Key words: Comparative analysis, Dermacentor steini, Mitochondrial genome, Phylogeny, Tick
Abstract
Ticks are a group of blood-sucking ectoparasites that play an important role in human health and livestock production development as vectors of zoonotic diseases. The phylogenetic tree of single genes cannot accurately reflect the true kinship between species. Based on the complete mitochondrial genome analysis one can help to elucidate the phylogenetic relationships among species. In this study, the complete mitochondrial genome of Dermacentor steini (isolate Longyan) was sequenced and compared with the mitochondrial genes of 3 other Chinese isolates (Nanchang, Jinhua and Yingtan). In Dermacentor steini 4 isolates had identical or similar mitochondrial genome lengths and an overall variation of 0.76% between sequences. All nucleotide compositions showed a distinct AT preference. The most common initiation and stop codons were ATG and TAA, respectively. Fewer base mismatches were found in the tRNA gene of D. steini (isolate Longyan), and the vicinity of the control region and tRNA gene was a hot rearrangement region of the genus Dermacentor. Maximum likelihood trees and Bayesian trees indicate that D. steini is most closely related to Dermacentor auratus. The results enrich the mitochondrial genomic data of species in the genus Dermacentor and provide novel insights for further studies on the phylogeographic classification and molecular evolution of ticks.
Introduction
Ticks are a group of blood-sucking ectoparasites that often parasitize on the body surfaces of mammals, birds, reptiles and amphibians (Burnard and Shao, 2019). Ticks have become the second largest vector of infectious diseases worldwide after mosquitoes because they can carry a variety of pathogens such as bacteria, fungi, viruses, nematodes and blood protozoans (Mead et al., 2015). Ticks can also cause severe allergic or toxic reactions, temporary paralysis and secondary infections in humans, livestock and other vertebrates through their bites, seriously endangering human health as well as livestock development (Chen et al., 2010).
The genus Dermacentor (Ixodida: Ixodidae) is a recently evolved genus with relatively limited species diversity compared to other genera (e.g. Ixodes and Haemaphysalis) in the family Ixodidae (Ernieenor et al., 2021). Forty species of the genus Dermacentor have been reported to be distributed worldwide, and 12 species have been found in China (Guo et al., 2016; Nava et al., 2017). All species of the genus Dermacentor have distinct external morphological characteristics and are classified in the subgenus Indocentor Schulze (Ernieenor et al., 2020). Hosts of the genus Dermacentor include horses, cattle, sheep and rodents, and can transmit a variety of pathogens, including Babesia bovis, B. caballi, B. canis, Theileria equi and other protozoa; Brucella, Rickettsia, Rocky Mountain spotted fever, Anaplasma and other bacteria, as well as tick-borne encephalitis virus (Parola et al., 2005; Swei et al., 2019; Ličková et al., 2020; Duncan et al., 2021; Eisen and Paddock, 2021; Yang et al., 2021). In addition, species of the genus Dermacentor are also most frequently associated with human intra-ear infections (Mariana et al., 2008). Among them, Dermacentor steini is considered to be the most abundant member of the subgenus Indocentor and is mainly parasitic on wild boars in Asian countries (Wassef and Hoogstraal, 1988; Lim et al., 2020). D. steini is morphologically most similar to D. auratus and D. atrosignatus and carries pathogens (Rickettsia sp., Anaplasma sp., Lanjan virus and Hepatozoon species) similar to those isolated from D. auratus (Wassef and Hoogstraal, 1986, 1988). Most importantly, D. steini feeds on human skin or other tissues and is an important reservoir host and vector of zoonotic diseases, which poses a greater threat to public health safety (Vongphayloth et al., 2016; Lim et al., 2020).
The mitochondrial genome (mitogenome) is a DNA molecule typically 15–20 kb in size, with 37 genes, including 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rRNAs) and usually longer non-coding regions (control regions) (Wang et al., 2018, 2020). The mitochondrial genome is widely considered as one of the most reliable and effective genetic markers in molecular phylogenetic studies because of its maternal inheritance, high mutation rate and lack of recombination (Yang et al., 2022a, 2022b). Meanwhile, the mitochondrial genome has been proven to be a useful tool for understanding the evolutionary, geographic and pathogenic relationships of tick species (Burger et al., 2013).
Given the wide distribution of ticks of the genus Dermacentor and their importance as vectors of zoonotic diseases, they have an important role in the development of human and livestock production. Thus, this study reports the mitochondrial genome sequence of D. steini (Longyan isolate) collected in Longyan, Fujian Province, China, and compared with those of 3 other D. steini (Jinhua, Nangchang and Yingtan isolates) from different regions of China.
Materials and methods
Sample collection
Rats were captured using mousetraps in Longyan, Fujian Province, China. Adult ticks were then collected from the body surface of the captured rats (Rattus andamanensis) and preserved in Eppendorf (EP) tubes with 95% ethanol, and the sample number was labelled P299.
Statement: The capture protocol and procedures for small mammals were approved by the Animal Ethics Committee of Dali University. The approval number is MECDU-201806-11.
DNA extraction and mitogenome sequencing
The adult ticks were taken out of the EP tubes containing 95% ethanol and immersed in sterile distilled water for 30 min to remove microorganisms from the surface, and then the ticks were cut with a sterile scalpel blade. Genomic DNA was extracted from ticks with the DNeasy Blood and Tissue Kit (Qiagen, Valencia, California, USA). The extracted DNA was used to construct Illumina PE libraries. The mitochondrial genome was then sequenced on the Illumina Novoseq 6000 platform (Winnerbio, Shanghai, China).
Genome annotation
MitoZ 2.3 (https://doi.org/10.1101/489955) was used to assemble the mitochondrial genome. The MITOS web server was used to predict genes (Bernt et al., 2013), and 13 PCGs were compared and edited using the BLAST tool in NCBI. The location and length of tRNA genes were further confirmed using tRNAscan-SE (Lowe and Eddy, 1997) and ARWEN (Laslett and Canbäck, 2008). Two rRNA genes were identified based on their putative secondary structures and previously sequenced mitochondrial genome sequences. The position of the control region was confirmed based on the boundaries of neighbouring genes. The mitochondrial sequence of D. steini (Longyan isolate) has been deposited in GenBank under the accession number OP383032.
Sequence analysis and phylogenetic reconstruction
Base composition and codon usage were calculated using GeneiousPrime (Kearse et al., 2012) and MEGA X (Kumar et al., 2016), respectively. Sliding window analysis was performed on the complete mitochondria of different D. steini isolates using DnaSP v5 (Librado and Rozas, 2009). The sliding window length was set to 100 bp with a step size of 25 bp to estimate the value of nucleotide diversity. Phylogenetic trees were constructed using Bayesian inference (BI) methods and maximum likelihood (ML) methods with Limulus polyphemus and Carcinoscorpius rotundicauda as outgroups. BI and ML analyses were performed in PhyloSuite (Zhang et al., 2020). For the Bayesian analysis, a total of 1 000 000 generations were run, with sampling every 1000 generations. The first 25% of the tree was burned to ensure sample independence and 4 Monte Carlo Markov chains were run. To estimate the support of the BI tree, posterior probabilities (PP) were calculated. For the ML tree, 5000 ultra-fast bootstrap replications were used to calculate branch reliability (bootstrap probability, BP). The constructed phylogenetic tree was viewed and edited with FigTree 1.4.1 (https://github.com/rambaut/figtree/). Species and accession numbers for the construction of the phylogenetic tree are shown in Supplementary Table 1.
Results
Mitochondrial genome characteristics of Dermacentor steini (Longyan isolate)
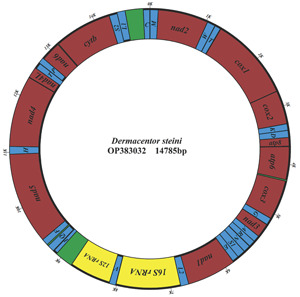
The mitochondrial genome of D. steini (Longyan isolate) is a circular double-stranded DNA molecule with a length of 14 785 bp. It includes 13 PCGs (cox1-3, cytb, nad1-6, nad4L, atp6 and atp8), 22 tRNA genes, 2 rRNA genes (rrnL and rrnS) and 2 control regions (Table 1), which is consistent with the reported mtgenomes of other species of the genus Dermacentor but not with the mitochondrial genomes of some metazoan, such as in trematodes, cestodes and Chromadorea nema-todes, which have been found to have lost the atp8 gene in their mitochondrial genomes (Yamasaki et al., 2012; Duan et al., 2015; Chang et al., 2016). Of the 37 genes, 23 genes (cox1-3, nad2-3, nad6, atp6, atp8, cytb, trnA, trnC, trnD, trnE, trnG, trnI, trnK, trnM, trnN, trnR, trnS1, trnS2, trnT, trnW) are encoded on the J-strand, and the remaining 12 genes (nad1, nad4, nad4L, nad5, trnF, trnH, trnL1, trnL2, trnP, trnQ, trnV, trnY) and 2 rRNA genes (16S rRNA and 12S rRNA) are encoded on the N-strand.
Table 1.
Dermacentor steini gene content, length, coding strand, initiation and stop codons of mitochondrial genomes of different isolate
| Gene/regions | Strand | Positions and lengths (bp) | Initiation and stop codons | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L.Y | N.C | J.H | Y.T | L.Y | N.C | J.H | Y.T | L.Y | N.C | J.H | Y.T | |
| trnM | J | J | J | J | 1–67 (67) | 1–67 (67) | 1–67 (67) | 1–67 (67) | ||||
| nad2 | J | J | J | J | 68–1027 (960) | 68–1027 (960) | 68–1027 (960) | 68–1027 (960) | ATT/TAA | ATT/TAA | ATT/TAA | ATT/TAA |
| trnW | J | J | J | J | 1026–1087 (62) | 1026–1087 (62) | 1027–1086 (62) | 1026–1087 (62) | ||||
| trnY | N | N | N | N | 1092–1154 (63) | 1092–1154 (63) | 1092–1154 (63) | 1092–1154 (63) | ||||
| cox1 | J | J | J | J | 1147–2685 (1539) | 1147–2685 (1539) | 1147–2685 (1539) | 1147–2685 (1539) | ATT/TAA | ATT/TAA | ATT/TAA | ATT/TAA |
| cox2 | J | J | J | J | 2690–3362 (673) | 2690–3362 (673) | 2690–3362 (673) | 2690–3362 (673) | ATG/T | ATG/T | ATG/T | ATG/T |
| trnK | J | J | J | J | 3363–3428 (66) | 3363–3427 (65) | 3363–3428 (66) | 3363–3428 (66) | ||||
| trnD | J | J | J | J | 3428–3487 (60) | 3429–3488 (60) | 3428–3488 (61) | 3427–3487 (61) | ||||
| atp8 | J | J | J | J | 3488–3646 (159) | 3488–3646 (159) | 3488–3646 (159) | 3488–3646 (159) | ATC/TAA | ATC/TAA | ATC/TAA | ATC/TAA |
| atp6 | J | J | J | J | 3640–4305 (666) | 3640–4305 (666) | 3640–4305 (666) | 3640–4305 (666) | ATG/TAA | ATG/TAG | ATG/TAG | ATG/TAA |
| cox3 | J | J | J | J | 4330–5108 (779) | 4330–5108 (779) | 4323–5100 (778) | 4323–5101 (779) | ATG/TA | ATG/TA | ATG/T | ATG/TA |
| trnG | J | J | J | J | 5107–5169 (63) | 5108–5168 (61) | 5101–5161 (61) | 5102–5161 (60) | ||||
| nad3 | J | J | J | J | 5166–5507 (342) | 5166–5507 (342) | 5162–5500 (339) | 5162–5500 (339) | ATA/TAA | ATA/TAA | ATT/TAA | ATT/TAA |
| trnA | J | J | J | J | 5507–5570 (64) | 5507–5569 (63) | 5500–5563 (64) | 5500–5563 (64) | ||||
| trnR | J | J | J | J | 5572–5639 (68) | 5572–5637 (66) | 5565–5632 (68) | 5565–5632 (68) | ||||
| trnN | J | J | J | J | 5634–5699 (66) | 5634–5699 (66) | 5638–5691 (54) | 5628–5690 (63) | ||||
| trnS1 | J | J | J | J | 5702–5759 (58) | 5702–5759 (58) | 5696–5751 (56) | 5696–5756 (61) | ||||
| trnE | J | J | J | J | 5759–5819 (61) | 5758–5820 (63) | 5752–5812 (61) | 5752–5813 (62) | ||||
| nad1 | N | N | N | N | 5818–6756 (939) | 5818–6756 (939) | 5811–6749 (939) | 5811–6749 (939) | ATT/TAA | ATT/TAA | ATT/TAA | ATT/TAA |
| trnL2 | N | N | N | N | 6757–6818 (62) | 6757–6818 (61) | 6750–6811 (62) | 6750–6811 (62) | ||||
| 16S rRNA | N | N | N | N | 6819–8018 (1200) | 6819–8018 (1200) | 6812–8004 (1193) | 6812–8004 (1193) | ||||
| trnV | N | N | N | N | 8015–8075 (61) | 8017–8077 (61) | 8004–8064 (61) | 8004–8064 (61) | ||||
| 12S rRNA | N | N | N | N | 8077–8788 (712) | 8079–8786 (708) | 8006–8775 (710) | 8066–8771 (706) | ||||
| CR1 | 8789–9100 (312) | 8787–9100 (314) | 8776–9088 (313) | 8772–9084 (313) | ||||||||
| trnI | J | J | J | J | 9101–9163 (63) | 9101–9163 (63) | 9089–9151 (63) | 9085–9147 (63) | ||||
| trnQ | N | N | N | N | 9172–9237 (66) | 9172–9237 (66) | 9160–9225 (66) | 9156–9221 (66) | ||||
| trnF | N | N | N | N | 9264–9327 (64) | 9264–9327 (64) | 9252–9315 (64) | 9248–9309 (62) | ||||
| nad5 | N | N | N | N | 9327–10 985 (1659) | 9327–10 985 (1659) | 9315–10 973 (1659) | 9311–10 969 (1659) | ATT/TAA | ATT/TAA | ATT/TAA | ATT/TAA |
| trnH | N | N | N | N | 10 986–11 048 (63) | 10 986–11 048 (63) | 10 974–11 036 (63) | 10 970–11 032 (63) | ||||
| nad4 | N | N | N | N | 11 049–12 363 (1315) | 11 049–12 363 (1315) | 11 037–12 351 (1315) | 11 033–12 347 (1315) | ATG/T | ATG/T | ATG/T | ATG/T |
| nad4L | N | N | N | N | 12 357–12 632 (276) | 12 357–12 632 (276) | 12 345–12 620 (276) | 12 341–12 616 (276) | ATG/TAA | ATG/TAA | ATG/TAA | ATG/TAA |
| trnT | J | J | J | J | 12 634–12 697 (64) | 12 635–12 698 (64) | 12 623–12 683 (61) | 12 619–12 681 (64) | ||||
| trnP | N | N | N | N | 12 695–12 757 (63) | 12 696–12 756 (61) | 12 684–12 744 (61) | 12 680–12 740 (61) | ||||
| nad6 | J | J | J | J | 12 759–13 193 (435) | 12 759–13 193 (435) | 12 732–13 181 (450) | 12 728–13 177 (450) | ATT/TAA | ATT/TAA | ATA/TAA | ATA/TAA |
| cytb | J | J | J | J | 13 208–14 290 (1083) | 13 208–14 290 (1083) | 13 196–14 278 (1083) | 13 190–14 272 (1083) | ATG/TAG | ATG/TAG | ATG/TAG | ATG/TAG |
| trnS2 | J | J | J | J | 14 289–14 355 (67) | 14 290–14 355 (66) | 14 277–14 344 (68) | 14 272–14 337 (66) | ||||
| trnL1 | N | N | N | N | 14 355–14 417 (63) | 14 355–14 417 (63) | 14 343–14 405 (63) | 14 337–14 399 (63) | ||||
| CR2 | 14 418–14 724 (307) | 14 418–14 725 (308) | 14 406–14 713 (308) | 14 400–14 706 (307) | ||||||||
| trnC | J | J | J | J | 14 725–14 781 (57) | 14 725–14 781 (57) | 14 714–14 767 (54) | 14 707–14 767 (61) | ||||
L.Y, Longyan; N.C, Nanchang; J.H, Jinhua; Y.T, Yingtan
The mitochondrial genome of D. steini (Longyan isolate) is relatively compact with 12 gene intergenic regions and 16 gene overlap regions (Table 1), with total lengths of 90 and 52 bp, respectively. The intergenic region occurred at 23 gene junctions. The longest intergenic sequence (26 bp) was found between trnQ and trnF, and the shortest intergenic sequence (1 bp) was found between trnA and trnR, trnV and 12S rRNA, nad4L and trnT, and trnP and nad6. Overlap regions occurred at 29 gene junctions. The longest overlap sequence, 8 bp, was found between trnY and cox1. The shortest overlap region, 1 bp, was found between nad3 and trnA, trnS1 and trnE, trnF and nad5, and trnS2 and trnL1. Among them, the 7 bp overlap between the nad4 and nad4L genes has also been widely reported in the mitochondrial genome of insects (Wang and Tang, 2017).
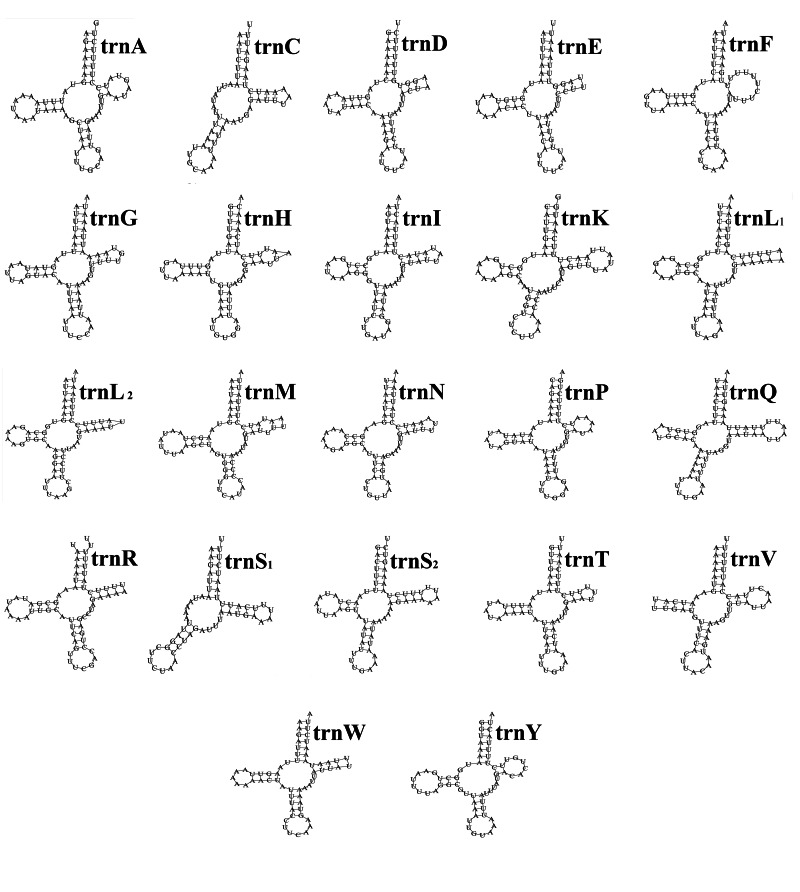
The typical secondary structure of tRNA genes is a cloverleaf structure with 4 arms: amino acid acceptor arm (AA-arm), DHC arm (D-arm), anticodon arm (AC-arm) and TΨC arm (T-arm) (Yuan et al., 2010). The 22 tRNA genes of D. steini (Longyan isolate) have a total length of 1391 bp. The length of individual tRNA genes ranged from 57 bp (trnC) to 68 bp (trnR), and their secondary structures are shown in Fig. 1. Except for trnC and trnS1, which lacked the D-arm, the other 20 tRNA genes were able to form a typical cloverleaf structure. Among these tRNA genes, 14 tRNA genes are encoded in the J-strand and the other 8 tRNA genes are encoded in the N-strand. Besides the typical Waston–Crick pairings (A–U, G–C), 12 non-canonical pairings, 9 G–U pairings and 3 U–U mismatches occurred.
Fig. 1.
Dermacentor steini (Longyan isolate) tRNA gene putative secondary structure.
Comparative analysis between mitochondrial genomes of D. steini (Longyan, Nanchang, Jinhua and Yingtan isolates)
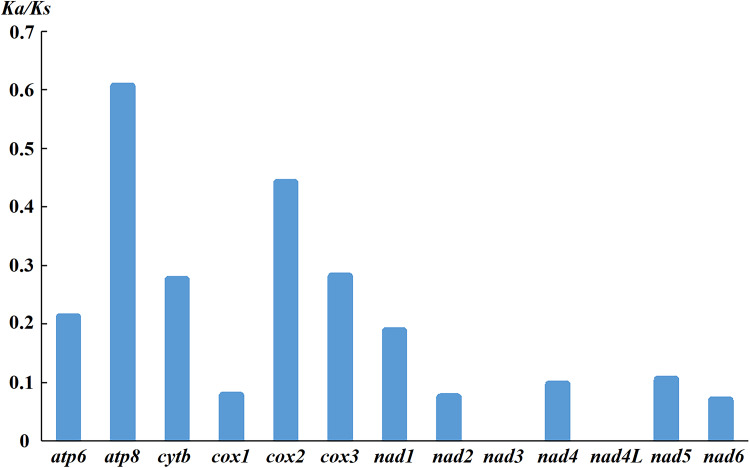
The mitochondrial genomes of D. steini isolated from Longyan and Nanchang were of the same length, 12 and 18 bp longer than those of Jinhua and Yingtan isolates, respectively. The 37 genes and 2 non-coding regions of the D. steini from Longyan, Nanchang, Jinhua and Yingtan isolate mitochondrial genomes were identical or similar in length, and the overall variation between sequences was 0.76%. From Table 2, the nucleotide composition of the mitochondrial genomes of the 4 isolates (Longyan, Nanchang, Jinhua and Yingtan) showed a distinct AT preference. All PCGs used the typical ATN as the initiation codon and no rare initiation codons (GTG and TTG) were present; of which; ATT and ATG were used up to 5 and 6 times, respectively. With the use of stop codons, most PCGs ended with the complete stop codon TAA. The truncated stop codon T/TA was found in the cox2, cox3 and nad4 genes of 4 isolates (Longyan, Nanchang, Jinhua and Yingtan). Figure 2 shows an analysis of the selection pressure of each protein-coding gene, which revealed that the ratio of non-synonymous mutation to synonymous mutation (Ka/Ks) for each protein-coding gene was less than 1, and the ratio of nad3 and nad4L was 0.
Table 2.
Dermacentor steini variation in nucleotide and predicted amino acid sequences of the mitochondrial genome of different isolates
| D. steini | Nucleotide sequence length | Nucleotide variance (%) | Number of amino acids | Amino acids variance (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Longyan | Jinhua | Nangchang | Yingtan | Longyan | Jinhua | Nanchang | Yingtan | |||
| Mitogenome | 14 785 | 14 773 | 14 785 | 14 767 | 0.76 | – | – | – | – | – |
| AT% | 76.9 | 76.9 | 76.9 | 76.8 | – | – | – | – | – | – |
| cox1 | 1539 | 1539 | 1539 | 1539 | 0.97 | 512 | 512 | 512 | 512 | 0.20 |
| cox2 | 673 | 673 | 673 | 673 | 0.74 | 224 | 224 | 224 | 224 | 0.13 |
| cox3 | 779 | 778 | 779 | 779 | 0.90 | 259 | 259 | 259 | 259 | 0.77 |
| nad1 | 939 | 939 | 939 | 939 | 0.53 | 312 | 312 | 312 | 312 | 0.32 |
| nad2 | 960 | 960 | 960 | 960 | 0.94 | 319 | 319 | 319 | 319 | 1.56 |
| nad3 | 342 | 339 | 342 | 339 | 0.00 | 113 | 112 | 113 | 112 | 0.88 |
| nad4 | 1315 | 1315 | 1315 | 1315 | 0.61 | 438 | 438 | 438 | 438 | 0.46 |
| nad4L | 276 | 276 | 276 | 276 | 0.36 | 91 | 91 | 91 | 91 | 0.00 |
| nad5 | 1659 | 1659 | 1659 | 1659 | 0.42 | 552 | 552 | 552 | 552 | 0.36 |
| nad6 | 435 | 450 | 435 | 450 | 1.11 | 144 | 149 | 144 | 149 | 0.67 |
| atp6 | 666 | 666 | 666 | 666 | 0.45 | 221 | 221 | 221 | 221 | 0.00 |
| atp8 | 159 | 159 | 159 | 159 | 1.89 | 53 | 53 | 53 | 53 | 3.85 |
| cytb | 1083 | 1083 | 1083 | 1083 | 0.55 | 360 | 360 | 360 | 360 | 0.83 |
| rrnL | 1200 | 1193 | 1200 | 1193 | 1.00 | – | – | – | – | – |
| rrnS | 712 | 710 | 708 | 706 | 1.54 | – | – | – | – | – |
Fig. 2.
Selection pressure analysis for different isolates in Dermacentor steini.
The nucleotide and amino acid variations of the 13 PCGs in Longyan, Nanchang, Jinhua and Yingtan isolates are shown in Table 2. The highest variation in nucleotides between homologous genes was found for atp8 (1.89%), and with lowest variation gene was nad3 (0.00%). For amino acid sequences, the lowest variation genes were nad4 and atp6 (0.00%), and the atp8 gene had the highest variation (5.66%). These results suggest that the atp8 gene was the most variable. The 2 rRNAs (rrnL and rrnS) of the Longyan, Nanchang, Jinhua and Yingtan isolates were similar in length to other ticks (Burger et al., 2014; Williams-Newkirk et al., 2015). rrnL was located between trnL and trnV with a 1.00% nucleotide variation; rrnS was located between trnV and NCR1 with a nucleotide variation of 1.54%.
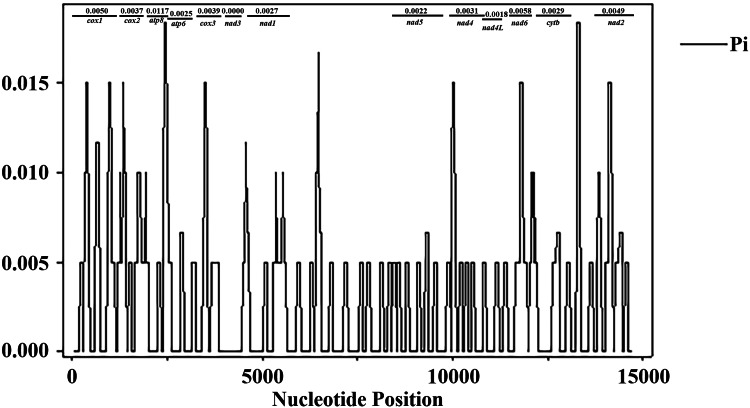
The nucleotide diversity (Pi) of the 13 PCGs of D. steini (Longyan, Nanchang, Jinhua and Yingtan isolates) was analysed using DnaSP (Fig. 3). It was found that the nucleotide variation of D. steini within and between genes was low, in the range of 0.0000–0.0117. According to the window analysis, the most variable sequences were atp8 (0.0117), nad6 (0.0058), cox1 (0.0050) and nad2 (0.0049), while the most conserved sequences were nad3 (0.000).
Fig. 3.
Sliding window analysis of the complete mitochondrial genomes of Dermacentor steini different isolates. The black line indicates the nucleotide diversity values (Pi) in the sliding window analysis with a window size of 100 bp and a step size of 25; the gene boundaries are indicated by the variation rate of each gene.
Rearrangement and phylogenetic analysis of the mitochondrial genome of D. steini (Longyan, Nanchang, Jinhua and Yingtan isolates)
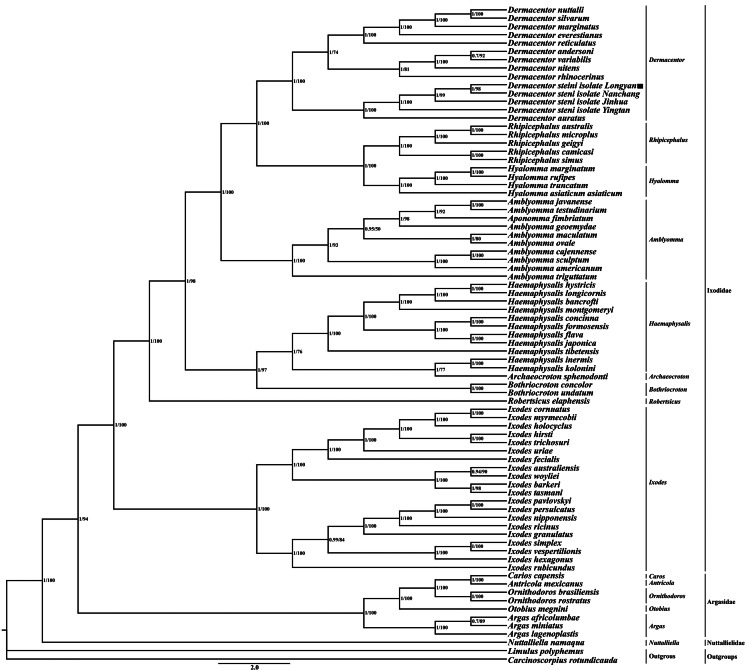
Although the mitochondrial genome evolves at a rapid rate, the order of the 37 genes is very conservative, and the rearrangement characteristics of mitochondrial genes are generally used to resolve controversial phylogenetic questions (Babbucci et al., 2014). Compared to the mitochondrial genome the arrangement pattern of arthropod hypothetical ancestors, smaller rearrangements occurred in the mitochondrial genomes of D. steini (Longyan, Nanchang, Jinhua and Yingtan isolates) and other species of the genus Dermacentor (Fig. 4). Rearrangements occurred mainly in 3 regions, namely, nad1-trnL2, trnI-trnQ-trnF, underwent translocations, as well as trnC underwent translocations and inversions, and all rearrangements occurred in the vicinity of control regions and tRNA genes. Phylogenetic trees were constructed using the maximum likelihood and Bayesian methods based on 13 PCGs with L. polyphemus (NC003057) and C. rotundicauda (JX437074) as outgroups (Fig. 5). The topology of the phylogenetic tree formed by the 2 methods was almost identical, except for slight differences in the support rates of branch nodes: the support rate of the Bayesian tree (BI) was generally higher than that of the maximum likelihood tree (ML), with the majority of nodes having a support rate of 1; while the support rates of the maximum likelihood tree (ML) were all in the range of 80–100, except for 3 nodes with a support rate of less than 80. The phylogenetic tree shows that the family Ixodidae is a monophyletic group, except for the genus Archaeocroton, in which all other genera are clustered separately into a single group. D. steini isolated from Longyan and Nanchang clustered preferentially together and then formed sister branches with high node support (BP > 95, PP > 0.95) to D. steini isolated from Jinhua and Yingtan. The present phylogenetic tree results show that the genus Dermacentor is most closely related to 2 genera (Rhipicentor and Hyalomma) in the family Ixodidae (BP > 95, PP > 0.95).
Fig. 4.
Mitochondrial genome rearrangement evolution in the genus Dermacentor. Green indicates translocations, blue indicates translocations and inversions.
Fig. 5.
Phylogenetic analysis based on the nucleotide sequences of the 13 PCGs in the mitogenome. The numbers beside the nodes are posterior probabilities (BI) and bootstrap (ML) indicates the species in this study.
Discussion
Most species of the genus Dermacentor are 3-host ticks, with larval and nymph ticks mainly parasitizing small mammals and adult ticks mainly parasitizing cattle, sheep and wild medium and large mammals, and are capable of transmitting a variety of diseases to humans and animals (Hajdušek et al., 2013; Ernieenor et al., 2020). Molecular biology studies are essential for the surveillance, prevention and control of tick-borne diseases.
In this study, the complete mitochondrial genome of D. steini (Longyan isolate) was sequenced and compared with the Nanchang, Jinhua and Yingtan isolates. The mitochondrial genome of D. steini (Longyan, Nanchang, Jinhua and Yingtan isolates) was found to be shorter than that of most species of the genus Dermacentor. The size of the mitochondrial genome varied among species, mainly due to the differences in the number and length of non-coding regions (Mo et al., 2019). The nucleotide composition of D. steini (Longyan, Nanchang, Jinhua and Yingtan isolates) showed a distinct AT preference, ranging from 76.8% to 76.9%, which is within the reported range of the Dermacentor genus ticks (Xin et al., 2018). It has been suggested that the higher AT content than GC content is a common derivative of arthropod mitochondrial genomes (Ikemura, 1985; Simon et al., 1994). All 37 mitochondrial genes of D. steini (Longyan, Nanchang, Jinhua and Yingtan isolates) were non-homogeneously distributed. Of the 23 genes located in the J-strand, the remaining genes were in the N-strand. In general, mitochondrial genes located on the J-strand are more susceptible to hydrolysis and oxidation (Brown et al., 1982). Among the 13 PCGs of D. steini (Longyan, Nanchang, Jinhua and Yingtan isolates), 4 genes (nad1, nad4, nad4L, nad5) were located on the N-strand, indicating that these 4 genes have relative stability.
The initiation codon and stop codon differ between species, and variations in the genetic code can be used as specific markers of the mitochondrial genome (Murrell et al., 2003). For example, ATN is considered to be the most common initiation codon in Insecta, while AGA and AGG are the most common stop codons in vertebrate mitochondrial genomes (Wolstenholme, 1992; Wang et al., 2019). The most common initiation and stop codons of D. steini (Longyan, Nanchang, Jinhua and Yingtan isolates) are ATG and TAA, respectively, which are also found in the mitochondrial genomes of other ticks (Liu et al., 2018). Three PCGs (cox2, cox3 and nad4) have incomplete stop codons, and those incomplete stop codons can be modified by post-transcriptional polyadenylation to eventually form complete stop codons (Donath et al., 2019). The mitochondrial protein-encoding genes have been conserved (Chen et al., 2012). The Ka/Ks of the 13 PCGs of D. steini (Longyan, Nanchang, Jinhua and Yingtan isolates) were all less than 1, indicating that these 13 PCGs were subject to purifying selection and did not undergo rapid evolution. The Ka/Ks of nad3 and nad4L were 0, further indicating that these 2 genes are highly conserved.
As the mitochondrial tRNA genes are approximately the same size, in comparison to nuclear tRNA genes, mitochondrial tRNA genes have a higher evolution rate (Saccone et al., 1999). The trnS1 gene lacks the D-arm and fails to form a typical cloverleaf secondary structure, which is considered to be an ancestral characteristic of metazoan (Lowe and Chan, 2016). The 22 tRNA genes of D. steini (Longyan isolate) also have the trnC gene lacking the D-arm. Dermauw et al. considered the trnC gene lacking D-arm as a common ancestral characteristic of Mesostigmata (Acari: Parasitiformes) species (Dermauw et al., 2010), but it has not been confirmed in the mitochondrial genome of ticks. The D. steini (Longyan isolate) tRNA gene underwent 9 G–U pairings and 3 U–U mismatches during the folding process. Compared with bacterial and cytoplasmic tRNA genes, mitochondrial tRNA genes contain more G–U pairing and mismatching, which is an important factor that makes mitochondrial tRNA genes less stable than cytoplasmic tRNA genes (Helm et al., 2000). The G–U pairing and mismatches that occur on mitochondrial tRNA genes are highly conserved in evolution or involved in the formation of higher structures that can be corrected by RNA editing, and their normal transport function is not affected, which is important for maintaining the stability and specificity of tRNA genes (Kunzmann et al., 1998; Helm et al., 2000). It has been thought that mismatches in tRNA genes may be linked to species evolution (Watanabe et al., 1994). Hence, whether the base mismatches occurring in the mitochondrial tRNA gene of D. steini (Longyan isolate) are related to its evolution needs to be further explored.
Geographic location is generally considered to be one of the factors influencing variation in the mitochondrial genome (Burnard and Shao, 2019). The overall variation among the mitochondrial genome sequences of D. steini (Longyan, Nanchang, Jinhua and Yingtan isolates) was 0.76%, which was less than 1%. It suggests that the variation in the mitochondrial genome of D. steini may be less affected by geographical location; it may also be due to the proximity (<1000 km) between the 4 locations (Longyan, Nanchang, Jinhua and Yingtan), which does not reflect the intra-species variation. The nucleotide and amino acid sequences of homologous genes as well as nucleotide diversity analysis showed that atp8 was the least conserved gene, which is consistent with the results of other tick species (Guo et al., 2016). It further suggests that the atp8 gene can be used as an alternative genetic marker for studying D. steini from different host body surfaces and different biogeographies.
The altered gene order of the mitochondrial genome is a well-known specific molecular marker that contains important phylogenetic information (Boore, 1999; Boore and Fuerstenberg, 2008). The study of mitochondrial genome rearrangement events is one of the hotspots for studying the evolutionary process of species. The mitochondrial genome arrangement is generally considered to be highly conserved at lower taxonomic levels (families and genera) and rearrangements occur much more frequently for short fragments than for long fragments (Dowton et al., 2009; Yuan et al. 2010). The mitochondrial genome rearrangements of species within the genus Dermacentor are relatively simple, not occurring in some very conserved regions (rrnL-trnV-rrnS), but all in 3 regions (nad1-trnL2, trnI-trnQ-trnF and trnC). These 3 regions are located in the vicinity of the control region and tRNA genes. It has been demonstrated in the mitochondrial genomes of sucking lice, insects and mites that the control region and the vicinity of tRNA genes are the hotspot regions in which rearrangements occur (Wei et al., 2009, 2010; Dong et al., 2021). It has been suggested that replication of the control region is responsible for the accelerated frequency of mitochondrial gene rearrangements, as in Thrips and Corrodentia (Shao and Barker, 2003; Shao et al., 2005; Yan et al., 2012). It remains to be verified whether the accelerated frequency of mitochondrial genome rearrangements in ticks is related to the replication of control regions. The BI and ML trees indicate that the family Ixodidae is a monophyletic group and the phylogenetic relationships within the family are relatively clear. D. steini isolated from Longyan clustered with 3 other isolates (Nanchang, Jinhua and Yingtan), and then formed a sister branch with D. auratus with high node support. It indicates that among the genus Dermacentor, D. steini is most closely related to D. auratus. It may be related to the similarity of these 2 species in morphology and in the pathogens they carry. Two phylogenetic trees showed that the genus Dermacentor is most closely related to 2 genera (Rhipicentor and Hyalomma), which is consistent with previous studies (Burnard and Shao, 2019; Uribe et al., 2020; Kelava et al., 2021). The analysis of the affinities of the genus Dermacentor facilitates the subsequent understanding of the phylogenetic analysis of species in the genus Dermacentor.
Conclusion
In this study, the complete mitochondrial genome of D. steini (Longyan isolate) parasitized on R. andamanensis was sequenced and compared with Nanchang, Jinhua and Yingtan isolates. The results showed that the mitochondrial genomes of D. steini different isolates were similar in length and the overall variation between sequences was less than 1%; the atp8 gene may serve as an alternative genetic marker for studying D. steini species from different hosts and biogeography; the mitochondrial genome underwent minor rearrangements. Phylogenetic analyses showed that the genus Dermacentor is most closely related to 2 genera (Rhipicentor and Hyalomma) in the family Ixodidae. The results enrich the mitochondrial genomic data of species in the genus Dermacentor and have important implications for the phylogeographic classification and molecular evolution of ticks.
Acknowledgements
We thank Gangxian He for his assistance in the initial formation of the circular mitochondrial genome map
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0031182022001639.
click here to view supplementary material
Data availability
The complete mitochondrial genome sequence of D. steini is available at the National Center for Biotechnology Information (NCBI) at [https://www.ncbi.nlm.nih.gov/] under accession number OP383032.
Author's contributions
Huijuan Yang and Wenge Dong conceived and designed the study. Huijuan Yang and Ting Chen conducted data gathering. Huijuan Yang performed statistical analyses. Huijuan Yang and Ting Chen wrote the manuscript. Wenge Dong revised the manuscript.
Financial support
This work was supported by the National Natural Science Foundation of China [No. 32060143 to Wenge Dong] and Major Science and Technique Programs in Yunnan Province [No. 202102AA310055-X to Xianguo Guo].
Conflict of interest
The authors declare that they have no conflicts of interests.
Ethical standards
Small mammal capture protocols and procedures were approved by the Animal Ethics Committees at Dali University. The approval ID is MECDU-201806-11.
References
- Babbucci M, Basso A, Scupola A, Patarnello T and Negrisolo E (2014) Is it an ant or a butterfly? Convergent evolution in the mitochondrial gene order of Hymenoptera and Lepidoptera. Genome Biology and Evolution 6, 3326–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M and Stadler PF (2013) MITOS: improved de novo metazoan mitochondrial genome annotation. Molecular Phylogenetics and Evolution 69, 313–319. [DOI] [PubMed] [Google Scholar]
- Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Research 27, 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL and Fuerstenberg SI (2008) Beyond linear sequence comparisons: the use of genome-level characters for phylogenetic reconstruction. Philosophical Transactions of the Royal Society B: Biological Sciences 363, 1445–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WM, Prager EM, Wang A and Wilson AC (1982) Mitochondrial DNA sequences of primates: tempo and mode of evolution. Journal of Molecular Evolution 18, 225–239. [DOI] [PubMed] [Google Scholar]
- Burger TD, Shao RF and Barker SC (2013) Phylogenetic analysis of the mitochondrial genomes and nuclear rRNA genes of ticks reveals a deep phylogenetic structure within the genus Haemaphysalis and further elucidates the polyphyly of the genus Amblyomma with respect to Amblyomma sphenodonti and Amblyomma elaphense. Ticks and Tick-Borne Diseases 4, 265–274. [DOI] [PubMed] [Google Scholar]
- Burger TD, Shao RF and Barker SC (2014) Phylogenetic analysis of mitochondrial genome sequences indicates that the cattle tick, Rhipicephalus (Boophilus) microplus, contains a cryptic species. Molecular Phylogenetics and Evolution 76, 241–253. [DOI] [PubMed] [Google Scholar]
- Burnard D and Shao RF (2019) Mitochondrial genome analysis reveals intraspecific variation within Australian hard tick species. Ticks and Tick-Borne Diseases 10, 677–681. [DOI] [PubMed] [Google Scholar]
- Chang QC, Liu GH, Gao JF, Zheng X, Zhang Y, Duan H, Yue DM, Fu X, Su X and Wang CR (2016) Sequencing and characterization of the complete mitochondrial genome from the pancreatic fluke Eurytrema pancreaticum (Trematoda: Dicrocoeliidae). Gene 576, 160–165. [DOI] [PubMed] [Google Scholar]
- Chen Z, Yang X, Bu F, Yang X, Yang X and Liu J (2010) Ticks (Acari: Ixodoidea: Argasidae, Ixodidae) of China. Experimental and Applied Acarology 51, 393–404. [DOI] [PubMed] [Google Scholar]
- Chen M, Tian LL, Shi QH, Cao TW and Hao JS (2012) Complete mitogenome of the lesser purple emperor Apatura ilia (Lepidoptera: Nymphalidae: Apaturinae) and comparison with other nymphalid butterflies. Zoological Research 33, 191–201. [DOI] [PubMed] [Google Scholar]
- Dermauw W, Vanholme B, Tirry L and Van Leeuwen T (2010) Mitochondrial genome analysis of the predatory mite Phytoseiulus persimilis and a revisit of the Metaseiulus occidentalis mitochondrial genome. Genome 53, 285–301. [DOI] [PubMed] [Google Scholar]
- Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M and Bernt M (2019) Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Research 47, 10543–10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong WG, Dong Y, Guo XG and Shao RF (2021) Frequent tRNA gene translocation towards the boundaries with control regions contributes to the highly dynamic mitochondrial genome organization of the parasitic lice of mammals. BMC Genomics 22, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowton M, Cameron SL, Dowavic JI, Austin AD and Whiting MF (2009) Characterization of 67 mitochondrial tRNA gene rearrangements in the Hymenoptera suggests that mitochondrial tRNA gene position is selectively neutral. Molecular Biology and Evolution 26, 1607–1617. [DOI] [PubMed] [Google Scholar]
- Duan H, Gao JF, Hou MR, Zhang Y, Liu ZX, Gao DZ, Guo DH, Yue DM, Su X, Fu X and Wang CR (2015) Complete mitochondrial genome of an equine intestinal parasite, Triodontophorus brevicauda (Chromadorea: Strongylidae): the first characterization within the genus. Parasitology International 64, 429–434. [DOI] [PubMed] [Google Scholar]
- Duncan KT, Grant A, Johnson B, Sundstrom KD, Saleh MN and Little SE (2021) Identification of Rickettsia spp. and Babesia conradae in Dermacentor spp. collected from dogs and cats across the United States. Vector-Borne and Zoonotic Diseases 21, 911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen RJ and Paddock CD (2021) Tick and tickborne pathogen surveillance as a public health tool in the United States. Journal of Medical Entomology 58, 1490–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernieenor FCL, Apanaskevich DA, Ernna G and Mariana A (2020) Description and characterization of questing hard tick, Dermacentor steini (Acari: Ixodidae) in Malaysia based on phenotypic and genotypic traits. Experimental and Applied Acarology 80, 137–149. [DOI] [PubMed] [Google Scholar]
- Ernieenor FCL, Apanaskevich DA, Ernna G, Ellyncia BB, Mariana A and Yaakop S (2021) Morphological and molecular identification of medically important questing Dermacentor species collected from some recreational areas of Peninsular Malaysia. Systematic Parasitology 98, 731–751. [DOI] [PubMed] [Google Scholar]
- Guo DH, Zhang Y, Fu X, Gao Y, Liu YT, Qiu JH, Chang QC and Wang CR (2016) Complete mitochondrial genomes of Dermacentor silvarum and comparative analyses with another hard tick Dermacentor nitens. Experimental Parasitology 169, 22–27. [DOI] [PubMed] [Google Scholar]
- Hajdušek O, Šíma R, Ayllón N, Jalovecká M, Perner J, De La Fuente J and Kopáček P (2013) Interaction of the tick immune system with transmitted pathogens. Frontiers in Cellular and Infection Microbiology 3, 26–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Brule H, Friede D, Giege R, Puetz D and Florenta C (2000) Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA 6, 1356–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T (1985) Codon usage and tRNA content in unicellular and multicellular organisms. Molecular Biology and Evolution 2, 13–34. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thiere T, Ashton B, Meintjes P and Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics (Oxford, England) 28, 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelava S, Mans BJ, Shao RF, Moustafa MAM, Matsuno K, Takano A, Kawabata H, Sato K, Fujita H, Ze C, Plantard O, Hornok S, Gao S, Barker D, Barker SC and Nakao R (2021) Phylogenies from mitochondrial genomes of 120 species of ticks: insights into the evolution of the families of ticks and of the genus Amblyomma. Ticks and Tick-Borne Diseases 12, 101577.. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G and Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzmann A, Brennicke A and Marchfelder A (1998) 5′ end maturation and RNA editing have to precede tRNA 3′ processing in plant mitochondria. Proceedings of the National Academy of Sciences 95, 108–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D and Canbäck B (2008) ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics (Oxford, England) 24, 172–175. [DOI] [PubMed] [Google Scholar]
- Librado P and Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics (Oxford, England) 25, 1451–1452. [DOI] [PubMed] [Google Scholar]
- Ličková M, Havlíková SF, Sláviková M, Slovák M, Drexler JF and Klempa B (2020) Dermacentor reticulatus is a vector of tick-borne encephalitis virus. Ticks and Tick-Borne Diseases 11, 101414. [DOI] [PubMed] [Google Scholar]
- Lim FS, Khoo JJ, Tan KK, Zainal N, Loong SK, Khor CS and AbuBakar S (2020) Bacterial communities in Haemaphysalis, Dermacentor and Amblyomma ticks collected from wild boar of an Orang Asli Community in Malaysia. Ticks and Tick-Borne Diseases 11, 101352. [DOI] [PubMed] [Google Scholar]
- Liu ZQ, Liu YF, Kuermanali N, Wang DF, Chen SJ, Guo HL, Zhao L, Wang JW, Han T, Wang YZ, Wang J, Shen CF, Zhang ZZ and Chen CF (2018) Sequencing of complete mitochondrial genomes confirms synonymization of Hyalomma asiaticum asiaticum and kozlovi, and advances phylogenetic hypotheses for the Ixodidae. PLoS One 13, e0197524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM and Chan PP (2016) tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Research 44, W54–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM and Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariana A, Srinovianti N, Ho TM, Halimaton I, Hatikah A, Shaharudin MH, Rosmaliza I, Wan Ishlah L and Sathananthar KS (2008) Intra-aural ticks (Acari: Metastigmata: Ixodidae) from human otoacariasis cases in Pahang, Malaysia. Asian Pacific Journal of Tropical Medicine 1, 20–24. [Google Scholar]
- Mead P, Hinckley A, Hook S and Beard CB (2015) TickNET – a collaborative public health approach to tickborne disease surveillance and research. Emerging Infectious Diseases 21, 1574–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo W, Guo L, Xiao Y, Xin T, Xia B and Zou Z (2019) The complete mitochondrial genome of Stratiolaelaps scimitus (Acari: Laelapidae) and the comparison of mitochondrial gene rearrangement in the Mesostigmata. International Journal of Acarology 45, 421–427. [Google Scholar]
- Murrell A, Campbell NJ and Barker SC (2003) The value of idiosyncratic markers and changes to conserved tRNA sequences from the mitochondrial genome of hard ticks (Acari: Ixodida: Ixodidae) for phylogenetic inference. Systematic Biology 52, 296–310. [DOI] [PubMed] [Google Scholar]
- Nava S, Venzal JM, González-Acuña D, Martins TF and Guglielmone AA (2017) Ticks of the Southern Cone of America: Diagnosis, Distribution, and Hosts with Taxonomy, Ecology and Sanitary Importance. London, UK: Academic Press. [Google Scholar]
- Parola P, Paddock CD and Raoult D (2005) Tick-borne rickettsioses around the world: emerging diseases challenging old concepts. Clinical Microbiology Reviews 18, 719–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone C, De Giorgi C, Gissi C, Pesole G and Reyes A (1999) Evolutionary genomics in Metazoa: the mitochondrial DNA as a model system. Gene 238, 195–209. [DOI] [PubMed] [Google Scholar]
- Shao RF and Barker SC (2003) The highly rearranged mitochondrial genome of the plague thrips, Thrips imaginis (Insecta: Thysanoptera): convergence of two novel gene boundaries and an extraordinary arrangement of rRNA genes. Molecular Biology and Evolution 20, 362–370. [DOI] [PubMed] [Google Scholar]
- Shao RF, Barker SC, Mitani H, Aoki Y and Fukunaga M (2005) Evolution of duplicate control regions in the mitochondrial genomes of metazoa: a case study with Australasian Ixodes ticks. Molecular Biology and Evolution 22, 620–629. [DOI] [PubMed] [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H and Flook P (1994) Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Annals of the Entomological Society of America 87, 651–701. [Google Scholar]
- Swei A, O'Connor KE, Couper LI, Thekkiniath J, Conrad PA, Padgett KA, Burns J, Yoshimizu MH, Gonzales B, Munk B, Shirkey N, Konde L, Mamoun CB, Lane RS and Kjemtrup A (2019) Evidence for transmission of the zoonotic apicomplexan parasite Babesia duncani by the tick Dermacentor albipictus. International Journal for Parasitology 49, 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe JE, Nava S, Murphy KR, Tarragona EL and Castro LR (2020) Characterization of the complete mitochondrial genome of Amblyomma ovale, comparative analyses and phylogenetic considerations. Experimental and Applied Acarology 81, 421–439. [DOI] [PubMed] [Google Scholar]
- Vongphayloth K, Brey PT, Robbins RG and Sutherland IW (2016) First survey of the hard tick (Acari: Ixodidae) fauna of Nakai District, Khammouane Province, Laos, and an updated checklist of the ticks of Laos. Systematic and Applied Acarology 21, 166–180. [Google Scholar]
- Wang Q and Tang G (2017) Genomic and phylogenetic analysis of the complete mitochondrial DNA sequence of walnut leaf pest Paleosepharia posticata (Coleoptera: Chrysomeloidea). Journal of Asia-Pacific Entomology 20, 840–853. [Google Scholar]
- Wang Z, Wang Z, Shi X, Wu Q, Tao Y, Guo H, Ji CY and Bai Y (2018) Complete mitochondrial genome of Parasesarma affine (Brachyura: Sesarmidae): gene rearrangements in Sesarmidae and phylogenetic analysis of the Brachyura. International Journal of Biological Macromolecules 118, 31–40. [DOI] [PubMed] [Google Scholar]
- Wang T, Zhang S, Pei T, Yu Z and Liu J (2019) Tick mitochondrial genomes: structural characteristics and phylogenetic implications. Parasites and Vectors 12, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Shi X, Guo H, Tang D, Bai Y and Wang Z (2020) Characterization of the complete mitochondrial genome of Uca lacteus and comparison with other Brachyuran crabs. Genomics 112, 10–19. [DOI] [PubMed] [Google Scholar]
- Wassef HY and Hoogstraal H (1986) Dermacentor (Indocentor) steini (Acari: Ixodoidea: Ixodidae): identity of male and female. Journal of Medical Entomology 23, 532–537. [DOI] [PubMed] [Google Scholar]
- Wassef HY and Hoogstraal H (1988) Dermacentor (Indocentor) steini (Acari: Ixodoidea: Ixodidae): hosts, distribution in the Malay Peninsula, Indonesia, Borneo, Thailand, the Philippines, and New Guinea. Journal of Medical Entomology 25, 315–320. [DOI] [PubMed] [Google Scholar]
- Watanabe YI, Kawai G, Yokogawa T, Hayashi N, Kumazawa Y, Ueda T, Nishikawa K, Hirao I, Miura KI and Watanabe K (1994) Higher-order structure of bovine mitochondrial tRNASerUGA: chemical modification and computer modeling. Nucleic Acids Research 22, 5378–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SJ, Shi M, He JH, Sharkey M and Chen XX (2009) The complete mitochondrial genome of Diadegma semiclausum (Hymenoptera: Ichneumonidae) indicates extensive independent evolutionary events. Genome 52, 308–319. [DOI] [PubMed] [Google Scholar]
- Wei SJ, Shi M, Sharkey MJ, van Achterberg C and Chen XX (2010) Comparative mitogenomics of Braconidae (Insecta: Hymenoptera) and the phylogenetic utility of mitochondrial genomes with special reference to Holometabolous insects. BMC Genomics 11, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Newkirk AJ, Burroughs M, Changayil SS and Dasch GA (2015) The mitochondrial genome of the lone star tick (Amblyomma americanum). Ticks and Tick-Borne Diseases 6, 793–801. [DOI] [PubMed] [Google Scholar]
- Wolstenholme DR (1992) Animal mitochondrial DNA: structure and evolution. International Review of Cytology 141, 173–216. [DOI] [PubMed] [Google Scholar]
- Xin ZZ, Liu Y, Zhang DZ, Wang ZF, Tang BP, Zhang HB, Zhou CL, Chai XY and Liu QN (2018) Comparative mitochondrial genome analysis of Spilarctia subcarnea and other noctuid insects. International Journal of Biological Macromolecules 107, 121–128. [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Ohmae H and Kuramochi T (2012) Complete mitochondrial genomes of Diplogonoporus balaenopterae and Diplogonoporus grandis (Cestoda: Diphyllobothriidae) and clarification of their taxonomic relationships. Parasitology International 61, 260–266. [DOI] [PubMed] [Google Scholar]
- Yan DK, Tang YX, Xue XF, Wang MH, Liu FQ and Fan JQ (2012) The complete mitochondrial genome sequence of the western flower thrips Frankliniella occidentalis (Thysanoptera: Thripidae) contains triplicate putative control regions. Gene 506, 117–124. [DOI] [PubMed] [Google Scholar]
- Yang Y, Christie J, Köster L, Du AF and Yao CQ (2021) Emerging human babesiosis with “ground zero” in North America. Microorganisms 9, 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HJ, Chen T and Dong WG (2022a) The complete mitochondrial genome of Parasitus fimetorum (Berlese, 1904) (Arachnida: Parasitiformes: Parasitidae). Mitochondrial DNA Part B 7, 1044–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HJ, Yang ZH and Dong WG (2022b) Morphological identification and phylogenetic analysis of Laelapin Mite Species (Acari: Mesostigmata: Laelapidae) from China. The Korean Journal of Parasitology 60, 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan ML, Wei DD, Wang BJ, Dou W and Wang JJ (2010) The complete mitochondrial genome of the citrus red mite Panonychus citri (Acari: Tetranychidae): high genome rearrangement and extremely truncated tRNAs. BMC Genomics 11, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX and Wang GT (2020) PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources 20, 348–355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0031182022001639.
click here to view supplementary material
Data Availability Statement
The complete mitochondrial genome sequence of D. steini is available at the National Center for Biotechnology Information (NCBI) at [https://www.ncbi.nlm.nih.gov/] under accession number OP383032.