Abstract
The Slack channel (KCNT1, Slo2.2) is a sodium-activated and chloride-activated potassium channel that regulates heart rate and maintains the normal excitability of the nervous system. Despite intense interest in the sodium gating mechanism, a comprehensive investigation to identify the sodium-sensitive and chloride-sensitive sites has been missing. In the present study, we identified two potential sodium-binding sites in the C-terminal domain of the rat Slack channel by conducting electrophysical recordings and systematic mutagenesis of cytosolic acidic residues in the rat Slack channel C terminus. In particular, by taking advantage of the M335A mutant, which results in the opening of the Slack channel in the absence of cytosolic sodium, we found that among the 92 screened negatively charged amino acids, E373 mutants could completely remove sodium sensitivity of the Slack channel. In contrast, several other mutants showed dramatic decreases in sodium sensitivity but did not abolish it altogether. Furthermore, molecular dynamics (MD) simulations performed at the hundreds of nanoseconds timescale revealed one or two sodium ions at the E373 position or an acidic pocket composed of several negatively charged residues. Moreover, the MD simulations predicted possible chloride interaction sites. By screening predicted positively charged residues, we identified R379 as a chloride interaction site. Thus, we conclude that the E373 site and the D863/E865 pocket are two potential sodium-sensitive sites, while R379 is a chloride interaction site in the Slack channel.
SIGNIFICANCE STATEMENT The research presented here identified two distinct sodium and one chloride interaction sites located in the intracellular C-terminal domain of the Slack (Slo2.2, KCNT1) channel. Identification of the sites responsible for the sodium and chloride activation of the Slack channel sets its gating property apart from other potassium channels in the BK channel family. This finding sets the stage for future functional and pharmacological studies of this channel.
Keywords: chloride binding site, gating mechanism, KCNT1 channel, Slo2.2, sodium binding site
Introduction
The Slack channel (KCNT1, Slo2.2) is a sodium and chloride-activated potassium channel, which belongs to the Slo channel family composed of the Slo1, Slo2, and Slo3 channels (Schreiber et al., 1998; Xia et al., 2002; Yuan et al., 2003; Santi et al., 2006). The Slack channel plays an essential role in many physiological and pathologic processes, including the control of slow spike frequency regulation of neurons, pain, itch sensation (Lu et al., 2015; Martinez-Espinosa et al., 2015; Liu et al., 2022), anxiety (Bausch et al., 2015; Quraishi et al., 2020; Zhang et al., 2022) epilepsy, and arrhythmia (Gertler et al., 1993; Heron et al., 2012; Kingwell, 2012; Ishii et al., 2013; Juang et al., 2014; Møller et al., 2015; Ohba et al., 2015; Tang et al., 2016; Alsaleem et al., 2019; Barcia et al., 2019; Cataldi et al., 2019; Kholin et al., 2019; Kuchenbuch et al., 2019; Yoshitomi et al., 2019). Despite extensive functional studies, the mechanism by which sodium activates the Slack channel remains unclear.
Since the Slack channel is gated by intracellular sodium, identifying the sodium-sensitive site of the Slack channel is necessary to elucidate the gating property of this channel. In earlier research, we identified the mutation D818N that decreased the sodium sensitivity of the rat Slack (rSlack) channel by approximately fourfold (Zhang et al., 2010). However, the mutant channel remained sensitive to intracellular sodium. Furthermore, the recently available cryo-EM structure of the chicken Slack (chSlack) channel obtained in a high sodium concentration argued against D818 and His823 forming a sodium binding motif in the Slack channel that would be analogous to that in GIRK channels (Rosenhouse-Dantsker et al., 2008; Hite and MacKinnon, 2017). Thus, we extended our efforts toward identifying novel sodium-sensitive sites in the Slack channel.
In addition to being gated by intracellular sodium, the rSlack channel (KCNT1, Slo2.2) has also been shown to be gated by chloride ions (Yuan et al., 2003; Santi et al., 2006). In contrast, the Caenorhabditis elegans Slo2 channel, a homolog of the Slack channel, is gated by intracellular calcium rather than chloride ions (Zhang et al., 2013), despite a shared 42% amino acid sequence identity. Although our prior work demonstrated a bidirectional dependence of channel activity between the Slack channel Cl– and Na+ ions (Zhang et al., 2010), the Cl–-sensitive site has remained elusive. Furthermore, the recent electron cryomicroscopy (cryo-EM) structure of the Slack channel did not identify the Cl– binding site (Hite and MacKinnon, 2017).
In this study, we performed a series of mutagenesis screening and electrophysiology recordings to identify and characterize the sodium-sensitive sites of the Slack channel. First, a broad neutralization mutagenesis screening of all acidic residues in the cytoplasmic C terminus of the rSlack channel revealed nine effective mutants: five decreasing sodium sensitivity >15-fold; and four abolishing the current even under high intracellular sodium concentrations. Second, by generating double mutations of the four mutants that abolished the current in the background of the M335A mutated Slack channel that exhibits activity in the absence of cytosolic sodium and enhances the ability of sodium to activate the Slack channel, we found that three of the four double mutants showed robust macroscopic current while two of them greatly decreased the sodium sensitivity of the Slack channel. One double mutation completely removed the sodium sensitivity of the channel, leaving residual single-channel activity in high cytosolic sodium concentration. Subsequently, molecular dynamics (MD) simulations revealed that one or two sodium ions could stay around the E373 sodium-sensitive site for a long time during the simulation process. Meanwhile, several other residues that vastly decreased the sodium sensitivity of the Slack channel also attracted sodium ions to stay, which implied that another sodium-sensitive site might exist. But no sodium ions could reside in the E373 and D863N/E865Q sites when we used the E373A/D863N/E865Q mutated channel model to perform MD simulations. The two potential sodium-sensitive sites in the interface of RCK1 and RCK2 domains of the same subunit differ from the Ca2+-sensitive sites located in the interface of different α subunits in the Slo1 channel, suggesting that conformations of RCK-based gating rings in the Slack channel are modulated differently from those in the Slo1 channel.
In addition, the MD simulations predicted the potential Cl– interaction sites of the Slack channel. Thus, we screened the predicted basic residues and identified the Cl–-sensitive site of the channel by comparing the sodium sensitivity with or without cytosolic Cl– ions. Together, we have identified two potential Na+-sensitive and one Cl–-sensitive sites of the Slack channel, providing the foundation for future studies on the relationship between physiological channel function and gating properties.
Materials and Methods
Mutagenesis and molecular biology
Slack channel mutations were generated using the QuickChange Site-Directed Mutagenesis Kit and verified by sequencing (Zhang et al., 2010; Tang et al., 2016). RNA was transcribed in vitro with T3 polymerase (Ambion). Transcribed cRNAs of each mutant were prepared at a concentration of ∼1–2 μg/μl. Each Xenopus oocyte was injected with 18.6–46.6 nl of cRNA. Macroscopic currents were typically recorded within 3–5 d.
Oocyte preparation
Stage VI Xenopus laevis oocytes were collected according to the standard protocol as previously described. All protocols and animal experiments that comply with ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines and regulations were approved by the Institutional Animal Care and Use Committee of Xuzhou Medical University (Tang et al., 2016). Before surgery, adult female X. laevis frogs were anesthetized by tricaine for 10 min. Under aseptic conditions, a small incision was made on one side of the abdomen to remove ovarian lobes. The lobes were immersed in sterile OR2 solution (85 mm NaCl, 5 mm KCI, 5 mm HEPES-NaOH, and 1 mm MgCl2, at pH 7.0) supplemented with tetracycline. Oocytes were defolliculated by shaking in OR2 solution containing 2 mg/ml collagenase (type II; Boehringer) at room temperature for 1–2 h and washed in ND96 solution (NaCl 96 mm, KCl 2 mm, MgCl2 1 mm, and HEPES 5 mm, at pH 7.5) at room temperature for another hour on a rotator (30 rpm).
Electrophysiology and data analysis
Macroscopic channel currents were recorded from an inside-out patch configuration with either an A-M 2400 patch-clamp amplifier (A-M Systems) or an EPC10 patch-clamp amplifier (HEKA). Data were filtered at 10 kHz. All currents were recorded within 5 min after patch excision to minimize significant rundown. Patch-clamp recording pipettes were made from borosilicate capillary tubes (catalog #B15023F, Vital Sense Scientific Instruments) with resistances of 2–4 MΩ. The standard pipette/extracellular solution for the inside-out patch without Cl– was the following ND96K-EGTA solution (in mm): 90 KGlu, 10 HEPES-K, 2 MgCl2, 5 EGTA, and 5 KOH, at pH 7.4. Bath/intracellular solutions contained the following (in mm): 90 KGlu, 10 HEPES-K, 2 MgCl2, 5 EGTA, 5 KOH, and 0–2000 NaGlu. The bath solution in the presence of Cl– using NaCl to replace NaGlu. Gigaohm seals were formed in the ND96K-EGTA solution with 0 Na+.
For testing the Cl– sensitivity of the Slack channel, the following perfused intracellular solutions were used: (1) 0 Na+ solution (in mm): 90 KGlu, 10 HEPES-K, 5 EGTA, and 5 KOH; (2) 250 Na+ solution (in mm): 90 KGlu, 10 HEPES-K, 5 EGTA, 5 KOH, and 250 NaGlu; (3) 250 Na+ solution with choline chloride: 250 Na solution with 0, 10, 25, 50, 75, and 100 mm choline chloride, respectively, and all solutions were titrated to pH 7.4 using KOH/MeS; and (4) 100 Na+ solution with choline chloride (in mm): 90 KGlu, 10 HEPES-K, 5 EGTA, 5 KOH, and 100 NaGlu with 0, 25, and 50 mm choline chloride.
Experiments were performed at room temperature (22–25°C). Salts of the compounds mentioned were purchased from Sigma-Aldrich or the Shanghai Shenggong Company. Data acquisition and analysis were conducted using pClamp9 (Molecular Devices), Clampfit 10.4, and Origin 7.5 software. The macroscopic currents recorded in different concentrations of [Na+]i were normalized to the maximal current in each patch. The EC50 values were calculated by fitting with the Hill equation. For the mutants that cannot reach the maximum open probability (po), even with the highest sodium concentration, the coefficient factor (Slope) of the Hill equation was fixed to 4 to estimate the minimum EC50 value. Data in all figures were expressed as the mean ± SEM. EC50 is the half maximal effective concentration that inducing channel open by the sodium or chloride ions.
The fitting function is as follows:
| (1) |
The n is the coefficient factor, while the k is the EC50. One-way ANOVA evaluated statistical significance. p < 0.05 was considered a significant difference. The n value in the tables gives the number of sampled patches for a given construct. The E373Q, M335A/E373Q, and M335A/E373A mutants were injected in parallel five times for single-channel recordings, and the oocytes were selected in a double-blind manner to test for possible contamination by endogenous currents.
Amplitude histograms of single-channel opening events were measured, idealized, and fitted with a Gaussian function using the software ANA.EXE provided by Michael Pusch (http://users.ge.ibf.cnr.it/pusch/programs-mik.htm), in which the single-channel traces could be selected, and histograms were calculated based on the bin setting as 1 pA. The histogram distribution values were calculated by setting an equidistant mean in the software algorithm. Then the channel values were counted based on the observed histogram peaks. Then the area under each peak was calculated by the Gaussian fit program. The po of the M335A mutant was calculated using the area under each peak (aj) at each current level (j) in the histogram, along with the number of channels (N) as follows:
| (2) |
The npo of various mutants in the absence of Na+ was estimated by setting N as the largest number of channels observed at 0 Na+ solutions. But, in calculating the po of E373Q/M335A and the E373A/M335A, the largest po cannot represent the actual channel numbers on the plasma membrane. Thus, the calculated po was defined as npo.
The Cl– dependent po was fitted by a dose–response function, as follows:
| (3) |
A1 is the minimal po in 0 Cl–, A2 is the maximal po at 50 Cl–, and the LogX0 is the EC50 of Cl– activation.
Confocal imaging
The HEK293T-cell cryotube was placed in a 37°C water bath for recovery. Once the frozen solution was thawed, the cells were mixed with 5 ml of the culture solution (10% fetal bovine serum in DMEM culture solution) and seeded into cell culture dishes specifically for confocal imaging. EGFP-N1 plasmids were prepared, including an open reading frame-encoded wild-type (WT) Slack channel-EGFP fused protein and its mutant (E373Q and E373A). Following the standard protocol, the plasmids were transfected into HEK293T cells with Lipofectamine 2000. After 48 h, wheat germ agglutinin (WGA) staining was performed under conditions avoiding light. Before staining, the HEK293T cells were washed three times with PBS buffer. Then cells were incubated with 5 µg/ml Alexa Fluor 568-labeled WGA for 20 min (Thermo Fisher Scientific). After washing with PBS three times, the cells were fixed with 4% PFA for 10 min and rewashed three times. Using a confocal microscope (model LSM 880, Zeiss), cells were observed while pictures were taken and analyzed using zen2.3 software. The colocalization rate was analyzed using ImageJ software with the plugin colocalization finder. The percentage of colocalization of EGFP (green) and WGA staining imaging (red) was calculated and demonstrated as overlapping rates using the ImageJ software. In brief, the images of the red and green channels were merged, and the color thresholds were set beside the RGB color shown in yellow. Then the overlap rate was calculated as yellow areas divided by green areas.
Immunoblotting and quantification of membrane expression of Slack channel mutants
According to the manufacturer protocol, membrane proteins were extracted from HEK293T cells transfected by rSlack-WT or rSlack-E373Q with a kit (KeyGEN). The process was as follows. First, the plates of HEK293T cells were washed twice with ice-cold PBS. Next, 1 ml of lysis buffer containing 1% protease inhibitor cocktail and 1 mm DTT was added to the plates. The cell lysates were harvested using a scraper and were pipetted into a 1.5 ml Eppendorf tube. The cell lysates were briefly vortexed and incubated for 1 min on ice and was repeated five times. Cytosolic proteins were separated by centrifugation at 12,000 rpm for 10 min at 4°C. The pellet was resuspended in cold Extraction Buffer, vortexed for 30 s, and placed on ice for 5 min, a process repeated five times. Last, membrane proteins were collected from the supernatant after centrifugation of the resuspended pellet at 12,000 rpm for 10 min at 4°C. Protein concentration and yield were determined using the BCA Protein Assay Kit (Beyotime Biotechnology). This method did not distinguish the internal membrane from the plasma membrane. Two hundred micrograms of membrane protein were electrophoresed and transferred by SDS-PAGE using 12.5% (w/v) acrylamide gels and blotted onto PVDF membranes (Immobilon-P). The protein blots were blocked with 5% nonfat milk in TBST. An appropriate number of the following primary antibodies was added and incubated overnight at 4°C: Na+/K+-ATPase α1 polyclonal antibody (1:1000; Abbkine); and mouse anti-GFP-Tag antibody (1:1000; ABclonal). After washing, the membrane was incubated for another 1 h at room temperature with the following secondary antibodies: HRP goat anti-mouse IgG (H + L; 1:2000; ABclonal), HRP goat anti-rabbit IgG (H + L; 1:2000; ABclonal). The membrane was rewashed in TBST, developed with clarity Western ECL substrate (Beyotime), and imaged on the Alliance Q9 Imager (UVITEC). Band intensities were determined using ImageJ software.
Molecular dynamics simulations
Modeling and simulation of the WT channel using the PDB file 5U70 as a template.
The molecular model of the rSlack tetramer was generated using a homology cryo-EM structure of the open state of the chSlo2.2 channel (Protein Data Bank ID:5U70) by the online SWISS-MODEL tool (https://swissmodel.expasy.org). The sequence alignment between chSlack and rSlack reveals ∼87.73% of homology. The constructed protein model was then put in a water box with 250 mm NaCl and hydrated with 446,706 water molecules.
GROMACS 5.1.4 was used for a free 500 ns dynamic simulation after energy minimization and equilibrium balancing. The amber 99-SB force field and the SPC/E model for water were used for the system. The temperature was set at 298.15K with Nosé–Hoover coupling. The Parrinello–Rahman method controlled the pressure at 1 atmosphere. Periodic boundary conditions were used, and the particle-mesh Ewald (PME) method was used to compute long-range electrostatic interactions. The bond lengths were constrained with the LINCS algorithm, while trajectories were saved every 50 ps for analysis. Molecular graphics were generated using VMD software. The simulation was conducted at the National Supercomputer Center (Lvliang City, People's Republic of China). Related calculations were performed on a TianHe-2 Supercomputer. To test whether the results were repeatable, the system was improved by using a dodecahedron water box to replace the cube water box to reduce the total number of molecules in the system; an additional 350 ns simulation was performed with 72,692 water molecules in the system.
Modeling and simulation of the WT channel and mutated channel using the PDB file 5U76 as a template.
Homology models of rSlack tetramer were built based on a crystal structure of the chSlack channel (Protein Data Bank ID:5U76). Ten initial rSlack channel homology models were generated using the MODELLER program based on the chSlack structure template, and the one with the best internal DOPE (Discrete Optimized Protein Energy) score of the program was selected for predicting the channel interactions. The H++ website server (http://biophysics.cs.vt.edu/) was used to add the hydrogen atoms. The pKa calculations determined the protonated states of the titratable residues at neutral physiological conditions. The CHARMM-GUI Membrane Builder webserver (http://www.charmmgui.org/?doc=input/membrane) was used to immerse the rSlack channel in the explicit lipid bilayer of POPC (1-palmitoyl-2-oleoylphosphatidylcholine), POPE (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine), POPS (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine), and cholesterol with a molecular ratio of 25:5:5:1, then the complexes were put in a water box of dimensions 178 × 181 × 195 Å, then 250 mm NaCl was added into the system. The system involved ∼481,000 atoms in the MD simulations. The Amber 16 program was used for MD simulations. The Antechamber module of AmberTools was used to generate the system parameters using the general AMBER (Assisted Model Building and Energy Refinement) force field. The tleap module neutralized the rSlack channel by adding additional Na+ or Cl– ions. The FF14SB and LIPID17 force fields were chosen for protein and mixed lipid membranes.
The simulations were conducted with the PMEMD.CUDA program in AMBER16 for 500 ns MD simulations. The MD simulations were performed with periodic boundary conditions to produce isothermal–isobaric ensembles (NPT, constant Number of atoms, Pressure, and Temperature). The pressure was regulated using the isotropic position scaling algorithm with the pressure relaxation time set to 2.0 ps. Long-range electrostatics were calculated using the PME method with a 10 Å cutoff. Before the program ran, the system conducted energy minimization using 2000 steps of the steepest descent followed by 3000 steps of conjugate gradient descent. Subsequently, the temperature of the system was increased from 0 to 303 K using Langevin dynamics with a collision frequency of 1 ps. During the increasing process, the protein was position restrained using an initial constant force of 500 kcal/mol/Å2 and weakened to 10 kcal/mol/Å2, allowing lipid and water molecules to move freely. Then, the system went through 5 ns equilibrium MD simulations. Finally, 500 ns production MD simulations were conducted, and coordinates were saved every 100 ps for analysis. A 4 fs timestep was used to accelerate the MD simulations by using a hydrogen mass repartition algorithm for system solutes.
The fractional occupancy was calculated as the sodium-bound site divided by the total site for a given binding site. When the distance between the sodium ion and oxygen atom of the given binding residue (E373 or D863) is shorter than 8 Å, the site is considered a sodium-bound site. Molecular graphics were generated using VMD. VMD is developed with NIH support by the Theoretical and Computational Biophysics group at the Beckman Institute, the University of Illinois at Urbana-Champaign.
Results
Neutralization of all acidic residues in the C terminus of the rSlack channel revealed mutants that dramatically decreased the sodium sensitivity of the slack channel
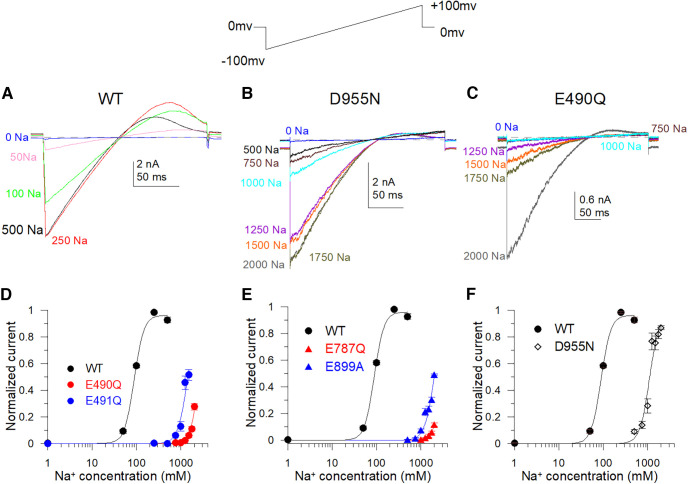
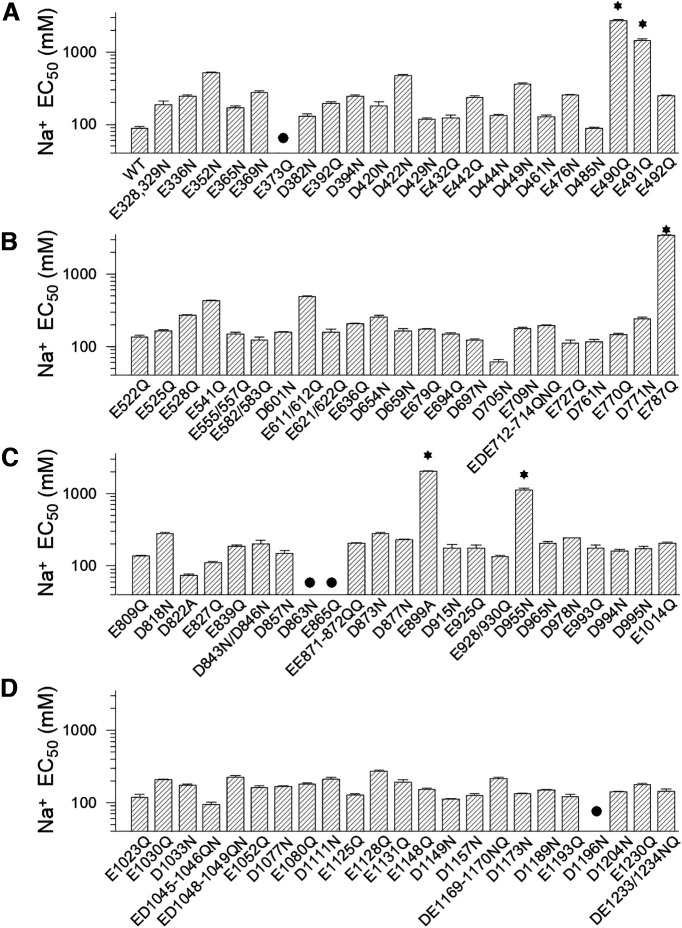
To test which acidic residues are responsible for sodium sense, we first individually neutralized all acidic residues in the cytosolic C terminus of the Slack channel by mutagenesis. Then a ramp protocol from −100 to 100 mV was used to measure sodium-dependent currents generated from channel mutants. The current levels at −90 mV in different cytosolic Na+ concentrations were used to estimate the sensitivity to Na+ of the wild-type and mutant Slack channels. The current levels measured in different Na+ concentrations were normalized to the current level at the highest Na+ concentration of each construct (Fig. 1A–C). The normalized currents were plotted and fitted by the Hill equation to generate the sodium dose–response curves and to calculate the EC50 or minimum EC50 of sodium dependence (concentration of sodium inducing a half-maximal effect for activation of the Slack channel, and in cases where the highest concentration used did not thoroughly saturate, we refer to the EC50 as a minimal EC50). Of the total 92 mutants tested (including double and triple mutants), some mutants mildly or moderately decreased Na+ sensitivity of the Slack channel (EC50 value increased twofold to fivefold). In contrast, the D955N mutant dramatically reduced channel sensitivity to Na+ by >15-fold (Fig. 1F). Moreover, the E787Q and E899A mutants (Fig. 1E) and the E490Q and E491Q mutants (Fig. 1D) decreased channel sensitivity to Na+ even more, because even when the cytosolic Na+ concentration reached 2 m, a saturated po had not been achieved. Thus, we estimated the minimum EC50 values of such mutants by fitting with the Hill equation using a coefficient factor of “4”. The minimum EC50 values were >1700 mm, which was >20-fold more elevated than the EC50 value of the WT Slack channel (Fig. 1D,E, Table 1). The EC50 values of all tested mutants of the Slack channel are summarized in Figure 2. Among the mutants tested, E373Q, D863N, E865Q, and D1196N did not elicit any current, even with 2 m cytosolic sodium ions (Fig. 2, Table 1). Thus, since these mutants could not be activated, one could not judge whether they lost sensitivity to Na+ or were functionally compromised because of a detrimental allosteric effect.
Figure 1.
Sodium-sensitive dependence of negatively charged mutants of the rSlack channel. A–C, Typical macroscopic current traces of the WT Slack channel (A), D955N (B), and E490Q (C) mutants were recorded by the inside-out patch configuration. Currents were elicited by a ramp protocol from −100 to +100 mV. D, Sample Hill equation fitted data of the Na+ dose–response generated from the macroscopic current of WT [EC50 = 89 ± 3.7 mm, coefficient factor (cf) = 4], E490Q (minimum EC50 = 2716 ± 84.7 mm, cf = 4), and E491Q (minimum EC50 = 1440 ± 64.3 mm, cf = 4) mutants. E, Hill equation fitted data of the Na+ dose–response in E787Q (minimum EC50 = 3391 ± 60.3 mm, cf = 4) and E899A (minimum EC50 = 2027 ± 38.4 mm, cf = 4) mutants. F, Hill equation fitted data of the Na+ dose response of D955N (minimum EC50 = 1114.3 ± 68.1 mm, cf = 4).
Table 1.
Characterization of the EC50 of slack channels negatively charged residue mutations
| Channels | EC50 or minimum EC50 | Patch numbers |
|---|---|---|
| WT | 89 ± 3.7 | 6 |
| E328/329N | 188 ± 23 | 6 |
| E336N | 245 ± 8 | 7 |
| D365N | 169 ± 910 | 7 |
| D369N | 277 ± 11 | 8 |
| E373Q | ||
| D382N | 129 ± 10 | 5 |
| E392Q | 195 ± 10 | 6 |
| D394N | 244 ± 9 | 8 |
| D420N | 181 ± 3 | 7 |
| D422N | 470 ± 21 | 9 |
| D429N | 117 ± 6 | 5 |
| E432Q | 123 ± 10 | 5 |
| E442Q | 236 ± 12 | 8 |
| D444N | 133 ± 5 | 5 |
| D449N | 358 ± 17 | 9 |
| D461N | 127 ± 9 | 5 |
| E476N | 254 ± 9 | 8 |
| D485N | 87 ± 5 | 6 |
| E490Q | 2716 ± 84.7 | 8 |
| E491Q | 1440 ± 64.3 | 8 |
| E492Q | 247 ± 8 | 8 |
| E522Q | 135 ± 9 | 4 |
| E525Q | 164 ± 6 | 5 |
| E528Q | 268 ± 5 | 8 |
| E541Q | 428 ± 6 | 8 |
| E555/557Q | 149 ± 10 | 4 |
| E582/583Q | 123 ± 12 | 5 |
| D601N | 156 ± 4 | 5 |
| E611/612Q | 487 ± 15 | 8 |
| E621/622Q | 158 ± 16 | 4 |
| E636Q | 205 ± 4 | 5 |
| D654N | 256 ± 17 | 6 |
| D659N | 164 ± 12 | 4 |
| E679Q | 173 ± 3 | 5 |
| E694Q | 149 ± 5 | 4 |
| D697N | 123 ± 5 | 4 |
| D705N | 61 ± 6 | 4 |
| E709N | 179 ± 7 | 4 |
| EDE712-714QNQ | 197 ± 2 | 6 |
| E727Q | 112 ± 11 | 4 |
| D761N | 115 ± 11 | 4 |
| E770Q | 145 ± 8 | 4 |
| D771N | 242 ± 13 | 7 |
| E787Q | 3391 ± 60.3 | 8 |
| E809Q | 136 ± 3 | 4 |
| D818N | 276 ± 14 | 8 |
| D822A | 74 ± 3 | 5 |
| E827Q | 111 ± 5 | 4 |
| E839Q | 185 ± 8 | 4 |
| D843-846N | 201 ± 25 | 6 |
| D857N | 148 ± 13 | 4 |
| D863N | ||
| E865Q | ||
| EE871-872QQ | 207 ± 3 | 6 |
| D873N | 278 ± 13 | 8 |
| D877N | 229 ± 4 | 8 |
| E899A | 2027.5 ± 38.4 | 8 |
| D915N | 177 ± 22 | 5 |
| E925Q | 176 ± 17 | 5 |
| E928-930Q | 135 ± 6 | 5 |
| D955N | 1114.3 ± 68.1 | 8 |
| D965N | 207 ± 10 | 6 |
| D978N | 244 ± 2 | 8 |
| E993Q | 177 ± 16 | 5 |
| D994N | 161 ± 8 | 5 |
| D995N | 173 ± 13 | 5 |
| E1014Q | 206 ± 6 | 8 |
| E1023Q | 119 ± 12 | 5 |
| E1030Q | 207 ± 5 | 8 |
| D1033N | 175 ± 5 | 6 |
| ED1045/1046QN | 94 ± 8 | 5 |
| ED1048/1049QN | 222 ± 13 | 8 |
| E1052Q | 160 ± 12 | 5 |
| D1077N | 169 ± 4 | 6 |
| E1080Q | 180 ± 8 | 6 |
| D1111N | 213 ± 10 | 8 |
| E1125Q | 129 ± 5 | 6 |
| E1128Q | 271 ± 9 | 8 |
| E1131Q | 191 ± 15 | 8 |
| E1148Q | 152 ± 6 | 6 |
| D1149N | 111 ± 3 | 8 |
| D1157N | 126 ± 6 | 8 |
| DE1169/1170NQ | 215 ± 9 | 6 |
| D1173N | 134 ± 1 | 6 |
| D1189N | 148 ± 4 | 6 |
| E1193Q | 120 ± 11 | 6 |
| D1196N | ||
| D1204N | 141 ± 3 | 5 |
| E1230Q | 179 ± 5 | 6 |
| DE1233/1234NQ | 143 ± 12 | 6 |
Figure 2.
Summary of Na+ sensitivity EC50 values for rSlack mutants. A–D, The sodium dependence EC50 values of 91 mutant constructs (including some double or triple amino acid mutants) are summarized. The Na+ sensitivity EC50 values were estimated. *Labels the minimum EC50 values of E490Q, E787Q, and E899Q mutants. The comparison of the EC50 of the WT channel with the EC50 of E490Q (p = 0.0009), E491Q (p = 0.001), E787Q (p = 0.0009), E899Q (p = 0.001), and D955 (p = 0.01) shows significant difference. The mutants (E373Q, D863N, E865Q, and D1196N) that did not show macroscopic currents were labeled with black circles.
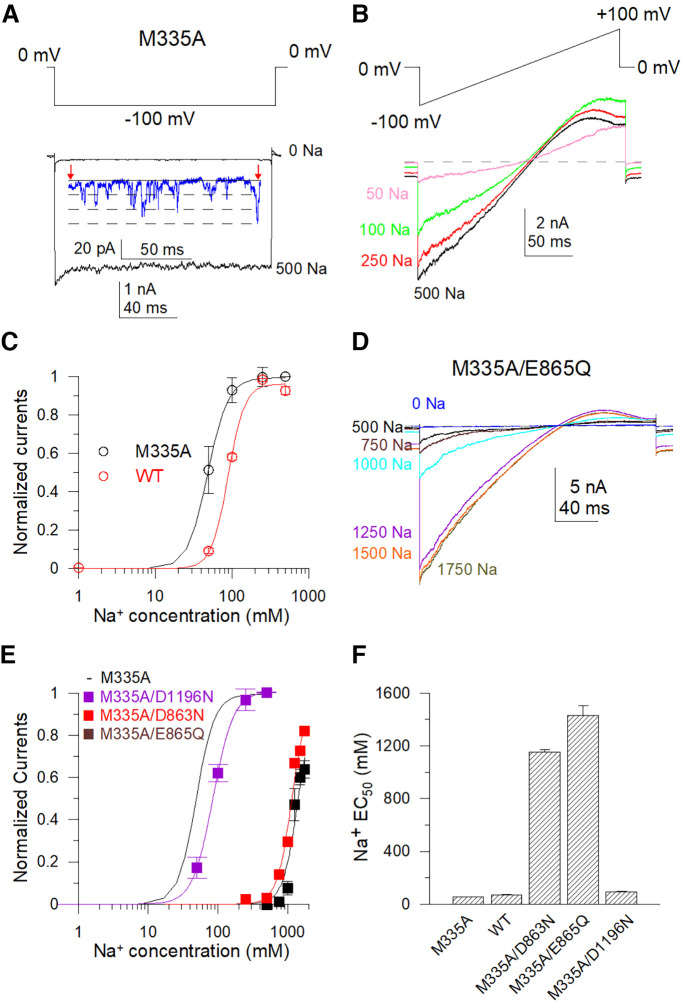
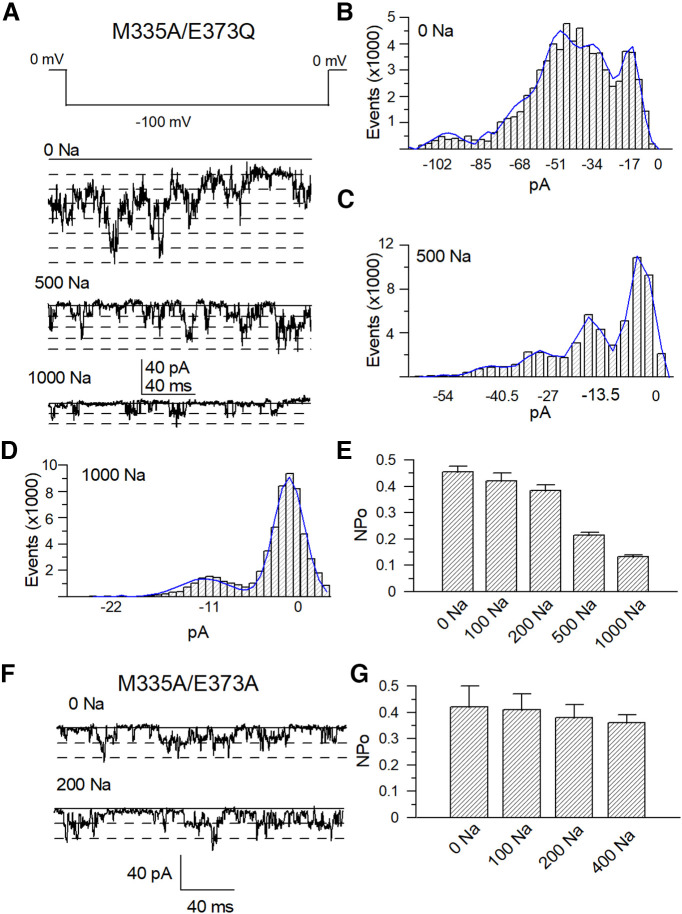
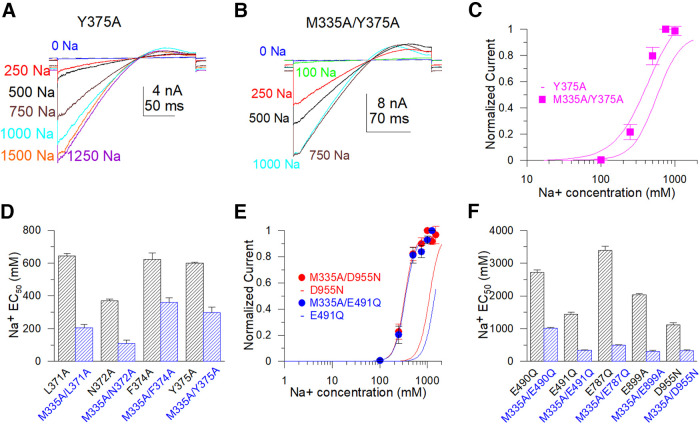
To assess the role of these “nonfunctional” mutants, we took advantage of the mutant M335A, corresponding to M333A in the chSlo2.2 channel, which shows channel activity even in zero cytosolic Na+, as it is located at the narrowest segment of the potassium ion permeation pathway (S6 helices gate) just below the ion selectivity filter (Hite and MacKinnon, 2017). The M335A mutant demonstrated single-channel level currents in the absence of cytosolic Na+, while ∼2.5 nA macroscopic currents could be measured in 500 mm [Na+]i (Fig. 3A). The calculated po was 0.0002 in the absence of [Na+]i. The EC50 value of the sodium dose dependence of M335A mutant is 50% lower than the WT Slack channel, suggesting that this mutant also enhanced the ability of sodium to activate the Slack channel (Fig. 3A,C). We generated the following double mutants of the four nonfunctional mutants in the background of the M335A mutant: M335A/E373Q, M335A/D863N, M335A/E865Q, and M335A/D1196N. Using this approach, we successfully rescued macroscopic currents from three double mutants, except for the M335A/E373Q mutant. The M335A/D863N and M335A/E865Q mutants exhibited dramatically decreased sodium sensitivity (EC50 values of these double mutants were >20-fold higher than the EC50 value of the M335A mutant), whereas the M335A/D1196N mutant only marginally reduced the sodium sensitivity of the Slack channel (Fig. 3D–F). However, although the M335A/E373Q mutant did not exhibit macroscopic current, it did maintain a single channel-level current in either the absence or presence of a high concentration of [Na+]i (Fig. 4A). The open probability of this mutant was not increased by increases in cytosolic Na+ concentration. Sample traces of the M335A/E373Q mutant show a total of seven discrete observed channel levels in the absence of Na+ and a total of four-channel levels in the presence of 500 mm Na+ (Fig. 4B,C). The averaged npo from different patches was calculated using the number of the most simultaneously opened channels seen without [Na+]i (n = 10). The npo of the M335A/E373Q mutant gradually declined in the presence of 0–1000 mm cytosolic Na+ rather than increased (Fig. 4D, Table 2). Then, we tested another mutant, M335A/E373A, which also demonstrated single channel-level currents in 0–400 mm cytosolic Na+ but without sodium dependence (Fig. 4F,G, Table 2). Thus, the M335A/E373Q and M335A/E373A mutants lost sensitivity to Na+ but were still functionally expressed on the cell membrane. These results suggest that the E373 and D863/E865 may form two different sodium-sensitive sites.
Figure 3.
Double mutants with M335A show functional expression but decreased sodium sensitivity. A, Sample single-channel current of M335A mutant with 0 mm intracellular sodium and macroscopic typical current traces with 500 mm cytosolic Na+ were recorded in the inside-out patch configuration. A, −100 mV hyperpolarization generated the currents. B, Sample current traces of M335A with 50–500 mm cytosolic Na+ are shown. Currents were generated by a −100 to +100 mV ramp protocol. C, Sodium sensitivity of rSlack channel and M335A mutants were estimated by Hill equation fitted data. D, Sample current traces of the M335A/E865Q mutant were recorded in the inside-out patch configuration. E, Hill equation fitted the data of the Na+ dose response of D863N/M335A [minimum EC50 = 1154 ± 17.5 mm, coefficient factor (cf) = 4], E865Q/M335A (minimum EC50 = 1430 ± 73.2 mm, cf = 4), and D1196N/M335A (EC50 = 82.6 ± 2.6 mm, cf = 4) mutants. F, Summary of EC50 and minimum EC50 values for the wild-type Slack channel, M335A (EC50 = 49 ± 3 mm, cf = 4), M335A/D863N, M335A/E865Q, and M335A/D1196N.
Figure 4.
Double mutant M335A/E373Q of the Slack channel can be functionally expressed but displays a complete loss of sodium sensitivity. A, Sample single-channel current traces of M335A/E373Q mutant with different cytosolic Na+ concentrations. B–D, Total amplitude histogram of single-channel currents with 0−1000 mm [Na+]i was fitted with a Gaussian function. E, Comparison of averaged po values of M335A/E373Q in different cytosolic Na+ concentrations (Table 3). F, Sample traces of M335A/E373A in 0 and 200 mm [Na+]i, respectively. G, Total amplitude histogram of single-channel currents with 0–400 mm [Na+]i were fitted with a Gaussian function, respectively. The open probabilities of the M335A/E373A in different cytosolic sodium concentrations (Table 3).
Table 2.
The single-channel recording po of the slack channel mutants
| Sodium channel (mm) | 10 po | 25 po | 50 po | 100 po | 200 po | Patch numbers |
|---|---|---|---|---|---|---|
| M335A | 0.002 ± 0.001 | 0.08 ± 0.02 | 0.53 ± 0.05 | 0.9 ± 0.05 | 0.92 ± 0.05 | 4 |
| Sodium channel (mM) | 50 npo | 100 npo | 200 npo | 500/400 npo | 1000 npo | Patch numbers |
|---|---|---|---|---|---|---|
| M335A/E373Q | 0.45 ± 0.022 | 0.42 ± 0.031 | 0.385 ± 0.020 | 0.21 ± 0.01 | 0.132 ± 000.6 | 10 |
| M335A/E373A | 0.42 ± 0.08 | 0.41 ± 0.06 | 0.38 ± 0.050 | 0.35 ± 0.03 | 8 |
To further test whether the lack of sodium sensitivity of the M335A/E373Q mutant was because of allosteric regulation, we mutated several other amino acid residues in the vicinity of E373 to alanine, including L371A, N372A, F374A, and Y375A, and examined their sensitivity to sodium. These mutants mildly altered the sodium sensitivity of the rSlack channel but still generated macroscopic currents (Fig. 5A,C, Table 3). This result indicates that adjacent residues of E373 seem not to be critical in disrupting the sodium sensitivity of this channel. Additional experiments also confirmed the role of M335A in enhancing the ability of the sodium interaction to activate the channel because the sodium EC50 values of all tested double mutants were >50% lower than the EC50 values of the single mutants without M335A (Fig. 5B,D,F).
Figure 5.
M335A increases the sodium sensitivity of Slack channel mutants. A, Sample current traces recorded in the inside-out patch configuration for Y375A. The ramp protocol ran from −100 to +100 mV. B, Sample current traces recorded in the inside-out patch configuration for the Y375A/M335A mutant with a ramp protocol ran from −100 to +100 mV. C, Hill equation fits the Na+ dose dependence data of Y375A and Y375A/M335A mutants. D, Comparison of EC50 values of the sodium dependence of L371A to Y375A with the EC50 values of the sodium dependence of double mutants of M335A plus corresponding mutants from L371A to Y375A. E, Hill equation fits the Na+ dose dependence data of M335A/D955N and M335A/E491Q. F, Comparison of minimum EC50 values of sodium dependence of E490Q, E491Q, E787Q, E899A, and D955N with the minimum EC50 values of sodium dependence of M335A/E490Q, M335A/E491Q, M335A/E787Q, M335A/E899A, and M335A/D955N.
Table 3.
Characterization of EC50 of sodium dependence of the rSlack channel mutations
| Channels | EC50 or minimum EC50 (in mm) | Patch numbers |
|---|---|---|
| M335A | 49 ± 3 | 4 |
| M335A/E490Q | 1072 ± 36 | 6 |
| M335A/E491Q | 341 ± 21 | 6 |
| M335A/E787Q | 480 ± 34 | 6 |
| M335A/D863N | 1154 ± 17.5 | 7 |
| M335A/E865Q | 1430 ± 73.2 | 8 |
| M335A/E899A | 982 ± 56 | 6 |
| M335A/D955N | 331 ± 12 | 6 |
| M335A/D1196N | 82.6 ± 2.6 | 5 |
| L371A | 643 ± 21 | 6 |
| N372A | 369 ± 20 | 5 |
| F374A | 623 ± 30 | 5 |
| Y375A | 600 ± 34 | 6 |
| M335A/L371A | 204 ± 17 | 5 |
| M335A/N372A | 110 ± 10 | 5 |
| M335A/F374A | 359 ± 38 | 6 |
| M335A/Y375A | 297 ± 6 | 5 |
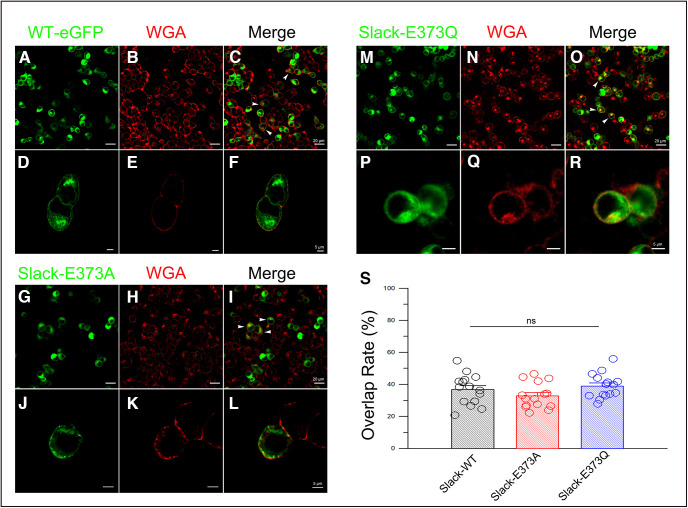
To test whether the membrane expression level of M335A/E373Q is altered, we generated expression constructs in which the EGFP protein was fused to the distal end of the C terminus of the WT rSlack channel or the rSlack E373Q, and E373A mutants (carried by the pEGFPN1 vector). Then the plasmids were transfected into HEK293T cells to assess fluorescence expression on the plasma membrane. The EGFP-fused Slack channel and the E373Q mutant were expressed in the cytosol but also colocalized with WGA, a plasma membrane marker labeled with Alexa Fluor 568. The fluorescence intensities on the membrane generated from EGFP fused with WT channel or E373Q/E373A mutants did not show significant differences (Fig. 6A–R). The colocalization rates of EGFP and WGA on the plasma membrane did not exhibit a significant difference among the WT Slack channel (0.37 ± 0.02), E373Q (0.38 ± 0.02), and E373A (0.33 ± 0.02) transfected cells (Fig. 6S). Furthermore, we purified the membrane proteins from transfected HEK293T cells and performed a Western blot analysis using an anti-GFP tag antibody. The results showed that the membrane expression levels of the WT Slack channel, E373Q, and E373A mutations were not significantly different (Fig. 7A). Although this method did not distinguish plasma membranes from internal membranes, these data suggested that the E373Q and E373A mutants do not alter the membrane expression levels of the channel.
Figure 6.
The E373Q mutant is expressed on the cell membrane in transfected HEK293 cells. A, Green fluorescence showing the expression of WT rSlack-EGFP in rSlack-EGFPN1 transfected HEK293 cells. B, The WGA staining on the cell membrane of the fixed HEK293 cells (red). C, The merged image shows the colocalization of WT rSlack-EGFP and WGA staining on the cell membrane of the HEK293 cells. D–F, The magnified picture of one cell shows the colocalization of the EGFP and WGA staining on the cell membrane. G–I, Green fluorescence shows the rSlackE373Q-EGFP expression in transfected HEK293 cells (G), while WGA stains show rSlackE373Q-EGFP expression in the HEK293 plasma membrane (H), and the merged expression of rSlackE373Q-EGFP and WGA in the transfected HEK293 cells (I). J–L, The magnified picture of merged fluorescence signals of the expression of E373Q-EGFP mutant and WGA staining on the plasma membrane in the HEK293 cells. M–R, The EGFP expression and WGA staining image of the R373A mutant. S, The overlap rates of EGFP and WGA staining of the WT, E373Q, and E373A mutants of the Slack channel.
Figure 7.
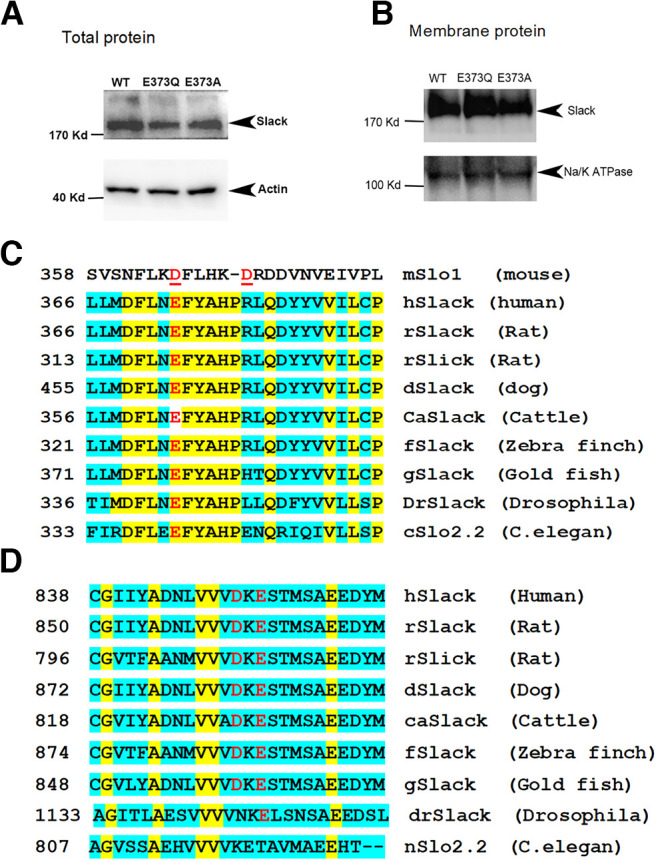
Western blot (WB) analysis of WT Slack channel, E373Q, and E373A membrane expression in transfected HEK293 cells. A, Western blot results of the expression of WT Slack channel, E373Q, and E373A mutants in total proteins. B, Western blot results of the expression of WT Slack channel, E373Q, and E373A mutants in the membrane protein fractions. C, Alignment of the context segments around the E373 sodium-sensitive site of Slack channel and Slo1 channel in different species. The species protein sequence IDs are as follows: mSlo1 human Slack, XP_011517182.1; rSlack, NP_068625.1; dog Slack, XP_005625191.1; cattle Slack, XP_027412510.1; zebra finch Slack, XP_027412510.1; Drosophila Slo2, EDW47242.1; nSLo2, NP_001024527.1; rat Slack, Q6UVM4.1; and goldfish Slack, XP_026052475.1. D, The alignment of the context segments around the D863/E865 sodium-sensitive site of the Slack channel.
Next, we aligned the neighboring amino acid sequences flanking the E373 and D863/E865 residues among Slack channels in different species. The rE373 site is conserved in almost all species, including the Ca2+-activated C. elegans Slo2 channel, while the D863/E865 is conserved in most aligned Slack channel sequences, except the C. elegans Slo2 channel (cSlo2) and the Drosophila Slack channels (Fig. 7B,C). The high conservation of E373 and D863/E865 in Slack channels supports a role for these residues as candidates for sodium-sensitive sites.
We compared the single-channel conductance of M335A with M335A/E373Q mutants to test whether their single-channel properties are similar when Na+ activates them. Single-channel currents of M335A were first measured at −100 mV membrane potential in different cytosolic Na+ concentrations (Fig. 8A). The open probability of M335A was plotted and fitted. The sodium dependence of the po of the M335A from single-channel recordings gave an EC50 of sodium at 46.5 ± 2.4 mm (Fig. 8B), consistent with the sodium dependence EC50 of M335A calculated by the G/Gmax in macroscopic current recordings (Fig. 3C). Single-channel currents of the M335A and M335A/E373Q mutants were measured from −100 to −200 mV in 0 mm cytosolic Na+ and plotted to calculate single-channel conductances (Fig. 8C,D, Table 2). In the absence of cytosolic sodium, the single-channel conductance of the M335A was 170 ± 5 pS, comparable to the conductance of M335A/E373Q of 163 ± 10 pS. These conductance estimates were 25 pS larger than the single conductance of the WT Slack channel measured in the presence of 50 mm cytosolic sodium (144 ± 4 pS), comparable to the single-channel conductance of the M335A mutant in 50 mm cytosolic sodium (145 ± 3 pS), suggesting a sodium ion-dependent blocking effect (Fig. 8A). In double-blinded recording experiments, the E373Q-injected oocytes were assessed in parallel injections of the M335A and M335A/E373Q mutants (at least five times). Uninjected oocytes under our recording conditions did not yield single-channel currents.
Figure 8.
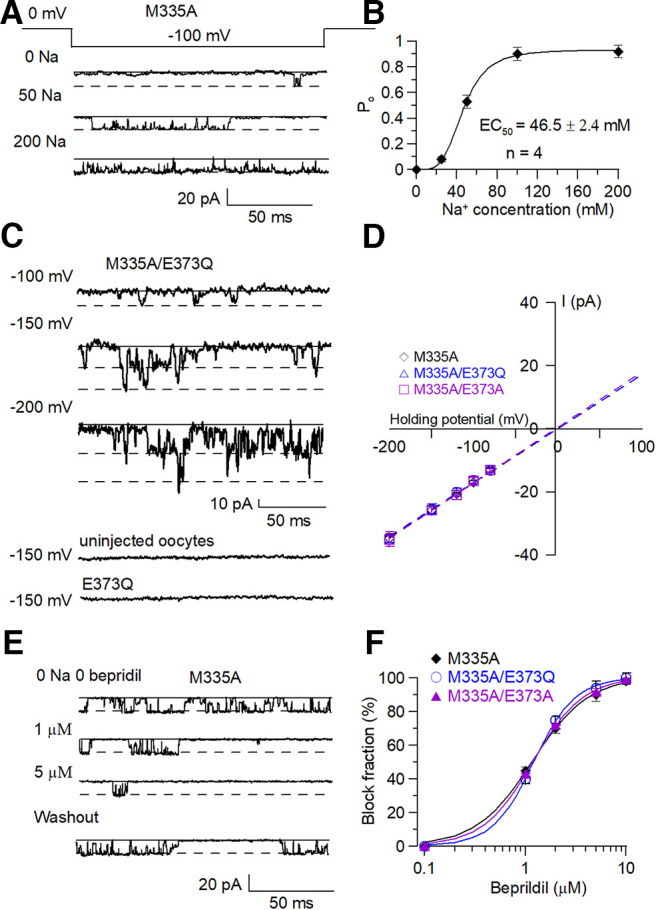
M335A/E373A and M335A/E373Q mutants show similar single-channel characteristics with the WT rSlack channel. A, The sample traces of M335A mutant in the presence of different cytosolic sodium ions at −100 mV membrane potential. B, The Hill equation fitted the Slack channel open probability versus sodium concentration data. The EC50 value is 46.5 ± 2.3 mm. C, The sample single-channel traces of M335A/E373Q mutant at indicated voltages in intracellular solution with 0 mm cytosolic Na+. D, The plotted traces show M335A single-channel conductance is 170 ± 5 pS while M335A/E373Q and M335A/E373A single-channel conductance is 163 ± 10 and 167 ± 4 pS at 0 mm cytosolic sodium, respectively. E, The sample single-channel traces of the M335A mutant were blocked by bepridil at indicated concentrations. F, The Hill equation fitted the dose–response curve of the bepridil blocking on M335A, M335A/E373A, and M335A/E373Q mutants. The blocking EC50 values of bepridil on these mutants are 1.55 ± 0.17, 1.2 ± 0.03, and 1.18 ± 0.04 μm, respectively.
To further confirm that the recorded single-channel currents were arising from the Slack channel, we tested whether the M335A, M335A/E373Q, and M335A/E373A mutants could be blocked by the Slack channel-specific blocker bepridil (Fig. 8E). The results indicated the po of these three mutants was inhibited by bepridil in a dose-dependent manner. The dose response and IC50 of the blocking effect were consistent with previous reports (Yang et al., 2006). All these results collectively led us to conclude that E373 is a Na+ interaction site in the Slack channel.
Molecular simulations revealed that besides the E373/D955 site, Na+ could interact stably with the acidic pocket residues in multiple conformations exemplified by the D863/E865 residues
Although we had experimentally identified sites that control Na+ sensitivity, how the sodium ions interact with the E373 and D863/E865 residues remained unclear. To test whether sodium ions could reside at E373 or D863/E865 sites, we set up a tetrameric rSlack homology model using the chSlack channel structure as a template (PDB:5U70; Hite and MacKinnon, 2017). We then performed a 500 ns molecular simulation (MD) of the rSlack model in a water box with 250 mm NaCl added. The stability of the channel was examined by calculating the root mean square deviations (rmsDs) of the initial structure of the track. The rmsD for the rSlack channel conformation in a water environment relative to the starting structure reached equilibration and fluctuated at ∼1 nm after 100 ns simulation time, indicating high stability for the MD simulation system (Fig. 9A). Then we analyzed the trajectory and observed whether the sodium ions resided in the residues whose mutants decreased the sodium sensitivity of the Slack channel. The 2195 Na+ ions were randomly placed in an initial position in the modeling system. The average traveled distance of Na+ ions from their initial position in the system to the end of the simulation was ∼100 Å (Fig. 9B). Ions localizing near residues E373 and D863/E865 were stable in their position. During the simulation process, two sodium ions moved to a place close to the E373/D955 residues on subunits b and c of the modeled rSlack channel, respectively, in which the sodium ions stably resided in the sensitive site for >250 ns until the end of the MD simulation run. After traveling close to the residing position, the distances of the sodium ion to residues E373/D955 were 2-8 Å during the simulation process (Fig. 9C,D). Interestingly, on subunit c of the modeled channel, an additional sodium ion traveled to the position close to residues E373/D955 and resided at the site until the end of the simulation (Fig. 9E,F). Thus, two sodium ions could simultaneously stay at E373/D955 for >200 ns. Another sodium ion resided in a position close to residues E490, E491, E787, D863, and E865 (Fig. 9G). The five residues were spatially close to one another, forming an acidic pocket that could trap sodium ions inside the pocket (Fig. 9H). On subunits b, c, and d of the rSlack model, the time of sodium ions resided in the pocket is >250 ns of the 500 ns run (Fig. 10A). The distances from the resident sodium ion to the residues of the pocket ranged between 0.5 and 8 Å (Fig. 9C,E,H). These simulation results revealed two distinct sodium ion interacting sites forming on the rSlack channel model. The average distance from the resident sodium ion to residue E373 was shorter than the distance to residue D955 (Fig. 9C). This result is consistent with the electrophysiological data that the sodium sensitivity of the M335A/E373Q mutant is lower than that of the D955N mutant. Similarly, the averaged distances from the resident sodium ion to residues D863 and E865 were also shorter than those to residues E490, E491, E787, and E899 (Fig. 9G). This result is consistent with the electrophysiological data that the D863N and E865Q mutants' sodium sensitivities are lower than those of the E490, E491, E787, and E899 mutants. The sodium on/off events from the acidic residues of this pocket during the 500 ns simulation of each tetramer Slack channel model subunit were summarized by a color-coded bar (Fig. 10A). To further test that the residence of the sodium ions near the residues we identified did not happen by chance, the simulation was repeated in a minor water box system for another 350 ns. The results of this second run reproduced the findings of the first run, namely that the sodium resided closest to the E373/D955 site and within an acidic pocket close to D863/865 (Fig. 10B). In this model, we also observed stable interactions of the Na+ ions with the E373 residue. When the sodium ions reached the E373 residue, the rmsD of the ions became fixed values, indicating that they stopped moving (Fig. 10C,D). Meanwhile, the distance between the E373 and D955 became short after sodium bound stably (Fig. 10E). The distance between E373 to D955 could be reduced from 7.7 to 2.3 Å as the sodium ions bound to the site (Fig. 10F). On another binding pocket, we measured the distances between residue D863 and the residues E899, E787, E490, and E491 during the simulation process (Fig. 11A). The distances from D863 to residue E787 and E891 were relatively large before the sodium binding (∼8 and 6 Å, respectively, at 140 ns). Still, they became somewhat shorter and fixed after sodium binding (∼5 and 3 Å, respectively, at 210 ns following sodium binding), indicating the stable sodium resident state is maintained when the sodium binds stably in the pocket (Fig. 11B).
Figure 9.
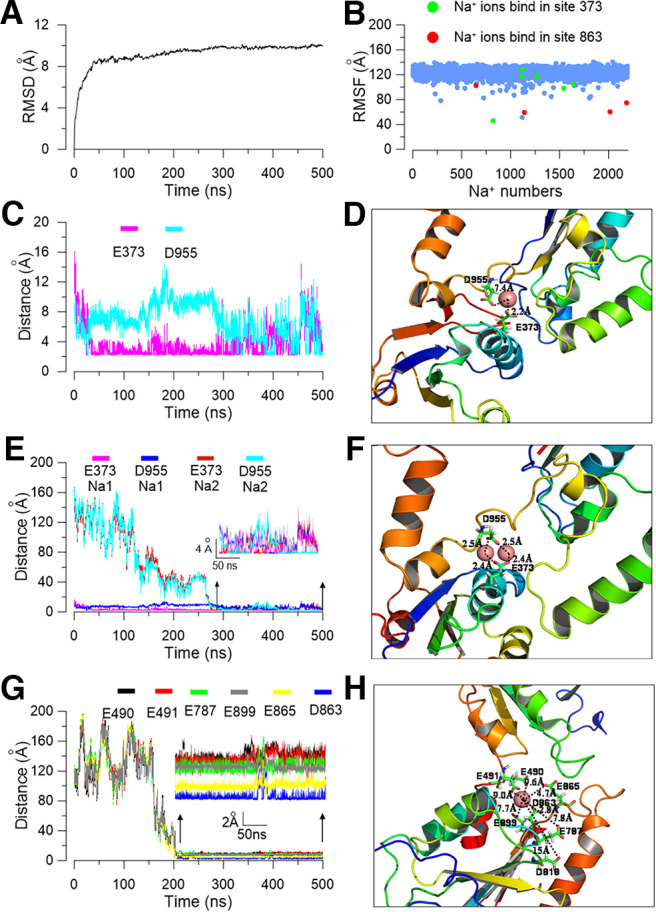
Molecular simulation shows two potential sodium binding sites in the C terminus of the Slack channel. A, The rmsD of the backbone of the Slack channel during MD simulation. B, The rmsF of total Na+ ions in the simulation system of a Slack channel homology model; the red-labeled sodium ions are in the E373 residue, while the green-labeled sodium ions are in the D863 position. C, The distances from the residing sodium ions to residues E373 and D955 in subunit b during the MD simulation process. D, A snapshot of the E373 sodium interaction site with only one sodium ion located between the E373 and D955 residues. E, During the MD simulation, the distances from two simultaneously resident sodium ions to residues E373 and D955 in the subunit c of the Slack channel. F, A snapshot of the simulated local structure of equilibrated coordinated sites with a conformation that two sodium ions simultaneously bind close to the E373 and D955 residues. G, The sodium ion distances to residues D863, E865, E787, E899, E490, and E491 in the acidic sodium pocket during the MD simulation in the Slack channel model. H, Simulated local structure of the acidic pocket formed by E490, E491, E787, D818, D863, and E865 amino acids of the Slack channel.
Figure 10.
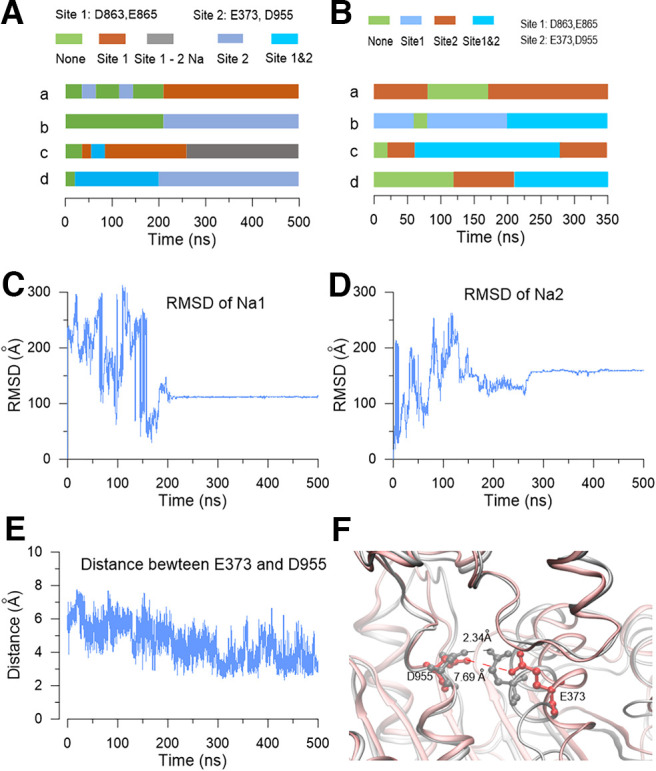
The time of the resident sodium ions staying in the sodium-sensitive site of the rSlack channel during the MD simulation. A, The time of four subunits with one or two sodium sites occupied or vacant in the homology model during the 500 ns MD simulation was indicated by different colors. B, The time of the four subunits with one or two sodium sites bound by one or two sodium ions and one vacant site during the 350 ns MD simulation. C, D, The rmsD of the two sodium ions (Na1 and Na2) binding to the E373/D955 site during the 500 ns MD simulation. E, The distance between the E373 and D955 site of one subunit during the 500 ns MD simulation. F, The snapshots of the local structure of the E373/D955 site with (gray) or without (red) sodium binding.
Figure 11.
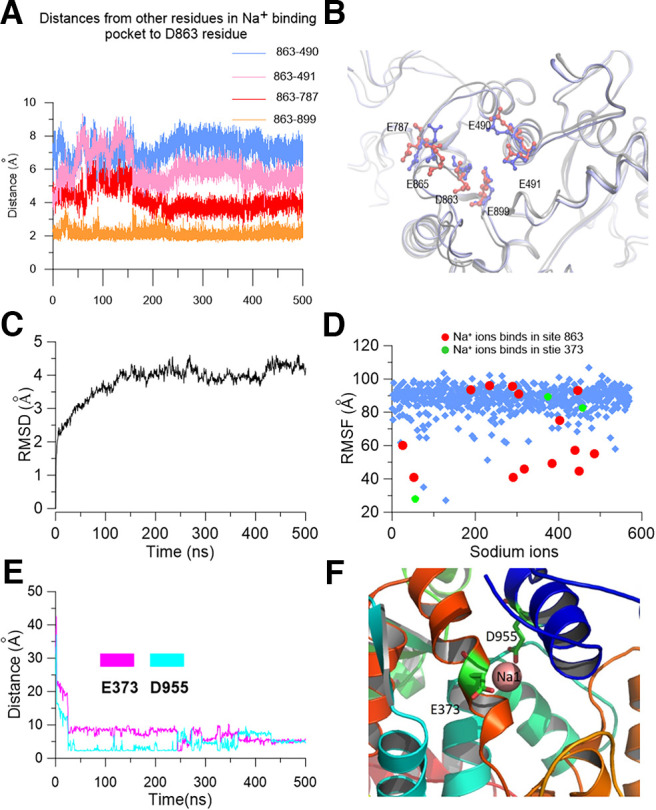
The location of the sodium binding site on the RCK1 and RCK2 interface in the Slack channel. A, Distances from residue D863 to residues E899, E787, E490, and E491, respectively, during a 500 ns MD simulation. B, The local structure of the sodium binding pocket around D863/E865. The blue color shows residues being closer to residue D863 at 210 ns. The red color shows residues relatively far away from D863 at 140 ns when the site is vacant. C, The rmsD of the backbone of the Slack channel model using 5U76 as the template. D, The rmsF of Na+ ions in the Slack channel homology model using the structure of the chSlack channel (PDBID:5U76). The red-labeled sodium ions resided in the D863/E865 pocket, while the green-labeled sodium ions resided in the E373 position. E, Distances from the resident sodium ion to residues E373 and D955 in subunit 1 during the MD simulation. F, The local structure of the sodium ion (Na1) resides in the E373/D955 position.
To test whether the sodium can bind with the channel in a closed-state conformation, we also ran an MD simulation using the template (PDB:5U76), which was acquired in a solution without sodium and probably represented a closed state of the KCNT1 channel. Thus, it likely can provide the initial steps of the sodium ion binding to the KCNT1 channel. The system stability and the sodium ion rms fluctuation (rmsF) are shown (Fig. 11C,D). In addition to consistently observing sodium ions stayed for a time at the position close to the E373/D955 coordination site (Fig. 11E,F), we also observed different sodium ions binding conformations of D863/E865 site from the local sodium ions binding conformation at this site obtained using the (PDB:5U70) template (Fig. 12A–D). For example, in one subunit (subunit 1) of the tetramer, we observed that at most four sodium ions could simultaneously interact with the D863/E865/E787/D818 residues (Fig. 12D). One sodium ion (Na3) resided in the position close to the D863/E865 site during the entire 500 ns simulation process, perhaps coordinated by three acidic residues of the acidic pocket, D863, E865, and D818. In contrast, three other sodium ions stayed for 134, 135, and 213 ns, respectively, and finally (Na2, Na4, and Na5) evaded the site (Fig. 12A,C). In another subunit (subunit 4), two sodium ions (Na6 and Na7) could reside in the interaction pocket, in which the E787/D818/D863/865 residues formed a conformation different from that in the subunit 1 (Fig. 12E,F). These results suggested that the sodium interaction pocket around 863/865 residues could form multiple conformations in a sodium solution, consistent with the various structures obtained by cryo-EM (Hite and MacKinnon, 2017).
Figure 12.
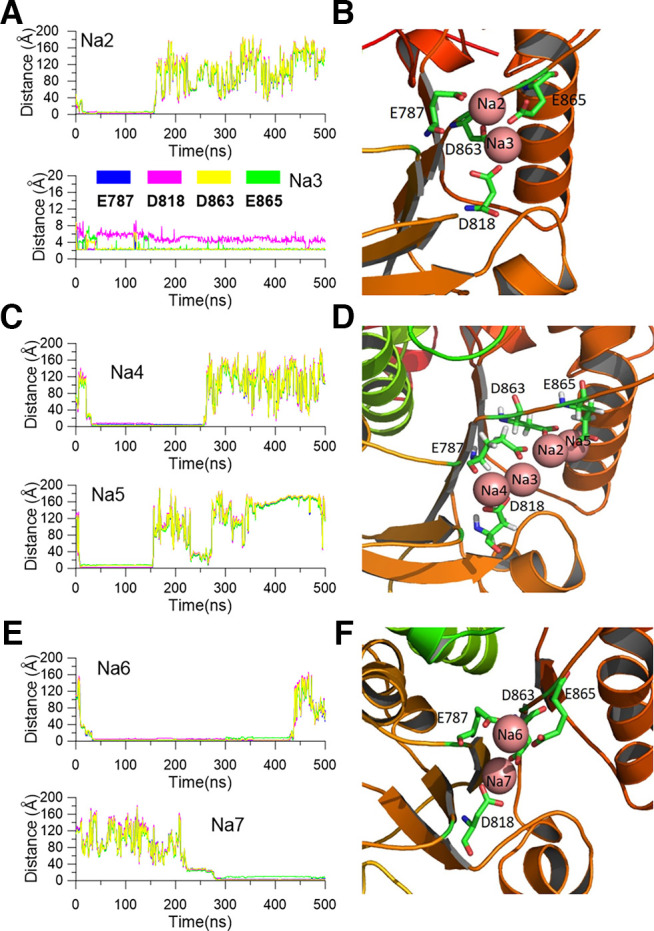
The distances from sodium ions to the interacting residues and the snapshot of the local structure of sodium interaction sites. A, The distances from the resident sodium ions (top, Na2; bottom, Na3) to the residues D863/E865/E787/D818 in subunit 1. B, The snapshot of the local structure when two sodium ions resided in the D863/E865 pocket. C, The distances from the Na4 (top) and Na5 (bottom) to D863/E865/E787/D818 residues in subunit 1. D, The snapshot of the local structure D863/E865 pocket when four sodium ions are located in the site. E, The distances from the sodium ions (Na6 top and Na7 bottom) to the residues D863/E865/E787/D818 in subunit 4. F, The local structure of the D863/E865 pocket in subunit 4 with two resident sodium ions.
To test whether mutation of these residues could prevent sodium binding with the channel, we subsequently did a molecular simulation using the sodium interaction site mutated template (D863N/E865Q and E373Q) to test whether the sodium ions still can bind with the channel (Fig. 13A,B). The result showed that in addition to a sodium ion momently staying in a position that is close to one subunit of the channel (Fig. 13C), with a distance 7–9 Å beyond the space of interaction with the mutated residue, the sodium ions cannot bind with the mutated interaction sites anymore in the 500 ns simulation process (Fig. 13D,E).
Figure 13.
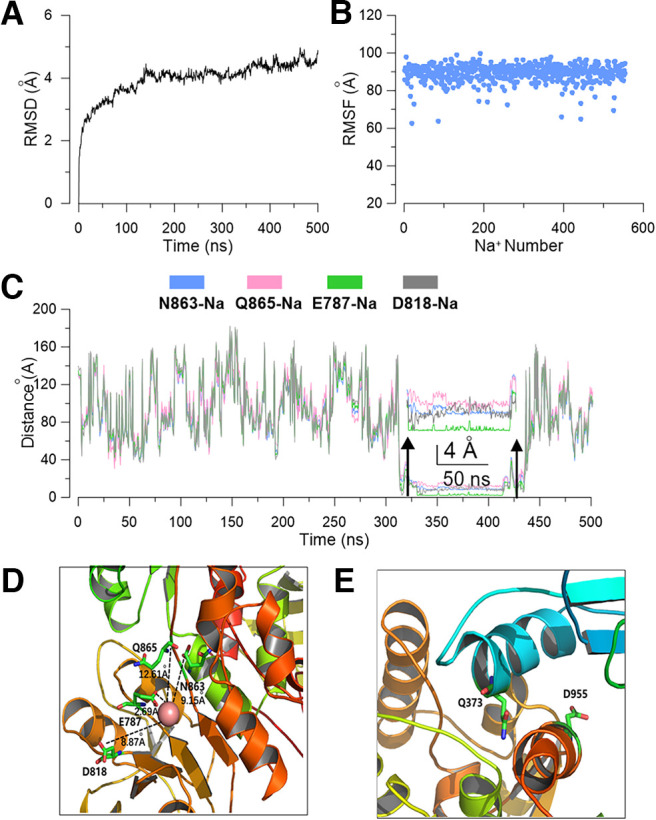
Molecular simulation shows that the D863/E865 and E373 mutated channels can no longer bind sodium. A, The rmsD of the backbone of the mutated Slack channel during MD simulation (5U76 as a template). B, The rmsF of total Na+ ions in the simulation system of a Slack channel homology model. C, During the MD simulation process, the distances from a sodium ion to residues D863/E865/E787/D818 pocket in subunit b. D, E, Snapshots of the simulated local structure of N863/Q865 (D) and the E373 (E) residues.
Determinants of Cl– sensitivity of the slack channel
To address how the Cl– ions modulate the gating of the Slack channel, we took advantage of the trajectories of the MD simulations to analyze the behavior of the Cl– ions in the system. All Cl– rmsFs were analyzed and sorted from small to large for every 100 ns (Fig. 14A). Then, the trajectories of possible bound Cl– were observed with VMD software. Basic amino acids <10 Å to stable (nonmobile) Cl– ions were considered a possible Cl– interaction site. The candidates of Cl–-interacting sites were listed in a table (Fig. 14B). Basic residues of the rSlack channel conserved in the cSlo2 were excluded as possible Cl– binding sites, as the cSlo2 channel is not sensitive to Cl– (Zhang et al., 2013). The sequence numbers and positions of the amino acids in the Slack channel are shown (Fig. 14C).
Figure 14.
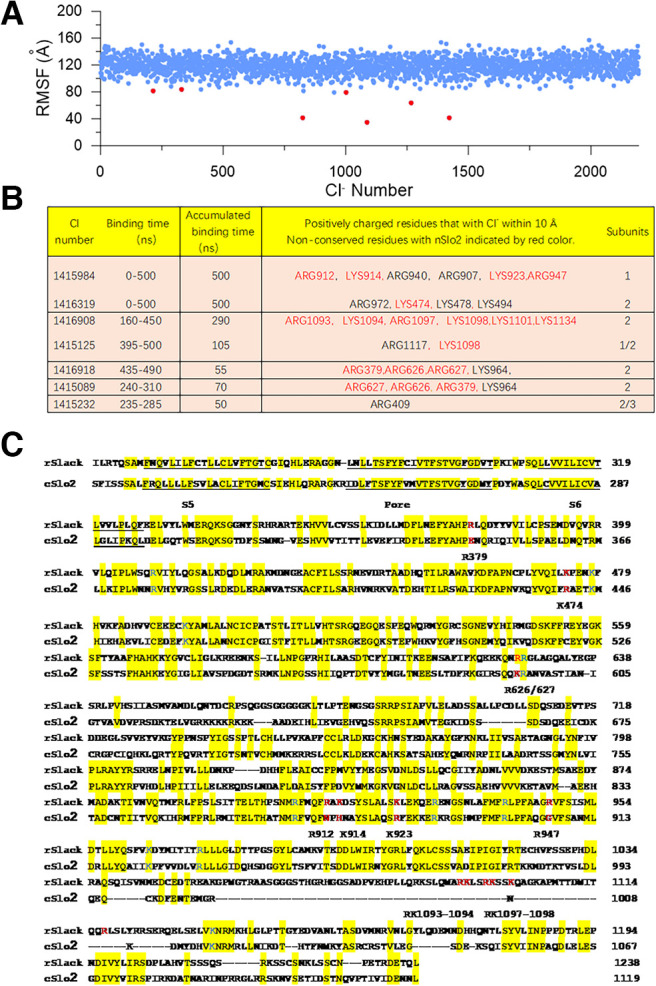
The simulation predicted the Cl–-sensitive site candidates in the Slack channel. A, The root-mean-square fluctuations (RMSF) of total Cl- ions in the system during the 500 ns MD simulation. B, The predicted amino acid candidate of the Cl- sensitive site of the Slack channel. C, The sequence alignment between the rSlack channel and C. elegans Slo2 channel. The candidate amino acids as the Cl−-sensitive site were labeled red.
Then, we neutralize these basic residues and express them in Xenopus oocytes to test whether these mutations alter the Cl– sensitivity. Electrophysiological recordings were performed in the inside-out patch configuration. Since the Slack channel sensitivity to Cl– is weaker than that to Na+, and the sensitivities to the two ions are interdependent (Zhang et al., 2010), it is difficult to directly compare the Cl– sensitivity of these mutants alone. Thus, we examined whether intracellular Cl– ions would enhance sodium sensitivity by comparing dose–response curves of sodium-dependent current in the absence or presence of Cl–. Therefore, we used two Na+ solutions, one not containing Cl– (NaGlu) and another containing Cl– (NaCl). A mutant example illustrated this enhancing effect, where the sodium sensitivity Kd value of K1098Q in NaCl solution shows a statistically significant difference of 30 mm lower value than the Kd value of this mutant in NaGlu solution (p < 0.05; Fig. 15A,B). Among the mutants tested, only R379Q removed the ability of Cl– to promote the Na+ sensitivity of the Slack channel (Fig. 15C–E). To exclude the allosteric effect of R379Q, we continued to mutate R379 to different amino acids. The R379A, R379C, R379N, and R379T mutants were generated to test the sodium dependence curve with different intracellular NaGlu and NaCl solution concentrations. The results indicated that all these mutants possessed similar sodium dose–response curves in intracellular sodium gluconate and sodium chloride solutions (Fig. 16A,B). These data showed that Cl– ions could not promote sodium binding in these mutants, suggesting the R379 is a chloride-sensitive site for the rSlack channel.
Figure 15.
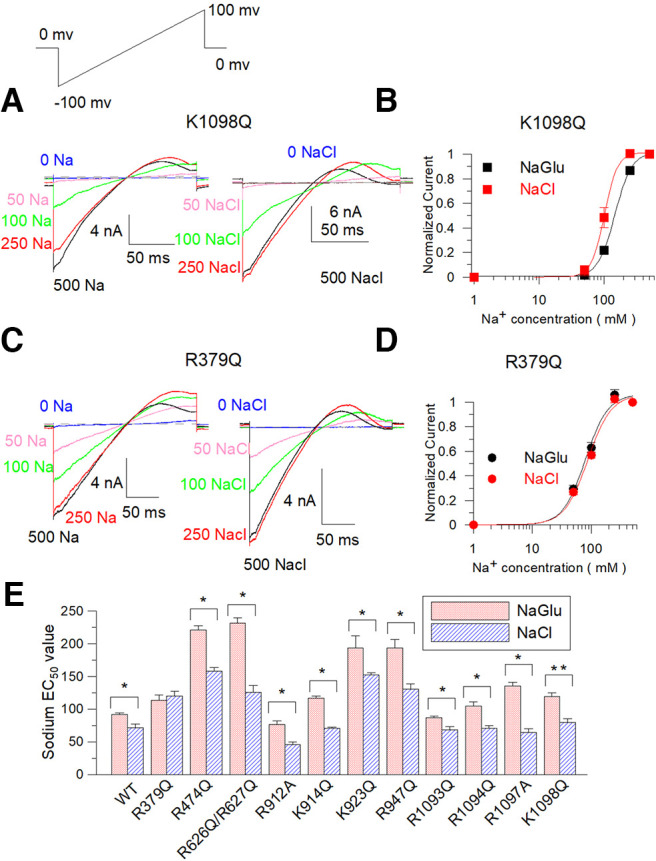
The R379Q mutant removes the chloride sensitivity of the Slack channel. A, Typical current traces of K1098Q were recorded in the inside-out configuration perfused with 0–500 mm NaGlu and 0–500 mm NaCl, respectively. B, Hill equation fitted data of the NaGlu (EC50 = 113.5 ± 8.3 mm, n = 3.9) and the NaCl (EC50 = 119.6 ± 7.6 mm, n = 3.8) dose response of K1098Q mutant. C, Typical currents of R379Q were recorded in an inside-out configuration bathed in 0–500 mm NaGlu and NaCl, respectively. D, Hill equation fitted data of the NaGlu (EC50 = 119.1 ± 5.9 mm, n = 3.9) and NaCl (EC50 = 79.4 ± 5.8 mm, n = 3.8) dose response of R379Q mutant, respectively. E, Comparison of candidate mutants of Cl–-sensitive sites of EC50 values of NaGlu and NaCl dose dependence. EC50 values of all mutants with NaGlu are as follows (in mm): WT, EC50 = 91.6 ± 2.7; R379Q, EC50 = 113.5 ± 8.3; R474Q, EC50 = 220.8 ± 6.6; R626/627Q, EC50 = 231.7 ± 7.5; R912A, EC50 =76 ± 6.1; K914Q, EC50 = 116.8 ± 3.0; K923Q, EC50 = 193.0 ± 18; R947Q, EC50 = 193.6 ± 13; R1093Q, EC50 = 87.1 ± 2.0; R1094Q, EC50 = 104.3 ± 7.0; R1097Q, EC50 = 134.9 ± 6.0; and K1098Q, EC50 = 119.1 ± 5.9. EC50 values of all mutants in NaCl solution are as follows (in mm): WT, EC50 = 71.6 ± 5.1; R379Q, EC50 = 119.6 ± 7.6; R474Q, EC50 = 151.6 ± 6.1; R626/627Q EC50 = 125.2 ± 11; R912A, EC50 = 46 ± 3.8; K914Q, EC50 = 70.7 ± 1.9; K923Q, EC50 = 152.2 ± 3.5; R947Q, EC50 = 130.8 ± 7.4; R1093Q, EC50 = 68 ± 5.6; R1094Q, EC50 = 71 ± 3.8; R1097Q, EC50 = 64.6 ± 5.6; and K1098Q, EC50 = 79.4 ± 5.8.
Figure 16.
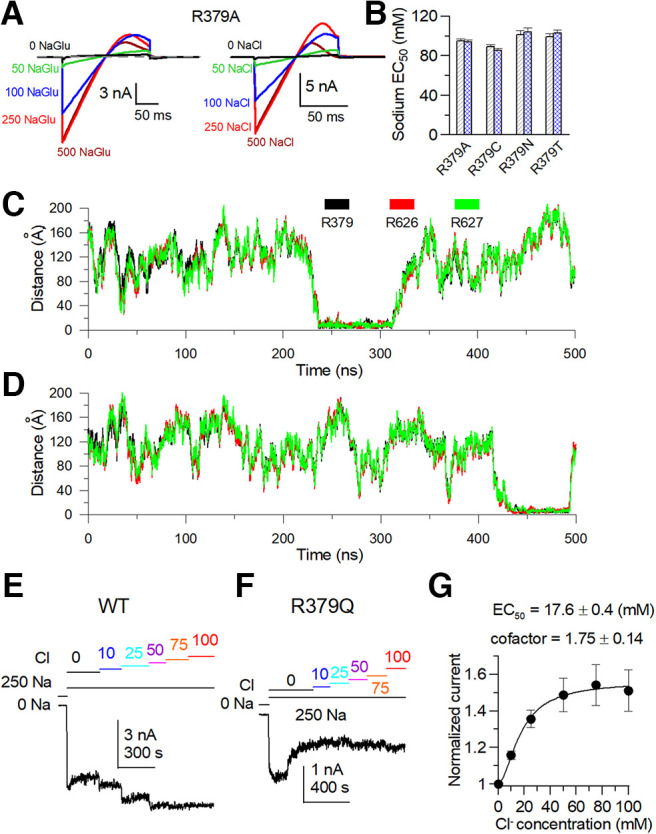
Distances of Cl– to positively charged amino acids R379Q and R626/627Q of rSlack channel in the simulated local structural model. A, Sample traces of R379A in NaGlu and NaCl solution. B, Comparison of sodium sensitivity of the R379 mutant EC50 values in NaGlu and NaCl solution. The sodium dependence of the R379 mutant EC50 value in NaGlu are as follows (in mm): R379A, EC50 = 95 ± 2; R379N, EC50 = 89 ± 2.1; R379C, EC50 = 101.3 ± 4.2; and R379T, EC50 = 99.3 ± 3.3. The EC50 values in NaCl are as follows (in mm): R379A, EC50 = 94 ± 1.6; R379N, EC50 = 85.4 ± 4.4; R379C, EC50 = 104 ± 7.3; and R379T, EC50 = 102.8 ± 4.2. C, During the simulation process, distances of the bound Cl– ion to residues R379, R626, and R627 amino acids. D, Distances of another bound Cl– ion to residues R379, R626, and R627 amino acids during the simulation process. E, F, The time course of the current change of the WT Slack channel and R379Q mutant were perfused in different Cl– and 250 mm sodium solution concentrations. G, The Cl– dose-dependent curve for the activation of the Slack channel.
The MD trajectories revealed that the Cl– ion stayed at this site and close to R626/R627 residues in two subunits of the Slack channel for >70 ns during the MD simulation (Fig. 16C,D). However, the R626/627Q mutant did not alter the sensitivity of the Slack channel to sodium (Fig. 15E). To further confirm that the R379 residue confers Cl– sensitivity to the rSlack channel, we assessed the Cl–-dependent activity of the rSlack channel in the presence of 250 mm intracellular sodium, which is a saturating concentration of sodium for activation of the Slack channel. The data showed that the WT rSlack channel still shows chloride dose-dependent activity in saturating sodium concentrations (Fig. 16E). In contrast, the R379Q mutant channel did not exhibit chloride dose-dependent activity. Still, it showed apparent time-dependent rundown (Fig. 16F). The Kd value of the Cl– activation of the rSlack channel is ∼18 mm with a coefficient factor of 1.75, which is twofold higher than the Kd in 100 mm intracellular sodium (Fig. 16G; Zhang et al., 2010). These results suggest the R379 is a chloride-sensitive site of the Slack channel.
To further test the role of R379 in sensing Cl–, we compared the single-channel po of the WT Slack channel and R379Q mutation in 100 mm intracellular sodium and 0–50 mm choline chloride. We found that the R379Q mutant showed higher po in 100 mm intracellular sodium but did not show Cl–-dependent enhancement of the po, while the WT Slack channel po increased from ∼0.16 to 0.49 as the Cl– concentration increased from 0 to 50 mm (Fig. 17A–F). The po comparison showed a statistically significant difference between the WT channel and the R379 mutant in 0 and 25 mm Cl– (Fig. 17G). The EC50 of the Cl– dependency of the WT channel is ∼23.7 mm, whereas the po of the R379Q did not show Cl– dependency (Fig. 17H). These results indicate that the R379 is critical for chloride sensing of the Slack channel.
Figure 17.
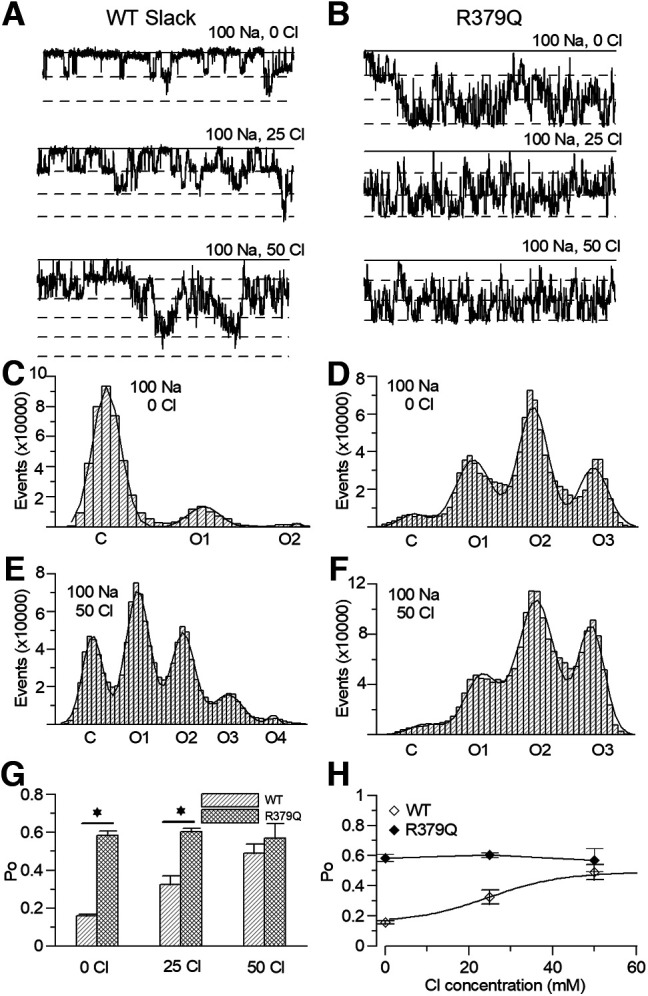
The single channel recording of Cl- dependence of the WT Slack and R379Q mutation. A-B, The sample traces of single channel recording traces of the WT Slack channel (A) and the R379Q mutant (B) under the 0 Cl, 25 Cl, and 50 Cl (in mM). C-F, The histogram analysis of the Po of the WT channel (C and E) and R379Q mutant (D and F), fitted by a Gaussian function. G, Comparison of the Po of the WT channel and the R379Q mutant under 100 Na and 0 Cl (WT, 0.158 ± 0.011; R379Q, 0.585 ± 0.022. t-test, p < 0.05, n = 5), 25 Cl (WT, 0.325 ± 00.046; R379Q, 0.604 ± 0.017. t-test, p < 0.05, n = 5), 50 Cl (WT, 0.489 ± 0.051; R379Q, 0.57 ± 0.076). H, The dose response curve of Cl− with 100 mM intracellular sodium. The Cl− EC50 = 23.74 ± 2.1 mM.
Discussion
Sodium is the known effective physiological stimulus that activates Slack channels. Studying how Na+ activates a Slack channel advances our understanding of the physiological function of this channel. Although we previously identified D818 as partly responsible for the Na+ sensitivity of the channel as neutralization mutations decreased the sodium sensitivity of the Slack channel more than fourfold, the D818 mutant remained sensitive to sodium. Thus, to search for additional sodium-sensitive sites in this study, we performed a comprehensive screening of acidic residues in the C terminus of the Slack channel. Unlike dissecting Ca2+-sensitive sites of the voltage and Ca2+-activated Slo1 channel, one challenge of this screening strategy is that the Slack channel activity can no longer be observed once the sodium sensitivity is removed. Thus, it is difficult to judge whether a mutant that kills activity altogether removes the sodium sensitivity of the Slack channel or the channel just cannot be functionally expressed. The M335A mutant of the rSlack channel, which corresponds to the M333A mutant in the chSlo2.2 channel, allows one to observe channel activity in zero cytosolic Na+. Although the M335A could also produce an allosteric effect that could alter the gating conformation change of the Slack channel, it did not affect sodium-dependent gating. By taking advantage of this mutation, we identified eight neutralization mutants that increased the EC50 of the Na+ sensitivity of the channel by >15-fold. The extent of sodium sensitivity reduction of these mutants was much greater than the fourfold decrease in EC50 seen with the D818N. Furthermore, several mutants caused fourfold to fivefold reductions in sodium sensitivity among the residues we screened. From electrophysiological data, we concluded that the E373 is the only residue, its mutants that completely removed the sodium sensitivity of the rSlack channel in contrast to other mutants that more or less maintained some sodium sensitivity. Subsequently, the sodium binding possibility of other acidic residues that decrease the sodium sensitivity was examined by the MD simulation.
We turned into a homology model based on recent structures and MD simulations in the hundreds of nanoseconds timescale a structural understanding of the Na+ binding sites. The simulations revealed that sodium ions could bind to a site composed of the two acidic residues E373/D955 and secondarily to an acidic pocket coupled to the first primary site consisting of the following six acidic residues: D863, E865, E490, E491, E899, and E787 residues. The simulation using the (PDB:5U70) template, which represents the open state conformation of the KCNT1 channel, showed sodium ions interacting with the D863/E865/E490/E491 residues, whereas the simulation using the (PDB:5U76) template, which represents the closed-state conformation, showed the D863/E865/E787/D818 residues interacting with sodium ions in different conformations. The D863/E865 pair was common to simulations based on both templates. In contrast, the simulation used D863N/E865Q and E373Q mutated channel model did not show sodium binding on both sites. These results suggest that D863/E865 may cooperate with E787/E490/E491 to form another sodium-sensitive site. In general, these simulation results are consistent with the electrophysiological data that these mutants dramatically decrease sodium sensitivity, suggesting a primary role for E373 and a secondary role for the D863/E865 pair. Thus, the two sodium-sensitive site activation mechanisms seem more plausible for explaining the sodium sensitivity of Slack channels. Furthermore, the average sodium fractional occupancy of the two sodium binding sites is 0.315 (373/955 site) and 0.43 (863/865 site), respectively (Fig. 10), which generally matched the maximum po of the Slack channel by ∼0.43. However, the fractional occupancy is not equal in all four subunits. If channel opening requires all four subunits to be occupied by sodium, the channel-opening ratio in the simulation is much less than the actual recorded po. Thus, a longer simulation time may be needed to estimate the simultaneous occupancy fraction in all four subunits. Also, our MD simulations did not provide the dynamic process of sodium binding activating the channel and the affinity of the sodium binding in different conformations, which needs a more refined model and probably a different algorithm. We plan to explore this further in future studies.
This is in line with the design of another member of this family, the Slo1 channel, which possesses three different calcium-binding sites (Schreiber et al., 1999; Shi et al., 2002; Xia et al., 2002). In the Slo1 channel, the activating effect of the three different calcium-binding sites on the Slo1 channel are additive and relatively independent (Zeng et al., 2005). But, different from the Slo1 channel, neutralization of either E373 or D863, E865 led to the deactivation of the Slack channel, indicating the activation effect of the two sites is interdependent. It suggests a unique activation style of the gating ring in the Slack channel. But on the M335A mutant background, the E373Q/M335A mutant showed Na+-insensitive activity, whereas the D863N or E865Q mutants could still elicit sodium-dependent macroscopic current in the presence of high concentrations of Na+. This result suggests distinct roles of sodium regulation of activity through the E373 site versus the D863/E865 site, ranking E373 higher than the D863/E865 in terms of critical importance.
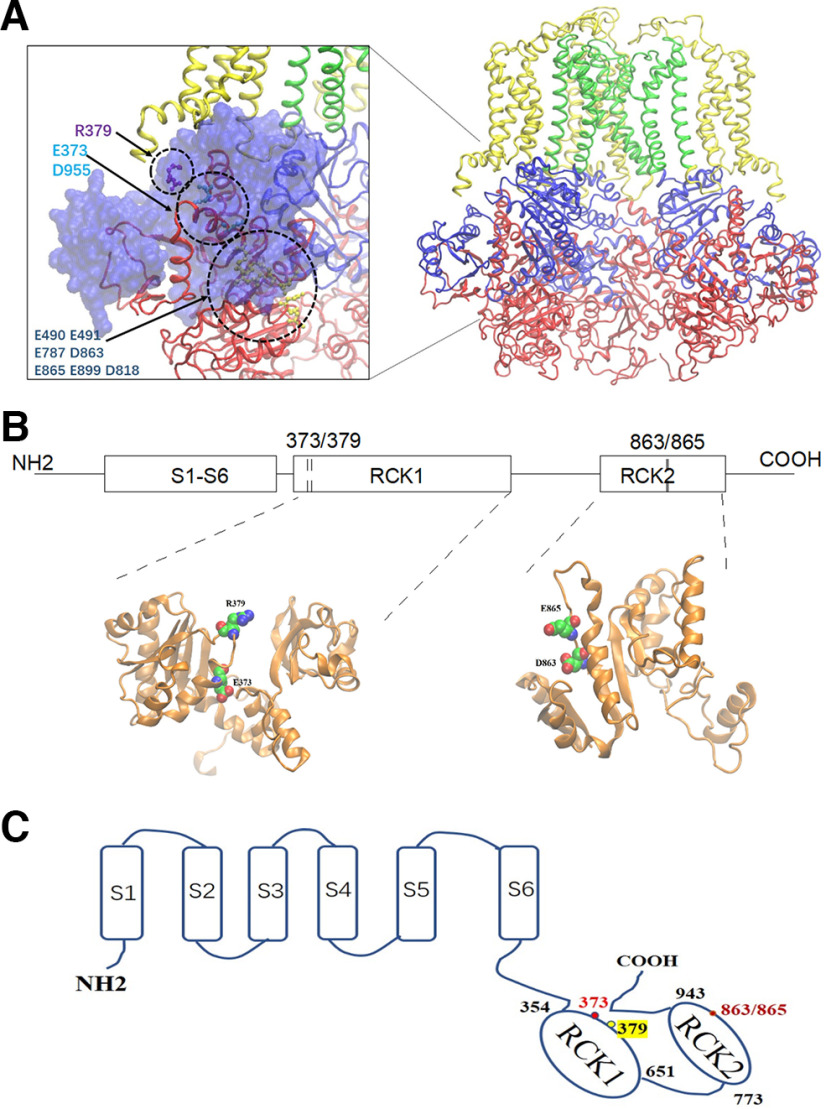
How could the sodium interaction control the gating of the Slack channel? We first considered insights offered by the structural character of the sodium interaction site. Unlike the Ca2+ bowl located in the assembly interface of different subunits in the BK (Slo1) channels, the E373/D955 residues in the Slack (Slo2.2) channel, and other residues in the acidic pocket (e.g., D863/E865) that control sodium sensitivity fall in the flexible interface between the RCK1 and RCK2 domains of the same α subunit (Fig. 18A). The E373 residue is located on the N lobe of the RCK1 domain of the Slack channel (Fig. 18B,C). Thus, because of sodium interaction with E373, the N lobe of the RCK1 domain may undergo a conformational change to its expanded conformation. As a previous crystallization study suggested, the expanded conformation may reposition the S6 helix through the polypeptide linker or interface interaction with the TMD domain (Wu et al., 2010; Hite et al., 2017).
Figure 18.
The sodium-sensitive and Cl–-sensitive sites are located in the same subunit in the RCK1 and RCK2 domain interface. A, The E373 and D863/E865 sensitive sites are located on the interface of RCK1 and RCK2 domains in the model of the Slack channel. Blue indicates the RCK1 domain, while red indicates the RCK2 domain. The green labels the pore domain, while the yellow labels the transmembrane domain. B, The linear scheme of the Slack channel with the location of the sodium-sensitive and chloride-sensitive sites. The local structure of the RCK1 and RCK2 domains. C, The cartoon structure of the Slack channel shows the location of the sodium-sensitive and Cl–-sensitive sites.
Although sodium ions are effective activators of the Slack channel, the EC50 value is much higher than the physiological intracellular sodium concentration (10−20 mm), and even in the case of spike firing of mitral cells (40 mm; Zylbertal et al., 2017). Despite the activation effect of Cl– ions with an EC50 of 23.7 mm (single-channel data) in a 100 mm cytosolic sodium solution is within the range of average neuronal intracellular concentration (Delpire and Staley, 2014; Kim et al., 2015), the Cl– may have only a trivial enhancing effect on the po of the Slack channel in physiological condition because the Cl– activation effect is sodium binding dependent. Interestingly, the R379Q mutant showed substantial rundown in the 200 mm [Na+]i, which was not observed in the WT channel under the same condition with Cl– ions, suggesting that the Cl– binding prevented rundown.
A previous study suggested that a “chloride bowl” existed at the far end of the C terminus (Yuan et al., 2000, 2003). Our simulation also permitted a chloride ion to stay in that position. However, the electrophysiological data generated from the mutant of these “Cl– bowl” residues did not support this hypothesis. Instead of the Cl– bowl, we found the R379Q mutant removed the Cl– sensitivity of the Slack channel. However, the simulation showed a relatively short time for a Cl– ion staying in this site during the simulation process. The structure of the 621–725 fragment of the Slack channel is missing in the cryo-EM structure. Electrophysiological data did not support residues R626/627, which are spatially closest to the R379 residue, as contributors to the Cl– ion sensitivity. The R474Q and R626/627Q mutations also altered the sodium dependence of the Slack channel. They could be an allosteric regulation of the sodium ion interaction with the Slack channel.
The R379Q mutant is also an epilepsy-related mutation identified in patients with seizures. The maximal po of this mutant increases, but its sodium sensitivity is somewhat decreased, probably because of a significant rundown. The macroscopic current of this mutant did not show a substantial decrease, probably because of the enhanced po. The R379 residue is also located in the interface of the RCK1 and RCK2 domains of the Slack channel (Fig. 18A and 18B), which may provide a basis for Cl– binding to promote sodium binding and prevent rundown. The R379 is also conserved in the Slick channel, which exhibits more robust Cl– dependence (Bhattacharjee et al., 2003). Since the Slick channel is also sensitive to chloride, the functional property of the Slack channel conferred by sodium binding may also apply to the Slick channel. Our results identify a Cl–-sensitive site of the Slack channel, which provides molecular insight for the Slack channel playing a role in response to the Cl– influx, such as during GABAA receptor activation, induced by anesthetics.
Our comprehensive survey of all acidic residues and predicted charged residues identified two sodium-sensitive sites and one chloride site. Our study also provides a reasonable model explaining the interaction of the Slack channel with sodium. The identification of sodium and chloride interaction sites in the Slack channel will guide us to understand better the diverse gating regulation mechanism of the Slack channel as well as the sodium signaling-related physiological and pathologic function of the Slack channel in various species.
Footnotes
This work was supported by The Important Project of Natural Science in Colleges and Universities in Jiangsu Province (Grant 14KJA320002, to Z.Z.), Jiangsu specially appointed professorships to Z.Z. and Q.-Y.T., the Natural Science Foundation of China (NSFC; Grants 81471314 and 81671090, to Z.Z.), the Natural Science Foundation of Jiangsu Province (Grant SBK201502515, to Z.Z.), the Xuzhou Science and Technology Program (Grant KC19036, to Z.Z.; and Grant KC16H0230, to Q.-Y.T.), and NSFC Grant 31671212 (to Q.-Y.T.). Support was also received from the Priority Academic Program Development of Jiangsu Higher Education Institutions and the Jiangsu Provincial Special Program of Medical Science (Grant BL2014029). D.E.L. was supported by National Institutes of Health Grant R01-HL059949-24.
The authors declare no competing financail interests.
References
- Alsaleem M, Carrion V, Weinstock A, Chandrasekharan P (2019) Infantile refractory seizures due to de novo KCNT 1 mutation. BMJ Case Rep 12:e231178. 10.1136/bcr-2019-231178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcia G, et al. (2019) Epilepsy with migrating focal seizures: KCNT1 mutation hotspots and phenotype variability. Neurol Genet 5:e363. 10.1212/NXG.0000000000000363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bausch AE, Dieter R, Nann Y, Hausmann M, Meyerdierks N, Kaczmarek LK, Ruth P, Lukowski R (2015) The sodium-activated potassium channel Slack is required for optimal cognitive flexibility in mice. Learn Mem 22:323–335. 10.1101/lm.037820.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee A, Joiner WJ, Wu M, Yang Y, Sigworth FJ, Kaczmarek LK (2003) Slick (Slo2.1), a rapidly-gating sodium-activated potassium channel inhibited by ATP. J Neurosci 23:11681–11691. 10.1523/JNEUROSCI.23-37-11681.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldi M, Nobili L, Zara F, Combi R, Prato G, Giacomini T, Capra V, De Marco P, Ferini-Strambi L, Mancardi MM (2019) Migrating focal seizures in autosomal dominant sleep-related hypermotor epilepsy with KCNT1 mutation. Seizure 67:57–60. 10.1016/j.seizure.2019.02.019 [DOI] [PubMed] [Google Scholar]
- Delpire E, Staley KJ (2014) Novel determinants of the neuronal Cl(-) concentration. J Physiol 592:4099–4114. 10.1113/jphysiol.2014.275529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertler T, Bearden D, Bhattacharjee A, Carvill G, (1993) KCNT1-related epilepsy. In: GeneReviews (Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A, eds). Seattle, WA: University of Washington. [PubMed] [Google Scholar]
- Heron SE, Smith KR, Bahlo M, Nobili L, Kahana E, Licchetta L, Oliver KL, Mazarib A, Afawi Z, Korczyn A, Plazzi G, Petrou S, Berkovic SF, Scheffer IE, Dibbens LM (2012) Missense mutations in the sodium-gated potassium channel gene KCNT1 cause severe autosomal dominant nocturnal frontal lobe epilepsy. Nat Genet 44:1188–1190. 10.1038/ng.2440 [DOI] [PubMed] [Google Scholar]
- Hite RK, MacKinnon R (2017) Structural titration of Slo2.2, a Na(+)-dependent K(+) channel. Cell 168:390–399.e11. 10.1016/j.cell.2016.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hite RK, Tao X, MacKinnon R (2017) Structural basis for gating the high-conductance Ca(2+)-activated K(+) channel. Nature 541:52–57. 10.1038/nature20775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii A, Shioda M, Okumura A, Kidokoro H, Sakauchi M, Shimada S, Shimizu T, Osawa M, Hirose S, Yamamoto T (2013) A recurrent KCNT1 mutation in two sporadic cases with malignant migrating partial seizures in infancy. Gene 531:467–471. 10.1016/j.gene.2013.08.096 [DOI] [PubMed] [Google Scholar]
- Juang JM, Lu TP, Lai LC, Ho CC, Liu YB, Tsai CT, Lin LY, Yu CC, Chen WJ, Chiang FT, Yeh SF, Lai LP, Chuang EY, Lin JL (2014) Disease-targeted sequencing of ion channel genes identifies de novo mutations in patients with non-familial Brugada syndrome. Sci Rep 4:6733. 10.1038/srep06733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholin AA, Zavadenko NN, Fedonyuk ID, Antonets AV, Mukhin KY, Malov AG, Vshivkov MI, Anisimov GV, Il'ina ES (2019) Early infantile epileptic encephalopathy type 14: three cases of epilepsy in infancy with migrating focal seizures due to KCNT1 mutations. Z Nevrol Psikhiatr Im SS Korsakova 119:74–82. 10.17116/jnevro201911907274 [DOI] [PubMed] [Google Scholar]
- Kim S, Ma L, Unruh J, McKinney S, Yu CR (2015) Intracellular chloride concentration of the mouse vomeronasal neuron. BMC Neurosci 16:90. 10.1186/s12868-015-0230-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingwell K (2012) Genetics: mutations in potassium channel KCNT1-a novel driver of epilepsy pathogenesis. Nat Rev Neurol 8:658. 10.1038/nrneurol.2012.229 [DOI] [PubMed] [Google Scholar]
- Kuchenbuch M, et al. (2019) KCNT1 epilepsy with migrating focal seizures shows a temporal sequence with poor outcome, high mortality and SUDEP. Brain 142:2996–3008. 10.1093/brain/awz240 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang FF, Song Y, Wang R, Zhang Q, Shen ZS, Zhang FF, Zhong DY, Wang XH, Guo Q, Tang QY, Zhang Z (2022) The slack channel deletion causes mechanical pain hypersensitivity in mice. Front Mol Neurosci 15:811441. 10.3389/fnmol.2022.811441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Bausch AE, Kallenborn-Gerhardt W, Stoetzer C, Debruin N, Ruth P, Geisslinger G, Leffler A, Lukowski R, Schmidtko A (2015) Slack channels expressed in sensory neurons control neuropathic pain in mice. J Neurosci 35:1125–1135. 10.1523/JNEUROSCI.2423-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Espinosa PL, Wu J, Yang C, Gonzalez-Perez V, Zhou H, Liang H, Xia XM, Lingle CJ (2015) Knockout of Slo2.2 enhances itch, abolishes KNa current, and increases action potential firing frequency in DRG neurons. Elife 4:e10013. 10.7554/eLife.10013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller RS, et al. (2015) Mutations in KCNT1 cause a spectrum of focal epilepsies. Epilepsia 56:e114–e120. 10.1111/epi.13071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba C, et al. (2015) De novo KCNT1 mutations in early-onset epileptic encephalopathy. Epilepsia 56:e121–e128. 10.1111/epi.13072 [DOI] [PubMed] [Google Scholar]
- Quraishi IH, Mercier MR, McClure H, Couture RL, Schwartz ML, Lukowski R, Ruth P, Kaczmarek LK (2020) Impaired motor skill learning and altered seizure susceptibility in mice with loss or gain of function of the Kcnt1 gene encoding Slack (KNa1.1) Na(+)-activated K(+) channels. Sci Rep 10:3213. 10.1038/s41598-020-60028-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenhouse-Dantsker A, Sui JL, Zhao Q, Rusinova R, Rodríguez-Menchaca AA, Zhang Z, Logothetis DE (2008) A sodium-mediated structural switch that controls the sensitivity of Kir channels to PtdIns(4,5)P(2). Nat Chem Biol 4:624–631. 10.1038/nchembio.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi CM, Ferreira G, Yang B, Gazula VR, Butler A, Wei A, Kaczmarek LK, Salkoff L (2006) Opposite regulation of Slick and Slack K+ channels by neuromodulators. J Neurosci 26:5059–5068. 10.1523/JNEUROSCI.3372-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber M, Wei A, Yuan A, Gaut J, Saito M, Salkoff L (1998) Slo3, a novel pH-sensitive K+ channel from mammalian spermatocytes. J Biol Chem 273:3509–3516. 10.1074/jbc.273.6.3509 [DOI] [PubMed] [Google Scholar]
- Schreiber M, Yuan A, Salkoff L (1999) Transplantable sites confer calcium sensitivity to BK channels. Nat Neurosci 2:416–421. 10.1038/8077 [DOI] [PubMed] [Google Scholar]
- Shi J, Krishnamoorthy G, Yang Y, Hu L, Chaturvedi N, Harilal D, Qin J, Cui J (2002) Mechanism of magnesium activation of calcium-activated potassium channels. Nature 418:876–880. 10.1038/nature00941 [DOI] [PubMed] [Google Scholar]
- Tang Q-Y, Zhang F-F, Xu J, Wang R, Chen J, Logothetis Diomedes E, Zhang Z (2016) Epilepsy-related slack channel mutants lead to channel over-activity by two different mechanisms. Cell Rep 14:129–139. 10.1016/j.celrep.2015.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Yang Y, Ye S, Jiang Y (2010) Structure of the gating ring from the human large-conductance Ca(2+)-gated K(+) channel. Nature 466:393–397. 10.1038/nature09252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia XM, Zeng X, Lingle CJ (2002) Multiple regulatory sites in large-conductance calcium-activated potassium channels. Nature 418:880–884. 10.1038/nature00956 [DOI] [PubMed] [Google Scholar]
- Yang B, Gribkoff VK, Pan J, Damagnez V, Dworetzky SI, Boissard CG, Bhattacharjee A, Yan Y, Sigworth FJ, Kaczmarek LK (2006) Pharmacological activation and inhibition of Slack (Slo2.2) channels. Neuropharmacology 51:896–906. 10.1016/j.neuropharm.2006.06.003 [DOI] [PubMed] [Google Scholar]
- Yoshitomi S, Takahashi Y, Yamaguchi T, Oboshi T, Horino A, Ikeda H, Imai K, Okanishi T, Nakashima M, Saitsu H, Matsumoto N, Yoshimoto J, Fujita T, Ishii A, Hirose S, Inoue Y (2019) Quinidine therapy and therapeutic drug monitoring in four patients with KCNT1 mutations. Epileptic Disord 21:48–54. [DOI] [PubMed] [Google Scholar]
- Yuan A, Dourado M, Butler A, Walton N, Wei A, Salkoff L (2000) SLO-2, a K+ channel with an unusual Cl- dependence. Nat Neurosci 3:771–779. 10.1038/77670 [DOI] [PubMed] [Google Scholar]
- Yuan A, Santi CM, Wei A, Wang ZW, Pollak K, Nonet M, Kaczmarek L, Crowder CM, Salkoff L (2003) The sodium-activated potassium channel is encoded by a member of the Slo gene family. Neuron 37:765–773. 10.1016/s0896-6273(03)00096-5 [DOI] [PubMed] [Google Scholar]
- Zeng XH, Xia XM, Lingle CJ (2005) Divalent cation sensitivity of BK channel activation supports the existence of three distinct binding sites. J Gen Physiol 125:273–286. 10.1085/jgp.200409239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Gao SH, Shen ZS, Wang Y, Hu SW, Duan GB, Liu Y, Zhong DY, Liu J, Sun MH, Zhang X, Cao TY, Cao JL, Tang QY, Zhang Z (2022) The Slack channel regulates anxiety-like behaviors via basolateral amygdala glutamatergic projections to ventral hippocampus. J Neurosci 42:3049–3064. 10.1523/JNEUROSCI.2027-21.2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Rosenhouse-Dantsker A, Tang QY, Noskov S, Logothetis DE (2010) The RCK2 domain uses a coordination site present in Kir channels to confer sodium sensitivity to Slo2.2 channels. J Neurosci 30:7554–7562. 10.1523/JNEUROSCI.0525-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Tang QY, Alaimo JT, Davies AG, Bettinger JC, Logothetis DE (2013) SLO-2 isoforms with unique Ca(2+) - and voltage-dependence characteristics confer sensitivity to hypoxia in C. elegans. Channels (Austin) 7:194–205. 10.4161/chan.24492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylbertal A, Yarom Y, Wagner S (2017) The slow dynamics of intracellular sodium concentration increase the time window of neuronal integration: a simulation study. Front Comput Neurosci 11:85. [DOI] [PMC free article] [PubMed] [Google Scholar]