Abbreviations
- ADP
adenosine diphosphate
- ARG1
arginase 1
- ASL
argininosuccinate lyase
- ASS1
argininosuccinate synthetase 1
- BIC
bicarbonate phosphorylation domain of CPS1
- CMP
cytidine monophosphate
- CPS1
carbamoyl‐phosphate synthetase 1
- CPS1B
CPS1Z, fibroblast cells from CPS1‐deficient patients
- GIC50
half‐maximal growth inhibition
- IC50
half maximal inhibitory concentration
- ID
integrating domain of CPS1
- iPSCs
induced pluripotent stem cells
- IR
ionizing radiation
- KRAS
Kirsten rat sarcoma virus proto‐oncogene
- LKB1
liver kinase B1
- NAG
N‐acetylglutamate
- NAGS
N‐acetylglutamate synthase
- NSCLC
non‐small cell lung carcinoma
- ORNT1
ornithine transporter 1
- OTC
ornithine transcarbamylase
- PDB
Protein Data Bank
- siRNA
small interfering RNA
- UDP
uridine diphosphate
- UMP
uridine monophosphate
Dear Editor,
Metabolically induced cancer heterogeneity creates a large source of novel potential targets towards an enhanced therapeutic window alone and in combination with classic chemo‐ and radiotherapy [1, 2]. This is of particular interest for non‐small cell lung cancer (NSCLC), which accounts for more than 80% of all lung tumor types characterized by limited responses to current treatment options [3, 4, 5]. Genetically‐defined KRAS/LKB1‐mutant NSCLC tumors exploit the proximal urea cycle enzyme carbamoyl phosphate synthetase 1 (CPS1) as an intermediate step for pyrimidine biosynthesis, thereby contrasting its crucial hepatic role in ammonia detoxification [6, 7, 8]. Thus, CPS1 could serve as a novel target for NSCLC susceptibility (Figure 1A). In this study, we investigated the metabolic changes observed in both NSCLC and healthy hepatic systems following the identification, characterization and application of a small molecule inhibitor of ectopic CPS1 functionality.
FIGURE 1.
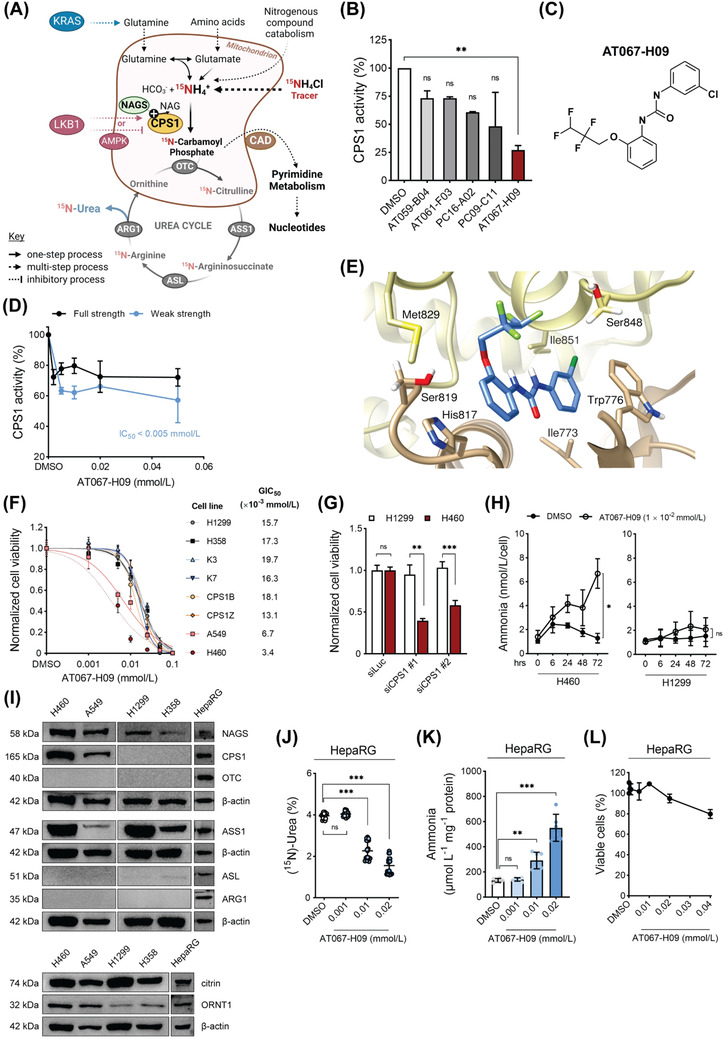
AT067‐H09 is a novel CPS1 inhibitor for KRAS/LKB1‐mutant NSCLC tumors. (A) Scheme of the metabolic utilization of the urea cycle and CPS1 towards pyrimidine biosynthesis in KRAS/LKB1‐mutant NSCLCs. Absence of at least two urea cycle enzymes abrogates (15N)‐Urea production and tracing. (B) CPS1 activity towards citrulline formation was determined using recombinant human CPS1 (rhCPS1) in the presence of 0.08 mg/mL of five out of more than 11,000 compounds. These five compounds were identified as potential CPS1 inhibitory ligands using a thermal stability assay‐based high‐throughput screening approach (Supplementary Figure S1). (C) Chemical structure of AT067‐H09. (D) Effect of AT067‐H09 on rhCPS1 activity, monitored continuously as ADP production, in saturating (“full strength”) and in 20% strength (“weak strength”) assay solution. Data points are means ±SD of two independent experiments in duplicates normalized to DMSO control. (E) AT067‐H09 predicted docking site. AT067‐H09 (sticks, cornflower blue) is localized in an allosteric pocket between the integrating (ID; yellow chain) and bicarbonate phosphorylation (BIC; tan chain) domains of CPS1 (PDB file, 6UEL), with its perfluorinated chain (green atoms) facing outwards from the protein core. Important amino acid residues that facilitate stabilizing interactions (hydrophobic and stacking) with AT067‐H09 are labeled. (F) Cell viability of NSCLC cell lines, which either express CPS1 (H460, A549) or lack CPS1‐expression (H1299, H358), of fibroblasts from healthy donors (K3, K7) and CPS1‐deficient patients (CPS1B, CPS1Z), all treated with increasing concentrations of AT067‐H09. GIC50 values are indicated. (G) Cell viability of H1299 and H460 cells 48 h after transfection with CPS1‐silencing siRNAs or control luciferase siRNA. (H) Ammonia accumulation in supernatants of H460 and H1299 cells upon treatment with 1 × 10−2 mmol/L AT067‐H09. (I) Western blotting of all urea cycle enzymes, grouped in those located intra‐mitochondrially (upper panel), namely NAGS, CPS1, and OTC, those located in the cytosol (middle panel), namely ASS1, ASL and ARG1, and the transporters citrin and ORNT1 (lower panel) in the studied NSCLC cell lines. Comparative immunoblots of urea cycle‐capable HepaRG cells are included. (J) Isotopically labeled urea (%) as a measure of urea cycle flux in HepaRG cells treated with AT067‐H09. 15NH4Cl (1 mmol/L) was used as the stable isotope tracer. (K) Ammonia accumulation in HepaRG cells when treated with 1 × 10−2 mmol/L AT067‐H09 for 48 h. (L) HepaRG cell viability with increasing AT067‐H09 concentrations. For all experiments, significance was obtained with one‐way ANOVA applying Dunnett's or Holm‐Sidak's multiple comparisons test, Kruskal‐Wallis test followed by Dunn's multiple comparisons test or unpaired t‐test. *P < 0.05. **P < 0.01, ***P < 0.001. ns = non‐significant. For all the panels, results are means ±SD of three or four independent experiments in triplicates unless otherwise indicated. Where shown, dose‐response curves were obtained by hyperbolic non‐linear regressions. Abbreviations: ADP, adenosine diphosphate; AMPK, adenosine monophosphate‐activated protein kinase; ARG1, arginase 1; ASL, argininosuccinate lyase; ASS1, argininosuccinate synthetase 1; BIC, bicarbonate phosphorylation domain of CPS1; CAD, protein complex comprised by carbamoyl‐phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase; CPS1, carbamoyl‐phosphate synthetase 1; CPS1B, CPS1Z, fibroblast cells from CPS1‐deficient patients; DMSO, dimethylsulfoxide; ID, integrating domain of CPS1; KRAS, Kirsten rat sarcoma virus proto‐oncogene; LKB1, liver kinase B1; NAGS, N‐acetylglutamate synthase; NSCLC, non‐small cell lung carcinoma; ORNT1; ornithine transporter 1; OTC, ornithine transcarbamylase; PDB, Protein Data Bank; rhCPS1, recombinant human CPS1 protein; siCPS1; CPS1‐targeting siRNA; siLuc, luciferase‐targeting siRNA; siRNA, small interfering RNA
Identification of AT067‐H09 as CPS1‐inhibitor
To search for CPS1 ligands, we performed a high‐throughput screening approach as part of a thermal stability assay with recombinant human CPS1 and identified 5 out of more than 11,000 compounds as potential CPS1 inhibitory ligands (Figure 1B, Supplementary Figure S1 and Supplementary Table S1; see also Supplementary Information for all assays). The compound 1‐(3‐chlorophenyl)‐3‐(2‐[2,2,3,3‐tetrafluoropropoxy]phenyl)urea (herein referred to as AT067‐H09, Figure 1C) exhibited the highest inhibitory effect on recombinant CPS1 activity towards citrulline synthesis and an IC50 below 5 × 10−3 mmol/L in the CPS1 activity assay (with full and weak strength concentrations of relevant cofactors) at which adenosine diphosphate (ADP) production was monitored at increasing concentrations of this compound (Figure 1D).
Molecular docking of AT067‐H09
Here, we aimed to predict the binding site for AT067‐H09 on the crystal structure of CPS1 (apo form) reported thus far at the highest resolution (PDB file: 6UEL). SwissDock identified 39 clusters with 256 docked conformers in this apo form of CPS1 and confidently positioned AT067‐H09 within the same pocket of the previously identified CPS1 inhibitor H3B‐120 [9] (Supplementary Table S2). This binding pocket was located between the integrating and bicarbonate phosphorylation domains of CPS1, where hydrophobic interactions of AT067‐H09 with residues Trp776, Leu778, His817 and Ile851 occurred, while its highly polar perfluorinated chain faced outwards, away from the protein backbone (Figure 1E). The propoxyphenyl and chlorophenyl groups of AT067‐H09 facilitate stacking interactions with amino acids His817 and Ile851 of the apoenzyme, further stabilizing the protein‐ligand conformation. The proximity of this binding pocket to the bicarbonate phosphorylation domain of CPS1 may concur with CPS1‐inhibition exhibiting a low IC50 value.
AT067‐H09 specific effects on KRAS/LKB1 mutant NSCLC cells
To validate metabolic rewiring and enhanced dependence of KRAS/LKB1‐mutant NSCLC tumors on CPS1, cell viability was probed in several NSCLC cell lines, which either expressed CPS1 (H460, A549, based on a perturbed KRAS/LKB1 genetic background) or lacked CPS1‐expression (H1299, H358) (Supplementary Table S3). H460 and A549 cells exhibited a strong concentration‐dependent reduction in cell viability with half‐maximal growth inhibition (GIC50) of 3.4 × 10−3 mmol/L and 6.7 × 10−3 mmol/L, respectively (Figure 1F), which was significantly lower than the GIC50 for H1299 wildtype and H358 KRAS mutant‐only NSCLC cells. Likewise, primary fibroblasts from CPS1‐deficient (CPS1B/Z, CPS1mut) or healthy individuals (K3/7, CPS1wt) exhibited considerably increased resistance to AT067‐H09 treatment. Furthermore, siRNA‐mediated CPS1 knockdown reproduced this effect in KRAS/LKB1‐mutant NSCLC cells but not on siRNA transfection of H1299 cells, also supporting CPS1‐driven tropism of KRAS/LKB1‐mutated tumor cells for AT067‐H09 (Figure 1G). Combination treatment with ionizing radiation (IR) was examined; however, no relevant synergism was observed for the CPS1‐expressing H460 cells (Supplementary Figure S2). Of note, the ammonia concentration escalated over time in the supernatant of the CPS1‐expressing H460 cells on incubation with 1 × 10−2 mmol/L AT067‐H09, while AT067‐H09 had little if any effect on ammonia content measured in the culture medium derived from H1299 cells (Figure 1H). In agreement with this, transfection of both cell types with siRNAs targeting CPS1 resulted in significantly increased ammonia levels only in H460 cells (Supplementary Figure S3), suggesting that CPS1 is an important element for ammonia homeostasis in H460 cells. A mechanism supporting CPS1 involvement in ammonia utilization in KRAS/LKB1‐mutant NSCLCs may be the incorporation of CPS1‐derived carbamoyl phosphate into the pyrimidine biosynthetic pathway [6]. In line, metabolic profiling on AT067‐H09‐treated H460 cells identified several pyrimidine pathway intermediates that were significantly downregulated on cellular incubation with AT067‐H09, including (S)‐dihydroorotate, uridine monophosphate (UMP), uridine‐5’‐diphosphate (UDP) and cytidine‐5’‐monophosphate (CMP) (Supplementary Figure S4).
Lack of complete urea cycle in KRAS/LKB1‐mutant NSCLCs
A previous study suggested the complete absence of the urea cycle in various NSCLC subtypes independent of the KRAS/LKB1‐mutant status [6]. We characterized the expression levels of all urea cycle enzymes and transporters in our cultured NSCLCs. Ornithine transcarbamylase (OTC) and arginase 1 (ARG1) were consistently undetectable in the four lung carcinoma cell lines, with argininosuccinate lyase (ASL) being expressed in very low abundance (Figure 1I). Surprisingly, N‐acetylglutamate synthase (NAGS) was strongly expressed in all cultured NSCLCs. The expression level of this enzyme has not been previously studied in lung carcinomas, but it is highly relevant for CPS1 ectopic activation since it provides the essential CPS1 allosteric cofactor NAG. The two transporters, ornithine transporter 1 (ORNT1) and citrin, were consistently expressed in all NSCLCs tested, with higher abundance in KRAS/LKB1‐mutant cells (Figure 1I). Lastly, argininosuccinate synthetase 1 (ASS1) was the most abundant urea cycle protein noted in H460 and H1299 cell lines, also supported by previous arginine and citrulline depletion studies [6]. Collectively, these results corroborate previous indications that a functional urea cycle as naturally occurring in hepatic tissue is not present in NSCLC.
AT067‐H09 specific effects on metabolically active hepatic cells
To investigate the potential impact of CPS1 inhibition on native hepatic metabolism and towards a therapeutic window, ureagenesis was investigated in differentiated HepaRG cells that faithfully model metabolic processes found in human hepatocytes [10]. When exposed to 1 × 10−2 mmol/L AT067‐H09, differentiated HepaRG cells showed impaired stable isotope incorporation reflected on the significantly reduced (15N)‐Urea enrichment value (Figure 1J) and a dose‐dependent increase in ammonia accumulation compared to vehicle‐treated cells (Figure 1K). Importantly, HepaRG viability was unaltered for AT067‐H09 concentrations up to at least 2 × 10−2 mmol/L, suggesting that the observed metabolic changes are not a toxicity‐related outcome (Figure 1L). Likewise, human control iPSCs, which were differentiated into hepatic‐like cells and treated with increasing concentrations of AT067‐H09, well‐tolerated this compound up to concentrations of at least 1 × 10−2 mmol/L (Supplementary Figure S5). No significant alterations in mitochondrial membrane potential were detectable in control experiments with HepG2 cells exposed to the same AT067‐H09 concentration range (Supplementary Figure S6). These findings indicate that liver metabolism and specifically the urea cycle were sensitive to transient pharmacological cues imposed by AT067‐H09 but that the viability of hepatocytes was less affected by AT067‐H09 and that hepatocytes with functionally active CPS1 tolerated AT067‐H09 at higher concentrations than KRAS/LKB1‐mutant NSCLC tumor cells.
In conclusion, compound AT067‐H09 could inhibit CPS1 activity and exert robust antiproliferative effects in CPS1‐expressing NSCLC cells. AT067‐H09 led to ammonia accumulation in CPS1‐expressing KRAS/LKB1‐mutated tumor cells and untransformed hepatocyte‐like normal cells without causing intolerable urea cycle flux impairment in hepatocytes. Metabolic interference via inhibition of CPS1 in tumor cells demonstrates enhanced cytotoxicity compared to untransformed cells, which might indicate differential oncogenic signaling or additionally affected downstream metabolic pathways in these targeted tumor cells. In this regard, our investigation is of great relevance for common malignant tumors caused by concurrent mutations in KRAS and LKB1, including NSCLC, where AT067‐H09 can act as a novel therapeutic approach, alone or in combination with state‐of‐the‐art treatment modalities, namely the immune checkpoint inhibitors. Future experiments focusing on preclinical testing could further explore the in vivo efficacy of AT067‐H09 toward CPS1‐dependent tumor sensitization.
DECLARATIONS
AUTHOR CONTRIBUTIONS
Georgios Makris, Carmen Diez‐Fernandez, Martin Pruschy and Johannes Häberle designed the study. Georgios Makris, Semih Kayhan, Marvin Kreuzer, Véronique Rüfenacht, Erica Faccin, Jarl Underhaug, Carmen Diez‐Fernandez, Philip A. Knobel, Martin Poms and Nadine Gougeard performed experiments and analyzed data. Georgios Makris, Vicente Rubio, Aurora Martinez, Martin Pruschy and Johannes Häberle analyzed data and revised the manuscript. Georgios Makris, Martin Pruschy and Johannes Häberle wrote the manuscript. All authors read and approved the final manuscript.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
FUNDING
This work was supported by the Swiss National Science Foundation, Switzerland (grant 320030_176088 to Johannes Häberle) and the Wolfermann‐Nägeli‐Stiftung, Switzerland (grant 2020/28 to Martin Pruschy). The high‐throughput screening was supported by the Research Council of Norway (NOR‐OPENSCREEN 245922/F50). The part of the work done by Vicente Rubio and Nadine Gougeard was supported by The Fundación Ramón Areces (grant CIVP20A6610). The work was also supported by a grant from European Union's Framework Program for Research and Innovation Horizon 2020 (2014‐2020) under Marie Skłodowska‐Curie (Grant Agreements No. 860245 (ITN THERADNET) to Marvin Kreuzer and Martin Pruschy.
AVAILABILITY OF DATA AND MATERIALS
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study protocol was approved by the Swiss Ethics Committee (permit number: BASEC‐No. 2021‐02314). The patients’ skin fibroblasts were obtained with the written informed consent of the respective individuals or their guardians.
CONSENT FOR PUBLICATION
Consent for publication was obtained from Swissethics (BASEC‐No. 2021‐02314).
Supporting information
Supporting Information
ACKNOWLEDGMENTS
We acknowledge the Functional Genomics Center Zurich (FGCZ) facility for the support with the metabolomic analysis and the Biophysics, Structural Biology and Screening (BiSS) core facility, University of Bergen, for the support with the high‐throughput screening.
REFERENCES
- 1. Schmidt DR, Patel R, Kirsch DG, Lewis CA, Vander Heiden MG, Locasale JW. Metabolomics in cancer research and emerging applications in clinical oncology. CA Cancer J Clin. 2021;71(4):333‐58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim J, DeBerardinis RJ. Mechanisms and Implications of Metabolic Heterogeneity in Cancer. Cell Metab. 2019;30(3):434‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Memmott RM, Wolfe AR, Carbone DP, Williams TM. Predictors of Response, Progression‐Free Survival, and Overall Survival in Patients With Lung Cancer Treated With Immune Checkpoint Inhibitors. J Thorac Oncol. 2021;16(7):1086‐98. [DOI] [PubMed] [Google Scholar]
- 4. Walsh RJ, Soo RA. Resistance to immune checkpoint inhibitors in non‐small cell lung cancer: biomarkers and therapeutic strategies. Ther Adv Med Oncol. 2020;12:1758835920937902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co‐occurring genomic alterations define major subsets of KRAS‐mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5(8):860‐77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim J, Hu Z, Cai L, Li K, Choi E, Faubert B, et al. CPS1 maintains pyrimidine pools and DNA synthesis in KRAS/LKB1‐mutant lung cancer cells. Nature. 2017;546(7656):168‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pierson DL, Brien JM. Human carbamylphosphate synthetase I. Stabilization, purification, and partial characterization of the enzyme from human liver. J Biol Chem. 1980;255(16):7891‐5. [PubMed] [Google Scholar]
- 8. Ratner S. Enzymes of arginine and urea synthesis. In: Grisolia S, Báguena R, Mayor F, editors. The Urea Cycle. New York: John Wiley and Sons; 1976. p. 181‐219. [Google Scholar]
- 9. Yao S, Nguyen TV, Rolfe A, Agrawal AA, Ke J, Peng S, et al. Small Molecule Inhibition of CPS1 Activity through an Allosteric Pocket. Cell Chem Biol. 2020;27(3):259‐68 e5. [DOI] [PubMed] [Google Scholar]
- 10. Guillouzo A, Corlu A, Aninat C, Glaise D, Morel F, Guguen‐Guillouzo C. The human hepatoma HepaRG cells: a highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem Biol Interact. 2007;168(1):66‐73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its supplementary information files).


