Keywords: Drosophila model, Icerguastat/IFB-088, OPMD, unfolded protein response, GADD34, PEK/PERK
Abstract
Oculopharyngeal muscular dystrophy (OPMD) is an autosomal dominant disease characterized by the progressive degeneration of specific muscles. OPMD is due to a mutation in the gene encoding poly(A) binding protein nuclear 1 (PABPN1) leading to a stretch of 11 to 18 alanines at N-terminus of the protein, instead of 10 alanines in the normal protein. This alanine tract extension induces the misfolding and aggregation of PABPN1 in muscle nuclei. Here, using Drosophila OPMD models, we show that the unfolded protein response (UPR) is activated in OPMD upon endoplasmic reticulum stress. Mutations in components of the PERK branch of the UPR reduce muscle degeneration and PABPN1 aggregation characteristic of the disease. We show that oral treatment of OPMD flies with Icerguastat (previously IFB-088), a Guanabenz acetate derivative that shows lower side effects, also decreases muscle degeneration and PABPN1 aggregation. Furthermore, the positive effect of Icerguastat depends on GADD34, a key component of the phosphatase complex in the PERK branch of the UPR. This study reveals a major contribution of the ER stress in OPMD pathogenesis and provides a proof-of-concept for Icerguastat interest in future pharmacological treatments of OPMD.
1. Introduction
Oculopharyngeal muscular dystrophy (OPMD) is an autosomal dominant disease that appears in the fifth decade. The disease is characterized by the progressive degeneration of specific muscles leading to ptosis (eyelid drooping), dysphagia (swallowing difficulties) and proximal limb weakness [1–3]. OPMD is caused by an expansion of GCG repeat in the gene encoding the poly(A) binding protein nuclear 1 (PABPN1) [4]. The mutation leads to a stretch of 11 to 18 alanines at the N-terminus of the protein, instead of 10 alanines in normal PABPN1. This extension induces misfolding and aggregation of PABPN1 in up to 10% of nuclei in affected muscles [5]. Aggregates contain the insoluble mutated-PABPN1, ubiquitin-proteasome system proteins, chaperones, mRNAs and RNA binding proteins [6–13].
PABPN1 has multiple functions in RNA metabolism. PABPN1 was discovered through its role in pre-mRNA cleavage and polyadenylation [14]. In this reaction, PABPN1 stimulates poly(A) polymerase activity and controls poly(A) tail lengths [14–18]. PABPN1 was then shown to be involved in alternative polyadenylation where it prevents utilization of weak proximal poly(A) sites by binding to non-canonical poly(A) signals [19,20]. PABPN1 also plays a role in polyadenylation-dependent decay or processing of nuclear RNAs by the exosome, including small nucleolar RNAs, long non-coding RNAs and abnormal mRNAs retained in the nucleus [21–25].
Defects in any of these molecular functions could participate in OPMD. Indeed, PABPN1 aggregates were reported to sequester the normal protein, leading to reduced levels of soluble PABPN1 that would contribute to OPMD pathogenesis through a loss-of-function mechanism [20,26,27]. In addition, PABPN1 aggregates or the process of aggregation are likely to substantially contribute to the pathology since decreasing the aggregation load, for example following oral treatments with small compounds, improve muscle function in Drosophila and mouse models of OPMD [28–32]. In line with this, it was recently shown using a Drosophila OPMD model that increased proteasome activity arises during OPMD progression, potentially in part through proteasome plugging with mutant PABPN1 oligomers, leading to muscle atrophy by degradation of myofibrillar proteins [33]. Thus, protein homeostasis appears to play a key role in OPMD.
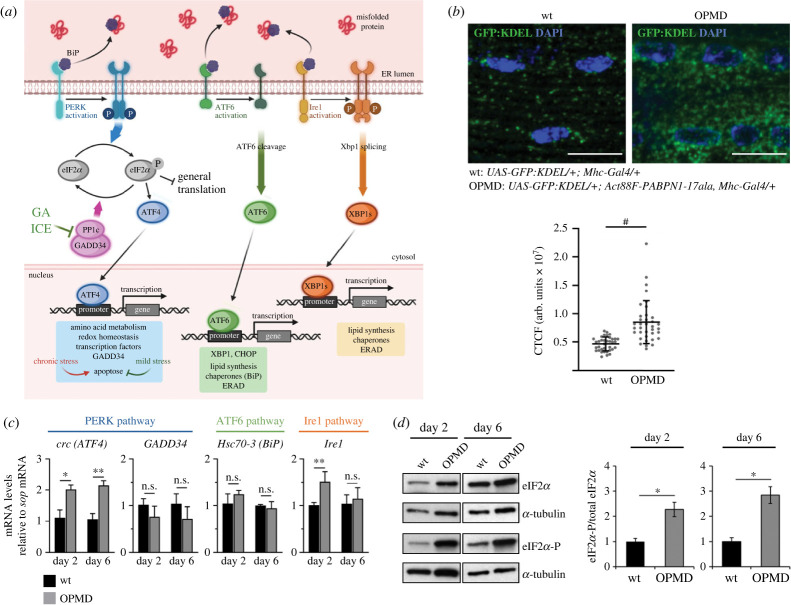
It has been established that reduced mitochondrial activity is one of the earliest defects in OPMD [27,34]. This defect results from an altered PABPN1 function in polyadenylation, which leads to shorter poly(A) tails of mRNAs encoding mitochondrial proteins involved in oxidative phosphorylation [34]. Decreased mitochondrial activity causes oxidative stress, which in turn produces oxidized proteins. These oxidized proteins can contribute to endoplasmic reticulum (ER) stress through accumulation of misfolded or unfolded proteins. The unfolded protein response (UPR) is initiated upon ER stress to restore ER homeostasis. The UPR is mediated by three transmembrane sensors: protein kinase R-like endoplasmic reticulum kinase (PERK, PEK in Drosophila), activating transcription factor 6 (ATF6) and inositol-requiring protein 1α (Ire1) [35–37]. The Binding Immunoglobulin Protein (BiP, Hsc70-3 in Drosophila) binds PERK, Ire1 and ATF6 in normal conditions. Upon ER stress, BiP recognizes and binds misfolded proteins and dissociates from the three sensors (figure 1a). PERK activation depends on its oligomerization and autophosphorylation. Following its activation, PERK phosphorylates eukaryotic translation initiation factor 2α (eIF2α). This leads to global inhibition of cap-dependent translation and translational activation of mRNAs encoding stress response proteins, such as activating transcription factor 4 (ATF4) and growth arrest and DNA damage-inducible protein (GADD34, also known as protein phosphatase 1 regulatory subunit 15A (PPP1R15A)). Translation activation depends on regulatory upstream ORFs (uORFs) present in the 5'-UTR of these mRNAs [38,39]. ATF4 translocates to the nucleus and activates the transcription of genes involved in the stress response, whereas GADD34 binds the catalytic subunit (PP1c) of the protein phosphatase 1 holoenzyme, which leads to dephosphorylation of eIF2α and recovery of translation, and thus ends the UPR [40–42]. In the two other branches of the UPR, activated ATF6 moves to the Golgi apparatus to be cleaved [43], while Ire1 is activated by oligomerization and autophosphorylation [44,45]. Ire1 promotes the splicing of the X-box binding protein 1 (XBP1) that translocates to the nucleus and activates genes involved in protein folding or degradation [46–48]. Cleaved ATF6 also translocates to the nucleus and together with XBP1 enhances through transcriptional activation, the production of chaperones such as BiP, to restore protein folding or increase protein degradation [47].
Figure 1.
The UPR is activated in the OPMD Drosophila model. (a) Schematic representation of UPR pathway in Drosophila where the three branches are represented. (b) Immunostaining of wild-type and OPMD (Act88F-PABPN1-17ala/+) GFP:KDEL-expressing thoracic muscles with anti-GFP antibody. DNA was revealed with DAPI. The quantification of GFP:KDEL fluorescence was performed using imageJ and represented as the CTCF (corrected total cell fluorescence) in arbitrary units. Quantifications were from 6 wild-type muscles (n = 36) and 12 OPMD muscles (n = 36), from three biological replicates. The bars represent the means with standard deviations. #p < 0.0001, using the unpaired-t test. Scale bars: 10 µm. (c) Quantification of UPR mRNA levels in wild-type and OPMD (Act88F-PABPN1-17ala/+) thoraxes at day 2 and day 6, using RT-qPCR. mRNA levels were normalized to sop mRNA and set to 1 in the wild-type. Means are from three to five biological replicates quantified in triplicates, error bars represent standard deviations. (d) Western blots of protein extracts from wild-type and OPMD (Act88F-PABPN1-17ala/+) thoraxes revealed with anti-eIF2α and anti-phosphorylated eIF2α. α-Tubulin was used as a loading control. Quantification of the ratio phosphorylated eIF2α/total eIF2α is shown. Means are from four biological replicates, error bars represent standard deviations. (c, d) *p < 0.05, **p < 0.01, n.s.: non-significant, using the unpaired-t test.
Among the small molecules identified using animal models to be active in OPMD, Guanabenz acetate (GA) is an FDA-approved antihypertensive drug that acts as an agonist of α2-adrenergic receptors. GA was shown to reduce muscle degeneration and PABPN1 aggregation upon oral treatment in a Drosophila model of OPMD [29]. GA was demonstrated to display an anti-aggregation activity by inhibiting the protein folding activity of the ribosome (PFAR) [49–51]. Synergistic positive effect between deletion of the ribosomal DNA locus and 6-aminophenanthridine, another anti-aggregative drug with the same mode of action as GA led to propose that both 6-aminophenanthridine and GA reduce OPMD phenotypes in the OPMD Drosophila model by preventing the PFAR activity of the ribosome [29]. More recently GA was found to show a positive effect in the A17 mouse model of OPMD (expression of PABPN1-17ala in skeletal muscles [32]), improving muscle strength and decreasing PABPN1 aggregation [52]. GA is also known to target the PERK branch of the UPR by inhibiting the phosphatase activity of the GADD34-PP1c phosphatase complex, through binding to GADD34 (figure 1a). This leads to prolonged eIF2α phosphorylation and translation inhibition, and extended protection of the ER by reducing the accumulation of misfolded proteins [53,54]. The ER stress is activated in muscles of the A17 OPMD mouse model and GA was shown to increase the levels of UPR markers in a cellular model of OPMD. Therefore, GA was proposed to reduce OPMD defects in the A17 mouse model through the UPR [52]. Together, data in the Drosophila and mouse models of OPMD suggest that the beneficial effect of GA on OPMD might involve both its effects in blocking the PFAR activity of the ribosome and extending the UPR.
Icerguastat (ICE), previously called IFB-088 and Sephin1, is a GA derivative with a similar effect on the UPR [54] (figure 1a), but devoid of its α2-adrenergic activity. As such, it has lower adverse effects and might be a better option for pharmacological treatments of proteinopathies. Indeed, no hypotensive effect of ICE was recorded during a phase 1 clinical trial on healthy volunteers (NCT03610334). Here, using an OPMD Drosophila model, we show that the UPR is activated in OPMD muscles. Using mutants of the PERK branch of the UPR, we functionally validate that this branch of the UPR participates in OPMD. We further show that ICE has a positive effect when provided orally to OPMD flies, in decreasing muscle weakness and reducing the PABPN1 aggregation load. ICE acts synergistically with reduced GADD34 gene dosage, suggesting that its effect in OPMD depends on its known role in preventing eIF2α dephosphorylation by the GADD34–PP1c phosphatase complex. Together these data reveal the functional implication of the ER stress response in OPMD and identify ICE as a potential candidate for future pharmacological treatments of OPMD and other proteinopathies involving the ER stress response.
2. Results
2.1. The PERK pathway is activated in the Drosophila model of OPMD
We previously developed Drosophila models of OPMD by expressing mammalian alanine-expanded PABPN1 (PABPN1-17ala) in muscles. PABPN1-17ala is specifically expressed in muscles either from larval stages using the UAS/Gal4 system and the Myosin heavy chain-Gal4 (Mhc-Gal4) driver or in adult thoracic muscles using the Act88F-PABPN1-17ala transgene [33,34,55]. Both models lead to a number of flies with abnormal wing position resulting from defective muscle function and progressive muscle degeneration [33,34,55]. To record ER stress in the OPMD Drosophila model, we monitored ER changes in thoracic muscles of Act88F-PABPN1-17ala/+ flies using the GFP:KDEL reporter protein that marks the ER lumen thanks to ER targeting and retention signals [56,57]. Immunostaining of GFP:KDEL-expressing muscles showed that GFP:KDEL labelled sites were substantially enlarged in OPMD muscles, particularly around nuclei (figure 1b) indicating a high level of ER reorganization consistent with ER stress.
We then used RT-qPCR to quantify mRNA expression of UPR pathway components in the Act88F-PABPN1-17ala model, at days 2 and 6 of adulthood. In the PERK branch, ATF4 is known to be regulated at transcriptional level, in addition to its translational regulation by eIF2α [58]. Drosophila ATF4 is encoded by the cryptocephal (crc) gene [59]. We found that crc mRNA levels were higher in OPMD muscles compared to wild-type at both days 2 and 6 (figure 1c). By contrast, GADD34 that is only regulated at the translation level in Drosophila [60] was not overexpressed in OPMD muscles using RT-qPCR (figure 1c). We quantified Hsc70-3 (BiP) and Ire1 mRNAs to assess activation of the ATF6 and Ire1 branches of the UPR, respectively. Hsc70-3 is known to be transcriptionally activated by ATF6 (figure 1a) and Drosophila Ire1 is also activated at transcriptional level [61]. Hsc70-3 mRNA levels were not higher than wild-type in OPMD muscles, suggesting a lack of activation of the ATF6 pathway, whereas Ire1 mRNA levels were higher at day 2, possibly consistent with its reported early activation under mild ER stress [62,63]. We next quantified eIF2α phosphorylation, a key marker of the PERK branch of the UPR, using western blots (figure 1d; electronic supplementary material, figure S1). eIF2α phosphorylation was higher in OPMD muscles compared to wild-type, at both day 2 and day 6 of adulthood.
These results reveal ER stress and activation of the PERK branch of the UPR in the OPMD Drosophila model.
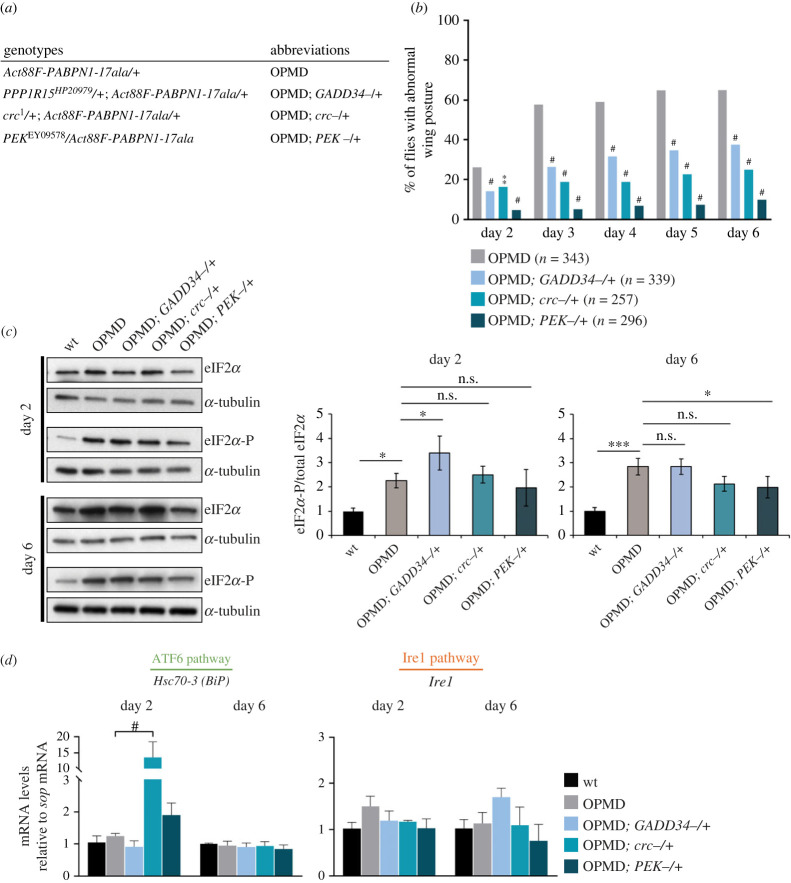
2.2. PERK pathway mutants reduce muscle defects in OPMD flies
Activation of the PERK branch of the UPR has a protective role under acute ER stress through the repression of general translation. However, PERK activation is detrimental under chronic stress through its role in activating apoptosis [64,65]. We used mutants of the Drosophila PERK pathway, namely of GADD34, crc and PEK genes (electronic supplementary material, figure S2a) to analyse the functional role of this pathway in OPMD pathogenesis. We analysed the effect of heterozygous GADD34, crc or PEK mutants on the percentage of Act88F-PABPN1-17ala flies showing wing position defects. The crc1 mutant is a known point mutant allele [66], however, GADD34 and PEK mutants were uncharacterized P-element insertion alleles (electronic supplementary material, figure S2a). We, therefore, validated that the corresponding mRNA levels were reduced in these two mutants (electronic supplementary material, figure S2b, c). The number of flies expressing Act88F-PABPN1-17ala (Act88F-PABPN1-17ala/+) with abnormal wing posture increased from 26% to 65% between day 2 and day 6 of adulthood (figure 2a,b). The presence of all three heterozygous GADD34, crc or PEK mutants led to a significant decrease in the number of flies with abnormal wing posture (figure 2b). The positive effects obtained with GADD34 and PEK heterozygous mutants were confirmed using UAS-RNAi transgenes specifically expressed in muscles with the Mhc-Gal4 driver (electronic supplementary material, figure S2d, e). PEK and GADD34 have antagonist activities on eIF2α phosphorylation and thus translational regulation, therefore, the decrease of wing posture defects in the presence of both mutants was surprising. We analysed whether this positive phenotypic effect was linked to eIF2α phosphorylation by quantifying the levels of phosphorylated eIF2α in thoracic muscles, in the presence or absence of the three heterozygous mutants using western blots. A decrease of GADD34 function is expected to result in increased levels of phosphorylated eIF2α that might reduce the ER stress through enhanced UPR response. By contrast, the levels of phosphorylated eIF2α are expected to decrease in the PEK heterozygous mutant, which might contribute to restart cap-dependent translation and increase the ER stress. It should be noted though, that variations in eIF2α phosphorylation levels might be difficult to record in vivo as quantifications were not performed under acute ER stress, but rather in flies several days old, which might blur the response. Quantification of eiF2α phosphorylation in thoracic muscles showed that indeed the presence of the GADD34−/+ mutant in Act88F-PABPN1-17ala/+ flies led to higher levels of phosphorylated eIF2α at day 2, whereas PEK−/+ mutant reduced these levels with a significant decrease at day 6, and crc−/+ mutant had no significant effect on eIF2α phosphorylation (figure 2c; electronic supplementary material, figure S3a). These results show that the level of eIF2α phosphorylation alone cannot explain the phenotypic improvement of OPMD flies by mutants of the PERK pathway, since reduced eIF2α phosphoryation in the PEK−/+ mutant is not consistent with the positive effect of this mutant through the UPR. Therefore, the decrease of muscle defects by PERK pathway mutants involves different mechanisms. This is consistent with data in mouse models of neuropathy showing that reduced function of PERK or GADD34 have similar effects in improving neuronal function [67,68].
Figure 2.
Half dosage of UPR genes reduces OPMD wing position defects. (a) Genotypes and abbreviations used in figures 2, 3 and 5. (b) Wing position defects were scored in OPMD (Act88F-PABPN1-17ala/+) flies in the absence or presence of UPR heterozygous mutants, each day between days 2 and 6. The numbers of scored flies are indicated (n). Quantifications were from of three biological replicates. **p < 0.01, #p < 0.0001, using the Chi2-test. (c) Western blots of protein extracts from OPMD thoracic muscles in the absence or presence of UPR hererozygous mutants, revealed with anti-eiF2α and anti-phosphorylated eiF2α. α-Tubulin was used as a loading control. Quantification of the ratio phosphorylated eiF2α/total eiF2α is shown. Means are from four biological replicates, error bars represent standard deviations. (d) Quantification of UPR mRNA levels in OPMD thoracic muscles in the absence or presence of UPR hererozygous mutants, at day 2 and day 6, using RT-qPCR. mRNA levels were normalized to sop mRNA and set to 1 in the wild-type. Means are from three biological replicates quantified in triplicates, error bars represent standard deviations. (c,d) *p < 0.05, ***p < 0.001, #p < 0.0001, n.s. or no significancy indicated: non-significant, using the one-way ANOVA test.
Because of the crosstalk between the three branches of the UPR, we investigated whether the ATF6 and Ire1 branches might be activated in response to reduced activity of the PERK branch in heterozygous mutants. The expression levels of Hsc70-3 and Ire1 mRNAs were quantified in thoracic muscles using RT-qPCR. Hsc70-3 mRNA highly accumulated in Act88F-PABPN1-17ala/+ thoracic muscles in the presence of the crc−/+ mutant at day 2 (figure 2d). This increased levels of Hsc70-3 mRNA might contribute to reduce muscle defects in Act88F-PABPN1-17ala/+; crc−/+ flies through higher synthesis of the chaperone Hsc70-3/BiP. Ire1 mRNA levels were not significantly affected in any of the three PERK pathway mutants (figure 2d).
These results reveal that targeting the PERK branch of the UPR strongly reduces muscle defects in the Drosophila OPMD model. This positive effect involves the UPR and increased eIF2α phosphorylation for GADD34−/+ mutant. In addition, the PEK kinase acts in OPMD through a distinct mechanism.
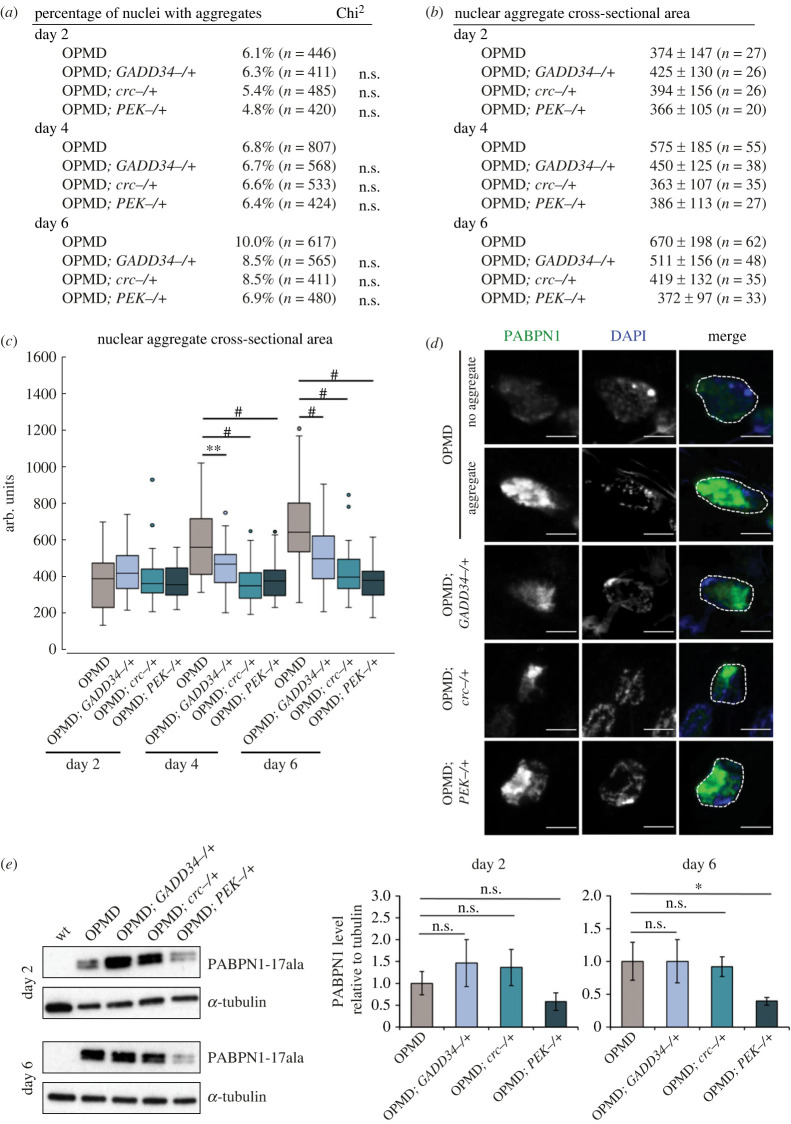
2.3. Targeting the PERK pathway decreases PABPN1-17ala aggregation in Drosophila
Aggregation of alanine-expanded PABPN1 is a hallmark of OPMD and genetic or pharmacological improvement of muscle function often correlates with reduced aggregation load [29,31,32,69], with some exceptions [33,34]. We, therefore, asked whether the improvement of muscle function in the presence of PERK pathway heterozygous mutants was associated with variations in PABPN1-17ala nuclear aggregates. Nuclear aggregates were visualized using anti-PABPN1 and DAPI staining in Act88F-PABPN1-17ala/+ thoracic muscles at days 2, 4 and 6 of adulthood. As previously reported [29,55], the number and size of PABPN1-17ala nuclear aggregates increased with time in OPMD muscles (figure 3a–d). Strikingly, in the presence of all three heterozygous GADD34, crc and PEK mutants, PABPN1-17ala aggregation was reduced at days 4 and 6. Although the percentage of nuclei with aggregates was not significantly reduced, the size of aggregates was reduced at days 4 and 6, with all three mutants (figure 3a–d). Because muscle defects and PABPN1-17ala aggregate formation are highly dependent on the levels of mutant PABPN1 [55], we quantified its levels in the absence and presence of the heterozygous mutants, in thoracic muscles using western blots. GADD34−/+ and crc−/+ mutants did not affect PABPN1-17ala levels, however, the PEK−/+ mutant reduced its levels, with a significant decrease at day 6 (figure 3e; electronic supplementary material, figure S3b). This low level of PABPN1-17ala in the presence of the PEK heterozygous mutant is likely the molecular basis of the reduced muscle defects in the presence of this mutant (figure 2b).
Figure 3.
PERK pathway heterozygous mutants reduce the size of PABPN1-17ala nuclear aggregates. (a) Percentage of nuclei with aggregates in OPMD thoracic muscles (Act88F-PABPN1-17ala/+) in the absence or presence of UPR heterozygous mutants. Adult thoracic muscles were dissected at days 2, 4 and 6 and stained with anti-PABPN1 and DAPI. Nuclear aggregates were visualized and scored using both staining. n.s. = non significant, using the chi2 test. (b,c) Quantification of nuclear aggregate areas. Each nuclear aggregate was delimited in a focal plan and the surface area was calculated using Image J. Mean values of the surface areas are shown in arbitrary units (b). Distribution of nuclear aggregate surface areas are shown as boxplots (c). The boxes represent 50% of the values, horizontal lines correspond to the medians (50% of the values on each side of the line), and vertical bars correspond to the range. Extreme values are in open circles. **p < 0.01, #p < 0.0001, no significancy indicated: non-significant, using the one-way ANOVA test. (d) Confocal images of nuclear aggregates visualized with anti-PABPN1 staining. DNA was revealed with DAPI. In OPMD individuals, nuclear aggregates are large and can occupy most of the nucleus. An example of nucleus without aggregate is shown for comparison (no aggregate). Nuclear aggregates were smaller in the presence of UPR heterozygous mutants. Nuclei are delimited with a white dotted line in the merge. Scale bars: 5 µm. (e) Western blots of protein extracts from OPMD thoracic muscles in the absence or presence of UPR heterozygous mutants revealed with anti-PABPN1 antibody. α-Tubulin was used as a loading control. Quantification of PABPN1 levels is shown. Means are from three biological replicates, error bars represent standard deviations. *p < 0.05, n.s.: non-significant, using the one-way ANOVA test.
Intriguingly, the decrease in the PABPN1-17ala aggregation load in the presence of the three heterozygous mutants (figure 3c) appears proportional to the positive effect of these mutants on the wing position phenotype (figure 2b), suggesting that these mutants might act primarily through PABPN1-17ala aggregation.
2.4. ICE is active in OPMD Drosophila models
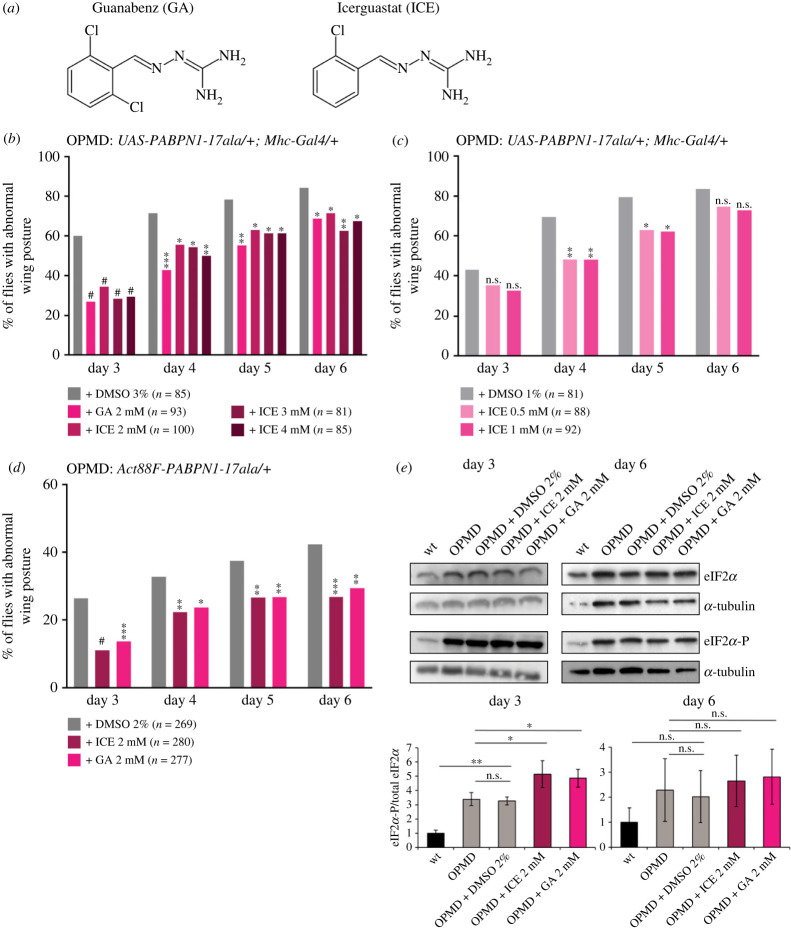
ICE is a GA derivative (figure 4a) that, as GA, was reported to prolong the UPR response by binding GADD34 and preventing eIF2α dephosphorylation [54]. ICE is lacking the α2-adrenergic activity present in GA and has therefore less side effects when used in proteinopathies. We tested the effect of ICE on two different OPMD Drosophila models.
Figure 4.
ICE reduces wing posture defects in OPMD Drosophila models. (a) Representation of GA and ICE molecules. (b–d) OPMD flies were transferred onto fresh instant Drosophila medium supplemented with ICE, GA or DMSO alone at day 2 after birth and then transferred to fresh medium with the drugs every day. Wing position defects were scored each day, between day 3 and day 6. Drug concentrations are indicated on the graphs. The numbers of scored flies are indicated (n). Quantifications were from one biological replicate in (b,c) and three biological replicates in (d). *p < 0.05, **p < 0.01, ***p < 0.001, #p < 0.0001, n.s.: non-significant, using the Chi2-test. The genotypes were UAS-PABPN1-17ala/+ ; Mhc-Gal4/+ in (b,c) and Act88F-PABPN1-17ala/+ in (d,e). (e) Western blots of protein extracts from thoracic muscles of OPMD flies fed with the drugs or DMSO alone, revealed with anti-eiF2α and anti-phosphorylated eiF2α. α-Tubulin was used as a loading control. Quantification of the ratio phosphorylated eiF2α/total eiF2α is shown. Means are from three biological replicates, error bars represent standard deviations. *p < 0.05, **p < 0.01, n.s.: non- significant, using the one-way ANOVA test.
First, we used the UAS/Gal4 system to express PABPN1-17ala in muscles with the Mhc-Gal4 driver. Flies expressing PABPN1-17ala were fed with ICE at 2 to 4 mM in the food from day 1 of adulthood and the number of flies with wing position defects were quantified at days 3 to 6. GA that has a positive effect on OPMD in Drosophila and mouse models [29,52] was used as a positive control. ICE had a beneficial effect in decreasing the number of OPMD flies with abnormal wing posture at the three concentrations of 2, 3 and 4 mM, compared to DMSO alone (figure 4b). ICE efficiency was stronger at day 3 when the wing position defects was lower, and was comparable between the three concentrations and comparable to GA efficiency at 2 mM. This indicated that the positive effect of the drug had already reached a plateau at the 2 mM concentration. We therefore analysed the effect of ICE at the lower concentrations of 1 and 0.5 mM in the food (figure 4c). The drug conserved a positive effect on the wing posture phenotype at these concentrations, but its efficiency decreased with a significant reduction of wing position defects at days 4 and 5 only (figure 4c).
Second, to confirm the positive effect of ICE on OPMD, we analysed its efficiency on the Act88F-PABPN1-17ala model. Similarly, Act88F-PABPN1-17ala/+ flies were fed with 2 mM ICE in the food and wing position defects were recorded at days 3 to 6. ICE showed a positive effect in this model also, with reduced numbers of flies showing wing position defects at all time points, compared to DMSO alone (figure 4d). As with the UAS-PABPN1-17ala model, the drug efficiency was higher at day 3 and was comparable to that of GA. We then asked whether the positive effect of ICE involves its known activity in inhibiting eIF2α dephosphorylation. We used western blots to quantify the levels of phosphorylated eIF2α in thoracic muscles of Act88F-PABPN1-17ala/+ flies fed with 2 mM ICE or GA in the food. eIF2α phosphorylation was higher following treatment with both ICE and GA at day 3 (figure 4e; electronic supplementary material, figure S4a). At day 6, eIF2α phosphorylation tended to be higher following ICE or GA treatment, but the increase was not significant (figure 4e; electronic supplementary material, figure S4a), possibly consistent with the lower efficiency of the compounds with time (figure 4d). These data suggest that ICE efficiency on muscle defects depends on its activity in the UPR in favouring translational repression. In addition, the same activity contributes to the beneficial effect of GA in OPMD.
We conclude that ICE is active in reducing OPMD muscle defects in Drosophila, with an efficiency similar to that of GA. These data also suggest that the positive effect of ICE involves its activity in targeting the PERK pathway, lengthening the UPR response.
2.5. ICE acts through GADD34 to reduce OPMD muscle defects
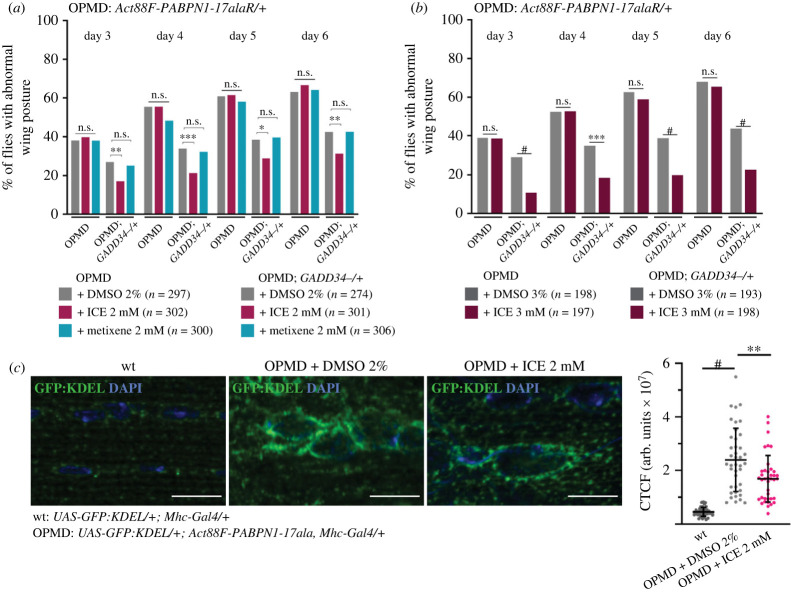
We used the genetic approach to confirm that ICE positive phenotypic effect depended on its ability to prevent GADD34-PP1c phosphatase complex activity. The Act88F-PABPN1-17alaR strain contains the same transgene as the Act88F-PABPN1-17ala strain, but its genetic background was changed by consecutive backcrosses. The resulting Act88F-PABPN1-17alaR/+ individuals showed a stronger OPMD phenotype, namely more individuals had wing position defects than Act88F-PABPN1-17ala/+ (compare figure 4d and figure 5a), and importantly, they were not sensitive to oral treatment with 2 mM ICE (figure 5a). We took advantage of this condition where 2 mM ICE concentration was suboptimal for Act88F-PABPN1-17alaR/+ flies to address a potential synergy with the GADD34 heterozygous mutant. We found that the GADD34−/+ mutant was able to reduce the percentage of flies with abnormal wing posture of Act88F-PABPN1-17alaR/+ flies. When ICE at 2 mM was provided to GADD34−/+; Act88F-PABPN1-17alaR/+ flies, the small molecule acted synergistically with GADD34−/+ in decreasing the percentage of flies with abnormal wing posture to a higher level than GADD34−/+ alone (figure 5a). A similar synergistic effect with GADD34−/+ but more pronounced was observed when ICE was provided at 3 mM (figure 5b). To confirm that the observed synergy depended on a direct link between ICE and GADD34−/+ mutant, we used another small molecule, metixene that is active in the OPMD Drosophila model but acts by reducing protein aggregation through the PFAR activity of the ribosome, not through the UPR [28]. As ICE, metixene at 2 mM was not active on Act88F-PABPN1-17alaR/+ flies (figure 5a), whereas it shows positive effect on Act88F-PABPN1-17ala/+ flies [28]. However, in contrast to ICE, it did not act synergistically with GADD34−/+ as the number of GADD34−/+; Act88F-PABPN1-17alaR/+ flies with abnormal wing posture did not decrease in the presence of metixene (figure 5a).
Figure 5.
ICE acts on OPMD through its GADD34 target. (a,b) Wing position defects were scored for OPMD (Act88F-PABPN1-17alaR/+) and OPMD; GADD34−/+ flies fed with drugs or DMSO alone. Flies were transferred onto fresh instant Drosophila medium supplemented with ICE, metixene or DMSO alone at day 2 after birth and then transferred to fresh medium with the drugs every day. Wing position defects were scored each day, from day 3 to day 6. Drug concentrations are indicating on the graphs. The numbers of scored flies are indicated (n). Quantifications were from three biological replicates in (a) and two biological replicates in (b). *p < 0.05, **p < 0.01, ***p < 0.001, #p < 0.0001, n.s.: non-significant, using the Chi2-test. (c) Immunostaining of wild-type and OPMD (Act88F-PABPN1-17ala/+) GFP:KDEL-expressing thoracic muscles with anti-GFP antibody. DNA was revealed with DAPI. OPMD flies were fed with 2 mM ICE or 2% DMSO alone. The quantification of GFP:KDEL fluorescence was performed using imageJ and represented as the CTCF (corrected total cell fluorescence) in arbitrary units. Quantifications were from 9 wild-type muscles (n = 40), 12 OPMD muscles from flies on 2% DMSO (n = 41) and 13 OPMD muscles from flies on 2 mM ICE (n = 39), from two biological replicates. The bars represent the means with standard deviations. **p < 0.01; #p < 0.0001, using the unpaired t-test. Scale bars: 10 µm.
We, next analysed a direct effect of ICE on the ER by recording changes in the ER structure using immunostaining of muscles expressing the GFP:KDEL reporter. We confirmed the enlargement of the ER lumen in OPMD muscles compared to wild-type (figure 5c). Strikingly, GFP:KDEL staining was significantly reduced when OPMD flies were fed with food containing 2 mM ICE, compared to DMSO alone, revealing a reduction of the ER, and thus decreased ER stress in the presence of the small molecule (figure 5c).
Together, these results are consistent with ICE acting in OPMD through its known function in inhibiting eIF2α dephosphorylation by the GADD34-PP1c phosphatase complex and maintaining translation inhibition to reduce ER stress.
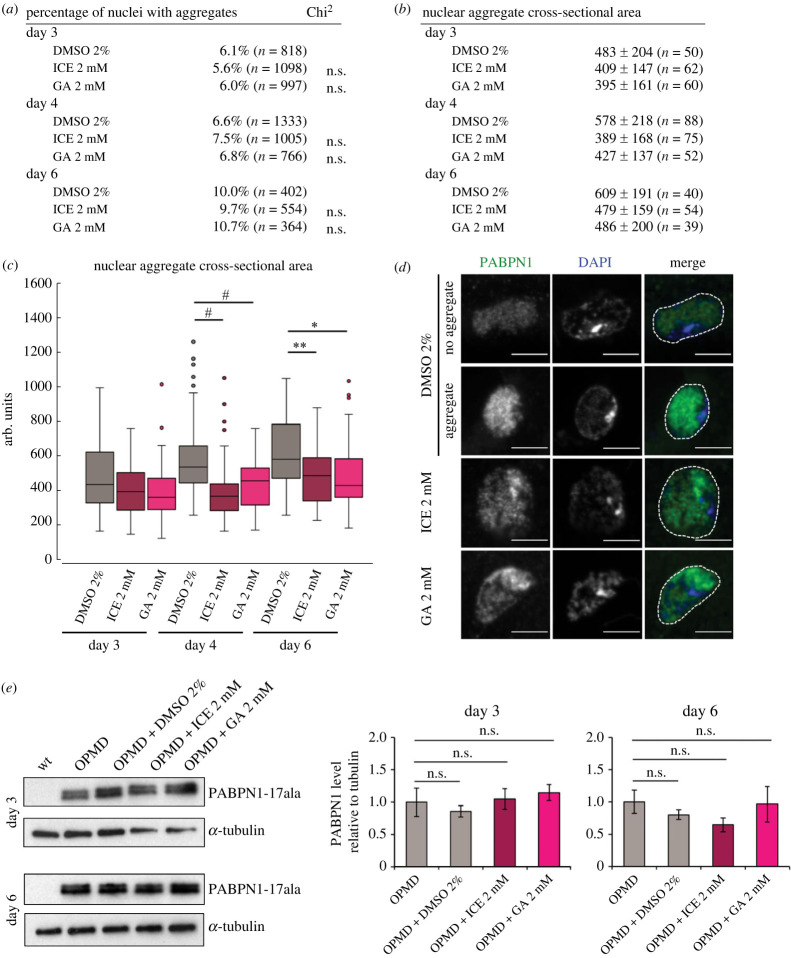
2.6. ICE reduces PABPN1-17ala aggregation in Drosophila
The decrease of muscle defects with PERK pathway heterozygous mutants correlates with reduced PABPN1-17ala aggregation in muscle nuclei. We, therefore, analysed whether ICE oral treatment also led to reduced PABPN1-17ala aggregate formation. Similarly to GA treatment [29] (figure 6a–d), the number of nuclei containing aggregates was not affected following oral treatment with 2 mM ICE (figure 6a), but the size of aggregates was significantly reduced over time (figure 6b–d). This smaller size of PABPN1-17ala aggregates was not due to lower levels of PABPN1-17ala protein in thoracic muscles following ICE treatment (figure 6e, electronic supplementary material, figure S4b).
Figure 6.
Oral treatment with ICE reduces PABPN1-17ala nuclear aggregates. (a) Percentage of nuclei with aggregates in OPMD thoracic muscles (Act88F-PABPN1-17ala/+). Adult flies were fed with the drugs that were provided in fresh medium every day from day 2. Adult thoracic muscles were dissected at days 3, 4 and 6 and stained with anti-PABPN1 and DAPI. Nuclear aggregates were visualized and scored using both staining. n.s.: non significant, using the Chi2-test. (b,c) Quantification of nuclear aggregate areas. Each nuclear aggregate was delimited in a focal plan and the surface area was calculated using ImageJ. Mean values of the surface areas are shown in arbitrary units (b). Distribution of nuclear aggregate surface areas are shown as boxplots (c). Legend is as in figure 3b, c. *p < 0.05, **p < 0.01, #p < 0.0001, no significancy indicated: non-significant, using the one-way ANOVA test. (d) Confocal images of nuclear aggregates visualized with anti-PABPN1 staining. DNA was revealed with DAPI. Examples of nuclei with or without an aggregate in OPMD individuals fed with DMSO alone. Nuclear aggregates were smaller when flies were raised on drug-supplemented medium. Examples of small aggregates at day 4 in the presence of 2 mM ICE or GA. Nuclei are delimited with a white dotted line in the merge. Scale bars: 5 µm. (e) Western blots of protein extracts from thoracic muscle of OPMD flies fed with the drugs or DMSO alone revealed with anti-PABPN1 antibody. α-Tubulin was used as a loading control. Quantification of PABPN1 levels is shown. Means are from three biological replicates, error bars represent standard deviations. n.s.: non-significant, using the one-way ANOVA test.
These results show that oral treatment with ICE decreases PABPN1-17ala aggregation in Drosophila OPMD muscles, similarly to GADD34 heterozygous mutant. They reinforce the notion that ICE would act positively on OPMD muscles through GADD34-PP1c complex activity.
3. Discussion
Several molecular pathways have previously been described to participate in OPMD pathogenesis. Here, we show that the ER stress and the downstream UPR pathway play a functional role in OPMD defects using the Drosophila model. ER stress and activation of the UPR have been recently reported in the A17 mouse model of OPMD [52]. Here we use the genetic approach to demonstrate that modulation of the PERK branch of the UPR has a positive effect in reducing OPMD phenotypes, in part through the repression of translation.
We find that the PERK branch is the main pathway activated in the Drosophila model of OPMD and maintained with time. In particular, eIF2α phosphorylation that prevents cap-dependent translation is increased in Drosophila OPMD muscles. Analysis of mRNA levels indicates that the Ire1 branch also appears to be activated, however, only at early stages. The Ire1 branch has been reported to be activated upon mild ER stress and beneficial [70–73]. By contrast, activation of the PERK pathway can be beneficial or detrimental for cells depending of the type of stress [63]. Upon mild stress, activation of the PERK pathway reduces the accumulation of misfolded proteins by inhibiting global translation through eIF2α phosphorylation. However, during chronic stress the PERK branch can activate an apoptotic cascade followed by cell death [64,65]. Both these mechanisms downstream of the PERK branch of the UPR might modulate OPMD defects. Quantification of eIF2α phosphorylation shows that it increases in OPMD Drosophila muscles in the presence of GADD34 heterozygous mutant, at early stages. This is consistent with a beneficial role of the PERK branch for OPMD at this early stage, through translation inhibition that would decrease the load of misfolded proteins in the ER. This beneficial effect would be enhanced by reducing the level of active GADD34-PP1c phosphatase in GADD34 heterozygous mutant. By contrast, the positive effect of PEK heterozygous mutant on OPMD muscle defects, associated with the decrease of eIF2α phosphorylation at later stages (day 6) is not consistent with a positive role of PEK through preventing translation. Thus, PEK would also act through an additional mechanism in OPMD pathogenesis. PEK function could involve apoptosis as it is the case during chronic stress, since apoptosis has been described to participate in OPMD defects [55]. In addition, PABPN1-17ala levels are reduced in OPMD Drosophila muscles in the presence of PEK heterozygous mutant. Although the molecular mechanisms leading to this decrease remain unknown, a lower amount of PABPN1-17ala is expected to substantially contribute to the decrease of muscle defects since the strength of these defects correlates with PABPN1-17ala levels in the Drosophila model [55]. Despite being counterintuitive, similar positive effects resulting from targeting both PERK and GADD34 have been reported for other diseases. For example, targeting either PERK or GADD34 in a mouse model of Parkinson's disease results in protection against neuronal degeneration [74,75]. Similarly, in a mouse model of Charcot–Marie–Tooth disease, both ablation of PERK and inactivation of GADD34 lead to improvement of motor performance [67,68]. Although the mechanisms underlying the beneficial effect of PERK inactivation in these diseases have not been identified, further analyses would be of interest in the future, and might lead to identifying additional therapeutic targets.
In this study, we identify a new molecule, ICE with a positive effect when provided orally to OPMD flies. This small molecule is a GA derivative and has the same property as GA in inactivating the GADD34–PP1c phosphatase complex, which leads to the maintenance of global translation repression [53,54,76]. We find that ICE, similarly to GA, is beneficial for OPMD by increasing eIF2α phosphorylation levels. Moreover, our genetics data indicate that GADD34 heterozygous mutant acts synergistically with ICE in lessening OPMD muscle defects. This provides functional evidence that ICE mechanism of action in OPMD involves GADD34-PP1c targeting. ICE was described to bind GADD34 and increase eIF2α phosphorylation levels from analyses in cell cultures [54]. However, more recent studies questioned this mode of action since ICE was found unable to disrupt the GADD34-PP1c complex in an in vitro set up [77,78]. Our data in the Drosophila model reinforce the conclusion that ICE can target the PERK branch of the UPR in vivo, although it could do so through a different mechanism than a direct interaction with GADD34. Interestingly, ICE has lower adverse effects than GA since it is devoid of the α2-adrenergic activity and thus has no hypotensive effect. Therefore, this small molecule has a higher potential for future development of pharmacological approaches to treat OPMD.
Intriguingly, both the genetic modulation of the PERK pathway and oral treatment of flies with ICE strongly reduces the size of PABPN1-17ala aggregates. Although reduction of muscle defects in OPMD animal models is not systematically associated with reduced PABPN1-17ala aggregation [33], this is nonetheless often the case. Indeed, treatments with diverse anti-aggregation drugs lead to both improvement of muscle function and reduction of aggregate formation [28,29,31,32,52]. These results have led to the conclusion that either the process of aggregation, or the presence of oligomeric forms of PABPN1-17ala play a key role in OPMD pathogenesis. Therefore, the decrease of PABPN1-17ala aggregation with PERK pathway heterozygous mutants or following ICE treatment might largely contribute to improving muscle function. The mechanisms underlying the reduction of PABPN1-17ala aggregation by alleviating the ER stress remain unknown. A direct link between PABPN1-17ala aggregation and the ER is unlikely since PABPN1 is a nuclear protein that does not transit through the ER. A possible hypothesis might involve the release of chaperone proteins upon reducing the ER stress; these chaperones would act in impeding PABPN1-17ala aggregation. In addition, although ICE was found to be less active as an anti-aggregation molecule [79], this activity might also contribute to reducing PABPN1-17ala aggregation as is the case for GA.
The positive effect of ICE in an animal model of OPMD along with its lower adverse effects make of this drug a promising therapeutic molecule for future treatments of OPMD. ICE was also shown to improve neuronal function in neurodegenerative diseases, such as Charcot–Marie–Tooth disease and amyotrophic lateral sclerosis [54,80]. Therefore, this molecule has strong potential for pharmacological approaches to diseases involving ER stress.
4. Material and methods
4.1. Drosophila stocks
w1118 was used as a control. OPMD was induced either by using the transgenic line Act88F-PABPN1-17ala to express PABPN1-17ala in adult indirect flight muscles [33,34] or the transgenic line UAS-PABPN1-17ala crossed with the Gal4 driver line Mhc-Gal4 to express PABPN1-17ala in all muscles from larval stages [55]. Mutant alleles were PEKEY0957 (#17582 at Bloomington Drosophila Stock Center (BDSC)), crc1/SM5 [66] (#266 at BDSC) and PPP1R15HP20979/CyO (#22330 at BDSC). Stocks containing UAS-RNAi constructs were TRiP.GL00030-attP2 (PEK-UAS-RNAi, #35162 at BDSC) and TRiP.HMS00811-attP2 (GADD34-UAS-RNAi, #33011 at BDSC); the attP2 stock (#36303 at BDSC) was used as a negative control. The UAS-KDEL:GFP stock [56] (#9898 at BDSC) was used to visualize the ER. All crosses were performed at 25°C, except crosses between UAS-PABPN1-17ala and Mhc-Gal4 that were maintained at 18°C; flies from these crosses were maintained at 18°C during the whole experiments.
4.2. Drug-supplemented medium and analysis of wing posture phenotype
Drug-supplemented medium was prepared with Instant Drosophila medium (Carolina Biological Supply Company) reconstituted with 2 ml of 1% yeast in water and supplemented with various concentrations of either Guanabenz acetate salt (Sigma, G110), metixene (a gift from Dr Cécile Voisset) or ICE (InFlectis BioScience) diluted in DMSO (Sigma, D2650). DMSO alone was used as control. Note that the concentration of 2 mM ICE in the food corresponds to an exposure of 0.23 mg kg−1/12 h, considering that a Drosophila ingests 20 nanoL h−1 [81]. This quantity of drug is in the range of that used for the treatment of OPMD mice with GA (1.7 mg kg−1 day−1) [52]. Adult males were grouped by twenty per vial. They were fed on drug-supplemented medium one day after birth and then changed to fresh drug-supplemented medium every day. Abnormal wing position was scored from day 3 to day 6 by direct observation through the vial after pooling five males in an empty vial every day. For mutant conditions, five males were pooled per vial containing regular Drosophila medium. Wing posture phenotype was scored every day from day 2 to day 6.
4.3. RNA extraction and RT-qPCR
Total RNA was prepared from five thoraxes per genotype using Trizol (Invitrogen) following recommendations of the manufacturer. DNA was digested using RQ1 RNase-Free DNase (Promega) for 30 min at 37°. Total RNA concentration was determined with nanodrop ND-1000 spectrophotometer. 1 µg of RNA was reverse transcribed for RT-qPCR using SuperScript III (Invitrogen) and random hexamers (Invitrogen). Quantitative PCR was performed on LightCycler LC480 (Roche) using LightCycler 480 SYBR Green I Master (Roche), and normalized with the Drosophila sop mRNA that encodes a ribosomal protein. Primers used are listed in table 1.
Table 1.
List of primers used in this study.
| gene | forward primer | reverse primer |
|---|---|---|
| sop | CACCCCAATAAAGTTGATAGACCT | ACCACCACGAGAGCCAAAT |
| GADD34 | TCGAGGAAGACGAGGATGA | AGGCGAGCATTACGTGAACT |
| crc (ATF4) | TTCCGCTCACTGACTCAAAC | AGGGTCAAGGCTTCATCATC |
| Ire1 | AAATCACAACAGGGCGACTC | GTGCGGAACCTGAACATTCT |
| Hsc70-3 (BiP) | AGGAAAAGGACAAGGAGCTG | GCGTCCGTTCTTGTACACAC |
| PEK | GAATTATCTGCGCCTTCAGC | CTCCGTGTCATTTTCGTCCT |
4.4. Immunostaining and western blots
Dissection and immunostaining of adult indirect flight muscles (IFMs) were performed as described previously [55] with some modifications. Following dissection, thoraxes were fixed for 45 min in 4% paraformaldehyde, 1× PBS. For immunostaining, IFMs were incubated in 1× PBS, 0.1% Triton X-100 four times for 15 min, then blocked in 1× PBS, 0.1% Triton X-100, 1% BSA for 1 h at room temperature. Antibodies diluted in 1× PBS, 0.1% Triton X-100, 1% BSA were as follows: rabbit anti-PABPN1 at 1:1000 [82]; mouse anti-GFP at 1:1000 (Roche, #11814 460 001). DNA was revealed by 3 min staining with 1 µg ml−1 DAPI (Sigma-Aldrich) in 1× PBS, 0.1% Triton X-100. Fluorescent images were acquired with a Leica SP8 confocal microscope. Quantification of KDEL:GFP staining was performed using ImageJ and calculated with the CTCF (corrected total cell fluorescence) as follows: CTCF = integrated density – (selection area* mean fluorescence of the background), where integrated density = selection area × mean fluorescence of the selection. The area of nuclei was subtracted from the area of the selection. Surface of aggregates was measured and quantified using the ImageJ software. For western blots, protein extracts were obtained from five thoraxes per genotype crushed in 50 µl of 2× Laemmli buffer supplemented with 10% β-mercaptoethanol and boiled for 10 min at 95°. Samples were loaded onto 4–15% or 10% SDS-PAGE gels and transferred to nitrocellulose membranes. Each membrane was blocked for 2 h in 5% milk diluted in 1× PBS, 0.1% Tween 20. Primary antibody incubation was performed overnight at 4°C in 1× PBS, 0.1% Triton X-100, 1% BSA, 0.02% azide. Antibodies used were: rabbit anti-eIF2α at 1:1000 (Abcam, Ab26197), rabbit anti-phosphorylated eIF2α at 1:1000 (Cell Signaling, #3398S), rabbit anti-PABPN1 at 1:1000 [82], mouse anti-α-Tubulin at 1:5000 (Sigma, T9026).
Acknowledgements
We are grateful to S. Marciniak and B. Mollereau for their gifts of Drosophila stocks, and to C. Voisset for her gift of metixene. We thank Bloomington Drosophila Stock Center for keeping and sending Drosophila stocks. We thank C. Salembier for producing the data shown in figure 4b,c.
Data accessibility
The data are provided in electronic supplementary material [83].
Authors' contributions
R.N.-S.: data curation, formal analysis, methodology, writing—original draft; A.C.: data curation, formal analysis; E.A.: resources; P.G.: resources; M.S.: conceptualization, funding acquisition, supervision, writing—original draft, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
E.A. is a full-time employee of InFlectis BioScience and P.G. is a full-time employee and founder of InFlectis BioScience.
Funding
This work was supported by the CNRS-University of Montpellier UMR9002, AFM-Téléthon eOPMD project 17110, FRM (Equipe FRM 2013 DEQ20130326534) and ANR (ANR-17-CE12-0011-01). R.N.S. held a PhD fellowship from the AFM-Téléthon.
References
- 1.Abu-Baker A, Rouleau GA. 2007. Oculopharyngeal muscular dystrophy: recent advances in the understanding of the molecular pathogenic mechanisms and treatment strategies. Biochim. Biophys. Acta BBA - Mol. Basis Dis. 1772, 173-185. ( 10.1016/j.bbadis.2006.10.003) [DOI] [PubMed] [Google Scholar]
- 2.Davies JE, Berger Z, Rubinsztein DC. 2006. Oculopharyngeal muscular dystrophy: potential therapies for an aggregate-associated disorder. Int. J. Biochem. Cell Biol. 38, 1457-1462. ( 10.1016/j.biocel.2006.01.016) [DOI] [PubMed] [Google Scholar]
- 3.Harish P, Malerba A, Dickson G, Bachtarzi H. 2015. Progress on gene therapy, cell therapy, and pharmacological strategies toward the treatment of oculopharyngeal muscular dystrophy. Hum. Gene Ther. 26, 286-292. ( 10.1089/hum.2015.014) [DOI] [PubMed] [Google Scholar]
- 4.Brais B, et al. 1998. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat. Genet. 18, 164-167. ( 10.1038/ng0298-164) [DOI] [PubMed] [Google Scholar]
- 5.Tomé FM, Fardeau M. 1980. Nuclear inclusions in oculopharyngeal dystrophy. Acta Neuropathol. (Berl.) 49, 85-87. ( 10.1007/BF00692226) [DOI] [PubMed] [Google Scholar]
- 6.Abu-Baker A. 2003. Involvement of the ubiquitin-proteasome pathway and molecular chaperones in oculopharyngeal muscular dystrophy. Hum. Mol. Genet. 12, 2609-2623. ( 10.1093/hmg/ddg293) [DOI] [PubMed] [Google Scholar]
- 7.Askanas V, Serdaroglu P, Engel WK, Alvarez RB. 1991. Immunolocalization of ubiquitin in muscle biopsies of patients with inclusion body myositis and oculopharyngeal muscular dystrophy. Neurosci. Lett. 130, 73-76. ( 10.1016/0304-3940(91)90230-q) [DOI] [PubMed] [Google Scholar]
- 8.Calado A, Tome FMS, Brais B, Rouleau GA, Kuhn U, Wahle E, Carmo-Fonseca M. 2000. Nuclear inclusions in oculopharyngeal muscular dystrophy consist of poly(A) binding protein 2 aggregates which sequester poly(A) RNA. Hum. Mol. Genet. 9, 2321-2328. ( 10.1093/oxfordjournals.hmg.a018924) [DOI] [PubMed] [Google Scholar]
- 9.Corbeil-Girard L-P, et al. 2005. PABPN1 overexpression leads to upregulation of genes encoding nuclear proteins that are sequestered in oculopharyngeal muscular dystrophy nuclear inclusions. Neurobiol. Dis. 18, 551-567. ( 10.1016/j.nbd.2004.10.019) [DOI] [PubMed] [Google Scholar]
- 10.Fan X, Messaed C, Dion P, Laganiere J, Brais B, Karpati G, Rouleau GA. 2003. hnRNP A1 and A/B interaction with PABPN1 in oculopharyngeal muscular dystrophy. Can. J. Neurol. Sci. 30, 244-251. ( 10.1017/S0317167100002675) [DOI] [PubMed] [Google Scholar]
- 11.Klein P, et al. 2016. Nuclear poly(A)-binding protein aggregates misplace a pre-mRNA outside of SC35 speckle causing its abnormal splicing. Nucleic Acids Res. 44, 10 929-10 945. ( 10.1093/nar/gkw703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leclerc A, Tomé FM, Fardeau M. 1993. Ubiquitin and beta-amyloid-protein in inclusion body myositis (IBM), familial IBM-like disorder and oculopharyngeal muscular dystrophy: an immunocytochemical study. Neuromuscul. Disord. NMD 3, 283-291. ( 10.1016/0960-8966(93)90021-b) [DOI] [PubMed] [Google Scholar]
- 13.Tavanez JP, Bengoechea R, Berciano MT, Lafarga M, Carmo-Fonseca M, Enguita FJ. 2009. Hsp70 chaperones and type I PRMTs are sequestered at intranuclear inclusions caused by polyalanine expansions in PABPN1. PLoS ONE 4, e6418. ( 10.1371/journal.pone.0006418) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wahle E. 1991. A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell 66, 759-768. ( 10.1016/0092-8674(91)90119-J) [DOI] [PubMed] [Google Scholar]
- 15.Benoit B, Nemeth A, Aulner N, Kühn U, Simonelig M, Wahle E, Bourbon HM. 1999. The Drosophila poly(A)-binding protein II is ubiquitous throughout Drosophila development and has the same function in mRNA polyadenylation as its bovine homolog in vitro. Nucleic Acids Res. 27, 3771-3778. ( 10.1093/nar/27.19.3771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerwitz Y. 2003. Stimulation of poly(A) polymerase through a direct interaction with the nuclear poly(A) binding protein allosterically regulated by RNA. EMBO J. 22, 3705-3714. ( 10.1093/emboj/cdg347) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kühn U, Buschmann J, Wahle E. 2017. The nuclear poly(A) binding protein of mammals, but not of fission yeast, participates in mRNA polyadenylation. RNA 23, 473-482. ( 10.1261/rna.057026.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kühn U, Gündel M, Knoth A, Kerwitz Y, Rüdel S, Wahle E. 2009. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J. Biol. Chem. 284, 22 803-22 814. ( 10.1074/jbc.M109.018226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Klerk E, et al. 2012. Poly(A) binding protein nuclear 1 levels affect alternative polyadenylation. Nucleic Acids Res. 40, 9089-9101. ( 10.1093/nar/gks655) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenal M, et al. 2012. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149, 538-553. ( 10.1016/j.cell.2012.03.022) [DOI] [PubMed] [Google Scholar]
- 21.Beaulieu YB, Kleinman CL, Landry-Voyer A-M, Majewski J, Bachand F. 2012. Polyadenylation-dependent control of long noncoding RNA expression by the poly(A)-binding protein nuclear 1. PLoS Genet. 8, e1003078. ( 10.1371/journal.pgen.1003078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bresson SM, Hunter OV, Hunter AC, Conrad NK. 2015. Canonical poly(A) polymerase activity promotes the decay of a wide variety of mammalian nuclear RNAs. PLOS Genet. 11, e1005610. ( 10.1371/journal.pgen.1005610) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bresson SM, Conrad NK. 2013. The human nuclear poly(A)-binding protein promotes RNA hyperadenylation and decay. PLoS Genet. 9, e1003893. ( 10.1371/journal.pgen.1003893) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemay J-F, Lemieux C, St-André O, Bachand F. 2010. Crossing the borders: Poly(A)-binding proteins working on both sides of the fence. RNA Biol. 7, 291-295. ( 10.4161/rna.7.3.11649) [DOI] [PubMed] [Google Scholar]
- 25.Meola N, et al. 2016. Identification of a nuclear exosome decay pathway for processed transcripts. Mol. Cell 64, 520-533. ( 10.1016/j.molcel.2016.09.025) [DOI] [PubMed] [Google Scholar]
- 26.Anvar SY, et al. 2013. A decline in PABPN1 induces progressive muscle weakness in Oculopharyngeal muscle dystrophy and in muscle aging. Aging 5, 412-426. ( 10.18632/aging.100567) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vest KE, et al. 2017. Novel mouse models of oculopharyngeal muscular dystrophy (OPMD) reveal early onset mitochondrial defects and suggest loss of PABPN1 may contribute to pathology. Hum. Mol. Genet. 26, 3235-3252. ( 10.1093/hmg/ddx206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bamia A, et al. 2021. Anti-prion drugs targeting the protein folding activity of the ribosome reduce PABPN1 aggregation. Neurother. J. Am. Soc. Exp. Neurother. 18, 1137-1150. ( 10.1007/s13311-020-00992-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barbezier N, Chartier A, Bidet Y, Buttstedt A, Voisset C, Galons H, Blondel M, Schwarz E, Simonelig M. 2011. Antiprion drugs 6-aminophenanthridine and guanabenz reduce PABPN1 toxicity and aggregation in oculopharyngeal muscular dystrophy. EMBO Mol. Med. 3, 35-49. ( 10.1002/emmm.201000109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chartier A, Simonelig M. 2013. Animal models in therapeutic drug discovery for oculopharyngeal muscular dystrophy. Drug Discov. Today Technol. 10, e103-e108. ( 10.1016/j.ddtec.2012.07.002) [DOI] [PubMed] [Google Scholar]
- 31.Davies JE, Sarkar S, Rubinsztein DC. 2006. Trehalose reduces aggregate formation and delays pathology in a transgenic mouse model of oculopharyngeal muscular dystrophy. Hum. Mol. Genet. 15, 23-31. ( 10.1093/hmg/ddi422) [DOI] [PubMed] [Google Scholar]
- 32.Davies JE, Wang L, Garcia-Oroz L, Cook LJ, Vacher C, O'Donovan DG, Rubinsztein DC. 2005. Doxycycline attenuates and delays toxicity of the oculopharyngeal muscular dystrophy mutation in transgenic mice. Nat. Med. 11, 672-677. ( 10.1038/nm1242) [DOI] [PubMed] [Google Scholar]
- 33.Ribot C, Soler C, Chartier A, Al Hayek S, Naït-Saïdi R, Barbezier N, Coux O, Simonelig M. 2022. Activation of the ubiquitin-proteasome system contributes to oculopharyngeal muscular dystrophy through muscle atrophy. PLOS Genet. 18, e1010015. ( 10.1371/journal.pgen.1010015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chartier A, et al. 2015. Mitochondrial dysfunction reveals the role of mRNA Poly(A) tail regulation in oculopharyngeal muscular dystrophy pathogenesis. PLOS Genet. 11, e1005092. ( 10.1371/journal.pgen.1005092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hetz C. 2012. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 13, 89-102. ( 10.1038/nrm3270) [DOI] [PubMed] [Google Scholar]
- 36.Ma K, Vattem KM, Wek RC. 2002. Dimerization and release of molecular chaperone inhibition facilitate activation of eukaryotic initiation factor-2 kinase in response to endoplasmic reticulum stress. J. Biol. Chem. 277, 18 728-18 735. ( 10.1074/jbc.M200903200) [DOI] [PubMed] [Google Scholar]
- 37.Wang W-A, Groenendyk J, Michalak M. 2014. Endoplasmic reticulum stress associated responses in cancer. Biochim. Biophys. Acta 1843, 2143-2149. ( 10.1016/j.bbamcr.2014.01.012) [DOI] [PubMed] [Google Scholar]
- 38.Lee Y-Y, Cevallos RC, Jan E. 2009. An upstream open reading frame regulates translation of GADD34 during cellular stresses that induce eIF2α phosphorylation. J. Biol. Chem. 284, 6661-6673. ( 10.1074/jbc.M806735200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vattem KM, Wek RC. 2004. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl Acad. Sci. USA 101, 11 269-11 274. ( 10.1073/pnas.0400541101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harding HP, Zhang Y, Scheuner D, Chen J-J, Kaufman RJ, Ron D. 2009. Ppp1r15 gene knockout reveals an essential role for translation initiation factor 2 alpha (eIF2alpha) dephosphorylation in mammalian development. Proc. Natl Acad. Sci. USA 106, 1832-1837. ( 10.1073/pnas.0809632106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma Y, Hendershot LM. 2003. Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J. Biol. Chem. 278, 34 864-34 873. ( 10.1074/jbc.M301107200) [DOI] [PubMed] [Google Scholar]
- 42.Novoa I, Zeng H, Harding HP, Ron D. 2001. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J. Cell Biol. 153, 1011-1022. ( 10.1083/jcb.153.5.1011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen J, Chen X, Hendershot L, Prywes R. 2002. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3, 99-111. ( 10.1016/s1534-5807(02)00203-4) [DOI] [PubMed] [Google Scholar]
- 44.Cox JS, Shamu CE, Walter P. 1993. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73, 1197-1206. ( 10.1016/0092-8674(93)90648-a) [DOI] [PubMed] [Google Scholar]
- 45.Mori K, Ma W, Gething MJ, Sambrook J. 1993. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 74, 743-756. ( 10.1016/0092-8674(93)90521-q) [DOI] [PubMed] [Google Scholar]
- 46.Korennykh AV, Egea PF, Korostelev AA, Finer-Moore J, Zhang C, Shokat KM, Stroud RM, Walter P. 2009. The unfolded protein response signals through high-order assembly of Ire1. Nature 457, 687-693. ( 10.1038/nature07661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamamoto K, Yoshida H, Kokame K, Kaufman RJ, Mori K. 2004. Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J. Biochem. (Tokyo) 136, 343-350. ( 10.1093/jb/mvh122) [DOI] [PubMed] [Google Scholar]
- 48.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881-891. ( 10.1016/s0092-8674(01)00611-0) [DOI] [PubMed] [Google Scholar]
- 49.Blondel M, et al. 2016. Protein folding activity of the ribosome is involved in yeast prion propagation. Sci. Rep. 6, 32117. ( 10.1038/srep32117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reis SD, Pang Y, Vishnu N, Voisset C, Galons H, Blondel M, Sanyal S. 2011. Mode of action of the antiprion drugs 6AP and GA on ribosome assisted protein folding. Biochimie 93, 1047-1054. ( 10.1016/j.biochi.2011.03.002) [DOI] [PubMed] [Google Scholar]
- 51.Tribouillard-Tanvier D, et al. 2008. Protein folding activity of ribosomal RNA is a selective target of two unrelated antiprion drugs. PLoS ONE 3, e2174. ( 10.1371/journal.pone.0002174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malerba A, et al. 2019. Pharmacological modulation of the ER stress response ameliorates oculopharyngeal muscular dystrophy. Hum. Mol. Genet. 28, 1694-1708. ( 10.1093/hmg/ddz007) [DOI] [PubMed] [Google Scholar]
- 53.Carrara M, Sigurdardottir A, Bertolotti A. 2017. Decoding the selectivity of eIF2α holophosphatases and PPP1R15A inhibitors. Nat. Struct. Mol. Biol. 24, 708-716. ( 10.1038/nsmb.3443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das I, Krzyzosiak A, Schneider K, Wrabetz L, D'Antonio M, Barry N, Sigurdardottir A, Bertolotti A. 2015. Preventing proteostasis diseases by selective inhibition of a phosphatase regulatory subunit. Science 348, 239-242. ( 10.1126/science.aaa4484) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chartier A, Benoit B, Simonelig M. 2006. A Drosophila model of oculopharyngeal muscular dystrophy reveals intrinsic toxicity of PABPN1. EMBO J. 25, 2253-2262. ( 10.1038/sj.emboj.7601117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okajima T, Reddy B, Matsuda T, Irvine KD. 2008. Contributions of chaperone and glycosyltransferase activities of O-fucosyltransferase 1 to Notch signaling. BMC Biol. 6, 1. ( 10.1186/1741-7007-6-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okajima T, Xu A, Lei L, Irvine KD. 2005. Chaperone activity of protein O-fucosyltransferase 1 promotes notch receptor folding. Science 307, 1599-1603. ( 10.1126/science.1108995) [DOI] [PubMed] [Google Scholar]
- 58.Dey S, Baird TD, Zhou D, Palam LR, Spandau DF, Wek RC. 2010. Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response. J. Biol. Chem. 285, 33 165-33 174. ( 10.1074/jbc.M110.167213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gauthier SA, Hewes RS. 2006. Transcriptional regulation of neuropeptide and peptide hormone expression by the Drosophila dimmed and cryptocephal genes. J. Exp. Biol. 209, 1803-1815. ( 10.1242/jeb.02202) [DOI] [PubMed] [Google Scholar]
- 60.Malzer E, Szajewska-Skuta M, Dalton LE, Thomas SE, Hu N, Skaer H, Lomas DA, Crowther DC, Marciniak SJ. 2013. Coordinate regulation of eIF2α phosphorylation by PPP1R15 and GCN2 is required during Drosophila development. J. Cell Sci. 126, 1406-1415. ( 10.1242/jcs.117614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walter F, O'Brien A, Concannon CG, Düssmann H, Prehn JHM. 2018. ER stress signaling has an activating transcription factor 6α (ATF6)-dependent ‘off-switch’. J. Biol. Chem. 293, 18 270-18 284. ( 10.1074/jbc.RA118.002121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, LaVail MM, Walter P. 2007. IRE1 signaling affects cell fate during the unfolded protein response. Science 318, 944-949. ( 10.1126/science.1146361) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rutkowski DT, et al. 2006. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 4, e374. ( 10.1371/journal.pbio.0040374) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de la Cadena SG, Hernández-Fonseca K, Camacho-Arroyo I, Massieu L. 2014. Glucose deprivation induces reticulum stress by the PERK pathway and caspase-7- and calpain-mediated caspase-12 activation. Apoptosis Int. J. Program. Cell Death 19, 414-427. ( 10.1007/s10495-013-0930-7) [DOI] [PubMed] [Google Scholar]
- 65.Demay Y, Perochon J, Szuplewski S, Mignotte B, Gaumer S. 2014. The PERK pathway independently triggers apoptosis and a Rac1/Slpr/JNK/Dilp8 signaling favoring tissue homeostasis in a chronic ER stress Drosophila model. Cell Death Dis. 5, e1452. ( 10.1038/cddis.2014.403) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hewes RS, Schaefer AM, Taghert PH. 2000. The cryptocephal gene (ATF4) encodes multiple basic-leucine zipper proteins controlling molting and metamorphosis in Drosophila. Genetics 155, 1711-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.D'Antonio M, Musner N, Scapin C, Ungaro D, Del Carro U, Ron D, Feltri ML, Wrabetz L. 2013. Resetting translational homeostasis restores myelination in Charcot-Marie-Tooth disease type 1B mice. J. Exp. Med. 210, 821-838. ( 10.1084/jem.20122005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Musner N, Sidoli M, Zambroni D, Del Carro U, Ungaro D, D'Antonio M, Feltri ML, Wrabetz L. 2016. Perk ablation ameliorates myelination in S63del-Charcot-Marie-Tooth 1B Neuropathy. ASN Neuro 8, 1759091416642351. ( 10.1177/1759091416642351) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chartier A, Raz V, Sterrenburg E, Verrips CT, van der Maarel SM, Simonelig M. 2009. Prevention of oculopharyngeal muscular dystrophy by muscular expression of Llama single-chain intrabodies in vivo. Hum. Mol. Genet. 18, 1849-1859. ( 10.1093/hmg/ddp101) [DOI] [PubMed] [Google Scholar]
- 70.Cheng K-C, Chiang H-C. 2018. XBP1 and PERK have distinct roles in Aβ-induced pathology. Mol. Neurobiol. 55, 7523-7532. ( 10.1007/s12035-018-0942-y) [DOI] [PubMed] [Google Scholar]
- 71.Credle JJ, Forcelli PA, Delannoy M, Oaks AW, Permaul E, Berry DL, Duka V, Wills J, Sidhu A. 2015. α-Synuclein-mediated inhibition of ATF6 processing into COPII vesicles disrupts UPR signaling in Parkinson's disease. Neurobiol. Dis. 76, 112-125. ( 10.1016/j.nbd.2015.02.005) [DOI] [PubMed] [Google Scholar]
- 72.Sado M, et al. 2009. Protective effect against Parkinson's disease-related insults through the activation of XBP1. Brain Res. 1257, 16-24. ( 10.1016/j.brainres.2008.11.104) [DOI] [PubMed] [Google Scholar]
- 73.Valdés P, et al. 2014. Control of dopaminergic neuron survival by the unfolded protein response transcription factor XBP1. Proc. Natl Acad. Sci. USA 111, 6804-6809. ( 10.1073/pnas.1321845111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mercado G, Castillo V, Soto P, López N, Axten JM, Sardi SP, Hoozemans JJM, Hetz C. 2018. Targeting PERK signaling with the small molecule GSK2606414 prevents neurodegeneration in a model of Parkinson's disease. Neurobiol. Dis. 112, 136-148. ( 10.1016/j.nbd.2018.01.004) [DOI] [PubMed] [Google Scholar]
- 75.Sun X, Aimé P, Dai D, Ramalingam N, Crary JF, Burke RE, Greene LA, Levy OA. 2018. Guanabenz promotes neuronal survival via enhancement of ATF4 and parkin expression in models of Parkinson disease. Exp. Neurol. 303, 95-107. ( 10.1016/j.expneurol.2018.01.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tsaytler P, Harding HP, Ron D, Bertolotti A. 2011. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science 332, 91-94. ( 10.1126/science.1201396) [DOI] [PubMed] [Google Scholar]
- 77.Crespillo-Casado A, Claes Z, Choy MS, Peti W, Bollen M, Ron D. 2018. A Sephin1-insensitive tripartite holophosphatase dephosphorylates translation initiation factor 2α. J. Biol. Chem. 293, 7766-7776. ( 10.1074/jbc.RA118.002325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Crespillo-Casado A, Chambers JE, Fischer PM, Marciniak SJ, Ron D. 2017. PPP1R15A-mediated dephosphorylation of eIF2α is unaffected by Sephin1 or Guanabenz. eLife 6, e26109. ( 10.7554/eLife.26109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nguyen PH, et al. 2014. Structure-activity relationship study around guanabenz identifies two derivatives retaining antiprion activity but having lost α2-adrenergic receptor agonistic activity. ACS Chem. Neurosci. 5, 1075-1082. ( 10.1021/cn5001588) [DOI] [PubMed] [Google Scholar]
- 80.Bai Y, et al. 2022. Treatment with IFB-088 improves neuropathy in CMT1A and CMT1B mice. Mol. Neurobiol. 59, 4159-4178. ( 10.1007/s12035-022-02838-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Plaçais P-Y, de Tredern É, Scheunemann L, Trannoy S, Goguel V, Han K-A, Isabel G, Preat T. 2017. Upregulated energy metabolism in the Drosophila mushroom body is the trigger for long-term memory. Nat. Commun. 8, 15510. ( 10.1038/ncomms15510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krause S, Fakan S, Weis K, Wahle E. 1994. Immunodetection of poly(A) binding protein II in the cell nucleus. Exp. Cell Res. 214, 75-82. ( 10.1006/excr.1994.1235) [DOI] [PubMed] [Google Scholar]
- 83.Naït-Saïdi R, Chartier A, Abgueguen E, Guédat P, Simonelig M. 2023. The small compound Icerguastat reduces muscle defects in oculopharyngeal muscular dystrophy through the PERK pathway of the unfolded protein response. Figshare. ( 10.6084/m9.figshare.c.6476390) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data are provided in electronic supplementary material [83].