Abstract
OBJECTIVE
The plasma proteome preceding diabetes can improve our understanding of diabetes pathogenesis.
RESEARCH DESIGN AND METHODS
In 8,923 Atherosclerosis Risk in Communities (ARIC) Study participants (aged 47–70 years, 57% women, 19% Black), we conducted discovery and internal validation for associations of 4,955 plasma proteins with incident diabetes. We externally validated results in the Singapore Multi-Ethnic Cohort (MEC) nested case-control (624 case subjects, 1,214 control subjects). We used Cox regression to discover and validate protein associations and risk-prediction models (elastic net regression with cardiometabolic risk factors and proteins) for incident diabetes. We conducted a pathway analysis and examined causality using genetic instruments.
RESULTS
There were 2,147 new diabetes cases over a median of 19 years. In the discovery sample (n = 6,010), 140 proteins were associated with incident diabetes after adjustment for 11 risk factors (P < 10−5). Internal validation (n = 2,913) showed 64 of the 140 proteins remained significant (P < 0.05/140). Of the 63 available proteins, 47 (75%) were validated in MEC. Novel associations with diabetes were found for 22 the 47 proteins. Prediction models (27 proteins selected by elastic net) developed in discovery had a C statistic of 0.731 in internal validation, with ΔC statistic of 0.011 (P = 0.04) beyond 13 risk factors, including fasting glucose and HbA1c. Inflammation and lipid metabolism pathways were overrepresented among the diabetes-associated proteins. Genetic instrument analyses suggested plasma SHBG, ATP1B2, and GSTA1 play causal roles in diabetes risk.
CONCLUSIONS
We identified 47 plasma proteins predictive of incident diabetes, established causal effects for 3 proteins, and identified diabetes-associated inflammation and lipid pathways with potential implications for diagnosis and therapy.
Graphical Abstract
Introduction
Type 2 diabetes pathogenesis involves an interplay of behaviors and genes over many years (1). Detailed characterization of the plasma proteome may provide insights into the dynamic changes preceding diabetes (2). Previous studies on the proteomics of diabetes risk have included a select number of proteins and generally focused on White populations (3–8). Broader proteomics coverage in larger, multiethnic samples should provide additional insights and broader generalizability. Furthermore, a robust design using discovery, followed by independent validation, should enhance the identification of novel plasma biomarkers, underlying pathways, clues for new treatment targets, and potentially improved risk prediction.
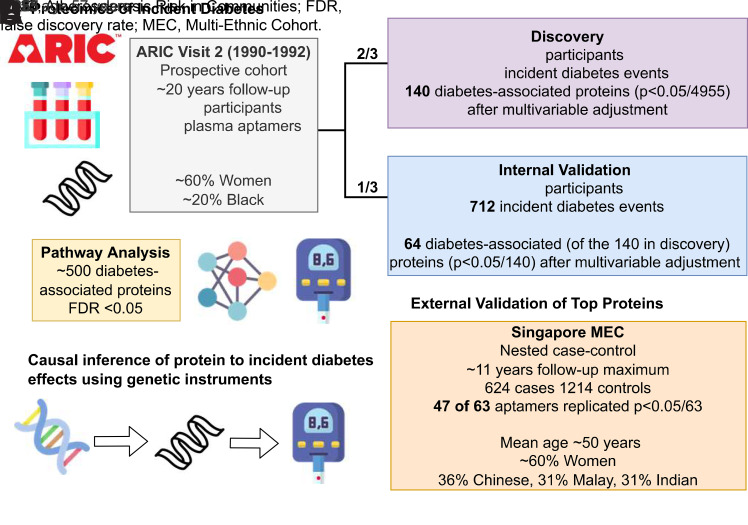
Using data from the Atherosclerosis Risk in Communities (ARIC) Study, we examined associations of nearly 5,000 aptamer measurements of the plasma proteome in midlife with incident diabetes among Black and White adults. We divided the ARIC Study data into discovery (2/3) and validation (1/3) samples, which permitted an internal validation of the proteome-diabetes associations, and we conducted an external replication of the top proteins in the Singapore Multi-Ethnic Cohort (MEC). Additionally, we 1) assessed the improvement in risk provided by proteome biomarkers beyond traditional risk factors, 2) performed a pathway analysis to identify biological pathways and upstream regulators relevant to diabetes development, and 3) used genetic models of plasma proteins to examine support for causal links between top proteins with diabetes risk.
Research Design and Methods
Study Design
The ARIC Study
The ARIC Study is a community-based cohort study that began in 1987–1989 when the 15,792 participants were aged 45–64 years (9). Participants were recruited from four U.S. communities: Forsyth County, North Carolina; suburban Minneapolis, Minnesota; Jackson, Mississippi; and Washington County, Maryland. Visit 2 occurred in 1990–1992 when participants were aged 46–70 years and was the first visit with HbA1c measurements. Participants were asked to fast >8 h and underwent phlebotomy during a clinic visit. Details relevant to covariate data collection are provided in the Supplementary Methods. Participants provided written informed consent. Institutional review boards at participating study centers approved the protocols.
Among the 14,348 ARIC participants who attended visit 2, we excluded 2,537 participants missing SomaScan visit 2 data (1,148 did not consent for industry-sponsored studies; 1,389 had no available sample or failed quality control [QC]), 382 participants who fasted <8 h, 1,590 individuals with prevalent diabetes (self-report diagnosis, glucose-lowering medication use, fasting blood glucose ≥126 mg/dL or HbA1c ≥6.5%), and 783 missing covariates of interest. Owing to small numbers, we excluded 42 participants who were neither White nor Black and 49 Black participants at the Maryland and Minnesota centers. To conduct protein discovery and then internal replication of the identified proteins, we randomly divided the data set into a 2/3 discovery (n = 6,010) and 1/3 validation (n = 2,913) sample.
Protein Measurements
The relative abundance of 5,284 plasma proteins was quantified using a highly multiplexed aptamer-based assay (SomaScan version 4; SomaLogic, Boulder, CO). Plasma samples (never thawed) from ARIC visit 2 were stored at −80°C until shipment on dry ice to the ARIC Study central laboratory, where specimens were thawed and underwent aliquoting into plates. These plates were sent to SomaLogic for protein measurements and were analyzed at SomaLogic in 2021. As described previously (10), the SomaScan uses Slow Offrate Modified Aptamer (SOMAmer) reagents to quantify the relative abundance of thousands of proteins and involves multiple QC steps with normalization, scaling, and calibration to minimize assay biases and batch effects. Protein abundances were standardized by SomaLogic using adaptive normalization by maximum likelihood techniques. After standard ARIC QC exclusions (see Supplementary Methods), we considered 4,955 SOMAmers (4,712 unique proteins) in the current analyses.
Ascertainment of Incident-Diagnosed Diabetes
Incident diabetes was based on a self-reported diagnosis by a health-care provider or glucose-lowering medication use (self-reported or medication brought to visits) reported by participants (or their proxies) during annual follow-up telephone calls through 2011–2013 or during ARIC visits 3 (1993–1995) or 4 (1996–1998) (11). We censored follow-up on the date the participant attended visit 5 (2011–2013) when the participants were all aged >65 years. If the participant was alive and did not attend visit 5, we censored follow-up on 31 December 2013. The date of the call or the visit where diabetes (diagnosis or medication) was first reported was used as the proxy for the date of diagnosis.
Statistical Analysis
We log2-transformed and then standardized the protein abundance to facilitate comparisons across proteins. We used a random number generator to divide the data set into a 2/3 discovery and 1/3 validation sample. In discovery, the Bonferroni-corrected two-sided P value was <10−5 (<0.05/4,955). We used Cox regression to examine associations of 4,955 proteins with incident diagnosed diabetes. Person-time accrued between visit 2 (baseline) to the date of diagnosed diabetes, death, loss to follow-up, or administrative censoring at visit 5, whichever came first. We used a set of hierarchical models, including model 0 (unadjusted). Model 1 adjusted for age (continuous), sex, race-center (White-Minnesota, White-Maryland, Black-Mississippi, White-North Carolina, Black-North Carolina), and estimated glomerular filtration rate (eGFR, continuous). We included eGFR in model 1 because kidney function is so strongly linked with the circulating proteome (12). Model 2 further added current smoking, physical activity (sport index), total cholesterol, HDL-cholesterol, systolic blood pressure, and hypertension medication use. Model 3 further added BMI. We further adjusted model 3 for HbA1c and fasting glucose (both modeled continuously, model 4) as complementary biomarkers of hyperglycemia. We considered model 3 as our main model for the discovery analysis. In a sensitivity analysis, we further adjusted all models for 10 protein principal components (PCsprotein) to account for correlations between proteins, including batch effects.
We explored the consistency of proteomic associations by baseline age (<55, ≥55 years), sex (male, female), race (Black, White), BMI (<25, ≥25 kg/m2), and HbA1c (<5.7%, 5.7 to <6.5%) in model 3. We calculated P interactions by adding a protein ∗ subgroup cross-product term in the models and a statistical significance threshold (Bonferroni P < 0.05/4,955/5) for testing interactions.
We used elastic net (13) with Cox regression models to examine whether proteins improved prediction of 20-year risk of diabetes beyond each of the adjustment models, quantified using the C statistics. Elastic net uses a machine learning approach to select a combination of proteins that optimize the predictive model for incident diabetes. This process was conducted for models including 1) covariate adjustment and 2) covariate adjustment + proteins (100 aptamers with the smallest P value were considered regardless of Bonferroni statistical significance). Covariates were forced in the elastic net models. We developed a predictive model (covariates and proteins) in discovery and tested model discrimination and calibration in the internal validation sample.
We used R version 4.1.2 software for all analyses.
Internal and External Validation
For the internal replication analysis within the ARIC Study, we examined whether proteins that were Bonferroni significant in the 2/3 discovery sample were also significant in the 1/3 internal validation sample at P < 0.05/no. of hits tested for each adjustment model.
We then externally replicated our proteomic findings in a nested matched case-control study (n = 1,858, mean age 50 [SD 11] years, 624 incident diabetes cases) within the Singapore MEC, a population-based cohort that included individuals of Chinese, Malay, and Indian ethnicity (14). Plasma proteins were quantified in MEC using SomaScan version 4. Of the 64 diabetes-associated proteins that were internally validated, 63 plasma proteins were available in MEC. Conditional logistic models were adjusted for a comparable set of covariates to our model 3 (see Supplementary Methods for details). For external validation, statistical significance was based on a Bonferroni threshold of P < 0.05/63. In a sensitivity analysis, we considered a less conservative threshold based on a false discovery rate (FDR) of q < 0.05.
Ingenuity Pathway Analysis
To explore mechanisms and upstream factors of diabetes-associated proteins (based on ARIC model 3), we conducted a pathway analysis using Ingenuity Pathway Analysis (IPA) (QIAGEN) based on published relationships between genes (or gene products) and regulators. Details regarding IPA have been previously published (15). Of the 4,955 proteins, 4,910 were mapped to genes in the IPA base, which we used to characterize canonical (biological) pathways and upstream regulators.
Genetic Instruments of Top Proteins with Diabetes
To characterize a causal nature of the diabetes-associated proteins, we performed a proteome-wide association study (PWAS) by combining genetic models from individuals of European ancestry for top proteins using previously developed elastic net regression models (16) with BMI-adjusted summary statistics from a genome-wide association study (GWAS) of diabetes among individuals of European ancestry performed by the DIAMANTE (DIAbetes Meta-ANalysis of Trans-Ethnic association Studies) consortium (17). This PWAS approach is equivalent to a two-sample Mendelian randomization (MR) that is restricted to the cis-region of the respective protein (i.e., ± 500 kilobase [kb] of the gene-encoding region) (18). The restriction to the cis-region leads to more conservative genetic models of SomaScan proteins than genome-wide modeling to increase support of the MR assumptions (e.g., pleiotropy). Moreover, this analysis is restricted to genetic models with significant genetic heritability (16). For the causal analyses, we considered the 64 internally validated proteins identified in model 3, of which 50 proteins had genetic instrument models. We used Bonferroni correction to determine statistical significance P < 0.05/50 (<0.001). In a sensitivity analysis, we performed colocalization analysis using a Bayesian framework (19). We estimated the posterior probability of the same causal variant underlying the protein GWAS and the diabetes GWAS (hypothesis 4 [H4]) for each protein with a significant PWAS finding in the encoding region ± 500 kb.
Results
Untargeted Discovery of the Proteomics of Incident Diabetes
In the discovery sample, there were 6,010 participants: mean age 56.8 (SD 5.7) years, 57% were women, and 19% self-identified their race as Black (Table 1). Over the median 19 (quartile 1, quartile 3: 13, 21) years of follow-up, we identified 1,435 cases of diagnosed diabetes.
Table 1.
Baseline participant characteristics in the ARIC Study (discovery and internal validation; 1990–1992) and in the Singapore MEC nested-case control study (external validation; 2004–2010)
| ARIC characteristics | MEC characteristics | ||||
|---|---|---|---|---|---|
| ARIC discovery | ARIC validation | MEC overall | MEC case subjects | MEC control subjects | |
| n = 6,010 | n = 2,913 | n = 1,838 | n = 624 | n = 1,214 | |
| Age, years | 56.8 (5.7) | 56.7 (5.7) | 50.6 (11.3) | 51.2 (11.6) | 50.3 (11.1) |
| Sex, n (%) | |||||
| Female | 3,406 (57) | 1,644 (56) | 1,043 (57) | 352 (56) | 691 (57) |
| Male | 2,604 (43) | 1,269 (44) | 795 (43) | 272 (44) | 523 (43) |
| Race/ethnicity, n (%) | |||||
| Black | 1,129 (19) | 543 (19) | — | — | — |
| White | 4,881 (81) | 2,370 (81) | — | — | — |
| Chinese | — | — | 667 (36) | 245 (39) | 422 (35) |
| Indian | — | — | 585 (32) | 185 (30) | 400 (33) |
| Malay | — | — | 586 (32) | 194 (31) | 392 (32) |
| eGFR, mL/min/1.73 m2 | 98.5 (15.7) | 99.0 (15.2) | — | — | — |
| SomaScan log2(cystatin-C), RFU | — | — | 11.1 (11.0, 11.2) | 11.1 (10.9, 11.3) | 11.1 (11.0, 11.3) |
| Physical activity (sports index) | 2.48 (0.8) | 2.46 (0.8) | — | — | — |
| Leisure-time physical activity, MET-h/week | — | — | 644 (967) | 643 (950) | 645 (976) |
| Current smoking, n (%) | 1,320 (22) | 650 (22) | 244 (13) | 87 (14) | 157 (13) |
| Total cholesterol, mg/dL | 210 (38) | 208 (39) | 208 (36) | 211 (36) | 207 (36) |
| HDL-cholesterol, mg/dL | 51 (17) | 51 (17) | 50 (14) | 46 (12) | 51 (15) |
| Systolic BP, mmHg | 120.0 (18.3) | 120.0 (18.0) | 131.5 (21.7) | 136.9 (21.9) | 128.7 (21.0) |
| BP-lowering medication use | 1,396 (23) | 623 (21) | 250 (14) | 125 (20) | 125 (10) |
| BMI, kg/m2 | 27.3 (5.0) | 27.3 (5.0) | 25.6 (4.8) | 27.2 (5.0) | 24.8 (4.5) |
| HbA1c, % | 5.4 (0.4) | 5.4 (0.4) | 5.7 (0.4) | 5.9 (0.4) | 5.6 (0.4) |
| Fasting glucose, mg/dL | 101.3 (9.4) | 101.4 (9.3) | 89.1 (9.9) | 94.3 (10.8) | 86.8 (8.5) |
Categorical variables are presented as indicated, and continuous variables are presented as mean (SD) or as median (quartile 1, quartile 3). In MEC, the case subjects were diabetes-free when baseline characteristics were measured and in MEC, for fasting glucose and HbA1c, data were available only for 91.24% and 82.54% of participants, while the data completeness of all other variables was 100%. BP, blood pressure; RFU, relative florescence intensities.
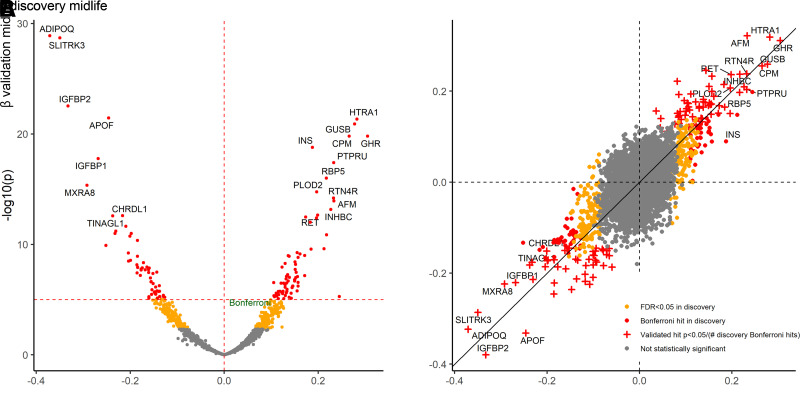
There were 596 proteins associated (Bonferroni P < 0.05/4,955) with an ∼20-year risk of diabetes in unadjusted analyses. There were 544 proteins associated with incident diabetes after adjustment for demographics and eGFR (model 1) and 312 proteins after additional adjustment for lifestyle and cardiometabolic risk factors (model 2). With further adjustment for BMI (model 3), 140 proteins were associated with an ∼20-year risk of diabetes with ADIPOQ, SLITRK3, IGFBP2, APOF, and HTRA1 as top proteins (Fig. 1A, Supplementary Table 1). After adjustment for baseline fasting glucose and HbA1c (model 4), there remained 53 statistically significant proteins (Supplementary Table 2).
Figure 1.
Volcano plots for ∼20-year risk of diabetes and scatterplot comparing discovery and validation: the ARIC study. Estimates adjusted for age, sex, race-center, eGFR, smoking status, physical activity, total cholesterol, HDL-cholesterol, systolic blood pressure, hypertension medication use, and BMI. Bonferroni-corrected P value in discovery based on P < 0.05/4,955.
After adjusting for the first 10 PCsprotein, the number of proteins associated with incident diabetes in model 3 was reduced (71 vs. 140 proteins). However, the top protein hits remained similar to our primary analysis without PCsproteins adjustment (Supplementary Table 3).
Results were generally consistent across age, sex, race, BMI, and HbA1c subgroups in discovery. There was a statistically significant P value for interaction (based on <2.0 × 10−6) for NTRK3 by race (P interaction = 1.9 × 10−6). In analyses stratified by race, NTRK3 was inversely associated with diabetes risk among White participants (hazard ratio [HR], 0.81; 95% CI 0.76, 0.87; P = 1 × 10−9) but not among Black participants (HR 1.03; 95% CI 0.91, 1.15; P = 0.68).
Internal and External Validation of Proteins Identified in Discovery
Participant characteristics in the internal validation sample (n = 2,913) were similar to the discovery sample (Table 1). In internal validation, there were 712 cases of diagnosed diabetes during a median of 19 years of follow-up. Of the 140 proteins identified in discovery (model 3), 64 proteins were also statistically significantly (P < 0.05/140) associated with incident diabetes in the internal validation sample, including 25 known (e.g., adiponectin [ADIPOQ], sex-hormone-binding globulin [SHBG], growth hormone receptor [GHR], aminoacylase-1 [ACY1]) and 39 novel hits (e.g., glutathione S-transferase A1 [GSTA1] and sodium/potassium-transporting ATPase subunit β-2 [ATP1B2]) (Supplementary Table 4). For the 64 proteins, the direction and magnitude of the effect estimates were similar across discovery and internal validation (Fig. 1B). NTRK3, the one protein with a statistical interaction identified in discovery, had a similar pattern in internal validation (P interaction = 0.03, P likelihood ratio test = 0.03; HRWhites 0.85 [95% CI 0.77, 0.93], HRBlacks 1.01 [0.88, 1.17]).
Using the 64 proteins that were internally validated in ARIC, we then externally replicated the proteins in the MEC. Characteristics of the MEC participants are provided in Table 1. There were 624 case subjects with incident diabetes and 1,214 matched control subjects (mean age 50 [SD 11] years, maximum follow-up 11 years). Of the 63 proteins available in the MEC, 47 (75%) were associated with incident diabetes (based on P < 0.05/63; in a sensitivity analysis, 60 proteins based on FDR q < 0.05) comprising 22 novel and 25 known diabetes-associated proteins (Supplementary Table 5). All 47 effect estimates for protein-diabetes associations were in the same direction in ARIC and MEC (Supplementary Fig. 1). The remaining 16 proteins that did not validate in MEC had effect estimates in the same direction as in ARIC.
Prediction
In the discovery elastic net regression models for 20-year diabetes risk, the protein-only model had a C statistic of 0.718 (95% CI 0.704, 0.732) (Table 2). Inclusion of the proteins selected by elastic net led to improved discrimination of incident diabetes beyond demographics and cardiometabolic factors (model 3: ΔC statistic 0.064; P < 10−300). The C statistic from the model with the 27 proteins selected by elastic net and model 4 covariates, including fasting glucose and HbA1c, was 0.770 (95% CI 0.758, 0.783). For comparison, the C statistic for a model adjusted for model 4 covariates and the 47 proteins that we internally validated was 0.763 (95% CI 0.750, 0.776) and led to improved discrimination beyond model 4 covariates (ΔC statistic 0.017, P < 2 × 10−8) in discovery.
Table 2.
Prediction of 20-year risk of diabetes in the discovery (n = 6,010 with 1,330 events) and internal validation (n = 2913 with 671 events) sample in ARIC
| Discovery | Proteins associated*with 20-year diabetes risk (n) | Proteins selected by elastic net** (n) | C statistic without any proteins | C statistic including proteins selected by elastic net model | ΔC statistic | P value for ΔC statistic | Validation | C statistic without any proteins | C statistic including proteins selected by discovery elastic net model | ΔC statistic | P value for ΔC statistic |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Model 0 | 601 | 26 | — | 0.718 (0.704, 0.732) | — | Model 0 | 0.691 (0.671, 0.711) | ||||
| Model 1 | 551 | 20 | 0.579 (0.563, 0.594) | 0.718 (0.705, 0.732) | 0.139 | <1E−300 | Model 1 | 0.592 (0.571, 0.613) | 0.694 (0.674, 0.714) | 0.102 | 7E−16 |
| Model 2 | 308 | 21 | 0.647 (0.633, 0.662) | 0.722 (0.708, 0.735) | 0.075 | <1E−300 | Model 2 | 0.639 (0.619, 0.660) | 0.695 (0.675, 0.715) | 0.055 | 2E−09 |
| Model 3 | 140 | 23 | 0.676 (0.662, 0.690) | 0.724 (0.711, 0.738) | 0.048 | <1E−300 | Model 3 | 0.663 (0.643, 0.683) | 0.694 (0.674, 0.714) | 0.031 | 6E−05 |
| Model 4 | 49 | 27 | 0.746 (0.733, 0.759) | 0.770 (0.758, 0.783) | 0.025 | 7E−13 | Model 4 | 0.720 (0.700, 0.740) | 0.731 (0.711, 0.750) | 0.011 | 0.04 |
Data are presented with the 95% CI. Model 0, unadjusted (protein only model). Model 1, age, sex, race-center, eGFR. Model 2, variables in model 1 plus physical activity, current smoking, total cholesterol, HDL-cholesterol, systolic blood pressure, and hypertension medication use. Model 3, variables in model 2 plus BMI. Model 4, variables in model 3 plus fasting glucose, HbA1c.
The number of proteins in the 20-year risk prediction models in Table 2 differ slightly from the results reported for ∼20-year risk (full follow-up) associations in the text (e.g., 49 proteins for 20-year risk in model 4 vs 53 proteins for ∼20-year risk in model 4) based on Bonferroni cutoff P value <1.01E−5.
Top 100 proteins considered in the elastic net models regardless of Bonferroni statistical significance (e.g., top 100 proteins considered in the elastic net regression along with the model 4 covariates when 49 proteins met threshold for statistical significance).
In the internal validation sample, the discrimination of the prediction model, including model 4 covariates plus the 27 proteins selected in discovery, was 0.731 (95% CI 0.711, 0.750), which improved discrimination beyond the model 4 covariates (ΔC statistic 0.011, P = 0.04) (Table 2). The discrimination in internal validation (C statistic 0.731) was lower than found in discovery (C statistic 0.770).
We provide the deciles of predicted versus observed risk in discovery and in validation in Supplementary Table 6 and visually inspected calibration plots (Supplementary Fig. 2). In discovery, as expected, calibration was excellent (Supplementary Fig. 2A). In internal validation, the protein-based model overestimated the observed diabetes risk across all deciles (Supplementary Fig. 2B). In the highest risk decile, the observed 20-year risk was 79% in discovery and 66% in internal validation (Supplementary Table 6).
Pathway Analysis
Pathway analysis identified acute-phase response signaling as the top biological pathway (Supplementary Table 7). Other inflammatory pathways, such as complement activation, STAT3 signaling, and interleukin 15 (IL15) production, were also among the top overrepresented biological pathways, as was lipid metabolism (liver X receptor/retinoid X receptor and farnesoid X receptor/retinoid X receptor activation). The proinflammatory cytokine IL17a was identified as the top upstream regulator of diabetes-associated proteins for a network of proteins associated with incident diabetes in an upstream regulator analysis (Supplementary Fig. 3 and Supplementary Table 8). Another top upstream regulator included tumor necrosis factor (Supplementary Fig. 3), which is a cytokine and master regulator of inflammatory signaling.
Genetic Instruments of Proteins with Diabetes
For the causal analyses, we considered the 50 proteins with genetic instrument models (the number of single nucleotide polymorphisms [SNPs] per protein models ranged from 16 to 170, genetic heritability from 1 to 37%) of the 64 proteins identified in model 3 discovery (140 “hits” based on P < 0.05/4,955) that were internally validated (P < 0.05/140 hits in model 3). The genetic instrument analyses supported causal roles for SHBG, ATP1B2, and GSTA1 in the development of diabetes (P < 0.05/50) (Table 3). The direction of the protein-diabetes association was consistent between the genetic instrument analyses and the untargeted analyses of diabetes proteomics for SHBG (inverse) and GSTA1 (positive), but the direction was opposite for ATP1B2 (positive in genetic instrument analyses vs. inverse in proteomic discovery and validation). Results for the remaining genetic instruments for proteins with diabetes risk are shown in Supplementary Table 9. For the three proteins with causal support, colocalization analyses provided further independent support of a causal link for GSTA1 (posterior probability H4 = 67.4%) and ATP1B2 (posterior probability H4 = 99.7%). Owing to the close proximity of SHBG and ATP1B2 on chromosome 17 (Supplementary Fig. 4), the potential SHBG signal in the diabetes GWAS is hidden by the stronger ATP1B2 signal, such that the posterior probability of H4 is only 3.6% for SHBG. When excluding the region corresponding to the ATP1B2 signal by visual inspection and restricting the colocalization analysis to the secondary peak (chromosome 17: 7,500,600–7,600,000) we do observe support for a shared causal variant (posterior probability H4 = 86%).
Table 3.
Causal support for selected proteins that validated internally and externally in association with diabetes risk
| Protein name | Gene | SNPs (n) | Heritability of the gene (%) | R2 for protein model (%) | Z statistic for genetic model association with diabetes risk | P value for the genetic model association with diabetes risk | ARIC Discovery SomaScan aptamer HR* (95% CI) | MEC SomaScan aptamer OR* (95% CI) |
|---|---|---|---|---|---|---|---|---|
| Sex hormone-binding globulin | SHBG | 100 | 6 | 7 | −5.32 | 1E−07 | 0.82 (0.76, 0.87) | 0.71 (0.63, 0.81) |
| Sodium/potassium-transporting ATPase subunit β-2 | ATP1B2 | 47 | 11 | 14 | 4.63 | 4E−06 | 0.83 (0.78, 0.88) | 0.78 (0.69, 0.87) |
| Glutathione S-transferase A1 | GSTA1 | 64 | 11 | 28 | 3.57 | 4E−04 | 1.14 (1.09, 1.19) | 1.37 (1.23, 1.52) |
Proteins with P < 0.05/50 or <0.001 shown here. Remaining proteins are provided in Supplementary Table 9. Of the 64 proteins considered, 50 had genetic models identified. OR, odds ratio.
Adjusted for age, sex, race (race-center in ARIC, ethnicity in MEC), eGFR (eGFR plus creatinine and cystatin C in ARIC, log2[SomaScan cystatin C] in MEC), physical activity (sport index in ARIC, leisure time METs per week in MEC), current smoking, total cholesterol, HDL-cholesterol, systolic blood pressure, hypertension medication use, BMI, and modeled per 1 SD of the log2(protein relative fluorescent units). The MEC case-control design used a risk-set sampling design such that the OR can approximate an HR.
Conclusions
In this large community-based study of Black and White adults, we identified numerous novel and established proteins associated with diabetes incidence in midlife by using an untargeted approach. The identified proteins were internally and externally validated. Furthermore, pathway analyses identified key biological mechanisms, including systemic inflammation and altered lipid metabolism, underlying the pathogenesis of diabetes. Three proteins were found to have a direct causal link to diabetes in genetic instrument analyses. Of particular clinical importance, we provide novel and known proteins that may help improve risk prediction for incident diabetes beyond current approaches for identifying individuals at high-risk for diabetes.
Comparisons with Other Proteomic Studies
Our study comprehensively assessed associations between 4,955 proteins and incident diabetes in >10,000 participants. Studies of the proteomics of diabetes have generally been smaller (∼880 to 3,000 participants), had ∼80 to 4,100 proteins quantified, and included predominantly White populations. Previous studies of proteomic alterations linked to incident diabetes identified proteins that related to inflammation and other metabolic processes related to glycemic regulation (Supplementary Table 10) (3–8). Of the 47 validated protein hits in this study, 25 were also identified by prior studies, including a recent 3-protein signature of isolated impaired glucose tolerance for diabetes risk (20). The known biological role in the regulation of glycemia of proteins, such as adiponectin, insulin-growth factor binding proteins, leptin, and insulin, support the potential importance of our novel hits. Interestingly, for adiponectin levels, our PWAS analysis did not find evidence for causality differing from a prior study (21). This may be due to our analysis conservatively focusing on only cis SNPs (i.e., ± 500 kb from the protein coding genes), whereas older MR analyses included trans SNPs. For adiponectin, trans SNPs include the SIAH2 (siah E3 ubiquitin protein ligase 2), which has an even stronger association with diabetes than adiponectin SNPs and several other genes (21). Our study expands on prior knowledge on the proteomic markers of diabetes with the identification of >20 novel proteins (Supplementary Table 4) that were rigorously associated with diabetes risk in 2 racially and ethnically diverse cohorts.
Mechanisms
Our analyses implicated inflammation as a top pathway leading to diabetes, consistent with existing understanding of diabetes pathogenesis. Inflammatory pathways have long been discussed as a mechanism contributing to diabetes risk arising from obesity (22). While inflammation could be a marker of obesity, in experimental models, proinflammatory cytokines produced by adipose tissue have been shown to cause insulin resistance (23). The cytokine, IL17a, was identified as a top upstream regulator in our pathway analysis. IL17 could be involved in insulin resistance development through adipose tissue disruption (24). IL17 has been shown to promote β-cell death in mouse islets (25), thereby adversely affecting insulin production. In mice with induced type 1 diabetes, IL17a promoted oxidative stress and apoptosis of β-cells (26), thereby leading to reductions in insulin secretion. Interestingly, there are pharmacological agents that can reduce IL17. Our results, if confirmed in other studies, suggest that such therapies might be investigated as repurposed therapies for prevention or management of diabetes.
Altered lipid metabolism was another key pathway identified in our pathway analysis. In the setting of obesity, excess lipids are stored in the liver and muscle rather than adipose tissue (27). Accumulation of lipids can lead to a proinflammatory state (28). Indeed, activation of liver X receptor/retinoid X receptor—one of our top pathways—has been shown to induce apoptosis of insulin-secreting cells (29). These prior laboratory data, in addition to our results, suggest inflammation and alterations in lipid metabolism may be important and perhaps intertwined processes underlying diabetes pathogenesis.
We found causal support for an inverse association between SHBG with diabetes risk. SHBG may play a role in diabetes through interaction of sex hormones with fat, muscle, and other peripheral tissues involved in glucose homeostasis. In our untargeted proteomic discovery analysis, the association of plasma SHBG with diabetes did not differ by sex (HRmen 0.72, HRwomen 0.77; P interaction > 0.05). Findings from mouse models indicate that monosaccharides, including glucose, slow SHBG transcriptional activity in the liver via downregulation of hepatocyte nuclear factor-4α (HNF-4α) (30). Our findings are in line with prior research implicating low SHBG as a risk factor for diabetes in observational (31,32), and MR (33,34) studies. Indeed, SHBG has been shown to decrease following weight gain (35). Our genetic instrument results model a life-long difference in SHBG levels and hence support a causal link for SHBG with diabetes.
Our genetic instrument results also supported a causal role for ATP1B2 and GSTA1. To our knowledge, links between ATP1B2 and GSTA1 with diabetes risk in humans are novel. With regard to ATP1B2, ATP and ADP have important role in intracellular respiration and could be posited to have a role in glucose trafficking (36). It is plausible that altered expression of ATP1B2 may reflect a state of systemic insulin resistance activity in the peripheral tissue (37). This notion is supported by rat models indicating alterations to ATPase sodium-potassium regulation within skeletal muscle in the presence of insulin-resistant states (high-fat diet, sedentary) (37). Mechanisms linking GSTA1 with diabetes are not well documented; however, laboratory-based studies suggest that GSTA1 and other glutathione S-transferase (GST) genes may play a role in inflammatory pathways. In vitro studies have suggested GSTA1 involvement in suppressing c-Jun N-terminal kinase–associated (inflammatory) cellular apoptosis (38). Tissue specimens from humans and mice have substantially lower expression of GSTA1 in pancreatic β-cells in the setting of type 2 diabetes (vs. cells exposed to nondiabetic conditions) (39). Future studies are needed to characterize these mechanisms in humans and determine whether these proteins may be modifiable via lifestyle or pharmacological intervention.
Clinical and Public Health Implications
We identified proteins that improved risk stratification for diabetes, with implications for clinical and public health practice. We used a machine learning approach to identify proteins associated with diabetes risk beyond demographics, BMI, fasting glucose, HbA1c, and other diabetes risk factors. Risk stratification for diabetes was improved when we added 27 top diabetes-associated proteins (e.g., SLITRK3, APOF, ADIPOQ, CPM, IGFBP1, RBP5, and RTN4R). Prior prediction models for diabetes risk have not included such an extensive list of proteins beyond common clinically used parameters. Importantly, these proteins improved prediction even beyond fasting glucose and HbA1c—biomarkers that are used to clinically define prediabetes and diabetes. Our findings suggest that novel proteins may help refine definitions of prediabetes and improve our ability to identify individuals at high risk for diabetes, eligible for targeted diabetes prevention strategies.
Strengths and Limitations
Strengths of this study include the rigorous assessment of the plasma proteome with incident diabetes, including internal validation, in a community-based population with Black and White adults, and external validation in a multiethnic Asian cohort. Other strengths include the long follow-up in ARIC, large sample size, and quantification of ∼5,000 proteins.
Limitations include, first, genetic models were created only among individuals of European ancestry due to the current paucity of summary GWAS data from genetically diverse populations. The genetic instruments for the proteins were also derived from GWAS data in ARIC. Analyses rely upon the currently available characterization of the genetic architecture for each protein, and statistical power can differ for proteins according to heritability, particularly when the heritability of the protein is low.
Second, we conservatively accounted for multiple comparisons using Bonferroni correction. We recognize that some proteins that did not meet the conservative Bonferroni threshold but met the FDR threshold may offer additional information on diabetes risk in future research.
Third, plasma samples underwent long-term storage; however, prior work in ARIC indicates excellent stability of proteins in ARIC samples (40).
Fourth, we used self-report to capture incident diabetes events in ARIC, but we previously showed in the ARIC Study that self-reported diabetes is highly specific (11).
Last, in ARIC, the type of diabetes was not known. However, given the age range of participants, it is likely that the vast majority of diabetes cases were type 2.
Conclusion
In conclusion, we identified 22 novel and 25 established proteins associated with incident diabetes after rigorous adjustment in two diverse cohorts. We identified a set of proteins that improved risk prediction for diabetes beyond traditional risk factors and the prevailing glycemia (assessed by both fasting glucose and HbA1c). In genetic analyses, we found causal evidence to support links of SHBG, ATP1B2, and GSTA1 with incident diabetes. Our findings suggest novel potential biological mechanisms, including some proteins (e.g., IL17a) that may be targetable by existing drugs, which may predict diabetes pathogenesis.
Article Information
Acknowledgments. The authors thank all ARIC Study and MEC participants, the study team, and investigators for their contributions to research.
Funding. The ARIC study has been funded in whole or in part with federal funds from the National Institutes of Health (NIH) National Heart, Lung, and Blood Institute (NHLBI), Department of Health and Human Services contract numbers 75N92022D00001, 75N92022D00002, 75N92022D00003, 75N92022D00004, 75N92022D00005, R01HL087641, and R01HL086694, National Human Genome Research Institute, contract U01HG004402, and NIH contract HHSN268200625226C. J.B.E.-T. was supported by NIH/National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant K23 HL153774. E.S. was supported by NIH/NHLBI grant K24 HL152440 and NIH/NIDDK grant R01 DK089174. O.T. is funded by NIH/NHLBI T32 HL007024. M.E.G. was supported by NIH/NHLBI K24 HL155861. This research was also supported by NIH/NIDDK R01 DK124399. The Singapore MEC study is supported by individual research and clinical scientist award schemes from the Singapore National Medical Research Council (NMRC), including MOH-000271-00, and the Singapore Biomedical Research Council (BMRC), the Singapore Ministry of Health (MOH), the National University of Singapore (NUS), and the Singapore National University Health System (NUHS). K.A.W. is funded by the National Institute on Aging’s Intramural Research Program. This study was funded, in part, by the National Institute on Aging’s Intramural Research Program.
Duality of Interest. Johns Hopkins University has signed a collaboration agreement with SomaLogic to conduct SomaScan of ARIC stored samples at no charge in exchange for the rights to analyze linked ARIC phenotype data. Jo.C. is a scientific advisor to SomaLogic. E.S. is a Deputy Editor at Diabetes Care. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. M.R.R. wrote the first draft of the manuscript. M.R.R. and Jo.C. were involved in the conception, design, conduct, and methodology of the study. Jing.C., K.A.W., P.S., Jiny.C., Y.L., and X.S. were involved in the analysis and interpretation of the results. J.B.E.-T., K.A.W., P.S., A.S., O.T., C.M.B., E.B., C.E.N., R.T.D., J.S.P., P.L.L., L.E.W., R.v.D., E.S.T., M.E.G., E.S., and Jo.C. edited and reviewed the manuscript. All authors approved the final version of the manuscript. M.R.R. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.21801163.
References
- 1. Ingelsson E, McCarthy MI. Human genetics of obesity and type 2 diabetes mellitus: past, present, and future. Circ Genom Precis Med 2018;11:e002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen ZZ, Gerszten RE. Metabolomics and proteomics in type 2 diabetes. Circ Res 2020;126:1613–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nowak C, Sundström J, Gustafsson S, et al. Protein biomarkers for insulin resistance and type 2 diabetes risk in two large community cohorts. Diabetes 2016;65:276–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gudmundsdottir V, Zaghlool SB, Emilsson V, et al. Circulating protein signatures and causal candidates for type 2 diabetes. Diabetes 2020;69:1843–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beijer K, Nowak C, Sundström J, Ärnlöv J, Fall T, Lind L. In search of causal pathways in diabetes: a study using proteomics and genotyping data from a cross-sectional study. Diabetologia 2019;62:1998–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Molvin J, Pareek M, Jujic A, et al. Using a targeted proteomics chip to explore pathophysiological pathways for incident diabetes–the Malmö Preventive Project. Sci Rep 2019;9:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elhadad MA, Jonasson C, Huth C, et al. Deciphering the plasma proteome of type 2 diabetes. Diabetes 2020;69:2766–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gou W, Yue L, Tang XY, et al. Circulating proteome and progression of type 2 diabetes. J Clin Endocrinol Metab 2022;107:1616–1625 [DOI] [PubMed] [Google Scholar]
- 9. Wright JD, Folsom AR, Coresh J, et al. The ARIC (Atherosclerosis Risk In Communities) Study: JACC Focus Seminar 3/8. J Am Coll Cardiol 2021;77:2939–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Candia J, Cheung F, Kotliarov Y, et al. Assessment of variability in the SOMAscan Assay. Sci Rep 2017;7:14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schneider AL, Pankow JS, Heiss G, Selvin E. Validity and reliability of self-reported diabetes in the Atherosclerosis Risk in Communities Study. Am J Epidemiol 2012;176:738–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang J, Brody EN, Murthy AC, et al. Impact of kidney function on the blood proteome and on protein cardiovascular risk biomarkers in patients with stable coronary heart disease. J Am Heart Assoc 2020;9:e016463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Williams SA, Kivimaki M, Langenberg C, et al. Plasma protein patterns as comprehensive indicators of health. Nat Med 2019;25:1851–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tan KHX, Tan LWL, Sim X, et al. Cohort profile: the Singapore Multi-Ethnic Cohort (MEC) study. Int J Epidemiol 2018;47:699–699j [DOI] [PubMed] [Google Scholar]
- 15. Krämer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014;30:523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang J, Dutta D, Köttgen A, et al.; CKDGen Consortium . Plasma proteome analyses in individuals of European and African ancestry identify cis-pQTLs and models for proteome-wide association studies. Nat Genet 2022;54:593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet 2018;50:1505–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu H, Zhou X. Transcriptome-wide association studies: a view from Mendelian randomization. Quant Biol 2021;9:107–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Giambartolomei C, Vukcevic D, Schadt EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 2014;10:e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carrasco-Zanini J, Pietzner M, Lindbohm JV, et al. Proteomic signatures for identification of impaired glucose tolerance. Nat Med 2022;28:2293–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nielsen MB, Çolak Y, Benn M, Nordestgaard BG. Low plasma adiponectin in risk of type 2 diabetes: observational analysis and one- and two-sample Mendelian randomization analyses in 756,219 individuals. Diabetes 2021;70:2694–2705 [DOI] [PubMed] [Google Scholar]
- 22. Duncan BB, Schmidt MI, Pankow JS, et al.; Atherosclerosis Risk in Communities Study . Low-grade systemic inflammation and the development of type 2 diabetes: the atherosclerosis risk in communities study. Diabetes 2003;52:1799–1805 [DOI] [PubMed] [Google Scholar]
- 23. Hotamisligil GS, Spiegelman BM. Tumor necrosis factor α: a key component of the obesity-diabetes link. Diabetes 1994;43:1271–1278 [DOI] [PubMed] [Google Scholar]
- 24. Abdel-Moneim A, Bakery HH, Allam G. The potential pathogenic role of IL-17/Th17 cells in both type 1 and type 2 diabetes mellitus. Biomed Pharmacother 2018;101:287–292 [DOI] [PubMed] [Google Scholar]
- 25. Catterall T, Fynch S, Kay TWH, Thomas HE, Sutherland APR. IL-17F induces inflammation, dysfunction and cell death in mouse islets. Sci Rep 2020;10:13077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qiu AW, Cao X, Zhang WW, Liu QH. IL-17A is involved in diabetic inflammatory pathogenesis by its receptor IL-17RA. Exp Biol Med (Maywood) 2021;246:57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shulman GI. Cellular mechanisms of insulin resistance. J Clin Invest 2000;106:171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lumeng CN, Saltiel AR. Inflammatory links between obesity and metabolic disease. J Clin Invest 2011;121:2111–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wente W, Brenner MB, Zitzer H, Gromada J, Efanov AM. Activation of liver X receptors and retinoid X receptors induces growth arrest and apoptosis in insulin-secreting cells. Endocrinology 2007;148:1843–1849 [DOI] [PubMed] [Google Scholar]
- 30. Selva DM, Hogeveen KN, Innis SM, Hammond GL. Monosaccharide-induced lipogenesis regulates the human hepatic sex hormone-binding globulin gene. J Clin Invest 2007;117:3979–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ding EL, Song Y, Manson JE, et al. Sex hormone-binding globulin and risk of type 2 diabetes in women and men. N Engl J Med 2009;361:1152–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ngo D, Benson MD, Long JZ, et al. Proteomic profiling reveals biomarkers and pathways in type 2 diabetes risk. JCI Insight 2021;6:e144392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yuan S, Wang L, Sun J, et al. Genetically predicted sex hormone levels and health outcomes: phenome-wide Mendelian randomization investigation. Int J Epidemiol 2022;51:1931–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perry JR, Weedon MN, Langenberg C, et al.; MAGIC . Genetic evidence that raised sex hormone binding globulin (SHBG) levels reduce the risk of type 2 diabetes. Hum Mol Genet 2010;19:535–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Singh P, Covassin N, Sert-Kuniyoshi FH, et al. Overfeeding-induced weight gain elicits decreases in sex hormone-binding globulin in healthy males—implications for body fat distribution. Physiol Rep 2021;9:e15127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Toyoda Y, Saitoh S. Adaptive regulation of glucose transport, glycolysis and respiration for cell proliferation. Biomol Concepts 2015;6:423–430 [DOI] [PubMed] [Google Scholar]
- 37. Galuska D, Kotova O, Barrès R, Chibalina D, Benziane B, Chibalin AV. Altered expression and insulin-induced trafficking of Na+-K+-ATPase in rat skeletal muscle: effects of high-fat diet and exercise. Am J Physiol Endocrinol Metab 2009;297:E38–E49 [DOI] [PubMed] [Google Scholar]
- 38. Romero L, Andrews K, Ng L, O’Rourke K, Maslen A, Kirby G. Human GSTA1-1 reduces c-Jun N-terminal kinase signalling and apoptosis in Caco-2 cells. Biochem J 2006;400:135–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Matsuoka TA, Kaneto H, Kawashima S, et al. Preserving Mafa expression in diabetic islet β-cells improves glycemic control in vivo. J Biol Chem 2015;290:7647–7657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tin A, Yu B, Ma J, et al. Reproducibility and variability of protein analytes measured using a multiplexed modified aptamer assay. J Appl Lab Med 2019;4:30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]