Significance
For over 100 years, bacteria have been studied in simplified conditions. During infection, bacteria colonize complex environments containing fluid flow, exemplified by the bloodstream. Here, we use microfluidics to examine how flow impacts bacterial pathogens. We discover that flow triggers an effect like “wind-chill” that makes cells sensitive to chemical concentrations 100 to 1,000 times lower than traditionally used. Using flow intensities and chemical concentrations found in the human bloodstream, we demonstrate that host-relevant flow impacts bacterial pathogens. Demonstrating the importance of our findings, we establish that flow sensitizes both Pseudomonas aeruginosa and Staphylococcus aureus to chemical stress. Overall, our research highlights the need to study flow to capture the full complexity of how bacterial pathogens operate in realistic contexts.
Keywords: microfluidics, shear flow, mechanosensing, hydrogen peroxide, Pseudomonas aeruginosa
Abstract
Cells regularly experience fluid flow in natural systems. However, most experimental systems rely on batch cell culture and fail to consider the effect of flow-driven dynamics on cell physiology. Using microfluidics and single-cell imaging, we discover that the interplay of physical shear rate (a measure of fluid flow) and chemical stress trigger a transcriptional response in the human pathogen Pseudomonas aeruginosa. In batch cell culture, cells protect themselves by quickly scavenging the ubiquitous chemical stressor hydrogen peroxide (H2O2) from the media. In microfluidic conditions, we observe that cell scavenging generates spatial gradients of H2O2. High shear rates replenish H2O2, abolish gradients, and generate a stress response. Combining mathematical simulations and biophysical experiments, we find that flow triggers an effect like “wind-chill” that sensitizes cells to H2O2 concentrations 100 to 1,000 times lower than traditionally studied in batch cell culture. Surprisingly, the shear rate and H2O2 concentration required to generate a transcriptional response closely match their respective values in the human bloodstream. Thus, our results explain a long-standing discrepancy between H2O2 levels in experimental and host environments. Finally, we demonstrate that the shear rate and H2O2 concentration found in the human bloodstream trigger gene expression in the blood-relevant human pathogen Staphylococcus aureus, suggesting that flow sensitizes bacteria to chemical stress in natural environments.
Classically, research on bacterial stress responses has focused on chemical stressors such as nutrient availability (1), pH (2), antibiotics (3), and oxidative stress (4). Bacteria respond to chemical perturbations with well-studied physiological responses to survive and grow in stressful situations (5–8). For simplicity, bacterial stress responses have been largely studied in batch cell culture, which has allowed researchers to identify and characterize many of the important signaling pathways associated with stress. However, simplified experimental systems neglect the dynamic mechanical features of natural and host systems (9).
Recently, a surge of research on bacterial mechanosensing has revealed that fluid flow impacts virulence (10, 11), biofilm formation (12), and gene expression (13). While bacterial transcriptional responses to flow were assumed to require the cellular measurement of forces (10, 12), one report challenged this assumption and revealed that flow can trigger bacterial gene expression in a force-independent manner (13). This report proposed naming flow-sensitive transcriptional responses “rheosensitive” (as rheo- is Greek for flow), due to lack of direct evidence that bacteria respond to flow by measuring forces (13). Thus, it is currently debated how flow generates bacterial transcriptional responses.
Here, we discover that flow generates transcriptional responses in the human pathogens Pseudomonas aeruginosa and Staphylococcus aureus by sensitizing them to H2O2 stress. We quantitatively establish that physical shear rate (a measure of fluid flow) triggers bacterial responses by abolishing spatial gradients of H2O2 generated by bacterial scavenging. Thus, flow triggers bacterial responses through a biophysical mechanism that it is highly reminiscent of “wind chill.” We also observe that bacteria in flow are sensitive to concentrations of the host-generated antimicrobial H2O2 that are 100–1,000 times less than those traditionally used in lab conditions. Notably, the incorporation of flow explains the long-standing mystery of why H2O2 concentrations required to generate responses in simplified lab conditions are orders of magnitude higher than those found in hosts.
Results
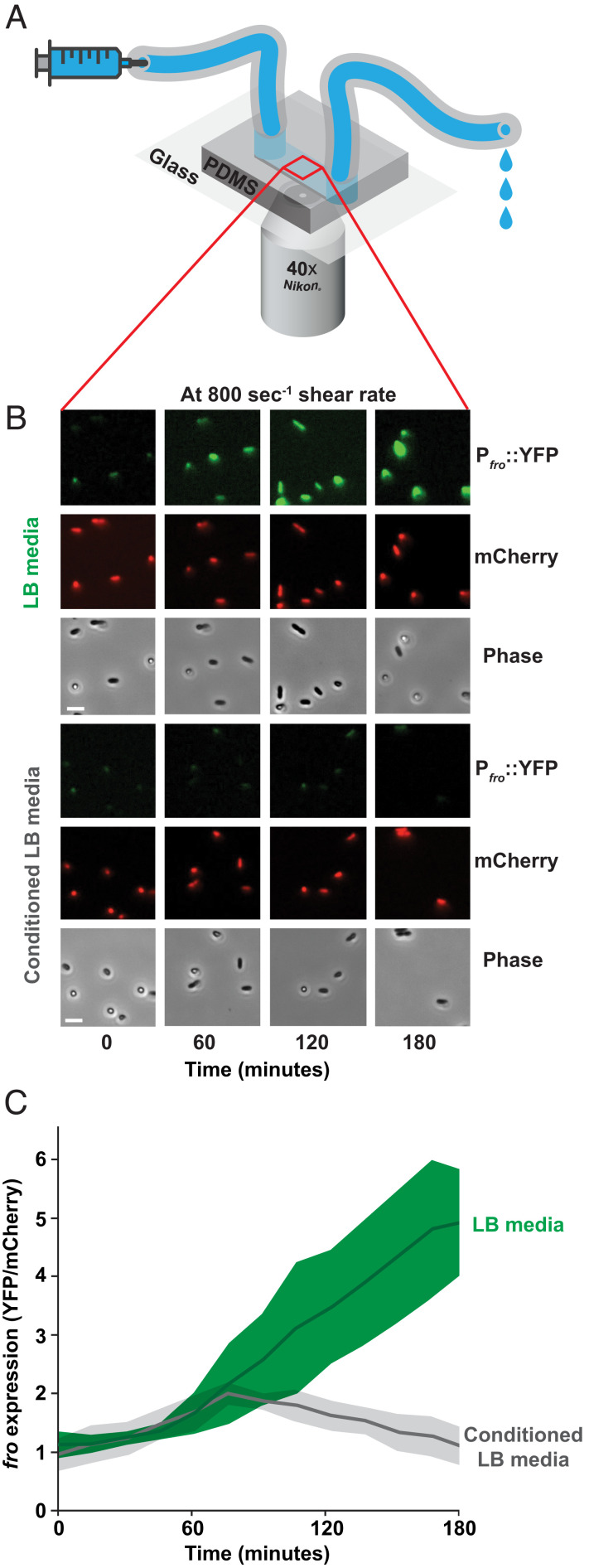
To understand how flow affects gene expression, we focused on the flow-sensitive transcriptional response in the bacterium P. aeruginosa. P. aeruginosa responds to flow by upregulating a suite of genes (13), including many genes upregulated during human infection (14). As a representative example of the larger flow-sensitive response, we focused our efforts on the fro operon. The fro operon is rapidly and robustly upregulated by flow (13) and required for full infection in multiple animal models (15, 16). For single-cell analysis, we used a fro reporter strain that reports on fro expression with yellow fluorescent protein (YFP) and encodes a constitutively expressed mCherry for normalization. To confirm that flow induces fro expression, cells in a microfluidic device were simultaneously subjected to flow from a syringe pump and imaged with a fluorescence microscope (Fig. 1A). Throughout this study, we represent the intensity of flow using shear rate, which is calculated using flow rate and channel dimensions (13). Consistent with previous results (13), fro is strongly induced after 3 h of exposure to a shear rate of 800 s−1 in an LB medium (Fig. 1B). Thus, the fro reporter represents a valuable tool to dissect how flow generates transcriptional responses in bacteria.
Fig. 1.
Conditioning media suppresses flow-sensitive gene expression in P. aeruginosa. (A) Microfluidic setup used in this study. Microfluidic devices are custom fabricated with polydimethylsiloxane and glass coverslips. Channels are 50 µm tall × 500 µm wide. Bacteria adhere to the channel wall. Microfluidic devices are simultaneously subjected to flow from a syringe pump and imaged 1 cm into the channel by a fluorescence microscope. (B) Fluorescence and phase images of the P. aeruginosa fro reporter strain in flow (at a shear rate of 800 s−1) over 180 min. Cell density in channels remains relatively constant due to the combinatorial effects of cell growth and cell departure. Images are representative of three biological replicates. (Scale bars, 5 µm.) (C) Quantification of fro expression by dividing YFP intensity by mCherry intensity as described in SI Appendix, Fig. S1. Green represents cells exposed to LB media, while gray represents cells exposed to conditioned LB. Shaded regions show SD of three biological replicates.
How does flow generate a transcriptional response? One possibility is that flow affects a rate-dependent biophysical process such as chemical transport. To test this hypothesis, we flowed fresh LB medium and LB medium that had been conditioned by P. aeruginosa into independent channels of the device. A conditioned medium was generated by exposing fresh, sterile LB to a culture of P. aeruginosa for 60 min, followed by filter-sterilizing the media to remove cells. We determined that cells exposed to flow with LB induced fro expression fivefold, but fro was not induced after 3 h of exposure to flow in conditioned LB (Fig. 1 B and C). Thus, our results suggest that chemical transport of a molecule into or out of cells underlies how bacteria respond to flow.
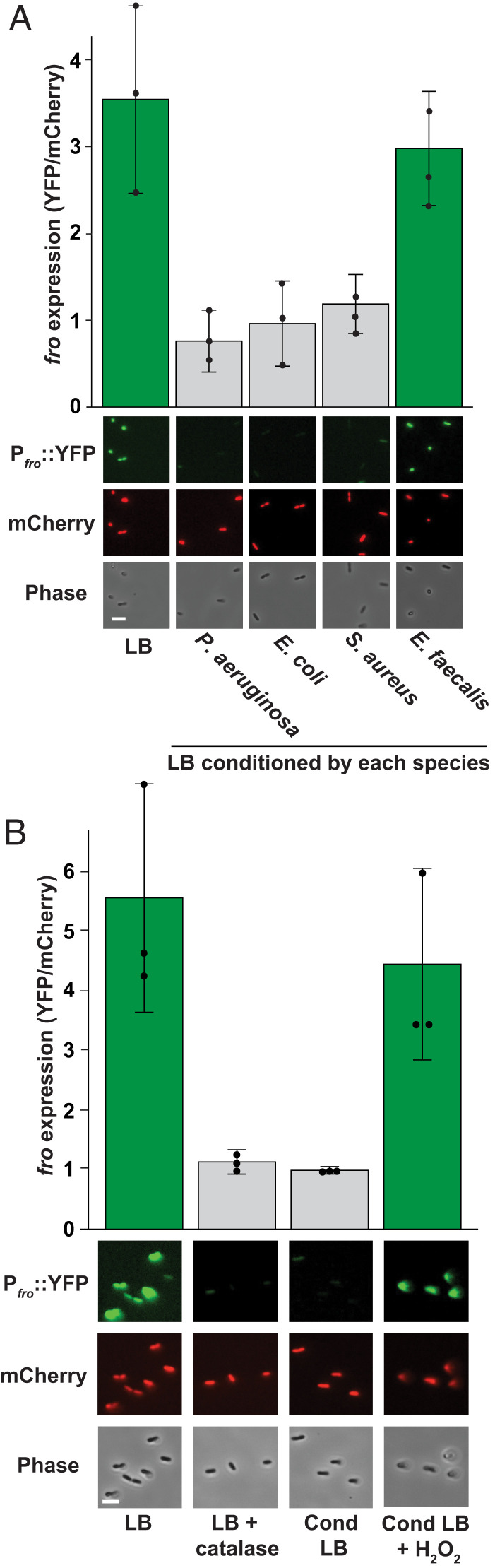
What is the identity of the flow-sensitive molecule? To examine whether the molecule was conserved, we tested the effect of conditioning media with Escherichia coli, S. aureus, and Enterococcus faecalis. Conditioning media with E. coli or S. aureus led to complete loss of fro induction in flow, while conditioning media with E. faecalis resulted in high fro induction (Fig. 2A). P. aeruginosa, E. coli, and S. aureus are catalase-positive (17), while E. faecalis is catalase-negative (18). Catalase specifically degrades H2O2 (19), supporting the hypothesis that the flow-sensitive molecule is H2O2. Consistent with the literature (17, 18), we confirm that P. aeruginosa, E. coli, and S. aureus can deplete H2O2 from media in 30 min, while E. faecalis cannot (SI Appendix, Fig. S2). To test the role of H2O2 scavenging enzymes during the conditioning of media, we compared the effect of conditioning media with wild-type E. coli and an E. coli ∆ahpC ∆katG ∆katE ∆ccp mutant known to lack scavenging ability (20). While media conditioned with wild-type E. coli prevented fro induction, media conditioned with the E. coli ∆ahpC ∆katG ∆katE ∆ccp mutant (20) was still capable of inducing fro expression (SI Appendix, Fig. S3). Taken together, our results support the hypothesis that the flow-sensitive molecule is H2O2.
Fig. 2.
H2O2 is a flow-sensitive molecule. (A) P. aeruginosa fro expression after 180 min in flow (at a shear rate of 800 s−1) with LB media or media conditioned by P. aeruginosa, E. coli, S. aureus, or E. faecalis for 60 min. Quantification shows the average and SD of three biological replicates. Images show YFP and mCherry fluorescence, as well as phase contrast. (B) fro expression after 180 min in flow (at a shear rate of 800 s−1) with LB media, LB treated with catalase, media conditioned by P. aeruginosa for 60 min, and conditioned media resupplied with 9 μM H2O2. Quantification shows the average and SD of three biological replicates. Images are taken 1 cm into the channel and show YFP and mCherry fluorescence, as well as phase contrast. (Scale bars, 5 µm.) Channels are 50 µm tall × 500 µm wide.
We reasoned that if H2O2 was the flow-sensitive molecule, it must be present in our media. We tested H2O2 levels with a peroxidase assay (21) and determined that our laboratory LB stocks contained approximately 9 μM H2O2 (SI Appendix, Fig. S4). The presence of H2O2 in LB is a reproducible but underappreciated detail (22, 23). The chemical production of H2O2 in LB is mediated by a light-dependent reaction involving riboflavin (23). To test if H2O2 in our media was required to trigger fro induction, we repeated our flow experiment with M9 minimal media lacking H2O2. Minimal media was incapable of inducing fro expression, supporting our hypothesis that H2O2 is required for fro induction (SI Appendix, Fig. S5). To directly test the role of catalase, we treated LB with purified catalase, which depleted H2O2 concentrations in LB to effectively zero (SI Appendix, Fig. S6). Catalase-treated media did not induce fro expression in flow, further supporting the hypothesis that the flow-sensitive molecule is H2O2. (Fig. 2B). Finally, we tested fro expression with conditioned media resupplied with 9 μM H2O2. Reintroduction of H2O2 restored robust fro induction (Fig. 2B), demonstrating that the flow-sensitive molecule is H2O2.
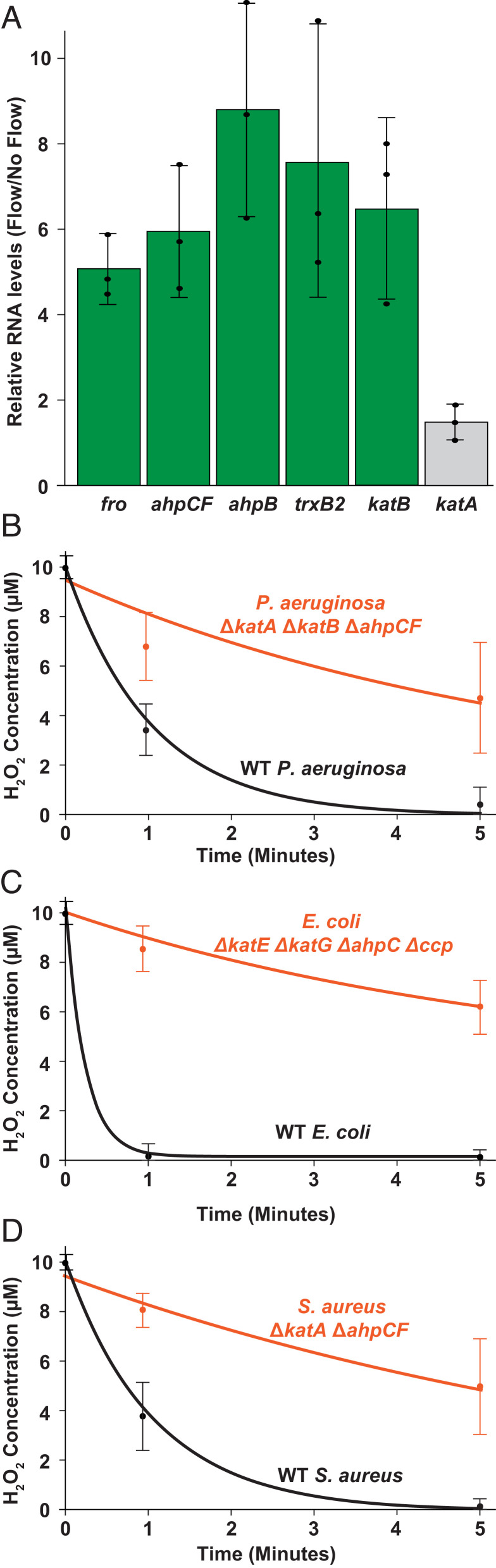
Armed with our understanding that H2O2 is the flow-sensitive molecule, we reanalyzed RNA-sequencing data from a previous microfluidic-based transcriptomic experiment (13). In addition to the fro operon, we identified many flow-induced genes implicated in H2O2 scavenging, such as ahpCF, ahpB, and katB (Fig. 3A). AhpCF and AhpB are NADH peroxidases that scavenge H2O2 (24–26). KatB is a catalase that converts H2O2 into water and oxygen (19, 24). AhpCF, AhpB, and KatB are induced by H2O2 and regulated by OxyR, a H2O2 sensor located in the cytoplasm (24, 27–30). Thus, our results suggest that shear rate leads to the intracellular accumulation of H2O2. In support of this model, an additional H2O2-regulated gene trxB2, which encodes thioredoxin reductase (27), was also upregulated in response to shear rate (Fig. 3A). Together, our results suggest the interplay of shear rate and H2O2 generate cellular stress and trigger a transcriptional response.
Fig. 3.
Fluid flow upregulates multiple enzymes that scavenge H2O2. (A) P. aeruginosa RNA sequencing data (13) showing relative transcript levels comparing flow conditions to no flow conditions. fro represents an average of the froABCD operon and ahpCF represents an average of ahpC and ahpF. Quantification shows the average and SD of three biological replicates. (B) H2O2 concentration of LB media over time when treated with wild-type P. aeruginosa and ΔkatA ΔkatB ΔahpCF mutant cells at an OD of 0.4 to 0.5. (C) H2O2 concentration of LB media over time when treated with wild-type E. coli and ΔkatE ΔkatG ΔahpC Δccp mutant cells at an OD of 0.4 to 0.5. (D) H2O2 concentration of LB media over time when treated with wild-type S. aureus and ΔkatA ΔahpCF mutant cells at an OD of 0.4 to 0.5. All H2O2 concentrations were measured using a peroxidase assay (21) and quantification shows the average and SD of three biological replicates.
What is the relationship between flow and H2O2? We hypothesize that shear rate replenishes H2O2 to overcome rapid scavenging by cells. To explore the kinetics of H2O2 scavenging, we used a peroxidase assay (21) to measure H2O2 concentrations of LB media treated with P. aeruginosa, E. coli, or S. aureus over 60 min. Additionally, we compared wild-type and mutant strains of P. aeruginosa, E. coli, and S. aureus. At a cell density of 0.5 OD, wild-type P. aeruginosa scavenged about 50% of the available H2O2 in 30 s, while a ∆katA ∆katB ∆ahpCF mutant was significantly impaired at scavenging H2O2 (Fig. 3B). Wild-type E. coli scavenges about 90% of the available H2O2 in 30 s, while an ∆ahpC ∆katG ∆katE ∆ccp mutant had significantly diminished scavenging ability (20) (Fig. 3C). Finally, wild-type S. aureus scavenged about 25% of the available H2O2 in 30 s and a ∆katA ∆ahpCF mutant was also significantly impaired at H2O2 scavenging (Fig. 3D). Together, our batch cell culture experiments reveal that bacteria rapidly scavenge H2O2, providing support to our hypothesis that flow triggers a biological response by replenishing H2O2.
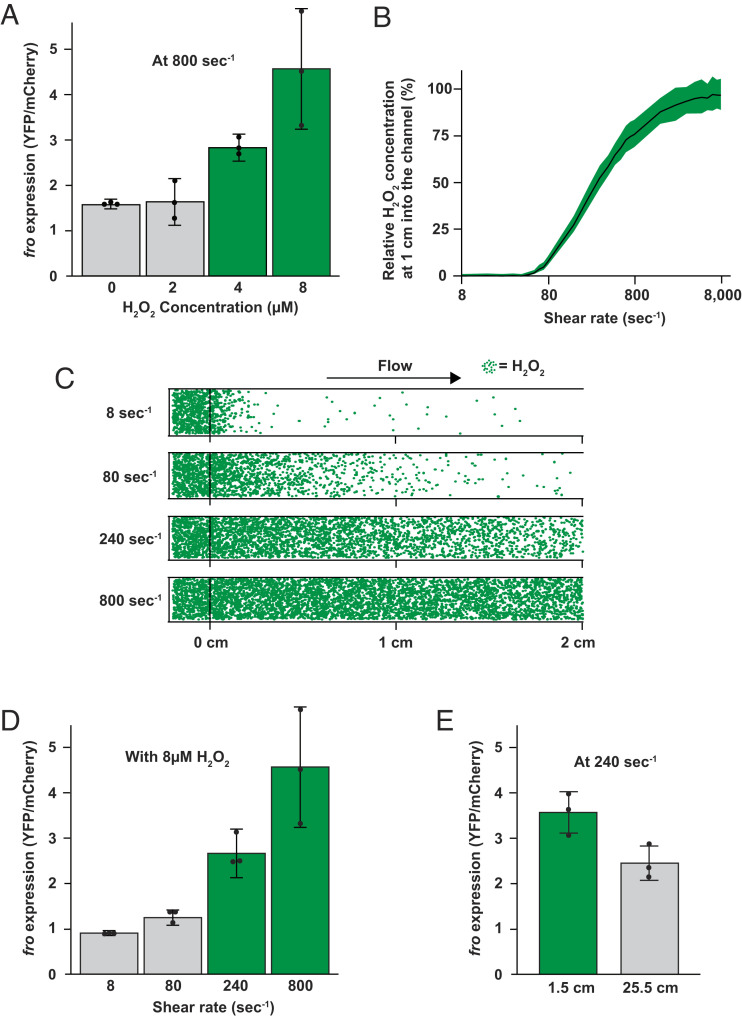
How sensitive are bacterial cells to H2O2 in flow? We measured fro induction at a range of H2O2 concentrations, while maintaining a constant shear rate of 800 s−1. We observed that 2 µM H2O2 did not induce fro expression. In contrast, 4 µM H2O2 led to a threefold induction and 8 µM H2O2 led to a fivefold induction in fro expression (Fig. 4A and SI Appendix, Fig. S7A). Thus, the minimum concentration necessary to elicit a flow-sensitive response at a shear rate of 800 s−1 is approximately 4 µM H2O2 (Fig. 4A). Surprisingly, 4 µM is 100 to 1,000 times lower than the H2O2 concentration traditionally studied in batch cell culture (24, 31–35). Moreover, the H2O2 concentration in the human bloodstream is thought to be between 1 and 5 µM (36). Together, these results support the hypothesis that bacteria in flow are highly sensitive to H2O2 and suggest that flow generates stress in natural environments.
Fig. 4.
Simulation and experiment determine the minimal shear rate to trigger flow-sensitive gene expression. (A) P. aeruginosa fro expression after 180 min in flow (at a shear rate of 800 s−1) with LB media with varied H2O2 concentrations imaged 1 cm into the channel. (B) Relationship between shear rate and the relative H2O2 concentrations 1 cm into a channel from our simulations. Quantification shows the average and SD of multiple bins in the middle of the channel. (C) Visual representation of H2O2 molecules in simulated microfluidic channels from the start (0 cm) to the end (2 cm) of a 2-cm channel at different shear rates. Green dots represent individual H2O2 molecules experiencing diffusion and fluid flow from left to right. To represent bacterial scavenging, 1 of every 100 molecules that hit the lower surface is removed. This assumption is based on calculations that use the scavenging kinetics defined in Fig. 3. (D) fro expression after 180 min in flow (at 8 μM H2O2) with varied shear rates imaged 1 cm into the channel. (E) fro expression near the start (1.5 cm) and end (25.5 cm) of a 27-cm long channel after 180 min in flow at 8 μM H2O2 with a shear rate of 240 s−1. Quantification in panels A, D, and E shows the average and SD of three biological replicates. Green and gray in panels A, D, and E signify expression levels that are statistically different with P < 0.05.
To understand the mathematical relationship between flow and H2O2, we calculated the Péclet number for our experimental system. The Péclet number describes if shear rate or diffusion is dominant in a particular regime and is proportional to the shear rate divided by diffusion. When shear rate dominates diffusion, the Péclet number is greater than one. When diffusion dominates shear rate, the Péclet number is less than one. To solve for the minimal shear rate required to overtake diffusion, we set the Péclet equation equal to one and solved for shear rate (SI Appendix, Fig. S8). Our calculations show that a shear rate of approximately 166 s−1 should dominate diffusion in our experimental system (SI Appendix, Fig. S8).
To generate testable mathematical predictions, we developed a simulation for the advection-diffusion transport of individual H2O2 molecules in a channel. The simulation uses experimental values for shear rate and the estimated diffusion coefficient of H2O2 (37). To simulate H2O2 removal by cells, we included a feature where 1 of every 100 molecules that contacted the channel surface was removed. We used the value of 1 in 100 based on calculations (described in Materials and Methods) that use the scavenging kinetics defined in Fig. 3. Our simulation shows that as the shear rate increases from 8 to 8,000 s−1, the H2O2 concentration at the middle of the channel increases (Fig. 4B). For low shear rates, H2O2 molecules are rapidly depleted. In contrast, higher shear rates replenish depleted molecules and maintain the concentration of H2O2 close to the initial value. Our simulated results highlight how rapid H2O2 scavenging generates gradients of H2O2, which are abolished by shear rates of at least 240 s−1. Together, our mathematical calculations and simulations predict that a shear rate of 166 s−1 to 240 s−1 will trigger flow-sensitive gene expression.
To test our mathematical predictions, we measured fro induction at a range of shear rates, while maintaining a constant H2O2 concentration of 8 μM. We observed that shear rates of 8 s−1 and 80 s−1 did not trigger fro expression. In contrast, 240 s−1 led to a threefold induction and 800 s−1 led to a fivefold induction in fro expression (Fig. 4D and SI Appendix, Fig. S7B). Thus, the minimum shear rate necessary to elicit a flow-sensitive response at a H2O2 concentration of 8 μM is approximately 240 s−1 (Fig. 4D). For perspective, human veins have shear rates of approximately 80 s−1 and human arteries have shear rates of approximately 800 s−1 (38).
To test the prediction that cell scavenging generates spatial H2O2 gradients, we examined fro expression at the beginning and end of a long microfluidic channel (SI Appendix, Fig. S9). For this experiment, we used a shear rate of 240 s−1 and a H2O2 concentration of 8 μM. Cells at the beginning of the channel induced fro expression 3.5-fold, while cells at the end of channel experienced lower induction levels (Fig. 4E). To further examine how flow transports H2O2 molecules, we repeated our long channel experiment at a shear rate of 80 s−1. As predicted by our simulation, a shear rate of 80 s−1 results in low fro expression 1.5 cm into the channel and at 25.5 cm into the channel (SI Appendix, Fig. S10A). Repeating our long channel experiment at a shear rate of 800 s−1 resulted in high fro expression at 1.5 cm into the channel and at 25.5 cm, which further validates our model and shows that flow can mediate long-range transport of H2O2 (SI Appendix, Fig. S10B). Based on our simulation, we hypothesized that cell density would affect H2O2 levels. We tested this hypothesis by performing an experiment where cell density was altered while shear rate was held constant at 80 s−1 and H2O2 concentration was held constant at 8 µM H2O2. Supporting our hypothesis and further validating our simulation, higher cell density led to lower fro expression and lower cell density led to higher fro expression (SI Appendix, Fig. S11). Together, our biophysical experiments support our mathematical predictions, establish that flow modulates chemical gradients, and demonstrate that physiological levels of flow trigger a transcriptional response in P. aeruginosa.
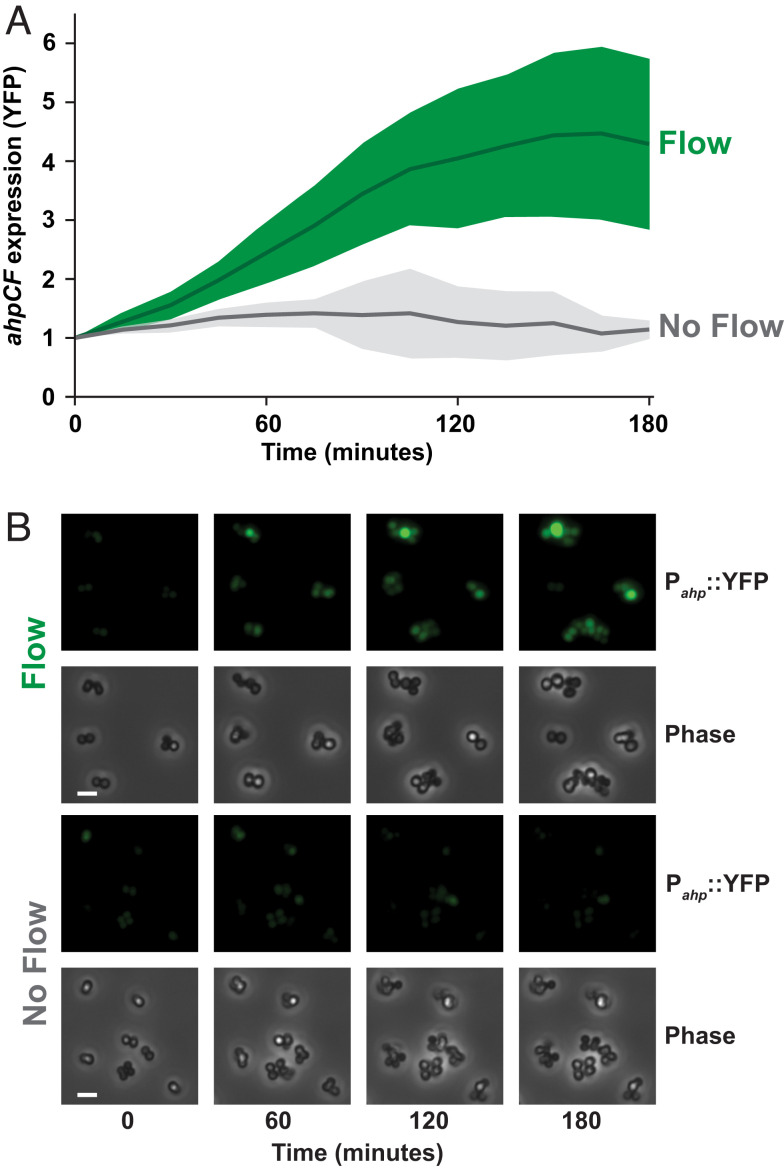
Does flow generate stress in other bacteria? The biological requirements for a flow-sensitive response are a permeable membrane, H2O2 scavenging, and H2O2 sensing capacity. As these three features are widespread in bacteria, we hypothesized that the human pathogen S. aureus would also exhibit flow-sensitive gene expression. We focused our efforts on S. aureus as it infects the human bloodstream, which contains high shear rates (38) and low micromolar H2O2 concentrations (36). We generated a S. aureus YFP fluorescent reporter to the promoter of ahpCF. In S. aureus, ahpCF expression is induced by H2O2 through the function of the H2O2-sensing transcriptional regulator PerR (39, 40) (SI Appendix, Fig. S12). When we subjected S. aureus cells to a shear rate of 800 s−1 and a H2O2 concentration of 8 μM, ahpCF was induced fourfold compared to no flow conditions (Fig. 5). Thus, flow triggers gene expression in S. aureus, suggesting that the flow-sensitive stress response we have discovered is widely conserved.
Fig. 5.
Shear rate triggers flow-sensitive gene expression in S. aureus. (A) S. aureus ahpCF expression over 180 min in the presence of flow (green) or no flow (gray). Cells contained a Pahp::YFP transcriptional reporter. Both treatments had 8 µM H2O2. Flow treatment was at a shear rate of 800 s−1. Quantification shows the average and SD of at least three biological replicates. (B) YFP fluorescence and phase contrast images taken 1 cm into the channel, representative of at least three biological replicates. (Scale bars, 5 µm.) Channels are 50 µm tall × 500 µm wide.
Discussion
Collectively, our results provide a molecular mechanism of how flow generates stress and triggers a transcriptional response (SI Appendix, Fig. S12). We show that our flow-sensitive transcriptional response hinges on the quantitative relationships between shear rate, H2O2, and cell scavenging. First, H2O2 from the environment diffuses into cells, where it is scavenged by catalases and NADH peroxidases (Fig. 3). Second, H2O2 scavenging results in a zone of depletion and H2O2 spatial gradients in the environment (Fig. 4). Third, shear rates above 240 s−1 replenish H2O2 in the environment, abolish spatial gradients, and lead to increased accumulation of H2O2 in cells (Fig. 4). Fourth, cells sense intracellular H2O2 levels and activate a transcriptional response that induces factors to mitigate H2O2 damage (Fig. 5). Thus, flow triggers a biological response by countervailing the ability of cells to remove a chemical stressor from the environment.
Reports have classified bacterial responses to flow as mechanosensing (10, 12) or rheosensing (13). Classification of bacterial flow responses as mechanosensing was influenced by a few well-established examples of force-sensing in eukaryotes (41, 42). Based on this classification, the study of bacterial flow responses focused on shear force and largely neglected the effects of flow on chemical transport. In contrast, the field of fluid dynamics has long appreciated that flow is a major driver of chemical transport (43). The attempt to classify bacterial flow responses as rheosensing (13) was intended to provide a more conservative view, as the mechanisms underlying these responses were unknown. Now, with the data presented here, it is clear that flow can trigger a bacterial response by affecting chemical transport of small molecules such as H2O2. Our results provide proof-of-principle that flow could amplify or nullify the effect of a wide variety of molecules, such as amino acids, carbon sources, oxygen, and antimicrobials. Thus, in the current manuscript, we refrain from classifying the flow-sensitive response as mechanosensing or rheosensing, as we prefer the more generalizable explanation that flow has a critical role in generating dynamics of chemical transport.
As an analogy to our findings, we note that flow-sensitive gene expression is conceptually like wind chill. In our system, shear rate generates a biological response by abolishing a chemical gradient. Wind chill describes how wind speed generates a biological response by abolishing a temperature gradient. Historically, the concept of wind chill did not exist until 1945 (44). However, it is now widely reported by the National Weather Service due to the critical importance of its effect on human health. By analogy, we propose that in order to understand how cells interact with their environment in natural systems, it is essential to include flow in experimental systems.
Our discovery that bacteria in flow are sensitive to H2O2 concentrations that are 100 to 1,000 times lower than traditionally used should provoke a paradigm shift in the way we think about H2O2 stress. For simplicity, studies on H2O2 stress in bacteria have primarily used batch cell cultures and millimolar amounts of H2O2 (24, 31–35). However, natural environments are unlikely to contain such high H2O2 concentrations (9). Additionally, it has been noted that H2O2 sensors OxyR and PerR are likely sensitive to much lower concentrations (23, 29, 45). Thus, the H2O2 concentrations required to generate bacterial responses in laboratory conditions do not reflect the environments where bacteria live. Our observation that bacteria in flow are sensitive to low-micromolar levels of H2O2 likely reconciles this discrepancy. Natural bacterial environments, such as the human bloodstream, contain low-micromolar levels of H2O2 (36). Without considering the effect of flow, these levels are insufficient to generate bacterial responses. In light of our study, it is apparent that H2O2 in flowing blood is sufficient to affect bacteria and potentially has a role in the restriction of bacterial growth.
Materials and Methods
The bacterial strains used in this paper are described in SI Appendix, Table S1. The primers used in this study are listed in SI Appendix, Table S2. The plasmids used are described in SI Appendix, Table S3. Information on the generation of mutant strains, H2O2 experiments, microfluidic experiments, microscopy, image quantification, and mathematical simulations are provided in Materials and Methods section of SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Satish Nair, Nicholas Wu, Ido Golding, Raven Huang, Wilfred van der Donk, Nick Martin, Andrian Gutu, and Lisa Wiltbank for helpful discussions and comments on the manuscript. We would also like to thank Noah Miller for helpful discussions and comments regarding image analysis. This work was supported by NIH grant R01AI155611 to T.E.K.-F. This work was also supported by start-up funds from the University of Illinois at Urbana-Champaign and NIH grant K22AI151263 to J.E.S.
Author contributions
G.C.P., A.M.S., A.S., M.D.K., J.N.R., T.E.K.-F., J.A.I., and J.E.S. designed research; G.C.P., A.M.S., A.S., M.D.K., J.-J.S.P., and J.N.R. performed research; G.C.P., A.M.S., A.S., M.D.K., J.A.I., and J.E.S. analyzed data; and G.C.P. and J.E.S. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix. The raw data and code used to support this study are available in Github (https://github.com/padrong/ImageAnalysis-Adaptations) (46).
Supporting Information
References
- 1.Boutte C. C., Crosson S., Bacterial lifestyle shapes stringent response activation. Trends Microbiol. 21, 174–180 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanjee U., Houry W. A., Mechanisms of acid resistance in Escherichia coli. Annu. Rev. Microbiol. 67, 65–81 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Liu Y., Imlay J. A., Cell death from antibiotics without the involvement of reactive oxygen species. Science 339, 1210–1213 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imlay J. A., The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol 11, 443–454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imlay J. A., Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77, 755–776 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potrykus K., Cashel M., (p)ppGpp: Still magical? Annu. Rev. Microbiol. 62, 35–51 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Raivio T. L., Silhavy T. J., Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55, 591–624 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Battesti A., Majdalani N., Gottesman S., The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65, 189–213 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imlay J. A., Where in the world do bacteria experience oxidative stress?: Oxidative stress in natural environments. Environ. Microbiol. 21, 521–530 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alsharif G., et al. , Host attachment and fluid shear are integrated into a mechanical signal regulating virulence in Escherichia coli O157:H7. Proc. Natl. Acad. Sci. U.S.A. 112, 5503–5508 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sirisaengtaksin N., Odem M. A., Bosserman R. E., Flores E. M., Krachler A. M., The E. coli transcription factor GrlA is regulated by subcellular compartmentalization and activated in response to mechanical stimuli. Proc. Natl. Acad. Sci. U.S.A. 117, 9519–9528 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodesney C. A., et al. , Mechanosensing of shear by Pseudomonas aeruginosa leads to increased levels of the cyclic-di-GMP signal initiating biofilm development. Proc. Natl. Acad. Sci. U.S.A. 114, 5906–5911 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanfilippo J. E., et al. , Microfluidic-based transcriptomics reveal force-independent bacterial rheosensing. Nat. Microbiol. 4, 1274–1281 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornforth D. M., et al. , Pseudomonas aeruginosa transcriptome during human infection. Proc. Natl. Acad. Sci. U.S.A. 115, E5125–E5134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skurnik D., et al. , A comprehensive analysis of in vitro and in vivo genetic fitness of Pseudomonas aeruginosa using high-throughput sequencing of transposon libraries. PLoS Pathogens 9, e1003582 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potvin E., et al. , In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ. Microbiol. 5, 1294–1308 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Yuan F., et al. , The richness and diversity of catalases in bacteria. Front. Microbiol. 12, 645477 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher K., Phillips C., The ecology, epidemiology and virulence of Enterococcus. Microbiology 155, 1749–1757 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Alfonso-Prieto M., Biarnés X., Vidossich P., Rovira C., The molecular mechanism of the catalase reaction. J. Am. Chem. Soc. 131, 11751–11761 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Khademian M., Imlay J. A., Escherichia coli cytochrome c peroxidase is a respiratory oxidase that enables the use of hydrogen peroxide as a terminal electron acceptor. Proc. Natl. Acad. Sci. U.S.A. 114, E6922–E6931 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohanty J. G., Jaffe J. S., Schulman E. S., Raible D. G., A highly sensitive fluorescent micro-assay of H2O2 release from activated human leukocytes using a dihydroxyphenoxazine derivative. J. Immunol. Methods 202, 133–141 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Ezraty B., Henry C., Hérisse M., Denamur E., Barras F., Commercial Lysogeny Broth culture media and oxidative stress: A cautious tale. Free Radical Biol. Med. 74, 245–251 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Li X., Imlay J. A., Improved measurements of scant hydrogen peroxide enable experiments that define its threshold of toxicity for Escherichia coli. Free Radical Biol. Med. 120, 217–227 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ochsner U. A., Vasil M. L., Alsabbagh E., Parvatiyar K., Hassett D. J., Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA Repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J. Bacteriol. 182, 4533–4544 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cosgrove K., et al. , Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J. Bacteriol. 189, 1025–1035 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seaver L. C., Imlay J. A., Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183, 7173–7181 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salunkhe P., Töpfer T., Buer J., Tümmler B., Genome-wide transcriptional profiling of the steady-state response of Pseudomonas aeruginosa to hydrogen peroxide. J. Bacteriol. 187, 2565–2572 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng M., Åslund F., Storz G., Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279, 1718–1722 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Åslund F., Zheng M., Beckwith J., Storz G., Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol—disulfide status. Proc. Natl. Acad. Sci. U.S.A. 96, 6161–6165 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Storz G., Imlayt J. A., Oxidative stress. Curr. Opin. Microbiol. 2, 188–194 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Panmanee W., Hassett D. J., Differential roles of OxyR-controlled antioxidant enzymes alkyl hydroperoxide reductase (AhpCF) and catalase (KatB) in the protection of Pseudomonas aeruginosa against hydrogen peroxide in biofilm vs. planktonic culture. FEMS Microbiol. Lett. 295, 238–244 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buvelot H., et al. , Hydrogen peroxide affects growth of S. aureus through downregulation of genes involved in pyrimidine biosynthesis. Front. Immunol. 12, 673985 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quillin S. J., Hockenberry A. J., Jewett M. C., Seifert H. S., Neisseria gonorrhoeae exposed to sublethal levels of hydrogen peroxide mounts a complex transcriptional response. mSystems 3, e00156–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang W., Small D. A., Toghrol F., Bentley W. E., Microarray analysis of Pseudomonas aeruginosa reveals induction of pyocin genes in response to hydrogen peroxide. BMC Genomics 6, 115 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng M., et al. , DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183, 4562–4570 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forman H. J., Bernardo A., Davies K. J. A., What is the concentration of hydrogen peroxide in blood and plasma? Arch. Biochem. Biophys. 603, 48–53 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Abdekhodaie M. J., Cheng J., Wu X. Y., Effect of formulation factors on the bioactivity of glucose oxidase encapsulated chitosan–alginate microspheres: In vitro investigation and mathematical model prediction. Chem. Eng. Sci. 125, 4–12 (2015). [Google Scholar]
- 38.Sakariassen K. S., Orning L., Turitto V. T., The impact of blood shear rate on arterial thrombus formation. Future Sci. OA 1, FSO30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horsburgh M. J., Clements M. O., Crossley H., Ingham E., Foster S. J., PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 69, 3744–3754 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee J.-W., Helmann J. D., The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440, 363–367 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Vollrath M. A., Kwan K. Y., Corey D. P., The micromachinery of mechanotransduction in hair cells. Annu. Rev. Neurosci. 30, 339–365 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansen C. E., Qiu Y., McCarty O. J. T., Lam W. A., Platelet mechanotransduction. Annu. Rev. Biomed. Eng. 20, 253–275 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Stone H. A., Stroock A. D., Ajdari A., Engineering flows in small devices: Microfluidics toward a lab-on-a-chip. Annu. Rev. Fluid Mech. 36, 381–411 (2004). [Google Scholar]
- 44.Siple P. A., Passel C. F., Measurements of dry atmospheric cooling in subfreezing temperatures. Proc. Am. Philosophical Soc. 89, 177–199 (1945). [DOI] [PubMed] [Google Scholar]
- 45.Helmann J. D., et al. , The global transcriptional response of Bacillus subtilis to peroxide stress is coordinated by three transcription factors. J. Bacteriol. 185, 243–253 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Padron G. C., Fluorescence intensity quantification script. Github. https://github.com/padrong/ImageAnalysis-Adaptations. Deposited 23 February 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix. The raw data and code used to support this study are available in Github (https://github.com/padrong/ImageAnalysis-Adaptations) (46).