Abstract
Centromeres are known to cluster around nucleoli in Drosophila and mammalian cells, but the significance of the nucleoli–centromere interaction remains underexplored. To determine whether the interaction is dynamic under different physiological and pathological conditions, we examined nucleolar structure and centromeres at various differentiation stages using cell culture models and the results showed dynamic changes in nucleolar characteristics and nucleoli–centromere interactions through differentiation and in cancer cells. Embryonic stem cells usually have a single large nucleolus, which is clustered with a high percentage of centromeres. As cells differentiate into intermediate states, the nucleolar number increases and the centromere association decreases. In terminally differentiated cells, including myotubes, neurons, and keratinocytes, the number of nucleoli and their association with centromeres are at the lowest. Cancer cells demonstrate the pattern of nucleoli number and nucleoli–centromere association that is akin to proliferative cell types, suggesting that nucleolar reorganization and changes in nucleoli–centromere interactions may play a role in facilitating malignant transformation. This idea is supported in a case of pediatric rhabdomyosarcoma, in which induced differentiation reduces the nucleolar number and centromere association. These findings suggest active roles of nucleolar structure in centromere function and genome organization critical for cellular function in both normal development and cancer.
INTRODUCTION
Nucleoli are multifunctional nuclear organelles beyond being the centers of ribosome synthesis. An increasing number of cellular functions are found associated with this prominent organelle, including signal recognition particle assembly, cell cycle regulation, p53 metabolism, stress sensing, gene regulation, and miRNA metabolism (Pederson, 2011; Bersaglieri and Santoro, 2019; Iarovaia et al., 2019). More recently, it has been discovered that nucleolar size and area are related to aging in vitro and in vivo (Pederson, 1998, 2011; Boisvert et al., 2007; Iarovaia et al., 2019), further demonstrating the multifunctionality of nucleoli.
Over the past decade, increasing evidence illustrates the spatial interaction of nucleoli with specific domains of chromosomes (Dillinger et al., 2017; Bersaglieri and Santoro, 2019; Cerqueira and Lemos, 2019). Several studies, including next-generation sequencing of nucleolar DNA and HiC experiments demonstrate the association between nucleoli/rDNA and many parts of all chromosomes. These chromosome regions were termed nucleoli-associated domains (NADs; Nemeth et al., 2010; van Koningsbruggen et al., 2010; Yu and Lemos, 2016; Yu and Lemos, 2018; Diesch et al., 2019). NADs are predominantly constitutive and/or facultative heterochromatin, both in human and mice (Vertii et al., 2019; Bizhanova et al., 2020; Bizhanova and Kaufman, 2021). In general, NAD association with the nucleoli correlates with lower levels of gene expression (Bersaglieri and Santoro, 2019; Bizhanova and Kaufman, 2021).
Centromeres are often found clustering around nucleoli in Drosophila and mammalian cells (Stahl et al., 1976; Ochs and Press, 1992). In addition, several findings indicate functional connections between centromeres and nucleoli. Centromeres are defined by the presence of a distinct class of nucleosomes containing the centromere-specific histone H3 variant, CENP-A (centromere protein-A; Sharma et al., 2019). Nucleolar protein NPM1 interacts with CENP-A (Foltz et al., 2006, 2009), HJURP, a nucleolar localized CENP-A chaperone (Dunleavy et al., 2009; Foltz et al., 2009; Shuaib et al., 2010), and plays a role in centromere assembly. A constitutive centromere-associated network protein, CENP-C, requires an interaction with the nucleolus and RNA for centromere assembly (Wong et al., 2007). Furthermore, in Drosophila, the clustering of centromeres around the nucleolus depends on NPM1 and Modulo (Chen et al., 2012; Padeken et al., 2013). These findings underscore the potential functional significance of the nucleoli–centromere association.
To determine the importance of nucleolar–centromere associations in cellular function, we examined the dynamics of these interactions in cell culture models that define multiple distinct developmental stages, including human embryonic stem cell (hESC), myoblast–myotube, neuroblast–neuron, and epidermal keratinocyte differentiation models. To analyze the pathological redistribution of centromeres, we also compared a panel of normal and cancer cells. Our data suggest that nucleolar structure and nucleoli–centromere interactions are regulated as cells experience different physiological and pathological conditions.
RESULTS AND DISCUSSION
A majority of centromeres associate with nucleoli in pluripotent embryonic stem cells
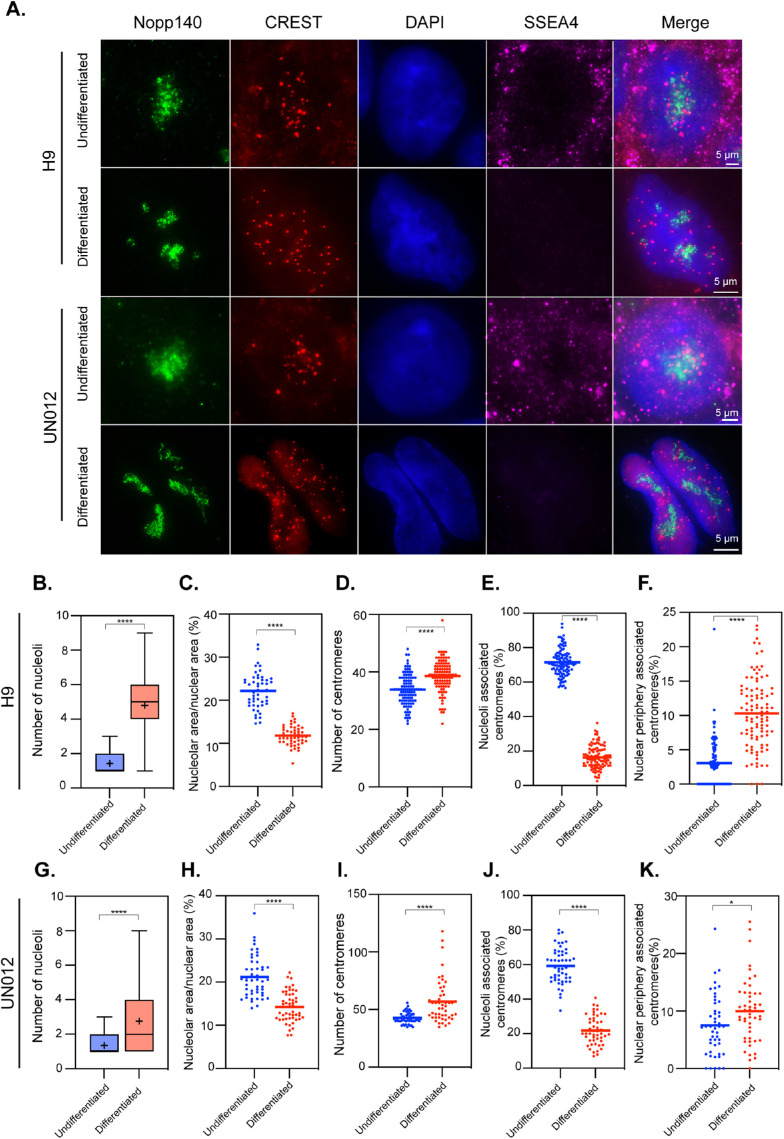
Embryonic stem cells represent a pluripotent stage of human embryonic development. The H9 embryonic stem cell line was expanded according to established protocols and allowed to differentiate by switching to 10% serum-supplemented DMEM over a 10-d period. Differentiated cells underwent morphological changes, spreading out and dispersing away from the original stem cell colonies. Cells were analyzed by immunofluorescence using antibodies that label nucleoli (Nopp140), centromeres (CREST), and a pluripotent stem cell marker, SSEA4. Cells were imaged through Z sectioning covering entire nuclei and the images presented (Figure 1A) are the maximal projection of the Z stacks to capture all the centromeres within cells. The number of detectable centromeres (individuals with clear separation from one another) and their associations with nucleoli were measured. As shown in Figure 1A, the undifferentiated H9 cells generally contained one to two large nucleoli, similar to previous observations (Meshorer and Misteli, 2006; Gupta and Santoro, 2020). These cells often displayed packed nucleoli-associated centromere clusters (Figure 1A, consider “CREST” and “Merge” panels). The detected number of centromeres was less than the total number of chromosomes (Figure 1D) perhaps due to their highly clustered nature around the nucleoli, leading to some partial visual overlap of individual centromeres. On average, 71% of centromeres are spatially associated with nucleoli in the undifferentiated H9 cells (Figure 1E). To determine whether this is a general property of the pluripotent state, we examined induced pluripotent cells: UN012. UN012 cells showed 59% of centromeres associated with nucleoli in the undifferentiated state (Figure 1, A and J).
FIGURE 1:
Nucleolus–centromere interactions decrease as H9 human embryonic stem cells and SSL012 iPS cells differentiate. Nucleoli are immunolabeled with anti-Nopp140 antibody (green) and centromeres by CREST (red) antibodies. Pluripotency was verified using SSEA4 antibody, which is lost in differentiating cells (A). The number of nucleoli (B, G) increases as nucleolar area (ratio of nucleolar area over nuclear area) decreases (C, H). The number of countable centromeres increases as cells differentiate (D, I). The average percentage of centromeres associated with nucleoli reduces significantly as cells differentiate (E, J). In comparison, the percentage of centromeres associating with the nuclear periphery is increased (F, K). Box plots indicate 25th–75th percentile values, with whiskers indicating minimum and maximum observed values. Scatter plots indicate the value of individual cellular measurements, with the horizontal line indicating the mean. Statistical significance was computed as two-tailed t tests of means with unequal variance. Bar = 5 μm. n > 50 per cell type. *: p < 0.05; ****: p < 0.0001.
When H9 and UN012 cells were induced to differentiate, they lost the stem marker, SSEA4 expression (Figure 1A). In these cells, the number of nucleoli increased (Figure 1, A, B, and G) while nucleolar area reduced (Figure 1, C and H). The association of centromeres with nucleoli was also significantly reduced (Figure 1, E and J). Conversely, the association of centromeres with the nuclear periphery (the edge of DAPI staining) increased (Figure 1, F and K). These findings demonstrate major nucleoli–centromeres reorganization during hESC or iPSC differentiation consistent with findings of changes in nucleoli (Meshorer and Misteli, 2006) and in chromatin organization during hESC differentiation (Bersaglieri and Santoro, 2019; Gupta and Santoro, 2020; Bersaglieri et al., 2022). Although nucleolar proteins such as NPM1 and HJURP have been shown to play a role in CENP-A deposition (Foltz et al., 2009; Padeken et al., 2013; Fujimura et al., 2020), the extent of the association of centromeres with nucleoli seems unlikely to be explained by the cell cycle–specific (early G1) process of centromere assembly, suggesting additional significance for the nucleoli–centromere interactions at the pluripotent stage. As hESCs or IPSCs represent the earliest stage of development, we examined the nucleoli and nucleoli–centromere relationship using other cell differentiation models from intermediate to terminal differentiation states.
The nucleoli–centromere association is reduced as cells differentiate toward the terminally differentiated state
We utilized three types of differentiation models: myoblasts to myotubes, neuroblasts to neurons, and three-dimensional (3D) human skin equivalent tissue development.
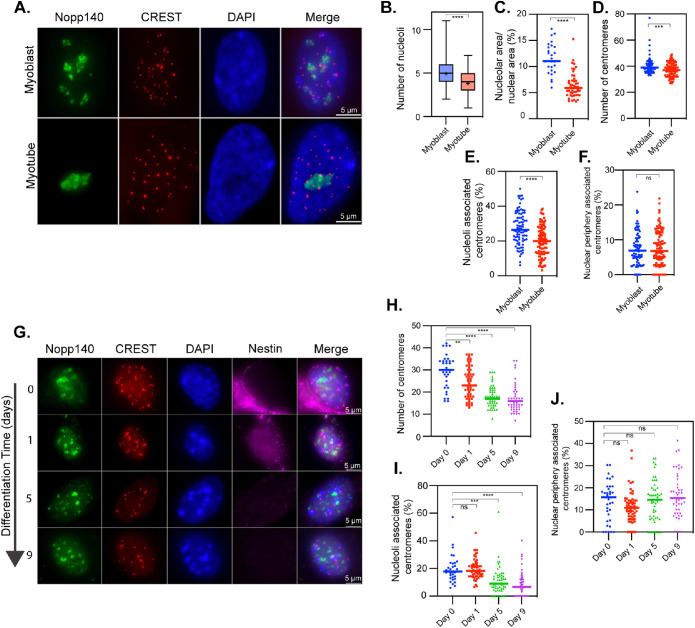
Primary human myoblasts were induced to differentiate into myotubes (see Materials and Methods). Cells were similarly prepared and immunolabeled as for the embryonic stem (ES) cells. To ensure cells were fully differentiated in the myotube analysis, only multinucleated cells were assessed (Supplemental Figure S1). As myoblasts differentiated into myotubes, there was a reduction in the number of nucleoli (Figure 2, A and B), nucleolar area (Figure 2C), and nucleoli-associated centromeres (Figure 2, A and E), but there was no obvious increase in association of centromeres with the nuclear periphery (Figure 2F). The evaluation of nucleoli and centromere localization showed a reduction of the total number of countable centromeres (Figure 2, A and D). Such a reduction is consistent with a reduction in CENP protein retention in differentiated myotubes as previously reported (Swartz et al., 2019) and with our previously published expression data comparing myoblasts and myotubes, which demonstrated significant down-regulation of multiple genes related to centromere structure and function (Supplemental Table S1; MacQuarrie et al., 2013). These findings demonstrate a dynamic reorganization of nucleolar structure and nucleoli–centromere association as cells progress through the myo-differentiation process. Compared to those changes seen in the ES cell system, the same pattern of decreasing nucleolar/nuclear area ratio and nucleoli–centromere association was observed with differentiation.
FIGURE 2:
Nucleolus–centromere interactions decrease as myoblasts and neuroblasts differentiate into myotubes and neurons. Nucleoli are immunolabeled with anti-Nopp140 antibody (green) and centromeres by CREST (red) antibodies (A, G). Only the nuclei in multinucleated myotubes (>1 nucleus per cell) were considered differentiated. Nucleoli number (B) and area (C) decreases as myoblasts differentiate into myotubes. In addition, the number of countable centromeres (D) and the percentage of centromeres associated with nucleoli (E) reduce significantly as cells differentiate into myotubes. However, the percentage of centromeres associating with the nuclear periphery is unchanged (F). (G) Nestin (purple) is a marker of undifferentiated neuroblasts. Countable centromeres (H) and the percentage of centromeres associated with nucleoli (I) reduces as cells differentiate into neurons, while the percentage of centromeres associated with the nuclear periphery is unchanged (J). The boxes in B indicate the 25th–75th percentile values, with the whiskers indicating minimum and maximum observed values. Scatter plots indicate the value of individual cellular measurements, with the horizontal line indicating the mean. Statistical significance was computed as two-tailed t tests of means with unequal variance. Bar = 5 μm. n > 75 per cell type for myocytes and 30–58 per time point for neurons. ns: not significant; *: p < 0.05; **: p < 0.01; ***: p < 0.001; ****: p < 0.0001.
The second system utilizes the differentiation of mouse neuroblasts to neurons. Neuroblasts were isolated directly from mouse brain and were induced to differentiate. Cells were analyzed at day 0 (neuroblast), 1, 5, and 9 to examine changes throughout differentiation. Nestin expression was used as a marker for undifferentiated neuroblasts. As cells differentiated, judged by the loss of nestin signals (Figure 2G), the number of detectable centromeres and the percentage of nucleolar-associated centromeres reduced significantly (Figure 2, H and I). There were no significant changes in centromeres’ association with the nuclear periphery (Figure 2J).
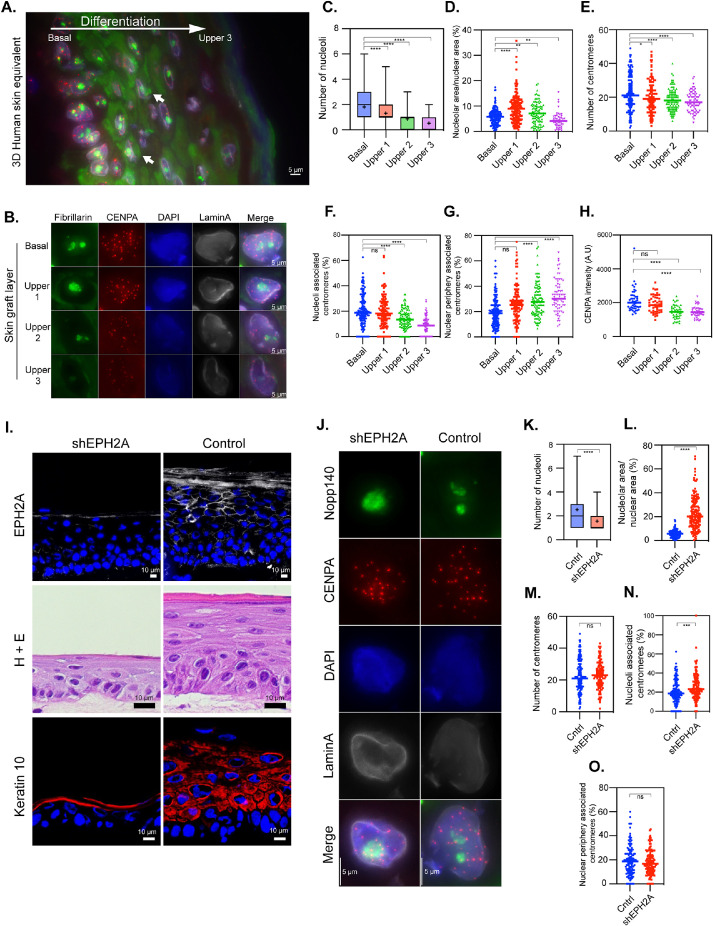
To further confirm the dynamics of nucleoli–centromere interactions through differentiation, we employed 3D human skin equivalents (3D HSE) derived from primary human keratinocytes. In this model, the morphogenesis and differentiation closely recapitulate that of normal human epidermis (Freeman et al., 1976; Arnette et al., 2016). In addition, mature 3D HSE provides a unique advantage because a single tissue cross-section encompasses cells at all stages of epidermal differentiation. This allows for an unbiased view of nucleoli and centromere characteristics through the differentiation process within a single image. In the basal layer, where epidermal stem cells reside, keratinocytes proliferate. Cells exit the cell cycle and move from the basal layer into the spinous, then the granular layer, where the cells are increasingly differentiated. Finally, cells terminally differentiate and undergo specialized cell death and enucleation in the cornified layer. This process of differentiation is initiated by culture at the air–liquid interface and occurs over the course of up to 12 d (Supplemental Figure S2). As shown in Figure 3A, the immunolabeling of a 6-d 3D HSE culture demonstrated clearly definable layers of cells at different stages of epidermal differentiation. As seen in the myocyte system, the number of nucleoli (Figure 3C) and the nucleolar area (Figure 3D) decreased as cellular differentiation progressed, except for layer 2 where the nucleolar area increased compared with the basal layer. It is interesting to note that fibrillarin (Figure 3A; green signal, arrows), a nucleolar preribosomal processing factor, displayed a significant cytoplasmic presence when cells departed from the basal layer and exited the cell cycle, suggesting potential alterations of nucleolar function as cells no longer divided from this layer on.
FIGURE 3:
Nucleolus–centromere interactions decrease as keratinocytes differentiate from basal cells toward cornified layers. Nucleoli are immunolabeled with anti-fibrillarin antibody (green), centromeres by anti-CENPA (red) antibodies, and nuclear lamin by anti-lamin A (grayscale) antibodies. (A) A cross-section of a three-dimensional (3D) human skin equivalent containing all layers in the same image demonstrates the clear changes in centromere numbers and intensity of the signals as well as their association with nucleoli as they develop into cornified layers. The large white arrow indicates the direction of differentiation, and small white arrows indicate examples of extranuclear fibrillarin staining. (B) Typical results of stains in cells from layers ranging from basal to upper layers are as shown. The number of nucleoli (C) and nucleolar area (D) decrease in parallel with the differentiation. Countable centromeres (E) and the percentage of centromeres associated with nucleoli (F) also reduces as cells differentiate. Correspondingly, the percentage of centromeres associating with nuclear periphery is increased (G). (H) CENPA fluorescent intensity is reduced progressively as cells differentiate. (I) Immunolabeling and a hematoxylin and eosin (H+E) staining demonstrate that transduction of a short hairpin construct targeting EPH2A (shEPH2A) reduces EPH2A expression (top row) and impairs differentiation in a 3D human skin equivalent compared with cells transduced with a construct with a scrambled sequence and no target (control). (J) Representative stains of transduced basal layer keratinocytes demonstrate a lower number of nucleoli (K), an increase in nucleolar area relative to nuclear area (L), no difference in centromere number (M), an increase in overlap between centromeres and nucleoli (N), and no difference in centromere overlap with the nuclear periphery (O). Box plots indicate the 25th–75th percentile values, with the whiskers indicating minimum and maximum observed values. Scatter plots indicate the value of individual cellular measurements, with the horizontal line indicating the mean. Statistical significance was computed as two-tailed t tests of means with unequal variance. n ranges from 72 to 154 per cell type (top) and >130 per condition (bottom). Bar = 5 μm. A.U. = arbitrary units; ns: not significant; *: p < 0.05; **: p < 0.01; ***: p < 0.001; ****: p < 0.0001.
CENP-A antibody staining (red) showed that the number of centromeres (Figure 3, B and E) and nucleoli–centromere interactions decreased (Figure 3, B and F) as keratinocytes differentiated toward the cornified layer. Interestingly, the percentage of centromeres associated with nuclear periphery increased (Figure 3, B and G). When all the differentiation layers were viewed on the same section, it is notable that the labeling signal of CENP-A (red) of each centromere was significantly reduced as cells differentiated (Figure 3, A and H). The reduction of centromere numbers is observed across all three terminal differentiation systems and is consistent with the findings in myoblast differentiation where CENP-A deposition to centromeres was reduced (Swartz et al., 2019) in addition to reduction in CENP gene expression (Supplemental Table S1).
To further evaluate the relationship between nucleoli–centromere association and differentiation, we asked whether this association changes when differentiation is inhibited. EPHA2, a receptor tyrosine kinase, plays an important role in epidermal differentiation and cancer prevention (Menges and McCance, 2008; Lin et al., 2010; Gordon et al., 2013; Perez White et al., 2017). To block keratinocyte differentiation, we knocked down EPHA2 in primary human keratinocytes using lentiviral shRNA and generated 3D HSE. Compared to the control culture, the EPHA2-depleted keratinocytes showed a lack of differentiation and stratification (Figure 3I) as judged by the H&E staining of the sections and reduction of a differentiation marker keratin 10. The nucleolar number reduced (Figure 3, J and K) but the nucleolar area increased (Figure 3, J and L) as cells remained undifferentiated. While there was no significant difference in the centromere number between control and EPHA2-deficient 3D HSE in the basal layer (Figure 3, J and M), the nucleoli–centromere association increased when EPHA2 is depleted (Figure 3, J and N), while the association of centromeres with the nuclear periphery did not change (Figure 3O). These data further corroborate the idea that differentiation couples with the reduction of nucleoli–centromere interactions.
Nucleoli–centromere interactions increase when cells are transformed into immortalized or cancerous cells
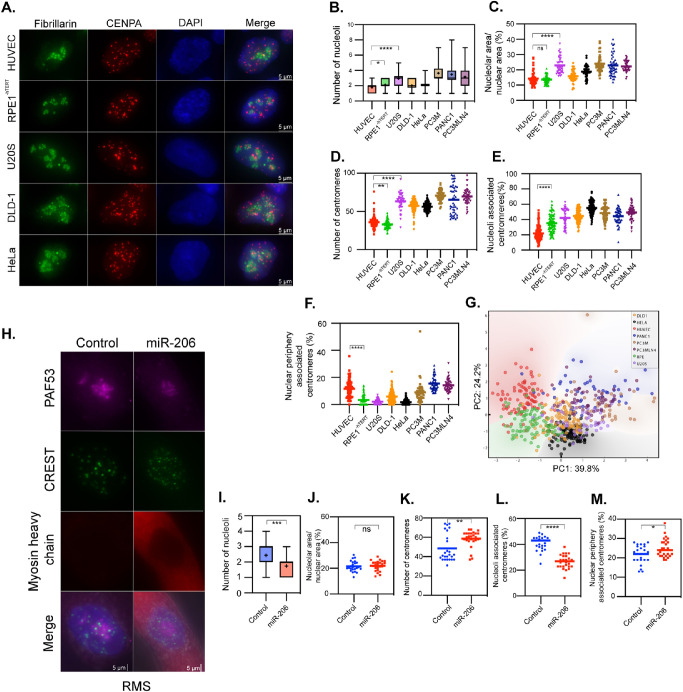
To further evaluate the dynamics of nucleoli–centromere interactions during differentiation, we examined a cohort of human cancer cells and a normal cell line because a key feature of cancer is the disruption of differentiated characteristics of the normal tissues of origin (Cao, 2017; Hanahan, 2022). If nucleoli–centromere interaction decreases as cells differentiate, we would expect the interaction to increase in cancer cells. We examined nucleoli and centromere dynamics in noncancerous primary human umbilical vein endothelial cells (HUVECs), hTERT-immortalized retinal pigment epithelial cells (RPE1-hTERT), colorectal adenocarcinoma cells (DLD-1), metastatic cervical adenocarcinoma (HeLa), metastatic prostate cancer (PC3M and PC3M LN4), metastatic pancreatic cancer (PANC1), and osteosarcoma cells (U2OS). The results (Figure 4A) showed that the number of nucleoli (Figure 4B) and nucleolar areas (Figure 4C) varied among the cancer cells, with cancer cells frequently having more nucleoli and increased area relative to normal cells (Figure 4, B and C). The number of centromeres (Figure 4D) was increased in all cancer cell lines compared with diploid cells, consistent with established aneuploidy in these cell lines. The percentage of nucleoli-associated centromeres was higher in the immortalized retinal pigment epithelium (RPE) cells compared with HUVECs, and typically higher still in cancer cells (Figure 4E). In addition, the percentage of nuclear periphery–associated centromeres decreased in the hTERT-immortalized and some of the cancer cells (Figure 4F). Principal component analysis (Figure 4G) was performed with the inclusion of all the variables from Figure 4, B–F, and demonstrated that nonmalignant cells (HUVEC, RPE) clustered separately from malignant cells (all tumor cell lines), and accounted for 64% of the variability in the data. These observations are consistent with a model in which increased proliferative and/or malignant capacity coincides with an increase in nucleoli number, area, and nucleoli-associated centromeres.
FIGURE 4:
Nucleolus–centromere interactions are increased in cancer cells compared with normal cells and reduce upon differentiation. Nucleoli were immunolabeled either with anti-fibrillarin antibody (green) and centromeres by anti-CenpA (red) antibodies or anti-PAF53 (purple) and CREST (green). (A) Representative stains for a subset of the tested cell lines quantified in panels B–G. For panels B–F, given the number of cell lines tested, only a subset of the potential pairwise statistical tests are shown. Compared to normal HUVEC cells and RPE1 cells with an hTERT-knockout, cancer cells frequently demonstrate increases in the number (B) and area (C) of nucleoli. (D) As expected, the number of centromeres varies in cancer cells, but is typically increased. The percentage of nucleolar-associated centromeres is frequently increased in the cancer cells (E), frequently occurring with a decreased spatial association of centromeres with the nuclear periphery (F). (G) A principal component analysis demonstrates that the nonmalignant HUVEC and RPE1 cells (red and green) largely cluster separately from cancer cells, while the cancer cells tend to group together. Percentage of variability in the data explained by PC1 and PC2 are indicated on the respective axes. When pediatric rhabdomyosarcoma cells were induced to differentiate upon transfection with miR-206 (H), the number of nucleoli (I) was reduced, while the nucleolar area did not change (J). Differentiated cells demonstrated a significant increase in countable centromeres (K), a reduction in nucleolus–centromere overlap (L), and an increase in centromere–nuclear periphery overlap (M). Box plots indicate the 25th–75th percentile values, with the whiskers indicating minimum and maximum observed values. Scatter plots indicate the value of individual cellular measurements, with the horizontal line indicating the mean. Statistical significance was computed as two-tailed t tests of means with unequal variance. Bar = 5 μm. n ranges from 49 to 105 per cell type (top) and 25 to 30 per condition (bottom). ns: not significant; *: p < 0.05; **: p < 0.01; ***: p < 0.001; ****: p < 0.0001.
To further confirm that the dynamics of nucleolar structure and nucleoli–centromere association paralleled the differentiation process, we asked whether induction of differentiation in a cancer cell line would restore the state of nucleolar structure and the nucleoli–centromere association in differentiated cells. RD cells are a cell culture model derived from a pediatric rhabdomyosarcoma (RMS). Increased expression of the promyogenic miRNA miR-206 has been demonstrated to induce differentiation of RMS cells and inhibit RMS tumor growth in vivo (Taulli et al., 2009; MacQuarrie et al., 2012). We examined and compared RMS cells transiently transfected with a miR-206 mimetic or a negative control mimetic and then kept in myogenic differentiation media for 48 h. The differentiation of these tumor cells into a more myotube-like state was determined by the expression of sarcomeric myosin heavy chain, a structural protein up-regulated during normal myogenic differentiation (Figure 4H; Schiaffino et al., 2015). The number of nucleoli was reduced upon differentiation (Figure 4, H and I), but the nucleolar area did not change within 48 h (Figure 4J). Furthermore, the nucleoli–centromere association (Figure 4, H and L) significantly reduced in spite of the increases in the number of centromeres (Figure 4, H and K) in myosin-expressing cells, and a small but statistically significant increase in the association of centromeres with the nuclear periphery was observed (Figure 4, H and M). These data are consistent with our observations of the reduction of nucleoli–centromere association in terminally differentiated cells. Examination of previously published gene expression data comparing RD cells to myotubes also demonstrated multiple centromere-related genes expressed at significantly higher levels in the RD cells compared with myotubes (Supplemental Table S2).
In summary, we report the dynamic nature of nucleolar number, area, and nucleoli–centromere interactions throughout cellular differentiation in several cell types. Nucleolar number and area do not always correspond to each other. The average number of nucleoli in the pluripotent state is 1.5, but the nucleolar area is the largest with an average nucleoli/nucleus area ratio of 21%, whereas in intermediate states, while nucleolar numbers increased to three to five in human cell models, the nucleolar area decreased. In terminally differentiated human cells, the nucleolar number and areas decrease further. These findings demonstrate that the nucleolar number and area are unlikely to be related to each other. Do they then correspond to ribosome synthesis? Previous reports have shown that ribosome synthesis and translation increase as cells differentiate and exit the pluripotent state (Tahmasebi et al., 2018; Gupta and Santoro, 2020; Li and Wang, 2020). Our data are consistent with the idea that nucleolar areas do not directly correspond with ribosome biogenesis, suggesting the possibility that nucleolar structure plays additional roles during differentiation.
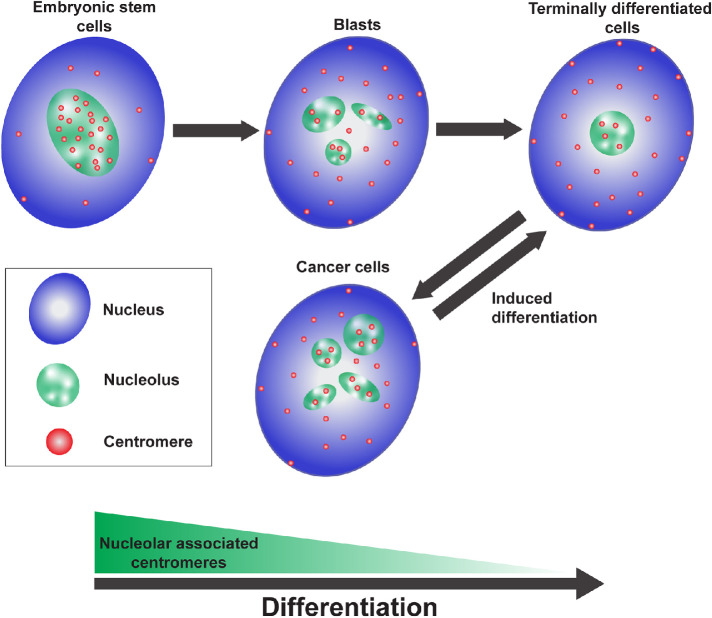
Many studies have previously reported on nucleolar-associated genomic domains (van Koningsbruggen et al., 2010; Bersaglieri and Santoro, 2019; Vertii et al., 2019; Bizhanova et al., 2020; Gupta and Santoro, 2020; Bersaglieri et al., 2022) and our findings of the dynamic nature of the nucleoli–centromere association through differentiation further supports a role for nucleoli in genome organization. The nucleoli–centromere association is the highest in pluripotent stem cells and lowest in the terminally differentiated state (Figure 5), in which possibly only the NOR containing chromosome centromeres still associate with nucleoli. The association is directly linked to the differentiation state as either the block of differentiation in a 3D HSE graft by shRNA against an essential epidermal differentiation factor EPHA2 (Figure 3) or the loss of differentiated characteristics (Cao, 2017) during carcinogenesis (Figure 4) is associated with an increase in nucleoli–centromere association. Furthermore, when a rhabdomyosarcoma cell line was induced to differentiate by the promyogenic miRNA miR-206, the association decreased as cells increased expression of myosin heavy chain, a marker of myogenic differentiation, an event associated with exit from the cell cycle (MacQuarrie et al., 2012).
FIGURE 5:
Schematic model demonstrating that the association of centromeres with nucleoli decreases as cells differentiate, is increased in cancer cells, and decreases once more as cancer cells are induced to differentiate.
Centromeres are special areas of chromosomes that enable equal segregation of chromosomes into the resulting daughter cells during cell division. Centromeres are composed with highly repetitive DNA sequences occupied by centromere-specific CENP-A nucleosomes flanked by histone H3–containing nucleosomes that form the pericentric heterochromatin (Morris and Moazed, 2007; Westhorpe and Straight, 2013). The spatial link of heterochromatin with nucleoli has been considered a regulatory mechanism that links to the suppression of gene expression (Bersaglieri and Santoro, 2019; Gupta and Santoro, 2020; Bizhanova and Kaufman, 2021). From pluripotent to terminally differentiated cells, the perinucleolar heterochromatin increases (Bersaglieri and Santoro, 2019; Gupta and Santoro, 2020; Bizhanova and Kaufman, 2021). ES cells generally have the least amount of heterochromatin (Savic et al., 2014; Politz et al., 2016). As the association of centromeres with nucleoli is the highest in ES cells, it is difficult to imagine that the changes of nucleoli–centromere interactions through differentiation stages are simply due to spatial shifting of bulk heterochromatin. Rather, this suggests a model (Figure 5) in which the shifting nucleoli–centromere association reflects a differentiation state–specific role for centromere function and/or genome organization in stem cells. In summary, our data from a variety of model systems demonstrates a dynamic, yet relatively consistent, pattern of interaction between nucleoli and centromeres in both normal differentiating and cancer cells, suggestive of potential functional significance to be further explored.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Immunostaining
The cells were fixed with 4% (wt/vol) paraformaldehyde in phosphate-buffered saline (PBS) for 15 min, washed with PBS, solubilized in 0.5% (vol/vol) Triton X-100 (Sigma) in PBS for 10 min, and washed again before being incubated in primary antibodies that included human CREST (a gift from R. Goldman, Feinberg School of Medicine, Northwestern University, Chicago, IL) at a dilution of 1:10, Nopp140 (a gift from T. Meier, Albert Einstein College of Medicine, Bronx, NY), SSEA4 (Abbomax; 620-500), Nestin (Cell Signaling; mAb33475), LaminA (a gift from R. Goldman), CENPA (Thermofisher; MA1-20832), Keratin10 (Abcam; Ab76318), fibrillarin (Sigma; AnA-N), mouse myosin heavy chain MF-20 antibody (DSHB; MF-20 concentrate) at 1:100, or rabbit POLRIE/PAF53 antibody at 1:100 (Proteintech; 16145-1-AP). Incubations were for 1 h before cells were washed, and then finally incubated in anti-human, anti-mouse, or anti-rabbit Alexa Fluor secondary antibodies at 1:200 (Invitrogen; A-11013; A-11014; A-11001; A-11005) for 1 h. The cells were then mounted using Vectashield antifade mounting media with DAPI (Vector Laboratories; H-1200).
Imaging, measurement, and statistical testing
Immunolabeled cells were imaged using a Nikon Eclipse Ti fluorescence microscope with 60× objective 1.4 NA and four channels of fluorescence. Cells were imaged through Z stack with 500-nm intervals. The top and bottom limits were set to include the entire nucleus. The Z stack of images was maximally projected and centromeres physically associated with nucleoli or the nuclear periphery in those projected images were hand scored and counted. The hand tracing tool in Element software was used to outline the extent of individual nucleoli and their respective nucleus. Nucleoli were visualized through nucleolar antibody staining, and nuclei were visualized using either nuclear outline via phase contrast, DAPI staining, or lamin A staining, depending on the cell system. Element software tools were then used to calculate the areas of traced nucleoli and nuclei and their ratio computed. Centromere intensity in the skin model was evaluated using the Element imaging tool. The intensity of each centromere was recorded and plotted.
Pairwise comparison testing for statistical significance was performed using two-tailed t tests of means with unequal variance in Prism 9. PCA (principal component analysis) was performed using the program Orange3 (Demsar et al., 2013) based on the data from Figure 4, B–F, and the variance that PC1 and PC2 account for individually is listed on each axis as indicated.
ES cells growth and differentiation
The National Institutes of Health (NIH)–approved and registered embryonic stem cell line NIHhESC-09-0022 (H9 ESCs) were acquired from WiCell and were grown on hESC-qualified Matrigel- (Corning; 354277) coated plates and expanded in serum-free mTeSR-plus stem cell medium (Stem Cell Technologies; 100-0276) without antibiotics. For imaging studies, cells were plated on Matrigel-coated glass coverslips that were pretreated with an acid wash, sterilized with ethanol, and cleaned using PBS washes before coating. ESCs were characterized based on morphology to grow in flat and tightly packed colonies with well-defined borders comprised of cells with high nuclear-to-cytoplasmic ratios and the ability to expand and differentiate. Cells were certified to have a normal karyotype and free from contaminants. For differentiation, cells were passaged, seeded, and briefly expanded to reach 25% confluence before being cultured in 15% serum-supplemented DMEM. Cells were allowed to differentiate for 10 d before fixation and staining.
Human-induced pluripotent stem cells (iPSCs; NU012) were derived from a healthy female. Peripheral blood mononuclear cells (PBMCs) were isolated from blood using SepMate tubes (Stem Cell Technologies; 85415). PBMCs were expanded in StemSpan SFEM II media (Stem Cell Technologies; 09605) with an erythroid expansion supplement (Stem Cell Technologies; 02692). Expanded PBMCs were reprogrammed using CytoTune 2.0 sendai vectors. Briefly, PBMCs were infected with Yamanaka factors encoded in sendai vectors and cultured overnight in StemSpan SFEM II supplemented media. Media was changed the next day and on day 3, cells were transferred to ReproTeSR medium (Stem Cell Technologies; 05926) for reprogramming. Reprogrammed colonies were selected and clonally expanded. Quality control testing was performed to confirm cells are undifferentiated, free of contaminants, and have a normal karyotype and short tandem repeats (STR) profile. For differentiation, cells were passaged, seeded, and briefly expanded to reach 25% confluence before being cultured in 15% serum-supplemented DMEM. Cells were allowed to differentiate for 10 d.
3 D HSEs
3D HSEs were established as previously described using primary neonatal foreskin-derived keratinocytes (Arnette et al., 2016). Cells were transduced with a lentiviral construct containing a short hairpin sequence targeting EPHA2 or a scrambled sequence (pLKO.EV and pLKO.shEPHA2; gifts of B. Wang, Case Western Reserve University, Cincinnati, OH) as previously described (Perez White et al., 2017).
Myoblast differentiation
Primary human skeletal muscle cells were acquired from ATCC (PCS-950-010) and cultured in mesenchymal stem cell basal medium (ATCC; PCS-500-030) supplemented with the Primary Skeletal Muscle Cell Growth Kit (ATCC; PCS-950-040) components (fetal bovine serum [FBS], dexamethasone, l-glutamine, epidermal growth factor [EGF], fibroblast growh factor b [FGF-b], and insulin) per manufacturer’s recommendations, as well as 1% penicillin–streptomycin (Life Technologies). Cells were kept at low density (<50% confluency) during propagation, and 0.25% trypsin-EDTA (Hyclone) used when passaging cells.
For myoblast samples, cells were split onto untreated coverslips at a low density, cultured in growth media for 16 h, and were then washed with PBS twice before being fixed in 4% paraformaldehyde in PBS for 10 min at room temperature. Before fixation, all myoblast samples were inspected to ensure confluency was <50%. For differentiated myotube samples, cells were split at a sufficient density onto untreated coverslips to reach 100% confluency after 16 h in growth media, then washed three times with PBS before being changed to Skeletal Muscle Differentiation Tool media (ATCC; PCS-950-050) and cultured for a further 96 h. Cells were then fixed as above for myoblast samples.
Rhabdomyosarcoma cells
The RD human rhabdomyosarcoma cell culture line was acquired from ATCC (CCL-136) and cultured in DMEM (Life Technologies) with 10% FBS (Hyclone) and 1% penicillin–streptomycin (Life Technologies). One day before transfection, cells were trypsinized (Hyclone) and plated onto untreated glass coverslips (VWR) at sufficient density to reach 50–60% confluency the following day. Cells were transfected using either a miR-206 miRNA mimetic (Thermofisher; AM17100) or negative control mimetic #1 (Thermofisher; 4464058) using RNAiMax (Thermofisher; 13778100). Transfections were performed according to the manufacturer’s protocol with the following modifications: the mimetic’s final concentration was 16 nM, no antibiotics were present in the media, and 1.5 μl of RNAiMax was used per well for a 12-well dish. After 24 h, cells were washed twice with PBS, and media was changed to low serum differentiation media (DMEM +1% horse serum [HyClone] + 1x insulin-transferrin-selenium [Corning; 25-800-CR] + 1% penicillin–streptomycin). Cells were fixed after 48 h in differentiation media as noted above for myoblasts.
Neuroblast differentiation
Neuroblasts/neural precursor cells are isolated and grown as described (Edens et al., 2019). Dorsal forebrains from timed-pregnant E13.5 mouse embryos were digested with accutase (Fisher; NC9839010). Neuroblasts were carried out on plates coated with Matrigel (Corning) at 80 μg/ml and maintained in DMEM-F12 medium (Life Technologies; 11320033) supplemented with B27 (Life Technologies; 17504044), N2 (Life Technologies; 17502048), and GlutaMax (Life Technologies; 35050079). A growth factor cocktail containing 20 ng/ml EGF (PeproTech; AF-100-15) and 20 ng/ml basic FGF (PeproTech; 100-18B) in heparin (5 μg/ml) was added to the medium fresh. Cells were carried at densities not exceeding 80%, and all experiments were performed on density- and passage-matched cultures. Cells were incubated in standard conditions: 37°C with 5% CO2.
Differentiation.
Neuroblasts were seeded onto coverslips or tissue culture dishes coated with 20 μg/ml poly-l-lysine and 4 μg/ml laminin in neural stem cell medium without growth factors (DMEM-F12 supplemented with N2, B27, and GlutaMax [all Life Technologies as above], but without bFGF and EGF). For 24-well plates, 300,000 neuroblasts were seeded into each well. The following day, day in vitro 1, cells were changed into fresh medium containing 50 ng/ml BDNF (brain derived neurotrophic factor; Life Technologies; PHC7074), 25 ng/ml GDNF (glial cell–derived neurotrophic factor; Life Technologies; PHC7041), and 10 uM forskolin (Sigma; F3917). Half of the culture media in each well was replaced with fresh medium containing BDNF, GDNF, and forskolin every 3 days until cells were fully differentiated into neurons.
Supplementary Material
Acknowledgments
D.R.F. is funded by GM1111907 and U10CA260699. Y.C.M. was supported by NIH R01NS-094564 and R21NS-106307. B.E.P.W. was supported by NIH NIAMS Grants no. K01AR-072773 and no. P30AR-075049. S.H. was supported by U10CA260699. K.L.M. is supported, in part, by the NIH’s National Center for Advancing Translational Sciences, Grant no. KL2TR-001424, as well as the Hyundai Hope on Wheels Scholar Hope Grant, the Stanley Manne Children’s Research Institute, and the Ann & Robert H. Lurie Children’s Hospital of Chicago. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We would also like to thank Robert Goldman for the CREST and lamin A antibodies, Thomas Meier for Nopp140 antibodies, and Joe Ibarra for technical assistance as well as Steven Kosak and Kurt Leano. We would also like to thank Stephen Adam, Paul Kaufman, and Anastassiia Vertii for critical reading.
Abbreviations used:
- ESC
embryonic stem cell
- HSE
human skin equivalent
- iPS
induced pluripotent cells
- NAD
nucleloi-associated domains
- RMS
rhabdomyosarcoma
- STR
short tandem repeats.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-06-0237) on February 8, 2023.
REFERENCES
- Arnette C, Koetsier JL, Hoover P, Getsios S, Green KJ (2016). In vitro model of the epidermis: connecting protein function to 3D structure. Methods Enzymol 569, 287–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersaglieri C, Kresoja-Rakic J, Gupta S, Bär D, Kuzyakiv R, Panatta M, Santoro R (2022). Genome-wide maps of nucleolus interactions reveal distinct layers of repressive chromatin domains. Nat Commun 13, 1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersaglieri C, Santoro R (2019). Genome organization in and around the nucleolus. Cells 8, 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizhanova A, Kaufman PD (2021). Close to the edge: heterochromatin at the nucleolar and nuclear peripheries. Biochim Biophys Acta Gene Regul Mech 1864, 194666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizhanova A, Yan A, Yu J, Zhu LJ, Kaufman PD (2020). Distinct features of nucleolus-associated domains in mouse embryonic stem cells. Chromosoma 129, 121–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI (2007). The multifunctional nucleolus. Nat Rev Mol Cell Biol 8, 574–585. [DOI] [PubMed] [Google Scholar]
- Cao Y (2017). Tumorigenesis as a process of gradual loss of original cell identity and gain of properties of neural precursor/progenitor cells. Cell Biosci 7, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerqueira AV, Lemos B (2019). Ribosomal DNA and the nucleolus as keystones of nuclear architecture, organization, and function. Trends Genet 35, 710–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Greene E, Bowers SR, Mellone BG (2012). A role for the CAL1-partner Modulo in centromere integrity and accurate chromosome segregation in Drosophila. PLoS One 7, e45094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demsar J, Curk T, Erjavec A, Gorup C, Hocevar T, Milutinovic M, Mozina M, Polajnar M, Toplak M, Staric A, et al. (2013). Orange: data mining toolbox in python. J Mach Learn Res 14, 2349–2353. [Google Scholar]
- Diesch J, Bywater MJ, Sanij E, Cameron DP, Schierding W, Brajanovski N, Son J, Sornkom J, Hein N, Evers M, et al. (2019). Changes in long-range rDNA-genomic interactions associate with altered RNA polymerase II gene programs during malignant transformation. Commun Biol 2, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillinger S, Straub T, Nemeth A (2017). Nucleolus association of chromosomal domains is largely maintained in cellular senescence despite massive nuclear eorganization. PloS One 12, e0178821. [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G (2009). HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137, 485–497. [DOI] [PubMed] [Google Scholar]
- Edens BM, Vissers C, Su J, Arumugam S, Xu Z, Shi H, Miller N, Rojas Ringeling F, Ming GL, He C, et al. (2019). FMRP modulates neural differentiation through m(6)A-dependent mRNA nuclear export. Cell Rep 28, 845–854. E845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW (2009). Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137, 472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW (2006). The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol 8, 458–469. [DOI] [PubMed] [Google Scholar]
- Freeman AE, Igel HJ, Herrman BJ, Kleinfeld KL (1976). Growth and characterization of human skin epithelial cell cultures. In Vitro 12, 352–362. [DOI] [PubMed] [Google Scholar]
- Fujimura A, Hayashi Y, Kato K, Kogure Y, Kameyama M, Shimamoto H, Daitoku H, Fukamizu A, Hirota T, Kimura K (2020). Identification of a novel nucleolar protein complex required for mitotic chromosome segregation through centromeric accumulation of Aurora B. Nucleic Acids Res 48, 6583–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon K, Kochkodan JJ, Blatt H, Lin SY, Kaplan N, Johnston A, Swindell WR, Hoover P, Schlosser BJ, Elder JT, et al. (2013). Alteration of the EphA2/Ephrin-A signaling axis in psoriatic epidermis. J Invest Dermatol 133, 712–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Santoro R (2020). Regulation and roles of the nucleolus in embryonic stem cells: from ribosome biogenesis to genome organization. Stem Cell Rep 15, 1206–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D (2022). Hallmarks of cancer: new dimensions. Cancer Discov 12, 31–46. [DOI] [PubMed] [Google Scholar]
- Iarovaia OV, Minina EP, Sheval EV, Onichtchouk D, Dokudovskaya S, Razin SV, Vassetzky YS (2019). Nucleolus: a central hub for nuclear functions. Trends Cell Biol 29, 647–659. [DOI] [PubMed] [Google Scholar]
- Li D, Wang J (2020). Ribosome heterogeneity in stem cells and development. J Cell Biol 219, e202001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Gordon K, Kaplan N, Getsios S (2010). Ligand targeting of EphA2 enhances keratinocyte adhesion and differentiation via desmoglein 1. Mol Biol Cell 21, 3902–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQuarrie KL, Yao Z, Fong AP, Diede SJ, Rudzinski ER, Hawkins DS, Tapscott SJ (2013). Comparison of genome-wide binding of MyoD in normal human myogenic cells and rhabdomyosarcomas identifies regional and local suppression of promyogenic transcription factors. Mol Cell Biol 33, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQuarrie KL, Yao Z, Young JM, Cao Y, Tapscott SJ (2012). miR-206 integrates multiple components of differentiation pathways to control the transition from growth to differentiation in rhabdomyosarcoma cells. Skeletal Muscle 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges CW, McCance DJ (2008). Constitutive activation of the Raf-MAPK pathway causes negative feedback inhibition of Ras-PI3K-AKT and cellular arrest through the EphA2 receptor. Oncogene 27, 2934–2940. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Misteli T (2006). Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol 7, 540–546. [DOI] [PubMed] [Google Scholar]
- Morris CA, Moazed D (2007). Centromere assembly and propagation. Cell 128, 647–650. [DOI] [PubMed] [Google Scholar]
- Nemeth A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Peterfia B, Solovei I, Cremer T, Dopazo J, Langst G (2010). Initial genomics of the human nucleolus. PLoS Genet 6, e1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs RL, Press RI (1992). Centromere autoantigens are associated with the nucleolus. Exp Cell Res 200, 339–350. [DOI] [PubMed] [Google Scholar]
- Padeken J, Mendiburo MJ, Chlamydas S, Schwarz HJ, Kremmer E, Heun P (2013). The nucleoplasmin homolog NLP mediates centromere clustering and anchoring to the nucleolus. Mol Cell 50, 236–249. [DOI] [PubMed] [Google Scholar]
- Pederson T (1998). The plurifunctional nucleolus. Nucleic Acids Res 26, 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T (2011). The nucleolus. Cold Spring Harb Perspect Biol 3, 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez White BE, Ventrella R, Kaplan N, Cable CJ, Thomas PM, Getsios S (2017). EphA2 proteomics in human keratinocytes reveals a novel association with afadin and epidermal tight junctions. J Cell Sci 130, 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JCR, Scalzo D, Groudine M (2016). The redundancy of the mammalian heterochromatic compartment. Curr Opin Genet Dev 37, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savic N, Bar D, Leone S, Frommel SC, Weber FA, Vollenweider E, Ferrari E, Ziegler U, Kaech A, Shakhova O, et al. (2014). lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell 15, 720–734. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Rossi AC, Smerdu V, Leinwand LA, Reggiani C (2015). Developmental myosins: expression patterns and functional significance. Skeletal Muscle 5, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma AB, Dimitrov S, Hamiche A, Van Dyck E (2019). Centromeric and ectopic assembly of CENP-A chromatin in health and cancer: old marks and new tracks. Nucleic Acids Res 47, 1051–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib M, Ouararhni K, Dimitrov S, Hamiche A (2010). HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci USA 107, 1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl A, Hartung M, Vagner-Capodano AM, Fouet C (1976). Chromosomal constitution of nucleolus-associated chromatin in man. Hum Genet 35, 27–34. [DOI] [PubMed] [Google Scholar]
- Swartz SZ, McKay LS, Su KC, Bury L, Padeganeh A, Maddox PS, Knouse KA, Cheeseman IM (2019). Quiescent cells actively replenish CENP-A nucleosomes to maintain centromere identity and proliferative potential. Dev Cell 51, 35–48. e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahmasebi S, Amiri M, Sonenberg N (2018). Translational control in stem cells. Front Genet 9, 709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, Tuschl T, Ponzetto C (2009). The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest 119, 2366–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Koningsbruggen S, Gierlinski M, Schofield P, Martin D, Barton GJ, Ariyurek Y, den Dunnen JT, Lamond AI (2010). High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell 21, 3735–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertii A, Ou J, Yu J, Yan A, Pages H, Liu H, Zhu LJ, Kaufman PD (2019). Two contrasting classes of nucleolus-associated domains in mouse fibroblast heterochromatin. Genome Res 29, 1235–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhorpe FG, Straight AF (2013). Functions of the centromere and kinetochore in chromosome segregation. Curr Opin Cell Biol 25, 334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, Choo KH (2007). Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res 17, 1146–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Lemos B (2016). A portrait of ribosomal DNA contacts with Hi-C reveals 5S and 45S rDNA anchoring points in the folded human genome. Genome Biol Evol 8, 3545–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Lemos B (2018). The long-range interaction map of ribosomal DNA arrays. PLoS Genet 14, e1007258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.