Summary
Behavior is shaped by both the internal state of an animal and its individual behavioral biases. Rhythmic variation in gonadal hormones during the estrous cycle is a defining feature of female internal state, one that regulates many aspects of sociosexual behavior. However, it remains unclear whether estrous state influences spontaneous behavior, and if so how these effects might relate to individual behavioral variation. Here we address this question by longitudinally characterizing the open field behavior of female mice across different phases of the estrous cycle, using unsupervised machine learning to decompose spontaneous behavior into its constituent elements1–4. We find that each female mouse exhibits a characteristic pattern of exploration that uniquely identifies it as an individual across many experimental sessions; in contrast, estrous state only negligibly impacts behavior, despite its known effects on neural circuits that regulate action selection and movement. Like female mice, male mice exhibit individual-specific patterns of behavior in the open field; however, the exploratory behavior of males is significantly more variable than that expressed by females both within and across individuals. These findings suggest an underlying functional stability to the circuits that support exploration in female mice, reveal a surprising degree of specificity in individual behavior, and provide empirical support for the inclusion of both sexes in experiments querying spontaneous behaviors.
Keywords: estrous, mouse, sex differences, exploration, spontaneous, behavior, individuality, variability, computational ethology
eTOC blurb
Levy et al. track open field behavior of female mice over weeks and find that behavior reflects individual identity far more than estrous state. Open field exploration by males is also much more variable than in females, arguing for inclusion of both sexes in studies of spontaneous behaviors.
Results
Mice exhibit a stereotyped pattern of behavior when exploring an open field1,5. This pattern is altered when the shape of the arena is changed, when sensory cues are introduced, when mice are hungry or in pain, or when brain dynamics are reconfigured through optogenetic perturbations or treatment with neuro- or psychoactive drugs1,3,4,6–8. Importantly, each of these different experimental manipulations evokes a characteristic change in behavior that is similar across mice, such that e.g., the identity and dose of an administered drug can be accurately predicted from behavior in the open field alone2. These observations support the broad proposal that the state of a mouse at any given moment determines how it explores the world9.
However, mouse behavior is also individualized. For example, different (and yet genetically identical and similarly housed) mice explore the same arena by expressing distinct patterns of behavior that are characteristic of each mouse4,10,11; these inter-mouse differences likely reflect stochastic aspects of gene expression and development as well as differences in experience, which collectively impinge upon neural circuit structure and function12. Thus, exploratory behavior — and likely other types of naturalistic, spontaneous behavior — reflects a balance between the tendency of mice to generate state-dependent behaviors appropriate for a given context, and their tendency to behave as unique individuals.
The day-to-day internal state of female rodents rhythmically varies due to the estrous cycle, in which levels of circulating gonadal hormones systematically rise and fall13–15. These hormones alter gene expression, connectivity, and synaptic function across the brain, serving as powerful neuromodulators that can ultimately affect behavior16–19. While estrous state clearly influences sex-specific behaviors, it remains uncertain whether female gonadal hormones more pervasively influence other forms of behavior expressed by both sexes, like exploration16,17,20–26. On the one hand, several studies have reported significant estrous-related changes in rodent open field behavior (e.g. time in center27, center entries28, distance traveled21); furthermore, the striatal and dopaminergic circuits that control the structure and dynamics of spontaneous exploration have been shown to be sensitive to gonadal hormones5,29,30. On the other hand, researchers have also reported the absence of cycle-related differences in exploratory behaviors, or behavioral changes that are linked to interactions between estrous phase and strain, age, or context22,23,25,31,32.
Inconsistencies across these studies may derive, at least in part, from the widespread use of low-dimensional summary statistics (e.g. average velocity) to measure complex and dynamic patterns of exploratory behavior. In addition, studies querying the behavioral effects of the estrous cycle often include only a small number of behavioral sessions, grouping the data by estrous phase and averaging across individual mice. Because this approach focuses on the estrous cycle as the only source of behavioral variability, it by design neglects other sources of inter-mouse behavioral variability — like individual behavioral biases — that are only measurable with repeated sampling of behavior over time.
To directly quantify the relative contribution of estrous state and individual identity to behavior, here we perform a longitudinal assessment of mouse spontaneous behavior using Motion Sequencing (MoSeq). MoSeq is a well-validated unsupervised machine learning algorithm that decomposes behavior into sub-second component modules (e.g., rears, runs, grooms), which are referred to as “syllables”1,2. This approach yields a high-dimensional description of behavior that captures the use of behavioral modules and sequences, while allowing simultaneous measurements of conventional behavioral parameters such as velocity and position1,3,4.
We first explored the naturalistic behavior of adult C57BL/6J female mice exploring an open field arena over multiple consecutive days while tracking their estrous cycle (Figures 1, S1). Commonly used summary statistics failed to identify a significant effect of estrous phase on open field exploration (Figure S2A–C). In addition, overall syllable distribution was not different between estrous phases (Figures 2A, and S2A,D,F), and phase-dependent patterns of behavior were not clustered in a low-dimensional embedding of syllable space (Figure 2B). Decoding analysis revealed that MoSeq-identified behavioral syllables and sequences could not predict the specific phase of the estrous cycle associated with any given behavioral session (Figures 2C, 2G and S2H). We noted, however, that averaging all instances of a particular estrous phase expressed by an individual mouse (rather than considering each session separately) enabled decoders to predict estrous phase based upon syllable use at a level modestly above chance (Figures 2C and 2G). The handful of syllables that conveyed the most information about estrous phase were associated with active exploration (e.g., running and rearing, Table S1, Figure S2G). Together, these findings suggest that subtle changes in exploratory behavior are associated with each estrous phase, but that these changes are masked by substantial session-to-session variability.
Figure 1: Weighing the relative influence of estrous state and individual variation on spontaneous behavior.
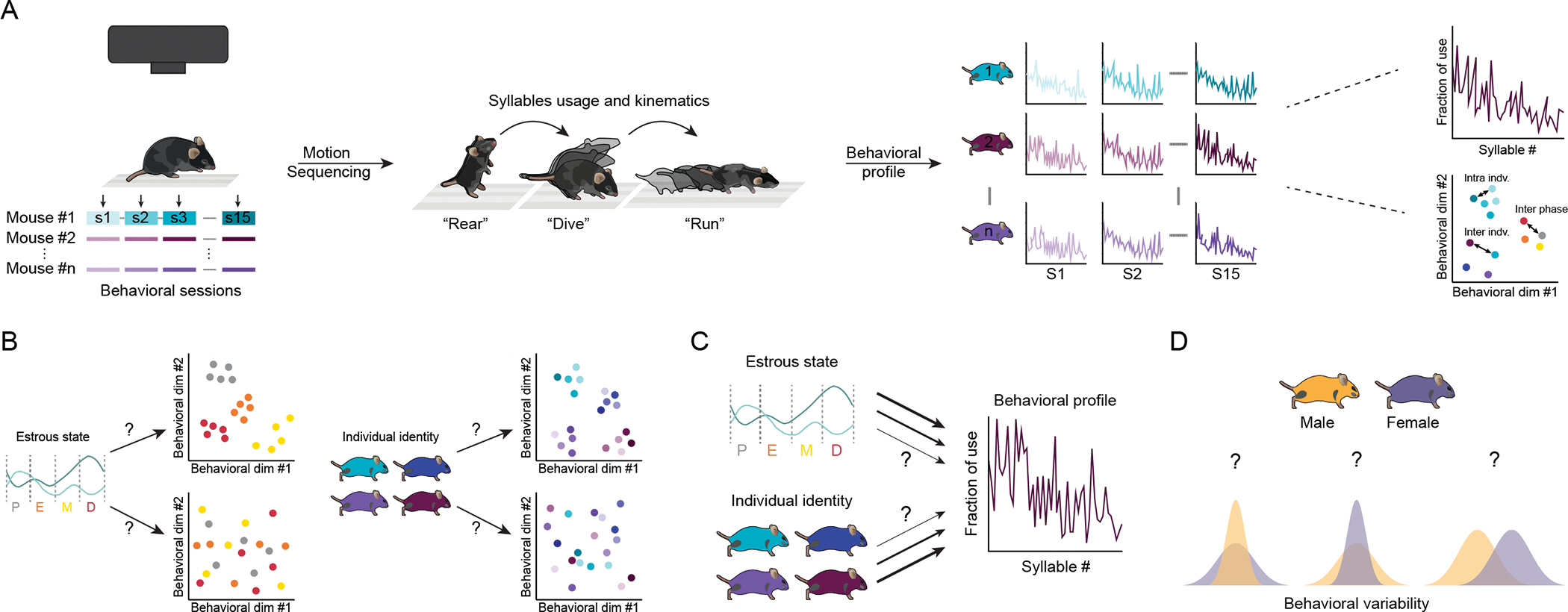
A) Individual mice (n=16 females, n=16 males) were recorded in an open field arena using a depth camera for 15 consecutive sessions, and the estrous phase of female mice was examined each day. Depth video recordings were analyzed using the Motion Sequencing algorithm (see Methods) which segments behavior into sub-second behavioral modules (termed “syllables”, see examples). The fraction of use of each syllable is then calculated for each session, and differences in behavior assessed between individuals (inter-indv.), between sessions for each individual (intra-indv.), and between estrous phases within each individual (inter-phase). Here and throughout, D = diestrus, E = estrus, M = metestrus, P = proestrus. B) The effect of estrous phase (adapted from14) and individual mouse identity on female mouse behavior (as captured by how frequently each behavioral syllable was used) was explored C) as was the relative contribution of each to the use of behavioral syllable and the variability in their use. D) Differences in the magnitude of intra-individual and inter-individual behavioral variability between males and females were also assessed. See also Figure S1.
Figure 2: Female spontaneous behavior reflects individual variation rather than estrous phase.
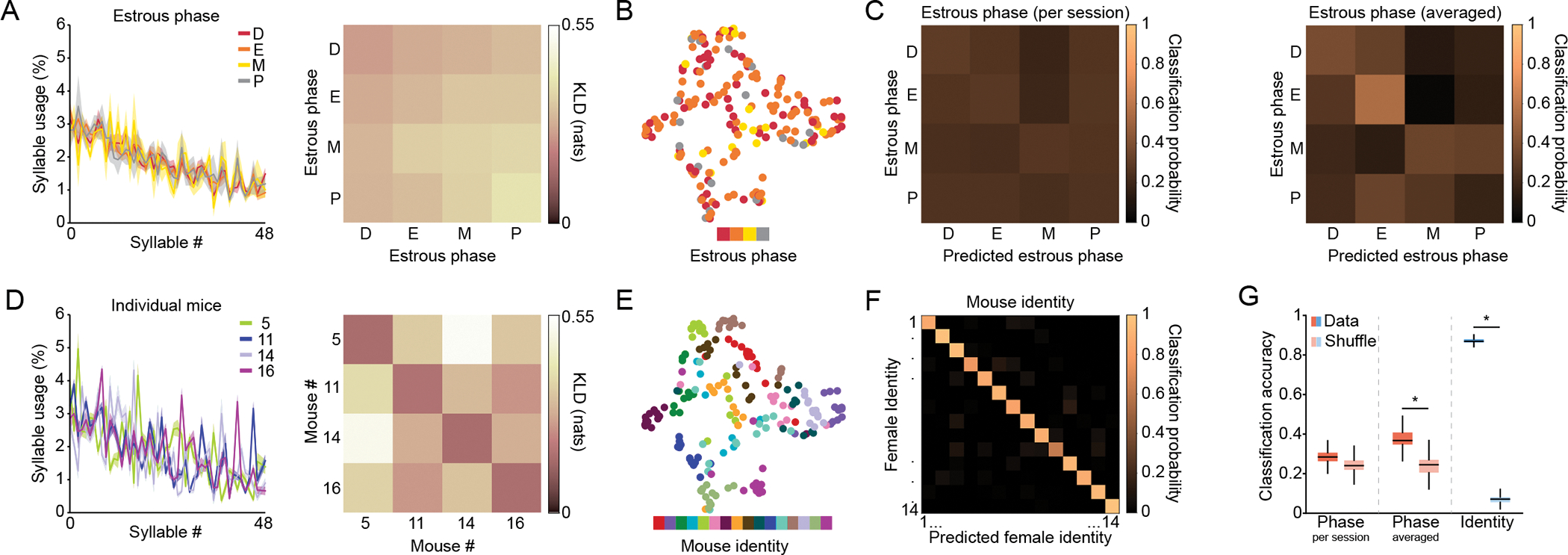
A) Left: syllable usage distribution for each of the estrous phases averaged across four representative mice (presented also in panel D). Mean ± standard error of the mean (shaded area) is presented. MoSeq assigns each syllable a unique identifier (Syllable #) based upon how often that syllable was used across all the data subject to modeling (in this case across all female mice and sessions in our dataset), such that Syllable “0” is the most often used and subsequent syllables are less used; we maintain this syllable ordering across all syllable usage plots herein, but vary how the data are aggregated (for example, by estrous phase or individual identity). See Figures S2D for a similar analysis across all mice. Right: Kullback-Leibler divergence (KLD) values of pairwise comparisons between phase-related data presented in left panel. Here we compute a phase-dependent KLD by pooling all the sessions corresponding to a particular phase across mice, calculating the pairwise KLD between all sessions corresponding to the same or a different phase (as labeled), and then plotting the average value of those pairwise comparisons in each cell. These values quantify how similar syllable usage distributions are across the different estrous phases, with lower values indicating greater similarity. It is important to note that because phase-based comparisons incorporate data from different animals, if different individual mice exhibit distinct patterns of behavior those differences will be incorporated into this metric. B) Uniform Manifold Approximation and Projection (UMAP) plot depicting syllable usage in females for each session color coded by estrous phase. To assess cluster quality, K-means clustering analysis was performed on high-dimensional data, and clustering quality was quantified using the Adjusted Rand Index (ARI), where higher values indicate a greater match between clustering and data labels; for phase (number of clusters = 4) ARI = 0.03. C) Confusion matrix for classification accuracy of a decoder trained to predict estrous phase based on syllable usages. Decoder was trained on data from individual sessions, left; or averaged data per phase per mouse, right (see Methods). D) Left: same data as in A, but syllable use is now averaged across all sessions corresponding to the four representative mice. See Figures S2E for a similar analysis across all mice. Right: same as the right panel in A, but KLD values are now computed among the four representative mice depicted in the left panel. Note that values on the diagonal indicate intra-individual variability. E) UMAP plot depicting syllable usage in females for each session color coded by mouse identity. Clustering analysis was performed as described in panel B. For individuals (number of clusters = 16) ARI = 0.43. F) Same as C but for prediction of individual mouse identity. G) Quantification of overall decoder performance (n=1000 restarts for data and shuffled data). Asterisk (*) denotes statistical significance, here indicating that the mean decoding distribution exceeds the 95th percentile shuffle threshold. For all relevant panels, Box plots depict median, interquartile range, and upper/lower adjacent values (black lines). See also Figure S2 and table S1.
In contrast, decoders accurately predicted individual mouse identity more than 85 percent of the time across all behavioral sessions (Figure 2F,G); these data demonstrate that each of the 16 female mice exhibits a characteristic pattern of behavior that is sufficiently stable over time to enable individual identification based upon behavior expressed within a single experimental session. Consistent with this observation, individual patterns of behavior were well clustered in a low-dimensional embedding of syllable space (Figures 2D,E and S2A,E). Importantly, most behavioral syllables identified by MoSeq conveyed at least some information about individual identity, suggesting that inter-individual differences in behavior reflect broad changes in syllable use, rather than the modulation of a small set of individualized syllables (Table S1 and Figure S2G). Taken together, these data reveal that female mice express an individual-specific pattern of behavior when exploring the open field, and that this individualized behavioral profile is relatively invariant across multiple experimental sessions encompassing different estrous phases.
To further explore the relative effect of estrous state and individual identity on behavior, we quantified the degree to which each contributed to the overall variability observed in our data. This analysis demonstrates that inter-individual differences in syllable use are far greater than phase-related differences in behavior expressed by each mouse (Figure 3A). Furthermore, modeling reveals that estrous state contributes little to the overall behavioral variation observed across the dataset, while the identity of each mouse accounted for almost all the explained variability (Figure 3B).
Figure 3: Individual identity, not estrous phase, accounts for variability in female behavior.
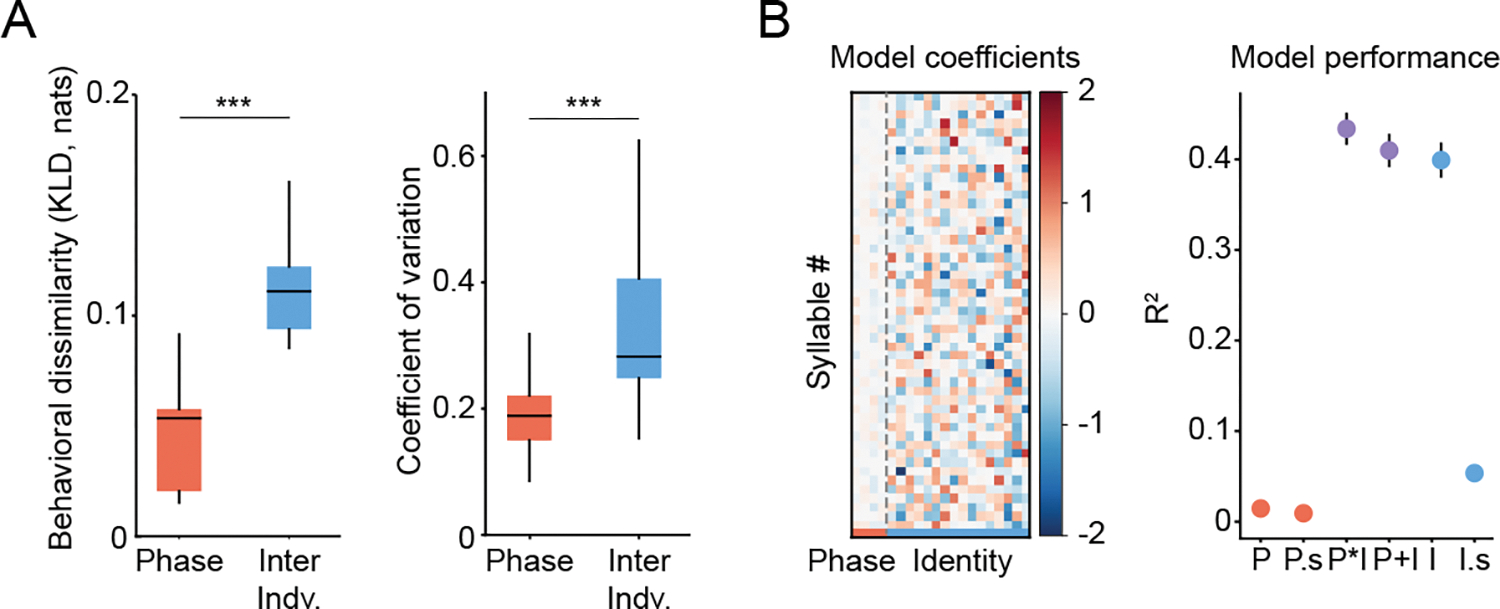
A) Behavioral dissimilarity among phases (within each mouse) and individuals (inter indv. = inter individual) as measured by Kullback-Leibler divergence (KLD) of syllable usage distributions (left) and by the coefficient of variation (CV) of the usage of each syllable (right). Wilcoxon signed-rank test was performed. For KLD: n=16 mice, T=1.0, p=6.10*10−5. For CV: n=49 syllables, T=32, p=7.72*10−9. B) Left: Weights from linear models fit to model syllable usage as a function of estrous phase and mouse identity. Right: Model performance as quantified by R2 (mean±SEM) for models trained to predict syllable usage as a function of phase (P); phase with phase labels shuffled within a mouse (P.s, n=100); phase, identity and their interaction (P*I); phase and identity without interaction (P+I, weights are shown in left panel); mouse identity (I); mouse identity, with shuffled identity labels (I.s, n=100). No significant differences in model performance were identified between P*I, P+I and I models (assessed via ANOVA). For all relevant panels, Box plots depict median, interquartile range, and upper/lower adjacent values (black lines). *p<0.05, **p<0.01, ***p<0.001.
Given these findings, we wondered whether the amount of variation observed in female open field behavior (either within individual mice or between mice) was greater or less than that observed in male behavior. To address this question, we longitudinally characterized open field behavior in age-matched male mice, and similarly found that male behavior largely reflected mouse identity (Figure S3A,B and Table S2). However, both intra- and inter-individual variability in male behavior was substantially greater than that observed in females (Figure 4A,B, S3 and S4); furthermore, variability in syllable use significantly increased over time in male but not female mice (Figure 4A,B, right panels). These findings demonstrate that despite hormonal fluctuations associated with the estrous cycle, female open field behavior — within individual mice, between different mice, and over time — is significantly more stable than that expressed by males.
Figure 4: Female behavioral variability is lower than that of males during spontaneous open field behavior.
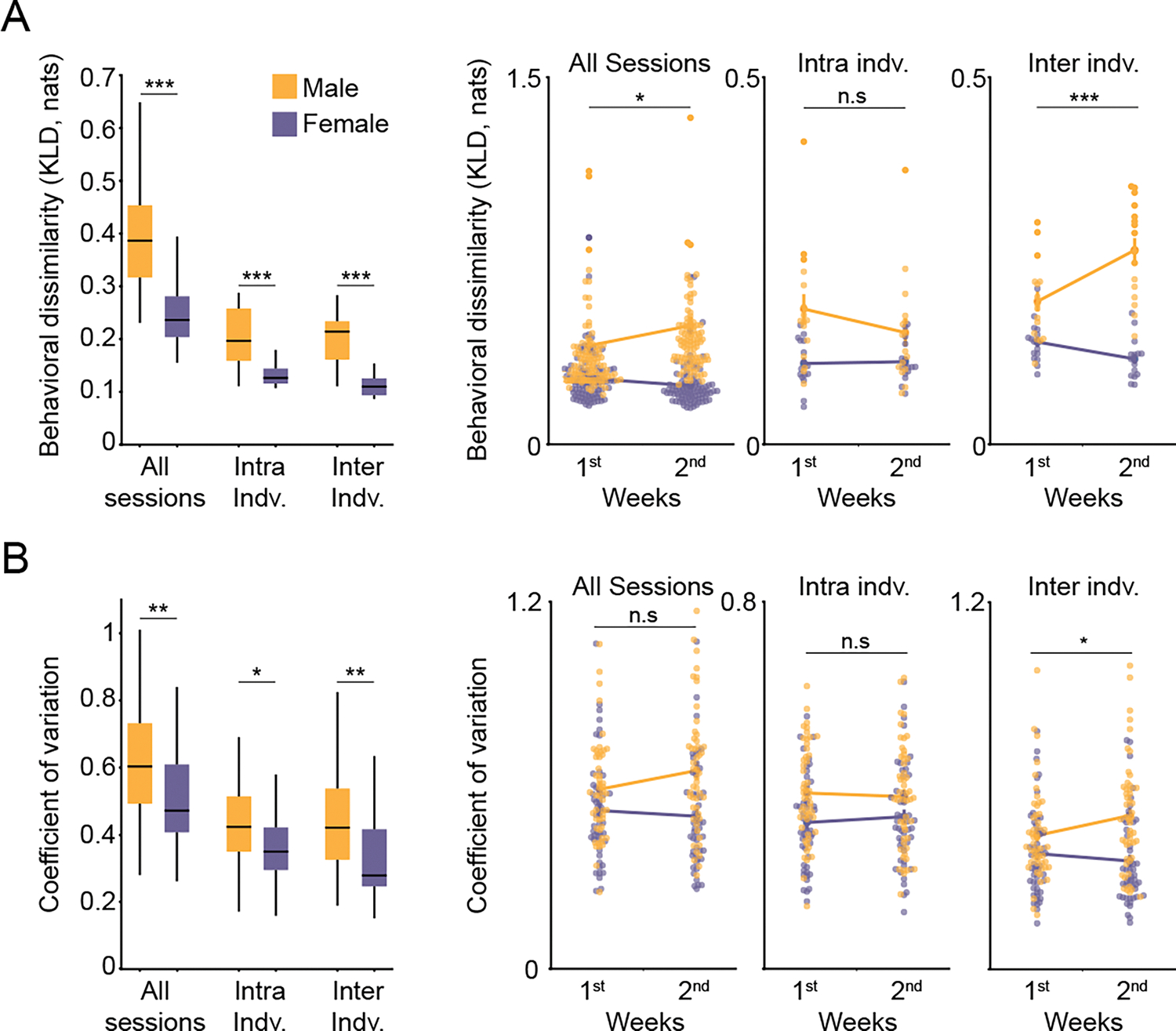
A) Left: Comparison of male and female behavioral variability, where variability is quantified across all behavioral sessions, across sessions within each individual (intra indv. = intra-individual variability), and across individuals (inter indv. = inter-individual variability) by measuring the KLD of syllable usage distributions. 2-way ANOVA for sex and experimental conditions (exp) as main factors was performed: Fsex(1,438)=55.46, p=5.10*10−13; Fexp(2,438)=16.12, p=1.74*10−7, Fsex*exp(2,438)=1.40, p=0.246. Individual contrasts were performed using student’s t-test with Bonferroni correction: pall sessions=6.42*10−11, nfemale=188 sessions, nmale=192 sessions; pintra-indv=3.3*10−4; pinter-indv=1.28*10−5, n=16 mice for male and females. Right: Variability across the first and second weeks of behavioral assessment (1st: day 1–7, nfemale=93 sessions, nmale=94 sessions; 2nd: day 8–14 nfemale=95 sessions, nmale=98 sessions, excluding the first experimental day in both sexes). Each point represents a single session (for “all sessions” analysis) or a single mouse (for “intra indv.” and “inter indv.” analysis), with mean±sem is presented as lines. 2-way ANOVA for sex and time as main factors was performed for each condition, with individual contrasts using student’s t-test and Bonferroni correction. Significant interaction effects are marked on figure. For all sessions: Fsex(1,376)=46.2613, p=4.08*10−11; Ftime(1,376)=0.90, p=0.34; Fsex*time(1,376)=4.01, p=0.045; For intra-individual: Fsex(1,60)=14.07, p=3.9*10−4; Ftime(1,60)=0.96, p=0.32; Fsex*time(1,60)=1.33, p=0.25; For inter-individual: Fsex(1,60)=73.03, p=5.77*10−12; Ftime(1,60)=3.8, p=0.054; Fsex*time(1,60)=15.23, p=2.4*10−4. Male significantly increased their inter-individual variation between the first and second week of behavioral recordings with p=0.006, all other comparisons were not significant when corrected for multiple comparisons. For visualization purposes (but not analytical purposes) 3 of the 784 data points were excluded because they lie outside the range of the graph. B) Same as in C, but depicted is the coefficient of variation of the usage of each syllable. Each point represents a single syllable, with mean±sem presented as lines. For left panel: 2-way ANOVA: Fsex(1,288)=27.97, p=2.43−7; Fexp(2,288)=42.56, p=6.38*10−17, Fsex*exp(2,288)=0.52, p=0.59. Individual contrasts were performed using student’s t-test with Bonferroni correction: pall sessions=0.0063; pintra-indv=0.015; pinter-indv=0.0051. Right panel: 2-way ANOVA: For all sessions: Fsex(1,192)=17.80, p=3.8*10−−4; Ftime(1,192)=0.7, p=0.39; Fsex*time(1,192)=2.52, p=0.11; For intra-individual: Fsex(1,192)=11.47, p=8.5*10−.4; Ftime(1,192)=0.038, p=0.85; Fsex*time(1, 192)=0.4, p=0.52; For inter-individual: Fsex(1,192)=21.63, p=6*10−6; Ftime(1,192)=0.86, p=0.35; Fsex*time(1,192)=4.04, p=0.045. nmice=16 and nsyllables=49 for both male and females. All individual contrasts were not significant when corrected for multiple comparisons. For all relevant panels, Box plots depict median, interquartile range, and upper/lower adjacent values (black lines). *p<0.05, **p<0.01, ***p<0.001. See also Figure S3, S4 and Table S2.
Discussion
The potential for estrous-driven variation to alter experimental outcomes has been the primary motivation for the widespread exclusion of female rodents in behavioral neuroscience research33–35. This concern has been previously considered in several meta-analyses, which have found that the overall distribution of behavioral variation (assessed by aggregating hundreds of behavioral metrics) is no greater in female rodents than in male rodents36–39. Similarly, one recent study found that bulk movement in the home cage is less variable across females than males40. However, these papers do not make clear whether the observed behavioral variation in females is primarily the consequence of estrous or individual variation.
Our granular, high-dimensional and longitudinal analysis reveals that the behavioral effects of estrous state are modest (at best) compared to that of individual variation during spontaneous open field behavior. For example, stable individual differences in behavior are apparent in single experimental sessions, whereas stereotyped estrous phase-dependent behavioral differences are not. To give an intuitive sense of the scale of the effects of the estrous cycle on exploratory behavior, in previous experiments we have demonstrated that mice treated with even small doses of neuro- or -psychoactive drugs exhibit far greater changes in behavior than those observed when we measure inter-individual variability4. Thus from a practical perspective the impact of estrous state on exploratory behavior is likely to be negligible.
Given the well-documented synaptic, cellular and network level effects of gonadal hormones on neural circuits implicated in exploration, the fact that estrous minimally influences spontaneous behavior suggests the existence of neural mechanisms that stabilize individual behavior in female mice across different estrous phases29,30. Furthermore, our data clearly demonstrate that female exploratory behavior is less variable (regardless of estrous state) than that of males. Taken together, our findings argue for the inclusion of both sexes in experiments querying behavior, and support the perspective that females — rather than males — should be the default sex used in studies of exploratory behavior in circumstances in which both sexes cannot be tested.
Estrous variation has been proposed to manifest in a species, strain, age and context-dependent manner22,23,31. Thus a main caveat of our work is that we only explore female and male mouse behavior in a single strain and experimental setting, one that does not impose an explicit task or goal; we have also not made manipulations to explore the causal mechanisms that underlie the striking degree of inter-individual variation we observe. Future work will be required to assess how general our findings are, to identify features of development or experience that are particularly important to defining individualized patterns of open field behavior, and to explain why behavioral variability is greater in males than females. We speculate that the use of dense, unsupervised behavioral characterization methods, as we have done here, will be useful for unveiling specific relationships between internal states and individual behavioral variation across a variety of contexts.
STAR Methods
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sandeep Robert Datta (srdatta@hms.harvard.edu)
Materials availability
This study did not generate new unique reagents.
Data and Code availability
Raw behavioral videos have been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
The behavioral analysis code and Moseq software used to model and analyze the data is freely available to all academic researchers online: http://www.moseq4all.org/.
All original code has been deposited at Github and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical commercial assays | ||
| JorVet Dip Quick Stain | Fischer Scientific | NC9581034 |
| Black Polyethylene Tank (OFA) | US plastics | #14317 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | #000664 |
| Software and algorithms | ||
| Motion Sequencing (Moseq) | The Datta lab | http://www.moseq4all.org/ |
| OLYMPUS OlyVIA 2.9 | Olympus Software Imaging Solutions | https://www.olympus-sis.com/ |
| Python version 3.6 | Python Software Foundation | https://www.python.org |
| Numpy | Harris el al42 | https://numpy.org/ |
| Seaborn | Waskom et al43 | https://seaborn.pydata.org/ |
| Pandas | Mckinney et al44 | https://pandas.pydata.org/ |
| Scikit-learn | Pedregosaet al45 | https://scikit-learn.org/stable/ |
| Scipy | Virtanen et al46 | https://scipy.org/ |
| Statsmodels | Seabold et al47 | https://www.statsmodels.org/stable/index.html |
| Matplotlib | Hunter et al48 | https://matplotlib.org/ |
| Deposited data | ||
| Analyzed data and related scripts | This paper | https://github.com/dattalab/spontaneous-behavior-reflects-individuality-not-estrous |
| Post-MoSeq behavioral analysis | This paper | 10.5281/zenodo.7622958 |
| Raw behavioral videos | This paper | 10.5281/zenodo.7622958 |
Experimental model
Animals
Animals used for this study were adult (3 months old) male and female C57BL/6J mice obtained from Jackson laboratories (stock number #000664, n=16 for each sex for all experiments). Mice were kept on a reverse 12-h light–dark cycle with food and water ad libitum and tested during the dark phase under dim red light. All mice were grouped-housed (four mice in a cage, randomly assigned). Mouse husbandry and experiments were performed following institutional and federal guidelines and were approved by Harvard Medical School’s Institutional Animal Care and Use Committee.
Method details
Behavioral procedure
At the day of the testing, mice were brought to the behavioral room and allowed to habituate in their home cage for a minimum of 30 minutes under dim red light. Following habituation, each individual mouse was placed in the middle of a circular 17″ diameter open-field arena (OFA) enclosed with 14″-high opaque walls (US Plastics 14317), immediately after which video recording began. The arena walls were sanded to eliminate reflective artifacts in the depth video. Depth videos of mouse behavior were acquired at 30 Hz using a Kinect v2 camera and app for Windows (Microsoft Corp.) Mice were allowed to freely explore the enclosure for a 20-min period after which they were returned to their home cage. Arenas were thoroughly cleaned using 70% Ethanol solution and allowed to air dry between trials and at the end of each experimental day. All animals were tested for 15 consecutive days. Female and male experiments were performed on consecutive weeks. To focus our conclusions on exploration rather than novelty responses, we excluded the first encounter of each mouse with the behavioral apparatus from analysis. For variability analysis, number of female experimental days was matched to males.
To control for possible handling and habituation related effects when comparing male and female data variability, an additional experiment was performed in which a novel batch of aged-matched (3 months old) males and females (n=16 for either sex) were habituated to the behavioral apparatus for 10 consecutive days, and then recorded for additional 7 days. Here, females were not tested for estrous phase, and thus male and female mice were matched for handling; further, providing an extensive habituation minimized any novelty/stress related behavioral effects in these control cohorts. Female and male experiments were performed on the same day to control for batch effects. Open field arenas were thoroughly cleaned using 70% ethanol and left to vent for 30mins between sexes. Behavioral results were analyzed as for the main experiments and reported in Figure S3.
Behavioral modeling
Depth videos of all mice used in this study were preprocessed, extracted, modeled, and analyzed using the Motion Sequencing (MoSeq) algorithm as previously described1,2, using the pipeline available here: http://www.moseq4all.org/. In brief, MoSeq is an unsupervised, ethologically inspired machine learning algorithm which automatically segments animal spontaneous behavior into sub-second motifs (termed “syllables”, e.g., rear, run, pause). Moseq labels each frame with a syllable label and identifies how often within a given session each syllable is used. Only those syllables used in more than one percent of frames across all sessions were used for analysis (n=49 for females and matched for the males and for the control experiment presented in Figure S3). Syllable number are sorted by fraction of use across all sessions, as seen in Figures 2A,B and S2D,E. The behavior depicted by each syllable was later manually annotated and presented in Table S1 (for females) and Table S2 (for males). In addition, MoSeq identifies the transition statistics that governs how often every syllable transitions into every other syllable (referred to in Figure S2H as “bigrams”). The MoSeq pipeline also quantified more traditional scalar metrics used to describe behavior (e.g., velocity, size, location, distance to center). Total distance traveled during each session was computed by adding the between-frame Euclidean distance between the mouse centroid coordinates. Time in center was calculated as total time mouse center point was present within 11cm circular radius around the center of open field arena. Sessions which could not be extracted due to technical errors or insufficient recording quality (e.g. reflections on OFA walls, sessions where mouse size was at 2.5% top/bottom of mouse size area distribution) were excluded from analysis prior to modeling; overall 9/240 of female sessions and 32/224 sessions in male data were excluded. For the handling/stress control experiment, 18/112 of female sessions and 12/112 sessions in male data were excluded.
Cytological evaluation of mouse estrous cycle
At the end of each experimental day a vaginal swab was collected from each female mouse and used to determine its estrous state as previously described13–15. Briefly, swabs were gently collected form the vaginal opening using a saline-dipped cotton swab at a similar time each day, spread on a glass slide and left to dry in room temperature, after which they were immediately stained (Stain Kit JorVet™ Dip Quick). Slides were then imaged at 10X magnification (Olympus VS120 Virtual Slide Microscope). Relative presence and proportion of leukocytes, cornified epithelial cells and nucleated epithelial cells were used to determine the estrous phase: Diestrus (D), Proestrus (P), Estrus (E) and Metestrus (M) (see Figure S1 for representative examples. Image contrast was adjusted for visualization purposes). Contaminated slides (i.e., significant urine residue or cellular debris) were removed from the analysis and corresponding behavioral sessions were excluded as well. Labeling was done by two independent observers and compared; behavioral trials corresponding to images in which labels were in disagreement were removed from analysis. Overall, 31/231 sessions were excluded. In accordance with the established distribution of phases across the cycle (in which P and M phases are brief and more difficult to detect13,15,41), the final number of included sessions per phase was D=62, E=99, M=20, P=19; the final number of included phases per mouse (after applying technical exclusion criteria described above for both behavioral recording and cytological assessment): 7/16 mice - four phases, 8/16 - three phases, one mouse was included only in the E and D phases and was therefore excluded from all decoder and associated analysis.
Phase and identity classifications
To predict estrous phase (out of the four estrous phases) and individual mouse identity (out of all same-sex mice) a random forest classifier was trained on session-based behavioral syllable usages (the usage of each syllable was used as an input feature, ntrees = 250, class_weights=balanced, so that weights are inversely proportional to class frequencies (i.e phase/mouse identity) in the input data). To maintain a balanced training set, mice that had less than ten total behavioral sessions or fewer than three of the four estrous phases in the data were excluded from all classification analysis in the manuscript (2/16 of females and 2/16 males).
All classifications were performed on all behavioral sessions, other than the classifier presented in right panel of Figure 2C and corresponding middle panel of Figure 2G, in which syllable usages were averaged over repetitions of the same phase within a mouse (e.g., two estrus days of a single mouse were averaged to give the mean behavioral “estrus” signature in that mouse, and similarly for all other estrous phases in that mouse, for all mice). To maintain a balanced training set, training data was bootstrapped by subsampling 9 sessions per phase (in the case of the phase decoding) or 9 sessions per mouse (in the case of the individual decoders) for each decoder restart. In the case of both average and per-session phase decoders, tests were performed on a single held-out mouse; in the case of the individual identity decoders, tests were performed on a single held-out session. The distribution of classification accuracy across 1000 bootstrapped restarts is presented and compared to classification accuracy of shuffled data.
Decoders presented in Figure S2B were trained on scalar quantiles (0.2, 0.4, 0.6, 0.8) calculated in each sessions: 2D velocity, 3D velocity, and distance to center, as well as total distance traveled and total time in center (each scalar was considered separately in Figure S2B and all measurements were aggregated together as one input vector in Figure S2C). Decoder presented in Figure S2H used syllable bigrams (the probability of a two syllables sequence) observed above chance in behavioral data as input. Here, syllable sequence was extracted for each session, and bigram probabilities were defined as the probability of two syllables to appear one after the other in a specific order, for each pair of syllables in the dataset (so that number of possible bigrams = 492 – 49, since self-transition were excluded). The probability for each bigram was calculated across all sessions. To identify bigrams that are observed above chance levels, syllable sequence was then permuted (n=2,000) and bigrams probabilities were calculated for each permutation resulting in a probability distribution for each bigram across the permutated data set. Decoder analysis included only bigrams which probability was greater than |±2SD| from the mean of the permutated bigram probability distribution (n=2185).
Identification of putative mouse size-related syllables
To identify candidate syllables whose usage might be correlated with mouse size, a linear model (ElasticNet with α=0.01) was fit to model mouse size-related features as a function of log-transformed, z-scored syllable usage data calculated per mouse. Size features included height, length, width and body area of the mouse as measured by the MoSeq algorithm from the recorded 3D videos. Each measure was calculated as the robust range per mouse. i.e. the differences between 95th and 5th percentile values of that measure per animal. Model weights were compared against shuffled data (n=1000 shuffled per measurement). To provide the most rigorous analysis, syllables with weights surpassing two-sided top 2.5 percentile of the shuffle weight distribution for any of the measurements were identified as correlating with mouse size. These syllables (n=8 for males, n=15 for females) were excluded from the decoder presented in Figure S4A,B, and from the variability analysis presented in Figure S4C,D.
Assessing the influence of size
To explore the relationship between mouse size and individual identity, we compared the degree of confusion between pairs of mice (as assessed via a classifier) with their size differences. To perform this analysis, we designed “held-out” decoders that excluded all sessions from a single mouse from the training data, and then used those sessions as the test data (decoder input features were behavioral syllables, and the overall design was as described above). This approach identified the individual mouse or mice that the classifier most confused with the held-out query mouse (Figures S4E and S4F). We also measured the size differences between individual mice, using the z-scored height, length, width and body area of the mouse as measured by the MoSeq algorithm from the recorded 3D videos. Each measure was calculated as the robust range per mouse. i.e. the differences between 95th and 5th percentile values of that measure per animal, and the overall size difference was determined by the Euclidian distance between measurement vectors for each pair of mice. In order to facilitate direct comparisons between classifier confusions and size, both datasets were normalized to a 0-to-1 scale. To allow for positive relationships, size distances were converted to size similarity such that similarity = 1-distance. Classification confusion and size similarity were then directly compared and fitted using a linear model as described in figure legend. Self-distances (both in size analysis and in the classification confusion analysis) were excluded from the final correlation analysis. As described above, and to allow evaluation of these results versus all other decoder results described in the manuscript, mice that had less than ten total behavioral sessions or fewer than three of the four estrous phases in the data were excluded from classification analysis and from all related analysis described here (2/16 of females and 2/16 males).
UMAP visualization
UMAP (Uniform Manifold Approximation and Projection) based dimensionality reduction was performed for visualization purposes only, with n_neighbors=8, min_dist=0.1, n_components=2 and metric = Euclidean. Syllable usages in all sessions were used and z-scored before UMAP analysis was applied.
Clustering analysis
Clustering was performed using a k-means algorithm applied on z-scored sessions-based syllable usage with 10 random seed initiations and 300 iterations. Clustering analysis was performed twice: i) with nclusters=4, the results of which were compared to labeling of sessions by phase, and ii) with nclusters=16, the results of which were compared to labeling of session by individual identity (using adjusted Rand index. 40 initial random seeds values were compared and maximum ARI is presented).
General linear model
A linear regression model (ElasticNet with α=0.01) was fit to model log-transformed, z-scored, session based, syllable usage for each syllable as a function of either): i) phase; ii) phase, with phase labels shuffled within a mouse (n=100); iii) phase, identity and their interaction; iv) phase and identity without interaction; v) mouse identity; vi) mouse identity, with shuffled identity labels (n=100). Coefficient of determination (R2) for each model are presented in Figure 3B as well as parameters for model iv. Performance of models iii,iv and v was compared using ANOVA for each syllable in order to explore the influence of adding phase information on explained variability.
Kullback-Leibler Divergence (KLD) analysis
Kullback-Leibler Divergence (KLD) analysis (relative entropy) was performed to calculate the pairwise dissimilarity of syllable usage distributions (P,Q) using the scipy.stats.entropy module such that .
For phase, syllable usage distributions were averaged per phase, per mouse. KLD between phases was then calculated for all possible pairwise combinations of phases within each mouse, and then averaged to give a single value indicating the overall dissimilarity of estrous phases within a single mouse. Dissimilarity of sessions within each individual (referred to as “intra-individual” across figures) was calculated as KLD for all possible pairwise combinations of sessions recorded for that individual mouse, and then averaged to give a single value indicating the overall dissimilarity of behavior across sessions for each single mouse. Dissimilarity of individuals (referred to as “inter-individual” across figures) was measured by calculating the pairwise KLDs of average syllable usage distribution of each mouse against all other mice, and then averaged to produce a single value for each mouse indicating its overall dissimilarity from all other mice. For “all sessions” condition presented across figures, KLD was calculated for each behavioral session against all other sessions (in all pairwise combinations) and then averaged per session, to give a single number indicating the dissimilarity of behavior observed in that session in comparison to all other sessions. For analysis of dynamics of variability presented in Figure 4A,B, the above was performed separately for each half of the experimental days (first 7 days – 1st week, versus second 7 days – 2nd week). For variability analysis presented in Figure S3C,D, measurements from the first week of female data was compared to those from the second week of male data. For analysis in Figure S3E,F, an independent experimental data was used controlling for mouse handling as described above. Analysis in Figure S4C,D excludes putative size-correlated syllables as described above. KLD values of self-distances (KLD=0) were removed from all analysis.
Coefficient of Variation (CV) analysis
Coefficient of variation analysis was performed to estimate the variation in usage of each syllable across phases and individuals such that . For phases, CV for each syllable was calculated across phases within each mouse (using the mean syllable usage across repetitions per phase), and then averaged across mice to give a single CV value per syllable. For intra-individual analysis, CV was calculated per syllable across repetition for each mouse, and then averaged across mice to give a single measurement per syllables. For inter-individual condition, CV for each syllable was calculated across mice, using the mean syllable usage across sessions per mouse. For “all sessions” analysis, CV for each syllable was calculated across all behavioral sessions. Details of specific dataset used for CV analysis in each figure is as detailed for KLD analysis above, and mentioned in the appropriate figure legends.
CV analysis shown in Figure S2F was calculated to test for the dynamic of syllable use throughout a single session. First, each 20 minute-long behavioral session was divided into five minute non-overlapping bins, and the syllable usage distribution was calculated for each bin. The CV across these bins was calculated for each syllable per session, and then averaged per estrous phase to give a single number indicating the variation in usage of a single syllable throughout the 20 minutes session for each phase.
Mutual information (MI) analysis
Mutual information was calculated to evaluate the information each syllable holds regarding the differences between phases or individuals using the sklearn.feature_selection.mutual_info_classif module, to evaluate MI between continuous data set (syllable usage) and discrete target (either phase or identity).
Quantification and statistical analysis
All quantification and statistical analysis were done in Python 3.6, using the following modules: Numpy42,43, Pandas44, Scikit-learn45, Scipy46, Statsmodels47, Seaborn43 and Matplotlib48. Specific details of statistical test, statistics, number of samples and p-values are described in the figure legends. Null hypothesis was rejected with α>0.05. When appropriate, p-values were adjusted for multiple comparisons using the Bonferroni method.
Supplementary Material
Highlights.
Spontaneous behavior of female mice is only negligibly affected by estrous state
Females and males exhibit strongly individualized patterns of exploration
Female spontaneous behavior is less variable than male behavior
Acknowledgments
SRD is supported by NIH grants U19NS113201, RF1AG073625, R01NS114020, the Brain Research Foundation, the Simons Collaboration on the Global Brain and the Simons Collaboration for Plasticity in the Aging Brain. DRL is supported by the Human Frontier Science Program fellowship LT000838/2020 and the Zuckerman STEM Leadership Program, and is an awardee of the Women’s Postdoctoral Career Development Award. Portions of this research were conducted on the O2 High Performance Compute Cluster at Harvard Medical School. Slide imaging was performed in the HMS/BCH center for neuroscience research #NS072030. We thank members of the Datta lab as well as Dr. Meital Oren-Suissa, Dr. Susana Q. Lima and Dr. Vanessa Ruta for useful comments on the manuscript.
Footnotes
Declaration of Interests
SRD sits on the scientific advisory boards of Neumora, Inc. and Gilgamesh Therapeutics.
Inclusion and Diversity
We worked to ensure sex balance in the selection of non-human subjects. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors of this paper self-identifies as a gender minority in their field of research. One or more of the authors of this paper self-identifies as a member of the LGBTQIA+ community. One or more of the authors of this paper received support from a program designed to increase minority representation in their field of research. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wiltschko AB, Johnson MJ, Iurilli G, Peterson RE, Katon JM, Pashkovski SL, Abraira VE, Adams RP, and Datta SR (2015). Mapping Sub-Second Structure in Mouse Behavior. Neuron 88, 1121–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin S, Gillis WF, Weinreb C, Zeine A, Jones SC, Robinson EM, Markowitz J, and Datta SR (2022). Characterizing the structure of mouse behavior using Motion Sequencing. arXiv preprint arXiv:2211.08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Markowitz JE, Gillis WF, Beron CC, Neufeld SQ, Robertson K, Bhagat ND, Peterson RE, Peterson E, Hyun M, and Linderman SW (2018). The striatum organizes 3D behavior via moment-to-moment action selection. Cell 174, 44–58. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiltschko AB, Tsukahara T, Zeine A, Anyoha R, Gillis WF, Markowitz JE, Peterson RE, Katon J, Johnson MJ, and Datta SR (2020). Revealing the structure of pharmacobehavioral space through motion sequencing. Nat Neurosci 23, 1433–1443. 10.1038/s41593-020-00706-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Markowitz JE, Gillis WF, Jay M, Wood J, Harris RW, Cieszkowski R, Scott R, Brann D, Koveal D, Kula T, et al. (2023). Spontaneous behaviour is structured by reinforcement without explicit reward. Nature. 10.1038/s41586-022-05611-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z, Roberson DP, Kotoda M, Boivin B, Bohnslav JP, González-Cano R, Yarmolinsky DA, Turnes BL, Wimalasena NK, Neufeld SQ, et al. (2022). Automated preclinical detection of mechanical pain hypersensitivity and analgesia. PAIN 163, 2326–2336. 10.1097/j.pain.0000000000002680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li C, Hou Y, Zhang J, Sui G, Du X, Licinio J, Wong M-L, and Yang Y (2019). AGRP neurons modulate fasting-induced anxiolytic effects. Translational Psychiatry 9, 111. 10.1038/s41398-019-0438-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burnett CJ, Li C, Webber E, Tsaousidou E, Xue Stephen Y., Brüning Jens C., and Krashes Michael J. (2016). Hunger-Driven Motivational State Competition. Neuron 92, 187–201. 10.1016/j.neuron.2016.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tinbergen N (1951). The Study of Instinct (Clarendon Press; ). [Google Scholar]
- 10.Freund J, Brandmaier AM, Lewejohann L, Kirste I, Kritzler M, Kruger A, Sachser N, Lindenberger U, and Kempermann G (2013). Emergence of individuality in genetically identical mice. Science 340, 756–759. 10.1126/science.1235294. [DOI] [PubMed] [Google Scholar]
- 11.Forkosh O, Karamihalev S, Roeh S, Alon U, Anpilov S, Touma C, Nussbaumer M, Flachskamm C, Kaplick PM, Shemesh Y, and Chen A (2019). Identity domains capture individual differences from across the behavioral repertoire. Nat Neurosci 22, 2023–2028. 10.1038/s41593-019-0516-y. [DOI] [PubMed] [Google Scholar]
- 12.Honegger K, and Bivort B.d. (2018). Stochasticity, individuality and behavior. Current Biology 28, R8–R12. 10.1016/j.cub.2017.11.058 [DOI] [PubMed] [Google Scholar]
- 13.Ajayi AF, and Akhigbe RE (2020). Staging of the estrous cycle and induction of estrus in experimental rodents: an update. Fertil Res Pract 6, 5. 10.1186/s40738-020-00074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLean AC, Valenzuela N, Fai S, and Bennett SAL (2012). Performing vaginal lavage, crystal violet staining, and vaginal cytological evaluation for mouse estrous cycle staging identification. J. Vis. Exp, e4389. 10.3791/4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Byers SL, Wiles MV, Dunn SL, and Taft RA (2012). Mouse estrous cycle identification tool and images. PLoS One 7, e35538. 10.1371/journal.pone.0035538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knoedler JR, Inoue S, Bayless DW, Yang T, Tantry A, Davis C-H, Leung NY, Parthasarathy S, Wang G, Alvarado M, et al. (2022). A functional cellular framework for sex and estrous cycle-dependent gene expression and behavior. Cell 0. 10.1016/j.cell.2021.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue S (2022). Neural basis for estrous cycle-dependent control of female behaviors. Neurosci. Res. 176, 1–8. 10.1016/j.neures.2021.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Jennings KJ, and de Lecea L (2020). Neural and Hormonal Control of Sexual Behavior. Endocrinology 161. 10.1210/endocr/bqaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutierrez-Castellanos N, Husain BF, Dias IC, and Lima SQ (2022). Neural and behavioral plasticity across the female reproductive cycle. Trends in Endocrinology & Metabolism. [DOI] [PubMed] [Google Scholar]
- 20.Chari T, Griswold S, Andrews NA, and Fagiolini M (2020). The Stage of the Estrus Cycle Is Critical for Interpretation of Female Mouse Social Interaction Behavior . Front. Behav. Neurosci. 14, 113. 10.3389/fnbeh.2020.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burke AW, and Broadhurst PL (1966). Behavioural Correlates of the Oestrous Cycle in the Rat. Nature 209, 223–224. 10.1038/209223a0. [DOI] [PubMed] [Google Scholar]
- 22.Lovick TA, and Zangrossi H Jr. (2021). Effect of Estrous Cycle on Behavior of Females in Rodent Tests of Anxiety. Front. Psychiatry 12, 711065. 10.3389/fpsyt.2021.711065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meziane H, Ouagazzal AM, Aubert L, Wietrzych M, and Krezel W (2007). Estrous cycle effects on behavior of C57BL/6J and BALB/cByJ female mice: implications for phenotyping strategies. Genes Brain Behav. 6, 192–200. 10.1111/j.1601-183X.2006.00249.x. [DOI] [PubMed] [Google Scholar]
- 24.Finger FW (1969). Estrus and general activity in the rat. Journal of Comparative and Physiological Psychology 68, 461–466. 10.1037/h0027490. [DOI] [PubMed] [Google Scholar]
- 25.Ogawa S, Chan J, Gustafsson J.-A.k., Korach KS, and Pfaff DW (2003). Estrogen Increases Locomotor Activity in Mice through Estrogen Receptor α: Specificity for the Type of Activity. Endocrinology 144, 230–239. 10.1210/en.2002-220519. [DOI] [PubMed] [Google Scholar]
- 26.Mullenix P (1981). Structure analysis of spontaneous behavior during the estrous cycle of the rat. Physiol. Behav. 27, 723–726. 10.1016/0031-9384(81)90246-8. [DOI] [PubMed] [Google Scholar]
- 27.Rocks D, Cham H, and Kundakovic M (2022). Why the estrous cycle matters for neuroscience. Biology of Sex Differences 13. 10.1186/s13293-022-00466-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koonce CJ, Walf AA, and Frye CA (2012). Type 1 5α-reductase may be required for estrous cycle changes in affective behaviors of female mice. Behavioural Brain Research 226, 376–380. 10.1016/j.bbr.2011.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krentzel AA, and Meitzen J (2018). Biological Sex, Estradiol and Striatal Medium Spiny Neuron Physiology: A Mini-Review. Front Cell Neurosci 12, 492. 10.3389/fncel.2018.00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zachry JE, Nolan SO, Brady LJ, Kelly SJ, Siciliano CA, and Calipari ES (2021). Sex differences in dopamine release regulation in the striatum. Neuropsychopharmacology 46, 491–499. 10.1038/s41386-020-00915-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller CK, Halbing AA, Patisaul HB, and Meitzen J (2021). Interactions of the estrous cycle, novelty, and light on female and male rat open field locomotor and anxiety-related behaviors. Physiology & Behavior 228, 113203. 10.1016/j.physbeh.2020.113203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scimonelli T, Marucco M, and Celis ME (1999). Age-related changes in grooming behavior and motor activity in female rats. Physiol. Behav. 66, 481–484. 10.1016/s0031-9384(98)00314-x. [DOI] [PubMed] [Google Scholar]
- 33.Shansky RM, and Murphy AZ (2021). Considering sex as a biological variable will require a global shift in science culture. Nat. Neurosci. 24, 457–464. 10.1038/s41593-021-00806-8. [DOI] [PubMed] [Google Scholar]
- 34.Beery AK, and Zucker I (2011). Sex bias in neuroscience and biomedical research. Neurosci Biobehav Rev 35, 565–572. 10.1016/j.neubiorev.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarthy MM, Arnold AP, Ball GF, Blaustein JD, and De Vries GJ (2012). Sex differences in the brain: the not so inconvenient truth. Journal of Neuroscience 32, 2241–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Becker JB, Prendergast BJ, and Liang JW (2016). Female rats are not more variable than male rats: a meta-analysis of neuroscience studies. Biol. Sex Differ. 7, 34. 10.1186/s13293-016-0087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prendergast BJ, Onishi KG, and Zucker I (2014). Female mice liberated for inclusion in neuroscience and biomedical research. Neurosci. Biobehav. Rev. 40, 1–5. 10.1016/j.neubiorev.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Beery AK (2018). Inclusion of females does not increase variability in rodent research studies. Curr Opin Behav Sci 23, 143–149. 10.1016/j.cobeha.2018.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaluve AM, Le JT, and Graham BM (2022). Female rodents are not more variable than male rodents: a meta-analysis of preclinical studies of fear and anxiety. Neurosci Biobehav Rev 143, 104962. 10.1016/j.neubiorev.2022.104962 [DOI] [PubMed] [Google Scholar]
- 40.Smarr B, and Kriegsfeld LJ (2022). Female mice exhibit less overall variance, with a higher proportion of structured variance, than males at multiple timescales of continuous body temperature and locomotive activity records. Biology of Sex Differences 13. 10.1186/s13293-022-00451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cora MC, Kooistra L, and Travlos G (2015). Vaginal Cytology of the Laboratory Rat and Mouse:Review and Criteria for the Staging of the Estrous Cycle Using Stained Vaginal Smears. Toxicologic Pathology 43, 776–793. 10.1177/0192623315570339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris CR, Millman KJ, Van Der Walt SJ, Gommers R, Virtanen P, Cournapeau D, Wieser E, Taylor J, Berg S, and Smith NJ (2020). Array programming with NumPy. Nature 585, 357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waskom M (2021). seaborn: statistical data visualization. J. Open Source Softw. 6, 3021. 10.21105/joss.03021. [DOI] [Google Scholar]
- 44.McKinney W, and Others (2010). Data structures for statistical computing in python. Proceedings of the 9th Python in Science Conference. conference.scipy.org. [Google Scholar]
- 45.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, and Dubourg V (2011). Scikit-learn: Machine learning in Python. the Journal of machine Learning research 12, 2825–2830. [Google Scholar]
- 46.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, and Bright J (2020). SciPy 1.0: fundamental algorithms for scientific computing in Python. Nature methods 17, 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seabold S, and Perktold J (2010). Statsmodels: Econometric and statistical modeling with python. In 61. (Austin, TX: ), pp. 10.25080. [Google Scholar]
- 48.Hunter JD (2007). Matplotlib: A 2D Graphics Environment. Computing in Science Engineering 9, 90–95. 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw behavioral videos have been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
The behavioral analysis code and Moseq software used to model and analyze the data is freely available to all academic researchers online: http://www.moseq4all.org/.
All original code has been deposited at Github and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical commercial assays | ||
| JorVet Dip Quick Stain | Fischer Scientific | NC9581034 |
| Black Polyethylene Tank (OFA) | US plastics | #14317 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | #000664 |
| Software and algorithms | ||
| Motion Sequencing (Moseq) | The Datta lab | http://www.moseq4all.org/ |
| OLYMPUS OlyVIA 2.9 | Olympus Software Imaging Solutions | https://www.olympus-sis.com/ |
| Python version 3.6 | Python Software Foundation | https://www.python.org |
| Numpy | Harris el al42 | https://numpy.org/ |
| Seaborn | Waskom et al43 | https://seaborn.pydata.org/ |
| Pandas | Mckinney et al44 | https://pandas.pydata.org/ |
| Scikit-learn | Pedregosaet al45 | https://scikit-learn.org/stable/ |
| Scipy | Virtanen et al46 | https://scipy.org/ |
| Statsmodels | Seabold et al47 | https://www.statsmodels.org/stable/index.html |
| Matplotlib | Hunter et al48 | https://matplotlib.org/ |
| Deposited data | ||
| Analyzed data and related scripts | This paper | https://github.com/dattalab/spontaneous-behavior-reflects-individuality-not-estrous |
| Post-MoSeq behavioral analysis | This paper | 10.5281/zenodo.7622958 |
| Raw behavioral videos | This paper | 10.5281/zenodo.7622958 |


