Abstract
Background
Exosomes are nanosized bio vesicles formed when multivesicular bodies and the plasma membrane merge and discharge into bodily fluids. They are well recognized for facilitating intercellular communication by transporting numerous biomolecules, including DNA, RNAs, proteins, and lipids, and have been implicated in varied diseases including cancer. Exosomes may be altered to transport a variety of therapeutic payloads, including as short interfering RNAs, antisense oligonucleotides, chemotherapeutic drugs, and immunological modulators, and can be directed to a specific target. Exosomes also possess the potential to act as a diagnostic biomarker in cancer, in addition to their therapeutic potential.
Conclusion
In this review, the physiological roles played by exosomes were summarized along with their biogenesis process. Different isolation techniques of exosomes including centrifugation-based, size-based, and polymer precipitation-based techniques have also been described in detail with a special focus on cancer therapeutic applications. The review also shed light on techniques of incubation of drugs with exosomes and their characterization methods covering the most advanced techniques. Myriad applications of exosomes in cancer as diagnostic biomarkers, drug delivery carriers, and chemoresistance-related issues have been discussed at length. Furthermore, a brief overview of exosome-based anti-cancer vaccines and a few prominent challenges concerning exosomal delivery have been concluded at the end.
Graphical abstract
Keywords: Exosomes, cancer, Biomarkers, Isolation, Biogenesis
Introduction
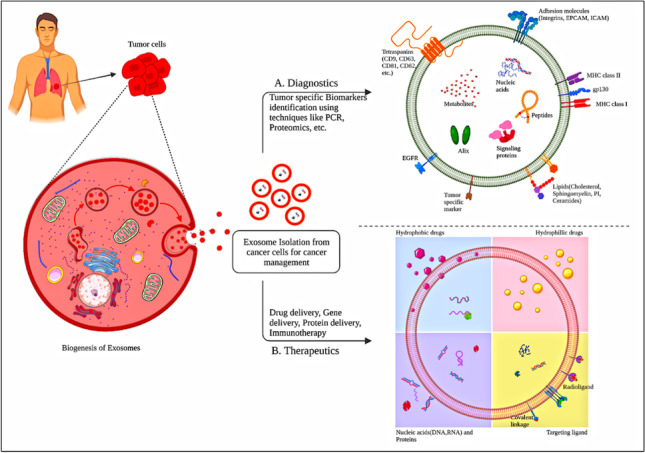
Cells communicate with each other in a coordinated manner to carry out their functions effectively. This intercellular communication is facilitated by connections such as gap junctions or by cell signaling processes. In cell signaling processes, chemical mediators are released by cells that alter or regulate the other cells that are present either in their vicinity or at distant locations. Extracellular vehicles (EVs), i.e., exosomes, have a similar role in intercellular communication and serve as cargo transporters in transporting proteins, lipids, and genetic material to the recipient cells [1]. These vesicles are formed in the endosomes or on the plasma membrane [2]. Exosomes are small, single-membrane EVs of a size ranging from 30 to 150 nm which are present in various biological fluids and are linked to a variety of biological processes and diseases, suggesting their potential role as biomarkers. A significant number of exosomes are found in most body fluids and they are frequently associated with multiple physiological and pathological processes [2–5]. For instance, exosomes play a crucial part in the regulation of gene expression in a recipient cell by delivering specific mRNA, controlling immune stimulation or repression, increasing organ excretion, and eliminating waste from the brain [1]. They also hold great potential as natural therapeutic agents and drug delivery vehicles [6]. Recent research has shown the pathophysiological consequences of exosomes on illnesses, particularly cancer. Cancer cells use exosome-mediated processes to create a favorable microenvironment that promotes tumor growth by boosting cell proliferation and preventing apoptosis. Exosomes can also induce the development of new vessels, ensuring access to nutrition, oxygen, and waste elimination, as well as assist in contributing to cancer cell metabolic reprogramming, allowing for their long-term proliferation. The invasive and disseminated capacity of tumors is highly boosted by cancer exosomes that carry information contributing to cancer cell migration, invasion, and extracellular matrix (ECM) [7].
Exosomes can be isolated from varied sources like cancer cells, immune cells, stem cells, and even food or plant cells. Although many isolation approaches have been utilized including ultracentrifugation, filtration, size exclusion chromatography, immunoaffinity capture, etc., there has not been a single optimized method for isolating exosomes from diversified sources. Many physicochemical factors like gene expression, elevated calcium concentration within the cells, and stimulation by drugs have been associated with the generation of exosomes. These are characterized by techniques like western blotting, microscopic techniques, nanoparticle tracking analysis, etc. [8].
In comparison with other delivery platforms, exosomes exhibit overwhelming advantages when used in the oncology sector such as long-term stability, targeting ability (active or passive targeting) culminating in a reduction in the frequency of administrations by improving efficiency, high drug loading with a capacity to load different cargoes like nucleic acids, drugs, peptides, proteins, etc. The aforementioned benefits have strengthened the position of exosomes in the biomedical field, oncology in particular. Although several advances in exosome-based research have been made in recent years, many challenges still remain, such as standard preparation and quality control procedures, as well as effective quantification methods for their comprehensive, and simultaneous inclusion. Several research activities are going under the umbrella of exosomes, ranging from manufacturing, purification, storage, quality control, modification, and their biological applications yet many are to be explored [8]. In this review, we aim to provide a comprehensive overview of exosomes, covering their composition, biogenesis, and isolation methods. The role of exosomes in the progression of cancer and the characteristics of cancer-derived exosomes have been elaborated at length. In addition, we have emphasized several anti-cancer drug loading techniques and diversified uses of exosomes in the context of diagnostics and therapeutics delivery for the treatment of cancer.
Biogenesis of exosomes
Exosomes are nano-sized EVs of endosomal origin. They are secreted by many cells and play a key role in cell-cell communication and in maintaining cellular homeostasis [9]. Along with cell surface proteins, extracellular constituents like lipids, proteins, metabolites, ions, small molecules, and fluid enter the cells through endocytosis. Plasma membrane invagination eventually culminates in the generation of multivesicular bodies, which may collide with other intracellular vesicles and organelles, adding to the variety of exosomal contents [10]. Exosomes constitute different proteins like heat shock proteins (HSP70, HSP90), GTPase, tetraspanins CD63, CD81, CD9, CD82, cytoskeletal heat shock nuclear enzyme RNA binding apoptotic signal transducers, different lipids like cholesterol, sphingomyelin, ceramides, different RNA like mRNA, miRNA, pre-miRNA, Y-RNA, circRNA, tRNA, snRNA, piRNA and different types of DNA like viral DNA, MtDNA, ssDNA and ds DNA [10–13].
The key factors involved in the biogenesis are as follows: (i) Rab GTPase proteins which control endosomal trafficking (ii) Endosomal Sorting Complex Required for Transport (ESCRT), which mainly includes (ESCRT) 0–3. Multiple proteins regulate intraluminal vesicles (ILV) formation, of which ESCRT-0 is involved in ubiquitin-dependent cargo cluster formation, ESCRT-1 and 2 induce the formation of buds, and ESCRT-3 has a role in vesicle scission (iii) Tetraspanins are transmembrane proteins that allow vesicles to form by causing membrane curvatures (iv) Sphingomyelinase, like other lipid-modifying enzymes, produces ceramides that assist in vesicle formation [14–16]. The presence of high amounts of the ceramide lipid has been found to help multivesicular endosome contents escape lysosomal digestion and be released as exosomes [17]. Exosome formation starts with the invagination of the outer membrane through the ubiquitination of surface receptors which results in the formation of early endosomes [18]. Early endosomes contain intraluminal vesicles (ILVs) that get matured and are known as multivesicular bodies (MVBs). These MVBs have two fates; one is fusion with lysosomes resulting in degradation, and the other being, fusion with the plasma membrane to release ILVs as exosomes by exocytosis [19]. It is important to note that not all MVEs produce exosomes; MVEs cargo can also be degraded by fusion with lysosomes [17]. The cells might be able to recycle the degraded products [10]. The plasma membrane-mediated exocytosis releases the exosomes with a lipid bilayer identical to that of the plasma membrane and is carried out by CD3, Lysosomal associated membrane protein (LAMP)-LAMP1, and LAMP2, which are present on few MVBs (Fig. 1) [10, 20].
Fig. 1.
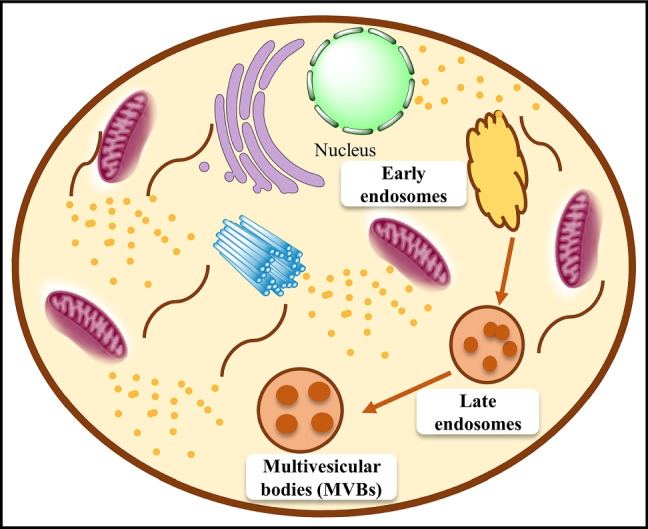
Biogenesis and secretion of exosomes: Exosomes are generated through endocytic membrane invagination and ILVs development inside the cell. During maturation, payloads (RNAs, proteins, and lipids) are integrated into ILVs through ESCRT-dependent or ESCRT-independent processes, and early endosomes mature to become MVBs. MVBs may be transported to the trans-Golgi network (TGN) for endosome recycling, to lysosomes for destruction.MVBs fusion with the cellular membrane is a precise process requiring several critical components such as Rab GTPases and SNARE complexes
Physiological functions of exosomes
Immune response
The immune system comprises ordered structures and biological processes that recognize and respond to extracellular environmental stimuli. Immune surveillance in cancer is a process in which pre-cancerous and cancerous cells activate an immune response that destroys altered malignant cells. Immune responses are regulated by numerous elements such as proteins, lipids, and RNAs in a process known as immunomodulation, which involves both innate and adaptive immunity. Exosomes have recently received great attention during tumor growth and genesis, with a focus on cancer immune surveillance and tumor escape responses at various stages. Cytotoxic T lymphocytes and natural killer (NK) cells mediated anti-tumor responses are directly suppressed by exosomes, which results in filiation of angiogenesis and induction of immune suppressor cell subsets leading to loss of immune surveillance [21].
One of the mechanisms by which cancer cells evade the immune system is the formation of membrane-covered vesicles or exosomes, originating from tumor cells, which can change the activity of the acquired immune system, including activated human T-cells [22].
Tumor-derived exosomes in cancer
Oncosomes are large microvesicles that are about 5 μm in diameter and are generated from tumor cells. They can carry oncogenic molecules that can modify the phenotype of receiving cells to encourage tumor growth [23]. They act as extracellular organelles which facilitate the growth of tumors and metastasis. They also play a role in the remodeling of the tumor microenvironment and take part in the transport of proteins and nucleic acids between tumor cells and neighboring cells [24]. As they are endogenous in origin, they provide a promising tool for cancer treatment with some key advantages in drug delivery [25]. Tumor cell-secreted exosomes play a role in paracrine signaling during tumor progression, tumor-stromal interactions, proliferative pathway activation, and immunosuppression [26]. However, the biological actions of tumor-derived exosomes differ from those of normal cell-derived exosomes. Exosomes from tumors enter cells by a variety of methods, depending on the target cells and the cancer cells that secrete them. For example, exosomes derived from glioblastoma utilize lipid Raft-mediated endocytosis for their uptake, which further depends on undisturbed ERK1/2eHSP27 signaling. Similarly, brain-metastatic breast cancer-derived exosomes utilize transcytosis to cross the brain endothelial cells, while the “CDC42-dependent clathrin-independent carrier/GPI-AP-enriched compartment (CLIC/GEEC) endocytic pathway” was utilized to enter astrocytes [27].
Isolation methods of exosomes
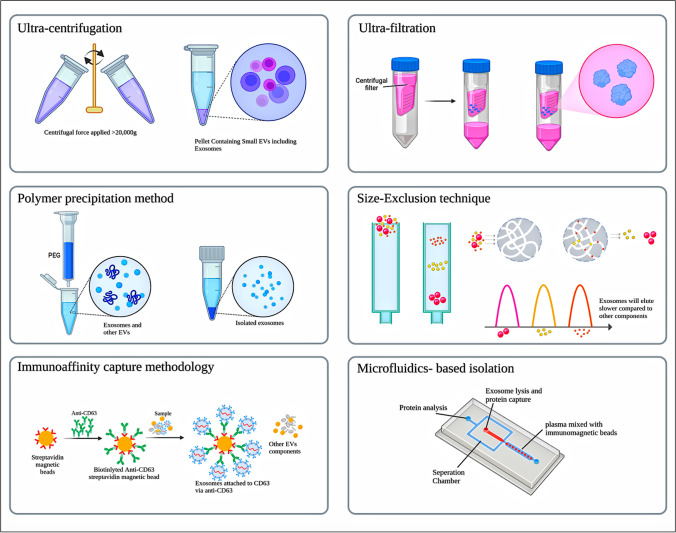
Though exosomes have several advantages, the isolation of high purity exosomes is a great challenge. There is a need for a robust and reproducible technique for the isolation of exosomes of higher purity. Exosomes have to be isolated from cellular debris and other intracellular components. As of now, some strategies of exosome isolation are commonly used, including ultrafiltration, ultracentrifugation, polymer precipitation, size-exclusion chromatography, and microfluidics immunoaffinity capture methodology [28]. Commercial exosome isolation kits are available based on the above principles, with some advantages and limitations [29]. The selection of the method depends on the objective and applications of isolated exosomes. These isolation techniques are discussed here in brief and represented diagrammatically in Fig. 2.
Fig. 2.
Different isolation techniques of exosomes
Centrifugation based techniques
Ultracentrifugation is the most prevalent technique and is regarded as the gold standard for the isolation of biological objects, which holds the potential for the isolation of exosomes even at an industrial scale [30]. Generally, these are divided into two types: differential centrifugation and density gradient centrifugation [31].
Johnston et al. established the differential centrifugation technique to separate exosomes from reticulocyte tissue culture in 1987 [32, 33]. In this method, sequential centrifugal forces with varying durations are required to separate exosomes and other components based on their differential size and density [34]. The process is performed at 4˚C. Before initiation, cleaning is carried out to get rid of larger biological material in a sample. Firstly, living cells are removed at 300 g, followed by the removal of dead cells, cellular debris, and large-sized EVs with low and medium speed centrifugation, i.e., at 2000 g and 10,000 g, respectively. Exosomes are then separated by high-speed centrifugation at 20,000 to 100,000 g. The isolated exosome pellet is suspended in phosphate buffer saline (PBS) which eliminates any leftover proteins. The pellet is then kept at -80˚C [35]. It is, however, susceptible to contamination from other lipoproteins and aggregates that are similarly pelleted at high speeds.
Density gradient centrifugation is yet another method which is used to improve purity and recovery rate. It is based on both ultracentrifugation and density-gradient medium. Principally, they are of two types, based on (i) medium-sucrose gradient media and (ii) iodixanol gradient media [36, 37], wherein the former is mostly used for biomolecules, including exosomes. Following ultracentrifugation, a sucrose cushion (10–40%) is used to purify isolated exosomes. Despite its great uses, it has some limitations, viz., it requires more time and is unsuitable in the case of low-volume samples like aqueous humor. It also has less mechanical stability which increases the chances of vesicle rupture [38]. Some studies report that the yield and purity of isolated exosomes mostly depend on factors like centrifugation time, force, rotor type, and sample viscosity [34]. Wei et al. performed a comparative analysis of isolation methods (ultra-high speed centrifugation vs. precipitation) of exosomes obtained from the serum of lung cancer patients. The particle size and its distribution were analyzed by nanoparticle tracking analysis (NTA) while transmission electron microscopy and cryo-electron microscopy were utilized for studying the morphological characteristics of exosomes. The results indicated that exosomes obtained by ultra-high-speed centrifugation were superior in terms of morphology, production, and smaller particle size (30 nm for centrifugation vs. 150 nm for precipitation) [39].
Size based techniques
Size-based techniques are rapid and do not require special equipment [40]. These are of the following types: ultrafiltration, sequential filtration, size-exclusion chromatography, and size-based microfluidics [34, 35]. In the ultrafiltration technique, exosomes are concentrated from large volumes into small volumes of biofluids using ultrafine nanomembranes with varied molecular weight cut-off (MWCO) from the sample. The separation is based on their size followed by sequential filtration of three continuous steps: normal dead-end filtration which removes cells, cell debris, and larger EVs; tangential flow filtration, in which free proteins are filtered out using a 500 kDa MWCO dialysis bag; and finally track-etched membrane filtration to isolate exosomes by passing samples from a 100 nm membrane filter [40]. Lobb et al. made a comparison between two exosome isolation methods in diagnosing lung cancer i.e., ultracentrifugation and ultrafiltration methods, and found that exosomes isolated from the ultrafiltration method were superior in terms of speed, number, and size of particles (less than 100 nm) [41]. The drawbacks of this approach include its low recovery rate and purity. In addition, the filters could clog up with outside debris [40].
Size exclusion chromatography (SEC) is another technique that separates biomaterials based on their size and molecular weight. In SEC, a liquid sample is allowed to pass through a porous stationary phase column and molecules with different hydrodynamic radii elute at a different rate. Smaller molecules with small hydrodynamic radii enter into the gel pores and thus they tend to elute slowly. Larger molecules having larger hydrodynamic radii, including exosomes, are known to elute faster because they cannot enter into the pores [29]. It is suitable for smaller quantities (as small as 15 µL) of biofluid and it gives highly resolved and reproducible exosome isolation using commercially available SEC columns. A fine adjustment in the pore size of the column yields a variety of biomaterials of different sizes. Despite many advantages, it has some challenges as we may get exosomes with a wide size distribution, which may correspond to some contaminants like lipoproteins or protein aggregates [33]. Some commercially available size exclusion-based kits include qEV original separation columns (Izon Science Ltd), EVs second purification columns (GL Sciences), PURE-EVs (Hansa Biomed), Exospin (Cell Guidance System, USA), ExoLutE (Rosetta Exosome, Korea), ExoMir (Bio Scientific, Austin, Texas, USA), Tangential flow filter-EV concentrator (Novus biologicals), Exosure (GeneCopoeia) [42].
Polymer precipitation method
In this technique, hydrophilic polymers affect the solubility and dispersibility of exosomes in the solvent and precipitate them out [36]. Polyethylene glycol (PEG) is one of the most commonly used fractional precipitating agents for the purification of proteins from a variety of sources [43]. The initial treatment is required to remove cells, after which a sample is incubated with a precipitating agent. After incubation at 4˚C overnight, precipitated exosomes are collected by centrifugation or filtration method [33]. This technique is easy and scalable for large sample sizes, so many companies are focusing on the development of isolation kits. Cho et al. presented a study in which a comparison has been made among the electrophoretic migration method, ultracentrifugation, and PEG precipitation for the isolation of exosomes. EVs were isolated from mouse plasma using ExoQuick solution via the PEG precipitation method. It was observed that PEG precipitation and the electro-migration system had a higher recovery rate than ultracentrifugation. However, the ultracentrifugation method was superior among all enlisted methods in terms of purity. This comparison revealed that particles containing contaminants such as proteins and other debris were recovered utilizing the precipitation approach [44]. The same precipitation method was utilized for the isolation of exosomes from 30 plasma samples of lung cancer patients. Further microRNA analysis was performed with real time-polymerase chain reaction (RT-PCR) method and its potential role as a biomarker for lung cancer was established [45]. Despite its speed, non-complexity and harmless nature without damaging exosomes which could be beneficial in clinical applications, this method is prone to contamination from samples while coisolating exosomes which could potentially interfere with sample analysis. However, this can be overcome by performing one additional step of post -precipitation purification or prefiltration [46]. Some of the precipitation-based products are ExoQuick (System BioSciences, USA), ExoPrep (Hansa Biomed Life Sciences, Estonia), Exosome purification kit (Norgen Biotek, Canada), Ex-Spin Isolation kit (Cell Guidance Systems, USA), PureExo Exosome Isolation kit (Biopalo Alto, CA, USA), miRCURY exosome isolation kit (Exiqon, Denmark), Total Exosome isolation reagent (Invitrogen, Carlsbad, CA, USA), RIBO Exosome Isolation reagent (RIBO, Guangzhou, China) [42].
Immunoaffinity capture chromatography (ICC)
ICC is a technique that uses specific exosome surface biomarkers for capturing exosomes that can couple with antibodies that are covalently connected to magnetic beads, chromatographic matrices, plates, or microfluidic devices. They can further bind to specific antigens or membrane proteins like CD9, CD63, ALIX, and Ep-CAM on the target particles’ unbound sites [30, 47]. Based on ICC, Enzyme-Linked immunosorbent Assay (ELISA), Magneto-immunoprecipitation, and Western blot are some of the techniques which are used for quantifying exosomes in biological fluids such as plasma, serum, and urine [48]. Zhang and co-workers prepared Tim4@ILI-01 immunoaffinity flake material with a view of enriching exosomes from the serum of patients suffering from adenocarcinoma which showed capturing efficiency of 85% which is around 5 times greater as compared to ultracentrifugation method [49]. In other instance, a magnetic bead-based approach was utilized by Shih et al. [50] for capturing circulating exosomes in analyzing human lung carcinoma wherein phosphatidylserine-binding protein and annexin A5 coating on magnetic beads were employed. The findings stated that this method was able to capture as much as 60% of induced apoptotic bodies. Overall, this technique is suitable for qualitative and quantitative analysis of exosomes, however; some constraints, such as its heavy cost, low yield, rough use, and storage conditions, limit its applicability on a large scale [47]. Some of the commercially available ICC-based products include exoRNeasy Serum/plasma kit (Qiagen, Hilden, Germany), Exosome Isolation kit CD63 isolation reagent (Thermofisher, USA), Exosome Isolation kitCD81/CD63 (Miltenyi Biotec, USA), Exosome-Human EpCAM isolation reagent (Thermofisher, USA), MagCapture Exosome Isolation kit (Qiagen, Germany), Capturem exosome isolation kit (Takara Bio, Europe), CD63 Immunobeads for exosome isolation (Novus Biologicals, Canada) [42].
Microfluidic techniques
Microfluidic techniques also referred to as a lab-on-a-chip-type microfluidic system, is the rapid microscale separation technique that deals with the physical and biochemical properties of exosomes such as size, density, and immunoaffinity for the isolation and detection of exosomes on a single chip [30]. In addition, some innovative mechanisms like acoustic, electrophoretic, and electromagnetic aspects can be integrated for the efficient capturing of exosomes [30]. The microfluidic techniques can be either active sorting or passive sorting. If the exosomes are captured by using externally applied control, such as acoustic standing waves, then it is considered active separation. If the exosomes are captured by means of microchannels, pores, and traps, then it is called passive separation. Size-exclusion, immunoaffinity, and flow-induced methods are considered to be passive sorting techniques, while acoustofluidics, electromagnetic, and electrophoretic are active sorting techniques. Both the active and passive modes can be integrated into a single microfluidic chip [51]. With an increase in the surface area of the chip, the performance also gets better.
Size-based microfluidics devices, fitted with filters of different pore sizes are used to sort exosomes. Exosomes are preserved in the devices when fluid flows over the channel. These devices include nano filters, nanoporous membranes, or nanoarrays [48]. Recently, an exosome track-etched magnetic nanopore (ExoTENPO) chip has been developed which incorporates several immune-labeled magnetic beads onto a single chip.
As an example of integrated technology, Wu et al. developed a unique automated on-chip technology, which separates desired EVs like exosomes directly from the undiluted whole blood samples. They have reported a separation method that was based on acousto-fluidics (integration of acoustics and microfluidics). By using the acoustic microfluidic chip, the exosomes were isolated as a basis of differential sizes. The whole blood is subjected to acoustic waves. The acoustic radiation force generated by acoustic standing waves is directly proportional to the size of the components. Thus, components of different sizes were moved to different recovery zones, which resulted in the isolation of exosomes. Two modules of this platform contained a cell removal module, in which larger blood components were removed with a 99% yield. Secondly, there was an exosome isolation module in which exosomes were isolated from the EVs mixture with 98.4% purity. By adjusting the input power of the radio frequency signal and the rate of fluid flow, the cut-off size for each module can be maintained. It offered the advantages of being contact-free, biocompatible, rapid, and able to achieve higher purity isolation of exosomes. It also preserved the desired characteristics and structure of exosomes [52].
Similarly, integrated electromagnetic beads and electrophoretic approaches with microfluidics were used to isolate EVs by automated continuous separation from biological fluids (e.g., plasma) with higher purity and yield [44, 53]. Cho et al. presented an electrophoretic migration method for the isolation of EVs from the plasma of melanoma mice and the results were compared with PEG precipitation and ultracentrifugation. An electric field was applied across a semi-permeable membrane with a suitable pore diameter (30 nm) for protein migration. In comparison to conventional procedures, this method recovered up to 65% of EVs (7.9 times better than ultracentrifugation, but EVs isolated by ultracentrifugation have been assumed to have the maximum purity compared to electrophoretic migration method and protein precipitation method) and this method can remove around 83.6% of proteins in 30 min (9 times faster than ultracentrifugation). It was observed that the sample isolated using polymer precipitation had some impurities, but the precipitation could collect the majority of the EVs (34.7% RNA recovery rate). After optimizing the working range of voltage, suitable device geometry, and appropriate buffers were used, and isolated EVs were fully compatible with biological processes and assays [44, 53].
Methods of loading anti-tumor cargo in exosomes
Exosomes are emerging as a potential cargo delivery platform, and identifying a strategy for effective cargo loading has become a necessity for using EVs as drug carriers. Methodologies to load cargoes into exosomes can be divided into two broad approaches: exogenous loading after EVs isolation and endogenous loading during EVs biogenesis and fusion method [38, 39].
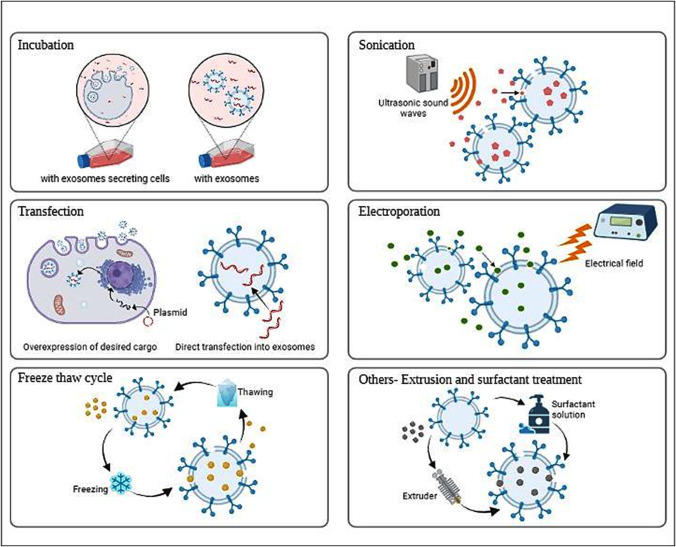
Incubation in exosomes, electroporation, sonication, and other techniques such as freeze-thaw cycles, saponin-assisted permeabilization, and extrusion are examples of exogenous cargo loading strategies. Endogenous loading is a technique of packaging particular materials into exosomes by influencing donor cells; strategies include donor cell incubation and transfection (Fig. 3) [54].
Fig. 3.
Different methods of loading cargo into exosomes
Coincubation
Exosomes (exogenous loading) or exosome-secreting cells (endogenous loading) are co-incubated with desired cargoes so that cargo diffuses across exosomal or cell membranes with the concentration gradient and gets enclosed into exosomes [55]. Principally, lipophilic drugs can be loaded via passive diffusion. Recent studies show that this method is simple as well as inexpensive compared to others [56].
Gong et al. studied the synergistic efficacy by co-delivering miR159 and doxorubicin using targeted exosomes for triple-negative breast cancer (TNBC) therapy. In this study, researchers used human macrophage-derived exosomes (A15-Exo). Doxorubicin hydrochloride (Dox) was packaged into A15-Exo by incubating 200 µg/mL of Dox, which showed maximal drug loading. These drug-loaded exosomes (A15-Exo/Dox) were then co-incubated with cholesterol-modified mi159 (Cho-miR159) to form an effective co-delivery system [57]. Munagala et al. investigated that bovine milk-derived exosomes loaded with chemotherapeutic drugs by direct co-incubation at room temperature showed significantly greater efficacy compared to free drug against cell line studies and lung tumor xenografts in vivo [58]. By co-incubating at 37˚C for an hour, nucleic acids like miRNA and proteins can be loaded into tumor-derived exosomes [59]. Incubation can be utilized either as a pre-loading or as a post-loading method [60]. Lin et al. produced hybrid exosomes by simply co-incubating with liposomes in the case of such a large nucleic acid. These hybrid exosomes have been used successfully to deliver plasmids like CRISPR-associated protein 9 (Cas9) expression vectors to MSCs designed to allow in vivo gene manipulation of the target gene [61]. Shaban et al. studied exosomal angiogenic cargo of endothelial cells in the senescence model which was induced by hydrogen peroxide. Exosomes were obtained from human umbilical vein cells (HUVECs) and were treated with complete media (Nor-HUVECs) and hydrogen peroxide (H2O2-HUVECs). It was then subsequently subjected to western blotting analysis of P53, P21, and P16, and gene expression of FMR1, miR-21, and miR-126 were analyzed by real time-PCR. The results indicated that the rate of migration of endothelial cells coincubated with exosomes (treated by H2O2) was decreased and under the influence of H2O2, endothelial cells generated exosomes with distinct cargo which could be utilized as biomarkers for age-related disorders [62]. Dysfunctioning of endothelial cells and senescence have also been implicated in disorders like cancer where cell proliferation is unregulated and poses a great threat worldwide. Further autophagy flux and exosome generation have been interlinked with each other in maintaining cell homeostasis. This autophagy pathway was studied by Mahbubfam et al. [63] in exosomes derived from HUVECs on incubation with H2O2. After performing molecular analysis, a significant elevation of CD63, CD81, TSAP6, and Rab11 was observed on exposure to H2O2 which indicated exosomal pathway induction in tandem with the autophagy process fostering senescence. Any modification in autophagy could change the further differentiation ability of CD146+ to mature into endothelial cells as evidenced by the research conducted by Hassanpour et al. [64]. A similar study was reported by Feghhi et al. [65] wherein HUVECs were incubated with polyhydroxylated polyhedral oligomeric silsesquioxane nanoparticles reinstating the key role played by exosomes in the process of angiogenesis.
Electroporation
Electroporation is another widely used technique, especially for hydrophilic molecules like doxorubicin [66–68]. Under short, high-voltage electric pulses, electroporation can form transient gaps (micropores) in the exosomal membrane, allowing drug loading with enhanced permeability. Some factors, such as voltage, pulses, pulse duration, time interval, and condenser capacity, need to be optimized [56].
Gomari et al. worked on “Targeted delivery of doxorubicin to HER2 positive tumor models.” Here, exosomes were isolated from transduced mesenchymal stem cells. Specified quantities of purified exosomes and doxorubicin were gently mixed with electroporation buffer at 4˚C and electroporation was performed. They reported that delivery of targeted doxorubicin-loaded exosomes efficiently reduced the growth rate of a breast cancer tumor model as well as the dosage of the drug [66]. Taffoli et al. reported that, compared to simple diffusion, electroporation boosted the doxorubicin loading efficiency threefold [56, 69]. However, exosomes directly loaded with nucleic acid via electroporation, on the other hand, have been demonstrated in certain instances to exhibit impaired functionality or even become inactive in recipient cells. Therefore, it has been concluded that electroporation might have a role in the formation of insoluble aggregates of nucleic acids [70].
Sonication
Sonication is a physical strategy based on the sonoporation phenomenon that uses low-frequency ultrasound waves to induce cavitation bubble formation. With the bursting of microbubbles, it produces cellular membrane pores by weakening membrane integrity, which allows cargo to penetrate cells [55, 71].
Li et al. mixed exosomes from pancreatic cancer cells with gemcitabine and sonicated the mixture. Exosomes loaded with gemcitabine using sonication were collected and found to have a higher loading capacity, i.e., 11.68 ± 3.68%, than exosomes incubated with gemcitabine, i.e., 2.79 ± 0.72% [72]. However, membrane alteration caused by sonication may reduce the loading effectiveness of hydrophobic drugs [73]. It has been observed that sonication when used to load siRNA into EVs causes less siRNA aggregation than electroporation; nonetheless, the amount of siRNA internalized into recipient cells by exosomes is still limited [74]. Kim et al. loaded exosomes with paclitaxel using three different methods viz., incubation, electroporation, and sonication. Loading capacity for incubation was found to be 1.4%, 5.3% for electroporation, and 28% with the sonication approach [73]. Recently, Sun et al. worked on ultrasound-based exosomal delivery of tissue-specific microRNA (miRNA) to boost efficacy while minimizing off-target effects [75].
Transfection
Transfecting cells or exosomes with protein-expressing plasmids or nucleic acids is a typical approach to enhance the loading of nucleic acids and proteins into exosomes [55]. For direct transfection of exosomes, some commercial transfecting reagents are available, such as HiPerFect reagent and Lipofectamine 2000, but efficiency was found to be very low rendering this method inappropriate for therapeutic purposes [59]. Transfection of exosome donor cells is an endogenous loading technique. The desired cargoes (e.g., RNA or proteins) as well as other genes of interest are engineered into exosome donor cells. Donor cells may overexpress the inserted gene and pack it into exosomes. This endogenous sorting of inserted cargo occurs during EVs biogenesis, followed by the release of the same EVs. Engineered EVs are then separated and purified [59, 71, 76]. For example, miR584-5p genes were transfected in mesenchymal stem cells (MSCs) before the isolation of exosomes from the supernatant. These exosomes were designed to deliver miR584 to glioma cells, resulting in the lower expression of matrix metalloproteinase-2 (MMP-2). MMP-2 has a crucial contribution in the progression of cancer. The extracellular matrix is significantly degraded by matrix metalloproteinase-2 (MMP-2), which increases the propensity for cancer to invade, proliferate, and spread [77]. Severic et al. studied “Genetically-engineered anti-PSMA exosome mimetics targeting advanced prostate cancer in vitro and in vivo”, in which the PSMA targeting peptide was expressed on the surface of monoblastic U937 cells by nucleofection. The cells were then extruded to generate PSMA-targeted exosome mimetics with active targeting characteristics against PSMA-expressing malignancies [78].
Physicochemical and morphological characteristics of exosomes or their cargoes are subsequently altered as a result of the aggregation caused by electroporation, sonication, or another physical approach, which promotes the loading of cargoes endogenously. [76]. However, this process is time-consuming and costly, and it is not ideal for large-scale manufacturing; also, transfection reagent application might harm or contaminate cells and exosomes. [55, 78].
Extrusion
An extrusion is a physical approach that causes membrane recombination when exosomes and cargoes are extruded together. Exosome membranes split and homogeneously blend with payloads during multiple extrusion cycles, resulting in cargo-loaded exosomes. The extrusion technique allows for the formation of an exosome-like nanostructure by repeatedly running donor cells or pure exosomes through polycarbonate membranes with different pore sizes under controlled pressure and temperature by using a mini-extruder. During the extrusion process, the structure of cells or exosomes is disaggregated into free lipid and protein molecules [55]. Exosomes can also be produced by multiple extrusions, which has a 100-fold higher production yield of cargo-loaded EVs [79]. Lunavat et al. proposed naturally released exosomes as RNAi carriers to develop RNA-based therapeutics, they developed exosome-mimetic extra vesicles by multiple extrusion of cells through filters followed by loading of these vesicles with the specific siRNA by electroporation [80]. Similarly, Jhan et al. used an extrusion technique to overcome the major obstacle to mass production. Using a membrane extrusion approach, they produced modified EVsby combining EVs surface composition with lipid-based components on a large scale. Following this approach, the number of vesicles post-isolation increased 6 to 43 times [81].
Kalimuthu and colleagues extracted exosomes from mesenchymal stem cells (MSCs). Cells were mixed with various concentrations of paclitaxel (25 µg, 50 µg, and 100 µg) and subjected to extrusion using polycarbonate membrane filters of various sizes (10 μm, 5 μm, and 1 μm) using a mini-extruder, with the optimum concentration (50 µg/mL) attaining a loading efficiency of 76% [82].
Fuhrmann et al. used a variety of passive and active ways to load porphyrins into exosomes, including electroporation, surfactant, extrusion, and dialysis. They observed variations in zeta potential compared to other loading methods due to the repeated and vigorous extrusion processes; moreover, extruded exosomes promote cytotoxicity, probably due to the surface modifications of the EVs membrane. Secondly, they observed that the viability of MDA-MB231 breast cancer cells treated with extrusion-loaded exosomes was considerably reduced compared to electroporation-loaded exosomes. In these studies, extrusion appears to have a high payload efficiency. However, recombination of exosomal surface structure might change the immunological aspects of exosomes, making them detectable to mononuclear phagocytes [83].
Freeze-thaw cycles
Freeze and thaw cycles are the most effective method to load cargo into exosomes. Cargoes and exosomes are incubated at room temperature for a specific time and then rapidly subjected to liquid nitrogen at -80˚C for freezing followed by thawing at room temperature. The cycle is repeated at least three times. The principle behind this approach is that after a few cycles of freezing and thawing, the lipid bilayer of exosomes is slightly disrupted, allowing cargo to pass into them [84].
Haney et al. worked on the design of a new exosomal-based drug delivery method for catalase, a powerful antioxidant, for the treatment of Parkinson’s disease (PD). Catalase was added into exosomes ex-vivo via a variety of methods, including room-temperature incubation, saponin permeabilization, sonication, freeze-thaw cycles, and extrusion. Then the catalase solution was loaded into exosomes as discussed above, incubated for 30 min, immediately frozen at -80˚C, and thawed at room temperature. The cycle of freeze-thaw was done three times. However, they observed that the loading significance of this process was generally moderate when compared to other techniques [85].
Kalani et al. designed a combined nanoformulation of curcumin (a neuroprotective molecule) and mouse embryonic stem cell exosomes (MESC-exo) to cure ischemia-reperfusion injury in mice. Curcumin was mixed with MESC-exosomes in a fixed proportion (1:4). After further incubation for 15 min at RT, rapid freeze-thawing was done for 2–3 times. The free drug was removed by centrifugation, and the nanoformulation was precipitated by ultracentrifugation [86]. Sato et al. engineered hybrid exosomes by fusing exosomal membranes with liposomes using the freeze-thaw method to improve the performance of exosomal nanocarriers for use in advanced drug delivery. In this, results suggest that this new strategy can be used to transport exogenous hydrophobic lipids as well as hydrophilic cargoes [87].
Recently, a study has been conducted to compare different methods to entrap hydrophilic low molecular weight compounds in stem cell-derived small EVs and to assess the impact of these loading methods on vesicle integrity. Pyranine and pentoxifylline were selected as probe hydrophilic models. When compared, freeze-thawing and osmotic shock have both shown to encapsulate small EVs better and keep their structure and biological functions intact [88]. Besides, repeated freeze-thaw cycles may cause protein inactivation and exosome clumping.
Other techniques
Saponin-assisted loading, hypotonic dialysis, and several innovative procedures are also used for loading antitumor cargoes into exosomes. Surfactants like saponin and triton, which may denature membrane components like cholesterol, are also used to generate pores on the surface of exosomes without disrupting the lipid bilayer membrane which ultimately leads to greater membrane permeability. However, since saponin is hemolytically active in higher quantities in vivo, the saponin utilized for drug loading should be employed in a regulated way, and a further purification step to nullify saponin is necessary. The hemolytic impact of saponin on blood cells is a significant concern, which might limit its therapeutic use [55]. Compared to other loading procedures (electroporation, extrusion, saponin, and dialysis), the saponin-assisted approach encapsulated hydrophilic porphyrins 11 times more effectively [83]. Recently, Kwon et al. developed an exosome-based hybrid nanostructure (EHN) with better targeting capacity and therapeutic efficiency against colorectal cancer. Metallic nanoparticles containing doxorubicin (Dox), and folic acid which act as a tumor targeting ligand are bound to EHN. In addition to incubation, the EHN were loaded with 0.2% saponin as a permeation enhancer [89].
Additionally, hypotonic dialysis has also been reported as a method for loading cargo into extracellular vesicles. As discussed in the above techniques, Fuhrmann et al. used hypotonic dialysis for the loading of porphyrins having intermediate hydrophobicity. To produce drug-loaded exosomes, the mixture of exosomes and porphyrins was poured onto dialysis membranes, and then dialyzed by stirring in 10 mM phosphate buffer. When compared to incubation at room temperature, this technique was reported to improve drug loading. It did, however, seem to change the size distribution pattern of exosomes. On the other hand, Fuhrmann et al. reported that porphyrin-loaded exosomes using hypotonic dialysis showed poor cellular uptake and thus had no impact on the photodynamic effect [62]. In addition, Wei et al. developed a nano drug combining doxorubicin and exosomes extracted from mesenchymal stem cells, which was investigated in vitro against osteosarcoma. Here, exosomes were extracted from mesenchymal stem cells using an isolation kit. Exosomes were blended with doxorubicin hydrochloride and dialyzed against phosphate buffer saline (PBS) overnight to load the drug into them. Dialyzed exosomes were taken up by cancer cells and released doxorubicin to inhibit cancer development in vitro [90].
Yang et al. recently published a novel cellular nano poration approach for mass-producing exosomes containing therapeutic mRNAs and targeting peptides. Cellular nanoporation produced 50-fold more exosomes and 1000-fold more exosomal mRNA transcripts compared to electroporation and other exosome production methods. In orthotropic PTEN-deficient mouse models, these mRNA-containing exosomes suppressed tumors and inhibited tumor development [91]. Recently, a new strategy called, “exosomes for protein loading via optically reversible protein-protein interaction” (EXPLORs) has been revealed for effective protein loading via exosomes which can also address the shortcomings of prior strategies. [59, 71]. Through this mechanism, cargo proteins are able to be actively loaded into exosomes employing endogenous biogenesis processes. This enables efficient cytosol delivery by means of regulated and reversible protein-protein interactions. To achieve the same goal, the protein modules photoreceptor cryptochrome 2 (CRY2) and CRY-interacting basic-helix-loop-helix 1 (CIB1) were selected to regulate floral initiation by phosphorylation in response to blue light. In order to bind CRY2-conjugated cargo proteins into exosomes, the CIB1 protein was first conjugated with the exosome-associated protein CD9, and then blue light illumination was employed. After the cargo protein was injected into exosomes and then separated from the CD9-conjugated CIBN by turning off the light source, it was released into the intraluminal area of the exosomes [92].
Characterization of loaded exosomes
There has been a great deal of advancement in the detection and characterization of EVs. Some biophysical techniques, including nanoparticle tracking analysis (NTA), dynamic light scattering (DLS), flow cytometry, and imaging-based techniques, including electron microscopy, atomic force microscopy (AFM), and biochemical characterization such as blotting techniques have been used for the characterization of exosomes.
Nanoparticle tracking analysis
Nanoparticle tracking analysis (NTA) has become the gold standard for the characterization of exosomes in the recent past. This fluorescence readout method has been used for the determination of the concentration and size of exosomes, capturing diameters within the range of 50-1000 nm. In this method, a laser beam is passed through the exosome-containing solution. Scattered light from the particles is then captured in the light-sensitive CCD camera and analyzed by image processing software. The software monitors each vesicle in Brownian motion and uses the Stokes-Einstein equation to relate Brownian motion to particle size [31, 36]. As compared to flow cytometry, NTA has greater resolution. Dragovic et al. demonstrated using human placental exosomes that NTA can measure EVs as small as 50 nm and with more sensitivity than conventional flow cytometry, having a lower limit of 300 nm [93]. Malvern has produced a commercial NTA instrument, branded as ‘Nanosight’, to measure a size range of 10 nm-2 μm and concentration within the range of 106 to 109 particles per mL [94]. Many researchers have used Nano sight (Malvern Instruments Ltd) to analyze the size distribution of isolated exosomes from various sources [58, 70]. However, NTA is having some challenges too; it needs a sample volume of 0.5 mL and involves a lengthy data acquisition procedure. So, it may cause photo-bleaching of fluorescent dye, as well as similar sizes of dye aggregates can be confounded with the obtained results of quantification. It needs many parameters for the optimization of data collection [40, 95].
Dynamic light scattering
Dynamic light scattering (DLS) is another method similar to NTA. Incident illumination is passed through the solution, and fluctuations in the intensity of scattered light from particles due to Brownian motion are detected at a certain angle to determine particle size and concentration. DLS needs sample volume in a small quantity (70µL) and is easy to use, compared to NTA [96]. The main disadvantage of DLS is that it has lower sensitivity and specificity in heterogeneous mixtures. Because the accuracy of the DLS can be distorted by the presence of only a few large particles, sample preparation needs to be done carefully [94]. DLS requires a higher concentration of the sample, which is challenging for exosomes [47, 84]. The Stokes-Einstein equation can then be used to obtain the size as well as the polydispersity index of particles present in the solution [29].
Flow cytometry
Flow cytometry is a typical method in which individual particles are passed in front of a laser beam and the scattered light or emitted fluorescence is measured at a certain angle using a forward angle light scatter detector, a side-scatter detector, and multiple fluorescence emission detectors [31, 84]. This method is a high-throughput analytical technique. Unlike NTA, it is a faster method, and unlike DLS, it requires less sample concentration [47]. Flow cytometers can distinguish exosomes based on their proteins, but it is not useful to detect particles smaller than 300 nm in diameter. Furthermore, the equipment is also expensive [48]. Fluorescence-activated cell sorting (FACS) is a specialized flow cytometry that captures and sorts exosomes based on fluorescent labeling using specific antibodies [31]. Flow cytometry has also been used for the analysis of exosome markers. Exosomes were incubated with fluorescein-conjugated specific antibodies (which are against a specific exosomal membrane marker) at 4˚C. The fluorescein-stained beads were then suspended in FACS buffer for further analysis by the BD FACSCalibur flow cytometer. FlowJo software was used for the data analysis. Thus, the presence of CD9 and CD63 protein markers on the membrane of exosomes confirmed the exosome identity [67].
Electron microscopy
Electron microscopy provides higher resolution and greater magnification than optical microscopy. Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) are the most commonly used microscopic techniques for the imaging of exosomes. An electron beam is used to create high-resolution images of submicron level. SEM analysis detects scattered electrons and TEM analysis detects electrons that pass through the sample [96]. It was observed that the determination of sample concentration and size distribution by electron microscopy can be risky. It has been proposed that electron microscopy can be used to identify exosomes, assess the quality of isolated exosomes, and ensure that exosomes are not ruptured [29, 84].
Cryogenic-Transmission Electron Microscopy (Cryo-TEM) is a gold standard for characterizing the morphology of the exosome. But it is a very costly instrument as well as it requires expertise in imaging and analysis. Atomic force microscopy (AFM) analysis is a technique alternatively used for qualitative analysis of extracted exosomes, i.e., evaluation of size, morphology, and homogeneity of isolated exosomes. This technique produces data on the 3D geometry of EVs. The AFM is advantageous over other techniques in terms of height measurement and other features like compression and deformation of particles can be captured. AFM imaging may be performed on either air or liquid samples [84]. The force between the probe and the sample is measured in this approach, producing an image from which height and diameter may be computed. Vesicle sizing via AFM was found to be consistent with Cryo-SEM [97].
Western blotting is an effective technique to characterize specific marker proteins of exosomes. Exosomes from different sources often contain different proteins intraluminal or on the membrane. Thus, marker-based exosomal characterization was recommended by the International Society for Extracellular Vesicles. The basic principle behind western blotting is simply affinity binding of target proteins and specific antibodies. Characterization of transmembrane proteins (such as CD9, CD63, CD81, etc.) or intraluminal proteins (such as TSG101) can be done by western blotting or enzyme-linked immunosorbent assay (ELISA). However, it just confirmed the presence of these protein biomarkers and not necessarily their colloidal form, which may get ruptured during isolation [84]. The process is complicated as well as time-consuming, too [47].
Exosomes in clinical applications
As diagnostic biomarkers
When tumor cells undergo cellular stress, the exosome release also increases to meet the overgrowing needs of tumor cells. In the process, the cancer cells shed exosomes into biological fluids such as serum, plasma, and urine. Exosomes have also been known to get labeled with fluorescent molecules like Di dyes, PKH67 and CFSE to better understand the pathophysiology of the concerned disease [98]. Notably, exosome-mediated cell-cell communication leads to an exchange of information by delivering nucleic acids, proteins, and lipids between cells, which further promotes tumor progression, proliferation, and metastasis. Therefore, the identification of exosomal nucleic acids, proteins, and lipids within distinct sites of tumor stages provides a key diagnostic tool for clinicians to evaluate and monitor tumor stage and progression. Thus, the exosomes derived from tumor cells highlight their pivotal role in the diagnosis and treatment of tumor disease [99].
Diagnosis using exosomal proteins
The researchers identified several proteins that reflect the alterations that occurred during various stages of cancer. Several exosomal proteins were identified, such as CD24 and EpCAM, in serum and ascites fluid and served as circulating biomarkers in the early stages of breast cancer [100]. EDIL3 and fibronectin were also found to be potential circulating biomarkers in early breast cancer stages along with treatment response markers [101, 102]. Survivin, a protein of the inhibitor of apoptosis (IAP) isolated from plasma-derived exosomes, was observed as a biomarker in breast and prostate cancer patients [103]. Zhao et al. used the ExoSearch chip for blood-based ovarian cancer detection. His group measured the potential biomarkers of CD24, EpCAM, and CA-125 proteins in the plasma samples of ovarian cancer patients [104]. Pancreatic cancer is a deadly disease with a poor prognosis and a high mortality rate. Pancreatic cancer detection is a serious clinical problem due to its poor prognosis and late detection. According to their proteomic profile, exosomes released from pancreatic lesions have the potential to be beneficial as a diagnostic tool for early instances of pancreatic cancer. GTP-binding proteins and glycoproteins, which are membrane-related proteins, were discovered to be the most abundant proteins in pancreatic tissues [105, 106]. The overexpression of epidermal growth factor receptor isoforms has also been found to serve as a biomarker tool for the diagnosis of pancreatic cancer [106]. Melo et al. used mass spectrometry to identify the biomarker protein Glypican-1 (GLP-1) and flow cytometry was used to isolate it from the serum of pancreatic cancer patients in both early and late stages, with absolute specificity and sensitivity [107]. Niu et al. carried out a proteomic analysis and demonstrated significant expression of alpha-2-HS-glycoprotein (AHSG) and extracellular matrix protein 1 (ECM1) in non-small cell lung cancer patients compared to the healthy group [108]. Based on the various biological characteristics of lung carcinoma, it is classified into lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD). In the clinical study, Cao et al. demonstrated an overexpressed level of tumor protein 63 (TP 63) and keratin 5 protein in lung squamous cell carcinoma. While in the case of lung adenocarcinoma, the levels of cell adhesion molecule 6 and surfactant protein were increased when compared to healthy individuals. These LUAD and LUSC carcinoma-specific exosomal protein biomarkers have been proven as effective tools for the diagnosis and identifying efficient treatment strategies [109].
Diagnosis using exosomal nucleic acids
The exosomal miRNA is potentially involved in the regulation of the tumor microenvironment and stimulation of various tumor-related pathways through the transfer of miRNA from the parent tumor cell to other neighboring cells through intercellular communication. Thus, the exosomal miRNA has been considered a biomarker for tumor diagnosis as it is indicative of tumor progression, aggressiveness, and severity. Exosomal miRNA has been reported to be significantly expressed in squamous cell carcinoma, lung, ovarian, colorectal, and pancreatic cancer. Exosomal miRNA 17-5p has been found to be overexpressed in colorectal cancer and pancreatic cancer, which has defined a strategy for identifying metastasis and stages of colorectal cancer [77]. Exosomes enriched in miR-224-5p provided novel potential targets for the suppression and therapy of non-small cell lung cancer (NSCLC) [110]. Huang et al. reported that the low level of miR-34c-3p extracted from exosomes assisted in NSCLC development. Therefore, miRNA has significant potential as a prognostic biomarker in NSCLC [111]. Sun et al. showed miRNA-3607-3p was enriched in the exosomes of natural killer cells. In pancreatic cancer, it suppresses pancreatic cancer cell proliferation, invasion, migration, as well as a malignant transformation of pancreatic cells by directly targeting IL-26 through in vitro and in vivo studies. Thus, miRNA-3607-3p has been proven to be used as a treatment therapy in pancreatic cancer [112]. Further, scientists demonstrated that the overexpression of exosomal miRNA-23b-3p [113] and miRNA 339-5p [114] resulted in pancreatic cancer cell growth, migration, and invasion activities. So, both these miRNAs were suggested as promising agents in the diagnosis of pancreatic cancer. Zhou et al. identified upregulated levels of miRNA-217 and miRNA-23b-3p via high-throughput sequencing. In vitro and in vivo studies revealed that upregulated miRNA-217 levels have promoted cell proliferation and invasion in prostate cancerous cells. Upregulated miRNA-23b-3p levels, on the other hand, supported the inhibition of cell proliferation and invasion in a high throughput screening study, indicating that both miRNAs can be used in the diagnosis and treatment of prostate cancer [115]. In breast cancer, the exosomal miRNA-21 was overexpressed in the late stage of cancer, which was considered a progress indicator for the late stage of breast cancer [116]. In the early stage of breast cancer, high levels of miRNA-105 [117] and overexpression of miRNA-373 in triple-negative breast cancer patients have been proven as effective biomarkers [118]. In the metastatic stage of breast cancer, exosomal miRNA-1246 is highly expressed and is implicated in cell proliferation, drug resistance, and cell migration [119]. In addition, there were reports demonstrating that an increase in exosomal miRNA-222 expression was associated with Adriamycin resistance in breast cancer patients [120].
Diagnosis using exosomal lipids
The exosomal lipid bilayer membrane contains a variety of lipids that help to preserve the exosomal shape and safeguard the proteins and nucleic acid contents [121]. Abnormal lipid metabolism is closely related to cancer progression and metastasis [122]. The exosomal lipid composition was found in prostate, breast, and pancreatic cancer cells, as well as hepatocellular carcinoma, glioblastoma, melanoma, B-lymphocytes, oligodendroglia precursor cells, mast cells, adipocytes, reticulocytes, and platelets. Exosomes contain a variety of lipids, including cholesterol, sphingomyelin, phosphatidylcholine, phosphatidylethanolamine, phosphatidylinositol, and ceramide [123–125]. Min et al. demonstrated potential lipid biomarkers such as phosphatidylcholines, phosphatidylethanolamines, phosphatidylserines, and phosphatidylinositol in prostate cancer patients [126]. Lea et al. investigated the endogenous lipid phosphatidylserine secreted by exosomes in the plasma of prostate cancer patients and healthy individuals. They found that the levels of phosphatidylserine in benign and malignant patients were higher than the healthy individuals [127]. The glycolipids such as hexosylceramides and lactosylceramides were abundantly found in prostate cancer-derived exosomes [128]. Lea et al. showed that the abundance of exosomal phosphatidylserine in ovarian cancer was higher than the healthy individuals. They carried out ELISA which could quantify even a picogram of phosphatidylserine (PS) and further based on a statistical test, the amount of phosphatidylserine was found higher in malignant women than in benign women. Thus, this study revealed the detection of exosomal phosphatidylserine in the blood of women and it has been proven as a biomarker for ovarian cancer [127]. In another instance, Skotland et al. established the potential utilization of lipids in urine exosomes as a prostate cancer biomarker. The quantitative lipidomic analysis using high throughput mass spectrometry revealed the overall composition of lipids in exosomes in urine samples of both prostate cancer patients and healthy individuals. The results warranted its use as a biomarker in prostate cancer [124].
As therapeutic target
Exosomes secreted by tumor cells can be used as a carrier of tumor-associated antigens carrying therapeutic targets, that have potential value in antitumor vaccination. Shi et al. evaluated the anticancer effects of a new exosomal vaccination which is comprised of an interferon-modified exosomal vaccine in an effort to counteract prostate cancer [128].
The exosomal vaccination was prepared to utilize a protein anchoring approach with cancer cell-derived exosomes. The immunogenicity and therapeutic efficiency of exosomes were assessed by evaluating the effects of the exosomal vaccine on M1 macrophage differentiation, macrophage capacity to ingest exosomes, antibody generation against exosomes, tumor angiogenesis, metastasis, and tumor development. The exosomal vaccination reduced the expression of vascular endothelial growth factor receptor 2 and reduced exosomes’ ability to promote tumor metastasis. The exosomal vaccination effectively reduced tumor development and increased survival time in mice with prostate cancer. As a result, this study demonstrated the utilization of exosomes as a therapeutic tool in immunotherapy for human prostate cancer [129]. Hartman et al. used tumor-associated antigens and targeted exosomes. The results demonstrated enhanced expression of tumor-associated antigens such as carcinoembryonic antigen and HER2, which were coupled to the C1C2 domain of cadherin protein in exosomes in vitro. It was concluded that exosomal targeting supported the anti-tumor vaccination strategy [130].
As drug carrier
Based on the evidence of drug integration into exosomes, it is hypothesized that drug-loaded exosomes may be used as therapeutic formulations for tumor targeting [73, 131]. Pascucci et al. loaded paclitaxel in mesenchymal stromal cells (MSC) derived exosomes as MSC have a prominent capability of uptake and release of exosomal vesicles. They used murine SR4987 cell lines as mesenchymal stem cell models and further release of paclitaxel from SR4987 cell lines has been investigated by HPLC. Human pancreatic cell lines CFPAC-1 were used to test the antitumor activity. The results demonstrated that MSC-derived paclitaxel-loaded exosomes have been proven to be an effective drug delivery system with a higher cell-target specificity [132]. Yong et al. developed novel biomimetic nanoparticles involving exosome-sheathed doxorubicin-loaded porous silicon nanoparticles that achieved increased tumor accumulation, extravasation from blood vessels, and penetration into deep tumor parenchyma with intravenous treatment, considerable cellular absorption, and cytotoxicity in both bulk cancer cells and cancer stem cells. The outcomes of these biomimetic nanoparticles suggested that the proposed exosomal-biomimetic doxorubicin-loaded porous silicon nanoparticles exocytosed from tumor cells proved to be a promising novel drug delivery system for cancer chemotherapy [133]. Zang et al. studied the effect of cisplatin-loaded umbilical cord-derived macrophage exosomes on ovarian cancer cell growth and treatment resistance. They tested the efficiency of M1 exosomes isolated from umbilical cord blood (UCB) monocytes for cisplatin delivery to drug-resistant ovarian cancer cells. The results demonstrated that UCB-derived M1 exosomes carrying cisplatin inhibited the development of the epithelial ovarian cancer lines A2780 and the cisplatin-resistant cell lines A2780/DDP better than cisplatin alone, notably in A2780/DDP cell lines [134]. For the treatment of pancreatic cancer, Yong et al. loaded gemcitabine in autologous exosomes. They loaded gemcitabine into exosomes using a sonication method, and cell line studies revealed that cellular uptake of autologous exosomes to parent cancer cells was selective when compared to heterologous cellular uptake, with improved cellular uptake, a regulated drug release profile, and preferable targeting efficacy to the tumor site. It was shown that administering an injection of exosome-loaded gemcitabine resulted in considerable tumor clearance, decreased tumor development following therapy, and extended life in tumor-challenged mice in a dose-dependent manner. The suggested formulation demonstrated, virtually perfect biocompatibility with lower immunogenicity, and the toxicity of free gemcitabine was significantly reduced using an exosome-loaded gemcitabine formulation [72]. Elanz and co-workers formulated targeted doxorubicin-loaded mesenchymal stem cell-derived exosomes for the treatment of colorectal cancer. They incorporated doxorubicin through the electroporation method, separated by ultracentrifugation, and encapsulation efficiency was found to be 35%. They also did functionalization of exosomes; the exosomal surface amine groups were covalently bonded with carboxylic acid-end Mucin 1 (MUC1) aptamer to provide selective guided drug delivery. The results demonstrated that doxorubicin-loaded functionalized exosomes provided preferential doxorubicin transportation to MUC1-positive cancer cells via in vitro cell line study and in vivo study reflected that single-dose intravenous injection of doxorubicin-loaded functionalized exosomal formulation significantly suppressed tumor growth compared to non-functionalized doxorubicin-loaded exosomes and free doxorubicin. From the results, it was concluded that MUC1 aptamer-decorated exosomes can be implemented therapeutically for the safe and effective delivery of doxorubicin to colon adenocarcinoma and can offer a promising platform for cancer therapy.
Chemoresistance
In recent years, exosome-induced resistance to chemotherapy has evolved as a novel mechanism. Exosomes confer resistance through direct drug export, trafficking of drug efflux pumps, and cell-to-cell exchange of miRNAs [135]. In the case of direct export of drugs, several drugs have been reported to be expelled into exosomal vesicles via the physical binding of drugs to the exosomal membrane, thereby proving the association of chemoresistance with vesicular shedding kinetics [136]. Later, it was found that the anthracyclines class of molecules showed preferential export from the cytoplasm into exosomes which have been assigned a regulatory role of ATP-transporter A3(ABCA-3) protein in exosome shedding [137, 138]. Koch et al. revealed that reduced exosome biogenesis leads to enhanced intracellular retention of doxorubicin after blocking ABCA-3 expression [137]. Generally, exosome biogenesis is often unregulated in drug-resistant cancer cells compared to drug-sensitive cancer cells [139]. Furthermore, chemotherapeutic drug therapy and tumor microenvironment conditions such as low pH are known to enhance exosome formation [140]. The rationale for developing compounds that target exosome formation to increase cancer cell chemosensitivity has been outlined collectively. Safaei et al. demonstrated two mechanisms for cisplatin resistance in ovarian cancer cells. The former encompasses resistance developed via increased exosomal pathways and the latter via direct drug export in exosomes [141]. Another reported mechanism for the development of resistance to chemotherapy is via efflux pump transporters. Exosomes have been reported to horizontally transfer the P-glycoprotein (P-gp), multidrug-resistant protein-1 (MRP-1), ATP-binding cassette transporter A3 (ABCA-3), and ATP-binding cassette transporter G2 (ABCG-2). Among all efflux pump transporters, P-gp exosomal delivery has been widely implicated in the development of cancer chemotherapy resistance [142, 143]. In the instance of miRNA-mediated resistance, miRNA exchange among cancer cells leads to more complex chemotherapeutic heterogeneity within the tumor microenvironment. Exosomal miRNA exchange has been linked to the development of resistance in two ways: exosomal miRNA interchange between drug-resistant and drug-sensitive tumor cells and functional miRNA transfer between tumor microenvironment cells and cancer cells. The exosomes extracted from docetaxel and Adriamycin-resistant breast cancer cells carried miRNA that caused treatment resistance in MCF-7 cells. Here, particular miRNAs such as miR-100, miR-222, miR-30a, miR-24, miR-26a, and miR27a have been discovered to be associated with the formation of the resistant phenotype.
As anticancer vaccine
The concept of cancer vaccine dated back to the 1970s with tumor peptides eliciting assuring outcomes. In the early 2000s, scientists found the expression of major histocompatibility complex (MHC)-I and MHC-II molecules from exosomes derived from dendritic cells induced cytotoxic T lymphocyte priming and tumor reduction. With the advent of engineering techniques and antigen-presenting artificial models, exosome-based vaccine development has taken a giant leap and is reflected in many clinical trials (Table 1). The cells which have been infected with viruses release exosomes that possess the ability to induce anti-viral interferon response. This attribute makes exosomes an indomitable player in anticancer vaccine development [144]. Cancer vaccines can exploit the use of tumor-specific antigens such as HER-2, human papillomavirus (HPV), and melanoma-associated antigen peptides-1 (MAGE-1) present exclusively in cancer cells [145]. Scientists have established a link between toll-like receptors (TLR) and tumor-derived damage-associated molecular patterns (DAMPs) inducing the release of cytokines through the activation of T cells. Damo et al. investigated the effect of a vaccine derived from ovalbumin and TLR pulsed bone marrow dendritic cells and found that the vaccine was responsible for the induction of CD8+ T cell proliferation and increase in the level of TNF-α CD4+ T cells in lymph nodes of mice suffering from melanoma and slowed the progression of tumor in vaccinated mice [146]. In another instance, Hartmann et al. investigated recombinant adenoviral vectors which expressed the extracellular domain of carcinoembryonic antigen and found elevated levels of protein in exosomes of a transgenic murine model. This research finding highlighted the low immunogenicity of tumor-associated antigen in patients suffering from cancer [130].
Table 1.
Overview of exosome-based clinical trials in the treatment of cancer
| Sr. No. | Disease | Exosome source | Status | Remarks | Clinical Trial Identification |
|---|---|---|---|---|---|
| 1 | Pancreatic cancer | Ultrasound-guided portal venous blood exosomes | Recruiting | Safety of sampling portal venous blood, analyzing mRNA markers. | NCT03821909 |
| 2 | Colon cancer | Curcumin conjugated with plant exosomes | Active, Phase I | Comparing exosome loaded curcumin on immune modulation, phospholipid profile of normal and malignant patients. | NCT01294072 |
| 3 | Sarcoma | Blood samples | Recruiting | Evaluation of cancer pathogenesis, progression and treatment efficacy of exosomes | NCT03800121 |
| 4 | Prostate cancer | Urine exosomes | Completed | Validation of non-digital rectal examination (DRE) exosome gene expression test of prostate cancer in biopsy. | NCT02702856 |
| 5 | Pancreatic cancer | Blood samples from patients | Active, not recruiting | Exosome purification for RNA sequencing and proteomics | NCT02393703 |
| 6 | Lung metastasis osteosarcoma | Blood samples | Recruiting | Identification of levels of circulating exosomal RNA with or without lung metastasis | NCT03108677 |
| 7 | Gallbladder carcinoma | Exosomal blood samples | Recruiting | Establishing a correlation between exosome biomarkers and gallbladder carcinoma | NCT03581435 |
| 8 | Stage IV pancreatic adenocarcinoma | Mesenchymal stromal cells-derived exosomes with KRAS G12D siRNA | Phase-I, not recruiting | Mesenchymal derived exosomes with KRASG12D in treating individuals with pancreatic cancer with KRAS G12D mutation. | NCT03608631 |
| 9 | Pancreatic ductal adenocarcinoma | Portal vein blood | Completed | Test 3 CTC isolation methods and analyses for onco exosomes in pancreatic cell culture media by flow cytometry. | NCT03032913 |
| 10 | Oral mucositis, head and neck cancer | Grape extract exosomes | Active, Phase-I, not recruiting. | Ability of plant exosomes to prevent oral mucositis in head and neck cancer. | NCT01668849 |
| 11 | Non-small cell lung cancer (NSCLC) | Plasma exosomes | Not recruiting | New radiotherapy combined with immunotherapy | NCT02890849 |
| 12 | Non-small cell lung cancer (NSCLC) | Dendritic cells derived exosomes | Completed Phase-2 | No induction of T cells monitored in patients | NCT01159288 |
| 13 | Thyroid cancer | Urine exosomal thyroglobulin and galectin 3 | Active, not recruiting | Identifying urinary exosomal proteins (thyroglobulin and galectin 3) | NCT03488134 |
| 14 | Colon cancer | Blood sampling | Recruiting | Novel ways of diagnosing and predicting the spread to other organs such as liver | NCT03432806 |
| 15 | Prostate cancer | Urine samples | Active, not recruiting | Validated urine test to predict the incidence of high-grade prostate cancer in initial prostate biopsy | NCT03235687 |
| 16 | Triple-negative breast cancer | Serum exosomes | Phase-I, recruiting | Assessing response to pembrolizumab in the primary tumor, circulating lymphocytes, | NCT02977468 |
| 17 | Thyroid cancer | Urine exosomes | Active, not recruiting | Evaluation of new therapeutic mechanisms and medications for poorly differentiated or anaplastic thyroid cancer | NCT02862470 |
| 18 | Lung cancer | Blood samples | Recruiting | Drug efficacy, surgical effect evaluation, recurrence monitoring, prognosis judgement, molecular differentiation by analysing blood ctDNA | NCT03317080 |
Challenges of exosomal delivery
As it has been discussed earlier, there arises a growing interest in exosomal delivery for therapeutic interventions. Many research reports corroborated that exosome production could be leveraged by adopting different strategies like genetic modification of stem cells, co-culturing of exosomes with biomaterials, and incubating exosomes with hypoxia, lipopolysaccharides, and reduced pH. However, large-scale production of exosomes for clinical utility remains a major obstacle and so is the isolation of exosomes which is considerably dependent on the physicochemical properties of exosomes and their purity. The standardization of storage conditions also poses an important consideration [147]. The fate of exosomes inside the patient’s body (pharmacokinetics) needs to be given proper attention for the betterment of therapy. There are also uncertainties pertaining to the superiority of exosomes among plant, animal, and bacterial sources and the impact of pre-conditioning on the efficacy of exosomal delivery. Another challenge in delivering exosomes is a lack of a standard method for targeted delivery to the affected site as it may carry the risk of getting captured by the liver or lungs or exosomes may get damaged during the process of loading rendering the delivery less efficacious [148].
EVs derived from animal cells have beneficial qualities in the treatment of many illnesses; nevertheless, one of the major hurdles in this research is determining whether and how many numbers of human EVs can be generated in vitro or purified from biological fluids. The number of EVs produced per unit of original substantial will have an impact on the ultimate manufacturing cost and clinical applications. Thus, an appropriate selection of alternate sources of exosomes is warranted. Although plant-derived exosomes have their natural origin and can be separated from a large reservoir, they suffer from isolation and characterization challenges which vary to a great extent jeopardizing the safety and efficacy. The surface markers, particle size, and densities also add up to the complexity of systematic analysis [149]. Another growing concern for the exosomes field is its heterogeneity which was raised by Johnstone et al. in 1987 [150]. The tumor microenvironment is principally responsible for the heterogeneity of EVs of which cancer cells with varying metastatic ability, fibroblasts associated with cancer, adipocytes, immune cells, mesothelial cells, and stromal cells are key players. Any abnormal mechanical as well as chemical factors are also responsible for stimulating the secretion of exosomes with different molecular characteristics leading to heterogeneity. Some of the well-reported factors which contribute in heterogeneity include pH, hypoxic conditions, calcium concentration intra and extracellular, radiation and chemotherapy-based treatment, and mechanical stress [151]. The heterogeneity of exosomes can also be attributed to differences in their size, content, functional impact on receipient cells, and cellular origin. Size differences could lead to different amount of exosomal contents. Exosome marker variability has been identified by proteomic analysis of EVs, raising concerns about their relevance in experimental design employing marker-determined purification techniques. Nonetheless, the proteome of breast cancer cells and their exosomes can reveal whether the cell of origin was epithelial or mesenchymal, and distinct proteins and nucleic acids are enriched in exosomes in comparison with their cell of origin, implying a specific protein-sorting mechanism involved in exosome biogenesis and/or content loading. Heterogeneity may also be predicated on the organ and tissue of origin of the exosomes, including whether they originate from cancer cells, giving them different features such as tropism to certain organs and absorption by specific cell types [152].
Conclusion
The exchange of biological molecules across cell membranes is a very important step in attaining homeostasis of normal cells and is assisted by the EVs, exosomes, in particular. These exosomes play a crucial role in intercellular communication and provide insights into the progression and growth of cancerous cells by discharging cargos trapped inside exosomes. There have been many isolation methods for exosomes and with advanced characterization techniques, these exosomes can be suitably evaluated for therapeutic purposes. Past reports have suggested that exosomes have been at the forefront of diversified applications in cancer, viz., diagnostic biomarkers, in drug delivery systems. It has even been reported that exosomes confer chemoresistance in some of cancers. As a whole, it could rightly be said that a detailed study focusing on the potential of exosomes in counteracting cancer and understanding the metastasis and progression of cancer has and in the future would vitalize existing anti-cancer therapies and open new doors towards tackling the same.
Acknowledgements
The authors are grateful to the National Institute of Pharmaceutical Education and Research (NIPER)-Ahmedabad for providing literature search facilities.
Author contributions
GP, TGA and AJ conceived and discussed the outline of the review. GP, TGA, MG and TS wrote the draft. TGA and SSG modified, edited and rectified the draft. GP, TGA and MG made all figures and tables. AJ and RG edited and finalized the manuscript. All authors read and approved the final manuscript.
Data availability
Not applicable. It is a review article.
Declarations
Ethical approval
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Patil SM, Sawant SS, Kunda NK. Eur. J. Pharm. Biopharm. 2020;154:259. doi: 10.1016/j.ejpb.2020.07.026. [DOI] [PubMed] [Google Scholar]
- 2.Simons M, Raposo G. Curr. Opin. Cell. Biol. 2009;21:575. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Van Niel G, D’Angelo G, Raposo G. Nat. Rev. Mol. Cell. Biol. 2018;19:213. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 4.M. Logozzi, R. Di Raimo, D. Mizzoni, S. Fais, Semin. Cancer Biol. (2021) [DOI] [PubMed]
- 5.Almohammai A, Rahbarghazi R, Keyhanmanesh R, Rezaie J, Ahmadi M. J. Inflamm. (United Kingdom) 2021;18:1. doi: 10.1186/s12950-021-00275-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi H, Kim Y, Mirzaaghasi A, Heo J, Kim YN, Shin JH, Kim S, Kim NH, Cho ES, Yook JI, Yoo TH, Song E, Kim P, Shin EC, Chung K, Choi K, Choi C. Sci. Adv. 2020;6:1. doi: 10.1126/sciadv.aaz6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruivo CF, Adem B, Silva M, Melo SA. Cancer Res. 2017;77:6480. doi: 10.1158/0008-5472.CAN-17-0994. [DOI] [PubMed] [Google Scholar]
- 8.Chavda VP, Pandya A, Kumar L, Raval N, Vora LK, Pulakkat S, Patravale V, Salwa Y, Duo, Tang BZ. Nano Today. 2023;49:101771. doi: 10.1016/j.nantod.2023.101771. [DOI] [Google Scholar]
- 9.N. Carolina, 1 (2020)
- 10.R. Summary, 6977, (2020)
- 11.Yokoi A, Ochiya T. Semin. Cancer Biol. 2021;74:79. doi: 10.1016/j.semcancer.2021.03.032. [DOI] [PubMed] [Google Scholar]
- 12.Rahbarghazi R, Jabbari N, Sani NA, Asghari R, Salimi L, Kalashani SA, Feghhi M, Etemadi T, Akbariazar E, Mahmoudi M, Rezaie J. Cell. Commun. Signal. 2019;17:1. doi: 10.1186/s12964-019-0390-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vahabi A, Rezaie J, Hassanpour M, Panahi Y, Nemati M, Rasmi Y, Nemati M. Biochem. Pharmacol. 2022;200:115038. doi: 10.1016/j.bcp.2022.115038. [DOI] [PubMed] [Google Scholar]
- 14.Wortzel I, Dror S, Kenific CM, Lyden D. Dev. Cell. 2019;49:347. doi: 10.1016/j.devcel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 15.Kowal J, Tkach M, Théry C. Curr. Opin. Cell. Biol. 2014;29:116. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 16.H.S. Joo, J.H. Suh, H.J. Lee, E.S. Bang, J.M. Lee, Int. J. Mol. Sci. 21 (2020) [DOI] [PMC free article] [PubMed]
- 17.L.T. Brinton, H.S. Sloane, M. Kester, K.A. Kelly, (2014)
- 18.Rodrigues P, Melim C, Veiga F, Figueiras A. Processes. 2020;8:1. doi: 10.3390/pr8121561. [DOI] [Google Scholar]
- 19.Milman N, Ginini L, Gil Z. Drug Resist. Updat. 2019;45:1. doi: 10.1016/j.drup.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 20.J. Liu, L. Ren, S. Li, W. Li, X. Zheng, 1 (n.d.)
- 21.D.W. Greening, S.K. Gopal, R. Xu, R.J. Simpson, W. Chen, Semin. Cell Dev. Biol. 1 (2015) [DOI] [PubMed]
- 22.Janas AM, Sapoń K, Janas T, Stowell MHB, Janas T. Biochim. Biophys. Acta - Biomembr. 2016;1858:1139. doi: 10.1016/j.bbamem.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 23.Redzic JS, Balaj L, van der Vos KE, Breakefield XO. Semin. Cancer Biol. 2014;28:14. doi: 10.1016/j.semcancer.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang C, Ji Q, Yang Y, Li Q, Wang Z. Technol. Cancer Res. Treat. 2018;17:1. doi: 10.1177/1533033818763450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu D, Liu Z, Li Y, Huang Q, Xia L, Li K. Biomaterials. 2021;274:120894. doi: 10.1016/j.biomaterials.2021.120894. [DOI] [PubMed] [Google Scholar]
- 26.H.W. King, M.Z. Michael, J.M. Gleadle, BMC Cancer 12 (2012) [DOI] [PMC free article] [PubMed]
- 27.Chen H, Chengalvala V, Hu H, Sun D. Acta Pharm. Sin B. 2021;11:2136. doi: 10.1016/j.apsb.2021.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pirisinu M, Pham TC, Zhang DX, Hong TN, Nguyen LT, Le MT. Semin. Cancer Biol. 2022;80:340. doi: 10.1016/j.semcancer.2020.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Kurian TK, Banik S, Gopal D, Chakrabarti S, Mazumder N. Mol. Biotechnol. 2021;63:249. doi: 10.1007/s12033-021-00300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang XX, Sun C, Wang L, Guo XL. J. Control Release. 2019;308:119. doi: 10.1016/j.jconrel.2019.07.021. [DOI] [PubMed] [Google Scholar]
- 31.Boriachek K, Islam MN, Möller A, Salomon C, Nguyen NT, Hossain MSA, Yamauchi Y, Shiddiky MJA. Small. 2018;14:1. doi: 10.1002/smll.201702153. [DOI] [PubMed] [Google Scholar]
- 32.R.M. By, A. Bianchini 74, 1844 (1989) [PubMed]
- 33.Yang D, Zhang W, Zhang H, Zhang F, Chen L, Ma L, Larcher LM, Chen S, Liu N, Zhao Q, Tran PHL, Chen C, Veedu RN, Wang T. Theranostics. 2020;10:3684. doi: 10.7150/thno.41580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu L, Sun HT, Wang S, Huang SL, Zheng Y, Wang CQ, Hu BY, Qin W, Zou TT, Fu Y, Shen XT, Zhu WW, Geng Y, Lu L, liang Jia H, Qin LX, Dong QZ. J. Hematol. Oncol. 2020;13:1. doi: 10.1186/s13045-019-0838-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bu H, He D, He X, Wang K. ChemBioChem. 2019;20:451. doi: 10.1002/cbic.201800470. [DOI] [PubMed] [Google Scholar]
- 36.Zhang M, Jin K, Gao L, Zhang Z, Li F, Zhou F, Zhang L. Small Methods. 2018;2:1. [Google Scholar]
- 37.Nikfarjam S, Rezaie J, Zolbanin NM, Jafari R. J. Transl Med. 2020;18:1. doi: 10.1186/s12967-020-02622-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen BY, Sung CWH, Chen C, Cheng CM, Lin DPC, Huang CTe, Hsu MY. Clin. Chim. Acta. 2019;493:14. doi: 10.1016/j.cca.2019.02.021. [DOI] [PubMed] [Google Scholar]
- 39.Wei H, Qian X, Xie F, Cui D. Ann. Transl Med. 2021;9:882. doi: 10.21037/atm-21-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li P, Kaslan M, Lee SH, Yao J, Gao Z. Theranostics. 2017;7:789. doi: 10.7150/thno.18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lobb RJ, Becker M, Wen SW, Wong CSF, Wiegmans AP, Leimgruber A, Möller A. J. Extracell. Vesicles. 2015;4:1. doi: 10.3402/jev.v4.27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shirejini SZ, Inci F. Biotechnol. Adv. 2022;54:107814. doi: 10.1016/j.biotechadv.2021.107814. [DOI] [PubMed] [Google Scholar]
- 43.Atha DH, Ingham KC. J. Biol. Chem. 1981;256:12108. doi: 10.1016/S0021-9258(18)43240-1. [DOI] [PubMed] [Google Scholar]
- 44.Cho S, Jo W, Heo Y, Kang JY, Kwak R, Park J. Sens. Actuators B Chem. 2016;233:289. doi: 10.1016/j.snb.2016.04.091. [DOI] [Google Scholar]
- 45.R. Cazzoli, F. Buttitta, M. Di Nicola, S. Malatesta, H. Pass, 4, 1156 (2011) [DOI] [PMC free article] [PubMed]
- 46.W.A. Afridi, S. Strachan, S. Kasetsirikul, A.S. Pannu, N. Soda, D. Gough, N.T. Nguyen, M.J.A. Shiddiky, ACS Meas. Sci. Au (2022) [DOI] [PMC free article] [PubMed]
- 47.Zhang Y, Bi J, Huang J, Tang Y, Du S, Li P. Int. J. Nanomedicine. 2020;15:6917. doi: 10.2147/IJN.S264498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahgoub EO, Razmara E, Bitaraf A, Norouzi FS, Montazeri M, Behzadi-Andouhjerdi R, Falahati M, Cheng K, Haik Y, Hasan A, Babashah S. Mol. Biol. Rep. 2020;47:7229. doi: 10.1007/s11033-020-05715-w. [DOI] [PubMed] [Google Scholar]
- 49.Zhang L, Wang H, Zhao G, Li N, Wang X, Li Y, Jia Y, Qiao X. Anal. Chem. 2021;93:6534. doi: 10.1021/acs.analchem.1c00528. [DOI] [PubMed] [Google Scholar]
- 50.Shih CL, Chong KY, Hsu SC, Chien HJ, Ma CT, Chang JWC, Yu CJ, Chiou CC. N Biotechnol. 2016;33:116. doi: 10.1016/j.nbt.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 51.Ayala-Mar S, Donoso-Quezada J, Gallo-Villanueva RC, Perez-Gonzalez VH. and J. González-Valdez Electrophoresis. 2019;40:3036. doi: 10.1002/elps.201800526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.M. Wu, Y. Ouyang, Z. Wang, R. Zhang, P.H. Huang, C. Chen, H. Li, P. Li, D. Quinn, M. Dao, S. Suresh, Y. Sadovsky, T.J. Huang, Proc. Natl. Acad. Sci. U. S. A. 114, 10584 (2017) [DOI] [PMC free article] [PubMed]
- 53.Niu F, Chen X, Niu X, Cai Y, Zhang Q, Chen T, Yang H. Micromachines. 2020;11:1. doi: 10.3390/mi11050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao X, Wu D, Ma X, Wang J, Hou W, Zhang W. Biomed. Pharmacother. 2020;128:110237. doi: 10.1016/j.biopha.2020.110237. [DOI] [PubMed] [Google Scholar]
- 55.Fu S, Wang Y, Xia X, Zheng JC. NanoImpact. 2020;20:100261. doi: 10.1016/j.impact.2020.100261. [DOI] [Google Scholar]
- 56.Mehryab F, Rabbani S, Shahhosseini S, Shekari F, Fatahi Y, Baharvand H, Haeri A. Acta Biomater. 2020;113:42. doi: 10.1016/j.actbio.2020.06.036. [DOI] [PubMed] [Google Scholar]
- 57.Gong C, Tian J, Wang Z, Gao Y, Wu X, Ding X, Qiang L, Li G, Han Z, Yuan Y, Gao S. J Nanobiotechnol. 2019;17:1. doi: 10.1186/s12951-019-0526-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Munagala R, Aqil F, Jeyabalan J, Gupta RC. Cancer Lett. 2016;371:48. doi: 10.1016/j.canlet.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang X, Zhang H, Yang H, Bai M, Ning T, Li S, Li J, Deng T, Ying G, Ba Y. Curr. Cancer Drug Targets. 2018;18:347. doi: 10.2174/1568009617666170710120311. [DOI] [PubMed] [Google Scholar]
- 60.Pi YN, Xia BR, Jin MZ, Jin WL, Lou G. Biochem. Pharmacol. 2021;186:114487. doi: 10.1016/j.bcp.2021.114487. [DOI] [PubMed] [Google Scholar]
- 61.Y. Lin, J. Wu, W. Gu, Y. Huang, Z. Tong, L. Huang, J. Tan, 1700611, 1 (2018) [DOI] [PMC free article] [PubMed]
- 62.Shaban SA, Rezaie J, Nejati V. Cardiovasc. Toxicol. 2022;22:592. doi: 10.1007/s12012-022-09740-y. [DOI] [PubMed] [Google Scholar]
- 63.Mahbubfam S, Rezaie J, Nejati V. Tissue Cell. 2022;76:101803. doi: 10.1016/j.tice.2022.101803. [DOI] [PubMed] [Google Scholar]
- 64.M. Hassanpour, J. Rezaie, M. Darabi, A. Hiradfar, R. Rahbarghazi, 0, 1 (2020) [DOI] [PMC free article] [PubMed]
- 65.M. Feghhi, J. Rezaie, A. Akbari, N. Jabbari, H. Jafari, F. Seidi, 197 (2021)
- 66.Gomari H, Moghadam MF, Soleimani M, Ghavami M, Khodashenas S. Int. J. Nanomedicine. 2019;14:5679. doi: 10.2147/IJN.S210731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bagheri E, Abnous K, Farzad SA, Taghdisi SM, Ramezani M, Alibolandi M. Life Sci. 2020;261:118369. doi: 10.1016/j.lfs.2020.118369. [DOI] [PubMed] [Google Scholar]
- 68.Schindler C, Collinson A, Matthews C, Pointon A, Jenkinson L, Minter RR, Vaughan TJ, Tigue NJ. PLoS ONE. 2019;14:1. doi: 10.1371/journal.pone.0214545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toffoli G, Hadla M, Corona G, Caligiuri I, Palazzolo S, Semeraro S, Gamini A, Canzonieri V, Rizzolio F. Nanomedicine. 2015;10:2963. doi: 10.2217/nnm.15.118. [DOI] [PubMed] [Google Scholar]
- 70.Kooijmans SAA, Stremersch S, Braeckmans K, De Smedt SC, Hendrix A, Wood MJA, Schiffelers RM, Raemdonck K, Vader P. J. Control Release. 2013;172:229. doi: 10.1016/j.jconrel.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 71.A. Familtseva, N. Jeremic, S.C. Tyagi, Mol. Cell. Biochem (2019) [DOI] [PubMed]
- 72.Li YJ, Wu JY, Wang JM, Bin Hu X, Cai JX, Xiang DX. Acta Biomater. 2020;101:519. doi: 10.1016/j.actbio.2019.10.022. [DOI] [PubMed] [Google Scholar]
- 73.Kim MS, Haney MJ, Zhao Y, Mahajan V, Deygen I, Klyachko NL, Inskoe E, Piroyan A, Sokolsky M, Okolie O, Hingtgen SD, Kabanov AV, Batrakova EV. Nanomed. Nanatechnol. Biol Med. 2016;12:655. doi: 10.1016/j.nano.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lamichhane TN, Jeyaram A, Patel DB, Parajuli B, Livingston NK, Arumugasaamy N, Schardt JS, Jay SM. Cell. Mol. Bioeng. 2016;9:315. doi: 10.1007/s12195-016-0457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun W, Xing C, Zhao L, Zhao P, Yang G, Yuan L. Mol. Ther. - Nucleic Acids. 2020;20:558. doi: 10.1016/j.omtn.2020.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Elsharkasy OM, Nordin JZ, Hagey DW, de Jong OG, Schiffelers RM, Andaloussi SEL, Vader P. Adv. Drug Deliv Rev. 2020;159:332. doi: 10.1016/j.addr.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 77.Kim R, Lee S, Lee J, Kim M, Kim WJ, Lee HW, Lee MY, Kim J, Chang W. BMB Rep. 2018;51:406. doi: 10.5483/BMBRep.2018.51.8.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Severic M, Ma G, Pereira SGT, Ruiz A, Cheung CCL, Al-Jamal WT. J. Control Release. 2021;330:101. doi: 10.1016/j.jconrel.2020.12.017. [DOI] [PubMed] [Google Scholar]
- 79.Jang SC, Kim OY, Yoon CM, Choi DS, Roh TY, Park J, Nilsson J, Lötvall J, Kim YK, Gho YS. ACS Nano. 2013;7:7698. doi: 10.1021/nn402232g. [DOI] [PubMed] [Google Scholar]
- 80.T.R. Lunavat, S.C. Jang, L. Nilsson, H.T. Park, G. Repiska, C. Lässer, J.A. Nilsson, Y.S. Gho, J. Lötvall, Biomaterials (2016) [DOI] [PubMed]
- 81.Jhan YY, Prasca-Chamorro D, Palou Zuniga G, Moore DM, Arun Kumar S, Gaharwar AK, Bishop CJ. Int. J. Pharm. 2020;573:118802. doi: 10.1016/j.ijpharm.2019.118802. [DOI] [PubMed] [Google Scholar]
- 82.S. Kalimuthu, P. Gangadaran, R.L. Rajendran, L. Zhu, M. Oh, H.W. Lee, A. Gopal, S.H. Baek, S.Y. Jeong, J. Lee, B. Ahn (2018)
- 83.Fuhrmann G, Serio A, Mazo M, Nair R, Stevens MM. J. Control Release. 2015;205:35. doi: 10.1016/j.jconrel.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 84.Wang J, Chen D, Ho EA. J. Control Release. 2021;329:894. doi: 10.1016/j.jconrel.2020.10.020. [DOI] [PubMed] [Google Scholar]
- 85.Haney MJ, Klyachko NL, Zhao Y, Gupta R, Plotnikova EG, He Z, Patel T, Piroyan A, Sokolsky M, Kabanov AV, Batrakova EV. J. Control Release. 2015;207:18. doi: 10.1016/j.jconrel.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kalani A, Chaturvedi P, Kamat PK, Maldonado C, Bauer P, Joshua IG, Tyagi SC, Tyagi N. Int. J. Biochem. Cell. Biol. 2016;79:360. doi: 10.1016/j.biocel.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sato YT, Umezaki K, Sawada S, Mukai SA, Sasaki Y, Harada N, Shiku H, Akiyoshi K. Sci. Rep. 2016;6:1. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hettich BF, Bader JJ, Leroux JC. Adv. Healthc. Mater. 2021;2100047:1. doi: 10.1002/adhm.202100047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kwon SH, Al Faruque H, Kee H, Kim E, Park S. Colloids Surf. B Biointerfaces. 2021;205:111915. doi: 10.1016/j.colsurfb.2021.111915. [DOI] [PubMed] [Google Scholar]
- 90.Wei H, Chen J, Wang S, Fu F, Zhu X, Wu C, Liu Z, Zhong G, Lin J. Int. J. Nanomedicine. 2019;14:8603. doi: 10.2147/IJN.S218988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang Z, Shi J, Xie J, Wang Y, Sun J, Liu T, Zhao Y, Zhao X, Wang X, Ma Y, Malkoc V, Chiang C, Deng W, Chen Y, Fu Y, Kwak KJ, Fan Y, Kang C, Yin C, Rhee J, Bertani P, Otero J, Lu W, Yun K, Lee AS, Jiang W, Teng L, Kim BYS, Lee LJ. Nat. Biomed. Eng. 2020;4:69. doi: 10.1038/s41551-019-0485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yim N, Ryu S, Choi K, Lee KR, Lee S, Choi H, Kim J, Shaker MR, Sun W, Park J, Kim D, Heo WDo, Choi C. Nat. Publ Gr. 2016;7:1. doi: 10.1038/ncomms12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dragovic RA, Gardiner C, Brooks AS, Tannetta DS, Ferguson DJP, Hole P, Carr B, Redman CWG, Harris AL, Dobson PJ, Harrison P, Sargent IL. Nanomed. Nanatechnol. Biol Med. 2011;7:780. doi: 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ko J, Carpenter E, Issadore D. Analyst. 2016;141:450. doi: 10.1039/C5AN01610J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.R.H. Nicolas, G.H. Goodwin, Chromosom. Proteins 41 (1982)
- 96.Doyle LM, Wang MZ. Cells. 2019;8:727. doi: 10.3390/cells8070727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.M. Skliar, V.S. Chernyshev, J. Vis. Exp. 2019, 1 (2019) [DOI] [PubMed]
- 98.Srivastava A, Rathore S, Munshi A, Ramesh R. Semin. Cancer Biol. 2022;86:80. doi: 10.1016/j.semcancer.2022.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sundararajan V, Sarkar FH, Ramasamy TS. Cell. Oncol. 2018;41:463. doi: 10.1007/s13402-018-0396-2. [DOI] [PubMed] [Google Scholar]
- 100.Rupp AK, Rupp C, Keller S, Brase JC, Ehehalt R, Fogel M, Moldenhauer G, Marmé F, Sültmann H, Altevogt P. Gynecol. Oncol. 2011;122:437. doi: 10.1016/j.ygyno.2011.04.035. [DOI] [PubMed] [Google Scholar]
- 101.Moon PG, Lee JE, Cho YE, Lee SJ, Jung JH, Chae YS, Bae HI, Kim YB, Kim IS, Park HY, Baek MC. Clin. Cancer Res. 2016;22:1757. doi: 10.1158/1078-0432.CCR-15-0654. [DOI] [PubMed] [Google Scholar]
- 102.Moon PG, Lee JE, Cho YE, Lee SJ, Chae YS, Jung JH, Kim IS, Park HY, Baek MC. Oncotarget. 2016;7:40189. doi: 10.18632/oncotarget.9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.S. Khan, J.M.S. Jutzy, M.M.A. Valenzuela, D. Turay, J.R. Aspe, A. Ashok, S. Mirshahidi, D. Mercola, M.B. Lilly, N.R. Wall, PLoS One 7 (2012) [DOI] [PMC free article] [PubMed]
- 104.Zhao Z, Yang Y, Zeng Y, He M. Lab. Chip. 2016;16:489. doi: 10.1039/C5LC01117E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.S. Klein-Scory, M.M. Tehrani, C. Eilert-Micus, K.A. Adamczyk, N. Wojtalewicz, M. Schnölzer, S.A. Hahn, W. Schmiegel, and I. Schwarte-Waldhoff, Proteome Sci. 12, 1 (2014) [DOI] [PMC free article] [PubMed]
- 106.Nuzhat Z, Kinhal V, Sharma S, Rice GE, Joshi V, Salomon C. Oncotarget. 2017;8:17279. doi: 10.18632/oncotarget.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, Kalluri R. Nature. 2015;523:177. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Niu L, Song X, Wang N, Xue L, Song X, Xie L. Cancer Sci. 2019;110:433. doi: 10.1111/cas.13862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.B. Cao, P. Wang, L. Gu, J. Liu, Oncol. Lett. 21 (2021) [DOI] [PMC free article] [PubMed]
- 110.Zhou J, Wang H, Sun Q, Liu X, Wu Z, Wang X, Fang W, Ma Z. Mol. Ther. - Nucleic Acids. 2021;23:1217. doi: 10.1016/j.omtn.2021.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.W. Huang, Y. Yan, Y. Liu, M. Lin, J. Ma, W. Zhang, J. Dai, J. Li, Q. Guo, H. Chen, B. Makabel, H. Liu, C. Su, H. Bi, J. Zhang, Signal Transduct. Target. Ther. 5 (2020) [DOI] [PMC free article] [PubMed]
- 112.Sun H, Shi K, Qi K, Kong H, Zhang J, Dai S, Ye W, Deng T, He Q, Zhou M. Front. Immunol. 2019;10:1. doi: 10.3389/fimmu.2019.00001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 113.Chen D, Wu X, Xia M, Wu F, Ding J, Jiao Y, Zhan Q, An F. Oncol. Rep. 2017;38:2182. doi: 10.3892/or.2017.5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shang D, Xie C, Hu J, Tan J, Yuan Y, Liu Z, Yang Z. J. Cell. Mol. Med. 2020;24:588. doi: 10.1111/jcmm.14766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou C, Chen Y, He X, Zheng Z, Xue D. Onco. Targets. Ther. 2020;13:11595. doi: 10.2147/OTT.S272869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shi J. J. Clin. Med. 2016;5:1. doi: 10.3390/jcm5040042. [DOI] [Google Scholar]
- 117.Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O’Connor STF, Chin AR, Yen Y, Wang Y, Marcusson EG, Chu P, Wu J, Wu X, Li AX, Li Z, Gao H, Ren X, Boldin MP, Lin PC, Wang SE. Cancer Cell. 2014;25:501. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Eichelser C, Stückrath I, Müller V, Milde-Langosch K, Wikman H, Pantel K, Schwarzenbach H. Oncotarget. 2014;5:9650. doi: 10.18632/oncotarget.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li XJ, Ren ZJ, Tang JH, Yu Q. Cell. Physiol. Biochem. 2018;44:1741. doi: 10.1159/000485780. [DOI] [PubMed] [Google Scholar]
- 120.Yu D, Wu Y, hui Zhang X, meng Lv M, Chen W, Chen X, Yang S, Shen H, liang Zhong S. J. hai Tang and J. hua Zhao Tumor Biol. 2016;37:3227. doi: 10.1007/s13277-015-4161-0. [DOI] [PubMed] [Google Scholar]
- 121.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Biochim. Biophys. Acta - Gen. Subj. 2012;1820:940. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 122.Fernandis AZ, Wenk MR, Chromatogr J. B Anal. Technol. Biomed. Life Sci. 2009;877:2830. doi: 10.1016/j.jchromb.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 123.Gurung S, Perocheau D, Touramanidou L, Baruteau J. Cell. Commun. Signal. 2021;19:1. doi: 10.1186/s12964-021-00730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Skotland T, Sandvig K, Llorente A. Prog. Lipid Res. 2017;66:30. doi: 10.1016/j.plipres.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 125.Skotland T, Hessvik NP, Sandvig K, Llorente A. J. Lipid Res. 2019;60:9. doi: 10.1194/jlr.R084343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Min HK, Lim S, Chung BC, Moon MH. Anal. Bioanal Chem. 2011;399:823. doi: 10.1007/s00216-010-4290-7. [DOI] [PubMed] [Google Scholar]
- 127.Lea J, Sharma R, Yang F, Zhu H, Sally E, Ward, Schroit AJ. Oncotarget. 2017;8:14395. doi: 10.18632/oncotarget.14795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Llorente A, Skotland T, Sylvänne T, Kauhanen D, Róg T, Orłowski A, Vattulainen I, Ekroos K, Sandvig K. Biochim. Biophys. Acta - Mol. Cell. Biol. Lipids. 2013;1831:1302. doi: 10.1016/j.bbalip.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 129.Shi X, Sun J, Li H, Lin H, Xie W, Li J, Tan W. Prostate. 2020;80:811. doi: 10.1002/pros.23996. [DOI] [PubMed] [Google Scholar]
- 130.Hartman ZC, Wei J, Glass OK, Guo H, Lei G, Yang XY, Osada T, Hobeika A, Delcayre A, Le Pecq JB, Morse MA, Clay TM, Lyerly HK. Vaccine. 2011;29:9361. doi: 10.1016/j.vaccine.2011.09.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ha D, Yang N, Nadithe V. Acta Pharm. Sin B. 2016;6:287. doi: 10.1016/j.apsb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pascucci L, Coccè V, Bonomi A, Ami D, Ceccarelli P, Ciusani E, Viganò L, Locatelli A, Sisto F, Doglia SM, Parati E, Bernardo ME, Muraca M, Alessandri G, Bondiolotti G, Pessina A. J. Control Release. 2014;192:262. doi: 10.1016/j.jconrel.2014.07.042. [DOI] [PubMed] [Google Scholar]
- 133.T. Yong, X. Zhang, N. Bie, H. Zhang, X. Zhang, F. Li, A. Hakeem, J. Hu, L. Gan, H.A. Santos, X. Yang, Nat. Commun. 10 (2019) [DOI] [PMC free article] [PubMed]
- 134.Zhang X, Liu L, Tang M, Li H, Guo X, Yang X. Drug Dev. Ind. Pharm. 2020;46:1150. doi: 10.1080/03639045.2020.1776320. [DOI] [PubMed] [Google Scholar]
- 135.Sharma A. Nanomedicine. 2017;12:2137. doi: 10.2217/nnm-2017-0184. [DOI] [PubMed] [Google Scholar]
- 136.Shedden K, Xie XT, Chandaroy P, Chang YT, Rosania GR. Cancer Res. 2003;63:4331. [PubMed] [Google Scholar]
- 137.Koch R, Aung T, Vogel D, Chapuy B, Wenzel D, Becker S, Sinzig U, Venkataramani V, Von Mach T, Jacob R, Truemper L, Wulf GG. Clin. Cancer Res. 2016;22:395. doi: 10.1158/1078-0432.CCR-15-0577. [DOI] [PubMed] [Google Scholar]
- 138.Chapuy B, Koch R, Radunski U, Corsham S, Cheong N, Inagaki N, Ban N, Wenzel D, Reinhardt D, Zapf A, Schweyer S, Kosari F, Klapper W, Truemper L, Wulf GG. Leukemia. 2008;22:1576. doi: 10.1038/leu.2008.103. [DOI] [PubMed] [Google Scholar]
- 139.Lopes-Rodrigues V, Di Luca A, Sousa D, Seca H, Meleady P, Henry M, Lima RT, O’Connor R, Vasconcelos MH. Biochim. Biophys. Acta - Gen. Subj. 2016;1860:618. doi: 10.1016/j.bbagen.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 140.Aubertin K, Silva AKA, Luciani N, Espinosa A, Djemat A, Charue D, Gallet F, Blanc-Brude O, Wilhelm C. Sci. Rep. 2016;6:1. doi: 10.1038/srep35376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Safaei R, Larson BJ, Cheng TC, Gibson MA, Otani S, Naerdemann W, Howell SB. Mol. Cancer Ther. 2005;4:1595. doi: 10.1158/1535-7163.MCT-05-0102. [DOI] [PubMed] [Google Scholar]
- 142.A. Levchenko, B.M. Mehta, X. Niu, G. Kang, L. Villafania, D. Way, D. Polycarpe, M. Sadelain, S.M. Larson, Proc. Natl. Acad. Sci. U. S. A. 102, 1933 (2005) [DOI] [PMC free article] [PubMed]
- 143.Ambudkar SV, Sauna ZE, Gottesman MM, Szakacs G. Trends Pharmacol. Sci. 2005;26:385. doi: 10.1016/j.tips.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rezaie J, Etemadi T, Feghhi M. Eur. J. Pharmacol. 2022;933:175292. doi: 10.1016/j.ejphar.2022.175292. [DOI] [PubMed] [Google Scholar]
- 145.P. Santos, F. Almeida, 12, 1 (2021)
- 146.M. Damo, D.S. Wilson, E. Simeoni, J.A. Hubbell, Nat. Publ. Gr. 1 (2015) [DOI] [PMC free article] [PubMed]
- 147.Rezaie J, Feghhi M, Etemadi T. Cell. Commun. Signal. 2022;20:1. doi: 10.1186/s12964-022-00959-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rezaie J, Ahmadi M, Ravanbakhsh R, Mojarad B, Mahbubfam S, Shaban SA, Shadi K, Berenjabad NJ, Etemadi T. Life Sci. 2022;289:120216. doi: 10.1016/j.lfs.2021.120216. [DOI] [PubMed] [Google Scholar]
- 149.Nemati M, Singh B, Mir RA, Nemati M, Babaei A, Ahmadi M, Rasmi Y, Golezani AG, Rezaie J. Cell. Commun. Signal. 2022;20:1. doi: 10.1186/s12964-022-00889-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.E. Willms, C. Cabañas, I. Mäger, and M. J. A. Wood, 9 (2018) [DOI] [PMC free article] [PubMed]
- 151.Lee AH, Koh IL, Dawson MR. Adv. Cancer Biol. - Metastasis. 2022;4:100040. doi: 10.1016/j.adcanc.2022.100040. [DOI] [Google Scholar]
- 152.R. Kalluri, V.S. Lebleu, C. Biology, 367 (2020) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable. It is a review article.