Significance
Photoacclimation in photosynthetic organisms remains poorly understood. Here the diatom Phaeodactylum tricornutum was used to study long-term responses to low and high growth irradiances. Extreme light conditions lead to suboptimal photon absorption in the plastid, which triggers feedback responses that alter the expression of specific nuclear genes. Surprisingly, multiple essential photoacclimation genes that have been identified in land plants and green algae are absent in diatoms. It is therefore likely that these photoautotrophs, which have a complex evolutionary origin, have evolved distinct mechanistic responses to light variations in the marine environment. Our results suggest that the nuclear genes identified in this study play key roles in feedback signaling between the nucleus and the plastid in diatoms leading to photoacclimation
Keywords: chlorophyll, diatom, photoacclimation, transcriptome, photosynthesis
Abstract
Unlike most higher plants, unicellular algae can acclimate to changes in irradiance on time scales of hours to a few days. The process involves an enigmatic signaling pathway originating in the plastid that leads to coordinated changes in plastid and nuclear gene expression. To deepen our understanding of this process, we conducted functional studies to examine how the model diatom, Phaeodactylum tricornutum, acclimates to low light and sought to identify the molecules responsible for the phenomenon. We show that two transformants with altered expression of two putative signal transduction molecules, a light-specific soluble kinase and a plastid transmembrane protein, that appears to be regulated by a long noncoding natural antisense transcript, arising from the opposite strand, are physiologically incapable of photoacclimation. Based on these results, we propose a working model of the retrograde feedback in the signaling and regulation of photoacclimation in a marine diatom.
Although higher plants have retrograde and anterograde signaling during development and in physiological responses to the environment (1), how the signals are transferred from the plastid to the nucleus and vice versa remain poorly understood. To elucidate these signals, unicellular eukaryotic algae have been widely used as model organisms (e.g., ref. 2). To optimize photosynthetic rates, eukaryotic algae photoacclimate to changes in irradiance on time scales of hours to a few days (3). In contrast to higher plants, where light-harvesting pigment accumulation is tissue specific and dependent on developmental processes, we focus on a marine diatom where, in response to low light, photoacclimation is manifested by increases in cellular chlorophyll concentration.
The optimal photosynthetic rate in any photoautotroph is obtained only when light absorption is equal to its utilization (4). An imbalance occurs when the rate at which the energy absorbed by the reaction centers and the rate of electron flow exceeds the capacity of the metabolic electron sinks (5). Because unicellular planktonic algae experience continuous variations in spectral irradiance due to physical turbulence, the passage of clouds across the sky, as well as phytoplankton population density in the euphotic zone (3, 6), they have evolved a series of mechanisms that lead to changes in the absorption of light on time scales from seconds to a few days. These include state transitions (7, 8) and nonphotochemical quenching (NPQ) (9–11), which are signaled by the transthylakoid proton gradient and occur on time scales of seconds to minutes, leading to changes in the effective cross-section of photosystem II (12). On time scales of hours to days however, a third process, photoacclimation, leads to changes in the abundance of nuclear-encoded light-harvesting pigment–protein complexes and/or plastid-encoded reaction center proteins (13). Changes in the expression of light-harvesting pigment–protein complexes are also associated with changes in the redox state of the plastoquinone pool (2, 14), however, the signal transduction pathway facilitating this mode of acclimation remains enigmatic.
Using nuclear transcriptome data, we examined the photophysiological changes occurring under low light in a model diatom, Phaeodactylum tricornutum. To identify the molecules involved in the signal transduction pathway leading to photoacclimation, we selected the top six up-regulated genes to target for RNA interference (RNAi) by an antisense approach and studied the resulting phenotypes of the transformed lines. Our results strongly suggest that two genes, as well as a putative regulatory long noncoding natural antisense transcript (NAT), are involved in the nuclear–plastid signal transduction pathway of photoacclimation in this organism.
Results
Low- and High-Light-Acclimated Cells Show Distinct Phenotypes.
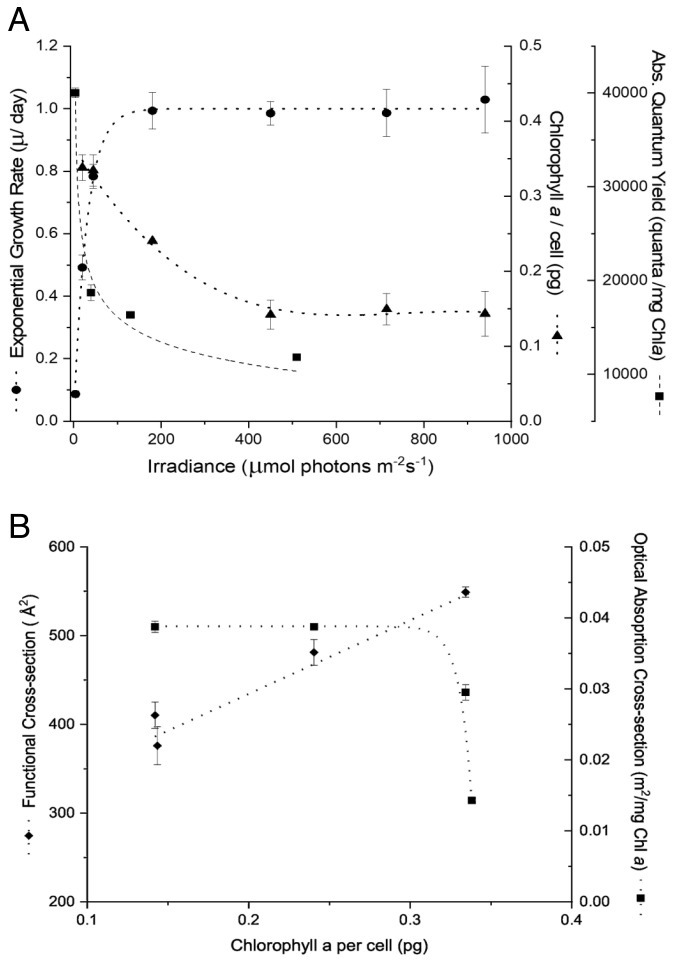
The growth rate of wild-type (WT) P. tricornutum was a hyperbolic function of light intensity (Fig. 1A). In the steady state, the exponential growth rate (μ) was accompanied by a reciprocal change in pigment content (e.g., Chl a/cell; Fig. 1A). The low light (LL)/high light (HL) Chl a ratio for this diatom was 2 to 2.5. In turn, the increased pigment content in cells fully adapted to LL led to self-shading within the plastids. This phenomenon was reflected in the decrease in the in vivo optical absorption cross-section per unit Chl, denoted a* (Fig. 1B). Thus, when the cells adapted to low light, the ensemble of pigments became less efficient in light absorption. However, the functional absorption cross-section of PSII (σPSII) increased with adaptation to low light (Fig. 1B). The ratio of σPSII to the optical absorption cross-section (σPSII /a*) is directly proportional to the absolute quantum yield of photosynthesis (12, 15). The results of this analysis show that in the WT diatom, the highest quantum yields were in cells with the highest chlorophyll content and reached a minimum as pigment content declined with increased growth irradiance (Fig. 1A).
Fig. 1.
(A). Effect of increasing incident light intensity on exponential growth rate (n = at least 12; Error bars represent ± SD); Chlorophyll a accumulation (n = 3; Error bars represent ± SD); and the absolute quantum yield of photosynthesis (σPSII/a*) (n = 3; Error bars represent ± SD) in wild-type P. tricornutum (B). Effect of change in concentration of Chlorophyll a per cell on the functional cross-section of PSII (σPSII) (n = 3; Error bars represent ± SD) and the optical cross-section or a* of wild-type P. tricornutum (n = 3; Error bars represent ± SD)
Nuclear Genes Showing Strong Differential Expression in Low- and High-Light-Acclimated Cells.
Light-harvesting complex genes.
In diatoms, the nuclear-encoded light-harvesting complex proteins (LHCs) are distinguished into three distinct clades: F, R and X (16). Based on the heat-map of our RNA-seq results (SI Appendix, Table S1), a greater number of the transcripts of these genes are up-regulated. Out of the 41 annotated lhc genes, 19 were significantly up-regulated under LL growth conditions, a majority of which are of the F-type (SI Appendix, Table S1). The gene transcript showing the most dynamic change in expression, however, is the nonphotochemical quenching (NPQ) gene lhcX1, which was up-regulated 32-fold. This was followed by lhcF15 and lhcF5 gene expression, both of which show approximately 20-fold change. Only four of the lhc genes were down-regulated more than twofold when comparing LL to HL, most of which are of the R-type.
Photosynthetic electron transport component genes.
There was an increase in transcript accumulation of all three nuclear-encoded PSII extrinsic subunit genes: psbU, psbQ/oee3, and psbO (SI Appendix, Table S2). This was accompanied by a twofold to 2.5-fold upregulation in PSII reaction center genes, specifically hcf136 and psb27, both of which encode proteins critical for PSII assembly and repair. Additionally, the gene coding for the mobile electron carrier, c553, which shuttles electrons between cytochrome b6/f and PSI in diatoms was up-regulated by 2.2-fold.
Chlorophyll biosynthesis genes.
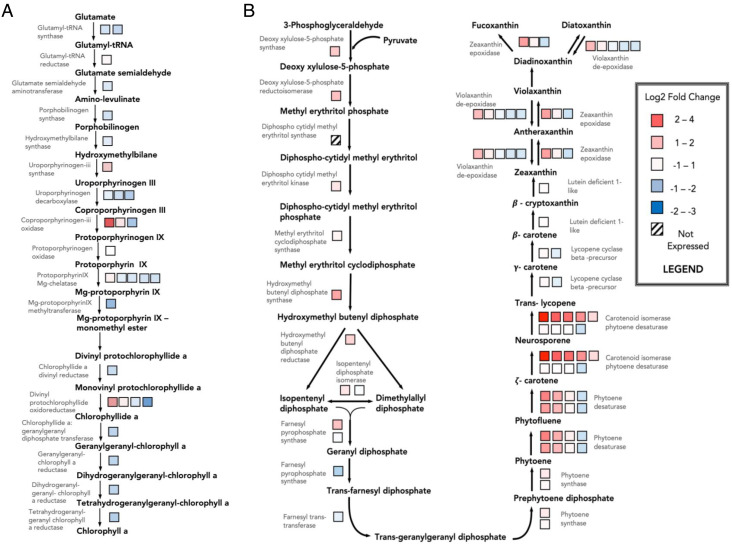
Almost all the enzymes catalyzing the chlorophyll a biosynthetic pathway are nuclear encoded. However, the actual synthesis of the pigment occurs in the plastid stroma, starting with the amino acid glutamate. Surprisingly, overlaying the transcript abundances for these genes from P. tricornutum on a schematic of the pathway, suggests that this process is down-regulated in steady-state LL growth conditions compared with HL grown cells (Fig. 2A). With the exception of one of the multigene families coding for coproporphyrinogen oxidase, and divinyl protochlorophyllide oxidoreductase, as well as the gene for uroporphyrinogen III synthase, the expression of all the other genes in this pathway generally decreased.
Fig. 2.
Heat map of transcript expression of wild-type P. tricornutum in LL as compared with HL of (A) Chlorophyll biosynthesis and (B) Carotenoid biosynthesis. The boxes represent the differential gene expression of multiple forms of the same enzyme or isozyme
Carotenoid biosynthesis genes.
The transcript levels of genes encoding for enzymes catalyzing the synthesis of carotenoids were up-regulated under LL growth conditions (Fig. 2B). The most strongly differentially expressed genes were multiple copies of crtiso_l, which encode for carotenoid isomerase-like phytoene dehydrogenases, and were up-regulated fivefold to 15-fold. On the other hand, crtiso2 was down-regulated ~threefold under LL. Another multigene family that was significantly up-regulated was the phytoene desaturase family, which precede CrtISOs in this pathway. The varied expression levels of the isozymes in these multigene families suggests that the individual genes are likely controlled by different regulatory factors, each likely responding to different stimuli to varying extents. It has recently been shown that zeaxanthin epoxidase-like (ZEP1) and violaxanthin deepoxidase-like enzymes (VDL1-2) (17) are involved in the synthesis of the light-harvesting carotenoid, fucoxanthin. Other ZEPs presumably epoxidize zeaxanthin to antheraxanthin and violaxanthin, as well as diatoxanthin (Dtx) to diadinoxanthin (Ddx). The reverse reactions catalyzed by vioxanthin deepoxidases (VDE) appear to have modified transcript levels to a similar extent, which suggests a form of coregulation. Based on the transcript profiles alone, more carotenoids (e.g., fucoxanthin) are synthesized under LL growth conditions.
Transcript Abundances of Low- and High-Light-Acclimated P. tricornutum Suggest the Involvement of Genes in Retrograde Signaling.
A comparison of transcriptomes of cells acclimated to LL versus HL showed an altered expression of 3,235 genes (i.e., 26% of the entire coding region of the genome). Of these, 1,749 were up-regulated and 1,486 down-regulated (log2fold change ≥ 2; NCBI GEO Accession # GSE133301). Of the eleven genes that showed a positive change of more than 30-fold, nine had uncharacterized open reading frames (ORFs). In silico analysis of the top six of these genes revealed the presence of potentially conserved functional domains and cellular location, as well as their hydrophobicity profiles (Table 1). Hypothetical roles of these six positively, differentially expressed genes could suggest their function in signal transduction. Each of these genes was targeted individually by an antisense RNAi strategy to examine their potential role in photoacclimation. Of these, the transformed strains with modifications to Phatr3_J43123 and Phatr3_J50052 expression showed the most dynamic phenotypes of photoacclimation disruption.
Table 1.
List of genes with the most significant difference in expression levels; i.e. those that showed more than 30-fold up-regulated in low-light-acclimated cells as compared with high light
| Gene Number (Phatr3) | Log2 Fold-change LL vs. HL | Fold-change in LL vs. HL | P-adj value (FDR) | NCBI BLAST Hit | NCBI Conserved Protein Domains | Signal Peptide (predicted) | Transmembrane (predicted) | Anchored to Membrane (predicted) | Targeted to Chloroplast (predicted) |
|---|---|---|---|---|---|---|---|---|---|
| EG01529 | 9.76 | 870 | 1.58 e-146 | Predicted Protein | - | + | + | - | inconclusive |
| J47715 | 7.67 | 203 | 2.34 e-146 | Predicted Protein | - | - | - | inconclusive | - |
| J43123 | 7.36 | 164 | 1.43 e-174 | Predicted Protein | - | + | + | - | + |
| EG00707 | 6.46 | 88 | 7.66 e-188 | Predicted Protein | - | + | + | - | + |
| J37655 | 6.45 | 87 | 7.43 e-113 | Predicted Protein | - | + | + | inconclusive | + |
| J50052 | 5.39 | 42 | 2.04 e-127 | Predicted Protein | Kinase (ABC1 family) | + | - | - | + |
All these genes were targeted for RNAi individually. Phatr3 – gene identification numbers from third annotation of genome (protists.ensembl.org); LL – low-light intensity of 20-μmol photons m−2s−1; HL – high-light intensity of 940-μmol photons m−2s−1; P-adj value (FDR) – P value of transcriptome results adjusted to false discovery rate <0.05; NCBI - The National Center for Biotechnology Information database; BLAST Hit – Results from The Basic Local Alignment Search Tool of NCBI; (+) indicates presence; (−) indicates absence. (n = 3; P < 0.05)
Phenotypic Characterization of Relevant Transformant Strains Shows Disruption in the Photoacclimation Response.
In silico sequence analysis identified Phatr3_J50052 as a nuclear-encoded hydrophilic kinase found only in unicellular algae and belonging to the AarF/ABC1/UbiB superfamily without any DNA-binding motifs (Table 1). Of the 70 resultant RNAi transformant lines, 67 strains showed more than a 25% increase/decrease in chlorophyll accumulation in the cell, with an average LL/HL Chl a ratio of 0.6 (n = 3). The phenotype of the most stable Phatr3_J50052 (light-specific kinase, LSK) transformant line #27 (LSK-27), was “locked” in a low-light state, with respect to chlorophyll content (Table 2). This transformant grew faster than the WT at LL and had a lower a* value consistent with increased thylakoid membrane packaging (Table 2). These changes did not affect σPSII (Table 2).
Table 2.
Phenotypic characterization of transformant strains (LSK-27 targeting kinase domain of Phatr3_J50052 and PTP-33 targeting Phatr3_J43123) in comparison to wild-type (WT) strains under high-light (HL) and low-light (LL) conditions
| WT (HL) | LSK-27 (HL) | PTP-33 (HL) | WT (LL) | LSK-27 (LL) | PTP-33 (LL) | |
|---|---|---|---|---|---|---|
| Exponential Growth Rate (μ d−1) | 1.1 ± 0.1 | 1.3 ± 0.3 | 1.3 ± 0.2 | 0.5 ± 0.04 | 1.0 ± 0.2 | 0.85 ± 0.2 |
| Chlorophyll a cell−1 (pg) | 0.27 ± 0.1 | 0.54 ± 0.1 | 0.28 ± 0.1 | 0.42 ± 0.1 | 0.87 ± 0.2 | 0.64 ± 0.1 |
| Optical Cross-section a* (m2/mg Chl a) | 41.1 ± 3.5 | 12.7 ± 0.7 | 16.6 ± 1.6 | 28.6 ± 1.4 | 26.6 ± 3.5 | 11.8 ± 1.3 |
| Functional Cross-section of PSII σPSII (Å2) | 364 ± 48 | 350 ± 48 | 363 ± 45 | 556 ± 22 | 562 ± 80 | 492 ± 83 |
| Quantum Efficiency of Photochemistry (Fv/Fm) | 0.53 ± 0.03 | 0.47 ± 0.08 | 0.49 ± 0.05 | 0.56 ± 0.01 | 0.55 ± 0.02 | 0.55 ± 0.03 |
| Relative Transcript Levels (qRT-PCR) | 1 ± n/a | 4.1 ± 0.9 | 22 ± 4.57 | 1 ± n/a | 12.9 ± 3.1 | 3.5 ± 0.34 |
HL, 750 to 800-μmol photons m−2s−1; LL, 20-μmol photons m−2s−1 (n = 3; P < 0.05; ±SD)
The physiological and photophysiological characteristics of the Phatr3_J43123 (plastid transmembrane protein or PTP) transformant line #33 (PTP-33) are presented in Table 2. In silico analysis of the annotated Phatr3_J43123 gene locus reveals that it encodes a 142 amino acid protein with no known functional domains, putatively targeted to the plastid outer-membranes or the thylakoids (Table 1). The significant differential expression of this “nonfunctional” gene in the WT in a LL-acclimated state (Table 1) indicates it could be an important regulator of the photoacclimation process. Of the 45 transformant lines generated from RNAi of Phatr3_J43123, 33 consistently showed abnormal cellular Chl a, with an average LL/HL ranging between 3 and 3.5 (n = 3). The PTP-33 line, which had the most stable phenotype, showed a 40% increase in Chl a content under LL as well as an increase in the exponential growth rate, when compared with WT LL (Table 2). However, changes were not observed between the WT and PTP-33 under HL. Despite Chl a accumulation occurring only under LL, there was an increase in thylakoid packaging or optical absorption cross-section irrespective of light intensity. LL acclimated PTP-33 cells also showed abnormal plastid division (SI Appendix, Figs. S1 and S2), where cells had enlarged plastids that appeared to undergo division independent of mitosis. Interestingly, both mutants grew faster under LL (even though σPSII and Fv/Fm are comparable to WT) implying that the mutants had better conversion of photosynthetically generated electrons to growth downstream of the light reactions.
A Long Noncoding Natural Antisense Transcript Putatively Regulates the Expression of Phatr3_J43123.
PTP-33 was designed to be a knockdown mutant of Phatr3_J43123 by an RNAi approach, with the full-antisense sequence of Phatr3_J43123 under the control of a strong constitutive promoter. However, instead of obtaining reduced accumulation of the Phatr3_J43123 transcript, we observed the opposite effect; an increased accumulation of this transcript in the PTP-33 mutant line suggesting increased expression (Table 2). Strand-specific RT-qPCR of Phatr3_J43123 revealed that PTP-33 had much higher levels of both sense and antisense transcripts, corresponding to the Phatr3-J43123 locus, than the WT (under both HL and LL conditions) but retained HL/LL differential regulation of the transcript (SI Appendix, Fig. S3). This result reveals that the overall concordant pattern of expression for the two transcripts is independent of light intensity. The result also strongly suggests that both transcripts form a sense–antisense concordant pair and that overexpression of the antisense transcript resulted in the overexpression of its cognate sense gene. Sequence analysis of the putative transcript originating from the opposite strand of Phatr3_J43123 using coding potential calculators, CPC and CPC2, which support vector machine-based classifiers that discriminate coding from noncoding transcripts with high accuracy (18, 19), revealed that it had a small unreliable putative ORF of 66 amino acids with a very low coding potential (CPC SVM score −1.48 with a CPC2 coding probability of 0.051), classifying it as a noncoding transcript. Since this putatively noncoding transcript is longer than 200 nucleotides, it falls in the category of long noncoding RNAs (lncRNAs) which have been previously characterized in P. tricornutum (20). Furthermore, the putative lncRNA transcript overlaps with the Phatr3_J43123 gene locus, which further classifies it as a long noncoding natural antisense transcript (NAT). Since the RNA-Seq datasets in this study were not strand specific, it was not possible to discriminate from the mapped reads which of the two strands (the protein coding gene or its cognate NAT) was up-regulated under LL. Our data strongly suggest that both transcripts are expressed in a concordant way.
Discussion
The results of this study reveal there are at least two nuclear-encoded genes involved in the light-dependent expression of light-harvesting complex proteins in P. tricornutum. One appears to be regulated by a putative lncRNA with cis-NAT regulatory functions. Changes in gene expression are signaled by the redox state of the plastoquinone pool (2), which effectively acts like a biophysical light sensor. The resultant nuclear as well as plastid gene products must be coordinated and tuned to bring about the physiological modifications in the plastid and the cell. Interfering with this signal transduction feedback loop of photoacclimation resulted in cells that lost their ability to alter the expression of nuclear-encoded light-harvesting pigment-binding proteins in response to changes in growth irradiance.
Photophysiology of Fully Acclimated P. tricornutum.
In P. tricornutum, 41 nuclear genes encode for three different families of LHC proteins. A subset of these genes was found to be responsive to changes in irradiance. Specifically, we observed a 32-fold change in expression of lhcX1 gene (SI Appendix, Table S1), which encodes for an LHC associated with NPQ (21, 22). Low-light-acclimated cells display a high-energy quenching component, qE, driven mainly by in vivo chlorophyll fluorescence quenching in close proximity to the PSII reaction center, where LHCX1 is primarily associated (23, 24). On the other hand, prolonged exposure to HL, enhances fluorescence quenching of fucoxanthin-chlorophyll protein (FCP), observed by the induction of other isoforms like LHCX2, which accumulate in the PSII antenna but not in the core (24). This can explain the negative change in expression of lhcX2 (SI Appendix, Table S1). Additionally, we observed a 20-fold increase in the expression of lhcF15 and lhcF5 genes, under LL in the WT (SI Appendix, Table S1). These lhc genes comprise major antenna complexes for both PSI and PSII (25, 26), which suggests an increase in antenna size, and/or number of antennae protein complexes leading to increased photon capture efficiency during photosynthesis under LL.
Despite the observed increase in expression of lhcs (SI Appendix, Table S1), there is no corresponding increase in transcript accumulation in the chlorophyll biosynthesis pathway (Fig. 2A). While this trend is clearly observed in photoautotrophs transitioning from HL to LL conditions, this does not appear to be the case once photoacclimation has stabilized (25). It has been suggested that regulating the activity of these biosynthesizing enzymes has a greater impact on chlorophyll biosynthesis than modifications at the transcript level (26). Therefore, it is likely that the observed levels of chlorophyll biosynthesis transcripts under LL (Fig. 2A) do not reflect the pigment content at the metabolite/protein level. In fact, exposure to LL could increase the half-life of the pigments embedded in the antenna, as a result of lower rates of ROS-associated chlorophyll degradation.
The functional absorption cross-section of PSII can also be altered by carotenoids in the LHCs, which are associated with NPQ (27). The ZEP and VDE reactions of the carotenoid biosynthesis pathway are responsible for the interconversions between the epoxidized and deepoxidized states of xanthophylls, respectively; however, paralogs of these enzymes have recently been reported to be involved in the synthesis of fucoxanthin, a light-harvesting carotenoid in diatoms (17). Although the ZEP can convert zeaxanthin to antheraxanthin, then to violaxanthin, in diatoms, this enzyme may also convert diadinoxanthin to diatoxanthin (28). The reverse reactions are catalyzed by vioxanthin deepoxidases (VDE). With the exception of zep3 and vdl2, these genes show almost no change in transcript abundances between LL and HL (Dataset S2). While upregulation of these genes would be necessary to increase the production of Ddx and Dtx for the xanthophyll cycle under HL stress, the same would also occur under LL, with the synthesis directed toward fucoxanthins for light harvesting instead. This can explain the low degree of relative change in transcript levels between LL and HL in the latter half of the carotenoid biosynthetic pathway (Fig. 2B). Taken together, these results suggest that, in a stable acclimated state, the up-regulated light-harvesting complex protein scaffolds preferentially bind fucoxanthins, rather than chlorophylls.
In the present study, only the nuclear transcriptomes were analyzed. For this reason, we can only extrapolate the possible change in stoichiometry of the photosystems based on the transcript profiles of the photosynthetic electron transport (PET)-component genes found in the nuclear genome (SI Appendix, Table S2). PSBU stabilizes the structure of PSII oxygen evolving complex (OEC) and protects the OEC against inactivation by reactive oxygen species (ROS) (29). The stoichiometric association of this subunit with the PSII core (30) leads us to propose that the changes in chloroplastic core subunit synthesis and assembly into functioning reaction centers in the thylakoids is highly coordinated in the retrograde signal transduction process. PSBO/OEE1 is another subunit that is required for structural stabilization of the Mn-cluster of the OEC (31). PSBQ functions as an oxygen-evolving enhancer protein, which is required for PSII assembly/stability (32) and is important for growth under low-light conditions in (e.g.) Arabidopsis thaliana (33). The ~twofold increase in transcript abundance of all these extrinsic PSII subunit genes suggests that these fully light-acclimated cells increase the synthesis of PSII reaction centers which optimizes light harvesting under LL growth conditions. We also observed a ~twofold upregulation in psb27 and hcf136 genes, both of which code for proteins associated with PSII assembly and repair. Given that these upregulations are under LL conditions where ROS-induced damage of PSII is very low, we propose that the expression of these genes is primarily for biogenesis of the reaction center rather than just its repair. Another electron transport component gene that is up-regulated in LL is the transcript of petJ, i.e., cytochrome c6/c553, which is the mobile electron carrier between Cytb6/f and PSI; unlike green algae, diatoms do not have plastocyanin. The differential expression of this gene as well as the other PET component genes, strongly suggests that LL acclimation in P. tricornutum is not only associated with an increased number of PSII subunits, but the entire photosynthetic apparatus.
Identification of Signaling Molecules in the Light-Intensity Acclimation Pathway.
The main body of information currently available on photosynthetic signal transduction is based on higher plants and green unicellular algae (34). However, there is no functional evidence for a number of these pathways in organisms of the secondary red algal lineage, such as diatoms. Indeed, homologs of various genes required for multiple photoacclimation pathways in green algae are not present in the P. tricornutum genome (35). This strongly suggests that organisms belonging to different endosymbiotic lineages have developed alternative signaling pathways that may be key to their ecological success. One major light-intensity-specific photoacclimation phenomenon occurring in the primary electron transport chain, is state transitions. First identified in a green alga (7), and a red, primary symbiotic microalga (36), state transitions are a rapid physiological mechanism that reversibly alters the effective absorption cross-sections of photosystems II and I in response to changes in light quality, thereby balancing the distribution of excitation energy between the two reaction centers. The phenomenon is driven by a protein kinase that is coupled to the redox state of the plastoquinone pool. The kinase responsible for this phosphorylation of LHCs, STT7 or STN7 (37, 38) is bound to thylakoid membranes and is conserved in higher plants and green algae (39, 40). However, there are no homologs to the state transition kinases such as STT7/STN7 in diatoms (40). Indeed, it has been reported previously that diatoms do not undergo state transitions (41). Hence, our finding of a soluble plastid kinase Phatr3_J50052/lsk (Table 1), which is only found in unicellular algae, is potentially significant. Overexpression of this gene resulted in a phenotype locked-in low light (Table 2). The lack of a DNA-binding region (based on sequence prediction) suggests that this kinase indirectly influences the expression of one or more lhc genes in the plastid or nuclear genomes, or could be involved in the feedback signaling cascade between the two. Comparing our transcriptome studies with transcriptomes of WT P. tricornutum in different stages of the cell cycle, redox-sensitive gene expression, as well as in the presence and absence of nitrogen, phosphorus, iron, cadmium, silica, revealed that this kinase is exclusively responsive to high-light-intensity changes (SI Appendix, Table S3). In Arabidopsis thaliana, the nonreceptor tyrosine kinase ABC1K1 (activity of bc1 complex kinase) is required for the stability of chlorophyll-binding proteins and prevents the accumulation of chlorophyll degradation products (42). We suggest that Phatr3_J50052, which has a ABC1-like kinase functional domain, may have a similar function by increasing the stability of the LHCs, thereby reducing the turnover of chlorophyll (Tables 1 and 2 and Fig. 3) (43).
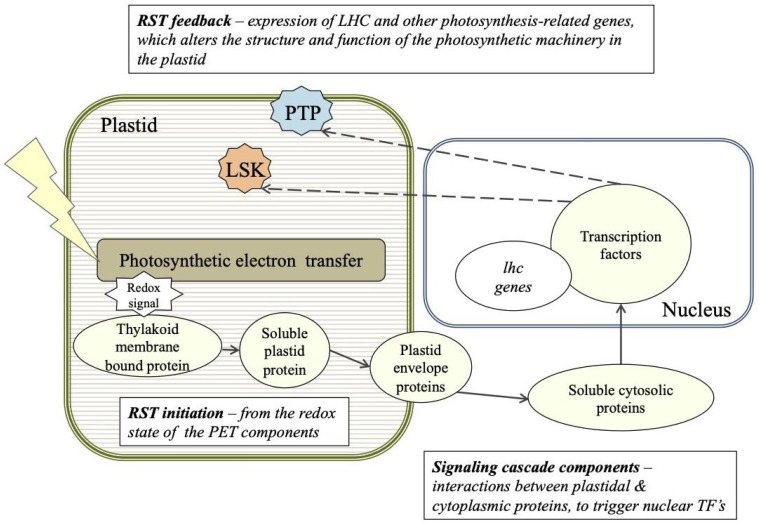
Fig. 3.
A working model of acclimation to irradiance and the retrograde signal transduction (RST) cascade between the nucleus and plastid in P. tricornutum. The putative cellular location of the genes of interest are based on sequence analysis predictions (refer to Table 1): PTP (blue) – Phatr3_J43123 plastid transmembrane protein; LSK (orange) – Phatr3_J50052 light-specific kinase.
In the case of PTP-33, a strong phenotype under LL as a result of its overexpression supports the hypothesis that this locus is an important regulator of light-harvesting antenna components in photoacclimation in diatoms. The RNAi-induced overexpression of the full-antisense Phatr3_ J43123 transcript resulted in the positive regulation of the cognate sense gene. This led us to investigate the expression patterns of both the coding and antisense strands from low- and high-light-intensity-acclimated PTP-33 cells, and further confirm the concordant expression patterns of both strands (SI Appendix, Fig. S3). Furthermore, the transcript expressed from the opposite strand of Phatr3_ J43123 shows a very low coding potential, classifying it as a putative long noncoding (lnc)RNA. LncRNAs are, by definition, transcripts longer than 200 nt that have no conserved ORF capable of producing proteins (44). For a long time, these molecules were considered transcriptional “noise;” however, lncRNAs are now recognized as being major regulators of gene activity throughout the Eukarya domain (44, 45). Instead of being expressed randomly, lncRNAs have specific expression patterns, generally responding to developmental and/or environmental cues (45, 46). In P. tricornutum, lncRNAs have been shown to be involved in responses to phosphate stress (20, 47) and elevated pCO2 (48). LncRNAs overlapping with protein-coding genes and expressed from the opposite strand are termed natural antisense transcripts (NATs) (44). NATs have been shown to act in cis- and/or in trans- and have an effect at the transcriptional level as well as the translational level of targeted genes or gene products (49). In P. tricornutum, NATs have been shown to be highly abundant, covering 21.5% of the annotated coding genome (47). Furthermore, several correlated NAT–mRNA pairs regulated by phosphate stress have been identified, the majority of which being concordant pairs (47).
Here, we show that a gene coding for a putative plastid transmembrane protein (Phatr3_J43123), as well as its putative NAT arising from the opposite strand, form a concordant NAT–mRNA pair highly up-regulated under LL. Although the nature of the regulation between these two transcripts remains unclear, our results strongly suggest that their expression is intrinsically correlated. Given the biplastid phenotype observed in the PTP-33 line under LL growth conditions (SI Appendix, Figs. S1-iii and S2), we also suggest that a potential trans-target for the regulatory lncRNA could be a putative ORF from the plastid genome, which shares a region of sequence complementarity with the NAT. It is predicted to have an FtsH-like conserved functional domain, likely to be involved in plastid division. This protein is known to accumulate in the mid-cell septum of dividing cells (50), along with conserved proteins FtsZ and FtsA, forming a complex (51) resulting in binary fission. The phenotype of the attempted RNAi line for J43123 (PTP-33) resulted in constitutive elevated expression of both the NAT and its cognate coding gene, leading to a marked phenotype characterized by an abnormal cell size and plastid formation under LL conditions, as seen in the PTP-33 line (SI Appendix, Figs. S1 and S2). Furthermore, PTP-33 also showed higher chlorophyll a content and higher exponential growth rate under LL in comparison to the WT (Table 2). Taken together, this suggests an important role for Phatr3_J43123 and its associated NAT in the regulation of plastid membrane integrity and pigment content during photoacclimation.
Based on sequence analysis predictions (Table 1), we propose a working model for the retrograde signaling process that enhances or suppresses gene expression of lhcs (Fig. 3). We suggest that a change in the redox state of photosynthetic electron transport between PSII and PSI (i.e., the PQ pool) triggers a retrograde signaling cascade. This cascade involves unknown cytosolic protein(s) that ultimately interact with nuclear transcription factors. The transcription factor(s) in turn alter the expression of nuclear-encoded LHC scaffold proteins, as well as leading to the induction of lsk and ptp whose products are then targeted to the plastid. Our experimental data suggest that LSK is a light-intensity-specific regulatory protein exclusive to diatoms. It appears to be essential to the stability of light-harvesting pigment–protein complexes within thylakoid membranes. Further, we have identified a plastid-targeted membrane-bound protein PTP (and its associated NAT), that is likely to be involved in the regulation of LHC content and plastid membrane division during photoacclimation. Neither of these proteins have been previously identified in any eukaryotic photoautotroph.
In conclusion, unicellular algae alter the expression of a myriad of nuclear-encoded light-harvesting proteins by light intensity via feedback signals originating in the photosynthetic electron transport chain (e.g., refs. 2, 21, and 33). Although several aspects of the retrograde signal in photoacclimation remain unknown, the data presented here strongly suggest that, in marine diatoms, the proteins LSK and PTP are critically involved.
Materials and Methods
Growth Conditions and Maintenance.
Phaeodactylum tricornutum (accession Pt1 8.6; CCMP632 in the Provasoli–Guillard National Center for Marine Algae and Microbiota) was maintained as described (52). To eliminate the effect of photoperiod and the cell cycle, cultures were grown in continuous white light from LEDs, and gradually acclimated to specific intensities of constant light ranging between 4- and 940-μmol photons m−2s−1. Cultures were maintained for at least thirty generations to ensure full acclimation.
RNAi Plasmid Design and Transformation in WT.
Sequences coding for RNA (iRNA) fragments were designed to target the functional domains of six specific genes (Table 1) based on NCBI BLAST annotations. These 325 to 425-bp sequence fragments were amplified with primers (SI Appendix, Table S4) into a pBlueScript-based plasmid, which was transformed into WT P. tricornutum biolistically as described (52). Single, independent transformation events of each plasmid into WT yielded 40 to 80 colonies. To screen for putative knockdown strains, each culture was grown under constant high light (HL) of ~800 to 950-μmol photons m−2s−1 and low light (LL) of 15 to 25-μmol photons m−2s−1. Of the 638 transformants isolated, the five strains that exhibited the most abnormal light acclimating phenotypes to either/both HL and LL were chosen for additional studies.
RNA Extraction, Sequencing and Analysis.
Samples for RNA-Seq were harvested and extracted from triplicate sets of individual cultures acclimated to 20- and 940-μmol photons m−2s−1. TruSeq RNA Library Prep Kit v2 (Illumina, CA) was used to prepare mRNA libraries for each of the six samples according to the manufacturer’s instructions. The 250-bp single-indexed libraries were multiplexed and sequenced on an Illumina MiSeq platform. The raw reads were trimmed for adaptor and low-quality sequences and then aligned to P. tricornutum version 3.0, which is the reannotation of 12,089 filtered gene models. After aligning the raw data to P. tricornutum’s version 3.0 set 12,089 filtered gene models (protists.ensembl.org), files were filtered to retrieve uniquely aligned reads with no more than three mismatches. Gene counts (unique aligned reads per gene) were used for differential expression (DE) analysis carried out using the DESeq R/Bioconductor package, which infers DE based on the negative binomial distribution. For this analysis, we used a cutoff of 5% to control for false detection rate (false positives) and considered only genes that had a log2 -fold change ≥ ±2, and a false detection rate < 0.05 to be DE. DESeq’s output for all 12,089 genes was submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus under accession no. GSE133301.
Quantification of Target Gene mRNA Copies Using Quantitative Real-time PCR (RT-qPCR).
Samples for RT-qPCR were extracted using TRIzol™ Reagent and measured as described in ref. 52. Strand-specific cDNA synthesis was carried out as above but with target-specific primers (SI Appendix, Table S4) also designed with Primer Express. At least three technical, as well as biological replicates were performed for each observation, and statistical significance was defined as P < 0.05.
Analytical Methods.
Relative chlorophyll fluorescence data, obtained from the Guava® easyCyte 12HT Sampling Flow Cytometer (0500-4012; EMD Millipore Sigma), were used for high-throughput screening. Based on significantly variable LL/HL ratios of Chl a/cell, the five most interesting transformants were analyzed. Chlorophyll a content per cell (Chl a), optical absorption cross-sections, referred to as a*, as well as PSII biophysical characteristics were measured as described in ref. 52. Functional metabolic assignments for identified gene transcripts into 16 relevant gene categories were done using DiatomCyc (www.diatomcyc.org), JGI Protist for Phatr2 (genome.jgi.doe.gov/Phatr2), Ensembl for Phatr3 (https://protists.ensembl.org/Phaeodactylum_tricornutum/Info/Index), and NCBI databases (www.ncbi.nlm.nih.gov). In silico gene localization predictions were obtained as described in ref. 52. Identification of putative diatom lncRNA candidates was based on coding potential calculators, CPC (18) and CPC2 (19) scores and then filtering for transcript length ≥ 200 nt and open reading frames (ORFs) <100 aa (20).
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
We thank Ehud Zelzion for the sequenced transcriptome analysis and Nicole Wagner for the transcriptome (both at Rutgers, The State University of New Jersey). This research was supported by the US NSF EAGER: Environmental signals in the marine diatom (Award # 1558128) for innovative research, the Rutgers University Professional Development Fund, the James G. Gibson (Biodiesel) Fund, and the Bennett L. Smith Endowment in Business and Natural Resources at Rutgers University to P.G.F.
Author contributions
A.A., O.L., and P.G.F. designed research; A.A. performed research; A.A., O.L., and H.C.d.C. analyzed data; and A.A., H.C.d.C., and P.G.F. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: E.-M.A., Turun yliopisto; and D.C., Mount Allison University.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Nott A., Jung H. S., Koussevitzky S., Chory J., Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 57, 739–759 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Escoubas J. M., Lomas M., LaRoche J., Falkowski P. G., Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc. Natl. Acad. Sci. U.S.A. 92, 10237–10241 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falkowski P. G., LaRoche J., Acclimation to spectral irradiance in algae. J. Phycol. 27, 8–14 (1991). [Google Scholar]
- 4.Falkowski P. G., Molecular ecology of phytoplankton photosynthesis in Primary Productivity and Biogeochemical Cycles in the Sea (Springer, US, 1992), pp. 47–67. [Google Scholar]
- 5.Huner N. P. A. Energy sensing and photostasis in photoautotrophs in Sensing, Signaling and Cell Adaptation, Storey K. B., Storey J. M., Eds. (Elsevier Science B.V., ed. 1, 2002), pp. 243–255. [Google Scholar]
- 6.Kirk J. T. O., Light and Photosynthesis in Aquatic Ecosystems (Cambridge University Press, 1994). 10.1017/CBO9780511623370. [DOI] [Google Scholar]
- 7.Bonaventura C., Myers J., Fluorescence and oxygen evolution from Chlorella pyrenoidosa. Biochim. Biophys. Acta 189, 366–383 (1969). [DOI] [PubMed] [Google Scholar]
- 8.Allen J. F., Bennett J., Steinback K. E., Arntzen C. J., Chloroplast protein phosphorylation couples plastoquinone redox state to distribution of excitation energy between photosystems. Nature 291, 25–29 (1981). [Google Scholar]
- 9.Müller P., Li X.-P., Niyogi K. K., Non-photochemical quenching. A response to excess light energy. Plant Physiol. 125, 1558–1566 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuzminov F. I., Gorbunov M. Y., Energy dissipation pathways in Photosystem 2 of the diatom, Phaeodactylum tricornutum, under high-light conditions. Photosynth Res. 127, 219–235 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Buck J. M., et al. , Lhcx proteins provide photoprotection via thermal dissipation of absorbed light in the diatom Phaeodactylum tricornutum. Nat. Commun. 10, 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falkowski P. G., Raven J. A., Aquatic Photosynthesis (Princeton University Press, ed. 2, 2007). [Google Scholar]
- 13.Falkowski P. G., Owens T. G., Light — Shade Adaptation. Plant Physiol. 66, 592–595 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maxwell D. P., Laudenbach D. E., Huner N. P. A., Redox regulation of light-harvesting complex II and cab mRNA abundance in Dunaliella salina. Plant Physiol. 109, 787–795 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ley A. C., Mauzerall D. C., Absolute absorption cross-sections for Photosystem II and the minimum quantum requirement for photosynthesis in Chlorella vulgaris. Biochim. Biophys. Acta 680, 95–106 (1982). [Google Scholar]
- 16.Büchel C., “Photosynthetic light reactions in diatoms. I. The lipids and light-harvesting complexes of the thylakoid membrane” in The Molecular Life of Diatoms (2022), pp. 397–422.
- 17.Bai Y., et al. , Green diatom mutants reveal an intricate biosynthetic pathway of fucoxanthin. Proc. Natl. Acad. Sci. U.S.A. 119, e2203708119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong L., et al. , CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35, W345–W349 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang Y. J., et al. , CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 45, W12–W16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cruz de Carvalho M. H., Sun H.-X., Bowler C., Chua N.-H., Noncoding and coding transcriptome responses of a marine diatom to phosphate fluctuations. New Phytol. 210, 497–510 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Bailleul B., et al. , An atypical member of the light-harvesting complex stress-related protein family modulates diatom responses to light. Proc. Nat. Acad. Sci. U.S.A. 107, 18214–18219 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lepetit B., et al. , High light acclimation in the secondary plastids containing diatom Phaeodactylum tricornutum is triggered by the redox state of the plastoquinone pool. Plant Physiol. 161, 853–865 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lepetit B., et al. , The diatom Phaeodactylum tricornutum adjusts nonphotochemical fluorescence quenching capacity in response to dynamic light via fine-tuned Lhcx and xanthophyll cycle pigment synthesis. New Phytol. 214, 205–218 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Taddei L., Dynamic changes between two LHCX-related energy quenching sites control diatom photoacclimation. Plant Physiol. 177, 00448.2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nymark M., et al. , An integrated analysis of molecular acclimation to high light in the marine diatom Phaeodactylum tricornutum. PLoS One 4, e7743 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka R., Tanaka A., Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58, 321–346 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Paulsen H., Carotenoids and the assembly of light-harvesting complexes in The Photochemistry of Carotenoids (1999), pp. 123–135. [Google Scholar]
- 28.Lohr M., Wilhelm C., Algae displaying the diadinoxanthin cycle also possess the violaxanthin cycle. Proc. Natl. Acad. Sci. U.S.A. 96, 8784–8789 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balint I., et al. , Inactivation of the extrinsic subunit of photosystem II, PsbU, in Synechococcus PCC 7942 results in elevated resistance to oxidative stress. FEBS Lett. 580, 2117–2122 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Shen J. R., Ikeuchi M., Inoue Y., Stoichiometric association of extrinsic cytochrome c550 and 12 kDa protein with a highly purified oxygen-evolving photosystem II core complex from Synechococcus vulcanus. FEBS Lett. 301, 145–149 (1992). [DOI] [PubMed] [Google Scholar]
- 31.Miyao M., Murata N., Role of the 33-kDa polypeptide in preserving Mn in the photosynthetic oxygen-evolution system and its replacement by chloride ions. FEBS Lett. 170, 350–354 (1984). [Google Scholar]
- 32.Roose J. L., Kashino Y., Pakrasi H. B., The PsbQ protein defines cyanobacterial Photosystem II complexes with highest activity and stability. Proc. Natl. Acad. Sci. U.S.A. 104, 2548–2553 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yi X., Hargett S. R., Frankel L. K., Bricker T. M., The PsbQ protein is required in Arabidopsis for photosystem II assembly/stability and photoautotrophy under low light conditions. J. Biol. Chem. 281, 26260–26267 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Pfannschmidt T., Chloroplast redox signals: How photosynthesis controls its own genes. Trends Plant Sci. 8, 33–41 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Falciatore A., Jaubert M., Bouly J. P., Bailleul B., Mock T., Diatom molecular research comes of age: Model species for studying phytoplankton biology and diversity. Plant Cell 32, 547–572 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murata N., Control of excitation transfer in photosynthesis I. Light-induced change of chlorophyll a fluoresence in Porphyridium cruentum. Biochim. Biophys. Acta 172, 242–251 (1969). [DOI] [PubMed] [Google Scholar]
- 37.Depège N., Bellafiore S., Rochaix J.-D., Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299, 1572–5 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Bellafiore S., Barneche F., Peltler G., Rochalx J. D., State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433, 892–895 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Rochaix J. D., et al. , Protein kinases and phosphatases involved in the acclimation of the photosynthetic apparatus to a changing light environment. Phil. Trans. R Soc. B 367, 3466–3474 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grouneva I., et al. , Phylogenetic viewpoints on regulation of light harvesting and electron transport in eukaryotic photosynthetic organisms. Planta 237, 399–412 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Owens T. G., Light-harvesting function in the diatom Phaeodactylum tricornutum. Plant Physiol. 80, 739–746 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang H., Yang M., Su Y., Qu L., Deng X. W., Arabidopsis atypical kinases ABC1K1 and ABC1K3 act oppositely to cope with photodamage under red light. Mol. Plant 8, 1122–1124 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riper D. M., Owens T. G., Falkowski P. G., Chlorophyll turnover in Skeletonema costatum, a marine plankton diatom. Plant Physiol. 64, 49–54 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rinn J. L., Chang H. Y., Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonasio R., Shiekhattar R., Regulation of transcription by long noncoding RNAs. Annu. Rev. Genet 48, 433–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ransohoff J. D., Wei Y., Khavari P. A., The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 19, 143–157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cruz de Carvalho M. H., Bowler C., Global identification of a marine diatom long noncoding natural antisense transcripts (NATs) and their response to phosphate fluctuations. Sci. Rep. 10, 14110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang R., et al. , A potential role for epigenetic processes in the acclimation response to elevated pCO2 in the model diatom Phaeodactylum tricornutum. Front Microbiol. 10, 3342 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deforges J., et al. , Control of cognate sense mrna translation by cis-natural antisense RNAs. Plant Physiol. 180, 305–322 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wehrl W., Niederweis M., Schumann W., The FtsH protein accumulates at the septum of Bacillus subtilis during cell division and sporulation. J. Bacteriol. 182, 3870–3873 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Addinall S. G., Lutkenhaus J., FtsA is localized to the septum in an FtsZ-dependent manner. J. Bacteriol. 178, 7167–72 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agarwal A., Di R., Falkowski P. G., Light-harvesting complex gene regulation by a MYB-family transcription factor in the marine diatom, Phaeodactylum tricornutum. Photosynth. Res. 1, 3 (2022). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
All study data are included in the article and/or SI Appendix.