Key words: Anthropogenic disturbance, β-specificity, DNA barcoding, habitat degradation, habitat restoration, host specialization, microsporidia, phylogenetic specificity
Abstract
The present study provides new insight into suitable microsporidian–host associations. It relates regional and continental-wide host specialization in microsporidians infecting amphipods to degraded and recovering habitats across 2 German river catchments. It provides a unique opportunity to infer the persistence of parasites following anthropogenic disturbance and their establishment in restored rivers. Amphipods were collected in 31 sampling sites with differing degradation and restoration gradients. Specimens were morphologically (hosts) and molecularly identified (host and parasites). Amphipod diversity and abundance, microsporidian diversity, host phylogenetic specificity and continental-wide β-specificity were investigated and related to each other and/or environmental variables. Fourteen microsporidian molecular operational taxonomic units (MOTUs), mainly generalist parasites, infecting 6 amphipod MOTUs were detected, expanding the current knowledge on the host range by 17 interactions. There was no difference in microsporidian diversity and host specificity among restored and near-natural streams (Boye) or between those located in urban and rural areas (Kinzig). Similarly, microsporidian diversity was generally not influenced by water parameters. In the Boye catchment, host densities did not influence microsporidian MOTU richness across restored and near-natural sites. High host turnover across the geographical range suggests that neither environmental conditions nor host diversity plays a significant role in the establishment into restored areas. Host diversity and environmental parameters do not indicate the persistence and dispersal of phylogenetic host generalist microsporidians in environments that experienced anthropogenic disturbance. Instead, these might depend on more complex mechanisms such as the production of resistant spores, host switching and host dispersal acting individually or conjointly.
Introduction
Freshwater ecosystems being particularly vulnerable to human activity and environmental changes are among the most impacted ecosystems in the world (Dudgeon et al., 2006). Impacts such as pollution, flow alteration and habitat destruction or degradation commonly result in biodiversity loss (Dudgeon et al., 2006; Birk et al., 2020). Fortunately, ecosystem restoration efforts to reverse biodiversity losses have soared in recent years (Fischer et al., 2021). However, the success of such initiatives is often hampered by complex interactions within living organisms and their responses to anthropogenic stressors (Lorenz et al., 2018; Birk et al., 2020). In this context, understanding less investigated aspects such as host–parasite interactions is of particular relevance.
Most parasites are dependent on their hosts. Some parasites are generalist and exploit a broad range of hosts, while others are specialized on one or a few hosts (Dobson et al., 2008; Lafferty, 2012). Heteroxenous parasites require multiple hosts to complete their life cycle, while monoxenous parasites need just one. Therefore, anthropogenic stressors can affect both hosts and parasites (Dobson et al., 2008; Moir et al., 2010). For instance, a reduction of hosts diversity and abundance caused by habitat degradation might trigger heteroxenous specialist parasite extinctions (Dobson et al., 2008; Moir et al., 2010). On the other hand, monoxenous generalist parasites are more likely to persist in degraded environments due to simple life cycles and host plasticity (Poulin and Morand, 2004; Lafferty, 2012). Hence, ubiquitous parasites with either simple or complex life cycles and various host specialization degrees such as microsporidians may reflect ecosystem degradation and recovery histories.
Microsporidians are a diverse and successful group of eukaryotic obligate intracellular parasites that exploit horizontal, vertical and mixed-mode transmissions (Dunn and Smith, 2001; Stentiford et al., 2013; Wadi and Reinke, 2020). Horizontal transmission occurs between related or unrelated hosts via spore ingestion, venereally or by direct invasion, and it is often associated with high host mortality (Wittner and Weiss, 1999). In contrast, vertical transmission occurs when spores are passed intergenerationally via transovarial transmission and is generally less virulent (Dunn and Smith, 2001). Despite being ubiquitous, microsporidians have received little attention, particularly those infecting aquatic organisms with little or no commercial value but high ecological importance, such as amphipods (Quiles et al., 2021). Microsporidian diversity in amphipods is high as evidenced by several studies (Grabner et al., 2015; Ironside and Wilkinson, 2018; Bojko and Ovcharenko, 2019; Park et al., 2020). Still, only a few common species have been characterized and described such as the horizontally transmitted Cucumispora spp. (Bojko et al., 2015, 2017) and vertically transmitted Nosema granulosis (Terry et al., 1999) and Dictyocoela spp. (Bacela-Spychalska et al., 2018). However, given the fragmentary nature of available information, knowledge of host specificity in microsporidians infecting amphipods and persistence following environmental disturbance remains scarce.
The present study provides new insight into microsporidian–host associations. It relates regional and continental-wide host specialization in microsporidians infecting amphipods to a gradient of degraded and recovering habitats across 2 German river catchments offering a unique opportunity to infer the persistence of parasites following anthropogenic disturbance and their establishment in restored rivers. One catchment, the Boye, has partly been used as an open sewer since the beginning of the last century and, starting from the 1990s, it was gradually restored (Winking et al., 2014, 2016). In contrast, the Kinzig catchment has suffered from moderate hydromorphological degradation, land use and wastewater overflow.
The first hypothesis is that microsporidian parasites in streams used as open sewers in the Boye catchment and those flowing in urbanized areas in the Kinzig catchment would have lower diversity and host specificity compared to sites in a near-natural state. Secondly, that to persist in such environments, microsporidian parasites should be able to use available amphipod hosts independently from their phylogenetic diversity. Thus, their presence is likely to depend on available host densities. Finally, if microsporidians have a wide geographical range and higher turnover of host species across their geographical distribution, both environmental conditions and host diversity play minor roles in their establishment in restored areas.
Materials and methods
Sampling
The sample consists of 519 amphipods collected in 31 sampling sites spread across the Boye (n = 13) and Kinzig (n = 18) catchments (Fig. 1). The source and upstream tributaries of the Boye are located in agricultural or forested areas and have never been used as open sewers, thus retaining near-natural conditions (Table 1). In contrast, the downstream section and tributaries which flow in urbanized areas were transformed into concrete channels at the beginning of the last century to transport domestic wastewater (Winking et al., 2014, 2016). These open sewers were lifeless except for rare occurrences of Oligochaeta. Still, starting from the 1990s, they have undergone partial restoration and subsequent colonization by pioneer invertebrate assemblages originating from upstream sections and neighbouring catchments (Winking et al., 2014, 2016). Tributaries of the river Kinzig, located in a rural landscape in the low mountain range of Hessen county, Germany, on the other hand, are in a near-natural state (Table 1). Samples were collected in March 2021 and immediately fixed in 96% ethanol. A standardized multi-habitat-sampling (Meier et al., 2006) was employed to estimate benthic invertebrate densities, while additional amphipod specimens were collected separately for parasitological analyses. Amphipods were morphologically identified to the lowest taxonomical level, dissected and screened for parasites under the microscope. Intestines were removed to avoid contamination between muscular tissues and the intestinal content. A small sample of muscular tissue was then used for molecular identification of both host and parasites.
Fig. 1.
Map showing sampling sites located in the (A) Boye and (B) Kinzig catchments (map created with QGIS v3.16.9).
Table 1.
Sampling locations in the Boye and Kinzig catchments, with coordinates, habitat description (Boye: near-natural, <5, 6–11 and >11 years since renaturation and Kinzig: urban or rural), host MOTUs, parasite MOTUs (in bracket the number of microsporidian MOTUs including those reported in other studies conducted in the same location), number of infected hosts (infected/total host individuals in the sample) and water parameters
| Catchment | Location | Coordinates | Habitat | Host MOTUs | Parasite MOTUs | Infected host | pH | EC (μs cm−1) | O2 (mg L−1) | T (°C) |
|---|---|---|---|---|---|---|---|---|---|---|
| Boye | BOYohBR | 51.569135; 6.927698 | R > 11 | 1 | 2 (4) | 2/17 | 7.53 | 593 | 9.5 | 8.7 |
| BOYohKI | 51.553107; 6.948059 | R < 5 | 2 | 1 | 2/10 | 7.80 | 754 | 11.1 | 7.6 | |
| BOYohSP | 51.564297; 6.930277 | R > 11 | 1 | 2 | 4/21 | 7.77 | 700 | 9.9 | 11.4 | |
| BOYuhHA | 51.534515; 6.99774 | R < 5 | 1 | 1 | 1/10 | 8.09 | 926 | 12 | 9.3 | |
| BOYuhSP | 51.561336; 6.932617 | R > 11 | 2 | 3 | 9/20 | 7.58 | 706 | 10.2 | 10 | |
| BRAob | 51.588333; 6.944944 | NN | 1 | 3 (3) | 6/25 | 7.54 | 873 | 9.1 | 11.1 | |
| HAAob | 51.570306; 6.960891 | R6–11 | 2 | 2 | 2/13 | 7.93 | 829 | 9.1 | 12.7 | |
| HAAun | 51.562638; 6.955581 | R6–11 | 1 | 1 | 1/10 | 7.37 | 1030 | 11.3 | 10.1 | |
| KIRob | 51.542223; 6.939196 | R > 11 | 1 | 1 | 7/10 | 8.05 | 1092 | 9.2 | 9.9 | |
| KIRun | 51.547455; 6.943704 | R > 11 | 1 | 2 | 7/15 | 7.65 | 979 | 10 | 8.2 | |
| SCHohVO | 51.539420; 6.908287 | NN | 1 | 2 | 3/11 | 7.65 | 584 | 10.3 | 7.5 | |
| VORohBOY | 51.554753; 6.932435 | R6–11 | 1 | 4 | 7/10 | 7.37 | 974 | 11.1 | 9 | |
| VORuhSC | 51.544809; 6.921403 | R6–11 | 1 | 1 (2) | 1/10 | 7.37 | 935 | 9.1 | 9 | |
| Kinzig | BIE1 | 50.163394; 9.294485 | Urban | 1 | 1 | 3/16 | 7.75 | 182 | 10.8 | 12.3 |
| BIE2 | 50.174435; 9.284504 | Rural | 1 | 2 | 2/17 | 8.15 | 160 | 11.1 | 10.2 | |
| FAL1 | 50.196262; 9.019494 | Rural | 3 | 3 | 6/13 | 8.15 | 805 | 12.0 | 9.6 | |
| FAL2 | 50.160807; 8.968476 | Urban | 2 | 4 | 11/34 | 8.29 | 771 | 11.5 | 8.6 | |
| GRU1 | 50.259856; 9.174480 | Rural | 1 | 3 | 6/15 | 7.72 | 146 | 12.2 | 8.2 | |
| GRU2 | 50.240705; 9.144059 | Urban | 3 | 3 | 5/21 | 7.19 | 166 | 11.3 | 8.4 | |
| KIN1 | 50.342661; 9.580566 | Rural | 2 | 4 | 5/26 | 9.04 | 357 | 12.1 | 8.8 | |
| KIN2 | 50.346310; 9.552487 | Urban | 2 | 3 | 5/24 | 8.51 | 431 | 13.3 | 6.9 | |
| KIN3 | 50.344639; 9.525316 | Urban | 2 | 2 | 4/22 | 8.83 | 402 | 12.1 | 8.4 | |
| KIN4 | 50.325129; 9.496401 | Rural | 2 | 3 | 4/23 | 8.50 | 450 | 13.6 | 6.4 | |
| KRE1 | 50.180280; 8.918023 | Urban | 2 | 2 | 5/16 | 8.17 | 841 | 15.3 | 12.9 | |
| KRE2 | 50.151738; 8.901119 | Rural | 1 | 1 | 3/22 | 8.21 | 852 | 13.9 | 12.3 | |
| ORB1 | 50.231196; 9.342359 | Urban | 2 | 1 | 8/29 | 7.21 | 111 | 11.6 | 9.3 | |
| ORB2 | 50.236741; 9.324385 | Rural | 1 | 3 | 7/13 | 7.03 | 132 | 11.3 | 10.7 | |
| SAL1 | 50.333618; 9.381166 | Rural | 2 | 2 | 5/6 | 9.54 | 153 | 13.2 | 10.1 | |
| SAL2 | 50.314300; 9.366880 | Urban | 2 | 2 | 3/14 | 9.45 | 169 | 14.2 | 9.4 | |
| STE1 | 50.332253; 9.466509 | Rural | 3 | 3 | 12/23 | 8.73 | 241 | 13.7 | 8.1 | |
| STE2 | 50.315518; 9.456635 | Urban | 1 | 2 | 3/3 | 9.10 | 248 | 12.1 | 8.7 |
DNA isolation and sequencing
DNA was isolated from muscular tissue following a modified salt precipitation protocol described by Grabner et al. (2015). Molecular identification of amphipods was obtained with the universal eukaryotic primers LCO1490 and HCO2198, which amplify the CO1 region, while that of microsporidians with the universal microsporidian primers V1 and Micuni3R, which amplify the SSU rDNA region (Weigand et al., 2016). If clear bands were visible, polymerase chain reaction products were sent to Microsynth Seqlab (Germany) for sequencing using LCO1490 and V1 primers, respectively.
Sequence editing and alignment
CO1 and SSU rDNA sequences were edited in Unipro UGENE version 40.0 (Okonechnikov et al., 2012). Only sequences with a minimum length of 200 bp were used for the analyses. Host and parasite sequences were separately aligned using the MAFFT 7 algorithm with a standard setting (Katoh et al., 2019). To identify hosts and their microsporidian parasites, sequences were blasted against records contained in GenBank. Haplotypes of amphipods and parasites were grouped in molecular operational taxonomic units (MOTUs) when the Kimura-2-parameter (K2p) corrected pairwise distances were below 2% (Supplementary file 1: Table S1). A threshold of 2% was chosen to account for potential intragenomic variation present in some microsporidians while remaining below commonly observed values of intraspecific variability in amphipods (Costa et al., 2009; Ironside, 2013; Grabner et al., 2015). Obtained MOTU sequences were then compared with highly similar sequences (minimum 98% identity match) retrieved from GenBank using K2p as described above to assess if microsporidians use a wider array of hosts than what was observed locally in the present study. When host and parasite sequences from the same individual were not available, we used the closest host sequences available (e.g. from the same host population or sequences available from different areas) to build a phylogenetic tree (Supplementary file 2: Dataset S1). Thus, resulting host specificity might be conservative as splitting these hosts in genetic clades within the same species was not feasible. Maximum likelihood phylogenetic trees with bootstrap support values (1000 replicates) for both amphipods and microsporidians were produced in IQ-Tree 2.0 (Minh et al., 2020). Based on Bayesian information criterion scores the TIM + F + R4 substitution model was selected for amphipods and TIM3 + F + G4 for microsporidians. Sequences of the amphipod Crangonyx islandicus (GenBank accession number HM015162) and the amphipod-infecting microsporidia Dictyocoela cavimanum (GenBank accession number KY073301) were used as outgroups. The naming of undescribed Microsporidium isolates with the exception of 2 Cucumispora isolates (sp01 and sp02) followed the classification used by previous studies (Bojko et al., 2015, 2017; Grabner, 2017; Bacela-Spychalska et al., 2018; Quiles et al., 2019, 2021).
Phylogenetic and geographic specificity analyses
Analyses of phylogenetic host specificity and subsequent statistical analyses were performed with the open-source software Rstudio (version 2021.09.0, Rstudio Inc.) based on R (version 4.1.1, R Core Team). The ape package (Paradis et al., 2004) was used to load and transform the phylogenetic tree in Newick format, while the picante package (Kembel et al., 2010) was used to calculate Faith's PD phylogenetic diversity index (Faith, 1992) as a measure of phylogenetic host specificity. Phylogenetic host specificity represents the total length of branches linking the host species to a parasite along the phylogenetic tree (Poulin et al., 2011). Thus, the higher the values, the less the parasite is species-specific. To compute β-diversity as a measure of geographic host specificity or β-specificity (Poulin et al., 2011), we used the extension of the Jaccard dissimilarity index for multiple-site using the betapart package (Baselga and Orme, 2012). β-Specificity ranges from zero for parasites that exploit the same host across all localities to one for parasites that use completely different hosts from one locality to another.
Statistical analyses
To assess if the number of microsporidian MOTUs was correlated with the number of amphipod MOTUs and water parameters across our sampling sites, the Spearman correlation coefficient was used, while differences in microsporidian MOTU richness across habitat type (Boye: near-natural, <5, 6–11 and >11 years since renaturation and Kinzig: urban or rural) were assessed with the Kruskal–Wallis rank-sum test for the Boye catchment and the Wilcoxon rank-sum test for the Kinzig catchment. General linear models followed by analysis of variance (ANOVA) were employed to assess if microsporidian host phylogenetic specificity and β-specificity are influenced by habitat type and host MOTU richness, using phylogenetic host specificity and β-specificity, respectively, as response variables and host MOTU richness and habitat type as predictors. Furthermore, the role of host densities (individuals/m2) in microsporidian MOTU richness and phylogenetic specificity among restored and near-natural sites in the Boye catchment was investigated using MOTU richness and phylogenetic specificity as response variables and host densities (pooled for each sampling site) and habitat type as predictors. Differences between continental- and regional-scale β-specificity were compared with the t-test.
Results
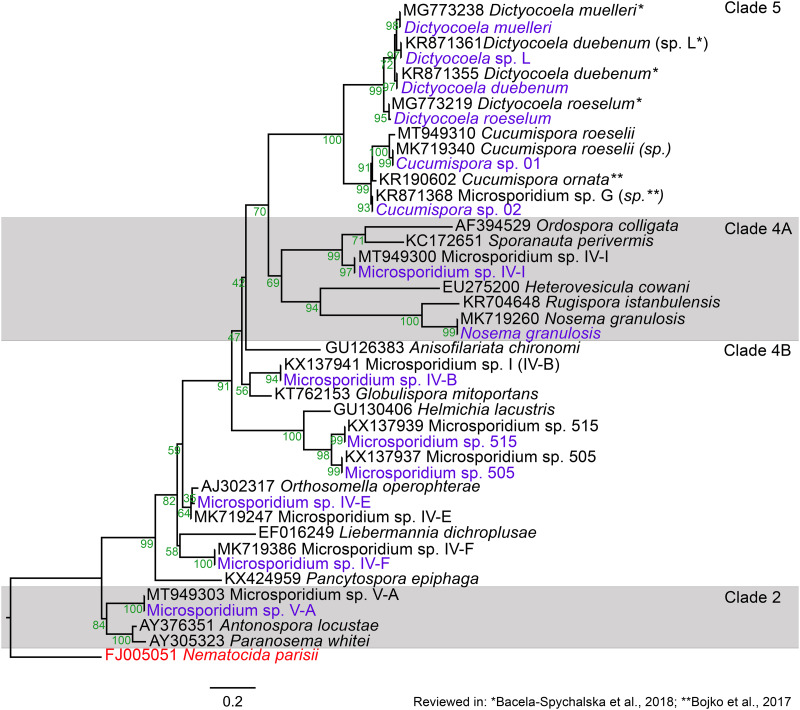
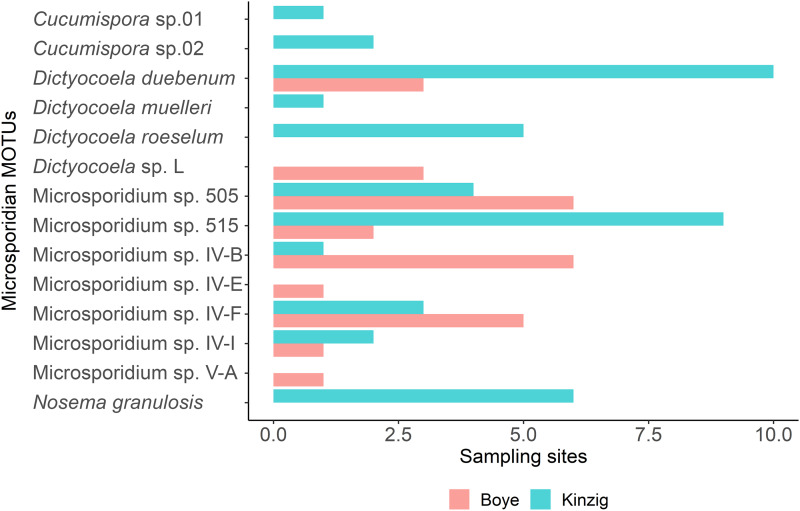
The sample comprised of 14 microsporidian MOTUs, infecting a total of 6 amphipods MOTUs (Fig. 2; Supplementary file 3: Fig. S1; Table 2), 3 in the Boye catchment (Gammarus pulex clade C, G. pulex clade E and Gammarus fossarum clade 2) and 5 in the Kinzig catchment (G. pulex clade D, G. pulex clade E, G. fossarum clade 1, G. fossarum clade 2 and Gammarus roeselii clade 2). Among microsporidian MOTUs 9 (Dictyocoela duebenum, D. sp. L., Microsporidium sp. 505, Microsporidium sp. 515, Microsporidium sp. IV-B, Microsporidium sp. IV-E, Microsporidium sp. IV-F, Microsporidium sp. IV-I and Microsporidium sp. V-A) were found in the Boye catchment and 11 (Cucumispora sp. 01, C. sp. 02, D. duebenum, Dictyocoela muelleri, Dictyocoela roeselum, Microsporidium sp. 505, Microsporidium sp. 515, Microsporidium sp. IV-B, Microsporidium sp. IV-F, Microsporidium sp. IV-I and N. granulosis) in the Kinzig catchment (Fig. 3). The most common microsporidians in the Boye catchment were Microsporidium sp. 505 and Microsporidium sp. IV-B with their presence being recorded in 6 sampling sites. In the Kinzig catchment, D. duebenum and Microsporidium sp. 515 were the most common with 10 and 9 observations, respectively (Fig. 3). Overall, 11 microsporidian MOTUs have been observed in new host MOTUs, further expanding the current knowledge on host range by 17 additional interactions (Table 2).
Fig. 2.
Maximum likelihood phylogenetic tree obtained with IQ-Tree 2.0 (Minh et al., 2020) using a TIM3 + F + G4 substitution model and based on partial small ribosomal subunit rDNA data (Supplementary file 2: Dataset S1). Labels with accession number are parasite sequences retrieved from GenBank. The name of described species and reviewed sequences are marked with asterisks. Bootstrap values (1000 replicates) are indicated in green. Outgroup is indicated in purple.
Table 2.
Microsporidian MOTU associations with amphipods host MOTUs
| Cucumispora sp. 01 | Cucumispora sp. 02 | Dictyocoela duebenum | Dictyocoela muelleri | Dictyocoela roeselum | Dictyocoela sp. L | Microsporidium sp. 505 | Microsporidium sp. 515 | Microsporidium sp. IV-B | Microsporidium sp. IV-E | Microsporidium sp. IV-F | Microsporidium sp. IV-I | Microsporidium sp. V-A | Nosema granulosis | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acanthogammarus lappaceus | a | c | ||||||||||||
| Acanthogammarus victorii | a | |||||||||||||
| Brandtia latissima | j | |||||||||||||
| Dikerogammarus haemobaphes | a/e | |||||||||||||
| Dikerogammarus villosus | a | s | s | |||||||||||
| Dorogostaiskia parasitica | a | |||||||||||||
| Echinogammarus berilloni | a/f/h | |||||||||||||
| Echinogammarus marinus | a/q | |||||||||||||
| Eulimnogammarus cyaneus | a | |||||||||||||
| Eulimnogammarus verrucosus | o | |||||||||||||
| Eulimnogammarus vittatus | j | |||||||||||||
| Gammarus balcanicus C1 | n | |||||||||||||
| G. balcanicus C2 | m | |||||||||||||
| G. balcanicus C3 | m | n | n | n | n | |||||||||
| G. balcanicus C4 | m | |||||||||||||
| G. balcanicus C5 | m | m | n | |||||||||||
| G. balcanicus C6 | m | |||||||||||||
| G. balcanicus C7 | m | m | ||||||||||||
| Gammarus chevreuxi | b/r | |||||||||||||
| Gammarus duebeni | a/k | k | k | k/i | ||||||||||
| Gammarus fossarum C1 | ** | ** | ** | |||||||||||
| G. fossarum C2 | ** | a/f | ** | e | e | ** | f | ** | ** | |||||
| Gammarus lacustris | d | j | ||||||||||||
| Gammarus locusta | a/h | |||||||||||||
| Gammarus pseudolimnaeus | a/p | |||||||||||||
| Gammarus pulex CC | a/e | f | f | f | f | ** | f | |||||||
| G. pulex CD | ** | ** | ** | ** | ** | |||||||||
| G. pulex CE | b/f | a/f | ** | f | f | f | ** | f | f | |||||
| Gammarus roeselii C1 | l | |||||||||||||
| G. roeselii C2 | e/l | l | a/f | a/l/r | a/l | a/e | f | e/n | e/l | l | l | ** | f/l | l |
| G. roeselii C3 | l | |||||||||||||
| Gammarus tigrinus | a/h | |||||||||||||
| Gammarus varsoviensis | a | |||||||||||||
| Gmelinoides fasciatus | a/j | u | ||||||||||||
| Linevichella vortex | u | |||||||||||||
| Niphagus aquilex | g | |||||||||||||
| Niphargus schellenbergi | g | t | ||||||||||||
| Pallasea cancellus | a | |||||||||||||
| Pontogammarus robustoides | a |
All parasite–host associations are indicated by either asterisks or letter corresponding to a literature reference. New associations are indicated by double asterisks and orange colour, while associations found in our sample that were previously reported by other study are indicated with blue colour. Associations without colour marking are only reported from literature.
**New host.
(a) Bacela-Spychalska et al. (2018); (b) Bojko et al. (2017); (c) Dimova et al. (2018); (d) Drozdova et al. (2020); (e) Grabner (2017); (f) Grabner et al. (2015); (g) Grabner et al. (2020); (h) Hogg et al. (2002); (i) Ironside (2013); (j) Ironside and Wilkinson (2018); (k) Krebes et al. (2010); (l) Quiles et al. (2019); (m) Quiles et al. (2020); (n) Quiles et al. (2021); (o) Madyarova et al. (2015); (p) Ryan and Kohler (2010); (q) Short et al. (2012); (r) Terry et al. (2004); (s) Wattier et al. (2007); (t) Weigand et al. (2016); (u) unpublished GenBank sequences.
Fig. 3.
Number of observed microsporidian MOTUs across sampling sites in the Boye and Kinzig catchments.
In both Boye and Kinzig catchments, microsporidian MOTU richness was neither correlated with host MOTU richness (the Spearman correlation coefficient, rs = −0.051, P = 0.872 and rs = 0.400, P = 0.100, respectively) nor significantly differed among habitat types (Kruskal–Wallis rank-sum test, χ2 = 3.6813, dƒ = 3, P = 0.298 and the Wilcoxon rank-sum test, W = 52.5, P = 0.2847, respectively). Similarly, no strong correlations among water parameters and microsporidian MOTU richness were detected. However, in 2 instances, our analyses revealed diverging patterns between catchments with microsporidian MOTU richness being negatively affected by pH in the Boye catchment and by temperature in the Kinzig catchment with their counterparts remaining unaffected (Table 3).
Table 3.
Correlations among water parameters of sampling sites located in the Boye (n = 13) and Kinzig (n = 18) catchments and microsporidian MOTU richness were calculated using the Spearman correlation coefficient
| Water parameters | Boye | Kinzig | ||
|---|---|---|---|---|
| rs | P | rs | P | |
| pH | −0.55 | 0.051 | 0.01 | 0.969 |
| Conductivity (μs cm−1) | −0.38 | 0.202 | 0.13 | 0.601 |
| Oxygen (mg L−1) | −0.33 | 0.271 | −0.05 | 0.853 |
| Temperature (°C) | 0.01 | 0.989 | −0.55 | 0.019 |
Microsporidian phylogenetic host specificity at a continental scale was higher than 1 in all cases except for C. roeselii, indicating that the vast majority of microsporidians are host generalist. This was also reflected by high β-specificity values, which pinpointed exploitation of completely different hosts from one locality to another, hence high host turnover across their geographical range (Table 4). Moreover, differences between regional- and continental-scale values (t-test, t₍₁₈₎ = −2.77, P = 0.013) highlighted the importance of a broader view when dealing with host-specialization measures (Table 4).
Table 4.
(A) Regional- and (B) continental-scale phylogenetic host specificity and β-specificity calculated with Faith's PD phylogenetic diversity index (Faith, 1992) and Jaccard dissimilarity index for multiple-site (Baselga and Orme, 2012), respectively
| Microsporidian MOTUs | Sampling sites | N host MOTUs | Phylogenetic specificity | β-Specificity |
|---|---|---|---|---|
| (A) Regional scale | ||||
| Cucumispora sp. 01 | 1 | 1 | – | – |
| Cucumispora sp. 02 | 2 | 2 | 1.92 | 0.50 |
| D. duebenum | 13 | 4 | 2.90 | 0.88 |
| D. muelleri | 1 | 1 | – | – |
| D. roeselum | 5 | 3 | 2.87 | 0.78 |
| Dictyocoela sp. L. | 3 | 2 | 1.22 | 0.80 |
| Microsporidium sp. 505 | 10 | 4 | 2.22 | 0.91 |
| Microsporidium sp. 515 | 11 | 5 | 2.25 | 0.93 |
| Microsporidium sp. IV-B | 7 | 2 | 1.01 | 0.85 |
| Microsporidium sp. IV-E | 1 | 2 | 1.91 | – |
| Microsporidium sp. IV-F | 8 | 3 | 2.87 | 0.92 |
| Microsporidium sp. IV-I | 3 | 2 | 1.92 | 0.80 |
| Microsporidium sp. V-A | 1 | 1 | – | – |
| N. granulosis | 6 | 4 | 2.90 | 0.84 |
| (B) Continental scale | ||||
| Cucumispora sp. 01 | 5 | 1 | 1 | 0 |
| Cucumispora sp. 02 | 5 | 4 | 3.72 | 0.90 |
| D. duebenum | 41 | 26 | 17.29 | 0.99 |
| D. muelleri | 23 | 8 | 5.55 | 0.94 |
| D. roeselum | 18 | 5 | 4.07 | 0.86 |
| Dictyocoela sp. L. | 5 | 5 | 3.51 | 0.92 |
| Microsporidium sp. 505 | 17 | 7 | 5.05 | 0.95 |
| Microsporidium sp. 515 | 18 | 10 | 6.11 | 0.97 |
| Microsporidium sp. IV-B | 11 | 6 | 4.17 | 0.95 |
| Microsporidium sp. IV-E | 2 | 3 | 2.91 | 1 |
| Microsporidium sp. IV-F | 11 | 5 | 4.05 | 0.94 |
| Microsporidium sp. IV-I | 4 | 3 | 2.91 | 0.91 |
| Microsporidium sp. V-A | 5 | 5 | 4.05 | 0.96 |
| N. granulosis | 26 | 9 | 6.08 | 0.96 |
In both Boye and Kinzig catchments, microsporidian phylogenetic specificity was not influenced by habitat type (ANOVA, F₍₃,₂₃₎ = 0.40, P = 0.754 and F₍₁,₄₁₎ = 0.12, P = 0.728, respectively) or host MOTU richness (ANOVA, F₍₁,₂₃₎ = 0.44, P = 0.515 and F₍₁,₄₁₎ = 0.15, P = 0.703 respectively). Similarly, microsporidian β-specificity was not influenced by habitat type (ANOVA, F₍₃,₂₃₎ = 0.55, P = 0. 650 and F₍₁,₄₁₎ = 0.86, P = 0.358 respectively) or host MOTU richness (ANOVA, F₍₁,₂₃₎ = 3.19, P = 0.09 and F₍₁,₄₁₎ = 0.18, P = 0.674 respectively). Furthermore, in the Boye catchment, no noticeable effect on microsporidian MOTU richness across restored and near-natural sites was observed when considering differences in host densities (ANOVA, F₍₁,₂₃₎ = 0.38, P = 0.546) and habitat type (ANOVA, F₍₃,₂₃₎ = 2.01, P = 0.141). Similarly, in the same catchment, neither host densities (ANOVA, F₍₁,₂₅₎ = 2.20, P = 0.150) nor host MOTU richness (ANOVA, F₍₁,₂₅₎ = 0.01, P = 0.996) affected microsporidian phylogenetic specificity.
Discussion
Most microsporidian species infect more hosts than previously reported, underlining the importance of parasitological studies using aquatic keystone species with little or no commercial value. Furthermore, the generalist nature of the observed microsporidians in terms of host uses highlights the importance of host specificity in parasite persistence and dispersal following anthropogenic disturbance.
Existing and novel associations between amphipods and microsporidian parasites
More than 30 microsporidian species from 12 genera and more than 150 undescribed isolates have been reported from amphipods globally (Bojko and Ovcharenko, 2019), with a still increasing trend. Among our samples, all microsporidian MOTUs found in G. fossarum clade 1 and G. pulex clade D account for 8 novel associations. New associations with microsporidians in these 2 amphipod clades are expected as they have been investigated less intensively than G. fossarum clade 2 (type B in literature) or G. pulex clades C and E (Grabner et al., 2015; Grabner, 2017; Bacela-Spychalska et al., 2018). However, we also report new parasite host associations in more commonly investigated amphipods. Gammarus fossarum clade 2 was infected with 9 microsporidian MOTUs of which 5 are new. Similarly, G. pulex clades C and E were infected with 6 and 5 microsporidian MOTUs, respectively, including 3 new associations. One new association was found even in G. roeselii clade 2 (group C in literature), which previously underwent a massive parasitological investigation (Grabner et al., 2015; Bojko et al., 2017; Grabner, 2017; Bacela-Spychalska et al., 2018; Quiles et al., 2019, 2021). The present finding highlights the importance of further parasitological studies in amphipods.
Variability between catchment degradation levels and microsporidians
Contrary to our expectations, both in the Boye and Kinzig catchments, there was no apparent difference in microsporidian diversity and host specificity among restored and near-natural streams or between those located in urban and rural areas. Similarly, microsporidian diversity was generally not influenced by water parameters. The sole exception is a moderate negative effect of pH in the Boye and temperature in the Kinzig catchments. However, the pH range in the Boye catchment (7.3–8.1) varied to a lower extent compared to its counterpart (7–9.5). Hence, we argue that if pH influences microsporidian diversity, this would have been more pronounced in the Kinzig catchment. Similarly, if temperature had an important effect on microsporidian diversity, this would be observed from both catchments given a similar temperature range. Hence other processes such as transmission mechanisms, host immune system, local adaptation, dispersal constraints and competitive interactions among parasites may play a more relevant role in shaping microsporidian communities.
Anthropogenic disturbance such as sewage discharge and reconstruction of natural watercourses, e.g. into concrete drainage channels, as in the case of the Boye catchment, may result in extinction of organisms, including parasites and their hosts. Thus, to persist, organisms should adapt to new conditions. Microsporidians may cope with similar situations by switching from a rapidly declining host to an alternative, more common host, even if that might entail reduced fitness benefits (Dunn et al., 2009; Moir et al., 2010). Accordingly, all microsporidians collected in the Boye catchment had low host phylogenetic specificity. In contrast, among those collected in the Kinzig catchment, Cucumispora sp. 01 showed the highest degree of host phylogenetic specificity, partially supporting our second hypothesis. However, phylogenetic specificity for these and other microsporidians might change over time as new hosts are discovered and it has been shown that for a single parasite species, different genotypes may be specialized on single host species or genotypes (Quiles et al., 2019, 2020). Differences in host diversity between the 2 catchments might also reflect different histories of colonization and anthropogenic disturbance which could have caused host–parasite coextinction. Accordingly, G. roeselii is absent from the Boye catchment but present in the surrounding watercourses (Grabner et al., 2015).
Unexpectedly, in the Boye catchment, host densities did not appear to influence microsporidian MOTU richness across restored and near-natural sites. Possible explanations are the release of resistant spores in the environment, which allow parasites to persist when hosts are present in low abundance or even absent (Dunn and Smith, 2001), host switching (Becnel and Andreadis, 1999) and different transmission mechanisms acting as confounding factors. Microsporidians might exploit horizontal, vertical or both transmission routes (Dunn and Smith, 2001; Haag et al., 2020). Among these transmission pathways, vertical and mixed transmission pathways may be relevant for parasite persistence and dispersal. For instance, a moth-infecting microsporidium, Orthosoma operoptherae, uses vertical transmission to over-winter in host eggs (Canning et al., 1985), while Octosporea bayeri utilizes vertical transmission to survive diapause in Daphnia magna during drought (Zbinden et al., 2008; Stentiford et al., 2013). Among the most common microsporidians collected in the Boye catchment, horizontal transmission is believed to be the only or the predominant mode of transmission of Microsporidium sp. 505 (Grabner et al., 2014), while Microsporidium sp. IV-B (also referred to as M3 or I in literature) is vertically transmitted (Terry et al., 2004). However, since the biology of most microsporidians found in the current study remains understudied, their primary transmission pathway is not known, preventing us from in-depth interpretations. Furthermore, given connectivity and the relative proximity among streams in the Boye catchment, the general lack of distinct patterns may be due to underlying mechanisms related to the dispersal of hosts and parasites across sites.
The high host turnover observed in microsporidians across their geographical range suggests that neither environmental condition nor host diversity plays a significant role in their establishment in restored areas, supporting our third hypothesis. Being internal parasites, microsporidians are less exposed to changes in the external environment. Moreover, given the wide geographical distribution of Microsporidia and their occurrence in a variety of environments, it is likely that they possess a certain degree of plasticity to environmental conditions. Host diversity is less relevant in generalist microsporidians, as they may use alternative and phylogenetically distant hosts. For instance, Enterocytozoon bieneusi can exploit birds and mammals (Wadi and Reinke, 2020), while Trachipleistophora hominis, whose natural host is an insect, may infect immunocompromised humans (Watson et al., 2015). This might be the case also for microsporidians infecting amphipods. For instance, Microsporidium sp. 1049 has been reported to infect both G. roeselii and chironomid larvae (Grabner, 2017). Thus, host switching in generalist parasites might overcome environmental impediments and favour persistence and dispersal. Microsporidians are most likely infecting a wider host spectrum than reported here, as current knowledge is based only on a few host species/MOTUs. Hence our estimates may be conservative, and further investigations on host specificity targeting alternative host species are required to understand persistence and dispersal mechanisms.
Conclusion
In conclusion, host diversity and environmental parameters do not dictate the persistence and dispersal of phylogenetic host generalist microsporidians in environments that experienced anthropogenic disturbance. Instead, these might depend on more complex mechanisms such as the production of resistant spores, host switching and host dispersal acting individually or conjointly.
Acknowledgements
We thank all collaborators of the Collaborative Research Center (CRC) RESIST for assistance during the fieldwork and in the laboratory. The Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) is gratefully acknowledged for funding this study. We acknowledge support by the Open Access Publication Fund of the University of Duisburg-Essen.
Author contributions
S. P. and B. S. conceived the study, and D. S. G. and B. S. supervised the project. S. M. P. and A. W. L. carried out the sampling and S. P. the data analyses. S. P. led the writing of the manuscript; D. S. G., S. M. P., A. W. L. and B. S. oversaw the analyses and writing. All authors contributed critically to the drafts and approved the final manuscript.
Financial support
This study was performed within the Collaborative Research Center (CRC) RESIST (A09) funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – CRC 1439/1 – project number: 426547801. We also acknowledge support from The Open Access Publication Fund of the University of Duisburg-Essen.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0031182022000452.
click here to view supplementary material
Data
The dataset supporting the conclusions of this article is included within the article (and its Supplementary material). Host and parasite sequences have been uploaded to GenBank and the following accession numbers obtained: amphipod hosts (ON093813, ON093814, ON093815, ON093816, ON093817 and ON093818) and microsporidian parasites (ON113505, ON113506, ON113507, ON113508, ON113509, ON113510, ON113511, ON113512, ON113513, ON113514, ON113515, ON113516, ON113517 and ON113518).
Conflict of interest
The authors declare there are no conflicts of interest.
References
- Bacela-Spychalska K, Wróblewski P, Mamos T, Grabowski M, Rigaud T, Wattier R, Rewicz T, Konopacka A and Ovcharenko M (2018) Europe-wide reassessment of Dictyocoela (Microsporidia) infecting native and invasive amphipods (Crustacea): molecular versus ultrastructural traits. Scientific Reports 8, 8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baselga A and Orme CDL (2012) Betapart: an R package for the study of beta diversity. Methods in Ecology and Evolution 3, 808–812. [Google Scholar]
- Becnel JJ and Andreadis TG (1999) Microsporidia in insects. In Wittner M and Weiss LM (eds), The Microsporidia and Microsporidiosis. Washington, DC: ASM Press, pp. 447–501. [Google Scholar]
- Birk S, Chapman D, Carvalho L, Spears BM, Andersen HE, Argillier C, Auer S, Baattrup-Pedersen A, Banin L, Beklioğlu M, Bondar-Kunze E, Borja A, Branco P, Bucak T, Buijse AD, Cardoso AC, Raoul-Marie Couture RM, Cremona F, de Zwart D, Feld CK, Ferreira MT, Feuchtmayr H, Gessner MO, Gieswein A, Globevnik L, Graeber D, Graf W, Gutiérrez-Cánovas C, Hanganu J, Işkın U, Järvinen M, Jeppesen E, Kotamäki N, Kuijper M, Lemm JU, Lu S, Solheim AL, Mischke U, Moe SJ, Nõges P, Nõges T, Ormerod SJ, Panagopoulos Y, Phillips G, Posthuma L, Pouso S, Prudhomme C, Rankinen K, Rasmussen JJ, Richardson J, Sagouis A, Santos JM, Schäfer RB, Schinegger R, Schmutz S, Schneider SC, Schülting L, Segurado P, Stefanidis K, Sures B, Thackeray SJ, Turunen J, Uyarra MC, Venohr M, von der Ohe PC, Willby N and Hering D (2020) Impacts of multiple stressors on freshwater biota across spatial scales and ecosystems. Nature Ecology & Evolution 4, 1060–1068. [DOI] [PubMed] [Google Scholar]
- Bojko J and Ovcharenko M (2019) Pathogens and other symbionts of the Amphipoda: taxonomic diversity and pathological significance. Diseases of Aquatic Organisms 136, 3–36. [DOI] [PubMed] [Google Scholar]
- Bojko J, Dunn AM, Stebbing PD, Ross SH, Kerr RC and Stentiford GD (2015) Cucumispora ornata n. sp. (Fungi: Microsporidia) infecting invasive ‘demon shrimp’ (Dikerogammarus haemobaphes) in the United Kingdom. Journal of Invertebrate Pathology 128, 22–30. [DOI] [PubMed] [Google Scholar]
- Bojko J, Bącela-Spychalska K, Stebbing PD, Dunn AM, Grabowski M, Rachalewski M and Stentiford GD (2017) Parasites, pathogens and commensals in the ‘low-impact’ non-native amphipod host Gammarus roeselii. Parasites & Vectors 10, 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canning EU, Barker RJ, Page AM and Nicholas JP (1985) Transmission of microsporidia, especially Orthosoma operophterae (Canning, 1960) between generations of winter moth Operophtera brumata (L) (Lepidoptera: Geometridae). Parasitology 90, 11–19. [Google Scholar]
- Costa FO, Henzler CM, Lunt DH, Whiteley NM and Rock J (2009) Probing marine Gammarus (Amphipoda) taxonomy with DNA barcodes. Systematics and Biodiversity 7, 365–379. [Google Scholar]
- Dimova M, Madyarova E, Gurkov A, Drozdova P, Lubyaga Y, Kondrateva E, Adelshin R and Timofeyev M (2018) Genetic diversity of microsporidia in the circulatory system of endemic amphipods from different locations and depths of ancient Lake Baikal. PeerJ 6, e5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson A, Lafferty KD, Kuris AM, Hechinger RF and Jetz W (2008) Homage to Linnaeus: how many parasites? How many hosts? Proceedings of the National Academy of Sciences of the USA 105, 11482–11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdova P, Madyarova E, Dimova M, Gurkov A, Vereshchagina K, Adelshin R and Timofeyev M (2020) The diversity of microsporidian parasites infecting the Holarctic amphipod Gammarus lacustris from the Baikal region is dominated by the genus Dictyocoela. Journal of Invertebrate Pathology 170, 107330. [DOI] [PubMed] [Google Scholar]
- Dudgeon D, Arthington AH, Gessner MO, Kawabata Z-I, Knowler DJ, Lévêque C, Naiman RJ, Prieur-Richard A-H, Soto D, Stiassny MLJ and Sullivan CA (2006) Freshwater biodiversity: importance, threats, status and conservation challenges. Biological Reviews 81, 163. [DOI] [PubMed] [Google Scholar]
- Dunn AM and Smith JE (2001) Microsporidian life cycles and diversity: the relationship between virulence and transmission. Microbes and Infection 3, 381–388. [DOI] [PubMed] [Google Scholar]
- Dunn RR, Harris NC, Colwell RK, Koh LP and Sodhi NS (2009) The sixth mass coextinction: are most endangered species parasites and mutualists? Proceedings of the Royal Society B: Biological Sciences 276, 3037–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith DP (1992) Conservation evaluation and phylogenetic diversity. Biological Conservation 61, 1–10. [Google Scholar]
- Fischer J, Riechers M, Loos J, Martin-Lopez B and Temperton VM (2021) Making the UN decade on ecosystem restoration a social-ecological endeavour. Trends in Ecology & Evolution 36, 20–28. [DOI] [PubMed] [Google Scholar]
- Grabner DS (2017) Hidden diversity: parasites of stream arthropods. Freshwater Biology 62, 52–64. [Google Scholar]
- Grabner DS, Schertzinger G and Sures B (2014) Effect of multiple microsporidian infections and temperature stress on the heat shock protein 70 (hsp70) response of the amphipod Gammarus pulex. Parasites & Vectors 7, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabner DS, Weigand AM, Leese F, Winking C, Hering D, Tollrian R and Sures B (2015) Invaders, natives and their enemies: distribution patterns of amphipods and their microsporidian parasites in the Ruhr Metropolis, Germany. Parasites & Vectors 8, 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabner D, Weber D and Weigand AM (2020) Updates to the sporadic knowledge on microsporidian infections in groundwater amphipods (Crustacea, Amphipoda, Niphargidae). Subterranean Biology 33, 71–85. [Google Scholar]
- Haag KL, Pombert J-F, Sun Y, de Albuquerque NRM, Batliner B, Fields P, Lopes TF and Ebert D (2020) Microsporidia with vertical transmission were likely shaped by nonadaptive processes. Genome Biology and Evolution 12, 3599–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JC, Ironside JE, Sharpe RG, Hatcher MJ, Smith JE and Dunn AM (2002) Infection of Gammarus duebeni populations by two vertically transmitted microsporidia; parasite detection and discrimination by PCR-RFLP. Parasitology 125, 59–63. [DOI] [PubMed] [Google Scholar]
- Ironside JE (2013) Diversity and recombination of dispersed ribosomal DNA and protein coding genes in microsporidia. PLoS ONE 8, e55878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ironside JE and Wilkinson TJ (2018) Accumulation and exchange of parasites during adaptive radiation in an ancient lake. International Journal for Parasitology 48, 297–307. [DOI] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J and Yamada KD (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20, 1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP and Webb CO (2010) Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. [DOI] [PubMed] [Google Scholar]
- Krebes L, Blank M, Frankowski J and Bastrop R (2010) Molecular characterisation of the microsporidia of the amphipod Gammarus duebeni across its natural range revealed hidden diversity, wide-ranging prevalence and potential for co-evolution. Infection, Genetics and Evolution 10, 1027–1038. [DOI] [PubMed] [Google Scholar]
- Lafferty KD (2012) Biodiversity loss decreases parasite diversity: theory and patterns. Philosophical Transactions of the Royal Society B: Biological Sciences 367, 2814–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz AW, Haase P, Januschke K, Sundermann A and Hering D (2018) Revisiting restored river reaches – assessing change of aquatic and riparian communities after five years. Science of the Total Environment 613–614, 1185–1195. [DOI] [PubMed] [Google Scholar]
- Madyarova EV, Adelshin RV, Dimova MD, Axenov-Gribanov DV, Lubyaga YA and Timofeyev MA (2015) Microsporidian parasites found in the hemolymph of four Baikalian endemic amphipods. PLoS ONE 10, e0130311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier C, Böhmer J, Biss R, Feld C, Haase P, Lorenz A, Rawer-Jost C, Rolauffs P, Schindehütte K and Schöll F (2006) Weiterentwicklung und Anpassung des nationalen Bewertungssystems für Makrozoobenthos an neue internationale Vorgaben. Retrieved from Gewaesser bewertung online website. Available at https://www.gewaesser-bewertung.de/files/kurzdarstellungen_mzb_coremetrics_2006.pdf (Accessed 14 November 2021).
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A and Lanfear R (2020) IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37, 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir ML, Vesk PA, Brennan KEC, Keith DA, Hughes L and McCarthy MA (2010) Current constraints and future directions in estimating coextinction. Conservation Biology 24, 682–690. [DOI] [PubMed] [Google Scholar]
- Okonechnikov K, Golosova O, Fursov M and the UGENE team (2012) Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28, 1166–1167. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J and Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. [DOI] [PubMed] [Google Scholar]
- Park E, Jorge F and Poulin R (2020) Shared geographic histories and dispersal contribute to congruent phylogenies between amphipods and their microsporidian parasites at regional and global scales. Molecular Ecology 29, 3330–3345. [DOI] [PubMed] [Google Scholar]
- Poulin R and Morand S (2004) Parasite Biodiversity. Washington, DC, USA: Smithsonian Institution Books. [Google Scholar]
- Poulin R, Krasnov BR and Mouillot D (2011) Host specificity in phylogenetic and geographic space. Trends in Parasitology 27, 355–361. [DOI] [PubMed] [Google Scholar]
- Quiles A, Bacela-Spychalska K, Teixeira M, Lambin N, Grabowski M, Rigaud T and Wattier RA (2019) Microsporidian infections in the species complex Gammarus roeselii (Amphipoda) over its geographical range: evidence for both host–parasite co-diversification and recent host shifts. Parasites & Vectors 12, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiles A, Wattier RA, Bacela-Spychalska K, Grabowski M and Rigaud T (2020) Dictyocoela microsporidia diversity and co-diversification with their host, a gammarid species complex (Crustacea, Amphipoda) with an old history of divergence and high endemic diversity. BMC Evolutionary Biology 20, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiles A, Rigaud T, Wattier RA, Grabowski M and Bacela Spychalska K (2021) Wide geographic distribution of overlooked parasites: rare microsporidia in Gammarus balcanicus, a species complex with a high rate of endemism. International Journal for Parasitology: Parasites and Wildlife 14, 121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JA and Kohler SL (2010) Virulence is context-dependent in a vertically transmitted aquatic host–microparasite system. International Journal for Parasitology 40, 1665–1673. [DOI] [PubMed] [Google Scholar]
- Short S, Guler Y, Yang G, Kille P and Ford AT (2012) Paramyxean–microsporidian co-infection in amphipods: is the consensus that microsporidia can feminise their hosts presumptive? International Journal for Parasitology 42, 683–691. [DOI] [PubMed] [Google Scholar]
- Stentiford GD, Feist SW, Stone DM, Bateman KS and Dunn AM (2013) Microsporidia: diverse, dynamic, and emergent pathogens in aquatic systems. Trends in Parasitology 29, 567–578. [DOI] [PubMed] [Google Scholar]
- Terry RS, Smith JE, Bouchon D, Rigaud T, Duncanson P, Sharpe RG and Dunn AM (1999) Ultrastructural characterisation and molecular taxonomic identification of Nosema granulosis n. sp., a transovarially transmitted feminising (TTF) microsporidium. Journal of Eukaryotic Microbiology 46, 492–499. [DOI] [PubMed] [Google Scholar]
- Terry RS, Smith JE, Sharpe RG, Rigaud T, Littlewood DTJ, Ironside JE, Rollinson D, Bouchon D, MacNeil C, Dick JTA and Dunn AM (2004) Widespread vertical transmission and associated host sex–ratio distortion within the eukaryotic phylum Microspora. Proceedings of the Royal Society of London. Series B: Biological Sciences 271, 1783–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadi L and Reinke AW (2020) Evolution of microsporidia: an extremely successful group of eukaryotic intracellular parasites. PLoS Pathogens 16, e1008276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson AK, Williams TA, Williams BAP, Moore KA, Hirt RP and Embley TM (2015) Transcriptomic profiling of host–parasite interactions in the microsporidian Trachipleistophora hominis. BMC Genomics 16, 983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattier RA, Haine ER, Beguet J, Martin G, Bollache L, Muskó IB, Platvoet D and Rigaud T (2007) No genetic bottleneck or associated microparasite loss in invasive populations of a freshwater amphipod. Oikos 116, 1941–1953. [Google Scholar]
- Weigand AM, Kremers J and Grabner DS (2016) Shared microsporidian profiles between an obligate (Niphargus) and facultative subterranean amphipod population (Gammarus) at sympatry provide indications for underground transmission pathways. Limnologica 58, 7–10. [Google Scholar]
- Winking C, Lorenz AW, Sures B and Hering D (2014) Recolonisation patterns of benthic invertebrates: a field investigation of restored former sewage channels. Freshwater Biology 59, 1932–1944. [Google Scholar]
- Winking C, Lorenz AW, Sures B and Hering D (2016) Start at zero: succession of benthic invertebrate assemblages in restored former sewage channels. Aquatic Sciences 78, 683–694. [Google Scholar]
- Wittner M and Weiss LM (1999) The Microsporidia and Microsporidiosis. Washington, DC: ASM Press. [Google Scholar]
- Zbinden M, Haag CR and Ebert D (2008) Experimental evolution of field populations of Daphnia magna in response to parasite treatment. Journal of Evolutionary Biology 21, 1068–1078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0031182022000452.
click here to view supplementary material
Data Availability Statement
The dataset supporting the conclusions of this article is included within the article (and its Supplementary material). Host and parasite sequences have been uploaded to GenBank and the following accession numbers obtained: amphipod hosts (ON093813, ON093814, ON093815, ON093816, ON093817 and ON093818) and microsporidian parasites (ON113505, ON113506, ON113507, ON113508, ON113509, ON113510, ON113511, ON113512, ON113513, ON113514, ON113515, ON113516, ON113517 and ON113518).