Abstract
Data System. The REal-time Assessment of Community Transmission-1 (REACT-1) Study was funded by the Department of Health and Social Care in England to provide reliable and timely estimates of prevalence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection over time, by person and place.
Data Collection/Processing. The study team (researchers from Imperial College London and its logistics partner Ipsos) wrote to named individuals aged 5 years and older in random cross-sections of the population of England, using the National Health Service list of patients registered with a general practitioner (near-universal coverage) as a sampling frame. We collected data over 2 to 3 weeks approximately every month across 19 rounds of data collection from May 1, 2020, to March 31, 2022.
Data Analysis/Dissemination. We have disseminated the data and study materials widely via the study Web site, preprints, publications in peer-reviewed journals, and the media. We make available data tabulations, suitably anonymized to protect participant confidentiality, on request to the study’s data access committee.
Public Health Implications. The study provided inter alia real-time data on SARS-CoV-2 prevalence over time, by area, and by sociodemographic variables; estimates of vaccine effectiveness; and symptom profiles, and detected emergence of new variants based on viral genome sequencing. (Am J Public Health. 2023;113(5):545–554. https://doi.org/10.2105/AJPH.2023.307230)
The REal-time Assessment of Community Transmission-1 (REACT-1) Study sought to provide reliable and timely estimates of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection prevalence and data on associated symptoms and covariates from random samples of the population of England.
DATA SYSTEM
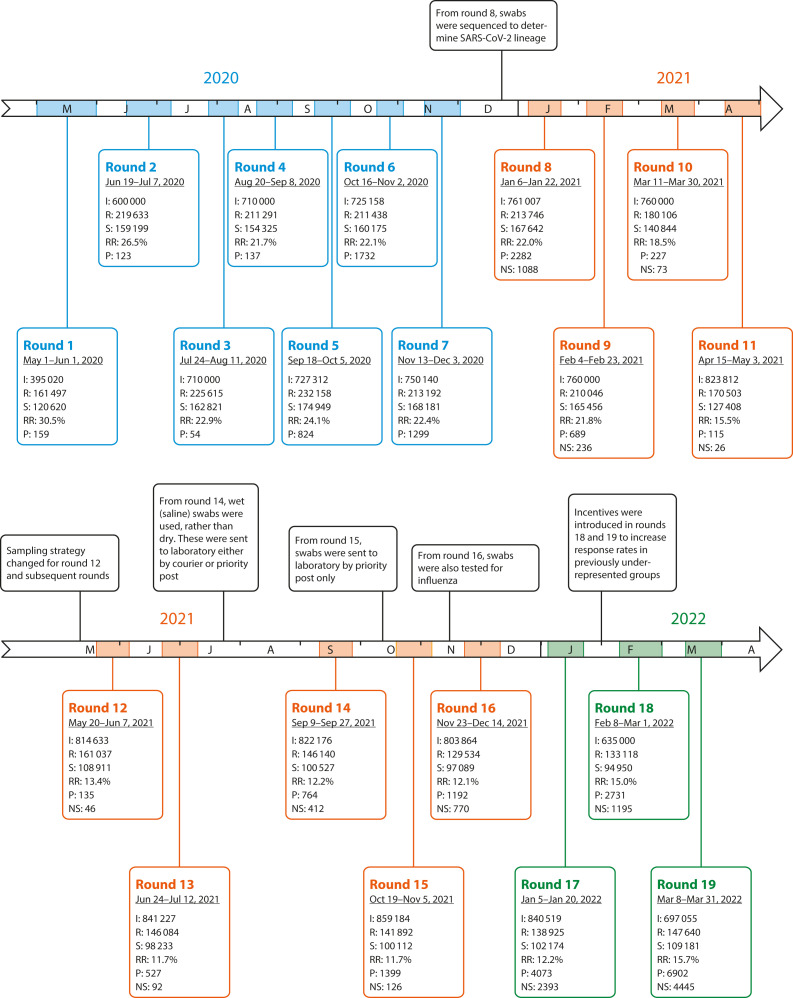
This study involved 19 distinct rounds of data collection, from May 1, 2020, to March 31, 2022 (Figure 1).
FIGURE 1—
REACT-1 Study Timeline Over 19 Rounds of Data Collection: England, May 1, 2020–March 31, 2022
Note. REACT-1 = REal-time Assessment of Community Transmission-1; SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2. For each round, we report the number of invitations sent (I), the number of participants registered (R), the number of valid swabs received (S), and response rate (RR), as defined by the number of valid swabs over the number of invitations sent and the total number of SARS-CoV-2‒positive swabs (P) these yielded. From round 8 onward (from January 6, 2021), viral sequencing was also available, and we report the number of SARS-CoV-2 (sub)-lineages determined per round (NS).
Name and Sponsor of the Program
The REACT-1 Study is funded by the Department of Health and Social Care in England.
Purpose
The aim of REACT-1 was to rapidly detect changes in SARS-CoV-2 transmission in England and to give early warning of any upturn in infections. As well as providing reliable estimates of prevalence of SARS-CoV-2 infection over time, by person and place, REACT-1 gave estimates of vaccine effectiveness against infection and identified new variants as they emerged in the population.
Public Health Significance
When REACT-1 was established during the first lockdown in England, little was known about the spread of SARS-CoV-2 in the population, who was most at risk, when infection rates were rising or falling, and where—for example, at national, regional, or subregional levels. REACT-1 was designed to provide rapid information on the time course of the infection, geographic spread (at high spatial resolution), and estimates of the reproduction number R.1,2 These data were given directly to the government as they were obtained to inform the public health response and enable timely implementation of public health interventions.
DATA COLLECTION/ PROCESSING
The study tested self-obtained throat and nose swabs from random samples of the population of England for presence of SARS-CoV-2, using reverse-transcriptase polymerase chain reaction (rt-PCR). Participants also completed a self-administered questionnaire, either online or by telephone. In later rounds, we carried out viral genome sequencing to obtain information on circulating variants.
Data Sources and Collection Mode
Source population
The study team (researchers from Imperial College London and its logistics partner Ipsos) invited random cross-sectional samples of individuals aged 5 years and older in England by mail to participate in REACT-1.
Registration
We invited named individuals on the National Health Service (NHS) list of patients to register for the study either online or by telephone. For participants, we obtained from the NHS register their name, sex, general practitioner practice code, address and residential postcode, mobile (cell) telephone number, e-mail address (for early rounds only), date of birth (to round 7, November 13 to December 3, 2020) or month and year of birth (subsequent rounds), and NHS number.
Questionnaires
Participants completed a brief questionnaire at registration either online or by telephone and a further, more detailed questionnaire for those who provided a swab. These included information on household composition; key worker status; social behaviors (e.g., mask wearing, commuting); contact with a person known or suspected to have COVID-19; whether, at time of survey, participants had experienced 1 or more of a list of symptoms and timing of the symptoms (participants not reporting symptoms may have developed symptoms later, but these were not captured); symptoms lasting more than 4 weeks; self-reported long COVID; and self-reported height and weight, smoking, vaccination history, and attitudes toward vaccination. Participants were asked for consent for recontact and for longer-term follow-up through linkage to their NHS records including data from the national immunization program. The questionnaires are available on the study Web site.3
Swabs
Individuals who registered for the study received a swab kit by mail, with both written and video instructions provided for collecting a self-obtained throat and nose swab (by parent or guardian for children aged 5–12 years). During the first round of data collection (May 1 to June 1, 2020), swabs were initially collected in viral transport medium and sent to 1 of 4 Public Health England laboratories for processing (n = 8595 swabs with reported result). All subsequent collections during round 1 to round 13 (June 24 to July 12, 2021) were obtained by using dry swabs. Participants were asked to obtain and refrigerate the sample and request a courier pick-up (same or next day) for sample collection. To maintain sample integrity, samples were then transported to a central hub before daily shipping to a commercial laboratory, all within a cold chain (4° to 8°C) for testing for presence of SARS-CoV-2.
For round 14 (September 9–27, 2021), we modified the way swab samples were handled, switching to wet swabs in saline solution. We randomized on a 1-to-1 basis whether samples were sent to the laboratory by priority mail or were picked up by courier without the cold chain (although samples were refrigerated on arrival at the depot before onward transportation to the laboratory). There was a slightly higher return rate for samples handled by mail, although prevalence estimates were slightly higher and cycle threshold (Ct) values among positives slightly lower (indicating slightly better preservation) in the samples sent by courier compared with samples shipped by mail.4 In subsequent rounds from round 15 (October 19 to November 5, 2021), we switched to shipment by priority mail in saline solution only for all swabs.
Ethical Procedures
Ethics
We obtained ethics approval from the South Central–Berkshire B Research Ethics Committee (IRAS ID: 283787). Participants gave individual consent to take part, either online or by telephone (parent or guardian for children aged 5–12 years and for those aged 13–17 years where parent or guardian completed registration on behalf of the child).
Public involvement
A public advisory panel provides input into the design, conduct, and dissemination of the REACT research program. We have a data access committee with lay representation.
Population and Geographic Coverage
Population coverage
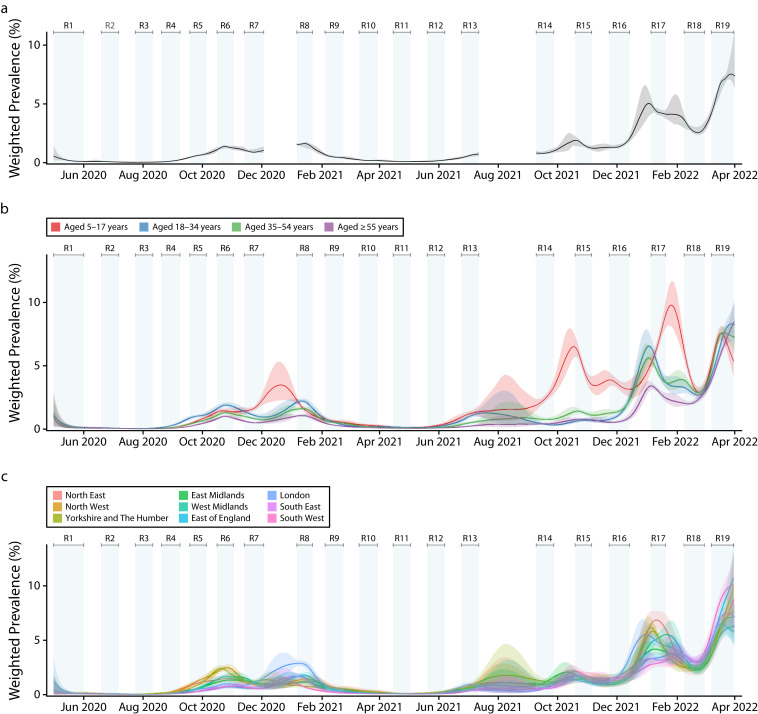
The target population was the population of England aged 5 years and older. We designed the study to provide robust estimates of prevalence at regional and subregional level, including estimates at lower-tier local authority (LTLA) level. We included data for 316 of the 317 LTLAs in England excluding the Isles of Scilly (after the first round), and by combining data for the City of London with Westminster, we report results across 315 LTLAs overall. At each round, we provided national and regional estimates of epidemic growth, and prevalence estimates for key demographic subgroups including by age, ethnicity, household size, occupation, and socioeconomic status. Smoothed estimates of weighted prevalence over the 19 rounds of the study (May 1, 2020, to March 31, 2022) by age and region, as well as overall, are shown in Figure 2.
FIGURE 2—
Smoothed Weighted SARS-CoV-2 Swab-Positivity Prevalence From All Rounds of the REACT-1 Study by (a) Overall Prevalence, (b) Age Groups, and (c) Region: England, May 1, 2020–March 31, 2022
Note. REACT-1 = REal-time Assessment of Community Transmission-1; SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2. We used the random iterative method (RIM) weighting approach to correct for possible nonresponse bias and ensure that our prevalence estimates were representative of the population of England as a whole. Using a Bayesian penalized-spline model fit to the daily swab-positivity data, we estimated the median (plain line) and 95% credible intervals (shaded regions) of weighted prevalence throughout the study period. Results are shown for the full population of England (A) with model fit on rounds 1–7, 8–13, and 14–19, separately, and for 4 broad age groups (B): 5–17 y, 18–34 y, 35–54 y, and 55 y and older, and by region (C), with model fit throughout the duration of the study. Ranking of weighted prevalence by age and region changed over the course of the epidemic in England. For example, there was particularly high prevalence in those aged 5 to 17 y in October 2021 and January 2022.
Sampling frame
We used the list of patients aged 5 years and older registered in the NHS with a general practitioner in England (near-universal population coverage). We obtained a new random sample at each round from data on the NHS register held centrally by NHS Digital.
Sampling strategy
For rounds 1 to 11 (between May 1, 2020, and May 3, 2021), we aimed to obtain LTLA-stratified random samples with approximately equal numbers of participants in each of the 315 LTLAs to enable local estimates of prevalence across England. From round 12 onward, we adjusted the sampling procedure to select the sample randomly in proportion to population at the LTLA level, because urban and inner-city areas (which tended to have higher infection rates) were relatively underrepresented in comparison with more sparsely populated rural areas.
Unit of Data Collection and Sample Size
Unit of data collection
Data collection was at the individual level. Named, randomly selected individuals were invited to take part; the invitation was nontransferable to other members of the family or household.
Sample size and response rates
Over the 19 rounds of the study, a total of 2 512 797 valid swabs were included from among 14 036 117 invitations, giving an overall response rate (number of valid swabs/number of invitations sent out) of 17.9%. Sample size by round varied from 94 950 in round 18 (February 8 to March 1, 2022, when prevalence rates were very high; Figure 2) to 168 181 in round 7 (November 13 to December 3, 2020). Response rates also varied by round, ranging from 11.7% in rounds 13 (June 24 to July 12, 2021) and 15 (October 19 to November 5, 2021) to 30.5% in round 1 (May 1 to June 1, 2020, during the first lockdown in England).
To maximize response rates, with our logistics partner Ipsos, we implemented a series of reminders to improve both registration rates and swab and survey return rates among those registering to take part. For registration, we included up to 4 reminders by letter, text, or (in 2 rounds) mobile phone contact. Among people who had requested a swab kit but not returned the swab or symptom survey, from round 3 (July 24 to August 11, 2020) Ipsos used a series of up to 3 reminders by either e-mail or text (or letter where an e-mail address or mobile phone number were not provided). A reminder by phone call was added from round 12 onward (May 20 to June 7, 2021) where a mobile phone number was provided at registration. Overall, a substantial proportion of those registering and those returning a swab did so following one of these reminders.
In addition, following a successful pilot in which different incentives (£10 to £30) were offered (in a randomized evaluation) to a subsample of people taking part in round 15, we added an incentive in rounds 18 (February 8 to March 1, 2022) and 19 (March 8–31, 2022) to increase response rates among underrepresented groups. For returning their completed test, those aged 13 to 17 and 35 to 44 years were offered a gift voucher worth £10 while those aged 18 to 34 years were offered a voucher worth £20. This had the effect of increasing the response rate in these groups, with the overall response rate in rounds 18 and 19 rising to 15.0% and 15.7%, respectively (Figure 1).
Completeness
We obtained sufficient data by round to be able to estimate prevalence by age, sex, region, and other key demographic groups including ethnicity, occupation, household size, and socioeconomic status (as determined by an area-level deprivation score).
Generalizability
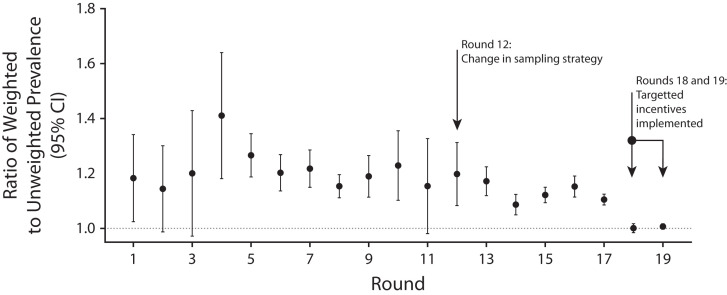
We weighted the data at each round (see subsequent “Prevalence estimates and random iterative method weighting” section) to be representative of England as a whole and to provide unbiased estimates of prevalence that were comparable across rounds, including with the change in sampling strategy to be in proportion to population size. The ratio of weighted to unweighted prevalence gives an indication as to the potential bias introduced by nonresponse in different demographic groups. This ratio was consistently above 1 for rounds 1 to 17 (i.e., unweighted prevalence was consistently below the expected rates for England). However, the variability around the ratio tended to decrease following the change in sampling strategy implemented in round 12, and the ratio itself fell to 1 (i.e., no bias on average) in both rounds 18 and 19, reflecting the use of incentives in those rounds among harder-to-reach younger people (Figure 3).
FIGURE 3—
Per-Round Ratio of Weighted Over Unweighted Prevalence of SARS-CoV-2 Swab Positivity in REACT-1 Over 19 Rounds: England, May 1, 2020–March 31, 2022
Note. SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2. We report the mean and 95% confidence intervals (CIs) for the ratio.
Surveillance Design and Data Collection Frequency
We designed the study to recruit a series of representative cross-sectional samples of the population of England aged 5 years and older. As noted, we carried out data collection over a 23-month period from May 1, 2020, to March 31, 2022, through a series of distinct rounds that took place over a 2- to 3-week period approximately monthly—exceptions were gaps between rounds 7 and 8 over Christmas 2020 and between rounds 13 and 14 during summer 2021 (Figure 2). The data were collected prospectively and reported in near-real-time. Where possible, we aimed to obtain independent samples at each round (i.e., sampling without replacement); however, given the scale of the sampling efforts (around one quarter of the population of England was invited) this could not always be achieved, and around 3% of the study population took part more than once.
Key Data Elements and Data Quality/Editing
Prevalence estimates and random iterative method weighting
We used random iterative method (RIM) weighting5 to correct for bias in (unweighted) prevalence estimates introduced by varying nonresponse in different demographic groups, taking account of age‒sex categories, deciles of an area-level index of multiple deprivation,6 LTLA population counts, and ethnicity. We obtained age, sex, and LTLA counts from the Office for National Statistics midyear population estimates,7 counts by ethnic group from the Labour Force Survey,8 and index of multiple deprivation deciles from linkage to residential postcodes using the NHS digital sampling frame. We based the RIM weighting on proportions rather than population totals, with age grouped into 9 categories: 5–12; 13–17; 18–24; 25–34; 35–44; 45–54; 55–64; 65–74; and 75 years or older, giving 18 age‒sex categories. Self-reported ethnicity was grouped into 9 categories: White, mixed/multiple ethnic groups, Indian, Pakistani, Bangladeshi, Chinese, any other Asian background, Black African/Caribbean/other, and any other ethnic group or missing.
For the RIM weighting (first stage) we weighted each sample to LTLA counts and age‒sex categories only, with adjustment to ensure that the final weighted estimates were as close as possible to the source population. Then, using the first stage weights as starting weights, we adjusted the RIM weighting for all 4 weighting variables, trimming the adjustment factor between the first and second stage weights at the first and 99th percentiles to dampen the extreme weights. We calculated the final weights as the first stage weights multiplied by the trimmed adjustment factor for the second stage, with credible intervals for weighted prevalence estimates calculated using the “survey” package in R.9
Time trends and smoothed prevalence
To analyze time trends in swab positivity, we used an exponential model of growth or decay assuming that the daily weighted number of positive samples (out of the daily weighted total number of samples) arose from a binomial distribution. The model is of the form I(t) = I0.ert, where I(t) is the swab positivity at time t, I0 is swab positivity at the beginning of data collection per round, and r is the growth rate. On a given day, the binomial likelihood for P (out of N) positive tests is then P ∼ B(N, I0.ert) based on day of swabbing or, if unavailable, day of sample collection. We used a bivariate No-U-Turn sampler to estimate posterior credible intervals assuming uniform prior distributions on I0 and r.10
To fit a smoothed trend (Figure 2), we used a Bayesian penalized-spline model11 to the daily data using a No-U-Turn Sampler in logit space, splitting the data into segments of approximately 5 days by regularly spaced knots, with further knots included beyond the period of the study to remove edge effects. We defined a system of 4th-order basis-splines (b-splines) over the knots, with a model comprising a linear combination of these b-splines. We guarded against overfitting through inclusion of a second-order random-walk prior distribution on the coefficients of the b-splines. This prior distribution takes the form bi = 2bi−1−bi−2 +ui, where bi is the ith b-spline coefficient and ui is normally distributed with ui ∼N(0,ρ2). The prior distribution assumes a constant first derivative and therefore penalizes against changes in growth rate unless supported by the data. The penalization of changes in growth rate is controlled by the parameter ρ, assigned an inverse γ prior distribution, ρ∼IG(0.001, 0.001). We assumed a uniform prior distribution on the first 2 b-spline coefficients.
Laboratory procedures
Swabs were sent by courier (earlier rounds) or by priority mail (later rounds) to a depot for onward transport to a single commercial laboratory for analysis by rt-PCR. The laboratory analyzed extracted nucleic acid for SARS-CoV-2 with the ViroBOAR 1·0 RT-qPCR kit (EuroFins Genomics, GmbH, Ebersberg, Germany) and on the Roche Lightcycler 480 II (Roche Diagnostics, Almere, The Netherlands) to detect in parallel 2 gene targets: N gene and E gene. The assay has a specificity close to 100% with limit of detection of 10 copies per microliter. In addition, the laboratory developed a multiplex for influenza A and B, which was added in November 2021 (Figure 1). Performance of the multiplex was tested against an established assay (Luminex NxTAG Respiratory Pathogen Panel, Luminex Corporation, Austin, Texas, USA) in an independent commercial laboratory, as well against sequencing of selected samples in the UK Health Security Agency laboratories.
Cycle threshold and laboratory calibration experiments
We used Ct values as a proxy for viral load for the 2 SARS-CoV-2 gene targets (N gene and E gene). The rt-PCR was considered positive if both gene targets were detected or if N gene was detected with Ct value less than 37. This Ct threshold was determined following 3 separate calibration experiments. In the first, we sent RNA extraction plates (n = 10) from the commercial laboratory to 2 UK Accreditation Service‒accredited laboratories for blinded reanalysis. We found concordant results for 919 negative samples and all 40 controls. We detected viral RNA in 11 of the 19 samples with a Ct value reported positive by the commercial laboratory (N gene Ct value ranging from 16.5 to 40.7); in 10 of these 11 samples, N gene Ct value was less than 37.
Second, in a serial dilution experiment of synthetic SARS-CoV-2 RNA, the commercial laboratory detected 2.5 copies at Ct 38; also, while following serial dilution of known positive samples with low viral load, the commercial laboratory identified an N gene signal at Ct greater than 37 in most instances.
Third, a Public Health England reference laboratory reanalyzed a further 40 unblinded positive samples with N gene Ct values greater than 35 (range = 35.7–46.8) and without a signal for E gene, detecting SARS-CoV-2 RNA in 15 of 40 (38%) samples (2 of 4 with N gene Ct value < 37). We then consolidated the results of all 3 calibration experiments to set the positivity criteria noted previously, which we used throughout each round of REACT-1.
Viral genome sequencing
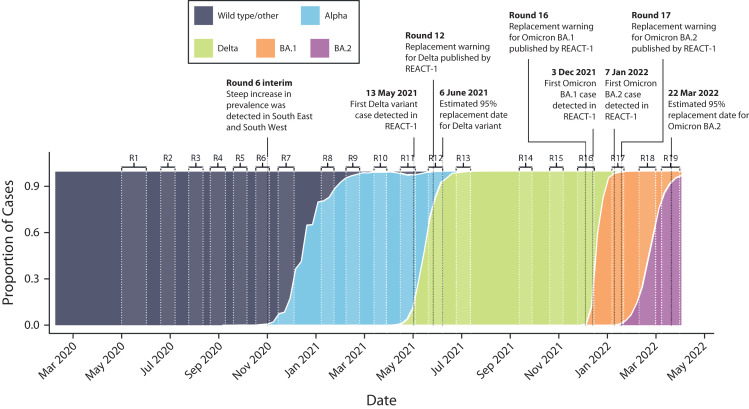
We undertook viral genome sequencing to provide information on specific variants from round 8 (January 6–22, 2021) onward. The Quadram Institute, in Norwich, UK, carried out the sequencing for samples that tested positive on rt-PCR with Ct value of (variously) 34 or below, or 32 or below, in either the N or E gene, and with sufficient volume of sample available. The Quadram Institute used the ARTIC protocol12 for viral RNA amplification, CoronaHiT for preparation of sequencing libraries,13 and the ARTIC bioinformatics pipeline,14 and assigned lineages by using PangoLEARN.15 Transitions of each of the variants during the course of the epidemic in the United Kingdom and their detection in England in the REACT-1 study are shown in Figure 4.
FIGURE 4—
Proportion of Wild Type, Alpha, and Delta Lineages and Omicron BA.1 and Omicron BA.2 Sublineages: GISAID, United Kingdom, April 1, 2022
Note. GISAID = Global Initiative on Sharing Avian Influenza Data; R = round; REACT-1 = REal-time Assessment of Community Transmission-1. For Delta lineage and Omicron BA.1 and Omicron BA.2 sublineages, we report the date of first detection in REACT-1 samples with determined lineage, the date at which REACT-1 reported a possible variant replacement, and the corresponding estimated date of replacement as defined by the date at which the estimated median proportion of the given variant was greater than 95%.
Managing disclosure risks
To protect confidentiality, individual data are not released, and tabular data are suppressed if there are fewer than 5 entries in a cell where 1 or more is a positive for SARS-CoV-2.
DATA ANALYSIS/DISSEMINATION
The data collected in REACT-1 were analyzed and reported in real-time to government and widely disseminated in timely fashion to the scientific community and the public.
Interpretation Issues
Over the course of the study, we observed a gradual decline in response rates, from a high of 30.5% in round 1 (May 1 to June 1, 2020), carried out during the first lockdown in England when it was very difficult to get a diagnostic test through other means. We were able to offset some of the decline in response by including a modest monetary incentive (via gift voucher) to people aged 13 to 44 years who are harder to reach in surveys. Moreover, our reported response rates are conservative estimates as we based them on numbers of swabs with a valid rt-PCR result compared with the numbers of invitation letters sent out, some of which may not have reached (or been opened by) the recipient. We used RIM weighting to correct the sample to be representative of the population of England as a whole. Nonetheless, to the extent that this relied on weighting factors, some bias may have been introduced into our estimates of prevalence, particularly when examining effects in specific groups (e.g., by occupation, ethnicity, or deprivation).
Changes in the way the swab samples were collected, transported, and tested may have introduced small differences across rounds, although these should not have affected within-round trends in prevalence. The switch from dry to wet (saline) swabs and from collection of samples with to without a cold chain might have affected diagnostic sensitivity. Because of financial and logistical constraints, we were not able to directly compare dry swab transported by courier on a cold chain, as was used before round 14, with wet swab sent by priority mail, which was used from round 15 on (Figure 1). However, we were reassured by the small differences between the samples collected by mail and courier in our randomized comparison during round 14.
More than 2 million people consented to data linkage to their NHS records, but for the remainder, we are unable to obtain hospitalizations or mortality data, or accurate vaccination data from the national COVID-19 immunization program. Although we do have self-report data on vaccination history, for those without linked data, dates of vaccination and vaccine type may be missing or less reliable than in the linked data, which may introduce bias. Furthermore, those who do and do not consent to data linkage might differ in important ways, such as social mixing patterns.
Linkage Ability
Participants were asked whether they consented to linkage to their health records. For participants with consent, data linkage (based on their unique NHS number) includes vaccine history (vaccine type and date) and outcome data (hospitalizations, deaths).
Data Release/Accessibility
Access to REACT-1 individual-level data are restricted to protect participants’ anonymity. Summary statistics, descriptive tables, and code from REACT-1 are available at https://github.com/mrc-ide/reactidd. REACT-1 study materials are available for each round at https://www.imperial.ac.uk/medicine/research-and-impact/groups/react-study/for-researchers/react-1-study-materials.
Key References/Other Information
We have published our protocol16 and on our findings during the 23 months of fieldwork, including methodological work on estimation of growth rates and reproduction number, on transitions between variants, and on the duration of swab-positivity after infection.1,2,4,17–25 We also published preprints giving results for each round. Links to all our publications are given on our Web site at https://www.imperial.ac.uk/medicine/research-and-impact/groups/react-study/real-time-assessment-of-community-transmission-findings.
PUBLIC HEALTH IMPLICATIONS
REACT-1 provided reliable and robust estimates of the prevalence of SARS-CoV-2 infection and of epidemic growth of COVID-19 in England. We provided prevalence estimates (and odds ratios) for different sociodemographic groups—for example, by age, sex, ethnicity, occupation, household size, and deprivation—and estimates of vaccine effectiveness against infection. The government used these data to inform the public health response across nearly 2 years of the epidemic in England.
Impact
Unlike the routine testing data, results from REACT-1 were not dependent on the availability of tests, which affected prevalence estimates in the routine surveillance data,22 nor were they biased by test-seeking behaviors, and importantly included asymptomatic as well as symptomatic infections, which is critical in understanding the infection dynamics.26 The results of REACT-1 were reported at least weekly to the government to provide situational awareness and inform policy and public health interventions. Thus, REACT-1 informed the timing of the second national lockdown in England, reinforced the need for the “rule of 6” (i.e., no more than 6 people allowed at social gatherings), provided key data underpinning school closure policy, and contributed to recommendations to protect those living in large households. REACT-1 was one of the first studies to detect the rapid rise of infections in southeast London, Kent, and Essex in September 202127 as the Alpha variant began to take hold and reported the rapid replacement of the Alpha variant by Delta in May 2021,24 Delta by Omicron BA.1 in December 2021,20 and Omicron BA.1 by Omicron BA.2 in February to March 202219 (Figure 4).
In addition to our role in providing situational awareness to government, we placed REACT-1 data into the public domain in near-real-time (through both preprints and press releases and the media), thus informing both the international scientific community and the public as to the current state of the epidemic in England.
ACKNOWLEDGMENTS
This study was funded by the Department of Health and Social Care in England. P. Elliott is director of the MRC Centre for Environment and Health (MR/L01341X/1 and MR/S019669/1). P. Elliott acknowledges support from Health Data Research UK; the National Institute for Health and Care Research (NIHR) Imperial Biomedical Research Centre; NIHR Health Protection Research Units in Chemical and Radiation Threats and Hazards, and Environmental Exposures and Health; the British Heart Foundation Centre for Research Excellence at Imperial College London (RE/18/4/34215); and the UK Dementia Research Institute at Imperial College London (MC_PC_17114). N. Steyn acknowledges support from the Oxford-Radcliffe Scholarship from University College, Oxford, and the Engineering and Physical Sciences Research Council Centre for Doctoral Training in Modern Statistics and Statistical Machine Learning (Imperial College London and University of Oxford). We thank A. Maslov for supporting this research through studentship support for N. Steyn. H. Ward acknowledges support from an NIHR Senior Investigator Award, the Wellcome Trust (205456/Z/16/Z), and the NIHR Applied Research Collaboration North West London. J. Elliott is an NIHR academic clinical fellow in infectious diseases. G. S. Cooke is supported by an NIHR professorship. C. A. Donnelly acknowledges support from the MRC Centre for Global Infectious Disease Analysis, the NIHR Health Protection Research Unit in Emerging and Zoonotic Infections, and the NIHR-funded Vaccine Efficacy Evaluation for Priority Emerging Diseases (PR-OD-1017-20007). M. Chadeau-Hyam and B. Bodinier acknowledge support from Cancer Research UK, Population Research Committee Project grant “Mechanomics” (grant 22184 to M. Chadeu-Hyam). M. Chadeu-Hyam acknowledges support from the H2020-EXPANSE (Horizon 2020 grant 874627) and H2020-LongITools (Horizon 2020 grant 874739).
We thank key collaborators on this work: K. Beaver, S. Clemens, G. Welch, N. Gilby, K. Ward, G. Pantelidou, and K. Pickering at Ipsos; G. Fontana and J. Alford at the Institute of Global Health Innovation at Imperial College London; E. Johnson, R. Elliott, and G. Blakoe at the School of Public Health, Imperial College London; and A. J. Page and N.-F. Alikhan at the Quadram Institute, Norwich, UK. We also thank North West London Pathology and Public Health England (now UK Health Security Agency) for help in calibration of the laboratory analyses, Patient Experience Research Centre at Imperial College London and the REACT Public Advisory Panel, NHS Digital for access to the NHS register, and the Department of Health and Social Care for logistical support.
Note. The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of this article.
CONFLICTS OF INTEREST
MC-H holds shares in the O-SMOSE company. Consulting activities conducted by the company are independent of the present work, and MC-H has no conflicts of interest to declare. All other authors have no competing interests to declare.
HUMAN PARTICIPANT PROTECTION
The REACT-1 study received ethical approval from the South Central–Berkshire B Research Ethics Committee, UK.
REFERENCES
- 1.Eales O, Ainslie KEC, Walters CE, et al. Appropriately smoothing prevalence data to inform estimates of growth rate and reproduction number. Epidemics. 2022;40:100604. doi: 10.1016/j.epidem.2022.100604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eales O, Walters CE, Wang H, et al. Characterising the persistence of RT-PCR positivity and incidence in a community survey of SARS-CoV-2. Wellcome Open Res. 2022;7:102. doi: 10.12688/wellcomeopenres.17723.1. [DOI] [Google Scholar]
- 3.REACT 1 study materials. Imperial College London. 2023. https://www.imperial.ac.uk/medicine/research-and-impact/groups/react-study/for-researchers/react-1-study-materials [Google Scholar]
- 4.Chadeau-Hyam M, Wang H, Eales O, et al. SARS-CoV-2 infection and vaccine effectiveness in England (REACT-1): a series of cross-sectional random community surveys. Lancet Respir Med. 2022;10(4):355–366. doi: 10.1016/S2213-2600(21)00542-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharot T.Weighting survey results. 2022. http://www.redresearch.com/wp/wp-content/uploads/2016/01/Weighting-Survey-Results.pdf
- 6.McLennan D, Noble S, Noble M, Plunkett E, Wright GN.The English Indices of Deprivation 2019. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/833951/IoD2019_Technical_Report.pdf2023
- 7.Population estimates for the UK, England and Wales; Wales: Office for National Statistics. 2021. [Google Scholar]
- 8.Labour Force Survey. Office for National Statistics. Available. 2023. https://www.ons.gov.uk/surveys/informationforhouseholdsandindividuals/householdandindividualsurveys/labourforcesurvey
- 9.Lumley T. Analysis of complex survey samples. J Stat Softw. 2004;9(8):1–19. doi: 10.18637/jss.v009.i08. [DOI] [Google Scholar]
- 10.Hoffman MD, Gelman A. The No-U-Turn sampler: adaptively setting path lengths in Hamiltonian Monte Carlo J Mach Learn Res. 2014151593–1623.https://www.jmlr.org/papers/volume15/hoffman14a/hoffman14a.pdf. Accessed January 1, 2023. [Google Scholar]
- 11.Lang S, Brezger A. Bayesian P-Splines. J Comput Graph Stat. 2004;13(1):183–212. doi: 10.1198/1061860043010. [DOI] [Google Scholar]
- 12.Quick J.2023. https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bp2l6n26rgqe/v3
- 13.Baker DJ, Aydin A, Le-Viet T, et al. CoronaHiT: high-throughput sequencing of SARS-CoV-2 genomes. Genome Med. 2021;13(1):21. doi: 10.1186/s13073-021-00839-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.A Nextflow Pipeline for Running the ARTIC Network’s Field Bioinformatics Tools. GitHub. Available at. 2023. https://github.com/connor-lab
- 15.Pangolin. 2022. https://github.com/cov-lineages/pangolin
- 16.Riley S, Atchison C, Ashby D, et al. REal-time Assessment of Community Transmission (REACT) of SARS-CoV-2 virus: study protocol. Wellcome Open Res. 2021;5:200. doi: 10.12688/wellcomeopenres.16228.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott P, Eales O, Bodinier B, et al. Dynamics of a national Omicron SARS-CoV-2 epidemic during January 2022 in England. Nat Commun. 2022;13(1):4500. doi: 10.1038/s41467-022-32121-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eales O, de Oliveira Martins L, Page AJ, et al. Dynamics of competing SARS-CoV-2 variants during the Omicron epidemic in England. Nat Commun. 2022;13(1):4375. doi: 10.1038/s41467-022-32096-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott P, Eales O, Steyn N, et al. Twin peaks: the Omicron SARS-CoV-2 BA.1 and BA.2 epidemics in England. Science. 2022;376(6600):eabq4411. doi: 10.1126/science.abq4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elliott P, Bodinier B, Eales O, et al. Rapid increase in Omicron infections in England during December 2021: REACT-1 study. Science. 2022;375(6587):1406–1411. doi: 10.1126/science.abn8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chadeau-Hyam M, Tang D, Eales O, et al. Omicron SARS-CoV-2 epidemic in England during February 2022: a series of cross-sectional community surveys. Lancet Reg Health Eur. 2022;21:100462. doi: 10.1016/j.lanepe.2022.100462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riley S, Ainslie KEC, Eales O, et al. Resurgence of SARS-CoV-2: detection by community viral surveillance. Science. 2021;372(6545):990–995. doi: 10.1126/science.abf0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chadeau-Hyam M, Eales O, Bodinier B, et al. Breakthrough SARS-CoV-2 infections in double and triple vaccinated adults and single dose vaccine effectiveness among children in autumn 2021 in England: REACT-1 study. EClinicalMedicine. 2022;48:101419. doi: 10.1016/j.eclinm.2022.101419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elliott P, Haw D, Wang H, et al. Exponential growth, high prevalence of SARS-CoV-2, and vaccine effectiveness associated with the Delta variant. Science. 2021;374(6574):eabl9551. doi: 10.1126/science.abl9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eales O, Wang H, Haw D, et al. Trends in SARS-CoV-2 infection prevalence during England’s roadmap out of lockdown, January to July 2021. PLOS Comput Biol. 2022;18(11):e1010724. doi: 10.1371/journal.pcbi.1010724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oran DP, Topol EJ. The proportion of SARS-CoV-2 infections that are asymptomatic. Ann Intern Med. 2021;174(9):1344–1345. doi: 10.7326/L21-0491. [DOI] [PubMed] [Google Scholar]
- 27.Riley S, Ainslie KEC, Eales O, et al. . medRxiv.2020 10.1101/2020.10.30.20223123 [DOI]