Abstract
Purpose
To explore the properties of short‐T2 signals in human brain, investigate the impact of various experimental procedures on these properties and evaluate the performance of three‐component analysis.
Methods
Eight samples of non‐pathological human brain tissue were subjected to different combinations of experimental procedures including D2O exchange and frozen storage. Short‐T2 imaging techniques were employed to acquire multi‐TE (33–2067 μs) data, to which a three‐component complex model was fitted in two steps to recover the properties of the underlying signal components and produce amplitude maps of each component. For validation of the component amplitude maps, the samples underwent immunohistochemical myelin staining.
Results
The signal component representing the myelin bilayer exhibited super‐exponential decay with T2,min of 5.48 μs and a chemical shift of 1.07 ppm, and its amplitude could be successfully mapped in both white and gray matter in all samples. These myelin maps corresponded well to myelin‐stained tissue sections. Gray matter signals exhibited somewhat different components than white matter signals, but both tissue types were well represented by the signal model. Frozen tissue storage did not alter the signal components but influenced component amplitudes. D2O exchange was necessary to characterize the non‐aqueous signal components, but component amplitude mapping could be reliably performed also in the presence of H2O signals.
Conclusions
The myelin mapping approach explored here produced reasonable and stable results for all samples. The extensive tissue and methodological investigations performed in this work form a basis for signal interpretation in future studies both ex vivo and in vivo.
Keywords: high‐performance gradient, super‐Lorentzian lineshape, tissue characterization, tissue preparation, ultrashort TE, white and gray matter
Short abstract
Click here for author‐reader discussions
1. INTRODUCTION
The central nervous system (CNS) relies heavily on myelin for efficient signal transmission along neural pathways. 1 , 2 Damage to the myelin sheath is an integral feature of several neuroinflammatory diseases such as multiple sclerosis and can result in severe disability. 3 , 4 The ability to non‐invasively map myelin content is highly desirable for monitoring of demyelinating disorders, pathomechanistic studies on demyelination and myelin repair, and development of putative remyelinating drug candidates.
CNS myelin is a compacted extension of the plasma membrane of oligodendrocytes. The membrane wraps concentrically around axons, trapping layers of intra‐ and extracellular water in the process, 5 and develops a characteristic lipid (70% of dry mass) and protein composition. 2 , 6 Consequently, the myelin sheath has two primary constituents: the trapped water collectively referred to as myelin water and the lipid‐protein bilayer.
CNS myelin is primarily present in white matter (WM), in which it comprises approximately 50% of dry mass, but oligodendrocytes and myelin‐wrapped axons exist also in gray matter (GM). 2
MRI lends itself well to myelin mapping due to its safe, non‐invasive application and its rich capacity for probing tissue properties. The majority of research toward myelin mapping with MRI is based on the analysis of signals stemming from aqueous protons, 7 for which myelin sensitivity is achieved either through the distinct relaxation properties of myelin water 8 or through interactions between non‐aqueous and aqueous magnetization pools. 9 Many techniques exploiting aqueous signals are readily available for use in vivo and have been shown to correlate with histological myelin stains and provide sensitivity to various CNS disorders. 5 , 10 , 11 These techniques represent a notable advancement in myelin specificity compared to conventional MRI, but yield inherently rather indirect measures of myelin content.
More direct myelin bilayer imaging methods are warranted to complement or possibly outperform the techniques relying on aqueous signals, but targeting signals from protons in the myelin bilayer has traditionally been impractical. Aqueous signals, with T2s in the range of tens to hundreds of milliseconds, are straightforward to capture with standard MRI systems. In contrast, due to the specific configuration of the myelin sheath, signals from bilayer‐bound protons exhibit a T2 spectrum ranging from a few microseconds to tens of milliseconds, 12 with around 75% of the signals exhibiting T2s below 100 μs. 13
As reviewed by Weiger et al., 14 research in recent years has demonstrated that dedicated short‐T2 imaging technology is capable of capturing and spatially resolving rapidly decaying signals from different tissues and materials, particularly for decay constants down to the hundreds of microseconds range. Using custom‐built, high‐performance RF and gradient hardware, even signals with ultrashort decay constants down to 10 μs have been imaged at high resolution. 15 Several studies have proposed the use of short‐T2 techniques for imaging of the myelin bilayer, 12 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 and a pilot myelin study using the imaging system described by Froidevaux et al. 15 demonstrated clear acquisition‐related advantages afforded by the advanced hardware. 13
Most reports of myelin bilayer imaging are based on ex vivo studies with large variations between experimental approaches. As such, it is yet unclear how best to interpret resulting data and whether the same interpretations would apply also to in vivo signals.
Much of the uncertainty in data interpretation relates to the selection, storage and preparation of imaging samples. Experimental approaches to myelin bilayer signal examination frequently involve the use of animal 12 , 13 , 21 , 22 , 23 , 26 , 27 or human 17 , 19 , 20 tissue samples, and it is common practice to freeze the tissue between dissection and imaging. However, it is unclear how well the signal properties of animal tissue match those of human tissue and whether freezing alters the MR signals like formalin fixation has been shown to do. 20 In addition, background water signals are often reduced by D2O exchange in order to isolate the contributions from non‐aqueous signals such as those stemming from the myelin bilayer. 12 , 13 , 19 , 20 , 21 , 22 , 23 , 26 Such chemical preparation is not feasible in vivo, and while an initial attempt showed promising results for myelin bilayer mapping also in non‐exchanged tissue, 13 key questions remain regarding which signal analysis approaches are valid in the presence of dominant background signals.
In this work, MRI signals from non‐aqueous and aqueous protons in non‐pathological human brain tissue (WM and GM) are observed and assigned to three signal components. The results are compared with literature findings in animal samples to explore the validity of animal tissue as a substitute for human tissue in MR‐based myelin research. Furthermore, the effects of frozen sample storage on the MRI signals are investigated. The performance of the three‐component analysis approach is discussed, with focus on the differences between employment in D2O‐exchanged samples and non‐exchanged counterparts. Finally, amplitude maps of the signal components are produced and qualitatively compared with myelin‐stained tissue sections, high‐resolution ultrashort‐T2 reference images and photographs of the samples.
2. METHODS
2.1. Tissue samples
Two tissue blocks from different regions of the cerebrum of a 56‐year‐old male donor were excised at autopsy (post‐mortem interval: six hour). The patient died of a cardiovascular event, and neuropathological workup did not suggest gross brain pathology. Written informed consent was obtained, and the study was approved by the regional ethics review board.
Each of the tissue blocks (labeled “1” and “2”, extracted from the temporal and frontal lobes, respectively) was divided into four samples on average 5 × 25 × 18 mm3. Two neighboring samples were imaged promptly after autopsy (referred to as fresh samples, labeled “s”), and the remaining two neighboring samples were stored at −80°C for five months prior to thawing and subsequent imaging (referred to as frozen samples, labeled “z”). For each set of two samples, one was subjected to D2O exchange (referred to as D2O samples, labeled “D”) performed in two steps over a total of 24 h (based on the procedures described by Wilhelm et al. 12 and Weiger et al. 13 ). The samples not subjected to D2O exchange were placed in water (referred to as H2O samples, labeled “H”) to avoid dehydration and imaged directly. Both the D2O and H2O solutions were saline (9 g/L NaCl). Samples were stored at 5°C while awaiting imaging, and all imaging was performed with the samples at room temperature. A schematic of the full procedure is provided in Figure 1.
FIGURE 1.
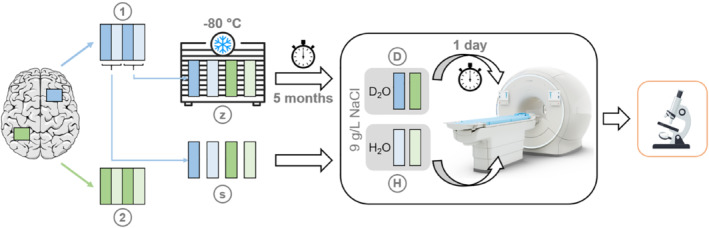
Schematic showing the processing steps for each sample. Tissue blocks from two regions (“1” and “2”) on the cerebrum were divided into four samples each, half of which were imaged directly and half of which were stored at −80°C for five months prior to imaging. Of the four samples that were arranged either fresh (“s”) or frozen–thawed (“z”), one from each tissue block underwent a 24‐h, two‐step D2O exchange (“D”) while the other was placed in H2O (“H”). After imaging, the samples were processed for myelin immunohistochemistry. Note that the placement of the tissue blocks on the cerebrum are for demonstration purposes only and are not meant to indicate the anatomical locations of the dissected tissue
2.2. Data acquisition
In order to capture the rapidly decaying signals from the myelin bilayer, dedicated ultrashort‐T2 techniques and hardware were employed. A 3 T Philips Achieva (Philips Healthcare, the Netherlands) system was equipped with a high‐performance gradient insert 28 capable of gradient strength in excess of 200 mT/m at 100% duty cycle, and the acquisition system was bypassed by a dedicated RF chain 29 including fast transmit/receive switches. 30 A 1H‐free loop coil of 40 mm diameter was used for both transmit and receive operations.
Two 3D imaging protocols were applied. The first protocol utilized single‐point imaging (SPI) 31 to acquire 14 images at TE between 33 and 2067 μs, which were used for model fitting. SPI captures the full k‐space at time TE, enabling unbiased voxel‐wise analysis because, with no time evolution across k‐space, each image accurately reflects the signal state at the respective TE. With the maximum gradient strength of 221 mT/m employed at the shortest TE, the nominal isotropic resolution was limited to 1.56 mm. The second protocol utilized zero‐TE imaging with hybrid filling of the dead‐time gap (HYFI) 32 to acquire a high‐resolution (0.39 mm isotropic) ultrashort‐T2 reference image. Further details on the protocols can be found in Table 1.
TABLE 1.
Protocols for MRI of the myelin bilayer
| Protocol | FOV [mm] | Δr [mm] | BW [kHz] | TE [μs] | RF pulse | TR [ms] | NSA | Scan time [m:s] | HYFI‐A | HYFI‐T2 [μs] |
|---|---|---|---|---|---|---|---|---|---|---|
| SPI | 50 | 1.56 | 7.5–470 | 2067–33 | 2 μs hard | 3 | 4 | 03:34 | N/A | N/A |
| HYFI | 50 | 0.39 | 425 | 12 | 2 μs hard | 1 | 32 | 27:41 | 0.17 | 50 |
Note: The first protocol uses an SPI 31 sequence with spherical k‐space support to acquire a multi‐TE image series. The second protocol uses a HYFI 32 sequence to acquire a high‐resolution ultrashort‐T2 reference image. Pulse power was adjusted for maximum steady‐state signal, calibrated in a D2O sample to specifically optimize non‐aqueous signals. The flip angles of approximately a few degrees were adjusted for the two protocols according to their different TRs to yield equivalent contrast. The scan time stated for the SPI protocol is per image, and the full multi‐TE series consisted of 14 images.
Abbreviations: BW, image bandwidth; HYFI‐A, HYFI amplitude coefficient; HYFI‐T2, HYFI target T2; NSA, number of signal averages; Δr, nominal 3D isotropic resolution.
2.3. Data analysis
The data analysis procedure consisted of three steps: (1) determining a signal model that approximates true tissue composition and facilitates constructive analysis; (2) evaluating the parameters of the signal components to gain knowledge of the underlying tissue and reduce the unknowns in the signal model; and (3) establishing a procedure for myelin mapping based on the formulated signal model.
2.3.1. Signal model
A three‐component signal model was employed, which splits the non‐aqueous signals into an ultrashort (U) and a short (S) component, both of super‐Lorentzian lineshape, 33 , 34 and represents the aqueous signals by one component (W) of Lorentzian lineshape. Following the interpretation by Weiger et al. 13 (for WM), the U‐component represents the myelin bilayer and the S‐component represents residual non‐aqueous content.
The signal evolution under this model is described by
where φ is an arbitrary global phase, Δω is a local resonance offset and ω 0 is the 1H Larmor frequency. A n , δ n and T2,n or T2,min,n are the amplitude, chemical shift and decay constant of component n, respectively, where T2 characterizes the Lorentzian lineshape of decay function D L and T2,min characterizes the super‐Lorentzian lineshape of decay function D SL. The full form of the decay functions D L and D SL are provided in Supporting Information Part 1.
A two‐component model with only one component representing the non‐aqueous signals was also explored. Unless otherwise stated, the three‐component model was used.
Note that decay constants as a tissue property are denoted by T2, while any measured signals technically decay by T2∗. However, T2′ is considered negligible here, and, therefore, the decay constants resulting from signal analysis retain the notation used to define the signal model.
Because water signals have decayed almost negligibly after the longest TE of the SPI series, the decay constant of the W‐component could not be reliably fitted and was instead fixed at 50 ms for all analyses: although water in the brain exhibits a range of decay constants, the exact value used to characterize the W‐component is of little practical impact in this study. Also the chemical shift of the W‐component was fixed (at 4.7 ppm) and was used to calibrate the chemical shifts of the non‐aqueous components.
2.3.2. Component analysis: open fits
To determine the parameter values of the signal model, least‐squares fitting was employed. For details on the algorithm, please consult the published analysis code (see Data Availability Statement). All parameters of the signal model – except those fixed for the W‐component – were treated as unknowns, and the bounds on the parameters were kept wide to avoid making assumptions on the underlying components; we refer to such fits as open fits.
Open fits were performed on complex signal values averaged over large regions of WM or GM, which stabilized the fitting with respect to noise and potential local signal variations. The regions were chosen based on tissue anatomy seen in the high‐resolution reference images, and were generally as large as possible while excluding voxels at tissue boundaries to limit partial volume effects. Such signal averaging is valid as long as variations in local resonance offset are small. Any mention of WM or GM regions or averaged signals for a given sample refers to the same data.
2.3.3. Myelin mapping: fixed fits
Applying the fitting procedure from analysis step 2 on a single‐voxel basis would yield maps of each free model parameter. With the interpretation that the U‐component represents the myelin bilayer, a map of the amplitude of the U‐component is a myelin map.
In order to enable useful interpretation, the amplitudes in the myelin map must be comparable between voxels, which can be achieved by defining the same signal components in each voxel (see the Discussion section). Reasonable component decay constants and chemical shifts were therefore determined from the open fits and fixed in the voxel‐wise fits. We refer to fits with fixed component decay constants and chemical shifts as fixed fits. Unless otherwise stated, the component parameters used in the fixed fits were the mean values found in WM; fixed fits based on the mean values found in GM were also explored.
Fixed fits were also run on the averaged WM and GM signals in order to compare component amplitudes in the two tissue types and across samples subjected to different experimental procedures. The absolute component amplitudes found for different datasets are not directly comparable due to variations in experimental factors such as coil loading and coil sensitivity. Therefore, amplitude normalization was performed, using different procedures for the aqueous and non‐aqueous components: the aqueous component amplitude was normalized by the sum of all component amplitudes, while the non‐aqueous component amplitudes were normalized by the sum of non‐aqueous component amplitudes to quantify their relative contributions to the rapidly decaying signals.
2.4. Processing
Tricubic interpolation was performed on all SPI data prior to analysis, reducing voxel size by a factor of two in each dimension. An additional in‐plane interpolation by a factor of three was applied to all MRI‐based images for display purposes.
Amplitude maps of the non‐aqueous components in H2O samples were masked based on the sample outline in the respective W‐component amplitude map.
For H2O samples, the two longest‐TE data points were discarded for fitting (i.e., ) because they exhibited unstable behavior, an effect that was not detected in D2O samples and is possibly related to the longer lifetime or larger presence of water signals. A linear fit to the phase of the remaining two longest‐TE data points was used to determine an approximate value for the local resonance offset in H2O datasets, which was corrected out of the data for WM and GM region fits. Using this correction avoids the assumption that the local resonance offset is equal for all voxels included in signal averages and ensures that the local resonance offset parameter stays well within fit boundaries. This correction is not valid for D2O data because the water component does not sufficiently dominate the longest‐TE data points.
2.5. Histological staining
After imaging, the brain samples were snap frozen at −80°C and subsequently cryosectioned into 20 μm thick sections. The sections underwent immunohistochemistry for myelin oligodendrocyte glycoprotein (MOG, primary antibody concentration 1:50), which is a key myelin component. For a complete overview of the histological processing, see Supporting Information Part 2.
3. RESULTS
The magnitude and phase behavior of the averaged WM signals for samples subjected to different experimental procedures (tissue block 1) are displayed in Figure 2. For the fresh D2O sample, the GM region is also included. H2O‐sample magnitudes converge to a higher level than D2O‐sample magnitudes, reflecting background water levels, and the phase curve shows characteristic behavior for D2O samples while for H2O samples the linear water phase dominates. Fresh samples exhibit a consistently higher magnitude than frozen samples, but the overall behavior is similar.
FIGURE 2.
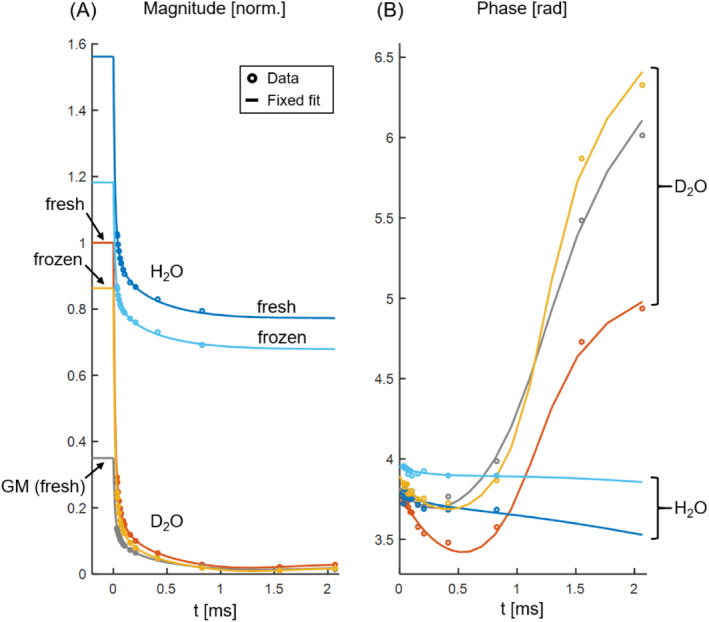
Magnitude (A) and phase (B) of the SPI data and associated fixed fits for the samples from tissue block 1. Data points represent an average over large WM regions unless specified as GM data. The magnitudes are normalized by the fit magnitude at time zero in WM for the fresh D2O sample. H2O and D2O samples are clearly distinguishable in both plots. In the absence of a dominant water pool, the phase curves exhibit a characteristic shape; in contrast, the phase of the H2O data is approximately linear and, because the local resonance offset has been corrected for, relatively flat. Frozen samples have reduced magnitude with respect to their fresh counterparts but otherwise exhibit similar signal behavior. The GM region has significantly lower magnitude than the WM region from the same sample, but the general signal behavior of the two tissue types is comparable
The decay constants and chemical shifts of the non‐aqueous components found by the open fits are given in Table 2 for the WM and GM regions in all D2O samples. There is a clear range of decay constant for each component: in WM, the U‐component averages 5.48 μs and the S‐component averages 102 μs, and in GM these values are prolonged to 9.42 μs and 171 μs, respectively. The chemical shifts are also quite consistent, averaging 1.07 ppm for the U‐component and 2.09 ppm for the S‐component in WM, and 0.58 ppm for the U‐component and 2.39 ppm for the S‐component in GM.
TABLE 2.
Non‐aqueous component parameters T2,min and chemical shift (δ) found by open fits to averaged WM and GM signals for all D2O (“D”) samples, as well as mean value and SD per tissue type
| Sample | T2,min,U [μs] | T2,min,S [μs] | δU [ppm] | δS [ppm] | |||||
|---|---|---|---|---|---|---|---|---|---|
| WM | 1Ds | 6.76 | (0.16) | 92.1 | (0.74) | 1.02 | (0.02) | 2.10 | (0.01) |
| 2Ds | 4.94 | (0.06) | 122 | (0.46) | 0.85 | (0.01) | 1.96 | (0.00) | |
| 1Dz | 5.53 | (0.16) | 88.1 | (0.48) | 1.30 | (0.02) | 2.06 | (0.01) | |
| 2Dz | 4.71 | (0.11) | 108 | (0.51) | 1.10 | (0.01) | 2.25 | (0.00) | |
| Mean | 5.48 | 102 | 1.07 | 2.09 | |||||
| SD | 0.92 | 15.5 | 0.18 | 0.12 | |||||
| GM | 1Ds | 10.3 | (0.19) | 157 | (1.28) | 0.77 | (0.02) | 2.47 | (0.00) |
| 2Ds | 8.88 | (0.08) | 219 | (1.07) | 0.10 | (0.01) | 2.20 | (0.00) | |
| 1Dz | 10.6 | (0.49) | 152 | (3.88) | 0.69 | (0.05) | 2.35 | (0.01) | |
| 2Dz | 7.92 | (0.34) | 155 | (2.35) | 0.75 | (0.04) | 2.51 | (0.01) | |
| Mean | 9.42 | 171 | 0.58 | 2.39 | |||||
| SD | 1.25 | 32.2 | 0.32 | 0.14 | |||||
Note: Standard errors obtained by propagation of the noise variance in the underlying images are given in parenthesis. T2,min is longer in GM than WM, but the signal components each exhibit a clear range of decay constant irrespective of tissue type. In WM and partly in GM, there is more variation between the two tissue blocks (“1” and “2”) than between fresh (“s”) and frozen (“z”) samples. This indicates that anatomical location affects the detectable signal components to some degree while freezing has little impact. The GM components found in sample 2Ds deviate somewhat from those found in the other samples, which is considered to be a manifestation of instability in the open fits. GM fits are generally less stable than WM fits due to the lower signal level.
Figure 3 shows a series of comparison plots of magnitude (top row) and phase (bottom row) behavior for different fits in the frozen D2O sample from tissue block 1. Note that the axis ranges are set individually for each plot. In Figure 3A, an open fit using the default three‐component signal model is compared with an open fit using the two‐component signal model, and it is clear that the two‐component model does not adequately represent the signal behavior, whereas the three‐component model fits the data well. Figure 3B illustrates the similarities between open and fixed fits to averaged WM signals, and the phase plot highlights the noise variations at low TE. Similarly, Figure 3C displays open and fixed fits to averaged GM signals, and, in addition, a fixed fit based on the average signal components found in GM is included. While there are differences between the components found by the open fits and the components used for the fixed fits (see Table 2), the differences in fit performance are marginal throughout Figure 3B–D. Figure 3D emphasizes the differences between WM and GM signal behavior. Overall, the presented details illustrate the suitability of the employed three‐component model and the use of fixed fits with equal component parameters for WM and GM.
FIGURE 3.
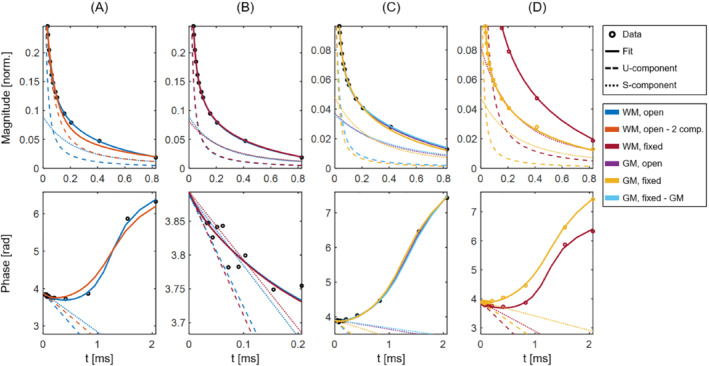
Comparison plots of magnitude (top row, same normalization as applied in Figure 2) and phase (bottom row) for different fits to averaged WM and GM signals in a representative sample (the frozen D2O sample from tissue block 1). Note the different axes used for the different plots. The underlying data points are shown as black circles in cases in which all fits are based on the same data, and as colored circles in cases in which the fits are based on different data. The W‐component is not shown for simplicity, but would manifest as a near‐horizontal line. A, Open fits in WM using both three‐component (blue) and two‐component (orange) signal models. B, Open (blue) and fixed (maroon) fits in WM. C, Open (purple) and fixed (yellow) fits in GM, as well as an additional fixed fit (light blue) based on the average signal components found in GM (see Table 2). D, Fixed fits in WM (maroon) and GM (yellow), highlighting the differences in signal behavior
Fixed fits applied to several averaged WM and GM signals are displayed in Figure 2, and normalized component amplitudes obtained from such fits for all samples are given in Table 3. The normalized W‐component amplitudes in D2O samples reflect the extent of D2O exchange and thus should not be directly compared, but comparing W‐component amplitudes within the same sample shows roughly twice as high a water fraction in GM as in WM. For H2O samples, the GM water increase averages around 30%. Both observations primarily reflect lower content of non‐aqueous components in GM than in WM. The normalized W‐component amplitudes in H2O samples are slightly lower than the brain water content expected from literature (approximately 70% in WM and 80% in GM) 35 ; although we urge caution when interpreting fitted component amplitudes (see the Discussion section), this apparent water reduction is likely a consequence of T1 weighting in the underlying data (see Supporting Information Part 3). In WM, the amplitude ratio between the non‐aqueous components AU/AS is roughly 90/10, except for the frozen H2O samples for which the ratio is rather 80/20. In GM, the AU/AS ratio is generally around 80/20, with deviations for samples 2Ds and 2Hs, which are closer to a 95/5 ratio, and the frozen H2O samples, for which the ratio is around 70/30. Differences between fresh and frozen H2O samples are also present in the W‐component, for which the amplitude is slightly higher for frozen than fresh samples.
TABLE 3.
Normalized amplitudes for each signal component in WM and GM regions for all samples, determined from fits in which the component decay constants and chemical shifts were fixed at the average WM values given in Table 2
| Sample |
|
|
|
||||
|---|---|---|---|---|---|---|---|
| WM | 1Ds | 0.03 | 0.90 | 0.10 | |||
| 2Ds | 0.03 | 0.93 | 0.07 | ||||
| 1Dz | 0.02 | 0.90 | 0.10 | ||||
| 2Dz | 0.05 | 0.90 | 0.10 | ||||
| 1Hs | 0.52 | 0.87 | 0.13 | ||||
| 2Hs | 0.47 | 0.94 | 0.06 | ||||
| 1Hz | 0.60 | 0.81 | 0.19 | ||||
| 2Hz | 0.58 | 0.81 | 0.19 | ||||
| Mean | 0.54 a | 0.88 | 0.12 | ||||
| SD | 0.06 a | 0.05 | 0.05 | ||||
| GM | 1Ds | 0.06 | 0.77 | 0.23 | |||
| 2Ds | 0.06 | 0.93 | 0.07 | ||||
| 1Dz | 0.04 | 0.81 | 0.19 | ||||
| 2Dz | 0.09 | 0.81 | 0.19 | ||||
| 1Hs | 0.71 | 0.75 | 0.25 | ||||
| 2Hs | 0.61 | 0.96 | 0.04 | ||||
| 1Hz | 0.76 | 0.73 | 0.27 | ||||
| 2Hz | 0.75 | 0.70 | 0.30 | ||||
| Mean | 0.71 a | 0.81 | 0.19 | ||||
| SD | 0.07 a | 0.09 | 0.09 |
Note: Standard errors are on the order of 10−3–10−5. The mean value and SD for each tissue type are also provided, which for the normalized W‐component amplitudes were determined only over the H2O samples. Relative water content is much higher in H2O samples (“H”) than in D2O samples (“D”) and higher in GM than in WM. The amplitude distribution of non‐aqueous signals AU/AS is around 90/10 in WM and 80/20 in GM. Frozen tissue (“z”) differs notably from fresh tissue (“s”) in H2O samples: in the frozen samples, the normalized amplitude of the W‐component is higher and the AU/AS ratio tends toward a lower U‐component contribution (80/20 in WM and 70/30 in GM) than in the fresh counterparts.
Calculated only over H2O samples.
Amplitude maps of the three signal components, obtained by performing voxel‐wise fixed fits, are displayed for all samples in Figures 4 (tissue block 1) and 5 (tissue block 2) along with photographs, myelin‐stained tissue sections, and high‐resolution reference images. Overall, good correspondence is seen between all image types (see for instance sample 1Dz in Figure 4), but due to the thickness and pliability of the samples and the resolution of the component amplitude maps, some topological sample features vary slightly between photographs, MRI slices and/or stained sections. Some of the component amplitude maps also suffer from partial volume effects. Certain myelin‐stained sections exhibit only weak WM/GM contrast, which likely relates to variability in the staining process. The similarities between U‐component amplitude maps and MOG staining density are of particular interest because they strengthen the interpretation of the U‐component as representing the myelin bilayer. The W‐component amplitude maps convincingly reflect water content, accounting for the T1 weighting in the underlying data. Also the S‐component amplitude maps exhibit some WM/GM contrast, but these maps are generally more uniform and harder to interpret than those of the U‐component. The component amplitude maps are of highest quality for the D2O samples but exhibit acceptable quality also for the H2O samples.
FIGURE 4.
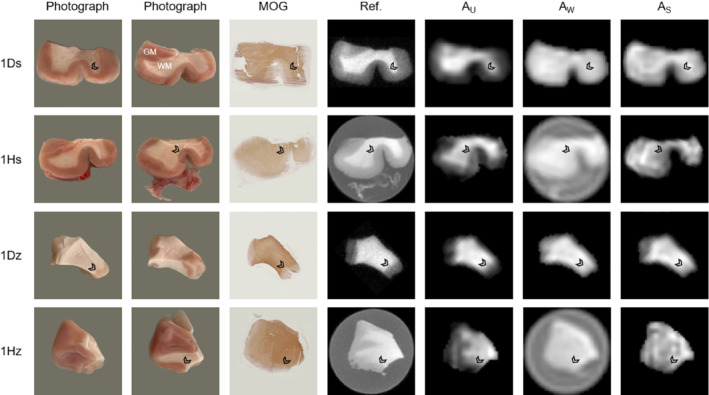
Comparison of (from left to right) photographs from both sides of the sample, tissue sections stained by myelin oligodendrocyte glycoprotein (MOG) immunohistochemistry, high‐resolution ultrashort‐T2 reference images, and fitted component amplitude maps for each sample from tissue block 1 (ordered according to Figure 1). Overall, good correspondence in terms of sample geometry and WM/GM boundaries (see arrowheads at comparable locations) is achieved for all image types. The component amplitude maps show the highest quality for D2O samples, although the map quality for H2O samples is also considered acceptable
FIGURE 5.
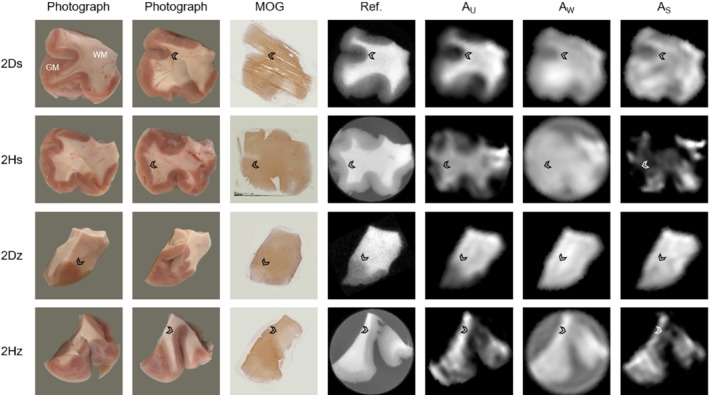
Identical results as presented in Figure 4 but for the samples in tissue block 2. Good correspondence between image types is seen also for these samples, and the component amplitude maps are of the same quality as those presented for the samples from tissue block 1 (see Figure 4). These results further verify the performance of the component amplitude mapping procedure, particularly considering the brain‐region dependence of the signal components seen in Table 2
4. DISCUSSION
In this work, MRI signals from non‐pathological human brain samples were observed using ultrashort‐T2 methods and analyzed by decomposition into one aqueous and two non‐aqueous components. The properties of these signal components were investigated for tissue subjected to different experimental procedures, specifically frozen storage and D2O exchange for reduction of background water signals. Signal analysis was performed separately for WM and GM, enabling comparison of the two tissue types and establishing a baseline for similar studies in diseased tissue.
4.1. Sample selection, storage, and preparation
Table 4 gives an overview of myelin bilayer signal components reported in literature. The components reported for ex vivo animal tissue are similar to those found in the present study for human tissue, with super‐Lorentzian analysis consistently placing the largest non‐aqueous signal component at T2,min of 5–10 μs and lipid range chemical shift. Additional components, where reported, also correspond well between animal and human tissue.
TABLE 4.
Overview of reported myelin bilayer signal components (ex vivo)
| Origin | Sample | Lineshape model | Component values | Component description | Reference |
|---|---|---|---|---|---|
| Human | Brain WM | Super‐Lorentzian |
T2,min,U = 5.48 μs T2,min,S = 102 μs δU = 1.07 ppm δS = 2.09 ppm |
U: Myelin S: Residual non‐aqueous content |
Current study |
| Porcine | Brain WM | Super‐Lorentzian |
T2,min,U = 7.5 μs T2,min,S = 101 μs δU = 1.38 ppm δS = 1.91 ppm |
U: Myelin S: Residual non‐aqueous content |
Weiger et al. 13 |
| Bovine | Myelin lipid extract | Super‐Lorentzian |
T2,min = 8 μs δ = 1.5 ppm |
Methylene, shortest and largest (74% of total myelin signal) | Wilhelm et al. 12 |
| Ovine | Cervical spinal cord | Super‐Lorentzian | T2,min = 10 μs | Methylene frequency, shortest and largest | Seifert et al. 21 |
| Bovine | Brain WM | Super‐Lorentzian (Gaussian‐based) |
σmin,U = 7.2 μs σmin,S = 87 μs δU = δS = 1.3 ppm |
U: Shortest and largest S: Remaining |
Manning et al. 27 |
| Rat | Optic nerve | Lorentzian | T2 = 80 μs | Shortest and largest | Horch et al. 26 |
Note: Lorentzian‐based super‐Lorentzian components are characterized (in the time‐domain) by decay constant T2,min, while Gaussian‐based super‐Lorentzian components are characterized (in the time‐domain) by the SD of the narrowest (i.e., shortest) constituent Gaussian, σmin. In studies in which multiple values were provided for the same parameter, the mean value is given. δ denotes the component chemical shift, which was calibrated with water at 4.7 ppm.
Freezing tissue had little effect on the parameters of the signal components, as evidenced by Table 2, in which there is greater variation between the two different regions of the brain (tissue blocks 1 and 2) than between fresh and frozen samples. However, for frozen samples, there were some differences in apparent water level and the amplitude relationship of the two non‐aqueous components, primarily for H2O samples (see Figure 2 and Table 3). A likely explanation for these differences is that freezing affects water content and T1 by altering the microstructural integrity of the tissue, which together with magnetisation transfer effects influences the component amplitudes. 36
D2O exchange is a useful procedure when attempting to characterize the non‐aqueous signal components because it largely removes the otherwise dominant signal contribution of mobile water. The procedure also diminishes any magnetisation transfer effects that can impact the evaluation of component amplitudes and, consequently, decay constants. In addition, the contributions of the non‐aqueous signals to the overall phase evolution are difficult to distinguish without D2O exchange (see Figure 2), which can lead to unreasonable chemical shift estimates and generally destabilize the fitting procedure. Employing open fits in H2O samples led to unstable fit behavior due to the accumulation of effects related to the considerable presence of water, and such fits were therefore not presented in this work.
We showed in Figures 4 and 5 that fixed fits provide acceptable component amplitude map quality also in H2O samples, removing the need for D2O exchange in cases in which the components of the signal model are known or can be reasonably assumed. This result has important implications for the applicability of the myelin mapping procedure to in vivo experiments with acquired data of similar information content, assuming that the underlying signal components in vivo are comparable to those found ex vivo (it is considered unlikely that biochemical changes directly post‐mortem should have a significant effect on signal components given the insignificance of changes introduced by the relatively aggressive procedure of frozen tissue storage).
4.2. Data analysis procedure
Data analysis was performed through a combination of two fitting approaches, namely open and fixed fits, in order to circumvent the inherent limitations of each approach. These limitations have important consequences regarding the interpretation and validity of the fitting results (especially for open fits) and are therefore discussed here in detail.
Open fits can provide valuable information about the parameters of the signal model but are prone to unstable behavior due to their many free parameters as well as interdependencies between the parameters. The instabilities can be explained separately for the magnitude and phase of the signal.
For the magnitude behavior, consider the relationship between component amplitude and decay constant in the case of only one signal component. Because TE1 > 0, the component amplitude is unbounded, and the signal magnitude at TE1 can be reached either with a smaller decay constant and a larger amplitude or with a larger decay constant and a smaller amplitude –that is, there is no unique solution. Naturally, the fitting process considers the data points at all TEs when determining the optimal parameter set, but component amplitudes and decay constants are still strongly dependent on each other, which corresponds to poor conditioning. With data subject to errors, it is therefore not instructive to compare component amplitudes (e.g., in different tissue types) unless the decay constants of the components are fixed; this caution also extends to comparisons with amplitudes expected from literature.
For the phase behavior, the explanation is more subtle. The component parameter governing the phase evolution is the chemical shift, which manifests as a linear change over time. At low TE for which the non‐aqueous components contribute significantly to the acquired signals, the chemical shifts have not had much time to influence the signal phase. Additionally, lower‐TE data points have lower SNR because the acquisition duration is shorter. Together, these effects may result in unstable chemical shift estimates, and even minor changes in the acquisition, reconstruction and analysis pipeline can influence the fitted chemical shifts. That said, using the same pipeline for multiple datasets often yielded similar chemical shift results, with values consistently within a reasonable range for primarily lipid signals.
Fixed fits, on the other hand, are comparatively stable and enable component amplitude comparisons, but impose assumptions on the signal components and, consequently, can bias the fitting results. It is therefore important that component characterization is performed with due care, which relates back to the informed use of open fits (or critical assessment of relevant literature results).
4.3. Component interpretation
The assignment of the fitted U‐component to myelin is based on a number of assumptions concerning the size and distinctiveness of the contribution to the rapidly decaying signals by myelin constituents, the validity of the approximations in the signal model, and the ability of the fitting procedure to correctly allocate signal properties to model parameters. As discussed by Weiger et al., 13 there are strong indications that WM ultrashort‐T2 signals indeed largely reflect myelin and that the U‐component primarily represents myelin lipids. In the present study, we have further validated the signal model and explored the limitations of the fitting procedure. Overall, we believe that the U‐component amplitude is the most quantitative direct measure of WM myelin bilayer content available with current MRI technology.
The situation for GM is fundamentally different because neuronal components such as somata and axons/dendrites (and, hence, non‐myelin plasma membranes) constitute a large portion of non‐aqueous material and quite likely exhibit similar signal characteristics as the myelin bilayer. Therefore, we cannot claim myelin specificity of the U‐component in GM, but contributions from the myelin bilayer to GM signals should nevertheless get assigned to the U‐component. It is therefore still likely that changes in myelin are partly reflected in the U‐component also for GM.
4.4. Signal components
GM exhibits different component parameters than WM (see Table 2), which likely reflects the different composition of the two tissue types. However, as demonstrated in Figures 4 and 5, fixing fit components based on WM values produced reasonable component amplitude maps also in GM, thus bypassing the need for pre‐analysis tissue segmentation. It is worth noting that the signal model was designed based on expected WM behavior, yet it appears to sufficiently represent also GM signals (see Figure 3D).
Considering the full spectrum of decay constants and chemical shifts presented in Table 2 together with the quality of component amplitude maps and region fits displayed in Figures 2, 4 and 5 based simply on fixed fits using average parameter values, we conclude that the exact values of the component parameters are not of great consequence as long as they are representative of the signal behavior. However, it is important to keep in mind that, for component amplitudes to be comparable, the same fixed values must be used across all analyses.
Although previous studies have reported multiple super‐Lorentzian myelin bilayer components, 12 it was not considered necessary here to resolve the rapidly decaying signals into more than two components due to the general similarity and lopsided amplitude distribution of the previously reported components. Nevertheless, signal models with more than three components were briefly explored but did not perform well, as expected for high‐dimensional multi‐exponential analysis.
4.5. Imaging protocols
The SPI protocol is limited by the minimum TE, which in turn is limited by image resolution and available gradient strength. The minimum TE of 33 μs raises a potential concern regarding the reliability of characterizing the U‐component, with T2,min of 5.5 μs, from the SPI data presented in this study; this concern ultimately relates to the SNR of the data used for fitting. Signals from a super‐Lorentzian component exhibit a T2 distribution for which T2,min describes the shortest‐lived signals; consequently, all other signals from the component exhibit T2 > T2,min. For the case of the U‐component, 20% of the total signal amplitude remains at TEmin, and, as can be seen in Figure 2, this signal level is significantly higher than the noise level. It is therefore reasonable to conclude that the U‐component can be characterized using the captured signals. This conclusion is supported by the reliability of the signal analysis procedure (demonstrated in Figure 3) and the good correspondence of the presented fitting results to those reported in literature (see Table 4).
The HYFI reference images provide high‐resolution depiction of ultrashort‐T2 signals, and reflect the contrast seen in the U‐component amplitude maps. This raises the question of whether the HYFI images are a sufficient representation of myelin content, which would bypass the resolution limitations of the SPI protocol and its associated signal analysis.
The most pressing argument against relying on single HYFI images concerns myelin specificity. The HYFI images depict the sum of all signal components, and while there are ways to attenuate the contribution of the water component, e.g., by long‐TE image subtraction or inversion recovery preparation, 18 , 37 the signals from the myelin bilayer cannot be separated from other non‐aqueous signals. In short, the SPI series facilitates greater myelin specificity at the cost of image resolution and SNR efficiency. We believe that the insights presented in this study will provide valuable information regarding how best to maintain myelin specificity when transitioning to in vivo application, for which the scan limitations are strict.
5. CONCLUSIONS
One aim of the present study was to explore the effects of different experimental procedures on the analysis of myelin bilayer signals, specifically regarding the selection, storage and preparation of imaging samples.
The human brain samples studied in this work exhibited similar component parameters to various animal samples reported in literature. These findings validate the use of animal tissue as a substitute for human tissue, although human tissue still offers obvious advantages depending on the particular purpose of the study.
Furthermore, it was demonstrated that tissue stored in a frozen state is qualitatively equivalent to fresh tissue, but quantitative comparison—in particular of signal amplitudes – between fresh and frozen tissue is limited.
Lastly, D2O exchange, which has been a popular method to reduce background water signals in previous short‐T2 myelin studies, was found also here to provide benefits for signal component investigation. However, when working with a fixed signal model, D2O exchange was not necessary to achieve acceptable myelin maps.
Another aim of this study was to investigate the rapidly decaying MRI signals from non‐pathological human brain. We showed that WM and GM can be described by the same signal model, which consists of two non‐aqueous components with super‐Lorentzian lineshape (T2,mins of around 5–10 μs and 100–200 μs, and chemical shifts in the lipid range) and one aqueous component with Lorentzian lineshape.
The employed myelin mapping procedure exhibited stable behavior, and the myelin maps corresponded well to sample anatomy and MOG staining density. The procedure can therefore be recommended for future use (contingent on the data being of comparable quality and structure to those presented in this study) and can be used without extensive prior signal analysis by fixing component parameter values reported here.
Overall, the results obtained in this work can act as a foundation for signal interpretation in further ex vivo and in vivo myelin studies.
CONFLICT OF INTEREST
Klaas Paul Pruessmann holds a research agreement with and receives research support from Philips and is a shareholder of Gyrotools LLC.
Supporting information
Appendix S1: Part 1: Decay functions in the signal model
Part 2: Immunohistochemistry procedure
Part 3: Demonstration of T1 weighting
ACKNOWLEDGMENTS
The authors thank Karl Frontzek from the Institute of Neuropathology, University Hospital Zurich, Switzerland, for providing the brain samples, and Richard Reynolds from the UK Multiple Sclerosis Tissue Bank for providing MOG antibody.
Baadsvik EL, Weiger M, Froidevaux R, Faigle W, Ineichen BV, Pruessmann KP. Mapping the myelin bilayer with short‐T2 MRI: Methods validation and reference data for healthy human brain. Magn Reson Med. 2023;89:665‐677. doi: 10.1002/mrm.29481
Click here for author‐reader discussions
DATA AVAILABILITY STATEMENT
Example data and MATLAB (MathWorks, USA) analysis code is provided at https://gitlab.ethz.ch/shortT2mri/myelin‐multi‐te‐fitting.
REFERENCES
- 1. Norton WT, Cammer W. Isolation and characterization of myelin. In: Morell P, ed. Myelin. 2nd ed. Springer; 1984:147‐195. [Google Scholar]
- 2. Rasband MN, Macklin WB. Myelin structure and biochemistry. In: Brady ST, Siegel GJ, Albers RW, Price DL, eds. Basic Neurochemistry. 8th ed. Elsevier; 2012:180‐199. [Google Scholar]
- 3. Popescu BFG, Lucchinetti CF. Pathology of demyelinating diseases. Annu Rev Pathol. 2012;7:185‐217. [DOI] [PubMed] [Google Scholar]
- 4. Love S. Demyelinating diseases. J Clin Pathol. 2006;59:1151‐1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Laule C, Vavasour IM, Kolind SH, et al. Magnetic resonance imaging of myelin. Neurotherapeutics. 2007;4:460‐484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baron W, Hoekstra D. On the biogenesis of myelin membranes: sorting, trafficking and cell polarity. FEBS Lett. 2010;584:1760‐1770. [DOI] [PubMed] [Google Scholar]
- 7. Piredda GF, Hilbert T, Thiran J‐P, Kober T. Probing myelin content of the human brain with MRI: a review. Magn Reson Med. 2021;85:627‐652. [DOI] [PubMed] [Google Scholar]
- 8. MacKay AL, Laule C. Magnetic resonance of myelin water: an in vivo marker for myelin. Brain Plast. 2016;2:71‐91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Varma G, Duhamel G, de Bazelaire C, Alsop DC. Magnetization transfer from Inhomogeneously broadened lines: a potential marker for myelin. Magn Reson Med. 2015;73:614‐622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Laule C, Moore GRW. Myelin water imaging to detect demyelination and remyelination and its validation in pathology. Brain Pathol. 2018;28:750‐764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heath F, Hurley SA, Johansen‐Berg H, Sampaio‐Baptista C. Advances in noninvasive myelin imaging. Dev Neurobiol. 2018;78:136‐151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wilhelm MJ, Ong HH, Wehrli SL, et al. Direct magnetic resonance detection of myelin and prospects for quantitative imaging of myelin density. Proc Natl Acad Sci. 2012;109:9605‐9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weiger M, Froidevaux R, Baadsvik EL, Brunner DO, Rösler MB, Pruessmann KP. Advances in MRI of the myelin bilayer. Neuroimage. 2020;217:116888. [DOI] [PubMed] [Google Scholar]
- 14. Weiger M, Pruessmann KP. Short‐T2 MRI: principles and recent advances. Prog Nucl Magn Reson Spectrosc. 2019;114‐115:237‐270. [DOI] [PubMed] [Google Scholar]
- 15. Froidevaux R, Weiger M, Rösler MB, et al. High‐resolution short‐T2 MRI using a high‐performance gradient. Magn Reson Med. 2020;84:1933‐1946. [DOI] [PubMed] [Google Scholar]
- 16. Du J, Ma G, Li S, et al. Ultrashort echo time (UTE) magnetic resonance imaging of the short T2 components in white matter of the brain using a clinical 3T scanner. Neuroimage. 2014;87:32‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nayak KS, Pauly JM, Gold GE, Nishimura DG. Imaging Ultra‐Short T2 Species in the Brain. Proceedings of the ISMRM 8th Scientific Meeting & Exhibition. International Society for Magnetic Resonance in Medicine; 2000:509. [Google Scholar]
- 18. Waldman A, Rees J, Brock C, Robson M, Gatehouse P, Bydder G. MRI of the brain with ultra‐short echo‐time pulse sequences. Neuroradiology. 2003;45:887‐892. [DOI] [PubMed] [Google Scholar]
- 19. Sheth V, Shao H, Chen J, et al. Magnetic resonance imaging of myelin using ultrashort Echo time (UTE) pulse sequences: phantom, specimen, volunteer and multiple sclerosis patient studies. Neuroimage. 2016;136:37‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seifert AC, Umphlett M, Hefti M, Fowkes M, Xu J. Formalin tissue fixation biases myelin‐sensitive MRI. Magn Reson Med. 2019;82:1504‐1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seifert AC, Li C, Wilhelm MJ, Wehrli SL, Wehrli FW. Towards quantification of myelin by solid‐state MRI of the lipid matrix protons. Neuroimage. 2017;163:358‐367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fan SJ, Ma Y, Zhu Y, et al. Yet more evidence that myelin protons can be directly imaged with UTE sequences on a clinical 3T scanner: Bicomponent T2∗ analysis of native and deuterated ovine brain specimens. Magn Reson Med. 2018;80:538‐547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fan SJ, Ma Y, Chang EY, Bydder GM, Du J. Inversion recovery ultrashort echo time imaging of ultrashort T2 tissue components in ovine brain at 3T: a sequential D2O exchange study. NMR Biomed. 2017;30:e3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boucneau T, Cao P, Tang S, et al. In vivo characterization of brain ultrashort‐T2 components. Magn Reson Med. 2018;80:726‐735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ercan E, Boernert P, Webb A, Ronen I. Whole‐brain tissue‐based assessment of the ultrashort T2 component using 3D UTE MRI Relaxometry. Proceedings of the ISMRM 20th Annual Meeting & Exhibition. International Society for Magnetic Resonance in Medicine; 2012:4279. [Google Scholar]
- 26. Horch RA, Gore JC, Does MD. Origins of the ultrashort‐T2 1H NMR signals in myelinated nerve: a direct measure of myelin content? Magn Reson Med. 2011;66:24‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Manning AP, Mackay AL, Michal CA. Understanding aqueous and non‐aqueous proton T1 relaxation in brain. J Magn Reson. 2021;323:106909. [DOI] [PubMed] [Google Scholar]
- 28. Weiger M, Overweg J, Rösler MB, et al. A high‐performance gradient insert for rapid and short‐T2 imaging at full duty cycle. Magn Reson Med. 2018;79:3256‐3266. [DOI] [PubMed] [Google Scholar]
- 29. Weiger M, Brunner DO, Dietrich BE, Müller CF, Pruessmann KP. ZTE Imaging in Humans. Magn Reson Med. 2013;70:328‐332. [DOI] [PubMed] [Google Scholar]
- 30. Brunner DO, Furrer L, Weiger M, et al. Symmetrically biased T/R switches for NMR and MRI with microsecond dead time. J Magn Reson. 2016;263:147‐155. [DOI] [PubMed] [Google Scholar]
- 31. Balcom BJ, MacGregor RP, Beyea SD, Green DP, Armstrong RL, Bremner TW. Single‐point ramped imaging with T1 enhancement (SPRITE). J Magn Reson Ser A. 1996;123:131‐134. [DOI] [PubMed] [Google Scholar]
- 32. Froidevaux R, Weiger M, Rösler MB, Brunner DO, Pruessmann KP. HYFI: hybrid filling of the dead‐time gap for faster zero echo time imaging. NMR Biomed. 2021;34:e4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wennerström H. Proton nuclear magnetic resonance lineshapes in lamellar liquid crystals. Chem Phys Lett. 1973;18:41‐44. [Google Scholar]
- 34. Bloom M, Burnell EE, Valic MI, Weeks G. Nuclear magnetic resonance line shapes in lipid bi‐layer model membranes. Chem Phys Lipids. 1975;14:107‐112. [DOI] [PubMed] [Google Scholar]
- 35. Van der Knaap MS, Valk J. Myelin and white matter. Magnetic Resonance of Myelination and Myelin Disorders. Springer Science & Business Media; 2005. [Google Scholar]
- 36. Evans SD, Nott KP, Kshirsagar AA, Hall LD. The effect of freezing and thawing on the magnetic resonance imaging parameters of water in beef, lamb and pork meat. Int J Food Sci Technol. 1998;33:317‐328. [Google Scholar]
- 37. Gatehouse P, Bydder G. Magnetic resonance imaging of short T2 components in tissue. Clin Radiol. 2003;58:1‐19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Part 1: Decay functions in the signal model
Part 2: Immunohistochemistry procedure
Part 3: Demonstration of T1 weighting
Data Availability Statement
Example data and MATLAB (MathWorks, USA) analysis code is provided at https://gitlab.ethz.ch/shortT2mri/myelin‐multi‐te‐fitting.


