Purpose of review
Here, we review recent findings on the role of long noncoding RNAs (lncRNAs) in cardiovascular disease (CVD). In addition, we highlight some of the latest findings in lncRNA biology, providing an outlook for future avenues of lncRNA research in CVD.
Recent findings
Recent publications provide translational evidence from patient studies and animal models for the role of specific lncRNAs in CVD. The molecular effector mechanisms of these lncRNAs are diverse. Overall, cell-type selective modulation of gene expression is the largest common denominator. New methods, such as single-cell profiling and CRISPR/Cas9-screening, reveal additional novel mechanistic principles: For example, many lncRNAs establish RNA-based spatial compartments that concentrate effector proteins. Also, RNA modifications and splicing features can be determinants of lncRNA function.
Summary
lncRNA research is passing the stage of enumerating lncRNAs or recording simplified on-off expression switches. Mechanistic analyses are starting to reveal overarching principles of how lncRNAs can function. Exploring these principles with decisive genetic testing in vivo remains the ultimate test to discern how lncRNA loci, by RNA motifs or DNA elements, affect CVD pathophysiology.
Keywords: cardiovascular, disease, long noncoding RNA, splicing, transcription
INTRODUCTION
Cardiovascular disease (CVD) is the leading cause of death globally. The vast majority of CVD death is due to myocardial infarction and stroke [1]. Here, we review recent studies from the last 1.5 years on long noncoding RNAs (lncRNAs) in CVD that did not stop with cellular in vitro analyses but stepped forward to investigate the pathophysiological roles of lncRNAs in vivo. As the lncRNA field is quickly evolving, we also review novel conceptual approaches for studying molecular effector mechanisms of lncRNAs.
Box 1.
no caption available
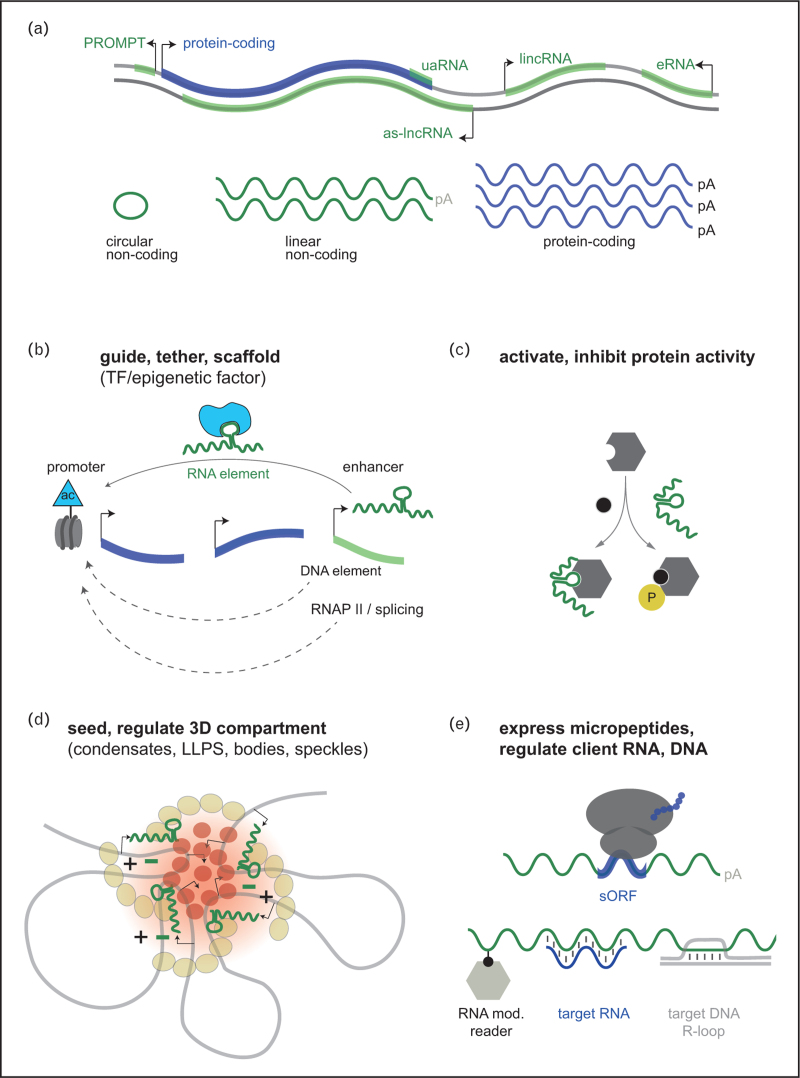
LncRNAs are a heterogeneous class of thousands of RNAs (Fig. 1a). Defining them is not trivial. Most commonly, lncRNAs refer to transcripts more than 200 nucleotides in length that do not appear to contain a protein-coding sequence and do not belong to ribosomal, transfer or small nuclear/nucleolar/microRNA-like species [2▪▪]. LncRNAs are predominantly transcribed at low levels by RNA polymerase II from epigenetically sculptured start sites. They are frequently encoded interleaved relative to protein-coding genes (Fig. 1a). An emerging view is that protein-coding genes and lncRNAs potentially represent a larger information continuum because many lncRNAs can regulate protein-coding genes [2▪▪]. For regulating genes, lncRNAs employ many effector mechanisms. They can tether, scaffold and modulate transcription regulatory complexes (Fig. 1b). Also the genomic orientation and relation to neighbouring genes may give first hints at the possible targets of a given lncRNA. On the basis of this, the large group of lncRNAs can be divided into subgroups, comprising lincRNAs (long intergenic noncoding RNAs), as-lncRNAs (antisense lncRNAs), eRNAs (enhancer RNAs), PROMPT (promoter-upstream transcripts) and uaRNA (noncoding RNA from 3’UTR of protein-coding genes) (Fig. 1a). The picture gets even more complex because, apart from linear lncRNAs, also circular forms of RNA (circRNAs) may be produced. CircRNAs may also be functional, as is reviewed separately in this issue.
FIGURE 1.
Classes and conceptual functions of lncRNAs. (a) Major classes of lncRNAs based on genomic orientation and relation to neighbouring genes. lincRNA (long intergenic noncoding RNA), as-lncRNA (antisense lncRNA), eRNA (enhancer RNA), PROMPT (promoter-upstream transcripts), uaRNA (noncoding RNA from 3’UTR of a protein-coding gene). (b--e) Major functions of lncRNAs. (b) Stable lincRNAs or more transient enhancer-like eRNAs as transcription and epigenetic regulators in cis and trans. Also, DNA elements or the processes of transcription and splicing can be functional determinants. (c) Role of low-abundance lncRNAs in seeding and maintaining different types of 3D-compartments. Shown is a nuclear condensate (nuclear body/ liquid-liquid phase separated structure) with IDRs/prion-like protein domains of an RBP and/or electrostatics (-/+) as building principles. (d) lncRNAs can affect protein activity. (e) A variety of other functions: lncRNAs may express peptides from previously undetected small ORFs, recruit effectors via RNA modifications, or bind other nucleic acids like RNAs (e.g. affecting stability, splicing or acting as competing endogenous RNA) or DNA (e.g. leading to R-loop formation and influencing transcription, replication, chromosome topology, damage repair). Ac, acetylation, example of activating histone tail posttranscriptional modification; IDR, intrinsically disordered domain; LLPS, liquid-liquid phase separated structure; P, phosphorylation as an example of protein activity change; TF, transcription factor.
Over the last years, several lncRNAs from these diverse classes have been found to be functional in physiology and disease [3▪▪]. In the cardiovascular system, dozens of lncRNAs have become known to affect heart and blood vessel development and pathophysiology, including ANRIL, MALAT, MIAT, Braveheart, Upperhand, Myheart, H19, Meg3, HOTAIR, Kcnq1ot1, Fendrr, Chaer, CARL, SENCR, SMILR or CARMEN, to name a few [4].
FUNCTIONS OF RECENTLY STUDIED LONG NONCODING RNAS IN CARDIOVASCULAR DISEASE
Several new lncRNAs have recently been functionally studied in a cardiovascular context in vivo (Table 1). Some of these lncRNAs affect the embryonic development of the heart and the vasculature. Others are expressed in cardiomyocytes, vascular endothelial cells or smooth muscle cells (VSMC) and modulate cell contractility, cell differentiation status, cell growth and cell survival (Table 1). Targeted gene knockouts and more fine-grained genetic manipulations were performed for most of the involved lncRNA loci, and effects on CVD were studied in animal CVD models (Table 1). Thereby, it was found that some cardiovascular lncRNAs aggravated, and others ameliorated CVD phenotypes, including atherosclerosis, myocardial pressure overload, infarction and congenital heart disease, as detailed in Table 1.
Table 1.
Recent in vivo studies on long noncoding RNAs in cardiovascular tissues
| lncRNA | Classification | Disease context | Species | Cell type | Cellular phenotype in vitro | Molecular function/mechanism in vitro | Effects in pathology/physiology models in vivo | Ref. |
| Caren | Pleiotropic translation inhibitor | Heart failure | Mouse | CM | CM hypertrophy potentially related to mitochondrial function | Primary effector unclear; Caren inhibits Hint 1 mRNA translation; mitochondrial respiratory function is decreased in KO; increased numbers of mitochondria in OE; Hint1 impairs mitochondrial respiration | Disease model TAC: more severe heart failure in KO; CAG-promoter-driven or CM-specific OE protects from hypertrophy; Hint1 heterozygous mutant with less severe heart defects | [17▪] |
| CARMN | TF tethering | Atherosclerosis | Mouse, rat, human | CM VSMC | Pro-differentiative and antiproliferative | Down in synthetic SMCs of atherosclerotic lesions (ScRNAseq); transactivates MYOCD/SRF complex during activation of target genes; in parallel, enhancer activity on the overlapping MIR143/145 locus | Disease model: conditional SMC-specific mouse knockout (Myh11-CreERT2) during left carotid artery ligation; adenoviral OE in the rat during left carotid artery balloon injury; CARMN protects from atherosclerotic neointima growth | [5] |
| CARMN | TF tethering | Atherosclerosis | Mouse and human | VSMC | Pro-proliferative and pro-migrative, pro-differentiation | Decreased in atherosclerotic plaque; CARMN interacts with SRF protein and increases SRF occupancy on promoters of SRF target genes | Disease model: systemic KD with ASOs in Ldlr–/– shows reduced atherosclerosis, arguing that WT CARMN is proatherogenic (which contrasts (5)) | [6] |
| CARMN | eRNA or host of miR | Atherosclerosis | Mouse and human | VSMC | Pro-differentiative and antiproliferative | Putatively enhances the expression of miR-143/14 (overlapping in the CARMN locus) | Disease model: liver AAV-PCSK9-induced hyperlipidemia-driven atherosclerosis; GapmeR and CRISPR/Cas9 KO of exon 1 cause more vulnerable plaques | [7] |
| circ-INSR | Unclear, affecting protein function | Cardiomyopathy | Human, mouse, rat | CM | Required for metabolism and survival after DNA replicative stress | Down during heart failure; BRCA1 is required for circRNA levels; effector mechanism potentially involves single-strand binding protein SSBP1 | Cardiotropic adenoviral OE of circRNA improves cardiac function in a model of doxorubicin-induced cardiotoxicity in mouse | [18] |
| CIRKIL | Regulator of protein in cytoplasm | Ischemia/reperfusion injury | Mouse and human | CM | Pro-apoptotic | CIRKIL expression increases in ischemic hearts; binds KU70 protein and inhibits nuclear import of KU70; unclear if cellular effects via DSB control or BAX cell death pathway | Disease models: (i) dox-inducible transgenic OE worsens heart function and CM damage and infarction area in I/R mice, which Ku70 OE antagonizes; (ii) CRISPR/Cas9 KO cardioprotection in ligation model | [16] |
| Ckip-1 3’ UTR | Ckip-1 has dual role as mRNA and as uaRNA | Cardiac hypertrophy and heart failure | Human and mouse | CM | Promotes mitochondrial respiration, reduces lipid accumulation | 3’UTR up in failing hearts; OE increases myocardial metabolism and ATP production independent of Ckip-1; 3’UTR interacts with Let-7f RNA and potentially affects AMPK-dependent metabolism |
Disease model: transgenic OE in the heart (α-MHC or TnT promoter) is cardioprotective during hypertrophy in TAC and confers partial rescue in a Ckip-1 KO mouse; Ckip-1 itself is a known cardioprotective gene | [23] |
| CPhar | TF tethering | Cardiac remodelling | Mouse | CM | Stimulates CM proliferation and size, antagonizes OGD/R- driven apoptosis | CPhar is up in the heart after endurance exercise and TAC-induced cardiac hypertrophy; sequesters C/EBPβ protein and inhibits target genes activation (e.g. ATF7) | Disease model: cardiotropic adenoviral OE in IR/I is cardioprotective; physiology model: cardiotropic KD with shRNA shows that the lncRNA is important for exercise-induced cardiac growth | [8] |
| H19 | Regulator of protein in cytoplasm and ceRNA for microRNAs | Progressive adult congenital heart dysfunction in Beckwith-Wiedemann syndrome | Mouse | EC | Loss of H19 leads to reactivation of mesenchymal genes in EC (endMT-like) | H19 lncRNA binds Let7 microRNAs and limits their bioavailability | Disease models: (i) Maternal deletion of imprinting control region at H19/Igf2 results in biallelic Igf2 and reduced H19 expression; progressive fibrosis and arterial dilation/ stiffening in adults are due to H19 loss; (ii) CRISPR/Cas9 deletion of Let-7 binding sites in H19 also causes heart fibrosis and cardiomegaly | [17▪] |
| HBL1 | Dual role as tether for epigenetic regulators and as miR suppressor | Cardiac development | Human and primates | Early CM lineage | lncRNA restrains the efficiency of cardiac differentiation by inhibiting cardiogenic genes in early mesoderm | lncRNA interacts with EED and JARID2 proteins, and these phenocopy HBL1; HBL1 targets JARID to cardiogenic and cardiovascular gene promoters for PRC2/EED complex to epigenetically repress them | CRISPR CAS9 KO of EED or JARID2 and in human stem cells and iPSCs and HBL1−/- iPSCs; disease link indirect by the genetic association of cardiogenic transcription factors (e.g., HAND2) with congenital heart diseases | [22] |
| lncExACT1 | Encoded as asRNA and functioning as eRNA | Cardiac hypertrophy and heart failure | Human, mouse, zebrafish | CM | KD is pro-proliferative and anti-apoptotic; OE increases CM size but not proliferation |
Down in exercised hearts but increased during heart failure; lincExACT1 transcriptionally activates neighbouring DCHS2 gene; DCHS2 phenocopies the lncRNA effects, DCHS2 KD blocks the effects of lncRNA OE | Disease models: (i) Gapmers protect against cardiac fibrosis during TAC and against myocardial I/RI jury; (ii) OE of human DCHS2 in CM leads to scarring and reduced proliferation in zebrafish heart regeneration; (iii) CRISPR/Cas9 KO in mouse heart with cardiotropic gRNAs causes cardiac hypertrophy and CM overproliferation; (iv) cardiotropic OE causes pathological hypertrophy | [11▪] |
| MIAT | Pleiotropic TF-regulator and trans-acting eRNA | Atherosclerosis | Human, mouse, pigs | VSMC, MΦ | Required for VSMC proliferation and survival and transdifferentiation of MΦ | Up in atherosclerosis; MIAT binds ELK1 protein in SMC and NFKB in macrophages and has predicted target sites in KLF4 promoter; MIAT affects oxLDL-induced KLF4 transcription | Disease models: (i) Miat KO during incomplete ligation and cuff placement in Apoe–/– mice; (ii) carotid ligation in Miat–/–Apoe–/– double KO mice; Knockout shows decreased SMC pathology in plaques, but increased fibrous cap thinning | [20▪] |
| OIP5-AS1 (Cyrano) | Unknown effector, maybe functioning as ceRNA | Cardiac hypertrophy and heart failure | Human and mouse | CM | Effector mechanism not clear | No effects on neighbouring genes; transcriptomic changes only under pressure overload; effector mechanism not clear | Disease model: knockout of locus is viable and cardioprotective in females, with more severe heart failure during TAC | [22] |
| Platr4 | TF scaffold | Cardiac development | Mouse | CM | Differentiation of cardiac-mesoderm-lineage | Platr4 interacts with the Hippo pathway effector TFs Yap and Tead4 to regulate downstream targets (e.g. Ctgf) | Physiology model: Platr4 is required for heart development; KO shows myocardial atrophy and mucinous degeneration of valves | [10] |
| RP11-728F11.4 | TF inhibiting | Atherosclerosis | Human and mouse | MΦ | Increases cholesterol uptake and inflammatory signalling via stimulating FXYD6 | Transcriptionally activates FXYD6 gene, potentially by binding FXYD6 mRNA and restraining the EWSR1 repressor | Disease model: pro-atherosclerotic, as deduced from hepatic and circulating dyslipidaemia and inflammatory signalling after systemic lentiviral OE in Apoe−/− background | [9] |
| TPRG1-AS1 | Regulator of protein in cytoplasm | Atherosclerosis | Human and mouse | VSMC | Negatively regulates transwell migration | Up in atherosclerotic plaques; lncRNA interacts with MYH9 protein and negatively affects MYH9 protein degradation, thereby impairing cytoskeletal function | Disease models: antiatherosclerotic, systemic adenoviral OE in rat carotid balloon injury model: VSMC-specific transgenic OE in carotid artery wire injury model or an Apoe-/- background. | [15] |
| Trdn-as | Encoded as asRNA, and affecting splicing of host RNA | Arrhythmia and heart failure | Human and mouse | CM | Modulates sarcoplasmic calcium release channels and, thus, contraction and CM beating | Down in failing hearts; lncRNA recruits SRSF1/10 splicing factors to the Trdn host premRNA; activates splicing of the major MT1 isoform (leading to normal levels of Triadin protein, a known regulator of Ca2+ handling). | Physiology model: Trdn-as maintains cardiac function, rhythmia, and exercise capacity resulting in premature death in exon2/3 KO mouse. Triadin KO also with cardiac arrhythmia | [13▪] |
| Veal2 VEAL2 | Regulator of protein enzyme activity | Diabetic retinopathy | Zebrafish and human | EC | Maintains EC permeability and angiogenesis independently of apoptosis effects | Up in diabetic retinopathy (also useful as a blood biomarker); Veal2/VEAL2 bind allosteric region in PRKCB2 and inhibit kinase activity; lncRNA motif is essential for the rescue of Veal2 mutant; VEAL2 does not impact the expression of neighbouring genes within 600kb | Physiology models: (i) Veal2 full KO shows essential function; (ii) blood vessel integrity defects leading to cranial hemorrhage in small 8bp mutant; (iii) lncRNA functions via restraining PRKCB2 activity in vivo (rescue with kinase inhibitor); (iv) human VEAL2 complements zebrafish mutant | [25▪▪] |
| ZNF593-AS | Affecting RNA level or stability | Cardiac hypertrophy and heart failure | Human and mouse | CM | Improves CM contractility (potentially by RYR2 in its known role of Ca2+ release channel from ER) | Down in DCM; 5’ motifs in lncRNA scaffold RYR2 mRNA and HNPNPC protein in the cytoplasm and increase RYR2 RNA level and RYR2 protein levels | Disease model: cardioprotective as deduced from systemic KD with Gapmers or adenoviral expression under troponin T promoter in transverse aortic constriction (TAC)-induced dilated cardiomyopathy | [14] |
List of reports that functionally studied long noncoding RNAs (linear and circular) in cardiovascular physiology or disease using in vivo modelling. The focus is on lncRNAs (alphabetically ordered) functioning in cardiomyocytes or cells of the vascular walls (EC, VSMC, macrophages). AAV, adeno-associated virus; ASO, antisense oligonucleotide; CM, cardiomyocyte; EC, vascular endothelial cell; IR/I, ischemia-reperfusion injury; KD, knockdown; KO, knockout; MΦ, macrophages; miR, microRNA; OE, overexpression; OGD/R, Oxygen glucose deprivation and reperfusion; TAC, Transverse Aortic Constriction; TF, transcription factor; UTR, untranslated region of mRNA; VSMC, vascular smooth muscle cell; WT, wildtype.
In the following, we describe the molecular functions of these recently identified cardiovascular lncRNAs. We group them by type of molecular effector mechanism to give an appreciation of the breadth and complexity of possible functions.
lncRNAs regulate transcription of target genes
The range of molecular effector mechanisms of lncRNAs is broad and cardiovascular lncRNAs are no exception. A recurrent theme is that lncRNAs guide, scaffold and regulate transcription factors or epigenetic regulatory complexes (Table 1, Fig. 1b). For example, CARMN, which is known for recruiting the PRC2 complex in cardiomyocytes, was recently found to transactivate the myocardin/SRF master regulator of vascular SMC differentiation. Thereby, CARMN maintained the contractile SMC state, and this function protected blood vessels from atherosclerotic neointima growth [5–7]. Another example is the cardiac CPhar, which sequestered the transcription regulator C/EBPβ. By inhibiting C/EBPβ from repressing targets such as ATF7, CPhar stimulated cardiomyocyte proliferation [8]. Conversely, the lncRNA RP11–728F11.4 turned out to be pro-atherosclerotic by preventing EWSR1 from repressing transcription in macrophages [9]. In a different study, the lncRNA Platr4 stimulated the Hippo pathway output transcription factors Yap1 and Tead4 during cardiogenic differentiation. Interestingly, Platr4-/- knockout mice developed myocardial and valve degeneration and sudden heart failure [10]. Within the same pathway, lncExACT1 exhibited enhancer activity towards its neighbouring gene, the protocadherin DCHS2, a regulator of Hippo signalling. Thereby, lncExACT1 protected against cardiac fibrosis and dysfunction during pressure-overload and dampened myocardial ischemia/reperfusion injury [11▪]. The last two reports merit specific attention in light of a large body of earlier work on the Hippo organ size control pathway in cardiovascular function and response to injury [12].
lncRNAs regulate splicing and mRNA stability
Apart from transcriptional regulation, lncRNAs also often control splicing and target mRNA stability. These processes are more complex to detect and quantify. For example, Trdn-as, stemming from a locus linked to heart arrhythmia, recruited the splicing factors SRSF1/10 to Trdn. Thereby, Trdn-as contributed to the efficient splicing of Triadin, and Triadin promoted cardiac function and exercise capacity [13▪]. In a different study, another asRNA exhibited a different function: ZNF593-AS scaffolded RYR2 mRNA and HNPNPC and increased the translation to RYR2 protein. By increasing RYR2 levels, ZNF593-AS beneficially influenced cardiac excitation-contraction coupling in cardiomyocytes in dilated cardiomyopathy [14].
lncRNAs regulate protein translation and transport
lncRNAs function not only in the nucleus but can also bind proteins in the cytoplasm: Cardiovascular TPRG1-AS1 accelerated the turnover of the cytoskeletal motor-protein MYH9 in VSMCs and curbed atherosclerotic neointima growth [15]. In another study, CIRKIL inhibited the nuclear import of Ku70, with potential effects on double-strand break control and cell death pathways. In disease models, CIRKIL worsened cardiomyocyte damage during myocardial infarction [16]. Loss of another well known lncRNA, H19, led to progressive heart fibrosis, heart failure and vascular abnormalities, as typically observed in patients with the Beckwith–Wiedemann syndrome. The authors mechanistically implicated H19 in this disease because H19 silenced mesenchymal genes in the endothelium, affected TGFβ-signalling and antagonized Mirlet7 microRNAs [17▪]. Finally, a circRNA, circ-INSR, was cardioprotective by prosurvival signalling during replicative stress in cells [18]. Whether other cytosolic or nuclear effector molecules exist for these four lncRNAs remains an open question.
lncRNAs regulate signalling pathways
In other cases, the function of the lncRNA is so pleiotropic that it is hard to pinpoint a single causative effector: For example, Caren knockout mice suffered heart failure during pressure overload. Mechanistically, Caren blocked the translation of the pleiotropic tumour suppressor Hint1, and engaged with other unknown factors to control mitochondrial respiration in cardiomyocytes [19▪]. A similarly complex lncRNA is MIAT. MIAT promoted pathological VSMC proliferation and transdifferentiation, which accelerated atherosclerotic plaque growth and vulnerability. This effect might involve NFKB pathway activation and KLF4 transcriptional induction [20▪]. As MIAT also promotes tumorigenesis by binding AEG-1, a protein that affects diverse pathways [21], it is possible that NFKB signalling is a central lncRNA effector across diseases and tissues. Finally, no clear effector mechanism was defined for OIP5-AS1 during its female-specific cardioprotective role [22].
lncRNAs can exhibit more than one role
lncRNA loci sometimes exhibit more than one role: For example, HBL1 targets JARID2 to repress cardiogenic gene promoters via PRC2. Independently, HBL1 also restrains miR-1's from targeting JARID[19▪]. Thus, HBL1 fine-tunes cardiogenesis twofold in the nucleus and the cytoplasm. HBL1 target genes are, thereby, relevant as possible genetic causes of congenital heart diseases [23]. A similarly complex scenario happens at the Ckip-1 locus. There, the 3’UTR of Ckip-1 mRNA takes on a life of its own as uaRNA (Fig. 1a) and sequesters Let-7f microRNAs. By antagonizing Let-7f, the Ckip-1 3’UTR activates cardioprotective metabolic remodelling [24]. These scenarios highlight the complexity of lncRNAs and show that care should be taken when attributing phenotypes to a single molecular effector mechanism (Table 1).
lncRNAs bind enzymes and regulate their activity
lncRNAs may also serve as selective regulators of protein enzyme activity, which speaks for specific structural interactions with clients (Fig. 1c) and goes far beyond simply tethering or scaffolding interacting partners (Fig. 1b): An instructive example is Veal2, which was found to be essential for the endothelial barrier function and normal vasculogenesis. This lncRNA inhibits the kinase activity of Prkcbb [25▪▪]. Specifically, the authors established that Veal2 inhibited lipid activators from access to an allosteric stretch in Prkcbb protein. To make this argument, the authors also provided corroborating structural insights from molecular dynamic simulations [25▪▪]. Astonishingly, this enzymatic regulation was conserved in humans, despite the lack of conservation in VEAL2 sequence. Finally, by rescuing the vascular defects of Veal2 mutants with chemical Prkcbb inhibitors, the study confirmed this effector mechanism in vivo. Moreover, reintroducing the lncRNA in trans into a lncRNA mutant provided formal evidence that the RNA was the functional moiety in vivo. Thus, at several levels, this work is a role model study. Importantly, from a translational perspective, re-expressing VEAL2 is a therapeutic avenue because VEAL2 strengthens cell-cell junctions and endothelial permeability in diabetic retinopathy [25▪▪].
Across the reviewed studies (Table 1), cardiovascular lncRNAs can bind DNA, RNA or proteins and affect many different molecular pathways. This makes it difficult to derive unifying features of lncRNA function. Also, given the paucity of bioinformatic methods to systematically predict effector mechanisms for lncRNAs, studying lncRNA loci remains often more complex than studying protein-coding genes. For this reason, it is important to consider potentially overarching concepts that can sharpen new experimental hypotheses and streamline the functional analysis of new lncRNAs, as discussed in the following two chapters.
NEW INSIGHTS FROM GENOME-WIDE STUDIES OF LONG NONCODING RNAs
Important new conceptual insights in lncRNA effector mechanisms have recently been obtained from new single-cell technologies and from CRISPR/Cas9-based genome-wide screens. They provide novel perspectives on how noncoding risk loci and lncRNAs expressed from these loci may be prioritized and functionally studied in CVD (Table 2).
Table 2.
Emerging concepts for long noncoding RNA functionality
| Emerging concept for lncRNA function | Specific lncRNA function | Ref. | |
| New insights from genome-wide studies of lncRNAs | Single-cell exploration of lncRNAs from enhancers and of noncoding risk variants from GWAS | - Single cell sequencing captures rare cell subtypes relevant to (CVD) pathophysiology. - Single cell eQTL, chromatin-accessibility caQTL mapping and allele-specific analysis are becoming feasible on cohort/population-scale. - Single cell profiling allows better than bulk analysis to pinpoint enhancer-target pairs and causal noncoding risk SNPs (e.g. lying in regulatory DNA elements), relevant cell types and contexts, and likely effector genes of noncoding risk SNPs. - Tiled multiplexed/combinatorial CRISPRi screening identifies regulatory regions in noncoding DNA and crosstalk between multiple enhancers. |
[26▪,27▪▪,28–31,32▪,33▪▪] |
| Unique features of lncRNAs compared with mRNAs | - The transcription burst frequency of lncRNAs is smaller than that of mRNAs, which contributes to higher variability in lncRNA abundance. - Often lncRNAs are expressed with < 1 burst per day. |
[34▪] | |
| Conservation and function of lncRNAs are not so directly related | - A similar association of conserved and nonconserved lincRNAs with cardiometabolic traits. - On the genomic scale, close to zero strong purifying selection in lncRNAs. - LncRNA function often lies in short conserved RNA features (possibly structural). |
[33▪▪,35–37] | |
| New insight on lncRNA numbers | - Discordance in lncRNAs numbers across databases; eRNAs often not included in databases. - Expected number of lncRNA transcripts is higher (400000) than number of lncRNA genes. - LncRNA exons amount to only 2.3% of the genome (similar to exons of protein-coding gene). - Unexpectedly many proteins (20% genome-wide) could be RNA-binding. |
[2▪▪,38▪▪] | |
| Expectations of lncRNA functionality on a genomic scale | - Minority (0.2%) of lncRNAs is essential for core cell functions (e.g. cell cycle control). - The hit rate for essential roles in genomic screens is 16x lower than for protein-coding genes. - Modulatory pro-differentiative roles of lncRNAs are often unique to a single/few cell types. |
[40▪▪,41] | |
| new concepts of lncRNA function with potential relevance to the experimental design in studying CVD | lncRNA-dependent spatial compartments and nuclear scaffolding | - Nascent RNA (primarily noncoding) bridges to an insoluble nuclear protein scaffold to maintain active expression states (by resisting inherent chromatin condensation). | [42] |
| nonstoichiometric amplification of lncRNA function | - A major function of lncRNAs is to seed and control RNA-based 3D compartments (also called bodies/condensates/speckles/paraspeckles/granules) in the nucleus and cytoplasm (affecting chromatin looping, transcription, splicing, polyadenylation). - LncRNA-based nuclear infrastructures serve to concentrate and spatially organize diffusible effector protein show different condensates and scaffolds relate to each other is not understood. |
[4,30] | |
| - LncRNAs are multivalent (e.g. multiple RBP binding motifs). - RBP protein features drive macro-assemblies (e.g. prion-like domains or IDRs), which can result in liquid-liquid phase separation (LLPS) and local reaction centres). |
[44▪▪] | ||
| - Transcription initiation (low RNA abundance) stimulates coactivator/RNAP II condensates, while RNA bursts during elongation (high RNA abundance) dissolve condensates. - Among others, this is a model of why regulatory lncRNA can be functional while low-abundant. |
[45▪] | ||
| lncRNA splicing can be the determining effector mechanism for lncRNA loci | - Programmed intron-retention drives nuclear retention of RNA. - Programmed intron-retention can control mRNA levels and indirectly impact translation rate. - 3‘UTR of mRNA acts as noncoding RNA (uaRNA) independent of mRNA. - Many more RNA isoforms of lncRNAs than thought, also among circRNAs. - RNA processing / RBP loading enhances lncRNA function (e.g. higher eRNA potency). |
[46–48] | |
| Functions of posttranscriptional modifications in RNA | - >100 RNA modifications known, a majority of which are functionally not understood. - m6A is common and can affect the splicing (and function) of lncRNAs. |
[49–51] | |
| Noncoding RNAs can express micropeptides | - LncRNAs can produce small peptides. - Around 7000 small ORFsare predicted genome-wide, only 10% of which are already quantifiable by peptidomics. |
[52–54] |
List of currently discussed mechanisms of how lncRNA can function, many of which still need to be specifically explored in the cardiovascular context.
eRNA, enhancer RNA; IDR, intrinsically disordered domain in a protein; PAS, promoter antisense RNA; RBP, RNA-binding protein.
Single-cell exploration of long noncoding RNAs from enhancers and of noncoding risk variants
The majority of genetic variation of disease risk identified in genome-wide association studies lies in the noncoding sequence space, most of which is transcribed at some point. Moreover, in genome-wide multi-tissue profiling, hundreds of lncRNA loci were associated with diseases/traits with a higher likelihood than the nearest protein-coding genes [26▪]. Nevertheless, it has been hard to predict or systematically test the function of disease-associated noncoding loci. Towards this goal, new approaches based on integrative single-cell methods are taking the lead in their systematic functional annotation: First, they aim to prioritize potentially causal noncoding disease risk SNPs by their regulatory potential. For example, many noncoding loci correspond to cis-regulatory elements, including enhancers expressing lncRNAs (eRNAs, Fig. 1b). Second, studies attempt to identify which cell type is relevant for a given risk SNP. Third, they try to unveil the target promoters of risk SNPs. Relevant potent approaches integrating single-cell transcriptomics and single-cell epigenomics data have recently been published. For example, studies have employed chromatin co-accessibility mapping of enhancers and their putative target promoters with single-cell/single-nuclear ATAC-seq to pinpoint CVD-relevant noncoding RNAs and their cellular context [27▪▪,28,29]. Similarly, allelic expression analysis in single cells is a powerful technology [30]. Moreover, regulatory gene network computation and machine learning helped infer the biological context for novel candidate CVD-driving target genes. Further fruitful approaches involved population-scale single-cell profiling and Mendelian randomization [31].
In parallel to correlative approaches, the function of noncoding risk loci has been tested experimentally with new systematic CRISPR/Cas9-based screening technologies: For example, it was possible to pinpoint regulatory DNA elements by tiled and multiplexed CRISPRi screening over noncoding loci that were hundreds of kilobases in size. The key to success was to systematically assay enhancer activity in these noncoding regions by molecular single-cell readouts, which can involve FlowFISH [32▪] or high-throughput sequencing [33▪▪].
For the cardiovascular field, the studies mentioned above revealed new cell-type--disease phenotype relations (e.g. in fibromyocytes in atherosclerosis), new lncRNAs with potential functionality in CVD and maps of epistatic interaction networks among CVD-relevant enhancers. Not last, millions of enhancer-gene interactions are now becoming known, many of which would have been invisible in bulk tissue analysis.
Unique features of long noncoding RNAs compared with mRNAs
Many lncRNAs are expressed at lower levels than mRNAs, show less well defined 3’-end processing and can stem from bidirectional promoters (eRNAs, PROMPTs/uaRNAs) (Fig. 1A). Recent single-cell sequencing and allele-specific measurements carved out further unique features that allowed new mechanistic insights: although protein-coding transcription occurs typically in periodic bursts (lasting 1–10 min and yielding 1–100 mRNA molecules per burst), lncRNAs have four-fold lower burst frequency and two-fold smaller burst size. One-third of lncRNAs burst less than once per day per allele. This feature has been directly linked to the typically higher cell-to-cell expression variability of lncRNAs compared with level-matched mRNAs [34▪]. This finding is relevant in the biomedical context, for example when using lncRNAs as biomarkers. These new methods may also allow researchers to address how the antisense orientation of an asRNA affects transcription initiation or elongation on its sense target gene. This task is relevant because several cardiovascular lncRNAs are encoded in antisense configuration to protein-coding genes, but there is no robust framework yet for testing their role (Table 1).
Conservation of long noncoding RNAs and impact for functional testing
Genetic conservation between species is often an indicator of biological function. However, lncRNAs evolve fast, and recent evolutionary studies confirmed that, globally, there is close to zero purifying selection against point mutations in lncRNA loci [35]. Also, nonconserved lincRNAs have a similar trend of association with cardiometabolic traits as conserved lincRNAs [33▪▪]. These findings suggest that simplified homology-based prioritization of conserved lncRNAs can skew which lncRNAs become the subject of study in disease modelling [36]. However, it is also evident that the ‘grammar’ of predicting lncRNA function from its sequence still needs improvement. Specifically, negative evidence for sequence conservation does not preclude the conservation of functionality in lncRNAs. Instead, one emerging view is that lncRNAs do not have recognizable orthologs across large species distances, partly because functionality may be due to short and scattered sequence motifs or hard-to-identify structural or topological features. Several technical approaches are evolving to identify such RNA motifs [37], and these may change how future studies evaluate lncRNA evolution and function.
New insight on numbers of long noncoding RNA genes and transcripts: is there an end in sight?
RNA sequencing has shown that up to 93% of the human genome is transcribed at some point, while protein-coding sequences account for only 1.5%. These numbers alone do, however, not mean that RNAs are much more important regulators than proteins [38▪▪]. Supporting this notion, the majority (90%) of all cellular RNAPII-dependent transcription stems from uncontrolled (spurious) starts at nucleosome-free foci and not from lncRNA genes. In contrast, lncRNA genes amount to only 2.3% of the human genome, which is on par with protein-coding genes [38▪▪].
Nevertheless, the number of lncRNA transcripts may still be more extensive than expected: Current databases list 107 039 (https://lncipedia.org) or 173 112 lncRNA transcripts (NONCODE). Moreover, experts estimate that up to 400 000 different lncRNA transcripts could exist when including the class of eRNAs [2▪▪] (Fig. 1a). Similarly, over the years, the number of proteins annotated as RBP in our genome increased to 4257 (amounting to ∼20% of the human proteome, which is a surprisingly large fraction) [39]. Although it is not predictable how many of these would engage noncoding RNA, the large numbers of RBPs and lncRNAs show the scale of work ahead.
Expectations of long noncoding RNA functionality on a genomic scale
Recent genome-scale functional screens give first hints about how many lncRNAs might eventually be functional, which is an important consideration also in a translational dimension: Recent comparative genome-wide CRISPR screening on lncRNAs and mRNAs showed that surprisingly few lncRNAs (0.2%) were essential for core cell functions such as cell cycle, cell growth or apoptosis. This fraction is 16-fold smaller than the hit rate of protein-coding genes [40▪▪]. Furthermore, not a single lncRNA was essential for robust cell growth across more cell types. This observation emphasizes the need to care about context-specificity when assaying lncRNAs in disease models [41] (Table 1). About 3.8% (200) of all lncRNAs scored as essential for differentiation in a typical stem cell based culture system (which is still two-fold less than protein-coding hits) [40▪▪]. Thereby, astonishingly, roles were rather pro-differentiative (87%) than antidifferentiative [40▪▪]. These findings and rates can guide how to test any new candidate lncRNA in the cardiovascular context most meaningfully. Hit rates might eventually be higher when assaying molecular effects of lncRNAs with broad and agnostic omics technologies, which was not achieved in the above genome-wide screens that used simplified cellular readouts for practical reasons.
NEW CONCEPTS OF LONG NONCODING RNAs FUNCTION WITH POTENTIAL RELEVANCE TO THE EXPERIMENTAL DESIGN IN STUDYING CARDIOVASCULAR DISEASE
Recent work suggests that there are some new overarching concepts for lncRNA function that have yet to be applied to the cardiovascular field but could simplify future research in CVD. In the following, we describe important concepts and point out their relevance for experimental design in studying CVD (Table 2).
Long noncoding RNA-dependent spatial compartments and nuclear scaffolding
Several insightful studies recently identified that many lncRNAs acquire previously unknown functionality by establishing three-dimensional (3D) infrastructures: One report found that many intron and repeat-containing RNAs and more than 80% of robustly expressed lncRNAs bridge to an insoluble proteinaceous material (e.g. SAF-A) in the nucleus. Importantly, these RNA-protein contacts served as a 3D scaffold that maintained gene activity by counteracting the inherent condensation properties of chromatin on a cytological scale [42] (Fig. 1d). Independent studies reported on similar but different roles of lncRNAs in controlling 3D chromatin topology: While physically attached to their DNA loci during RNA transcription, lncRNAs (including the cardiovascular Airn, Kcnq1ot1 or MALAT) seed and regulate 3D chromatin looping over megabases. These loops typically coalesce into reaction centres that allow to concentrate diffusible activator or repressor proteins [43▪]. Thus, in the face of the dazzling diversity of lncRNA functions in gene expression control (Table 1), a substantial number of these functions might be reducible to a few basic properties related to 3D reaction compartmentalization (Fig. 1d).
Nonstoichiometric amplification of long noncoding RNA function
Another riddle is how lncRNAs that decay fast and exist with less than 10 copies per cell could ever be significant regulators of biochemical reactions. Remarkably, some functional lncRNAs exist at much lower levels than their targets. An insightful recent review discusses conceptual models that reconcile this paradox [44▪▪]. First, lncRNAs such as cardiovascular NEAT1 contain multiple RNA-binding protein (RBP) binding sites, which helps to amplify their impact. Second, even with few RNA copies, lncRNAs use properties of the bound RBPs to amplify their function at the protein level. To this end, specific intrinsically disordered regions (IDRs) or prion-like domains in the RBPs propel protein structural state-switches or oligomerization. Surprisingly, it is these protein switches that drive liquid-liquid phase separation and other types of 3D compartmentalization that functionally support effective transcription and splicing and other RNA-based processes [44▪▪] (Fig. 1d). A nonexclusive model recently highlighted how noncoding RNAs could also gain impact by the effect of electrostatic forces from their backbones. In this model, the charge balance from noncoding RNA appeared to be critical to forming condensates of coactivators and RNA Polymerase II (RNAPII) during transcription initiation [45▪]. Thinking about these nonlinear reaction schemes is vital for future experimental design, for exploiting the therapeutic potential of lncRNAs, and not lastly, for reinterpreting published results that drew conclusions from nonphysiological overexpression of lncRNAs.
Long noncoding RNA splicing can be the determining effector mechanism
Although lncRNA sequence is generally not well conserved, splice-control sequences in lncRNAs surprisingly are. In addition, enhancers that produce well spliced lncRNAs (eRNAs, Fig. 1a) are more potent, and it was long unknown why. Recent studies show that splicing over a lncRNA locus has consequences independent of the actual lncRNA sequence content. Specifically, splicing the first exon of a lncRNA stimulated RNAPII progression along the noncoding locus. The transcription process then remodels the local chromatin landscape. For 50% of local eRNAs, this (and not the RNA moiety itself) might be the actual determinant for enhancer function [46]. This finding suggests that lncRNA processing and/or RBP-loading onto lncRNAs may be an underappreciated factor in how lncRNA loci become functional. Such considerations will undoubtedly impact the interpretation of studies that deleted entire lncRNA loci to assess the role of given lncRNAs for CVD (Table 1). Splicing of lncRNAs can also have additional consequences: For example, intron-retention in lncRNAs was identified as a physiological regulatory process to retain lncRNAs in the nucleus [47]. Moreover, if occurring in mRNAs, it can reduce the net translation of protein-coding genes, as shown for MYH7b in the heart [48]. New long-range RNA sequencing platforms such as nanopore sequencing are getting a much better grip on detecting lncRNA splice-isoforms and help to expand the number of noncoding transcript isoforms to be studied.
Functions of lncRNA modifications
Epitranscriptomics is an emerging new field. More than 150 posttranscriptional chemical RNA modifications are known to date, and these can affect the processing, stability or sorting of lncRNAs, yet with largely untapped therapeutic potential [49]. For example, conceptually alike histone tail modifications and chromatin reader-writer systems, region-selectively deposited m6A methylation in RNA not only has steric effects on RNA [50] but can also recruit readers such as the YTHDC1 protein (Fig. 1e). Among others, YTHDC1 attracts the splicing factor SRSF3 in the case of ANRIL, a major lncRNA also in cardiovascular context. Thereby, m6A implements a local splicing switch from the short to the long isoform, which determines ANRIL's repressor function [51].
Noncoding RNAs can express micropeptides
Synthetic screening, better IRES prediction and translational profiling revealed that microexons and small open reading frames (sORFs) are more common in lncRNAs than expected (Fig. 1e). Current measurements may be undersampling true micropeptides, as peptidomics is not sufficiently sensitive for lncRNAs that are typically present at low copy numbers. However, up to 0.3 micropeptides might exist per transcribed lncRNA, which is significant for a molecular species characterized by its noncoding character. Up to 7000 micropeptides can be expected to be expressed in the human genome [52]. Currently, only 10% of these are already quantifiable, and most need to be functionally tested [53]. An example in the cardiovascular system is lncRNA PSR, which produces a 117 amino acid long peptide (Arteridin) required for SMC phenotypic-switching. Surprisingly, PSR additionally functions as lncRNA because it binds Arteridin in complex with the atherosclerosis-driving transcription factor YBX-1 [54]. This scenario reveals yet again the multiplicity and repurposing of function among lncRNAs.
Gold standards for best practices in dissecting lncRNA functions are currently being formulated [3▪▪], and common pitfalls are highlighted [55], which are particularly important for translational attempts: First, special care is advised regarding RNAseq and PCR artefacts in CLIP experiments [56,57▪▪] or poor stringency in pulldowns. The latter is an issue even for well known and often reported lncRNA-interactors like PRC2 [57▪▪], which has also been implicated as an effector of many cardiovascular lncRNAs. Second, frameworks are emerging for describing best practice approaches in studying lncRNAs [3▪▪]: Brute force deletion of a lncRNA locus may still be a pragmatic entry point (as performed for most studies in Table 1), especially when coupled to deep in vivo phenotyping [3▪▪]. In many cases, further clarifying steps are needed and have not yet been reported for cardiovascular lncRNAs in typical CVD disease models (Table 1). Specifically, more fine-grained CRISPRi/a-based probing, deletions scanning, insertions of polyA-terminators and rescue experimentation (with wildtype and mutant RNA versions) can determine if a DNA region or the RNA product in a locus is relevant. This distinction is essential for identifying the therapeutic target in a lncRNA locus [3▪▪] (Fig. 1, Table 1). Such a distinction is specifically relevant because lncRNA loci can be active at the DNA level, while the lncRNA molecule itself may be without function. Synthetically tethering lncRNAs to their proposed effector molecule (client DNA, RNA or protein) can help to formally document that RNA is the functional moiety and that it engaged a specific effector [3▪▪,55]. Last, it is common sense that evidence for molecular effector mechanisms of lncRNAs should not solely involve phenocopying, especially not when making unwarranted use of epistasis experiments with indirect cellular readouts or when involving overexpression at nonphysiological levels [3▪▪,38▪▪,55].
CONCLUSION
Some lncRNAs play important roles in the pathophysiology of CVD. The functions of these lncRNAs are diverse, and new concepts still emerge. These are driven by advances in sequencing and genome engineering, interaction profiling and quantitative modelling of RNA-dependent gene regulatory networks. Some lncRNAs also show therapeutic potential for ameliorating the functions of cardiomyocytes, endothelial cells or VSMCs in the heart and vasculature (Table 1). However, the way to lncRNA-based therapies is still far. On this road, it is reasonable to expect that lncRNAs exhibit moderate, cell-type specific, modulatory or combinatorial effects. Therefore, interrogating lncRNA function in vivo, either during development or in animal disease models and in organoids, remains a central avenue toward unveiling lncRNA functions. Current frontiers for therapeutic exploitation are also related to remaining technical challenges. One is to pinpoint the role of lncRNAs from single cells in their spatial tissue context. Another is the systematic prediction, imaging and testing of RNA structural motifs as functional determinants and as therapeutic drug targets in vivo. Together, as the decades-long fruitful research on model lncRNAs like XIST can tell, studying lncRNAs is a challenging multistep and multidisciplinary endeavour that bears ever-new surprises and where decisive mechanistic work pays off.
Acknowledgements
The authors thank Dr Andreas Herbst for critically reading the manuscripts and for his input.
Financial support and sponsorship
This work was funded by the German Research Foundation within project A07 of the collaborative research centre TRR267.
Conflicts of interest
There are no conflicts of interest.
Footnotes
Lesca M. Holdt and Daniel Teupser contributed equally to this article.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
REFERENCES
- 1.Vaduganathan M, Mensah GA, Turco JV, et al. The global burden of cardiovascular diseases and risk: a compass for future health. J Am Coll Cardiol 2022; 80:2361–2371. [DOI] [PubMed] [Google Scholar]
- 2▪▪.Mattick JS, Amaral PP, Carninci P, et al. Long noncoding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol 2023; doi: 10.1038/s41580-022-00566-8. [Online ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]; A must-read review by a panel of authorities in the field on the definition, classification and broad functions of lncRNAs across different biological systems.
- 3▪▪.Andergassen D, Rinn JL. From genotype to phenotype: genetics of mammalian long noncoding RNAs in vivo. Nat Rev Genet 2022; 23:229–243. [DOI] [PubMed] [Google Scholar]; This is a must-read review, presenting the most relevant considerations and guidelines for functional in vivo testing of lncRNA loci. The authors lay out a practical step-by-step framework to hone in on the significant known functions of lncRNAs, and the review also describes necessary technologies and typical pitfalls.
- 4.Holdt LM, Kohlmaier A, Teupser D. Erdmann J, Moretti A. Long noncoding RNAs in cardiovascular disease. Genetic causes of cardiac disease. Cham: Springer International Publishing; 2019. 199–288. [Google Scholar]
- 5.Dong K, Shen J, He X, et al. CARMN is an evolutionarily conserved smooth muscle cell-specific LncRNA that maintains contractile phenotype by binding myocardin. Circulation 2021; 144:1856–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ni H, Haemmig S, Deng Y, et al. A smooth muscle cell-enriched long noncoding RNA regulates cell plasticity and atherosclerosis by interacting with serum response factor. Arterioscler Thromb Vasc Biol 2021; 41:2399–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vacante F, Rodor J, Lalwani MK, et al. CARMN loss regulates smooth muscle cells and accelerates atherosclerosis in mice. Circ Res 2021; 128:1258–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao R, Wang L, Bei Y, et al. Long noncoding RNA cardiac physiological hypertrophy-associated regulator induces cardiac physiological hypertrophy and promotes functional recovery after myocardial ischemia-reperfusion injury. Circulation 2021; 144:303–317. [DOI] [PubMed] [Google Scholar]
- 9.Dong XH, Lu ZF, Kang CM, et al. The long noncoding RNA RP11-728F11.4 promotes atherosclerosis. Arterioscler Thromb Vasc Biol 2021; 41:1191–1204. [DOI] [PubMed] [Google Scholar]
- 10.Hazra R, Brine L, Garcia L, et al. Platr4 is an early embryonic lncRNA that exerts its function downstream on cardiogenic mesodermal lineage commitment. Dev Cell 2022; 57:2450–2468. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11▪.Li H, Trager LE, Liu X, et al. lncExACT1 and DCHS2 regulate physiological and pathological cardiac growth. Circulation 2022; 145:1218–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study is relevant because it involves lncExACT1 in DCHS2 protocadherin regulation. Thereby, the authors identify a new regulator or the Hippo pathway, one the most central signalling nodes in cell proliferation control during tissue maintenance and regeneration, also in the cardiovascular system.
- 12.Wang J, Liu S, Heallen T, Martin JF. The Hippo pathway in the heart: pivotal roles in development, disease, and regeneration. Nat Rev Cardiol 2018; 15:672–684. [DOI] [PubMed] [Google Scholar]
- 13▪.Zhao Y, Riching AS, Knight WE, et al. Cardiomyocyte-specific long noncoding RNA regulates alternative splicing of the Triadin gene in the heart. Circulation 2022; 146:699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reports on the peculiarity that many 3’UTRs of protein-coding mRNAs are expressed separately from their mRNA, hypothetically after cleavage and also termed uaRNAs, a phenomenon reported first by the Mattick laboratory. Here, the 3’UTR of the protein-coding gene Ckip-1 was upregulated in failing hearts and was cardioprotective in a TAC mouse model.
- 14.Fan J, Li H, Xie R, et al. LncRNA ZNF593-AS alleviates contractile dysfunction in dilated cardiomyopathy. Circ Res 2021; 128:1708–1723. [DOI] [PubMed] [Google Scholar]
- 15.Ren X, Zhu H, Deng K, et al. Long noncoding RNA TPRG1-AS1 suppresses migration of vascular smooth muscle cells and attenuates atherogenesis via interacting with MYH9 protein. Arterioscler Thromb Vasc Biol 2022; 42:1378–1397. [DOI] [PubMed] [Google Scholar]
- 16.Xiao H, Zhang M, Wu H, et al. CIRKIL exacerbates cardiac ischemia/reperfusion injury by interacting with Ku70. Circ Res 2022; 130:e3–e17. [DOI] [PubMed] [Google Scholar]
- 17▪.Park KS, Rahat B, Lee HC, et al. Cardiac pathologies in mouse loss of imprinting models are due to misexpression of H19 long noncoding RNA. Elife 2021; 10:e67250. [DOI] [PMC free article] [PubMed] [Google Scholar]; This high-quality study shows how detailed the genetic work must be when dissecting the function of a lncRNA locus. Together with the careful phenotypic analyses, this is another role-model paper for cardiovascular research on lncRNAs.
- 18.Lu D, Chatterjee S, Xiao K, et al. A circular RNA derived from the insulin receptor locus protects against doxorubicin-induced cardiotoxicity. Eur Heart J 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19▪.Sato M, Kadomatsu T, Miyata K, et al. The lncRNA Caren antagonizes heart failure by inactivating DNA damage response and activating mitochondrial biogenesis. Nat Commun 2021; 12:2529. [DOI] [PMC free article] [PubMed] [Google Scholar]; The study showcases the importance of careful cell-type specific genetic dissection of lncRNAs. The authors use an SMC-specific knockout and overexpression, revealing that Caren maintained the mature, contractile SMC state. Caren partially protected from atherosclerotic neointima growth in disease modelling.
- 20▪.Fasolo F, Jin H, Winski G, et al. Long noncoding RNA MIAT controls advanced atherosclerotic lesion formation and plaque destabilization. Circulation 2021; 144:1567–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]; The study implicates MIAT in atherosclerotic plaque growth and vulnerability in a multifaceted study. The authors may touch on a unifying function of MIAT across tissue and disease contexts.
- 21.Peng KL, Vasudevan HN, Lockney DT, et al. Miat and interacting protein Metadherin maintain a stem-like niche to promote medulloblastoma tumorigenesis and treatment resistance. Proc Natl Acad Sci U S A 2022; 119:e2203738119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhuang A, Calkin AC, Lau S, et al. Loss of the long noncoding RNA OIP5-AS1 exacerbates heart failure in a sex-specific manner. iScience 2021; 24:102537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Liu S, Han L, et al. LncRNA HBL1 is required for genome-wide PRC2 occupancy and function in cardiogenesis from human pluripotent stem cells. Development 2021; 148:dev199628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Y, Ling S, Li J, et al. 3’ untranslated region of Ckip-1 inhibits cardiac hypertrophy independently of its cognate protein. Eur Heart J 2021; 42:3786–3799. [DOI] [PubMed] [Google Scholar]
- 25▪▪.Sehgal P, Mathew S, Sivadas A, et al. LncRNA VEAL2 regulates PRKCB2 to modulate endothelial permeability in diabetic retinopathy. EMBO J 2021; 40:e107134. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study is a role model for modern lncRNA research. The study is of high quality regarding controls, mechanistic depth of deciphering the effector mechanism in vivo and identifying a lncRNA homolog based on conserved biochemical interaction with an effector protein.
- 26▪.de Goede OM, Nachun DC, Ferraro NM, et al. Population-scale tissue transcriptomics maps long noncoding RNAs to complex disease. Cell 2021; 184:2633–2648. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]; This comprehensive study is an important study and resource. By combining GWAS, eQTL and sQTL data, the authors map associations of lncRNAs with traits on a genome-wide scale across dozens of tissues. The study lays out potentially disease-relevant lncRNAs and their functional context. Traits include BMI, diabetes and coronary artery disease.
- 27▪▪.Turner AW, Hu SS, Mosquera JV, et al. Single-nucleus chromatin accessibility profiling highlights regulatory mechanisms of coronary artery disease risk. Nat Genet 2022; 54:804–816. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study with a cardiovascular focus is a strong example of how single-cell approaches change the research on noncoding variants associated with diseases and traits. It presents a generalizable approach that draws from single-cell technologies, epigenetic principles, machine learning and regulatory gene network modelling and identifies new CVD driver genes.
- 28.Zhang K, Hocker JD, Miller M, et al. A single-cell atlas of chromatin accessibility in the human genome. Cell 2021; 184:5985–6001. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ameen M, Sundaram L, Shen M, et al. Integrative single-cell analysis of cardiogenesis identifies developmental trajectories and noncoding mutations in congenital heart disease. Cell 2022; 185:4937–4953. e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Findley AS, Monziani A, Richards AL, et al. Functional dynamic genetic effects on gene regulation are specific to particular cell types and environmental conditions. Elife 2021. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yazar S, Alquicira-Hernandez J, Wing K, et al. Single-cell eQTL mapping identifies cell type-specific genetic control of autoimmune disease. Science 2022; 376:eabf3041. [DOI] [PubMed] [Google Scholar]
- 32▪.Nasser J, Bergman DT, Fulco CP, et al. Genome-wide enhancer maps link risk variants to disease genes. Nature 2021; 593:238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors expand on a generalizable method they developed earlier. Here, they comprehensively link noncoding variants from 72 diseases and across more than 100 biosamples to so far unknown target genes and cell types. Highly relevant also to lncRNAs, the study yields more than 6 million enhancer-gene connections, showing the power of the approach for future application in CVD contexts.
- 33▪▪.Lin X, Liu Y, Liu S, et al. Nested epistasis enhancer networks for robust genome regulation. Science 2022; 377:1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work is the first time a study takes on the challenge of deciphering the complexity in the logic of how multiple enhancers regulate a target promoter, which is the norm and not the exception in gene structures. On the basis of multiplexed CRISPRi with tens of thousands of gRNAs (and their combinations), the study reveals the unexpectedly layered additive and synergistic effects between enhancers. Findings are relevant for future noncoding CVD risk locus fine-mapping.
- 34▪.Johnsson P, Ziegenhain C, Hartmanis L, et al. Transcriptional kinetics and molecular functions of long noncoding RNAs. Nat Genet 2022; 54:306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work is a prominent example of how modern work on lncRNAs is transforming. The study offers fundamental new insight into how lncRNAs are transcribed and represents a more broadly applicable approach for testing how genomic orientation may affect the yet unclear modes by which lncRNAs control transcriptional initiation or elongation of target genes.
- 35.Dukler N, Mughal MR, Ramani R, et al. Extreme purifying selection against point mutations in the human genome. Nat Commun 2022; 13:4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao T, O’Reilly ME, Selvaggi C, et al. Cis-regulated expression of nonconserved lincRNAs associates with cardiometabolic related traits. J Hum Genet 2022; 67:307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross CJ, Ulitsky I. Discovering functional motifs in long noncoding RNAs. Wiley Interdiscip Rev RNA 2022; 13:e1708. [DOI] [PubMed] [Google Scholar]
- 38▪▪.Ponting CP, Haerty W. Genome-wide analysis of human long noncoding RNAs: a provocative review. Annu Rev Genomics Hum Genet 2022; 23:153–172. [DOI] [PubMed] [Google Scholar]; This review is another must-read work, especially for translationally-oriented audiences. It addresses, with numbers and clear logic, some of the currently prominent questions in the lncRNA field. The work rightfully points out instances where many current treadmill studies surf on the lncRNA boom and propagate misconceptions and overstatements, skewing research directions and interpretations.
- 39.Gebauer F, Schwarzl T, Valcarcel J, Hentze MW. RNA-binding proteins in human genetic disease. Nat Rev Genet 2021; 22:185–198. [DOI] [PubMed] [Google Scholar]
- 40▪▪.Wu D, Poddar A, Ninou E, et al. Dual genome-wide coding and lncRNA screens in neural induction of induced pluripotent stem cells. Cell Genom 2022; 2: [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors perform comparative genome-wide lncRNA and mRNA CRISPRi screening in a prototypical iPSC differentiation model. Findings are likely relevant beyond the system they use. Overall hit rates for lncRNAs from the screen are in the low percentage range, which is in line with many other genome-wide screens of lncRNAs published over the years.
- 41.Liu SJ, Horlbeck MA, Cho SW, et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 2017; 355: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Creamer KM, Kolpa HJ, Lawrence JB. Nascent RNA scaffolds contribute to chromosome territory architecture and counter chromatin compaction. Mol Cell 2021; 81:3509–3525. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43▪.Quinodoz SA, Jachowicz JW, Bhat P, et al. RNA promotes the formation of spatial compartments in the nucleus. Cell 2021; 184:5775–5790. e30. [DOI] [PMC free article] [PubMed] [Google Scholar]; The study describes a new sequencing-based technology to map and explore RNA-dependent nuclear compartments, including lncRNA-based spatial 3D infrastructures. The study gives the reader a good appreciation of the different flavours of nuclear lncRNA-based bodies and the challenges ahead.
- 44▪▪.Unfried JP, Ulitsky I. Substoichiometric action of long noncoding RNAs. Nat Cell Biol 2022; 24:608–615. [DOI] [PubMed] [Google Scholar]; This review is highly insightful and transformative, written by authorities in the field. It describes a fundamental general concept: how low-abundant lncRNAs gain disproportionally significant functionality. Relevant mechanisms range from reaction centre compartmentalization in the nucleus to chaperone-like functions, to name a few.
- 45▪.Henninger JE, Oksuz O, Shrinivas K, et al. RNA-mediated feedback control of transcriptional condensates. Cell 2021; 184:207–225. e24. [DOI] [PMC free article] [PubMed] [Google Scholar]; The study is another fascinating example of currently published studies that achieve to reduce the seemingly so diverse roles of RNA in gene expression control to a smaller number of elegant ground principles. The authors offer evidence that low-abundant RNAs stimulate coactivator condensates during transcription initiation, while high levels of RNA dissolve condensates during ongoing full-blown elongation.
- 46.Engreitz JM, Haines JE, Perez EM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016; 539:452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dumbovic G, Braunschweig U, Langner HK, et al. Nuclear compartmentalization of TERT mRNA and TUG1 lncRNA is driven by intron retention. Nat Commun 2021; 12:3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee LA, Broadwell LJ, Buvoli M, Leinwand LA. Nonproductive splicing prevents expression of MYH7b protein in the mammalian heart. J Am Heart Assoc 2021; 10:e020965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer 2020; 20:303–322. [DOI] [PubMed] [Google Scholar]
- 50.Jones AN, Tikhaia E, Mourao A, Sattler M. Structural effects of m6A modification of the Xist A-repeat AUCG tetraloop and its recognition by YTHDC1. Nucleic Acids Res 2022; 50:2350–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang ZW, Pan JJ, Hu JF, et al. SRSF3-mediated regulation of N6-methyladenosine modification-related lncRNA ANRIL splicing promotes resistance of pancreatic cancer to gemcitabine. Cell Rep 2022; 39:110813. [DOI] [PubMed] [Google Scholar]
- 52.Patraquim P, Magny EG, Pueyo JI, et al. Translation and natural selection of micropeptides from long noncanonical RNAs. Nat Commun 2022; 13:6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chothani SP, Adami E, Widjaja AA, et al. A high-resolution map of human RNA translation. Mol Cell 2022; 82:2885–2899. e8. [DOI] [PubMed] [Google Scholar]
- 54.Yu J, Wang W, Yang J, et al. LncRNA PSR regulates vascular remodeling through encoding a novel protein arteridin. Circ Res 2022; 131:768–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo JK, Guttman M. Regulatory noncoding RNAs: everything is possible, but what is important? Nat Methods 2022; 19:1156–1159. [DOI] [PubMed] [Google Scholar]
- 56.Hafner M, Katsantoni M, Köster T, et al. CLIP and complementary methods. Nat Rev Methods Primers 2021; 1:20. [Google Scholar]
- 57▪▪.Rosenberg M, Blum R, Kesner B, et al. Motif-driven interactions between RNA and PRC2 are rheostats that regulate transcription elongation. Nat Struct Mol Biol 2021; 28:103–117. [DOI] [PMC free article] [PubMed] [Google Scholar]; The study is highly relevant for the entire lncRNA field and provides deep mechanistic insight into RNA-PRC2 contacts, claimed to be effector process for hundreds of earlier papers by often less than satisfactory evidence. The findings reconcile seemingly contradictory data from years of work on PRC2. The study also offers generalizable guidelines for state-fo-the art high-stringency biochemical pulldowns of RNA-protein complexes.