Human papillomavirus (HPV)–associated cancers of the anus, cervix, oropharynx, penis, vagina, and vulva constitute 5% of all cancers worldwide. In the United States, human papillomavirus–associated oropharyngeal squamous cell carcinoma (HPV+OPSCC) has been rapidly increasing in incidence, surpassing cervical cancer as the most common HPV-associated malignancy, with an estimated annual increase of 5% in White men aged 55 to 64 years from 1992 to 2015.1 By the year 2030, HPV+OPSCC will account for more than 30,000 new cancer diagnoses per year.1
Clinical practice guidelines from the College of American Pathologists currently recommend p16 immunohistochemistry as the diagnostic of choice for HPV+OPSCC because of its widespread availability, reproducibility of interpretation, low cost, and high correlation with high-risk human papillomavirus (HR-HPV) infection. Although p16 overexpression is a safe, effective, and widely used surrogate marker for HR-HPV in oropharyngeal squamous cell carcinoma (OPSCC), it suffers from a number of limitations. First, HPV+OPSCC classically presents with large cystic nodal metastases and small primary tumors; thus, the most common diagnostic approach is fine-needle aspiration (FNA) of a neck lymph node. FNA has intrinsic failure rates due to inadequate cellular material for evaluation in the range of 20% to 30% for the diagnosis of HPV+OPSCC.2–6 Even at high-volume institutions with dedicated head and neck radiologists and surgeons performing ultrasound-guided FNA and experienced head and neck cytopathologists, repeat biopsy is necessary in ~15% of cases (unpublished institutional data). Furthermore, the interpretation of p16 on FNA specimens lacks consensus guidelines and is more variable than tissue interpretation, and this has resulted in a lack of standardization and decreased sensitivity.2–6 Repeat FNA or subsequent tissue biopsy is often required to confirm a diagnosis of HPV+OPSCC, and this leads to increased costs, delays in diagnosis, patients being subjected to multiple invasive procedures, and increased diagnostic uncertainty. Second, although p16 is a relatively sensitive and specific surrogate marker for HR-HPV when used on tissue biopsies from oropharynx tumors in the United States, its performance and utility in other clinical settings are less clear. The most widely cited performance metrics for p16 are based on US studies examining staining patterns in comparison with direct HPV testing (DNA polymerase chain reaction (PCR) or RNA in situ hybridization/reverse transcription-PCR in OPSCC, where HPV infection rates range from 60% to 80%. However, HPV is significantly less prevalent in other parts of the world. When the HPV-associated fraction of OPSCC drops, p16 specificity decreases as well, and this limits the utility of p16 for much of the world. Similarly, HPV is known to be an oncogenic driver in additional subsites of the head and neck. For example, a subset of nasopharyngeal carcinomas are HPV-associated, as are up to approximately one-quarter of sinonasal squamous cell carcinomas.7 The utility of p16 as a surrogate marker for HPV at these sites appears to be much less reliable, likely for similar reasons. Lastly, widespread adoption of p16 as a surrogate marker for HPV has indirectly led to stagnation in the field of OPSCC predictive and prognostic biomarkers. p16 fails to provide information necessary for better understanding important relationships between the virus and its host. For example, a growing body of literature suggests that specifics of the viral genome directly affect tumor behavior at the genotype, sublineage, and single-nucleotide polymorphism levels in cervical cancer.8,9 Despite being the most common HPV-associated malignancy, HPV+OPSCC lags significantly behind cervical cancer in our understanding of even the most basic questions regarding host-viral interactions, such as the relationship between genotype and clinical outcomes. This information is unlikely to come to light in well-conducted, large, multi-institutional studies until direct HPV testing is routinely performed in clinical practice.
Tissue and cytology liquid-based direct HPV tests have improved, and RNA in situ hybridization for HR-HPV is an increasingly clinically available test for tissue specimens and cytology cell block preparations. These approaches are effective as cocktails and highly reliable for transcriptionally active HR-HPV. However, most do not provide individualized genotyping and, furthermore, require tumor tissue, an adequate cell block, and/or an aspirate specimen with liquid for testing, which may necessitate multiple procedures to acquire successfully.
Blood-based molecular diagnostics have emerged as diagnostic tools applicable to a broad spectrum of tumor types from early diagnosis to treatment monitoring and the detection of recurrent disease. Most liquid biopsy approaches focus on the detection of somatic mutations in cell-free DNA, which is released by cancer cells, or methylation. Many cell-free DNA cancer diagnostics, hindered by less than ideal sensitivity, remain in the research setting for now, although significant progress continues to be made. Virally driven tumors also release viral DNA fragments into the blood with the advantage of being more easily detected. The most robust literature comes from Epstein-Barr virus–associated nasopharyngeal carcinoma, with studies showing that circulating tumor Epstein-Barr virus DNA is detectable in >95% of patients at the time of diagnosis and that levels correlate with disease stage, the risk of recurrence after chemoradiotherapy, and overall survival.10–16 More recently, using quantitative (q) PCR and droplet digital polymerase chain reaction (ddPCR)–based approaches, a number of groups have demonstrated that most patients with HPV+OPSCC have detectable circulating tumor human papillomavirus DNA (ctHPVDNA) at presentation. A recent meta-analysis of ctHPVDNA showed an overall sensitivity of 0.81 and an overall specificity of 0.98 at the time of presentation.17 When studies were restricted to those performing ddPCR on plasma, which has emerged as the preferred methodology at many centers, including our own, the pooled sensitivity increased to 0.92. Our own experience suggests that sensitivity can be even higher with the incorporation of the addition of probes to capture less common genotypes and with an increased input quantity of cell-free DNA.
The potential of ctHPVDNA as a diagnostic tool is significant. ctHPVDNA is noninvasive and can provide direct HPV testing and genotyping. Early performance metrics of ctHPVDNA compare favorably with existing standard-of-care approaches mentioned previously, although these have not yet been compared head to head in a prospective, well-controlled fashion. Similarly, cost and time to diagnosis may favor ctHPVDNA, but this will need to be prospectively evaluated. ctHPVDNA has the added advantage of being not only a diagnostic test but also a tool for longitudinal monitoring of treatment responses and recurrence. Chera et al18 and others have demonstrated that ctHPVDNA levels decrease during chemoradiotherapy treatment and that those with early ctHPVDNA clearance have an improved prognosis. Furthermore, ctHPVDNA has been shown to detect recurrences earlier than traditional monitoring techniques such as cross-sectional imaging19 (Fig. 1). The potential for an integrated diagnostic and monitoring platform carries significant appeal.
Figure 1.
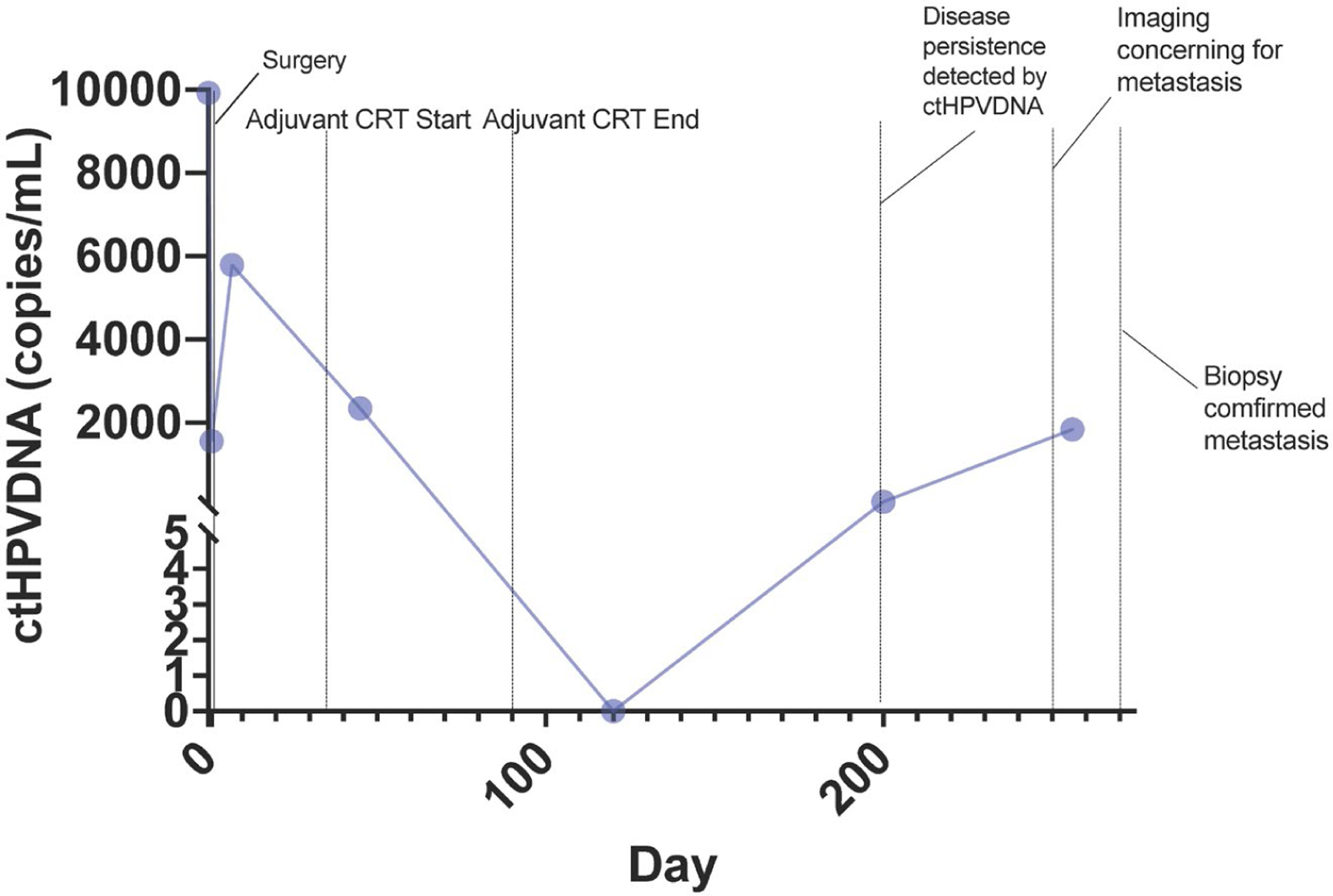
Longitudinal ctHPVDNA for a single patient treated with surgery followed by adjuvant chemoradiotherapy with early detection of distant metastatic disease on postoperative day 200. Routine imaging revealed lung nodules on day 250 with biopsy and pathology confirmation of metastatic HPV+OPSCC on day 260, 2 months later than ctHPVDNA detection.
ddPCR-based approaches to ctHPVDNA, however, have several limitations. Sample quality and quantity can be limiting factors. Processing blood for cell-free DNA detection requires specific protocols and can be disrupted by a number of easily made mistakes. Furthermore, probe multiplexing is limited, and this restricts the number of genotypes that can be tested at one time. Although most HPV+OPSCC cases are caused by HPV genotype 16 (followed by HPV-35 and HPV-33), a small percentage of cases are related to additional rare genotypes that would be missed with existing approaches if used alone. Furthermore, HPV-associated cancers outside the oropharynx have an even more diverse array of genotypes. Perhaps most significantly, a standalone blood-based diagnostic test would need to overcome traditional viewpoints of what constitutes proof of a cancer, which in head and neck oncology to date has equated to a tissue-based diagnosis.
In June 2021, Sastre-Garau et al20 published findings from a multi-national, prospective study evaluating a next-generation sequencing (NGS)–based HPV liquid biopsy called CaptHPV. CaptHPV uses probes designed for more than 200 HPV genotypes and variants followed by NGS library preparation and sequencing. Using this approach, they demonstrated 95% sensitivity and 98% specificity across an array of HPV-associated cancers. Although the sample size of HPV+OPSCC cases in this study was small (only 3 HPV+OPSCC cases were included in the cohort of 81 HPV+ cancers overall), the proof of principle remains. NGS-based liquid biopsies have the ability to overcome some of the limitations of ddPCR, such as the number of genotypes that can be detected. Furthermore, the flexibility of NGS to capture and interrogate other features of interest is superior. For example, Sastre-Garau et al also reported the integration status and captured significant portions of the viral genome, which could be used for subtyping and viral single-nucleotide polymorphism interrogation. NGS-based ctHPVDNA detection, however, carries with it a slew of challenges, including higher cost, more complex informatics, and higher minimum sample input quantities, among others.
Although much work remains to be done to rigorously assess ctHPVDNA as a standalone diagnostic test for HPV+OPSCC or as an adjunct to standard-of-care cytology or tissue-based diagnosis, the potential of a noninvasive, sensitive, specific, cost-effective, rapid, and longitudinally integrated diagnostic and monitoring blood-based test is apparent. Regardless of the platform used, the future of blood-based molecular diagnostics for HPV-associated cancers is bright, and the increasing incidence of HPV+OPSCC only makes the need even greater.
FUNDING SUPPORT
No specific funding was disclosed.
Footnotes
CONFLICT OF INTEREST DISCLOSURES
The author made no disclosures.
REFERENCES
- 1.Tota JE, Best AF, Zumsteg ZS, et al. Evolution of the oropharynx cancer epidemic in the United States: moderation of increasing incidence in younger individuals and shift in the burden to older individuals. J Clin Oncol 2019;37:1538–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rollo F, Dona MG, Pellini R, et al. Cytology and direct human papillomavirus testing on fine needle aspirates from cervical lymph node metastases of patients with oropharyngeal squamous cell carcinoma or occult primary. Cytopathology 2018;29:449–454. [DOI] [PubMed] [Google Scholar]
- 3.Wong KS, Krane JF, Jo VY. Heterogeneity of p16 immunohistochemistry and increased sensitivity of RNA in situ hybridization in cytology specimens of HPV-related head and neck squamous cell carcinoma. Cancer Cytopathol 2019;127:632–642. [DOI] [PubMed] [Google Scholar]
- 4.Xu B, Ghossein R, Lane J, et al. The utility of p16 immunostaining in fine needle aspiration in p16-positive head and neck squamous cell carcinoma. Hum Pathol 2016;54:193–200. [DOI] [PubMed] [Google Scholar]
- 5.Yang Z, Gomez-Fernandez C, Lora Gonzalez M, et al. HPV testing through p16 immunocytochemistry in neck-mass FNA and its correlation with tissue samples. Cancer Cytopathol 2019;127: 458–464. [DOI] [PubMed] [Google Scholar]
- 6.Conrad R, Yang SE, Chang S, et al. Comparison of cytopathologist-performed ultrasound-guided fine-needle aspiration with cytopathologist-performed palpation-guided fine-needle aspiration: a single institutional experience. Arch Pathol Lab Med 2018;142:1260–1267. [DOI] [PubMed] [Google Scholar]
- 7.Chang Sing Pang KJW, Mur T, Collins L, Rao SR, Faden DL. Human papillomavirus in sinonasal squamous cell carcinoma: a systematic review and meta-analysis. Cancers (Basel) 2020;13:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mirabello L, Yeager M, Cullen M, et al. HPV16 sublineage associations with histology-specific cancer risk using HPV whole-genome sequences in 3200 women. J Natl Cancer Inst 2016;108:djw100. [DOI] [PMC free article] [PubMed]
- 9.Mirabello L, Yeager M, Yu K, et al. HPV16 E7 genetic conservation is critical to carcinogenesis. Cell 2017;170:1164–1174.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hui EP, Ma BB, Chan KC, et al. Clinical utility of plasma Epstein-Barr virus DNA and ERCC1 single nucleotide polymorphism in nasopharyngeal carcinoma. Cancer 2015;121:2720–2729. [DOI] [PubMed] [Google Scholar]
- 11.Lo YM, Chan LY, Lo KW, et al. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res 1999;59:1188–1191. [PubMed] [Google Scholar]
- 12.Leung SF, Zee B, Ma BB, et al. Plasma Epstein-Barr viral deoxyribonucleic acid quantitation complements tumor-node-metastasis staging prognostication in nasopharyngeal carcinoma. J Clin Oncol 2006;24:5414–5418. [DOI] [PubMed] [Google Scholar]
- 13.Chan AT, Lo YM, Zee B, et al. Plasma Epstein-Barr virus DNA and residual disease after radiotherapy for undifferentiated nasopharyngeal carcinoma. J Natl Cancer Inst 2002;94:1614–1619. [DOI] [PubMed] [Google Scholar]
- 14.Analysis of plasma Epstein-Barr Virus DNA to screen for nasopharyngeal cancer. N Engl J Med 2018;378:973. [DOI] [PubMed] [Google Scholar]
- 15.Lin JC, Wang WY, Chen KY, et al. Quantification of plasma Epstein-Barr virus DNA in patients with advanced nasopharyngeal carcinoma. N Engl J Med 2004;350:2461–2470. [DOI] [PubMed] [Google Scholar]
- 16.Lam WKJ, Jiang P, Chan KCA, et al. Sequencing-based counting and size profiling of plasma Epstein-Barr virus DNA enhance population screening of nasopharyngeal carcinoma. Proc Natl Acad Sci U S A 2018;115:E5115–E5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wuerdemann N, Jain R, Adams A, et al. Cell-free HPV-DNA as a bio-marker for oropharyngeal squamous cell carcinoma—a step towards personalized medicine? Cancers (Basel) 2020;12:2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chera BS, Kumar S, Beaty BT, et al. Rapid clearance profile of plasma circulating tumor HPV type 16 DNA during chemoradiotherapy correlates with disease control in HPV-associated oropharyngeal cancer. Clin Cancer Res 2019;25:4682–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chera BS, Kumar S, Shen C, et al. Plasma circulating tumor HPV DNA for the surveillance of cancer recurrence in HPV-associated oropharyngeal cancer. J Clin Oncol 2020;38:1050–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sastre-Garau X, Diop M, Martin F, et al. A NGS-based blood test for the diagnosis of invasive HPV-associated carcinomas with extensive viral genomic characterization. Clin Cancer Res Published online June 9, 2021. doi: 10.1158/1078-0432.CCR-21-0293 [DOI] [PMC free article] [PubMed]


