Abstract
Breast cancer subtypes and their phenotypes parallel different stages of the mammary epithelial cell developmental hierarchy. Discovering mechanisms that control lineage identity could provide novel avenues for mitigating disease progression. Here we report that the transcriptional corepressor, TLE3, is a guardian of luminal cell fate in breast cancer and operates independently of estrogen receptor. In luminal breast cancer, TLE3 actively repressed the gene expression signature associated with highly aggressive basal-like breast cancers (BLBC). Moreover, maintenance of the luminal lineage depended on appropriate localization of TLE3 to its transcriptional targets, a process mediated by interactions with FOXA1. By repressing genes that drive BLBC phenotypes, including SOX9 and TGFβ2, TLE3 prevented the acquisition of a hybrid epithelial-mesenchymal state and reduced metastatic capacity and aggressive cellular behaviors. These results establish TLE3 as an essential transcriptional repressor that sustains the more differentiated and less metastatic nature of luminal breast cancers. Approaches to induce TLE3 expression could promote the acquisition of less aggressive, more treatable disease states to extend patient survival.
Statement of Significance
Transcriptional co-repressor TLE3, actively suppresses SOX9 and TGF-β transcriptional programs to sustain luminal lineage identity of breast cancer cells and inhibit metastatic progression.
Introduction
Over two decades ago, the transformational studies conducted by Perou and colleagues shifted our understanding of breast cancer by classifying this disease into subtypes using transcriptomic signatures that were also prognostic of patient outcomes (1). Subsequent studies identified key genetic drivers of each subtype, with luminal breast cancers driven by estrogen receptor (ER)-stimulated programs and HER2 amplification/overexpression promoting HER2-enriched disease (2,3). However, no single driver has been identified for basal-like breast cancers (BLBC), a subtype that lacks receptors for estrogen and progesterone as well as HER2 overexpression. The transcriptomes of BLBC dictate their stem-like and aggressive nature compared to the more differentiated luminal tumors (4). Recent single-cell sequencing studies have revealed the highly fluid nature of these subtypes, with individual cells residing along a continuum of epigenetic and transcriptomic profiles ranging from basal to luminal (5). Coupled with studies demonstrating that mammary cell fate can be altered by an imbalance in chromatin modifiers, these findings reveal remarkable plasticity, even in more differentiated cell states (6-8). However, mechanisms by which transcriptional regulators and chromatin modifiers cooperate to establish these fates, including the relatively differentiated luminal breast cancer phenotype, even in the absence of ER, are not well understood.
Chromatin modifiers play crucial roles in the maintenance of epigenetic barriers that control lineage-restrictive transcriptional programs. In breast cancer, these include the polycomb-repressive complex members BMI1 (9,10) and EZH2 (11,12), and histone modifiers PYGO2 (13) and JARID1B (14). Pioneering factors such as GATA3 and FOXA1 can also modulate chromatin accessibility and enable ER recruitment to drive programs associated with the luminal cell fate (15,16). Recently, a collaborative mechanism involving FOXA1 and the SWI/SNF chromatin remodeling complex has been identified that promotes the luminal fate in ER+ breast cancer cells (7,17). Specifically, recruitment of SWI/SNF and ER by FOXA1 at luminal-defining genes drives their transcription. Mutation or loss of the SWI/SNF subunit, ARID1A, decreases chromatin accessibility at luminal signature genes, desensitizing cells to endocrine therapies (7). Thus, the integrity of the FOXA1-ER-SWI/SNF complex is required to maintain the gene expression program within hormone-responsive cells. In addition to stimulating luminal gene expression, FOXA1 also prevents the acquisition of BLBC phenotypes by directly binding to and repressing BLBC genes in luminal breast cancer cells (18). Importantly, this function of FOXA1 is independent of ER. FOXA1 lacks a repression domain, thus previously uncharacterized molecular interactions must facilitate its repressive functions including the repression of genes characteristic of less-differentiated basal-like cells that drive pro-metastatic phenotypes.
Here we identify the transcriptional corepressor protein, TLE3, as a novel driver of both ER-positive and -negative luminal breast cancer lineages. Members of the Groucho/Grg/Transducin-like enhancer of split (TLE) family are key regulators of cellular identity, where they govern transcriptional programs to suppress stem cell features and drive differentiation (19). TLE proteins do not bind DNA directly; rather, they facilitate short- and long-range chromatin condensation through protein-protein interactions. These include direct interactions between TLE proteins and histone tails, as well as TLE-mediated recruitment of HDAC proteins or the Polycomb repressive complex (PRC) that results in chromatin condensation (20). TLE proteins are recruited to specific sites of DNA through interactions with sequence-specific transcription factors, including those of the FOX family (21-24), TCF/LEF (25), and HES (26) proteins. In breast cancer, TLE3 can partner with FOXA1 and ER at ER-target genes to prevent their expression in the absence of estrogens (21). Additionally, TLE3 and FOXA1 interact in ER-negative apocrine cells to repress the ErbB2 downstream signaling gene, RELB (27), indicating that the FOXA1/TLE3 interaction exhibits ER-dependent and -independent roles in this disease. Nonetheless, the full spectrum of ER-independent functions of TLE3 and its role in controlling breast cancer cell fate are not yet known.
By identifying ER-independent genes that are repressed by TLE3, we discovered that TLE3 is a critical modulator of breast cancer cell differentiation. In association with its transcriptional binding partner, FOXA1, TLE3 directly represses the expression of a set of genes associated with the BLBC subtype such as TGFβ2 and SOX9 that are associated with poor patient outcomes. Consequently, TLE3 averts the acquisition of aggressive phenotypes associated with BLBC, including migration, invasion, and metastasis. Together, these data establish TLE3 as a key governor of the luminal breast cancer lineage that suppresses breast cancer metastatic phenotypes.
METHODS
Statistical Methods
Statistical analyses were performed using two-tailed t-test for all in vitro data unless otherwise stated. Significance was concluded if the p-value was less than 0.05. Unless noted otherwise, all in vitro data are represented as a means with standard deviations of three independent experiments each completed in triplicate.
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ruth Keri (kerir@ccf.org).
Materials Availability
Cell lines generated in this study are available upon request.
REAGENTS
| Reagent | Supplier | Catalog number | RRID number |
|---|---|---|---|
| TLE3-antibody | Proteintech | 11372-1-AP | RRID:AB_2203743 |
| FOXA1-antibody | Santa Cruz | Sc-6553 | RRID:AB_2104865 |
| SOX9-antibody | Millipore Sigma | AB5535 | RRID:AB_2239761 |
| ER-antibody | Santa Cruz | Sc-542 | RRID:AB_631470 |
| B-actin antibody | Millipore Sigma | A2228 | RRID:AB_476697 |
| N-cadherin antibody | Cell Signaling | 13116 | RRID:AB_2687616 |
| Snai1 antibody | Cell Signaling | 3879 | RRID:AB_2255011 |
| IRDye 800CW Goat anti-mouse IgM | LI-COR Biosciences | 926-32280 | RRID:AB_2814919 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
MDA-MB-453, SKBR3, MDA-MB-468, and T47D cell lines were obtained from The American Type Culture Collection (ATCC). TLE3-WT and TLE3-KO MDA-MB-453 and T47D cells were generated by Synthego using CRISPR-Cas9 gene editing to target the 5th exon of TLE3. T47D and MDA-MB-453 TLE3-KO cells had 97% and 79% editing efficiency, respectively, and were maintained as cell pools.
MDA-MB-468 and T47D cells were cultured in RPMI 1640 cell culture medium containing 10% fetal bovine serum and 1% Pen-strep. T47D cells were additionally supplemented with 0.2 units/ml insulin. MDA-MB-453 cells were cultured in DMEM containing 2mM L-glutamine, 10% FBS, and 1% Pen-strep. SKBR3 cells were cultured in McCoy’s 5 medium with 1.5mM L-glutamine, 10% FBS, and 1% Pen-strep. All cells were incubated at 37 degrees Celsius in 5% CO2. Cells were monitored for mycoplasma contamination monthly and cell lines were authenticated using STR analysis in 2022 (Labcorp, Burlington, NC).
Animal Models
All in vivo experiments were performed with approval from the Institutional Animal Care and Use Committees at Case Western Reserve University or Cleveland Clinic. NOD Scid-γ (NSG) female mice were obtained from Jackson Laboratories. For metastasis studies using TLE3 overexpression, mice were anesthetized with 2-3% isoflurane. MDA-MB-468 stable TLE3-GFP and TLE3-OE cells were injected into the inguinal mammary fat pads at 1 million cells/pad (10 mice per group). Primary tumor volume was measured 1-2 times per week with calipers. Lungs were collected for histology 50 days after cells were injected into the fat pads.
METHOD DETAILS
Gene silencing and overexpression
RNAi
Transient silencing was achieved using reverse transfection. siRNA targeting TLE3, FOXA1, SOX9, or a nontargeting siRNA (siNS) directed to firefly luciferase mRNA were diluted in Opti-MEM medium at a final concentration of 100nM, mixed with Lipofectamine 2000 at a 1:100 dilution and incubated for 20 min at 37°C. Cells were resuspended in pen-strep-free complete medium and mixed with the siRNA/Opti-MEM/Lipofectamine mix at a concentration of cells that would achieve 70-80% confluency at endpoint. The following siRNAs were purchased from Horizon Discovery and used in this study: ON-TARGETplus Human TLE3 siRNA Smartpool (L-019929-00-0010); ON-TARGETplus Human TLE3 siRNA (J-019929-07); siGENOME Human FOXA1 siRNA Smartpool (M-010319-01-0010); siGENOME Human FOXA1 siRNA (D-010319-04-0010).
Overexpression
Adenovirus expressing TLE3 and GFP (Ad-hTLE3-Ef1aGFP cat# S1100) or GFP alone (cat# V1040), as a control, were obtained from Welgen Inc. MDA-MB-468 cells were plated in complete media containing 1x108 viral particles/mL at a concentration to achieve 60-70% confluency at endpoint. Virus-containing media was replaced with complete media at 24 hours and cells were collected at 48 hours. For stable overexpression of TLE3, we obtained lentiviral particles overexpressing TLE3 (pLenti-CMV-hTLE3-IRES-GFP-PGK-Puro; 1x1012 LP/mL) and GFP (pLenti-CMV-GFP-PGK-Puro; 1x1012 LP/mL) from Welgen Inc. MDA-MB-468 cells were plated at 20k/mL in a 24 well plate in pen-strep-free RPMI containing 10μg/mL polybrene (Millipore Sigma #TR-1003-G) solution. Lentiviral particles were added to the cell suspension at a 1:100 dilution for TLE3-expressing particles and 1:200 for GFP control particles. Medium was changed 24 hours later and selected and maintained in media containing 1.5μg/mL puromycin (Gibco #A1113803). Infection efficiency for both transient and stable TLE3 overexpression models was observed using an immunofluorescence microscope to determine the percent of cells expressing GFP at 48 hours post-transduction and western blot to confirm overexpression of TLE3 protein.
Western blots
Cells were lysed in RIPA buffer containing protease (Millipore Sigma #539138) and phosphatase (PhosSTOP; Millipore Sigma #4906845001) inhibitors for 30 min on ice and then cleared by spinning at 10,000 rpm for 10 min. DNA pellets were removed and lysates were stored at −80°C. Protein concentration was determined using BioRad Protein Assay and diluted to 2μg/μL in Laemmli buffer plus β-mercaptoethanol. After boiling, 80-100μg of protein lysate and molecular weight marker (LI-COR, #928-60000) were run in a 4-20% Tris-glycine gel (Invitrogen #XP04200BOX) and transferred onto an Immobilon-FL PVDF membrane (Millipore Sigma #IPFL00010) at 80 V for 120 min. Total protein was stained using REVERT total protein stain (LI-COR #926-11011) and imaged using a LI-COR Odyssey Fc. All membranes were blocked in 5% nonfat milk in TBS with 0.05% Tween-20, with the exception of those probed for TLE3 protein in which Intercept Blocking Buffer (LI-COR #927-60001) was used. The following antibodies were used for immunoblotting: anti-TLE3 (1:250), Proteintech #11372-1-AP; anti-FOXA1 (1:500) Santa Cruz #sc-6653; SOX9 (1:500) Millipore Sigma #AB5535, anti-ER-α (1:250) Santa Cruz #sc-542; β-Actin (1:10,000) Millipore Sigma #A2228; β-Catenin (1:500) Cell Signaling #8480; anti-Snai1 (1:500) Cell Signaling #3879. Following overnight incubation, membranes were incubated for 1 hr in fluorophore-bound secondary antibodies (IRDye 800CW IgM at 1:20,000) in the dark. Westerns were imaged using a LI-COR Odyssey Fc and quantified using Image Studio v5.2. Relative protein levels were quantified in relation to total protein levels or β-actin.
Tissue Staining and Pathology
Lung tissue was fixed overnight in 4% paraformaldehyde and embedded in paraffin. Tissue was serially sectioned at 5μm and collected every 30μm throughout the lungs. Sections were stained with H&E, de-identified, and submitted to two board-certified pathologists for quantitation of metastases.
Quantitative PCR and RNA-sequencing
RNA isolation and cDNA-synthesis
RNA was isolated using either TRIzol Reagent (ThermoFisher #15596026) followed by DNase treatment (Invitrogen DNA-free kit #AM1906) or RNeasy Plus minikit (Qiagen #74004). RNA concentration and quality was measured using a NanoDrop OneC (Thermo Scientific # 13-400-519). SuperScript IV reverse transcriptase (ThermoFisher #18090010) with random primers (ThermoFisher #48190011) was used to generate complementary DNA according to the manufacturer’s protocol.
qPCR
Quantitative real-time PCR (qRT-PCR) experiments were performed on a StepOnePlus real-time PCR machine (ThermoFisher 4376600). Gene expression was normalized to GAPDH. All experiments were performed in triplicate with two replicates. The following TaqMan realtime assays were purchased from ThermoFisher: SOX9 (Hs00165814_m1); TLE3 (Hs01032572_m1), EGFR (Hs01076090_m1), FOXA1 (Hs04187555_m1), KRT4 Hs00361611_m1), UBE2E3 (Hs00961488).
RNA-seq
MDA-MB-453, SKBR3, and T47D cells were reverse transfected with SMARTpool siRNAs against TLE3, FOXA1, or siNS for 48 hr as described above. RNA was isolated using RNAeasy Plus Minikit. Novogene Corporation Inc. conducted library preparation, sequencing, and analyses. Sequencing was performed using the Illumina NovaSeq 6000 platform with paired-end (150 bp) reads that were mapped to hg19. Reliability of sample variability was tested with correlation analysis of FPKM values and PCA. The DESeq2 R package was used to determine differential gene expression using a Benjamini-Hochberg adjusted p-value cutoff of 0.05.
Gene set enrichment
GSEA software was utilized to assess enrichment of basal, luminal, and mesenchymal gene sets using the following gene signatures deposited within the MSigDB database: SMID BREAST CANCER BASAL UP (28); CHARAFE BREAST CANCER LUMINAL VS BASAL DN (29); CHARAFE BREAST CANCER LUMINAL VS BASAL UP (29); CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN (29); KOINUMA_TARGETS_OF_SMAD2_OR_SMAD3 (30). The BL1 signature was downloaded from the supplemental files provided by Lehmann et al (31).
To assess whether loss of TLE3 enriches for SOX9 targets, we first generated a list of direct SOX9 gene targets using publicly available SOX9 ChIP-seq (32) data. We then filtered this list to contain those genes that are negatively correlated with TLE3 within the TCGA PanCancer database. This core set of 342 genes was then used as a signature to perform GSEA analysis.
To determine enrichment of Molecular Functions and Biological Processes, gene lists of interest were queried using Gene ontology (http://geneontology.org/) online software.
TLE3-Repressed Gene Set
Commonly upregulated genes in response to TLE3 silencing [determined by RNA-seq (p-adj < 0.05)] across all three cell lines were filtered to those with a Log2 fold change value >1. The 80 most highly upregulated genes were then queried within all breast cancer patients in the TCGA or METABRIC dataset using cBioPortal. A Pearson correlation coefficient was calculated for each breast cancer patient by comparing Log2 fold change values from the RNA-seq data to z-score expression levels reported within the TCGA or METABRIC. Patients were divided into those with high and low correlation coefficients as determined by quartiles (low = 25th percentile; high = 75th percentile) and progression-free survival curves were generated. To ensure the rigor of the TLE3-R signature, we randomly selected 80 genes expressed in the siNS samples from the RNA-seq dataset as a negative control. This gene set was evaluated in the same manner as the TLE3-R repressed gene set.
Evaluation of TLE3, FOXA1, and SOX9 expression in the Breast Cancer Atlas data
Raw counts data was obtained from the Broad Institute’s Single Cell Portal from Wu et al 2021 (33), and the metadata delineating cell identities was obtained from the publication. The data was filtered for cancer epithelial cells and processed using Seurat (Version 4.1.1) (34). The subset count data was normalized again using the SCTransform algorithm (Version 0.3.3) (35). Epithelial cells with a “cycling” profile were removed to eliminate confounding results. TSNE (t-distributed stochastic neighbor embedding) projection was used to visualize the data. The expression data for FOXA1, TLE3, and SOX9, and the expression differences of TLE3 and SOX9, and FOXA1 and SOX9 were visualized on the TSNE plots. The plots for the cell type subsets were also visualized to evaluate the differences between the expression of these genes.
ChIP-sequencing
ChIP
Parental MDA-MB-453, T47D, and SKBR3 cell lines were grown to 70% confluency and approximately 28 million cells were collected for each biological replicate. For siFOXA1 vs. siNS ChIP-seq, MDA-MB-453 cells were reverse transfected with siFOXA1 (Dharmacon #D-010319-04-0010) as described above for 48 hours. ChIP was performed using the iDeal ChIP-seq kit for Transcription Factors (Diagenode #C01010055) according to the manufacturer’s protocol. For TLE3-ChIP, cells were crosslinked using Crosslink Gold (Diagenode #C01019027) prior to fixation with formaldehyde. Chromatin shearing was performed using a QSonica Sonicator Ultrasonic Processor (PN Q125) at the following settings: Time = 04:10; Pulse = 25 on/59 off; Amp1 = 50%. Immunoprecipitation was carried out using the following antibodies: Proteintech (#11372-1AP) anti-TLE3 at 4.6μg/IP; Diagenode (#C15410231-100) anti-FOXA1 at 2μg/IP; Diagenode (#C15410206) Rabbit IgG at 1μg/IP). Input samples were pooled prior to sequencing. ChIP-PCR was used to confirm TLE3 and FOXA1 binding sites using the following primers for parental T47D, MDA-MB-453, and SKBR3 cells:
TRIM2: Left 5’ GTGTGCCCAGGGTAGTGTTT 3’
Right 5’ TGACTGCCACTCCTCAACAG 3’
SOX9: Left 5’ GCACCTTTGGCAATCCTAGA 3’
Right 5’ GCCAGAGAGGTTGGACTCTG 3’
For siFOXA1 vs. siNS MDA-MB-453 ChIP-PCR, the following primers were used:
SOX9: Left 5’ CTCTGCCTTCATCTTCACGC 3’
Right 5’ CTGGTTAGTCTTGCAGGCAC 3’
PAX7: Left 5’ CTAGCCAATCACAGAGCCCT 3’
Right 5’ ACCCTTCTCAATGACCCCAG 3’
Sequencing, alignment, and peak calling
Sequencing, alignment, and peak calling were completed by Diagenode. Briefly, paired-end sequencing was performed using a NovaSeq 6000 with NovaSeq Control Software version 1.6.0. FastQC was used for quality control of sequencing reads and reads were aligned using BWA to the hg38 reference genome deposited in the UCSC genome browser. PCR duplicates and multimapping reads were filtered using Samtools. Data was filtered to include only reads that aligned to human DNA. Next, ENCODE blacklisted regions were removed using BEDTools v.2.17 and peak calling was completed using MACS2.
Data analysis
Unless otherwise mentioned, all analyses were implemented in R (https://www.r-project.org, v4.1.1). The quality of the ChIP-seq data was assessed using ChIPQC (v1.25.1).
IDR analysis
To measure consistency between the ChIP replicates, we incorporated the irreproducibility discovery rate (IDR) framework (36), which evaluates a pair of ranked lists of peaks or ranges and assigns values that reflect its reproducibility. A single set of high confidence peaks from the two biological replicates was derived for each unique combination of cell line (MDA-MB-453, T47D or SKBR3) and transcription factor of interest (FOXA or TLE3). Peak detection was applied in the paired-end reads setting at IDR q < 0.01, using IDRfilter in the ChIPpeakAnno R package (v3.27.7). The resultant peaks were then used for overlaps between and across different cell lines and transcription factors.
Peak overlap
Peak overlaps between two or multiple peak sets were determined using the default settings of ChIPpeakAnno findOverlapsOfPeaks, namely, peaks are considered as overlapping if the maximum gap between peaks is −1 (gap is considered to be −1 if one range has its start or end strictly inside the other) and the minimum number of positions that overlap between them is 0 for any types of overlap between genomic ranges. If multiple peaks are involved in any group of connected/overlapping peaks in any input peak list, the “keepAll” option was set for the connected peaks, which will add the number of involved peaks for each peak list to the corresponding overlapping counts, but only add the minimal involved peaks in each group of connected/overlapped peaks to the overlapping counts.
Peak annotation
Peaks are annotated to the nearest genes and genomic features over the regions within 3 kb upstream and downstream from the transcription start sites (TSS) using the ChIPseeker R package (v1.29.2). The distribution of annotated genomic features is visualized in a bar plot, also generated by ChIPseeker. To compare functional annotations of the target genes between FOXA1 and TLE3, KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis was performed using the clusterProfiler R package (v4.1.4). The top 15 significantly enriched categories (Benjamini-Hochberg multiple testing correction; P-adj < 0.01) are visualized in a dot plot generated by the same package.
Motif discovery
For the de novo motif discovery of TLE3 ChIP-seq data in T47D, MDA-MB-453 and SKBR3, we applied the GADEM method(37) using the rGADEM R package (v2.42.0). Briefly, rGADEM is an unsupervised and stochastic motif discovery tool that combines sampling with subsequent enrichment analysis to find over-represented sequence motifs. The extracted motifs of interest are compared with JASPAR2020 database using TFBSTools (v1.32.0) and JASPAR2020 R packages. Putative binding sites were declared with P value < 0.0002 and motifs selected with a cutoff value at 0 for the logarithm of the E-value. The Pearson correlation coefficient between the position weight matrix (PWM) of the unknown motif and the PWM of the JASPAR motif was computed for each discovered motif, and used as its similarity index to the known motifs. For motif analysis of siNS vs. siFOXA1 TLE3-ChIP-seq data, we used HOMER (v4.11.1) de novo motif analysis with default parameters.
Differential binding affinity analysis
To determine differential binding between siNS and siFOXA1 TLE3-ChIP-seq samples, we utilized the DiffBind R package (v3.4.11). Only overlapping peaks present in all samples were used to generate the consensus peak set for analysis. Sites that were differentially bound between the siNS and siFOXA1 groups were identified using the DESeq2 method with an FDR cutoff of 0.05. The MA plot generated from these data indicate statistically significantly differentially bound sites as pink dots. Blue dots indicate binding sites with non-significant differences, and regions of the plot containing many blue dots close together are instead represented by a blue cloud to reduce clutter.
Summary plots and heatmaps of ChIP-seq data
To generate a visual representation of TLE3 binding density for siNS and siFOXA1 samples at basal signature genes, we utilized deepTools (v2.5.2). Briefly, bam files for each sample were normalized against matched input, filtered for ENCODE hg38 blacklisted regions, and converted into bigwig files, all using bamCompare. These normalized bigwig files were used with computeMatrix to generate a matrix comparing TLE3 binding intensity for each sample in a 4 kb window centered at the TSS of each basal signature gene identified by Smid and Charafe-Jauffret. Basal gene hg38 coordinates were downloaded in bed format from UCSC Genome Table Browser, using the ncbiRefSeqSelect table. Heatmaps and density plots were created from the binding matrix using plotHeatmap.
Overlap of ChIP-seq and RNA-seq data
The potential target genes that were bound and regulated were proposed from the subset of genes that are both significantly differentially expressed, as detected from RNA-seq analysis, and bound by transcription factors of interest, as detected from ChIP-seq analysis. The gene expression profile of the potential target genes was visualized in a volcano plot generated by the EnhancedVolcano R package (v1.12.0).
For certain analyses, the basal gene subsets from either the ChIP-seq data or from combined ChIP-seq/RNA-seq were filtered and evaluated. The cross tabulation of the overlap of TLE3 and FOXA1 peaks was generated by the use of gplots R package (v3.1.1). Other plots were generated using ggpubr package (v0.4.0), if not otherwise specified.
In vitro assays
Invasion and migration
Parental T47D and MDA-MB-453 cells were transiently transfected as described earlier with ON-TARGETplus Human TLE3 siRNA (J-019929-07) or siNS 48 hr prior to plating for migration and invasion assays. Cells with stable (TLE3-WT or TLE3-KO) or transient silencing were suspended in serum-free media and plated at a concentration of 8x105 cells/mL into modified Boyden chambers (Corning #3422) to assess migration or into matrigel-coated invasion chambers at a concentration of 4x105 cells/mL (Corning #354480). Cells were allowed to migrate or invade in response to serum as a chemoattractant for 72 hr. To assess migration and invasion of MDA-MB-468 cells with transient overexpression of TLE3, cells were infected with 1x108 Ad-GFP or Ad-TLE3 for 24 hr, at which point media was changed and cells were cultured for another 24 hr. The next day, cells were plated into migration or invasion chambers at 7.5x105 cells/mL and allowed to migrate for 24 hr in response to serum as a chemoattractant. Chamber inserts were fixed and stained with Diff-Quick and mounted onto slides. Migration and invasion were quantified by calculating the average number of cells per field of view (20x magnification) across five images/insert. Three independent experiments were conducted for each cell line with two or three replicates.
Cell growth
Parental T47D and MDA-MB-453 cells were transiently transfected as described earlier with siNS or ON-TARGETplus Human TLE3 siRNA (J-019929-07) in 24 well plates. Stable T47D and MDA-MB-453 TLE3-KO and TLE3-WT, and stable MDA-MB-468 TLE3OE and GFP cells were plated at equal concentrations in 24 well plates. Cells were stained with 0.05% crystal violet on days ranging between 2-7 post-plating. Crystal violet was solubilized using 10% acetic acid and absorbance was read on a GloMax Explorer (Promega #GM3500) plate reader at 600nm.
Analysis of publicly available data
Kaplan-Meier Curves
Kaplan-Meier curves using metadata from TCGA, GEO, and EGA were generated using KM plotter (https://kmplot.com/analysis/) for breast cancer. JetSet status was set to best probe set for all genes analyzed. Cutoff number for high vs. low expression was determined by lower and upper quartiles, and number of patients is noted in the figure legends for each plot. Kaplan-Meier curves for TCGA or METABRIC data were generated using cBioPortal to extract data, and GraphPad Prism was used to graph and run statistical analyses. Metastasis data from the METABRIC cohort was extracted using Oncomine. Patient metastatic data was overlaid with patient TLE3 expression data extracted from cBioPortal and TLE3-high vs. low expressing patients were divided at the median.
Multivariate analysis
Multivariate analysis for TLE3, ESR1, MK167, and ERBB2 expression was conducted using KM plotter (https://kmplot.com/analysis/) for breast cancer.
Correlation plots
Spearman correlation values for genes correlated with TLE3 within the Luminal A/B tumors of the TCGA dataset were downloaded from cBioPortal. Genes with correlation values between −0.01 and 0.01 were removed for visualization purposes. All negatively correlated genes with a Pearson correlation value of <−0.01 were queried within the MSigDB-GSEA website (https://www.gsea-msigdb.org/gsea/msigdb/) for enriched pathways. Spearman correlation values of TLE3 or known corepressors (38) with FOXA1, were obtained using cBioPortal. For FOXA1 and TLE3 correlation in cell lines, expression data was downloaded from Heiser 2012 (39) dataset using UCSC-Xenabrowser (https://xenabrowser.net/) website.
Data Code and Availability
The RNA-seq data generated in this study are publicly available at NCBI GEO (GEO accession number GSE205356).
The ChIP-seq data generated in this study are publicly available at NCBI GEO. TLE3 and FOXA1 ChIP-seq data in T47D, MDA-MB-453, and SKBR3 cells are available under GEO accession number GSE204667. TLE3 ChIP-seq data in siFOXA1 MDA-MB-453 cells are available under GEO accession number GSE205862.
The scRNA-seq data referenced in this study and published by Wu, et al. (33) are publicly available at NCBI GEO (GEO accession number GSE176078).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available upon request.
Results
TLE3 expression is associated with breast cancer differentiation status and increased recurrence-free survival
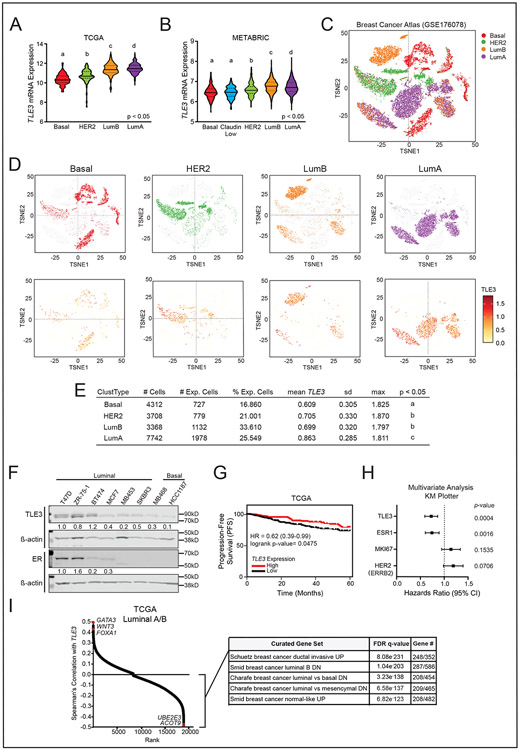
To discern the potential role of TLE3 in breast cancer, we first assessed its expression across subtypes by querying the TCGA (Fig. 1A) and METABRIC (Fig. 1B) datasets. TLE3 expression was significantly higher in more differentiated Luminal A and B tumor subtypes compared to all others. We also analyzed TLE3 expression patterns using publicly available scRNA-seq data of 130,246 single cells isolated from 26 primary patient tumors spanning ER+, HER2+ and TNBC subtypes (33) (Fig. 1C). Isolated tumor epithelial cells were segregated by intrinsic subtypes including Basal, HER2, Luminal A, and Luminal B. The most differentiated and highly treatable Luminal A tumors had the highest mean TLE3 expression and both luminal A and B tumors had the highest percentage of cells expressing this gene (Fig. 1D-E). In contrast, BLBCs exhibited the lowest expression levels in a smaller fraction of cells. Assessment of TLE3 protein expression across a panel of human breast cancer cell lines further revealed that TLE3 is generally more highly expressed in luminal compared to basal lines, and that TLE3 expression is not dependent on ER (Fig. 1F). We further found that TLE3 is prognostic of patient outcomes using the TCGA dataset (40) . Patients whose breast tumors had higher TLE3 expression had longer progression-free survival (PFS) compared to those with low TLE3 (Fig. 1G). These data support a previous study examining TLE3 protein expression and survival reporting that breast cancer patients with high TLE3-expressing tumors had longer RFS (41). Lastly, examination of subtype-specific outcomes using the KM plotter meta-analysis tool revealed that TLE3 levels were also prognostic in patients with Luminal A breast cancer (SFig. 1A). A similar, but non-significant, trend was observed in Luminal B patients (SFig. 1B).
Figure 1: TLE3 is highly expressed in luminal breast cancers and is associated with increased recurrence-free survival.
A-B) Violin plots for TLE3 expression in breast tumor subtypes within the A) TCGA and B) METABRIC datasets.
C) t-SNE visualization of cancer epithelial cell scRNA-seq data (GSE176078) (33). Cells are clustered and colored by intrinsic molecular subtype.
D) t-SNE visualization of cancer epithelial cells. Top Row: each plot depicts cells classified in distinct molecular subtypes. Bottom row: log-normalized expression of TLE3 in each subtype, visualized by color intensity and size of data point (larger point = higher expression).
E) TLE3 scRNA-seq expression levels in Basal, HER2, Luminal B, and Luminal A subtypes. Distinct letters under p<0.05 indicate differences in expression of TLE3 between groups.
F) Representative western blots for TLE3 and ER expression in breast cancer cell lines. Numbers represent protein quantitation relative to β-actin. There is no value where ER was undetectable.
G) Kaplan-Meier plot of PFS for all breast cancer patients in the TCGA dataset with high vs. low TLE3 expression, corresponding to the top and bottom quartiles, respectively.
H) Multivariate analysis accounting for TLE3, ESR1, MKI67, and ERBB2 expression in all breast cancer patients using KM plotter.
I) Spearman correlation of TLE3 with all genes expressed in Luminal A and B tumors in the TCGA dataset. Table inset shows the top five most highly enriched gene sets (C2) reported by MSigDB when analyzing all negatively correlated genes.
ER signaling is a major determinate of breast cancer outcomes (1,42) and the association of TLE3 with recurrence-free survival could be confounded by ER co-expression. Thus, we examined whether TLE3 is prognostic of patient outcomes independently of ER using a multivariate proportional hazard model of breast cancer patient metadata from KM plotter (Fig. 1H). Tumors with lower TLE3 levels were significantly associated with ~28% lower recurrence-free survival independently of ESR1 (HR=0.72, 95% CI: 0.59-0.86, p=0.0004). As MKI67 and ERBB2 levels are associated with poor patient outcomes in basal and HER2+ tumors, respectively, we also included these factors in the analysis and found that low TLE3 remained prognostic of worse outcomes when these factors were considered.
To begin discovering the functional role of TLE3 in luminal breast cancer, we identified the set of genes in Luminal A and B tumors whose expression is correlated with TLE3 using the TCGA dataset (40). Pathway analysis of genes negatively correlated with TLE3 (i.e., genes that may be repressed by TLE3) revealed enrichment for gene expression signatures associated with BLBC (Fig. 1I). These findings suggest that TLE3 may contribute to luminal lineage tumor phenotypes by repressing BLBC genes through its canonical function as a transcriptional corepressor.
TLE3 dictates breast cancer cell fate by directly repressing BLBC signature genes
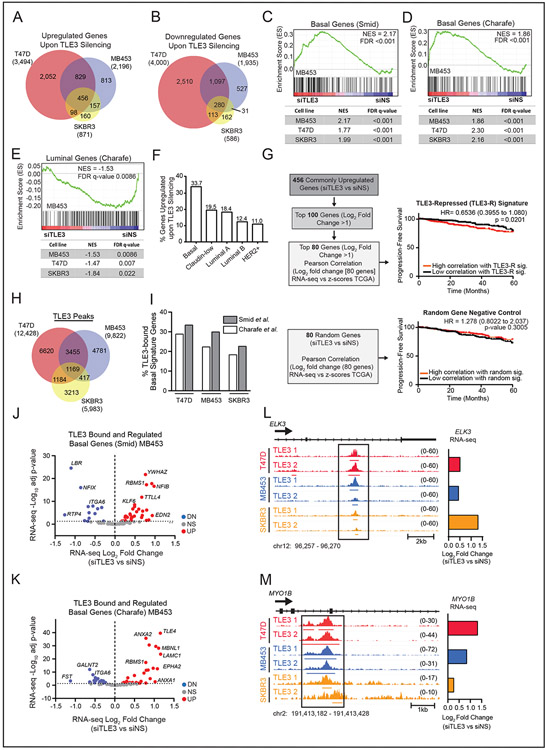
To determine the extent to which TLE3 may repress BLBC signature gene expression, we transiently silenced TLE3 in three luminal breast cancer cell lines (SFig. 2A), one of which expresses ER (T47D) and two that do not (MDA-MB-453 and SKBR3) (Fig. 1F). RNA-seq analysis revealed 456 upregulated (Fig. 2A), and 280 downregulated (Fig. 2B), genes conserved across all three cell lines (p-adj < 0.05). Upregulated genes were enriched for processes involved in transcriptional regulation and cellular adhesion (SFig. 2B). More importantly, gene set enrichment analysis (GSEA) revealed that TLE3 loss results in an induction of BLBC gene expression (28,29) (Fig. 2C-D and SFig. 2C). Hence, TLE3 suppresses basal gene expression in luminal cells. Silencing TLE3 also resulted in a coordinated decrease in luminal signature gene expression (Fig. 2E). Together, these data indicate that TLE3 is necessary to sustain the luminal, and prevent basal, cell fates as defined by their gene expression patterns. To further assess the role of TLE3 in sustaining the luminal fate, we evaluated the impact of TLE3 suppression on genes comprising the PAM50 classifier that identifies tumors by intrinsic subtype. TLE3 silencing broadly shifts the expression of basal and luminal genes, indicating partial subtype switching (SFig. 2D). To confirm these findings, we generated TLE3-null luminal breast cancer cell lines, MDA-MB-453 and T47D, using CRISPR-Cas9 (SFig. 2E). qPCR analysis of TLE3 knockout (TLE3-KO) and wildtype controls (TLE3-WT) confirmed that TLE3 loss induces the expression of genes associated with BLBCs, including EGFR, UBE2E3, SOX9, and KRT4 (SFig. 2F-G).
Figure 2. TLE3 directly represses basal signature genes.
A-B) Venn diagram of significantly A) upregulated and B) downregulated genes following TLE3 silencing for 48 hours in T47D, MDA-MB-453, and SKBR3 cells (p-adj < 0.05).
C-D) GSEA of changed gene expression from RNA-seq data using MDA-MB-453, T47D, and SKBR3 cells (siTLE3 vs. siNS) for basal signature genes derived from primary breast tumors C) SMID_BREAST_CANCER_BASAL_UP or from cell lines D) CHARAFE_BREAST_CANCER_LUMINAL_VS_BASAL_DN. GSEA for MDA-MB-453 are shown as representative enrichment plots.
E) GSEA of RNA-seq expression data from MDA-MB-453, T47D, and SKBR3 cell lines (siTLE3 vs. siNS) for CHARAFE_BREAST_CANCER_LUMINAL_VS_BASAL_UP MSigDB luminal signature gene set. GSEA of MDA-MB-453 data is shown as a representative enrichment plot.
F) Percent of genes upregulated with TLE3 silencing, classified as Basal, Claudin-Low, Luminal A, Luminal B, or HER2+ based on their highest expression level in the METABRIC dataset.
G) RNA-seq data from T47D, SKBR3, and MDA-MB-453 cells transiently transfected with siNS or siTLE3 for 48 hours was used to generate a TLE3-repressed (TLE3-R) gene signature. Expression of the TLE3-R gene signature was correlated with human tumor expression levels and progression-free survival up to 60 months (top) is reported. Survival curve for correlation with a random gene signature (negative control) (bottom).
H) Venn diagram of TLE3 ChIP-seq peak overlap in T47D, SKBR3, and MDA-MB-453 cells. The total number of peaks detected in each cell line is shown in parentheses.
I) Percent of TLE3 peaks that mapped to basal signature genes (defined by Smid and Charafe-Jauffret gene sets) in T47D, MDA-MB-453, and SKBR3 cells.
J-K) Volcano plot of basal genes within the Smid (J) and Charafe-Jauffret (K) signatures that are both bound (ChIP-seq) by TLE3 and significantly differentially expressed in response to TLE3 silencing (RNA-seq) in MDA-MB-453 cells. Red and blue symbols indicate statistically significant up- or downregulated expression, respectively (p-adj < 0.05).
L-M) ChIP-seq tracks of TLE3 binding in T47D, MDA-MB-453, and SKBR3 cells at basal signature genes, L) ELK3 and M) MYO1B. Height of peak tracks is shown in parentheses. Log2 fold changes as determined by RNA-seq (siTLE3 vs. siNS) are shown to the right.
The upregulation of BLBC signature genes in response to TLE3 loss suggested that TLE3 may actively repress an expression program that promotes aggressive disease. We assessed the association of the 456 genes that were upregulated in response to TLE3-silencing (i.e., TLE3-repressed genes) with different breast cancer subtypes in the METABRIC dataset. A large subset (33.7%) were most highly expressed in BLBCs compared to all other subtypes (Fig. 2F). By correlating the expression of the 100 most upregulated genes derived from the RNA-seq data (conserved across all three cell lines) to expression levels in patients in the TCGA dataset, we found that this gene set was associated with reduced progression-free survival (PFS) (SFig. 2H). Using just the top 80 of the most upregulated genes conserved across all three cell lines [‘TLE3-R’ (TLE3-repressed) signature (Fig. 2G Top, Sup.Table 1)] was still significantly associated with outcomes. This result contrasted with a negative control gene set comprised of 80 randomly-selected genes that are expressed in breast cancer (Fig. 2G Bottom). The association of the set of TLE3-repressed genes with worse outcomes was confirmed in the METABRIC dataset (SFig. 2I). Together, these data indicate that TLE3 represses a core set of genes in aggressive breast cancers. GO biological processes that are enriched in the TLE3-R gene set include “Protein Localization”, “Cell Migration”, and “Locomotion” (SFig. 2J), notable cellular features associated with BLBC.
As TLE3 is a canonical co-repressor (19), we postulated that it may directly bind to BLBC genes to inhibit their expression in luminal cells. Using ChIP-sequencing, we identified TLE3 binding sites in T47D, MDA-MB-453, and SKBR3 luminal breast cancer cell lines. Approximately 75% of global TLE3 binding sites mapped to intragenic regions, while ~25% mapped to distal intergenic regions (SFig. 3A), suggesting that TLE3 may have both short- and long-range corepressor activity. Consistent TLE3 binding sites were identified at 1169 regions across all three cell lines (Fig. 2H). Regarding associations of binding with gene regulation, T47D cells exhibited the greatest number of TLE3 peaks, with ~40% mapping to genes that were also differentially expressed upon TLE3 silencing (p-adj < 0.05, SFig. 3B). Genes that were both bound and regulated by TLE3 were also identified in MDA-MB-453 and SKBR3 cell lines, albeit at lower frequency (~22% and 11%, respectively). The lower percentage in these two cell lines is likely due to the ability of TLE3 to modulate ER-target genes in T47D cells (19,21), while TLE3 targets in MDA-MB-453 and SKBR3 cells are independent of ER as they lack this receptor (Fig. 1F) (43). Similar to previous reports, we also found that TLE3 peaks mapped to genes that were both up- and downregulated in response to TLE3 silencing, suggesting that it both represses and activates transcription (21,25) (SFig. 3C-E).
When examining TLE3 binding specifically to basal signature genes, we detected peaks within all three cell lines at basal signature genes defined by Smid (28) and Charafe-Jauffret (29) (18.4-28.9% and 22.7-33.5%, respectively, Fig. 2I). Many of these genes were upregulated upon TLE3 silencing (Fig. 2J-K and SFig. 3F-I). Hence, TLE3 binds to genes that drive BLBC phenotypes, such as ELK3 and MYO1B, and represses their transcription in luminal breast cancer cells (Fig. 2L-M) independently of ER. These data establish TLE3 as a transcriptional repressor of genes associated with poor outcomes from breast cancer.
TLE3 suppresses migration and invasion of luminal breast cancer cells
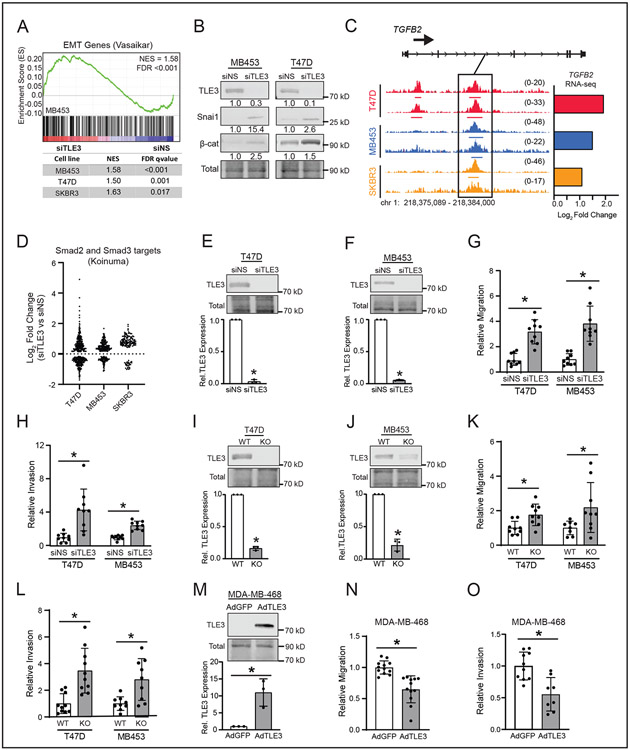
As a group, BLBCs are more migratory, invasive, and metastatic than their luminal counterparts (42,44). These phenotypes are often driven by an epithelial to mesenchymal transition (EMT). Several genes induced by TLE3 silencing are established EMT modulators, including TUFT1 and CGN (45) (SFig. 4A-B), suggesting that TLE3 may actively repress the mesenchymal phenotype of BLBCs. While no overt morphological changes were observed in response to TLE3 silencing or knock-out (data not shown), GSEA analysis of RNA-seq data (siTLE3 vs. siNS) revealed that TLE3-silenced cells increase the expression of EMT-associated genes (Fig. 3A). Moreover, protein markers of EMT, β-catenin and SNAIL, were induced in MDA-MB-453 and T47D cells following transient TLE3 silencing (Fig. 3B). TGF-β signaling drives EMT during development and carcinogenesis. TGF-β ligands (TGFβ1-3) bind their cognate receptors, induce downstream signaling, and activate SMAD transcription factors that then stimulate the expression of mesenchymal genes (46). Notably, TGFβ2 expression was upregulated following TLE3 silencing and consistent TLE3 binding sites were detected at the TGFβ2 locus in all three lines (Fig. 3C), suggesting that TLE3 represses TGFβ signaling in luminal cells. As an indicator of activated TGF-β signaling, we assessed the expression of SMAD2/3 target genes (30) following TLE3 silencing in the RNA-seq dataset. The majority of SMAD2/3 targets were upregulated in response to TLE3 silencing in both MDA-MB-453 and SKBR3 cell lines, indicating activation of this signaling pathway (Fig. 3D). In contrast, a similar proportion of SMAD targets were up- and downregulated in T47D cells, potentially reflecting a role of ER in this pathway (47). Together, these data suggest that TLE3 directly represses TGFβ2 expression, signaling, and EMT in luminal cells and upon TLE3 removal, they acquire a partial mesenchymal gene signature that is more typical of BLBC cells.
Figure 3. TLE3 suppresses cellular migration and invasion of luminal breast cancer cells.
A) GSEA of RNA-seq data from MDA-MB-453, T47D, and SKBR3 cell lines (siTLE3 vs. siNS) for EMT genes reported by Vasaikar, et al (45). GSEA of MDA-MB-453 is shown as a representative plot.
B) Representative western blots of mesenchymal markers, Snai1 and β-catenin, in siTLE3 vs. siNS T47D and MDA-MB-453 cells after 96 hr.
C) ChIP-seq tracks of TLE3 binding in T47D, MDA-MB-453, and SKBR3 cells at the TGFβ2 locus. Height of peak tracks is shown in parentheses. Log2 fold changes as determined by RNA-seq (siTLE3 vs. siNS) are shown to the right.
D) Bee swarm depicting siTLE3 vs. siNS RNA-seq log2 fold change values of Smad2 and Smad3 target genes [reported by Koinuma, et al (30)] in T47D, MB453, and SKBR3 cell lines (p-adj < 0.05).
E-F) Representative western blots and quantitation of TLE3 expression in siTLE3 vs. siNS E) T47D and F) MDA-MB-453 cells after five days. Westerns quantified relative to total protein (*p < 0.05). n=3 in triplicate.
G-H) Relative G) migration and H) invasion of TLE3-silenced cells compared to control after 72 hr (*p < 0.05). n=3 in triplicate.
I-J) Western blot images and quantitation for TLE3 expression in I) T47D and J) MDA-MB-453 TLE3-WT and TLE3-KO cells. Westerns quantified relative to total protein (*p < 0.05). n=3 in triplicate.
K-L) Relative K) migration and L) invasion of TLE3-KO vs. TLE3-WT cells after 72 hr (*p < 0.05). n=3 in triplicate.
M) Western blot images and quantitation of TLE3 in MDA-MB-468 cells infected with adenovirus expressing GFP (AdGFP) or TLE3 (AdTLE3) 72 hr after initial infection. Western blots were quantified relative to total protein (*p < 0.05). n=3 in triplicate.
N-O) Relative N) migration and O) invasion of AdTLE3 vs. AdGFP MDA-MB-468 cells after 24 hr (*p < 0.05). n=3 in triplicate.
The transcriptional shift to a more BLBC gene expression pattern and upregulation of EMT genes upon TLE3 silencing suggested that TLE3 may repress aggressive cell phenotypes associated with BLBC. Indeed, transient (Fig. 3E-H) and stable (Fig. 3I-L) suppression of TLE3 resulted in increased migration and invasion of cells relative to TLE3-intact controls. These changes were not due to differences in proliferation as TLE3 loss did not increase cell growth (SFig. 4C-G). As expected, orthogonal studies using a viral vector to overexpress TLE3 in basal MDA-MB-468 cells (Fig. 3M) revealed reduced migration (Fig. 3N) and invasion (Fig. 3O) compared to cells infected with an empty vector control (AdGFP). Similar to TLE3 silencing, transient TLE3 overexpression did not affect cell growth (SFig. 4H). Together, these data indicate that TLE3 represses the invasive and migratory capacity of luminal breast cancer cells in the absence of major changes in proliferation and that these phenotypes are associated with an enhanced EMT-like transcriptional program.
TLE3 suppresses breast cancer metastasis
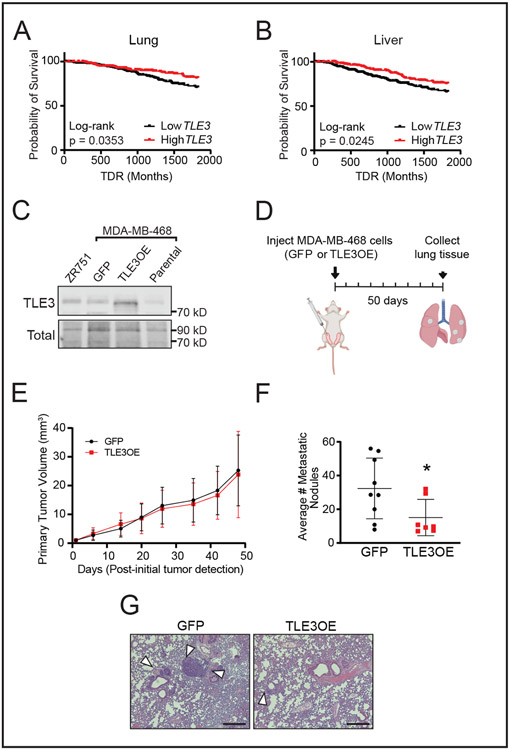
EMT gene signatures and elevated migration and invasion are commonly associated with metastatic progression. To determine if TLE3 expression is associated with breast cancer metastasis, we evaluated metastatic outcomes of patients in the METABRIC dataset. Consistent with the migration and invasion studies, patients whose tumors expressed high TLE3 had a reduced probability of metastatic relapse, both to the lung and liver (Fig. 4A-B). To directly determine if elevated TLE3 is sufficient to repress metastasis, TLE3 was stably overexpressed in MDA-MB-468 basal breast cancer cells, a line that readily metastasizes to the lungs in mice (48) (Fig. 4C). No difference in primary tumor growth was observed 50 days after TLE3-overexpressing (TLE3-OE) or GFP-expressing control cells were orthotopically injected into the mammary fat pads of female mice (Fig. 4D-E). In contrast, the lungs of mice with TLE3-overexpressing tumors had significantly reduced numbers of metastatic nodules compared to controls (Fig. 4F-G). Combined with the patient data, these results indicate that TLE3 is a metastasis suppressor in breast cancer.
Figure 4. TLE3 suppresses breast cancer metastasis.
A-B) Probability of distant recurrence to the A) lung and B) liver in patients with primary tumors with high vs. low TLE3 expression in the METABRIC dataset. (High vs. low expression determined by upper and lower 50% TLE3 expression).
C) Western blot for TLE3 in ZR751 (luminal) and MDA-MB-468 (basal) cells stably transduced with control GFP or TLE3 overexpressing (TLE3-OE) lentivirus. Parental MDA-MB-468 cells included for comparison.
D) Schematic of lung metastasis experiment using orthotopically injected tumor cells. Created using BioRender.com.
E) Primary tumor volumes of MDA-MB-468 cells with stable overexpression of GFP or TLE3.
F) Average number of metastatic lung nodules. Student’s t-test (*p = 0.04). n=10 mice.
G) Representative images of H&E stained lung sections from mice harboring MDA-MB-468 GFP or TLE3-OE primary tumors. Arrowheads indicate metastases.
TLE3 represses expression of the metastatic driver SOX9
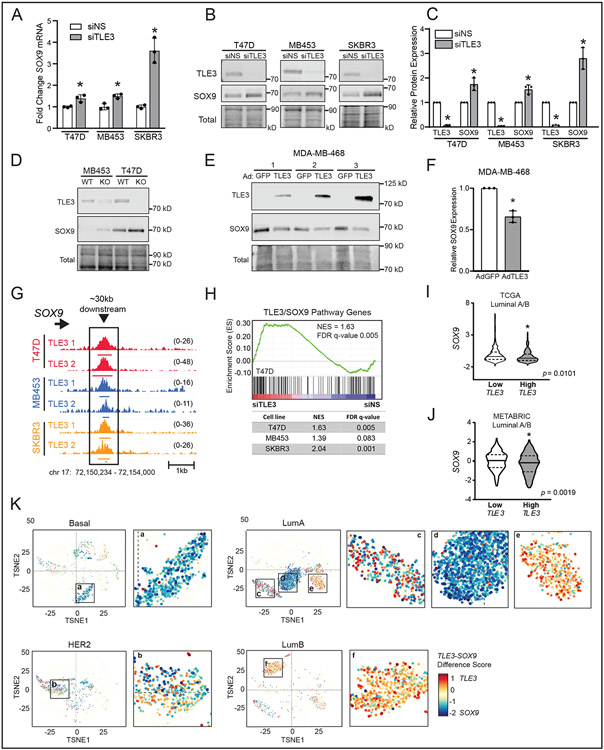
Interrogation of the RNA-seq dataset revealed that TLE3 silencing significantly upregulates SOX9 in all three cell models (Fig. 5A). In addition to being a basal signature gene, SOX9 is a transcription factor essential for mammary gland development that also drives breast cancer progression (28,49,50). Upregulation of SOX9 protein was confirmed following transient (Fig. 5B-C) and stable (Fig. 5D) loss of TLE3. Moreover, transient overexpression of TLE3 in the basal breast cancer cell line, MDA-MB-468, significantly suppressed SOX9 protein expression (Fig. 5E-F). Examination of TLE3 ChIP-seq data further showed that TLE3 directly binds to a region approximately 30kb downstream of the SOX9 gene locus in T47D, MDA-MB-453, and SKBR3 cell lines (Fig. 5G), indicating that TLE3 directly represses SOX9 expression. This is likely due to the ability of TLE3 to tether and condense surrounding chromatin (51).
Figure 5. TLE3 represses expression of the metastatic driver SOX9.
A) Fold change in SOX9 expression (RNA-seq) following transfection with a non-silencing siRNA (siNS) or siRNA targeting TLE3 (siTLE3) after 48 hr (*p-adj < 0.05).
B) Representative western blots of TLE3 and SOX9 expression following transient silencing of TLE3 after 96 hr in T47D, MDA-MB-453, and SKBR3 cells. n=3.
C) Quantitation of SOX9 and TLE3 expression shown in B, relative to total protein. (*p < 0.05). n=3.
D) Representative western blots for TLE3 and SOX9 in TLE3-WT and TLE3-KO T47D and MDA-MB-453 cell lines.
E) Western blots for TLE3 and SOX9 expression in MDA-MB-468 cells infected with adenovirus expressing GFP (as a control) or TLE3 after 48 hr. Numbers 1,2,3 indicate protein collected from three independent experiments. n=3.
F) Quantitation of SOX9 expression shown in E, relative to total protein (*p < 0.05). n=3.
G) ChIP-seq tracks of the conserved binding site for TLE3 approximately 30kb downstream of the SOX9 gene locus in T47D, MDA-MB-453, and SKBR3 cells. Height of peak tracks is shown in parentheses.
H) GSEA of RNA-seq data (siTLE3 vs. siNS) in T47D, MDA-MB-453, and SKBR3 cell lines for SOX9 direct targets (32) that were also negatively correlated with TLE3 in the TCGA dataset. GSEA of T47D is shown as a representative.
I-J) SOX9 levels in Luminal A and B tumors within the I) TCGA or J) METABRIC dataset that have low (bottom quartile) or high (top quartile) TLE3 expression.
K) t-SNE visualization of scRNA-seq data of individual cancer epithelial cells (GSE176078) classified as Basal, HER2, Luminal A (LumA), or Luminal B (LumB). The difference of TLE3 and SOX9 expression (TLE3-SOX9) is depicted by color intensity and size of the point (larger point = larger expression difference). High TLE3 relative to SOX9 expression is red while high SOX9 relative to TLE3 is blue.
To assess the functional impact of SOX9 repression by TLE3, we queried whether TLE3 silencing would result in the upregulation of SOX9 transcriptional targets in luminal breast cancer cells. Using a publicly available SOX9 ChIP-seq dataset, we identified a set of direct SOX9 gene targets (32). We then asked which of these genes were negatively correlated with TLE3 in the TCGA PanCancer dataset to develop a core set of genes that are downstream targets of the TLE3/SOX9 pathway in breast cancer. Notably, we found that this signature set of genes is enriched in the RNA-seq analysis of TLE3-silenced cells (Fig. 5H). Thus, TLE3 suppression of SOX9 in luminal breast cancer cells prevents the expression of SOX9 targets. Moreover, SOX9 target genes are highly dynamic and can be rapidly activated when TLE3 expression is blocked. Together, these data indicate that the direct repression of SOX9 by TLE3 in luminal cells as one mechanism for its ability to suppress basal gene expression and promote luminal lineage phenotypes.
To determine if the inverse relationship between TLE3 and SOX9 is conserved in patient tumors, we queried gene expression data for Luminal A and Luminal B breast cancers in the TCGA and METABRIC datasets and found that tumors with high TLE3 expression had significantly lower SOX9 expression while low TLE3 was associated with higher SOX9 (Fig. 5I-J). The inverse expression patterns of TLE3 and SOX9 were also observed at the single cell level. Basal breast cancer cells expressed high SOX9 and very low TLE3 (Fig. 5K a), whereas a large portion of Luminal B cells expressed high TLE3 and low SOX9 (Fig. 5K f). Of note, we observed three different populations of Luminal A cells (Fig. 5K c-e). The first was comprised of roughly equal populations of TLE3highSOX9low and TLE3lowSOX9high cells (Fig. 5K c). The second primarily contained cells that were TLE3lowSOX9high (Fig. 5K d). Lastly, a third cluster of cells was identified that is predominantly TLE3highSOX9low (Fig. 5K e). This aligns with the reported heterogeneity of Luminal A tumors when examined at the single cell level (33). Moreover, these data affirm the inverse relationship of TLE3 and SOX9 in many cells, supporting the conclusion that TLE3 is a core repressor of SOX9 in luminal breast cancer.
TLE3 and FOXA1 expression are highly correlated in breast cancer
Due to its lack of a DNA binding domain, TLE3 interacts with partnering transcription factors to impact gene expression (26,51). We noted that TLE3 loss in luminal breast cancer cells conveys a phenotype similar to that observed with FOXA1 silencing (18), specifically an increase in migration and invasion (Figure 3). FOXA1 is a pioneering transcription factor that promotes luminal breast cancer phenotypes by activating luminal as well as repressing BLBC genes (18,52). Importantly, FOXA1 and TLE3 have been reported to co-regulate subsets of genes in breast and prostate cancer models (51,53,54). Thus, we postulated that TLE3 repression of BLBC genes in luminal cells may require interactions with FOXA1. Supporting this possibility, TLE3 binding sites identified by ChIP-seq were enriched in forkhead box protein binding motifs (SFig. 5A). Analysis of the METABRIC dataset also revealed that, of the 43 corepressor that are expressed in breast cancer (38), TLE3 is in the top five that are positively correlated with FOXA1 across all breast cancer subtypes (SFig. 5B). Of these five corepressors, low TLE3 expression was also associated with the largest decrease in recurrence-free survival [HR=0.52 (0.42-0.65), metadata from KM plotter] (SFig. 5C-G). Interrogation of individual datasets also revealed that TLE3 and FOXA1 mRNA expression are positively correlated in breast tumors in the METABRIC (SFig. 5H) and TCGA (SFig. 5I) datasets. TLE3 and FOXA1 are also highly correlated in breast cancer cell lines (39) (SFig. 5J, r = 0.8137). These data suggest that FOXA1 may be a core mediator of TLE3 function in breast cancer.
TLE3 collaborates with FOXA1 to repress BLBC signature genes
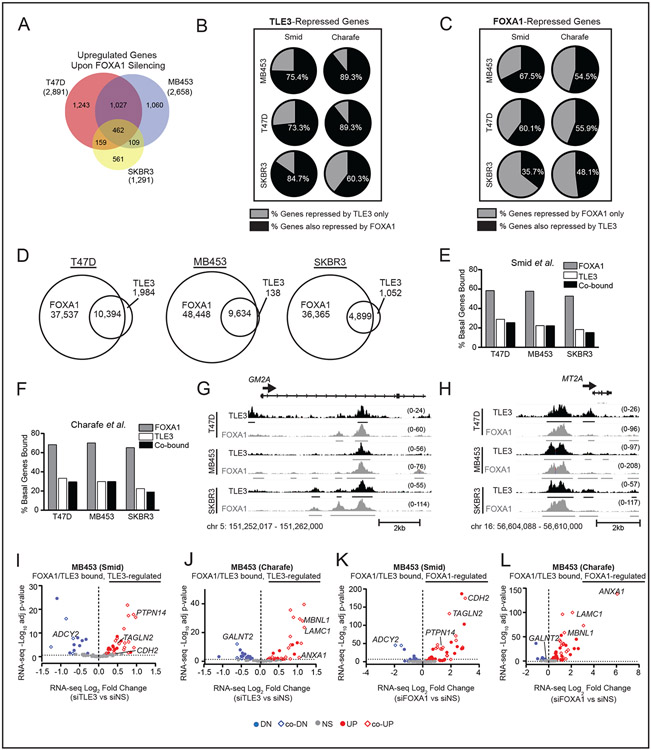
To assess the extent to which TLE3 and FOXA1 co-regulate target genes, we first identified the FOXA1 up-regulated transcriptome in T47D, MDA-MB-453, and SKBR3 cell lines following transient FOXA1 silencing (Fig. 6A). Similar to our prior report (18), luminal signature gene expression is reduced in T47D and MDA-MB-453 cells (SFig. 6A) while BLBC gene expression is increased (SFig. 6B-C) in all cell lines tested, when FOXA1 is lost. When comparing the TLE3- and FOXA1-regulated transcriptomes, ~50% of genes regulated by TLE3 were also modulated by FOXA1, in the same direction (SFig. 6D). Restricting the analysis to BLBC signature genes defined by Smid (28) or Charafe-Jauffret (29), revealed that the majority (60-89%) of TLE3-repressed genes were also repressed by FOXA1 (Fig. 6B). In contrast, <35% of FOXA1-regulated genes were co-regulated by TLE3 (with the exception of T47D cells where ~50% of genes were co-regulated) (SFig. 6E). When specifically examining BLBC genes, ~50% of genes repressed by FOXA1 were also inhibited by TLE3 (Fig. 6C). Together, these data indicate that FOXA1 co-regulates the majority of TLE3-regulated BLBC genes, whereas about half of FOXA1 BLBC gene targets are co-regulated by TLE3.
Figure 6. TLE3 collaborates with FOXA1 to repress BLBC signature genes.
A) Venn diagram of RNA-seq data for overlapping genes that are significantly upregulated in siFOXA1 vs. siNS T47D, SKBR3, and MDA-MB-453 cells (p-adj < 0.05). Total number of upregulated genes within each cell line are shown in parentheses.
B) Percent of TLE3-repressed (upregulated upon TLE3 silencing) BLBC signature genes defined by Smid or Charafe-Jauffret that are also repressed by FOXA1 (derived from RNA-seq data, p-adj < 0.05).
C) Percent of FOXA1-repressed BLBC genes that are also repressed by TLE3 (p-adj < 0.05).
D) Venn diagram showing the number of overlapping TLE3 and FOXA1 peaks in T47D, MB453, and SKBR3 cell lines.
E-F) Percent of basal signature genes bound by FOXA1, TLE3, or both in the E) Smid or F) Charafe-Jauffret signature gene lists.
G-H) Examples of TLE3 and FOXA1 overlapping peaks mapped to representative basal signature genes, G) GM2A and H) MT2A in T47D, MDA-MB-453, and SKBR3 cell lines. Peak height is shown in parentheses. n=2.
I-J) Volcano plot of basal signature genes defined by I) Smid or J) Charafe-Jauffret co-bound by TLE3 and FOXA1 that were differentially expressed in response to TLE3 silencing (RNA-seq data siTLE3 vs. siNS). Colored dots indicate significantly upregulated (red) or downregulated (blue) genes (p-adj < 0.05), while gray dots indicate genes not significantly changed.
K-L) Same as I-J but in response to FOXA1 silencing (RNA-seq data siFOXA1 vs. siNS, p-adj < 0.05).
To explore whether TLE3 and FOXA1 collaboratively and directly regulate BLBC genes, we assessed their overlapping binding patterns. Using ChIP-seq for FOXA1, we identified 16,748 conserved peaks in T47D, MDA-MB-453, and SKBR3 cells (SFig. 6F). Integrating TLE3 and FOXA1 peak sets revealed that the vast majority (82-99%) of TLE3 binding sites overlap with FOXA1 sites in each cell line (Fig. 6D and SFig. 6G). The overlap was lower when comparing across cell lines (≥54%), but still constituted a majority. Hurtado, et al. reported similar binding patterns between FOXA1 and ER, highlighting the cell line-specific binding activity of FOXA1 (55). In contrast to TLE3-bound sites, the majority of FOXA1 binding sites do not overlap with TLE3 bound regions (Fig. 6D and SFig. 6G). Thus, most TLE3-bound sites reside in regions that also bind FOXA1, while FOXA1 interacts with a much broader set of sites across the genome. A similar trend was observed at BLBC signature genes. Nearly all TLE3 peaks reside at sites that also bind FOXA1, whereas FOXA1 binds many additional sites in the absence of TLE3 (Fig. 6E-F; examples of genes that bind both FOXA1 and TLE3 are shown in Fig. 6G-H). Together, these data indicate that TLE3 binding almost always coincides with FOXA1, both globally and at basal signature genes, whereas the majority of FOXA1 binding sites are independent of TLE3.
The expression of many BLBC genes that were bound by TLE3 and FOXA1 was also regulated by both proteins, underscoring the functional significance of co-binding (Fig. 6I-L and SFig. 6H-O). For example, FOXA1 silencing increased SOX9 mRNA and protein expression in SKBR3 and MDA-MB-453 cells (SFig. 7A-B), similar to that observed with TLE3 loss (Fig. 5A-C). Moreover, TLE3 and FOXA1 binding sites at the SOX9 locus are nearly identical (SFig. 7C). As indicated above, overexpression of TLE3 in the MDA-MB-468 BLBC cell line represses SOX9 expression (Fig. 5E-F). Notably, these cells have very low, but detectable FOXA1 (SFig. 7D), raising the possibility that TLE3 repression of SOX9 in BLBC cells may require endogenous FOXA1. Transient FOXA1 silencing blocks the ability of TLE3 to suppress SOX9 expression in MDA-MD-468 cells that were engineered to overexpress TLE3 (SFig. 7D-G), indicating that low levels of FOXA1 are indeed necessary and sufficient to facilitate SOX9 repression by TLE3. Similar to the inverse expression of TLE3 with SOX9 that occurs in individual human tumor cells (Fig. 5K), FOXA1 and SOX9 are also inversely correlated (SFig. 7H), supporting the likelihood that TLE3 and FOXA1 co-repress SOX9 in human tumors.
TLE3 binding sites are dictated by FOXA1
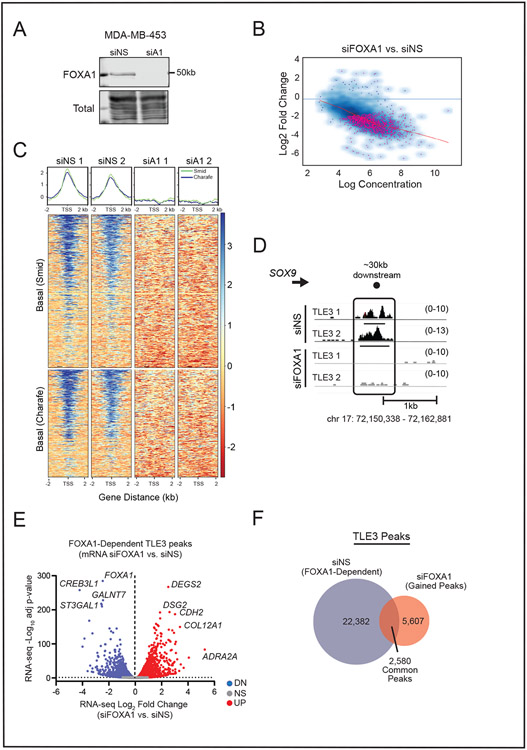
TLE3 associates with chromatin by interacting with histone tails or transcription factors (51). The extensive overlap of TLE3 binding sites with FOXA1 sites suggests that FOXA1 may be the primary binding partner of TLE3 at its target genes in luminal breast cancer. Indeed, FOXA1 is necessary for TLE3 recruitment to the ERα target gene, TFF1, in the ER+ breast cancer cell line, MCF-7 (21). However, the extent to which FOXA1 is required for genome-wide TLE3 binding in breast cancer cells, independently of ER, is currently unknown. We transiently silenced FOXA1 and quantified its impact on genome-wide binding of TLE3 using MDA-MB-453 cells that lack ER [Fig. 7A and (43)]. TLE3 binding was significantly changed at 1,461 genomic sites following FOXA1 silencing (Fig. 7B). The vast majority of TLE3 binding to BLBC genes was eliminated with the loss of FOXA1 (Fig. 7C), including TLE3 binding to the SOX9 locus (Fig. 7D). Thus, FOXA1 is an obligate partner required for TLE3 binding to most chromatin sites in luminal breast cancer cells that lack ER. This interaction is functionally significant as many genes that lose TLE3 binding in response to FOXA1 silencing also undergo changes in their transcription (Fig. 7E). Interestingly, a similar number of genes were up- and downregulated, suggesting that the TLE3/FOXA1 interaction may also have activating properties at a subset of sites.
Figure 7. TLE3 binding sites are dictated by FOXA1.
A) Representative western blot confirming FOXA1 silencing in MDA-MB-453 cells after 48 hr.
B) Affinity plot showing 1,461 significantly different TLE3-bound regions between siNS and siFOXA1 samples (FDR < 0.05). Pink dots denote sites with significantly changed binding.
C) Density plots and heatmaps of TLE3 binding at basal signature genes as defined by Smid or Charafe-Jauffret in siNS vs. siFOXA1 (siA1) samples.
D) ChIP-seq tracks of TLE3 binding in siNS or siFOXA1 MDA-MB-453 cells at the SOX9 locus. Height of peak tracks is shown in parentheses.
E) Volcano plot of peaks specific to the siNS group and their change in gene expression following FOXA1 silencing after 48 hr (RNA-seq, p-adj < 0.05).
F) Venn diagram depicting TLE3 peaks common and unique to siNS or siFOXA1 samples.
A relatively small number of shared/common peaks remained unchanged in response to FOXA1 silencing, indicating that TLE3 must utilize a FOXA1-independent mechanism for binding at these loci (Fig. 7F). In addition, 5,607 new TLE3-bound peaks were detected after FOXA1 loss, suggesting that TLE3 may utilize a different binding partner to interact with other sites in the genome when FOXA1 is absent. Motif analysis of these novel TLE3 binding sites identified the transcription factors AP-2γ, GATA, and other FOX factors as potential partners (SFig. 8). As a whole, these results demonstrate that TLE3 maintains luminal lineage fidelity of breast cancers by interacting with FOXA1 and repressing the expression of a cohort of BLBC genes.
Discussion
Cancer cell phenotypic plasticity facilitates the establishment of tumor subpopulations capable of driving therapeutic resistance and metastasis (56). Here, we report the discovery that TLE3 is a gatekeeper of luminal cell identity in breast cancer. We demonstrate that TLE3 promotes luminal breast cancer epithelial cell lineage fidelity through transcriptional repression of a cohort of BLBC genes that can drive aggressive tumor phenotypes including metastatic outgrowth. Additionally, TLE3 serves as a critical determinant of FOXA1 target gene repression in both ER-dependent and -independent contexts. This includes repression of SOX9, an established driver of breast cancer metastasis (49). Moreover, this study identifies a core set of genes coordinately repressed by TLE3 that are functionally significant in human tumor samples. Many of these genes have not been extensively studied in breast cancer. Thus, these results provide new targets for investigation relevant to tumor cell plasticity.
A well-studied mechanism underlying tumor cell plasticity is EMT. EMT is often considered a binary conversion where cells lose epithelial and gain mesenchymal traits, providing the ability to migrate and invade. The mesenchymal state has also been associated with cancer stem cells (57) and increased tumorigenesis, metastasis, and therapeutic resistance (58). More recent data have revealed that EMT can gradually progress through intermediate hybrid states (partial EMT) that generate heterogeneous populations in cancer and during normal mammary morphogenesis (59,60). Partial EMT states defined by the simultaneous expression of epithelial and mesenchymal markers have been associated with increased invasive and metastatic potential in multiple cancer types (56). Data presented herein demonstrates that TLE3 suppression results in increased expression of mesenchymal genes including CDH2, CTNNB1, and SNAI1, whereas expression of the epithelial markers, CDH1 and CLDN1, remained unchanged. Thus, TLE3 regulates partial/hybrid EMT states (61) that have been shown to be necessary for BLBC tumorigenicity (62) and have been identified in patient tumors (63). We also identified TGFβ2, an established activator of EMT and hybrid states (64), as a direct downstream target of TLE3. TLE3 silencing induced TGFβ2 expression and activation of SMAD2 and SMAD3 target genes, including SOX9. Together, these data implicate an EMT-regulatory switch involving opposing signals between TLE3 and SMADs on a subset of EMT-associated genes. TLE3 normally represses expression of these genes in luminal cells and its loss induces TGFβ2 gene transcription and subsequently, SMAD signaling. Thus, the ability of luminal breast cancer cells to undergo EMT may ultimately be defined by TLE3 expression. Future studies are needed to directly assess the extent to which TLE3 controls TGF-β signaling and expression of its downstream target genes, both in vitro and in vivo.
The FOX family of transcription factors are most well recognized for their pioneering functions. Upon DNA binding, FOX proteins relax surrounding chromatin, allowing for the binding of other transcriptional regulators, including corepressors (52). This enabling property permits control of gene expression programs by context-specific binding partners. We found that FOXA1 is essential for TLE3 to bind to its target genes. Given the ability of FOXA1 to open chromatin sites in breast cancer, it is conceivable that the loss of FOXA1 could induce the formation of heterochromatin and would indirectly prevent TLE3 binding. However, direct interactions between TLE and FOX proteins have previously been reported in multiple cell types (21-24,27). Moreover, the loss of TLE3 binding that occurs with FOXA1 silencing results in an induction of gene expression, indicative of open, rather than closed, chromatin states. As a specific example, silencing TLE3 or FOXA1 induces expression of the metastatic driver, SOX9. Affirming the ability of TLE3 and FOXA1 to bind and regulate the SOX9 gene, we previously reported that FOXA1 directly represses SOX9 expression in breast cancer cells (18). In addition, previously published TLE3-ChIP-seq data confirms that TLE3 binds to the SOX9 gene in MCF-7 breast cancer cells (22) as well as prostate cancer cells (65).
In addition to its ability to recruit TLE3, FOXA1 can also recruit ARID1A, a subunit of the SWI/SNF chromatin remodeling complex, to a set of target genes (7). Like TLE3, this represses BLBC signature gene expression. While FOXA1 can recruit ARID1A to sites prior to ER binding (17), the majority of FOXA1/ARID1A binding overlaps with ER upon exposure to ligand. Importantly, the effect of the FOXA1/ARID1A interaction in the context of luminal cells devoid of ER has not been explored. To our knowledge, the current study is the first to identify a genome-wide mechanism by which FOXA1 promotes luminal identity in the context of both ER-positive and -negative luminal breast cancer cells. The data reported herein utilized two ER− cell lines, leading us to conclude that a set of BLBC signature genes are repressed by TLE3/FOXA1 independently of ER. To further explore the potential for ER to mediate the effects of TLE3 on BLBC genes, we interrogated a dataset of ER-regulated genes in T47D cells (66). Only a minority (9.15%) of TLE3-regulated genes across all three cell lines are established ER targets (data not shown), further indicating that TLE3 repression of BLBC genes is independent of ER.
Because TLE3 has also been associated with ER transcriptional complexes (22), it is plausible that it may collaborate with FOXA1, ARID1A, and ER to repress BLBC targets in ER+ cells but interacts with alternate binding partners, such as androgen or glucocorticoid receptors (AR or GR, respectively) in ER− luminal cells. Supporting this possibility, TLE3 co-localizes with FOXA1 and AR to modulate target gene expression and anti-androgen responsiveness in prostate cancer cells (53). Significant overlap between AR and glucocorticoid receptor (GR) cistromes has also been reported and many of these sites include FOXA1 motifs (67). Lastly, it is notable that TLE3 can be expressed in non-luminal cells (Fig. 1F), suggesting that it has a different function in these cells. Indeed, TLE3 has been suggested to regulate the functions of EN1, a key modulator of triple-negative breast cancer cell growth (68). Together, these results suggest that the varying partners of TLE3 are critical for defining its role in regulating breast cancer cell fates.
Our study revealed that approximately 25% of conserved TLE3 binding sites in luminal breast cancer cells are intergenic (SFig. 3A). Of note, a greater percentage (~50%) of binding sites for other lineage determinants (i.e., FOXA1 and GATA3) are intergenic (69,70). This likely reflects the expanded ability of these proteins to collaborate with additional binding partners other than TLE3 (52). We also discovered that TLE3 silencing upregulates FOXA1 expression (GEO dataset GSE205356), an observation that was recently reported during β-cell development (71). These results suggest that TLE3 regulates FOXA1 activity through two distinct processes to ensure both the appropriate levels and function of FOXA1 in luminal breast cancer.
ChIP-seq analysis led to the unexpected finding that TLE3 becomes redistributed to sites containing AP-2 (TFAP) motifs following FOXA1 loss. TFAP2C can promote luminal identity of normal mammary epithelial cells as well as breast cancer cells (72) and loss of TFAP2C in luminal breast cancer induces BLBC gene expression. Additionally, AP-2 interacts with FOXA1 to facilitate long-range chromatin interactions (73). Thus, FOXA1, TLE3, and AP-2 may function in a coordinated fashion to maintain proper lineage fidelity. Although FOXA1 is highly expressed in most breast cancers, mutations in FOXA1 can alter its activity (24). FOXA1 is mutated in ~4% of all breast cancers and this incidence is increased in the metastatic setting (74). The majority of FOXA1 mutations in breast cancer occur within the C-terminal forkhead domain, the region required for interaction with TLE3 (24). Thus, mutations in this region (termed Class 2) may disrupt the interaction between FOXA1 and TLE3. If so, this could shift TLE3 to collaborate with AP-2 in the context of FOXA1 mutations. Additional studies examining the role of TLE3 in the context of FOXA1 mutations should reveal the extent to which a TLE3/AP-2 interaction may drive tumor cell phenotypes and uncover mechanisms by which disruption of normal FOXA1 activity contributes to tumor progression.
In summary, we identified the genome-wide regulatory landscape of TLE3 in luminal breast cancer cells and discovered its key role in sustaining luminal lineage specification, even in the absence of ER. This involves the definitive repression of basal signature genes that are associated with partial/hybrid EMT as well as metastasis. FOXA1 is required for the majority of TLE3 binding. These data reveal that that TLE3 works closely with FOXA1 as a key repressor of aggressive phenotypes. Future studies that identify upstream inducers of TLE3 expression may reveal novel pathways that could promote the acquisition of less aggressive, more treatable disease states that extend patient survival.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Vinay Varadan and Hannah Hill for their insightful input. We also acknowledge shared resources supported by the Case Comprehensive Cancer Center (NIH P30CA043703) including the Flow Cytometry and Histology Cores, as well as the Imaging and Small Animal Imaging Cores at the Cleveland Clinic.
This work was supported by the following funding:
DoD W81XWH-18-1-0455, NIH T32CA059366 (LJA)
NIH T32GM008803 (PRM)
NIH R01CA206505 (RAK)
DoD W81XWH-06-1-0712 (GMS)
NIH F31CA224809 (BMW)
NIH P30CA043703 (RAK)
NIH T32GM008056 (LC-V)
NIH R25CA225461 (MV)
NIH R25GM075207 (AA)
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
REFERENCES
- 1.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature 2000;406:747–52 [DOI] [PubMed] [Google Scholar]
- 2.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987;235:177–82 [DOI] [PubMed] [Google Scholar]
- 3.Williams C, Lin CY. Oestrogen receptors in breast cancer: basic mechanisms and clinical implications. Ecancermedicalscience 2013;7:370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marra A, Trapani D, Viale G, Criscitiello C, Curigliano G. Practical classification of triple-negative breast cancer: intratumoral heterogeneity, mechanisms of drug resistance, and novel therapies. NPJ Breast Cancer 2020;6:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anstine LJ, Keri R. A new view of the mammary epithelial hierarchy and its implications for breast cancer initiation and metastasis. J Cancer Metastasis Treat 2019;5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wainwright EN, Scaffidi P. Epigenetics and Cancer Stem Cells: Unleashing, Hijacking, and Restricting Cellular Plasticity. Trends Cancer 2017;3:372–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu G, Chhangawala S, Cocco E, Razavi P, Cai Y, Otto JE, et al. ARID1A determines luminal identity and therapeutic response in estrogen-receptor-positive breast cancer. Nat Genet 2020;52:198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giraddi RR, Chung CY, Heinz RE, Balcioglu O, Novotny M, Trejo CL, et al. Single-Cell Transcriptomes Distinguish Stem Cell State Changes and Lineage Specification Programs in Early Mammary Gland Development. Cell Rep 2018;24:1653–66 e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srinivasan M, Bharali DJ, Sudha T, Khedr M, Guest I, Sell S, et al. Downregulation of Bmi1 in breast cancer stem cells suppresses tumor growth and proliferation. Oncotarget 2017;8:38731–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res 2006;66:6063–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez ME, Moore HM, Li X, Toy KA, Huang W, Sabel MS, et al. EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proc Natl Acad Sci U S A 2014;111:3098–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Granit RZ, Gabai Y, Hadar T, Karamansha Y, Liberman L, Waldhorn I, et al. EZH2 promotes a bi-lineage identity in basal-like breast cancer cells. Oncogene 2013;32:3886–95 [DOI] [PubMed] [Google Scholar]
- 13.Saxena M, Kalathur RKR, Rubinstein N, Vettiger A, Sugiyama N, Neutzner M, et al. A Pygopus 2-Histone Interaction Is Critical for Cancer Cell Dedifferentiation and Progression in Malignant Breast Cancer. Cancer Res 2020;80:3631–48 [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto S, Wu Z, Russnes HG, Takagi S, Peluffo G, Vaske C, et al. JARID1B is a luminal lineage-driving oncogene in breast cancer. Cancer Cell 2014;25:762–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell 2006;127:1041–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Theodorou V, Stark R, Menon S, Carroll JS. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res 2013;23:12–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagarajan S, Rao SV, Sutton J, Cheeseman D, Dunn S, Papachristou EK, et al. ARID1A influences HDAC1/BRD4 activity, intrinsic proliferative capacity and breast cancer treatment response. Nat Genet 2020;52:187–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernardo GM, Bebek G, Ginther CL, Sizemore ST, Lozada KL, Miedler JD, et al. FOXA1 represses the molecular phenotype of basal breast cancer cells. Oncogene 2013;32:554–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agarwal M, Kumar P, Mathew SJ. The Groucho/Transducin-like enhancer of split protein family in animal development. IUBMB Life 2015;67:472–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel SR, Bhumbra SS, Paknikar RS, Dressler GR. Epigenetic mechanisms of Groucho/Grg/TLE mediated transcriptional repression. Mol Cell 2012;45:185–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jangal M, Couture JP, Bianco S, Magnani L, Mohammed H, Gevry N. The transcriptional co-repressor TLE3 suppresses basal signaling on a subset of estrogen receptor alpha target genes. Nucleic Acids Res 2014;42:11339–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohammed H, D'Santos C, Serandour AA, Ali HR, Brown GD, Atkins A, et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep 2013;3:342–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmes KA, Hurtado A, Brown GD, Launchbury R, Ross-Innes CS, Hadfield J, et al. Transducin-like enhancer protein 1 mediates estrogen receptor binding and transcriptional activity in breast cancer cells. Proc Natl Acad Sci U S A 2012;109:2748–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parolia A, Cieslik M, Chu SC, Xiao L, Ouchi T, Zhang Y, et al. Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature 2019;571:413–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Villanueva CJ, Waki H, Godio C, Nielsen R, Chou WL, Vargas L, et al. TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell Metab 2011;13:413–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher AL, Caudy M. Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev 1998;12:1931–40 [DOI] [PubMed] [Google Scholar]
- 27.Naderi A, Meyer M, Dowhan DH. Cross-regulation between FOXA1 and ErbB2 signaling in estrogen receptor-negative breast cancer. Neoplasia 2012;14:283–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JG, et al. Subtypes of breast cancer show preferential site of relapse. Cancer Res 2008;68:3108–14 [DOI] [PubMed] [Google Scholar]
- 29.Charafe-Jauffret E, Ginestier C, Monville F, Finetti P, Adelaide J, Cervera N, et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene 2006;25:2273–84 [DOI] [PubMed] [Google Scholar]
- 30.Koinuma D, Tsutsumi S, Kamimura N, Taniguchi H, Miyazawa K, Sunamura M, et al. Chromatin immunoprecipitation on microarray analysis of Smad2/3 binding sites reveals roles of ETS1 and TFAP2A in transforming growth factor beta signaling. Mol Cell Biol 2009;29:172–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011;121:2750–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larsimont JC, Youssef KK, Sanchez-Danes A, Sukumaran V, Defrance M, Delatte B, et al. Sox9 Controls Self-Renewal of Oncogene Targeted Cells and Links Tumor Initiation and Invasion. Cell Stem Cell 2015;17:60–73 [DOI] [PubMed] [Google Scholar]
- 33.Wu SZ, Al-Eryani G, Roden DL, Junankar S, Harvey K, Andersson A, et al. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet 2021;53:1334–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–87 e29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 2019;20:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qunhua Li JBB, Haiyan Huang, Bickel Peter J.. Measuring reproducibility of high-throughput experiments. 1752–79 p. [Google Scholar]
- 37.Li L. GADEM: a genetic algorithm guided formation of spaced dyads coupled with an EM algorithm for motif discovery. J Comput Biol 2009;16:317–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet 2010;11:109–23 [DOI] [PubMed] [Google Scholar]
- 39.Heiser LM, Sadanandam A, Kuo WL, Benz SC, Goldstein TC, Ng S, et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci U S A 2012;109:2724–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018;173:291–304 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kashiwagi S, Fukushima W, Asano Y, Goto W, Takada K, Noda S, et al. Identification of predictive markers of the therapeutic effect of eribulin chemotherapy for locally advanced or metastatic breast cancer. BMC Cancer 2017;17:604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 2001;98:10869–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vranic S, Gatalica Z, Wang ZY. Update on the molecular profile of the MDA-MB-453 cell line as a model for apocrine breast carcinoma studies. Oncol Lett 2011;2:1131–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prat A, Karginova O, Parker JS, Fan C, He X, Bixby L, et al. Characterization of cell lines derived from breast cancers and normal mammary tissues for the study of the intrinsic molecular subtypes. Breast Cancer Res Treat 2013;142:237–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vasaikar SV, Deshmukh AP, den Hollander P, Addanki S, Kuburich NA, Kudaravalli S, et al. EMTome: a resource for pan-cancer analysis of epithelial-mesenchymal transition genes and signatures. Br J Cancer 2021;124:259–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu S, Chen S, Zeng J. TGFbeta signaling: A complex role in tumorigenesis (Review). Mol Med Rep 2018;17:699–704 [DOI] [PubMed] [Google Scholar]
- 47.Band AM, Laiho M. Crosstalk of TGF-beta and estrogen receptor signaling in breast cancer. J Mammary Gland Biol Neoplasia 2011;16:109–15 [DOI] [PubMed] [Google Scholar]
- 48.Jin X, Demere Z, Nair K, Ali A, Ferraro GB, Natoli T, et al. A metastasis map of human cancer cell lines. Nature 2020;588:331–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma Y, Shepherd J, Zhao D, Bollu LR, Tahaney WM, Hill J, et al. SOX9 Is Essential for Triple-Negative Breast Cancer Cell Survival and Metastasis. Mol Cancer Res 2020;18:1825–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jana S, Madhu Krishna B, Singhal J, Horne D, Awasthi S, Salgia R, et al. SOX9: The master regulator of cell fate in breast cancer. Biochem Pharmacol 2020;174:113789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sekiya T, Zaret KS. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell 2007;28:291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seachrist DD, Anstine LJ, Keri RA. FOXA1: A Pioneer of Nuclear Receptor Action in Breast Cancer. Cancers (Basel) 2021;13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palit SA, Vis D, Stelloo S, Lieftink C, Prekovic S, Bekers E, et al. TLE3 loss confers AR inhibitor resistance by facilitating GR-mediated human prostate cancer cell growth. Elife 2019;8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang JC, Waltner-Law M, Yamada K, Osawa H, Stifani S, Granner DK. Transducin-like enhancer of split proteins, the human homologs of Drosophila groucho, interact with hepatic nuclear factor 3beta. J Biol Chem 2000;275:18418–23 [DOI] [PubMed] [Google Scholar]
- 55.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 2011;43:27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bakir B, Chiarella AM, Pitarresi JR, Rustgi AK. EMT, MET, Plasticity, and Tumor Metastasis. Trends Cell Biol 2020;30:764–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hong D, Fritz AJ, Zaidi SK, van Wijnen AJ, Nickerson JA, Imbalzano AN, et al. Epithelial-to-mesenchymal transition and cancer stem cells contribute to breast cancer heterogeneity. J Cell Physiol 2018;233:9136–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aggarwal V, Montoya CA, Donnenberg VS, Sant S. Interplay between tumor microenvironment and partial EMT as the driver of tumor progression. iScience 2021;24:102113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pastushenko I, Brisebarre A, Sifrim A, Fioramonti M, Revenco T, Boumahdi S, et al. Identification of the tumour transition states occurring during EMT. Nature 2018;556:463–8 [DOI] [PubMed] [Google Scholar]
- 60.Thong T, Wang Y, Brooks MD, Lee CT, Scott C, Balzano L, et al. Hybrid Stem Cell States: Insights Into the Relationship Between Mammary Development and Breast Cancer Using Single-Cell Transcriptomics. Front Cell Dev Biol 2020;8:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Campbell K, Casanova J. A common framework for EMT and collective cell migration. Development 2016;143:4291–300 [DOI] [PubMed] [Google Scholar]
- 62.Kroger C, Afeyan A, Mraz J, Eaton EN, Reinhardt F, Khodor YL, et al. Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. Proc Natl Acad Sci U S A 2019;116:7353–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013;339:580–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hao Y, Baker D, Ten Dijke P. TGF-beta-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis. Int J Mol Sci 2019;20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stelloo S, Nevedomskaya E, Kim Y, Hoekman L, Bleijerveld OB, Mirza T, et al. Endogenous androgen receptor proteomic profiling reveals genomic subcomplex involved in prostate tumorigenesis. Oncogene 2018;37:313–22 [DOI] [PubMed] [Google Scholar]
- 66.Williams C, Edvardsson K, Lewandowski SA, Strom A, Gustafsson JA. A genome-wide study of the repressive effects of estrogen receptor beta on estrogen receptor alpha signaling in breast cancer cells. Oncogene 2008;27:1019–32 [DOI] [PubMed] [Google Scholar]
- 67.Arora VK, Schenkein E, Murali R, Subudhi SK, Wongvipat J, Balbas MD, et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell 2013;155:1309–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peluffo G, Subedee A, Harper NW, Kingston N, Jovanovic B, Flores F, et al. EN1 Is a Transcriptional Dependency in Triple-Negative Breast Cancer Associated with Brain Metastasis. Cancer Res 2019;79:4173–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 2008;132:958–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cohen H, Ben-Hamo R, Gidoni M, Yitzhaki I, Kozol R, Zilberberg A, et al. Shift in GATA3 functions, and GATA3 mutations, control progression and clinical presentation in breast cancer. Breast Cancer Res 2014;16:464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Theis A, Singer RA, Garofalo D, Paul A, Narayana A, Sussel L. Groucho co-repressor proteins regulate beta cell development and proliferation by repressing Foxa1 in the developing mouse pancreas. Development 2021;148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cyr AR, Kulak MV, Park JM, Bogachek MV, Spanheimer PM, Woodfield GW, et al. TFAP2C governs the luminal epithelial phenotype in mammary development and carcinogenesis. Oncogene 2015;34:436–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tan SK, Lin ZH, Chang CW, Varang V, Chng KR, Pan YF, et al. AP-2gamma regulates oestrogen receptor-mediated long-range chromatin interaction and gene transcription. EMBO J 2011;30:2569–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arruabarrena-Aristorena A, Maag JLV, Kittane S, Cai Y, Karthaus WR, Ladewig E, et al. FOXA1 Mutations Reveal Distinct Chromatin Profiles and Influence Therapeutic Response in Breast Cancer. Cancer Cell 2020;38:534–50 e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.