Abstract
Bifidobacterium asteroides is considered the ancestor of the genus Bifidobacterium, which has evolved in close touch with the hindgut of social insects. However, recent studies revealed high intraspecies biodiversity within this taxon, uncovering the putative existence of multiple bifidobacterial species, thus, suggesting its reclassification. Here, a genomic investigation of 98 B. asteroides‐related genomes retrieved from public repositories and reconstructed from metagenomes of the hindgut of Apis mellifera and Apis cerana was performed to shed light on the genetic variability of this taxon. Phylogenetic and genomic analyses revealed the existence of eight clusters, of which five have been recently characterized with a representative type strain of the genus and three were represented by putative novel bifidobacterial species inhabiting the honeybee gut. Then, the dissection of 366 shotgun metagenomes of honeybee guts revealed a pattern of seven B. asteroides‐related taxa within A. mellifera that co‐exist with the host, while A. cerana microbiome was characterized by the predominance of one of the novel species erroneously classified as B. asteroides. A further glycobiome analysis unveiled a conserved repertoire of glycosyl hydrolases (GHs) reflecting degradative abilities towards a broad range of simple carbohydrates together with genes encoding specific GHs of each B. asteroides‐related taxa.
INTRODUCTION
Members of the genus Bifidobacterium are Gram‐positive bacteria subjected to growing interest due to their health‐promoting properties exerted on human health (Hidalgo‐Cantabrana et al., 2017; Milani et al., 2017; Ventura et al., 2014). They are mainly distributed in the gut of mammals (Turroni et al., 2018), but they can also be identified in the hindgut of social insects, oral cavities, sewage, blood and foods (Bunesova et al., 2014). The advent of the genomic era allowed to easily decode the genome sequences of bifidobacteria, providing a tool to shed light on genetics and phylogenetics of these microorganisms (Lugli et al., 2014; Milani et al., 2014). Furthermore, a combination of genome sequencing and bacterial isolation attempts expanded our knowledge regarding the genus Bifidobacterium through the identification and characterization of more than 100 different bifidobacterial species (Lugli, Alessandri, et al., 2021; Lugli, Calvete‐Torre, et al., 2021; Lugli, Duranti, et al., 2019; Lugli, Milani, et al., 2019). Remarkably, in the last 10 years, more bifidobacterial species have been described than in the previous century, starting from the identification of the first Bifidobacterium bifidum in 1899 by Henri Tissier from the intestinal microbiota of a breastfed infant (Tissier, 1900).
So far, several members of the genus Bifidobacterium have been characterized from the guts of different social insects, highlighting a rich bifidobacterial‐related environment second only to the gut of mammals (Bunesova et al., 2014). Four species were identified from the guts of bumble bees (Bombus lucorum, Bombus lapidarius, Bombus terrestris, and Bombus hypnorum), named after Bifidobacterium actinocoloniiforme, Bifidobacterium bohemicum, Bifidobacterium bombi and Bifidobacterium commune (Killer et al., 2009, 2011; Praet et al., 2015). Furthermore, Bifidobacterium aemilianum and Bifidobacterium xylocopae were isolated from the guts of Xylocopa violacea, the violet carpenter bee (Alberoni et al., 2019). In addition, among two of the eight species of honey bees, that is, Apis mellifera (the western honey bee) and Apis cerana (the eastern honey bee) (Danforth, 2007), three bifidobacterial species were characterized, that is, Bifidobacterium asteroides, Bifidobacterium coryneforme and Bifidobacterium indicum (Milani et al., 2014).
Among the species inhabiting the gut of social insects, B. asteroides was subjected to genomic investigation predicting its capability for a putative respiratory metabolism (Bottacini et al., 2012). This notion was also investigated from an evolutionary perspective highlighting that respiration is a common metabolic feature of this ancient bifidobacterial species, which has been lost in mammal‐derived Bifidobacterium species (Bottacini et al., 2012). Furthermore, the glycan‐degrading abilities of B. asteroides have been investigated in respect to those of the other members of the genus, highlighting a more extended adaptation history to their hosts that resulted in a different repertoire of glycosyl hydrolases (GHs) characterized by GH43 and GH3 enzymes (Milani et al., 2015).
More recently, four novel Bifidobacterium species were isolated from honey bees, named after Bifidobacterium apousia, Bifidobacterium choladohabitans, Bifidobacterium mizhiense and Bifidobacterium polysaccharolyticum, whose genomic investigation revealed a close genetic relatedness towards B. asteroides type strain DSM 20089 (Chen et al., 2021; Li et al., 2022). Furthermore, based on genomic investigation aiming at exploring the intraspecies genetic diversity of members of the genus Bifidobacterium, B. asteroides was fragmented into four putative distinct bifidobacterial species (Lugli et al., 2018), resulting in the most complex Bifidobacterium taxon identified to date. Lately, different B. asteroides clusters have been confirmed by strain‐level investigation of the honey bee gut microbiota (Wu et al., 2021), requiring further investigation to disentangle the high biodiversity of this widespread taxon in the gut of A. mellifera.
In this study, we report an exhaustive genomic and metagenomic dissection of B. asteroides genomes retrieved from public repositories as well as reconstructed from the metagenomes of their hosts, that is, A. mellifera and A. cerana, allowing to redesign the B. asteroides phylogeny. Furthermore, to explore the distribution of B. asteroides‐related taxa among their shared hosts, 366 honey bee metagenomes (Regan et al., 2018; Wu et al., 2021, 2022) were investigated to uncover their ecological and evolutionary dynamics.
EXPERIMENTAL PROCEDURES
Bifidobacterium genome sequence selection and quality control
Complete and partial genomes of 17 B. asteroides strains, and 170 unclassified bifidobacterial strains, were retrieved from the NCBI database (Tables 1 and S1). Then, the genome sequence of B. asteroides DSM 20089, together with those of bifidobacteria isolated from social insects (Table 1), was used to discard those strains showing an average nucleotide identity (ANI) lower than 94% employing the software fastANI (Jain et al., 2018). Amino acid sequences of coding genes were predicted using Prodigal (Hyatt et al., 2010) and then used for further genomic analyses, while the quality of genomes was estimated for completeness and contamination using CheckM (Parks et al., 2015) (Table S3).
TABLE 1.
Bifidobacterium asteroides‐related genomes.
| NCBI code | Strain name | Predicted species | Cluster | Host |
|---|---|---|---|---|
| GCA_003202695.1 | Bifidobacterium asteroides ESL0200 | Bifidobacterium sp. nov. | CL1 | Apis mellifera |
| GCA_019469425.1 | Bifidobacterium asteroides ESL0447 | Bifidobacterium sp. nov. | CL2 | Apis cerana |
| GCA_016102005.1 | Bifidobacterium choladohabitans JCM 34586 | Bifidobacterium choladohabitans | CL3 | Apis mellifera |
| GCA_000499285.1 | Bifidobacterium sp. 7101 | Bifidobacterium choladohabitans | CL3 | Apis mellifera |
| GCA_000967265.1 | Bifidobacterium asteroides Bin7 | Bifidobacterium choladohabitans | CL3 | Apis mellifera |
| GCA_009683175.1 | Bifidobacterium asteroides VRA_9sq_n | Bifidobacterium choladohabitans | CL3 | Apis mellifera |
| GCA_016100645.1 | Bifidobacterium sp. M0353 | Bifidobacterium choladohabitans | CL3 | Apis mellifera |
| GCA_016100655.1 | Bifidobacterium sp. W8107 | Bifidobacterium choladohabitans | CL3 | Apis mellifera |
| GCA_016100735.1 | Bifidobacterium sp. W8104 | Bifidobacterium choladohabitans | CL3 | Apis mellifera |
| GCA_016100765.1 | Bifidobacterium sp. W8105 | Bifidobacterium choladohabitans | CL3 | Apis mellifera |
| GCA_016100775.1 | Bifidobacterium sp. W8103 | Bifidobacterium choladohabitans | CL3 | Apis mellifera |
| GCA_020884755.1 | Bifidobacterium mizhiense JCM 34710 | Bifidobacterium mizhiense | CL4 | Apis mellifera |
| GCA_000499185.1 | Bifidobacterium sp. A11 | Bifidobacterium mizhiense | CL4 | Apis mellifera |
| GCA_003688305.1 | Bifidobacterium sp. wkB344 | Bifidobacterium mizhiense | CL4 | Apis mellifera |
| GCA_007559275.1 | Bifidobacterium apousia JCM 34587 | Bifidobacterium apousia | CL5 | Apis mellifera |
| GCA_000967185.1 | Bifidobacterium asteroides Bin2 | Bifidobacterium apousia | CL5 | Apis mellifera |
| GCA_002846895.1 | Bifidobacterium asteroides 1460B | Bifidobacterium apousia | CL5 | Apis mellifera |
| GCA_016100785.1 | Bifidobacterium sp. W8120 | Bifidobacterium apousia | CL5 | Apis mellifera |
| GCA_016101425.1 | Bifidobacterium sp. W8112 | Bifidobacterium apousia | CL5 | Apis mellifera |
| GCA_002715865.1 | Bifidobacterium asteroides DSM 20089 | Bifidobacterium asteroides | CL6 | Apis mellifera |
| GCA_000304215.1 | Bifidobacterium asteroides PRL2011 | Bifidobacterium asteroides | CL6 | Apis mellifera |
| GCA_003202855.1 | Bifidobacterium asteroides ESL0170 | Bifidobacterium asteroides | CL6 | Apis mellifera |
| GCA_016101165.1 | Bifidobacterium sp. W8114 | Bifidobacterium asteroides | CL6 | Apis mellifera |
| GCA_016101375.1 | Bifidobacterium sp. W8110 | Bifidobacterium asteroides | CL6 | Apis mellifera |
| GCA_016102155.1 | Bifidobacterium asteroides W8130 | Bifidobacterium asteroides | CL6 | Apis mellifera |
| GCA_016102205.1 | Bifidobacterium asteroides W8118 | Bifidobacterium asteroides | CL6 | Apis mellifera |
| GCA_016102215.1 | Bifidobacterium asteroides W8109 | Bifidobacterium asteroides | CL6 | Apis mellifera |
| GCA_003202755.1 | Bifidobacterium asteroides ESL0199 | Bifidobacterium sp. nov. | CL7 | Apis mellifera |
| GCA_016101585.1 | Bifidobacterium polysaccharolyticum JCM 34588 | Bifidobacterium polysaccharolyticum | CL8 | Apis mellifera |
| GCA_000970835.1 | Bifidobacterium asteroides Hma3 | Bifidobacterium polysaccharolyticum | CL8 | Apis mellifera |
| GCA_003202715.1 | Bifidobacterium asteroides ESL0198 | Bifidobacterium polysaccharolyticum | CL8 | Apis mellifera |
| GCA_003688325.1 | Bifidobacterium sp. wkB338 | Bifidobacterium polysaccharolyticum | CL8 | Apis mellifera |
| GCA_016100595.1 | Bifidobacterium sp. M0399 | Bifidobacterium polysaccharolyticum | CL8 | Apis mellifera |
| GCA_016101415.1 | Bifidobacterium sp. M0404 | Bifidobacterium polysaccharolyticum | CL8 | Apis mellifera |
| GCA_017349335.1 | Bifidobacterium asteroides wkB204 | Bifidobacterium polysaccharolyticum | CL8 | Apis mellifera |
| GCA_016100445.1 | Bifidobacterium sp. M0307 | Bifidobacterium indicum/coryneforme | ‐ | Apis mellifera |
| GCA_016102235.1 | Bifidobacterium sp. W8108 | Bifidobacterium indicum/coryneforme | ‐ | Apis mellifera |
| GCA_022484665.1 | Bifidobacterium sp. UW_MP_BIF13_2 | Bifidobacterium sp. nov. | ‐ | biological waste material |
| GCA_022486445.1 | Bifidobacterium sp. UW_MP_BIF13_1 | Bifidobacterium sp. nov. | ‐ | biological waste material |
| GCA_022647885.1 | Bifidobacterium sp. UW_MP_BIF3_1 | Bifidobacterium sp. nov. | ‐ | biological waste material |
| GCA_001263395.1 | Bifidobacterium actinocoloniiforme DSM 22766 | ‐ | ‐ | Bombus |
| GCA_003315635.1 | Bifidobacterium aemilianum DSM 104956 | ‐ | ‐ | Xylocopa violacea |
| GCA_000741525.1 | Bifidobacterium bohemicum DSM 22767 | ‐ | ‐ | Bombus |
| GCA_000737845.1 | Bifidobacterium bombi DSM 19703 | ‐ | ‐ | Bombus |
| GCA_900094885.1 | Bifidobacterium commune R‐52791 | ‐ | ‐ | Bombus |
| GCA_000737865.1 | Bifidobacterium coryneforme LMG 18911 | ‐ | ‐ | Apis mellifera |
| GCA_000706765.1 | Bifidobacterium indicum LMG 11587 | ‐ | ‐ | Apis cerana |
| GCA_003315615.1 | Bifidobacterium xylocopae DSM 104955 | ‐ | ‐ | Xylocopa violacea |
Phylogenomic comparison of B. asteroides‐related taxa
Collected genome sequences were subjected to a pangenome calculation using PGAP (Pan‐Genomes Analysis Pipeline) (Zhao et al., 2012). The proteome of all assessed genomes was organized into functional gene clusters using the GF (gene family) method, which involves a comparison of each protein to all other proteins using BLAST analysis, followed by clustering into clusters of orthologous genes (cut‐off E‐value of 1 × 10−5 and 50% identity across at least 80% of both protein sequences), using MCL (a graph‐theory‐based Markov cluster algorithm) (Enright et al., 2002). Protein families shared between all genomes, named core genes, were defined by selecting the families that contained at least one single protein member for each genome. The concatenated core genome sequences were aligned using MAFFT (multiple alignment using fast Fourier Transform) (Katoh et al., 2018), and the corresponding phylogenomic tree was constructed using the neighbour‐joining method in ClustalW version 2.1 (Li, 2003). The core genome tree was built using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). For each genome pair, the ANI value was calculated using the software fastANI (Jain et al., 2018). Then, hierarchical clustering analysis (HCA) based on a matrix of ANI values between Bifidobacterium strains was constructed using OriginPro graphing and analysis 2021 (https://www.originlab.com/2021).
Metagenome dataset selection
In this project, 366 publicly available datasets retrieved after 10 BioProject (listed in NCBI) from various geographical locations were obtained through honey bee hindgut sequencing literature (Table S3). In detail, we selected shotgun microbial profiling datasets, including microbiomes belonging to A. mellifera and A. cerana (Table S3).
Taxonomic classification of the honey bee microbiome reads and whole metagenome assembly
To analyse high‐quality sequenced data only, each dataset was subjected to a filtering step removing low‐quality reads (minimum mean quality score 20, window size 5, quality threshold 25 and minimum length 100) using the fastq‐mcf script (https://github.com/ExpressionAnalysis/ea-utils/blob/wiki/FastqMcf.md). Filtered reads were then collected and taxonomically classified through the METAnnotatorX2 pipeline (Milani et al., 2021), using the up‐to‐date RefSeq (genome) database retrieved from the NCBI (https://www.ncbi.nlm.nih.gov/refseq/). Filtered reads were then subjected to whole metagenome assembly using Spades v3.15 (Bankevich et al., 2012) with default parameters and the metagenomic flag option (‐meta) together with k‐mer sizes of 21, 33, 55 and 77. As mentioned above, for the short reads, reconstructed contig sequences were taxonomically classified based on their sequence identity using megablast against the same RefSeq database (Chen et al., 2015). ORFs of each assembled genome were then predicted with Prodigal (Hyatt et al., 2010) and annotated utilizing the MEGAnnotator pipeline (Lugli et al., 2016). In all, the METAnnotatorX2 pipeline was employed for various purposes, from read filtering to taxonomic classification of the assembled contigs (Milani et al., 2018, 2021).
Metagenome tracing of B. asteroides‐related taxa
Complete and partial genome sequences retrieved from NCBI and reclassified from CL1 to CL8 were used to trace their presence among the 366 publicly available datasets collected in this study. Previous filtered reads of each microbiome were then collected and taxonomically reclassified using a homemade RefSeq database with B. asteroides genomes subdivided into the eight CLs. Then, the reconstructed genome sequences obtained from the 366 samples after WMS assembly were reclassified following the updated database. Next, reconstructed genomes of B. asteroides‐related taxa were selected based on statistics (completeness >80% and contamination <3%) retrieved using the CheckM software (Parks et al., 2015). Each reconstructed genome was then validated using the 94% ANI threshold employing the software fastANI (Jain et al., 2018) and used for further phylogenomic and genetic analyses.
Bifidobacterium asteroides glycobiome
The proteome of each reconstructed bifidobacterial strain was screened for genes predicted to encode carbohydrate‐active enzymes based on sequence similarity to genes classified in the carbohydrate‐active enzyme (CAZy) database (Drula et al., 2022). Thus, each predicted proteome of a given bifidobacterial strain was screened for orthologues against the dbCAN2 meta server (Zhang et al., 2018) composed of 2,141,452 CDS using HMMER v3.3.2 (cut‐off E‐value of 1 × 10−15 and coverage >0.35).
Statistical analysis
Bacterial abundance at the species level between microbiomes was validated by ANOVA analysis. Furthermore, PERMANOVA analysis was performed using 1000 permutations to estimate p‐values of differences among samples in PCoA analyses. The hierarchical clustering analysis (HCA) of samples was performed using OriginPro graphing and analysis 2021 (https://www.originlab.com/2021), employing the Bray–Curtis matrix and Pearson correlation as a distance metric and the sum square of distances and furthest neighbour for clustering methods.
RESULTS AND DISCUSSION
Evaluation of intraspecies genomic variability of Bifidobacterium asteroides species
The analysis of the currently available Bifidobacterium genome sequences deposited in NCBI provided an exhaustive overview of the genomic diversity for each taxon. Seven species displayed more than a hundred sequenced genomes, representing to date the most studied bifidobacterial species, that is, B. longum (n = 674), B. breve (n = 204), B. bifidum (n = 178), B. pseudocatenulatum (n = 178), B. animalis (n = 123), B. adolescentis (n = 117), and B. pseudolongum (n = 107). Nonetheless, after the latter species, B. catenulatum (n = 57), B. dentium (n = 49) and finally B. asteroides (n = 17) showed the highest number of sequenced genomes, leaving behind additional 91 bifidobacterial species poorly characterized. This data highlighted an increasing interest of the scientific community about B. asteroides, probably due to the growing concerns regarding the health and behaviours of its common host, the honey bee.
The collection of 17 B. asteroides genome sequences from the NCBI database allowed us to investigate the genome variability of this taxon. Each genome was subjected to ANI evaluation against the other B. asteroides genome sequences. Such analyses revealed ANI values lower than 90% in respect to the B. asteroides type strain, that is, B. asteroides ESL0447 (ANI value of 89.9%) and B. asteroides ESL0200 (ANI value of 89.2%), which cast doubt on the correct taxonomic classification of these strains (Figure 1). In this regard, it should be noted that two strains displaying an ANI value of <95% are considered to belong to two distinct species (Konstantinidis et al., 2006; Richter & Rosselló‐Móra, 2009). Moreover, recent phylogenomics studies regarding the Bifidobacterium genus have demonstrated that 94% is an excellent ANI threshold to define bifidobacterial species boundaries (Lugli et al., 2017, 2018). Thus, 10 out of 17 strains were below this accepted threshold, showing that more than half of the submitted genomes may not represent members of the B. asteroides taxon (Figure 1).
FIGURE 1.
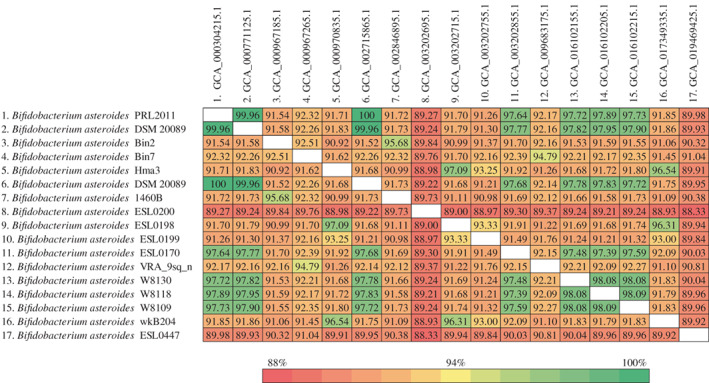
Average nucleotide identity (ANI) values between 17 Bifidobacterium asteroides strains retrieved from the NCBI database. Colour‐based scheme highlight those 10 strains with ANI values below the 94% threshold to be represented as members of the B. asteroides taxon.
The genome sequences of additional 170 bifidobacterial unclassified strains were collected from the NCBI repository to ensure an exhaustive screening of the genomic diversity of the B. asteroides taxon (Table S1). Thus, an ANI evaluation including genome sequences of these unclassified bifidobacteria altogether with 17 B. asteroides strains (n = 187) was performed to gather additional B. asteroides‐related genomes. In this context, a hierarchical clustering analysis employing the ANI values matrix of all 187 genome sequences allowed to identify 26 unclassified genomes related to the B. asteroides strains (Figure S1). Subsequently, sequence quality of the so‐gathered genomes was evaluated through the CheckM pipeline, discarding strains with completeness <85% and contamination >3% (Table S2). The process results in selecting 36 distinct strains related to B. asteroides with a suitable genome quality for further phylogenetic and metagenomic analyses (Table 1).
Phylogenetic dissection of B. asteroides‐related strains revealed the existence of eight possible novel different taxa
Based on the recent scientific literature about the bifidobacterial ecology, B. asteroides was only one of those Bifidobacterium species identified and isolated from social insects, such as A. mellifera, A. cerana, Bombus and Xylocopa violacea. Thus, the type strains of the closed phylogenetic species to B. asteroides, that is, Bifidobacterium actinocoloniiforme DSM 22766, B. aemilianum DSM 104956, B. apousia JCM 34587, B. bohemicum DSM 22767, B. bombi DSM 19703, B. choladohabitans JCM 34586, B. commune R‐52791, B. coryneforme LMG 18911, B. indicum LMG 11587, B. mizhiense JCM 34710, B. polysaccharolyticum JCM 34588 and B. xylocopae DSM 104955, were added to the analysis. Gathered genomes were then employed to study the phylogeny of the 36 strains recognized as B. asteroides‐related strains (Table 1). The relationship between bifidobacterial species isolated from social insects was initially evaluated by performing gene prediction among the 48 genome sequences, avoiding inconsistencies due to different gene prediction methods. Then, a pangenome analysis of the collected strains was undertaken to determine putative orthologous genes between the 48 strains. The analysis identified 10,698 clusters of orthologous genes (COGs), representing the pangenome of strains isolated from social insects. Collected COGs allowed the identification of 370 genes shared between all analysed genomes. Furthermore, after exclusion of paralogs, the concatenation of 318 core protein sequences was used to build a Bifidobacterium phylogenomic tree using Scardovia inopinata JCM 12537 gene sequences as outgroup (Figure 2).
FIGURE 2.
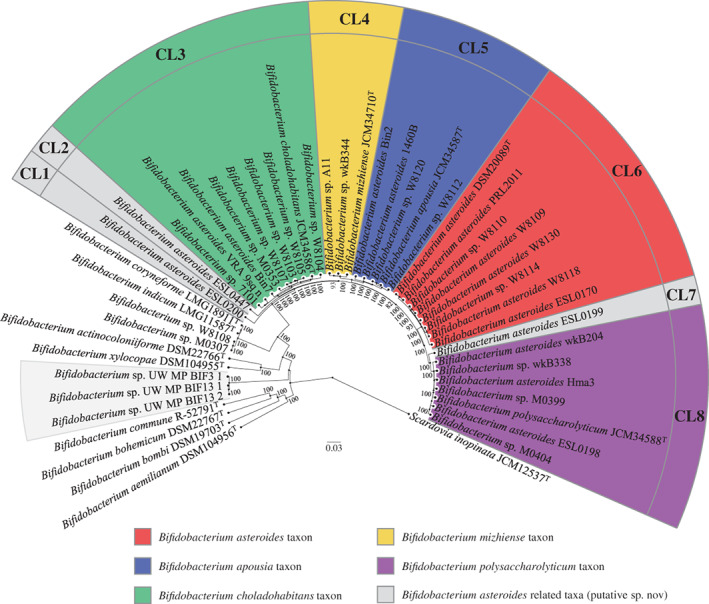
Phylogenomic tree of 48 Bifidobacterium asteroides‐related strains based on the concatenation of 318 core protein sequences. Different colours highlight the division into eight taxa (named from CL1 to CL8) between strains previously classified as B. asteroides or unclassified bifidobacterial species. The phylogenetic tree was constructed by the neighbour‐joining method, with the genome sequence of Scardovia inopinata JCM 12537 as an outgroup. Bootstrap percentages above 50 are shown at node points based on 1000 replicates of the phylogenetic tree.
The phylogenetic reconstruction of bifidobacteria isolated from insects allowed to define eight clusters from CL1 to CL8 containing Bifidobacterium strains previously classified as B. asteroides highlighting divergent evolutionary routes (Figure 2). B. asteroides type strain fell in CL6 with only seven other bifidobacteria, representing those strains that belong to this taxon (Table 1). In contrast, all the other related strains were distributed among the other seven clusters except for Bifidobacterium sp. UW_MP_BIF3_1, Bifidobacterium sp. UW_MP_BIF13_1, and Bifidobacterium sp. UW_MP_BIF13_2, which cluster apart from any classified taxon (Figure 2). Interestingly, these latter strains were genomically related to Bifidobacterium crudilactis (ANI between 79.4 % and 80.1%), representing a putative novel species of the genus Bifidobacterium. However, since these strains do not belong to a B. asteroides‐related taxon, they were excluded from further analyses, as well as Bifidobacterium (B). sp. W8108 and B. sp. M0307, which were found to be related to B. coryneforme LMG 18911 and B. indicum LMG 11587 (Figure 2).
Among the identified clusters, four of them were represented by Bifidobacterium taxa recently characterized, that is, B. choladohabitans JCM 34586 (CL3), B. mizhiense JCM 34710 (CL4), B. apousia JCM 34587 (CL5) and B. polysaccharolyticum JCM 34588 (CL8) (Chen et al., 2021; Li et al., 2022) (Figure 2). Thus, thanks to isolation attempts and genomic characterization of the isolates, the large part of the identified clusters were reclassified and a representative type strain defined. Nonetheless, three clusters (CL1, CL2 and CL7) still await an exhaustive characterization representing three putative novel bifidobacterial species inhabiting the honey bee's gut. Specifically, the latter clusters were composed of single strains, such as B. asteroides ESL0200 (CL1), B. asteroides ESL0447 (CL2) and B. asteroides ESL0199 (CL7). Thus, further investigation of the honey bee microbiome is necessary to better explore the B. asteroides biodiversity.
Altogether genomic and phylogenetic analyses highlighted that the intraspecies variability of the B. asteroides taxon is distributed among eight clusters. However, to date, only five clusters have been characterized with a representative type strain of the genus, while the remaining three clusters may represent putative novel bifidobacterial species inhabiting the gut of honey bees.
Taxonomic classification of the honey bee microbiome
Since each representative strain of the eight B. asteroides‐related clusters was isolated from the gut of honey bees, we decided to explore its distribution by collecting A. mellifera and A. cerana metagenomics data from the short read archive (SRA). Thus, sequenced DNA of 314 A. mellifera and 52 A. cerana microbiomes was selected from 10 sequencing projects publicly available in the NCBI repository (Table S3). Collected data were subjected to microbial profiling based on short‐read taxonomic classification down to species level after filtering steps based on DNA sequence quality and host DNA removal (Table S4).
In silico investigation uncovered that the most abundant species among honey bee microbiomes were represented by microorganisms typically associated with the host, such as Bartonella apis, B. asteroides, Frischella perrara, Gilliamella apicola, Gilliamella apis, Lactobacillus apis and Snodgrassella alvi. All the latter species were found with a prevalence higher than 45%, representing the core microbiota of honey bees (Figure 3). However, significant differences occurred by comparing the gut microbiota composition of A. mellifera in respect to that of A. cerana (PERMANOVA p‐value of <0.05), as depicted by a beta‐diversity investigation represented through principal coordinate analysis (PCoA) based on Bray–Curtis dissimilarity index (Figure 3). More in specific, with the exclusion of B. asteroides and Gilliamella apicola, the other five most prevalent taxa above reported were identified with a significant low prevalence in the gut of A. cerana (Figure 3). In contrast, Apibacter mensalis taxon was prevalent in 62% of the A. cerana samples and completely undetected in the microbiota of A. mellifera, highlighting a strict co‐evolutionary route of this microorganism and its host. In addition, the dissection of A. mellifera and A. cerana microbiomes revealed a number of putative unknown species that have not been classified so far with a prevalence >50% belonging to the genus Lactobacillus, Bifidobacterium, Gilliamella, Snodgrassella, and Bartonella, showing that the microbial biodiversity of the honey bee has not been fully explored (Figure 3).
FIGURE 3.
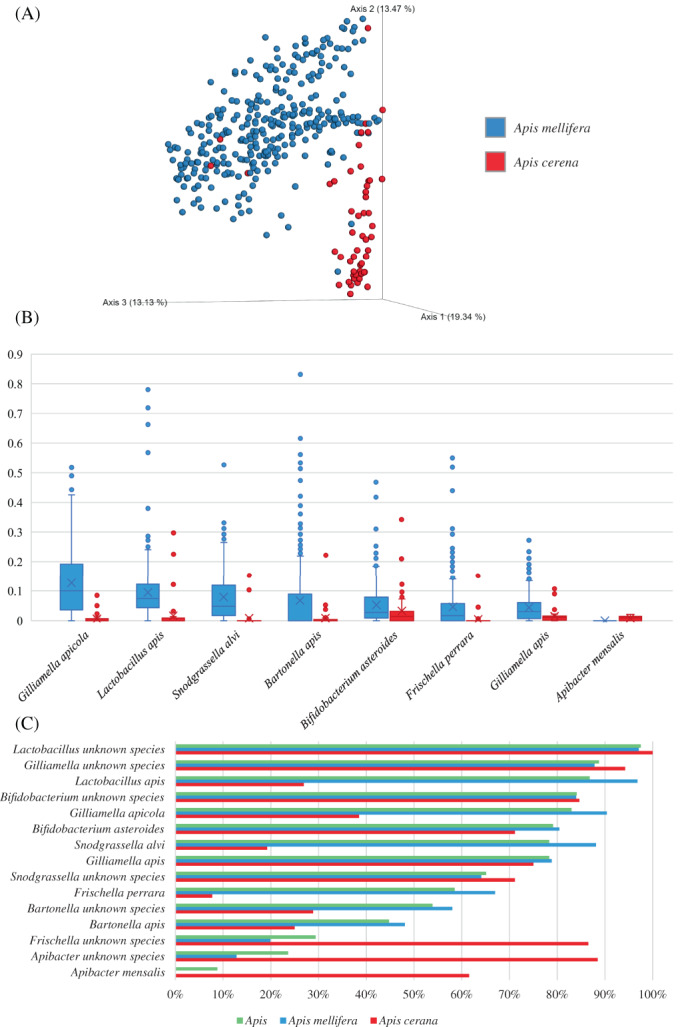
Taxonomic composition of 366 gut microbiomes of Apis mellifera and Apis cerana. (A) Shows a principal coordinate analysis (PCoA) based on microbial distribution using the Bray–Curtis index represented in different colours by means of A. mellifera (light blue) and A. cerana (red). (B) Displays a Whisker plot based on average abundances of main microbial constituents inhabiting the gut of A. mellifera (light blue) and A. cerana (red). The y‐axis shows the average abundance of each observation. Dots reflect the distribution of a data set, while the boxes represent 50% of the data set, distributed between the first and third quartiles. The median divides the boxes into the interquartile range, while the X represents the mean. The lines extending vertically outside the boxes show the outlier range. (C) Depicts the prevalent bacterial taxa identified among all processed samples (green), A. mellifera microbiomes (light blue), and A. cerana microbiomes (red).
Focusing on bifidobacteria, B. asteroides was the taxa with the highest average abundance of 5%, followed by B. indicum and B. coryneforme with average abundances of 0.26 and 0.25%, respectively. Notably, the near‐identical average abundance of the latter two taxa reflects the inability to discriminate between them since their ANI has been previously reported to be >98% (Lugli et al., 2014). Thus, based on this notion, it should be more accurate to state that B. indicum/coryneforme was identified with an average abundance of 0.5%. Furthermore, a portion of the honey bee microbiome was represented by unclassified species of the Bifidobacterium genus (average abundance of 6%), highlighting the gap in knowledge concerning bifidobacteria and honey bees (Figure 3). Interestingly, none of the bifidobacterial species isolated from other social insects was detected from the hindgut of A. mellifera and A. cerana, that is, B. actinocoloniiforme, B. aemilianum, B. bohemicum, B. bombi, B. commune and B. xylocopae. These findings highlighted a specialization of bifidobacteria for certain insects since no trace of the latter taxa were isolated from Bombus and Xylocopa violacea was identified in honey bees.
These findings confirmed that B. asteroides is the most common member of the genus Bifidobacterium harboured by A. mellifera and A. cerana. However, up‐to‐date bioinformatics pipelines cannot distinguish between the above predicted B. asteroides‐related clusters (CL1–CL8). For this reason, a further investigation was conducted to explore the actual distribution of all different putative species recognized as members of the B. asteroides taxon.
Bifidobacterial distribution among Apis hindgut revealed a complex Bifidobacterium‐based ecosystem
An in‐house database was built encompassing representative strain of each B. asteroides‐related cluster to investigate their distribution among A. mellifera and A. cerana. In this context, the chromosomal sequences of the 35 reclassified B. asteroides were subdivided from CL1 to CL8, representing both described and putative novel species, that is, B. sp. nov. CL1, B. sp. nov. CL2, B. choladohabitans (CL3), B. mizhiense (CL4), B. apousia (CL5), B. asteroides (CL6), B. sp. nov. CL7 and B. polysaccharolyticum (CL8) (Table 1). Therefore, 366 honey bee microbiomes were re‐analysed allowing to taxonomically classify the previous DNA widely assigned to B. asteroides to the proper B. asteroides CL.
Interestingly, collecting data taxonomically classified to members of the genus Bifidobacterium, the amount of predicted DNA belonging to unknown species of bifidobacteria was reduced from 55% to 9%, demonstrating that our in‐house database constituted by the eight B. asteroides CLs was able to disentangle 91% of bifidobacteria harboured by A. mellifera and A. cerana (Figure 4). Similarly, the amount of B. asteroides DNA was also reduced from 42% to 3% (B. asteroides [CL6]), revealing how limited was the contribution of the actual B. asteroides taxon to the microbiota variability of honey bees and uncovering a more complex bifidobacterial ecosystem. Instead, those bifidobacterial taxa highly distributed among honey bee microbiomes were represented by B. choladohabitans (CL3) (31%), B. polysaccharolyticum (CL8) (17%), B. apousia (CL5) (15%) and B. sp. nov. CL2 (10%) (Figure 4).
FIGURE 4.
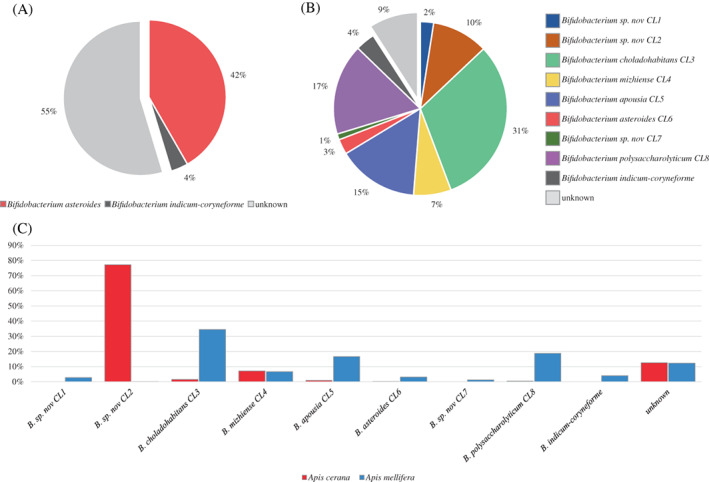
Distribution of Bifidobacterium asteroides‐related taxa among 366 gut microbiomes of Apis. mellifera and Apis cerana. (A) Exhibits the relative abundance of bifidobacteria at species level using a non‐up‐to‐date database for taxonomical classification of bifidobacteria, while panel B shows the distribution of B. asteroides‐related taxa using an updated database built using the reclassified genomes analysed in this project. (C) Displays the distribution of B. asteroides‐related taxa A. mellifera microbiomes (light blue) and A. cerana microbiomes (red).
Comparing taxonomic data between A. mellifera and A. cerana microbiomes, a very diverse distribution of bifidobacteria emerged, showing B. asteroides‐related taxa specialized in the colonization of a specific Apis species (Figure 4). As already depicted in Figure 3, the gut microbiota of A. mellifera and A. cerana showed significant differences in their microbiota composition, which were also confirmed by the intraspecies variability of B. asteroides (Figure 4). In this context, B. sp. nov. CL2 represented 77% of bifidobacterial DNA in A. cerana, highlighting the predominance of this single CL, while in A. mellifera, no data regarding CL2 was identified. Nevertheless, the distribution of B. asteroides‐related taxa in A. mellifera, with the exception of CL2, followed the data reported above, with most abundant CLs represented by B. choladohabitans (CL3) (34%), B. polysaccharolyticum (CL8) (19%), and B. apousia (CL5) (17%). Furthermore, B. mizhiense (CL4) was the only B. asteroides‐related taxa identified with the same abundance of 7% in both species of Apis, while B. sp. nov. CL7 was the rarest taxon with an abundance of 1% in A. mellifera.
A network based on their abundances was produced to unravel associations between the above‐described taxa (Figure 5), using data of negative and positive significant correlations retrieved from the taxonomic classification of Apis microbiomes. The analysis highlighted positive correlations between the most prevalent taxa represented by Frischella perrara, Gilliamella apicola, Gilliamella apis, Lactobacillus apis and Snodgrassella alvi, verifying an interconnection among the core microbiota of honey bee (Figures 3 and 5). Furthermore, positive correlations revealed complex interconnections between all B. asteroides‐related CLs except for CL2, constituting the core Bifidobacterium taxa of A. mellifera (Figure 5). Instead, CL2 displayed positive correlations only with CL4 and Apibacter mensalis, representing those taxa inhabiting the hindgut of A. cerana. Thus, the correlation analysis revealed a pattern of multi‐bifidobacterial taxa within the gut of A. mellifera that co‐exist with the host, while in A. cerana, a simple distribution characterized by members of CL2 (Figure 5). To understand the completely different pattern of bifidobacteria inhabiting the gut of the two species of Apis, further investigation on their genomic characteristics may unveil their specificity with the host.
FIGURE 5.
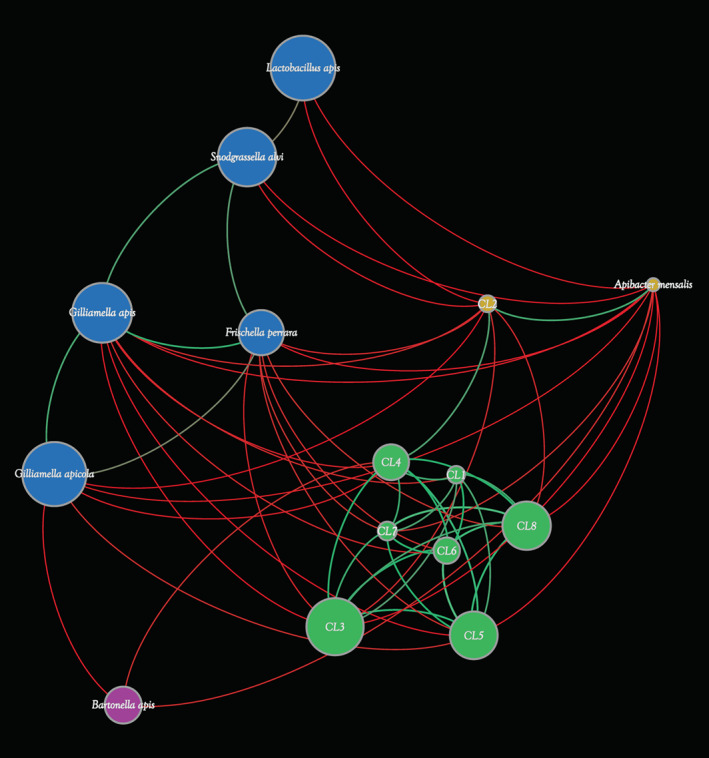
Network analysis based on the correlation of identified main microbial constituent of the honey bee gut microbiota. Circles represent the main taxa, of which Bifidobacterium asteroides‐related taxa are reported as CLs (CL1–CL8), and their diameter is proportional to their average abundance in the honey bee gut microbiomes. Each colour indicates a different cluster generated through positive interactions (green lines) and negative interactions (red lines).
Reconstruction of B. asteroides‐related species and their degradative capability
The shotgun metagenomic data of honey bee gut was submitted to genome reconstruction, allowing the collection of 990,385,248 bases of bifidobacterial DNA. The reconstruction of B. asteroides‐related chromosomal sequences revealed 391 partially reconstructed genomes whose size was >500 Kb, of which 151 belonged to members of CL3 (Table S5). On the contrary, five and seven partially reconstructed genomes were classified as members of CL7 and CL6, followed by 17 genomes of CL1, reflecting their lower average abundance and prevalence identified between samples (Table S5).
Partially reconstructed B. asteroides‐related genomes with completeness >80% and contamination <3% (n = 63) were then employed to validate the eight CLs defined through genomic analyses. A further pangenome analysis was performed based on B. asteroides‐related coding sequences allowing the generation of a phylogenetic tree extending the eight clusters except for CL6 and CL7 (Figure S2). More in detail, the analysis highlighted the predominance of specific CLs within the gut of A. mellifera and A. cerana, represented by B. choladohabitans (CL3), B. sp. nov. CL2, and B. polysaccharolyticum (CL8), which are in high abundance in certain metagenomes and thus allowed the reconstruction of nearly complete genome sequences. As reported in Figure S2, the existence of a B. sp. nov. CL2 was validated, including 30 genomes of high quality to the analysis, while no additional high‐quality genomes were reconstructed for CL7. Notably, since no near‐complete B. asteroides (CL6) genome sequences were reconstructed, the analysis uncovered again that the common ecological attribution of this species to honey bee had been an erroneous simplification of an actual more complex bifidobacterial ecosystem of A. mellifera.
Coding sequences of reclassified B. asteroides‐related taxa (n = 35) and assembled high‐quality genomes (n = 63) were used to access the metabolic signature of bifidobacteria inhabiting the gut of A. mellifera and A. cerana aiming to dissect their carbohydrate‐active enzyme repertoire. Identified genes encoding glycosyl hydrolases (GHs) were used to cluster together the 98 bifidobacterial strains resulting in the establishment of six enzymatic clusters (ECs) that resemble the distribution of predicted CLs (Figure 6). More in specific, members of CL1, CL2, CL3, CL6 and CL8 generated their own EC, highlighting similar GH profiles, while members of CL4 and CL5 clustered together in a common EC. Notably, the single strain representing B. sp. nov. CL7 was excluded from the analysis. Altogether a consistent GH core was identified between all B. asteroides‐related strains, encompassing GH43, GH3, GH31, GH2, GH42, GH30, GH13, GH32 and GH36 (Figure 6), reflecting metabolic activities towards a broad range of simple carbohydrates such as sucrose, glucose, fructose, galactose, ribose, mannose, melibiose, raffinose, xylose and cellobiose (Bottacini et al., 2012;Chen et al., 2021; Li et al., 2022). Recently, it has been demonstrated that an increase of sugars in flower nectar, the honey bee food, resulted in an increase in the relative abundance of bifidobacteria within the microbiome driven by their high‐grown performance on simple carbohydrates (Wang et al., 2020). Notably, B. sp. nov. CL1 was characterized by the lowest number of genes encoding GHs, that is, 27, and the unique presence of GH144 encoding for enzymes that hydrolyze glycosidic linkages in glucans. In contrast, B. polysaccharolyticum (CL8) and B. sp. nov. CL2 showed the highest repertoire of GHs, that is, 68 and 66, respectively (Figure 6). While members of CL8 were characterized by the unique presence of GH35 encoding galactosidase enzymes, members of CL2 were not characterized by the presence of any unique GH signature. In addition, members of B. choladohabitans (CL3) were characterized by the unique presence of GH77 encoding for an amylomaltase, while members of B. asteroides (CL6) possessed a unique repertoire of GHs, represented by GH4, GH67 and GH136 associated with genes encoding α‐glucosidase, α‐galactosidase, α‐glucuronidase and lacto‐N‐biosidase.
FIGURE 6.
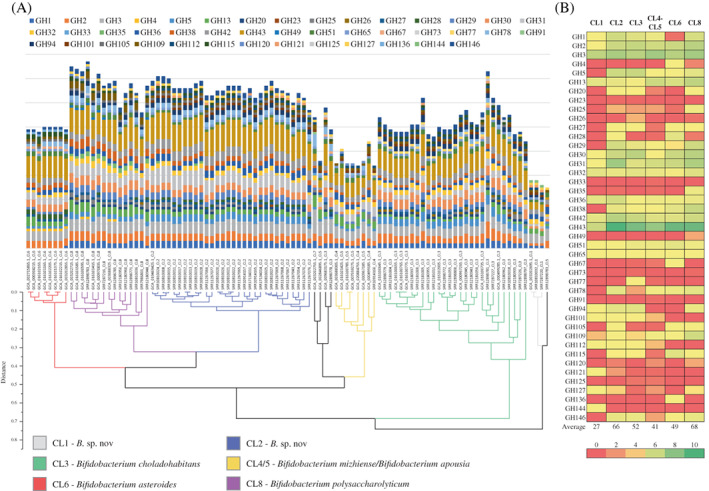
Predicted glycobiome of Bifidobacterium asteroides‐related taxa. (A) Depicts a hierarchical clustering based on predicted GH families of the 98 B. asteroides‐related strains retrieved from NCBI and reconstructed in this work through honey bee gut microbiomes. The predicted GH arsenal for every analysed bifidobacterial strain is represented by a bar plot, while B. asteroides‐related clusters are highlighted with different colours. (B) Displays a heat map with the average number of predicted genes encoding GHs in each CL.
Together with GHs, the genes encoding for polysaccharide lyases (PLs) and carbohydrate esterases (CEs) repertoire were also predicted from the genomes of the 98 B. asteroides‐related strains revealing the absence of any specific PLs and a conserved CEs encoding gene among CLs (Table S6). In detail, a single copy of CE1 and CE9 was shared between all genomes representing a core element of B. asteroides‐related strains, revealing no difference between the eight CLs. Moreover, the genomes of all members of CL2 and CL6 encompassed a copy of CE8 encoding gene for a pectin methylesterase, representing a species‐specific CE. Furthermore, only a slight difference between species was identified for CL3 encoding genes, where half of the genomes of the members harboured a copy of a CE2 encoding for an acetyl xylan esterase.
The hindgut of A. mellifera is distinguished by the coexistence of a plethora of different bifidobacterial species, each characterized by the usage of unique GHs to complement their ability to compete in an ecological niche rich in simple sugars. In contrast, members of CL2 that specifically colonize the gut of A. cerana did not exhibit any unique GH signature. Instead, they shared with CL6 members the specific EC8 encoding gene for a pectin methylesterase.
CONCLUSIONS
For years many bifidobacterial strains have been erroneously classified as belonging to the B. asteroides taxon, resulting in an overestimation of this species inhabiting the hindgut of honey bees through metagenomic taxonomic profiling. Here, we characterize the existence of eight species among the so‐called B. asteroides taxon by using genomics, metagenomics, and phylogenetics analyses. While five of them were validated by the characterization of isolates (Chen et al., 2021; Li et al., 2022), three are still missing a conventional classification. Interestingly, investigating their distribution among honey bee microbiomes, one unclassified species was found predominant in the gut of A. cerana and utterly absent in A. mellifera, opening a new insights regarding bifidobacterial species distribution among honey bees and their coevolution with the host. Notably, the host‐specificity of such B. asteroides‐related species may represent potential valuable novel candidates of next generation of probiotics for honey bees. Furthermore, a glycobiome analysis, encompassing reclassified B. asteroides‐related taxa and assembled high‐quality genomes, revealed a conserved repertoire of GHs reflecting their metabolic behaviours towards a broad range of simple carbohydrates abundant in the gut of honey bees.
AUTHOR CONTRIBUTIONS
Gabriele Andrea Lugli performed bioinformatics analyses, wrote the manuscript and designed the study; Federico Fontana and Chiara Tarracchini managed the metadata and data results; Leonardo Mancabelli performed the statistical analyses; Christian Milani and Francesca Turroni supervised the project and edited the manuscript; Marco Ventura supervised the project and designed the study.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
Supporting information
Figure S1. Selection of unclassified bifidobacterial species related to Bifidobacterium asteroides. The hierarchical clustering is based on an ANI values matrix of all 187 genome sequences collected from the NCBI database. The additional 26 genomes identified as related to the B. asteroides taxon are highlighted in green.
Figure S2. Phylogenomic tree of 98 Bifidobacterium asteroides‐related strains based on the concatenation of core protein sequences of reconstructed genomes retrieved from the metagenomes of the hindgut of honey bees. Different colours highlight the division into eight taxa (named from CL1 to CL8). The phylogenetic tree was constructed by the neighbour‐joining method, with the genome sequence of Bifidobacterium actinocoloniiforme DSM 22766 as an outgroup. Bootstrap percentages above 50 are shown at node points based on 1000 replicates of the phylogenetic tree.
Table S1. Unclassified bifidobacteria in the NCBI repository
Table S2. Quality control of Bifidobacterium asteroides‐related strains
Table S3. Shotgun metagenomic samples of Apis mellifera and Apis cerana
Table S4. Filtering table of shotgun metagenomics data
Table S5. Quality control of reconstructed Bifidobacterium asteroides‐related strains
Table S6. Carbohydrate esterase (CE) profiling of Bifidobacterium asteroides‐related strains
ACKNOWLEDGEMENTS
We thank GenProbio Srl for the financial support of the Laboratory of Probiogenomics. Part of this research is conducted using the high‐performance computing (HPC) facility of the University of Parma. Francesca Turroni is funded by Italian Ministry of Health through the Bando Ricerca Finalizzata (Grant Number GR‐2018‐12365988). Open Access Funding provided by Universita degli Studi di Parma within the CRUI‐CARE Agreement.
Lugli, G.A. , Fontana, F. , Tarracchini, C. , Mancabelli, L. , Milani, C. , Turroni, F. et al. (2022) Exploring the biodiversity of Bifidobacterium asteroides among honey bee microbiomes. Environmental Microbiology, 24(12), 5666–5679. Available from: 10.1111/1462-2920.16223
Funding information Ministero della Salute, Grant/Award Number: GR‐2018‐12365988; University of Parma; Ministry of Health
DATA AVAILABILITY STATEMENT
Raw sequences of shotgun metagenomics experiments are accessible through SRA study listed in Table S3. In addition, genome sequences used in this project have been reported in Tables 1 and S1.
REFERENCES
- Alberoni, D. , Gaggìa, F. , Baffoni, L. , Modesto, M.M. , Biavati, B. & Di Gioia, D. (2019) Bifidobacterium xylocopae sp. nov. and Bifidobacterium aemilianum sp. nov., from the carpenter bee (Xylocopa violacea) digestive tract. Systematic and Applied Microbiology, 42, 205–216. [DOI] [PubMed] [Google Scholar]
- Bankevich, A. , Nurk, S. , Antipov, D. , Gurevich, A.A. , Dvorkin, M. , Kulikov, A.S. et al. (2012) SPAdes: a new genome assembly algorithm and its applications to single‐cell sequencing. Journal of Computational Biology, 19, 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottacini, F. , Milani, C. , Turroni, F. , Sánchez, B. , Foroni, E. , Duranti, S. et al. (2012) Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PLoS One, 7, e44229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunesova, V. , Vlkova, E. , Rada, V. , Killer, J. & Musilova, S. (2014) Bifidobacteria from the gastrointestinal tract of animals: differences and similarities. Benef Microbes, 5, 377–388. [DOI] [PubMed] [Google Scholar]
- Chen, J. , Wang, J. & Zheng, H. (2021) Characterization of Bifidobacterium apousia sp. nov., Bifidobacterium choladohabitans sp. nov., and Bifidobacterium polysaccharolyticum sp. nov., three novel species of the genus Bifidobacterium from honey bee gut. Systematic and Applied Microbiology, 44, 126247. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Ye, W. , Zhang, Y. & Xu, Y. (2015) High speed BLASTN: an accelerated MegaBLAST search tool. Nucleic Acids Research, 43, 7762–7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danforth, B. (2007) Bees. Current Biology, 17, R156–R161. [DOI] [PubMed] [Google Scholar]
- Drula, E. , Garron, M.L. , Dogan, S. , Lombard, V. , Henrissat, B. & Terrapon, N. (2022) The carbohydrate‐active enzyme database: functions and literature. Nucleic Acids Research, 50, D571–D577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright, A.J. , Van Dongen, S. & Ouzounis, C.A. (2002) An efficient algorithm for large‐scale detection of protein families. Nucleic Acids Research, 30, 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo‐Cantabrana, C. , Delgado, S. , Ruiz, L. , Ruas‐Madiedo, P. , Sánchez, B. & Margolles, A. (2017) Bifidobacteria and their health‐promoting effects. Microbiology Spectrum, 5, BAD‐0010‐2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt, D. , Chen, G.L. , LoCascio, P.F. , Land, M.L. , Larimer, F.W. & Hauser, L.J. (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics, 11, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, C. , Rodriguez‐R, L.M. , Phillippy, A.M. , Konstantinidis, K.T. & Aluru, S. (2018) High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nature Communications, 9, 5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K. , Rozewicki, J. & Yamada, K.D. (2018) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics, 20, 1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killer, J. , Kopečný, J. , Mrázek, J. , Koppová, I. , Havlík, J. , Benada, O. et al. (2011) Bifidobacterium actinocoloniiforme sp. nov. and Bifidobacterium bohemicum sp. nov., from the bumblebee digestive tract. International Journal of Systematic and Evolutionary Microbiology, 61, 1315–1321. [DOI] [PubMed] [Google Scholar]
- Killer, J. , Kopecny, J. , Mrazek, J. , Rada, V. , Benada, O. , Koppova, I. et al. (2009) Bifidobacterium bombi sp. nov., from the bumblebee digestive tract. International Journal of Systematic and Evolutionary Microbiology, 59, 2020–2024. [DOI] [PubMed] [Google Scholar]
- Konstantinidis, K.T. , Ramette, A. & Tiedje, J.M. (2006) The bacterial species definition in the genomic era. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 361, 1929–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, K.B. (2003) ClustalW‐MPI: ClustalW analysis using distributed and parallel computing. Bioinformatics, 19, 1585–1586. [DOI] [PubMed] [Google Scholar]
- Li, T.T. , Zhang, H.X. & Gu, C.T. (2022) Bifidobacterium mizhiense sp. nov., isolated from the gut of honeybee (Apis mellifera). International Journal of Systematic and Evolutionary Microbiology, 5, 005390. [DOI] [PubMed] [Google Scholar]
- Lugli, G.A. , Alessandri, G. , Milani, C. , Viappiani, A. , Fontana, F. , Tarracchini, C. et al. (2021) Genetic insights into the dark matter of the mammalian gut microbiota through targeted genome reconstruction. Environmental Microbiology, 23, 3294–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli, G.A. , Calvete‐Torre, I. , Alessandri, G. , Milani, C. , Turroni, F. , Laiolo, P. et al. (2021) Phylogenetic classification of ten novel species belonging to the genus Bifidobacterium comprising B. phasiani sp. nov., B. pongonis sp. nov., B. saguinibicoloris sp. nov., B. colobi sp. nov., B. simiiventris sp. nov., B. santillanense sp. nov., B. miconi . Syst Appl Microbiol, 44, 126273. [DOI] [PubMed] [Google Scholar]
- Lugli, G.A. , Duranti, S. , Milani, C. , Mancabelli, L. , Turroni, F. , van Sinderen, D. et al. (2019) Uncovering bifidobacteria via targeted sequencing of the mammalian gut microbiota. Microorganisms, 7, 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli, G.A. , Milani, C. , Duranti, S. , Alessandri, G. , Turroni, F. , Mancabelli, L. et al. (2019) Isolation of novel gut bifidobacteria using a combination of metagenomic and cultivation approaches. Genome Biology, 20, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli, G.A. , Milani, C. , Duranti, S. , Mancabelli, L. , Mangifesta, M. , Turroni, F. et al. (2018) Tracking the taxonomy of the genus Bifidobacterium based on a phylogenomic approach. Applied and Environmental Microbiology, 84, e02249–e02217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli, G.A. , Milani, C. , Mancabelli, L. , Van Sinderen, D. & Ventura, M. (2016) MEGAnnotator: a user‐friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiology Letters, 363, fnw049. [DOI] [PubMed] [Google Scholar]
- Lugli, G.A. , Milani, C. , Turroni, F. , Duranti, S. , Ferrario, C. , Viappiani, A. et al. (2014) Investigation of the evolutionary development of the genus bifidobacterium by comparative genomics. Applied and Environmental Microbiology, 80, 6383–6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli, G.A. , Milani, C. , Turroni, F. , Duranti, S. , Mancabelli, L. , Mangifesta, M. et al. (2017) Comparative genomic and phylogenomic analyses of the Bifidobacteriaceae family. BMC Genomics, 18, 568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani, C. , Casey, E. , Lugli, G.A. , Moore, R. , Kaczorowska, J. , Feehily, C. et al. (2018) Tracing mother‐infant transmission of bacteriophages by means of a novel analytical tool for shotgun metagenomic datasets: METAnnotatorX. Microbiome, 6, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani, C. , Duranti, S. , Bottacini, F. , Casey, E. , Turroni, F. , Mahony, J. et al. (2017) The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiology and Molecular Biology Reviews, 81, e00036–e00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani, C. , Lugli, G.A. , Duranti, S. , Turroni, F. , Bottacini, F. , Mangifesta, M. et al. (2014) Genomic encyclopedia of type strains of the genus Bifidobacterium . Applied and Environmental Microbiology, 80, 6290–6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani, C. , Lugli, G.A. , Duranti, S. , Turroni, F. , Mancabelli, L. , Ferrario, C. et al. (2015) Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Scientific Reports, 5, 15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani, C. , Lugli, G.A. , Fontana, F. , Mancabelli, L. , Alessandri, G. , Longhi, G. et al. (2021) METAnnotatorX2: a comprehensive tool for deep and shallow metagenomic data set analyses. mSystems, 6, e00583–e00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks, D.H. , Imelfort, M. , Skennerton, C.T. , Hugenholtz, P. & Tyson, G.W. (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Research, 25, 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praet, J. , Meeus, I. , Cnockaert, M. , Aerts, M. , Smagghe, G. & Vandamme, P. (2015) Bifidobacterium commune sp. nov. isolated from the bumble bee gut. Antonie van Leeuwenhoek, 107, 1307–1313. [DOI] [PubMed] [Google Scholar]
- Regan, T. , Barnett, M.W. , Laetsch, D.R. , Bush, S.J. , Wragg, D. , Budge, G.E. et al. (2018) Characterisation of the British honey bee metagenome. Nature Communications, 9, 4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, M. & Rosselló‐Móra, R. (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proceedings of the National Academy of Sciences of the United States of America, 106, 19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier, H. (1900) Recherches sur la flore intestinale des nourrissons: état normal et pathologique.
- Turroni, F. , Milani, C. , Duranti, S. , Ferrario, C. , Lugli, G.A. , Mancabelli, L. et al. (2018) Bifidobacteria and the infant gut: an example of co‐evolution and natural selection. Cellular and Molecular Life Sciences, 75, 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura, M. , Turroni, F. , Lugli, G.A. & van Sinderen, D. (2014) Bifidobacteria and humans: our special friends, from ecological to genomics perspectives. Journal of the Science of Food and Agriculture, 94, 163–168. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Liu, C. , Liu, Z. , Wang, Y. , Ma, L. & Xu, B. (2020) The different dietary sugars modulate the composition of the gut microbiota in honeybee during overwintering. BMC Microbiology, 20, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Lang, H. , Mu, X. , Zhang, Z. , Su, Q. , Hu, X. et al. (2021) Honey bee genetics shape the strain‐level structure of gut microbiota in social transmission. Microbiome, 9, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y. , Zheng, Y. , Wang, S. , Chen, Y. , Tao, J. , Chen, Y. et al. (2022) Genetic divergence and functional convergence of gut bacteria between the eastern honey bee Apis cerana and the Western honey bee Apis mellifera . Journal of Advanced Research, 37, 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Yohe, T. , Huang, L. , Entwistle, S. , Wu, P. , Yang, Z. et al. (2018) DbCAN2: a meta server for automated carbohydrate‐active enzyme annotation. Nucleic Acids Research, 46, W95–W101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Wu, J. , Yang, J. , Sun, S. , Xiao, J. & Yu, J. (2012) PGAP: pan‐genomes analysis pipeline. Bioinformatics, 28, 416–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Selection of unclassified bifidobacterial species related to Bifidobacterium asteroides. The hierarchical clustering is based on an ANI values matrix of all 187 genome sequences collected from the NCBI database. The additional 26 genomes identified as related to the B. asteroides taxon are highlighted in green.
Figure S2. Phylogenomic tree of 98 Bifidobacterium asteroides‐related strains based on the concatenation of core protein sequences of reconstructed genomes retrieved from the metagenomes of the hindgut of honey bees. Different colours highlight the division into eight taxa (named from CL1 to CL8). The phylogenetic tree was constructed by the neighbour‐joining method, with the genome sequence of Bifidobacterium actinocoloniiforme DSM 22766 as an outgroup. Bootstrap percentages above 50 are shown at node points based on 1000 replicates of the phylogenetic tree.
Table S1. Unclassified bifidobacteria in the NCBI repository
Table S2. Quality control of Bifidobacterium asteroides‐related strains
Table S3. Shotgun metagenomic samples of Apis mellifera and Apis cerana
Table S4. Filtering table of shotgun metagenomics data
Table S5. Quality control of reconstructed Bifidobacterium asteroides‐related strains
Table S6. Carbohydrate esterase (CE) profiling of Bifidobacterium asteroides‐related strains
Data Availability Statement
Raw sequences of shotgun metagenomics experiments are accessible through SRA study listed in Table S3. In addition, genome sequences used in this project have been reported in Tables 1 and S1.


