Summary
Wnt and Rspondin (RSPO) signaling drives proliferation, and bone morphogenetic protein inhibitors (BMPi) impede differentiation, of intestinal stem cells (ISCs). Here we identify the mouse ISC niche as a complex, multi-layered structure that encompasses distinct mesenchymal and smooth muscle populations. In young and adult mice, diverse sub-cryptal cells provide redundant ISC-supportive factors; few of these are restricted to single cell types. Niche functions refine during postnatal crypt morphogenesis, in part to oppose dense aggregation of differentiation-promoting BMP+ sub-epithelial myofibroblasts at crypt-villus junctions. Muscularis mucosae, a specialized muscle layer, first appears during this period and supplements neighboring RSPO and BMPi sources. Components of this developing niche are conserved in human fetuses. In vivo ablation of mouse postnatal smooth muscle raises BMP signaling activity, potently limiting a pre-weaning burst of crypt fission. Thus, distinct and progressively specialized mesenchymal cells together create the milieu required to propagate crypts during rapid organ growth and to sustain adult ISCs.
Keywords: Intestinal stem cell niche, smooth muscle, trophocytes, intestinal sub-epithelial myofibroblasts, intestinal crypt morphogenesis, crypt fission, epithelial-mesenchymal cocultures, single-cell RNA profiles
Graphical Abstract
eTOC blurb
A mesenchymal niche sustains intestinal stem cells and McCarthy et al. describe the layered anatomy, molecular properties, overlapping functions, and postnatal maturation of this supportive sub-epithelial tissue. Cells and secreted factors that constitute the complex niche in young mice correspond to those identified in the mid-gestation human fetus.
Introduction
Lgr5+ intestinal stem cells (ISCs) at the base of adult small intestine (SI) crypts1 replicate in response to Wnt/RSPO2 and differentiate in response to bone morphogenetic protein (BMP) signals.3 Accordingly, the crypt base is a zone of high Wnt/RSPO and low BMP activity, the same conditions that favor ISC expansion ex vivo.4 Sub-epithelial mesenchyme is the principal source of these signals.5–7 At birth, the mouse SI lacks crypts. Over the ensuing ~2 weeks,8,9 crypts progressively form and lengthen,10 sequestering ISCs at the bottom alongside Paneth cells, and undergoing fission to expand epithelial mass.11–14 Crypt size,15 bifurcation,11,13 monoclonality,16 and Paneth cell numbers17 increase in the third week of life. The 3 weeks of substantial epithelial and ISC expansion between birth and weaning are likely associated with appearance of a stable ISC niche in the adjacent mesenchyme.
This mesenchyme, historically regarded as a collection of ‘myofibroblasts’ and smooth muscle (SM),18 has come into focus through identification of overlapping CD34+Gp38+ and PDGFRA+ cell sources of crucial niche factors.19–22 PDGFRAhi cells abutting the epithelium resemble telocytes, diverse cells with a distinctive morphology found in many organs;23,24 they are proposed as a key source of canonical Wnt ligands.25 However, careful review of their properties26,27 indicates that these are the same cells as intestinal sub-epithelial myofibroblasts (ISEMFs) long described in the literature.28,29 Because telocytes lack defined molecular or functional uniformity across tissues, here we use the ISEMF nomenclature, a longstanding and descriptive convention. In contrast, sub-cryptal PDGFRAlo cells that co-express Grem1 (a BMP inhibitor, BMPi) and Rspo3 support ISC expansion ex vivo with no added trophic factors; they are therefore called “trophocytes.”30 PDGFRA− smooth muscle (Acta2+Myh11+) and other PDGFRAlo cells in the trophocyte vicinity also express Grem1 or other trophic factors31,32 but anatomic expression domains are unresolved. Mouse villus morphogenesis has been studied in detail33,34 and fetal cell types were recently catalogued in mouse colon35 and human SI36–39 but it is unclear which cells contribute niche activity or how the ISC niche arises in concert with crypts.
Here we identify SM contributions toward the ISC niche in young and adult mice and show that distinct SM populations substitute in part for selected trophic factors in ex vivo ISC cultures. The niche matures in parallel with postnatal murine crypts, coincident with increasing ISEMF density at the villus base. Niche elements positioned to counteract the resulting concentrated BMP source include the muscularis mucosae (MM) and previously uncharacterized cells in the superficial muscularis propria (MP). MM is molecularly distinct from contiguous lamina propria SM, expresses high Rspo3 and Grem2, arises de novo from resident stromal cells, and delimits a distinct anatomic compartment for trophocytes. Human MM cells (called “myofibroblasts” in recent studies) express a similar repertoire of trophic factors at an analogous developmental stage. In vivo ablation of mouse SM during the pre-weaning peak in crypt fission augments local BMP activity and markedly attenuates crypt fission. Trophocytes provide the most potent RSPO ligand, RSPO2. Thus, a partially redundant, evolutionarily conserved, and multi-layer ISC niche develops step-wise, as distinct mesenchymal cells express overlapping trophic factors at defined distances from ISCs at the crypt base.
Results
Activity and formation of the isthmus ISEMF shelf, a potent BMP source
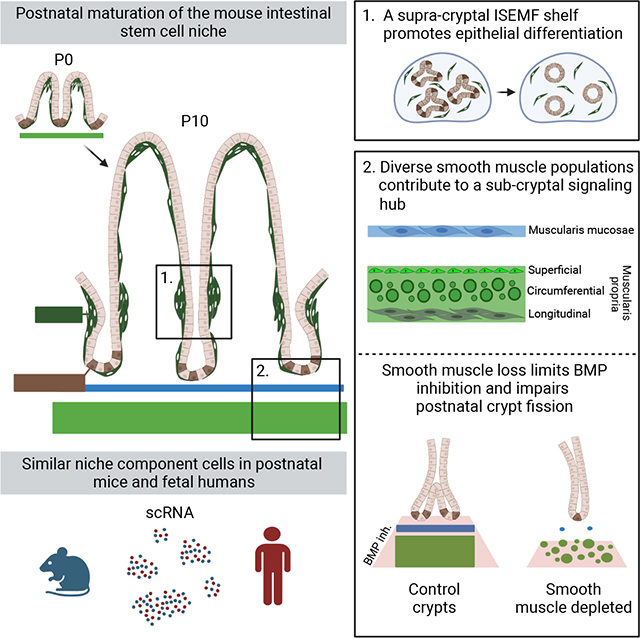
As BMPs drive cell differentiation and their inhibition promotes ISC self-renewal,3,40 phospho-SMAD1/5/9 (pSMAD, a marker of BMP signaling), is restricted to adult mouse villi and absent in crypt cells.41 The presumed function of ISEMFs as a potent BMP source30,35 has not been tested. We cultured adult mouse duodenal crypts in media containing recombinant (r) epidermal growth factor (EGF), rRSPO1, and the BMPi rNOG (ENR medium). After organoids had established, we replaced rNOG with various factors or with cells isolated by flow cytometry from PdgfraH2B-eGFP mice, where PDGFRAhi ISEMFs express high GFP30,42 (Figure 1A, Table S1). Without exogenous cells or in the presence of GFP− (PDGFRA−) cells, organoids enlarged into budding structures, while exposure to GFPhi ISEMFs or to rBMPs arrested organoid growth (Figure 1B). In these structures, ISCs occupy protruding buds.4 Organoids that survived ISEMF co-culture retracted those buds (Figure S1A) and exposure to BMPi rNOG or rGREM1 restored budding forms (Figures 1A and S1A). Proliferative ISCs in organoid buds take up the S-phase marker 5-ethynyl-2’-deoxyuridine (EdU) and this uptake was reduced in ISEMF co-cultures (Figures 1B and S1B). Furthermore, organoids co-cultured with ISEMFs reduced expression of ISC marker genes and increased expression of BMP target genes and differentiation markers (Figure 1C). Thus, ISEMF-derived BMPs tilt the balance from ISC self-renewal to differentiation; subtle morphologic differences between organoids exposed to rBMPs or ISEMFs may reflect disparate BMP levels or additional ISEMF products, e.g., non-canonical Wnts.
Figure 1. ISEMFs promote epithelial differentiation in vitro.
A) Two- to 3-day old established organoids cultured with recombinant (r) RSPO1, EGF, and indicated cell types from PdgfraH2B-eGFP mouse SI. The diagram depicts ISEMFs (green) and trophocytes (red) as BMP and BMPi sources, respectively. Representative organoids are shown 48 h later (scale bar 100 μm). Co-cultures without cells or with GFP− (PDGFRA−) cells produced budding structures, whereas crypts co-cultured with ISEMFs or rBMPs retracted their buds, a process that reversed in the presence of BMPi rNOG or rGREM1. The graph represents fractions of budding (white), unbranched (non-budding, grey), fully spheroidal (stippled), and non-viable (black) organoids after co-culture with the indicated cells or factors. Statistics for budding fractions were determined by one-way ANOVA followed by Dunnett’s posttest. *p <0.05, ****p <0.0001 (n=3–10 independent organoid cultures as indicated above each bar).
B) Representative images of organoid EdU uptake and quantitation (percent EdU+ organoids per condition). Cell replication is markedly reduced in organoids co-cultured with ISEMFs or rBMPs. White arrowheads: organoid buds with EdU+ cells, green arrowheads: live GFPhi ISEMFs, additional examples in Figure S1B. Scale bar 50 μm. Statistics, reported relative to crypt cultures without added cells, were determined by one-way ANOVA followed by Tukey’s multiple comparisons test. ****P <0.0001, ns: not significant.
C) qRT-PCR analysis of organoids after 48 h co-culture (n=3 or 4 biological replicates). BMP- or ISEMF-exposed organoids reduced expression of ISC markers and increased markers of BMP activation and epithelial differentiation. Transcript levels are represented relative to control crypt cultures with no added cells. Statistical comparisons use one-way ANOVA followed by Dunnett’s posttest. ****P <0.0001, ***P <0.001, **P <0.01, *P <0.05.
See also Figure S1.
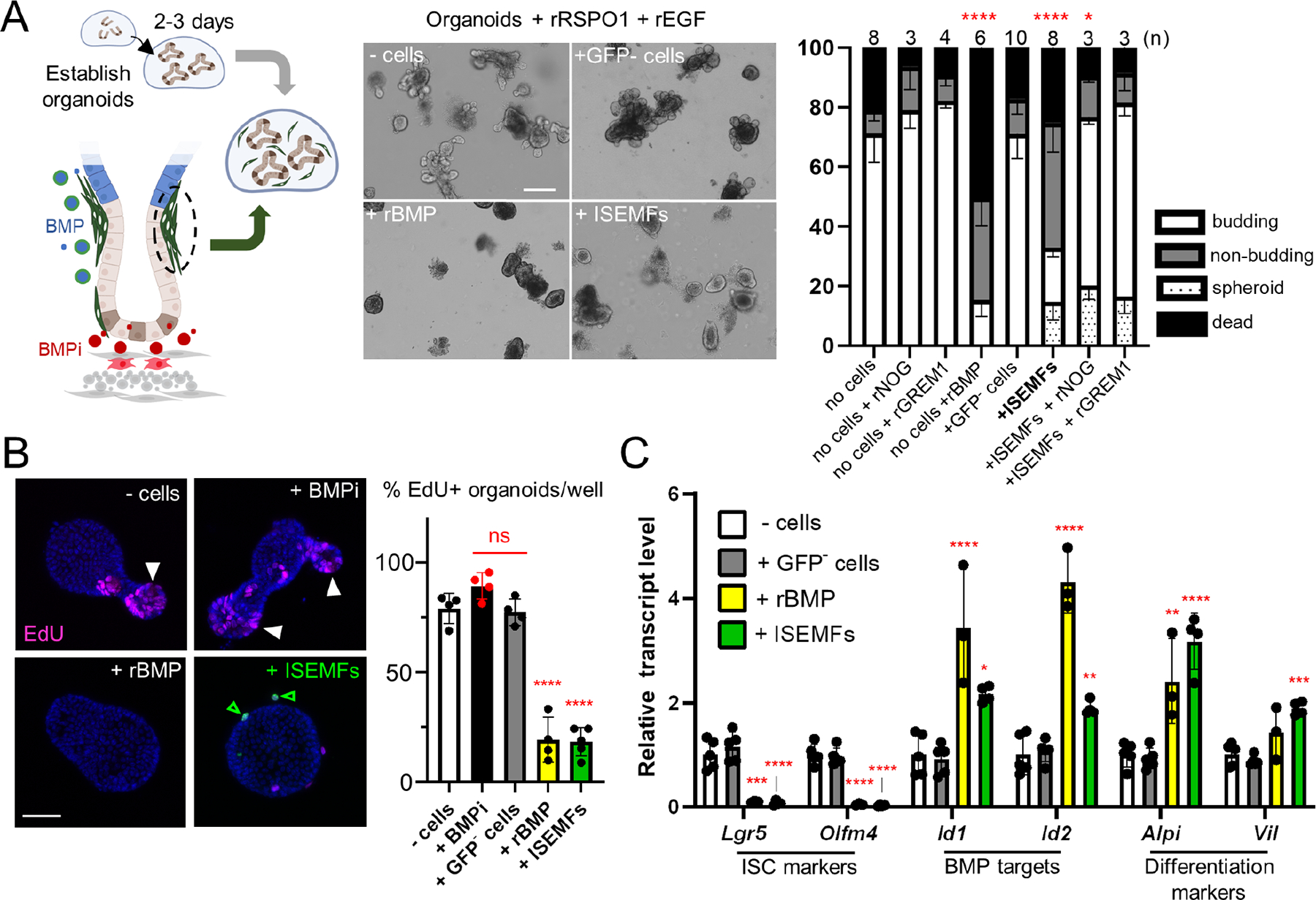
Before crypt formation, newborn mouse ISCs reside in inter-villus troughs9,43 and the organoids they generate differ from adult ISCs.44 A watershed period between P10 and P16 is marked by crypt deepening, emergence of adult ISC properties, and extensive crypt fission.10–15 PDGFRAhi precursors to ISEMFs constitute the leading edges of new villi during fetal life33,45 (Figure S1C) and to ask when adult levels of BMP signaling emerge, we tracked postnatal ISEMF distribution and epithelial pSMAD expression. Through the first week of life, high ISEMF density was evident only at villus tops, with negligible aggregation at the villus base at postnatal day (P) 1 (Figure 2A). Quantified relative to the villus trunk, isthmus aggregation increased progressively through P21 (Figure 2A–B). In line with these findings, epithelial pSMAD at P1 and P5 was confined to the top halves of emerging and enlarging villi, becoming steadily more pronounced at villus bottoms by P14 (Figures 2C and S1D; pSMAD signal is generally stronger in the stroma). ISEMFs express Bmp5 even at birth (Figure S1E) and organoids co-cultured with P14 ISEMFs did not thrive (Figure S1F), indicating that ISEMFs are functional by then. Epithelial expression of the BMP-responsive gene Id1 also increased by P21 (Figure S1G). As mouse46 and chick47 intestinal villus cells are responsive to BMP signaling and express BMP receptors 1a and 2 in embryos and neonates, respectively, new villus responsiveness is an unlikely basis for the cryptward shift in BMP signaling. We suggest, rather, that it reflects increased ISEMF density at the crypt-villus junction (isthmus, Figure 2D).
Figure 2. Postnatal epithelial BMP signaling coincides with isthmus ISEMF aggregation and young trophocytes provide limited support for organoid growth.
A-B) In PdgfraH2B-eGFP mice, GFPhi ISEMFs present on established and emerging (arrowhead) villi on postnatal day (P) 1 concentrate over the next 3 weeks at the crypt-villus isthmus shelf (white brackets). From representative images of proximal SI at the indicated ages (A, scale bars 50 μm) we quantified GFPhi ISEMF cell numbers (B) at the isthmus (white brackets, 5 lowest villus epithelial cells) in relation to villus trunks (red brackets, next 5 cells toward the villus tip); graph displays data from >25 villi/sample (n=3 animals at each age). Statistical differences are reported relative to P1 after two-way ANOVA followed by Tukey’s multiple comparisons test. ns: not significant, *P <0.0001.
C) Representative pSMAD1/5/9 (magenta) immunostaining (green: LAMININ/basement membrane, blue: DAPI) in mouse proximal SI on indicated postnatal days. cr, crypts; scale bars 50 μm; boxed regions are magnified in adjoining images; grey scale and extended images are shown in Figure S1D. pSMAD+ epithelial cell nuclei (filled arrowheads; pSMAD− cells indicated by empty arrowheads) are present at villus tops from birth, while pSMAD+ cells at villus bottoms increase over the ensuing 2 weeks. Graph displays pSMAD+ epithelial cell fractions at the bottoms (blue dots) and tops (grey dots) of >25 villi/sample (n=3 mice at each age). Statistical differences are reported relative to P1 villus bottoms after two-way ANOVA followed by Tukey’s multiple comparisons test. ns: not significant, *P <0.0001.
D) Increased epithelial pSMAD1/5 coincides with increasing isthmus ISEMF density.
E) PDGFRAlo mesenchymal cells include trophocytes (CD81+, red) and CD81− stromal cells (CD81−PRAlo, purple). Co-cultures of P14 (left graph) or adult (right graph) SI crypts with isolated adult (black) or P14 (blue) mesenchymal cells in factor-free medium. In contrast to adult trophocytes, which support robust organoid growth, P14 trophocytes are inactive. Graph depicts organoids per sample relative to control wells (- cells + EGF, NOG, and RSPO1 (ENR); 94.5 ±46.8 P14 organoids/well; 64.5 ±20.03 adult organoids/well; n=6 each). Statistical significance determined by Student’s t-test at **P <0.01, ***P <0.001 (n=3–6 biological replicates).
See also Figure S1.
To ask if CD81+PDGFRAlo trophocytes, a significant BMPi source in adult mice,19,20,22,30 are active in that period, we cultured SI crypts with trophocytes isolated from P14 or adult PdgfraH2B-eGFP mice (Figure S2A). In contrast to adult cells, P14 trophocytes did not support organoid growth from adult or P14 crypts in the absence of extrinsic factors (Figure 2E). Fluorescence and bright-field microscopy confirmed that P14 trophocytes were viable in co-culture and they showed modest activity in the presence of rEGF plus low levels of rBMPi or rRSPO that did not alone support organoid growth (Figure S2B). Lower trophocyte Grem1 expression at P14 than in adults (Figure S2C) may explain this limited ISC support or other stromal cells may provide niche activity prior to weaning and possibly beyond.
Delineation of a multi-component sub-cryptal signaling center
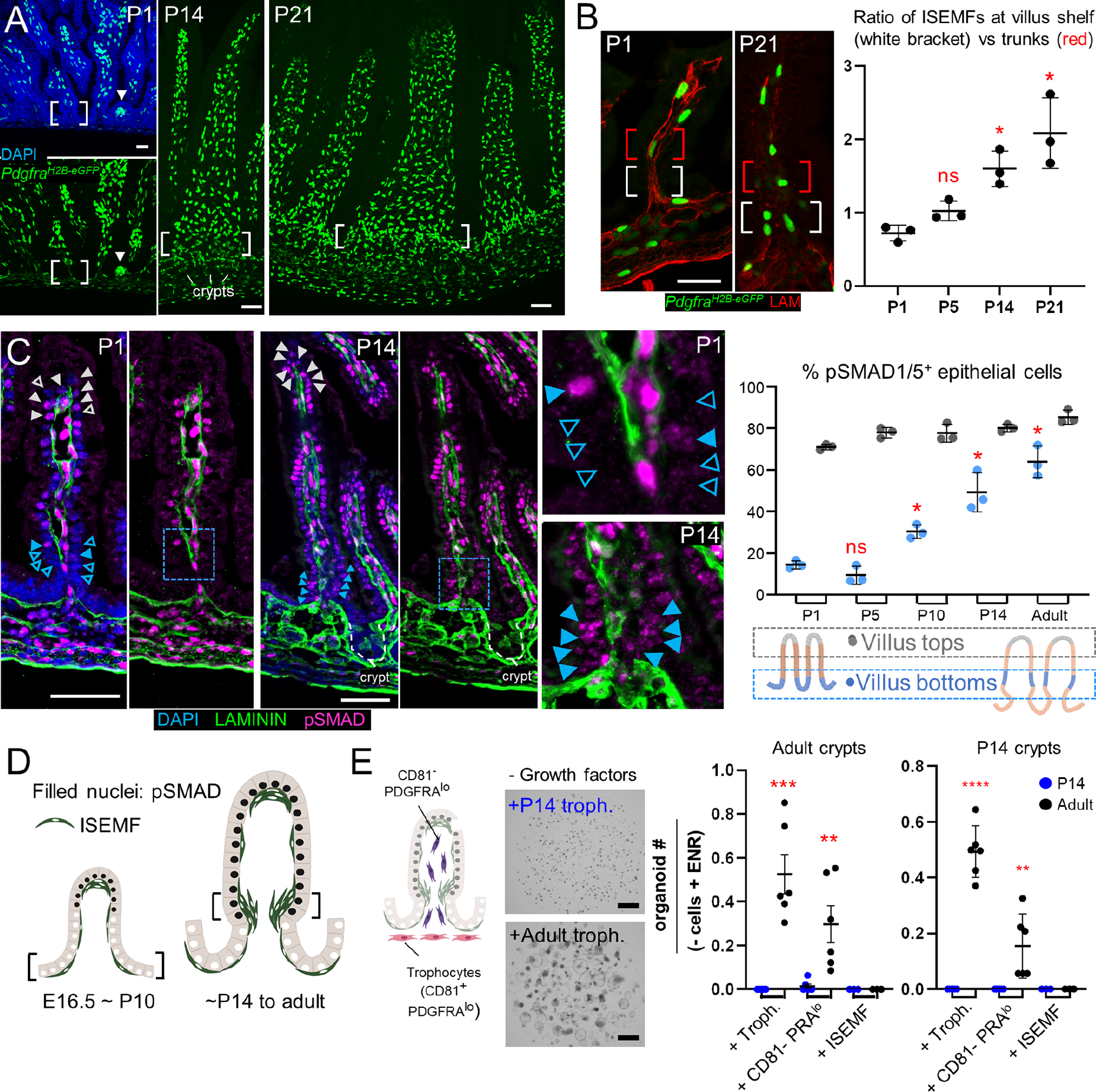
To identify such cells, we extracted SI mesenchyme at various times between P1 and P14 for single-cell (sc) RNA-seq. We profiled >61,000 cells, encompassing unfractionated wild-type mesenchyme and GFP+ cells from PdgfraH2B-eGFP mice (Figure S2D). After excluding replicating cells, leukocytes, and Peyer’s patch-associated follicular cells,48 >51,000 informative cells from all stages included all known adult populations (Figures 3A and S2D–E). One PDGFRA+ population marked by Gata6 and low Wt1 expression likely represents mesothelium-like cavity fibroblasts.49 Notably, dual Grem1 and Rspo3 expression was evident not only in trophocytes but also in diverse Myh11hiActa2hi smooth muscle populations (Figure 3B).
Figure 3. Diverse postnatal intestinal smooth muscle cell populations.
A) Uniform manifold approximation and projection (UMAP) of combined scRNA-seq data from whole mesenchyme and GFP+ cells from PdgfraH2B-eGFP mice, representing 51,084 non-lymphoid (Ptprc-) cells from P1/P2, P4/P5, P9, and P14. ISEMFs, intestinal subepithelial myofibroblasts; LECs, lymphatic endothelial cells; BECs, blood endothelial cells.
B) Overlay of aggregate trophic factor levels (Grem1 and Rspo3, red) on the global UMAP, showing high dual expression in distinct populations, including trophocytes. Aggregate smooth muscle marker expression (Myh11+Acta2, red) on the UMAP plot identifies distinct populations of smooth muscle corresponding to muscularis mucosae (MM), muscularis propria (MP), and pericytes. Interstitial cells of Cajal (ICC) form a distinct cluster (yellow arrow). Right: ACTA2-immunostained P14 proximal SI highlights key anatomic structures: LPM (dashed red outline), MM (dotted blue outline), MP (dashed green outline), and the epithelium (white dashed outline).
Scale bar 50 μm.
C) Top genes that discriminate each indicated smooth muscle population. MP segregates into Nog+Chrdl1+ superficial cells and the classic circumferential and longitudinal subpopulations. Gene names are listed in Table S2. Violin plots below show that superficial MP expresses markedly lower levels of genes for contractile proteins.
D) Relative expression of ICC markers and BMPi genes in each smooth muscle compartment and in ICCs. Superficial MP expresses high levels of Nog and Chrdl1, projected below onto the dashed green box within the UMAP plot from Fig. 3B. Images show RNAscope in situ hybridization (ISH) with the indicated probes (left: magenta dots, right: greyscale, green arrowheads: positive cells, green immunostain: LAMININ). Scale bar 50 μm. The findings place Nog+Chdl+ cells in the superficial MP, above the zone of circumferential fibers.
E) RNAscope ISH for Grem2 at P14 showing strong expression in MM (blue arrowheads) and lower levels in MP (dashed green outline). Top, 2-color fluorescence; bottom, Grem2 ISH in grey. Scale bar 50 μm.
F) Average expression of selected BMPi and Rspo genes across all cell types represented in the postnatal mesenchymal scRNA survey. Factors that support ISCs are largely restricted to sub-cryptal cell populations. Right: schematic representation of anatomic locations of smooth muscle populations and superficial (Sup.) Nog+Chrdl1+ MP cells.
See also Figures S2 and S3.
Intestinal SM includes lamina propria-associated myocytes (LPM) in villus stalks, muscularis mucosae (MM) beneath crypts, and the external muscularis propria (MP), which contains inner circumferential and outer longitudinal layers (Figure 3B, note: pericytes also express high Acta2 and Myh11). Graph-based clustering revealed molecular distinctions among these anatomically defined populations (Figure 3C), with several BMPi and Rspo factors prominent among differentially expressed genes (Table S2). One population that clustered near MP expresses low levels of classic SM markers Acta2, Myh11, and Actg2 (Figure 3C). It overlaps transcriptionally with interstitial cells of Cajal (ICC)50 and with MP but is distinct from either and it uniquely expresses two BMPi, Nog and Chrdl1 (Figures 3B–D and S2F; Table S2). In situ hybridization (ISH) of P14 SI localized these Nog+Chrdl1+ cells to the superficial MP surface, closest to crypts and distinct from deeper Pcp4hi circumferential or Osr1+ longitudinal MP (Figures 3D and S3A–C). Nog expression was previously noted in fetal circumferential MP;33,51 our findings in postnatal mice place this BMPi-expressing cell type in the vicinity of crypt base ISCs.
Although sub-cryptal MM is physically contiguous with villus LPM, the two SM populations are molecularly distinct (Figure 3C). Both MM and LPM express Hhip, as we verified by ISH (Figure S3D), but whereas other muscle cells express Grem1 and LPM lacks any BMPi, the MM uniquely expresses high Grem2 (Figures 3E and S3D–E; Table S2). Combined ISH for Nog and Grem2 distinguished Grem2+ MM from Nog+ cells (Figure S3F, note: Grem2+ cells deep in the MP are likely ICCs). Altogether, this previously unappreciated sub-cryptal SM cell and BMPi diversity reveals that, in addition to trophocytes, postnatal ISCs abut a milieu, including MM and superficial MP, that is rich in key trophic factors (Figure 3F).
Next we examined the adult sub-epithelium, where MM and MP are anatomically distinct (Figure S4A) and a myenteric plexus containing ETV1+ ICCs and PDGFRAhi interstitial cells (PRAICs) lies between longitudinal and circumferential MP.52,53 To better resolve these layers, we generated Etv1Cre(ER-T2);R26RTdTom;PdgfraH2B-eGFP mice (Table S1), where tdTomato marks axons projecting from ETV1+ ICCs54 and GFP marks PDGFRAhi (ISEMFs and PRAICs) and PDGFRAlo cells. Axonal tdTomato+ projections intercalate between circumferential and longitudinal MP, as expected, and demarcate a 1- to 2-cell layer positioned between circumferential MP and PDGFRAlo trophocytes (Figure 4A). Laminin immunostaining combined with Grem1 ISH (which marks various SM populations – Figure 3F) indicated that this thin layer houses Nog+Chdl1+ superficial MP and that PDGFRAlo trophocytes lie between it and MM (Figure 4B). ISH for Grem1, Grem2, and Rspo3 revealed the same arrangement of BMPi-expressing smooth muscle in Laminin-stained adult colon (Figure S4B) and that SI and colonic trophocytes consistently localize between MM and Nog+Chdl1+ superficial MP (Figure S4C–D). In turn, Grem2hi MM lies directly beneath Olfm4hi ISCs at the crypt base (Figure S4E). Thus, in addition to trophocytes, diverse cell types likely contribute to adult ISC niche activity from a layered signaling hub (Figure 4B).
Figure 4: A sub-cryptal signaling hub in adult mouse mesenchyme and conserved mesenchymal populations in postnatal mouse and fetal human SI.
A) Representative whole-mount image from Etv1Cre(ER-T2);R26RTdTom;PdgfraH2B-eGFP adult mouse ileum, highlighting the space beneath crypts (dashed white outlines) demarcated by axonal projections (red) from Etv1+ interstitial cells of Cajal (ICCs). Pdgfralo trophocytes lie between sub-cryptal MM (unstained) and Nog+Chrdl1+ superficial MP (also unstained; neural projections separate these cells from circumferential smooth muscle). GFPhi nuclei far from peri-cryptal ISEMFs are PDGFRA+ interstitial cells72, distinct from Etv1Cre-labeled TdTom+ projections ICCs.
Scale bar 50 μm.
B) Composite Grem1 ISH overlaid on LAMININ-stained PdgfraH2B-eGFP adult mouse duodenum (sections 10 μm apart), showing high Grem1 levels in MM (arrowhead) and superficial MP (between brackets; red: PDGFRA Ab stain; green: PdgfraH2B-eGFP nuclei; blue: DAPI). The sub-cryptal signaling center encompasses muscularis mucosae (MM), trophocytes, and Nog+Chrdl1+ superficial MP.
C) Developmental timing of major intestinal morphogenetic events and UMAP from integrated analysis of postnatal mouse scRNA-seq datasets (blue, Figure 3A) with 42,598 fetal human terminal ileal cells (red) from PCW 8 to PCW 2236. Table on right (continued in Figure S4F) shows relative expression of molecular markers in cells as annotated in each species. Anatomic positions of cells are illustrated on the right. FRC, follicle reticular cell; MFs, myofibroblasts; BECs, blood endothelial cells; LECs, lymphatic endothelial cells; MP, muscularis propria; ISEMF, intestinal subepithelial myofibroblast.
D) UMAP plot from (C) separated by species, revealing heterogeneity of the human S3 pool (trophocytes and their progenitors) and a seeming lack of human Nog+Chdl1+ superficial MP.
E) Correlations between human fetal and mouse postnatal mesenchymal cell populations, depicted with reference to area under the receiver operating characteristic (AUROC) scores.
F) Relative scRNA expression of the indicated BMPi and RSPO genes across mouse postnatal and human fetal mesenchymal cell types.
See also Figures S4 and S5.
Conserved mesenchymal composition in fetal mice and humans
Human crypt morphogenesis occurs in utero and the fetal human gut resembles the adult organ by post coitus week (PCW) 24, after villi first appear at PCW 8–9 and crypts at PCW 11–12 (Figure 4C).55–57 To study relations between human and mouse mesenchymal populations, we integrated our mouse SI survey from P1 to P14 with a dataset of human ileal cells isolated from 8 to 22 PCW.36 We assigned non-immune (PTPRC-), non-proliferating (KI67-), and non-epithelial (EPCAM-) cell clusters using established mouse (our study) and human36–39 molecular markers (Figure 4C). Cells identified in both species included muscularis propria, LPM and MM (classified together), trophocytes, CD81−Pdgfralo stroma, and ISEMFs; in the human study, the latter four cell types are designated, respectively, as “myofibroblasts” and S3, S1, and S2 “fibroblasts,” while follicular reticular cells (FRC) found in Peyer’s patches48 are called S4 (Figures 4C and S4F). Most cell types from both species clustered together (Figure 4D). Nog+ superficial mouse MP was not represented in the human fetal cell isolates, where S3 fibroblasts (corresponding to trophocytes) were more heterogenous than in mice, including a subpopulation of S3 “progenitors.” Unsupervised metaneighbor analysis, which quantifies the degree of molecular identity between cell types,58 confirmed high inter-species concordance (Figure 4E).
As in young mice, fetal human sub-cryptal cells express most ISC-trophic factors, with subtle differences. Human cells express little GREM1, NOG or CHRDL1, the three BMPi that dominate in sub-cryptal mouse cells; instead GREM2 and Follistatin (FST) are the dominant BMPi (Figure 4F). In contrast to high RSPO3 levels in mouse cells, the dominant counterpart in human fetal“myofibroblasts” and trophocytes is RSPO2 and mesothelium expresses more trophic factors in human embryos than in young mice (Figure 4F). In the absence of discrete cells matching mouse superficial MP (Figure 4D), fetal human ICCs express BMPi and RSPO genes (Figure 4F), while cells classified as circumferential MP and “myofibroblasts” express low levels of Nog and Chrdl1 (Figure S4G). Like mouse ISEMFs, fetal human S2 cells robustly express BMP genes, especially BMP4 and BMP5. Other factors pertinent to niche functions include secreted Wnt antagonists in multiple cell types (Figures 4C and S4H). Canonical WNT2B levels are high in human myofibroblasts (corresponding to mouse MM) and various cells express non-canonical WNT4 and WNT5A (Figure S4H). Thus, cell types and their molecular signatures are highly conserved in developing mouse and human SI mesenchyme, with subtle differences in expression of orthologous trophic factor genes.
The adult mouse niche represents a culmination of dynamic postnatal gene activity and de novo MM genesis
Trophic factor levels increase in diverse mouse sub-cryptal cells from birth to P14 (Figure S5A), thereby likely increasing ISC support. Additionally, our scRNA isolations recovered many more MM and LPM cells at P14 than earlier (Figure S5B), mirroring “myofibroblast” appearance after PCW 14 in human development (Figure S5C).36,37,39 Indeed, ACTA2 immunostaining revealed sparse MM in newborn mice, with a modest increase from P5 to P10; a continuous MM structure was present only by P14 (Figure 5A–B). Whereas adult smooth muscle lacks Pdgfra (or GFP in PdgfraH2B-eGFP mice),30 large fractions of MM and LPM cells carried GFP+ nuclei at P10 and P14 (Figures 5C and S5D). Moreover, in our temporal scRNA survey, the 8,030 Pdgfra+ (GFP+) cells from PdgfraH2B-eGFP mice included distinct pools of Acta2+Myh11+ cells corresponding to Hhip+ LPM and Grem2+Rspo3+ MM (Figures 5D and S5E). Together, these findings suggest that MM and LPM derive from resident PDGFRA+ precursors, retaining H2B-eGFP (which is stable59) for some duration. To test this hypothesis, we crossed PdgfraCre(ER-T2),60 and R26RL-S-L-TdTom mice,61 (Table S1) and induced Cre activity soon after compound heterozygotes were born (P0). The following day (P1), both PDGFRAlo stroma and PDGFRAhi ISEMFs were marked with TdTom (Figure S5F), indicating that the label could be followed from these cells into their derivatives. At P10, both MM and LPM expressed TdTom (Figures 5E and S5F). Thus, resident PDGFRAhi or PDGFRAlo stromal cells generate MM and LPM de novo, with MM arising almost exclusively after birth (Figure 5F).
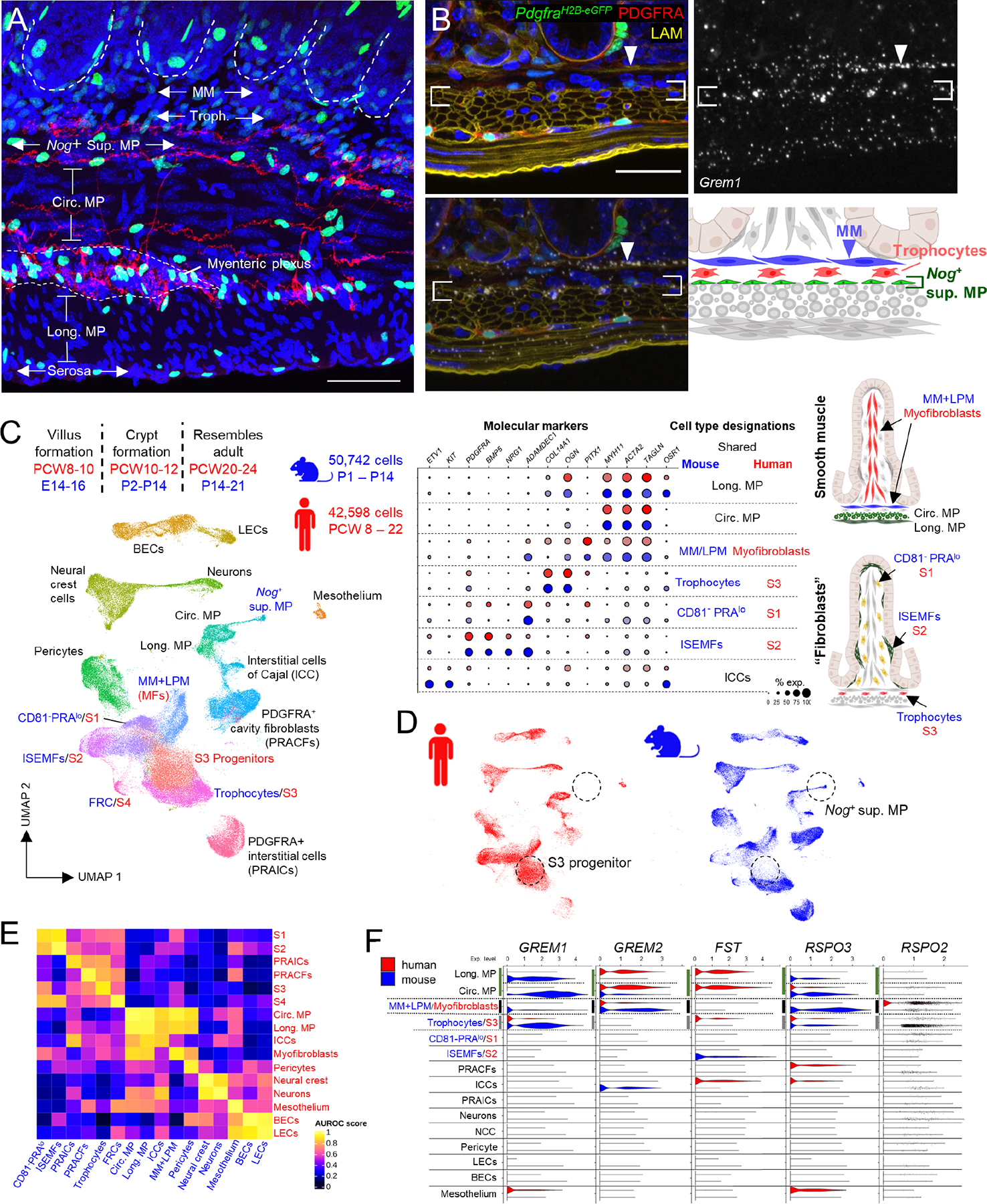
Figure 5. The MM arises postnatally from native PDGFRA+ precursor cells.
A) Representative ACTA2 immunostaining of proximal PdgfraH2B-eGFP SI at P1, P5, and P10. Boxed areas are magnified in the respective insets. While some LPM is present at P1 (arrow), MM arises between P5 and P10 (arrowheads). Scale bar 50 μm..
B) Graph depicting MM cells per mm of tissue at the indicated ages. Statistical differences with respect to P1 were determined by one-way ANOVA followed by Dunnett’s multiple comparisons test. **p <0.0001, *p <0.05, n=3 animals at each age..
C) GFP+ nuclei are evident in LPM (arrow) and MM (arrowheads) in PdgfraH2B-eGFP proximal SI at P14. Areas outlined with white dashes are shown to the right (for MP) or below (for MM) with GFP signals only in greyscale. Scale bar 50 μm..
D) Aggregate Acta2 and Myh11 expression (red) overlaid on a composite UMAP plot of GFP+ cells isolated from PdgfraH2B-eGFP mice at P14. Complementing the images in (C), the isolation shows that PDGFRA+ cells at P14 include a substantial smooth muscle fraction. Inset: Grem2 expression projected on the cells boxed in the composite UMAP plot distinguishes Grem2+ MM from Grem2- LPM..
E) PdgfraCre(ER-T2);R26RTdTom pups were treated with tamoxifen (TAM) at P0 and their intestines were examined with ACTA2 immunostaining at P10, when MM (arrowheads) showed lineage tracing (green) from Pdgfra+ precursors present at birth. Signals are merged in the left and separated in the right images. Scale bar 50 μm. tdtomato+ MM and LPM cell fractions are shown (n: number of cells counted) from >4 mm of proximal SI in each of 3 independent animals..
F) Thus, PDGFRA+ cells present at birth subsequently give rise to MM and LPM smooth muscle cells..
See also Figures S5 and S6.
To determine if adult MM retains key products expressed at P14, we generated Myh11Cre(ER-T2);PdgfraH2B-eGFP;R26RTdTom mice, where SM cells label red and PDGFRA+ cells are green (Figure S6A and Table S1). After excluding the MP, scRNA analysis of TdTom+GFP− cells from the SI yielded few Grem2+ MM cells; nevertheless, integration with previous data from MP-depleted adult SI mesenchyme30 revealed Grem2+Rspo3+ MM distinct from LPM (Figure S6B–C). Similarly, MP-depleted TdTom+GFP− cells from adult colon, where MM is thicker than in the SI (Figure S6D), identified a distinct Grem1+Grem2+Rspo3+ population corresponding to MM, separate from abundant Hhip+ LPM (Figure S6D). Thus, mouse MM sustains expression of ISC-trophic factors into adulthood.
Smooth muscle requirements for ISC support in vivo
Wide, duplicative expression of niche factors in distinctive sub-cryptal cells suggests their likely joint and redundant contributions to an ISC signaling center. Ablation of adult Grem1+ cells triggers epithelial BMP activity and rapid ISC attrition,30 but because many mesenchymal cells express Grem1 (Figure 3F), this finding does not implicate a defined single niche component. To isolate SM contributions to the ISC niche in vivo, we generated Myh11Cre-ER(T2);R26RL-S-L-DTA;PdgfraH2B-eGFP mice (Myh11;DTA, Table S1 – note: Cre activity in Myh11Cre-ER(T2);R26RL-S-L-tDtomato mice mirrors ACTA2 immunostaining, Figure S7A). Adult Myh11;DTA mice became moribund within 2 days of tamoxifen (TAM) exposure, likely owing to global SM attrition and precluding assessment of niche functions. However, administering TAM at P14, by when the MM is well formed, allowed us to follow animals for 1 week. Weight loss and proportionally reduced SI length indicated muscle attrition (Figure S7B). MM was substantially depleted at P16 and P21 (Figure 6A), circumferential MP was modestly reduced at P21 (Figure S7C), PDGFRAhi ISEMFs were preserved, and PDGFRAlo cells were slightly reduced (Figure 7D), likely reflecting loss of PDGFRA+ smooth muscle precursors. Thus, SM ablation was incomplete, as reported with R26RDTA and other Cre drivers62 or because TAM activity is limited in intestinal sub-epithelium63. Nevertheless, MM appeared especially vulnerable and TAM-treated Myh11;DTA pups were informative with respect to SM functions in the pre-weaning ISC niche.
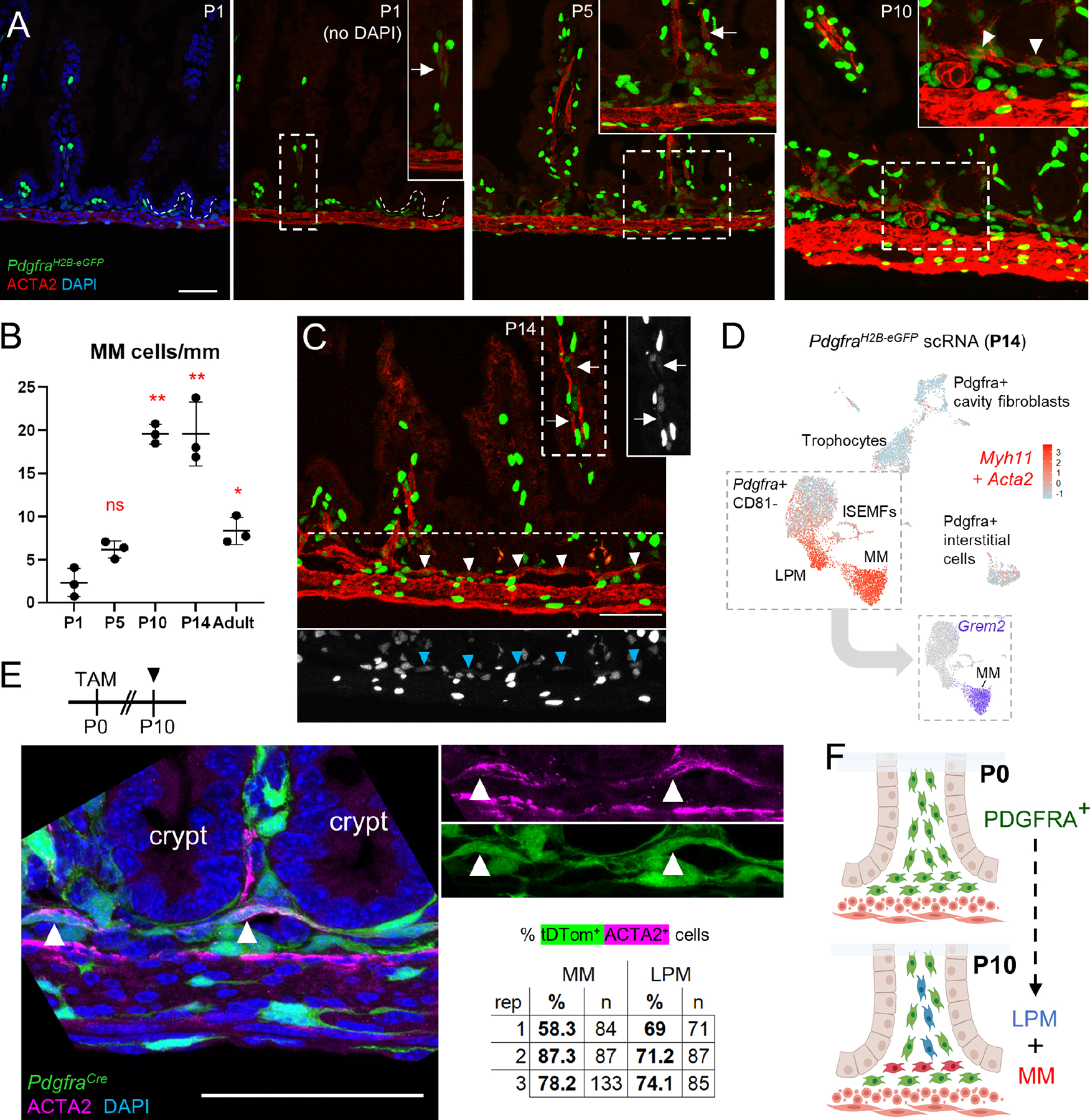
Figure 6. BMP-associated crypt attrition in mice with intestinal SM deficiency.
A) Intestines harvested at P16 and P21 from Myh11Cre(ER-T2);R26RDTA mice treated with TAM at P14 show reduced ACTA2-stained MM. Representative images at P21 are shown and boxed areas are magnified in the insets, where arrowheads point to the MM in the control (Cre− DTA+) specimen and dashed white lines mark the superficial MP border; cr: crypts. Scale bar 50 μm. Right: MM cells quantified from >4 mm tissue in each of 3 or 4 independent animals. Statistical significance was determined using unpaired Student’s t-test. **p <0.01, ***p <0.001.
B) F-actin (phalloidin)- stained epithelial whole-mount images showing considerably fewer bifid crypts (arrowheads) in Myh11Cre(ER-T2);R26RDTA compared to control pups treated with TAM at P14 and examined at P16 (Figure S7A) and P21. Right: Bifid crypts quantified from >100 crypts in each of 3–6 mice from each age. Statistical significance was determined using unpaired Student’s t-test. *p <0.05, ***p <0.001.
C) Crypt base cells expressing RNA for ISC markers Olfm4 (RNAscope ISH signal in magenta) and Axin2 (green) are decreased in TAM-treated Myh11;DTA animals at P21. n=4 mice per condition, >40 crypts per sample, statistics by unpaired Student’s t-test. *p <0.05, ***p <0.001.
D) Increased pSMAD1/5/9 immunostaining (arrowheads in bottom right image) in TAM-treated Myh11Cre(ER-T2);R26RDTA crypts, which lack pSMAD1/5/9 in controls (see also Figure 1A). Boxed areas are magnified in the two right images. pSMAD+ crypts were quantified in >80 crypts from 3 mice for each condition; statistical significance was determined using unpaired Student’s t-test. **p <0.01.
See also Figure S7.
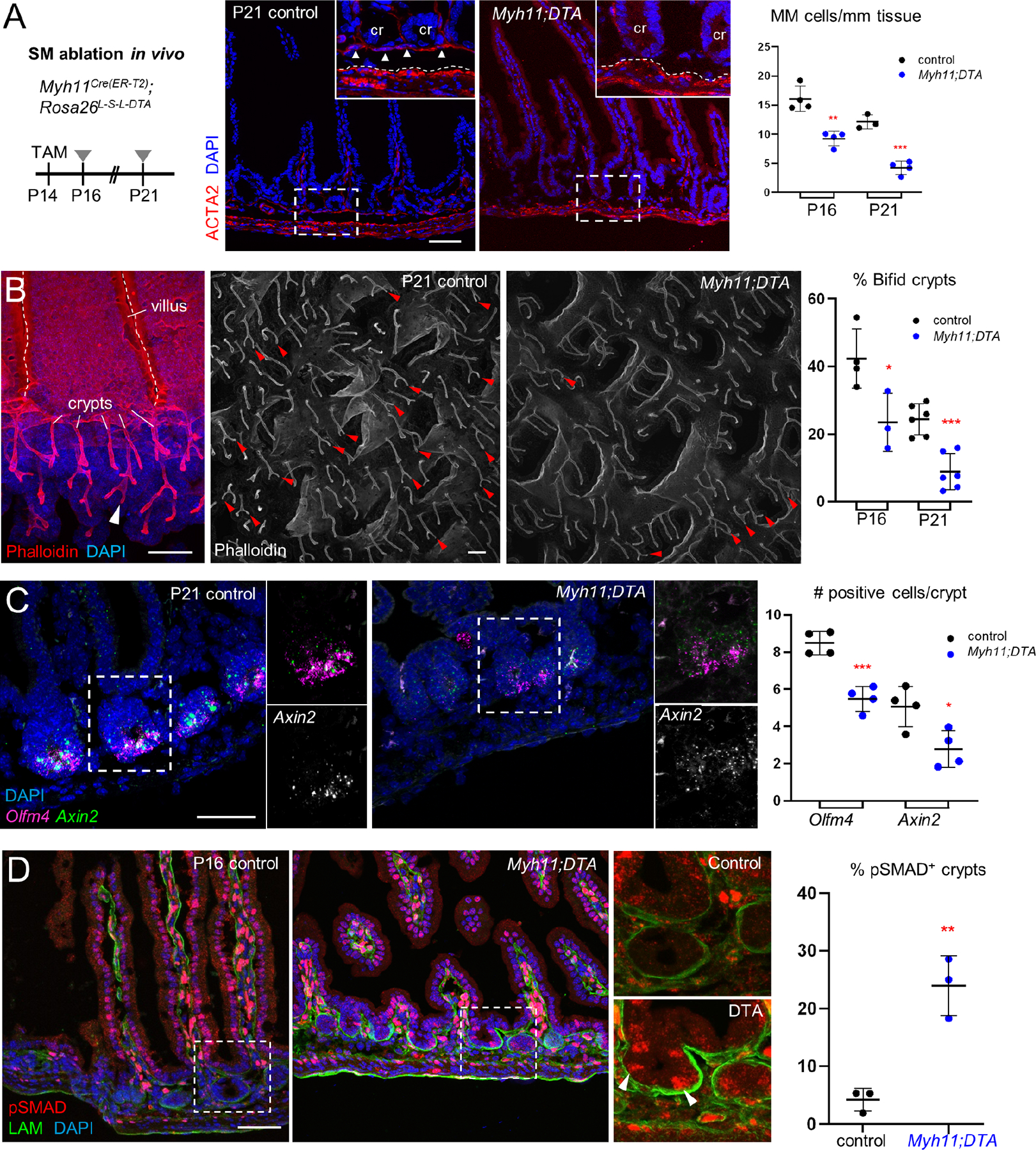
Figure 7. Adult MM and MP complement trophocyte activity in vitro by substituting for BMPi.
A) Adult SI crypts did not generate organoids when co-cultured with unfractionated P14 or adult MP (arrowheads) alone but did so when sub-optimal concentrations of rEGF and rRSPO were added to the co-cultures. In the graph, organoids are represented relative to the numbers generated in ENR medium (no added cells, 66 ±10.4 organoids, n=3). Significance determined by Student’s t-test, ****p <0.0001. -GF, no recombinant trophic factors.
B) Organoids arising from SI crypts after 5 days of culture in a sub-optimal concentration of rRSPO1 (see Methods) and 50 ng/mL or 100 ng/mL of rNOG, rGREM1, and rGREM2, which are roughly equipotent in this assay. n=3 biological replicates. One-way ANOVA with Dunnett’s test shows significance of all conditions to at least p<0.01 compared to control cultures with no added BMPi.
C) After MP removal, MM stripped from PdgfraH2B-eGFP colon retained GFP+ trophocytes on the undersurface (green arrowheads in inset) and flow cytometry separated GFP+ trophocytes from GFP− MM (see Figure S7G–H). Scale bars 50 μm. Right: In adult SI crypt co-cultures, the GFP+ trophocytes supported organoid growth without added factors, while GFP− MM provided support only in the presence of rEGF and rRSPO1 (ER medium). Significance assessed with respect to ENR-only controls (no cells, 31.7 ±5.5 organoids/well, n=5) using Student’s t-test. ***p <0.001, ****p <0.0001.
D) Top: Rspo2 ISH (green) and LAMININ immunostaining (red) in adult SI. Dashed lines demarcate the MM above and MP below. Trophocytes lie between these layers and express Rspo2; a greyscale version of the Rspo2 signal is shown below the color image without LAM; cr: crypts. Bottom: Rspo1 (red) ISH confirms high expression in the serosa as indicated by scRNA-seq (see Figure 3F). Dashed line: superficial MP, scale bars 50 μm.
E) Crypts cultured with as little as 1 ng/mL recombinant murine (m) RSPO2 form organoids with an efficiency that requires at least 10 ng/mL mRSPO3. Scale bar 50 μm. Graph depicts organoid-forming efficiency at 5 days relative to 100 ng/ml rRSPO3 (107.3 ±19.6 organoids/well, n=3). Statistical comparisons by one-way ANOVA followed by Tukey’s posttest, ****P <0.0001.
See also Figure S7.
Crypt cell proliferation, which occurs largely in transit amplifying cells, was intact (Figure S7E) but crypt fission, which peaks in the 3rd week of mouse life,15 was significantly attenuated by P21 and starting as early as P16 (Figures 6B and S7F). ISC markers Olfm4 and Axin2 were also reduced at P21 (Figure 6C). Crypts with pSMAD+ cells were increased ~5-fold (Figure 6D), pointing to BMPi deficiency as a basis for these defects. Thus, SM deficiency compromises ISC function, at least in part from unopposed BMP signaling.
Complementary ISC support from SM and trophocytes
Myh11;DTA mice implicate smooth muscle –but not a discrete cell type– in ISC support and because the above findings do not address the relative contributions of trophocytes and smooth muscle, we assessed smooth muscle niche activity in vitro. A lack of selective surface markers precludes purification of MM or Nog+Chdl1+ cells by flow cytometry. Therefore, first we cultured unfractionated P14 or adult MP with P14 or adult SI crypts. Neither young nor adult whole MP substituted for ENR medium in supporting organoid formation; however, in the presence of low doses of rEGF and rRSPO1 that were alone insufficient for organoid growth, MP co-cultures consistently generated organoids (Figures 7A and S7G). Thus, although MP shows low potency by itself in this assay, it supports organoids when RSPO levels are limiting.
Second, noting that MM is a significant source of Grem2, we found that rGREM2 supported organoid growth at least as potently as rGREM1 (Figure 7B); thus, MM is also well suited to complement trophocyte activity. Indeed, MM isolated as a membrane from adult PdgfraH2B-eGFP colon supported organoid growth from adult SI crypts in the absence of additional factors (Figure S7H). Because this effect could reflect trophocyte contamination, we digested the intact membrane, sorted GFP− (MM) from GFP+ (trophocyte) cells by flow cytometry (Figure 7C), and verified that GFP+ cells are CD81+ trophocytes (Figure S7I). Cell yields allowed us to co-culture only 10,000 cells from each fraction with crypts in small Matrigel droplets, with little flexibility in factor supplementation; nevertheless, the trophocyte fraction promoted organoid growth in the absence of soluble factors. MM was inert by itself, but in the presence of small amounts of rEGF and rRSPO1, which approached trophocyte activity in this assay, its activity consistently exceeded that of trophocytes alone (Figure 7C). Thus, MP and MM each reveal intrinsic and complementary niche activity when factor levels are sub-optimal. Organoid growth in the absence of rNOG implicates sub-cryptal SM as a functional source of BMPi.
Finally, because adult trophocytes alone support ISCs robustly in vitro30 (see Figs. 2E and 7C), these cells must harbor some distinctive activity not limited to BMPi. Several cell types express Rspo3 and serosal cells express Rspo1 (Rspo4 is absent from any cell type), but only trophocytes express Rspo2 (Figures 3F and 4F), a finding we verified by ISH (Figure 7D). RSPO3 is superior to RSPO1 in sustaining organoids20 and ISC attrition in vivo requires both RSPO2 and RSPO3 antibodies,64 but RSPO2 activity has not been examined in isolation. We found that adult mouse SI crypts required 10 ng/mL murine rRSPO3 to generate organoids at top efficiency and 2.5 ng/mL was ineffective, while 1 ng/mL rRSPO2 achieved near-peak efficiency (Figure 7E). Human RSPO2, the dominant homolog in fetal myofibroblasts (Figure 4F), is also more active than RSPO1 or RSPO3 in supporting mouse organoids (Figure S7J). Thus, redundant BMPi coupled with unique expression of RSPO2 may explain why trophocytes can support organoid growth alone.
Discussion
Immature ISC precursors located in inter-villus epithelium first generate shallow troughs, which deepen into the earliest crypts and, before mouse weaning, undergo extensive fission to expand crypt and epithelial mass.9,10,15,16,43 Classical11,13,15,17 and recent10 studies highlight the P10-P15 interval as a watershed, when crypts and ISCs acquire adult forms. The present study integrates mesenchymal scRNA profiles with high-resolution microscopy and functional assessment of the emerging ISC niche during this watershed period. Mesenchymal niche cells and their products are substantively similar between postnatal (P1 to P14) mice and fetal (PCW 12 to PCW 22) humans. In both species, each putative niche component expresses at least one BMPi and one RSPO gene, but mouse and human SM cells and trophocytes differ in expression of some orthologs; RSPO2, for example, is restricted to mouse trophocytes and to human myofibroblasts that correspond to mouse MM.39 These similarities and differences reinforce our conclusion that the functional ISC niche is a finely layered structure consisting of distinct cell populations that are strategically positioned near the crypt base and elaborate multiple, partially redundant trophic factors.
Our findings add developmental and fine spatial context to prior work showing that BMPi drive ISC properties,41,65–67 that BMP signaling restrains ISC pool size3 and crypt fission,68 and that human colonic mucosa is polarized for BMP and BMPi expression.21 ISEMF aggregation at the villus base coincides with BMP signaling in adjacent epithelium and, likely to counter that effect, sub-epithelial niche elements develop in parallel. MM, a sub-cryptal niche component not previously well appreciated, differs from contiguous and perpendicular LPM fibers in expressing Rspo3 and BMPi, and arises after birth from resident PDGFRAhi or PDGFRAlo cells. RNA levels of various trophic factors also increase after birth in trophocytes and in Acta2loMyh11lo cells that uniquely express the BMPi Nog and Chrdl1. The latter cells reside between circumferential MP fibers and trophocytes, and share transcriptional similarities with ICC and smooth muscle, but are distinct from all these cell types. Apparent absence of a matching population in human fetal cell isolates may reflect a species difference in niche cell composition, their attrition during the isolation protocol or, most likely, their bioinformatic clustering with smooth muscle or ICCs. Of note, ICCs arise from the resident mesenchyme69,70 and some authors suggest that ICC and selected SM cells derive from a common Kit+ precursor.71 The sum of these observations lends motivation to investigate the relationship and ontogeny of ICCs and Nog+Chrdl1+ superficial MP.
The layered niche of mouse RSPO- and BMPi-expressing cells achieves adult form and function between birth and weaning. Given that P14 trophocytes fail to support organoids without added BMPi, other cells with overlapping expression of trophic factors may first help drive postnatal crypt expansion and, subsequently, contribute toward lifelong ISC support. De novo emergence of MM may, for example, counteract concomitant ISEMF aggregation and enhanced BMP signaling at the villus base in the third week of mouse life. Our findings collectively imply that niche cell populations are redundant with one another and that some may be dispensable in resting tissue. However, trophocytes do support organoid growth in the absence of any recombinant factor or other non-epithelial cells and their unique expression of Rspo2, the most potent RSPO, likely accounts for some portion of this robust activity. In the future, molecular markers that allow better cell fractionation and precise in vivo perturbation will complete the emerging understanding of individual niche elements at rest and under conditions of high ISC attrition or stress, such as infectious enteritis and inflammatory bowel disease. The cell types and molecular markers identified in this study will aid in those endeavors.
Limitations of the study
Establishing the specific contribution and requirement of niche components is challenging because few surface markers allow isolation of a pure cell population and available Cre-driver mouse strains typically target multiple cell types. For example, previous ablation of Grem1+ cells affected ISCs profoundly30 but it is uncertain if that effect reflects loss of trophocytes, SM, or both. In the present study, depletion of Myh11+ cells also elicited ISC dysfunction, as reflected in markedly reduced crypt fission. However, Myh11Cre marks all SM, ablation was unavoidably incomplete, and our findings in Myh11Cre;DTA mice could reflect indirect effects of SM loss on other niche cells rather than direct consequences on ISCs. Our delineation of niche functions in crypt co-cultures mitigates some of these limitations, revealing that MM and MP independently provide BMPi but likely insufficient RSPO for organoid growth. These in vitro findings must be interpreted cautiously, as gene expression and cell behaviors likely drift in culture.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ramesh Shivdasani (ramesh_shivdasani@dfci.harvard.edu).
Materials availability
This study did not generate new unique reagents/materials.
Data and Code Availability
Data are deposited in the Gene Expression Omnibus (GSE184158). No new software was developed for this study. Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals.
PdgfraH2B-eGFP (Jackson Laboratories (JAX) strain 007669)42, Rosa26RLSL-TdTomato (JAX strain 007909), Myh11Cre(ER-T2) (JAX strain 019079), PdgfraCre(ER-T2) (JAX strain 032770), Etv1Cre(ER-T2) (JAX strain 013048) and Rosa26RLSL-DTA (JAX strain 009669) mouse lines were purchased from Jackson Laboratories (Table S1). Adult mice were more than 8 weeks of age at the time of treatments or cell isolations. Animals were housed in a specific pathogen-free barrier facility, maintained on a 12-hour light/dark cycle, and had ad libitum access to standard chow and water. All experiments used mice of both sexes and littermates as controls. All animal procedures and experiments were approved and monitored by the Animal Care and Use Committee at the Dana-Farber Cancer Institute.
METHOD DETAILS
Mouse treatments.
Postnatal PdgfraCre(ER-T2);R26RLSL-TdTom and Myh11Cre(ERT2);Rosa26 LSL-DTA mice received 1 dose of 4-OH tamoxifen (Sigma-Aldrich, 1 mg per 25 g body weight) by gastric gavage at P1 or intra-peritoneal (IP) injection at P14. Myh11Cre(ER-T2);R26RLSL-TdTom;PdgfraH2B-eGFP adults received 4-OH tamoxifen (1 mg) by IP injection on 2 consecutive days to allow recombination at LoxP sites and were harvested at the indicated times, usually 5 days later. BrDU (10 μg/g body weight) was administered by IP injection 1 h before euthanasia. Postnatal mice were harvested at the time points indicated in the figure legends.
Immunohistochemistry and quantitation.
Whole-mount tissue immunohistochemistry was performed as described.30,76 Briefly, proximal small intestines were harvested, pinned onto agarose plates, and fixed overnight in 4% paraformaldehyde (PFA); in this and all subsequent steps, the tissue was rocked gently. After rinsing in phosphate-buffered saline (PBS), the tissue was placed in 10%, then 20% sucrose over the course of 1 day, followed by blocking buffer (PBS containing 0.125% bovine serum albumin, 0.003% Triton X-100, 0.05% donkey serum, and 0.0005% NaN3) for 6 h and overnight in blocking buffer containing 4′,6-diamidino-2-phenylindole (DAPI). Tissue was rinsed with PBS, cut into 1-mm fragments, placed on glass slides with spacers (Grace Bio-Labs, 654002), and cleared using FocusClear (CelExplorer Labs, FC-101) for 30 min, before applying VectaShield mounting medium (Vector Laboratories) and a coverslip. To generate representative and comprehensive anatomic resolution, images were taken from at least 3 independent animals.
Proximal SI epithelial immunochemistry was performed as described.10 Fresh tissue was incubated in 5 mM EDTA in Hank’s Balanced Salt Solution (HBSS) at room temperature, rocked gently for 5 min, and washed briefly in PBS before separating the epithelium gently from underlying mesenchyme. Tissue was fixed in 4% PFA at 4oC overnight, with subsequent PBS washing and incubation with DAPI to stain nuclei and Phalloidin (Invitrogen, A12381) to visualize F-actin. Crypt bifurcation was quantified in 3D-rendered images of whole-mount phalloidin stained intestines, reported as a fraction of >100 intact crypts.
Routine immunohistochemistry was performed on tissues fixed as described above and placed in OCT compound (Tissue-Tek, 4583). 7 μm sections were prepared using a Leica CM3050 cryostat. The following antibodies (Ab, all at 1:1000 dilution unless indicated) were used: Laminin (Sigma, L9393); GFP (Abcam, ab6662); CD31 (BD Biosciences, 557355); bromodeoxyuridine (BrDU, Life Technologies, B23151); Alpha-smooth muscle actin (ACTA2; Abcam, ab5694); PDGFRA (R&D Systems, AF1062, 1:100); and Alexa Fluor-conjugated secondary goat anti-rat, goat anti-rabbit, or donkey anti-goat IgG (Invitrogen, A11081, A21071, A11058). Signals were amplified using biotin/streptavidin HRP-conjugated secondary Ab (Jackson Immuno, 111–065-003 and 016–030-084) and detected using Tyramide Signal Amplification Plus kit (Akoya Biosciences, NEL744001KT, 1:100). Images were taken using a Leica SP5X laser scanning confocal microscope and further processed using ImageJ Fiji software.75
pSMAD1/5/9 was immunostained as described77. Briefly, antigens were retrieved by boiling slides in citrate buffer (pH 6.0) for 1 min and pSMAD1/5 Ab (Cell Signaling, 41D10) was added overnight at 4oC. pSMAD1/5+ crypts were quantified on images acquired and processed as above. Epithelial compartments were delineated with respect to LAM Ab-stained basement membrane, and quantified by representing the lowest and highest 5 villus epithelial cells (10 cells per villus) as the fraction of pSMAD1/5+ cells in >25 villi per sample (Figure 2C) or crypts carrying ≥1 pSMAD1/5+ cell as a fraction of >80 crypts per sample (Figure 6D). To quantify PdgfraH2B-eGFP ISEMFs, 25 μm sections were co-stained with LAM Ab to demarcate the epithelium. Mesenchymal GFPhi cells abutting the bottom 5 villus epithelial cells were counted and are reported with respect to GFPhi cells abutting the next 5 epithelial cells along the villus trunk (Figure 2B). More than 25 villi were counted per sample.
To quantify SM populations (Figures 6A and S7C), ACTA2+ cells in the MM and in the circular and longitudinal layers of the MP were counted per length of proximal SI examined (average 5 mm/sample). Co-labeled ACTA2+Pdgfralo cells are reported as a ratio of the total number of ACTA2+ MM and LPM (Figures 5C and S5D). PdgfraCre(ER-T2);Rosa26DTA intestines exposed to TAM at P0 were harvested at P10 and stained with ACTA2 Ab to quantify various SM compartments. For quantitation of Pdgfralo cells in Myh11Cre;PdgfraH2B-eGFP;DTA experiments (Figure S7D), cells found in the submucosa, between crypts and MP, are reported as numbers/length of intestine (>4mm/sample). BrdU+ cells/crypt are reported as numbers per crypt (≥25 crypts/sample).
In situ RNA hybridization and quantitation.
mRNAs were localized by the RNAscope (Advanced Cell Diagnostics) method78 on intestines collected from at least 3 different animals per treatment. Probe sets were designed by Advanced Cell Diagnostics for Id1, Bmp5, Chrdl1, Grem1, Grem2, Hhip, Noggin, Olfm4, Axin2, Osr1, Pcp4, Rspo1, Rspo2, and Rspo3. After hybridization according to the manufacturer’s protocols, tissue sections were washed for 5 min in PBS containing 0.1% Tween-20, blocked for 1 h at room temperature in PBS containing 5% normal goat serum, and exposed overnight at 4oC to Laminin (Sigma, L9393, 1:1,000) or GFP (Abcam, ab6556, 1:100) Ab. After multiple 5-min washes in PBS and 90-min incubation with AlexaFluor-conjugated secondary Ab as above (Invitrogen) at room temperature, DAPI was applied and slides were mounted according to the RNAscope protocol. Images were taken using a Leica SP5X laser scanning confocal or a Leica Thunder Imager microscope and processed using ImageJ Fiji software.75 For quantitation in SM layers delineated by laminin (Figure S3B and S3E, 2 samples), each cell with at least 1 fluorescent ISH dot was counted as one and reported as a fraction of all SM cells present in the respective sub-compartment. Every cell with at least one Olfm4+ or Axin2+ ISH dot was reported per crypt (Figure 6C, >40 crypts per sample).
Schematic illustrations
were generated with BioRender.
Mesenchymal cell isolation and flow cytometry.
Mesenchymal cells were isolated from the pooled proximal halves of the small intestine from 3–6 wildtype or PdgfraH2B-eGFP pups. Timepoints harvested from wildtype tissue include P1, P4, P5, P9, and P14, and from GFP+ isolated PdgfraH2B-eGFP tissue include P2, P5, and P14. After manual stripping of external muscles (MP) and serosa, whole adult Myh11Cre(ER-T2);R26RL-L-TdTom; PdgfraH2B-eGFP SI or colon was processed as described.30 Epithelium was denuded by shaking the tissue for 20 min at 37oC in pre-warmed HBSS (Life Technologies) containing 10 mM EDTA. The remaining tissue was rinsed with HBSS, minced using a scalpel, and digested with gentle rocking for 1 h at 37oC in 3 mg/mL collagenase II (Worthington, LS004176) diluted in HBSS containing 5% fetal bovine serum (FBS). Extracted cells were centrifuged at 300 g for 5 min, washed with FACS buffer (PBS containing 0.1% BSA), and leukocytes were depleted in ACK Lysis buffer (Gibco) for 3 min. Washed cells were suspended in FACS buffer and, to deplete epithelial and immune populations, stained with conjugated EPCAM (BioLegends, 118214, 1:100) and CD45 (eBiosciences, 17–0451-82, 1:100) Ab for 20 min at 4°C. Cells were sorted on a FACSAria IIII flow cytometer, with gating against DAPI (BD Pharmingen) to identify live cells. Graphs of isolated cell fractions were generated using FlowJo software v10.
scRNA-seq library preparation, sequencing, alignment, quality control, and data analysis.
5,000 to 10,000 cells isolated by flow cytometry were loaded onto a Chromium Controller (10X Genomics), followed by library preparation according to the manufacturer’s recommendations (Single Cell 3’ V3 assay) and sequencing on a HiSeq4000 instrument (Illumina). Libraries were de-multiplexed, aligned to the mm10 mouse transcriptome, and unique molecular identifiers (UMIs) were counted using Cell Ranger (10X Genomics) v3.1.1. Data were analyzed using the Seurat package v4.0.3 in R.73 Cells with ≥1,000 and ≤ 4,000 detected genes, >2,500 and <15,000 total transcripts, and <10% mitochondrial transcripts were retained. Merged datasets utilized the “merge” function and data were normalized and log-transformed using the “SCTransform” function in Seurat, regressing out mitochondrial read fractions and other confounding variables. Datasets integrated after normalization used the “integrate” function in Seurat. Differential markers were identified in clusters using the “FindAllMarkers” function in Seurat, with parameters: min.pct 0.25 and logfc.threshold 0.25. To identify cell types in each collection, the data were queried for known mesenchymal cell-specific genes.30,79 UMAP plots for gene expression were generated using the “FeaturePlot” function.
For merged datasets (Figures 3A and S2D), the top 15 principal components were selected using the “FindNeighbors” function, followed by identifying clusters using the “FindClusters” (resolution: 2) function in Seurat. The “RunUMAP” function was used to reduce the top 15 principal components using the “uwot” method. Based on marker expression, clusters were merged to compile 20 mesenchymal cell populations; among these, we removed Mki67+ (proliferating), Ptprc+ (leukocytes), and Ccl19+ (follicle reticular, FRC) cells (Figure S2D) to generate the final merged dataset (Figure 3A). To generate correlation heatmaps (Figure 3C) and gene lists (Table S2), we separated SM and ICC cell types using the “FindAllMarkers” function in Seurat and plotted the top 10 and top 50 marker genes for each. Genes differentially expressed between P14 and P1/P2 (Figure S5A) were identified for each cell type using the “FindMarkers” function and dot plots depicting log2 fold-differences were generated using code based on the “DotPlot” function within the ggplot2 package in R. Integrated analysis (Figure S6C) included published adult scRNA-seq data30 (Gene Expression Omnibus, GSE130681).
Mouse-human cell comparisons.
We compared fetal human terminal Ileum data (Fawkner-Corbett et al, 2020) with our mouse data (Figure 3A). To integrate scRNA-seq data from the two species, we first used biomaRt74 to convert mouse gene names to their human orthologs (keeping only the ~15,000 genes with one-to-one mapping), then integrated datasets using the canonical correlation analysis-based workflow implemented in Seurat v4.80 Human cell annotations were generated by using marker genes specified in Table S1 of the Fawkner-Corbett study. Cross-species comparisons within and between cell types were performed using MetaNeighbor,58 which constructs a cell-cell correlation network based on expression of highly variable genes and computes an AUROC metric to characterize the similarity between groups of cells. Quantitation of myofibroblasts across human fetal development (Figure S5C) was generated from analysis of small intestinal mesenchymal data from GutAtlas.com.37
In vitro co-cultures, imaging, quantitation, and analysis.
Unfractionated mesenchyme extracted from P14 or adult PdgfraH2B-eGFP mice was plated on non-pyrogenic, gas plasma surface-treated polystyrene tissue culture plates (Falcon) in Dulbecco’s Modified Eagle and F12 media (Gibco, 12634–010) supplemented with penicillin, streptomycin, Glutamax, HEPES buffer, and 10% FBS (Basal media + FBS, see Ref. 30 for extraction methods, Figure S2A). Using forceps, we carefully peeled off SI muscularis propria (MP), and in the colon, the muscularis mucosae from PdgfraH2B-eGFP mice. SM was disaggregated for 10 min in HBSS containing 5%FBS and 3 mg/mL collagenase II, then plated as described above. The medium was replaced 24 h after plating; 2 to 3 days later, cells were removed using 0.25% Trypsin/EDTA (Corning), washed in FACS buffer, and GFPhi (ISEMFs), CD81+Pdgfralo (trophocytes), CD81−Pdgfralo stroma, and GFP− cell fractions were harvested by flow cytometry (Figure S2A) as described previously30 as unfractionated mesenchyme (Figure 1A) or SM populations (Figure 7C). MP was disaggregated using 0.25% trypsin/EDTA for 5 min, washed with PBS/0.1%BSA, and placed directly into co-cultures with isolated crypts.
For organoid co-cultures (Figures 1A and S1F), crypts were plated for 2 to 3 days in complete basal media supplemented with N2, B27 and N-acetylcysteine as described,81 EGF (Thermo Fisher, 50 ng/mL), RSPO1 (10% culture supernatant from 293T-HA-RspoI-Fc cells), and rNOG (Peprotech, 100 ng/mL), then removed from Matrigel in Cell Recovery Solution (Corning) for 10 min at 4°C and recovered by centrifugation at 200 g for 15 min. 50–100 crypts were replated in 20 μl Matrigel drops in 24-well tissue culture plates, together with 2×104 mesenchymal cells harvested, cultured, and purified as described above. Complete Basal Media was replaced containing EGF, RSPO1 (10% culture supernatant from 293T-HA-RspoI-Fc cells), and rNOG, rGREM1 (Thermo Fisher, 100 ng/ml), or BMP2 (Peprotech #120–02, 50 ng/mL) and BMP7 (R&D#5666-BP, 50 ng/mL). Bright field and fluorescent organoid images were captured using an Olympus CKX53 or Nikon Eclipse T/2 microscope, respectively. Organoid features were scored from 3 or 4 experiments with 2 or 3 technical replicates per group per experiment. RNA from co-cultured organoids after 48 hrs (Fig2) or from FACS-sorted mesenchymal populations prior to co-culture (Figure S2C) was harvested and qRT-PCR was performed on 3 or 4 replicates from each treatment group; mRNA expression values are represented relative to Gapdh (2−ΔCT) and relative to organoids cultured without additional cells. To quantify EdU uptake (Figure 1B), organoids were treated with 10 μM EdU working solution and harvested 2 h later, followed by EdU immunohistochemistry as described (Invitrogen #C10640). Organoids were imaged using a Leica Thunder Imager and scored based on appearance of EdU+ aggregates or clusters found in budding regions of organoids (n=3–5 replicates per group, >20 organoids scored per replicate).
For culture experiments with BMPi (Figure 7B), ~100 isolated crypts were plated in Matrigel drops in media containing a sub-optimal concentration of RSPO1 (0.5% culture supernatant from 293T-HA-Rspo1-Fc cells), EGF (50 ng/mL), and the indicated concentrations of rNOG, rGREM1, or rGREM2 (R&D#2069-PR). For culture experiments with RSPOs (Figures 7E and S7J), ~100 isolated crypts were plated in Matrigel drops in basal condition media containing rNOG (100 ng/mL), EGF (50 ng/mL), and the indicated concentrations of mRSPO2, mRSPO3 (R&D, 6946-RS and 4120-RS), hRSPO1, hRSPO2, or hRSPO3 (R&D, 4645-RS, 3266-RS, and 3500-RS). Organoid structures were counted with a Nikon Eclipse TS100 microscope 5 days after plating. Three biological replicates are reported per condition, each including 3 technical replicates. Figure S7H was compiled as a composite using Microsoft 365 ProPlus Office PowerPoint.
Quantification and statistical analysis.
Statistical analyses were performed in Prism software package v7.03 (GraphPad). Replicate numbers, statistical methods, and P values are given in the respective figure legends. No sample size estimations and no blinding were performed.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER | |
|---|---|---|---|
| Antibodies | |||
| CD31 | BD Pharmingen | 557355; RRID: AB_396660 | |
| pSMAD1/5/9 | Cell Signaling | 41D10; RRID:AB_491015 | |
| Laminin | Sigma | L9393; RRID: AB_477163 | |
| PDGFRA | R&D Systems | AF1062; RRID: AB_2236897 | |
| Alpha-smooth muscle actin (ACTA2) | Abcam | 5694; RRID: AB_2223021 | |
| CD81 | Invitrogen | 13–0811-81; RRID: AB 466514 | |
| GFP | Abcam | 6556/6662; RRIDs: AB 305564 and AB 305635 | |
| Lysozyme | Dako | A0099; RRID: AB_2341230 | |
| CD45 | eBiosciences | 17–0451-82; RRID:AB 469392 | |
| EPCAM | BioLegend | 118214; RRID:AB 1134102 | |
| Chemicals, Peptides, and Recombinant Proteins | |||
| 0.25%Trypsin-EDTA | Gibco | 25200–056 | |
| Collagenase II | Worthington | LS004176 | |
| RSPONDIN3 (HUMAN) | R&D Systems | 3500-RS | |
| RSPONDIN2 (HUMAN) | R&D Systems | 3266-RS | |
| RSPONDIN3 | R&D Systems | 4120-RS | |
| RSPONDIN2 | R&D Systems | 6946-RS | |
| GREM2 | R&D Systems | 2069-PR | |
| BMP2 | Peprotech | 120–02 | |
| BMP7 | R&D Systems | 5666-BP | |
| NOGGIN | Peprotech | 120–10C | |
| RSPONDIN1 (HUMAN) | R&D Systems | 4645-RS | |
| EGF | ThermoFisher | PHG0311 | |
| GREMLIN1 | ThermoFisher | 956GR050 | |
| Matrigel | Corning | 356231 | |
| Diphtheria toxin | Enzo Life Sciences | BML-G135–0001 | |
| 4-OH Tamoxifen | Sigma | T5648 | |
| DMEM/F12 tissue culture medium | Gibco | 12634–010 | |
| Fetal bovine serum | Corning | 35–010-CV | |
| Penicillin/Streptomycin | Life Technologies | 15140163 | |
| Glutamax | Gibco | 35050–661 | |
| N-2 Supplement (100X) | Life Technologies | 17502001 | |
| B-27 Supplement (50X) | Life Technologies | 17504001 | |
| NICOTINAMIDE | Sigma | 72340 | |
| N-Acetylcysteine | Sigma | A9165 | |
| Normal goat serum | Cell Signaling | 5425S | |
| DAPI | BD Pharmingen | 5649097 | |
| Dulbecco-s modified Eagle medium (DMEM) | Corning | 17–205-CV | |
| Hanks’ balanced salt solution (HBSS, 10X) | Gibco | 14065–056 | |
| Phosphate buffered saline (PBS, 10X) | Corning | 46–013-CM | |
| AlexaFluor goat 546 anti-rat IgG | Invitrogen | A11081 | |
| AlexaFluor goat 633 anti-rabbit IgG | Invitrogen | A21071 | |
| AlexaFluor 594 donkey anti-goat IgG | Invitrogen | A11058 | |
| OCT compound | Tissue-Tek | 4583 | |
| Biotin | Invitrogen | 17–4317-82 | |
| Bovine serum albumin | Sigma | A9647 | |
| Paraformaldehyde | EMS | 15714-S | |
| Picric acid | Fluka | 80456 | |
| FocusClear | CElExplorer | FC-101 | |
| Triton-X 100 | Sigma | T8787 | |
| Sodium azide | Sigma | S2002 | |
| HEPES | Sigma | H3375 | |
| VectaShield mounting medium | Vector Laboratories | H-1000 | |
| 5-bromo-2-deoxyuridine | Life Technologies | B23151 | |
| Power SYBR | Thermofisher | 4367659 | |
| Phalloidin | Invitrogen | A12381 | |
| Critical Commercial Assays | |||
| RNAscope Multiplex Fluorescent Reagent Kit v2 | Advanced Cell Diagnostics (ACD) | 323100 | |
| Single Cell 3’ V3.1 assay | 10X Genomics | PN-1000147 | |
| Deposited Data | |||
| Raw and analyzed data | This paper; Table S2 | GEO: GSE184158 | |
| Mouse reference genome Mm10, GRCm38 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/grc/mouse | |
| Adult mesenchymal RNA datasets | McCarthy, et al.30 | GEO: GSE130681 | |
| Experimental Models: Organisms/Strains | |||
| Mouse: PdgfraH2BeGFP | Jackson Laboratory | 007669 | |
| Mouse: ROSA26LSL-TdTomato | Jackson Laboratory | 007909 | |
| Mouse: Myh11Cre(ER-T2) | Jackson Laboratory | 019079 | |
| Mouse: Etv1Cre(ER-T2) | Jackson Laboratory | 013048 | |
| Mouse: PdgfraCre(ER-T2> | Jackson Laboratory | 032770 | |
| Mouse: ROSA26L-S-L-DTA | Jackson Laboratory | 009669 | |
| Oligonucleotides | |||
| Grem1 probes | Advanced Cell Diagnostics (ACD) | 314741 | |
| Grem2 probes | ACD | 473981 | |
| Hhip probes | ACD | 448441 | |
| Noggin probes | ACD | 467391 | |
| Olfm4 probes | ACD | 311831 | |
| Osr1 probes | ACD | 496281 | |
| Axin2 probes | ACD | 400331 | |
| Rspo3 probes | ACD | 483781 | |
| Rspo2 probes | ACD | 402008 | |
| Rspo1 probes | ACD | 479591 | |
| Id1 probes | ACD | 312221 | |
| Bmp5 probes | ACD | 401241 | |
| Pcp4 probes | ACD | 402311 | |
| Id1 | GAGGCGGCATGTGTTCCA | CTGGAGGCTGAAAGGTGGAG | |
| Id2 | AAAAACAGCCTGTCGGACCA | GGTGATGCAGGCTGACGATA | |
| Vil1 | GGGATCCCTTCAAGTGGAGTA | TGGTATTGGCAGT GAAAA CG | |
| Alpi | CAGCCCTTTCCAATTTCACTG | AGGACATCGCCACTCAACTC | |
| Lgr5 | CCTACTCGAAGACTTACCCAGT | GCATTGGGGTGAATGATAGCA | |
| Cd81 | ACACCTTCTACGTGGGCATC | TGCTTCACATCCTTGGCGAT | |
| Bmp5 | TAGATGTGGGCTGGCTTGTC | ACCTCGCTTGCCTTGAAGAA | |
| Grem1 | CTGGAGACCCAGAGTACCGT | CGGTGCGATTCATTCTGTCAC | |
| Rspo3 | ATGTGGCTTCAAAAGGGGGA | TGGGGTCTCAATGCTGGACT | |
| Olfm4 | CAGCCACTTTCCAATTTCACTG | GCTGGACATACTCCTTCACCTTA | |
| Software and Algorithms | |||
| GraphPad Prism v9.0.2 | GraphPad | ||
| R | R | https://cran.r-proiect.org/bin/windows/ | |
| Seurat package v4.1 | Butler et al.73 | https://github.com/satiialab/seurat | |
| Cell Ranger v3.1.1 | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation | |
| BioMart v2.38.0 | Durinck et al.74 | https://github.com/grimbough/biomaRt | |
| MetaNeighbor | Crow et al.58 | https://github.com/maggiecrow/MetaNeighbor | |
| ImageJ Fiji | Schindelin et al.75 | https://imagei.net/Fiii/ | |
| Biorender | BioRender.com | ||
| FlowJo Software v10 | FlowJo | ||
| Other | |||
| ACD HybEZ II Hybridization System | ACD | 321710 | |
| 10X Genomics Controller | 10X Genomics | ||
| EdU labeling kit | Invitrogen | C10640 | |
| Slide spacers | Grace Bio-Labs | 654002 | |
Highlights.
The mouse intestinal stem cell niche matures considerably between birth and weaning.
ISEMFs induce epithelial differentiation and concentrate at crypt tops over that time.
Diverse sub-cryptal smooth muscle cells provide BMP inhibitors and other niche factors.
Young mice share nearly all niche cell populations with the developing human fetus.
Acknowledgments
Supported by National Institutes of Health awards U01DK103152 and R01DK121540 (to R.A.S.) and K01DK125639 (to N.M.). We appreciate generous support from the Lind family, valuable assistance from E. Manieri and flow cytometry staff at the Dana-Farber Cancer Institute, advice on human fetal cell scRNA-seq data from A. Antanaviciute and A. Simmons, and microscopy and organoid services from the Harvard Digestive Diseases Center (P30 DK034854)
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests.
REFERENCES
- 1.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, and Clevers H (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007. [DOI] [PubMed] [Google Scholar]
- 2.Clevers H (2013). The intestinal crypt, a prototype stem cell compartment. Cell 154, 274–284. [DOI] [PubMed] [Google Scholar]
- 3.Qi Z, Li Y, Zhao B, Xu C, Liu Y, Li H, Zhang B, Wang X, Yang X, Xie W, et al. (2017). BMP restricts stemness of intestinal Lgr5(+) stem cells by directly suppressing their signature genes. Nat. Commun 8, 13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, and Clevers H (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. [DOI] [PubMed] [Google Scholar]
- 5.Farin HF, Van Es JH, and Clevers H (2012). Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 143, 1518–1529 e1517. [DOI] [PubMed] [Google Scholar]
- 6.McCarthy N, Kraiczy J, and Shivdasani RA (2020). Cellular and molecular architecture of the intestinal stem cell niche. Nat. Cell Biol 22, 1033–1041. [DOI] [PubMed] [Google Scholar]
- 7.Kabiri Z, Greicius G, Madan B, Biechele S, Zhong Z, Zaribafzadeh H, Edison, Aliyev J, Wu Y, Bunte R, et al. (2014). Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development 141, 2206–2215. [DOI] [PubMed] [Google Scholar]
- 8.Calvert R, and Pothier P (1990). Migration of fetal intestinal intervillous cells in neonatal mice. Anat. Rec 227, 199–206. [DOI] [PubMed] [Google Scholar]
- 9.Itzkovitz S, Blat IC, Jacks T, Clevers H, and van Oudenaarden A (2012). Optimality in the development of intestinal crypts. Cell 148, 608–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sumigray KD, Terwilliger M, and Lechler T (2018). Morphogenesis and Compartmentalization of the Intestinal Crypt. Dev. Cell 45, 183–197 e185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng H, and Bjerknes M (1985). Whole population cell kinetics and postnatal development of the mouse intestinal epithelium. Anat. Rec 211, 420–426. [DOI] [PubMed] [Google Scholar]
- 12.Maskens AP, and Dujardin-Loits RM (1981). Kinetics of tissue proliferation in colorectal mucosa during post-natal growth. Cell Tissue Kinet. 14, 467–477. [DOI] [PubMed] [Google Scholar]
- 13.St Clair WH, and Osborne JW (1985). Crypt fission and crypt number in the small and large bowel of postnatal rats. Cell Tissue Kinet. 18, 255–262. [DOI] [PubMed] [Google Scholar]
- 14.Langlands AJ, Almet AA, Appleton PL, Newton IP, Osborne JM, and Nathke IS (2016). Paneth Cell-Rich Regions Separated by a Cluster of Lgr5+ Cells Initiate Crypt Fission in the Intestinal Stem Cell Niche. PLoS Biol. 14, e1002491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Nafussi AI, and Wright NA (1982). Cell kinetics in the mouse small intestine during immediate postnatal life. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol 40, 51–62. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt GH, Winton DJ, and Ponder BA (1988). Development of the pattern of cell renewal in the crypt-villus unit of chimaeric mouse small intestine. Development 103, 785–790. [DOI] [PubMed] [Google Scholar]
- 17.Bry L, Falk P, Huttner K, Ouellette A, Midtvedt T, and Gordon JI (1994). Paneth cell differentiation in the developing intestine of normal and transgenic mice. Proc. Natl. Acad. Sci. USA 91, 10335–10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powell DW, Pinchuk IV, Saada JI, Chen X, and Mifflin RC (2011). Mesenchymal cells of the intestinal lamina propria. Annu. Rev. Physiol 73, 213–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stzepourginski I, Nigro G, Jacob JM, Dulauroy S, Sansonetti PJ, Eberl G, and Peduto L (2017). CD34+ mesenchymal cells are a major component of the intestinalstem cells niche at homeostasis and after injury. Proc. Natl. Acad. Sci. USA 114, E506–E513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greicius G, Kabiri Z, Sigmundsson K, Liang C, Bunte R, Singh MK, and Virshup DM (2018). PDGFRalpha(+) pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc. Natl. Acad. Sci. USA 115, E3173–E3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kosinski C, Li VS, Chan AS, Zhang J, Ho C, Tsui WY, Chan TL, Mifflin RC, Powell DW, Yuen ST, et al. (2007). Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc. Natl. Acad. Sci. USA 104, 15418–15423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim JE, Fei L, Yin WC, Coquenlorge S, Rao-Bhatia A, Zhang X, Shi SSW, Lee JH, Hahn NA, Rizvi W, et al. (2020). Single cell and genetic analyses reveal conserved populations and signaling mechanisms of gastrointestinal stromal niches. Nat. Commun 11, 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cretoiu D, Cretoiu SM, Simionescu AA, and Popescu LM (2012). Telocytes, a distinct type of cell among the stromal cells present in the lamina propria of jejunum. Histol. Histopathol 27, 1067–1078. [DOI] [PubMed] [Google Scholar]
- 24.Popescu LM, and Faussone-Pellegrini MS (2010). TELOCYTES - a case of serendipity: the winding way from Interstitial Cells of Cajal (ICC), via Interstitial Cajal-Like Cells (ICLC) to TELOCYTES. J. Cell. Mol. Med 14, 729–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shoshkes-Carmel M, Wang YJ, Wangensteen KJ, Toth B, Kondo A, Massasa EE, Itzkovitz S, and Kaestner KH (2018). Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature 557, 242–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Powell DW, Adegboyega PA, Di Mari JF, and Mifflin RC (2005). Epithelial cells and their neighbors I. Role of intestinal myofibroblasts in development, repair, and cancer. Am. J. Physiol 289, G2–7. [DOI] [PubMed] [Google Scholar]
- 27.Roulis M, and Flavell RA (2016). Fibroblasts and myofibroblasts of the intestinal lamina propria in physiology and disease. Differentiation 92, 116–131. [DOI] [PubMed] [Google Scholar]
- 28.Furuya S, and Furuya K (2007). Subepithelial fibroblasts in intestinal villi: roles in intercellular communication. Int. Rev. Cytol 264, 165–223. [DOI] [PubMed] [Google Scholar]
- 29.Joyce NC, Haire MF, and Palade GE (1987). Morphologic and biochemical evidence for a contractile cell network within the rat intestinal mucosa. Gastroenterology 92, 68–81. [DOI] [PubMed] [Google Scholar]
- 30.McCarthy N, Manieri E, Storm EE, Saadatpour A, Luoma AM, Kapoor VN, Madha S, Gaynor LT, Cox C, Keerthivasan S, et al. (2020). Distinct Mesenchymal Cell Populations Generate the Essential Intestinal BMP Signaling Gradient. Cell Stem Cell 26, 391–402 e395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koppens MAJ, Davis H, Valbuena GN, Mulholland EJ, Nasreddin N, Colombe M, Antanaviciute A, Biswas S, Friedrich M, Lee L, et al. (2021). Bone Morphogenetic Protein Pathway Antagonism by Grem1 Regulates Epithelial Cell Fate in Intestinal Regeneration. Gastroenterology 161, 239–254 e239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin-Alonso M, Iqbal S, Vornewald PM, Lindholm HT, Damen MJ, Martinez F, Hoel S, Diez-Sanchez A, Altelaar M, Katajisto P, et al. (2021). Smooth muscle-specific MMP17 (MT4-MMP) regulates the intestinal stem cell niche and regeneration after damage. Nat. Commun 12, 6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walton KD, Whidden M, Kolterud A, Shoffner SK, Czerwinski MJ, Kushwaha J, Parmar N, Chandhrasekhar D, Freddo AM, Schnell S, and Gumucio DL (2016). Villification in the mouse: Bmp signals control intestinal villus patterning. Development 143, 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shyer AE, Tallinen T, Nerurkar NL, Wei Z, Gil ES, Kaplan DL, Tabin CJ, and Mahadevan L (2013). Villification: how the gut gets its villi. Science 342, 212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brugger MD, Valenta T, Fazilaty H, Hausmann G, and Basler K (2020). Distinct populations of crypt-associated fibroblasts act as signaling hubs to control colon homeostasis. PLoS Biol. 18, e3001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fawkner-Corbett D, Antanaviciute A, Parikh K, Jagielowicz M, Geros AS, Gupta T, Ashley N, Khamis D, Fowler D, Morrissey E, et al. (2021). Spatiotemporal analysis of human intestinal development at single-cell resolution. Cell 184, 810–826 e823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elmentaite R, Kumasaka N, Roberts K, Fleming A, Dann E, King HW, Kleshchevnikov V, Dabrowska M, Pritchard S, Bolt L, et al. (2021). Cells of the human intestinal tract mapped across space and time. Nature 597, 250–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elmentaite R, Ross ADB, Roberts K, James KR, Ortmann D, Gomes T, Nayak K, Tuck L, Pritchard S, Bayraktar OA, et al. (2020). Single-Cell Sequencing of Developing Human Gut Reveals Transcriptional Links to Childhood Crohn’s Disease. Dev. Cell 55, 771–783 e775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holloway EM, Czerwinski M, Tsai YH, Wu JH, Wu A, Childs CJ, Walton KD, Sweet CW, Yu Q, Glass I, et al. (2021). Mapping Development of the Human Intestinal Niche at Single-Cell Resolution. Cell Stem Cell 28, 568–580 e564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen L, Toke NH, Luo S, Vasoya RP, Fullem RL, Parthasarathy A, Perekatt AO, and Verzi MP (2019). A reinforcing HNF4-SMAD4 feed-forward module stabilizes enterocyte identity. Nat. Genetics 51, 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haramis AP, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus GJ, and Clevers H (2004). De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science 303, 1684–1686. [DOI] [PubMed] [Google Scholar]
- 42.Hamilton TG, Klinghoffer RA, Corrin PD, and Soriano P (2003). Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol. and Cell. Biol 23, 4013–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim TH, Escudero S, and Shivdasani RA (2012). Intact function of Lgr5 receptor-expressing intestinal stem cells in the absence of Paneth cells. Proc. Natl. Acad. Sci. USA 109, 3932–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fordham RP, Yui S, Hannan NR, Soendergaard C, Madgwick A, Schweiger PJ, Nielsen OH, Vallier L, Pedersen RA, Nakamura T, et al. (2013). Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell 13, 734–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karlsson L, Lindahl P, Heath JK, and Betsholtz C (2000). Abnormal gastrointestinal development in PDGF-A and PDGFR-(alpha) deficient mice implicates a novel mesenchymal structure with putative instructive properties in villus morphogenesis. Development 127, 3457–3466. [DOI] [PubMed] [Google Scholar]
- 46.Blank U, Seto ML, Adams DC, Wojchowski DM, Karolak MJ, and Oxburgh L (2008). An in vivo reporter of BMP signaling in organogenesis reveals targets in the developing kidney. BMC Dev Biol 8, 86. 10.1186/1471-213X-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nerurkar NL, Mahadevan L, and Tabin CJ (2017). BMP signaling controls buckling forces to modulate looping morphogenesis of the gut. Proc. Natl. Acad. Sci. USA 114, 2277–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Link A, Vogt TK, Favre S, Britschgi MR, Acha-Orbea H, Hinz B, Cyster JG, and Luther SA (2007). Fibroblastic reticular cells in lymph nodes regulate the homeostasis of naive T cells. Nat. Immunol 8, 1255–1265. [DOI] [PubMed] [Google Scholar]
- 49.Buechler MB, Kim KW, Onufer EJ, Williams JW, Little CC, Dominguez CX, Li Q, Sandoval W, Cooper JE, Harris CA, et al. (2019). A Stromal Niche Defined by Expression of the Transcription Factor WT1 Mediates Programming and Homeostasis of Cavity-Resident Macrophages. Immunity 51, 119–130 e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee MY, Ha SE, Park C, Park PJ, Fuchs R, Wei L, Jorgensen BG, Redelman D, Ward SM, Sanders KM, and Ro S (2017). Transcriptome of interstitial cells of Cajal reveals unique and selective gene signatures. PloS One 12, e0176031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huycke TR, Miller BM, Gill HK, Nerurkar NL, Sprinzak D, Mahadevan L, and Tabin CJ (2019). Genetic and Mechanical Regulation of Intestinal Smooth Muscle Development. Cell 179, 90–105 e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kurahashi M, Nakano Y, Hennig GW, Ward SM, and Sanders KM (2012). Platelet-derived growth factor receptor alpha-positive cells in the tunica muscularis of human colon. J. Cell. Mol. Med 16, 1397–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ward SM, Burns AJ, Torihashi S, and Sanders KM (1994). Mutation of the proto-oncogene c-kit blocks development of interstitial cells and electrical rhythmicity in murine intestine. J. Physiol 480 ( Pt 1), 91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chi P, Chen Y, Zhang L, Guo X, Wongvipat J, Shamu T, Fletcher JA, Dewell S, Maki RG, Zheng D, et al. (2010). ETV1 is a lineage survival factor that cooperates with KIT in gastrointestinal stromal tumours. Nature 467, 849–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCracken VJ, and Lorenz RG (2001). The gastrointestinal ecosystem: a precarious alliance among epithelium, immunity and microbiota. Cell Microbiol. 3, 1–11. [DOI] [PubMed] [Google Scholar]
- 56.Spence JR, Lauf R, and Shroyer NF (2011). Vertebrate intestinal endoderm development. Dev. Dyn 240, 501–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Montgomery RK, Mulberg AE, and Grand RJ (1999). Development of the human gastrointestinal tract: twenty years of progress. Gastroenterology 116, 702–731. [DOI] [PubMed] [Google Scholar]
- 58.Crow M, Paul A, Ballouz S, Huang ZJ, and Gillis J (2018). Characterizing the replicability of cell types defined by single cell RNA-sequencing data using MetaNeighbor. Nat. Commun 9, 884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kanda T, Sullivan KF, and Wahl GM (1998). Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol 8, 377–385. [DOI] [PubMed] [Google Scholar]
- 60.Chung MI, Bujnis M, Barkauskas CE, Kobayashi Y, and Hogan BLM (2018). Niche-mediated BMP/SMAD signaling regulates lung alveolar stem cell proliferation and differentiation. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eilken HM, Dieguez-Hurtado R, Schmidt I, Nakayama M, Jeong HW, Arf H, Adams S, Ferrara N, and Adams RH (2017). Pericytes regulate VEGF-induced endothelial sprouting through VEGFR1. Nat. Commun 8, 1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chee YC, Pahnke J, Bunte R, Adsool VA, Madan B, and Virshup DM (2018). Intrinsic Xenobiotic Resistance of the Intestinal Stem Cell Niche. Dev. Cell 46, 681–695 e685. [DOI] [PubMed] [Google Scholar]
- 64.Storm EE, Durinck S, de Sousa e Melo F, Tremayne J, Kljavin N, Tan C, Ye X, Chiu C, Pham T, Hongo JA, et al. (2016). Targeting PTPRK-RSPO3 colon tumours promotes differentiation and loss of stem-cell function. Nature 529, 97–100. [DOI] [PubMed] [Google Scholar]
- 65.Batts LE, Polk DB, Dubois RN, and Kulessa H (2006). Bmp signaling is required for intestinal growth and morphogenesis. Dev. Dyn 235, 1563–1570. [DOI] [PubMed] [Google Scholar]
- 66.Davis H, Irshad S, Bansal M, Rafferty H, Boitsova T, Bardella C, Jaeger E, Lewis A, Freeman-Mills L, Giner FC, et al. (2015). Aberrant epithelial GREM1 expression initiates colonic tumorigenesis from cells outside the stem cell niche. Nat. Med 21, 62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He XC, Zhang J, Tong WG, Tawfik O, Ross J, Scoville DH, Tian Q, Zeng X, He X, Wiedemann LM, et al. (2004). BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat. Genetics 36, 1117–1121. [DOI] [PubMed] [Google Scholar]
- 68.Auclair BA, Benoit YD, Rivard N, Mishina Y, and Perreault N (2007). Bone morphogenetic protein signaling is essential for terminal differentiation of the intestinal secretory cell lineage. Gastroenterology 133, 887–896. [DOI] [PubMed] [Google Scholar]
- 69.Lecoin L, Gabella G, and Le Douarin N (1996). Origin of the c-kit-positive interstitial cells in the avian bowel. Development 122, 725–733. [DOI] [PubMed] [Google Scholar]
- 70.Young HM, Ciampoli D, Southwell BR, and Newgreen DF (1996). Origin of interstitial cells of Cajal in the mouse intestine. Dev. Biol 180, 97–107. [DOI] [PubMed] [Google Scholar]
- 71.Radenkovic G, Radenkovic D, and Velickov A (2018). Development of interstitial cells of Cajal in the human digestive tract as the result of reciprocal induction of mesenchymal and neural crest cells. J. Cell. Mol. Med 22, 778–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kurahashi M, Nakano Y, Hennig GW, Ward SM, and Sanders KM (2012). Platelet-derived growth factor receptor alpha-positive cells in the tunica muscularis of human colon. J. Cell. Mol. Med 16, 1397–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature Biotech. 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Durinck S, Spellman PT, Birney E, and Huber W (2009). Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc 4, 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bernier-Latmani J, and Petrova TV (2016). High-resolution 3D analysis of mouse small-intestinal stroma. Nat. Protoc 11, 1617–1629. [DOI] [PubMed] [Google Scholar]
- 77.Nerurkar NL, Mahadevan L, and Tabin CJ (2017). BMP signaling controls buckling forces to modulate looping morphogenesis of the gut. Proc. Natl. Acad. Sci. USA 114, 2277–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, and Luo Y (2012). RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn 14, 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kinchen J, Chen HH, Parikh K, Antanaviciute A, Jagielowicz M, Fawkner-Corbett D, Ashley N, Cubitt L, Mellado-Gomez E, Attar M, et al. (2018). Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell 175, 372–386 e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sato T, and Clevers H (2013). Primary mouse small intestinal epithelial cell cultures. Methods in Mol. Biol 945, 319–328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are deposited in the Gene Expression Omnibus (GSE184158). No new software was developed for this study. Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.


