Significance
Pathogens and their hosts are engaged in a molecular arms race whereby both parties continually develop new strategies for infection and immunity. Host immune surveillance systems reflect an important battleground in these arms races as the evolution of microbial factors that interfere with host sensors enables pathogens to modulate a network of inducible antimicrobial defenses. Here, it was found that the intracellular bacterial pathogen Coxiella burnetii blocks induction of antimicrobial responses by the immune sensor RIG-I. The C. burnetii protein EmcB was identified as a ubiquitin-specific protease that deactivates RIG-I to interfere with the host immune response. These data implicate a role for RIG-I in antibacterial defense and identify a mechanism by which C. burnetii interferes with this immunosurveillance pathway.
Keywords: bacterial effectors, type IV secretion, innate immunity
Abstract
Eukaryotes have cytosolic surveillance systems to detect invading microorganisms and initiate protective immune responses. In turn, host-adapted pathogens have evolved strategies to modulate these surveillance systems, which can promote dissemination and persistence in the host. The obligate intracellular pathogen Coxiella burnetii infects mammalian hosts without activating many innate immune sensors. The Defect in Organelle Trafficking/Intracellular Multiplication (Dot/Icm) protein secretion system is necessary for C. burnetii to establish a vacuolar niche inside of host cells, which sequesters these bacteria in a specialized organelle that could evade host surveillance systems. However, bacterial secretion systems often introduce agonists of immune sensors into the host cytosol during infection. For instance, nucleic acids are introduced to the host cytosol by the Dot/Icm system of Legionella pneumophila, which results in type I interferon production. Despite host infection requiring a homologous Dot/Icm system, C. burnetii does not induce type I interferon production during infection. Here, it was found that type I interferons are detrimental to C. burnetii infection and that C. burnetii blocks type I interferon production mediated by retionic acid inducible gene I (RIG-I) signaling. Two Dot/Icm effector proteins, EmcA and EmcB, are required for C. burnetii inhibition of RIG-I signaling. EmcB is sufficient to block RIG-I signaling and is a ubiquitin-specific cysteine protease capable of deconjugating ubiquitin chains from RIG-I that are necessary for signaling. EmcB preferentially cleaves K63-linked ubiquitin chains of three or more monomers, which represent ubiquitin chains that potently activate RIG-I signaling. Identification of a deubiquitinase encoded by C. burnetii provides insights into how a host-adapted pathogen antagonizes immune surveillance.
Coxiella burnetii is an obligate intracellular bacterial pathogen that manipulates eukaryotic signaling pathways to establish a vacuolar niche within the cytosol of mammalian host cells (1, 2). Intracellular replication of C. burnetii requires a specialized type IVB protein secretion apparatus called the Dot/Icm system, along with an arsenal of over 130 effector proteins that are translocated into the host cytosol (3, 4). These Dot/Icm effectors target mammalian signaling pathways to create a replication-permissive vacuole (5–8). However, the biochemical and molecular function of most Dot/Icm effectors remains unknown.
Host immune surveillance systems detect nonself molecules through sensors called pattern recognition receptors (PRRs) (9–13). Insight into a possible role for Dot/Icm effector proteins of C. burnetii may be gleaned from other host-adapted intracellular bacterial pathogens that encode specialized protein secretion systems and repertoires of effectors with sophisticated methods of subverting these receptors. Antagonism of PRRs by pathogen-encoded effectors is often necessary to maintain a replication-permissive niche within the host (11). Whether C. burnetii has effectors that interfere specifically with PRR function is not clear.
Upon infection of host cells, C. burnetii does not trigger robust immune activation (14–16). This is in contrast to the related pathogen Legionella pneumophila, which requires a homologous Dot/Icm secretion apparatus to replicate in eukaryotic host cell, and stimulates multiple innate immune pathways regulated by cytosolic sensors. For example, the double-stranded DNA (dsDNA) sensor cGAS is robustly activated upon infection of mammalian host cells by L. pneumophila, and there is also evidence that L. pneumophila is capable of activating the double-stranded RNA (dsRNA) sensing RIG-I-like receptors (RLRs), RIG-I and melanoma differentiation-associated gene 5 (MDA5) (17–21). Activation of these nucleic acid-sensing pathways results in the production of type I interferons (IFNs), which are cytokines that activate cell-autonomous defense pathways capable of restricting intracellular replication of L. pneumophila (17–20, 22). Importantly, the detection of L. pneumophila by most cytosolic innate immune sensors, including the nucleic acid sensors, requires a functional Dot/Icm system (17–19, 23, 24). Indeed, there are numerous examples of specialized bacterial protein secretion systems introducing agonists of immune sensors into the cytosol of infected cells and activating antimicrobial defenses (24–31). Intriguingly, C. burnetii infection does not induce type I IFN production (15, 16), raising the possibility that there may be pathogen-encoded mechanisms to subvert detection by sensors that induce this response during infection. Moreover, although type I IFNs are induced during infection by many bacterial pathogens, these cytokines have typically been considered an “antiviral” defense, and their role in most bacterial infections remains poorly understood (32). Thus, the goal of this study was to investigate the role type I IFN may play during C. burnetii infection and identify mechanisms by which C. burnetii may manipulate type I IFN production.
Results
The Dot/Icm Effectors EmcA and EmcB Are Necessary for Inhibition of RIG-I Signaling by C. burnetii.
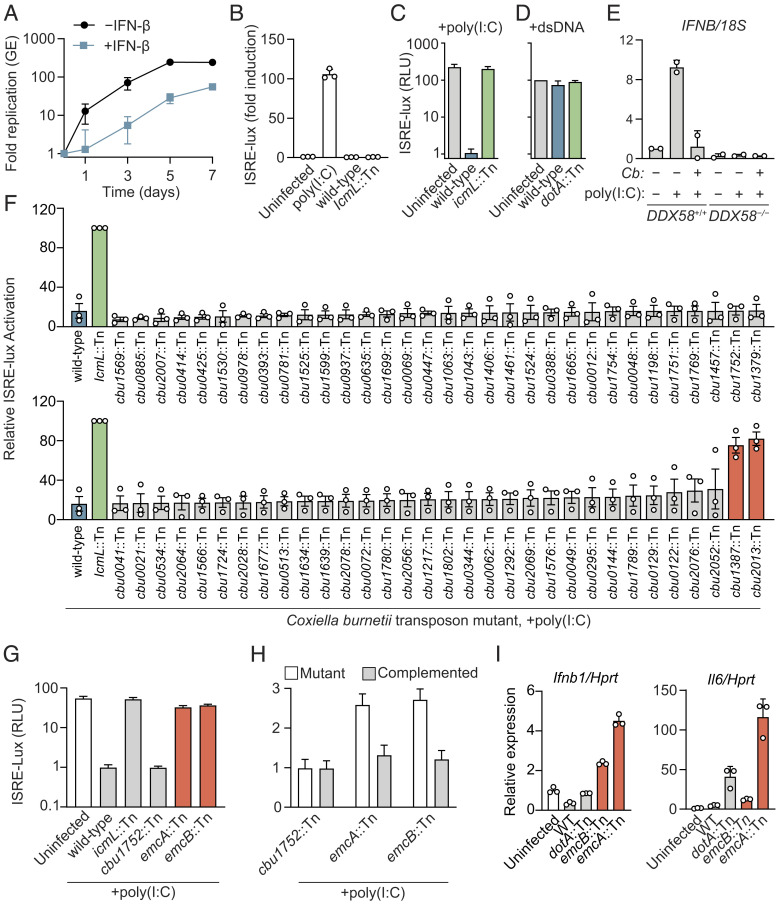
Type I IFNs are induced by a diverse array of bacterial pathogens; however, their effect on the host antibacterial response remains unclear (32). To investigate the role of type I IFNs on C. burnetii infection, THP-1 macrophages were treated with IFN-β prior to infection with C. burnetii, and intracellular replication of C. burnetii was measured for 7 d. There was a significant decrease in C. burnetii replication in THP-1 cells pretreated with type I IFN as compared with untreated cells (Fig. 1A). Because C. burnetii does not induce type I IFN during infection (15), the question arose as to whether C. burnetii could interfere with signaling of PRRs coupled to this signaling response to inhibit type I IFN production.
Fig. 1.
C. burnetii inhibits RIG-I signaling by a Dot/Icm-dependent mechanism. (A) qPCR analysis of C. burnetii genome equivalents (GE) during infection of PMA-differentiated THP-1 pretreated with IFN-β or untreated infected at an MOI of 20. Data are mean ± SD of three independent experiments. P < 0.05 untreated vs. IFN-β at D3 to D7 by unpaired t test. (B) ISRE reporter activation by C. burnetii infection. Human Embryonic Kidney 293 (HEK293) ISRE-luciferase cells were infected for 48 h with C. burnetii at an MOI of 200 or transfected with poly(I:C). Data are mean ± SD of three technical replicates representative of two independent experiments. P < 0.01 uninfected vs. poly(I:C) by one-way ANOVA with Dunnett’s post hoc test. (C) ISRE reporter induction by poly(I:C). Cells were infected for 24 h with indicated strain at an MOI of 200, transfected with poly(I:C), and incubated an additional 24 h before luminescence was measured. Data are mean ± SD of five technical replicates representative of three independent experiments. P < 0.01 wild type (WT) Cb vs. uninfected and icmL::Tn, uninfected vs. icmL::Tn n.s., by one-way ANOVA with Tukey’s post hoc test. (D) ISRE reporter induction by cGAS-STING pathway activation. Cells were infected for 24 h with indicated strain at an MOI of 200, transfected with plasmids encoding cGAS and stimulator of Interferon genes (STING), and incubated an additional 24 h before luminescence was measured. Data are mean ± SD of two independent experiments. n.s. for uninfected vs. WT and dotA::Tn by one-way ANOVA with Tukey’s post hoc test. (E) qPCR analysis of interferon Beta gene (IFNB) messenger RNA (mRNA) in HEK293T RIG-I (DDX58) WT and KO cells. Cells were infected for 24 h with wild-type Cb at an MOI of 1,000, transfected with poly(I:C) and incubated an additional 24 h. Data are relative to uninfected DDX58+/+ without poly(I:C) and normalized to 18S. Data are mean ± SD of two independent experiments. P < 0.01 poly(I:C) + no Cb vs. all other conditions; all other comparisons, n.s. by two-way ANOVA with Tukey’s post hoc test. (F) Screen for C. burnetii transposon insertion mutants unable to block ISRE-luciferase production by poly(I:C). HEK293-ISRE cells were infected at an multiplicity of infection (MOI) of 200 for 24 h before transfection with poly(I:C) for 24 h. Data are mean ± SD of three independent experiments. P < 0.0001 for WT vs. icmL::Tn, cbu1387::Tn and cbu2013::Tn; WT vs. all others n.s. by one-way ANOVA with Dunnett post hoc test. (G) ISRE reporter induction by poly(I:C) in cells infected with indicated C. burnetii mutant. Data are mean ± SD of five replicates representative of two independent experiments. P < 0.01 for WT vs. cbu1387::Tn, cbu2013::Tn, icmL::Tn and uninfected; WT vs. cbu1752::Tn n.s. by one-way ANOVA with Tukey’s post hoc test. (H) ISRE reporter induction by poly(I:C) in cells infected with indicated C. burnetii mutant or complemented strain. Data are mean ± SD of 10 replicates representative of two independent experiments. P < 0.01 for cbu1387::Tn vs. cbu1387::Tn complement and cbu2013::Tn mutant vs. cbu2013::Tn complement by one-way ANOVA with Šídák’s post hoc test. (I) qPCR analysis of cytokine mRNA in J774A.1 cells infected with indicated C. burnetii strain for 4 d at an MOI of 500. Results are relative to those of uninfected J774A.1. P < 0.05 for WT vs. emcB::Tn and emcA::Tn Ifnb1; WT vs. emcB::Tn Il6 n.s., WT vs. emcA::Tn Il6 <0.05 by one-way ANOVA with Dunnett’s post hoc test.
To determine if the lack of type I IFN production observed during C. burnetii infection is due to antagonism of type I IFN inducing pathways, an infection-based luciferase reporter assay was used to test whether C. burnetii can block RLR or cGAS-STING signaling. HEK293 cells encoding luciferase under the transcriptional control of an interferon-stimulated response element (ISRE-luciferase) were used. Activation of cGAS or RLR signaling induces type I IFN production, which drives transcription of the luciferase gene. Consistent with prior reports that C. burnetii does not induce type I IFN production, HEK293 cells infected with isogenic strains of C. burnetii Nine Mile phase II RSA439 showed no induction of ISRE-luciferase irrespective of the presence of a functional Dot/Icm system (Fig. 1B). When the RIG-I pathway was induced by transfection of polyinosinic:polycytidylic acid [poly(I:C)], a robust ISRE-luciferase response was detected for uninfected cells and cells infected with a Dot/Icm-deficient mutant of C. burnetii; however, ISRE-luciferase production was significantly lower after poly(I:C) transfection of cells infected with the isogenic wild-type strain of C. burnetii (Fig. 1C). Wild-type C. burnetii was unable to suppress ISRE-luciferase production in a HEK293 cell system after the cGAS/STING pathway was induced by ectopic production of these signaling factors (Fig. 1D), which suggests that suppression of the poly(I:C)-induced ISRE-luciferase response is not the result of C. burnetii acting on a downstream component shared by both the RIG-I and cGAS/STING pathways. Levels of IFNB mRNA were measured to confirm that C. burnetii can suppress the RIG-I pathway during infection. Similar to ISRE-luciferase data, IFNB expression was induced upon poly(I:C) stimulation of uninfected cells, and this response was blocked in cells infected with wild-type C. burnetii (Fig. 1E). Induction of IFNB by poly(I:C) was not detected in RIG-I-deficient cells (DDX58−/−), which indicates that the poly(I:C) response requires RIG-I signaling (Fig. 1E). Thus, C. burnetii has the capacity to antagonize RIG-I-dependent signaling during infection by a process that requires the Dot/Icm system.
To identify C. burnetii effectors required for inhibition of the RIG-I pathway, a library of C. burnetii transposon insertion mutants deficient in single effector proteins (33) was screened for the ability to block poly(I:C)-induced activation of the ISRE-luciferase reporter. Two effector mutants, cbu1387::Tn and cbu2013::Tn, had defects in blocking RIG-I signaling (Fig. 1F). These effectors were named EmcA and EmcB, respectively (effector that modulates cytokine signaling A and B). The ability of the C. burnetii emcA::Tn and emcB::Tn strains to block poly(I:C)-induced ISRE signaling was complemented upon introduction of the intact gene into the mutant (Fig. 1H). Thus, EmcA and EmcB are Dot/Icm-translocated effector proteins necessary for C. burnetii inhibition of RIG-I signaling.
The C. burnetii emcA::Tn mutant has a defect in intracellular replication (33). The transposon insertion library that was screened contained multiple mutants with defects in intracellular replication (33); however, the C. burnetii emcA::Tn strain was the only mutant with an identified defect in intracellular replication that also had a defect in RIG-I inhibition. Although previous screens for genes required for intracellular replication had not identified the C. burnetii emcB::Tn mutant (7, 33), this mutant was compared with an isogenic wild-type strain to test for any subtle replication defects. No measurable intracellular replication defect was detected for the emcB::Tn mutant in THP-1 cells (SI Appendix, Fig. S1A). Last, the C. burnetii cbu1752::Tn mutant has an intracellular replication defect comparable to the emcA::Tn mutant but retained the ability to efficiently suppress poly(I:C)-induced ISRE reporter activation (Fig. 1G), which indicates that intracellular replication is not necessary to suppress RIG-I signaling during infection.
Higher levels of Ifnb1 expression were detected in J774A.1 macrophage-like cells infected with the emcA::Tn mutant or the emcB::Tn mutant when compared with cells infected with wild-type C. burnetii (Fig. 1I), which suggests these effectors may interfere with type I IFN production induced by an endogenous agonist during infection. There was higher Il6 expression in cells infected with the EmcA::Tn mutant compared with cells infected with either wild-type C. burnetii or the EmcB::Tn mutant (Fig. 1I), which suggests that EmcA and EmcB have overlapping functions in terms of inhibiting the RIG-I pathway but may employ different mechanisms to block a host response to infection.
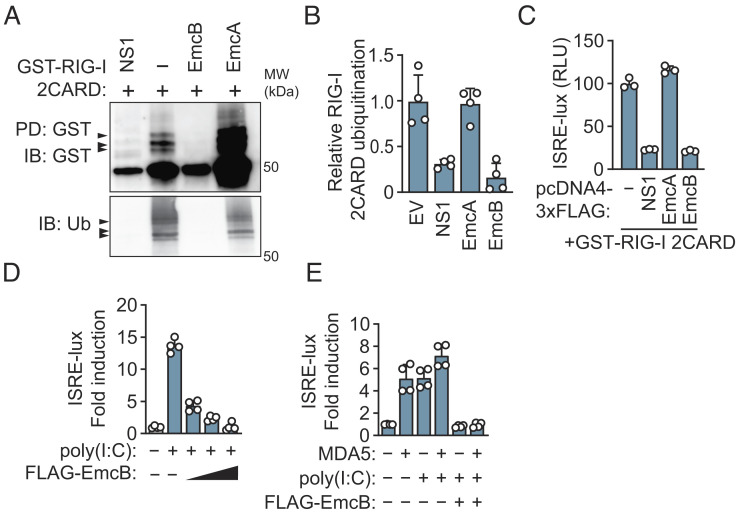
EmcB Is Sufficient to Block MDA5 and RIG-I by Inhibition of Ubiquitination.
RIG-I signaling requires ubiquitination of RIG-I after RNA detection (34). Many viral pathogens interfere with RIG-I signaling by modulating RIG-I ubiquitination. For example, the NS1 protein of Influenza A inhibits ubiquitination of RIG-I, and Open Reading Frame 64 protein (ORF64) of Kaposi Sarcoma Herpes Virus is a deubiquitinase that removes ubiquitin chains from RIG-I (35, 36). To test whether EmcA or EmcB could block ubiquitination of RIG-I, FLAG-tagged EmcA or EmcB were coexpressed in HEK293T cells with glutathione S-transferase (GST)-tagged RIG-I 2CARD (tandem caspase activation and recruitment domain). RIG-I 2CARD is a constitutively-active form of RIG-I that forms signaling-competent oligomers in the absence of dsRNA (37). RIG-I filaments are recognized by an E3 ubiquitin ligase and polyubiquitinated, which is necessary to nucleate oligomerization of the adaptor mitochondrial antiviral-signaling protein (MAVS) to induce transcription of interferon-stimulated genes and produce type I IFNs (34, 38–46). Isolation of GST-RIG-I 2CARD from HEK293T cells revealed polyubiquitinated species that migrated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) as multiple higher molecular weight bands (Fig. 2A). As expected, production of the Influenza A NS1 protein prevented the ubiquitination of GST-RIG-I 2CARD (Fig. 2A). Production of FLAG-EmcA did not interfere with ubiquitination of GST-RIG-I 2CARD; however, no ubiquitinated GST-RIG-I 2CARD was detected in cells producing FLAG-EmcB (Fig. 2 A and B). FLAG-EmcA and FLAG-EmcB were produced in similar amounts in the transfected cells (SI Appendix, Fig. S2A). Moreover, ectopic expression of FLAG-EmcB, but not FLAG-EmcA, reduced GST-RIG-I 2CARD-mediated activation of the ISRE-luciferase reporter to a similar level as cells producing NS1 protein (Fig. 2C). FLAG-EmcB also reduced GST-RIG-I 2CARD-mediated activation of a luciferase reporter fused to the IFNB promoter (SI Appendix, Fig. S2B), and blocked poly(I:C)-mediated ISRE-luciferase activation in a dose-dependent manner (Fig. 2D). Because RIG-I 2CARD is ubiquitinated by tripartite motif containing protein 25 (TRIM25), whereas, really interesting gene [RING] finger protein leading to RIG-I activation (RIPLET) (RNF135) is the E3 ligase that ubiquitinates full-length RIG-I (38–42, 47), the ability of EmcB to deubiquitinate full-length RIG-I was tested. These data demonstrated a defect in K63-linked ubiquitination of full-length GST-RIG-I purified under activating conditions in cells coexpressing FLAG-EmcB (SI Appendix, Fig. S2 C and D). Thus, EmcB is sufficient to block RIG-I signaling downstream of dsRNA recognition by influencing RIG-I ubiquitination.
Fig. 2.
EmcB is sufficient to inhibit RLR signaling and prevents ubiquitination of RIG-I 2CARD. (A) Western blot analysis of GST-RIG-I 2CARD ubiquitination. GST-RIG-I 2CARD affinity purified from HEK293T cells coexpressing indicated protein for 48 h. Blot representative of four independent experiments. (B) Densitometric analysis of ubiquitination of GST-RIG-I 2CARD from A. P < 0.01 for − vs. NS1, − vs. EmcB, n.s. for − vs. EmcA by one-way ANOVA with Tukey’s post hoc test. (C) ISRE-luciferase signal from HEK293T cells transfected with GST-RIG-I 2CARD and indicated construct for 60 h. Luciferase signal normalized to no-GST-RIG-I 2CARD condition. Data are mean ± SD of three technical replicates representative of three independent experiments. P < 0.01 for − vs. NS1, − vs. EmcB; NS1 vs. EmcB n.s. by one-way ANOVA with Tukey’s post hoc test. (D) ISRE-luciferase signal from HEK293T cells transfected with increasing amounts of indicated pcDNA4 construct and poly(I:C). Luciferase signal normalized to no-poly(I:C) condition. Data are mean ± SD of four technical replicates representative of two independent experiments. Comparison n.s. for poly(I:C) vs. poly(I:C) + EmcB high by two-way ANOVA with Tukey’s post hoc test. (E) ISRE-luciferase signal from cells transfected with pcDNA4-FLAG-EmcB, poly(I:C) and MDA5 for 60 h. Data are mean ± SD of four technical replicates representative of two independent experiments. P < 0.0001 for poly(I:C), MDA5 vs. poly(I:C), MDA5, EmcB by two-way ANOVA with Tukey’s post hoc test.
RIG-I belongs to a family of RNA helicases that includes MDA5, which is also a sensor of cytosolic dsRNA. MDA5 and RIG-I share a similar architecture with a N-terminal tandem CARD domain and helicase RNA-binding domain. Importantly, both require polyubiquitination to signal through the shared adaptor MAVS (48). Cotransfection of MDA5 with poly(I:C) was sufficient to induce ISRE luciferase production, which was reduced by coexpression of EmcB (Fig. 2E). Thus, EmcB is sufficient to block signaling by both RIG-I and MDA5.
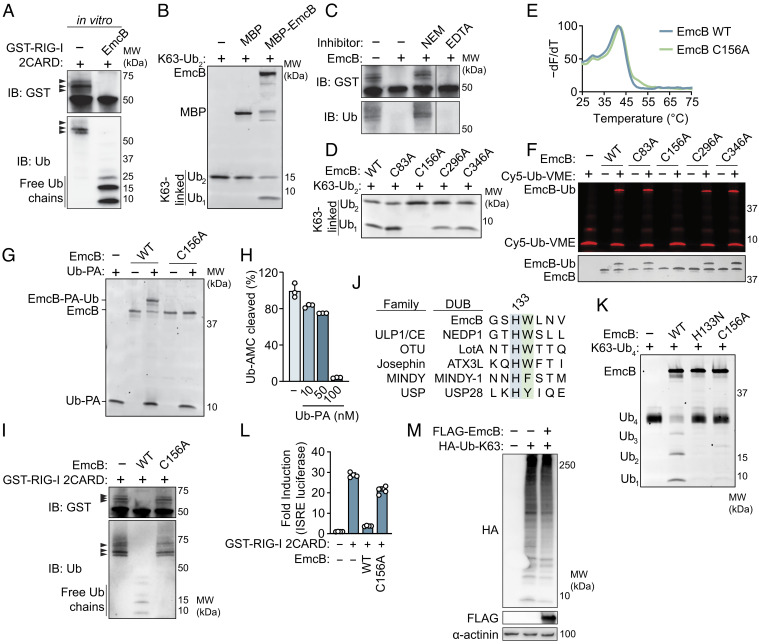
EmcB Is a Cysteine Protease That Deconjugates Ubiquitin from RIG-I.
These data suggested that EmcB could inhibit RIG-I signaling either by interfering with the ubiquitination of RIG-I or by deconjugating ubiquitin chains attached to RIG-I. To better understand how EmcB interferes with RIG-I signaling, ubiquitinated GST-RIG-I 2CARD was isolated from HEK293 cells and incubated with purified maltose-binding protein (MBP)-EmcB. This resulted in the removal of ubiquitin from GST-RIG-I 2CARD and the accumulation of free monomeric and dimeric ubiquitin (Fig. 3A). The accumulation of monoubiquitin suggested that EmcB might function as a deubiquitinase. The generation of K63-linked ubiquitin on RIG-I is necessary for signaling through MAVS (38, 42, 48–50). Thus, the ability of EmcB to cleave K63-linked ubiquitin was tested in vitro. Purified MBP-EmcB cleaved K63-linked di-ubiquitin into monoubiquitin (Fig. 3B), which indicates that the EmcB is a ubiquitin protease capable of interfering with RIG-I signaling by deconjugation of K63-linked ubiquitin chains.
Fig. 3.
EmcB is a cysteine protease that deconjugates ubiquitin chains from RIG-I. (A) Western blot analysis of in vitro reconstitution of EmcB-mediated deubiquitylation of RIG-I. Purified GST-RIG-I 2CARD (~200 μM) was incubated with recombinant EmcB (~40 μM) for 1 h. Blot representative of three independent experiments. (B) Reconstitution of EmcB DUB activity. Purified K63-linked di-ubiquitin (0.5 μM) was incubated with MBP, MBP-EmcB (1 μM) or buffer for 1 h. Gels were imaged with SYPRO Ruby. Data are representative of three independent replicates. (C) Western blot of in vitro RIG-I 2CARD deubiquitination of EmcB with protease inhibitors. Purified, ubiquitinated GST-RIG-I 2CARD (~200 μM) was coincubated with purified EmcB (~40 μM) for 1 h. EmcB was preincubated with 100 mM NEM, 20 mM etheylenediaminetetraacetic acid (EDTA) or buffer. Blot representative of two independent experiments. (D) SDS-PAGE analysis of in vitro deubiquitinase assay. Purified K63-linked diubiquitin chain (0.5 μM) was incubated with indicated EmcB protein (0.5 μM), resolved on an SDS-PAGE and stained with SYPRO Ruby. (E) Thermal denaturation assay of purified EmcB. Protein was incubated with SYPRO Orange and SYBR fluorescence was measured over increasing temperature to determine melting point of each protein. Data are representative of three independent experiments. Unpaired t test of WT EmcB vs. C156A Tm n.s. (F) SDS-PAGE analysis of activity-based, cysteine deubiquitinase probe reactions. Cy5-Ubiquitin-vinylmethylester (Cy5-Ub-VME) (2.4 μM) was incubated with EmcB (0.6 μM) for 1 h. Gel was imaged in Cy5 channel (Top) and after silver staining (Bottom). Gel representative of two independent experiments. (G) SDS-PAGE analysis of Ubiquitin-propargylamine (Ub-PA) covalent modification of EmcB; 2.4 μM Ub-PA or buffer was incubated for 1 h with 0.6 μM of indicated EmcB. Representative of two independent experiments. (H) Inhibition of EmcB DUB activity by Ub-PA; 2 nM EmcB was preincubated for 10 min with indicated concentration of Ub-PA before coincubation with Ubiquitin-AMC (250 nM), a fluorogenic DUB activity substrate. Cleavage dequenches AMC, a fluorescent molecule. Relative Fluorescence units (RFU) was measured after 1 h. Data are mean ± SD of three technical replicates representative of two independent experiments. P < 0.01 for mock vs. 10 nM, mock vs. 50 nM, mock vs. 100 nM, 10 nM vs. 100 nM, and 50 nM vs. 100 nM by one-way ANOVA with Tukey’s post hoc test. (I) Alignment of catalytic histidine (blue) of representative deubiquitinases from five families with histidine-tryptophan dyad of EmcB. The flanking aromatic residue that imbues substrate specificity for C-terminal di-glycine is green. (J) SDS-PAGE analysis of tetraubiquitin cleavage assay. Indicated EmcB construct (0.3 μM) was incubated with K63-linked tetraubiquitin (1 μM) for 1 h. Representative Sypro Ruby stained SDS-PAGE from two independent experiments. (K) Western blot analysis of purified, ubiquitinated RIG-I 2CARD with EmcB. Affinity purified GST-RIG-I 2CARD (~200 μM) was incubated with indicated EmcB (40 μM) for 1 h. Blot representative of two independent experiments. (L) ISRE-luciferase in cells expressing catalytically inactive EmcB. HEK293T cells were transfected with GST-RIG-I 2CARD and indicated EmcB construct for 60 h. Luciferase signal was normalized to untreated cells. Data are mean ± SD of four technical replicates representative of three independent experiments. P < 0.0001 for EmcB vs. EmcB C156A by one-way ANOVA with Tukey’s post hoc test. (M) Western blot analysis of global K63-linked ubiquitin in cells producing FLAG-EmcB. Cells expressing vector or EmcB, and human influenza hemagglutinin (HA)-tagged K63-only Ubiquitin for 48 h. Blot representative of three independent experiments.
Deubiquitinases are divided into two major biochemical classes—metalloproteases and cysteine proteases. Preincubation of EmcB with the cysteine protease inhibitor N-ethylmaleimide (NEM) blocked the deconjugation of ubiquitin from RIG-I 2CARD (Fig. 3C). In contrast, the addition of the divalent cation chelator EDTA did not alter the ability of EmcB to deconjugate ubiquitin from RIG-I 2CARD (Fig. 3C). These data suggest EmcB is a cysteine protease. No amino acid sequence or predicted structural similarities were observed between EmcB and other cysteine proteases. Thus, to identify a potential catalytic cysteine residue in EmcB, the four cysteines in this protein were individually changed to alanine. Each EmcB mutant was tested for ubiquitin protease activity, and Cys156 was found to be the only cysteine required for EmcB ubiquitin protease activity (Fig. 3D). The EmcB C156A protein displayed similar thermal denaturation properties when compared with wild-type EmcB (Tm 41.3 °C ± 0.3 vs. 41.7 °C ± 0.6) and eluted similarly by size exclusion chromatography (Fig. 3E and SI Appendix, Fig. S3A), which indicates mutant EmcB C156A protein folds similarly to wild-type protein. Thus, the Cys156 residue is likely to be the catalytic cysteine required for EmcB ubiquitin protease activity.
Activity-based probes were used to confirm that Cys156 is the catalytic cysteine required for EmcB ubiquitin protease activity. Incubation of EmcB with the specific, irreversible activity-based cysteine deubiquitinase targeting probe ubiquitin-vinyl methyl ester (Cy5-Ub-VME) (51) resulted in the covalent attachment of the probe to EmcB, and this covalent modification required Cys156 (Fig. 3F). In addition, a chemically unique cysteine deubiquitinase activity-based protein profiling probe that covalently modifies catalytic cysteines, ubiquitin-propargylamine (Ub-PA) (52, 53), modified EmcB but not EmcB C156A (Fig. 3G). Preincubation of EmcB with Ub-PA inhibited EmcB activity as measured by cleavage of a fluorogenic ubiquitin hydrolase activity probe (Ub-AMC) (Fig. 3H). Importantly, the cysteine protease activity of EmcB was required to deconjugate ubiquitin chains from RIG-I 2CARD (Fig. 3I).
Cysteine deubiquitinases contain conserved catalytic histidine-cysteine dyads and sometimes an additional acidic residue that hydrogen bonds with the basic residue. In all families of cysteine ubiquitin proteases, the catalytic histidine is followed by a bulky aromatic residue that maintains active site specificity for the penultimate residue of ubiquitin (G75), as R groups of any other amino acid would sterically clash with the aromatic residue, thus preventing cleavage of nonspecific substrates (Fig. 3J). In contrast, cysteine proteases closely related to deubiquitinases (DUBs) that cleave substrates lacking a C-terminal di-glycine lack this hydrophobic residue, which accommodates substrate binding within the active site pocket (54). EmcB contains a single histidine-tryptophan motif (HW133-134), and mutation of this histidine to asparagine resulted in a loss of deubiquitinase activity (Fig. 3K), consistent with EmcB being a ubiquitin-specific cysteine protease.
To test whether the proteolytic activity of EmcB is necessary to inhibit RIG-I signaling, cells expressing wild-type EmcB or EmcB C156A were transfected with GST-RIG-I 2CARD, and ISRE-luciferase reporter activity was measured. Cells expressing EmcB C156A exhibited a strong ISRE luciferase response compared with cells expressing wild-type EmcB, which indicates that ubiquitin protease activity is necessary for inhibition of RIG-I signaling (Fig. 3L). Importantly, production of FLAG-EmcB in HEK293T cells did not significantly alter the global abundance of K63-linked ubiquitin conjugates in the cell (Fig. 3M), which indicates that EmcB does not have promiscuous deubiquitinase activity when expressed ectopically in cells.
EmcB Preferentially Cleaves Long Ubiquitin Chains.
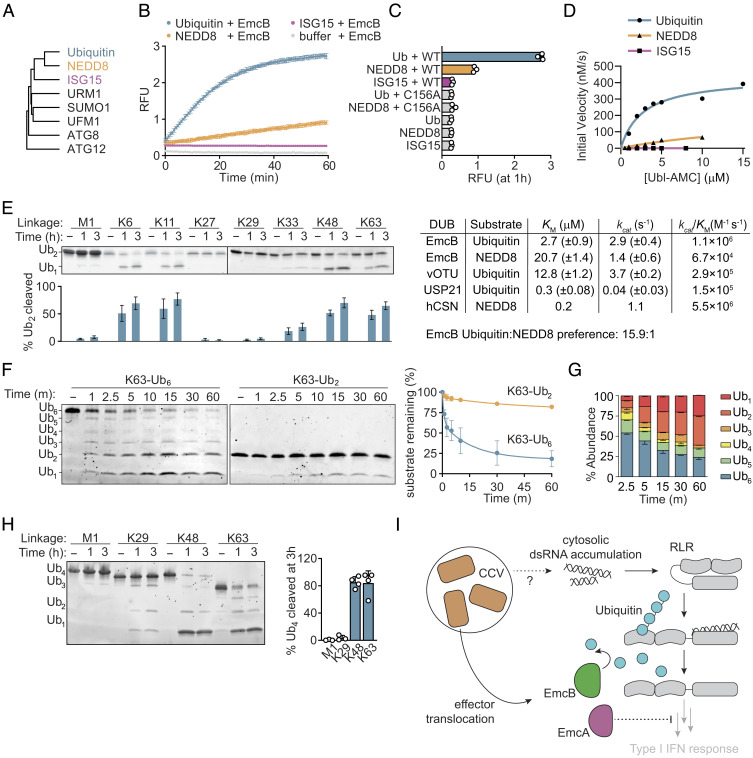
Ubiquitin is a member of a diverse family of widespread eukaryotic posttranslation modifiers that share a conserved beta-grasp fold and regulate diverse cellular activities (Fig. 4A) (55, 56). Notably, ubiquitin is the only one of these modifiers necessary for RIG-I activation. To better understand the ability of EmcB to target RIG-I, the breadth of ubiquitin-like modifiers cleaved by EmcB was measured with an in vitro protease activity assay using ubiquitin-like modifiers conjugated to the fluorophore 7-amido-4-methylcoumarin (AMC) as substrates. Cleavage of AMC from the ubiquitin-like protein results in fluorescence of the probe. EmcB cleaved Ubiquitin-AMC efficiently and had weak activity on the most closely related ubiquitin-like modifier neural precursor cell-expressed developmentally down-regulated 8 (NEDD8)-AMC (~60% identity to ubiquitin), but not interferon-stimulated gene 15 (ISG15)-AMC (~30% identity to ubiquitin) (Fig. 4B). Cleavage of Ubiquitin-AMC or NEDD8-AMC by the mutant EmcB C156A was not detectable, which demonstrates that activity is dependent on the catalytic cysteine in EmcB (Fig. 4C). The rate of NEDD8-AMC cleavage by EmcB was slow compared with cleavage of Ubiquitin-AMC (Fig. 4B). Consistent with this, EmcB exhibited more than a 10-fold greater preference toward ubiquitin compared with NEDD8 (Fig. 4D). Thus, ubiquitin is the preferred substrate for EmcB cleavage.
Fig. 4.
EmcB is a ubiquitin-specific protease that efficiently cleaves long ubiquitin chains. (A) Phylogenetic tree of ubiquitin-like modifiers. Tree was rooted on autophagy related gene 12 (ATG12). (B) Fluorometric cleavage assay of C-terminal ubiquitin-like AMC conjugates. Ub-AMC, NEDD8-AMC, and ISG15-AMC (400 nM) were incubated with 3.5 nM EmcB, and RFU was measured. Cleavage allows fluorescence of AMC. Data are mean ± SD of three technical replicates representative of two independent experiments. (C) Fluorometric cleavage assay of Ubiquitin and ubiquitin-like modifier-7-Amino-4-methylcoumarin (UBL-AMC) conjugates. RFU measurement at 1 h of incubation of indicated AMC conjugate with indicated protein. Data are mean ± SD representative of two independent experiments. P < 0.0001 for Ub vs. EmcB, Ub EmcB vs. Ub EmcB C156A, NEDD8 mock vs. NEDD8 EmcB; Ub vs. Ub EmcB C156A, NEDD8 mock vs. NEDD8 EmcB C156, ISG15 mock vs. ISG15 EmcB all n.s. by one-way ANOVA with Dunnett correction. (D) Kinetics of EmcB protease activity toward ubiquitin-like AMC conjugates; 150 nM EmcB was incubated with indicated concentration of indicated AMC conjugate, and initial velocity was determined. Comparison of activity to values of select deubiquitinases and deNEDDylase provided from refs. 57–59. (E) SDS-PAGE analysis of diubiquitin cleavage. Representative SYPRO Ruby stained SDS-PAGE of diubiquitin (3 μM) with EmcB (1 μM). Quantification of cleaved diubiquitin by densitometry below indicated lane are mean ± SD of three independent experiments. P value comparing cleavage for each chain at 0 h vs. indicated time, M1 1 h and 3 h n.s., K6 1 h and 3 h <0.0001, K11 and 3 h <0.0001, K27 1 h and 3 h n.s., K29 1 h and 3 h n.s., K33 1 h n.s., 3 h <0.05, K48 1 h and 3 h <0.0001, and K63 1 h and 3 h <0.0001 by one-way ANOVA with Dunnett post hoc test. (F) SDS-PAGE analysis of ubiquitin cleavage by chain length. Representative Sypro Ruby-stained gel of K63-linked ubiquitin (1 μM) of indicated length incubated with EmcB (0.25 μM). Quantification of cleavage of full-length hexaubiquitin (blue) or diubiquitin (green). Data are mean ± SEM of two independent experiments. P < 0.01 hexaubiquitin vs. diubiquitin at 2.5 min+ by one-way ANOVA with Šídák post hoc test. (G) Relative ubiquitin chain length abundance of hexaubiquitin from F. Data are mean ± SEM. (H) SDS-PAGE analysis of tetraubiquitin cleavage. Representative Sypro Ruby-stained SDS-PAGE of tetraubiquitin (1 μM) cleavage by EmcB (0.25 μM). Representative of ≥3 independent experiments. Quantification of cleavage at 3 h, bars are mean ± SD. (I) Model depicting how translocation of EmcA and EmcB by C. burnetti residing in host cells can inhibit RLR signaling at different stages which prevents type I IFN production.
Ubiquitin monomers can be linked into polymeric chains by either canonical peptide bonds between the C terminus of one monomer to the N terminus of the adjacent one (“M1-linked”), or by isopeptide bonds between the C terminus of one monomer with the side chain of one of seven internal lysines within an adjacent ubiquitin monomer (60). These linkages give ubiquitin chains unique topologies with distinct cellular functions. Deubiquitinases exhibit variable specificity toward different linkages ranging from specific cleavage of just one linkage to the promiscuous cleavage of all eight (61, 62). Importantly, RIG-I signaling requires only K63-linked ubiquitin. EmcB protease activity was detected for diubiquitin with K6, K11, K48 and K63 linkages. EmcB inefficiently cleaved K33-linked di-ubiquitin and minimal, if any, cleavage of M1-, K27-, or K29-linked di-ubiquitin was detected (Fig. 4E).
Ubiquitin chain length plays an important role in the regulation of diverse cellular functions. In the case of RIG-I signaling, ubiquitin chains comprising three or more monomers activate downstream signaling better than di-ubiquitin (48, 49). Moreover, deubiquitinases can display a preference for ubiquitin chain lengths. EmcB efficiently cleaved K63-linked hexaubiquitin as compared with diubiquitin (Fig. 4F). Hexaubiquitin cleavage by EmcB resulted in the accumulation of monomeric and dimeric ubiquitin, and a relative absence of trimeric, tetrameric, and pentameric ubiquitin, suggesting that EmcB may preferentially recognize ubiquitin chains of this length (Fig. 4G). The kinetics of EmcB cleavage of tetraubiquitin chains comprising the four most abundant linkages in cells was measured to determine how chain length influences EmcB activity on different ubiquitin linkages. EmcB efficiently cleaved K48- and K63-linked chains (Fig. 4H). EmcB activity against K29-linked tetraubiquitin was low, and there was no detectable cleavage of M1-linked tetraubiquitin by EmcB (Fig. 4H). Thus, EmcB protease activity is robust on long K63-linked ubiquitin chain, which are the ubiquitin chains required for assembly of the RIG-I signaling complex.
Discussion
Many cell-autonomous defenses are selectively activated by host sensors that detect microbial determinants introduced into the cell through pathogen-associated activities, which include protein secretion systems that can introduce agonists of these sensors into the cytosol of the host cell. Thus, it stands to reason that many of these pathogens will have evolved mechanisms to interfere with cell autonomous defense pathways that are deleterious for pathogen survival and replication in the host. This study showed that type I IFN can induce cell-autonomous restriction of C. burnetii replication and identified two different effector proteins, EmcA and EmcB, that enable C. burnetii to interfere with the signaling pathway activated by the cytosolic double-stranded RNA sensor RIG-I to prevent infected host cells from producing type I IFN (Fig. 4I). EmcB function as a ubiquitin-specific protease that deconjugates long, K63-linked ubiquitin chains from RIG-I, which are required for the formation of an active RIG-I signaling complex necessary that interacts with the adaptor protein MAVS. Thus, there has likely been selective pressure applied on C. burnetii to evolve mechanisms to prevent the induction of type I IFN production to enhance infection and persistence in mammalian hosts.
EmcA and EmcB block activation of RLR signaling by unique mechanisms. Endogenous levels of EmcA and EmcB translocated during infection by C. burnetii were required for blocking RLR-triggered type I IFN production stimulated by the addition of exogenous poly(I:C). The efficiency of RLR signaling inhibition during C. burnetii infection was dependent on both EmcA and EmcB, which indicates that the independent biological activities of these effectors may function cooperatively or synergistically. EmcA was necessary for C. burnetii to block RLR signaling but not sufficient when produced in cells. This could indicate that EmcA requires additional infection-derived factors not present during ectopic expression, or that EmcA may facilitate the function of another effector. Although these were the only two effectors identified in this screen, there is evidence that other C. burnetii effectors may have the capacity to modulate a type I IFN response downstream of RIG-I activation. For example, NopA (CBU1217) inhibits induction of type I IFNs during C. burnetii infection by preventing the nuclear translocation of the transcription factor interferon regulatory factor 3 (IRF3), and CBU1314 has recently been reported to block the polymerase associated factor 1 (PAF1) complex, an integral component of the transcriptional complex that mediates ISRE-dependent transcription (63–65). These independent studies indicate that C. burnetii has evolved multiple mechanisms to interfere with type I IFNs during infection, which could presumably be stimulated by a diverse number of innate immune sensors and is consistent with an important role of these cytokines in restricting C. burnetii replication and dissemination.
RIG-I and MDA5 agonists can be microbial-derived RNA or derived from the host (nuclear or mitochondrial RNA). Indeed, in bacterial infections where there is RIG-I-dependent type I IFN production, the particular RIG-I agonists are often not known (18). The cytosolic bacterial pathogen Listeria monocytogenes secretes an RNA-binding protein called Zea that presents bacterial RNA to RIG-I to potentiate type I IFN production (66). Although there is evidence that L. pneumophila infection can activate RIG-I by a Dot/Icm-dependent mechanism, it is unknown whether stimulation is mediated by bacterial RNA delivered by the pathogen or stimulation by a host-derived RNA (17–19, 23). Similarly, during C. burnetii infection there is the potential for RLR agonists to be directly translocated into the host cytosol by the Dot/Icm system, indirectly introduced from damage to the pathogen-containing vacuole, or arise from host sources such as the mitochondria (67, 68). The Dot/Icm system is related to type IV secretion systems that mediate DNA transfer by a process called conjugation, and the Dot/Icm machine retains the ability to transfer nucleic acids to bacterial and eukaryotic recipient cells (69–72). Intriguingly, stimulation of the cGAS-STING pathway during L. pneumophila infection requires the Dot/Icm system (20), although a specific DNA substrate stimulating this pathway has not been identified. Thus, how cytosolic nucleic acid sensor activation occurs during infection by bacterial pathogens remains an interesting question for future studies.
RIG-I-mediated induction of type I IFNs could induce the production of host proteins such as IDO1 and IRG1, which regulate cell-autonomous defense pathways that limit C. burnetii intracellular replication (73–75). RIG-I also activates cell-autonomous defenses by directly inducing expression of interferon-stimulated genes with antimicrobial functions that could potentially limit replication of C. burnetii (76). Thus, the evolutionary pressure on C. burnetii to antagonize RIG-I signaling may be multifaceted and the mechanisms of RLR-mediated restriction of C. burnetii merits further investigation.
Pathogen-encoded effectors must outcompete opposing cognate host enzymes to efficiently interfere with host signaling. The high catalytic efficiency of EmcB for ubiquitin and weak activity for NEDD8 are consistent with results obtained for other bacterial deubiquitinases, such as OtDUB from Orientia tsutsugamushi, or RickCE from Rickettsia spp. (77, 78). The catalytic activity displayed by EmcB for NEDD8 was ~100 times lower than the constitutive photomorphogenesis 9 (COP9)-signalosome deneddylase, which suggests that NEDD8 is not a biologically relevant target for EmcB cleavage (Fig. 4D) (57). In contrast, the catalytic efficiency exhibited by EmcB toward ubiquitin was comparable to that of known pathogen-encoded deubiquitinases such as viral Ovarian Tumor-associated protease (vOTU) from Crimean Congo Hemorrhagic Fever Virus, and greater than that of the known endogenous negative regulator of RIG-I ubiquitination, ubiquitin-specific protease 21 (USP21) (Fig. 4D) (58, 59, 79). These data are consistent with a model whereby EmcB could overcome E3 ligase activity to disassemble RIG-I filaments.
EmcB is capable of efficiently cleaving K63-linked ubiquitin, the known activating linkage for RIG-I, as well as other linkages of ubiquitin. These findings are typical of many deubiquitinases. EmcB may have additional substrate recognition features or may be recruited to discrete subcellular locations that enable specific targeting of RIG-I to direct the proteolytic activity against K63-linked chains appended to the tandem CARD domain. For instance, the ubiquitin-like [Ubl]-specific protease 1/Cysteine endopeptidase (ULP1/CE) clan protease RavZ efficiently recognizes substrates through accessory domains that bind PI3P and curved membranes, which concentrate RavZ at the preautophagosome (80, 81).
C. burnetii is a recent evolutionary branch of the Coxiella genus that likely originated from a tick endosymbiont and coevolved with mammals to become a successful pathogen (2). An emcB-like gene with conservation of the catalytic cysteine and histidine-tryptophan motif that enable ubiquitin protease activity can be found in an environmental Coxiella spp. metagenome (82). Although RLRs are present in invertebrates, they were lost in the evolution of Arthropoda (83, 84). Thus, it is possible that ancestral EmcB-like enzymes participate in the remodeling of conserved ubiquitin signaling pathways in arthropods. Indeed, other endosymbionts of arthropods such as Wolbachia and Orientia encode deubiquitinases, suggesting that there may be alternative pressures for Coxiella that are tick endosymbionts to modulate ubiquitin signaling (78, 85). Understanding how EmcB evolved to provide C. burnetii with a mechanism to interfere with RIG-I an interesting question for future studies.
Materials and Methods
Infection Assays.
C. burnetii Nine Mile phase II, strain RSA493 clone 4, and isogenic derivatives were grown in acidified citrate cysteine medium-2 (Sunrise Science Products) at 5% CO2, 2.5% O2 at 37 °C (86, 87). C. burnetii stationary phase cultures were quantified by qPCR for the dotA gene against a standard curve to enumerate genome equivalents (GEs) and determine the multiplicity of infection (88). Infected, phorbol 12-myristate 13-acetate (PMA)-differentiated THP-1 cell were lysed in nuclease-free water. Cell lysates were centrifuged to collect the bacteria. Genomic DNA was extracted using the Illustra Bacteria GenomicPrep Mini Prep Kit (Cytiva). GEs were quantified by qPCR and the fold replication was determined by dividing the number of Ges at the indicated time point by the number of GEs at 4 h postinfection (day 0).
Cell-Based Luciferase Reporter Assays.
HEK293 cells stably expressing an ISRE-firefly luciferase reporter were infected with indicated C. burnetii strain as above. Twenty-four hours later, cells were transfected with poly(I:C) (1 ng/μL) using X-tremegene 9 (Sigma-Aldrich) or TransIT-LT1 (Mirus) in Dulbecco's Modified Eagle Medium (DMEM). Twenty-four hours after transfection, cells were lysed, and luminescence was measured using a tris(hydroxymethyl)aminomethane-etheylenediaminetetraacetic acid (TE).
Protein Purification and Biochemical Assays.
Codon-optimized EmcB (Genscript) was cloned into a custom pET vector encoding an N-terminal 6×His-SUMO2-tagged fusion protein (89) and expressed in BL21 DE3 RIL Escherichia coli (Agilent). After induction with isopropyl β-D-1-thiogalactopyranoside (IPTG), cells were pelleted, snap frozen, and stored at −80 °C. Pellets were resuspended in ice cold lysis buffer (20 mM N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid (HEPES)-KOH pH 7.5, 250 mM NaCl, 20 mM imidazole, 10% glycerol, 1 mM Dithiothreitol (DTT)) and sonicated (10 s on, 20 s off, 70% power, total: 5 min) before clarification. Recombinant protein was purified from clarified lysates by gravity chromatography with Nickel-nitrilotriacetic acid (Ni-NTA) resin (QIAGEN). Protein was eluted with lysis buffer with 300 mM imidazole. Proteins were dialyzed and incubated with SUMO endopeptidase 2 (SENP2) overnight at 4 °C. Proteins were further purified by size-exclusion chromatography, concentrated to >10 mg/mL and frozen in small aliquots.
For diubiquitin cleavage assays, 0.5 µM EmcB was incubated with 0.5 µM K63-linked diubiquitin chains in 15 µL reactions of 50 mM Tris-HCl pH 7.5, 120 mM KCl, 5 mM MgCl2, and 5 mM DTT for 2 h, quenched with 5 µL of 4× Laemmli sample buffer without reducing agent and resolved on PROTEAN TGX gels (Bio-Rad). Gels were stained with Sypro™ Ruby and imaged with the Amersham ImageQuant 800 biomolecular imager (ultraviolet (UV) channel).
More detailed materials and methods can be found in SI Appendix text.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Michaela Gack for providing plasmids encoding GST-RIG-I 2CARD (pEBG-RIG-I 2CARD) and Influenza A virus NS1 proteins (pCAGGS-NS1), Dr. Kate Fitzgerald for the HEK293 ISRE-luciferase cells, and Saumendra Sakar for providing DDX58−/− HEK293T cells. pGL3-IFNB was a gift from Nicolas Manel (Addgene plasmid #102597). pEBG was a gift from David Baltimore (Addgene plasmid #22227). pRK5-HA-Ubiquitin-K63 was a gift from Ted Dawson (Addgene plasmid #17606). We thank Brianna Duncan–Lowey for providing the custom pET vector, guidance on protein purification and biochemistry, and feedback on the manuscript. Financial support for this work was provided by the NIH R01AI114760 (C.R.R.) and T32GM007205 (J.D.-L).
Author contributions
J.D.-L., E.C., A.J., and C.R.R. designed research; J.D.-L., E.C., and A.J. performed research; S.C.O.R. contributed new reagents/analytic tools; J.D.-L., E.C., A.J., and C.R.R. analyzed data; and J.D.-L., E.C., and C.R.R. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Hackstadt T., Williams J. C., Biochemical stratagem for obligate parasitism of eukaryotic cells by Coxiella burnetii. Proc. Natl. Acad. Sci. U.S.A. 78, 3240–3244 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duron O., et al. , The recent evolution of a maternally-inherited endosymbiont of ticks led to the emergence of the Q fever pathogen, Coxiella burnetii. PLoS Pathog. 11, e1004892 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carey K. L., Newton H. J., Lührmann A., Roy C. R., The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Pathog. 7, e1002056 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beare P. A., et al. , Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. mBio 2, e00175-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voth D. E., Heinzen R. A., Coxiella type IV secretion and cellular microbiology. Curr. Opin. Microbiol. 12, 74–80 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newton H. J., Roy C. R., The Coxiella burnetii dot/icm system creates a comfortable home through lysosomal renovation. mBio 2, e00226-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newton H. J., et al. , A screen of Coxiella burnetii mutants reveals important roles for Dot/Icm effectors and host autophagy in vacuole biogenesis. PLoS Pathog. 10, e1004286 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu J., Luo Z. Q., Legionella and Coxiella effectors: Strength in diversity and activity. Nat. Rev. Microbiol. 15, 591–605 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Janeway C. A., The immune system evolved to discriminate infectious nonself from noninfectious self. Immunol. Today 13, 11–16 (1992). [DOI] [PubMed] [Google Scholar]
- 10.Roy C. R., Mocarski E. S., Pathogen subversion of cell-intrinsic innate immunity. Nat. Immunol. 8, 1179–1187 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Reddick L. E., Alto N. M., Bacteria fighting back: How pathogens target and subvert the host innate immune system. Mol. Cell 54, 321–328 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anand I., Choi W., Isberg R. R., The vacuole guard hypothesis: How intravacuolar pathogens fight to maintain the integrity of their beloved home. Curr. Opin. Microbiol. 54, 51–58 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baxt L. A., Garza-Mayers A. C., Goldberg M. B., Bacterial subversion of host innate immune pathways. Science 340, 697–701 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Zamboni D. S., McGrath S., Rabinovitch M., Roy C. R., Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 49, 965–76 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Bradley W. P., et al. , Primary role for Toll-like receptor-driven tumor necrosis factor rather than cytosolic immune detection in restricting Coxiella burnetii phase II replication within mouse macrophages. Infect. Immun. 84, 998–1015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clemente T. M., et al. , Coxiella burnetii blocks intracellular interleukin-17 signaling in macrophages. Infect. Immun. 86, e00532-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Opitz B., et al. , Legionella pneumophila induces IFNβ in lung epithelial cells via IPS-1 and IRF3, which also control bacterial replication. J. Biol. Chem. 281, 36173–36179 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Monroe K. M., McWhirter S. M., Vance R. E., Identification of host cytosolic sensors and bacterial factors regulating the type I interferon response to Legionella pneumophila. PLoS Pathog. 5, e1000665 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiu Y. H., MacMillan J. B., Chen Z. J., RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138, 576–591 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lippmann J., et al. , Dissection of a type I interferon pathway in controlling bacterial intracellular infection in mice. Cell. Microbiol. 13, 1668–1682 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watson R. O., et al. , The cytosolic sensor cGAS detects mycobacterium tuberculosis DNA to induce type I interferons and activate autophagy. Cell Host Microbe 17, 811–819 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sumpter R., et al. , Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79, 2689–2699 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stetson D. B., Medzhitov R., Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24, 93–103 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Shin S., et al. , Type IV secretion-dependent activation of host MAP kinases induces an increased proinflammatory cytokine response to Legionella pneumophila. PLoS Pathog. 4, e1000220 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Case C. L., et al. , Caspase-11 stimulates rapid flagellin-independent pyroptosis in response to Legionella pneumophila. Proc. Natl. Acad. Sci. U.S.A. 110, 1851–1856 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zamboni D. S., et al. , The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat. Immunol. 7, 318–325 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Ivanov S. S., Roy C. R., Pathogen signatures activate a ubiquitination pathway that modulates the function of the metabolic checkpoint kinase mTOR. Nat. Immunol. 14, 1219–1228 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Case C. L., Shin S., Roy C. R., Asc and Ipaf inflammasomes direct distinct pathways for caspase-1 activation in response to Legionella pneumophila. Infect. Immun. 77, 1981–1991 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou P., et al. , Alpha-kinase 1 is a cytosolic innate immune receptor for bacterial ADP-heptose. Nature 561, 122–126 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Gaudet R. G., et al. , Cytosolic detection of the bacterial metabolite HBP activates TIFA-dependent innate immunity. Science 348, 251–255 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Wassermann R., et al. , Mycobacterium tuberculosis differentially activates cGAS- and inflammasome-dependent intracellular immune responses through ESX-1. Cell Host Microbe 17, 799–810 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Boxx G. M., Cheng G., The roles of type I interferon in bacterial infection. Cell Host Microbe 19, 760–769 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crabill E., Schofield W. B., Newton H. J., Goodman A. L., Roy C. R., Dot/Icm-translocated proteins important for biogenesis of the Coxiella burnetii-containing vacuole identified by screening of an effector mutant sublibrary. Infect. Immun. 86, e00758-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gack M. U., et al. , TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446, 916–920 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Inn K.-S., et al. , Inhibition of RIG-I-mediated signaling by Kaposi’s sarcoma-associated herpesvirus-encoded deubiquitinase ORF64. J. Virol. 85, 10899–10904 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gack M. U., et al. , Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5, 439–449 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saito T., et al. , Regulation of innate antiviral defenses through a shared repressor domain in RIG-1 and LGP2. Proc. Natl. Acad. Sci. U.S.A. 104, 582–587 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oshiumi H., Miyashita M., Matsumoto M., Seya T., A distinct role of riplet-mediated K63-linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog. 9, e1003533 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oshiumi H., Matsumoto M., Hatakeyama S., Seya T., Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-β induction during the early phase of viral infection. J. Biol. Chem. 284, 807–817 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Shi Y., et al. , Ube2D3 and Ube2N are essential for RIG-I-mediated MAVS aggregation in antiviral innate immunity. Nat. Commun. 8, 15138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oshiumi H., et al. , The ubiquitin ligase riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe 8, 496–509 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Cadena C., et al. , Ubiquitin-dependent and -independent roles of E3 ligase RIPLET in innate immunity. Cell 177, 1187–1200.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferrage F., et al. , Structure and dynamics of the second CARD of human RIG-I provide mechanistic insights into regulation of RIG-I activation. Structure 20, 2048–2061 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu L. G., et al. , VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol. Cell 19, 727–740 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Kawai T., et al. , IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6, 981–988 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Seth R. B., Sun L., Ea C. K., Chen Z. J., Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell 122, 669–682 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Hayman T. J., et al. , RIPLET, and not TRIM25, is required for endogenous RIG-I-dependent antiviral responses. Immunol. Cell Biol. 97, 840–852 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Jiang X., et al. , Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 36, 959–973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng W., et al. , Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141, 315–330 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peisley A., Wu B., Xu H., Chen Z. J., Hur S., Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature 508, 110–114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borodovsky A., et al. , Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 9, 1149–1159 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Ekkebus R., et al. , On terminal alkynes that can react with active-site cysteine nucleophiles in proteases. J. Am. Chem. Soc. 135, 2867–2870 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sommer S., Weikart N. D., Linne U., Mootz H. D., Covalent inhibition of SUMO and ubiquitin-specific cysteine proteases by an in situ thiol-alkyne addition. Bioorg. Med. Chem. 21, 2511–2517 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Hermanns T., et al. , An evolutionary approach to systematic discovery of novel deubiquitinases, applied to Legionella. Life Sci. Alliance 3, e202000838 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hochstrasser M., Origin and function of ubiquitin-like proteins. Nature 458, 422–429 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Isaacson M. K., Ploegh H. L., Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe 5, 559–570 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mosadeghi R., et al. , Structural and kinetic analysis of the COP9-signalosome activation and the cullin-RING ubiquitin ligase deneddylation cycle. Elife 5, e12102 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Akutsu M., Ye Y., Virdee S., Chin J. W., Komander D., Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains. Proc. Natl. Acad. Sci. U.S.A. 108, 2228–2233 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ye Y., et al. , Polyubiquitin binding and cross-reactivity in the USP domain deubiquitinase USP21. EMBO Rep. 12, 350–357 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Komander D., Rape M., The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Komander D., Mechanism, specificity and structure of the deubiquitinases. Subcell. Biochem. 54, 69–87 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Lange S. M., Armstrong L. A., Kulathu Y., Deubiquitinases: From mechanisms to their inhibition by small molecules. Mol. Cell 82, 15–29 (2022). [DOI] [PubMed] [Google Scholar]
- 63.Burette M., et al. , Modulation of innate immune signaling by a Coxiella burnetii eukaryotic-like effector protein. Proc. Natl. Acad. Sci. U.S.A. 117, 13708–13718 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fischer N. L., A Coxiella burnetii effector interacts with the host PAF1 complex and suppresses the innate immune response. bioRxiv [Preprint] (2022). 10.1101/2022.04.20.488957 (Accessed 14 October 2022). [DOI]
- 65.Shah P. S., et al. , Comparative flavivirus-host protein interaction mapping reveals mechanisms of dengue and zika virus pathogenesis. Cell 175, 1931–1945.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pagliuso A., et al. , An RNA-binding protein secreted by a bacterial pathogen modulates RIG-I signaling. Cell Host Microbe 26, 823–835.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dhir A., et al. , Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 560, 238–242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sadeq S., Al-Hashimi S., Cusack C. M., Werner A., Endogenous double-stranded RNA. Noncoding RNA 7, 15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guzmán-Herrador D. L., et al. , DNA delivery and genomic integration into mammalian target cells through type IV A and B secretion systems of human pathogens. Front. Microbiol. 8, 1503 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Segal G., Shuman H. A., Possible origin of the Legionella pneumophila virulence genes and their relation to Coxiella burnetii [2]. Mol. Microbiol. 33, 669–670 (1999). [DOI] [PubMed] [Google Scholar]
- 71.Zusman T., Yerushalmi G., Segal G., Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila. Infect. Immun. 71, 3714–3723 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Christie P. J., Vogel J. P., Bacterial type IV secretion: Conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8, 354–360 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ganesan S., Roy C. R., Host cell depletion of tryptophan by IFNγ-induced Indoleamine 2,3-dioxygenase 1 (IDO1) inhibits lysosomal replication of Coxiella burnetii. PLoS Pathog. 15, e1007955 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kohla L., Macrophages inhibit Coxiella burnetii by the ACOD1-itaconate pathway for containment of Q fever. EMBO Mol Med. 15, e15931 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen M., et al. , Itaconate is an effector of a Rab GTPase cell-autonomous host defense pathway against Salmonella. Science 369, 450–455 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dixit E., et al. , Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141, 668–681 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pruneda J. N., et al. , The molecular basis for ubiquitin and ubiquitin-like specificities in bacterial effector proteases. Mol. Cell 63, 261–276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Berk J. M., et al. , A deubiquitylase with an unusually high-affinity ubiquitin-binding domain from the scrub typhus pathogen Orientia tsutsugamushi. Nat. Commun. 11, 2343 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fan Y., et al. , USP21 negatively regulates antiviral response by acting as a RIG-I deubiquitinase. J. Exp. Med. 211, 313–328 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Choy A., et al. , The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science 338, 1072–1076 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Horenkamp F. A., et al. , The Legionella anti-autophagy effector RavZ targets the autophagosome via PI3P- and curvature-sensing motifs. Dev. Cell 34, 569–576 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anantharaman K., et al. , Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun. 7, 13219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mukherjee K., Korithoski B., Kolaczkowski B., Ancient origins of vertebrate-specific innate antiviral immunity. Mol. Biol. Evol. 31, 140–153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yao S., Chan J., Xu Y., Wu S., Zhang L., Divergences of the RLR gene families across lophotrochozoans: Domain grafting, exon-intron structure, expression, and positive selection. Int. J. Mol. Sci. 23, 3415 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beckmann J. F., Ronau J. A., Hochstrasser M., A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat. Microbiol. 2, 17007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Omsland A., et al. , Isolation from animal tissue and genetic transformation of Coxiella burnetii are facilitated by an improved axenic growth medium. Appl. Environ. Microbiol. 77, 3720–3725 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Omsland A., et al. , Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc. Natl. Acad. Sci. U.S.A. 106, 4430–4434 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Coleman S. A., Fischer E. R., Howe D., Mead D. J., Heinzen R. A., Temporal analysis of Coxiella burnetii morphological differentiation. J. Bacteriol. 186, 7344–7352 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lowey B., et al. , CBASS immunity uses CARF-related effectors to sense 3′–5′- and 2′–5′-linked cyclic oligonucleotide signals and protect bacteria from phage infection. Cell 182, 38–49.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.