Abstract
We present a genome assembly from an individual female Lysandra bellargus (the Adonis blue; Arthropoda; Insecta; Lepidoptera; Lycaenidae). The genome sequence is 529 megabases in span. The majority of the assembly (99.93%) is scaffolded into 46 chromosomal pseudomolecules with the W and Z sex chromosomes assembled. The complete mitochondrial genome was also assembled and is 15.6 kilobases in length. Gene annotation of this assembly on Ensembl has identified 13,249 protein coding genes.
Keywords: Lysandra bellargus, Adonis blue, genome sequence, chromosomal, Lepidoptera
Species taxonomy
Eukaryota; Metazoa; Ecdysozoa; Arthropoda; Hexapoda; Insecta; Pterygota; Neoptera; Endopterygota; Lepidoptera; Glossata; Ditrysia; Papilionoidea; Lycaenidae; Polyommatinae; Polyommatini; Polyommatina, Lysandra; Lysandra bellargus (Rottemburg, 1775) (NCBI:txid138070).
Background
The Adonis blue, Lysandra bellargus (Rottemburg, 1775), is a butterfly belonging to the gossamer-wing butterfly family Lycaenidae. It inhabits the western Palearctic region, being found most commonly in southern and central Europe, western Russia, Turkey, Transcaucasia, Caucasus, north Iraq and Iran. Its presence has been confirmed in northern Morocco and found occasionally as far north as southern Sweden ( Raper, 2021). While L. bellargus is listed as a species of Least Concern on the IUCN Red List of Europe ( van Swaay et al., 2010), it is considered a vulnerable species in the UK ( Fox et al., 2022). Within the UK, the Adonis blue is at its northern range limit and exists primarily in southern counties, such as Dorset, Wiltshire, Kent, Sussex and Surrey, as well as the Isle of Wight. Populations in the UK have been in general decline since the 1950s ( Thomas, 1983) and were severely reduced in the late 1970s after a drought caused widespread damage to the host plant ( Harper et al., 2003). However, there is evidence of colonies of less than 50 individuals recovering to populations of 60,000 over five years ( Bourn et al., 1998).
The Adonis blue is bivoltine, prefers calcareous grassland in sheltered, warm conditions and lays its eggs on horseshoe vetch ( Hippocrepis comosa). The Adonis blue is named after the sky-blue wings of the males, which are surrounded by a black rim with a white margin. The female is chocolate brown with a small number of blue scales near the base of the wings ( Still, 1996). The caterpillar is dark green with spines and has yellow stripes lining its back and sides ( Carter & Hargreaves, 1986).
The karyotype of the Adonis blue is unusual in that it has 45 chromosomes, while most butterflies in Lycaenidae have a conserved haploid chromosome number of either 23 or 24 ( Lorkovic, 1990). The genome sequence will offer further insight into lepidopteran genome evolution.
Genome sequence report
The genome was sequenced from a single female L. bellargus collected from El Brull, Catalunya, Spain ( Figure 1). A total of 52-fold coverage of Pacific Biosciences single-molecule HiFi long reads and 65-fold coverage of 10X Genomics read clouds were generated. Primary assembly contigs were scaffolded with chromosome conformation Hi-C data. Manual assembly curation corrected 207 missing/misjoins and removed 13 haplotypic duplications, reducing the assembly size by 0.53% and the scaffold number by 47.35%, and increasing the scaffold N50 by 72.56%.
Figure 1. Fore and hind wings of the female Lysandra bellargus specimen from which the genome was sequenced.
Dorsal (left) and ventral (right) surface view of wings from specimen EB_LB_191 (ilLysBell1) from El Brull, Catalunya, Spain, used to generate Pacific Biosciences, 10X genomics and Hi-C data.
The final assembly has a total length of 529 Mb in 139 sequence scaffolds with a scaffold N50 of 11.2 Mb ( Table 1). The majority, 99.93%, of the assembly sequence was assigned to 46 chromosomal-level scaffolds, representing 44 autosomes (numbered by sequence length) and the W and Z sex chromosomes ( Figure 2– Figure 5; Table 2). The putative W chromosome contains multiple separate regions with homology to microsporidian, insect and insect mitochondrial sequences, also displaying features consistent with its being a lepidopteran W chromosome (Hi-C interactions with lepidopteran chromosomes, telomeric repeats consistent with a lepidopteran origin, high repeat density, reduced coverage). We observed no evidence of extant infection with a microsporidian parasite. This suggests horizontal gene transfer from a microsporidian endosymbiont, which we have previously observed in other lepidopteran W chromosomes.
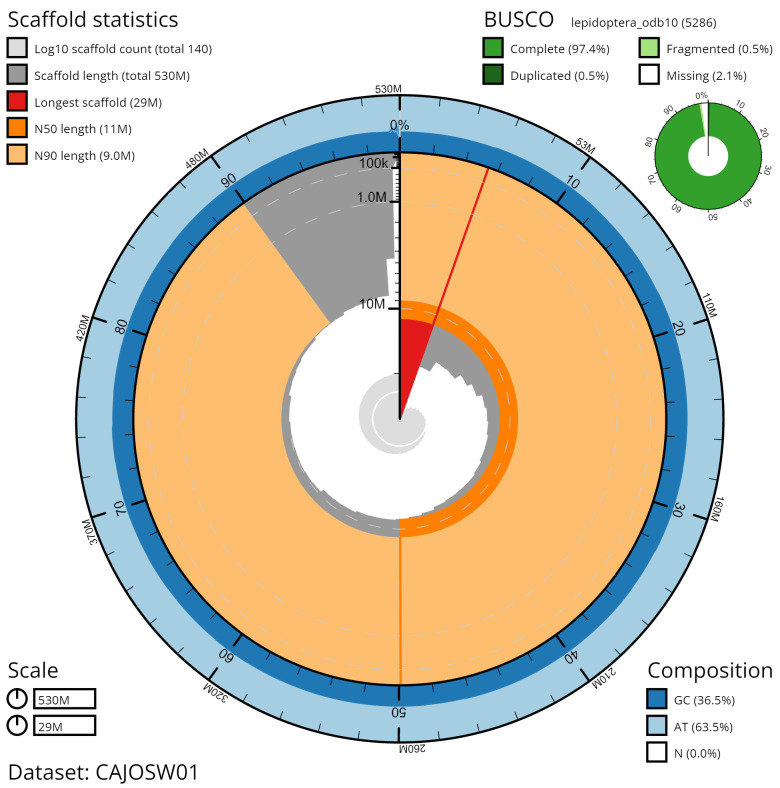
Figure 2. Genome assembly of Lysandra bellargus, ilLysBell1.1: metrics.
The BlobToolKit Snailplot shows N50 metrics and BUSCO gene completeness. The main plot is divided into 1,000 size-ordered bins around the circumference with each bin representing 0.1% of the 528,933,758 bp assembly. The distribution of chromosome lengths is shown in dark grey with the plot radius scaled to the longest chromosome present in the assembly (29,060,993 bp, shown in red). Orange and pale-orange arcs show the N50 and N90 chromosome lengths (11,205,580 and 8,974,749 bp), respectively. The pale grey spiral shows the cumulative chromosome count on a log scale with white scale lines showing successive orders of magnitude. The blue and pale-blue area around the outside of the plot shows the distribution of GC, AT and N percentages in the same bins as the inner plot. A summary of complete, fragmented, duplicated and missing BUSCO genes in the lepidoptera_odb10 set is shown in the top right. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/ilLysBell1.1/dataset/CAJOSW01/snail.
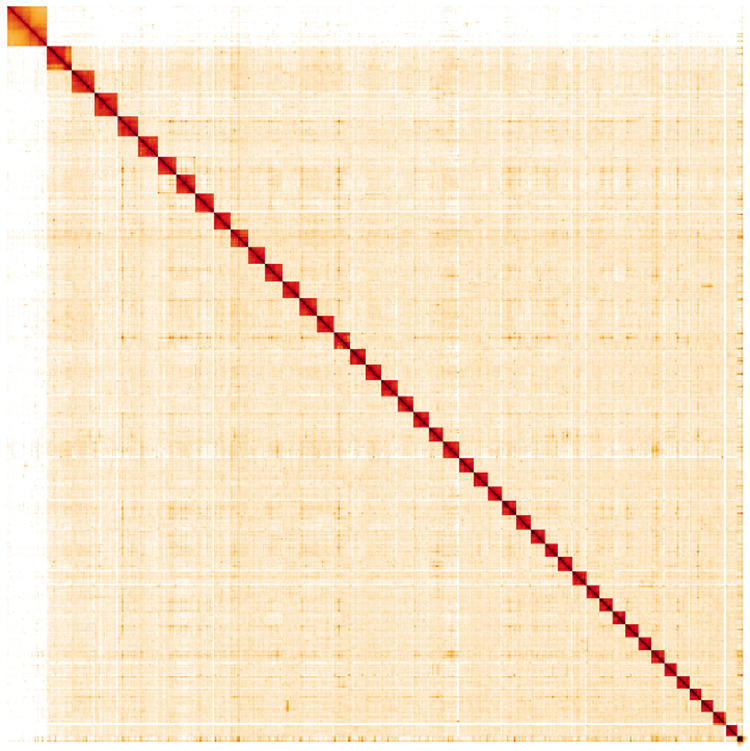
Figure 5. Genome assembly of Lysandra bellargus, ilLysBell1.1: Hi-C contact map.
Hi-C contact map of the ilLysBell1.1 assembly, visualised in HiGlass. Chromosomes are arranged in size order from left to right and top to bottom. The interactive Hi-C map can be viewed at https://genome-note-higlass.tol.sanger.ac.uk/l/?d=BZpgv8y6Q_2udwxedQCuYA.
Table 1. Genome data for Lysandra bellargus, ilLysBell1.1.
| Project accession data | |
|---|---|
| Assembly identifier | ilLysBell1.1 |
| Species | Lysandra bellargus |
| Specimen | ilLysBell1 (genome assembly, Hi-C); ilLysBell2 (RNA-Seq) |
| NCBI taxonomy ID | 138070 |
| BioProject | PRJEB43534 |
| BioSample ID | SAMEA7536572 |
| Isolate information | Female, whole organism (ilLysBell1); male, whole organism (ilLysBell2) |
| Raw data accessions | |
| PacificBiosciences SEQUEL II | ERR6576322 |
| 10X Genomics Illumina | ERR6054513-ERR6054516 |
| Hi-C Illumina | ERR6054517 |
| PolyA RNA-Seq Illumina | ERR6363262 |
| Genome assembly | |
| Assembly accession | GCA_905333045.1 |
| Accession of alternate haplotype | GCA_905332955.1 |
| Span (Mb) | 529 |
| Number of contigs | 361 |
| Contig N50 length (Mb) | 2.9 |
| Number of scaffolds | 139 |
| Scaffold N50 length (Mb) | 11.2 |
| Longest scaffold (Mb) | 18.03 |
| BUSCO * genome score | C:97.4%[S:96.9%,D:0.5%],F:0.5%,M:2.1%,n:5,286 |
| Genome annotation | |
| Number of protein-coding
genes |
13,249 |
| Average length of coding
sequence (bp) |
1,443.19 |
| Average number of exons per
transcript |
7.00 |
| Average intron size (bp) | 2,238.35 |
*BUSCO scores based on the lepidoptera_odb10 BUSCO set using v5.3.2. C= complete [S= single copy, D=duplicated], F=fragmented, M=missing, n=number of orthologues in comparison. A full set of BUSCO scores is available at https://blobtoolkit.genomehubs.org/view/ilLysBell1.1/dataset/CAJOSW01/busco.
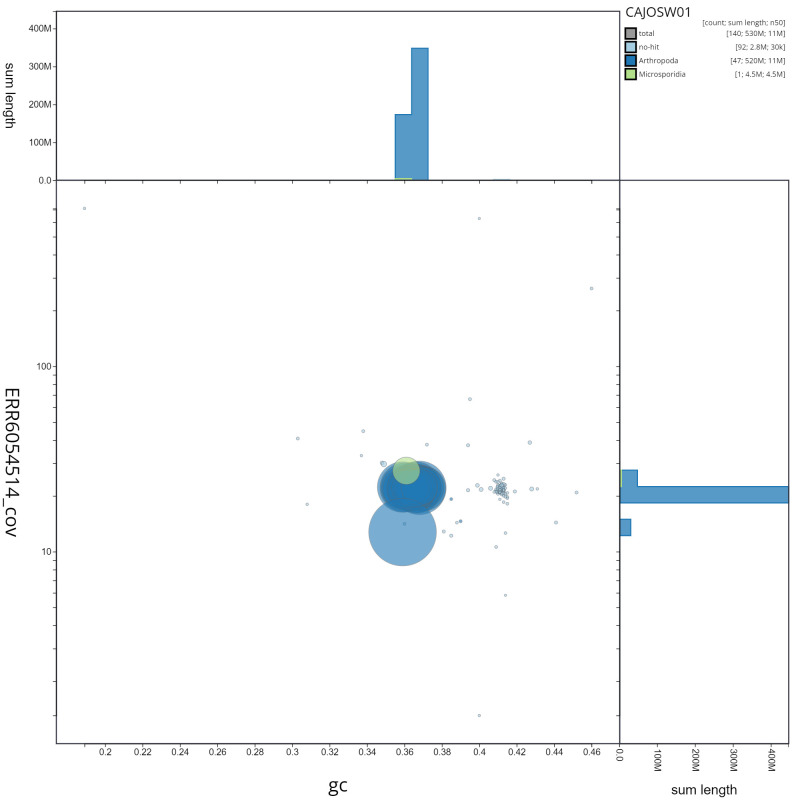
Figure 3. Genome assembly of Lysandra bellargus, ilLysBell1.1: GC coverage.
BlobToolKit GC-coverage plot. Scaffolds are coloured by phylum. Circles are sized in proportion to scaffold length. Histograms show the distribution of scaffold length sum along each axis. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/ilLysBell1.1/dataset/CAJOSW01/blob.
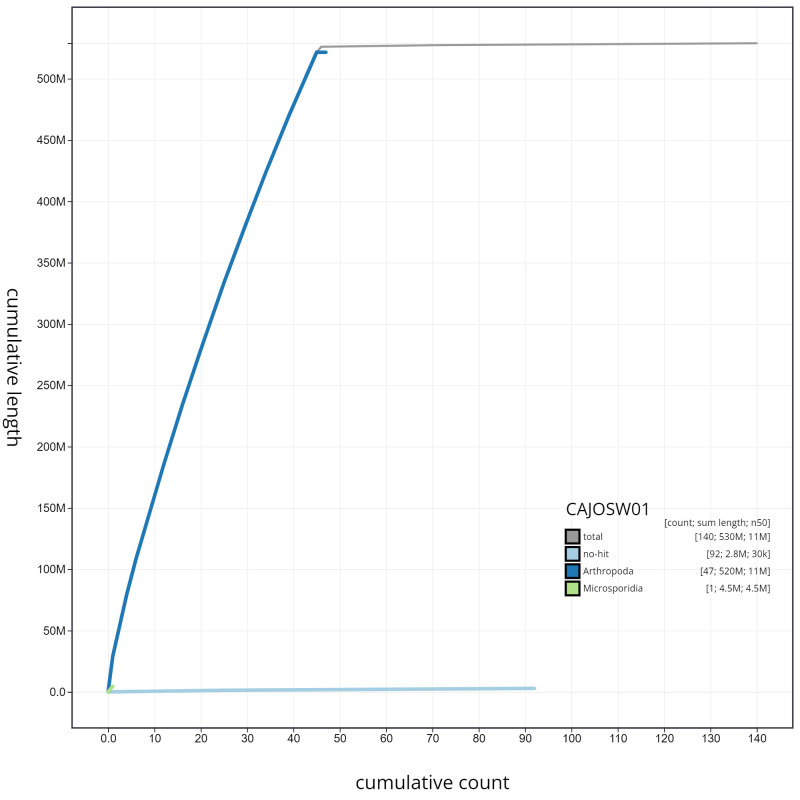
Figure 4. Genome assembly of Lysandra bellargus, ilLysBell1.1: cumulative sequence.
BlobToolKit cumulative sequence plot. The grey line shows cumulative length for all scaffolds. Coloured lines show cumulative lengths of scaffolds assigned to each phylum using the buscogenes taxrule. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/ilLysBell1.1/dataset/CAJOSW01/cumulative.
Table 2. Chromosomal pseudomolecules in the genome assembly of Lysandra bellargus, ilLysBell1.1.
| INSDC accession | Chromosome | Size (Mb) | GC% |
|---|---|---|---|
| HG995320.1 | 1 | 18.03 | 36.8 |
| HG995321.1 | 2 | 16.11 | 36.6 |
| HG995322.1 | 3 | 16 | 35.9 |
| HG995323.1 | 4 | 14.92 | 36 |
| HG995324.1 | 5 | 14.16 | 36.7 |
| HG995325.1 | 6 | 13.29 | 36.5 |
| HG995326.1 | 7 | 13.03 | 36.4 |
| HG995327.1 | 8 | 12.89 | 36.3 |
| HG995328.1 | 9 | 12.69 | 36.7 |
| HG995329.1 | 10 | 12.63 | 37 |
| HG995330.1 | 11 | 12.36 | 36.2 |
| HG995331.1 | 12 | 12.19 | 36.7 |
| HG995332.1 | 13 | 12.19 | 36.4 |
| HG995333.1 | 14 | 12.14 | 36.5 |
| HG995334.1 | 15 | 12.01 | 36.7 |
| HG995335.1 | 16 | 11.66 | 36.6 |
| HG995336.1 | 17 | 11.44 | 35.9 |
| HG995337.1 | 18 | 11.21 | 36.6 |
| HG995338.1 | 19 | 11.11 | 36.5 |
| HG995339.1 | 20 | 11.09 | 36.5 |
| HG995340.1 | 21 | 11.03 | 36.4 |
| HG995341.1 | 22 | 10.81 | 36.6 |
| HG995342.1 | 23 | 10.76 | 37 |
| HG995343.1 | 24 | 10.71 | 36.4 |
| HG995344.1 | 25 | 10.24 | 36.2 |
| HG995345.1 | 26 | 10.16 | 36.5 |
| HG995346.1 | 27 | 10.09 | 36.2 |
| HG995347.1 | 28 | 10.09 | 36.4 |
| HG995348.1 | 29 | 10.04 | 36.3 |
| HG995349.1 | 30 | 9.99 | 37 |
| HG995350.1 | 31 | 9.86 | 35.9 |
| HG995351.1 | 32 | 9.83 | 36.4 |
| HG995352.1 | 33 | 9.52 | 36.6 |
| HG995353.1 | 34 | 9.46 | 36.2 |
| HG995354.1 | 35 | 9.37 | 36.2 |
| HG995355.1 | 36 | 9.26 | 36.1 |
| HG995356.1 | 37 | 9.25 | 36.7 |
| HG995357.1 | 38 | 8.99 | 36.7 |
| HG995358.1 | 39 | 8.97 | 36.5 |
| HG995359.1 | 40 | 8.94 | 36.6 |
| HG995360.1 | 41 | 8.65 | 36.4 |
| HG995361.1 | 42 | 8.59 | 36.5 |
| HG995362.1 | 43 | 8.41 | 36.3 |
| HG995363.1 | 44 | 8.29 | 36.8 |
| HG995364.1 | W | 4.54 | 36.1 |
| HG995319.1 | Z | 29.06 | 35.9 |
| HG995365.1 | MT | 0.02 | 19.1 |
| - | Unplaced | 2.86 | 40.3 |
The assembly has a BUSCO v5.3.2 ( Manni et al., 2021) completeness of 97.4% (single 96.9%, duplicated 0.5%) using the lepidoptera_odb10 reference set (n=5,286). While not fully phased, the assembly deposited is of a single haplotype. Contigs corresponding to the second haplotype have also been deposited.
Genome annotation report
The ilLysBell1.1 genome was annotated using the Ensembl rapid annotation pipeline ( Table 1; Lysandra bellargus Ensembl page). The resulting annotation includes 24,348 transcribed mRNAs from 13,249 protein-coding and 2,895 non-coding genes. There is an average of 7.00 exons and 6.00 introns per canonical protein coding transcript, with an average intron length of 2,238.35 bases.
Methods
Sample acquisition and nucleic acid extraction
Two L. bellargus specimens (ilLysBell1, female; ilLysBell2, male) were collected using a net from El Brull, Catalunya, Spain (latitude 41.8103, longitude 2.3054) by Konrad Lohse (University of Edinburgh) and Alex Hayward (University of Exeter). The specimens were identified by Roger Vila (Institut de Biologia Evolutiva, Barcelona) and flash frozen from live in a dry shipper.
DNA was extracted at the Scientific Operations Core, Wellcome Sanger Institute. The ilLysBell1 sample was weighed and dissected on dry ice with head tissue set aside for Hi-C sequencing. Whole organism tissue was disrupted by manual grinding in lysis buffer with a disposable pestle. Fragment size analysis of 0.01–0.5 ng of DNA was then performed using an Agilent FemtoPulse. High molecular weight (HMW) DNA was extracted using the Qiagen MagAttract HMW DNA extraction kit. Low molecular weight DNA was removed from a 200-ng aliquot of extracted DNA using 0.8X AMpure XP purification kit prior to 10X Chromium sequencing; a minimum of 50 ng DNA was submitted for 10X sequencing. HMW DNA was sheared into an average fragment size between 12–20 kb in a Megaruptor 3 system with speed setting 30. Sheared DNA was purified by solid-phase reversible immobilisation using AMPure PB beads with a 1.8X ratio of beads to sample to remove the shorter fragments and concentrate the DNA sample. The concentration of the sheared and purified DNA was assessed using a Nanodrop spectrophotometer and Qubit Fluorometer and Qubit dsDNA High Sensitivity Assay kit. Fragment size distribution was evaluated by running the sample on the FemtoPulse system.
RNA was extracted from the whole organism tissue of ilLysBell2 in the Tree of Life Laboratory at the WSI using TRIzol, according to the manufacturer’s instructions. RNA was then eluted in 50 μl RNAse-free water and its concentration assessed using a Nanodrop spectrophotometer and Qubit Fluorometer using the Qubit RNA Broad-Range (BR) Assay kit. Analysis of the integrity of the RNA was done using Agilent RNA 6000 Pico Kit and Eukaryotic Total RNA assay.
Sequencing
Pacific Biosciences HiFi circular consensus and 10X Genomics Chromium read cloud sequencing libraries were constructed according to the manufacturers’ instructions. Sequencing was performed by the Scientific Operations core at the Wellcome Sanger Institute on Pacific Biosciences SEQUEL II (HiFi), Illumina HiSeq X Ten and Illumina HiSeq 4000 (RNA-Seq) instruments. Hi-C data were generated in the Tree of Life laboratory from remaining whole organism tissue of ilLysBell1 using the Arima v2 kit and sequenced on a NovaSeq 6000 instrument.
Genome assembly
Assembly was carried out with Hicanu ( Nurk et al., 2020); haplotypic duplication was identified and removed with purge_dups ( Guan et al., 2020). One round of polishing was performed by aligning 10X Genomics read data to the assembly with longranger align, calling variants with freebayes ( Garrison & Marth, 2012). The assembly was then scaffolded with Hi-C data ( Rao et al., 2014) using SALSA2 ( Ghurye et al., 2019). The assembly was checked for contamination and corrected using the gEVAL system ( Chow et al., 2016) as described previously ( Howe et al., 2021). Manual curation ( Howe et al., 2021) was performed using gEVAL, HiGlass ( Kerpedjiev et al., 2018) and Pretext. The mitochondrial genome was assembled using MitoHiFi ( Uliano-Silva et al., 2021), which performs annotation using MitoFinder ( Allio et al., 2020). The genome was analysed and BUSCO scores generated within the BlobToolKit environment ( Challis et al., 2020). Table 3 contains a list of all software tool versions used, where appropriate.
Table 3. Software tools used.
| Software
tool |
Version | Source |
|---|---|---|
| Hicanu | 2.1 | Nurk et al., 2020 |
| purge_dups | 1.2.3 | Guan et al., 2020 |
| SALSA2 | 2.2 | Ghurye et al., 2019 |
| longranger
align |
2.2.2 |
https://support.10xgenomics.
com/genome-exome/software/ pipelines/latest/advanced/ other-pipelines |
| freebayes | 1.3.1-17-
gaa2ace8 |
Garrison & Marth, 2012 |
| MitoHiFi | 1.0 | Uliano-Silva et al., 2021 |
| HiGlass | 1.11.6 | Kerpedjiev et al., 2018 |
| PretextView | 0.2.x |
https://github.com/wtsi-hpag/
PretextView |
| BlobToolKit | 3.2.6 | Challis et al., 2020 |
Genome annotation
The Ensembl gene annotation system ( Aken et al., 2016) was used to generate annotation for the Lysandra bellargus assembly (GCA_905333045.1). Annotation was created primarily through alignment of transcriptomic data to the genome, with gap filling via protein-to-genome alignments of a select set of proteins from UniProt ( UniProt Consortium, 2019).
Author information
Members of the Wellcome Sanger Institute Tree of Life programme are listed here: https://doi.org/10.5281/zenodo.6866293.
Members of Wellcome Sanger Institute Scientific Operations: DNA Pipelines collective are listed here: https://doi.org/10.5281/zenodo.5746904.
Members of the Tree of Life Core Informatics collective are listed here: https://doi.org/10.5281/zenodo.6125046.
Members of the Darwin Tree of Life Consortium are listed here: https://doi.org/10.5281/zenodo.6418363.
Funding Statement
This work was supported by Wellcome through core funding to the Wellcome Sanger Institute (206194) and the Darwin Tree of Life Discretionary Award (218328). KL and DL are supported by an ERC grant (ModelGenom Land 757648). KL was also supported by a NERC fellowship (NE/L011522/1). AH is supported by a Biotechnology and Biological Sciences Research Council (BBSRC) David Phillips Fellowship (BB/N020146/1). RV was supported by the Spanish government through grant PID2019-107078GB-I00/ MCIN/AEI/ 10.13039/501100011033.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 2 approved, 2 approved with reservations]
Data availability
European Nucleotide Archive: Lysandra bellargus (Adonis blue). Accession number PRJEB43534; https://identifiers.org/ena.embl/PRJEB43534
The genome sequence is released openly for reuse. The L. bellargus genome sequencing initiative is part of the Darwin Tree of Life (DToL) project. All raw sequence data and the assembly have been deposited in INSDC databases. Raw data and assembly accession identifiers are reported in Table 1.
References
- Aken BL, Ayling S, Barrell D, et al. : The Ensembl Gene Annotation System. Database (Oxford). 2016;2016:baw093. 10.1093/database/baw093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allio R, Schomaker-Bastos A, Romiguier J, et al. : MitoFinder: Efficient Automated Large-Scale Extraction of Mitogenomic Data in Target Enrichment Phylogenomics. Mol Ecol Resour. 2020;20(4):892–905. 10.1111/1755-0998.13160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourn NAD, Warren MS, Butterfly Conservation, et al. : ADONIS BLUE Lysandra Bellargus (Polyommatus Bellargus).Butterfly Conservation Society.1998. Reference Source [Google Scholar]
- Carter DJ, Hargreaves B: A Field Guide to Caterpillars of Butterflies and Moths in Britain and Europe.Collins,1986. Reference Source [Google Scholar]
- Challis R, Richards E, Rajan J, et al. : BlobToolKit - Interactive Quality Assessment of Genome Assemblies. G3 (Bethesda). 2020;10(4):1361–74. 10.1534/g3.119.400908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow W, Brugger K, Caccamo M, et al. : gEVAL - a web-based browser for evaluating genome assemblies. Bioinformatics. 2016;32(16):2508–10. 10.1093/bioinformatics/btw159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox R, Dennis EB, Brown AF, et al. : A Revised Red List of British Butterflies. Insect Conserv Divers. Royal Entomological Society of London,2022. 10.1111/icad.12582 [DOI] [Google Scholar]
- Garrison E, Marth G: Haplotype-Based Variant Detection from Short-Read Sequencing.2012; arXiv:1207.3907. 10.48550/arXiv.1207.3907 [DOI] [Google Scholar]
- Ghurye J, Rhie A, Walenz BP, et al. : Integrating Hi-C Links with Assembly Graphs for Chromosome-Scale Assembly. PLoS Comput Biol. 2019;15(8):e1007273. 10.1371/journal.pcbi.1007273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, McCarthy SA, Wood J, et al. : Identifying and Removing Haplotypic Duplication in Primary Genome Assemblies. Bioinformatics. 2020;36(9):2896–98. 10.1093/bioinformatics/btaa025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper GL, Maclean N, Goulson D: Microsatellite Markers to Assess the Influence of Population Size, Isolation and Demographic Change on the Genetic Structure of the UK Butterfly Polyommatus bellargus. Mol Ecol. 2003;12(12):3349–57. 10.1046/j.1365-294x.2003.02012.x [DOI] [PubMed] [Google Scholar]
- Howe K, Chow W, Collins J, et al. : Significantly Improving the Quality of Genome Assemblies through Curation. GigaScience. 2021;10(1):giaa153. 10.1093/gigascience/giaa153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerpedjiev P, Abdennur N, Lekschas F, et al. : HiGlass: Web-Based Visual Exploration and Analysis of Genome Interaction Maps. Genome Biol. 2018;19(1):125. 10.1186/s13059-018-1486-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic Z: The Butterfly Chromosomes and Their Application in Systematics and Phylogeny. Butterflies of Europe. 1990;332–96. [Google Scholar]
- Manni M, Berkeley MR, Seppey M, et al. : BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol Biol Evol. 2021;38(10):4647–54. 10.1093/molbev/msab199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Walenz BP, Rhie A, et al. : HiCanu: Accurate Assembly of Segmental Duplications, Satellites, and Allelic Variants from High-Fidelity Long Reads. Genome Res. 2020;30(9):1291–1305. 10.1101/gr.263566.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, et al. : A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159(7):1665–80. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper C: United Kingdom Species Inventory (UKSI).Natural History Museum,2021. 10.15468/RM6PM4 [DOI] [Google Scholar]
- Still J: Collins Wild Guide: Butterflies and Moths.Harper Collins, London,1996. [Google Scholar]
- Thomas JA: The Ecology and Conservation of Lysandra bellargus (Lepidoptera: Lycaenidae) in Britain. J Appl Ecol. 1983;20(1):59–83. 10.2307/2403376 [DOI] [Google Scholar]
- Uliano-Silva M, Nunes JGF, Krasheninnikova K, et al. : marcelauliano/MitoHiFi: mitohifi_v2.0.2021. 10.5281/zenodo.5205678 [DOI] [Google Scholar]
- UniProt Consortium: UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019;47(D1):D506–15. 10.1093/nar/gky1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Swaay C, Cuttelod A, Collins S, et al. : European Red List of Butterflies.2010. Reference Source [Google Scholar]