In her letter (1), Miglietta warns about the limitations in our knowledge of the biology of species in the Turritopsis genus and inquires about how those limitations affect our manuscript (2).
First, we state precisely throughout the manuscript “no reported evidence of postreproductive rejuvenation” in Turritopsis rubra—we use the expression “incapable of rejuvenation” once, and only because of space restrictions. The literature Miglietta mentioned refers to “subadult” (not mature) medusae (3, 4) or to undefined Turritopsis species (5). We cited Li et al. (6) because it is the only source that mentions explicitly T. rubra and its rejuvenation rate in the discussion. In addition, we explored ontogeny reversal to T. rubra medusae, and rejuvenation capabilities of both species after reproduction are clearly different (Fig. 1). In view of these data, T. rubra is unlikely to retain a functioning genomic complement for postreproductive rejuvenation. Excluding any possibility of sporadic postreproductive reversion in T. rubra is beyond the aims of this manuscript and irrelevant to our conclusions.
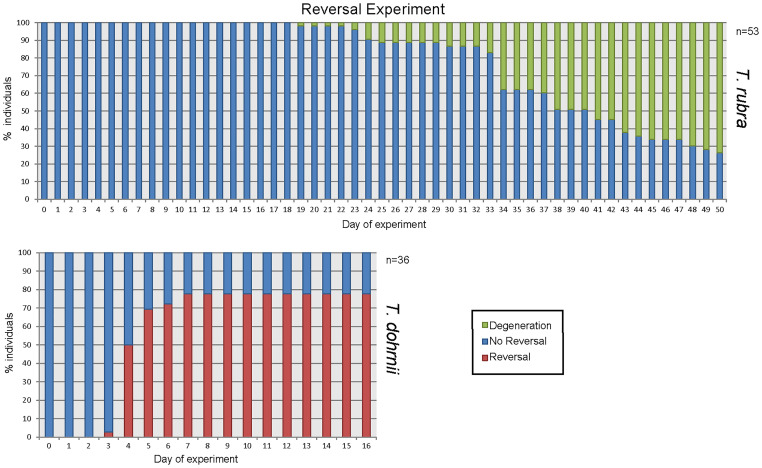
Fig. 1.
Results of ontogeny reversal trial in 53 T. rubra medusae induced by mechanical damage: None of them reverted and 74% degenerated by day 50 of the experiment (Top). These results, which were not included in the manuscript, are compared with life cycle reversal experiment of T. dohrnii (Bottom). We considered “reversal” once the sample reached the early stolon stage and “degeneration” when tissues were clearly degraded and did not recover from the stress challenge.
Our manuscript does not contain any claim that our transcriptomic experiment is novel, although we do state that underexpression of Polycomb repressive complex targets was previously undescribed in jellyfish. We appreciate all previous transcriptomic works, and we cite Matsumoto et al. (7) in supplementary material. However, the goal of our work was not to discuss the transcriptome in detail but to complement our genomic analysis. In addition, these studies differ from ours in that they compare only three stages, and they just use the Gene Ontology database for Gene set enrichment analysis (7, 8). Thus, any comparison with our work would have been inconsistent.
Regarding reversal, four samples analyzed had reverted spontaneously. We would expect this heterogeneity to slightly decrease sensitivity of the analysis and to increase robustness of the results, i.e., the highlighted expression differences are consistent independently of how the reversal starts. Importantly, transcription patterns of “spontaneous” and “CsCl” samples were similar [(2); SI Appendix, Fig. S2C].
As Miglietta indicates, whether “no-reversal” individuals would reverse later is difficult to know. By defining a priori the “no-reversal” group for “individuals that had not accomplished reversal at the end of the experiment,” we avoided any bias. Consistent with our hypothesis, “no reversal” and “medusa” stages had similar transcriptomes [(2); SI Appendix, Fig. S2C], therefore, they were unlikely to be in a reversal process.
Finally, we agree with Miglietta about the importance of species identification in understudied nonmodel organisms. In this sense, we are confident about the identity of the species studied since we sequenced the mitochondrial 16S gene prior to genome annotation. The results can now be found at https://github.com/vqf/turritopsis (2).
We are happy to exercise transparency in our work, and we would appreciate the same level of courtesy and good faith with which we address our peers. In summary, our conclusions are well founded: All the limitations outlined by Dr. Miglietta had been considered in our work, which we publish in the hope that it may help the study of Turritopsis species and the aging process in general.
Acknowledgments
Author contributions
M.P.-T. and V.Q. wrote the paper.
Competing interests
The authors declare no competing interest.
Contributor Information
Maria Pascual-Torner, Email: mariapascual1.618@gmail.com.
Víctor Quesada, Email: quesadavictor@uniovi.es.
References
- 1.Miglietta M. P., On the perils of working on non-model organisms.
- 2.Pascual-Torner M., et al. , Comparative genomics of mortal and immortal cnidarians unveil novel keys behind rejuvenation. Proc. Natl. Acad. Sci. U.S.A. 119, e2118763119 (2022), 10.1073/pnas.2118763119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kubota S., Difference of rejuvenation rate among three species of Turritopsis (Hydrozoa, Anthomedusae) from Japan. Bull. Biogeograph. Soc. Jpn. 68, 139–142 (2013). [Google Scholar]
- 4.Kubota S., Distinction of two morphotypes of Turritopsis nutricula medusae (Cnidaria, Hydrozoa, Anthomedusae) in Japan, with reference to their different abilities to revert to the hydroid stage and their distinct geographical distributions. Biogeography 7, 41–50 (2005). [Google Scholar]
- 5.Kubota S., Rejuvenation of senile medusa of Turritopsis sp. (Hydrozoa, Anthomedusae). Bull. Biogeogr. Soc. Jpn. 70, 189–191 (2015). [Google Scholar]
- 6.Li J.-Y., Guo D.-H., Wu P.-C., He L.-S., Ontogeny reversal and phylogenetic analysis of Turritopsis sp.5 (Cnidaria, Hydrozoa, Oceaniidae), a possible new species endemic to Xiamen, China. PeerJ. 6, e4225 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsumoto Y., Piraino S., Miglietta M. P., Transcriptome characterization of reverse development in Turritopsis dohrnii (Hydrozoa, Cnidaria). G3 9, 4127–4138 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsumoto Y., Miglietta M. P., Cellular reprogramming and immortality: Expression profiling reveals putative genes involved in Turritopsis dohrnii’s life cycle reversal. Genome Biol. Evol. 13, 136 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]