Wuxiang virus (WUXV), a new species of Phlebovirus from sandfly specimens (identified as Phlebotomus chinensis) collected in Wuxiang County, Shanxi Province, China, belongs to the order Bunyavirales, family Phenuiviridae[1]. Like other Bunyavirales, WUXV contains a negative-sense tripartite RNA genome comprising a small (S), a medium (M), and a large (L) segment. The S segment encodes nucleocapsid protein (NP) and a nonstructural protein (NSP). The M segment encodes a glycoprotein precursor (Gp), which is cleaved into mature N and C glycoproteins. The L segment encodes an RNA-dependent RNA polymerase (RdRp)[1].
WUXV is the first report of a Phlebovirus isolated from sandfly specimens collected from the natural environment in China. The virus was subsequently isolated three times in the same province[1–3]. Although there are no reports of human infection with WUXV, a study showed that it can cause cytopathic effects in mammalian cells, death in neonatal mice, and infection in mammals[1,2]. However, a relevant detection method has not been established. Therefore, it is imperative to create an efficient and rapid method to detect WUXV in sandflies in the environment.
Reverse-transcription recombinase-aided amplification (RT-RAA) is a new isothermal amplification technology using specific enzymes and proteins for the rapid detection of pathogens. Under constant temperature, the amplification process was repeated with high efficiency, and the amplification product of the target gene could reach detectable levels five minutes after the reaction. Its use has been widespread in the clinical detection of human and animal pathogens, such as coronavirus[4] and tick-borne encephalitis virus[5].
This study presents a rapid and sensitive RT-RAA assay to detect WUXV. We designed and synthesized specific primers and probes in the S segment of WUXV (Supplementary Table S1 available in www.besjournal.com). The amplification length was 220 bp. The titer of WUXV strain SXYQ1872–1 (GenBank accession no. MT786487.1) was determined using a plaque assay, and then serial 10-fold dilutions ranging from 103 to 101 pfu/μL were made, and the viral RNA was extracted for use as templates in RT-RAA.
The RT-RAA reaction was performed with an RT exo kit (Jiangsu Qitian Bio-technology, China), which comprised a mixture of all the enzymes (SSB, 800 ng/μL; UvsX, 120 ng/μL; DNA polymerase, 30 ng/μL) needed for reverse-transcription and DNA amplification in lyophilized form in a single tube. The reaction mixture contained the following components: 25 μL of rehydration buffer, 2.1 μL of each primer (10 μmol/L), 0.6 μL of probe (10 μmol/L), 1 μL of the RNA template, and diethylpyrocarbonate (DEPC) water 16.7 μL. Then, 47.5 μL of master mix/template solution was mixed and transferred to each lyophilized enzyme mix. Then, 2.5 μL of 280 mmol/L magnesium acetate was pipetted into each tube lid. The tube lids were closed carefully, vortexed briefly, and centrifuged simultaneously to trigger the RT-RAA reaction. The tube was then placed in a QT-F1620RAA fluorescence detection device (Jiangsu Qitian Bio-technology, China) at 39 °C for 30 minutes. A negative control (DEPC water) was included in each run.
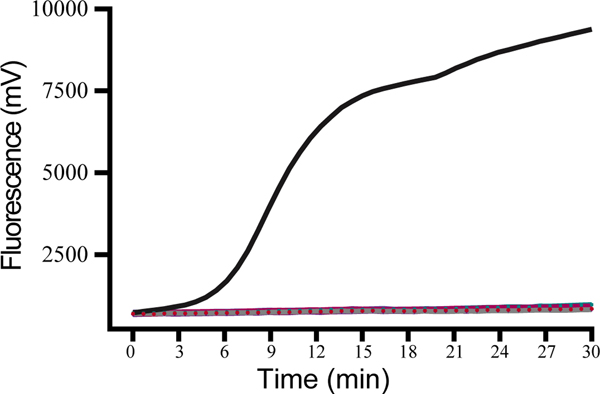
To test the specificity of this assay, the nucleotides of Japanese encephalitis virus, yellow fever virus, West Nile virus, Tahyna virus, severe fever with thrombocytopenia syndrome virus, Kadipiro virus (KDV), and Liaoning virus (LNV) were examined. Acquired experimental results showed that the RT-RAA assay was positive for WUXV and negative for the other seven arboviruses belonging to four families, four genera, and the negative control. No cross-reactivity of RNA from any control virus was observed (Figure 1).
Figure 1.
Specificity of the RT-RAA assay for Wuxiang virus (WUXV).
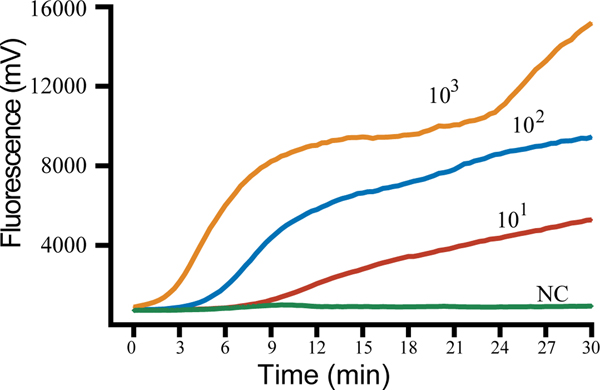
The sensitivity of the method was assessed using a dilution gradient of the WUXV virus ranging from 103 to 101 pfu/μL, with each dilution measured eight times to assess the reproducibility of the method (Table 1). The lower limit of detection was 10 pfu/reaction viral RNA. The higher the template concentration, the greater the fluorescence value and the earlier the peak time (Figure 2).
Table 1.
Assay data used for probit analysis to calculate the detection limit of RT-RAA for WUXV
| Pfu/reaction | No. of positive samples/No. of sample tested by the RT-RAA assays for detection of WUXV |
|---|---|
| 1 × 103 | 8/8 |
| 1 × 102 | 8/8 |
| 1 × 101 | 8/8 |
| 1 × 100 | 1/8 |
Note. Each dilution was tested in a total of eight replicates. WUXV: Wuxiang virus.
Figure 2.
Sensitivity of the RT-RAA assay for Wuxiang virus (WUXV). WUXV titers from 103 to 101 pfu per reaction were used to determine the sensitivity of the RT-RAA assay. NC, negative control.
A total of 30 pools of sandflies were used to perform the RT-RAA assay, and 7 of them were identified as positive for WUXV, which was consistent with the results of the sequencing assay (Supplementary Table S2 available in www.besjournal.com ). The results indicated the accuracy of the assay developed in this study.
WUXV is a sandfly-borne virus. Other such viruses include sandfly fever Sicilian virus (SFSV) and Toscana virus. The former can cause febrile disease after infection, referred to as “three-day fever”[6]; The latter can cause acute meningitis[7]. Using the plaque reduction neutralization test, it was found that humans (8.7%, 4/46) and chickens (100%, 4/4) in the virus isolation region contained WUXV-neutralizing antibodies, suggesting that they could be infected with WUXV[2]. Although there has been no clear report of human WUXV cases, these results indicate that humans are susceptible to the virus, and it may be a potential human disease pathogen.
According to our research, the WUXV is a stable viral population present in local sandflies[3]. Moreover, the sandfly species is distributed widely throughout the plain, mountainous, and Loess Plateau regions north of the Yangtze River. As we detected sandflies only in Shanxi Province, it is unclear whether they carry the virus in other regions. Hence, we need to expand the detection range. Moreover, with the improvement of the population mean standard of living and the development of transportation, tourist regions have been further expanded, which also creates favorable conditions for the contact between humans and sandflies, increasing the possibility of transmission of the sandfly-borne virus. Therefore, a rapid, sensitive, and convenient detection method is indispensable to the monitoring of WUXV among sandflies in the environment.
The current WUXV detection methods are based on virus isolation or rely on traditional reverse-transcription PCR. However, these methods are expensive and time-consuming and require highly specialized equipment, making them unsuitable for widespread application in the field.
As WUXV was only isolated in 2018, few sequences of its three segments are available. There are 23 full-length or partial sequences of the L segment in the NCBI database; however, there are only three full-length sequences, and the rest are short ones of only about 400 bp. Due to the limited number of known sequences, the specificity of the primers and probes designed for this segment may not meet the needs of the experiment. Moreover, the RdRp gene of phleboviruses is highly conserved, with more than 95% identity at the amino acid level[8]. High intergeneric homology will increase the probability of cross-reaction. As for the M segment, it presents on the surface of virions and is thus exposed to the selective pressures of the host. These sequences could vary widely among strains[9]. We selected the S segment to establish the detection method. First, there are 64 known reference sequences for the S segment, significantly more than for the M and L segments. Second, the sequences of the N proteins of the different phleboviruses exhibit low homology, but sequence data indicate high homology within one phlebovirus. Apart from this, some studies have also established detection methods for the S segment. Another study designed primers and probes targeting the NP gene for Congo Hemorrhagic Fever (CCHFV) and SFSV[10].
The sensitivity of the established RT-RAA assay reached 10 pfu per reaction, and there was no crossreaction with seven insect-borne viruses, including Flavivirus, Alphavirus, Orthobunyavirus, and Seadornavirus, which showed strong specificity and high sensitivity. Notably, our RT-RAA assay can detect WUXV in sandflies, requiring only 30 minutes at a constant temperature of 39 °C, producing a positive signal in approximately 10 minutes, and showing 100% consistency with sequencing results. Moreover, for this method, under the condition of constant temperature, the repeated and efficient amplification of nucleic acids can be accomplished. The detection results are determined by the instrument, which is more accurate and objective than human interpretation. The reaction is conducted in a single tube, and the cover is not opened during the process to avoid the possibility of contamination. Furthermore, the reaction reagents are stored in freeze-dried powder form. Compared with liquid reagents, these are easier to manipulate and reduce the loss caused by pipetting error. Meanwhile, they are convenient to transport, avoiding the wastage caused by repeated freezing and thawing during transportation. Although there are many advantages to the RT-RAA assay for virus detection, it also has some limitations. First, because long primers and probes form dimers more easily than short ones, it is difficult to achieve multiplex detection of pathogens. Second, RT-RAA cannot achieve quantitative detection of the nucleic acid of the detected virus. Furthermore, we only used 30 pools of sandflies to validate the assay. This method should be evaluated using more sandfly samples.
However, the advantages of this method make it more suitable for rapid detection in the general laboratory and in the field.
Supplementary Material
Acknowledgments
This research was funded by a development grant from the State Key Laboratory of Infectious Disease Prevention and Control [2015SKLID505 to HW]; the project of the Priority Academic Program Development of Jiangsu Higher Education Institutions [PAPD]; and the United States National Institutes of Health U01 [AI151810].
Footnotes
The authors disclose no potential conflicts of interest.
Biographical note of the first author: YAO Xiao Hui, female, born in 1995, PhD Candidate, majoring in animal infectious disease research.
REFERENCES
- 1.Wang J, Fu SH, Xu ZQ, et al. Emerging sand fly–borne phlebovirus in China. Emerg Infect Dis, 2020; 26, 2435–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang QY, Fu SH, Cheng JX, et al. Re-isolation of Wuxiang virus from wild sandflies collected from Yangquan county, China. Virol Sin, 2021; 36, 1177–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu XY, Cheng JX, Fu SH, et al. Wuxiang virus is a virus circulated naturally in Wuxiang county, China. Vector Borne Zoonotic Dis, 2021; 21, 289–300. [DOI] [PubMed] [Google Scholar]
- 4.Xue GH, Li SL, Zhang WW, et al. Reverse-transcription recombinase-aided amplification assay for rapid detection of the 2019 novel coronavirus (SARS-CoV-2). Anal Chem, 2020; 92, 9699–705. [DOI] [PubMed] [Google Scholar]
- 5.Wang QY, Li F, Shen XX, et al. A reverse-transcription recombinase-aided amplification assay for the rapid detection of the far-eastern subtype of tick-borne encephalitis virus. Biomed Environ Sci, 2019; 32, 357–62. [DOI] [PubMed] [Google Scholar]
- 6.Ayhan N, Baklouti A, Prudhomme J, et al. Practical guidelines for studies on sandfly-borne phleboviruses: Part I: important points to consider ante field work. Vector Borne Zoonotic Dis, 2017; 17, 73–80. [DOI] [PubMed] [Google Scholar]
- 7.Depaquit J, Grandadam M, Fouque F, et al. Arthropod-borne viruses transmitted by Phlebotomine sandflies in Europe: a review. Euro Surveill, 2010; 15, 19507. [PubMed] [Google Scholar]
- 8.Calisher CH, Calzolari M. Taxonomy of phleboviruses, emphasizing those that are sandfly-borne. Viruses, 2021; 13, 918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu DY, Tesh RB, Da Rosa APA, et al. Phylogenetic relationships among members of the genus Phlebovirus (Bunyaviridae) based on partial M segment sequence analyses. J Gen Virol, 2003; 84, 465–73. [DOI] [PubMed] [Google Scholar]
- 10.Ibrahim SM, Aitichou M, Hardick J, et al. Detection of CrimeanCongo hemorrhagic fever, hanta, and sandfly fever viruses by real-time RT-PCR. Methods Mol Biol, 2011; 665, 357–68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.