Significance
Nucleotide excision DNA repair (NER) removes a large variety of genomic lesions. NER can be initiated through two distinct pathways: global genome repair (GG-NER) and transcription-coupled repair (TC-NER). Both pathways subsequently funnel into a common pathway that involves recruitment of the transcription factor IIH (TFIIH) complex and the central scaffold protein XPA that enables full NER complex assembly. Although downstream steps after damage recognition are thought to be identical, we identify a disease-associated mutation in XPA that severely weakens the interaction with the TFIIH complex causing TC-NER to be affected to a greater extent than GG-NER. This differential impact on GG-NER and TC-NER suggests unanticipated mechanistic differences in the transition from lesion recognition to dual incision for the two pathways of NER.
Keywords: DNA repair, nucleotide excision repair, xeroderma pigmentosum protein A, transcription factor II H, transcription-coupled repair
Abstract
XPA is a central scaffold protein that coordinates the assembly of repair complexes in the global genome (GG-NER) and transcription-coupled nucleotide excision repair (TC-NER) subpathways. Inactivating mutations in XPA cause xeroderma pigmentosum (XP), which is characterized by extreme UV sensitivity and a highly elevated skin cancer risk. Here, we describe two Dutch siblings in their late forties carrying a homozygous H244R substitution in the C-terminus of XPA. They present with mild cutaneous manifestations of XP without skin cancer but suffer from marked neurological features, including cerebellar ataxia. We show that the mutant XPA protein has a severely weakened interaction with the transcription factor IIH (TFIIH) complex leading to an impaired association of the mutant XPA and the downstream endonuclease ERCC1-XPF with NER complexes. Despite these defects, the patient-derived fibroblasts and reconstituted knockout cells carrying the XPA-H244R substitution show intermediate UV sensitivity and considerable levels of residual GG-NER (~50%), in line with the intrinsic properties and activities of the purified protein. By contrast, XPA-H244R cells are exquisitely sensitive to transcription-blocking DNA damage, show no detectable recovery of transcription after UV irradiation, and display a severe deficiency in TC-NER-associated unscheduled DNA synthesis. Our characterization of a new case of XPA deficiency that interferes with TFIIH binding and primarily affects the transcription-coupled subpathway of nucleotide excision repair, provides an explanation of the dominant neurological features in these patients, and reveals a specific role for the C-terminus of XPA in TC-NER.
Nucleotide excision repair (NER) is a versatile DNA repair pathway that removes a large variety of genomic lesions, including those induced by UV light, environmental genotoxins, or chemotherapeutic agents. NER can be initiated through two distinct damage-recognition pathways: global genome repair (GG-NER) and transcription-coupled repair (TC-NER). GG-NER involves lesion recognition throughout the genome by the xeroderma pigmentosum complementation C (XPC) and DNA damage-binding 2 (DDB2) proteins (1–3). TC-NER is initiated when transcribing RNA polymerase II (RNAPII) encounters a lesion that blocks its progression and involves Cockayne syndrome A and B (CSA and CSB) proteins, UV-stimulated scaffold protein A (UVSSA), and ELOF1 (4–10).
Both damage-recognition modes subsequently funnel into a common pathway involving the recruitment of the 10-subunit transcription factor IIH (TFIIH) complex, which verifies the presence of the lesion with the help of XPA and RPA (2, 11–13). XPA functions as a central scaffold protein that interacts with many components of the NER preincision complex, including TFIIH (14–16), RPA (17–21), and ERCC1 (22–24), as well as with DNA through its zinc-finger domain (25, 26). The arrival of XPA at NER complexes stimulates the dissociation of TFIIH’s kinase module (consisting of Cdk7-cyclin H, and MAT1) thereby activating its XPD helicase subunit, allowing it to track along the damaged strand to verify the presence of a DNA lesion (16, 27). Subsequently, XPA recruits the ERCC1-XPF endonuclease to the NER preincision complex (2, 24), which, together with the XPG endonuclease, mediates dual incision around the DNA lesion (28). After dual incision and removal of a stretch of single-stranded DNA containing the lesion, the resulting gap is filled by DNA polymerases and sealed by DNA ligases (29, 30).
The importance of NER is underscored by the severity of clinical syndromes caused by inherited NER defects, including xeroderma pigmentosum (XP) and Cockayne syndrome (CS). XP is characterized by extreme sensitivity to UV light and an over 2,000-fold increased risk of skin cancer along with varying levels of neurological manifestations. In contrast, CS is characterized by severe neurodegeneration and developmental defects without cancer predisposition. The specific genes affected in patients, together with their role in distinct NER pathways, to a large extent explain the variability in clinical syndromes. CS patients have a defect in CSA or CSB and have a selective defect in TC-NER along with an associated defect in the processing of DNA damage stalled RNAPII (5, 10, 31). Patients with XP have a defect in any of the known XP genes, annotated XPA-XPG, and either have a selective defect in GG-NER (e.g., patient with defective XPC or XPE/DDB2) or in both TC-NER and GG-NER (e.g., patients with defects in XPA).
Patients with null-mutations in XPA typically present with classical XP, which is associated with a complete NER defect, severe UV sensitivity, cancer predisposition, and often neurological features (32). However, multiple XPA gene alterations, especially single amino acid substitutions, have been linked to varying clinical manifestations in patients. While a C108F amino acid substitution in the zinc-finger domain of XPA, and a homozygous Q185H mutation, causing missplicing of exon 4 to 5, have been associated with a severe clinical phenotype (33, 34), a previously described patient compound heterozygous for the Q185H and an H244R amino acid substitution in the last exon of XPA showed residual DNA repair and milder XP features (35, 36). Similarly, a nonsense mutation causing a premature stop-codon in the last exon of XPA (R228*), which is found in Tunisian and Japanese XP patients with mild clinical features (37, 38), resulted in moderate UV sensitivity (39). The data from patients with the R228* or H244R amino acid substitutions have led to the notion that mutations in the last exon of XPA cause mild features due to substantial levels of residual repair.
The relatively mild phenotype of the C-terminal mutations may be explained by the structure of the XPA protein (40). The central domain of XPA (residues 98 to 239), which includes the zinc finger, DNA- and RPA70-binding domains assumes a defined fold (41), and mutations that affect the folding of this domain appear to destabilize and inactivate the protein. The N- and C-terminal parts of XPA are disordered in the free protein and some domains undergo a disorder to order transition upon interaction with partner protein RPA32 (residues 29 to 24) (20), ERCC1 (residues 67 to 80) (24), and TFIIH (C-terminus, residues 240 to 273) (16). Mutations in these regions are therefore expected to only affect a part of the protein, in line with the milder phenotypes of the R228* and H244R mutations.
Here, we describe two Dutch patients with a homozygous H244R amino acid substitution in XPA who present with mild cutaneous manifestations of XP without developing skin cancer, but with marked neurological features including cerebellar ataxia. Functional analysis revealed that the XPAH244R mutant protein shows a severely weakened interaction with the TFIIH complex, which is sufficient to maintain intermediate levels of GG-NER (~50%) but results in a severe TC-NER defect. Our study suggests that C-terminal disease-associated XPA mutations can lead to interaction defects with TFIIH that affect TC-NER to a greater extent than GG-NER.
Results
Two Patients with Mild XP-A and Neurological Symptoms.
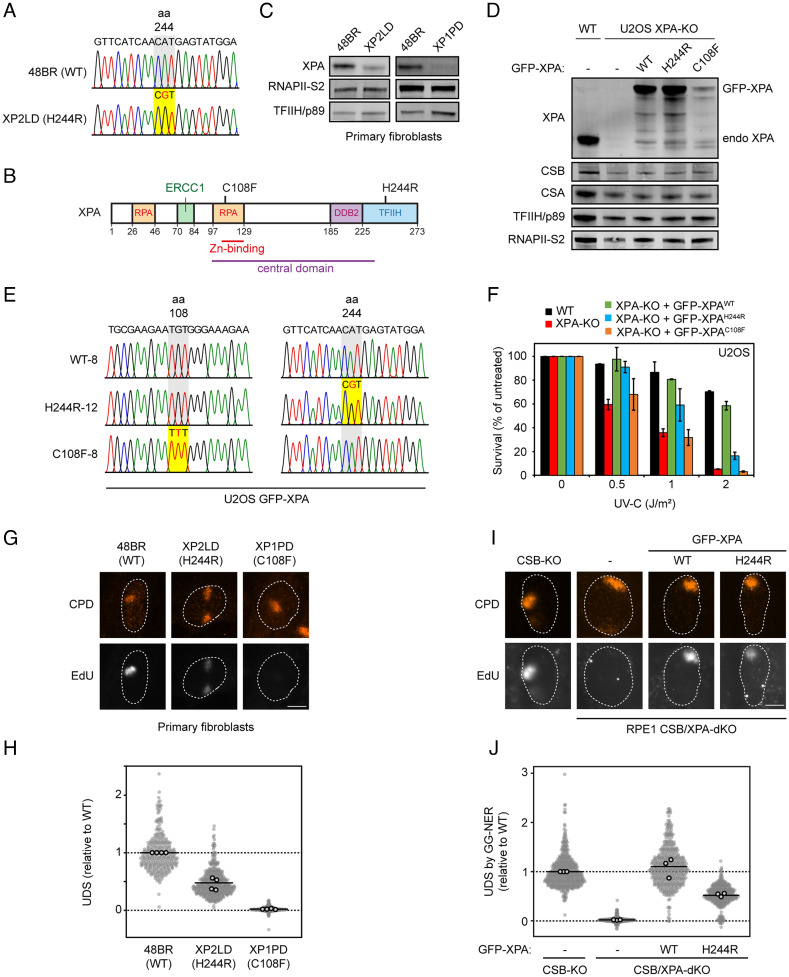
A 46-y-old female patient (XP2LD) and her 51-y-old brother (XP3LD) had experienced photosensitivity from early childhood, presenting with erythema and blistering of the skin following exposure to sunlight (SI Appendix, Fig. S1A). Both protected their skin against sun exposure by wearing a hat when outside and by applying sunscreen in the spring and summer months. The male patient suffered from cerebellar ataxia, dysarthria and mild axonal polyneuropathy, progressive cognitive impairment, and autism-spectrum disorder. Starting from adulthood, the younger sister developed similar but milder neurological features. MRI scanning of the brain revealed generalized atrophy of moderate severity in both patients (SI Appendix, Fig. S1B). On skin examination, both patients had some scattered lentigines on the upper back, shoulders, and nose, hypopigmented macules on the lower legs, but no further pigmentary changes or poikiloderma. The male patient had a few actinic keratoses on the face. Phototesting performed in the male patient showed that the minimal erythemal dose for broadband UVB was low but not significantly reduced, while the minimal erythemal dose for UVA was normal. Their family history was negative for photosensitivity, skin cancer, and neurological disorders. A diagnosis of xeroderma pigmentosum type A was made by diagnostic whole-exome sequencing analysis, which revealed a homozygous mutation in the XPA gene, an A to G nucleotide substitution at base 731 in the cDNA (c.731A>G; Fig. 1A), leading to a histidine to arginine amino acid substitution at position 244 of the protein, in both patients (p.His244Arg). Their parents, who were of Dutch descent and not consanguineous, are carriers of this pathogenic gene variant. At the age of 49 y, the male patient had not developed cutaneous squamous cell carcinoma, basal cell carcinoma, or melanoma. His sister at the age of 44 y had not developed any malignant skin tumors either. Although photosensitivity and neurological symptoms are common features of XP type A; the absence of skin carcinogenesis is unusual.
Fig. 1.
Patient or rescue cells expressing XPAH244R show residual UDS. (A) Sequence confirmation of the H244R amino acid substitution in genomic DNA of patient XP2LD compared to WT DNA (48BR). (B) Schematic representation of the XPA protein with its interaction domains. (C) XPA protein expression in primary fibroblasts from 48BR (WT), XP2LD (XPAH244R), and XP1PD (XPAC108F) by western blot. RNAPII-S2 and TFIIH/p89 levels are used as loading controls. (D) Expression levels of endogenous XPA in U2OS(FRT) WT and U2OS(FRT) XPA-KO cells, and GFP-XPA expression in U2OS(FRT) XPA-KO cells reconstituted with GFP-XPAWT, GFP-XPAH244R, or GFP-XPAC108F. CSB, CSA, TFIIH/p89, and RNAPII-S2 are used as loading controls. (E) Sequence confirmation of the H244R or C108F amino acid substitution in the U2OS(FRT) XPA-KO cells reconstituted with GFP-XPAWT, GFP-XPAH244R, or GFP-XPAC108F cDNA. (F) Clonogenic survival assay after irradiation with various doses of UV-C in U2OS(FRT) WT, XPA-KO, and GFP-XPA reconstituted cell lines. The bars are the average of two experiments. The error bars reflect the SD. (G) Representative images and (H) quantification of EdU incorporation in the indicated primary fibroblasts after 30 J/m2 UV-C irradiation through 5-µm pore membranes. Sites of local damage are identified by CPD staining. The EdU signal has been normalized for CPD levels at sites of local damage. (I) Representative images and (J) quantification of EdU incorporation in the indicated RPE1 cells deficient in TC-NER (CSB-KO cells) to specifically measure UDS by GG-NER after 30 J/m2 UV-C irradiation through 5-µm pore membranes. Sites of local damage are identified by CPD staining. The EdU signal has been normalized for CPD levels at sites of local damage. (H and J) All cells are depicted as individual data points with the bar representing the mean of all data points. The individual means of four biological replicates are depicted as gray points with black circles.
Reduced XPA Protein Expression Caused by an H244R Amino Acid Substitution.
We obtained primary fibroblasts of the female patient and annotated this primary fibroblast cell line as XP2LD. The nucleotide substitution in the XPA gene was confirmed by Sanger sequencing (Fig. 1A) and was located in the C-terminal TFIIH-binding domain of XPA (Fig. 1B). Notably, H244 is one of four conserved potential Zinc-binding residues (along with H242, C261, and C264; SI Appendix, Fig. S2A), although Zn binding of the C-terminal domain has not been experimentally detected. We also obtained primary fibroblasts from a previously described patient XP1PD, presenting with classical XP. This patient is compound heterozygous for a 5-nucleotide deletion (at nucleotides 349 to 353) inducing a frameshift on one allele, and a C108F amino acid substitution in the Zn finger of the DNA-binding domain of XPA on the other allele (Fig. 1B) (34). Cells of both XP1PD and XP2LD expressed XPA, but to a reduced level compared to that in control fibroblasts (48BR). While the XPAC108F protein was barely detectable by western blot, XPAH244R levels were reduced to about 25% of wild-type levels (Fig. 1C). These findings suggest that while the C108F amino acid substitution severely affected XPA protein stability, the effect of the H244R amino acid substitution was moderate.
Generation of Reconstituted Cell Lines for Functional Characterization of XPA Mutants.
To remove potential residual XPA activity, we generated U2OS cell lines expressing inducible versions of GFP-tagged XPA variants. We first knocked out endogenous XPA using CRISPR-Cas9-mediated genome editing. Successful knockout (KO) was confirmed by a lack of XPA protein by western blot analysis (Fig. 1D) and by sequencing genomic DNA (SI Appendix, Fig. S2B). Subsequently, we stably expressed wild-type (WT) GFP-tagged XPA or GFP-XPA containing the H244R or C108F amino acid substitutions in these XPA-KO cells. Successful genomic integration was confirmed by sequencing (Fig. 1E) and by the inducible expression of GFP-XPA to levels near endogenous XPA expression levels in wild-type cells (Fig. 1D). Note that the inducible expression of GFP-XPA proteins enabled us to achieve similar expression levels between GFP-XPAWT and GFP-XPAH244R, while GFP-XPAC108F was present at much lower levels, suggesting that the severe instability of this mutant due to disruption of the zinc-finger in the central domain could not be overcome by expression at high levels.
Expression of XPAH244R Confers Partial Protection against UV Irradiation.
To assess to what extent XPA mutant proteins provide protection against UV-induced DNA damage, we monitored UV sensitivity in XPA knockout cells expressing GFP-tagged wild-type XPA (GFP-XPAWT) or the two XPA mutants GFP-XPAC108F or GFP-XPAH244R in clonogenic survival experiments (Fig. 1F). As expected, knockout of XPA in U2OS cells resulted in severe UV sensitivity, which could be rescued to a large extent by reexpression of GFP-XPAWT. The expression of GFP-XPAH244R in XPA-deficient cells resulted in a partial, rescue of UV sensitivity, while the GFP-XPAC108F expressing cells were as sensitive as the XPA-KO cells. These results confirm the essential role of the central DNA-binding domain of XPA for its functionality and show that the XPAH244R protein is still partially functional in NER.
Cells Expressing XPAH244R Display About 50% Global Genome NER Activity.
To investigate the NER capacity in patient-derived fibroblasts (Fig. 1 G and H) and reconstituted U2OS cells (SI Appendix, Fig. S2 C and D), we measured unscheduled DNA synthesis (UDS) after UV irradiation. Cells were locally exposed to 30 J/m2 UV-C through 5-µm pore filters and incubated with the thymidine analog 5-ethynyl-deoxy-uridine (EdU) for 1 h. Local incorporation of EdU in nonreplicating cells is a measure for NER-associated DNA repair synthesis (42). At sites of local damage, identified by cyclobutane pyrimidine dimer (CPD) staining, clear DNA incorporation was detected in wild-type fibroblasts (48BR) (Fig. 1G). While cells from the patient XP1PD were completely impaired in EdU incorporation, patient-derived XP2LD cells (expressing XPAH244R) maintained ~50% residual EdU incorporation compared to control fibroblasts (Fig. 1 G and H).
Clear EdU incorporation at sites of local UV damage was also observed in parental U2OS cells, which was completely abolished upon knockout of XPA (SI Appendix, Fig. S2 C and D). Reexpression of XPAWT rescued DNA repair-associated EdU incorporation, while expression of XPAH244R showed unscheduled DNA synthesis at ~60% compared to wild-type cells. By contrast, reexpression of XPAC108F failed to restore UDS in XPA-deficient cells (SI Appendix, Fig. S2 C and D). To rule out any contribution from TC-NER to this residual NER activity and to confirm these findings in another cell-line, we generated double knockouts of CSB and XPA in RPE1-hTERT cells (SI Appendix, Fig. S2E). Robust UV-induced EdU incorporation caused exclusively by GG-NER was detected in CSB-KO cells, while GG-NER-associated DNA repair synthesis was virtually absent in CSB/XPA-dKO cells (Fig. 1 I and J). Stable expression of XPAWT restored EdU incorporation, while XPAH244R restored UDS levels to ~50% (Fig. 1 I and J and SI Appendix, Fig. S2E). Together, these results confirm that the XPAH244R protein supports up to ~50% global genome NER activity.
Purified XPAH244R Protein Tends to Aggregate and Has Reduced Intrinsic NER Activity.
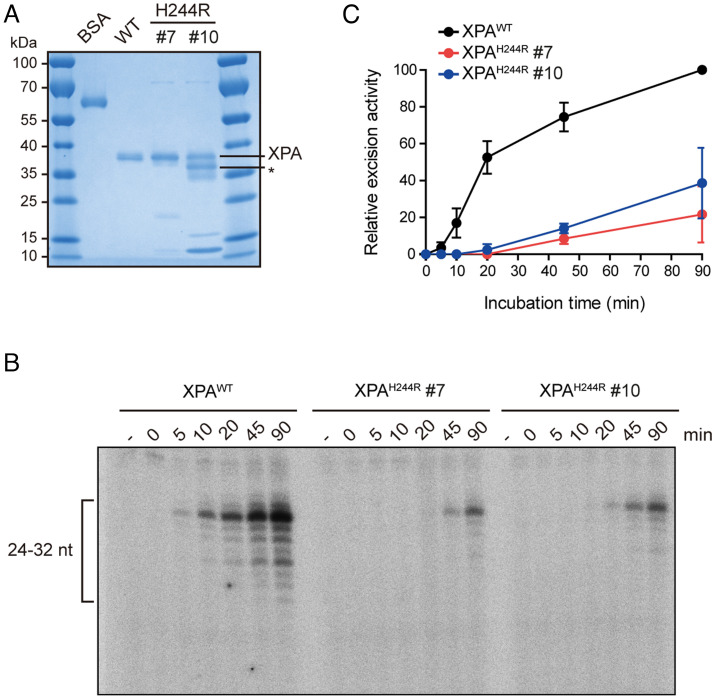
To gain more insight into the intrinsic properties of the XPAH244R protein, we expressed from E. coli and purified using Ni affinity, heparin, and gel-filtration columns and compared its activity to the XPAWT protein. While the WT protein eluted as one peak on the gel filtration column, representing a single band on an SDS PAGE gel, XPAH244R eluted as two peaks, designated fractions #7 and #10 (Fig. 2A and SI Appendix, Fig. S3A). While fraction #7 ran as a single band on an SDS-PAGE gel, #10 ran as two main bands, at the same and higher mobility (lower apparent MW) than the WT protein. We suspected that the lower band may be a degradation product, but electrospray ionization (ESI) mass spectrometry (MS) analysis revealed that both bands represent full-length XPAH244R, with the H244R mutation clearly present in a proteolytic fragment (SI Appendix, Fig. S3B). Treatment with iodoacetamide did not alter the relative mobility of the multiple bands, while it shifted the mobility of the mutant and WT protein to the same extent. These experiments show that incomplete reduction of cysteine residues does not cause the multiple bands seen with the purified XPAH244R protein (SI Appendix, Fig. S3C). While the exact origin of the two bands is unknown, our interpretation is that XPAH244R tends to aggregate.
Fig. 2.
The H244R mutation in XPA reduces in vitro NER activity. (A) Purification of XPAH244R protein. Two fractions from purification, #7 and #10 were tested for in vitro assays. (B) In vitro NER activity of XPAWT and XPAH244R mutant using purified NER proteins. A plasmid containing a site-specific AAF lesion was incubated with purified XPA (20 nM), XPA-RAD23B (5 nM), TFIIH (10 nM), RPA (42 nM), XPG (27 nM), and XPF-ERCC1 (13 nM) for 0 to 90 min. The excision products were detected by annealing to a complementary oligonucleotide with a 4dG overhang, which was labeled with [α-32P] dCTP. (C) Quantification of (B). Two independent experiments were performed.
We then tested the intrinsic DNA-binding activity of XPAH244R in an electrophoretic mobility shift assay (EMSA). We observed that both fraction #7 and #10 of XPAH244R displayed robust DNA-binding activity, albeit with lower affinity than the WT protein (SI Appendix, Fig. S3D). We have observed similar binding behavior of other mutant XPA proteins with mutations outside the central domain (21), and so believe that difference in binding affinity is not significant enough to suggest misfolding of the core domain. Interestingly, the XPA-DNA bands of fraction #10 were diffuse, in line with the multiple bands observed by SDS PAGE (Fig. 2A and SI Appendix, Fig. S3D).
We next compared the in vitro NER activity of XPAH244R and XPAWT by incubation of these proteins with a plasmid containing a site-specific dG-AAF lesion along with purified NER factors XPC-RAD23B, TFIIH, RPA, XPG, and ERCC1-XPF. The NER excision product was detected by radioactive labeling and analyzed on a sequencing gel. We found that both fractions #7 and #10 of XPAH244R displayed a three to fourfold reduction in NER activity at longer incubation times (Fig. 2 B and C), in line with the results showing partial NER activity of this XPA allele in cells.
Proteomic Interactome Mapping of GFP-XPAH244R Reveals a Weakened Interaction with TFIIH.
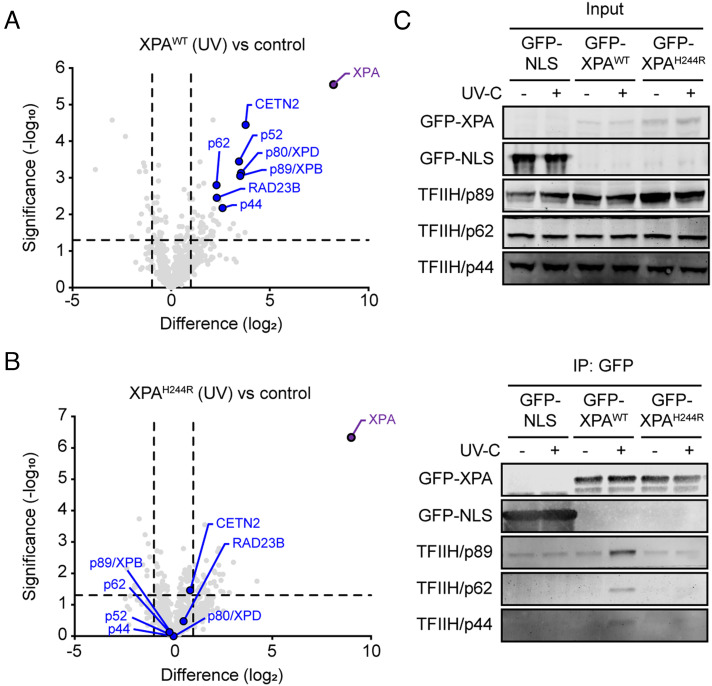
XPA has been extensively studied for its role as a scaffold for various core NER proteins and interaction domains have been identified for binding to RPA, ERCC1, and TFIIH (Fig. 1B)(16, 19, 20, 24). To map the interactome of both wild-type and mutant XPA in an unbiased manner, we performed pull-down experiments with GFP-XPAWT, GFP-XPAH244R, or a control GFP-protein tagged with a nuclear localization signal (GFP-NLS). Cells were either mock-treated or exposed to UV irradiation (20 J/m2), and interacting proteins were analyzed by label-free mass spectrometry (MS).
A comparison between GFP-XPAWT versus GFP-NLS revealed that, in the absence of UV irradiation, wild-type XPA associates with the RNAPII (POLR2A, POLR2B) and the PAF1 complexes (CDC73, CTR9, LEO1, and WDR61; SI Appendix, Fig. S4A; in green), which may reflect a previously reported role of GG-NER proteins in transcription unrelated to DNA repair (43). Following UV irradiation, XPAWT strongly associated with core NER proteins, including five subunits of the TFIIH complex (p44/GTF2H2, p52/GTF2H4, p62/GTF2H1, p80/XPD/ERCC2, and p89/XPB/ERCC3) and the XPC-associated proteins RAD23B and CETN2 (Fig. 3A and SI Appendix, Fig. S4B; in blue), reflecting the assembly of NER complexes involved in DNA repair.
Fig. 3.
XPAH244R has a weakened association with TFIIH after UV. (A) Volcano plot depicting the UV-specific enrichment of proteins after pull-down of GFP-XPAWT at 1 h after irradiation with 20 J/m2 UV-C versus (unirradiated) GFP-NLS analyzed by label-free MS. The enrichment (log2) is plotted on the x-axis and the significance (2-sided t test -log10 P-value) is plotted on the y-axis. Highlighted are XPA (purple) and NER proteins (blue). (B) As in a, but for pull-down of GFP-XPAH244R at 1 h after irradiation with 20 J/m2 UV-C compared to unirradiated, mock-treated GFP-NLS. (C) Coimmunoprecipitation of GFP-XPA or GFP-NLS from U2OS(FRT) WT cells expressing GFP-NLS or U2OS(FRT) XPA-KO cells reconstituted with GFP-XPAWT or GFP-XPAH244R, either without or after UV irradiation (20 J/m2 UV-C). The input is 0.75% of the total protein lysate.
Proteomic analysis of GFP-XPAH244R versus GFP-NLS revealed that in nonirradiated cells, the mutant XPA protein failed to associate with the RNAPII and PAF1 complex (SI Appendix, Fig. S4 C and D; in green). Following UV irradiation, the UV-induced association with TFIIH subunits and XPC complex members seen with wild-type XPA protein was not detectable after pull-down of the mutant XPA protein (Fig. 3B and SI Appendix, Fig. S4 E and F in blue). Immunoprecipitation experiments on GFP-XPAWT and GFP-XPAH244R followed by western blotting confirmed the UV-induced association of XPAWT with TFIIH subunits (p89, p62, p44), while we failed to detect any association of XPAH244R with the TFIIH complex (Fig. 3C). Our results show that histidine 244 in XPA is essential for a robust TFIIH-XPA interaction, in line with earlier findings showing that the last 48 C-terminal amino acids of XPA are essential for binding of TFIIH (14, 15). The observation that cells expressing XPAH244R maintain about 50% residual global genome NER activity suggests that a weakened XPA-TFIIH interaction, likely mediated by the central domain of XPA (16), is sufficient to support residual levels of DNA repair.
Impaired TFIIH Binding Reduces the Association of XPA with Sites of DNA Damage.
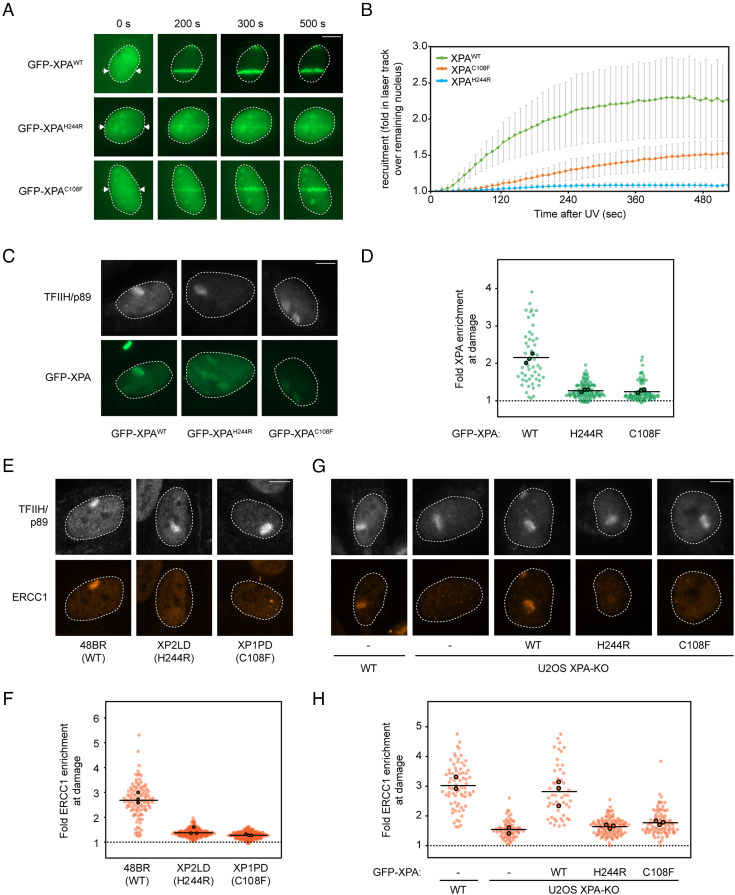
The recruitment of XPA to sites of DNA damage is essential for damage verification and the efficient assembly and positioning of NER proteins, including ERCC1-XPF, at damaged sites (2). To address whether the impaired TFIIH association leads to reduced localization of the mutant XPAH244R protein to sites of DNA damage, we locally irradiated U2OS cells expressing different variants of GFP-XPA using a 254-nm UV-C laser, followed by live-cell imaging to monitor GFP-XPA recruitment to laser tracks (44, 45). We observed the rapid and robust recruitment of GFP-XPAWT to sites of laser-induced DNA damage, reaching maximum binding within ~400 s with a t1/2 of 120 s (Fig. 4 A and B). In contrast, GFP-XPAH244R was barely visible at sites of UV-C laser irradiation, while recruitment of GFP-XPAC108F could be detected albeit much weaker than the wild-type protein (Fig. 4 A and B). To ensure that DNA damage was properly induced in these experiments, we cotransfected mCherry-tagged DDB2, which was robustly recruited to UV-C laser tracks, while GFP-XPAH244R failed to be efficiently recruited in the same cells (SI Appendix, Fig. S5A).
Fig. 4.
Reduced accumulation of XPAH244R impairs ERCC1-XPF recruitment at UV lesions. (A) Representative images of the recruitment of indicated GFP-tagged XPA variants to UV-C laser tracks, measured by live cell imaging. (B) Quantification of (A). Data represents average and SD of three independent experiments of 25 to 78 cells per experiment. (C) Representative images of the recruitment of indicated GFP-tagged XPA mutants reconstituted in U2OS(FRT) XPA-KO cells, to sites of damage after 30 J/m2 UV-C irradiation through 5-µm pore membranes. Sites of local damage are identified by TFIIH/p89 staining. (D) Quantification of (C). Quantification of the recruitment of TFIIH/p89 is shown in SI Appendix, Fig. S5B. (E) Representative images and (F) quantification of ERCC1 recruitment in the indicated primary fibroblasts after 30 J/m2 UV-C irradiation through 5-µm pore membranes. Sites of local damage are identified by TFIIH/p89 staining. Quantification of the recruitment of TFIIH/p89 is shown in SI Appendix, Fig. S5C. (G) Representative images and (H) quantification of ERCC1 recruitment in the indicated U2OS(FRT) cells after 30 J/m2 UV-C irradiation through 5-µm pore membranes. Sites of local damage are identified by TFIIH/p89 staining. Quantification of the recruitment of TFIIH/p89 is shown in SI Appendix, Fig. S5D. (D, F, and H) All cells are depicted as individual data points with the bar representing the mean of all data points. The individual means of three biological replicates are depicted as colored points with black circles.
To confirm these findings, we locally irradiated cells with a UV-C germicidal lamp through 5-µm pores, fixed cells after 20 min, and quantified GFP-XPA levels at sites of DNA damage marked by immunofluorescent staining of the TFIIH subunit p89/XPB (Fig. 4 C and D and SI Appendix, Fig. S5B). These data confirmed that GFP-XPAWT was clearly recruited to sites of TFIIH-marked DNA damage, whereas GFP-XPAH244R or GFP-XPAC108F could not be detected at this time point (Fig. 4D). Together, these results show that the TFIIH interaction mediated directly or indirectly through the H244 residue of XPA is essential for the stable association of XPA with sites of DNA damage.
ERCC1 Fails to Associate with UV-Damaged Sites in Cells Expressing XPAH244R.
Having established that the mutant XPAH244R protein is not efficiently recruited to NER preincision complexes, we next asked how this affects the association of the endonuclease ERCC1-XPF, considering the known role of XPA in bringing ERCC1-XPF to NER complexes (2, 24). While robust recruitment of endogenous ERCC1 was detected in wild-type primary fibroblasts (48BR), ERCC1 was not visible at sites of local damage marked by TFIIH in XP2LD (H244R) or XP1PD (C108F) primary fibroblasts at 20 min after local UV-C irradiation (Fig. 4 E and F and SI Appendix, Fig. S5C). Likewise, endogenous ERCC1 was clearly detectable at damaged sites in wild-type U2OS cells as well as XPA-KO cells rescued with GFP-XPAWT, while ERCC1 could not be observed at damaged sites in XPA-KO cells or in XPA-KO cells complemented with XPAH244R or XPAC108F (Fig. 4 G and H). The recruitment of TFIIH was not affected under these conditions (Fig. 4G and SI Appendix, Fig. S5D). These data indicate that the strongly reduced interaction between TFIIH and XPAH244R severely compromises the association of XPA with a preincision complex and the subsequent recruitment of ERCC1-XPF. Nonetheless, the mutant XPAH244R protein still supports ~50% levels of unscheduled DNA synthesis at sites of local UV damage in patient fibroblasts (Fig. 1H) and reconstituted XPA knockout cells in either U2OS (SI Appendix, Fig. S2 C and D) or RPE1 cells (Fig. 1 I and J). This suggests that even the transient association of mutant XPAH244R protein with NER complexes is sufficient to maintain residual levels of residual global genome NER, consistent with the absence of skin cancer in the patients described here.
Expression of XPAH244R Confers Sensitivity to Illudin S-Induced Transcription Stress.
While most UV-induced DNA lesions are removed by GG-NER, those on transcribed strands of genes can stall transcribing RNAPII enzymes and trigger TC-NER (31, 46). GG-NER and TC-NER are thought to converge in the DNA opening and damage verification steps initiated by the arrival of TFIIH (47). In GG-NER, TFIIH is recruited by association with XPC, while in TC-NER, TFIIH recruitment to DNA damage-stalled RNAPII is dependent on CSB, CSA, and UVSSA (7, 10). We have previously shown that XPA is not required for TFIIH to join TC-NER complexes (7), in line with the notion that XPA acts downstream of TFIIH in both GG-NER and TC-NER. However, it is currently not known whether the association of XPA with GG-NER and TC-NER complexes through TFIIH follows the same mechanism (16).
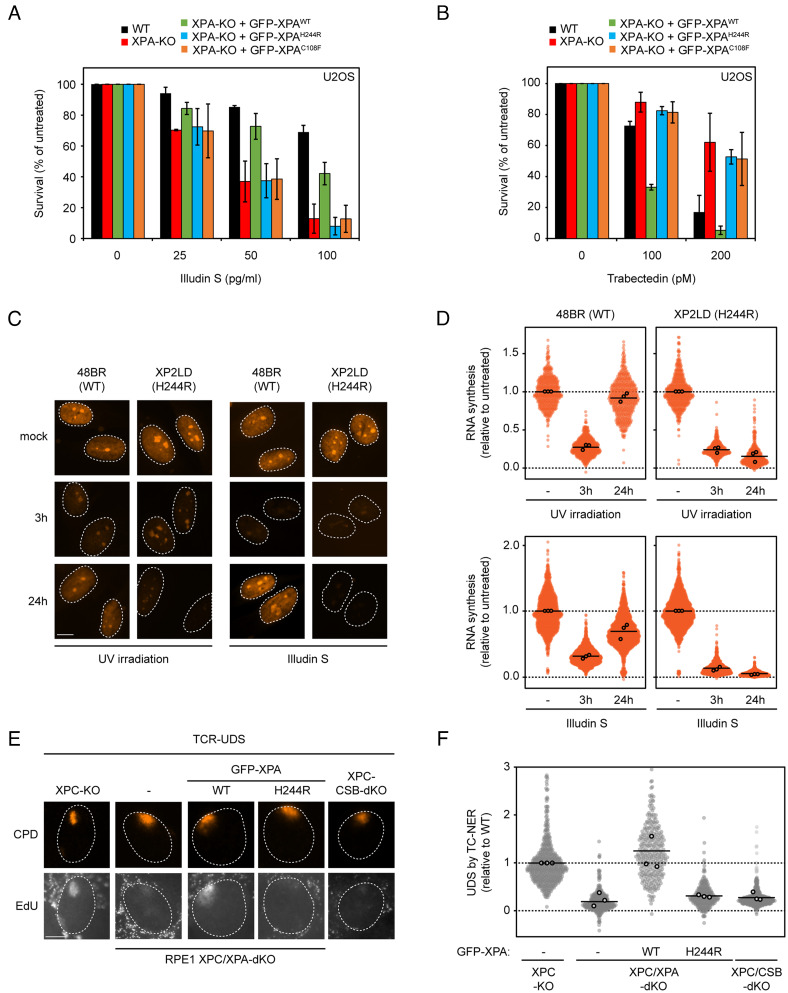
We therefore set out to investigate whether the XPAH244R allele affects TC-NER activity. We performed clonogenic survival assays with Illudin S, a compound that forms nondistorting DNA adducts that escape recognition by GG-NER and are exclusively repaired by TC-NER (8, 48). Sensitivity to Illudin S therefore specifically reflects a loss of TC-NER activity. XPA knockout U2OS cells displayed an approximately fivefold hypersensitivity to Illudin S compared to the parental cell lines, and reexpression of GFP-XPAWT in these knockout cells rescued Illudin S sensitivity to a large extent (Fig. 5A). By contrast, cells expressing either GFP-XPAH244R or GFP-XPAC108F showed a hypersensitivity that was indistinguishable from the XPA-KO cells (Fig. 5A). These findings show that XPAH244R is defective in TC-NER. Importantly, while XPAC108F expressing cells were fully deficient in both GG-NER and TC-NER, those expressing XPAH244R had a more pronounced TC-NER defect, suggesting that this XPA allele more dramatically affects TC-NER compared to GG-NER.
Fig. 5.
Patient or rescue cells expressing XPAH244R are defective in TC-NER. (A) Clonogenic survival assay after 72 h treatment with various doses of Illudin S in U2OS(FRT) WT, XPA-KO, and GFP-XPA reconstituted cell lines. The bars are the average of three to four experiments. The error bars reflect the SD. (B) Clonogenic survival assay after continuous treatment with various doses of Trabectedin in U2OS(FRT) WT, XPA-KO, and GFP-XPA reconstituted cell lines. The bars are the average of two to three experiments. The error bars reflect the SD. (C) Representative images of indicated primary fibroblasts pulse-labeled with 5-ethynyl-uridine (5-EU). Cells were either mock-treated, irradiated with 9 J/m2 UV-C, or exposed to 30 ng/mL Illudin S for 3 h. Cells were subsequently pulse-labelled with 5-EU pulse-labeled at the indicated timepoints. (D) Quantification of RNA synthesis from (C), relative to the mock condition per cell line. (E) Representative images and (F) quantification of EdU incorporation in the indicated RPE1 cells deficient in GG-NER (XPC-KO cells) to specifically measure UDS by TC-NER after 100 J/m2 UV-C irradiation through 5-µm pore membranes. Sites of local damage are identified by CPD staining. (D and F) All cells are depicted as individual data points with the bar representing the mean of all data points. The individual means of three to four biological replicates are depicted as points with black circles.
Cells Expressing XPAH244R Are Resistant to TC-NER-Induced Toxicity Caused by Trabectedin.
To further confirm the specificity of the TC-NER defect, we exposed cells to the trabectedin, a marine alkaloid isolated from colonial sea squirts and an approved anticancer drug (49). Trabectedin is highly toxic to cells and elicits toxic DNA breaks in a manner that depends on transcription and processing by TC-NER (50). Consistent with this fact, wild-type and GG-NER-deficient cells are highly sensitive to trabectedin, while cells deficient in TC-NER are resistant to this compound (51).
Clonogenic survival assays after treatment with increasing doses of trabectedin indeed revealed that wild-type cells did not form colonies at the highest dose (200 pM), while XPA-knockout cells were resistant and did form colonies under these conditions (Fig. 5B). Reexpression of GFP-XPAWT restored trabectedin sensitivity even beyond wild-type levels, while cells expressing either GFP-XPAH244R or GFP-XPAC108F showed a similar level of resistance to trabectedin as XPA-KO cells (Fig. 5B). These findings support the conclusion from our illudin S experiments that the XPAH244R specifically affects TC-NER, likely due to a weakened interaction between TFIIH and XPAH244R.
The XPAH244R Protein Fails to Support Transcription Recovery after UV Irradiation.
UV-induced DNA lesions in transcribed strands trigger the stalling of RNAPII leading to a strong transcriptional arrest. The restart of transcription is fully dependent on the activity of TC-NER. Therefore, the recovery of transcription is deficient in cells from individuals with the TC-NER-deficiency disorder Cockayne syndrome (52). To directly measure transcription restart after UV irradiation, we visualized nascent transcription by 5-ethynyl-uridine labeling following global UV irradiation in either reconstituted XPA knockout cells (SI Appendix, Fig. S6 A and B) or primary fibroblasts (Fig. 5 C and D) (53, 54). Nascent transcription was strongly reduced at 3 h after UV irradiation in XPA-KO cells and, as expected, did not recover within 24 h. While reexpression of XPAWT restored transcription recovery, expression of either XPAH244R or XPAC108F failed to do so, in line with their inability to support TC-NER (SI Appendix, Fig. S6 A and B). Similarly, wild-type primary fibroblasts recovered from the transcription arrest within 18 h after UV irradiation or Illudin S exposure, while XP2LD (H244R) patient cells failed to restore transcription (Fig. 5 C and D).
Cells Expressing XPAH244R Display Low Levels of Transcription-Coupled NER Activity.
To specifically measure unscheduled DNA synthesis (UDS) associated with TC-NER, we locally exposed serum-starved XPC-KO cells (RPE1-hTERT) with 100 J/m2 UV-C through 5-µm pore filters and incubated with EdU for 4 h (Fig. 5 E and F). The captured EdU incorporation was specific for TC-NER since the UDS signal was abolished by the additional knockout of either CSB or XPA in these cells (Fig. 5 E and F). We subsequently expressed either GFP-XPAWT or GFP-XPAH244R in the XPC/XPA-dKO cells and confirmed expression levels by western blot analysis (SI Appendix, Fig. S6C). This enabled us to specifically measure to what extent XPAH244R supports TC-NER in cells that are completely deficient in GG-NER. Reexpression of XPAWT fully restored EdU incorporation to the levels seen in XPC-KO cells, while XPAH244R showed limited residual TC-NER activity similar to XPC/XPA-dKO or XPC/CSB-dKO cells (Fig. 5 E and F).
Together, our data reveal that the XPAH244R mutant protein in XP2LD patient cells shows a severely weakened interaction with the TFIIH complex that affects TC-NER activity to a much greater degree than GG-NER.
Discussion
A New Case of Mild XP-A with Considerable Levels of Global Genome NER.
XPA is a central scaffold protein that interacts with NER repair proteins and DNA through distinct domains (16, 19, 20, 24, 26). XPA is thought to associate with the NER complex through interactions with TFIIH and help the lesion verification process by anchoring TFIIH at the NER bubble (16, 27). In addition, together with RPA, XPA orients the NER complex on the bubble and recruits the ERCC1-XPF endonuclease to the NER complex, licensing the 5′ incision reaction (2, 19, 21, 24). Inherited defects in XPA cause xeroderma pigmentosum (XP), which is characterized by severe clinical features including skin cancer susceptibility, UV sensitivity, and neurological abnormalities. However, the spectrum and severity of disease manifestations are strongly dependent on the residual activity of the mutant XPA protein.
In this study, we describe two new patients (XP2LD and XP3LD), currently in their late forties and early fifties, respectively, who express an XPAH244R mutant protein and display only moderate UV sensitivity, and did not develop skin cancer. The XPAH244R mutant protein displays a strongly reduced association with the TFIIH complex as shown by proteomics and coimmunoprecipitation experiments. Both endogenous and ectopically expressed mutant XPAH244R protein failed to stably associate with sites of UV damage and to facilitate the association of ERCC1-XPF with NER complexes.
Despite the reduced association with NER complexes, the XPAH244R allele maintained up to ~50% GG-NER activity when quantifying residual repair by measuring unscheduled DNA synthesis, UV survival, and in vitro NER assays. A previously described patient, XP8LO, who is compound heterozygous for the same XPAH244R allele and a splice mutation that is considered to be a null-allele, also presented with mild clinical symptoms and moderate sensitivity to UV light (33, 35). Similarly, we previously reported on ERCC1 patients with an ERCC1R156W allele that supported considerable NER levels despite seemingly abolishing the interaction between XPA and ERCC1 and a lack of detectable ERCC1-XPF association with NER complexes (44). Likewise, XPG deletion mutants with a weakened interaction with TFIIH that could not be detected at sites of local UV damage still supported considerable NER activity (55).
These findings suggest that even a transient interaction between mutant XPA and the NER complex that is not experimentally detectable can be sufficient for considerable residual NER activity and explain the relatively mild phenotype and absence of skin cancer development in these two XP-A patients.
The C-Terminus of XPA Mediates Its Association with TFIIH.
TFIIH consist of a core complex, containing XPB/p89, XPD/p80, p52, p44, p62, p34, and TTDA/p8 and a kinase module consisting of CDK7, Cyclin H, and MAT1. Crosslinking data suggests that XPA associates through its central domain primarily with XPB, while the protein also associates with XPD through an intercalating hairpin, located close to its Zn-finger that binds DNA (16). In this way, XPA clamps the XPB subunit to DNA, preventing its dissociation. Its C-terminus extends to and interacts primarily with TTDA/p8 and p52 (16). Even though XPA has contacts with several TFIIH subunits, our analysis of a single H244R amino acid substitution in the C-terminus of XPA reveals a severe reduction in association with TFIIH, likely by abolishing the interaction with the TTDA/p8 and p52 subunits. One possibility is that XPA initially associates with TFIIH through its C-terminus, after which the kinase CAK module dissociates, facilitating the engagement with DNA and the XPB and XPD subunits (16, 27). This observation is strengthened by the observation that a mutant XPA protein lacking its C-terminus failed to release CAK from the TFIIH core complex (27). Another possibility is that the binding of XPA is cooperative and depends on all interaction surfaces and that loss of any one of them would disrupt the interaction between these NER proteins. Our finding that a mutation in the zinc-finger (C108F), that is important for the folding of the XPA core domain and impairs NER, still allows residual recruitment of XPAC108F to sites of local damage suggests that the C-terminal interaction with TFIIH is sufficient for its association with NER complexes.
Selective Impact of the XPA Allele on NER Subpathways.
Despite residual GG-NER activity, both patient fibroblasts and reconstituted XPA knockout cells expressing XPAH244R show severely limited residual TC-NER activity. Cells with the XPAH244R allele showed strong sensitivity to illudin S, failed to recover transcription after UV, failed to support unscheduled DNA synthesis associated specifically with TC-NER, and had acquired resistance to trabectedin, all of which are hallmarks of TC-NER deficiency (8, 48, 51, 52). This is consistent with previous findings showing that TC-NER of the transcribed strand of the DHFR gene was reduced in XPAH244R-expressing cells (36).
Patients with defects in proteins essential for TC-NER, present with Cockayne syndrome (CS) or the milder UV sensitivity syndrome (UVSS), both of which are characterized by UV sensitivity, while only CS is characterized by severe neurological abnormalities. We have postulated before that CS is not caused by defective TC-NER per se but rather by a defect in the ability to process and degrade DNA damage-stalled RNAPII. Processing of RNAPII is impaired in cells defective in CSB and CSA (causing CS) but not in cells deficient in UVSSA (causing UVSS) (5, 10, 31, 46). Considering that the clinical features of the XP-A patients described here more closely resemble UV-sensitivity syndrome than the severe neurodegenerative CS, it is likely that the processing and degradation of RNAPII are fully intact in cells from these patients, like in cells from UVSS individuals.
How Do C-Terminal Mutations in XPA Cause a Defect in Transcription-Coupled Repair?
In GG-NER, the DDB2 lesion-recognition factor recognizes lesions in chromatin. XPC-RAD23B then binds to lesions that destabilize the DNA duplex and recruits the TFIIH complex to initiate the formation of a preincision complex. DDB2 may still be bound to the lesion at that time, and the arrival of TFIIH stimulates its dissociation from the lesion (56). The initiation of TC-NER involves the sequential association of CSB, CSA, and UVSSA with the DNA damage-stalled RNAPII. (7, 8, 31). Once positioned, UVSSA mediates the association of the TFIIH complex through interaction with its p62 subunit (47). In TC-NER, the recruitment of TFIIH and the formation of a preincision complex and subsequent DNA repair also requires the ELOF1-stimulated ubiquitylation of RNAPII at lysine 1268 and likely requires the removal of stalled RNAPII blocking access to the lesion buried within its active site (7, 8, 31).
While the p62 subunit of TFIIH has a key role for GG-NER (by interacting with XPC) and TC-NER (by interacting with UVSSA), the pathway for association of TFIIH and XPA must differ in other ways for the two pathways. These differences may explain the differential impact of the XPAH244R allele on the two NER subpathways. While the mutant XPAH244R protein has a severely weakened interaction with TFIIH, it is possible that during GG-NER, the association of mutant XPA with NER complexes is stabilized by the reported interactions with DDB2 (57) or XPC-RAD23B-CEN2 (58, 59), which are missing in TC-NER. These additional interactions may still allow for the assembly of a functional NER complex—albeit below our detection limit—that is proficient in ERCC1-XPF binding, dual incision, and residual repair activity in GG-NER.
In TC-NER by contrast, the presence of the stalled RNAPII might further complicate proper TFIIH recruitment and NER complex assembly. It is therefore possible that the initial association of XPA with TC-NER complexes is dependent on the full set of protein–protein interactions between XPA and TFIIH. In line with this notion, our pull-down experiments with RNAPII revealed detectable interactions with CSB, CSA, UVSSA, and TFIIH subunits after UV irradiation, while none of the downstream NER proteins XPA, XPG, and ERCC1-XPF could not be detected (7). This may indicate that XPA does not associate with the complex containing RNAPII and possibly other TC-NER-specific proteins but rather associates with TFIIH after RNAPII has been removed or degraded (31). The elucidation of how XPA contributes to TC-NER in conjunction with TFIIH binding will require the development of new approaches to monitor the transition from lesion recognition to dual incision.
Materials and Methods
Cell Lines.
All cell lines are listed in SI Appendix. Human U2OS-Flp-In/T-Rex, RPE1-hTERT, and primary fibroblasts were cultured at 37 °C in 5% CO2 in DMEM, supplemented with antibiotics, 10% fetal calf serum (FCS), and glutaMAX (Gibco).
Plasmids.
The pU6-gRNA:PGK-puro-2A-tagBFP plasmid containing a sgRNA against exon 2 of XPA (sequence: 5-GCCCCAAAGATAATTGACACAGG-3) was obtained from the Sigma-Aldrich library. Overlap PCR was used to generate GFP-XPA with either amino acid substitution H244R (GFP-XPAH244R) or C108F (GFP-XPAC108F) and cloned in pcDNA5/FRT/TO-puro (for U2OS cells) or pPGK-EGFP-IRES-PURO (for RPE1-hTERT cells). Further details are provided in SI Appendix.
Generation of TC-NER Knockout Cells.
Knockouts in U2OS-Flp-In/T-Rex or RPE1-hTERT were generated as described previously (7, 8). Stable integration of GFP-tagged proteins by Flp-In or random integration was described previously (7, 8). Details are provided in SI Appendix.
Immunoprecipitation for Co-IP.
Cell pellets were lysed in EBC-150 buffer I [50 mM Tris, pH 7.5, 150 mM NaCl, 0.5% NP-40, 2 mM MgCl2 supplemented with protease and phosphatase inhibitor cocktails (Roche)] and 500 U/mL Benzonase® Nuclease HC (Novagen) for 1 h at 4 °C under rotation. The lysates were cleared from insoluble chromatin and were subjected to immunoprecipitation with GFP-Trap beads (Chromotek) for 1.5 h at 4 °C. The beads were then washed 4 to 6 times with EBC-150 buffer II (50 mM Tris, pH 7.5, 150 mM NaCl, 0.5% NP-40, 1 mM EDTA) and boiled in Laemmli sample buffer. Bound proteins were resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
Western Blotting.
Cell extracts were generated by cell lysis and boiled in Laemmli sample buffer. Proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on 4 to 12% bis-tris Criterion Xt precast gels (Biorad) in NuPAGE™ MOPS SDS Running Buffer (Thermo Fisher) and transferred to 0.45-µm PVDF membranes (Immobilon). Protein expression was analyzed by immunoblotting with the indicated antibodies in Rockland buffer. Details are provided in SI Appendix.
Clonogenic Survival Assays.
Cells were plated in low density in culture dishes and allowed to attach. Following treatment, the cells were allowed to form clones for 7 to 10 d. To visualize clones, cells were subjected to methylene blue fixation and staining. Details are provided in SI Appendix.
Unscheduled DNA Synthesis.
UDS at site of local damage was measured as described in refs. 8 and 44. Details are provided in SI Appendix.
Quantification of ERCC1 and GFP-XPA Levels at Local Damage.
Cells were locally UV-irradiated and stained for XPA and ERCC1 as described previously (44). Details are provided in SI Appendix.
Nascent Transcript Level Measurements.
Transcription recovery after UV irradiation was measured by 5-ethynyl-uridine (EU) pulse labeling as described (7, 8, 54). Details are provided in SI Appendix.
UV-C Laser Microirradiation.
Laser tracks were generated using a diode-pumped solid-state 266-nm Yttrium Aluminum Garnet laser (average power 5 mW, repetition rate up to 10 kHz, pulse length 1 ns) as described before (44). Images were acquired in Zeiss ZEN and quantified in Image J. Details are provided in SI Appendix.
Generation of Mass Spectrometry Samples.
Individual mass spectrometry samples were subjected to coimmunoprecipitation using GFP-Trap beads as described above. After pulldown, the beads were washed two times with EBC-150 buffer II and two times with 50 mM NH4HCO3 followed by overnight digestion using 2.5 µg trypsin at 37 °C under constant shaking. All samples were desalted using a Sep-Pak tC18 cartridge by washing with 0.1% formic acid and concentrated on STAGE-tips as described previously (60). Details are provided in SI Appendix.
Mass Spectrometry Data Acquisition and Analysis.
Mass spectrometry was performed essentially as previously described (61). Raw mass spectrometry data were further analyzed in MaxQuant v 1.6.7.0 according to (62) using standard settings with modifications. Details are provided in SI Appendix.
Expression and Purification of XPAWT and XPAH244R.
Wild-type or mutant full-length XPA with an N-terminal His6 tag was expressed in Escherichia coli Rosetta pLysS cells as described in ref. 21. Details are provided in SI Appendix.
Electrophoretic Mobility Shift Assay (EMSA).
EMSA was conducted using three-way junction substrate as described in (19). The annealed three-way junction oligonucleotides (100 nM) with fluorescein-label were incubated with wild-type or mutant XPA (0, 20, 40, 60, and 80 nM) in a 10-µL mixture containing 25 mM Tris-HCl (pH 8.0), 1 mM DTT, 0.1 mg/mL BSA, 5% glycerol, and 1 mM EDTA at 25 °C for 30 min. The reaction mixture was loaded onto 8% native polyacrylamide gel and run at 4 °C for 2 h at 20 mA with 0.5% TBE buffer. Gels were scanned using an Amersham Typhoon RGB imager.
In Vitro NER Activity Assay with Purified Proteins.
A plasmid containing a site-specific acetylaminofluorene (AAF) lesion was incubated with purified NER protein as previously described: XPC-RAD23B (63), TFIIH (64), RPA (65), XPG (66), ERCC1-XPF (67). Details are provided in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
We acknowledge both siblings for contributing to this paper. We would also like thank Minyong Kee (Ulsan National Institute of Science and Technology, Republic of Korea) for providing dG-AAF-containing oligonucleotides, Chi-Lin Tsai (MD Anderson, United States) for providing purified TFIIH protein, Walter Chazin (Vanderbilt University, United States) for purified RPA protein and Miaw-Sheue Tsai (Lawrence Berkeley National Laboratory, United States) for help with expressing XPC-RAD23B, ERCC1-XPF, and XPG, and Walter Chazin for discussion of the properties of the H244R protein. This work was funded by a Leiden University Medical Center Research Fellowship, a VIDI grant (ALW.016.161.320) from the Dutch research council (Nederlandse Organisatie voor Wetenschappelijk Onderzoek) and an ERC (European research council) consolidator grant (101043815) to M.S.L., an ERC (European research council) starting grant (310913) to A.C.O.V., a Young Investigator Grant (11367) from the Dutch Cancer society (Koningin Wilhelmina fonds) and an EMERGIA 2020 program grant (EMERGIA20_00276) from the Andalusian Government (Junta de Andalucía) to R.G.-P., and by grants from the Korean Institute for Basic Science (IBS-R022-A1 to O.D.S.), and the National Cancer Institute (P01CA092584 and R01CA218315 to O.D.S.).
Author contributions
D.v.d.H., O.D.S., and M.S.L. designed research; D.v.d.H., M.K., A.P.W., P.J.v.d.M., M.W., F.L., H.-S.K., F.K., K.A., A.K., R.G.-P., J.-E.Y., B.-G.K., and M.S.L. performed research; D.v.d.H., M.K., F.K., A.C.O.V., R.v.D., O.D.S., and M.S.L. contributed new reagents/analytic tools; D.v.d.H., M.K., A.P.W., P.J.v.d.M., M.W., F.L., H.-S.K., F.K., K.A., A.K., R.G.-P., J.-E.Y., B.-G.K., R.v.D., O.D.S., and M.S.L. analyzed data; and D.v.d.H., R.v.D., O.D.S., and M.S.L. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. R.D.W. is a guest editor invited by the Editorial Board.
Contributor Information
Orlando D. Schärer, Email: orlando.scharer@gmail.com.
Martijn S. Luijsterburg, Email: m.luijsterburg@lumc.nl.
Data, Materials, and Software Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (68) partner repository with the dataset identifier PXD033520. All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Sugasawa K., et al. , UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121, 387–400 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Volker M., et al. , Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8, 213–224 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Sugasawa K., et al. , Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell 2, 223–232 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Mellon I., Spivak G., Hanawalt P. C., Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51, 241–249 (1987). [DOI] [PubMed] [Google Scholar]
- 5.Nakazawa Y., et al. , Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nat. Genet. 44, 586–592 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Schwertman P., et al. , UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat. Genet. 44, 598–602 (2012). [DOI] [PubMed] [Google Scholar]
- 7.van der Weegen Y., et al. , The cooperative action of CSB, CSA, and UVSSA target TFIIH to DNA damage-stalled RNA polymerase II. Nat. Commun. 11, 2104 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Weegen Y., et al. , ELOF1 is a transcription-coupled DNA repair factor that directs RNA polymerase II ubiquitylation. Nat. Cell Biol. 23, 595–607 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geijer M. E., et al. , Elongation factor ELOF1 drives transcription-coupled repair and prevents genome instability. Nat. Cell Biol. 23, 608–619 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakazawa Y., et al. , Ubiquitination of DNA damage-stalled RNAPII promotes transcription-coupled repair. Cell 180, 1228–1244.e1224 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Missura M., et al. , Double-check probing of DNA bending and unwinding by XPA-RPA: An architectural function in DNA repair. EMBO J. 20, 3554–3564 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans E., Moggs J. G., Hwang J. R., Egly J. M., Wood R. D., Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 16, 6559–6573 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riedl T., Hanaoka F., Egly J. M., The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 22, 5293–5303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park C. H., Mu D., Reardon J. T., Sancar A., The general transcription-repair factor TFIIH is recruited to the excision repair complex by the XPA protein independent of the TFIIE transcription factor. J. Biol. Chem. 270, 4896–4902 (1995). [DOI] [PubMed] [Google Scholar]
- 15.Nocentini S., Coin F., Saijo M., Tanaka K., Egly J. M., DNA damage recognition by XPA protein promotes efficient recruitment of transcription factor II H. J. Biol. Chem. 272, 22991–22994 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Kokic G., et al. , Structural basis of TFIIH activation for nucleotide excision repair. Nat. Commun. 10, 2885 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L., Lu X., Peterson C. A., Legerski R. J., An interaction between the DNA repair factor XPA and replication protein A appears essential for nucleotide excision repair. Mol. Cell Biol. 15, 5396–5402 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuda T., et al. , DNA repair protein XPA binds replication protein A (RPA). J. Biol. Chem. 270, 4152–4157 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Topolska-Wos A. M., et al. , A key interaction with RPA orients XPA in NER complexes. Nucleic Acids Res. 48, 2173–2188 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mer G., et al. , Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell 103, 449–456 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Kim M., et al. , Two interaction surfaces between XPA and RPA organize the preincision complex in nucleotide excision repair. Proc. Natl. Acad. Sci. U.S.A. 119, e2207408119. (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L., Elledge S. J., Peterson C. A., Bales E. S., Legerski R. J., Specific association between the human DNA repair proteins XPA and ERCC1. Proc. Natl. Acad. Sci. U.S.A. 91, 5012–5016 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L., Peterson C. A., Lu X., Legerski R. J., Mutations in XPA that prevent association with ERCC1 are defective in nucleotide excision repair. Mol. Cell Biol. 15, 1993–1998 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsodikov O. V., et al. , Structural basis for the recruitment of ERCC1-XPF to nucleotide excision repair complexes by XPA. EMBO J. 26, 4768–4776 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka K., et al. , Analysis of a human DNA excision repair gene involved in group A xeroderma pigmentosum and containing a zinc-finger domain. Nature 348, 73–76 (1990). [DOI] [PubMed] [Google Scholar]
- 26.Kuraoka I., et al. , Identification of a damaged-DNA binding domain of the XPA protein. Mutat. Res. 362, 87–95 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Coin F., et al. , Nucleotide excision repair driven by the dissociation of CAK from TFIIH. Mol. Cell 31, 9–20 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Staresincic L., et al. , Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 28, 1111–1120 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogi T., et al. , Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell 37, 714–727 (2010). [DOI] [PubMed] [Google Scholar]
- 30.van Toorn M., et al. , Active DNA damage eviction by HLTF stimulates nucleotide excision repair. Mol. Cell 82, 1343–1358.e8 (2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van den Heuvel D., van der Weegen Y., Boer D. E. C., Ogi T., Luijsterburg M. S., Transcription-coupled DNA repair: From mechanism to human disorder. Trends Cell Biol. 31, 359–371 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Kraemer K. H., Lee M. M., Scotto J., Xeroderma pigmentosum. Cutaneous, ocular, and neurologic abnormalities in 830 published cases. Arch. Dermatol. 123, 241–250 (1987). [DOI] [PubMed] [Google Scholar]
- 33.Satokata I., Tanaka K., Yuba S., Okada Y., Identification of splicing mutations of the last nucleotides of exons, a nonsense mutation, and a missense mutation of the XPAC gene as causes of group A xeroderma pigmentosum. Mutat. Res. 273, 203–212 (1992). [DOI] [PubMed] [Google Scholar]
- 34.Satokata I., Tanaka K., Okada Y., Molecular basis of group A xeroderma pigmentosum: A missense mutation and two deletions located in a zinc finger consensus sequence of the XPAC gene. Hum. Genet. 88, 603–607 (1992). [DOI] [PubMed] [Google Scholar]
- 35.Weerd-Kastelein E. A., Keijzer W., Sabour M., Parrington J. M., Bootsma D., A xeroderma pigmentosum patient having a high residual activity of unscheduled DNA synthesis after UV is assigned to complementation group A. Mutat. Res. 37, 307–312 (1976). [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi T., et al. , Mutational analysis of a function of xeroderma pigmentosum group A (XPA) protein in strand-specific DNA repair. Nucleic Acids Res. 26, 4662–4668 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishigori C., et al. , High prevalence of the point mutation in exon 6 of the xeroderma pigmentosum group A-complementing (XPAC) gene in xeroderma pigmentosum group A patients in Tunisia. Am. J. Hum. Genet. 53, 1001–1006 (1993). [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi Y., et al. , XPA gene mutations resulting in subtle truncation of protein in xeroderma pigmentosum group A patients with mild skin symptoms. J. Invest. Dermatol. 130, 2481–2488 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Satokata I., et al. , Three nonsense mutations responsible for group A xeroderma pigmentosum. Mutat Res. 273, 193–202 (1992). [DOI] [PubMed] [Google Scholar]
- 40.Sugitani N., Sivley R. M., Perry K. E., Capra J. A., Chazin W. J., XPA: A key scaffold for human nucleotide excision repair. DNA Repair 44, 123–135 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugitani N., Shell S. M., Soss S. E., Chazin W. J., Redefining the DNA-binding domain of human XPA. J. Am. Chem. Soc. 136, 10830–10833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Limsirichaikul S., et al. , A rapid non-radioactive technique for measurement of repair synthesis in primary human fibroblasts by incorporation of ethynyl deoxyuridine (EdU). Nucleic Acids Res. 37, e31 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le May N., et al. , NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol. Cell 38, 54–66 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Apelt K., et al. , ERCC1 mutations impede DNA damage repair and cause liver and kidney dysfunction in patients. J. Exp. Med. 218, e20200622 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Apelt K., et al. , Human HMGN1 and HMGN2 are not required for transcription-coupled DNA repair. Sci. Rep. 10, 4332 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jia N., et al. , Dealing with transcription-blocking DNA damage: Repair mechanisms, RNA polymerase II processing and human disorders. DNA Repair 106, 103192 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Okuda M., Nakazawa Y., Guo C., Ogi T., Nishimura Y., Common TFIIH recruitment mechanism in global genome and transcription-coupled repair subpathways. Nucleic Acids Res. 45, 13043–13055 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaspers N. G., et al. , Anti-tumour compounds illudin S and Irofulven induce DNA lesions ignored by global repair and exclusively processed by transcription- and replication-coupled repair pathways. DNA Repair 1, 1027–1038 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Erba E., et al. , Ecteinascidin-743 (ET-743), a natural marine compound, with a unique mechanism of action. Eur. J. Cancer 37, 97–105 (2001). [DOI] [PubMed] [Google Scholar]
- 50.Guirouilh-Barbat J., Redon C., Pommier Y., Transcription-coupled DNA double-strand breaks are mediated via the nucleotide excision repair and the Mre11-Rad50-Nbs1 complex. Mol. Biol. Cell 19, 3969–3981 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takebayashi Y., et al. , Antiproliferative activity of ecteinascidin 743 is dependent upon transcription-coupled nucleotide-excision repair. Nat. Med. 7, 961–966 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Mayne L. V., Lehmann A. R., Failure of RNA synthesis to recover after UV irradiation: An early defect in cells from individuals with Cockayne’s syndrome and xeroderma pigmentosum. Cancer Res. 42, 1473–1478 (1982). [PubMed] [Google Scholar]
- 53.Nakazawa Y., Yamashita S., Lehmann A. R., Ogi T., A semi-automated non-radioactive system for measuring recovery of RNA synthesis and unscheduled DNA synthesis using ethynyluracil derivatives. DNA Repair 9, 506–516 (2010). [DOI] [PubMed] [Google Scholar]
- 54.van den Heuvel D., et al. , A CSB-PAF1C axis restores processive transcription elongation after DNA damage repair. Nat. Commun. 12, 1342 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dunand-Sauthier I., et al. , The spacer region of XPG mediates recruitment to nucleotide excision repair complexes and determines substrate specificity. J. Biol. Chem. 280, 7030–7037 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Ribeiro-Silva C., et al. , Ubiquitin and TFIIH-stimulated DDB2 dissociation drives DNA damage handover in nucleotide excision repair. Nat. Commun. 11, 4868 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wakasugi M., et al. , Physical and functional interaction between DDB and XPA in nucleotide excision repair. Nucleic Acids Res. 37, 516–525 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.You J. S., Wang M., Lee S. H., Biochemical analysis of the damage recognition process in nucleotide excision repair. J. Biol. Chem. 278, 7476–7485 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Bunick C. G., Miller M. R., Fuller B. E., Fanning E., Chazin W. J., Biochemical and structural domain analysis of xeroderma pigmentosum complementation group C protein. Biochemistry 45, 14965–14979 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rappsilber J., Mann M., Ishihama Y., Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Kumar R., Gonzalez-Prieto R., Xiao Z., Verlaan-de Vries M., Vertegaal A. C. O., The STUbL RNF4 regulates protein group SUMOylation by targeting the SUMO conjugation machinery. Nat. Commun. 8, 1809 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tyanova S., Temu T., Cox J., The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Cheon N. Y., Kim H. S., Yeo J. E., Scharer O. D., Lee J. Y., Single-molecule visualization reveals the damage search mechanism for the human NER protein XPC-RAD23B. Nucleic Acids Res. 47, 8337–8347 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gradia S. D., et al. , MacroBac: New technologies for robust and efficient large-scale production of recombinant multiprotein complexes. Methods Enzymol. 592, 1–26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brosey C. A., et al. , NMR analysis of the architecture and functional remodeling of a modular multidomain protein, RPA. J. Am. Chem. Soc. 131, 6346–6347 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hohl M., Thorel F., Clarkson S. G., Scharer O. D., Structural determinants for substrate binding and catalysis by the structure-specific endonuclease XPG. J. Biol. Chem. 278, 19500–19508 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Enzlin J. H., Scharer O. D., The active site of the DNA repair endonuclease XPF-ERCC1 forms a highly conserved nuclease motif. EMBO J. 21, 2045–2053 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perez-Riverol Y., et al. , The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (68) partner repository with the dataset identifier PXD033520. All study data are included in the article and/or SI Appendix.