular Vesicles. Nat. Rev. Mol. Cell Biol 2018, 19, 213–228. [DOI] [PubMed] [Google Scholar]
- (143).Tkach M; Théry C Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [DOI] [PubMed] [Google Scholar]
- (144).Pitt JM; Kroemer G; Zitvogel L Extracellular Vesicles: Masters of Intercellular Communication and Potential Clinical Interventions. J. Clin. Invest 2016, 126, 1139–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (145).Zhang D; Lee H; Wang X; Rai A; Groot M; Jin Y Exosome-Mediated Small RNA Delivery: A Novel Therapeutic Approach for Inflammatory Lung Responses. Mol. Ther 2018, 26, 2119–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (146).El Andaloussi S; Mäger I; Breakefield XO; Wood MJA Extracellular Vesicles: Biology and Emerging Therapeutic Opportunities. Nat. Rev. Drug Discovery 2013, 12, 347–357. [DOI] [PubMed] [Google Scholar]
- (147).Johnsen KB; Gudbergsson JM; Skov MN; Pilgaard L; Moos T; Duroux M A Comprehensive Overview of Exosomes as Drug Delivery Vehicles — Endogenous Nanocarriers for Targeted Cancer Therapy. Biochim. Biophys. Acta, Rev. Cancer 2014, 1846, 75–87. [DOI] [PubMed] [Google Scholar]
- (148).Wiklander OPB; Brennan MÁ; Lötvall J; Breakefield XO; El Andaloussi S Advances in Therapeutic Applications of Extracellular Vesicles. Sci. Transl. Med 2019, 11, No. eaav8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (149).Ha D; Yang N; Nadithe V Exosomes as Therapeutic Drug Carriers and Delivery Vehicles Across Biological Membranes: Current Perspectives and Future Challenges. Acta Pharm. Sin. B 2016, 6, 287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (150).Hessvik NP; Llorente A Current Knowledge on Exosome Biogenesis and Release. Cell. Mol. Life Sci 2018, 75, 193–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (151).Zhou Y; Zhou G; Tian C; Jiang W; Jin L; Zhang C; Chen X Exosome-Mediated Small RNA Delivery for Gene Therapy. Wiley Interdiscip. Rev. RNA 2016, 7, 758–771. [DOI] [PubMed] [Google Scholar]
- (152).Wahlgren J; Karlson TDL; Brisslert M; Vaziri Sani F; Telemo E; Sunnerhagen P; Valadi H Plasma Exosomes Can Deliver Exogenous Short Interfering RNA to Monocytes and Lymphocytes. Nucleic Acids Res 2012, 40, No. e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (153).El Andaloussi S; Lakhal S; Mäger I; Wood MJA Exosomes for Targeted siRNA Delivery Across Biological Barriers. Adv. Drug Delivery Rev 2013, 65, 391–397. [DOI] [PubMed] [Google Scholar]
- (154).Alvarez-Erviti L; Seow Y; Yin H; Betts C; Lakhal S; Wood MJA Delivery of siRNA to the Mouse Brain by Systemic Injection of Targeted Exosomes. Nat. Biotechnol 2011, 29, 341–345. [DOI] [PubMed] [Google Scholar]
- (155).Yang Z; Xie J; Zhu J; Kang C; Chiang C; Wang X; Wang X; Kuang T; Chen F; Chen Z; et al. Functional Exosome-Mimic for Delivery of siRNA to Cancer: In Vitro and In Vivo Evaluation. J. Controlled Release 2016, 243, 160–171. [DOI] [PubMed] [Google Scholar]
- (156).Pegtel DM; Cosmopoulos K; Thorley-Lawson DA; van Eijndhoven MAJ; Hopmans ES; Lindenberg JL; de Gruijl TD; Würdinger T; Middeldorp JM Functional Delivery of Viral miRNAs via Exosomes. Proc. Natl. Acad. Sci. U. S. A 2010, 107, 6328–6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (157).Yuan A; Farber EL; Rapoport AL; Tejada D; Deniskin R; Akhmedov NB; Farber DB Transfer of MicroRNAs by Embryonic Stem Cell Microvesicles. PLoS One 2009, 4, No. e4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (158).Mittelbrunn M; Gutiérrez-Vázquez C; Villarroya-Beltri C; González S; Sánchez-Cabo F; González MÁ; Bernad A; Sánchez-Madrid F Unidirectional Transfer of microRNA-Loaded Exosomes From T Cells to Antigen-Presenting Cells. Nat. Commun 2011, 2, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (159).Valadi H; Ekström K; Bossios A; Sjöstrand M; Lee JJ; Lötvall JO Exosome-Mediated Transfer of mRNAs and microRNAs is a Novel Mechanism of Genetic Exchange Between Cells. Nat. Cell Biol 2007, 9, 654–659. [DOI] [PubMed] [Google Scholar]
- (160).Lu M; Xing H; Xun Z; Yang T; Ding P; Cai C; Wang D; Zhao X Exosome-Based small RNA Delivery: Progress and Prospects. Asian J. Pharm. Sci 2018, 13, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (161).Shahabipour F; Barati N; Johnston TP; Derosa G; Maffioli P; Sahebkar A Exosomes: Nanoparticulate Tools for RNA Interference and Drug Delivery. J. Cell. Physiol 2017, 232, 1660–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (162).Darband SG; Mirza-Aghazadeh-Attari M; Kaviani M; Mihanfar A; Sadighparvar S; Yousefi B; Majidinia M Exosomes: Natural Nanoparticles as Bio Shuttles for RNAi Delivery. J. Controlled Release 2018, 289, 158–170. [DOI] [PubMed] [Google Scholar]
- (163).Yang Z; Shi J; Xie J; Wang Y; Sun J; Liu T; Zhao Y; Zhao X; Wang X; Ma Y; et al. Large-Scale Generation of Functional mRNA-Encapsulating Exosomes via Cellular Nanoporation. Nat. Biomed. Eng 2020, 4, 69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (164).Usman WM; Pham TC; Kwok YY; Vu LT; Ma V; Peng B; Chan YS; Wei L; Chin SM; Azad A; et al. Efficient RNA Drug Delivery Using Red Blood Cell Extracellular Vesicles. Nat. Commun 2018, 9, 2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (165).Shi J; Kundrat L; Pishesha N; Bilate A; Theile C; Maruyama T; Dougan SK; Ploegh HL; Lodish HF Engineered Red Blood Cells as Carriers for Systemic Delivery of a Wide Array of Functional Probes. Proc. Natl. Acad. Sci. U. S. A 2014, 111, 10131–10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (166).Siljander PRM Platelet-Derived Microparticles - an Updated Perspective. Thromb. Res 2011, 127, S30–S33. [DOI] [PubMed] [Google Scholar]
- (167).Michael JV; Wurtzel JGT; Mao GF; Rao AK; Kolpakov MA; Sabri A; Hoffman NE; Rajan S; Tomar D; Madesh M; et al. Platelet Microparticles Infiltrating Solid Tumors Transfer miRNAs That Suppress Tumor Growth. Blood 2017, 130, 567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (168).Li S; Feng S; Ding L; Liu Y; Zhu Q; Qian Z; Gu Y Nanomedicine Engulfed by Macrophages for Targeted Tumor Therapy. Int. J. Nanomed 2016, 11, 4107–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (169).Yi L; Xiao H; Xu M; Ye X; Hu J; Li F; Li M; Luo C; Yu R; Bian X; et al. Glioma-Initiating Cells: A Predominant Role in Microglia/Macrophages Tropism to Glioma. J. Neuroimmunol 2011, 232, 75–82. [DOI] [PubMed] [Google Scholar]
- (170).Wayne EC; Long C; Haney MJ; Batrakova EV; Leisner RM; Parise LV; Kabanov AV Targeted Delivery of siRNA Lipoplexes to Cancer Cells Using Macrophage Transient Horizontal Gene Transfer. Adv. Sci 2019, 6, 1900582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (171).Zhang Y; Qin Y; Li H; Peng Q; Wang P; Yang L; Chen S; Li M; Fu J; Yu X; et al. Artificial Platelets for Efficient siRNA Delivery to Clear “Bad Cholesterol. ACS Appl. Mater. Interfaces 2020, 12, 28034–28046. [DOI] [PubMed] [Google Scholar]
- (172).Jiang Z; Thayumanavan S Noncationic Material Design for Nucleic Acid Delivery. Adv. Ther 2020, 3, 1900206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (173).Ni R; Feng R; Chau Y Synthetic Approaches for Nucleic Acid Delivery: Choosing the Right Carriers. Life 2019, 9, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (174).Lostalé-Seijo I; Montenegro J Synthetic Materials at the Forefront of Gene Delivery. Nat. Rev. Chem 2018, 2, 258–277. [Google Scholar]
- (175).Ozpolat B; Sood AK; Lopez-Berestein G Liposomal siRNA Nanocarriers for Cancer Therapy. Adv. Drug Delivery Rev 2014, 66, 110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (176).Allen TM; Cullis PR Liposomal Drug Delivery Systems: From Concept to Clinical Applications. Adv. Drug Delivery Rev 2013, 65, 36–48. [DOI] [PubMed] [Google Scholar]
- (177).Wan C; Allen TM; Cullis PR Lipid Nanoparticle Delivery Systems for siRNA-Based Therapeutics. Drug Delivery Transl. Res 2014, 4, 74–83. [DOI] [PubMed] [Google Scholar]
- (178).Mitchell MJ; Billingsley MM; Haley RM; Wechsler ME; Peppas NA; Langer R Engineering Precision Nanoparticles for Drug Delivery. Nat. Rev. Drug Discovery 2021, 20, 101–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (179).Khalil IA; Yamada Y; Harashima H Optimization of siRNA Delivery to Target Sites: Issues and Future Directions. Expert Opin. Drug Delivery 2018, 15, 1053–1065. [DOI] [PubMed] [Google Scholar]
- (180).Khalil IA; Younis MA; Kimura S; Harashima H Lipid Nanoparticles for Cell-SpecificI In Vivo Targeted Delivery of Nucleic Acids. Biol. Pharm. Bull 2020, 43, 584–595. [DOI] [PubMed] [Google Scholar]
- (181).Midoux P; Pichon C Lipid-Based mRNA Vaccine Delivery Systems. Expert Rev. Vaccines 2015, 14, 221–234. [DOI] [PubMed] [Google Scholar]
- (182).Grijalvo S; Puras G; Zárate J; Sainz-Ramos M; Qtaish NAL; López T; Mashal M; Attia N; Díaz Díaz D; Pons R; et al. Cationic Niosomes as Non-Viral Vehicles for Nucleic Acids: Challenges and Opportunities in Gene Delivery. Pharmaceutics 2019, 11, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (183).Xue HY; Guo P; Wen W-C; Lun Wong H Lipid-Based Nanocarriers for RNA Delivery. Curr. Pharm. Des 2015, 21, 3140–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (184).Bangham AD; Horne RW Negative Staining of Phospholipids and Their Structural Modification by Surface-Active Agents As Observed in the Electron Microscope. J. Mol. Biol 1964, 8, 660–668. [DOI] [PubMed] [Google Scholar]
- (185).Bangham AD; Standish MM; Watkins JC Diffusion of Univalent Ions Across the Lamellae of Swollen Phospholipids. J. Mol. Biol 1965, 13, 238–252. [DOI] [PubMed] [Google Scholar]
- (186).Barenholz Y Doxil® — The first FDA-approved nano-drug: Lessons learned. J. Controlled Release 2012, 160, 117–134. [DOI] [PubMed] [Google Scholar]
- (187).Dimitriadis GJ Entrapment of Plasmid DNA in Liposomes. Nucleic Acids Res 1979, 6, 2697–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (188).Fraley RT; Fornari CS; Kaplan S Entrapment of a Bacterial Plasmid in Phospholipid Vesicles: Potential for Gene Transfer. Proc. Natl. Acad. Sci. U. S. A 1979, 76, 3348–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (189).Dimitriadis GJ Translation of Rabbit Globin mRNA Introduced by Liposomes Into Mouse Lymphocytes. Nature 1978, 274, 923–924. [DOI] [PubMed] [Google Scholar]
- (190).Wilson T; Papahadjopoulos D; Taber R The Introduction of Poliovirus RNA Into Cells via Lipid Vesicles (Liposomes). Cell 1979, 17, 77–84. [DOI] [PubMed] [Google Scholar]
- (191).Malone RW; Felgner PL; Verma IM Cationic Liposome-Mediated RNA Transfection. Proc. Natl. Acad. Sci. U. S. A 1989, 86, 6077–6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (192).Hatakeyama H; Akita H; Harashima H The Polyethyleneglycol Dilemma: Advantage and Disadvantage of PEGylation of Liposomes for Systemic Genes and Nucleic Acids Delivery to Tumors. Biol. Pharm. Bull 2013, 36, 892–899. [DOI] [PubMed] [Google Scholar]
- (193).Fang Y; Xue J; Gao S; Lu A; Yang D; Jiang H; He Y; Shi K Cleavable PEGylation: a Strategy for Overcoming the “PEG Dilemma” in Efficient Drug Delivery. Drug Delivery 2017, 24, 22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (194).Cheng X; Lee RJ The Role of Helper Lipids in Lipid Nanoparticles (LNPs) Designed for Oligonucleotide Delivery. Adv. Drug Delivery Rev 2016, 99, 129–137. [DOI] [PubMed] [Google Scholar]
- (195).Kulkarni JA; Witzigmann D; Leung J; Tam YYC; Cullis PR On the Role of Helper Lipids in Lipid Nanoparticle Formulations of siRNA. Nanoscale 2019, 11, 21733–21739. [DOI] [PubMed] [Google Scholar]
- (196).Karnik R; Gu F; Basto P; Cannizzaro C; Dean L; Kyei-Manu W; Langer R; Farokhzad OC Microfluidic Platform for Controlled Synthesis of Polymeric Nanoparticles. Nano Lett 2008, 8, 2906–2912. [DOI] [PubMed] [Google Scholar]
- (197).Maeki M; Fujishima Y; Sato Y; Yasui T; Kaji N; Ishida A; Tani H; Baba Y; Harashima H; Tokeshi M Understanding the Formation Mechanism of Lipid Nanoparticles in Microfluidic Devices With Chaotic Micromixers. PLoS One 2017, 12, No. e0187962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (198).Maeki M; Kimura N; Sato Y; Harashima H; Tokeshi M Advances in Microfluidics for Lipid Nanoparticles and Extracellular Vesicles and Applications in Drug Delivery Systems. Adv. Drug Delivery Rev 2018, 128, 84–100. [DOI] [PubMed] [Google Scholar]
- (199).Cullis PR; Hope MJ Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther 2017, 25, 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (200).Guevara ML; Persano S; Persano F Lipid-Based Vectors for Therapeutic mRNA-Based Anti-Cancer Vaccines. Curr. Pharm. Des 2019, 25, 1443–1454. [DOI] [PubMed] [Google Scholar]
- (201).Barros SA; Gollob JA Safety Profile of RNAi Nanomedicines. Adv. Drug Delivery Rev 2012, 64, 1730–1737. [DOI] [PubMed] [Google Scholar]
- (202).Sato Y; Hatakeyama H; Sakurai Y; Hyodo M; Akita H; Harashima H A pH-sensitive cationic lipid facilitates the delivery of liposomal siRNA and gene silencing activity in vitro and in vivo. J. Controlled Release 2012, 163, 267–276. [DOI] [PubMed] [Google Scholar]
- (203).Rossi JJ RNAi Therapeutics: SNALPing siRNAs In Vivo. Gene Ther 2006, 13, 583–584. [DOI] [PubMed] [Google Scholar]
- (204).Tam YYC; Chen S; Cullis PR Advances in Lipid Nanoparticles for siRNA Delivery. Pharmaceutics 2013, 5, 498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (205).Rizk M; Tüzmen Ş Update on the Clinical Utility of an RNA Interference-Based Treatment: Focus on Patisiran. Pharmgenomics Pers. Med 2017, 10, 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (206).Urban-Klein B; Werth S; Abuharbeid S; Czubayko F; Aigner A RNAi-Mediated Gene-Targeting Through Systemic Application of Polyethylenimine (PEI)-Complexed siRNA In Vivo. Gene Ther 2005, 12, 461–466. [DOI] [PubMed] [Google Scholar]
- (207).Démoulins T; Milona P; Englezou PC; Ebensen T; Schulze K; Suter R; Pichon C; Midoux P; Guzmán CA; Ruggli N; et al. Polyethylenimine-Based Polyplex Delivery of Self-Replicating RNA Vaccines. Nanomedicine 2016, 12, 711–722. [DOI] [PubMed] [Google Scholar]
- (208).Bettinger T; Carlisle RC; Read ML; Ogris M; Seymour LW Peptide-Mediated RNA Delivery: a Novel Approach for Enhanced Transfection of Primary and Post-Mitotic Cells. Nucleic Acids Res 2001, 29, 3882–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (209).Miyazaki T; Uchida S; Nagatoishi S; Koji K; Hong T; Fukushima S; Tsumoto K; Ishihara K; Kataoka K; Cabral H Polymeric Nanocarriers With Controlled Chain Flexibility Boost mRNA Delivery In Vivo Through Enhanced Structural Fastening. Adv. Healthcare Mater 2020, 9, 2000538. [DOI] [PubMed] [Google Scholar]
- (210).Koji K; Yoshinaga N; Mochida Y; Hong T; Miyazaki T; Kataoka K; Osada K; Cabral H; Uchida S Bundling of mRNA Strands Inside Polyion Complexes Improves mRNA Delivery Efficiency In Vitro and In Vivo. Biomaterials 2020, 261, 120332. [DOI] [PubMed] [Google Scholar]
- (211).Zhu D; Shen H; Tan S; Hu Z; Wang L; Yu L; Tian X; Ding W; Ren C; Gao C; et al. Nanoparticles Based on Poly (β-Amino Ester) and HPV16-Targeting CRISPR/shRNA as Potential Drugs for HPV16-Related Cervical Malignancy. Mol. Ther 2018, 26, 2443–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (212).Su X; Fricke J; Kavanagh DG; Irvine DJ In Vitro and In Vivo mRNA Delivery Using Lipid-Enveloped pH-Responsive Polymer Nanoparticles. Mol. Pharmaceutics 2011, 8, 774–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (213).Dosta P; Ramos V; Borrós S Stable and Efficient Generation of Poly(β-amino ester)s for RNAi Delivery. Mol. Syst. Des. Eng 2018, 3, 677–689. [Google Scholar]
- (214).Siahmansouri H; Somi MH; Babaloo Z; Baradaran B; Jadidi-Niaragh F; Atyabi F; Mohammadi H; Ahmadi M; Yousefi M Effects of HMGA2 siRNA and Doxorubicin Dual Delivery by Chitosan Nanoparticles on Cytotoxicity and Gene Expression of HT-29 Colorectal Cancer Cell Line. J. Pharm. Pharmacol 2016, 68, 1119–1130. [DOI] [PubMed] [Google Scholar]
- (215).Corbet C; Ragelle H; Pourcelle V; Vanvarenberg K; Marchand-Brynaert J; Préat V; Feron O Delivery of siRNA Targeting Tumor Metabolism Using Non-Covalent PEGylated Chitosan Nanoparticles: Identification of an Optimal Combination of Ligand Structure, Linker and Grafting Method. J. Controlled Release 2016, 223, 53–63. [DOI] [PubMed] [Google Scholar]
- (216).Nascimento AV; Gattacceca F; Singh A; Bousbaa H; Ferreira D; Sarmento B; Amiji MM Biodistribution and Pharmacokinetics of Mad2 siRNA-Loaded EGFR-Targeted Chitosan Nanoparticles in Cisplatin Sensitive and Resistant Lung Cancer Models. Nanomedicine 2016, 11, 767–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (217).Son S; Nam J; Zenkov I; Ochyl LJ; Xu Y; Scheetz L; Shi J; Farokhzad OC; Moon JJ Sugar-Nanocapsules Imprinted With Microbial Molecular Patterns for mRNA Vaccination. Nano Lett 2020, 20, 1499–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (218).Kozlowska AK; Florczak A; Smialek M; Dondajewska E; Mackiewicz A; Kortylewski M; Dams-Kozlowska H Functionalized Bioengineered Spider Silk Spheres Improve Nuclease Resistance and Activity of Oligonucleotide Therapeutics Providing a Strategy for Cancer Treatment. Acta Biomater 2017, 59, 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (219).Yan Y; Zhou K; Xiong H; Miller JB; Motea EA; Boothman DA; Liu L; Siegwart DJ Aerosol Delivery of Stabilized Polyester-siRNA Nanoparticles to Silence Gene Expression in Orthotopic Lung Tumors. Biomaterials 2017, 118, 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (220).Yan Y; Liu L; Xiong H; Miller JB; Zhou K; Kos P; Huffman KE; Elkassih S; Norman JW; Carstens R; et al. Functional Polyesters Enable Selective siRNA Delivery to Lung Cancer Over Matched Normal Cells. Proc. Natl. Acad. Sci. U. S. A 2016, 113, E5702–E5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (221).Ghosh P; Han G; De M; Kim CK; Rotello VM Gold Nanoparticles in Delivery Applications. Adv. Drug Delivery Rev 2008, 60, 1307–1315. [DOI] [PubMed] [Google Scholar]
- (222).Giljohann DA; Seferos DS; Prigodich AE; Patel PC; Mirkin CA Gene Regulation with Polyvalent siRNA-Nanoparticle Conjugates. J. Am. Chem. Soc 2009, 131, 2072–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (223).Braun GB; Pallaoro A; Wu G; Missirlis D; Zasadzinski JA; Tirrell M; Reich NO Laser-Activated Gene Silencing via Gold Nanoshell-siRNA Conjugates. ACS Nano 2009, 3, 2007–2015. [DOI] [PubMed] [Google Scholar]
- (224).Trewyn BG; Giri S; Slowing II; Lin VSY Mesoporous Silica Nanoparticle Based Controlled Release, Drug Delivery, and Biosensor Systems. Chem. Commun 2007, 31, 3236–3245. [DOI] [PubMed] [Google Scholar]
- (225).Xia T; Kovochich M; Liong M; Meng H; Kabehie S; George S; Zink JI; Nel AE Polyethyleneimine Coating Enhances the Cellular Uptake of Mesoporous Silica Nanoparticles and Allows Safe Delivery of siRNA and DNA Constructs. ACS Nano 2009, 3, 3273–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (226).Tang F; Li L; Chen D Mesoporous Silica Nanoparticles: Synthesis, Biocompatibility and Drug Delivery. Adv. Mater 2012, 24, 1504–1534. [DOI] [PubMed] [Google Scholar]
- (227).Lee MS; Lee JE; Byun E; Kim NW; Lee K; Lee H; Sim SJ; Lee DS; Jeong JH Target-Specific Delivery of siRNA by Stabilized Calcium Phosphate Nanoparticles Using Dopa-Hyaluronic Acid Conjugate. J. Controlled Release 2014, 192, 122–130. [DOI] [PubMed] [Google Scholar]
- (228).Roy I; Mitra S; Maitra A; Mozumdar S Calcium Phosphate Nanoparticles as Novel Non-Viral Vectors for Targeted Gene Delivery. Int. J. Pharm 2003, 250, 25–33. [DOI] [PubMed] [Google Scholar]
- (229).Li J; Yang Y; Huang L Calcium Phosphate Nanoparticles With an Asymmetric Lipid Bilayer Coating for siRNA Delivery to the Tumor. J. Controlled Release 2012, 158, 108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (230).Mahmoudi M; Sant S; Wang B; Laurent S; Sen T Superparamagnetic Iron Oxide Nanoparticles (SPIONs): Development, Surface Modification and Applications in Chemotherapy. Adv. Drug Delivery Rev 2011, 63, 24–46. [DOI] [PubMed] [Google Scholar]
- (231).Medarova Z; Pham W; Farrar C; Petkova V; Moore A In Vivo Imaging of siRNA Delivery and Silencing in Tumors. Nat. Med 2007, 13, 372–377. [DOI] [PubMed] [Google Scholar]
- (232).Lee J-H; Lee K; Moon SH; Lee Y; Park TG; Cheon J All-in-One Target-Cell-Specific Magnetic Nanoparticles for Simultaneous Molecular Imaging and siRNA Delivery. Angew. Chem., Int. Ed 2009, 48, 4174–4179. [DOI] [PubMed] [Google Scholar]
- (233).Seeman NC Nucleic Acid Junctions and Lattices. J. Theor. Biol 1982, 99, 237–247. [DOI] [PubMed] [Google Scholar]
- (234).Storhoff JJ; Mirkin CA Programmed Materials Synthesis with DNA. Chem. Rev 1999, 99, 1849–1862. [DOI] [PubMed] [Google Scholar]
- (235).Guo P The Emerging Field of RNA Nanotechnology. Nat. Nanotechnol 2010, 5, 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (236).Nishikawa M; Tan M; Liao W; Kusamori K Nanostructured DNA for the Delivery of Therapeutic Agents. Adv. Drug Delivery Rev 2019, 147, 29–36. [DOI] [PubMed] [Google Scholar]
- (237).Jones MR; Seeman NC; Mirkin CA Programmable Materials and the Nature of the DNA Bond. Science 2015, 347, 1260901. [DOI] [PubMed] [Google Scholar]
- (238).Jasinski D; Haque F; Binzel DW; Guo P Advancement of the Emerging Field of RNA Nanotechnology. ACS Nano 2017, 11, 1142–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (239).Kim J; Yin P; Green AA Ribocomputing: Cellular Logic Computation Using RNA Devices. Biochemistry 2018, 57, 883–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (240).Krishnan Y; Seeman NC Introduction: Nucleic Acid Nanotechnology. Chem. Rev 2019, 119, 6271–6272. [DOI] [PubMed] [Google Scholar]
- (241).Wang X; Chandrasekaran AR; Shen Z; Ohayon YP; Wang T; Kizer ME; Sha R; Mao C; Yan H; Zhang X; et al. Paranemic Crossover DNA: There and Back Again. Chem. Rev 2019, 119, 6273–6289. [DOI] [PubMed] [Google Scholar]
- (242).Zuo H; Mao C A Minimalist’s Approach for DNA Nanoconstructions. Adv. Drug Delivery Rev 2019, 147, 22–28. [DOI] [PubMed] [Google Scholar]
- (243).Hu Q; Li H; Wang L; Gu H; Fan C DNA Nanotechnology-Enabled Drug Delivery Systems. Chem. Rev 2019, 119, 6459–6506. [DOI] [PubMed] [Google Scholar]
- (244).Dong Y; Yao C; Zhu Y; Yang L; Luo D; Yang D DNA Functional Materials Assembled from Branched DNA: Design, Synthesis, and Applications. Chem. Rev 2020, 120, 9420–9481. [DOI] [PubMed] [Google Scholar]
- (245).Samanta D; Ebrahimi SB; Mirkin CA Nucleic-Acid Structures as Intracellular Probes for Live Cells. Adv. Mater 2020, 32, 1901743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (246).Wu X; Wu T; Liu J; Ding B Gene Therapy Based on Nucleic Acid Nanostructure. Adv. Healthcare Mater 2020, 9, 2001046. [DOI] [PubMed] [Google Scholar]
- (247).Tan X; Jia F; Wang P; Zhang K Nucleic acid-based drug delivery strategies. J. Controlled Release 2020, 323, 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (248).Liu J; Wang Z; Zhao S; Ding B Multifunctional nucleic acid nanostructures for gene therapies. Nano Res 2018, 11, 5017–5027. [Google Scholar]
- (249).Hu Q; Wang S; Wang L; Gu H; Fan C DNA Nanostructure-Based Systems for Intelligent Delivery of Therapeutic Oligonucleotides. Adv. Healthcare Mater 2018, 7, 1701153. [DOI] [PubMed] [Google Scholar]
- (250).Huang J; Ma W; Sun H; Wang H; He X; Cheng H; Huang M; Lei Y; Wang K Self-Assembled DNA Nanostructures-Based Nanocarriers Enabled Functional Nucleic Acids Delivery. ACS Appl. Bio Mater 2020, 3, 2779–2795. [DOI] [PubMed] [Google Scholar]
- (251).Seeman NC; Sleiman HF DNA Nanotechnology. Nat. Rev. Mater 2018, 3, 17068. [Google Scholar]
- (252).Afonin KA; Viard M; Kagiampakis I; Case CL; Dobrovolskaia MA; Hofmann J; Vrzak A; Kireeva M; Kasprzak WK; KewalRamani VN; et al. Triggering of RNA Interference with RNA-RNA, RNA-DNA, and DNA-RNA Nanoparticles. ACS Nano 2015, 9, 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (253).Shu Y; Pi F; Sharma A; Rajabi M; Haque F; Shu D; Leggas M; Evers BM; Guo P Stable RNA Nanoparticles as Potential New Generation Drugs for Cancer Therapy. Adv. Drug Delivery Rev 2014, 66, 74–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (254).Roberts TC; Langer R; Wood MJA Advances in Oligonucleotide Drug Delivery. Nat. Rev. Drug Discovery 2020, 19, 673–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (255).Eckstein F Phosphorothioates, Essential Components of Therapeutic Oligonucleotides. Nucleic Acid Ther 2014, 24, 374–387. [DOI] [PubMed] [Google Scholar]
- (256).Monia BP; Johnston JF; Sasmor H; Cummins LL Nuclease Resistance and Antisense Activity of Modified Oligonucleotides Targeted to Ha-ras. J. Biol. Chem 1996, 271, 14533–14540. [DOI] [PubMed] [Google Scholar]
- (257).Iwamoto N; Butler DCD; Svrzikapa N; Mohapatra S; Zlatev I; Sah DWY; Meena Standley SM; Lu G; Apponi LH; et al. Control of Phosphorothioate Stereochemistry Substantially Increases the Efficacy of Antisense Oligonucleotides. Nat. Biotechnol 2017, 35, 845–851. [DOI] [PubMed] [Google Scholar]
- (258).Veedu RN; Wengel J Locked Nucleic Acid as a Novel Class of Therapeutic Agents. RNA Biol 2009, 6, 321–323. [DOI] [PubMed] [Google Scholar]
- (259).Nair JK; Willoughby JLS; Chan A; Charisse K; Alam MR; Wang Q; Hoekstra M; Kandasamy P; Kel’in AV; Milstein S; et al. Multivalent N-Acetylgalactosamine-Conjugated siRNA Localizes in Hepatocytes and Elicits Robust RNAi-Mediated Gene Silencing. J. Am. Chem. Soc 2014, 136, 16958–16961. [DOI] [PubMed] [Google Scholar]
- (260).Benizri S; Gissot A; Martin A; Vialet B; Grinstaff MW; Barthélémy P Bioconjugated Oligonucleotides: Recent Developments and Therapeutic Applications. Bioconjugate Chem 2019, 30, 366–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (261).Soutschek J; Akinc A; Bramlage B; Charisse K; Constien R; Donoghue M; Elbashir S; Geick A; Hadwiger P; Harborth J; et al. Therapeutic Silencing of an Endogenous Gene by Systemic Administration of Modified siRNAs. Nature 2004, 432, 173–178. [DOI] [PubMed] [Google Scholar]
- (262).Nishina T; Numata J; Nishina K; Yoshida-Tanaka K; Nitta K; Piao W; Iwata R; Ito S; Kuwahara H; Wada T; et al. Chimeric Antisense Oligonucleotide Conjugated to α-Tocopherol. Mol. Ther.– Nucleic Acids 2015, 4, No. e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (263).Nishina K; Unno T; Uno Y; Kubodera T; Kanouchi T; Mizusawa H; Yokota T Efficient In Vivo Delivery of siRNA to the Liver by Conjugation of α-Tocopherol. Mol. Ther 2008, 16, 734–740. [DOI] [PubMed] [Google Scholar]
- (264).Eguchi A; Meade BR; Chang Y-C; Fredrickson CT; Willert K; Puri N; Dowdy SF Efficient siRNA Delivery Into Primary Cells by a Peptide Transduction Domain-dsRNA Binding Fomain Fusion Protein. Nat. Biotechnol 2009, 27, 567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (265).Ämmälä C; Drury WJ; Knerr L; Ahlstedt I; Stillemark-Billton P; Wennberg-Huldt C; Andersson EM; Valeur E; Jansson-Löfmark R; Janzén D; et al. Targeted Delivery of Antisense Oligonucleotides to Pancreatic β-cells. Sci. Adv 2018, 4, No. eaat3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (266).McNamara JO; Andrechek ER; Wang Y; Viles KD; Rempel RE; Gilboa E; Sullenger BA; Giangrande PH Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol 2006, 24, 1005–1015. [DOI] [PubMed] [Google Scholar]
- (267).Kruspe S; Giangrande PH Aptamer-siRNA Chimeras: Discovery, Progress, and Future Prospects. Biomedicines 2017, 5, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (268).Ni S; Yao H; Wang L; Lu J; Jiang F; Lu A; Zhang G Chemical Modifications of Nucleic Acid Aptamers for Therapeutic Purposes. Int. J. Mol. Sci 2017, 18, 1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (269).Song E; Zhu P; Lee S-K; Chowdhury D; Kussman S; Dykxhoorn DM; Feng Y; Palliser D; Weiner DB; Shankar P; et al. Antibody Mediated In Vivo Delivery of Small Interfering RNAs via Cell-Surface Receptors. Nat. Biotechnol 2005, 23, 709–717. [DOI] [PubMed] [Google Scholar]
- (270).Satake N; Duong C; Yoshida S; Oestergaard M; Chen C; Peralta R; Guo S; Seth PP; Li Y; Beckett L; et al. Novel Targeted Therapy for Precursor B-Cell Acute Lymphoblastic Leukemia: Anti-CD22 Antibody-MXD3 Antisense Oligonucleotide Conjugate. Mol. Med 2016, 22, 632–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (271).Zhou J; Rossi J Cell-Type-Specific Aptamer and Aptamer-Small Interfering RNA Conjugates for Targeted Human Immunodeficiency Virus Type 1 Therapy. J. Invest. Med 2014, 62, 914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (272).Matsuda S; Keiser K; Nair JK; Charisse K; Manoharan RM; Kretschmer P; Peng CGV; Kel’in A; Kandasamy P; Willoughby JLS; et al. siRNA Conjugates Carrying Sequentially Assembled Trivalent N-Acetylgalactosamine Linked Through Nucleosides Elicit Robust Gene Silencing In Vivo in Hepatocytes. ACS Chem. Biol 2015, 10, 1181–1187. [DOI] [PubMed] [Google Scholar]
- (273).Thomas M; Kularatne SA; Qi L; Kleindl P; Leamon CP; Hansen MJ; Low PS Ligand-Targeted Delivery of Small Interfering RNAs to Malignant Cells and Tissues. Ann. N. Y. Acad. Sci 2009, 1175, 32–39 [DOI] [PubMed] [Google Scholar]
- (274).Ray KK; Landmesser U; Leiter LA; Kallend D; Dufour R; Karakas M; Hall T; Troquay RPT; Turner T; Visseren FLJ; et al. Inclisiran in Patients at High Cardiovascular Risk with Elevated LDL Cholesterol. N. Engl. J. Med 2017, 376, 1430–1440. [DOI] [PubMed] [Google Scholar]
- (275).Willoughby JLS; Chan A; Sehgal A; Butler JS; Nair JK; Racie T; Shulga-Morskaya S; Nguyen T; Qian K; Yucius K; et al. Evaluation of GalNAc-siRNA Conjugate Activity in Pre-clinical Animal Models with Reduced Asialoglycoprotein Receptor Expression. Mol. Ther 2018, 26, 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (276).Nikam RR; Gore KR Journey of siRNA: Clinical Developments and Targeted Delivery. Nucleic Acid Ther 2018, 28, 209–224. [DOI] [PubMed] [Google Scholar]
- (277).Shen X; Corey DR Chemistry, Mechanism and Clinical Status of Antisense Oligonucleotides and Duplex RNAs. Nucleic Acids Res 2018, 46, 1584–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (278).Stockert RJ; Morell AG; Scheinberg IH Mammalian Hepatic Lectin. Science 1974, 186, 365–366. [DOI] [PubMed] [Google Scholar]
- (279).Brown MD; Schätzlein AG; Uchegbu IF Gene Delivery With Synthetic (Non Viral) Carriers. Int. J. Pharm 2001, 229, 1–21. [DOI] [PubMed] [Google Scholar]
- (280).de Ilarduya CT; Sun Y; Düzgüneş N Gene Delivery by Lipoplexes and Polyplexes. Eur. J. Pharm. Sci 2010, 40, 159–170. [DOI] [PubMed] [Google Scholar]
- (281).Felgner PL; Gadek TR; Holm M; Roman R; Chan HW; Wenz M; Northrop JP; Ringold GM; Danielsen M Lipofection: a Highly Efficient, Lipid-Mediated DNA-Transfection Procedure. Proc. Natl. Acad. Sci. U. S. A 1987, 84, 7413–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (282).Zhi DF; Zhang SB; Cui SH; Zhao YA; Wang YH; Zhao DF The Headgroup Evolution of Cationic Lipids for Gene Delivery. Bioconjugate Chem 2013, 24, 487–519. [DOI] [PubMed] [Google Scholar]
- (283).Niculescu-Duvaz D; Heyes J; Springer CJ Structure-Activity Relationship in Cationic Lipid Mediated Gene Transfection. Curr. Med. Chem 2003, 10, 1233–1261. [DOI] [PubMed] [Google Scholar]
- (284).Zhi D; Zhang S; Wang B; Zhao Y; Yang B; Yu S Transfection Efficiency of Cationic Lipids with Different Hydrophobic Domains in Gene Delivery. Bioconjugate Chem 2010, 21, 563–577. [DOI] [PubMed] [Google Scholar]
- (285).Kohli AG; Kierstead PH; Venditto VJ; Walsh CL; Szoka FC Designer lipids for drug delivery: From heads to tails. J. Controlled Release 2014, 190, 274–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (286).Kanasty R; Dorkin JR; Vegas A; Anderson D Delivery materials for siRNA therapeutics. Nat. Mater 2013, 12, 967–977. [DOI] [PubMed] [Google Scholar]
- (287).Mahato RI Water Insoluble and Soluble Lipids for Gene Delivery. Adv. Drug Delivery Rev 2005, 57, 699–712. [DOI] [PubMed] [Google Scholar]
- (288).Wasungu L; Hoekstra D Cationic Lipids, Lipoplexes and Intracellular Delivery of Genes. J. Controlled Release 2006, 116, 255–264. [DOI] [PubMed] [Google Scholar]
- (289).Kulkarni JA; Cullis PR; van der Meel R Lipid Nanoparticles Enabling Gene Therapies: From Concepts to Clinical Utility. Nucleic Acid Ther 2018, 28, 146–157. [DOI] [PubMed] [Google Scholar]
- (290).Hsu W-L; Chen H-L; Liou W; Lin H-K; Liu W-L Mesomorphic Complexes of DNA with the Mixtures of a Cationic Surfactant and a Neutral Lipid. Langmuir 2005, 21, 9426–9431. [DOI] [PubMed] [Google Scholar]
- (291).Semple SC; Akinc A; Chen J; Sandhu AP; Mui BL; Cho CK; Sah DW; Stebbing D; Crosley EJ; Yaworski E; et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol 2010, 28, 172–176. [DOI] [PubMed] [Google Scholar]
- (292).Hafez IM; Maurer N; Cullis PR On the Mechanism Whereby Cationic Lipids Promote Intracellular Delivery of Polynucleic Acids. Gene Ther 2001, 8, 1188–1196. [DOI] [PubMed] [Google Scholar]
- (293).Hafez IM; Cullis PR Roles of Lipid Polymorphism in Intracellular Delivery. Adv. Drug Delivery Rev 2001, 47, 139–148. [DOI] [PubMed] [Google Scholar]
- (294).Filion MC; Phillips NC Toxicity and Immunomodulatory Activity of Liposomal Vectors Formulated With Cationic Lipids Toward Immune Effector Cells. Biochim. Biophys. Acta, Biomembr 1997, 1329, 345–356. [DOI] [PubMed] [Google Scholar]
- (295).Lappalainen K; Jääskeläinen I; Syrjänen K; Urtti A; Syrjänen S Comparison of Cell Proliferation and Toxicity Assays Using Two Cationic Liposomes. Pharm. Res 1994, 11, 1127–1131. [DOI] [PubMed] [Google Scholar]
- (296).Dokka S; Toledo D; Shi X; Castranova V; Rojanasakul Y Oxygen Radical-Mediated Pulmonary Toxicity Induced by Some Cationic Liposomes. Pharm. Res 2000, 17, 521–525. [DOI] [PubMed] [Google Scholar]
- (297).Soenen SJH; Brisson AR; De Cuyper M Addressing the Problem of Cationic Lipid-Mediated Toxicity: The Magnetoliposome Model. Biomaterials 2009, 30, 3691–3701. [DOI] [PubMed] [Google Scholar]
- (298).Solodin I; Brown CS; Bruno MS; Chow C-Y; Jang E-H; Debs RJ; Heath TD A Novel Series of Amphiphilic Imidazolinium Compounds for in Vitro and in Vivo Gene Delivery. Biochemistry 1995, 34, 13537–13544. [DOI] [PubMed] [Google Scholar]
- (299).van der Woude I; Wagenaar A; Meekel AAP; ter Beest MBA; Ruiters MHJ; Engberts JBFN; Hoekstra D Novel Pyridinium Surfactants for Efficient, Nontoxic In Vitro Gene Delivery. Proc. Natl. Acad. Sci. U. S. A 1997, 94, 1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (300).Ilies MA; Johnson BH; Makori F; Miller A; Seitz WA; Thompson EB; Balaban AT Pyridinium Cationic Lipids in Gene Delivery: an In Vitro and In Vivo Comparison of Transfection Efficiency Versus a Tetraalkylammonium Congener. Arch. Biochem. Biophys 2005, 435, 217–226. [DOI] [PubMed] [Google Scholar]
- (301).Yingyongnarongkul B-E; Howarth M; Elliott T; Bradley M Solid-Phase Synthesis of 89 Polyamine-Based Cationic Lipids for DNA Delivery to Mammalian Cells. Chem. - Eur. J 2004, 10, 463–473. [DOI] [PubMed] [Google Scholar]
- (302).Christensen D; Korsholm KS; Rosenkrands I; Lindenstrøm T; Andersen P; Agger EM Cationic Liposomes as Vaccine Adjuvants. Expert Rev. Vaccines 2007, 6, 785–796. [DOI] [PubMed] [Google Scholar]
- (303).Mintzer MA; Simanek EE Nonviral Vectors for Gene Delivery. Chem. Rev 2009, 109, 259–302. [DOI] [PubMed] [Google Scholar]
- (304).Stamatatos L; Leventis R; Zuckermann MJ; Silvius JR Interactions of Cationic Lipid Vesicles With Negatively Charged Phospholipid Vesicles and Biological Membranes. Biochemistry 1988, 27, 3917–3925. [DOI] [PubMed] [Google Scholar]
- (305).Floch V; Loisel S; Guenin E; Hervé AC; Clément JC; Yaouanc JJ; des Abbayes H; Férec C Cation Substitution in Cationic Phosphonolipids: A New Concept To Improve Transfection Activity and Decrease Cellular Toxicity. J. Med. Chem 2000, 43, 4617–4628. [DOI] [PubMed] [Google Scholar]
- (306).Mahidhar YV; Rajesh M; Chaudhuri A Spacer-Arm Modulated Gene Delivery Efficacy of Novel Cationic Glycolipids: Design, Synthesis, and in Vitro Transfection Biology. J. Med. Chem 2004, 47, 3938–3948. [DOI] [PubMed] [Google Scholar]
- (307).Kranz LM; Diken M; Haas H; Kreiter S; Loquai C; Reuter KC; Meng M; Fritz D; Vascotto F; Hefesha H; et al. Systemic RNA Delivery to Dendritic Cells Exploits Antiviral Defence for Cancer Immunotherapy. Nature 2016, 534, 396–401. [DOI] [PubMed] [Google Scholar]
- (308).Krienke C; Kolb L; Diken E; Streuber M; Kirchhoff S; Bukur T; Akilli-Öztürk Ö; Kranz LM; Berger H; Petschenka J; et al. A Noninflammatory mRNA Vaccine for Treatment of Experimental Autoimmune Encephalomyelitis. Science 2021, 371, 145–153. [DOI] [PubMed] [Google Scholar]
- (309).Reinhard K; Rengstl B; Oehm P; Michel K; Billmeier A; Hayduk N; Klein O; Kuna K; Ouchan Y; Wöll S; et al. An RNA Vaccine Drives Expansion and Efficacy of Claudin-CAR-T Cells Against Solid Tumors. Science 2020, 367, 446–453. [DOI] [PubMed] [Google Scholar]
- (310).Sahin U; Oehm P; Derhovanessian E; Jabulowsky RA; Vormehr M; Gold M; Maurus D; Schwarck-Kokarakis D; Kuhn AN; Omokoko T; et al. An RNA Vaccine Drives Immunity in Checkpoint-Inhibitor-Treated Melanoma. Nature 2020, 585, 107–112. [DOI] [PubMed] [Google Scholar]
- (311).Cheng X; Liu Q; Li H; Kang C; Liu Y; Guo T; Shang K; Yan C; Cheng G; Lee RJ Lipid Nanoparticles Loaded with an Antisense Oligonucleotide Gapmer Against Bcl-2 for Treatment of Lung Cancer. Pharm. Res 2017, 34, 310–320. [DOI] [PubMed] [Google Scholar]
- (312).Truong B; Allegri G; Liu X-B; Burke KE; Zhu X; Cederbaum SD; Häberle J; Martini PGV; Lipshutz GS Lipid Nanoparticle-Targeted mRNA Therapy as a Treatment for the Inherited Metabolic Liver Disorder Arginase Deficiency. Proc. Natl. Acad. Sci. U. S. A 2019, 116, 21150–21159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (313).Lonez C; Vandenbranden M; Ruysschaert J-M Cationic Lipids Activate Intracellular Signaling Pathways. Adv. Drug Delivery Rev 2012, 64, 1749–1758. [DOI] [PubMed] [Google Scholar]
- (314).Kedmi R; Ben-Arie N; Peer D The Systemic Toxicity of Positively Charged Lipid Nanoparticles and the Role of Toll-Like Receptor 4 in Immune Activation. Biomaterials 2010, 31, 6867–6875. [DOI] [PubMed] [Google Scholar]
- (315).Maruggi G; Chiarot E; Giovani C; Buccato S; Bonacci S; Frigimelica E; Margarit I; Geall A; Bensi G; Maione D Immunogenicity and Protective Efficacy Induced by Self-Amplifying mRNA Vaccines Encoding Bacterial Antigens. Vaccine 2017, 35, 361–368. [DOI] [PubMed] [Google Scholar]
- (316).Baeza Garcia A; Siu E; Sun T; Exler V; Brito L; Hekele A; Otten G; Augustijn K; Janse CJ; Ulmer JB; et al. Neutralization of the Plasmodium-Encoded MIF Ortholog Confers Protective Immunity Against Malaria Infection. Nat. Commun 2018, 9, 2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (317).Brito LA; Chan M; Shaw CA; Hekele A; Carsillo T; Schaefer M; Archer J; Seubert A; Otten GR; Beard CW A Cationic Nanoemulsion for the Delivery of Next-Generation RNA Vaccines. Mol. Ther 2014, 22, 2118–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (318).Bogers WM; Oostermeijer H; Mooij P; Koopman G; Verschoor EJ; Davis D; Ulmer JB; Brito LA; Cu Y; Banerjee K Potent Immune Responses in Rhesus Macaques Induced by Nonviral Delivery of a Self-Amplifying RNA Vaccine Expressing HIV Type 1 Envelope With a Cationic Nanoemulsion. J. Infect. Dis 2015, 211, 947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (319).Brazzoli M; Magini D; Bonci A; Buccato S; Giovani C; Kratzer R; Zurli V; Mangiavacchi S; Casini D; Brito LM; et al. Induction of Broad-Based Immunity and Protective Efficacy by Self-amplifying mRNA Vaccines Encoding Influenza Virus Hemagglutinin. J. Virol 2016, 90, 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (320).Kaneda MM; Sasaki Y; Lanza GM; Milbrandt J; Wickline SA Mechanisms of Nucleotide Trafficking During siRNA Delivery to Endothelial Cells Using Perfluorocarbon Nanoemulsions. Biomaterials 2010, 31, 3079–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (321).Dave V; Tak K; Sohgaura A; Gupta A; Sadhu V; Reddy KR Lipid-Polymer Hybrid Nanoparticles: Synthesis Strategies and Biomedical Applications. J. Microbiol. Methods 2019, 160, 130–142. [DOI] [PubMed] [Google Scholar]
- (322).Mukherjee A; Waters AK; Kalyan P; Achrol AS; Kesari S; Yenugonda VM Lipid-Polymer Hybrid Nanoparticles as a Next-Generation Drug Delivery Platform: State of the Art, Emerging Technologies, and Perspectives. Int. J. Nanomed 2019, 14, 1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (323).Zhang L; Chan JM; Gu FX; Rhee J-W; Wang AZ; Radovic-Moreno AF; Alexis F; Langer R; Farokhzad OC Self-Assembled Lipid-Polymer Hybrid Nanoparticles: A Robust Drug Delivery Platform. ACS Nano 2008, 2, 1696–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (324).Gao L-Y; Liu X-Y; Chen C-J; Wang J-C; Feng Q; Yu M-Z; Ma X-F; Pei X-W; Niu Y-J; Qiu C; et al. Core-Shell Type Lipid/rPAA-Chol Polymer Hybrid Nanoparticles for In Vivo siRNA Delivery. Biomaterials 2014, 35, 2066–2078. [DOI] [PubMed] [Google Scholar]
- (325).Felgner JH; Kumar R; Sridhar CN; Wheeler CJ; Tsai YJ; Border R; Ramsey P; Martin M; Felgner PL Enhanced Gene Delivery and Mechanism Studies With a Novel Series of Cationic Lipid Formulations. J. Biol. Chem 1994, 269, 2550–2561. [PubMed] [Google Scholar]
- (326).Bennett MJ; Aberle AM; Balasubramaniam RP; Malone JG; Malone RW; Nantz MH Cationic Lipid-Mediated Gene Delivery to Murine Lung: Correlation of Lipid Hydration With in Vivo Transfection Activity. J. Med. Chem 1997, 40, 4069–4078. [DOI] [PubMed] [Google Scholar]
- (327).Anderson DM; Hall LL; Ayyalapu AR; Irion VR; Nantz MH; Hecker JG Stability of mRNA/Cationic Lipid Lipoplexes in Human and Rat Cerebrospinal Fluid: Methods and Evidence for Nonviral mRNA Gene Delivery to the Central Nervous System. Hum. Gene Ther 2003, 14, 191–202. [DOI] [PubMed] [Google Scholar]
- (328).Bennett MJ; Aberle AM; Balasubramaniam RP; Malone JG; Nantz MH; Malone RW Considerations for the Design of Improved Cationic Amphiphile-Based Transfection Reagents. J. Liposome Res 1996, 6, 545–565. [Google Scholar]
- (329).Kikuchi A; Aoki Y; Sugaya S; Serikawa T; Takakuwa K; Tanaka K; Suzuki N; Kikuchi H Development of Novel Cationic Liposomes for Efficient Gene Transfer into Peritoneal Disseminated Tumor. Hum. Gene Ther 1999, 10, 947–955. [DOI] [PubMed] [Google Scholar]
- (330).Sato Y; Murase K; Kato J; Kobune M; Sato T; Kawano Y; Takimoto R; Takada K; Miyanishi K; Matsunaga T; et al. Resolution of Liver Cirrhosis Using Vitamin A-Coupled Liposomes to Deliver siRNA Against a Collagen-Specific Chaperone. Nat. Biotechnol 2008, 26, 431–442. [DOI] [PubMed] [Google Scholar]
- (331).Soule B; Tirucherai G; Kavita U; Kundu S; Christian R LBP-015 - Safety, Tolerability, and Pharmacokinetics of BMS-986263/ND-L02-s0201, a Novel Targeted Lipid Nanoparticle Delivering HSP47 siRNA, in Healthy Participants: A Randomised, Placebo-Controlled, Double-Blind, Phase 1 Study. J. Hepatol 2018, 68, S112. [Google Scholar]
- (332).Gorman CM; Aikawa M; Fox B; Fox E; Lapuz C; Michaud B; Nguyen H; Roche E; Sawa T; Wiener-Kronish JP Efficient in Vivo Delivery of DNA to Pulmonary Cells Using the Novel Lipid EDMPC. Gene Ther 1997, 4, 983–992. [DOI] [PubMed] [Google Scholar]
- (333).MacDonald RC; Rakhmanova VA; Choi KL; Rosenzweig HS; Lahiri MK O-ethylphosphatidylcholine: A Metabolizable Cationic Phospholipid Which is a Serum-Compatible DNA Transfection Agent. J. Pharm. Sci 1999, 88, 896–904. [DOI] [PubMed] [Google Scholar]
- (334).MacDonald RC; Ashley GW; Shida MM; Rakhmanova VA; Tarahovsky YS; Pantazatos DP; Kennedy MT; Pozharski EV; Baker KA; Jones RD; et al. Physical and Biological Properties of Cationic Triesters of Phosphatidylcholine. Biophys. J 1999, 77, 2612–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (335).Das A; Niven R Use of Perfluorocarbon (Fluorinert) to Enhance Reporter Gene Expression Following Intratracheal Instillation Into the Lungs of Balb/c Mice: Implications for Nebulized Delivery of Plasmids. J. Pharm. Sci 2001, 90, 1336–1344. [DOI] [PubMed] [Google Scholar]
- (336).McDonald RJ; Liggitt HD; Roche L; Nguyen HT; Pearlman R; Raabe OG; Bussey LB; Gorman CM Aerosol Delivery of Lipid:DNA Complexes to Lungs of Rhesus Monkeys. Pharm. Res 1998, 15, 671–679. [DOI] [PubMed] [Google Scholar]
- (337).Faneca H; Simões S; Pedroso de Lima MC Association of Albumin or Protamine to Lipoplexes: Enhancement of Transfection and Resistance to Serum. J. Gene Med 2004, 6, 681–692. [DOI] [PubMed] [Google Scholar]
- (338).Faneca H; Faustino A; Pedroso de Lima MC Synergistic Antitumoral Effect of Vinblastine and HSV-Tk/GCV Gene Therapy Mediated by Albumin-Associated Cationic Liposomes. J. Controlled Release 2008, 126, 175–184. [DOI] [PubMed] [Google Scholar]
- (339).Tenchov B; Sugimoto Y; Koynova R; Brueggemeier RW; Lee RJ Highly Efficient Cationic Ethylphosphatidylcholine siRNA Carrier for GFP Suppression in Modified Breast Cancer Cells. Anticancer Res 2012, 32, 2563–2566. [PMC free article] [PubMed] [Google Scholar]
- (340).Persano S; Guevara ML; Li Z; Mai J; Ferrari M; Pompa PP; Shen H Lipopolyplex Potentiates Anti-Tumor Immunity of mRNA-Based Vaccination. Biomaterials 2017, 125, 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (341).Zhang J; Shrivastava S; Cleveland RO; Rabbitts TH Lipid-mRNA Nanoparticle Designed to Enhance Intracellular Delivery Mediated by Shock Waves. ACS Appl. Mater. Interfaces 2019, 11, 10481–10491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (342).Pierrat P; Laverny G; Creusat G; Wehrung P; Strub JM; VanDorsselaer A; Pons F; Zuber G; Lebeau L Phospholipid-Detergent Conjugates as Novel Tools for siRNA Delivery. Chem. - Eur. J 2013, 19, 2344–2355. [DOI] [PubMed] [Google Scholar]
- (343).Pierrat P; Kereselidze D; Wehrung P; Zuber G; Pons F; Lebeau L Bioresponsive Deciduous-Charge Amphiphiles for Liposomal Delivery of DNA and siRNA. Pharm. Res 2013, 30, 1362–1379. [DOI] [PubMed] [Google Scholar]
- (344).Skjørringe T; Gjetting T; Jensen TG A modified Protocol for Efficient DNA Encapsulation Into Pegylated Immunoliposomes (PILs). J. Controlled Release 2009, 139, 140–145. [DOI] [PubMed] [Google Scholar]
- (345).Manosroi A; Thathang K; Werner RG; Schubert R; Manosroi J Stability of Luciferase Plasmid Entrapped in Cationic Bilayer Vesicles. Int. J. Pharm 2008, 356, 291–299. [DOI] [PubMed] [Google Scholar]
- (346).Li P; Zhang L; Ai K; Li D; Liu X; Wang E Coating Didodecyldimethylammonium Bromide Onto Au Nanoparticles Increases the Stability of its Complex With DNA. J. Controlled Release 2008, 129, 128–134. [DOI] [PubMed] [Google Scholar]
- (347).Zhen G; Hinton TM; Muir BW; Shi S; Tizard M; McLean KM; Hartley PG; Gunatillake P Glycerol Monooleate-Based Nanocarriers for siRNA Delivery in Vitro. Mol. Pharmaceutics 2012, 9, 2450–2457. [DOI] [PubMed] [Google Scholar]
- (348).Hattori Y; Nakamura A; Arai S; Kawano K; Maitani Y; Yonemochi E siRNA Delivery to Lung-Metastasized Tumor by Systemic Injection With Cationic Liposomes. J. Liposome Res 2015, 25, 279–286. [DOI] [PubMed] [Google Scholar]
- (349).Zheng X; Vladau C; Zhang X; Suzuki M; Ichim TE; Zhang Z-X; Li M; Carrier E; Garcia B; Jevnikar AM; et al. A Novel in Vivo siRNA Delivery System Specifically Targeting Dendritic Cells and Silencing CD40 Genes for Immunomodulation. Blood 2009, 113, 2646–2654. [DOI] [PubMed] [Google Scholar]
- (350).Mével M; Montier T; Lamarche F; Delépine P; Le Gall T; Yaouanc J-J; Jaffres̀ P-A; Cartier D; Lehn P; Clément J-C Dicationic Lipophosphoramidates as DNA Carriers. Bioconjugate Chem 2007, 18, 1604–1611. [DOI] [PubMed] [Google Scholar]
- (351).Bell PC; Bergsma M; Dolbnya IP; Bras W; Stuart MCA; Rowan AE; Feiters MC; Engberts JBFN Transfection Mediated by Gemini Surfactants: Engineered Escape from the Endosomal Compartment. J. Am. Chem. Soc 2003, 125, 1551–1558. [DOI] [PubMed] [Google Scholar]
- (352).Rosenzweig HS; Rakhmanova VA; MacDonald RC Diquaternary Ammonium Compounds as Transfection Agents. Bioconjugate Chem 2001, 12, 258–263. [DOI] [PubMed] [Google Scholar]
- (353).Gaucheron J; Wong T; Wong KF; Maurer N; Cullis PR Synthesis and Properties of Novel Tetraalkyl Cationic Lipids. Bioconjugate Chem 2002, 13, 671–675. [DOI] [PubMed] [Google Scholar]
- (354).Heden TD; Neufer PD; Funai K Looking Beyond Structure: Membrane Phospholipids of Skeletal Muscle Mitochondria. Trends Endocrinol. Metab 2016, 27, 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (355).Kasireddy K; Ali SM; Ahmad MU; Choudhury S; Chien P-Y; Sheikh S; Ahmad I Synthesis of Cationic Cardiolipin Analogues. Bioorg. Chem 2005, 33, 345–362. [DOI] [PubMed] [Google Scholar]
- (356).Chien PY; Wang JK; Carbonaro D; Lei S; Miller B; Sheikh S; Ali SM; Ahmad MU; Ahmad I Novel Cationic Cardiolipin Analogue-Based Liposome for Efficient DNA and Small Interfering RNA Delivery in Vitro and in Vivo. Cancer Gene Ther 2005, 12, 321–328. [DOI] [PubMed] [Google Scholar]
- (357).Pal A; Ahmad A; Khan S; Sakabe I; Zhang C; Kasid UN; Ahmad I Systemic Delivery of RafsiRNA Using Cationic Cardiolipin Liposomes Silences Raf-1 Expression and Inhibits Tumor Growth in Xenograft Model of Human Prostate Cancer. Int. J. Oncol 2005, 26, 1087–1091. [PubMed] [Google Scholar]
- (358).Fabio K; Gaucheron J; Di Giorgio C; Vierling P Novel Galactosylated Polyamine Bolaamphiphiles for Gene Delivery. Bioconjugate Chem 2003, 14, 358–367. [DOI] [PubMed] [Google Scholar]
- (359).Maslov MA; Morozova NG; Chizhik EI; Rapoport DA; Ryabchikova EI; Zenkova MA; Serebrennikova GA Synthesis and Delivery Activity of New Cationic Cholesteryl Glucosides. Carbohydr. Res 2010, 345, 2438–2449. [DOI] [PubMed] [Google Scholar]
- (360).Ivanova EA; Maslov MA; Kabilova TO; Puchkov PA; Alekseeva AS; Boldyrev IA; Vlassov VV; Serebrennikova GA; Morozova NG; Zenkova MA Structure-Transfection Activity Relationships in a Series of Novel Cationic Lipids With Heterocyclic Head-Groups. Org. Biomol. Chem 2013, 11, 7164–7178. [DOI] [PubMed] [Google Scholar]
- (361).Moreau L; Grinstaff MW; Barthélémy P Vesicle Formation From a Synthetic Adenosine Based Lipid. Tetrahedron Lett 2005, 46, 1593–1596. [Google Scholar]
- (362).Bestel I; Campins N; Marchenko A; Fichou D; Grinstaff MW; Barthélémy P Two-Dimensional Self-Assembly and Complementary Base-Pairing Between Amphiphile Nucleotides on Graphite. J. Colloid Interface Sci 2008, 323, 435–440. [DOI] [PubMed] [Google Scholar]
- (363).Milani S; Baldelli Bombelli F; Berti D; Baglioni P Nucleolipoplexes: A New Paradigm for Phospholipid Bilayer-Nucleic Acid Interactions. J. Am. Chem. Soc 2007, 129, 11664–11665. [DOI] [PubMed] [Google Scholar]
- (364).Moreau L; Barthélémy P; Li Y; Luo D; Prata CAH; Grinstaff MW Nucleoside Phosphocholine Amphiphile for in Vitro DNA Transfection. Mol. BioSyst 2005, 1, 260–264. [DOI] [PubMed] [Google Scholar]
- (365).Ceballos C; Prata CA; Giorgio S; Garzino F; Payet D; Barthelemy P; Grinstaff MW; Camplo M Cationic Nucleoside Lipids Based on a 3-Nitropyrrole Universal Base for siRNA Delivery. Bioconjugate Chem 2009, 20, 193–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (366).Ceballos C; Khiati S; Prata CAH; Zhang X-X; Giorgio S; Marsal P; Grinstaff MW; Barthélémy P; Camplo M Cationic Nucleoside Lipids Derived from Universal Bases: A Rational Approach for siRNA Transfection. Bioconjugate Chem 2010, 21, 1062–1069. [DOI] [PubMed] [Google Scholar]
- (367).Luvino D; Khiati S; Oumzil K; Rocchi P; Camplo M; Barthélémy P Efficient Delivery of Therapeutic Small Nucleic Acids to Prostate Cancer Cells Using Ketal Nucleoside Lipid Nanoparticles. J. Controlled Release 2013, 172, 954–961. [DOI] [PubMed] [Google Scholar]
- (368).Oumzil K; Benizri S; Tonelli G; Staedel C; Appavoo A; Chaffanet M; Navailles L; Barthelemy P pH-Cleavable Nucleoside Lipids: A New Paradigm for Controlling the Stability of Lipid-Based Delivery Systems. ChemMedChem 2015, 10, 1797–1801. [DOI] [PubMed] [Google Scholar]
- (369).Stewart AO; Williams RM C-Glycosidation of Pyridyl Thioglycosides. J. Am. Chem. Soc 1985, 107, 4289–4296. [Google Scholar]
- (370).Harusawa S; Imazu T; Takashima S; Araki L; Ohishi H; Kurihara T; Sakamoto Y; Yamamoto Y; Yamatodani A Synthesis of 4(5)-[5-(Aminomethyl)tetrahydrofuran-2-yl- or 5-(Aminomethyl)-2,5-dihydrofuran-2-yl]imidazoles by Efficient Use of a PhSe Group: Application to Novel Histamine H3-Ligands1. J. Org. Chem 1999, 64, 8608–8615. [Google Scholar]
- (371).Bray BL; Mathies PH; Naef R; Solas DR; Tidwell TT; Artis DR; Muchowski JM N-(Triisopropylsilyl)pyrrole. A Progenitor “Par Excellence” of 3-Substituted Pyrroles. J. Org. Chem 1990, 55, 6317–6328. [Google Scholar]
- (372).Scaringe SA RNA Oligonucleotide Synthesis via 5′-Silyl-2′-Orthoester Chemistry. Methods 2001, 23, 206–217. [DOI] [PubMed] [Google Scholar]
- (373).Li S; Gao X; Son K; Sorgi F; Hofland H; Huang L DC-Chol Lipid System in Gene Transfer. J. Controlled Release 1996, 39, 373–381. [Google Scholar]
- (374).Tang F; Hughes JA Use of Dithiodiglycolic Acid as a Tether for Cationic Lipids Decreases the Cytotoxicity and Increases Transgene Expression of Plasmid DNA in Vitro. Bioconjugate Chem 1999, 10, 791–796. [DOI] [PubMed] [Google Scholar]
- (375).Arakawa K; Eguchi T; Kakinuma K Tightly Packed Membranes Composed of 36-Membered Macrocyclic Diether Phospholipid Found in Archaea Growing under Deep-sea Hydrothermal Vents. Chem. Lett 1998, 27, 901–902. [Google Scholar]
- (376).Pungente MD; Jubeli E; Opstad CL; Al-Kawaz M; Barakat N; Ibrahim T; Abdul Khalique N; Raju L; Jones R; Leopold PL; et al. Synthesis and Preliminary Investigations of the siRNA Delivery Potential of Novel, Single-Chain Rigid Cationic Carotenoid Lipids. Molecules 2012, 17, 3484–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (377).Matsumoto A; Cabral H; Sato N; Kataoka K; Miyahara Y Assessment of Tumor Metastasis by the Direct Determination of Cell-Membrane Sialic Acid Expression. Angew. Chem., Int. Ed 2010, 49, 5494–5497. [DOI] [PubMed] [Google Scholar]
- (378).Büll C; Stoel MA; den Brok MH; Adema GJ Sialic Acids Sweeten a Tumor’s Life. Cancer Res 2014, 74, 3199–3204. [DOI] [PubMed] [Google Scholar]
- (379).Liu H; Li Y; Sun K; Fan J; Zhang P; Meng J; Wang S; Jiang L Dual-Responsive Surfaces Modified with Phenylboronic Acid-Containing Polymer Brush To Reversibly Capture and Release Cancer Cells. J. Am. Chem. Soc 2013, 135, 7603–7609. [DOI] [PubMed] [Google Scholar]
- (380).Matsumoto A; Sato N; Kataoka K; Miyahara Y Noninvasive Sialic Acid Detection at Cell Membrane by Using Phenylboronic Acid Modified Self-Assembled Monolayer Gold Electrode. J. Am. Chem. Soc 2009, 131, 12022–12023. [DOI] [PubMed] [Google Scholar]
- (381).Otsuka H; Uchimura E; Koshino H; Okano T; Kataoka K Anomalous Binding Profile of Phenylboronic Acid with N-Acetylneuraminic Acid (Neu5Ac) in Aqueous Solution with Varying pH. J. Am. Chem. Soc 2003, 125, 3493–3502. [DOI] [PubMed] [Google Scholar]
- (382).Tang Q; Liu J; Jiang Y; Zhang M; Mao L; Wang M Cell-Selective Messenger RNA Delivery and CRISPR/Cas9 Genome Editing by Modulating the Interface of Phenylboronic Acid-Derived Lipid Nanoparticles and Cellular Surface Sialic Acid. ACS Appl. Mater. Interfaces 2019, 11, 46585–46590. [DOI] [PubMed] [Google Scholar]
- (383).Wang M; Alberti K; Varone A; Pouli D; Georgakoudi I; Xu Q Enhanced Intracellular siRNA Delivery Using Bioreducible Lipid-Like Nanoparticles. Adv. Healthcare Mater 2014, 3, 1398–1403. [DOI] [PubMed] [Google Scholar]
- (384).Audouy SAL; de Leij LFMH; Hoekstra D; Molema G In vivo characteristics of cationic liposomes as delivery vectors for gene therapy. Pharm. Res 2002, 19, 1599–1605. [DOI] [PubMed] [Google Scholar]
- (385).Scheule RK; George JAS; Bagley RG; Marshall J; Kaplan JM; Akita GY; Wang KX; Lee ER; Harris DJ; Jiang C; et al. Basis of Pulmonary Toxicity Associated with Cationic Lipid-Mediated Gene Transfer to the Mammalian Lung. Hum. Gene Ther 1997, 8, 689–707. [DOI] [PubMed] [Google Scholar]
- (386).Ruysschaert JM; Elouahabi A; Willeaume V; Huez G; Fuks R; Vandenbranden M; Distefano P A Novel Cationic Amphiphile for Transfection of Mammalian Cells. Biochem. Biophys. Res. Commun 1994, 203, 1622–1628. [DOI] [PubMed] [Google Scholar]
- (387).Bayer TS; Booth LN; Knudsen SM; Ellington AD Arginine-rich Motifs Present Multiple Interfaces for Specific Binding by RNA. RNA 2005, 11, 1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (388).Rothbard JB; Kreider E; VanDeusen CL; Wright L; Wylie BL; Wender PA Arginine-Rich Molecular Transporters for Drug Delivery: Role of Backbone Spacing in Cellular Uptake. J. Med. Chem 2002, 45, 3612–3618. [DOI] [PubMed] [Google Scholar]
- (389).Nakase I; Niwa M; Takeuchi T; Sonomura K; Kawabata N; Koike Y; Takehashi M; Tanaka S; Ueda K; Simpson JC; et al. Cellular Uptake of Arginine-Rich Peptides: Roles for Macropinocytosis and Actin Rearrangement. Mol. Ther 2004, 10, 1011–1022. [DOI] [PubMed] [Google Scholar]
- (390).Kosuge M; Takeuchi T; Nakase I; Jones AT; Futaki S Cellular Internalization and Distribution of Arginine-Rich Peptides as a Function of Extracellular Peptide Concentration, Serum, and Plasma Membrane Associated Proteoglycans. Bioconjugate Chem 2008, 19, 656–664. [DOI] [PubMed] [Google Scholar]
- (391).Santel A; Aleku M; Keil O; Endruschat J; Esche V; Fisch G; Dames S; Loffler K; Fechtner M; Arnold W; et al. A Novel siRNA-Lipoplex Technology for RNA Interference in the Mouse Vascular Endothelium. Gene Ther 2006, 13, 1222–1234. [DOI] [PubMed] [Google Scholar]
- (392).Santel A; Aleku M; Keil O; Endruschat J; Esche V; Durieux B; Löffler K; Fechtner M; Röhl T; Fisch G; et al. RNA Interference in the Mouse Vascular Endothelium by Systemic Administration of siRNA-Lipoplexes for Cancer Therapy. Gene Ther 2006, 13, 1360–1370. [DOI] [PubMed] [Google Scholar]
- (393).Aleku M; Schulz P; Keil O; Santel A; Schaeper U; Dieckhoff B; Janke O; Endruschat J; Durieux B; Röder N; et al. Atu027, a Liposomal Small Interfering RNA Formulation Targeting Protein Kinase N3, Inhibits Cancer Progression. Cancer Res 2008, 68, 9788–9798. [DOI] [PubMed] [Google Scholar]
- (394).Santel A; Aleku M; Röder N; Möpert K; Durieux B; Janke O; Keil O; Endruschat J; Dames S; Lange C; et al. Atu027 Prevents Pulmonary Metastasis in Experimental and Spontaneous Mouse Metastasis Models. Clin. Cancer Res 2010, 16, 5469–5480. [DOI] [PubMed] [Google Scholar]
- (395).Schultheis B; Strumberg D; Santel A; Vank C; Gebhardt F; Keil O; Lange C; Giese K; Kaufmann J; Khan M; et al. First-in-Human Phase I Study of the Liposomal RNA Interference Therapeutic Atu027 in Patients With Advanced Solid Tumors. J. Clin. Oncol 2014, 32, 4141–4148. [DOI] [PubMed] [Google Scholar]
- (396).Schultheis B; Strumberg D; Kuhlmann J; Wolf M; Link K; Seufferlein T; Kaufmann J; Gebhardt F; Bruyniks N; Pelzer U A phase Ib/IIa study of combination therapy with gemcitabine and Atu027 in patients with locally advanced or metastatic pancreatic adenocarcinoma. J. Clin. Oncol 2016, 34, 385–385. [Google Scholar]
- (397).Chen Y; Sen J; Bathula SR; Yang Q; Fittipaldi R; Huang L Novel Cationic Lipid That Delivers siRNA and Enhances Therapeutic Effect in Lung Cancer Cells. Mol. Pharmaceutics 2009, 6, 696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (398).Chen Y; Bathula SR; Li J; Huang L Multifunctional Nanoparticles Delivering Small Interfering RNA and Doxorubicin Overcome Drug Resistance in Cancer. J. Biol. Chem 2010, 285, 22639–22650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (399).Mevel M; Kamaly N; Carmona S; Oliver MH; Jorgensen MR; Crowther C; Salazar FH; Marion PL; Fujino M; Natori Y; et al. DODAG; a Versatile New Cationic Lipid that Mediates Efficient Delivery of pDNA and siRNA. J. Controlled Release 2010, 143, 222–232. [DOI] [PubMed] [Google Scholar]
- (400).Bernatowicz MS; Wu Y; Matsueda GR 1H-Pyrazole-1-carboxamidine Hydrochloride an Attractive Reagent for Guanylation of Amines and its Application to Peptide Synthesis. J. Org. Chem 1992, 57, 2497–2502. [Google Scholar]
- (401).Adami RC; Seth S; Harvie P; Johns R; Fam R; Fosnaugh K; Zhu TY; Farber K; McCutcheon M; Goodman TT; et al. An Amino Acid-based Amphoteric Liposomal Delivery System for Systemic Administration of siRNA. Mol. Ther 2011, 19, 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (402).Sanchez-Arribas N; Martinez-Negro M; Villar EM; Perez L; Aicart E; Taboada P; Guerrero-Martinez A; Junquera E Biocompatible Nanovector of siRNA Consisting of Arginine-Based Cationic Lipid for Gene Knockdown in Cancer Cells. ACS Appl. Mater. Interfaces 2020, 12, 34536–34547. [DOI] [PubMed] [Google Scholar]
- (403).Metwally AA; Blagbrough IS Self-Assembled Lipoplexes of Short Interfering RNA (siRNA) Using Spermine-Based Fatty Acid Amide Guanidines: Effect on Gene Silencing Efficiency. Pharmaceutics 2011, 3, 406–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (404).Metwally AA; Pourzand C; Blagbrough IS Efficient Gene Silencing by Self-Assembled Complexes of siRNA and Symmetrical Fatty Acid Amides of Spermine. Pharmaceutics 2011, 3, 125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (405).Brown AJ; Ikonen E; Olkkonen VM Cholesterol Precursors: More than Mere Markers of Biosynthesis. Curr. Opin. Lipidol 2014, 25, 133–139. [DOI] [PubMed] [Google Scholar]
- (406).Puras G; Mashal M; Zárate J; Agirre M; Ojeda E; Grijalvo S; Eritja R; Diaz-Tahoces A; Martínez Navarrete G; Avilés-Trigueros M; et al. A Novel Cationic Niosome Formulation for Gene Delivery to the Retina. J. Controlled Release 2014, 174, 27–36. [DOI] [PubMed] [Google Scholar]
- (407).Bertrand JR; Lucas C; Pham NM; Durieu C; Couvreur P; Malvy CP; Desmaele D Turning Squalene into Cationic Lipid Allows a Delivery of siRNA in Cultured Cells. Nucleic Acid Ther 2015, 25, 121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (408).Lee J; Saw PE; Gujrati V; Lee Y; Kim H; Kang S; Choi M; Kim JI; Jon S Mono-arginine Cholesterol-based Small Lipid Nanoparticles as a Systemic siRNA Delivery Platform for Effective Cancer Therapy. Theranostics 2016, 6, 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (409).Bang EK; Cho H; Jeon SS; Tran NL; Lim DK; Hur W; Sim T Amphiphilic Small Peptides for Delivery of Plasmid DNAs and siRNAs. Chem. Biol. Drug Des 2018, 91, 575–587. [DOI] [PubMed] [Google Scholar]
- (410).Torres AG; Gait MJ Exploiting Cell Surface Thiols to Enhance Cellular Uptake. Trends Biotechnol 2012, 30, 185–190. [DOI] [PubMed] [Google Scholar]
- (411).Kowalski PS; Lintermans LL; Morselt HWM; Leus NGJ; Ruiters MHJ; Molema G; Kamps JAAM Anti-VCAM-1 and Anti-E-selectin SAINT-O-Somes for Selective Delivery of siRNA into Inflammation-Activated Primary Endothelial Cells. Mol. Pharmaceutics 2013, 10, 3033–3044. [DOI] [PubMed] [Google Scholar]
- (412).Kowalski PS; Zwiers PJ; Morselt HWM; Kuldo JM; Leus NGJ; Ruiters MHJ; Molema G; Kamps JAAM Anti-VCAM-1 SAINT-O-Somes Enable Endothelial-Specific Delivery of siRNA and Downregulation of Inflammatory Genes in Activated Endothelium in Vivo. J. Controlled Release 2014, 176, 64–75. [DOI] [PubMed] [Google Scholar]
- (413).Leus NGJ; Morselt HWM; Zwiers PJ; Kowalski PS; Ruiters MHJ; Molema G; Kamps JAAM VCAM-1 Specific PEGylated SAINT-Based Lipoplexes Deliver siRNA to Activated Endothelium in Vivo but do not Attenuate Target Gene Expression. Int. J. Pharm 2014, 469, 121–131. [DOI] [PubMed] [Google Scholar]
- (414).Kirby AJ; Camilleri P; Engberts JBFN; Feiters MC; Nolte RJM; Söderman O; Bergsma M; Bell PC; Fielden ML; García Rodríguez CL; et al. Gemini Surfactants: New Synthetic Vectors for Gene Transfection. Angew. Chem., Int. Ed 2003, 42, 1448–1457. [DOI] [PubMed] [Google Scholar]
- (415).Sharma VD; Ilies MA Heterocyclic Cationic Gemini Surfactants: A Comparative Overview of Their Synthesis, Self-assembling, Physicochemical, and Biological Properties. Med. Res. Rev 2014, 34, 1–44. [DOI] [PubMed] [Google Scholar]
- (416).Ilies MA; Seitz WA; Johnson BH; Ezell EL; Miller AL; Thompson EB; Balaban AT Lipophilic Pyrylium Salts in the Synthesis of Efficient Pyridinium-Based Cationic Lipids, Gemini Surfactants, and Lipophilic Oligomers for Gene Delivery. J. Med. Chem 2006, 49, 3872–3887. [DOI] [PubMed] [Google Scholar]
- (417).Meekel AAP; Wagenaar A; Šmisterová J; Kroeze Jessica E; Haadsma P; Bosgraaf B; Stuart Marc CA; Brisson A; Ruiters Marcel HJ; Hoekstra D; et al. Synthesis of Pyridinium Amphiphiles Used for Transfection and Some Characteristics of Amphiphile/DNA Complex Formation. Eur. J. Org. Chem 2000, 2000, 665–673. [Google Scholar]
- (418).Muñoz-Úbeda M; Misra SK; Barrán-Berdón AL; Datta S; Aicart-Ramos C; Castro-Hartmann P; Kondaiah P; Junquera E; Bhattacharya S; Aicart E How Does the Spacer Length of Cationic Gemini Lipids Influence the Lipoplex Formation with Plasmid DNA? Physicochemical and Biochemical Characterizations and their Relevance in Gene Therapy. Biomacromolecules 2012, 13, 3926–3937. [DOI] [PubMed] [Google Scholar]
- (419).Bhadani A; Singh S Novel Gemini Pyridinium Surfactants: Synthesis and Study of Their Surface Activity, DNA Binding, and Cytotoxicity. Langmuir 2009, 25, 11703–11712. [DOI] [PubMed] [Google Scholar]
- (420).Wang H; Kaur T; Tavakoli N; Joseph J; Wettig S Transfection and Structural Properties of Phytanyl Substituted Gemini Surfactant-Based Vectors for Gene Delivery. Phys. Chem. Chem. Phys 2013, 15, 20510–20516. [DOI] [PubMed] [Google Scholar]
- (421).Barbero N; Magistris C; Quagliotto P; Bonandini L; Barolo C; Buscaino R; Compari C; Contardi L; Fisicaro E; Viscardi G Synthesis, Physicochemical Characterization, and Interaction with DNA of Long-Alkyl-Chain Gemini Pyridinium Surfactants. ChemPlusChem 2015, 80, 952–962. [DOI] [PubMed] [Google Scholar]
- (422).Satyal U; Draghici B; Dragic LL; Zhang Q; Norris KW; Madesh M; Brailoiu E; Ilies MA Interfacially Engineered Pyridinium Pseudogemini Surfactants as Versatile and Efficient Supramolecular Delivery Systems for DNA, siRNA, and mRNA. ACS Appl. Mater. Interfaces 2017, 9, 29481–29495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (423).Sharma VD; Aifuwa EO; Heiney PA; Ilies MA Interfacial Engineering of Pyridinium Gemini Surfactants for the Generation of Synthetic Transfection Systems. Biomaterials 2013, 34, 6906–6921. [DOI] [PubMed] [Google Scholar]
- (424).Dobbs W; Heinrich B; Bourgogne C; Donnio B; Terazzi E; Bonnet ME; Stock F; Erbacher P; Bolcato-Bellemin AL; Douce L Mesomorphic Imidazolium Salts: New Vectors for Efficient siRNA Transfection. J. Am. Chem. Soc 2009, 131, 13338–13346. [DOI] [PubMed] [Google Scholar]
- (425).Dobbs W; Douce L; Allouche L; Louati A; Malbosc F; Welter R New Ionic Liquid Crystals Based on Imidazolium Salts. New J. Chem 2006, 30, 528–532. [Google Scholar]
- (426).Perche F; Benvegnu T; Berchel M; Lebegue L; Pichon C; Jaffrès P-A; Midoux P Enhancement of dendritic cells transfection in vivo and of vaccination against B16F10 melanoma with mannosylated histidylated lipopolyplexes loaded with tumor antigen messenger RNA. Nanomedicine 2011, 7, 445–453. [DOI] [PubMed] [Google Scholar]
- (427).Pichon C; Midoux P In Synthetic Messenger RNA and Cell Metabolism Modulation: Methods and Protocols; Rabinovich PM, Ed.; Humana Press: Totowa, NJ, 2013; pp 247–274; DOI: 10.1007/978-1-62703-260-5_16. [DOI] [Google Scholar]
- (428).Gonçalves C; Berchel M; Gosselin M-P; Malard V; Cheradame H; Jaffres̀ P-A; Guégan P; Pichon C; Midoux P Lipopolyplexes Comprising Imidazole/Imidazolium Lipophosphoramidate, Histidinylated Polyethyleneimine and siRNA as Efficient Formulation for siRNA Transfection. Int. J. Pharm 2014, 460, 264–272. [DOI] [PubMed] [Google Scholar]
- (429).Pal A; Datta S; Aswal VK; Bhattacharya S Small-Angle Neutron-Scattering Studies of Mixed Micellar Structures Made of Dimeric Surfactants Having Imidazolium and Ammonium Headgroups. J. Phys. Chem. B 2012, 116, 13239–13247. [DOI] [PubMed] [Google Scholar]
- (430).Barrán-Berdón AL; Misra SK; Datta S; Muñoz-Úbeda M; Kondaiah P; Junquera E; Bhattacharya S; Aicart E Cationic Gemini Lipids Containing Polyoxyethylene Spacers as Improved Transfecting Agents of Plasmid DNA in Cancer Cells. J. Mater. Chem. B 2014, 2, 4640–4652. [DOI] [PubMed] [Google Scholar]
- (431).Datta S; Biswas J; Bhattacharya S How Does Spacer Length of Imidazolium Gemini Surfactants Control the Fabrication of 2D-Langmuir Films of Silver-Nanoparticles at the Air-Water Interface? J. Colloid Interface Sci 2014, 430, 85–92. [DOI] [PubMed] [Google Scholar]
- (432).In M; Zana R Phase Behavior of Gemini Surfactants. J. Dispersion Sci. Technol 2007, 28, 143–154. [Google Scholar]
- (433).Zana R Dimeric and oligomeric surfactants. Behavior at interfaces and in aqueous solution: a review. Adv. Colloid Interface Sci 2002, 97, 205–253. [DOI] [PubMed] [Google Scholar]
- (434).Kamboj R; Singh S; Bhadani A; Kataria H; Kaur G Gemini Imidazolium Surfactants: Synthesis and Their Biophysiochemical Study. Langmuir 2012, 28, 11969–11978. [DOI] [PubMed] [Google Scholar]
- (435).Pietralik Z; Kołodziejska Ż; Weiss M; Kozak M Gemini Surfactants Based on Bis-Imidazolium Alkoxy Derivatives as Effective Agents for Delivery of Nucleic Acids: A Structural and Spectroscopic Study. PLoS One 2015, 10, No. e0144373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (436).Andrzejewska W; Pietralik Z; Skupin M; Kozak M Structural studies of the formation of lipoplexes between siRNA and selected bis-imidazolium gemini surfactants. Colloids Surf., B 2016, 146, 598–606. [DOI] [PubMed] [Google Scholar]
- (437).Martinez-Negro M; Kumar K; Barran-Berdon AL; Datta S; Kondaiah P; Junquera E; Bhattacharya S; Aicart E Efficient Cellular Knockdown Mediated by siRNA Nanovectors of Gemini Cationic Lipids Having Delocalizable Headgroups and Oligo-Oxyethylene Spacers. ACS Appl. Mater. Interfaces 2016, 8, 22113–22126. [DOI] [PubMed] [Google Scholar]
- (438).Tousignant JD; Gates AL; Ingram LA; Johnson CL; Nietupski JB; Cheng SH; Eastman SJ; Scheule RK Comprehensive analysis of the acute toxicities induced by systemic administration of cationic lipid: plasmid DNA complexes in mice. Hum. Gene Ther 2000, 11, 2493–2513. [DOI] [PubMed] [Google Scholar]
- (439).Scheule RK; George JAS; Bagley RG; Marshall J; Kaplan JM; Akita GY; Wang KX; Lee ER; Harris DJ; Jiang C Basis of Pulmonary Toxicity Associated with Cationic Lipid-Mediated Gene Transfer to the Mammalian Lung. Hum. Gene Ther 1997, 8, 689–707. [DOI] [PubMed] [Google Scholar]
- (440).Zhang J-S; Liu F; Huang L Implications of Pharmacokinetic Behavior of Lipoplex for its Inflammatory Toxicity. Adv. Drug Delivery Rev 2005, 57, 689–698. [DOI] [PubMed] [Google Scholar]
- (441).Jayaraman M; Ansell SM; Mui BL; Tam YK; Chen J; Du X; Butler D; Eltepu L; Matsuda S; Narayanannair JK; et al. Maximizing the Potency of siRNA Lipid Nanoparticles for Hepatic Gene Silencing In Vivo. Angew. Chem., Int. Ed 2012, 51, 8529–8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (442).Whitehead KA; Dorkin JR; Vegas AJ; Chang PH; Veiseh O; Matthews J; Fenton OS; Zhang Y; Olejnik KT; Yesilyurt V; et al. Degradable Lipid Nanoparticles with Predictable in Vivo siRNA Delivery Activity. Nat. Commun 2014, 5, 4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (443).Maurer N; Wong KF; Stark H; Louie L; McIntosh D; Wong T; Scherrer P; Semple SC; Cullis PR Spontaneous Entrapment of Polynucleotides upon Electrostatic Interaction with Ethanol-Destabilized Cationic Liposomes. Biophys. J 2001, 80, 2310–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (444).Semple SC; Klimuk SK; Harasym TO; Dos Santos N; Ansell SM; Wong KF; Maurer N; Stark H; Cullis PR; Hope MJ; et al. Efficient Encapsulation of Antisense Oligonucleotides in Lipid Vesicles Using Ionizable Aminolipids: Formation of Novel Small Multilamellar Vesicle Structures. Biochim. Biophys. Acta, Biomembr 2001, 1510, 152–166. [DOI] [PubMed] [Google Scholar]
- (445).Guo X; Wang H; Li Y; Leng X; Huang W; Ma Y; Xu T; Qi X Transfection Reagent Lipofectamine Triggers Type I Interferon Signaling Activation in Macrophages. Immunol. Cell Biol 2019, 97, 92–96. [DOI] [PubMed] [Google Scholar]
- (446).Miller AD Cationic Liposomes for Gene Therapy. Angew. Chem., Int. Ed 1998, 37, 1768–1785. [Google Scholar]
- (447).Dalby B; Cates S; Harris A; Ohki EC; Tilkins ML; Price PJ; Ciccarone VC Advanced Transfection with Lipofectamine 2000 Reagent: Primary Neurons, siRNA, and High-Throughput Applications. Methods 2004, 33, 95–103. [DOI] [PubMed] [Google Scholar]
- (448).Santel A; Aleku M; Keil O; Endruschat J; Esche V; Fisch G; Dames S; Löffler K; Fechtner M; Arnold W; et al. A Novel siRNA-Lipoplex Technology for RNA Interference in the Mouse Vascular Endothelium. Gene Ther 2006, 13, 1222–1234. [DOI] [PubMed] [Google Scholar]
- (449).Zhang Y; Cristofaro P; Silbermann R; Pusch O; Boden D; Konkin T; Hovanesian V; Monfils PR; Resnick M; Moss SF; et al. Engineering Mucosal RNA Interference in Vivo. Mol. Ther 2006, 14, 336–342. [DOI] [PubMed] [Google Scholar]
- (450).Ewert K; Ahmad A; Evans HM; Schmidt H-W; Safinya CR Efficient Synthesis and Cell-Transfection Properties of a New Multivalent Cationic Lipid for Nonviral Gene Delivery. J. Med. Chem 2002, 45, 5023–5029. [DOI] [PubMed] [Google Scholar]
- (451).Schulze U; Schmidt H-W; Safinya CR Synthesis of Novel Cationic Poly(Ethylene Glycol) Containing Lipids. Bioconjugate Chem 1999, 10, 548–552. [DOI] [PubMed] [Google Scholar]
- (452).Ewert KK; Zidovska A; Ahmad A; Bouxsein NF; Evans HM; McAllister CS; Samuel CE; Safinya CR In Nucleic Acid Transfection; Bielke W, Erbacher C, Eds.; Springer Berlin Heidelberg: Berlin, Heidelberg, 2010; pp 191–226; DOI: 10.1007/128_2010_70. [DOI] [Google Scholar]
- (453).Leal C; Ewert KK; Shirazi RS; Bouxsein NF; Safinya CR Nanogyroids Incorporating Multivalent Lipids: Enhanced Membrane Charge Density and Pore Forming Ability for Gene Silencing. Langmuir 2011, 27, 7691–7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (454).Ghonaim HM; Li S; Blagbrough IS Very Long Chain N-4, N-9 -Diacyl Spermines: Non-Viral Lipopolyamine Vectors for Efficient Plasmid DNA and siRNA Delivery. Pharm. Res 2009, 26, 19–31. [DOI] [PubMed] [Google Scholar]
- (455).Ahmed OAA; Adjimatera N; Pourzand C; Blagbrough I . N4,N9-Dioleoyl Spermine Is a Novel Nonviral Lipopolyamine Vector for Plasmid DNA Formulation. Pharm. Res 2005, 22, 972–980. [DOI] [PubMed] [Google Scholar]
- (456).Ahmed OAA; Pourzand C; Blagbrough IS Varying the Unsaturation in N4,N9-Dioctadecanoyl Spermines: Nonviral Lipopolyamine Vectors for More Efficient Plasmid DNA Formulation. Pharm. Res 2006, 23, 31–40. [DOI] [PubMed] [Google Scholar]
- (457).O’Sullivan MC; Dalrymple DM A One-Step Procedure for the Selective Trifluoroacetylation of Primary Amino Groups of Polyamines. Tetrahedron Lett 1995, 36, 3451–3452. [Google Scholar]
- (458).Bergeron RJ; McManis JS Total Synthesis of (±)-15-Deoxyspergualin. J. Org. Chem 1987, 52, 1700–1703. [Google Scholar]
- (459).Metwally AA; Reelfs O; Pourzand C; Blagbrough IS Efficient Silencing of EGFP Reporter Gene with siRNA Delivered by Asymmetrical N4,N9-diacyl Spermines. Mol. Pharmaceutics 2012, 9, 1862–1876. [DOI] [PubMed] [Google Scholar]
- (460).Blagbrough IS; Metwally AA; Ghonaim HM Asymmetrical N4,N9-diacyl Spermines: SAR Studies of Nonviral Lipopolyamine Vectors for Efficient siRNA Delivery with Silencing of EGFP Reporter Gene. Mol. Pharmaceutics 2012, 9, 1853–1861. [DOI] [PubMed] [Google Scholar]
- (461).Moazed D; Noller HF Interaction of Antibiotics with Functional Sites in 16S Ribosomal RNA. Nature 1987, 327, 389–394. [DOI] [PubMed] [Google Scholar]
- (462).Wang H; Tor Y Electrostatic Interactions in RNA Aminoglycosides Binding. J. Am. Chem. Soc 1997, 119, 8734–8735. [Google Scholar]
- (463).François B; Russell RJM; Murray JB; Aboul-Ela F; Masquida B; Vicens Q; Westhof E Crystal Structures of Complexes Between Aminoglycosides and Decoding A Site Oligonucleotides: Role of the Number of Rings and Positive Charges in the Specific Binding Leading to Miscoding. Nucleic Acids Res 2005, 33, 5677–5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (464).Desigaux L; Sainlos M; Lambert O; Chevre R; Letrou-Bonneval E; Vigneron JP; Lehn P; Lehn JM; Pitard B Self-Assembled Lamellar Complexes of siRNA with Lipidic Aminoglycoside Derivatives Promote Efficient siRNA Delivery and Interference. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 16534–16539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (465).Pitard B; Oudrhiri N; Vigneron J-P; Hauchecorne M; Aguerre O; Toury R; Airiau M; Ramasawmy R; Scherman D; Crouzet J; et al. Structural Characteristics of Supramolecular Assemblies Formed by Guanidinium-Cholesterol Reagents for Gene Transfection. Proc. Natl. Acad. Sci. U. S. A 1999, 96, 2621–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (466).Habrant D; Peuziat P; Colombani T; Dallet L; Gehin J; Goudeau E; Evrard B; Lambert O; Haudebourg T; Pitard B Design of Ionizable Lipids To Overcome the Limiting Step of Endosomal Escape: Application in the Intracellular Delivery of mRNA, DNA, and siRNA. J. Med. Chem 2016, 59, 3046–3062. [DOI] [PubMed] [Google Scholar]
- (467).Obata Y; Suzuki D; Takeoka S Evaluation of Cationic Assemblies Constructed with Amino Acid Based Lipids for Plasmid DNA Delivery. Bioconjugate Chem 2008, 19, 1055–1063. [DOI] [PubMed] [Google Scholar]
- (468).Kim HS; Song IH; Kim JC; Kim EJ; Jang DO; Park YS In Vitro and in Vivo Gene-Transferring Characteristics of Novel Cationic Lipids, DMKD (O,O’-dimyristyl-N-lysyl aspartate) and DMKE (O,O’-dimyristyl-N-lysyl glutamate). J. Controlled Release 2006, 115, 234–241. [DOI] [PubMed] [Google Scholar]
- (469).Suh MS; Shim G; Lee HY; Han SE; Yu YH; Choi Y; Kim K; Kwon IC; Weon KY; Kim YB; et al. Anionic Amino Acid-Derived Cationic Lipid for siRNA Delivery. J. Controlled Release 2009, 140, 268–276. [DOI] [PubMed] [Google Scholar]
- (470).Xiao H; Altangerel A; Gerile G; Wu Y; Baigude H Design of Highly Potent Lipid-Functionalized Peptidomimetics for Efficient in Vivo siRNA Delivery. ACS Appl. Mater. Interfaces 2016, 8, 7638–7645. [DOI] [PubMed] [Google Scholar]
- (471).Wang X-L; Ramusovic S; Nguyen T; Lu Z-R Novel Polymerizable Surfactants with pH-Sensitive Amphiphilicity and Cell Membrane Disruption for Efficient siRNA Delivery. Bioconjugate Chem 2007, 18, 2169–2177. [DOI] [PubMed] [Google Scholar]
- (472).Malamas AS; Gujrati M; Kummitha CM; Xu RZ; Lu ZR Design and Evaluation of New pH-Sensitive Amphiphilic Cationic Lipids for siRNA Delivery. J. Controlled Release 2013, 171, 296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (473).Gujrati M; Malamas A; Shin T; Jin EL; Sun YL; Lu ZR Multifunctional Cationic Lipid-Based Nanoparticles Facilitate Endosomal Escape and Reduction-Triggered Cytosolic siRNA Release. Mol. Pharmaceutics 2014, 11, 2734–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (474).Wang X-L; Nguyen T; Gillespie D; Jensen R; Lu Z-R A Multifunctional and Reversibly Polymerizable Carrier for Efficient siRNA Delivery. Biomaterials 2008, 29, 15–22. [DOI] [PubMed] [Google Scholar]
- (475).Parvani JG; Gujrati MD; Mack MA; Schiemann WP; Lu Z-R Silencing β3 Integrin by Targeted ECO/siRNA Nanoparticles Inhibits EMT and Metastasis of Triple-Negative Breast Cancer. Cancer Res 2015, 75, 2316–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (476).Gujrati M; Vaidya AM; Mack M; Snyder D; Malamas A; Lu Z-R Targeted Dual pH-Sensitive Lipid ECO/siRNA Self-Assembly Nanoparticles Facilitate In Vivo Cytosolic sieIF4E Delivery and Overcome Paclitaxel Resistance in Breast Cancer Therapy. Adv. Healthcare Mater 2016, 5, 2882–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (477).Vaidya AM; Sun Z; Ayat N; Schilb A; Liu X; Jiang H; Sun D; Scheidt J; Qian V; He S; et al. Systemic Delivery of Tumor-Targeting siRNA Nanoparticles against an Oncogenic LncRNA Facilitates Effective Triple-Negative Breast Cancer Therapy. Bioconjugate Chem 2019, 30, 907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (478).Asai T; Matsushita S; Kenjo E; Tsuzuku T; Yonenaga N; Koide H; Hatanaka K; Dewa T; Nango M; Maeda N; et al. Dicetyl Phosphate-Tetraethylenepentamine-Based Liposomes for Systemic siRNA Delivery. Bioconjugate Chem 2011, 22, 429–435. [DOI] [PubMed] [Google Scholar]
- (479).Sako M; Song F; Okamoto A; Koide H; Dewa T; Oku N; Asai T Key Determinants of siRNA Delivery Mediated by Unique pH-Responsive Lipid-Based Liposomes. Int. J. Pharm 2019, 569, 118606. [DOI] [PubMed] [Google Scholar]
- (480).Okamoto A; Koide H; Morita N; Hirai Y; Kawato Y; Egami H; Hamashima Y; Asai T; Dewa T; Oku N Rigorous Control of Vesicle-Forming Lipid pK(a) by Fluorine-Conjugated Bioisosteres for Gene-Silencing with siRNA. J. Controlled Release 2019, 295, 87–92. [DOI] [PubMed] [Google Scholar]
- (481).Siegemund G; Schwertfeger W; Feiring A; Smart B; Behr F; Vogel H; McKusick B Ullmann’s Encyclopedia of Industrial Chemistry; Wiley-VCH Verlag GmbH & Co. KGaA, 2000; DOI: 10.1002/14356007.a11_349. [DOI] [Google Scholar]
- (482).Chang RS; Suh MS; Kim S; Shim G; Lee S; Han SS; Lee KE; Jeon H; Choi HG; Choi Y; et al. Cationic Drug-Derived Nanoparticles for Multifunctional Delivery of Anticancer siRNA. Biomaterials 2011, 32, 9785–9795. [DOI] [PubMed] [Google Scholar]
- (483).Zimmermann TS; Lee ACH; Akinc A; Bramlage B; Bumcrot D; Fedoruk MN; Harborth J; Heyes JA; Jeffs LB; John M; et al. RNAi-Mediated Gene Silencing in Non-Human Primates. Nature 2006, 441, 111–114. [DOI] [PubMed] [Google Scholar]
- (484).Wang X; Yu BO; Wu YUN; Lee RJ; Lee LJ Efficient Down-Regulation of CDK4 by Novel Lipid Nanoparticle-Mediated siRNA Delivery. Anticancer Res 2011, 31, 1619–1626. [PubMed] [Google Scholar]
- (485).Hsu S-H; Yu B; Wang X; Lu Y; Schmidt CR; Lee RJ; Lee LJ; Jacob ST; Ghoshal K Cationic Lipid Nanoparticles for Therapeutic Delivery of siRNA and miRNA to Murine Liver Tumor. Nanomedicine 2013, 9, 1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (486).Li L; Wang R; Wilcox D; Sarthy A; Lin X; Huang X; Tian L; Dande P; Hubbard RD; Hansen TM; et al. Developing Lipid Nanoparticle-Based siRNA Therapeutics for Hepatocellular Carcinoma Using an Integrated Approach. Mol. Cancer Ther 2013, 12, 2308–2318. [DOI] [PubMed] [Google Scholar]
- (487).Niculescu-Duvaz D; Heyes J; Springer C Structure-Activity Relationship in Cationic Lipid Mediated Gene Transfection. Curr. Med. Chem 2003, 10, 1233–1261. [DOI] [PubMed] [Google Scholar]
- (488).Ferrari ME; Rusalov D; Enas J; Wheeler CJ Synergy Between Cationic Lipid and Co-lipid Determines the Macroscopic Structure and Transfection Activity of Lipoplexes. Nucleic Acids Res 2002, 30, 1808–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (489).Heyes JA; Niculescu-Duvaz D; Cooper RG; Springer CJ Synthesis of Novel Cationic Lipids: Effect of Structural Modification on the Efficiency of Gene Transfer. J. Med. Chem 2002, 45, 99–114. [DOI] [PubMed] [Google Scholar]
- (490).Balasubramaniam RP; Bennett MJ; Aberle AM; Malone JG; Nantz MH; Malone RW Structural and Functional Analysis of Cationic Transfection Lipids: the Hydrophobic Domain. Gene Ther 1996, 3, 163–172. [PubMed] [Google Scholar]
- (491).Tao W; Davide JP; Cai M; Zhang GJ; South VJ; Matter A; Ng B; Zhang Y; Sepp-Lorenzino L Noninvasive Imaging of Lipid Nanoparticle-Mediated Systemic Delivery of Small-Interfering RNA to the Liver. Mol. Ther 2010, 18, 1657–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (492).Zhang J; Fan H; Levorse DA; Crocker LS Interaction of Cholesterol-Conjugated Ionizable Amino Lipids with Biomembranes: Lipid Polymorphism, Structure-Activity Relationship, and Implications for siRNA Delivery. Langmuir 2011, 27, 9473–9483. [DOI] [PubMed] [Google Scholar]
- (493).Abrams MT; Koser ML; Seitzer J; Williams SC; DiPietro MA; Wang W; Shaw AW; Mao X; Jadhav V; Davide JP; et al. Evaluation of Efficacy, Biodistribution, and Inflammation for a Potent siRNA Nanoparticle: Effect of Dexamethasone Co-treatment. Mol. Ther 2010, 18, 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (494).Tao W; Mao X; Davide JP; Ng B; Cai M; Burke PA; Sachs AB; Sepp-Lorenzino L Mechanistically Probing Lipid-siRNA Nanoparticle-associated Toxicities Identifies Jak Inhibitors Effective in Mitigating Multifaceted Toxic Responses. Mol. Ther 2011, 19, 567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (495).Zhang J; Fan H; Levorse DA; Crocker LS Ionization Behavior of Amino Lipids for siRNA Delivery: Determination of Ionization Constants, SAR, and the Impact of Lipid pKa on Cationic Lipid-Biomembrane Interactions. Langmuir 2011, 27, 1907–1914. [DOI] [PubMed] [Google Scholar]
- (496).Zhang J; Fan H; Levorse DA; Crocker LS Interaction of Cholesterol-Conjugated Ionizable Amino Lipids with Biomembranes: Lipid Polymorphism, Structure-Activity Relationship, and Implications for siRNA Delivery. Langmuir 2011, 27, 9473–9483. [DOI] [PubMed] [Google Scholar]
- (497).Huang Z; Szoka FC Sterol-Modified Phospholipids: Cholesterol and Phospholipid Chimeras with Improved Biomembrane Properties. J. Am. Chem. Soc 2008, 130, 15702–15712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (498).Maurer N; Fenske DB; Cullis PR Developments in Liposomal Drug Delivery Systems. Expert Opin. Biol. Ther 2001, 1, 923–947. [DOI] [PubMed] [Google Scholar]
- (499).Semple SC; Klimuk SK; Harasym TO; Dos Santos N; Ansell SM; Wong KF; Maurer N; Stark H; Cullis PR; Hope MJ Efficient Encapsulation of Antisense Oligonucleotides in Lipid Vesicles Using Ionizable Aminolipids: Formation of Novel Small Multilamellar Vesicle Structures. Biochim. Biophys. Acta, Biomembr 2001, 1510, 152–166. [DOI] [PubMed] [Google Scholar]
- (500).Bailey AL; Cullis PR Modulation of Membrane Fusion by Asymmetric Transbilayer Distributions of Amino Lipids. Biochemistry 994, 33, 12573–12580. [DOI] [PubMed] [Google Scholar]
- (501).Conceição M; Mendonça L; Nóbrega C; Gomes C; Costa P; Hirai H; Moreira JN; Lima MC; Manjunath N; Pereira de Almeida L Intravenous Administration of Brain-Targeted Stable Nucleic Acid Lipid Particles Alleviates Machado-Joseph Disease Neurological Phenotype. Biomaterials 2016, 82, 124–137. [DOI] [PubMed] [Google Scholar]
- (502).Semple SC; Akinc A; Chen J; Sandhu AP; Mui BL; Cho CK; Sah DWY; Stebbing D; Crosley EJ; Yaworski E; et al. Rational Design of Cationic Lipids for siRNA Delivery. Nat. Biotechnol 2010, 28, 172–176. [DOI] [PubMed] [Google Scholar]
- (503).Lee JB; Zhang K; Tam YYC; Tam YK; Belliveau NM; Sung VYC; Lin PJC; LeBlanc E; Ciufolini MA; Rennie PS; et al. Lipid nanoparticle siRNA systems for silencing the androgen receptor in human prostate cancer in vivo. Int. J. Cancer 2012, 131, E781–E790. [DOI] [PubMed] [Google Scholar]
- (504).Lin PJC; Tam YYC; Hafez I; Sandhu A; Chen S; Ciufolini MA; Nabi IR; Cullis PR Influence of Cationic Lipid Composition on Uptake and Intracellular Processing of Lipid Nanoparticle Formulations of siRNA. Nanomedicine 2013, 9, 233–246. [DOI] [PubMed] [Google Scholar]
- (505).Basha G; Novobrantseva TI; Rosin N; Tam YYC; Hafez IM; Wong MK; Sugo T; Ruda VM; Qin J; Klebanov B; et al. Influence of Cationic Lipid Composition on Gene Silencing Properties of Lipid Nanoparticle Formulations of siRNA in Antigen-Presenting Cells. Mol. Ther 2011, 19, 2186–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (506).Zhang X; Goel V; Robbie GJ Pharmacokinetics of Patisiran, the First Approved RNA Interference Therapy in Patients With Hereditary Transthyretin-Mediated Amyloidosis. J. Clin. Pharmacol 2020, 60, 573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (507).Nabhan JF; Wood KM; Rao VP; Morin J; Bhamidipaty S; LaBranche TP; Gooch RL; Bozal F; Bulawa CE; Guild BC Intrathecal Delivery of Frataxin mRNA Encapsulated in Lipid Nanoparticles to Dorsal Root Ganglia as a Potential Therapeutic for Friedreich’s Ataxia. Sci. Rep 2016, 6, 20019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (508).Rungta RL; Choi HB; Lin PJ; Ko RW; Ashby D; Nair J; Manoharan M; Cullis PR; Macvicar BA Lipid Nanoparticle Delivery of siRNA to Silence Neuronal Gene Expression in the Brain. Mol. Ther.–Nucleic Acids 2013, 2, No. e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (509).Chen S; Tam YYC; Lin PJC; Sung MMH; Tam YK; Cullis PR Influence of Particle Size on the in Vivo Potency of Lipid Nanoparticle Formulations of siRNA. J. Controlled Release 2016, 235, 236–244. [DOI] [PubMed] [Google Scholar]
- (510).Bahl K; Senn JJ; Yuzhakov O; Bulychev A; Brito LA; Hassett KJ; Laska ME; Smith M; Almarsson Ö; Thompson J; et al. Preclinical and Clinical Demonstration of Immunogenicity by mRNA Vaccines against H10N8 and H7N9 Influenza Viruses. Mol. Ther 2017, 25, 1316–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (511).Liang F; Lindgren G; Lin A; Thompson EA; Ols S; Röhss J; John S; Hassett K; Yuzhakov O; Bahl K; et al. Efficient Targeting and Activation of Antigen-Presenting Cells In Vivo after Modified mRNA Vaccine Administration in Rhesus Macaques. Mol. Ther 2017, 25, 2635–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (512).Feldman RA; Fuhr R; Smolenov I; Ribeiro A; Panther L; Watson M; Senn JJ; Smith M; Almarsson Ö; Pujar HS; et al. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine 2019, 37, 3326–3334. [DOI] [PubMed] [Google Scholar]
- (513).Richner JM; Himansu S; Dowd KA; Butler SL; Salazar V; Fox JM; Julander JG; Tang WW; Shresta S; Pierson TC; et al. Modified mRNA Vaccines Protect against Zika Virus Infection. Cell 2017, 168, 1114–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (514).Richner JM; Jagger BW; Shan C; Fontes CR; Dowd KA; Cao B; Himansu S; Caine EA; Nunes BTD; Medeiros DBA; et al. Vaccine Mediated Protection Against Zika Virus-Induced Congenital Disease. Cell 2017, 170, 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (515).John S; Yuzhakov O; Woods A; Deterling J; Hassett K; Shaw CA; Ciaramella G Multi-antigenic human cytomegalovirus mRNA vaccines that elicit potent humoral and cell-mediated immunity. Vaccine 2018, 36, 1689–1699. [DOI] [PubMed] [Google Scholar]
- (516).Maier MA; Jayaraman M; Matsuda S; Liu J; Barros S; Querbes W; Tam YK; Ansell SM; Kumar V; Qin J; et al. Biodegradable Lipids Enabling Rapidly Eliminated Lipid Nanoparticles for Systemic Delivery of RNAi Therapeutics. Mol. Ther 2013, 21, 1570–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (517).Miao L; Lin J; Huang Y; Li L; Delcassian D; Ge Y; Shi Y; Anderson DG Synergistic Dipid Compositions for Albumin Receptor Mediated Delivery of mRNA to the Liver. Nat. Commun 2020, 11, 2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (518).Dong Y; Love KT; Dorkin JR; Sirirungruang S; Zhang Y; Chen D; Bogorad RL; Yin H; Chen Y; Vegas AJ; et al. Lipopeptide Nanoparticles for Potent and Selective siRNA Delivery in Rodents and Nonhuman Primates. Proc. Natl. Acad. Sci. U. S. A 2014, 111, 3955–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (519).Rajappan K; Tanis SP; Mukthavaram R; Roberts S; Nguyen M; Tachikawa K; Sagi A; Sablad M; Limphong P; Leu A; et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med 2020, 383, 2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (520).Joseph E; Payne PC, Priya K; Steven P Tanis Ionizable Cationic Lipid for RNA Delivery, US10526284 B2, Jan. 7, 2020. [Google Scholar]
- (521).de Alwis R; Gan ES; Chen S; Leong YS; Tan HC; Zhang SL; Yau C; Matsuda D; Allen E; Hartman P; et al. A Single Dose of Self-Transcribing and Replicating RNA Based SARS-CoV-2 Vaccine Produces Protective Adaptive Immunity In Mice. Mol. Ther 2021, 29, 1970–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (522).Ramishetti S; Hazan-Halevy I; Palakuri R; Chatterjee S; Gonna SN; Dammes N; Freilich I; Shmuel LK; Danino D; Peer D A Combinatorial Library of Lipid Nanoparticles for RNA Delivery to Leukocytes. Adv. Mater 2020, 32, 1906128. [DOI] [PubMed] [Google Scholar]
- (523).Liu Y; Liu J; Xia H; Zhang X; Fontes-Garfias CR; Swanson KA; Cai H; Sarkar R; Chen W; Cutler M; et al. Neutralizing Activity of BNT162b2-Elicited Serum — Preliminary Report. N. Engl. J. Med 2021, 384, 1466–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (524).Walsh EE; Frenck RW; Falsey AR; Kitchin N; Absalon J; Gurtman A; Lockhart S; Neuzil K; Mulligan MJ; Bailey R; Desmosterol in Parenteral Liposomal Pharmaceutical Formulations. Int. J. Pharm 2019, 569, 118576. [DOI] [PubMed] [Google Scholar]
- (525).Polack FP; Thomas SJ; Kitchin N; Absalon J; Gurtman A; Lockhart S; Perez JL; Pérez Marc G; Moreira ED; Zerbini C; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med 2020, 383, 2603–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (526).Widge AT; Rouphael NG; Jackson LA; Anderson EJ; Roberts PC; Makhene M; Chappell JD; Denison MR; Stevens LJ; Pruijssers AJ; et al. Durability of Responses after SARS-CoV-2 mRNA-1273 Vaccination. N. Engl. J. Med 2021, 384, 80–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (527).Baden LR; El Sahly HM; Essink B; Kotloff K; Frey S; Novak R; Diemert D; Spector SA; Rouphael N; Creech CB; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med 2021, 384, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (528).Anderson EJ; Rouphael NG; Widge AT; Jackson LA; Roberts PC; Makhene M; Chappell JD; Denison MR; Stevens LJ; Pruijssers AJ; et al. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N. Engl. J. Med 2020, 383, 2427–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (529).Ansell SM; Du X Novel Lipids and Lipid Nanoparticle Formulations for Delivery of Nucleic Acids WO2017075531 A1, May 4, 2017. [Google Scholar]
- (530).FDA. Interim Clinical Considerations for Use of mRNA COVID-19 Vaccines Currently Authorized in the United States https://www.cdc.gov/vaccines/covid-19/info-by-product/clinical-considerations.html (accessed 2021-05-19).
- (531).Hassett KJ; Benenato KE; Jacquinet E; Lee A; Woods A; Yuzhakov O; Himansu S; Deterling J; Geilich BM; Ketova T; et al. Optimization of Lipid Nanoparticles for Intramuscular Administration of mRNA Vaccines. Mol. Ther.–Nucleic Acids 2019, 15, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (532).Witzigmann D; Kulkarni JA; Leung J; Chen S; Cullis PR; van der Meel R Lipid Nanoparticle Technology for Therapeutic Gene Regulation in the Liver. Adv. Drug Delivery Rev 2020, 159, 344–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (533).Gruner SM; Cullis PR; Hope MJ; Tilcock CPS Lipid Polymorphism: the Molecular basis of Nonbilayer Phases. Annu. Rev. Biophys. Biophys. Chem 1985, 14, 211–238. [DOI] [PubMed] [Google Scholar]
- (534).Sabnis S; Kumarasinghe ES; Salerno T; Mihai C; Ketova T; Senn JJ; Lynn A; Bulychev A; McFadyen I; Chan J; et al. A Novel Amino Lipid Series for mRNA Delivery: Improved Endosomal Escape and Sustained Pharmacology and Safety in Non-human Primates. Mol. Ther 2018, 26, 1509–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (535).Fabre A-L; Colotte M; Luis A; Tuffet S; Bonnet J An Efficient Method for Long-Term Room Temperature Storage of RNA. Eur. J. Hum. Genet 2014, 22, 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (536).Pogocki D; Schöneich C Chemical Stability of Nucleic Acid Derived Drugs. J. Pharm. Sci 2000, 89, 443–456. [DOI] [PubMed] [Google Scholar]
- (537).Jaeger J; Sorensen K; Wolff SP Peroxide Accumulation in Detergents. J. Biochem. Biophys. Methods 1994, 29, 77–81. [DOI] [PubMed] [Google Scholar]
- (538).Wang C; Siriwardane DA; Jiang W; Mudalige T et al. Property-Driven Design and Development of Lipids for Efficient Quantitative Analysis of Cholesterol Oxidation Products and Delivery of siRNA. J. Med. Chem 2020, 63, 12992–13012. [DOI] [PubMed] [Google Scholar]
- (539).Ayat NR; Sun Z; Sun D; Yin M; Hall RC; Vaidya AM; Liu X; Schilb AL; Scheidt JH; Lu Z-R Formulation of Biocompatible Targeted ECO/siRNA Nanoparticles with Long-Term Stability for Clinical Translation of RNAi. Nucleic Acid Ther 2019, 29, 195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (540).Ball RL; Bajaj P; Whitehead KA Achieving Long-Term Stability of Lipid Nanoparticles: Examining the Effect of pH, Temperature, and Lyophilization. Int. J. Nanomed 2017, 12, 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (541).Ryals RC; Patel S; Acosta C; McKinney M; Pennesi ME; Sahay G The Effects of PEGylation on LNP Based mRNA Delivery to the Eye. PLoS One 2020, 15, No. e0241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (542).Crommelin DJA; Anchordoquy TJ; Volkin DB; Jiskoot W; Mastrobattista E Addressing the Cold Reality of mRNA Vaccine Stability. J. Pharm. Sci 2021, 110, 997–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (543).Schoenmaker L; Witzigmann D; Kulkarni JA; Verbeke R; Kersten G; Jiskoot W; Crommelin DJA mRNA-Lipid Nanoparticle COVID-19 Vaccines: Structure and Stability. Int. J. Pharm 2021, 601, 120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (544).Finn JD; Smith AR; Patel MC; Shaw L; Youniss MR; van Heteren J; Dirstine T; Ciullo C; Lescarbeau R; Seitzer J; et al. A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Rep 2018, 22, 2227–2235. [DOI] [PubMed] [Google Scholar]
- (545).Dahlman JE; Kauffman KJ; Xing Y; Shaw TE; Mir FF; Dlott CC; Langer R; Anderson DG; Wang ET Barcoded Nanoparticles for High Throughput in Vivo Discovery of Targeted Therapeutics. Proc. Natl. Acad. Sci. U. S. A 2017, 114, 2060–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (546).Lokugamage MP; Sago CD; Gan Z; Krupczak BR; Dahlman JE Constrained Nanoparticles Deliver siRNA and sgRNA to T Cells In Vivo without Targeting Ligands. Adv. Mater 2019, 31, 1902251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (547).Akita H; Ishiba R; Hatakeyama H; Tanaka H; Sato Y; Tange K; Arai M; Kubo K; Harashima H A Neutral Envelope-Type Nanoparticle Containing pH-Responsive and SS-Cleavable Lipid-Like Material as a Carrier for Plasmid DNA. Adv. Healthcare Mater 2013, 2, 1120–1125. [DOI] [PubMed] [Google Scholar]
- (548).Ukawa M; Akita H; Hayashi Y; Ishiba R; Tange K; Arai M; Kubo K; Higuchi Y; Shimizu K; Konishi S; et al. Neutralized nanoparticle composed of SS-cleavable and pH-activated lipid-like material as a long-lasting and liver-specific gene delivery system. Adv. Healthcare Mater 2014, 3, 1222–1229. [DOI] [PubMed] [Google Scholar]
- (549).Akita H; Ishiba R; Togashi R; Tange K; Nakai Y; Hatakeyama H; Harashima H A Neutral Lipid Envelope-Type Nanoparticle Composed of a pH-Activated and Vitamin E-scaffold Lipid-Like Material as a Platform for a Gene Carrier Targeting Renal Cell Carcinoma. J. Controlled Release 2015, 200, 97–105. [DOI] [PubMed] [Google Scholar]
- (550).Tanaka H; Takahashi T; Konishi M; Takata N; Gomi M; Shirane D; Miyama R; Hagiwara S; Yamasaki Y; Sakurai Y; et al. Self-Degradable Lipid-Like Materials Based on “Hydrolysis Accelerated by the Intra-Particle Enrichment of Reactant (HyPER)” for Messenger RNA Delivery. Adv. Funct. Mater 2020, 30, 1910575. [Google Scholar]
- (551).Liang C; Chang J; Jiang Y; Liu J; Mao L; Wang M Selective RNA Interference and Gene Silencing Using Reactive Oxygen Species-Responsive Lipid Nanoparticles. Chem. Commun 2019, 55, 8170–8173. [DOI] [PubMed] [Google Scholar]
- (552).Tanaka H; Sakurai Y; Anindita J; Akita H Development of Lipid-Like Materials for RNA Delivery Based on Intracellular Environment-Responsive Membrane Destabilization and Spontaneous Collapse. Adv. Drug Delivery Rev 2020, 154–155, 210–226. [DOI] [PubMed] [Google Scholar]
- (553).Ma XF; Sun J; Qiu C; Wu YF; Zheng Y; Yu MZ; Pei XW; Wei L; Niu YJ; Pang WH; et al. The role of Disulfide-Bridge on the Activities of H-Shape Gemini-Like Cationic Lipid Based siRNA Delivery. J. Controlled Release 2016, 235, 99–111. [DOI] [PubMed] [Google Scholar]
- (554).Akita H; Noguchi Y; Hatakeyama H; Sato Y; Tange K; Nakai Y; Harashima H Molecular Tuning of a Vitamin E-Scaffold pH-Sensitive and Reductive Cleavable Lipid-like Material for Accelerated in Vivo Hepatic siRNA Delivery. ACS Biomater. Sci. Eng 2015, 1, 834–844. [DOI] [PubMed] [Google Scholar]
- (555).Toriyabe N; Sakurai Y; Kato A; Yamamoto S; Tange K; Nakai Y; Akita H; Harahsima H The Delivery of Small Interfering RNA to Hepatic Stellate Cells Using a Lipid Nanoparticle Composed of a Vitamin A-Scaffold Lipid-Like Material. J. Pharm. Sci 2017, 106, 2046–2052. [DOI] [PubMed] [Google Scholar]
- (556).Watanabe A; Tanaka H; Sakurai Y; Tange K; Nakai Y; Ohkawara T; Takeda H; Harashima H; Akita H Effect of Particle Size on Their Accumulation in an Inflammatory Lesion in a Dextran Sulfate Sodium (DSS)-Induced Colitis Model. Int. J. Pharm 2016, 509, 118–122. [DOI] [PubMed] [Google Scholar]
- (557).Tanaka H; Watanabe A; Konishi M; Nakai Y; Yoshioka H; Ohkawara T; Takeda H; Harashima H; Akita H The Delivery of mRNA to Colon Inflammatory Lesions by Lipid-Nano-Particles Containing Environmentally-Sensitive Lipid-Like Materials with Oleic Acid Scaffolds. Heliyon 2018, 4, No. e00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (558).Tateshita N; Miura N; Tanaka H; Masuda T; Ohtsuki S; Tange K; Nakai Y; Yoshioka H; Akita H Development of a Lipoplex-Type mRNA Carrier Composed of an Ionizable Lipid with a Vitamin E Scaffold and the KALA Peptide for Use as an Ex Vivo Dendritic Cell-Based Cancer Vaccine. J. Controlled Release 2019, 310, 36–46. [DOI] [PubMed] [Google Scholar]
- (559).Sato Y; Hatakeyama H; Hyodo M; Harashima H Relationship Between the Physicochemical Properties of Lipid Nanoparticles and the Quality of siRNA Delivery to Liver Cells. Mol. Ther 2016, 24, 788–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (560).Yamamoto N; Sato Y; Munakata T; Kakuni M; Tateno C; Sanada T; Hirata Y; Murakami S; Tanaka Y; Chayama K; et al. Novel pH-Sensitive Multifunctional Envelope-Type Nanodevice for siRNA-Based Treatments for Chronic HBV Infection. J. Hepatol 2016, 64, 547–555. [DOI] [PubMed] [Google Scholar]
- (561).Warashina S; Nakamura T; Sato Y; Fujiwara Y; Hyodo M; Hatakeyama H; Harashima H A Lipid Nanoparticle for the Efficient Delivery of siRNA to Dendritic Cells. J. Controlled Release 2016, 225, 183–191. [DOI] [PubMed] [Google Scholar]
- (562).Sato Y; Hashiba K; Sasaki K; Maeki M; Tokeshi M; Harashima H Understanding Structure-Activity Relationships of pH-Sensitive Cationic Lipids Facilitates the Rational Identification of Promising Lipid Nanoparticles for Delivering siRNAs In Vivo. J. Controlled Release 2019, 295, 140–152. [DOI] [PubMed] [Google Scholar]
- (563).Sakurai Y; Hatakeyama H; Sato Y; Hyodo M; Akita H; Harashima H Gene Silencing via RNAi and siRNA Quantification in Tumor Tissue Using MEND, a Liposomal siRNA Delivery System. Mol. Ther 2013, 21, 1195–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (564).Watanabe T; Hatakeyama H; Matsuda-Yasui C; Sato Y; Sudoh M; Takagi A; Hirata Y; Ohtsuki T; Arai M; Inoue K; et al. In Vivo Therapeutic Potential of Dicer-Hunting siRNAs Targeting Infectious Hepatitis C Virus. Sci. Rep 2014, 4, 4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (565).Younis MA; Khalil IA; Abd Elwakil MM; Harashima H A Multifunctional Lipid-Based Nanodevice for the Highly Specific Codelivery of Sorafenib and Midkine siRNA to Hepatic Cancer Cells. Mol. Pharmaceutics 2019, 16, 4031–4044. [DOI] [PubMed] [Google Scholar]
- (566).Gindy ME; Feuston B; Glass A; Arrington L; Haas RM; Schariter J; Stirdivant SM Stabilization of Ostwald Ripening in Low Molecular Weight Amino Lipid Nanoparticles for Systemic Delivery of siRNA Therapeutics. Mol. Pharmaceutics 2014, 11, 4143–4153. [DOI] [PubMed] [Google Scholar]
- (567).Suzuki Y; Hyodo K; Tanaka Y; Ishihara H siRNA-Lipid Nanoparticles with Long-Term Storage Stability Facilitate Potent Gene-Silencing In Vivo. J. Controlled Release 2015, 220, 44–50. [DOI] [PubMed] [Google Scholar]
- (568).Suzuki Y; Ishihara H Structure, activity and uptake mechanism of siRNA-lipid nanoparticles with an asymmetric ionizable lipid. Int. J. Pharm 2016, 510, 350–358. [DOI] [PubMed] [Google Scholar]
- (569).Suzuki Y; Hyodo K; Suzuki T; Tanaka Y; Kikuchi H; Ishihara H Biodegradable Lipid Nanoparticles Induce a Prolonged RNA Interference-Mediated Protein Knockdown and Show Rapid Hepatic Clearance in Mice and Nonhuman Primates. Int. J. Pharm 2017, 519, 34–43. [DOI] [PubMed] [Google Scholar]
- (570).Viricel W; Poirier S; Mbarek A; Derbali RM; Mayer G; Leblond J Cationic Switchable Lipids: pH-Triggered Molecular Switch for siRNA Delivery. Nanoscale 2017, 9, 31–36. [DOI] [PubMed] [Google Scholar]
- (571).Miyaura N; Yamada K; Suzuki A A New Stereospecific Cross-Coupling by the Palladium-Catalyzed Reaction of 1-Alkenylboranes with 1-Alkenyl or 1-Alkynyl Halides. Tetrahedron Lett 1979, 20, 3437–3440. [Google Scholar]
- (572).Miyaura N; Suzuki A Stereoselective Synthesis of Arylated (E)-Alkenes by the Reaction of Alk-1-enylboranes with Aryl halides in the Presence of Palladium Catalyst. J. Chem. Soc., Chem. Commun 1979, 19, 866–867. [Google Scholar]
- (573).Sonogashira K; Tohda Y; Hagihara N A Convenient Synthesis of Acetylenes: Catalytic Substitutions of Acetylenic Hydrogen with Bromoalkenes, Iodoarenes and Bromopyridines. Tetrahedron Lett 1975, 16, 4467–4470. [Google Scholar]
- (574).Viricel W; Mbarek A; Leblond J Switchable Lipids: Conformational Change for Fast pH-Triggered Cytoplasmic Delivery. Angew. Chem., Int. Ed 2015, 54, 12743–12747. [DOI] [PubMed] [Google Scholar]
- (575).Wijagkanalan W; Kawakami S; Takenaga M; Igarashi R; Yamashita F; Hashida M Efficient Targeting to Alveolar Macrophages by Intratracheal Administration of Mannosylated Liposomes in Rats. J. Controlled Release 2008, 125, 121–130. [DOI] [PubMed] [Google Scholar]
- (576).Zhang C; Zhang X; Zhao W; Zeng C; Li W; Li B; Luo X; Li J; Jiang J; Deng B; et al. Chemotherapy Drugs Derived Nanoparticles Encapsulating mRNA Encoding Tumor Suppressor Proteins to Treat Triple-Negative Breast Cancer. Nano Res 2019, 12, 855–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (577).Zhang C; Zhao W; Bian C; Hou X; Deng B; McComb DW; Chen X; Dong Y Antibiotic-Derived Lipid Nanoparticles to Treat Intracellular Staphylococcus aureus. ACS Appl. Bio Mater 2019, 2, 1270–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (578).Semple SC; Chonn A; Cullis PR Influence of Cholesterol on the Association of Plasma Proteins with Liposomes. Biochemistry 1996, 35, 2521–2525. [DOI] [PubMed] [Google Scholar]
- (579).Briuglia M-L; Rotella C; McFarlane A; Lamprou DA Influence of Cholesterol on Liposome Stability and on In Vitro Drug Release. Drug Delivery Transl. Res 2015, 5, 231–242. [DOI] [PubMed] [Google Scholar]
- (580).Lee J; Saw PE; Gujrati V; Lee Y; Kim H; Kang S; Choi M; Kim J-I; Jon S Mono-arginine Cholesterol-based Small Lipid Nanoparticles as a Systemic siRNA Delivery Platform for Effective Cancer Therapy. Theranostics 2016, 6, 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (581).Gao X; Huang L A novel cationic liposome reagent for efficient transfection of mammalian cells. Biochem. Biophys. Res. Commun 1991, 179, 280–285. [DOI] [PubMed] [Google Scholar]
- (582).Zuidam NJ; Barenholz Y Electrostatic Parameters of Cationic Liposomes Commonly Used for Gene Delivery as Determined by 4-Heptadecyl-7-Hydroxycoumarin. Biochim. Biophys. Acta, Biomembr 1997, 1329, 211–222. [DOI] [PubMed] [Google Scholar]
- (583).Ajmani PS; Hughes JA 3β [N-(N′, N′-Dimethylaminoethane)-Carbamoyl] Cholesterol (DC-Chol)-Mediated Gene Delivery to Primary Rat Neurons: Characterization and Mechanism. Neurochem. Res 1999, 24, 699–703. [DOI] [PubMed] [Google Scholar]
- (584).Zhang Y; Li H; Sun J; Gao J; Liu W; Li B; Guo Y; Chen J DC-Chol/DOPE Cationic Liposomes: A Comparative Study of the Influence Factors on Plasmid pDNA and siRNA Gene Delivery. Int. J. Pharm 2010, 390, 198–207. [DOI] [PubMed] [Google Scholar]
- (585).Lee J; Ahn HJ PEGylated DC-Chol/DOPE Cationic Liposomes Containing KSP siRNA as a Systemic siRNA Delivery Carrier for Ovarian Cancer Therapy. Biochem. Biophys. Res. Commun 2018, 503, 1716–1722. [DOI] [PubMed] [Google Scholar]
- (586).Michel T; Luft D; Abraham M-K; Reinhardt S; Salinas Medina ML; Kurz J; Schaller M; Avci-Adali M; Schlensak C; Peter K; et al. Cationic Nanoliposomes Meet mRNA: Efficient Delivery of Modified mRNA Using Hemocompatible and Stable Vectors for Therapeutic Applications. Mol. Ther.–Nucleic Acids 2017, 8, 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (587).Zhao YC; Zhang L; Feng SS; Hong L; Zheng HL; Chen LL; Zheng XL; Ye YQ; Zhao MD; Wang WX; et al. Efficient Delivery of Notch1 siRNA to SKOV3 Cells by Cationic Cholesterol Derivative-Based Liposome. Int. J. Nanomed 2016, 11, 5485–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (588).Dong Y; Zhang X Biomimetic Nanomaterials and Uses Thereof WO2019027999 A1, Jul. 2, 2019. [Google Scholar]
- (589).Akinc A; Zumbuehl A; Goldberg M; Leshchiner ES; Busini V; Hossain N; Bacallado SA; Nguyen DN; Fuller J; Alvarez R; et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol 2008, 26, 561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (590).Love KT; Mahon KP; Levins CG; Whitehead KA; Querbes W; Dorkin JR; Qin J; Cantley W; Qin LL; Racie T; et al. Lipid-Like Materials for Low-Dose, In Vivo Gene Silencing. Proc. Natl. Acad. Sci. U. S. A 2010, 107, 1864–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (591).Ball RL; Knapp CM; Whitehead KA Lipidoid Nanoparticles for siRNA Delivery to the Intestinal Epithelium: In Vitro Investigations in a Caco-2 Model. PLoS One 2015, 10, No. e0133154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (592).Altınoglu S; Wang M; Xu Q Combinatorial Library Strategies for Synthesis of Cationic Lipid-Like Nanoparticles and Their Potential Medical Applications. Nanomedicine 2015, 10, 643–657. [DOI] [PubMed] [Google Scholar]
- (593).Cho SW; Goldberg M; Son SM; Xu QB; Yang F; Mei Y; Bogatyrev S; Langer R; Anderson DG Lipid-Like Nanoparticles for Small Interfering RNA Delivery to Endothelial Cells. Adv. Funct. Mater 2009, 19, 3112–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (594).Mahon KP; Love KT; Whitehead KA; Qin J; Akinc A; Leshchiner E; Leshchiner I; Langer R; Anderson DG Combinatorial Approach to Determine Functional Group Effects on Lipidoid-Mediated siRNA Delivery. Bioconjugate Chem 2010, 21, 1448–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (595).Chen D; Love KT; Chen Y; Eltoukhy AA; Kastrup C; Sahay G; Jeon A; Dong Y; Whitehead KA; Anderson DG Rapid Discovery of Potent siRNA-Containing Lipid Nanoparticles Enabled by Controlled Microfluidic Formulation. J. Am. Chem. Soc 2012, 134, 6948–6951. [DOI] [PubMed] [Google Scholar]
- (596).Li Y; Yang T; Yu Y; Shi N; Yang L; Glass Z; Bolinger J; Finkel IJ; Li W; Xu Q Combinatorial Library of Chalcogen-Containing Lipidoids for Intracellular Delivery of Genome-Editing Proteins. Biomaterials 2018, 178, 652–662. [DOI] [PubMed] [Google Scholar]
- (597).Zhang X; Zhao W; Nguyen GN; Zhang C; Zeng C; Yan J; Du S; Hou X; Li W; Jiang J; et al. Functionalized Lipid-Like Nanoparticles for In Vivo mRNA Delivery and Base Editing. Sci. Adv 2020, 6, No. eabc2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (598).Li B; Luo X; Deng B; Wang J; McComb DW; Shi Y; Gaensler KML; Tan X; Dunn AL; Kerlin BA; et al. An Orthogonal Array Optimization of Lipid-like Nanoparticles for mRNA Delivery in Vivo. Nano Lett 2015, 15, 8099–8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (599).Zhang X; Li B; Luo X; Zhao W; Jiang J; Zhang C; Gao M; Chen X; Dong Y Biodegradable Amino-Ester Nanomaterials for Cas9 mRNA Delivery in Vitro and in Vivo. ACS Appl. Mater. Interfaces 2017, 9, 25481–25487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (600).Alabi CA; Love KT; Sahay G; Yin H; Luly KM; Langer R; Anderson DG Multiparametric Approach for the Evaluation of Lipid Nanoparticles for siRNA Delivery. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 12881–12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (601).Li L; Zahner D; Su Y; Gruen C; Davidson G; Levkin PA A Biomimetic Lipid Library for Gene Delivery Through Thiol-yne Click Chemistry. Biomaterials 2012, 33, 8160–8166. [DOI] [PubMed] [Google Scholar]
- (602).Molla MR; Chakraborty S; Munoz Sagredo L; Drechsler M; Orian Rousseau V; Levkin PA Combinatorial Synthesis of a Lipidoid Library by Thiolactone Chemistry: In Vitro Screening and In Vivo Validation for siRNA Delivery. Bioconjugate Chem 2020, 31, 852–860. [DOI] [PubMed] [Google Scholar]
- (603).Miao L; Li L; Huang Y; Delcassian D; Chahal J; Han J; Shi Y; Sadtler K; Gao W; Lin J; et al. Delivery of mRNA Vaccines with Heterocyclic Lipids Increases Anti-Tumor Efficacy by STING-Mediated Immune Cell Activation. Nat. Biotechnol 2019, 37, 1174–1185. [DOI] [PubMed] [Google Scholar]
- (604).Molla MR; Böser A; Rana A; Schwarz K; Levkin PA One-Pot Parallel Synthesis of Lipid Library via Thiolactone Ring Opening and Screening for Gene Delivery. Bioconjugate Chem 2018, 29, 992–999. [DOI] [PubMed] [Google Scholar]
- (605).Nguyen DN; Chen SCY; Lu J; Goldberg M; Kim P; Sprague A; Novobrantseva T; Sherman J; Shulga-Morskaya S; de Fougerolles A; et al. Drug Delivery-mediated Control of RNA Immunostimulation. Mol. Ther 2009, 17, 1555–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (606).Svensson RU; Shey MR; Ballas ZK; Dorkin JR; Goldberg M; Akinc A; Langer R; Anderson DG; Bumcrot D; Henry MD Assessing siRNA Pharmacodynamics in a Luciferase-expressing Mouse. Mol. Ther 2008, 16, 1995–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (607).Akinc A; Goldberg M; Qin J; Dorkin JR; Gamba-Vitalo C; Maier M; Jayaprakash KN; Jayaraman M; Rajeev KG; Manoharan M; et al. Development of Lipidoid-siRNA Formulations for Systemic Delivery to the Liver. Mol. Ther 2009, 17, 872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (608).Frank-Kamenetsky M; Grefhorst A; Anderson NN; Racie TS; Bramlage B; Akinc A; Butler D; Charisse K; Dorkin R; Fan Y; et al. Therapeutic RNAi Targeting PCSK9 Acutely Lowers Plasma Cholesterol in Rodents and LDL Cholesterol in Nonhuman Primates. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 11915–11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (609).Whitehead KA; Dorkin JR; Vegas AJ; Chang PH; Veiseh O; Matthews J; Fenton OS; Zhang Y; Olejnik KT; Yesilyurt V; et al. Degradable Lipid Nanoparticles with Predictable In Vivo siRNA Delivery Activity. Nat. Commun 2014, 5, 4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (610).Knapp CM; Guo PH; Whitehead KA Lipidoid Tail Structure Strongly Influences siRNA Delivery Activity. Cell. Mol. Bioeng 2016, 9, 305–314. [Google Scholar]
- (611).Ball RL; Hajj KA; Vizelman J; Bajaj P; Whitehead KA Lipid Nanoparticle Formulations for Enhanced Co-delivery of siRNA and mRNA. Nano Lett 2018, 18, 3814–3822. [DOI] [PubMed] [Google Scholar]
- (612).Hajj KA; Ball RL; Deluty SB; Singh SR; Strelkova D; Knapp CM; Whitehead KA Branched-Tail Lipid Nanoparticles Potently Deliver mRNA In Vivo due to Enhanced Ionization at Endosomal pH. Small 2019, 15, 1805097. [DOI] [PubMed] [Google Scholar]
- (613).Hajj KA; Melamed JR; Chaudhary N; Lamson NG; Ball RL; Yerneni SS; Whitehead KA A Potent Branched-Tail Lipid Nanoparticle Enables Multiplexed mRNA Delivery and Gene Editing In Vivo. Nano Lett 2020, 20, 5167–5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (614).Zhao X; Chen J; Qiu M; Li Y; Glass Z; Xu Q Imidazole-Based Synthetic Lipidoids for In Vivo mRNA Delivery into Primary T Lymphocytes. Angew. Chem., Int. Ed 2020, 59, 20083–20089. [DOI] [PubMed] [Google Scholar]
- (615).Ma F; Yang L; Sun Z; Chen J; Rui X; Glass Z; Xu Q Neurotransmitter-Derived Lipidoids (NT-lipidoids) for Enhanced Brain Delivery Through Intravenous Injection. Sci. Adv 2020, 6, No. eabb4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (616).Liu F; Yang L; Li Y; Junier A; Ma F; Chen J; Han H; Glass Z; Zhao X; Kumamoto CA; et al. In Vitro and In Vivo Study of Amphotericin B Formulation with Quaternized Bioreducible Lipidoids. ACS Biomater. Sci. Eng 2020, 6, 1064–1073. [DOI] [PubMed] [Google Scholar]
- (617).Li Y; Jarvis R; Zhu K; Glass Z; Ogurlu R; Gao P; Li P; Chen J; Yu Y; Yang Y; et al. Protein and mRNA Delivery Enabled by Cholesteryl-Based Biodegradable Lipidoid Nanoparticles. Angew. Chem., Int. Ed 2020, 59, 14957–14964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (618).Liu J; Chang J; Jiang Y; Meng X; Sun T; Mao L; Xu Q; Wang M Fast and Efficient CRISPR/Cas9 Genome Editing In Vivo Enabled by Bioreducible Lipid and Messenger RNA Nanoparticles. Adv. Mater 2019, 31, 1902575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (619).Sui L; Wang M; Han Q; Yu L; Zhang L; Zheng L; Lian J; Zhang J; Valverde P; Xu Q; et al. A Novel Lipidoid-MicroRNA Formulation Promotes Calvarial Bone Regeneration. Biomaterials 2018, 177, 88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (620).Wang M; Alberti K; Sun S; Arellano CL; Xu Q Combinatorially Designed Lipid-like Nanoparticles for Intracellular Delivery of Cytotoxic Protein for Cancer Therapy. Angew. Chem., Int. Ed 2014, 53, 2893–2898. [DOI] [PubMed] [Google Scholar]
- (621).Qiu M; Glass Z; Chen J; Haas M; Jin X; Zhao X; Rui X; Ye Z; Li Y; Zhang F; et al. Lipid Nanoparticle-Mediated Codelivery of Cas9 mRNA and Single-Guide RNA Achieves Liver-Specific In Vivo Genome Editing of Angptl3. Proc. Natl. Acad. Sci. U. S. A 2021, 118, No. e2020401118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (622).Wang M; Zuris JA; Meng FT; Rees H; Sun S; Deng P; Han Y; Gao X; Pouli D; Wu Q; et al. Efficient Delivery of Genome-Editing Proteins Using Bioreducible Lipid Nanoparticles. Proc. Natl. Acad. Sci. U. S. A 2016, 113, 2868–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (623).Yang L; Ma F; Liu F; Chen J; Zhao X; Xu Q Efficient Delivery of Antisense Oligonucleotides Using Bioreducible Lipid Nanoparticles In Vitro and In Vivo. Mol. Ther.–Nucleic Acids 2020, 19, 1357–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (624).Paley EL Tryptamine-Induced Tryptophanyl-tRNA trp Deficiency in Neurodifferentiation and Neurodegeneration Interplay: Progenitor Activation with Neurite Growth Terminated in Alzheimer’s Disease Neuronal Vesicularization and Fragmentation. J. Alzheimer’s Dis 2011, 26, 263–298. [DOI] [PubMed] [Google Scholar]
- (625).Mosnaim AD; Callaghan OH; Hudzik T; Wolf ME Rat Brain-Uptake Index for Phenylethylamine and Various Monomethylated Derivatives. Neurochem. Res 2013, 38, 842–846. [DOI] [PubMed] [Google Scholar]
- (626).Zhou K; Nguyen LH; Miller JB; Yan Y; Kos P; Xiong H; Li L; Hao J; Minnig JT; Zhu H; et al. Modular Degradable Dendrimers Enable Small RNAs to Extend Survival in an Aggressive Liver Cancer Model. Proc. Natl. Acad. Sci. U. S. A 2016, 113, 520–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (627).Cheng Q; Wei T; Jia Y; Farbiak L; Zhou K; Zhang S; Wei Y; Zhu H; Siegwart DJ Dendrimer-Based Lipid Nanoparticles Deliver Therapeutic FAH mRNA to Normalize Liver Function and Extend Survival in a Mouse Model of Hepatorenal Tyrosinemia Type I. Adv. Mater 2018, 30, 1805308. [DOI] [PubMed] [Google Scholar]
- (628).Sahay G; Querbes W; Alabi C; Eltoukhy A; Sarkar S; Zurenko C; Karagiannis E; Love K; Chen D; Zoncu R; et al. Efficiency of siRNA Delivery by Lipid Nanoparticles is Limited by Endocytic Recycling. Nat. Biotechnol 2013, 31, 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (629).Kauffman KJ; Dorkin JR; Yang JH; Heartlein MW; DeRosa F; Mir FF; Fenton OS; Anderson DG Optimization of Lipid Nanoparticle Formulations for mRNA Delivery in Vivo with Fractional Factorial and Definitive Screening Designs. Nano Lett 2015, 15, 7300–7306. [DOI] [PubMed] [Google Scholar]
- (630).DeRosa F; Smith L; Shen Y; Huang Y; Pan J; Xie H; Yahalom B; Heartlein MW Improved Efficacy in a Fabry Disease Model Using a Systemic mRNA Liver Depot System as Compared to Enzyme Replacement Therapy. Mol. Ther 2019, 27, 878–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (631).Turnbull IC; Eltoukhy AA; Fish KM; Nonnenmacher M; Ishikawa K; Chen J; Hajjar RJ; Anderson DG; Costa KD Myocardial Delivery of Lipidoid Nanoparticle Carrying modRNA Induces Rapid and Transient Expression. Mol. Ther 2016, 24, 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (632).Xu X; Xie K; Zhang X-Q; Pridgen EM; Park GY; Cui DS; Shi J; Wu J; Kantoff PW; Lippard SJ; et al. Enhancing Tumor Cell Response to Chemotherapy Through Nanoparticle-Mediated Codelivery of siRNA and Cisplatin Prodrug. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 18638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (633).Kong N; Tao W; Ling X; Wang J; Xiao Y; Shi S; Ji X; Shajii A; Gan ST; Kim NY; et al. Synthetic mRNA Nanoparticle-Mediated Restoration of p53 Tumor Suppressor Sensitizes p53-Deficient Cancers to mTOR Inhibition. Sci. Transl. Med 2019, 11, No. eaaw1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (634).Islam MA; Rice J; Reesor E; Zope H; Tao W; Lim M; Ding J; Chen Y; Aduluso D; Zetter BR; et al. Adjuvant-Pulsed mRNA Caccine Nanoparticle for Immunoprophylactic and Therapeutic Tumor Suppression in Mice. Biomaterials 2021, 266, 120431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (635).Busscher GF; Rutjes FPJT; van Delft FL 2-Deoxystreptamine: Central Scaffold of Aminoglycoside Antibiotics. Chem. Rev 2005, 105, 775–792. [DOI] [PubMed] [Google Scholar]
- (636).Fourmy D; Recht MI; Blanchard SC; Puglisi JD Structure of the A Site of Escherichia Coli 16S Ribosomal RNA Complexed with an Aminoglycoside Antibiotic. Science 1996, 274, 1367–1371. [DOI] [PubMed] [Google Scholar]
- (637).Arya DP; Xue L; Willis B Aminoglycoside (Neomycin) Preference Is for A-Form Nucleic Acids, Not Just RNA: Results from a Competition Dialysis Study. J. Am. Chem. Soc 2003, 125, 10148–10149. [DOI] [PubMed] [Google Scholar]
- (638).Ecker DJ; Griffey RH RNA as a small-molecule drug target: doubling the value of genomics. Drug Discovery Today 1999, 4, 420–429. [DOI] [PubMed] [Google Scholar]
- (639).Mingeot-Leclercq M-P; Glupczynski Y; Tulkens PM Aminoglycosides: Activity and Resistance. Antimicrob. Agents Chemother 1999, 43, 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (640).Zhang Y; Pelet JM; Heller DA; Dong Y; Chen D; Gu Z; Joseph BJ; Wallas J; Anderson DG Lipid-Modified Aminoglycoside Derivatives for In Vivo siRNA Delivery. Adv. Mater 2013, 25, 4641–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (641).Yu X; Liu S; Cheng Q; Wei T; Lee S; Zhang D; Siegwart DJ Lipid-Modified Aminoglycosides for mRNA Delivery to the Liver. Adv. Healthcare Mater 2020, 9, No. e1901487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (642).Dong Y; Eltoukhy AA; Alabi CA; Khan OF; Veiseh O; Dorkin JR; Sirirungruang S; Yin H; Tang BC; Pelet JM; et al. Lipid-Like Nanomaterials for Simultaneous Gene Expression and Silencing In Vivo. Adv. Healthcare Mater 2014, 3, 1392–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (643).Li B; Luo X; Deng B; Giancola JB; McComb DW; Schmittgen TD; Dong Y Effects of Local Structural Transformation of Lipid-Like Compounds on Delivery of Messenger RNA. Sci. Rep 2016, 6, 22137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (644).Yin H; Song C-Q; Suresh S; Wu Q; Walsh S; Rhym LH; Mintzer E; Bolukbasi MF; Zhu LJ; Kauffman K; et al. Structure-Guided Chemical Modification of Guide RNA Enables Potent Non-Viral In Vivo Genome Editing. Nat. Biotechnol 2017, 35, 1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (645).Lokugamage MP; Gan Z; Zurla C; Levin J; Islam FZ; Kalathoor S; Sato M; Sago CD; Santangelo PJ; Dahlman JE Mild Innate Immune Activation Overrides Efficient Nanoparticle-Mediated RNA Delivery. Adv. Mater 2020, 32, 1904905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (646).Bartke N; Hannun YA Bioactive Sphingolipids: Metabolism and Function. J. Lipid Res 2009, 50, S91–S96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (647).Hannun YA; Obeid LM Principles of Bioactive Lipid Signalling: Lessons from Sphingolipids. Nat. Rev. Mol. Cell Biol 2008, 9, 139–150. [DOI] [PubMed] [Google Scholar]
- (648).Fenton OS; Kauffman KJ; McClellan RL; Appel EA; Dorkin JR; Tibbitt MW; Heartlein MW; DeRosa F; Langer R; Anderson DG Bioinspired Alkenyl Amino Alcohol Ionizable Lipid Materials for Highly Potent In Vivo mRNA Delivery. Adv. Mater 2016, 28, 2939–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (649).Halland N; Braunton A; Bachmann S; Marigo M; Jørgensen KA Direct Organocatalytic Asymmetric α-Chlorination of Aldehydes. J. Am. Chem. Soc 2004, 126, 4790–4791. [DOI] [PubMed] [Google Scholar]
- (650).Fenton OS; Kauffman KJ; Kaczmarek JC; McClellan RL; Jhunjhunwala S; Tibbitt MW; Zeng MD; Appel EA; Dorkin JR; Mir FF; et al. Synthesis and Biological Evaluation of Ionizable Lipid Materials for the In Vivo Delivery of Messenger RNA to B Lymphocytes. Adv. Mater 2017, 29, 1606944. [DOI] [PubMed] [Google Scholar]
- (651).Geng Y; Discher DE Hydrolytic Degradation of Poly-(ethylene oxide)-block-Polycaprolactone Worm Micelles. J. Am. Chem. Soc 2005, 127, 12780–12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (652).Fenton OS; Kauffman KJ; McClellan RL; Kaczmarek JC; Zeng MHD; Andresen JL; Rhym LH; Heartlein MW; DeRosa F; Anderson DG Customizable Lipid Nanoparticle Materials for the Delivery of siRNAs and mRNAs. Angew. Chem., Int. Ed 2018, 57, 13582–13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (653).Billingsley MM; Singh N; Ravikumar P; Zhang R; June CH; Mitchell MJ Ionizable Lipid Nanoparticle-Mediated mRNA Delivery for Human CAR T Cell Engineering. Nano Lett 2020, 20, 1578–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (654).Fairbanks BD; Scott TF; Kloxin CJ; Anseth KS; Bowman CN Thiol-Yne Photopolymerizations: Novel Mechanism, Kinetics, and Step-Growth Formation of Highly Cross-Linked Networks. Macromolecules 2009, 42, 211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (655).Hoogenboom R Thiol-Yne Chemistry: A Powerful Tool for Creating Highly Functional Materials. Angew. Chem., Int. Ed 2010, 49, 3415–3417. [DOI] [PubMed] [Google Scholar]
- (656).Hulme C; Gore V Multi-component Reactions: Emerging Chemistry in Drug Discovery“‘From Xylocain to Crixivan’. Curr. Med. Chem 2003, 10, 51–80. [DOI] [PubMed] [Google Scholar]
- (657).Koopmanschap G; Ruijter E; Orru RVA Isocyanide-Based Multicomponent Reactions Towards Cyclic Constrained Peptidomimetics. Beilstein J. Org. Chem 2014, 10, 544–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (658).Tanaka Y; Hasui T; Suginome M Acid-Free, Aminoborane-Mediated Ugi-Type Reaction Leading to General Utilization of Secondary Amines. Org. Lett 2007, 9, 4407–4410. [DOI] [PubMed] [Google Scholar]
- (659).Kazmaier U; Ackermann S A Straightforward Approach Towards Thiazoles and Endothiopeptides via Ugi Reaction. Org. Biomol. Chem 2005, 3, 3184–3187. [DOI] [PubMed] [Google Scholar]
- (660).Pan SC; List B Catalytic Three-Component Ugi Reaction. Angew. Chem., Int. Ed 2008, 47, 3622–3625. [DOI] [PubMed] [Google Scholar]
- (661).Sonoke S; Ueda T; Fujiwara K; Kuwabara K; Yano J Galactose-Modified Cationic Liposomes as a Liver-Targeting Delivery System for Small Interfering RNA. Biol. Pharm. Bull 2011, 34, 1338–1342. [DOI] [PubMed] [Google Scholar]
- (662).Walsh CL; Nguyen J; Szoka FC Synthesis and Characterization of Novel Zwitterionic Lipids with pH-Responsive Biophysical Properties. Chem. Commun 2012, 48, 5575–5577. [DOI] [PubMed] [Google Scholar]
- (663).Patel S; Ryals RC; Weller KK; Pennesi ME; Sahay G Lipid Nanoparticles for Delivery of Messenger RNA to the Back of the Eye. J. Controlled Release 2019, 303, 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (664).Walsh CL; Nguyen J; Tiffany MR; Szoka FC Synthesis, Characterization, and Evaluation of Ionizable Lysine-Based Lipids for siRNA Delivery. Bioconjugate Chem 2013, 24, 36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (665).Miller JB; Zhang S; Kos P; Xiong H; Zhou K; Perelman SS; Zhu H; Siegwart DJ Non-Viral CRISPR/Cas Gene Editing In Vitro and In Vivo Enabled by Synthetic Nanoparticle Co-Delivery of Cas9 mRNA and sgRNA. Angew. Chem., Int. Ed 2017, 56, 1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (666).Hirai Y; Saeki R; Song F; Koide H; Fukata N; Tomita K; Maeda N; Oku N; Asai T Charge-Reversible Lipid Derivative: A Novel Type of pH-Responsive Lipid for Nanoparticle-Mediated siRNA Delivery. Int. J. Pharm 2020, 585, 119479. [DOI] [PubMed] [Google Scholar]
- (667).Liu S; Cheng Q; Wei T; Yu X; Johnson LT; Farbiak L; Siegwart DJ Membrane-Destabilizing Ionizable Phospholipids for Organ-Selective mRNA Delivery and CRISPR-Cas Gene Editing. Nat. Mater 2021, 20, 701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (668).Wang D; Tu C; Su Y; Zhang C; Greiser U; Zhu X; Yan D; Wang W Supramolecularly Engineered Phospholipids Constructed by Nucleobase Molecular Recognition: Upgraded Generation of Phospholipids for Drug Delivery. Chem. Sci 2015, 6, 3775–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (669).Menger FM; Peresypkin AV A Combinatorially-Derived Structural Phase Diagram for 42 Zwitterionic Geminis. J. Am. Chem. Soc 2001, 123, 5614–5615. [DOI] [PubMed] [Google Scholar]
- (670).Wei T; Cheng Q; Min Y-L; Olson EN; Siegwart DJ Systemic Nanoparticle Delivery of CRISPR-Cas9 Ribonucleoproteins for Effective Tissue Specific Genome Editing. Nat. Commun 2020, 11, 3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (671).Cheng Q; Wei T; Farbiak L; Johnson LT; Dilliard SA; Siegwart DJ Selective Organ Targeting (SORT) Nanoparticles for Tissue-Specific mRNA Delivery and CRISPR-Cas Gene Editing. Nat. Nanotechnol 2020, 15, 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (672).Kim JB; Lee YM; Ryu J; Lee E; Kim WJ; Keum G; Bang EK Coordinative Amphiphiles as Tunable siRNA Transporters. Bioconjugate Chem 2016, 27, 1850–1856. [DOI] [PubMed] [Google Scholar]
- (673).Tai WY; Gao XH Noncovalent Tagging of siRNA with Steroids for Transmembrane Delivery. Biomaterials 2018, 178, 720–727. [DOI] [PubMed] [Google Scholar]
- (674).Thewalt JL; Bloom M Phosphatidylcholine: Cholesterol Phase Diagrams. Biophys. J 1992, 63, 1176–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (675).Reynolds A; Leake D; Boese Q; Scaringe S; Marshall WS; Khvorova A Rational siRNA Design for RNA Interference. Nat. Biotechnol 2004, 22, 326–330. [DOI] [PubMed] [Google Scholar]
- (676).Landen CN; Kinch MS; Sood AK EphA2 as a Target for Ovarian Cancer Therapy. Expert Opin. Ther. Targets 2005, 9, 1179–1187. [DOI] [PubMed] [Google Scholar]
- (677).Landen CN; Chavez-Reyes A; Bucana C; Schmandt R; Deavers MT; Lopez-Berestein G; Sood AK Therapeutic EphA2 Gene Targeting In Vivo Using Neutral Liposomal Small Interfering RNA Delivery. Cancer Res 2005, 65, 6910. [DOI] [PubMed] [Google Scholar]
- (678).Maric S; Thygesen MB; Schiller J; Marek M; Moulin M; Haertlein M; Forsyth VT; Bogdanov M; Dowhan W; Arleth L Biosynthetic Preparation of Selectively Deuterated Phosphatidylcholine in Genetically Modified Escherichia Coli. Appl. Microbiol. Biotechnol 2015, 99, 241–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (679).Kim J; Jozic A; Sahay G Naturally Derived Membrane Lipids Impact Nanoparticle-Based Messenger RNA Delivery. Cell. Mol. Bioeng 2020, 13, 463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (680).Trang P; Wiggins JF; Daige CL; Cho C; Omotola M; Brown D; Weidhaas JB; Bader AG; Slack FJ Systemic Delivery of Tumor Suppressor microRNA Mimics Using a Neutral Lipid Emulsion Inhibits Lung Tumors in Mice. Mol. Ther 2011, 19, 1116–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (681).Sugrue RJ; Hay AJ Structural Characteristics of the M2 Protein of Influenza a Viruses: Evidence That it Forms a Tetrameric Channe. Virology 1991, 180, 617–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (682).Liu J; Obando D; Liao V; Lifa T; Codd R The Many Faces of the Adamantyl Group in Drug Design. Eur. J. Med. Chem 2011, 46, 1949–1963. [DOI] [PubMed] [Google Scholar]
- (683).Štimac A; Šekutor M; Mlinarić-Majerski K; Frkanec L; Frkanec R Adamantane in Drug Delivery Systems and Surface Recognition. Molecules 2017, 22, 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (684).Gan Z; Lokugamage MP; Hatit MZC; Loughrey D; Paunovska K; Sato M; Cristian A; Dahlman JE Nanoparticles Containing Constrained Phospholipids Deliver mRNA to Liver Immune Cells In Vivo Without Targeting Ligands. Bioeng. Transl. Med 2020, 5, No. e10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (685).Urban P; Pritzl SD; Konrad DB; Frank JA; Pernpeintner C; Roeske CR; Trauner D; Lohmüller T Light-Controlled Lipid Interaction and Membrane Organization in Photolipid Bilayer Vesicles. Langmuir 2018, 34, 13368–13374. [DOI] [PubMed] [Google Scholar]
- (686).Neises B; Steglich W Simple Method for the Esterification of Carboxylic Acids. Angew. Chem., Int. Ed. Engl 1978, 17, 522–524. [Google Scholar]
- (687).Sago CD; Lokugamage MP; Paunovska K; Vanover DA; Monaco CM; Shah NN; Gamboa Castro M; Anderson SE; Rudoltz TG; Lando GN; et al. High-Throughput In Vivo Screen of Functional mRNA Delivery Identifies Nanoparticles for Endothelial Cell Gene Editing. Proc. Natl. Acad. Sci. U. S. A 2018, 115, E9944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (688).Ciani L; Ristori S; Salvati A; Calamai L; Martini G DOTAP/DOPE and DC-Chol/DOPE Lipoplexes for Gene Delivery: Zeta Potential Measurements and Electron Spin Resonance Spectra. Biochim. Biophys. Acta, Biomembr 2004, 1664, 70–79. [DOI] [PubMed] [Google Scholar]
- (689).Hattori Y; Suzuki S; Kawakami S; Yamashita F; Hashida M The role of Dioleoylphosphatidylethanolamine (DOPE) in Targeted Gene Delivery with Mannosylated Cationic Liposomes via Intravenous Route. J. Controlled Release 2005, 108, 484–495. [DOI] [PubMed] [Google Scholar]
- (690).Harvey RD; Ara N; Heenan RK; Barlow DJ; Quinn PJ; Lawrence MJ Stabilization of Distearoylphosphatidylcholine Lamellar Phases in Propylene Glycol Using Cholesterol. Mol. Pharmaceutics 2013, 10, 4408–4417. [DOI] [PubMed] [Google Scholar]
- (691).Fasbender A; Marshall J; Moninger TO; Grunst T; Cheng S; Welsh MJ Effect of Co-lipids in Enhancing Cationic Lipid-Mediated Gene Transfer In Vitro and In Vivo. Gene Ther 1997, 4, 716–725. [DOI] [PubMed] [Google Scholar]
- (692).Farhood H; Serbina N; Huang L The role of Dioleoyl Phosphatidylethanolamine in Cationic Liposome Mediated Gene Transfer. Biochim. Biophys. Acta, Biomembr 1995, 1235, 289–295. [DOI] [PubMed] [Google Scholar]
- (693).Harper PE; Mannock DA; Lewis RNAH; McElhaney RN; Gruner SM X-Ray Diffraction Structures of Some Phosphatidylethanolamine Lamellar and Inverted Hexagonal Phases. Biophys. J 2001, 81, 2693–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (694).Sato Y; Okabe N; Note Y; Hashiba K; Maeki M; Tokeshi M; Harashima H Hydrophobic Scaffolds of pH-sensitive Cationic Lipids Contribute to Miscibility with Phospholipids and Improve the Efficiency of Delivering Short Interfering RNA by Small-sized Lipid Nanoparticles. Acta Biomater 2020, 102, 341–350. [DOI] [PubMed] [Google Scholar]
- (695).Zhang R; El-Mayta R; Murdoch TJ; Warzecha CC; Billingsley MM; Shepherd SJ; Gong N; Wang L; Wilson JM; Lee D; et al. Helper Lipid Structure Influences Protein Adsorption and Delivery of Lipid Nanoparticles to Spleen and Liver. Biomater. Sci 2021, 9, 1449–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (696).Kapoor M; Burgess DJ Efficient and Safe Delivery of siRNA Using Anionic Lipids: Formulation Optimization Studies. Int. J. Pharm 2012, 432, 80–90. [DOI] [PubMed] [Google Scholar]
- (697).Olton D; Li J; Wilson ME; Rogers T; Close J; Huang L; Kumta PN; Sfeir C Nanostructured Calcium Phosphates (Nano-CaPs) for Non-viral Gene Delivery: Influence of the Synthesis Parameters on Transfection Efficiency. Biomaterials 2007, 28, 1267–1279. [DOI] [PubMed] [Google Scholar]
- (698).Zhang Y; Peng L; Mumper RJ; Huang L Combinational Delivery of c-myc siRNA and Nucleoside Analogs in a Single, Synthetic Nanocarrier for Targeted Cancer Therapy. Biomaterials 2013, 34, 8459–8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (699).Zelphati O; Szoka FC Mechanism of Oligonucleotide Release from Cationic Liposomes. Proc. Natl. Acad. Sci. U. S. A 1996, 93, 11493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (700).Xu Y; Szoka FC Mechanism of DNA Release from Cationic Liposome/DNA Complexes Used in Cell Transfection. Biochemistry 1996, 35, 5616–5623. [DOI] [PubMed] [Google Scholar]
- (701).Bhattacharya S; Mandal SS Evidence of Interlipidic Ion-Pairing in Anion-Induced DNA Release from Cationic Amphiphile-DNA Complexes. Mechanistic Implications in Transfection. Biochemistry 1998, 37, 7764–7777. [DOI] [PubMed] [Google Scholar]
- (702).Wang J; Megha London E Relationship between Sterol/Steroid Structure and Participation in Ordered Lipid Domains (Lipid Rafts): Implications for Lipid Raft Structure and Function. Biochemistry 2004, 43, 1010–1018. [DOI] [PubMed] [Google Scholar]
- (703).Lu JJ; Langer R; Chen J A Novel Mechanism is Involved in Cationic Lipid-Mediated Functional siRNA Delivery. Mol. Pharmaceutics 2009, 6, 763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (704).Pozzi D; Marchini C; Cardarelli F; Amenitsch H; Garulli C; Bifone A; Caracciolo G Transfection Efficiency Boost of Cholesterol-Containing Lipoplexes. Biochim. Biophys. Acta, Biomembr 2012, 1818, 2335–2343. [DOI] [PubMed] [Google Scholar]
- (705).Rodrigueza WV; Haydn Pritchard P; Hope MJ The Influence of Size and Composition on the Cholesterol Mobilizing Properties of Liposomes In Vivo. Biochim. Biophys. Acta, Biomembr 1993, 1153, 9–19. [DOI] [PubMed] [Google Scholar]
- (706).Tenchov BG; MacDonald RC; Siegel DP Cubic Phases in Phosphatidylcholine-Cholesterol Mixtures: Cholesterol as Membrane “Fusogen. Biophys. J 2006, 91, 2508–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (707).Arteta MY; Kjellman T; Bartesaghi S; Wallin S; Wu X; Kvist AJ; Dabkowska A; Székely N; Radulescu A; Bergenholtz J Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc. Natl. Acad. Sci. U. S. A 2018, 115, E3351–E3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (708).Zhigaltsev IV; Maurer N; Wong KF; Cullis PR Triggered Release of Doxorubicin Following Mixing of Cationic and Anionic Liposomes. Biochim. Biophys. Acta, Biomembr 2002, 1565, 129–135. [DOI] [PubMed] [Google Scholar]
- (709).Ikonen E Cellular Cholesterol Trafficking and Compartmentalization. Nat. Rev. Mol. Cell Biol 2008, 9, 125–138. [DOI] [PubMed] [Google Scholar]
- (710).Paunovska K; Gil CJ; Lokugamage MP; Sago CD; Sato M; Lando GN; Gamboa Castro M; Bryksin AV; Dahlman JE Analyzing 2000 in Vivo Drug Delivery Data Points Reveals Cholesterol Structure Impacts Nanoparticle Delivery. ACS Nano 2018, 12, 8341–8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (711).Kuai R; Li D; Chen YE; Moon JJ; Schwendeman A High-density Lipoproteins: Nature’s Multifunctional Nanoparticles. ACS Nano 2016, 10, 3015–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (712).Paunovska K; Da Silva Sanchez AJ; Sago CD; Gan Z; Lokugamage MP; Islam FZ; Kalathoor S; Krupczak BR; Dahlman JE Nanoparticles Containing Oxidized Cholesterol Deliver mRNA to the Liver Microenvironment at Clinically Relevant Doses. Adv. Mater 2019, 31, No. e1807748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (713).Sago CD; Lokugamage MP; Paunovska K; Vanover DA; Monaco CM; Shah NN; Castro MG; Anderson SE; Rudoltz TG; Lando GN High-Throughput In Vivo Screen of Functional mRNA Delivery Identifies Nanoparticles for Endothelial Cell Gene Editing. Proc. Natl. Acad. Sci. U. S. A 2018, 115, E9944–E9952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (714).Dong Y; Love KT; Dorkin JR; Sirirungruang S; Zhang Y; Chen D; Bogorad RL; Yin H; Chen Y; Vegas AJ Lipopeptide Nanoparticles for Potent and Selective siRNA Delivery in Rodents and Nonhuman Primates. Proc. Natl. Acad. Sci. U. S. A 2014, 111, 3955–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (715).Eygeris Y; Patel S; Jozic A; Sahay G Deconvoluting Lipid Nanoparticle Structure for Messenger RNA Delivery. Nano Lett 2020, 20, 4543–4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (716).Patel S; Ashwanikumar N; Robinson E; Xia Y; Mihai C; Griffith JP 3rd; Hou S; Esposito AA; Ketova T; Welsher K; et al. Naturally-Occurring Cholesterol Analogues in Lipid Nanoparticles Induce Polymorphic Shape and Enhance Intracellular Delivery of mRNA. Nat. Commun 2020, 11, 983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (717).Herrera M; Kim J; Eygeris Y; Jozic A; Sahay G Illuminating Endosomal Escape of Polymorphic Lipid Nanoparticles That Boost mRNA Delivery. Biomater. Sci 2021, DOI: 10.1039/D0BM01947J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (718).Hung W-C; Lee M-T; Chung H; Sun Y-T; Chen H; Charron NE; Huang HW Comparative Study of the Condensing Effects of Ergosterol and Cholesterol. Biophys. J 2016, 110, 2026–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (719).Prades J; Vögler O; Alemany R; Gomez-Florit M; Funari SS; Ruiz-Gutiérrez V; Barceló F Plant Pentacyclic Triterpenic Acids as Modulators of Lipid Membrane Physical Properties. Biochim. Biophys. Acta, Biomembr 2011, 1808, 752–760. [DOI] [PubMed] [Google Scholar]
- (720).Halling KK; Slotte JP Membrane Properties of Plant Sterols in Phospholipid Bilayers as Determined by Differential Scanning Calorimetry, Resonance Energy Transfer and Detergent-Induced Solubilization. Biochim. Biophys. Acta, Biomembr 2004, 1664, 161–171. [DOI] [PubMed] [Google Scholar]
- (721).Subramanian K; Balch WE NPC1/NPC2 Function as a Tag Team Duo to Mobilize Cholesterol. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 15223–15224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (722).Eygeris Y; Patel S; Jozic A; Sahay G Deconvoluting Lipid Nanoparticle Structure for Messenger RNA Delivery. Nano Lett 2020, 20, 4543–4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (723).Yanez Arteta M; Kjellman T; Bartesaghi S; Wallin S; Wu W; Kvist AJ; Dabkowska A; Székely N; Radulescu A; Bergenholtz J; et al. Successful Reprogramming of Cellular Protein Production through mRNA Delivered by Functionalized Lipid Nanoparticles. Proc. Natl. Acad. Sci. U. S. A 2018, 115, E3351–E3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (724).Imam ZI; Kenyon LE; Ashby G; Nagib F; Mendicino M; Zhao C; Gadok AK; Stachowiak JC Phase-Separated Liposomes Enhance the Efficiency of Macromolecular Delivery to the Cellular Cytoplasm. Cell. Mol. Bioeng 2017, 10, 387–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (725).Hafez IM; Ansell S; Cullis PR Tunable pH-Sensitive Liposomes Composed of Mixtures of Cationic and Anionic Lipids. Biophys. J 2000, 79, 1438–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (726).Shi G; Guo W; Stephenson SM; Lee RJ Efficient Intracellular Drug and Gene Delivery Using Folate Receptor-Targeted pH-Sensitive Liposomes Composed of Cationic/Anionic Lipid Combinations. J. Controlled Release 2002, 80, 309–319. [DOI] [PubMed] [Google Scholar]
- (727).Yu B; Hsu S-H; Zhou C; Wang X; Terp MC; Wu Y; Teng L; Mao Y; Wang F; Xue W Lipid nanoparticles for hepatic delivery of small interfering RNA. Biomaterials 2012, 33, 5924–5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (728).Wang X; Yu B; Ren W; Mo X; Zhou C; He H; Jia H; Wang L; Jacob ST; Lee RJ Enhanced Hepatic Delivery of siRNA and microRNA Using Oleic Acid Based Lipid Nanoparticle Formulations. J. Controlled Release 2013, 172, 690–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (729).Wang XM; Yu B; Ren W; Mo XK; Zhou CG; He HY; Jia HL; Wang L; Jacob ST; Lee RJ; et al. Enhanced Hepatic Delivery of siRNA and microRNA Using Oleic Acid Based Lipid Nanoparticle Formulations. J. Controlled Release 2013, 172, 690–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (730).Thomas G; Loriette C; Pepin D; Chambaz J; Bereziat G Selective Channelling of Arachidonic and Linoleic Acids into Glycerolipids of Rat Hepatocytes in Primary Culture. Biochem. J 1988, 256, 641–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (731).Schürer NY; Stremmel W; Grundmann J-U; Schliep V; Kleinert H; Bass NM; Williams ML Evidence for a Novel Keratinocyte Fatty Acid Uptake Mechanism with Preference for Linoleic Acid: Comparison of Oleic and Linoleic Acid Uptake by Cultured Human Keratinocytes, Fibroblasts and a Human Hepatoma Cell Line. Biochim. Biophys. Acta, Lipids Lipid Metab 1994, 1211, 51–60. [DOI] [PubMed] [Google Scholar]
- (732).Angeletti C; Tacconi de Alaniz MJ Fatty Acid Uptake and Metabolism in Hep G2 Human-Hepatoma Cells. Mol. Cell. Biochem 1995, 143, 99–105. [DOI] [PubMed] [Google Scholar]
- (733).Li L; Wang H; Ong ZY; Xu K; Ee PLR; Zheng S; Hedrick JL; Yang Y-Y Polymer- and Lipid-Based Nanoparticle Therapeutics for the Treatment of Liver Diseases. Nano Today 2010, 5, 296–312. [Google Scholar]
- (734).Yu B; Hsu SH; Zhou CG; Wang XM; Terp MC; Wu W; Teng LS; Mao YC; Wang F; Xue WM; et al. Lipid Nanoparticles for Hepatic Delivery of Small Interfering RNA. Biomaterials 2012, 33, 5924–5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (735).Voutila J; Reebye V; Roberts TC; Protopapa P; Andrikakou P; Blakey DC; Habib R; Huber H; Saetrom P; Rossi JJ; et al. Development and Mechanism of Small Activating RNA Targeting CEBPA, a Novel Therapeutic in Clinical Trials for Liver Cancer. Mol. Ther 2017, 25, 2705–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (736).Russo L; Berardi V; Tardani F; La Mesa C; Risuleo G Delivery of RNA and Its Intracellular Translation into Protein Mediated by SDS-CTAB Vesicles: Potential Use in Nanobiotechnology. BioMed Res. Int 2013, 2013, 734596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (737).He S; Fan W; Wu N; Zhu J; Miao Y; Miao X; Li F; Zhang X; Gan Y Lipid-Based Liquid Crystalline Nanoparticles Facilitate Cytosolic Delivery of siRNA via Structural Transformation. Nano Lett 2018, 18, 2411–2419. [DOI] [PubMed] [Google Scholar]
- (738).Zhou C; Mao Y; Sugimoto Y; Zhang Y; Kanthamneni N; Yu B; Brueggemeier RW; Lee LJ; Lee RJ SPANosomes as Delivery Vehicles for Small Interfering RNA (siRNA). Mol. Pharmaceutics 2012, 9, 201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (739).Taratula O; Kuzmov A; Shah M; Garbuzenko OB; Minko T Nanostructured Lipid Carriers as Multifunctional Nanomedicine Platform for Pulmonary Co-delivery of Anticancer Drugs and siRNA. J. Controlled Release 2013, 171, 349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (740).Silva JPN; Oliveira IMSC; Oliveira ACN; Lućio M; Gomes AC; Coutinho PJG; Oliveira MECDR Structural Dynamics and Physicochemical Properties of pDNA/DODAB:MO Lipoplexes: Effect of pH and Anionic Lipids in Inverted Non-lamellar Phases Versus Lamellar Phases. Biochim. Biophys. Acta, Biomembr 2014, 1838, 2555–2567. [DOI] [PubMed] [Google Scholar]
- (741).Leal C; Bouxsein NF; Ewert KK; Safinya CR Highly Efficient Gene Silencing Activity of siRNA Embedded in a Nanostructured Gyroid Cubic Lipid Matrix. J. Am. Chem. Soc 2010, 132, 16841–16847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (742).Oliveira ACN; Raemdonck K; Martens T; Rombouts K; Simón-Vázquez R; Botelho C; Lopes I; Lućio M; González-Fernández Á; Real Oliveira MECD; et al. Stealth Monoolein-Based Nanocarriers for Delivery of siRNA to Cancer Cells. Acta Biomater 2015, 25, 216–229. [DOI] [PubMed] [Google Scholar]
- (743).Oliveira ACN; Martens TF; Raemdonck K; Adati RD; Feitosa E; Botelho C; Gomes AC; Braeckmans K; Real Oliveira MECD Dioctadecyldimethylammonium:Monoolein Nanocarriers for Efficient in Vitro Gene Silencing. ACS Appl. Mater. Interfaces 2014, 6, 6977–6989. [DOI] [PubMed] [Google Scholar]
- (744).Erel-Akbaba G; Carvalho LA; Tian T; Zinter M; Akbaba H; Obeid PJ; Chiocca EA; Weissleder R; Kantarci AG; Tannous BA Radiation-Induced Targeted Nanoparticle-Based Gene Delivery for Brain Tumor Therapy. ACS Nano 2019, 13, 4028–4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (745).Silva JPN; Oliveira ACN; Casal MPPA; Gomes AC; Coutinho PJG; Coutinho OP; Oliveira MECDR DODAB: Monoolein-Based Lipoplexes as Non-viral Vectors for Transfection of Mammalian Cells. Biochim. Biophys. Acta, Biomembr 2011, 1808, 2440–2449. [DOI] [PubMed] [Google Scholar]
- (746).Silva JPN; Oliveira ACN; Gomes AC; Oliveira MR Development of Dioctadecyldimethylammonium Bromide/Monoolein Liposomes for Gene Delivery. Cell Interaction 2012, 245–272. [Google Scholar]
- (747).Ruktanonchai U; Limpakdee S; Meejoo S; Sakulkhu U; Bunyapraphatsara N; Junyaprasert V; Puttipipatkhachorn S The Effect of Cetyl palmitate Crystallinity on Physical Properties of Gamma-Oryzanol Encapsulated in Solid Lipid Nanoparticles. Nanotechnology 2008, 19, 095701. [DOI] [PubMed] [Google Scholar]
- (748).Montana G; Bondì ML; Carrotta R; Picone P; Craparo EF; San Biagio PL; Giammona G; Di Carlo M Employment of Cationic Solid-Lipid Nanoparticles as RNA Carriers. Bioconjugate Chem 2007, 18, 302–308. [DOI] [PubMed] [Google Scholar]
- (749).Béduneau A; Saulnier P; Hindré F; Clavreul A; Leroux J-C; Benoit J-P Design of Targeted Lipid Nanocapsules by Conjugation of Whole Antibodies and Antibody Fab’ Fragments. Biomaterials 2007, 28, 4978–4990. [DOI] [PubMed] [Google Scholar]
- (750).Martinez-Pomares L The Mannose Receptor. J. Leukocyte Biol 2012, 92, 1177–1186. [DOI] [PubMed] [Google Scholar]
- (751).White KL; Rades T; Furneaux RH; Tyler PC; Hook S Mannosylated Liposomes as Antigen Delivery Vehicles for Targeting to Dendritic Cells. J. Pharm. Pharmacol 2010, 58, 729–737. [DOI] [PubMed] [Google Scholar]
- (752).Irache JM; Salman HH; Gamazo C; Espuelas S Mannose-Targeted Systems for the Delivery of Therapeutics. Expert Opin. Drug Delivery 2008, 5, 703–724. [DOI] [PubMed] [Google Scholar]
- (753).Carrillo-Conde B; Song E-H; Chavez-Santoscoy A; Phanse Y; Ramer-Tait AE; Pohl NLB; Wannemuehler MJ; Bellaire BH; Narasimhan B Mannose-Functionalized “Pathogen-like” Polyanhydride Nanoparticles Target C-Type Lectin Receptors on Dendritic Cells. Mol. Pharmaceutics 2011, 8, 1877–1886. [DOI] [PubMed] [Google Scholar]
- (754).Markov OV; Mironova NL; Shmendel EV; Maslov MA; Zenkova MA Systemic Delivery of Complexes of Melanoma RNA with Mannosylated Liposomes Activates Highly Efficient Murine Melanoma-Specific Cytotoxic T cells In Vivo. Mol. Biol 2017, 51, 102–107. [DOI] [PubMed] [Google Scholar]
- (755).Markov OV; Mironova NL; Shmendel EV; Serikov RN; Morozova NG; Maslov MA; Vlassov VV; Zenkova MA Multicomponent Mannose-Containing Liposomes Efficiently Deliver RNA in Murine Immature Dendritic Cells and Provide Productive Anti-tumour Response in Murine Melanoma Model. J. Controlled Release 2015, 213, 45–56. [DOI] [PubMed] [Google Scholar]
- (756).Goswami R; Chatzikleanthous D; Lou G; Giusti F; Bonci A; Taccone M; Brazzoli M; Gallorini S; Ferlenghi I; Berti F; et al. Mannosylation of LNP Results in Improved Potency for Self-Amplifying RNA (SAM) Vaccines. ACS Infect. Dis 2019, 5, 1546–1558. [DOI] [PubMed] [Google Scholar]
- (757).Vogel G; Eichenberger W Betaine Lipids in Lower Plants. Biosynthesis of DGTS and DGTA in Ochromonas Danica (Chrysophyceae) and the Possible Role of DGTS in Lipid Metabolism. Plant Cell Physiol 1992, 33, 427–436. [Google Scholar]
- (758).Kotlova ER; Sinyutina NF Advanced Research on Plant Lipids; Springer, 2003; pp 373–376; DOI: 10.1007/978-94-017-0159-4. [DOI] [Google Scholar]
- (759).Mochizuki S; Kanegae N; Nishina K; Kamikawa Y; Koiwai K; Masunaga H; Sakurai K The role of the Helper Lipid Dioleoylphosphatidylethanolamine (DOPE) for DNA Transfection Cooperating with a Cationic Lipid Bearing Ethylenediamine. Biochim. Biophys. Acta, Biomembr 2013, 1828, 412–418. [DOI] [PubMed] [Google Scholar]
- (760).Grijalvo S; Puras G; Zarate J; Sainz-Ramos M; Qtaish NAL; Lopez T; Mashal M; Attia N; Diaz D; Pons R; et al. Cationic Niosomes as Non-Viral Vehicles for Nucleic Acids: Challenges and Opportunities in Gene Delivery. Pharmaceutics 2019, 11, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (761).Hui SW; Langner M; Zhao YL; Ross P; Hurley E; Chan K The Role of Helper Lipids in Cationic Liposome-Mediated Gene Transfer. Biophys. J 1996, 71, 590–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (762).Ojeda E; Puras G; Agirre M; Zarate J; Grijalvo S; Eritja R; DiGiacomo L; Caracciolo G; Pedraz J-L The Role of Helper Lipids in the Intracellular Disposition and Transfection Efficiency of Niosome Formulations for Gene Delivery to Retinal Pigment Epithelial Cells. Int. J. Pharm 2016, 503, 115–126. [DOI] [PubMed] [Google Scholar]
- (763).Mashal M; Attia N; Puras G; Martínez-Navarrete G; Fernández E; Pedraz JL Retinal Gene Delivery Enhancement by Lycopene Incorporation into Cationic Niosomes Based on DOTMA and Polysorbate 60. J. Controlled Release 2017, 254, 55–64. [DOI] [PubMed] [Google Scholar]
- (764).Di Mascio P; Kaiser S; Sies H Lycopene as the Most Efficient Biological Carotenoid Singlet Oxygen Quencher. Arch. Biochem. Biophys 1989, 274, 532–538. [DOI] [PubMed] [Google Scholar]
- (765).Kauffman KJ; Webber MJ; Anderson DG Materials for Non-Viral Intracellular Delivery of Messenger RNA Therapeutics. J. Controlled Release 2016, 240, 227–234. [DOI] [PubMed] [Google Scholar]
- (766).Zhai J; Hinton TM; Waddington LJ; Fong C; Tran N; Mulet X; Drummond CJ; Muir BW Lipid-PEG Conjugates Sterically Stabilize and Reduce the Toxicity of Phytantriol-Based Lyotropic Liquid Crystalline Nanoparticles. Langmuir 2015, 31, 10871–10880. [DOI] [PubMed] [Google Scholar]
- (767).Biswas A; Chakraborty K; Dutta C; Mukherjee S; Gayen P; Jan S; Mallick AM; Bhattacharyya D; Sinha Roy R Engineered Histidine-Enriched Facial Lipopeptides for Enhanced Intracellular Delivery of Functional siRNA to Triple Negative Breast Cancer Cells. ACS Appl. Mater. Interfaces 2019, 11, 4719–4736. [DOI] [PubMed] [Google Scholar]
- (768).Zhu Q-L; Zhou Y; Guan M; Zhou X-F; Liu Y; Chen W-L; Zhang C-G; Yuan Z-Q; Liu C; Zhu A-J Low-Density Lipoprotein-Coupled N-Succinyl Chitosan Nanoparticles Co-delivering siRNA and Doxorubicin for Hepatocyte-Targeted Therapy. Biomaterials 2014, 35, 5965–5976. [DOI] [PubMed] [Google Scholar]
- (769).Mintzer MA; Simanek EE Nonviral Vectors for Gene Delivery. Chem. Rev 2009, 109, 259–302. [DOI] [PubMed] [Google Scholar]
- (770).Singha K; Namgung R; Kim WJ Polymers in Small-Interfering RNA Delivery. Nucleic Acid Ther 2011, 21, 133–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (771).Nogueira SS; Schlegel A; Maxeiner K; Weber B; Barz M; Schroer MA; Blanchet CE; Svergun DI; Ramishetti S; Peer D Polysarcosine-Functionalized Lipid Nanoparticles for Therapeutic mRNA Delivery. ACS Appl. Nano Mater 2020, 3, 10634–10645. [Google Scholar]
- (772).Gao L-Y; Liu X-Y; Chen C-J; Wang J-C; Feng Q; Yu M-Z; Ma X-F; Pei X-W; Niu Y-J; Qiu C Core-Shell Type Lipid/rPAA-Chol Polymer Hybrid Nanoparticles for In Vivo siRNA Delivery. Biomaterials 2014, 35, 2066–2078. [DOI] [PubMed] [Google Scholar]
- (773).Chahal JS; Khan OF; Cooper CL; McPartlan JS; Tsosie JK; Tilley LD; Sidik SM; Lourido S; Langer R; Bavari S Dendrimer-RNA Nanoparticles Generate Protective Immunity Against Lethal Ebola, H1N1 Influenza, and Toxoplasma Gondii Challenges with a Single Dose. Proc. Natl. Acad. Sci. U. S. A 2016, 113, E4133–E4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (774).Tam YYC; Chen S; Cullis PR Advances in Lipid Nanoparticles for siRNA Delivery. Pharmaceutics 2013, 5, 498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (775).Suzuki T; Suzuki Y; Hihara T; Kubara K; Kondo K; Hyodo K; Yamazaki K; Ishida T; Ishihara H PEG Shedding-Rate-Dependent Blood Clearance of PEGylated Lipid Nanoparticles in Mice: Faster PEG Shedding Attenuates Anti-PEG IgM Production. Int. J. Pharm 2020, 588, 119792. [DOI] [PubMed] [Google Scholar]
- (776).Li R; Li Y; Zhang J; Liu Q; Wu T; Zhou J; Huang H; Tang Q; Huang C; Huang Y Targeted Delivery of Celastrol to Renal Interstitial Myofibroblasts Using Fibronectin-Binding Liposomes Attenuates Renal Fibrosis and Reduces Systemic Toxicity. J. Controlled Release 2020, 320, 32–44. [DOI] [PubMed] [Google Scholar]
- (777).Li M; Yu H; Wang T; Chang N; Zhang J; Du D; Liu M; Sun S; Wang R; Tao H Tamoxifen Embedded in Lipid Bilayer Improves the Oncotarget of Liposomal Daunorubicin In Vivo. J. Mater. Chem. B 2014, 2, 1619–1625. [DOI] [PubMed] [Google Scholar]
- (778).Kulkarni JA; Darjuan MM; Mercer JE; Chen S; Van Der Meel R; Thewalt JL; Tam YYC; Cullis PR On the Formation and Morphology of Lipid Nanoparticles Containing Ionizable Cationic Lipids and siRNA. ACS Nano 2018, 12, 4787–4795. [DOI] [PubMed] [Google Scholar]
- (779).Wahane A; Waghmode A; Kapphahn A; Dhuri K; Gupta A; Bahal R Role of Lipid-Based and Polymer-Based Non-Viral Vectors in Nucleic Acid Delivery for Next-Generation Gene Therapy. Molecules 2020, 25, 2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (780).Akinc A; Maier MA; Manoharan M; Fitzgerald K; Jayaraman M; Barros S; Ansell S; Du X; Hope MJ; Madden TD The Onpattro Story and the Clinical Translation of Nanomedicines Containing Nucleic Acid-Based Drugs. Nat. Nanotechnol 2019, 14, 1084–1087. [DOI] [PubMed] [Google Scholar]
- (781).Nosova A; Koloskova O; Nikonova A; Simonova V; Smirnov V; Kudlay D; Khaitov M Diversity of PEGylation Methods of Liposomes and Their Influence on RNA Delivery. MedChemComm 2019, 10, 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (782).Sonoke S; Ueda T; Fujiwara K; Sato Y; Takagaki K; Hirabayashi K; Ohgi T; Yano J Tumor Regression in Mice by Delivery of Bcl-2 Small Interfering RNA with Pegylated Cationic Liposomes. Cancer Res 2008, 68, 8843–8851. [DOI] [PubMed] [Google Scholar]
- (783).Mui BL; Tam YK; Jayaraman M; Ansell SM; Du X; Tam YYC; Lin PJ; Chen S; Narayanannair JK; Rajeev KG Influence of Polyethylene Glycol Lipid Desorption Rates on Pharmacokinetics and Pharmacodynamics of siRNA Lipid Nanoparticles. Mol. Ther.–Nucleic Acids 2013, 2, No. e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (784).Oberli MA; Reichmuth AM; Dorkin JR; Mitchell MJ; Fenton OS; Jaklenec A; Anderson DG; Langer R; Blankschtein D Lipid Nanoparticle Assisted mRNA Delivery for Potent Cancer Immunotherapy. Nano Lett 2017, 17, 1326–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (785).Zhu X; Tao W; Liu D; Wu J; Guo Z; Ji X; Bharwani Z; Zhao L; Zhao X; Farokhzad OC Surface De-PEGylation Controls Nanoparticle-Mediated siRNA Delivery In Vitro and In Vivo. Theranostics 2017, 7, 1990–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (786).Islam MA; Xu Y; Tao W; Ubellacker JM; Lim M; Aum D; Lee GY; Zhou K; Zope H; Yu M; et al. Restoration of Tumour-Growth Suppression In Vivo via Systemic Nanoparticle-Mediated Delivery of PTEN mRNA. Nat. Biomed. Eng 2018, 2, 850–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (787).Kulkarni A; DeFrees K; Hyun S-H; Thompson DH Pendant Polymer: Amino-β-Cyclodextrin: siRNA Guest: Host Nanoparticles as Efficient Vectors for Gene Silencing. J. Am. Chem. Soc 2012, 134, 7596–7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (788).Chen C-W; Lu D-W; Yeh M-K; Shiau C-Y; Chiang C-H Novel RGD-Lipid Conjugate-Modified Liposomes for Enhancing siRNA Eelivery in Human Retinal Pigment Epithelial Cells. Int. J. Nanomed 2011, 6, 2567–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (789).Hashiba K; Sato Y; Harashima H pH-Labile PEGylation of siRNA-Loaded Lipid Nanoparticle Improves Active Targeting and Gene Silencing Activity in Hepatocytes. J. Controlled Release 2017, 262, 239–246. [DOI] [PubMed] [Google Scholar]
- (790).Li S-D; Chono S; Huang L Efficient Gene Silencing in Metastatic Tumor by siRNA Formulated in Surface-Modified Nanoparticles. J. Controlled Release 2008, 126, 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (791).Akinc A; Querbes W; De S; Qin J; Frank-Kamenetsky M; Jayaprakash KN; Jayaraman M; Rajeev KG; Cantley WL; Dorkin JR Targeted Delivery of RNAi Therapeutics with Endogenous and Exogenous Ligand-Based Mechanisms. Mol. Ther 2010, 18, 1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (792).Zang X; Ding H; Zhao X; Li X; Du Z; Hu H; Qiao M; Chen D; Deng Y; Zhao X Anti-EphA10 Antibody-Conjugated pH-Sensitive Liposomes for Specific Intracellular Delivery of siRNA. Int. J. Nanomed 2016, 11, 3951–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (793).Krzysztoń R; Salem B; Lee D; Schwake G; Wagner E; Rädler J Microfluidic Self-Assembly of Folate-Targeted Monomolecular siRNA-Lipid Nanoparticles. Nanoscale 2017, 9, 7442–7453. [DOI] [PubMed] [Google Scholar]
- (794).Santiwarangkool S; Akita H; Nakatani T; Kusumoto K; Kimura H; Suzuki M; Nishimura M; Sato Y; Harashima H PEGylation of the GALA Peptide Enhances the Lung-Targeting Activity of Nanocarriers That Contain Encapsulated siRNA. J. Pharm. Sci 2017, 106, 2420–2427. [DOI] [PubMed] [Google Scholar]
- (795).Qiao J-B; Fan Q-Q; Zhang C-L; Lee J; Byun J; Xing L; Gao X-D; Oh Y-K; Jiang H-L Hyperbranched Lipoid-Based Lipid Nanoparticles for Bidirectional Regulation of Collagen Accumulation In Liver Fibrosis. J. Controlled Release 2020, 321, 629–640. [DOI] [PubMed] [Google Scholar]
- (796).Zhang L; Lu Z; Zhao Q; Huang J; Shen H; Zhang Z Enhanced Chemotherapy Efficacy by Sequential Delivery of siRNA and Anticancer Drugs Using PEI-Grafted Graphene Oxide. Small 2011, 7, 460–464. [DOI] [PubMed] [Google Scholar]
- (797).Ke X; Shelton L; Hu Y; Zhu Y; Chow E; Tang H; Santos JL; Mao H-Q Surface-Functionalized PEGylated Nanoparticles Deliver Messenger RNA to Pulmonary Immune Cells. ACS Appl. Mater. Interfaces 2020, 12, 35835–35844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (798).Zhupanyn P; Ewe A; Büch T; Malek A; Rademacher P; Müller C; Reinert A; Jaimes Y; Aigner A Extracellular Vesicle (ECV)-Modified Polyethylenimine (PEI) Complexes for Enhanced siRNA Delivery In Vitro and In Vivo. J. Controlled Release 2020, 319, 63–76. [DOI] [PubMed] [Google Scholar]
- (799).Alshamsan A; Haddadi A; Incani V; Samuel J; Lavasanifar A; Uludag H Formulation and Delivery of siRNA by Oleic Acid and Stearic Acid Modified Polyethylenimine. Mol. Pharmaceutics 2009, 6, 121–133. [DOI] [PubMed] [Google Scholar]
- (800).Dahlman JE; Barnes C; Khan OF; Thiriot A; Jhunjunwala S; Shaw TE; Xing Y; Sager HB; Sahay G; Speciner L In Vivo Endothelial siRNA Delivery Using Polymeric Nanoparticles with Low Molecular Weight. Nat. Nanotechnol 2014, 9, 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (801).Navarro G; Essex S; Sawant RR; Biswas S; Nagesha D; Sridhar S; de ILarduya CT; Torchilin VP Phospholipid-modified polyethylenimine-based nanopreparations for siRNA-mediated gene silencing: Implications for transfection and the role of lipid components. Nanomedicine 2014, 10, 411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (802).Fan X; Zhao X; Su W; Tang X Triton X-100-Modified Adenosine Triphosphate-Responsive siRNA Delivery Agent for Antitumor Therapy. Mol. Pharmaceutics 2020, 17, 3696–3708. [DOI] [PubMed] [Google Scholar]
- (803).Cao Y; Liu X; Peng L Molecular Engineering of Dendrimer Nanovectors for siRNA Delivery and Gene Silencing. Front. Chem. Sci. Eng 2017, 11, 663–675. [Google Scholar]
- (804).Shen W; Liu H; Ling-Hu Y; Wang H; Cheng Y Enhanced siRNA Delivery of a Cyclododecylated Dendrimer Compared to Its Linear Derivative. J. Mater. Chem. B 2016, 4, 5654–5658. [DOI] [PubMed] [Google Scholar]
- (805).Han S; Ganbold T; Bao Q; Yoshida T; Baigude H Sugar Functionalized Synergistic Dendrimers for Biocompatible Delivery of Nucleic Acid Therapeutics. Polymers 2018, 10, 1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (806).Baigude H; McCarroll J; Yang C-S; Swain PM; Rana TM Design and Creation of New Nanomaterials for Therapeutic RNAi. ACS Chem. Biol 2007, 2, 237–241. [DOI] [PubMed] [Google Scholar]
- (807).Yu T; Liu X; Bolcato-Bellemin AL; Wang Y; Liu C; Erbacher P; Qu F; Rocchi P; Behr JP; Peng L An Amphiphilic Dendrimer for Effective Delivery of Small Interfering RNA and Gene Silencing In Vitro and In Vivo. Angew. Chem., Int. Ed 2012, 51, 8478–8484. [DOI] [PubMed] [Google Scholar]
- (808).Khan OF; Zaia EW; Yin H; Bogorad RL; Pelet JM; Webber MJ; Zhuang I; Dahlman JE; Langer R; Anderson DG Ionizable Amphiphilic Dendrimer-Based Nanomaterials with Alkyl-Chain-Substituted Amines for Tunable siRNA Delivery to the Liver Endothelium In Vivo. Angew. Chem., Int. Ed 2014, 53, 14397–14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (809).Khan OF; Zaia EW; Jhunjhunwala S; Xue W; Cai W; Yun DS; Barnes CM; Dahlman JE; Dong Y; Pelet JM Dendrimer-Inspired Nanomaterials for the In Vivo Delivery of siRNA to Lung Vasculature. Nano Lett 2015, 15, 3008–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (810).Xiong Q; Li Y; Zhou K; Chen P; Guo H; Chen L; Ding J; Song T; Shi J Optimized Fluorodendrimer-Incorporated Hybrid Lipid-Polymer Nanoparticles for Efficient siRNA Delivery. Biomater. Sci 2020, 8, 758–762. [DOI] [PubMed] [Google Scholar]
- (811).Benner NL; McClellan RL; Turlington CR; Haabeth OA; Waymouth RM; Wender PA Oligo (Serine Ester) Charge-Altering Releasable Transporters: Organocatalytic Ring-Opening Polymerization and Their Use for In Vitro and In Vivo mRNA Delivery. J. Am. Chem. Soc 2019, 141, 8416–8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (812).Haabeth OA; Blake TR; McKinlay CJ; Waymouth RM; Wender PA; Levy R mRNA Vaccination with Charge-Altering Releasable Transporters Elicits Human T Cell Responses and Cures Established Tumors in Mice. Proc. Natl. Acad. Sci. U. S. A 2018, 115, E9153–E9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (813).Hao J; Kos P; Zhou K; Miller JB; Xue L; Yan Y; Xiong H; Elkassih S; Siegwart DJ Rapid Synthesis of a Lipocationic Polyester Library via Ring-Opening Polymerization of Functional Valerolactones for Efficacious siRNA Delivery. J. Am. Chem. Soc 2015, 137, 9206–9209. [DOI] [PubMed] [Google Scholar]
- (814).Dong Y; Dorkin JR; Wang W; Chang PH; Webber MJ; Tang BC; Yang J; Abutbul-Ionita I; Danino D; DeRosa F Poly (Glycoamidoamine) Brushes Formulated Nanomaterials for Systemic siRNA and mRNA Delivery In Vivo. Nano Lett 2016, 16, 842–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (815).Luo X; Wang W; Dorkin JR; Veiseh O; Chang P-H; Abutbul-Ionita I; Danino D; Langer R; Anderson DG; Dong Y Poly (Glycoamidoamine) Brush Nanomaterials for Systemic siRNA Delivery In Vivo. Biomater. Sci 2017, 5, 38–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (816).McKinlay CJ; Vargas JR; Blake TR; Hardy JW; Kanada M; Contag CH; Wender PA; Waymouth RM Charge-Altering Releasable Transporters (CARTs) for the Delivery and Release of mRNA in Living Animals. Proc. Natl. Acad. Sci. U. S. A 2017, 114, E448–E456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (817).Zhao Z; Li Y; Liu H; Jain A; Patel PV; Cheng K Co-Delivery of IKBKE siRNA and Cabazitaxel by Hybrid Nanocomplex Inhibits Invasiveness and Growth of Triple-Negative Breast Cancer. Sci. Adv 2020, 6, No. eabb0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (818).Tai Z; Wang X; Tian J; Gao Y; Zhang L; Yao C; Wu X; Zhang W; Zhu Q; Gao S Biodegradable Stearylated Peptide with Internal Disulfide Bonds for Efficient Delivery of siRNA In Vitro and In Vivo. Biomacromolecules 2015, 16, 1119–1130. [DOI] [PubMed] [Google Scholar]
- (819).Li Y; Li Y; Wang X; Lee RJ; Teng L Fatty Acid Modified Octa-Arginine for Delivery of siRNA. Int. J. Pharm 2015, 495, 527–535. [DOI] [PubMed] [Google Scholar]
- (820).Jing X; Foged C; Martin-Bertelsen B; Yaghmur A; Knapp KM; Malmsten M; Franzyk H; Nielsen HM Delivery of siRNA Complexed with Palmitoylated α-Peptide/β-Peptoid Cell-Penetrating Peptidomimetics: Membrane Interaction and Structural Characterization of a Lipid-Based Nanocarrier System. Mol. Pharmaceutics 2016, 13, 1739–1749. [DOI] [PubMed] [Google Scholar]
- (821).Koloskova O; Nikonova A; Budanova U; Shilovskiy I; Kofiadi I; Ivanov A; Smirnova O; Zverev V; Sebaykin YL; Andreev S Synthesis and Evaluation of Novel Lipopeptide as a Vehicle for Efficient Gene Delivery and Gene Silencing. Eur. J. Pharm. Biopharm 2016, 102, 159–167. [DOI] [PubMed] [Google Scholar]
- (822).Kim WJ; Christensen LV; Jo S; Yockman JW; Jeong JH; Kim Y-H; Kim SW Cholesteryl Oligoarginine Delivering Vascular Endothelial Growth Factor siRNA Effectively Inhibits Tumor Growth in Colon Adenocarcinoma. Mol. Ther 2006, 14, 343–350. [DOI] [PubMed] [Google Scholar]
- (823).Cline LL; Janout V; Fisher M; Juliano RL; Regen SL A Molecular Umbrella Approach to the Intracellular Delivery of Small Interfering RNA. Bioconjugate Chem 2011, 22, 2210–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (824).Sharma M; El-Sayed NS; Do H; Parang K; Tiwari RK; Aliabadi HM Tumor-Targeted Delivery of siRNA Using Fatty Acyl-CGKRK Peptide Conjugates. Sci. Rep 2017, 7, 6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (825).Busatto S; Walker SA; Grayson W; Pham A; Tian M; Nesto N; Barklund J; Wolfram J Lipoprotein-Based Drug Delivery. Adv. Drug Delivery Rev 2020, 159, 377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (826).Feingold KR; Grunfeld C Introduction to lipids and lipoproteins; Endotext; MDText.com, Inc., 2015. [Google Scholar]
- (827).Dong Y; Love KT; Dorkin JR; Sirirungruang S; Zhang Y; Chen D; Bogorad RL; Yin H; Chen Y; Vegas AJ Lipopeptide Nanoparticles for Potent and Selective siRNA Delivery in Rodents and Nonhuman Primates. Proc. Natl. Acad. Sci. U. S. A 2014, 111, 3955–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (828).Ding Y; Wang Y; Zhou J; Gu X; Wang W; Liu C; Bao X; Wang C; Li Y; Zhang Q Direct Cytosolic siRNA Delivery by Reconstituted High Density Lipoprotein for Target-Specific Therapy of Tumor Angiogenesis. Biomaterials 2014, 35, 7214–7227. [DOI] [PubMed] [Google Scholar]
- (829).Jiang G; Chen H; Huang J; Song Q; Chen Y; Gu X; Jiang Z; Huang Y; Lin Y; Feng J Tailored Lipoprotein-Like miRNA Delivery Nanostructure Suppresses Glioma Stemness and Drug Resistance Through Receptor-Stimulated Macropinocytosis. Adv. Sci 2020, 7, 1903290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (830).Han Y; Ding B; Zhao Z; Zhang H; Sun B; Zhao Y; Jiang L; Zhou J; Ding Y Immune Lipoprotein Nanostructures Inspired Relay Drug Delivery for Amplifying Antitumor Efficiency. Biomaterials 2018, 185, 205–218. [DOI] [PubMed] [Google Scholar]
- (831).Guo Y; Yuan W; Yu B; Kuai R; Hu W; Morin EE; Garcia-Barrio MT; Zhang J; Moon JJ; Schwendeman A Synthetic High-Density Lipoprotein-Mediated Targeted Delivery of Liver X Receptors Agonist Promotes Atherosclerosis Regression. EBioMedicine 2018, 28, 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (832).Jin H; Lovell JF; Chen J; Lin Q; Ding L; Ng KK; Pandey RK; Manoharan M; Zhang Z; Zheng G Mechanistic Insights into LDL Nanoparticle-Mediated siRNA Delivery. Bioconjugate Chem 2012, 23, 33–41. [DOI] [PubMed] [Google Scholar]
- (833).Yang M; Jin H; Chen J; Ding L; Ng KK; Lin Q; Lovell JF; Zhang Z; Zheng G Efficient Cytosolic Delivery of siRNA Using HDL-Mimicking Nanoparticles. Small 2011, 7, 568–573. [DOI] [PubMed] [Google Scholar]
- (834).Cruz W; Huang H; Barber B; Pasini E; Ding L; Zheng G; Chen J; Bhat M Lipoprotein-Like Nanoparticle Carrying Small Interfering RNA Against Spalt-Like Transcription Factor 4 Effectively Targets Hepatocellular Carcinoma Cells and Decreases Tumor Burden. Hepatol. Commun 2020, 4, 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (835).McMahon KM; Plebanek MP; Thaxton CS Properties of Native High-Density Lipoproteins Inspire Synthesis of Actively Targeted In Vivo siRNA Delivery Vehicles. Adv. Funct. Mater 2016, 26, 7824–7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (836).Huang J-L; Jiang G; Song Q-X; Gu X; Hu M; Wang X-L; Song H-H; Chen L-P; Lin Y-Y; Jiang D; et al. Lipoprotein-Biomimetic Nanostructure Enables Efficient Targeting Delivery of siRNA to Ras-Activated Glioblastoma Cells via Macropinocytosis. Nat. Commun 2017, 8, 15144. [DOI] [PMC free article] [PubMed] [Google Scholar]


