Significance
Vaccines and repurposed drugs have been critical in preventing and treating SARS-CoV-2 infections; however, the virus’s ability to evolve rapidly requires alternative strategies that can keep pace with this pandemic pathogen. We performed the first comprehensive screening of siRNA chemical architectures and identified an effective multimeric platform for local lung delivery. Using this platform, we designed and screened over 200 siRNAs and antisense oligonucleotides targeting highly conserved regions within the SARS-CoV-2 genome. We identified four compounds targeting all clinically relevant variants, including delta and omicron, and are fully protective in a mouse model of SARS-CoV-2 infection. Our agile platform is adaptable for multiple diseases by changing the nucleic acid sequence, promising broad and deep impact in pulmonary medicine.
Keywords: SARS-COV-2, antisense oligos, small interfering RNAs, mouse model, lung
Abstract
The continuous evolution of SARS-CoV-2 variants complicates efforts to combat the ongoing pandemic, underscoring the need for a dynamic platform for the rapid development of pan-viral variant therapeutics. Oligonucleotide therapeutics are enhancing the treatment of numerous diseases with unprecedented potency, duration of effect, and safety. Through the systematic screening of hundreds of oligonucleotide sequences, we identified fully chemically stabilized siRNAs and ASOs that target regions of the SARS-CoV-2 genome conserved in all variants of concern, including delta and omicron. We successively evaluated candidates in cellular reporter assays, followed by viral inhibition in cell culture, with eventual testing of leads for in vivo antiviral activity in the lung. Previous attempts to deliver therapeutic oligonucleotides to the lung have met with only modest success. Here, we report the development of a platform for identifying and generating potent, chemically modified multimeric siRNAs bioavailable in the lung after local intranasal and intratracheal delivery. The optimized divalent siRNAs showed robust antiviral activity in human cells and mouse models of SARS-CoV-2 infection and represent a new paradigm for antiviral therapeutic development for current and future pandemics.
The COVID-19 pandemic is currently in its third year, with at least seven major variants of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) driving the disease (1–5). Despite the development and deployment of multiple vaccines which have been successful in reducing disease severity and hospitalizations (6–10), vaccine breakthrough infections have been reported across multiple strains (11, 12). Additionally, SARS-CoV-2 variants of concern including delta and omicron show increased resistance to neutralizing antibodies, immune evasion, and transmissibility (13–18). Given the continued emergence of vaccine and antibody-resistant variants, the identification and development of drugs targeting SARS-CoV-2 is a pressing need (19). The RNA genome of coronaviruses makes RNA-targeted therapeutics a natural treatment modality.
Small interfering RNAs (siRNAs) are a class of RNA-targeted therapeutics that recruit endogenous cellular proteins to degrade a specific RNA target (20, 21). The design of siRNAs is modular in nature: The chemical architecture, conjugated ligand, and/or delivery vehicle determines the siRNA’s tissue distribution and stability in a way that is largely independent of the siRNA sequence. Thus, siRNA scaffolds that provide desired pharmacokinetic properties in a given tissue can be repurposed for new targets by adapting the targeting sequence of the molecule while retaining the same chemical architecture (22–24). Advanced modification of siRNAs (20, 25–29) has allowed clinical application with efficacy in the liver lasting up to 6 months after a single dose (30). Nevertheless, the lung has seen limited success to date. Antisense oligonucleotides (ASOs) represent a second class of RNA-targeted therapeutics that bind to target mRNA and result in degradation or inhibition of mRNA function. While the mechanisms of siRNAs and ASOs are different, they represent two attractive strategies for the inhibition of viral RNA (31). In this work, we show that multivalent, fully chemically modified asymmetric siRNAs are bioavailable and effective in the lung after local delivery. We also identify siRNA target sequences, conserved across coronavirus variants of concern, which allow robust inhibition of viral replication and pathology in cells and in vivo.
Results
Identification of Highly Conserved SARS-CoV-2 Regions for siRNA-Based Targeting.
SARS coronaviruses have caused multiple severe outbreaks in the last two decades (32, 33). Like any virus, SARS-CoV-2 accumulates mutations that are naturally selected to enhance infectivity, adaptation to new hosts, escape from acquired natural, or vaccine-based immunity or drug resistance (4, 34). The infectivity in terms of receptor engagement and entry of the virus is mainly defined by epitopes on the surface of the viral particle. Most antigens targeted for neutralizing antibody-mediated immunity are from the same regions (3, 13, 14, 35).
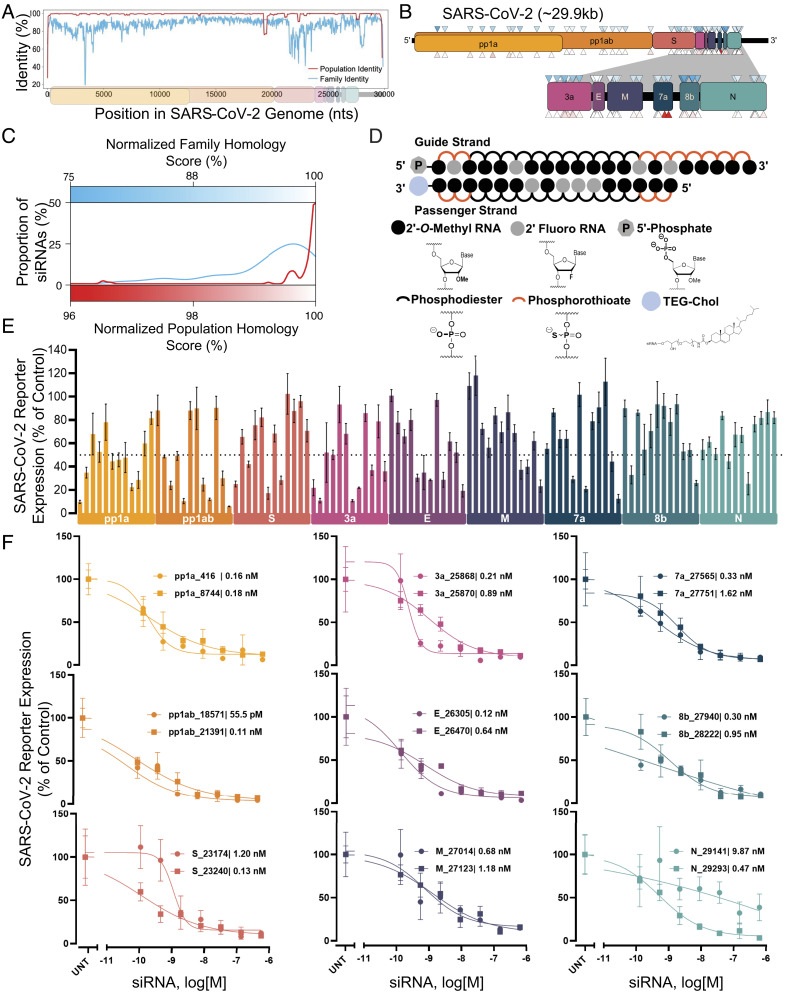
In contrast to neutralizing antibodies, targeting viral RNA allows access to the entire viral genome. We performed genomic alignments of SARS-CoV-2 variants from patients to the SARS-CoV-2 reference genome (population identity), as well as alignments of the SARS-CoV-2 reference genome to that of other coronaviruses (family identity). (Fig. 1A and SI Appendix, Fig. S1). We designed 108 siRNA sequences using a modified siRNA design algorithm similar to what was reported previously (36). Targeting sites were distributed throughout the SARS-CoV-2 genome with twelve siRNAs per open reading frame (ORF) (Fig. 1B). A strong preference was given to siRNAs targeting regions with high family and population homology scores since highly conserved regions are less likely to tolerate mutation. More than 95% of designed siRNAs had homology scores of >85% at the family level and >99% at the population level (Fig. 1C).
Fig. 1.
Design and screening of fully chemically modified siRNAs targeting conserved SARS-CoV-2 gene regions. (A) Population identity indicates that from the alignment of the SARS-CoV-2 target genome with SARS-CoV-2 genomes from the COVID-19 patient. Family identity indicates that from the alignment of the SARS-CoV-2 target genome with the most closely related SARS-CoVs. All identities are averaged over a sliding window of fifty bases to facilitate visualization. (B) siRNAs were designed to target the open reading frame of the SARSCoV-2 genome indicated by triangles. Triangle colors correspond to the homology scores in (C)-upper triangles for family homology score and lower triangles for population homology score. (C) Each siRNA sequence was scored based on its homology to the SARS-CoV-2 population (red) and family (blue). (D) Schematic of the fully chemically modified siRNA. (E and F) Percent expression of SARS-CoV-2 reporter in HeLa cells 72 h after uptake of siRNA (n = 3; E: 1.5 µM, F: concentration indicated). Reporter expressions were assayed using the psiCHECK-2 reporter system for SARS-CoV-2. Data presented relative to UNT (mean ± SD of independent biological replicates). Target regions of SARS-CoV-2 are indicated in each graph. The dotted line indicates 50% silencing (E). IC50 values of each siRNA are indicated on each graph (F). UNT, untreated control.
Durable in vivo siRNA activity depends on full chemical stabilization (25) with the tolerance of chemical modification patterns showing significant sequence dependence (36). To enable a seamless transition to in vivo studies, all the screens were done in the context of fully chemically modified siRNAs (Fig. 1D). All 2′-positions of ribose sugars of nucleotides were modified with 2′-O-methyl (2′-O-Me) or 2′-fluoro (2′-F groups), and terminal internucleotide linkages were metabolically stabilized with phosphorothioates. For in vitro screening, compounds were conjugated to cholesterol, which enables efficient uptake in all cell types (37). To allow testing and comparison of an independent mechanism of gene silencing, we also designed and synthesized 53 LNA gapmer ASOs (five or six ASOs per viral ORF). Full sequences and chemical modification patterns of all oligonucleotides used are shown in SI Appendix, Table S1.
Identification of Functional siRNAs and ASOs Targeting the SARS-CoV-2 Genome.
The 108 siRNAs and 53 ASOs were synthesized, and their identity was confirmed by LC–MS. To rapidly screen sequences, we engineered a panel of SARS-CoV-2 genome psiCHECK-2 reporter plasmids to contain the siRNA target sites within the mRNA for a luciferase reporter. Fig. 1E and SI Appendix, Fig. S2 show the results of the primary siRNA screens. 38% (41 siRNAs) induced more than 50% reporter silencing, while 8% (9 siRNAs) induced more than 80% reporter silencing and dose-dependent activity (Fig. 1 E and F). For ASOs, most of the compounds were highly active and of similar efficacy to the siRNAs (SI Appendix, Fig. S3).
There were no distinct differences in efficacy across regions of the viral genome. Top hits per target region showed subnanomolar IC50 in uptake without any delivery vehicle, indicating highly potent, clinical quality leads. Thus, the screen identified plentiful, highly conserved, fully chemically stabilized siRNAs and ASOs spanning the entire SARS-CoV-2 genome. While the reporter assays allowed for rapid screening and identification of hits, the lack of target sequence context (neighboring nucleotides, mRNA secondary structure, and localization) may have reduced the relevance of the siRNA efficacy predictions to the authentic viral sequence (see next paragraph). Future antiviral screens may be aided by the incorporation of target flanking regions in the reporter plasmid to understand the role of sequence context in the efficacy of siRNAs.
Antiviral siRNA Activity Shows a Poor Correlation with Reporter Assays.
To evaluate the ability of lead siRNAs and ASOs to modulate virus infection, we engineered A549 cells to overexpress human angiotensin-converting enzyme 2 (ACE2) and TMPRSS2 (transmembrane serine protease 2; see Methods) to promote SARS-CoV-2 entry and viral processing. Cells were pretreated with 41 siRNAs (10 nM) and 45 ASOs (25 nM), all of which showed more than 50% efficacy in the SARS-CoV-2 reporter assay. After 36 h, cells were challenged with SARS-CoV-2, and qRT-PCR was used to evaluate viral RNA levels 48 h postinfection. Remdesivir (5 µM) was used as a positive control. Untreated cells and a nontargeting control siRNA/ASO of identical chemical configuration were used as negative controls. Uninfected cells were used for background measurement.
Surprisingly, only 12% (5 out 41) of reporter-active siRNAs showed efficacy against virus infection (Fig. 2A and SI Appendix, Fig. S4). Moreover, multiple highly potent compounds with reporter IC50 values in the subnanomolar range were inactive against the live virus (SI Appendix, Fig. S6). Thus, factors other than functional RNA-induced silencing complex (RISC) assembly critically define antiviral siRNA efficacy. Similarly, only 13% (6 out of 45) of reporter-active ASOs showed anti-viral efficacy (SI Appendix, Figs. S5 and S6). Since ASO activity is dependent on target binding and RNase H recruitment and not the assembly and loading of a multisubunit RISC complex, the poor performance of both siRNAs and ASOs against live virus indicates viral-specific factors that hinder gene silencing efficacy. It is therefore crucial for future antiviral efforts to characterize viral mRNA localization and viral mRNA-associated protein factors to better understand the susceptibility of the viral transcriptome to RNAi and RNase H-mediated gene silencing. Moreover, it is essential that any future antiviral oligonucleotide discovery pipelines emphasize testing/validation in the context of live viral infection and do not rely exclusively on in silico design or reporter assays.
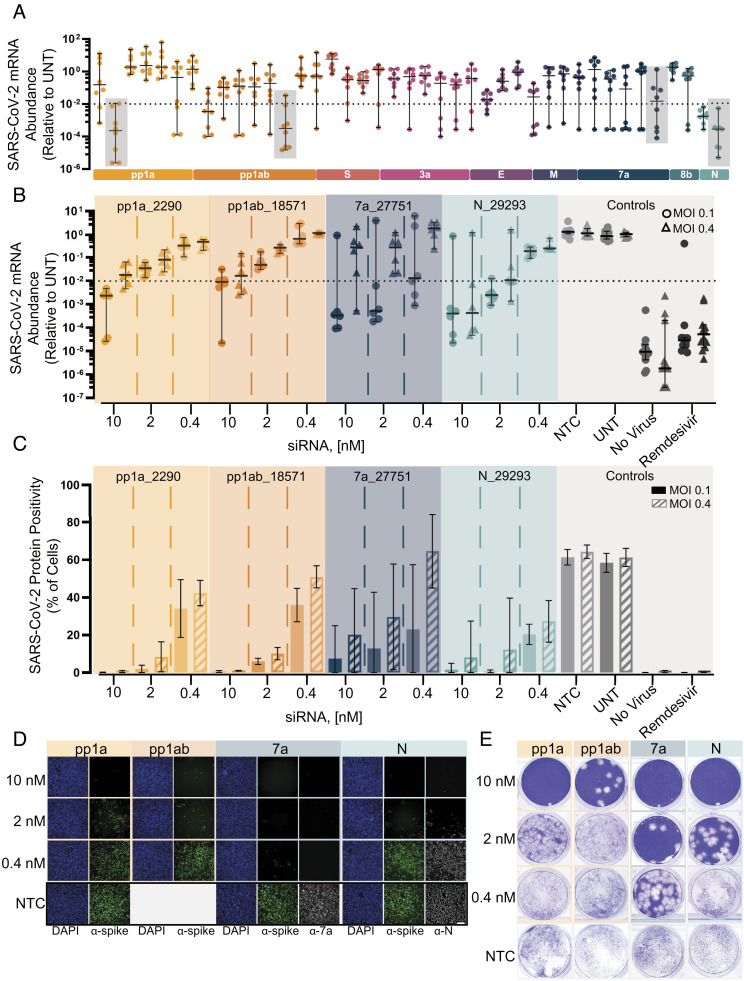
Fig. 2.
Screening and lead characterization of fully chemically modified siRNAs in SARSCoV-2 infection model in human cells identify several lead candidates. (A) Relative abundance of viral mRNA in supernatant of A549ACE2/TMPRSS2 cells 48 h postinfection of SARS-CoV-2 with MOI 0.1. siRNAs were transfected 36 h prior to the infection (siRNA: n = 8, controls: n = 20; 10 nM). Abundance of viral mRNA was measured by qRT-PCR. Data presented relative to UNT (median ± 95% CI of independent biological replicates). Lead siRNAs selected in screening were further characterized (gray box). The dotted line indicates 99% reduction in mRNA abundance. (B–E) Samples collected and analyzed from A549ACE2plus cells 48 h postinfection of SARS-CoV-2 with MOI 0.1 or 0.4. siRNAs were transfected 36 h prior to the infection (siRNA: n = 6, controls: n = 12; concentration indicated). Remdesivir with 5 µM was used as a positive control for antiviral activity. No virus represents cells which were not infected with virus. (B) Abundance of viral mRNA was measured by qRT-PCR. Data presented relative to UNT (median ± 95% CI of independent biological replicates). The dotted line indicates 99% reduction in mRNA abundance. (C) Percent of spike protein-positive cells was determined by immunofluorescence staining (mean ± SD of independent biological replicates). (D) Immunofluorescence images of the spike protein (α-spike; green), Orf7a protein (α-7a; gray), nucleocapsid protein (α-N; gray), and DAPI (blue). Original magnification, ×10. (Scale bar: 100 μm.) (E) Image of the viral plaque-forming assay conducted with supernatant. (Scale bar: 100 μm.) MOI, multiplicity of infection; NTC, nontargeting control; UNT, untreated control.
Oligonucleotides with robust activity against replicating virus were identified in four out of nine genomic regions (pp1a, pp1a/b, ORF7a, and nucleocapsid) with top siRNAs and ASO achieving three logs of viral reduction (Fig. 2A, SI Appendix, Figs. S4 and S5). The failure to identify antiviral oligonucleotides targeting other parts of the SARS-CoV-2 genome might be simply due to insufficient sequence space sampling rather than inherent biological limitations.
Four Fully Chemically Stabilized siRNAs Show Robust anti-SARS-CoV-2 Efficacy In Vitro.
We selected four siRNA lead compounds from different viral regions for more detailed characterization (pp1a_2290, pp1ab_18571, orf7a_27751, and nucleocapsid_29293). We first measured viral mRNA and protein levels as a proxy for viral replication after SARS-CoV-2 infection in A549ACE2/TMPRSS2 cells. The mRNA assay showed dose-responsive inhibition across all dose levels. The overall performance was similar between all four siRNAs with the highest siRNA dose tested (10 nM) resulting in two log (99%) inhibition of viral load. The top two compounds, orf7a_27751 and nucleocapsid_29293, achieved 99% reduction in viral mRNA at lower concentrations of 0.4 nM and 2 nM, respectively (Fig. 2B, circles and SI Appendix, Fig. S7A). However, orf7a_27751 was less effective when we repeated the experiment at fourfold higher multiplicity of infection (MOI) (Fig. 2B, triangles and SI Appendix, Fig. S7C).
At the protein level, siRNA treatment at 10 nM resulted in robust reduction of spike-positive cells for three out of the four siRNAs (Fig. 2C and SI Appendix, Fig. S7 B and D). To confirm that we observe silencing of targeted proteins, the orf7a_27751-treated and nucleocapsid_29293-treated cells were stained for antibodies for ORF7 and nucleocapsid protein correspondence (Fig. 2D), confirming efficient protein knockdown and again demonstrating a higher level of biochemical potency of orf7a_27751 at MOI 0.1. It is worth noting the most potent compound identified at lower MOI (orf7a_27751) was the least effective at higher MOI. Thus, the MOI selected for viral challenge experiments is an important consideration for future antiviral lead optimization. To confirm the observed biochemical readouts (mRNA and protein), the amount of live virus in cellular supernatants was quantified using a plaque-forming assay. Fig. 2E shows dose-dependent viral inhibition for all four lead siRNAs with orf7a_27751 showing the highest potency.
Multivalent, Fully Chemically Stabilized siRNA Configurations Support Robust Lung Delivery, Distribution, and Target Gene Silencing.
Optimization of the chemical scaffold for target tissue delivery is key to the clinical translation of siRNAs, with conjugate-mediated delivery of fully chemically modified siRNAs currently dominating the clinic. There are four major approaches for nonformulated siRNA delivery: GalNAc conjugates (38), antibody conjugates (39, 40), hydrophobic conjugates (41, 42), and divalent siRNA compounds (43). GalNAc-mediated delivery is hepatocyte-specific and has no impact on lung accumulation (44). Hydrophobic conjugates, in general, enhance tissue retention and enable broad extrahepatic distribution (41, 42). The divalent approach has been successfully employed to significantly enhance brain distribution and efficacy with local cerebrospinal fluid administration (43).
To identify the optimal scaffold for lung delivery, we systematically evaluated the impact of siRNA multimerization and hydrophobic conjugation on lung delivery and distribution. A panel of fully chemically stabilized siRNAs varying in valency (monovalent to tetravalent) or hydrophobic conjugate (eicosapentaenoic acid [EPA] or docosanoic acid [DCA]) was synthesized, purified by HPLC, and characterized by MS (Fig. 3 A and B). The divalent scaffold used here was structurally identical to that previously reported (43).
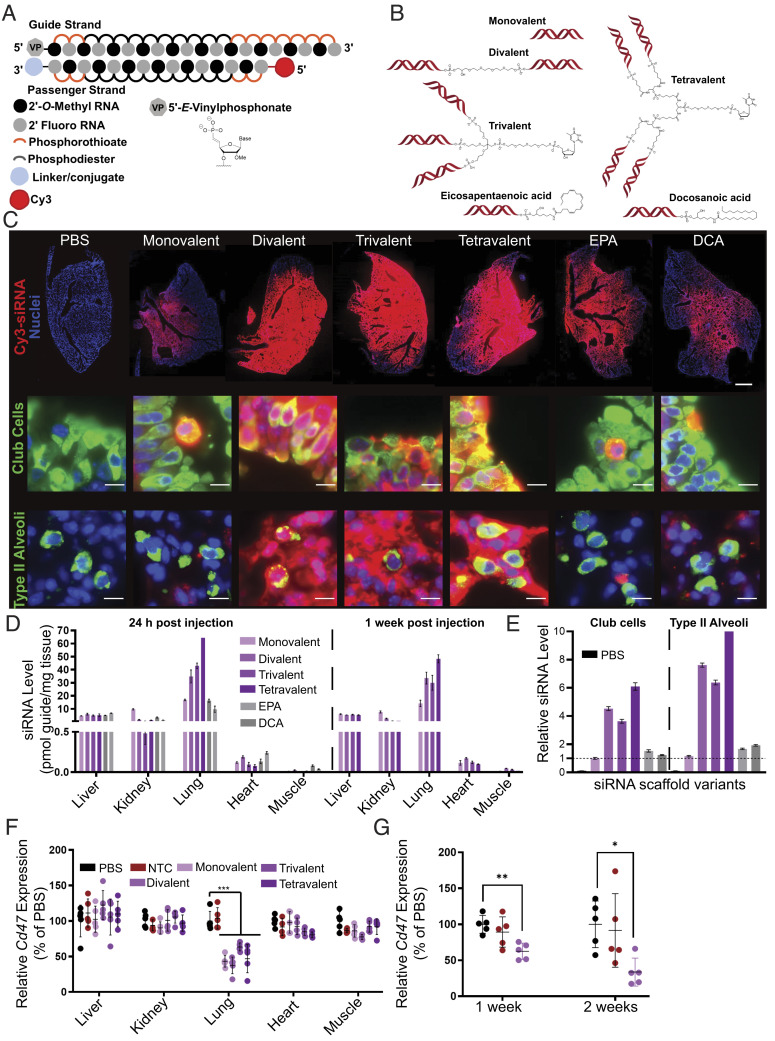
Fig. 3.
Local administration of multivalent fully chemically modified siRNA supports robust lung accumulation and safe and efficacious target gene silencing. (A) Schematic of the fully chemically modified siRNA. Multivalent linker or hydrophobic moiety is conjugated to the 3′ end of the passenger strand. Cy3 dye labeled to the 5′ end of the passenger strand. (B) Schematic of multivalent and hydrophobic siRNA scaffold. (C) Representative fluorescence images of lung after intratracheal injection of siRNA in mice sacrificed at 24 h postinjection (n = 2 mice; 3.75 mg/kg). (Top) Distribution of siRNA (red) with staining of DAPI (blue). Original magnification, ×5. (Scale bar: 1 mm.) (Middle and Bottom) Distribution of siRNA (red) with staining of club cells (green), type II alveoli (green), and DAPI (blue). Original magnification, ×40. (Scale bar: 10 µm.) Images collected at the same laser intensity and acquisition time. d, Tissue siRNA levels after intratracheal injection of siRNA in mice sacrificed at 24 h (Left) and 1 wk (Right) postinjection measured by PNA hybridization assay (n=2 mice; 3.75 mg/kg; mean ± SD of independent biological replicates). (E) siRNA uptake in club cells and type II alveoli after intratracheal injection of siRNA in mice sacrificed at 24 h quantified from microscope image (2 mice; 3.75 mg/kg). Bar colors same as D, with PBS group also shown in black on the left of each set. Data presented relative to monovalent siRNA of club cells (mean ± SEM of individual cells, n = 501 (PBS); 622 (monovalent); 798 (divalent); 946 (trivalent); 325 (tetravalent); 742 (EPA); and 545 (DCA) for club cells, n = 137 (PBS); 598 (monovalent); 753 (divalent); 796 (trivalent); 325 (tetravalent); 503 (EPA); and 326 (DCA) for type II alveoli). (F) Percent of Cd47 mRNA levels after intratracheal injection of siRNA targeting Cd47 in mice sacrificed at 1 wk postinjection (n = 5 mice; 7.5 mg/kg). mRNA levels were measured using QuantiGene assay, normalized to a housekeeping gene, Hprt, and presented relative to PBS (mean ± SD of independent biological replicates). (G) Percent of Cd47 mRNA level in bulk lung tissue after intratracheal injection of siRNA targeting Cd47 in mice sacrificed at 1 wk and 2 wk postinjection (3.25 mg/kg, n = 5 mice). mRNA levels were measured using QuantiGene assay, normalized to Hprt, and presented relative to PBS (mean ± SD of independent biological replicates).
The compounds were fluorescently labeled with Cy3 on the passenger strand to enable spatial and cell type–specific evaluation of distribution and accumulation. The mice were dosed intratracheally, and lung distribution was evaluated 48 h after administration. Dosage was adjusted such that all treatment groups received the same molar equivalent of siRNA guide strand. Fully chemically stabilized, nonlipid-conjugated asymmetric monovalent siRNA showed some distribution to proximal lung structures. Increasing the siRNA valency improved lung accumulation and overall distribution, while the increase in accumulation afforded by hydrophobic conjugation was less pronounced (Fig. 3C and SI Appendix, Figs. S8 and S9). The results from histology were further verified using whole lung flow cytometry analysis, where multivalency increased siRNA accumulation in multiple cell types (SI Appendix, Fig. S10), with epithelial (including alveolar type I, alveolar type II, and ciliated epithelia) and endothelial cells showing quantitative uptake.
To complement the accumulation data derived from measuring Cy3-labeled siRNA passenger strands, we quantified guide strand accumulation in five tissues at 24 h and 1-wk post-administration using PNA hybridization (Fig. 3D)(45), an assay that is independent of the presence of a fluorescent label. Consistent with the imaging results, guide strand quantification confirmed lung-specific enhancement of siRNA accumulation with the multivalent scaffolds. While the difference between monovalent and divalent siRNAs was profound, further enhancement in accumulation with increased valency was limited. In this set of five tissues, most of the nonlung accumulation was detected in the liver and kidney, the primary siRNA clearance tissues.
Consistent with the high metabolic stability of chemically modified siRNA scaffolds, the levels of lung accumulation were similar at one and 7 d postadministration (Fig. 3D). The accumulation of compounds was confirmed in club cells and type II alveolar epithelial cells, both cell types essential for SARS-CoV-2 infection (46, 47). The quantification of club cells and alveolar type accumulation were consistent with overall lung distribution with multivalency substantially enhancing uptake (four to eightfold) compared to monovalent compounds. DCA and EPA conjugates show no significant increase in club cell accumulation but demonstrate twofold enhancement in type II alveolar cells (Fig. 3E).
To test whether this uptake was functional in the bulk lung, we administered siRNAs targeting the housekeeping gene CD47 intratracheally at a dose of 7.5 mg/kg and found lung-specific target silencing (~45 to 65%) 1 wk posttreatment for all multivalent scaffolds. While the accumulation was significantly enhanced by multivalency, the observed levels of lung gene silencing at a time points tested were similar between monovalent and multivalent siRNAs (Fig. 3F). The divalent scaffold was further evaluated for efficacy 2 wk post treatment at 7.5 mg/kg (51% silencing) as well as with a lower dose of 3.25 mg/kg with 37% silencing observed at 1 wk and 66% silencing 2 wk posttreatment (Fig. 3G). Nontargeting control siRNA (NTC) in the divalent scaffold did not induce any significant changes in gene expression. The observed silencing was localized to the lung, consistent with the distribution profiles of these scaffolds. We next evaluated the tolerability of different scaffolds. There was no detectable toxicity at functional doses for all configurations, and animals were healthy with no observable adverse effects. With divalent siRNAs, a dose as high as 50 mg/kg (highest tested) was well tolerated with no detectable serum cytokine elevation (SI Appendix, Fig. S11).
While trivalent and tetravalent siRNA did show enhanced accumulation compared to the divalent scaffold, which is expected to enhance the duration of effect, they significantly increase the complexity of manufacturing, with lower yields and duplexing efficiency compared to the divalent scaffold. As the enhancement of activity using trivalent and tetravalent scaffolds compared to the divalent scaffold is relatively small and divalent siRNA manufacturing is relatively straightforward and has already been synthetically optimized for gene silencing in the central nervous system (43), the divalent siRNA was selected as a lead scaffold for local lung delivery. Interestingly, multivalency also improved extra-hepatic accumulation of systemically administered siRNA compared to monovalent siRNA at 24 h and 1 wk post administration (SI Appendix, Fig. S12). However, the absolute levels in the lung achieved by systemic administration were far lower than those achieved by local administration. Therefore, local administration was selected as the route of delivery in subsequent experiments.
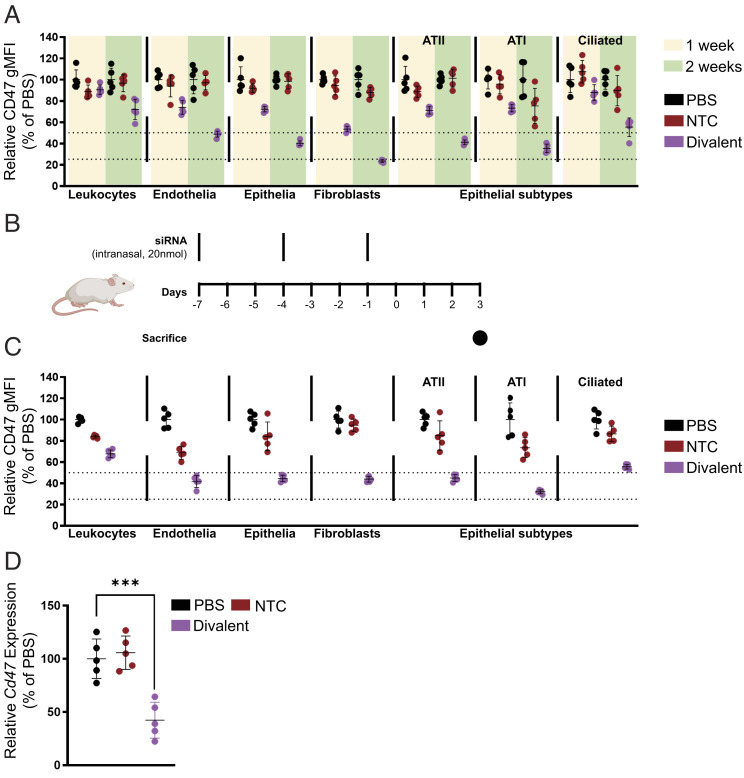
In order to examine the distribution of locally administered divalent siRNA in the lung more closely, we decided to measure CD47 silencing in specific lung cell types using flow cytometry(48) 1 wk and 2 wk post administration of 7.5 mg/kg divalent siRNA (Fig. 4A). We found that silencing improved at 2 wk post injection compared to 1 wk with maximum silencing observed in lung fibroblasts (47% at 1 wk and 77% at 2 wk). Total epithelial cell and endothelial cell silencing levels were similar (28% at 1 wk and 51 to 60% at 2 wk) and leukocyte silencing was minimal (9% at 1 wk and 28% at 2 wk). The epithelial cells were further gated into ATI, ATII, and ciliated subtypes with silencing levels in ATI and ATII cells similar to total epithelial cells (29% at 1 wk and 60 to 65% at 2 wk). Silencing in ciliated epithelia was slightly lower (12% at 1 wk and 45% at 2 wk).
Fig. 4.
Local administration of divalent fully chemically modified siRNA elicits potent gene silencing in multiple lung cell subtypes following intratracheal or intranasal injection. (A) Percentage CD47 geometric mean fluorescence intensity presented relative to PBS in leukocytes (CD45+), endothelial cells (CD45–CD31+CD326–), fibroblasts (CD45–CD31–CD326–CD140a+), type II alveoli (CD45–CD31–CD326medMHCII+), type I alveoli (CD45–CD31–CD326lowMHCII–), and ciliated cells (CD45–CD31–CD326hiMHCII–) after a single intratracheal injection of siRNA of 7.5 mg/kg sacrificed at 1 wk and 2 wk followed by flow cytometry analysis (n = 5 mice). (B) Schematic of intranasal multidose regimen optimized for expedited silencing. (C) Percentage CD47 geometric mean fluorescence intensity presented relative to PBS in leukocytes (CD45+), endothelial cells (CD45– D31+CD326–), fibroblasts (CD45–CD31–CD326–CD140a+), type II alveoli (CD45–CD31–CD326medMHCII+), type I alveoli (CD45–CD31–CD326lowMHCII–), and ciliated cells (CD45–CD31–CD326hiMHCII–) after intranasal injection of siRNA (20 nmol or ~10 mg/kg ×3) sacrificed at day 7 post first injection followed by flow cytometry analysis (n = 5 mice; ~10 mg/kg ×3). (D) Percent of Cd47 mRNA levels after intranasal injection of siRNA (20 nmol or ~10 mg/kg ×3) sacrificed at day 7 post first injection (n = 5 mice). mRNA levels were measured using QuantiGene assay, normalized to a housekeeping gene, Hprt, and presented relative to PBS (mean ± SD of independent biological replicates).
While robust silencing was achieved using divalent siRNA scaffolds at a duration of 2 wk post injection, we wanted to develop a dosing regimen that would allow us to achieve sufficiently high silencing levels in a shorter period of time. Additionally, while intratracheal dosing is the most efficient method for local lung delivery, the invasiveness of this route of administration prevents repeat dosing and has the potential to produce significant inflammation at the injection site, potentially confounding antiviral efficacy testing. Thus, we developed an intranasal administration model involving three doses of 10 mg/kg divalent siRNA administered three days apart (Fig. 4B). This dosing regimen produced robust levels of silencing in leukocyte (33%), endothelial (60%), epithelial (66%), ATI (78%), ATII (55%), ciliated epithelia (45%), and fibroblast (66%) silencing in 1 wk (Fig. 4C). Bulk lung expression measurements showed 58% silencing of Cd47 (Fig. 4D) using this dosing scheme. This expedited dosing regimen allowed us to quickly test the efficacy of SARS-CoV-2 targeting siRNA in an infection model and reduce the time required for lead identification.
Treatment with Antiviral siRNA Shows Robust Efficacy in a SARS-CoV-2 Animal Infection Model.
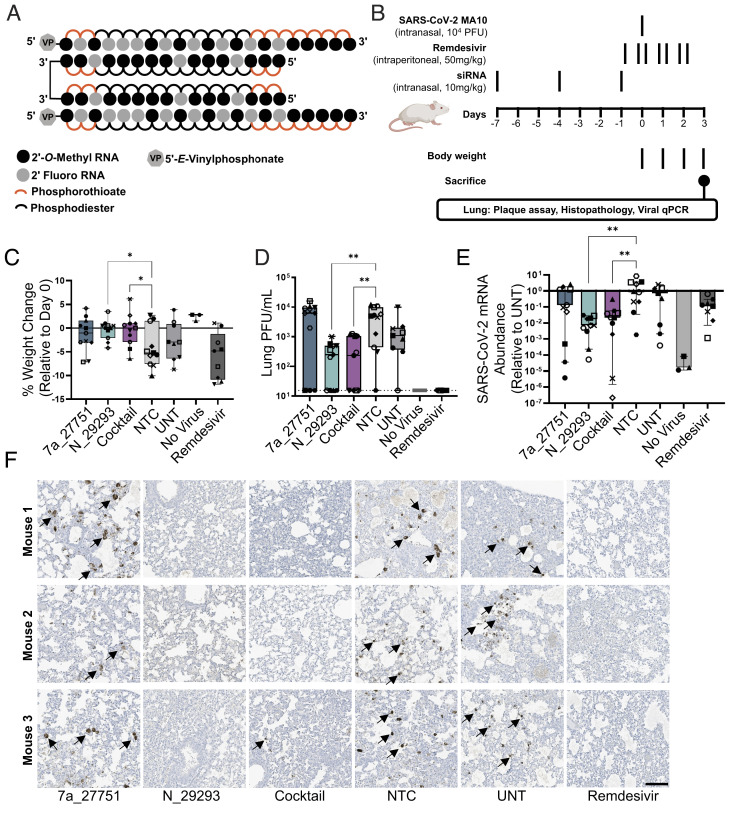
To look at the impact of lead siRNA on infection in vivo, we used a SARS-CoV-2 mouse model based on intranasal delivery of the mouse-adapted SARS-CoV-2 strain MA10 in BALB/c mice (47). This animal model allows for measuring the efficacy of drugs that impair viral replication, with lung viral titers, lung pathology, and body weight loss being used as primary readouts (49). Two anti-viral lead compounds, orf7a_27751 and N_29293, were synthesized in a divalent scaffold (Fig. 5A). Mice were treated intranasally with 10 mg/kg siRNAs (nontargeting control or NTC, orf7a_27751, N_29293) or a mixture of 5 mg/kg orf7a_27751 plus 5 mg/kg N_29293 (cocktail) prior to infection (Fig. 5B). All siRNAs were administered in the divalent scaffold. As a positive control, mice were treated with seven doses (twice a day, 50 mg/kg) of remdesivir 1 d before and throughout the course of infection.
Fig. 5.
Intranasal administration of divalent siRNA is protective in a mouse model of SARS-CoV-2 infection. (A) Schematic of the fully chemically modified divalent siRNA. (B) Study design describing a mouse model of SARS-CoV-2 infection. Divalent siRNAs, 7a_27751, N_29293, and cocktail (1:1 mixture of 7a_27751 and N_29293), were given as pretreatment at days 7, 4, and 1 before viral infection with the SARS-CoV-2 strain MA10 (day 0) by intranasal administration (10 mg/kg/injection). Remdesivir was administered twice a day from day -1 to day 2 by intraperitoneal injection (50 mg/kg/injection) as a positive control. After viral infection, body weight was measured every day, and mice were sacrificed on day 3 (siRNA: n = 11, UNT: n = 9, no virus: n = 3, remdesivir: n = 9 mice). (C) Percent weight change on day 3 relative to day 0 (box plot: center line, median; box limits, upper and lower quartiles; whiskers, min to max; points, independent biological replicates). (D) Viral load in lung quantified by plaque-forming assay (box plot: center line, median; box limits, upper and lower quartiles; whiskers, min to max; points, independent biological replicates). The dotted line indicates the limit of detection (15.6 PFU/mL). (E) Abundance of viral mRNA in lung was measured by qRT-PCR, normalized to a housekeeping gene, Hprt. Data presented relative to UNT (median ± 95% CI of independent biological replicates). (F) Representative immunostaining images of mice lungs. Arrows indicate positive staining of nucleocapsid protein of SARS-CoV-2. Original magnification, ×20. (Scale bar: 100 µm.) C–E, Individual mice are identified by a unique symbol. One-way ANOVA with Dunnett test for multiple comparisons (**P < 0.01, *P < 0.05). MOI, multiplicity of infection; NTC, nontargeting control; UNT, untreated control; PFU, plaque-forming units.
We observed that SARS-CoV-2 infection resulted in significant weight loss over 3 d in animals treated with NTC siRNA, while both the siRNA N_29293 and the cocktail efficiently prevented weight loss (Fig. 5C). We also observed more than 10-fold reduction of lung viral titers (plaque-forming assay, Fig. 5D) and 25 to 100-fold reduction in viral RNA levels (qRT-PCR, Fig. 5E) at 3 d postinfection in animals treated with siRNA N_29293 or the cocktail. The lungs of siRNA N_29293 or cocktail-treated mice also showed a clear reduction in the presence of viral nucleocapsid by immunostaining (Fig. 5F). By each of these metrics, mice treated with the orf7a_27751 siRNA alone did not significantly differ from the NTC siRNA-treated or untreated mice. The ORF7a viral protein is an accessory protein of unclear function and may not be essential for SARS-CoV-2 replication in vitro and in vivo (50, 51). It is still largely unclear whether the lack of ORF7a affects some unknown aspects of SARS-CoV-2 pathogenesis. The minimal activity of orf7a_27751 on mouse lung viral titers may be in part related to the dispensability of the ORF7a protein in vivo. We also note that the ORF7a siRNA, while highly potent, lost efficacy at higher viral titer in cellular experiments (Fig. 2 B and C). In summary, intranasal administration of divalent siRNA N_29293 demonstrated a profound impact on SARS-CoV-2 infection as evidenced by protection from weight loss, decreased viral plaques and viral mRNA abundance, and viral nucleocapsid detection in the lung by immunohistochemistry.
Discussion
Here, we report the development of siRNA architectures that support robust silencing of targets in the lung after local administration. We further show that local administration of a SARS-CoV-2 genome-targeted divalent siRNA silences viral replication and pathology in mice. While previous studies have demonstrated in vivo antiviral efficacy of siRNA using various formulation or adeno-associated viral delivery(52–54), this study is the first demonstration of unformulated fully chemically stabilized siRNA showing broad lung distribution and antiviral activity after local administration. During the course of revision of our manuscript, LNA-modified ASOs were also demonstrated to suppress SARS-CoV-2 replication in the lungs of infected mice, with both prophylactic and therapeutic effects (55). Since the lead LNA-modified ASO evaluated in vivo in that work interrupts the secondary structure, while the lead siRNAs in our study elicit the degradation of viral RNA, these results highlight the utility of RNA-targeted technologies in combating respiratory viruses by multiple mechanisms.
Complete chemical stabilization in the context of an asymmetric siRNA architecture design was sufficient to support some gene silencing in the lung after local administration. However, multimeric siRNAs showed significantly enhanced tissue retention with up to 30 to 40% of the administered dose retained in the respiratory tract. Interestingly, the level of lung accumulation was much better with the multivalent strategy than with hydrophobic conjugation (Fig. 3 D and E). In particular, multivalent architectures support a more uniform delivery with a 5- to 10-fold increased accumulation in endothelial and alveolar cells and robust silencing in multiple lung cell types (Fig. 4). The accumulation afforded by multivalency was about twofold greater than that of monovalent siRNA (Fig. 3C, SI Appendix, Figs. S9 and S10). The increase in accumulation with an increase in valency could be due to increased numbers of single-stranded, phosphorothioate-modified tails, which are known to bind cell-surface proteins and promote tissue accumulation; larger molecules also have slower tissue clearance kinetics because of their larger size. Therefore, while short-term silencing (1 wk) studies did not show significant differences in efficacy between monovalent and multivalent siRNAs (Fig. 3F), the increased accumulation of multivalent siRNA is likely to extend the duration of silencing in the lung.
The uptake and silencing activity of multivalent siRNAs in nasal tissue has not been evaluated in this study for either CD47 silencing or viral infection. Given that viral load in the nasal turbinate is an important characteristic of certain SARS-CoV-2 variants of concern, this will be an important question to address in future studies. The recent study mentioned above showed antiviral activity in the nasal wash after intranasal administration of an LNA-modified ASO which implicates the activity of the oligonucleotide in nasal turbinate (55). Divalent siRNAs were also well-tolerated at doses up to 50 mg/kg. Thus, for local lung administration, multivalent siRNAs provide a safe and productive way to enable functional silencing. While we used intranasal and intratracheal administration in the rodent experiments, to achieve uniform distribution beyond the upper respiratory tract in larger animal models and clinical trials, the oligonucleotides would need to be administered using liquid nebulizers. Many clinically approved nebulizers are widely used to treat various respiratory conditions. Due to complete chemical stabilization, siRNAs and ASOs are stable in solution for extended periods, making liquid nebulization clinically feasible.
We observed a lack of correlation between siRNA/ASO efficacy in the psiCHECK reporter screen and the compounds’ impact on viral infection in cells (correlation plot in SI Appendix, Fig. S6). Usually, the correlation between siRNA efficacy in reporter assays and endogenous silencing is relatively high. However, in the case of virus infection in cell culture, the correlation was low, with only a small fraction of RISC-competent siRNAs showing antiviral efficacy. The lack of functional correlation may result from multiple factors, including the limited accessibility of highly complex viral RNA structures, the biology of viral replication cycles, or both. A previous in vitro study using recombinant SARS-CoV-2 has shown that siRNAs targeting ORF1, where only genomic RNA is present, can effectively suppress viral replication—perhaps by avoiding binding to highly abundant subgenomic RNAs (56). Nevertheless, in our hands, an siRNA targeting the nucleocapsid region, which is found in both genomic and subgenomic RNAs, showed high silencing efficacy in cells and in vivo, implying that both genomic and subgenomic viral RNAs can be good targets (Fig. 2). In selecting targets, the role and dispensability of the target genes should be considered: for example, ORF7a-, ORF7b-, or ORF8-deficient SARS-CoV-2 induced similar pathology compared to WT SARS-CoV-250, which might explain the minimal activity of our ORF7a-targeted siRNA, orf7a_27751 (Fig. 5). Regardless of the contributing factors, for future antiviral oligonucleotide selection, in vitro and in vivo experiments should be conducted using replicating virus rather than reporter assays as early as possible to reduce the number of false positives.
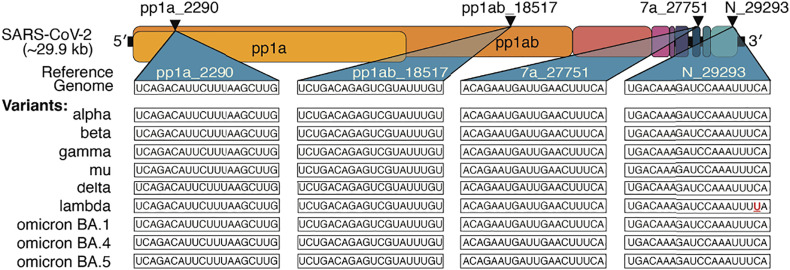
As SARS-CoV-2 continues to evolve, variants including delta and omicron have wrought significant damage to economic and healthcare infrastructures worldwide, with more variants likely to emerge. In siRNA selection, we specifically targeted regions of high conservation in the contexts of SARS-CoV-2 and the SARS family as a whole. This paper’s original screen of 162 bioinformatically optimized oligonucleotides generated four siRNAs with >2-log antiviral activity against replicating virus in human cells and a mouse model of infection, which show high conservation across variants of concern (Fig. 6). As remdesivir was more effective in blocking viral replication (Fig. 5), dosing regimens need to be optimized to maximize safety and efficacy prior to clinical application. We note that the siRNAs were well-tolerated at the doses employed while remdesivir caused some weight loss at the selected dose. To further reduce the chances of escape mutants, advancing a cocktail of two to three individual sequences into the clinic would be advisable. Additionally, and especially for emerging viruses and viral variants, targeting multiple genes may be advantageous where information about gene function and essentiality is limited. As the pharmacokinetics and pharmacodynamics of siRNAs are mainly driven by their chemical architecture (22), this is feasible, while a combinatorial small-molecule approach would have no guarantee of compatible biodistribution and retention properties.
Fig. 6.
Lead siRNA compounds target all current SARS-CoV-2 variants of clinical concern. Triangles indicate target location in the coding regions of the SARS-CoV-2 genome. Sequences targeted by lead siRNA compounds and comparison with current SARS-CoV-2 variants of clinical concern. The mismatched base is denoted in red and underlined.
There are currently multiple lines of pandemic defense, including a range of highly effective vaccines (1), monoclonal antibodies, and small molecules (57). Conceptually, RNAi-based drugs lie between vaccines and small molecule therapeutics, providing rapid-onset, multimonth but nonpermanent modulation of target activity (30). Moreover, siRNA-based approaches offer the added advantages of rapid sequence modification to target emerging variants and the ability to focus targeting on conserved regions outside the S protein, which has become a mutational hotspot allowing immune evasion and drug resistance (34). In conclusion, we have described the development and optimization of an RNAi-based approach for gene silencing in the lung, by showing that multivalent, fully chemically stabilized siRNAs provide broad delivery to the lung following local administration. We screened and identified a battery of fully chemically modified siRNAs spanning the SARS-CoV-2 genome and identified candidates that efficiently block viral replication in human cells and in a mouse model of the disease.
Materials and Methods
Methods for Designing SARS-CoV-2-Targeting siRNAs Considering Both Macro and Microevolution of the Virus.
Phylogenetic tree construction to identify related coronavirus species was performed with the Augur package. siRNA sequences were selected based on standard rules of optimal GC content and minimization of off-targets with seed region homology. Sequences were constrained to targeting regions of high family and population homology based on multiple sequence alignment. Details of phylogenetic tree construction, siRNA selection, and homology score calculations can be found in the Supporting Information.
Oligonucleotide Synthesis, Purification, and Quality Control.
Oligonucleotides were synthesized by phosphoramidite solid-phase synthesis with methylamine gas deprotection. Oligonucleotides for in vitro screens were precipitated on support, washed with ethanol, and dissolved in water. Oligonucleotides for in vivo experiments were purified by HPLC. Oligonucleotides were confirmed using LC-MS analysis. A detailed description of the methods is provided in the Supporting Information.
Cell Culture.
HeLa cells (ATCC, #CCL-2) were maintained in Dulbecco′s Modified Eagle′s Medium (DMEM) (Cellgro, #10-013CV). The media were supplemented with 9% fetal bovine serum (FBS) (Gibco, #26140), and cells were grown at 37 °C and 5% CO2. Cells were split every 3 to 7 d and discarded after 15 passages.
Direct (i.e., Vehicle-Free) Delivery of Oligonucleotides.
cDNA sequences corresponding to 20-nucleotide long, unique regions of target mRNAs were cloned into a psiCHECK-2 vector (Promega; C8021) according to the manufacturer′s protocol. HeLa cells were transfected with reporter plasmid using Lipofectamine 2000 (Invitrogen, Carlsbad, CA; 11668019) according to the manufacturer′s protocol. Twenty-four hours later, cells were diluted in DMEM containing 6% FBS and transferred to 96-well culture plates. siRNAs were diluted in OptiMEM (Gibco; 31985-088) to create seven-point dose responses and added to cells. Treated cells were incubated for 72 h at 37 °C and 5% CO2.
Method for Quantitative Analysis of SARS-CoV-2 Reporter Expression.
mRNA was quantified using the Dual-Glo® Luciferase Assay System according to the manufacturer′s protocol (Promega, #E2940). Luminescence was detected on a Veritas Luminometer (Promega, #998-9100) or a Tecan M1000 (Tecan). For each cell treatment plate, data were normalized to control reporter (fLuc) and plotted as a percentage of the mean results from untreated cells.
SARS-CoV-2 In Vitro Infection.
A549ACE2/TMPRSS2 cells were seeded at 70 to 80% confluency. Cells were transfected with siRNA and Lipofectamine RNAiMAX. After 36 h, low-passage SARS-CoV-2 (USA-WA1/2020 strain, BEI Resources) in MEM media with 3% FBS was added and incubated for 1 h. The cells were washed twice with phosphate-buffered saline and plated in MEM with 3% FBS for 48 h. All work with live SARS-CoV-2 was performed in a biosafety level 3 laboratory facility (BSL3) by personnel trained to handle BSL3 agents and adhering to necessary Standard Operating Procedures. Details of A549ACE2/TMPRSS2 cell line generation can be found in SI Appendix.
Animal Administrations.
Animal experiments were conducted as approved by the UMass Chan Medical School Institutional Animal Care and Use Committee (IACUC), protocols A-2551 and PROTO201900346. Female FVB (Charles River Laboratories) and BALB/c mice (Jackson Laboratory), 8 wk of age, were administered 50 µL of PBS or siRNA as previously described (58). Mice were anesthetized during the procedure using ketamine/xylazine or isoflurane for intratracheal or intranasal instillation, respectively. For intratracheal instillation, the solution was administered through the catheter intubated using the Endotracheal Tube Introducer (Hallowell). For intranasal instillation, the solution was slowly administered via the nostril in a dropwise manner.
siRNA Distribution Analysis Using Fluorescence Microscopy.
Formalin-fixed, paraffin-embedded tissue sections were deparaffinized in xylene, rehydrated, and stained with antiuteroglobin antibody for club cells (1:1,600) (Abcam, ab40873) or antiprosurfactant protein C antibody for alveoli type II (1:1,000) (Abcam, ab40879). Goat anti-rabbit IgG H&L (Alexa Fluor® 488) (Abcam, ab150077) was used as secondary antibody (1:1,000). The nucleus was stained with DAPI (Molecular Probes). Images of three sections per mouse, 100 µm apart, were acquired with Leica DMi8 inverted tiling microscope (Leica Microsystems) and processed using LAS X.
Peptide Nucleic Acid (PNA) Hybridization Assay for Tissue siRNA Quantification.
Quantification of antisense strands in tissues was performed using a PNA hybridization assay as described (45). Briefly, tissues were lysed in MasterPure tissue lysis solution (EpiCentre) containing 0.2 mg/mL proteinase K (Invitrogen). Sodium dodecyl sulfate (SDS) was precipitated from lysates by adding 3 M potassium chloride and pelleted centrifugation at 4,000 × g for 15 min. siRNA in the supernatant were hybridized to a Cy3-labeled PNA probe fully complementary to the antisense strand (PNABio). Samples were analyzed by HPLC (Agilent) over a DNAPac PA100 anion-exchange column (ThermoFisher). Cy3 fluorescence was monitored, and peaks were integrated. The final concentrations were ascertained using calibration curves.
Flow Cytometry for Lung siRNA Distribution Analysis.
Mice were treated with Cy3-labeled siRNA and euthanized as per IACUC protocol A-2551 after 24 h; lungs were harvested and dissociated to single-cell suspension. Dissociated cells were then stained with selected antibodies for cell-type identification (SI Appendix, Fig. S13) at 4 °C for 30 min. Cells were analyzed using a MACSQuant VYB flow cytometer (Miltenyi Biotec), and data including Cy3 fluorescence intensity were analyzed using FlowJo software (BD Biosciences, v10.6). Cell dissociation protocols and antibody details can be found in the SI Appendix.
Serum Cytokine Analysis.
ELISA quantification of secreted cytokine levels was performed using customized 36-plex ProcartaPlex Immunoassays (ThermoFisher) following the manufacturer’s instructions. Detailed methods can be found in the SI Appendix.
RNA Isolation from Cell Culture Supernatants and Lung Tissue.
Cell culture supernatants or lung homogenates were diluted with TRIzol LS Reagent (ThermoFisher), following which RNA was isolated using chloroform separation and ethanol precipitation according to the manufacturer’s recommended protocol.
qRT-PCR.
Viral RNA from in vitro assays was measured with CDC probe 2019-nCoV_N2 (IDT, 10006713, Coralville, IA, USA) using the QuantiFast Pathogen RT-PCR +IC Kit (Qiagen). The reaction was performed under the following condition: 50 °C for 15 min, 95 °C for 5 min, followed by 45 cycles of 95 °C for 15 s and 60 °C for 30 s. To measure viral RNA from in vivo studies, isolated RNA from lung tissue was used for cDNA generation with a High-Capacity cDNA Reverse Transcription Kit (ThermoFisher) according to the manufacturer′s protocol. Viral RNA was detected with CDC probe 2019-nCoV_N2 and normalized with Hprt detected with Hprt-specific probe (IDT, Mm.PT.39a.22214828). qPCR reactions were performed using iTaq Super Mix (BioRad) under the following conditions: 95 °C for 3 min, followed by 50 cycles of 95 °C for 10 s and 60 °C for 30 s. All reactions were performed on the CFX96 Real-Time Systems (BioRad).
In Vivo mRNA Silencing Experiments.
Tissues were collected and stored in RNAlater (Sigma) at 4 °C overnight. mRNA was quantified using the QuantiGene 2.0 Assay (Affymetrix). Tissue punches were lysed in 300 μL Homogenizing Buffer (Affymetrix) containing 0.2 mg/mL proteinase K (Invitrogen). Diluted lysates and probe sets (mouse CD47 or mouse Hprt) were added to the bDNA capture plate and the signal was amplified and detected as described previously (59). Luminescence was detected on a Tecan M1000 (Tecan). Flow cytometry-based CD47 expression analysis was performed as described previously (48).
Plaque Assays.
Approximately 2 × 105 Vero E6 cells (ATCC, cat#CRL-1586) were seeded to each well of 12-well plates and cultured at 37 °C, 5% CO2 for 18 h. Virus was serially diluted in MEM with 3% FBS for in vitro-generated samples and transferred to the monolayers. The viruses were incubated with the cells at 37 °C with 5% CO2 for 1 h. After that, the virus-containing medium was removed and overlay medium was added to the infected cells per well. The overlay medium contained MEM with 0.42% BSA, 20 mM HEPES, 0.24% NaHCO3, and 0.7% agarose (Oxoid, LP0028). After a 3-d incubation, plates were fixed with 4% paraformaldehyde overnight and stained with crystal violet solution (Sigma-Aldrich) the next day. Plaques were counted on a light box.
Immunofluorescence Staining of Cells.
Cells were plated on 96-well tissue culture plates (black polystyrene microplates, Corning) and infected with the low passage of SARS-CoV2 at indicated MOI and infection period. Infected cells were fixed with 4% paraformaldehyde for 15 min at room temperature, gently washed 2× in PBS, and permeabilized with 1% Triton X-100 in PBS and blocked with 5% BSA. Fixed cells were either labeled with either a human monoclonal antibody against the spike antigen (1:200) (60), ORF7a mouse monoclonal antibody (1:500; [3C9] GTX632602; GeneTex), or a mouse monoclonal antibody that recognizes the NP antigen (1:1,000; 40143-MM05; Sinobiological) conjugated with AlexaFluor-488 by incubation for 2 h at 4 °C; after washing with saline-Tween 20 (0.05% [vol/vol]), these cells were labeled with goat anti-mouse secondary antibody conjugated with AlexaFluor-594 (1:500) by incubation for 1 h at 4 °C. Nuclei were stained with DAPI (1 μg/mL, Abcam) for 15 min at 4 °C. Images were acquired with the ImageXpress Micro-XL system and processed by MetaXpress Software.
Mouse-Adapted SARS-CoV-2 Infection and Treatment Studies.
The in vivo infection studies were performed in an animal biosafety level 3 (ABSL3) facility in at UMass Chan Medical School. The study procedures were conducted with approval by the IACUC at UMass Chan Medical School. Briefly, mice were anesthetized and inoculated intranasally with 1 × 104 plaque-forming units of SARS-CoV-2 MA10 followed by compound administration. Mice were weighed daily and at the end of the study, lungs were harvested for virus titer assays, immunohistochemistry staining, and virus qRT-PCR. Detailed descriptions can be found in the SI Appendix.
Mouse Infection Experiment: Lung Image Acquisition.
Images were acquired with a TissueFAXS SL tissue cytometer (TissueGnostics GmbH) built on a Zeiss AxioImager.Z2 microscope base with Märzhäuser stage, Lumencor SOLA light engine, Zeiss EC Plan-Neofluar 20x/0.5 NA objective, and Pixelink PL-D674CU camera.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This project was funded by the Mathers Foundation (to A.K., J.P.W., and R.W.F.), the Harrington Discovery Institute (Harrington Scholar Awards to A.K. and K.A.F.), the NIH (R35 GM131839 and R01 NS104022 to A.K. and S10 OD020012), the Ellison Foundation of Lynn, MA (grant to J.K.W.), the Ono Pharmaceutical Foundation (Breakthrough Science Award to J.K.W.), the Massachusetts Consortium on Pathogen Readiness (to R.W.F. and K.A.F.), the Department of Defense COVID-19 Expansion for AIRe Program, W81XWH2110029 (to R.W.F., K.A.F., J.P.W., W.M.M., and R.M.), and by UMass Chan Medical School, including via the COVID19 Pandemic Fund Pilot (to R.M.), and the Dean’s Strategic Development Fund. BioRender was used to generate Fig. 4B. We thank the Sanderson Center for Optical Experimentation (SCOPE) light microscopy core facility at UMass Chan Medical School (Worcester, MA), which provided access to the TissueGnostics TissueFAXS SL tissue cytometer. The SCOPE TissueFAXS SL was funded by a Massachusetts Life Science Center Bits to Bytes award to Drs. Christina Baer and Dorothy Schafer.
Author contributions
V.N.H., M. Shin, C.-W.C., D.O., K.Y., M. Somasundaran, S.H., S.M.D., C.F., B.M.D.C.G., S.L., G.G., R.W.F., K.A.F., J.P.W., J.K.W., and A.K. designed research; V.N.H., M. Shin., C.-W.C., D.O., A.B., K.Y., Z.G., M. Somasundaran., Q.T., P.M.K., J. Cruz, I.L.C., P.L., S.-Y.L., J.M., S.P.S., B.M.D.C.G., J. Caiazzi, M.M., K.K., F.H., K.M.P., and W.M.M. performed research; D.O., K.Y., G.D., N.M., D.A.C., D.E., J.S., Y.T., A.C., K.M.P., Q.L., Y.W., R.M., and D.K. contributed new reagents/analytic tools; V.N.H., M. Shin, A.B., Q.T., K.M., and Z.K. analyzed data; K.M. and D.K. performed bioinformatic sequence design; G.D., N.M., D.A.C., D.E., J.S., and Y.T. synthesized and purified compounds; Z.K. contributed to microscopy image analysis; R.M., G.G., W.M.M., R.W.F., K.A.F., J.P.W., J.K.W., and A.K. supervised elements of the project; and V.N.H., M. Shin., J.P.W., J.K.W., and A.K. wrote the paper.
Competing interests
The authors have filed a patent application related to this work. G. Harold and Leila Y. Mathers Foundation (Mathers Foundation), UH | Harrington Discovery Institute, University Hospitals (HDI), HHS | NIH, HHS | NIH, HHS | NIH, Larry Ellison Foundation (LEF), Ono Pharmaceutical Foundation, Massachusetts Consortium on Pathogen Readiness (MCPR), U.S. Department of Defense (DOD), University of Massachusetts Chan Medical School, University of Massachusetts Chan Medical School.
Footnotes
Reviewers: S.C., University of Pennsylvania Perelman School of Medicine; and J.R., Beckman Research Institute of City of Hope.
Contributor Information
Katherine A. Fitzgerald, Email: kate.fitzgerald@umassmed.edu.
Jennifer P. Wang, Email: jennifer.wang@umassmed.edu.
Jonathan K. Watts, Email: jonathan.watts@umassmed.edu.
Anastasia Khvorova, Email: anastasia.khvorova@umassmed.edu.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Hadj Hassine I., Covid-19 vaccines and variants of concern: A review. Rev. Med. Virol. 32, e2313 (2021), 10.1002/rmv.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chakraborty C., Bhattacharya M., Sharma A. R., Present variants of concern and variants of interest of severe acute respiratory syndrome coronavirus 2: Their significant mutations in S-glycoprotein, infectivity, re-infectivity, immune escape and vaccines activity. Rev. Med. Virol. 32, e2270. [Google Scholar]
- 3.Lauring A. S., Hodcroft E. B., Genetic variants of SARS-CoV-2-what do they mean? JAMA 325, 529–531 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Chakraborty C., et al. , Mode of transmission, and mutational landscape of newly emerging SARS-CoV-2 variants. mBio 12, e0114021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tao K., et al. , The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet 22, 757–773 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baden L. R., et al. , Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 384, 403–416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson M. G., et al. , Prevention and attenuation of Covid-19 with the BNT162b2 and mRNA-1273 vaccines. N. Engl. J. Med. 385, 320–329 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine-Tiefenbrun M., et al. , Initial report of decreased SARS-CoV-2 viral load after inoculation with the BNT162b2 vaccine. Nat. Med. 27, 790–792 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Chemaitelly H., mRNA-1273 COVID-19 vaccine effectiveness against the B.1.1.7 and B.1.351 variants and severe COVID-19 disease in Qatar. Nat. Med. 27, 1614–1621 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Tang L., et al. , Asymptomatic and symptomatic SARS-CoV-2 infections after BNT162b2 vaccination in a routinely screened workforce. JAMA 325, 2500–2502 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Servellita V., et al. , Predominance of antibody-resistant SARS-CoV-2 variants in vaccine breakthrough cases from the San Francisco Bay Area, California. Nat. Microbiol. 7, 277–288 (2022), 10.1038/s41564-021-01041-4. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Beltran W. F., et al. , Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 184, 2523 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kannan S. R., et al. , Omicron SARS-CoV-2 variant: Unique features and their impact on pre-existing antibodies. J. Autoimmun. 126, 102779 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann M., et al. , SARS-CoV-2 variant B.1.617 is resistant to bamlanivimab and evades antibodies induced by infection and vaccination. Cell Rep. 36, 109415 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang P., et al. , Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 29, 747–751.e744 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kannan S. R., et al. , Evolutionary analysis of the delta and delta plus variants of the SARS-CoV-2 viruses. J. Autoimmun. 124, 102715 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Planas D., et al. , Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 596, 276–280 (2021). [DOI] [PubMed] [Google Scholar]
- 18.VanBlargan L. A., et al. , An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 28, 490–495 (2022), 10.1038/s41591-021-01678-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozlov M., Why scientists are racing to develop more COVID antivirals. Nature 601, 496 (2022), 10.1038/d41586-022-00112-8. [DOI] [PubMed] [Google Scholar]
- 20.Hu B., et al. , Therapeutic siRNA: State of the art. Signal Transduct. Target. Ther. 5, 101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crooke S. T., Witztum J. L., Bennett C. F., Baker B. F., RNA-targeted therapeutics. Cell Metab. 27, 714–739 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Khvorova A., Watts J. K., The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 35, 238–248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levin A. A., Treating disease at the RNA level with oligonucleotides. N. Engl. J. Med. 380, 57–70 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Shen X., Corey D. R., Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 46, 1584–1600 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hassler M. R., et al. , Comparison of partially and fully chemically-modified siRNA in conjugate-mediated delivery in vivo. Nucleic Acids Res. 46, 2185–2196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foster D. J., et al. , Advanced siRNA designs further improve in vivo performance of GalNAc-siRNA conjugates. Mol. Ther. 26, 708–717 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parmar R., et al. , 5’-(E)-Vinylphosphonate: A stable phosphate mimic can improve the RNAi Activity of siRNA-GalNAc conjugates. ChemBioChem 17, 985–989 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Haraszti R. A., et al. , 5-Vinylphosphonate improves tissue accumulation and efficacy of conjugated siRNAs in vivo. Nucleic Acids Res. 45, 7581–7592 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biscans A., et al. , The chemical structure and phosphorothioate content of hydrophobically modified siRNAs impact extrahepatic distribution and efficacy. Nucleic Acids Res. 48, 7665–7680 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khvorova A., Oligonucleotide therapeutics - a new class of cholesterol-lowering drugs. N. Engl. J. Med. 376, 4–7 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Watts J. K., Corey D. R., Silencing disease genes in the laboratory and the clinic. J. Pathol. 226, 365–379 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piret J., Boivin G., Pandemics throughout history. Front. Microbiol. 11, 631736 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graham R. L., Donaldson E. F., Baric R. S., A decade after SARS: Strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 11, 836–848 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harvey W. T., et al. , SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 19, 409–424 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Beltran W. F., et al. , Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 184, 2523 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shmushkovich T., et al. , Functional features defining the efficacy of cholesterol-conjugated, self-deliverable, chemically modified siRNAs. Nucleic Acids Res. 46, 10905–10916 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alterman J. F., et al. , Hydrophobically modified siRNAs silence huntingtin mRNA in primary neurons and mouse brain. Mol. Ther. Nucleic Acids 4, e266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuda S., et al. , siRNA conjugates carrying sequentially assembled trivalent N-acetylgalactosamine linked through nucleosides elicit robust gene silencing in vivo in hepatocytes. ACS Chem Biol. 10, 1181–1187 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Song E., et al. , Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat. Biotechnol. 23, 709–717 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Baumer S., et al. , Antibody-mediated delivery of anti-KRAS-siRNA in vivo overcomes therapy resistance in colon cancer. Clin Cancer Res. 21, 1383–1394 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Biscans A., et al. , Diverse lipid conjugates for functional extra-hepatic siRNA delivery in vivo. Nucleic Acids Res. 47, 1082–1096 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osborn M. F., Khvorova A., Improving siRNA delivery in vivo through lipid conjugation. Nucleic Acid Ther. 28, 128–136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alterman J. F., et al. , A divalent siRNA chemical scaffold for potent and sustained modulation of gene expression throughout the central nervous system. Nat. Biotechnol. 37, 884–894 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Janas M. M., et al. , The nonclinical safety profile of GalNAc-conjugated RNAi therapeutics in subacute studies. Toxicol. Pathol. 46, 735–745 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Godinho B., et al. , Pharmacokinetic profiling of conjugated therapeutic oligonucleotides: A high-throughput method based upon serial blood microsampling coupled to peptide nucleic acid hybridization assay. Nucleic Acid Ther. 27, 323–334 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bridges J. P., Vladar E. K., Huang H., Mason R. J., Respiratory epithelial cell responses to SARS-CoV-2 in COVID-19. Thorax 77, 203–209 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leist S. R., et al. , A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell 183, 1070–1085.e1012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shin M., et al. , Intratracheally administered LNA gapmer antisense oligonucleotides induce robust gene silencing in mouse lung fibroblasts. Nucleic Acids Res. 50, 8418–8430 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Owen D. R., et al. , An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science 374, 1586–1593 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Silvas J. A., et al. , Contribution of SARS-CoV-2 accessory proteins to viral pathogenicity in K18 human ACE2 transgenic mice. J. Virol. 95, e0040221 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Addetia A., et al. , Identification of multiple large deletions in ORF7a resulting in in-frame gene fusions in clinical SARS-CoV-2 isolates. J. Clin. Virol. 129, 104523 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khaitov M., et al. , Silencing of SARS-CoV-2 with modified siRNA-peptide dendrimer formulation. Allergy 76, 2840–2854 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Idris A., et al. , A SARS-CoV-2 targeted siRNA-nanoparticle therapy for COVID-19. Mol. Ther. 29, 2219–2226 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Becker J., et al. , Ex vivo and in vivo suppression of SARS-CoV-2 with combinatorial AAV/RNAi expression vectors. Mol. Ther. 30, 2005–2023 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu C., et al. , An intranasal ASO therapeutic targeting SARS-CoV-2. Nat. Commun. 13, 4503 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ambike S., et al. , Targeting genomic SARS-CoV-2 RNA with siRNAs allows efficient inhibition of viral replication and spread. Nucleic Acids Res. 50, 333–349 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takashita E., et al. , Efficacy of antibodies and antiviral drugs against Covid-19 omicron variant. N. Engl. J. Med. (2022), 10.1056/NEJMc2119407. [DOI] [PMC free article] [PubMed]
- 58.Gruntman A. M., Mueller C., Flotte T. R., Gao G., Gene transfer in the lung using recombinant adeno-associated virus. Curr. Protoc. Microbiol. 14, Unit14D.2 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coles A. H., et al. , A high-throughput method for direct detection of therapeutic oligonucleotide-induced gene silencing in vivo. Nucleic Acid Ther. 26, 86–92 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.ter Meulen J., et al. , Human monoclonal antibody combination against SARS coronavirus: Synergy and coverage of escape mutants. PLoS Med. 3, e237 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.